SUMMARY
Active DNA demethylation via Ten-eleven Translocation (TET) family enzymes is essential for epigenetic reprogramming in cell state transitions. TET enzymes catalyze up to three successive oxidations of 5-methylcytosine (5mC), generating 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), or 5-carboxycytosine (5caC). Although these bases are known to contribute to distinct demethylation pathways, the lack of tools to uncouple these sequential oxidative events has constrained our mechanistic understanding of the role of TETs in chromatin reprogramming. Here, we describe the first application of biochemically-engineered TET mutants that unlink 5mC oxidation steps, examining their effects on somatic cell reprogramming. We show that only TET enzymes proficient for oxidation to 5fC/5caC can rescue the reprogramming potential of Tet2-deficient mouse embryonic fibroblasts. This effect correlated with rapid DNA demethylation at reprogramming enhancers and increased chromatin accessibility later in reprogramming. These experiments demonstrate that DNA demethylation through 5fC/5caC has roles distinct from 5hmC in somatic reprogramming to pluripotency.
Keywords: epigenetics, reprogramming, Ten-eleven Translocation (TET), DNA demethylation, induced pluripotent stem cells (iPSCs), 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), 5-carboxycytosine (5caC), bACE-seq
INTRODUCTION
As a key regulator of tissue-specific gene expression patterns and chromatin organization, DNA methylation presents a significant epigenetic barrier in the reprogramming of somatic cells to induced pluripotent stem cells (iPSCs) (Gao et al., 2013; Nashun et al., 2015; Takahashi & Yamanaka, 2006). Erasure of DNA methylation proceeds through one of two distinct mechanisms: (1) passive loss of 5mC during DNA replication via suppression of DNA methyltransferase (DNMT) activity, or (2) active demethylation by TET enzymes (Figure 1A) (Hill et al., 2014; Tahiliani et al., 2009). Active demethylation is initiated through the progressive oxidation of 5mC to 5hmC, 5fC, or 5caC (Ito et al., 2011), after which demethylation is achieved through one of two potential pathways. First, 5hmC is not well-recognized by the DNMT1 maintenance methylation machinery (Hashimoto et al., 2012), allowing for its passive loss over several rounds of DNA replication and cellular division (termed here the “hmC pathway”, Figure 1A). Although 5fC and 5caC are also subject to passive loss (Inoue et al., 2011), their steady state levels are many orders of magnitude lower than 5hmC, suggesting that their involvement in this pathway is limited (Wagner et al., 2015; Wu et al., 2014). Instead, 5fC and 5caC can be targeted for base excision by thymine DNA glycosylase (TDG) (He et al., 2011; Maiti & Drohat, 2011; Zhang et al., 2012). In this mode of active demethylation (termed here the “fC/caC pathway”, Figure 1A), subsequent steps of base-excision repair (BER) restore an unmodified cytosine at the former abasic site (Kohli & Zhang, 2013).
Figure 1 – Characterization of mouse TET fC/caC catalytic mutants.
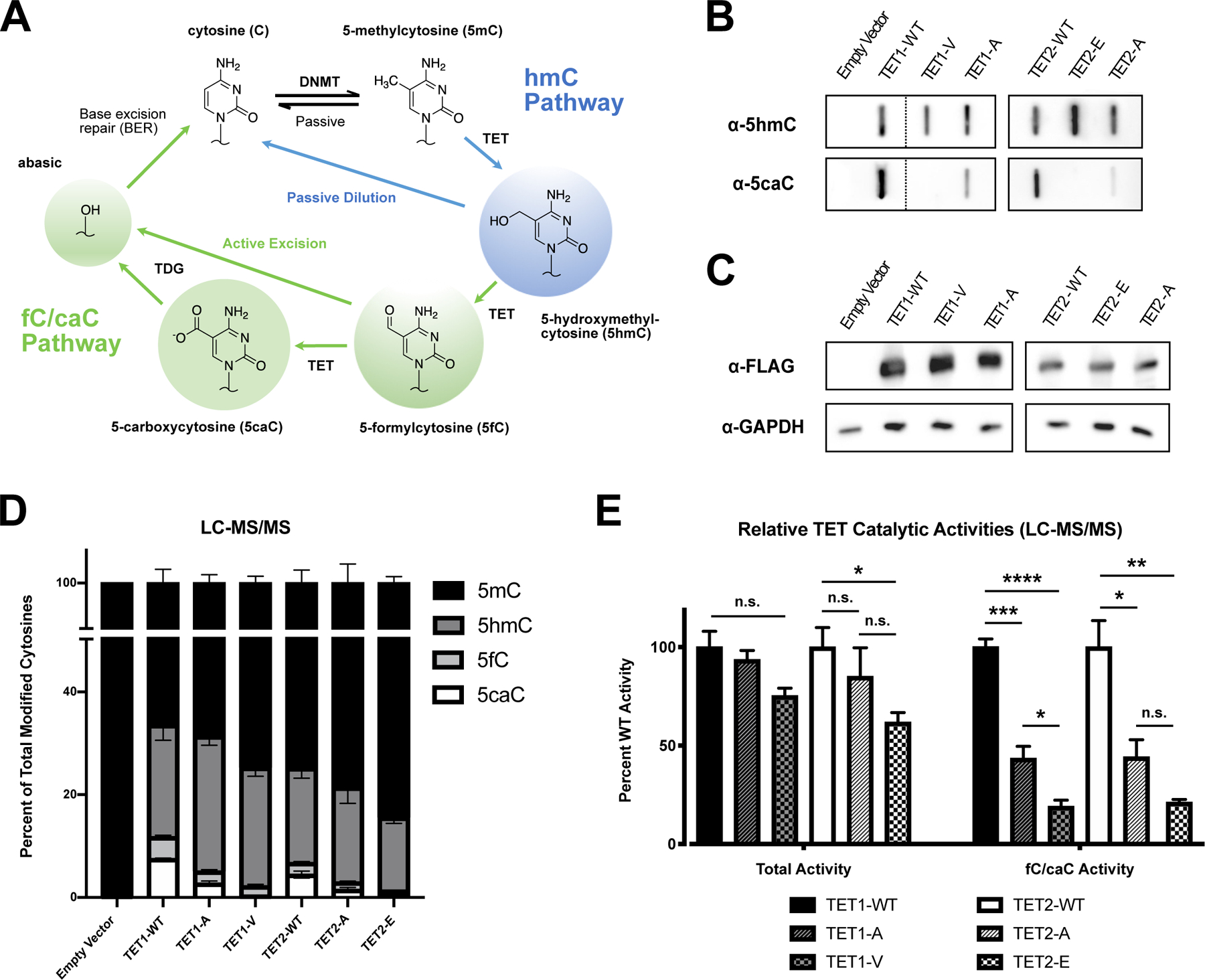
(A) Schematic representation of DNA demethylation pathways. (B) Representative slot blots for 5hmC and 5caC in DNA from transfected HEK293T cells (n=5; note: Tet1 results spliced from full blot in S1B). (C) Western blot for FLAG-tagged TET catalytic variant expression. Variants tested included wild-type (WT), 5hmC-stalling (T>V or T>E), and low-efficiency (T>A) TET1 and TET2, respectively, and empty vector control. GAPDH served as a loading control. (D) Genomic levels of modified cytosines quantified by LC-MS/MS and expressed as the percentage of total modified cytosines present in each sample (mean ± SEM; n=3–4). (E) Relative catalytic activities of TET1 and TET2 variants. Modified cytosines were normalized to their mean levels in cells transfected with WT Tet1 or Tet2. Relative catalytic activities are presented as total TET activity (5hmC + 5fC + 5caC) or specific fC/caC activity (mean ± SEM; n=3–4; one-way ANOVA with Tukey multiple comparisons; n.s. = not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001).
TET enzymes and TDG are critical for iPSC reprogramming. TET1 has been proposed to have a vitamin C-dependent role in promoting iPSC formation through a positive feedback loop with Pou5f1 (Oct4) and Nanog (Chen et al., 2013; Chen et al., 2015; Costa et al., 2013; Olariu et al., 2016). TET2, furthermore, can directly interact with KLF4 or PARP1 to drive site-specific demethylation of reprogramming enhancers and promoters, and Tet2-depleted cells have reduced reprogramming potential (Doege et al., 2012; Sardina et al., 2018). Importantly, both Tet triple-knockout and Tdg null mouse embryonic fibroblasts (MEFs) fail to undergo iPSC reprogramming, suggesting the fC/caC pathway may be essential to the process (Hu et al., 2014). However, given the multifunctional roles of TDG in DNA repair, transcriptional activation, and histone modification, as well as the relatively early stage in reprogramming at which Tdg-null MEFs arrest, the contribution of the hmC and fC/caC pathways to epigenetic reprogramming remains poorly defined (Nedderman et al., 1996; Tini et al., 2002; Um et al., 1998; Cortázar et al., 2007; Cortázar et al., 2011; Cortellino et al., 2011). Furthermore, although differential accumulation of 5hmC and 5fC/5caC across the genome suggests that these pathways might have distinct roles, functional studies into differences in their epigenetic reprogramming potential have been hindered by a lack of molecular tools to distinguish between the pathways in vivo (Shen et al., 2013; Wu et al., 2011).
We recently identified a threonine residue (T1372) in the active site of the human TET2 catalytic domain (CD) that can be mutated to alter the enzyme’s catalytic processivity (Liu et al., 2017). While some substitutions reduced the overall activity in each oxidative step, others elicited a “5hmC-stalling” phenotype whereby 5hmC is efficiently generated but not further oxidized, thereby depleting the cell of downstream 5fC and 5caC. We posited that mutants with altered processivity can be used to inform the importance of the 5hmC-driven mode of DNA demethylation versus the fC/caC pathway. Here, we introduce TET2 mutants into Tet2-depleted cells, in concert with a novel chemoenzymatic sequencing approach, to investigate the specific role of the fC/caC pathway in iPSC formation and chromatin reorganization.
RESULTS
Characterization of mouse TET fC/caC catalytic mutants
To determine the role of the fC/caC pathway in epigenetic reprogramming, we developed an allelic series of mouse TET mutants exhibiting diverse catalytic capacities. Because the human TET2 T1372 residue is conserved in mouse TET1 (T1642) and TET2 (T1285) (Figure S1A), we first tested whether corresponding mutations elicited similar changes in catalytic activity. We transfected HEK293T cells with candidate FLAG-Tet1-CDT1642 or FLAG-Tet2-CDT1285 mutants and collected DNA after 48 hours to measure the effect on global modified cytosine levels. Slot blot analysis of 5hmC and 5caC levels recapitulated the previously reported 5hmC-stalling phenotype through T1642V or T1285E substitution of mouse TET1-CD and TET2-CD, respectively (Figure 1B, S1B). Furthermore, T>A substitutions resulted in a phenotype whereby the enzymes produced both 5hmC and 5caC, but at a reduced rate compared to their wild-type (WT) counterparts, which has previously been termed as a “low efficiency” variant (Liu et al., 2017). Importantly, Western blots of transfected cellular lysates indicated that TET protein levels were unaffected by T1642 or T1285 substitution in HEK293T cells (Figure 1C).
To quantify the catalytic activity of our TET mutants more rigorously, we next analyzed DNA from transfected HEK293T cells by liquid chromatography-tandem mass spectrometry (LC-MS/MS). In the absence of transfected TET, 5mC accounted for the majority of total modified cytosines (99.74 ± 0.02%; n=4) (Figure 1D). When WT TET1-CD or TET2-CD was transfected, however, we observed a robust increase in 5hmC, 5fC, and 5caC levels. By contrast, TET 5hmC-stalling and low efficiency mutants exhibited a range of oxidative potential that largely matched their expected activity based on slot blot analysis. From the LC-MS/MS data, we quantified TET catalytic activity in two ways: “Total activity,” referring to the combined percentage of 5hmC, 5fC, and 5caC genome-wide, and “fC/caC activity,” referring to the combined percentage of 5fC/5caC (Figure 1E). Because total activity is driven largely by 5hmC production, these levels were only modestly affected in our catalytic mutants. Conversely, fC/caC activity was strongly affected in all catalytic mutants, with low efficiency mutants exhibiting approximately 45% of WT fC/caC activity vs. 20% for 5hmC-stalling mutants.
To confirm that the observed phenotypes were reproducible in additional cell types, we repeated our mouse TET1-CDT1642 mutant transfections in mouse NIH3T3 fibroblasts, and found that low efficiency and 5hmC-stalling phenotypes were recapitulated (Figures S1C–D). Together, these experiments demonstrate that the human TET2-CDT1372 mutant phenotypes were conserved for the mouse TET2-CD ortholog as well as its TET1-CD isoform.
fC/caC-proficient TET activity rescues iPSC reprogramming in Tet2−/− MEFs
To elucidate the mechanism by which TET enzymes promote epigenetic reprogramming in cell state transitions, we performed iPSC induction on Tet2−/− MEFs transduced with our TET2 catalytic mutants. These Tet2−/− MEFs carry a single-copy insertion of the STEMCCA OKSM reprogramming cassette in a Rosa26:M2rtTA background (OKSM-rtTA), allowing for doxycycline (Dox)-inducible expression of the four Yamanaka factors (Figure S2A–D) (Stadtfeld et al., 2010).
Prior to Dox induction, Tet2−/−; OKSM-rtTA MEFs were retrovirally transduced with either empty vector or one of four Tet2-CD constructs: WT Tet2-CD (Tet2-WT), low efficiency Tet2-CDT1285A (Tet2-A), 5hmC-stalling Tet2-CDT1285E (Tet2-E), or catalytically inactive Tet2-CDH1295Y,D1297A (Tet2-HxD) (Ko et al., 2010). Two days after infection, MEFs were seeded onto feeder cells and placed in 2i/LIF media + Dox for iPSC induction (Figure 2A). After 5 days of Dox treatment, we evaluated samples from each group by qRT-PCR to confirm proper Tet2 expression levels (Figure S2E). Furthermore, we verified that expression of Tet1, Tet3, Dnmt1, Dnmt3a, and Dnmt3b was unaffected by Tet2 depletion or mutant overexpression (Figure S2E).
Figure 2 – Rescue of Tet2−/− iPSC reprogramming efficiency is dependent on TET2 fC/caC activity.
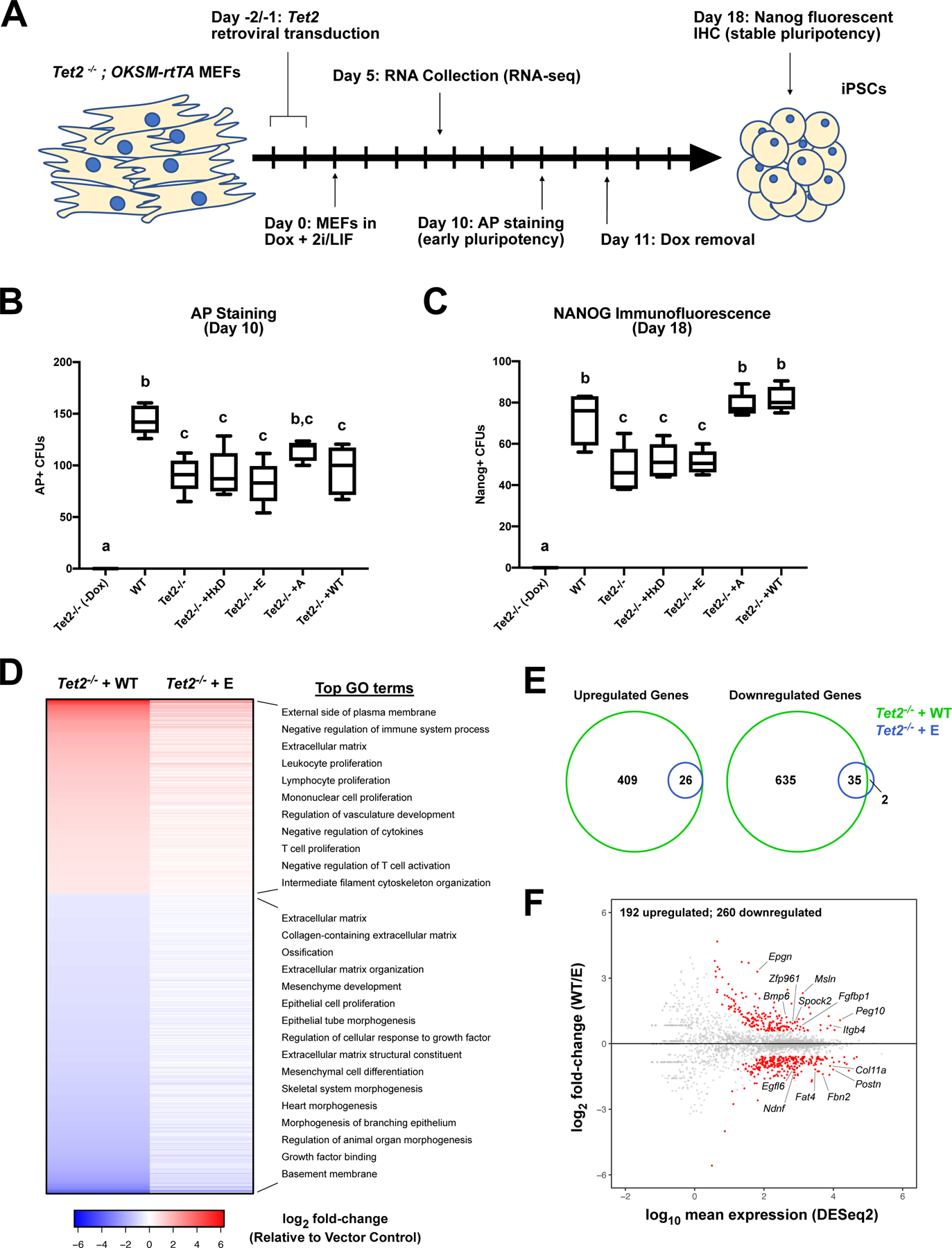
(A) Schematic illustration of Tet2−/− MEF reprogramming paradigm. (B) Quantification of AP staining of pluripotent colonies at day 10. Box plots indicate median AP+ colony forming units (CFUs) (one-way ANOVA with Tukey multiple comparisons; n=5; groups with different letters denote significant differences between groups, while shared letters indicate no difference was detected). (C) Quantification of immunohistochemical staining for NANOG-positive stable pluripotent colonies at day 18 (median NANOG+ CFU; one-way ANOVA with Tukey multiple comparisons; n=5). (D) Heat map of differentially expressed genes in Tet2-WT or Tet2-E transduced cells relative to vector control at day 5, with enriched GO terms indicated on right (RNA-seq; n=3–4; FDR < 0.05; fold-change > 1.5). (E) Venn overlap of significantly altered transcripts in Tet2-WT and Tet2-E transduced cells relative to vector control. (F) MA plot for Tet2-WT vs. Tet2-E gene expression. Red dots represent differentially expressed genes, with biologically relevant genes designated (FDR < 0.05; fold-change > 1.5).
After 10 days of Dox treatment, we performed alkaline phosphatase (AP) staining to determine the relative proportion of iPSC-like colonies prior to Dox withdrawal (e.g. while exogenous OKSM factors are still expressed). Interestingly, all Tet2−/− cultures, regardless of Tet2 overexpression, exhibited ~30% fewer AP+ colonies relative to WT reprogramming MEFs (Figure 2B). Because cellular proliferation rates are positively correlated with iPSC reprogramming efficiency, we performed 5-ethnyl-2’-deoxyuridine (EdU) staining (Hanna et al., 2009). Although untreated Tet2−/− MEFs exhibited a slightly slower rate of EdU incorporation relative to WT MEFs, this effect disappeared after 3 days of Dox treatment (Figure S2F). Likewise, we observed no differences in rate of apoptosis or cell death between WT and Tet2−/− MEFs as assayed by Annexin V and propidium iodide staining (Figure S2G).
As a more rigorous assay of stable iPSC formation, we removed Dox from the media after 11 days of treatment, requiring cells to utilize endogenous OKSM expression to maintain pluripotency. One week after Dox withdrawal, we examined NANOG expression as a marker of stable iPSC colonies. In agreement with previous literature, reprogramming Tet2−/− cultures exhibited on average 40% fewer NANOG+ colonies relative to WT cultures (Figure 2C) (Doege et al., 2012; Sardina et al., 2018). Strikingly, this reduced reprogramming efficiency was fully rescued by either Tet2-WT or Tet2-A overexpression, but not Tet2-E or Tet2-HxD. Taken together with the AP staining results, these data suggest that although entry into the early iPSC state is generally reduced in a Tet2−/− background, TET2’s fC/caC activity can rescue reprogramming efficiency during the later maturation phase as iPSC colonies transition to stable pluripotency. Furthermore, given the comparable potential of TET2-E and TET2-A to generate 5hmC, we can exclude that this effect might be due to a general loss of activity relative to TET2-WT.
To test the generality of our results, we repeated our experiments in Tet2−/− MEFs with retrovirally transduced WT Tet1-CD (Tet1-WT), 5hmC-stalling Tet1-CDT1642V (Tet1-V), or catalytically inactive Tet1-CDH1672Y,D1674A (Tet1-HxD), and observed that only TET1-WT rescued NANOG+ iPSC colony counts to WT levels (Figure S2H–I). These experiments suggest that fC/caC generative potential, as opposed to TET isoform identity, is more critical for the rescue of Tet2−/− reprogramming.
Previous research identified the role of TET proteins in regulating early transcriptional changes in MEF reprogramming, particularly the mesenchymal-to-epithelial transition (MET) (Hu et al. 2014). To test the influence of fC/caC activity on the transcriptome, we performed RNA-seq on Tet2−/− MEFs retrovirally transduced with empty vector, Tet2-WT, or Tet2-E after 5 days of Dox treatment. Relative to untreated Tet2−/− MEFs, there was a 72% overlap in differentially regulated genes (2956 upregulated, 3165 downregulated) among the three conditions, indicating the general reprogramming trajectory is unaltered by TET2 expression (Figure S2J; Table S4). However, we identified 1044 genes (409 upregulated, 635 downregulated) with significantly altered expression in Tet2-WT cells relative to vector control at iPSC day 5 (Figure 2D). Gene ontology (GO) enrichment analysis identified several pathways critical for early MEF reprogramming, including cell proliferation, extracellular matrix (ECM) reorganization, immune regulation, and mesenchymal fate repression. This effect was strongly attenuated in Tet2-E cells (26 upregulated, 35 downregulated relative to vector control), suggesting that TET2’s fC/caC activity is essential to promote rapid transcriptional changes during early MEF reprogramming (Figure 2D–E). Indeed, direct comparison of the Tet2-WT and Tet2-E day 5 transcriptomes identified 452 genes with significantly altered expression (fold-change > 1.5; false discovery rate < 0.05) (Figure 2F). Upregulated genes included several signaling factors known to promote cell growth and survival, (Peg10, Itgb4, Epgn, Spock2, Fgfbp1, Bmp6), as well as Zfp961, a KRAB-zinc finger protein implicated in retrotransposon silencing that may influence iPSC reprogramming efficiency (Friedli et al., 2014; Wolf et al., 2020). Among downregulated genes were several involved in mesenchymal ECM organization, cell adhesion, and motility (Ndnf, Postn, Egfl6, Fbn2, Fat4, and Coll11a1). Aberrant expression of these key factors may contribute to the diminished reprogramming potential of Tet2-E cells relative to Tet2-WT.
DNA demethylation at KLF4 reprogramming enhancers correlates with TET2 fC/caC activity
Because rescue of Tet2−/− iPSC reprogramming was dependent on TET fC/caC activity, we next tested whether this result correlated with increased DNA demethylation. Bisulfite (BS) sequencing is commonly used to assess DNA demethylation; treatment of DNA with BS deaminates unmodified C, 5fC, and 5caC, giving a readout of 5mC + 5hmC. However, because the hmC-stalling variants were unable to rescue reprogramming, it was necessary to distinguish 5hmC alone. We therefore used BS-assisted APOBEC-Coupled Epigenetic (bACE) pyrosequencing (Schutsky et al., 2018). Briefly, DNA is treated with BS and then the DNA deaminase APOBEC3A. The enzyme deaminates residual 5mC, but cytosine 5-methylenesulfonate (CMS), the product of 5hmC reaction with BS, is resistant to deamination, providing a specific readout of 5hmC. Pyrosequencing of a control oligonucleotide confirmed sensitive detection of 5mC + 5hmC by standard BS and 5hmC alone by bACE pyrosequencing (Figure S3A).
We focused our analysis on TET2 target enhancers. A recent study identified a subset of enhancers targeted by the TET2-KLF4 complex whose active DNA demethylation early in reprogramming (iPSC day 2–4) contributes to increased chromatin accessibility at later stages (Sardina et al., 2018). Selecting four enhancers highlighted by the authors as representative of this effect (Sall4 intragenic, Smarcd2, Tet2 intragenic, and Ebf3), we measured changes in 5mC and 5hmC levels relative to untreated control Tet2−/− MEFs after 5 days of Dox treatment (Figure 3A–D). 5hmC levels at all four enhancers were significantly augmented by overexpression of catalytically active Tet2. Most notably, Tet2-E generated levels of 5hmC at least as high as Tet2-WT, yet this was insufficient to rescue reprogramming.
Figure 3 – DNA demethylation at Klf4 reprogramming enhancers correlates with TET2 fC/caC activity.
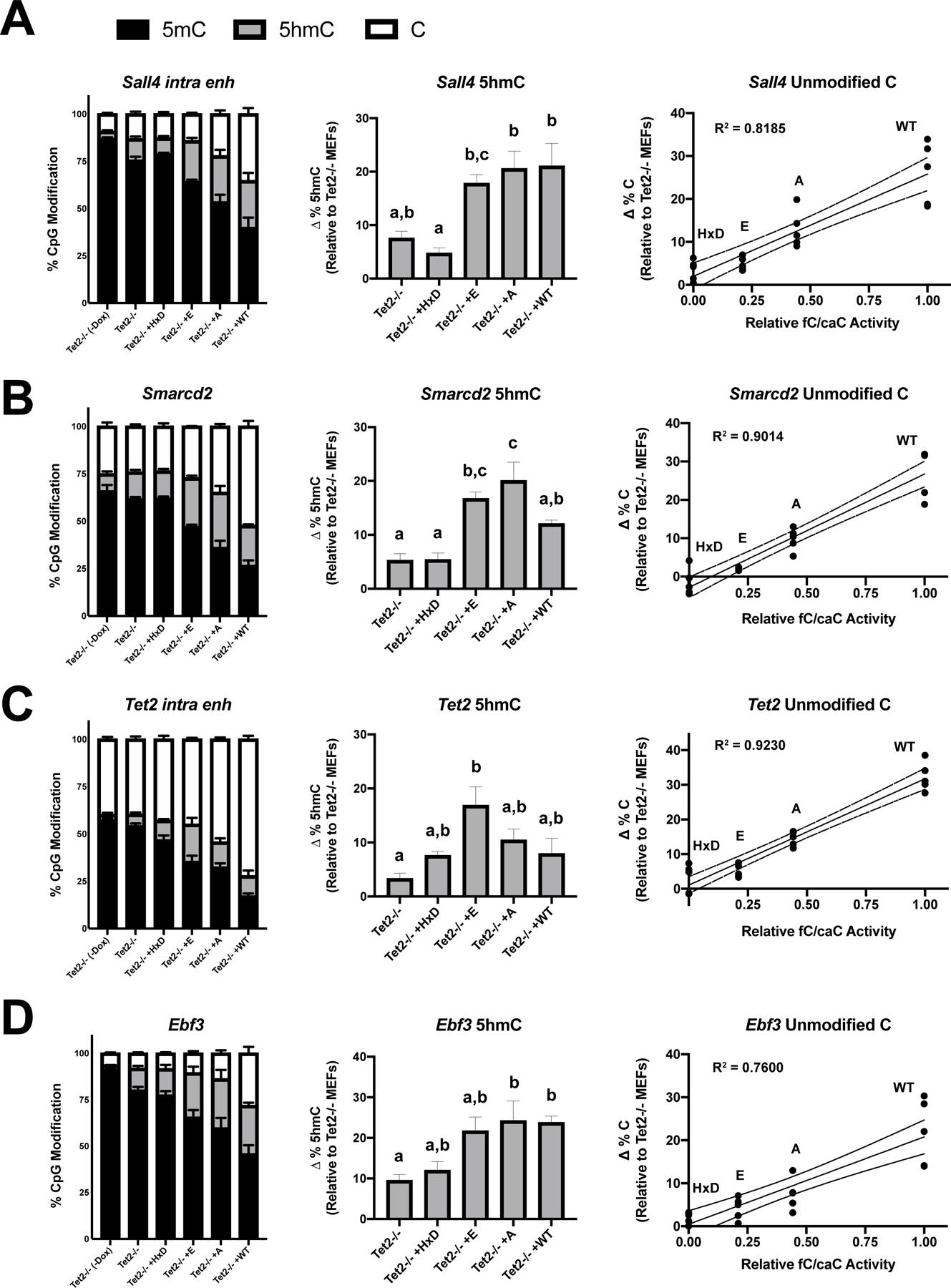
Relative levels of 5mC, 5hmC, and unmodified cytosine at (A) Sall4, (B) Smarcd2, (C) Tet2, and (D) Ebf3 proximal enhancers in untreated Tet2−/− MEFs or retrovirally transduced cells at day 5, as measured by combined BS-, bACE-, and MAB-pyrosequencing. For the central and right panels, change in 5hmC and unmodified cytosine levels were calculated by subtracting mean levels in untreated Tet2−/− MEFs. Bar graphs represent mean ± SEM, with letters designating statistically distinct groups (one-way ANOVA with Tukey multiple comparisons). For the right panel, data points represent independent experiments plotted against relative fC/caC activity values determined from HEK overexpression experiments (Fig. 1E). Simple linear regressions were performed for each enhancer, with the line of best fit (solid) and 95% confidence intervals (dotted) indicated (n=4–5).
Given that unmodified cytosine and 5fC/5caC are indistinguishable by BS, we also performed M.SssI Methylation-Assisted pyrosequencing (MAB-seq) of these four loci. However, 5fC/5caC levels were below our detection limit (<5% of cytosines) for all tested conditions, suggesting these bases are still efficiently removed by TDG in reprogramming Tet2−/− MEFs (Figure S3A; Table S5). Thus, by subtracting the BS and bACE signals, we can attribute the remaining signal to unmodified cytosine. For all four enhancers, we observed a strong linear correlation between the fC/caC activity of a given TET2 mutant (defined in Figure 1E) and the proportion of unmodified cytosine generated (lack of fit F-test p > 0.05; n = 4–5). This relationship was not observed when unmodified cytosine levels were instead plotted against total TET2 catalytic activity (Figure S3B; lack of fit F-test p < 0.05; n = 4–5). A similar effect was also observed at the miR200b cluster promoter, whose activities are thought to regulate MET during iPSC reprogramming (Figure S3C–D) (Hu et al., 2014). Our data therefore suggest that the degree of DNA demethylation at TET2 target regions during iPSC reprogramming correlated most strongly with the ability of the TET mutant to generate the higher order oxidation products 5fC and 5caC.
To test the effect of TET2 catalytic activity on the methylation status of known pluripotency loci, we performed bisulfite sequencing of the Oct4 promoter and Nanog intron 1 (Figure S3E). Although the average DNA methylation at these loci was unaffected after 5 days of Dox treatment, we observed a trending increase in lowly methylated clones among Tet2-WT cells. This effect likely reflects an expansion of rare, stably pluripotent cells in the Tet2-WT population, consistent with increased reprogramming efficiency.
Influence of TET2 catalytic activity on local chromatin accessibility
In addition to DNA demethylation, MEFs undergo significant alterations in chromatin accessibility during reprogramming. To examine how TET2 activity might influence local chromatin accessibility, we performed ATAC-seq on reprogramming Tet2−/− MEFs transduced with empty vector, Tet2-WT, or Tet2-E. Cells collected at days 5 and 10 of reprogramming were sorted for Stage-Specific Embryonic Antigen-1 (SSEA-1) to enrich for reprogramming intermediates on trajectory to complete reprogramming (Brambink et al., 2008). The ATAC-seq datasets were then cross-referenced with a previously published iPSC day 4 hydroxymethylated DNA immunoprecipitation (hMeDIP) dataset to identify iPSC-specific ATAC peaks overlapping regions enriched for 5hmC, which marks putative sites of TET2 activity (Figure 4A) (Sardina et al., 2018).
Figure 4 – Differential requirements for fC/caC activity for chromatin opening during reprogramming.
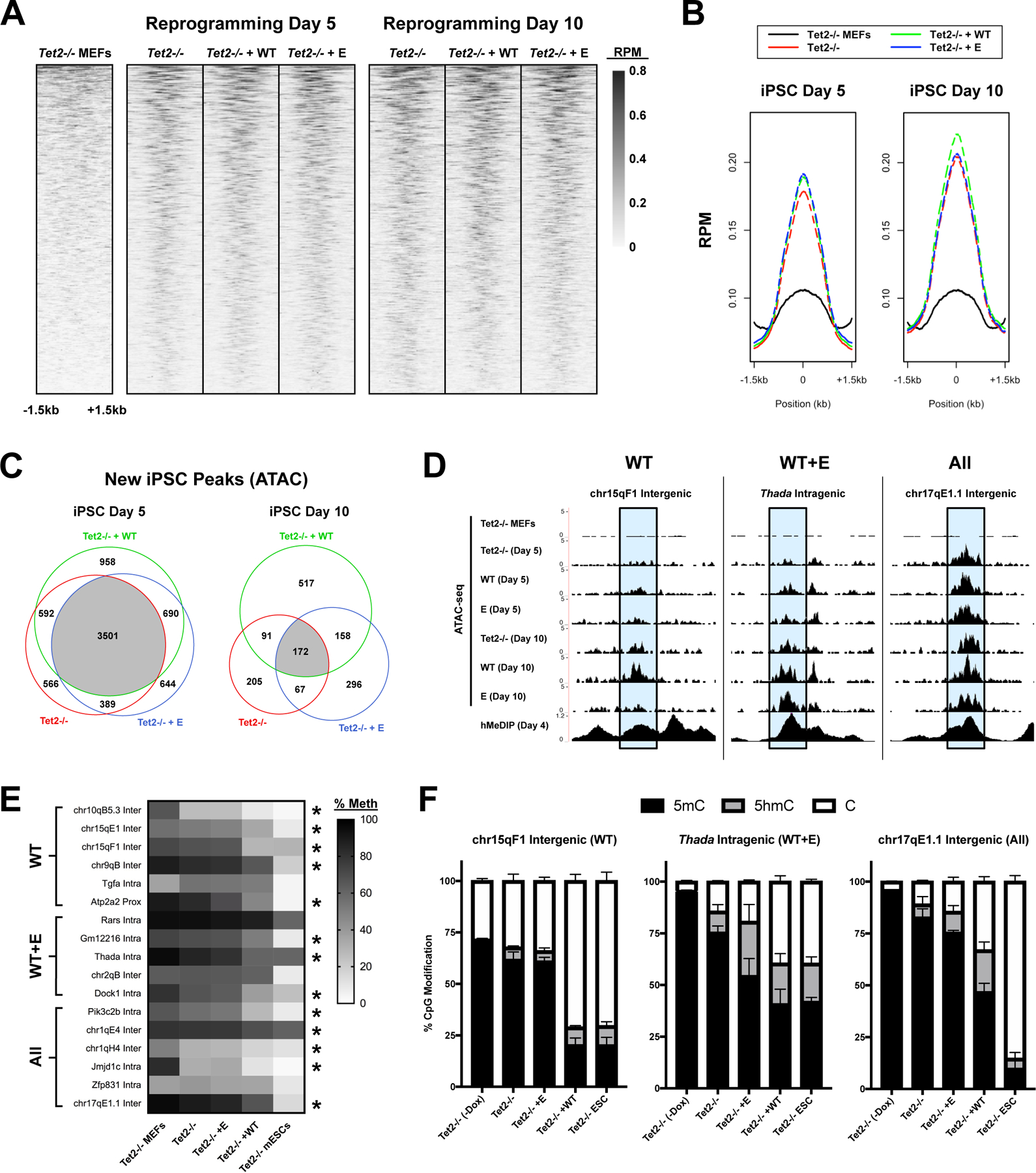
(A) Heatmaps of ATAC-seq signal at iPSC-specific accessible regions overlapping 5hmC peaks (GEO: GSE103470) (Sardina et al., 2018), measured in reads per million (RPM). Plots are individually sorted, centered at 5hmC peak summits, and represent the average of 4 biological replicates. (B) Metaplot of ATAC-seq signal at 5hmC peaks. (C) Venn diagrams of highly reproducible ATAC-5hmC peaks (present in ≥ 3/4 replicates) between different conditions. Peaks are subdivided based on the time point at which the ATAC peak was first observed. (D) Representative examples of WT-specific (WT), WT- and E-specific (WT+E), and shared (All) ATAC peaks overlapping 5hmC peaks (shaded in blue). UCSC genome browser snapshots include ATAC signal (merge of 4 biological replicates) for each condition at day 5 or 10, as well as hMeDIP signal at iPSC day 4 (Sardina et al., 2018). (E) Heatmap of mean DNA methylation (5mC+5hmC) at ATAC-5hmC peaks (n=3–4; * significantly reduced in WT vs. MEFs / Tet2−/− / E; one-way ANOVA with Tukey multiple comparisons). (F) Relative level of unmodified cytosine, 5hmC, and 5mC at representative ATAC-5hmC peaks from (D) in untreated Tet2−/− MEFs, untreated Tet2−/− mESCs, or transduced Tet2−/− cells at day 5 (n=3–4).
At day 5 of reprogramming, Tet2-WT or Tet2-E expression increased the average ATAC signal at 5hmC peaks as compared to empty vector control (Figure 4B). Focusing on reproducible ATAC peaks (present in ≥ 3/4 replicates), we noted that nearly half of all iPSC-specific peaks (48%) are shared by all three conditions, suggesting accelerated opening in the Tet2-WT and Tet2-E conditions rather than unique opening events (Figure 4C). Based on proximity to nearest gene promoters, only 5 ATAC peaks unique to Tet2-WT cells were linked to upregulated genes at day 5 (Adh7, AU018091, Avil, Rap1gap, and Scng), indicating early transcriptional differences are driven primarily by DNA demethylation at shared regions rather than WT-specific chromatin decompaction.
By day 10 of reprogramming, however, the average ATAC signal was higher at 5hmC peaks in Tet2-WT cells relative to empty vector and Tet2-E (Figure 4B). Consistently, 34% of new ATAC peaks are unique to Tet2-WT cells, suggesting an increased reliance on the fC/caC pathway to promote chromatin opening at later time points (Figure 4C). GO enrichment analysis of genes proximal to WT-specific peaks identified several key pathways involved in iPSC reprogramming, including Wnt signaling and epithelial cell proliferation (Table S6) (Marson et al., 2008). Notably, we observed a WT-specific increase in chromatin accessibility at two distal Sox2 enhancers, likely representing an expansion of the stably pluripotent iPSC population (Figure S4A).
To investigate the relationship between chromatin accessibility and DNA demethylation, we defined three groups of 5hmC-ATAC peaks: 1) Increased accessibility with Tet2-WT expression (≥1.5-fold increase in signal relative to Tet2-E and vector control), 2) Increased accessibility with Tet2-WT or Tet2-E expression, and 3) Accessible in all three conditions (i.e. Tet2-independent) (Figure 4D). We then selected 17 candidate regions exhibiting these features and measured the change in C, 5mC, and 5hmC levels at day 5 of reprogramming relative to levels in untreated Tet2−/− MEFs and mouse embryonic stem cells (mESCs) (Figure 4E–F, S4B–D). Surprisingly, regardless of their pattern of chromatin accessibility, all regions with appreciable demethylation by day 5 (13/17 regions) experienced increased DNA demethylation in Tet2-WT cells relative to Tet2-E or vector. Only one region (Atp2a2 proximal) showed evidence of accelerated demethylation by Tet2-E relative to vector. Additionally, 5hmC-ATAC regions where opening appeared to be independent of TET2 also tended to exhibit TET2-independent DNA demethylation (4/6 regions). Collectively, these data suggest that while Tet2-WT expression is associated with increased chromatin accessibility at 5hmC-enriched regions, chromatin accessibility in these regions isn’t necessarily coupled to TET2-dependent DNA demethylation. Instead, our results support the involvement of the fC/caC pathway as a shared driver of DNA demethylation during iPSC reprogramming (Figure S4E).
DISCUSSION
Since the discovery of TET enzymatic activity more than a decade ago (Tahiliani et al., 2009), the relative contribution of the fC/caC pathway to active DNA demethylation has been vigorously debated. In part, this stems from the steady-state abundance of genomic 5hmC compared to 5fC and 5caC, even under conditions of TDG depletion (Wagner et al., 2015; Wu et al., 2014; Xia et al., 2015). Research has also been limited by a lack of tools to uncouple TET’s oxidative products (Crawford et al., 2016). Here, we leveraged our biochemically defined TET catalytic mutants to establish that 5hmC generation alone is insufficient to drive the epigenetic changes observed in iPSC reprogramming. Instead, the fC/caC pathway promotes rapid DNA demethylation at reprogramming loci, thereby supporting a more efficient cell state transition.
Why would the fC/caC pathway more effectively contribute to chromatin reorganization? One tangible advantage is that its independence from cell division could enable a more rapid and robust transition to the unmethylated state. Through stable, transcription factor-mediated recruitment (as with TET2-KLF4), multiple proximal CpG dyads could be converted within a single cell cycle, thereby generating an unmethylated platform for subsequent epigenetic alterations. Alternatively, BER of 5fC/5caC may promote chromatin reorganization beyond 5mC erasure. TDG can recruit histone modifiers such as p300/CBP to generate active H3K27ac marks, and because TET-mediated 5mC oxidation is least efficient for nucleosome-bound DNA, chromatin remodeling would also be expected to have a positive feed-forward effect on DNA demethylation (Kizaki et al., 2016; Lake et al., 2016; Tini et al., 2002).
We also used a novel approach to resolve 5hmC signal from 5mC. In examining 5hmC patterns, it is notable that even during the early stages of iPSC reprogramming, MEFs undergo at least one cell cycle per day (Hanna et al., 2009). Despite having undergone numerous cellular divisions by iPSC reprogramming day 5, however, we observe 5hmC accumulation at reprogramming regions in the 5hmC-stalling condition rather than DNA demethylation, suggesting the hmC pathway is unable to promote significant levels of DNA demethylation. Interestingly, a recent study on the substrate preferences of maintenance methyltransferase DNMT1 found that its catalytic activity on an unmodified CpG is reduced only 3-fold when positioned opposite a 5hmCpG relative to 5mCpG, highlighting the potential inefficiency of 5hmC-driven passive dilution (Seiler et al., 2018).
In summary, our results demonstrate that the fC/caC pathway is a major driver of epigenetic changes during iPSC reprogramming. Importantly, this study represents the first evidence of biological activity specifically attributable to one mode of TET oxidative activity, and suggests that the hmC and fC/caC demethylation pathways may have distinct functions in regulating epigenetic identity and cell fate.
LIMITATIONS
Tet2−/− iPSC reprogramming rescue experiments were performed through retroviral overexpression of the Tet1 or Tet2 catalytic domains, as opposed to the full-length protein. Because the N-terminal domain has been proposed to regulate protein-protein interactions, it is possible that loss of these non-catalytic interactions may contribute to discrepancies between our reprogramming system and that of WT MEFs. Furthermore, because we focused our 5mC/hmC/fC/caC sequencing analyses on a discrete set of regions implicated in iPSC reprogramming, high-throughput sequencing approaches will be required in the future to determine how the entire genome is influenced by TET2-WT vs. stalling TET2-E expression. Finally, because RNA-seq was performed only on reprogramming day 5 cells, we are unable to evaluate whether WT-specific changes in ATAC signal at day 10 are reflected in transcriptional changes at later time points. Understanding the interplay of DNA methylation and chromatin accessibility at these sites and how they mediate the establishment of poised or active gene states will be an important topic for future study.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Marisa S. Bartolomei (bartolom@pennmedicine.upenn.edu).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
The accession number for raw and processed RNA- and ATAC-sequencing data reported in this paper is GEO: GSE158334. Source data and image files for all figures in the paper are available at Mendeley Data (DOI: 10.17632/42kc23sjy9.1).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture
HEK293T, NIH3T3, PLAT-E, and primary MEFs were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with 2mM GlutaMAX (Gibco) 10% fetal bovine serum (FBS), and 1x Pen/Strep (Gibco) at 37°C in atmospheric oxygen. Mouse ESCs and reprogramming iPSCs were maintained on Mito-C feeder cells and cultured in DMEM supplemented with 15% heat-inactivated FBS (Gibco), 2mM GlutaMAX, 0.1mM non-essential amino acids (Gibco), 1mM sodium pyruvate (Gibco), 0.1mM 2-mercaptoethanol (Gibco), 1x Pen/Strep, and 1000U/mL ESGRO recombinant mouse LIF (Sigma-Aldrich). Reprogramming iPSCs were supplemented with 1 µM PD0325901 (Stemolecule) and 3 µM CHIR99021 (Sigma-Aldrich), as well as 2 µg/mL doxycycline (Dox) (Stem Cell) prior to Dox withdrawal. Primary MEFs and reprogramming iPSCs were kept in a low (5%) oxygen 37°C incubator for improved cell growth and survival.
Derivation of Tet2−/− OKSM; rtTA MEFs
Wild-type KH2-OKSM; rosa26:M2rtTA (OKSM; rtTA) mESCs were previously described (Stadtfeld et al. 2010). These are male XY cells in a V6.5 129XBI/6 hybrid background. In order to generate Tet2−/− OKSM; rtTA mESCs, we co-transfected wild-type OKSM; rtTA mESCs with pX330 vectors encoding guide RNAs for introns 8 and 10 of Tet2, as well as the ploxPneo-1 selection marker, using Lipofectamine 3000. Transfected mESCs were grown in the presence of 200 µg/mL geneticin (Gibco) for 5 days, at which point individual clones were picked for expansion. We screened for homozygous Tet2−/− OKSM; rtTA mESC clones using a PCR surveyor assay, followed by sequencing of candidates to confirm excision of Tet2 exons 9 and 10.
Wild-type or homozygous Tet2−/− OKSM; rtTA mESCs were injected into mouse E3.5 blastocysts to generate chimeric embryos. MEFs were prepared from E12.5 chimeric embryos and grown in the presence of 2 µg/mL puromycin (Takara Bio USA) for 48 hrs to select for Tet2−/− OKSM; rtTA MEFs.
METHOD DETAILS
Plasmid construction
The wild-type and HxD Tet2-catalytic domain (CD) pMXs retroviral vectors were a gift from Dr. Guo-Liang Xu (Hu et al. 2014). To generate the Tet2-CD T1285E/A pMXs vectors, we performed site-directed mutagenesis using the Agilent QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent). N-terminal FLAG tags were added with PCR primer adapters to each of these Tet2-CD mutants followed by restriction cloning to integrate them into pLEXm mammalian expression vectors. Tet1-CD was PCR amplified from mouse embryonic stem cell (mESC) cDNA and cloned into pFastBac1. We next performed site-directed mutagenesis to generate the Tet1-CD T1642E/V/A and HxD catalytic mutants, introduced N-terminal FLAG tags as with Tet2-CD, and used restriction cloning to integrate them into pLEXm expression and pMXs retroviral vectors.
For CRISPR/Cas9 mutagenesis, the ploxPneo-1 selection marker was a gift from Dr. Shirley Tilghman (Nagy et al. 1998). pX330 hSpCas9 CRISPR targeting vectors for Tet2 introns 8 and 10 were generated from restriction cloning of T4-phosphorylated (NEB) annealed oligos. All vector sequences were confirmed by sequencing.
TET overexpression in HEK293T and NIH3T3 cells
We transfected HEK293T and NIH3T3 cells in 6-well plates with 2.5 µg of Tet1/2-CD mutant pLEXm vector or empty vector control using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s protocol. 24 hrs after transfection, we changed media in each of the wells, and harvested cells by trypsinization 48 hrs after transfection. Cells were washed in phosphate-buffered saline and split into one of two downstream treatments: 1.) Cells were lysed using CytoBuster Protein Extraction Reagent (Millipore) for Western blots, or 2.) Genomic DNA was extracted by phenol-chloroform extraction.
Slot blots for cytosine modifications in genomic DNA
We performed slot blots for cytosine modifications in genomic DNA as previously described (Liu et al. 2016). Membranes were blotted at 4°C overnight with primary antibodies against 5hmC (1:10,000 rabbit anti-5hmC, Active Motif) or 5caC (1:5,000 anti-5caC, Active Motif), washed, and incubated with secondary goat anti-rabbit-HRP (Santa Cruz Biotechnology) for 2 hrs at room temperature. Notably, the instability of 5fC limits the ability to characterize this modification well by slot blot analysis. The membranes were then washed, incubated with Immobilon HRP Chemiluminescent Substrate (Millipore) according to the manufacturer’s instructions, and imaged using an Amersham Imager 600.
LC-MS/MS analysis of DNA
Briefly, to quantify genomic levels of modified cytosines in transfected HEK293T or NIH3T3 cells, we concentrated 1 – 1.5 µg of purified DNA by ethanol precipitation and degraded samples to component nucleosides with Nucleoside Digestion Mix (NEB) at 37°C overnight. The mixture was diluted 10-fold into 0.1% formic acid. LC-MS/MS was performed as previously described in DeNizio and Liu et al. 2019. Standard curves were generated from individual nucleosides (Berry & Associates), and sample peak areas were fit to the standard curve to determine amounts of each modified cytosine in the DNA sample. Each cytosine modification was expressed as the percent of total cytosine modifications in each sample.
Western blots for FLAG-tagged TET1 and TET2 mutants.
Transfected cells were lysed using CytoBuster Protein Extraction Reagent. The lysates were then diluted 100-fold into 10 µg protein in 20 µL of CytoBuster and run on a 4–12% SDS-PAGE gel, with empty vector-transfected samples as a control. The gel was transferred onto a PVDF membrane at 200 mAmps for 120 min. After transfer, the membrane was cut at the 50-kDa marker such that the upper half contained the TET1- and TET2-CD mutant protein bands and the bottom half contained the α-GAPDH loading control. Membranes were blocked separately for 1 hr at room temperature with 5% non-fat dry milk in Tris-buffered saline with 0.1% Tween-20 (TBST). The membranes were then blotted in 5% non-fat dry milk in TBST with either 1:10,000 mouse anti-FLAG M2 (Sigma-Aldrich) or 1:10,000 rabbit anti-GAPDH (Cell Signaling Technology) primary antibodies at 4 °C overnight. The following day, membranes were washed 3x in TBST, blotted with 1:10,000 mouse-IgGκ BP (Santa Cruz Biotechnology) or 1:2,500 goat anti-rabbit IgG (Cell Signaling Technology) secondary antibodies for 2 hrs at room temperature, washed 3x in TBST, and imaged with Immobilon Western Chemiluminescent HRP Substrate on an Amersham Imager 600.
Retroviral transduction and iPSC reprogramming
Plat-E retroviral packaging cells were seeded at a density of 6.5 × 105 cells / well on 6-well plates and, after 24 hrs, transfected with 1.5 µg of pMXs retroviral construct using Lipofectamine 3000. Cell media was replaced 10 hrs after initial transfection. Retroviral media was collected 48 and 72 hrs after transfection and filtered through a 0.45-µm PES filter. Primary wild-type or Tet2−/− OKSM; rtTA MEFs (seeded 48 hrs prior at a density of 5 × 104 cells / well on 6-well plates) were incubated for 24 hrs with 200 µL of retroviral solution and 1.8 mL MEF media, supplemented with 4 µg/mL polybrene (American Bio). Following two rounds of infection, cells were seeded on Mito-C-treated MEF feeder cells at a density of 5 × 103 cells / well on 6-well plates.
After 24 hrs, the retrovirus-transduced MEFs were moved into iPSC media (2i/LIF+Dox) to initiate reprogramming. Cell media was changed every day for the duration of reprogramming. After 3 days of Dox treatment, rates of cell proliferation and apoptosis were determined using the Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay Kit (Invitrogen) and Dead Cell Apoptosis Kit with Annexin V FITC and PI (Invitrogen), respectively, on a BD Accuri C6 Plus flow cytometer. To assay early pluripotency, we performed alkaline phosphatase (AP) staining on day 10 of reprogramming using the StemAb Alkaline Phosphatase Staining Kit II (Stemgent). AP+ colonies were counted using ImageJ. After 11 days of treatment, reprogramming cells were moved into iPSC media without Dox for one week to test for stable pluripotency.
NANOG fluorescent immunohistochemistry
Reprogramming day 18 6-well cultures were washed in 1x phosphate-buffered saline, 0.1% Tween-20 (PBST) and fixed in 4% paraformaldehyde for 15 mins at room temperature. Cells were then washed 3x in ice cold PBST, and permeabilized for 10 mins in 0.2% Triton X-100 (Supelco). Permeabilized cells were washed 2x in PBST and blocked for 1 hr in 10 mg/mL bovine serum albumin (BSA, Sigma-Aldrich). Cells were next incubated overnight at 4°C with primary antibodies against Nanog (1:100 rabbit anti-Nanog, Abcam), washed in PBST, and incubated for 1 hr at room temperature with goat anti-rabbit IgG H&L Alexa Fluor 488 (1:500, Abcam). The cells were then washed and imaged on an Amersham Imager 600. Nanog+ colonies were counted manually using Fiji.
RNA extraction, reverse transcription, and qRT-PCR
Total RNA from reprogramming day 5 cells was extracted using the RNeasy Mini Kit (QIAGEN). RNA samples were treated with DNaseI recombinant (Roche) and reverse transcribed with Superscript III Reverse Transcriptase (Invitrogen) according to the manufacturer’s protocol. Quantitative real-time PCR (qRT-PCR) was performed using Power SYBR Green Master Mix (Applied Biosystems) on a QuantStudio 7 Flex Real-Time PCR system. Relative expression levels were determined using the Pffafl method normalized to the housekeeping gene Nono.
RNA sequencing (RNA-seq)
Total RNA (4 µg) extracted from reprogramming day 5 cells was processed for RNA-seq using a KAPA Stranded mRNA-Seq Kit (Roche) according to the manufacturer’s protocol, with the following alterations. For step 3.1, mRNA was fragmented at 85°C for 6 min to generate 301–400 bp fragments. For step 8, indexing was performed using a KAPA Single-Indexed Adaptor Kit (Roche), with 8 cycles of library amplification. All purification steps were completed using Ampure XP beads (Beckman). Sequencing libraries were prepped in two sets, with the RNA in each set randomized. Prior to sequencing, each library was analyzed with a Bioanalyzer High-Sensitivity DNA chip (Agilent) and KAPA Library Quantification Kit (Roche) to establish library quality. Libraries were sequenced on an Illumina NextSeq 500 using 75 bp paired-end sequencing (40 × 40 cycles).
Bisulfite-assisted APOBEC3A-Coupled Enzymatic (bACE) pyrosequencing
Genomic DNA from reprogramming day 5 cells was collected by phenol-chloroform extraction. Relative 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) levels at genomic loci of interest were determined using a modified APOBEC-Coupled Enzymatic (ACE) sequencing methodology (Schutsky et al. 2018). Validation of bACE pyrosequencing was also performed with a spike-in oligonucleotide as described in Figure S3A with 5hmC at sites 5, 6, and 7. For each sample, we bisulfite treated 200 ng of genomic DNA, with 100 pg each of CpG methylated (meth) lambda and T4 phage spike-in DNA controls added, using the EpiTect Bisulfite Kit (QIAGEN). Samples were eluted in 20 µL 0.1x EB (1 mM Tris-Cl, pH 8.5), and 2 µL of bisulfite treated DNA were incubated at 37°C for 2 hrs in a 50 µL reaction of 0.5 µM recombinant MBP-APOBEC3A (A3A)-His in 35 mM SPG (2:7:7 succinic acid:sodium dihydrogen phosphate:glycine) pH 5.5, 0.1% Tween-20, 2% DMSO. This MBP-A3A-His fusion protein was purified with a C-terminal His tag as previously described in Wang et al. 2020. The A3A-treated bisulfite DNA was then purified using the Zymo Oligo Clean & Concentrator kit (Zymo Research) and eluted in 15 µL 1x EB. The bisulfite and bisulfite+A3A DNA fractions for each sample were separately processed for pyrosequencing as previously described (de Waal et al. 2014). Briefly, 1 µL of bisulfite or bisulfite+A3A DNA was used as template for PCR amplification of regions of interest using the PyroMark PCR Kit (QIAGEN) and sequenced on a PyroMark Q96 MD instrument. Relative 5mC levels at a given locus were determined by subtracting percent 5hmC levels determined by bACE pyrosequencing from percent 5mC/5hmC levels determined by standard bisulfite pyrosequencing. To confirm the efficiency of A3A-mediated 5mC conversion and protection of bisulfite-converted 5hmC CMS adducts, we performed pyrosequencing of meth lambda and T4 phage bACE-treated spike-in DNA for each sample. bACE treatment and pyrosequencing was repeated for any samples returning percent cytosine values ≥ 10% for meth lambda phage or ≤ 90% for T4 phage.
Methylase-assisted bisulfite (MAB) pyrosequencing
Relative 5fC and 5caC levels at genomic loci of interest were determined using a modified MAB sequencing methodology (Wu et al. 2016). Efficiency of CpG methylation by the methyltransferase M.SssI (NEB) was assessed using unmethylated lambda phage spike-in (Promega) and a spike-in oligonucleotide as described in Figure S3A with 5caC at site 5, 6, and 7. For each sample, 500 ng of genomic DNA, 1.25 ng of unmethylated lambda spike-in (0.25% w/w), and 1.25 ng oligonucleotide spike-in (0.25% w/w) was incubated at 37°C for 4 hrs in a 50 µL reaction containing 20 units of M.SssI, 32 µM S-adenosylmethionine (SAM) in 1X NEBuffer 2. An additional 5 units of M.SssI and 32 µM of SAM were added, followed by a second 4 hrs incubation at 37°C. M.SssI treated gDNA was purified using 1.8X AMPure XP bead and eluted in 40 µL of 0.1X EB. M.SssI treatment was followed by bisulfite mutagenesis using EpiTect Bisulfite Kit (QIAGEN). PCR amplification and pyrosequencing are as described in bACE pyrosequencing method.
Fast-ATAC of SSEA1+ reprogramming MEFs
To isolate SSEA1+ reprogramming MEFs, cells were harvested by trypsinization after 5 or 10 days of Dox treatment, filtered through a Falcon 100 µm strainer, and incubated for 30 mins on ice in 1.2 ng/µL SSEA1 eFluor 660 Monoclonal antibody (Invitrogen). Cells were then washed twice and resuspended in 1x PBS + 0.1% BSA for fluorescence activated cell sorting (FACS) on a BD FACSJazz cell sorter.
SSEA1+ reprogramming cells were next applied directly to the Fast-ATAC optimized ATAC-seq workflow (Corces et al. 2016), with the following modifications. Post-FACS cells were washed 1x in MEF media, and then 75,000 cells were resuspended in 1 mL warm RPMI media (Gibco). Following a 5 min spin at 4°C (500 x g), cells were washed with 1 mL ATAC-RSB (10 mM Tris-HCl pH 7.4, 10 mM NaCl, and 3 mM MgCl2 + 0.1% Tween), and then resuspended in TDE1 transposase solution (Illumina) + 0.01% digitonin. The transposase mix was incubated at 37°C with shaking for 30 mins, after which the transposed DNA was purified using a MinElute Reaction Cleanup Kit (QIAGEN). Transposed fragments were amplified for 8 cycles using indexed primers according to Buenostro et al 2015, and then purified by AMPure XP bead selection (Beckman Coulter). ATAC libraries were quantified using the NEBNext Library Quant Kit (NEB) and assayed using a Bioanalyzer High Sensitivity DNA chip (Agilent) to establish library quality. Pooled libraries were sequenced on an Illumina NextSeq 500 using a High Output Kit v2.5 (75 cycles) (Illumina).
bACE next-generation sequencing
To supplement our bACE pyrosequencing analysis of 5mC and 5hmC levels in day 5 reprogramming MEFs, we performed targeted next-generation sequencing of select reprogramming loci using a modified BisPCR2 workflow (Bernstein et al. 2015). Genomic DNA was subjected to bisulfite +/− A3A as described above and then used as a template for target enrichment using the PyroMark PCR Kit. Amplified regions were pooled for column purification (4–6 regions per pool, for a total of 150ng), and the purified pools were barcoded with indexing primers using a Multiplex PCR Kit (QIAGEN). All indexed pools for a given sample were then pooled once more for column purification, after which library quality was determined using a Bioanalyzer DNA 1000 chip (Agilent). Finally, all indexed libraries were pooled and sequenced on an Illumina MiSeq using a MiSeq Reagent Nano Kit v2 (500 cycles) (Illumina).
QUANTIFICATION AND STATISCIAL ANALYSIS
Statistics were performed using GraphPad Prism. Comparisons of > 2 groups was performed using one-way ANOVA adjusted for Tukey’s multiple comparisons, with the exception of EdU and Annexin-V flow cytometry data, which was corrected using Sidak’s multiple comparisons test based on the assumption that each comparison is independent of the other. In all figures, statistical significance in comparisons between > 3 groups is indicated by lowercase letters; groups with different letters denote significant differences between groups, while shared letters indicate no difference was detected. Information on statistical tests performed, exact values of n, and how significance was defined is available in the figure legends. For cell culture experiments, n is defined as either cultures treated with separately prepared transfection or retroviral solutions, or cultures derived from different frozen stocks (EdU and Annexin-V staining experiments). All experiments were performed on at least three separate days.
Flow cytometry
Data were manually analyzed using standard BD Accuri C6 software (https://www.bdbiosciences.com/instruments/accuri/features/software.jsp).
AP staining and Nanog immunofluorescence image quantitation
AP staining and Nanog immunofluorescence image quantitation was performed using Fiji (https://imagej.net/Fiji/Downloads). AP staining images were background subtracted (rolling ball radius = 40 pixels), binarized using threshold values to match colonies patterns of the brightest and dimmest samples, watershedded, and counted using the Analyze Particles tool (size = 15 pixel units to infinity; circularity = 0–1.00). Nanog colony counts were manually performed using the Grid feature.
RNA-seq analysis
Paired-end reads were aligned against reference genome mm9 with STAR (Dobin et al. 2013; version 2.6.1a), using default parameters and allowing maximum fragment lengths of 2000 bp. Only correctly paired primary aligned pairs were retained for downstream analysis, which were filtered with SAMtools (Li et al., 2009; version 1.7). Alignment BAM files were converted to BED files and mitochondria reads were removed with BEDtools (Quinlan and Hall 2010; version 2.27.1). Bedgraph files were generated using BEDtools, and each library was normalized to 10 million reads for visualization. Read count for each RefSeq gene was quantified with featureCounts (Liao et al. 2014; version 1.6.2) using default parameters. Differential expression analysis was performed with using the R package DESeq2 (Love et al., 2014; version 1.26.0). GO analysis of selected gene sets was performed with the R package clusterProfiler (Yu et al., 2012; version 3.14.3).
ATAC-seq analysis
ATAC-seq reads were aligned using Bowtie2 (Langmead and Salzberg 2012) (default parameters, paired-end mode). Duplicate reads were removed using SAMTools (Li et al. 2009) (function: rmdup), along with mitochondrial reads and reads in blacklist regions. Bedgraph files normalized to reads per million (RPM) were prepared using BEDTools (Quinlan and Hall 2010) (function: genomecov; parameters: -bg -scale 1000000/READ_COUNT), and then converted to Bigwig tracks for UCSC browser visualization (Kent et al. 2010; http://genome.ucsc.edu).
Peak calling was performed using macs2 (Zhang et al. 2008) (function: callpeak; parameters: -f bed -g mm --qvalue 0.01 --bdg --SPMR) (function: bdgcmp -c control_lambda -m subtract). Peaks from reprogramming day 5 and 10 were merged as one set of non-overlapping 500bp windows (+/−250bp from peak summit) using Bedops (Neph et al. 2012). iPSC-specific peaks were determined by identifying non-overlapping regions between this merged list and untreated Tet2−/− MEF narrowPeak replicates (n=4) using BEDTools (function: intersect; parameters: -c -F 0.25). From this iPSC-specific peak list, regions overlapping 5hmC peaks (Sardina et al. 2018; GSE117919; iPSC day 4 5hMeDIP signal ≥ 0.2 RPKM across 2 replicates) were identified using BEDTools (function: intersect; parameters: -wa). Highly reproducible 5hmC-ATAC peaks for each condition and time point were defined as regions with ≥ 3 overlapping narrowPeak replicates (function: intersect; -c -F 0.5). GO analysis of genes proximal to 5hmC-ATAC peaks was performed with the R packages ChIPSeeker (Yu et al., 2015; version 1.22.1) and clusterProfiler (Yu et al., 2012; version 3.14.3).
To generate heatmaps for ATAC signal at 5hmC peaks, 5hmC-ATAC regions were expanded to non-overlapping 3kb windows (+/−1.5kb from 5hmC peak center) with ≥ 2 overlapping ATAC peaks in any iPSC day 5 or 10 sample (22,108 windows total) and used to calculate read coverage with the Rpackage GenomicRanges (Lawrence et al. 2013) (function: peak.coverage; parameters: bins=3000, RPM=T). We then generated heatmaps using the Rpackage pheatmap (https://www.rdocumentation.org/packages/pheatmap), as well as metaplots for average ATAC signal at 5hmC-ATAC peaks across replicates for each condition and timepoint.
bACE next-generation sequencing analysis
Sequenced reads were trimmed using Trim Galore (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore) and mapped with Bismark (version 0.22.3; https://www.bioinformatics.babraham.ac.uk/projects/bismark) in paired-end mode. As with our previous ACE pipeline, non-deaminated reads were filtered out based on the presence of ≥ 3 consecutive instances of non-CG methylation, although this was not observed in any of our tested samples (function: filter_non_conversion; parameters: --paired --consecutive). Bedgraph files were prepared using the Bismark Methylation Extractor to calculate percent methylation at each CpG with ≥ 30x coverage.
Supplementary Material
Table S3. Bisulfite analysis regions, related to STAR Methods.
Table S2. Primer list, related to STAR Methods.
Table S5. MAB pyrosequencing of spike-in control and Sall4, Smarcd2, Tet2, and EBf3 enhancers, related to Figure 3.
Table S6. ATAC-seq GO terms, related to Figure 4.
Table S4. RNA-seq and GO terms, related to Figure 2.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-5hmC | Active Motif | Cat # 39769; RRID:AB_10013602 |
| Rabbit polyclonal anti-5caC | Active Motif | Cat # 61225; RRID:AB_2793557 |
| Goat anti-rabbit IgG-HRP | Santa Cruz Biotechnology | Cat # sc-2004; RRID:AB_631746 |
| Mouse monoclonal anti-FLAG M2 | Sigma-Aldrich | Cat # F1804; RRID:AB_262044 |
| Rabbit monoclonal anti-GAPDH | Cell Signaling Technology | Cat # 2118S; RRID:AB_561053 |
| Mouse IgGκ BP-HRP | Santa Cruz Biotechnology | Cat # sc-516102; RRID:AB_2687626 |
| Goat anti-rabbit IgG-HRP | Cell Signaling Technology | Cat # 7074S; RRID:AB_2099233 |
| Rabbit polyclonal anti-Nanog | Abcam | Cat # ab80892; RRID:AB_2150114 |
| Goat anti-rabbit IgG H&L Alexa Fluor 488 | Abcam | Cat # 150081; AB_2734747 |
| SSEA1 eFluor 660 monoclonal antibody | Invitrogen | Cat # 50–8813-41; RRID:AB_11217669 |
| Biological Samples | ||
| Unmethylated lambda phage DNA | Promega | Cat # D152A |
| Methylated lambda phage DNA | Schutsky et al., 2018 | N/A |
| T4 phage DNA | Schutsky et al., 2018 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| T4 Polynucleotide Kinase | NEB | Cat # M0201S |
| GlutaMAX | Gibco | Cat # 35050061 |
| Fetal bovine serum (FBS) | TCB | Cat # 101 |
| Penicilin / Streptomycin | Gibco | Cat # 15140122 |
| Heat-inactivated FBS | Gibco | Cat # 10082147 |
| MEM non-essential amino acids | Gibco | Cat # 11140050 |
| Sodium pyruvate (cell culture) | Gibco | Cat # 11360070 |
| 2-Mercaptoethanol (cell culture) | Gibco | Cat # 21985023 |
| ESGRO recombinant mouse LIF | Sigma-Aldrich | Cat # ESG1107 |
| PD0325901 | Stemolecule | Cat # 04–0006 |
| CHIR99021 | Sigma-Aldrich | Cat # SML1046 |
| Doxycycline (cell culture) | Stem Cell | Cat # 72742 |
| Lipofectamine 3000 Transfection Reagent | Invitrogen | Cat # L3000001 |
| Cytobuster Protein Extraction Reagent | Millipore | Cat # 71009 |
| Immobilon HRP Chemiluminescent Substrate | Millipore | Cat # WBKLS0100 |
| Nucleoside Digestion Mix | NEB | Cat # M0649S |
| Geneticin selective antibiotic | Gibco | Cat # 10131035 |
| Puromycin selective antibiotic | Takara Bio | Cat # 631305 |
| Polybrene | American Bio | Cat # AB01643–00001 |
| Triton X-100 | Supelco | Cat # TX1568–1 |
| Bovine serum albumin, fraction V | Sigma-Aldrich | Cat # 12659 |
| DNase I recombinant | Roche | Cat # 4716728001 |
| Superscript III Reverse Transcriptase | Invitrogen | Cat # 18080–044 |
| RPMI media | Gibco | Cat # 11875119 |
| TDE1 transposase | Illumina | Cat # 15027865 |
| 2’-Deoxycytidine-5-Carboxylic acid sodium salt (LC/MS-MS) | Berry & Associates | Cat # PY 7593 |
| 5-Formyl-2’-deoxycytidine (LC/MS-MS) | Berry & Associates | Cat # PY 7589 |
| 5-Hydroxymethyl-2’-deoxycytidine (LC/MS-MS) | Berry & Associates | Cat # PY 7588 |
| 5-Methyl-2′-deoxycytidine Hydrochloride (LC/MS-MS) | Berry & Associates | Cat # PY 7635 |
| Critical Commercial Assays | ||
| Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay Kit | Invitrogen | Cat # C10424 |
| Dead Cell Apoptosis Kit with Annexin V FITC and PI | Invitrogen | Cat # V13242 |
| StemAb Alkaline Phosphatase Staining Kit II | Stemgent | Cat # 00–0055 |
| Power SYBR Green Master Mix (qRT-PCR) | Applied Biosystems | Cat # 4367659 |
| Recombinant MBP-APOBEC3A (A3A)-His | Schutsky et al., 2018; This paper | N/A |
| CpG Methyltransferase (M.SssI) | NEB | Cat # M0226S |
| NEBNext Library Quant Kit | NEB | Cat # E7630 |
| Bioanalyzer High-Sensitivity DNA chip | Agilent | Cat # 5067–4626) |
| High Output Kit v2.5 (75 cycles) | Illumina | Cat # 20024906 |
| Bioanalyzer DNA 1000 chip | Agilent | Cat # 5067–1504 |
| MiSeq Reagent Nano Kit v2 (500 cycles) | Illumina | Cat # MS-103–1003 |
| Deposited Data | ||
| 5hMeDIP-seq (iPSC day 4) | Sardina et al., 2018 | GSM3315344; GSM3315345 |
| RNA-seq (iPSC day 5) | This paper | GSE158334 |
| ATAC-seq (iPSC day 5 and 10) | This paper | GSE158334 |
| Caldwell et al., 2021 Original Data | This paper | DOI: 10.17632/42kc23sjy9.1 |
| Experimental Models: Cell Lines | ||
| Human: HEK293T cells | ATCC | Cat # CRL-11268; RRID:CVCL_1926 |
| Mouse: NIH/3T3 cells | ATCC | Cat # CRL-1658; RRID:CVCL_0594 |
| Human: PLAT-E retroviral packaging cells | Cell Bio Labs | Cat # RV-101; RRID:CVCL_B488 |
| Mouse: CF1 Mito-C feeder cells | This paper | N/A |
| Mouse: KH2-OKSM; rosa26:M2rtTA (OKSM; rtTA) mESCs | Stadtfeld et al., 2010 | N/A |
| Mouse: WT and Tet2−/− OKSM; rtTA primary MEFs | This paper | N/A |
| Oligonucleotides | ||
| Mouse Tet1 / Tet2 cloning primers (See Table S2 for primer sequences) | This paper | N/A |
| iPSC reprogramming qRT-PCR primers (See Table S2 for primer sequences) | This paper; Hu et al., 2014 | N/A |
| bACE-pyroseq/MiSeq primers (See Table S3 for primer sequences and genomic coordinates) | This paper | N/A |
| bACE 5mC/5hmC spike-in oligonucleotide (See Figure S3A for oligo info) | This paper | N/A |
| Recombinant DNA | ||
| Tet1-CD WT and T1642 / Y2049 / HxD mutant pLEXm expression vectors (See Table S1 for full list) | This paper | N/A |
| Tet2-CD WT and T1285 / HxD pLEXm expression vectors (See Table S1 for full list) | This paper | N/A |
| Tet1-CD WT and T1642V / HxD pMXs retrovirus vectors | This paper | N/A |
| Tet2-CD WT and HxD pMXs retrovirus vectors | Hu et al., 2014 | N/A |
| Tet2-CD T1285A/E mutant pMXs retrovirus vectors | This paper | N/A |
| Tet2 Int8a pX330 CRISPR vector | This paper | N/A |
| Tet2 Int10a pX330 CRISPR vector | This paper | N/A |
| ploxPneo-1 | Nagy et al., 1998 | N/A |
| Software and Algorithms | ||
| Fiji | Schinderlin et al., 2012 | https://imagej.net/Fiji/Downloads |
| STAR (version 2.6.1a) | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| Bowtie2 (version 2.2.5) | Langmead & Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| SAMTools (version 1.7) | Li et al., 2009 | http://samtools.sourceforge.net/ |
| BEDTools (version 2.27.1) | Quinlan & Hall, 2010 | https://github.com/arq5x/bedtools2/releases |
| featureCounts (version 1.6.2) | Liao et al., 2014 | https://www.rdocumentation.org/packages/Rsubread/versions/1.22.2/topics/featureCounts |
| DESeq2 (version 1.26.0) | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| clusterProfiler (version 3.14.3) | Yu et al., 2012 | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| ChIPseekers (version 1.22.1) | Yu et al., 2015 | https://www.bioconductor.org/packages/release/bioc/html/ChIPseeker.html |
| Bigwig tracks | Kent et al., 2010 | http://genome.ucsc.edu |
| macs2 (version 2.2.7.1) | Zhang et al., 2008 | https://github.com/taoliu/MACS/wiki/Install-macs2 |
| Bedops (version 2.4.39) | Neph et al., 2012 | https://bedops.readthedocs.io/en/latest/content/installation.html |
| GenomicRanges (R package) | Lawrence et al., 2013 | https://bioconductor.org/packages/release/bioc/html/GenomicRanges.html |
| Pheatmap (R package) | Raivo Kolde | https://www.rdocumentation.org/packages/pheatmap |
| Trim Galore (version 0.6.4) | Felix Krueger | http://www.bioinformatics.babraham.ac.uk/projects/trim_galore) |
| Bismark (version 0.22.3) | Felix Krueger | https://www.bioinformatics.babraham.ac.uk/projects/bismark |
| GraphPad Prism 8 (version 8.4.1) | GraphPad Software | www.graphpad.com |
| Other | ||
| Agilent QuikChange Lightning Site-Directed Mutagenesis Kit | Agilent | Cat # 210519 |
| RNeasy Mini Kit | Qiagen | Cat # 74104 |
| KAPA Stranded mRNA-Seq Kit (KK8420) | Roche | Cat # 07962193001 |
| KAPA Single-Indexed Adaptor Kit (KK8701) | Roche | Cat # 0800570200 |
| KAPA Library Quantification Kit (KK4924) | Roche | Cat # 07960140001 |
| EpiTect Bisulfite Kit | Qiagen | Cat # 59104 |
| Zymo Oligo Clean & Concentrator Kit | Zymo Research | Cat # D4060 |
| Pyromark PCR Kit | Qiagen | Cat # 978703 |
| MinElute Reaction Clean-up Kit | Qiagen | Cat # 28204 |
| AMPure XP | Beckman Coulter | Cat # A63881 |
| Multiplex PCR Kit | Qiagen | Cat # 206143 |
ACKNOWLEDGMENTS
We thank William Jones and Emily Fabyanic for help developing the bACE-Seq methodology, and Joanne Thorvaldsen, Aimee Juan, and Suhee Chang for tissue culture assistance. This work was supported by the National Institutes of Health: GM051279 (M.S.B.), GM118501 (R.M.K.), GM127408 (R.B.), T32GM008216 (B.A.C.) and 1F31HD098764 (B.A.C.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Bernstein DL, Kameswaran V, Le Lay JE, Sheaffer KL & Kaestner KH (2015). The BisPCR2 method for targeted bisulfite sequencing. Epigenetics Chromatin 8, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambrink T, Foreman R, Welstead GG, Lengner CJ, Wernig M, Suh H & Jaenisch R (2008). Sequential Expression of Pluripotency Markers during Direct Reprogramming of Mouse Somatic Cells. Cell Stem Cell 2, 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J, Wu B, Chang H & Greenleaf W (2015). ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr. Protoc. Mol. Biol 109, 21.29.1–21.29.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Guo L, Zhang L, Wu H, Yang J, Liu H, Wang X, Hu X, Gu T, Zhou Z, Liu J, Liu J, Wu H, Mao SQ, Mo K, Li Y, Lai K, Qi J, Yao H, et al. (2013). Vitamin C modulates TET1 function during somatic cell reprogramming. Nat. Genet 45, 1504–1509 [DOI] [PubMed] [Google Scholar]
- Chen J, Gao S, Gao Y, Huang H, Xu K, Chen X, Jiang Y, Li H, Gao S, Tao Y, Wang H, Zhang Y, Wang H & Cai T (2015). The combination of Tet1 with Oct4 generates high-quality mouse-induced pluripotent stem cells. Stem Cells 33, 686–698 [DOI] [PubMed] [Google Scholar]
- Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, Snyder MP, Pritchard JK, Kundaje A, Greenleaf WJ, Majeti R & Chang HY (2016). Lineage-specific and single cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet 48, 1193–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortázar D, Kunz C, Saito Y, Steinacher R & Schär P (2007). The enigmatic thymine DNA glycosylase. DNA Repair (Amst) 6, 489–504 [DOI] [PubMed] [Google Scholar]
- Cortázar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, Wirz A, Schuermann D, Jacobs AL, Siegrist F, Steinacher R, Jiricny J, Bird A & Schär P (2011). Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature 470, 419–423 [DOI] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, Abramowitz LK, Bartolomei MS, Rambow F, Bassi MR, Bruno T, Fanciulli M, Renner C, Klein-Szanto AJ, Matsumoto Y, et al. (2011). Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146, 67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Y, Ding J, Theunissen TW, Faiola F, Hore T. a, Shliaha PV, Fidalgo M, Saunders A, Lawrence M, Dietmann S, Das S, Levasseur DN, Li Z, Xu M, Reik W, Silva JCR & Wang J (2013). NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature 495, 370–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford DJ, Liu MY, Nabel CS, Cao XJ, Garcia BA & Kohli RM (2016). Tet2 Catalyzes Stepwise 5-Methylcytosine Oxidation by an Iterative and de novo Mechanism. J. Am. Chem. Soc 138, 730–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Waal E, Mak W, Calhoun S, Stein P, Ord T, Krapp C, Coutifaris C, Schultz RM & Bartolomei MS (2014). In Vitro Culture Increases the Frequency of Stochastic Epigenetic Errors at Imprinted Genes in Placental Tissues from Mouse Concepti Produced Through Assisted Reproductive Technologies. Biol. Reprod 90, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denizio JE, Liu MY, Leddin EM, Cisneros GA & Kohli RM (2019). Selectivity and Promiscuity in TET-Mediated Oxidation of 5-Methylcytosine in DNA and RNA. Biochemistry 58, 411–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M & Gingeras TR (2013). STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doege CA, Inoue K, Yamashita T, Rhee DB, Travis S, Fujita R, Guarnieri P, Bhagat G, Vanti WB, Shih A, Levine RL, Nik S, Chen EI & Abeliovich A (2012). Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature 488, 652–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedli M, Turelli P, Kapopoulou A, Rauwel B, Castro-Diáz N, Rowe HM, Ecco G, Unzu C, Planet E, Lombardo A, Mangeat B, Wildhaber BE, Naldini L & Trono D (2014). Loss of transcriptional control over endogenous retroelements during reprogramming to pluripotency. Genome Res 24, 1251–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Chen J, Li K, Wu T, Huang B, Liu W, Kou X, Zhang Y, Huang H, Jiang Y, Yao C, Liu X, Lu Z, Xu Z, Kang L, Chen J, Wang H, Cai T & Gao S (2013). Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell Stem Cell 12, 453–469 [DOI] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, Van Zon J, Lengner CJ, Creyghton MP, Van Oudenaarden A & Jaenisch R (2009). Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 463, 595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X & Cheng X (2012). Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res 40, 4841–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li N, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C & Xu GL (2011). Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill PWS, Amouroux R & Hajkova P (2014). DNA demethylation, Tet proteins and 5-hydroxymethylcytosine in epigenetic reprogramming: An emerging complex story. Genomics 104, 324–333 [DOI] [PubMed] [Google Scholar]
- Hu X, Zhang L, Mao SQ, Li Z, Chen J, Zhang RR, Wu HP, Gao J, Guo F, Liu W, Xu GF, Dai HQ, Shi YG, Li X, Hu B, Tang F, Pei D & Xu GL (2014). Tet and TDG mediate DNA demethylation essential for mesenchymal-to-epithelial transition in somatic cell reprogramming. Cell Stem Cell 14, 512–522 [DOI] [PubMed] [Google Scholar]
- Inoue A, Shen L, Dai Q, He C & Zhang Y (2011). Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res 21, 1670–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C & Zhang Y (2011). Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Zweig AS, Barber G, Hinrichs AS & Karolchik D (2010). Data and text mining BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26, 2204–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizaki S, Zou T, Li Y, Han YW, Suzuki Y, Harada Y & Sugiyama H (2016). Preferential 5-Methylcytosine Oxidation in the Linker Region of Reconstituted Positioned Nucleosomes by Tet1 Protein. Chem. - A Eur. J 22, 16598–16601 [DOI] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, Liu XS, Aravind L, Agarwal S, Maciejewski JP & Rao A (2010). Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 468, 839–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli RM & Zhang Y (2013). TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502, 472–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake RJ, Boetefuer EL, Won K-J & Fan H-Y (2016). The CSB chromatin remodeler and CTCF architectural protein cooperate in response to oxidative stress. Nucleic Acids Res 44, 2125–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B & Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P & Carlson M (2013). Software for Computing and Annotating Genomic Ranges. PLoS Comput Biol 9, e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R & 1000 Genome Project Data Processing Subgroup. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK & Shi W (2014). FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 [DOI] [PubMed] [Google Scholar]
- Liu MY, Denizio JE & Kohli RM (2016). Quantification of oxidized 5-methylcytosine bases and TET enzyme activity In Methods in Enzymology 573, Marmostein R, ed. (Academic Press Inc; ), pp. 365–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MY, Torabifard H, Crawford DJ, DeNizio JE, Cao X, Garcia BA, Cisneros GA, & Kohli RM (2017). Mutations along a TET2 active site scaffold stall oxidation at 5-hydroxymethylcytosine. Nat. Chem. Biol 13, 181–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W & Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti A & Drohat AC (2011). Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: Potential implications for active demethylation of CpG sites. J. Biol. Chem 286, 35334–35338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Foreman R, Chevalier B, Bilodeau S, Kahn M, Young RA & Jaenisch R (2008). Wnt Signaling Promotes Reprogramming of Somatic Cells to Pluripotency. Cell Stem Cell 3, 132–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy KN, Sonkodi I, Szöke I, Nagy E, & Newman HN The microflora associated with human oral carcinomas. Oral Oncol 34, 304–308 [PubMed] [Google Scholar]
- Nashun B, Hill PW & Hajkova P (2015). Reprogramming of cell fate: epigenetic memory and the erasure of memories past. EMBO J 34, 1296–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neddermann P, Gallinari P, Lettieri T, Schmid D, Truong O, Hsuan JJ, Wiebauer K & Jiricny J (1996). Cloning and Expression of Human G/T Mismatch-specific Thymine-DNA Glycosylase. J. Biol. Chem 271, 12767–12774 [DOI] [PubMed] [Google Scholar]
- Neph S, Kuehn MS, Reynolds AP, Haugen E, Thurman RE, Johnson AK, Rynes E, Maurano MT, Vierstra J, Thomas S, Sandstrom R, Humbert R & Stamatoyannopoulos JA (2012). Genome analysis BEDOPS: high-performance genomic feature operations. Bioinformatics 28, 1919–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olariu V, Lövkvist C & Sneppen K (2016). Nanog, Oct4 and Tet1 interplay in establishing pluripotency. Sci. Rep 6, 25438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR & Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardina JL, Collombet S, Tian TV, Gomez A, Stefano B. Di, Berenguer C, Brumbaugh J, Stadhouders R, Segura-Morales C, Gut M, Gut IG, Heath S, Aranda S, Croce L. Di, Hochedlinger K, Thieffry D & Graf T (2018). Transcription Factors Drive Tet2-Mediated Enhancer Demethylation to Reprogram Cell Fate Article Transcription Factors Drive Tet2-Mediated Enhancer Demethylation to Reprogram Cell Fate. Cell Stem Cell 23, 1–1529979984 [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J, White DJ, Hartenstein V, Eliceiri K, Tomancak P & Cardona A (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutsky EK, DeNizio JE, Hu P, Liu MY, Nabel CS, Fabyanic EB, Hwang Y, Bushman FD, Wu H & Kohli RM (2018). Nondestructive, base-resolution sequencing of 5-hydroxymethylcytosine using a DNA deaminase. Nat. Biotechnol 36, 1083–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler CL, Fernandez J, Koerperich Z, Andersen MP, Kotandeniya D, Nguyen ME, Sham YY & Tretyakova NY (2018). Maintenance DNA Methyltransferase Activity in the Presence of Oxidized Forms of 5-Methylcytosine: Structural Basis for Ten Eleven Translocation-Mediated DNA Demethylation. Biochemistry 57, 6061–6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Wu H, Diep D, Yamaguchi S, D’Alessio AC, Fung HL, Zhang K & Zhang Y (2013). Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell 153, 692–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Borkent M & Hochedlinger K (2010). A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nat Methods. 7, 53–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L & Rao A (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K & Yamanaka S (2006). Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- Tini M, Benecke A, Um SJ, Torchia J, Evans RM & Chambon P (2002). Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol. Cell 9, 265–277 [DOI] [PubMed] [Google Scholar]
- Um S, Harbers M, Beneckeʈ A, Pierrat B, Losson RG & Chambon P (1998). Retinoic Acid Receptors Interact Physically and Functionally with the T:G Mismatch-specific Thymine-DNA Glycosylase. J. Biol. Chem 273, 20728–20736 [DOI] [PubMed] [Google Scholar]
- Wagner M, Steinbacher J, Kraus TFJ, Michalakis S, Hackner B, Pfaffeneder T, Perera A, Müller M, Giese A, Kretzschmar HA & Carell T (2015). Age-Dependent Levels of 5-Methyl-, 5-Hydroxymethyl-, and 5-Formylcytosine in Human and Mouse Brain Tissues. Angew. Chemie Int. Ed 54, 12511–12514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Luo M, Berrios KN, Schutsky EK, Wu H & Kohli RM (2021). Bisulfite-Free Sequencing of 5-Hydroxymethylcytosine with APOBEC-Coupled Epigenetic Sequencing (ACE-Seq). Methods Mol. Biol 2198, 349–367 [DOI] [PubMed] [Google Scholar]
- Wolf G, de Iaco A, Sun MA, Bruno M, Tinkham M, Hoang D, Mitra A, Ralls S, Trono D & Macfarlan TS (2020). Krab-zinc finger protein gene expansion in response to active retrotransposons in the murine lineage. Elife 9, 1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Wang Z, Cui K & Zhao K (2011). Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev 25, 679–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Wu X, Shen L & Zhang Y (2014). Single-base resolution analysis of active DNA demethylation using methylase-assisted bisulfite sequencing. Nat. Biotechnol 32, 1231–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Wu X & Zhang Y (2016). Base-resolution profiling of active DNA demethylation using MAB-seq and caMAB-seq. Nat. Protoc 11, 1081–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Han D, Lu X, Sun Z, Zhou A, Yin Q, Zeng H, Liu M, Jiang X, Xie W, He C & Yi C (2015). Bisulfite-free, base-resolution analysis of 5-formylcytosine at the genome scale. Nat. Methods 12, 1047–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y & He QY (2012). ClusterProfiler: An R package for comparing biological themes among gene clusters. Omi. A J. Integr. Biol 16, 284–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG & He QY (2015). ChIP seeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W & Shirley Liu X (2008). Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu GL, Luo C, Jiang H & He C (2012). Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat. Chem. Biol 8, 328–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S3. Bisulfite analysis regions, related to STAR Methods.
Table S2. Primer list, related to STAR Methods.
Table S5. MAB pyrosequencing of spike-in control and Sall4, Smarcd2, Tet2, and EBf3 enhancers, related to Figure 3.
Table S6. ATAC-seq GO terms, related to Figure 4.
Table S4. RNA-seq and GO terms, related to Figure 2.
Data Availability Statement
The accession number for raw and processed RNA- and ATAC-sequencing data reported in this paper is GEO: GSE158334. Source data and image files for all figures in the paper are available at Mendeley Data (DOI: 10.17632/42kc23sjy9.1).


