Abstract
MicroRNAs (miRNAs) are noncoding small RNAs that regulate various pathophysiological cellular processes. Here, we report that expression of the miR-378 family was significantly induced by metabolic inflammatory inducers, a high-fructose diet, and inflammatory cytokine tumor necrosis factor-α. Hepatic miRNA profiling revealed that expression of miR-378a was highly upregulated, which, in turn, targeted the 3′-untranslated region of PPARα mRNA, impaired mitochondrial fatty acid β-oxidation, and induced mitochondrial and endoplasmic reticulum stress. More importantly, the upregulated miR-378a can directly bind to and activate the double-strand RNA (dsRNA)–dependent protein kinase R (PKR) to sustain the metabolic stress. In vivo, genetic depletion of miR-378a prevented PKR activation and ameliorated inflammatory stress and insulin resistance. Counterbalancing the upregulated miR-378a using nanoparticles encapsulated with an anti-miR-378a oligonucleotide restored PPARα activity, inhibited PKR activation and ER stress, and improved insulin sensitivity in fructose-fed mice. Our study delineated a novel mechanism of miR-378a in the pathogenesis of metabolic inflammation and insulin resistance through targeting metabolic signaling at both mRNA (e.g., PPARα) and protein (e.g., PKR) molecules. This novel finding of functional interaction between miRNAs (e.g., miR-378a) and cellular RNA binding proteins (e.g., PKR) is biologically significant because it greatly broadens the potential targets of miRNAs in cellular pathophysiological processes.
Introduction
The development of metabolic syndrome is usually accompanied by the activation of metabolic inflammation, which is triggered by the activation of inflammatory pathways and increased secretion of inflammatory cytokines. The double-strand RNA (dsRNA)–dependent protein kinase R (PKR) is a key component in metabolic inflammation, which directly interacts with several inflammatory kinases, including IκB kinase (IKK) and c-Jun N-terminal kinase (JNK), insulin signaling molecule insulin receptor substrate 1 (IRS-1), and the endoplasmic reticulum (ER) stress marker eIF2α (1–3). Although originally identified as an intracellular sensor of viral dsRNA and regulator of interferon inflammatory signaling (4), studies have delineated the pivotal role of PKR in nutrient surplus–associated metabolic stress (5–7) and insulin resistance (1–3). Activation of PKR has been found in the liver, muscle, and adipose tissues of obese human subjects with insulin resistance. Bariatric surgery that reduces human body weight ameliorates activation of PKR, IKK, JNK, and IRS-1-Ser312 in subcutaneous adipose tissue of obese patients (8). Furthermore, inhibition of PKR kinase activity by disrupting the PKR C-terminal kinase domain protects mice from high-fat diet–induced insulin resistance (6,9,10). This evidence demonstrates the close association between PKR activation and the onset of insulin resistance. However, the mechanism of how PKR is activated in metabolic disease remains unknown.
It has been well accepted that PKR is activated by the binding of viral dsRNAs or RNA molecules with specific three-dimensional structures to the two dsRNA-binding domains located in the N-terminus of PKR (11–13). However, short RNA duplexes of 16-nucleotide (nt) segments, small nucleolar RNAs, and siRNAs with lengths ranging from 21 to 23 nt were also reported to activate PKR under different conditions (14–17). Mutation in the dsRNA-binding motifs of PKR disrupts the integrity of the binding domain and compromises the kinase activity (18), which is associated with amelioration of metabolic inflammation (6). However, endogenous molecules that can act as PKR activators in the context of metabolic stress have not been identified. Studies have reported that aberrant expression of microRNAs (miRNAs) plays a role in overnutrition-induced metabolic diseases, including obesity, nonalcoholic fatty liver disease, and insulin resistance (19,20). These findings raise an intriguing question: Can the upregulated miRNAs act as PKR activators?
miRNAs are endogenous, noncoding small RNAs between 18 and 25 nt long that regulate multiple cellular processes by complementary base pairing between their seed sequences and the 3′- or 5′-untranslated region (UTR) of the target mRNAs. This action results in decay and/or translational repression of the target mRNA (21,22). About 60% of mRNAs in the genome are targets of miRNAs, which regulate diverse pathophysiological cellular activities, including metabolic inflammation and insulin signaling (23,24). Furthermore, miRNA mimics and anti-miRNAs (inhibitors) have been used to specifically modulate the actions of miRNAs in animal model research and clinical trials. However, the question of whether miRNAs can directly bind to cellular RNA-binding proteins and exert their regulatory impacts on biological processes remains unclear.
Peroxisome proliferator–activated receptor-γ (PPARγ) coactivator-1β (PGC-1β) and PPARα are partner transcription factors that regulate expression of genes encoding mitochondrial fatty acid oxidation enzymes, including CPT1α, ACOX1, LCAD, and MCAD, and play an essential role in regulating mitochondrial metabolic homeostasis and oxidative stress (25–30). miR-378 and miR-378* are two miRNAs that are coexpressed with PGC-1β to counterbalance the functional activity of PGC-1β in metabolic signaling. Mice with genetic depletion of miR-378 and miR-378* are resistant to high-fat diet–induced obesity (31–34). Previously, our group reported that high-fructose diets compromised mitochondrial fatty acid β-oxidation and induced ER stress and hepatic steatosis by impairing PPARα signaling (35). However, an unanswered question in that study is what is the molecular mechanism that compromised PPARα signaling upon exposure to high-fructose diet? In the current study, we demonstrate that miR-378a targeted the 3′-UTR of PPARα mRNA at multiple sites, leading to reduced PPARα expression and the associated oxidative stress. miR-378a further bound to the protein molecules of PKR and activated the kinase to sustain metabolic inflammation and insulin resistance.
Research Design and Methods
Animal Protocols
All animal experiments were approved by the University of Nebraska-Lincoln institutional animal care and use committee and were performed according to National Institutes of Health guidelines (Guide for the Care and Use of Laboratory Animals). Mice used in this study were 12–24 weeks old. Male mice were used unless otherwise specified. miR-378a knockout (miR-378a-KO) mice and the wild-type (WT) littermates were a gift from Dr. Vincent Giguere (McGill University, Montreal, Quebec, Canada). Animals were housed on alternating 12-h light and dark cycles with free access to food and water. For the dietary trial, two groups of mice were fed either a chow (ID: 2016; Envigo, Madison, WI) or a high-fructose (60% fructose, ID: 161.506; Dyets, Inc., Bethlehem, PA) diet for 4 weeks. Detailed dietary composition has been described previously (35). At the end of the feeding trial, mice were fasted for 12 h followed by anesthetization using isoflurane. Livers were excised, frozen in liquid nitrogen, and stored at −80°C for further analysis. For tumor necrosis factor-α (TNF-α) treatment, two groups of WT and miR-378a-KO mice (female) were fasted for 8 h with free access to water followed by treatment with recombinant mouse TNF-α (8 μg/200 g body weight) via intraperitoneal injection. Liver tissue samples were collected at 5 h posttreatment. To determine the therapeutic effect of miR-378a inhibitor (anti-miR-378a), mice were fed a high-fructose diet for 4 weeks. At the beginning of week 3 of the feeding trial, mice were randomly divided into two groups and subjected to treatment with a dose of either nanoparticles (NPs) carrying the anti-miR-378a (3 mg/kg) or NPs carrying scramble miRNA as control (3 mg/kg) every 3 days through tail vein injection. In total, four doses of NPs were given to mice before livers, muscles, and adipose tissues were collected for further analysis.
Statistical Analyses
The Anderson-Darling test on the residuals was used to test normality, and Levene test was used to test for equal variances. The two-tailed Student t test was used for statistical analyses of two-group comparisons. All results are presented as mean ± SD. Statistically significant differences of P < 0.05 or P < 0.01 compared with controls are reported. An additional description of materials and methods can be found in the Supplementary Material.
Data and Resource Availability
The data sets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Results
Upregulation of miR-378a Induced by High-Fructose Diet Targets the PPARα Signaling Pathway
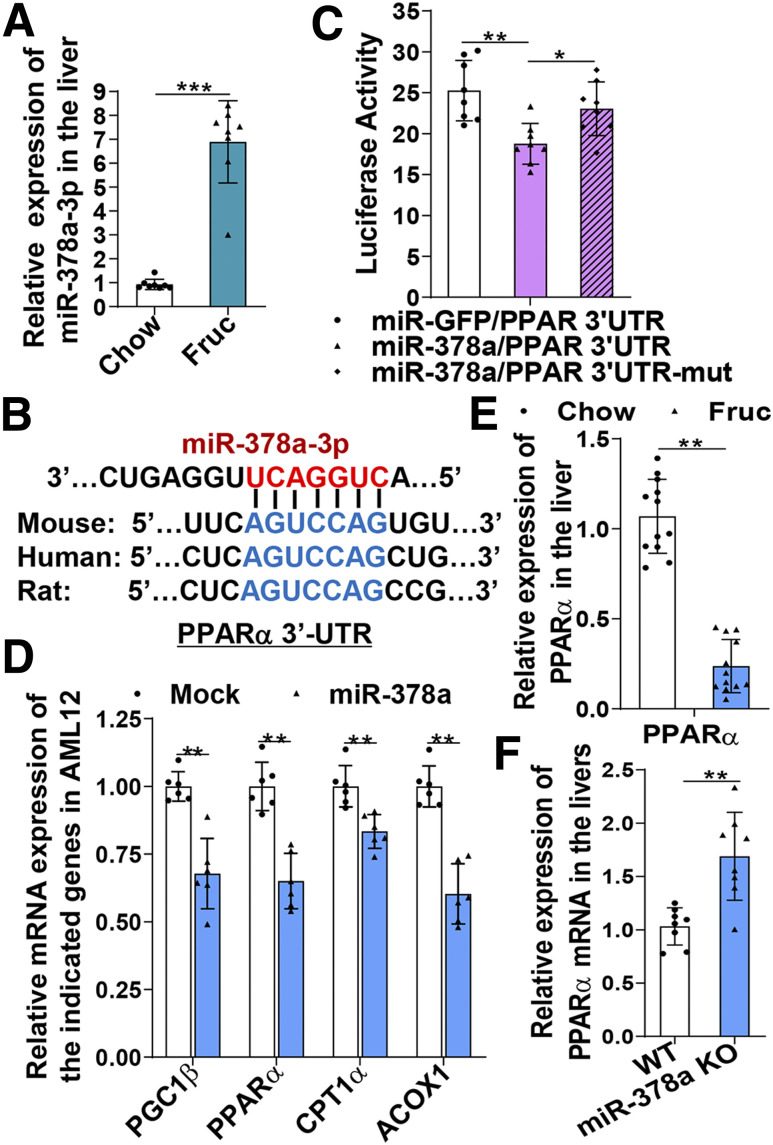
Fructose is a highly lipogenic sugar that has a profound impact on metabolic signaling and disorders. We previously reported that a high-fructose diet induced significant changes in hepatic miRNA expression, which targets multiple metabolic pathways in the liver and induces hepatic steatosis and insulin resistance (20). Bioinformatics analysis of the hepatic miRNA expression profile further revealed that expression of the miR-378 family, including miR-378a, b, c, and d, was significantly increased upon exposure to the fructose diet (Supplementary Fig. 1A and B). Among them, the abundance of miR-378a-3p is at the top of the family members (Supplementary Fig. 1A and B), indicating the potentially significant biological impact of this miRNA. Upregulation of miR-378a by fructose was further verified by a TaqMan miRNA probe specific for miR-378a (Fig. 1A). The use of TargetScan to search for potential targets of miR-378a identified multiple target sites of miR-378a within the PPARα 3′-UTR (Fig. 1B and Supplementary Fig. 1C). More importantly, the targeted sequences are conserved among humans, rats, and mice, implicating the evolutional significance of these targeted sites. To investigate whether PPARα is a true target of miR-378a, we generated a luciferase construct containing a WT or mutated miR-378a binding site of the PPARα 3′-UTR. Coexpression of the luciferase constructs with a plasmid expressing either miR-378a or a mock control (green fluorescent protein [GFP]) in COS-7 cells for 48 h revealed that expression of miR-378a inhibited luciferase activity of PPARα 3′-UTR WT compared with mock-transfected cells (Fig. 1C). In contrast, mutation at the targeted site abolished the inhibitory effect of miR-378a as indicated by the recovered luciferase activity in the PPARα 3′-UTR mutant transfected cells (Fig. 1C), suggesting that PPARα is a true target of miR-378a. The inhibitory effect of miR-378a on PPARα was further investigated by expressing miR-378a in AML12, a mouse hepatocyte cell line, and testing alterations on the mRNA expression of PPARα and its downstream genes. Successful overexpression of miR-378a in the transfected cells was verified by a TaqMan probe specific for miR-378a (Supplementary Fig. 1D). PPARα mRNA and its target genes involved in mitochondrial fatty acid β-oxidation, including CPT1α and ACOX1, were significantly reduced in the miR-378a–expressing cells (Fig. 1D). mRNA of PGC-1β, a gene that has been reported as a true target of miR-378a, was also significantly reduced (Fig. 1D). A similar inhibitory effect of miR-378a on PPARα signaling was also observed in McA cells, a rat hepatoma cell line (Supplementary Fig. 1E). Consistently, in vivo, upregulation of miR-378a induced by a high-fructose diet was accompanied by significant reduction of hepatic PPARα mRNA (Fig. 1E). In contrast, genetic depletion of miR-378a in mice markedly upregulated hepatic PPARα mRNA expression to a level even higher than in the WT mice (Fig. 1F). Together, these results demonstrate that miR-378a induced by a high-fructose diet targets PPARα signaling and compromises mitochondrial metabolic homeostasis, which might contribute to the initiation of mitochondrial oxidative stress.
Figure 1.
Upregulation of miR-378a induced by a high-fructose diet targets the PPARα signaling pathway. Mice (C57BL/6J) were subjected to chow or fructose diets for 4 weeks. Total miRNAs were extracted from the livers. A: Hepatic miRNA-378a-3p detected by a TaqMan miRNA probe specific for miR-378a and normalized to small nucleolar RNA202 (n = 8/group). B: miR-378a-3p response elements within the 3′-UTR of human, rat, and mouse PPARα mRNAs predicted by TargetScan. C: COS-7 cells were cotransfected with miR-GFP or miR-378a plus the firefly luciferase reporter of WT PPARα 3′-UTR or mutant PPARα 3′-UTR for 48 h. Cell lysates were examined for firefly luciferase activities and normalized to Renilla luciferase activities. D: AML12 cells were transfected with miR-378a or a mock vector (GFP) for 48 h. mRNAs of the indicated genes were detected by quantitative RT-PCR (qRT-PCR). E: mRNA of PPARα detected by qRT-PCR in the livers of 4-week chow-fed or fructose-fed mice (C57BL/6J, n = 12/group). F: mRNA of PPARα detected by qRT-PCR in the livers of WT and miR-378a-KO mice (n = 8/group). Data are mean ± SD. The two-tailed Student t test was used for statistical analyses of two-group comparisons. *P < 0.05, **P < 0.01, ***P < 0.001 vs. controls. Fruc, fructose; mut, mutant.
Elevated miR-378a Induces Mitochondrial and ER Stress via Mediation of PPARα Signaling
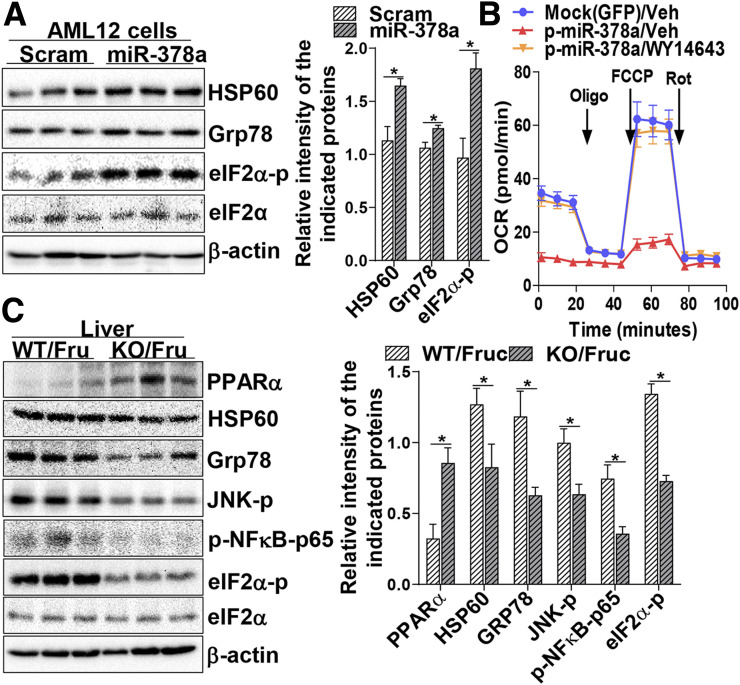
To determine the biological consequence of increased miR-378a and compromised PPARα signaling, the metabolic homeostasis of mitochondria and ER was investigated. Immunoblotting analysis revealed that the mitochondrial and ER stress markers HSP60 and phosphorylation of eIF2α (eIF2α-p) were activated in the livers of fructose-fed mice (Supplementary Fig. 2A). To investigate the direct impact of miR-378a on mitochondrial and ER stress, we transiently expressed miR-378a in AML12, and scrambled miRNA was used as control. Increased expression of miR-378a was found to be associated with the activation of mitochondrial and ER stress markers HSP60, Grp78, and eIF2α-p (Fig. 2A), indicating that increased miR-378a alone is sufficient to activate the stress signaling. The impact of miR-378a on metabolic stress was further determined by a Seahorse (FX24) assay, which showed that the oxygen consumption rate of mitochondria was reduced upon exposure to high abundances of miR-378a, leading to a reduction of mitochondrial ATP production and compromised oxidative capacity (Supplementary Fig. 2B). The mechanism of miR-378a–induced oxidative stress was further investigated by determining the association between mitochondrial respiratory reaction and PPARα activity. We reasoned that if miR-378a–induced oxidative stress is mediated by PPARα signaling, restoration of PPARα activity will recover the integrity of mitochondria and ATP production. To test this hypothesis, the miR-378a–expressing cells were treated or untreated with a PPARα agonist, WY14,643 (30 μmol/L), for 24 h. Seahorse analysis showed that treatment with WY14,643 significantly recovered mitochondrial oxidative capacity, which reached a comparable level of the control (GFP/vehicle) (Fig. 2B), demonstrating the involvement of PPARα in miR-378a–induced mitochondrial oxidative stress. Furthermore, miR-378a-KO abolished the inhibitory effect of high-fructose diet on PPARα expression, which was associated with less activation of mitochondrial and ER stress markers HSP60, Grp78, phosphorylated JNK (JNK-p), eIF2α-p, and phosphorylated nuclear factor-κB-p65 (p-NFκB-p65) at Ser536 (Fig. 2C). Together, these results demonstrated that upregulation of miR-378a compromised PPARα signaling and contributed to the induction of metabolic inflammation, oxidative stress, and ER stress.
Figure 2.
Elevated miR-378a induces mitochondrial and ER stress via mediation of PPARα signaling. A: AML12 cells were transfected with miR-378a or scrambled (Scram) miRNA (10 nmol/L) as control for 60 h and subjected to protein extraction. Mitochondrial and ER stress markers HSP60, Grp78, and eIF2α-p as well as total eIF2α and β-actin were determined by immunoblotting analysis. B: Oxygen consumption rate (OCR) was determined in McA cells transfected with miR-378a or mock (GFP) for 12 h followed by treatment with PPARα agonist WY14,643 (30 μmol/L) or vehicle (Veh) (DMSO) for an additional 24 h using Seahorse (FX24) analysis in the presence of 1 μmol/L oligomycin (Oligo), 1 μmol/L carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP), and 0.5 μmol/L rotenone (Rot). C: Protein expression of PPARα and mitochondrial and ER stress markers HSP60, Grp78, JNK-p, p-NFκB-p65, and eIF2α-p in the livers of WT or miR-378a-KO mice fed a high-fructose diet for 4 weeks were determined by immunoblotting analysis (n = 5/group). For cell treatment, two independent experiments were performed in triplicate. Data are mean ± SD. The two-tailed Student t test was used for statistical analyses of two-group comparisons. *P < 0.05 vs. controls. Fruc, fructose.
miR-378a Induced by Inflammatory Cytokine TNF-α Disturbs Metabolic Homeostasis of Mitochondria and ER
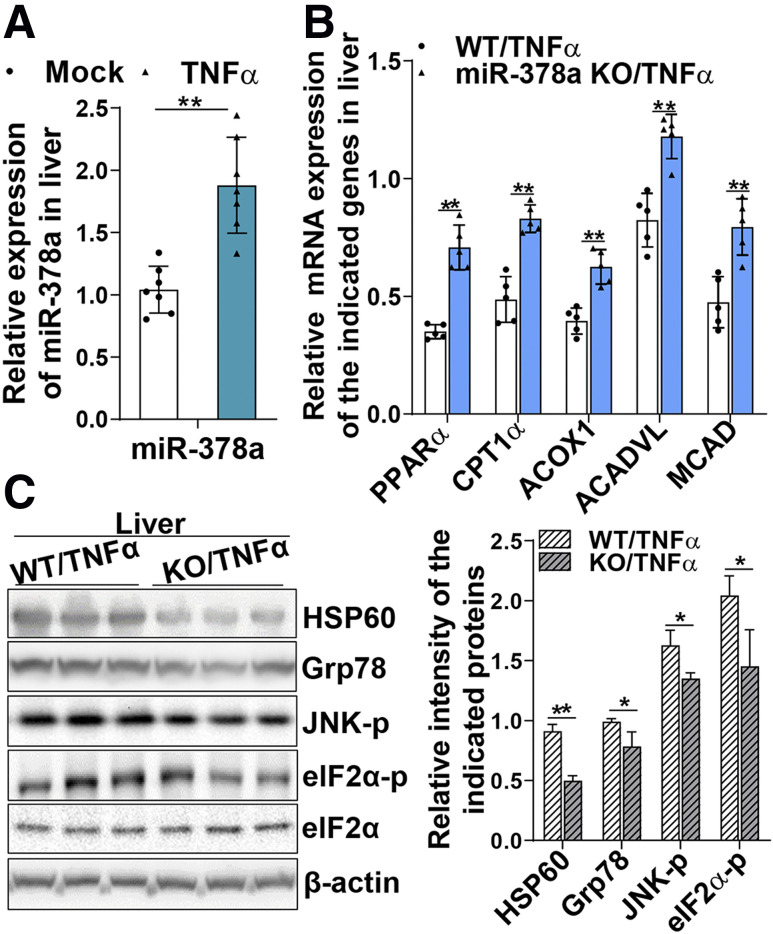
To investigate whether inflammatory cytokine TNF-α can induce miR-378a expression, two groups of WT and miR-378a-KO mice were treated with recombinant mouse TNF-α (8 μg/200 g body weight) for 5 h via intraperitoneal injection. Hepatic miR-378a expression was found to be significantly induced by TNF-α treatment (Fig. 3A). Furthermore, depletion of miR-378a released the inhibitory effect of TNF-α on PPARα, as indicated by the significantly higher expression of PPARα mRNA and its target genes CPT1α, ACOX1, ACADVL, and MCAD (Fig. 3B). Depletion of miR-378a further protected mice from TNF-α–induced hepatic mitochondrial and ER stress, as evidenced by less activation of stress makers HSP60, Grp78, JNK-p, and eIF2α-p (Fig. 3C). Together, these results demonstrate that expression of miR-378a can be induced by inflammatory cytokine TNF-α, which is engaged in the trigger of mitochondrial oxidative and ER stress.
Figure 3.
miR-378a induced by inflammatory cytokine TNF-α disturbs metabolic homeostasis of mitochondria and ER. miR-378a-KO mice and their WT littermates (n = 5–7/group) were fasted for 8 h followed by treatment with TNF-α (8 μg/200 g body weight) for 5 h. Liver tissues were collected for protein and RNA extractions. A: miR-378a detected in the livers of mice treated with mock (saline) or TNF-α by a TaqMan miRNA probe specific for miR-378a and normalized to small nucleolar RNA202. B: mRNAs of the indicated genes in the TNF-α–treated WT and miR-378a-KO mice. C: Mitochondrial and ER stress markers HSP60, Grp78, JNK-p, and eIF2α-p as well as total eIF2α and β-actin in the livers of TNF-α–treated WT and KO mice were detected by immunoblotting analysis. Data are mean ± SD. The two-tailed Student t test was used for statistical analyses of two-group comparisons. *P < 0.05, **P < 0.01 vs. controls.
miR-378a Interacts With and Activates PKR to Sustain Metabolic Inflammation
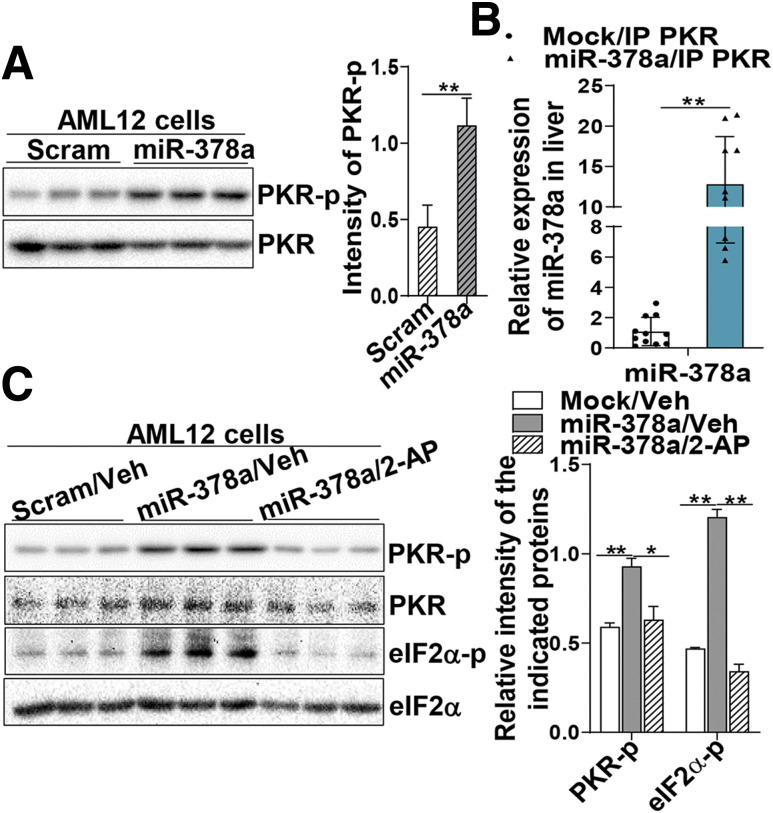
The inflammatory protein kinase PKR is activated by the binding of dsRNAs to its dsRNA-binding motifs and plays an important role in metabolic inflammation. We found that high-fructose diet induced activation of PKR and JNK, two key kinases in conveying metabolic inflammation, in the liver (Supplementary Fig. 3A). To investigate whether high-fructose–induced miR-378a is involved in PKR activation, we transfected miR-378a and scramble miRNAs in AML12 cells and determined the activation of PKR by immunoblotting analysis using a phosphorylation-specific antibody against PKR. Figure 4A shows that upregulation of miR-378a activated PKR (Fig. 4A). To determine whether miR-378a directly interacted with PKR and activated the kinase, cellular PKR proteins were immunoprecipitated from the cells transfected with miR-378a or mock (GFP) using an anti-PKR antibody. Bound RNAs were released from the PKR immune complex and further subjected to an assay using a TaqMan miRNA probe specific for miR-378a. Figure 4B shows that a substantial amount of miR-378a was pulled down in the PKR immune complexes compared with the mock (Fig. 4B), suggesting the interaction between miR-378a and PKR. To test whether other miRNAs were also involved in PKR activation, miR-203 that was also induced by high-fructose diet (20) and miR-125a that has been reported in metabolic inflammation were selected as additional candidates for the PKR binding assay. Data in Supplementary Fig. 3B reveal that miR-203 presented at a very low level in the PKR immune complexes and that no changes were observed between the control and the treated groups. In contrast, miR-125a was significantly presented in the immune complexes, although at less abundance compared with miR-378a (Supplementary Fig. 3B), suggesting that in addition to miR-378a, certain miRNAs (e.g., miR-125a) may also contribute to PKR activation. Next, the role of miR-378a as a PKR activator was further verified by using a PKR inhibitor, 2-aminopurine (2-AP). miR-378a–expressing cells were treated with either 2-AP or vehicle (PBS:glacial acetic acid [200:1]) as control for 24 h. Significantly less PKR activation (phosphorylation) was detected in the 2-AP–treated cells compared with the vehicle-treated cells (Fig. 4C). Consistently, less phosphorylation was observed in the PKR substrate eIF2α upon 2-AP treatment (Fig. 4C). These results demonstrate that increased miR-378a in metabolic inflammation can activate PKR via direct interaction with the protein molecules of the kinase.
Figure 4.
miR-378a interacts with and activates the dsRNA-dependent protein kinase PKR to sustain metabolic inflammation. A: Phosphorylation (active form) of PKR in AML12 cells transfected with scrambled (Scram) miRNA (10 nmol/L) as control or miR-378a for 48 h were determined by immunoblotting analysis using a phosphorylation-specific antibody against PKR-Thr446. B: Relative abundance of miR-378a bound to PKR was determined in the immune complex of PKR using a TaqMan miRNA probe specific for miR-378a and normalized to small nucleolar RNA202. The detailed procedure is provided in the Supplementary Material. C: AML12 cells were transfected with miR-378a or Scram miRNA (10 nmol/L) as control for 24 h and followed by treatment with 2-AP (0.5 mmol/L) (Sigma) or vehicle (Veh) (PBS:glacial acetic acid [200:1]) for an additional 24 h. Phosphorylation of PKR and its substrate eIF2α were determined by immunoblotting analysis. Two independent experiments were performed in triplicate. Data are mean ± SD. The two-tailed Student t test was used for statistical analyses of two-group comparisons. *P < 0.05, **P < 0.01 vs. controls. IP, immunoprecipitated; PKR-p, phosphorylated protein kinase R.
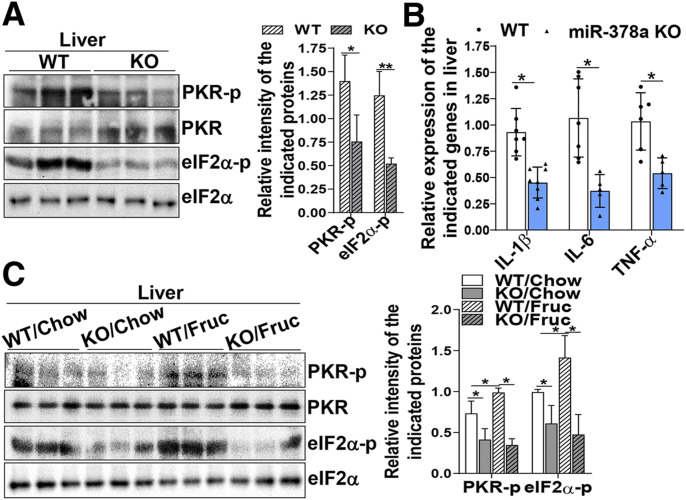
To further investigate whether miR-378a can act as an activator of PKR in vivo, the miR-378a PKR activation status was determined in WT and miR-378a-KO mice. Consistently, genetic depletion of miR-378a resulted in a significant decrease in the phosphorylation of PKR and its substrate eIF2α in the mouse livers (Fig. 5A). This phenotype was associated with substantially reduced expression of inflammatory cytokine genes interleukin-1β (IL-1β), IL-6, and TNF-α (Fig. 5B), implicating an overall amelioration of metabolic inflammation in the KO mice. Challenging the WT and KO mice with a high-fructose diet for 4 weeks showed that miR-378a-KO prevented fructose-induced PKR activation and its substrate eIF2α (Fig. 5C). Together, these data strongly demonstrate that upregulation of miR-378a activates PKR to sustain, at least partially, the metabolic inflammation induced by a high-fructose diet.
Figure 5.
Depletion of miR-378a by genetic intervention or anti-miR-378a oligonucleotides improves fructose-induced insulin resistance. A and B: miR-378a-KO mice and their WT littermates were fasted for 12 h. Liver tissues were collected for the following analyses: immunoblotting analysis of PKR phosphorylation (active form) and its substrate eIF2α using phosphorylation-specific antibodies against PKR and eIF2α (n = 6/group) (A) and mRNA expression of the indicated inflammatory cytokine genes determined by quantitative RT-PCR (n = 5–7/group) (B). C: Immunoblotting analysis of PKR phosphorylation and its substrate eIF2α in the liver tissues of WT and miR-378a-KO mice fed a chow or high-fructose diet for 4 weeks (n = 5/group). Data are mean ± SD. The two-tailed Student t test was used for statistical analyses of two-group comparisons. *P < 0.05, **P < 0.01 vs. controls. Fruc, fructose; PKR-p, phosphorylated protein kinase R.
Depletion of miR-378a by Genetic Intervention or Anti-miR-378a Oligonucleotides Improves Fructose-Induced Insulin Resistance
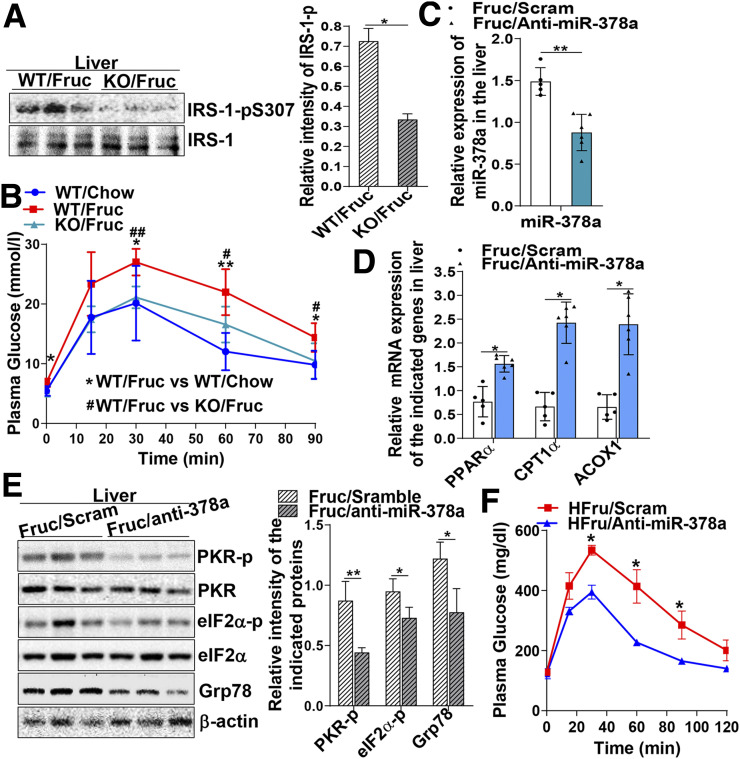
Activation of PKR has been shown to activate JNK and contribute to insulin resistance by inducing phosphorylation of IRS-1 at an inhibitory residue Ser307 (IRS-1-pS307) (6). To investigate the impact of miR-378a-PKR signaling on hepatic insulin sensitivity, IRS-1-pS307 was determined in the livers of fructose-fed mice and found to be increased (Supplementary Fig. 4A). In contrast, miR-378a-KO released the inhibitory effect of fructose on IRS-1 as less IRS-1-pSer307 was detected in the fructose-fed KO mice (Fig. 6A). A glucose tolerance test further verified the improvement of insulin sensitivity in the KO mice as significantly lower blood glucose levels were observed at 30, 60, and 90 min after administrating a dose of glucose (2 g/kg) into the mice (Fig. 6B). miR-378a-KO recovered the insulin sensitivity to a comparable level of WT mice fed a regular chow diet (Fig. 6B). A similar result was obtained by insulin tolerance test (0.5 units/kg) (Supplementary Fig. 4B).
Figure 6.
Depletion of miR-378a by genetic intervention or anti-miR-378a oligonucleotides improves fructose-induced insulin resistance. A: Immunoblotting analysis of IRS-1-pS307 and total protein levels in the livers of miR-378a-KO mice fed a high-fructose diet for 4 weeks (n = 5/group). B: Plasma glucose levels in a glucose tolerance test of the chow- or fructose-fed WT and miR-378a-KO mice (n = 5–7/group). C–F: Mice (C57BL/6J) were subjected to a high-fructose diet for 4 weeks. At week 3 of the feeding trial, mice were randomly divided into two groups (n = 5–6/group) and subjected to treatment with either a dose of NPs carrying the anti-miR-378a (3 mg/kg) or NPs carrying scrambled (Scram) miRNA as control (3 mg/kg) every 3 days through tail vein injection. In total, four doses of NPs were given to mice by the end of the feeding trial. C: miR-378a determined in the mouse liver tissues by a TaqMan miRNA probe specific for miR-378a and normalized to small nucleolar RNA202. D: Hepatic mRNA expression of PPARα, CPT1α, and ACOX1 measured by quantitative RT-PCR. E: Activation of PKR and ER stress markers Grp78 and eIF2α-p determined by immunoblotting analysis. F: Blood glucose levels determined by a glucose tolerance test conducted at the end point of the scram miRNA– or anti-miR-378a–treated mice. Data are mean ± SD. The two-tailed Student t test was used for statistical analyses of two-group comparisons. *P < 0.05, **P < 0.01 vs. controls; #P < 0.05, ##P < 0.01 vs. controls. Fruc, fructose; HFru, high fructose.
Next, a second mouse model that more resembles the physiological condition was established to further explore the therapeutic potential of miR-378a in insulin resistance. We formulated an NP encapsulating an anti-miR-378a oligonucleotide (anti-miR-378a) with an average particle size <100 nm. The stability and uptake efficiency of the NPs were characterized as reported in a previous publication (36). The fructose-induced insulin resistance mouse model was established as described in the Research Design and Methods and the NP treatment started at week 3 of the feeding trial. In total, four doses of NPs were given to mice by the end of the feeding trial. Body weights were monitored and found to be comparable between the anti-miR-378a– and scramble RNA–treated groups (Supplementary Fig. 5A). Analyzing the abundance of miR-378a in the livers of treated mice revealed that the anti-miR-378a oligonucleotide successfully reduced the abundance of hepatic miR-378a in the fructose-fed mice (Fig. 6C). Inhibition of miR-378a led to increased expression of PPARα mRNA and its target genes CPT1α and ACOX1 (Fig. 6D). More importantly, reduced miR-378a further prevented the activation of PKR and ER stress markers Grp78 and eIF2α in the fructose-fed mice (Fig. 6E). Improved insulin sensitivity was detected in the anti-miR-378a–treated mice, as indicated by the lower blood glucose levels at 30, 60, and 90 min on glucose tolerance test (Fig. 6F). Increased insulin sensitivity was further detected in liver, muscle, and adipose tissue, the major insulin-responsive organs and tissues, as indicated by the increased AKT-pS473 and reduced IRS-1-pS307 detected in the anti-miR-378a–treated mice (Supplementary Fig. 5B–D). Together, these results strongly indicate that inhibition of miR-378a in fructose-fed mice improves PPARα activity and suppresses PKR activation and the associated metabolic inflammation, leading to the improvement of insulin sensitivity.
Discussion
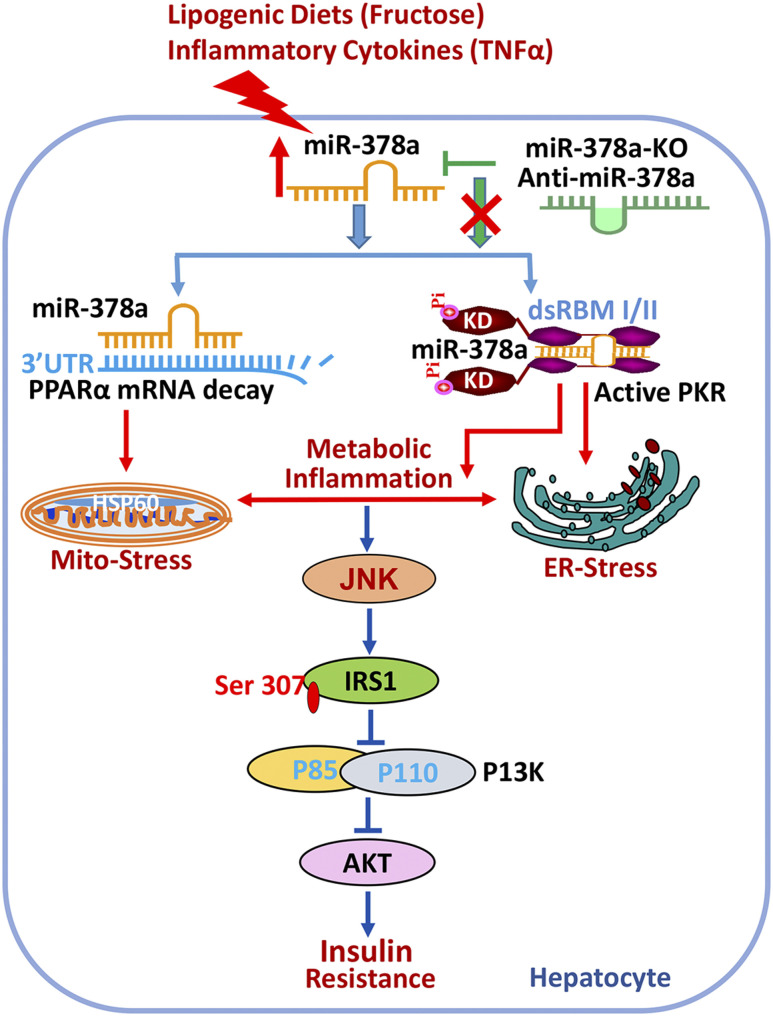
Emerging evidence has revealed that aberrant expression of miRNAs is associated with the development of insulin resistance and diabetes in both human subjects and animal models (37). In this study, we characterized a novel regulatory mechanism of miR-378a in metabolic inflammation and insulin resistance. Specifically, in the fructose- and TNF-α–induced metabolic inflammation, hepatic miR-378a was significantly increased, which targeted the miRNA response element within the 3′-UTR of PPARα mRNA and compromised PPARα activity, leading to the induction of oxidative and ER stress. More importantly, miR-378a further directly interacted with the protein molecule of PKR and activated the kinase to sustain metabolic inflammation and induce insulin resistance (Fig. 7). This finding, for the first time, unveiled a novel mechanism of miRNA that can directly bind to and activate the dsRNA-dependent protein kinase PKR in the context of metabolic stress.
Figure 7.
miR-378a targets the 3′-UTR of PPARα mRNA and activates the dsRNA-dependent PKR in metabolic inflammation. Metabolic inflammatory inducers, such as a high-fructose diet and inflammatory cytokine TNF-α, induce expression of miR-378a, which targets multiple miRNA response elements within the 3′-UTR of PPARα mRNA and induces mitochondrial and ER stress. Upregulated miR-378a further directly binds to the dsRNA-binding domains in PKR and activates the kinase to sustain the metabolic inflammation and induces IRS-1-pS307, which blunts hepatic insulin signaling. Genetic depletion of miR-378a (miR-378a-KO) or treatment with an anti-miR-378a oligonucleotide that counterbalances miR-378a expression can ameliorate inflammatory stress and improve insulin resistance. dsRBM, double-stranded RNA-binding motif; KD, kinase domain; Pi, phosphorylation.
Intensive studies have demonstrated PKR to be a key determinant in metabolic inflammation and the associated metabolic disease (e.g., insulin resistance) (1,6,8). However, the mechanism of metabolic inflammation initiation and the associated kinase activation (e.g., PKR) remains unclear. PKR was originally identified as a protein kinase that is activated by viral dsRNAs and plays a pivotal role in antiviral response. However, other types of dsRNAs or RNA molecules can also activate the kinase (38). dsRNA duplex is produced during the process of pre-miRNAs to miRNAs (39). This makes miRNAs the potential candidates of PKR activators. In this study, we demonstrated that miR-378a interacts with the protein molecule of PKR and activates the kinase. Genetic depletion of miR-378a or counterbalancing the upregulated miR-378a induced by fructose using an anti-miR-378a oligonucleotide inhibited PKR activation and improved insulin sensitivity. Although other noncoding RNAs, such as miR-182 and noncoding RNA886 (ncRNA886), have also been reported to regulate PKR-interferon signaling (40,41), regulation of PKR by miR-182 is through the canonical mechanism that directly targets the 3′-UTR of PKR mRNA (41) whereas ncRNA886 is through binding to PKR in CD4+ T cells. Interestingly, instead of activating the kinase, interaction of ncRNA886 with PKR prevents phosphorylation of the kinase by dsRNA, and the mechanism is currently unknown (40). Identification that miR-378a binds to and activates a protein kinase not only unveils a molecular mechanism of PKR activation in metabolic syndrome but also further delineates an unprecedented regulatory mechanism of miRNAs in cellular pathophysiological processes via interaction with certain RNA-binding proteins.
During metabolic stress, PKR activates the stress kinase JNK, which further induces IRS-1-pSer307 and blunts the downstream insulin signaling (6,42). Genetic depletion of PKR protects mice from high-fat diet–induced insulin resistance and diabetes (9,43). In this study, we found that activation of PKR by miRNA-378a was associated with JNK activation and subsequent inhibition of IRS-1 activity, leading to reduced AKT phosphorylation and decreased insulin sensitivity. A study has reported that miR-378a can inhibit insulin sensitivity by targeting the p110α subunit of PI3K (44). Thus, one may argue that insulin resistance in the fructose-fed mice may be a consequence of upregulated miR-378a targeting the p110α subunit of PI3K. Although we cannot completely rule out the possible involvement of the miR-378a-PI3K axis in this regulation, it is important to point out that in the insulin signaling pathway, IRS-1 is upstream of PI3K (Fig. 7) such that inhibition of IRS-1 by PKR and JNK contributes to the development of insulin resistance. Moreover, the fact that counterbalancing miR-378a by an anti-miR-378a oligonucleotide suppressed PKR activation, ameliorated ER stress, and improved insulin sensitivity further supports the critical role of the miR-378a-PKR axis in regulating insulin sensitivity.
A study has reported that increased expression of miR-378 in mouse liver triggered hepatic inflammation and facilitated the development of nonalcoholic steatohepatitis and liver fibrosis via the mediation of NF-κB and TNF-α (45). This study, from another angle, demonstrates the role of miR-378a in inflammatory-associated liver diseases. It would be interesting to investigate whether PKR was activated in this scenario. Of note, inconsistent observations have been reported from studies using PKR-KO mouse models (6,45). One study shows that a high-fat diet only induced modest inflammation in adipose tissue and failed to induce obesity and hepatic steatosis in a PKR-KO mouse model (46). At a glance, these observations may challenge the proposed role of PKR in lipogenic diet–induced metabolic inflammation (6). However, this inconsistency could be due to the different PKR-KO mouse models used in the studies. There are two PKR-KO mouse models: an N-terminal PKR-KO that depleted the N-terminal regulatory domain but retained the C-terminal kinase domain and a C-terminal PKR-KO that depleted the C-terminal kinase domain but retained the N-terminal regulatory domain (47). The use of different PKR-KO mouse models may contribute to the different observations in the two studies. It is also possible that the inconsistent observations were associated with the differences in gut bacterial community and the consequent changes in metabolic behavior because the two PKR-KO mouse colonies were housed in different facilities.
A unique mechanism in miRNA regulation is that a single miRNA has the potential to target multiple relevant genes in a signaling pathway, enabling the miRNA to amplify its regulatory impact on the cellular processes (48). Indeed, miR-378 has been found to target multiple molecules in the mitochondrial respiratory chain, energy homeostasis, and hepatosteatosis (34,44,49,50). Our study adds novel knowledge to this regulatory network by demonstrating that PPARα, a key transcription factor that regulates genes involved in mitochondrial fatty acid β-oxidation, is a true target of miR-378a that regulates mitochondrial oxidative stress and metabolic inflammation.
In conclusion, our study unveils a novel regulatory mechanism of miR-378a by showing that in addition to targeting PPARα at the mRNA molecule, miR-378a can interact with the RNA-binding protein kinase PKR and activate the kinase in metabolic stress. This novel finding not only advances our knowledge of miRNA physiological function but also might further lend support to new therapeutic strategies for the prevention and treatment of insulin resistance and type 2 diabetes.
Article Information
Acknowledgments. The authors thank Dr. Vincent Giguere in the Department of Medicine and Oncology at McGill University for sharing the miR-378-KO mice and the WT littermates.
Funding. This work was supported by British Heart Foundation grant PG/19/86/34788 and National Institutes of Health grant P20GM-104320-01A to Q.S.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. H.W., Y.S., Y.W., and V.K. performed experiments and analyzed the data. H.W., Y.S., Y.W., and Q.S. designed the study. Y.S., V.K., and Q.S. wrote and edited the manuscript. R.I.M. edited the manuscript. Q.S. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.13491342.
H.W., Y.S., and Y.W. contributed equally to the work.
References
- 1.Hotamisligil GS Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell 2010;140:900–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su Q, Wang S, Baltzis D, Qu LK, Wong AH, Koromilas AE. Tyrosine phosphorylation acts as a molecular switch to full-scale activation of the eIF2alpha RNA-dependent protein kinase. Proc Natl Acad Sci U S A 2006;103:63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hummasti S, Hotamisligil GS. Endoplasmic reticulum stress and inflammation in obesity and diabetes. Circ Res 2010;107:579–591 [DOI] [PubMed] [Google Scholar]
- 4.Sud N, Rutledge AC, Pan K, Su Q. Activation of the dsRNA-activated protein kinase PKR in mitochondrial dysfunction and inflammatory stress in metabolic syndrome. Curr Pharm Des 2016;22:2697–2703 [DOI] [PubMed] [Google Scholar]
- 5.Hsu LC, Park JM, Zhang K, et al. . The protein kinase PKR is required for macrophage apoptosis after activation of Toll-like receptor 4. Nature 2004;428:341–345 [DOI] [PubMed] [Google Scholar]
- 6.Nakamura T, Furuhashi M, Li P, et al. . Double-stranded RNA-dependent protein kinase links pathogen sensing with stress and metabolic homeostasis. Cell 2010;140:338–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett RL, Blalock WL, Abtahi DM, Pan Y, Moyer SA, May WS. RAX, the PKR activator, sensitizes cells to inflammatory cytokines, serum withdrawal, chemotherapy, and viral infection. Blood 2006;108:821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carvalho BM, Oliveira AG, Ueno M, et al. . Modulation of double-stranded RNA-activated protein kinase in insulin sensitive tissues of obese humans. Obesity (Silver Spring) 2013;21:2452–2457 [DOI] [PubMed] [Google Scholar]
- 9.Nakamura T, Kunz RC, Zhang C, et al. . A critical role for PKR complexes with TRBP in immunometabolic regulation and eIF2α phosphorylation in obesity. Cell Rep 2015;11:295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carvalho-Filho MA, Carvalho BM, Oliveira AG, et al. . Double-stranded RNA-activated protein kinase is a key modulator of insulin sensitivity in physiological conditions and in obesity in mice. Endocrinology 2012;153:5261–5274 [DOI] [PubMed] [Google Scholar]
- 11.Manche L, Green SR, Schmedt C, Mathews MB. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol Cell Biol 1992;12:5238–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmedt C, Green SR, Manche L, Taylor DR, Ma Y, Mathews MB. Functional characterization of the RNA-binding domain and motif of the double-stranded RNA-dependent protein kinase DAI (PKR). J Mol Biol 1995;249:29–44 [DOI] [PubMed] [Google Scholar]
- 13.Bevilacqua PC, Cech TR. Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry 1996;35:9983–9994 [DOI] [PubMed] [Google Scholar]
- 14.Youssef OA, Safran SA, Nakamura T, Nix DA, Hotamisligil GS, Bass BL. Potential role for snoRNAs in PKR activation during metabolic stress. Proc Natl Acad Sci U S A 2015;112:5023–5028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol 2003;5:834–839 [DOI] [PubMed] [Google Scholar]
- 16.Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet 2003;34:263–264 [DOI] [PubMed] [Google Scholar]
- 17.Marques JT, Devosse T, Wang D, et al. . A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat Biotechnol 2006;24:559–565 [DOI] [PubMed] [Google Scholar]
- 18.Dabo S, Meurs EF. dsRNA-dependent protein kinase PKR and its role in stress, signaling and HCV infection. Viruses 2012;4:2598–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Latorre J, Moreno-Navarrete JM, Mercader JM, et al. . Decreased lipid metabolism but increased FA biosynthesis are coupled with changes in liver microRNAs in obese subjects with NAFLD. Int J Obes 2017;41:620–630 [DOI] [PubMed] [Google Scholar]
- 20.Sud N, Zhang H, Pan K, Cheng X, Cui J, Su Q. Aberrant expression of microRNA induced by high-fructose diet: implications in the pathogenesis of hyperlipidemia and hepatic insulin resistance. J Nutr Biochem 2017;43:125–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasquinelli AE MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 2012;13:271–282 [DOI] [PubMed] [Google Scholar]
- 22.Krist B, Florczyk U, Pietraszek-Gremplewicz K, Józkowicz A, Dulak J. The role of miR-378a in metabolism, angiogenesis, and muscle biology. Int J Endocrinol 2015;2015:281756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chakraborty C, Doss CG, Bandyopadhyay S, Agoramoorthy G. Influence of miRNA in insulin signaling pathway and insulin resistance: micro-molecules with a major role in type-2 diabetes. Wiley Interdiscip Rev RNA 2014;5:697–712 [DOI] [PubMed] [Google Scholar]
- 24.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mello T, Materozzi M, Galli A. PPARs and mitochondrial metabolism: from NAFLD to HCC. PPAR Res 2016;2016:7403230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gardner OS, Dewar BJ, Graves LM. Activation of mitogen-activated protein kinases by peroxisome proliferator-activated receptor ligands: an example of nongenomic signaling. Mol Pharmacol 2005;68:933–941 [DOI] [PubMed] [Google Scholar]
- 27.Gardner OS, Shiau CW, Chen CS, Graves LM. Peroxisome proliferator-activated receptor gamma-independent activation of p38 MAPK by thiazolidinediones involves calcium/calmodulin-dependent protein kinase II and protein kinase R: correlation with endoplasmic reticulum stress. J Biol Chem 2005;280:10109–10118 [DOI] [PubMed] [Google Scholar]
- 28.Irukayama-Tomobe Y, Miyauchi T, Kasuya Y, Sakai S, Goto K, Yamaguchi I. Activation of peroxisome proliferator-activated receptor-alpha decreases endothelin-1-induced p38 mitogen-activated protein kinase activation in cardiomyocytes. J Cardiovasc Pharmacol 2004;44(Suppl. 1):S358–S361 [DOI] [PubMed] [Google Scholar]
- 29.Lin J, Tarr PT, Yang R, et al. . PGC-1beta in the regulation of hepatic glucose and energy metabolism. J Biol Chem 2003;278:30843–30848 [DOI] [PubMed] [Google Scholar]
- 30.LoVerme J, Russo R, La Rana G, et al. . Rapid broad-spectrum analgesia through activation of peroxisome proliferator-activated receptor-alpha. J Pharmacol Exp Ther 2006;319:1051–1061 [DOI] [PubMed] [Google Scholar]
- 31.John E, Wienecke-Baldacchino A, Liivrand M, Heinäniemi M, Carlberg C, Sinkkonen L. Dataset integration identifies transcriptional regulation of microRNA genes by PPARγ in differentiating mouse 3T3-L1 adipocytes. Nucleic Acids Res 2012;40:4446–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knezevic I, Patel A, Sundaresan NR, et al. . A novel cardiomyocyte-enriched microRNA, miR-378, targets insulin-like growth factor 1 receptor: implications in postnatal cardiac remodeling and cell survival. J Biol Chem 2012;287:12913–12926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eichner LJ, Perry MC, Dufour CR, et al. . miR-378(∗) mediates metabolic shift in breast cancer cells via the PGC-1β/ERRγ transcriptional pathway. Cell Metab 2010;12:352–361 [DOI] [PubMed] [Google Scholar]
- 34.Carrer M, Liu N, Grueter CE, et al. . Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378*. Proc Natl Acad Sci U S A 2012;109:15330–15335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su Q, Baker C, Christian P, et al. . Hepatic mitochondrial and ER stress induced by defective PPARα signaling in the pathogenesis of hepatic steatosis. Am J Physiol Endocrinol Metab 2014;306:E1264–E1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar V, Mondal G, Dutta R, Mahato RI. Co-delivery of small molecule hedgehog inhibitor and miRNA for treating liver fibrosis. Biomaterials 2016;76:144–156 [DOI] [PubMed] [Google Scholar]
- 37.Nigi L, Grieco GE, Ventriglia G, et al. . MicroRNAs as regulators of insulin signaling: research updates and potential therapeutic perspectives in type 2 diabetes. Int J Mol Sci 2018;19:3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nallagatla SR, Hwang J, Toroney R, Zheng X, Cameron CE, Bevilacqua PC. 5′-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science 2007;318:1455–1458 [DOI] [PubMed] [Google Scholar]
- 39.Lund E, Dahlberg JE. Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs. Cold Spring Harb Symp Quant Biol 2006;71:59–66 [DOI] [PubMed] [Google Scholar]
- 40.Golec E, Lind L, Qayyum M, Blom AM, King BC. The noncoding RNA nc886 regulates PKR signaling and cytokine production in human cells. J Immunol 2019;202:131–141 [DOI] [PubMed] [Google Scholar]
- 41.Inoue K, Deng Z, Chen Y, et al. . Bone protection by inhibition of microRNA-182. Nat Commun 2018;9:4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su Q, Wang S, Baltzis D, et al. . Interferons induce tyrosine phosphorylation of the eIF2alpha kinase PKR through activation of Jak1 and Tyk2. EMBO Rep 2007;8:265–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su Q, Tsai J, Xu E, et al. . Apolipoprotein B100 acts as a molecular link between lipid-induced endoplasmic reticulum stress and hepatic insulin resistance. Hepatology 2009;50:77–84 [DOI] [PubMed] [Google Scholar]
- 44.Liu W, Cao H, Ye C, et al. . Hepatic miR-378 targets p110α and controls glucose and lipid homeostasis by modulating hepatic insulin signalling. Nat Commun 2014;5:5684. [DOI] [PubMed] [Google Scholar]
- 45.Zhang T, Hu J, Wang X, et al. . MicroRNA-378 promotes hepatic inflammation and fibrosis via modulation of the NF-κB-TNFα pathway. J Hepatol 2019;70:87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lancaster GI, Kammoun HL, Kraakman MJ, Kowalski GM, Bruce CR, Febbraio MA. PKR is not obligatory for high-fat diet-induced obesity and its associated metabolic and inflammatory complications. Nat Commun 2016;7:10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baltzis D, Li S, Koromilas AE. Functional characterization of pkr gene products expressed in cells from mice with a targeted deletion of the N terminus or C terminus domain of PKR. J Biol Chem 2002;277:38364–38372 [DOI] [PubMed] [Google Scholar]
- 48.Ivey KN, Srivastava D. microRNAs as developmental regulators. Cold Spring Harb Perspect Biol 2015;7:a008144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang T, Zhao X, Steer CJ, Yan G, Song G. A negative feedback loop between microRNA-378 and Nrf1 promotes the development of hepatosteatosis in mice treated with a high fat diet. Metabolism 2018;85:183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeon TI, Park JW, Ahn J, Jung CH, Ha TY. Fisetin protects against hepatosteatosis in mice by inhibiting miR-378. Mol Nutr Food Res 2013;57:1931–1937 [DOI] [PubMed] [Google Scholar]