Abstract
Developmental exposure to endocrine disrupting chemicals (EDCs), e.g., bisphenol A (BPA) or genistein (GEN), causes longstanding epigenome effects. MicroRNAs (miRs) regulate which mRNAs will be translated to proteins and thereby serve as the final checkpoint in epigenetic control. Scant amount is known, however, whether EDCs affect neural miRNA (miR) patterns. We aimed to test the hypothesis that developmental exposure of California mice (Peromyscus californicus) to GEN, BPA, or both chemicals influences hypothalamic miR/small RNA profiles and ascertain the extent such biomolecular alterations correlate with behavioral and metabolic changes. California mice were developmentally exposed to GEN (250 mg/kg feed weight, FW), GEN (250 mg/kg FW) + BPA (5 mg/kg FW), low dose (LD) BPA (5 mg/kg FW), or upper dose BPA (50 mg/kg FW). Adult offspring were tested in a battery of behavioral and metabolic tests; whereupon, mice were euthanized, frozen brains collected, small RNAs were isolated from hypothalamic punches, and subsequent small RNA sequencing. California mice exposed to one or both EDCs engaged in one or more repetitive behaviors. GEN, LD BPA, and UD BPA altered aspects of ultrasonic and audible vocalizations. Each EDC exposure led to sex-dependent differences in differentially expressed miR/small RNAs with miR7-2, miR146, and miR148a being increased in all female and male EDC exposed groups. Current findings reveal that developmental exposure to GEN and/or BPA affects hypothalamic miR/small RNA expression patterns, and such changes correlate with EDC-induced behavioral and metabolic alterations. miR146 is likely an important mediator and biomarker of EDC exposure in mammals, including humans.
Keywords: Bisphenol A (BPA), EDC, DOHaD, Epigenetics, Genistein, Small RNA, Phytoestrogens, Neurobehavior, Transcription, Translation, Cognition, Anxiety, Stereotypical Behaviors, Socio-communication behaviors
1. Introduction
Endocrine disrupting chemicals (EDCs) are widely prevalent in food products, everyday household items, and in the environment. Two main EDCs that have received considerable attention are bisphenol A (BPA), plasticizer, and the phytoestrogen, genistein (GEN). Developmental exposure to both chemicals disturbs normal brain programming that manifest in later neurobehavioral changes (Butler et al., 2020; Cheong et al., 2018; Jasarevic et al., 2011; Jasarevic et al., 2013; Johnson et al., 2016; Johnson et al., 2015a; Marshall et al., 2019; Rosenfeld, 2015, 2017; Rosenfeld and Trainor, 2014; Williams et al., 2013).
We have previously tested the effects of such chemicals in California mice (Peromyscus californicus) who may serve as a better model of most human societies as they monogamous and biparental (Rosenfeld et al., 2013). We found several behaviors vulnerable to BPA and other EDC exposure. Developmental exposure to BPA reduced territorial marking behavior by males, a behavior needed to protect him and his mate from potential intruders, and suppressed biparental care (Johnson et al., 2015a; Williams et al., 2013). We have recently shown that California mice developmentally exposed to low dose BPA (5 mg/kg feed weight) and GEN (250 mg/kg feed weight) show impairments at weaning and adulthood in socio-communication behaviors (Kaur et al., 2020; Marshall et al., 2019). Moreover, such EDC-induced behavioral deficits are associated with select micro (mi)RNAs and coding genes in the hypothalamus and hippocampus (Butler et al., 2020) and gut microbiome/metabolome changes (Kaur et al., 2020; Marshall et al., 2019).
We have also found that early exposure of California mice to BPA or ethinyl estradiol (E2, estrogen in birth control pills) altered the hypothalamic transcriptome profile (Johnson et al., 2017b). Even if a gene is transcribed, miRNAs (miRs) can act to block their translation. miR are initially transcribed as transcripts that can span several kilobases in length (Moreno-Moya et al., 2014; Van Wynsberghe et al., 2011). Such transcripts are then cleaved by RNase III enzymes, Drosha in the nucleus and Dicer in the cytoplasm to produce 70 nucleotide (nt) long precursor miR and then 19–22 nt long mature miRs, respectively. miR govern post transcriptional expression by associating with the multiprotein RNA induced silencing complex (RISC) and binding to the 3’ untranslated region of mRNA. This binding results in mRNA degradation or suppression of translation of the mRNA to a protein, such that miRs epigenetically silence certain genes.
In other tissues and organs, BPA and other EDC have been shown to induce shifts in miR patterns (Chou et al., 2017; De Felice et al., 2015; Gao et al., 2018; Li et al., 2015; Reed et al., 2018; Verbanck et al., 2017). In the hypothalamus, prenatal exposure to estradiol benzoate or polychlorinated biphenyls (PCBs) affects miR expression in a sex, age, developmental, and region-specific (medial preoptic nucleus, MPN and ventromedial nucleus, VMN) manner (Topper et al., 2015). However, effects of BPA on miR profiles in the mammalian brain remain an understudied area. To our knowledge, the only study in this area was our aforementioned study that examined select miRs in the hypothalamus and hippocampus of California mice (Butler et al., 2020). Given the importance of miRs in potentially understanding epigenetic-based disorders and as potential biomarkers, we sought to examine how developmental exposure to a lower dose of BPA, upper dose of BPA, GEN, and the combination of BPA and GEN affect global miR and other small RNA patterns in adult California mice. Moreover, we tested various behavioral domains in male and female offspring to then link potential biomolecular and behavioral changes. Our underlying hypothesis was that developmental exposure to one or both EDCs would lead to behavioral disturbances that associated with shifts in miR patterns in the hypothalamus, a brain region that regulates various traits, especially socio-sexual behaviors. At the outset, it was not clear if the combination of GEN and BPA would lead to enhanced effects on these parameters or whether concurrent GEN exposure may mitigate the potential harmful effects of BPA. We chose to examine these two EDCs because both lead to other epigenetic changes, in particular DNA methylation (Dolinoy et al., 2007; Dolinoy et al., 2006). It has been suggested co-exposure to both BPA and GEN results in the latter reversing harmful hypomethylation effects of the former (Dolinoy et al., 2007), which we speculated may be the case as well for miRs. BPA has been shown to induce miR changes in other organs (Chou et al., 2017; De Felice et al., 2015; Gao et al., 2018; Li et al., 2015; Reed et al., 2018; Verbanck et al., 2017).To test these hypotheses, California mice females were exposed for two weeks prior to conception to one of the two doses of BPA through the diet, GEN, GEN+BPA, or a phytoestrogen-free control (AIN93G) diet. Behavioral and metabolic testing was performed in adult male and female offspring followed by miR isolation and small RNA-seq with the hypothalamus of these same mice. mixOmics analyses (Rohart et al., 2017) was then used to integrate the small RNA-seq and phenotypic results.
2. Material and Methods
2.1. California Mice and Treatments
Female and male adult California mice were purchased from the Peromyscus Genetic Stock Center (PGSC) at the University of South Carolina (Columbia, SC). Breeder pairs were 60–90 days of age and free of common rodent pathogens. To ensure the mice did not carry any transmittable diseases, they were quarantined for 8 weeks after arrival at the University of Missouri. After this period and testing of sentinel mice who, the breeder pairs were transported to the Animal Sciences Research Center (ASRC). Additional breeder pairs are routinely purchased from the PGSC to maintain the outbred status of the colony. All experiments were approved by the University of Missouri Animal Care and Use Committee (Protocol Numbers 8693 and 9590) and performed in accordance with the recommendations in the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
All weaned mice were housed in polypropylene cages (10.5 inches × 6 inches, Allentown, NJ) to reduce exposure to bisphenol A (BPA). Polypropylene cages are considered to have minimal BPA (Howdeshell et al., 2003). Breeder pairs and parents with pups were housed in polysulfone cages (17 inches × 8.25 inches, Allentown, NJ), as our facility does not have large size polypropylene cages. Newer polysulfone cages have detectable amount of BPA, however, older cages do not, which were used in this study. Water was provided in glass bottles.
Virgin males and females (8–12 weeks of age) were randomly assigned to one of five diets: 1) a phytoestrogen-free AIN 93G diet (control-CTL diet, TD.95092; Envigo, Madison, WI) supplemented with 7% corn oil by weight to minimize phytoestrogenic contamination that would otherwise come from using soy oil as the primary fatty acid source; 2) AIN base diet supplemented with 250 mg/kg feed weight of genistein (GEN, TD.10691); 3) AIN base diet supplemented with a “lower dose- LD” of BPA (5 mg/kg feed weight, TD.170514); or 4) AIN base diet supplemented with a “upper dose-UD” of BPA (50 mg/kg feed weight, TD.09518), which have been documented to lead to internal serum concentrations in rodents (Jasarevic et al., 2013; Sieli et al., 2011) approximating those previously measured in pregnant women and the human population in general that is unknowingly exposed to this chemical (Padmanabhan et al., 2008; Teeguarden et al., 2011; Vandenberg et al., 2010; Vandenberg et al., 2007); or 5) AIN base diet supplemented with a combination LD-BPA and GEN. Diets 2–5 are identical to the control AIN diet in terms of nutrient composition. BPA and GEN were shipped to Envigo, where their laboratory added the chemicals into the base diet. These chemicals do not show to decrease diet palatability. The P0 females were exposed to one of the five diets for a two-week period before mating, throughout gestation, and lactation.
This treatment regimen and dosages were used to replicate maternal diet exposure of fetuses and neonates to GEN and BPA, which can be transmitted to the offspring both across the placenta and through the milk. While the past study that tested the combination of BPA and GEN used the upper dose of BPA (50 mg BPA/kg feed weight) (Dolinoy et al., 2007), we instead chose to use the lower dose of BPA for this combination group. The reason for this choice is that in our past California mice studies, the lower dose of BPA tends to lead to greater neurobehavioral differences (Kaur et al., 2020). Further, our goal in this study was also more so to examine low dose BPA effects alone and in combination with GEN. Moreover, in rodents, brain development has been shown to extend throughout the post-natal period (Howdeshell, 2002; Rice and Barone, 2000). Offspring were weaned at 30 days of age, which is standard for this species, and placed on the AIN 93G phytoestrogen-free diet.
From each litter, one male and one female pup were randomly selected for neurobehavioral and miR analyses. This was done to avoid litter bias effects. The remaining pups from the breeder pairs were selected to be strangers and not used in the testing procedures. California mice have an average litter of approximately three pups and female mice have 4 nipples. Five dams were included in each treatment groups for a total of 25 dams. One male and one female pup were selected from each litter. Thus, the experiments consisted of 5 CTL females, 5 CTL males, 5 GEN females, 5 GEN males, 5 LD-BPA females, 5 LD-BPA males, 5 UD-BPA females, 5 UD-BPA males, 5 GEN+BPA females, and 5 GEN+BPA males. After the completion of all the behavioral and metabolic testing detailed below, mice were humanely euthanized in accordance with AVMA euthanasia panel guidelines, brain and other tissues was frozen in liquid nitrogen, and then stored at −80°C until processing.
2.2. Barnes Maze Testing
At PND 90, test mice underwent Barnes maze testing to assess spatial learning and memory, as done previously (Jasarevic et al., 2011; Jasarevic et al., 2013; Rosenfeld, 2013; Williams et al., 2013). Briefly, the Barnes apparatus is a circular platform with 12 holes along the perimeter. Four intra-maze visual cues were placed above holes 3, 6, 9, and 12. A bright light was positioned above the maze, as a non-painful stimulus, to encourage test individuals to seek the escape hole.
Mice were randomly assigned to an escape hole at the beginning of the test week. The assigned escape hole was constant throughout the five-day testing period. Mice were placed in the testing room for 30 minutes prior to testing to allow for habituation. The test individual was then placed under a clean polypropylene box in the center of the maze. At the start of the trial, the box was removed, and timer started. The test concluded once the test individual entered the escape hole or when five minutes had elapsed. If the test individual did not enter escape hole on first day of testing, the animal was guided to the escape hole. After a 30-minute interval, a second trial was performed. The maze platform, polypropylene box, and escape hole were cleaned with 70% ethanol between each trial and each animal. This procedure also removed odor cues that could affect the performance of subsequent individuals. The tests were recorded with a Sony Handycam HDR-CX440 (San Diego, CA). Videos were analyzed using ANY-maze software version 6.2 (Stoelting, Wood Dale, IL). The program tracked the movement of the mouse and analyzed various parameters, including latency to find the correct escape hole, number of entries into correct and incorrect quadrants, total distance traveled, and velocity.
2.3. Elevated Plus Maze (EPM)
At PND97, individuals were then tested in the elevated plus maze (EPM), which measures anxiety-like behavior, exploratory, and repetitive behaviors, as described previously (Fountain et al., 2008; Jasarevic et al., 2011; Jasarevic et al., 2013; Williams et al., 2013). The apparatus consists of two open and two closed arms connected at the center and perpendicular to each other. The arms were raised 100 cm off the floor. The closed arms had walls that enclosed the space from the surroundings. Test individuals were acclimated in the testing room for 30 minutes prior to testing. The test began when the experimental animal was placed at the center and concluded after 5 minutes. The apparatus was cleaned with 70% ethanol between each trial, which also removed previous odor cues. The test was recorded with a Sony Handycam HDR-CX440 (San Diego, CA) and then analyzed by using the Observer software version 11.5 (Noldus, Leesberg, VA). Behaviors and duration of behaviors were analyzed including grooming, rearing, head dipping, time spent in center, open arms, and closed arms.
2.4. Reverse Barnes Maze Testing
At PND104, the mice underwent reverse Barnes maze testing, which tests ability to extinguish a learned response in that the escape hole was moved directly opposite of the one randomly selected for the initial Barnes maze test. One of the behaviors in rodents that mimics clinical signs observed in patients with autism spectrum disorders (ASD) is failure to extinguish a learned response (Crawley, 2012; Silverman et al., 2010). The mice underwent five days of consecutive testing to assess their ability to learn the new escape hole. All other procedures for reverse Barnes maze trials and parameters used for analyses were the same as for the initial Barnes maze testing.
2.5. Social Behavior Testing
At PND180, the test individuals underwent Crawley’s sociability and preference for social novelty three-chambered test, as detailed previously by (Marshall et al., 2019; Moy et al., 2004). This was done to examine for potential social deficits. The apparatus allows the test individual to move freely between the three chambers using two openings. The left and right chamber contained wire mesh cups that held stranger (novel) individuals that were similar in age, sex, and parental diet to the test animal. The stranger individuals though had different parental lineage relative to the test animal. Test and stranger individuals were acclimated to the testing room for 30 minutes. The test consisting of three trials. In the first trials, the test animal was placed in the center chamber and could roam freely within the chamber for five minutes. This allowed the animal to acclimate to the test apparatus. No strangers were placed in their wire cups in the first trial. Between trial 1 and 2, novel strangers were acclimated to the wire mesh cups for five minutes. In trial 2, stranger 1 was placed under the wire cup on the left chamber while the test individual was placed in the center of the maze. In trial 3, the test individual was placed in the center chamber and stranger 1 and 2 individuals were placed under the cup in the left and the right chambers. Trial 2 and 3 lasted for 10 minutes. This allowed for sufficient time to observe novel behaviors while avoiding habituation that might impact the results. The trials were video recorded using a Logitech Carl Zeiss Tessar HD 1080P (Newark, CA) camera mounted to a Joby Gorilla Pod Original Tripod (Daymen US Inc., Petaluma, CA). Video data were analyzed using Observer software version 11.5 (Noldus, Leesberg, VA). Behaviors such as rearing, grooming, and nose-to-nose contacts with the stranger mice (in Trials 2 and 3) were determined, along with the location of the mouse within the three-chambered apparatus.
2.6. Vocalizations
Following trial 3, ultrasonic and audible vocalizations produced in isolation were measured, as described previously (Johnson et al., 2018b; Marshall et al., 2019). The test individual was placed in a clean polypropylene box, which was then placed in chamber lined with 2 inches of acoustic foam paneling (Soundproof Cow, Chambersburg, PA). The box contained a light source (20-watt LED puck lights, Intertek, London, UK) and an Avisoft Bioacoustics CM16/CMPA40–5V microphone (Glienicke, Germany) that was 33 cm from the floor of the box. A microphone was attached to National Instruments USB 6351 data collection board, which was connected to a Dell OptiPlex 7010 (Dell Incorporated, Roundrock, TX). Vocalizations were recorded for 5 minutes. Recordings consist of audible calls and ultrasonic vocalizations (USVs) that were above 20 kHz and out of the hearing range for humans. After each test individual, the box was cleaned with 70% ethanol, which also removed previous odor cues. Data were collected by using code written by Dr. Katrin Schenk (Randolph College) and MATLAB 2015a.Ink version 8.5.0.197613 (R2015a) software (MathWorks, Natick, MA). Categories evaluated included number of syllables, syllable duration, syllable median frequency, average syllable power, and power percent above and below 20 kHz, as detailed previously (Johnson et al., 2018b; Marshall et al., 2019).
2.7. Indirect Calorimetry Testing
After the behavioral tests detailed above were completed, test individuals underwent indirect calorimetric testing for 3 days. Mice were tested in the Promethion continuous measurement indirect calorimetry system (Sable Systems International, Las Vegas, NV), as we have described previously (Butler et al., 2020; Johnson et al., 2017a; Johnson et al., 2015b). Data were divided into three 12-hour light cycles and three 12-hour dark cycles. Categories evaluated included general activity level, energy expenditure, respiratory quotient, body mass, and food and water intake.
2.8. EchoMRI
After completion of the indirect calorimetry testing, mice were tested in the EchoMRI-1100 (EchoMRI LLC, Houston, TX) to analyze body composition, as we have described previously (Butler et al., 2020; Johnson et al., 2017a; Johnson et al., 2015b). Parameters measured include lean tissue, fat tissue, and water content.
2.9. Hypothalamic Punches and RNA Isolation
Upon completion of all assessments at PND180, mice were humanely euthanized. Brains were removed from the cranium and rapidly frozen on dry ice. The frozen brains were stored at −80°C until further processing. The hypothalamic region, MPN, supraoptic nucleus (SON), and paraventricular nucleus (PVN),was micro-punched on dry ice, as we have done previously (Butler et al., 2020; Johnson et al., 2018a; Johnson et al., 2017b). A Harris Micro-Punch 1 mm in diameter and 1 mm in depth (Catalogue # 15091, Ted Pella, Redding, CA, USA) was used to obtain two bilateral punches that spanned these nuclei. RNA and miRs/small RNAs were isolated by using the AllPrep DNA/RNA/miRNA Universal Kit (Catalogue #80224; Qiagen, Hilden, Germany). After RNA isolation, RNA was quantified by spectrophotometrical analysis (Nanodrop 2000, ThermoFisher Scientific, Waltham, MA, USA). The results were further confirmed by analyzing the RNA on the Fragment Analyzer (Agilent, Santa Clara, CA). Only those samples that had an RNA integrity number (RIN) score above 7 were used for small RNA-seq analyses. For the small RNA sequencing studies, the number of replicates included 5 CTL females, 5 CTL males, 5 GEN females, 5 GEN males, 5 LD-BPA females, 5 LD-BPA males, 5 UD-BPA females, 5 UD-BPA males, 5 GEN+BPA females, and 5 GEN+BPA males.
2.10. Illumina TruSeq Small RNA Library Preparation
Small RNA sequencing was provided by the University of Missouri DNA Core Facility. Libraries were constructed by following the manufacturer’s protocol with reagents supplied in Illumina’s TruSeq small RNA sample preparation kit. Briefly, a two-step ligation process is performed with total RNA to add adapters to the 3’and 5’ ends of the small RNA, respectively. The adapter added small RNA was then reverse transcribed using SuperScript II Reverse Transcriptase and followed by PCR reaction to amplify and enrich the small RNA library. The amplified small RNA libraries were pooled and purified by gel electrophoresis on a 6% polyacrylamide gel as described by Illumina. Bands corresponding to the desired DNA insert size of 19 – 45 bases were excised from the gel. The DNA fragments (libraries) were recovered from the gel slice by elution with water and ethanol precipitation. Purified library pool was quantified with the Qubit HS DNA kit and the fragment size analyzed by an Agilent Fragment Analyzer Automated CE system. Libraries were diluted according to Illumina’s standard sequencing protocol for sequencing on the NextSeq 500 (Illumina).
2.11. Bioinformatics Analyses of Small RNA-seq Data
All reads were trimmed to remove adapters and low-quality sequences by using cutadapt program (Version 2.8) (Martin, 2011). Trimmed reads were used for miR/small RNA identification by using bowtie2 v2.2.7 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml), which aligns small RNA reads to a genome sequence. In this case, reads were aligned to both the Mus musculus genome (UCSC mm10) and separately to the Peromyscus maniculatus bairdii genome (GenBank GCA_000500345.1). The number of reads that aligned to each gene were counted with the featureCount tool from Subread software suite (Liao et al., 2014). The P. maniculatus bairdii annotation was published in 2013 ((https://www.ncbi.nlm.nih.gov/assembly/GCF_000500345.1/), and it resulted in much less features in comparison to using the Mus musculus GRCm38.99 annotation. While the sequencing and annotation of the California mouse is still in the draft stage, it has been shown with using 62,094 orthologous gene pairs for related cousins, P. leucopus, to Mus musculus that there is conserved synteny for species in the cricetine, includes Peromyscus species, and murid, includes Mus species, families in the Rodentia order (Long et al., 2019). In the current study, count data from GRCm38.99 annotation were thus used for final data analysis.
2.12. Principal Component Analysis (PCA) Analysis
Principal Component Analysis (PCA) for each sample was carried by using r package of mixOmics 6.12.1 (https://bioconductor.org/packages/release/bioc/vignettes/mixOmics/inst/doc/vignette.html#pca). 3D scatter plot was performed by pca3d r program v0.10.1 (https://rdrr.io/cran/pca3d/man/pca3d-package.html).
2.13. Differential Gene Expression Analysis (DGEA): DESeq2
The raw read counts were used to carry out differential gene expression analysis (DGEA) by means of DESeq2 to study the effects of maternal GEN ± BPA exposure on the hypothalamic small RNA expression (Love et al., 2014). The genes with an average of less than 5 read counts in at least one group were filtered out before carrying out DGEA. Genes were considered upregulated if they had an absolute fold-change ≥ 2 and adjusted p-value ≤ 0.05. To create sample-to-sample distance heatmap, raw counts data were transformed by rlog function in DEseq2 and transformed count matrix was transposed to get heatmap by DEseq2 built in dist function. Volcano plot was created by using plot function of DEseq2.
2.14. mRNA Target Predictions
To predict the potential mRNA targets for those miRs that were differentially expressed with the eight treatment comparisons, the miRror 2.0 tool (Friedman et al., 2010) was employed. Eleven databases were used for searching the targets. These included PITA_TOP, PicTar_4way, TargetRank-all (TRnk all), TargetScan Conserved (TScan), microCosm (mCosm), miRanda Conserved (microRNA.org), DIANA-microT (microT), EIMMO-MirZ (MirZ), miRDB, RNA22, MAMI, and Map2 (miRNAMap2). Based on the predicted mRNAs, we then performed functional enrichment analyses with the WEB-based Gene SeT AnaLysis Toolkit (WebGestalt) (Wang et al., 2013) and GO biological terms to determine which pathways might be affected.
2.15. qPCR Analyses
Total RNA including both mRNA and miR were isolated as described above. One μg of total RNA was reverse transcribed to cDNA with miScript II RT kit (Catalogue #218161; Qiagen) and miScript HiFlex buffer. The resulting cDNA was used for quantitative real-time polymerase chain reaction (qPCR). For qPCR analyses, the Bio-Rad SYBR Green Master Mix (Catalogue # 1725121, Hercules, CA) was used according to the manufacturer’s protocol. Primers were ordered from Integrated DNA Technologies, Inc. (IDT, Coralville, IA). qPCR analyses were performed for miR146a and ten mRNAs potentially affected by this miR. The ten mRNAs included Syt1, Syt14, Synpo2, Grid1, Nptn, Kdm2b, Cdk5, Ptpra, Klf4, and Ywhab. These ten mRNAs were chosen based on select ones previously identified to be affected by miR146a, as listed in in Table S2 for the following paper (https://www.pnas.org/content/suppl/2009/01/16/0812591106.DCSupplemental/ST2_PDF.pdf) and our own analyses as detailed above.
In pilot studies, we tested a range of housekeeping genes, including B2m1, Rpl7, and Actb, and found that B2m1 provides the most consistent results in that it is not affected in California mice by maternal exposure to BPA (Wright et al., 2017). Table S1 provides information on the primer sequences and efficiency. Primers for miR146a were designed based on a previous study (Hou et al., 2018). To calculate the amplification efficiency of primers, pooled sample was serially diluted to 1:10, 1:100, 1:1000, and 1:10000. Their resulting Ct values were plotted on a logarithmic scale along with corresponding concentrations. Next, a linear regression curve was generated through the data points, and the slope of the trend line was calculated. Finally, efficiency was calculated by using the equation of E = −1+10(−1/slope). For the actual assays, two replicates were tested for each sample. The Bio-Rad CFX Connect Real-Time PCR Detection System (Bio-Rad) was used for qPCR analysis. The cycling conditions for cDNA derived from coding genes were 1) 95°C for 5 min for polymerase activation 2) 40 cycles of; denaturation 15 seconds at 95 °C, annealing and extension 30 seconds at 56 °C, and 3) dissociation melt curve analysis of denaturation at 95°C, complete annealing at 65 °C, followed by a gradual increase in temperature up to 95°. For cDNA derived from miR, cycling conditions used were 1) 95°C for 3 min for polymerase activation, 2) 40 cycles of; denaturation 15 seconds at 95 °C, annealing and extension 30 seconds at 59 °C, and 3) dissociation melt curve analysis of denaturation at 95°C, complete annealing at 65 °C, followed by a gradual increase in temperature up to 95°.
2.16. Statistical Analyses of Behavioral, Metabolic, and qPCR Data
Data were analyzed by using SAS version 9.4 Software (SAS Institute, Cary, NC). A split plot in space and time (an ANOVA-based method) was used to analyze the data, as detailed by (Steel, 1996). The main plot consisted of maternal treatment, and the subplot was F1 pup sex and maternal treatment × F1 pup sex. Behavioral assessments listed above were first recorded with a camera, with the manners noted above, timed using a stopwatch, and recorded. All dependent variables including social behaviors, ultrasonic vocalization, and elevated plus maze tests were analyzed by using the general linear model (GLM) procedure of SAS. Sources of variation considered were parent treatment, test mouse sex, and interaction between treatment and sex. Treatment effects were determined with dam as the error term (experimental unit). Differences between treatment groups and control were determined by Fisher’s protected least-significant difference (LSD). The LSD was only calculated if the overall F test was significant.
Dependent variables assessed in the Barnes maze, including distance traveled and velocity were analyzed as a split plot in space and time (Steel, 1996). The linear statistical model contained the fixed effects of maternal treatment, sex, day and all possible interactions with treatment, sex and day. To determine whether there were litter effects, source (dam) within treatment was used as the denominator of F for maternal treatment, source within day × sex was used as the denominator for sex and interaction of treatment × sex, source within day was used as the denominator of F for day and the remaining interaction used the residual mean square as the denominator of F.
Latency data for the Barnes and reverse Barnes maze testing were further analyzed by using the PROC PHREG and Proportional Hazard Ratio functions in the SAS. These analyses adjust for right-censoring (defined here as not locating the escape hole within the allotted time of 300 seconds) while still accommodating the study design of 300 s/trial. Data are reported as a hazard ratio that signifies the odds of a subject in a treatment group locating the correct escape hole compared with the other groups tested. A significant result indicates the odds are not 1:1. A result >1 indicates the test group was more likely to locate the correct escape hole than all other groups tested. A result <1 indicates that the treatment group is less likely to locate the correct escape hole compared with the other study groups. The litter was used as the denominator of F for the effects of maternal treatment, offspring sex and test day, and potential interactions between maternal treatment, offspring sex and test day. Latency data are reported as the mean and 95% lower and upper confidence limits.
Indirect calorimetric testing data were analyzed as a repeated measurement analysis in which the main plot contained the effects of the maternal treatment and offspring sex. The denominator of F for the main plot was litter within maternal diet and offspring sex. The subplot contained the time series of both day and cycle. The day and cycle were factorial arranged in which the cycle contained two cycles (dark and light) and day contained the two days in which mice were measured in this unit. The subplot effect of day and cycle and day × cycle and the interactions of day and cycle with the main plot effect were tested using litter within maternal diet, offspring sex, day, and cycle as the denominator of F.
EchoMRI data were analyzed as a complete randomized design in which treatments were arranged as a five by two factorial (five maternal treatment and two offspring sexes). Dam within maternal treatment was used as error term to determine maternal treatment effects. If the overall F was significant, then differences were determined using Fisher’s protected LSD. All data are presented as actual means and standard error of the mean (SEM).
mRNA and miR expression data as determined by qPCR analyses were normalized relative to housekeeping genes (B2m1 for mRNA and U6 for miRs). They were then analyzed by using the PROC GLM procedure of SAS version 9.4 Software (SAS Institute, Cary, NC). Graphs are based though on 2-ΔΔCt values relative to the CTL values, whose mean value was set to 1 for graphing purposes.
2.16. Integrative Correlation Analyses
We used the mixOmics R package (Rohart et al., 2017) to correlate the behavioral and metabolic outcomes with small RNA-seq profiles in the hypothalamus. We conducted sparse discriminant analysis with partial least square regression with function ‘block.splsda’. The circos plot was generated by using the ‘circosPlot’ function with correlations calculated by the method described by González, et al. (González et al., 2012). A correlation coefficient ≥ 0.85 was used as the cutoff.
3. Results
3.1. Barnes Maze and Reverse Barnes Maze Testing
For the various parameters analyzed in the Barnes maze, no treatment X sex interactions were evident. Thus, the male and female data for all categories were considered together. The only significant difference for latency was California mice developmentally exposed to GEN showed reduced latency to enter the correct escape hole compared to CTL (GEN: 112.81 ± 9.27 seconds-s vs. CTL: 156.95 ± 10.58 s, p < 0.001, Fig. S1A). For speed within the maze, those exposed to GEN or UD BPA had increased velocity relative to CTL (GEN: 0.11 ± 0.006 meters- m/s; UP BPA: 0.12 ± 0.007 m/s; CTL: 0.10 ± 0.005 m/s, p < 0.01, Fig. S1B).
When we further analyzed the Barnes maze latency data with a hazard ratio approach, it is only on day 3 of Barnes maze testing are differences between treatment groups evident. On this day, GEN exposed individuals show an increase likelihood of locating the escape hole relative to CTL (GEN to CTL Ratio: 3.86, lower 95% CI: 1.85, and upper 95% CI 13.77; p = 0.01, Fig. S2A). In contrast, LD BPA individuals showed reduced likelihood of locating the escape hole on this day (LD BPA to CTL ratio: 0.25, lower 95% CI 0.06, and upper 95% CI: 1.04; p = 0.03, Fig. S2B). Reverse Barnes maze latency data was analyzed with this same approach and no treatment differences were observed on any of the days (Fig. S2B).
3.2. EPM testing
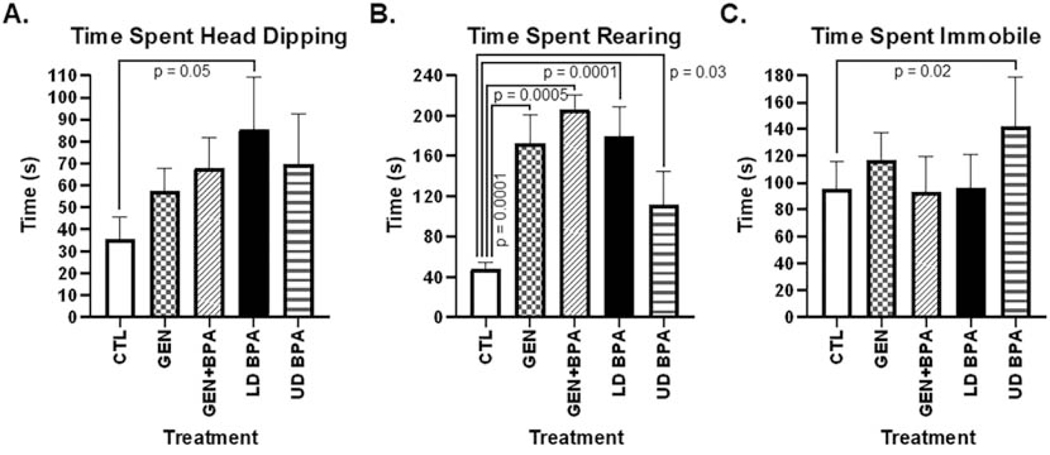
While no differences were observed in frequency or duration spent in the open or closed arms, treatment differences were noted for rearing behaviors and time spent immobile. Those exposed to LD BPA spent more time engaged in head-dipping behaviors relative to controls (LD BPA: 85.61 ± 23.61 s vs. CTL: 35.71 ± 9.75 s, p = 0.05, Fig. 1A). Those developmentally exposed to GEN, GEN + BPA, and LD BPA had increased duration of rearing behaviors relative to CTL (GEN: 172.50 ± 28.73 s, GEN + BPA: 206.33 ± 14.63 s, LD BPA: 179.92 ± 29.34 s vs. CTL: 48.30 ± 6.00 s; p = 0.0001 to 0.0005; Fig. 1B). In contrast, UD BPA spent less time rearing than CTL (111.04 ± 33.72 s; p = 0.03). However, California mice offspring developmentally exposed to UD BPA spent more time immobile than controls (UD BPA: 141.80 ± 37.16 s vs. CTL: 95.66 ± 19.87 s; p = 0.02; Fig. 1C).
Fig. 1.
EPM results. This test measures anxiogenic, exploratory and stereotypical behaviors, and the duration of the test was five minutes. Anxiogenic behaviors are determined based on greater time spent in the closed arms and generally remaining immobile or freezing. Exploratory behaviors are indicated by amount of time spent in the open arms and mobile. Rearing behaviors includes head dipping and rearing. The duration of the test is five minutes. While all parameters were analyzed, only those that showed differences are included. A) Time spent engaging in head dipping behaviors. B) Time spent rearing. C) Time spent immobile. The p value differences are included above the bar graphs.
3.3. Socio-Communication Behaviors
When mice were tested in the social testing at PND 180 days of age, UD BPA showed a trend to spend less time with the stranger 2 relative to stranger 1 in the third trial (25.2 ± 12.3 s vs. 78.1 ± 28.3 s; p =0.06, Fig. S3).
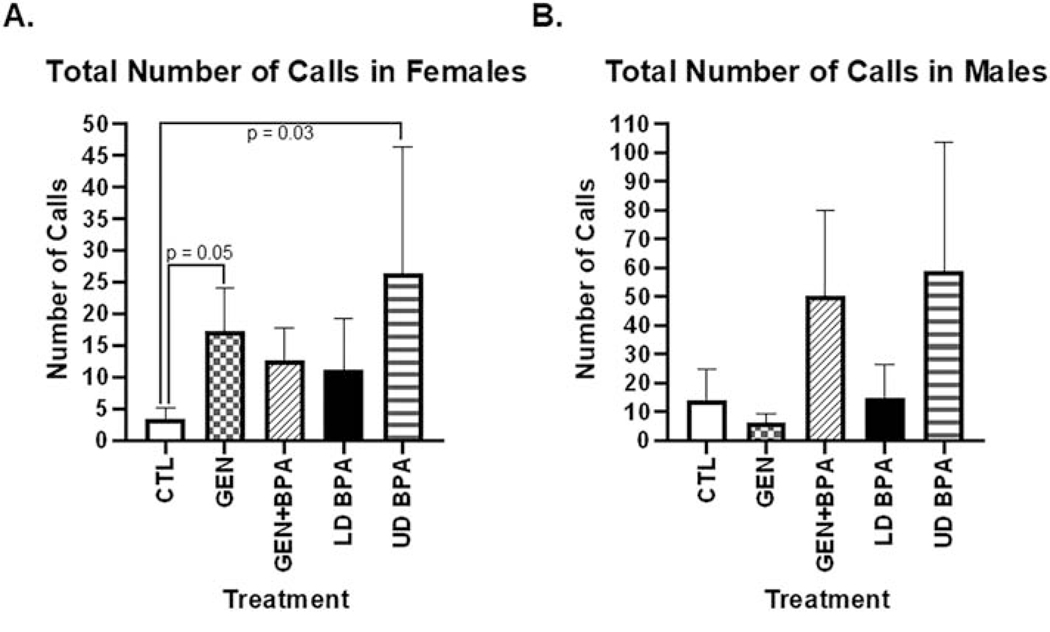
For some of the vocalization parameters, treatment X sex interactions were observed. For total number of calls in isolation, females exposed to GEN and UD BPA exhibited greater number of calls than CTL females (GEN: 17.4 ± 6.68; UD BPA: 26.4 ± 19.96 vs CTL: 3.4 ± 1.75; p =0.03 to 0.05; Fig. 2A). No differences in total number of calls was observed in any of the male groups (Fig. 2B), However, males exposed to LD BPA showed greater duration of calls relative to CTL males (LD BPA: 0.009 ± 0.001 s vs. CTL: 0.007 ± 0.0009 s; p = 0.03; Fig. S4); whereas, no differences were detected in the female groups for this category.
Fig. 2.
Total number of calls emitted during vocalization testing. This test was conducted for five minutes. All parameters listed in the methods were anlayzed, bu only total number of calls showed maternal treatment X sex differences. A) Females. B) Males. The p value differences are included above the bar graphs.
For fraction of calls above 20 kHz, i.e. in the ultrasonic range, females exposed to GEN, GEN + BPA, and LD BPA had increased number of calls in this range compared to controls (GEN: 46.15 ± 27.43; GEN + BPA: 286.41 ± 246.98; LD BPA: 173.35 ± 89.95 vs. CTL: 15.38 ± 10.34; p = 0.02 to 0.03; Fig. S5). Similarly, males exposed to GEN and LD BPA showed greater number of calls above 20 kHz than CTL males (GEN: 93.52 ± 48.07; LD BPA: 190.30 ± 111.61 vs. CTL: 12.23 ± 10.29; p = 0.02 to 0.03; Fig. S5).
Bursts are a collection of calls, and for this category only a significant treatment effect was detected. California mice offspring exposed to UD BPA had greater number of calls in a given burst than CTL (UD BPA: 2.5 ± 1.08 vs. CTL: 0.5 ± 0.40; p = 0.03; Fig. S6).
3.4. Indirect Calorimetry Testing
In the indirect calorimetry testing, GEN individual had reduced RQ value compared to CTL (GEN: 1.73 ± 0.08 CO2 released/O2 absorbed vs. CTL: 1.84 ± 0.08 CO2 released/O2 absorbed; p = 0.01; Fig. S7). This group also had reduced energy expenditure vs. CTL (GEN: 5.87 ± 0.31 kcal/day vs CTL: 6.42 ± 0.30 kcal/day p = 0.004; Fig. S7). Average body mass was reduced in all EDC exposed groups but GEN + BPA relative to CTL (GEN: 33.40 ± 0.57 g; LD BPA: 33.46 ± 0.68 g; UD BPA: 29.57 ± 1.23 g vs CTL: 35.37 ± 1.0g; p = 0.001 to 0.03; Fig. S7). No other parameters analyzed with indirect calorimetry testing differed based on treatment.
3.5. EchoMRI
In this assessment, the only category that differed according to treatment and sex was lean mass. Females exposed to GEN + BPA and UD BPA had increased lean mass relative to CTL (GEN + BPA: 28.30 ± 2.02g; UD BPA: 28.62 ± 2.89g vs. CTL: 30.58 ± 1.52g; p = 0.01 to 0.05; Fig. S8). In contrast, males exposed to GEN or GEN + BPA had reduced lean mass relative to CTL (GEN: 26.14 ± 0.59g; GEN + BPA: 26.41 ± 1.12g vs. CTL: 30.58 ± 1.52g; p = 0.01 to 0.03; Fig. S8).
3.6. Differentially Expressed miRs and Other Small RNAs
Small RNA-seq was performed on hypothalamuses from all individuals in the five treatment groups. The average number of small RNA reads was 12,603,667, the average number of mapped reads was 10,189,234, and the average mapping rate percentage was 81.28% (Table S2).
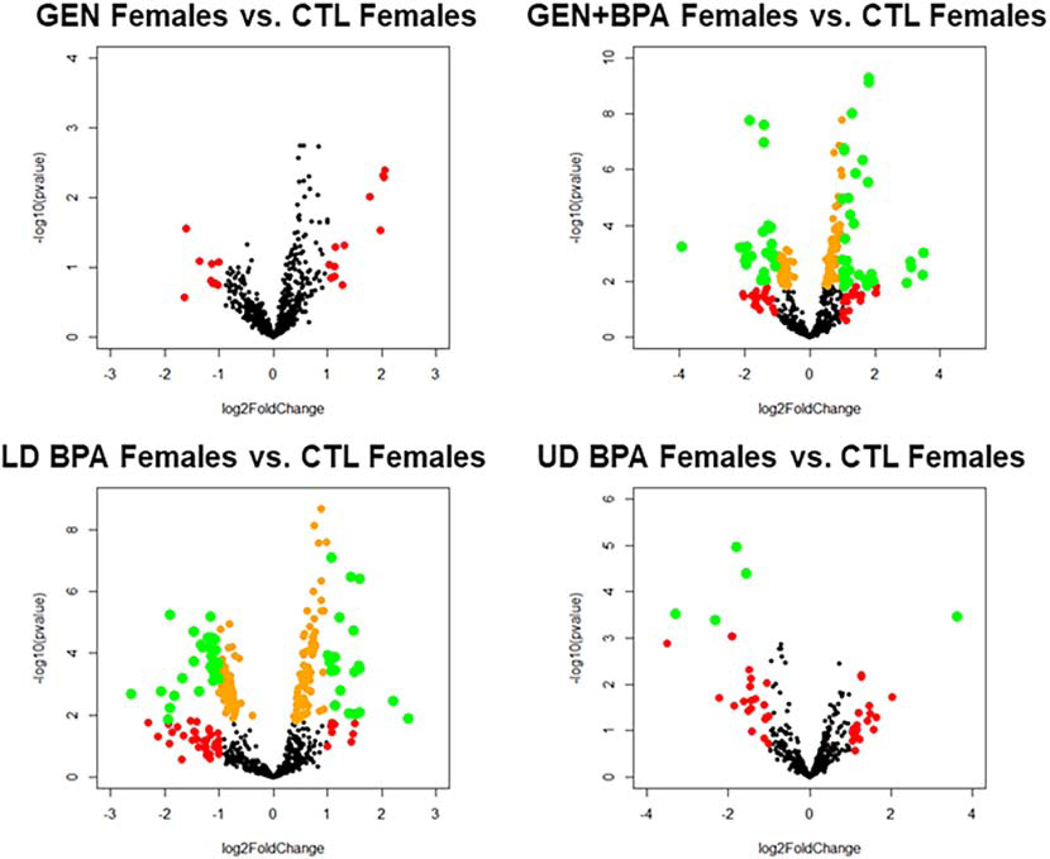
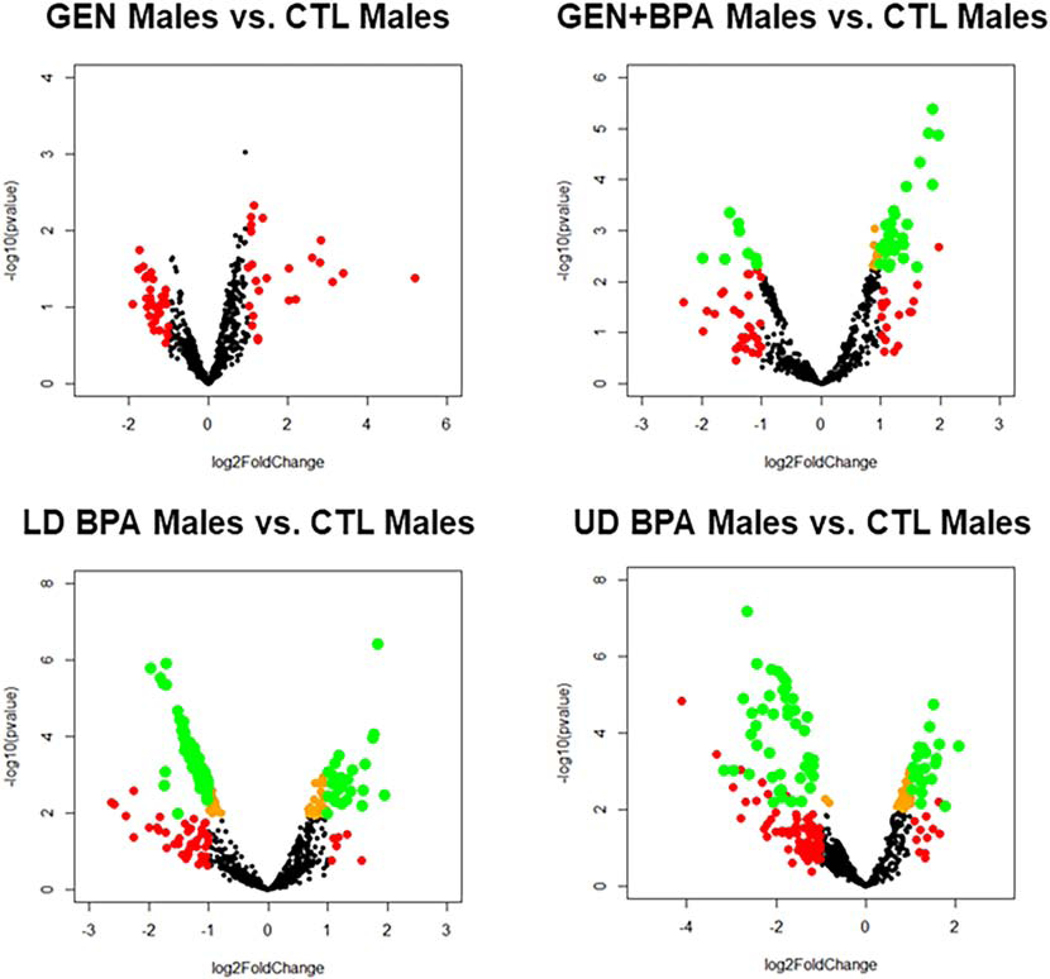
As shown in the 3D PCA plots and heatmaps for the female groups, for all four comparisons (GEN females vs. CTL females, GEN+BPA females vs. CTL females; LD BPA females vs. CTL females, UD BPA females vs. CTL females), there is separation based on treatment group (Fig. S9). Volcano plots indicates for all four comparisons, several small RNAs were differentially expressed (DE), but for GEN females vs. CTL females, none were DE based on an adjusted p value (FDR, Fig. 3). The 3D PCA plots and heatmaps for the male groups (GEN males vs. CTL males, GEN+BPA males vs. CTL males, LD BPA males vs. CTL males, and UD BPA males vs. CTL males), separation between treatment groups is evident. Similar to the female groups, volcano plots for male groups shows that all four comparisons have small RNAs that are DE, but the GEN males vs. CTL males do not have any that are significant based on a FDR (Fig. 4).
Fig. 3.
Volcano plots for female comparison groups. Transcripts represented by Orange: p < 0.05; Transcripts represented by Red: log2 Fold Change < −1 or > 1; Transcripts represented by Green: p < 0.05 AND log2 Fold Change < −1 or > 1.
Fig. 4.
Volcano plots for male comparison groups. Transcripts represented by Orange: p < 0.05; Transcripts represented by Red: log2 Fold Change < −1 or > 1; Transcripts represented by Green: p < 0.05 AND log2 Fold Change < −1 or > 1.
Supplemental Files 1 and 2 provides the small RNA that are differentially expressed in the four female and four male treatment group comparisons, respectively. As noted in the volcano plot, no small RNAs were DE in GEN females vs. CTL females and GEN males vs. CTL males based on FDR. However, several were DE based on p value alone. Tables 1–8 list up to the 25 DE miRs and small RNAs for all the female and male comparisons. As the GEN female and male groups did not have any DE miRs or small RNAs based on FDR relative to CTL counterparts, Tables 1 and 5 are instead based on p value. The UD BPA females vs. CTL females only demonstrated 6 DE (based on FDR) miR or small RNA compared to CTL females (Table 4).
Table 1.
The top 25 miR and other small RNA expression differences in GEN females vs. CTL females.
| Gene Symbol | Fold Change GEN Females vs. CTL Females | P value* | Directionality in GEN Females vs. CTL females |
|---|---|---|---|
| miR1298 | 1.4034354 | 0.001781 | ↑ |
| Gm23751 | 1.47135797 | 0.00179 | ↑ |
| miR146 | 1.78836542 | 0.001875 | ↑ |
| miR421 | 1.36924469 | 0.002702 | ↑ |
| miR96 | 4.14480504 | 0.004023 | ↑ |
| miR182 | 4.0615682 | 0.004832 | ↑ |
| Snord88c | 1.56719727 | 0.004985 | ↑ |
| Gm23287 | 4.09486096 | 0.005109 | ↑ |
| miR222 | 1.46465536 | 0.005765 | ↑ |
| miR298 | 1.38872638 | 0.005994 | ↑ |
| miR148a | 1.59102935 | 0.007445 | ↑ |
| miR383 | 1.76688733 | 0.009317 | ↑ |
| miRlet7f-2 | 1.48499733 | 0.009686 | ↑ |
| miR183 | 3.40486272 | 0.009734 | ↑ |
| AC161763.1 | 1.3584389 | 0.012621 | ↑ |
| miRlet7a-2 | 1.39544508 | 0.017995 | ↑ |
| miR7j | 1.36856277 | 0.019257 | ↑ |
| miR7-1 | 1.99505005 | 0.020599 | ↑ |
| miR335 | 1.38574501 | 0.020992 | ↑ |
| miR184 | 1.63480121 | 0.022064 | ↑ |
| miR7-2 | 1.99626025 | 0.022486 | ↑ |
| miR211 | 1.80300333 | 0.022902 | ↑ |
| Gm26016 | 0.32753953 | 0.027425 | ↓ |
| miR200b | 3.89660695 | 0.029901 | ↑ |
| miR98 | 1.35469043 | 0.031589 | ↑ |
For this comparison, no miRs or other small RNAs were different based on FDR, and thus, this table is based on p value.
Table 8.
The top 25 miR and other small RNA expression differences in UD BPA males vs. CTL males.
| Gene Symbol | Fold Change UD BPA Males vs. CTL Males | Padj (FDR) | Directionality in UD BPA Males vs. CTL Males |
|---|---|---|---|
| Gm25106 | 0.16061368 | 4.00E–05 | ↓ |
| Gm26316 | 0.2322568 | 0.00036661 | ↓ |
| Gm23744 | 0.18674006 | 0.00036661 | ↓ |
| Gm24497 | 0.26131741 | 0.00036661 | ↓ |
| Gm23444 | 0.28925899 | 0.00038949 | ↓ |
| Rnu2–10 | 0.29219499 | 0.00038949 | ↓ |
| Gm23472 | 0.27816116 | 0.00038949 | ↓ |
| Gm24950 | 0.27975825 | 0.00049597 | ↓ |
| Rnu5g | 0.29202826 | 0.00049597 | ↓ |
| Gm23971 | 0.32552228 | 0.00057419 | ↓ |
| Gm23849 | 0.29380949 | 0.00057419 | ↓ |
| Rnu1b6 | 0.15089703 | 0.00057419 | ↓ |
| Gm23143 | 0.22787631 | 0.00057419 | ↓ |
| miR199f-2 | 2.84535906 | 0.00076347 | ↑ |
| Gm25899 | 0.20408402 | 0.00091136 | ↓ |
| Gm22068 | 0.29977858 | 0.00091136 | ↓ |
| Gm25813 | 0.33605557 | 0.00093013 | ↓ |
| Gm23510 | 0.17360752 | 0.00099051 | ↓ |
| Gm22122 | 0.23975897 | 0.00100378 | ↓ |
| Rnu1a1 | 0.30158987 | 0.00100378 | ↓ |
| Gm25360 | 0.40432493 | 0.00108967 | ↓ |
| Gm25939 | 0.34247169 | 0.00162679 | ↓ |
| miR10b | 2.71404529 | 0.00170746 | ↑ |
| Gm25189 | 0.18492581 | 0.00170746 | ↓ |
| Gm25313 | 0.39075083 | 0.00213088 | ↓ |
Table 5.
The top 25 miR and other small RNA expression differences in GEN males vs. CTL males.
| Gene Symbol | Fold Change GEN Males vs. CTL Males | P value* | Directionality in GEN Males vs. CTL Males |
|---|---|---|---|
| miR448 | 1.89053139 | 0.00093793 | ↑ |
| miR1298 | 2.18946998 | 0.00468859 | ↑ |
| miR1264 | 2.09720073 | 0.00672952 | ↑ |
| miR346 | 2.57542128 | 0.00691474 | ↑ |
| miR221 | 2.10156859 | 0.00832021 | ↑ |
| miR212 | 1.90630374 | 0.00955776 | ↑ |
| miR148a | 2.10082364 | 0.01031741 | ↑ |
| miR34c | 1.60263051 | 0.01166418 | ↑ |
| miRlet7f-2 | 1.75260685 | 0.01227667 | ↑ |
| Gm24463 | 1.75358135 | 0.01327666 | ↑ |
| Gm44347 | 7.12243107 | 0.01352464 | ↑ |
| miR224 | 1.89818484 | 0.01431426 | ↑ |
| miR365-2 | 1.86560704 | 0.01473562 | ↑ |
| Gm22962 | 1.6830436 | 0.01617742 | ↑ |
| Rnu1b6 | 0.30060178 | 0.01786225 | ↓ |
| Gm22946 | 6.0923221 | 0.02270571 | ↑ |
| Gm24265 | 0.53147352 | 0.0229547 | ↓ |
| miRlet7g | 1.68401428 | 0.02332439 | ↑ |
| Gm25313 | 0.52589828 | 0.02438234 | ↓ |
| miR195a | 1.78748994 | 0.02509915 | ↑ |
| miR335 | 1.65120366 | 0.02625555 | ↑ |
| Gm44469 | 6.98777892 | 0.02653117 | ↑ |
| Snord98 | 1.60248761 | 0.02686947 | ↑ |
| miR7j | 1.6218154 | 0.02727921 | ↑ |
| miR344b | 1.6851915 | 0.02740922 | ↑ |
For this comparison, no miRs or other small RNAs were different based on FDR, and thus, this table is based on P value.
Table 4.
Differentially expressed miR and other small RNA in UD BPA females vs. CTL females.
| Gene Symbol | Fold Change UD BPA Females vs. CTL Females | Padj (FDR) | Directionality in UD BPA Females vs. CTL Females |
|---|---|---|---|
| Gm25106 | 0.28808677 | 0.00810887 | ↓ |
| Gm23744 | 0.3386575 | 0.01466976 | ↓ |
| miR351 | 95.4802705 | 0.04999373 | ↑ |
| Snora47 | 0.10231611 | 0.04999373 | ↓ |
| Gm44347 | 12.3950411 | 0.04999373 | ↑ |
| Gm49697 | 0.20162595 | 0.04999373 | ↓ |
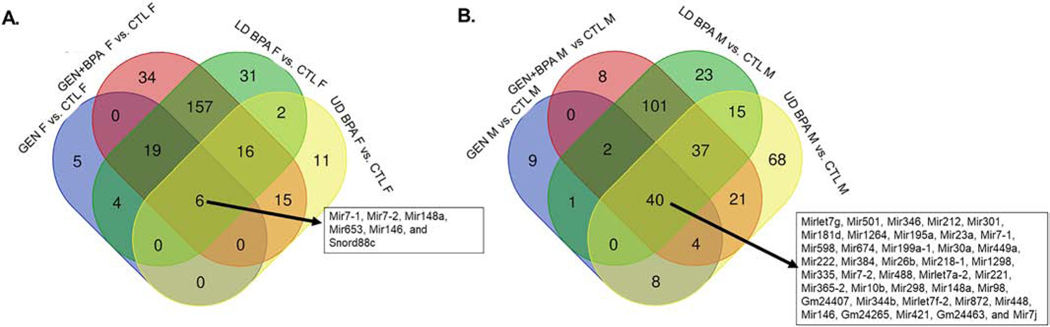
After determining the miRs and other small RNAs that were DE for the female and male comparisons, Venn diagram analyses was then used to determine which miRs and other small RNAs overlapped in the female and male group comparisons. To include the GEN comparison groups, small RNAs that were DE based on p value rather than FDR were considered. As shown in Fig. 5A, six miRs/small RNAs (miR7-1, miR7-2, Mir148a, Mir653, Mir146, and Snord88c) were DE in the female comparisons. Forty (Mirlet7g, Mir501, Mir346, Mir212, Mir301, Mir181d, Mir1264, Mir195a, Mir23a, miR7-1, Mir598, Mir674, Mir199a-1, Mir30a, Mir449a, Mir222, Mir384, Mir26b, miR218-1, Mir1298, Mir335, miR7-2, Mir488, Mirlet7a-2, Mir221, miR365-2, Mir10b, Mir298, Mir148a, Mir98, Gm24407, Mir344b, Mirlet7f-2, Mir872, Mir448, Mir146, Gm24265, Mir421, Gm24463, and Mir7j) overlapped in the male comparisons (Fig. 5B). The three miRs that overlap and were upregulated in all female and male EDC exposed group comparisons are miR7-2, miR146, and miR148a. One or more of these miRs are among the top DE miRs/small RNAs in GEN females vs. CTL females (bolded in Table 1), GEN+BPA females vs. CTL females (bolded in Table 2), LD BPA females vs. CTL females vs. CTL females (bolded in Table 3), GEN males vs. CTL males (bolded in Table 5), GEN+BPA males vs. CTL males (bolded in Table 6), and LD BPA males vs. CTL males (bolded in Table 7).
Fig. 5.
Venn diagrams of differentially expressed (DE) miRs/small RNAs. These diagrams were put together to determine which if any miRs and other small RNAs overlapped for the various comparisons with female and male groups. As the GEN comparison group only showed DE miRs/small RNAs based on p value, genes were selected based on p ≤ 0.05. A) Female comparison groups. B) Male comparison groups. Six and 40 miR/small RNAs overlapped respectively in the female and male comparisons. For all comparisons, miR146 and miR148 were DE and greater in the EDC-treated group.
Table 2.
The top 25 miR and other small RNA expression differences in GEN+BPA females vs. CTL females.
| Gene Symbol | Fold Change GEN+BPA Females vs. CTL Females | Padj (FDR) | Directionality in GEN+BPA Females vs. CTL Females |
|---|---|---|---|
| miR7-1 | 3.51464988 | 3.02E–07 | ↑ |
| miR7-2 | 3.52656827 | 3.02E–07 | ↑ |
| miR148a | 2.4768534 | 2.67E–06 | ↑ |
| miR501 | 1.99363376 | 2.85E–06 | ↑ |
| Gm25106 | 0.28093969 | 2.85E–06 | ↓ |
| Gm25099 | 0.38252947 | 3.48E–06 | ↓ |
| Gm25313 | 0.38261344 | 1.22E–05 | ↓ |
| miR335 | 1.8787968 | 1.35E–05 | ↑ |
| miR384 | 2.10328996 | 1.60E–05 | ↑ |
| miR212 | 2.09361049 | 1.72E–05 | ↑ |
| Gm24195 | 1.68188942 | 1.82E–05 | ↑ |
| miR146 | 3.08513212 | 3.05E–05 | ↑ |
| miR421 | 1.93443652 | 6.43E–05 | ↑ |
| miR211 | 2.68126249 | 7.66E–05 | ↑ |
| miR1264 | 1.97334103 | 8.35E–05 | ↑ |
| miR449a | 3.45553117 | 0.00013993 | ↑ |
| miR218-1 | 1.84718134 | 0.00042566 | ↑ |
| miR152 | 2.30855259 | 0.00046593 | ↑ |
| miR346 | 2.0275343 | 0.00047363 | ↑ |
| miR135b | 1.84348865 | 0.00066893 | ↑ |
| miR218-2 | 1.86308793 | 0.00066893 | ↑ |
| miR362 | 1.74362186 | 0.00072305 | ↑ |
| miR199a-1 | 2.36645446 | 0.00144546 | ↑ |
| miR301 | 1.63232824 | 0.00186302 | ↑ |
| miR10b | 2.59065821 | 0.00279282 | ↑ |
Table 3.
The top 25 miR and other small RNA expression differences in LD BPA females vs. CTL females.
| Gene Symbol | Fold Change LD BPA Females vs. CTL Females | Padj (FDR) | Directionality in LD BPA Females vs. CTL Females |
|---|---|---|---|
| miR7-1 | 2.921183 | 1.84E–12 | ↑ |
| miR7-2 | 2.91071 | 1.84E–12 | ↑ |
| miR146 | 3.130154 | 1.73E–10 | ↑ |
| miR98 | 1.838709 | 4.08E–07 | ↑ |
| miR92b | 1.687184 | 1.11E–06 | ↑ |
| miR384 | 1.773278 | 3.04E–06 | ↑ |
| miR212 | 1.969491 | 3.04E–06 | ↑ |
| miR148a | 2.099285 | 8.07E–06 | ↑ |
| miR211 | 2.705332 | 2.92E–05 | ↑ |
| miR10a | 3.03354 | 2.96E–05 | ↑ |
| miR1264 | 1.842555 | 3.07E–05 | ↑ |
| miR421 | 1.668585 | 6.47E–05 | ↑ |
| miRlet7f-2 | 1.842017 | 0.00010982 | ↑ |
| miR221 | 1.915698 | 0.00020568 | ↑ |
| miR26a-1 | 1.542192 | 0.00020568 | ↑ |
| Gm24195 | 1.836472 | 0.00020568 | ↑ |
| Gm25106 | 0.269411 | 0.00025998 | ↓ |
| Gm25313 | 0.452563 | 0.00029023 | ↓ |
| miR199a-1 | 2.345227 | 0.00029557 | ↑ |
| miR222 | 1.685525 | 0.00029959 | ↑ |
| Rnu2-10 | 0.57459 | 0.00039922 | ↓ |
| miR92-2 | 1.590633 | 0.0004749 | ↑ |
| Gm25099 | 0.508079 | 0.00056575 | ↓ |
| miR10b | 2.801688 | 0.0006132 | ↑ |
| miRlet7a-2 | 1.699544 | 0.0006132 | ↑ |
Table 6.
The top 25 miR and other small RNA expression differences in GEN+BPA males vs. CTL males.
| Gene Symbol | Fold Change GEN+BPA Males vs. CTL Males | Padj (FDR) | Directionality in GEN+BPA Males vs. CTL Males |
|---|---|---|---|
| miR871 | 0.09598443 | 0.00028475 | ↓ |
| miR7-2 | 3.65982139 | 0.00098684 | ↑ |
| miR7-1 | 3.51835965 | 0.00161711 | ↑ |
| miR146 | 3.91892362 | 0.00161711 | ↑ |
| miR10b | 3.17095407 | 0.00438947 | ↑ |
| miR148a | 2.70972643 | 0.0093332 | ↑ |
| miR216a | 3.67078923 | 0.0093332 | ↑ |
| Gm25107 | 0.34818425 | 0.02299567 | ↓ |
| miR1264 | 2.33325956 | 0.02299567 | ↑ |
| miR181d | 2.35970004 | 0.02299567 | ↑ |
| Gm26498 | 0.38651663 | 0.02483923 | ↓ |
| miR7j | 2.12294488 | 0.02483923 | ↑ |
| miR98 | 2.22478561 | 0.02483923 | ↑ |
| miRlet7f-2 | 2.24456301 | 0.02483923 | ↑ |
| miR346 | 2.74250356 | 0.02483923 | ↑ |
| miR448 | 1.86272197 | 0.02639755 | ↑ |
| Gm26230 | 0.39012788 | 0.02813473 | ↓ |
| miR384 | 2.30263221 | 0.02912914 | ↑ |
| miR212 | 2.20964577 | 0.03036753 | ↑ |
| miR184 | 2.59085414 | 0.03346464 | ↑ |
| miR221 | 2.20270513 | 0.03553398 | ↑ |
| n-TSaga9 | 1.85140606 | 0.03646631 | ↑ |
| miRlet7g | 2.11179824 | 0.03646631 | ↑ |
| miR298 | 2.16286301 | 0.03646631 | ↑ |
| miR501 | 2.36086026 | 0.03646631 | ↑ |
Table 7.
The top 25 miR and other small RNA expression differences in LD BPA males vs. CTL males.
| Gene Symbol | Fold Change LD BPA Males vs. CTL Males | Padj (FDR) | Directionality in LD BPA Males vs. CTL Males |
|---|---|---|---|
| miR146 | 3.59785964 | 0.00025617 | ↑ |
| Gm25107 | 0.25615114 | 0.00036562 | ↓ |
| Gm24407 | 0.30642514 | 0.00036562 | ↓ |
| Gm26498 | 0.29822392 | 0.00047675 | ↓ |
| Gm26230 | 0.28601463 | 0.00047675 | ↓ |
| Gm24265 | 0.30665545 | 0.00047675 | ↓ |
| Gm23297 | 0.35120726 | 0.00203618 | ↓ |
| Gm22510 | 0.35871764 | 0.00292911 | ↓ |
| Gm25121 | 0.37748401 | 0.00294054 | ↓ |
| Gm24566 | 0.37187679 | 0.00330348 | ↓ |
| Gm25229 | 0.36851196 | 0.0040413 | ↓ |
| Gm26389 | 0.38065228 | 0.00417328 | ↓ |
| miR7-1 | 3.37753124 | 0.00429822 | ↑ |
| miR7-2 | 3.4316525 | 0.00429822 | ↑ |
| Gm26390 | 0.37666111 | 0.00429822 | ↓ |
| Gm22111 | 0.37831251 | 0.00429822 | ↓ |
| Gm23305 | 0.38005448 | 0.00429822 | ↓ |
| Gm25017 | 0.40771793 | 0.00523512 | ↓ |
| Gm22393 | 0.38645527 | 0.00582397 | ↓ |
| Gm22130 | 0.39145192 | 0.00582397 | ↓ |
| Gm23682 | 0.42511949 | 0.00616365 | ↓ |
| Gm26374 | 0.42426062 | 0.00667177 | ↓ |
| Gm26336 | 0.37899441 | 0.00667177 | ↓ |
| Gm25085 | 0.40527951 | 0.00720853 | ↓ |
| miRlet7f-2 | 2.27826272 | 0.00809886 | ↑ |
If only genes that are DE based on FDR, 246, 275, and 6 are DE in the GEN+BPA Females vs. CTL Females, LD BPA Females vs. CTL Females, and UD BPA Females vs. CTL Females, respectively. The only small RNAs that overlap in these three groups include Gm25106 and Gm23744. For the males, those genes that are DE based on FDR include 52, 158, and 118 for GEN+BPA Males vs. CTL males, LD BPA Males vs. CTL Males, and UD BPA Males vs. CTL Males, respectively. Of these 29 overlap in all three groups miR7-2, miR7-1, Mir146, Mir10b, Mir148a, Mir1264, Mir181d, Mir7j, Mir98, Mirlet7f-2, Mir346, Mir448, Mir384, Mir212, Mir221, Mirlet7g, Mir501, Mirlet7a-2, Mir421, Mir488, Mir30a, Mir186, Mir378c, Mir26a-1, miR9-3, Mir195a, Gm24195, Mir199a-1, and Mir1983. The collective results thus suggest that BPA induces more small RNA changes then GEN exposure in females and males. The combination of GEN+ BPA in females reduces those that are seen otherwise with just LD BPA exposure, suggesting that GEN may at least partially mitigate the effects of BPA on small RNAome.
3.7. mRNAs Potentially Targeted by Differentially Expressed miRs
After determining miRs that were differentially expressed for these eight comparisons, we next determined those mRNAs that would likely be affected based on the collection of differentially expressed miRs for the various comparisons. Supplemental Files 3 and 4 provides the entire list of mRNAs predicated to be affected in the female and male group comparisons, respectively.
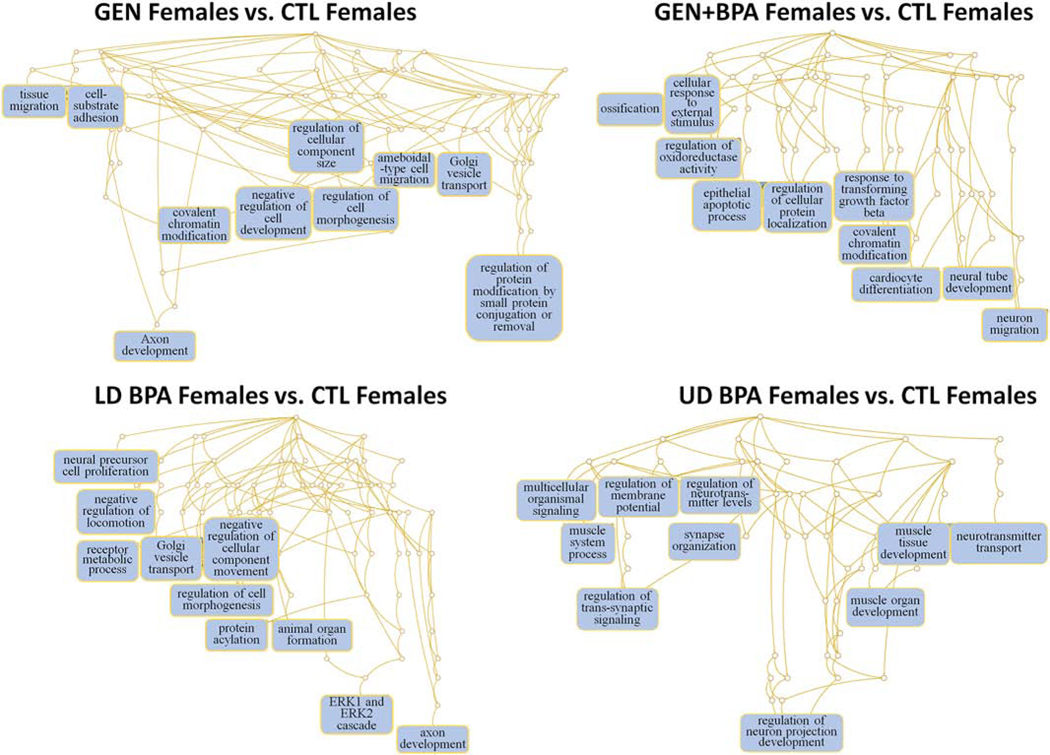
Based on the target mRNAs, we then determined which pathways would likely be impacted by changes in expression for miR profiles in the eight comparisons. In GEN Females vs. CTL females, the main pathways predicted to be affected are ameboidal-type cell migration and regulation of protein modification by small protein modification or removal (Fig. 6). For GEN+BPA Females vs. CTL Females, notable pathways likely to be affected are neural tube development and neuron migration (Fig. 6). Neural precursor cell, negative regulation of locomotion, receptor metabolic process, Golgi vesicle transport, regulation of cell morphogenesis, and axon development are some of the pathways likely affected in LD BPA females vs. CTL females (Fig. 6). For UD BPA Females vs. CTL Females, notable pathways that are predicted to be affected include regulation of trans-synaptic signaling, regulation of membrane potential, regulation of neurotransmitter levels, synapse organization, and regulation of neuron projection development (Fig. 6).
Fig. 6.
Pathways predicted to be affected based on mRNAs targeted by DE miRs in GEN Females vs. CTL Females, GEN+BPA Females vs. CTL Females, LD BPA, Females vs. CTL Females, and UD BPA Females vs. CTL Females. Based on the predicted target mRNAs, we performed functional enrichment analyses with the WEB-based Gene SeT AnaLysis Toolkit (WebGestalt) (Wang et al., 2013) and GO biological terms to determine which pathways might be affected.
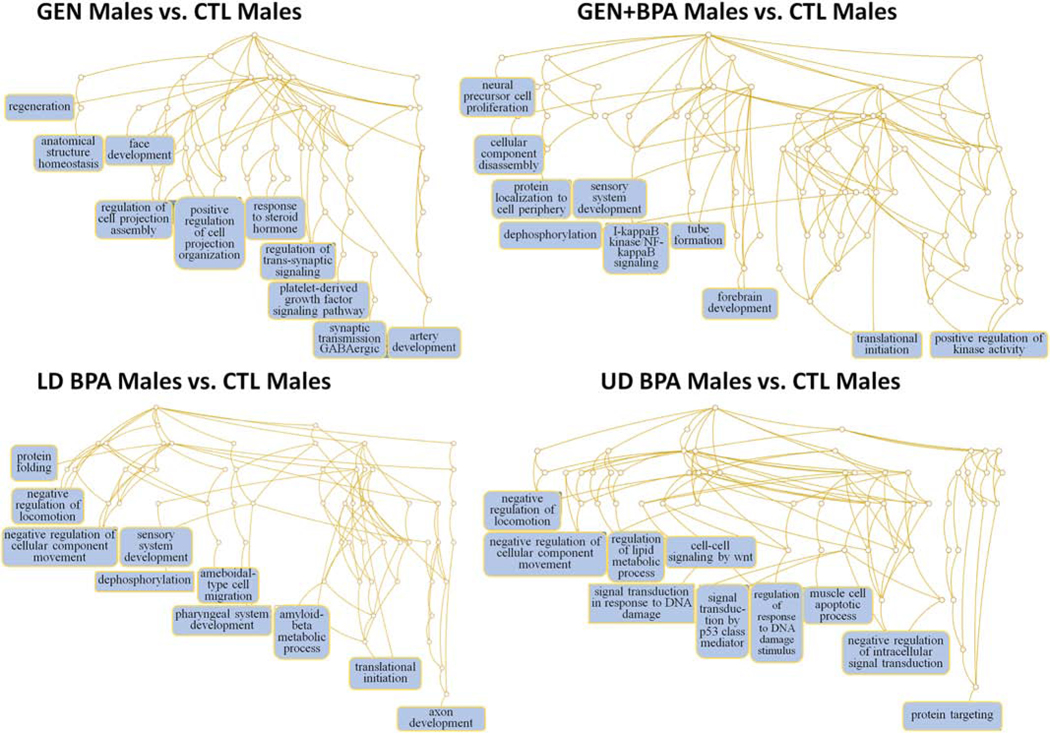
Pathways likely to be affected in GEN Males vs. CTL Males include positive regulation of cell projection assembly, response to steroid hormone, regulation of trans-synaptic signaling, and synaptic transmission, GABAeric (Fig. 7). Those potentially affected in GEN+BPA Males vs. CTL Males include dephosphorylation, tube formation, forebrain development, translation initiation, and positive regulation of kinase activity (Fig. 7). Negative regulation of locomotion, sensory system development, dephosphorylation, and ameboidal-type cell migration, and axon development are example pathways predicted to be altered in LD BPA Males vs. CTL Males (Fig. 7). In UD BPA Males vs. CTL Males, possible pathways affected include negative regulation of locomotion, negative regulation of cellular component movement, signal transduction in response to DNA damage, and protein targeting (Fig. 7).
Fig. 7.
Pathways predicted to be affected based on mRNAs targeted by DE miRs in GEN Males vs. CTL Males, GEN+BPA Males vs. CTL Males, LD BPA, Males vs. CTL Males, and UD BPA Males vs. CTL Males. Based on the predicted target mRNAs, we performed functional enrichment analyses with the WEB-based Gene SeT AnaLysis Toolkit (WebGestalt) (Wang et al., 2013) and GO biological terms to determine which pathways might be affected.
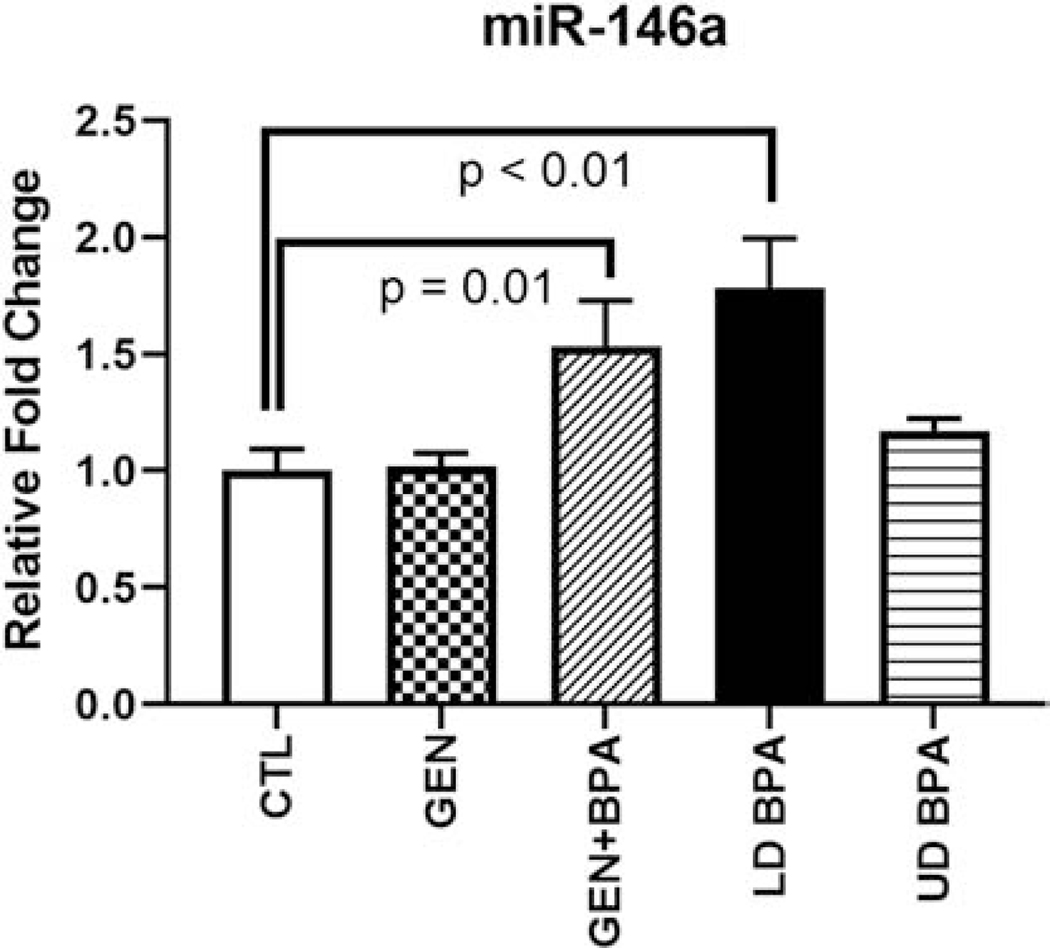
3.8. qPCR Analyses for miR146 and potential target mRNAs
For these analyses, no treatment X sex interactions were observed. Thus, female and male data within the same group were combined. qPCR analyses for miR146 revealed that this miR showed increased expression in those exposed to GEN+BPA or LD BPA relative to CTL (Fig. 8). However, no differences were detected in the other two groups, GEN or UD BPA, which could be due to a sensitivity issue. Our prediction at the outset was that increased expression of miR146 in EDC groups would result in reduced expression of target mRNA. However, several of the predicted mRNAs, including Cdk5, Grid1, Klf4, Ptpra, and Syt14, showed increased expression in the LD BPA group relative to CTL (Fig. S11). Nptn was the only gene down-regulated in one of the EDC-exposed groups, GEN+BPA.
Fig. 8.
qPCR analyses for miR146 in EDC-exposed and CTL groups. Graph is based on 2-ΔΔCt values relative to the CTL values, whose mean value was set to 1 for graphing purposes. P value differences relative to CTL group are indicated above the respective bars.
3.8. Integrative Correlation Analyses
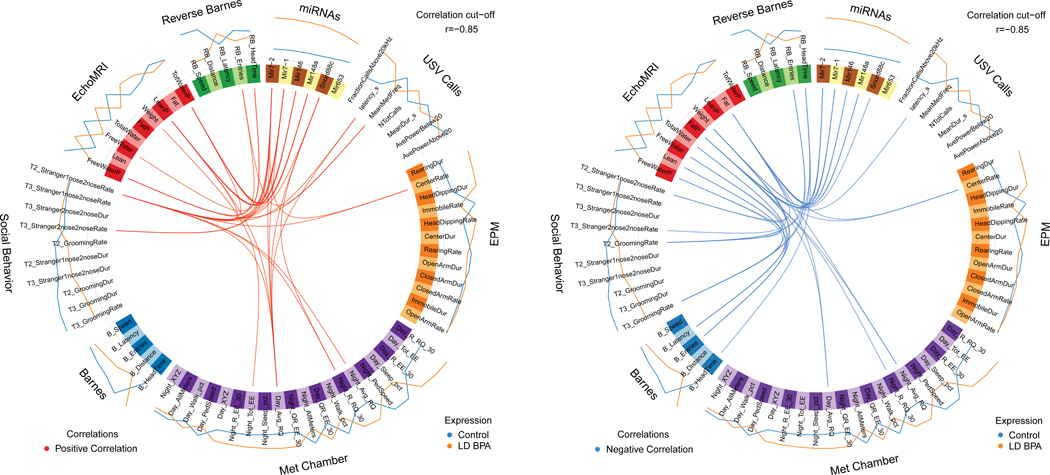
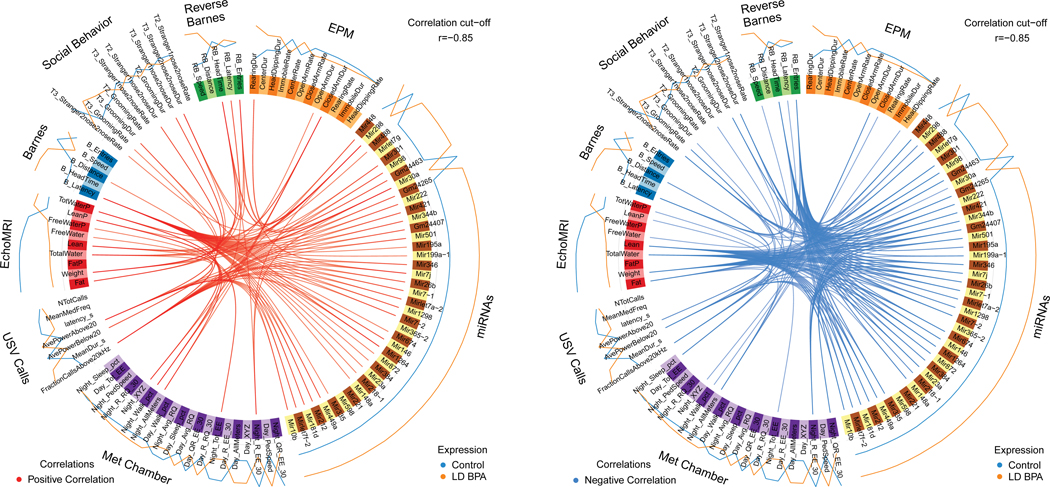
miR/small RNA changes that overlapped in the female (6 altogether) and male (40 altogether) group comparisons were correlated with behavioral and metabolic results by using mixOmics R package (Rohart et al., 2017) with an r value set to 0.85, which is considered highly stringent, For all eight group comparisons, several positive and negative correlations were evident between the miR/small RNA and phenotypic assessments. Results from LD BPA Female vs CTL Females and LD BPA Males vs. CTL Males are discussed below (Figs 9 and 10). The other mixOmics results are included as Supplemental Materials (Figs. S12 through S17).
Fig. 9.
Circos plot correlations between DE miRs/small RNAs and behavioral/metabolic parameters in LD BPA females vs. Control females. Red lines in the center indicate a positive correlation. In contrast, blue lines indicate a negative correlation. Results for Control females are indicated with a blue line outside of the circle. Orange line indicates results for LD BPA females. The color of the line further from the circle indicates the treatment group where these results are greater.
Fig. 10.
Circos plot correlations between DE miRs/small RNAs and behavioral/metabolic parameters in LD BPA males vs. Control males. Red lines in the center indicate a positive correlation. In contrast, blue lines indicate a negative correlation. Results for Control males are indicated with a blue line outside of the circle. Orange line indicates results for LD BPA males. The color of the line further from the circle indicates the treatment group where these results are greater.
In LD BPA Females vs. CTL Females, miR7-1, miR7-2, miR146, miR148, and Snord88c positively correlated with time spent with stranger 2 in trial 1 of social testing (Fig. 9). Snord88c also correlated with various RQ parameters. In these same group comparisons, miR7-1, miR7-2, miR146, miR148, and Snord88c negatively correlated with fat (Fig. 9). Snord88c also inversely correlated with body weight.
In LD BPA Males vs. CTL Males, all of the 40 in common DE miRs/small RNAs positively correlated with latency in the Barnes maze except for Mirlet7g, miR365-2, miR221, and miR598 (Fig. 10). miR346, miR7j, miR26b, miR7-1, miRlet7a-2, miR1298, miR7-2, miR449a, miR212, miR181d, miRlet7f2, and miR10b positively associated with fraction of calls above 20 kHz. For the same comparison, all miRs but miRlet7g, miR365-2, miR221, and miR598 negatively correlated with lean fat, total water, body weight, and fat. miR674, miR146, miR1264, miR872, miR384, miR218-1, and miR148a inversely correlated with parameters in the reverse Barnes maze. A few of the DE miRs inversely associated with social behaviors shown to stranger 2 in trial 3 of social behavioral testing. Collectively the findings suggest that many of the DE miRs are positively associated with aspects of communication, specifically calling within the ultrasonic range, whereas, these same ones in many cases are negatively associated with metabolic parameters, cognition, and social behaviors.
For all female and male group comparisons, similar correlations were evident as detailed for LD BPA vs. CTL groups (Figs. S12 through S17).
4. Discussion
The goals of the current study were to determine whether developmental exposure to GEN, GEN+BPA, LD BPA, and UD BPA affect miRs/small RNA profiles in the hypothalamus of male and female adult California mice. Correspondingly, we sought to determine how early exposure to these chemicals influences behavioral and metabolic parameters and whether miR/small RNA changes correlate with such phenotypic changes. For the combination group, we hypothesized at the outset that either developmental exposure to both chemicals would lead to exacerbated responses or GEN might mitigate some of the alterations induced by BPA. While we have studied how these chemicals affect select miRs in the hypothalamus and hippocampus (Butler et al., 2020), this the first study to our knowledge that has examined the global changes EDCs might induce on the miR/small RNA patterns in any brain region.
For Barnes maze testing, those exposed to GEN surprisingly showed improved spatial learning and memory, as evidenced by reduced latency to find the correct escape hole, especially on Day 3 of testing. This finding though is consistent with our previous studies showing early GEN exposure improves Barnes maze performance (Kaur et al., 2020). Rats perinatally exposed to GEN (1 or 10 mg/kg/day administered to the dam from gestational day 10 to postnatal day 14) demonstrated improved spatial learning and memory but impaired passive avoidance learning and memory (Kohara et al., 2014). GEN improves spatial and placement learning and memory in other adult rodent models (Alonso et al., 2010; Bagheri et al., 2011; Kohara et al., 2015; Pisani et al., 2012).
Both doses of BPA led to minimal effects on spatial learning and memory in California mice. This finding is consistent with our previous studies (Kaur et al., 2020; Williams et al., 2013), and presumably relates to the fact that this trait is not sexually selected in monogamous California mice. In contrast, developmental exposure of polygynous deer mice (P. maniculatus bairdii), who rely on this behavior to locate prospective breeding partners that are widely dispersed in the environment, to the same doses of BPA results in impaired spatial learning and memory (Jasarevic et al., 2011; Jasarevic et al., 2013).
Our past studies though show that socio-communication behaviors in California mice at various ages are vulnerable to developmental exposure to GEN or BPA (Butler et al., 2020; Kaur et al., 2020; Marshall et al., 2019; Williams et al., 2013). Current findings are consistent with these previous results. At adulthood, exposure to the UD BPA results in less social behaviors to a novel individual. Females exposed to GEN or UD BPA elicit more calls when placed in isolation. The average duration of calls becomes more pronounced in males exposed to LD BPA. Females exposed to GEN, GEN+BPA, and LD BPA are more likely to call in the ultrasonic range, and the same is the case for males exposed to GEN or LD BPA. Males and females exposed to UD BPA produced more calls in a single burst than controls. The results thus suggest that all EDCs alter varying aspects of the calls with many of these features being sex dependent.
California mice exposed to one or both EDCs are also more likely to engage in one or more stereotypical behaviors, such as head-dipping or rearing. Those exposed to UD BPA also show hints of anxiogenic behaviors by remaining immobile for greater time than controls when tested in the EPM. These findings are consistent with other rodent models and Drosophila melanogaster studies that suggests that BPA exposure, especially, can result in exposed individuals engaging in repetitive (Kaur et al., 2015) and anxiety-like behaviors (Harris et al., 2018; Jasarevic et al., 2011; Jasarevic et al., 2013; Kumar and Thakur, 2017; Luo et al., 2014; Matsuda et al., 2012; Xu et al., 2012).
The other notable finding was that GEN, GEN+BPA, LD BPA, and UD BPA exposure groups showed reduced body mass. These results were surprising given the fact that BPA and other EDCs are considered obesogens (Schug et al., 2011; Thayer et al., 2012). In previous studies with younger California mice, we did not detect any differences in body mass (Johnson et al., 2015b). A previous mouse study found that developmental exposure to BPA (50 ng 50 mg, or 50 mg/kg diet) resulted in a lean body composition in females at certain ages (Anderson et al., 2013). The underlying mechanisms of how BPA and/or GEN promotes a lean body mass phenotype at adulthood merit further pursuit. As detailed below, one likely mediating factor is miR146a.
Developmental exposure to GEN, GEN+BPA, and BPA led to many sex-dependent differences in miRs. Exposure to LD BPA or UD BPA resulted in the greatest number of miR and other small RNA expression changes. In contrast, GEN altered some miRs/small RNAs, but in both female and males, none were significant after applying an FDR. The reason for this is not clear, as sufficient replicates were tested. Those that were changed in GEN+BPA group likely reflect more the effects of BPA exposure. The fact that this combination treatment did not result in greater number of miR/small RNA changes suggest that these two xenoestrogens do not exert synergistic effects.
Three miRs that were increased in all female and male treatment groups, when based on those that differed based on p value, include miR7-2, miR146, and miR148a. Of the three miRs, miR146 and its primary form, miR146a, has been well studied in the brain. miR-146a exerts anti-inflammatory activity by targeting tumor necrosis factor receptor (TNFR)-associated factor 6 (TRAF6) and interleukin-1 receptor-associated kinase 1 (IRAK1) (Taganov et al., 2006). Mice deficient in miR146a show altered microglial function and proteome (Martin et al., 2020). In this same study, investigators showed that miR146 is upregulated in brain lesions with multiple sclerosis patients (Martin et al., 2020). An earlier study showed that this same miR was increased in the peripheral blood mononculear cells (PBMC) of MS patients, suggestive that it might serve as a potential biomarker for this disease (Fenoglio et al., 2011). In autism spectrum disorder (ASD) patients, miR146a is upregulated in the brain and in other non-neuronal cells, and it may be considered one of the most common miR deregulation events in ASD (Fregeac et al., 2016; Mor et al., 2015; Nguyen et al., 2016; Talebizadeh et al., 2008). This miR has also been implicated in other neurobehavioral disorders, such as epiplepsy and intellectual disability (Aronica et al., 2010; Iyer et al., 2012; Nguyen et al., 2016). In stable human neural stem cell line (H9 hNSC), over-expression of miR146a result in deregulation of many distinct markers of neuronal cell lineages (Nguyen et al., 2018), thereby, indicating proper expression of this miR is essential for normal segregation of neuronal cell populations.
While this study is the first to our knowledge to show EDCs can affect miR146 expression in the brain, other reports have shown that these compounds can increase its expression in other organs. In the testes, BPA-induced miR-146a-5p impairs steroidogenesis through negative regulation of Metastatic tumor antigen 3 (MTA3) signaling (Gao et al., 2018). Similarly, BPA exposure is associated with over-expression of miR146a in human placenta (De Felice et al., 2015). BPA treatment of two immortalized human cytotrophoblast cell lines strongly induces miR146a (Avissar-Whiting et al., 2010). Taken together, the findings provide robust evidence that BPA exposure stimulates miR146 expression across multiple organs. Consequently, miR146 may be the ideal biomarker of exposure to BPA and possibly other EDCs. If BPA is acting through miR146 to induce disease across a wide variety of tissues, it also suggests that mice lacking this gene may be resistant to BPA exposure. The fact that developmental exposure to GEN also upregulates expression of miR146 suggests that it may be inducible by a wide variety of xenoestrogens. The fact that BPA trigged up-regulation of it in the past mouse studies and in the current California mice studies further suggests that xenoestrogens may induce this miR in a variety of mammalian species, including humans.
We predicted at the outset that qPCR analyses would validate the small RNA-seq results in relation to miR146 expression. This miR was upregulated in the GEN+BPA and LD BPA exposed groups, but not the two other EDC groups (Fig. 8). It should be noted that these two groups showed the most robust upregulation of miR146, as noted by being in the top 25 DE miRs/small RNAs based on FDR for these comparisons (Tables 2, 3, 6, and 7). Thus, the contrasting findings might suggest small RNA-seq is a more sensitive assay than qPCR. We further hypothesized that potential targets of miR146 would be downregulated in one or more of the EDC treated groups. However, several of the predicted target mRNAs, including Cdk5, Grid1, Klf4, Ptpra, and Syt14, showed increased expression in the LD BPA group relative to CTL (Fig. S11). Nptn was the only gene down-regulated in one of the EDC-exposed groups, GEN+BPA. It is uncertain if such gene expression changes are due to alterations in miR expression, especially in the GEN+BPA and LD BPA groups where this miR was upregulated based on both RNA-seq and qPCR analyses. If so, it is not clear how miR146 would act to upregulate these four mRNAs that are important in neural function. The main actions currently ascribed to miR146 are involvement in innate immunity and carcinogenesis (Paterson and Kriegel, 2017; Testa et al., 2017).
miR148a was also upregulated in all treatment groups. Not much is known, however, about this miR. One report showed that this miR, along with miR145, is upregulated in the brain by congenital Zika virus infection (Castro et al., 2019). miR148a has been implicated in targeting estrogen receptors (ESRs) (Nguyen-Dien et al., 2014). The expression of other miRs was affected by both maternal treatment and offspring sex, suggesting potential sexual dimorphism in miR responses in the hypothalamus. Intriguingly, an upcoming publication revealed that social stress in California mice induced sex-specific miR responses in the bed nucleus of the stria terminalis (Luo et al., In Review). Specifically, the researchers found more transcripts increase in females than males, including several in the let7 family.
The DE miRs in each of the current group comparisons were then used to determine those mRNAs that would likely be affected. Based on these mRNAs, functional enrichment was performed to determine the pathways that would likely be impacted by developmental exposure to GEN, BPA, or both compounds. Major pathways that were affected in one or more EDC groups include axon development, cell migration, neural precursor cell formation, negative regulation of locomotion, protein modifications, neuron migration, regulation of ERK1 and ERK2 cascade, regulation of neuronal projection development, regulation of neurotransmitter levels, synapse organization and signaling, response to steroid hormone, synaptic transmission, and forebrain development. To examine pathways predicted to be affected by upregulation of miR146a alone, we performed the same analyses with just those mRNAs predicted to be affected by miR146a as listed in Table S2 for the following paper (https://www.pnas.org/content/suppl/2009/01/16/0812591106.DCSupplemental/ST2_PDF.pdf). Pathways likely affected by deregulation of mRNAs regulated by miR146a include regulation of cell-cell adhesion, response to nerve growth factor, regulation of innate immune response, positive regulation of defense response, ERK1 and ERK2 cascade, and protein polyubiquitination (Fig. S18). Taken together, these analyses suggest that miR146a acts as a major contributor to the ultimate pathways predicted to be affected by the collection of BPA/GEN altered miRs. Pathways predicted to be affected by miR146a upregulation are consistent with previous reports suggests a role for this miR in suppressing inflammation and influencing neuronal cell differentiation.
MixOmics analyses revealed several both positive and negative correlations between DE miRs common across female and male groups and socio-vocalization behaviors and metabolic parameters. miR146 showed several positive and negative associations with behavioral and metabolic parameters in the different group comparisons. For instance, in GEN Females vs. CTL Females, this miR positively associated with total energy expenditure during the day but inversely associated with reverse Barnes parameters (Fig. S12). In GEN+BPA Females vs. CTL Females, miR146 positively correlated with total energy expenditure during the day and showed no negative associations (Fig. S13). For LD BPA Females vs. CTL Females, this miR was positively linked to social behaviors shown to Stranger 1 in Trial 2 of social testing and inversely with body fat (Fig. 9). In UD BPA Females vs. CTL Females, miR146 positively correlated with lean percentage but negatively associated with Barnes maze latency (Fig. S14). In GEN Males vs. CTL Males, miR146 and fraction of calls above 20 kHz were positively associated, but this miR inversely correlated with fat, body weight, lean and total water (Fig. S15). For GEN+BPA Males vs. CTL Males, no positive associations were observed for miR and the other indices, but it was negatively correlated with aspects of energy expenditure, especially during the night (Fig. S16). In LD BPA Males vs. CTL Males, miR146 and fraction of calls above 20 kHz were positively linked, whereas, it negatively correlated with body weight and fat (Fig. 10). In UD BPA Males vs. CTL Males, this miR positively associated with free water and free water percentage, and no negative associations were observed with this miR (Fig. S17).
miR146 and other DE miRs associated with various aspects of California mice socio-vocalizations and metabolic parameters. In zebra finches (Taeniopygia guttata), several miRs have been identified to regulate song-responsive mRNAs (Gunaratne et al., 2011). In this same species, miR9 overexpression results in variable song production at adulthood and eliminates social context-dependent modulation of song variability (Shi et al., 2018). A previous study with miR146a deficient mice suggest that besides acting as an important anti-inflammatory factor, it also suppresses diet-induced obesity and regulates various metabolic processes as placing these transgenic mice on a high fat diet results in exaggerated weight gain, increased adiposity, hepatosteatosis, and dysregulated blood glucose concentrations (Runtsch et al., 2019). miR146a and 148 are dysregulated in human patients with obesity, prediabetes, and type 2 diabetes (Nunez Lopez et al., 2016). Current results and combined findings suggest that EDC-induced upregulation of miR146 might reduce body weight and fat content but may detrimentally impact aspects of socio-communication behaviors.
In conclusion, current studies show that developmental exposure to GEN and/or BPA alter aspects of socio-communication behaviors. These EDCs also increase the likelihood of engaging in repetitive and anxiety-like behaviors at adulthood. All EDC treated groups demonstrate a reduced body mass phenotype at adulthood. Developmental exposure to GEN, BPA, or the combination generally results in sex-dependent differences in miR/small RNA signature patterns. Three miRs that are increased in all EDC groups are miR7-2, miR146, and miR148a. Of these, miR146 or one of its forms, miR146a, is associated with various neurobehavioral disorders, including ASD. In other organs, BPA increases miR146(a). Overexpression of this antiflammatory miR may impact various neurogenic and synaptogenic pathways. Integrative correlation analyses suggests that miR146 and other DE miRs female and male groups associate with socio-communication and metabolic traits. miR146 might be an important modulator and biomarker of EDC exposure in mammals, including humans.
Supplementary Material
Highlights.
Small RNA-seq was used to examine endocrine disruptor effects on neural miRNA.
Bisphenol A (BPA) and genistein (GEN) caused behavioral and metabolic changes.
Both EDCs affected miR profiles in the hypothalamus, including increasing miR146.
Hypothalamic miR changes, namely miR146, correlated with phenotypic alterations.
miRNA146 may be a biomarker of BPA and other endocrine disruptor exposure.
Acknowledgements
The studies were supported by NIEHS 1R01ES025547 (CSR). We appreciate all the undergraduate students who assisted with animal husbandry and general care of the California mice colonies.
Footnotes
Conflict of Interest Statement: The authors declare they have no competing financial or other interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alonso A, Gonzalez-Pardo H, Garrido P, Conejo NM, Llaneza P, Diaz F, Del Rey CG, Gonzalez C, 2010. Acute effects of 17 beta-estradiol and genistein on insulin sensitivity and spatial memory in aged ovariectomized female rats. Age 32, 421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson OS, Peterson KE, Sanchez BN, Zhang Z, Mancuso P, Dolinoy DC, 2013. Perinatal bisphenol A exposure promotes hyperactivity, lean body composition, and hormonal responses across the murine life course. FASEB J 27, 1784–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronica E, Fluiter K, Iyer A, Zurolo E, Vreijling J, van Vliet EA, Baayen JC, Gorter JA, 2010. Expression pattern of miR-146a, an inflammation-associated microRNA, in experimental and human temporal lobe epilepsy. Eur J Neurosci 31, 1100–1107. [DOI] [PubMed] [Google Scholar]
- Avissar-Whiting M, Veiga KR, Uhl KM, Maccani MA, Gagne LA, Moen EL, Marsit CJ, 2010. Bisphenol A exposure leads to specific microRNA alterations in placental cells. Reprod Toxicol 29, 401–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagheri M, Joghataei MT, Mohseni S, Roghani M, 2011. Genistein ameliorates learning and memory deficits in amyloid beta(1–40) rat model of Alzheimer’s disease. Neurobiol Learn Mem 95, 270–276. [DOI] [PubMed] [Google Scholar]
- Butler MC, Long CN, Kinkade JA, Green MT, Martin RE, Marshall BL, Willemse TE, Schenk AK, Mao J, Rosenfeld CS, 2020. Endocrine disruption of gene expression and microRNA profiles in hippocampus and hypothalamus of California mice: Association of gene expression changes with behavioural outcomes. J Neuroendocrinol 32, e12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro FL, Geddes VEV, Monteiro FLL, Gonçalves R, Campanati L, Pezzuto P, Paquin-Proulx D, Schamber-Reis BL, Azevedo GS, Gonçalves AL, Cunha DP, Moreira MEL, Vasconcelos ZFM, Chimeli L, Melo A, Tanuri A, Nixon DF, Ribeiro-Alves M, Aguiar RS, 2019. MicroRNAs 145 and 148a are ppregulated during congenital Zika virus Infection. ASN Neuro 11, 1759091419850983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong A, Johnson SA, Howald EC, Ellersieck MR, Camacho L, Lewis SM, Vanlandingham MM, Ying J, Ho SM, Rosenfeld CS, 2018. Gene expression and DNA methylation changes in the hypothalamus and hippocampus of adult rats developmentally exposed to bisphenol A or ethinyl estradiol: a CLARITY-BPA consortium study. Epigenetics 13, 704–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou WC, Lee PH, Tan YY, Lin HC, Yang CW, Chen KH, Chuang CY, 2017. An integrative transcriptomic analysis reveals bisphenol A exposure-induced dysregulation of microRNA expression in human endometrial cells. Toxicol In Vitro 41, 133–142. [DOI] [PubMed] [Google Scholar]
- Crawley JN, 2012. Translational animal models of autism and neurodevelopmental disorders. Dialogues Clin Neurosci 14, 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Felice B, Manfellotto F, Palumbo A, Troisi J, Zullo F, Di Carlo C, Di Spiezio Sardo A, De Stefano N, Ferbo U, Guida M, Guida M, 2015. Genome-wide microRNA expression profiling in placentas from pregnant women exposed to BPA. BMC Med Genomics 8, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinoy DC, Huang D, Jirtle RL, 2007. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci USA 104, 13056–13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL, 2006. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Persp 114, 567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenoglio C, Cantoni C, De Riz M, Ridolfi E, Cortini F, Serpente M, Villa C, Comi C, Monaco F, Mellesi L, Valzelli S, Bresolin N, Galimberti D, Scarpini E, 2011. Expression and genetic analysis of miRNAs involved in CD4+ cell activation in patients with multiple sclerosis. Neurosci Lett 504, 9–12. [DOI] [PubMed] [Google Scholar]
- Fountain ED, Mao J, Whyte JJ, Mueller KE, Ellersieck MR, Will MJ, Roberts RM, Macdonald R, Rosenfeld CS, 2008. Effects of diets enriched in omega-3 and omega-6 polyunsaturated fatty acids on offspring sex-ratio and maternal behavior in mice. Biol. Reprod 78, 211–217. [DOI] [PubMed] [Google Scholar]
- Fregeac J, Colleaux L, Nguyen LS, 2016. The emerging roles of microRNAs in autism spectrum disorders. Neurosci Biobehav Rev 71, 729–738. [DOI] [PubMed] [Google Scholar]
- Friedman Y, Naamati G, Linial M, 2010. MiRror: a combinatorial analysis web tool for ensembles of microRNAs and their targets. Bioinformatics 26, 1920–1921. [DOI] [PubMed] [Google Scholar]
- Gao GZ, Zhao Y, Li HX, Li W, 2018. Bisphenol A-elicited miR-146a-5p impairs murine testicular steroidogenesis through negative regulation of Mta3 signaling. Biochem Biophys Res Commun 501, 478–485. [DOI] [PubMed] [Google Scholar]
- González I, Lê Cao K-A, Davis MJ, Déjean S, 2012. Visualising associations between paired ‘omics’ data sets. BioData Mining 5, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaratne PH, Lin YC, Benham AL, Drnevich J, Coarfa C, Tennakoon JB, Creighton CJ, Kim JH, Milosavljevic A, Watson M, Griffiths-Jones S, Clayton DF, 2011. Song exposure regulates known and novel microRNAs in the zebra finch auditory forebrain. BMC Genomics 12, 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EP, Allardice HA, Schenk AK, Rissman EF, 2018. Effects of maternal or paternal bisphenol A exposure on offspring behavior. Horm Behav 101, 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou T, Liao J, Zhang C, Sun C, Li X, Wang G, 2018. Elevated expression of miR-146, miR-139 and miR-340 involved in regulating Th1/Th2 balance with acute exposure of fine particulate matter in mice. Int Immunopharmacol 54, 68–77. [DOI] [PubMed] [Google Scholar]
- Howdeshell KL, 2002. A model of the development of the brain as a construct of the thyroid system. Environ Health Perspect 110 Suppl 3, 337–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howdeshell KL, Peterman PH, Judy BM, Taylor JA, Orazio CE, Ruhlen RL, Vom Saal FS, Welshons WV, 2003. Bisphenol A is released from used polycarbonate animal cages into water at room temperature. Environ Health Persp 111, 1180–1187. https://www.ncbi.nlm.nih.gov/assembly/GCF_000500345.1/. https://www.pnas.org/content/suppl/2009/01/16/0812591106.DCSupplemental/ST2_PDF.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer A, Zurolo E, Prabowo A, Fluiter K, Spliet WG, van Rijen PC, Gorter JA, Aronica E, 2012. MicroRNA-146a: a key regulator of astrocyte-mediated inflammatory response. PloS One 7, e44789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasarevic E, Sieli PT, Twellman EE, Welsh TH Jr., Schachtman TR, Roberts RM, Geary DC, Rosenfeld CS, 2011. Disruption of adult expression of sexually selected traits by developmental exposure to bisphenol A. Proc Natl Acad Sci USA 108, 11715–11720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasarevic E, Williams SA, Vandas GM, Ellersieck MR, Liao C, Kannan K, Roberts RM, Geary DC, Rosenfeld CS, 2013b. Sex and dose-dependent effects of developmental exposure to bisphenol A on anxiety and spatial learning in deer mice (Peromyscus maniculatus bairdii) offspring. Horm Behav 63, 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SA, Ellersieck MR, Rosenfeld CS, 2018a. Hypothalamic gene expression changes in F1 California mice (Peromyscus californicus) parents developmentally exposed to bisphenol A or ethinyl estradiol. Heliyon 4, e00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SA, Farrington MJ, Murphy CR, Caldo PD, McAllister LA, Kaur S, Chun C, Ortega MT, Marshall BL, Hoffmann F, Ellersieck MR, Schenk AK, Rosenfeld CS, 2018b. Multigenerational effects of bisphenol A or ethinyl estradiol exposure on F2 California mice (Peromyscus californicus) pup vocalizations. PLoS One 13, e0199107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SA, Javurek AB, Painter MS, Ellersieck MR, Welsh TH Jr., Camacho L, Lewis SM, Vanlandingham MM, Ferguson SA, Rosenfeld CS, 2016. Effects of developmental exposure to bisphenol A on spatial navigational learning and memory in rats: A CLARITY-BPA study. Horm Behav 80, 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SA, Javurek AB, Painter MS, Murphy CR, Conard CM, Gant KL, Howald EC, Ellersieck MR, Wiedmeyer CE, Vieira-Potter VJ, Rosenfeld CS, 2017a. Effects of a maternal high-fat diet on offspring behavioral and metabolic parameters in a rodent model. J Dev Orig Health Dis 8, 75–88. [DOI] [PubMed] [Google Scholar]
- Johnson SA, Javurek AB, Painter MS, Peritore MP, Ellersieck MR, Roberts RM, Rosenfeld CS, 2015a. Disruption of parenting behaviors in california mice, a monogamous rodent species, by endocrine disrupting chemicals. PloS One 10, e0126284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SA, Painter MS, Javurek AB, Ellersieck MR, Wiedmeyer CE, Thyfault JP, Rosenfeld CS, 2015b. Sex-dependent effects of developmental exposure to bisphenol A and ethinyl estradiol on metabolic parameters and voluntary physical activity. J Dev Orig Health Dis 6, 539–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SA, Spollen WG, Manshack LK, Bivens NJ, Givan SA, Rosenfeld CS, 2017b. Hypothalamic transcriptomic alterations in male and female California mice (Peromyscus californicus) developmentally exposed to bisphenol A or ethinyl estradiol. Physiol Rep 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur K, Simon AF, Chauhan V, Chauhan A, 2015. Effect of bisphenol A on Drosophila melanogaster behavior--a new model for the studies on neurodevelopmental disorders. Behav Brain Res 284, 77–84. [DOI] [PubMed] [Google Scholar]
- Kaur S, Sarma SJ, Marshall BL, Liu Y, Kinkade JA, Bellamy MM, Mao J, Helferich WG, Schenk AK, Bivens NJ, Lei Z, Sumner LW, Bowden JA, Koelmel JP, Joshi T, Rosenfeld CS, 2020. Developmental exposure of California mice to endocrine disrupting chemicals and potential effects on the microbiome-gut-brain axis at adulthood. Sci Rep 10, 10902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y, Kawaguchi S, Kuwahara R, Uchida Y, Oku Y, Yamashita K, 2015. Genistein improves spatial learning and memory in male rats with elevated glucose level during memory consolidation. Physiol Behav 140, 15–22. [DOI] [PubMed] [Google Scholar]
- Kohara Y, Kuwahara R, Kawaguchi S, Jojima T, Yamashita K, 2014. Perinatal exposure to genistein, a soy phytoestrogen, improves spatial learning and memory but impairs passive avoidance learning and memory in offspring. Physiol Behav 130, 40–46. [DOI] [PubMed] [Google Scholar]
- Kumar D, Thakur MK, 2017. Anxiety like behavior due to perinatal exposure to Bisphenol-A is associated with decrease in excitatory to inhibitory synaptic density of male mouse brain. Toxicology 378, 107–113. [DOI] [PubMed] [Google Scholar]
- Li Q, Kappil MA, Li A, Dassanayake PS, Darrah TH, Friedman AE, Friedman M, Lambertini L, Landrigan P, Stodgell CJ, Xia Y, Nanes JA, Aagaard KM, Schadt EE, Murray JC, Clark EB, Dole N, Culhane J, Swanson J, Varner M, Moye J, Kasten C, Miller RK, Chen J, 2015. Exploring the associations between microRNA expression profiles and environmental pollutants in human placenta from the National Children’s Study (NCS). Epigenetics 10, 793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W, 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Long AD, Baldwin-Brown J, Tao Y, Cook VJ, Balderrama-Gutierrez G, Corbett-Detig R, Mortazavi A, Barbour AG, 2019. The genome of Peromyscus leucopus, natural host for Lyme disease and other emerging infections. Sci Adv 5, eaaw6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S, 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G, Wang S, Li Z, Wei R, Zhang L, Liu H, Wang C, Niu R, Wang J, 2014. Maternal bisphenol a diet induces anxiety-like behavior in female juvenile with neuroimmune activation. Toxicol Sci 140, 364–373. [DOI] [PubMed] [Google Scholar]
- Luo P, Manning CE, Fass JN, Williams AV, Hao R, Campi KL, Trainor BC, In Review. Sex-specific effects of social defeat stress on miRNA expression in the anterior BNST. Behav Neurosci, Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall BL, Liu Y, Farrington MJ, Mao J, Helferich WG, Schenk AK, Bivens NJ, Sarma SJ, Lei Z, Sumner LW, Joshi T, Rosenfeld CS, 2019. Early genistein exposure of California mice and effects on the gut microbiota-brain axis. J Endocrinol 242, 139–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal; Vol 17, No 1: Next Generation Sequencing Data AnalysisDO - 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- Martin NA, Hyrlov KH, Elkjaer ML, Thygesen EK, Wlodarczyk A, Elbaek KJ, Aboo C, Okarmus J, Benedikz E, Reynolds R, Hegedus Z, Stensballe A, Svenningsen Å F, Owens T, Illes Z, 2020. Absence of miRNA-146a Differentially Alters Microglia Function and Proteome. Front Immunol 11, 1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S, Matsuzawa D, Ishii D, Tomizawa H, Sutoh C, Nakazawa K, Amano K, Sajiki J, Shimizu E, 2012. Effects of perinatal exposure to low dose of bisphenol A on anxiety like behavior and dopamine metabolites in brain. Prog Neuropsychopharmacol Biol Psychiatry 39, 273–279. [DOI] [PubMed] [Google Scholar]
- Mor M, Nardone S, Sams DS, Elliott E, 2015. Hypomethylation of miR-142 promoter and upregulation of microRNAs that target the oxytocin receptor gene in the autism prefrontal cortex. Mol Autism 6, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Moya JM, Vilella F, Simon C, 2014. MicroRNA: key gene expression regulators. Fertil. Steril 101, 1516–1523. [DOI] [PubMed] [Google Scholar]
- Moy SS, Nadler JJ, Perez A, Barbaro RP, Johns JM, Magnuson TR, Piven J, Crawley JN, 2004. Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes Brain Behav 3, 287–302. [DOI] [PubMed] [Google Scholar]
- Nguyen-Dien GT, Smith RA, Haupt LM, Griffiths LR, Nguyen HT, 2014. Genetic polymorphisms in miRNAs targeting the estrogen receptor and their effect on breast cancer risk. Meta Gene 2, 226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LS, Fregeac J, Bole-Feysot C, Cagnard N, Iyer A, Anink J, Aronica E, Alibeu O, Nitschke P, Colleaux L, 2018. Role of miR-146a in neural stem cell differentiation and neural lineage determination: relevance for neurodevelopmental disorders. Mol Autism 9, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LS, Lepleux M, Makhlouf M, Martin C, Fregeac J, Siquier-Pernet K, Philippe A, Feron F, Gepner B, Rougeulle C, Humeau Y, Colleaux L, 2016. Profiling olfactory stem cells from living patients identifies miRNAs relevant for autism pathophysiology. Mol Autism 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez Lopez YO, Garufi G, Seyhan AA, 2016. Altered levels of circulating cytokines and microRNAs in lean and obese individuals with prediabetes and type 2 diabetes. Mol Biosyst 13, 106–121. [DOI] [PubMed] [Google Scholar]
- Padmanabhan V, Siefert K, Ransom S, Johnson T, Pinkerton J, Anderson L, Tao L, Kannan K, 2008. Maternal bisphenol-A levels at delivery: a looming problem? J Perinatol 28, 258–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson MR, Kriegel AJ, 2017. MiR-146a/b: a family with shared seeds and different roots. Physiol Genomics 49, 243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisani SL, Neese SL, Doerge DR, Helferich WG, Schantz SL, Korol DL, 2012. Acute genistein treatment mimics the effects of estradiol by enhancing place learning and impairing response learning in young adult female rats. Horm Behav 62, 491–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed BG, Babayev SN, Chen LX, Carr BR, Word RA, Jimenez PT, 2018. Estrogen-regulated miRNA-27b is altered by bisphenol A in endometrial stromal cells. Reproduction 156, 559–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice D, Barone S Jr., 2000. Critical periods of vulnerability for the developing nervous system: evidence from humans and animal models. Environ Health Perspect 108 Suppl 3, 511–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohart F, Gautier B, Singh A, Le Cao K-A, 2017. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comp Biol 13, e1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld CS, 2015. Bisphenol A and phthalate endocrine disruption of parental and social behaviors. Front Neurosci 9, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld CS, 2017. Neuroendocrine disruption in animal models due to exposure to bisphenol A analogues. Front Neuroendocrinol 47, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld CS, Ferguson SA, 2013. Barnes maze testing strategies with small and large rodent models. J Vis Exp 84,e51194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld CS, Johnson SA, Ellersieck MR, Roberts RM, 2013. Interactions between parents and parents and pups in the monogamous California mouse (Peromyscus californicus). PloS One 8, e75725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld CS, Trainor BC, 2014. Environmental health factors and sexually dimorphic differences in behavioral disruptions. Curr Environ Health Rep 1, 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runtsch MC, Nelson MC, Lee SH, Voth W, Alexander M, Hu R, Wallace J, Petersen C, Panic V, Villanueva CJ, Evason KJ, Bauer KM, Mosbruger T, Boudina S, Bronner M, Round JL, Drummond MJ, O’Connell RM, 2019. Anti-inflammatory microRNA-146a protects mice from diet-induced metabolic disease. PLoS Genet 15, e1007970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug TT, Janesick A, Blumberg B, Heindel JJ, 2011. Endocrine disrupting chemicals and disease susceptibility. J Steroid Biochem Mol Biol 127, 204–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Piccus Z, Zhang X, Yang H, Jarrell H, Ding Y, Teng Z, Tchernichovski O, Li X, 2018. miR-9 regulates basal ganglia-dependent developmental vocal learning and adult vocal performance in songbirds. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieli PT, Jasarevic E, Warzak DA, Mao J, Ellersieck MR, Liao C, Kannan K, Collet SH, Toutain PL, Vom Saal FS, Rosenfeld CS, 2011. Comparison of serum bisphenol A concentrations in mice exposed to bisphenol A through the diet versus oral bolus exposure. Environ Health Perspect 119, 1260–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Yang M, Lord C, Crawley JN, 2010. Behavioural phenotyping assays for mouse models of autism. Nature reviews. Neuroscience 11, 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel RG, 1996. Principles and Procedures of Statistics: A Biometrical Approach, 3rd edition. McGraw-Hill Higher Education, New York, New York. [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D, 2006. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA 103, 12481–12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talebizadeh Z, Butler MG, Theodoro MF, 2008. Feasibility and relevance of examining lymphoblastoid cell lines to study role of microRNAs in autism. Autism Res 1, 240–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeguarden JG, Calafat AM, Ye X, Doerge DR, Churchwell MI, Gunawan R, Graham MK, 2011. Twenty-four hour human urine and serum profiles of bisphenol a during high-dietary exposure. Toxicol Sci 123, 48–57. [DOI] [PubMed] [Google Scholar]
- Testa U, Pelosi E, Castelli G, Labbaye C, 2017. miR-146 and miR-155: Two key modulators of immune response and tumor development. Noncoding RNA 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer KA, Heindel JJ, Bucher JR, Gallo MA, 2012. Role of environmental chemicals in diabetes and obesity: a National Toxicology Program workshop review. Environ Health Persp 120, 779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topper VY, Walker DM, Gore AC, 2015. Sexually dimorphic effects of gestational endocrine-disrupting chemicals on microRNA expression in the developing rat hypothalamus. Mol Cell Endocrinol 414, 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wynsberghe PM, Chan SP, Slack FJ, Pasquinelli AE, 2011. Analysis of microRNA expression and function. Methods Cell Biol 106, 219–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg LN, Chahoud I, Heindel JJ, Padmanabhan V, Paumgartten FJ, Schoenfelder G, 2010. Urinary, circulating, and tissue biomonitoring studies indicate widespread exposure to bisphenol A. Environ Health Perspect 118, 1055–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg LN, Hauser R, Marcus M, Olea N, Welshons WV, 2007. Human exposure to bisphenol A (BPA). Reprod Toxicol 24, 139–177. [DOI] [PubMed] [Google Scholar]
- Verbanck M, Canouil M, Leloire A, Dhennin V, Coumoul X, Yengo L, Froguel P, Poulain-Godefroy O, 2017. Low-dose exposure to bisphenols A, F and S of human primary adipocyte impacts coding and non-coding RNA profiles. PLoS One 12, e0179583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Duncan D, Shi Z, Zhang B, 2013. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acid Res 41, W77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SA, Jasarevic E, Vandas GM, Warzak DA, Geary DC, Ellersieck MR, Roberts RM, Rosenfeld CS, 2013. Effects of developmental bisphenol A exposure on reproductive-related behaviors in California mice (Peromyscus californicus): a monogamous animal model. PloS One 8, e55698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright EC, Johnson SA, Hao R, Kowalczyk AS, Greenberg GD, Ordones Sanchez E, Laman-Maharg A, Trainor BC, Rosenfeld CS, 2017. Exposure to extrinsic stressors, social defeat or bisphenol A, eliminates sex differences in DNA methyltransferase expression in the amygdala. J. Neuroendocrinol 29(6):10.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Hong X, Xie L, Li T, Yang Y, Zhang Q, Zhang G, Liu X, 2012. Gestational and lactational exposure to bisphenol-A affects anxiety- and depression-like behaviors in mice. Horm Behav 62, 480–490. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.