Abstract
Background and aim
SARS-CoV-2 quick testing is relevant for the containment of new pandemic waves. Antigen testing in self-collected saliva might be useful. We compared salivary and naso-pharyngeal swab (NPS) SARS-CoV-2 antigen detection by a rapid chemiluminescent assay (CLEIA) and two different point-of-care (POC) immunochromatographic assays, with results of molecular testing.
Methods
234 patients were prospectively enrolled. Paired self-collected saliva (Salivette) and NPS were obtained to perform rRT-PCR, chemiluminescent (Lumipulse G) and POC (NPS: Fujirebio and Abbott; saliva: Fujirebio) for SARS-CoV-2 antigen detection.
Results
The overall agreement between NPS and saliva rRT-PCR was 78.7%, reaching 91.7% at the first week from symptoms. SARS-CoV-2 CLEIA antigen was highly accurate in distinguishing positive and negative NPS (ROC-AUC = 0.939, 95%CI:0.903–0.977), with 81.6% sensitivity and 93.8% specificity. This assay on saliva reached the optimal value within 7 days from symptoms onset (Sensitivity: 72%; Specificity: 97%). Saliva POC antigen was limited in sensitivity (13%), performing better in NPS (Sensitivity: 48% and 66%; Specificity: 100% and 99% for Espline and Abbott respectively), depending on viral loads.
Conclusions
Self-collected saliva is a valid alternative to NPS for SARS-CoV-2 detection by molecular, but also by CLEIA antigen testing, which is therefore potentially useful for large scale screening.
Keywords: COVID-19, Point-of-care, Naso-pharyngeal swab, Chemiluminescence
Abbreviations: (NPS), naso-pharyngeal swab; (CLEIA), chemiluminescent assay; (POC), point-of-care; (ROC), receiver operating characteristic; (AUC), area under the receiver operating characteristic curve; (SD), standard deviation
1. Introduction
Saliva testing for SARS-CoV-2, one of the strategies for COVID-19 diagnosis and monitoring, is advocated mainly for screening asymptomatic subjects in order to rapidly detect and isolate infected individuals and their contacts, thus limiting viral spread and containing further waves of the pandemic [1], [2], [3], [4], [5], [6]. Although the molecular detection of SARS-CoV-2 RNA in naso-pharyngeal swabs (NPS) is considered the “gold standard” technique for identifying symptomatic or asymptomatic individuals [7], it has limitations in both the analytical and the healthcare settings. From the analytical viewpoint, it is widely agreed that the sensitivity of rRT-PCR of NPS ranges from 70 to 90% [5], [8], reaching values around 50% after the first two weeks of disease [9], [10]. Therefore, COVID-19 disease cannot be ruled out when a NPS result is negative, but the patient has clinical symptoms, and biochemical data and radiological findings that evidence a clinical scenario typical of the disease [11]. In this context anti-SARS-CoV-2 antibodies should also be taken into account [12]. From the healthcare organizational viewpoint, NPS testing calls for the involvement of healthcare workers and services for sample collection. This pre-requisite might be promptly met by the healthcare system when the demand is low, but not necessarily when it is high, as occurs in a pandemic. Any delay in testing puts individuals without a diagnosis at risk, consequently exposing the community to viral contagion.
Saliva testing might not only have the advantage of relieving health care resources, but also of reducing hazard exposure to healthcare workers during sampling; it might also limit the risk of viral spread incurred when numerous individuals queue for a long time waiting for testing, since saliva can be self-collected at home. However, none of these advantages support the use of saliva testing if its results are not as reliable as those of NPS. When rRT-PCR is used for SARS-CoV-2 testing, the reliability of saliva testing is reportedly equal to, or even higher than, NPS [5], [10], [13], [14], although some studies report contradictory findings (i.e. saliva as less sensitive) probably depending on salivary viral load kinetic [3], [9], [14], [15], [16] .
Saliva testing by rRT-PCR is reliable, but time consuming, calling for dedicated laboratory equipment for nucleic acid extraction and amplification, and personnel trained in molecular techniques [17]. These requirements, which might not be fulfilled by all laboratories, compromise the advantage of using the saliva sample. In front of a safe and rapid collection procedure, the overall testing process remains long not only because molecular testing takes time, but also because molecular laboratories might be limited in number, especially in low resource countries, thus increasing the turnaround time due to sample transportation and processing. In order to speed up testing while maximizing the number of tested individuals, the search for SARS-Co-2 antigens rather than RNA, by immunometric techniques is now emerging, including point-of-care (POC) rapid immunochromatographic assays based on lateral flow technology [18]. The market now offers a number of SARS-CoV-2 antigen detection immunometric assays, which are high throughput but require laboratory instrumentation [19], and ultra-rapid POC devices suggested for use in medical cabinets by general practitioners and nurses. Since they are simple to use, these devices are also considered potentially employable in auto-testing. However, simplicity is not synonymous with accuracy [20]. To date, no exhaustive data are present in the literature on antigen detection using saliva, an approach that might maximize an effective and timely COVID-19 diagnosis, encompassing the advantages of a) saliva self-collection and b) rapid viral protein detection, thus making wide-scale screening possible in many parts of the world.
The aim of this prospective study was to compare in saliva and NPS the diagnostic accuracy of molecular testing with SARS-CoV-2 antigen detection by a rapid chemiluminescent assay and two different point of care ultra-rapid immunochromatographic assays.
2. Materials and methods
2.1. Patients and samples
A total of 234 subjects were enrolled between 1st August and 30th November 2020. One hundred thirty-eight (52 females, 86 males, mean age ± SD: 56 ± 17 years) were COVID-19 inpatients, and 96 (47 females, 49 males, mean age ± SD: 42 ± 15 years) were outpatients screened for suspected SARS-CoV-2 (i.e. contact with a SARS-CoV-2 positive subject or with typical symptoms). After giving fully informed consent in writing (Local Ethic Committee Nr. 27444), patients were asked to collect a morning saliva sample (Salivette device, SARSTEDT AG & Co, Nümbrecht, Germany). After saliva sampling, trained nurses collected three NPS from each patient. A subset of 32 inpatients repeated saliva collection and testing 7 days after enrollment and, among them, 23 repeated also NPS.
Clinical data on each inpatient were also retrieved from the hospital information system (HIS).
This research did not receive any specific grant from funding agencies in the public, commercial, or not-profit sectors.
2.2. Laboratory testing
For molecular testing, saliva and NPS were analyzed by TaqPath COVID-19 RT-PCR kit (Applied Biosystems, USA) as detailed in Supplementary materials and methods [21].
Chemiluminescence immunoassay (CLEIA) was performed using a LUMIPULSE SARS-CoV-2 Ag kit on a LUMIPULSE G1200 automated analyzer (Fujirebio, Tokjo, Japan), following the manufacturer’s instructions. After the first result, available in 30 min, the diagnostic system provides 120 results per hour. ESPLINE rapid test and PanbioTM COVID-19 Ag Rapid Test (ABBOTT, Chicago, Illinois, USA) were the two evaluated POC lateral flow immunochromatographic assays.
2.3. Statistical analysis of data
The statistical analysis of data was made with Stata software ver. 13.1 (Lakeway drive, TX, US) by Wilcoxon rank test, Kruskal-Wallis rank test, Student’s t test, Fisher’s exact test, and multiple linear regression as detailed in Supplementary materials and methods.
3. Results
3.1. Clinical data
The clinical characteristics of the 138 inpatients are shown in Table 1 .
Table 1.
Clinical characteristics of inpatients.
| Inpatients n = 138 (%) |
||
|---|---|---|
| Days since onset of symptoms | ≤ 7 | 38 (27.6) |
| 7–14 | 74 (53.6) | |
| greater than 14 | 26 (18.8) | |
| Symptoms at onset | Pneumonia | 93 (67.4) |
| Fever greater than 37.5 °C | 97 (70.3) | |
| Dyspnea | 21 (15.2) | |
| Cough | 46 (33.3) | |
| Gastrointestinal | 25 (18.1) | |
| Other, minor | 55 (39.9) | |
| Therapy | Steroids only | 33 (23.9) |
| Steroids and oxygen | 13 (9.4) | |
| Steroids, oxygen and Remdesivir | 57 (41.3) | |
| Steroids, oxygen, Remdesivir and convalescent plasma | 35 (25.4) |
While no significant differences were found between in- and outpatients (n = 96) for gender distribution (Fisher’s exact test: p = 0.106), the mean age of outpatients was significantly lower than that of inpatients (Student’s t test for unpaired data: t = 6.51, p < 0.001).
3.2. NPS and saliva molecular testing
At enrollment, NPS rRT-PCR results were positive among 84/138 (60.9%) inpatients and 3/96 (3.1%) outpatients, while saliva in 67/127 (52.8%) inpatients and in 4/96 (4.2%) outpatients. Positive NPS and saliva were more frequent among inpatients with a time lapse from symptoms of <7 days (37/38, 97.4% for NPS; 32/36, 88.9% for saliva), with respect to those with 7 to 14 days’ (40/74, 54.1% for NPS; 31/69, 44.9% for saliva) and those with more than 14 days’ (7/26, 26.9% for NPS, 4/22, 18.2% for saliva) (Fisher’s exact test: p < 0.001 for both NPS and saliva). Accordingly, the lowest median Ct values of NPS and saliva were found among inpatients with a time lapse from symptoms of <7 days (Supplementary Table S1).
Among the 96 outpatients, four had positive findings at NPS and/or saliva testing (Supplementary Table S2). The patient who was negative at NPS but positive at saliva (n. 4), repeated NPS two and ten days after enrollment, being positive in both cases.
On considering the overall inpatient population, agreement of 78.7% was found for rRT-PCR among paired NPS and saliva samples (n = 127, Cohen k = 0.569, SE = 0.086, p < 0.001). Agreement was 91.67% in patients tested <7 days after onset of symptoms (n = 36, Cohen k = 0.372, SE = 0.130, p < 0.001), 71.0% in those tested after 7 to 14 days (n = 69, Cohen k = 0.427, SE = 0.117, p < 0.001) and 81.8% in those after more than 14 days (n = 22, Cohen k = 0.488, SE = 0.206, p = 0.009).
3.3. NPS and saliva antigen testing
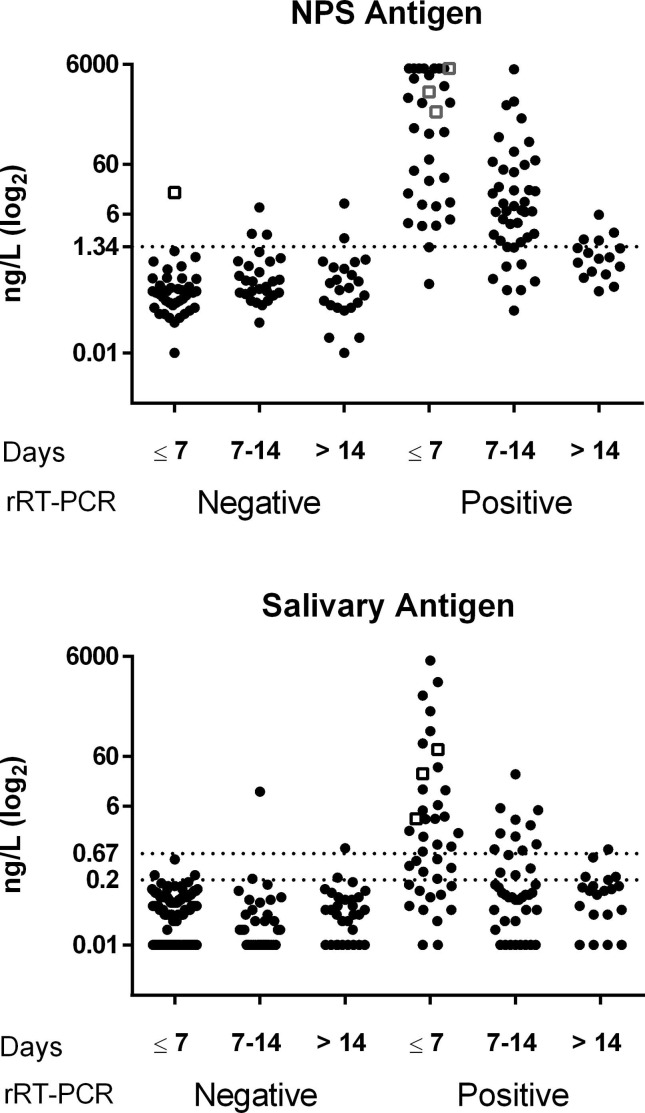
Fig. 1 shows the individual CLEIA antigen levels measured in NPS and saliva after subdividing patients on the basis of the time lapse between symptom onset and testing, data on analytical reproducibility being reported in Supplementary Table S3. SARS-CoV-2 antigen measured in NPS enabled distinction between positive and negative swabs classified on the basis of rRT-PCR with a high diagnostic accuracy (area under the ROC curve = 0.939, 95% CI: 0.903–0.977). Based on the threshold reported by the manufacturer (1.34 ng/L), the overall sensitivity and specificity were 81.6% (95% CI: 71.0–89.5%) and 93.8 (95% CI: 86.2–98.0%), respectively, with a positive likelihood ratio (LR) and negative LR of 13.2 (95% CI: 5.62–31.1) and 0.19 (95% CI: 0.12–0.32), respectively.
Fig. 1.
SARS-CoV-2 antigen in NPS and saliva. The antigen was assayed by CLEIA in subjects classified as negative or positive on the basis of NPS rRT-PCR and subdivided on the basis of the time lapse between symptoms onset and testing (Days). The upper graph shows the results obtained in NPS samples. The dotted line (1.34 ng/L) is the cut-off recommended by the manufacturer. The lower graph shows the results obtained in salivary samples. The dotted lines are the cut-off (0.67 ng/L) and the limit of detection (0.2 ng/L) recommended by the manufacturer. In both graphs, patients enrolled within 7 days and classified as negative are all but one (open square) outpatients. Among rRT-PCR positive results, open squares represent the three outpatients who were found to be positive.
Salivary SARS-CoV-2 antigen allowed to distinguish between positive and negative samples classified on the basis of NPS rRT-PCR with a good diagnostic accuracy (area under the ROC curve = 0.805, 95% CI: 0.740–0.870) (Supplementary Table S4). For salivary antigen, the manufacturer’s suggested cut-off is 0.67 ng/L and the limit of quantification, 0.2 ng/L. With these two thresholds, sensitivity, specificity, positive and negative LR were calculated considering the patients overall and after subdividing them on the basis of duration of symptoms (Table 2 ).
Table 2.
Sensitivity, specificity, positive and negative likelihood ratio (LR) with 95% confidence intervals (95% CI) of salivary CLEIA antigen. The manufacturer’s suggested cut-off (0.67 ng/L) and the manufacturer’s declared limit of quantification (0.20 ng/L) were considered as thresholds.
| Cut-off | Sensitivity % (95% CI) | Specificity % (95% CI) | Pos LR (95% CI) | Neg LR (95% CI) | |
|---|---|---|---|---|---|
| Overall | ≧0.67 ng/L | 41.3 (30.4–52.8) | 98.6 (95.0–99.8) | 29.1 (7.17–118.0) | 0.59 (0.49–0.71) |
| ≧0.20 ng/L | 53.8 (42.2–65.0) | 95.7 (91.0–98.4) | 12.6 (5.6–28.4) | 0.48 (0.38–0.61) | |
| ≤ 7 days | ≧0.67 ng/L | 56.4 (39.6–72.2) | 100 (96.2–100) | 107.0 (6.6–1719.0) | 0.44 (0.31–0.62) |
| ≧0.20 ng/L | 71.8 (55.1–85.0) | 96.8 (91.0–99.3) | 22.5 (7.3–69.7) | 0.29 (0.18–0.48) | |
| 7–14 days | ≧0.67 ng/L | 35.1 (20.2–52.5) | 99.2 (95.6–100) | 29.4 (5.7–153.0) | 0.65 (0.52–0.83) |
| ≧0.20 ng/L | 38.2 (22.2–56.4) | 93.3 (77.9–99.2) | 5.7 (1.4–23.4) | 0.66 (0.50–0.87) | |
| More than 14 days | ≧0.67 ng/L | 40.0 (12.2–73.8) | 99.1 (95.0–100) | 30.3 (5.3–173.0) | 0.60 (0.36–0.98) |
| ≧0.20 ng/L | 28.6 (3.7–71.0) | 94.1 (71.3–99.9) | 4.8 (0.5–45.3) | 0.76 (0.47–1.23) |
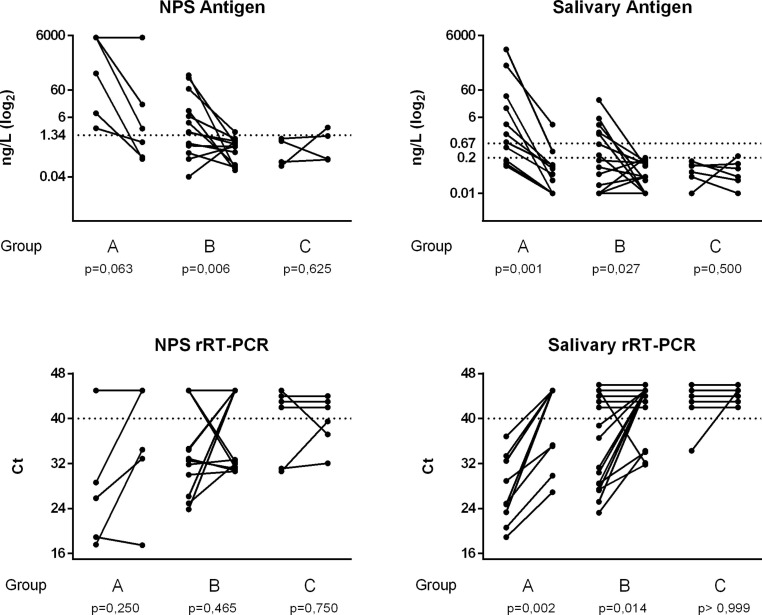
Fig. 2 shows the individual levels of SARS-CoV-2 antigen measured in NPS (n = 23) and saliva (n = 32) and the corresponding Ct values obtained in a series of inpatients for whom two consecutive samples were available: one at enrollment and the other, after 7 days.
Fig. 2.
Kinetics of SARS-CoV-2 antigen and Ct values in NPS and saliva. Two consecutive samples (seven days apart) were available in a series of inpatients, who were subdivided on the basis of days from symptoms onset to enrollment in three groups: within 7 days (group A), between 7 and 14 days (group B), after 14 (group C). The p values reported were obtained after Wilcoxon rank test for paired data.
Multiple linear regression analyses were performed considering the Ct values (Orf1ab) of NPS and saliva as dependent variables, while antigen levels, clinical and demographic data as predictor variables (Supplementary Table S5). The Ct vales were significantly correlated with antigen values independently from all clinical and demographic characteristics (r2 = 0.793 for NPS and r2 = 0.598 for saliva).
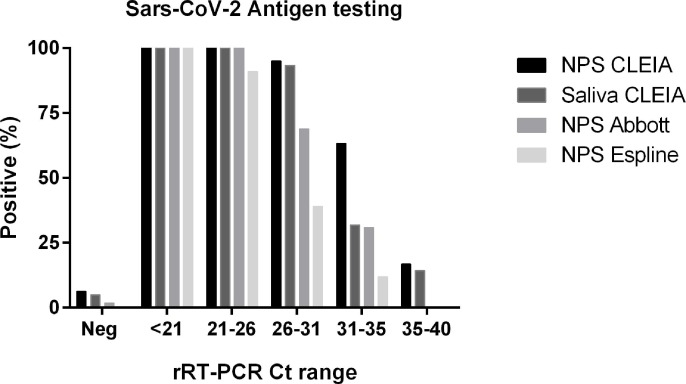
Fig. 3 shows the percentages of positive results for SARS-CoV-2 antigen by POC and CLEIA in NPS and saliva after subdividing samples based on viral load estimated with the Ct values at molecular analyses. POC sensitivity was satisfactory only for NPS with high viral loads (i.e. Ct values < 25), while CLEIA enabled the detection of viral particles with a good sensitivity also for Ct values above 25.
Fig. 3.
Comparison of NPS and saliva SARS-CoV-2 rapid antigen testing. Percentages of positive results for SARS-CoV-2 antigen testing by means of rapid immunochromatographic assays (Abbott and Espline) and CLEIA in NPS and saliva after subdividing samples on the basis of viral load (Ct ranges) at molecular analyses.
4. Discussion
There is a pressing need for novel strategies for the effective containment of a third wave of SARS-Cov-2 infection, particularly while awaiting vaccines. The early, reliable identification of SARS-CoV-2 infection appears to be the key to reducing community transmission yet the recommended diagnosis based on rRT-PCR analysis of NPS, although accurate, does not enable an early and prompt diagnosis. Moreover, it calls personnel trained in NPS collection, analysis and interpretation. SARS-CoV-2 antigen determination in NPS by point-of-care immunochromatographic assays or by chemiluminescent assays developed with laboratory instrumentation has been proposed in order to overcome these limitations, and to facilitate large scale analyses [22]. This approach allows a reduction in the analytical time but does not obviate the need to perform NPS by trained personnel within dedicated medical cabinets. This bottle-neck could be overcome by using self-collected saliva. To identify the best possible strategy for detecting the infection by antigenic rapid testing, in this second wave pandemic we studied two series of subjects representing real world scenarios: inpatients with COVID-19 disease and outpatients screened for SARS-CoV-2 due to a history of positive contact or suspect clinical signs. NPS and saliva, simultaneously collected from all patients enrolled in the study, were used as matrices for antigen detection employing immunochromatographic assays and rapid CLEIA, and for viral sequences identification by rRT-PCR. As expected, the prevalence of positive rRT-PCR findings in NPS was lower among outpatients (3%) than inpatients (61%); in this latter group, it progressively declined in parallel with duration of symptoms, as the viral load, assessed by the Ct value, declined. Based on this observation, our patients’ series was subdivided according to the duration of symptoms before enrollment: less than one week, one to two weeks, and more than two weeks. The molecular detection of viral sequences in NPS and saliva gave concordant results in a high percentage of cases at the onset (92%) and in the late phases (82%) of the infection. Conversely, patients enrolled one to two weeks after symptoms onset were more likely to have positive NPS results than positive saliva rRT-PCR results, in agreement with previous data in the literature [16]. These findings corroborate the hypothesis that buccal mucosa and salivary glands are the first sites of viral colonization, according to previous data the highest load occurring in the first week of symptom onset, followed by a progressive decline during the course of the disease [3], [9], [14]. As saliva is the first route for viral dissemination, it represents a suitable matrix for screening asymptomatic subjects, who are known to carry a viral load comparable to that of symptomatic patients [6], [23]. CLEIA antigen testing in NPS enabled a highly accurate distinction between positive and negative swabs. The area under the ROC curve was higher than 0.9, and better than that reported earlier in a Japanese series by Hirotsu et al. [20]. Unlike the approach used in the cited study, we evaluated a large number of infected patients and for any patient one single sample, while Hirotsu et al. included serial samples from a limited number of infected patients. Moreover, CLEIA antigenic testing in NPS had a very high sensitivity not only in the first week, but also in the second week from symptoms onset paralleling molecular findings. With respect to CLEIA, rapid immunochromatographic assays in NPS appeared very reliable for high viral loads but much less so in the presence of less abundant viral loads (i.e. Ct values ranging from 25 to 30) or even worse, for higher Ct values (greater than 30), in agreement with data reported in the literature [19], [24].
This must be considered a limitation of these rapid immunochromatographic assays taking also into account that Ct values ranging from 20 to 30 are considered normal findings [25]. We therefore suggest that CLEIA antigenic testing in NPS should be preferred to rapid immunochromatographic assays for obtaining accurate and fast results in the emergency setting to rapidly identify infected symptomatic patients before their hospitalization, on considering that in this context NPS collection does not represent a limitation. CLEIA antigenic testing, which requires a simple laboratory instrumentation usable by minimally trained personnel, enables the result to be obtained in 30 min and processes more than 100 samples per hour and can therefore be used by any emergency laboratory.
In the setting of screening asymptomatic subjects, salivary antigen might make the difference. In self-collected saliva, CLEIA antigen allowed subjects with positive to be differentiated from those with negative NPS with a good overall accuracy (0.81), which was even better in the early infection phase (accuracy 0.88) [26]. The antigen levels in saliva declined more rapidly that in NPS paralleling the decline in viral load [27]. Interestingly considering the patients with repeated sampling, the Ct of NPS especially in the time frame of one-two weeks from symptom onset exerted a higher variable pattern than saliva findings. This discrepancy might depend on saliva collection being more standardized than NPS collection, but also on the possibility that viral RNA fragments in the absence of infectivity could be detected in NPS by molecular testing [28]. We focused our analyses mainly in this early phase because it is more representative of the possible screening scenario. By using the manufacturer’s suggested cut-off of 0.67 ng/L for CLEIA salivary antigen, in the presence of specificity close to 100%, the highest sensitivity was achieved within the first week of onset of symptoms. However, the 56.4% sensitivity observed is too low for screening programs, especially when the prevalence of disease is low, as might occur in the near future following the effects of lockdown policies. Therefore we evaluated whether an increase in sensitivity, without decreasing specificity, could be achieved by lowering the cut-off to the limit of detection suggested by the manufacturer (0.2 ng/L), and achieved 72% sensitivity and 96% specificity. We believe that CLEIA salivary testing could be applied in screening asymptomatic cohorts (e.g. in schools or farms) according to the following strategic plan: 1. Values above 0.67 ng/L to be considered positive for SARS-CoV-2; values below 0.2 ng/L to be considered negative for SARS-CoV-2; values ranging from 0.2 to 0.67 ng/L to be considered a grey zone and, for these samples, a reflex rRT-PCR activated. To be effective, this strategy should be used on a large scale, and undertaken at least once a week, in agreement with the proposal for less sensitive point of care tests with a quick return suggested as useful in surveillance when repeated at least every three days [29]. However, immunochromatographic saliva testing had an unacceptable overall sensitivity of about 13%, in line with previous data obtained with the same [26], or different diagnostic systems [25]; this renders it unhelpful, even if repeated daily.
The limitation of our study is represented by the low number of subjects enrolled for screening. Feasibility of saliva testing for screening programs is under evaluation at the University of Padova since October 2020, aiming to test saliva samples from about 4000 asymptomatic employees every 15 days.
4.1. Conclusion
SARS-CoV-2 antigen testing by CLEIA is fast enough to meet requirements for the early detection of the infection by any laboratory. CLEIA antigen testing in NPS might be suggested in the emergency setting, while CLEIA antigen testing with molecular reflex testing for the grey zone results in saliva is suggested by the authors of this study for large scale screening of asymptomatic subjects in risk cohorts.
Author's contributions: DB and AA designed the study and drafted the manuscript. AA and NC collected the samples and clinical data. CC, FN, SM and CFZ performed the analyses. DB, AA and AP analyzed and summarized all the data. AMC and MP revised the final manuscript. All authors approved the final version of the manuscript.
Acknowledgements
We thank all personnel of the Tropical and Infectious Disease Unit for their valuable assistance in collection of biomaterials. We thank Mrs Tamara Avellone, Cinzia Centobene, Monica Razetti and Daniela Rinaldi for their technical assistance in laboratory testing.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cca.2021.02.014.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Aita A., Basso D., Cattelan A.M., Fioretto P., Navaglia F., Barbaro F., Stoppa A., Coccorullo E., Farella A., Socal A., Vettor R., Plebani M. SARS-CoV-2 identification and IgA antibodies in saliva: One sample two tests approach for diagnosis. Clinica Chimica Acta. 2020;510:717–722. doi: 10.1016/j.cca.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.K.K.-W. To, O.T.-Y. Tsang, C. Chik-Yan Yip, K.-H. Chan, T.-C. Wu, J.M.C. Chan, et al., Consistent detection of 2019 novel coronavirus in saliva, Clin. Infect. Dis. (2020). https://doi:10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed]
- 3.D. Sapkota, T.M. Søland, H.K. Galtung, L.P. Sand, S. Giannecchini, K.K.W. To, et al., COVID-19 salivary signature: diagnostic and research opportunities, J. Clin. Pathol. (2020). https://doi:10.1136/jclinpath-2020-206834. [DOI] [PubMed]
- 4.Migueres M., Mengelle C., Dimeglio C., Didier A., Alvarez M., Delobel P., Mansuy J.-M., Izopet J. Saliva sampling for diagnosing SARS-CoV-2 infections in symptomatic patients and asymptomatic carriers. Journal of Clinical Virology. 2020;130:104580. doi: 10.1016/j.jcv.2020.104580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.I. Yokota, P.Y. Shane, K. Okada, Y. Unoki, Y. Yang, T. Inao, et al., Mass screening of asymptomatic persons for SARS-CoV-2 using saliva, Clin. Infect. Dis. (2020). https://doi:10.1093/cid/ciaa1388. [DOI] [PMC free article] [PubMed]
- 6.Hamid H., Khurshid Z., Adanir N., Zafar M.S., Zohaib S. COVID-19 Pandemic and Role of Human Saliva as a Testing Biofluid in Point-of-Care Technology. Eur J Dent. 2020;14(S 01):S123–S129. doi: 10.1055/s-0040-1713020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanson K.E., Caliendo A.M., Arias C.A., Englund J.A., Lee M.J., Loeb M., et al. Clin. Infect; Dis: 2020. Infectious Diseases Society of America Guidelines on the Diagnosis of COVID-19. https://doi:10.1093/cid/ciaa760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.L. Comber, K.A. Walsh, K. Jordan, K.K. O'Brien, B. Clyne, C. Teljeur, et al., Alternative clinical specimens for the detection of SARS-CoV-2: A rapid review, Rev. Med. Virol. (2020). https://doi:10.1002/rmv.2185. [DOI] [PubMed]
- 9.S. Zheng, J. Fan, F. Yu, B. Feng, B. Lou, Q. Zou, et al., Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study, Bmj. 369 (2020) m1443. https://doi:10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed]
- 10.Sakanashi D., Asai N., Nakamura A., Miyazaki N., Kawamoto Y., Ohno T., Yamada A., Koita I., Suematsu H., Hagihara M., Shiota A., Kurumiya A.i., Sakata M., Kato S., Muramatsu Y., Koizumi Y., Kishino T., Ohashi W., Yamagishi Y., Mikamo H. Comparative evaluation of nasopharyngeal swab and saliva specimens for the molecular detection of SARS-CoV-2 RNA in Japanese patients with COVID-19. Journal of Infection and Chemotherapy. 2021;27(1):126–129. doi: 10.1016/j.jiac.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin Y.-H., Cai L., Cheng Z.-S., Cheng H., Deng T., Fan Y.-P., Fang C., Huang D.i., Huang L.-Q., Huang Q., Han Y., Hu B.o., Hu F., Li B.-H., Li Y.-R., Liang K.e., Lin L.-K., Luo L.-S., Ma J., Ma L.-L., Peng Z.-Y., Pan Y.-B., Pan Z.-Y., Ren X.-Q., Sun H.-M., Wang Y., Wang Y.-Y., Weng H., Wei C.-J., Wu D.-F., Xia J., Xiong Y., Xu H.-B., Yao X.-M., Yuan Y.-F., Ye T.-S., Zhang X.-C., Zhang Y.-W., Zhang Y.-G., Zhang H.-M., Zhao Y., Zhao M.-J., Zi H., Zeng X.-T., Wang Y.-Y., Wang X.-H. A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version) Military Med Res. 2020;7(1) doi: 10.1186/s40779-020-0233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plebani M. SARS-CoV-2 antibody-based SURVEILLANCE: New light in the SHADOW. EBioMedicine. 2020;61:103087. doi: 10.1016/j.ebiom.2020.103087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rao M., Rashid F.A., Sabri F.S.A.H., Jamil N.N., Zain R., Hashim R., et al. Clin. Infect; Dis: 2020. Comparing nasopharyngeal swab and early morning saliva for the identification of SARS-CoV-2. https://doi:10.1093/cid/ciaa1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uwamino Y., Nagata M., Aoki W., Fujimori Y., Nakagawa T., Yokota H., Sakai-Tagawa Y., Iwatsuki-Horimoto K., Shiraki T., Uchida S., Uno S., Kabata H., Ikemura S., Kamata H., Ishii M., Fukunaga K., Kawaoka Y., Hasegawa N., Murata M. Accuracy and stability of saliva as a sample for reverse transcription PCR detection of SARS-CoV-2. J Clin Pathol. 2021;74(1):67–68. doi: 10.1136/jclinpath-2020-206972. [DOI] [PubMed] [Google Scholar]
- 15.Y.C. Manabe, C. Reuland, T. Yu, R. Azamfirei, T. Church, D.M. Brown, et al., Variability of Salivary and Nasal Specimens for SARS-CoV-2 Detection, medRxiv. (2020). https://doi:10.1101/2020.10.07.20208520.
- 16.A.L. Wyllie, J. Fournier, A. Casanovas-Massana, M. Campbell, M. Tokuyama, P. Vijayakumar, et al., Saliva or Nasopharyngeal Swab Specimens for Detection of SARS-CoV-2, N. Engl. J. Med. 383 (2020) 1283–1286. https://doi:10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed]
- 17.C.B.E.M. Reusken, E.K. Broberg, B. Haagmans, A. Meijer, V.M. Corman, A. Papa, et al., Laboratory readiness and response for novel coronavirus (2019-nCoV) in expert laboratories in 30 EU/EEA countries, January 2020, Euro Surveill. 25 (2020). https://doi:10.2807/1560-7917.ES.2020.25.6.2000082. [DOI] [PMC free article] [PubMed]
- 18.L. Azzi, Saliva is the Key Element for SARS-CoV-2 Mass Screening, Clin. Infect. Dis. (2020). https://doi:10.1093/cid/ciaa1440. [DOI] [PMC free article] [PubMed]
- 19.J. Dinnes, J.J. Deeks, A. Adriano, S. Berhane, C. Davenport, S. Dittrich, et al., Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection, Cochrane Database Syst Rev. 8 (2020) CD013705. https://doi:10.1002/14651858.CD013705. [DOI] [PMC free article] [PubMed]
- 20.Hirotsu Yosuke, Maejima Makoto, Shibusawa Masahiro, Nagakubo Yuki, Hosaka Kazuhiro, Amemiya Kenji, Sueki Hitomi, Hayakawa Miyoko, Mochizuki Hitoshi, Tsutsui Toshiharu, Kakizaki Yumiko, Miyashita Yoshihiro, Yagi Shintaro, Kojima Satoshi, Omata Masao. Comparison of automated SARS-CoV-2 antigen test for COVID-19 infection with quantitative RT-PCR using 313 nasopharyngeal swabs, including from seven serially followed patients. International Journal of Infectious Diseases. 2020;99:397–402. doi: 10.1016/j.ijid.2020.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D. Basso, A. Aita, F. Navaglia, E. Franchin, P. Fioretto, S. Moz, et al., SARS-CoV-2 RNA identification in nasopharyngeal swabs: issues in pre-analytics, Clin. Chem. Lab. Med. 58 (2020) 1579–1586. doi:10.1515/cclm-2020-0749. [DOI] [PubMed]
- 22.Diao Bo, Wen Kun, Zhang Ji, Chen Jian, Han Chao, Chen Yongwen, Wang Shufeng, Deng Guohong, Zhou Hongwei, Wu Yuzhang. Accuracy of a nucleocapsid protein antigen rapid test in the diagnosis of SARS-CoV-2 infection. Clinical Microbiology and Infection. 2021;27(2):289.e1–289.e4. doi: 10.1016/j.cmi.2020.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Ruoshi, Cui Bomiao, Duan Xiaobo, Zhang Ping, Zhou Xuedong, Yuan Quan. Saliva: potential diagnostic value and transmission of 2019-nCoV. Int J Oral Sci. 2020;12(1) doi: 10.1038/s41368-020-0080-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porte Lorena, Legarraga Paulette, Vollrath Valeska, Aguilera Ximena, Munita José M, Araos Rafael, Pizarro Gabriel, Vial Pablo, Iruretagoyena Mirentxu, Dittrich Sabine, Weitzel Thomas. Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. International Journal of Infectious Diseases. 2020;99:328–333. doi: 10.1016/j.ijid.2020.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mak Gannon CK, Cheng Peter KC, Lau Stephen SY, Wong Kitty KY, Lau CS, Lam Edman TK, Chan Rickjason CW, Tsang Dominic NC. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. Journal of Clinical Virology. 2020;129:104500. doi: 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.M. Nagura-Ikeda, K. Imai, S. Tabata, K. Miyoshi, N. Murahara, T. Mizuno, et al., Clinical Evaluation of Self-Collected Saliva by Quantitative Reverse Transcription-PCR (RT-qPCR), Direct RT-qPCR, Reverse Transcription-Loop-Mediated Isothermal Amplification, and a Rapid Antigen Test To Diagnose COVID-19, J. Clin. Microbiol. 58 (2020) 497. https://doi:10.1128/JCM.01438-20. [DOI] [PMC free article] [PubMed]
- 27.Mao Ming-Hui, Guo Jing-Jing, Qin Li-Zheng, Han Zheng-Xue, Wang Ya-Jie, Yang Di. Serial semiquantitative detection of SARS-CoV-2 in saliva samples. Journal of Infection. 2020 doi: 10.1016/j.jinf.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Y. Sohn, S.J. Jeong, W.S. Chung, J.H. Hyun, Y.J. Baek, Y. Cho, et al., Assessing Viral Shedding and Infectivity of Asymptomatic or Mildly Symptomatic Patients with COVID-19 in a Later Phase, J Clin Med. 9 (2020). https://doi:10.3390/jcm9092924. [DOI] [PMC free article] [PubMed]
- 29.D.B. Larremore, B. Wilder, E. Lester, S. Shehata, J.M. Burke, J.A. Hay, et al., Test sensitivity is secondary to frequency and turnaround time for COVID-19 surveillance, medRxiv. (2020). https://doi:10.1101/2020.06.22.20136309. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.