Abstract
The open reading frame 8 (orf8) is an accessory protein of SARS-CoV-2. It has 121 amino acids with two genotypes, orf8L and orf8S. In this study, we overexpressed the orf8L and orf8S of SARS-CoV-2 as well as the orf8b of SARS-CoV to investigate their roles in the regulation of endoplasmic reticulum (ER) stress and the inhibition of interferon beta (IFNß) production. We found that the two genotypes of SARS-CoV-2 orf8 are capable of inducing ER stress without significant difference by triggering the activating transcription factor 6 (ATF6) and inositol-requiring enzymes 1 (IRE1) branches of the ER stress pathway. However, the third branch of ER stress pathway, i.e. the protein kinase-like ER kinase (PERK), was unaffected by the overexpression of SARS-CoV-2 orf8L or orf8S. Moreover, both orf8L and orf8S of SARS-CoV-2 are capable of down regulating the production of IFNß and interferon-stimulated genes (ISG), ISG15 and ISG56 induced by polyinosinic-polycytidylic acid (poly (I:C)). Moreover, we also found decreased nuclear translocation of Interferon regulatory factor 3 (IRF3), after overexpressing orf8L and orf8S induced by poly (I:C). Our data demonstrated that SARS-CoV-2 orf8 protein could induce ER stress by activating the ATF6 and IRE1 pathways, but not the PERK pathway, and functions as an interferon antagonist to inhibit the production of IFNß. However, these functions appeared not to be affected by the genotypes of SARS-CoV-2 orf8L and orf8S.
Keywords: SARS-CoV-2, Open reading frame 8, Open reading frame 8 genotypes, Endoplasmic reticulum stress, Interferon beta
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causal agent of the COVID-19 pandemic and is phylogenetically related to the 2002–2003 severe acute respiratory syndrome coronavirus (SARS-CoV) and other bat-related SARS-CoVs (Ceraolo and Giorgi, 2020; Chan et al., 2020). Several proteins of SARS-CoV can trigger endoplasmic reticulum (ER) stress and unfolded protein responses (UPR) to maintain cellular homeostasis (Minakshi et al., 2009; Sung et al., 2009; Shi et al., 2019). The suppression of primary interferons (IFNs) production and signaling has been observed in subjects infected with SARS-CoV-2 (Yuen et al., 2020). However, studies that investigate the role of the individual viral proteins of SARS-CoV-2 in the regulation of ER stress and innate immune suppression need to be conducted.
ER is responsible for the correct folding and maturation of a large number of proteins (Rashid et al., 2017). Protein folding is both sensed and regulated by ER resident chaperones, i.e. GRP78 and GRP94 (Stevens et al., 2010). When the proteins are not folded correctly, ER stress is induced and causes cell toxicity (Rashid et al., 2017). Cells respond to ER stress by activating a protective response termed the UPR (Rashid et al., 2017). SARS-CoV and SARS-CoV-2 have developed mechanisms to modulate UPR (Chan et al., 2006; Minakshi et al., 2009; Köseler et al., 2020; Sung et al., 2009; Shi et al., 2019). Various individual proteins of SARS-CoV including the spike, orf3a, and orf8b have been shown to induce ER stress pathways (Shi et al., 2019; Minakshi et al., 2009; Sung et al., 2009). Also, the activation of the ER stress pathways has important immunoregulatory roles in mediating IFNß production (Martinon et al., 2010; Mitzel et al., 2014).
Viral pathogen-associated molecular patterns (PAMPs), such as viral genomes or viral mRNAs, are first recognized by host pattern recognition receptors (PRR) (Lim et al., 2016; Chen et al., 2014). These are followed by the activation of transcription factors such as IRF3 and adaptor proteins to control the production of type I/III IFNs (Kawai and Akira, 2007). The above responses activate the interferon-stimulated genes (ISGs) and are critical for the control of early coronavirus infection (Schneider et al., 2014; Ivashkiv and Donlin, 2014).
Viruses such as coronaviruses have evolved mechanisms to suppress IFN production by targeting different aspects of the IFN signaling cascade (Lim et al., 2016). Coronaviruses use their structural proteins and non-structural or accessory proteins to suppress the host innate immune system (Kamitani et al., 2009; Huang et al., 2011; Wathelet et al., 2007; Liu et al., 2014; Lim et al., 2016; Zhong et al., 2012). Moreover, RNA viruses including SARS-CoV-2 have the capability of gaining rapid mutation, which is correlated with virus virulence and its dissemination (Lu et al., 2020). Although the viral genes of SARS-CoV-2 are relatively conserved, the orf8 gene is prone to mutation (Laha et al., 2020). Previous studies have identified two genotypes of SARS-CoV-2 orf8 protein i.e. orf8L and orf8S from China during the early stage of COVID-19 pandemic, which are characterized by the presence of either Leucine (orf8L) or Serine (orf8S) at amino acid 84 (Ceraolo and Giorgi, 2020; Tang et al., 2020). Several important functions have been attributed to SARS-CoV orf8b, including apoptosis (Chen et al., 2007), down regulation of the envelope protein (Keng et al., 2006), induction of the ER stress pathways (Shi et al., 2019), and antagonizing the IFN signaling pathway. (Lee et al., 2019; Wong et al., 2020). Moreover, the presence of N-terminal signal sequences in the orf8 of SARS-CoV-2, which are responsible for protein residence in ER, prompted us to investigate whether this protein could induce ER stress (Mohammad et al., 2020). SAR-CoV-2 is poor inducer of IFN-I responses. IFN-I levels in the sera of COVID-19 patients are not detectable by commonly used assays (Ribero et al., 2020). It was also found that that SARS-CoV-2 orf8 protein could down regulate the major histocompatibility complex class I (MHC-I) (Zhang et al., 2020), which in turn compelled us to investigate whether this protein could evade the immune system by suppressing IFNß production.
Therefore, in this study, we investigated the role of SARS-CoV-2 orf8 in regulating ER stress pathways and IFNß production by overexpressing the two SARS-CoV-2 orf8 genotypes orf8L and orf8S. We found that both orf8L and orf8S of SARS-CoV-2 induced ER stress by activating ATF6 and IRE1 pathways of ER stress, but not the PERK pathway. Moreover, we also observed that SARS-CoV-2 orf8 antagonized INFß production by decreasing the nuclear translocation of IRF3 and suppressed interferon-stimulated genes (ISG) ISG15 and ISG56. The study will further contribute to the understanding of viral pathogenesis of SARS-CoV-2.
2. Materials and methods
2.1. Sequence alignment of orf8L and orf8S
The primary amino acid sequences of SARS-CoV-2 orf8L and orf8S were retrieved from NCBI (https://www.ncbi.nlm.nih.gov/) (NCBI Resource Coordinators, 2018) using the accession numbers MN908947.3 and MN975262.1, respectively. Sequence alignments of the two isolates of orf8 were performed using clustal Omega.
2.2. Construction of plasmids
The full-length constructs of SARS-CoV-2 orf8L and orf8b were cloned into pCDNA3.1-EGFP-HA-C vector (kindly provided by Dr. Yi-Ping Li of Institute of Human Virology, Sun Yat-sen University, Guangdong, China) where the HA tag is present at the C-terminus. The pCDNA3.1-EGFP-HA orf8S plasmid was constructed by PCR-based point mutation to create a mismatch mutation in the primers using pCDNA3.1-EGFP-HA-orf8L as template. All constructs were confirmed by sequencing. The primer sequences are included in Supplementary table.
For the functional analysis of orf8L and orf8S, the promoter regions of IFNß (−309 to +9), GRP78 (−304 to +1), and GRP94 (−363 to +34) were cloned into pGL3 vector. The promoter regions were amplified by PCR from the genomic DNA of HEK-293 T cells. The PCR product was then sub-cloned into the XhoI site of the vector. All the primers used for plasmid construction are given in supplementary table. For immunostaining experiments, the full-length constructs of SARS-CoV-2 orf8L/S and SARS-CoV-orf8b were cloned into pCAGGS-FLAG-N (kindly provided by Dr. Jianzhong Wang, College of veterinary Medicine, Jilin Agricultural University, China), where the FLAG tag is present at the N terminus.
2.3. Cell culture and transfection
HEK-293 T cells were cultured under the standard conditions with DMEM (GE Healthcare, South Logan, UT, USA) plus 10 % heat inactivated fetal bovine serum (FBS) at 37 °C and 5% CO2. Plasmid transfections were performed with Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the supplier’s protocol. For 12 well plate, each well was incubated with 2ug poly (I:C) and 5 μL of Lipofectamine 3000. For TG treatments, actively growing cells were incubated for 8 h with 300 nM TG.
2.4. RT-PCR reactions
Total RNA was isolated using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. DNA was eliminated with nuclease-free DNase (Promega, Madison, WI, USA). For RT–PCR, complementary DNA was synthesized from RNA with the corresponding primers and a GoScript Reverse Transcription System (Promega) according to the manufacturer’s protocol. About 500 ng RNA were used for cDNA synthesis. Quantitative real-time PCR was performed with GoTaq SYBR Green qPCR Master Mix (Promega) on a PikoReal 96 real-time PCR system (Thermo Scientific, Waltham, MA, USA) according to standard procedures. All PCR products were sequenced for validation. All the primers are given in supplementary table.
2.5. Luciferase assay
HEK-293 T cells (1 × 105) were transfected with 500 ng of expression plasmids for SARS-CoV orf8b and SARS-CoV-2 or8L or orf8S as well as empty vector control, 500 ng of pGL3 Basic Vector carrying the corresponding promoter region of IFNß, GRP78 or GRP94, and 10 ng pRL-TK renilla plasmid. Poly (I:C) was transfected 6 h before cells lysis. Luciferase assays were performed 48 h after transfection using Dual Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocol. Briefly, cells were lysed with passive lysis buffer at 4 °C for 20 min. The luciferase assay buffer II was then added, and firefly luciferase (F-luc) activity was immediately read using a Fluoroskan Ascent FL microplate reader (Thermo Scientific). Next, Stop & Glo Buffers and Stop & Glo substrates were added and mixed briefly. Renilla luciferase (R-luc) activity was immediately read. F-luc activity was normalized to R-luc activity to avoid the variation of transfection efficiency.
2.6. Western Blotting and immunostaining
The cells were washed twice with ice-cold PBS and resuspended in RIPA lysis buffer (150 mM NaCl, 50 mM Tris pH 8.0, 1 % Nonidet P-40, 0.5 % deoxycholate) with protease inhibitor mixture (Roche Applied Science, Mannheim, Germany). Cells were incubated on ice for 20 min and rotated at 4 °C for 10 min. The lysates were clarified by centrifugation for 20 min at 14,000 rpm at 4 °C. Samples were separated on 10 % SDS–PAGE gels and then transferred to PVDF membranes (Millipore, Billerica, MA, USA). The membranes were blocked with 5 % non-fat milk for 1 h at room temperature, and then incubated with primary antibodies in 5 % non-fat milk over night at 4 °C. After washing the membranes three times with PBST, the membranes were incubated with the corresponding secondary antibodies, and processed according to the ECL Western blotting protocol (GE Healthcare). For immunostaining, HEK-293 T cells seeded in 12 well culture plates containing clean sterile cover slips (15 mm) were washed twice with PBS, and fixed with 4 % paraformaldehyde, treated with 0.1 % triton X-100 and blocked with 1%BSA followed by staining with the indicated antibodies. Images were captured using confocal microscope (ZEISS). The following primary antibodies were used in Western blotting: anti-ATF6 (Origene, cat#TA336753) and anti-GAPDH (cell signaling, cat#2118), anti-eIF2α (cell signaling, cat#2103), peIF2α (cell signaling, cat#3597), anti-ERdj4 (Abcam, cat#ab118282). The antibodies used in immune staining are: anti-FLAG (abcam, cat#ab45766), anti-IRF3 (CST, 1cat#1904).
3. Results
3.1. Sequence alignment of two genotypes of SARS-CoV-2 orf8 protein
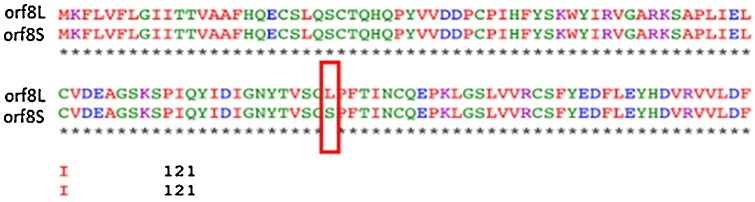
The SAR-CoV-2 orf8 protein is 121 amino acids and emerged due to deletion of 382 nucleotides, including 40-nt deletion at the 3’ end of orf7b, 6-nt deletion within the intergenic region of orf7b/8, and 336-nt deletion at the 5’ end of orf8 (Su et al., 2020; Gordon et al., 2020). In this study, we aligned the two genotypes of orf8 protein, i.e. orf8L and orf8S, which are caused by the mutation of C/U at nt 251 and the change of Serine to Leucine at the amino acid 84 (Fig. 1 ).
Fig. 1.
Sequence alignment of orf8L and orf8S. Amino acid sequences of orf8L and orf8S. The only difference between orf8L and orf8S is at amino acid 84 where Leucine in orf8L was substituted with Serine orf8S.
3.2. Induction of ER stress by orf8L and orf8S of SARS-CoV-2
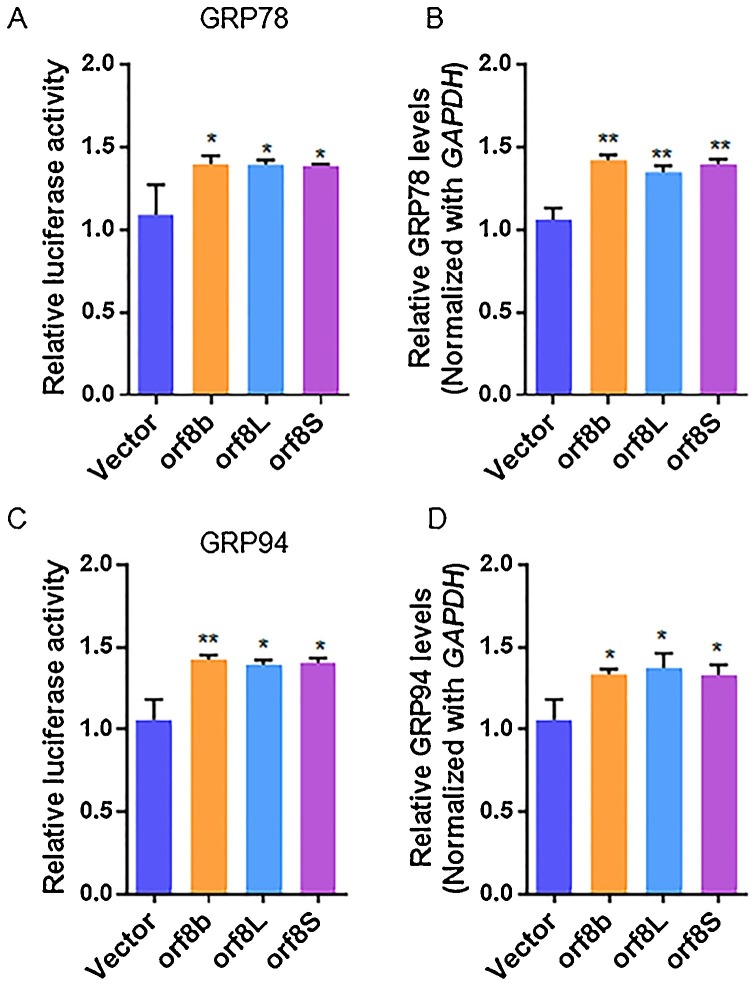
The orf8b protein of SARS-CoV was able to induce ER stress and UPR (Shi et al., 2019). In this study, we investigated the role of SARS-CoV-2 orf8 protein in inducing ER stress and UPR, and also ascertained any differential roles of the individual SARS-CoV-2 orf8 genotypes in inducing UPR. We first analyzed the transcriptional activation of GRP78 and GRP94 genes, which are the molecular chaperones and sensitive biomarkers of ER stress in cells expressing orf8L and orf8S of SARS-CoV-2, and used orf8b of SARS-CoV as a positive control. The relative luciferase activity showed a significant increase by about 1.4 folds in cells co-transfected with the plasmid DNAs of luciferase reporter constructs driven by the promoters of GRP78 (Fig. 2 A) or GRP94 (Fig. 2C) genes, and orf8L or orf8S when compared with the negative control of the vector DNA. Furthermore, the levels of luciferase activity were comparable between the cells expressing SARS-CoV orf8b and SARS-CoV-2 orf8L or orf8S. We also assessed the mRNA levels of GRP78 and GRP94 in the cells overexpressing orf8L and orf8S. As expected, the mRNA levels of GRP78 (Fig. 2B) and GRP94 (Fig. 2D) were upregulated. These results suggest that both orf8L and orf8S of SARS-CoV-2 were capable of upregulating ER-resident chaperones with the same level of intensity.
Fig. 2.
ER stress was induced in HEK-293 T cells after overexpressing SARS-CoV-2 orf8. (A and C) HEK-293 T cells were co-transfected with firefly luciferase plasmids harboring the corresponding promoter of GRP78 (A) or GRP94 (C), pRL-null (Renilla plasmid), and either of orf8L, orf8S, orf8b or vector control. The ratio of the reporter (Firefly) to control (Renilla) in relative luminescence units were plotted. (B and D) mRNA levels of GRP78 (B) and GRP94 (D) were examined by qRT-PCR in HEK-293 T cells after treatment with orf8L, orf8S, orf8b and vector control. Orf8L and orf8S referred to SARS-CoV-2, orf8b referred to SARS-CoV * p < 0.05; ** p < 0.01; *** p < 0.001. p values were determined with two-tailed student’s t test. Error bars represent standard deviation S.D.
3.3. SARS-CoV-2 orf8L and orf8S activate the ATF6/XBP1 branches of UPR
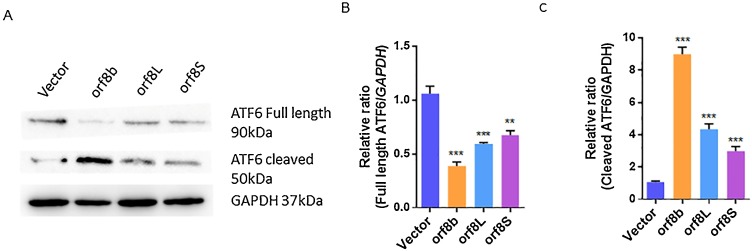
ER stress activates three major signaling pathways of the UPR, which are mediated through PERK kinase, IRE1, and ATF6, respectively (Hou et al., 2019; Rashid et al., 2017). We first investigated whether the ATF6 branch of UPR is activated when orf8L or orf8S are overexpressed. We observed that the levels of full-length ATF6 was significantly reduced by about 56 % for orf8L and 52 % for orf8S of the negative control (Fig. 3 A and B). In contrast, the levels of cleaved ATF6 significantly increased when orf8b of SARS-CoV, a previously known activator of ATF6, and SARS-CoV-2 orf8L and orf8S were overexpressed. The levels of cleaved ATF6 were increased by 40 % for orf8L and 35 % for orf8S (Fig. 3A and C). These results indicate that the ATF6 branch of UPR was induced by orf8L and orf8S of SARS-CoV-2 although the induction was weaker than the SARS-CoV orf8b protein.
Fig. 3.
SARS-CoV-2 orf8L and orf8S regulated the ATF6 pathway. (A) Levels of full length and cleaved ATF6 were examined by Western blots. GAPDH was used as loading control. (B-C) The full length and cleaved ATF6 densitometry analysis of Western blot treated with orf8L, orf8S, orf8b, or vector control. GAPDH was used as loading control. Orf8L and orf8S referred to SARS-CoV-2, orf8b referred to SARS-CoV * p < 0.05; ** p < 0.01; *** p < 0.001. p values were determined with two-tailed student’s t test. Error bars represent standard deviation S.D.
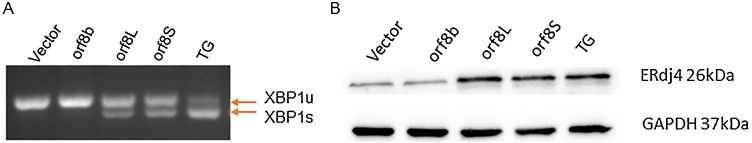
Next, we investigated the effect of orf8L and orf8S of SARS-CoV-2 on the IRE1 pathway by examining the XBP1 splicing (XBP1s) and used cells treated with Thapsigargin (TG) as a positive control (Rashid et al., 2017). The XBP1 splicing was observed in the cells treated with TG, and the cells transfected with both orf8L and orf8S of SARS-CoV-2 (Fig. 4 A). Since XBP1s is essential for the expression of ERdj4, we wonder if over expression of orf8L and orf8S could also upregulate the ERdj4 levels. Our results showed that orf8L or orf8S over expression and TG treatment significantly increased the levels of ERdj4 (Fig. 4B). However, orf8b of SARS-CoV, which failed to induce the splicing of XBP1 (Fig. 4A), was incapable to induce ERdj4 (Fig. 4B). These results indicate that the IRE1 branch of UPR was upregulated by orf8L and orf8S of SARS-CoV-2 with similar efficiency, but not by SARS-CoV orf8b.
Fig. 4.
Regulation of the IRE1 pathway by orf8L and orf8S. (A) The cytoplasmic splicing of XBP-1 mRNA response to orf8L and orf8S and TG was detected by separating the RT-PCR product on an agarose gel. (B) Levels of ERdj4 were examined by Western blots. GAPDH was used as loading control.
3.4. PERK pathway was not induced by orf8L and orf8S of SARS-CoV-2
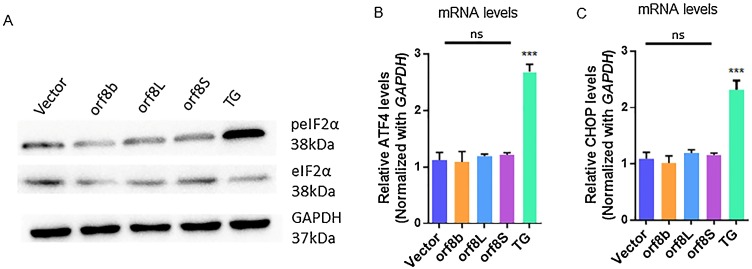
Under ER stress, the kinase activity of PERK is activated, which in turn phosphorylates its downstream target, the eukaryotic translation initiation factor 2 subunit α (eIF2α). To investigate if orf8L and orf8S of SARS-CoV-2 regulate the PERK pathway of UPR, we first examined the phosphorylation status of eIF2α (peIF2α) using Western blotting. The levels of peIF2α were unaffected in the cells expressing orf8b of SARS-CoV as reported previously (Sung et al., 2009). Similarly, no change in the phosphorylation levels of eIF2α was found in the cells expressing orf8L and orf8S of SARS-CoV-2. However, the levels of peIF2α increased in the cells treated with TG (Fig. 5 A). Furthermore, the mRNA levels of ATF4 (Fig. 5B) and CHOP (Fig. 5C) were similar in all the cells transfected with the vector-only control and SARS-CoV orf8b or SARS-CoV-2 orf8L and orf8S. In contrast, in the presence of TG, the mRNA levels of ATF4 and CHOP dramatically increased (Fig. 5B and C). These results confirm that the orf8 proteins of both SARS-CoV and SARS-CoV-2 do not regulate the PERK pathway under ER stress conditions.
Fig. 5.
The PERK arm of UPR was not regulated by orf8L and orf8S. (A) Levels of phosphorylated eIF2α (top panel) and eIF2α (middle panel) were examined by western blots treated with orf8L, orf8S, orf8b, vector control and TG. GAPDH was used as loading control. (B-C) mRNA levels of ATF4 (B) and CHOP (C) were examined by qRT-PCR in HEK-293 T cells after treatment with orf8L, orf8S, orf8b, vector control and TG. Orf8L and orf8S referred to SARS-CoV-2, orf8b referred to SARS-CoV. Orf8L and orf8S referred to SARS-CoV-2, orf8b referred to SARS-CoV.* p < 0.05; ** p < 0.01; *** p < 0.001. p values were determined with two-tailed student’s t test. Error bars represent standard deviation S.D.
3.5. SARS-CoV-2 orf8L and orf8S are interferon antagonist
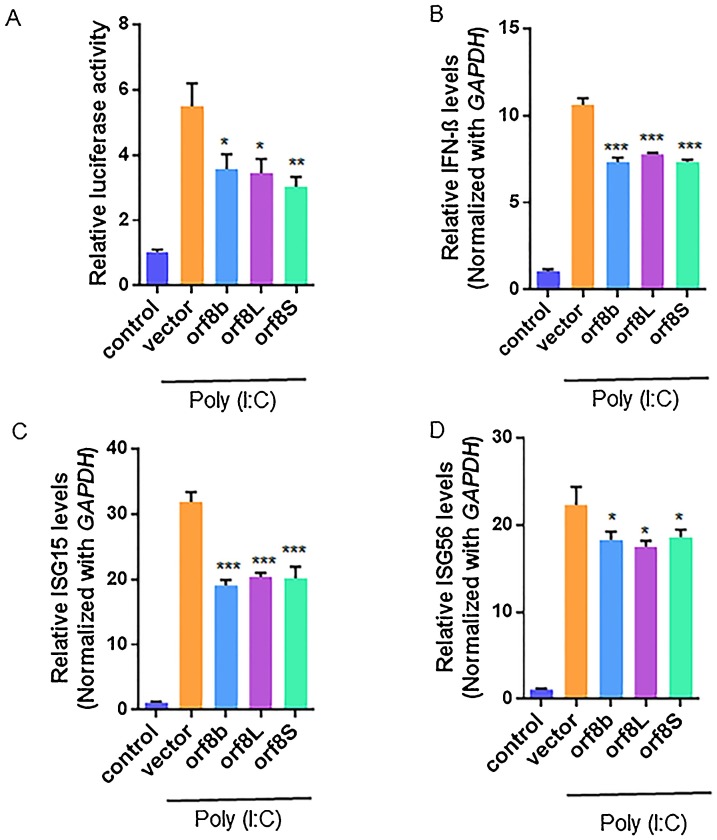
Next, we asked whether orf8L and orf8S of SARS-CoV-2 influenced the induction of INFß. For this purpose, a well-known immune stimulant, Poly (I:C) was used to induce INFß. The levels of INFß were assessed by measuring its promoter activity (Fig. 6 A). As expected, the levels of INFß significantly increased in the cells treated with Poly (I:C). However, we found that the levels of INFß dramatically decreased in the cells expressing both SARS-CoV orf8b and SARS-CoV-2 orf8L/orf8S, and there were no significant differences among the three proteins (Fig. 6A). Furthermore, the mRNA levels of INFß also significantly decreased in the cells expressing orf8L, orf8S, and orf8b, representing about 80 % of the vector control (Fig. 6B). Similarly, the mRNA levels of the two interferon-stimulated genes (ISGs), ISG15 and ISG56, were down regulated in the cells expressing orf8L and orf8S as well as orf8b compared to the control cells. The mRNA levels of ISG15 and ISG56 decreased by about 26% and 23%, respectively (Fig. 6C and D).
Fig. 6.
orf8L and orf8S are IFNß antagonist. (A) HEK-293 T cells were co-transfected with firefly luciferase plasmids harboring the corresponding promoter of IFNß, pRL-null (Renilla plasmid) and either of orf8L, orf8S, orf8b or vector control. Cells were also transfected with Poly (I:C) to activate the IFN. The ratio of the reporter (Firefly) to control (Renilla) in relative luminescence units were plotted. (B-D) mRNA levels of IFNß (B) ISG15 (C) and ISG56 (D) were examined by qRT-PCR in HEK-293 T cells after treatment with orf8L, orf8S, orf8b and vector control. Cells were also transfected with Poly (I:C) to activate the IFN expression pathway. * p < 0.05; ** p < 0.01; *** p < 0.001. p values were determined with two-tailed student’s t test. Error bars represent standard.
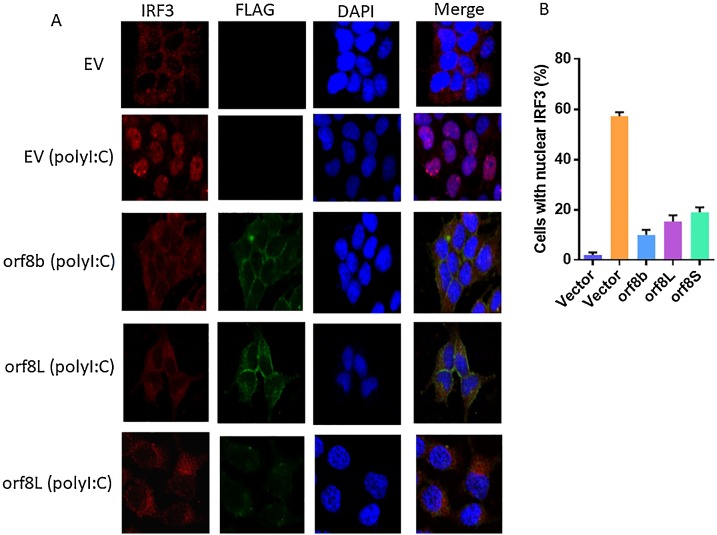
To determine the possible inhibitory mechanism of SARS-CoV-2 orf8 in inhibiting IFN induction, the nuclear translocation of IRF3 was determined after overexpressing orf8L and orf8S, orf8b or empty vector in the presence or absence of poly (I:C). As expected, orf8b of SARS-CoV, which is previously known as inhibitor of IRF3 nuclear localization, could inhibit nuclear translocation of IRF3 induced by poly (I:C). Both orf8L and orf8S could also decrease the nuclear translocation of IRF3 although less efficiently than orf8b of SARS-CoV in our experiment (Fig. 7 A and B).
Fig. 7.
Suppression of IRF3 nuclear translocation by SARS-CoV-2 proteins. (A) HEK-293 T Cells were transfected with pCAGGS-FLAG-N (EV) or pCAGGS-FLAG-N orf8b/orf8L/S for 20 h, and then transfected with or without poly (I:C) for more 12 h. At 32 h post infection. Cells were analyzed to detect IRF3 (red) pCAGGS-FLAG-N orf8b/orf8L/S proteins (green) singals. (B) Percentage of nuclear IRF3-positive transfected cells were counted from three fields of view.
4. Discussion
SARS-CoV orf8b protein has been shown to induce ER stress in cells (Sung et al., 2009). Our study showed that SARS-CoV-2 orf8 protein could also induce ER stress by activating ER stress markers, GRP78 and GRP94, in the cells expressing SARS-CoV-2 orf8L or orf8S proteins (Fig. 2). Previous studies and our results indicate that the orf8 protein of SARAS-CoV and SARS-CoV-2 may share common functions in regulating ER stress although there are only 54.4 % nucleotides identities between the SARS-CoV orf8b and SARS-CoV-2 orf8 (Su et al., 2020). Of note, the N-terminal signal sequence from 2-nt to 16-nt are highly conserved between the SARS-CoV-2 and SARS-CoV and is responsible for locating the viral proteins to reside in ER (Oostra et al., 2007; Mohammad et al., 2020). Besides, both SARS-CoV-2 orf8 and SARS-CoV orf8b have multiple cysteine residues, which are important for inducing ER stress through the formation of disulfide bridges. It is believed that the ER functions to form intra- or intermolecular disulfide bonds between these cysteine residues (Mohammad et al., 2020). Our results therefore provided evidence that SARS-CoV-2 orf8 induce ER stress.
There are three well-known ER stress-related pathways, i.e. ATF6, IRE1/XBP1, and PERK (Rashid et al., 2017). Previous studies and our results indicated that SARS-CoV orf8b mainly activated the ATF6 pathway under ER stress conditions (Sung et al., 2009). However, the SARS-CoV-2 orf8 protein appeared to modulate ER stress by stimulating both ATF6 and IRE1 pathways, but not the PERK pathway (Fig. 3, Fig. 4, Fig. 5). Furthermore, the two genotypes of SARS-CoV-2, orf8L and orf8S, showed similar functions in regulating ER stress and its related pathways (Fig. 2, Fig. 3, Fig. 4, Fig. 5).
In our study, we also demonstrated that both SARS-CoV-2 orf8 and SARS-CoV orf8b could antagonize IFNß production (Fig. 6), which is one of the important strategies employed by viruses for the successful infection of host cells by overcoming host immune responses (Wong et al., 2018). We also determined that the decrease nuclear translocation of IRF3 after overexpressing orf8L orf8S in the presence of poly I:C (Fig. 7). It has recently been reported that the overexpression of SARS-CoV-2 orf8 resulted in the down regulation of MHC-I in different human cell lines to mediate immune evasion (Zhang et al., 2020). In our study, we found that SARS-CoV-2 orf8 could antagonize IFNß and inhibit ISGs (Fig. 6). These results suggested that SARS-CoV-2 adapted multiple mechanisms to evade the host immune system, such as the orf8-mediated down regulation of MHC-I (Zhang et al., 2020) and inhibition of IFNß production (Fig. 6).
In this preliminary study, SARS-CoV-2 orf8 was found to play a role in regulating the ER stress pathway and antagonizing IFNß production. However, the detailed mechanism remains to be elucidated about how the regulation of the ER stress pathways influences the inhibition of IFNß and ISGs (Mitzel et al., 2014). Moreover, it will be interesting to find out whether SARS-CoV-2 orf8 physically interacts with transcription factors to impede IFNß production. In our study, we observed that the two genotypes of SARS-CoV-2 orf8 protein, orf8L and orf8S, function similarly in inducing ER stress pathways and antagonizing IFNß production. The MHC-I gene was also found to be down regulated by both orf8L and orf8S to the same level in a recent study (Zhang et al., 2020). These results indicate that this mutation does not affect the crucial role played by orf8 in the induction of ER stress and immune evasion. However, the role of other latest identified genotypes of SARS-CoV-2 orf8, V62 L and S24 L need to be analyzed (Laha et al., 2020). Thus, although the most commonly observed mutation of the SARS-CoV-2 orf8 is the L84S, the recent identification of the V62 L and S24 L mutations may have clinical implications (Laha et al., 2020). Furthermore, it has been observed that the V62 L mutation is usually accompanied by L84S mutation, hence it will be important to study the orf8 protein with double mutations (V62 L and L84S) and also explore the roles of the individual genotype to induce ER stress pathways and to evade innate immune in future investigations (Laha et al., 2020).
5. Conclusions
During viral infection, host cells recruit multiple mechanisms to fight virus invasion and to maintain cellular homeostasis. In this study, we demonstrated that both SARS-CoV orf8b and SARS-CoV-2 orf8 protein activate ER stress pathways. Furthermore, both SARS-CoV orf8b and SARS-CoV-2 orf8 proteins suppress IFNß production. We also determined the decreased nuclear translocation of IRF3 in orf8L and orf8S overexpressing cells induced by poly (I:C). The identification of SARS-CoV-2 orf8 protein in the regulation of UPR and its antagonizing role in modulating the production of IFNß will provide better insight into the pathogenesis of SARS-CoV-2 infection.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Farooq Rashid: Conceptualization, Methodology, Writing - original draft. Emmanuel Enoch Dzakah: Writing - review & editing. Shixing Tang: Supervision, Conceptualization, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.virusres.2021.198350.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Ceraolo C., Giorgi F.M. Genomic variance of the 2019-nCoV coronavirus. J. Med. Virol. 2020;92:522–528. doi: 10.1002/jmv.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.-P., Siu K.-L., Chin K.-T., Yuen K.-Y., Zheng B., Jin D.-Y. Modulation of the unfolded protein response by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2006;80:9279–9287. doi: 10.1128/JVI.00659-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.-W., Kok K.-H., Zhu Z., Chu H., To K.K.-W., Yuan S., Yuen K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-Y., Ping Y.-H., Lee H.-C., Chen K.-H., Lee Y.-M., Chan Y.-J., Lien T.-C., Jap T.-S., Lin C.-H., Kao L.-S., Chen Y.-M.A. Open reading frame 8a of the human severe acute respiratory syndrome coronavirus not only promotes viral replication but also induces apoptosis. J. Infect. Dis. 2007;196:405–415. doi: 10.1086/519166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Yang X., Zheng Y., Yang Y., Xing Y., Chen Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell. 2014;5:369–381. doi: 10.1007/s13238-014-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Hüttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.-P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O’Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.-Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L., Dong J., Zhu S., Yuan F., Wei L., Wang J., Quan R., Chu J., Wang D., Jiang H., Xi Y., Li Z., Song H., Guo Y., Lv M., Liu J. Seneca valley virus activates autophagy through the PERK and ATF6 UPR pathways. Virology. 2019;537:254–263. doi: 10.1016/j.virol.2019.08.029. [DOI] [PubMed] [Google Scholar]
- Huang C., Lokugamage K.G., Rozovics J.M., Narayanan K., Semler B.L., Makino S. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv L.B., Donlin L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani W., Huang C., Narayanan K., Lokugamage K.G., Makino S. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol. 2009;16:1134–1140. doi: 10.1038/nsmb.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T., Akira S. Antiviral signaling through pattern recognition receptors. J. Biochem. 2007;141:137–145. doi: 10.1093/jb/mvm032. [DOI] [PubMed] [Google Scholar]
- Keng C.T., Choi Y.W., Welkers M.R.A., Chan D.Z.L., Shen S., Gee Lim S., Hong W., Tan Y.J. The human severe acute respiratory syndrome coronavirus (SARS-CoV) 8b protein is distinct from its counterpart in animal SARS-CoV and down-regulates the expression of the envelope protein in infected cells. Virology. 2006;354:132–142. doi: 10.1016/j.virol.2006.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köseler A., Sabirli R., Gören T., Türkçüer I., Kurt Ö. Endoplasmic reticulum stress markers in SARS-COV-2 infection and pneumonia: case-control study. In Vivo (Brooklyn) 2020;34:1645–1650. doi: 10.21873/invivo.11956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laha S., Chakraborty J., Das S., Manna S.K., Biswas S., Chatterjee R. Characterizations of SARS-CoV-2 mutational profile, spike protein stability and viral transmission. Infect. Genet. Evol. 2020;85 doi: 10.1016/j.meegid.2020.104445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Bae S., Myoung J. Middle East respiratory syndrome coronavirus-encoded ORF8b strongly antagonizes IFN-β promoter activation: its implication for vaccine design. J. Microbiol. 2019;57:803–811. doi: 10.1007/s12275-019-9272-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y.X., Ng Y.L., Tam J.P., Liu D.X. Human coronaviruses: a review of virus-host interactions. Dis. (Basel, Switzerland) 2016;4 doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Fung T.S., Chong K.K.-L., Shukla A., Hilgenfeld R. Accessory proteins of SARS-CoV and other coronaviruses. Antiviral Res. 2014;109:97–109. doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet (Lond., Engl.) 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F., Chen X., Lee A.-H., Glimcher L.H. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat. Immunol. 2010;11:411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakshi R., Padhan K., Rani M., Khan N., Ahmad F., Jameel S. The SARS coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS One. 2009;4:1–10. doi: 10.1371/journal.pone.0008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitzel D.N., Lowry V., Shirali A.C., Liu Y., Stout-Delgado H.W. Age-enhanced endoplasmic reticulum stress contributes to increased Atg9A inhibition of STING-mediated IFN-β production during Streptococcus pneumoniae infection. J. Immunol. 2014;192:4273–4283. doi: 10.4049/jimmunol.1303090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad S., Bouchama A., Alharbi B.M., Rashid M. 2020. SARS-CoV-2 ORF8 and SARS-CoV ORF8ab: Genomic Divergence and Functional Convergence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Resource Coordinators Database resources of the national center for biotechnology information. Nucleic Acids Res. 2018;46:D8–D13. doi: 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostra M., de Haan C.A.M., Rottier P.J.M. The 29-nucleotide deletion present in human but not in animal severe acute respiratory syndrome coronaviruses disrupts the functional expression of open reading frame 8. J. Virol. 2007;81:13876–13888. doi: 10.1128/JVI.01631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid F., Awan H.M., Shah A., Chen L., Shan G. Induction of miR-3648 upon ER stress and its regulatory role in cell proliferation. Int. J. Mol. Sci. 2017;18 doi: 10.3390/ijms18071375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribero M.S., Jouvenet N., Dreux M., Nisole S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020;16:1–22. doi: 10.1371/journal.ppat.1008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C.-S., Nabar N.R., Huang N.-N., Kehrl J.H. SARS-Coronavirus Open Reading Frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discov. 2019;5:101. doi: 10.1038/s41420-019-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens R.C., Sancho J., Martinez A. Protein misfolding and cellular stress in disease and aging. Methods Mol. Biol. 2010;648:313–324. doi: 10.1007/978-1-60761-756-3. [DOI] [Google Scholar]
- Su Y.C.F., Anderson D.E., Young B.E., Linster M., Zhu F., Jayakumar J., Zhuang Y., Kalimuddin S., Low J.G.H., Tan C.W., Chia W.N., Mak T.M., Octavia S., Chavatte J.-M., Lee R.T.C., Pada S., Tan S.Y., Sun L., Yan G.Z., Maurer-Stroh S., Mendenhall I.H., Leo Y.-S., Lye D.C., Wang L.-F., Smith G.J.D. Discovery and genomic characterization of a 382-nucleotide deletion in ORF7b and ORF8 during the early evolution of SARS-CoV-2. MBio. 2020;11 doi: 10.1128/mBio.01610-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S.-C., Chao C.-Y., Jeng K.-S., Yang J.-Y., Lai M.M.C. The 8ab protein of SARS-CoV is a luminal ER membrane-associated protein and induces the activation of ATF6. Virology. 2009;387:402–413. doi: 10.1016/j.virol.2009.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Wu C., Li X., Song Y., Yao X., Wu X., Duan Y., Zhang H., Wang Y., Qian Z., Cui J., Lu J. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020;7:1012–1023. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathelet M.G., Orr M., Frieman M.B., Baric R.S. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 2007;81:11620–11633. doi: 10.1128/JVI.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H.H., Fung T.S., Fang S., Huang M., Le M.T., Liu D.X. Accessory proteins 8b and 8ab of severe acute respiratory syndrome coronavirus suppress the interferon signaling pathway by mediating ubiquitin-dependent rapid degradation of interferon regulatory factor 3. Virology. 2018;515:165–175. doi: 10.1016/j.virol.2017.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L.-Y.R., Ye Z.-W., Lui P.-Y., Zheng X., Yuan S., Zhu L., Fung S.-Y., Yuen K.-S., Siu K.-L., Yeung M.-L., Cai Z., Woo P.C.-Y., Yuen K.-Y., Chan C.-P., Jin D.-Y. Middle east respiratory syndrome coronavirus ORF8b accessory protein suppresses type I IFN expression by impeding HSP70-dependent activation of IRF3 kinase IKKε. J. Immunol. 2020 doi: 10.4049/jimmunol.1901489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen C.K., Lam J.Y., Wong W.M., Mak L.F., Wang X., Chu H., Cai J.P., Jin D.Y., To K.K.W., Chan J.F.W., Yuen K.Y., Kok K.H. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microbes Infect. 2020;9:1418–1428. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang J., Chen Y., Luo B., Yuan Y., Huang F., Yang T., Yu F., Liu J., Liu B., Song Z., Chen J., Pan T., Zhang X., Li Y., Li R., Huang W., Xiao F., Zhang H. bioRxiv; 2020. The ORF8 Protein of SARS-CoV-2 Mediates Immune Evasion Through Potently Downregulating MHC-I. 2020.05.24.111823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y., Tan Y.W., Liu D.X. Recent progress in studies of arterivirus- and coronavirus-host interactions. Viruses. 2012;4:980–1010. doi: 10.3390/v4060980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.