Abstract
The endonuclease activity within the influenza virus cap-snatching process is a proven therapeutic target. The anti-influenza drug baloxavir is highly effective, but is associated with resistance mutations that threaten its clinical efficacy. The endonuclease resides within the N-terminal domain of the PA subunit (PAN) of the influenza RNA dependent RNA polymerase, and we report here complexes of PAN with RNA and DNA oligonucleotides to understand its specificity and the structural basis of baloxavir resistance mutations. The RNA and DNA oligonucleotides bind within the substrate binding groove of PAN in a similar fashion, explaining the ability of the enzyme to cleave both substrates. The individual nucleotides occupy adjacent conserved pockets that flank the two-metal active site. However, the 2′ OH of the RNA ribose moieties engage in additional interactions that appear to optimize the binding and cleavage efficiency for the natural substrate. The major baloxavir resistance mutation at position 38 is at the core of the substrate binding site, but structural studies and modeling suggest that it maintains the necessary virus fitness via compensating interactions with RNA. These studies will facilitate the development of new influenza therapeutics that spatially match the substrate and are less likely to elicit resistance mutations.
INTRODUCTION
The influenza virus RNA dependent RNA polymerase (RdRp) is a trimeric complex comprising the PB1, PB2 and PA subunits. The RdRp performs transcription to generate the viral mRNA, and replication to generate the viral genomic RNA, during the infection cycle (1). To synthesize viral mRNAs that can be processed by the host ribosome, the RdRp employs a ‘cap-snatching’ mechanism by which host mRNAs are sequestered and trimmed down to 5′-capped RNA primers (2–4). Over the past decade, crystallographic studies on components of the RdRp and the complete trimer have provided key insights into how the complex functions and how the three subunits interact to coordinate these activities (5,6). Early studies had suggested that the PB2 subunit contains the cap-binding functionality (7) while the PA subunit harbors the endonuclease activity (8), and these were confirmed by structural studies of subdomains of the two subunits (9–11). The structure of the trimer subsequently revealed that the two subdomains are perfectly positioned to both process bound host mRNAs and to allow the resulting 5′-capped primers into the active site of the central PB1 polymerase subunit (12–15). Recent studies by cryoelectron microscopy have revealed the entire transcription cycle (16). The RdRp physically binds to the host RNA polymerase II to facilitate the cap-snatching mechanism (17) and structural studies have provided key insights into the nature of this interaction (18).
Cap-snatching is an essential process in the influenza infection cycle, and was quickly recognized as a potential therapeutic target. This was particularly true of the endonuclease activity and the discovery that the active site within the N-terminal domain of the PA subunit (PAN) contains a two-metal center that can be targeted by chelating scaffolds (19–23). Structure-assisted drug discovery using the crystal structure of PAN has been pursued by several groups, including ours (24–30). The active metabolite (baloxavir) of the anti-influenza drug XOFLUZA or Baloxavir marboxil that was recently approved for therapeutic use (31) is a particularly potent endonuclease inhibitor (32–34).
There is little information on how the PAN endonuclease binds its RNA substrate, although structures are available for complexes with single nucleotides bound at the two-metal center (25,35,36). The large open cavity that surrounds the two-metal center together with conserved distal patches within the cavity (24) suggest that the RNA binding surface is quite extensive. Also, it has been shown that the RNA-binding locale imparts sequence specificity for the cleavage reaction (37). To investigate this, we have determined the crystal structure of a short RNA oligomer in complex with PAN. The structure reveals that the binding surface is indeed quite extensive and encompasses conserved pockets both 5′ and 3′ to the central nucleotide. We also determined the complex structures with short DNA oligomers that explain the observed activity with both RNA and DNA and reveal additional 5′ and 3′ binding pockets. The complex structures reveal that the single nucleotide complexes do not reflect the true interaction with the two-metal center, and that a dinucleotide should be considered as the core element of the substrate that engages the active site.
MATERIALS AND METHODS
Chemicals
DNA and RNA substrates were obtained from Integrated DNA Technologies, Inc. (USA). Crystallization solutions were obtained from Qiagen (USA). All other chemicals were of research grade, obtained from Sigma-Aldrich Corp. (USA).
Cloning, expression and purification of PAN endonuclease
The PAN endonuclease construct comprised residues 1–209 from the PA subunit of the pandemic H1N1 influenza virus A/California/04/2009 in which a flexible loop comprising residues 51–72 was replaced with a three-residue GGS linker. This choice of construct was based on previous work (24) and routinely produces high quality crystals suitable for analysis of protein-ligand complexes (29). The construct was cloned with an N-terminal His-tag into the pET28a+ expression vector. The vector was transformed into BL21(DE3) cells and the protein was expressed in LB medium overnight at 18°C after induction at an OD600 of 0.8 with 0.2 mM isopropyl-β-thiogalactopyranoside (IPTG). The protein was initially purified from cell lysates by HisTrap affinity chromatography and final purification was achieved by size-exclusion chromatography using a Superdex 75 column in 20 mM Tris pH 8.0, 150 mM NaCl and 1 mM TCEP. Although a thrombin cleavable site was included to remove the N-terminal His-tag, this step proved unnecessary because the protein crystallized well with the His-tag present. Three versions of the protein were used in these studies, wild type and the E119D and I38T point mutants, and all three were purified as described above.
Crystallization and X-ray structure analysis
To obtain the PAN–oligonucleotide complexes, we first screened a number of DNA oligonucleotides using the catalytically impaired active site PAN mutant E119D. The oligonucleotides varied between pentamers to octamers based on the length of the conserved active site cleft (24), and several contained a single GC pair based on the observed specificity of the endonuclease (37). The screening resulted in seven complexes at ∼2.0 Å resolution. Based on these structures, we then tested several RNA oligonucleotides and succeeded in obtaining one structure with a non-cleavable sulfate linkage between the GC pair to further increase the chances of obtaining an intact bound oligonucleotide. Finally, two DNA oligonucleotides yielded complex structures with wild type PAN. We obtained a total of 10 complex structures, and the four reported here represent those that are structurally unique with the highest resolution. The detailed procedures are as follows.
The purified proteins were concentrated to ∼10 mg/ml and crystallizations were performed using the hanging drop vapor diffusion method. Crystals appeared in 1–4 days and grew to their maximum size in a week. The crystallization conditions for the various complexes were as follows. (1) E119D PAN–AG*CAUC (RNA oligo where * represents a non-cleavable sulfate in place of the phosphate group) complex: the protein was incubated with 20 mM oligo for an hour prior to crystallization with 0.1 M HEPES pH 7.5, and 1.0 M ammonium sulfate. The cryo solution consisted of the crystallization solution supplemented with 5 mM MnCl2 and 33% glycerol. (2) E119D PAN–TTAGCATT (DNA oligo) complex: apo crystals grown in 0.1 M HEPES pH 7.5 and 1.0 M ammonium sulfate were soaked overnight in the same crystallization solution containing 5 mM oligo. The cryo solution consisted of the crystallization solution supplemented with 5 mM MnCl2 and 33% glycerol. (3) Wild type PAN–TTAGCATT (DNA oligo) complex: apo crystals were grown in 0.1 M HEPES pH 7.5, 1.0 M ammonium sulfate, 2% (w/v) PEG 400, 5 mM MnCl2 and 5 mM MgCl2, and soaked overnight in the same crystallization solution supplemented with 10 mM oligo. The cryo solution consisted of the crystallization solution supplemented with 5 mM oligo and 33% glycerol. (4) Wild type PAN–GTGAGCAGTG (DNA oligo) complex: the protein was incubated with 20 mM oligo for an hour prior to crystallization with 0.1 M HEPES pH 7.5, and 1.0 M ammonium sulfate. The cryo solution consisted of the crystallization solution supplemented with 2 mM MnCl2 and 33% glycerol. (5) Wild type, E119D and I38T PAN in complex with baloxavir acid: 0.1 M HEPES pH 7.8, 1.0 M ammonium sulfate, 10 mM MnCl2, 10 mM MgCl2, 0.5% PVP K15 and 1 mM baloxavir acid. The cryo solution consisted of 0.1 M HEPES pH 7.8, 1.3 M ammonium sulfate, 10 mM MnCl2, 10 mM MgCl2, 0.5% PVP K15, 1 mM baloxavir acid and 33% glycerol. All crystals were flash frozen in liquid nitrogen.
X-ray diffraction data were collected on beam lines 22-ID and 22-BM at the Southeast Regional Collaborative Access Team (SERCAT) at the Advanced Photon Source (APS), Argonne National Laboratory. Data sets were indexed, integrated, and scaled using HKL2000 (38). The structures were solved by molecular replacement using Phaser (39) and pdb coordinates 5CZN as the search model. Refinement was performed using iterative rounds of Phenix.refine (40) and rebuilding using COOT (41). The data collection, refinement statistics and Protein DataBank accession codes are provided in Table 1 (oligonucleotide complexes) and Table 2 (baloxavir complexes). All the figures in this article were generated and edited using PyMOL and GIMP, respectively.
Table 1.
Data collection and refinement statistics for the PAN–RNA and PAN–DNA crystal structures
| PAN complex | ||||
| PAN species | E119D | E119D | WT | WT |
| Oligonucleotide | RNA: AG*CAUCa | DNA: TTAGCATT | DNA: TTAGCATT | DNA: GTGAGCAGTG |
| Data collection b | ||||
| Beamline (SERCAT) | APS/22-ID | APS/22-ID | APS/22-ID | APS/22-ID |
| Wavelength (Å) | 1.000 | 1.000 | 1.000 | 1.000 |
| Space group | I422 | I422 | I422 | I422 |
| Unit cell (Å) | ||||
| a, b, c | 89.20, 89.20, 134.25 | 89.30, 89.30, 134.02 | 89.89, 89.89, 134.30 | 89.65, 89.65, 133.78 |
| Resolution range (Å) | 50.00–1.99 | 50.00–2.08 | 50.00–1.92 | 50.00–2.20 |
| R merge c | 0.047 (0.843) | 0.080 (0.485) | 0.049 (0.779) | 0.078 (0.790) |
| R meas | 0.071 (0.804) | 0.108 (0.594) | 0.061 (0.740) | 0.104 (0.969) |
| R pim | 0.024 (0.426) | 0.030 (0.219) | 0.023 (0.308) | 0.042 (0.486) |
| Unique reflections | 18 911 (1369) | 16 288 (1435) | 21 245 (2048) | 25 628 (2145) |
| Multiplicity | 4.9 (4.7) | 12.3 (5.9) | 7.3 (5.4) | 3.2 (1.9) |
| Mean I/σ(I) | 30.5 (2.0) | 25.0 (1.8) | 46.4 (2.0) | 20.8 (1.4) |
| CC1/2 | 0.999 (0.807) | 0.999 (0.865) | 0.999 (0.786) | 0.996 (0.684) |
| Completeness (%) | 99.9 (100.0) | 98.9 (89.6) | 99.4 (95.5) | 99.0 (92.1) |
| Wilson B-factor (Å−2) | 39.80 | 37.68 | 39.4 | 50.80 |
| Refinement | ||||
| R work (%) | 17.76 | 17.46 | 17.72 | 18.99 |
| R free (%) | 20.98 | 20.49 | 21.67 | 21.54 |
| R.m.s. deviations | ||||
| Bonds (Å) | 0.017 | 0.016 | 0.009 | 0.008 |
| Angles (°) | 1.43 | 1.93 | 0.87 | 0.80 |
| Coordinate error (Å)d | 0.25 | 0.21 | 0.23 | 0.28 |
| Protein residues | 177 | 178 | 179 | 179 |
| Average B factor (Å2) | ||||
| All atoms | 56.61 | 57.93 | 59.12 | 77.80 |
| Protein atoms | 55.51 | 57.29 | 57.84 | 77.20 |
| Ligand atoms | 93.64 | 86.79 | 106.94 | 117.80 |
| Solvent atoms | 59.56 | 62.27 | 64.67 | 77.50 |
| Ramachandran plot | ||||
| Favored (%) | 96.57 | 94.89 | 97.18 | 98.31 |
| Allowed (%) | 2.86 | 3.41 | 1.69 | 1.69 |
| Outliers (%) | 0.57 | 1.70 | 1.13 | 0.00 |
| Clash score | 6.0 | 4.3 | 5.4 | 2.7 |
| PDB accession code | 7KL3 | 6W7A | 6WHM | 6WS3 |
a‘*’ indicates a non-cleavable sulfate group in place of the phosphate group at this position.
bValues in parentheses refer to the highest resolution shell.
cRmerge = ∑|(I–<I>)|/ ∑(I), where I is the observed intensity.
dCalculated by maximum likelihood.
Table 2.
Data collection and refinement statistics for the PAN–baloxavir crystal structures
| PAN complex | |||
| PAN species | Wild type | I38T | E119D |
| Bound compound | Baloxavir (BLX) | Baloxavir (BLX) | Baloxavir (BLX) |
| Data collection a | |||
| Beamline (SERCAT) | APS/22-BM | APS/22-BM | APS/22-BM |
| Wavelength (Å) | 1.000 | 1.000 | 1.000 |
| Space group | I422 | I422 | I422 |
| Unit cell (Å) | |||
| a, b, c | 89.72, 89.72, 133.76 | 89.82, 89.82, 133.77 | 89.96, 89.96, 133.94 |
| Resolution range (Å) | 46.03–2.09 | 46.06–2.25 | 44.98–2.25 |
| R merge b | 0.069 (0.500) | 0.064 (0.556) | 0.048 (0.618) |
| R meas | 0.096 (0.619) | 0.104 (0.744) | 0.086 (0.845) |
| R pim | 0.031 (0.251) | 0.032 (0.332) | 0.027 (0.377) |
| Unique reflections | 15263 (762) | 12419 (671) | 12694 (768) |
| Multiplicity | 4.8 (2.7) | 9.3 (3.8) | 5.1 (2.4) |
| Mean I/σ(I) | 12.0 (2.1) | 12.0 (1.4) | 12.7 (1.4) |
| CC1/2 | 1.000 (0.835) | 1.000 (0.679) | 0.998 (0.692) |
| Completeness (%) | 92.5 (57.5) | 93.1 (56.9) | 94.9 (65.3) |
| Wilson B-factor (Å−2) | 38.80 | 46.00 | 47.9 |
| Refinement | |||
| R work (%) | 21.90 | 19.25 | 19.59 |
| R free (%) | 25.07 | 23.59 | 22.94 |
| R.m.s. deviations | |||
| Bonds (Å) | 0.008 | 0.003 | 0.006 |
| Angles (°) | 1.3 | 0.51 | 0.84 |
| Coordinate error (Å)c | 0.31 | 0.26 | 0.31 |
| Protein residues | 180 | 183 | 182 |
| Average B factor (Å2) | |||
| All atoms | 50.5 | 60.2 | 63.8 |
| Protein atoms | 50.6 | 59.2 | 62.8 |
| Ligand atoms | 51.4 | 76.6 | 77.5 |
| Solvent atoms | 45.0 | 61.0 | 61.1 |
| Ramachandran plot | |||
| Favored (%) | 96.61 | 98.32 | 96.63 |
| Allowed (%) | 3.39 | 1.12 | 2.81 |
| Outliers (%) | 0 | 0.56 | 0.56 |
| Clash score | 4.4 | 11.3 | |
| PDB accession code | 7K0W | 7KAF | 7KBC |
aValues in parentheses refer to the highest resolution shell.
bRmerge = ∑|(I–<I>)|/ ∑(I), where I is the observed intensity.
cCalculated by maximum likelihood.
Modeling studies
The models were constructed based on the PAN E119D/RNA complex reported here (pdb code: 7KL3) and the wild type PAN complex with baloxavir also reported here (pdb code: 7K0W). Wherever necessary, side chains were manipulated and adjusted using COOT (41). Preprocessing of the coordinates was performed in SCHRODINGER/MAESTRO. Hydrogen atoms and missing side chains were added using PRIME. Only water molecules within 5 Å of the complex in the crystal structures were included in the model. The modeled structures were energy minimized using OPLS_2005 Force Field in SCHRODINGER/MAESTRO, and all atoms were allowed to move to avoid clashes. Overall, the changes in the conformations of the protein and the RNA were minimal.
RESULTS
The structure of the PAN(E119D)–RNA complex
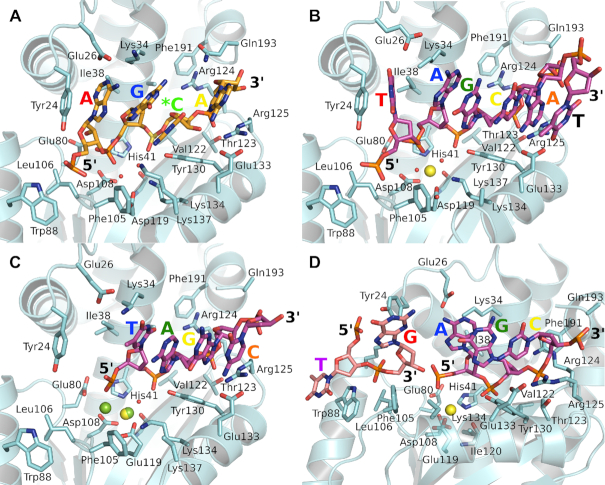
To increase the likelihood of crystallizing the complex and to attempt to understand the basis of the PAN specificity (37), we used the catalytically compromised point mutant E119D (35) and an RNA oligomer (5′-AGCAUC-3′) that contained a single preferred GC cleavage sequence and a sulfate connection between the GC pair in place of the phosphate. The complex structure was determined at 2.0 Å resolution (Table 1), although only four (AGCA) of the six nucleotides are visible in the structure (Figure 1A). Each of the four nucleotides (defined as the 5′ phosphate nucleotide) engages a distinct pocket that we refer to as P-1, P0, P1 and P2, where P0 contains the two-metal catalytic site and P-1 and P1/P2 are 5′ and 3′, respectively, to P0. The nucleotides in these four pockets are A (P-1), G (P0), C (P1) and A (P2). This register is not consistent with the observed GC specificity of the endonuclease (37) that would have G and C in the P-1 and PO pockets with the connecting phosphate group occupying the two-metal catalytic center. The interactions within each of the four pockets are as follows.
Figure 1.
RNA and DNA oligonucleotides bound to the PAN endonuclease. RNA oligomers have orange carbon atoms and DNA oligomers have magenta carbon atoms. (A) Active site mutant E119D bound to RNA oligomer AG*CA, where * indicates the location of the uncleavable sulfate linkage. (B) Mutant E119D bound to DNA oligomer TAGCAT. One metal is visible at the active site and was refined as a Mn2+ (yellow sphere). (C) Wild type PAN endonuclease bound to DNA oligomer TAGC. Two metal sites are visible at the active site; a Mg2+ (green sphere), and a shared Mg2+/Mn2+ (overlapping green/yellow spheres) with 70% and 30% occupancies, respectively. (D) Wild type PAN endonuclease bound to DNA oligomer TGAGC. In this structure, the oligomer is cleaved into a 5′ TG (light pink carbon atoms) and a 3′ AGC. There is a single Mn2+ (yellow sphere) at the active site. In each figure, the letter indicating the bound 5′ nucleotide is colored according to the pocket it occupies: P-2, magenta; P-1, red; P0 (5′ phosphate at the two-metal site), blue; P1, green; P2, yellow; and P3, orange. The orientation of each figure is the same apart from D to more clearly show the cleaved DNA. In each structure, the interacting amino acids are in very similar orientations except in D where Tyr24 is rotated as the guanine base leaves the P-1 pocket.
In the P-1 pocket, the base (A) stacks with Tyr24 and forms hydrogen bonding interactions with Glu26 that, in turn, forms a salt bridge with Lys34. The ribose group has no significant interaction and the 5′ phosphate interacts with main chain amide group of Leu106 and potentially with the OH group of Tyr24. In the P0 pocket, the base (G) is flanked on one side by the P-1 base and acyl chain of Lys34, and by the P1 base on the other side. The ribose group forms a van der Waals interaction with Ile38 and the 5′ phosphate engages in hydrogen bonding interactions with several water molecules. There are no metal atoms visible in the electron density map, although one or two of the water molecules may actually be low occupancy metal ions. In the P1 pocket, the base (C) stacks on the P0 base, forms van der Waal interactions with the P2 base and Tyr130, and a hydrogen-bonding interaction with Lys137. The ribose forms van der Waal interactions with Tyr130 and the 2′ OH forms a hydrogen bond with the guanidinium group of Arg125. The 5′ phosphate interacts with Tyr130, the main chain amide of Val122 and two water molecules. Finally, in the P2 pocket, the base (A) forms van der Waal interactions with Arg125 that in turn forms a salt bridge with Glu133. The ribose also interacts with Arg125 and the 2′ OH appears to interact with the main chain carbonyl oxygen of Gln193. The 5′ phosphate is tightly sequestered between and interacts with Arg124, Arg125, Thr123 and the main chain amide of Arg124. The orientation of Arg124 is fixed by Phe191, specifically by a stacking interaction and a hydrogen bond with the main chain carbonyl oxygen.
The structure of the PAN(E119D)–DNA complex
We and others have previously demonstrated that PAN can also use DNA oligonucleotides as substrates (10,24). To investigate how PAN binds DNA, we determined the complex structure at 2.1 Å resolution (Table 1) using the oligonucleotide 5′-TTAGCATT-3′ and the E119D mutant (Figure 1B). The complex is similar to the RNA complex, but six nucleotides (5′-TAGCAT-3′) are visible instead of four and the GC pair now occupies the P1 and P2 pockets rather than the P0 and P1 pockets. A key difference is the presence of a catalytic Mn2+ ion at the active site coordinated to His41, Asp108, Asp119 (normally Glu119), the main chain carbonyl oxygen of Ile120 and two water molecules. The two water molecules replace the P0 phosphate oxygen atom that normally coordinates the Mn2+ ion, and the phosphate group and the P-1 ribose group are ∼1.5 Å further away from the catalytic center than in the RNA structure. Apart from this difference, the sugar-phosphate backbones of the RNA and DNA complexes overlap well (Supplementary Figure S1A).
The residues that create the four pockets are largely unperturbed when compared to the RNA complex, but the orientations of the individual bases within the pockets are quite different (Supplementary Figure S1B), which suggests that the bases make minimal contributions to the substrate binding. However, one key difference is in the P-1 pocket where the movement of the P0 phosphate and the P-1 ribose appears to reorient the base (T) that, in turn, leads to a shift in the adjacent loop that contains Tyr24, Gly25 and Glu26. The five nucleotides 3′ to the P-1 site (AGCAT) create a stacked structure that is not present in the RNA structure. The base stacking appears to be initiated by a ‘book end’ interaction between the P0 base (A) and Lys34. The 3′ A interacts closely with the Arg125/Glu133 salt bridge and we therefore designate this as the P3 pocket.
The structure of the PAN(WT)–DNA complex
Assuming that DNA is a poorer substrate for PAN than RNA, we attempted to obtain the complex structure of wild type PAN with an intact DNA oligonucleotide. We obtained two structures, but the DNA is cleaved in both. However, one structure only contains the bound 3′ product (we call this structure ‘cleaved’) whereas the second structure contains both the 5′ and the 3′ products (‘product’). The cleaved structure is at 1.9 Å resolution using the sequence 5′-TTAGCATT-3′ (Table 1), and the product structure is at 2.2 Å using the sequence 5′-GTGAGCAGTG-3′ (Table 1). The catalytic GC dinucleotide is once again not at the two-metal center in either complex. The sugar-phosphate backbones of the bound DNA generally align well with the bound RNA and DNA in the E119D complexes (Supplementary Figure S1A), although the orientation of the bases show significant variations (Supplementary Figure S1C and Supplementary Figure S1D).
In the ‘cleaved’ complex (Figure 1C), only four nucleotides are visible (TAGC) and occupy the P0, P1, P2 and P3 pockets, respectively. Consistent with the catalytic mechanism, the P0 phosphate is present and adjacent to the two-metal site that contains the two metal ions in this wild type Glu119 structure. The refinement is consistent with one site being occupied by a mixture of Mn2+ and Mg2+ ions. Although there is no nucleotide in the P-1 pocket and no base to stack onto Tyr24, this residue and its associated loop region maintain the same conformation observed in the E119D intact DNA complex structure. In the ‘product’ structure (Figure 1D), five nucleotides are visible, TG on the 5′ side of the cut and AGC on the 3′ side. The latter occupy the P0, P1 and P2 pockets and superimpose well on the TAG nucleotides present in the cleaved structure. In contrast, the 5′ TG nucleotides are in new locations; the G is flipped out of the P-1 pocket and the terminal T base stacks onto the exposed Trp88 that we designate as the P-2 pocket. The 5′ phosphate of the flipped out G remains in its P-1 pocket but the sidechain of Tyr24 has rotated by ∼40°. Again, consistent with the catalytic mechanism, the P0 phosphate is present and the P-1 nucleotide has a free 3′ OH, although there is only one metal ion (refined as Mn2+) at the two-metal site.
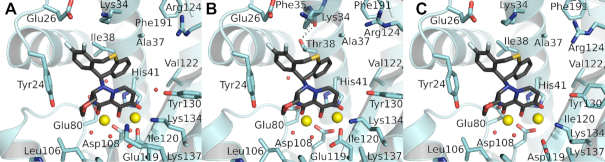
The structures of PAN-–baloxavir complexes
We recently demonstrated that the point mutant I38T consistently appears in PAN mutants that are resistant to compound RO-7, an analog of the approved drug baloxavir (42). Consistent with this finding, the same drug resistance mutation has now emerged upon baloxavir pressure (42,43). To investigate this, we determined the complex structures of baloxavir bound to wild type PAN (Figure 2A) and PAN-I38T (Figure 2B). These structures have previously been reported (43) but we wanted to confirm that our PAN construct does not introduce any significant differences. The two structures are very similar, at 2.09 and 2.25 Å, respectively (Table 2), and contain two tightly bound active site Mn2+ ions coordinated to the compound. Unsurprisingly, the structures are very similar to those of the RO-7 complex (42) and to the previously determined baloxavir complexes. Ile38 forms a tight and extensive apolar interaction with baloxavir and its mutation to the polar threonine would significantly impact binding affinity as observed (43). Reduced susceptibility to baloxavir has also been reported for the mutant E119D (43), although not to the extent of I38T, and we therefore determined the structure of baloxavir bound to PAN-E119D (Figure 2C) at 2.25 Å (Table 2). The complex is very similar to that of the wild type PA although the shorter aspartic acid reduces the interaction with the oxazino-pyridotriazin-dione head-group, which may explain the reduced susceptibility.
Figure 2.
Baloxavir bound to the PAN endonuclease. (A) Wild type PAN. (B) I38T PAN. (C) E119D PAN. In each figure, baloxavir is represented as sticks with black carbon atoms, the PAN cartoon and key residues are colored cyan, the active site Mn2+ ions are large yellow spheres and the water molecules are small red spheres. Note in B that the side chain of the Thr38 point mutant is oriented such that the hydroxyl group can make hydrogen bond interactions with the main chain carbonyl oxygens of Lys34 and Phe35 (dotted black lines) while the methyl group optimizes its apolar interaction with baloxavir.
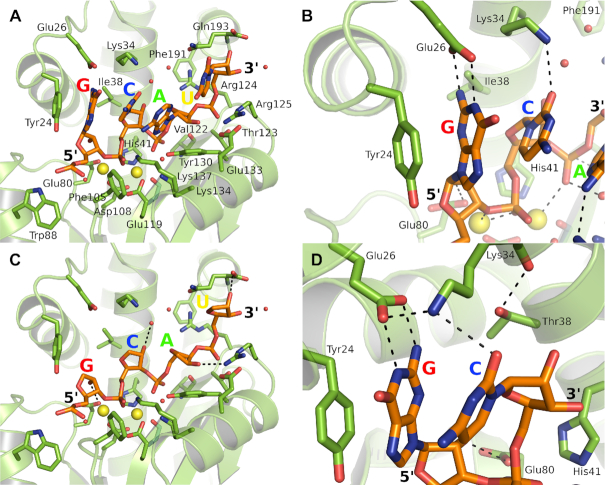
The model structure of the cognate PAN–RNA complex
Despite using specifically designed oligonucleotides, we have not been able to determine the wild type structure of the bound substrate with the preferred GC pair at the P-1 and P0 positions that would provide insights into the structural basis for the specificity. Each of our structures is variably distorted by the use the E119D active site mutant, the DNA substrate and the lack of two metal ions at their known locations in the active site. We therefore modeled the wild type complex and performed energy minimization. The model was created in four steps. First, two Mn2+ ions and conserved water molecules were placed at the active site locale according to the high resolution wild type baloxavir complex structure described above. Second, the RNA oligonucleotide GCAU was placed according to the PAN(E119D)-RNA complex structure (Figure 1A) but with the G and C placed at the P-1 and P0 positions respectively. Third, the P0 phosphate was placed at the active site and coordinating two Mn2+ ions according to the two-metal catalytic mechanism (44). Finally, the wild type Glu119 was restored at the active site.
In the energy minimized model (Figure 3A), the GC pair in the P-1 and P0 positions do indeed make ideal interactions with Tyr24, Glu26, Lys34 and the active site metal ions (Figure 3B). Of particular note are the hydrogen bonding interactions between the guanine base and Glu26, the salt bridge between Glu26 and Lys34, and the accommodation of the single ring cytosine base in a tight pocket bounded by the 5′ and 3′ bases and the sidechain of Lys34. The model suggests that the GC pair serves to accurately place the bridging phosphate at the two-metal active site. Thus, the larger purine moiety in the P-1 pocket is necessary to simultaneously maintain the optimal interactions with Tyr24 and Glu26 and the correct placement of the phosphate adjacent to the two catalytic Mn2+ ions. The role of the preferred cytosine in the P0 pocket is less clear, but a larger purine base would appear to be sterically disfavored and a uracil may have less ideal interactions with the surrounding residues. The interactions of the AU nucleotides are very similar to those of the actual RNA crystal structure described above. The model suggests that the 2′ OH groups are important in the RNA complex and provide additional binding affinity and specificity compared to DNA (Figure 3C). Thus, the P-1 2′ OH interacts with Glu80, the P0 2′ OH potentially with Arg124 and/or the main chain carbonyl oxygen of Lys34 via water molecules, the P1 2′ OH with Arg125 and the P2 2′ OH potentially with the main chain carbonyl oxygen of Gln193.
Figure 3.
Energy minimized models of RNA oligonucleotide GCAU bound to PAN endonuclease. (A) Overview of the wild type model in the same orientation as Figure 1. (B) Close up of the GC dinucleotide at the two-metal active site center of the wild type model showing the key hydrogen bonding interactions. (C) The interactions of the 2′ OH groups of the oligonucleotide sugar-phosphate backbone in the wild type model. (D) Close up of the Thr38 interactions in the I38T mutant model. To distinguish these as models rather than actual crystal structures, the RNA oligomer has orange carbon atoms and the protein and side chains are in light green. The metals are modeled as Mn2+ (yellow) and water molecules are shown as red spheres.
The model structure of the PAN(I38T)–RNA complex
In the RNA and DNA complexes, Ile38 interacts with the P0 ribose group and the I38T mutation should therefore also compromise binding. Indeed, it has been observed that PAN-I38T is significantly compromised in terms of endonuclease activity and that viruses bearing this mutation display impaired replicative capacity (43). To support this, we have failed to obtain RNA or DNA complex structures with PAN-I38T. While a number of mutations have been identified that have impacted the susceptibility of influenza viruses to baloxavir-like inhibitors, changes at position 38 are the most commonly observed clinically and the I38T mutation, in particular, has been described as the marker of baloxavir resistance (45). One would therefore predict that it retains sufficient fitness in terms of RNA binding to support the cap-snatching process. To investigate this, we repeated the modeling using the mutant protein. The model suggests that the threonine side chain is oriented to maximize its interaction with the P0 ribose (Figure 3D). The side chain hydroxyl group forms a hydrogen bond interaction with the main chain carbonyl oxygen of Lys34 but may also form a hydrogen bond with the ribose 2′ OH. The N2 nitrogen of the P-1 guanine is also within 5Å of the side chain hydroxyl of Thr38. Water molecules are not present in the model structure and may contribute to these potential hydrogen bonding interactions.
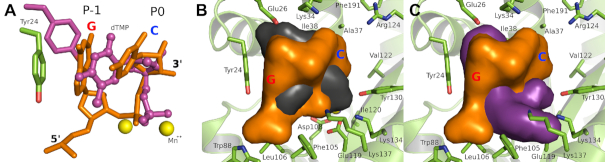
Comparison with single nucleotide PAN complexes
We and others have previously determined the structures of single nucleotides bound to the PAN endonuclease (25,35,36), which have provided key information for our drug discovery studies (29). However, comparison of the RNA and DNA complexes presented here with these single nucleotide complex structures reveals that the latter are artifactual and do not accurately reflect the mode of binding at the P0 pocket. Although the single nucleotide complexes overlap well between themselves, they actually represent a compromise between the oligonucleotide binding at the P-1 and P0 pockets (Figure 4A). Thus, while the phosphate and ribose correctly occupy the P0 pocket, the base reaches over to occupy the P-1 pocket location adjacent to Tyr24. The key roles of the P-1 and P0 pockets is consistent with the GC specificity of the endonuclease (37) residing in these pockets.
Figure 4.
Implications for PAN endonuclease inhibitor development. (A) Overlap of the bound P-1/P0 RNA dinucleotide with the previously determined bound mononucleotide (pdb file 5CCY). Note that the mononucleotide actually spans the P-1 and P0 sites. The dinucleotide is colored orange and the mononucleotide is colored magenta and shown in ball-and-sticks. The Mn2+ ions at the active site are shown as yellow spheres. Note that Tyr24 in the mononucleotide complex (magenta sticks) moves to maintain the stacking interaction with the base. (B) Spatial comparison of baloxavir and the RNA dinucleotide bound at the P-1 and P0 sites. (C) Spatial comparison of L-742,001 and the RNA dinucleotide bound at the P-1 and P0 sites. In (B) and (C), the RNA dinucleotide is shown as an orange van der Waal surface, the PAN cartoon and key residues are colored lime green, and baloxavir and L-742,001 are shown as black and magenta van der Waal surfaces, respectively. In all panels, the RNA dinucleotide and the PAN endonuclease are derived from the wild type RNA-bound model shown in Figure 3B with GC occupying the P-1 and P0 pockets.
In terms of drug discovery, the dinucleotide that occupies the P-1 and P0 sites is the structure that should be spatially mimicked to reduce the potential for resistance (Figure 3B). Comparison of baloxavir (Figure 4B) with the bound dinucleotide reveals that there is significant spatial overlap but there are opportunities for the emergence of resistance as observed. We have also previously demonstrated that F105S consistently appears in PAN mutants that are resistant to compound L-742,001 and predicted that this mutation would have minimal impact on substrate binding (35). The RNA complex confirms this (Figure 1A) and shows that Phe105 is relatively distant from the bound substrate and free to mutate without affecting activity. Comparison of L-742,001 with the bound dinucleotide reveals poor spatial overlap (Figure 4C).
DISCUSSION
Our four complex structures comprising uncleaved RNA and DNA oligonucleotides bound to the catalytically compromised E119D mutant, and cleaved DNA oligonucleotides bound to the wild type enzyme reveal six binding pockets that we refer to as P-2, P-1, P0, P1, P2 and P3. The central three pockets, in which P0 contains the two-metal center that engages the catalytically processed phosphate group, are consistent in the four structures, and the peripheral P-2, P2 and P3 sites show some variability and associated weaker electron density. Although the catalytically preferred GC dinucleotide (37) is not bound appropriately in the P-1 and P0 sites, other features of the complexes support the validity of these structures. Thus, the amino acids that create the six nucleotide binding pockets are conserved, the correct products with the phosphate group in the P0 site and the free 3′ OH in the P-1 site are present in the cleaved DNA complexes, and the 5′-3′ orientation of the bound oligonucleotides is consistent with the overall cap-snatching mechanism and the position of the PB2 cap-binding domain in the RdRp complex (12,13).
Although the endonuclease is active with both RNA and DNA substrates, comparison of the two complexes reveals that the 2′ OH in the RNA complex is involved in additional interactions compared to the DNA complex. Although the RNA and the DNA occupy the same binding pockets, there are small but significant differences between their backbone conformations (Supplementary Figure S1A) that suggest that these 2′ OH interactions contribute to the expected preference for the RNA substrate. This is supported by the modeling studies where the 2′ OH interactions are maintained. As regards the preferred GC specificity of the endonuclease (37), the structure of our RNA complex fails to explain this because the GC pair does not occupy the P-1 and P0 pockets. However, the modeling study suggests that the GC pair serves to simultaneously place the bridging phosphate at the two-metal active site for optimal catalysis and maintain the base interactions with Tyr24, Glu26 and Lys34 on the opposite side of the substrate binding groove. In the PAN(E119D)–DNA complex, where a smaller pyrimidine (T) occupies the P-1 pocket, the P0 phosphate is forced to move away from the two-metal site by ∼1.5 Å in order to maintain the interactions with Tyr24 and Glu26. The model also suggests that G is preferred over A because of more favorable hydrogen bonding interactions with Glu26.
There are a number of potential explanations for why the GC dinucleotide is not bound at the P-1 and P0 sites in our structures. First and foremost, it has been established that the GC specificity derives not only from the endonuclease itself but also from the spacing of the GC pair from the 5′ cap and its interaction with the cap-binding domain in the PB2 subunit in the RdRp complex (37). We provide compelling evidence that there is GC specificity built into the endonuclease active site locale, in particular for G at the P-1 site, but this is apparently insufficient in the absence of the cap binding domain. Indeed, previous studies have strongly suggested that the spacing is the dominant specificity factor (37). Second, although GC specificity is clear, it is not absolute and the endonuclease can cleave other sequences albeit less efficiently (37). Our short oligonucleotides, the positions of the GC pair within the oligonucleotides and other features of the sequence may prevent the preferred binding in the isolated endonuclease within the crystal. Third, although the model structure supports the preferred binding of guanine at the P-1 site, the bases at other positions have no obvious specific interactions with the binding pockets and have variable conformations (Supplementary Figure S1). This is consistent with the requirement that the cap-snatching mechanism operates on multiple host mRNAs. Finally, the use of the inactive E119D mutant to obtain the uncleaved RNA complex and the presence of the non-cleavable sulfate group between the G and the C are likely to impact the binding of the GC pair to the two-metal active site. Unfortunately, we currently do not have a complex structure with an unmodified RNA oligonucleotide.
The successful deployment of baloxavir as an effective influenza therapeutic (31) has confirmed the early predictions that the essential cap-snatching mechanism of the influenza RdRp and the endonuclease component within the PAN subunit would be excellent targets for drug discovery (19–21). However, we have previously shown that resistance can rapidly arise for inhibitors that target the two-metal active site of the PAN endonuclease (35,42) and baloxavir resistance has already emerged (43). An important goal of the structural studies presented here is to facilitate the development of novel PAN endonuclease inhibitors less prone to resistance. It has been established that drug resistance can be minimized when the drug occupies the same space within the active site as the actual substrate because resistance mutations are likely to also have a significant negative impact on activity (46). Our finding that the P-1/P0 dinucleotide represents the ‘core’ feature of the bound substrate will now facilitate future drug discovery efforts that minimize resistance potential. The stacking interaction of the P-1 base with Tyr24 and the P0 interaction of the ribose with Ile38 are important stabilizing features in the substrate complex. These interactions are maintained in the artifactual single nucleotide complexes and would be predicted to be key features of an effective drug. Indeed, Ile38 features prominently in the baloxavir complex but in such a manner that allows resistance mutations. The importance of the Tyr24 interaction is confirmed in the ‘product’ structure (Figure 1D) where the side chain has rotated out of position to presumably allow the release of the 5′ oligonucleotide. The Influenza Research Database (47) lists 32820 PAN sequences from H1N1 and H3N2 human viruses, and of these, 32798 contain tyrosine at position 24, 18 have histidine, 2 have cysteine, and 2 have phenylalanine. Also, it has been shown (48) that 10–25% of swine influenza viruses contain the Y24H variant. Apart from cysteine, the aromatic side chains of these residues are consistent with the importance of the stacking interaction with the P-1 base.
DATA AVAILABILITY
Crystal structures (PDB database): 7KL3, 6W7A, 6WHM, 6WS3, 7K0W 7KAF, 7KBC. Modelled structures (modelarchive.org): ma-a4act, ma-nf0st.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Darcie Miller for crystallographic advice and assistance with data collection, and Jeremy Jones for helpful discussions. The diffraction data were collected at Southeast Regional Collaborative Access Team (SERCAT) beam lines 22-ID and 22-BM at the Advanced Photon Source, Argonne National Laboratory, and we thank SERCAT staff for their assistance. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Notes
Present address: Gyanendra Kumar, Research & Development Department, BioMarin Pharmaceutical Inc., 105 Digital Drive, Novato, CA 94949, USA.
Contributor Information
Gyanendra Kumar, Department of Structural Biology, Memphis, TN 38105, USA.
Maxime Cuypers, Department of Structural Biology, Memphis, TN 38105, USA.
Richard R Webby, Department of Infectious Diseases, St. Jude Children's Research Hospital, Memphis, TN 38105, USA.
Thomas R Webb, Wildflower Biopharma, Inc., 8650 Genesee Ave. STE 214, San Diego, CA 92122, USA; Department of Chemistry and Biochemistry, University of California, Santa Cruz, Santa Cruz, CA 95064, USA.
Stephen W White, Department of Structural Biology, Memphis, TN 38105, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH [AI098757 to T.R.W., in part]; Comprehensive Cancer Center [CA021765]; American Lebanese Syrian Associated Charities (ALSAC); U.S. Department of Energy [W‐31‐109‐Eng‐38]; supporting SERCAT institutions may be found at www.ser-cat.org/members.html. Funding for open access charge: St. Jude Children's Research Hospital, Department of Structural Biology.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ortin J., Martin-Benito J.. The RNA synthesis machinery of negative-stranded RNA viruses. Virology. 2015; 479–480:532–544. [DOI] [PubMed] [Google Scholar]
- 2. Bouloy M., Plotch S.J., Krug R.M.. Globin mRNAs are primers for the transcription of influenza viral RNA in vitro. Proc. Natl. Acad. Sci. U.S.A. 1978; 75:4886–4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Krug R.M., Bouloy M., Plotch S.J.. RNA primers and the role of host nuclear RNA polymerase II in influenza viral RNA transcription. Philos. Trans. R. Soc. Lond. B Biol Sci. 1980; 288:359–370. [DOI] [PubMed] [Google Scholar]
- 4. Plotch S.J., Bouloy M., Ulmanen I., Krug R.M.. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981; 23:847–858. [DOI] [PubMed] [Google Scholar]
- 5. Te Velthuis A.J., Fodor E. Influenza virus RNA polymerase: insights into the mechanisms of viral RNA synthesis. Nat. Rev. Microbiol. 2016; 14:479–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wandzik J.M., Kouba T., Cusack S.. Structure and function of influenza polymerase. Cold Spring Harb. Perspect. Med. 2020; 10:a038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fechter P., Mingay L., Sharps J., Chambers A., Fodor E., Brownlee G.G.. Two aromatic residues in the PB2 subunit of influenza A RNA polymerase are crucial for cap binding. J. Biol. Chem. 2003; 278:20381–20388. [DOI] [PubMed] [Google Scholar]
- 8. Hara K., Schmidt F.I., Crow M., Brownlee G.G.. Amino acid residues in the N-terminal region of the PA subunit of influenza A virus RNA polymerase play a critical role in protein stability, endonuclease activity, cap binding, and virion RNA promoter binding. J. Viro.l. 2006; 80:7789–7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guilligay D., Tarendeau F., Resa-Infante P., Coloma R., Crepin T., Sehr P., Lewis J., Ruigrok R.W., Ortin J., Hart D.J.et al.. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat. Struct. Mol. Biol. 2008; 15:500–506. [DOI] [PubMed] [Google Scholar]
- 10. Dias A., Bouvier D., Crepin T., McCarthy A.A., Hart D.J., Baudin F., Cusack S., Ruigrok R.W.. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature. 2009; 458:914–918. [DOI] [PubMed] [Google Scholar]
- 11. Yuan P., Bartlam M., Lou Z., Chen S., Zhou J., He X., Lv Z., Ge R., Li X., Deng T.et al.. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature. 2009; 458:909–913. [DOI] [PubMed] [Google Scholar]
- 12. Pflug A., Guilligay D., Reich S., Cusack S.. Structure of influenza A polymerase bound to the viral RNA promoter. Nature. 2014; 516:355–360. [DOI] [PubMed] [Google Scholar]
- 13. Reich S., Guilligay D., Pflug A., Malet H., Berger I., Crepin T., Hart D., Lunardi T., Nanao M., Ruigrok R.W.et al.. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature. 2014; 516:361–366. [DOI] [PubMed] [Google Scholar]
- 14. Pflug A., Gaudon S., Resa-Infante P., Lethier M., Reich S., Schulze W.M., Cusack S.. Capped RNA primer binding to influenza polymerase and implications for the mechanism of cap-binding inhibitors. Nucleic Acids Res. 2018; 46:956–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kouba T., Drncova P., Cusack S.. Structural snapshots of actively transcribing influenza polymerase. Nat. Struct. Mol. Biol. 2019; 26:460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wandzik J.M., Kouba T., Karuppasamy M., Pflug A., Drncova P., Provaznik J., Azevedo N., Cusack S.. A structure-based model for the complete transcription cycle of influenza polymerase. Cell. 2020; 181:877–893. [DOI] [PubMed] [Google Scholar]
- 17. Engelhardt O.G., Smith M., Fodor E.. Association of the influenza A virus RNA-dependent RNA polymerase with cellular RNA polymerase II. J. Virol. 2005; 79:5812–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lukarska M., Fournier G., Pflug A., Resa-Infante P., Reich S., Naffakh N., Cusack S.. Structural basis of an essential interaction between influenza polymerase and Pol II CTD. Nature. 2017; 541:117–121. [DOI] [PubMed] [Google Scholar]
- 19. Tomassini J., Selnick H., Davies M.E., Armstrong M.E., Baldwin J., Bourgeois M., Hastings J., Hazuda D., Lewis J., McClements W.et al.. Inhibition of cap (m7GpppXm)-dependent endonuclease of influenza virus by 4-substituted 2,4-dioxobutanoic acid compounds. Antimicrob. Agents Chemother. 1994; 38:2827–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tomassini J.E., Davies M.E., Hastings J.C., Lingham R., Mojena M., Raghoobar S.L., Singh S.B., Tkacz J.S., Goetz M.A.. A novel antiviral agent which inhibits the endonuclease of influenza viruses. Antimicrob. Agents Chemother. 1996; 40:1189–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hastings J.C., Selnick H., Wolanski B., Tomassini J.E.. Anti-influenza virus activities of 4-substituted 2,4-dioxobutanoic acid inhibitors. Antimicrob. Agents Chemother. 1996; 40:1304–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Doan L., Handa B., Roberts N.A., Klumpp K.. Metal ion catalysis of RNA cleavage by the influenza virus endonuclease. Biochemistry. 1999; 38:5612–5619. [DOI] [PubMed] [Google Scholar]
- 23. Parkes K.E., Ermert P., Fassler J., Ives J., Martin J.A., Merrett J.H., Obrecht D., Williams G., Klumpp K.. Use of a pharmacophore model to discover a new class of influenza endonuclease inhibitors. J. Med. Chem. 2003; 46:1153–1164. [DOI] [PubMed] [Google Scholar]
- 24. DuBois R.M., Slavish P.J., Baughman B.M., Yun M.K., Bao J., Webby R.J., Webb T.R., White S.W.. Structural and biochemical basis for development of influenza virus inhibitors targeting the PA endonuclease. PLoS Pathog. 2012; 8:e1002830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kowalinski E., Zubieta C., Wolkerstorfer A., Szolar O.H., Ruigrok R.W., Cusack S.. Structural analysis of specific metal chelating inhibitor binding to the endonuclease domain of influenza pH1N1 (2009) polymerase. PLoS Pathog. 2012; 8:e1002831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sagong H.Y., Bauman J.D., Patel D., Das K., Arnold E., LaVoie E.J.. Phenyl substituted 4-hydroxypyridazin-3(2H)-ones and 5-hydroxypyrimidin-4(3H)-ones: inhibitors of influenza A endonuclease. J. Med. Chem. 2014; 57:8086–8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carcelli M., Rogolino D., Gatti A., De Luca L., Sechi M., Kumar G., White S.W., Stevaert A., Naesens L.. N-acylhydrazone inhibitors of influenza virus PA endonuclease with versatile metal binding modes. Sci. Rep. 2016; 6:31500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fudo S., Yamamoto N., Nukaga M., Odagiri T., Tashiro M., Hoshino T.. Two distinctive binding modes of endonuclease inhibitors to the N-Terminal region of influenza virus polymerase acidic subunit. Biochemistry. 2016; 55:2646–2660. [DOI] [PubMed] [Google Scholar]
- 29. Beylkin D., Kumar G., Zhou W., Park J., Jeevan T., Lagisetti C., Harfoot R., Webby R.J., White S.W., Webb T.R.. Protein-structure assisted optimization of 4,5-Dihydroxypyrimidine-6-Carboxamide inhibitors of influenza virus endonuclease. Sci. Rep. 2017; 7:17139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Credille C.V., Dick B.L., Morrison C.N., Stokes R.W., Adamek R.N., Wu N.C., Wilson I.A., Cohen S.M.. Structure-activity relationships in metal-binding pharmacophores for influenza endonuclease. J. Med. Chem. 2018; 61:10206–10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. O’Hanlon R., Shaw M.L.. Baloxavir marboxil: the new influenza drug on the market. Curr. Opin. Virol. 2019; 35:14–18. [DOI] [PubMed] [Google Scholar]
- 32. Jones J.C., Marathe B.M., Lerner C., Kreis L., Gasser R., Pascua P.N., Najera I., Govorkova E.A.. A novel endonuclease inhibitor exhibits broad-spectrum anti-influenza virus activity in vitro. Antimicrob. Agents. Chemother. 2016; 60:5504–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jones J.C., Marathe B.M., Vogel P., Gasser R., Najera I., Govorkova E.A.. The PA endonuclease inhibitor RO-7 protects mice from lethal challenge with influenza A or B viruses. Antimicrob. Agents Chemother. 2017; 61:e02460-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Noshi T., Kitano M., Taniguchi K., Yamamoto A., Omoto S., Baba K., Hashimoto T., Ishida K., Kushima Y., Hattori K.et al.. In vitro characterization of baloxavir acid, a first-in-class cap-dependent endonuclease inhibitor of the influenza virus polymerase PA subunit. Antiviral. Res. 2018; 160:109–117. [DOI] [PubMed] [Google Scholar]
- 35. Song M.S., Kumar G., Shadrick W.R., Zhou W., Jeevan T., Li Z., Slavish P.J., Fabrizio T.P., Yoon S.W., Webb T.R.et al.. Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:3669–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhao C., Lou Z., Guo Y., Ma M., Chen Y., Liang S., Zhang L., Chen S., Li X., Liu Y.et al.. Nucleoside monophosphate complex structures of the endonuclease domain from the influenza virus polymerase PA subunit reveal the substrate binding site inside the catalytic center. J. Virol. 2009; 83:9024–9030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Datta K., Wolkerstorfer A., Szolar O.H., Cusack S., Klumpp K.. Characterization of PA-N terminal domain of Influenza A polymerase reveals sequence specific RNA cleavage. Nucleic Acids Res. 2013; 41:8289–8299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 39. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W.et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jones J.C., Kumar G., Barman S., Najera I., White S.W., Webby R.J., Govorkova E.A.. Identification of the I38T PA substitution as a resistance marker for next-generation influenza virus endonuclease inhibitors. MBio. 2018; 9:e00430-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Omoto S., Speranzini V., Hashimoto T., Noshi T., Yamaguchi H., Kawai M., Kawaguchi K., Uehara T., Shishido T., Naito A.et al.. Characterization of influenza virus variants induced by treatment with the endonuclease inhibitor baloxavir marboxil. Sci. Rep. 2018; 8:9633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang W., Lee J.Y., Nowotny M.. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell. 2006; 22:5–13. [DOI] [PubMed] [Google Scholar]
- 45. Gubareva L.V., Fry A.M.. Baloxavir and treatment-emergent resistance: public health insights and next steps. J. Infect. Dis. 2020; 221:337–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Romano K.P., Ali A., Royer W.E., Schiffer C.A.. Drug resistance against HCV NS3/4A inhibitors is defined by the balance of substrate recognition versus inhibitor binding. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:20986–20991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang Y., Aevermann B.D., Anderson T.K., Burke D.F., Dauphin G., Gu Z., He S., Kumar S., Larsen C.N., Lee A.J.et al.. Influenza Research Database: An integrated bioinformatics resource for influenza virus research. Nucleic Acids Res. 2017; 45:D466–D474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stevaert A., Dallocchio R., Dessi A., Pala N., Rogolino D., Sechi M., Naesens L.. Mutational analysis of the binding pockets of the diketo acid inhibitor L-742,001 in the influenza virus PA endonuclease. J. Virol. 2013; 87:10524–10538. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Crystal structures (PDB database): 7KL3, 6W7A, 6WHM, 6WS3, 7K0W 7KAF, 7KBC. Modelled structures (modelarchive.org): ma-a4act, ma-nf0st.