Abstract
Mammalian circRNAs can influence different cellular processes by interacting with proteins and other nucleic acids. Here, we used ribonucleoprotein immunoprecipitation (RIP) analysis to identify systematically the circRNAs associated with the cancer-related protein AUF1. Among the circRNAs interacting with AUF1 in HeLa (human cervical carcinoma) cells, we focused on hsa_circ_0032434 (circPCNX), an abundant target of AUF1. Overexpression of circPCNX specifically interfered with the binding of AUF1 to p21 (CDKN1A) mRNA, thereby promoting p21 mRNA stability and elevating the production of p21, a major inhibitor of cell proliferation. Conversely, silencing circPCNX increased AUF1 binding to p21 mRNA, reducing p21 production and promoting cell division. Importantly, eliminating the AUF1-binding region of circPCNX abrogated the rise in p21 levels and rescued proliferation. Therefore, we propose that the interaction of circPCNX with AUF1 selectively prevents AUF1 binding to p21 mRNA, leading to enhanced p21 mRNA stability and p21 protein production, thereby suppressing cell growth.
INTRODUCTION
AU-rich element RNA-binding factor 1 (AUF1), also known as heterogeneous nuclear ribonucleoprotein D (HNRNPD), is an RNA-binding protein (RBP) consisting of four different isoforms (p37, p40, p42 and p45). AUF1 was among the first AU-rich element RNA-binding proteins to be purified, cloned and studied for its complex regulation of mRNA targets (1–4). Although AUF1 is predominantly known to promote decay of mRNAs encoding cell cycle-regulatory proteins, inflammatory cytokines, oncoproteins and apoptosis-related proteins [CCND1 (cyclin D1), CDKN1A (p21), CDKN1B (p27), CDKN2A (p16), RB, TNF (TNFα), IL1B (interleukin-1β), IL6, IL8, FOS, JUND, MYC, BCL2, BAX], AUF1 was also found to stabilize some mRNA targets, including PTH and VHL mRNAs (3), and to promote the translation of MYC and Mef2c mRNAs (5,6). Besides binding RNAs to form ribonucleoprotein (RNP) complexes, AUF1 can also bind to DNA and affect gene transcription, as shown for TERT (7,8). Through these pleiotropic functions, AUF1 has been implicated in controlling the gene expression programs that drive processes such as proliferation, senescence, carcinogenesis and the immune response (3,9). Notably, previous identification of AUF1 target RNAs en masse using PAR-CLIP (photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation) analysis revealed many noncoding (nc)RNA targets of AUF1 (10); however, this search was not designed to find AUF1 target circular (circ)RNAs.
CircRNAs belong to a vast category of RNA molecules generally lacking coding potential, covalently closed into a circle, and usually generated through a backsplice event during RNA splicing (11). Although circRNAs were first discovered in 1979 by electron microscopy (12), only recent advances in RNA sequencing and computational analysis have permitted the accurate and systematic study of circRNAs, as well as investigation of their function in cellular processes (13). A growing body of evidence finds that many circRNAs are expressed in tissue-specific patterns and can govern cellular processes including proliferation, senescence and differentiation (13–19). Accordingly, circRNAs have been implicated in the pathogenesis of diseases including cancer, autoimmunity and neurological disorders (20–23). Given that circRNAs lack 5′ and 3′ ends, they are protected from exonucleolytic decay and are believed to be long-lived molecules; hence, circRNAs are emerging as attractive therapeutic agents and targets (24). Despite this remarkable progress, the role of circRNAs in most cellular processes remains largely unknown.
CircRNAs have emerged as promising potential regulators of biological processes including neurogenesis, myogenesis, immune function and cancer (14,18,20,25). Here, we sought to identify systematically the circRNA targets of AUF1. We employed RNP immunoprecipitation (RIP) analysis followed by microarray analysis to identify bound circRNAs, and we validated a subset of them using reverse transcription (RT) followed by quantitative (q)PCR analysis. One of the highly enriched circRNA ligands in the AUF1 RIP cohort, hsa_circ_0032434 (hsa_circRNA_101387), which we renamed circPCNX because it is generated from the host gene PCNX, was selected for further investigation. An abundant RNA in HeLa cells, circPCNX levels were not affected by binding to AUF1, but instead circPCNX caused a robust and selective reduction in AUF1 binding to p21 mRNA, in turn leading to a rise in both p21 mRNA and p21 protein levels and suppression of cell growth.
MATERIALS AND METHODS
Cell culture and modulation of circPCNX levels
Human cervical carcinoma HeLa cells were cultured in DMEM (Gibco) supplemented with 10% fetal bovine serum (Gibco) and the antibiotics penicillin and streptomycin (Gibco) at 37°C in a humidified atmosphere. For AUF1 ribonucleoprotein (RNP) immunoprecipitation (RIP), RNA and protein analyses, 6 × 105 or 4 × 105 cells were seeded to either overexpress or silence circPCNX, respectively, in 60-mm dishes, unless otherwise indicated. In overexpression experiments, 500 ng of empty vector [pcDNA3 (EV)], pcDNA3-circPCNX, pcDNA3-circPCNX(m1b), pcDNA3-AUF1 (a pool of four vectors to express Flag-tagged p37, p40, p42, p45), 300 ng of pcDNA3-circPCNX(Δ1b), 100 ng of empty vector [pCMV6 (EV)], pCMV6-p21-3′wt or pCMV6-p21-3′del were transfected using Lipofectamine® 2000 (Thermo Fisher Scientific) following the manufacturer's instructions. For silencing, 100 ng of control (Ctrl), circPCNX or AUF1 siRNAs (Integrated DNA Technologies) were transfected using Lipofectamine® 2000. CircInteractome was employed to design siRNAs spanning the circPCNX junction (26).
RNA isolation, reverse transcription (RT), quantitative (q)PCR analysis and droplet digital (dd)PCR analysis
Total RNA was isolated using the Direct-zolTM RNA MiniPrep kit (Zymo Research). For reverse transcription (RT) followed by quantitative PCR (qPCR) analysis, 1 μg of total RNA was employed. For qPCR analysis, 0.1 μl cDNA was used with 250 nM of primers (Supplementary Table S1) and KAPA SYBR® FAST qPCR Master Mix (KAPA Biosystems). To assess the levels of circRNAs, divergent primers spanning the respective junctions were designed using CircInteractome (26). RT-qPCR analysis was carried out on a QuantStudio 5 Real-Time PCR System (Thermo Fisher Scientific) with a cycle setup of 3 min at 95°C, 40 cycles of 5 s at 95°C and 20 s at 60°C. Relative RNA levels were calculated after normalizing to GAPDH mRNA using the 2−ΔΔCt method.
Droplet digital PCR (ddPCR) analysis was employed to quantify the number of circPCNX copies per cell. Total RNA (1 μg) was reverse-transcribed and droplets were generated using EvaGreen Supermix (Bio-Rad) on the QX200™ AutoDG™ Droplet Digital™ PCR System (Bio-Rad Laboratories). The PCR reaction contained 5 μl of cDNA and 250 nM of divergent primers. The PCR amplification setup was: 5 min at 95°C, 30 s at 95°C followed by 60 s at 60°C for 40 cycles, 5 min at 95°C, and then held at 4°C. The absolute copy numbers were assessed using QX200 Droplet Digital PCR System and calculated as described (27). In addition, RNA copy number was estimated using a reference mRNA of known abundance as described (28). To estimate mRNA half-life, cells were treated with Actinomycin D (5 μg/ml) for up to 4 h.
Plasmids
Initially, circPCNX (pcDNA3-circPCNX) was cloned into the vector pcDNA3.1, a kind gift from Dr Sebastian Kadener (29). Briefly, genomic DNA was used to amplify sequences upstream and downstream of the circPCNX sequence. The pcDNA3-circPCNX plasmid was then employed as a template to generate the circPCNX variants: the mutant plasmid circPCNX [pcDNA3-circPCNX(m1b)] and the truncated plasmid circPCNX [pcDNA3-circPCNX(Δ1b)].
The full-length 3′UTR of p21 mRNA was cloned into psiCHECK2 vector (Promega), downstream of the Renilla luciferase (RL) open reading frame, using as template the p21 3′UTR described earlier (30) (psiCHECK2-p21-3′wt); this template was also employed to generate the truncated p21 3′UTR (psiCHECK2-p21-3′del). A plasmid expressing the coding region of p21 mRNA was obtained from OriGene (Cat. # SC119947) and the aforementioned p21 3′UTRs were then ligated; the resulting plasmids were named pCMV6-p21-3′wt or pCMV6-p21-3′del harboring the entire p21 mRNA coding sequence and either an intact or truncated 3′UTR, respectively. The primer pairs employed to generate the plasmids are listed in Supplementary Table S1. All plasmids were verified by sequencing.
Ribonucleoprotein (RNP) immunoprecipitation (RIP) analysis and microarray analysis
The association of endogenous AUF1 with circRNAs and mRNAs was analyzed by RIP analysis as described (31,32). Briefly, HeLa cells were lysed in polysome extraction buffer (PEB; 20 mM Tris–HCl at pH 7.5, 100 mM KCl, 5 mM MgCl2 and 0.5% NP-40) supplemented with 1% protease inhibitor cocktail and 100 units of RiboLOCK RNase inhibitor (Thermo Fisher Scientific), for 10 min on ice. Following a 30-min centrifugation at 10 000 × g, the supernatants were collected, and 1 mg of cell lysate was incubated with protein A sepharose beads (GE Healthcare) coated with 2 μg of either anti-AUF1 (Millipore, Cat. No. 07-260) or control IgG (BD Biosciences, Cat. No. 557273) antibodies for 2 h at 4°C. The beads were then washed with ice-cold NT2 buffer (50 mM Tris–HCl [pH 7.5], 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40)]; the bound RNA was extracted from the beads using Direct-zol™ RNA MiniPrep kit (Zymo Research) and subjected to circRNA microarray analysis (Arraystar; Supplementary Table S2). To validate the microarray analysis or perform additional AUF1 RIP assays for the circRNA variants or the p21 mRNA, the isolated RNA was subjected to RT-qPCR analysis as described above.
Cell counting, BrdU incorporation and cell cycle
To assess population growth, 4 × 104 or 2 × 104 cells/well were plated to either overexpress or silence circPCNX, respectively, and then either counted or subjected to BrdU incorporation analysis. For overexpression, 30 ng of pcDNA3 (EV), pcDNA3-circPCNX, pcDNA3-circPCNX(m1b), pcDNA3-AUF1 or 10 ng of pcDNA3-circPCNX(Δ1b) were transfected using Lipofectamine® 2000, following the manufacturer's instructions. For silencing, 50 ng of Ctrl, circPCNX or AUF1 siRNAs were transfected using Lipofectamine® 2000. For circPCNX-p21 mRNA cotransfection, 50 ng of Ctrl or circPCNX siRNA were cotransfected with 100 ng of pCMV6 (EV), pCMV6-p21-3′wt or pCMV6-p21-3′del using Lipofectamine® 2000, following the manufacturer's instructions. Cell proliferation was assessed using the BrdU Cell Proliferation Assay (Cell Signaling Technology). Briefly, BrdU was added one day after transfection and the incorporation was detected for up to four or five days by reading at 450 nm on a Victor 3V plate reader (Perkin Elmer). Total cells and dead cells were counted on an automatic cell counter (Bio-Rad) after staining with 0.4% Trypan Blue (Gibco).
To trigger cell cycle arrest, 5 × 104 cells were treated with two 18-h cycles of 2 mg/ml of Thymidine (33), whereupon RNA was isolated for RT-qPCR analysis and cell cycle distribution was assessed by flow cytometry. For cell cycle distribution analysis, cells were permeabilized using 70% ethanol for 30 min at 4°C, washed with PBS, and then treated with 10 μg/ml of RNase A for 15 min at 37°C; cells were then washed with PBS and stained with 2 μg/ml of propidium iodide (PI) for 15 min at 25°C in the dark. PI intensity per particle was assessed using BD FACS Canto™ II.
Western blot analysis
Cells were lysed in RIPA buffer (10 mM Tris–HCl, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.1% SDS) supplemented with 1× protease and phosphatase inhibitor cocktail (Thermo Scientific) for 10 min on ice. Following a 10-min centrifugation at 10 000 × g, the supernatants were collected and whole-cell extracts were separated by SDS-PAGE and transferred onto nitrocellulose membranes (Trans-Blot® Turbo™ RTA Transfer Kit, Bio-Rad). Membranes were then blocked in 5% milk in TBS at 25°C for 1 h. Incubation with antibodies recognizing AUF1 (Millipore Sigma, Cat. No. 07-260), GAPDH (Santa Cruz Biotechnology, Cat. No. sc-32233), HDAC3 (Santa Cruz Biotechnology, Cat. No. sc-11417), HSP90 (Santa Cruz Biotechnology, Cat. No. sc-69703) and p21 (Santa Cruz Biotechnology, Cat. No. sc-53870), diluted in 1% milk in TBS-Tween, was carried out for 16 h at 4°C; membranes were then washed three times with TBS–Tween for 5 min and incubated for 1 h with secondary antibodies conjugated with horseradish peroxidase (KwikQuant) diluted in 1% milk in TBS-Tween, then washed three times with TBS-Tween for 5 min, at 25°C. Membranes were developed using Enhanced Chemiluminescence (ECL), and digitized images were captured using KwikQuant Imager (Kindle Biosciences). ImageJ software was employed to assess the band densities.
Biotin pulldown analysis
HeLa cells were lysed in PEB buffer for 10 min on ice, and 200 ng of whole-cell extracts were incubated with 1 μg of either (i) custom-designed partially overlapping biotinylated fragments 1–5, or (ii) a biotinylated fragment spanning the circPCNX junction, or (iii) a pool of fragments spanning p21 mRNA for 30 min at 25°C. The reaction was supplemented with TENT binding buffer (10 mM Tris–HCl, 250 mM NaCl, 1 mM EDTA, 0.5% Triton X100, pH), 200 units of RiboLOCK, and protease inhibitors. Biotinylated complexes were pulled down with prewashed Streptavidin-coupled Dynabeads (Invitrogen) following a 30-min incubation at 25°C. Beads were then washed three times for 5 min at 25°C with TENT buffer, whereupon sample buffer was added to the beads and western blot analysis was performed to assess AUF1 enrichment. TriPure Isolation Reagent (Sigma) was added to the beads and RNA was extracted using the Direct-zol™ RNA MiniPrep kit to assess the enrichment in p21 mRNA and circPCNX.
Luciferase reporter analysis
To test the impact of the p21 3′UTR, dual reporter plasmids psiCHECK2-p21-3′wt and psiCHECK2-p21-3′del were generated. The p21 full-length 3′UTR (wild-type, 3′wt) or 3′UTR with a deletion of the AUF1 binding site (3′del) were cloned downstream of the renilla luciferase (RL) open reading frame in psiCHECK2. Plasmid psiCHECK2 also constitutively expresses firefly luciferase (FL) to normalize for transfection efficiency. Cell cultures (4 × 105 cells/well) were cotransfected with 250 ng of the dual reporter constructs and either 250 ng of pcDNA3-AUF1 or 50 ng of circPCNX siRNA or their respective controls using Lipofectamine® 2000 (Thermo Fisher Scientific). Cells were lysed and luciferase activities were assessed using a dual-luciferase assay kit (Promega) on a Victor 3V plate reader (Perkin Elmer). In experiments measuring RL and FL activities, the levels of expressed RL mRNA (without or with ectopic 3′UTR) and FL mRNA were routinely measured by RT-qPCR analysis.
Immunofluorescence
AUF1 immunofluorescence was carried out as described (34). Briefly, 500 cells/well were plated in chamber slides (Thermo Scientific Nunc), fixed 24 h later with 4% formaldehyde, permeabilized with 0.2% Triton X-100, and blocked in 10% normal goat serum for 1 h at 37°C (Life Technologies). After blocking, cells were incubated with anti-AUF1 antibody in 10% normal goat serum for 1 h at 37°C and then with a secondary antibody linked to Alexa Fluor 568 (Life Technologies) in 10% normal goat serum for 30 min at 37°C. Slides were then washed three times with 1× PBS, the nuclei were stained with DAPI (Dojindo Laboratories) for 15 min at 25°C, and mounting media (Life Technologies) was employed to cover the cells. Visualization of the staining was performed with Zeiss LSM 880.
Statistical analysis
All experiments were carried out a minimum of three times unless otherwise stated. Quantitative data are represented as the means ± SD, and compared statistically by unpaired Student's t test, using SigmaPlot (12.0). Statistical significance was indicated as follows: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. Graphs were generated using GraphPad Prism (8.0) or FlowJO (10.0).
RESULTS
Circular RNA targets of AUF1
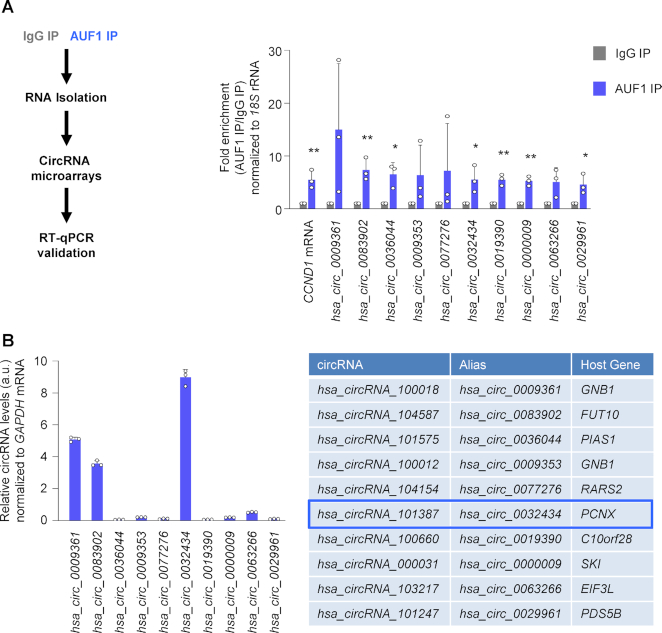
To identify circRNAs interacting with AUF1, an RBP linked to many conditions exacerbated with age, such as cancer, muscle loss and cell senescence, we employed human cervical carcinoma HeLa cells. We performed ribonucleoprotein immunoprecipitation (RNP IP or RIP) analysis using an anti-AUF1 antibody under IP conditions that preserved native interactions. RNA isolated from the RIP complexes was reverse-transcribed and studied by circRNA microarray (chip) analysis (Figure 1A, schematic, Supplementary File S2). The circRNAs that were at least 3-fold enriched in one or more of the AUF1 RIP-chip analyses were then validated by reverse transcription (RT) followed by real-time quantitative (qPCR) analysis, using divergent primers that amplified the junction sequences of the circRNAs. Of the top 10 most highly enriched circRNAs in the AUF1 RIP (Figure 1A, right), we initially focused on three abundant circRNAs in HeLa cells, hsa_circ_0009361, hsa_circ_0083902 and hsa_circ_0032434 (Figure 1B, left). Given that we were subsequently unable to overexpress or silence hsa_circ_0009361 and hsa_circ_0083902 (not shown), these circRNAs were not analyzed further. We tested an additional 10 AUF1-associated circRNAs, but all were less enriched by AUF1 RIP analysis and were less abundant than hsa_circ_0032434, while 10 circRNAs that did not associate with AUF1 in RIP microarray were confirmed not to be enriched by AUF1 RIP analysis (not shown). In sum, hsa_circ_0032434 was selected for deeper analysis; we renamed it circPCNX as it was generated from a linear RNA transcribed from the gene PCNX; PCNX is an evolutionarily conserved transmembrane protein.
Figure 1.
circPCNX is an abundant circRNA target of AUF1. (A) Schematic of the RIP assay and the downstream analysis (left). Validation of the circRNA targets of AUF1 in HeLa cells, as detected by RIP followed by microarray analysis (RIP-chip). The top 10 enriched circRNAs in the AUF1 IP are depicted (right). All circRNA values in the RIP were initially normalized to 18S rRNA and then the levels of the individual circRNAs in AUF1 RIP were normalized to the levels of those circRNAs in control IgG IP. CCND1 mRNA, a target of AUF1, was included as a positive control. The data are plotted as ‘enrichment’ in AUF1 IP relative to IgG IP. (B) Basal expression levels of the top 10 circRNAs highly enriched by AUF1 RIP in HeLa cells (left); data were normalized to GAPDH mRNA levels. Table listing the top 10 candidates including the circRNA ID, the alias name and the host gene (right). a.u., arbitrary units. Data in (A, B) represent the mean values ± SD from four biological replicates. Significance was established using Student's t-test. * P ≤ 0.05; ** P ≤ 0.01.
Distribution and abundance of AUF1 and circPCNX
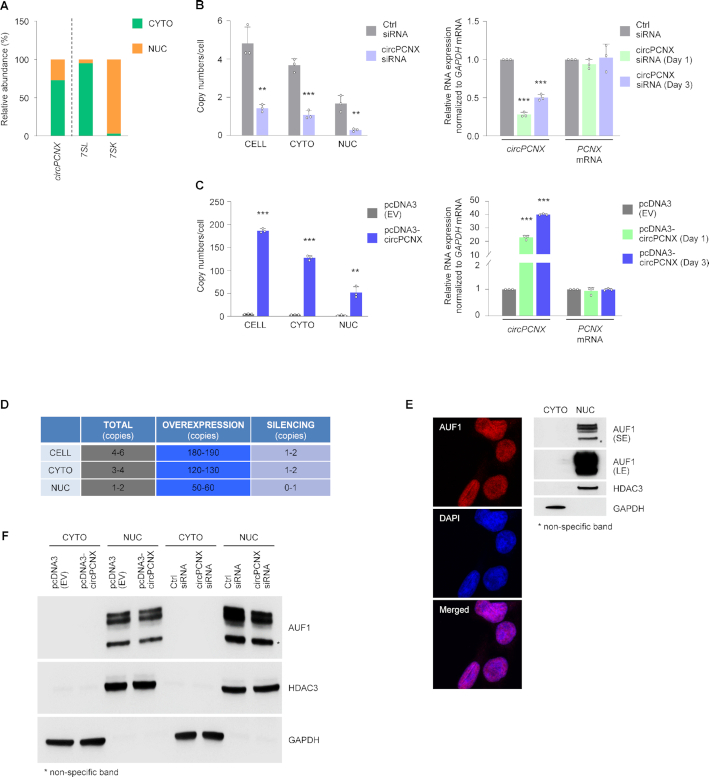
circPCNX is 203 nucleotides (nts) in length and arises from exon 11 of the PCNX mRNA (NM_014982.3, positions 3186–3388). RT-qPCR analysis of RNA from fractionated lysates revealed that circPCNX was predominantly (70%) cytoplasmic (Figure 2A); 7SL and 7SK RNAs were included as controls for the cytoplasmic and nuclear fractions, respectively (Figure 2A) (35). After fractionating HeLa cells into nuclear and cytoplasmic lysates, AUF1 RIP followed by RT-qPCR analysis revealed that both nuclear and cytoplasmic circPCNX associated with AUF1 (not shown). To further study the impact of circPCNX, we modulated its levels by silencing and overexpressing it. To silence circPCNX, we tested three different small interfering (si)RNAs spanning the circPCNX junction (Figure 2B, Supplementary Figure S1, Materials and Methods) and used siRNA #1 (hereafter siRNA) for subsequent studies. Under basal conditions circPCNX was present in 4–6 copies per cell as assessed by droplet digital PCR (ddPCR) and qPCR analysis (28) (Materials and Methods); ∼3–4 copies and ∼1–2 copies in the cytoplasm (CYTO) and nucleus (NUC), respectively (Figure 2B, left). Silencing circPCNX significantly reduced its abundance to 1–2 copies per cell, almost exclusively cytoplasmic (Figure 2B, left); as measured by RT-qPCR analysis, silencing reduced circPCNX expression levels by 75% and 50% at days 1 and 3 after transfection, respectively (Figure 2B, right).
Figure 2.
Stoichiometric analysis and modulation of the levels of AUF1 and circPCNX. (A) Subcellular localization of circPCNX. The relative levels of circPCNX in each subcellular compartment, cytoplasm (CYTO) and nucleus (NUC), fractionated as explained in Materials and Methods, were assessed by RT-qPCR analysis. 7SL (a cytoplasmic RNA) and 7SK (a nuclear RNA) were included as fractionation controls. (B) The numbers of circPCNX copies per cell or cell compartment were assessed by ddPCR analysis 3 days after transfection with control (Ctrl) siRNA or an siRNA directed at circPCNX (left). The relative levels of circPCNX and PCNX mRNA one or three days later in each siRNA transfection group were assessed by RT-qPCR analysis (right). (C) The numbers of circPCNX per cell or cell compartment were assessed by ddPCR analysis 3 days after transfection with an empty vector [pcDNA3(EV)] or with a vector to overexpress circPCNX (pcDNA3-circPCNX) (left). The relative levels of circPCNX and PCNX mRNA one or three days after transfection were assessed by RT-qPCR analysis (right). (D) Table summarizing copy numbers per cell in the cultures described in panels B and C, as estimated by ddPCR analysis at day 3 following transfection. (E) Subcellular localization of AUF1 in HeLa cells using microscopy (left) and western blot analysis (right). DAPI-stained cell nuclei; the levels of GAPDH and HDAC3 served to monitor the cytoplasmic and nuclear fractionation, respectively (right). SE, short exposure; LE, long exposure. (F) Western blot analysis of the subcellular localization of AUF1 two days after transfection to either overexpress or silence circPCNX. GAPDH and HDAC3 were assayed to monitor loading and the quality of the cytoplasmic and nuclear fractionation procedures. Data in (B, C) represent the mean values ± SD from three biological replicates. Significance was established using Student's t-test. * P ≤ 0.05, ** P ≤ 0.01 or *** P ≤ 0.001.
To perform gain-of-function experiments, we designed vector pcDNA3-circPCNX to overexpress circPCNX (Materials and Methods). After transfection, RT followed by ddPCR and qPCR analyses revealed that overexpression of pcDNA3-circPCNX increased circPCNX levels overall, reaching ∼190 copies/cell as estimated by ddPCR analysis (Figure 2C, left), 120–130 copies in the cytoplasm, 50–60 copies in the nucleus, corresponding to ∼20- to 40-fold increases at days 1 and 3 after transfection, respectively (Figure 2C, right). These results are summarized in Figure 2D. Importantly, neither overexpression nor silencing of circPCNX significantly altered the levels of PCNX mRNA, enabling further analysis of circPCNX function (Figure 2B, C).
We then sought to explore the stoichiometry of the AUF1-circPCNX interaction more thoroughly by investigating the subcellular localization of AUF1 (Figure 2E). In line with previous observations in HeLa cells (36,37), AUF1 was predominantly localized in the nucleus as shown by microscopy and western blot analysis (Figure 2E and Supplementary Figure S1B). We monitored the levels of GAPDH and HDAC3 as controls for cytoplasmic and nuclear lysates, respectively (Figure 2E, right). Notably, the lower abundance of AUF1 in the cytoplasm—the compartment where circPCNX was predominantly localized—permits a stoichiometry that favors circPCNX interactions with AUF1. However, it was challenging to establish an accurate number of AUF1 molecules per cell due to a number of factors. Briefly, AUF1 has 4 isoforms (p37, p40, p42, p45), but establishing which endogenous isoform associated with circPCNX was not possible given the present lack of commercial AUF1 isoform-specific antibodies. In addition, while AUF1 phosphorylation can affect binding (38,39), it was not possible to determine whether HeLa AUF1 was phosphorylated in the cytoplasm or whether phosphorylation influenced binding to circPCNX; the impact of AUF1 phosphorylation on the fate of target RNAs remains unknown. Furthermore, the relative concentration of AUF1 and circPCNX in different subcytoplasmic areas has not been studied, even though their relative concentration in specific regions of the cell may help explain the influence of circPCNX on AUF1 actions. Here too, the required detection tools are not yet available. Other factors influencing the number of AUF1 molecules associating with circPCNX are mentioned below (Discussion). Given the limitations of quantifying circPCNX and AUF1 molecules in the cell, we focused on the empirical consequences of lowering or overexpressing each one, the ‘functional stoichiometry’ of circPCNX and AUF1.
To begin testing if there was a functional consequence of the binding of AUF1 and circPCNX, we first examined if silencing or overexpressing circPCNX influenced AUF1 levels and/or subcellular localization. As shown, neither AUF1 levels (Supplementary Figure S1C) nor its subcellular distribution (Figure 2F) changed significantly by altering circPCNX abundance. In sum, circPCNX is a moderately abundant circRNA that associates with AUF1 and does not modulate AUF1 levels or subcellular localization.
circPCNX suppresses cell proliferation
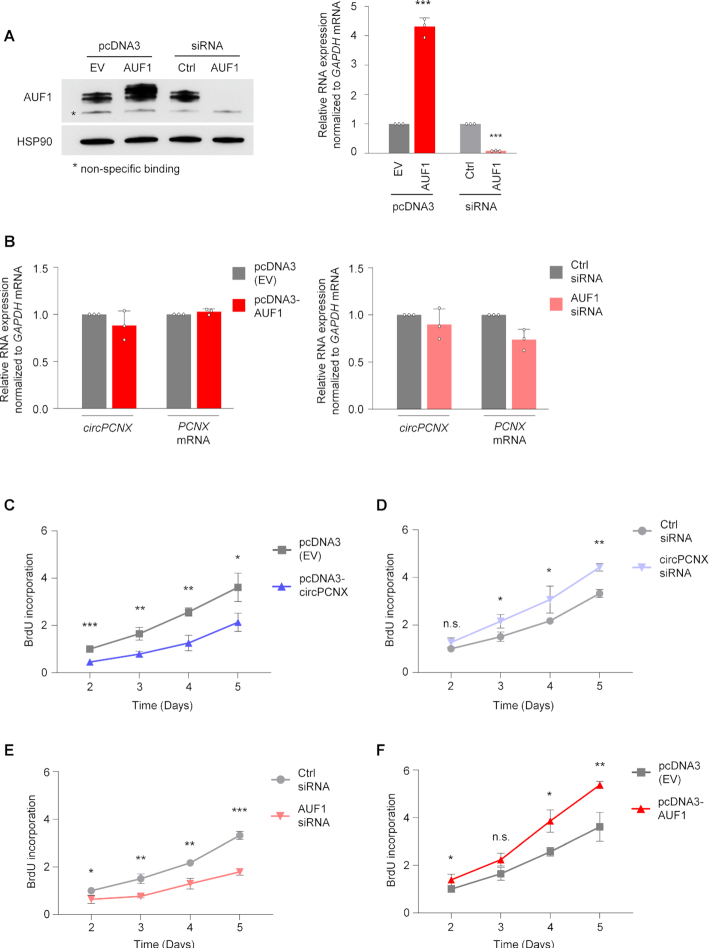
We then tested whether changing AUF1 levels might alter circPCNX levels. AUF1 was overexpressed in HeLa cells using a pool of four plasmids (pcDNA3-AUF1), each expressing one AUF1 isoform, p37, p40, p42, p45 and it was silenced using a pool of siRNAs (AUF1 siRNA) (Materials and Methods). Forty-eight h after transfection with plasmids or siRNAs, the levels of AUF1 mRNA and AUF1 protein were assessed by RT-qPCR and western blot analyses, respectively (Figure 3A). Despite the fact that AUF1 decreases the stability of subsets of mRNAs (4), overexpression or silencing of AUF1 did not significantly affect the levels of circPCNX, other circRNAs or the respective host mRNAs (Figure 3B and Supplementary Figure S2A, B).
Figure 3.
circPCNX negatively affects cell proliferation. (A) Overexpression or silencing of AUF1. Three days after transfection of either empty plasmid pcDNA3(EV) or plasmids expressing each of the four Flag-tagged AUF1 isoforms, as well as following transfection of control (Ctrl) siRNA or siRNAs directed at all four AUF1 isoforms (AUF1 siRNA), the levels of AUF1 protein as well as loading control HSP90 were assessed by western blot analysis (left) and the levels of AUF1 mRNA (all isoforms, right) by RT-qPCR analysis. (B) Effect of AUF1 overexpression (left) or silencing (right) on the levels of circPCNX and PCNX mRNA as determined by RT-qPCR analysis three days after transfection. (C, D) Proliferation of HeLa cells was measured at the indicated times following circPCNX overexpression (C) or silencing (D) as described in Figure 2, by assaying BrdU incorporation. (E, F) Proliferation of HeLa cells was measured at the indicated times following AUF1 silencing (E) or overexpression (F) as described in Figure 2, by assessing BrdU incorporation. Data in (A–F) represent the mean values ± SD from three biological replicates. Significance was established using Student's t-test. n.s. not significant; * P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.001.
Given that altering AUF1 levels did not change circPCNX abundance, we asked the converse question, whether overexpressing or silencing circPCNX modulated AUF1 function. In the course of these experiments, we observed that modulating circPCNX levels changed the rates of cell proliferation. To examine this phenotype more directly, 2 days after transfection, we counted cells each day for the following 5 days. Overexpression of circPCNX led to a prominent decrease in cell numbers, whereas circPCNX silencing caused a significant increase in cell numbers compared to the respective controls (Supplementary Figure S3A, B). Interestingly, the effects of altering AUF1 levels on cell number mirrored the effects of modulating circPCNX: silencing AUF1 decreased cell counts while overexpressing AUF1 increased cell numbers over the ensuing five days (Supplementary Figure S3C, D). Importantly, the observed changes in cell numbers were not due to changes in cell viability (Supplementary Figure S3E–H).
In light of the findings that cell numbers changed while cell viability appeared unaltered, we hypothesized that circPCNX affected cell proliferation. To investigate this possibility, we assayed BrdU incorporation (Materials and Methods) in cells in which AUF1 and circPCNX levels were altered. Briefly, BrdU incorporation was measured 2 days after transfection and once daily for the following 5 days. Akin to the results from cell counting, circPCNX overexpression significantly inhibited cell proliferation ∼40–50% throughout a 5-day span (Figure 3C), while circPCNX silencing accelerated cell proliferation (Figure 3D). In further agreement with the cell counting data, AUF1 inhibition lowered cell proliferation rate (by ∼40–50%), whereas AUF1 overexpression modestly accelerated cell proliferation (Figure 3E, F). Taken together, these findings indicate that overexpressing circPCNX or silencing AUF1 in HeLa cells reduced population growth, while silencing circPCNX or overexpressing AUF1 promoted cell proliferation.
circPCNX attenuates AUF1 binding to p21 mRNA
AUF1 binds and affects the stability of many RNAs implicated in cell proliferation (10,40), including coding transcripts (CCNB1, CCND1, IL6, IL8, p21, p16, TERT and AUF1 mRNAs) and long noncoding (lnc)RNAs (NEAT1 and MALAT1). To investigate if the effects on population growth by modulating circPCNX and AUF1 levels were linked, we studied whether circPCNX modulated the impact of AUF1 on these target RNAs. Overexpressing or silencing circPCNX altered the levels of CCNB1, IL6 and IL8 mRNAs in only one of these conditions, whereas lncRNA NEAT1 and TERT mRNA changed moderately in both conditions (Supplementary Figure S4A, B). Interestingly, the levels of AUF1 target p21 (CDKN1A) mRNA, encoding a potent suppressor of proliferation (40–42), were significantly affected by changes in circPCNX levels. AUF1 was previously shown to bind to the p21 mRNA 3′ untranslated region (UTR) and promoted p21 mRNA decay (10,40).
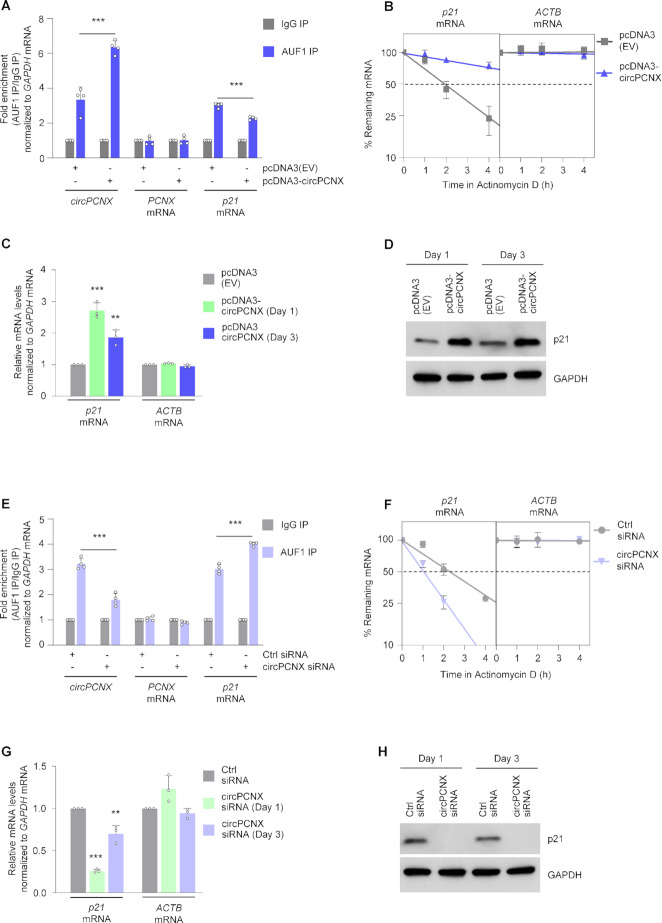
After overexpressing circPCNX for 24 h, RIP analysis indicated that AUF1 binding to p21 mRNA was significantly attenuated (25% lower), while AUF1 binding to circPCNX increased ∼ twofold; there was no significant enrichment of the linear host (PCNX mRNA) in the AUF1 RIP (Figure 4A and Supplementary Figure S5A). Of note, pcDNA3-circPCNX did not appear to give rise to substantial levels of linear RNA products, and only circularized RNA (ectopic circPCNX) was found to bind AUF1 (not shown). Given that AUF1 is known to destabilize p21 mRNA (10,40), we then assessed p21 mRNA stability following circPCNX overexpression. Using Actinomycin D to block de novo transcription in order to assess mRNA half-life, we found that overexpressing circPCNX for 24 h increased p21 mRNA half-life; the stable transcript ACTB mRNA and the labile, cell cycle-related transcript activating transcription factor 1 (ATF1) mRNA were included as control transcripts that were not affected by changes in circPCNX levels (Figure 4B and Supplementary Figure S5C) (43,44). To examine the impact of this enhanced p21 mRNA stability, we performed both RT-qPCR and western blot analyses 1 and 3 days after transfection to assess changes in p21 mRNA and p21 protein levels, respectively. In line with the decline in AUF1 binding to p21 mRNA, there was a 2- to 3-fold rise in p21 mRNA levels (Figure 4C) and a robust increase in p21 protein levels (Figure 4D) at both time points after transfection.
Figure 4.
Effects of circPCNX on p21 mRNA and p21 protein. (A–D) AUF1 binding to p21 mRNA (A), p21 mRNA stability (B), p21 mRNA levels (C), and p21 protein levels (D) were assessed after transfection with pcDNA3(EV) or with pcDNA-circPCNX to overexpress circPCNX. (A) AUF1 binding to circPCNX, PCNX mRNA and p21 mRNA was assessed one day after transfection; values were first normalized to the levels of a transcript (GAPDH mRNA) that encodes a housekeeping protein, and afterwards normalized to the RNA levels in the IgG IP. (B) One day after transfection, cells were treated with Actinomycin D to block de novo transcription for the times indicated, and the time required for p21 mRNA to reach 50% of its initial abundance (the half-life) in each transfection group was assessed by RT-qPCR analysis. A stable transcript (ACTB mRNA) was included as control. At one and three days after transfection of plasmids, changes in the levels of p21 mRNA (C) or p21 protein (D) were assessed by RT-qPCR and western blot analyses, respectively. (E–H) AUF1 binding to p21 mRNA (E), p21 mRNA stability (F), p21 mRNA levels (G), and p21 protein levels (H) were assessed after transfection with Ctrl siRNA or circPCNX siRNA to silence circPCNX. (E) AUF1 binding to circPCNX, to PCNX mRNA and to p21 mRNA was assessed one day after transfection; as above, values were first normalized to GAPDH mRNA, and afterwards normalized to RNAs in the IgG IP. (F) One day after transfection, cells were treated with Actinomycin D to block de novo transcription for the times indicated, and the time required for p21 mRNA to reach 50% of its initial abundance (the half-life) in each transfection group was assessed by RT-qPCR analysis. A stable transcript (ACTB mRNA) was included as control. At one and three days after siRNA transfections, changes in the levels of p21 mRNA (G) or p21 protein (H) were assessed by RT-qPCR and western blot analyses, respectively. Data in (A–C, E–G) represent the mean values ± SD from three biological replicates. Significance was established using Student's t-test. ** P ≤ 0.01 or *** P ≤ 0.001.
We then examined the effects of silencing circPCNX for 24 h. RIP analysis indicated that silencing circPCNX reduced the association of AUF1 with circPCNX (by ∼50%), but it significantly increased the binding of AUF1 to p21 mRNA by up to 1.5-fold compared to the control (Ctrl) siRNA transfection group (Figure 4E and Supplementary Figure S5B). In line with this finding, increased binding of AUF1 to p21 mRNA promoted p21 mRNA decay, whereas the levels of ACTB and ATF1 mRNAs were unchanged (Figure 4F and Supplementary Figure S5D). Along with enhanced decay, p21 mRNA levels decreased by 75% or 30% by 1 and 3 days after transfection, respectively, as shown by RT-qPCR (Figure 4G). Consequently, there was a robust reduction in p21 protein levels, as identified by western blot analysis (Figure 4H); as anticipated, this reduction was reversed and p21 levels were restored by silencing AUF1 (not shown). In conclusion, increasing circPCNX levels reduced AUF1 binding to p21 mRNA and elevated p21 production, while conversely, decreasing circPCNX increased AUF1 binding to p21 mRNA and reduced p21 levels.
AUF1 binding site in circPCNX
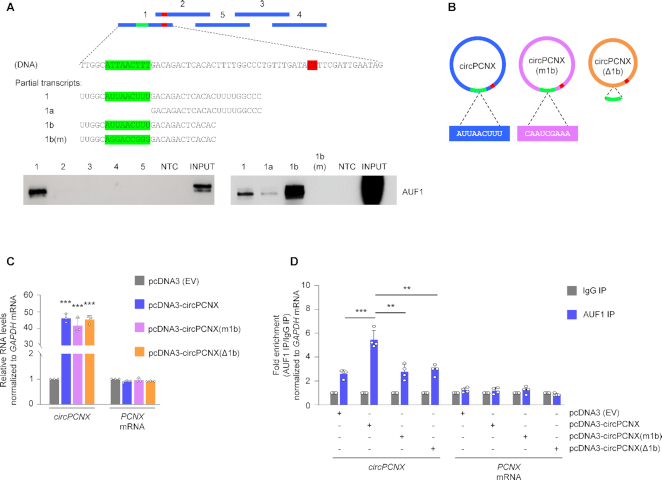
To continue to ascertain if the impact of circPCNX on proliferation was dependent on the control of p21 expression by AUF1, we set out to map the site(s) of AUF1 interaction on circPCNX. We generated five biotinylated RNAs spanning the length of circPCNX (Figure 5A, schematic), incubated them with HeLa whole-cell protein lysates, pulled down the biotinylated RNP complexes with streptavidin beads, and performed western blot analysis to detect AUF1 in the RNPs. After finding that AUF1 bound RNA segment 1 (Figure 5A), we generated additional biotinylated RNAs to narrow down the binding site. Although both segments 1a and 1b were AU-rich, the classic sequences of interaction with AUF1, segment 1a bound only weakly while segment 1b had strong affinity for AUF1. Within fragment 1b, we identified the AU-rich sequence AUUAACUUU resembling an AUF1 binding site. Interestingly, this sequence (green in Figure 5A) is proximal to, but does not include, the circPCNX junction (red ‘GA’ dinucleotide in Figure 5A).
Figure 5.
Site of AUF1 binding to circPCNX. (A) Schematic of the biotinylated circPCNX fragments (blue) used in biotin pulldown experiments. NTC, negative control; m, mutant. The green and red colors indicate the AUF1 binding site and the junction, respectively. Purified fragments 1–5 as well as the ‘Partial transcripts’ shown were incubated with HeLa whole-cell lysates and the interaction with AUF1 was revealed after pulldown using streptavidin beads, followed by western blot analysis to identify AUF1 in the pulldown material. (B) Schematic of the three circPCNX variants expressed from engineered plasmid vectors, employed in the study. In circPCNX(m1b) the 9-mer AUF1-binding segment was mutated whereas in circPCNX(Δ1b) it was deleted. (C) Relative levels of circPCNX or PCNX mRNA in cells transfected with the plasmids shown in panel B, three days following transfection. (D) Twenty-four hours after transfecting HeLa cells with the plasmids shown, the association of circPCNX (or PCNX mRNA in control reactions) with AUF1 was measured by RIP followed by RT-qPCR analysis and represented as ‘fold enrichment’. Data in (C, D) represent the mean values ± SD from four biological replicates. Significance was established using Student's t-test. ** P ≤ 0.01 or *** P ≤ 0.001.
Further evidence that AUF1 bound to region 1b was sought by mutating [m1b] or deleting [Δ1b], the AU-rich nucleotides in 1b from the pcDNA-circPCNX expression vector (Figure 5B). As shown, transfection of these plasmids led to comparable expression levels of circPCNX (intact), circPCNX(m1b) or circPCNX(Δ1b) and did not affect PCNX mRNA levels (Figure 5C). We then assessed whether modifying site 1b influenced AUF1 binding to circPCNX by AUF1 RIP followed by RT-qPCR analysis 24 h after plasmid transfection. We found that AUF1 showed enhanced binding only to ectopic circPCNX, but not to circPCNX(m1b) or circPCNX(Δ1b), where binding was found to be at basal levels as observed in cells transfected with the control pcDNA3 (EV) (Figure 5D). These findings confirmed that AUF1 bound specifically to circPCNX segment 1b.
Interfering with the circPCNX-AUF1 complex restores cell proliferation
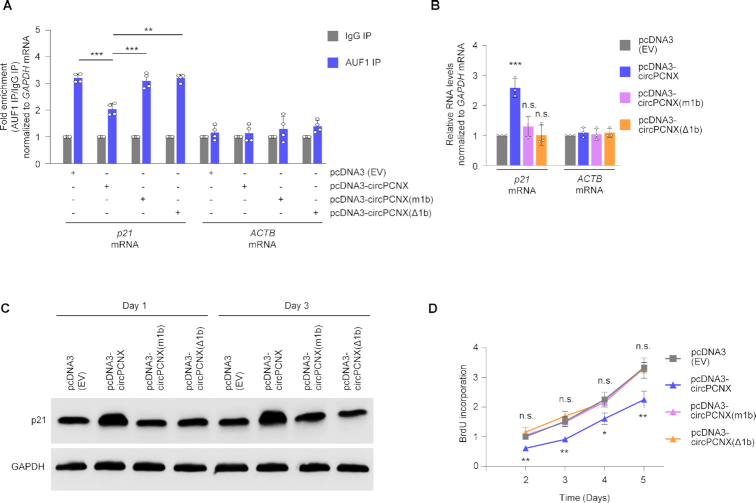
Following the identification of AUF1 binding to circPCNX, we sought to investigate whether overexpression of the truncated or mutated circPCNX might influence the ability of AUF1 to bind p21 mRNA. We transfected all four plasmids and tested the ability of AUF1 to bind p21 mRNA in cells that overexpressed circPCNX, circPCNX(m1b) or circPCNX(Δ1b), relative to cells transfected with pcDNA3 (EV). Briefly, 24 h after transfection, we lysed cells and performed AUF1 RIP followed by RT-qPCR analysis to assess the enrichment of p21 mRNA in AUF1 IP. As seen earlier, overexpressing circPCNX reduced significantly AUF1 binding to p21 mRNA as compared to similar binding in control cells [pcDNA3 (EV)] (Figure 6A). Interestingly, however, expression of either circPCNX(m1b) or circPCNX(Δ1b) did not reduce the AUF1 binding to p21 mRNA, indicating that the region of interaction of circPCNX with AUF1 was required to prevent AUF1 binding to p21 mRNA (Figure 6A and Supplementary Figure S6A). In line with these observations, the rise in p21 mRNA levels observed when circPCNX was overexpressed (and consequently AUF1 binding to p21 mRNA was reduced) was lost after expressing circRNAs circPCNX(m1b) or circPCNX(Δ1b), which did not bind AUF1, further supporting the view that preventing AUF1 binding to p21 mRNA through possible competition with circPCNX led to a rise in p21 mRNA expression (Figure 6B). In turn, overexpressing either circPCNX(m1b) or circPCNX(Δ1b) did not cause the rise in p21 protein levels seen after overexpressing circPCNX (Figure 6C).
Figure 6.
AUF1 binding to p21 mRNA is attenuated by circPCNX, but not by mutant circPCNX unable to bind AUF1. (A) Twenty-four hours after transfecting HeLa cells with the plasmids shown, the association of p21 mRNA (or ACTB mRNA, in control reactions) with AUF1 was measured by RIP followed by RT-qPCR analysis and represented as ‘fold enrichment’. (B, C) In HeLa cells transfected with the plasmids indicated, the levels of p21 mRNA (B) were assessed 3 days after transfection by RT-qPCR analysis, and the levels of p21 protein (C) were assessed one and three days after transfection by western blot analysis, including GAPDH as loading control. (D) At the times shown following transfection of HeLa cells with the plasmids indicated, cell proliferation in each transfection group was assessed by using BrdU incorporation analysis. Data in (A, B, D) represent the mean values ± SD from four biological replicates. Significance was established using Student's t-test. * P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.001.
Finally, we monitored cell proliferation in populations that expressed different p21 levels resulting from intact or modified circPCNX. As measured by cell counting and BrdU incorporation analysis over 5 days following transfection, proliferation progressed robustly in control cells [pcDNA3 (EV)] and in cells expressing circPCNX(m1b) or circPCNX(Δ1b), but declined when the intact circPCNX was overexpressed (Figure 6D and Supplementary Figure S6B, C). Taken together, these findings indicate that overexpression of circPCNX, a circRNA capable of binding AUF1 and thus preventing AUF1 binding to p21 mRNA, elevated p21 expression and reduced cell proliferation; by contrast, overexpression of circPCNX mutants unable to bind AUF1 led to the preservation of AUF1-p21 mRNA complexes, retaining low levels of p21 and active cell division.
circPCNX affects AUF1 binding to the 3′UTR of p21 mRNA
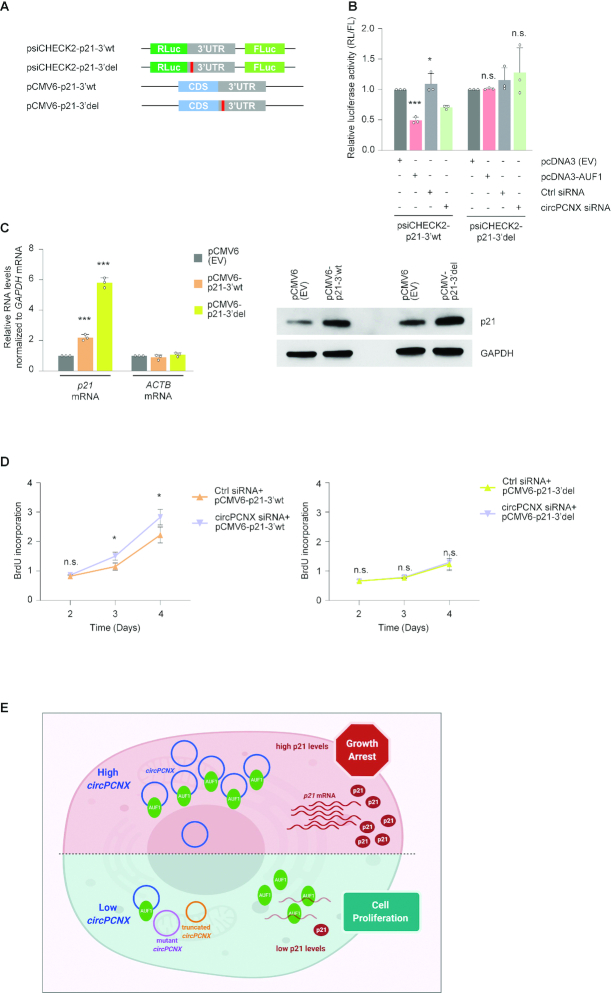
AUF1 controls p21 mRNA stability by binding the p21 3′UTR (40). Therefore, we sought further validation that the circPCNX-AUF1 complex governed the changes in p21 mRNA and p21 protein levels through the AUF1-binding site on the p21 3′UTR. To this end, we cloned in plasmid psiCHECK2, immediately downstream of the reporter renilla luciferase (RL) open reading frame, an intact [wild-type full-length (3′wt)] 3′UTR of p21 mRNA (psiCHECK2-p21-3′wt) or a truncated (deletion) 3′UTR of p21 mRNA (psiCHECK2-p21-3′del) lacking the AUF1-binding site identified by AUF1 PAR-CLIP analysis (10) (Materials and Methods). To further validate the importance of the AUF1-binding site in the effect of circPCNX on cell proliferation, we constructed plasmids encoding a full-length (pCMV6-p21-3′wt) or truncated (pCMV6-p21-3′del) p21 mRNA, lacking the AUF1-binding site (Figure 7A).
Figure 7.
Expression levels of p21 mRNA and p21 protein are attenuated by circPCNX, but not by a mutant circPCNX unable to bind AUF1. (A) Reporter plasmids were created using the backbone of the psiCHECK2 dual luciferase reporter by inserting the p21 3′UTR (gray) with either the full-length wild-type (3′wt) sequence or with a mutation (3′del) in the AUF1-binding site (red). Vectors using the pCMV6 backbone were also created to express p21 protein from mRNAs that had the intact 3′UTR (3′wt) or had a mutation in the AUF1-binding site (3′del). (B) Forty-eight hours after transfecting HeLa cells with either pcDNA3(EV) or pcDNA3-AUF1 or with the siRNAs shown, the reporter plasmids psiCHECK2-p21-3′wt or psiCHECK2-p21-3′del were transfected and 24 h after that, luciferase activities (RL/FL) in each transfection group were assessed. (C) Twenty-four hours after transfecting HeLa cells with the plasmids indicated, the levels of p21 mRNA (with 3′wt or 3′del 3′UTRs) were measured by RT-qPCR analysis and normalized to neo mRNA expressed from the same plasmids; the levels of control ACTB mRNA were quantified in the same reactions; the levels of p21 protein in each transfection group were assessed by western blot analysis. (D) Ctrl or circPCNX siRNAs were transfected along with the plasmids shown (left, p21 3′wt; right, p21 3′del), and the rates of proliferation were assessed by measuring BrdU incorporation at the times indicated. (E) Summary model. Top, when circPCNX accumulates in cells, it can sequester AUF1 away from its target mRNAs, notably p21 mRNA, in turn allowing p21 mRNA stabilization, increased p21 expression, and growth suppression. Bottom, when circPCNX levels decline, or if circPCNX is unable to bind AUF1 (mutated or truncated), then AUF1 associates with p21 mRNA, reducing p21 mRNA stability and p21 levels, and enhancing cell proliferation. Image was created using BioRender. Data in (B–D) represent the mean values ± SD from four biological replicates. Significance was established using Student's t-test. * P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.001.
Initially, we cotransfected the reporter constructs with either pcDNA3-AUF1 or circPCNX siRNA or the respective controls, and assessed luciferase activity 24 h after transfection. In cells cotransfected with psiCHECK2-p21-3′wt and pcDNA3-AUF1, luciferase activity decreased by approximately 50% compared to cells co-overexpressing pcDNA3 (EV); this reduction was associated with a corresponding reduction in RL mRNA but not FL mRNA (Supplementary Figure S7A). In line with our previous observations, circPCNX silencing mimicked the effect of pcDNA3-AUF1 on the intact p21 3′UTR. To further validate whether AUF1 was the downstream effector of circPCNX-mediated p21 reduction, we employed a reporter bearing a truncated p21 3′UTR (psiCHECK2-p21-3′del) lacking the predicted AUF1 binding site. The observation that co-overexpression of psiCHECK2-p21-3′del with pcDNA3-AUF1 did not significantly alter luciferase signals supported the accuracy of the predicted AUF1-binding site (Figure 7B). Moreover, the circPCNX siRNA-mediated effect on the activity of the psiCHECK2-p21-3′wt construct was completely abrogated when we overexpressed psiCHECK2-p21-3′del. We thus proposed that circPCNX effectively sequestered AUF1, and thus by reducing circPCNX, AUF1 binding to the 3′UTR of p21 mRNA increased, in turn promoting p21 mRNA decay.
Finally, to study if AUF1 and p21 were effectors of the influence of circPCNX on population growth (Figure 3C, D), we assessed cell proliferation in cells overexpressing pCMV6-p21-3′wt or pCMV6-p21-3′del followed by circPCNX silencing. Initially, we confirmed that the p21 variants were moderately overexpressed using RT-qPCR and western blot analyses (Figure 7C). Given that pCMV6-p21-3′del lacked the AUF1 binding site, we hypothesized that the deletion rendered p21 mRNA more stable than the full-length p21 mRNA. To compare mRNA expression levels across transfection groups, we normalized to neo mRNA, expressed from the vector backbone to confer resistance to neomycin (Supplementary Figure S7B). We then assessed the effect of expressing pCMV6-p21-3′wt or pCMV6-p21-3′del on proliferation by cell counting and BrdU incorporation analysis until day 4. As expected, overexpression of the intact p21 mRNA inhibited cell proliferation, while expression of p21-3′del mRNA caused an even greater reduction of cell proliferation (Supplementary Figures S7C, D). Importantly, whereas circPCNX silencing modestly but significantly rescued the p21-3′wt-mediated effect on cell proliferation (Figure 7D, left and Supplementary Figure S7E, left), circPCNX levels had no significant effect on the inhibition of cell proliferation elicited by p21-3′del, which was refractory to AUF1-mediated repression (Figure 7D, right and Supplementary Figure S7E, right). We therefore propose that circPCNX bound AUF1 away from the 3′UTR of p21 mRNA, leading to a rise in p21 production and to inhibition of cell proliferation (Figure 7E).
DISCUSSION
CircRNAs have recently drawn immense interest as part of ribonucleoprotein or RNA-RNA complexes implicated in different processes affecting cell function, physiology and disease states (45,46). Here, we investigated circRNAs associated with AUF1, a protein with pleiotropic cellular functions, including cell senescence, division and the immune and stress responses, as well as in muscle metabolism and cancer. We focused on a previously uncharacterized circRNA, hsa_circ_0032434 or hsa_circRNA_101387, later named circPCNX, which was both highly enriched in AUF1 RIP and moderately abundant in HeLa cells (Figure 1). Our results led us to propose a model in which elevated circPCNX directly binds AUF1 and prevents AUF1 from interacting with p21 mRNA, enabling increases in p21 mRNA and p21 protein levels, and in turn inhibiting proliferation (Figures 3 and 4). Conversely, a reduction in circPCNX levels has the opposite effect, freeing AUF1 to bind p21 mRNA, in turn enhancing p21 mRNA decay and reducing p21 expression levels (Figures 3 and 4). In support of this model, circPCNX variants lacking the AUF1-binding site [circPCNX(m1b) and circPCNX(Δ1b)] reversed the circPCNX-mediated effects on AUF1 binding and restored cell proliferation (Figure 6). It is important to recognize that additional circRNAs might contain PCNX exon 11; two out of five circRNAs identified with PCNX exon 11 flanking the junction were found moderately enriched in AUF1 RIP (albeit less than circPCNX). The composition of other circRNAs in which exon 11 might be present in the circRNA body, away from the junction, could not be established with confidence and hence these circRNAs were not studied. As our ability to reconstruct the bodies of circRNAs improves, studying their influence on AUF1 actions is warranted.
Although we initially hypothesized that circPCNX might influence the levels of many of the mRNA targets of AUF1 encoding proliferation-associated proteins, we were surprised to find that only p21 mRNA among the cohort tested appeared to be preferentially elevated when circPCNX was overexpressed and reduced when circPCNX was silenced (Figure 4; Supplementary Figure S4). We dedicated extensive efforts to finding the mechanism responsible for this selectivity. We hypothesized that perhaps circPCNX might bind to p21 mRNA, thereby facilitating the competition of the two RNAs for AUF1. However, as shown by pulldown of these RNAs in native conditions as well as after crosslinking (Supplementary Figure S8A-C), p21 mRNA and circPCNX did not appear to interact physically. Other possible factors contributing to this selective competition, including colocalization of p21 mRNA, circPCNX and AUF1 in specific subcytoplasmic spaces or in distinct macromolecular complexes, await further analysis. However, it was clear that the effects of circPCNX-AUF1 on cell proliferation were carried out through AUF1 modulating p21 levels, as circPCNX variants lacking the AUF1 binding site were unable to compete with AUF1 for binding to p21 mRNA, in turn lowering p21 levels and enabling proliferation (Figure 6). In line with this observation, circPCNX silencing derepressed AUF1 to bind full-length p21 mRNA leading to reduced p21 production and enhanced proliferation. These effects on p21 mRNA stability and proliferation were lost in the presence of mutations in the p21 mRNA or circPCNX RNA that eliminated the AUF1-binding sites (Figures 5-7 and Supplementary Figures S6, S7). Understanding why a circRNP influences the binding of the RBP to some mRNA targets but not others is a question of increasing interest. In a recent report, the interaction of another RBP, the insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3), with the circRNA CDR1as affected only a subset of IGF2BP3 target mRNAs (47). Although for this circRNP the selectivity of IGF2BP3 was attributed to cellular context, the determinants of the circRNP associations and the impact upon the RBP’s ability to bind its target mRNAs remain to be studied systematically for virtually all circRNAs and RBPs.
Another critical question in circRNP biology is stoichiometry—that is, the relative levels of the circRNA studied and the associated RBP. In this cell system, circPCNX was predominantly cytoplasmic, and AUF1, while likely present in thousands of copies per HeLa cell, was primarily nuclear, and only a small fraction of AUF1 was localized in the cytoplasm (Figure 2). We thus focused on the circRNP complexes (circPCNX-AUF1) affecting cytoplasmic AUF1 target mRNAs. Reaching an accurate estimation of the levels of circPCNX-AUF1 was challenging due to several factors. First, AUF1 has 4 isoforms (p37, p40, p42, p45); while all four appeared to have affinity for the biotinylated circPCNX segment of interaction (Figure 4A), establishing which endogenous isoform was capable of associating with circPCNX was not possible given a lack of AUF1 isoform-specific antibodies. Second, AUF1 phosphorylation was previously proposed to influence RNA binding (38,39). In this study, the extent of AUF1 phosphorylation in cells or cell compartments was not determined, nor was it established whether phosphorylation influenced binding to circPCNX or p21 mRNA; in fact, the impact of AUF1 phosphorylation on the binding and fate of target mRNAs has not been investigated until now. Third, the relative concentration of AUF1 in different subcytoplasmic domains has not been studied for this RBP or for circPCNX or p21 mRNA. The relative abundance of circPCNX in regions where AUF1 binds p21 mRNA may help explain the selective manner in which circPCNX reduces AUF1 binding to p21 mRNA, but the necessary detection tools are not available at present. Finally, while circPCNX harbors a single AUF1 binding site (Figure 5A), AUF1 was previously shown to multimerize, and thus one circPCNX copy could potentially interact with multiple AUF1 molecules (48–50). Given the limitations of quantifying the relative ratios of circPCNX and AUF1 molecules in the cell, we focused on the empirical consequences of lowering or overexpressing each one. Through careful analysis, we discovered that elevating circPCNX caused derepression of p21 mRNA by AUF1, in turn raising p21 levels and decreasing cell division.
Given that circPCNX suppressed cell proliferation, we explored its possible influence in cell processes and diseases with a key component of cell division, particularly those implicating p21, such as cell senescence and cancer. Disappointingly, we did not detect significant changes in circPCNX levels in human diploid fibroblasts rendered senescent by replicative exhaustion (not shown) or in HeLa cells arrested in the G1 phase of the cell cycle (Supplemental Figures S8D, S8E), despite the fact that a rise in p21 levels contributes to senescence-associated growth arrest. Hence, these findings support the view that an elevation in circPCNX can trigger growth arrest, but not all growth arrest paradigms induce circPCNX levels. We then hypothesized that circPCNX might be a tumor suppressor, given that its elevation increased p21 levels and arrested cell division. Microarray analysis of circRNA expression profiles in bladder cancer, acute myeloid leukemia and hepatocellular carcinoma revealed low albeit insignificant levels of circPCNX (51–53), while in colorectal cancer, gastric cancer and cutaneous squamous cell carcinoma circPCNX levels were elevated (54–56). Whether high levels of circPCNX are functionally linked to these cancer phenotypes remains to be tested. Perhaps the anti-apoptotic function of p21 could in some cases promote tumor growth (57–59), and thus the possibility that circPCNX could help confer resistance to tumor growth awaits further study. Finally, one study showed that circPCNX levels were significantly higher in intervertebral disc degeneration (60). The function of circPCNX in this pathology warrants future investigation, perhaps through analysis that might include other nervous system dysfunctions. To close, the knowledge of circRNAs expressed in different developmental processes and diseases is accumulating rapidly. Elucidating the molecular partners, including RBPs, through which circRNAs influence cell function, as shown here for circPCNX, AUF1 and p21, will be critical for developing effective interventions.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the NIA-IRP and the NCI-IRP, NIH. We thank Dr R. Moaddel and C. Dunn (NIA) for assisting with this manuscript.
Contributor Information
Dimitrios Tsitsipatis, Laboratory of Genetics and Genomics, National Institute on Aging (NIA) Intramural Research Program (IRP), National Institutes of Health (NIH), Baltimore, MD, USA.
Ioannis Grammatikakis, Regulatory RNAs and Cancer Section, Genetics Branch, Center for Cancer Research, National Cancer Institute IRP, NIH, Bethesda, MD, USA.
Riley K Driscoll, Laboratory of Genetics and Genomics, National Institute on Aging (NIA) Intramural Research Program (IRP), National Institutes of Health (NIH), Baltimore, MD, USA.
Xiaoling Yang, Laboratory of Genetics and Genomics, National Institute on Aging (NIA) Intramural Research Program (IRP), National Institutes of Health (NIH), Baltimore, MD, USA.
Kotb Abdelmohsen, Laboratory of Genetics and Genomics, National Institute on Aging (NIA) Intramural Research Program (IRP), National Institutes of Health (NIH), Baltimore, MD, USA.
Sophia C Harris, Laboratory of Genetics and Genomics, National Institute on Aging (NIA) Intramural Research Program (IRP), National Institutes of Health (NIH), Baltimore, MD, USA.
Jen-Hao Yang, Laboratory of Genetics and Genomics, National Institute on Aging (NIA) Intramural Research Program (IRP), National Institutes of Health (NIH), Baltimore, MD, USA.
Allison B Herman, Laboratory of Genetics and Genomics, National Institute on Aging (NIA) Intramural Research Program (IRP), National Institutes of Health (NIH), Baltimore, MD, USA.
Ming-Wen Chang, Laboratory of Genetics and Genomics, National Institute on Aging (NIA) Intramural Research Program (IRP), National Institutes of Health (NIH), Baltimore, MD, USA.
Rachel Munk, Laboratory of Genetics and Genomics, National Institute on Aging (NIA) Intramural Research Program (IRP), National Institutes of Health (NIH), Baltimore, MD, USA.
Jennifer L Martindale, Laboratory of Genetics and Genomics, National Institute on Aging (NIA) Intramural Research Program (IRP), National Institutes of Health (NIH), Baltimore, MD, USA.
Krystyna Mazan-Mamczarz, Laboratory of Genetics and Genomics, National Institute on Aging (NIA) Intramural Research Program (IRP), National Institutes of Health (NIH), Baltimore, MD, USA.
Supriyo De, Laboratory of Genetics and Genomics, National Institute on Aging (NIA) Intramural Research Program (IRP), National Institutes of Health (NIH), Baltimore, MD, USA.
Ashish Lal, Regulatory RNAs and Cancer Section, Genetics Branch, Center for Cancer Research, National Cancer Institute IRP, NIH, Bethesda, MD, USA.
Myriam Gorospe, Laboratory of Genetics and Genomics, National Institute on Aging (NIA) Intramural Research Program (IRP), National Institutes of Health (NIH), Baltimore, MD, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute on Aging (NIA) [Z01-AG000511-20]; National Cancer Institute (NCI) [ZIA BC 011646]. Funding for open access charge: NIA [Z01-AG000511-20].
Conflict of interest statement. None declared.
REFERENCES
- 1. Brewer G. An A + U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol. Cell. Biol. 1991; 11:2460–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang W., Wagner B.J., Ehrenman K., Schaefer A.W., DeMaria C.T., Crater D., DeHaven K., Long L., Brewer G.. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol. 1993; 13:7652–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. White E.J., Brewer G., Wilson G.M.. Post-transcriptional control of gene expression by AUF1: mechanisms, physiological targets, and regulation. Biochim. Biophys. Acta. 2013; 1829:680–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gratacos F.M., Brewer G.. The role of AUF1 in regulated mRNA decay. Wiley Interdiscip. Rev. RNA. 2010; 1:457–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liao B., Hu Y., Brewer G.. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat. Struct. Mol. Biol. 2007; 14:511–518. [DOI] [PubMed] [Google Scholar]
- 6. Panda A.C., Abdelmohsen K., Yoon J.H., Martindale J.L., Yang X., Curtis J., Mercken E.M., Chenette D.M., Zhang Y., Schneider R.J.et al.. RNA-binding protein AUF1 promotes myogenesis by regulating MEF2C expression levels. Mol. Cell. Biol. 2014; 34:3106–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pont A.R., Sadri N., Hsiao S.J., Smith S., Schneider R.J.. mRNA decay factor AUF1 maintains normal aging, telomere maintenance, and suppression of senescence by activation of telomerase transcription. Mol. Cell. 2012; 47:5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tolnay M., Baranyi L., Tsokos G.C.. Heterogeneous nuclear ribonucleoprotein D0 contains transactivator and DNA-binding domains. Biochem. J. 2000; 348:151–158. [PMC free article] [PubMed] [Google Scholar]
- 9. Abdelmohsen K., Kuwano Y., Kim H.H., Gorospe M.. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol. Chem. 2008; 389:243–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yoon J.H., De S., Srikantan S., Abdelmohsen K., Grammatikakis I., Kim J., Kim K.M., Noh J.H., White E.J., Martindale J.L.et al.. PAR-CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity. Nat. Commun. 2014; 5:5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hombach S., Kretz M.. Non-coding RNAs: classification, biology and functioning. Adv. Exp. Med. Biol. 2016; 937:3–17. [DOI] [PubMed] [Google Scholar]
- 12. Hsu M.T., Coca-Prados M.. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979; 280:339–340. [DOI] [PubMed] [Google Scholar]
- 13. Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M.et al.. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013; 495:333–338. [DOI] [PubMed] [Google Scholar]
- 14. Yu C.Y., Kuo H.C.. The emerging roles and functions of circular RNAs and their generation. J. Biomed. Sci. 2019; 26:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O.. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012; 7:e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Panda A.C., Grammatikakis I., Kim K.M., De S., Martindale J.L., Munk R., Yang X., Abdelmohsen K., Gorospe M.. Identification of senescence-associated circular RNAs (SAC-RNAs) reveals senescence suppressor CircPVT1. Nucleic Acids Res. 2017; 45:4021–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O.. Cell-type specific features of circular RNA expression. PLoS Genet. 2013; 9:e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Panda A.C., Grammatikakis I., Munk R., Gorospe M., Abdelmohsen K.. Emerging roles and context of circular RNAs. Wiley Interdiscip. Rev. RNA. 2017; 8:e1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pandey P.R., Yang J.H., Tsitsipatis D., Panda A.C., Noh J.H., Kim K.M., Munk R., Nicholson T., Hanniford D., Argibay D.et al.. circSamd4 represses myogenic transcriptional activity of PUR proteins. Nucleic Acids Res. 2020; 48:3789–3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vo J.N., Cieslik M., Zhang Y., Shukla S., Xiao L., Zhang Y., Wu Y.M., Dhanasekaran S.M., Engelke C.G., Cao X.et al.. The landscape of circular RNA in cancer. Cell. 2019; 176:869–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xia X., Tang X., Wang S.. Roles of CircRNAs in autoimmune diseases. Front. Immunol. 2019; 10:639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Floris G., Zhang L., Follesa P., Sun T.. Regulatory role of circular RNAs and neurological disorders. Mol. Neurobiol. 2017; 54:5156–5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Akhter R. Circular RNA and Alzheimer's disease. Adv. Exp. Med. Biol. 2018; 1087:239–243. [DOI] [PubMed] [Google Scholar]
- 24. Holdt L.M., Kohlmaier A., Teupser D. Circular RNAs as therapeutic agents and targets. Front Physiol. 2018; 9:1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kristensen L.S., Hansen T.B., Veno M.T., Kjems J.. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018; 37:555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dudekula D.B., Panda A.C., Grammatikakis I., De S., Abdelmohsen K., Gorospe M.. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016; 13:34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wojciechowska M., Sobczak K., Kozlowski P., Sedehizadeh S., Wojtkowiak-Szlachcic A., Czubak K., Markus R., Lusakowska A., Kaminska A., Brook J.D.. Quantitative methods to monitor RNA biomarkers in myotonic dystrophy. Sci. Rep. 2018; 8:5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kanno J., Aisaki K., Igarashi K., Nakatsu N., Ono A., Kodama Y., Nagao T.. “Per cell” normalization method for mRNA measurement by quantitative PCR and microarrays. BMC Genomics. 2006; 7:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S.. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014; 56:55–66. [DOI] [PubMed] [Google Scholar]
- 30. Wang W., Furneaux H., Cheng H., Caldwell M.C., Hutter D., Liu Y., Holbrook N., Gorospe M.. HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 2000; 20:760–769.10629032 [Google Scholar]
- 31. Abdelmohsen K., Panda A.C., Munk R., Grammatikakis I., Dudekula D.B., De S., Kim J., Noh J.H., Kim K.M., Martindale J.L.et al.. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017; 14:361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Penalva L.O., Tenenbaum S.A., Keene J.D.. Gene expression analysis of messenger RNP complexes. Methods Mol. Biol. 2004; 257:125–134. [DOI] [PubMed] [Google Scholar]
- 33. Chen G., Deng X.. Cell synchronization by double thymidine block. Bio Protoc. 2018; 8:e2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Indig F.E., Diaz-Gonzalez F., Ginsberg M.H.. Analysis of the tetraspanin CD9-integrin alphaIIbbeta3 (GPIIb-IIIa) complex in platelet membranes and transfected cells. Biochem. J. 1997; 327:291–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Abdelmohsen K., Panda A., Kang M.J., Xu J., Selimyan R., Yoon J.H., Martindale J.L., De S., Wood W.H. 3rd, Becker K.G.et al.. Senescence-associated lncRNAs: senescence-associated long noncoding RNAs. Aging Cell. 2013; 12:890–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cathcart A.L., Rozovics J.M., Semler B.L.. Cellular mRNA decay protein AUF1 negatively regulates enterovirus and human rhinovirus infections. J. Virol. 2013; 87:10423–10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gao X., Dong H., Lin C., Sheng J., Zhang F., Su J., Xu Z.. Reduction of AUF1-mediated follistatin mRNA decay during glucose starvation protects cells from apoptosis. Nucleic Acids Res. 2014; 42:10720–10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wilson G.M., Lu J., Sutphen K., Suarez Y., Sinha S., Brewer B., Villanueva-Feliciano E.C., Ysla R.M., Charles S., Brewer G.. Phosphorylation of p40AUF1 regulates binding to A + U-rich mRNA-destabilizing elements and protein-induced changes in ribonucleoprotein structure. J. Biol. Chem. 2003; 278:33039–33048. [DOI] [PubMed] [Google Scholar]
- 39. Blum J.L., Samarel A.M., Mestril R.. Phosphorylation and binding of AUF1 to the 3′-untranslated region of cardiomyocyte SERCA2a mRNA. Am. J. Physiol. Heart Circ. Physiol. 2005; 289:H2543–2550. [DOI] [PubMed] [Google Scholar]
- 40. Lal A., Mazan-Mamczarz K., Kawai T., Yang X., Martindale J.L., Gorospe M.. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004; 23:3092–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Niculescu A.B. 3rd, Chen X., Smeets M., Hengst L., Prives C., Reed S.I.. Effects of p21(Cip1/Waf1) at both the G1/S and the G2/M cell cycle transitions: pRb is a critical determinant in blocking DNA replication and in preventing endoreduplication. Mol. Cell. Biol. 1998; 18:629–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ogryzko V.V., Wong P., Howard B.H.. WAF1 retards S-phase progression primarily by inhibition of cyclin-dependent kinases. Mol. Cell. Biol. 1997; 17:4877–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tani H., Mizutani R., Salam K.A., Tano K., Ijiri K., Wakamatsu A., Isogai T., Suzuki Y., Akimitsu N.. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 2012; 22:947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hao Q., Zhao X., Zhang Y., Dong Z., Hu T., Chen P.. Targeting overexpressed activating transcription factor 1 (ATF1) inhibits proliferation and migration and enhances sensitivity to paclitaxel in esophageal cancer cells. Med. Sci. Monit. Basic Res. 2017; 23:304–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Du W.W., Zhang C., Yang W., Yong T., Awan F.M., Yang B.B.. Identifying and characterizing circRNA-Protein interaction. Theranostics. 2017; 7:4183–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Panda A.C. Circular RNAs act as miRNA sponges. Adv. Exp. Med. Biol. 2018; 1087:67–79. [DOI] [PubMed] [Google Scholar]
- 47. Hanniford D., Ulloa-Morales A., Karz A., Berzoti-Coelho M.G., Moubarak R.S., Sanchez-Sendra B., Kloetgen A., Davalos V., Imig J., Wu P.et al.. Epigenetic silencing of CDR1as drives IGF2BP3-mediated melanoma invasion and metastasis. Cancer Cell. 2020; 37:55–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wilson G.M., Sun Y., Lu H., Brewer G.. Assembly of AUF1 oligomers on U-rich RNA targets by sequential dimer association. J. Biol. Chem. 1999; 274:33374–33381. [DOI] [PubMed] [Google Scholar]
- 49. DeMaria C.T., Sun Y., Long L., Wagner B.J., Brewer G.. Structural determinants in AUF1 required for high affinity binding to A + U-rich elements. J. Biol. Chem. 1997; 272:27635–27643. [DOI] [PubMed] [Google Scholar]
- 50. DeMaria C.T., Brewer G.. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J. Biol. Chem. 1996; 271:12179–12184. [DOI] [PubMed] [Google Scholar]
- 51. Fu L., Yao T., Chen Q., Mo X., Hu Y., Guo J.. Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget. 2017; 8:58405–58416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li W., Zhong C., Jiao J., Li P., Cui B., Ji C., Ma D. Characterization of hsa_circ_0004277 as a new biomarker for acute myeloid leukemia via circular RNA profile and bioinformatics analysis. Int. J. Mol. Sci. 2017; 18:597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhong Z., Lv M., Chen J.. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci. Rep. 2016; 6:30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen Z., Ren R., Wan D., Wang Y., Xue X., Jiang M., Shen J., Han Y., Liu F., Shi J.et al.. Hsa_circ_101555 functions as a competing endogenous RNA of miR-597-5p to promote colorectal cancer progression. Oncogene. 2019; 38:6017–6034. [DOI] [PubMed] [Google Scholar]
- 55. Shao Y., Li J., Lu R., Li T., Yang Y., Xiao B., Guo J.. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017; 6:1173–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sand M., Bechara F.G., Gambichler T., Sand D., Bromba M., Hahn S.A., Stockfleth E., Hessam S.. Circular RNA expression in cutaneous squamous cell carcinoma. J. Dermatol. Sci. 2016; 83:210–218. [DOI] [PubMed] [Google Scholar]
- 57. Abbas T., Dutta A.. p21 in cancer: intricate networks and multiple activities. Nat. Rev. Cancer. 2009; 9:400–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Roninson I.B. Oncogenic functions of tumour suppressor p21(Waf1/Cip1/Sdi1): association with cell senescence and tumour-promoting activities of stromal fibroblasts. Cancer Lett. 2002; 179:1–14. [DOI] [PubMed] [Google Scholar]
- 59. Georgakilas A.G., Martin O.A., Bonner W.M.. p21: a two-faced genome guardian. Trends Mol. Med. 2017; 23:310–319. [DOI] [PubMed] [Google Scholar]
- 60. Liu X., Che L., Xie Y.K., Hu Q.J., Ma C.J., Pei Y.J., Wu Z.G., Liu Z.H., Fan L.Y., Wang H.Q.. Noncoding RNAs in human intervertebral disc degeneration: An integrated microarray study. Genom Data. 2015; 5:80–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.