Abstract
Argonaute (Ago) proteins are conserved nucleic acid-guided proteins present in all domains of life. Eukaryotic Argonaute proteins (eAgos) are key players in RNA interference pathways and function as RNA-guided RNA endonucleases at physiological temperatures. Although eAgos are considered to evolve from prokaryotic Argonaute proteins (pAgos), previously studied pAgos were unable to catalyze RNA-guided RNA cleavage at physiological temperatures. Here, we describe a distinctive pAgo from mesophilic bacteria Kurthia massiliensis (KmAgo). KmAgo utilizes DNA guides to cleave single-stranded DNA (ssDNA) and RNA targets with high activity. KmAgo also utilizes RNA guides to cleave ssDNA and RNA targets at moderate temperatures. We show that KmAgo can use 5′ phosphorylated DNA guides as small as 9-mers to cut ssDNA and RNA, like Clostridium butyricum Ago. Small DNA binding confers remarkable thermostability on KmAgo, and we can suppress the guide-independent plasmid processing activity of empty KmAgo by elevating the DNA guide loaded temperature. Moreover, KmAgo performs programmable cleavage of double-stranded DNA and highly structured RNA at 37°C. Therefore, KmAgo can be regarded as a DNA-guided programmable omnipotent nuclease for cleaving most types of nucleic acids efficiently. This study broadens our understanding of Ago proteins and could expand the pAgo-based DNA and RNA manipulation toolbox.
INTRODUCTION
Argonaute (Ago) proteins, present in all three domains of life, use small DNA or RNA guides to recognize and sometimes cleave complementary nucleic acid targets (1), which are similar to CRISPR–Cas proteins. As the catalytic engine of eukaryotic RNA interference (RNAi) machinery, eukaryotic Argonaute proteins (eAgos) bind small non-coding RNA molecules as guides to direct the RNA-induced silencing complex (RISC) toward complementary RNA targets (2,3). Accordingly, eAgos in eukaryotes participate in antivirus defense systems and regulatory processes (4). Despite recent studies showing that prokaryotic Argonaute proteins (pAgos) may function in the DNA replication and provide cell defense against foreign genetic elements in vivo (5–8), the role of pAgos in bacteria and archaea (which lack RNAi pathways) remains poorly understood.
Genomic studies showed that the diversity of pAgos is far greater than that of eAgos (9). pAgos can be classified into long pAgo clade (further subdivided into two clades, long-A and long-B), short pAgo clade and PIWI-RE clade (9,10). All pAgos characterized so far belong to the long pAgo clade and they share a high degree of structural homology with eAgos (11). Both eAgos and long pAgos adopt the same four-domain architecture, including the N-terminal (N), PIWI-Argonaute-Zwille (PAZ), middle (MID) and catalytic RNase H-like P element-induced wimpy testis (PIWI) domains [except for AfAgo (Archaeoglobus fulgidus) that only has MID and PIWI domains] (6). The 5′ and 3′ ends of a nucleic acid guide molecule are anchored in the MID and PAZ domains, respectively. Agos with complementary DEDX (X = N, D or H) catalytic tetrad in PIWI domains possess endonuclease activity (12) and cleave the complementary target at a site between the 10′ and 11′ nucleotides starting from the 5′ end of the guide (13–15). Despite their structural homology, the functional roles and guide and/or target preferences of characterized pAgos are more divergent compared to those of eAgos.
While eAgos prefer to bind small RNA guides and exclusively cleave RNA targets, early structural and biochemical studies of thermophilic pAgos showed that they utilize DNA guides to cleave single-stranded DNA (ssDNA) or RNA targets, including AaAgo (Aquifex aeolicus) (16), TtAgo (Thermus thermophilus) (17), PfAgo (Pyrococcus furiosus) (18) and MjAgo (Methanocaldococcus jannaschii) (19,20). The subsequent studies demonstrated that both TtAgo and PfAgo can be programmed with DNA guides to cleave double-stranded DNA (dsDNA) targets at elevated temperatures (≥65°C) (18,21). However, MpAgo (Marinitoga piezophila) was reported to use small RNA guides to cleave DNA and RNA substrates (12). In analogy with the Clustered Regularly Interspaced Short Palindromic Repeats-CRISPR associated (CRISPR–Cas) enzymes, pAgos may potentially be used in nucleic acid detection (22,23), molecular cloning (24) and genome editing applications (10). However, all the pAgos mentioned above are from thermophilic prokaryotes and they only have low levels of DNA-guided ssDNA and RNA cleavage activity at moderate temperatures (20–50°C) (16,18,21), which limits the application of pAgos. Recent studies focus on the pAgos of mesophilic prokaryotes to search for a pAgo that can cleave dsDNA at moderate temperatures. Some pAgos from mesophilic prokaryotes can catalyze DNA-guided dsDNA cleavage at 37°C, including CbAgo (Clostridium butyricum), LrAgo (Limnothrix rosea), CpAgo (Clostridium perfringens) and IbAgo (Intestinibacter bartlettii) (25–27). Furthermore, both thermophilic pAgos and mesophilic pAgos perform guide-independent ‘chopping’ of dsDNA, including TtAgo, MjAgo, LrAgo, CbAgo and SeAgo (Synechococcus elongatus) (19,25,28,29). Except for NgAgo (Natronobacterium gregory) that can only catalyze DNA-dependent RNA target cleavage and CbAgo that can utilize RNA guides to cleave DNA targets at 37°C, the characterized mesophilic pAgos catalyze preferentially DNA-guided DNA target cleavage at moderate temperatures (25–27,29–31).
Although eAgos are considered to originate from pAgos, eAgos catalyze preferentially RNA-guided RNA cleavage at moderate temperature (1,5). The quest for a pAgo that can utilize RNA guides to cleave RNA targets at moderate temperatures is intriguing. We here report the characterization of the Ago protein from the mesophilic bacteria Kurthia massiliensis (KmAgo) that can utilize short DNA and RNA as guides to cleave both ssDNA and RNA targets at moderate temperatures. In addition, we demonstrate that KmAgo can perform precise guide-dependent cleavage of both dsDNA and highly structured RNA at 37°C. The programmable DNA and RNA endonuclease activity of KmAgo broadens our understanding of Ago proteins and could advance the development of pAgo-based applications at moderate temperatures.
MATERIALS AND METHODS
Protein expression and purification
The KmAgo gene (WP_010289662.1; K. massiliensis) and KmAgo double mutant (KmAgo_DM) (D527A, D596A) gene were synthesized by Wuhan Genecreate Biotechnology Co., Ltd and cloned into pET28a expression vectors in frame with the N-terminal 6× His tag. The KmAgo and KmAgo_DM proteins were expressed in Escherichia coli BL21(DE3) (Novagen). Cultures were grown at 37°C in Luria-Bertani (LB) medium containing 50 μg/ml kanamycin and until OD600 reached 0.8. KmAgo expression was induced by addition of isopropyl-β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. During the expression, cells were incubated at 18°C for 20 h with continuous shaking. The cells were collected by centrifugation and stored at −80°C for further protein purification.
Cells were lysed by sonication (SCIENTZ-IID, 400 W, 2 s on/4 s off for 15 min) in Buffer A containing 20 mM Tris–HCl, pH 7.5, 500 mM NaCl, 10 mM imidazole, supplemented with an ethylenediaminetetraacetic acid (EDTA)-free protease inhibitor cocktail tablet (Roche). The lysate was clarified by centrifugation at 18 000 rpm for 50 min and the supernatant was bound to Ni-NTA agarose resin, washed with 9 column volumes Buffer A and subsequently extensively washed with Buffer A containing 60 mM imidazole. Bound protein was eluted in Buffer A containing 300 mM imidazole. The eluted protein was concentrated against Buffer B [20 mM N-2-Hydroxyethylpiperazine-N-2-Ethane Sulfonic Acid (HEPES), pH 7.5, 0.5 M NaCl and 1 mM dithiothreitol (DTT)] by ultrafiltration using an Amicon 50K filter unit (Millipore). Next, the protein was diluted in 20 mM HEPES pH 7.5 to lower the final salt concentration to 125 mM NaCl. The diluted protein was applied to a Heparin column (HiTrap Heparin HP, GE Healthcare) equilibrated with Buffer C (20 mM HEPES pH 7.5, 125 mM NaCl and 1 mM DTT), washed with at least 10 column volumes of the same buffer and eluted with a linear NaCl gradient (0.125–2 M). Fractions containing pAgos were concentrated by ultrafiltration using an Amicon 50K filter unit (Millipore). Next, the heparin-purified protein was loaded onto a size exclusion column (Superdex 200 16/600 column, GE Healthcare) and eluted with Buffer D (20 mM HEPES, pH 7.5, 500 mM NaCl and 1 mM DTT). Purified KmAgo was diluted in Buffer D to a final concentration of 8 μM. Aliquots were flash-frozen in liquid nitrogen and stored at −80°C.
Preparation of HIV-1 ΔDIS 5′UTR transcript and its guide DNAs
Dayeh et al. reported that HIV-1 ΔDIS 5′UTR is a highly structured RNA (see Supplementary Table S2) (32). The HIV-1 ΔDIS 5′UTR was in vitro transcribed using T7 RNA polymerase and synthetic DNA templates carrying a T7 promoter sequence. Transcripts used for cleavage assays was DNase I-treated and gel-purified. Guide DNAs (gDNAs) for experiments targeting the HIV-1 ΔDIS 5′UTR sequence (Supplementary Table S3) were designed as described in (32) with minor modifications. Briefly, Dayeh et al. designed 14 gDNAs (length was 23 nt) for targeting the different regions of HIV-1 ΔDIS 5′UTR. We selected the target regions of gDNA_4 to gDNA_14 and designed 11 gDNAs (length was 18 nt) to guide KmAgo cleavage at the corresponding sites.
Single-stranded activity assays
Most cleavage assays were performed at the 4:2:1 pAgo:guide:target molar ratio at 37°C, unless otherwise indicated. 800 nM KmAgo was mixed with 400 nM guide in reaction buffer containing 10 mM HEPES–NaOH pH 7.5, 100 mM NaCl, 5 mM MnCl2, 5% glycerol and incubated at 37°C for 10 min for guide loading. Nucleic acid target was added to 200 nM. The reaction was stopped after indicated time intervals by mixing the samples with 2× RNA loading dye (95% formamide, 18 mM EDTA, 0.025% SDS and 0.025% bromophenol blue) and heating it for 5 min at 95°C. The cleavage products were resolved by 20% denaturing polyacrylamide gel electrophoresis (PAGE), stained with SYBR Gold (Invitrogen), visualized with Gel Doc™ XR+ (Bio-Rad), and analyzed by the ImageJ and Origin software. For analysis of temperature dependence of target cleavage, all samples were incubated at indicated temperatures simultaneously using a polymerase chain reaction (PCR) thermocycler (T100, Bio-Rad).
Fluorescently labeled ssDNA or RNA targets (synthesized by Genscript) were used to analyze the specificity of the nucleic acid of KmAgo. For activity assays, 800 nM purified KmAgo, 400 nM ssDNA or RNA guide were mixed in reaction buffer and incubated at 37°C for 10 min. Then 200 nM fluorescently labeled ssDNA or RNA target was added to the mixture. After incubation at 37°C for 60 min, the reactions were stopped via the addition of 2× RNA Loading dye at a 1:1 ratio (v/v). The samples were resolved on 20% denaturing PAGE, visualized with Gel Doc™ XR+ (Bio-Rad). All the nucleic acids used in this study are listed in Supplementary Table S1.
Electrophoretic mobility shift assay (EMSA)
To examine the loading of guide onto KmAgo, KmAgo and 3′ end FAM-labeled guide were incubated in 20 μl of reaction buffer containing 10 mM HEPES–NaOH pH 7.5, 100 mM NaCl, 5 mM metal (MnCl2, MgCl2) or EDTA, respectively, for 30 min at 37°C. The concentration of the 3′ end FAM-labeled guide was fixed as 128 nM, whereas the concentration of KmAgo varied. Then the samples were mixed with 2.2 μl 10× loading buffer (250 mM Tris–HCl pH 7.5, 40% glycerol) and resolved by 10% native PAGE with 0.5× Tris-Borate-EDTA (TBE) buffer. Nucleic acids were visualized using Gel Doc™ XR+. All the nucleic acids used in this study are listed in Supplementary Table S1.
To analyze the loading of the guide-target duplex, reactions contained KmAgo_DM, 128 nM 3′ end FAM-labeled guide, 64 nM 5′ end FAM-labeled target, HEPES–NaOH pH 7.5, 100 mM NaCl, 5 mM metal (MnCl2, MgCl2) or EDTA, respectively, were combined for 20 μl and incubated for 30 min at 37°C. The concentration of KmAgo_DM varied. Then the samples were mixed with 2.2 μl 10× loading buffer (250 mM Tris–HCl pH 7.5, 40% glycerol) and resolved by 10% native PAGE with 0.5× TBE. Nucleic acids were visualized using Gel Doc™ XR+. All the nucleic acids used in this study are listed in Supplementary Table S1.
Thermostability assay
Nucleic acids isolated experiments demonstrated that the purified KmAgo is essentially free of associated nucleic acids. To analyze thermostability of the KmAgo-guide duplex, 8 pmol KmAgo and 10 pmol guide were preincubated at 37°C for 10 min before treated at 37, 45, 50, 55, 60, 65 or 70°C for 30 min, respectively. Following incubated at 37°C for 10 min, the KmAgo-guide duplex were used to cleave 4 pmol DNA target at 37°C for 15 min. To analyze thermostability of the empty KmAgo, 8 pmol KmAgo was preincubated at 37°C for 10 min before treated at different temperatures for 30 min. Then the treated KmAgo was incubated with 10 pmol guide at 37°C for 10 min to form KmAgo-guide duplex, and the KmAgo-guide duplex was used to cleave 4 pmol DNA target at 37°C for 15 min. The cleavage products were assayed by 20% denaturing PAGE.
Double-stranded DNA and highly structured RNA activity assay
In two half-reactions, 8 pmol of KmAgo was loaded with either 10 pmol of forward or reverse DNA guide in reaction buffer containing 5 mM HEPES–NaOH pH 7.5, 50 mM NaCl, 0.5 mM MnCl2, 2.5% glycerol. The half-reactions were incubated for 30 min at 55°C. Next, both half-reactions were mixed and 200 ng target plasmid was added, after which the mixture was incubated for 2 h of 37°C. After the incubation, NdeI (NEB) or ScaI-HF (NEB) and Cutsmart buffer (NEB) were added and incubation for 2 h at 37°C. A 6× DNA loading dye (NEB) was added to the plasmid sample prior to resolving it on a 1.0% agarose gel stained with ethidium bromide. In the double-stranded assays in which linear plasmid was targeted, 200 ng pUC19, linearized by NdeI, was added to both half-reactions and incubated for 2 h at 37°C. The reaction products were resolved on a 1.0% agarose gel.
For KmAgo–gDNA duplex formation and HIV-1 ΔDIS 5′UTR cleavage, KmAgo (800 nM) was pre-mixed with gDNA (400 nM) for 10 min at 37°C in a 9 μl reaction followed by addition 1 μl of HIV-1 ΔDIS 5′UTR substrate (final concentration: 250 nM) and incubation at 37°C for 15 min. Reactions were quenched with 2× RNA loading dye and heating it for 5 min at 95°C. The cleavage products were resolved by 20% denaturing PAGE, stained with SYBR Gold, and visualized with Gel Doc™ XR+ (Bio-Rad).
RESULTS
KmAgo uses small DNA and RNA guides for endonucleolytic cleavage of ssDNA and ssRNA targets at ambient temperature
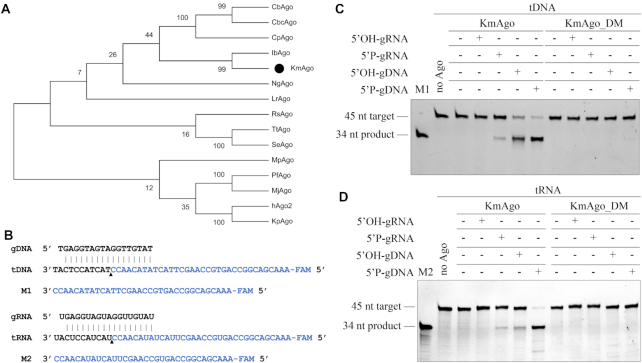
As PfAgo was reported to one of the pAgos most closely related to eAgos (18), we chose the protein sequence of PfAgo (WP_011011654.1) as the query and used the web interface of the BLASTp program to search for mesophilic pAgos. Considering most characterized eAgos come from aerobic organisms, pAgo from aerobic mesophilic bacillus, K. massiliensis (KmAgo), was chosen as the candidate for its low identity (18.30%) compared with PfAgo. Furthermore, despite KmAgo is phylogenetically closest related to the IbAgo (Figure 1A) which from anaerobic bacillus, the sequence identity is only 29.67%. A multiple sequence alignment of KmAgo with other pAgos (Supplementary Figure S1A) suggests that KmAgo contains the conserved DEDD catalytic residues which are essential for nuclease activity in ‘slicing’ Agos (26). In the case of KmAgo, this concerns residues D527, E562, D596 and D713. We obtained the catalytically inactive variants of KmAgo (KmAgo_DM) with substitutions of two out of four catalytic tetrad residues (D527A/D596A, Supplementary Figure S1A).
Figure 1.
KmAgo exhibits DNA-guided and RNA-guided RNA cleavage activity at 37°C. (A) Maximum likelihood phylogenetic tree analysis of KmAgo based on amino acid sequences. The numbers at the nodes indicate the bootstrap values for maximum likelihood analysis of 1000 resampled data sets. (B) Sequence of the synthetic let7 miRNA-based guide and target sequences that were used for the in vitro cleavage assays. Black triangle indicates the cleavage site. (C) Cleavage activity assay with FAM-labeled ssDNA target. (D) Cleavage activity assay with FAM-labeled RNA target. KmAgo, guides and targets were mixed in a 4:2:1 molar ratio (800 nM KmAgo preloaded with 400 nM guide, plus 200 nM target) and incubated for 1 h at 37°C. Catalytic mutant KmAgo_DM was used as a control. Lanes M1 and M2 contain chemically synthesized 5′-end, FAM-labeled, 34-nt ssDNA and RNA corresponding to the cleavage products of target DNA and target RNA, respectively.
To study the biochemical properties of KmAgo, KmAgo and KmAgo_DM were successfully expressed in E. coli using a T7-based pET expression system and purified (Supplementary Figure S1B). To determine if there were copurified nucleic acids, we measured the A260/A280 ratio of the purified KmAgo with Thermo Scientific NanoDrop 8000 Spectrophotometer and the ratio was 0.56. Moreover, we carried out the isolation of copurified nucleic acids as described in (26), and no copurified nucleic acids were detected (data not shown). Both the A260/A280 ratio and nucleic acids isolation results indicated that the purified KmAgo was essentially nucleic-acid free. Then we studied the nucleic acid specificity of KmAgo with in vitro cleavage assay using synthetic fluorescently labeled ssDNA or RNA targets (Figure 1B). KmAgo was loaded with 18 nt DNA or RNA guides containing a 5′ phosphate (5′P) or 5′ hydroxyl (5′OH) group followed by the addition of complementary 5′ end FAM-labeled 45-nt long ssDNA or RNA targets. After incubation for 1 h at 37°C in reaction buffer containing 5 mM Mn2+, the products were resolved on 20% denaturing gel. In reactions containing DNA target (tDNA) and DNA guide (gDNA) or RNA guide(gRNA), KmAgo used 5′ phosphorylated DNA guide (5′P-gDNA) and 5′ phosphorylated RNA guide (5′P-gRNA) to cleave complementary single-stranded DNA targets at a single site between nucleotides 10 and 11 on DNA guide, identical to all previously studied Ago proteins (11), resulting in the appearance of the 34 nt long 5′-fragment of the DNA target (Figure 1C and Supplementary Figure S1C). Strikingly, in reactions containing RNA target (tRNA), KmAgo also used 5′P-gDNA and 5′P-gRNA to cleave complementary single-stranded RNA target between nucleotides 10 and 11 on RNA guide, resulting in the 34 nt long 5′-fragment of the RNA target (Figure 1D and Supplementary Figure S1D). This 5′ phosphorylated RNA-mediated RNA cleavage activity at ambient temperature is same as eAgos (eAgos bind small RNA guides and cleave RNA targets at moderate temperatures) (12,33) and has not been observed in other pAgo homologs (6,25–27,29–31). Surprisingly, in contrast to observations from other eukaryotic and prokaryotic Ago homologs, the efficiency of DNA and RNA cleavage guided with 5′ phosphorylated DNA were almost similar high in ambient temperature. The efficiency of DNA and RNA cleavage guided with 5′ phosphorylated RNA was lower than with 5′ phosphorylated DNA. No cleavage products were observed in the absence of KmAgo protein or guides (Figure 1C and D).
KmAgo cleavage required the intact catalytic tetrad in the PIWI domain, and point mutations in the tetrad eliminated the activity of KmAgo (Figure 1C and D). Though no substrate cleavage was observed with a 5′ hydroxylated RNA guide (5′OH-gRNA), we observed the DNA and RNA cleavage with a 5′hydroxylated DNA guide (5′OH-gDNA) (Figure 1C and D). For DNA target, cleavage not only occurred between target position 10′-11′ relative to the guide 5′ end but also occurred at several positions around the canonical cleavage site (Figure 1C and Supplementary Figure S1C). Changes in the cleavage position were also observed for some other Ago proteins with non-phosphorylated guides, including hAgo2 (Homo sapiens) (33), MjAgo (19), LrAgo (25), CpAgo and IbAgo (27). The absence of 5′P-MID interactions might cause the guide-target duplex sliding in the active site, thus lead to such cleavage site changes (25).
Divalent cations and temperature affect the cleavage activity of KmAgo
Since divalent cations are required for Ago activity (34), we next tested whether divalent metal ion contributes to the discrimination between 5′ phosphorylated DNA guides and 5′phosphorylated RNA guides for guide-dependent target cleavage. In the presence of different divalent metal ions (Mg2+, Ca2+, Mn2+, Fe2+, Co2+, Ni2+, Cu2+ and Zn2+), KmAgo was active to DNA target and RNA target with both Mn2+ and Mg2+ when the guides were 5′ phosphorylated DNA (Supplementary Figure S2A and B). Titration of Mn2+ ions showed that KmAgo was equally active between 0.05 and 10 mM Mn2+, while titration of Mg2+ ions showed that KmAgo required increased Mg2+ concentrations > 0.5 mM (Supplementary Figure S2E and F). When the guides were 5′ phosphorylated RNA, KmAgo was able to utilize Mn2+ as cation to cleave both DNA target and RNA target (Supplementary Figure S2C and D). In contrast to eAgos such as hAgo2 (35), KmAgo was unable to use Mg2+ as cation for its RNA-guided activity. Titration of Mn2+ showed that KmAgo was active when the Mn2+ concentrations increased to 0.5 mM (Supplementary Figure S2G). Considering most pAgos use divalent cation for interaction with the first guide phosphate of 5′ phosphorylated guides (6,26), we performed Electrophoretic Mobility Shift Assay (EMSA) to determine whether KmAgo can load RNA guides with Mg2+. As shown in Supplementary Figure S3, KmAgo was able to bind both 5′P-gRNA and 5′P-gRNA/tRNA duplex in the presence of EDTA, Mn2+ or Mg2+, which suggested that the reason why KmAgo had no RNA-guided catalytic activity with Mg2+ might be that Mg2+ could not activate RNA-guided cleavage activity (6). Furthermore, KmAgo could cleave DNA target with 5′OH-gDNAs when the concentrations of Mg2+ or Mn2+ increased to 0.5 mM (Supplementary Figure S2H, I and J).
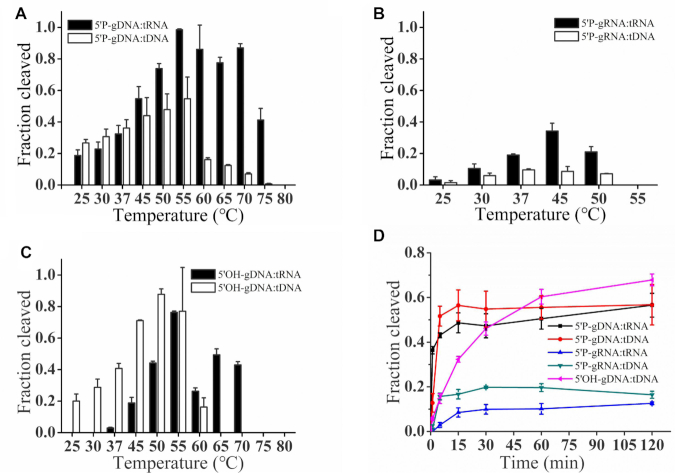
To further explore the full temperature range at which KmAgo is active, we tested the influence of temperature on DNA and RNA cleavage activity mediated by three different kinds of guides at temperatures ranging from 25 to 80°C (Figure 2). The optimum temperatures of KmAgo were significantly different when directed by varied guides: the activity of 5′P-gDNA-mediated DNA cleavage was enhanced with increasing temperature from 25 to 55°C and then decreased rapidly with higher temperature; the 5′P-gDNA-mediated RNA cleavage was most active in the range from 50 to 70°C (Figure 2A). However, distinct to highly stable DNA target, the RNA target could be degraded partially under high temperature (Supplementary Figure S4A). Therefore, we assumed that the robust RNA cleavage activity over 55°C might be caused by the RNA target degradation together with the residual activity of KmAgo under high temperatures. As shown in Figure 2B, the activity of 5′P-gRNA-mediated target cleavage increased at the temperature range from 25 to 45°C, then decreased at higher temperatures and lost at 55°C. Figure 2C showed that the 5′OH-gDNA-mediated DNA cleavage increased at the temperature range from 25 to 55°C, then decreased rapidly at 60°C and lost at higher temperatures. Though there was no activity <37°C when KmAgo used 5′OH-gDNA to cleave RNA, the activity increased rapidly at the temperature range from 37 to 55°C and maintained relatively high activity until 70°C. Similarly, the RNA target in 5′OH-gDNA-mediated reaction could also be degraded partially under high temperature (Supplementary Figure S4), which might contribute to the cleavage activity.
Figure 2.
Effects of temperature on KmAgo activity. (A) Effects of temperature on KmAgo activity mediated by 5′P-gDNA. (B) Effects of temperature on KmAgo activity mediated by 5′P-gRNA. (C) Effects of temperature on KmAgo activity mediated by 5′OH-gDNA. (D) Cleavage kinetics of ssDNA and ssRNA targets using DNA-guided and RNA-guided KmAgo. The assay in (A), (B), (C) was performed in reaction buffer containing Mn2+ ions for 15 min at indicated temperatures. The assay in (D) was performed for indicated times (1, 5, 15, 30, 60, 120 min) at 37°C. Results from three independent experiments were quantified. Error bars represent SD of three independent experiments.
To investigate the catalytic properties of KmAgo mediated by three different guides, we performed a cleavage kinetics assay at 37°C with 5 mM Mn2+ (Figure 2D and Supplementary Figure S5). The fastest reaction rates were observed for 5′ phosphorylated DNA–mediated ssDNA cleavage followed by 5′ phosphorylated DNA-mediated RNA cleavage. The reaction rates of DNA cleavage guided with 5′ hydroxylated DNA was only slightly lower in comparison to 5′ phosphorylated DNA and higher in comparison to 5′ phosphorylated RNA. The cleavage rates of KmAgo using the 5′ phosphorylated RNA guides were the lowest. Therefore, KmAgo prefers to use 5′ phosphorylated DNA as a guide strand.
The sequence of the nucleic acid guide affects KmAgo activity
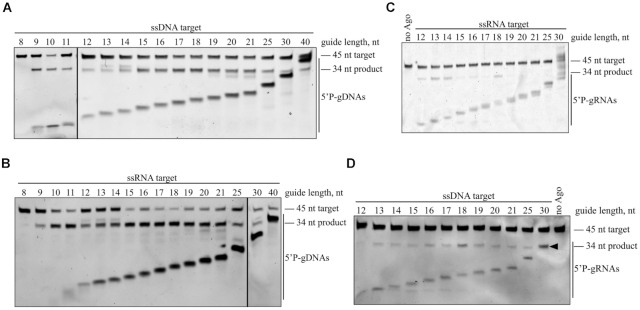
We first investigated the role of the guide length by testing a series of 5′P-gDNAs from 8 to 40 nt long that shared identical sequences at their 5′ ends, so that the predicted cleavage site was the same for all guides. Notably, 9–40 nt guides led to efficient DNA cleavage within 15 min (Figure 3A), like C. butyricum Ago (31). For RNA target, 9–30 nt guides also led to efficient cleavage within 15 min (Figure 3B). Then we tested a series of 5′P-gRNAs from 12 to 30 nt, revealing that gRNAs between 13 and 25 nt long supported similar rates of DNA target cleavage (Figure 3D), but only 12–18 nt long RNA guides led to RNA target cleavage (Figure 3C).
Figure 3.
Effects of the guide length on KmAgo activity. (A) Effects of the 5′P-gDNA length on DNA cleavage activity. (B) Effects of the 5′P-gDNA length on RNA cleavage activity. (C) Effects of the 5′P-gRNA length on RNA cleavage activity. (D) Effects of the 5′P-gRNA length on DNA cleavage activity. Black triangle indicates the 30 nt 5′P-gRNA. All experiments were performed at the 4:2:1 KmAgo:guide:target molar ratio in reaction buffer containing Mn2+ ions for 15 min at 37°C.
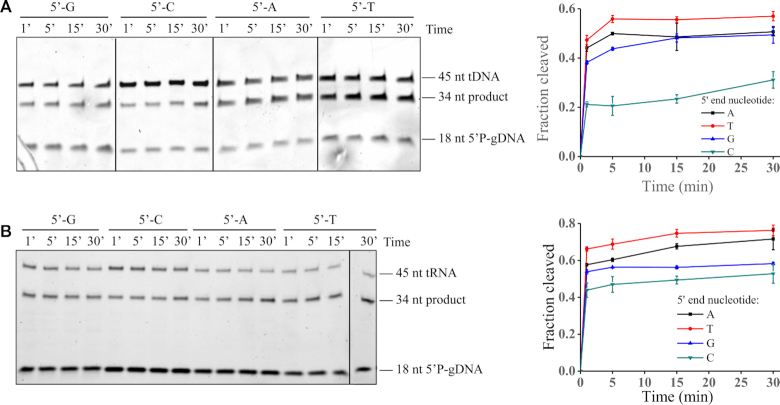
To determine if KmAgo has a preference for the first nucleotide of the guide, we also tested four guide variants with different 5′-terminal nucleotides but otherwise identical sequences (Supplementary Table S1). Firstly, we tested the 45 nt DNA and RNA target cleavage guided with four 5′P-gDNAs. Despite slightly lower cleavage rates were observed when KmAgo was loaded with DNA guides containing a 5′-C, KmAgo loaded with DNA guides containing 5′-T, 5′-A or 5′-G cleaved the target comparably (Figure 4A and B). Then we tested the 45 nt DNA and RNA cleavage guided with four 5′P-gRNAs. Guides beginning with U or A cleaved the RNA comparably whereas RNA guides with a 5′-C displayed lower cleavage (Supplementary Figure S6A). Furthermore, RNA guides with a 5′-G displayed no activity to RNA. In conclusion, KmAgo has no obvious preference for the 5′end nucleotide of the DNA guides, but has a preference for the 5′-U and 5′-A nucleotides of the RNA guides.
Figure 4.
Effects of the 5′-end nucleotide identity on KmAgo activity. (A) Effects of the 5′-end nucleotide of 5′P-gDNA on DNA cleavage activity. (B) Effects of the 5′-end nucleotide of 5′P-gDNA on RNA cleavage activity. Quantification of the cleavage efficiencies is shown on the right. All experiments were performed at the 4:2:1 KmAgo:guide:target molar ratio in reaction buffer containing Mn2+ ions at 37°C. Means and standard deviations from three independent measurements are shown.
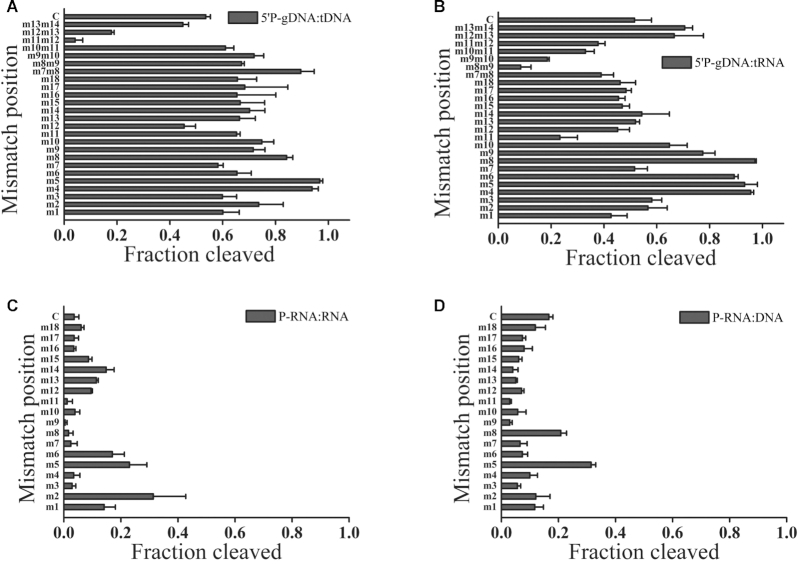
Previous studies of eAgos and several mesophilic pAgos showed that mismatches between the guide and target strands may have large effects on the cleavage efficiency and precision (25,32). To study the effect of mismatches on target cleavage, we designed a set of DNA and RNA guides, each containing a single mismatched nucleotide at a certain position (Supplementary Table S1), and tested them in the cleavage reaction with KmAgo (Figure 5 and Supplementary Figure S7). Similar to LrAgo, mismatches in the seed region (guide positions 2–8) of guides substantially stimulated the activity of KmAgo (Figure 5A) (25). In contrast to the majority of Ago proteins studied to date, mismatches at the site of cleavage (guide positions 10–11) and the 3′-supplementary region of DNA guides (guide positions 13–15) had no significant effect on DNA cleavage, while mismatches at positions 7 and 12 slightly decreased the efficiency of DNA cleavage. However, though mismatches at 3′-supplementary region of DNA guides also had no significant effect on RNA cleavage, a mismatch at the site of cleavage (guide position 11) decreased the efficiency of RNA cleavage (Figure 5B). To further evaluate the effect of mismatches between 5′P-gDNA and target strands, we systematically introduced dinucleotide mismatches to the gDNA spanning an 8-nt window (guide positions 7–14) flanking the cleavage site (Supplementary Table S1). Despite dinucleotide mismatches to positions between g7 and g11 stimulated the DNA cleavage activity of KmAgo, dinucleotide mismatches to positions between g11 and g13 dramatically reduced DNA cleavage (Figure 5A). Dinucleotide mismatches to positions between g8 and g12 dramatically reduced RNA cleavage, but dinucleotide mismatches to positions between g12 and g14 slightly increased the RNA cleavage activity (Figure 5B). Interestingly, single-nucleotide mismatch to positions between g10 and g15 of 5′P-gRNAs strongly decreased the DNA cleavage activity of KmAgo (Figure 5D). Moreover, despite mismatches at the cleavage site (guide positions 11) and guide positions 7–9 of 5′P-gRNAs almost lost the RNA cleavage activity, mismatches in the 3′-supplementary region (guide positions 12–15) increased the RNA cleavage activity (Figure 5C).
Figure 5.
Effects of mismatches in the guide-target duplex on the slicing activity of KmAgo. (A) Effects of single- and dinucleotide mismatches in the 5′P-gDNA:tDNA duplex on the slicing activity of KmAgo. (B) Effects of single- and dinucleotide mismatches in the 5′P-gDNA–tRNA duplex on the slicing activity of KmAgo. (C) Effects of single-nucleotide mismatches in the 5′P-gRNA:tRNA duplex on the slicing activity of KmAgo. (D) Effects of single-nucleotide mismatches in the 5′P-gRNA:tDNA duplex on the slicing activity of KmAgo. All experiments were performed at the 4:2:1 KmAgo:guide:target molar ratio in reaction buffer containing Mn2+ ions at 37°C. Error bars represent SD of three independent experiments.
KmAgo is stabilized by small DNA binding
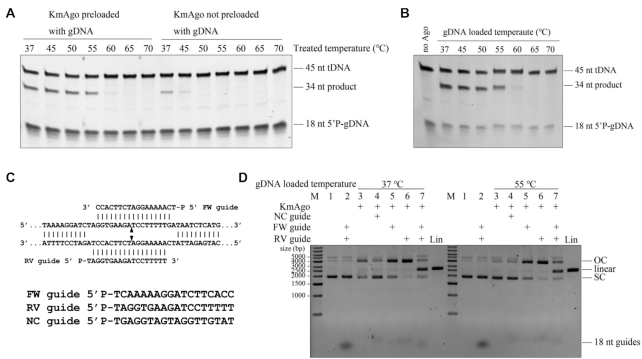
Small RNA binding confers remarkable stability on hAgo2 (36). We have successfully purified KmAgo essentially without endogenous nucleic acids. To investigate the thermostability of empty KmAgo and KmAgo–gDNA duplex, we treated them at different temperatures for 30 min before tested their DNA target cleavage activity respectively. Increasing treated temperature up to 50°C resulted in KmAgo not preloaded with gDNA lost its DNA cleavage activity (Figure 6A). In contrast, KmAgo preloaded with gDNA was still able to efficiently cleave DNA target when treated temperature increased to 55°C (Figure 6A). The obviously enhanced thermostability of KmAgo loaded with gDNA indicated that KmAgo is stabilized by small DNA binding. We further investigated the effects of temperature of KmAgo–gDNA duplex formation stage (gDNA loaded temperature) on DNA target cleavage activity (Figure 6B). KmAgo–gDNA duplex could efficiently cleave ssDNA target when gDNA loaded temperature increased to 55°C, which suggested that the duplex formed very fast.
Figure 6.
Small DNAs stabilize KmAgo. (A) KmAgo preloaded with gDNA tolerates higher treated temperature. KmAgo preloaded with gDNA, KmAgo and gDNA were incubated at 37°C for 10 min before treated at indicated temperature for 30 min, and then incubated at 37°C for 10 min; KmAgo not preloaded with gDNA, KmAgo was incubated at 37°C for 10 min before treated at indicated temperature for 30 min, and then incubated with gDNA at 37°C for 10 min. (B) Effects of gDNA loaded temperatures on KmAgo activity. All cleavage experiments in (A) and (B) were performed at the 4:5:1 KmAgo:guide:target molar ratio in reaction buffer containing Mn2+ ions at 37°C for 15 min. (C) Schematic of the sequences of the 5′ phosphorylated DNA guides. (D) The effects of KmAgo–gDNA duplex loaded temperature on plasmid cleavage activity. Plasmid cleavage assay performed by loading KmAgo with the indicated 5′P-gDNAs at 37°C or 55°C for 30 min, followed by incubation with the target plasmid at 37°C for 2 h, and analysis of the target plasmid by electrophoresis. gDNA loaded temperature, the temperature of KmAgo–gDNA duplex formation stage. M, molecular weight marker; Lin, linearized plasmid; SC, supercoiled plasmid; OC, open circular plasmid. See also Supplementary Figure S8A.
Previous studies showed that the ‘chopping’ activity of empty CbAgo and empty LrAgo can result in plasmid linearization or degradation, respectively (25). We designed a pair of 5′ phosphorylated 18 nt ssDNA guides (‘FW’ and ‘RV’ guides) which were corresponded to the same target site in plasmid pUC19 and designed an irrelevant guide, which had a random sequence without overlap with pUC19 (noncomplementary (‘NC’) guide) (Figure 6C). We observed that empty KmAgo treated at 37°C for 30 min converted the plasmid substrate from a supercoiled to open-circular state possibly by nicking one of the strands, but this nonspecific plasmid cleavage activity was suppressed when KmAgo was loaded with NC guide at 37°C for 30 min before adding plasmid (Figure 6D and Supplementary Figure S8A). We therefore searched for conditions that would eliminate the ‘chopping’ activity of empty pAgos and keep the ability to use DNA guides for specific cleavage of dsDNA by increasing the temperature of the gDNA loaded step. Importantly, we observed no plasmid processing by KmAgo in the absence of guide molecules at 37°C when KmAgo was pre-incubated at 55°C for 30 min. Thus, we can inactive the empty KmAgo by elevating the gDNA loaded temperature to suppress the guide-independent plasmid relaxation activity.
KmAgo could generate dsDNA breaks in double-stranded DNA with a GC-content of 53% or lower and cleave highly structured RNA
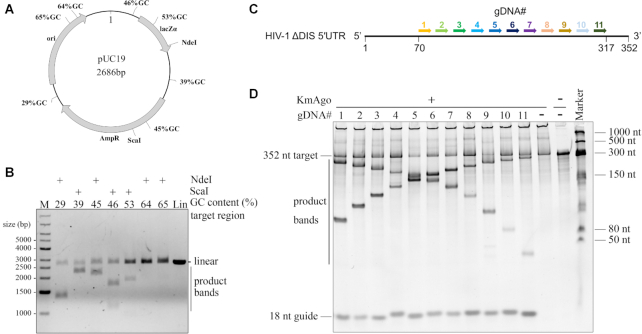
Mesophilic pAgos have successfully been used to generate dsDNA breaks in plasmid DNA and their cleavage ability are affected by the GC-content of dsDNA target (25–27). KmAgo was able to specifically generate dsDNA breaks in plasmid DNA when loaded with FW guide and RV guide at 55°C for 30 min (Figure 6D). To investigate whether the GC-content of the dsDNA target plays a role during dsDNA cleavage by KmAgo, we designed seven guide sets responding to different 80 bp target regions of the pUC19 plasmid with increasing GC-content at the sites of guide binding (Supplementary Table S4). The target plasmid was digested with a restriction enzyme (NdeI or ScaI) either before or after the incubation with two KmAgo–gDNA complexes (Figure 7B and Supplementary Figure S8B). Interestingly, KmAgo could cleave the negatively supercoiled plasmid, but not the linearized plasmid, suggesting that, like CbAgo, KmAgo relies on the negative supercoiled state of the target plasmid to facilitate local DNA melting before cleavage can take place (26). Furthermore, KmAgo–gDNA complexes were able to efficiently generate dsDNA breaks in regions with a GC-content of 46% or lower (Figure 7B and Supplementary S8C). The cleavage efficiency was significantly decreased at 53% GC and lost at 64% GC (Figure 7B and Supplementary S8C). Taken together, this suggests that, like other mesophilic pAgos, the cleavage efficiency of double-strand DNA depends on the GC-content at the cleavage site.
Figure 7.
Double stranded plasmid DNA and highly structured RNA cleavage by KmAgo. (A) Schematic overview of the pUC19 target plasmid. Black polylines indicate target sites while percentages indicate the GC-content of the 80 bp segments in which these target sites are located. (B) Pre-assembled KmAgo-gDNA complexes targeting various pUC19 segments were incubated with pUC19. Cleavage products were incubated with NdeI or ScaI and were further analysed by agarose gel electrophoresis. M, molecular weight marker; Lin, linearized plasmid. (C) Schematic overview of the HIV-1 ΔDIS 5′UTR. Arrows with different colors indicate the target region and the corresponding gDNAs are numbered from 1 to 11 with the corresponding colours. (D) Substrates and products generated by the assay described in (C) were resolved by denaturing PAGE (8%) revealing cleavability of the highly structured RNA by KmAgo–gDNA complex.
To the best of our knowledge, only KpAgo (Kluyveromyces polysporus) could cleave highly structured RNA (32), cleavage of highly structured RNA with pAgos has not been reported. Considering KmAgo guided with 5′ phosphorylated DNA could cleave RNA with high efficiency, we next examined whether KmAgo-guide complex can cleave target sequences in a highly structured RNA that contains a diverse set of conformational features such as varied lengths of bulges, hairpin loops, and single-stranded regions. Dayeh et al. have predicted the secondary structure of HIV-1 ΔDIS 5′UTR with SHAPE and designed a set of gDNAs to span the HIV-1 ΔDIS 5′UTR sequence in 23-nt increments (Supplementary Figure S7D) (32). Since 45-nt unstructured RNA targets are cleaved best when gDNAs are 15–20 nt long (Figure 3B), we designed 11 gDNAs (length was 18 nt) targeting the target regions of gDNA_4 to gDNA_14 in Dayeh's research to examine if KmAgo could cleave at the corresponding same sites (Figure 7C). Cleavage products were detected at the expected position for all sites, albeit to different extents (Figure 7D), demonstrating that KmAgo-gDNA duplex cleaves the target sequences even in highly structured RNAs.
DISCUSSION
eAgos utilize preferentially RNA guides to cleave RNA targets at moderate temperature and they are considered to originate from pAgos (1,5,32,34). Previously characterized pAgo proteins were unable to catalyze RNA guide-mediated RNA target cleavage at moderate temperature (11). Here, we present a detailed characterization of pAgo protein from mesophilic bacteria K. massiliensis. We show that KmAgo is an omnipotent endonuclease that can be programmed with DNA guides to cleave most types of nucleic acid substrates efficiently and precisely, including ssDNA, dsDNA, unstructured RNA and highly structured RNA. KmAgo can also be programmed with RNA guides to cleave both ssDNA and RNA targets precisely at 37°C. This discovery demonstrates that pAgos can also utilize RNA guides to cleave RNA targets at moderate temperatures like eAgos. Like most of Ago proteins, KmAgo utilizes Mn2+ and Mg2+ as cations, with Mn2+ being a better cation than Mg2+. Furthermore, consistent with eAgos, KmAgo has a preference for the 5′-U nucleotides of the RNA guides (6). Therefore, the guide–target duplex binding channel and guide 5′ end binding pockets of KmAgo may be similar to those of eAgos, allowing RNA guide and target binding. However, further investigations are required to demonstrate this hypothesis.
The activity and specificity of KmAgo are mainly affected by the type of guides. Similar to the majority of pAgos, KmAgo utilizes preferentially 5′ phosphorylated DNA guides. Unexpectedly, in contrast to the majority of studied pAgos that were reported to have a strong preference for DNA targets (11), KmAgo can cleave both DNA and RNA targets with almost the same high efficiency at moderate temperature when guided with 5′P DNA guides. Not only that, KmAgo is shown to use 9–30 nt 5′P DNA guides for efficient cleavage of DNA and RNA targets. Our results for minimum 5′P DNA guide requirements are similar to the 7 nt described for CbcAgo and the 9 nt described for TtAgo (31,37). In these pAgos, small 5′P DNA guides (i.e. 9 nt) could be effectively recognized by the MID domain of pAgos. These small DNA guides may function as seed to position the target nucleic acids near the active site of PIWI domain and result in the observed cleavage activity (31). Furthermore, KmAgo performs precise 5′P DNA-guided slicing of the target at a wide range of temperatures (from 25 to 75°C), while KmAgo loaded with 5′P RNA guides could only utilize Mn2+ as cation to cleave target at a narrow temperature range (from 25 to 50°C). This indicates that KmAgo loaded with DNA guide may be more stable than that with RNA guide. Previous studies of Ago proteins demonstrated that the complementarity between guide and target is important for cleavage. Analysis of LrAgo and zebrafish Argonaute2 demonstrated that mismatches in the seed region stimulated target cleavage (25,38). For KmAgo, mismatches in the seed region (guide positions 2–8) of guide stimulate target cleavage, and mismatches in the 3′-supplementary region (guide positions 13–16) of 5′P DNA guides do not significantly affect target cleavage. For 5′P RNA guides, mismatches in the 3′-supplementary region of guides stimulate the RNA cleavage activity but inhibit the DNA cleavage activity. Dinucleotide mismatches in positions 9–12 of 5′P DNA guides or single-nucleotide mismatches in positions 9–12 of 5′P RNA guides strongly inhibit target cleavage by KmAgo. Mismatches in the seed region may affect target positioning and/or conformational changes in the active site of KmAgo during catalysis. Future studies will be necessary to determine the structural basis of the apparent preference for these nucleotides at these positions.
The biochemical characterization of KmAgo reported herein is the first example of a pAgo that catalyzes RNA-guided RNA cleavage and owns high DNA-guided RNA and DNA cleavage activity at moderate temperatures, which may help us improve the understanding of the evolution of Ago protein. Research of PfAgo suggested that ancestors of Ago proteins might have utilized DNA guides and/or DNA targets (18). Phylogenetic analyses indicated that the Ago of the last eukaryotic common ancestor was an RNA-guided RNA-interfering protein (39). pAgos like KmAgo may be key intermediators in the evolution journey of Ago proteins. On the one hand, KmAgo has evolved a high DNA-guided RNA cleavage activity possessed by a small amount of characterized pAgos, and maintains high DNA-guided DNA cleavage activity like that of the possible Ago ancestor. On the other hand, KmAgo has evolved a RNA-guided RNA cleavage activity owned by the Ago protein of the last eukaryotic common ancestor. Further exploration of the mesophilic pAgos, which prefer RNA guide to cleave RNA or DNA, may be necessary to understand the evolution of Ago proteins.
Similar to the majority of reported pAgos, KmAgo shows guide-independent plasmid relaxation activity. Considering the obviously enhanced thermostability of KmAgo–gDNA duplex compared to the empty KmAgo, we inactivate the empty KmAgo by elevating the guide loaded temperature to 55°C to suppress the guide-independent plasmid processing activity of empty KmAgo. In addition, KmAgo could generate dsDNA breaks in dsDNA with a GC content up to 53% at 37°C when programmed with corresponding 5′P DNA guides. This means KmAgo might be a promising tool used for genome editing. Moreover, our data indicate that KmAgo–gDNAs duplex can be readily programmed to cleave both structured and unstructured RNA sequences at moderate temperatures. KmAgo may be developed into many convenient and efficient RNA manipulation tools in vitro for its unexpensive and easy synthesized DNA guides. Furthermore, KmAgo or other RNA targeting pAgos may not interfere with the cellular RNAi pathways (6). We propose that the guide-dependent efficient RNase activity of KmAgo can be applied for biomedical research, specifically degrading mRNA or functional RNAs in eukaryotic cells. Alternatively, the RNA targeting capabilities of KmAgo could be selectively harnessed by combination of KmAgo_DM and other effector components. Fusing various effector protein domains to KmAgo_DM to expand the functionality of KmAgo beyond RNA cleavage like RNase-defective dCas13 may be an effective strategy (40). In conclusion, this study advances our understanding of the evolutionary journey of Ago proteins and could expand the Ago-based DNA and RNA manipulation toolbox.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Prof. Shihui Yang for valuable revised advice of the manuscript.
Contributor Information
Yang Liu, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Collaborative Innovation Center for Green Transformation of Bio-resources, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Wenqiang Li, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Collaborative Innovation Center for Green Transformation of Bio-resources, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Xiaoman Jiang, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Collaborative Innovation Center for Green Transformation of Bio-resources, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Yaping Wang, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Collaborative Innovation Center for Green Transformation of Bio-resources, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Zhiwei Zhang, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Collaborative Innovation Center for Green Transformation of Bio-resources, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Qi Liu, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Collaborative Innovation Center for Green Transformation of Bio-resources, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Ruyi He, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Collaborative Innovation Center for Green Transformation of Bio-resources, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Quan Chen, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Collaborative Innovation Center for Green Transformation of Bio-resources, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Jun Yang, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Collaborative Innovation Center for Green Transformation of Bio-resources, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Longyu Wang, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Collaborative Innovation Center for Green Transformation of Bio-resources, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Fei Wang, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Collaborative Innovation Center for Green Transformation of Bio-resources, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
Lixin Ma, State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Collaborative Innovation Center for Green Transformation of Bio-resources, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan, Hubei 430062, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Hubei Province [2017ACA174]; Open Research Fund Program of State Key Laboratory of Biocatalysis and Enzyme Engineering [SKLBEE2018003]. Funding for open access charge: Department of Science and Technology, Hubei Province.
Conflict of interest statement. None declared.
REFERENCES
- 1. Swarts D.C., Makarova K., Wang Y., Nakanishi K., Ketting R.F., Koonin E.V., Patel D.J., van der Oost J. The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol. 2014; 21:743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Meister G. Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 2013; 14:447–459. [DOI] [PubMed] [Google Scholar]
- 4. Koonin E.V. Evolution of RNA- and DNA-guided antivirus defense systems in prokaryotes and eukaryotes: common ancestry vs convergence. Biol. Direct. 2017; 12:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Makarova K.S., Wolf Y.I., van der Oost J., Koonin E.V. Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol. Direct. 2009; 4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lisitskaya L., Aravin A.A., Kulbachinskiy A. RNA interference and beyond: structure and functions of prokaryotic Argonaute proteins. Nat. Commun. 2018; 9:5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kuzmenko A., Oguienko A., Esyunina D., Yudin D., Petrova M., Kudinova A., Maslova O., Ninova M., Ryazansky S., Leach D. et al. DNA targeting and interference by a bacterial Argonaute nuclease. Nature. 2020; 587:632–637. [DOI] [PubMed] [Google Scholar]
- 8. Jolly S.M., Gainetdinov I., Jouravleva K., Zhang H., Strittmatter L., Bailey S.M., Hendricks G.M., Dhabaria A., Ueberheide B., Zamore P.D. Thermus thermophilus Argonaute functions in the completion of DNA replication. Cell. 2020; 182:1545–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ryazansky S., Kulbachinskiy A., Aravin A.A. The expanded universe of prokaryotic Argonaute proteins. mBio. 2018; 9:e01935-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hegge J.W., Swarts D.C., van der Oost J. Prokaryotic Argonaute proteins: novel genome-editing tools?. Nat. Rev. Microbiol. 2018; 16:5–11. [DOI] [PubMed] [Google Scholar]
- 11. Wu J., Yang J., Cho W. C., Zheng Y. Argonaute proteins: structural features, functions and emerging roles. J. Adv. Res. 2020; 24:317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nakanishi K., Weinberg D.E., Bartel D.P., Patel D.J. Structure of yeast Argonaute with guide RNA. Nature. 2012; 486:368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sheng G., Zhao H., Wang J., Rao Y., Tian W., Swarts D.C., van der Oost J., Patel D.J., Wang Y. Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc. Natl Acad. Sci. U.S.A. 2014; 111:652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kaya E., Doxzen K.W., Knoll K.R., Wilson R.C., Strutt S.C., Kranzusch P.J., Doudna J.A. A bacterial Argonaute with noncanonical guide RNA specificity. Proc. Natl Acad. Sci. U.S.A. 2016; 113:4057–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miyoshi T., Ito K., Murakami R., Uchiumi T. Structural basis for the recognition of guide RNA and target DNA heteroduplex by Argonaute. Nat. Commun. 2016; 7:11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yuan Y.R., Pei Y., Ma J.B., Kuryavyi V., Zhadina M., Meister G., Chen H.Y., Dauter Z., Tuschl T., Patel D.J. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell. 2005; 19:405–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Y., Juranek S., Li H., Sheng G., Wardle G. S., Tuschl T., Patel D.J. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009; 461:754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Swarts D.C., Hegge J.W., Hinojo I., Shiimori M., Ellis M.A., Dumrongkulraksa J., Terns R.M., Terns M.P., van der Oost J. Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA. Nucleic Acids Res. 2015; 43:5120–5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zander A., Willkomm S., Ofer S., van Wolferen M., Egert L., Buchmeier S., Stockl S., Tinnefeld P., Schneider S., Klingl A. et al. Guide-independent DNA cleavage by archaeal Argonaute from Methanocaldococcus jannaschii. Nat. Microbiol. 2017; 2:17034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Willkomm S., Oellig C.A., Zander A., Restle T., Keegan R., Grohmann D., Schneider S. Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein. Nat. Microbiol. 2017; 2:17035. [DOI] [PubMed] [Google Scholar]
- 21. Swarts D.C., Jore M.M., Westra E.R., Zhu Y., Janssen J.H., Snijders A.P., Wang Y., Patel D.J., Berenguer J., Brouns S.J.J.J. et al. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014; 507:258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Song J., Hegge J.W., Mauk M.G., Chen J., Till J.E., Bhagwat N., Azink L.T., Peng J., Sen M., Mays J. et al. Highly specific enrichment of rare nucleic acid fractions using Thermus thermophilus argonaute with applications in cancer diagnostics. Nucleic Acids Res. 2020; 48:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. He R., Wang L., Wang F., Li W., Liu Y., Li A., Wang Y., Mao W., Zhai C., Ma L. Pyrococcus furiosus Argonaute-mediated nucleic acid detection. Chem. Commun. (Camb.). 2019; 55:13219–13222. [DOI] [PubMed] [Google Scholar]
- 24. Enghiad B., Zhao H. Programmable DNA-guided artificial restriction enzymes. ACS Synth. Biol. 2017; 6:752–757. [DOI] [PubMed] [Google Scholar]
- 25. Kuzmenko A., Yudin D., Ryazansky S., Kulbachinskiy A., Aravin A.A. Programmable DNA cleavage by Ago nucleases from mesophilic bacteria Clostridium butyricum and Limnothrix rosea. Nucleic Acids Res. 2019; 47:5822–5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hegge J.W., Swarts D.C., Chandradoss S.D., Cui T.J., Kneppers J., Jinek M., Joo C., van der Oost J. DNA-guided DNA cleavage at moderate temperatures by Clostridium butyricum Argonaute. Nucleic Acids Res. 2019; 47:5809–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cao Y., Sun W., Wang J., Sheng G., Xiang G., Zhang T., Shi W., Li C., Wang Y., Zhao F. et al. Argonaute proteins from human gastrointestinal bacteria catalyze DNA-guided cleavage of single- and double-stranded DNA at 37 °C. Cell Discov. 2019; 5:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Swarts D.C., Szczepaniak M., Sheng G., Chandradoss S.D., Zhu Y., Timmers E.M., Zhang Y., Zhao H., Lou J., Wang Y. et al. Autonomous generation and loading of DNA guides by bacterial Argonaute. Mol. Cell. 2017; 65:985–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Olina A., Kuzmenko A., Ninova M., Aravin A.A., Kulbachinskiy A., Esyunina D Genome-wide DNA sampling by Ago nuclease from the cyanobacterium Synechococcus elongatus. RNA Biol. 2020; 17:677–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ye S., Bae T., Kim K., Habib O., Lee S.H., Kim Y.Y, Lee K.-I., Kim S., Kim J.-S. DNA-dependent RNA cleavage by the Natronobacterium gregoryi. 2017; bioRxiv doi:20 January 2017, preprint: not peer reviewed 10.1101/101923. [DOI]
- 31. García-Quintans N., Bowden L., Berenguer J., Mencía M. DNA interference by a mesophilic Argonaute protein, CbcAgo. F1000Research. 2019; 8:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dayeh D.M., Cantara W.A., Kitzrow J.P., Musier-Forsyth K., Nakanishi K. Argonaute-based programmable RNase as a tool for cleavage of highly-structured RNA. Nucleic Acids Res. 2018; 46:e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rivas F.V., Tolia N.H., Song J.J., Aragon J.P., Liu J., Hannon G.J., Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005; 12:340–349. [DOI] [PubMed] [Google Scholar]
- 34. Song J.J., Smith S.K., Hannon G.J., Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004; 305:1434–1437. [DOI] [PubMed] [Google Scholar]
- 35. Lima W.F., Wu H., Nichols J.G., Sun H., Murray H.M., Crooke S.T. Binding and cleavage specificities of human Argonaute2. J. Biol. Chem. 2009; 284:26017–26028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Elkayam E., Kuhn C.D., Tocilj A., Haase A.D., Greene E.M., Hannon G.J., Joshua-Tor L. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012; 150:100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Y., Juranek S., Li H., Sheng G., Tuschl T., Patel D.J. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008; 456:921–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen G.R., Sive H., Bartel D.P. A seed mismatch enhances Argonaute2-catalyzed cleavage and partially rescues severely impaired cleavage found in fish. Mol. Cell. 2017; 68:1095–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shabalina S.A., Koonin E.V. Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 2008; 23:578–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Terns M.P. CRISPR-based technologies: impact of RNA-targeting systems. Mol. Cell. 2018; 72:404–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.