Abstract
Approximately 13% of the human genome can fold into non-canonical (non-B) DNA structures (e.g. G-quadruplexes, Z-DNA, etc.), which have been implicated in vital cellular processes. Non-B DNA also hinders replication, increasing errors and facilitating mutagenesis, yet its contribution to genome-wide variation in mutation rates remains unexplored. Here, we conducted a comprehensive analysis of nucleotide substitution frequencies at non-B DNA loci within noncoding, non-repetitive genome regions, their ±2 kb flanking regions, and 1-Megabase windows, using human-orangutan divergence and human single-nucleotide polymorphisms. Functional data analysis at single-base resolution demonstrated that substitution frequencies are usually elevated at non-B DNA, with patterns specific to each non-B DNA type. Mirror, direct and inverted repeats have higher substitution frequencies in spacers than in repeat arms, whereas G-quadruplexes, particularly stable ones, have higher substitution frequencies in loops than in stems. Several non-B DNA types also affect substitution frequencies in their flanking regions. Finally, non-B DNA explains more variation than any other predictor in multiple regression models for diversity or divergence at 1-Megabase scale. Thus, non-B DNA substantially contributes to variation in substitution frequencies at small and large scales. Our results highlight the role of non-B DNA in germline mutagenesis with implications to evolution and genetic diseases.
Graphical Abstract
Graphical Abstract.
Guiblet et al. show that loci capable of forming non-canonical (non-B) DNA structures are a major driver of variation in nucleotide substitution levels across the genome. Image credit: Wilfried Guiblet.
INTRODUCTION
Mutation rates vary across the genome (1,2), and this phenomenon contributes to differences in the levels of intra- and interspecific genetic variation (henceforth called ‘diversity’ and ‘divergence’, respectively). As a result, certain genomic regions may be at a higher (or at a lower) risk of acquiring mutations important for adaptation and/or genetic diseases (1–3). In a broad sense, deciphering the causes of regional variation in mutation rates is essential to understanding both evolution and diseases (1,2).
Numerous genomic features contribute to regional variation in mutation rates, but those identified to date cannot account for all such variation. Some features are directly related to DNA sequence and usually act at the scale of single nucleotides, e.g. guanines and cytosines are more mutable than adenines and thymines (4,5). Neighboring nucleotides also have an effect, e.g. methylated cytosines in CpG dinucleotides are 10 times more mutable than other sites because of their spontaneous deamination (6), and several other contexts leading to guanine holes and increased mutagenesis were previously identified (7). Other genomic features—such as recombination rate (8), replication timing (9), chromatin accessibility (10,11), histone modifications (12,13), and Lamina Associated Domains (14)—contribute to regional variation in mutation rates through the variable activity of different enzymatic processes along the genome. These frequently act at larger scales, from several hundreds of kilobases to several megabases (Mbs). The magnitude of regional variation in mutation rates decreases with the increase in the genomic scale considered; most such regional variation in fact occurs at the single-nucleotide scale (1). At the 1-Mb scale, which is considered the natural long-range variation scale for mammalian genomes (15), most regions have mutation rates deviating by ∼2-fold (1). Notably, at this scale, several analyses indicated that the genomic features listed above explain only ∼50% of the regional variation in mutation rates (12,16,17). The correlation in regional variation in mutation rates between human and great apes (18) suggests that the unexplained portion of this variation is not random, and that additional factors remain to be discovered. Non-B DNA may be one such factor.
Certain DNA sequence motifs have the ability to fold (at least over part of their length) into secondary conformations that differ from the canonical right-handed B-DNA helix that has 10 bp per turn (19–21). Such non-B DNA sequence motifs (henceforth called ‘non-B DNA motifs’) range in length from a few dozen to a few hundred nucleotides and are non-randomly distributed across the genome (22–24). A genomic locus harboring a non-B DNA motif is usually referred to as a non-B DNA locus, and the same non-B DNA motif is frequently present at multiple loci in the genome.
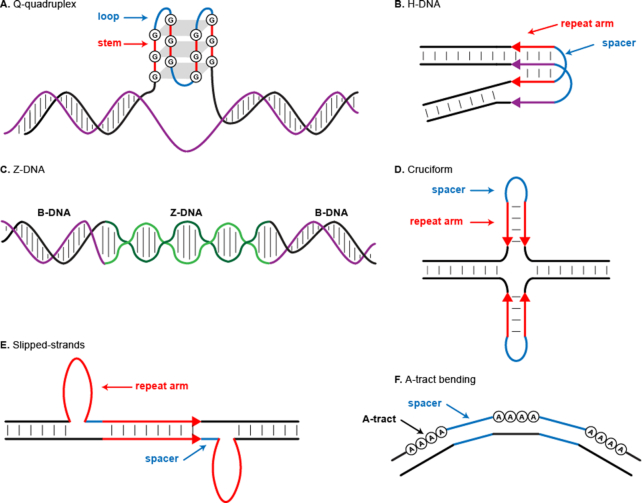
Several types of non-B DNA have been identified based on the structures they can form (Figure 1), which in turn depend on their motif sequences (25,26). The G-quadruplex (henceforth called ‘G4’) structure (Figure 1A) alternates stems consisting of guanines and loops consisting of unspecified nucleotides, with the canonical sequence of G3+N1−12G3+N1−12G3+N1−12G3+ (27). Alternating purine and pyrimidine sequences can form Z-DNA (Figure 1C)—a left-handed double-stranded helix with 12 bases per turn (28). Each mirror, inverted and direct repeat locus contains two repeat arms, usually separated by a non-repetitive spacer. Homopurines and homopyrimidines organized in mirror repeats with or without a ≤100-nucleotide spacer can form H-, or triplex, DNA (Figure 1B) (29). Inverted sequence repeats with or without a ≤100-nucleotide spacer can form DNA cruciforms (Figure 1D) (30,31). Direct sequence repeats with or without a ≤10-nucleotide spacer can form slipped-strand structures (Figure 1E) (32). Finally, A-phased repeats—three or more segments composed of three to nine adenines and/or thymines (A-tracts) and whose centers are separated by 10 bp—can bend, or create a curvature in, the double helix (Figure 1F) (26,33,34).
Figure 1.
Schematic of different types of non-B DNA structures. (A) G-quadruplex, (B) H-DNA, (C) Z-DNA, (D) cruciform, (E) slipped strands and (F) A-tract bending.
Such non-B DNA loci have been implicated in a myriad of cellular functions (reviewed in (35)) and associated with multiple human diseases. These loci regulate gene expression (3,36–40), contribute to telomere maintenance (41,42), participate in the life cycle of transposable elements (43), serve as direct protein-binding DNA targets (44), and are likely involved in recombination (44–48) and hypomethylation of CpG islands (49–51). Because certain non-B DNA loci are functional, mutations interfering with their ability to form structures may be harmful for the organism. Relatedly, non-B DNA structures have been linked to cancer (44,52–56) and several neurological diseases (57–64), although the mechanisms by which these structures contribute to diseases are not completely understood.
In the cell, non-B DNA loci can affect several DNA metabolic processes, and both replication-dependent and independent pathways contribute to elevated mutation rates at such loci. Replicative polymerases encounter many non-B DNA structures that act as natural impediments to DNA synthesis elongation. Specialized DNA polymerases associated with the replication fork, including Pols eta and kappa, perform highly efficient synthesis through non-B DNA structures (reviewed in (56,65,66)), and can take over synthesis from stalled replicative polymerases (67). DNA damage-induced mutagenesis has been associated with non-B structure formation (reviewed in (68)), and damage induced by reactive oxygen species is affected by the inherent structure and local sequence of DNA (reviewed in (69)). Non-B DNA structures are recognized by DNA repair pathways, such as nucleotide excision repair (70–73), and error-prone repair processing can result in mutations (74–76). In addition, non-B DNA structures are associated with the formation of double-strand breaks (DSBs), which lead to increased genomic instability (71,77–84). Even though non-B DNA loci have the potential to influence mutations by affecting multiple processes in the cell, their effects on mutation rates have not been studied in detail.
While an elevated density of single nucleotide polymorphisms (SNPs) at non-B DNA was previously reported (24), little is known about where mutations concentrate within the non-B DNA motifs, and whether the increase in mutations is limited to the motifs themselves or extends into their flanking sequences. Furthermore, no studies have reported whether the effect of non-B DNA structures on SNP density depends on their stability. In all, genetic variation at non-B DNA and its flanking sequences remains understudied.
We recently demonstrated that, across the human genome, both the speed and accuracy of the modified bacteriophage phi29 polymerase used in Pacific Biosciences (PacBio) sequencing are modulated by the presence of non-B DNA loci and that, for G4 loci, this effect depends on their stability (85). We also showed G4 loci that are most divergent between human and orangutan or are highly diverse among human populations have more pronounced polymerization slowdown and error rates (85). These results suggest that both sequencing errors and germline mutations are elevated at G4 motifs and may have similar mechanisms, i.e. result from polymerization slowdown.
Here, we extend this study, and test a hypothesis that non-B DNA motifs contribute to regional variation in mutation rates. We use the frequency of human SNPs as a measure of intraspecific variation or ‘diversity’, and the frequency of fixed nucleotide substitutions (FNSs) between human and orangutan reference genomes as a measure of interspecific variation or ‘divergence’. These measures proxy germline mutation rates because we minimize selection effects by limiting our analysis to putatively neutrally evolving non-coding regions of the genome. We analyze the influence of non-B DNA on nucleotide substitution frequencies at two different scales: the small scale of single nucleotides and the large scale of 1-Mb genomic windows. Using statistical methods from the functional data analysis domain, which investigate shapes of signals (86,87), we test whether diversity and divergence levels differ between each type of non-B DNA and randomly selected control sequences genome-wide. Furthermore, we test whether the predicted stability of G4 loci affects the levels of diversity and divergence. Moreover, we describe distortions in the nucleotide substitution spectrum surrounding G4s. Lastly, we evaluate whether adding non-B DNA loci to statistical models that include known contributing genomic features (1,12,16,88,89) increases the explained share of regional variation in diversity and divergence. Overall, our study measures the contributions of non-B DNA to germline variation in nucleotide substitution frequencies across the human genome.
MATERIALS AND METHODS
Non-coding non-repetitive (NCNR) subgenome
To obtain the NCNR subgenome of the hg19 version of the human genome, we excluded (i) all repeats as annotated with RepeatMasker (90) (rmsk track at the UCSC Genome Browser (91)); (ii) NCBI RefSeq (92) genes (exons and their 1-kb up- and downstream flanking regions, and 5 kb upstream of the 5′ UTRs and downstream of the 3′ UTRs); (iii) conserved elements as annotated with phastConsElements100way (93) and (iv) enhancers as annotated with GeneHancerRegElementsDoubleElite (94,95) (Supplementary Figure S1). The hg19 version of the human genome was used because it has a larger number of annotations of genomic features than the more recent hg38 version.
Divergence and diversity nucleotide substitutions datasets
69 329 877 FNSs that occurred between human and orangutan were retrieved from the 100-way Vertebrate Multiz Alignment (96) obtained from the UCSC Genome Browser (91). SNPs from the Simons Genome Diversity Project (97) were acquired from the Seven Bridges Cancer Genomic Cloud (https://cgc.sbgenomics.com/). 44 833 480 SNPs from all individuals in this project were merged into a single BED file. We only considered variants (26 875 194 FNSs and 15 555 617 SNPs) located in the NCNR subgenome in subsequent analyses.
Non-B DNA annotations
G4 loci were annotated in the human genome reference (version hg19) using Quadron software (98). Annotations of the other non-B DNA loci (direct, mirror, and inverted repeats, as well as Z-DNA and A-phased motifs) were downloaded from the non-B Database (non-B DB, https://nonb-abcc.ncifcrf.gov/, as assessed on September 8, 2020) (99).
Small-scale variation in nucleotide substitution frequency
We compared SNP (diversity) and FNS (divergence) frequencies inside non-B DNA loci and their 2-kb up- and downstream flanking regions. We only considered non-B DNA located within the NCNR subgenome (Supplementary Table S1). Non-B DNA loci longer than 100 bp were discarded, to minimize the possibility that longer (and more rare) motifs and their flanking sequences may overlap with multiple shorter (and more common) loci. Even with this filter, some loci did overlap; when such overlaps occurred for loci of the same non-B DNA type, we retained only one locus (and its flanking regions) selected at random (Supplementary Figure S2). Overlaps between non-B DNA loci of different types are rare (23) and do not interfere with statistical analyses, thus we did not filter them out. For each SNP or FNS located in the flanking region of a non-B DNA locus, we computed the distance from the closest annotated motif end. For each SNP or FNS located inside a motif, we computed a position scaled between 0 and 1. This scaling was performed for each section of the non-B locus (stems and loops in G4s, repeat arms and spacers in inverted, direct, and mirror repeats) in order to ‘align’ all non-B loci of the same type while retaining the information about sections. 180 bins were considered in total, for all sections of the scaled interval to be larger than the ones observed at non-B DNA loci. The sections’ maximal sizes were 30 bins for stems and 20 bins for loops in G4s; 50 bins for repeat arms and 80 bins for spacers in inverted, direct, and mirror repeats; 40 bins for A-tracts and 30 bins for spacers in A-phased repeats. Because the resolution in the scaled intervals was higher than in the original one, each SNP or FNS could occupy several scaled positions (Supplementary Figure S3). For each non-B DNA locus and its flanking regions, a non-overlapping control interval of matching length and chromosome was created using bedtools shuffle (100).
IWTomics
IWTomics (Interval-Wise Testing for Omics data; (101)) was employed to compare SNP and FNS frequencies between non-B DNA and control sequences. For each non-B DNA type and each nucleotide substitution frequency, we performed three separate tests: one for each 2-kb flanking sequence and one for the non-B DNA locus itself. IWTomics performs a functional permutation test for the null hypothesis that two groups of curves have the same distribution, versus the alternative hypothesis that their distributions differ (two-sided test). Here, the two groups are represented by non-B DNA and control sets, while each curve consists of the substitution counts measured in contiguous nucleotides (for flanking sequences) or in contiguous bins (for non-B DNA loci). Although substitution counts are discrete measurements and each of these curves might look quite noisy, the substitution frequency difference between the two groups—employed in the test statistics of the test—is a rather smooth curve (e.g. Supplementary Figure S4, bottom panels). This makes functional methods suitable for our analysis and guarantees that IWTomics has good power and accuracy. For each comparison, we based the test on 1,000 permutations and on the test statistic
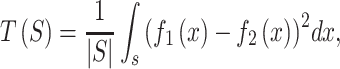 |
where 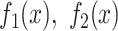 are the substitution frequencies in the two groups and S is the subinterval where the test is performed. We computed an adjusted P-value curve—controlling the interval-wise error rate—for any scale ranging from the single nucleotide to the entire 2000-bp interval (for flanking regions) or from the single bin to the entire 180-bin interval (for non-B DNA loci). An example of a complete IWTomics output is shown in Supplementary Figure S4.
are the substitution frequencies in the two groups and S is the subinterval where the test is performed. We computed an adjusted P-value curve—controlling the interval-wise error rate—for any scale ranging from the single nucleotide to the entire 2000-bp interval (for flanking regions) or from the single bin to the entire 180-bin interval (for non-B DNA loci). An example of a complete IWTomics output is shown in Supplementary Figure S4.
Substitution spectrum at the first flanking position
To account for the potential sequence context bias introduced by the non-B DNA motif annotations, SNP and FNS frequencies were computed in their trinucleotide context (by considering a substituted base with its two flanking bases). For each trinucleotide, we counted total occurrences surrounding non-B DNA loci and their controls, and identified and counted those harbouring a SNP or an FNS in the central base. Next, we added up substitution counts in trinucleotides for each substitution type (for instance, all NAN→NCN were summed as A→C substitutions), and computed substitution frequencies dividing these by the counts of total corresponding trinucleotides. We tested whether substitution frequencies surrounding non-B DNA loci differed from those of their controls using two-sided Fisher's exact tests. P-values were adjusted for multiple testing using a Bonferroni correction.
Building 1-Mb genomic windows
Our multiple regression analysis utilized 1-Mb consecutive non-overlapping windows obtained partitioning the hg19 human genome. We excluded windows that overlapped with gaps in the hg19 assembly using bedtools (100), resulting in 2511 1-Mb autosomal windows. The coverage of NCNR subgenome in each 1-Mb window was computed with bedtools annotate (100).
Genomic features
For each of our 1-Mb windows, we extracted genomic features from various sources (Supplementary Table S2). GC content was computed directly from the genomic DNA sequence. RNA polymerase II and replication origins were measured as ‘coverages’, computing the proportion of the window covered by the feature using bedtools (100). Replication timing and recombination rates were measured as weighted averages, i.e. the sum of all annotated intervals’ length multiplied by their score. DNA methylation coverages (for CHH, CHG and CPG methylation (102)) were also measured as weighted averages, but using a different method implemented through the Galaxy tool ‘Assign Weighted Average Values’ (103,104). Specifically, a window partially or totally overlapping multiple feature intervals was assigned the average of the values of the features weighted by the corresponding number of overlapping bases. DNA binding profiles of H2AFZ, H3K9me3, H3K9ac, H3K27ac, H3K36me3, H4K20me1, H3K79me2, H3K4me1, as well as CTCF motifs, were collected from the ENCODE project portal (105). The results of ChIP-seq assays performed on hESC H1 cell lines were downloaded (provided on GitHub) and histone modification features were measured as ‘signals’—the average number of reads aligned in each window. DNase Hypersensitive Sites were also collected from the ENCODE project portal (105) and measured as ‘coverage’, i.e. as the proportion of the window covered by DHS peaks. Mappability (106), CpG islands (107), and Lamina Associated Domains (108) tracks were downloaded from the UCSC Genome Browser (91). The distance of each window to the closest centromere and telomere was computed as described in the code on the GitHub. Finally, the telomeric hexamer TTAGGG was annotated on hg19 using fastaRegexFinder (https://github.com/dariober).
Multiple linear regressions
Multiple regression models were built for both SNP frequency and FNS frequency using the NCNR subgenome (see above). Since these frequencies are not reliable in windows with small NCNR coverage (this ranged from 4.8% to 66.6%, Supplementary Figure S5A), we filtered out all windows where this coverage was <20%. Moreover, since SNP and FNS frequencies reliability increases (their variance decreases) with NCNR coverage, we employed weighted least squares—weighing each retained window with its NCNR coverage. As predictors, we used all genomic features listed above—measured as coverage, count, or signal (Supplementary Table S2)—as well as the coverage of each non-B DNA type (Supplementary Table S1), in the same 1-Mb genomic windows. For G4 loci, instead of simple coverage, we used a weighted coverage, computed weighing each G4 locus in a 1-Mb window with its predicted stability score provided by Quadron (98). Windows with average mappability score <0.8 (Supplementary Figure S5B) were also filtered out. Finally, we identified and filtered out a total of 12 windows that represented strong outliers, which may have been influential points for the regressions (in particular, three windows with no G4s, one with unusually large inverted repeats coverage, one with unusually large mirror repeats coverage, eight windows with very small RNA Pol II coverage, one with very large H2AFZ signal and one with very large H3K79me2 signal). A total of 2203 windows were retained for the regression analysis. Several genomic features, as well as SNP frequency, were log-transformed to obtain more symmetric distributions, while other variables were scaled (see Supplementary Table S3 and the GitHub for details). Telomere hexamer coverage was binarized (0 when the coverage was 0 and 1 when it was >0). In order to avoid strong multicollinearity in the models, we used hierarchical clustering (with 1 – |Spearman's correlation| as dissimilarity and complete linkage) to group predictors (Supplementary Figure S6A). Predictors with Spearman's correlations higher than 0.8 in absolute value were clustered, and only one predictor from each cluster was included in the regression models (Supplementary Figure S6). A total of 15 genomic features and all six non-B DNA predictors were retained through this exercise (using Pearson's correlation produced similar clustering; Supplementary Figure S6B). Mappability was not included in the clustering, but it was forced to be in the regression models as a predictor, in order to control for it.
A two-step procedure was then employed to include the relevant quadratic terms in the models and perform variable selection. First, for each variable (all non-B DNA and genomic features except telomere hexamer, which was binarized), we compared a weighted linear model comprising the variable and mappability as predictors, with a weighted quadratic model comprising the variable, its squared values, and mappability as predictors. The quadratic term was retained if the P-value of the ANOVA test comparing the linear and quadratic models was <10−10. Second, we employed a weighted regression model with Elastic Net regularization (109) to select relevant (linear and/or quadratic) terms. The elastic net mixing parameter was set to 0.5 (equal mixture of Lasso and Ridge penalties). The regularization parameter λ was selected through repeated 10-fold cross-validation (five repetitions) to minimize mean squared error (we obtained the same results maximizing R-squared). Mappability was forced to be in the final models, hence it was not included in the Elastic Net penalty. Finally, we re-fitted the final models comprising the terms selected by the Elastic Net and mappability (Supplementary Table S3), we evaluated their R-squared and we computed the coefficient of partial determination of each selected variable (non-B DNA and genomic feature) as
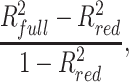 |
where 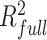 is the R-squared of the full model and
is the R-squared of the full model and 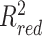 is the R-squared of the reduced model obtained by removing all linear and/or quadratic terms involving the variable.
is the R-squared of the reduced model obtained by removing all linear and/or quadratic terms involving the variable.
RESULTS
To study the effects of non-B DNA loci on regional variation in mutation rates, we used sequence-motif-based annotations of Z-DNA loci, and of direct, inverted, mirror, and A-phased repeats (Supplementary Table S1) in the hg19 version of the human genome available in the non-B DNA DataBase (99). G4 loci (Supplementary Table S1) were annotated with Quadron (98)—which, compared with other G4 annotation tools (110), has the advantage of assigning a predicted stability score to each G4 locus by using a machine-learning algorithm trained on the experimental output of G4-seq (111).
To minimize the effects of selection, regional variation in mutation rates was assessed by evaluating the distribution of nucleotide substitutions in putatively neutrally evolving non-coding regions of the genome (see Materials and Methods). From these non-coding regions, we additionally excluded repetitive elements as annotated by RepeatMasker (90) because of difficulties in sequencing, read mapping, and subsequent variant calling, and because of the demonstrated role of some non-B DNA motifs in the life cycle of transposable elements (43). This resulted in ∼900 Mb of the Non-Coding Non-Repetitive (NCNR) subgenome (i.e. approximately one third of the genome). We assumed that these sequences evolve largely neutrally (14,88,89,112) and thus, the levels of diversity and divergence reflect variation in mutation frequencies. Within the NCNR subgenome, we were able to analyze 171 653 direct repeats, 2 017 399 inverted repeats, 213 899 mirror repeats, 146 174 A-phased repeats, 108 279 Z-DNA loci and 178 312 G4 loci (Supplementary Table S1).
Always within the NCNR subgenome, we gathered nucleotide substitutions, the most common type of genetic variants (2), from two independent datasets. First, we considered the frequency of FNSs (the number of fixed differences per site per locus) obtained from whole-genome alignments of human and orangutan genomes (96). Second, we considered the frequency of SNPs (the number of SNPs per site per locus) from the Simons Genome Diversity Project (SGDP) (97). This dataset was generated from 279 human genomes sequenced at a relatively high depth (∼30×). We did not use low-depth sequencing datasets, e.g. the 1000 Genomes Project (113), as they have high probability of false SNPs arising from sequencing errors (85).
Non-B DNA loci affect small-scale variation in nucleotide substitution frequency
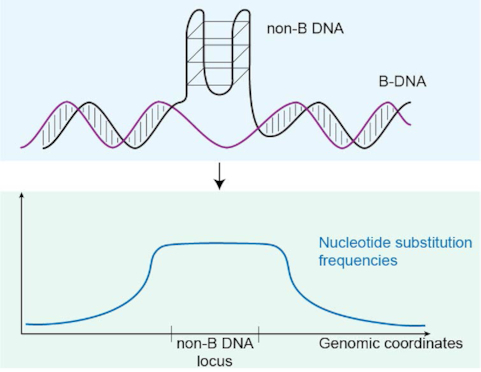
We studied small-scale variation in nucleotide substitution frequency in non-B DNA loci and their immediate flanking regions (2 kb upstream and 2 kb downstream), as compared to NCNR subgenome control sequences matching the non-B loci in number, length and chromosome of origin, and chosen at random. Previous studies suggested a mutagenic effect of non-B DNA extending into adjacent hundreds of bases (80,114); we increased this distance to ensure we can detect potential effects even at larger distances. Whenever the flanking regions of non-B DNA loci overlapped, only one of the overlapping loci was chosen (at random) for analysis (Supplementary Table S1; see Materials and Methods). To include non-B DNA loci of different lengths (from 11 to 100 bp) in a unified statistical analysis, we scaled each locus to a common length interval split in 180 bins (Supplementary Figure S3). Such scaling was chosen based on the number and maximum length of different sections within non-B DNA loci (e.g. stems and loops in G4s, and repeat arms and potential spacers in inverted, direct, mirror, and A-phased repeats; see Materials and Methods). After scaling the loci, we aggregated nucleotide substitutions per bin for each non-B DNA type genome-wide, and computed the corresponding fixed and polymorphic nucleotide substitution frequencies dividing the respective per-site nucleotide substitution counts by the number of loci. Differences in polymorphic and fixed nucleotide substitution frequencies between non-B DNA loci and control sequences were evaluated using IWTomics (115), a functional data analysis method. IWTomics compares two groups of curves (e.g. the substitution frequencies along non-B DNA loci, or their up- and downstream flanking regions versus the respective control sequences), performs permutation tests on all possible subintervals, and produces adjusted P-value curves for any possible subinterval length (from the single bin to the whole 180 bin interval for non-B loci, and from the single nucleotide to the whole 2000-bp interval for each flanking region). This approach identifies locations of significant differences between the two groups of curves.
G4 loci
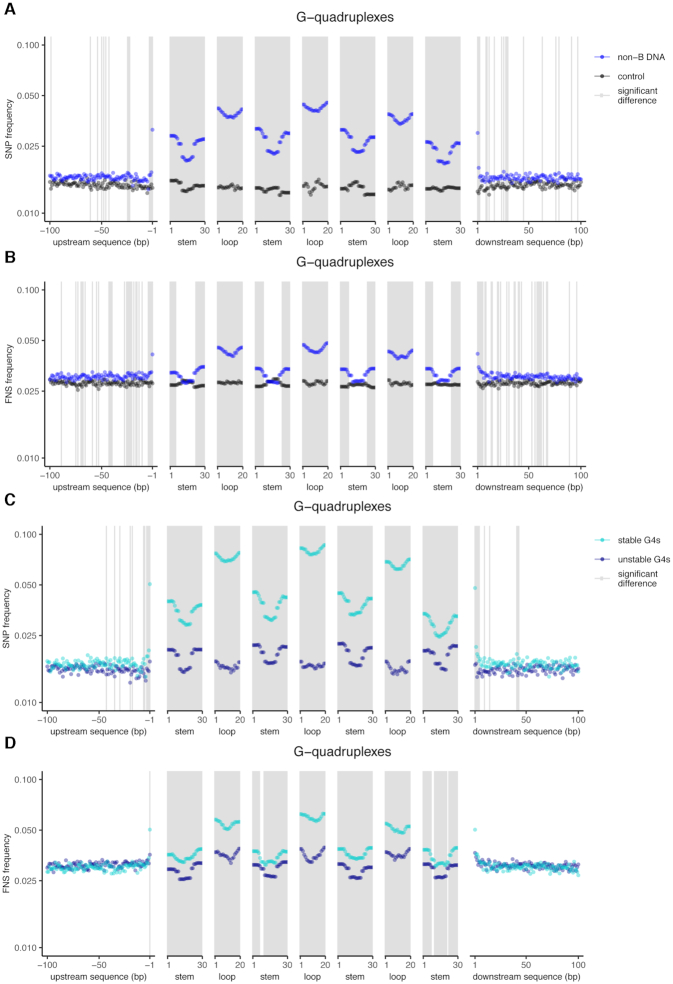
We studied variation in nucleotide substitution frequency within G4 loci, separating each locus into its stems (consisting of guanines) and loops (consisting of any nucleotides). In order to more easily parse patterns in stems and loops, we focused on G4s with four stems and three loops. These are the most abundant in the genome; they constitute 49.7% of the G4s we considered in the NCNR subgenome. We found that G4 loci exhibited significantly elevated polymorphic and fixed nucleotide substitution frequencies compared to control sequences (Figure 2A and B). This elevation was evident in both stems and loops, but was particularly striking in loops, which had up to 3.2-fold SNP frequency increase and up to 1.7-fold FNS frequency increase compared to control sequences.
Figure 2.
Genome-wide nucleotide substitution frequencies at G4 loci and their flanking sequences. The positions of nucleotide substitutions within motifs were scaled based on motif size (see Materials and Methods for details). Stems are runs of guanines and loops are unspecified nucleotides between stems. Flanking regions are the 2 kb up- and downstream from the loci. For clarity of visualization, only the first 100 bps are shown (the full 2 kb are shown in Supplementary Figure S8) and the Y-axes are displayed on a log scale. Gray areas indicate significantly different rates between groups (IWTomics adjusted P-value curve <0.01). A comparison between all G4 loci and control sequences for (A) single-nucleotide polymorphism (SNP) frequencies and (B) fixed nucleotide substitution (FNS) frequencies. A comparison between stable and unstable G4 loci for (C) SNP and (D) FNS frequencies.
We next divided G4 loci into stable and unstable based on their predicted stability. Following published recommendations (98), we labeled loci with Quadron score >19 as stable (a total of 20 156 loci) and loci with Quadron score ≤19 as unstable (a total of 30 212 loci). We found that SNP and FNS frequencies were significantly higher at stable versus unstable G4 loci, and again this difference was more pronounced in loops (up to 5.1- and 1.8-fold for SNP and FNS frequencies, respectively) than in stems (up to 2.1- and 1.3-fold for SNP and FNS frequencies, respectively; Figure 2C and D).
To test whether mutations at G4 loci tend to stabilize or destabilize G4 structures, we analyzed the effects of rare variants on G4 stability (we considered SNPs with derived allele frequency ≤0.01, excluding those supported by 1–2 reads only to minimize the effect of sequencing errors). Because genetic drift dominates over natural selection when allele frequencies are low (116), the distribution of rare variants is likely to reflect mutational processes rather than natural selection. Interestingly, we found that, on average, there are as many mutations leading to an increase as there are mutations leading to a decrease in predicted G4 stability. Specifically, we substituted positions in the reference human genome by the corresponding rare variants, creating an alternative human genome, and reran G4 annotation with Quadron (98). A total of 609 013 G4s were present at the same or similar coordinates (requiring ≥90% overlap) in both alternative and reference human genomes. Among these shared G4s, the proportion predicted to change from stable to unstable (201/(201 + 179)) was not significantly different from proportion predicted to change from unstable to stable (179/(201 + 179); P = 0.281, 1-sample test with continuity correction for proportion of G4s with increased stability equal to 50%), based on the Quadron stability threshold of 19 (98).
Other non-B DNA loci
Other types of non-B DNA loci also affected variation in both SNP and FNS frequencies in NCNR (Figures 3–4), with patterns that were specific to the different types of loci. For inverted repeats (Figures 3A and 4A), SNP and FNS frequencies were both significantly elevated in the spacer (up to 1.3- and 1.2-fold, respectively), but significantly depressed in most repeat arms (up to 1.2-fold for both), as compared to control sequences (the comparisons in the rest of the paragraph are also against control sequences). For direct repeats (Figures 3B and 4B), SNP and FNS frequencies were both significantly elevated in the spacer (up to 3.0- and 1.8-fold, respectively); however, SNP frequency was significantly elevated (up to 1.4-fold), whereas FNS frequency was significantly depressed (up to 1.5-fold), in the repeat arms. For mirror repeats (Figures 3C and 4C), both SNP and FNS frequencies were significantly elevated in the spacer (up to 1.4- and 1.2-fold, respectively); however, SNP frequency was significantly elevated (up to 1.3-fold), whereas FNS frequency was significantly depressed (up to 1.2-fold), in the repeat arms. For Z-DNA (Figures 3D and 4D), both SNP and FNS frequencies were significantly elevated (up to 3.3- and 2.0-fold, respectively). A-phased repeats can be separated into A-tracts, which are required for structure formation, and spacers, which can vary in sequence without affecting structure formation. We focused on those with three A-tracts and two spacers (the most abundant in the genome). Both SNP and FNS frequency in repeat arms were significantly depressed (up to 1.7- and 1.6-fold lower, respectively; Figures 3E and 4E). In spacers, SNP and FNS frequencies were both significantly elevated (up to 1.2- and 1.3-fold, respectively; Figures 3E and 4E).
Figure 3.
Genome-wide single-nucleotide polymorphism (SNP) frequencies at non-G4 non-B DNA loci and their flanking sequences. The positions of SNPs within motifs were scaled based on motif size (see Materials and Methods for details). Inverted, direct, and mirror repeats are split into spacers and repeat arms, and A-phased repeats are split into A-tracts and spacers. For clarity of visualization, only the first 100 bps are shown (the full 2 kb are shown in Supplementary Figure S8) and the Y-axes are displayed on a log scale. Gray areas indicate significantly different SNP frequency in non-B DNA vs. control sequences (IWTomics adjusted P-value curve < 0.01).
Figure 4.
Genome-wide fixed nucleotide substitution (FNS) frequencies at non-G4 non-B DNA loci and their flanking sequences. The positions of FNSs within motifs were scaled based on motif size (see Materials and Methods for details). Inverted, direct, and mirror repeats are split into spacers and repeat arms, and A-phased repeats are split into A-tracts and spacers. Flanking regions are the 2 kb up- and downstream from the loci. For clarity of visualization, only the first 100 bp are shown (the full 2 kb are shown in Supplementary Figure S8) and the Y-axes are displayed on a log scale. Gray areas indicate significantly different FNS frequency in non-B DNA versus controls (IWTomics adjusted P-value curve < 0.01).
We also discovered that spacer length influences variation in SNP frequencies for inverted and mirror repeats (Supplementary Figure S7). Namely, inverted and mirror repeats with <15-bp-long spacers exhibited a more pronounced elevation in SNP frequency. This result suggests that inverted and mirror repeats with short spacers have a higher probability of forming stable structures as compared to the ones with long spacers.
Nucleotide substitution frequencies at flanking sequences of non-B DNA loci
For most types of non-B DNA loci, the alteration in nucleotide substitution frequencies continued into their flanking regions. Even though we analyzed 2 kb up- and downstream of flanking regions, most of the effect was observed in the first 100 bp, and thus we present them in Figures 2–4 (full 2-kb flanking sequences are presented in Supplementary Figure S8). For all NCNR G4 loci (with four stems and three loops), we measured significantly elevated SNP and FNS frequencies extending into the flanking regions. Contiguous regions of significant elevation extended into the neighboring 3–6 bp, and more scattered regions of smaller, but still significant, elevation existed up to ∼500 bp. However, this flanking region elevation was smaller than that within the loci themselves and was the highest at the immediate first flanking nucleotide position upstream and downstream. Indeed, the first flanking nucleotide position experienced up to 2.2- and 1.5-fold increase in SNP and FNS frequency, respectively, compared to control sequences (Figure 2A and B). In contrast, the other flanking nucleotides analyzed experienced only up to 1.4- and 1.3-fold increase in SNP and FNS frequency, respectively, compared to control sequences (Figure 2A and B). Importantly, the elevation in SNP and FNS frequency was more pronounced in the flanking regions of stable than unstable G4 loci (Supplementary Figure S9A–D).
In the flanking regions of direct repeats, SNP frequency was significantly elevated (up to 2.5-fold), although gradually decreasing over a distance of ∼450 bp (Figure 3B), whereas FNS frequency was elevated at the first flanking position only (up to 1.7-fold; Figure 4B), as compared with control sequences. Both SNP and FNS frequencies were significantly elevated (up to 1.4- and 1.1-fold, respectively) in the flanking regions of mirror repeats, mainly in the first few (one to eight) flanking nucleotides (Figures 3C and 4C). The influence of inverted repeats on SNP and FNS frequency did not extend into their flanking regions (Figures 3A and 4A). The elevated SNP and FNS frequencies at Z-DNA loci extended over a distance of ∼175 and ∼12 bp, respectively (Figures 3D and 4D), in the surrounding flanking regions. In the flanking regions of A-phased repeats, the decrease in SNP and FNS frequency was concentrated at the first flanking nucleotide (Figures 3E and 4E).
SNP frequencies at the first flanking nucleotide positions (immediately adjacent to non-B motif annotations), both up- and downstream, were particularly high for all types of non-B DNA loci except for inverted repeats (Figures 2–4). In an effort to explain this observation, we asked whether this increase was driven by a particular substitution type (A→T, A→C, etc.), thus skewing the substitution spectrum. To investigate this possibility, we compared the frequencies of different substitutions at the first flanking nucleotide position, separately for each type of non-B DNA.
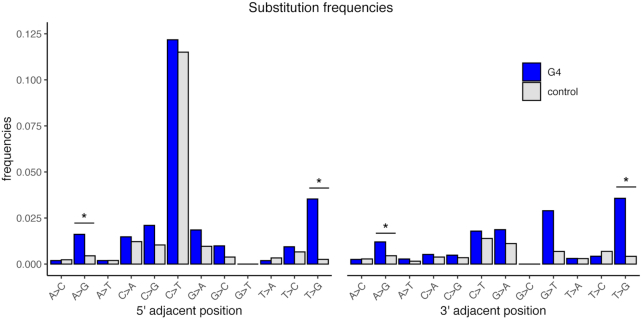
The substitution spectra were indeed different for the first flanking positions of G4 loci as compared to control sequences (Figure 5; Supplementary Tables S4A and B). Because G4 structures form only on one DNA strand, they might skew the substitution spectrum at the first flanking base in a strand-specific fashion. To account for this, we studied the substitution spectrum at G4 loci annotated on the reference strand only (i.e. restricting G4 motifs to the G-rich strand and not considering C-rich strand motifs; G-quadruplexes annotated on the non-reference strand were analyzed in Supplementary Figure S10, and the results were similar). The requirement of the first and the last bases at G4 loci to be a guanine might lead to a nucleotide substitution spectrum bias at the first flanking positions. For instance, a cytosine in the upstream flanking base followed by a guanine in the annotated G4 can form a CpG dinucleotide, which, if methylated, has elevated rates of CpG→TpG substitutions (117). To correct for this potential bias at the immediate flanking bases of G4 loci, we computed nucleotide substitution frequencies in their trinucleotide context, i.e. the ratio of the number of occurrences of a trinucleotide with the mutated middle base and the total number of occurrences of this trinucleotide. This procedure was performed separately for G4 loci (with four stems and three loops and located in the NCNR subgenome) and control sequences, and effectively corrected for the differences in trinucleotide composition between these two groups of sequences. After applying such corrections, we observed that the T→G substitution frequency was significantly elevated at the first up- and downstream flanking positions of stable G4 loci (odds ratios 14.04 and 8.51, Bonferroni-corrected P-values 1.09 × 10−30 and 1.60 × 10−13, respectively, Fisher's exact test; Figure 5 and Supplementary Table S4A). The A→G substitution frequency was also significantly elevated at the first up- and downstream positions of stable G4 loci, albeit to a smaller extent (odds ratios 3.61 and 2.66, Bonferroni-corrected P-values 1.34 × 10−6 and 3.97 × 10−3, respectively, Fisher's exact test). The substitution frequencies at the first flanking positions of unstable G4 loci were not significantly different from those at control sequences (Supplementary Table S4B). These results suggest that the first flanking nucleotides of stable G4s have a tendency to acquire additional guanines, leading to G4 elongation.
Figure 5.
Frequencies of polymorphic substitutions at the immediate first 5′ and 3′ flanking positions of stable G4 loci annotated on the reference strand. Only the frequencies of trinucleotides present at the immediate flanking positions of stable G4 loci were compared with those present at control sequences (trinucleotides present only in control sequences were not considered). A correction for the trinucleotide context was applied (see Materials and Methods). Two-sided Fisher's exact test was used to evaluate significant differences, and P-values were adjusted for multiple testing using Bonferroni correction. An asterisk (*) marks significant differences between G4 and control sequences (adjusted P-value < 0.05).
Whereas other types of non-B DNA we examined also displayed differences in their nucleotide substitution frequencies at the first flanking positions compared to those of control sequences (Supplementary Table S4C–G), none demonstrated a mutation spectrum as skewed as that observed in G4s. In Z-DNA, all substitution frequencies were elevated except for C→T and G→A (Supplementary Table S4F). All substitution frequencies were evenly (2–3-fold) elevated at both immediate flanking positions of direct repeats (Supplementary Table S4D).
Non-B DNA loci explain a substantial portion of large-scale variation in nucleotide substitution frequencies
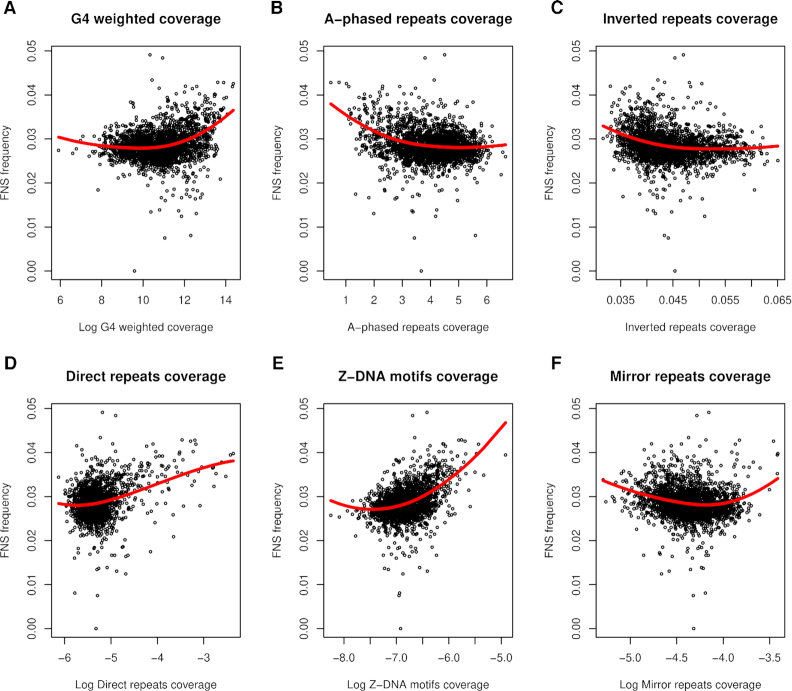
To evaluate whether non-B DNA loci contribute to large-scale regional variation in nucleotide substitution frequencies across the genome, we studied how divergence, as measured with human-orangutan FNS frequency, and diversity, as measured with human SNP frequency from the SGDP (97), vary with the coverage of each of the six non-B DNA types measured in 1-Mb genomic windows (see Materials and Methods; for G4 loci, we computed a weighted average of per-window coverage, weighting with predicted stability as provided by Quadron scores (98)). Nucleotide substitution frequencies and non-B DNA coverages were measured in the NCNR portion of each 1-Mb window. Non-linear (quadratic) relationships were observed between nucleotide substitution frequencies and coverage of each non-B DNA type studied (Figures 6 and Supplementary Figure S9); however, there were some differences among non-B DNA types. When considering divergence, G4, direct repeat and Z-DNA coverage exhibited an almost flat trend followed by positive associations, A-phased repeat and inverted repeat coverage exhibited negative associations followed by a flat trend, whereas mirror repeat coverage exhibited a negative trend followed by a positive trend, depending on their values (Figure 6). Similar patterns were observed considering diversity (Supplementary Figure S11).
Figure 6.
Relationships between fixed nucleotide substitution (FNS) frequency and non-B DNA. (A) G-quadruplexes coverage weighted by stability, (B) A-phased repeats coverage, (C) inverted repeats coverage, (D) direct repeats coverage, (E) Z-DNA motifs coverage, and (F) mirror repeats coverage. Red curves represent loess (locally estimated scatterplot smoothing) fits superimposed to the scatterplots to visualize trends. See Supplementary Figure S11 for an analogous analysis performed using SNP data.
We next examined how non-B DNA contributes to regional variation in nucleotide substitution frequencies in the context of 26 additional genomic features that were previously shown to affect such variation (Supplementary Table S2). To minimize multicollinearity, the overall set of potential predictors was reduced by clustering features based on pairwise Spearman's correlations and retaining only one feature per cluster (Supplementary Figure S6), similar to the procedure used in (118,119). We retained all six non-B DNA types and 15 additional genomic features. Next, we fitted two multiple regressions (separately for divergence and diversity) on this reduced set of predictors and, when needed, their quadratic terms, performing further variable selection with the Elastic Net and measuring the contribution of each retained predictor with a coefficient of partial determination (partial R2; see Materials and Methods). Average mappability score (106) was included as a predictor in all three models to control for its effect.
The resulting models explained 58.0% and 55.6% of variation in divergence and diversity, respectively (Table 1 and Supplementary Table S3). Importantly, all types of non-B DNA were retained as significant predictors in both models. Among different types of non-B DNA, Z-DNA, direct repeats, and G4 loci were the strongest predictors of divergence (explaining 2.32%, 1.63% and 1.04%, respectively), whereas direct repeats and G4 loci were the strongest predictors of diversity (explaining 8.01% and 1.08% of variation, respectively). Even when considered in the context of other relevant genomic features, as a group, different types of non-B DNA explained as much as 15.94% and 19.95% of variation not explained by the other features in the two regressions, respectively. If we were to exclude non-B DNA, our models would explain 50.0% and 44.5% of variation in divergence and diversity, respectively (note that partial R2 values usually do not add up because of correlations among predictors). In our full models (Table 1), among other genomic features, distance to telomere, replication timing, H3K9me3, and lamina-associated domains were strong predictors of divergence (explaining 15.3%, 2.08%, 1.72% and 1.35% of variation, respectively), whereas distance to telomere, recombination rate, distance to centromere, H3K9me3, and DNase hypersensitive sites were strong predictors of diversity (explaining 14.2%, 6.12%, 3.45%, 1.52% and 1.08% of variation, respectively). Notably, the contribution of all non-B DNA types taken as a group exceeded that of any other genomic feature in both divergence and diversity regressions. Thus, our results strongly suggest that non-B DNA is a key contributor to large-scale regional variation in nucleotide substitution frequencies.
Table 1.
Coefficients of partial determination of regional variation in nucleotide substitution and the proportion of variation explained by each model (at a 1-Megabase scale)
| Predictors/models | Fixed nucleotide substitution (FNS) frequency | Single nucleotide polymorphism (SNP) frequency |
|---|---|---|
| G4 weighted coveragea | 1.04% | 1.08% |
| A-phased repeats coverage | 0.50% | 0.10% |
| Inverted repeats coverage | 0.48% | 0.68% |
| Direct repeats coverage | 1.63% | 8.01% |
| Z-DNA motifs coverage | 2.32% | 0.66% |
| Mirror repeats coverage | 0.47% | 0.27% |
| All non-B | 15.94% | 19.95% |
| Replication timing | 2.08% | 0.54% |
| Recombination rate | 0.65% | 6.12% |
| DNase hypersensitive sites coverage | 0.12% | 1.08% |
| RNA polymerase II binding sites coverage | 0.49% | 0.66% |
| CHH methylation coverage | 0.29% | 0.01% |
| CpG island coverage | 0.67% | 0.35% |
| Lamina Associated Domains coverage | 1.35% | 0.72% |
| Distance to telomere | 15.25% | 14.19% |
| Distance to centromere | 0.76% | 3.45% |
| H2AFZ signal | 0.29% | 0.39% |
| H3K27me3 signal | 0.00% | 0.14% |
| H3K36me3 signal | 0.38% | 0.12% |
| H3K9ac signal | 0.15% | 0.27% |
| H3K9me3 signal | 1.72% | 1.52% |
| Telomere hexamer presence | 0.28% | 0.02% |
| R-squared | 58.00% | 55.61% |
aCoverage of G4 loci was weighted by their stability.
DISCUSSION
Harnessing publicly available datasets that span different evolutionary times (human-orangutan divergence and human diversity) and sophisticated statistical methods, our genome-wide study illuminates the relationship between non-B DNA loci and variation in nucleotide substitution frequencies. Using IWTomics, we delineated distinct patterns of small-scale variation in and around different types of non-B DNA loci. Using multiple regression analysis, we determined the contribution of non-B DNA to explaining regional variation in nucleotide substitution frequencies at the 1-Mb scale. Our results compellingly argue for a pivotal role of non-B DNA in shaping the mutation dynamics of the genome, which has important biological implications.
On the one hand, our genome-wide study supports a ‘unifying hypothesis’, whereby non-B DNA plays a role linking genomic architecture and local DNA environment with mutations that underlie human inherited disease (120). Thus, future studies should consider the potential of a locus to form non-B DNA when evaluating candidate genetic variants for human genetic diseases. On the other hand, because of its high mutability, non-B DNA likely represents an inexhaustible source of novel genetic variation in natural populations. In agreement with this, a recent study in stickleback fish found that Z-DNA forming in an enhancer of the Pitx1 gene increases the probability of deletions that lead to the loss of pelvic hindfins, and are repeatedly utilized for adaptation to a freshwater environment (3). Based on our findings, we expect that many analogous examples of the use and re-use of non-B DNA in adaptation will be discovered in the near future.
Small-scale variation in nucleotide substitution frequency
We have shown that non-B DNA loci often have nucleotide substitution frequencies significantly different from those characterizing B DNA (usually higher). This corroborates an earlier report of increased SNP density at non-B DNA in the 1000 Genomes Project (24), even though this study might have been biased by a potentially increased Illumina sequencing error rate at non-B DNA that is expected to elevate false positives for genomes sequenced at low depth (85). The present study does not suffer from this limitation as it uses deeply sequenced genomes. The altered nucleotide substitution frequencies at non-B DNA loci we detected here are in agreement with another study from our group, in which we found that PacBio sequencing errors are affected by non-B DNA (85). Thus, non-B DNA loci likely affect both errors in the PacBio sequencer and mutations in germline cells. Other recent studies found non-B DNA loci to be mutation hotspots in cancer genomes (23,64), suggesting that such loci also drive mutagenesis in somatic cells.
G4 loci
Aggregating variants at tens of thousands of G4 loci, we observed an increase in both diversity and divergence, consistent with previous reports of elevated 1000 Genome Project SNP frequency at G4 loci (24) and of G4 motif enrichment at cancer mutations (23). Potential mechanisms underlying heightened mutagenesis at G4 loci include DNA polymerase errors during replication or repair, and/or damage-induced mutations. Importantly, these mechanisms are not mutually exclusive. Due to their G-rich sequence, G4 motifs are subject to oxidative DNA damage (reviewed in (121)), which, if unrepaired, can lead to mutations. Eukaryotic replicative DNA polymerases are inhibited at some G4 structures (reviewed in (122)). Possibly, G4s compromise the high fidelity of replicative DNA polymerases, leading to errors during DNA synthesis. This hypothesis is based on our previously published genome-wide results showing a direct link between polymerization errors during PacBio sequencing and germline mutations at G4 loci (85). Indeed, we found levels of divergence and diversity to be negatively correlated with polymerization speed and accuracy in the sequencer at such loci (85). Alternatively, error-prone polymerases may be engaged at a slowed or stalled fork to ensure complete G4 replication. This mechanism is supported by evidence suggesting that specialized polymerases eta, kappa, and Rev1 are important for G4 stability (122).
We demonstrated that stable G4 loci have higher nucleotide substitution frequencies as compared to unstable G4 loci (Figure 2C and D). Because stability reflects the probability and strength of structure formation, this result suggests that stable G4 structures are a more serious and/or more common obstacle for polymerization progression leading to increased errors, which frequently result in mutations. In support of this claim, we previously found positive relationships between polymerization slowdown and error rates, and between polymerization slowdown and G4 stability, as measured by circular dichroism (85). Potential mechanisms include replication fork restart at G4 motifs by PrimPol, an error prone polymerase (123), or fork degradation and breakage followed by polymerase theta-mediated, error-prone DNA end joining (124). The effect of stability on polymerization accuracy was also found for other types of non-B DNA—Z-DNA and triplex DNA (125).
Stable and unstable G4 loci differed more in divergence comparisons than in diversity comparisons, suggesting that the stability of G4 structures evolves over time. While the classification of individual G4 loci into stable versus unstable likely remains valid for human populations, there is a probability that extrapolating the predicted stability of some human G4 loci to orangutan loci might be incorrect, due to accumulated mutations in the latter. Thus, for instance, some G4 loci annotated as stable in the human genome might be unstable in the orangutan genome. As a consequence, they may be a weaker obstacle for polymerization or accrue less oxidative damage, leading to fewer mutations, and resulting in lower fixed nucleotide substitution frequencies.
Within G4 loci, we found that nucleotide substitution frequencies were more elevated in loops, which can vary without impeding G4 structures, than in guanine stems, which are critical for G4 formation. This suggests that stems and loops differ in mutability and/or selective pressure. Guanines located in both stem tetrads and loop sequences are subject to oxidative damage when folded into quadruplex structures, and the 5′ guanine within a tetrad can be a hotspot of damage (126). If G4 stems had low mutability, we would expect guanines to harbor few polymerization errors. However, this was not the case for the polymerase used in PacBio sequencing, which exhibited many polymerase errors at guanines (85). Therefore, the precise mechanisms leading to the observed differences in stem versus loop substitutions remain to be elucidated.
To assess potential selection in stems, we tested whether the site frequency spectra differed between stems and loops in the SGDP SNPs (97). Although the two distributions were similar visually, we observed a significantly higher proportion of SNPs with lower minor allele frequency in stems than in loops (P-value < 2.2e–16, Kolmogorov–Smirnov test; Supplementary Figure S12). Assuming the latter evolve neutrally, this suggests purifying selection acting on the former. Such an observation is consistent with a study in S. cerevisiae, proposing that stems are conserved to safeguard G4 structure formation (127), and contradicts another study, proposing that the underrepresentation of SNPs in G4 guanine tracts has neutral causes (128). Notably, we found evidence of selection acting on G4 stems even though our analyses focused on G4 loci found in the NCNR subgenome, which ought to have a high probability of evolving neutrally. We also found the centers of G4 stems to be more conserved than their edges (Figure 2), consistent with previous reports of the guanine at the center of stems being the most important nucleotide for the formation of a G4 structure (128–130). In summary, our results suggest that even though they drive mutagenesis and genome instability interfering with DNA replication (131,132), and even when they are located in the NCNR subgenome, some stable G4 loci might be beneficial for other essential processes and thus preserved in the genome.
A-phased repeats
In contrast to G4 loci, A-phased repeats had depressed overall nucleotide substitution frequencies. This is consistent with slightly lowered rates of polymerization errors observed for A-phased repeats during PacBio sequencing (85). Interestingly, these loci showed a periodic pattern that was in some ways similar to that of G4 loci. In our analysis, nucleotide substitution frequencies were depressed in A-tracts but similar to controls in spacers. Lowered nucleotide substitution frequencies in A-tracts may be explained in part by the previously reported low mutability of adenines (1,4,5); PacBio polymerization error rates at adenines were also reported to be low (85).
Direct, inverted, and mirror repeats
We previously reported elevated PacBio sequencing errors at mirror, direct, and (to a lower extent) inverted repeats (85). Elevated SNP frequencies (1000 Genomes Project, (24)) and cancer somatic mutations (23) were also reported for these loci. Intriguingly, we observed both increases and decreases in nucleotide substitution frequencies at mirror, inverted and direct repeats. For all three repeat types, frequencies were higher in spacers than in repeat arms. This is reminiscent of observations for cancer mutations (23), and consistent with previous experimental studies demonstrating that spacers are more mutable in hairpin structures (84,133). We also found that nucleotide substitution patterns within repeat arms differed among repeat types. (84,133). Non-B DNA loci composed of repeats can pose an obstacle to DNA replicative polymerases, and the observed nucleotide substitution frequencies could reflect processing by low-fidelity, specialized DNA polymerases (65,66,134–136) gene conversion and increased DNA damage. Gene conversion is common at cruciform and slipped-strand structures (137), and could explain the decreased nucleotide substitution frequencies in repeat arms of inverted and direct repeats—particularly for fixed nucleotide substitutions, where multiple rounds of gene conversion might have taken place. Oxidative DNA damage was proposed as an important mechanism of increased mutagenesis at mirror repeats (125,138).
Z-DNA
At Z-DNA loci, the elevated diversity and divergence we detected are consistent with previous reports of increased germline and somatic mutations (23,24), but contradict our previous study, in which such loci showed decreased levels of PacBio polymerization errors (85). A potential resolution of this contradiction might lie in the importance of compromised DNA repair (125,138) and of environmental factors (139,140) for Z-DNA mutagenesis. We found that the edges of Z-DNA loci were particularly enriched in nucleotide substitutions (Figures 3D and 4D), potentially because they coincide with the B–Z junctions (125,138).
Nucleotide substitution frequencies in regions flanking non-B DNA loci
Our results suggest that the effect of most types of non-B DNA loci on diversity and divergence is not restricted to their sequences but extends into their flanking regions—albeit at a smaller magnitude and limited range. We found an elevation in substitution frequencies extending up to ∼500 bp in the flanking regions of G4 loci, which is further than the range of elevated indel frequencies previously reported for humans (23) and C. elegans (80) (150 and 200 bp, respectively). We also found substitution frequencies to be elevated up to ∼300 bp from direct repeats, which is further than the 50 bp previously reported for microsatellites, a similar type of loci (141). While Z-DNA and H-DNA-forming mirror repeats have been reported to increase mutation frequencies in their neighboring regions (125), we found that this effect is limited to at most ∼100 bp and 4 bp, respectively. Several molecular mechanisms have been proposed to explain the influence of non-B DNA loci on nucleotide substitutions in their flanking regions. Double-strand breaks associated with non-B DNA structures and the resulting DNA repair can be mutagenic (124,138). Alternatively, folding into non-B DNA structures may expose downstream sequences to oxidative damage through long-range hole migration, creating hotspots for mutagenesis in the surrounding sequences (125).
We also found a marked elevation in the nucleotide substitution frequencies at the first flanking nucleotides of some types of non-B DNA loci. Diversity and divergence were particularly elevated at the first flanking nucleotides of stable G4 loci, and showed a skewed substitution spectrum favoring T→G and A→G substitutions. This specificity is intriguing, and the mechanism(s) responsible are not clear at this time. Oxidative damage to dGTP precursor pools and subsequent incorporation of 8-oxo-dGTP opposite template A causes T→G substitutions (69). Possibly, the alternative folding of G4 distorts the immediate flanking template bases, and promotes this misincorporation by DNA polymerases during replication. Alternatively, the skewed spectrum may reflect the error specificity of specialized polymerases that are engaged at G4 motifs (122). Among the nuclear DNA polymerases thus far identified for this function, only DNA Pol eta produces T→G and A→G substitutions at measurable frequencies (142). Notably, these substitutions result in the elongation of G4 loci, which should further increase their stability—an observation that is in line with the potential functionality of some G4 loci in the NCNR portion of the human genome. The first flanking nucleotide of Z-DNA loci also showed a skewed substitution spectrum, but this was not the case for other non-B DNA loci considered in our study. These results echo findings of Bacolla et al. (143), who found that SNP frequency and spectrum at the first flanking nucleotides of mononucleotide microsatellites is affected by microsatellite repeat identity and length.
Large-scale variation in nucleotide substitution frequency
Our multiple regression models indicate that non-B DNA loci across the human genome are prominent contributors to regional variation in divergence and diversity measured at the 1-Mb scale. Each type of non-B DNA loci contributed significantly, with particularly strong contributions of direct repeats and G4 loci to variation in diversity, and of direct repeats and Z-DNA loci to variation in divergence (Table 1). Although we analyzed the NCNR genome, many of the studied non-B DNA loci do have a repetitive nature and could elevate mutation rates up to 10 kilobases away from the repetitive tract via repeat-induced mutagenesis (144). When taken together, non-B DNA loci explained ∼20% of the variation in divergence and ∼16% of the variation in diversity—greater than that of any previously reported predictors, which we confirmed to have substantial contributions—e.g. distance to telomeres, distance to centromeres (89,145), recombination rate (89,145,146), replication timing (9,17), histone modification H3K9me3 (12), and DNase hypersensitive sites (11,112,147).
Though comparisons are hindered by differences in methodologies, data sources, and overall sets of predictors considered, including non-B DNA does appear to increase the proportion of regional variation explained in our models as compared to that in prior modeling efforts. We explained 58.0% of variation in human-orangutan divergence, as compared to 52.6% and 52% obtained in prior models for human-chimpanzee (145) and human-macaque (89) divergence, respectively. We explained 55.6% of variation in SNP frequency from SGDP (97), as compared to 35% obtained in prior models of SNP frequency from dbSNP (12).
Limitations of the study and future directions
At any given time, only a subset of annotated non-B DNA loci are expected to fold into non-B DNA structures in vivo (148). The preferential conformation of a motif depends on the precise sequence and stability of B versus non-B DNA (35) and on conditions within the cell. Supercoiling, transcriptional activity, and conditions in the nucleus (e.g. ionic concentrations) can impact transitions between B and non-B DNA conformations at a locus. Due to this transient nature, the effects of non-B DNA structures at individual loci are difficult to study and must be aggregated into genome-wide trends. Thus, the results of our study cannot be extrapolated to individual loci. At present, it remains challenging to identify individual loci that form non-B DNA structures at a given time in vivo. However, new methods such as permanganate sequencing (148), kethoxal-assisted single-stranded DNA sequencing (149), and G4-ChIP-seq (36,150) will increase our ability to study the effects of individual non-B loci on nucleotide substitution frequencies in the near future.
While including non-B DNA loci as predictors in our models of large-scale variation in substitution frequencies provided a substantial improvement, we did not evaluate the interaction terms. Including interactions between predictors will complicate the analysis, but may bolster our predictions and further increase the amount of explained variation. For instance, the distribution of non-B DNA is non-uniform along the genome and may correlate with other genomic features used in the models (e.g. recombination rate; note that multicollinearity in our models was controlled by clustering and eliminating some correlated predictors at the outset). Co-occurrence of non-B DNA loci and other features creates an opportunity for joint effects on nucleotide substitution frequencies, which ought to be evaluated in future studies.
Despite our best efforts to restrict our study to neutrally evolving regions of the genome by limiting it to the NCNR subgenome, we captured footprints of purifying selection at G4 loci. Because we are aggregating multiple loci, it may be challenging to distinguish local changes in mutability from selective pressure. However, the possibility of natural selection acting upon some loci previously considered as neutral is an exciting prospect; the potential functions of such loci should be characterized in future work. In fact, natural selection at some non-B DNA loci has been reported previously (24,151)—but the issue remains underexplored to date.
Finally, our study offers only glimpses into the molecular mechanisms underlying our findings. Our results are consistent with DNA polymerase errors being an important mechanism behind increased mutagenesis at non-B DNA loci, and it will therefore be critical to deepen the investigation through further genome-wide studies. Additionally, future work evaluating the role of DNA repair and the effect of single-strandedness on oxidative damage-induced mutagenesis genome-wide will be important for a complete understanding of the mutability of non-B DNA loci and their flanking sequences.
DATA AVAILABILITY
All computational tools used in this study are available as a GitHub repository at the following URL: https://github.com/makovalab-psu/nonB-RegVar.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Monika Cechova for discussions about studying substitution types in the flanking sequences of G4 loci, and Suzanne Hile and MaryElizabeth Stein for their help in revising the manuscript.
Contributor Information
Wilfried M Guiblet, Bioinformatics and Genomics Graduate Program, Penn State University, UniversityPark, PA 16802, USA.
Marzia A Cremona, Department of Statistics, The Pennsylvania State University, University Park, PA 16802, USA; Department of Operations and Decision Systems, Université Laval, Canada; CHU de Québec – Université Laval Research Center, Canada.
Robert S Harris, Department of Biology, Penn State University, University Park, PA 16802, USA.
Di Chen, Intercollege Graduate Degree Program in Genetics, Huck Institutes of the Life Sciences, Penn State University, UniversityPark, PA 16802, USA.
Kristin A Eckert, Department of Pathology, Penn State University, College of Medicine, Hershey, PA 17033, USA; Center for Medical Genomics, Penn State University, University Park and Hershey, PA, USA.
Francesca Chiaromonte, Department of Statistics, The Pennsylvania State University, University Park, PA 16802, USA; Center for Medical Genomics, Penn State University, University Park and Hershey, PA, USA; EMbeDS, Sant’Anna School of Advanced Studies, 56127 Pisa, Italy.
Yi-Fei Huang, Department of Biology, Penn State University, University Park, PA 16802, USA; Center for Medical Genomics, Penn State University, University Park and Hershey, PA, USA.
Kateryna D Makova, Department of Biology, Penn State University, University Park, PA 16802, USA; Center for Medical Genomics, Penn State University, University Park and Hershey, PA, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01GM136684 and R01CA23715]; Clinical and Translational Sciences Institute; Institute of Computational and Data Sciences; Huck Institutes of the Life Sciences; Eberly College of Science of the Pennsylvania State University; Pennsylvania Department of Health using Tobacco Settlement and CURE Funds (the department specifically disclaims any responsibility for any analysesy or conclusions); CBIOS Predoctoral Training Program awarded to Penn State by the National Institutes of Health (W.M.G. is a trainee). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Hodgkinson A., Eyre-Walker A.. Variation in the mutation rate across mammalian genomes. Nat. Rev. Genet. 2011; 12:756–766. [DOI] [PubMed] [Google Scholar]
- 2. Makova K.D., Hardison R.C.. The effects of chromatin organization on variation in mutation rates in the genome. Nat. Rev. Genet. 2015; 16:213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xie K.T., Wang G., Thompson A.C., Wucherpfennig J.I., Reimchen T.E., MacColl A.D.C., Schluter D., Bell M.A., Vasquez K.M., Kingsley D.M.. DNA fragility in the parallel evolution of pelvic reduction in stickleback fish. Science. 2019; 363:81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gojobori T., Li W.H., Graur D.. Patterns of nucleotide substitution in pseudogenes and functional genes. J. Mol. Evol. 1982; 18:360–369. [DOI] [PubMed] [Google Scholar]
- 5. Bulmer M. Neighboring base effects on substitution rates in pseudogenes. Mol. Biol. Evol. 1986; 3:322–329. [DOI] [PubMed] [Google Scholar]
- 6. Coulondre C., Miller J.H., Farabaugh P.J., Gilbert W.. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978; 274:775–780. [DOI] [PubMed] [Google Scholar]
- 7. Bacolla A., Temiz N.A., Yi M., Ivanic J., Cer R.Z., Donohue D.E., Ball E.V., Mudunuri U.S., Wang G., Jain A.et al.. Guanine holes are prominent targets for mutation in cancer and inherited disease. PLoS Genet. 2013; 9:e1003816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lercher M.J., Hurst L.D.. Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 2002; 18:337–340. [DOI] [PubMed] [Google Scholar]
- 9. Stamatoyannopoulos J.A., Adzhubei I., Thurman R.E., Kryukov G.V., Mirkin S.M., Sunyaev S.R.. Human mutation rate associated with DNA replication timing. Nat. Genet. 2009; 41:393–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boulikas T. Evolutionary consequences of nonrandom damage and repair of chromatin domains. J. Mol. Evol. 1992; 35:156–180. [DOI] [PubMed] [Google Scholar]
- 11. Ying H., Epps J., Williams R., Huttley G.. Evidence that localized variation in primate sequence divergence arises from an influence of nucleosome placement on DNA repair. Mol. Biol. Evol. 2010; 27:637–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schuster-Böckler B., Lehner B.. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012; 488:504–507. [DOI] [PubMed] [Google Scholar]
- 13. Polak P., Karlić R., Koren A., Thurman R., Sandstrom R., Lawrence M., Reynolds A., Rynes E., Vlahoviček K., Stamatoyannopoulos J.A.et al.. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature. 2015; 518:360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ananda G., Chiaromonte F., Makova K.D.. A genome-wide view of mutation rate co-variation using multivariate analyses. Genome Biol. 2011; 12:R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gaffney D.J., Keightley P.D.. The scale of mutational variation in the murid genome. Genome Res. 2005; 15:1086–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hodgkinson A., Chen Y., Eyre-Walker A.. The large-scale distribution of somatic mutations in cancer genomes. Hum. Mutat. 2012; 33:136–143. [DOI] [PubMed] [Google Scholar]
- 17. Agarwal I., Przeworski M.. Signatures of replication timing, recombination, and sex in the spectrum of rare variants on the human X chromosome and autosomes. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:17916–17924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Terekhanova N.V., Seplyarskiy V.B., Soldatov R.A., Bazykin G.A.. Evolution of local mutation rate and its determinants. Mol. Biol. Evol. 2017; 34:1100–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Watson J.D., Crick F.H.C.. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953; 171:964–967. [DOI] [PubMed] [Google Scholar]
- 20. Wilkins M.H.F., Stokes A.R., Wilson H.R.. Molecular structure of nucleic acids: molecular structure of deoxypentose nucleic acids. Nature. 1953; 171:738–740. [DOI] [PubMed] [Google Scholar]
- 21. Franklin R.E., Gosling R.G.. Molecular configuration in sodium thymonucleate. Nature. 1953; 171:740–741. [DOI] [PubMed] [Google Scholar]
- 22. Huppert J.L., Balasubramanian S.. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005; 33:2908–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Georgakopoulos-Soares I., Morganella S., Jain N., Hemberg M., Nik-Zainal S.. Noncanonical secondary structures arising from non-B DNA motifs are determinants of mutagenesis. Genome Res. 2018; 28:1264–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Du X., Gertz E.M., Wojtowicz D., Zhabinskaya D., Levens D., Benham C.J., Schäffer A.A., Przytycka T.M.. Potential non-B DNA regions in the human genome are associated with higher rates of nucleotide mutation and expression variation. Nucleic Acids Res. 2014; 42:12367–12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mirkin S.M., Others Discovery of alternative DNA structures: a heroic decade (1979–1989). Front. Biosci. 2008; 13:1064–1071. [DOI] [PubMed] [Google Scholar]
- 26. Cer R.Z., Bruce K.H., Mudunuri U.S., Yi M., Volfovsky N., Luke B.T., Bacolla A., Collins J.R., Stephens R.M.. Non-B DB: a database of predicted non-B DNA-forming motifs in mammalian genomes. Nucleic Acids Res. 2011; 39:D383–D391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sen D., Gilbert W.. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988; 334:364–366. [DOI] [PubMed] [Google Scholar]
- 28. Rich A., Nordheim A., Wang A.H.. The chemistry and biology of left-handed Z-DNA. Annu. Rev. Biochem. 1984; 53:791–846. [DOI] [PubMed] [Google Scholar]
- 29. Mirkin S.M., Lyamichev V.I., Drushlyak K.N., Dobrynin V.N., Filippov S.A., Frank-Kamenetskii M.D.. DNA H form requires a homopurine-homopyrimidine mirror repeat. Nature. 1987; 330:495–497. [DOI] [PubMed] [Google Scholar]
- 30. Panayotatos N., Wells R.D.. Cruciform structures in supercoiled DNA. Nature. 1981; 289:466–470. [DOI] [PubMed] [Google Scholar]
- 31. Lilley D.M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc. Natl. Acad. Sci. U.S.A. 1980; 77:6468–6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sinden R.R., Pytlos-Sinden M.J., Potaman V.N.. Slipped strand DNA structures. Front. Biosci. 2007; 12:4788–4799. [DOI] [PubMed] [Google Scholar]
- 33. Barbič A., Zimmer D.P., Crothers D.M.. Structural origins of adenine-tract bending. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:2369–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Neidle S. Oxford Handbook of Nucleic acid Structure. 1999; Oxford University Press. [Google Scholar]
- 35. Zhao J., Bacolla A., Wang G., Vasquez K.M.. Non-B DNA structure-induced genetic instability and evolution. Cell. Mol. Life Sci. 2010; 67:43–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hänsel-Hertsch R., Beraldi D., Lensing S.V., Marsico G., Zyner K., Parry A., Di Antonio M., Pike J., Kimura H., Narita M.et al.. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016; 48:1267–1272. [DOI] [PubMed] [Google Scholar]
- 37. Baral A., Kumar P., Halder R., Mani P., Yadav V.K., Singh A., Das S.K., Chowdhury S.. Quadruplex-single nucleotide polymorphisms (Quad-SNP) influence gene expression difference among individuals. Nucleic Acids Res. 2012; 40:3800–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hizver J., Rozenberg H., Frolow F., Rabinovich D., Shakked Z.. DNA bending by an adenine–thymine tract and its role in gene regulation. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:8490–8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Belotserkovskii B.P., Liu R., Tornaletti S., Krasilnikova M.M., Mirkin S.M., Hanawalt P.C.. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:12816–12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wittig B., Dorbic T., Rich A.. Transcription is associated with Z-DNA formation in metabolically active permeabilized mammalian cell nuclei. Proc. Natl. Acad. Sci. U.S.A. 1991; 88:2259–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Parkinson G.N., Lee M.P.H., Neidle S.. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002; 417:876–880. [DOI] [PubMed] [Google Scholar]
- 42. Moye A.L., Porter K.C., Cohen S.B., Phan T., Zyner K.G., Sasaki N., Lovrecz G.O., Beck J.L., Bryan T.M.. Telomeric G-quadruplexes are a substrate and site of localization for human telomerase. Nat. Commun. 2015; 6:7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sahakyan A.B., Murat P., Mayer C., Balasubramanian S.. G-quadruplex structures within the 3′ UTR of LINE-1 elements stimulate retrotransposition. Nat. Struct. Mol. Biol. 2017; 24:243. [DOI] [PubMed] [Google Scholar]
- 44. Brázda V., Laister R.C., Jagelská E.B., Arrowsmith C.. Cruciform structures are a common DNA feature important for regulating biological processes. BMC Mol. Biol. 2011; 12:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mani P., Yadav V.K., Das S.K., Chowdhury S.. Genome-wide analyses of recombination prone regions predict role of DNA structural motif in recombination. PLoS One. 2009; 4:e4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van Wietmarschen N., Merzouk S., Halsema N., Spierings D.C.J., Guryev V., Lansdorp P.M.. BLM helicase suppresses recombination at G-quadruplex motifs in transcribed genes. Nat. Commun. 2018; 9:271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maizels N., Gray L.T.. The G4 genome. PLoS Genet. 2013; 9:e1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Aranda A., Pérez-Ortín J.E., Benham C.J., del Olmo M.L.Í.. Analysis of the structure of a natural alternating d (TA) n sequence in yeast chromatin. Yeast. 1997; 13:313–326. [DOI] [PubMed] [Google Scholar]
- 49. Mao S.-Q., Ghanbarian A.T., Spiegel J., Cuesta S.M., Beraldi D., Di Antonio M., Marsico G., Hänsel-Hertsch R., Tannahill D., Balasubramanian S.. DNA G-quadruplex structures mold the DNA methylome. Nat. Struct. Mol. Biol. 2018; 25:951–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Halder R., Halder K., Sharma P., Garg G., Sengupta S., Chowdhury S.. Guanine quadruplex DNA structure restricts methylation of CpG dinucleotides genome-wide. Mol. Biosyst. 2010; 6:2439–2447. [DOI] [PubMed] [Google Scholar]
- 51. Jara-Espejo M., Peres Line S.R.. DNA G-quadruplex stability, position and chromatin accessibility are associated with CpG island methylation. FEBS J. 2020; 287:483–495. [DOI] [PubMed] [Google Scholar]
- 52. Yuan L., Tian T., Chen Y., Yan S., Xing X., Zhang Z., Zhai Q., Xu L., Wang S., Weng X.et al.. Existence of G-quadruplex structures in promoter region of oncogenes confirmed by G-quadruplex DNA cross-linking strategy. Sci. Rep. 2013; 3:1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H.. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Simonsson T., Pecinka P., Kubista M.. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998; 26:1167–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Miller D.M., Thomas S.D., Islam A., Muench D., Sedoris K.. c-Myc and cancer metabolism. Clin. Cancer Res. 2012; 18:5546–5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bochman M.L., Paeschke K., Zakian V.A.. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012; 13:770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Haeusler A.R., Donnelly C.J., Periz G., Simko E.A.J., Shaw P.G., Kim M.-S., Maragakis N.J., Troncoso J.C., Pandey A., Sattler R.et al.. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014; 507:195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Maizels N. G4-associated human diseases. EMBO Rep. 2015; 16:910–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wolfe A.L., Singh K., Zhong Y., Drewe P., Rajasekhar V.K., Sanghvi V.R., Mavrakis K.J., Jiang M., Roderick J.E., Van der Meulen J.et al.. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014; 513:65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bacolla A., Wells R.D.. Non-B DNA conformations, genomic rearrangements, and human disease. J. Biol. Chem. 2004; 279:47411–47414. [DOI] [PubMed] [Google Scholar]
- 61. Mirkin S.M. Expandable DNA repeats and human disease. Nature. 2007; 447:932–940. [DOI] [PubMed] [Google Scholar]
- 62. Orr H.T., Zoghbi H.Y.. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007; 30:575–621. [DOI] [PubMed] [Google Scholar]
- 63. Pearson C.E., Sinden R.R.. Alternative structures in duplex DNA formed within the trinucleotide repeats of the myotonic dystrophy and fragile X loci. Biochemistry. 1996; 35:5041–5053. [DOI] [PubMed] [Google Scholar]
- 64. Bacolla A., Tainer J.A., Vasquez K.M., Cooper D.N.. Translocation and deletion breakpoints in cancer genomes are associated with potential non-B DNA-forming sequences. Nucleic Acids Res. 2016; 44:5673–5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bournique E., Dall’Osto M., Hoffmann J.-S., Bergoglio V.. Role of specialized DNA polymerases in the limitation of replicative stress and DNA damage transmission. Mutat. Res. 2018; 808:62–73. [DOI] [PubMed] [Google Scholar]
- 66. Tsao W.-C., Eckert K.A.. Detours to Replication: Functions of specialized DNA polymerases during oncogene-induced replication stress. Int. J. Mol. Sci. 2018; 19:3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Barnes R.P., Hile S.E., Lee M.Y., Eckert K.A.. DNA polymerases eta and kappa exchange with the polymerase delta holoenzyme to complete common fragile site synthesis. DNA Repair (Amst.). 2017; 57:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang G., Vasquez K.M.. Impact of alternative DNA structures on DNA damage, DNA repair, and genetic instability. DNA Repair (Amst.). 2014; 19:143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Poetsch A.R. The genomics of oxidative DNA damage, repair, and resulting mutagenesis. Comput. Struct. Biotechnol. J. 2020; 18:207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Khristich A.N., Mirkin S.M.. On the wrong DNA track: molecular mechanisms of repeat-mediated genome instability. J. Biol. Chem. 2020; 295:4134–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang G., Vasquez K.M.. Naturally occurring H-DNA-forming sequences are mutagenic in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:13448–13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rodriguez R., Miller K.M., Forment J.V., Bradshaw C.R., Nikan M., Britton S., Oelschlaegel T., Xhemalce B., Balasubramanian S., Jackson S.P.. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 2012; 8:301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. De S., Michor F.. DNA secondary structures and epigenetic determinants of cancer genome evolution. Nat. Struct. Mol. Biol. 2011; 18:950–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rodgers K., McVey M.. Error-prone repair of DNA double-strand breaks. J. Cell. Physiol. 2016; 231:15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang Y., Rohde L.H., Wu H.. Involvement of nucleotide excision and mismatch repair mechanisms in double strand break repair. Curr. Genomics. 2009; 10:250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhao J., Wang G., Del Mundo I.M., McKinney J.A., Lu X., Bacolla A., Boulware S.B., Zhang C., Zhang H., Ren P.et al.. Distinct mechanisms of nuclease-directed DNA-structure-induced genetic instability in cancer genomes. Cell Rep. 2018; 22:1200–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Paeschke K., Capra J.A., Zakian V.A.. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011; 145:678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Krasilnikova M.M., Mirkin S.M.. Replication stalling at Friedreich's Ataxia (GAA)n repeats in vivo. Mol. Cell. Biol. 2004; 24:2286–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Mirkin E.V., Mirkin S.M.. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 2007; 71:13–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cheung I., Schertzer M., Rose A., Lansdorp P.M.. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat. Genet. 2002; 31:405–409. [DOI] [PubMed] [Google Scholar]
- 81. Wang G., Christensen L.A., Vasquez K.M.. Z-DNA-forming sequences generate large-scale deletions in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:2677–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Collins N.S., Bhattacharyya S., Lahue R.S.. Rev1 enhances CAG·CTG repeat stability in Saccharomyces cerevisiae. DNA Repair (Amst.). 2007; 6:38–44. [DOI] [PubMed] [Google Scholar]
- 83. Marcadier J.L., Pearson C.E.. Fidelity of primate cell repair of a double-strand break within a (CTG)·(CAG) tract. J. Biol. Chem. 2003; 278:33848–33856. [DOI] [PubMed] [Google Scholar]
- 84. Vasquez K.M., Wang G.. The yin and yang of repair mechanisms in DNA structure-induced genetic instability. Mutat. Res. 2013; 743-744:118–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Guiblet W.M., Cremona M.A., Cechova M., Harris R.S., Kejnovská I., Kejnovsky E., Eckert K., Chiaromonte F., Makova K.D.. Long-read sequencing technology indicates genome-wide effects of non-B DNA on polymerization speed and error rate. Genome Res. 2018; 28:1767–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ramsay J., Silverman B.W.Henry Overton Wills Professor of Mathematics B W Silverman . Functional Data Analysis. 2005; Springer Science & Business Media. [Google Scholar]
- 87. Cremona M.A., Xu H., Makova K.D., Reimherr M., Chiaromonte F., Madrigal P.. Functional data analysis for computational biology. Bioinformatics. 2019; 35:3211–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chiaromonte F., Makova K.D.. Paganoni A.M., Secchi P.. Using Statistics to Shed Light on the Dynamics of the Human Genome: A Review. Advances in Complex Data Modeling and Computational Methods in Statistics. 2015; Cham: Springer International Publishing; 69–85. [Google Scholar]
- 89. Tyekucheva S., Makova K.D., Karro J.E., Hardison R.C., Miller W., Chiaromonte F.. Human-macaque comparisons illuminate variation in neutral substitution rates. Genome Biol. 2008; 9:R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. SMIT A., F.A.. 2004; Repeat-Masker Open-3.0.
- 91. Haeussler M., Zweig A.S., Tyner C., Speir M.L., Rosenbloom K.R., Raney B.J., Lee C.M., Lee B.T., Hinrichs A.S., Gonzalez J.N.et al.. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. 2019; 47:D853–D858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Pruitt K.D., Brown G.R., Hiatt S.M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C.M., Hart J., Landrum M.J., McGarvey K.M.et al.. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014; 42:D756–D763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A.. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010; 20:110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Fishilevich S., Nudel R., Rappaport N., Hadar R., Plaschkes I., Iny Stein T., Rosen N., Kohn A., Twik M., Safran M.et al.. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database. 2017; 2017:bax028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Stein T.I., Nudel R., Lieder I., Mazor Y.et al.. The GeneCards Suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinformatics. 2016; 54:1.30.1–1.30.33. [DOI] [PubMed] [Google Scholar]
- 96. Miller W., Rosenbloom K., Hardison R.C., Hou M., Taylor J., Raney B., Burhans R., King D.C., Baertsch R., Blankenberg D.et al.. 28-way vertebrate alignment and conservation track in the UCSC Genome Browser. Genome Res. 2007; 17:1797–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Mallick S., Li H., Lipson M., Mathieson I., Gymrek M., Racimo F., Zhao M., Chennagiri N., Nordenfelt S., Tandon A.et al.. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature. 2016; 538:201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sahakyan A.B., Chambers V.S., Marsico G., Santner T., Di Antonio M., Balasubramanian S.. Machine learning model for sequence-driven DNA G-quadruplex formation. Sci. Rep. 2017; 7:14535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Cer R.Z., Donohue D.E., Mudunuri U.S., Temiz N.A., Loss M.A., Starner N.J., Halusa G.N., Volfovsky N., Yi M., Luke B.T.et al.. Non-B DB v2.0: a database of predicted non-B DNA-forming motifs and its associated tools. Nucleic Acids Res. 2013; 41:D94–D100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Quinlan A.R. BEDTools: the Swiss-Army Tool for genome feature analysis. Curr. Protoc. Bioinformatics. 2014; 47:11.12.1–11.12.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Cremona M.A., Pini A., Chiaromonte F., Vantini S.. 2017; IWTomics: Interval-Wise testing for omics data. [Google Scholar]
- 102. Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q.-M.et al.. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009; 462:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Goecks J., Nekrutenko A., Taylor J., Team G.. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010; 11:R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Afgan E., Baker D., Batut B., van den Beek M., Bouvier D., Cech M., Chilton J., Clements D., Coraor N., Grüning B.A.et al.. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018; 46:W537–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. ENCODE Project Consortium The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004; 306:636–640. [DOI] [PubMed] [Google Scholar]
- 106. Derrien T., Estellé J., Marco Sola S., Knowles D.G., Raineri E., Guigó R., Ribeca P.. Fast computation and applications of genome mappability. PLoS One. 2012; 7:e30377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Gardiner-Garden M., Frommer M.. CpG islands in vertebrate genomes. J. Mol. Biol. 1987; 196:261–282. [DOI] [PubMed] [Google Scholar]
- 108. Guelen L., Pagie L., Brasset E., Meuleman W., Faza M.B., Talhout W., Eussen B.H., de Klein A., Wessels L., de Laat W.et al.. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008; 453:948–951. [DOI] [PubMed] [Google Scholar]
- 109. Zou H., Hastie T.. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Series B Stat. Methodol. 2005; 67:301–320. [Google Scholar]
- 110. Hon J., Martínek T., Zendulka J., Lexa M.. pqsfinder: an exhaustive and imperfection-tolerant search tool for potential quadruplex-forming sequences in R. Bioinformatics. 2017; 33:3373–3379. [DOI] [PubMed] [Google Scholar]
- 111. Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S.. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015; 33:877–881. [DOI] [PubMed] [Google Scholar]
- 112. Don P.K., Ananda G., Chiaromonte F., Makova K.D.. Segmenting the human genome based on states of neutral genetic divergence. Proc. Natl. Acad. Sci. USA. 2013; 110:14699–14704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. 1000 Genomes Project Consortium Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A.et al.. A global reference for human genetic variation. Nature. 2015; 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Lemmens B., van Schendel R., Tijsterman M.. Mutagenic consequences of a single G-quadruplex demonstrate mitotic inheritance of DNA replication fork barriers. Nat. Commun. 2015; 6:8909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Cremona M.A., Pini A., Cumbo F., Makova K.D., Chiaromonte F., Vantini S.. IWTomics: testing high-resolution sequence-based ‘Omics’ data at multiple locations and scales. Bioinformatics. 2018; 34:2289–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Cvijović I., Good B.H., Jerison E.R., Desai M.M.. Fate of a mutation in a fluctuating environment. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E5021–E5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Hwang D.G., Green P.. Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:13994–14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Fungtammasan A., Walsh E., Chiaromonte F., Eckert K.A., Makova K.D.. A genome-wide analysis of common fragile sites: what features determine chromosomal instability in the human genome. Genome Res. 2012; 22:993–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Campos-Sánchez R., Kapusta A., Feschotte C., Chiaromonte F., Makova K.D.. Genomic landscape of human, bat, and ex vivo DNA transposon integrations. Mol. Biol. Evol. 2014; 31:1816–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Cooper D.N., Bacolla A., Férec C., Vasquez K.M., Kehrer-Sawatzki H., Chen J.-M.. On the sequence-directed nature of human gene mutation: the role of genomic architecture and the local DNA sequence environment in mediating gene mutations underlying human inherited disease. Hum. Mutat. 2011; 32:1075–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Fleming A.M., Burrows C.J.. Interplay of Guanine Oxidation and G-Quadruplex Folding in Gene Promoters. J. Am. Chem. Soc. 2020; 142:1115–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Estep K.N., Butler T.J., Ding J., Brosh R.M.. G4-interacting DNA helicases and polymerases: potential therapeutic targets. Curr. Med. Chem. 2019; 26:2881–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Schiavone D., Jozwiakowski S.K., Romanello M., Guilbaud G., Guilliam T.A., Bailey L.J., Sale J.E., Doherty A.J.. PrimPol is required for replicative tolerance of G Quadruplexes in vertebrate cells. Mol. Cell. 2016; 61:161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Koole W., van Schendel R., Karambelas A.E., van Heteren J.T., Okihara K.L., Tijsterman M.. A polymerase theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat. Commun. 2014; 5:3216. [DOI] [PubMed] [Google Scholar]
- 125. Bacolla A., Wang G., Jain A., Chuzhanova N.A., Cer R.Z., Collins J.R., Cooper D.N., Bohr V.A., Vasquez K.M.. Non-B DNA-forming sequences and WRN deficiency independently increase the frequency of base substitution in human cells. J. Biol. Chem. 2011; 286:10017–10026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Fleming A.M., Zhou J., Wallace S.S., Burrows C.J.. A role for the fifth G-Track in G-quadruplex forming oncogene promoter sequences during oxidative stress: do these ‘Spare Tires’ have an evolved function. ACS Cent Sci. 2015; 1:226–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Capra J.A., Paeschke K., Singh M., Zakian V.A.. G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comput. Biol. 2010; 6:e1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Nakken S., Rognes T., Hovig E.. The disruptive positions in human G-quadruplex motifs are less polymorphic and more conserved than their neutral counterparts. Nucleic Acids Res. 2009; 37:5749–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Gros J., Rosu F., Amrane S., De Cian A., Gabelica V., Lacroix L., Mergny J.-L.. Guanines are a quartet's best friend: impact of base substitutions on the kinetics and stability of tetramolecular quadruplexes. Nucleic Acids Res. 2007; 35:3064–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Lee J.Y., Kim D.S.. Dramatic effect of single-base mutation on the conformational dynamics of human telomeric G-quadruplex. Nucleic Acids Res. 2009; 37:3625–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Valton A.-L., Prioleau M.-N.. G-quadruplexes in DNA replication: a problem or a necessity. Trends Genet. 2016; 32:697–706. [DOI] [PubMed] [Google Scholar]
- 132. Piazza A., Adrian M., Samazan F., Heddi B., Hamon F., Serero A., Lopes J., Teulade-Fichou M.-P., Phan A.T., Nicolas A.. Short loop length and high thermal stability determine genomic instability induced by G-quadruplex-forming minisatellites. EMBO J. 2015; 34:1718–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Saini N., Zhang Y., Nishida Y., Sheng Z., Choudhury S., Mieczkowski P., Lobachev K.S.. Fragile DNA motifs trigger mutagenesis at distant chromosomal loci in saccharomyces cerevisiae. PLoS Genet. 2013; 9:e1003551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Voineagu I., Narayanan V., Lobachev K.S., Mirkin S.M.. Replication stalling at unstable inverted repeats: interplay between DNA hairpins and fork stabilizing proteins. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:9936–9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Lai P.J., Lim C.T., Le H.P., Katayama T., Leach D.R.F., Furukohri A., Maki H.. Long inverted repeat transiently stalls DNA replication by forming hairpin structures on both leading and lagging strands. Genes Cells. 2016; 21:136–145. [DOI] [PubMed] [Google Scholar]
- 136. Shastri N., Tsai Y.-C., Hile S., Jordan D., Powell B., Chen J., Maloney D., Dose M., Lo Y., Anastassiadis T.et al.. Genome-wide identification of Structure-Forming repeats as principal sites of fork collapse upon ATR inhibition. Mol. Cell. 2018; 72:222–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Chuzhanova N., Chen J.-M., Bacolla A., Patrinos G.P., Férec C., Wells R.D., Cooper D.N.. Gene conversion causing human inherited disease: Evidence for involvement of non-B-DNA-forming sequences and recombination-promoting motifs in DNA breakage and repair. Hum. Mutat. 2009; 30:1189–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. McKinney J.A., Wang G., Mukherjee A., Christensen L., Subramanian S.H.S., Zhao J., Vasquez K.M.. Distinct DNA repair pathways cause genomic instability at alternative DNA structures. Nat. Commun. 2020; 11:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Tartier L., Michalik V., Spotheim-Maurizot M., Rahmouni A.R., Sabattier R., Charlier M.. Radiolytic signature of Z-DNA. Nucleic Acids Res. 1994; 22:5565–5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Ribeiro D.T., Madzak C., Sarasin A., Di Mascio P., Sies H., Menck C.F.. Singlet oxygen induced DNA damage and mutagenicity in a single-stranded SV40-based shuttle vector. Photochem. Photobiol. 1992; 55:39–45. [DOI] [PubMed] [Google Scholar]
- 141. Vowles E.J., Amos W.. Evidence for widespread convergent evolution around human microsatellites. PLoS Biol. 2004; 2:E199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Hile S.E., Wang X., Lee M.Y.W.T., Eckert K.A.. Beyond translesion synthesis: polymerase κ fidelity as a potential determinant of microsatellite stability. Nucleic Acids Res. 2012; 40:1636–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Bacolla A., Zhu X., Chen H., Howells K., Cooper D.N., Vasquez K.M.. Local DNA dynamics shape mutational patterns of mononucleotide repeats in human genomes. Nucleic Acids Res. 2015; 43:5065–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Shah K.A., Mirkin S.M.. The hidden side of unstable DNA repeats: mutagenesis at a distance. DNA Repair (Amst.). 2015; 32:106–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Hellmann I., Prüfer K., Ji H., Zody M.C., Pääbo S., Ptak S.E.. Why do human diversity levels vary at a megabase scale. Genome Res. 2005; 15:1222–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Duret L., Arndt P.F.. The impact of recombination on nucleotide substitutions in the human genome. PLos Genet. 2008; 4:e1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Drillon G., Audit B., Argoul F., Arneodo A.. Evidence of selection for an accessible nucleosomal array in human. BMC Genomics. 2016; 17:526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Kouzine F., Wojtowicz D., Baranello L., Yamane A., Nelson S., Resch W., Kieffer-Kwon K.-R., Benham C.J., Casellas R., Przytycka T.M.et al.. Permanganate/S1 nuclease footprinting reveals non-B DNA structures with regulatory potential across a mammalian genome. Cell Syst. 2017; 4:344–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Wu T., Lyu R., You Q., He C.. Kethoxal-assisted single-stranded DNA sequencing captures global transcription dynamics and enhancer activity in situ. Nat. Methods. 2020; 17:515–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Hänsel-Hertsch R., Spiegel J., Marsico G., Tannahill D., Balasubramanian S.. Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat. Protoc. 2018; 13:551–564. [DOI] [PubMed] [Google Scholar]
- 151. Lee D.S.M., Ghanem L.R., Barash Y.. Integrative analysis reveals RNA G-quadruplexes in UTRs are selectively constrained and enriched for functional associations. Nat. Commun. 2020; 11:527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All computational tools used in this study are available as a GitHub repository at the following URL: https://github.com/makovalab-psu/nonB-RegVar.