Abstract
In most bacteria, efficient use of carbohydrates is primarily mediated by the phosphoenolpyruvate (PEP):carbohydrate phosphotransferase system (PTS), which concomitantly phosphorylates the substrates during import. Therefore, transcription of the PTS-encoding genes is precisely regulated by transcriptional regulators, depending on the availability of the substrate. Fructose is transported mainly through the fructose-specific PTS (PTSFru) and simultaneously converted into fructose 1-phosphate (F1P). In Gammaproteobacteria such as Escherichia coli and Pseudomonas putida, transcription of the fru operon encoding two PTSFru components, FruA and FruB, and the 1-phosphofructokinase FruK is repressed by FruR in the absence of the inducer F1P. Here, we show that, contrary to the case in other Gammaproteobacteria, FruR acts as a transcriptional activator of the fru operon and is indispensable for the growth of Vibrio cholerae on fructose. Several lines of evidence suggest that binding of the FruR-F1P complex to an operator which is located between the –35 and –10 promoter elements changes the DNA structure to facilitate RNA polymerase binding to the promoter. We discuss the mechanism by which the highly conserved FruR regulates the expression of its target operon encoding the highly conserved PTSFru and FruK in a completely opposite direction among closely related families of bacteria.
INTRODUCTION
Most bacteria rely on the phosphoenolpyruvate (PEP): carbohydrate phosphotransferase system (PTS) to transport sugars into the cell efficiently by simultaneously phosphorylating them (1–3). The PTS is composed of two general cytoplasmic proteins, enzyme I (EI) and histidine-containing phosphocarrier protein (HPr), which are commonly used to transport most PTS sugars and various sugar-specific enzyme II (EII) complexes. Each EII complex usually consists of two cytosolic domains (EIIA and EIIB) and one transmembrane domain (EIIC). EI autophosphorylates with PEP, and HPr mediates phosphoryl transfer from EI to EIIA. Then the phosphoryl group is transferred to EIIB, which leads to the phosphorylation of a PTS sugar during its transport into the cell through EIIC.
Fructose is considered an important sugar in the early evolution of the carbohydrate metabolic pathway in bacteria since it feeds directly into glycolysis without isomerization or epimerization, and it is the first hexose synthesized through gluconeogenesis (4,5). It is also noteworthy that the fructose PTS (PTSFru) is more widespread among bacterial species than any other carbohydrate PTS including the glucose PTS (PTSGlc) (4). The uniqueness of the PTSFru is that it possesses its own HPr-like domain, FPr, fused to an EIIA domain via a central M domain of unknown function to constitute FruB. FruB catalyzes the phosphoryl transfer reaction from autophosphorylated EI to FruA (EIIB′BCFru), which finally phosphorylates fructose to fructose 1-phosphate (F1P) during its translocation across the membrane. F1P is then converted to fructose 1,6-bisphosphate (FBP) by the 1-phosphofructokinase FruK (Figure 1A) (6). In most bacteria belonging to Gammaproteobacteria such as Escherichia coli and Pseudomonas putida, FruA, FruB and FruK are encoded in a single operon (fru operon).
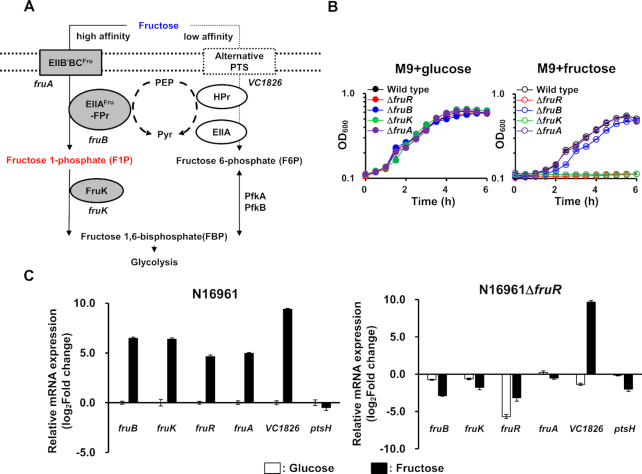
Figure 1.
Vibrio cholerae FruR activates fru operon transcription in the presence of fructose. (A) The schematic representation of fructose metabolism in V. cholerae. Fructose is mainly transported through the fructose-specific PTS (PTSFru) which consists of FruB (FPr-IIAFru) and FruA (EIIB′BCFru), and concomitantly phosphorylated to fructose 1-phosphate (F1P), which is then channeled towards glycolysis following the activity of 1-phosphofructokinase (FruK) to yield fructose 1,6-bisphosphate (FBP). VC1826 was also shown to transport fructose (28). (B) Growth curves of in-frame deletion mutants of fruB, fruK, fruA, and fruR in M9 medium supplemented with 0.2% glucose (left panel) or fructose (right panel). Growth was measured by absorbance at 600 nm using a multimode microplate reader (TECAN). (C) The relative mRNA expression of fru operon genes, fruR, and VC1826 in the wild-type V. cholerae N16961 (left) and a fruR mutant (right) grown on fructose. The mRNA expression levels of indicated genes are shown as relative values (log2 scale) to that of the wild-type strain grown on glucose. The HPr-encoding gene (ptsH) was used as a negative control. The means and standard deviations of three independent measurements are shown in (B) and (C).
To cope with the metabolic needs of the cell, genes encoding the PTS are precisely regulated by the concerted actions of diverse mechanisms in bacteria (7). Unlike other carbohydrate PTSs, PTSFru is transcribed independently of cyclic AMP and its receptor CRP (8), which is one of the major transcriptional regulators of carbon metabolism in Gammaproteobacteria (9,10). Instead, the expression of the fru operon is primarily regulated by FruR, which has been characterized as a fructose repressor. In enteric bacteria and P. putida, FruR directly binds to its cognate binding site(s) located downstream of the transcription start site (TSS) of the fru operon, and F1P is an inducer of this operon (11–13). However, comparative genomic analysis revealed that the FruR of Vibrio cholerae is phylogenetically distinct from its orthologs in E. coli or P. putida, which implies that its FruR regulatory mechanism could be different from these two species (14).
To gain selective advantage in their host environment, pathogens often use metabolic cues to regulate virulence genes as well as metabolic genes (15). According to the transcriptome analysis of V. cholerae, the transcriptional level of genes coding for the PTSFru and FruR rose during host infection (16), implying that fructose could be an important nutrient for V. cholerae in the host. However, the transcriptional regulatory mechanism of the fru operon has not been explored in Vibrio species. Here, we reveal that, in V. cholerae, FruR is indispensable for the expression of the fru operon and thus for growth on fructose. In the presence of F1P, FruR activates transcription of the fru operon by facilitating the binding of RNA polymerase to the promoter, in contrast to what is observed in other Gammaproteobacteria.
MATERIALS AND METHODS
Bacterial strains, plasmids and culture conditions
Details of strains, plasmids and oligonucleotides used in this study are listed in Supplementary Tables S1 and S2. All V. cholerae N16961 strains were cultured in Luria-Bertani (LB) medium or M9 minimal medium supplemented with the indicated sugars at 37°C. All Escherichia coli strains were grown in LB medium at 37°C. All plasmids were constructed using standard PCR-based cloning procedures and verified by sequencing. The following supplements were added if necessary: ampicillin, 100 μg ml−1; chloramphenicol, 2 μg ml−1; tetracycline, 2 μg ml−1 for V. cholerae and ampicillin, 100 μg ml−1; chloramphenicol, 20 μg ml−1 for E. coli; isopropyl-β-d-1-thiogalactopyranoside (IPTG), 1 mM; 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal), 80 μg ml−1. In-frame deletion mutants of V. cholerae were generated by allelic exchange using a pDM4-based plasmid as described previously (17).
The E. coli SM10 λpir strain carrying pDM4-based plasmids was conjugated to V. cholerae, and all transconjugants were confirmed by PCR as previously described (18). The plasmids, except for pDM4-based plasmids, were directly transformed into V. cholerae by electroporation, as previously described (19). For duplication of the fruR-fruB intergenic region on chromosome, the 1 kb PCR products encompassing 338 bp of the fruR–fruB intergenic region and 662 bp DNA region from the initiation codon of fruB or fruR were cloned into pDM4. After single homologous recombination, these constructs were chromosomally integrated into fruB or fruR. To construct the fruB::lacZ and fruR::lacZ transcriptional fusion vector, the fruR-fruB intergenic region was amplified by PCR using the appropriate primer pairs (Supplementary Table S2). The PCR product was digested with SalI (New England Biolabs, Beverley, MA, USA, #R3138S) and inserted into the corresponding site of the pJK1113-based expression vector carrying promoter-less E. coli lacZ (pJK-LacZ). To replace the nucleotide sequences of the FruR-binding sites in the fruR–fruB intergenic region with the mutated sequences, site-directed mutagenesis PCR was performed using the appropriate primer pairs (Supplementary Table S2). Each FruR-binding site was replaced with 5′-AGATCGTGAGTATTCG-3′, which corresponds to the nucleotide sequences of –162 to –147 relative to the initiation codon of fruB. To construct the FruR expression plasmid pACYC-FruR, the fruR gene was amplified by PCR and linked to the cat promoter by cloning into an appropriate site in pACYC-184 using the Gibson assembly method (20). Amino acid substitution mutants of FruR were generated by site-directed mutagenesis PCR using appropriate primer pairs (Supplementary Table S2).
Measurement of bacterial growth in sugar supplemented medium
Overnight-grown V. cholerae N16961 cultures were diluted 100-fold into fresh LB medium and cultured at 37°C until OD600 reached 1.0. The cell pellets were rinsed twice with M9 medium lacking a carbon source followed by incubation in the same medium for 30 min as previously described (3). The cells were then inoculated into a 96-well plate containing M9 medium supplemented with 0.2% glucose or fructose. The optical density of all cultures was measured at 600nm using a multimode microplate reader (Spark TM 10 M multimode microplate reader, Tecan Group Ltd, Männedorf, Switzerland) (21).
RNA extraction and quantitative real-time reverse transcription-PCR (qRT-PCR)
RNA extraction and qRT-PCR were performed as previously described (17). The V. cholerae strains were grown in LB medium and resuspended in M9 medium in the same manner as for the growth measurement. Each culture was divided into two aliquots, one supplemented with 0.2% fructose, the other with 0.2% glucose. Both samples were cultured for 30 min at 37°C. After fixing the cells with the same volume of 100% methanol for 1 h at -20°C, total RNA was isolated using the TaKaRa MiniBEST Universal RNA Extraction Kit (Takara Bio, Inc., Otsu, Japan, #9767) according to the manufacturer's instructions. Total RNA (2500 ng) from each sample was converted into cDNA using the RNA to cDNA EcoDry Premix (Random Hexamers) (Clontech Laboratories, Inc., Mountain View, CA, USA #639545). The 30-fold diluted cDNA was subjected to real-time PCR amplification using a FAST SYBR™ green master mix (Thermo Fisher Scientific, Inc., Waltham, MA, USA, # 4385616) with specific primers in a CFX96™ Real-Time System (BioRad, Hercules, CA, USA).
Electrophoretic mobility shift assay (EMSA)
Non-radioisotope-labeled EMSA was performed as previously described (12). A 338-bp fruB probe was amplified by PCR using the V. cholerae N16961 chromosome as a template with the primers fruBR/E-F and fruBR/E-R (Supplementary Table S2). The 338-bp fruB probes containing the mutated FruR-binding sites were amplified by PCR using the pJK-PfruBMut::LacZ as templates with the same primers. The probes were incubated with the indicated proteins and metabolites in TGED buffer (10 mM Tris–HCl, pH 8.0; 5% v/v glycerol; 0.1 mM EDTA and 1 mM DTT), and 200 μg ml–1 of bovine serum albumin (BSA) as non-specific protein competitor. Fructose 1-phosphate (F1P) was purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA, #sc-285345) and other metabolites from Sigma-Aldrich (Waltham, MA, USA) unless otherwise specified. Each sample was incubated at 37°C for 10 min and then analysed on a 6% polyacrylamide gel (acrylamide/bisacrylamide ratio of 29:1) in TBE (89 mM Tris, 89 mM boric acid, 2 mM EDTA) followed by EtBr staining. DNA bands were visualized using a gel documentation system (GDS-200C, KBT, Seongnam, Korea) and their intensities were quantified using ImageJ software (NIH Image, National Institutes of Health, Bethesda, MD; online at: http://rsbweb.nih.gov/ij/).
DNase I footprinting
DNase I footprinting experiments were performed as described in a previous study (22). The 338-bp fruB probe covering the entire fruR–fruB intergenic region and 538-bp fruB probe covering from –199 to +339 bp relative to the TSS were prepared by PCR using the V. cholerae N16961 chromosome as a template with the 5′ 6-carboxyfluorescein (6-FAM)-labeled forward primer fruBR/F(6FAM) and fruBR/LF(6FAM), respectively, and the reverse primer fruBR/E-R and fruBR/LR, respectively (Bionics, Korea) (Supplementary Table S2). The purified PCR product was incubated with the indicated amounts of proteins and metabolites at 37°C for 10 min prior to digestion with 0.02 U DNase I (New England Biolabs, Beverley, MA, USA, # M0303S) for 1 min. The cleavage reaction was stopped by adding the same volume of stop solution (200 mM NaCl, 30 mM EDTA, 1% SDS) followed by phenol extraction and EtOH precipitation. DNase I digestion reactions were analysed by capillary electrophoresis in an ABI 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA) with Peak Scanner software v1.0 (Applied Biosystems, Foster City, CA).
β-galactosidase assay
A V. cholerae N16961 ΔlacZ strain was transformed with the plasmid carrying E. coli lacZ transcriptionally fused with the wild-type or mutated fruB promoter and grown on glucose or fructose to measure the β-galactosidase activities as previously described (23). Cultured cells (80 μl) were 10-fold diluted in Z-buffer (0.06 M Na2HPO4, 0.04 M NaH2PO4, 0.01 M KCl, 1 mM MgSO4 and 0.04 M β-mercaptoethanol) and lysed with 20 μl 0.1% SDS and 40 μl chloroform at 37°C for 10 min. The β-galactosidase activity was then measured as described by Miller (24).
Purification of proteins
Non-tagged FruR proteins were overexpressed in E. coli BL21(DE3)/pLysSRARE (Novagen). Harvested cells were resuspended in buffer A (50 mM Tris–HCl, pH 8.0; 10 mM DTT; 10 mM EDTA; and 10% glycerol) containing 50 mM NaCl and disrupted by three passages through a French pressure cell at 9000 psi. After centrifugation at 100 000 × g at 4°C for 60 min, the supernatant was applied to a HiTrap Heparin HP affinity column (GE Healthcare Life Sciences). Protein elution was performed using a 20-column volume 0.5–1 M NaCl gradient in buffer A at a flow rate of 2 ml min–1. The fractions containing FruR were concentrated using Amicon Ultracel-3K centrifugal filters (Merck Millipore), and then chromatographed on a HiLoad 16/60 Superdex 200 prep grade column (GE Healthcare Life Sciences) equilibrated with buffer A containing 200 mM NaCl to achieve higher purity (>95%). His-tagged RpoD was overexpressed in E. coli ER2566 and purified by immobilized metal affinity chromatography (IMAC) using TALON metal affinity resin according to the manufacturer's instructions (Clontech Laboratories, Inc). Proteins bound to the resin were eluted with buffer B containing 200 mM imidazole. To remove the imidazole and achieve higher purity (>98%), further chromatography with a HiLoad 16/600 Superdex 200 prep grade column was performed on the concentrated sample.
RESULTS
Vibrio cholerae FruR activates fru operon transcription in the presence of fructose
In V. cholerae, there are 25 PTS components responsible for the utilization of 10 types of carbohydrates (25–28). The fructose PTS (PTSFru) consists of the membrane-spanning transporter FruA (VCA0516) and cytosolic FruB (VCA0518). It phosphorylates fructose to fructose 1-phosphate (F1P) during its translocation across the membrane. F1P is then converted to fructose 1,6-bisphosphate (FBP) by the 1-phosphofructokinase FruK (VCA0517). In V. cholerae, the transcription of the fruBKA (fru) operon was shown to increase approximately 80-fold in minimal medium supplemented with fructose in contrast to in minimal medium lacking a carbon source (27). While transcriptional repression of the fru operon by FruR is reported to be relieved in the presence of fructose in other Gammaproteobacteria such as E. coli and P. putida (11–13), the transcriptional regulatory mechanism of the fru operon has not been investigated in V. cholerae. To determine the fructose-mediated transcriptional regulation of the fru operon, we constructed strains lacking one of the fru operon genes or fruR (VCA0519), which is contiguous to but divergently transcribed from the operon, and examined the growth of each strain on glucose and fructose. All mutants showed growth rates similar to those of the wild-type (WT) N16961 strain on glucose (Figure 1B). As it is known that another EIIC-containing protein (VC1826) is capable of supporting the growth on fructose (Figure 1A) (28) and HPr also partly contributes to fructose transport (27), the fruA mutant exhibited a growth rate similar to that of the WT and the fruB mutant showed only a slight growth retardation on fructose (Figure 1B). However, the fruK mutant showed a severe growth defect on fructose as previously reported in other bacteria (Figure 1B) (29,30). Interestingly, the fruR mutant also showed a growth defect as severe as the fruK mutant. This led us to assume that FruR might be absolutely required for the transcription of the fru operon in V. cholerae as a transcription activator, contrary to what has been reported in E. coli and P. putida (11–13). To validate our assumption, qRT-PCR experiments were performed to assess the transcription levels of the fru operon genes and fruR in WT and the fruR mutant in the presence of glucose or fructose. We found that the transcription of fruR and the fru operon was dramatically activated in WT cells in the presence of fructose compared to that in glucose-grown cells (Figure 1C, left panel), whereas these genes were hardly expressed in the fruR mutant cells regardless of the sugar source (Figure 1C, right panel). It is noteworthy that the transcription of VC1826 significantly increases in the presence of fructose regardless of the presence of FruR, implying that the fructose-induced transcriptional activation of VC1826 is independent of FruR. To further confirm that the transcription of the fru operon is activated in the presence of fructose in a VcFruR-dependent manner, we constructed a ΔfruR strain harboring a plasmid carrying the fruBKA operon under control of its own promoter or the constitutive cat promoter and examined the growth of each strain on glucose and fructose (Supplementary Figure S1). An exogenous supply of the fruBKA operon under control of its own promoter did not rescue the growth defect of the ΔfruR strain, whereas the constitutive expression of fruBKA restored the growth of the mutant on fructose to a wild-type level. These data validate that FruR is essential for the transcription of the fru operon, and that FruR-mediated transcriptional activation of fruK is absolutely required for the growth on fructose.
There are three FruR-binding sites in the fruR-fruB intergenic region
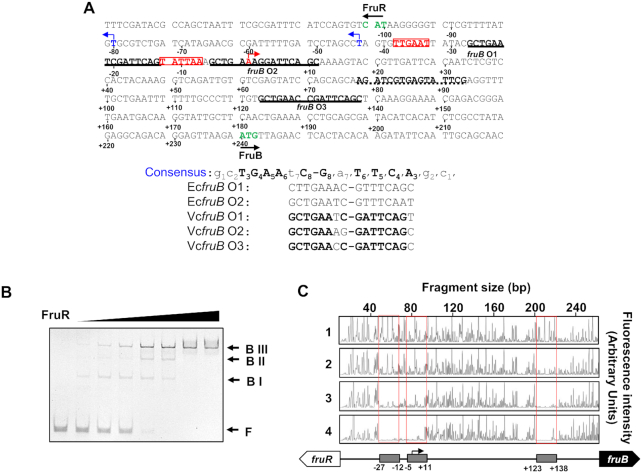
In E. coli, FruR is known to belong to the GalR-LacI family of transcriptional regulators and directly binds to the 16-bp imperfect palindromic sequence (5′-GCTGAAnC/GnTTCAGC-3′) (31). We searched for these nucleotide sequences in two V. cholerae chromosomes using PRODORIC software (http://prodoric.tu-bs.de) (32,33). As a result, three binding sites were identified in the fruR–fruB intergenic region, herein referred to as fruB O1, O2 and O3, centered at 258.5, 236.5 and 109.5 bp upstream of the initiation codon of fruB, respectively (Figure 2A). To confirm the binding of FruR to the fruB promoter, an electrophoretic mobility shift assay (EMSA) was performed with a 338-bp probe containing the entire fruR-fruB intergenic region (Figure 2B). When we incubated the probe with increasing amounts of FruR, we detected three shifted bands (BI, BII, and BIII), which could be inferred from the presence of three FruR-binding consensus sequences in the intergenic region. In the presence of an excess amount of FruR, however, only the uppermost band of the probe bound to three FruR molecules (BIII) could be seen. To confirm the specificity of FruR interaction with the fruR-fruB intergenic region we included a DNA probe encompassing +121 to +618 relative to the initiation codon of fruB (Supplementary Figure S2). It was not bound by FruR and had no effect on the FruR band shift and so confirmed the specific binding of FruR to the fruB promoter. To determine the precise FruR-binding sites, DNase I footprinting assays were performed with 5′-6-carboxyfluorescein (6-FAM)-labeled probes containing the fruR–fruB intergenic region. As expected from the EMSA data, the DNA sequences corresponding to the predicted fruB O1, O2 and O3 were protected by FruR from DNase I digestion (Figure 2C). It is known that the GalR-LacI family of transcription factors including E. coli FruR (EcFruR) bind to tandem binding sites and induce DNA looping as tetramers (34). However, V. cholerae FruR (VcFruR) lacks the leucine-mini-zipper domain that structurally determines the tetrameric state of EcFruR (Supplementary Figure S3A) (35–37). Consequently, gel filtration experiments revealed that VcFruR exists as a dimer in the solution (Supplementary Figure S3B). The regulatory effect of a transcription factor is usually determined by the location of its binding site(s) relative to the transcription start site (TSS) (4,31). To map the TSS(s) of the fru operon, RNA isolated from WT N16961 cells grown on fructose or glucose were subjected to primer extension analysis. A fruB transcript starting 239 bp upstream of the initiation codon of fruB was detected in fructose-grown cells, but not in glucose-grown cells (Supplementary Figure S4A). The promoter region contains the consensus sequence elements associated with σ70 binding in V. cholerae (–35 element: TTGnnn, –10 element: TAnAAT) (boxed in Figure 2A) (38). Interestingly, fruB O1 is located between the –35 and –10 elements, while the sixth nucleotide of fruB O2 corresponds to the TSS (marked with a bent red arrow in Figure 2A). The fructose-dependent transcription from this promoter was further confirmed by RT-PCR experiments: while the fructose-activated transcript could be detected using the forward primers annealing to sequences downstream of the TSS, with ∼1000-fold activation regardless of the primer set used, it was not detected using a forward primer annealing to the sequence immediately upstream of the TSS (Supplementary Figure S4B). Therefore, we concluded that the TSS of the fru operon is located 239 bp upstream of the initiation codon of fruB.
Figure 2.
There are three FruR-binding sites in the fruR-fruB intergenic region. (A) Nucleotide sequence in the fruR–fruB intergenic region. The numbers refer to the nucleotide position relative to the fruB TSS. The three binding sites of FruR are underlined with solid lines. The sequence used to mutate the FruR-binding sites is underlined with a dotted line. The transcription start sites (TSSs) of fruB and fruR are marked with red and blue arrows, respectively. The –35 and –10 elements of the fruB promoter were colored and boxed in red. The initiation codons of fruB and fruR were colored in green and marked with arrows. Shown below are the FruR-binding sequences aligned in comparison to two FruR-binding sequences in the E. coli fru promoter, with conserved bases in bold. (B) VcFruR binding to operators was examined using electrophoretic mobility shift assay (EMSA). The 338-bp fruB probe (60 ng) containing the entire fruR-fruB intergenic region was incubated with increasing amounts of VcFruR (0–180 ng) and analysed on a 6% polyacrylamide gel in TBE as described under ‘Materials and methods.’ (C) DNase I footprinting of binding sites of VcFruR in the fruR-fruB intergenic region. The 338-bp DNA fragment (200 ng; 14.6 nM) was labeled with 6-carboxyfluorescein (6-FAM) and incubated with increasing amounts of VcFruR (0, 100, 200 and 400 ng; 69.4, 138.8 and 277.6 nM in the lanes 1–4, respectively) prior to digestion with DNase I. The fluorescence intensity of the 6-FAM-labeled fragments is shown on the each electropherogram and fragment sizes were determined by comparison to the internal molecular weight standards. The DNA regions corresponding to fruB O1, O2 and O3 were marked with red boxes. Schematic diagram of the fruR–fruB intergenic region is shown below, with the three binding sites depicted in gray rectangles. A bent arrow indicates the TSS of the fru operon and nucleotide positions relative to the TSS are indicated.
FruR binding to fruB O1 is indispensable for fruBKA transcription in V. cholerae
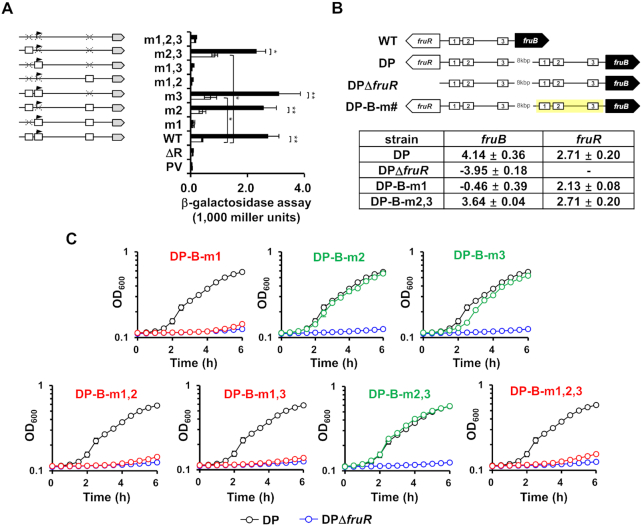
It is known that, in E. coli and P. putida, the transcription of the fru operon is negatively regulated by FruR which binds to specific DNA sequences located downstream of the TSS in the absence of F1P (11,12). Since the fruB TSS is located downstream of fruB O1, overlaps with fruB O2, and located upstream of fruB O3 in V. cholerae, we sought to investigate how each of the three VcFruR-binding sites contributes to the transcriptional activation of the fru operon. For this, we deleted the lacZ gene from WT V. cholerae N16961 (fruR+) and its otherwise isogenic ΔfruR derivative, and transformed these strains with a set of eight plasmids, each carrying E. coli lacZ transcriptionally fused with the V. cholerae fruB promoter (PfruB) with the three FruR-binding sites either wild type or mutated in all possible combinations (Figure 3A). Then we measured the PfruB activity in all strains grown either on glucose or fructose by measuring β-galactosidase activity. In the case of the fruR+ strain harboring the plasmid carrying the wild-type PfruB, cells grown on fructose exhibited a significantly higher β-galactosidase activity than cells grown on glucose. However, the fruR− strain harboring the same plasmid exhibited little β-galactosidase activity regardless of the sugar source (Figure 3A). Interestingly, all fruR+ strains harboring plasmids carrying the PfruB with wild-type fruB O1 showed a fructose-induced increase in β-galactosidase activity regardless of the mutation in fruB O2 and O3, whereas fruR+ strains harboring plasmids carrying the mutated fruB O1 exhibited little β-galactosidase activity in fructose medium, indicating that the binding of FruR at fruB O1 is necessary and sufficient for the fructose-induced transcriptional activation of the fru operon. Interestingly, a strain harboring the plasmid carrying the mutated fruB O3 exhibited a small but significant increase in β-galactosidase activity in glucose medium compared to the strain carrying the wild-type PfruB, indicating that fruB O3 could be involved in glucose-induced transcriptional repression of the fru operon. Surprisingly, however, the mutation at fruB O2 did not have any significant effect on the transcription of the fru operon regardless of the sugar source. To further confirm that fruB O1 is solely responsible for, and fruB O2 and O3 are not involved in, the transcriptional activation of the fru operon in the presence of fructose, we constructed strains in which the fruR-fruB intergenic sequence was duplicated and separated with a large distance (DP) and mutations were introduced into the FruR-binding sites on the fruB-side promoter (DP-B-m#) so that we could minimize the perturbation in the expression level of FruR induced by the mutations (Figure 3B). We then compared the transcriptional levels of fruR and the fru operon and the growth rate on fructose of these strains to those of the DP strain. As expected from the β-galactosidase assays, strains having mutated fruB O1 on the fruB-side promoter showed no transcription of the fru operon and accordingly exhibited a severe growth defect on fructose as the ΔfruR strains did (Figure 3B and 3C). Strains carrying mutated fruB O2 and O3 on the fruB-side promoter exhibited a similar transcriptional activation of the fru operon, and thus a similar growth rate to the DP strain on fructose (Figure 3B and 3C). Since fruB O1 is located more proximal to the initiation codon of fruR than that of fruB, we wondered if the fruB O1 is also involved in the transcriptional activation of fruR itself, thus forming a feedforward loop. To address this question, we first mapped the TSS(s) of fruR. Primer extension analysis using RNA isolated from WT V. cholerae N16961 cells grown on fructose revealed two TSSs of the fruR gene, which are located 23.5 and 60.5 bp downstream of the center of fruB O1 (indicated in Supplementary Figure S5A and marked with blue arrows in Figure 2A). We predicted the putative –35 and –10 promoter elements based on the TSSs identified by the primer extension experiments. In particular, we found that the minor promoter elements of fruR overlapped with those of fruB. The fructose-dependent transcription from the two promoters was further confirmed by RT-PCR experiments (Supplementary Figure S5B). We then measured fruR promoter (PfruR) activity using the fruR+lacZ− and fruR−lacZ− strains harboring the plasmid carrying E. coli lacZ transcriptionally fused to the wild-type PfruR. Interestingly, both strains exhibited a fructose-induced increase in β-galactosidase activity (Supplementary Figure S6A). To further confirm that VcFruR is not responsible for the transcriptional activation of fruR in the presence of fructose, we constructed mutant derivatives of the DP strain in which mutations were introduced into the FruR-binding sites on the fruR-side promoter (DP-R-m#) (Supplementary Figure S6B). As expected from the β-galactosidase assays, DP-R-m1 and DP-R-m1,2,3 strains exhibited a similar transcriptional activation of fruR and thus a similar growth rate to the DP strain on fructose (Supplementary Figure S6B). Therefore, we assume that the fructose-dependent transcriptional activation of fruR might be accomplished by an unknown transcription factor. Although the transcriptional level of fruR was shown to decrease in a V. cholerae strain lacking the global transcriptional regulator H-NS in a previous study (39), hns transcription was not significantly affected by fructose (Supplementary Figure S7). Therefore, a separate study would be necessary to assess how fruR expression responds to the presence of fructose. Based on these results, we concluded that the binding of VcFruR to fruB O1 is a prerequisite for the transcriptional activation of the fru operon, but binding of VcFruR to fruB O2 and O3 could be involved in fruR repression, in the presence of fructose (Supplementary Figure S6B).
Figure 3.
FruR binding to fruB O1 is indispensable for fruBKA transcription in V. cholerae. (A) Effect of VcFruR binding to each operator on the transcriptional activation of the fru operon was measured by the lacZ reporter assay using a fruR+lacZ− strain harboring a plasmid carrying E. coli lacZ transcriptionally fused with the wild-type or mutated fruB promoter (mutated site indicated as m#). Schematic representation of plasmids is shown left, with the lacZ gene and mutation site depicted by a grey arrow and X, respectively. The fruR+lacZ− strain harboring the plasmid containing promoter-less lacZ served as a negative control (PV). Indicated strains were grown on glucose (open bars) or fructose (filled bars) and then lysed to measure the β-galactosidase activity as described under ‘Materials and Methods.’ (right panel). Statistical significance was determined using the Student's t-test. (*P< 0.05; **P< 0.01). (B) The transcriptional activation by FruR binding to fruB O1 was further confirmed by measuring the fruB expression in fructose-grown strains carrying the chromosomal duplication of the wild-type or mutated fruR-fruB intergenic sequence. Schematic representation of the strains carrying duplicated regions is shown in the upper panel, with FruR-binding sites and mutated areas depicted by white rectangles and yellow shadings, respectively. The mRNA expression levels of fruB and fruR in the indicated strains grown on fructose were analyzed by qRT-PCR and shown as log2 values relative to that in the DP strain grown on glucose (lower panel). (C) Growth of DP-B-m# strains on fructose was compared with the DP (black) and DPΔfruR (blue) strains and the strains showing the same growth defect as the DPΔfruR strain were indicated in red. The means and standard deviations of three independent measurements are shown in (A) and (C).
F1P does not release VcFruR from the operators but affects their binding affinity
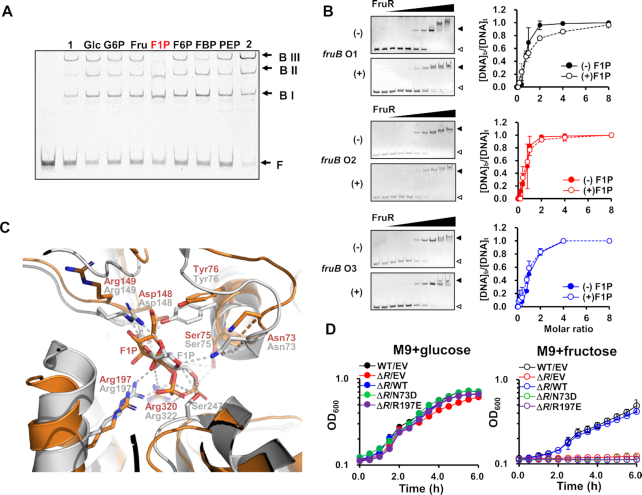
Metabolic intermediates of fructose, such as F1P and FBP, have been reported to play important roles as metabolite effectors in the transcriptional regulation of metabolic genes in diverse bacterial species (9,10,40). Recent studies have shown that, within the physiological concentration range, F1P is the only metabolite effector of EcFruR and P. putida FruR (PpFruR) (13,40,41). In these species, F1P is known to release FruR from the operators and induce transcription of the fru operon. It is known that the intracellular concentration of F1P is ∼0.15 mM, which is ∼10-fold less than that of FBP, in bacterial cells grown on fructose (42). Thus, we first examined whether F1P can also function as an allosteric effector of VcFruR. EMSA was performed in the presence of 2 mM sugar phosphates using a molar ratio of VcFruR and the fruB probe which showed three shifted bands as well as the free probe (Figure 2B). The electrophoretic mobility pattern of the VcFruR-probe complexes was not influenced by any sugar phosphates tested including fructose 6-phosphate (F6P) and FBP except F1P, which is in accordance with earlier findings in other species (Figure 4A) (11–13,40). Interestingly, however, F1P slightly decreased the band intensity of BIII while slightly increasing that of BI and BII, but did not change the band intensity of the free probe, which is contrary to what is expected from earlier observations in other species. Therefore, we wondered if F1P may alter the binding affinity of VcFruR for operators without dissociating VcFruR from the operators. To verify this assumption, we performed EMSA with probes containing one wild-type and two mutated binding sites in the presence of increasing concentrations of FruR and measured the binding affinity of VcFruR for each binding site in the absence or presence of F1P (Figure 4B). Despite the existence of slight sequence differences (Figure 2A), the three binding sites exhibited similar affinities for VcFruR and the Kd values were determined to be 3.81, 2.93 and 4.04 nM, respectively. While F1P did not significantly change the binding affinities of VcFruR for fruB O2 and O3, the Kd value of FruR for fruB O1 increased from 3.81 to 11.62 nM in the presence of F1P (Figure 4B). We further confirmed that F1P weakens the binding affinity of VcFruR for fruB O1 without affecting that for fruB O2 and fruB O3 by EMSA using the fruB probe, where only one of the three binding sites was mutated in the presence of increasing concentrations of F1P (Supplementary Figure S8). When VcFruR was mixed with the Mut 1 probe in which only fruB O1 was mutated, F1P did not change the band intensity of the BI, BII, and free probe. Interestingly, however, when VcFruR was mixed with Mut 2 or Mut 3, which contained wild-type fruB O1 with mutated fruB O2 or fruB O3, respectively, F1P decreased the band intensity of BIIs and notably increased the band intensity of free probes (Supplementary Figure S8). Considering the effect of F1P on the binding affinities of FruR for the binding sites, we speculated that F1P may not dissociate FruR from the operators in fructose-grown cells, where VcFruR usually exists in a large molar excess to its binding sites (9,14,43). As predicted, when we performed EMSA with an excess molar ratio of VcFruR to the probe (∼18:1) in the presence of increasing concentrations of F1P, F1P did not release VcFruR from its binding sequences even at 5 mM (Supplementary Figure S9). Considering the physiological levels of F1P and FruR, we assumed that a significant proportion of the FruR-F1P complex might be bound to fruB O1 in vivo in the presence of fructose. Therefore, we suggest that F1P may affect the binding of VcFruR to DNA, but not as a negative allosteric effector. A structural and biophysical study on PpFruR revealed a cavity where F1P binds (12) and the amino acids lining the cavity are well conserved in VcFruR. Therefore, we generated a docking model of VcFruR with F1P to determine the amino acids essential for the binding of F1P to VcFruR. The structure of the full-length VcFruR was modeled using the crystal structure of E. coli PurR, which also belongs to the GalR-LacI family of transcriptional regulators, as a template (44), and the modeled structure was docked with F1P (Figure 4C). By superimposing the structural model of the VcFruR-F1P complex with the crystal structure of the PpFruR–F1P complex (13), we found that the side chains of all residues in the F1P binding cavity of VcFruR were similarly oriented as the corresponding residues in the PpFruR–F1P complex, except for Arg149 (Figure 4C). Site-directed mutagenesis showed that, while the fruR mutant harboring a plasmid bearing wild-type fruR exhibited a growth rate similar to that of the WT N16961 strain carrying the empty vector, the mutant strain harboring a plasmid bearing fruR mutated at a conserved F1P-binding residue (N73D or R197E) exhibited a similar growth defect on fructose as the fruR mutant carrying the empty vector (Figure 4D). From these results, we conclude that the binding of F1P is essential for the transcriptional regulatory activity of VcFruR by altering the binding of VcFruR to fruB O1.
Figure 4.
F1P does not release VcFruR from the operators but affects its binding affinity. (A) The effect of metabolites on DNA binding of FruR was examined by EMSA. The 338-bp fruB probe (60 ng) was incubated with FruR (60 ng) in the presence of 2 mM metabolite in TGED buffer and analyzed on a 6% polyacrylamide gel in TBE. Lanes 1 and 2 contained 45 and 90 ng of FruR, respectively, in the absence of metabolite. Shifted bands (BI, BII and BIII) and free probe (F) are indicated with arrows. (B) The affinities of FruR for binding sites were measured in the absence or presence of F1P. Sixty nanograms of each probe (4.4 nM) having only one of the three binding sites remaining intact was incubated with increasing amounts of VcFruR (0–240 ng; 0–58.4 nM) in the absence (–) and presence (+) of F1P and subjected to EMSA (left panels). Then free and FruR-bound DNA (open and closed triangle, respectively) were quantified using ImageJ software and the fraction of FruR-bound DNA ([DNA]b/[DNA]t) was plotted against the molar ratio of VcFruR to each probe (right panels). The means and standard deviations of three measurements are shown. (C) Structural modeling of full-length VcFruR was performed using the SWISS Model server (60), with the crystal structure of E. coli PurR (PDB code: 1JFS) as a template (44). The modeled structure of VcFruR (colored in orange) was docked with F1P using PatchDock server (61) and superimposed with the PpFruR-F1P complex structure (PDB code: 3O75, colored in grey) (13). F1P and interacting residues are shown as sticks. Polar interactions in the PpFruR-F1P complex are shown as dotted lines. The superimposition of the modeled VcFruR-F1P complex structure with the crystal structure of the PpFruR-F1P complex had a root‐mean‐square deviation of 2.380 Å for 249 amino acids. (D) Comparison of growth of wild type and the fruR mutant harboring a plasmid carrying wild-type (WT) or mutant (N73D and R197E) FruR on glucose or fructose. The means and standard deviations of three independent measurements are shown. EV, empty vector.
The FruR–F1P complex facilitates RNA polymerase binding to DNA
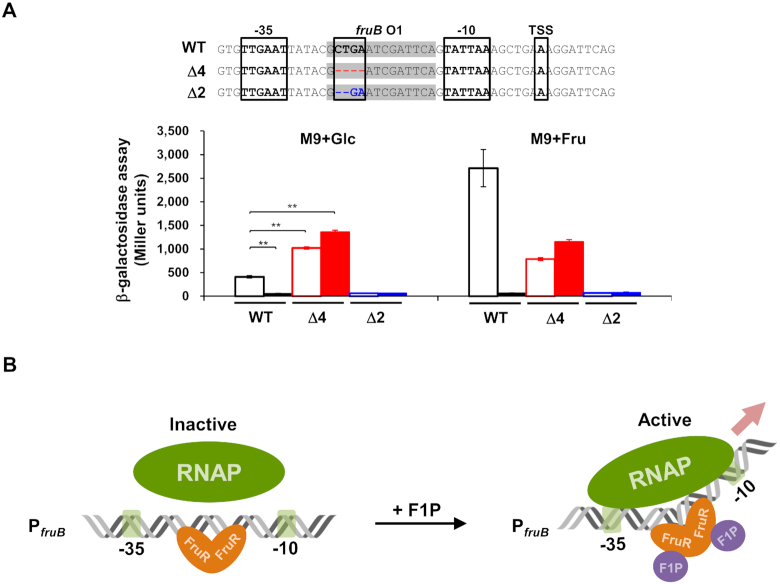
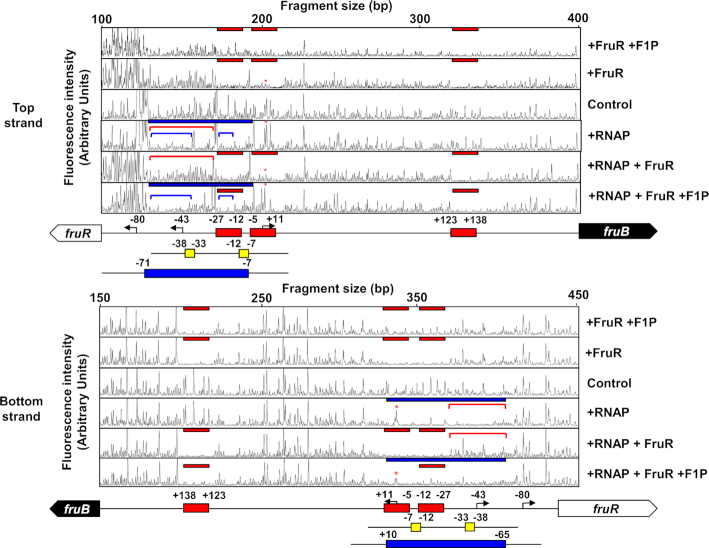
Bacterial transcription activators are classified into three types according to the position of the binding sites relative to the –35 and –10 promoter elements (45). Since the functional fruB O1 is located between –35 and –10 promoter elements, we assumed that VcFruR could have a similar mode of action as E. coli MerR (45). MerR is known to regulate transcription of the merTP(C/F)AD(E) operon (mer operon), which is contiguous to but divergently transcribed from merR, by binding to a site between –35 and –10 elements of the merT promoter (PmerT). While the optimal spacer length between –35 and –10 elements in E. coli promoters is known to be 17 ± 1 bases (46), the mer operon has 19 bp, which requires modification of the DNA structure for optimal transcription initiation (47). MerR acts as a transcription activator in the presence of an effector, Hg (II). In the presence of Hg (II), MerR–Hg (II) binds to the mer promoter region and assists the RNA polymerase (RNAP) complex to form an open initiation complex (48,49). We wondered if this regulatory mechanism might also be operating in the VcFruR-mediated transcriptional regulation of the fru operon because the spacer length between –35 and –10 elements in the fru promoter of V. cholerae is also unusually long (20 bp) (Figure 2A), compared to that in E. coli and P. putida (18 and 15 bp, respectively) (12,50). It is known that deletion of 1 or 2 bp in the spacer region between –35 and –10 promoter elements increases PmerT activity regardless of the presence of MerR or effector Hg (II) (51). To examine the effect of the spacer length between –35 and –10 elements of PfruB on its activity, we constructed V. cholerae N16961 lacZ−fruR− and lacZ−fruR+ strains harboring plasmids carrying E. coli lacZ transcriptionally fused with the PfruB with a 2 or 4 bp deletion in the fruB O1 sequence (Δ2 and Δ4, respectively, in Figure 5A). In both strains harboring the plasmid carrying the Δ2 promoter, β-galactosidase activity was not observed regardless of the presence of fructose, whereas lacZ was strongly expressed in strains carrying the Δ4 promoter, regardless of the presence of FruR or fructose. This result implies that the VcFruR-mediated transcriptional activation of the fru operon can be achieved by the facilitation of open complex formation by RNAP through optimization of the spacer length of the fruB promoter (Figure 5B). To further verify our assumption, we conducted footprinting assays with the hybrid RNAP holoenzyme consisting of E. coli core enzyme and V. cholerae σ70 and VcFruR in the absence or presence of F1P, using a DNA probe spanning from –199 to +339 relative to the TSS (Figure 6, the TSS is marked with a red asterisk in each panel). As expected from the previous Chip-seq data of V. cholerae σ70 (38), the hybrid RNAP holoenzyme protected the DNA region from –76 to –7 and –65 to +10 (marked with blue bars in Figure 6) from DNase I digestion in the top strand and bottom strand, respectively. It should be noted that the DNA region spanning from –27 to –12 corresponds to the FruR-binding site fruB O1 (see Figure 2A), while the region from –55 to –40 corresponds to the UP element, which is the binding site of the C-terminal domains of the α subunits of RNAP; it further increases the association of RNAP with DNA and thereby stimulates transcription (52). When the RNAP was added to the DNase I reaction mixture together with VcFruR; however, the protection by the RNAP holoenzyme in the region encompassing –71 to –28 and –65 to –28 were abolished in the top and bottom strands, respectively (compare the regions marked with red lines in Figure 6), while the protection by FruR at fruB O1, O2 and O3 occurred regardless of RNAP (marked with red bars). These data indicate that FruR efficiently competes with RNAP for the promoter, thus inhibiting the initiation of transcription in the absence of F1P. Interestingly, however, when F1P was added to the DNase I reaction mixture containing both RNAP and FruR, the protection of the regions encompassing –71 to –40 and –27 to –21 became stronger (compare the regions marked with blue lines in Figure 6), while the region between –40 and –27 became more sensitive to DNase I digestion (compare the region between blue lines in Figure 6), than when RNAP was added alone (Figure 6). Based on the increased sensitivity to DNase I digestion, we speculate that the binding of FruR-F1P at fruB O1 may induce a structural change in the spacer region between –35 and –10 elements. It is also worth noting that, while the protection of fruB O3 from DNase I digestion was still observed, the protection of fruB O2 including the TSS was abolished in the presence of RNAP, FruR and F1P. Together with the data above, this footprinting experiment suggests that, in the presence of F1P, FruR bound to fruB O1 strengthens the association of the RNAP holoenzyme to the promoter and displaces another FruR molecule from fruB O2 so that the transcription of the fru operon can be initiated. Furthermore, we performed KMnO4 footprinting assays to examine the open complex formation of RNA polymerase in the absence or presence of VcFruR and F1P. We found that in the presence of VcFruR and F1P, the KMnO4 reactivity at –10 thymine was increased. This result implies that VcFruR–F1P complex may facilitate the open complex formation (Supplementary Figure S10). Although it is still not possible to explain the exact mechanism by which the FruR–F1P complex affects the association of RNAP to the promoter, we hypothesize that the FruR–F1P complex bound to fruB O1 optimizes the accessibility of RNAP to the fru promoter.
Figure 5.
Optimization of the spacer length between –35 and –10 promoter elements increased the fruB promoter activity independently of the presence of VcFruR. (A) The effect of the spacer length between –35 and –10 elements of the fruB promoter on its activity was measured by the lacZ reporter assay using a V. cholerae N16961 lacZ−fruR− (filled bars) or lacZ−fruR+ (open bars) strain harboring plasmids carrying E. coli lacZ transcriptionally fused with the modified V. cholerae fruB promoter with 2 or 4 bp deletion in the fruB O1 sequence (Δ2 and Δ4, marked as blue and red, respectively) as well as the wild-type fruB promoter (WT). Nucleotide sequences corresponding to fruB O1 are shaded in grey. The –35 and –10 promoter elements and TSS are boxed. Deleted nucleotides in the modified promoters are shown in blue and red, respectively, and marked as ‘-’ (upper panel). Indicated strains were grown on glucose or fructose and then lysed to measure the β-galactosidase activity (lower panel). Statistical significance was determined using the Student's t-test (**P< 0.01). (B) A schematic view of the proposed mechanism of transcriptional activation of the fru operon by FruR in V. cholerae. In the absence of F1P, FruR binds to the binding site fruB O1 located between –27 to –12 relative to the TSS in the fruB promoter, which has a suboptimal spacer length of 20 bp, and interferes with the binding of RNAP to the promoter. Accordingly, transcriptional activation of the fru operon does not occur. In the presence of fructose, however, it is transported through the PTSFru and concomitantly phosphorylated to F1P. Then, the FruR–F1P complex binds to fruB O1 and induces a structural change in the DNA spacer region between the -35 and -10 elements, thereby facilitating RNAP binding to the promoter to trigger the transcriptional activation of the fru operon.
Figure 6.
The effect of VcFruR and F1P on RNAP binding to the top and bottom strands of the fruB promoter was assessed by DNase I footprinting assays. A 6-FAM-labeled 538-bp fruB probe (200 ng; 30.6 nM) was incubated with either hybrid RNAP holoenzyme (0.7 μg of E. coli core RNAP and 1.4 μg V. cholerae σ70) or VcFruR (650 ng; 451 nM) in the absence or presence of 2 mM F1P as indicated. The DNA regions protected from DNase I digestion by VcFruR (red bars) and RNAP (blue bars) were then determined. The DNA regions encompassing –76 to –28, –76 to –40 and –27 to –21 relative to the TSS are indicated with red and blue lines, respectively. The TSS of the fru operon is marked with red asterisks. Schematic diagrams of the fruR-fruB intergenic region are shown below, and FruR-binding sites, –35 and –10 elements and RNAP-binding sites are depicted in red, yellow, and blue rectangles, respectively. The bent arrows indicate the TSSs of the fru operon and fruR with nucleotide positions relative to the fruB TSS are indicated. The fluorescence intensity of the 6-FAM-labeled fragments is shown on the y-axis of each electropherogram and fragment sizes were determined by comparison with the internal molecular weight standards.
DISCUSSION
In this study, we propose a molecular mechanism of the FruR-mediated transcriptional activation of the fru operon in V. cholerae which is completely different from that in other Gammaproteobacteria (Figure 5B). In E. coli, FruR binds to two sites centered at 7.5 and 73.5 bp downstream of the TSS of the fru operon, which is located far from the fruR gene (11). In P. putida, FruR binds to a site centered at 19.5 bp downstream of the TSS of the fru operon, which is adjacent to but divergently transcribed from fruR (12). In both Enterobacteriaceae and Pseudomonadaceae, a fruR mutant exhibited an increased expression level of the fru operon (4,12,31,53) and thereby an increased uptake rate of fructose compared to the wild-type strain (54,55). Therefore, FruR is known as a transcriptional repressor of the fru operon in Gammaproteobacteria. However, in this study, we found that a V. cholerae fruR mutant cannot grow on fructose as the sole carbon and energy source, due to a lack of expression of the fru operon (Figure 1 and Supplementary Figure S1). In the presence of F1P, VcFruR activated the transcription of the fru operon by binding to an operator (fruB O1) located between –35 and –10 elements. While there are three FruR-binding sites in the promoter region of the V. cholerae fru operon, the other two binding sites do not seem to play a role in the fructose-mediated transcriptional activation of the operon (Figure 3). Meanwhile, the binding of VcFruR to fruB O3 is likely to be involved in the repression of the fru operon by FruR in the presence of glucose (Figure 3A). F1P alters the affinity of VcFruR for the binding site fruB O1, which facilitates the binding of RNAP to the promoter. We assumed that this transcriptional regulatory mechanism of VcFruR might be similar to that of E. coli MerR (EcMerR) since they share some important common characteristics even though VcFruR lacks structural features typical of the MerR family transcription factors, such as three cysteine residues involved in metal binding (56). First, both VcFruR and EcMerR are homodimers (Supplementary Figure S3) (57), while other GalR-LacI family of transcriptional regulators including EcFruR are usually homo-tetramers. Second, EcMerR and VcFruR bind to an operator located between –35 and –10 elements of mer and fru operons, respectively (Figure 2B) (48). Third, both the E. coli mer operon and the V. cholerae fru operon have suboptimal spacer lengths (19 and 20 bp, respectively) between –35 and –10 elements (Figure 2A) (47), so that structural rearrangement would be required to optimize the binding of RNAP to the promoter. Fourth, while MerR and FruR outcompete RNAP for binding to the operator in the absence of their effectors, both facilitate the association of RNAP to the promoter in the presence of their effectors (Figure 6) (58). Consequently, we hypothesized that VcFruR also induces a structural change at fruB O1 to enhance the binding of RNAP to the promoter, as EcMerR is known to distort the DNA to activate transcription of the mer operon in the presence of Hg(II) (48,49). Indeed, we found that RNAP bound to the UP element more tightly, while the region near –30 became more sensitive to DNase I digestion, in the presence of VcFruR and F1P compared to when RNAP was present alone (Figure 6). Furthermore, when VcFruR and F1P were added together with RNAP, the region near the TSS also became more sensitive to DNase I digestion than when FruR was present alone. However, we were unable to detect a direct interaction between VcFruR and RNAP, regardless of the presence of F1P (Supplementary Figure S11). Thus, we assume that VcFruR-F1P induces a structural change in the DNA spacer region between –35 and –10 elements (Figure 6).
It is known that EcFruR acts as a transcription activator of ppsA, whose mechanism though differs from that of the transcriptional activation of the fru operon by VcFruR, based on the following reasons: first, EcFruR is a Class I transcription activator (45) that binds to the operator centered at –45.5 relative to the TSS and strengthens the binding of the C-terminal domain of the α subunit of RNAP to DNA (59). In contrast, the regulatory mechanism of VcFruR is similar to that of the MerR-type transcriptional regulator that induces DNA distortion by binding to the region between the -35 and -10 promoter elements and facilitates RNAP binding to the promoter. Second, the presence of FruR is not essential for the transcription of ppsA. According to a previous study, the addition of EcFruR strongly stimulated the transcription of ppsA, but a significant amount of transcript was generated even without EcFruR (59). However, in the absence of VcFruR, the fru operon is hardly expressed due to the abnormally longer than optimal spacer length between –35 and –10 elements in the promoter. Third, F1P is not essential for EcFruR-mediated transcriptional activation. According to a previous study (59), EcFruR alone elicits a conformational change in the ppsA promoter. Although it is known that F1P interferes with the binding of EcFruR to the ppsA promoter (11), there are no studies on how F1P affects the EcFruR-mediated transcriptional activation of ppsA. In this study, however, we showed that F1P is essential for the transcriptional activation of the fru operon by VcFruR and facilitates the binding of RNAP to the fru promoter.
It is generally considered that F1P interacts with FruR to prevent its binding to the fru promoter in Enterobacteriaceae and Pseudomonadaceae (11–13,41). In this study, however, we show that F1P does not block VcFruR binding to DNA but weakens the binding affinity of FruR for fruB O1 by ∼3-fold without affecting the affinity for fruB O2 and fruB O3 (Figure 4B and Supplementary Figure S8). A previous study demonstrated that F1P inhibits FruR binding to an operator that contains symmetrical nucleotide sequences in a stronger manner than to a binding site containing asymmetrical nucleotide sequences (11). Since fruB O1 has a more symmetrical nucleotide sequence at central positions of the operator palindrome than fruB O2 and O3 (Figure 2A), we speculated that F1P specifically reduces FruR binding to fruB O1. Despite the decreased affinity, the binding of FruR–F1P to fruB O1 is essential for fructose-dependent transcriptional activation of the fru operon in V. cholerae (Figures 3 and 4). Since our findings are in contrast to those obtained in other species, we examined previous studies on the effect of F1P on the binding of FruR to DNA in E. coli and P. putida (11–13,33,40). Interestingly, in all of the EMSA data in these previous reports, a significant fraction of FruR remained bound to the DNA probe even in the presence of a very high concentration of F1P (up to 5 mM), suggesting that F1P may not block FruR binding to DNA but rather decrease its binding affinity for the fru promoter in these species. Furthermore, in a recent study on the transcriptional regulation of central carbon metabolism by FruR using a chromatin immuno-precipitation method with exonuclease treatment (ChIP-exo) in E. coli (9), FruR still remained bound to the promoter of the fru operon in cells grown on fructose as well as in cells grown on glucose. Therefore, the molecular mechanism underlying transcriptional activation by VcFruR revealed in this study could be more comprehensive than previously thought. In conclusion, this study provides a new perspective on the transcriptional regulation mechanism of the fru operon by FruR, which needs to be re-examined in other Gammaproteobacterial species.
DATA AVAILABILITY
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request
Supplementary Material
Contributor Information
Chang-Kyu Yoon, School of Biological Sciences and Institute of Microbiology, Seoul National University, Seoul 08826, Korea.
Deborah Kang, School of Biological Sciences and Institute of Microbiology, Seoul National University, Seoul 08826, Korea.
Min-Kyu Kim, Radiation Research Division, Korea Atomic Energy Research Institute, Jeongeup 56212, Korea.
Yeong-Jae Seok, School of Biological Sciences and Institute of Microbiology, Seoul National University, Seoul 08826, Korea.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation (NRF) of Korea [NRF-2018R1A5A1025077, NRF-2019R1A2C2004143 to Y.-J.S., NRF-2020R1F1A1057780 to M.-K.K.]. Funding for open access charge: National Research Foundation of Korea.
Conflict of interest statement. None declared.
REFERENCES
- 1. Deutscher J., Francke C., Postma P.W.. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 2006; 70:939–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Park S., Park Y.H., Lee C.R., Kim Y.R., Seok Y.J.. Glucose induces delocalization of a flagellar biosynthesis protein from the flagellated pole. Mol. Microbiol. 2016; 101:795–808. [DOI] [PubMed] [Google Scholar]
- 3. Choe M., Park Y.H., Lee C.R., Kim Y.R., Seok Y.J.. The general PTS component HPr determines the preference for glucose over mannitol. Sci. Rep. 2017; 7:43431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Saier M.H. Jr, Ramseier T.M. The catabolite repressor/activator (Cra) protein of enteric bacteria. J. Bacteriol. 1996; 178:3411–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Granot D., David-Schwartz R., Kelly G.. Hexose kinases and their role in sugar-sensing and plant development. Front Plant Sci. 2013; 4:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kornberg H.L. Routes for fructose utilization by Escherichia coli. J. Mol. Microbiol. Biotechnol. 2001; 3:355–359. [PubMed] [Google Scholar]
- 7. Plumbridge J. Regulation of PTS gene expression by the homologous transcriptional regulators, Mlc and NagC, in Escherichia coli (or how two similar repressors can behave differently). J. Mol. Microbiol. Biotechnol. 2001; 3:371–380. [PubMed] [Google Scholar]
- 8. Feldheim D., Chin A., Nierva C., Feucht B., Cao Y.W., Xu Y.F., Sutrina S., Saier M.. Physiological consequences of the complete loss of phosphoryl-transfer proteins HPr and FPr of the phosphoenolpyruvate: sugar phosphotransferase system and analysis of fructose (fru) operon expression in Salmonella typhimurium. J. Bacteriol. 1990; 172:5459–5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim D., Seo S.W., Gao Y., Nam H., Guzman G.I., Cho B.K., Palsson B.O.. Systems assessment of transcriptional regulation on central carbon metabolism by Cra and CRP. Nucleic Acids Res. 2018; 46:2901–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kochanowski K., Gerosa L., Brunner S.F., Christodoulou D., Nikolaev Y.V., Sauer U.. Few regulatory metabolites coordinate expression of central metabolic genes in Escherichia coli. Mol. Syst. Biol. 2017; 13:903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ramseier T.M., Negre D., Cortay J.C., Scarabel M., Cozzone A.J., Saier M.H. Jr. In vitro binding of the pleiotropic transcriptional regulatory protein, FruR, to the fru, pps, ace, pts and icd operons of Escherichia coli and Salmonella typhimurium. J. Mol. Biol. 1993; 234:28–44. [DOI] [PubMed] [Google Scholar]
- 12. Chavarria M., Santiago C., Platero R., Krell T., Casasnovas J.M., de Lorenzo V.. Fructose 1-phosphate is the preferred effector of the metabolic regulator Cra of Pseudomonas putida. J. Biol. Chem. 2011; 286:9351–9359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chavarria M., Durante-Rodriguez G., Krell T., Santiago C., Brezovsky J., Damborsky J., de Lorenzo V.. Fructose 1-phosphate is the one and only physiological effector of the Cra (FruR) regulator of Pseudomonas putida. FEBS Open Bio. 2014; 4:377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ravcheev D.A., Khoroshkin M.S., Laikova O.N., Tsoy O.V., Sernova N.V., Petrova S.A., Rakhmaninova A.B., Novichkov P.S., Gelfand M.S., Rodionov D.A.. Comparative genomics and evolution of regulons of the LacI-family transcription factors. Front Microbiol. 2014; 5:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rohmer L., Hocquet D., Miller S.I.. Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol. 2011; 19:341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mandlik A., Livny J., Robins W.P., Ritchie J.M., Mekalanos J.J., Waldor M.K.. RNA-Seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe. 2011; 10:165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Park S., Yoon J., Lee C.R., Lee J.Y., Kim Y.R., Jang K.S., Lee K.H., Seok Y.J.. Polar landmark protein HubP recruits flagella assembly protein FapA under glucose limitation in Vibrio vulnificus. Mol. Microbiol. 2019; 112:266–279. [DOI] [PubMed] [Google Scholar]
- 18. Kim H.M., Park Y.H., Yoon C.K., Seok Y.J.. Histidine phosphocarrier protein regulates pyruvate kinase A activity in response to glucose in Vibrio vulnificus. Mol. Microbiol. 2015; 96:293–305. [DOI] [PubMed] [Google Scholar]
- 19. Hamashima H., Iwasaki M., Arai T.. A simple and rapid method for transformation of Vibrio species by electroporation. Methods Mol. Biol. 1995; 47:155–160. [DOI] [PubMed] [Google Scholar]
- 20. Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A. 3rd, Smith H.O.. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009; 6:343–345. [DOI] [PubMed] [Google Scholar]
- 21. Kim H.M., Yoon C.K., Ham H.I., Seok Y.J., Park Y.H.. Stimulation of Vibrio vulnificus pyruvate kinase in the presence of glucose to cope with H2O2 stress generated by its competitors. Front Microbiol. 2018; 9:1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Choi S.H., Lee K.L., Shin J.H., Cho Y.B., Cha S.S., Roe J.H.. Zinc-dependent regulation of zinc import and export genes by Zur. Nat. Commun. 2017; 8:15812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee J.W., Park Y.H., Seok Y.J.. Rsd balances (p)ppGpp level by stimulating the hydrolase activity of SpoT during carbon source downshift in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E6845–E6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miller J. Experiments in Molecular Genetics. 1972; NY: Cold Spring Harbor Laboratory Press; . [Google Scholar]
- 25. Houot L., Watnick P.I.. A novel role for enzyme I of theVibrio cholerae phosphoenolpyruvate phosphotransferase system in regulation of growth in a biofilm. J. Bacteriol. 2008; 190:311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Houot L., Chang S., Pickering B.S., Absalon C., Watnick P.I.. The phosphoenolpyruvate phosphotransferase system regulatesVibrio cholerae biofilm formation through multiple independent pathways. J. Bacteriol. 2010; 192:3055–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Houot L., Chang S., Absalon C., Watnick P.I.. Vibrio cholerae phosphoenolpyruvate phosphotransferase system control of carbohydrate transport, biofilm formation, and colonization of the germfree mouse intestine. Infect. Immun. 2010; 78:1482–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hayes C.A., Dalia T.N., Dalia A.B.. Systematic genetic dissection of PTS in Vibrio cholerae uncovers a novel glucose transporter and a limited role for PTS during infection of a mammalian host. Mol. Microbiol. 2017; 104:568–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ferenci T., Kornberg H.L.. The utilization of fructose byEscherichia coli. Properties of a mutant defective in fructose 1-phosphate kinase activity. Biochem. J. 1973; 132:341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Van Dijken J.P., Quayle J.R.. Fructose metabolism in four Pseudomonas species. Arch. Microbiol. 1977; 114:281–286. [DOI] [PubMed] [Google Scholar]
- 31. Shimada T., Yamamoto K., Ishihama A.. Novel members of the Cra regulon involved in carbon metabolism in Escherichia coli. J. Bacteriol. 2011; 193:649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Munch R., Hiller K., Grote A., Scheer M., Klein J., Schobert M., Jahn D.. Virtual footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics. 2005; 21:4187–4189. [DOI] [PubMed] [Google Scholar]
- 33. Nègre D., Bonod-Bidaud C., Geourjon C., Deléage G., Cozzone A.J., Cortay J.C.. Definition of a consensus DNA-binding site for the Escherichia coli pleiotropic regulatory protein, FruR. Mol. Microbiol. 1996; 21:257–266. [DOI] [PubMed] [Google Scholar]
- 34. Swint-Kruse L., Matthews K.S.. Allostery in the LacI/GalR family: variations on a theme. Curr. Opin. Microbiol. 2009; 12:129–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Weickert M.J., Adhya S.. A family of bacterial regulators homologous to Gal and Lac repressors. J. Biol. Chem. 1992; 267:15869–15874. [PubMed] [Google Scholar]
- 36. Cortay J.C., Negre D., Scarabel M., Ramseier T.M., Vartak N.B., Reizer J., Saier M.H. Jr, Cozzone A.J.. In vitro asymmetric binding of the pleiotropic regulatory protein, FruR, to the ace operator controlling glyoxylate shunt enzyme synthesis. J. Biol. Chem. 1994; 269:14885–14891. [PubMed] [Google Scholar]
- 37. Chen J., Matthews K.S.. Deletion of lactose repressor carboxyl-terminal domain affects tetramer formation. J. Biol. Chem. 1992; 267:13843–13850. [PubMed] [Google Scholar]
- 38. Manneh-Roussel J., Haycocks J.R.J., Magan A., Perez-Soto N., Voelz K., Camilli A., Krachler A.M., Grainger D.C.. cAMP receptor protein controls Vibrio cholerae gene expression in response to host colonization. mBio. 2018; 9:e00966-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang H., Ayala J.C., Benitez J.A., Silva A.J.. RNA-seq analysis identifies new genes regulated by the histone-like nucleoid structuring protein (H-NS) affecting Vibrio cholerae virulence, stress response and chemotaxis. PLoS One. 2015; 10:e0118295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bley Folly B., Ortega A.D., Hubmann G., Bonsing-Vedelaar S., Wijma H.J., van der Meulen P., Milias-Argeitis A., Heinemann M.. Assessment of the interaction between the flux-signaling metabolite fructose-1,6-bisphosphate and the bacterial transcription factors CggR and Cra. Mol. Microbiol. 2018; 109:278–290. [DOI] [PubMed] [Google Scholar]
- 41. Chavarria M., de Lorenzo V.. The imbroglio of the physiological Cra effector clarified at last. Mol. Microbiol. 2018; 109:273–277. [DOI] [PubMed] [Google Scholar]
- 42. Chavarria M., Fuhrer T., Sauer U., Pfluger-Grau K., de Lorenzo V.. Cra regulates the cross-talk between the two branches of the phosphoenolpyruvate: phosphotransferase system of Pseudomonas putida. Environ. Microbiol. 2013; 15:121–132. [DOI] [PubMed] [Google Scholar]
- 43. Chavarria M., Goni-Moreno A., de Lorenzo V., Nikel P.I.. A metabolic widget adjusts the phosphoenolpyruvate-dependent fructose influx in Pseudomonas putida. mSystems. 2016; 1:e00154-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huffman J.L., Lu F., Zalkin H., Brennan R.G.. Role of residue 147 in the gene regulatory function of the Escherichia coli purine repressor. Biochemistry. 2002; 41:511–520. [DOI] [PubMed] [Google Scholar]
- 45. Browning D.F., Busby S.J.. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004; 2:57–65. [DOI] [PubMed] [Google Scholar]
- 46. Singh S.S., Typas A., Hengge R., Grainger D.C.. Escherichia coli σ 70 senses sequence and conformation of the promoter spacer region. Nucleic Acids Res. 2011; 39:5109–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lund P.A., Brown N.L.. Regulation of transcription in Escherichia coli from the mer and merR promoters in the transposon Tn501. J. Mol. Biol. 1989; 205:343–353. [DOI] [PubMed] [Google Scholar]
- 48. Ansari A.Z., Chael M.L., O’Halloran T.V.. Allosteric underwinding of DNA is a critical step in positive control of transcription by Hg-MerR. Nature. 1992; 355:87–89. [DOI] [PubMed] [Google Scholar]
- 49. Ansari A.Z., Bradner J.E., O’Halloran T.V.. DNA-bend modulation in a repressor-to-activator switching mechanism. Nature. 1995; 374:370–375. [DOI] [PubMed] [Google Scholar]
- 50. Huerta A.M., Collado-Vides J.. Sigma70 promoters in Escherichia coli: specific transcription in dense regions of overlapping promoter-like signals. J. Mol. Biol. 2003; 333:261–278. [DOI] [PubMed] [Google Scholar]
- 51. Parkhill J., Brown N.L.. Site-specific insertion and deletion mutants in the mer promoter-operator region of Tn501; the nineteen base-pair spacer is essential for normal induction of the promoter by MerR. Nucleic Acids Res. 1990; 18:5157–5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ross W., Ernst A., Gourse R.L.. Fine structure of E. coli RNA polymerase-promoter interactions: α subunit binding to the UP element minor groove. Genes Dev. 2001; 15:491–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sarkar D., Siddiquee K.A., Arauzo-Bravo M.J., Oba T., Shimizu K.. Effect of cra gene knockout together with edd andiclR genes knockout on the metabolism in Escherichia coli. Arch. Microbiol. 2008; 190:559–571. [DOI] [PubMed] [Google Scholar]
- 54. Yao R., Kurata H., Shimizu K.. Effect of cra gene mutation on the metabolism of Escherichia coli for a mixture of multiple carbon sources. Adv Biosci Biotechnol. 2013; 4:477. [Google Scholar]
- 55. Ramseier T.M., Bledig S., Michotey V., Feghali R., Saier M.H. Jr. The global regulatory protein FruR modulates the direction of carbon flow in Escherichia coli. Mol. Microbiol. 1995; 16:1157–1169. [DOI] [PubMed] [Google Scholar]
- 56. Brown N.L., Stoyanov J.V., Kidd S.P., Hobman J.L.. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 2003; 27:145–163. [DOI] [PubMed] [Google Scholar]
- 57. Helmann J.D., Ballard B.T., Walsh C.T.. The MerR metalloregulatory protein binds mercuric ion as a tricoordinate, metal-bridged dimer. Science. 1990; 247:946–948. [DOI] [PubMed] [Google Scholar]
- 58. O’Halloran T.V., Frantz B., Shin M.K., Ralston D.M., Wright J.G.. The MerR heavy metal receptor mediates positive activation in a topologically novel transcription complex. Cell. 1989; 56:119–129. [DOI] [PubMed] [Google Scholar]
- 59. Nègre D., Oudot C., Prost J.F., Murakami K., Ishihama A., Cozzone A.J., Cortay J.C.. FruR-mediated transcriptional activation at the ppsA promoter of Escherichia coli. J. Mol. Biol. 1998; 276:355–365. [DOI] [PubMed] [Google Scholar]
- 60. Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L.et al.. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018; 46:W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H.J.. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005; 33:W363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request