Summary
Universal red blood cells (RBCs) differentiated from O-negative human induced pluripotent stem cells (hiPSCs) could find applications in transfusion medicine. Given that each transfusion unit of blood requires 2 trillion RBCs, efficient bioprocesses need to be developed for large-scale in vitro generation of RBCs.
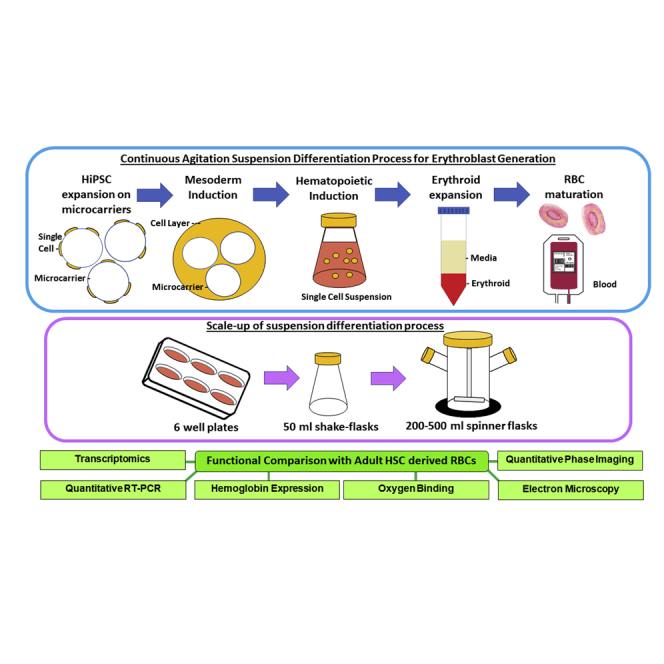
We have developed a scalable suspension agitation culture platform for differentiating hiPSC-microcarrier aggregates into functional RBCs and have demonstrated scalability of the process starting with 6 well plates and finally demonstrating in 500 mL spinner flasks. Differentiation of the best-performing hiPSCs generated 0.85 billion erythroblasts in 50 mL cultures with cell densities approaching 1.7 × 107 cells/mL. Functional (oxygen binding, hemoglobin characterization, membrane integrity, and fluctuations) and transcriptomics evaluations showed minimal differences between hiPSC-derived and adult-derived RBCs.
The scalable agitation suspension culture differentiation process we describe here could find applications in future large-scale production of RBCs in controlled bioreactors.
Keywords: iPSC, scalable, red blood cell, spinner flasks, enucleation
Graphical Abstract
Highlights
-
•
Scalable process for differentiating hiPSC-microcarrier aggregates into RBCs
-
•
Erythroid differentiation potential of multiple hiPSC lines was evaluated
-
•
hiPSC RBCs and adult RBCs revealed minor differences functionally and transcriptionally
-
•
Co-culture of hiPSC RBCs with OP9 cells (2D and 3D) promoted improved enucleation
In this article, Dr. Oh and colleagues show that agitation microcarrier suspension cultures can be used as a scalable platform for differentiating O-negative hiPSCs into RBCs. Following screening, differentiation of the best-performing hiPSCs was scaled up in stirred spinner flasks. Co-culture of erythroblasts with OP9 cells significantly improved enucleation. Functional and transcriptomics analysis revealed minimal differences between hiPSC-derived versus adult-derived RBCs.
Introduction
O-negative rhesus factor D-negative (O-neg) blood, the universal donor blood type, is considered a limited and valuable source of red blood cells (RBCs) for emergency transfusion applications (Hirani et al., 2017). Anticipated supply shortages in the future due to an aging population and risks from emerging viruses and pathogens (Alter et al., 2007) have driven initiatives to develop alternate and ready sources of universal donor blood. Differentiation of RBCs from human induced pluripotent stem cells (hiPSCs) is one such approach actively being investigated. The unlimited proliferation potential of hiPSCs coupled with their potential to differentiate into hematopoietic lineages (Paes et al., 2017) has made these cells appealing as limitless sources of starting materials for generating universal RBCs. It has been postulated that as few as 10 hiPSC clones derived from patients with rare blood phenotypes would be sufficient to cover the necessary blood types for 99% of the population with recurrent transfusion needs (Peyrard et al., 2011).
Each transfusion unit of blood consists of 2 trillion RBCs. In vitro derivation of such large numbers of RBCs requires overcoming a few unmet challenges. First is the lack of efficient bioprocesses that can be scaled up from laboratory to industrial scale for RBC manufacture. Although several groups have shown the potential for efficient differentiation of hiPSCs toward RBCs (Dorn et al., 2015; Mao et al., 2016; Olivier et al., 2016), most of these may not be favorable for clinical development, either due to the use of undefined or xenogenic components or due to the lack of scalability of the process. Other groups have also immortalized adult erythroblasts to produce RBCs (Trakarnsanga et al., 2017; Kurita et al., 2013; Hirose et al., 2013). Yet a second challenge that needs to be overcome is the lack of cost-effective means to achieve ultra-high-density cultures of RBCs. Given that each unit of blood requires 2 × 1012 RBCs, one would have to achieve cell densities of at least 1 × 108 cells/mL in order to generate the desired cell numbers in a minimal medium volume. Thus far, the highest reported cell density for RBC culture appears to be in the range of 1 × 107 cells/mL (Ying Wang et al., 2016).
Development of a scalable process that can eventually be transferred to large-scale stirred bioreactors would require the entire process to be performed in continuous agitation suspension culture. We have previously described means to scale up the pluripotent expansion stage by culturing hiPSCs on Laminin-521 (LN-521)-coated microcarriers (MCs) (Lam et al., 2016; Sivalingam et al., 2018). We have also shown that hiPSC-MC aggregates in suspension culture can efficiently differentiate into T-Bra+ and KDR+ mesodermal cells (Sivalingam et al., 2018), demonstrating that hiPSC-MC aggregates could differentiate as embryoid bodies (EBs) in a scalable manner.
The current study was undertaken to develop an agitation suspension culture bioprocess for differentiation of hiPSCs to erythroid cells with prospects of transferring the process to larger-scale controlled bioreactors for future manufacture of RBCs. Using process optimization, we show that hiPSC-MC aggregates can be efficiently differentiated into mature and functional RBCs. We demonstrate the scalability of the process starting from 6 well plates all the way to 500 mL spinner flasks. We show that it is possible to differentiate hiPSC-MC aggregates into high-density cultures of erythroid cells approaching concentrations of 1.7 × 107 cells/mL in spinner flasks. More importantly, we show that functional and transcriptomics evaluation revealed minimal differences between hiPSC-derived RBCs and adult derived RBCs. The scalable agitation suspension culture differentiation process we describe could serve as a platform for developing large-scale blood differentiation processes in controlled bioreactors.
Results
Continuous Agitation Suspension Culture Differentiation of hiPSC-MC Aggregates in 6 Well-ULA Plates
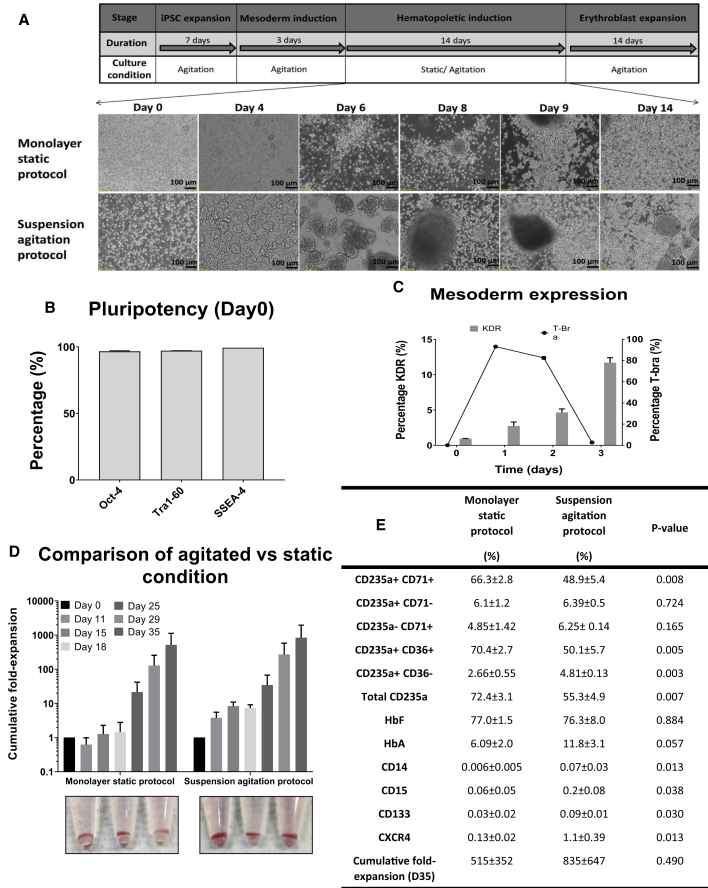
To develop a continuous agitation suspension culture differentiation platform, we first evaluated whether an hiPSC line expanded under agitation on MC during the pluripotent expansion and mesoderm stages could be further differentiated through the hematopoietic and erythroid induction stages under continuous agitation in suspension culture in 6 well ULA plates (suspension agitation protocol). A control experiment was performed whereby the hematopoietic induction stage was achieved on normal tissue culture plates under static conditions (monolayer static protocol) (Figure 1A). An O-neg hiPSC line, D5, was used for both of these experiments. hiPSC-MC aggregates maintained high levels of pluripotency (>98% Oct-4, Tra1-60, and SSEA-4 positive by fluorescence-activated cell sorting [FACS]) following 7 days of expansion under continuous agitation (Figure 1B). Following hematopoietic mesoderm induction for 3 days under agitation, high levels of the primitive streak/early mesoderm marker T-bra (>83%) were observed at day 1 post differentiation. KDR expression (12 ± 4.9%) was enriched by day 3 of differentiation (Figure 1C). Single cells derived by enzymatically detaching cells from MCs were seeded into either 6 well tissue culture plates (monolayer static protocol) or ULA plates (suspension agitation protocol) for hematopoietic induction. In tissue culture plates, seeded cells initially attached and expanded as adherent hemogenic endothelial cells (days 4–9) that subsequently gave rise to suspension culture hematopoietic cells with great proliferation observed by day 14 post seeding. In ULA plates, seeded single cells initially formed small clusters (150–200 μm in diameter) in suspension by day 4. Single cells were subsequently observed to emerge from these small clusters followed by expansion from day 8 to 14. Following induction of erythroid differentiation (day 14–35), extensive expansion of single suspension cells was noted, with day 35 cumulative fold expansion of continuous agitation (suspension agitation protocol: 835 ± 647) being comparable (p > 0.05) to static (monolayer static protocol: 515 ± 352) (Figure 1D). Both continuous agitation and static culture differentiation protocols resulted in formation of hemoglobinized erythroid cells with 55.3 ± 4.9% and 72.4 ± 3.1% expressing the CD235a erythroid marker, respectively (Figures 1D and 1E). Erythroid cells mainly expressed fetal hemoglobin, although adult hemoglobin expression at a lower level was also noted (Figure 1E).
Figure 1.
Development of Agitation Suspension Culture Differentiation of O-Neg hiPSCs
(A–C) (A) Schematic of differentiation process from hiPSC to erythroblast stage. Bright-field images of cells from an O-neg hiPSC line, D5, undergoing hematopoietic differentiation in tissue culture plates under static conditions (monolayer static protocol) or in 6 well ULA plates under continuous agitation (suspension agitation protocol) from day 0 to 14 of the hematopoietic induction stage. Flow cytometry evaluation of (B) pluripotency markers following 7 days of agitation expansion in 6 well ULA plates and (C) T-Bra and KDR cell populations during mesoderm differentiation.
(D) Cumulative fold expansion of total viable cells following differentiation using monolayer static protocol or suspension agitation protocol and corresponding hemoglobinized cell pellets observed at day 35 of differentiation. There is no significant difference in expansion fold between static and agitated conditions, p > 0.05.
(E) Table summarizing FACS characterization of differentiated cells from day 35 of differentiation. Percentages of erythroid-specific markers CD235a, CD71, and CD36 and fetal/adult hemoglobin expression as well as markers for myelomonocytic cells (CD14, CD15) and hematopoietic stem cells (CD133, CXCR4) are shown together with cumulative fold expansion on day 35. All data represent the mean ± SD with at least three independent replicates. Representative experiment shown.
Interestingly, in the agitation condition, extensive expansion of single cells in suspension from day 8 to 14 was observed only after the formation of cell clusters/aggregates in suspension culture (Figure S2A). Co-expression of CD34/CD144/CD31 and CD43 by FACS (Figure S2B) and immunofluorescence imaging (CD34/CD144 co-staining) (Figure S2C) established the presence of hemogenic endothelial cells (Angelos et al., 2018) (CD34+ CD144+ CD31+ or CD43+) within these cell aggregates. In contrast, single cells obtained from day 5 of differentiation did not express markers of hemogenic endothelial populations but differentiated further into CD34/CD45-expressing primitive hematopoietic progenitors (Figure S2B).
To establish if the expanding cells in suspension culture were derived from differentiation of the hemogenic endothelial cell aggregates or from primitive hematopoietic progenitors already present in suspension, we separated out the “hemogenic endothelial cell aggregates” from single cells by sieving through 40 μm sieves on day 5 of differentiation and proceeded to differentiate these different cell populations (Figure S2A). The control group was differentiated without any additional sieving (mixture of aggregates and single cells). Interestingly, the majority of expansion was observed in the groups that had the hemogenic endothelial cell aggregates (control group I, no sieving, 30.2 ± 4.3-fold expansion; group II, sieved cell aggregates, 28.7 ± 21.4-fold expansion on day 21) rather than single cells (group III, sieved non-aggregated single cells, 1.6 ± 1.5-fold expansion). Erythroid differentiation from sieved hemogenic endothelial aggregates (aggregated population) was similar to that of the control group (no sieving), while differentiation of the sieved single cells (non-aggregated population) failed to result in erythroid differentiation (Figure S2D). Taken together, these data suggest that the hematopoietic differentiation reported in our continuous suspension agitation protocol was via differentiation from hemogenic endothelial cell clusters rather than from expansion of primitive hematopoietic progenitor cells.
Differentiation of Multiple hiPSC Lines in 50 mL Shaker Flasks Using Continuous Agitation Suspension Differentiation Approach
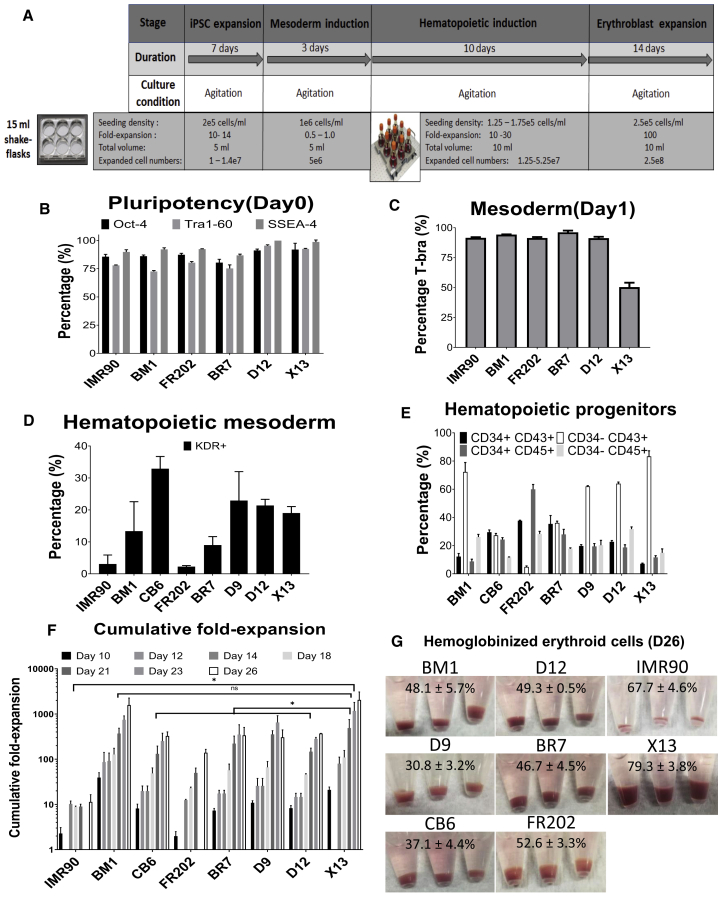
Having established that erythroid differentiation from hiPSCs can be entirely performed in suspension under continuous agitation conditions, we next proceeded to evaluate differentiation of hiPSCs derived from O-neg erythroid progenitors (D9, D12, X13, BR7) and of cord-blood-derived hiPSCs (CB6), bone-marrow-derived hiPSCs (BM1), and dermal-fibroblast-derived hiPSCs (FR202 and IMR90). For each hiPSC line, pluripotent expansion (7 days) and mesoderm differentiation (3 days) of hiPSC-MC aggregates were first established in 6 well ULA plates. After 3 days of hematopoietic mesoderm development, single cells derived following trypsinization of hiPSC-MC aggregates were further differentiated in 50 mL shaker flasks (Figure 2A). hiPSC-MC aggregates maintained their pluripotency during the expansion stage (Oct-4, 80.4%–92.0%; Tra1-60, 72.5%–95.4%; and SSEA-4, 86.8%–99.9%) (Figure 2B), differentiated into T-Bra+ primitive streak/mesoderm (50.4%–96.2%) on day 1 of differentiation (Figure 2C), and had evidence of hematopoietic-fated mesoderm marker KDR+ (3.1%–32.9%) by day 3 of differentiation (Figure 2D). Among these eight lines, the mesoderm differentiated from CB6 showed the highest KDR+ hematopoietic mesoderm marker, which was 32.9% ± 3.8%. By day 12 of differentiation, CD34+ CD45+ (8.8%–59.9%) and CD34+ CD43+ (7.2%–37.6%) hematopoietic progenitor cells, as well as more mature CD34− CD45+ (11.6%–31.7%) and CD34− CD43+ (4.9%–83.3%) hematopoietic-committed cells, were detected (Figure 2E). CD34− CD43+ hematopoietic-committed cells from X13 displayed the highest levels (83.3% ± 6.7%) compared with the other seven lines (Figure 2E). Following erythroid differentiation, BM1 (1,580 ± 675) and X13 (2,049 ± 1,019) achieved the highest cumulative fold expansion by day 26 of differentiation (Figure 2F). All eight hiPSC lines were able to differentiate into hemoglobinized erythroblasts (Figure 2G), with CD235a+ expression ranging from 30.8%–79.3%. X13 hiPSC differentiated erythroblasts yielded 79.3 ± 3.8% CD235a+ erythroblasts, highest among the eight lines differentiated (Figure 2G). To investigate whether there was correlation between D26 CD235a+ erythroblast production and the different hematopoietic markers, we performed a heatmap analysis (Figure S3B). Among all the parameters, only the marker CD34− CD43+ had an R2 value of 0.486 (Figure S3A), which indicates that these different hematopoietic markers do not have a strong correlation with final RBC production.
Figure 2.
Agitation Suspension Culture Differentiation of Multiple hiPSC Lines in Shaker Flasks
(A–F) (A) Schematic showing initial expansion of cell-MC aggregates in 6 well ULA plates during iPSC expansion and mesoderm induction stages. Subsequent stages of differentiation were achieved in 50 mL shaker flasks under continuous agitation. FACS evaluation of (B) pluripotency markers (insufficient samples for CB6 and D9), (C) mesoderm/primitive-streak marker (T-bra) (day 1 of differentiation; insufficient samples for CB6 and D9), (D) hematopoietic-fated mesoderm markers (KDR+) (day 3 of differentiation), and (E) hematopoietic progenitor markers (CD34+CD43+, CD34+CD45+)/committed hematopoietic cells (CD34− CD43+, CD34− CD45+) (day 12 of differentiation) for multiple hiPSC lines (insufficient samples for IMR90).
(F) Cumulative fold expansion of total viable cells following differentiation of multiple human iPSC lines on shaker flasks. ∗p < 0.05 comparing X13 with other lines except for BM1.
(G) Corresponding hemoglobinized cell pellets at day 26 of differentiation with percentage of CD235a+ cells (determined by FACS) indicated. All data represent the mean ± SD with at least three independent replicates. Representative experiment shown.
Both fetal hemoglobin and adult hemoglobin expression was verified by immunoblotting (Figure S3C). Expression of embryonic hemoglobin (Epsilon in the figure) was detected in BM1, D12, and X13 hiPSCs, suggesting the presence of erythroid cells derived from primitive erythropoiesis. Interestingly, erythroid cells differentiated from CB6, BR7, and D9 hiPSCs had little or no embryonic hemoglobin, suggesting that these cells may have been differentiated via definitive erythropoiesis. Oxygen equilibrium profiles of erythroblasts differentiated from hiPSCs were left-shifted (p50: 12.4 to 14.7) compared with adult RBCs (p50: 20.7 ± 1.0) (Figure S3D). Following erythroblast expansion, from day 18 to 26, maximal cell concentrations ranged from 4 to 12.6 × 106 cells/mL (Figure S3E). Complete medium replenishment was critical for cell viability and expansion, especially when cell densities were high (>8 × 106 cells/mL). For instance, at day 23, both D9 (▼) and X13 (▼) differentiated erythroblasts had high cell densities and viabilities (D9 9.64 × 106 cells/mL, 97.2 ± 1.2% viability; X13 8.63 × 106 cells/mL, 97.1 ± 1.0% viability). However, without medium renewal for 24 h, D9 erythroblast cell concentrations dropped to 7.35 × 106 cells/mL and 70.8 ± 8.1% viability (day 24), whereas with complete medium renewal, X13 erythroblasts continued to proliferate to 1.25 × 107 cells/mL and maintained a high viability of 96.5 ± 0.7% (day 26). Analysis of the conditioned medium from day 24 cultures showed that lactate and ammonia levels had spiked to 2.49 ± 0.03 g/L and 4.65 ± 0.2 mM, respectively, for D9 erythroblast cultures without medium renewal for 24 h. For X13 erythroblasts that received medium renewal on day 23, lactate and ammonia levels were significantly lower (p < 0.05) at 0.96 ± 0.2 g/L and 1.05 ± 0.2 mM, respectively (Figure S3F). Thus, the X13 hiPSC line was chosen as the best-performing line for erythroid cell differentiation with the highest cumulative fold expansion. High-resolution digital karyotyping performed on genomic DNA of X13 differentiated erythroid cells showed normal copy numbers (defined as between 1.6 and 2.4 copies) for all chromosomes evaluated (Figures S3G and S3H).
Scale-Up of Continuous Agitation Suspension Culture Differentiation to 125 mL Spinner Flasks
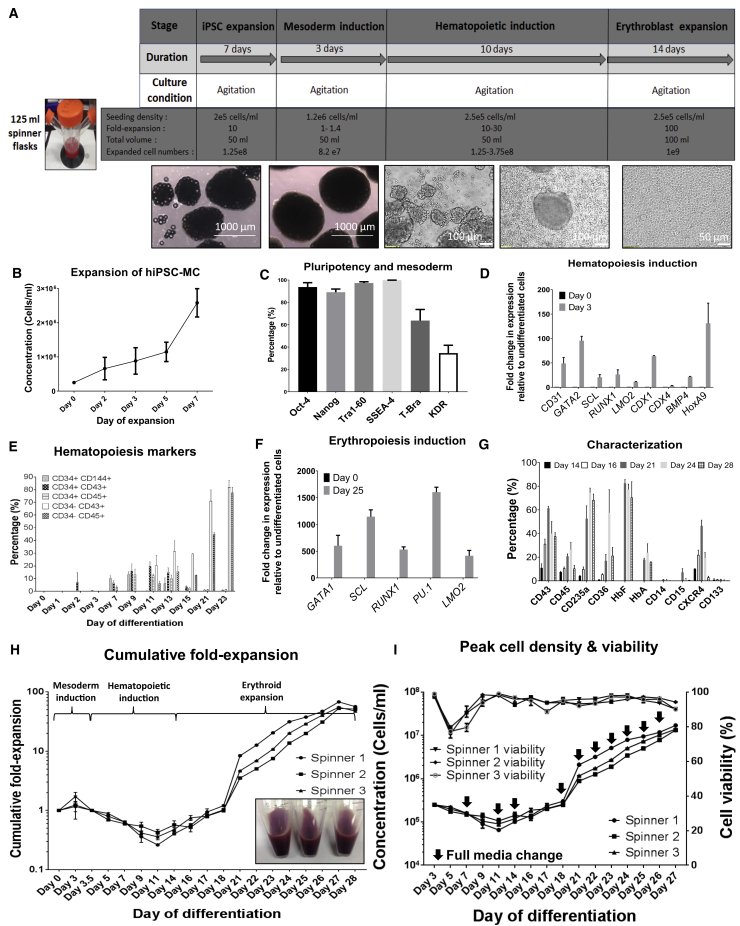
Following the demonstration that multiple hiPSC lines can be differentiated under constant agitation condition in 50 mL shaker flasks, we next chose to demonstrate scale-up of differentiation and reproducibility in 125 and 500 mL spinner flasks (Figure 3A). X13 hiPSCs, which had the best differentiation outcome in shaker flasks, were used for this study. In 125 mL spinner flasks, X13-MC aggregates achieved approximately 10-fold expansion during the pluripotent stage (Figure 3B) while maintaining pluripotency levels (Figure 3C). Upon onset of differentiation of X13 hiPSC-MC aggregates, the primitive streak/early mesoderm marker T-bra (63.7 ± 10%) and hematopoietic-fated mesoderm KDR+ marker (34.6 ± 6.9%) were detected at days 1 and 3 post differentiation, respectively (Figure 3C). KDR+ CD235a−, another marker indicative of definitive hematopoiesis (Sturgeon et al., 2014), was also detected on day 3 (15.6 ± 2%). Key markers and transcription factors involved in early hematopoiesis were found to be upregulated by day 3 of differentiation (fold upregulation compared with undifferentiated cells; Figure 3D). CD34+ CD144+ hemogenic endothelial cells (10.3 ± 3.6%) were detected as early as 7 days post differentiation. CD34+ CD43+ (19.7 ± 6.2%) and CD34+ CD45+ (12.2 ± 2.3%) hematopoietic progenitors peaked at around day 11 post differentiation and continued to mature into CD34− CD43+ (81.9 ± 9.5%) and CD34− CD45+ (77.8 ± 7.1%)-committed hematopoietic cells until day 23 post differentiation (Figure 3E). During erythroid differentiation, key markers and transcription factors associated with erythropoiesis were upregulated (Figure 3F). A significant increase in CD235a+ erythroid cells was noted starting from day 16 (9.9 ± 1.9%) to day 24 (74.7 ± 3.6%) of differentiation (Figure 3G). Day 24 differentiated erythroblasts had high levels of HbF (77.1 ± 7.5%) and lower levels of HbA (23.8 ± 7.4%). By day 28 of differentiation, the cultures had very few hematopoietic stem cells (<3% CXCR4 and <0.5% CD133) and a negligible amount of (<1%) myelomonocytic cells (CD14/CD15) (Figure 3G and Table 1). X13 hiPSC-MC aggregates expanded 1.1- to 1.7-fold during mesoderm induction. Following single-cell seeding after 3 days of mesoderm induction, from day 5 to day 11, no expansion was observed, possibly due to the seeded single cells forming hemogenic endothelial cell aggregates. From day 11 onward, cell expansion was noted, with exponential increase in cell numbers observed during the erythroid expansion stage (day 18 onward). All three spinner cultures yielded hemoglobinized erythroid cells with an average cumulative fold expansion of 58.6 ± 8.1-fold on day 27 of differentiation (Figure 3H). With complete medium replenishment during the erythroblast expansion stage, we were able to achieve mean peak cell densities of 1.47 × 107 ± 2.04 × 106 cells/mL with 91.4 ± 2.4% viability (Figure 3I), generating an average of 7.35 × 108 cells with a total medium usage of 575 mL from start to end of differentiation. Metabolic profiles from Table 2 suggest that at cell densities above 1 × 107 cells/mL, lactate levels could reach inhibitory levels >15 mM or 1.36 g/L (Bayley et al., 2018; Hassell et al., 1991) within 1.5 days without medium replenishment. However, glucose consumption was found to be non-limiting at these cell densities.
Figure 3.
Scale-Up of Differentiation of X13 hiPSC in 125 mL Spinner Flasks
(A) Schematic of suspension culture continuous agitation differentiation process from hiPSC to erythroblast stage in 125 mL spinner flasks. Corresponding images of cell aggregates/cells during differentiation are shown below.
(B and C) (B) Concentration of viable cells during pluripotent expansion stage. (C) Flow cytometry evaluation of pluripotency markers (Oct-4, Tra1-60, SSEA-4), mesoderm/primitive streak marker (T-bra), and hematopoietic-fated mesoderm marker (KDR+) (representative experiment shown).
(D) RT-PCR quantified fold expression of early markers (GAPDH normalized) involved in hematopoietic induction at day 3 of differentiation.
(E) Time course of marker expression for hemogenic endothelial cells (CD34+ CD144+), hematopoietic progenitor markers (CD34+ CD43+; CD34+ CD45+), and pan-hematopoietic markers (CD34− CD43+; CD34− CD45+) during the course of differentiation.
(F) RT-PCR quantified fold expression of markers upregulated during erythropoiesis at day 25 of differentiation relative to undifferentiated cells.
(G) Time course of marker expression (determined by FACS) during the differentiation process from day 14 to 28. Data in (B, D, E, F, G) represent the mean ± SD with at least three independent replicates. Representative experiment shown.
(H) Cumulative fold expansion of cells during pluripotent expansion (day −8 to day 0), mesoderm induction (day 0 to day 3), hematopoietic induction (day 5 to day 14), and erythroid expansion (day 15 to day 28) stages, in three independent spinner flasks (p > 0.05 indicates three independent spinner flasks are equivalent). Corresponding hemoglobinized cell pellets at day 28 of differentiation are shown.
(I) Concentration of viable cells and corresponding viability in three independent spinner flask experiments during differentiation are shown. Black arrows indicate complete medium change. Statistical tests were performed on the pooled data from at least three independent replicates.
Table 1.
Expression Profile of X13 hiPSC-Derived Erythroblasts from Spinner Culture Differentiation
| Expression Profile | Marker | Spinner 1 | Spinner 2 | Spinner 3 | Mean ± SD |
|---|---|---|---|---|---|
| Erythroid | CD235a | 63.5 | 73.5 | 68.1 | 68.4 ± 5.0 |
| Early erythroid | CD235a+ CD71+ | 44.3 | 58.5 | 51.4 | 51.4 ± 7.1 |
| Late erythroid | CD235a+ CD71− | 19.2 | 15 | 16.7 | 16.9 ± 2.1 |
| Thrombospondin receptor | CD36 | 16.8 | 29.4 | 17.5 | 21.2 ± 7.1 |
| Hematopoietic progenitor | CD45 | 13.1 | 7.6 | 10.5 | 10.4 ± 2.8 |
| Hematopoietic progenitor | CD43 | 40.7 | 34.3 | 37.8 | 37.6 ± 3.3 |
| Hematopoietic stem cell | CXCR4 | 3.74 | 3.21 | 1.84 | 2.9 ± 1.0 |
| Hematopoietic stem cell | CD133 | 0.17 | 0.09 | 0.08 | 0.12 ± 0.05 |
| Myelomonocytic cell | CD15 | 0.47 | 0.58 | 0.51 | 0.5 ± 0.05 |
| Myelomonocytic cell | CD14 | 0.17 | 0.12 | 0.12 | 0.14 ± 0.03 |
| Fetal hemoglobin | HbF | 59.6 | 85.4 | 66.3 | 70.4 ± 13.3 |
| Adult hemoglobin | HbA | 16.3 | 15.2 | 15.3 | 15.6 ± 0.69 |
Table 2.
Peak Cell Density, Glucose Consumption, and Metabolite Production in Deriving Erythroblasts at Day 27 of Differentiation
| Day 27 | Spinner 1 | Spinner 2 | Spinner 3 |
|---|---|---|---|
| Concentration (cells/mL) | 1.7 × 107 ± 5.2 × 105 | 1.32 × 107 ± 5.5 × 105 | 1.38 × 107 ± 5.5 × 105 |
| Total cells/50 mL | 8.5 × 108 ± 2.6 × 107 | 6.6 × 108 ± 2.7 × 107 | 6.9 × 108 ± 2.7 × 107 |
| Total medium used (mL) | 575 | 575 | 575 |
| Lactate production (mg/L/per 24 h/million cells) | 1.76 | 1.73 | 1.81 |
| Ammonia production (mM/per 24 h/million cells) | 0.0016 | 0.0014 | 0.0018 |
| Glucose consumption (mg/L/per 24 h/million cells) | 1.7 | 1.1 | 1.3 |
The aforementioned spinner flask differentiation approach was applied to another hiPSC line, FR202. Figure S4A shows the FR202 hiPSCs expanded and differentiated into the mesoderm lineage on MCs in spinner flasks under continuous agitation. Following hematopoietic induction and erythroid differentiation in 125 mL spinner flasks, cells successfully formed hemoglobinized erythroblasts. Like X13 hiPSCs, expression of the mesoderm marker T-Bra, hematopoietic mesoderm marker KDR+, and hematopoietic markers CD34+ CD43+ and CD34+ CD45+ on cells was noted during the mesoderm and hematopoietic induction stages, respectively (Figure S4B). Erythroid cells differentiated from FR202 hiPSCs were able to reach a cell density between 5.44 × 106 and 9.88 × 106 cells/mL by day 29 of differentiation (Figure S4C), achieving a cumulative fold expansion between 206- and 805-fold (Figure S4D) and a total yield of 3.3–5.9 × 108 erythroid cells in a final volume of 60 mL. Differentiated cells were 50% CD235a+ erythroblasts, which had high expression of fetal hemoglobin (Figure S4E). As with X13 differentiated erythroblasts, oxygen equilibration curves were left-shifted (p50: 10.5) compared with adult RBCs (p50: 16.3) (Figure S4F). We were able to scale up the erythroid differentiation process to 500 mL spinner flasks, generating approximately 1.6 × 109 erythroid cells in 200 mL volumes at densities of 8.2 × 106 ± 5.9 × 105 cells/mL.
Transcriptome Analysis of hiPSC Erythroblasts versus Adult Erythroblasts
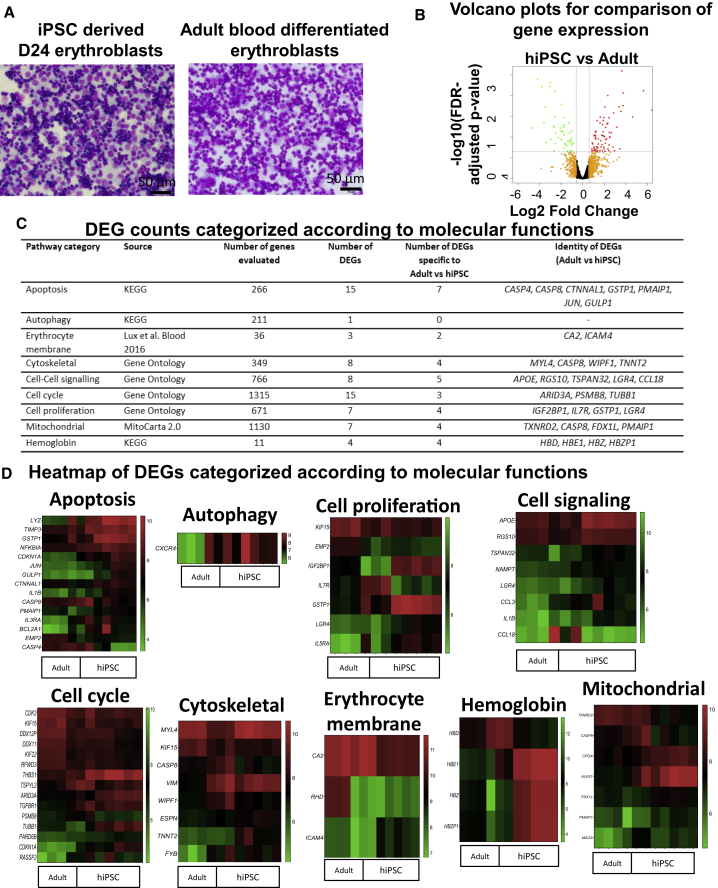
Having achieved scale-up of erythroid differentiation from hiPSCs, it is important to determine whether erythroblasts thus generated are similar in terms of gene expression to counterparts differentiated from adult peripheral blood cells. To this end, we performed microarray-based transcriptome profiling of D24 erythroblasts derived from X13 hiPSCs and the adult peripheral blood CD34+ cells from the same donor (X13). Representative Giemsa staining of X13 iPSC-derived and adult blood-differentiated erythroblasts showed similar morphologies, reflecting mainly erythroblast cells, and a portion of more mature erythroblasts with condensed nuclei and reduced cell size (Figure 4A). Average linkage clustering of expressed genes (n = 10,342) and volcano plots revealed minimal changes in gene expression across two groups (Figure 4B). Comparison of erythroblasts differentiated from X13 hiPSCs with those differentiated from adult CD34 cells identified 119 differentially expressed genes (DEGs) (73 upregulated; 46 downregulated) (Figure 4C). We found that most of them, 64 of 73 upregulated DEGs (Figure S5A) and 40 of 46 downregulated DEGs (Figure S5B), had less than 10-fold change in expression. Pathway mapping of DEGs showed that only 3 of 73 upregulated genes mapped significantly to a single identified pathway (metabolism of xenobiotics by cytochrome p450; p = 1.7 × 10−2) (Figure S5C) and 9 of 46 downregulated genes mapped significantly to three different pathways (tuberculosis, hematopoietic cell lineage, and nitrogen metabolism) (Figure S5D). Gene ontology classification of DEGs revealed that a majority were categorized as being involved in biological regulation (upregulated 17%, downregulated 22%) and metabolic processes (upregulated 22%, downregulated 18%) (Figures S5E and S5F). To ascertain if erythroblasts differentiated from hiPSCs versus adult CD34+ cells differed in certain biological processes, we next focused on genes involved in growth (cell-cell signaling, cell cycle, cell proliferation, mitochondria), maturation (apoptosis, autophagy), and functional characteristics (erythrocyte membrane, cytoskeleton, hemoglobin) (Figure 4D). Comparison of DEGs between hiPSCs and adult erythroid cells revealed significant differences in expression of only a small number of genes involved in apoptosis, cytoskeleton, cell proliferation, cell signaling, mitochondria, cell cycle, and hemoglobin (Figure 4C). qRT-PCR was used to verify changes in gene expression of selected DEGs (14 of 28 DEGs) from the above-mentioned categories, and we observed concordance between qRT-PCR and microarray results for adult versus hiPSC erythroid cells (Figure S5G). Previous transcriptome (Merryweather-Clarke et al., 2016) and proteomic (Wilson et al., 2016) studies highlighted differences in the expression of key erythropoiesis and other genes between hiPSCs and adult erythroblasts. These included genes involved in heme biosynthesis, erythropoiesis, autophagy, cytoskeleton, and biological regulation by intracellular enzymes. Our microarray analysis revealed significant differences (fold change ≥1.5; p < 0.05) in the expression of only two of the previously identified genes (ARID3A and BCL11A) between hiPSCs and adult erythroblasts (Table S1 and Figure S5H), suggesting that erythroblasts generated from hiPSCs using our scale-up method are substantially more similar in gene expression to adult erythroblasts compared with what others have previously accomplished.
Figure 4.
Transcriptome Comparison of Erythroid Cells Differentiated from X13 hiPSC and X13 Donor Adult Peripheral Blood
(A) Representative Giemsa staining of X13 iPSC-derived and adult CD34 blood-differentiated erythroblasts for transcriptome study.
(B) Volcano plots for comparison of gene expression (log2 fold change) between the day 24 erythroid cells differentiated from hiPSC (n = 5 samples) and the adult CD34 cells (n = 3 samples). Red and green dots indicate significantly up- and downregulated genes, respectively (absolute fold change ≥1.5 and false discovery rate [FDR]-adjusted p < 0.05), while orange dots denote the remaining genes that have absolute fold change ≤1.5. The dotted lines indicate the fold change and FDR-adjusted p value thresholds that were applied.
(C) Table showing number of DEGs belonging to the different categories. Genes evaluated for each category were derived from the indicated sources.
(D) Heatmaps (normalized expression signal intensity) of significant DEGs categorized according to molecular functions (apoptosis, autophagy, cytoskeletal, cell proliferation, cell signaling, cell cycle, erythrocyte membrane, mitochondrial, and hemoglobin) for comparison.
Functional Evaluation and Terminal Maturation of Differentiated Erythroblasts
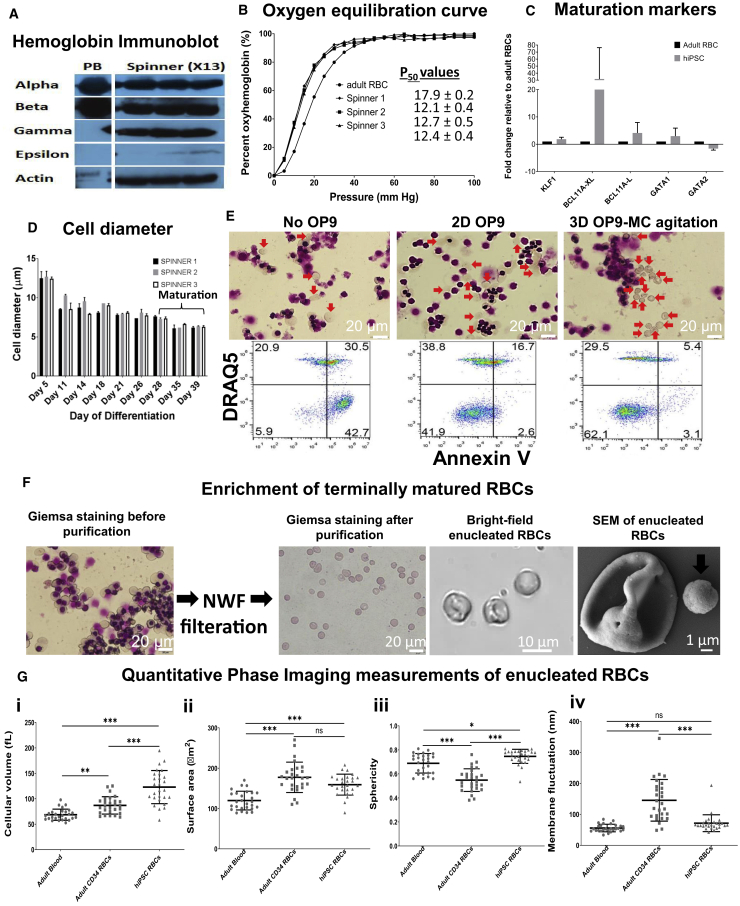
Having demonstrated differentiation of hiPSCs toward high-density cultures of erythroid cells, we next performed functional validation of the erythroblasts. Expression of hemoglobin protein subtypes was confirmed by immunoblots (Figures 5A and S6A). Erythroblasts differentiated from hiPSCs in spinner flasks (spinner X13) expressed alpha, beta, and gamma globins and had low-level expression of the epsilon hemoglobin subtype. On the other hand, adult RBCs from peripheral blood expressed only alpha and beta hemoglobin, consistent with the expression pattern of adult hemoglobin. Consistent with the expression of gamma hemoglobin, which has higher affinity for oxygen, the P50 values for oxygen equilibration curves of erythroblasts differentiated from hiPSCs in spinner flasks (P50: 12.1–12.7) were significantly lower than those of adult RBCs (P50: 17.9) (Figure 5B). A critical process in erythroid development is the terminal maturation of erythroblasts into enucleated erythrocytes. Using qRT-PCR we first evaluated the expression of genes involved in hemoglobin switching (HbF to HbA) and the terminal maturation process. Following terminal maturation, KLF1 (1.8-fold), a key transcription factor necessary for HbF to HbA switching, and BCL11A (variants BCL11A-L, 4.1-fold, and BCL11A-XL, 31.7-fold), another transcription factor implicated in repression of HbF (Trakarnsanga et al., 2014), were found to be expressed at significantly higher levels in hiPSC-derived erythroid cells compared with RBCs differentiated from adult CD34 cells (adult RBC) (Figure 5C). GATA-1, a master regular of erythropoiesis, was 2.9-fold higher, while GATA-2 was 1.6-fold lower compared with adult RBCs, consistent with the expected decline in GATA-2 levels during the onset of terminal maturation (Leonard et al., 1993).
Figure 5.
Terminal Maturation of X13 hiPSC-Derived Erythroblasts from Spinner Differentiation
(A) Cell lysates from peripheral blood (PB) and X13 hiPSC-derived erythroblasts from spinner differentiation (day 27 post differentiation) were immunoblotted with antibodies specific to alpha, beta, gamma, and epsilon human hemoglobin subtypes and the housekeeping control human β-actin. White lines demarcate regions of gel images that were merged.
(B) Oxygen equilibrium curves (percentage oxyhemoglobin versus oxygen pressure [mm Hg]) for adult RBCs (•) and erythroblasts differentiated from X13 hiPSCs (day 27 post differentiation) from spinners 1 (▪), 2 (▲), and 3 (▼) are shown.
(C) qRT-PCR quantified fold expression (GAPDH-normalized and relative to adult erythroblasts) of genes involved in erythroid maturation from X13 hiPSC-derived erythroblasts (day 27 post differentiation) terminally matured for 11 days (data represent the mean ± SD of at least three independent replicates). Representative experiment shown.
(D) Change in cell diameter during differentiation in three independent spinner flask experiments. Day 28 erythroblasts were further matured until day 39.
(E) (Top) Giemsa staining and (bottom) flow cytometry evaluation for non-apoptotic (Annexin V−) enucleated RBCs (DRAQ5−) of X13 hiPSC erythroblasts after 21 days of terminal maturation without OP9 co-culture (No OP9), with monolayer OP9 co-culture (2D OP9), and with OP9-MC co-culture under agitation (3D OP9-MC). Red arrows indicate enucleated erythroid cells. Representative data from n = 3 independent experiments.
(F) Giemsa staining of terminally matured erythroblasts before and after processing through NWF filters is shown. Scale bar, 20 μm. Bright-field images highlighting enucleated RBCs with biconcave appearance and SEM image of a single enucleated RBC are shown. Scale bar, 1 μm. Arrows indicate the expelled pyrenocyte.
(G) Quantitative phase imaging data (mean ± SD) for (i) cell volume, hiPSC RBCs were significantly greater compared with adult CD34 and adult blood; (ii) surface area, hiPSC RBCs were significantly higher compared with adult blood; (iii) sphericity, hiPSC RBCs were significantly greater compared with adult CD34 (∗∗∗p < 0.0001) and adult blood (∗p < 0.05); and (iv) refractive index tomography measurements for dynamic membrane fluctuations of RBCs, hiPSC RBCs were significantly lower compared with adult CD34 RBCs (∗∗p < 0.001). RBCs were differentiated from adult CD34 cells (n = 27 cell counts) and hiPSCs (n = 27 cell counts) and adult blood (n = 27 cell counts).
Reduction in cell size was noted during the course of differentiation, with cells having a mean diameter of 12.6 ± 0.14 μm at the start of differentiation and 7.42 ± 0.16 μm at the end of erythroblast expansion (day 28) and finally 6.28 ± 0.07 μm following 11 days of terminal maturation (day 39) (Figure 5D).
To demonstrate the feasibility of scale-up of erythroblasts co-cultured with stromal cell lines, we show that a murine stromal cell line, OP9, can be effectively grown on MCs under agitation. Intriguingly, in the absence of OP9 co-culture, X13 hiPSC-derived erythroblasts (no OP9) succumbed to massive cell death and apoptosis, whereas with OP9 co-culture (as a monolayer or on MC under agitation) efficient enucleation and reduced apoptosis were observed after 21 days of maturation (Figure 5E). In three independent experiments, the enucleation efficiencies (DRAQ5−, Annexin V− population) of X13 hiPSC-differentiated erythroblasts after 21 days of maturation were noted to be low for conditions without OP9 co-culture (no OP9, 6.27%–6.42%) and were much higher in the presence of OP9 co-culture either as a monolayer (2D OP9, 34.1%–42.2%) or as 3D-MC with agitation (3D OP9-MC, 18.1%–59.3%).
Enucleated RBCs could effectively be enriched (Figure 5F) by passing through non-woven fabric (NWF) filters or leukocyte reduction filters (Pall). Figure 5F shows bright-field and scanning electron microscopy (SEM) images of enucleated RBCs having a biconcave appearance. Giemsa staining shows that adult blood-derived RBCs measure less than 10 μm in diameter, whereas in vitro-differentiated RBCs from adult and hiPSCs are slightly larger than 10 μm in diameter (Figure S6B). Quantitative phase imaging and refractive index tomography were used to compare cellular volume, surface area, sphericity, hemoglobin content, hemoglobin concentration, and dynamic membrane fluctuations of purified populations of enucleated RBCs (Figures 5G and S6C). Cellular volume and surface area were found to be significantly greater (p < 0.0001) in both in vitro-differentiated cells (adult CD34 and hiPSC) compared with adult blood. hiPSC-differentiated cells generally had a higher cellular volume and surface area (Figures 5G i and 5G ii), possibly due to a higher proportion of immature reticulocytes compared with adult CD34 and adult blood-derived cells (33.6%, Figure S7A). Adult blood- and adult CD34-differentiated RBCs and hiPSC RBCs differed in their sphericity index (ranging from 0.55 to 0.75), with hiPSC RBCs displaying the least discoid profile (sphericity of 0.75), again possibly due to a greater proportion of reticulocyte cells (Figure 5G iii). Comparison of hemoglobin concentrations showed that adult blood generally had slightly higher levels compared with adult CD34- and hiPSC-differentiated RBCs (Figure S6C i). On the other hand, measurement of mean Hb content showed that hiPSC-differentiated RBCs, due to their larger cellular volume, had significantly higher Hb content compared with adult blood and adult CD34 RBCs (Figure S6C ii). Last, dynamic membrane fluctuations were measured as an indication of membrane deformability of the RBCs. The dynamic membrane fluctuations of in vitro-differentiated RBCs from adult CD34 (145.2 nm) were found to be significantly higher than those of adult blood (55.7 nm) and hiPSC (71.4 nm) (Figure 5G iv). Three-dimensional tomography imaging of adult blood-, adult CD34-, and hiPSC-differentiated RBCs identified cells with discoid appearance (Figure S7B).
Finally, high-resolution electron microscopy imaging of enucleated RBCs derived from in vitro differentiation of hiPSCs and adult CD34 cells revealed that RBCs from both groups had good surface morphology (Figure S7C).
Discussion
hiPSCs have been investigated as a potentially limitless source of cells for generating RBCs (Dorn et al., 2015; Mao et al., 2016; Olivier et al., 2016; Sivalingam et al., 2018). However, to date, scale-up of the differentiation process starting from the hiPSC stage to the EB stage and subsequent hematopoietic and erythroblast stages have not been clearly demonstrated.
Olivier et al. (Olivier et al., 2016) had previously described an EB-based differentiation process that allowed for generating 50,000–200,000 erythroid cells per hiPSC seeded. Although the process allowed for generation of large numbers of erythroid cells, it required the initial differentiation of EB-differentiated cells as a monolayer in tissue culture flasks, thereby making the process restrictive to scale-up.
In this work, by modifying the protocol described by Olivier et al. (Olivier et al., 2016) and combining with our previously reported hiPSC-MC differentiation protocol (Sivalingam et al., 2018), we were able to demonstrate an entirely suspension-culture-based differentiation process performed under continuous agitation. We successfully scaled up the differentiation process, starting from 6 well ULA plates to shaker flasks and finally demonstrating in stirred spinner flasks as a prelude to scaling up to controlled stirred-tank bioreactors. Using the MC-hiPSC aggregates has an added advantage of generating consistent and evenly sized MC-EBs, which have been demonstrated to be scalable in suspension culture platforms (Lam et al., 2016; Sivalingam et al., 2018). Furthermore, in our spinner culture differentiation platform, by keeping the levels of lactate in culture below their inhibitory threshold, we show peak cell densities that are at least 4-fold higher (1.7 × 107 cells/mL) than the 4 × 106 cells/mL that was reported previously (Ying Wang et al., 2016). To generate one transfusion unit of RBCs (1 × 1012 cells) in a minimum operation volume of a 20 L bioreactor, one would ideally have to reach cell densities exceeding 1 × 108 cells/mL. Data suggest that even at erythroid cell concentrations of 1.7 × 107 cells/mL, lactate (inhibitory level >15 mM) and ammonia (inhibitory level >4 mM) can reach inhibitory levels (Bayley et al., 2018) in less than 1.5 days without medium replenishment. As such, further process refinements such as a dilution feeding strategy (Zhang et al., 2017) or perfusion strategy (Allenby et al., 2018) may be necessary in order to replenish depleted cytokines/growth factors/nutrients, as well as preventing the accumulation of culture metabolites from reaching inhibitory levels in order to allow for cell densities exceeding 1 × 108 cells/mL to be achieved.
In performing high-density cultures, it is pertinent to demonstrate that the final cell product is healthy and functional. In this study, high-resolution electron microscopy imaging and quantitative phase imaging provided additional evidence that hiPSC-derived RBCs (in comparison with adult-derived RBCs) do have good membrane morphology and similar dynamic membrane fluctuation profiles (Kim et al., 2018) indicative of their membrane deformability capabilities, which would be important for passing through narrow capillaries in the human body.
Comparison of the gene expression of adult and hiPSC erythroblasts identified <1.2% of expressed genes (119 of 10,342) with expression fold change greater than or equal to 1.5-fold. None of these DEGs mapped significantly to erythroid cell-specific signaling pathways as evaluated by DAVID pathway mapping analysis. Further query of these 119 DEGs against a database of genes directing important molecular functions of erythroid cells (4,755 genes) such as apoptosis, autophagy, erythrocyte membrane, cytoskeleton, cell-cell signaling, cell cycle, cell proliferation, mitochondrial, and hemoglobin biogenesis produced only 33 hits, suggesting a high degree of similarity between hiPSC- and adult-derived erythroblasts. In terms of the erythropoiesis induction-related genes examined in Figure 3F, there were no significant differences between the adult- and the hiPSC-derived erythroblasts (Figure S4G).
A potential issue with the use of hiPSC-derived erythroid cells for transfusion applications would be the lack of reduced adult beta hemoglobin expression relative to adult-derived RBCs. By immunoblot analysis, we show that the hiPSC-differentiated erythroid cells do express both adult and fetal hemoglobin, with little or no embryonic hemoglobin, although the levels of adult beta hemoglobin from hiPSC-erythroid cells are significantly less compared with adult RBCs. Differences observed in hemoglobin expression could lead to altered affinity of the RBCs for oxygen binding and release. With regard to this, it should be pointed out that individuals with hereditary persistence of fetal hemoglobin who have high levels of gamma fetal hemoglobin (up to 30% of total hemoglobin) are asymptomatic and pathologically normal (Christaki et al., 2019). Previous studies have shown that overexpression of KLF1 and BCL11a could bring about hemoglobin switching from fetal to adult subtype (Trakarnsanga et al., 2014). Such strategies could be incorporated into genetically engineered hiPSCs to improve beta hemoglobin expression in the differentiated RBCs.
A major bottleneck for generation of hiPSC-RBCs is the rather low efficiency of enucleation of hiPSC-derived erythroblasts. Previously, transcriptomics and proteomics studies have raised the possibility of poor enucleation of hiPSC-derived erythroid cells due to defects in autophagy and lysosomal-mediated organelle clearance (Merryweather-Clarke et al., 2016) during terminal maturation and even possibly due to defects in cytoskeletal organization and remodeling (Trakarnsanga et al., 2019; Wilson et al., 2016). However, our transcriptomics analysis did not reveal any significant expression differences in some of these implicated genes (VIM, TRIM58, VCPIP1) between hiPSC- and adult-derived erythroid cells, at least at the erythroblast stage. Of genes whose expression differed significantly between adult and hiPSC erythroid cells, only two coincided with a list of 40 genes previously reported (Merryweather-Clarke et al., 2016; Trakarnsanga et al., 2019; Wilson et al., 2016). ARID3A, an erythroid transcription factor involved in cell cycle and transcriptional regulation was found to be significantly upregulated in hiPSC compared with adult erythroid cells. Second, BCL11A, a repressor of HbF, was found to be downregulated 3.5-fold in hiPSC compared with adult erythroid cells under conditions that favor erythroid expansion. This might explain the continued expression of HbF in hiPSC-derived erythroid cells. We found no differences in expression of genes involved in autophagy or cytoskeletal remodeling, as previously suggested (Merryweather-Clarke et al., 2016; Trakarnsanga et al., 2019; Wilson et al., 2016). These differences may partly be due to the different protocols used for differentiating hiPSC erythroid cells. Erythroid cells differentiated from our protocol had evidence of definitive erythropoiesis, such as adult beta hemoglobin expression, which was not demonstrated in the previous publications.
Here, we show that co-culture with stromal cells may help prevent erythroblast cell death and promote enucleation. Previous studies have reiterated this point with the use of OP9 and other stromal cell cultures (Giarratana et al., 2005; Lu et al., 2008; Suzuki et al., 1992). However, all these co-culture studies so far have been performed only in monolayer, making the scalability of this approach challenging. We show that a co-culture system using OP9 stromal cells can potentially be scaled up by culturing them on MCs, allowing the entire process to be performed in suspension culture and paving the way for future scale-up.
In conclusion, we have developed a suspension agitation culture differentiation process for differentiating hiPSCs toward erythroblasts that can be volumetrically scaled up. hiPSC-derived erythroid cells were capable of undergoing enucleation, had expression of adult beta hemoglobin, albeit at reduced levels (in comparison with adult RBCs), were highly similar to adult-derived erythroid cells at the molecular level, and had similar membrane morphologies and dynamic membrane fluctuations/membrane deformability profiles. Further development would be necessary to adapt our differentiation processes into bioreactors for large-scale generation of high-density functional RBCs.
Experimental Procedures
For details of this section, please also refer to the Supplemental Experimental Procedures.
Briefly, all O-neg hiPSC lines were cultured on LN-521-coated MCs (Sivalingam et al., 2018) under static or agitation conditions for 7 days and subjected to hematopoietic mesoderm differentiation for 2 days using a combination of BMP4, Activin A, VEGF-165, B-FGF, and small-molecule CHIR-99021, as detailed in the Supplemental Experimental Procedures. On day 3 of differentiation, single cells derived from MC aggregates following treatment with TrypLE Express and sieving of MCs were subjected to hematopoietic induction using a cocktail of cytokines (based on a protocol adapted from Olivier et al., 2016) for a further 8 days, under agitation. Then, the cells were further differentiated toward the erythroid lineage and expanded by fresh medium replenishment with cytokines and maintenance of cell seeding density at 5 × 105 to 1 × 106 cells/mL, under continuous agitation. Terminal maturation of erythroid cells was performed for a period of 2–3 weeks with or without co-culture of stromal cells, using IMDM supplemented with 10% human AB plasma, Holo-Transferrin, cytokines, and growth factors. The detailed protocol can be found in the Supplemental Experimental Procedures.
Requests for materials should be addressed to the corresponding author.
Data and Code Availability
The authors declare that all data supporting the findings of this study are available within the article and its Supplemental Information files. Microarray data are available at Gene Expression Omnibus under accession no. GSE137917.
Author Contributions
S.K.W.O. was the principal investigator and takes primary responsibility for the article. J.S. performed all experiments, analyzed data, and wrote the manuscript. Y.S., H.Y.C., Z.R.L., H.K.T., and A.T.L.M. contributed toward the laboratory work. A.P.L., L.H.L., and A.H.M.T. performed the microarray study. T.W. performed bioinformatics. H.J.W., N.B.N., and B.M. performed the electron microscopy experiments. J.S., Y.S., B.M., L.R., S.R., Y.H.L., and S.K.W.O. wrote the manuscript. Y.H.L. provided hiPSC lines for the study.
Conflicts of Interests
Dr. Steve Oh has patents on microcarrier technology for stem cell cultivation filed by A∗STAR. He is also a founder of Zenzic Labs and SingCell. The rest of the authors declare no competing interests.
Acknowledgments
This work was supported by a Joint Council Office grant from the Agency for Science, Technology and Research, Singapore (A∗STAR) (1331AFG075), Singapore Immunology Network Core funding, A∗STAR, and NUHS Start-Up funding (NUHSRO/2018/006/SU/01). The authors would like to thank Mickey Koh and team (Health Sciences Authority, Singapore) for useful discussions and Benedict and Dennis (EINST Technology Pte Ltd) for performing quantitative phase imaging using the Tomocube HT-2S system, Electron Microscopy Unit, National University of Singapore, and particularly Professor Ong Wei Yi, Miss Tan Suat Hoon, and Mister Lu Thong Beng. The authors would like to thank Naresh Waran Gnanasegaran for providing a peripheral blood sample for this study.
Published: December 10, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.11.008.
Supporting Citations
The following references appear in the supplemental information: Babicki et al., 2016; Chen et al., 2016; Clincke et al., 2013; Friebel and Meinke, 2006; Hong et al., 2010; Lee et al., 2017; Liu et al., 2013; Livak and Schmittgen, 2001; Lux, 2016; Malleret et al., 2013; Park et al., 2009; Sivalingam et al., 2016; Tan et al., 2014; Tao et al., 2011; Yu et al., 2007; Zhang et al., 2015.
Supplemental Information
References
- Allenby M.C., Tahlawi A., Morais J.C.F., Li K., Panoskaltsis N., Mantalaris A. Ceramic hollow fibre constructs for continuous perfusion and cell harvest from 3D hematopoietic organoids. Stem Cells Int. 2018;2018:6230214. doi: 10.1155/2018/6230214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter H.J., Stramer S.L., Dodd R.Y. Emerging infectious diseases that threaten the blood supply. Semin. Hematol. 2007;44:32–41. doi: 10.1053/j.seminhematol.2006.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelos M.G., Abrahante J.E., Blum R.H., Kaufman D.S. Single cell resolution of human hematoendothelial cells defines transcriptional signatures of hemogenic endothelium. Stem Cells. 2018;36:206–217. doi: 10.1002/stem.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babicki S., Arndt D., Marcu A., Liang Y., Grant J.R., Maciejewski A., Wishart D.S. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44:W147–W153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayley R., Ahmed F., Glen K., McCall M., Stacey A., Thomas R. The productivity limit of manufacturing blood cell therapy in scalable stirred bioreactors. J. Tissue Eng. Regener. Med. 2018;12:e368–e378. doi: 10.1002/term.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.Y., Tan H.K., Loh Y.H. Derivation of transgene-free induced pluripotent stem cells from a single drop of blood. Curr. Protoc. Stem Cell Biol. 2016;38:4A 9 1–4A 9 10. doi: 10.1002/cpsc.12. [DOI] [PubMed] [Google Scholar]
- Christaki E.E., Politou M., Antonelou M., Athanasopoulos A., Simantirakis E., Seghatchian J., Vassilopoulos G. Ex vivo generation of transfusable red blood cells from various stem cell sources: a concise revisit of where we are now. Transfus. Apher. Sci. 2019;58:108–112. doi: 10.1016/j.transci.2018.12.015. [DOI] [PubMed] [Google Scholar]
- Clincke M.F., Molleryd C., Zhang Y., Lindskog E., Walsh K., Chotteau V. Very high density of CHO cells in perfusion by ATF or TFF in WAVE bioreactor. Part I. Effect of the cell density on the process. Biotechnol. Prog. 2013;29:754–767. doi: 10.1002/btpr.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn I., Klich K., Arauzo-Bravo M.J., Radstaak M., Santourlidis S., Ghanjati F., Radke T.F., Psathaki O.E., Hargus G., Kramer J. Erythroid differentiation of human induced pluripotent stem cells is independent of donor cell type of origin. Haematologica. 2015;100:32–41. doi: 10.3324/haematol.2014.108068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebel M., Meinke M. Model function to calculate the refractive index of native hemoglobin in the wavelength range of 250-1100 nm dependent on concentration. Appl. Opt. 2006;45:2838–2842. doi: 10.1364/ao.45.002838. [DOI] [PubMed] [Google Scholar]
- Giarratana M.C., Kobari L., Lapillonne H., Chalmers D., Kiger L., Cynober T., Marden M.C., Wajcman H., Douay L. Ex vivo generation of fully mature human red blood cells from hematopoietic stem cells. Nat. Biotechnol. 2005;23:69–74. doi: 10.1038/nbt1047. [DOI] [PubMed] [Google Scholar]
- Hassell T., Gleave S., Butler M. Growth inhibition in animal cell culture. The effect of lactate and ammonia. Appl. Biochem. Biotechnol. 1991;30:29–41. doi: 10.1007/BF02922022. [DOI] [PubMed] [Google Scholar]
- Hirani R., Wong J., Diaz P., Mondy P., Hogan C., Dennington P.M., Pink J., Irving D.O. A national review of the clinical use of group O D- red blood cell units. Transfusion. 2017;57:1254–1261. doi: 10.1111/trf.14012. [DOI] [PubMed] [Google Scholar]
- Hirose S., Takayama N., Nakamura S., Nagasawa K., Ochi K., Hirata S., Yamazaki S., Yamaguchi T., Otsu M., Sano S. Immortalization of erythroblasts by c-MYC and BCL-XL enables large-scale erythrocyte production from human pluripotent stem cells. Stem Cell Reports. 2013;1:499–508. doi: 10.1016/j.stemcr.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.H., Werbowetski-Ogilvie T., Ramos-Mejia V., Lee J.B., Bhatia M. Multiparameter comparisons of embryoid body differentiation toward human stem cell applications. Stem Cell Res. 2010;5:120–130. doi: 10.1016/j.scr.2010.04.007. [DOI] [PubMed] [Google Scholar]
- Kim G., Lee M., Youn S., Lee E., Kwon D., Shin J., Lee S., Lee Y.S., Park Y. Measurements of three-dimensional refractive index tomography and membrane deformability of live erythrocytes from Pelophylax nigromaculatus. Sci. Rep. 2018;8:9192. doi: 10.1038/s41598-018-25886-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita R., Suda N., Sudo K., Miharada K., Hiroyama T., Miyoshi H., Tani K., Nakamura Y. Establishment of immortalized human erythroid progenitor cell lines able to produce enucleated red blood cells. PLoS One. 2013;8:e59890. doi: 10.1371/journal.pone.0059890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam A.T., Chen A.K., Ting S.Q., Reuveny S., Oh S.K. Integrated processes for expansion and differentiation of human pluripotent stem cells in suspended microcarriers cultures. Biochem. Biophys. Res. Commun. 2016;473:764–768. doi: 10.1016/j.bbrc.2015.09.079. [DOI] [PubMed] [Google Scholar]
- Lee S., Park H., Kim K., Sohn Y., Jang S., Park Y. Refractive index tomograms and dynamic membrane fluctuations of red blood cells from patients with diabetes mellitus. Sci. Rep. 2017;7:1039. doi: 10.1038/s41598-017-01036-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard M., Brice M., Engel J.D., Papayannopoulou T. Dynamics of GATA transcription factor expression during erythroid differentiation. Blood. 1993;82:1071–1079. [PubMed] [Google Scholar]
- Liu F., Bhang S.H., Arentson E., Sawada A., Kim C.K., Kang I., Yu J., Sakurai N., Kim S.H., Yoo J.J. Enhanced hemangioblast generation and improved vascular repair and regeneration from embryonic stem cells by defined transcription factors. Stem Cell Reports. 2013;1:166–182. doi: 10.1016/j.stemcr.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu S.J., Feng Q., Park J.S., Vida L., Lee B.S., Strausbauch M., Wettstein P.J., Honig G.R., Lanza R. Biologic properties and enucleation of red blood cells from human embryonic stem cells. Blood. 2008;112:4475–4484. doi: 10.1182/blood-2008-05-157198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux S.E.t. Anatomy of the red cell membrane skeleton: unanswered questions. Blood. 2016;127:187–199. doi: 10.1182/blood-2014-12-512772. [DOI] [PubMed] [Google Scholar]
- Malleret B., Xu F., Mohandas N., Suwanarusk R., Chu C., Leite J.A., Low K., Turner C., Sriprawat K., Zhang R. Significant biochemical, biophysical and metabolic diversity in circulating human cord blood reticulocytes. PLoS One. 2013;8:e76062. doi: 10.1371/journal.pone.0076062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao B., Huang S., Lu X., Sun W., Zhou Y., Pan X., Yu J., Lai M., Chen B., Zhou Q. Early development of definitive erythroblasts from human pluripotent stem cells defined by expression of glycophorin A/CD235a, CD34, and CD36. Stem Cell Reports. 2016;7:869–883. doi: 10.1016/j.stemcr.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merryweather-Clarke A.T., Tipping A.J., Lamikanra A.A., Fa R., Abu-Jamous B., Tsang H.P., Carpenter L., Robson K.J., Nandi A.K., Roberts D.J. Distinct gene expression program dynamics during erythropoiesis from human induced pluripotent stem cells compared with adult and cord blood progenitors. BMC Genomics. 2016;17:817. doi: 10.1186/s12864-016-3134-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier E.N., Marenah L., McCahill A., Condie A., Cowan S., Mountford J.C. High-efficiency serum-free feeder-free erythroid differentiation of human pluripotent stem cells using small molecules. Stem Cells Transl. Med. 2016;5:1394–1405. doi: 10.5966/sctm.2015-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paes B., Moco P.D., Pereira C.G., Porto G.S., de Sousa Russo E.M., Reis L.C.J., Covas D.T., Picanco-Castro V. Ten years of iPSC: clinical potential and advances in vitro hematopoietic differentiation. Cell Biol. Toxicol. 2017;33:233–250. doi: 10.1007/s10565-016-9377-2. [DOI] [PubMed] [Google Scholar]
- Park Y., Yamauchi T., Choi W., Dasari R., Feld M.S. Spectroscopic phase microscopy for quantifying hemoglobin concentrations in intact red blood cells. Opt. Lett. 2009;34:3668–3670. doi: 10.1364/OL.34.003668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrard T., Bardiaux L., Krause C., Kobari L., Lapillonne H., Andreu G., Douay L. Banking of pluripotent adult stem cells as an unlimited source for red blood cell production: potential applications for alloimmunized patients and rare blood challenges. Transfus. Med. Rev. 2011;25:206–216. doi: 10.1016/j.tmrv.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Sivalingam J., Chen H.Y., Yang B.X., Lim Z.R., Lam A.T.L., Woo T.L., Chen A.K., Reuveny S., Loh Y.H., Oh S.K. Improved erythroid differentiation of multiple human pluripotent stem cell lines in microcarrier culture by modulation of Wnt/beta-Catenin signaling. Haematologica. 2018;103:e279–e283. doi: 10.3324/haematol.2017.180919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivalingam J., Lam A.T., Chen H.Y., Yang B.X., Chen A.K., Reuveny S., Loh Y.H., Oh S.K. Superior red blood cell generation from human pluripotent stem cells through a novel microcarrier-based embryoid body platform. Tissue Eng. C Methods. 2016;22:765–780. doi: 10.1089/ten.TEC.2015.0579. [DOI] [PubMed] [Google Scholar]
- Sturgeon C.M., Ditadi A., Awong G., Kennedy M., Keller G. Wnt signaling controls the specification of definitive and primitive hematopoiesis from human pluripotent stem cells. Nat. Biotechnol. 2014;32:554–561. doi: 10.1038/nbt.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki J., Fujita J., Taniguchi S., Sugimoto K., Mori K.J. Characterization of murine hemopoietic-supportive (MS-1 and MS-5) and non-supportive (MS-K) cell lines. Leukemia. 1992;6:452–458. [PubMed] [Google Scholar]
- Tan H.K., Toh C.X., Ma D., Yang B., Liu T.M., Lu J., Wong C.W., Tan T.K., Li H., Syn C. Human finger-prick induced pluripotent stem cells facilitate the development of stem cell banking. Stem Cells Transl. Med. 2014;3:586–598. doi: 10.5966/sctm.2013-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z.Y., Xia H., Cao J., Gao Q. Development and evaluation of a prototype non-woven fabric filter for purification of malaria-infected blood. Malar. J. 2011;10:251. doi: 10.1186/1475-2875-10-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trakarnsanga K., Ferguson D., Daniels D.E., Griffiths R.E., Wilson M.C., Mordue K.E., Gartner A., Andrienko T.N., Calvert A., Condie A. Vimentin expression is retained in erythroid cells differentiated from human iPSC and ESC and indicates dysregulation in these cells early in differentiation. Stem Cel. Res. Ther. 2019;10:130. doi: 10.1186/s13287-019-1231-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trakarnsanga K., Wilson M.C., Lau W., Singleton B.K., Parsons S.F., Sakuntanaga P., Kurita R., Nakamura Y., Anstee D.J., Frayne J. Induction of adult levels of beta-globin in human erythroid cells that intrinsically express embryonic or fetal globin by transduction with KLF1 and BCL11A-XL. Haematologica. 2014;99:1677–1685. doi: 10.3324/haematol.2014.110155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trakarnsanga K., Griffiths R.E., Wilson M.C., Blair A., Satchwell T.J., Meinders M. An immortalized adult human erythroid line facilitates sustainable and scalable generation of functional red cells. Nat. Commun. 2017;8:14750. doi: 10.1038/ncomms14750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M.C., Trakarnsanga K., Heesom K.J., Cogan N., Green C., Toye A.M., Parsons S.F., Anstee D.J., Frayne J. Comparison of the proteome of adult and cord erythroid cells, and changes in the proteome following reticulocyte maturation. Mol. Cell Proteomics. 2016;15:1938–1946. doi: 10.1074/mcp.M115.057315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Wang Y.G., He C., Ye Z., Gerecht S., Cheng L. Scalable production of human erytrhocytes from induced pluripotent stem cells. BioRxiv. 2016 doi: 10.1101/050021. [DOI] [Google Scholar]
- Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Stobbe P., Silvander C.O., Chotteau V. Very high cell density perfusion of CHO cells anchored in a non-woven matrix-based bioreactor. J. Biotechnol. 2015;213:28–41. doi: 10.1016/j.jbiotec.2015.07.006. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wang C., Wang L., Shen B., Guan X., Tian J., Ren Z., Ding X., Ma Y., Dai W. Large-scale ex vivo generation of human red blood cells from cord blood CD34(+) cells. Stem Cells Transl. Med. 2017;6:1698–1709. doi: 10.1002/sctm.17-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the article and its Supplemental Information files. Microarray data are available at Gene Expression Omnibus under accession no. GSE137917.