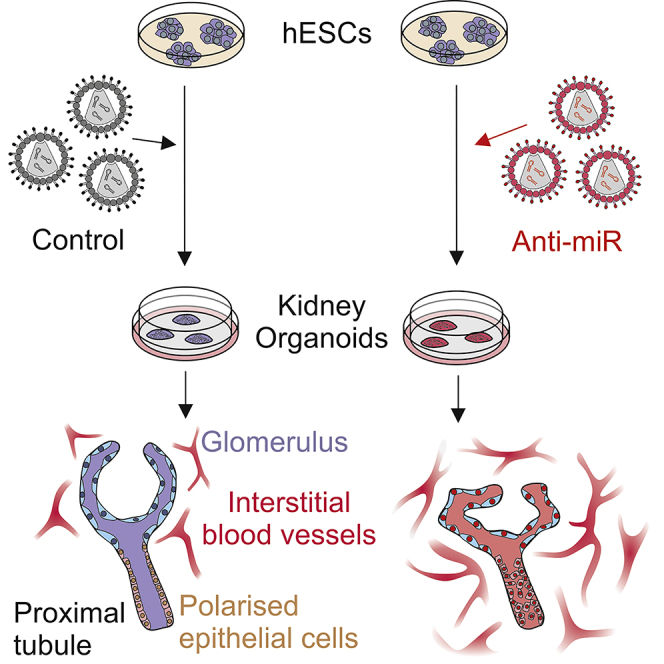
Summary
MicroRNAs (miRNAs) are gene expression regulators and they have been implicated in acquired kidney diseases and in renal development, mostly through animal studies. We hypothesized that the miR-199a/214 cluster regulates human kidney development. We detected its expression in human embryonic kidneys by in situ hybridization. To mechanistically study the cluster, we used 2D and 3D human embryonic stem cell (hESC) models of kidney development. After confirming expression in each model, we inhibited the miRNAs using lentivirally transduced miRNA sponges. This reduced the WT1+ metanephric mesenchyme domain in 2D cultures. Sponges did not prevent the formation of 3D kidney-like organoids. These organoids, however, contained dysmorphic glomeruli, downregulated WT1, aberrant proximal tubules, and increased interstitial capillaries. Thus, the miR-199a/214 cluster fine-tunes differentiation of both metanephric mesenchymal-derived nephrons and kidney endothelia. While clinical implications require further study, it is noted that patients with heterozygous deletions encompassing this miRNA locus can have malformed kidneys.
Keywords: kidney, development, human pluripotent stem cells, microRNA, organoids, miR-199a, miR-214
Graphical Abstract
Highlights
-
•
miR-199a/214 is expressed in the developing human kidney
-
•
Its function can be modeled using hESC-derived kidney organoids
-
•
Inhibiting these miRNAs results in dysmorphic glomeruli and proximal tubules
-
•
Their inhibition also leads to a denser vascular network between tubules
Bantounas and colleagues studied the role of the miR-199a/214 cluster in hESC-kidney development because they found it was expressed in native embryonic human kidneys. Using hESC-derived 3D kidney organoids, they found that blocking the miRNAs' function resulted in aberrations in glomeruli and proximal tubules and increased vascular density between tubules. Thus, the cluster fine-tunes kidney differentiation.
Introduction
MicroRNAs (miRNAs) are small (20–25 nucleotides) RNAs that regulate gene expression by binding to the 3′ UTRs of their target gene mRNA and inhibiting their translation and/or causing their degradation (Bartel, 2009; Filipowicz et al., 2008). Diverse studies, mostly in animal models, are implicating miRNAs in acquired kidney diseases and in renal development (Jones et al., 2018; Nakagawa et al., 2015; Sankrityayan et al., 2019; Shaffi et al., 2018; Trionfini et al., 2015; Zhao et al., 2019). MiR-199a and miR-214 are transcribed as part of the long non-coding RNA dynamin 3 opposite strand (DNM3OS), from which they are excised to yield their mature forms (Lee et al., 2009) and they have been found to be involved in the development and disease of various tissues. As examples, miR-199a is involved in osteogenesis, chondrogenesis, and adipogenesis, and in the stress response to hypoxia in the heart, lung, and brain (Gu and Chan, 2012), while miR-214 participates in muscle, bone, pancreatic, and nervous system development (Chen et al., 2010; Flynt et al., 2007; Joglekar et al., 2007; Shi et al., 2013; Sun et al., 2018; Watanabe et al., 2008) and protects against heart ischemia (Aurora et al., 2012). More recently, expression and possible roles for miRNAs have begun to be investigated in the kidney. Indeed, a number of groups have recently implicated miRNAs in genetic diseases of the kidney (Trionfini et al., 2015). The miR-17-92 cluster has been shown to be important in the regulation of nephron progenitors in mice. miR-19b in the cluster suppresses the cystic fibrosis transmembrane conductance regulator (Cftr) gene, in murine nephron progenitors, and misregulation of this gene causes changes in proliferation of the progenitors (Marrone et al., 2014; Phua et al., 2019). Knockout studies revealed a requirement for miR-210 through its regulation of Wnt signaling in the development of normal nephron numbers in male mice (Hemker et al., 2020). An anti-inflammatory/anti-cystic role for miR-214 in autosomal dominant polycystic kidney disease has also been identified (Lakhia et al., 2020). MiR-214 also promotes kidney fibrosis in experimental animals (Denby et al., 2014), which may be linked to its ability to promote epithelial-to-mesenchymal transition (EMT) in tubular epithelial cells (Liu et al., 2018c). MiR-199a expression is downregulated in human renal cell carcinoma (He et al., 2015; Liu et al., 2018b) and deregulated in rodent genetic models of polycystic kidney disease (Dweep et al., 2013), in lupus nephritis (Ye et al., 2018), and in ischemia/reperfusion injury (Godwin et al., 2010).
While the role and mechanisms of action of miR-199a/214 in the kidney have begun to be elucidated, functional studies that focus on human kidney development have yet to be undertaken. We hypothesized that the miR-199a/214 cluster regulates human kidney development and elected to use human embryonic stem cell (hESC)-based models (Bantounas et al., 2018; Takasato et al., 2014, 2016) to investigate its function. Following confirmation of miR-199a/214 expression in histological sections of human embryonic kidneys and in hESC-derived kidney tissues, we differentiated hESCs in 2D and 3D in the presence or absence of competitive inhibitors of the miRNAs (miRNA “sponges”), which bind to and mask miRNAs thus preventing the latter from accessing their targets (Ebert et al., 2007; Ebert and Sharp, 2010). We found that inhibiting this miRNA cluster resulted in aberrations of the metanephric mesenchyme (MM) domain in 2D cultures. Sponges did not prevent the formation of 3D kidney-like organoids. Histological and molecular analyses of these organoids, however, revealed glomerular dysmorphology and an increase of the capillary network between tubules. Thus, the miR-199a/214 cluster fine-tunes the differentiation of the human kidney.
Results
Expression of the miR-199a/214 Cluster in the Developing Human Kidney
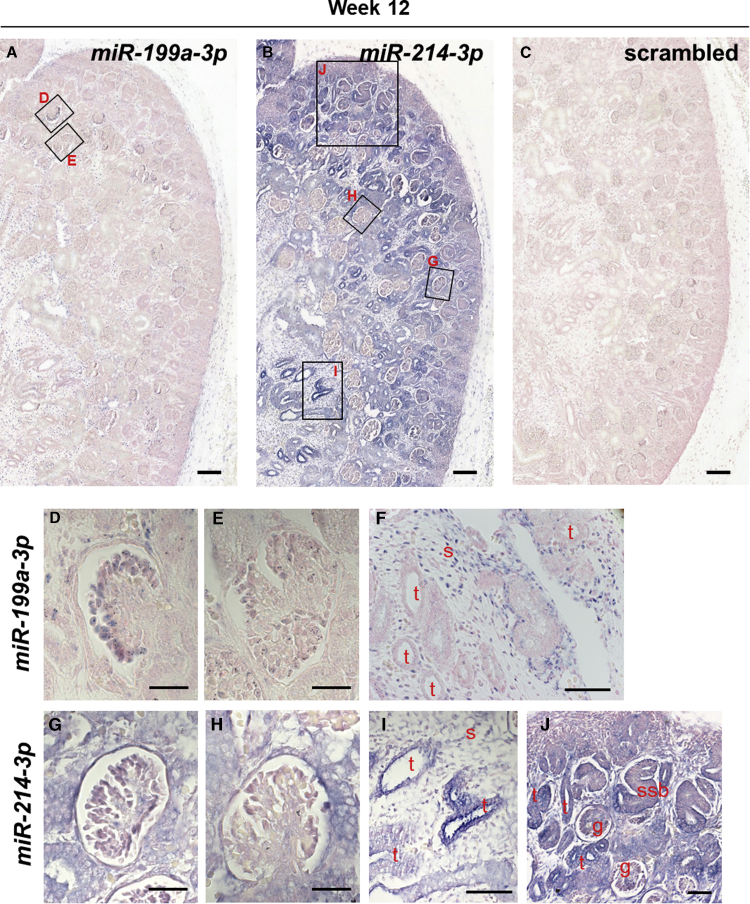
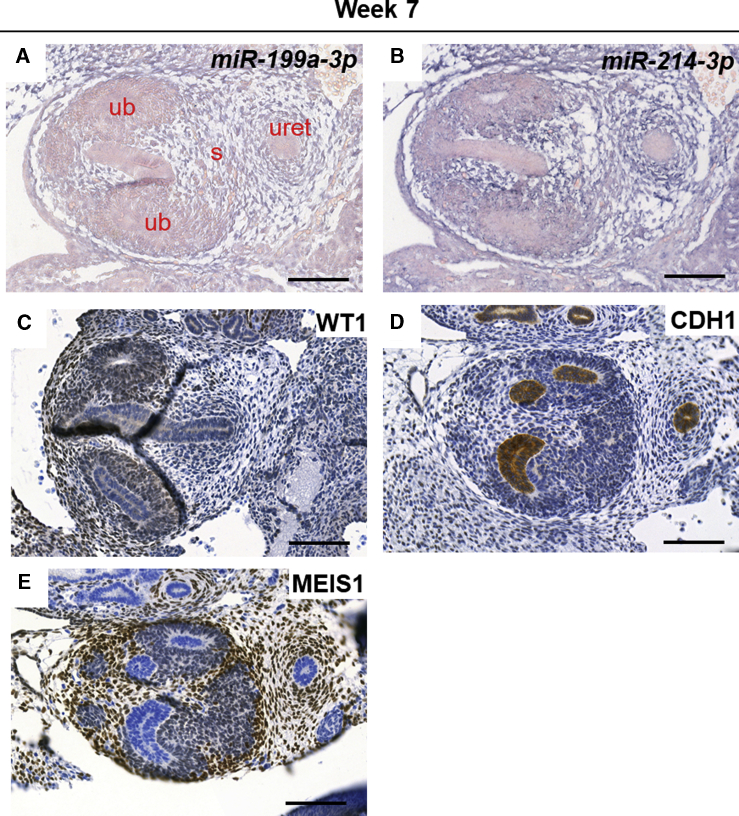
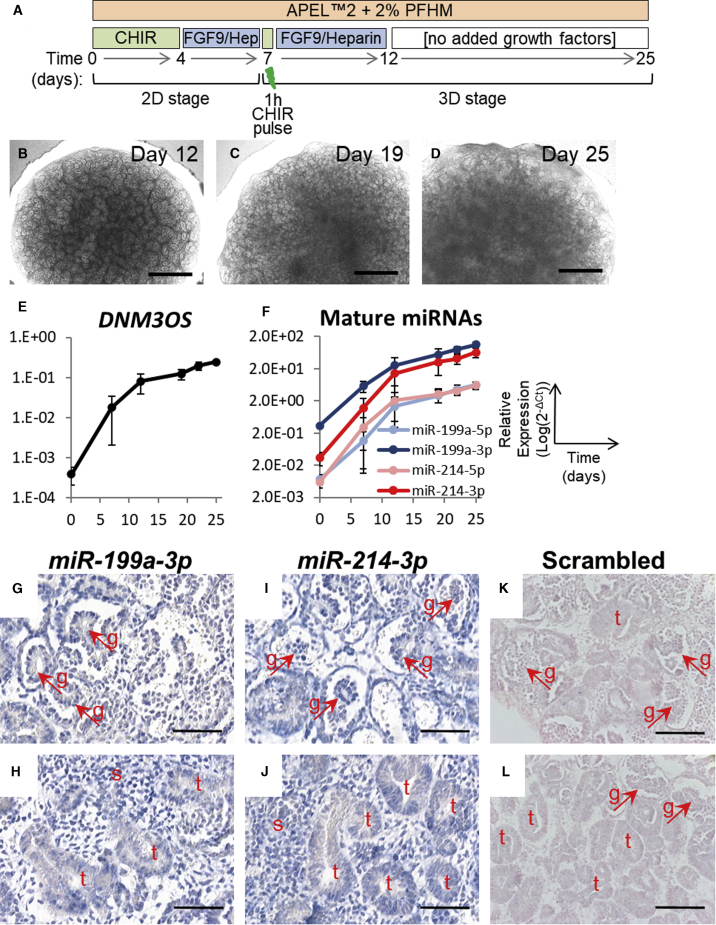
To determine expression of the miR-199a/214 cluster in the developing kidney, we performed in situ hybridization (ISH) experiments, using digoxigenin-labeled locked nucleic acid (LNA) probes, firstly on 12-week gestation human embryonic kidney sections (Figure 1). We focused on miR-199a-3p and miR-214-3p because these are the dominant mature strands stemming from their corresponding pre-miRNA hairpins in most tissues, according to the miRBase database (Kozomara and Griffiths-Jones, 2011). MiR-214-3p expression appeared more widespread than that of miR-199a-3p (Figures 1A–1C). On higher-power images, both were expressed in less mature more superficial glomerular tufts, including in presumptive podocytes (Figures 1D and 1G), but they appeared downregulated in the most mature (i.e., deepest) glomeruli (Figures 1E and 1H). In addition, both miRNAs were expressed in the interstitium between tubules (Figures 1F and 1I). MiR-214-3p but not miR-199a-3p was expressed in cortical tubules (Figures 1F and 1I) and in the nephrogenic zone, where it was prominent in S-shaped bodies, nephron precursors (Figure 1J). We also examined expression of the miRNAs in week 7 human embryonic sections (Figures 2A and 2B) and stained for WT1, CDH1, and MEIS1 to identify MM, tubules, and interstitium, respectively (Figures 2C–2E). We detected both miR-214-3p and miR-199a-3p in the interstitium within the rudimentary metanephros and around the ureter stalk at 7 weeks gestation, in a similar location to MEIS1 immunostaining.
Figure 1.
In Situ Hybridization for the miR-199a/214 Cluster in Human Embryonic Kidney at Week 12 of Gestation
(A–C) Low-magnification images showing part of the cortical region of week 12 human embryonic kidney sections probed with a miR-199a-3p, miR-214-3p, or scrambled control probe. Squares show the positions of the high-magnification images in (D, E, and G–J).
(D–F) MiR-199a-3p was present in outer cortical, maturing (D), but not in more mature, deeper layer (E) glomeruli, and was also present in interstitial cells (F).
(G–I) MiR-214-3p was also present in early maturing (G) but not more mature (H) glomeruli and was additionally expressed in interstitial cells and some tubules (I).
(J) Higher-magnification view of the nephrogenic zone showing strong miR-214-3p expression. Scale bars, 100 μm (A–C); 50 μm (D, E, G, H); 200 μm (F, I); and 40 μm (J). t, tubule; s, stroma; g, glomerulus; ssb, S-shaped body. Stained areas appear purple, over Nuclear Fast Red (pink) counterstain.
Figure 2.
Expression of Members of the miR-199a/214 Cluster in Human Embryonic Kidney at Week 7 of Gestation
(A and B) In situ hybridization with LNA probes against miR-199a-3p and miR-214-3p, revealing expression of both miRNAs mainly in the stroma. Stained areas appear purple, over Nuclear Fast Red (pink) counterstain.
(C–E) Sections of week 7 human embryonic kidneys were stained for different lineage markers (WT1 for MM; CDH1 for tubules; MEIS1 for stroma). Scale bars, 100 μm. s, stroma; ub, ureteric bud tip; uret, ureter. Stained areas are brown, over hematoxylin (blue) counterstain.
Expression of the miR-199a/214 Cluster in hESC-Kidney Development
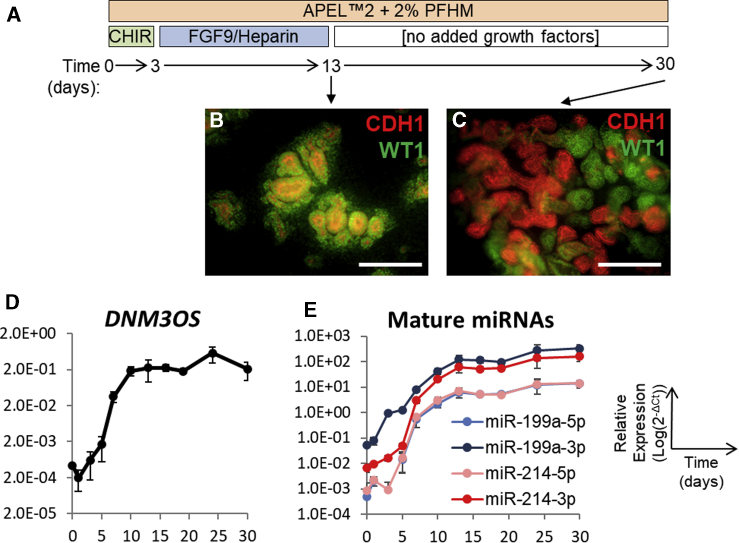
Having confirmed expression of miR-199a and miR-214 in vivo, we wanted to investigate their potential roles in the developing kidney. To this end, we first used a previously established method of hESC differentiation to kidney in 2D culture, capable of modeling early events in kidney development (Bantounas et al., 2018; Takasato et al., 2014) (Figure 3A). In this model, stem cells are first differentiated, through intermediate mesoderm, to precursor cells forming multilayered structures emanating from the cell monolayer. Cells within these immature structures initially express both epithelial (CDH1 [E-cadherin]) and MM (WT1 [Wilms tumor 1]) markers (Figure 3B). As the cells mature, these structures are divided into mutually exclusive WT1+ and CDH1+ domains (Figure 3C). We traced the expression of the primary transcript (DNM3OS) as well as individual mature miRNAs of the cluster by qPCR in three different hESC lines: MAN13, MAN11, and HUES1 during kidney differentiation (Figures 3D, 3E and S1). In each line, we observed a similar pattern of expression, with both the primary and mature transcripts being expressed simultaneously, starting between days 7 and 10 of differentiation and increasing thereafter, reaching a plateau in HUES1 and MAN13 during the second half of the differentiation protocol. MiR-199a-3p and miR-214-3p were the dominant mature species for miR-199a and miR-214, respectively, while expression of the 5p strands was at least 10-fold lower in both cases.
Figure 3.
Expression of the miR-199a/214 Cluster in MAN13 hESC Cultures Differentiating to Kidney in 2D
(A) Schematic representation of the 2D differentiation protocol.
(B and C) 2D cultures immunostained for CDH1 and WT1 at day 13 (B) and day 30 (C) of the differentiation protocol, showing the eventual separation of CDH1+ (epithelial cells) and WT1+ (MM) to discrete areas of the developing structures. Scale bars, 120 μm.
(D and E) Time course of the primary transcript (D) and mature miRNAs (E) of the miR-199a/214 cluster during 2D differentiation, assessed by qPCR (mean ± SEM; n = 3 independent differentiation experiments).
See also Figure S1.
Inhibition of miR-199a/214 Decreases the Amount of MM and Alters Epithelial Morphology in Differentiating hESC Cultures
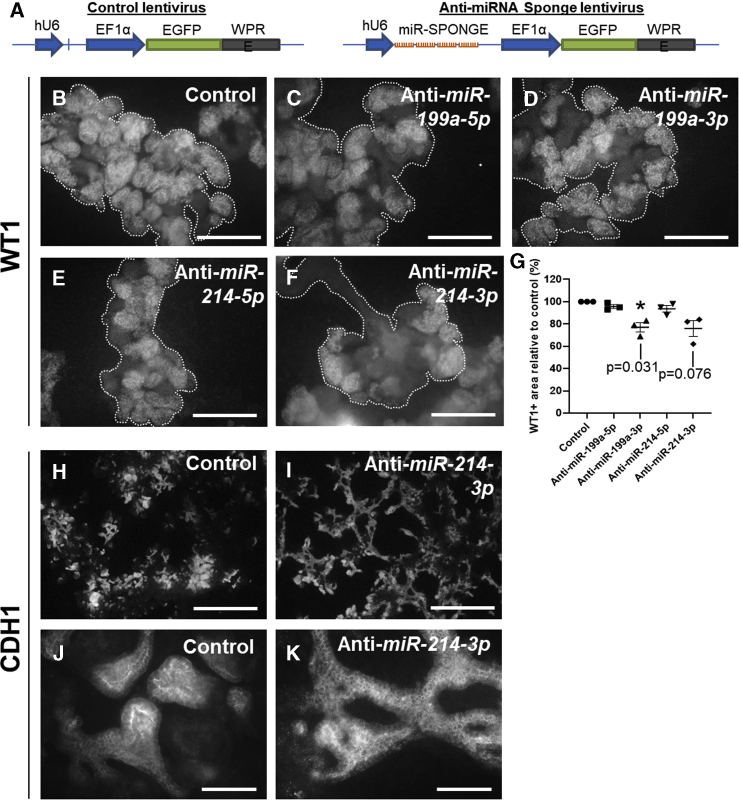
We next wanted to know whether inhibiting either of these miRNAs would have an effect on the differentiating cultures. We constructed lentiviral vectors expressing miRNA inhibitors (sponges) against each of the four mature species of the miRNA cluster (miR-199a-5p, miR-199a-3p, miR-214-5p, and miR-214-3p), previously shown to be effective by Tsujimura et al. (2015), under the control of a U6 promoter (Figure 4A). These inhibitors act by binding to the mature miRNAs, thereby preventing the latter from accessing their intracellular targets (Ebert et al., 2007). MAN13 hESCs were transduced with the sponge-expressing lentiviral vectors (or with control lentivirus expressing only EGFP) on the day of plating and then differentiated. At the end of the 2D protocol (day 30), the cultures were immunostained for WT1, as a marker of MM in the developing multilayered kidney tissue, and also for CDH1 to assess the extent of epithelialization. Inhibiting miR-199a-3p significantly decreased the extent of the MM domain, as judged by the proportion of multilayered kidney tissue masses that immunostained positive for WT1 (Figures 4B–4G). Moreover, inhibition of miR-214-3p (but not of miR-199a-3p) appeared to change the morphology of the epithelia making these structures more elongated compared with controls (Figures 4H–4K and S2).
Figure 4.
Inhibition of Members of the miR-199a/214 Cluster by miRNA Sponges in Differentiating 2D Cultures
MAN13 hESCs transduced either with control (EGFP-only) lentiviral vectors or with vectors expressing “sponge” inhibitors against miR-199a-5p, miR199a-3p, miR-214-5p, or miR-214-3p schematically shown in (A) were differentiated in 2D and immunostained with WT1 or CDH1 antibodies at the end (day 30) of the protocol.
(B–G) Inhibition of miR-199a-3p caused a significant decrease in the extent of the WT1+-stained area (MM) within developing cell aggregates (examples outlined) in differentiating cultures. Quantified in (G): mean ± SEM; n = 3 independent differentiation experiments; ∗p < 0.05, one-sample t test; individual p values are also shown for anti-miR-199a-3p and anti-miR-214-3p). Scale bars, 120 μm.
(H and I) Low-magnification images showing that inhibition of miR-214-3p results in more elongated epithelial structures (CDH1+) in the cultures. Scale bars, 600 μm.
(J and K) Examples of individual CDH1+ tubules showing normal (control) versus elongated (anti-miR-214-3p) phenotype. Scale bars, 60 μm.
See also Figures S2 and S3.
Following this, we tested if overexpressing miR-199a, using a lentiviral vector in a different hESC line, MAN11, which normally generates a lower proportion of the WT1+ (MM) domain than MAN13, had opposite effects. Indeed, overexpression of miR-199a increased the extent of the WT1+ domain, while by the end of the differentiation protocol, it made CDH1+ tubules appear less thin/elongated; the opposite of the effects elicited by the sponge (Figure S3).
Effects of Downregulating miR-199a/214 in Organoids
We next investigated whether the perturbation of the WT1+ domain observed in 2D culture correlated with defects in nephron maturation in more mature tissues. As the 2D differentiation method that we employed does not progress beyond the condensation of MM around branching tubules (Bantounas et al., 2018), we differentiated hESCs into 3D kidney organoids (Figures 5A–5D). We and others have previously shown that this method predominantly yields MM-derived nephrons with immature glomeruli and proximal tubules, as well as minor populations of distal-like tubules and collecting duct-like structures (Bantounas et al., 2018; Takasato et al., 2016). To ascertain that this was a suitable model for our investigation, we first differentiated MAN13 hESCs using this method and determined the level of both the pre-miRNA (DNM3OS) and the mature miRNAs of the miR-199a/214 cluster during differentiation by qPCR. Similarly to the 2D model, the expression of all molecular species increased with time and miR-199a-3p and miR-214-3p were the dominant strands (Figures 5E and 5F). We also performed LNA probe ISH with probes against miR-199a-3p and miR-214-3p, finding that interstitial cells, tubules, and glomeruli expressed these miRNAs (Figures 5G–5L).
Figure 5.
Endogenous Expression of miR199a/214 Cluster Members during Differentiation of MAN13 hESC-Derived 3D Kidney Organoids
(A) Schematic representation of the differentiation protocol. On day 7, the cells are pelleted and transferred to Transwell membranes to continue developing as 3D organoids.
(B–D) Phase contrast images showing the development of the organoids during the 3D part of the differentiation protocol; clear morphogenesis is visible in the periphery of the organoids. Scale bars, 600 μm.
(E and F) Time course of the primary transcript (D) and mature miRNAs (E) of the miR-199a/214 cluster during 3D organoid differentiation, assessed by qPCR (mean ± SEM; n = 3 independent differentiation experiments, with three organoids pooled per time point in each experiment).
(G–L) In situ hybridization with LNA probes against miR-199a-3p and miR-214-3p on day 30 organoids shows strong interstitial and glomerular expression as well as some often patchy expression in the tubules. Scale bars, 50 μm. t, tubules; g, glomeruli (shown by arrows). Stained areas appear purple, over Nuclear fast red (pink) counterstain.
Inhibition of miR-199a-3p or miR-214-3p Results in Formation of Dysplastic Nephrons and a Denser Vascular Network
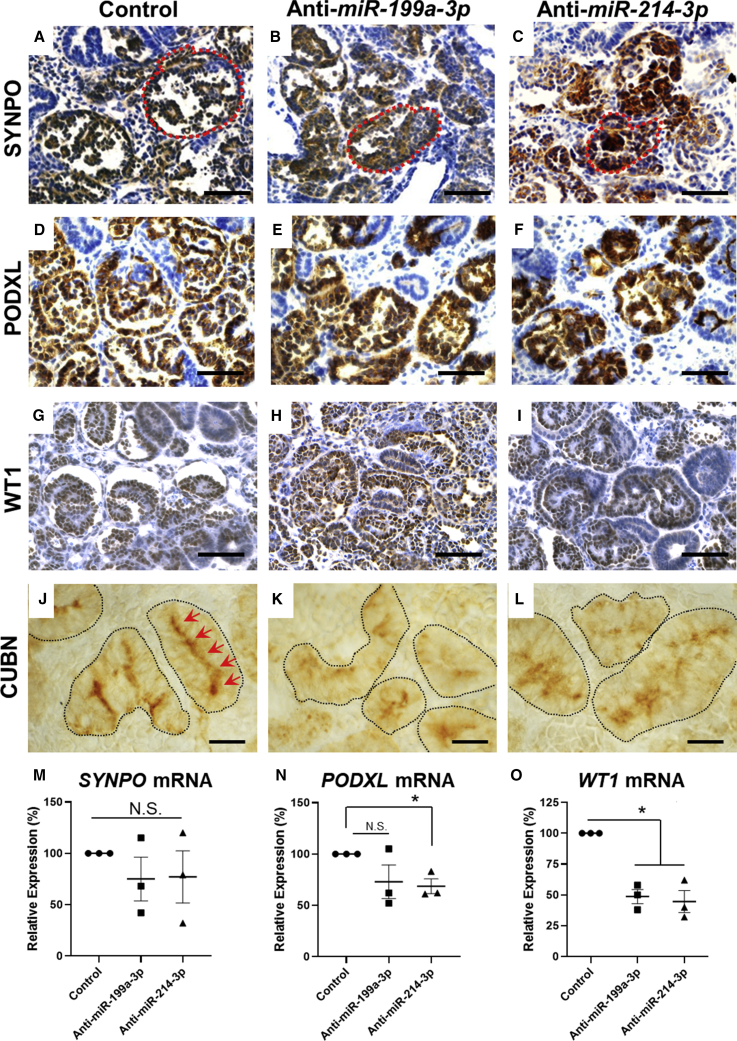
After verifying expression of the miRNAs in the 3D organoid model, we proceeded to inhibit their activity in this system. We transduced MAN13 hESCs with lentiviruses expressing sponges against miR-199a-3p or miR-214-3p, or a control EGFP lentivirus (Figure 4A), then differentiated them as 3D organoids. We confirmed transgene expression from the vectors through to the end of the differentiation protocol (day 25), as shown by the persistence of EGFP signal (Figure S4), at which point we undertook immunohistochemical and molecular analyses. In control organoids, we detected round glomerular profiles with delicate fronds in their tufts, and these glomeruli immunostained for SYNPO (Synaptopodin), PODXL (Podocalyxin), and WT1. In contrast, subsets of glomeruli were dysmorphic in organoids in which miR-199a-3p or miR-214-3p was inhibited (Figures 6A–6I). These glomeruli appeared to have irregular, sometimes square-shaped outlines and lacked delicate fronds in their tufts, and it was difficult to discern discrete junctions between the Bowman space and the adjacent tubule. There was, however, no difference in proportions of each organoid that immunostained for the three glomerular markers, suggesting that the total mass of glomeruli was not affected. Interestingly, as measured by qPCR, transcripts for PODXL were significantly lower than in controls when miR-214-3p was inhibited, and WT1 transcripts were significantly downregulated when either miRNA was inhibited (Figures 6M–6O). Numerous other kidney differentiation transcripts were not affected by miRNA inhibition (Figure S5), suggesting a specific effect. Moreover, our manipulations had no significant effect on the average size of the organoids, as assessed by measuring the areas of their transverse mid-sections on histology (Figure S6A).
Figure 6.
Characterization of Glomerular and Proximal Tubule Morphology following Inhibition of miR-199a/214 Activity
(A–I) MAN13-derived organoids treated with anti-miRNA sponges and control organoids were fixed at the end of differentiation (day 25) and immunostained using antibodies against glomerular markers: (A–C) SYNPO, with a typical glomerulus circled in each condition. Note the circular profiles in controls with fine fronds in the glomerular tufts, whereas the glomeruli are dysmorphic in the sponge-treated organoids; (D–F) PODXL; (G–I) WT1. Scale bars, 100 μm. Immunostaining is brown over hematoxylin (blue) counterstain.
(J–L) Organoids immunostained for CUBN (brown; no counterstain), marking proximal tubules (individual tubules outlined) revealed a continuous linear apical pattern in control tubules (J) (marked by arrows), which is disorganized in the anti-miRNA sponge organoids (K, L). Scale bars, 20 μm.
(M–O) Quantification of transcripts encoding SYNPO, PODXL, and WT1 by qPCR at day 25 of differentiation (mean ± SEM; n = 3 independent differentiation experiments; each dot represents a pooled RNA sample from three organoids; N.S., not significant; ∗p < 0.05, one-sample t test).
See also Figures S4–S7.
Organoid sections were then immunostained for the proximal tubule apical protein Cubilin (CUBN) (Nielsen et al., 2016) to determine whether miRNA inhibition had any adverse effects on proximal tubules (Figures 6J–6L). The proportions of organoid cross-sections with positive CUBN staining did not differ significantly between the three groups (Figure S6B). On close inspection, however, the normal apical linear pattern of CUBN seen in control organoids (Figure 6J) was difficult to discern in sponge-treated organoids, in which the pattern appeared more diffuse, sometimes granular and cytoplasmic, pointing to a loss of its proper cellular polarity (Figures 6K and 6L). To examine more distal parts of the nephron and presumptive collecting ducts, we immunostained for CDH1, but observed no differences between control and sponge-treated organoids (Figures S6C–S6H).
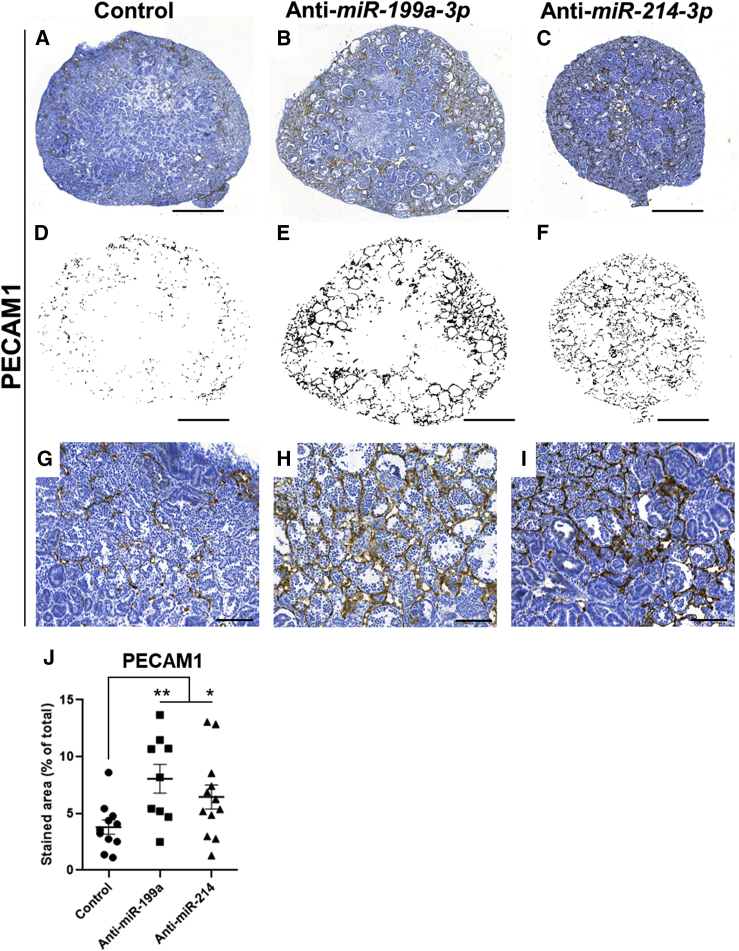
We further asked if our miRNA manipulations had an effect on the development of blood vessels in the organoids. Compared with controls, PECAM1 immunostaining (marking vascular endothelial cells) revealed a significantly denser network of vessels in both anti-miR-199a-3p- and anti-miR-214-3p-treated organoids compared with controls (Figure 7). Vasculature was observed surrounding the developing Bowman capsule but in no condition did we observe glomerular tufts with capillary loops. Thus, the over-abundant capillaries remain confined to the interstitial space.
Figure 7.
Inhibition of miR-199a or miR-214 Increases Vascular Density in Kidney Organoids
(A–C) MAN13-derived organoid sections were immunostained for PECAM1 (marking vascular endothelial cells). Scale bars, 500 μm.
(D–F) PECAM1+ areas of the organoids in (A–C) are highlighted in each case showing the increased extent of staining in the anti-miRNA organoids.
(G–I) High-power images showing the denser network of blood vessels in the anti-miRNA organoids compared with controls. Scale bars, 100 μm. Immunostaining is brown over hematoxylin (blue) counterstain.
(J) Quantification of the extent PECAM1 staining in organoids (mean ± SEM; each dot represents a separate organoid; organoids from three independent experiments were used ∗p < 0.05, ∗∗p < 0.01 ANOVA, followed by post-hoc t tests).
See also Figures S4–S7.
Molecular Sponges that Target the miR-199a/214 Cluster Result in a Tendency for Increases in Key Kidney Developmental Transcripts
Finally, to gain insight into the mechanism of action of the miR-199a/214 cluster, we measured the levels of several miR-199a/214 target transcripts that have been shown to be destabilized by these miRNAs in other (non-renal) models and which have also been shown to play roles in kidney development (reviewed in Little and McMahon, 2012; Papakrivopoulou et al., 2013). We predicted that, if the activity of the miRNAs were to be inhibited by the molecular sponges, then the levels of the target transcripts should increase. Indeed, we observed tendencies for several of these targets to increase at the later time points of our organoid differentiation protocol. FZD4, VANGL2, and MAPK8 tended to increase when the anti-miR-199a-3p sponge was applied (Figure S7A), while levels of FZD4 transcripts tended to increase in the presence of the anti-miR-214-3p sponge (Figure S7B). As assessed by the DIANA-miRPath software (Vlachos et al., 2012), these three species of transcripts are not only targets of the miR-199a/214 cluster, but they also participate in the same WNT signaling pathway that controls planar cell polarity. Indeed, an aberration in planar cell polarity may help explain the loss of uniform apical CUBN immunostaining in tubules in the organoids when either of the miRNAs was inhibited (Figures 6J–6L). Furthermore, WNT9b from the developing ureteric bud signals to the surrounding MM, binding to FZD4 to synchronize the maturation of both tissues (Carroll et al., 2005) and this is in agreement with our observation that the balance between MM and tubules is deregulated in our sponge-treated 2D cultures (Figures 4B–4G). In addition to this, levels of TWIST1 and JAG1 transcripts, both previously shown to be downstream of WNT (Howe et al., 2003; Katoh and Katoh, 2006), also showed a tendency to increase at certain points of the organoid differentiation protocol after exposure to the molecular sponges.
Discussion
In this study, we showed that a particular miRNA locus, encoding the miR-199a/214 cluster, is expressed both in human embryonic kidneys in vivo, and in kidney tissues differentiated from cultured hESCs. We inhibited these miRNAs using molecular sponges, and we overexpressed them in human stem cell-derived kidney tissues. Neither manipulation prevented the formation of kidney-like tissues, either in 2D or in 3D organoid cultures. Critically, however, we report here that the miR-199a/214 cluster has demonstrable effects in fine-tuning the development of human nephrons, and the extent of the capillary network between tubules. These effects are also consistent with the fine-tuning roles played by miRNAs in other, non-kidney, examples of mammalian development and differentiation (e.g., Martinez and Walhout, 2009; Schratt, 2009; Xiao and Dudley, 2017). Indeed, we noted that the expression level of the miR-199a/214 cluster tended to increase during the stem cell-to-kidney differentiation protocol. This, again, is in accord with the idea that these molecules do not control, for example, whether a specific organ, such as a kidney, will initiate but instead will modify its tissue components as its nephron and vascular lineages continue to mature.
Using ISH of human embryonic kidneys we detected both miRNAs in the interstitium within the rudimentary metanephros and around the ureter stalk at 7 weeks gestation. At 12 weeks, both miRNAs were expressed in the interstitium, with miR-214 also detected in the nephrogenic zone and in deeper tubules. Next, hESCs were used to investigate the role of the cluster in kidney development. miR-199a/214 was upregulated during 2D kidney differentiation in several wild-type lines. In 3D organoids, interstitial cells, tubules, and avascular glomeruli expressed these miRNAs as assessed by ISH. It should be noted that, as previously observed (Bantounas et al., 2018), glomeruli that differentiate in organoid culture remain immature compared with in vivo counterparts. This may explain why we observed marked miRNA expression in glomeruli within organoids yet only transient expression of the miRNAs in glomeruli in native embryonic human kidneys, i.e., the outer layers of glomeruli, that would have recently formed from MM, were positive but the deeper, more mature, glomeruli appeared to have downregulated the miRNAs. In 2D cultures, transduction of lentiviral vectors overexpressing sponge inhibitors into hESCs decreased the MM domain, as assessed by WT1 immunostaining, and altered the morphology of adjacent tubules, as assessed by CDH1 immunostaining. To understand the repercussions in more mature developing kidneys, we differentiated control or miRNA sponge-transduced hESCs into 3D organoids. Here, we documented dysmorphic glomeruli and a more prominent interstitial vascular network. Thus, the miR-199a/214 cluster fine-tunes differentiation of both MM-derived nephrons and capillaries between tubules.
While clinical implications of these observations require further study, it is noted that a subset of people with heterozygous deletions encompassing the DNM3OS locus that encodes the miR-199a/214 cluster, have malformed kidneys and renal tracts (Chatron et al., 2015). Importantly, these patients did not completely lack kidneys but instead had organs that were, for example, abnormal in shape (Chatron et al., 2015). Although there is no published information about kidney physiology or histology in these individuals, based on our ex vivo organoid experiments we predict that the following may be found in the patients on further investigation: a reduced glomerular filtration rate, because of dysmorphic glomeruli; and a Fanconi-like syndrome of proximal tubule dysfunction, because of the combination of disturbed polarity demonstrated by CUBN mislocalization, as well as aberrations in peritubular capillaries. In future, it would be informative to generate induced pluripotent stem cells from such individuals; we predict that the kidney organoids that would form from them would have similar phenotypic aberrations to those in the current study. To our knowledge, patients with biallelic deletions of the DNM3OS locus have yet to be reported, so their potential renal phenotype is unknown.
In the 2D differentiating cultures, inhibiting miR-199a or miR-214 resulted in a proportionally smaller WT1+ domain (representing MM) at the end of the protocol, which could be linked to nephron abnormalities that were observed in our 3D organoid experiments. A similar effect in terms of the MM domain was observed by Takasato et al. (2016), whereby increasing the duration of Wnt pathway activation at the beginning of hESC differentiation to kidney tissue resulted in a higher proportion of MM in the cultures, at the expense of presumptive ureteric bud-derived epithelium, whereas decreasing it had the opposite effect. Wnt signaling is central to nephron development (Carroll et al., 2005; Kobayashi et al., 2008; Park et al., 2007); so, mechanistically, the two observations could be linked, given that the miR-199a/214 cluster has been shown to control members of the Wnt pathway (including β-catenin, GSK3β, and Frizzled-4 [Alexander et al., 2013; Ghatak and Raha, 2015; Wang et al., 2012; Xia et al., 2012]) in a variety of tissues. Notably, although in 2D there was a reduction of the WT1+ cell population when the miRNAs were inhibited, we did not observe the same in our 3D organoids, where the overall area stained with an anti-WT1 antibody was the same across all conditions. It must be noted, however, that our 3D organoids reach a later stage in kidney development, when the majority of WT1+ cells are confined to glomeruli and, thus, WT1 staining represents immature podocytes rather than MM. Nonetheless, in miRNA sponge-treated organoids, we observed a trend for increased levels of target transcripts of genes involved in WNT signaling, implying the deregulation of the pathway and potentially linking our 2D with our 3D organoid results, as well as mechanistically explaining our observed phenotype. In future studies, detailed proteomic analyses of control versus sponge-treated organoids should be informative to both confirm the effects on these target molecules and to identify further targets. The latter is especially important given that levels of many target transcripts do not decrease upon miRNA binding, but instead the levels of encoded proteins are reduced due to miRNA-induced block in translation.
In addition, despite the extent of PODXL and WT1 immunostaining being unchanged compared with controls, transcripts encoding these proteins were actually lower in the sponge-treated organoids. Since immunohistochemistry is not quantitative, the amount of WT1 protein in tissues may therefore actually be less, contributing to the abnormal glomerulogenesis observed: indeed, it is known that WT1 drives expression of PODXL (Palmer et al., 2001), which may in turn lead to the glomerular abnormalities observed in our organoids.
Another abnormality that was observed in organoids treated with the miRNA sponges was in proximal tubules that, like glomeruli, are derived from WT1+ MM. In the sponge-treated organoids, proximal tubules lacked the normal apical linear pattern of CUBN, perhaps suggesting aberrant polarity. This phenotype is in keeping with the expression of the miRNAs we observed in control organoid tubules, as well as miR-214-3p expression in human embryonic kidney tubules in vivo. Moreover, we observed tendencies for increased transcript levels of FZD4, VANGL2, and MAPK8, mRNAs encoding planar cell polarity molecules implicated in kidney tubule morphogenesis and differentiation (Papakrivopoulou et al., 2013). An aberration of CUBN would be predicted to compromise the processing of glomerular ultrafiltrate (Nielsen et al., 2016) and we note that an important role for miRNAs in regulating ion transport in the kidney has already been suggested (Elvira-Matelot et al., 2011; Liu et al., 2017).
Epithelial structures were clearly affected in 2D cultures transduced with the anti-miR-214 sponge vector: CDH1+ epithelia became more elongated and connected up to form a network. These branched CDH1+ tubules in 2D cultures are more likely to represent ureteric bud derivatives and inhibiting miR-214-3p could, thus, favor ureteric bud epithelium stabilization, while restricting MM (as shown by the reduction in WT1+ cells). In agreement with this interpretation, CDH1 itself has been previously reported as a direct target of miR-214 (Wang et al., 2016), while in some cancers miR-214 was found to promote EMT (Liu et al., 2018a; Long et al., 2015; Zhao et al., 2018) and therefore inhibiting it would be expected to result in epithelial stabilization.
We have also shown that inhibition of miR-199a/214 resulted in an increased vascular network in kidney organoids. This is in agreement with previous studies in non-kidney tissues indicating that miR-199a and miR-214 are mainly antiangiogenic. For instance, miR-214 is expressed in vascular smooth muscle cells, where it targets proangiogenic QKI (van Mil et al., 2012) and it is upregulated in heart failure patients (Duan et al., 2015), in both cases preventing vascular sprouting. The cluster also targets vascular endothelial growth factor (VEGF) and its receptors in endometrial cell hypoxia, coronary heart disease, and cancer (Dai et al., 2015; Ghosh et al., 2017; Jin et al., 2015). In fact, inhibition of these miRNA is emerging as a mechanism that allows tumor angiogenesis and survival (He et al., 2013; Lombardo et al., 2018; Orso et al., 2020; Wang et al., 2012). In the light of these findings, it is, therefore, not surprising that we observed a similar “contra-vascular” function for the cluster in the kidney. Our data reinforce the idea that considering regulation of the vascularization as predominantly reliant on growth factor (e.g., VEGFA)-receptor interactions is a considerable simplification. This is supported by the recent data from Wang et al. (2020) showing that, although miR-218-2 homozygous knockout mice were not viable, in the heterozygote knockout the density of capillaries was found to be reduced around kidney tubules postnatally.
Finally, given the very strong stromal expression of the miRNA cluster in both human tissue sections and organoids, it may appear surprising that abnormalities following miRNA inhibition were mainly in nephrons. However, it must be stressed that the different cell populations in the developing kidney depend on each other to co-develop and differentiate and that miRNA may play a role in this communication. Furthermore, a significant change occurred in the vasculature throughout the interstitium. In keeping with the results of our study, when Dicer1 (the main nuclease effecting miRNA maturation) was knocked out in the kidney stroma (Foxd1+ cells of mice), severe glomerular, tubular, and angiogenic abnormalities were observed (Nakagawa et al., 2015).
In summary, our data strongly suggest the importance of the miR-199a/214 cluster in fine-tuning human kidney development, as assessed by a human stem cell model. Further work is necessary to both define the precise molecular mechanisms of the observed aberrations of glomerular, tubule, and capillary differentiation, and to determine the potential physiological effects of these phenotypes.
Experimental Procedures
Human Tissues
Human tissues, collected after maternal consent and ethical approval (REC 08/H0906/21+5), were provided by the Medical Research Council and Wellcome Trust Human Developmental Biology Resource (http://www.hdbr.org/). Kidneys were fixed, processed, and sections prepared as described previously (Lopes et al., 2019).
hESC Culture and Renal Differentiation
ESCs were grown on 24-well or 6-well plates coated with 5 μg mL−1 recombinant human Vitronectin (Life Technologies, no. A14700) in the case of MAN11 and MAN13 (Ye et al., 2017) or with Matrigel (BD Biosciences, no. 734-1440) in the case of HUES1, in mTeSR1 (STEMCELL Technologies, no. 5850) or TeSR2 medium (STEMCELL Technologies, no. 5860), with the medium changed once daily. The cells were passaged by treatment of the cultures with 0.5 mM EDTA solution (pH 8) (Invitrogen, no. 15575-038; diluted in PBS) and replating the cells in mTeSR1 or TeSR2 medium, containing 5 nM ROCK inhibitor, Y-27632 (Tocris, no. 1254) for 24 h.
For differentiation, stem cells were plated on Vitronectin-coated plates, at a density of 18,000 cells cm−2 when untransduced, or at 20,500 cells cm−2 when transduced with lentivirus (to account for additional cell death due to some viral toxicity) in mTeSR1 or TeSR2 medium containing 10 nM Y-27632. The following day the medium was replaced with STEMdiff APEL (STEMCELL Technologies, no. 05210), containing 8 μM CHIR-99021 (Tocris, no. 4423) for 3 days, followed by APEL supplemented with 200 ng mL−1 FGF9 (PeproTech, no. 100-23) and 1 μg mL−1 heparin (Sigma, no. 3149) for a further 10 days. Subsequently, the cells were cultured in basal APEL medium, which was changed daily.
3D hESC-kidney organoid culture was performed according to Takasato et al. (2016) and Bantounas et al. (2018). In brief, hESCs were seeded as above and, beginning the following day (day 0), they were incubated in STEMdiff APEL 2 medium (STEMCELL Technologies, no. 05270) with 2% PFHM-II (protein-free hybridoma medium II) (Thermo Fisher Scientific, no. 12040077), supplemented with 8 μM CHIR-99021 for a period of 4 days, before switching to APEL 2/2% PFMH with FGF9/heparin, as above. At day 7, the cells were collected by treatment with TrypLE (Life Technologies, no. 12605-028) and divided into 200,000 cell aliquots. The aliquots were then centrifuged at 400 × g in a microcentrifuge and the resulting cell pellets were placed on Millicell Cell Culture inserts (pore size, 0.4 μm; diameter, 30 mm; no. PICM03050), which were placed into 6-well plates with 1.2 mL of APEL 2/2% PFMH/5 μM CHIR-99021 medium per well. After 1 h, the medium was changed back to APEL 2/PFMH/FGF9/heparin until day 12 and then to APEL 2/PFMH for the remainder of the protocol (day 25). At the 3D stage, the medium was changed once every 2 days.
RNA Extraction and qPCR
RNA was extracted using the mirVana miRNA isolation kit (Thermo Fisher Scientific, AM1560) according to the manufacturer's instructions. Mature miRNA PCR was conducted using the appropriate TaqMan miRNA Assays (Thermo Fisher Scientific) and longer transcripts were detected using the TaqMan RNA-to-Ct 1-Step Kit (Thermo Fisher Scientific, no. 4392653). For a more detailed description and primer sequences, see Supplemental Experimental Procedures and Table S2.
ISH of miRNA in Kidney Tissue and 3D Organoid Sections
Digoxigenin-labeled miRCURY LNA probes were used to detect mature miRNAs on paraffin-embedded sections of kidney organoids. See Supplemental Experimental Procedures for details.
Immunostaining of 2D Cultures and Immunohistochemistry on 3D Organoid Sections
See Supplemental Experimental Procedures. Primary antibodies used are listed in Table S1.
Construction of miRNA Sponge-Expressing Lentiviral Shuttle Plasmids, Lentiviral Vector Production, and Transduction of Stem Cells
Previously published miRNA sponge-expressing plasmids: pLLX (empty vector), pLLX-sponge-miR-199a-5p, pLLX-sponge-miR-199a-3p, pLLX-sponge-miR-214-5p, and pLLX-sponge-miR-214-5p were a gift from Professor Nakashima's group (Kyushu University, Japan) (Tsujimura et al., 2015). Each of these five plasmids was digested with ApaI/XhoI to excise the pU6 sponge expression cassette, which was then cloned into ApaI/XhoI-digested pLL3.7 vector (Addgene, no. 11795). Lentiviral vectors were then produced as described previously in Bantounas et al. (2018) (see Supplemental Experimental Procedures). hESCs were transduced with lentivirus at a multiplicity of infection of 5 IU/cell.
Author Contributions
I.B., A.S.W., and S.J.K. designed the study. I.B., F.M.L., and K.M.R. conducted the research. I.B., F.M.L., A.S.W., and S.J.K. analyzed the data. I.B., A.S.W., and S.J.K. wrote the paper. All authors approved the final paper.
Acknowledgments
We are most grateful to the following funding bodies: UK Research and Innovation/Medical Research Council (MRC) UK Regenerative Medicine Platform hub grant MR/K026739/1; Kidney Research UK John Feehally-Stoneygate Project and Innovation award JFS/RP/008/20160916; Kidneys for Life pump priming grant; Horizon 2020 Marie Skłodowska-Curie Actions Initial Training Network RENALTRACT (642937) grant, EPSRC/MRC Center for Doctoral Training grant EP/L014904/1. The authors declare no conflict of interest. We thank Paul A. Humphreys for assistance with the graphical abstract art.
Published: December 10, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.11.007.
Contributor Information
Ioannis Bantounas, Email: ioannis.bantounas@manchester.ac.uk.
Susan J. Kimber, Email: sue.kimber@manchester.ac.uk.
Supplemental Information
References
- Alexander M.S., Kawahara G., Motohashi N., Casar J.C., Eisenberg I., Myers J.A., Gasperini M.J., Estrella E.A., Kho A.T., Mitsuhashi S. MicroRNA-199a is induced in dystrophic muscle and affects WNT signaling, cell proliferation, and myogenic differentiation. Cell Death Differ. 2013;20:1194–1208. doi: 10.1038/cdd.2013.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurora A.B., Mahmoud A.I., Luo X., Johnson B.A., van Rooij E., Matsuzaki S., Humphries K.M., Hill J.A., Bassel-Duby R., Sadek H.A. MicroRNA-214 protects the mouse heart from ischemic injury by controlling Ca(2)(+) overload and cell death. J. Clin. Invest. 2012;122:1222–1232. doi: 10.1172/JCI59327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantounas I., Ranjzad P., Tengku F., Silajdzic E., Forster D., Asselin M.C., Lewis P., Lennon R., Plagge A., Wang Q. Generation of functioning nephrons by implanting human pluripotent stem cell-derived kidney progenitors. Stem Cell Reports. 2018;10:766–779. doi: 10.1016/j.stemcr.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll T.J., Park J.S., Hayashi S., Majumdar A., McMahon A.P. Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev. Cell. 2005;9:283–292. doi: 10.1016/j.devcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Chatron N., Haddad V., Andrieux J., Desir J., Boute O., Dieux A., Baumann C., Drunat S., Gerard M., Bonnet C. Refinement of genotype-phenotype correlation in 18 patients carrying a 1q24q25 deletion. Am. J. Med. Genet. A. 2015;167A:1008–1017. doi: 10.1002/ajmg.a.36856. [DOI] [PubMed] [Google Scholar]
- Chen H., Shalom-Feuerstein R., Riley J., Zhang S.D., Tucci P., Agostini M., Aberdam D., Knight R.A., Genchi G., Nicotera P. miR-7 and miR-214 are specifically expressed during neuroblastoma differentiation, cortical development and embryonic stem cells differentiation, and control neurite outgrowth in vitro. Biochem. Biophys. Res. Commun. 2010;394:921–927. doi: 10.1016/j.bbrc.2010.03.076. [DOI] [PubMed] [Google Scholar]
- Dai L., Lou W., Zhu J., Zhou X., Di W. MiR-199a inhibits the angiogenic potential of endometrial stromal cells under hypoxia by targeting HIF-1alpha/VEGF pathway. Int. J. Clin. Exp. Pathol. 2015;8:4735–4744. [PMC free article] [PubMed] [Google Scholar]
- Denby L., Ramdas V., Lu R., Conway B.R., Grant J.S., Dickinson B., Aurora A.B., McClure J.D., Kipgen D., Delles C. MicroRNA-214 antagonism protects against renal fibrosis. J. Am. Soc. Nephrol. 2014;25:65–80. doi: 10.1681/ASN.2013010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q., Yang L., Gong W., Chaugai S., Wang F., Chen C., Wang P., Zou M.H., Wang D.W. MicroRNA-214 is upregulated in heart failure patients and suppresses XBP1-mediated endothelial cells angiogenesis. J. Cell Physiol. 2015;230:1964–1973. doi: 10.1002/jcp.24942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dweep H., Sticht C., Kharkar A., Pandey P., Gretz N. Parallel analysis of mRNA and microRNA microarray profiles to explore functional regulatory patterns in polycystic kidney disease: using PKD/Mhm rat model. PLoS One. 2013;8:e53780. doi: 10.1371/journal.pone.0053780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert M.S., Neilson J.R., Sharp P.A. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert M.S., Sharp P.A. MicroRNA sponges: progress and possibilities. RNA. 2010;16:2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvira-Matelot E., Jeunemaitre X., Hadchouel J. Regulation of ion transport by microRNAs. Curr. Opin. Nephrol. Hypertens. 2011;20:541–546. doi: 10.1097/MNH.0b013e328348b4aa. [DOI] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Flynt A.S., Li N., Thatcher E.J., Solnica-Krezel L., Patton J.G. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat. Genet. 2007;39:259–263. doi: 10.1038/ng1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghatak S., Raha S. Micro RNA-214 contributes to proteasome independent downregulation of beta catenin in Huntington's disease knock-in striatal cell model STHdhQ111/Q111. Biochem. Biophys. Res. Commun. 2015;459:509–514. doi: 10.1016/j.bbrc.2015.02.137. [DOI] [PubMed] [Google Scholar]
- Ghosh A., Dasgupta D., Ghosh A., Roychoudhury S., Kumar D., Gorain M., Butti R., Datta S., Agarwal S., Gupta S. MiRNA199a-3p suppresses tumor growth, migration, invasion and angiogenesis in hepatocellular carcinoma by targeting VEGFA, VEGFR1, VEGFR2, HGF and MMP2. Cell Death Dis. 2017;8:e2706. doi: 10.1038/cddis.2017.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin J.G., Ge X., Stephan K., Jurisch A., Tullius S.G., Iacomini J. Identification of a microRNA signature of renal ischemia reperfusion injury. Proc. Natl. Acad. Sci. U S A. 2010;107:14339–14344. doi: 10.1073/pnas.0912701107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S., Chan W.Y. Flexible and versatile as a chameleon-sophisticated functions of microRNA-199a. Int. J. Mol. Sci. 2012;13:8449–8466. doi: 10.3390/ijms13078449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., Wang L., Zhou W., Zhang Z., Wang L., Xu S., Wang D., Dong J., Tang C., Tang H. MicroRNA expression profiling in clear cell renal cell carcinoma: identification and functional validation of key miRNAs. PLoS One. 2015;10:e0125672. doi: 10.1371/journal.pone.0125672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Jing Y., Li W., Qian X., Xu Q., Li F.S., Liu L.Z., Jiang B.H., Jiang Y. Roles and mechanism of miR-199a and miR-125b in tumor angiogenesis. PLoS One. 2013;8:e56647. doi: 10.1371/journal.pone.0056647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemker S.L., Cerqueira D.M., Bodnar A.J., Cargill K.R., Clugston A., Anslow M.J., Sims-Lucas S., Kostka D., Ho J. Deletion of hypoxia-responsive microRNA-210 results in a sex-specific decrease in nephron number. FASEB J. 2020;34:5782–5799. doi: 10.1096/fj.201902767R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe L.R., Watanabe O., Leonard J., Brown A.M. Twist is up-regulated in response to Wnt1 and inhibits mouse mammary cell differentiation. Cancer Res. 2003;63:1906–1913. [PubMed] [Google Scholar]
- Jin Y., Yang C.J., Xu X., Cao J.N., Feng Q.T., Yang J. MiR-214 regulates the pathogenesis of patients with coronary artery disease by targeting VEGF. Mol. Cell Biochem. 2015;402:111–122. doi: 10.1007/s11010-014-2319-5. [DOI] [PubMed] [Google Scholar]
- Joglekar M.V., Parekh V.S., Hardikar A.A. New pancreas from old: microregulators of pancreas regeneration. Trends Endocrinol. Metab. 2007;18:393–400. doi: 10.1016/j.tem.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Jones T.F., Bekele S., O'Dwyer M.J., Prowle J.R. MicroRNAs in acute kidney injury. Nephron. 2018;140:124–128. doi: 10.1159/000490204. [DOI] [PubMed] [Google Scholar]
- Katoh M., Katoh M. Notch ligand, JAG1, is evolutionarily conserved target of canonical WNT signaling pathway in progenitor cells. Int. J. Mol. Med. 2006;17:681–685. [PubMed] [Google Scholar]
- Kobayashi A., Valerius M.T., Mugford J.W., Carroll T.J., Self M., Oliver G., McMahon A.P. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell. 2008;3:169–181. doi: 10.1016/j.stem.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhia R., Yheskel M., Flaten A., Ramalingam H., Aboudehen K., Ferre S., Biggers L., Mishra A., Chaney C., Wallace D.P. Interstitial microRNA miR-214 attenuates inflammation and polycystic kidney disease progression. JCI Insight. 2020;5:133785. doi: 10.1172/jci.insight.133785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.B., Bantounas I., Lee D.Y., Phylactou L., Caldwell M.A., Uney J.B. Twist-1 regulates the miR-199a/214 cluster during development. Nucleic Acids Res. 2009;37:123–128. doi: 10.1093/nar/gkn920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little M.H., McMahon A.P. Mammalian kidney development: principles, progress, and projections. Cold Spring Harb. Perspect. Biol. 2012;4:a008300. doi: 10.1101/cshperspect.a008300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Edinger R.S., Klemens C.A., Phua Y.L., Bodnar A.J., LaFramboise W.A., Ho J., Butterworth M.B. A microRNA cluster miR-23-24-27 is upregulated by aldosterone in the distal kidney nephron where it alters sodium transport. J. Cell Physiol. 2017;232:1306–1317. doi: 10.1002/jcp.25599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Luo J., Zhao Y.T., Wang Z.Y., Zhou J., Huang S., Huang J.N., Long H.X., Zhu B. TWIST1 upregulates miR-214 to promote epithelial-to-mesenchymal transition and metastasis in lung adenocarcinoma. Int. J. Mol. Med. 2018;42:461–470. doi: 10.3892/ijmm.2018.3630. [DOI] [PubMed] [Google Scholar]
- Liu J., Liu B., Guo Y., Chen Z., Sun W., Gao W., Wu H., Wang Y. MiR-199a-3p acts as a tumor suppressor in clear cell renal cell carcinoma. Pathol. Res. Pract. 2018;214:806–813. doi: 10.1016/j.prp.2018.05.005. [DOI] [PubMed] [Google Scholar]
- Liu M., Liu L., Bai M., Zhang L., Ma F., Yang X., Sun S. Hypoxia-induced activation of Twist/miR-214/E-cadherin axis promotes renal tubular epithelial cell mesenchymal transition and renal fibrosis. Biochem. Biophys. Res. Commun. 2018;495:2324–2330. doi: 10.1016/j.bbrc.2017.12.130. [DOI] [PubMed] [Google Scholar]
- Lombardo G., Gili M., Grange C., Cavallari C., Dentelli P., Togliatto G., Taverna D., Camussi G., Brizzi M.F. IL-3R-alpha blockade inhibits tumor endothelial cell-derived extracellular vesicle (EV)-mediated vessel formation by targeting the beta-catenin pathway. Oncogene. 2018;37:1175–1191. doi: 10.1038/s41388-017-0034-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long H., Wang Z., Chen J., Xiang T., Li Q., Diao X., Zhu B. microRNA-214 promotes epithelial-mesenchymal transition and metastasis in lung adenocarcinoma by targeting the suppressor-of-fused protein (Sufu) Oncotarget. 2015;6:38705–38718. doi: 10.18632/oncotarget.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes F.M., Roberts N.A., Zeef L.A., Gardiner N.J., Woolf A.S. Overactivity or blockade of transforming growth factor-beta each generate a specific ureter malformation. J. Pathol. 2019;249:472–484. doi: 10.1002/path.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrone A.K., Stolz D.B., Bastacky S.I., Kostka D., Bodnar A.J., Ho J. MicroRNA-17~92 is required for nephrogenesis and renal function. J. Am. Soc. Nephrol. 2014;25:1440–1452. doi: 10.1681/ASN.2013040390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez N.J., Walhout A.J.M. The interplay between transcription factors and microRNAs in genome-scale regulatory networks. Bioessays. 2009;31:435–445. doi: 10.1002/bies.200800212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa N., Xin C., Roach A.M., Naiman N., Shankland S.J., Ligresti G., Ren S., Szak S., Gomez I.G., Duffield J.S. Dicer1 activity in the stromal compartment regulates nephron differentiation and vascular patterning during mammalian kidney organogenesis. Kidney Int. 2015;87:1125–1140. doi: 10.1038/ki.2014.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Christensen E.I., Birn H. Megalin and cubilin in proximal tubule protein reabsorption: from experimental models to human disease. Kidney Int. 2016;89:58–67. doi: 10.1016/j.kint.2015.11.007. [DOI] [PubMed] [Google Scholar]
- Orso F., Quirico L., Dettori D., Coppo R., Virga F., Ferreira L.C., Paoletti C., Baruffaldi D., Penna E., Taverna D. Role of miRNAs in tumor and endothelial cell interactions during tumor progression. Semin. Cancer Biol. 2020;60:214–224. doi: 10.1016/j.semcancer.2019.07.024. [DOI] [PubMed] [Google Scholar]
- Palmer R.E., Kotsianti A., Cadman B., Boyd T., Gerald W., Haber D.A. WT1 regulates the expression of the major glomerular podocyte membrane protein Podocalyxin. Curr. Biol. 2001;11:1805–1809. doi: 10.1016/s0960-9822(01)00560-7. [DOI] [PubMed] [Google Scholar]
- Papakrivopoulou E., Dean C.H., Copp A.J., Long D.A. Planar cell polarity and the kidney. Nephrol. Dial. Transpl. 2013;29:1320–1326. doi: 10.1093/ndt/gft484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.S., Valerius M.T., McMahon A.P. Wnt/beta-catenin signaling regulates nephron induction during mouse kidney development. Development. 2007;134:2533–2539. doi: 10.1242/dev.006155. [DOI] [PubMed] [Google Scholar]
- Phua Y.L., Chen K.H., Hemker S.L., Marrone A.K., Bodnar A.J., Liu X., Clugston A., Kostka D., Butterworth M.B., Ho J. Loss of miR-17~92 results in dysregulation of Cftr in nephron progenitors. Am. J. Physiol. Ren. Physiol. 2019;316:F993–F1005. doi: 10.1152/ajprenal.00450.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankrityayan H., Kulkarni Y.A., Gaikwad A.B. Diabetic nephropathy: the regulatory interplay between epigenetics and microRNAs. Pharmacol. Res. 2019;141:574–585. doi: 10.1016/j.phrs.2019.01.043. [DOI] [PubMed] [Google Scholar]
- Schratt G. Fine-tuning neural gene expression with microRNAs. Curr. Opin. Neurobiol. 2009;19:213–219. doi: 10.1016/j.conb.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Shaffi S.K., Galas D., Etheridge A., Argyropoulos C. Role of microRNAs in renal parenchymal diseases—a new dimension. Int. J. Mol. Sci. 2018;19:1797. doi: 10.3390/ijms19061797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi K., Lu J., Zhao Y., Wang L., Li J., Qi B., Li H., Ma C. MicroRNA-214 suppresses osteogenic differentiation of C2C12 myoblast cells by targeting Osterix. Bone. 2013;55:487–494. doi: 10.1016/j.bone.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Sun Y., Kuek V., Liu Y., Tickner J., Yuan Y., Chen L., Zeng Z., Shao M., He W., Xu J. MiR-214 is an important regulator of the musculoskeletal metabolism and disease. J. Cell Physiol. 2018;234:231–245. doi: 10.1002/jcp.26856. [DOI] [PubMed] [Google Scholar]
- Takasato M., Er P.X., Becroft M., Vanslambrouck J.M., Stanley E.G., Elefanty A.G., Little M.H. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat. Cell Biol. 2014;16:118–126. doi: 10.1038/ncb2894. [DOI] [PubMed] [Google Scholar]
- Takasato M., Er P.X., Chiu H.S., Maier B., Baillie G.J., Ferguson C., Parton R.G., Wolvetang E.J., Roost M.S., Lopes S.M. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2016;536:238. doi: 10.1038/nature17982. [DOI] [PubMed] [Google Scholar]
- Trionfini P., Benigni A., Remuzzi G. MicroRNAs in kidney physiology and disease. Nat. Rev. Nephrol. 2015;11:23–33. doi: 10.1038/nrneph.2014.202. [DOI] [PubMed] [Google Scholar]
- Tsujimura K., Irie K., Nakashima H., Egashira Y., Fukao Y., Fujiwara M., Itoh M., Uesaka M., Imamura T., Nakahata Y. miR-199a links MeCP2 with mTOR signaling and its dysregulation leads to Rett syndrome phenotypes. Cell Rep. 2015;12:1887–1901. doi: 10.1016/j.celrep.2015.08.028. [DOI] [PubMed] [Google Scholar]
- van Mil A., Grundmann S., Goumans M.J., Lei Z., Oerlemans M.I., Jaksani S., Doevendans P.A., Sluijter J.P. MicroRNA-214 inhibits angiogenesis by targeting Quaking and reducing angiogenic growth factor release. Cardiovasc. Res. 2012;93:655–665. doi: 10.1093/cvr/cvs003. [DOI] [PubMed] [Google Scholar]
- Vlachos I.S., Kostoulas N., Vergoulis T., Georgakilas G., Reczko M., Maragkakis M., Paraskevopoulou M.D., Prionidis K., Dalamagas T., Hatzigeorgiou A.G. DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. 2012;40:W498–W504. doi: 10.1093/nar/gks494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chen J., Li F., Lin Y., Zhang X., Lv Z., Jiang J. MiR-214 inhibits cell growth in hepatocellular carcinoma through suppression of beta-catenin. Biochem. Biophys. Res. Commun. 2012;428:525–531. doi: 10.1016/j.bbrc.2012.10.039. [DOI] [PubMed] [Google Scholar]
- Wang X., Liu J., Yin W., Abdi F., Pang P.D., Fucci Q.A., Abbott M., Chang S.L., Steele G., Patel A. miR-218 expressed in endothelial progenitor cells contributes to the development and repair of the kidney microvasculature. Am. J. Pathol. 2020;190:642–659. doi: 10.1016/j.ajpath.2019.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Shen E., Wang Y., Li J., Cheng D., Chen Y., Gui D., Wang N. Cross talk between miR-214 and PTEN attenuates glomerular hypertrophy under diabetic conditions. Sci. Rep. 2016;6:31506. doi: 10.1038/srep31506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Sato T., Amano T., Kawamura Y., Kawamura N., Kawaguchi H., Yamashita N., Kurihara H., Nakaoka T. Dnm3os, a non-coding RNA, is required for normal growth and skeletal development in mice. Dev. Dyn. 2008;237:3738–3748. doi: 10.1002/dvdy.21787. [DOI] [PubMed] [Google Scholar]
- Xia H., Ooi L.L., Hui K.M. MiR-214 targets beta-catenin pathway to suppress invasion, stem-like traits and recurrence of human hepatocellular carcinoma. PLoS One. 2012;7:e44206. doi: 10.1371/journal.pone.0044206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L., Dudley A.C. Fine-tuning vascular fate during endothelial-mesenchymal transition. J. Pathol. 2017;241:25–35. doi: 10.1002/path.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H., Su B., Ni H., Li L., Chen X., You X., Zhang H. microRNA-199a may be involved in the pathogenesis of lupus nephritis via modulating the activation of NF-kappaB by targeting Klotho. Mol. Immunol. 2018;103:235–242. doi: 10.1016/j.molimm.2018.10.003. [DOI] [PubMed] [Google Scholar]
- Ye J., Bates N., Soteriou D., Grady L., Edmond C., Ross A., Kerby A., Lewis P.A., Adeniyi T., Wright R. High quality clinical grade human embryonic stem cell lines derived from fresh discarded embryos. Stem Cell Res. Ther. 2017;8:128. doi: 10.1186/s13287-017-0561-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Diao C., Wang X., Xie Y., Liu Y., Gao X., Han J., Li S. LncRNA BDNF-AS inhibits proliferation, migration, invasion and EMT in oesophageal cancer cells by targeting miR-214. J. Cell. Mol. Med. 2018;22:3729–3739. doi: 10.1111/jcmm.13558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Ma S.X., Shang Y.Q., Zhang H.Q., Su W. MicroRNAs in chronic kidney disease. Clin. Chim. Acta. 2019;491:59–65. doi: 10.1016/j.cca.2019.01.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.