Summary
Burgeoning applications of human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) in disease modeling, regenerative medicine, and drug screening have broadened the usage of hiPSC-CMs and entailed their long-term storage. Cryopreservation is the most common approach to store hiPSC-CMs. However, the effects of cryopreservation and recovery on hiPSC-CMs remain poorly understood. Here, we characterized the transcriptome, electro-mechanical function, and drug response of fresh hiPSC-CMs without cryopreservation and recovered hiPSC-CMs from cryopreservation. We found that recovered hiPSC-CMs showed upregulation of cell cycle genes, similar or reduced contractility, Ca2+ transients, and field potential duration. When subjected to treatment of drugs that affect electrophysiological properties, recovered hiPSC-CMs showed an altered drug response and enhanced propensity for drug-induced cardiac arrhythmic events. In conclusion, fresh and recovered hiPSC-CMs do not always show comparable molecular and physiological properties. When cryopreserved hiPSC-CMs are used for assessing drug-induced cardiac liabilities, the altered drug sensitivity needs to be considered.
Keywords: hiPSC-CMs, cryopreservation, recovery, microelectrode arrays, drug response, drug safety assessment, cardiac arrhythmia
Highlights
-
•
We compare fresh and recovered hiPSC-CMs from cryopreservation
-
•
Recovered hiPSC-CMs show altered transcriptome and electro-mechanical function
-
•
Recovered hiPSC-CMs show altered drug response
-
•
Recovered hiPSC-CMs show enhanced propensity for drug-induced arrhythmic events
In this article, Wu and colleagues show that recovered hiPSC-CMs from cryopreservation and fresh hiPSC-CMs show different transcriptome profiling and electro-mechanical function in a line-dependent manner. When hiPSC-CMs are treated with drugs that affect electrophysiology, recovered hiPSC-CMs show altered drug response and enhanced propensity for drug-induced arrhythmic events.
Introduction
Significant advances in human induced pluripotent stem cell (hiPSC) technology in the past decade have led to a paradigm shift in the field of cardiovascular research. Using well-defined differentiation protocols, hiPSCs can be robustly differentiated into cardiomyocytes (hiPSC-CMs) that are now widely applied in cardiovascular disease modeling, regenerative medicine, and large-scale screens for novel therapeutics or assessing cardiotoxicity (see reviews by Chen et al., 2016; Zhang et al., 2018). With the rapidly expanding usage of hiPSC-CMs, long-term storage of amassed hiPSC-CMs with the desired phenotype has important practical and financial advantages and is considered a prerequisite for large-scale clinical biobanking initiatives. In parallel with efforts to optimize protocols for hiPSC-CM cryopreservation, several studies have also demonstrated that the recovered hiPSC-CMs display typical genetic and electro-mechanical characteristics of cardiomyocytes (Chen et al., 2015; Hwang et al., 2015; Xu et al., 2011). Additionally, cryopreservation has a negligible effect on hiPSC-CM survival, engraftment size, and host-graft electro-mechanical integration in injured animal hearts after cell transplantation (Shiba et al., 2012; Xu et al., 2011; Zhao et al., 2018), indicating the feasibility of using cryopreserved hiPSC-CMs in future clinical trials. While encouraging, there is still little knowledge as to whether recovered hiPSC-CMs are functionally equivalent to fresh hiPSC-CMs and whether recovered hiPSC-CMs can fully recapitulate the pharmacological response to drug treatments, a cornerstone of cardiac safety assessment.
Recently, hiPSC-CMs have offered a superior in vitro model for assessing drug-induced cardiac liabilities such as arrhythmias because of their greater resemblance to human cardiomyocytes in physiology and pharmacological response than animal cardiomyocytes. Under the Comprehensive In Vitro Proarrhythmia Assay (CiPA) initiative, hiPSC-CMs represent a significant arm of the new concerted effort for improving the specificity and accuracy of evaluating drug-induced proarrhythmic risks such as QT prolongation and torsades de pointes (TdP) (Colatsky et al., 2016; Sager et al., 2014). Indeed, large datasets of drug toxicity have been converted into integrated scoring models to predict the proarrhythmic risks of compounds (Ando et al., 2017; Blinova et al., 2018; Kopljar et al., 2018). In such large-scale studies, the use of recovered hiPSC-CMs from cryopreservation is unavoidable. However, cryopreservation has been reported to cause cell microstructural damage, including partial lesions to the nucleus and cell membrane and loss of mitochondrial integrity (Kim et al., 2011). Moreover, cryopreservation has been associated with activation of cellular stress response pathways and accumulation of reactive oxygen species (ROS) in recovered cells (Pegg, 2015; Xu et al., 2011), which may affect ion channel function. Recovered hiPSC-CMs can exhibit the pharmacological response to ion channel modulators (Blinova et al., 2018; Hortigon-Vinagre et al., 2016), but, to achieve the goal of the CiPA initiative, it is essential to assess whether cryopreservation affects the drug response of hiPSC-CMs.
To understand the effects of cryopreservation on hiPSC-CMs, we examined the transcriptome and electro-mechanical function of fresh (without cryopreservation) and recovered hiPSC-CMs from cryopreservation. Furthermore, to assess the pharmacological response of recovered hiPSC-CMs to drugs affecting electrophysiology, we assessed the effects of four drugs from three different categories (antihypertensive, nifedipine; class III antiarrhythmics, ibutilide and E4031; and tyrosine kinase inhibitor, sunitinib) on hiPSC-CM electrophysiological properties. Overall, the significance of this study is in performing the comparability analysis between fresh and recovered hiPSC-CMs, and the findings may provide a frame of reference for future applications of cryopreserved hiPSC-CMs in large-scale drug testing studies and facilitate investigators to determine whether fresh hiPSC-CMs are necessary for cardiac drug safety studies.
Results
Cell Viability Is Independent of the Duration of Cryopreservation
We acquired five healthy control hiPSC lines with different genetic backgrounds from the Stanford Cardiovascular Institute (CVI) Biobank and differentiated hiPSCs into hiPSC-CMs. Day 23 hiPSC-CMs were kept in culture as fresh control or cryopreserved in liquid nitrogen for further use (Figure 1A). Both fresh and recovered groups had over 90% cardiac troponin T (cTNT)-positive hiPSC-CMs (Figure S1A and S1B and Video S1). Cell viability is one of the major concerns of cryopreservation, as ice crystallization during the freezing process may damage the cell membrane and lead to diminished cell recovery. Therefore, we sought to determine whether the duration of cryopreservation affects cell viability. We examined the recovered hiPSC-CMs after 6-month or 18-month cryopreservation using calcein and ethidium homodimer-1 (EthD-1) (Figure S1C). The percentage of live (calcein-positive) or dead (EthD-1-positive) hiPSC-CMs was comparable between 6-month and 18-month groups, suggesting that the longer 18-month duration of cryopreservation has a negligible effect on cell viability (Figures S1D and S1E).
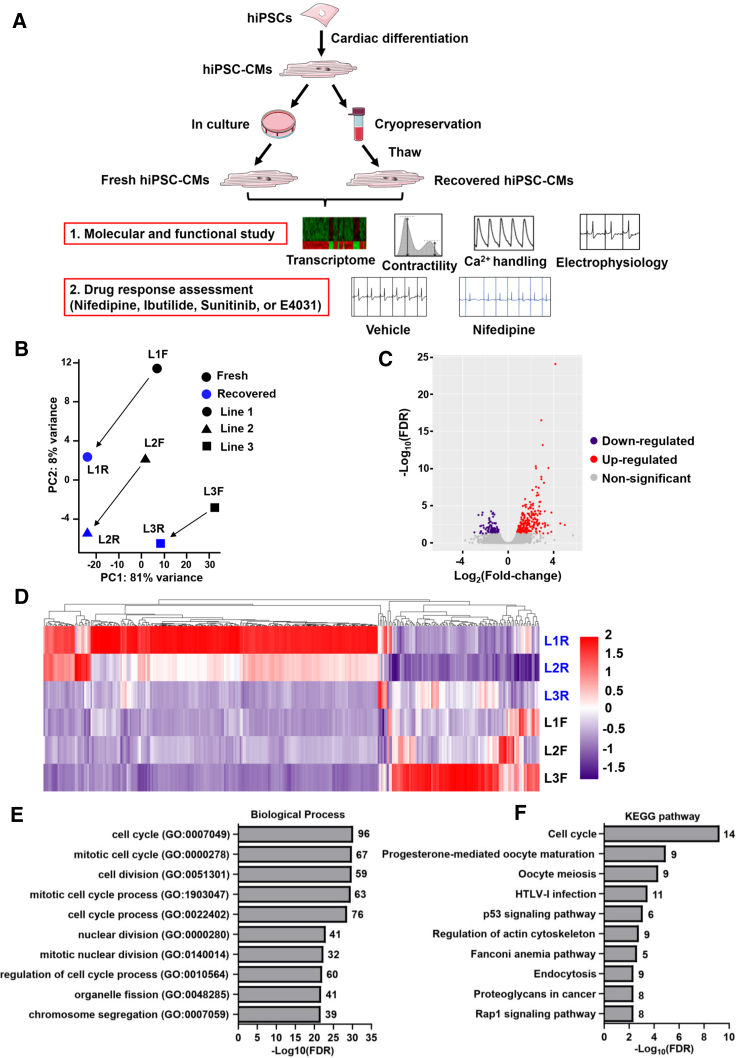
Figure 1.
Molecular Signature of Fresh and Recovered hiPSC-CMs
(A) Schematic outline of the study design. Healthy control hiPSC lines were differentiated into hiPSC-CMs that were either continuously cultured for further experiments or cryopreserved and recovered later. Transcriptome and functionality of fresh and recovered hiPSC-CMs were characterized. Moreover, the drug response of fresh and recovered hiPSC-CMs was assessed.
(B) PCA showed the transcriptomic differences between fresh and recovered hiPSC-CMs from three lines.
(C) Volcano plot showed the downregulated, non-significant, and upregulated genes in recovered hiPSC-CMs.
(D) Heatmap showed the hierarchical clustering of DEGs.
(E and F) Gene Ontology (GO) and KEGG analysis revealed the top 10 cellular processes that were enriched in recovered hiPSC-CMs. Numbers indicate the number of genes found in each pathway. L, line; F, fresh; R, recovered.
Recovered hiPSC-CMs Show Upregulation of Cell Cycle Genes
To better understand the effects of cryopreservation on hiPSC-CMs, we performed RNA sequencing (RNA-seq) analysis on both fresh and recovered hiPSC-CMs in three hiPSC lines. Principal component analysis (PCA) revealed that recovered hiPSC-CMs from individual lines had the same variation shift on the first two PC axes compared with their corresponding fresh hiPSC-CMs, suggesting that differentially expressed genes (DEGs) may have coordinated variations after cryopreservation (Figure 1B). Using the fold change over fresh control, we identified 316 DEGs with 97 downregulated and 219 upregulated genes (Figure 1C). Heatmap of the DEGs showed that recovered hiPSC-CMs from lines 1 and 2 exhibited more fold change in the upregulated genes than line 3, suggesting a line-dependent effect (Figure 1D). When we examined the DEGs, we found that GJA5 encoding CONNEXIN 40 was upregulated in recovered hiPSC-CMs, but not at the protein level, and the expression of genes associated with other ion channels, Ca2+ handling, and sarcomere structure was not statistically different between fresh and recovered hiPSC-CMs (Figures S2A–S2C and Table S1). Additionally, the MYH7/MYH6 ratio, an indicator of hiPSC-CM maturation (Veerman et al., 2015), was not statistically different between fresh and recovered hiPSC-CMs (Figure S2D). Furthermore, pathway analysis indicated that upregulated genes in recovered hiPSC-CMs were primarily associated with the cell cycle (Figures 1E and 1F and Table S1). To quantify proliferating hiPSC-CMs, cells were incubated with 5-bromo-2′-deoxyuridine (BrdU) overnight followed by an examination of BrdU immunostaining (Figures S2E and S2F). The data indicated that recovered hiPSC-CMs exhibited an enhanced cell proliferation compared with age-matched fresh hiPSC-CMs (Figure S2G).
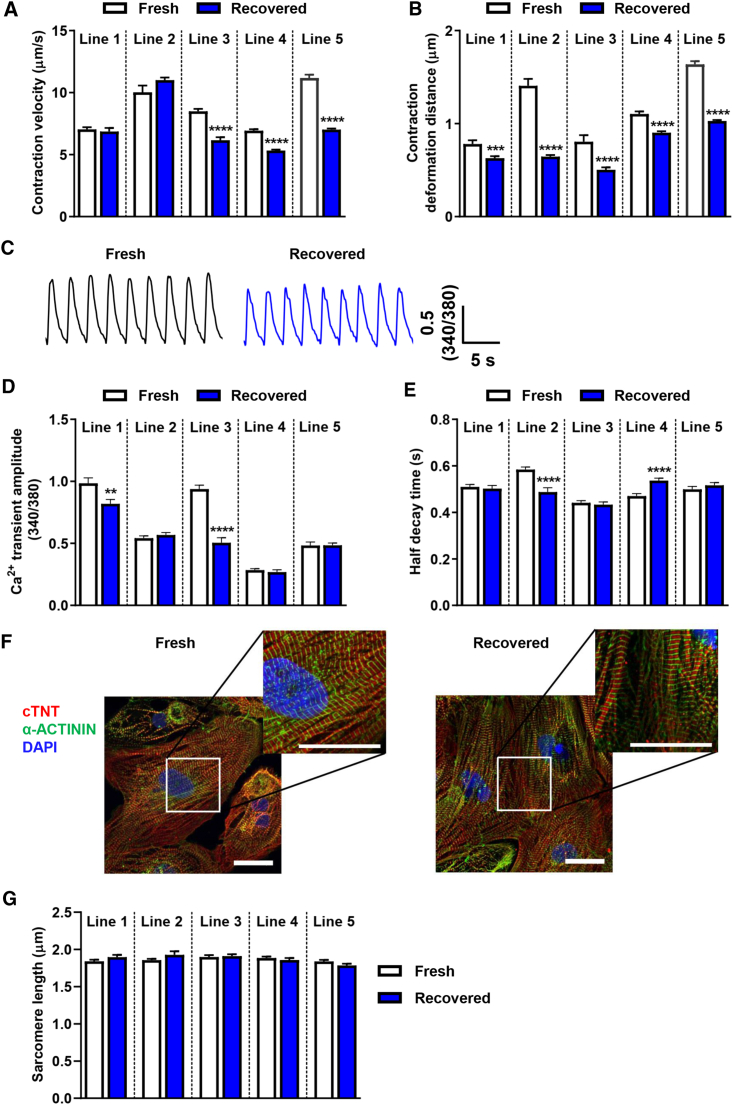
Recovered hiPSC-CMs Show Reduced Contractility
To understand whether the functionality of recovered hiPSC-CMs is comparable with fresh hiPSC-CMs, we examined the contraction of hiPSC-CMs using a motion tracking system (Kitani et al., 2019). Recovered hiPSC-CMs showed a significantly reduced contraction velocity in three out of five lines, while recovered hiPSC-CMs from all five lines showed a significant decrease in contraction deformation distance (Figures 2A and 2B). Because Ca2+ acts as the second messenger during excitation-contraction coupling, it plays an important role in regulating cardiomyocyte contractile function. Thus, we compared the Ca2+ handling of field-stimulated (0.5-Hz) fresh hiPSC-CMs with that of recovered hiPSC-CMs using the Ca2+ fluorescent indicator, Fura-2 (Figure 2C). Consistent with the contractility findings, cryopreservation had a line-dependent effect on Ca2+ handling. For example, recovered hiPSC-CMs from lines 1 and 3 showed a significant decrease in Ca2+ transient amplitude compared with their fresh controls (Figure 2D). However, cryopreservation had a mixed impact on the half-decay time (Figure 2E). These data show that cryopreservation has a line-dependent effect on Ca2+ handling, and the reduced Ca2+ transient amplitude may partially contribute to the reduced contractile function. Immunostaining of the sarcomere structure using anti-cTNT and anti-α-ACTININ antibody revealed that the sarcomere length of recovered hiPSC-CMs was similar to that of fresh hiPSC-CMs (Figures 2F and 2G).
Figure 2.
Recovered hiPSC-CMs Show Altered Contractility and Ca2+ Kinetics in a Line-Dependent Manner
(A and B) Recovered hiPSC-CMs from five lines showed reduced contraction velocity and contraction deformation distance (line 1, n = 20 and 35; line 2, n = 15 and 30; line 3, n = 30 and 30; line 4, n = 50 and 50; line 5, n = 50 and 50 independent measurements in fresh and recovered groups, respectively). Data were collected from three independent differentiations for each line and the assay was repeated three times.
(C) Representative Ca2+ transients of line 1 fresh (black) and recovered hiPSC-CMs (blue).
(D and E) Ca2+ transient amplitude and half-decay time of Ca2+ transients were altered in recovered hiPSC-CMs in a line-dependent manner (line 1, n = 42 and 48; line 2, n = 75 and 82; line 3, n = 55 and 52; line 4, n = 44 and 51; line 5, n = 48 and 58 cells in fresh and recovered groups, respectively). Data were collected from three to five independent coverslip preparations from three independent differentiations for each line.
(F) Immunostaining of line 1 fresh and recovered hiPSC-CMs showed the expression of cTNT (red), α-ACTININ (green), and nuclear stain DAPI (blue). Scale bars represent 30 μm.
(G) Comparable sarcomere length between fresh and recovered groups across cell lines (n = 36 per group from six randomly selected regions).
All data represented as mean ± SEM, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 compared with their respective fresh samples.
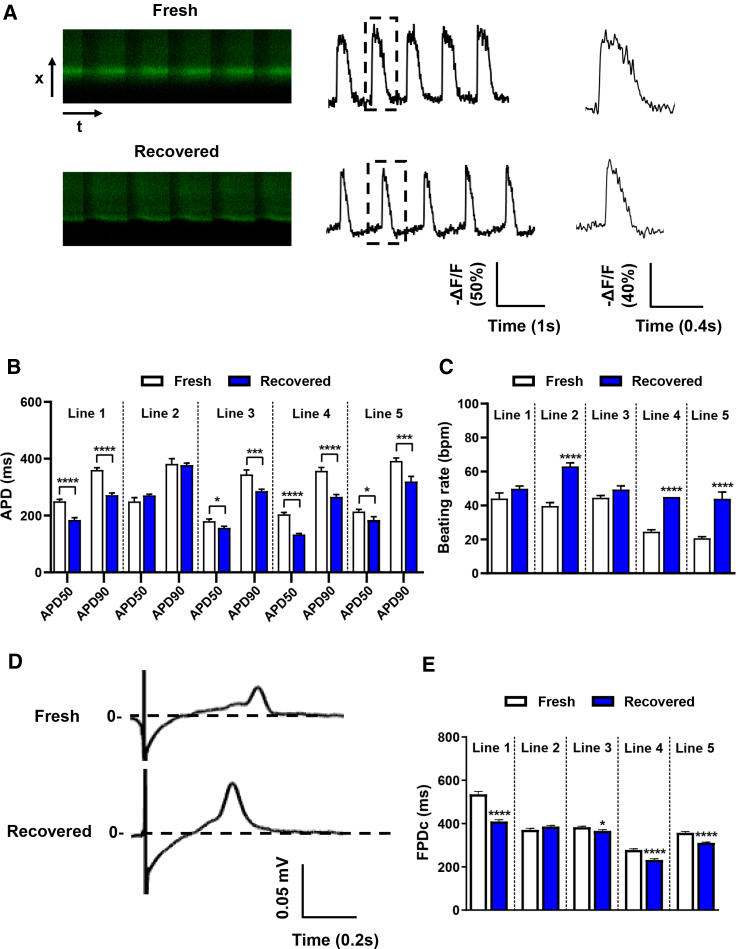
Recovered hiPSC-CMs Show Altered Electrophysiological Properties at Both Single-Cell and Monolayer Levels
To understand how cryopreservation affects hiPSC-CM electrophysiological properties, we next examined the electrical activity at the single-cell level using optical imaging and at the monolayer level using microelectrode arrays (MEAs). Previously, we demonstrated the usefulness of the voltage sensor Allosteric Sensor for Action Potentials 2 (ASAP2) for optical imaging of single-cell cardiac action potentials (APs) (Ma et al., 2018; Zhang et al., 2019). Using the same strategy, we examined the cardiac APs in fresh and recovered hiPSC-CMs (Figure 3A). Compared with fresh hiPSC-CMs, recovered hiPSC-CMs from four lines showed a shorter AP duration 50 (APD50) and 90 (APD90) with a range of reduction from ∼20 to 80 ms, whereas only line 2 hiPSC-CMs exhibited similar APD50 and APD90 between fresh and recovered groups (Figure 3B). Recovered hiPSC-CMs also showed a trend toward increased beating rate (Figure 3C). These findings suggest that recovered hiPSC-CMs may have different electrophysiological properties.
Figure 3.
Recovered hiPSC-CMs Show Altered Electrophysiology
(A) Representative single-cell AP recordings of line 1 fresh and recovered hiPSC-CMs using ASAP2.
(B) APD50 and APD90 were altered in recovered hiPSC-CMs from four out of five lines (line 1, n = 50 and 50; line 2, n = 59 and 48; line 3, n = 33 and 25; line 4, n = 14 and 21; line 5, n = 22 and 21 cells in fresh and recovered groups, respectively). Data were collected from three independent dish preparations from three independent differentiations for lines 1, 2, and 3, and two independent differentiations for lines 4 and 5.
(C) Beating rate of fresh and recovered hiPSC-CMs measured using ASAP2.
(D) Representative MEA recordings showed that FPD was shorter in recovered hiPSC-CMs.
(E) Recovered hiPSC-CMs from four out of five lines showed altered FPDc (line 1, n = 101 and 114; line 2, n = 84 and 78; line 3, n = 94 and 76; line 4, n = 93 and 92; line 5, n = 94 and 93 independent measurements in fresh and recovered groups, respectively).
Data were collected from two independent differentiations for each line. All data represented as mean ± SEM, ∗p < 0.05, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 compared with their respective fresh samples.
To further examine hiPSC-CM electrical activity at the monolayer level, we conducted an MEA study that is commonly used for investigating drug-induced cardiotoxicity (Blinova et al., 2017, 2018; Nozaki et al., 2017). Consistent with the single-cell optical imaging study, recovered hiPSC-CMs had a shorter rate-corrected field potential duration (FPDc) in four out of five lines (lines 1, 3, 4, and 5), whereas line 2 showed a comparable FPDc between fresh and recovered cells (Figures 3D and 3E). The cell viability of fresh and recovered hiPSC-CMs was comparable (Figures S2H–S2J), thus these results indicate that recovered hiPSC-CMs can generate an electrical influx propagating through a monolayer, but that electrical influx can be influenced by the cryopreservation/thaw process.
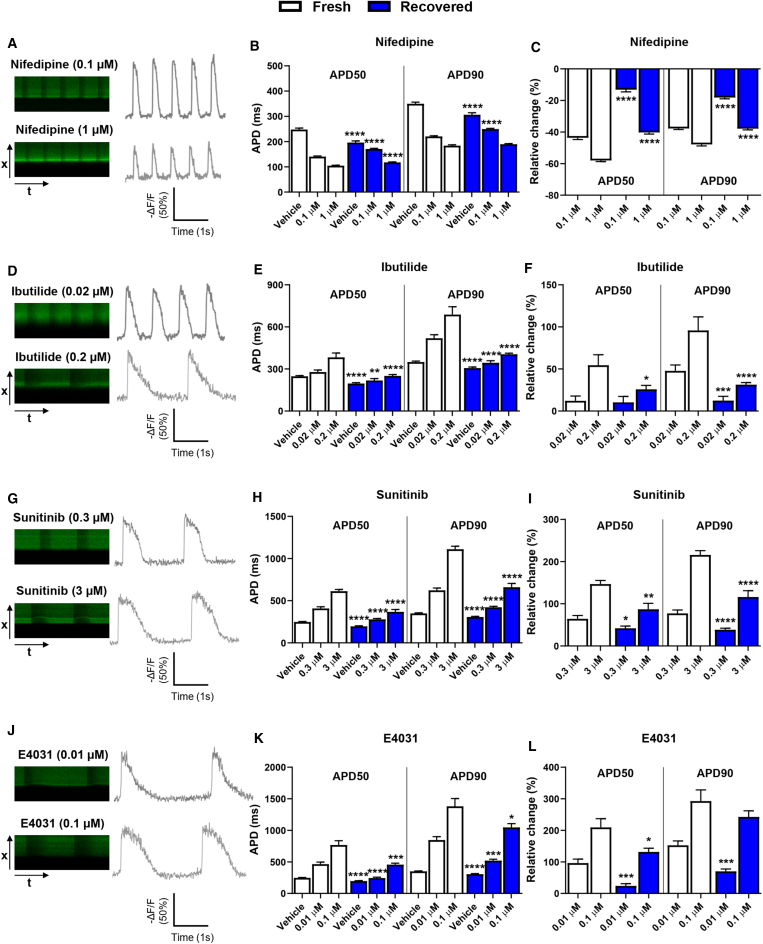
Recovered hiPSC-CMs Show Altered Drug Response at the Cellular Level
hiPSC-CMs have emerged as a promising platform for assessing cardiac safety liabilities (see review by Paik et al., 2020). However, it remains unclear whether recovered hiPSC-CMs have a comparable drug sensitivity with their fresh counterparts. Therefore, we tested four drugs (nifedipine, ibutilide, sunitinib, and E4031) that affect the QT interval and are associated with different levels of TdP risk (Colatsky et al., 2016; Guo et al., 2013). We first used the single-cell optical imaging approach to investigate the drug-induced APD changes with two different doses for each drug. Due to the low-throughput nature of single-cell imaging, we investigated the hiPSC-CM drug response using only lines 1, 2, and 3. Nifedipine, an L-type Ca2+ channel blocker, is known to shorten APD by reducing the L-type Ca2+ current (ICaL). Both fresh and recovered hiPSC-CMs showed a dose-dependent effect on the decrease in APD50 and APD90 (Figures 4A–4C). Recovered hiPSC-CMs from lines 1 and 2 had a blunted response (Figures 4A–4C, Figures S3A and S3B), whereas recovered hiPSC-CMs from line 3 appeared to show a greater drug response than fresh cells at the high dose (Figures S4A and S4B).
Figure 4.
Recovered hiPSC-CMs Show Altered Drug Response at the Cellular Level
Representative single-cell AP recordings using ASAP2 in line 1 fresh hiPSC-CMs treated with (A) nifedipine, (D) ibutilide, (G) sunitinib, and (J) E4031 with two different doses. Line 1 recovered hiPSC-CMs showed blunted changes in APD50 and APD90 compared with fresh hiPSC-CMs when cells treated with (B and C) nifedipine (fresh, n = 33 [0.01 μM], n = 19 [0.1 μM]; recovered, n = 18 [0.01 μM], n = 21 [0.1 μM]), (E and F) ibutilide (fresh, n = 14 [0.02 μM], n = 15 [0.2 μM]; recovered, n = 23 [0.02 μM], n = 30 [0.2 μM]), (H and I) sunitinib (fresh, n = 13 [0.3 μM], n = 15 [3 μM]; recovered, n = 18 [0.3 μM], n = 15 [3 μM]), and (K and L) E4031 (fresh, n = 22 [0.01 μM], n = 13 [0.1 μM]; recovered, n = 12 [0.01 μM], n = 11 [0.1 μM]). Raw APD50 and APD90 data are presented in (B), (E), (H), and (K). Data of relative change to vehicle are presented in (C), (F), (I), and (L). The cell number of vehicle was 93 and 89 in fresh and recovered groups, respectively, and the same dataset was used for plotting graphs in four drug treatments. Data were collected from three independent dish preparations from two independent differentiations. All data represented as mean ± SEM, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 compared with their respective fresh samples at the same dose.
Ibutilide, sunitinib, and E4031 have been reported to prolong APD and increase the risk of TdP (Blinova et al., 2018; Guo et al., 2013; Hortigon-Vinagre et al., 2016). Indeed, all three drugs caused APD prolongation in a dose-dependent manner in line 1 and again recovered hiPSC-CMs showed a blunted response compared with fresh hiPSC-CMs (Figures 4D–4L). Line 2 recovered hiPSC-CMs had a reduced drug response to ibutilide and E4031, but a comparable response to sunitinib compared with their fresh control (Figures S3C–S3H). Line 3 recovered hiPSC-CMs appeared to show a greater drug response than fresh cells (Figures S4C–S4H). Taken together, these findings suggest that recovered hiPSC-CMs tend to have different drug responses.
Recovered hiPSC-CMs Have an Altered Drug Sensitivity with an Enhanced Propensity for Drug-Induced Arrhythmias
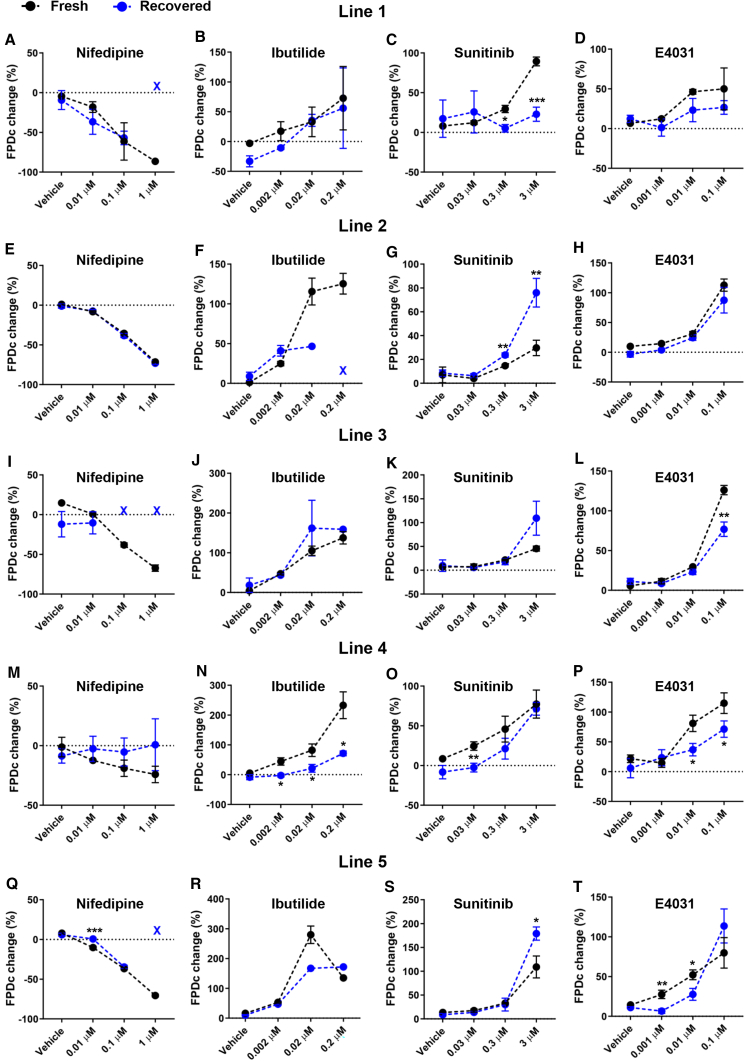
Next, we investigated whether the altered drug response in recovered hiPSC-CMs can also be observed using MEAs that were previously used in the CiPA study (Blinova et al., 2018). For each condition, the FPDc and the arrhythmic events were evaluated using five cell lines. Fresh hiPSC-CMs from line 1 showed a gradual decrease in FPDc with increasing doses of nifedipine, and the dose-dependent effect was consistently recapitulated in fresh cells of the other four cell lines (Figures 5A, 5E, 5I, 5M, 5Q, and S5A). However, lines 1, 3, and 5 recovered hiPSC-CMs showed no signal at the highest dose (Figures 5A, I, and Q). Only line 2 fresh and recovered hiPSC-CMs showed a comparable drug response to nifedipine (Figure 5E). It should be noted that the extent of nifedipine-induced FPDc shortening is comparable with that previously reported (Blinova et al., 2018), demonstrating the consistency across different experimental sites.
Figure 5.
Recovered hiPSC-CMs Show Altered Drug Responses at the Monolayer Level
(A–D) Drug-induced changes in FPDc in line 1 fresh and recovered hiPSC-CMs.
(E–H) Drug-induced changes in FPDc in line 2 fresh and recovered hiPSC-CMs.
(I–L) Drug-induced changes in FPDc in line 3 fresh and recovered hiPSC-CMs.
(M–P) Drug-induced changes in FPDc in line 4 fresh and recovered hiPSC-CMs.
(Q–T) Drug-induced changes in FPDc in line 5 fresh and recovered hiPSC-CMs. Color-coded marks (X) indicate no measurements under that drug treatment condition due to drug-induced cardiac arrest.
All data represented as mean ± SEM, from three to six independent measurements, ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 compared with their respective fresh samples at the same dose.
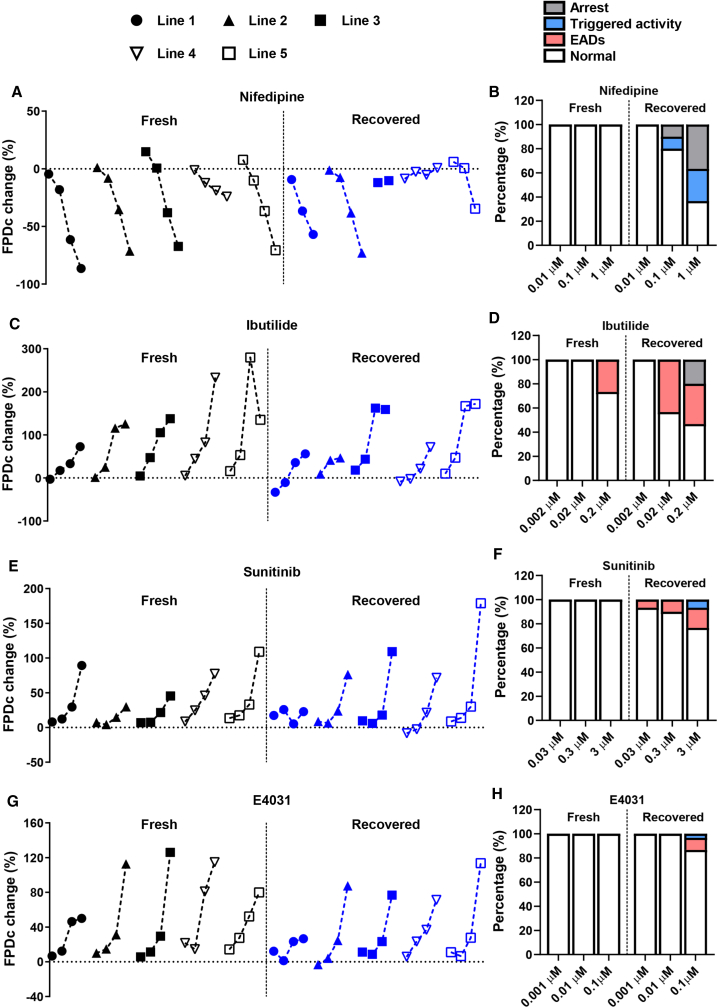
Consistent with the results observed for nifedipine, fresh hiPSC-CMs showed a dose-dependent increase in FPDc with increasing doses of ibutilide across five lines except for the highest dose in line 5 (Figures 5B, 5F, 5J, 5N, and 5R and Figure S5B). Fresh hiPSC-CMs showed either similar or larger FPDc prolongation caused by ibutilide than recovered hiPSC-CMs. Similarly, fresh and recovered hiPSC-CMs showed a dose-dependent sunitinib-induced FPDc prolongation except for line 1 recovered hiPSC-CMs (Figures 5C, 5G, 5K, 5O, 5S, and S5C). Recovered hiPSC-CMs from lines 1, 3, and 4 had a similar or smaller drug response to sunitinib (Figures 5C, 5K, and 5O), whereas recovered hiPSC-CMs from lines 2 and 5 had a statistically larger response at the highest dose (3 μM) (Figures 5G and 5S). The trend of drug response to E4031 was relatively consistent between fresh and recovered hiPSC-CMs, but the extent of response was generally smaller in recovered hiPSC-CMs (Figures 5D, 5H, 5L, 5P, and 5T and Figure S5D). To further investigate whether the altered drug response is attributed to changes in the proportion of cardiomyocyte subtypes in recovered hiPSC-CMs, we conducted single-cell patch clamp experiments using three lines due to its low-throughput feature. Consistent with multiple studies (Argenziano et al., 2018; Burridge et al., 2014; Hwang et al., 2015), the majority of subtype was ventricular in both fresh and recovered groups (Figures S6A and S6B and Table S2). We did not observe consistent changes in the proportion of certain subtypes in the recovered group (Figure S6B), suggesting that the altered drug response is not attributable to the proportion of cardiomyocyte subtypes. Only maximum dV/dt (derivative of voltage with respect to time) consistently decreased in recovered hiPSC-CMs from all three lines (Figure S6C), but the overall maximum diastolic potential (MDP) and AP amplitude were comparable between fresh and recovered groups, despite a decrease in MDP in line 4 recovered hiPSC-CMs (Figures S6D and S6E). Overall, fresh hiPSC-CMs consistently showed dose-dependent FPDc changes across different cell lines; however, recovered hiPSC-CMs showed the altered drug sensitivity (Figures 6A, 6C, 6E, and 6G).
Figure 6.
Recovered hiPSC-CMs Are More Susceptible to Drug-Induced Arrhythmic Events
Summary of (A) nifedipine (vehicle, 0.01, 0.1, and 1 μM), (C) ibutilide (vehicle, 0.002, 0.02, and 0.2 μM), (E) sunitinib (vehicle, 0.03, 0.3, and 3 μM), and (G) E4031 (vehicle, 0.001, 0.01, and 0.1 μM)-induced changes in FPDc in fresh and recovered hiPSC-CMs from five lines. For individual lines, symbols from left to right indicate vehicle and the three tested doses for each drug as shown above, respectively. Doses that induce cardiac arrest in some lines are not shown (please refer to Figure 5). Pooled percentage of (B) nifedipine, (D) ibutilide, (F) sunitinib, and (H) E4031-induced arrhythmic events in fresh and recovered hiPSC-CMs. Data were collected from three to six independent measurements from five cell lines.
During the same experiments, we evaluated the arrhythmic susceptibility of fresh and recovered hiPSC-CMs under each drug treatment condition. Three types of arrhythmic events were identified by MEA: arrest (no electrical signal), triggered activity, and early after-depolarizations (EADs) (Figures S5E and S5F). Interestingly, fresh hiPSC-CMs did not show any nifedipine-induced arrhythmic events, whereas recovered hiPSC-CMs exhibited a higher frequency of arrhythmic events with increasing doses of nifedipine (Figure 6B and Table S3). Consistent with the high-risk prediction of TdP for ibutilide, the highest dose induced EADs in fresh hiPSC-CMs, and this effect was further exacerbated in recovered hiPSC-CMs (Figure 6D and Table S3). For sunitinib and E4031, we only observed arrhythmic events in recovered hiPSC-CMs but not fresh hiPSC-CMs (Figures 6F and 6H and Table S3). The enhanced propensity for drug-induced arrhythmic events was also observed at the single-cell level (Figures S6F–S6H). Moreover, some recovered but not fresh hiPSC-CMs from all three lines consistently showed AP alternans that are a risk factor predisposing to drug-induced arrhythmias (Grabowski et al., 2004) (Figures S6I and S6J). These findings indicate that recovered hiPSC-CMs are more susceptible to drug-induced cardiac arrhythmias compared with fresh hiPSC-CMs: they exhibit a lower threshold for triggering arrhythmias under drug doses that still maintain normal rhythm in fresh cells.
Discussion
Accumulating evidence has demonstrated that hiPSC-CMs are a reliable cellular platform for assessing acute drug-induced proarrhythmic risk (Blinova et al., 2017, 2018; Cerignoli et al., 2012; Hortigon-Vinagre et al., 2016) and chronic drug-induced contractile and structural liabilities (Kitani et al., 2019; Kopljar et al., 2017; Pointon et al., 2013). To achieve the assessment goals, a large number of “off-the-shelf” hiPSC-CMs are usually necessary. Cryopreservation has been the standard approach for long-term storage of hiPSC-CMs that can be readily thawed for use; however, it remains unclear how different fresh and recovered hiPSC-CMs are in terms of genetic signature, functionality, and (more importantly) the pharmacological response. To address these gaps in knowledge, we designed our study to systemically characterize the comparability between fresh and recovered hiPSC-CMs from cryopreservation. Here, we report that (1) transcriptome profiling shows enhanced cell proliferative capacity in recovered hiPSC-CMs; (2) recovered hiPSC-CMs appear to have reduced contractile properties and Ca2+ transient amplitude, as well as shortened APD or FPDc in a line-dependent manner; (3) recovered hiPSC-CMs show an altered drug response in some cell lines; and (4) recovered hiPSC-CMs exhibit a lower threshold for drug-induced arrhythmic events.
The cryopreservation/thaw process not only disrupts the cell-cell and cell-matrix adhesion leading to anoikis but also increases the generation of ROS and activates multiple cellular stress pathways that enhance apoptosis (Preininger et al., 2016). One study showed an enhanced apoptotic activity of human embryonic stem cell-derived cardiomyocytes 5 days after thawing (Kim et al., 2011), thus improving the viability of recovered cells has been one of the major focuses in cryobiology. Recently, a small molecule cocktail has been shown to improve the survival of hiPSC-CMs and hiPSC-derived motor neurons by 36% and 63%, respectively, after re-plating (Chen et al., 2019). Despite using different cryoprotectants, the viability of cryopreserved hiPSC-CMs after thawing still ranges from ∼50% to 90% (Chen et al., 2015; Kim et al., 2011; Xu et al., 2011). Here, we showed ∼60% viability of hiPSC-CMs even after 18-month cryopreservation. This observation could be underestimated due to the approach we used here, as the percentage of dead cells was ∼20% and there was a small percentage of calcein/EthD-1 double-negative cells. Although cryopreservation caused cell death, the remaining live cells exhibited an upregulation of cell cycle and division genes and were more proliferative compared with their fresh counterparts. This finding is consistent with a previous report showing an enhanced proliferative capacity of recovered human atrial tissues from cryopreservation (Yokomuro et al., 2010). It should also be noted that the enhanced proliferation of recovered hiPSC-CMs could reflect their less mature status, which is contrary to a recent study showing cryopreservation may actually promote maturation of hiPSC-CMs (Brink et al., 2020). At both genetic (Figure S2) and functional levels (Figure S6), we did not observe improved maturation in recovered hiPSC-CMs. The exact reasons for the inconsistencies between the two studies are not clear, but could be attributed to the different number of lines studied, different genetic background of donor sources, different seeding densities, and different cryopreservation approaches used. Therefore, a standardized protocol of culturing, differentiation, cryopreservation, and re-plating may be necessary for the field to minimize variations.
Interestingly, the recent study also reported that cryopreservation does not adversely affect cardiomyocyte function (Brink et al., 2020). However, only one cell line was used in the contraction study, and the measured parameters (CD90 [contraction duration at 90% peak amplitude], time to peak, and relaxation time) only partially reflected the contractile performance. By contrast, here we used high-speed video microscopy with motion vector analysis to precisely measure the contraction velocity (Hayakawa et al., 2014; Kitani et al., 2019; Kopljar et al., 2017; Lam et al., 2019). The reduced contractility could not be associated with the overt sarcomere structure change, which was also not identified by the high-resolution electron micrograph in a previous study (Kim et al., 2011). In two lines (lines 1 and 3), we found that Ca2+ transient amplitude was reduced, which may lead to reduced contractility. Other factors such as myofilament Ca2+ sensitivity may also affect contractile machinery function. Moreover, it should be noted that Ca2+ handling was measured at the single-cell level. Despite the comparable Ca2+ transient amplitude between fresh and recovered hiPSC-CMs in the other lines, whether changes in Ca2+ propagation in monolayer contribute to the reduced contraction will need further investigation. In addition to the mechanical force, electrophysiological properties were also changed in recovered hiPSC-CMs, which were unlikely caused by changes in the expression of ion channel genes. This suggests that ion channel trafficking, membrane targeting, and ion channel gating may play a more important role in determining the electrophysiological properties in recovered hiPSC-CMs.
Drug-induced cardiovascular liability remains one of the major contributors to the attrition of new chemical entities and is associated with 16% of withdrawn or discontinued drugs (Siramshetty et al., 2016). Such liability presents in various forms, from acute cardiac repolarization leading to arrhythmias to chronic mechanical dysfunction resulting in heart failure. According to crediblemeds.org, 15% of drugs on the market can cause QT prolongation and 4% are associated with TdP risk. Moreover, it has been estimated that up to 60% of drug candidates may have potential cardiac repolarization liability (Munawar et al., 2018). Increasingly, hiPSC-CMs are being used for cardiac risk assessment to better predict drug-induced arrhythmias with fewer false-positives under the CiPA program (Authier et al., 2017). The primary goal of our study is to investigate whether recovered hiPSC-CMs show the drug response comparable with their fresh counterparts. We observed that recovered cells tend to have lesser drug-induced repolarization changes in most tested drugs and doses. This can be partially explained by the shorter APD in recovered cells at baseline, as it has been suggested that drug-induced changes in APD are correlated with baseline APD values, and it also highlights the lack of electrical plasticity of recovered cells (Barandi et al., 2010a, 2010b; Cummins et al., 2014).
Another significant finding of this study is that recovered hiPSC-CMs are more susceptible to drug-induced arrhythmic events. This observation seems to contradict the premise that prolonged repolarization increases the arrhythmic risk since recovered hiPSC-CMs appear to have a shorter APD or FPDc. This paradox suggests a shift in the drug sensitivity of recovered hiPSC-CMs and a lack of gradual increment in the drug response that could result in a lower threshold for arrhythmias, which may impair the prediction accuracy of drug toxicity when cryopreserved hiPSC-CMs are used in pre-clinical drug testing. Furthermore, the increased frequency of alternans in recovered hiPSC-CMs may also explain the enhanced propensity for drug-induced arrhythmias as it is well known that alternans create a substrate conducive to arrhythmias (Rosenbaum et al., 1994; Weiss et al., 2011).
In summary, we systematically compared the genetic and functional characteristics between fresh and recovered hiPSC-CMs from cryopreservation and determined their drug responses. Our data provide additional insights but should not be interpreted as showing the categorical unsuitability of using cryopreserved hiPSC-CMs for drug screening and toxicity testing. Instead, the results raise awareness on potential limitations and the necessity of establishing a sensible frame of reference to take into account changes in drug responses induced by cryopreservation, which could help improve the prediction accuracy of drug toxicity in the future.
Experimental Procedures
Differentiation of hiPSCs to hiPSC-CMs
When hiPSCs reached a confluency of 85%, the E8 medium was replaced with RPMI supplemented with B27 without insulin (Life Technologies) and 6 μM CHIR-99021 (Selleck Chemicals), and cells were transferred to a humidified incubator with 5% CO2 and 21% O2 at 37°C for 48 h. The medium was changed to RPMI-B27 without insulin for 24 h, and then to RPMI-B27 without insulin supplemented with 5 μM IWR-1 (Selleck Chemicals) for 48 h. On day 5, the medium was changed back to RPMI-B27 without insulin for 48 h, and then switched to RPMI-B27 for 72 h. On day 11, the medium was changed to RPMI-B27 without D-glucose (Life Technologies) for 72 h to allow metabolic purification of hiPSC-CMs. Cells were then dissociated after a 6-min incubation with Accutase (Sigma Aldrich) at 37°C followed by seeding into 6-well plates cultured with RPMI-B27 supplemented with 5% KnockOut Serum Replacement (KOSR) (Thermo Fisher Scientific). After 24 h, the medium was changed to RPMI-B27, followed by a second metabolic purification as described above. After the second purification, hiPSC-CMs were continuously cultured in RPMI-B27 for further experiments and described as fresh hiPSC-CMs.
Cryopreservation of hiPSC-CMs
On day 23 post differentiation, hiPSC-CMs were dissociated using Accutase as described above. Approximately 2 million hiPSC-CMs were re-suspended in 1 mL of freezing medium that contains 90% fetal bovine serum (FBS) (Life Technologies) and 10% DMSO (Sigma Aldrich) and stored in cryovials in isopropanol-filled containers at −80°C overnight. Cryovials were then transferred to liquid nitrogen for long-term storage.
Thawing Frozen hiPSC-CMs
Cryovials were removed from liquid nitrogen and quickly placed in a 37°C water bath to thaw the cells until ∼5-mm diameter ice crystal was left. Thawed cells were then transferred into 9 mL of cold RPMI-B27 supplemented with 5% KOSR followed by centrifugation at 300 × g at room temperature. Cells were re-suspended in 1 mL of RPMI-B27 with 5% KOSR and re-plated in a well of the 6-well plate. After 24 h, the medium was changed to RPMI-B27. Cells that had been recovered for 1 week were used for experiments except for single-cell patch clamp experiments (recovered for 2 weeks).
Phase-Contrast Video-Image-Based Contraction Assays
Phase-contrast cell motion movies of cardiomyocyte contraction were captured using a high-speed digital CMOS (complementary metal oxide semiconductor) camera (KP-FM400WCL, Hitachi Kokusai Denki Engineering) mounted on an inverted microscope (Eclipse Ti, Nikon). Video images of the hiPSC-CM monolayer (50,000 cells per well) in 96-well plates (37°C and 5% CO2) were recorded as sequential phase-contrast images using SI8000C View Software (Sony) with a 10× objective (NA [numerical aperture]: 0.3) at a frame rate of 75 frames per second (fps) and a resolution of 1,024 × 1,024 pixels. Data analysis was performed using SI8000C Analyzer software (Sony).
Field Potential Recordings
The field potential recordings were performed using the MEA platform (Maestro, Axion Biosystems). Before recording, hiPSC-CMs were enzymatically dissociated using TrypLE Select Enzyme (10X) (Thermo Fisher Scientific) for 6 min at 37°C and 50,000 cells were plated on the electrode area in 48-well MEA plates (Axion Biosystems) that were previously coated with Matrigel. Thawed hiPSC-CMs were directly seeded on plates. MEA experiments were performed 7 days after seeding at 37°C in a 5% CO2 environment. Three concentrations of each drug were used. Concentrated (2×) testing solutions for each concentration were prepared freshly on the day of testing by diluting DMSO stocks into the RPMI-B27 medium, then the prepared dilution was warmed at 37°C to be at the same temperature as the cells. The targeted concentration was attained when drugs were added to the MEA plate to avoid removing the medium that will modify the temperature in the well and apply unwanted shear stress on the cells. The same amount of DMSO was used as vehicle controls. Each concentration of drugs was tested in individual MEA plate wells, so no cells were exposed to multiple concentrations of drugs. Data acquisition was performed using AxIS Navigator software (Axion Biosystem). Data analysis was achieved using the Cardiac Analysis Tool (Axion Biosystems), and FPD was rate-corrected (FPDc) using the Frederica formula, which is a built-in function in the Cardiac Analysis Tool. Data were presented as percentage change between baseline and drug testing recordings. Arrhythmic events such as arrest, triggered activity, and EADs were identified manually.
Statistical Analysis
Data were analyzed and graphed using Prism (GraphPad). Data were presented as mean ± SEM. Comparisons were conducted via unpaired two-tailed Student's t test with significance defined by p < 0.05 (∗), p < 0.01 (∗∗), p < 0.001 (∗∗∗), and p < 0.0001 (∗∗∗∗).
Data and Code Availability
The accession number for the RNA-seq data in this paper is GSE146266.
Author Contributions
J.Z.Z. designed the study; conducted contractility, optical imaging, and biochemical studies; and wrote the manuscript. N.B. conducted MEA experiments, single-cell patch clamp, and wrote the manuscript. T.Z. differentiated cells and conducted flow cytometry. Y.L. analyzed RNA-seq data. R.S. differentiated hiPSC-CMs. J.C.W. designed the study, wrote the manuscript, and provided funding support.
Conflicts of Interest
J.C.W. is a cofounder of Khloris Biosciences but has no competing interests, as the work presented here is completely independent. The other authors declare no competing interests.
Acknowledgments
This work was supported by American Heart Association 17MERIT3361009, Burroughs Wellcome Fund IRSA 1015009, Leducq Fondation 18CVD05, and National Institutes of Health R01 HL130020 and R01 HL133272 (J.C.W.).
Published: December 17, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.11.010.
Supplemental Information
References
- Ando H., Yoshinaga T., Yamamoto W., Asakura K., Uda T., Taniguchi T., Ojima A., Shinkyo R., Kikuchi K., Osada T. A new paradigm for drug-induced torsadogenic risk assessment using human iPS cell-derived cardiomyocytes. J. Pharmacol. Toxicol. Methods. 2017;84:111–127. doi: 10.1016/j.vascn.2016.12.003. [DOI] [PubMed] [Google Scholar]
- Argenziano M., Lambers E., Hong L., Sridhar A., Zhang M., Chalazan B., Menon A., Savio-Galimberti E., Wu J.C., Rehman J. Electrophysiologic characterization of calcium handling in human induced pluripotent stem cell-derived atrial cardiomyocytes. Stem Cell Reports. 2018;10:1867–1878. doi: 10.1016/j.stemcr.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Authier S., Pugsley M.K., Koerner J.E., Fermini B., Redfern W.S., Valentin J.-P., Vargas H.M., Leishman D.J., Correll K., Curtis M.J. Proarrhythmia liability assessment and the comprehensive in vitro proarrhythmia assay (CiPA): an industry survey on current practice. J. Pharmacol. Toxicol. Methods. 2017;86:34–43. doi: 10.1016/j.vascn.2017.02.021. [DOI] [PubMed] [Google Scholar]
- Barandi L., Harmati G., Horvath B., Szentandrassy N., Magyar J., Varro A., Nanasi P.P., Banyasz T. Drug-induced changes in action potential duration are proportional to action potential duration in rat ventricular myocardium. Gen. Physiol. Biophys. 2010;29:309–313. doi: 10.4149/gpb_2010_03_309. [DOI] [PubMed] [Google Scholar]
- Barandi L., Virag L., Jost N., Horvath Z., Koncz I., Papp R., Harmati G., Horvath B., Szentandrassy N., Banyasz T. Reverse rate-dependent changes are determined by baseline action potential duration in mammalian and human ventricular preparations. Basic Res. Cardiol. 2010;105:315–323. doi: 10.1007/s00395-009-0082-7. [DOI] [PubMed] [Google Scholar]
- Blinova K., Dang Q., Millard D., Smith G., Pierson J., Guo L., Brock M., Lu H.R., Kraushaar U., Zeng H. International multisite study of human-induced pluripotent stem cell-derived cardiomyocytes for drug proarrhythmic potential assessment. Cell Rep. 2018;24:3582–3592. doi: 10.1016/j.celrep.2018.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blinova K., Stohlman J., Vicente J., Chan D., Johannesen L., Hortigon-Vinagre M.P., Zamora V., Smith G., Crumb W.J., Pang L. Comprehensive translational assessment of human-induced pluripotent stem cell derived cardiomyocytes for evaluating drug-induced arrhythmias. Toxicol. Sci. 2017;155:234–247. doi: 10.1093/toxsci/kfw200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink L.v.d., Brandão K.O., Yiangou L., Mol M.P.H., Grandela C., Mummery C.L., Verkerk A.O., Davis R.P. Cryopreservation of human pluripotent stem cell-derived cardiomyocytes is not detrimental to their molecular and functional properties. Stem Cell Res. 2020;43:101698. doi: 10.1016/j.scr.2019.101698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge P.W., Matsa E., Shukla P., Lin Z.C., Churko J.M., Ebert A.D., Lan F., Diecke S., Huber B., Mordwinkin N.M. Chemically defined generation of human cardiomyocytes. Nat. Methods. 2014;11:855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerignoli F., Charlot D., Whittaker R., Ingermanson R., Gehalot P., Savchenko A., Gallacher D.J., Towart R., Price J.H., McDonough P.M. High throughput measurement of Ca2+ dynamics for drug risk assessment in human stem cell-derived cardiomyocytes by kinetic image cytometry. J. Pharmacol. Toxicol. Methods. 2012;66:246–256. doi: 10.1016/j.vascn.2012.08.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.Y., Matsa E., Wu J.C. Induced pluripotent stem cells: at the heart of cardiovascular precision medicine. Nat. Rev. Cardiol. 2016;13:333–349. doi: 10.1038/nrcardio.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Tristan C.A., Chen L., Jovanovic V.M., Malley C., Chu P.-H., Ryu S., Deng T., Ormanoglu P., Tao D. A versatile polypharmacology platform promotes cytoprotection and viability of human pluripotent and differentiated cells. bioRxiv. 2019:815761.3. doi: 10.1038/s41592-021-01126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V.C., Ye J., Shukla P., Hua G., Chen D., Lin Z., Liu J.C., Chai J., Gold J., Wu J. Development of a scalable suspension culture for cardiac differentiation from human pluripotent stem cells. Stem Cell Res. 2015;15:365–375. doi: 10.1016/j.scr.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colatsky T., Fermini B., Gintant G., Pierson J.B., Sager P., Sekino Y., Strauss D.G., Stockbridge N. The comprehensive in vitro proarrhythmia assay (CiPA) initiative- update on progress. J. Pharmacol. Toxicol. Methods. 2016;81:15–20. doi: 10.1016/j.vascn.2016.06.002. [DOI] [PubMed] [Google Scholar]
- Cummins M.A., Dalal P.J., Bugana M., Severi S., Sobie E.A. Comprehensive analyses of ventricular myocyte models identify targets exhibiting favorable rate dependence. PLoS Comput. Biol. 2014;10:e1003543. doi: 10.1371/journal.pcbi.1003543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski M., Karpinski G., Filipiak K.J., Opolski G. Images in cardiovascular medicine. Drug-induced long-QT syndrome with macroscopic T-wave alternans. Circulation. 2004;110:e459–e460. doi: 10.1161/01.CIR.0000147541.49575.15. [DOI] [PubMed] [Google Scholar]
- Guo L., Coyle L., Abrams R.M., Kemper R., Chiao E.T., Kolaja K.L. Refining the human iPSC-cardiomyocyte arrhythmic risk assessment model. Toxicol. Sci. 2013;136:581–594. doi: 10.1093/toxsci/kft205. [DOI] [PubMed] [Google Scholar]
- Hayakawa T., Kunihiro T., Ando T., Kobayashi S., Matsui E., Yada H., Kanda Y., Kurokawa J., Furukawa T. Image-based evaluation of contraction-relaxation kinetics of human-induced pluripotent stem cell-derived cardiomyocytes: correlation and complementarity with extracellular electrophysiology. J. Mol. Cell. Cardiol. 2014;77:178–191. doi: 10.1016/j.yjmcc.2014.09.010. [DOI] [PubMed] [Google Scholar]
- Hortigon-Vinagre M.P., Zamora V., Burton F.L., Green J., Gintant G.A., Smith G.L. The use of ratiometric fluorescence measurements of the voltage sensitive dye di-4-ANEPPS to examine action potential characteristics and drug effects on human induced pluripotent stem cell-derived cardiomyocytes. Toxicol. Sci. 2016;154:320–331. doi: 10.1093/toxsci/kfw171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H.S., Kryshtal D.O., Feaster T.K., Sánchez-Freire V., Zhang J., Kamp T.J., Hong C.C., Wu J.C., Knollmann B.C. Comparable calcium handling of human iPSC-derived cardiomyocytes generated by multiple laboratories. J. Mol. Cell. Cardiol. 2015;85:79–88. doi: 10.1016/j.yjmcc.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.Y., Ku S.Y., Liu H.C., Cho H.J., Oh S.K., Moon S.Y., Choi Y.M. Cryopreservation of human embryonic stem cells derived-cardiomyocytes induced by BMP2 in serum-free condition. Reprod. Sci. 2011;18:252–260. doi: 10.1177/1933719110385130. [DOI] [PubMed] [Google Scholar]
- Kitani T., Ong S.G., Lam C.K., Rhee J.W., Zhang J.Z., Oikonomopoulos A., Ma N., Tian L., Lee J., Telli M.L. Human-induced pluripotent stem cell model of trastuzumab-induced cardiac dysfunction in patients with breast cancer. Circulation. 2019;139:2451–2465. doi: 10.1161/CIRCULATIONAHA.118.037357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopljar I., De Bondt A., Vinken P., Teisman A., Damiano B., Goeminne N., Van den Wyngaert I., Gallacher D.J., Lu H.R. Chronic drug-induced effects on contractile motion properties and cardiac biomarkers in human induced pluripotent stem cell-derived cardiomyocytes. Br. J. Pharmacol. 2017;174:3766–3779. doi: 10.1111/bph.13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopljar I., Lu H.R., Van Ammel K., Otava M., Tekle F., Teisman A., Gallacher D.J. Development of a human iPSC cardiomyocyte-based scoring system for cardiac hazard identification in early drug safety de-risking. Stem Cell Reports. 2018;11:1365–1377. doi: 10.1016/j.stemcr.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam C.K., Tian L., Belbachir N., Wnorowski A., Shrestha R., Ma N., Kitani T., Rhee J.W., Wu J.C. Identifying the transcriptome signatures of calcium channel blockers in human induced pluripotent stem cell-derived cardiomyocytes. Circ. Res. 2019;125:212–222. doi: 10.1161/CIRCRESAHA.118.314202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N., Zhang J.Z., Itzhaki I., Zhang S.L., Chen H., Haddad F., Kitani T., Wilson K.D., Tian L., Shrestha R. Determining the pathogenicity of a genomic variant of uncertain significance using CRISPR/Cas9 and human-induced pluripotent stem cells. Circulation. 2018;138:2666–2681. doi: 10.1161/CIRCULATIONAHA.117.032273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munawar S., Windley M.J., Tse E.G., Todd M.H., Hill A.P., Vandenberg J.I., Jabeen I. Experimentally validated pharmacoinformatics approach to predict hERG inhibition potential of new chemical entities. Front. Pharmacol. 2018;19:1035. doi: 10.3389/fphar.2018.01035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki Y., Honda Y., Watanabe H., Saiki S., Koyabu K., Itoh T., Nagasawa C., Nakamori C., Nakayama C., Iwasaki H. CSAHi study-2: validation of multi-electrode array systems (MEA60/2100) for prediction of drug-induced proarrhythmia using human iPS cell-derived cardiomyocytes: assessment of reference compounds and comparison with non-clinical studies and clinical information. Regul. Toxicol. Pharmacol. 2017;88:238–251. doi: 10.1016/j.yrtph.2017.06.006. [DOI] [PubMed] [Google Scholar]
- Paik D.T., Chandy M., Wu J.C. Patient and disease–specific induced pluripotent stem cells for discovery of personalized cardiovascular drugs and therapeutics. Pharmacol. Rev. 2020;72:320–342. doi: 10.1124/pr.116.013003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg D.E. Principles of cryopreservation. Methods Mol. Biol. 2015;1257:3–19. doi: 10.1007/978-1-4939-2193-5_1. [DOI] [PubMed] [Google Scholar]
- Pointon A., Abi-Gerges N., Cross M.J., Sidaway J.E. Phenotypic profiling of structural cardiotoxins in vitro reveals dependency on multiple mechanisms of toxicity. Toxicol. Sci. 2013;132:317–326. doi: 10.1093/toxsci/kft005. [DOI] [PubMed] [Google Scholar]
- Preininger M.K., Singh M., Xu C. Cryopreservation of human pluripotent stem cell-derived cardiomyocytes: strategies, challenges, and future directions. Adv. Exp. Med. Biol. 2016;951:123–135.2. doi: 10.1007/978-3-319-45457-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum D.S., Jackson L.E., Smith J.M., Garan H., Ruskin J.N., Cohen R.J. Electrical alternans and vulnerability to ventricular arrhythmias. N. Engl. J. Med. 1994;330:235–241. doi: 10.1056/NEJM199401273300402. [DOI] [PubMed] [Google Scholar]
- Sager P.T., Gintant G., Turner J.R., Pettit S., Stockbridge N. Rechanneling the cardiac proarrhythmia safety paradigm: a meeting report from the Cardiac Safety Research Consortium. Am. Heart J. 2014;167:292–300. doi: 10.1016/j.ahj.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Shiba Y., Fernandes S., Zhu W.Z., Filice D., Muskheli V., Kim J., Palpant N.J., Gantz J., Moyes K.W., Reinecke H. Human ES-cell-derived cardiomyocytes electrically couple and suppress arrhythmias in injured hearts. Nature. 2012;489:322–325. doi: 10.1038/nature11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siramshetty V.B., Nickel J., Omieczynski C., Gohlke B.-O., Drwal M.N., Preissner R. WITHDRAWN--a resource for withdrawn and discontinued drugs. Nucleic Acids Res. 2016;44:D1080–D1086. doi: 10.1093/nar/gkv1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerman C.C., Kosmidis G., Mummery C.L., Casini S., Verkerk A.O., Bellin M. Immaturity of human stem-cell-derived cardiomyocytes in culture: fatal flaw or soluble problem? Stem Cells Dev. 2015;24:1035–1052. doi: 10.1089/scd.2014.0533. [DOI] [PubMed] [Google Scholar]
- Weiss J.N., Nivala M., Garfinkel A., Qu Z., Winslow R. Alternans and arrhythmias. Circ. Res. 2011;108:98–112. doi: 10.1161/CIRCRESAHA.110.223586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Police S., Hassanipour M., Li Y., Chen Y., Priest C., O'Sullivan C., Laflamme M.A., Zhu W.Z., Van Biber B. Efficient generation and cryopreservation of cardiomyocytes derived from human embryonic stem cells. Regen. Med. 2011;6:53–66. doi: 10.2217/rme.10.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomuro H., Shiono N., Ozawa T., Fujii T., Watanabe Y., Koyama N., Okada M. Effect of cryopreservation on cell proliferation and immunogenicity of transplanted human heart cells. Ann. Thorac. Cardiovasc. Surg. 2010;16:105–112. [PubMed] [Google Scholar]
- Zhang J.Z., Guo H., Wu J.C. Applications of genetically engineered human pluripotent stem cell reporters in cardiac stem cell biology. Curr. Opin. Biotechnol. 2018;52:66–73. doi: 10.1016/j.copbio.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.Z., Termglinchan V., Shao N.Y., Itzhaki I., Liu C., Ma N., Tian L., Wang V.Y., Chang A.C.Y., Guo H. A human iPSC double-reporter system enables purification of cardiac lineage subpopulations with distinct function and drug response profiles. Cell Stem Cell. 2019;24:802–811.e5. doi: 10.1016/j.stem.2019.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Chen H., Xiao D., Yang H., Itzhaki I., Qin X., Chour T., Aguirre A., Lehmann K., Kim Y. Comparison of non-human primate versus human induced pluripotent stem cell-derived cardiomyocytes for treatment of myocardial infarction. Stem Cell Rep. 2018;10:422–435. doi: 10.1016/j.stemcr.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNA-seq data in this paper is GSE146266.