Summary
Mammary gland ductal morphogenesis depends on the differentiation of mammary stem cells (MaSCs) into basal and luminal lineages. The AP-2γ transcription factor, encoded by Tfap2c, has a central role in mammary gland development but its effect in mammary lineages and specifically MaSCs is largely unknown. Here, we utilized an inducible, conditional knockout of Tfap2c to elucidate the role of AP-2γ in maintenance and differentiation of MaSCs. Loss of AP-2γ in the basal epithelium profoundly altered the transcriptomes and decreased the number of cells within several clusters of mammary epithelial cells, including adult MaSCs and luminal progenitors. AP-2γ regulated the expression of genes known to be required for mammary development, including Cebpb, Nfkbia, and Rspo1. As a result, AP-2γ-deficient mice exhibited repressed mammary gland ductal outgrowth and inhibition of regenerative capacity. The findings demonstrate that AP-2γ can regulate development of mammary gland structures potentially regulating maintenance and differentiation of multipotent MaSCs.
Keywords: mammary gland, development, Tfap2c, AP-2γ
Highlights
-
•
AP-2γ-deficient mice exhibit repressed ductal outgrowth and regenerative capacity
-
•
Loss of AP-2γ reduced cells within mammary stem and luminal progenitor clusters
-
•
AP-2γ target genes, including Cebpb, Nfkbia, and Rspo1, regulate mammary development
-
•
AP-2γ potentially regulates maintenance of multipotent mammary stem cells
Gu and colleagues utilized a conditional knockout of Tfap2c to examine transcriptional effects of AP-2γ on mammary development. AP-2γ-deficient mice have decreased numbers of cell populations harboring mammary stem cells and have altered expression of genes required for mammary development, including Cebpb, Nfkbia, and Rspo1. They demonstrate that AP-2γ is implicated in maintenance of multipotent mammary stem cells.
Introduction
The ductal system of the mouse mammary gland develops postnatally from a single rudimentary duct that extends from the nipple during the first 3 weeks postpartum (Ball, 1998; Silberstein, 2001). With the start of the prepubertal period occurring at age 4–5 weeks in mice, the mammary gland becomes responsive to reproductive hormones that initiate a growth period with terminal end bud (TEB) structures driving ductal morphogenesis. TEBs are club-shaped structures at the tip of growing ducts, which are composed of two cell types—an inner layer of body cells that give rise to the luminal mammary epithelial cells and a single layer of undifferentiated cap cells that develop into the myoepithelial layer (Morris and Stein, 2017; Williams and Daniel, 1983). By age 9 weeks, invasion of the TEBs into the stroma generates a mammary tree that fills the mammary fat pad resulting in a mammary gland characteristic of mature virgin animals.
Adult tissue stem cells are classically characterized as undifferentiated multipotent cells capable of regenerating the structure of the organ or tissue through cell division (Post and Clevers, 2019). The existence of mammary stem cells (MaSCs) was first demonstrated by showing that adult mammary tissue could reproduce the mammary gland when transplanted into the cleared fat pad of 3-week-old syngeneic mice (Deome et al., 1959). The frequency of MaSCs defined by mammary repopulating units (MRUs) in transplantation assays was estimated to be approximately 2% of basal cells (BCs) (Fu et al., 2019; Shackleton et al., 2006; Stingl et al., 2006). The mouse MaSC population has the expression profile of Lin− CD24+ CD29high CD49fhigh and expresses cytokeratins 5 and 14 (encoded by the Krt5 and Krt14 genes, respectively). Consistent with the hypothesis that multipotent stem cells reside within the basal layer, BC-derived organoids are capable of forming structures comprised of inner cuboidal luminal cells capable of milk production surrounded by a myoepithelial layer with elongated morphology, whereas luminal cell-derived organoids lack a myoepithelial layer (Jamieson et al., 2017).
Mammary gland developmental programs are characterized by changes in the transcriptome orchestrated by several transcription factors (Pal et al., 2017; Wuidart et al., 2018). The AP-2 transcription factor family is required for normal embryogenesis and proper embryonic development of neural crest derivatives, and epidermal and urogenital tissues (Hilger-Eversheim et al., 2000; Winger et al., 2006). AP-2 factors also play an important role in regulating mammary gland development. In the mouse mammary gland, AP-2α and AP-2γ are expressed within the nuclei of basal and luminal mammary epithelial cells by age 7 weeks and both factors are expressed within the body and cap cells of the TEB (Jager et al., 2010; Zhang et al., 2003). Studies have examined the effects of AP-2α overexpression driven by a transgene using the MMTV promoter (Zhang et al., 2003). The initial phase of mammary gland development was normal with mature virgin 8-week-old mice expressing the transgene, demonstrating normal mammary tree structures; however, at 6 months mice expressing the transgene demonstrated a sparser ductal network with reduced alveolar buds. During pregnancy lobuloalveolar tissue in mice expressing the transgene had reduced proliferation and increased apoptosis. Parallel studies in mice overexpressing AP-2γ demonstrated hyperproliferation, as determined by increased Ki-67 expression and bromodeoxyuridine incorporation, as well as increased apoptosis, with the overall effect resulting in hypoplasia of the alveolar epithelium during pregnancy (Jager et al., 2003).
Genetic knockout (KO) of Tfap2c results in early embryonic lethality due to loss of AP-2γ expression in extra-embryonic membranes (Auman et al., 2002). Conditional knockout (CKO) of Tfap2c using the Sox2 promoter to drive Cre recombinase showed that loss of AP-2γ impaired mammary gland branching during the prepubertal period (Jager et al., 2010). Loss of AP-2γ resulted in a reduction in the number of branch points and maximal ductal length but did not impair the generation of TEBs. By 8 months, the mammary ducts had completely filled the fat pad in virgin Tfap2c CKO mice; however, there was a reduction in the degree of branching, showing that AP-2γ controls the speed of ductal elongation and the development of tertiary branches and lateral buds. In another study, CKO of Tfap2c using the MMTV promoter to drive Cre resulted in loss of AP-2γ expression in the luminal epithelial cells only (Cyr et al., 2015). Mammary gland branching was delayed in Tfap2c CKO mice with a reduction in branch points and an increase in the relative proportion of BCs compared with luminal cells.
The previous experimental models offered a limited ability to form conclusions on the role of AP-2γ in the basal and MaSC populations and were not able to identify AP-2γ target genes necessary for mammary gland development. To advance our understanding of the role of AP-2γ in mammary gland development, here we used a transgene that uses the Krt5 promoter to drive expression of a tamoxifen-inducible Cre-ER, allowing control over the timing of Tfap2c KO and also directing the loss of AP-2γ to the BC population that includes MaSCs.
Results
Loss of AP-2γ Impaired Mammary Gland Ductal Outgrowth
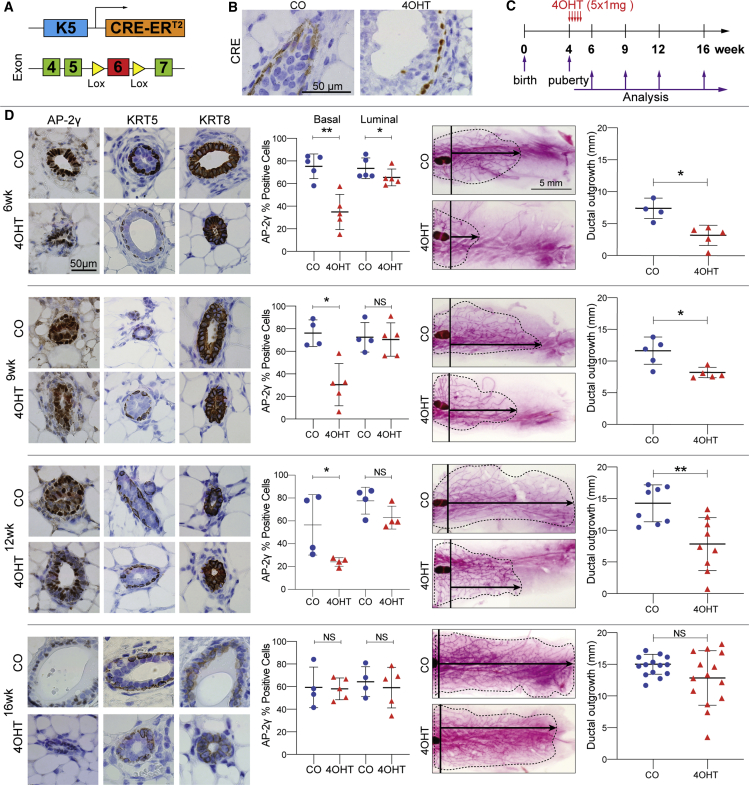
To control the timing of Tfap2c KO in mouse MaSCs, we utilized FVB mice expressing a tamoxifen-inducible Cre recombinase fused to the estrogen receptor with expression driven by the bovine keratin 5 promoter (Indra et al., 1999) (Figure 1A). We confirmed that localization of CRE was restricted to the cytoplasm in the basal layer with nuclear localization induced by exposure to 4-hydroxy-tamoxifen (4OHT) (Figure 1B). The floxed Tfap2c allele was designed to delete exon 6, which disrupts expression of functional AP-2γ protein. FVB/Tfap2cfl/fl/Krt5-Cre-ERT2 mice were pulsed with corn oil (CO) or 4OHT at a dose previously shown not to interfere with morphogenesis (Scheele et al., 2017). Genetically identical female littermates were treated for 5 days beginning in early puberty at age 4 weeks and analyzed for AP-2γ expression with immunohistochemistry and mammary gland branching was assessed by whole mount at 6, 9, 12, and 16 weeks (Figures 1C and 1D). Significantly reduced AP-2γ expression was demonstrated in the BCs at 6, 9, and 12 weeks with a slight reduction of expression in the luminal cells only at 6 weeks. Differences in AP-2γ expression resolved by 16 weeks. Loss of exon 6 within the Tfap2c mRNA was detected in the basal mammary epithelial cells (MMECs) but not in the luminal cells within 1 week of treatment with 4OHT (Figure S1A). A delay in mammary gland ductal elongation was demonstrated with CKO of Tfap2c at 6, 9, and 12 weeks. However, no significant differences in mammary gland ductal structure were found at 16 weeks comparing the 4OHT- and CO-treated mice. A parallel set of experiments were performed in FVB/Krt5-Cre-ERT2 mice demonstrating that tamoxifen treatment using an identical protocol had no effect on mammary gland ductal morphogenesis (Figures S1B and S1C). The findings suggest that CKO of Tfap2c was either mosaic or failed to target a precursor cell population, so that wild-type cells could outcompete mutant cells and lead to an eventual recovery of ductal outgrowth. Collectively, the findings indicate that AP-2γ plays a critical role in mammary gland ductal elongation.
Figure 1.
Loss of AP-2γ Represses Mammary Ductal Outgrowth
(A) Krt5 promoter was used to drive expression of Cre recombinase to eliminate expression of AP-2γ through Lox sites designed to remove exon 6 of the Tfap2c gene.
(B) FVB/Tfap2cfl/fl/Krt5-Cre-ERT2 mice were pulsed with corn oil (CO) or 4-hydroxy-tamoxifen (4OHT) and Cre expression was examined by immunohistochemistry; treatment with 4OHT-induced nuclear CRE expression.
(C) Schematic showing treatment of mice with CO or 4OHT starting at age 4 weeks for 5 days; analysis was performed at age 6, 9, 12, and 16 weeks.
(D) Analysis of protein expression and whole mounts. Immunohistochemistry (left side) was used to examine loss of AP-2γ protein in MMECs with CKO of Tfap2c. Expression of KRT5 and KRT8 demonstrates basal and luminal MMECs, respectively. There was a significant reduction of AP-2γ expression noted at 6, 9, and 12 weeks in the (BC) compartment. Luminal cells had a significant reduction in AP-2γ expression noted at 6 weeks but this reduction did not persist at 9 and 12 weeks. At least three representative microscopic fields (40× magnification) were quantified per animal. The percentages were calculated by comparing stained cells to total epithelial cells. Results are expressed as mean ± SEM; number for CO/4OHT: n = 5/5 (6 weeks), n = 4/5 (9 weeks), n = 4/4 (12 weeks), n = 4/5 (16 weeks). ∗p < 0.05, ∗∗p < 0.01; n.s., not significant; statistical significance by Mann-Whitney U test. Representative whole mounts of mammary glands from mice at 6, 9, 12, and 16 weeks (right side); black arrows illustrate distances measured. Corresponding graphs show growth of the mammary tree measured from the lymph node to the most distant mammary branching demonstrating a significant reduction in growth of the mammary tree with CKO of Tfap2c, which resolves by 16 weeks. Results are represented as mean ± SEM; number for CO/4OHT mice: n = 4/5 (6 weeks), n = 5/5 (9 weeks), n = 8/9 (12 weeks), n = 14/14 (16 weeks); ∗p < 0.05, ∗∗p < 0.01; n.s., not significant; statistical significance by Mann-Whitney U test.
CKO of Tfap2c Alters MMEC Clusters and Patterns of Gene Expression
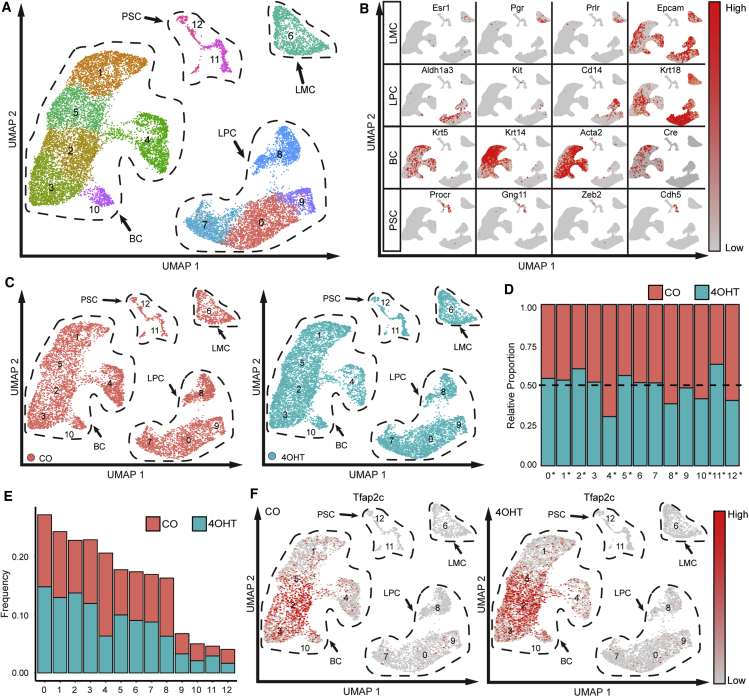
Single-cell RNA sequencing (scRNA-seq) was used to examine changes in the transcriptome in MMECs with loss of AP-2γ. Four-week-old FVB/Tfap2cfl/fl/Krt5-Cre-ERT2 mice were similarly pulsed with 4OHT or CO and mammary glands were harvested at age 9 weeks (Figure S1D). Single cells were generated and flow sorted to enrich luminal and basal mammary epithelial populations, which were combined 1:1 and analyzed with scRNA-seq (Figure 2). Of note, we did not find a statistically significant difference in the total numbers of basal and luminal cells harvested comparing CO- and 4OHT-treated FVB/Tfap2cfl/fl/Krt5-Cre-ERT2 animals. The RNA-seq data from the combined population of 7,866 MMECs from CO-treated animals and 10,350 MMECs from 4OHT-treated animals were analyzed using uniform manifold approximation and projection (UMAP), which identified 13 distinct MMEC clusters, referred to as clusters 0–12 (Figure 2A). The pattern of expression was used to identify the major MMEC groups—luminal mature cells, luminal progenitor cells (LPCs), BCs, and protein C receptor (PROCR+) stem cells (PSCs) (Wang et al., 2015; Sun et al., 2018) (Figures 2B and S2A). The pattern of gene expression for the PSC cluster matched the pattern of expression for the Procr+ population of multipotent MaSCs reported by Wang et al. (2015) (Figure S2B). CKO of Tfap2c significantly reduced the relative proportion of cells within clusters 4 and 8, belonging to the basal and luminal clusters, respectively (Figures 2C–2E). Less profound decreases were also identified in several other clusters, including cluster 10 (BC cluster) and 12 (PSC cluster). In addition, there was a significant increase in the relative proportion of cells in cluster 11, belonging to the PSC subgroup (Figures 2D and 2E). The Tfap2c gene was most highly expressed in the basal compartment with the highest percentage of Tfap2c-expressing cells in cluster 2 (Figure 2F). Scattered cells in LPC clusters 0, 7, 8, and 9 were also demonstrated to have relatively high Tfap2c expression. Although CKO eliminated AP-2γ protein expression in the basal layer (Figure 1), the mRNA with exon 6 deleted was expressed, as expected (Figure 2F).
Figure 2.
scRNA-Seq Analysis
(A) Experiments were performed in two independent replicates and in each experiment two CO- and two 4OHT-treated mice were used for mammary gland harvest for a total of n = 4 mice per condition. UMAP cluster analysis identified 13 mammary cell clusters; the transcriptome defined closely related clusters, including the BC, luminal progenitor cluster (LPC), mature luminal cluster (LMC), and the Procr+ stem cell cluster (PSC).
(B) Examples of gene expression used to classify the major groups of mammary cell clusters.
(C) UMAP clusters shown separately for CO-treated (orange) and 4OHT-treated (teal) animals.
(D) Relative proportion of mammary cells in each cluster for CO- and 4OHT-treated animals, where 0.50 would indicate no change in relative number of cells; ∗p < 0.05.
(E) Frequency of cells from CO- and 4OHT-treated mice shown by the relative amount of cells by cluster.
(F) Expression pattern of Tfap2c in CO and 4OHT-treated mice.
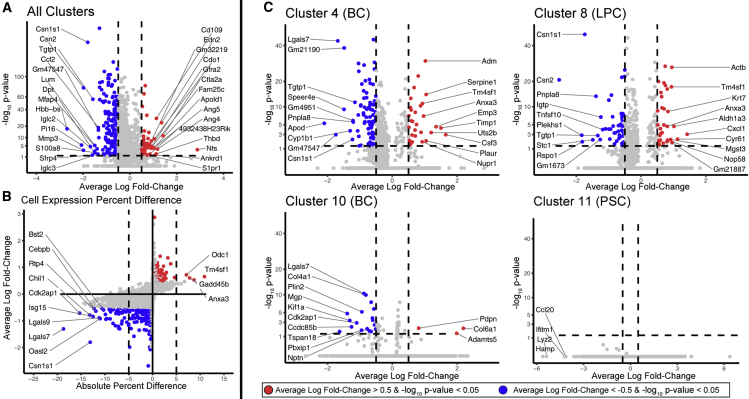
The data were analyzed to identify genes with significant changes in expression with loss of AP-2γ and this gene list was subsequently subjected to Ingenuity Pathway Analysis (IPA). IPA indicated that many genes altered by loss of AP-2γ were involved in cell cycle, migration, and differentiation (Table S1). There was a total of 644 genes that demonstrated significant changes in expression with CKO of Tfap2c (Table S2). Using a difference based on a log-fold change of ±0.5 and significance with p < 0.05, there was a greater number of genes repressed compared with those induced with CKO of Tfap2c (Figure 3A). When limited to genes with an absolute percentage difference of 5% of cells in the cluster, there were 30 genes repressed and 4 genes induced by loss of AP-2γ (Figures 3B and S3). When examined by cluster, changes in gene expression were noted in every cluster, except cluster 11 (Figures 3C and S4). When considering genes that demonstrated significant changes in four or more clusters, Cdk2ap1, Cebpb, Csn1s1, Hspa1a, Krt15, Lgals7, Plin2, Pnpla8, and Sp140 were repressed and Anxa3, Gadd45b, Krt18, Nfkbia, Odc1, and Tm4sf1 were induced with CKO of Tfap2c (Table S2). For all of these genes, changes in expression always included basal clusters (1, 2, 3, 4, 5, and 10), and for Lgals7 and Krt15, changes in expression were restricted to the basal clusters. Expression analysis of selected genes from bulk luminal and basal MMECs isolated at age 5 weeks confirmed the regulation by AP-2γ (Figure S5).
Figure 3.
Changes in Gene Expression with CKO of Tfap2c
(A) Volcano plot with vertical axis showing the −log10 p value and horizontal axis average log-fold change for genes altered across all MMECs with loss of AP-2γ; colored dots represent genes with p < 0.05 and average log-fold change >±0.5; blue is used for genes repressed and red is used for genes induced with KO of Tfap2c.
(B) Same data as shown in (A) plotted with average log-fold change versus the difference in the percentage of cells demonstrating expression of the genes. Labeled genes had an adjusted p value < 0.05 and ±5% difference.
(C) Volcano plots as presented in (A) for changes in gene expression for clusters 4, 8, 10, and 11.
CKO of Tfap2c Repressed Multipotency of MaSCs
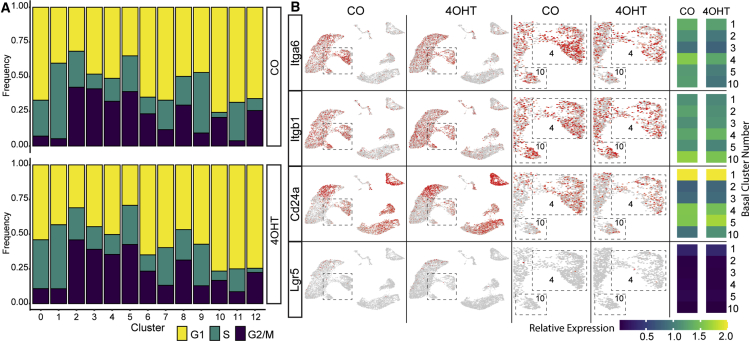
Previous studies (Fu et al., 2019; Visvader and Stingl, 2014) have defined MaSCs by expression of the markers Lin− KRT5+, CD49high, CD29high, and CD24+. LGR5 has been reported as another potential MaSC marker, but studies have been inconsistent showing repopulating activity in LGR5+ and LGR5− subsets (Visvader and Stingl, 2014). An examination of mouse mammary glands using scRNA-seq did not identify a single, unique cluster representing MaSCs, consistent with previous reports that a functional MaSC state may be generated from cells with different transcriptional signatures (Giraddi et al., 2018). The current analysis of mammary gland transcriptome using scRNA-seq did not define a MaSC cluster specifically (Figures 2 and 4); however, based on the pattern of expression of MaSC markers, a small subset of cells in clusters 4 and 10 may represent the MaSC population. Within the basal clusters, cluster 10 had the highest percentage of cells in G1/G0 and lowest percentage of cells in S phase (Figure 4A). CKO of Tfap2c reduced the number of cells within basal clusters 4 and 10 (Figures 2D and 2E) and the pattern of expression was consistent with cell populations harboring an MaSC phenotype; cells in clusters 4 and 10 were noted to be Itga6/CD49f+, Itgb1/CD29High, Cd24a/CD24+, and Lgr5Low (Figure 4B). Cells with the MaSC marker expression were reduced with KO of Tfap2c. The impact of AP-2γ deficiency on MaSC signature and on mammary gland ductal outgrowth suggested that CKO of Tfap2c potentially reduced the frequency and/or function of MaSCs.
Figure 4.
Changes in MaSCs with Loss of AP-2γ
(A) Cell cycle shown as proportion of cells in G1, S, and G2/M by mammary cell cluster for CO- and 4OHT-treated mice.
(B) Expression pattern for the Itga6, Itgb1, Cd24a, and Lgr5 genes from CO- and 4OHT-treated mice. Middle panel: magnified view of boxed area focused on basal clusters 4 and 10; right panel: heatmap showing relative expression of Itga6, Itgb1, Cd24a, and Lgr5 in basal MMEC clusters in CO- versus 4OHT-treated mice.
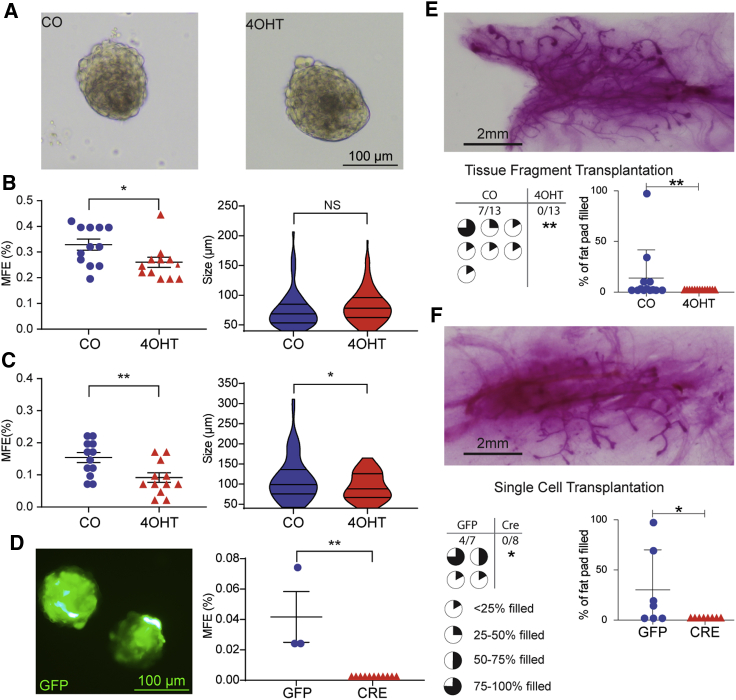
To examine changes in the functional properties of MaSCs, MMECs isolated from FVB/Tfap2cfl/fl/Krt5-Cre-ERT2 mice treated with CO versus 4OHT were examined for the ability to form mammospheres. MMECs isolated from 4OHT-treated mice demonstrated a significant reduction in the ability to form mammospheres, as measured by mammosphere-forming efficiency, compared with CO-treated mice (Figures 5A and 5B). The formation of secondary mammospheres has been advocated as a more reliable method to assess self-renewal of MaSC activity (Shaw et al., 2012). Secondary mammospheres were generated from primary mammospheres recovered from 4OHT- versus CO-treated animals; primary mammospheres from 4OHT-treated mice demonstrated a reduced capacity to form secondary mammospheres (Figure 5C). Secondary mammospheres recovered from mice with CKO of Tfap2c were also noted to be smaller compared with control mammospheres. To avoid potential confounding effects of 4OHT, basal MMECs from FVB/Tfap2cfl/fl mice were transduced with adenovirus vectors expressing green fluorescent protein (Ad-GFP) or GFP and Cre recombinase (Ad-Cre), subjected to fluorescence-activated cell sorting (FACS) to recover GFP+ cells and used to form mammospheres; whereas Ad-GFP-transduced cells formed mammospheres, Ad-Cre-transduced cells failed to form mammospheres (Figure 5D), consistent with the notion that KO of Tfap2c resulted in a loss of mammosphere-forming capability.
Figure 5.
Loss of AP-2γ Reduces Mammospheres and Mammary Gland Reconstitution
Mammospheres formed from MMECs harvested from 9-week-old FVB/Tfap2cfl/fl/Krt5-Cre-ERT2 mice that had been treated starting at 4 weeks with CO versus 4OHT.
(A) Examples of primary mammospheres after 14 days of culture.
(B) Data for mammosphere-forming efficiency (MFE) and size of mammospheres; n = 12 plating replicates of 4,000 cells per condition; ∗p < 0.05; n.s., not significant, Student's t test.
(C) Secondary mammospheres formed from primary mammospheres described in (A and B) with data for MFE and size; n = 12 plating replicates of 4,000 cells per condition ∗p < 0.05, ∗∗p < 0.01, Student’s t test.
(D) Examples of GFP+ mammospheres in Ad5CMVeGFP-transduced MMECs cultured for 14 days and MFE for Ad5CMVeGFP and Ad5CMVCre-eGFP transduction; ∗∗p < 0.01, Student’s t test.
(E) Top panel shows example of reconstituted mammary gland branching from CO-treated animals; data demonstrated that mammary tree reconstitution was found with transplantation of mammary tissue from CO-treated but not 4OHT-treated mice, 7/13 versus 0/13, p < 0.01, Fisher's exact test; graph of percent filled results are mean ± SEM, ∗p < 0.05, Mann-Whitney U test.
(F) Top panel shows example of reconstituted mammary gland branching from Ad-GFP-transduced cells; data demonstrate reconstitution of the mammary tree with cells transduced with Ad-GFP but not with Ad-Cre, 4/7 versus 0/8, p < 0.05, Fisher's exact test; graph of percent filled results are mean ± SEM; ∗p < 0.05, Mann-Whitney U test.
Mammary gland transplantation was used to further demonstrate the effect of CKO of Tfap2c on functional MaSCs. Mammary tissue was isolated from 9-week-old mice that had been treated with CO or 4OHT for 5 days starting at age 4 weeks. Mammary tissue isolated from CO-treated animals was capable of reconstituting the mammary gland when transplanted into a cleared fat pad of 3-week-old syngeneic FVB mice, whereas, mammary tissue harvested from 4OHT-treated animals was not able to regenerate the mammary gland structures in cleared fat pads (Figure 5E). To further examine the effect of AP-2γ on mammary gland regenerative capacity, MMEC BCs were harvested from FVB/Tfap2cfl/fl mice and transduced with Ad-Cre or Ad-GFP. Basal MMECs transduced with Ad-GFP demonstrated the capacity to regenerate the mammary tree; however, cells transduced with Ad-Cre failed to demonstrate the capacity to regenerate mammary gland structures (Figure 5F). These data demonstrate a critical role for AP-2γ in maintenance of multipotent, functional MaSCs.
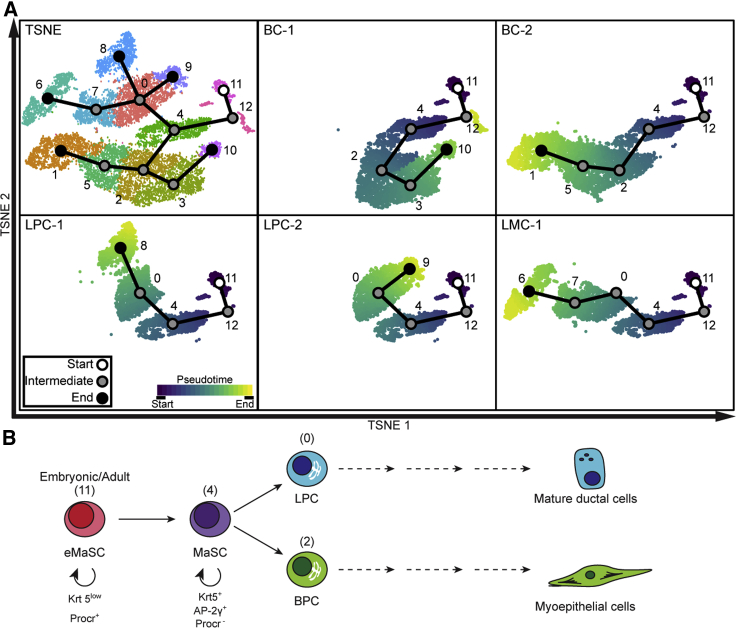
Pseudotime Analysis
Pseudotime analysis of scRNA-seq data was used to generate a model of mammary gland development within the context of Tfap2c expression (Figure 6). The earliest MMEC precursor was represented by the Procr+ cell clusters (11 and 12). Gene expression in the Procr+ cells include expression of known MaSC markers, including Zeb2, Gng11, Etv5, and Cdh5, but with low expression of basal lineage marker Krt5 (Figure 2), consistent with earlier studies indicating that the Procr+ cell population is a multipotent embryonic MaSC population (Wang et al., 2015). Our data showed that Krt5low/Procr+ embryonic MaSCs persist in adult mammary glands during puberty but were not affected by Tfap2c CKO (Figure 3C). The pseudotime trajectory starting at Procr+ clusters 11/12 differentiates to cluster 4, and provides additional evidence for this cluster containing KRT5+ MaSCs (Figure 6). From cluster 4, the differentiation pathway diverged into luminal and basal lineages terminating in either mature luminal or mature BC populations. Unsupervised pseudotime trajectory analysis using p-Creode further supported a stem cell role for the PSC cluster and close developmental relationship to cells in cluster 4 (Figure S6).
Figure 6.
Analysis of Pseudotime and Model of Mammary Cell Development
(A) Pseudotime analysis from scRNA-seq with Slingshot; first panel showing pseudotime overlying tSNE plot with same clustering as previous figures and remaining panels showing each individual lineage inferred from Slingshot.
(B) Model of mammary development supported by results with CKO of Tfap2c; numbers in parentheses refer to cluster number.
Discussion
This study provides important insight into the role of AP-2γ during the prepubertal stage of mammary gland development. CKO of Tfap2c within the KRT5+ BC compartment repressed mammary gland ductal outgrowth and inhibited the ability of mammary epithelial cells to form mammospheres and regenerate mammary gland structures. Where they can be compared, the current findings are consistent with previous reports that examined the role of AP-2γ in the mouse mammary gland (Cyr et al., 2015; Jager et al., 2010). As noted previously, Igfbp5 was identified as an AP-2γ target gene in the mammary gland (Jager et al., 2003) and we noted a significant repression of Igfbp5 expression in basal cluster 4 and luminal cluster 7 (Table S2). The current findings extend previous reports by clarifying the role of AP-2γ in the basal/MaSC compartment and by identifying AP-2γ target genes that regulate mammary gland development.
There is considerable debate concerning the potential role of multipotent KRT5+ MaSCs within the basal compartment in mammary gland outgrowth during the pubertal period (Lee et al., 2019). Lineage-tracing experiments using Krt5-CreER/Rosa-YFP mice treated with tamoxifen at age 4 weeks demonstrated labeling of the basal compartment exclusively (Van Keymeulen et al., 2011). Similarly, lineage-tracing experiments designed to label KRT14+ basal stem cells reinforced support for unipotent MaSCs in postpubertal mice (Wuidart et al., 2016). Additional studies have provided evidence that the bulk of mammary embryonic growth proceeds from lineage-restricted unipotent cells after embryonic day 15.5 (Lilja et al., 2018). On the other hand, transplantation experiments demonstrated that CD29highCD24+ BCs were capable of generating normal mammary gland structures. Lineage-tracing experiments reported by Rios et al. (2014), using high-resolution 3D imaging, demonstrated labeling of both the luminal and basal populations of the ductal tree when driven by the Krt5 promoter, supporting the model of a multipotent KRT5+ MaSC giving rise to mature basal and luminal cells during pubertal mammary gland development. Key differences in results may be influenced by techniques used for processing tissues and the use of 3D versus 2D imaging to visualize labeled cells (Rios et al., 2016).
One possibility for finding alteration of the transcriptome of some luminal cells is that CKO of Tfap2c in KRT5+ MaSCs gave rise to luminal cells before the loss of AP-2γ protein; however, once function AP-2γ protein was lost, the MaSCs with a deleted Tfap2c gene lost their multipotent ability. Previous studies in luminal breast cancer showed that knockdown of TFAP2C induced epithelial-mesenchymal transition with repression of luminal gene expression and induction of basal gene expression (Bogachek et al., 2016; Cyr et al., 2015). One likely hypothesis is that AP-2γ is necessary for multipotent MaSCs to generate luminal progenitors, which could explain both the findings in mammary gland development and luminal breast cancer models (Figure 6B). If CKO of Tfap2c in MaSCs blocked their ability to give rise to luminal progenitors, then all subsequent luminal epithelial cells would have to be derived from MaSCs with an intact Tfap2c gene, and would explain why decreased AP-2γ expression is not detected at later time points (Figure 1). Alternatively, loss of AP-2γ may decrease viability of BCs that could account for effects on mammosphere formation and mammary ductal reconstitution. Furthermore, it is possible that tissue dissociation in combination with loss of AP-2γ reduces the viability or proliferation of BCs and could account for the experimental findings. There was a relative reduction in basal clusters 4 and 10 but a relative increase in KRT5+ basal clusters 1, 2, 3, and 5; hence, the effects on BC viability might be specific to cells in clusters 4 and 10, which could include the MaSC population. Further questions remain as to whether loss of AP-2γ leads to proliferative arrest of BCs, to a reduced ability to invade the extracellular matrix, to an alteration in the expression of genes that affect BC identity, or to defective signaling between basal and hormone-responsive luminal cells. Whether loss of AP-2γ affects the viability of MaSCs or alters their transcriptome leading to altered function or identity is a complex issue that is not easily resolved definitively.
Cluster 11, composed of PROCR+ KRT5− cells, was the only cluster without significant changes in the pattern of gene expression with CKO of Tfap2c, suggesting that cluster 11 represents a multipotent embryonic MaSC population at the top of the developmental hierarchy; we also confirmed that the expression pattern in our PROCR+ cell clusters matched the signature profile described by Wang et al. (2015) (Figure S2). The lack of changes in expression of AP-2γ target genes in cluster 11 is consistent with the hypothesis that the PROCR+ cells in cluster 11 precede expression of Krt5 and therefore have an intact Tfap2c gene. Other investigators have identified a PROCR+ mammary epithelial cell population, although they did not conclude that this population represented an MRU (Bach et al., 2017). Pseudotime analysis suggested that the PROCR+ cells lead to cluster 4, which likely includes the KRT5+ MaSCs (Figures 6 and S9). The data suggest that following the pulse with 4OHT, PROCR+ cells give rise to KRT5+ MaSCs with an intact Tfap2c gene, which are multipotent resulting in normal mammary morphogenesis by 16 weeks.
Several genes known to regulate mammary gland development were altered by CKO of Tfap2c. Mice deficient for Cebpb demonstrate disrupted mammary gland morphogenesis with reduced ductal growth, secondary branching, and loss of secretory cells during pregnancy (Robinson et al., 1998; Seagroves et al., 1998). C/EBPβ-deficient mice also lack expression of β-casein, and, in addition to Cebpb being repressed in several clusters, the Csn2 gene (encoding β-casein) was significantly repressed in luminal clusters 0, 7, and 8 with CKO of Tfap2c (Table S2). IκBα encoded by the Nfkbia gene is a major inhibitor of the nuclear factor κB (NF-κB) transcription factor (Henkel et al., 1993). Mammary epithelial cells deficient in IκBα when transplanted into syngeneic animals develop an increase in lateral ductal branching and intraductal hyperplasia (Brantley et al., 2001). CKO of Tfap2c increased expression of Nfkbia in several basal clusters and is consistent with loss of AP-2γ inhibiting ductal growth through repression of NF-κB. Loss of Rspo1 expression resulted in an absence of mammary side-branching and loss of alveolar development during pregnancy (Chadi et al., 2009) and CKO of Tfap2c resulted in repression of Rspo1 expression in several of the luminal epithelial cell clusters. Taken together our findings indicate that loss of AP-2γ leads to repression of mammary gland branching and normal alveolar development, at least in part, through the regulation of Cebpb, Nfkbia, and Rspo1.
In summary, AP-2γ plays an important role in mammary gland ductal outgrowth during the prepubertal period. We hypothesize that loss of AP-2γ reduced the number of functional MaSCs and significantly repressed the capability for MaSCs to regenerate mammary gland structures. AP-2γ regulates the expression of several genes known to be necessary for mammary gland development, including Cebpb, Nfkbia, and Rspo1. The model system described will further allow an examination of AP-2γ influence during other stages of mammary gland development and lactation.
Experimental Procedures
Mice
All animal studies conformed to guidelines set by the Office of the Institutional Animal Care and Use Committee (IACUC) and performed with IACUC approval. FVB/NJ and KRT5-Cre-ERT2 (FVB.Cg-Tg(Krt5-cre/ERT2)2Ipc/JeldJ) mice were purchased from Jackson Laboratory (stock no. 001800 and 018394). The Tfap2cfl/fl mouse strain was described previously (Auman et al., 2002) and was backcrossed >15 generations on FVB/NJ background to limit genetic variability. Knockout mice were generated by crossing Tfap2cfl/fl with Krt5-Cre-ERT2 mice, and the offspring were back crossed with Tfap2cfl/fl to generate FVB/Tfap2cfl/fl/Krt5-Cre-ERT2. Pulsing with 4OHT was performed starting at age 4 weeks at 30 mg/kg/day intraperitoneal injection for 5 days. All experimental analyses were performed in female mice. Genotype analysis was conducted with qPCR analysis of tail DNA by Transnetyx (Cordova, TN).
Mammary Cell Preparation, Labeling, and Flow Cytometry
Mammary glands of CO (vehicle)-treated and 4OHT-treated 8- to 9-week-old virgin littermate female FVB/Tfap2cfl/fl/Krt5-Cre-ERT2 mice were digested in a mixture of gentle collagenase/hyaluronidase (STEMCELL, cat. no. 07919) with 5% fetal bovine serum (FBS) in complete EpiCult-B (STEMCELL, cat. no. 05611) media for 15 h at 37°C. The red blood cells were lysed in NH4Cl. The cell suspension was further digested in 0.25% trypsin-EDTA for 1 min then 5 U/mL dispase (STEMCELL, cat. no. 07913) plus 0.1 mg/mL DNase I (STEMCELL, cat. no. 07900) for 1 min followed by filtration through a 40-μm cell strainer. After washing in Hank’s balanced salt solution, cell viability was assessed by trypan blue. Single-cell isolation procedure was performed as described previously (Borcherding et al., 2015). FACS was carried out using FACS Aria (Becton Dickinson). The Lin– population was defined as TER119–, CD31−, and CD45–. FACS data were analyzed using FlowJo software (v.10.1r7, Tree Star). The following antibodies were used: CD31-Biotin (BioLegend, 102404, 1:250), TER-119-Biotin (BioLegend, 116204, 1:250), CD45-Biotin (BioLegend, 103014, 1:250), Strep-PE-Cy7 (Invitrogen, 25-4317-82, 1:250), CD24-PE (BioLegend, 101807, 1:250), CD49f-FITC (BioLegend, 313606, 1:200).
Whole-Mount Analysis of Mammary Branching
Mammary glands were collected and whole mounts performed on pair 4 mammary glands as described previously (Plante et al., 2011). To analyze branching morphogenesis, whole-mount mammary gland images were taken on a light box, and branching distance was measured from lymph node to the furthest TEBs. For mammary gland transplantation experiments, the branching area was measured. All images were quantified by ImageJ.
Immunohistochemistry
Immunohistochemical analysis was accomplished on formalin-fixed, paraffin-embedded samples from the fourth mammary gland. The following antibodies were used for analysis: anti-AP-2γ, 1:100 (Santa Cruz Biotechnology, no. sc-12762); anti-CRE, 1:50 (Cell Signaling Technology, no. 15036); anti-CK5, 1:100 (Abcam, ab5235); and anti-CK8/18, 1:500 (Abcam, ab53280). Standard protocols were followed, and Vector ABC kits used for amplification. Bright-field imaging was collected on an Olympus BX-51 microscope.
Quantitative Real-Time RNA Analysis
RNA was isolated from basal and luminal cells recovered from flow cytometry using the RNeasy Mini Kit (QIAGEN, cat. no. 74104). Using the random hexamers method (Thermo Fisher Scientific), mRNA was converted to cDNA by quantitative real-time PCR. Relative expression of exon 2/3 of Tfap2c (TFS, cat. no. 4351372; ID: Mm00493470_g1), exon 6 of Tfap2c (TFS, cat. no. 4351372; ID: Mm00493474_m1), Cebpb (TFS, Mm00843434_s1), Lgals7 (TFS, Mm00456135_m1), Rspo1 (TFS, Mm00507077_m1), and Csn2 (TFS, Mm04207885_m1) was determined using TaqMan primers and the ΔΔCt method of qPCR and compared using CO- versus 4OHT-treated mice. Amplification of Gapdh (TFS, cat. no. 4331182; ID: Mm99999915_g1) was used as an endogenous control.
Single-Cell Capture and Library Preparation for Sequencing
Following the same flow cytometry protocol as above, freshly sorted basal and luminal epithelial cells were submitted in a 1:1 ratio for sequencing using the manufacturer’s recommended Chromium (10× Genomics) and Illumina technologies. Two separate submissions of littermate-controlled, mammary epithelial cells were submitted to 10× Genomics. In each of the two submissions, two CO-treated MMECs were combined and two 4OHT-treated mice MMECs were combined and then were each submitted for sequencing. This resulted in a total of four CO-treated and four 4OHT-treated, 9-week-old mice used for downstream analysis. The amplified cDNA was constructed into 3′ expression libraries and pooled together in separate lanes of a 150-bp, paired-end chemistry flow cell. The Illumina HiSeq 4000 was used with an Agilent Bioanalyzer, and average library sizes ranged from 329 to 432 bp in length. The adapter sequence of the library was 124 bases in length. Basecalls were converted into FASTQ files by the University of Iowa Genomics Division using the Illumina bcl2fastq software. These each captured an estimated 3,546 to 5,545 cells per sample, which were submitted for sequencing with mean reads per cell ranging from 63,842 to 94,776.
scRNA-Seq Data Processing and Quality Control
Transcripts were mapped using Cell Ranger v.3.0.1 and mm10 reference genome (GRCm38.91). Using R, the filtered gene count matrix was imported, and quality control for all samples was completed before downstream analysis using the standard workflow put forth by the Seurat R package, v.3.1 (Butler et al., 2018; Stuart et al., 2019). All cells with unique RNA feature counts <200, or percent mitochondrial DNA >5% were removed.
scRNA-Seq Clustering and Visualization
scRNA-seq data normalization using Seurat was completed to account for feature expression measurements in each cell in relation to total expression values and the data were log transformed using a 1 × 104 factor (Butler et al., 2018; Stuart et al., 2019). The subsets of features showing high cell-to-cell variability were identified in the dataset. The four individual samples were combined using mutual nearest neighbor approach, as described previously, into a singular “integrated” Seurat object (Stuart et al., 2019), enabling further analysis to allow for identification of common cell types between datasets and to control for batch effect between samples. Linear scaling was performed and principal-component analysis calculations were completed. UMAP was calculated using the RunUmap function with default settings and principal components input set to 20 (Becht et al., 2018). The FindClusters Seurat function with default Louvain algorithm and a resolution of 0.5 clustered cells was completed, identifying 13 clusters. The sequence for Cre transgene was concatenated into the mm10 reference genome and all four samples were rerun through Cell Ranger. The single-cell expression data for the transgene was extracted and added to the previous data to allow for its visualization while keeping clustering consistent with the accepted mm10 reference genome. Differential gene expression was calculated using the Wilcoxon rank-sum test with a pseudocount of 0.01 and without thresholds for log-fold change or percentage of cells. The p values for differential gene expression were adjusted using the Bonferroni method for multiple hypotheses comparison.
Pathway Analysis
Differential gene expression results derived from the scRNA-seq quantifications were analyzed using IPA (QIAGEN, Hilden, Germany). Each cluster was individually analyzed to examine the relationship between its highly significantly up- or downregulated genes between CO versus 4OHT. Analysis was completed with an adjusted p < 0.05 and log-fold change ≥ and ≤0.5 to isolate the top networks altered in each cluster by KO of Tfap2c.
Cell-Cycle Analysis
Cell-cycle phase was calculated with scRNA-seq data in both CO- and 4OHT-treated mice on the previously generated integrated object. Scores were determined based on previously published markers (Nestorowa et al., 2016). Utilizing the CellCycleScoring function in Seurat on the integrated metadata of the four samples, S and G2/M scores were stored in the object metadata along with classification of each cell as G2M, S, and G1 phase based on the previously published markers. Cell-cycle distribution based on cluster was visualized by creating a frequency table and graphs using the ggplot2 R package v.3.2.1.
Mammosphere Assay
Freshly sorted basal MMECs from CO- and 4OHT-treated FVB/Tfap2cfl/fl/Krt5-CreERT2 littermate-controlled mice were cultured at 4,000 cells per well at a density of 8,000 viable cells/mL on low attachment plates in EpiCult-B basal medium supplemented with EpiCult-B proliferation supplement, 10 ng/mL epidermal growth factor, 10 ng/mL fibroblast growth factor β, 4 μg/mL, 10% FBS, and 50 μg/mL gentamicin in a low-oxygen incubator at 37°C for 14 days (Dong et al., 2013; Shaw et al., 2012). The above sorting was completed in triplicate. Approximately 48,000 BCs were sorted for each of the two conditions, which allowed for 12 wells to be plated in total. At 21 days, secondary mammospheres were generated by introduction of trypsin and mechanical disruption of primary mammospheres via gentle aspiration through a 20G syringed needle. Again, 48,000 cells per condition were harvested allowing for 12 replicates of each condition to be plated on low-adherent plates. Secondary mammospheres were incubated in a low-oxygen incubator at 37°C for 14 days. Mammosphere analysis and data collection were performed in a blinded manner to treatment of the mammospheres. All images for comparative analysis were taken at 14 days. Total mammosphere count was evaluated by ImageJ software using 40 μm minimum diameter and evidence of 3D structure to differentiate from singular BCs, cellular aggregates, and other debris. Mammosphere formation efficiency was determined by percentage of mammospheres formed per plated basal epithelial cell. For adenovirus mammosphere assays, the BCs were harvested from Tfap2cfl/fl mice and immediately transduced with adenovirus after initial sorting for MMECs. The following day, all BCs were reharvested and, after filtering for single cells, both adenovirus-GFP and GFP-tagged Cre recombinase were sorted for GFP positivity by FACS. These cells were plated in nonadherent plates as described previously and were evaluated for mammosphere formation at 14 days.
Adenovirus Transduction
An enhanced adenovirus-GFP (Ad5CMVeGFP) and enhanced GFP-tagged CRE recombinase adenovirus (Ad5CMVCre-eGFP) were both purchased from the University of Iowa Viral Vector Core (Iowa City, IA) at a viral titer of 5 × 1010 plaque-forming units per mL. Ad5CMVeGFP expresses the enhanced GFP by the CMV promoter and Ad5CMVCre-eGFP has GFP downstream of an IRES sequence. All transductions were performed at a multiplicity of infection of 200 in serum-free Opti-MEM for 1 h. All downstream adenovirus experiments were performed in triplicate with MMECs acquired from cell sorting as described previously.
Mouse Mammary Gland Transplantation
Three-week-old virgin FVB/NJ recipient females were anesthetized, and the fat pad of the right fourth mammary gland was cleared up to the lymph node and complete removal of the mammary gland was confirmed histologically. In one set of experiments, 1-mm3 sections of mammary gland were harvested from 9-week-old FVB/Tfap2cfl/fl/Krt5-Cre-ERT2 mice that had been previously treated with 4OHT or CO from 4 to 5 weeks to conditionally knock out Tfap2c, and then were transplanted into the cleared fat pads following the identical clearing method as above. In another congruent experiment, freshly FACS-sorted MMECs were harvested from FVB/Tfap2cfl/fl mouse mammary glands, transduced with either Ad5CMVeGFP or Ad5CMVCre-eGFP, and 9,000–10,000 GFP+ MMECs were transplanted into the newly cleared fat pad of recipient mice. After 8 weeks, transplanted mammary glands were collected for whole-mount staining and analysis, conducted by a blinded investigator.
Pseudotime Analysis
Slingshot was used to infer and verify cell lineages (Street et al., 2018). The Slingshot object was generated using the previously generated integrated scRNA object. The tSNE plot for pseudotime inference was generated using dims = 20, max_iter = 2,000, seed.use = 2, and the remainder of the settings to default. Clusters were labeled per previous Seurat cluster labeling for consistency and Slingshot was run with the tSNE identifier for reducedDim, start.clus set to cluster 11, and maximum iterations set to 20. Five cell lineages were identified using the slingPseudotime function. Unsupervised trajectory analysis using p-Creode was performed on the previously generated Seurat integrated object (Herring et al., 2018). Default parameters were utilized as per program recommendations with the representative trajectory of n = 200 runs displayed. For consistency, nodes of inferred trajectory from p-Creode were colored according to previous assigned Seurat cluster color.
Statistical Analysis
For whole-mount analysis of branching and immunohistochemical data, statistical significance was assessed using the non-parametric Mann-Whitney U test, for small samples not normally distributed. For mammary gland transplantation experiments, the Mann-Whitney U test and Fisher's exact test were performed. p values of less than 0.05 were considered significant. Mammosphere counts and sizes were analyzed using the Student’s t test as they were continuous, unpaired data. The chi-square test was used to determine significance between scRNA-seq cluster cell points from Seurat as the cell count in each cluster was categorical with either CO or 4OHT treatment. All scRNA-seq statistical analyses were performed in R version 3.6.0. All remaining statistical analyses were performed in GraphPad Prism v.8.0.1.
Data Availability
RNA-seq data are available and can be downloaded from the Gene Expression Omnibus (GEO: GSE143159).
Author Contributions
Conceptualization, V.W.G., E.C., T.W., W.Z., and R.J.W.; Methodology, V.W.G., E.C., D.T.T., T.W., W.Z., and R.J.W.; Software, E.C., D.T.T., N.B., and M.V.K.; Validation, V.W.G., E.C., D.T.T., V.C.C., N.B., K.E.K., V.T.W., A.W.L., D.M.v.d.H., J.R.W., and M.V.K.; Formal Analysis, V.W.G., E.C., D.T.T., V.C.C., N.B., V.T.W., A.W.L., and M.V.K.; Investigation, V.W.G., E.C., D.T.T., V.C.C., K.E.K., V.T.W., A.W.L., D.M.v.d.H., and J.R.W.; Resources, V.W.G., E.C., D.T.T., and K.E.K.; Data Curation, E.C., D.T.T., N.B., and M.V.K.; Writing – Original Draft, V.W.G. and R.J.W.; Writing – Review & Editing, V.W.G., E.C., D.T.T., V.C.C., N.B., K.E.K., V.T.W., A.W.L., D.M.v.d.H., J.R.W., M.V.K., T.W., W.Z., and R.J.W.; Visualization, V.W.G., E.C., D.T.T., and K.E.K.; Supervision, V.W.G., W.Z., and R.J.W.; Project Administration, W.Z. and R.J.W.; Funding Acquisition, T.W., W.Z., and R.J.W.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by NIH grants R01CA183702 (PI: R.J.W.), CA200673 (PI: W.Z.), CA203834 (PI: W.Z.) T32CA148062 (P.I.: R.J.W.), DOD/CDMRP grant BC180227 (PI: W.Z.) and by generous gifts from the Kristen Olewine Milke Breast Cancer Research Fund (to R.J.W.), from J.D. and Jill Thoreson (to R.J.W.), and from Dr. and Mrs. James Robert Spencer Family Cancer Research Fund (to W.Z.). E.C., D.T.T., K.E.K., V.T.W., A.W.L., and D.M.v.d.H. were supported by NIH grant T32CA148062. N.B. was supported by an F30 fellowship CA206255.
Published: December 30, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.12.002.
Contributor Information
Weizhou Zhang, Email: zhangw@ufl.edu.
Ronald J. Weigel, Email: ronald-weigel@uiowa.edu.
Supplemental Information
References
- Auman H.J., Nottoli T., Lakiza O., Winger Q., Donaldson S., Williams T. Transcription factor AP-2gamma is essential in the extra-embryonic lineages for early postimplantation development. Development. 2002;129:2733–2747. doi: 10.1242/dev.129.11.2733. [DOI] [PubMed] [Google Scholar]
- Bach K., Pensa S., Grzelak M., Hadfield J., Adams D.J., Marioni J.C., Khaled W.T. Differentiation dynamics of mammary epithelial cells revealed by single-cell RNA sequencing. Nat. Commun. 2017;8:2128. doi: 10.1038/s41467-017-02001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball S.M. The development of the terminal end bud in the prepubertal-pubertal mouse mammary gland. Anat. Rec. 1998;250:459–464. doi: 10.1002/(SICI)1097-0185(199804)250:4<459::AID-AR9>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Becht E., McInnes L., Healy J., Dutertre C.A., Kwok I.W.H., Ng L.G., Ginhoux F., Newell E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2018;3 doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- Bogachek M.V., Park J.M., De Andrade J.P., Lorenzen A.W., Kulak M.V., White J.R., Gu V.W., Wu V.T., Weigel R.J. Inhibiting the SUMO pathway represses the cancer stem cell population in breast and colorectal carcinomas. Stem Cell Reports. 2016;7:1140–1151. doi: 10.1016/j.stemcr.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borcherding N., Bormann N., Kusner D., Kolb R., Zhang W. Transcriptome analysis of basal and luminal tumor-initiating cells in ErbB2-driven breast cancer. Genom Data. 2015;4:119–122. doi: 10.1016/j.gdata.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantley D.M., Chen C.L., Muraoka R.S., Bushdid P.B., Bradberry J.L., Kittrell F., Medina D., Matrisian L.M., Kerr L.D., Yull F.E. Nuclear factor-kappaB (NF-kappaB) regulates proliferation and branching in mouse mammary epithelium. Mol. Biol. Cell. 2001;12:1445–1455. doi: 10.1091/mbc.12.5.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadi S., Buscara L., Pechoux C., Costa J., Laubier J., Chaboissier M.C., Pailhoux E., Vilotte J.L., Chanat E., Le Provost F. R-Spondin1 is required for normal epithelial morphogenesis during mammary gland development. Biochem. Biophys. Res. Commun. 2009;390:1040–1043. doi: 10.1016/j.bbrc.2009.10.104. [DOI] [PubMed] [Google Scholar]
- Cyr A.R., Kulak M.V., Park J.M., Bogachek M.V., Spanheimer P.M., Woodfield G.W., White-Baer L.S., O'Malley Y.Q., Sugg S.L., Olivier A.K. TFAP2C governs the luminal epithelial phenotype in mammary development and carcinogenesis. Oncogene. 2015;34:436–444. doi: 10.1038/onc.2013.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deome K.B., Faulkin L.J., Jr., Bern H.A., Blair P.B. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res. 1959;19:515–520. [PubMed] [Google Scholar]
- Dong Q., Wang D., Bandyopadhyay A., Gao H., Gorena K.M., Hildreth K., Rebel V.I., Walter C.A., Huang C., Sun L.Z. Mammospheres from murine mammary stem cell-enriched basal cells: clonal characteristics and repopulating potential. Stem Cell Res. 2013;10:396–404. doi: 10.1016/j.scr.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu N., Nolan E., Lindeman G.J., Visvader J.E. Stem cells and the differentiation hierarchy in mammary gland development. Physiol. Rev. 2019;100:489–523. doi: 10.1152/physrev.00040.2018. [DOI] [PubMed] [Google Scholar]
- Giraddi R.R., Chung C.Y., Heinz R.E., Balcioglu O., Novotny M., Trejo C.L., Dravis C., Hagos B.M., Mehrabad E.M., Rodewald L.W. Single-cell transcriptomes distinguish stem cell state changes and lineage specification programs in early mammary gland development. Cell Rep. 2018;24:1653–1666 e1657. doi: 10.1016/j.celrep.2018.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkel T., Machleidt T., Alkalay I., Kronke M., Ben-Neriah Y., Baeuerle P.A. Rapid proteolysis of I kappa B-alpha is necessary for activation of transcription factor NF-kappa B. Nature. 1993;365:182–185. doi: 10.1038/365182a0. [DOI] [PubMed] [Google Scholar]
- Herring C.A., Banerjee A., McKinley E.T., Simmons A.J., Ping J., Roland J.T., Franklin J.L., Liu Q., Gerdes M.J., Coffey R.J. Unsupervised trajectory analysis of single-cell RNA-seq and imaging data reveals alternative tuft cell origins in the gut. Cell Syst. 2018;6:37–51 e39. doi: 10.1016/j.cels.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilger-Eversheim K., Moser M., Schorle H., Buettner R. Regulatory roles of AP-2 transcription factors in vertebrate development, apoptosis and cell-cycle control. Gene. 2000;260:1–12. doi: 10.1016/s0378-1119(00)00454-6. [DOI] [PubMed] [Google Scholar]
- Indra A.K., Warot X., Brocard J., Bornert J.M., Xiao J.H., Chambon P., Metzger D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999;27:4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager R., Schafer S., Hau-Liersch M., Schorle H. Loss of transcription factor AP-2gamma/TFAP2C impairs branching morphogenesis of the murine mammary gland. Dev. Dyn. 2010;239:1027–1033. doi: 10.1002/dvdy.22239. [DOI] [PubMed] [Google Scholar]
- Jager R., Werling U., Rimpf S., Jacob A., Schorle H. Transcription factor AP-2gamma stimulates proliferation and apoptosis and impairs differentiation in a transgenic model. Mol. Cancer Res. 2003;1:921–929. [PubMed] [Google Scholar]
- Jamieson P.R., Dekkers J.F., Rios A.C., Fu N.Y., Lindeman G.J., Visvader J.E. Derivation of a robust mouse mammary organoid system for studying tissue dynamics. Development. 2017;144:1065–1071. doi: 10.1242/dev.145045. [DOI] [PubMed] [Google Scholar]
- Lee E., Piranlioglu R., Wicha M.S., Korkaya H. Plasticity and potency of mammary stem cell subsets during mammary gland development. Int. J. Mol. Sci. 2019;20:2357. doi: 10.3390/ijms20092357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilja A.M., Rodilla V., Huyghe M., Hannezo E., Landragin C., Renaud O., Leroy O., Rulands S., Simons B.D., Fre S. Clonal analysis of Notch1-expressing cells reveals the existence of unipotent stem cells that retain long-term plasticity in the embryonic mammary gland. Nat. Cel. Biol. 2018;20:677–687. doi: 10.1038/s41556-018-0108-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J.S., Stein T. Pubertal ductal morphogenesis: isolation and transcriptome analysis of the terminal end bud. Methods Mol. Biol. 2017;1501:131–148. doi: 10.1007/978-1-4939-6475-8_5. [DOI] [PubMed] [Google Scholar]
- Nestorowa S., Hamey F.K., Pijuan Sala B., Diamanti E., Shepherd M., Laurenti E., Wilson N.K., Kent D.G., Gottgens B. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood. 2016;128:e20–e31. doi: 10.1182/blood-2016-05-716480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal B., Chen Y., Vaillant F., Jamieson P., Gordon L., Rios A.C., Wilcox S., Fu N., Liu K.H., Jackling F.C. Construction of developmental lineage relationships in the mouse mammary gland by single-cell RNA profiling. Nat. Commun. 2017;8:1627. doi: 10.1038/s41467-017-01560-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante I., Stewart M.K., Laird D.W. Evaluation of mammary gland development and function in mouse models. J. Vis. Exp. 2011;21:2828. doi: 10.3791/2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post Y., Clevers H. Defining adult stem cell function at its simplest: the ability to replace lost cells through mitosis. Cell Stem Cell. 2019;25:174–183. doi: 10.1016/j.stem.2019.07.002. [DOI] [PubMed] [Google Scholar]
- Rios A.C., Fu N.Y., Cursons J., Lindeman G.J., Visvader J.E. The complexities and caveats of lineage tracing in the mammary gland. Breast Cancer Res. 2016;18:116. doi: 10.1186/s13058-016-0774-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios A.C., Fu N.Y., Lindeman G.J., Visvader J.E. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014;506:322–327. doi: 10.1038/nature12948. [DOI] [PubMed] [Google Scholar]
- Robinson G.W., Johnson P.F., Hennighausen L., Sterneck E. The C/EBPbeta transcription factor regulates epithelial cell proliferation and differentiation in the mammary gland. Genes Dev. 1998;12:1907–1916. doi: 10.1101/gad.12.12.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheele C.L., Hannezo E., Muraro M.J., Zomer A., Langedijk N.S., van Oudenaarden A., Simons B.D., van Rheenen J. Identity and dynamics of mammary stem cells during branching morphogenesis. Nature. 2017;542:313–317. doi: 10.1038/nature21046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seagroves T.N., Krnacik S., Raught B., Gay J., Burgess-Beusse B., Darlington G.J., Rosen J.M. C/EBPbeta, but not C/EBPalpha, is essential for ductal morphogenesis, lobuloalveolar proliferation, and functional differentiation in the mouse mammary gland. Genes Dev. 1998;12:1917–1928. doi: 10.1101/gad.12.12.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton M., Vaillant F., Simpson K.J., Stingl J., Smyth G.K., Asselin-Labat M.L., Wu L., Lindeman G.J., Visvader J.E. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Shaw F.L., Harrison H., Spence K., Ablett M.P., Simoes B.M., Farnie G., Clarke R.B. A detailed mammosphere assay protocol for the quantification of breast stem cell activity. J. Mammary Gland Biol. Neoplasia. 2012;17:111–117. doi: 10.1007/s10911-012-9255-3. [DOI] [PubMed] [Google Scholar]
- Silberstein G.B. Postnatal mammary gland morphogenesis. Microsc. Res. Tech. 2001;52:155–162. doi: 10.1002/1097-0029(20010115)52:2<155::AID-JEMT1001>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Stingl J., Eirew P., Ricketson I., Shackleton M., Vaillant F., Choi D., Li H.I., Eaves C.J. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- Street K., Risso D., Fletcher R.B., Das D., Ngai J., Yosef N., Purdom E., Dudoit S. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 2018;19:477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902 e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Miao Z., Zhang X., Chan U.I., Su S.M., Guo S., Wong C.K.H., Xu X., Deng C.X. Single-cell RNA-seq reveals cell heterogeneity and hierarchy within mouse mammary epithelia. J. Biol. Chem. 2018;293:8315–8329. doi: 10.1074/jbc.RA118.002297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Keymeulen A., Rocha A.S., Ousset M., Beck B., Bouvencourt G., Rock J., Sharma N., Dekoninck S., Blanpain C. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- Visvader J.E., Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev. 2014;28:1143–1158. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Cai C., Dong X., Yu Q.C., Zhang X.O., Yang L., Zeng Y.A. Identification of multipotent mammary stem cells by protein C receptor expression. Nature. 2015;517:81–84. doi: 10.1038/nature13851. [DOI] [PubMed] [Google Scholar]
- Williams J.M., Daniel C.W. Mammary ductal elongation: differentiation of myoepithelium and basal lamina during branching morphogenesis. Dev. Biol. 1983;97:274–290. doi: 10.1016/0012-1606(83)90086-6. [DOI] [PubMed] [Google Scholar]
- Winger Q., Huang J., Auman H.J., Lewandoski M., Williams T. Analysis of transcription factor AP-2 expression and function during mouse preimplantation development. Biol. Reprod. 2006;75:324–333. doi: 10.1095/biolreprod.106.052407. [DOI] [PubMed] [Google Scholar]
- Wuidart A., Ousset M., Rulands S., Simons B.D., Van Keymeulen A., Blanpain C. Quantitative lineage tracing strategies to resolve multipotency in tissue-specific stem cells. Genes Dev. 2016;30:1261–1277. doi: 10.1101/gad.280057.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuidart A., Sifrim A., Fioramonti M., Matsumura S., Brisebarre A., Brown D., Centonze A., Dannau A., Dubois C., Van Keymeulen A. Early lineage segregation of multipotent embryonic mammary gland progenitors. Nat. Cel. Biol. 2018;20:666–676. doi: 10.1038/s41556-018-0095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Brewer S., Huang J., Williams T. Overexpression of transcription factor AP-2alpha suppresses mammary gland growth and morphogenesis. Dev. Biol. 2003;256:127–145. doi: 10.1016/s0012-1606(02)00119-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data are available and can be downloaded from the Gene Expression Omnibus (GEO: GSE143159).