Abstract
Background
Radiation with platinum-based chemotherapy is the standard of care for unresectable stage III non-small cell lung cancer (NSCLC). Despite aggressive treatment, progression-free survival and overall survival remain poor. It is unclear whether any tumor genetic mutations are associated with response to chemoradiation therapy.
Methods
We retrospectively reviewed clinical outcomes of patients with stage III NSCLC treated with definitive radiation who had undergone tumor molecular profiling through a next-generation DNA sequencing platform. Cox proportional hazards model was used to investigate associations between clinical outcomes and genetic mutations detected by next-generation sequencing.
Results
110 patients were identified with stage III NSCLC and underwent definitive radiation between 2013 and 2017 and tumor molecular profiling. Concurrent or sequential chemotherapy was given in 104 patients (95%). Unbiased genomic analyses revealed a significant association between AKT2 mutations and decreased local-regional tumor control and overall survival (hazard ratios [HR] 12.5 and 13.7, P = .003 and P = .003, respectively). Analyses restricted to loss-of-function mutations identified KMT2C and KMT2D deleterious mutations as negative prognostic factors for overall survival (HR 13.4 and 7.0, P < .001 and P < .001, respectively). Deleterious mutations in a panel of 38 DNA damage response and repair pathway genes were associated with improved local-regional control (HR 0.32, P = .049).
Conclusions
This study coupled multiplexed targeted sequencing with clinical outcome and identified mutations in AKT2, KMT2C, and KMT2D as negative predictors of local-regional control and survival, and deleterious mutations in damage response and repair pathway genes were associated with improved local-regional disease control after chemoradiation therapy. These findings will require validation in a larger cohort of patients with prospectively collected and detailed clinical information.
Introduction
Approximately 30% of patients with non-small cell lung cancer (NSCLC) present with stage III (locally advanced) disease at diagnosis. The standard of care for unresectable stage III NSCLC has been concurrent chemoradiation, based on several randomized clinical trials.1, 2, 3 More recently, the addition of consolidative immunotherapy with durvalumab was associated with improved 2-year progression-free survival and overall survival in stage III patients compared with chemoradiation alone.4,5 Despite such improvements in outcome, more than 50% of patients still experience progression of disease within 24 months. Among patients who developed progression of disease in the PACIFIC study, 75% to 80% of patients had intrathoracic progression as the first site of progression,6 which highlights the challenge of local-regional tumor control in this patient population.
Lung adenocarcinoma and squamous cell carcinoma exhibit high rates of somatic mutation. Whole exome sequencing of lung adenocarcinoma and squamous cell carcinoma has revealed recurring somatic mutations in known oncogenes and tumor suppressions.7,8 However, the functional significance of majority of the somatic mutations remains unknown. It is unclear whether any genetic mutations are associated with local-regional response to radiation and survival after chemoradiotherapy.
Recent years have seen the advent of efforts to expand tumor genomics testing in the clinic, with the goal of identifying mutations that could guide therapy and predict outcome. At Memorial Sloan Kettering Cancer Center (MSKCC), a targeted, next-generation sequencing (NGS) platform named Memorial Sloan Kettering-Integrated Mutational Profiling of Actionable Cancer Targets (MSK-IMPACT) was developed to detect mutations and copy number changes in 468 frequently mutated cancer genes. Since its inception in January 2014, this sequencing platform was rapidly integrated into routine clinical practice.9 The sequencing test was used in patients with recurrent and metastatic disease, which include a subset of patients who had stage III NSCLC. The sequencing test was also performed in patients with de novo diagnosis of stage III NSCLC to aid in diagnosis and assess candidacy for clinical trials. In comparison to untreated primary tumors from the Cancer Genome Atlas Project, primary tumors and distant metastases sequenced on our NGS platform showed strong concordance in identities and frequencies of mutations detected in the Cancer Genome Atlas.10 We undertook this study to survey the landscape of genetic mutations in stage III NSCLC and use an unbiased approach to investigate any associations between tumor somatic mutations and clinical outcomes after definitive chemoradiation.
Methods
Patient selection
An institutional database was queried with the approval of the institutional review board (MSKCC-IRB #16-142) to identify stage III (American Joint Committee on Cancer [AJCC] 7th edition) NSCLC patients treated with definitive radiation therapy from 2013 to 2017 who underwent tumor genetic panel testing between 2014 and 2017. Patients were identified based on International Classification of Diseases (ICD)-9 and ICD-10 codes for malignant neoplasm of the trachea, bronchus, and lung. Patients were excluded if the radiation dose was less than 50 Gy or number of fractions was less than 25 (as these were considered palliative treatment), underwent definitive surgical resection, or were not clinically stage III. The cohort of patients was cross-referenced with MSKCC NGS database and those patients with at least 1 identified tumor genetic abnormality were included.
Tumor genomic testing
Patients were selected and consented for tumor genomic testing to identify genetic alterations that are readily actionable or candidates for clinical trial as part of routine clinical care. Tumor genomic testing was performed with MSK-IMPACT, which is a US Food and Drug Administration (FDA)–approved assay that detects genetic alterations involving 341 (version 1), 410 (version 2), or 468 (version 3) cancer-associated genes.9 The assay was performed on tissue samples taken either from a primary or metastatic site, depending on the availability of tissue. If multiple samples were taken from the patient, the first sample was chosen for analysis. The definition of a deleterious mutation included either a frameshift, splice site, or nonsense mutation that often leads to a loss of function effect.
Radiation treatment
Simulation was performed with patients in the supine and arms-up position in a customized immobilization cradle. Four-dimensional (4D)-computed tomography (CT) simulation was included and an internal target volume (ITV) approach was used to account for respiratory motion. The ITV was expanded by 5 to 7 mm to create the clinical target volume, which was then expanded by 5 mm to create the planning target volume. Patients were treated with conventional fractionation with 1.8 to 2 Gy per fraction using either 3-dimensional (3D)-conformal radiation therapy or intensity modulated radiation therapy. Elective nodal radiation (other than the ipsilateral hilar nodes) was not done. After completion of radiation therapy, patients were followed every 3 months for years 1 to 2 and every 6 months for years 3 to 4 in a clinic with surveillance CT scans.
Clinical outcome assessment
Patient clinical outcome was retrospectively collected with institutional review board approval. Patient clinical and tumor characteristics were collected, and response after definitive radiation therapy was assessed based on CT imaging. Local-regional failure after radiation was defined by progression of disease within the radiation field, including irradiated lymph nodes. Distant failure was defined by progression of disease in the contralateral thorax, or outside the thoracic cavity.
Statistical analysis
Cox proportional hazards model was used to assess hazard ratio for local-regional recurrence and death associated with the presence of any or deleterious mutation in each mutated gene. The models have been tested to confirm there is no violation of standard proportional hazards assumptions. P values were corrected for multiple hypothesis testing using false discovery rate (FDR) method.11 Overall and local-regional recurrence-free survivals were estimated using the Kaplan-Meier method. Multivariate Cox regression analysis was performed to estimate associations between factors of interest, including tumor mutational status, age, sex, smoking history, tumor stage, tumor histology, and receipt of chemotherapy. DNA damage response and repair pathway included 48 genes from MSK-IMPACT as previously described.12,13 All analyses were performed with R software (version 3.6).
Results
Patient characteristics
We identified a total of 110 patients who had stage III NSCLC and underwent tumor genomic testing with MSK-IMPACT (Table 1). The median age was 67 years (range, 29-88). 53% of patients were male and 47% were female. 90% of patients had prior smoking history. 73 patients (66%) had adenocarcinoma, 18 patients (16%) had squamous cell carcinoma, 12 patients (11%) had mixed squamous and adenocarcinoma histology, and 7 patients (6%) had rare histologic types including large cell neuroendocrine carcinoma (2%), sarcomatoid carcinoma of the lung (2%), and combined small cell carcinoma and adenocarcinoma of the lung (1%).
Table 1.
Baseline patient clinical and tumor profiling characteristics
| Characteristics | No. (%) |
|---|---|
| Age | |
| Median (range) | 67 (29-88) |
| Sex | |
| Male | 58 (53%) |
| Female | 52 (47%) |
| Smoking status | |
| Current smoker | 29 (26%) |
| Former smoker | 70 (64%) |
| Never smoker | 11 (10%) |
| Stage (AJCC 7th edition) | |
| IIIA | 48 (44%) |
| IIIB | 62 (56%) |
| Histology | |
| Adenocarcinoma | 73 (66%) |
| Squamous cell carcinoma | 18 (16%) |
| Mixed adenosquamous | 12 (11%) |
| Other | 7 (6%) |
| Radiation dose (Gy) | |
| Median (range) | 60 (50.4 - 74) |
| Chemotherapy | |
| Yes | 104 (95%) |
| Concurrent | 83 (75%) |
| Sequential | 21 (19%) |
| No | 6 (5%) |
| MSK-IMPACT gene panel | |
| IMPACT-468 | 40 (36%) |
| IMPACT-410 | 55 (50%) |
| IMPACT-341 | 15 (14%) |
| IMPACT sample site | |
| Primary | 47 (43%) |
| Metastatic | 63 (57%) |
Abbreviations: AJCC = American Joint Committee On Cancer; MSK-IMPACT = Memorial Sloan Kettering-Integrated Mutational Profiling of Actionable Cancer Targets.
Radiation treatment and clinical outcome
Median radiation dose received was 60 Gy (range, 50.4 Gy-74 Gy), delivered in 1.8 Gy or 2 Gy per fraction. One patient received 50.4 Gy due to bilateral hilar nodal (N3) disease making it difficult to meet lung dose constraints. A total of 104 patients (95%) received chemotherapy either concurrently (75%) or sequentially (19%). No patients received adjuvant immunotherapy as it was not standard of care at that time.
The median follow-up time was 15.3 months (range, 0.8-58.7 months) from the end of RT. Median overall survival of all patients was 24.7 months (Figure E1A). One-year and 2-year overall survival rates were 80% and 51%, respectively. In total, 36 patients developed local-regional failure, 69 patients developed distant metastases, and 22 patients had both local-regional and distant failures. Median time to local-regional failure was 40.3 months (Figure E1B). Median time to developing distant metastasis was 12.0 months.
Tumor molecular profiling
A total of 110 samples were profiled using MSK-IMPACT during the study period; 47 samples were collected from the primary site and 63 samples were collected from a metastatic site including lymph node. Among metastatic sites, the most common site of collection was lymph node (76%), followed by brain (8%), bone (5%), and liver (5%). Among different gene panels used, 50% of the tumor samples were profiled with the 410-gene panel, 40% of samples used the 468-gene panel, and the remaining 15% of samples used the 341-gene panel (Table 1). The average sample sequencing coverage was 778×, which provided sensitivity to detect mutations at low allele frequencies.
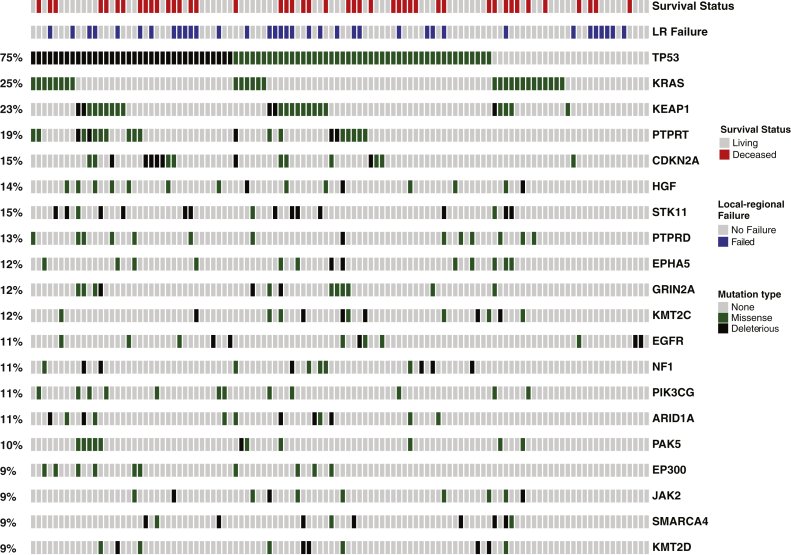
The most frequently mutated genes were TP53 and KRAS, with mutations found in 75% and 25% of the tumors, respectively (Fig. 1). For actionable mutations in adenocarcinoma, 18% of tumors had EGFR mutations and ALK mutations were found in 11% of adenocarcinoma tumors. We also found frequent mutations in tumor suppressor genes aside from TP53, including KEAP1 (23%), CDK2NA (19%), STK11 (15%),14 NF1 (11%), and SMARCA4 (9%).14, 15, 16, 17, 18, 19, 20
Figure 1.
Mutational profile of stage III non-small cell lung cancer. Oncoprint of clinical outcomes (survival status and local-regional failure) and 20 most frequently mutated genes identified by Memorial Sloan Kettering-Integrated Mutational Profiling of Actionable Cancer Targets clinical tumor mutational profiling in 110 patients with stage III non-small cell lung cancer. Genes are ordered based on mutational frequency. Mutational types are color-coded (black, deleterious mutations defined as either frameshift or nonsense mutation; green, missense mutation; gray, no alterations).
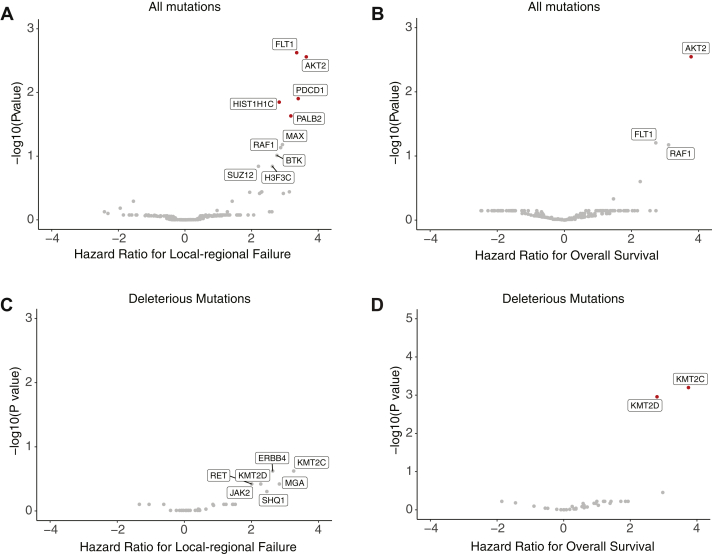
Genomic analyses for mutations associated with differential response after chemoradiation
Unbiased genomics analyses revealed somatic mutations that were associated with differential local-regional disease control and survival after definitive chemoradiation (Figs. 2A and 2B). To investigate the association of loss of function mutations and clinical outcome, we restricted our analysis to include only deleterious mutations. There was no individual deleterious mutation that was associated with worse local-regional disease control (Fig 2C). However, deleterious mutations in KMT2C and KMT2D were both independently associated with worse overall survival (HR 13.4 and 7.0, P < .001 and .001, respectively) (Fig 2D). KMT2C and KMT2D mutations tended to co-occur (Fisher's exact test, P = .002) and mutations in both genes were found in 5 of 13 patients with KMT2C mutations and 5 of 10 patients with KMT2D mutations.
Figure 2.
Genomic analyses for mutations associated with differential response after chemoradiation. (A) Volcano plots of mutations associated with differential local-regional failure. Statistical significance (log10-transformed P values from log-rank test) was plotted against risk of local-regional failure (log2-fold change of Cox proportional hazard ratios). P values were corrected for multiple hypothesis testing. Mutations with significant P values after transformation (>-log10[0.05]) were labeled in red. (B) Volcano plots of any mutations associated with differential survival probability. (C) Volcano plots of deleterious mutations associated with differential local-regional failure. (D) Volcano plots of deleterious mutations associated with differential survival probability.
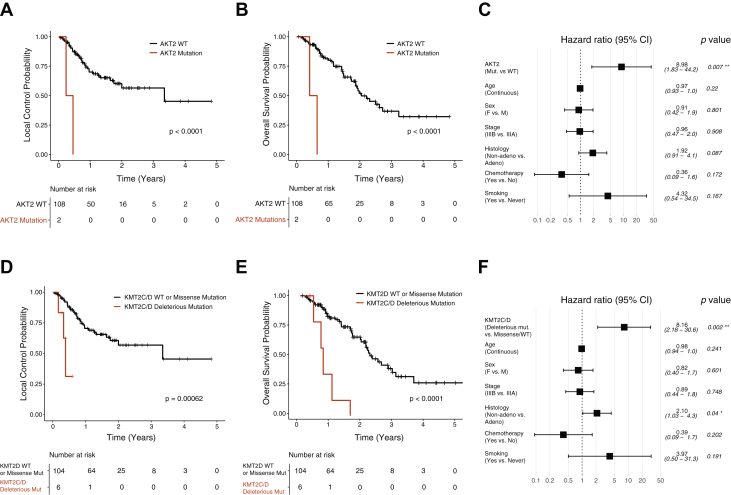
Somatic mutations in AKT2 (n = 2) were significantly associated with worse local-regional control (HR 12.5 and 13.7, P = .003 and .003, respectively) (Figs. 3A and 3B). Multivariable Cox regression analysis showed that AKT2 mutations independently increased the likelihood of local-regional failure (Fig. 3C). Two patients with AKT2 mutations had 2 missense mutations, P115L and W334L. P115L resides between the Pleckstrin homology (PH) domain and the protein kinase domain and W334L is in the protein kinase domain. Patients with deleterious mutations in either KMT2C or KMT2D exhibited worse local-regional tumor control and survival (Figs. 3D and 3E). Multivariable Cox regression analysis showed KMT2C or KMT2D mutations are independently associated with worse overall survival after chemoradiation (Fig. 3F).
Figure 3.
Association of missense mutations in AKT2 and deleterious mutations in KMT2C and KMT2D with clinical outcome after definitive chemoradiation. (A) Kaplan-Meier estimate of local-regional control probability for tumors without versus with AKT2 mutations (n = 2). (B) Kaplan-Meier estimate of overall survival probability for patients with tumor harboring AKT2 mutations (n = 2). (C) Multivariate analysis of AKT2 mutation and patient factors associated with local-regional failure. (D) Kaplan-Meier survival curves of local-regional control probability for tumors without versus with KMT2C or KMT2D mutations (n = 6). (E) Kaplan-Meier survival curves of overall survival probability for patients with tumor containing KMT2C or KMT2D mutations (n = 6). (F) Multivariate analysis of KMT2C or KMT2D mutations and patient factors associated with overall survival.
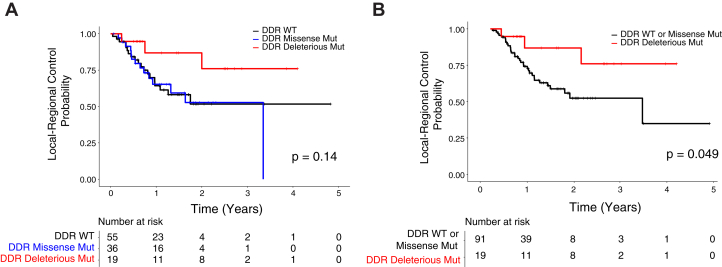
Association between mutations in DNA damage response and repair genes and local-regional disease control
Because an impairment of DNA damage repair pathways may sensitize tumor cells to DNA damage induced by radiation, we investigated the association of mutations in damage response and repair (DDR) genes with outcome after chemoradiation. Tumors were classified based on the presence of mutations in 38 DDR genes included in the MSK-IMPACT panel.12 Kaplan-Meier analysis revealed similar local-regional control between tumors with wild-type and missense mutations in DDR genes (Fig 4A), and tumors with deleterious mutations in DDR genes had significantly improved local-regional control compared with those with missense mutations or wild-type DDR genes (Fig 4B). There is a nonsignificant improvement in overall survival in patients with deleterious DDR mutations compared with wild type DDR genes (Figure E4B). Frequencies of deleterious mutations in DDR genes ranged from 0% to 3%, and DDR genes from multiple pathways such as homologous recombination, mismatch repair, Fanconi anemia, and nuclear excision repair were represented in the mutated cohort (Fig E4C).
Figure 4.
Local-regional disease control in tumors with DNA damage response and repair (DDR) pathway mutations. (A) Local-regional control probability after definitive chemoradiation in patients with wild-type (n = 55) versus missense mutations (n = 36) versus deleterious mutations (n = 19) in 38 DDR genes from Memorial Sloan Kettering-Integrated Mutational Profiling of Actionable Cancer Targets. (B) Local-regional control probability comparing patients with or without deleterious mutations in DDR genes.
Discussion
The current study used an unbiased genomic approach to identify predictors of response to chemoradiation in stage III NSCLC. To date, limited evidence has been published that supports the existence of genetic predictors of radiation response in NSCLC. Prior studies have investigated the effect of individual gene mutations in various stages of NSCLC. In early stage NSCLC, KRAS mutations have been reported to correlate with worse local control after stereotactic body radiation therapy (SBRT); however, the studies were limited by sample sizes of 10 to 45 genotyped patients.21,22 Our institution had reported higher rate of local recurrence after SBRT in tumors harboring PIK3CA mutations in 166 tumors of primary lung (93%) or metastatic origin (7%).23 In addition, mutations in KEAP1/NFE2L2 (HR 2.19, 95% CI 1.41-3.38) and STK11 (HR 2.36, 95% CI 1.19-5.08) have been linked to worse overall survival in advanced NSCLC.24, 25, 26
Our study identified AKT2 mutation as a potential predictor of poor response after definitive chemoradiation in stage III NSCLC. Although mutational frequency of AKT2 was only 1.8%, the 2 patients with tumors containing AKT2 mutations had significantly worse local-regional control and overall survival after radiation, and this effect is independent from potential confounding factors. AKT2 encodes 1 of the 3 closely related serine/threonine-protein kinases (AKT1, AKT2, and AKT3) that are expressed in different tissue types and regulate processes critical to oncogenesis including cell survival, proliferation, and metabolism. AKT2 has been implicated as an oncogene and found to be amplified in 3% to 12% of ovarian carcinoma, breast carcinoma, and pancreatic carcinoma.27, 28, 29 The 2 mutations seen in AKT2 (P115L and W334L) in our study were both considered pathogenic in the Catalog of Somatic Mutations in Cancer (COSMIC) database. FATHMM scores for P115L and W334 were 0.87 and 0.96, respectively (scores range from 0-1 and values greater than 0.5 are predicted to be pathogenic). It is possible that these particular missense mutations in AKT2 may lead to more aggressive biology and radioresistant phenotype, though it is difficult to draw firm conclusions from the small number of tumors that harbored such mutations.
In our analysis limited to deleterious mutations that presumably demonstrate a loss of function phenotype, deleterious mutations in KMT2C and KMT2D were significantly associated with worse overall survival. KMT2C and KMT2D are members of the histone-lysine N-methyltransferase 2 (KMT2) family of proteins.30 KMT2 proteins methylate lysine 4 on the histone H3 tail (H3K4) and change chromatin structures to promote genomic accessibility and transcription of specific genes. KMT2C and KMT2D are 2 of the most frequently mutated genes in cancer and have been described in multiple cancer types including non-small cell7 and small cell lung cancer,31 prostate cancer,32 breast cancer,33 renal cell carcinoma,34 and non-Hodgkin lymphoma.35 Most cancer-associated KMT2C and KMT2D mutations are frameshift or nonsense mutations, and it is postulated that they function as tumor suppressor genes. In our study, 12% of tumors had mutations in KMT2C and 9% had mutations in KMT2D, and significant fractions were deleterious mutations (33% of KMT2C mutations and 55% of KMT2D mutations). Furthermore, mutations in KMT2D were previously shown to be associated with worse survival in locally advanced and metastatic NSCLC. In a study of 194 patients with stage III (23%) and stage IV (77%) NSCLC, 34 patients with KMT2D mutations had worse survival with lower median overall survival of 10 months compared with 30 months in patients with wild-type KMT2D.36 In the context of prior molecular, sequencing, and clinical studies, our finding supports loss of function mutations in KMT2C and KMT2D as potential prognostic factors for worse response after chemoradiation in stage III NSCLC.
It has been hypothesized that impaired DNA damage response and repair mechanisms may sensitize the tumor cells to genotoxic therapies such as radiation. There is emerging clinical evidence suggesting mutations in DDR genes can predict better response. Loss-of-function mutations in the ATM gene, which plays a central role in DNA damage response, was found in 8 exceptional responders after radiation.37 In a larger cohort of 95 patients with NSCLC who received radiation therapy to either thoracic or extrathoracic lesions, the 2-year cumulative incidence of local failure was 6.7% for pathogenic ATM mutations compared with 19.9% for missense ATM mutations.38 It is likely that mutations in other DDR genes are also important in determining the response after radiation. The presence of somatic mutations in a panel of 34 DDR genes is associated with improved response after platinum-based chemotherapy in patients with locally advanced and metastatic urothelial carcinoma.12 Interestingly, our analysis showed that only deleterious mutations, not missense mutations, in DDR genes correlate with improved local-regional tumor control. This is in contrast with the previous study showing any type of mutations in the DDR gene can correlate with improved outcome in urothelial carcinoma.12 Due to the rare frequencies of individual DDR gene mutations and insufficient number of patients, our study did not have the power to detect a difference in outcome at an individual gene or pathway level.
It is important to address several limitations of the study. First, it is important to keep in mind the exploratory nature of the study due to the limited number of patients available for analysis. Insufficient power due to small patient cohort may limit the detection of many potential genomic predictors with low prevalence. In the stage III NSCLC population, genomic testing is only recently being integrated into more routine practice; therefore, the study cohort did not capture all of the patients with stage III NSCLC treated with definitive radiation at our institution. Patients included in this cohort were chosen for gene panel testing to look for actionable mutations or clinical trial eligibility, and therefore may reflect a bias toward more aggressive disease. Second, the gene panel only includes a select group of up to 468 genes that were previously known to be involved in cancer. It does not include the rest of the exome or noncoding regions of the genome that may be implicated in cancer. Lastly, the study did not account for staging and treatment heterogeneity within the stage III NSCLC cohort. The study included both stage IIIA and IIIB patients who were treated with concurrent or sequential chemotherapy, and a small number of patients (5%) with definitive radiation alone.
Although our study represents a step toward understanding the genomic determinants of chemoradiation response, more studies are needed to validate our findings. These studies will require a much larger cohort of patients, prospectively collected and detailed clinical information, and deep tumor sequencing to fully investigate the relationship between each mutation and clinical outcome.
Conclusion
The study used an unbiased genomic approach and identified mutations in several genes including AKT2, KMT2C, and KMT2D as negative predictors of local-regional control and survival in stage III NSCLC patients. Deleterious mutations in DNA damage response and repair pathway genes were associated with improved local-regional control after chemoradiation.
Footnotes
Sources of support: This research was funded in part by NCI Cancer Center Support Grant P30 CA008748.
Disclosures: Dr Samstein has a patent for tumor mutational burden as a predictor of response to immunotherapy with royalties paid to Personal Genome Diagnostics. Dr Paik reports personal fees from Celgene, EMD Serono, Takeda, AbbVie, Boehringer Ingelheim, and Calithera, outside the submitted work. Dr Chaft reports grants and personal fees from AstraZeneca Pharmaceuticals LP and Bristol-Myers Squibb Company, grants from Merck & Co, Inc and Genentech, Inc, outside the submitted work. Dr Gomez reports grants from Merck, BMS, Varian, and AstraZeneca, personal fees from Merck, BMS, Varian, AstraZeneca, Reflexion, Vindico, Medscape, WebMD, and Reflexion, outside the submitted work. Dr Rimner reports grants from Boehringer Ingelheim and Pfizer, grants and personal fees from AstraZeneca and Merck, nonfinancial support from Philips/Elekta, personal fees from Cybrexa, MoreHealth, and ResearchToPractice, and grants from Varian Medical Systems, outside the submitted work. Dr Wu reports grants and nonfinancial support from CivaTech Oncology, Inc, personal fees from MORE Health and AstraZeneca, and nonfinancial support from AlphaTau Medical, outside the submitted work. Other authors do not have any conflicts of interest to disclose.
Research data are stored in an institutional repository and will be shared upon request to the corresponding author.
Supplementary material for this article can be found at https://doi.org/10.1016/j.adro.2020.10.027.
Supplementary Materials
References
- 1.Auperin A., Le Pechoux C., Rolland E. Meta-analysis of concomitant versus sequential radiochemotherapy in locally advanced non-small-cell lung cancer. J Clin Oncol. 2010;28:2181–2190. doi: 10.1200/JCO.2009.26.2543. [DOI] [PubMed] [Google Scholar]
- 2.Bradley J.D., Paulus R., Komaki R. Standard-dose versus high-dose conformal radiotherapy with concurrent and consolidation carboplatin plus paclitaxel with or without cetuximab for patients with stage IIIA or IIIB non-small-cell lung cancer (RTOG 0617): a randomised, two-by-two factorial phase 3 study. Lancet Oncol. 2015;16:187–199. doi: 10.1016/S1470-2045(14)71207-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dillman R.O., Seagren S.L., Propert K.J. A randomized trial of induction chemotherapy plus high-dose radiation versus radiation alone in stage III non-small-cell lung cancer. N Engl J Med. 1990;323:940–945. doi: 10.1056/NEJM199010043231403. [DOI] [PubMed] [Google Scholar]
- 4.Antonia S.J., Villegas A., Daniel D. Overall survival with durvalumab after chemoradiotherapy in stage III NSCLC. N Engl J Med. 2018 doi: 10.1056/NEJMoa1809697. [DOI] [PubMed] [Google Scholar]
- 5.Antonia S.J., Villegas A., Daniel D. Durvalumab after chemoradiotherapy in stage III non-small-cell lung cancer. N Engl J Med. 2017;377:1919–1929. doi: 10.1056/NEJMoa1709937. [DOI] [PubMed] [Google Scholar]
- 6.Raben D., Rimner A., Senan S. Patterns of disease progression with durvalumab in stage III non-small cell lung cancer (PACIFIC) Int J Radiat Oncol Biol Phys. 2019;105:683. [Google Scholar]
- 7.Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng D.T., Mitchell T.N., Zehir A. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zehir A., Benayed R., Shah R.H. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benjamini Y., Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Meth. 1995;57:289–300. [Google Scholar]
- 12.Teo M.Y., Bambury R.M., Zabor E.C. DNA damage response and repair gene alterations are associated with improved survival in patients with platinum-treated advanced urothelial carcinoma. Clin Cancer Res. 2017;23:3610–3618. doi: 10.1158/1078-0432.CCR-16-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teo M.Y., Seier K., Ostrovnaya I. Alterations in DNA damage response and repair genes as potential marker of clinical benefit from PD-1/PD-L1 blockade in advanced urothelial cancers. J Clin Oncol. 2018;36:1685–1694. doi: 10.1200/JCO.2017.75.7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez-Cespedes M., Parrella P., Esteller M. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res. 2002;62:3659–3662. [PubMed] [Google Scholar]
- 15.Goeman F., De Nicola F., Scalera S. Mutations in the KEAP1-NFE2L2 pathway define a molecular subset of rapidly progressing lung adenocarcinoma. J Thorac Oncol. 2019;14:1924–1934. doi: 10.1016/j.jtho.2019.07.003. [DOI] [PubMed] [Google Scholar]
- 16.Jeong Y., Hoang N.T., Lovejoy A. Role of KEAP1/NRF2 and TP53 mutations in lung squamous cell carcinoma development and radiation resistance. Cancer Discov. 2017;7:86–101. doi: 10.1158/2159-8290.CD-16-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Medina P.P., Romero O.A., Kohno T. Frequent BRG1/SMARCA4-inactivating mutations in human lung cancer cell lines. Hum Mutat. 2008;29:617–622. doi: 10.1002/humu.20730. [DOI] [PubMed] [Google Scholar]
- 18.Redig A.J., Capelletti M., Dahlberg S.E. Clinical and molecular characteristics of NF1-mutant lung cancer. Clin Cancer Res. 2016;22:3148–3156. doi: 10.1158/1078-0432.CCR-15-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shapiro G.I., Edwards C.D., Kobzik L. Reciprocal Rb inactivation and p16INK4 expression in primary lung cancers and cell lines. Cancer Res. 1995;55:505–509. [PubMed] [Google Scholar]
- 20.Singh A., Misra V., Thimmulappa R.K. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3 doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cassidy R.J., Zhang X., Patel P.R. Next-generation sequencing and clinical outcomes of patients with lung adenocarcinoma treated with stereotactic body radiotherapy. Cancer. 2017;123:3681–3690. doi: 10.1002/cncr.30794. [DOI] [PubMed] [Google Scholar]
- 22.Mak R.H., Hermann G., Lewis J.H. Outcomes by tumor histology and KRAS mutation status after lung stereotactic body radiation therapy for early-stage non-small-cell lung cancer. Clin Lung Cancer. 2015;16:24–32. doi: 10.1016/j.cllc.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lockney N.A., Yang T.J., Barron D. PIK3CA mutation is associated with increased local failure in lung stereotactic body radiation therapy (SBRT) Clin Transl Radiat Oncol. 2017;7:91–93. doi: 10.1016/j.ctro.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai J., Yang H., Shi S. Behavioral, morphological, and gene expression changes induced by (60)Co-gamma ray irradiation in Bactrocera tau (Walker) Front Physiol. 2018;9:118. doi: 10.3389/fphys.2018.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeong Y., Hellyer J.A., Stehr H. Role of KEAP1/NFE2L2 mutations in the chemotherapeutic response of patients with non-small cell lung cancer. Clin Cancer Res. 2020;26:274–281. doi: 10.1158/1078-0432.CCR-19-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.La Fleur L., Falk-Sorqvist E., Smeds P. Mutation patterns in a population-based non-small cell lung cancer cohort and prognostic impact of concomitant mutations in KRAS and TP53 or STK11. Lung Cancer. 2019;130:50–58. doi: 10.1016/j.lungcan.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Bellacosa A., de Feo D., Godwin A.K. Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. Int J Cancer. 1995;64:280–285. doi: 10.1002/ijc.2910640412. [DOI] [PubMed] [Google Scholar]
- 28.Cheng J.Q., Ruggeri B., Klein W.M. Amplification of AKT2 in human pancreatic cells and inhibition of AKT2 expression and tumorigenicity by antisense RNA. Proc Natl Acad Sci U S A. 1996;93:3636–3641. doi: 10.1073/pnas.93.8.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan Z.Q., Sun M., Feldman R.I. Frequent activation of AKT2 and induction of apoptosis by inhibition of phosphoinositide-3-OH kinase/Akt pathway in human ovarian cancer. Oncogene. 2000;19:2324–2330. doi: 10.1038/sj.onc.1203598. [DOI] [PubMed] [Google Scholar]
- 30.Rao R.C., Dou Y. Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat Rev Cancer. 2015;15:334–346. doi: 10.1038/nrc3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peifer M., Fernandez-Cuesta L., Sos M.L. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grasso C.S., Wu Y.M., Robinson D.R. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sjoblom T., Jones S., Wood L.D. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 34.Dalgliesh G.L., Furge K., Greenman C. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morin R.D., Mendez-Lago M., Mungall A.J. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ardeshir-Larijani F., Bhateja P., Lipka M.B. KMT2D mutation is associated with poor prognosis in non-small-cell lung cancer. Clin Lung Cancer. 2018;19:e489–e501. doi: 10.1016/j.cllc.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 37.Ma J., Setton J., Morris L. Genomic analysis of exceptional responders to radiotherapy reveals somatic mutations in ATM. Oncotarget. 2017;8:10312–10323. doi: 10.18632/oncotarget.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pitter K.L., Casey D.L., Setton J. Pathogenic mutations in ATM as determinants of local control in non-small cell lung cancers treated with radiation therapy. Int J Radiat Oncol Biol Phys. 2018;102:S226. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.