Abstract
This study was performed to investigate the role of interleukin-1 receptor accessory protein (IL1RAP) in stomach carcinoma in vitro and in vivo, determine whether IL1RAP knockdown could regulate the development of stomach carcinoma, and elucidate the relationship between IL1RAP knockdown and inflammation by tumor microenvironment-related inflammatory factors in stomach carcinoma. We first used TCGA and GEPIA systems to predict the potential function of IL1RAP. Second, western blot and RT-PCR were used to analyze the expression, or mRNA level, of IL1RAP at different tissue or cell lines. Third, the occurrence and development of stomach carcinoma in vitro and in vivo were observed by using IL1RAP knockdown lentivirus. Finally, the inflammation of stomach carcinoma in vitro and in vivo was observed. Results show that in GEPIA and TCGA systems, IL1RAP expression in STAD tumor tissue was higher than normal, and high expression of IL1RAP in STAD patients had a worse prognostic outcome. Besides, GSEA shown IL1RAP was negative correlation of apopopsis, TLR4 and NF-κB signaling pathway. We also predicted that IL1RAP may related to IL-1 s, IL-33, and IL-36 s in STAD. The IL1RAP expression and mRNA level in tumor, or MGC803, cells were increased. Furthermore, IL1RAP knockdown by lentivirus could inhibit stomach carcinoma development in vitro and in vivo through weakening tumor cell proliferation, migration, invasion, therefore reducing tumor volume, weight, and biomarker levels, and increasing apoptotic level. Finally, we found IL1RAP knockdown could increase inflammation of tumor microenvironment-related inflammatory factors of stomach carcinoma, in vitro and in vivo. Our study demonstrates that IL1RAP is possibly able to regulate inflammation and apoptosis in stomach carcinoma. Furthermore, TLR4, NF-κB, IL-1 s, IL-33, and IL-36 s maybe the downstream target factor of IL1RAP in inflammation. These results may provide a new strategy for stomach carcinoma development by regulating inflammation.
Keywords: IL1RAP, stomach carcinoma, TCGA, inflammation, apoptosis
Introduction
Gastric cancer remains one of the major causes of malignant disease morbidity and mortality.1 Helicobacter pylori virulence factors can induce an inflammatory response in the gastric mucosa and play a vital role in the development of stomach carcinoma.2
Epithelial cells, as well as macrophages and neutrophils, infiltrate into the gastric mucosa and release antimicrobial compounds, such as interleukin (IL), tumor necrosis factor, and matrix metalloproteinases.3,4 Inflammatory stimulation is required for the induction of the nuclear factor kappa B (NF-κB) and Toll-like receptor 4 (TLR4) signaling pathways, which could lead to the expression of the abovementioned pro-inflammatory cytokines.5,6
The tumor microenvironment harbors multiple immunosuppressive mechanisms, many of which involve suppressing the immune cells that can release ILs and affect tumor progression7; therefore, analysis of the markers of inflammation, with respect specifically to cell of origin and growth regulation, will be useful for treatment, prophylaxis, and prevention of stomach carcinomas.
The surface molecule IL-1 receptor accessory protein (IL1RAP; also IL-1R3, IL-RAcP) has been reported to be consistently overexpressed across multiple genetic subtypes of acute myeloid leukemia and has emerged as a novel therapeutic target.8 IL1RAP is involved in 3 signaling pathways that affect many IL-1 family cytokines (IL-1α, IL-1β, IL-33, IL-36β, and IL-36γ) in several diseases9; however, the mechanism of IL1RAP in the inflammation of stomach carcinoma is not fully understood.
In this study, we focused on the role of IL1RAP in stomach carcinoma and used a lentivirus to knockdown IL1RAP to boost inflammation progression and tumor development in vitro and in vivo. Our findings indicate that IL1RAP may affect inflammation and tumor development in stomach carcinoma. These results may provide a new strategy to research stomach carcinoma development by inflammation progress.
Materials and Methods
The Cancer Genome Atlas Stomach Adenocarcinoma (TCGA-STAD) genomic datasets were analyzed using gene expression profiling interactive analysis (GEPIA, https://gepia.cancer-pku.cn/). The results were automatically generated by selecting different modules. Specific quantified values and expression profiles in different cancer types were generated in the “general information” module. The expression profile assay and isoform analysis in the boxplot for IL1RAP expression were generated using the “profile” and “box plot” functions in the “expression DIY” module. Survival analysis was performed in the “survival analysis” module.
GSEA (4.0.3) was used to analyze TCGA-STAD data. According to the median expression of IL1RAP in TCGA, patients were divided into IL1RAP high expression group and IL1RAP low expression group. Analysis parameters—Gene set “c2. Cp. Reactome. Version. Symbols. GMT”—were downloaded from the molecular signature database (http://software.broadinstitute.org/gsea/msigdb/index.jsp) and used for analysis of enrichment, and 1000 permutations were performed for each gene set analysis to obtain a normalized enrichment score (NES). FDR <0.25 and P <0.05 were considered as significant enrichment.
We selected 25 patients pathologically diagnosed with stomach carcinoma by the gastrointestinal-surgery department in our hospital from July 2018 to July 2019 and used the para-carcinoma tissue as a control. All patients provided informed consent, and the experimental design was approved by the Ethics Committee of our hospital.
GES-1 is a human normal gastric mucosa cell line, and AGS, SGC-7901, and MGC803 are stomach carcinoma cell lines. Cells were cultured in RPMI-1640 medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% FBS and PenStrep (100 U/mL penicillin and 100 mg/mL streptomycin) in an incubator maintained at 5% CO2 and 37°C and trypsinized in a 0.25% trypsin solution containing 0.02% EDTA, as described previously.10
Healthy, 2-day- or 4-week-old, male BALB/c mice were supplied by Liaoning Changsheng Biotechnology Co., Ltd. (Shenyang, Liaoning, China). Mice were housed in a temperature- and light-controlled environment under pathogen-free conditions and provided with unlimited access to food and water. Mice were cared for in strict accordance with the Guide for the Care and Use of Laboratory Animals (NIH Publication No. 85–23, revised 1996), and the experimental design was approved by the Ethics Committee of our hospital.
The cell lines used in this study were as follows: GES-1 (CL-0563, Procell, Wuhan, China), AGS (CL-0022), SGC-7901, and MGC803 (ML-CS-0276, ATCC, Manassas, VA, USA).
The following primary antibodies were obtained from Abcam Inc. (Cambridge, MA, USA): Anti-IL1RAP (ab8110), Anti-CEA (ab207718), Anti-CA199 (ab3982), Anti-c-Met (ab51067), Anti-TLR4 (ab13556), Anti-IL-1α (ab227482), Anti-IL-1β (ab2105), Anti-IL-33 (ab207737), Anti-IL-36β (ab180890), and Anti-IL-36γ (ab180894). The primary antibodies Anti-NF-κB (BM3940) and Anti-β-actin (M01263-2) were purchased from Boster (Wuhan, Chin). The secondary antibodies conjugated to horseradish peroxidase—anti-rabbit IgG (H+L) (AS014) and anti-mouse IgG (H+L) (AS003)—were purchased from ABclonal (Wuhan, China).
IL1RAP knockdown was achieved by using an active lentivirus (sh-IL1RAP) designed and chemically synthesized by GenePharma Corporation (Shanghai, China). Lentiviral vectors were stored at −80°C. Stomach carcinoma cells were inoculated into 6-well plates at 2 × 105 cells/well, and the serum-free medium was replaced after the cell density passed 70%. Next, 10 µL of the lentivirus was diluted in the cell medium and incubated for 6 h. Protein or mRNA was collected after 72 h. Cells transfected with lentivirus were used to establish a stomach carcinoma mouse model after the passage. The sequences of the IL1RAP lentiviruses are listed in Table 1.
Table 1.
The Name and Sequence of IL1RAP Lentivirus.
| Name | Sequence |
|---|---|
| sh-IL1RAP-421 | 5′-TTTTATGGAATCCTGCAAAG-3′ |
| sh-IL1RAP-662 | 5′-TTAGTAAGGAGAAAGATGTG-3′ |
| sh-IL1RAP-1146 | 5′-CTATGAGAAAGAACCAGGAG-3′ |
| Negative Control | 5′-TTGCCCCTCCCTTTAATATC-3′ |
| control-shIL1RAP | 5′-CATCGTCATGTGATCATCAC-3′ |
The subjects were randomly divided into 3 groups (n = 6 in each group): (1) control group: untreated MGC803 cells or mice that received MGC803 injection; (2) sh-IL1RAP group: MGC803 cells treated with sh-IL1RAP or mice that received MGC803 injection and were then treated with sh-IL1RAP injection; (3) control-sh-IL1RAP group: MGC803 cells treated with IL1RAP control lentivirus or mice that underwent MGC803 injection and were treated with IL1RAP control lentivirus injection.
Four-week-old male BALB/c mice were fed in an SPF environment, and MGC803 cells were injected subcutaneously. After 60 h, vital signs and inoculation sites in the mice were observed. Tumor tissue was collected after modeling for 28 d, and the tumor volume was calculated using the formula: 0.5 × length × width2 (mm3).
A total of 40 µg of protein from each sample was subjected to 12% SDS-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. All blots were imaged using the ChemiDocTM Touch Imaging System and analyzed with the Image Lab software (version 3.0; Bio-Rad, Hercules, California, USA).
Total RNA was extracted from the frozen tissue of subject animals using a RNAiso reagent kit (TaKaRa Biotechnology, Dalian, China). A total of 40 µL of RNA was reverse transcribed into cDNA. Quantitative PCR was performed as previously described.11 The IL1RAP level was calculated as a ratio normalized to β-actin.
For immunohistochemical (IHC) staining, 4-μm-thick tissue sections were incubated with the anti-IL1RAP (1:100) primary antibody at 4°C overnight. The sections were then incubated with the secondary antibody (1:200) at room temperature for 2 h. Staining were measured using a commercial streptavidin-biotin-peroxidase staining kit (Golden Bridge Biotechnology Co., Beijing, China).
Cells were visualized using the propidium iodide (PI)-Hoechst assay (40755ES64, Qcbio Science&Technologies Co., Ltd, Shanghai, China) according to the manufacturer’s instructions. Images were captured through fluorescence microscopy at 400× magnification. Images were acquired with a Nikon Eclipse Ni inverted microscope (TE2000; Nikon, Tokyo, Japan).
The tissue fraction was determined using the TUNEL assay. The TUNEL assay was performed according to the manufacturer’s instructions (ab66108; Abcam, Inc.). Images were captured using fluorescence microscopy at 400× magnification.
After stirring the solution for 30 min, the bacteria were removed with a microporous (diameter 0.22 m) filter and stored at 4°C away from light, after packaging. Cells in the logarithmic growth stage were inoculated into 96-well plates in groups, as previously described.12
For cell migration assays, a transwell chamber was pretreated with Matrigel (Corning, Corning, NY, USA) and dried at 37°C for 1 h, and the assay was conducted following the manufacturer’s instructions. The results of the transwell migration and invasion assays were calculated according to the number of transferred cells.
For cell cycle determination, cells were plated in six-well plates at a density of 2 × 105 cells per well. After 12 h, various concentrations (0, 25, 50, and 100 mM) of eriodyctiol were added to each well, and the cells were incubated for an additional 48 h. Detailed procedures were performed as described previously.13
Data are expressed as mean ± standard deviation. Statistical analysis was performed using GraphPad Prism 6.0 (GraphPad Software, San Diego, CA, USA) and SPSS18.0 (IBM, New York, NY, USA). Differences were analyzed using one-way ANOVA, and multiple comparisons were analyzed using the Sidak test. Differences were considered statistically significant at P <0.05.
Results
Bioinformatic Analysis of IL1RAP in STAD Using TCGA and GEPIA
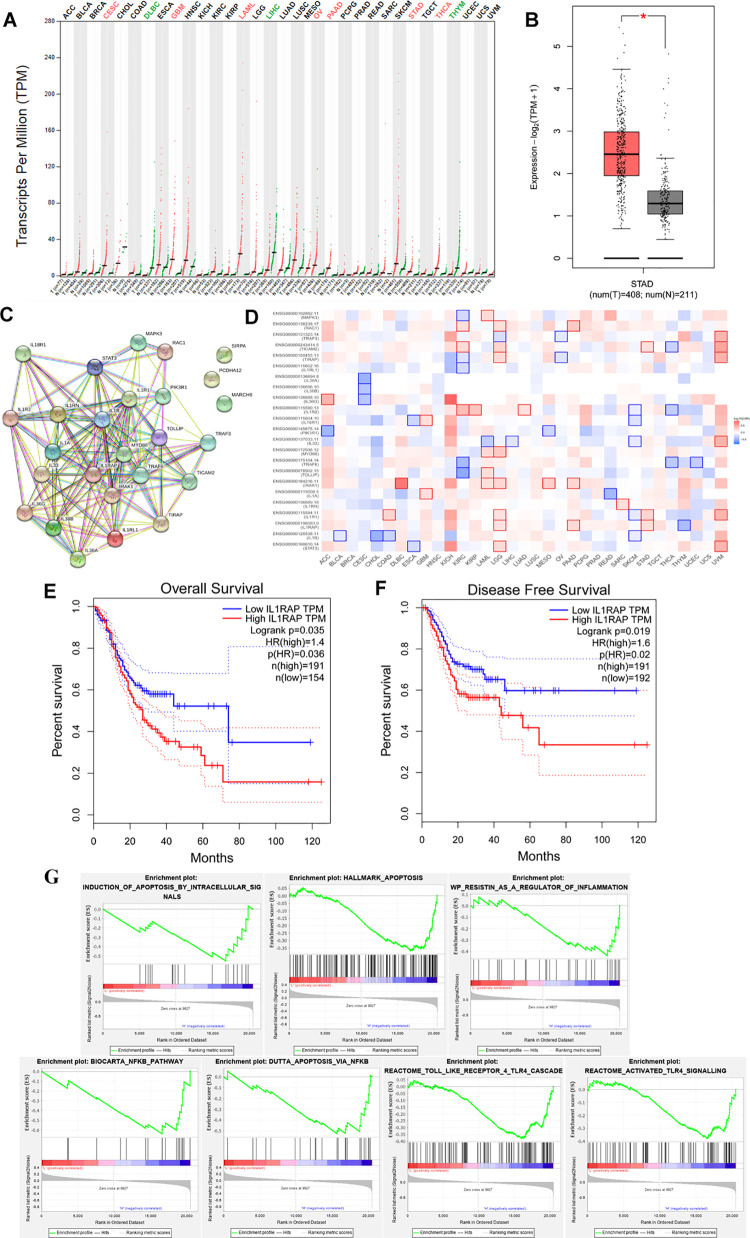
Predictions of IL1RAP function in STAD through GEPIA and TCGA are shown in Figure 1. As shown in Figure 1A, the predicted expression of IL1RAP as transcripts per million was higher in STAD tissue than that in normal tissue. The expression profile of IL1RAP in STAD tissue was similar as it was shown in Figure 1B (P < 0.05). The interacting proteins for IL1RAP, as determined using string assay, were MAPK3, RAC1, TRAF3, TICAM2, TLRAP, IL1RL1, IL-36A, IL-36B, IL-36G, IL18R1, PI3KR1, IL-33, MYD88, TRAF6, TOLLIP, IRAK1, IL-1A, IL1RN, IL1R1, IL-1B, and STAT3 (Figure 1C). The survival map for IL1RAP and its interacting proteins is shown in Figure 1D, which shows significant results in STAD of IL1RAP. Through analysis of overall survival and disease-free survival (Figure 1E-F), we can confirm that given the high expression of IL1RAP isoform, the patients with STAD had a worse prognostic outcome, especially in the disease-free survival assay (P < 0.05). GSEA showed that IL1RAP in STAD was negatively correlated with apoptosis, TLR4, and NF-κB signaling pathways.
Figure 1.
(A) Transcripts per million of IL1RAP in different cancers by GEPIA and TCGA system. (B) Expression profile assay of IL1RAP in STAD and normal tissue. (C) Interacting proteins of String assay and (D) Survival map for IL1RAP and its interacting proteins in STAD. (E) Overall survival analyses and (F) Disease-free survival analyses of high-level and low-level IL1RAP in STAD patients. (G) GSEA assay of IL1RAP in STAD patients.
Expression of IL1RAP in Different Tissues and STAD
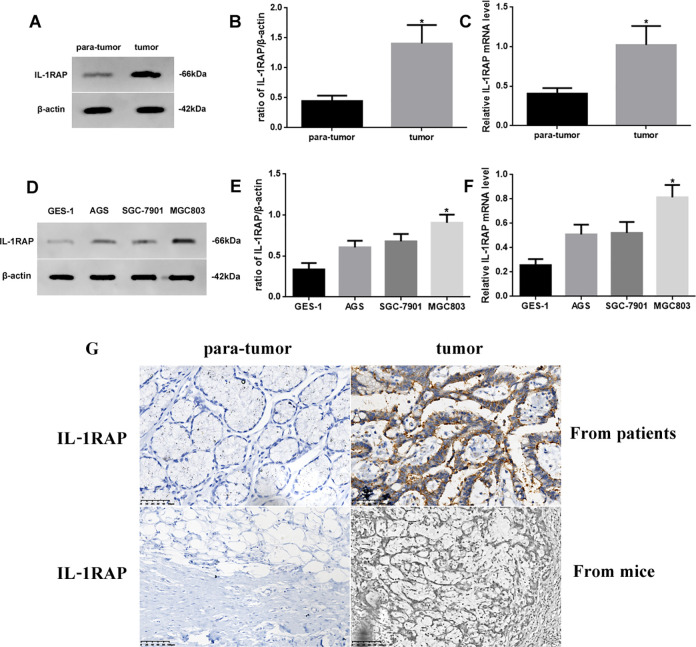
As bioinformatic analysis predicted that IL1RAP was related to STAD, we measured the difference in protein and mRNA expression of IL1RAP in different tissues or cell lines using western blotting and RT-PCR, respectively (Figure 2). IL1RAP protein and mRNA expression in stomach tumor tissue was increased (P < 0.05) compared to para-tumor tissue (Figure 2A-C). In addition, the IL1RAP protein and mRNA expression in MGC803 cells was most significantly increased (P < 0.05) compared to GES-1, AGS, and SGC-7901 cells (Figure 2D-F). We used MGC803 as the model stomach carcinoma cell line for follow-up experiments. The approximate localization of IL1RAP as determined through IHC staining is shown in Figure 2G; the fluorescence signals in IL1RAP-positive cells in the IHC sections were observed outside the cytoplasm and membrane.
Figure 2.
(A) Western blot assay of IL1RAP expression in tumor and para-tumor tissue, (B) Quantification, and (C) Quantitative RT-PCR assay for mRNA level of IL1. RAP in different tissues. (D) Western blot assay of IL1RAP expression in GES-1, AGS, SGC-7901, and MGC803 cells, (E) Quantification, and (F) Quantitative RT-PCR assay for mRNA level of IL1RAP in different cells. (G) IHC assay of IL1RAP expression of para-tumor and tumor from patients (×400) and mice (×200). Protein and mRNA levels were normalized to β-actin. (Tumor or MGC803 cells vs. para-tumor or GES-1 cells, *P < 0.05, n = 6 per group, all data was represented as Mean ± Standard error).
IL1RAP Knockdown Could Regulate the Development of Stomach Carcinoma In Vitro
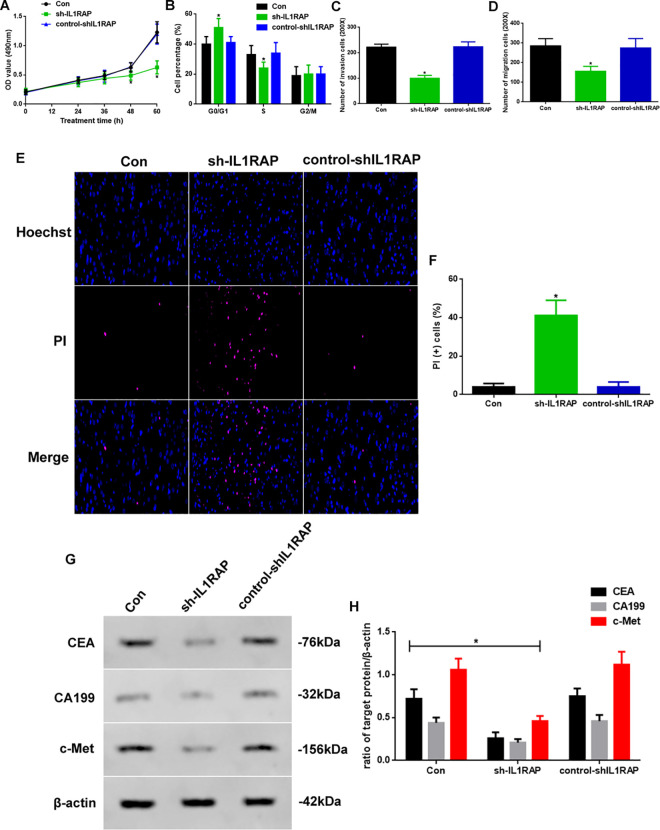
As IL1RAP expression was shown to be increased in tumor tissue, we were prompted to determine the role of this protein in stomach carcinoma progression. We developed a method for decreasing the IL1RAP level: IL1RAP was downregulated through lentivirus (sh-IL1RAP)-mediated knockdown, as shown in Figure 3. We measured proliferation, cell cycle, migration, and invasion of control-, sh-IL1RAP-, and control-shIL1RAP-treated MGC803 cells. The optical density at 48 and 60 h, as measured in the MTT assay, was significantly lower in the sh-IL1RAP group than that in the other 2 groups (Figure 3A). The distribution in the G0/G1 phase in sh-IL1RAP cells was significantly higher than that in the other 2 groups, while the opposite was observed in the S phase (Figure 3B, P < 0.05). Additionally, the migration and invasion abilities of the sh-IL1RAP group were decreased (Figure 3C-D, P < 0.05). To investigate the function of IL1RAP in apoptosis, we used PI staining to observe apoptotic changes (Figure 3E). Apoptosis in the sh-IL1RAP group was increased (Figure 3F, P < 0.05). In addition, we detected biomarkers of stomach carcinoma through western blotting (Figure 3G). The expression of CEA, CA199, and c-Met in the sh-IL1RAP group was reduced (P < 0.05) compared to other groups (Figure 3H).
Figure 3.
(A) Proliferation assay in 72 hours, (B) Cell cycle assay, (C) Migration assay and (D) Invasion assay at 60 hours of Con, sh-IL1RAP, and control-shIL1RAP group. (E) PI-Hoechst staining assay (×400) of apoptosis and (F) PI (+) cell assay. (G) Western blot assay of CEA, CA199 and c-Met expression in vitro. (H) Quantification of CEA, CA199, and c-Met expression. Protein levels were normalized to β-actin (sh-IL1RAP vs. other group, * P < 0.05, n = 6 per group).
IL1RAP Knockdown Could Regulate the Development of Stomach Carcinoma In Vivo
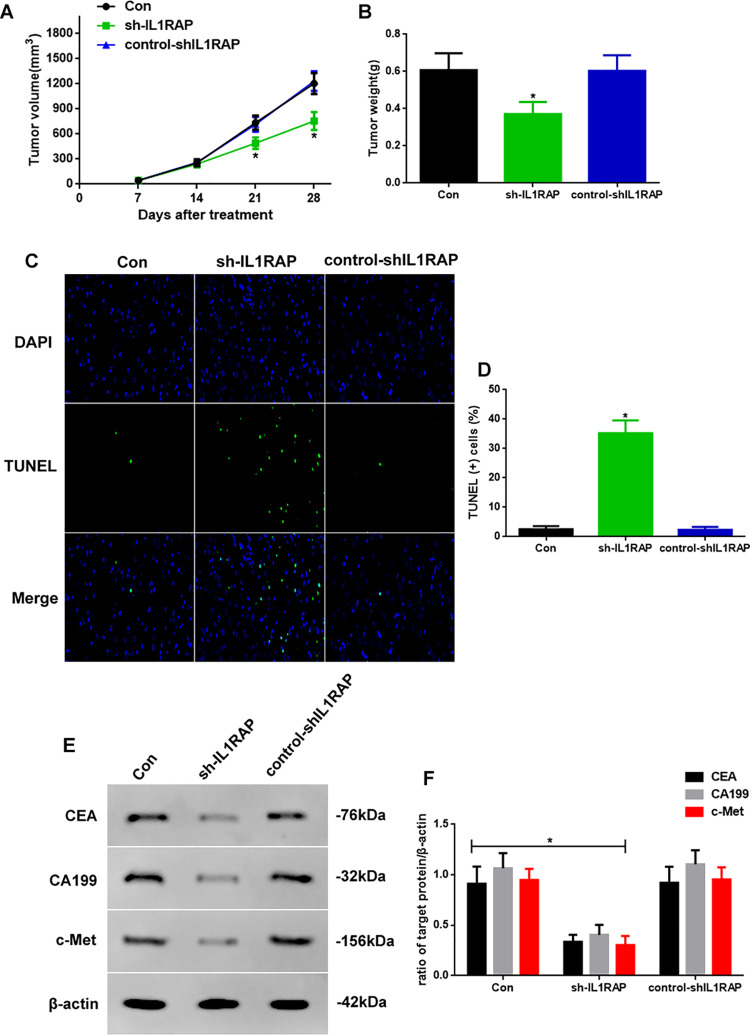
As knockdown of IL1RAP regulated the development of stomach carcinoma in vitro, we next determined if it also regulated this process in vivo (Figure 4). First, we measured the tumor volume and weight in control, sh-IL1RAP, and control-shIL1RAP mice. The tumor volumes at 21 and 28 d in sh-IL1RAP mice were significantly lower than those in the other 2 groups (Figure 4A, P < 0.05); additionally, the tumor weights in sh-IL1RAP mice were reduced (Figure 4B, P < 0.05). Second, to investigate the apoptotic function of IL1RAP, we performed TUNEL assay (Figure 4C) and found that the apoptotic levels in sh-IL1RAP mice were high (Figure 4D, P < 0.05). Finally, we detected the biomarkers of stomach carcinoma through western blotting (Figure 4C). The expression of CEA, CA199, and c-Met in the sh-IL1RAP group was decreased (P < 0.05) compared to the other groups (Figure 3D), which was consistent with the in vitro results.
Figure 4.
(A) Tumor volume in 28-day period and (B) Tumor weight at 28 days of Con, sh-IL1RAP, and control-shIL1RAP mice. (C) TUNEL-DAPI staining assay (×400) of apoptosis and (D) TUNEL (+) cell assay. (E) Western blot assay of CEA, CA199, and c-Met expression in vivo. (F) Quantification of CEA, CA199, and c-Met expression. Protein levels were normalized to β-actin (sh-IL1RAP vs. other group, * P < 0.05, n = 6 per group).
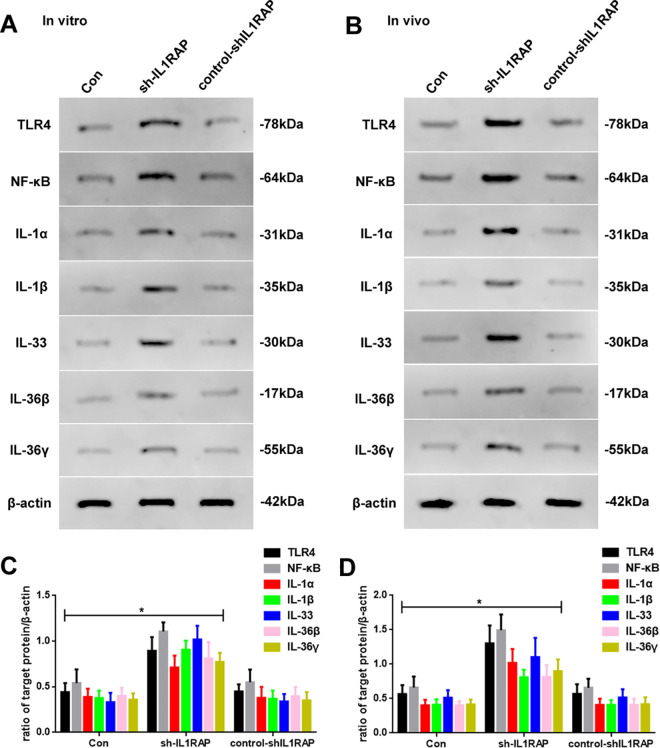
IL1RAP Knockdown Could Increase Inflammatory Marker Levels In Vitro and In Vivo
To further confirm whether the tumor-regulatory effect of IL1RAP was mediated through inflammation, we determined the expression of the inflammatory promoter and inflammation-related proteins that were related to IL1RAP, through western blotting (Figure 5). As shown in Figure 5A, IL1RAP knockdown enhanced protein expression of TLR4, NF-κB, IL-1α, IL-1β, IL-33, IL-36β, and IL-36γ (Figure 5C). The in vivo outcomes were thus consistent with our in vitro observations (Figure 5B and D).
Figure 5.
(A) Western blot assay of TLR4, NF-κB, IL-1α, IL-1β, IL-33, IL-36β, and IL-36γ expression of Con, sh-IL1RAP, and control-shIL1RAP groups in vitro. (B) Western blot assay of TLR4, NF-κB, IL-1α, IL-1β, IL-33, IL-36β, and IL-36γ expression of Con, sh-IL1RAP, and control-shIL1RAP mice in vivo. (C) Quantification of TLR4, NF-κB, IL-1α, IL-1β, IL-33, IL-36β, and IL-36γ of Con, sh-IL1RAP, and control-shIL1RAP groups in vitro. (D) Quantification of TLR4, NF-κB, IL-1α, IL-1β, IL-33, IL-36β, and IL-36γ of Con, sh-IL1RAP, and control-shIL1RAP groups in vivo. Protein levels were normalized to β-actin (sh-IL1RAP vs. other group, * P < 0.05, n = 6 per group).
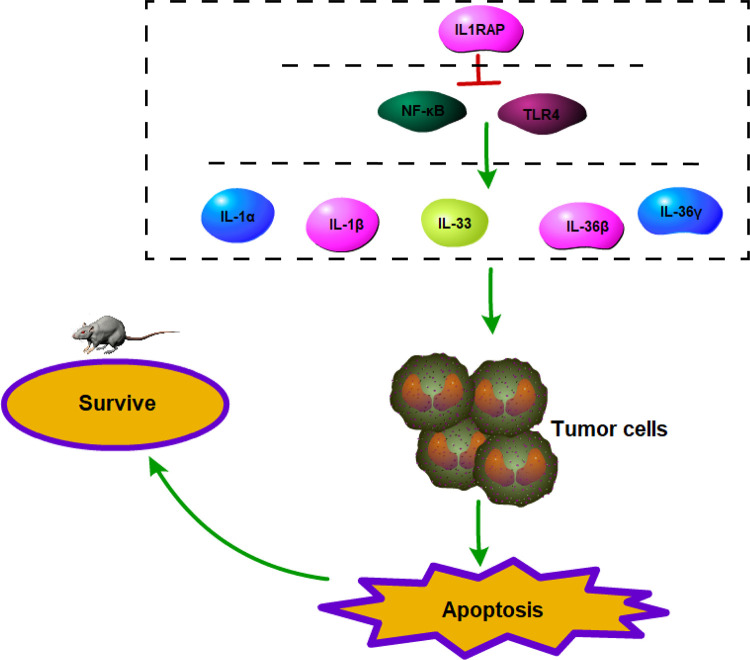
The potential mechanisms of how IL1RAP induces inflammation in stomach carcinoma are shown in Figure 6. IL1RAP probably regulates the inflammatory promoter, similar to TLR4 and NF-κB, and the IL1RAP-related proteins such as IL-1, IL-33, or IL-36, to restrain the inflammatory progress and apoptosis, enhance the development of stomach carcinoma, and become fatal for mice with stomach carcinoma.
Figure 6.
The potential mechanisms of inflammation by IL1RAP mediated in stomach carcinoma. IL1RAP probably could reduce the related proteins of inflammation expression just like TLR4, NF-κB, IL-1α, IL-1β, IL-33, IL-36β, and IL-36γ, weaken the inflammatory progress and apoptosis progress to tumor cells, and cut down lives of mice of stomach carcinoma.
Discussion
Gastrointestinal malignancies rank fifth in terms of cancer morbidity and third in cancer-related death, have high incidence and mortality rates, and impose enormous worldwide health and economic burdens.14 The unique physiology of tumor cells creates a hostile and nutrient-poor microenvironment, and chemical and metabolic adaptations that occur within and among heterogeneous cell types of tumors facilitate growth.15 The innate immune system is evolutionarily conserved and plays a role in immune surveillance via specific cells, such as macrophages, dendritic cells, neutrophils, and natural killer cells, as well as through soluble factors such as the interleukin system, to withstand the tumor microenvironment.16 Briefly, the inflammatory microenvironment plays a crucial role in stomach carcinoma.
ILs are cytokines involved in inflammation and development of many types of cancer.17 IL-1 is an inflammatory cytokine that plays a key role in carcinogenesis, tumor progression, and the inflammatory tumor microenvironment.18 IL1RAP has been shown to be an important inflammatory regulator in the IL-1 pathway and is related to the TLRs.19 IL1RAP has been reported to be consistently overexpressed in some cancer types and has emerged as a novel therapeutic target20,21; however, the mechanism of IL1RAP in the inflammation of stomach carcinoma is not fully understood. In our study, we used GEPIA and TCGA to predict IL1RAP function in STAD. The results showed that IL1RAP expression in STAD tissues was higher than that in noncancerous tissues. As demonstrated via the string assay, the proteins interacting with IL1RAP were IL-1, IL-33, and IL-36, which belong to the IL family. In addition, the high expression of IL1RAP isoform in patients with STAD had a worse prognostic outcome, especially in the disease-free survival assay. These results suggest that high levels of IL1RAP may cause the development of STAD and may regulate the inflammatory process.
To verify the bioinformatic analysis of IL1RAP, we determined the protein and mRNA levels of IL1RAP in different tissues and cell lines and found that IL1RAP in tumor or MGC803 cells was increasingly expressed, suggesting that IL1RAP may be related to the development of stomach carcinoma.
To further investigate the function of IL1RAP in the development of stomach carcinoma, IL1RAP knockdown was performed to reduce IL1RAP expression. Our results showed that the knockdown of IL1RAP, both in vivo and in vitro, inhibited stomach carcinoma development by weakening tumor proliferation, migration, and invasion; enhancing apoptosis; and reducing tumor volume, weight, and expression of stomach carcinoma biomarkers such as CEA, CA199, and c-Met. These results suggest that lowered levels of IL1RAP could improve stomach carcinoma outcomes; however, the specific mechanism of IL1RAP in stomach carcinoma remains unclear.
IL1RAP has been reported as a tumor-associated antigen for cell-targeted immunotherapy and is involved in different signaling pathways.22,23 IL1RAP is involved in 3 signaling pathways that affect many cytokines of the IL-1 family (IL-1α, IL-1β, IL-33, IL-36β, and IL-36γ) in many diseases9; however, the relationship between IL1RAP and IL-1α, IL-1β, IL-33, IL-36β, and IL-36γ in the inflammation process of stomach carcinoma has not been fully reported.
To investigate the inflammatory function of IL1RAP in stomach carcinoma, we measured TLR4, NF-κB, IL-1α, IL-1β, IL-33, IL-36β, and IL-36γ levels and found that the levels of these proteins were increased by IL1RAP knockdown, which could further describe the immunomodulatory function of IL1RAP. These results suggest that IL1RAP may be involved in the regulation of tumor microenvironment-related inflammatory factors and plays a crucial role in the inflammation process of stomach carcinoma. As shown in Figure 1G, the GSEA shows that IL1RAP expression in STAD was negatively correlated with apoptosis, TLR4, and NF-κB signaling pathways, which is consistent with our results.
Our findings indicate that IL1RAP knockdown significantly activated inflammation and inhibited the development of stomach carcinoma and then improved the level of apoptosis, reducing the level of biomarkers of gastrointestinal tumors. Taken together, these findings may provide a new strategy for the treatment of stomach carcinoma development by regulating inflammation.
Supplemental Material
Supplemental Material, sj-pdf-1-tct-10.1177_1533033821995282 for The Potential Role of IL1RAP on Tumor Microenvironment-Related Inflammatory Factors in Stomach Adenocarcinoma by Qing Lv, Qinghua Xia, Anshu Li and Zhiyong Wang in Technology in Cancer Research & Treatment
Footnotes
Authors’ Note: QL and QX designed the study and write the first draft; Polished the first draft and confirmed the methodology and material parts; AL & ZW analyzed the data, write, and revised the paper. All authors read and approved the final manuscript. All animals were cared for in strict accordance with the Guide for the Care and Use of Laboratory Animals (NIH Publication No. 85-23, revised 1996), and the experimental design was applied by the Ethics Committee of Wuhan Union Hospital.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was funded by the Foundational fund of Wuhan Union hospital under grant number 0203201915.
ORCID iD: Zhiyong Wang  https://orcid.org/0000-0002-4596-2655
https://orcid.org/0000-0002-4596-2655
Supplemental Material: Supplemental material for this article is available online.
References
- 1. de Martel C, Forman D, Plummer M. Gastric cancer: epidemiology and risk factors. Gastroenterol Clin North Am. 2013;42(2):219–240. doi:10.1016/j.gtc.2013.01.003 [DOI] [PubMed] [Google Scholar]
- 2. Waldum HL, Fossmark R. Types of gastric carcinomas. Int J Mol Sci. 2018;19(12):4109 doi:10.3390/ijms19124109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Serizawa T, Hirata Y, Hayakawa Y, et al. Gastric metaplasia induced by Helicobacter pylori is associated with enhanced SOX9 expression via interleukin-1 signaling. Infect Immun. 2016;84(2):562–572. doi:10.1128/IAI.01437-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Torres-Martinez AC, Gallardo-Vera JF, Lara-Holguin AN, Montano LF, Rendon-Huerta EP. Claudin-6 enhances cell invasiveness through claudin-1 in AGS human adenocarcinoma gastric cancer cells. Exp Cell Res. 2017;350(1):226–235. doi:10.1016/j.yexcr.2016.11.025 [DOI] [PubMed] [Google Scholar]
- 5. Backert S, Naumann M. What a disorder: proinflammatory signaling pathways induced by Helicobacter pylori . Trends Microbiol. 2010;18(11):479–486. doi:10.1016/j.tim.2010.08.003 [DOI] [PubMed] [Google Scholar]
- 6. Chen G, Xu M, Chen J, et al. Clinicopathological features and increased expression of toll-like receptor 4 of gastric cardia cancer in a high-risk Chinese population. J Immunol Res. 2018;2018:7132868 doi:10.1155/2018/7132868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu HT, Ai X, Lu M, Song Z, Li H. Characterization of intratumoral and circulating IL-10-producing B cells in gastric cancer. Exp Cell Res. 2019;384(2):111652 doi:10.1016/j.yexcr.2019.111652 [DOI] [PubMed] [Google Scholar]
- 8. Mitchell K, Barreyro L, Todorova TI, et al. IL1RAP potentiates multiple oncogenic signaling pathways in AML. J Exp Med. 2018;215(6):1709–1727. doi:10.1084/jem.20180147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hojen JF, Kristensen MLV, McKee AS, et al. IL-1R3 blockade broadly attenuates the functions of six members of the IL-1 family, revealing their contribution to models of disease. Nat Immunol. 2019;20(9):1138–1149. doi:10.1038/s41590-019-0467-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fan L, Tan B, Li Y, et al. Upregulation of miR185 promotes apoptosis of the human gastric cancer cell line MGC803. Mol Med Rep. 2018;17(2):3115–3122. doi:10.3892/mmr.2017.8206 [DOI] [PubMed] [Google Scholar]
- 11. Kong Q, Yu M, Zhang M, et al. Conditional Dnmt3b deletion in hippocampal dCA1 impairs recognition memory. Mol Brain. 2020;13(1):42 doi:10.1186/s13041-020-00574-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen J, He GM, Xian GY, Su XQ, Yu LL, Yao F. Mechanistic biosynthesis of SN-38 coated reduced graphene oxide sheets for photothermal treatment and care of patients with gastric cancer. J Photochem Photobiol B. 2020;204:111736 doi:10.1016/j.jphotobiol.2019.111736 [DOI] [PubMed] [Google Scholar]
- 13. Cikutovic-Molina R, Herrada AA, Gonzalez W, Brown N, Zuniga L. TASK-3 gene knockdown dampens invasion and migration and promotes apoptosis in KATO III and MKN-45 human gastric adenocarcinoma cell lines. Int J Mol Sci. 2019;20(23):6077 doi:10.3390/ijms20236077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xiang T, Jiang HS, Zhang BT, Liu G. CircFOXO3 functions as a molecular sponge for miR-143-3p to promote the progression of gastric carcinoma via upregulating USP44. Gene. 2020;753:144798 doi:10.1016/j.gene.2020.144798 [DOI] [PubMed] [Google Scholar]
- 15. Halbrook CJ, Lyssiotis CA. Employing metabolism to improve the diagnosis and treatment of pancreatic cancer. Cancer Cell. 2017;31(1):5–19. doi:10.1016/j.ccell.2016.12.006 [DOI] [PubMed] [Google Scholar]
- 16. Wang M, Busuttil RA, Pattison S, Neeson PJ, Boussioutas A. Immunological battlefield in gastric cancer and role of immunotherapies. World J Gastroenterol. 2016;22(28):6373–6384. doi:10.3748/wjg.v22.i28.6373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hong JT, Son DJ, Lee CK, Yoon DY, Lee DH, Park MH. Interleukin 32, inflammation and cancer. Pharmacol Ther. 2017;174:127–137. doi:10.1016/j.pharmthera.2017.02.025 [DOI] [PubMed] [Google Scholar]
- 18. Mantovani A, Barajon I, Garlanda C. IL-1 and IL-1 regulatory pathways in cancer progression and therapy. Immunol Rev. 2018;281(1):57–61. doi:10.1111/imr.12614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. O’Neill LA. The interleukin-1 receptor/Toll-like receptor superfamily: 10 years of progress. Immunol Rev. 2008;226:10–18. doi:10.1111/j.1600-065X.2008.00701.x [DOI] [PubMed] [Google Scholar]
- 20. He J, Li X, Zhu W, Yu Y, Gong J. Research of differential expression of sIL1RAP in low-grade gliomas between children and adults. Brain Tumor Pathol. 2018;35(1):19–28. doi:10.1007/s10014-017-0304-x [DOI] [PubMed] [Google Scholar]
- 21. Smallridge RC, Chindris AM, Asmann YW, et al. RNA sequencing identifies multiple fusion transcripts, differentially expressed genes, and reduced expression of immune function genes in BRAF (V600E) mutant vs BRAF wild-type papillary thyroid carcinoma. J Clin Endocrinol Metab. 2014;99(2):E338–E347. doi:10.1210/jc.2013-2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Warda W, Larosa F, Neto Da Rocha M, et al. CML hematopoietic stem cells expressing IL1RAP can be targeted by chimeric antigen receptor-engineered T cells. Cancer Res. 2019;79(3):663–675. doi:10.1158/0008-5472.CAN-18-1078 [DOI] [PubMed] [Google Scholar]
- 23. Chen R, Li M, Zhang Y, Zhou Q, Shu HB. The E3 ubiquitin ligase MARCH8 negatively regulates IL-1beta-induced NF-kappaB activation by targeting the IL1RAP coreceptor for ubiquitination and degradation. Proc Natl Acad Sci U S A. 2012;109(35):14128–14133. doi:10.1073/pnas.1205246109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, sj-pdf-1-tct-10.1177_1533033821995282 for The Potential Role of IL1RAP on Tumor Microenvironment-Related Inflammatory Factors in Stomach Adenocarcinoma by Qing Lv, Qinghua Xia, Anshu Li and Zhiyong Wang in Technology in Cancer Research & Treatment