Abstract
Expression of the GFAP gene has attracted considerable attention because its onset is a marker for astrocyte development, its upregulation is a marker for reactive gliosis, and its predominance in astrocytes provides a tool for their genetic manipulation. The literature on GFAP regulation is voluminous, as almost any perturbation of development or homeostasis in the CNS will lead to changes in its expression. In this review, we limit our discussion to mechanisms proposed to regulate GFAP synthesis through a direct interaction with its gene or mRNA. Strengths and weaknesses of the supportive experimental findings are described, and suggestions made for additional studies. This review covers 15 transcription factors, DNA and histone methylation, and microRNAs. The complexity involved in regulating the expression of this intermediate filament protein suggests that GFAP function may vary among both astrocyte subtypes and other GFAP-expressing cells, as well as during development and in response to perturbations.
Keywords: GFAP, astrocyte, regulation of transcription and translation, gliogenesis, gene expression, gene structure
Introduction
GFAP encodes the predominant intermediate filament protein present in mature astrocytes. The biological roles of this protein are described in the companion review (Messing and Brenner, 2020). Here, we review the gene’s expression, which has attracted considerable attention because its onset is a marker for astrocyte development, its upregulation is a marker for reactive gliosis, and its predominant activity in astrocytes is a tool for their genetic manipulation. It also has clinical relevance, because the ability to inhibit GFAP synthesis may provide a treatment for Alexander disease (Hagemann et al., 2018), a usually fatal astrogliopathy caused by GFAP mutations (Messing et al., 2012). The literature on GFAP transcription is voluminous, as almost any perturbation of development or homeostasis in the CNS will lead to changes in GFAP expression. This review is limited to regulatory factors shown to bind to the GFAP gene or its mRNA. As examples of this limitation, both Sox9 (Wang X et al., 2018) and histone acetylation (Kanski et al., 2014; de Menezes et al., 2018) are important for GFAP expression, but neither is discussed because the mechanisms of these effects have not been studied. Factors affecting GFAP stability are described in the Messing and Brenner (2020) review. Despite these stringent selection criteria, a large number of regulatory factors qualified for inclusion. These are discussed in two main sections, the first being devoted to mechanisms regulating the developmental onset of GFAP expression, and the second to transcription factors more generally in play.
Nomenclature
In the discussion that follows, the nucleotide (nt) positions of sites are relative to +1 being the transcription start site. Note that the starting RNA sequence determined for human GFAP, AGAGCCAGAGCA, (Brenner et al., 1990) commences 1 nt 5’ of that for rat and mouse (Feinstein et al., 1992), despite their sequences being highly homologous in this region. Positions for the human gene are in accordance with the NCBI Reference Sequence NC_000017.11, found at https://www.ncbi.nlm.nih.gov/gene/2670. Positions upstream of –512 may be 1 nt less than numbers previously published from our laboratories, because our sequence of the promoter has a string of 19 T’s proceeding upstream from –494, whereas the NCBI sequence has 18 T’s. Mouse and rat numberings are from NCBI Reference Sequences NC_000077.6 and NC_005109.4, respectively. We use the standard notation of GFAP to refer specifically to the human gene, Gfap for the rat or mouse gene, and GFAP (no italics) when not referring to a specific species. However, since we found mechanistic studies of GFAP expression only for mouse, rat and humans, the non-italicized GFAP refers to the gene of these three species; it is also used for designating the protein itself.
Methodology and Caveats
Transient Transfection
In this review, we present the experimental evidence for findings, and caveats for their interpretation. Primary among the methods used is transient transfection assays. Although attractive for its rapidity and low cost (and used extensively by the authors), in many instances results obtained for expression of GFAP promoters are not supported by subsequent studies in transgenic mice. Among these are multiple reports that GFAP promoter segments extending no further upstream than –250 support the same activity as ones extending further than –1600. For example, Sun et al. (2001) found that a rat Gfap promoter commencing at –106 yielded the same reporter activity as one commencing at –1876. As will be discussed below, sequences upstream of –250, such as the STAT binding site, are critical for GFAP expression. Furthermore, a mouse transgene commencing at –256 was inactive (personal communication of K. Ikenaka, K. Nakahira, and K. Mikoshiba, cited in Brenner, 1994). Use of a luciferase reporter is a feature common to the multiple instances in which a proximal GFAP promoter yielded substantial activity, suggesting it may contribute to the expression. Consistent with this possibility, K. Ikenaka reported that use of the luciferase reporter masks the contribution to activity of the upstream region of the GFAP promoter (personal communication cited in Brenner and Messing, 1996). Another example of a misleading result from transient transfection is the finding of Lee et al. (2006) that a human GFAP promoter consisting of bp –1756 to –1488 joined to –132 to +47 (the gfaABD promoter) has 10 times higher activity in transfected cells than a promoter spanning –2162 to +47 (the gfa2 promoter), but is not more active in transgenic mice (details about this and other studies cited in this paragraph are provided in the appropriate sections below). Also, whereas a gfaABD-nlacZ transgene expressed strongly in the cerebellum and weakly in cerebral cortex, and this difference was maintained in astrocytes cultured from the transgenic mice, the construct expressed equally well when transiently transfected into primary astrocytes isolated from these two regions. Consequently, any result derived from transient transfection should be confirmed by another method.
Electrophoretic Mobility Shift and Chromatin Immunoprecipitation Assays
Two other techniques commonly used to study transcriptional regulation of GFAP whose results should be interpreted with caution are electrophoretic mobility shift assays (EMSAs) and chromatin immunoprecipitation (ChIP) assays. EMSAs show that a sequence of interest can bind a transcription factor, but do not establish that such binding occurs in vivo. Conversely, ChIP assays can demonstrate in vivo binding, but do not define the sequence bound. ChIP typically uses sonicated DNA whose average length is between 500 and 700 bp, and likely contains some fragments considerably longer. Thus the sequence actually bound by the transcription factor could be 500 bp or more away from the target amplified (none of the papers reviewed reported size fractionating the sonicated DNA).
Cell Cultures
Nearly all the studies of developmental expression of GFAP use progenitor cells isolated from cerebral cortices of embryonic mouse or rat. These are used within a few days of culture, or after several passages to obtain a population that is both more uniform and more susceptible to transfection. Different terms have been used by different laboratories for each of these cell populations. Here we will refer to the former as neuroepithelial cells (NECs), and the passaged cells as neural stem cells (NSCs).
Transgenic Mice
As a final note of caution, studies in genetically altered mice are presently the standard for investigating the mechanisms of GFAP regulation, but may fall short of illuminating the actual circuitry in mice or man. Alteration of the endogenous GFAP gene has yet to be performed to identify transcriptional regulatory regions, and the biology of astrocytes in mice may differ markedly from that in primates, including humans (Oberheim et al., 2009). As an illustration of this latter possibility, Shinohara et al. (2016) observed that when a 300 bp marmoset Gfap promoter-driven GFP reporter was packaged into lentivirus and injected into the cerebral cortex of mice, only about 20% of the expressing cells were astrocytes; but when packaged into AAV and injected into the cerebral cortex of marmosets, about 90% of the expressing cells were astrocytes.
Developmental Time Course of GFAP Expression
GFAP expression can be detected in mice and rats by E14 (Teter et al., 1996; Fan et al., 2005), and in humans as early as about 6 weeks post conception (Holst et al., 2019). Although GFAP has served as a marker of astrocyte development, this first detection is in radial glial cells, which give rise to neurons and oligodendrocytes as well as to astrocytes; the first detection of GFAP in human cells with the morphological appearance of astrocytes has been reported to be at about 15 weeks post conception (Holst et al., 2019).
We are aware of only two studies following GFAP expression after birth in young animals. Using northern blotting, Mokuno et al. (1989) detected Gfap mRNA in rat sciatic nerve at P21, but not at P3, P6 or P10. The mRNA level appeared to increase severalfold between P21 and adulthood. An earlier onset of Gfap expression was observed in mouse brain by Riol et al. (1992), who performed mRNA run off experiments using nuclei isolated from mice at ages 3, 15, 22, 34, 55 and 217 days. They observed maximum activity for the 3-day old mice, followed by about a 35% decrease at day 15 to a level that remained relatively constant through day 55. Comparisons between 55-day old mice and 217-day old mice were inconclusive, yielding either a slight increase of about 20%, or decrease of about 35%, depending on whether labeled transcripts or labeled probe was used for the assay. Multiple other investigators have reported clearly elevated levels of GFAP expression in older individuals. In humans, GFAP mRNA levels increase from three to fourfold between middle age (25–59 years) and older age (60–79 years) (Nichols et al., 1993). In mice, about a 2-fold increase in Gfap mRNA was reported between 12 and 29 months (Goss et al., 1991); and GFAP immunostaining also increases in aging mice, although the change has not been quantified (Kohama et al., 1995). In rats, about a 2-fold increase in both Gfap mRNA and protein between 3 months and 24 months has been reported (Morgan et al., 1999). These comparisons between groups of younger and older animals may yield the conclusion that the increase in GFAP expression is restricted to advanced aging. However, a recent study using quantitative PCR to analyze Gfap mRNA levels in mice spanning the ages of 1.1 months to 16.4 months found a gradual increase of about 7% per month (Brenner et al., 2019), which yields fold-changes similar to those reported previously (e.g., about 2-fold in 10 months). These GFAP increases with aging have been suggested to be a consequence of subclinical brain pathology, perhaps due to oxidative damage (Goss et al., 1991; Morgan et al., 1999) or chronic inflammation (Clarke et al., 2018). Another possibility, that they are simply due to an increased fraction of brain volume being occupied by astrocytes, is unlikely based on current evidence. Although Hansen et al. (1987) did observe about a 2-fold increase with age in fibrous astrocytes in the human mid-frontal cortex cellular layer (the volume actually occupied was not determined), no change was present in the more astrocyte-rich molecular layer. Studies in other brain regions of humans and rodents have found no change or a decrease in astrocyte numbers with age (reviewed in Palmer and Ousman, 2018).
GFAP Promoter Regions
This section summarizes studies that identified general regions of the GFAP promoter required for its activity. The possible roles of specific transcription factor binding sites are described in the subsequent two sections. Discovery of regions of the GFAP gene contributing to its expression is a first step for deciphering the mechanisms of its regulation by directing the search for specific transcription factor binding sites. However, it also has the immediate utility of producing astrocyte-specific expression cassettes for transgenes. Descriptions of these promoters and their properties are provided in Messing and Brenner (2020).
Initial Identification of a Functional GFAP Promoter
Initial studies seeking to identify elements of the GFAP gene contributing to its transcriptional regulation used transient transfection. Conflicting results were obtained from different laboratories for the contribution of an upstream region around –1500, a promoter proximal region extending from about –120 to +55 and segments in the first intron and at the 3’ end of the gene (reviewed in Brenner, 1994). Results supporting the importance of the upstream and promoter proximal regions, and the inactivity of the downstream regions, were subsequently obtained using transgenic mice expressing a lacZ reporter. These initial transgenic studies identified extended segments of the mouse and human GFAP genes that direct strong reporter activity in astrocytes. The mouse segment commences at –1980 and extends through the entire coding region, terminating about 1.1 kb 3’ of its polyadenylation site (Mucke et al., 1991; Johnson et al., 1995). The lacZ reporter replaced a segment between +49 to +86 (these cloning coordinates are updates corrected for sequencing errors—see Brenner et al., 1990). Follow-up studies indicated that the region downstream of this insertion site could be deleted with little, if any, effect on expression (Johnson et al., 1995). The human GFAP sequence, gfa2, extends from –2162 to +47 (Brenner et al., 1994).
Finer Mapping of the Human GFAP Promoter
Subregions of the gfa2 Promoter
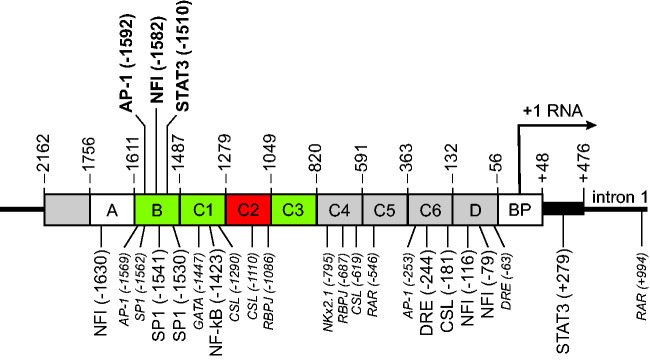
Based on deletion mapping and footprinting, the gfa2 promoter was divided into an upstream region (–2162 to –1757), an A region (–1756 to –1612), B region (–1611 to –1488), C region (–1487 to –133), D region (–132 to –57) and basal promoter (BP) (–56 to +47) (Besnard et al., 1991) (these coordinates are illustrated in Figure 1, and tabulated in Table 1 together with the contribution of each region to activity). Finer mapping studies determined that deletion of the upstream region was without effect (Lee et al., 2008; Brenner et al., 2019), whereas elimination of the C region resulted in expression being restricted to distinct areas of the CNS and occurring in several neuronal populations (Lee et al., 2006). The dichotomy between astrocytes revealed by the regional expression of the C region-deleted promoter (gfa28, renamed gfaABD, but note that it also contains the basal promoter) could be traced back to the developing embryo.
Figure 1.
Transcription Factor Binding Sites in the Human GFAP Gene. The rectangle shows the regions of the human GFAP gene included in the gfa2 promoter (not drawn to scale). Position numbers above the gene refer to the start of the section to the right. The fill colors indicate the result of testing the activity of the section in transgenic mice as described in the section on GFAP Promoter Regions: white = not tested, grey = no significant effect, green = required, red = strongly inhibits. Position numbers for the transcription factors are for the 5’ end of the binding site. Although discussed in the text, PAX3 is not shown because no site was identified in the human gene (see Table 2). Font size indicates the level of evidence supporting a role in human GFAP transcription: large & bold = strong; medium & standard = moderate; small & italic = weak.
Table 1.
Human GFAP Promoter Regions.
| gfa2 Regions | |||
|---|---|---|---|
| Region | Locationa | Transcription factor sites | Effect on transgene activity |
| Upstream | −2162 to −1757 | an alu sequence spans -2069 to −1757 | No effect |
| A | −1756 to −1612 | NFI | Not tested |
| B | −1611 to −1488 | See subregions below | Required |
| B1 | −1611 to −1581 | consensus AP−1, NFI | Mutation in ABC1D reduces activity >99% |
| B2 | −1580 to −1548 | AP−1, NFI, Sp1 | Mutation in ABC1D reduces activity ∼84% and contributes to spatial distribution, and suppression in neurons |
| B3 | −1547 to −1519 | Sp1 (x2) | Mutation in ABC1D reduces activity ∼87% and delays developmental onset |
| B4 | −1518 to −1488 | STAT3 | Mutation in ABC1D reduces activity ∼58% and delays developmental onset. Also contributes to spatial distribution and suppression in neurons |
| C | −1487 to −133 | See subregions below | |
| C1 | −1487 to −1255 | CSL, GATA, NF-κB (see C1.1 and C1.2 subregions below) | Required for general expression throughout the CNS |
| C1.1 | −1487 to −1433 | GATA | Contributes to general CNS expression |
| C1.2 | −1442 to −1398 | NF-κB | Contributes to silencing neuronal GFAP expression |
| C2 | −1279 to −1025 | CSL, RBPJ | Strongly represses activity |
| C3 | −1049 to −796 | Contributes to general CNS expression and silencing expression in neurons | |
| C4 | −820 to −567 | RBPJ, CSL, Nkx2.1 | Little if any effect |
| C5 | −591 to −339 | RAR | Little if any effect |
| C6 | −363 to −132 | AP−1, CSL, DRE | Little if any effect |
| D | −132 to −57 | DRE, NFI (x2) | No effect (but see text for caveats) |
| BP | −56 to +47 | Not tested | |
|
hGFAP Transgene Promotersb | |||
| Name | GenBank coordinates | Expression characteristics | |
| gfa2 | −2162 to +47 | Similar to endogenous GFAP except weak response to injury | |
| ABCD | −1756 to +47 | Similar to gfa2 | |
| ABC1D | −1756 to −1255/−132 to +47 | Similar to gfa2 | |
| ABD | −1756 to −1488/−132 to +47 | Expression in astrocytes largely limited to the dorsal and caudal cortical regions, hippocampus, and caudal vermis of the cerebellum; expresses in several populations of neurons | |
aLocations are relative to the RNA start site, using NCBI Reference Sequence NC_000017.11.bThe basal promoter (BP) is present in each of these promoters, and has the protein initiating ATG at +15 changed to TTG.
The C Region
Subregions within the C segment that contribute to general expression throughout the CNS and prevent activity in neurons were identified by Lee et al. (2008). Both properties were restored by including the sequence from –1487 to –1255 (the C1 fragment) to produce the gfaABC1D promoter (this also contains the basal promoter region). Other subfragments of the C region restored a weaker level of activity, potently repressed activity, or had no effect (see Table 1).
The B Region
Yeo et al. (2013) examined the contribution of the B region by dividing it into four contiguous segments and block mutating them one at a time in the context of the gfaABC1D-nlac transgene. All four block mutations resulted in substantially reduced activity: >99% reduction for B1, about 84% for B2, 87% for B3 and 58% for B4. That each of the reductions was greater than 50% indicates that the regions act cooperatively. In addition, the B2 and B4 block mutants expressed in neurons in the hippocampus, and had regionally restricted expression patterns in the brain differing from that of gfaABD-nLac, suggesting they could be used to define further heterogeneity among astrocytes. The restricted pattern of expression was evident in the embryo, as had been observed for the gfaABD promoter. In addition, the B3 and B4 block mutants showed developmental delay in transgene expression.
The D Region
The D region (–132 to –57) has about 80% sequence identity among human, mouse and rat, suggesting functional importance. Shinohara et al. (2016) obtained evidence for the D region having promoter activity from deletion mapping of the marmoset Gfap promoter by lentiviral transfection of mouse cerebella. They observed that a promoter segment encompassing bp –200 to +14 supported about half the activity of one commencing at –1991. Although this activity could be attributed to the sequence from –200 to –133 rather than to the D region (–132 to –57), the analysis of subfragments of the C-region described above found that the sequence from –820 to –133 did not contribute to promoter function. A contrary conclusion, that the D region does not contribute to activity, was reached by Pignataro et al. (2017), who found no difference between the ability of the ABC1 segment with or without the D region to drive expression of GFP packaged in AAV that was then injected into mouse striatum. A possible explanation of this negative finding of Pignataro et al. is that their constructs likely lack the GFAP basal promoter, which provides a TATA box and the RNA start site. Although the exact sequence of their ABC1D construct was not described, its stated size of 587 nt (in the text) or 543 nt (in their Figure 1) is well short of the 681 nt that would be present were the GFAP basal promoter present. Thus, the conflicting results of Shinohara et al. and Pignataro et al. suggest that the activity of the D region is dependent on its context. Evidence for this was provided by Besnard et al. (1991), whose results indicated that the D region is critical when the upstream enhancer segments (e.g., A and B) are at their normal location, but not when they are brought close to the basal promoter, as in the ABC1D construct.
Regulatory Regions Yet to Be Discovered
Analysis of the GFAP promoter has focused on promoters extending no further than about –2000, since transgenic analyses have shown them to closely mimic the development and cell specificity of endogenous GFAP expression, and to increase activity in response to injury. However, a recent quantitative comparison found that the response of the gfa2 promoter to injury in transgenic mice was only about 25% that of the endogenous Gfap gene, indicating that important regulatory elements lie outside of the –2162 to +47 segment (Brenner et al., 2019). A similar conclusion was reached previously by Verderber et al. (1995) in a study comparing the expression of endogenous GFAP to that of a lacZ reporter in the mouse retina. The lacZ reporter was embedded in the first exon of a mouse genomic clone extending from –1980 to about 1.1 kb past its polyadenylation site (see the Initial Identification of a Functional GFAP Promoter section above for a description of this construct). As expected, both endogenous GFAP and β-galactosidase were expressed in astrocytes in the ganglion cell layer of the retina, and neither in resting Müller cells. However, upon injury, endogenous GFAP but not β-galactosidase was observed in the Müller cells, suggesting that injury-responsive elements present in the endogenous gene were absent in the transgene.
Developmental Regulation of GFAP Expression
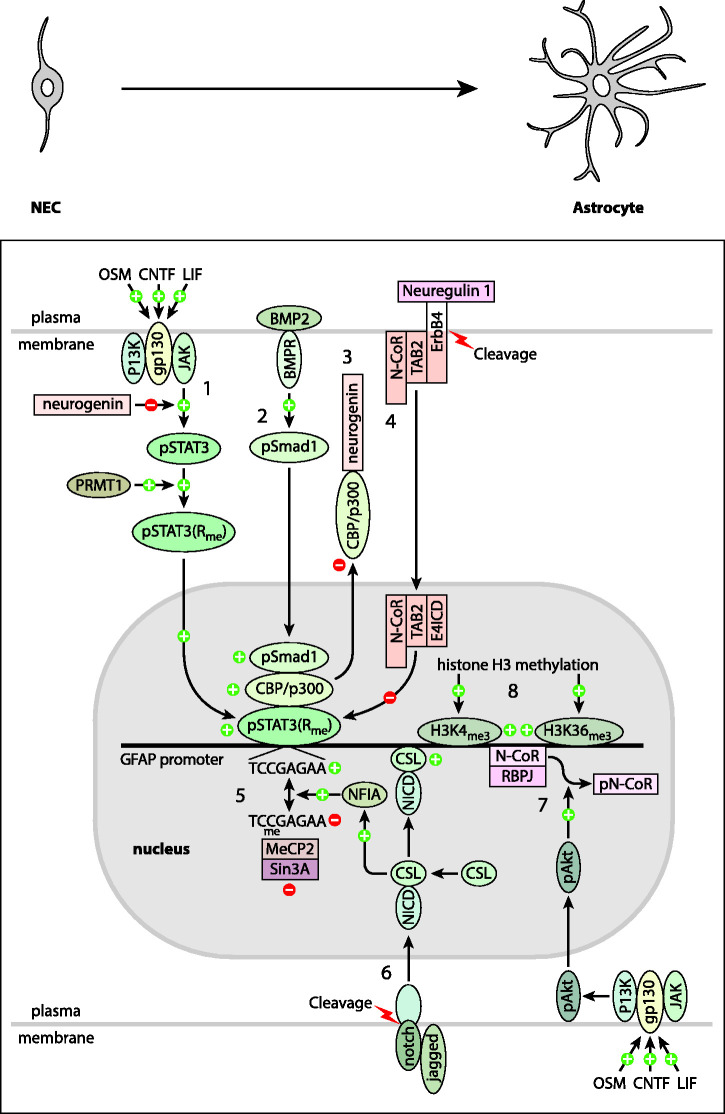
Figure 2 depicts pathways proposed to impinge directly on the GFAP gene to either keep it repressed early in development, or to activate it as astrocytes begin to differentiate. Numbers in the figure are keyed to the subdivisions of this section.
Figure 2.
Developmental Regulation of GFAP Expression. Shown are mechanisms proposed to regulate the developmental timing of GFAP expression that are discussed in the text. Plus signs (+), and ovoid shapes indicate processes that stimulate GFAP expression, whereas minus signs (–) and rectangular boxes indicate those that are inhibitory. Numbers in the figure are keyed both to this legend and the subdivisions of the section Developmental Regulation of GFAP Expression. Foremost among the positive factors is STAT3 [1]. It is activated by cytokines such as CNTF, LIF and oncostatin M (OSM) binding to their receptors complexed with gp130, resulting in the activation of Janus kinase (JAK). JAK then activates STAT3 by phosphorylation. pSTAT is further activated by methylation by protein arginine methyltransferase 1 (PRMT1) (it is not known if this occurs in the cytoplasm or nucleus). pSTAT3(Rme) (hereafter STAT3) enters the nucleus and attaches to its consensus binding sequence, TCCGAGAA, in the GFAP promoter. Assisting STAT3 in its stimulation of GFAP expression is Smad1, which is activated following binding of BMP2 to its plasma membrane receptor [2]. Smad1 is believed to form a complex with STAT3 via mutual interaction with CBP/p300. Several mechanisms prevent premature expression of GFAP, many targeting the activity of STAT3. Neurogenin, which is present at high levels early in development, inhibits the phosphorylation of STAT3 by JAK, and also suppresses GFAP expression by sequestering CBP/p300 [3]. STAT3 activity is also inhibited by a complex of TAB2, N-CoR, and the intracellular domain of the receptor ErbB4 (E4ICD), which is generated by cleavage of ErbB4 following neuregulin 1 binding [4]. In addition, binding of STAT3 to its consensus sequence is inhibited by methylation of the CpG within the consensus sequence [5]. The CpG methylation also contributes to preventing GFAP expression by attracting the transcriptional repressor Sin3A via its binding to MeCP2. Another inhibitory mechanism is binding of the transcriptional repression complex RBPJ/N-CoR to the GFAP promoter [7]. Inhibition is relieved by developmental decreases in the levels of Sin3A, neurogenin and neuregulin. Notch signaling also contributes to activation of GFAP expression through generation of the Notch intracellular domain (NICD) by proteolytic cleavage following binding of a ligand such as jagged [6]. The NICD then forms a complex with CSL, a transcriptional activator, which binds to the GFAP promoter. In addition, by unknown mechanisms, the NICD/CSL complex increases levels of NFIA, which facilitates demethylation of the CpG in the STAT3 binding site. Also contributing to GFAP expression is activation of Akt by phosphorylation via the PI3K pathway (a single arrow is shown, but multiple steps are involved) [7]. pAkt in turn phosphorylates the transcriptional repressor N-CoR, causing it to exit the nucleus. Developmental increases in methylation of lysines 4 and 36 of histone 3 are also associated with increased GFAP transcription [8]. A stimulatory mechanism not illustrated, but discussed in the text in the section BRG1 and Gene Clustering, is association of the GFAP promoter with other STAT-activated genes via the bridging protein BRG1.
STAT3 (pathway 1 in Figure 2)
A critical role for STAT3 in GFAP developmental expression was discovered in the course of investigating astrogenesis. These studies were primarily performed using cultured embryonic rat or mouse neuroepithelial cells (NECs) that respond to several signaling molecules by differentiating into astrocytes and synthesizing GFAP. These signaling molecules include cytokines that act through the gp130 receptor to activate the JAK-STAT and MAPK pathways (Takizawa et al., 2001b), such as ciliary neurotrophic factor (CNTF), leukemia inhibitory factor (LIF) and oncostatin M (OSM); and bone morphogenetic proteins (BMPs), which act through BMP receptors to activate Smads. In their study of the role of CNTF in astrogenesis, Bonni et al. (1997) found that CNTF increased expression of a luciferase reporter driven by a rat Gfap promoter through activation of the JAK-STAT pathway. A conserved consensus STAT binding site, TTCCGAGAA, was identified in this promoter at –1421 (see Table 2 for a listing of all transcription factor binding sites described in this review, their consensus sequence, and their positions in the human, mouse and rat GFAP genes, the three major species studied). Mutation of this site showed it to be essential for CNTF-dependent activation. Gel mobility supershift experiments documented binding of STAT1 and STAT3 to this DNA segment. Rajan and McKay (1998) also used gel mobility supershift assays to identify STAT1 and STAT3 as the STAT isoforms activated when astrogenesis is stimulated by CNTF, finding STAT3 to be the predominant species. Additional evidence for STAT3 being the principle, if not exclusive, STAT isoform acting at the GFAP promoter was presented by Hong and Song (2014). This includes severe loss of astrogenesis in Stat3 null mice but inconsequential effects of a Stat1 null, and response of a 2.5 kb mouse Gfap promoter to STAT3 but not to STAT1. Further supporting the role of STAT3 in GFAP expression, Herrmann et al. (2008) observed that conditional knockout (KO) of STAT3 reduced GFAP levels severalfold in the spinal cord of both uninjured mice and mice with a spinal cord crush injury. In a complementary study, Yeo et al. (2013) observed that mutating the consensus STAT binding site in a gfaABC1D-nlac transgene decreased reporter activity over 90%.
Table 2.
Transcription Factor Consensus Sequences and Binding Sites.
| Site |
Consensus |
Human |
Mouse |
Rat |
||||
|---|---|---|---|---|---|---|---|---|
| Sequence | ID | Loca | GFAP sequence | Loc | Gfap sequence | Loc | Gfap sequence | |
| AP−1 | (G/C/A)TGA(C/G/T)TCA(T/C) | MA0476.1 | −1592 −1569 −253 |
ATGACTCAC AatgTTCgg tTGAtTCAg or tTGctTCAg |
−1599 −1576 −355 |
GTGACTCAC AatgGTCAg AaGtaTgca |
−1504 −1481 −378 |
ATGACTCAC AatgGTCAg AaGtaTgca |
| CSL | TTCCCA b | MA1116.1 | −1290 −1110 −619 −181 |
TTCCCA TTCCCA TTCCCA TgCCCA |
−1304 −1162 −664 −184 |
gTtCCA aTatCc TTCCtA TTCCCA |
−1201 −1067 −735 −184 |
TTtCCA aTatCc aTCCtA TTCCCA |
| DRE | (A/G)NGTCA(A/G)(A/G)G | Carrion et al., 1998 | −244 −63 |
cGGTCAGGG GGcaCccGG |
−346 −65 |
cTGTCAAcc AAGTCAGGG |
−369 −65 |
cTGTCAAAc AAGTCAGGG |
| GATA | AGATAA(G/A)(A/G/C) | MA0037.2 | −1447 | gGATAAAA | −1453 | gGATAAAA | −1358 | gGATgAAAG |
| NFI | (T/C)TGGCN4(T/C)GCCA(A/G/T) | MA1527.1 | −1630 −1582 −116 −79 |
CTGGCN4CcCCAG TTGGCN4acaCAA TgGGCN4gcCCAA agGGgN4TGCCAG |
−1635 −1589 −119 −81 |
CTGGtN4CcCCAG TTGGCN4acatAA TgGGCN4gcCCAG TgGGgN4TtCCAG |
−1544 −1494 −118 −81 |
CTGGCN4CcCCtc TTGGCN4acatAA TgGGCN4gcCCAG aTGGgN4TGCCAG |
| NF-κB | GGG(A/G)(A/C/T)(T/A/G)T(T/C/A)CC | MA0105.1 | −1423 | GGGGCTgCCC | −1436 | GGGGCTgCCC | −1341 | GGGGCTaCCC |
| Nkx2.1 | (A/T/G)(A/T/G)N(T/G/C)(T/A)(C/T/G)(A/G)AGT(G/A)(G/C)N(T/C/A)b | PH0171.1 | −795 | GGGCTGGtGTcCAA | −842 | AGGCTCAAGTtGAC | −922 | AGGCTCAAaTtGAT |
| PAX3 | G(T/A)(C/T)A(C/T)(G/A)(C/G)NN(A/G/C)T(T/C)(A/T) | MA1546.1 | N/A N/A |
no homology no homology |
−2021 −411 |
GTCtCACAAATCT cTCtCACTAATTg |
−1937 −433 |
GTCACACAAGcCT GTCACACTAATTgb |
| RAR | AGGTCA(A/C/T)NN(A/T)(A/G)AGGTCAc | MA0858.1 | N/A −546 N/A +994 |
no homology AGGTCANAGGTCA no homology tGGTCAN10caTttc |
−2502 N/A N/A +928 |
AGtTCAAGGTCA no homology no homology AGGTCAN10TGtCCT |
−2498 N/A −508 +924 |
AGtTCAAGGTCA no homology AGGcCANAGGTCA AGGTCAN10TGtCCT |
| RBPJ | TGGGAA | MA1116.1 | N/A −1086 −687 |
no homology TGGGAA TGGGAA |
−1909 −1139 −731 |
TGGcAA TaGGAA TGaGAA |
−1822 −1044 −802 |
TGGGAA TGaaAA TGaGAA |
| Sp1 | (C/T/G)(C/T)CC(G/T/A)CC(C/T)(C/TA) | MA0079.3 | −1562 −1541 −1530 |
GCCCACCCCb TCCCGCCgC CCCaGCCCC |
−1568 −1547 −1536 |
GCCCACCCC TCCCGCtgT TCCaGgCCT |
−1473 −1452 −1441 |
GCCCGCCCC
TCCCGCtgC TCCaGgCCC |
| STAT3 | TTCCN(G/A)(G/T)AA | MA0137.2 | −1510 +279 |
TTCCGAGAA
TTCCTGGAA |
−1516 +269 |
TTCCGAGAA
TTCCTGGAA |
−1421 +272 |
TTCCGAGAA
TTCCTGGAA |
The sequences and positions of transcription factor binding sites discussed in the text are shown for human, mouse and rat, together with a consensus sequence. Consensus sequences are from the 2020 addition of the JASPAR database (Khan et al., 2018) (http://jaspar.genereg.net/), except for DRE (the binding site for DREAM), which was not in the database and was obtained from Carrión et al. (1998). When multiple, differing sequences were present in the JASPAR database, the one best matching that in the GFAP gene was selected. A nt is included in the listed consensus if it was present at least 10 times more often in the frequency matrix than at least one of the other three nts. Additional nts are included for a position in their order of frequency if they were present at least 10% as often as the most common nt. A lower case letter in the GFAP sequences indicates a mismatch with the consensus sequence. Nt positions are relative to +1 being the transcription start site, and are in accordance with NCBI Reference Sequences NC_000017.11, NC_000077.6, and NC_005109.4 for human, mouse and rat, respectively. N/A = not applicable because no homology is present.
a5’ end of the sequence (3’ end if the sequence is the reverse complement). Numbering is relative to the RNA start site (see text).
bSequence is the reverse complement
cMatches to the consensus are given only for the AGGTCA repeats, because the space between them can vary from 1 to 5 nucleotides. The mouse sequence at +928 and the rat sequence at +924 are inverted repeats. The absence of a spacer for the −2502 mouse and -2498 rat sequences and the N10 spacers for the mouse +928 and rat +924 sequences suggests binding will be weak (Rastinejad et al., 2000).
Honda et al. (2017) found that the ability of STAT3 to stimulate GFAP synthesis is augmented by its arginine methylation by protein arginine methyltransferase 1 (PRMT1). Expression of short hairpin RNA (shRNA) directed against PRMT1 curtailed acquisition of Gfap mRNA expression in mouse E14.5 NECs induced to differentiate by LIF. Immunoblotting showed that the shRNA strongly reduced arginine methylation of STAT3 in this system. That this reduction in STAT3 methylation was responsible for the inhibition of Gfap expression by a direct effect on the Gfap promoter was indicated by use of a mouse 2.5kb Gfap-luc reporter. Whereas the increase in luciferase activity following LIF stimulation was inhibited about 70% by the PRMT1 shRNA, no significant inhibition by PRMT1 shRNA was observed when the STAT3 binding site in the reporter was mutated. A caveat is that the STAT site mutation itself lowered reporter expression to an extent that it is not clear that further inhibition by PRMT1 knockdown could be readily discerned. Whether the STAT3 methylation is constitutive or regulated was not discussed.
Smad1 (pathway 2 in Figure 2)
It is generally accepted that Smad1 synergistically interacts with BMP2 to stimulate GFAP expression, but alternative possible mechanisms for its contribution remain to be explored. The Smad1/BMP2 cooperativity model arose from the observation by Nakashima et al. (1999) that differentiation of astrocytes from E14 mouse NECs was synergistically augmented by activation of STAT3 by LIF and of Smads by BMP2. Neither LIF nor BMP2 at concentrations up to 200 ng/ml induced astrogenesis, yet GFAP-positive cells appeared when LIF and BMP2 were added together at 80 ng/ml each. An interaction between LIF and BMP2 was also observed for the activity of a 2.5 kb mouse Gfap promoter driving a luciferase reporter transfected into the NECs. Mutation of the consensus STAT binding site attenuated the increase in activity produced by either LIF or BMP. When a Smad binding sequence could not be identified in the Gfap promoter, the authors pursued the possibility that the interaction results from Smad binding to STAT3. No direct interaction was observed, but the transcriptional activator CBP/p300 was found capable of linking the two; STAT3 was shown to bind to the N-terminal region of CBP/p300, and Smad1 to the C-terminal region. A complex containing these three proteins was detected by immunoprecipitation, but only when CBP/p300 was over-expressed in COS-7 cells, raising the possibility that the association was an artifact of the elevated CBP/p300 levels. However, Sun et al. (2001) subsequently reported detection of an interaction between CBP and Smad1 in rat E14 NECs, and between CBP and STAT3 in P3 cells from the subventricular zone.
Although the bridging model provides an explanation for the synergistic interaction of LIF and BMP2 in stimulating astrogenesis, the biological relevance of the interaction of STAT3 and Smad1 at the GFAP promoter is unclear. When measured by the increase in units of luciferase reporter activity produced by addition of BMP2 alone (0.17 unit increase) or LIF alone (1.03 unit increase), or in combination (3.00 unit increase), there is indeed a more than additive effect (data taken from Figure 2A of Nakashima et al.). But transcription factors acting independently may increase activity multiplicatively rather than additively; for example, if they bind to different sites on the basal transcription machinery (He et al., 2010). By this criterion there is no synergy between BMP2 and LIF for Gfap promoter activity. Expression of BMP2 resulted in a 5.0-fold increase, LIF in a 25.5-fold increase, and the combination in a 72.5-fold increase. This is less, rather than more, than the 127.5-fold increase expected if LIF and BMP2 acted independently and multiplicatively. A similar result pertains to the experiment in which a dominant negative STAT was used to investigate dependence of activation by BMP2 on the presence of STAT3. Expression of the dominant negative STAT3 did result in a 2-fold decrease in the units of BMP2-induced luciferase activity, but there was no change in the fold-increase, since the control displayed a similar 2-fold decrease in activity units (why BMP2 had any effect on the reporter activity in the absence of LIF was not addressed). A final consideration is that Smad1 binding to CBP would occur not only for CBP that had bound to STAT, but also for CBP bound to any other transcription factor that occupied a region of CBP different from the Smad1 binding site. The prospect that the independent effects of STAT3 and Smad1 are multiplicative rather than additive, and thus that the criterion for transcriptional synergy may not be met by the Nakashima et al. experiments, leaves open the possibility that BMP2 activates GFAP transcription through association with factors other than STAT3. Dore et al. (2009) have suggested that BMP2 acts indirectly on GFAP transcription through increasing Sp1 levels. Thus, the mechanism by which Smads contribute to GFAP expression remains open to further investigation.
Neurogenin (pathway 3 in Figure 2)
Another proposed timing mechanism for STAT-dependent GFAP expression is changes in the levels of neurogenin, a protein previously identified as a transcription factor contributing to the differentiation of neurons. Levels of neurogenin are high during neurogenesis, but become very low during astrogenesis. Sun et al. (2001) investigated whether neurogenin might have a dual role of inhibiting astrogenesis in addition to promoting neurogenesis. For these studies, they analyzed the effects of neurogenin in rat E14 NECs, which are unresponsive to stimulation to produce astrocytes, and either E17 or E18 NECs or NSCs, which are responsive (the NSCs were isolated from E14 cortices and passaged 2 or 3 times). Confirmation came from observing that raising the level of neurogenin in cultured rat astrocytes or NSCs by viral transfection reduced GFAP levels. That this effect was not simply an indirect result of inhibiting astrocyte maturation was indicated by over-expression of neurogenin inhibiting activity of a 1.9 kb rat Gfap promoter-luciferase reporter construct transiently transfected into rat E18 cortical astrocytes. However, the mechanism of inhibition did not involve DNA binding, because it occurred even in the absence of the DNA binding domain of the expressed neurogenin protein. Instead, co-immunoprecipitation in E14 NECs found association of CBP with neurogenin, but not with STAT3, leading to the suggestion that neurogenin inhibits GFAP transcription by competing with STAT3 for CBP. Similarly, association of CBP with STAT3 was attenuated in NSCs by over-expression of neurogenin. This competition model was supported by finding that the inhibition of GFAP production in NSCs by over-expression of neurogenin could be prevented by over-expression of CBP. Evidence was also presented for another mechanism by which neurogenin contributes to GFAP silencing—by inhibition of the JAK-STAT pathway. When over-expressed in LIF-stimulated NSCs, neurogenin markedly reduced activation of STAT3. Finally, as pointed out by the authors, CBP mediates the activity of multiple transcription factors, so its sequestration by neurogenin could provide a global switch in gene activity.
A key finding in this study is that association of STAT3 with CBP was undetectable in E14 NECs. The inference that the absence of this interaction is due to sequestration of CBP by neurogenin would be strengthened by showing that STAT3 is capable of binding CBP in the E14 cells. Perhaps STAT3 must be activated, translocated to the nucleus and/or bound to DNA for the interaction with CBP to occur. Possible confirmatory experiments include testing the effect of over-expressing CBP in the E14 cells; or more directly to the point, knockdown of neurogenin.
N-CoR, Neuregulin and ErbB4 (pathway 4 in Figure 2)
Another mechanism proposed to regulate developmental control of GFAP transcription is the formation of a transcriptional repressor complex following binding of a ligand such as neuregulin 1 to the ErbB4 receptor. Subsequent to such binding, the cytoplasmic tail of ErbB4 (E4ICD) was shown by Sardi et al. (2006) to be liberated by proteolytic cleavage, and to then form a complex with TAB2 (TAK1 binding protein 2) and the transcriptional repressor N-CoR, which translocates to the nucleus where it represses genes involved in astrogenesis. Association of E4ICD with TAB2 was detected using a yeast 2-hybrid system, and confirmed by co-immunoprecipitation following transfection of HEK293 cells. Co-immunoprecipitation also identified an association between N-CoR and E4ICD that was dependent on the presence of TAB2, indicating that TAB2 serves as a bridging molecule between E4ICD and N-CoR. Of greater biological relevance, co-immunoprecipitation of E4ICD with both TAB2 and N-CoR was detected in E14 rat NECs exposed to neuregulin 1. A ChIP assay showed binding of E4ICD to the Gfap promoter that was dependent on neuregulin 1 treatment, although the DNA sequence required for the binding was not determined. Manipulation of E4ICD/TAB2/N-CoR complex components by knockdown and overexpression provided evidence that the complex partially inhibits astrogenesis in vivo, as well as the ability of CNTF to both induce GFAP synthesis in cultured NECs and to increase expression of a luciferase reporter driven by a rat Gfap promoter. Since CNTF is believed to stimulate GFAP synthesis through recruitment of STAT3, these latter observations suggest that the complex interferes with the ability of STAT3 to stimulate GFAP transcription. A puzzling finding, however, was that although neuregulin 1 inhibited the ability of CNTF to stimulate expression from the Gfap promoter, it had no effect on the basal level of promoter expression. This is unexpected, because STAT3 signaling is considered essential for GFAP basal promoter activity as well as for its activation by CNTF. A possible explanation, not examined by the authors, is that the E4ICD/TAB2/N-CoR complex inhibits the CNTF signaling pathway rather than acting locally at the GFAP promoter.
DNA Structure of the STAT Binding Site (pathway 5 in Figure 2)
Methylation
Takizawa et al. (2001a) observed that methylation of the CpG in the endogenous mouse Gfap gene STAT3 consensus binding site is high in E11.5 NECs, but low in E14.5 cells. Using EMSAs, they found that this methylation prevented STAT3 binding, thus providing an explanation for why E11.5 NECs do not initiate GFAP synthesis in response to LIF, despite STAT3 being activated, whereas E14.5 cells do. Two experiments established a causal relationship between CpG methylation of the STAT3 site and gene activity. In the first, transient transfection was used to determine the effect of CpG methylation on the activity of a construct consisting of 8 STAT3 binding sites joined to a minimal Gfap promoter driving a luciferase reporter. Prior in vitro CpG methylation of this construct reduced its activity about 4-fold. Conversely, inhibition of CpG methylation by 5-aza-2’-deoxycytidine in E11.5 NECs resulted in some activation of GFAP synthesis. A caveat for both of these experiments is that the methylation status of CpG’s outside the STAT3 binding site could be responsible for the observed effects.
Nucleosome Positioning
Although demethylation of the STAT binding site may be necessary for activation of the GFAP gene, it is not sufficient. Urayama et al. (2013) found that LIF failed to induce GFAP synthesis in mouse embryonic stem cells carrying a triple KO of DNA methyltransferases, despite the STAT site being demethylated. Furthermore, although the STAT signaling pathway was active, a ChIP assay found that STAT3 failed to bind to the demethylated Gfap promoter. Simultaneous DNA demethylation by 5-aza-2’-deoxycytidine and reduction of the levels of the inhibitory histone 3 modifications using a conditional KO were also insufficient to allow LIF induction of GFAP (the effect on STAT binding was not reported). A micrococcal nuclease assay suggested that failure of LIF to induce GFAP synthesis in the triple KO embryonic stem cells was due to sequestration of the STAT site by nucleosomes. Nuclease accessibility to DNA in the region of the STAT site in the triple KO embryonic stem cells was similar to that in the wild type embryonic stem cells and in E11.5 NECs, both of which also failed to initiate GFAP synthesis in response to LIF; whereas this region was more susceptible to micrococcal nuclease cleavage in the LIF-responsive E14.5 NECs. Over-expression of NFIA in E11.5 NECs rendered Gfap susceptible to LIF induction, and also rendered the region of the STAT binding site more susceptible to micrococcal nuclease, suggesting a role for NFIA in reordering of chromatin. It would have been of interest to perform a similar NFIA over-expression experiment in the embryonic stem cells in order to demonstrate that they, like the NECs, are capable of Gfap transcription. In the absence of such a positive control, it remains uncertain whether a repressive chromatin structure in these cells is indeed responsible for their unresponsiveness to LIF.
Cell Specificity of STAT Binding Site Methylation
In addition to contributing to developmental regulation of GFAP expression, methylation of the STAT site has been invoked as an explanation for the gene being silent in non-astrocytic cells. For example, Takizawa et al. (2001a) found that methylation of the STAT site was high in neurons and in non-CNS tissues such as heart, femoral muscle and liver. However, Fan et al. (2005) observed that the STAT site was demethylated in neurons in mice with a conditional KO of the DNA methylase Dnmt1, yet their GFAP gene remained silent despite an active JAK/STAT pathway. Thus, consistent with the findings of Urayama et al. (2013) described above, mechanisms in addition to CpG methylation act to suppress GFAP synthesis in non-expressing cells.
Alternative Findings for STAT3 Binding Site Methylation
Contrary data for CpG methylation preventing STAT3 binding have been presented by Cheng et al. (2011). They found that the consensus STAT3 binding site at –1510 remained almost fully methylated when human teratocarcinoma NTera-2 cells were differentiated into astrocytes, yet a ChIP assay showed increased binding of STAT3 to this region. Another confounding finding of Cheng et al. is the absence of recruitment of CBP or p300 to the upstream STAT site, an unexpected result given the association of STAT3 with CBP/p300 shown by several other investigators. These results suggest that there may be multiple, independent, routes to GFAP gene activation and astrogenesis. Perhaps contributing to an alternative pathway is a possible STAT3 binding site observed by Cheng et al. in exon 1, which does not contain a CpG methylation site. This sequence, TTCCTGGAA, differs from the –1510 sequence of TTCCGAGAA, but also matches the consensus (Table 2). It starts at +279 in human GFAP, and is conserved in mouse and rat. During differentiation of the NTera-2 cells into astrocytes, binding of STAT3 to this region increased, and unlike the –1510 site, increased binding of CBP and p300 was also observed. The biological relevance of this putative STAT3 binding site remains to be determined. Absence of an effect on transcription is suggested by the finding of Johnson et al. (1995) of similar expression of mouse transgenes that do or do not contain the entire Gfap coding region downstream of +86. However, displacement of possible downstream regulatory elements from the RNA start site by the 3 kb length of the inserted lacZ reporter could attenuate their effects.
Sin3A as a Mediator of Inhibitory Methylation
Another mechanism by which the CpG methylation inhibits GFAP expression during development involves Sin3A, a transcriptional repressor without DNA binding capacity, but known to interact with the CpG binding protein MeCP2. Cheng et al. (2011) observed that levels of Sin3A decreased when NTera-2 human embryonal carcinoma cells were differentiated into cells resembling astrocytes. In undifferentiated NTera-2 cells, ChIP assays showed that Sin3A had about a 2-fold and MeCP2 about a 1.5-fold greater association with a region around the STAT site at –1510 compared to a control region in GFAP intron 6. Following differentiation, Sin3A binding was reduced to about 1.5-fold that of the control, and MeCP2 binding was reduced to about half that of the control. Surprisingly, these changes in binding occurred despite the STAT binding region remaining almost fully methylated. These findings prompted the suggestion that the reduction of Sin3A during differentiation of NTera-2 cells is responsible for unmasking the GFAP gene for transcription, but intervention experiments remain to be done to establish causality.
Notch (pathway 6 in Figure 2)
Upon binding of a ligand to Notch, its intracellular domain is clipped off, migrates to the nucleus, and binds to the transcription factor CSL (CBF1, Suppressor of hairless, Lag-1), converting it from a repressor to an activator. Namihira et al. (2009) found that ectopic expression of the Notch intracellular domain in mouse E11.5 NECs results in demethylation of the Gfap promoter, including the STAT binding site, and enables LIF to induce Gfap expression in these cells. Similarly, activation of Notch signaling in the E11.5 cells by incubation with the Notch ligand JAGGED1 renders them responsive to LIF for GFAP synthesis. A role for NFIA in the pathway from Notch to Gfap gene demethylation was suggested by finding that over-expression of the Notch intracellular domain in the E11.5 cells increased the level of NFIA; and in a ChIP assay, that NFIA bound to the Gfap promoter. Furthermore, transfection of the E11.5 cells with a dominant negative NFIA prevented both the LIF responsiveness and Gfap demethylation, and similar effects were observed in cells isolated from NFIA null mice. Conversely, over-expression of NFIA in the telencephalon of developing mouse embryos by exo utero electroporation resulted in precocious GFAP synthesis. The mechanisms by which Notch signaling increases NFIA levels, and by which NFIA induces demethylation, remain to be discovered.
A more direct role for Notch signaling in regulating GFAP transcription was proposed by Ge et al. (2002). These authors found that over-expression of a constitutively activated Notch increased expression of a –1876 rat Gfap promoter-driven luciferase reporter about 6-fold when transiently transfected into E13 rat NSCs. Sequence analysis revealed a consensus binding site for its CSL interaction partner of TTCCCA in the rat Gfap promoter at –184. A 40-mer centered on this site, but not a mutated version of the sequence, bound recombinant CSL in an EMSA assay. ChIP assays to show binding in vivo were not reported. Mutation of the site in the rat Gfap promoter that commences at –1876 reduced the Notch-induced increase in activity of the transiently transfected luciferase reporter by about half. However, raising uncertainty about the contribution of the –184 site was the finding that Notch signaling produced a similar level of stimulation of a rat promoter commencing at –106, which lacks the putative CSL binding site. Based on the differential responses of –384 and –106 promoters to mutated versions of Notch, the authors suggested that Notch may affect the activity of the –106 promoter by a mechanism different from that of the –384 and longer promoters. However, the similarity in the fold-increases (5.5 for –384, 5.2 for –106) leaves this question open. The CSL binding sequence is conserved perfectly in mouse, also starting at –184, and imperfectly in humans (TgCCCA) (a lower case letter indicates a mismatch to the consensus), starting at –181. Perfect matches are present in human at –1290, –1110 and –619, but none of these is conserved in mouse or rat. The latter two sites, as well as that at –181, are absent in the gfaABC1D human promoter, which expresses in transgenic mice without discernible deficit compared to the gfa2 promoter (Lee et al., 2008).
N-CoR, RBPJ and Akt (pathway 7 in Figure 2)
In addition to activating the JAK-STAT pathway, another role attributed to developmental cytokines such as CNTF is relief of transcriptional repression by an RBPJ/N-CoR complex. RBPJ is a DNA binding protein that participates in inhibition or activation of transcription, depending on its binding partner, and N-CoR is known to bind to RBPJ (Kao et al., 1998) (both papers cited in this paragraph use the RBPJ alias CBF1). Involvement in GFAP transcription was demonstrated by finding that over-expression of N-CoR in mouse E13 NECs inhibited by about 60% the expression of a luciferase reporter driven by a proximal rat Gfap promoter (Hermanson et al., 2002). These authors state that this proximal sequence, extending from –384 to +13, contains a conserved RBPJ consensus binding site, but the consensus sequence they adopted was not stated. Ling et al. (1993), Tun et al. (1994) and the JASPAR database (see Table 2) all report TGGGAA as the consensus core RBPJ binding sequence, but this sequence is not present between –384 and +13 in the rat Gfap promoter. Nevertheless, Hermanson et al. found association of both RBPJ and N-CoR with the rat Gfap promoter proximal region by ChIP assay. (As noted in Table 2, a search of the rat Gfap promoter from –3000 to +13 did find the TGGGAA consensus at –1822, but it is unlikely that this is the binding site detected by the ChIP assay, because the DNA was sonicated to produce fragments of 500 to 800 bp, and the antisense ChIP primer ended over 1500 nt distant from this site at –166. A similar search of the mouse Gfap promoter yielded no sites, whereas human GFAP had matches at –1086 and –687.) CNTF relieves N-CoR repression by activating the Akt pathway, resulting in phosphorylation of N-CoR, and its translocation from the nucleus to the cytoplasm (Hermanson et al., 2002).
Histone Methylation (pathway 8 in Figure 2)
The possibility that the onset of STAT binding to the GFAP promoter is regulated by changes in histone methylation arose from an investigation by Song and Ghosh (2004) of how FGF2 potentiates the ability of CNTF to induce astrogenesis. They found that incubation of rat E18 NECs with FGF2 resulted in loss of the transcriptionally repressive lysine 9 methylation of histone 3 (H3K9me3), and gain of the permissive lysine 4 methylation (H3K4me). ChIP assays showed that these changes in histone methylation occurred in the region of the rat Gfap consensus STAT3 binding site, and correlated with increases in STAT3 and CBP binding to this region. Two experiments supported a causal contribution of histone methylation to Gfap expression in astrocytes. Over-expression of a lysine 4 methytransferase in NECs had a potentiating effect on astrocyte differentiation similar to that of FGF2. This suggests that it is an increase in lysine 4 methylation, rather than a decrease in lysine 9 methylation, that potentiates Gfap expression, but this was not confirmed by actual measurements of the effect of the methytransferase over-expression on the levels of these methylations. In a reciprocal experiment, a methyl transferase inhibitor prevented methylation of lysine 4 of histone 3 in NECs, and also prevented FGF2 + CNTF from activating GFAP production. A caveat is that this experiment lacked a control to establish that the inhibitor was not simply having a toxic effect. In this experiment also, the effect on lysine 9 was not reported; presumably lysine 9 would also be demethylated, suggesting again that its methylation status is not a factor in GFAP gene activity. A similar conclusion was reached by Urayama et al. (2013) using embryonic stem cells deficient in lysine 4 methylation. Further support for a positive role for H3K4 methylation in GFAP transcription was obtained by Kong et al. (2018) through experiments manipulating the level of KDM5A, which demethylates H3K4me2 and H3K4me3. Expression in HEK293T cells of a luciferase reporter driven by the human gfa2 promoter was reduced by about half by over-expression of KDM5A; and conversely, transfection of rat NSCs with KDM5A siRNA increased GFAP levels. In contrast to the observations of Song and Ghosh, however, ChIP assays found that differentiation of the rat NSCs with CNTF produced no change in association of H3K4me3 with the Gfap promoter in the region of the STAT binding site, whereas association with a region encompassing the RNA start site was markedly increased. The participation of methylation of H3K4 but not of H3K9 is consistent with the finding of Song and Ghosh that the level of lysine 9 methylation is low in neurons as well as in astrocytes, whereas the level of lysine 4 methylation is high in astrocytes but low in neurons. The overall conclusion is that lysine 4 methylation of histone 3 is a key determinant for both the timing and cell specificity of GFAP expression. However, since H3K4me3 is generally associated with transcriptionally active promoters (reviewed in Park et al., 2020), it cannot be ruled out that its effects on GFAP expression are indirect via activation of other genes.
An investigation by Cascante et al. (2014) also found no evidence for a contribution from changes in the repressive H3K9me3, but observed a positive effect from increased levels of another H3 methylation, trimethylated lysine 36 (H3K36me3). They used siRNA to knockdown KDM4A, which demethylates H3K36me3 and was found by ChIP assay to associate with the rat Gfap gene. When combined with treatment with the histone deacetylase inhibitor valproic acid, knockdown of KDM4A in rat E15.5 NECs increased production of GFAP positive cells and the levels of Gfap mRNA and protein. ChIP assays showed that this knockdown increased association of H3K36me3 with the Gfap exon 6 and exon 8 regions, as well as association of Pol II with both these regions and the promoter (the region targeted in the promoter was not specified). There was no change in the association of either H3K36me3 or the repressive H3K9me3 with the promoter region (neither the effect on STAT3 binding nor the association of H3K9me3 with exons 6 and 8 was determined). The increased presence of H3K36me3 with the downstream regions exon 6 and exon 8, but not with the promoter, is consistent with its known association with transcriptional elongation rather than initiation (Li et al., 2019). Nevertheless, their findings prompted Cascante et al. to suggest that the downstream methylation status could influence promoter activity; although as they pointed out, a less direct mechanism could be in play, such as formation of a previously described KDM4A/N-CoR repressor complex. Experiments using a reporter driven by a GFAP promoter lacking exons 6 and 8 might be instructive in this regard.
BRG1 and Gene Clustering
The role in GFAP expression of gene clustering, the physical association of unlinked but possibly functionally related genes, has been investigated by Ito et al. (2018) using astrocyte induction of mouse NECs by LIF. Double label fluorescent in situ hybridization (FISH) had shown that as NPCs differentiate into astrocytes, the Gfap gene clusters with several other newly activated genes. The association with the oncostatin M receptor gene, Osmr, was selected by Ito et al. for further study (Osmr is on mouse chromosome 15, and Gfap on chromosome 11). ChIP assay showed that a protein that mediates both chromatin remodeling and gene clustering, BRG1, bound to the STAT binding regions of both the Gfap and Osmr promoters following LIF induction. A critical finding was that shRNA knockdown of BRG1 in NPCs by viral transfection reduced LIF-induced clustering of Gfap with Osmr by about 50% and Gfap mRNA levels by about 75%, but this is a puzzling result given that the stated transfection efficiency of the NPCs was only 25%. In reciprocal experiments, inhibition of STAT activation inhibited BRG1 binding to the Gfap promoter. These findings led the authors to suggest that a synergistic interaction between BRG1 and STAT3 contributes to Gfap transcription. They suggest that STAT3 initially binds to its site, resulting in histone modification that enables BRG1 binding. BRGI then further modifies chromatin structure to facilitate increased STAT3 binding. A critical component of this model, yet to be demonstrated, is that BRG1 actually affects STAT3 binding. In addition, it was not determined whether the gene clustering contributes to gene activation or is simply an epiphenomenon. A possibility we suggest is that the association of like-regulated genes increases the local concentration of shared transcription factors (in this case, STAT3), and thus the occupancy of their binding sites.
Transcription Factors Regulating GFAP Expression in Mature Astrocytes
Figure 1 displays the binding sites of transcription factors proposed to regulate human GFAP transcription in mature astrocytes, and Table 2 presents their consensus DNA binding sequences and the location and sequence match of possible sites in the human, rat and mouse GFAP genes. Evidence for the role of each of these transcription factors is discussed below in alphabetical order, except that CSL, RBPJ and STAT were described above.
AP-1
A consensus AP-1 binding site is present in the human promoter at –1592, and is conserved in mouse and rat. Since its role in GFAP transcription has been recently reviewed (Brenner et al., 2019), only a brief summary will be provided here. Masood et al. (1993) used gel mobility supershift assays to show binding of both Fos and Jun family members to this sequence, and that its mutation in a gfaABD-CAT construct reduced activity of the chloramphenicol acetyltransferase reporter by over 90% when transiently transfected into U251 cells. These results were confirmed and extended by Gopalan et al. (2006a, 2006b), who observed that the activity of a GFAP-CAT reporter transiently transfected into U373 cells was reduced about 4-fold when the AP-1 site was mutated, and that expression of a dominant negative c-Jun in these cells markedly lowered the level of GFAP mRNA. Consistent with these findings, Yeo et al. (2013) found that block mutation of the B1 segment (–1611 to –1581), which contains the AP-1 binding site, reduced the activity of the gfaABC1D-nlac transgene in mice by over 99%. Surprisingly, however, when only the AP-1 site was mutated, transgene activity was essentially unchanged. This indicated that regions yet to be identified in the B1 segment are critical for GFAP expression, and that the AP-1 site contributes little, if at all, to the basal level of activity. Instead, as described below in the Response to Injury section, Brenner et al. (2019) found that the AP-1 site is essential for the upregulation of a gfa2-nlac transgene in response to injury.
A different AP-1 site was reported by Dore et al. (2009) at –1481 in the rat Gfap promoter. The basis for considering it an AP-1 site is unclear, as its sequence, AatgGTCAg, is a poor match to the consensus. The corresponding sequence in the human promoter, starting at –1569, is an even poorer match (AatgTTCgg). Mutation of this site in a rat promoter driving expression of a luciferase reporter had no effect on activity as measured by transient transfection into a neural crest-derived cell line.
Bachetti et al. (2010) identified yet another putative AP-1 binding site in the course of surveying the human GFAP promoter out to –2123 for single nucleotide polymorphisms (SNPs) whose alleles might differentially affect transcription. They focused on a C/A SNP at –250 because the “A” allele was predicted to yield an AP-1 binding site (tTGATTCAg), whereas the “C” allele was not (tTGcTTCAg). Surprisingly, in EMSAs the C allele yielded a stronger signal than the A allele. Evidence that the signal was indeed due to AP-1 was its inhibition by curcumin and production of a supershift by a c-Jun antibody. However, several aspects of the data leave uncertain the conclusion that in this instance AP-1 binding is strengthened by a mismatch within its core consensus sequence. One is that negative controls were not reported that would help establish specificity of the binding, such as use of unrelated oligonucleotides as competitors. Another is that although the reference cited by the authors for curcumin being able to “specifically inhibit formation of the AP-1/DNA complex in vitro” does state that curcumin inhibits AP-1 binding, it does not claim specificity. Instead, it remarks that curcumin “is known to have diverse biological functions.” Finally, contrary to what the authors state, the c-Jun antibody used for the supershift increases, rather than decreases, the unshifted binding signal.
Regardless of what actually binds to the –250 site, transient transfection using a GFAP promoter segment spanning –2076 to +48 linked to a luciferase reporter showed 60% greater activity for the C allele than the A allele. A caveat for this experiment is that the promoter used for the transient transfections retains the GFAP translation initiation codon, which precedes that for luciferase. Although this seems unlikely to affect the relative contributions of the two alleles, it could compromise results if it is not in frame with the reporter.
To obtain evidence for an effect of the C/A SNP at –250 on human GFAP levels, Bachetti et al. examined a database linking SNPs to brain gene expression. The database did not contain the –250 SNP, but was interrogated using an included SNP strongly linked to it. On this basis, the C/C alleles corresponded to about a 17% higher level of GFAP mRNA than A/A, and a 3% higher level than A/C. The authors state in the Discussion that this “showed higher GFAP expression in the presence of the C allele than with the A allele,” although in Results they note that these differences were not statistically significant. Thus, additional studies are needed to determine whether the –250 site actually binds AP-1, and whether GFAP expression is affected by the allele present.
Nevertheless, prompted by the suggestion that the –250 C allele results in higher GFAP levels than the A allele, Takahashi et al. (2020) correlated the –250 genotype of 1,212 Japanese subjects with various brain structural properties obtained by MRI. The authors speculated that higher GFAP levels would reflect a greater contribution of astrocytes to brain volume and function. Contrary to expectations, the associations found for the C/C genotype that were statistically significant were lower volumes of gray matter, white matter and intracranial space, lower cerebral blood flow, and greater diffusivity. The magnitudes of the differences between the A/A and C/C genotypes for these properties were small, ranging from 0.3% for diffusivity to 2.6% for cerebral blood flow. The false positive report probability was calculated to be 0.33. No measurements of GFAP levels were performed, so this publication does not provide information about the effect of the –250 allele on GFAP transcription. However, it does suggest that it has biological consequences, although it could be indirect through linkage to other genes, or simply be a false positive.
Also in the belief that the –250 allele may influence GFAP expression, Sereika et al. (2018) correlated the allele genotype with glioma grade. They observed that GFAP mRNA levels were lower in the more deadly grade IV gliomas than in grade II, suggesting that reduced GFAP expression correlates with poorer prognosis. But contrary to expectation, longevity for grade IV patients was greater for those with the presumptively lower expressing A/A genotype than those with the C/C genotype. Actual measurement in 45 glioma patients found no effect of the –250 genotype on GFAP mRNA levels.
In contrast to this negative result, Yoshida et al. (2013) observed a correlation between the –250 SNP genotype and the severity of Alexander disease, a usually fatal astrogliopathy caused by GFAP mutations, and exacerbated by increased expression of the protein (Messing et al., 2012). Three of the 4 patients with a C/C genotype, but none of 6 patients with a C/A or A/A genotype, had lost the ability to walk by age 50, consistent with the prediction that outcome will be more severe with the C/C alleles. More importantly, 3 of the patients had the same disease-causing R79H mutation, but different –250 alleles. Of these, the one with the C/C alleles experienced disease onset at 5 years, whereas onsets with the C/A and A/A alleles were at ages 38 and 36, respectively. Analyses with larger sample sizes are required to solidify these findings for Alexander disease, and more generally, to determine if the –250 SNP actually has a role in GFAP expression.
DREAM
Cebolla et al. (2008) presented evidence that Downstream Regulatory Element Antagonist Modulator (DREAM) mediates the stimulation of astrogenesis by pituitary adenylate cyclase-activating polypeptide (PACAP). Progressive deletions of a rat Gfap promoter driving luciferase expression in E17 rat NECs revealed that a region between –384 and –106 was responsible for PACAP stimulation, and two DREAM binding sites (DRE) were identified within this segment, one at –369 (cTGTCAAAc) and the other at –65 (AAGTCAGGG)(the authors give the consensus as GTCA, but the Carrión et al. (1998) reference they cite gives the more extensive sequence used here, which includes several degenerate positions (see Table 2)). The –369 sequence is conserved in the human gene, present at –244, but the –65 site is not (Table 2). ChIP assays showed binding of DREAM to the proximal region of the rat promoter, and gel mobility supershift assays showed binding to Gfap oligonucleotides centered on either sequence, but not to mutated versions. Mutation of either site in a promoter commencing at –1546 caused almost complete loss of both stimulated and basal activity. Finally, NECs isolated from DREAM null mice did not respond to PACAP by onset of GFAP synthesis.
There are several aspects of these findings that are problematical. A promoter deleted to –35 had the same basal activity as one starting at –1546, a finding contrary to a previous report from the same laboratory (Cebolla and Vallejo, 2006); and as detailed above, almost certainly not reflective of the actual promoter requirements for GFAP transcription. This makes uncertain how to interpret the finding that mutation of the DREAM sites in the –1546 promoter reduces its activity below that of the –35 promoter, which lacks these sites. Also, although NECs isolated from DREAM null mice no longer responded to PACAP, they continued to increase GFAP synthesis in response to CNTF, and by P7 the GFAP level in DREAM null mice was equal to or greater than that in wild type controls. Thus further studies are needed to determine the biological relevance of DREAM binding to the GFAP promoter.
GATA
As noted in Table 1, sequences within the C1.1 region (–1487 to –1433) of the human GFAP promoter contribute to the gfaABC1D-nlac transgene being expressed throughout the CNS, rather than being spatially restricted as observed for gfaABD-nlac. Yeo et al. (2013) identified a GATA binding site within this sequence at –1447, which is perfectly conserved in mouse and imperfectly in rat (Table 2). Mutation of the sequence reduced transgene activity by about half, but did not restrict its spatial expression, indicating that other C1.1 sequences contribute to this latter property. The ability of the GATA transcription factor to bind to this site was not tested.
NFI
A possible role for NFI in controlling the methylation status of the STAT binding site during development was discussed above in the section on Notch. Cebolla and Vallejo (2006) proposed an additional role for NFI in developmental activation of the GFAP gene based on their observation that NFI levels increase in rat cerebral cortex at the same time as GFAP synthesis. Using transient transfection of a rat Gfap-luciferase reporter into RC2.E10 cells, a cell line established from rat E16 cortex, they found that a region between –106 and –35 was critical for activity, and pursued the involvement of an NFI-like binding sequence at –81, aTGGgN4TGCCAG (see Table 2). This site had been previously implicated by Krohn et al. (1999) in mediating the rat Gfap response to transforming growth factor-β1 (TGB-β1) and interleukin-1β (IL-1β), but they had been unsuccessful in detecting binding of NFI to this sequence by EMSA. Cebolla and Vallejo did observe such an NFI-specific supershift, and also detected NFI binding to this region of the endogenous Gfap gene by ChIP assay of E17 rat NECs. Mutation of the site resulted in about an 80% reduction in activity of a –1546 Gfap-luciferase reporter when transfected into unstimulated E17 rat NECs, or cells stimulated with either PACAP or CNTF. A caveat for the studies of Cebolla and Vallejo is that in the transient transfection system used, a promoter commencing at –106 had activity similar to one commencing at –1546, whereas other studies have indicated binding sites in the upstream region (e.g., for STAT) are critical for expression. Thus, the transfection system may not correctly report biological activity. In addition, it remains to be demonstrated that NFI binds to the imperfectly conserved homologous sites in the GFAP promoters of mouse (TgGGgN4TtCCAG, also at –81) and human (agGGgN4TGCCAG, at –79).
Binding of NFI to other candidate sites in the human GFAP gene was investigated in the astrocytoma cell line U373 by the Kordula laboratory (Gopalan et al., 2006a; Singh et al., 2011) and in the U251 line by Brun et al. (2009). The three sites studied were those previously identified by Besnard et al. (1991), CTGGCN4CcCCAG at –1630, TTGGCN4acaCAA at –1582 and TgGGCN4gcCCAA at –116 (note that this site differs from the one above identified by Cebolla and Vallejo (2006), which starts at –79 in the human sequence). In EMSAs, Gopalan et al. observed an NFI supershift for binding at the –1630 site, but not for the other two; however, Brun et al. detected NFI binding to all three sites. Brun et al. also observed binding by ChIP assays in U251 cells to both an upstream region that would include –1630 and –1582, and to a promoter proximal region that would include –116. Both groups performed transient transfection experiments in which the NFI sites were mutated, finding that mutation of each site reduced reporter activity, and that mutation of the –116 site produced the greatest reduction, about 5-fold. The contribution of each NFI isoform (NFIA, NFIB, NFIC and NFIX) to GFAP expression was also examined by both laboratories using knockdown by small interfering RNAs. Although both found that knockdown of NFIX reduced GFAP mRNA about 70%, results for the other isoforms differed between the two laboratories. Reductions in GFAP mRNA levels found by Gopalan et al. in U373 cells and by Brun et al. in U251 cells were, respectively, no effect and 50% for knockdown of NFIA, no effect and 75% for knockdown of NFIB, and 40% and 85% for knockdown of NFIC. These differences indicate exquisite sensitivity to the experimental system. The Kordula laboratory followed up their observations that NFIX had primacy for regulating GFAP expression in their system by determining that its activity was attributable to the X3 splice form (Singh et al., 2011).
The above studies performed in astrocytoma cell lines were extended to the mouse by Yeo et al. (2013), who investigated whether the approximately 84% reduction in ABC1D-nlac transgene activity resulting from mutation of the B2 block (–1580 to –1548) was due to the presence of the NFI site beginning at –1582. Mutation specifically of this site reduced activity by about 55%, only partially accounting for the effect of the block mutation. However, its mutation also led to expression in neurons, suggesting that NFI contributes to silencing GFAP expression in neurons as well as stimulating it in astrocytes.
Reduced levels of GFAP in NFIA null and NFIB null mice have been cited as further evidence for a role for NFI in GFAP transcription. However, the authors of these studies point out that the effect on GFAP could be indirect, because only about 5% of the NFIA null mice and none of the NFIB null mice survive the perinatal period, and the mice that do survive have significant non-astrocytic defects in the CNS and other organs (das Neves et al., 1999; Steele-Perkins et al., 2005). NFIC null mice have been studied solely for defects in tooth formation. If fed a diet to compensate for this defect, they live a normal lifespan (Steele-Perkins et al., 2003). Effects on GFAP expression have not been examined. Two lines of NFIX null mice have been produced. One line dies at about three weeks of age (Driller et al., 2007). Its Gfap mRNA levels were stated to be unaffected, but neither the age of the mice nor anatomical location examined was specified. Mice of the other line also die at about three weeks of age when fed a normal lab chow, but can live to adulthood if provided a modified diet, albeit with some persistent defects in CNS architecture (Campbell et al., 2008). Developmental studies of these mice suggest that NFIX acts after NFIA and NFIB to regulate the timing of CNS differentiation (Piper et al., 2011; Matuzelski et al., 2017). In an analysis of the spinal cord, generation of astrocytes and Gfap expression was delayed in the Nfix null mice, but reached wild type levels by P10, indicating that NFIX is not required for Gfap expression in mature astrocytes (Matuzelski et al., 2017). These findings with Nfi null mice indicate very different roles for GFAP synthesis for each isoform in vivo from those suggested by the cell culture studies described above.
NF-κB
Bae et al. (2006) investigated the role of nuclear factor kappa B (NF-κB) in regulation of GFAP transcription in cultured cells. Aspirin, which can block NF-κB activity, reduced GFAP mRNA and protein levels about 4-fold in both an A172 human glioblastoma cell line and in human primary brain astrocytes. A possible GGGGCTgCCC NF-κB binding site in the human promoter was noted conserved in both mouse and rat (the locations stated in their publication differ somewhat from those in the NCBI sequences; it starts at bp –1423 in the human). Mutation of this site in a 2.5 kb mouse promoter reduced activity of a linked luciferase reporter about 4-fold when transiently transfected into the A172 cells, and this reduced activity was no longer inhibited by aspirin. Further evidence for a positive transcriptional role for NF-κB came from decreased levels of GFAP protein in A172 cells transfected with an IκBα super repressor. Participation of NF-κB in reactive upregulation of GFAP expression was suggested by finding that treatment of human primary brain astrocytes with aspirin prevented the 3-fold increase in GFAP mRNA and protein levels produced by hypoxia. A caveat for this study is that a direct effect on the GFAP gene was tested only in a cell line and by transient transfection with a luciferase reporter. The primary astrocytes were used solely to test the effect of aspirin, which this group subsequently found to inhibit STAT3 activity (Kim et al., 2009).
An in vivo test of the contribution of the NF-κB site was made by Yeo et al. (2013), who analyzed mice carrying a gfaABC1D-nlac transgene with the NF-κB site mutated. Both expressing lines obtained had a normal level of reporter activity, but showed strong neuronal expression throughout the hippocampus, prompting the suggestion that the site is not important for basal activity in astrocytes, but serves to suppress GFAP expression in neurons. Caveats for this study are that the contribution of neuronal expression was not considered in the overall report that the level of expression was unaffected by the NF-κB site mutation, and the effect of the mutation on the reactive response was not tested.
Nkx2.1
Evidence that the homeobox transcription factor Nkx2.1 contributes directly to GFAP expression has been obtained from studies in mice and cultured cells, but its role in humans is uncertain. Nkx2.1 is expressed in progenitor cells in the ventral telencephalic region that give rise to GABAergic interneurons, oligodendrocytes, and astrocytes (Minocha et al., 2017). Studies with Nkx2.1 null mice showed that it contributes to the proliferation of the progenitors, and in its absence the number of astrocytes in the dorsal telencephalic region derived from normally Nkx2.1 positive cells is severely reduced. A role for Nkx2.1 in differentiation as well as proliferation was suggested by its effect on Gfap expression. When HEK293 cells were transfected with a lacZ reporter driven by a mouse Gfap promoter, the number of cells staining for lacZ was greatly increased by cotransfection with an Nkx2.1 expression vector. Analysis of the mouse Gfap promoter identified a segment starting at –839 that matches the core binding sequence of Nkx2.1, CTCAAGT. This site was thus present in the promoter sequence used for the HEK293 transfections, which commenced at –1666 (the beginning position of –1679 given in the paper used the translation start site as its reference point). ChIP analysis found binding of Nkx2.1 to a region centered on this CTCAAGT sequence. The findings with HEK293 cell transfection demonstrate that Nkx2.1 has the capability to stimulate Gfap promoter activity, but do not establish that it does so under physiological conditions; e.g., in astrocytes and with normal levels of Nkx2.1. More importantly, the putative CTCAAGT binding site is imperfectly conserved in rat (CTCAAaT, starting at –919), and not at all in human (CTggtGT, present at –792) (these positions differ by 3 places from those presented in Table 2, because the Table presents the full 14 nt Nkx2.1 consensus binding sequence, which includes 3 additional nucleotides of low specificity at its 5’ end). When compared to the full 14 nt Nkx2.1 consensus binding sequence, and ignoring the 2 positions at which any nt may be present (the N’s), the human sequence does have 10 of 12 matches; however, most of these are to nucleotides infrequently present at each position; matches to the most frequent nucleotides occur in only 3 of the 12 positions, whereas the mouse sequence has matches for 8 of the 12 positions. Thus, the biological relevance of regulation of GFAP expression by Nkx2.1 requires further study.
PAX3
PAX3 is a transcription factor with both paired box and homeodomain DNA binding domains. It functions early in development in formation of the neural tube and differentiation of skeletal muscle and neural crest cells. Liu et al. (2011) suggested that PAX3 may also have a role in preventing premature astrogenesis, based on observations that over-expression of PAX3 in rat E16 NECs inhibited astrogenesis stimulated by fetal bovine serum, whereas knockdown of PAX3 stimulated astrogenesis even in the absence of serum. A possible direct role for PAX3 in inhibiting GFAP synthesis was suggested by the presence of candidate paired box binding sites in the rat Gfap promoter at –1937 and –433 (a search for homeodomain binding sites was not reported). The ability of PAX3 to bind to these sequences was verified by EMSAs that included both wild type and mutant versions of the putative binding sequences as well as a PAX3 antibody-induced supershift. It was also verified by ChIP assays, although the latter lacked a negative control of a non-binding DNA region. As a functional test, a rat Schwann cell line was transfected with a luciferase reporter driven by a segment of the rat Gfap promoter extending from –1987 to +13. Deletion of the –1937 PAX3 binding site increased reporter activity by 60-fold, and deletion of the –433 binding site increased activity 110-fold. Whether knockdown of the endogenous PAX3 would similarly increase activity of the wild type promoter was not reported.
Together, these observations by Liu et al. make a compelling case for PAX3 acting directly to inhibit Gfap transcription in this model system. However, the biological relevance is uncertain. Kioussi et al. (1995) observed a positive, rather than a negative, correlation between Pax3 mRNA levels and GFAP protein in mouse Schwann cells in vivo and in culture during development, and also in response to injury and upon microinjection of a Pax3 expression vector. Similarly, although Wei et al. (2018) did observe an inverse relationship between PAX3 and GFAP during differentiation of mouse NECs, levels of both were increased in NECs following exposure to a microRNA inhibitor. A positive correlation between PAX3 and GFAP levels was also observed by Yang et al. (2016) during differentiation of mouse NECs adapted to grow in low glucose. In humans, mutations in the PAX3 gene result in Waardenburg syndrome, which is characterized by defects in skeletal and neural crest development, but without any reported effect on astrocytes. None of the 1,078 papers mentioning Waardenburg syndrome in PubMed contained either GFAP or astrocyte in any search field. Finally, as noted by Liu et al., neither of the putative rat Gfap PAX3 binding sites is conserved in mouse or man. There is strong homology between the corresponding rat and mouse sequences, but at both mouse sites the core TCAC is TCtC (Table 2). There is no homology for either site in the human sequence. The authors do point out that PAX3 binding sites may be present at –6.5 kb in the mouse sequence and –8.5 kb in human.
Retinoic Acid
Asano et al. (2009) found that retinoic acid about doubled the induction of GFAP synthesis by LIF in mouse E14.5 NECs. Retinoic acid receptors (RARs) bind to DNA as dimers, each subunit binding to an AGGTCA core, with the cores being either direct or indirect repeats separated by a spacer of 1 to 5 nts (Rastinejad et al., 2000). In the absence of retinoic acid, the bound receptors complex with transcriptional repressors such as N-Cor. Association with retinoic acid switches the receptors from complexing with repressors to binding to activators, such as p300/CBP. Asano et al. (2009) identified a putative RAR binding site, AGtTCAAGGTCA (but note that there is no spacer between the two core sequences), in the mouse promoter at –2502, and a ChIP assay showed binding of RARs to a corresponding upstream region of the mouse Gfap gene. Mutation of the binding site in a Gfap-luciferase reporter eliminated the ability of retinoic acid to augment LIF-induced activity. The mutation also increased LIF-induced reporter activity to the same level as obtained with the wild type reporter in the presence of retinoic acid, suggesting that retinoic acid acts solely to remove an inhibitory effect. Despite this finding, the authors suggest that retinoic acid acts not only to relieve repression due to RAR binding, but also has a positive effect. This was prompted by finding increased acetylation of histone H3 bound to the promoter, which was attributed to presumed binding of p300/CBP to the RAR. As just noted, such a positive contribution is inconsistent with their transient transfection results, which found that the activity of the wild type promoter in the presence of LIF and retinoic acid was the same, and not greater, than that of the mutated promoter in the presence of LIF.
A possible negative effector acting at the RAR binding site is peroxisome proliferator-activated receptor gamma (PPARγ), which can form a heterodimer with RARs. Rai et al. (2014) observed that treatment of either rats or rat primary astrocytes with a mixture of arsenic, cadmium and lead resulted in both an increase in activation of PPARγ via phosphorylation and about a 50% decrease in GFAP protein levels. A causal role for PPARγ for the GFAP decrease was indicated by inhibition of both of these changes by a PPARγ antagonist. Two putative RAR binding sites were identified in the rat Gfap gene, a direct repeat starting at –508 with a 1 nt spacer, and an inverted repeat in intron 1 starting at +924 with a 10 nt spacer (the start point of the –508 site was erroneously stated to be +507, and the DNA segment containing it was presented as if it were the sense strand, but it is actually the reverse complement). ChIP and antibody supershift EMSAs were used as evidence of binding of PPARγ to these DNA regions, and transfection of cultured rat astrocytes with 3 copies of each binding site driving expression of luciferase showed the predicted decrease in activity upon exposure to the mixture of arsenic, cadmium and lead.
Although the ChIP, EMSA and reporter transfection assays performed by Rai et al. appear to make a strong case for direct inhibition of Gfap expression by PPARγ, each of the observations has caveats. Technical issues with the EMSAs include the claimed antibody supershift bands not being visible, absence of controls for non-specific DNA binding or for non-specific effects of the added antibody, the amount of free probe in some lanes being markedly reduced without compensating signal appearing elsewhere, and it being unclear whether single or double stranded DNA was used as the probe. The Methods section states that Gapdh was targeted as a negative control for the ChIP assay, but those data are not mentioned in the Results. The cell transfection experiments lack an appropriate control for a general toxicity of the added metals mixture, and the responses of the putative PPARγ binding sites were tested in isolation rather than in the context of the Gfap promoter. An additional concern for both studies is the authenticity of the putative RAR binding sites. Binding of RAR to the mouse site identified by Asano et al. at –2502 is expected to be weak because it lacks a spacer between the core repeats. Although this site is conserved in rat, it is not present in the human GFAP promoter. The mouse Gfap gene has strong homology to the putative rat RAR binding site identified by Rai et al. at +924, but binding is expected to be weak also for this site due to the unusually long 10 nt spacer. More importantly, the human sequence has homology to only the first segment of the duplex site (Table 2). There is no homology between mouse and rat in the region of the putative rat binding site at –508. In a search of the human sequence, we did find two core AGGTCA binding sequences separated by 1 nt starting at –546 (this was the only candidate site with a 1 to 5 nt spacer found in a search from –1 to –3,000). However, it is unlikely that this corresponds to the rat site, as there is no homology to rat on either side of this sequence.
Sp1
In their analysis of the human B region, Yeo et al. (2013) noted the presence of two possible Sp1 binding sites in the B3 segment, one at –1541 and the other at –1530 (Table 2). Both sites are conserved with 2 mismatches in mouse and rat. EMSA or ChIP assays to demonstrate binding to these sequences was not performed. However, mutation of two nts within each site in a gfaABC1D-nlac transgene accounted for the entire 87% reduction in activity produced by the B3 block mutation.
A different Sp1 site was identified by Dore et al. (2009) in the rat Gfap promoter at –1473, just upstream of the ones investigated by Yeo et al. (2013). The corresponding mouse and human sequences both have a single nt difference from the rat sequence, but still match the consensus (Table 2). Here also, binding to the site was not demonstrated. In contrast to the results obtained by Yeo et al. (2013) in transgenic mice for mutation of the Sp1 sites in the B3 region, mutation of this site in a rat promoter driving expression of a luciferase reporter increased activity as measured by transient transfection into a neural crest-derived cell line. This suggested that under these conditions the site binds a repressor. However, consistent with Sp1 having a net positive effect, Hung et al. (2020) observed that astrocyte-specific knockout of Sp1 reduced Gfap expression.
Response to Injury
Correlation Versus Causality
Several hundred papers have been published investigating the injury-induced signals that result in astrogliosis. Since GFAP levels are commonly used as a marker of this process, each of these papers potentially provides information about signaling pathways that activate this gene (for an extensive compilation of changes in GFAP levels in disease or injury, see Li et al., 2020). In most of these studies, however, the focus is on the effect of drugs or signaling molecules on astrogliosis and injury outcome; experiments are usually not designed to trace the pathway linking the injury-induced signal to GFAP expression. Here we discuss those investigations which did seek to determine causal links.
One such study was performed by O’Callaghan et al. (2014) to determine how neurotoxins lead to Gfap activation in mice. They observed that treatment with a striatal toxin, MPTP, or with a hippocampal toxin, kainic acid, had a similar chain of responses: levels of the mRNAs encoding the cytokines TNF-α, OSM and LIF rapidly increased, followed by a rise in pSTAT3, and then an increase in Gfap mRNA and protein. To determine if the observed temporal progression is a causal chain, the cytokine increases were inhibited by corticosterone. Unexpectedly, this did not prevent either the rise in pSTAT3 or Gfap expression, leading the authors to suggest that despite the strong correlation, the neuroinflammatory response occurs independently of the GFAP increase. However, there are alternatives to this striking conclusion. One possibility is that the corticosterone did not sufficiently reduce the cytokine levels; although described as being “suppressed,” their data show that significant increases still occurred. As an extreme case, corticosterone reduced the MPTP-induced level of Osm mRNA by only about 15%. Another possibility is that the measured mRNA levels of the cytokines do not correspond to their protein levels. Yet another alternative is that the GFAP increase may be independently mediated by several different signaling pathways, so that an elevation in cytokines is sufficient, but not necessary, for the rise in GFAP. However, sufficiency of a cytokine increase appears to be ruled out by another experiment performed by the group, in which they examined events during the acute phase following acute lipopolysaccharide (LPS) administration. At 6 hours after s.c. injection of LPS, increases in the mRNA levels of Tnf, Osm and Lif were observed that were similar to those produced by MPTP, yet there was no change in GFAP amounts over a 72 h period. This indicates that increased levels of these cytokines are not sufficient to trigger GFAP synthesis (with the caveat that cytokine mRNA levels, not protein, were measured).
O’Callaghan et al. (2014) also investigated whether activated STAT3 necessarily causes increased Gfap transcription. Two different experiments produced seemingly conflicting outcomes. An astrocyte-specific knockout of STAT3 markedly attenuated the MPTP-induced increase in GFAP protein, indicating that pSTAT3 is essential for Gfap expression. On the other hand, although LPS did not increase GFAP levels, it did stimulate STAT3 activation, suggesting that this activation does not necessarily result in increased Gfap expression. Possible explanations for this latter finding are that the level of pSTAT is already saturating for GFAP synthesis (but not for other gliotic changes, which it does initiate); or perhaps more likely, that the LPS-induced pSTAT increase occurred in a cell type other than astrocytes, like microglia (it was shown that the pSTAT increase in response to MPTP and kainic acid occurred in astrocytes, but this analysis was not performed for LPS).
Like the O’Callaghan et al. (2014) study, Kirsch et al. (2010) observed strong correlative evidence for a signaling pathway that proved to be misleading. They noted that following optic nerve crush in the mouse, levels of CNTF, pSTAT3 and GFAP all increased severalfold in Müller cells, strongly suggesting a regulatory pathway. However, when a Cntf null mouse was used to test causality, they found that instead of the knockout attenuating STAT3 phosphorylation and Gfap expression, they were instead increased. Experiments with a Lif null mouse indicated that LIF was the actuating signal in this injury model, and that the effects of the Cntf null were due to it unleashing increased injury-induced production of LIF. That LIF had its effect by activating the JAK/STAT pathway was indicated by finding that knockout of STAT3 in astrocytes reduced the GFAP increase following optic nerve crush (this effect was only measured by immunostaining). However, a finding escaping comment was that heterozygosity for the Lif null allele was sufficient to prevent the GFAP increase in response to injury in the Cntf null mice, but the increase in pSTAT3 remained intact. This suggests that in this injury model, STAT3 phosphorylation is necessary, but not sufficient, for the Gfap response.
In addition to neurotoxins and optic nerve crush, LIF has been implicated as the signaling molecule following a cortical stab wound in mice. Sugiura et al (2000) found that the increase in GFAP staining following such an injury is reduced in Lif null mice. However, it was not determined whether this was a direct or indirect effect; e.g., it could simply be due to the observed reduction in the number of astrocytes in the injury region.
Despite LIF being implicated in each of the three injury models just described, it is not a universal messenger for CNS injury. For example, Kirsch et al. (2010) noted in their study that activation of GFAP synthesis in Müller cells by kainic acid was unaffected by knockout of either Cntf or Lif. Evidence for other mediators is presented below.
Role of STAT3
Both O’Callaghan et al. (2014) and Kirsch et al. (2010) used an astrocyte-specific knockout of STAT3 to evaluate its participation in GFAP expression. This null mouse, as well as other genetic manipulations of STAT3 activity, has been used for the same purpose by several other laboratories, with it generally being found that interference with STAT3 activity greatly attenuates an injury-induced GFAP increase (for a review, see the Discussion section of Brenner et al., 2019). However, this finding does not necessarily identify STAT3 as a mediator of the GFAP reactive response, but could instead be due to it having a general role for GFAP expression irrespective of physiological state. As previously discussed in the Smad1 section of Developmental Regulation of GFAP Expression, the critical question for interpreting these results is whether the relevant parameter is the increase in activity units, or the fold-increase of expression over the basal level. For example, in the O’Callaghan et al. (2014) study, the increase in units of GFAP protein following MPTP treatment was about 9 times greater for the control than for the astrocytic STAT3 null; but because the conditional knockout depresses the basal level of GFAP as well as its upregulation, the fold-changes in activity were similar at 3.2 for the control and 3.4 for the null (data taken from their Figure 6). As noted previously (Brenner et al., 2019), if the fold-change criterion were adopted, then only a modest contribution of JAK/STAT to GFAP upregulation following injury would be evident in some studies, and no effect in others.
The same consideration applies to the report of Sun et al. (2008) that ischemia produces reactive gliosis through purinergic signaling via astrocytic P2Y1 receptors. At 24 hours after transient right middle cerebral artery occlusion in rats, levels of Gfap mRNA increased much less in rats treated with a P2Y1 inhibitor than in untreated rats; but because the inhibitor also lowered the basal Gfap mRNA level, the injury-induced fold-change was actually several times greater in the presence of the inhibitor.
Roles of AP-1 and Calcium
The possible involvement of AP-1 in GFAP upregulation following injury was investigated by Brenner et al. (2019) using transgenic mice. They compared the injury-induced induction of a lacZ reporter driven by either the human gfa2 or ABCD promoter to that driven by the same promoter with the consensus AP-1 binding site at –1592 mutated. Unlike the case for pSTAT3, mutation of the AP-1 binding site had no effect on Gfap expression in uninjured mice, but abolished the response for each of the three injury models examined—kainic acid-induced excitotoxicity, cryoinjury, and Alexander disease-associated gliosis. These observations suggest that this AP-1 binding site is essential for the GFAP reactive response to multiple types of CNS injury.
Prior publications indicating a role for AP-1 transcription factors in the injury-induced upregulation of GFAP expression are reviewed in the Discussion section of Brenner et al, 2019. Three papers particularly pertinent are Raivich et al. (2004), Gadea et al. (2008), and Gao et al. (2013).
Raivich et al. (2004) observed that the knockout of Jun had no effect on GFAP levels in uninjured mice, but reduced its increase in response to facial nerve axotomy, consistent with AP-1 having a critical role for injury-induced GFAP upregulation, but not for its basal expression.
Gadea et al. (2008) also identified AP-1 as key to injury-induced GFAP upregulation in their study of endothelin-1 (ET-1) and reactive gliosis. This study was prompted by prior observations that ET-1 levels are increased in response to a variety of brain injuries. Finding that ET-1 elevated Gfap mRNA and protein levels in primary cultures of rat astrocytes, they used various inhibitors to determine the signaling pathway. ET-1 was found to act through its ETB-R receptor, leading to phosphorylation of JNK and c-Jun, with these activations being dependent on the mixed lineage kinase pathway. Surprisingly, no effect on Gfap expression resulted from treatment with a JNK inhibitor or a MEK inhibitor, but together they completely prevented the GFAP response. Since an endpoint of the MEK pathway is activation of c-Fos, these findings implicate an AP-1 Jun/Fos dimer in the injury response of the Gfap gene.
A related pathway leading to AP-1 activation, but differing by the absence of a role for MEK signaling, was described by Gao et al. (2013). They found that the increase in Gfap mRNA and protein in a scratch model of primary mouse astrocytes was triggered by uptake of extracellular calcium, followed by activation of JNK and then AP-1. Increased AP-1 activity was demonstrated by transfection with a luciferase reporter driven by AP-1 binding sites, and by ChIP assays. Inhibitors of the MEK/ERK and JAK/STAT pathways had no effect, indicating that these signaling systems do not participate in this injury response. Consistent with these cell culture findings that indicated a primary role for calcium uptake, the in vivo GFAP increase following a cortical stab wound was markedly reduced by injection of the calcium chelator BAPTA-AM. The mechanism by which elevated intracellular calcium activated JNK was not determined in this investigation. However, a prior examination of calcium activation of JNK in bladder smooth muscle cells indicated participation of calmodulin and calcineurin, but not ERK (Kushida et al., 2001).
Another GFAP-activating pathway involving increased intracellular calcium was described by Brahmachari et al. (2006). Building on observations that increased nitric oxide (NO) levels are often associated with CNS injury, they investigated whether NO mediated the GFAP increase associated with reactive gliosis. Gfap expression was indeed elevated by exposure of mouse primary astrocytes to multiple activators of the inducible nitric oxide synthase (iNOS). These activators included LPS, interferon-γ, IL-1β, HIV-1 gp120, amyloid β1–42, and double-stranded RNA. Use of an iNOS inhibitor and an NO scavenger confirmed that the GFAP increase was dependent on NO production, and use of an NO donor established that NO was sufficient to trigger the increase. For LPS, the findings were extended to the mouse in vivo: striatal injection of LPS resulted in increased Gfap mRNA and protein, which was reduced by co-injection of the iNOS inhibitor or NO scavenger. [The LPS-induced increase in GFAP described here contrasts with the absence of an increase observed by O’Callaghan et al. (2014) as reported in the section Correlation versus Causality in Response to Injury, above. The different outcomes are likely due to the LPS being injected systemically by O’Callaghan et al., but intracranially by Brahmachari et al. Systemic LPS is reported to minimally enter the brain, and likely produce neuroinflammation indirectly (Banks and Robinson, 2010).] NF-κB, previously shown to mediate induction of iNOS by LPS, interferon-γ and IL-1 (Nomura, 2001), was positioned prior to NO production in the pathway by finding that an inhibitor of its activation prevented the GFAP increase stimulated by any of the above-mentioned activators, but not that by the NO donor. This participation by NF-κB is consistent with the report by Bae et al. (2006) described above (section Transcription Factors Regulating GFAP Expression in Mature Astrocytes, which see also for caveats) that aspirin, an NF-κB inhibitor, prevented the increase in GFAP mRNA seen in untreated human primary astrocytes subjected to hypoxia. The GFAP-activating pathway was further probed by Brahmachari et al. (2006) through use of a guanylate cyclase inhibitor, which placed that enzyme downstream of NO production. Lastly, reduction of the LPS-induced GFAP increase by an inhibitor of cGMP-activated protein kinase indicated that this enzyme mediates the effect of cGMP.
The linkage between cGMP-activated protein kinase and increased GFAP synthesis was not pursued by Brahmachari et al. (2006), but a subsequent study by Sticozzi et al. (2020) of the role of guanylate cyclase in GFAP upregulation implicated increased calcium levels. By treating the human glioma cell line U373 with inhibitors of iNOS and guanylate cyclase, they deduced that these two enzymes were required for the induction of GFAP by TNF-α or IL-6. A severalfold elevation in intracellular calcium that accompanied the GFAP increase suggested that it was a participant in the pathway, and this was supported by finding that the GFAP increase was blocked by inhibitors of intracellular calcium channels. The calcium increase was placed downstream of iNOS and guanylate cyclase, because it was blocked by their inhibitors. Caveats for these findings are that the induced increases in GFAP were only about 20% above the control level, they were only measured by immunostaining, and the U373 cell line likely has aberrant physiology. However, they were supported by experiments performed in vivo. Injection of TNF-α or IL-6 into rat striatum resulted in increased cGMP and GFAP, and a similar response was produced by injection of an NO donor. These increases were abrogated by prior treatment with the inhibitors of iNOS, guanylate cyclase or calcium channels.
Combining the findings of these latter three papers yields an injury-induced pathway for GFAP expression that begins with production of cytokines, their binding to astrocyte membrane receptors, and activation of NF-κB. NF-κB in turn induces iNOS synthesis, with the resultant NO activating soluble guanylate cyclase, producing cGMP that triggers an increase in intracellular calcium, perhaps via a cGMP-dependent protein kinase. Finally, the calcium acts through calmodulin and calcineurin to activate AP-1, which binds to the GFAP promoter. Whether this stitched-together pathway is indeed correct is yet to be determined. For example, could progression through the multiple steps be sufficiently rapid to account for the increases in calcium that were observed by Sticozzi et al. (2020) to occur in cultured cells and mouse striatum as soon as 15 minutes after IL-6 or TNF-α application?
Roles of Other Signaling Systems
IL-1β was mentioned in the above section as a candidate for signaling for reactive gliosis. This was tested by Lin et al. (2006) by examining the effect on the GFAP response to a cortical stab wound of knockout of the interleukin 1 receptor type 1 (Il1r1), which serves both IL-1α and IL-1β. Following such an injury, the level of GFAP increased more slowly in the Il1r1 null mouse than the wild type, although it eventually reached the wild type level. This suggests that IL-1α and/or IL-1β acts as an extracellular messenger to increase GFAP expression during the first few days following the trauma. Which of the multiple possible downstream mediators of the GFAP increase were involved was not pursued, nor was it established whether the primary effect of the IL-1 receptor type 1 deficiency was in astrocytes or in some other cell type, like microglia.
Yet another signaling system responding to a cortical stab wound involves the Protease Activated Receptor (PAR) family of G-protein coupled receptors, which as their name states, are activated by various proteases. Nicole et al. (2005) found that the GFAP increase induced by this injury was markedly curtailed in F2r null mice (F2r is the gene encoding PAR-1) compared to the wild type. The pathway from PAR-1 activation to Gfap expression was not pursued. It may be indirect, perhaps via other cell types, since activation of PAR-1 in cultured wild type astrocytes had no effect on GFAP levels.
Another PAR family member, PAR-2 (encoded by F2rl1), was studied by Radulovic et al. (2015) for its role in the gliotic response to a spinal cord crush injury. Suggestive of a role was the observation that the mRNAs encoding both PAR-2 and neurosin, a PAR-2 agonist, increased about 2-fold following such an injury. Knockout of PAR-2 completely prevented the increase in GFAP present in wild type mice 3 days post-lesion. The F2rl1 null mice also had attenuated increases compared to the wild type in the levels of TNF-α, IL-1β, TGF-β, IL-6 and IL-10. These observations, together with results from experiments with primary astrocyte cultures, led the authors to propose an indirect mechanism for injury-induced activation of Gfap by PAR-2. The elevated levels of neurosin and PAR-2 were proposed to activate the MEK pathway, resulting in increased production and release of IL-6 from astrocytes and immune cells. The IL-6 then acts through the gp130 receptor to increase STAT3 phosphorylation, and thereby stimulate Gfap expression.
Key findings with the primary cultures of astrocytes contributing to this model were that neurosin elicited about a 2-fold increase in both IL-6 secretion and pSTAT, and that IL-6 caused about a 2-fold elevation in its own mRNA as well as those for neurosin and Gfap, and over a 1,000-fold increase in PAR-2 mRNA. The stimulation of IL-6 secretion by neurosin was partially reduced by a MEK1/2 inhibitor, and blocked by a pSTAT inhibitor, but no experiment was performed to demonstrate that the ability of IL-6 to increase Gfap expression was dependent on pSTAT3. When F2rl1 null astrocytes were used, the stimulation of IL-6 release by neurosin was reduced by half compared to wild type astrocytes, and there was no increase in pSTAT (the effect of the PAR-2 knockout on the responses to IL-6 was not tested).
Many of these observations fit the proposed model, but some do not. One seeming inconsistency is that the wild type and knockout had similar increases in pSTAT3 in the injury core after 3 days, but only the wild type had an elevated GFAP level. Another is that despite primary wild type astrocytes increasing both IL-6 release and STAT3 phosphorylation in response to neurosin, neurosin did not increase Gfap mRNA levels. Also, the model predicts that IL-6 secretion should be completely prevented by inhibition of MEK1/2, but only partially by inhibition of pSTAT3, yet the opposite was observed. Finally, the model does not address the apparently transitory effect of PAR-2 on Gfap expression: at 30 days post-injury, Gfap mRNA levels in the injury epicenter for the wild type and the knockout were both about 2-fold greater than in the uninjured controls. Thus, there is more to learn about the contribution of PAR-2 to the gliotic response.
BMP is another candidate proposed for signaling for reactive gliosis following a spinal cord compression injury. Its participation was investigated by Sahni et al. (2010) by developing an astrocyte-specific KO of the BMP receptor BMPR1a, prompted by finding that the mRNA for this receptor as well as that for BMP4 increased about 3-fold following a compression injury. They were rewarded by the null mouse displaying an attenuated gliosis response compared to the wild type, including a reduced increase in GFAP, and by astrocytes cultured from the null also having reduced Gfap mRNA levels. When the KO was found not to affect the increased phosphorylation of STAT3 or SMADs following spinal cord injury, a possible role for the microRNA miR-21 was investigated, because its level was known to be regulated by BMP signaling (see below in the section Regulation of mRNA Translation for a description of microRNAs). The level of miR-21 was indeed found to be elevated in astrocytes cultured from the BMPR1a conditional KO. Furthermore, over-expression of miR-21 in cultured astrocytes by lentiviral transfection resulted in decreased Gfap mRNA and protein. However, the authors suggest that miR-21 is unlikely to have a direct effect on Gfap expression, because its over-expression also resulted in the astrocytes having smaller processes and cell size, effects unrelated to GFAP reduction (reviewed in Brenner, 2014), and because an miR-21 binding site could not be identified in the mouse Gfap gene.
Expression of GFAP Isoforms
GFAP is encoded by a single gene, but multiple mRNA isoforms exist (reviewed in Messing and Brenner, 2020). Regulatory mechanisms affecting their expression likely function at the transcriptional level for GFAPα, GFAPβ and GFAPγ, which have different RNA start points, and at the post-transcriptional level for the other isoforms, GFAPδ, GFAPκ and GFAPλ, which arise from alternative splicing. None of the transcriptional studies to date has discriminated among GFAPα, GFAPβ and GFAPγ, but because they were performed in the CNS, they can be assumed to apply to the GFAPα isoform, which accounts for greater than 90% of CNS GFAP mRNA. However, substantial variations in the ratios between levels of some of the isoform mRNAs and GFAPα mRNA have been documented, revealing that they can be differentially regulated. For example, GFAPβ mRNA, which initiates upstream of GFAPα mRNA, accounts for about 75% of the GFAP mRNA in Schwann cells but only about 5% in astrocytes (Galea et al., 1995). Levels of GFAP mRNA splice variants that have the same start point as GFAPα generally vary in parallel with GFAPα, but their ratios can also differ between tissues or physiological conditions. For example, the relative level of GFAPδ mRNA, an alternatively spliced mRNA in which exons 8 and 9 are replaced by exon 7a, is about 5-fold higher in vanishing white matter disease compared to controls (Bugiani et al., 2011); and GFAPκ has been reported to be the primary isoform expressed in human enteric glial cells (Clairembault et al., 2014). Possible mechanisms for altering the relative levels of a GFAP splice form include global changes in efficiency of mRNA splicing, or targeted regulation exploiting their sequence differences, such as microRNAs binding to 3’UTR sequences differentially present among the splice variants (see the microRNA section below).
Non-CNS Expression
Although GFAP’s primary claim to fame is as a marker of astrocytes, it is also expressed in cell types outside the CNS (reviewed in Messing and Brenner, 2020). In several instances, non-CNS cells that contain GFAP have been observed to express GFAP promoter-driven transgenes used for astrocytes, suggesting that these promoters also contain elements sufficient for non-CNS activity. Examples include expression of thymidine kinase in enteric glia (Bush et al., 1998) and hepatic stellate cells (Puche et al., 2013) driven by the mouse Gfap genomic sequence extending from –1980 to about +10,874, and expression of gfa2-driven transgenes in Schwann cells and support cells in the ear (Rio et al., 2002), pancreatic stellate cells (Ding et al., 2009) and hepatic stellate cells (Chen et al., 2008). In the absence of quantitative comparisons between transgene and endogenous GFAP expression, however, it is possible that elements contributing to non-CNS expression reside outside the transgene promoter sequences.
We are aware of only one study seeking to identify a regulatory element contributing to non-CNS GFAP expression. Using a rat hepatic stellate cell line stably transfected with a gfa2-lacZ reporter, Zhang and Zhuo (Zhang and Zhuo, 2006) (2006) observed inhibition of expression of both lacZ and the endogenous Gfap gene by epigallocatechin gallate and genistein, compounds known to antagonize the fibrogenic response of these cells. Immunoblotting of nuclear extracts revealed that each inhibitor reduced the level of c-Jun and c-Fos by about 50%, and EMSAs demonstrated binding of nuclear extracts from untreated cells to sequences corresponding to the AP-1 binding site present in the rat and human GFAP promoters. Caveats are that the EMSAs lacked a negative control of a mutated or unrelated oligonucleotide binding target, the effect of the inhibitor treatments on binding activity was not determined, the decreases in c-Jun and c-Fos were not shown to affect Gfap transcription, and the properties of the cell line may differ from those of stellate cells in vivo. Nevertheless, the suggestion that AP-1 participates in GFAP upregulation in the fibrogenic response of hepatic stellate cells is consistent with findings that it is critical for increased GFAP expression in astrogliosis. It is also consistent with findings of Ding et al. (2009) with a pancreatic stellate cell line stably transfected with a gfa2-lacZ transgene. They observed that lacZ activity was reduced by inhibitors of JNK and the MEK1/2 pathway, which are known to activate fibrogenesis by these cells as well as c-Jun and c-Fos.
Although Zhang and Zhuo (2006) found that epigallocatechin gallate reduced the expression of both endogenous Gfap and the gfa2-lacZ transgene, there were remarkable differences in the timing, dose-response, and extent of the reductions. Inhibition of the transgene was rapid and transient, peaking at 2 h with about a 40% reduction in activity and gone by 4 h, whereas inhibition of the endogenous gene peaked at 24 h with about a 70% reduction, and was still present at 48 h. A dose of 1.25 μM had no effect on transgene expression, but reduced expression of the endogenous Gfap gene by the 70% maximum amount. These large differences in the responses of the two genes suggest that regulation of the endogenous gene includes regulatory sequences not present in the transgene. However, because the integration site of a GFAP-driven transgene can strongly affect its activity, especially outside the CNS (reviewed in Su et al., 2004), additional studies are required to establish this point.
Regulation of mRNA Translation
microRNA
MicroRNAs (miRNAs) are small (∼22 nt) non-coding RNAs that affect mRNA translation or stability, usually reducing expression. After associating with an Argonaute protein, the seed sequence in the microRNA, corresponding to nts 2-8, binds to its complement in an mRNA, usually located in the 3’UTR. After binding, additional proteins that modulate expression of the targeted mRNA are recruited to the miRNA/Argonaute complex (Gebert and MacRae, 2019).
Several reports have linked microRNAs to GFAP gene activity. In a search for targets of miR-3099, Abidin et al. (2017) identified the mouse Gfap gene based on sequence analysis of mouse 3’UTRs by a battery of algorithms, and by an inverse correlation between the presence of Gfap mRNA and miR-3099 in various mouse brain regions. For a functional assay, a segment of the Gfap gene containing the miR-3099 binding site was cloned downstream of a luciferase reporter. Co-transfection of this reporter together with an miR-3099 expression vector into HEK293FT cells resulted in 4-fold lower activity than co-transfection with a miR-3099 scrambled sequence vector.
Neither the miR-3099 binding sequence nor its location within the mouse Gfap 3’UTR was provided in this report, but the oligonucleotide sequences used to amplify the 3’UTR fragments tested point to a region extending from +5,980 to +6,768. In a subsequent publication, Abidin et al. (2019) give the seed sequence as AGGCUAG, corresponding to a binding site (reverse complement) of CTAGCCT, which is present 8 times in the amplified segment due to multiple direct repeats of a larger sequence in which it is embedded. The authors appear unaware that the amplified sequence is located in the 3’UTR of Gfapδ, a minor GFAP splice variant, which accounts for only about 4% of the total Gfap mRNA in mouse brain (Thomsen et al., 2013). In the predominant Gfapα isoform, this sequence is in intron 7, and thus not present in mature Gfapα mRNA. Accordingly, a significant contribution of these miR-3099 sites to negative regulation of GFAP levels in the mouse seems unlikely. It is even more problematic for human GFAP, since miR-3099 is not present in humans (although a possible human homolog has been identified; Abidin et al., 2019), and none of the 8 mouse binding sites is conserved in the corresponding region of the human gene.
Choi et al. (2016) provide evidence that miR-326, miR-330, and miR-3099 act to increase the efficiency of Gfap mRNA translation in mouse. Data include positive correlations between the levels of these three miRNAs and GFAP protein during mouse brain development, during CNTF-induced differentiation of mouse E13.5 NSCs, and in response to knockout of PINK1. Like Abidin et al., they also used a functional assay of transient transfection into HEK293T cells of a luciferase reporter linked to either wild type or mutant forms of the Gfap 3’UTR. However, in contrast to the findings of Abidin et al., inclusion of mimics for each of the three microRNAs increased, rather than decreased, reporter activity, with the miR-3099 mimic producing a 2-fold increase. It is unclear why such similar assay systems produced such discrepant results.
Although Choi et al. also did not explicitly specify the positions of the miRNA binding sites, sufficient information was provided to establish that the miR-3099 binding sites were the same 8 sites studied by Abidin et al., and that the shared binding site for miR-326 and miR-330 is slightly 3’ of those, starting at +6,889. Thus the same concerns apply about biological relevance of these sites, given that they are all in the non-conserved mouse 3’UTR of the Gfapδ isoform.
Wang et al. (2015) observed that the level of another miRNA, miR-145, decreased as that of GFAP increased following spinal cord injury in rats. To test for a causal relationship between miR-145 and GFAP levels, a lentivirus carrying a GFAP promoter-driven miR-145 was transfected into the rat spinal cord immediately after injury. The astrocyte hypertrophy and GFAP increase induced by the injury were markedly reduced when the miR-145 sequence was expressed compared to expression of a scrambled sequence. The presence of a perfect match to the AACUGGA binding site for miR-145 in the 3’UTR of the rat Gfapα mRNA suggested that Gfap expression might be a direct target of the miRNA. This site is located 249 nt from the translation terminating UGA codon, corresponding to +7,541 of the rat genomic sequence. Its role was tested by inserting a segment of the 3’UTR containing this binding site downstream of a luciferase reporter, and transfecting the construct into U373 cells. Cotransfection with the lentivirus expressing miR-145 reduced both reporter expression and endogenous GFAP levels by about 50%. Treatment with an miR-145 antagonist prevented both of these reductions.
Although strongly suggestive that the identified miR-145 binding site regulates rat Gfap expression, in the absence of an experiment in which the site was mutated it remains possible that the effect of miR-145 is indirect. It is also possible that the inhibition is an artifact of supraphysiological levels of miR-145 produced by its over-expression. Treating cells with the miR-145 antagonist in the absence of over-expression could have been informative for biological significance. Whether miR-145 has a direct effect on human GFAP expression is also questionable. Although miR-145 is present in humans, and its over-expression in U373 cells reduced the level of endogenous (human) GFAP, its putative binding site is not conserved at the corresponding position in the human 3’UTR (AtgaGGA), nor is a match present anywhere in the entire GFAPα mRNA.
Involvement of another miRNA, miR-139, was pursued by Wang J et al. (2018). In a search for SNPs associated with risk for glioblastoma, they identified a C/G polymorphism in the 3’UTR of GFAPα at +8182. Reduced disease prevalence was associated with the G allele, which was present at a frequency of 33% in their population of 406 individuals. A computer analysis predicted that the G allele, but not the C allele, would produce a binding site for miR-139, and thus might reduce GFAP expression. Consistent with this possibility, GFAP protein levels were about 8-fold lower in glioblastoma cells freshly isolated from patients homozygous for the G allele (3 samples) compared to patients homozygous for the C allele (6 samples), and levels from heterozygous patients (3 samples) were intermediate. Suggesting that these differences were indeed due to miR-139, transfection of U251 cells with an miR-139 mimic decreased GFAP levels up to 7-fold, and transfection with an miR-139 antagonist increased GFAP levels up to 6-fold. However, these results do not necessarily implicate the GFAP SNP, because the allele genotype of the U251 cells was not stated. Were it C/C, the findings would imply an indirect effect of miR-139. As a more direct test of the role of miR-139 binding to the putative GFAP 3’UTR site, a 100 bp segment of the UTR containing either the C or G allele, or a mutated version of the presumed binding site, was inserted downstream of a luciferase reporter. These constructs were then transfected into U251 cells either alone or with an miR-139 mimic or antagonist. Consistent with miR-139 not binding to the C allele, no statistical difference was found between the activities supported by the C allele and mutant constructs, and neither the miR-139 mimic nor antagonist affected their activity. On the other hand, consistent with miR-139 binding to the G allele, it supported about 45% less reporter activity than the C allele, the miR-139 mimic further reduced activity by about 30%, and the miR-139 antagonist increased activity to near that of the C allele.
These results appear to provide strong evidence for miR-139 binding to the G allele and producing a significant suppression of GFAP expression. However, there are several curiosities not addressed by the authors. It is considered a requirement for miRNA function that at least 6 of the 7 nts in its seed sequence (nts 2-8) form Watson-Crick base pairs with the mRNA target; and the preceding mRNA nt, which is recognized by the Argonaute protein and not by nt 1 of the miRNA, is usually an A (Agarwal et al., 2015). In this instance, only 4 of the seed nts are predicted to form base pairs with the mRNA target, one of these being a G:U pair, and the preceding mRNA nt is a G rather than an A. Functional binding of a miRNA to such a poorly matched mRNA target is thus highly unlikely, especially since complementarity between the remainder of the miR-139 sequence and the mRNA is also poor (8 base pairs among the remaining 15 positions, with 2 being G:U). Another peculiarity is that the GFAP 3’UTR segments analyzed were apparently cloned downstream of an efficient SV40 polyadenylation sequence that follows the luciferase coding sequence in the pGL3-basic vector used. Thus, the SNP allele variants are not expected to be included in the mature luciferase mRNA. Possibly miR-139 is instead affecting reporter activity by a transcriptional mechanism. The absence of a promoter in the pGL3-basic vector used makes it particularly sensitive to such an effect.
In addition to association of the G allele with lower risk of glioblastoma, using cells isolated from glioblastoma patients, Wang et al. found it associated with lower levels of vimentin, a lower rate of migration, and less resistance to the chemotherapeutic drug imatinib. None of these properties has previously been attributed to a reduced level of GFAP, and studies of Gfap null mice found no effect on either migration or vimentin levels (Brenner, 2014). This raises the possibility that the GFAP SNP is serving as a linkage marker to other, causative, genetic differences. As noted by the authors, their study was limited to a particular ethnic group in eastern China, requiring that their intriguing findings be confirmed by a more extensive study.
Quaking
Quaking (QKI) is a sequence-specific RNA-binding protein expressed in oligodendrocytes and astrocytes that is associated with the dysmyelinating quaking phenotype in mice (Hardy et al., 1996). Functions of QKI include contributing to mRNA splicing, transport, translation and/or stability. To investigate a possible role for QKI in astrocytes, Radomska et al. (2013) transfected siRNA into human primary astrocytes to knockdown either all 4 known QKI isoforms (QKI5, QKI6, QKI7 and QKI7b), or specifically QKI7 and QKI7b (hereafter referred to as QKI7/7b). Genome-wide RNA sequencing (RNA-seq) showed that each treatment caused up or down changes in a small group of mRNAs. GFAP mRNA was among these for the QKI7/7b knockdown, being reduced about 50%. Although it was not among the mRNAs changed by total QKI knockdown (surprisingly, no mRNA was common to both groups), qPCR found that total QKI knockdown also produced about a 50% decrease in GFAP mRNA. A possible QKI binding site was identified in the 3’UTR of GFAPα mRNA, consisting in part of a consensus ACUAAC core sequence starting at +8,371, which is 217 nts downstream of the UGA translation termination codon. Also part of the site is a CAAC partial core sequence (a “half sequence”) beginning 8 nts upstream, at +8,363. The CAAC conforms to the YAAY half sequence consensus stated by Radomska et al., but not to the UAAY consensus in the source they cite (Galarneau and Richard, 2005); thus, the GFAP site might not provide optimal binding. The core sequence, but not the half sequence, is conserved in both rat and mouse.
The presence of this putative QKI binding site suggests a direct, positive, contribution of QKI7/7b to GFAP expression. However, no manipulation of the site was performed, leaving open whether it is functional. Since the site is present in GFAPα mRNA, but not in GFAPδ or GFAPκ mRNA, knockdown of QKI7/7b would be expected to differentially affect their levels. Unfortunately, an experiment performed to investigate this possibility using RNA-seq was inconclusive. Use of qPCR rather than RNA-seq might have yielded more quantitatively definitive data.
In summary, although there is suggestive evidence for post-transcriptional regulation of GFAP expression by miRNA and QKI, compelling data are yet to be obtained.
Concluding Remarks
The above studies reveal that GFAP expression is highly regulated, multifaceted, and responsive to changes in both the intracellular and extracellular environments, yet much remains to be learned. Important roles for several transcription factor binding sites have been established, but many others await more definitive demonstration, or are still to be discovered (see Yeo et al., 2013, for a listing of several additional candidate transcription factors). As one example, the alteration of multiple regions within the commonly used gfa2 promoter (–2162 to +47) produces effects on expression that are yet to be explored, and important regulatory elements responsible for increased transcription in response to injury remain to be discovered that lie outside of this region (Brenner et al., 2019). As a second example, a proposed mechanism in Alexander disease is that expression of mutant protein invokes a positive feedback loop that increases promoter activity and exacerbates disease (Jany et al., 2013), but the specific factors and pathways that mediate this effect have yet to be defined. The purpose of this complex regulatory machinery for expression of an intermediate filament protein is mysterious. The lifespan and behavior of GFAP null mice is little different from that of their wild type littermates, at least in the confines of a laboratory cage (see Brenner, 2014, for a review of functions attributed to GFAP). Perhaps the complexity of its regulatory system is telling us that in the wild, GFAP has multiple important functions that differ depending on the physiological state and the cell type in which it’s expressed.
Abbreviations
Akt= Protein kinase B; AP-1= Activator protein 1; BMP= Bone morphogenetic protein; BRG1= Brahma-related gene 1; CBF1= Centromere-binding protein 1; CBP= CREB-binding protein; ChIP= Chromatin immunoprecipitation; CNTF= Ciliary neurotrophic factor; CSL= CBF1, Suppressor of hairless, Lag-1; Dnmt1= DNA methyltransferase 1; DREAM= Downstream Regulatory Element Antagonist Modulator; E4ICD= Cytoplasmic tail of ErbB4 that is cleaved off following ligand binding; EMSAs= Electrophoretic mobility shift assays; ErbB4= Erb-B2 receptor tyrosine kinase 4; ET-1= Endothelin-1; FISH= Fluorescent in situ hybridization; gp130= Glycoprotein 130; H3K9me3= Trimethylated lysine 9 of histone 3; H3K36me3= Trimethylated lysine 36 of histone 3; IL= interleukin; iNOS= Inducible nitric oxide synthase; JAK= Janus kinase; KO= Knockout; LIF= Leukemia inhibitory factor; LPS= Lipopolysaccharide; MAPK= Mitogen-activated protein kinase; MeCP2= Methyl CpG binding protein 2; miRNA= MicroRNA; N-CoR= Nuclear receptor co-repressor; NECs= Neuroepithelial cells; NF-κB= Nuclear factor kappa B; NFIA= Nuclear factor I A; NICD= Notch intracellular domain; NO= Nitric oxide; NSCs= Neural stem cells; nt= Nucleotide; OSM= Oncostatin M; Osmr= Oncostatin M receptor gene; p300= Histone acetyltransferase p300; PACAP= Pituitary adenylate cyclase-activating polypeptide; PAR= Protease activated receptor; PI3K= Phosphatidylinositol 3-kinase; PPARγ= Peroxisome proliferator-activated receptor gamma; PRMT1= Protein arginine methyltransferase 1; pSTAT3=STAT3 phosphorylated on tyrosine 705; RAR= Retinoic Acid Receptor; RBPJ= Recombination signal binding protein for immunoglobulin kappa J region; shRNA= Short hairpin RNA; Sin3A= SIN3 transcription regulator family member A; Smad= Mothers against decapentaplegic homolog; SNP= Single nucleotide polymorphism; STAT= Signal transducer and activator of transcription; TAB2= TAK1 binding protein 2; TGF-β= Transforming growth factor beta; TNF-α= Tumor necrosis factor-α.
Acknowledgments
The authors thank John Svaren for his insightful comments on the manuscript.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Work in our laboratories has been funded by the National Institutes of Health (HD076892, NS110719, NS42803, NS39055, NS060120), the United Leukodystrophy Association, CLIMB, the Palamaro Fund, the Juanma Fund, the Jelte Rijkaart Fund, and Elise’s Corner.
ORCID iD: Albee Messing https://orcid.org/0000-0001-6049-0521
References
- Abidin S. Z., Fam S. Z., Chong C. E., Abdullah S., Cheah P. S., Nordin N., Ling K. H. (2019. ) miR-3099 promotes neurogenesis and inhibits astrogliogenesis during murine neural development. Gene, 697, 201–212. [DOI] [PubMed] [Google Scholar]
- Abidin S. Z., Leong J. W., Mahmoudi M., Nordin N., Abdullah S., Cheah P. S., Ling K. H. (2017) In silico prediction and validation of Gfap as an miR-3099 target in mouse brain. Neurosci Bull, 33, 373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal V., Bell G. W., Nam J. W., Bartel D. P. (2015) Predicting effective microRNA target sites in mammalian mRNAs. Elife, 4, 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano H., Aonuma M., Sanosaka T., Kohyama J., Namihira M., Nakashima K. (2009) Astrocyte differentiation of neural precursor cells is enhanced by retinoic acid through a change in epigenetic modification. Stem Cells, 27, 2744–2752. [DOI] [PubMed] [Google Scholar]
- Bachetti T., Di Zanni E., Lantieri F., Caroli F., Regis S., Filocamo M., Rainero I., Gallone S., Cilia R., Romano S., Savoiardo M., Pareyson D., Biancheri R., Ravazzolo R., Ceccherini I. (2010) A novel polymorphic AP-1 binding element of the GFAP promoter is associated with different allelic transcriptional activities. Ann Hum Genet, 74, 506–515. [DOI] [PubMed] [Google Scholar]
- Bae M. K., Kim S. R., Lee H. J., Wee H. J., Yoo M. A., Ock Oh S., Baek S. Y., Kim B. S., Kim J. B., Sik Y., Bae S. K. (2006) Aspirin-induced blockade of NF-kappaB activity restrains up-regulation of glial fibrillary acidic protein in human astroglial cells. Biochim Biophys Acta, 1763, 282–289. [DOI] [PubMed] [Google Scholar]
- Banks W. A., Robinson S. M. (2010). Minimal penetration of lipopolysaccharide across the murine blood-brain barrier. Brain Behav Immun, 24, 102–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard F., Brenner M., Nakatani Y., Chao R., Purohit H. J., Freese E. (1991) Multiple interacting sites regulate astrocyte-specific transcription of the human gene for glial fibrillary acidic protein. J. Biol Chem, 266, 18877–18883. [PubMed] [Google Scholar]
- Bonni A., Sun Y., Nadal-Vicens M., Bhatt A., Frank D. A., Rozovsky I., Stahl N., Yancopoulos G. D., Greenberg M. E., 1997. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science, 278, 477–483. [DOI] [PubMed] [Google Scholar]
- Brahmachari S., Fung Y. K., Pahan K. (2006) Induction of glial fibrillary acidic protein expression in astrocytes by nitric oxide. J Neurosci, 26, 4930–4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner M. (1994) Structure and transcriptional regulation of the GFAP gene. Brain Pathol, 4, 245–257. [DOI] [PubMed] [Google Scholar]
- Brenner M. (2014) Role of GFAP in CNS injuries. Neurosci Lett, 565, 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner M., Messing A. (1996) GFAP transgenic mice. Methods, 10, 351–364. [DOI] [PubMed] [Google Scholar]
- Brenner M., Kisseberth W. C., Su Y., Besnard F., Messing A. (1994) GFAP promoter directs astrocyte-specific expression in transgenic mice. J Neurosci, 14, 1030–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner M., Lampel K., Nakatani Y., Mill J., Banner C., Mearow K., Dohadwala M., Lipsky R., Freese E. (1990) Characterization of human cDNA and genomic clones for glial fibrillary acidic protein. Brain Res Mol Brain Res, 7, 277–286. [DOI] [PubMed] [Google Scholar]
- Brenner M., Messing A., Olsen M. L. (2019) AP-1 and the injury response of the GFAP gene. J Neurosci Res, 97, 149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun M., Coles J. E., Monckton E. A., Glubrecht D. D., Bisgrove D., Godbout R. (2009) Nuclear factor I regulates brain fatty acid-binding protein and glial fibrillary acidic protein gene expression in malignant glioma cell lines. J Mol Biol, 391, 282–300. [DOI] [PubMed] [Google Scholar]
- Bugiani M., Boor I., van Kollenburg B., Postma N., Polder E., van Berkel C., van Kesteren R. E., Windrem M. S., Hol E. M., Scheper G. C., Goldman S. A., van der Knaap M. S. (2011) Defective glial maturation in vanishing white matter disease. J Neuropathol Exp Neurol, 70, 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush T. G., Savidge T. C., Freeman T. C., Cox H. J., Campbell E. A., Mucke L., Johnson M. H., Sofroniew M. V. (1998) Fulminant jejuno-ileitis following ablation of enteric glia in adult transgenic mice. Cell, 93, 189–201. [DOI] [PubMed] [Google Scholar]
- Campbell C. E., Piper M., Plachez C., Yeh Y. T., Baizer J. S., Osinski J. M., Litwack E. D., Richards L. J., Gronostajski R. M. (2008) The transcription factor Nfix is essential for normal brain development. BMC Dev Biol, 8, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrión A. M., Mellström B., Naranjo J. R. (1998) Protein kinase A-dependent derepression of the human prodynorphin gene via differential binding to an intragenic silencer element. Mol Cell Biol, 18, 6921–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascante A., Klum S., Biswas M., Antolin-Fontes B., Barnabe-Heider F., Hermanson O. (2014) Gene-specific methylation control of H3K9 and H3K36 on neurotrophic BDNF versus astroglial GFAP genes by KDM4A/C regulates neural stem cell differentiation. J Mol Biol, 426, 3467–3477. [DOI] [PubMed] [Google Scholar]
- Cebolla B., Vallejo M., 2006. Nuclear factor-I regulates glial fibrillary acidic protein gene expression in astrocytes differentiated from cortical precursor cells. J Neurochem, 97, 1057–1070. [DOI] [PubMed] [Google Scholar]
- Cebolla B., Fernandez-Perez A., Perea G., Araque A., Vallejo M. (2008) DREAM mediates cAMP-dependent, Ca2+-induced stimulation of GFAP gene expression and regulates cortical astrogliogenesis. J Neurosci, 28, 6703–6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. W., Chen Y. X., Zhang X. R., Qian H., Chen W. Z., Xie W. F. (2008) Targeted inhibition of platelet-derived growth factor receptor-beta subunit in hepatic stellate cells ameliorates hepatic fibrosis in rats. Gene Ther, 15, 1424–1435. [DOI] [PubMed] [Google Scholar]
- Cheng P. Y., Lin Y. P., Chen Y. L., Lee Y. C., Tai C. C., Wang Y. T., Chen Y. J., Kao C. F., Yu J. (2011) Interplay between SIN3A and STAT3 mediates chromatin conformational changes and GFAP expression during cellular differentiation. PLoS One, 6, e22018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi I., Woo J. H., Jou I., Joe E. H. (2016) PINK1 deficiency decreases expression levels of mir-326, mir-330, and mir-3099 during brain development and neural stem cell differentiation. Exp Neurobiol, 25, 14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clairembault T., Kamphuis W., Leclair-Visonneau L., Rolli-Derkinderen M., Coron E., Neunlist M., Hol E. M., Derkinderen P. (2014) Enteric GFAP expression and phosphorylation in Parkinson's disease. J Neurochem, 130, 805–815. [DOI] [PubMed] [Google Scholar]
- Clarke L. E., Liddelow S. A., Chakraborty C., Munch A. E., Heiman M., Barres B. A. (2018) Normal aging induces A1-like astrocyte reactivity. Proc Natl Acad Sci USA, 115, E1896–E1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- das Neves L., Duchala C. S., Tolentino-Silva F., Haxhiu M. A., Colmenares C., Macklin W. B., Campbell C. E., Butz K. G., Gronostajski R. M. (1999) Disruption of the murine nuclear factor I-A gene (Nfia) results in perinatal lethality, hydrocephalus, and agenesis of the corpus callosum. Proc Natl Acad Sci USA, 96, 11946–11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Menezes M. F., Nicola F., Vital da Silva I. R., Vizuete A., Elsner V. R., Xavier L. L., Goncalves C. A. S., Netto C. A., Mestriner R. G. (2018) Glial fibrillary acidic protein levels are associated with global histone H4 acetylation after spinal cord injury in rats. Neural Regen Res, 13, 1945–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Maubach G., Masamune A., Zhuo L. (2009) Glial fibrillary acidic protein promoter targets pancreatic stellate cells. Dig Liver Dis, 41, 229–236. [DOI] [PubMed] [Google Scholar]
- Dore J. J., DeWitt J. C., Setty N., Donald M. D., Joo E., Chesarone M. A., Birren S. J. (2009) Multiple signaling pathways converge to regulate bone-morphogenetic-protein-dependent glial gene expression. Dev Neurosci, 31, 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driller K., Pagenstecher A., Uhl M., Omran H., Berlis A., Grunder A., Sippel A. E. (2007) Nuclear factor I X deficiency causes brain malformation and severe skeletal defects. Mol Cell Biol, 27, 3855–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G., Martinowich K., Chin M. H., He F., Fouse S. D., Hutnick L., Hattori D., Ge W., Shen Y., Wu H., ten Hoeve J., Shuai K., Sun Y. E. (2005) DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development, 132, 3345–3356. [DOI] [PubMed] [Google Scholar]
- Feinstein D. L., Weinmaster G. A., Milner R. J. (1992) Isolation of cDNA clones encoding rat glial fibrillary acidic protein: Expression in astrocytes and in Schwann cells. J Neurosci Res, 32, 1–14. [DOI] [PubMed] [Google Scholar]
- Gadea A., Schinelli S., Gallo V. (2008) Endothelin-1 regulates astrocyte proliferation and reactive gliosis via a JNK/c-Jun signaling pathway. J Neurosci, 28, 2394–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galarneau A., Richard S. (2005) Target RNA motif and target mRNAs of the Quaking STAR protein. Nat Struct Mol Biol, 12, 691–698. [DOI] [PubMed] [Google Scholar]
- Galea E., Dupouey P., Feinstein D. L. (1995) Glial fibrillary acidic protein mRNA isotypes: Expression in vitro and in vivo. J Neurosci Res, 41, 452–461. [DOI] [PubMed] [Google Scholar]
- Gao K., Wang C. R., Jiang F., Wong A. Y., Su N., Jiang J. H., Chai R. C., Vatcher G., Teng J., Chen J., Jiang Y. W., Yu A. C. (2013) Traumatic scratch injury in astrocytes triggers calcium influx to activate the JNK/c-Jun/AP-1 pathway and switch on GFAP expression. Glia, 61, 2063–2077. [DOI] [PubMed] [Google Scholar]
- Ge W., Martinowich K., Wu X., He F., Miyamoto A., Fan G., Weinmaster G., Sun Y. E. (2002) Notch signaling promotes astrogliogenesis via direct CSL-mediated glial gene activation. J Neurosci Res, 69, 848–860. [DOI] [PubMed] [Google Scholar]
- Gebert L. F. R., MacRae I. J. (2019) Regulation of microRNA function in animals. Nat Rev Mol Cell Biol, 20, 21–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalan S. M., Wilczynska K. M., Konik B. S., Bryan L., Kordula T. (2006. a) Nuclear factor-1-X regulates astrocyte-specific expression of the alpha1-antichymotrypsin and glial fibrillary acidic protein genes. J Biol Chem, 281, 13126–13133. [DOI] [PubMed] [Google Scholar]
- Gopalan S. M., Wilczynska K. M., Konik B. S., Bryan L., Kordula T. (2006. b) Astrocyte-specific expression of the {alpha}1-antichymotrypsin and glial fibrillary acidic protein genes requires activator protein-1. J Biol Chem, 281, 1956–1963. [DOI] [PubMed] [Google Scholar]
- Goss J. R., Finch C. E., Morgan D. G., 1991. Age-related changes in glial fibrillary acidic protein mRNA in the mouse brain. Neurobiol Aging, 12, 165–170. [DOI] [PubMed] [Google Scholar]
- Hagemann T. L., Powers B., Mazur C., Kim A., Wheeler S., Hung G., Swayze E., Messing A. (2018) Antisense suppression of glial fibrillary acidic protein as a treatment for Alexander disease. Ann Neurol, 83, 27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen L. A., Armstrong D. M., Terry R. D. (1987) An immunohistochemical quantification of fibrous astrocytes in the aging human cerebral cortex. Neurobiol Aging, 8, 1–6. [DOI] [PubMed] [Google Scholar]
- Hardy R. J., Loushin C. L., Friedrich V. L., Jr., Chen Q., Ebersole T. A., Lazzarini R. A., Artzt K. (1996) Neural cell type-specific expression of QKI proteins is altered in quaking viable mutant mice. J Neurosci, 16, 7941–7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Samee M. A., Blatti C., Sinha S. (2010) Thermodynamics-based models of transcriptional regulation by enhancers: the roles of synergistic activation, cooperative binding and short-range repression. PLoS Comput Biol, 6, e1000935 10.1371/journal.pcbi.1000935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanson O., Jepsen K., Rosenfeld M. G. (2002) N-CoR controls differentiation of neural stem cells into astrocytes. Nature, 419, 934–939. [DOI] [PubMed] [Google Scholar]
- Herrmann J. E., Imura T., Song B., Qi J., Ao Y., Nguyen T. K., Korsak R. A., Takeda K., Akira S., Sofroniew M. V. (2008) STAT3 is a critical regulator of astrogliosis and scar formation after spinal cord injury. J Neurosci, 28, 7231–7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst C. B. Brochner C. B. Vitting-Seerup K., andMollgard K. (2019) Astrogliogenesis in human fetal brain: complex spatiotemporal immunoreactivity patterns of GFAP, S100, AQP4 and YKL-40. J Anat, 235, 590–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda M., Nakashima K., Katada S. (2017) PRMT1 regulates astrocytic differentiation of embryonic neural stem/precursor cells. J Neurochem, 142, 901–907. [DOI] [PubMed] [Google Scholar]
- Hong S., Song M. R. (2014) STAT3 but not STAT1 is required for astrocyte differentiation. PLoS One, 9, e86851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung C. Y., Hsu T. I., Chuang J. Y., Su T. P., Chang W. C., Hung J. J. (2020) Sp1 in astrocyte is important for neurite outgrowth and synaptogenesis. Mol Neurobiol, 57, 261–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Noguchi A., Uosaki Y., Taga T., Arakawa H., Takizawa T. (2018) Gfap and Osmr regulation by BRG1 and STAT3 via interchromosomal gene clustering in astrocytes. Mol Biol Cell, 29, 209–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jany P. L., Hagemann T. L., Messing A. (2013) GFAP expression as an indicator of disease severity in mouse models of Alexander disease. ASN Neuro, 5, art:e00109 10.1042/AN20130003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W. B., Ruppe M. D., Rockenstein E. M., Price J., Sarthy V. P., Verderber L. C., Mucke L. (1995) Indicator expression directed by regulatory sequences of the glial fibrillary acidic protein (GFAP) gene: In vivo comparison of distinct GFAP-lacZ transgenes. Glia, 13, 174–184. [DOI] [PubMed] [Google Scholar]
- Kanski R., Sneeboer M. A., van Bodegraven E. J., Sluijs J. A., Kropff W., Vermunt M. W., Creyghton M. P., De Filippis L., Vescovi A., Aronica E., van Tijn P., van Strien M. E., Hol E. M. (2014) Histone acetylation in astrocytes suppresses GFAP and stimulates a reorganization of the intermediate filament network. J Cell Sci, 127, 4368–4380. [DOI] [PubMed] [Google Scholar]
- Kao H. Y., Ordentlich P., Koyano-Nakagawa N., Tang Z., Downes M., Kintner C. R., Evans R. M., Kadesch T. (1998) A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev, 12, 2269–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., Fornes O., Stigliani A., Gheorghe M., Castro-Mondragon J. A., van der Lee R., Bessy A., Cheneby J., Kulkarni S. R., Tan G., Baranasic D., Arenillas D. J., Sandelin A., Vandepoele K., Lenhard B., Ballester B., Wasserman W. W., Parcy F., Mathelier A. (2018) JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res, 46, D260–D266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. R., Bae M. K., Kim J. Y., Wee H. J., Yoo M. A., Bae S. K. (2009) Aspirin induces apoptosis through the blockade of IL-6-STAT3 signaling pathway in human glioblastoma A172 cells. Biochem Biophys Res Commun, 387, 342–347. [DOI] [PubMed] [Google Scholar]
- Kioussi C., Gross M. K., Gruss P. (1995) Pax3: a paired domain gene as a regulator in PNS myelination. Neuron, 15, 553–562. [DOI] [PubMed] [Google Scholar]
- Kirsch M., Trautmann N., Ernst M., Hofmann H. D. (2010) Involvement of gp130-associated cytokine signaling in Muller cell activation following optic nerve lesion. Glia, 58, 768–779. [DOI] [PubMed] [Google Scholar]
- Kohama S. G., Goss J. R., Finch C. E., McNeill T. H. (1995) Increases of glial fibrillary acidic protein in the aging female mouse brain. Neurobiol Aging, 16, 59–67. [DOI] [PubMed] [Google Scholar]
- Kong S. Y., Kim W., Lee H. R., Kim H. J. (2018) The histone demethylase KDM5A is required for the repression of astrocytogenesis and regulated by the translational machinery in neural progenitor cells. FASEB J, 32, 1108–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krohn K., Rozovsky I., Wals P., Teter B., Anderson C. P., Finch C. E. (1999) Glial fibrillary acidic protein transcription responses to transforming growth factor-beta1 and interleukin-1beta are mediated by a nuclear factor-1-like site in the near-upstream promoter. J Neurochem, 72, 1353–1361. [DOI] [PubMed] [Google Scholar]
- Kushida, N.Kabuyama, Y., Yamaguchi, O., & Homma, Y. (2001). Essential role for extracellular Ca(2+) in JNK activation by mechanical stretch in bladder smooth muscle cells. Am J Physiol Cell Physiol, 281, C1165–C1172. [DOI] [PubMed] [Google Scholar]
- Lee Y., Messing A., Su M., Brenner M. (2008) GFAP promoter elements required for region-specific and astrocyte-specific expression. Glia, 56, 481–493. [DOI] [PubMed] [Google Scholar]
- Lee Y., Su M., Messing A., Brenner M. (2006) Astrocyte heterogeneity revealed by expression of a GFAP-LacZ transgene. Glia, 53, 677–687. [DOI] [PubMed] [Google Scholar]
- Li D., Liu X., Liu T., Liu H., Tong L., Jia S., Wang Y. F. (2020) Neurochemical regulation of the expression and function of glial fibrillary acidic protein in astrocytes. Glia, 68, 878–897. [DOI] [PubMed] [Google Scholar]
- Li J., Ahn J. H., Wang G. G. (2019) Understanding histone H3 lysine 36 methylation and its deregulation in disease. Cell Mol Life Sci, 76, 2899–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H. W., Basu A., Druckman C., Cicchese M., Krady J. K., Levison S. W. (2006) Astrogliosis is delayed in type 1 interleukin-1 receptor-null mice following a penetrating brain injury. J Neuroinflammation, 3, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling P. D., Rawlins D. R., Hayward S. D. (1993) The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc Natl Acad Sci USA, 90, 9237–9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhu H., Liu M., Du J., Qian Y., Wang Y., Ding F., Gu X. (2011) Downregulation of Pax3 expression correlates with acquired GFAP expression during NSC differentiation towards astrocytes. FEBS Lett, 585, 1014–1020. [DOI] [PubMed] [Google Scholar]
- Masood K., Besnard F., Su Y., Brenner M. (1993) Analysis of a segment of the human glial fibrillary acidic protein gene that directs astrocyte-specific transcription. J Neurochem, 61, 160–166. [DOI] [PubMed] [Google Scholar]
- Matuzelski E., Bunt J., Harkins D., Lim J. W. C., Gronostajski R. M., Richards L. J., Harris L., Piper M. (2017) Transcriptional regulation of Nfix by NFIB drives astrocytic maturation within the developing spinal cord. Dev Biol, 432, 286–297. [DOI] [PubMed] [Google Scholar]
- Messing A., Brenner M. (2020) GFAP at 50, ASN Neuro, 12 10.1177/1759091420949680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing A., Brenner M., Feany M. B., Nedergaard M., Goldman J. E. (2012) Alexander disease. J Neurosci, 32, 5017–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minocha S., Valloton D., Arsenijevic Y., Cardinaux J. R., Guidi R., Hornung J. P., Lebrand C. (2017) Nkx2.1 regulates the generation of telencephalic astrocytes during embryonic development. Sci Rep, 7, 43093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokuno K., Kamholz J., Behrman T., Black C., Sessa M., Feinstein D., Lee V., Pleasure D. (1989) Neuronal modulation of Schwann cell glial fibrillary acidic protein (GFAP). J Neurosci Res, 23, 396–405. [DOI] [PubMed] [Google Scholar]
- Morgan T. E., Xie Z., Goldsmith S., Yoshida T., Lanzrein A. S., Stone D., Rozovsky I., Perry G., Smith M. A., Finch C. E., 1999. The mosaic of brain glial hyperactivity during normal ageing and its attenuation by food restriction. Neuroscience, 89, 687–699. [DOI] [PubMed] [Google Scholar]
- Mucke L., Oldstone M. B., Morris J. C., Nerenberg M. I. (1991) Rapid activation of astrocyte-specific expression of GFAP-lacZ transgene by focal injury. New Biol, 3, 465–474. [PubMed] [Google Scholar]
- Nakashima K., Yanagisawa M., Arakawa H., Kimura N., Hisatsune T., Kawabata M., Miyazono K., Taga T. (1999) Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science, 284, 479–482. [DOI] [PubMed] [Google Scholar]
- Namihira M., Kohyama J., Semi K., Sanosaka T., Deneen B., Taga T., Nakashima K. (2009) Committed neuronal precursors confer astrocytic potential on residual neural precursor cells. Dev Cell, 16, 245–255. 10.1016/j.devcel.2008.12.014 [DOI] [PubMed] [Google Scholar]
- Nichols N. R., Day J. R., Laping N. J., Johnson S. A., Finch C. E., (1993). GFAP mRNA increases with age in rat and human brain. Neurobiol Aging, 14, 421–429. [DOI] [PubMed] [Google Scholar]
- Nicole O., Goldshmidt A., Hamill C. E., Sorensen S. D., Sastre A., Lyuboslavsky P., Hepler J. R., McKeon R. J., Traynelis S. F. (2005) Activation of protease-activated receptor-1 triggers astrogliosis after brain injury. J Neurosci, 25, 4319–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura Y. (2001) NF-kappaB activation and IkappaB alpha dynamism involved in iNOS and chemokine induction in astroglial cells. Life Sci, 68, 1695–1701. [DOI] [PubMed] [Google Scholar]
- Oberheim N. A., Takano T., Han X., He W., Lin J. H., Wang F., Xu Q., Wyatt J. D., Pilcher W., Ojemann J. G., Ransom B. R., Goldman S. A., Nedergaard M. (2009) Uniquely hominid features of adult human astrocytes. J Neurosci, 29, 3276–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Callaghan J. P., Kelly K. A., VanGilder R. L., Sofroniew M. V., Miller D. B. (2014) Early activation of STAT3 regulates reactive astrogliosis induced by diverse forms of neurotoxicity. PLoS One, 9, e102003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer A. L., Ousman S. S. (2018) Astrocytes and aging. Front Aging Neurosci, 10, 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Kim G. W., Kwon S. H., Lee J. S. (2020) Broad domains of histone H3 lysine 4 trimethylation in transcriptional regulation and disease. FEBS J. 10.1111/febs.15219. [DOI] [PubMed] [Google Scholar]
- Pignataro D., Sucunza D., Vanrell L., Lopez-Franco E., Dopeso-Reyes I. G., Vales A., Hommel M., Rico A. J., Lanciego J. L., Gonzalez-Aseguinolaza G. (2017) Adeno-associated viral vectors serotype 8 for cell-specific delivery of therapeutic genes in the central nervous system. Front Neuroanat, 11, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M., Harris L., Barry G., Heng Y. H., Plachez C., Gronostajski R. M., Richards L. J., 2011. Nuclear factor one X regulates the development of multiple cellular populations in the postnatal cerebellum. J Comp Neurol, 519, 3532–3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puche J. E., Lee Y. A., Jiao J., Aloman C., Fiel M. I., Munoz U., Kraus T., Lee T., Yee H. F., Jr., Friedman S. L. (2013) A novel murine model to deplete hepatic stellate cells uncovers their role in amplifying liver damage in mice. Hepatology, 57, 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radomska K. J., Halvardson J., Reinius B., Lindholm Carlstrom E., Emilsson L., Feuk L., Jazin E. (2013) RNA-binding protein QKI regulates glial fibrillary acidic protein expression in human astrocytes. Hum Mol Genet, 22, 1373–1382. [DOI] [PubMed] [Google Scholar]
- Radulovic M., Yoon H., Wu J., Mustafa K., Fehlings M. G., Scarisbrick I. A. (2015) Genetic targeting of protease activated receptor 2 reduces inflammatory astrogliosis and improves recovery of function after spinal cord injury. Neurobiol Dis, 83, 75–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai A., Tripathi S., Kushwaha R., Singh P., Srivastava P., Sanyal S., Bandyopadhyay S. (2014) CDK5-induced p-PPARgamma(Ser 112) downregulates GFAP via PPREs in developing rat brain: effect of metal mixture and troglitazone in astrocytes. Cell Death Dis, 5, e1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raivich G., Bohatschek M., Da Costa C., Iwata O., Galiano M., Hristova M., Nateri A. S., Makwana M., Riera-Sans L., Wolfer D. P., Lipp H. P., Aguzzi A., Wagner E. F., Behrens A. (2004) The AP-1 transcription factor c-Jun is required for efficient axonal regeneration. Neuron, 43, 57–67. [DOI] [PubMed] [Google Scholar]
- Rajan P., McKay R. D. (1998) Multiple routes to astrocytic differentiation in the CNS. J Neurosci, 18, 3620–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastinejad F., Wagner T., Zhao Q., Khorasanizadeh S. (2000) Structure of the RXR-RAR DNA-binding complex on the retinoic acid response element DR1. EMBO J, 19, 1045–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio C., Dikkes P., Liberman M. C., Corfas G. (2002) Glial fibrillary acidic protein expression and promoter activity in the inner ear of developing and adult mice. J Comp Neurol, 442, 156–162. [DOI] [PubMed] [Google Scholar]
- Riol H., Fages C., Tardy M. (1992) Transcriptional regulation of glial fibrillary acidic protein (GFAP)-mRNA expression during postnatal development of mouse brain. J Neurosci Res, 32, 79–85. [DOI] [PubMed] [Google Scholar]
- Sahni V, Mukhopadhyay A, Tysseling V, Hebert A, Birch D, McGuire TL, Stupp SI, Kessler JA. (2010) BMPR1a and BMPR1b signaling exert opposing effects on gliosis after spinal cord injury. J Neurosci, 30, 1839–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardi S. P. Murtie J. Koirala S. Patten B. A., andCorfas G. (2006) Presenilin-dependent ErbB4 nuclear signaling regulates the timing of astrogenesis in the developing brain. Cell, 127, 185–197. [DOI] [PubMed] [Google Scholar]
- Sereika M., Urbanaviciute R., Tamasauskas A., Skiriute D., Vaitkiene P. (2018) GFAP expression is influenced by astrocytoma grade and rs2070935 polymorphism. J Cancer, 9, 4496–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara Y., Konno A., Takahashi N., Matsuzaki Y., Kishi S., Hirai H. (2016) Viral vector-based dissection of marmoset GFAP promoter in mouse and marmoset brains. PLoS One, 11, e0162023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S. K., Wilczynska K. M., Grzybowski A., Yester J., Osrah B., Bryan L., Wright S., Griswold-Prenner I., Kordula T. (2011) The unique transcriptional activation domain of nuclear factor-I-X3 is critical to specifically induce marker gene expression in astrocytes. J Biol Chem, 286, 7315–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M. R., Ghosh A. (2004) FGF2-induced chromatin remodeling regulates CNTF-mediated gene expression and astrocyte differentiation. Nat Neurosci, 7, 229–235. [DOI] [PubMed] [Google Scholar]
- Steele-Perkins G., Butz K. G., Lyons G. E., Zeichner-David M., Kim H. J., Cho M. I., Gronostajski R. M. (2003) Essential role for NFI-C/CTF transcription-replication factor in tooth root development. Mol Cell Biol, 23, 1075–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele-Perkins G. Plachez C. Butz K. G. Yang G. Bachurski C. J. Kinsman S. L. Litwack E. D. Richards L. J., andGronostajski R. M. (2005) The transcription factor gene Nfib is essential for both lung maturation and brain development. Mol Cell Biol, 25, 685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sticozzi C., Belmonte G., Frosini M., Pessina F. (2020) Nitric oxide/cyclic gmp-dependent calcium signalling mediates IL-6- and TNF-alpha-induced expression of glial fibrillary acid protein. J Mol Neurosci. 10.1007/s12031-020-01708-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M., Hu H., Lee Y., d'Azzo A., Messing A., Brenner M. (2004) Expression specificity of GFAP transgenes. Neurochem Res, 29, 2075–2093. [DOI] [PubMed] [Google Scholar]
- Sugiura S. Lahav R. Han J. Kou S.Y. Banner L.R. de Pablo F., andPatterson P.H. (2000). Leukaemia inhibitory factor is required for normal inflammatory responses to injury in the peripheral and central nervous systems in vivo and is chemotactic for macrophages in vitro. Eur J Neurosci, 12, 457–466. [DOI] [PubMed] [Google Scholar]
- Sun J. J., Liu Y., Ye Z. R. (2008) Effects of P2Y1 receptor on glial fibrillary acidic protein and glial cell line-derived neurotrophic factor production of astrocytes under ischemic condition and the related signaling pathways. Neurosci Bull, 24, 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Nadal-Vicens M., Misono S., Lin M. Z., Zubiaga A., Hua X., Fan G., Greenberg M. E. (2001) Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell, 104, 365–376. [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Takeuchi H., Sakai M., Yu Z., Kikuchi Y., Ito F., Matsuoka H., Tanabe O., Yasuda J., Taki Y., Kawashima R., Tomita H. (2020) A single nucleotide polymorphism (-250 A/C) of the GFAP gene is associated with brain structures and cerebral blood flow. Psychiatry Clin Neurosci, 74, 49–55. [DOI] [PubMed] [Google Scholar]
- Takizawa T., Nakashima K., Namihira M., Ochiai W., Uemura A., Yanagisawa M., Fujita N., Nakao M., Taga T. (2001. a) DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell, 1, 749–758. [DOI] [PubMed] [Google Scholar]
- Takizawa T., Yanagisawa M., Ochiai W., Yasukawa K., Ishiguro T., Nakashima K., Taga T. (2001. b) Directly linked soluble IL-6 receptor-IL-6 fusion protein induces astrocyte differentiation from neuroepithelial cells via activation of STAT3. Cytokine, 13, 272–279. [DOI] [PubMed] [Google Scholar]
- Teter B., Rozovsky I., Krohn K., Anderson C., Osterburg H., Finch C. (1996) Methylation of the glial fibrillary acidic protein gene shows novel biphasic changes during brain development. Glia, 17, 195–205. [DOI] [PubMed] [Google Scholar]
- Thomsen R., Daugaard T. F., Holm I. E., Nielsen A. L. (2013) Alternative mRNA splicing from the glial fibrillary acidic protein (GFAP) gene generates isoforms with distinct subcellular mRNA localization patterns in astrocytes. PLoS One, 8, e72110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tun T., Hamaguchi Y., Matsunami N., Furukawa T., Honjo T., Kawaichi M. (1994) Recognition sequence of a highly conserved DNA binding protein RBP-J kappa. Nucleic Acids Res, 22, 965–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urayama S., Semi K., Sanosaka T., Hori Y., Namihira M., Kohyama J., Takizawa T., Nakashima K. (2013) Chromatin accessibility at a STAT3 target site is altered prior to astrocyte differentiation. Cell Struct Funct, 38, 55–66. [DOI] [PubMed] [Google Scholar]
- Verderber L., Johnson W., Mucke L., Sarthy V. (1995) Differential regulation of a glial fibrillary acidic protein-LacZ transgene in retinal astrocytes and Muller cells. Invest Ophthalmol Vis Sci, 36, 1137–1143. [PubMed] [Google Scholar]
- Wang C. Y., Yang S. H., Tzeng S. F. (2015) MicroRNA-145 as one negative regulator of astrogliosis. Glia, 63, 194–205. [DOI] [PubMed] [Google Scholar]
- Wang J., Wang M. L., Wang C. H., Sun S. Y., Zhang H. B., Jiang Y. Y., Xu Q. W., Wang Y., Gu S. X. (2018) A novel functional polymorphism of GFAP decrease glioblastoma susceptibility through inhibiting the binding of miR-139. Aging (Albany NY ), 10, 988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Shu Q., Ni Y., Xu G. (2018) CRISPR-mediated SOX9 knockout inhibits GFAP expression in retinal glial (Muller) cells. Neuroreport, 29, 1504–1508. [DOI] [PubMed] [Google Scholar]
- Wei C., Ren L., Li K., Lu Z. (2018) The regulation of survival and differentiation of neural stem cells by miR-124 via modulating PAX3. Neurosci Lett, 683, 19–26. [DOI] [PubMed] [Google Scholar]
- Yang P., Shen W. B., Reece E. A., Chen X., Yang P. (2016) High glucose suppresses embryonic stem cell differentiation into neural lineage cells. Biochem Biophys Res Commun, 472, 306–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo S., Bandyopadhyay S., Messing A., Brenner M. (2013) Transgenic analysis of GFAP promoter elements. Glia, 61, 1488–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Mizuta I., Saito K., Ohara R., Kurisaki H., Ohnari K., Riku Y., Hayashi Y., Suzuki H., Shii H., Fujiwara Y., Yonezu T., Nagaishi A., Nakagawa M. (2013) Effects of a polymorphism in the GFAP promoter on the age of onset and ambulatory disability in late-onset Alexander disease. J Hum Genet, 58, 635–638. [DOI] [PubMed] [Google Scholar]
- Zhang, C., & , Zhuo, L. (2006). Epigallocatechin gallate and genistein attenuate glial fibrillary acidic protein elevation lnduced by fibrogenic cytokines in hepatic stellate cells. Int. J. Mol, 18, 1141–1151. [PubMed] [Google Scholar]