Abstract
Pseudorabies (PR), also called Aujeszky’s disease, is a highly infectious disease caused by pseudorabies virus (PRV). Without specific host tropism, PRV can infect a wide variety of mammals, including pig, sheep, cattle, etc., thereby causing severe clinical symptoms and acute death. PRV was firstly reported in China in 1950s, while outbreaks of emerging PRV variants have been documented in partial regions since 2011, leading to significant economic losses in swine industry. Although scientists have been devoting to the design of diagnostic approaches and the development of vaccines during the past years, PR remains a vital infectious disease widely prevalent in Chinese pig industry. Especially, its potential threat to human health has also attracted the worldwide attention. In this review, we will provide a summary of current understanding of PRV in China, mainly focusing on PRV history, the existing diagnosis methods, PRV prevalence in pig population and other susceptible mammals, molecular characteristics, and the available vaccines against its infection. Additionally, promising agents including traditional Chinese herbal medicines and novel inhibitors that may be employed to treat this viral infection, are also discussed.
Keywords: Pseudorabies virus (PRV), Diagnosis methods, Prevalence, Multiple-species infection, Prevention and control measures
Introduction
The great disruption of viral infection, such as currently circulating novel coronavirus to the health of human beings, African swine fever virus (ASFV) to the benign development of swine industry, is being witnessed, thereby appealing for more attention to the virus induced diseases. Since the first document of pseudorabies virus (PRV) in Hungary in 1902 (Lee and Wilson 1979), this pathogen has been regarded as one of the most significant causative agents, leading to fatal losses to pig industry worldwide (Xia et al. 2018). PRV, also called suid herpesvirus (SuHV-1) or Aujeszky’s disease virus (ADV), is an enveloped and double-stranded linear DNA virus which belongs to the subfamily Alphaherpesvirinae, within the family Herpesviridae (Sun et al. 2016). PRV can infect a wide variety of mammals, including pigs, wild boars (Minamiguchi et al. 2019), bears (Zanin et al. 1997), ruminants (e.g. sheep, goat, and cattle) (Kong et al. 2013; Cheng et al. 2019), carnivores (e.g. minks and foxes) (Jin et al. 2016), and rodents, etc. In particular, the latest researches revealed that human beings might also be another potential host of this pathogen (Ai et al. 2018; Liu Q et al. 2020). Although PRV has a wide range of hosts, pigs (including other wild boars) are the unique natural host and reservoir (Freuling et al. 2017). The clinical symptoms of PRV-infected pigs are characterized by diarrhea, vomiting, nervous system disorders (such as tremor, dyskinesia, and lethargy), with high morbidity and mortality for sucking piglets less than two weeks, dyspnea and slow growth for fattening pigs, reproductive disorder for breeding pigs (Sun et al. 2016).
The first identification of pseudorabies (PR) occurred in cat was reported in 1947 in China (Zhou and Sun 1957), thereafter, PRV infection in diverse species has been extensively documented. Given its significant impact on Chinese swine industry (Sun et al. 2016), China has accordingly made a lot of efforts to investigate the epidemiology and molecular biology, and develop the detection methods and commercialized vaccines for the control and prevention of PRV infection. Though significant progress in this field has been made, the threat posed by PRV does not vanish, mainly due to numerous factors such as huge pig herds with distinct management levels and various breeding conditions, the occurrence of PRV variants, and voluntary policy of vaccination. In the present review, an update understanding of PRV in China, with an emphasis on the diagnosis methods, prevalence, clinical conditions of PR in different hosts, molecular characteristics, and the prevention of this disease, is presented. The potential threat of PRV infection to public health is highlighted, meanwhile, the effective agents against PRV infection is also described.
History of Pseudorabies in China
In 1947, the first case of PRV infection in a domestic cat was reported in eastern China (Zhou and Sun 1957), and those in pigs, cattle, goats, and other hosts were subsequently documented. Owing to the lack of accurate detection technology and poor biosafety measures in pig industry, PR had been widely prevalent in most regions of China in the early 1980s. With the frequent communication among pig raising markets and the increasing intensity of pig herds in China, the outbreaks of PRV infection with higher morbidity and mortality had frequently occurred in the following years. Fortunately, the prevalence of PR in China has been largely controlled, mainly via the wide application of the commercial Bartha-K61 vaccine (a live-attenuated vaccine based on the PRV Bartha-K61 strain) which was introduced from Hungary. However, it seems that the battle to control PR has not came to the end, since this infectious disease currently remains the major disaster influencing the development of the swine industry in China (An et al. 2013; Sun et al. 2016).
Since 2011, the outbreaks of PR have frequently been reported in many pig farms with a fatality rate up to 50% in infected newborn piglets in China, though these pig herds had been immunized with Bartha-K61 vaccine (An et al. 2013; Tong et al. 2015; Wang et al. 2014). Further molecular biological analysis confirmed that the pathogenic agents were PRV variants which displayed distinct genetic variations compared to the classical PRV strains. Moreover, growing evidence indicated that Bartha-K61 vaccine could not confer complete protection for piglets against these new variants (An et al. 2013; Luo et al. 2014; Tong et al. 2015). More recently, this pathogen has received increasing attention since the first case of human endophthalmitis caused by PRV infection was reported in 2017 (Ai et al. 2018).
Development of Diagnostic Methods
Traditional clinical and pathological conditions based diagnostic approaches fail to accurately diagnose PR. In recent years, numerous diagnostic approaches for PRV detection have been invented and applied. They can be divided into two categories: serological technologies detecting PRV specific antibodies and molecular biology methods probing PRV nucleic acids. Several representative PRV detecting approaches of these two types are listed in Table 1 and briefly introduced as below.
Table 1.
Various diagnostic approaches developed in China.
| Target | Methods | Sensitivity and specificity | Reference |
|---|---|---|---|
| PRV antibody |
Blocking ELISA: targeting to the gB antibody |
High sensitivity (80.9%) and specificity (96.4%) compared with the commercial ELISA kit (IDEXX) | Sun HF et al. (2018) |
|
Indirect ELISA: targeting to the gE antibody |
Total 89.1% positive coincidence rate compared with the commercial ELISA kit (IDEXX) (allowing DIVA) | Zheng et al. (2017) | |
|
Indirect ELISA: targeting to the gE antibody |
High 88.0% sensitivity and 91.5% specificity compared with the commercial ELISA kit (IDEXX) (allowing DIVA) | Kou et al. (2018) | |
|
Indirect ELISA: targeting to the gB antibody |
Highly total positive coincidence rate (97.8%) compared with the commercial ELISA kit. and the lowest detection limit was 1: 128 dilution of the positive serum | Liu MM et al. (2019) | |
|
ICA: targeting to the gE antibody |
The lowest detection limit was 1:1280 dilution of the positive serum, and 95.3% positive coincidence rate compared with the commercial ELISA kit (IDEXX) (allowing DIVA) | Lei and Zhang (2016) | |
|
DFM: 2 targeting to the gE antibody |
Total 74.7% positive coincidence rate compared with the conventional PCR, while which was more sensitive than the later one (allowing DIVA) | Xu LH et al. (2017) | |
|
Liquid chip technology: targeting to the gE antibody |
This method could be applied to detect both PRV and PRRSV antibodies with higher sensitivity than the commercial ELISA kits (allowing DIVA) | Xiao et al. (2018) | |
|
IFAT: targeting to the gE antibody |
High sensitivity (93.8%) and specificity (91.7%) compared with the commercial ELISA kit (IDEXX) (allowing DIVA) | Zhu et al. (2019) | |
| PRV antigen |
LFA: targeting to the gB antigen |
Detection limit of inactivated PRV antigens were lower than 1 × 106.6 TCID50/0.1 mL with 86.7% positive coincidence rate compared with the conventional PCR | Wang TH et al. (2018) |
| PRV DNA |
NanoPCR: targeting to the gB, gE, and gG genes |
This test could be used for the differentiation of wild PRV and gene-deleted vaccine strains with higher sensitivity than the conventional PCR (allowing DIVA) | Ma et al. (2013) |
|
DdPCR: targeting to the gE gene |
Higher sensitivity (6.1 copies/μL) than qPCR with high specify (96.2%) compared with viral isolation (allowing DIVA) | Chen et al. (2017) | |
|
LAMP: targeting to the gB gene |
Showed 100 times higher sensitivity than conventional PCR with the detection limit of 1 fg/μL | Xu ZL et al. (2017) | |
|
PCR combined with nucleic acid probe spot hybridization: targeting to the gE gene |
The sensitivity was 100 times higher than conventional PCR, being 10 pg/μL and 1 ng/ul, respectively (allowing DIVA) | Wunaerhan et al. (2017) | |
|
PCR: targeting to the gE, gB and TK genes |
This approach could be used to distinguish between PRV wild virus, SA215 and Bartha-K61 vaccine strains with detection limit of 1.8 × 106 copies/μL (allowing DIVA) | Jiang et al. (2018) | |
|
RPA: targeting to the gE and gB genes |
Both the sensitivities of gE and gB gene were 100 copies/μL, this approach could be used to distinguish PRV wild and attenuated virus with 100% specify compared with qPCR (allowing DIVA) | Liu LB et al. (2018) | |
|
TaqMan qPCR: targeting to the gE and gD genes |
The detection limits of gE and gB genes were 12.1 and 39.4 copies, respectively, which was more sensitive than qPCR and conventional PCR (allowing DIVA) | Lan et al. (2018) | |
|
QPCR: targeting to the gB gene |
The detection limit was lower than 1000 copies/μL, showing higher sensitivity than conventional PCR | Hua et al. (2019) | |
|
QPCR: targeting to the gE gene |
The detection limit was 10 copies/μl with high specificity (100%) compared with conventional PCR and commercial kits | Wen et al. (2019) | |
|
Multiple RT-PCR: targeting to the gE gene |
The approach could detect the nucleic acids of PRV, PCV2, PPV, PPRSV, JEV, and CSFV with the detection limit pf 10–3 ng/μL for PRV (allowing DIVA) | Li et al. (2019) | |
|
QIAxcel CGE: targeting to the gE gene |
This method could be applied for the detection of PRV, CSFV, JEV, PCV2, PRRSV, PPV and ASFV, with the detection limit of 4.53 × 103 copies/μL for PRV (allowing DIVA) | Wu et al. (2019) | |
|
Multiple PCR: targeting to the gE gene |
This method could be applied for the detection of PRV, PCV2 and PPV, with the detection limit of 72 pg/μL for PRV (allowing DIVA) | Xin et al. (2019) | |
| NGS | This approach could be used for detecting undetermined pathogens with high sensitivity | Ai et al. (2018) |
DIVA: Distinction between the infected and vaccinated animals; ICA: Gold immunochromatographic assay; DFM: Direct immunofluorescence method; IFAT: Indirect immunofluorescent antibody detection method; LFA: Lateral flow assay; NanoPCR: Nanoparticle-assisted PCR assay; DdPCR: Droplet digital PCR assay; LAMP: Loop-mediated isothermal amplification; RPA: Recombinase polymerase amplification; QIAxcel CGE: QIAxcel capillary gel electrophoresis; NGS: Next generation sequencing.
Serological Approaches for the Detection of PRV Infection
Owing to the extensive application of PRV glycoprotein E (gE)-deleted vaccines in China, a lot of PRV-gE antibody based serological tests have been developed for rapid and effective differentiation of infected and vaccinated animals (DIVA), while gB antibody based serological approaches have been designed for evaluating the immunological levels induced by vaccine immunization. Various serological methods including enzyme-linked immunosorbent assays (ELISA) (Zheng et al. 2017; Kou et al. 2018; Sun HF et al. 2018; Sun et al. 2018b; Liu MM et al. 2019), serum neutralization test (SNT), and direct-immunofluorescence method (DFM) (Xu LH et al. 2017) have been also applied in recent years. However, the most widely applied approach is the ELISA based one, with higher sensitive and more simple properties relative to others. Among these distinct ELISA tests, competitive ELISAs (cELISA) targeting to the gB or gE antibody are the most widely used in China (Hu D et al. 2015; Liu Y et al. 2018; Wu et al. 2018; Xia et al. 2018).
Besides cELISAs, other types of serological approaches have also been developed for reducing cost and increasing sensitivity and specificity (shown in Table 1). Notably, no unitary standard has been established for comparing the features between these various formats of serological approaches. Therefore, the sensitivity and specificity of the newly designed ELISA tests including blocking ELISA (Sun HF et al. 2018), indirect ELISA (Zheng et al. 2017; Kou et al. 2018; Liu MM et al. 2019), indirect fluorescence antibody test (Zhu et al. 2019), are usually compared to these of commercial cELISA kits. Fantastically, some of them, such as immunochromatogragphic assay (Lei and Zhang 2016) and liquid chip technology (Xiao et al. 2018), display higher sensitivity than commercial cELISA kits. In particular, a novel serological approach based on blocking fluorescent lateral flow immunoassay exhibits advantages of less time and high sensitivity in differently diagnosing wild PRV-infected and vaccinated pigs relative to a commercial gE-ELISA kit (Chen H et al. 2020). In addition, one serological test targeting to the inactivated PRV gB antigen presents relatively high positive coincidence rate (86.67%) with the conventional PCR, while it is not universal thus far (Wang JH et al. 2018).
Molecular Biology Approaches for the Detection of PRV Infection
To further improve the sensitivity and specificity for PRV detection, increasing molecular biology approaches targeting to the specific sequences of PRV genes (e.g. gE, gC, gD, gB, and gG) have been created, such as polymerase chain reaction (PCR), real-time PCR (RT-PCR), nano PCR, loop-mediated isothermal amplification (LAMP) (Table 1). In general, PCR and RT-PCR approaches are the most widely applied ones for rapidly detecting the existence of PRV or distinguishing between the PRV wild type and gE-gene deleted strains in China (Jiang et al. 2018; Ren et al. 2018). Especially, multiplex PCR assays have also been developed for synchronous detection of PRV and other pathogens (Li et al. 2019). Among other approaches, next generation sequencing (NGS) is the most powerful and supersensitive method for detecting the presence of PRV in humans (Ai et al. 2018; Yang H et al. 2019), while this approach might be not suitable for large-scale clinical detection due to its high cost. Relative to conventional PCR, the features of higher sensitivity and no requirement for expensive facilities of LAMP approach make it more suitable for rapid and on-site PRV detection in the field (Zhang et al. 2010), while the easily contaminated property of it remains to be overcome in the future.
Apart from the approaches described above, direct pathogen detection is another effective method, which also facilitates vaccine design for the prevention of PR greatly. This method mainly consists of viral isolation and subsequent laboratory diagnosis involving serological, molecular biology, and electron microscopy technologies. Viral isolation is regarded as “gold standard” in pathogen diagnosis, and a mass of PRV strains from different hosts have successfully been isolated in China via this way (Liu et al. 2017; Cheng et al. 2019; Lian et al. 2020), whereas this approach is only suitable for professional laboratories.
Current Prevalence of Pseudorabies in China
Current Prevalence of Pseudorabies Virus in Pigs
In order to better control this infectious disease, the serological and molecular epidemiology of PRV infection in Chinese pig population have been extensively performed in recent years, while its prevalent status is affected by numerous factors including regions, sample sizes and seasons, etc.
Sero-prevalence of Pseudorabies Virus in Pigs
A series of sero-prevalence approaches, mainly targeting the gE antibody, have been developed to comprehensively assess wild PRV prevalence in pigs and explore potentially affecting elements in China. To provide a comprehensive and update view on these, we collect a total of 108 representative studies investigating PRV sero-prevalence in pigs across distinct regions covering 29 provinces of China, which are available in Chinese and English databases from 2011 to the present. Thus, 256,326 serum samples are included and analyzed in this review. Among them, 76,553 specimens manifested positive for PRV gE antibody, yielding the average 29.87% positive rate of PRV infection in pigs, as supported by two latest reports (Chen C et al. 2020; Ning et al. 2020). The sero-prevalence rate of PR in the three regions (Jiangsu, Henan, and Jiangxi) from 2018 to 2019 was over 35% (1398/3675) (Chen C et al. 2020), while that for 24 provinces from 2016 to 2018 was 34.2% (32083/93912) (Ning et al. 2020). These demonstrate that in spite of the improved feeding and management levels and widely applied effective vaccines, the prevalence of wild PRV in pig population is still severe in recent years, indicating the continuous threat of PR to the pig industry in China.
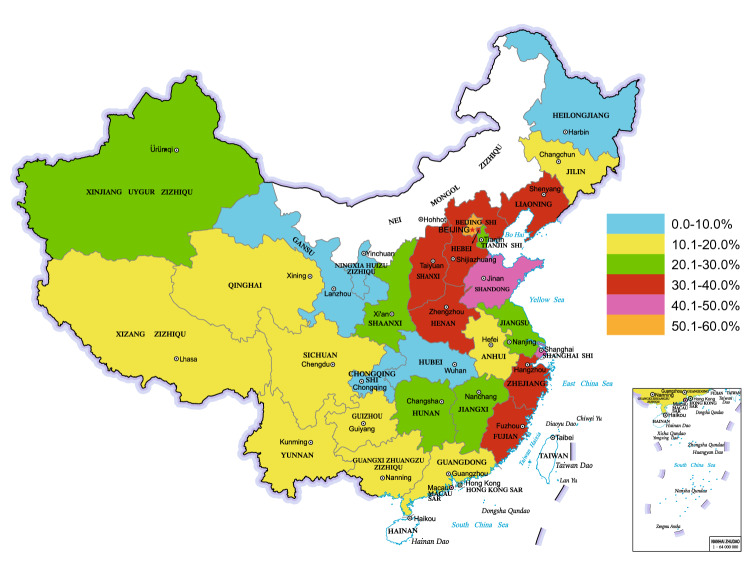
The sero-prevalence results of wild PRV infection in pigs from different regions of China are summarized and shown in Fig. 1, which shows an over 30% PRV-gE antibody positive rate in 9 provinces/regions (31.03%, 9/29) (red, purple and orange marked). Furthermore, the sero-prevalence in northern China, eastern China, and central and southern China are higher than those in northeastern China, northwestern China, and southwestern China. Several risk factors including feeding patterns, seasons, and categories of pigs, are considerably associated with the occurrence of PR in pig population. For example, the sero-prevalence of PR in free-range or small pig farms is much higher than that in middle or extensive pig farms. Additionally, other factors, like times of vaccine immunization, altitudes of pig farms, also have significant impact (Wu et al. 2018), while sex and variety of pigs exhibit less influence (Wu et al. 2018).
Fig. 1.
Current sero-prevalence of pseudorabies virus in pigs. Results of 108 representative studies investigating PRV sero-prevalence in pigs across distinct regions covering 29 provinces of China. These data collected from Chinese and English databases from 2011 to the present were analyzed. Data of the regions marked with white were not available.
Epidemiology of Pseudorabies in Pigs
Recently, PCR and RT-PCR methods have been widely employed to investigate the PRV epidemiology of pigs in China (Sun Y et al. 2018; Ma et al. 2020). A total of 27 representative articles available in Chinese or English database from 2011 to the present are chosen and summarized. The analytic results showed that the overall nucleic acid positive rate of PRV in pigs reached 11.5% (4742/41237), as also supported by a recent study, which indicated a nearly 8.0% of PRV-positive rate among 16,256 tissue specimens collected from 27 provinces in China. Furthermore, the average rate in eastern and central China was over 10.0%, while that of northwestern China was below 7.0%, suggesting that the occurrence of this disease is markedly related to regions (Sun Y et al. 2018). Similarly, another recent study focusing on the PRV prevalence in the years of 2015, 2017, and 2018 in Shandong Province revealed an average positive rate of 11.8% (147/1243) (Ma et al. 2020), as consistent with ours. Notably, the general epidemiological trend of PRV infection is similar to that of the sero-prevalence in different regions (Sun Y et al. 2018). Additionally, pigs showing nervous symptoms or reproductive failures are at high risk of PRV infection, although these consequences may be caused by the infection of various pathogens.
Pseudorabies in Other Animals
In China, a variety of animals (including some wild species) are raised for satisfying the demands of consumers, while some of these animal species are susceptible to PRV. Accordingly, growing reports of PR in different mammals (apart from pigs) have been documented in recent years. For example, the outbreak of PRV infection has caused the death of nearly 8000 minks in Shandong Province, China (Liu et al. 2017). Similarly, a morbidity rate of 80% in foxes has led to the death of 1200 animals due to PRV infection (Jin et al. 2016).
Representative characteristics of PR in non-nature host species are summarized in Table 2. In general, the clinical and pathological signs of these PRV infected species (cow/cattle, dog, wolf, goat/sheep, etc.) have some common features, including pruritus, hyperemia or hemorrhage in some organs (Zhang L et al. 2015; Jin et al. 2016; Liu et al. 2016; Zhang et al. 2016; Sun Y et al. 2018; Cheng et al. 2019; Lian et al. 2020). Additionally, reports of PRV infection with other animals (tigers and wild pigs) have also been documented in China (Li et al. 2014; Qin et al. 2015). Overall, the major causes for PRV infection in plant-eating animals (i.e. cow/cattle and goat/sheep) can be ascribed to living spaces adjacent to pig breeding sites with low feeding management (severe rat plague) (Cheng et al. 2019), while the reasons for carnivorous animals (i.e. wolf, mink, dog, and fox, etc.) may be stemmed from feeding raw or undercooked PRV-infected pork meat or offal to these animals (Liu et al. 2017; Jin et al. 2016; Lian et al. 2020).
Table 2.
Clinical signs and pathological characteristics of different PRV-infected species in China.
| Species | Case numbers | Clinical signs | Pathological characteristics | Reference |
|---|---|---|---|---|
| Cattle/cow | 9 | Pruritus, nervous symptoms, being excited and manic, etc | Leptomeningeal hyperemia, consolidation of lung lobes, etc | Cheng et al. (2019) |
| Dog | 12 | Pruritus, hypersalivation, broken winded, etc | Endocardial and thymic hemorrhage, pulmonary hemorrhage and/or congestion, etc | Zhang L et al. (2015) |
| Wolf | 1 | Pruritus, vomiting, quadriplegia, etc |
hemorrhagic spots and edema in the meninges Hemorrhagic spots and necrosis in the liver, etc |
Lian et al. (2020) |
| Goat/sheep | 11 | Pruritus, nervous symptoms, muscle spasm, etc | Leptomeningeal hyperemia, consolidation of lung lobes, etc | Zhang et al. (2016) |
| Fox | 2 | Pruritus, vomiting, broken winded, etc | Sugillation in the lung, hemorrhage in the spleen, thymus and liver, etc | Jin et al. (2016) |
| Mink | 9 | Pruritus, diarrhea, muscle spasm, etc | Hemorrhage in the thymus and submandibular lymph node, liver and spleen tumefaction, etc | Liu et al. (2017) |
| Raccoon | 3 | Pruritus, vomiting, etc | Not mentioned | Liu et al. (2016) |
Though PRV infection is fatal to these animal species, studies on the prevalence of this disease remain limited. A report performed by Wang et al. indicated that 38 (5.8%) among 566 dead mink specimens collected in Shandong province displayed PRV-positive (Wang GS et al. 2018). Another study revealed that PRV infection led to 21 of 300 (7.0%) cows suffering from abortion related diseases (Gu et al. 2012). These scant information still hint that these animal industries should pay attention to the occurrence of PRV infection.
PRV Infection as a Potential Threat to Humans
Since the first identification of PRV in 1902, whether this pathogen can infect humans remains controversial (Wong et al. 2019). Though several reports indicating high possibility of PRV infection in humans with different symptoms (such as fever, weakness and dysphagia) have been documented, the causes of these cases were not determined due to the lack of substantial evidences (Mravak et al. 1987; Anusz et al. 1992; Skinner et al. 2001).
In July 2017, the first case of human endophthalmitis with clinical symptoms of fever, visual impairment and headaches, etc. in China was proved to be PRV positive by NGS combining with PCR analysis (Ai et al. 2018). Further phylogenetic analysis confirmed that it was caused by an emerging PRV variant, suggesting that in spite of the case occurred in China, the potential threat of PRV infection to humans should receive more attention worldwide (Ai et al. 2018). The latest research showed that 1.27% (6/472) patients with clinically suspected central nervous system (CNS) infection were PRV-positive (Fan et al. 2020). Likewise, another retrospective seroepidemiological survey conducted in nine provinces of China indicated that 12.16% (75/617), 14.25% (63/442) and 6.25% (18/276) patients with encephalitis exhibited positive for PRV infection in 2012, 2013, and 2017, respectively (Li et al. 2020). Up to now, increasing cases of PRV infection in humans have been recorded in several provinces of China, and some of patients were eventually blind or dead (Ai et al. 2018; Fan et al. 2018; Zhao et al. 2018; Wang Y et al. 2019; Yang H et al. 2019; Yang X et al. 2019; Fan et al. 2020; Liu Q et al. 2020; Wang et al. 2020). The detailed information of these cases are summarized in Table 3. Notably, almost all patients in these reports were engaged in the works associated with pigs (e.g. slaughterer, veterinarian, and swine herder). Moreover, most of them had been injured when dealing with (sick) pigs or pork. In addition, all infected patients displayed similar clinical symptoms, including fever, headache, respiratory failure, seizure, and even visual impairment or loss, while the occurred clinical symptoms mainly depended on the development stages of the disease. It should be mentioned that most of the cases above were diagnosed as PR via detecting the presence of PRV DNA or specific IgM antibodies. Live viruses (namely hSD-1/2019) have successfully been isolated only from one case (i.e. the cerebrospinal fluid sample) (Liu Q et al. 2020). Further analysis showed its close phylogenetic relationship with those emerging PRV variants prevalent in China. Moreover, the in vivo pathogenicity of this isolated virus in experimental mice or pigs was higher than that of other variants (such as JX2/CHN2015, HuBXY/2018, and HeN1/CHN2012 strains) (Liu Q et al. 2020). However, no experiments investigating the mechanisms of PRV infecting humans have been carried out thus far.
Table 3.
Human PRV infection cases in China.
| Date | Age/Sex | Occupation | Injured at work | Interval after injury | Symptoms | Diagnostic methods | Treatment | Reference |
|---|---|---|---|---|---|---|---|---|
| NA | 43/male | Veterinarian | Yes | 4 days | Fever, headache, tonic–clonic seizures and coma, etc | NGS; PCR and ELISA methods | Acyclovir treatment for 2 weeks; antibiotics | Yang H et al. (2019) |
| NA | 59/male | Swineherd | Yes | 6 days | Fever, seizures, tonic–clonic seizures and coma, etc | NGS, ELISA method | Antimicrobial therapy, penciclovir and foscarnet sodium treatment for 17 days | Zheng et al. (2019) |
| NA | 44/male | Sick pig handler | NA | NA | Fever, seizures, visual loss, etc | NGS and PCR methods | Acyclovir and other treatments | Wang Y et al. (2019) |
| 2011 | A young female | NA | NA | Least 5 days | Fever, chotic behavior, recurrent seizures, etc | NGS (CSF) | Acyclovir, foscarnet, VPA treatment, etc. (death) | Fan et al. (2020) |
| Nov 2016 | 42/female | Pork cutter/cooker | NA | 4 days | Fever, tonic–clonic seizures, coma, blindness, etc | NA | Acyclovir and other treatments | Zhao et al. (2018) |
| Jun 2017 | 46/female | Swineherd | Exposed to the sewage | 3 days | Fever, visual impairment, headaches, etc | NGS, real-time PCR and PCR methods | NA | Ai et al. (2018) |
| Dec 2017 | 55/male | Pork cutter/cooker | NA | 4 days | Fever, headaches, cough, coma, etc | NGS (CSF) | NA | Zhao et al. (2018) |
| Dec 2017 | 51/man | Pork cutter/cooker | NA | NA | Fever, headache, coma, tonic–clonic seizures and death, etc | NGS (CSF) | Acyclovir and other treatments | Zhao et al. (2018) |
| Nov 2017 | 38/male | Pork cutter/cooker | NA | 4 days | Fever, headache, coma, tonic–clonic seizures, etc | NA | Acyclovir and other treatments | Zhao et al. (2018) |
| 2018 | A mid-aged male | NA | NA | NA | Fever, bilateral retinal necrosis, consciousness loss, etc | NGS (CSF) | Acyclovir treatment (death) | Fan et al. (2020) |
| 2018 | A mid-aged male | Pig farmer | NA | One month | Fever, cognitive decline, consciousness loss, etc | NGS (CSF) | Acyclovir, foscarnet, and methylprednisolone treatment, etc | Fan et al. (2020) |
| 2018 | A young male | NA | NA | Least 3 days | Fever, seizures, consciousness loss, etc | NGS (CSF) | Continues renal replacement, clonazepam, VPA, acyclovir, etc | Fan et al. (2020) |
| Before Mar 2018 | 59/male | Pig farmer | Yes | 40 days | Fever, weakness, tonic–clonic seizures, respiratory failure, etc | NGS (CSF) | Penciclovir treatment combined with Sodium phosphonate | Fan et al. (2018) |
| Feb 2018 | 50/male | Pig slaughterer | Yes | 5 days | Fever, headache, visual disturbance, coma, etc | NGS | IVIG treatment for 5 days, Glucocorticoids, antiviral and other treatments | Yang X et al. (2019) |
| Mar 2018 | 50/female | Pork cutter | Yes | 7 days | Fever, coma, respiratory failure, seizure, etc | NGS | Yang X et al. (2019) | |
| Mar 2018 | 43/male | Sick pig handler | Yes | 7 days | Fever, extremity tremors, respiratory failure, etc | NGS | Yang X et al. (2019) | |
| Apr 2018 | 59/male | Pork cutter | Yes | 10 days | Fever, seizures, respiratory failure, etc | NGS | Yang X et al. (2019) | |
| Apr 2018 | 50/male | Pork cutter | NA | NA | Fever, seizure, respiratory failure, etc | NGS | Yang X et al. (2019) | |
| Apr 2018 | 25/male | Veterinarian | Exposed to the PRV infected pigs | 6 days | Fever, headache, cognitive impairment, seizures, consciousness loss, and meningeal irritation sign | NGS (CSF), DdPCR, etc | Ganciclovir and foscarnet treatment | Liu Q et al. (2020) |
| Aug 2018 | 43/male | Veterinarian | Exposed to the diseased pigs | 11 days | Fever, respiratory failure, headache, cognitive impairment, seizures, consciousness loss, and meningeal irritation sign | Virus isolation, NGS (CSF), ddPCR, etc | Acyclovir treatment | Liu Q et al. (2020) |
| Mar 2019 | 44/male | Pork cutter | Yes | 14 days | Fever, seizure, tonic–clonic seizures and coma, etc | NGS and PCR methods | Acyclovir, dexamethasone and other treatments | Wang et al. (2020) |
| Mar 2019 | 35/male | Pig butcher | Exposed to the pigs and pork | 7 days | Fever, respiratory failure, headache, cognitive impairment, seizures, and consciousness loss | NGS (CSF), ddPCR, etc | Acyclovir and foscarnet treatment | Liu Q et al. (2020) |
| Feb 2019 | 49/male | Pig butcher | Exposed to the pork | 3 days | Fever, respiratory failure, headache, cognitive impairment, seizures, and consciousness loss | NGS (CSF), ddPCR, etc | Ganciclovir and foscarnet treatment | Liu Q et al. (2020) |
NA: no available; NGS: next generation sequencing; CSF: cerebral spinal fluid; DdPCR: droplet digital PCR.
In spite of these, the potential factors contributing to the PRV cross-species transmission events from animals to human are mainly derived from the speculations. The natural recombination events in some PRV variants circulating in Chinese pig population may alter viral virulence and immune response to novel hosts (He et al. 2019; Liu J et al. 2020). In addition, it has been confirmed that multiple alphaherpesviruses including PRV, herpes simplex virus type 1 (HSV-1) and type 2 (HSV-2), and bovine herpesvirus 1 (BHV-1) could utilize nectin-1 as an efficient cellular receptor for cell entry (Geraghty et al. 1998; Campadelli-Fiume et al. 2000; Milne et al. 2001; Li et al. 2017; Menotti et al. 2000; Yue et al. 2020). Moreover, the amino acid residues in nectin-1 among distinct species (e.g. human, swine, sheep, dog, cat, etc.) are relatively conserved (Wong et al. 2019). However, whether nectin-1 plays an important role in the cross-species infection of PRV needs further investigation.
Molecular Characteristics of Current Circulating PRV Strains in China
Genotypical Features of Prevalent PRV Strains in China
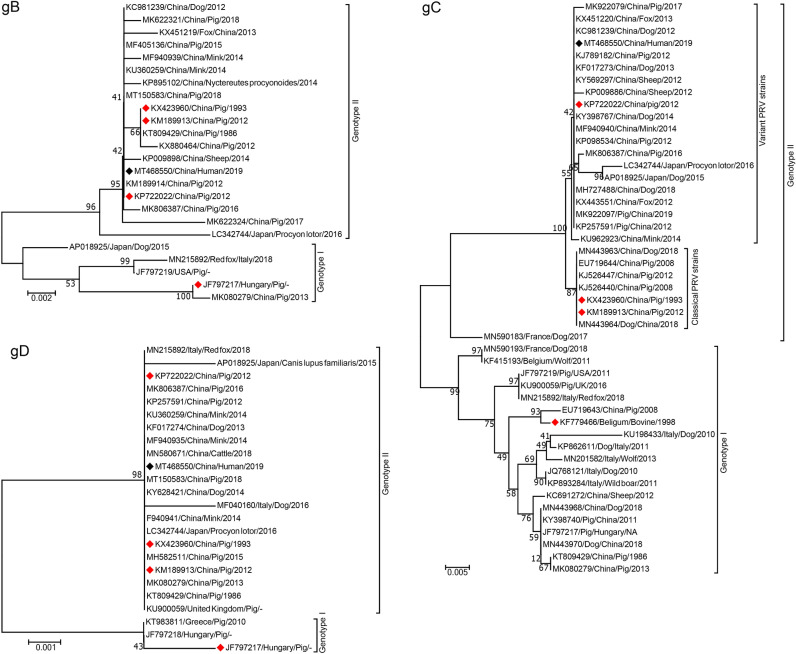
Undeniably, PRV remains the major pathogen that posts huge threats to pig industry worldwide. Currently, PRV isolates from the world could be divided into two genotypes, genotype I strains have been widely prevalent in different countries from Europe, America, and parts of China, while most of genotype II PRV isolates are from Asian countries, mainly in China (He et al. 2019a). Since China has suffered from two PR outbreaks, which were caused by PRV classical strains in 1990s and variant PRV strains in 2011 respectively, PRV isolates circulated in China represent emerging variant strains and classical local strains (Sun et al. 2016). Due to the high genetic variations of PRV gC, gB and gD gene sequences, they are often employed to analyze the genetic evolution of PRV strains (Ye C et al. 2015; Zhai et al. 2019). To investigate genetic characteristics of Chinese PRV strains from various species, we construct neighbor-joining phylogenetic trees based on the partial gC, gB, and gD gene sequences of the representative PRV strains isolated after 1970s from China and other countries. The results indicate that all PRV strains from different species including humans can be clustered into two major genotypes (Fig. 2). As shown in Fig. 2A, a tiny minority of PRV Chinese isolates and isolates from Europe and USA, including Bartha vaccine strain from Hungary (accession number: KF779466), are the members of genotype I group. While the genotype II group is composed of other Chinese strains and Japan strains, which can be further subdivided into two groups, namely classical and variant PRV strains, the former group mainly includes vaccine strains Ea (accession number: KX423960) and Fa (accession number: KM189913) which are widely used in China, and other PRV strains, the latter group is composed of a recently licensed PRV vaccine (accession number: KP722022) and other strains isolated after 2011. In line with our findings, numerous studies have shown that PRV strains in genotype II but not genotype I were more prevalent in China (He et al. 2019a; Zhai et al. 2019). This finding partially explains why currently available vaccines derived from Bartha or Bartha-K61 strains (genotype I) cannot provide complete protection against PRV variants (genotype II). Furthermore, PRV strains from other species including fox, mink, sheep, dog, and raccoon are randomly distributed with these from pigs in China in genotype I or II, together with the latest research conducted by Liu J et al. (2020), suggesting that PRV strains from the above species including humans may have the similar origin with those of pigs (Moreno et al. 2015; He et al. 2019a).
Fig. 2.
Phylogenetic trees based on the different gene sequences, showing the genetic relationships among PRV strains from different hosts/regions, were constructed by the neighbor-joining method with a bootstrap test of 1000 replicates using MEGA 7.0 software. The hosts, countries, years, and GenBank accession numbers of the reference strains employed in this phylogenetic tree are labeled, Black and red diamonds represent isolates from human and vaccines generated.
Mutation and Recombination of PRV Strains in China
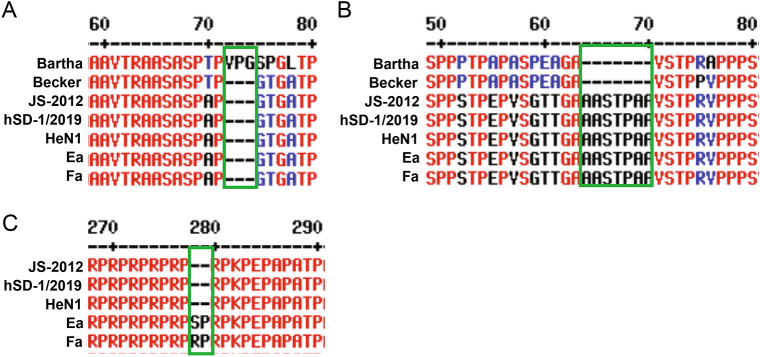
According to the analysis results of He et al. an emerging PRV variant (Qihe547 strain) in China showed that the average amino acid (aa) differences with PRV genotype I strains, classical and variant strains in genotype II are 4.94%, 1.16%, and 0.46%, respectively (He et al. 2019a). Compared with PRV genotype I strains such as Bartha and Becker, currently circulating genotype II PRV strains in China displayed high genetic variations in some hypervariable regions, such as internal and terminal repeat regions (Ye C et al. 2015). Furthermore, unique aa insertions and deletions (indels), and mutations are frequently found in various protein-coding gene sequences of PRV strains in genotype II. Thus, there are many unique genetic features to differentiate PRV genotype I and II strains. For example, both classical (e.g. Ea and Fa) and emerging (e.g. JS-2012, HeN1, and hSD/2019) PRV strains in genotype II show a 9-nucleotide deletion (encoding 75VPG77) in UL27 (gB) gene, and a 21-nucleotide insertion (encoding 63AASTPAA69) in UL44 (gC) gene compared with PRV strains in genotype I (Sun et al. 2018b). Also, there are some key features of PRV variants that are different from the classical local strains, such as a 6-nucleotide insertion (encoding 288SP299) in US6 (gD) gene of classical local strains compared to emerging PRV variants (Fig. 3).
Fig. 3.
Alignment of partial amino acid sequence of gB (A), gC (B), and gD (C) genes of representative PRV strains (genotype I strains (eg. Bartha and Becker), classical (eg. Fa and Ea) and variant strains (JS-2012, hSD-1/2019, and HeN1) in the genotype II). Unique aa deletions/insertions existed between different PRV strains are shown in the green box.
Besides aa mutations and indels, recombinant events also accelerate the evolution of PRV genome and the change of viral virulence. He et al. has confirmed that inter- and intra-clade recombinant events occurred among various PRV strains using SimPlot software (He et al. 2019a). Recently, two inter-clade recombinant PRV strains (FJ-W2 and FJ-ZXF) have been reported, the gB genes of these two strains belonged to the genogroup I, whereas the gD, gE, and gC genes of which were located in the genogroup II (Zhai et al. 2019). Similarly, the gB gene of another PRV strain FJ62 from Sichuan Province of China had a 100.0% sequence identity with the MY-1 strain isolated from a Japanese wild boar, while its gC, gD, and gE sequences displayed higher homology with Chinese strains relative to others, namely 99.5%, 99.9%, and 99.9%, respectively (Huang et al. 2020). This suggests that the variant strain FJ62 may be generated from the natural recombination of PRV strains from genotype I (from Japan) and genotype II (from China) (Huang et al. 2020).
What Has Been Done for the Prevention of PR in China
To better control and prevent PR in China, the Chinese Government has issued a series of policies with the intent to eradicate PR in pig breeding farms by the end of 2020 (Sun et al. 2016). Accordingly, lots of efforts have been made for the development of effective means to control PRV infection, mainly including vaccines and other novel viral inhibitors.
Main Vaccines against PRV Infection
As a major challenge, PRV has in fact been prevalent in pig farms of China for nearly seventy years, vaccination is one of the most effective approaches for preventing it and minimizing the economic losses caused by PR (Freuling et al. 2017). The first live gene-modified vaccine (attenuated Bartha-K61 strain) widely applied in China was imported from Hungary in 1970s, immunization with it has effectively controlled PR in some regions in China (Sun et al. 2016; Zhou et al. 2017). However, outbreaks of PR in farms where pigs had been vaccinated with Bartha-K61 strain based live modified vaccines have frequently occurred in most of regions in China since 2011, which were mainly caused by emerging PRV variants with high sequence divergence comparing with classical PRV strains in China (Ye et al. 2016). These suggest that the existing vaccines derived from Bartha-K61 strain cannot confer complete protection for pigs against the current PRV variants (Hu D et al. 2015; Tong et al. 2015). With the rapid development of biotechnologies and deep understanding of the biological functions of PRV encoding genes, some gene-modified and other types of vaccines derived from local virulent PRV strains have also been generated.
A series of novel gene-modified vaccines based on classical or variant PRV strains have been created in recent years (Summarized in Table 4). It has been confirmed that gE, gI, and TK genes are close to the virulence of PRV strains, while they do not affect viral immunogenicity, therefore these genes are ideal targets for researchers to create genetic engineering vaccines against PR (Cong et al. 2016; Zhao et al. 2020). Before the emergence of variant PRV strains in 2011, some gene-modified vaccines based on classical PRV strains had been licensed in China, including the triple-gene-deleted (gE/gI/TK) vaccine generated based on the PRV Fa strain licensed in 2003, which was regarded as the first genetically modified vaccine against PR in China (Zhu et al. 2004). Likewise, another TK/gG-deleted vaccine developed based on the PRV Ea strain was licensed in 2006 (He et al. 2006). These two licensed vaccines have been widely applied in Chinese pig population against PR until now, however, whether those two vaccines could provide complete protection against variant PRV strains are undefined (Sun et al. 2016). Additionally, a large number of researchers have developed novel genetic engineering vaccines based on emerging PRV variants since 2011, but only two types of vaccines have been licensed so far, including the gE-gene-deleted inactivated vaccine based on the PRV HeN1201 strain in 2019 (Wang T et al. 2015), and another natural four-gene-deleted (gI/gE/Us9/Us2) vaccine based on the PRV C strain in 2017 (Gao et al. 2015). Though other candidates including killed and live attenuated vaccines based on the variant PRV strains have been proved to offer effective protection against PR caused by variant strains, considering the safety issue of these candidates applied in the field, further clinical trials need to be conducted.
Table 4.
List of genetic modified vaccines against PRV infection.
| Gene-deleted vaccines | Strain | Features | Technologies | Main advantages | Status | References |
|---|---|---|---|---|---|---|
| Single gene-deleted vaccine (inactivated) | AH02LA (variant) | gE-deleted | BCA | High safe without virulence reversion; Allowing DIVA with more complete protection than Bartha K61 vaccines | Not available | Wang J et al. (2016) |
| HN1201 (variant) | gE-deleted | HR | Licensed | Wang T et al. (2015) | ||
| Double gene-deleted vaccine (inactivated) | ZJ01 (variant) | gE/gI-deleted | BCA | Not available | Gu et al. (2015) | |
| Single gene-deleted vaccine | TJ (variant) | gE-deleted | HR | Safe to piglets without visible gross pathological lesions; Effective immune response; Completely provides protection against emerging PRV variants; Allowing DIVA; | Not available | Wang et al. (2014) |
| Double gene-deleted vaccine | Ea (classical) | TK/gG deleted | NA | Licensed | He et al. (2006) | |
| JS-2012 (variant) | gE/gI-deleted | HR | Not available | Tong et al. (2016) | ||
| XJ (variant) | Yin et al. (2017) | |||||
| AH02LA (variant) | TK/gE deleted | HR | Not available | Wang J et al. (2018) | ||
| JS-2012 (variant) | gE/US2 deleted | High-temperature passage | Not available | Liang et al. (2017) | ||
| Triple gene-deleted vaccine | Fa (classical) | gE/gI/TK-deleted | HR | Safer than double gene-deleted vaccines to piglets and growing pigs; Effective immune response; Completely provides protection against emerging PRV variants; Allowing DIVA; | Licensed | Zhu et al. (2004) |
| SA215 (classical) | ||||||
| ZJ01 (variant) | Not available | Dong et al. (2017) | ||||
| SMX (variant) | Not available | Hu RM et al. (2015) | ||||
| HN1201 (variant) | Not available | Zhang C et al. (2015) | ||||
| HeN1 (variant) | gE/gI/TK-deleted | CRISPR/Cas9 | Not available | Tang et al. (2016) | ||
| NY (variant) | Not available | Zhao et al. (2020) | ||||
| 201,715 (variant) | gE/gC/TK-deleted | CRISPR/Cas9 | Not available | Lin et al. (2020) | ||
| Four gene-deleted vaccine | C (variant) | gI/gE/Us9/Us2-deleted | Natural losses | Licensed | Gao et al. (2015) |
HR: homologous DNA recombination; DIVA: distinction between the infected and vaccinated animals; BCA: bacterial artificial chromosome; CRISPR/Cas9: clustered regularly interspaced short palindromic repeats/Cas9.
Considering the large genome of PRV, the property of tolerating gene modification can also be used as an excellent vector for expressing other foreign protein(s), meanwhile, the infectivity and immunogenicity of PRV itself are not affected (Kimman et al. 1992). Some live attenuated PRV recombinant vaccines which could encode key antigens of other animal pathogens have been created and described in a previous review (Dong et al. 2014). As updated in Table 5, researchers usually selected the gene regions encoding nonessential glycoproteins (eg. gG, gI, gE, and TK) to insert exogenous sequences (Chen et al. 2011; Tong W et al. 2020; Zheng et al. 2020). To date, various exogenous genes encoding key antigens of animal pathogens have been successfully inserted into PRV genome, such as the VP2 gene of Porcine parvovirus (Zheng et al. 2020), the E2 gene of Classical swine fever virus (Wang YM et al. 2015; Lei et al. 2016), as well as the Bp26 gene of Brucella melitensis (Yao et al. 2015). In general, most of PRV recombinant vaccines could provide effective protection against multiple infectious disease including PR, but these candidate vaccines are not available in the markets yet. Fantastically, the latest research showed that using Bacillus subtilis as a recombinant vaccine expressing PRV gC and gD proteins could efficiently induce mucosal immune responses against this disease (Wang J et al. 2019), and the efficient protection provided by a gD-based subunit vaccine against PRV variant infection in pig models was also confirmed recently (Zhang et al. 2020).
Table 5.
List of live attenuated recombinant vaccines against PRV infection.
| Insertion sites in PRV genome |
Parental PRV strains | Insertion genes | Function (animal model) | References |
|---|---|---|---|---|
| gG gene | HB-98 strain | Porcine IL6 gene and VP2 gene of PPV | Provided partial protection against the virulent PPV and PRV challenges (mice) | Zheng et al. (2020) |
| gI gene | gE/gI/TK-deleted SA 215 strain | VP2 gene of PPV | Completely protected pigs against maternal PRV infection and significantly reduced the death rate (1/28) after PPV challenge compared with the control (7/31) (pig) | Chen et al. (2011) |
| gG gene | HB-98 strain | Porcine IL18 gene and Cap gene of PCV2 | Protected mice against PRV variants infection and significantly reduced the amount of PCV2 viremia (mice) | Zheng et al. (2015) |
| Between gE and gI gene | gE/TK-deleted strain | prM and E genes of JEV | Provided 100% and 80% protection against PRV and JEV infection, respectively (mice) | Qian et al. (2015) |
| Between gE and gI gene | gE/gI-deleted strain | E2 gene of CSFV | Provided complete protection against maternal PRV and CSFV infection (pig) | Wang YM et al. (2015) |
| gE/gI/TK-deleted TJ strain | Lei JL et al. (2016) | |||
| Between gG and gD gene | gE/gI-deleted JS-2012 strain | Provided complete protection against maternal PRV and CSFV infection without MDAs (pig) | Tong W et al. (2020) | |
| Between gG and US9 gene | gE/gI/TK-deleted TJ strain | Cap gene of PCV2 and E2 gene of CSFV | Only protected pigs against PRV infection (pig) | Abid et al. (2019) |
| gG gene | gG deleted strain | SiRNA targeting to the N gene of HP-PRRSV | Safe to pigs and efficiently inhibited HP-PRRSV replication in vivo (pig) | Cao et al. (2015) |
| Between gE and gI gene | gE/TK-deleted strain | Bp26 gene of Brucella melitensis | Induced good humoral and cell-mediated immune response in mice (mice) | Yao et al. (2015) |
| gG gene | HB-98 strain | SAG1 and MIC genes of Toxoplasma gondii | Induced partial protection against a lethal challenge with Toxoplasma gondii strain (mice) | Nie et al. (2011) |
| Between gE and gI gene | gE/gI/TK-deleted TJ strain | P12A and 3C genes of FMDV | Significantly increased the survival rate (3/5) after FMDV challenge compared with the control (0/5) (pig) | Zhang et al. (2011) |
PPV: porcine parvovirus; PCV2: porcine circovirus type 2; JEV: Japanese encephalitis virus; CSFV: classical swine fever virus; HP-PRRSV: high pathogenic porcine reproductive and respiratory syndrome virus.
In China, various types of vaccines against PR have been developed recently, mainly including inactivated vaccines, live attenuated vaccines, and live virus-vectored vaccines. Most of vaccines aforementioned could generate high level of neutralizing and gB antibodies, and offer effective protection against PRV challenge. Furthermore, these candidate vaccines also allow DIVA, thereby accelerating the eradication of PR in China. As shown in Table 4, the inactivated vaccines display high safety to the vaccinated animals without viral virulence reversion, while they are generally less efficacious compared to genetically modified-live vaccines (the live attenuated and virus-vectored vaccines) (Freuling et al. 2017). Notably, genetically modified-live vaccines summarized in the Tables 4 and 5, have their own unignorable disadvantages. The important one is their safety to pig populations should be confirmed via lengthy testing (Wang J et al. 2016). Moreover, the safety issues of these PRV modified-live vaccines to non-target species has also attracted widespread concern. For example, vaccination with attenuated Bartha-K61 strain could cause PR in sheep with severe clinical signs (Kong et al. 2013). Similarly, immunization with Bartha-K61 and HB98 (another commercial live attenuated PRV vaccine generated based on Ea strain in China) vaccines also posed potential health threat to dogs (Lin et al. 2019). In addition, the latest report verified that infection with a gE-deleted PRV strain could cause PR in an adult red fox in Italy (Moreno et al. 2020). Finally, as the origin of variant PRV strains circulating since 2011 in China remains undefined, potential contribution of these genetically modified-live vaccine to the evolution of this pathogen should also be considered (Wang T et al. 2015; Bo et al. 2020).
Traditional Chinese Herbal Medicines as Potential Anti-PRV Drugs
Considering the great impact of PR on pig industry, Chinese scientists have also devoted to identifying inhibitory agents against PRV infection. The antiviral activities of Chinese herbal medicines have received comprehensive attention, as summarized in Table 6. For instance, the druggable components derived from leaves of Panax ginseng exhibited effective anti-PRV activity in vitro, while the detailed antiviral mechanisms remain undetermined (Hou et al. 1998). Likewise, resveratrol, as a polyphenolic stilbenoid initially exacted from Veratrum grandiforum and leaves of vines (Pezzuto, 2019), has a variety of bioactivities, such as high anti-oxidative and anti-inflammatory properties (Malaguarnera et al. 2020). In particular, the antiviral activities of resveratrol against PRV infection have well been recognized in a series of observations. Resveratrol showed efficient anti-PRV activities with a low 50% inhibition concentration (EC50) of 17.17 μmol/L in vitro, the underlying mechanisms of it in suppressing PRV replication were at least partially associated with the inhibition of the IκB kinase activation (Zhao et al. 2017). Its anti-PRV and immune-adjuvant abilities were also confirmed in both mice and pig models (Yao et al. 2015; Su et al. 2016; Zhao et al. 2018). Additionally, the ability of resveratrol to inhibit ASFV replication in vitro previously described (Galindo et al. 2011), together with interesting observations as stated above, suggest that this natural herb should warrant intensive investigations in the future, thereby pushing it forward to be an efficient agent for protecting animals or humans from PRV infection.
Table 6.
Different types of compounds with anti-PRV infection activity.
| Source | Extracts | Mechanism | IC50 | CC50 | In vitro | In vivo | PRV strain | MOI | References |
|---|---|---|---|---|---|---|---|---|---|
| Resveratrol | Ethanol | Inhibition of viral replication; Inhibition of IKB kinase activation | > 262.87 μmol/L | 17.17 ± 0.35 μmol/L | √ | √ | Rong A | 0.01 | Chen et al. (2019), Zhao et al. (2017) |
| Germacrone | Dimethyl sulfoxide | Inhibition of viral replication | 233.5 μmol/L for Vero, 184.1 μmol/L for PK15 | 54.51 μmol/L for Vero, 88.78 mol/L for PK15 | √ | × | PRV variant Bartha K61 vaccine | 0.1–10 | He et al. (2019b) |
| Isatis indigotica (leaf) | Ethanol | Inhibition of viral replication | 226 μg/mL | 11 μg/mL | √ | × | TNL | 100 pfu/well | Hsuan et al. (2009) |
| Radix isatidis | Ethanol and water | Inhibition of viral replication; Killing virus directly | Not mentioned | Not mentioned | √ | × | MinA | 100 TCID50 | Tong C et al. (2020) |
| Marine Bacillus S-12–86 lysozyme | Water | Inhibition of viral replication | 100 μg/mL | 0.46 μg/mL | √ | × | Attenuated | Not mentioned | Zhu et al. (2013) |
| Diammonium glycyrrhizin | Not mentioned | Killing virus directly | 1.25 mg/mL | Not mentioned | √ | √ | Bartha K61 vaccine | 104 pfu/mL | Sui et al. (2010) |
| Vanadium-substituted Heteropolytungstate | DMEM | Killing virus directly | 400 μg/mL | 5 μg/mL | √ | × | Bartha | 200 TCID50/mL | Liu et al. (1998) |
| Graphene Oxide | DMEM | Killing virus directly | Not mentioned | Not mentioned | √ | × | HNX variant | 0.01 | Ye S et al. (2015) |
| Ivermectin | Dimethyl sulfoxide | Blocking the nuclear translocation of viral DNA polymerase UL42 | Not mentioned | Not mentioned | √ | √ | Not mentioned | 0.01 | Lv et al. (2018) |
| Phosphonoformate sodium | Not mentioned | Inhibition of viral DNA polymerase | 480 μg/mL | Nearly 60 μg/mL | √ | × | Kaplan | 2 | Ren et al. (2011) |
Other Compounds of Anti-PRV Infection
Other compounds with antiviral properties also receive extensive attention, as described in Table 6. For example, Vanadium-substituted Heteropolytungstate should be the first compound exhibiting anti-PRV activity through directly killing the virus (Liu et al. 1998). Similarly, Graphene Oxide has been confirmed its antiviral function against PRV infection (Ye S et al. 2015). Therefore, these agents might hold the promise to be natural disinfectants in the future, if their killing characteristics of multiple viruses prevalent in pig farms are further verified. Additionally, ivermectin, an effectively anti-parasitic drugs, also manifested a nearly 100% inhibitory activity against PRV replication under the concentration of 2.0 or 2.5 μmol/L in vitro. The molecular mechanism of it was ascribed to blocking the nuclear translocation of viral DNA polymerase UL42 (Lv et al. 2018). Moreover, further in vivo study revealed that ivermectin treatment could decrease the death rate of PRV infected mice, and relieve the lesions in mouse brains caused by PRV infection (Lv et al. 2018). However, the anti-PRV activity of ivermectin in pig models warrants further investigations.
Novel small RNAs
Small RNAs including small interfering RNAs (siRNAs) and microRNAs (miRNAs) are widely used to explore gene functions due to their features of targeting mRNA degradation (Tan et al. 2019), and also to be useful tools for efficiently inhibiting virus replication and interfering protein synthesis (Du et al. 2009; Wong et al. 2020). For example, application of siRNA targeting to PRV DNA polymerase processivity factor UL42 could efficiently suppress viral replication in cultured cells (Wang YP et al. 2016). Similarly, over-expression of miRNA-21 significantly inhibited PRV replication via directly targeting to interferon-γ inducible protein-10 gene in PK15 cells (Huang et al. 2014). Additionally, another research group proved the capability of PRV-miRNA-LLTTa to reduce the numbers of PRV genomic copies in vitro, while the potential mechanism requires further investigations (Liu H et al. 2019).
Conclusions and Future Directions
PRV remains the major infectious pathogen which greatly affects the development of pig industry in China. Chinese Government and scientists have spent nearly seventy years in fighting PR. The past efforts devoted to investigating the prevalence of PR in China, and developing diagnostic approaches for detecting PRV, vaccines and other novel inhibitors against this virus, are well summarized as described above.
In terms of PRV prevalence in China, the literatures collected from English and Chinese databases clearly indicate that this virus has widely spread in different provinces/regions of this country with intense epidemic. The factors contributing to the severe PRV infection in China are complex. Overall, the biosecurity levels in Chinese large-scale pig farms have been greatly improved, and PR in some breeding pig farms has been successfully eradicated via implementing effective elimination strategies. However, pigs in these PR purified farms are still faced with high risks of being re-infected upon introducing PRV infected breeding pigs, even PRV infected semen (Song et al 2017) and animal feeds (Dee et al. 2018) from the outside. Additionally, as a voluntary policy, vaccination for PR eradication in some pig farms (especially free-range and small farms) in remote regions of China is not always performed (Sun et al. 2016), thus, poor biosecurity leads to more severe prevalence of PR in these farms.
Also, severe prevalence of PRV in Chinese pigs could increase the possibility of other susceptible animals to get infected with this pathogen, which in turn increases the opportunities of human infection. Though the cases of human infection with PRV currently are still limited, potential threat of its infection to humans should truly receive widespread concerns. Common characteristics in these cases are summarized in Table 3. In particular, it has been proved that PRV may infect humans via wound (Yang H et al. 2019, Yang X et al. 2019) and eyes (Ai et al. 2018). Additionally, considering these facts that intranasal immunization is the main approach for vaccination and PRV genome is detected in the air specimens from pig farms (Li et al. 2019), whether airborne route is another pathway for PRV infection in humans needs further investigations. It is therefore suggested that self-protection for these susceptible population is very essential upon working. Moreover, molecular analysis revealed the occurrence of PR in other animals and humans might be caused by PRV variants circulating in Chinese pig population (Ai et al. 2018; Chen et al. 2019; Jin et al. 2016; Liu Q et al. 2020), while the understanding of PRV transmission from pigs to other species (especially humans) should further be explored.
In recent years, different types of diagnostic approaches for PRV diagnosis have been created. It should be noted that despite not reaching to commercialization, some diagnostic products designed by veterinary detection institutes have widely been applied for the diagnosis of PR or the investigation of the PRV prevalence in China. Notably, available approaches for determining PRV infection in humans remain very limited. The confirmation of PRV infection in humans by most hospitals mainly relies on NGS, while this approach is time-consuming and relatively expensive compared with others. Furthermore, considering that PRV infection may be fatal to humans, NGS seems not to be suitable for patients with severe infection. In particular, diagnostic standards for detection of PRV infection in humans are not available in Chinese hospitals, including clinical and laboratory tests, which are essential for early diagnosis of this fatally infectious disease in humans. Therefore, these should be the emphases for future efforts.
Additionally, various types of gene-modified vaccine candidates have been generated with efficient protection for pigs against PRV infection. However, they have not been commercial for application mainly due to multiple factors, such as their safety to pig populations and the possibility of viral virulence reversion. Additionally, as different species show various susceptibility and immune response to PRV infection, safer vaccines with efficient protection against PRV infection in other species including goat, dog, mink, also need to be developed in the future. Furthermore, several antiviral drugs treating other herpesvirus such as HSV-1 can be introduced for curing human PRV infection, such as valacyclovir, acyclovir (Wang Y et al. 2019), ribavirin (Pancheva 1991). Meanwhile, some newly developed therapies including monoclonal antibodies, small molecule inhibitors, are also available approaches for the treatment of PR in humans.
In conclusion, though effective vaccines have widely been used in pig population against PRV infection, this pathogen is still prevalent in Chinese pig herds. The degree of PR elimination in pig population seems to rely heavily on whether the mandatory vaccination policy is conducted or not. In addition, PRV infection in other susceptible species including humans should receive more concerns. Some effectively diagnostic and treatment approaches urgently need to be exploited, especially in term of human infection.
Acknowledgements
This work was supported by the General Program of National Natural Science Foundation of China (Grants No. 31571432/31802252), the Hunan Provincial Natural Science Foundation of China (Grant No. 2015JC3097/2020JJ4041), and the Postgraduate Scientific Research Innovation Project of Hunan Province (CX20200659). Support was also provided by “Shennong” Scholar funding to Aibing Wang. Additionally, we sincerely thank Fan Liao and Kuan-kuan Xiong for the assistance in data collection.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no competing interests.
Animal and Human Right Statement
The present manuscript does not include any animal or human experiments by any of the authors.
Contributor Information
Lingchen Yang, Email: lcyang@hunau.edu.cn.
Aibing Wang, Email: bingaiwang@hunau.edu.cn.
References
- Abid M, Teklue T, Li Y, Wu H, Wang T, Qiu HJ, Sun Y. Generation and immunogenicity of a recombinant pseudorabies virus co-expressing classical swine fever virus E2 protein and porcine circovirus type 2 capsid protein based on fosmid library platform. Pathogens. 2019;8:279. doi: 10.3390/pathogens8040279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai JW, Weng SS, Cheng Q, Cui P, Li YJ, Wu HL, Zhu YM, Xu B, Zhang WH. Human endophthalmitis caused by pseudorabies virus infection, China, 2017. Emerg Infect Dis. 2018;24:1087–1090. doi: 10.3201/eid2406.171612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An TQ, Peng JM, Tian ZJ, Zhao HY, Li N, Liu YM, Chen JZ, Leng CL, Sun Y, Chang D, Tong GZ. Pseudorabies virus variant in bartha-K61-vaccinated pigs, China, 2012. Emerg Infect Dis. 2013;19:1749–1755. doi: 10.3201/eid1911.130177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anusz Z, Szweda W, Popko J, Trybała E. Is Aujeszky's disease a zoonosis? Przegl Epidemiol. 1992;46:181–186. [PubMed] [Google Scholar]
- Bo Z, Miao Y, Xi R, Gao X, Miao D, Chen H, Jung YS, Qian Y, Dai J. Emergence of a novel pathogenic recombinant virus from Bartha vaccine and variant pseudorabies virus in China. Transbound Emerg Dis. 2020 doi: 10.1111/tbed.13813. [DOI] [PubMed] [Google Scholar]
- Campadelli-Fiume G, Cocchi F, Menotti L, Lopez M. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev Med Virol. 2000;10:305–319. doi: 10.1002/1099-1654(200009/10)10:5<305::AID-RMV286>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Cao SF, Guo QY, Wang Y. Inhibition of highly pathogenic porcine reproductive and respiratory syndrome virus replication by recombinant pseudorabies virus-mediated RNA interference in piglets. Vet Microbiol. 2015;181:212–220. doi: 10.1016/j.vetmic.2015.10.016. [DOI] [PubMed] [Google Scholar]
- Chen Y, Guo W, Xu Z, Yan Q, Luo Y, Shi Q, Chen D, Zhu L, Wang X. A novel recombinant pseudorabies virus expressing parvovirus VP2 gene: immunogenicity and protective efficacy in swine. Virol J. 2011;8:307. doi: 10.1186/1743-422X-8-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YN, Wang J, Zi ZC, Kang WH, Wang BY, Ni JQ, Yuan L. Development of droplet digital PCR for the detection of pseudorabies virus. Acta Veterinaria et Zootechnica Sinica. 2017;48:1705–1710. [Google Scholar]
- Chen M, Chen X, Song X, Muhammad A, Jia R, Zou Y, Yin L, Li L, He C, Ye G, Lv C, Zhang W, Yin Z. The immune-adjuvant activity and the mechanism of resveratrol on pseudorabies virus vaccine in a mouse model. Int Immunopharmacol. 2019;76:105876. doi: 10.1016/j.intimp.2019.105876. [DOI] [PubMed] [Google Scholar]
- Chen C, Cao MZ, Lyu L, Bai J, Wang XW, Jiang P. Epidemiological investigation of the antibody against gE of pseudorabies virus in swine in three provinces in mid-eastern China from 2018 to 2019. Anim Husb Vet Med. 2020;52:122–124. [Google Scholar]
- Chen H, Zhang XL, Jin Z, Huang LP, Dan H, Xiao W, Liang J, Zou S, Tang Y. Differential diagnosis of PRV-infected versus vaccinated pigs using a novel EuNPs-virus antigen probe-based blocking fluorescent lateral flow immunoassay. Biosens Bioelectron. 2020;155:112101. doi: 10.1016/j.bios.2020.112101. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Kong Z, Liu P, Fu Z, Zhang J, Liu M, Shang Y. Natural infection of a variant pseudorabies virus leads to bovine death in China. Transbound Emerg Dis. 2019;67:518–522. doi: 10.1111/tbed.13427. [DOI] [PubMed] [Google Scholar]
- Cong X, Lei JL, Xia SL, Wang YM, Li Y, Li S, Luo Y, Sun Y, Qiu HJ. Pathogenicity and immunogenicity of a gE/gI/TK gene-deleted pseudorabies virus variant in susceptible animals. Vet Microbiol. 2016;182:170–177. doi: 10.1016/j.vetmic.2015.11.022. [DOI] [PubMed] [Google Scholar]
- Dee SA, Bauermann FV, Niederwerder MC, Singrey A, Clement T, de Lima M, Long C, Patterson G, Sheahan MA, Stoian AMM, Petrovan V, Jones CK, De Jong J, Ji J, Spronk GD, Minion L, Christopher-Hennings J, Zimmerman JJ, Rowland RRR, Nelson E, Sundberg P, Diel DG. Survival of viral pathogens in animal feed ingredients under transboundary shipping models. PLoS ONE. 2018;13:e0194509. doi: 10.1371/journal.pone.0194509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Zarlenga DS, Ren X. An overview of live attenuated recombinant pseudorabies viruses for use as novel vaccines. J Immunol Res. 2014;2014:824630. doi: 10.1155/2014/824630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Bai J, Sun T, Gu Z, Wang J, Sun H, Jiang P. Comparative pathogenicity and immunogenicity of triple and double gene-deletion pseudorabies virus vaccine candidates. Res Vet Sci. 2017;115:17–23. doi: 10.1016/j.rvsc.2017.01.011. [DOI] [PubMed] [Google Scholar]
- Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Zheng LL, He JY, Hui HX, Zou YL, Xin ZK. A case of human viral encephalitis caused by pseudorabies virus infection. Chin J Nerv Ment Dis. 2018;44:747–748. [Google Scholar]
- Fan S, Yuan H, Liu L, Li H, Wang S, Zhao W, Wu Y, Wang P, Hu Y, Han J, Lyu Y, Zhang W, Chen P, Wu H, Gong Y, Ma Z, Li Y, Yu J, Qiao X, Li G, Zhao Y, Wang D, Ren H, Peng B, Cui L, Wang J, Guan H. Pseudorabies virus encephalitis in humans: a case series study. J Neurovirol. 2020;26:556–564. doi: 10.1007/s13365-020-00855-y. [DOI] [PubMed] [Google Scholar]
- Freuling CM, Müller TF, Mettenleiter TC. Vaccines against pseudorabies virus (PrV) Vet Microbiol. 2017;206:3–9. doi: 10.1016/j.vetmic.2016.11.019. [DOI] [PubMed] [Google Scholar]
- Galindo I, Hernáez B, Berná J, Fenoll J, Cenis JL, Escribano JM, Alonso C. Comparative inhibitory activity of the stilbenes resveratrol and oxyresveratrol on African swine fever virus replication. Antiviral Res. 2011;91:57–63. doi: 10.1016/j.antiviral.2011.04.013. [DOI] [PubMed] [Google Scholar]
- Gao JF, Lai Z, Shu YH, Qi SH, Ma JJ, Wu BQ, Gong JP. Isolation and identification of porcine pseudorabies virus (PRV) C strain. Acta Agric Shanghai. 2015;31:32–36. [Google Scholar]
- Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- Gu WJ, Fan F, Mao AM. Correlation investigation of pseudorabies virus infection and abortion in cattle. Chin J Anim Husb and Vet Med. 2012;4:21. [Google Scholar]
- Gu Z, Dong J, Wang J, Hou C, Sun H, Yang W, Bai J, Jiang P. A novel inactivated gE/gI deleted pseudorabies virus (PRV) vaccine completely protects pigs from an emerged variant PRV challenge. Virus Res. 2015;195:57–63. doi: 10.1016/j.virusres.2014.09.003. [DOI] [PubMed] [Google Scholar]
- He QG, Chen HC, Fang LR, Wu B, Liu ZF, Xiao SB, Jin ML. The safety, stability and immunogenicity of double gene-negative mutant of pseudorabies virus strain (PRV HB-98) Chin J Vet Med. 2006;26:165–168. [Google Scholar]
- He W, Auclert LZ, Zhai X, Wong G, Zhang C, Zhu H, Xing G, Wang S, He W, Li K, Wang L, Han GZ, Veit M, Zhou J, Su S. Interspecies transmission, genetic diversity, and evolutionary dynamics of pseudorabies virus. J Infect Dis. 2019;219:1705–1715. doi: 10.1093/infdis/jiy731. [DOI] [PubMed] [Google Scholar]
- He W, Zhai X, Su J, Ye R, Zheng Y, Su S. Antiviral activity of germacrone against pseudorabies virus in vitro. Pathogens. 2019;8:258. doi: 10.3390/pathogens8040258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou SK, Zhang ML, Liu J, Gou XK, Tian HY, Feng SZ, Jin NY. Investigation of the antiviral activities of six Chinese herbal medicines. Chin J Vet Sci. 1998;18:596–598. [Google Scholar]
- Hsuan SL, Chang SC, Wang SY, Liao TL, Jong TT, Chien MS, Lee WC, Chen SS, Liao JW. The cytotoxicity to leukemia cells and antiviral effects of Isatis indigotica extracts on pseudorabies virus. J Ethnopharmacol. 2009;123:61–67. doi: 10.1016/j.jep.2009.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Zhang Z, Lv L, Xiao Y, Qu Y, Ma H, Niu Y, Wang G, Liu S. Outbreak of variant pseudorabies virus in Bartha-K61-vaccinated piglets in central Shandong Province, China. J Vet Diagn Invest. 2015;27:600–605. doi: 10.1177/1040638715593599. [DOI] [PubMed] [Google Scholar]
- Hu RM, Zhou Q, Song WB, Sun EC, Zhang MM, He QG, Chen HC, Wu B, Liu ZF. Novel pseudorabies virus variant with defects in TK, gE and gI protects growing pigs against lethal challenge. Vaccine. 2015;33:5733–5740. doi: 10.1016/j.vaccine.2015.09.066. [DOI] [PubMed] [Google Scholar]
- Hua T, Tang B, Huang J, Chang C, Liu GY, Zhang XH, Hou JB, Zhang DH. Establishment of real-time PCR assay of pseudorabies virus and viral load detection of PRV attenuated vaccine. Acta Agric Jiangxi. 2019;31:73–78. [Google Scholar]
- Huang J, Ma G, Fu L, Jia H, Zhu M, Li X, Zhao S. Pseudorabies viral replication is inhibited by a novel target of miR-21. Virology. 2014;456–457:319–328. doi: 10.1016/j.virol.2014.03.032. [DOI] [PubMed] [Google Scholar]
- Huang J, Zhu L, Zhao J, Yin X, Feng Y, Wang X, Sun X, Zhou Y, Xu Z. Genetic evolution analysis of novel recombinant pseudorabies virus strain in Sichuan, China. Transbound Emerg Dis. 2020;67:1428–1432. doi: 10.1111/tbed.13484. [DOI] [PubMed] [Google Scholar]
- Jiang ZY, Li B, Cai Y, Yan JQ, Gong SY, Fan Y, Huang JB, Zhu L, Xu ZW. Multiplex PCR identification of pseudorabies vaccine strains and wild viruses. Jiangsu J Agric Sci. 2018;34:1307–1311. [Google Scholar]
- Jin HL, Gao SM, Liu Y, Zhang SF, Hu RL. Pseudorabies in farmed foxes fed pig offal in Shandong province, China. Arch Virol. 2016;161:445–448. doi: 10.1007/s00705-015-2659-9. [DOI] [PubMed] [Google Scholar]
- Kimman TG, de Wind N, Oei-Lie N, Pol JM, Berns AJ, Gielkens AL. Contribution of single genes within the unique short region of Aujeszky's disease virus (suid herpesvirus type 1) to virulence, pathogenesis and immunogenicity. J Gen Virol. 1992;73:243–251. doi: 10.1099/0022-1317-73-2-243. [DOI] [PubMed] [Google Scholar]
- Kong HJ, Zhang KS, Liu YJ, Shang YJ, Wu B, Liu XT. Attenuated live vaccine (Bartha-K16) caused pseudorabies (Aujeszky's disease) in sheep. Vet Res Commun. 2013;37:329–332. doi: 10.1007/s11259-013-9568-8. [DOI] [PubMed] [Google Scholar]
- Kou XJ, Gao F, Guo HL, Liu JF. Establishment of indirect ELISA antibody detection kit for gE protein of porcine pseudorabies virus. China J Anim Quar. 2018;35:91–94. [Google Scholar]
- Lan DS, Gu GB, Hou ZZ. Development of a Taqman real-time PCR for differentiation of wild-type strain from gE-deleted vaccine strain of pseudorabies virus. China Anim Husb Vet Med. 2018;45:1804–1812. [Google Scholar]
- Lee JY, Wilson MR. A review of pseudorabies (Aujeszky's disease) in pigs. Can Vet J. 1979;20:65–69. [PMC free article] [PubMed] [Google Scholar]
- Lei YL, Zhang W. Development and preliminary application of a colloidal gold immunochromatographic test strip for the detection of antibody against pseudorabies virus. Chin J Vet Med. 2016;52:35–37. [Google Scholar]
- Lei JL, Xia SL, Wang Y, Du M, Xiang GT, Cong X, Luo Y, Li LF, Zhang L, Yu J, Hu Y, Qiu HJ, Sun Y. Safety and immunogenicity of a gE/gI/TK gene-deleted pseudorabies virus variant expressing the E2 protein of classical swine fever virus in pigs. Immunol Lett. 2016;174:63–71. doi: 10.1016/j.imlet.2016.04.014. [DOI] [PubMed] [Google Scholar]
- Li YG, Gao XL, Sang XY, Wang HJ, Zhang CL, Wang AR, Yu ZJ, Ren ZG, Zheng XX, Feng N, Wang TC, Yang ST, Huang G, Gao YW, Xia XZ, Hu YH. Identification of pseudorabies virus (PRV) and analysis of the gB gene from tigers in China. J Pathog Biol. 2014;9:1057–1060. [Google Scholar]
- Li A, Lu GW, Qi JX, Wu LL, Tian KG, Luo TR, Shi Y, Yan JH, Gao GF. Structural basis of nectin-1 recognition by pseudorabies virus glycoprotein D. PLoS Pathog. 2017;13:e1006314. doi: 10.1371/journal.ppat.1006314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wei XB, Zhang XL, Xu H, Zhao XS, Zhou SF, Huang SB, Liu XY. Establishment of a multiplex RT-PCR assay for identification of atmospheric virus contamination in pig farms. Environ Pollut. 2019;253:358–364. doi: 10.1016/j.envpol.2019.07.028. [DOI] [PubMed] [Google Scholar]
- Li XD, Fu SH, Chen LY, Fan Li, Deng JH, Lu XC, Wang HY, Tian KG. Detection of pseudorabies virus antibodies in human encephalitis cases. Biomed Environ Sci. 2020;33:444–447. doi: 10.3967/bes2020.059. [DOI] [PubMed] [Google Scholar]
- Lian KQ, Zhang ML, Zhou LL, Song YW, Wang GD, Wang SS. First report of a pseudorabies-virus-infected wolf (Canis lupus) in China. Arch Virol. 2020;165:459–462. doi: 10.1007/s00705-019-04502-8. [DOI] [PubMed] [Google Scholar]
- Liang C, Tong W, Zheng H, Liu F, Wu JQ, Li GX, Zhou EM, Tong GZ. A high-temperature passaging attenuated Pseudorabies vaccine protects piglets completely against emerging PRV variant. Res Vet Sci. 2017;112:109–115. doi: 10.1016/j.rvsc.2017.02.008. [DOI] [PubMed] [Google Scholar]
- Lin WC, Shao YY, Tan C, Shen Y, Zhang XH, Xiao JF, Wu YT, He LL, Shao GM, Han MZ, Wang H, Ma JY, Xie QM. Commercial vaccine against pseudorabies virus: a hidden health risk for dogs. Vet Microbiol. 2019;233:102–112. doi: 10.1016/j.vetmic.2019.04.031. [DOI] [PubMed] [Google Scholar]
- Lin JX, Li ZL, Feng ZH, Fang Z, Chen JH, Chen WZ, Liang WW, Chen Q. Pseudorabies virus PRV strain with defects in gE, gC, and TK genes protects piglets against an emerging PRV variant. J Vet Med Sci. 2020;82:846–855. doi: 10.1292/jvms.20-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Tu CC, Zhang ML, Hou SK, Wang EB. Activity of vanadium-substituted heteropolytungstate to antipseudorabies virus. Chin J Appl Chem. 1998;15:41–44. [Google Scholar]
- Liu YZ, Rui P, Ma ZJ, Ma R, Yang WJ, Zhang ZZ. Isolation and identification of nyctereutes pseudorabies virus Rac 1 strain. Chin J Prev Vet Med. 2016;38:202–205. [Google Scholar]
- Liu H, Li XT, Hu B, Deng XY, Zhang L, Lian SZ, Zhang HL, Lv S, Xue XH, Lu RG, Shi N, Yan MH, Xiao PP, Yan XJ. Outbreak of severe pseudorabies virus infection in pig-offal-fed farmed mink in Liaoning Province, China. Arch Virol. 2017;162:863–866. doi: 10.1007/s00705-016-3170-7. [DOI] [PubMed] [Google Scholar]
- Liu LB, Wang JC, Geng YY, Wang JF, Yuan WZ. Development and application of the dual PRA different detection method for wide virus and vaccine strain of pseudorabies virus. Chin J Prev Vet Med. 2018;40:306–310. [Google Scholar]
- Liu Y, Zhang S, Xu Q, Wu JJ, Zhai X, Li S, Wang J, Ni J, Yuan L, Song X, Zhao B, Zhou Z, Wang C, Yang L. Investigation on pseudorabies prevalence in Chinese swine breeding farms in 2013–2016. Trop Anim Health Prod. 2018;50:1279–1285. doi: 10.1007/s11250-018-1555-1. [DOI] [PubMed] [Google Scholar]
- Liu H, Yang L, Shi Z, Lv R, Yang X, Wang C, Chen L, Chang H. Functional analysis of prv-miR-LLT11a encoded by pseudorabies virus. J Vet Sci. 2019;20:e68. doi: 10.4142/jvs.2019.20.e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MM, Ding GM, Fu YG, Liu AH, Chen JN, Li BY, Zeng QY, Liu GL. Establishment of an indirect ELISA antibody detection method for porcine pseudorabies virus. Chin J Prev Vet Med. 2019;41:484–488. [Google Scholar]
- Liu J, Chen C, Li X. Novel Chinese pseudorabies virus variants undergo extensive recombination and rapid interspecies transmission. Transbound Emerg Dis. 2020 doi: 10.1111/tbed.13784.10.1111/tbed.13784. [DOI] [PubMed] [Google Scholar]
- Liu Q, Wang X, Xie C, Ding S, Yang H, Guo S, Li J, Qin L, Ban F, Wang D, Wang C, Feng L, Ma H, Wu B, Zhang L, Dong C, Xing L, Zhang J, Chen H, Yan R, Wang X, Li W. A novel human acute encephalitis caused by pseudorabies virus variant strain. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa987. [DOI] [PubMed] [Google Scholar]
- Luo Y, Li N, Cong X, Wang CH, Du M, Li L, Zhao B, Yuan J, Liu DD, Li S, Li YF, Sun Y, Qiu HJ. Pathogenicity and genomic characterization of a pseudorabies virus variant isolated from Bartha-K61-vaccinated swine population in China. Vet Microbiol. 2014;174:107–115. doi: 10.1016/j.vetmic.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Lv C, Liu W, Wang B, Dang R, Qiu L, Ren J, Yan C, Yang Z, Wang X. Ivermectin inhibits DNA polymerase UL42 of pseudorabies virus entrance into the nucleus and proliferation of the virus in vitro and vivo. Antivir Res. 2018;159:55–62. doi: 10.1016/j.antiviral.2018.09.010. [DOI] [PubMed] [Google Scholar]
- Ma X, Cui Y, Qiu Z, Zhang B, Cui S. A nanoparticle-assisted PCR assay to improve the sensitivity for rapid detection and differentiation of wild-type pseudorabies virus and gene-deleted vaccine strains. J Virol Methods. 2013;193:374–378. doi: 10.1016/j.jviromet.2013.07.018. [DOI] [PubMed] [Google Scholar]
- Ma Z, Han Z, Liu Z, Meng F, Wang H, Cao L, Li Y, Jiao Q, Liu S, Liu M. Epidemiological investigation of porcine pseudorabies virus and its coinfection rate in Shandong Province in China from 2015 to 2018. J Vet Sci. 2020;21:e36. doi: 10.4142/jvs.2020.21.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaguarnera M, Khan H, Cauli O. Resveratrol in autism spectrum disorders: behavioral and molecular effects. Antioxidants. 2020;9:188. doi: 10.3390/antiox9030188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menotti L, Lopez M, Avitabile E, Stefan A, Cocchi F, Adelaide J, Lecocq E, Dubreuil P, Campadelli-Fiume G. The murine homolog of human Nectin1delta serves as a species nonspecific mediator for entry of human and animal alpha herpesviruses in a pathway independent of a detectable binding to gD. Proc Natl Acad Sci USA. 2000;97:4867–4872. doi: 10.1073/pnas.97.9.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne RS, Connolly SA, Krummenacher C, Eisenberg RJ, Cohen GH. Porcine HveC, a member of the highly conserved HveC/nectin 1 family, is a functional alphaherpesvirus receptor. Virology. 2001;281:315–328. doi: 10.1006/viro.2000.0798. [DOI] [PubMed] [Google Scholar]
- Minamiguchi K, Kojima S, Sakumoto K, Kirisawa R. Isolation and molecular characterization of a variant of Chinese gC-genotype II pseudorabies virus from a hunting dog infected by biting a wild boar in Japan and its pathogenicity in a mouse model. Virus Genes. 2019;55:322–331. doi: 10.1007/s11262-019-01659-x. [DOI] [PubMed] [Google Scholar]
- Moreno A, Sozzi E, Grilli G, Gibelli LR, Gelmetti D, Lelli D, Chiari M, Prati P, Alborali GL, Boniotti MB, Lavazza A, Cordioli P. Detection and molecular analysis of pseudorabies virus strains isolated from dogs and a wild boar in Italy. Vet Microbiol. 2015;177:359–365. doi: 10.1016/j.vetmic.2015.04.001. [DOI] [PubMed] [Google Scholar]
- Moreno A, Chiapponi C, Sozzi E, Morelli A, Silenzi V, Gobbi M, Lavazza A, Paniccià M. Detection of a gE-deleted pseudorabies virus strain in an Italian red fox. Vet Microbiol. 2020;244:108666. doi: 10.1016/j.vetmic.2020.108666. [DOI] [PubMed] [Google Scholar]
- Mravak S, Bienzle U, Feldmeier H, Hampl H, Habermehl KO. Pseudorabies in man. Lancet. 1987;1:501–502. doi: 10.1016/s0140-6736(87)92105-2. [DOI] [PubMed] [Google Scholar]
- Nie H, Fang R, Xiong BQ, Wang LX, Hu M, Zhou YQ, Zhao JL. Immunogenicity and protective efficacy of two recombinant pseudorabies viruses expressing toxoplasma gondii SAG1 and MIC3 proteins. Vet Parasitol. 2011;181:215–221. doi: 10.1016/j.vetpar.2011.04.039. [DOI] [PubMed] [Google Scholar]
- Ning HB, Li C, Zhang TT, Su J, Yang HC. Serological investigation of pseudorabies virus infection and its transmission risk factors on large scale pig farms in China during the 2016 to 2018 period. Anim Husb Vet Med. 2020;53:97–106. [Google Scholar]
- Pancheva SN. Potentiating effect of ribavirin on the anti-herpes activity of acyclovir. Antivir Res. 1991;16:151–161. doi: 10.1016/0166-3542(91)90021-I. [DOI] [PubMed] [Google Scholar]
- Pezzuto JM. Resveratrol: twenty years of growth, development and controversy. Biomol Ther. 2019;27:1–14. doi: 10.4062/biomolther.2018.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian P, Zhi XW, Wang B, Zhang HW, Chen HC, Li XM. Construction and immune efficacy of recombinant pseudorabies virus expressing PrM-E proteins of Japanese encephalitis virus genotype I. Virol J. 2015;12:214. doi: 10.1186/s12985-015-0449-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin SM, Liu JF, Chen FL, Mo YS, Li CT, Wu JM. Survey of infection with pseudorabies virus in wild animals in Guangxi and sequence analysis of gE gene. Chin J Prev Vet Med. 2015;37:344–347. [Google Scholar]
- Ren X, Li G, Sui X. Antiviral activities of phosphonoformate sodium to pseudorabies herpesvirus infection in vitro. Pharm Biol. 2011;49:608–613. doi: 10.3109/13880209.2010.538416. [DOI] [PubMed] [Google Scholar]
- Ren M, Lin H, Chen S, Yang M, An W, Wang Y, Xue C, Sun Y, Yan Y, Hu J. Detection of pseudorabies virus by duplex droplet digital PCR assay. J Vet Diagn Invest. 2018;30:105–112. doi: 10.1177/1040638717743281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner GR, Ahmad A, Davies JA. The infrequency of transmission of herpesviruses between humans and animals; postulation of an unrecognised protective host mechanism. Comp Immunol Microbiol Infect Dis. 2001;24:255–269. doi: 10.1016/S0147-9571(01)00014-5. [DOI] [PubMed] [Google Scholar]
- Song C, Gao L, Bai W, Zha X, Yin G, Shu X. Molecular epidemiology of pseudorabies virus in Yunnan and the sequence analysis of its gD gene. Virus Genes. 2017;53:392–399. doi: 10.1007/s11262-017-1429-z. [DOI] [PubMed] [Google Scholar]
- Su D, Wu S, Guo J, Wu X, Yang Q, Xiong X. Protective effect of resveratrol against pseudorabies virus-induced reproductive failure in a mouse model. Food Sci Biotechnol. 2016;25:103–106. doi: 10.1007/s10068-016-0105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui X, Yin J, Ren X. Antiviral effect of diammonium glycyrrhizinate and lithium chloride on cell infection by pseudorabies herpesvirus. Antivir Res. 2010;85:346–353. doi: 10.1016/j.antiviral.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Luo Y, Wang CH, Yuan J, Li N, Song K, Qiu HJ. Control of swine pseudorabies in China: opportunities and limitations. Vet Microbiol. 2016;183:119–124. doi: 10.1016/j.vetmic.2015.12.008. [DOI] [PubMed] [Google Scholar]
- Sun HF, Xun X, Dong J, Sun T, Bai J, Jiang P. Development of a blocking ELISA for the detection of gB proteinantibodies against pseudorabies virus in swine. Chin Vet Sci. 2018;48:712–716. [Google Scholar]
- Sun Y, Liang W, Liu QY, Zhao TT, Zhu HC, Hua L, Peng Z, Tang XB, Stratton CW, Zhou DN, Tian YX, Chen HC, Wu B. Epidemiological and genetic characteristics of swine pseudorabies virus in mainland China between 2012 and 2017. Peer J. 2018;6:e5785. doi: 10.7717/peerj.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Yuan XM, Liu YS, Cai X, Guo SY, Wang AB. Non-muscle myosin II: role in microbial infection and its potential as a therapeutic target. Front Microbiol. 2019;10:401. doi: 10.3389/fmicb.2019.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YD, Liu JT, Wang TY, An TQ, Sun MX, Wang SJ, Fang QQ, Hou LL, Tian ZJ, Cai XH. Live attenuated pseudorabies virus developed using the CRISPR/Cas9 system. Virus Res. 2016;225:33–39. doi: 10.1016/j.virusres.2016.09.004. [DOI] [PubMed] [Google Scholar]
- Tong W, Liu F, Zheng H, Liang C, Zhou YJ, Jiang YF, Shan TL, Gao F, Li GX, Tong GZ. Emergence of a pseudorabies virus variant with increased virulence to piglets. Vet Microbiol. 2015;181:236–240. doi: 10.1016/j.vetmic.2015.09.021. [DOI] [PubMed] [Google Scholar]
- Tong W, Li GX, Liang C, Liu F, Tian Q, Cao YY, Li L, Zheng XC, Zheng H, Tong GZ. A live, attenuated pseudorabies virus strain JS-2012 deleted for gE/gI protects against both classical and emerging strains. Antivir Res. 2016;130:110–117. doi: 10.1016/j.antiviral.2016.03.002. [DOI] [PubMed] [Google Scholar]
- Tong C, Chen Z, Liu F, Qiao Y, Chen T, Wang X. Antiviral activities of radix isatidis polysaccharide against pseudorabies virus in swine testicle cells. BMC Complement Med Ther. 2020;20:48. doi: 10.1186/s12906-020-2838-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong W, Zheng H, Li GX, Gao F, Shan TL, Zhou YJ, Yu H, Jiang YF, Yu LX, Li LW, Kong N, Tong GZ, Li JC. Recombinant pseudorabies virus expressing E2 of classical swine fever virus (CSFV) protects against both virulent pseudorabies virus and CSFV. Antivir Res. 2020;173:104652. doi: 10.1016/j.antiviral.2019.104652. [DOI] [PubMed] [Google Scholar]
- Wang CH, Yuan J, Qin HY, Luo Y, Cong X, Li Y, Chen J, Li S, Sun Y, Qiu HJ. A novel gE-deleted pseudorabies virus (PRV) provides rapid and complete protection from lethal challenge with the PRV variant emerging in Bartha-K61-vaccinated swine population in China. Vaccine. 2014;32:3379–3385. doi: 10.1016/j.vaccine.2014.04.035. [DOI] [PubMed] [Google Scholar]
- Wang T, Xiao Y, Yang Q, Wang Y, Sun Z, Zhang C, Yan S, Wang J, Guo L, Yan H, Gao Z, Wang L, Li X, Tan F, Tian K. Construction of a gE-deleted pseudorabies virus and its efficacy to the new-emerging variant PRV challenge in the form of killed vaccine. Biomed Res Int. 2015;2015:684945. doi: 10.1155/2015/684945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YM, Yuan J, Cong X, Qin HY, Wang CH, Li YF, Li S, Luo YZ, Sun Y, Qiu HJ. Generation and efficacy evaluation of a recombinant pseudorabies virus variant expressing the E2 protein of classical swine fever virus in pigs. Clin Vaccine Immunol. 2015;22:1121–1129. doi: 10.1128/CVI.00383-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Guo R, Qiao Y, Xu M, Wang Z, Liu Y, Gu Y, Liu C, Hou J. An inactivated gE-deleted pseudorabies vaccine provides complete clinical protection and reduces virus shedding against challenge by a Chinese pseudorabies variant. BMC Vet Res. 2016;12:277. doi: 10.1186/s12917-016-0897-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YP, Huang LP, Du WJ, Wei YW, Wu HL, Feng L, Liu CM. Targeting the pseudorabies virus DNA polymerase processivity factor UL42 by RNA interference efficiently inhibits viral replication. Antivir Res. 2016;132:219–224. doi: 10.1016/j.antiviral.2016.06.010. [DOI] [PubMed] [Google Scholar]
- Wang GS, Du Y, Wu JQ, Tian FL, Yu XJ, Wang JB. Vaccine resistant pseudorabies virus causes mink infection in China. BMC Vet Res. 2018;14:20. doi: 10.1186/s12917-018-1334-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JH, Ban FG, Zhao XL, Xie CH, Yan RQ, Wang SJ, Ma ZY, Wang DF, Wang C. Preparation of a Lateral flow assay for detection of giycoprotein B of pseudorabies virus in swine herds. China J Ani Quare. 2018;35:85–90. [Google Scholar]
- Wang J, Song Z, Ge A, Guo R, Qiao Y, Xu M, Wang Z, Liu Y, Zheng Y, Fan H, Hou J. Safety and immunogenicity of an attenuated Chinese pseudorabies variant by dual deletion of TK & gE genes. BMC Vet Res. 2018;14:287. doi: 10.1186/s12917-018-1536-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang Y, Zhang E, Zhou M, Lin J, Yang Q. Intranasal administration with recombinant bacillus subtilis induces strong mucosal immune responses against pseudorabies. Microb Cell Fact. 2019;18:103. doi: 10.1186/s12934-019-1151-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wang L, Zhao Q. Efficacy evaluation of two live virus vaccines against an emerging pseudorabies virus variant. Pol J Vet Sci. 2019;22:639–645. doi: 10.24425/pjvs.2019.129975. [DOI] [PubMed] [Google Scholar]
- Wang Y, Nian H, Li Z, Wang W, Wang X, Cui Y. Human encephalitis complicated with bilateral acute retinal necrosis associated with pseudorabies virus infection: a case report. Int J Infect Dis. 2019;89:51–54. doi: 10.1016/j.ijid.2019.09.019. [DOI] [PubMed] [Google Scholar]
- Wang D, Tao XG, Fei MM, Chen J, Guo W, Li P, Wang JQ. Human encephalitis caused by pseudorabies virus infection: a case report. J Neurovirol. 2020;26:442–448. doi: 10.1007/s13365-019-00822-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen SX, An LM, Zhao X, Zhang J, Pan YY, Ge WX. Establishment and application of Real-time PCR method for detection of pseudorabies virus. Jiangsu Agric Sci. 2019;47:50–53. [Google Scholar]
- Wong G, Lu JH, Zhang WH, Gao GF. Pseudorabies virus: a neglected zoonotic pathogen in humans? Emerg Microbes Infect. 2019;8:150–154. doi: 10.1080/22221751.2018.1563459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong RR, Abd-Aziz N, Affendi S, Poh CL. Role of microRNAs in antiviral responses to dengue infection. J Biomed Sci. 2020;27:4. doi: 10.1186/s12929-019-0614-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu QX, Zhang H, Dong HL, Mehmood K, Chang ZY, Li K, Liu SZ, Rehman MU, Nabi F, Javed MT, Zhu HY, Li JK. Seroprevalence and risk factors associated with pseudorabies virus infection in Tibetan pigs in Tibet. BMC Vet Res. 2018;14:25. doi: 10.1186/s12917-018-1347-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XL, Xiao L, Lin H, Yang ZX, Yao XP, Wang Y, Zhang PF, Jiang RJ. Establishment and preliminary application of multiplex PCR based on QIAxcel capillary gel electrophoresis to detect seven porcine viral pathogens. Chin J Prev Vet Med. 2019;41:824–829. [Google Scholar]
- Wunaerhan JSH, Ren YB, Meng FF, Tian SB, Fu YJ, Zhang JH, Li D, Meiremuguli Ren J, Lin HL, Cui ZZ, Chang S, Zhao P. A combined PCR with dot blot hybridization for pseudorabies virus. Chin J Anim Infect Dis. 2017;27:86–89. [Google Scholar]
- Xia LM, Sun QY, Wang JJ, Chen Q, Liu PH, Shen CJ, Sun JH, Tu YP, Shen SF, Zhu JC, Zhao HJ, Wang QZ, Li BL, Tao J, Soares Magalhaes RJ, Yan YX, Cai C. Epidemiology of pseudorabies in intensive pig farms in Shanghai, China: herd-level prevalence and risk factors. Prev Vet Med. 2018;159:51–56. doi: 10.1016/j.prevetmed.2018.08.013. [DOI] [PubMed] [Google Scholar]
- Xiao L, Gong F, Liu XN, Li XZ, Rao D, Lian XX, Huang BH, Zhang Y, Huang R, Guo PJ, Chen ML. Simultaneous detection of antibodies against porcine pseudorabies virus and porcine reproductive and respiratory syndrome virus with Liquid chip technology. Progress Vet Med. 2018;39:32–35. [Google Scholar]
- Xin CX, Zheng SX, Shi JL, Peng Z, Wu XY, Gao M, Liu YW, Wang S, Wang JB, Li J. Establishment and application of a multiplex PCR for detecting porcine circovirus type 2, Porcine pseudorabies virus and Porcine parvovirus. J Domest Anim Ecol. 2019;40:65–68. [Google Scholar]
- Xu LH, Peng Z, Zhao TT, Xu SX, Chen HC, Wu B. Development and preliminary application of a direct immunofluorescence method for the detection of pseudorabies virus. Chin J Pev Vet Med. 2017;39:993–997. [Google Scholar]
- Xu ZL, Huang YH, Li ZY, Liu ZL, Zhou SS, Qin YR, Qiu J, Pan MN. Establishment of a visualized LAMP method for detection of pseudorabies virus. China Anim Health Insp. 2017;34:79–82. [Google Scholar]
- Yang H, Han H, Wang H, Cui Y, Liu H, Ding S. A Case of human viral encephalitis caused by pseudorabies virus infection in China. Front Neurol. 2019;10:534. doi: 10.3389/fneur.2019.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Guan H, Li C, Li Y, Wang S, Zhao X, Zhao Y, Liu Y. Characteristics of human encephalitis caused by pseudorabies virus: a case series study. Int J Infect Dis. 2019;87:92–99. doi: 10.1016/j.ijid.2019.08.007. [DOI] [PubMed] [Google Scholar]
- Yao L, Wu CX, Zheng K, Xu XJ, Zhang H, Chen CF, Liu ZF. Immunogenic response to a recombinant pseudorabies virus carrying bp26 gene of brucella melitensis in mice. Res Vet Sci. 2015;100:61–67. doi: 10.1016/j.rvsc.2015.03.030. [DOI] [PubMed] [Google Scholar]
- Ye C, Zhang QZ, Tian ZJ, Zheng H, Zhao K, Liu F, Guo JC, Tong W, Jiang CG, Wang SJ, Shi M, Chang XB, Jiang YF, Peng JM, Zhou YJ, Tang YD, Sun MX, Cai XH, An TQ, Tong GZ. Genomic characterization of emergent pseudorabies virus in China reveals marked sequence divergence: evidence for the existence of two major genotypes. Virology. 2015;483:32–43. doi: 10.1016/j.virol.2015.04.013. [DOI] [PubMed] [Google Scholar]
- Ye S, Shao K, Li Z, Guo N, Zuo Y, Li Q, Lu Z, Chen L, He Q, Han H. Antiviral activity of graphene oxide: how sharp edged structure and charge matter. ACS Appl Mater Interfaces. 2015;7:21571–21579. doi: 10.1021/acsami.5b06876. [DOI] [PubMed] [Google Scholar]
- Ye C, Guo JC, Gao JC, Wang TY, Zhao K, Chang XB, Wang Q, Peng JM, Tian ZJ, Cai XH, Tong GZ, An TQ. Genomic analyses reveal that partial sequence of an earlier pseudorabies virus in China is originated from a Bartha-vaccine-like strain. Virology. 2016;491:56–63. doi: 10.1016/j.virol.2016.01.016. [DOI] [PubMed] [Google Scholar]
- Yin Y, Xu ZW, Liu XW, Li P, Yang F, Zhao J, Fan Y, Sun XG, Zhu L. A live gI/gE-deleted pseudorabies virus (PRV) protects weaned piglets against lethal variant PRV challenge. Virus Genes. 2017;53:565–572. doi: 10.1007/s11262-017-1454-y. [DOI] [PubMed] [Google Scholar]
- Yue D, Chen ZJ, Yang FL, Ye F, Lin S, He B, Cheng YW, Wang JC, Chen ZM, Lin X, Yang J, Chen H, Zhang ZL, You Y, Sun HL, Wen A, Wang LL, Zheng Y, Cao Y, Li YH, Lu GW. Crystal structure of bovine herpesvirus 1 glycoprotein D bound to nectin-1 reveals the basis for its low-affinity binding to the receptor. Sci Adv. 2020;6:eaba5147. doi: 10.1126/sciadv.aba5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai X, Zhao W, Li K, Zhang C, Wang C, Su S, Zhou J, Lei J, Xing G, Sun H, Shi Z, Gu J. Genome characteristics and evolution of pseudorabies virus strains in Eastern China from 2017 to 2019. Virol Sin. 2019;34:601–609. doi: 10.1007/s12250-019-00140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CF, Cui SJ, Zhu C. Loop-mediated isothermal amplification for rapid detection and differentiation of wild-type pseudorabies and gene-deleted virus vaccines. J Virol Methods. 2010;169:239–243. doi: 10.1016/j.jviromet.2010.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Huang J, Wang Q, He Y, Xu Z, Xiang M, Wu B, Chen H. Recombinant pseudorabies virus expressing P12A and 3C of FMDV can partially protect piglets against FMDV challenge. Res Vet Sci. 2011;91:90–94. doi: 10.1016/j.rvsc.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Zhang C, Guo L, Jia X, Wang T, Wang J, Sun Z, Wang L, Li X, Tan F, Tian K. Construction of a triple gene-deleted Chinese pseudorabies virus variant and its efficacy study as a vaccine candidate on suckling piglets. Vaccine. 2015;33:2432–2437. doi: 10.1016/j.vaccine.2015.03.094. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhong C, Wang JS, Lu Z, Liu L, Yang W, Lyu Y. Pathogenesis of natural and experimental pseudorabies virus infections in dogs. Virol J. 2015;12:44. doi: 10.1186/s12985-015-0274-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZZ, Wang GW, Hu DF, Niu YJ, Zhang MX, Lv CW, Lv L, Zhang QL, Xiao YH, Liu SQ. The occurrence of sheep pseudorabies and variation analysis of the glycoprotein E gene of the virus. Chin J Vet Med. 2016;52:46–48. [Google Scholar]
- Zhang T, Liu Y, Chen Y, Wang A, Feng H, Wei Q, Zhou E, Zhang G. A single dose glycoprotein D-based subunit vaccine against pseudorabies virus infection. Vaccine. 2020;38:6153–6161. doi: 10.1016/j.vaccine.2020.07.025. [DOI] [PubMed] [Google Scholar]
- Zhao XH, Cui QK, Fu QT, Song X, Jia RY, Yang Y, Zou YF, Li LX, He CL, Liang XX, Yin LZ, Lin JC, Ye G, Shu G, Zhao L, Shi F, Lv C, Yin ZQ. Antiviral properties of resveratrol against pseudorabies virus are associated with the inhibition of IκB kinase activation. Sci Rep. 2017;7:8782. doi: 10.1038/s41598-017-09365-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao WL, Wu YH, Li HF, Li SY, Fan SY, Wu HL, Li YJ, Lü YL, Han J, Zhang WC, Zhao Y, Li GL, Qiao XD, Ren HT, Zhu YC, Peng B, Cu LY, Guan HZ. Clinical experience and next-generation sequencing analysis of encephalitis caused by pseudorabies virus. Zhonghua yi xue za zhi. 2018;98:1152–1157. doi: 10.3760/cma.j.issn.0376-2491.2018.15.006. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Wang LQ, Zheng HH, Yang YR, Liu F, Zheng LL, Jin Y, Chen HY. Construction and immunogenicity of a gE/gI/TK-deleted PRV based on porcine pseudorabies virus variant. Mol Cell Probes. 2020;53:101605. doi: 10.1016/j.mcp.2020.101605. [DOI] [PubMed] [Google Scholar]
- Zheng LL, Guo XQ, Zhu QL, Chao AJ, Fu PF, Wei ZY, Wang SJ, Chen HY, Cui BA. Construction and immunogenicity of a recombinant pseudorabies virus co-expressing porcine circovirus type 2 capsid protein and interleukin 18. Virus Res. 2015;201:8–15. doi: 10.1016/j.virusres.2015.02.010. [DOI] [PubMed] [Google Scholar]
- Zheng H, Zhang Q, Zou M, Dong YQ, Liu S, Fan GC, Wu FX, Li XC. Prokaryotic expression on major epitope domain of gE for PRV SD and establishment of indirect ELISA for PRV gE antibody. China J Anim Quar. 2017;34:83–89. [Google Scholar]
- Zheng LH, Liu XJ, Yuan DQ, Li RX, Lu JH, Li XD, Tian KG, Dai EH. Dynamic cerebrospinal fluid analyses of severe pseudorabies encephalitis. Transbound Emerg Dis. 2019;66:2562–2565. doi: 10.1111/tbed.13297. [DOI] [PubMed] [Google Scholar]
- Zheng HH, Wang LQ, Fu PF, Zheng LL, Chen HY, Liu F. Characterization of a recombinant pseudorabies virus expressing porcine parvovirus VP2 protein and porcine IL-6. Virol J. 2020;17:19. doi: 10.1186/s12985-020-1292-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou SW, Sun LS. The diagnostic report of Pseudorabies (Aujeszky’s disease) J Anim Husb Vet Med. 1957;2:77–86. [Google Scholar]
- Zhou J, Li S, Wang X, Zou M, Gao S. Bartha-k61 vaccine protects growing pigs against challenge with an emerging variant pseudorabies virus. Vaccine. 2017;35:1161–1166. doi: 10.1016/j.vaccine.2017.01.003. [DOI] [PubMed] [Google Scholar]
- Zhu L, Guo WZ, Xu ZW. Fluctuant rule of colostral antibodies and the date of initial immunization for the piglets from sows inoculated with pseudorabies virus gene-deleted vaccine SA215. Chin J Vet Med. 2004;24:320–322. [Google Scholar]
- Zhu WJ, Zou M, Sun M, Liu JZ, Wang YJ. The anti-PRV and anti-PRRSV effects of Marine Bacillus S-12 low temperature lysozyme in vitro. Chin J Marine Drugs. 2013;32:26–33. [Google Scholar]
- Zhu HC, Liang W, Fan GF, Peng Z, Hua L, Tang XB, Chen HC, Wu B. Establishment of an indirect immunofluorescent antibody (IDF) detection method for PRV gE protein by using baculovirus expression system. Chin J Vet Sci. 2019;39:1428–1434. doi: 10.1007/s11430-018-9354-y. [DOI] [Google Scholar]