Abstract
The human gastrointestinal (GI) tract harbors gut microbiome, which plays a crucial role in preserving homeostasis at the intestinal host‐microbial interface. Conversely, specific gut microbiota may be altered during various pathological conditions and produce a number of toxic compounds and oncoproteins, in turn, to induce both inflammatory response and carcinogenesis. Recently, promising findings have been documented toward the implementation of certain intestinal microbiome in the next era of cancer biology and cancer immunotherapy. Notably, intestinal microbiota can cooperate with immune checkpoint inhibitors (ICIs) of its host, especially in enhancing the efficacy of programmed death 1 (PD‐1) protein and its ligand programmed death ligand 1 (PD‐L1) blockade therapy for cancer. Herein, we review the dual function of gut microbiota in triggering GI cancers, its association with host immunity and its beneficial functions in modulation of cancer immunotherapy responses. Furthermore, we consider the significance of gut microbiota as a potential biomarker for predicting the efficacy of cancer immunotherapy. Finally, we summarize the relevant limitations that affect the effectiveness and clinical applications of gut microbiome in response to immunotherapy.
Keywords: cancer immunotherapy, gut microbiomarkers, gut microbiota, immune checkpoint inhibitors, oncomicrobes, PD‐1/PD‐L1 inhibitors
In this review, we discuss the dual role of gut microbiome in triggering GI cancers, its association with host immunity and its beneficial functions in modulation of cancer immunotherapy responses. Furthermore, we consider the significance of gut microbiota as a potential biomarker for predicting the response to immunotherapy.
1. INTRODUCTION
The human gut microbiome is composed of a complex community of microbes, approximately 1013–1014 cells, which plays critical task in disease and health status. 1 The intestinal microbiota consists of different microorganism types including archaea, bacteria, viruses, fungi, and protozoa that live on and inside various humans’ organs. 2 , 3 Different physiological acts can be attributed to gut microbiome, particularly inflammation, metabolism and immunity. 4 , 5
The immune system exploits different effector responses, cells and factors to eliminate pathogenic microbes and cancerous cells. 6 Notably, gut microbiota destruction, identified as “dysbiosis,” has been correlated with a number of inflammatory conditions. 4
Intestinal dysbiosis of healthy gut microbiota results in deterioration of mutualistic relationship and may associate with many diseases like metabolic syndrome, type 1 and type 2 diabetes, obesity, inflammatory bowel disease (IBD), irritable bowel syndrome (IBS), different types of cancers particularly gastrointestinal (GI) cancers (Table 1). 7 , 8
TABLE 1.
Potential gut microbiomarkers associated with different types of GI cancers
| Implicated microbiota | Type of cancer | Mode of action | Ref. |
|---|---|---|---|
| Helicobacter pylori ↑ | Gastric cancer |
|
16 |
| Fusobacterium nucleatum↑ | Colon cancer |
|
7, 17, 18 |
| Streptococcus gallolyticus ↑ | Colon cancer |
|
7, 17 |
| Enterococcus faecalis ↑ | Colon cancer |
|
7, 17 |
| Enterotoxogenic Bacteroides fragilis (ETBF) ↑ | Colon cancer |
|
7, 17 |
| Genotoxic Escherichia coli ↑ | Colon cancer |
|
7, 17 |
| Porphyromonas gingivalis ↑ | Pancreatic cancer |
|
18 |
| Clostridium spp.↑ | Liver cancer |
|
19 |
Through changes in the intestinal lumen, certain commensal microbiota can quickly proliferate and acquire pathogenic features, such as vancomycin‐resistant Enterococcus or Clostridium difficile. 9 , 10 Gut microbiome complies all the prerequisites for representation as an endocrine body structure due to its plasticity and capability of producing various biologically functional components. 11 , 12 These metabolic by‐products and biologically active compounds like hormones that are released from this so‐called endocrine organ may circulate and disseminate to other body sites, and affect different pivotal biological procedures. 11 Recent evidence strongly supports the important role of gut microbiota as a new therapeutic option in cancer treatment. 13 Moreover, gut microbiota and their released metabolites have profound impacts on the development and response of peripheral immune system, and also it was demonstrated that can improve the therapeutic effectiveness of immune checkpoint inhibitors (ICIs) against cancerous cells. 14 , 15 Herein, we aimed to review the relationship between gut microbiota, host immune response and cancer immunotherapy, with a focus on the interaction of gut microbiota and ICIs. Also, we brought up the related pitfalls and challenges that may potentially affect the therapeutic capacity of microbiota in cancer immunotherapy. Furthermore, we discussed the possible role of chronic infections or inflammation that may interfere with cancer immunotherapy.
1.1. Gut microbiome and host immune system
Recent studies have suggested critical roles for the gut microbiome in the educating and development of major players of the host immunity through a complex microbiota‐immunity crosstalk in both homeostatic conditions and diseases. 20 , 21 These multifaceted dialogs not only authorize the immunological tolerance of commensal bacteria, but also enable the host immune cells to identify and begin an assault against microbial pathogens. Disturbance in the gut microbiome equilibrium is termed dysbiosis, which can result in considerable alterations in the taxonomical composition as well as the metagenomic functions of the gut microbiota and induce the overgrowth (blooming) of otherwise less abundant or potentially deleterious microbiota such as pathobionts. 22 , 23 , 24 Once the dysbiosis occurred, it can directly or indirectly result in functional impairment of local, locoregional, and systemic immune responses leading to disintegration of epithelial barriers, and subsequently delivery of mucosa‐associated microbes and their components into the mesenteric lymph nodes (MLNs) and into the peripheral circulation. 23 Moreover, dysbiosis‐associated inflammation can recruit neutrophils to the intestinal epithelium, alter the inflammatory cytokine and chemokine profiles, activate the T helper 17 (Th17) and effector T cells, which in turn may cause a negative feedback control of the gut microbiota. 25 , 26 , 27
It has been well established that intestinal microbiota remarkably modulates and controls the development and operation of both the innate and adaptive immune systems. The microbial components and biomolecules, called microbe‐ or pathogen‐associated molecular patterns (MAMPs or PAMPs), and also their sensors named pattern recognition receptors (PRRs), are the key players which mediate the conversation between microbiota and host innate immune cells such as monocytes/macrophages, dendritic cells (DCs), and natural killer (NK) cells. 28 , 29 , 30
In homeostatic conditions, the immune system orchestrates tolerance to beneficial intestinal microbiota such as Bifidobacterium and Lactobacillus species, while strongly reacts against the virulent microorganisms and opportunistic pathogens or pathobionts mainly through induction of the profound pro‐inflammatory responses. 31 , 32 , 33 , 34 Hence, there is a natural and prudent immunosurveillance system in the intestinal lumen which carefully monitors the microbial communities for maintaining the host‐microbiota mutualism and host defense. Moreover, normal intestinal flora can generate and synthesize various immunomodulatory compounds and metabolites such as short‐chain fatty acids (SCFAs) like propionate, acetate, and butyrate, and also secondary bile acids and ubiquitous bacterial fermentation products. 12 , 35 Of note, SCFAs act as effective inhibitors of histone deacetylases (HDACs) and lysine deacetylase (KDAC) in innate immune cells such as macrophages and DCs. 36 , 37 , 38 , 39 Furthermore, these bioactive agents are capable to interact with the over‐mentioned receptors on the immune cells and adjust their size, metabolic processes and functions which may result in host health benefits. 32 , 40 Thus, understanding the involved mechanisms behind the interactions between gut microbiome and immune system can be utilized to design and develop novel therapies to treat immune‐mediated and immune‐associated diseases.
1.2. Gut microbiota and NK cells
NK cells are key players of the innate immune system, and are characterized by the surface expression of marker CD56 and the lack of CD3 expression. 41 This group of innate immune cells represents a heterogeneous subset of large granular lymphocytes, and constitutes nearly 5%–20% of all peripheral lymphocytes which are engaged in the clearance of virus‐infected cells and lysis of tumor cells. 42 , 43 , 44 Beside their cytotoxic effector functions, NK cells are significantly involved in regulating the immune response by producing several cytokines and chemokines, mainly interferon‐γ (IFN‐γ) and tumor necrosis factor (TNF)‐α upon stimulation, to modulate other types of cells related to both the adaptive and innate immune responses. 44 , 45 , 46 It has been shown that NK cells are not normally active killers but rather require to be completely activated in a process known as NK cell priming. 47
Regarding the prominent function of NK cells in the biology of cancer, they obviously represented as forthcoming immunotherapeutic targets for the treatment of different malignancies, and a rising number of studies and ongoing clinical trials support the use of various therapeutic agents that target NK cell‐related pathways as cell‐based cancer immunotherapies. 48 , 49 The continuous existence of metabolites/products/ligands (e.g., LPS, peptidoglycan, SCFAs, and AhR ligands) originated from gut microbiota can induce the differentiation and activity of myeloid (monocytes/macrophages) lineage, including NK cells, and bone marrow progenitors, and also various groups of innate lymphoid cells (ILCs) through interacting with PRRs. 21 , 50 , 51 Totally, NK cells play a critical role in response to gut microbial invasion, mainly via secretion of IFN‐γ, which can provoke recruitment of further NK cells from peripheral blood to augment the antimicrobial immune responses. 52 These immune cells encounter a great number of antigens derived from commensal or potentially pathogenic microbes or pathobionts shaping the gut microbiome. Moreover, the crosstalk between NK cells and gut microbiota can lead to induction of adaptive T cell‐mediated immunity through interacting with professional antigen presenting cells (APCs) such as DCs. 52 , 53 It is also suggested that NK cells can evoke an intestinal inflammatory response during microbial invasions in the gut, which is irrespective of viral and tumor elimination. Also, these innate cells can exploit different toll‐like receptors (TLRs) to interact with various bacterial components like PAMPs, MAMPs, LPS, peptidoglycans, viral dsRNA, and DNA with CpG motifs to elicit inflammatory responses. 54
NK cells have crucial roles in early defense against viral infections and a variety of tumors, and are involved in DC maturation, indicating a DC‐NK interplay which is of vital significance in antitumor immunity, and emphasizes the rationale for inspecting this crosstalk in the expansion of more efficacious cancer immunotherapies. 55 , 56 , 57 On the contrary, certain strains of gut microbiota have been observed to modulate gut‐associated lymphoid tissue (GALT), enhancing the functional capability of innate immune response, activating DCs, and promoting NK cells though a direct cytochemical pathway by pathogens which invade the epithelial layer of the host gut. 42 , 52 It has been documented that NK cell priming and antiviral immune response were seriously compromised in germ‐free (GF) mice, which suggests that the presence of commensal microbiota is required to calibrate the function and priming of NK cells in GF mice. 47 Furthermore, lactic acid bacteria (LAB) have been demonstrated to have considerable impact on maturation of DCs, therefore, activating NK cells. 58 It has been shown that some strains of gut‐derived interleukin (IL)‐12‐inducing LAB can stimulate various subsets of DCs such as blood DCs and lymph node (LN) DCs, and activate NK cells to secrete IFN‐γ. 42 , 59 , 60 Also, certain strains of probiotic bacteria that originated from a healthy gut microbiome, in particular lactobacilli and bifidobacteria, were reported to be involved in activation of NK cells, their functionality and cytotoxicity as a result of DC‐NK interplay. 42 , 61 , 62 Taken together, these observations should represent a convincing rationale to explore the ligand‐receptor interactions between NK cells and healthy gut microbiota, which can be exploited as innovative targeted immunotherapies to help those with different conditions of intestinal inflammatory diseases associated with the gut immune system.
1.3. Role of oncomicrobes in cell proliferation and cancer initiation
Oncomicrobes contain microorganisms that induce direct DNA mutations and change host cellular signal transduction pathways. Until recently, oncomicrobes were mostly recognized to be viral agents such as human papillomavirus (HPV) that integrate their oncogenes inside the genetic content and frequently target the genes associated in various cancers. 63 However, a few numbers of microbes are known as true oncomicrobes partially because of restrictions in recognizing microorganisms as irregular causes of cancers. The responsible microorganism may be depleted in the cancerous locations because it may have launched cellular injury via a “hit‐and‐run” strategy after a quick exposure to host cells. 63 , 64 In spite of lacking sufficient information associating cancer with specific bacterial species, various direct, and indirect plans are proposed by which they can induce different carcinogenesis pathways. Certain microbial species have evolved competitive approaches that contain the capacity to cause DNA damage of competing microorganisms. Also, such strategies can change host DNA material by forcing genetic alterations that may be involved in tumorigenesis. In addition, microbial DNA may be inserted into the host cellular genomes, especially the mitochondrial genetic content, via RNA intermediate molecules. These events occur mostly in cancerous tissues than normal adjacent tissues. 63 Certain bacterial proteins are documented to induce signaling pathways involved in the host cellular cascades that modulate cell proliferation and stemness. For instance, Wnt/β‐catenin pathway, is aberrantly regulated via components generated by a number of bacteria, consisting of Salmonella typhi, Fusobacterium nucleatum, and Helicobacter pylori. 7 , 63 , 65 DNA damage may also occur by bacterial toxins. For example, Escherichia coli producing colibactin, a newly identified substituted spirobicyclic molecule, induces crosslinking of double‐stranded DNA, 66 , 67 and cytolethal distending toxin (CDT) expressed by ƹ‐ and γ‐proteobacteria, demonstrates DNase activity and can directly induce DNA breaks 68 (Figure 1).
FIGURE 1.
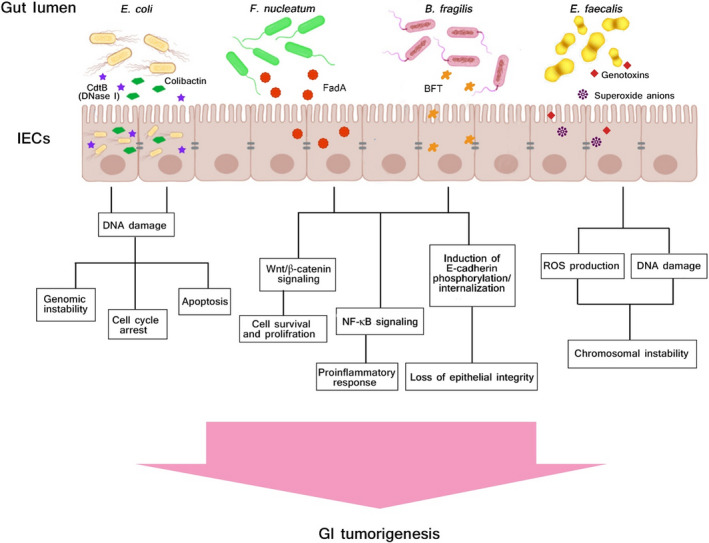
Schematic illustration of host‐microbiome interplay as potential trigger of gastrointestinal (GI) cancer. The mechanisms underlying the effects of certain gut microbiota and microbiome‐derived toxins and metabolites as potential triggers of GI tumorigenesis are described. Moreover, a series of pathways and process of carcinogenesis by which the gut microbiota may be involved in the genesis and development of GI tumorigenesis are depicted in the picture and mentioned in this review
2. CANCER IMMUNOTHERAPY
Cancer immunotherapy has recently attracted a great attention in the next era of cancer treatment. This new therapeutic strategy employs the host immune system to render antitumor effects against cancerous cells. 69 Recently, ICIs are introduced as promising immunotherapeutic biomolecules, which have shown hopeful clinical outcomes in treatment of various cancers, as shown by monoclonal antibodies (mAbs) blocking cytotoxic T‐lymphocyte antigen‐4 (CTLA‐4), programmed cell death ligand 1 (PD‐L1) and programmed cell death protein 1 (PD‐1). 69 , 70 However, development of primary and acquired resistance throughout the duration of treatment period may decrease the ubiquitous clinical use of ICIs. 71 Of note, selection of appropriate cases is critical to prevent subsequent resistance to such drugs and increase the efficacy of ICIs. 72 Thus, robust attempts to combat the resistance to immunotherapy are extremely required.
It was observed that tumor cells expressing PD‐L1 induced apoptosis of co‐cultured activated effector T cells, and this process was inhibited by an antihuman PD‐L1 mAb. 73 , 74 In addition, the growth of murine tumors expressing PD‐L1 was blocked in syngeneic mice by the antimurine PD‐L1 mAb. Furthermore, similar findings were achieved through the examination of a variety cancer cells using animal models. 75 , 76 , 77 These important findings opened the way to run several clinical trials exploiting mAbs targeting PD‐1, PD‐L1, and CTLA‐4 in cancer immunotherapy for different kinds of cancers. Presently, the U.S. Food and Drug Administration (FDA) have authorized the consumption of some mAbs including: cemiplimab (Libtayo), pembrolizumab (Keytruda), avelumab (Bavencio), atezolizumab (Tecentriq), durvalumab (Imfinzi), and nivolumab (Opdivo) for targeting PD‐1 and PD‐L1 in cancer immunotherapy. 78 , 79 , 80 Also, ipilimumab (Yervoy) that targets the CTLA‐4 was demonstrated to function synergistically with nivolumab to induce T‐cell antitumor activity in melanoma and small lung cell carcinoma. 81 Despite the obvious efficacy of PD‐L1, PD‐1, and CTLA‐4 suppression in cancer therapy, not all patients responded to these treatments. Therefore, practical strategies to enhance the effectiveness of cancer immunotherapy are demanded. 6 , 82
2.1. Interaction of PD‐1 and PD‐L1 in tumor microenvironment
In anticancer immunity, the immune system recognizes the tumor‐specific antigens expressed through gene mutations, and specific CD8+ cytotoxic T lymphocytes (CTLs) are recruited to the sites of tumor targeting the corresponding antigens. 83 This certain cluster of effector CTLs identify the target tumor cells and induce programmed cell death of cancerous cells. Surprisingly, cancerous cells exploit different tactics to escape immune surveillance. For instance, they resist neutralizing effects of the antitumor CTLs by enhancing the expression level of PD‐L1 in tumor ecosystem. 75 , 84 Healthy host cells normally do not produce noticeable level of PD‐L1 on their surfaces, while PD‐L1 is significantly produced by tumor cells, immune, and nonimmune cells. 6 , 85 Interferon gamma (IFN‐γ) cytokine that is secreted by the infiltrating antitumor CTLs into tumor microenvironment, plays a key role in induction of PD‐L1 expression. 85 , 86 Moreover, some other cytokines like IL‐4, IL‐10, and TNF‐α can also upregulate the PD‐L1 expression. 87 , 88 The interplay between PD‐1 and PD‐L1 in tumor ecosystem capacitates the tumor cells to withstand the endogenous antitumor functions excreted from the host immune response. 87 The interaction of PD‐L1 in tumor tissues with expressed PD‐1 on the activated T cells impairs the normal functions of effector T cells via multiple strategies, like induction of T cell programmed cell death, exhaustion, and anergy. 6 , 69 , 85 , 86 Recently, it was shown that crosstalk of PD‐1 with PD‐L1 expressed on tumor‐related macrophages prohibits the phagocytic capacity of macrophages against tumor cells. 89 The significance of PD‐1 and PD‐L1 interplay in cancer cell escape promoted the utilization of such biomolecules as prominent therapeutic agents in immunotherapy of cancer. 87 , 88 , 89
2.2. Gut microbiome and immunotherapy responses
Today, growing evidence has revealed that gut microbiome can play a key role in the modulation of immunotherapy responses in patients under treatment by immunotherapeutic drugs such as ICIs. 90 , 91 , 92 The host response to ICIs, PD‐1/PD‐L1 blockade or CTLA‐4 inhibition, could be affected by the composition of intestinal microbiome. 93 , 94 , 95 Upon PD‐1/PD‐L1 inhibition, mice with various intestinal microbial compositions have been shown to exert different responses to the anticancer immunotherapy. 93 Gut microbiota analysis depicted that bifidobacteria were enhanced in mice with slow tumor growth, and exhibited promising responses to anti‐PD‐1 therapy. These favorable influences from mice having a more beneficial microbiome may be transported to other mice through fecal microbial transplantation (FMT). 92 FMT is an effective strategy to normalize the intestinal microbiota which has already been employed in various clinical indications such as IBD, IBS, multiple sclerosis (MS), different type of cancers, and particularly in treatment of recurrent Clostridioides (formerly, Clostridium) difficile (rCDI) infection that do not response to conventional antimicrobial therapies. 96 , 97 , 98 , 99 , 100 , 101 , 102 , 103 FMT is defined as a therapeutic procedure that involves transplantation of the entire intestinal microbiota from a healthy donor into the intestinal tract of a patient to completely rebuild and normalize the structure and functionality of gut microbiome. 104 , 105 , 106 In recent years, FMT also has attracted great interest to be applied along with cancer immunotherapy for solid tumor malignancies, specifically for improving the efficacy of ICIs. 92 Together, due to enhancing the systemic and antitumor immune response in cancer patients, FMT could be administrated as a dramatic tool for the treatment of patients receiving ICIs.
Furthermore, the antitumor activity of PD‐L1 inhibition was increased when mice having an unpleasant gut microbiota were provided with oral probiotics containing Bifidobacterium bacteria. 107 Such effects mostly arose from the maturation induction of dendritic cells DCs that lead to enhancement of cellular function of the tumor‐specific CD8+ T cells. 107 Following CTLA‐4 blockade treatment, the richness of intestinal microbiota clearly differed in mice, as indicated by the relative enrichment of Burkholderiales and Bacteroidales and reduction of Clostridiales. 93 Furthermore, mice oral feeding with Bacteroides thetaiotaomicron, Bacteroides fragilis, and Burkholderia cepacia increased the effectiveness of anti‐CTLA‐4 treatment by inducing T helper 1 (Th1) response and improving DC maturation. Nevertheless, consumption of broad‐spectrum antimicrobials in GF and specific‐pathogen‐free (SPF) mice significantly reduced the activity of anti‐CTLA‐4 treatment. This effect might be restored via FMT from individuals having predominant species of Bacteroides. 93
Recent investigations have also confirmed the significance of intestinal microbiome in improving the effectiveness of cancer immunotherapy. 69 , 92 With PD‐1/PD‐L1 blockade therapies, the overall survival and the progression‐free survival (PFS) rates were significantly elevated in cases with epithelial tumors whom did not consume antimicrobials for routine purposes compared to cases with tumor that received antibiotics. 108 This phenomenon declares that antibiotic usage may cause intestinal dysbiosis, hence, hindering the antitumor immunity and immune checkpoint blockade responses. Data obtained from the comprehensive metagenomic sequencing of fecal specimens from such cases demonstrated that responder participants to anti‐PD‐1 treatment had various compositions of intestinal microbiota, which were enriched in Alistipes and Akkermansia. 69 Before PD‐1 blockade therapy, FMT was exploited in GF mice using fecal samples from responder donors that strengthened the immunity, while immune response of GF mice taking FMT from nonresponder donors was restored using Akkermansia muciniphila alone or in combination with Enterococcus hirae. 69 Importantly, A. muciniphila was associated with enhanced infiltration of immune cells in tumor sites as CCR9+CXCR3+CD4+ T cells were migrated to the site of tumor, and CD4+ T cells to CD4+FoxP3+ T cells (Tregs) ratio was elevated. 91 , 107 In subjects with metastatic melanoma, the gut microbiome diversity was remarkably enhanced in responder cases to PD‐1 blockade treatment, and specific bacterial species were relatively more enriched, like Faecalibacterium, Ruminococcaceae, and Clostridiales. 63 , 66 However, nonresponder patients had less diverse population of gut microbiota and higher abundance of Bacteroidales. 92 Analysis of the intestinal microbiome composition and the immunological patterns in the cancerous tissue showed that the expression of specific markers of cytotoxic T cells and antigen display were enhanced in individuals with beneficial intestinal microbiome in comparison with subjects having inappropriate gut microbiome. 92 It was reported that tumor microenvironment of cases who responded to anti‐PD‐1 was abundant in Collinsella aerofaciens, Bifidobacterium longum, and Enterococcus faecium. 70 Moreover, transfer of responder fecal specimens to GF mice positively reproduced the dominant phenotype, lower rate of tumor growth and promoted therapeutic impacts compared with mice that received nonresponder fecal samples. Consequently, these restorations of gut microbiota led to a rise in the overall population of CD8+ T cells and a reduction in Tregs in the tumor site. 95
3. COMMENSAL MICROBIOTA AS POTENTIAL CONTROLLER OF CANCER IMMUNOTHERAPY
3.1. Beneficial microbiota
Results obtained from metagenomic studies using 16S ribosomal RNA (16S rRNA) sequencing revealed that Bifidobacterium adolescentis, Bifidobacterium breve, and Bifidobacterium longum were associated with increased efficacy of drugs used for cancer immunotherapy. 6 , 107 The function of these microbes in increasing defensive immune responses against tumors were subsequently evaluated by administering mice having solid tumors with B. longum and B. breve cocktail via oral feeding. 107 In this experiment, Bifidobacterium‐treated mice demonstrated significant improvement in controlling tumor outgrowth as compared to untreated mice. It is hypothesized that Bifidobacterium cocktail can cooperate with immune checkpoint blockade to promote and activate antitumor immunity as depicted in Figure 2. Since Bifidobacterium species enhanced the anti‐melanoma effects by induction of innate immunity, the application of Bifidobacterium cocktail against tumor growth can be expanded to other types of cancers. Some of the typical bacterial species and viral agents that have been proposed to be positively or negatively linked to anti‐PD‐1 and anti‐PD‐L1 therapies are presented in Table 2.
FIGURE 2.
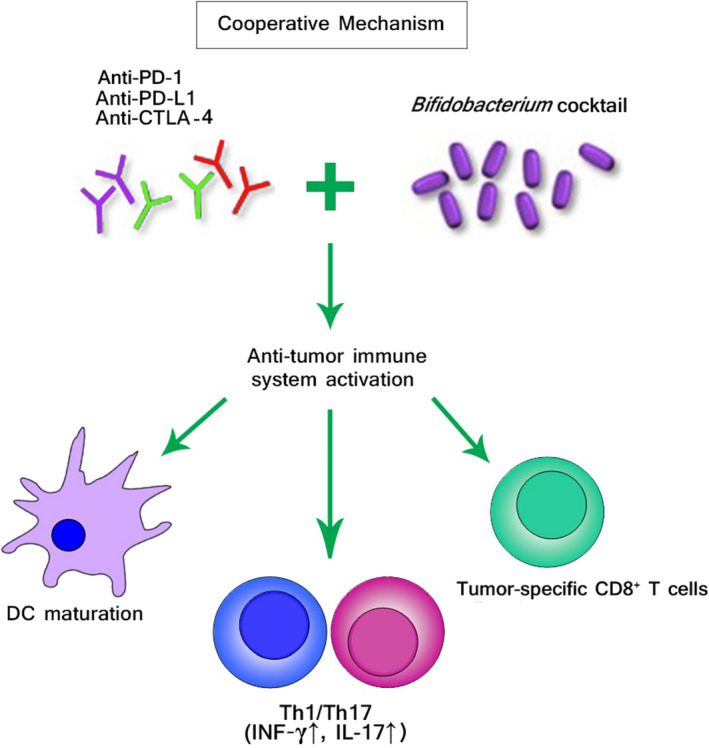
The putative effects of commensal microbiota on cancer immunotherapy. Certain beneficial microbial species are known to have a range of effects on host antitumor immune responses, and cancer immunotherapy. Bifidobacterium cocktail cooperates with immune checkpoint inhibitors (ICIs) blockade to promote and activate antitumor immunity. The identification of such cooperative mechanism may provide a novel and promising prospect for probiotic‐based therapies that could be integrated with cancer immunotherapy to ameliorate patient outcomes and even convert nonresponders
TABLE 2.
Microbial species and viral agents that are positively and negatively associated with PD‐1 and PD‐L1 blockade immunotherapy
| Microbiota | Main effects on immunity | Potential effect on immunotherapy | Ref. |
|---|---|---|---|
| Beneficial microbiota | Enhanced the antitumor efficacy of PD‐L1 blockade, enhancement of DC maturation, improving activity of the tumor‐specific CD8+ T cells, increased IFN‐γ production | Effective | 90, 107 |
| Bifidobacterium | |||
| Bacteroides fragilis, Bacteroides thetaiotaomicron, Burkholderia cepacia | Increased the efficacy of anti‐CTLA‐4 therapy by inducing Th1 response and promoting DC maturation, an increase in CD8+ T cells and a decrease in Tregs in the tumor environment | Effective | 93 |
| Akkermansia muciniphila | Enhanced the infiltration of immune cells in tumor site, as CCR9+CXCR3+CD4+ T cells were recruited to the tumor microenvironment and the ratio of CD4+ T cells to CD4+FoxP3+ T cells (Tregs) was enhanced | Effective | 69 |
| Enterococcus hirae | Enhanced IL‐12 secretion by DCs | Effective | 110 |
| Harmful microbiota | Increased host PD‐1 and PD‐L1 expression, higher level of pro‐inflammatory cytokines (TNF‐α), suppressed the proliferation of CD4+ T cells, the inhibitory effect can be blocked using antibodies PD‐L1 | Ineffective | 111 |
| Helicobacter pylori | |||
| HBV, HCV, HPV, EBV | Established chronic infections in humans and increased host PD‐1 or PD‐L1 expression | Ineffective | 6 |
Abbreviations: CTLA‐4, cytotoxic T‐lymphocyte‐associated protein 4; DC, dendritic cell; EBV, Epstein‐Barr virus; HBV, hepatitis B virus; HCV, hepatitis C virus; HPV, human papillomavirus; IFN‐γ, interferon gamma; PD‐1, programmed death 1; PD‐L1, programmed death ligand 1; Th1, T helper type 1; TNF‐α, tumor necrosis factor‐α; Tregs, regulatory T cells.
Recently, another study examined the fecal specimens of metastatic melanoma cases prior to PD‐1 blockade cancer treatment, and demonstrated that abundance of C. aerofaciens, E. faecium, and B. longum were higher in the PD‐1 blockade immunotherapy responders, underpinning the antitumor actions of such microbes. 70 Also, Frankel et al. proved that patients bearing melanoma who responded to ICIs were populated with Bacteroides caccae. 95 Moreover, they presented that the kind of bacterial species which are increased within responders are most probably to be associated on the type of antibodies used against cancer immunotherapy. The gut microbiota of cases who responded to nivolumab (targeting PD‐1) were abundant with Holdemania filiformis, Faecalibacterium prausnitzii and Bacteroides thetaiotaomicron, while cases who responded to pembrolizumab (targeting PD‐1) were populated with Dorea formicogenerans. Nevertheless, the exact mechanisms behind these alterations are not well understood. 95 Wargo et al. examined the human gut microbiome in participants with PD‐1 blockade therapy by whole genome shotgun sequencing and 16S rRNA metagenomics, and discovered that composition and diversity of bacteria in participants who responded to the immunotherapy were notably varied from that in participants who did not respond to the immunotherapy. The responders showed more diverse bacterial composition and higher number of Clostridiales, while the nonresponders were enriched with Bacteroidales. 109 In another study, the effects of gut microbiome in anti‐PD‐1 therapy were investigated in patients bearing different cancers consisting of lung cancer, urothelial carcinoma, and renal cell carcinoma. They showed that cases who received antimicrobials prior or soon after beginning the anti‐PD‐1 treatment had diminished rate of survival, unless responders were enriched by A. muciniphila. They also observed that administration of A. muciniphila to SPF or GF mice was capable to rebuild the antitumor effects of anti‐PD‐1 therapy which was prevented by antibiotic usage. 69 However, the precise mechanisms by which A. muciniphila enhances anti‐PD‐1 immunotherapy needs to be clarified.
3.2. Harmful microbiota
In recent studies, unmethylated CpG oligodeoxynucleotides that are frequently found in bacterial chromosomes, were documented to increase the antitumor function of CD8+ T cells by reducing PD‐1 expression through the IL‐12 cascade, proposing that intestinal microbiota that are positively related to anti‐PD‐1 and anti‐PD‐L1 immunotherapy may produce some metabolites which directly suppress PD‐1 and PD‐L1 expression. 111 , 112 Also, it seems likely that gut microbiota indirectly affect PD‐L1/PD‐1 expression via both systematically or locally mediating immune functions, thus, impacting the efficacy of anti‐PD‐1 and anti‐PD‐L1 treatment. 6 For instance, polysaccharide A from B. fragilis was shown to stimulate Th1 cell responses. 113 In addition, it was shown that oral feeding therapy with neomycin resulted in compromised immunity to infection by respiratory influenza virus, which was associated with significant reduction in the population of Gram‐positive bacteria in the intestine but not the nasal tract. 114 Furthermore, there are well‐known microbial agents that are directly responsible for chronic infections in humans, some of them are identified to enhance PD‐1/PD‐L1 expression in host tissues. 115 , 116 For instance, H. pylori infection is one of the most prevalent human infections that can develop chronic active gastritis, peptic ulcers, and gastric adenocarcinoma. 117 , 118 Actually, H. pylori‐infected patients have a considerable secretion level of pro‐inflammatory cytokines, like TNF‐α 119 , 120 , 121 and higher production of PD‐L1 in gastric tissue as observed in a gastric cell line model of epithelial cells. 115 , 119 , 120 Furthermore, H. pylori suppressed the proliferation of human CD4+ T cells originated from blood sample, however, such repressive impact can be inhibited by using antibodies against PD‐L1. 116 Moreover, enhanced level of PD‐L1 expression was observed in gastric tissues of H. pylori‐infected patients, and also coculture of H. pylori‐infected primary gastric epithelial cells with T cells resulted in overexpression of PD‐L1 on gastric epithelial cells, which eventually led to induction of apoptosis in T cells. Taken together, these findings propose that H. pylori infection could induce a nonspecific suppression of circulating T cells, more importantly tumor‐specific T cells. Additionally, many viruses such as hepatitis B virus (HBV), hepatitis C virus (HCV), human papillomavirus (HPV), and Epstein‐Barr virus (EBV) are also capable to cause chronic infections and enhance human PD‐L1/PD‐1 expression. 121 , 122 , 123 , 124
3.3. Significance of gut microbiota as a promising biomarker to predict ICI efficacy
In the past few years, there has been rapidly rising interest in identifying potential biomarkers for predicting drug response to checkpoint blockade and providing prognostic data, basically in relation to cancer immunotherapy. 125 , 126 Along with the progress of the high‐throughput sequencing (HTS) technology, microarray tools and large‐scale analysis methods, a great number of biomarker identification strategies have been profoundly explored and have already resulted in promising outcomes. 127 , 128 Recent evidence conveys the potential application of intestinal microbiota as a predictive biomarker predicting the effectiveness of hematopoietic stem cell transplantation (HSCT), chemotherapy, and antitumor immunotherapy. 129 , 130 , 131 It has also been shown that modulation of the intestinal microbiota may abolish inflammatory complications caused by ICI blockade therapy, thus, supporting the importance of microbial biomarkers and signatures in predicting the inflammatory adverse events (IAE) caused by cancer immunotherapy. 132
Currently, the number of gut microbiome signatures as potential biomarkers that predict host response, and acquired resistance to ICI blockade treatment is rapidly expanding. In the recent years, substantial researches documented the synergistic cooperation of the certain gut microbiota with PD‐1/PD‐L1/CTLA‐4 inhibitors. For instance, A. muciniphila, Alistipes indistinctus, Bacteroides, B. cepacia, D. formicigenerans, Parabacteroides merdae/distasonis, C. aerofaciens, Eubacterium spp., Veillonella parvula, Klebsiella pneumoniae, Bifidobacterium spp., Lactobacillus spp., Streptococcus parasanguinis, Blautia spp., E. hirae, E. faecium, H. filiformis, Faecalibacterium prausnitzii, and Gemmiger formicilis as well as Ruminococcaceae family have been positively associated with response to checkpoint inhibition in the preclinical and clinical studies. 69 , 70 , 92 , 94 , 95 , 130 However, baseline enrichment in B. thetaiotaomicron, Roseburia intestinalis, Anaerotruncus colihominis, Blautia obeum, and some combination of antibiotics have been negatively correlated with response to anti‐PD‐1 and anti‐CTLA‐4 blockade and compromised the efficacy of immunotherapy. 69 , 129 , 133 Furthermore, incorporation of gut microbiota‐derived proteomics, metabolomics, and genomics data paired with composition profiling of intestinal microbiota may lead to identification of unique metabolic signatures, which can be exploited as comprehensive biomarkers predicting the response to cancer immunotherapy. 134 However, there remain several critical issues such as inaccuracies in predicting the response to immunotherapy, that have to be conveyed in order to validate the application and efficacy of the intestinal microbiota as a prognostic and predictive biomarker for immunotherapy in the clinical practice.
3.4. Limitations and possible suggestions to enhance gut microbiota efficacy in cancer immunotherapy
In spite of promising exploitation of gut microbiota in the era of immunotherapy for cancer, there are as yet some issues and challenges which need to be considered. For example, the existence of unpleasant bacterial species in the intestinal tract can negatively influence the effectiveness of immunotherapy. Commonly, antibiotics are consumed to eliminate pathogenic bacterial species, but at the same time they may cause important risks owing to lack of specificity, particularly intestinal dysbiosis. On the contrary, use of probiotics in combination with prebiotics can synergistically help the intestinal colonization and augmentation of useful microbial species, and may have a booster effect to strengthen the host antitumor immune response. Moreover, the dietary fiber components may be metabolized and converted to biomolecules with immunomodulatory effects such as butyrate as a well‐known SCFA. 135 Alternatively, bacteriophages (viruses that attack bacteria) have been mainly exploited in food industry to demolish pathogenic bacteria owing to their notable selectivity for certain bacterial agents. 136
Recently, several studies have demonstrated that commensal gut microbiota can provide protection against the invasion of pathogenic microbes via colonization resistance mechanism, and also induction of the native or adaptive immune response though the immunomodulatory effects. This beneficial microbiota advocates colonization resistance through direct competing for nutrients and cellular attachment sites, and also produces various inhibitory metabolites which can restrict the overgrowth of the harmful microorganisms. 137 Furthermore, despite the brilliant outcomes of FMT in the treatment of rCDI, administration of FMT could be a promising supplementary option beside immunotherapy against various types of human cancers. 96 , 138 However, application of FMT in cancer immunotherapy needs addressing several important issues, particularly the selection of an ideal donor, administration route, immune status of the recipient, and the types of cancer immunotherapeutic agents used. Moreover, it is noteworthy that still there are inconsistent findings between different studies regarding the impact of gut microbiota on cancer treatment. 69 , 92 , 93 , 94 , 102 Hence, further studies including large cohorts, and clinical trials should be performed to assess the impact of gut microbiota on the effectiveness of ICIs.
4. CONCLUSIONS
The era of microbiota and cancer immunotherapy has recently been introduced and is still in its infancy. Currently, some primary reports of preclinical and clinical investigations on the function of gut microbiota in cancer immunotherapy have proposed it as an appropriate and alternative approach in war on cancer. It is worth noting to identify the specific microbiota and clarify their underlying mechanisms in the context of immune checkpoint blockade. Importantly, supplementation with specific probiotics or prebiotics and restoring the favorable intestinal microbiome by applying FMT or the prevention of the unfavorable bacteria by narrow‐spectrum antibiotics may improve the effectiveness of ICIs in tumor control. However, some problems and challenges stand to be addressed about how and when to manipulate intestinal microbiome to increase the potency of cancer immunotherapy. Finally, studying the potential variations in response to ICIs and exploring the possible hypotheses behind this therapeutic strategy should be accurately addressed by performing future studies in the setting of cancer immunotherapy, gut microbiome, metabolomics, proteomics, and genomics.
CONFLICT OF INTEREST
The authors have no conflict of interest to disclose.
AUTHOR CONTRIBUTIONS
SR and AY contributed to the literature review and wrote the draft of the manuscript. AY worked on concept and design of the study and interpreted the collected information. HAA and MRZ provided clinical advice and guidance for improving of the manuscript. AY critically revised the final version of the manuscript. All authors approved the final version of the manuscript and the authorship list.
Funding information
This work was supported by Foodborne and Waterborne Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
ACKNOWLEDGEMENT
The authors would express sincere thankfulness to Mrs. Raha Rezasoltani for drawing the figures for the manuscript. We also would like to thank the staff of Foodborne and Waterborne Diseases Research Center in Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
DATA AVAILABILITY STATEMENT
All data generated or analyzed during this study are included in this published article.
REFERENCES
- 1. Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258‐1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McQuade JL, Daniel CR, Helmink BA, Wargo JA. Modulating the microbiome to improve therapeutic response in cancer. Lancet Oncol. 2019;20:e77‐e91. [DOI] [PubMed] [Google Scholar]
- 3. Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell. 2016;164:337‐340. [DOI] [PubMed] [Google Scholar]
- 4. Rezasoltani S, Dabiri H, Aghdaie HA, Sepahi AA, Modaresi MH, Mojarad EN. The gut microflora assay in patients with colorectal cancer: in feces or tissue samples? Iran J Microbiol. 2019;11:1‐6. [PMC free article] [PubMed] [Google Scholar]
- 5. Rezasoltani S, Bashirzadeh DA, Mojarad EN, Aghdaei HA, Norouzinia M, Shahrokh S. Signature of gut microbiome by conventional and advanced analysis techniques: advantages and disadvantages. Middle East J Dig Dis. 2020;12:5‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang Y, Ma R, Liu F, Lee SA, Zhang L. Modulation of gut microbiota: a novel paradigm of enhancing the efficacy of programmed death‐1 and programmed death ligand‐1 blockade therapy. Front Immunol. 2018;9:374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rezasoltani S, Aghdaei HA, Mojarad EN, Dabiri H, Ghanbari R, Zali MR. Gut microbiota, epigenetic modification and colorectal cancer. Iran J Microbiol. 2017;9:55‐63. [PMC free article] [PubMed] [Google Scholar]
- 8. Rezasoltani S, Mojarrad EN, Norouzinia M, Aghdaei HA. The necessity of gut microbiome characterization in diseases prevention and therapy. Gastroenterol Hepatol Bed Bench. 2017;10:150‐151. [PMC free article] [PubMed] [Google Scholar]
- 9. Chow J, Tang H, Mazmanian SK. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr Opin Immunol. 2011;23:473‐480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zechner EL. Inflammatory disease caused by intestinal pathobionts. Curr Opin Microbiol. 2017;35:64‐69. [DOI] [PubMed] [Google Scholar]
- 11. Tang WHW, Hazen SL. The contributory role of gut microbiota in cardiovascular disease. J Clin Invest. 2014;124:4204‐4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242‐249. [DOI] [PubMed] [Google Scholar]
- 13. Fong W, Li Q, Yu J. Gut microbiota modulation: a novel strategy for prevention and treatment of colorectal cancer. Oncogene. 2020;39:4925‐4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li W, Deng Y, Chu Q, Zhang P. Gut microbiome and cancer immunotherapy. Cancer Lett. 2019;447:41‐47. [DOI] [PubMed] [Google Scholar]
- 15. Yi M, Jiao D, Xu H, et al. Biomarkers for predicting efficacy of PD‐1/PD‐L1 inhibitors. Mol Cancer. 2018;17:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Noto JM, Peek RM Jr. The gastric microbiome, its interaction with Helicobacter pylori, and its potential role in the progression to stomach cancer. PLoS Pathog. 2017;13:e1006573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rezasoltani S, Sharafkhah M, Aghdaei HA, et al. Applying simple linear combination, multiple logistic and factor analysis methods for candidate fecal bacteria as novel biomarkers for early detection of adenomatous polyps and colon cancer. J Microbiol Methods. 2018;155:82‐88. [DOI] [PubMed] [Google Scholar]
- 18. Wei MY, Shi S, Liang C, et al. The microbiota and microbiome in pancreatic cancer: more influential than expected. Mol Cancer. 2019;18:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mima K, Baba H. The gut microbiome, antitumor immunity, and liver cancer. Hepatobiliary Surg Nutr. 2019;8:67‐68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75‐84. [DOI] [PubMed] [Google Scholar]
- 21. Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30:492‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stecher B, Maier L, Hardt WD. ‘Blooming’ in the gut: how dysbiosis might contribute to pathogen evolution. Nat Rev Microbiol. 2013;11:277‐284. [DOI] [PubMed] [Google Scholar]
- 23. Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol. 2017;17:219‐232. [DOI] [PubMed] [Google Scholar]
- 24. Nadalian B, Yadegar A, Houri H, et al. Prevalence of the pathobiont adherent‐invasive Escherichia coli and inflammatory bowel disease: a systematic review and meta‐analysis. J Gastroenterol Hepatol. 2020. [DOI] [PubMed] [Google Scholar]
- 25. Zhang D, Frenette PS. Cross talk between neutrophils and the microbiota. Blood. 2019;133:2168‐2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zeng MY, Inohara N, Nuñez G. Mechanisms of inflammation‐driven bacterial dysbiosis in the gut. Mucosal Immunol. 2017;10:18‐26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Luo A, Leach ST, Barres R, Hesson LB, Grimm MC, Simar D. The microbiota and epigenetic regulation of T helper 17/regulatory T cells: in search of a balanced immune system. Front Immunol. 2017;8:417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Negi S, Das DK, Pahari S, Nadeem S, Agrewala JN. Potential role of gut microbiota in induction and regulation of innate immune memory. Front Immunol. 2019;10:2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chu H, Mazmanian SK. Innate immune recognition of the microbiota promotes host‐microbial symbiosis. Nat Immunol. 2013;14:668‐675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hug H, Mohajeri MH, Fata GL. Toll like receptors: regulators of the immune response in the human gut. Nutrients. 2018;10:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489:231‐241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Belkaid Y, Harrison OJ. Homeostatic immunity and the microbiota. Immunity. 2017;46:562‐576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Swiatczak B, Cohen IR. Gut feelings of safety: tolerance to the microbiota mediated by innate immune receptors. Microbiol immunol. 2015;59:573‐585. [DOI] [PubMed] [Google Scholar]
- 34. Luu M, Monning H, Visekruna A. Exploring the molecular mechanisms underlying the protective effects of microbial SCFAs on intestinal tolerance and food allergy. Front immunol. 2020;11:1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Levy M, Thaiss CA, Elinav E. Metabolites: messengers between the microbiota and the immune system. Genes Dev. 2016;30:1589‐1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Park J, Kim M, Kang SG, et al. Short‐chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR–S6K pathway. Mucosal Immunol. 2015;8:80‐93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schilderink R, Verseijden C, Seppen J, et al. The SCFA butyrate stimulates the epithelial production of retinoic acid via inhibition of epithelial HDAC. Am J Physiol Gastrointest Liver Physiol. 2016;310:G1138‐G1146. [DOI] [PubMed] [Google Scholar]
- 38. Goverse G, Molenaar R, Macia L, et al. Diet derived short chain fatty acids stimulate intestinal epithelial cells To induce mucosal tolerogenic dendritic cells. J Immunol. 2017;198:2172‐2181. [DOI] [PubMed] [Google Scholar]
- 39. Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci USA. 2014;111:2247‐2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jia W, Xie G, Jia W. Bile acid–microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol. 2018;15:111‐128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu SY, Fu T, Jiang YZ, Shao ZM. Natural killer cells in cancer biology and therapy. Mol Cancer. 2020;19:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Aziz N, Bonavida B. Activation of natural killer cells by probiotics. For Immunopathol Dis Therap. 2016;7:41‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9:503‐510. [DOI] [PubMed] [Google Scholar]
- 44. Abel AM, Yang C, Thakar MS, Malarkannan S. Natural killer cells: development, maturation, and clinical utilization. Front Immunol. 2018;9:1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Eversloh ML, Cicek BB, Siracusa F, et al. NK cells gain higher IFN‐γ competence during terminal differentiation. Eur J Immunol. 2014;44:2074‐2084. [DOI] [PubMed] [Google Scholar]
- 46. Vivier E, Raulet DH, Moretta A, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ganal SC, Sanos SL, Kallfass C, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37:171‐186. [DOI] [PubMed] [Google Scholar]
- 48. Hu W, Wang G, Huang D, Sui M, Xu Y. Cancer immunotherapy based on natural killer cells: current progress and new opportunities. Front Immunol. 2019;10:1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Miller JS, Lanier LL. Natural killer cells in cancer immunotherapy. Annu Rev Cancer Biol. 2019;3:77‐103. [Google Scholar]
- 50. Dzutsev A, Badger JH, Chanona EP, et al. Microbes and cancer. Annu Rev Immunol. 2017;35:199‐228. [DOI] [PubMed] [Google Scholar]
- 51. McCoy KD, Burkhard R, Geuking MB. The microbiome and immune memory formation. Immunol Cell Biol. 2019;97:625‐635. [DOI] [PubMed] [Google Scholar]
- 52. Poggi A, Benelli R, Venè R, et al. Human gut‐associated natural killer cells in health and disease. Front Immunol. 2019;10:961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Spits H, Bernink JH, Lanier L. NK cells and type 1 innate lymphoid cells: partners in host defense. Nat Immunol. 2016;17:758‐764. [DOI] [PubMed] [Google Scholar]
- 54. Guimaraes FSF, Parlato M, Philippart F, Misset B, Cavaillon JM, Conquy MA. Toll‐like receptors expression and interferon‐γ production by NK cells in human sepsis. Crit Care. 2012;16:R206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fernandez NC, Lozier A, Flament C, et al. Dendritic cells directly trigger NK cell functions: cross‐talk relevant in innate anti‐tumor immune responses in vivo. Nat Med. 1999;5:405‐411. [DOI] [PubMed] [Google Scholar]
- 56. Ferlazzo G, Tsang ML, Moretta L, Melioli G, Steinman RM, Münz C. Human dendritic cells activate resting natural killer (NK) cells and are recognized via the NKp30 receptor by activated NK cells. J Exp Med. 2002;195:343‐351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Andoniou CE, van Dommelen SL, Voigt V, et al. Interaction between conventional dendritic cells and natural killer cells is integral to the activation of effective antiviral immunity. Nat Immunol. 2005;6:1011‐1019. [DOI] [PubMed] [Google Scholar]
- 58. Rizzello V, Bonaccorsi I, Dongarrà ML, Fink LN, Ferlazzo G. Role of natural killer and dendritic cell crosstalk in immunomodulation by commensal bacteria probiotics. J Biomed Biotechnol. 2011;2011:473097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fink LN, Frøkiaer H. Dendritic cells from peyer's patches and mesenteric lymph nodes differ from spleen dendritic cells in their response to commensal gut bacteria. Scand J Immunol. 2008;68:270‐279. [DOI] [PubMed] [Google Scholar]
- 60. Konstantinov SR, Smidt H, de Vos WM, et al. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc Natl Acad Sci USA. 2008;105:19474‐19479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gui Q, Wang A, Zhao X, et al. Effects of probiotic supplementation on natural killer cell function in healthy elderly individuals: a meta‐analysis of randomized controlled trials. Eur J Clin Nutr. 2020;74(12):1630‐1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Medina M, Izquierdo E, Ennahar S, Sanz Y. Differential immunomodulatory properties of Bifidobacterium logum strains: relevance to probiotic selection and clinical applications. Clin Exp Immunol. 2007;150:531‐538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hamm AK, Weir TL. Editorial on “Cancer and the microbiota” published in Science. Ann Transl Med. 2015;3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014;15:317‐328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lu R, Wu S, Zhang YG, et al. Enteric bacterial protein AvrA promotes colonic tumorigenesis and activates colonic beta‐catenin signaling pathway. Oncogenesis. 2014;3(6):e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vizcaino MI, Crawford JM. The colibactin warhead crosslinks DNA. Nat Chem. 2015;7:411‐417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Xue M, Kim CS, Healy AR, et al. Structure elucidation of colibactin and its DNA cross‐links. Science. 2019;365(6457):eaax2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Battaglia KB, Alexander D, Dlakić M, Shenker BJ. A journey of cytolethal distending Ttoxins through cell membranes. Front Cell Infect Microbiol. 2016;6:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD‐1‐based immunotherapy against epithelial tumors. Science. 2018;359:91‐97. [DOI] [PubMed] [Google Scholar]
- 70. Matson V, Fessler J, Bao R, et al. The commensal microbiome is associated with anti–PD‐1 efficacy in metastatic melanoma patients. Science. 2018;359:104‐108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. O'Donnell JS, Long GV, Scolyer RA, Teng MW, Smyth MJ. Resistance to PD1/PDL1 checkpoint inhibition. Cancer Treat Rev. 2017;52:71‐81. [DOI] [PubMed] [Google Scholar]
- 72. Sharma P, Lieskovan SH, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168:707‐723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Dai Z, Zhang J, Wu Q, et al. Intestinal microbiota: a new force in cancer immunotherapy. Cell Commun Signal. 2020;18:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dong H, Strome SE, Salomao DR, et al. Tumor‐associated B7–H1 promotes T‐cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8:793‐800. [DOI] [PubMed] [Google Scholar]
- 75. Iwai Y, Ishida M, Tanaka Y, Okazaki T, Honjo T, Minato N. Involvement of PD‐L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD‐L1 blockade. Proc Natl Acad Sci USA. 2002;99:12293‐12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Blank C, Brown I, Peterson AC, et al. PD‐L1/B7H‐1 inhibits the effector phase of tumor rejection by T cell receptor (TCR) transgenic CD8+ T cells. Cancer Res. 2004;64:1140‐1145. [DOI] [PubMed] [Google Scholar]
- 77. Hirano F, Kaneko K, Tamura H, et al. Blockade of B7–H1 and PD‐1 by monoclonal antibodies potentiates cancer therapeutic immunity. Cancer Res. 2005;65:1089‐1096. [PubMed] [Google Scholar]
- 78. Bhandaru M, Rotte A. Monoclonal antibodies for the treatment of melanoma: present and future strategies. Methods Mol Biol. 2019;1904:83‐108. [DOI] [PubMed] [Google Scholar]
- 79. Weiss SA, Wolchok JD, Sznol M. Immunotherapy of melanoma: facts and hopes. Clin Cancer Res. 2019;25:5191‐5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Morgado M, Plácido A, Morgado S, Roque F. Management of the adverse effects of immune checkpoint inhibitors. Vaccines. 2020;8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Temraz S, Nassar F, Nasr R, Charafeddine M, Mukherji D, Shamseddine A. Gut microbiome: A promising biomarker for immunotherapy in colorectal cancer. Int J Mol Sci. 2019;20:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Gebologlu IK, Iz SG, Avci CB. Monoclonal antibodies in cancer immunotherapy. Mol Biol Rep. 2018;45:2935‐2940. [DOI] [PubMed] [Google Scholar]
- 83. Engels B, Engelhard VH, Sidney J, et al. Relapse or eradication of cancer is predicted by peptide‐major histocompatibility complex affinity. Cancer Cell. 2013;23:516‐526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Pacheco NV, Pizano LA, Osorio EF, et al. PD‐L1 expression induced by the 2009 pandemic influenza A(H1N1) virus impairs the human T cell response. Clin Dev Immunol. 2013;2013:989673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Dong H, Zhu G, Tamada K, Chen L. B7–H1, a third member of the B7 family, co‐stimulates T‐cell proliferation and interleukin‐10 secretion. Nat Med. 1999;5:1365‐1369. [DOI] [PubMed] [Google Scholar]
- 86. Gebauer NS, Majdic O, Szekeres A, et al. B7–H1 (programmed death‐1 ligand) on dendritic cells is involved in the induction and maintenance of T cell anergy. J Immunol. 2003;170:3637‐3644. [DOI] [PubMed] [Google Scholar]
- 87. Sznol M, Chen L. Antagonist antibodies to PD‐1 and B7–H1 (PD‐L1) in the treatment of advanced human cancer. Clin Cancer Res. 2013;19:1021‐1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kondo A, Yamashita T, Tamura H, et al. Interferon‐gamma and tumor necrosis factor‐alpha induce an immunoinhibitory molecule, B7–H1, via nuclear factor‐kappaB activation in blasts in myelodysplastic syndromes. Blood. 2010;116:1124‐1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Gordon SR, Maute RL, Dulken BW, et al. PD‐1 expression by tumour‐associated macrophages inhibits phagocytosis and tumour immunity. Nature. 2017;545:495‐499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Johnson CH, Spilker ME, Goetz L, Peterson SN, Siuzdak G. Metabolite and microbiome interplay in cancer immunotherapy. Cancer Res. 2016;76:6146‐6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Gharaibeh RZ, Jobin C. Microbiota and cancer immunotherapy: in search of microbial signals. Gut. 2019;68:385‐388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti–PD‐1 immunotherapy in melanoma patients. Science. 2018;359:97‐103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Vétizou M, Pitt JM, Daillère R, et al. Anticancer immunotherapy by CTLA‐4 blockade relies on the gut microbiota. Science. 2015;350:1079‐1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Chaput N, Lepage P, Coutzac C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. 2017;28:1368‐1379. [DOI] [PubMed] [Google Scholar]
- 95. Frankel AE, Coughlin LA, Kim J, et al. Metagenomic shotgun sequencing and unbiased metabolomic profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia. 2017;19:848‐855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Levy AN, Allegretti JR. Insights into the role of fecal microbiota transplantation for the treatment of inflammatory bowel disease. Therap Adv Gastroenterol. 2019;12:1756284819836893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Salhy ME, Hatlebakk JG, Gilja OH, Kristoffersen AB, Hausken T. Efficacy of faecal microbiota transplantation for patients with irritable bowel syndrome in a randomised, double‐blind, placebo‐controlled study. Gut. 2020;69:859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Johnsen PH, Hilpüsch F, Cavanagh JP, et al. Faecal microbiota transplantation versus placebo for moderate‐to‐severe irritable bowel syndrome: a double‐blind, randomised, placebo‐controlled, parallel‐group, single‐centre trial. Lancet Gastroenterol Hepatol. 2018;3:17‐24. [DOI] [PubMed] [Google Scholar]
- 99. Schepici G, Silvestro S, Bramanti P, Mazzon E. The gut microbiota in multiple sclerosis: an overview of clinical trials. Cell transplant. 2019;28:1507‐1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Chen D, Wu J, Jin D, Wang B, Cao H. Fecal microbiota transplantation in cancer management: current status and perspectives. Int J Cancer. 2019;145:2021‐2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wang Z, Hua W, Li C, et al. Protective role of fecal microbiota transplantation on colitis and colitis‐associated colon cancer in mice is associated with Treg cells. Front Microbiol. 2019;10:2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Azimirad M, Yadegar A, Aghdaei HA, Kelly CR. Enterotoxigenic Clostridium perfringens infection as an adverse event after faecal microbiota transplantation in two patients with ulcerative colitis and recurrent Clostridium difficile infection: A neglected agent in donor screening. J Crohns Colitis. 2019;13:960‐961. [DOI] [PubMed] [Google Scholar]
- 103. Azimirad M, Yadegar A, Gholami F, et al. Treatment of recurrent Clostridioides difficile infection using fecal microbiota transplantation in Iranian patients with underlying inflammatory bowel disease. J Inflamm Res. 2020;13:563‐570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Siedlecka KK, Daca A, Fic M, van de Wetering T, Folwarski M, Makarewicz W. Therapeutic methods of gut microbiota modification in colorectal cancer management‐fecal microbiota transplantation, prebiotics, probiotics, and synbiotics. Gut Microbes. 2020;11:1518‐1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Cammarota G, Ianiro G, Tilg H, et al. European consensus conference on faecal microbiota transplantation in clinical practice. Gut. 2017;66:569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Ianiro G, Mullish BH, Kelly CR, et al. Reorganisation of faecal microbiota transplant services during the COVID‐19 pandemic. Gut. 2020;69:1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti–PD‐L1 efficacy. Science. 2015;350:1084‐1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Borody TJ, Warren EF, Leis SM, Surace R, Ashman O, Siarakas S. Bacteriotherapy using fecal flora: toying with human motions. J clin gastroenterol. 2004;38:475‐483. [DOI] [PubMed] [Google Scholar]
- 109. Andrews MC, Wargo JA. Cancer evolution during immunotherapy. Cell. 2017;171:740‐742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Daillère R, Vétizou M, Waldschmitt N, et al. Enterococcus hirae and Barnesiella intestinihominis facilitate cyclophosphamide‐induced therapeutic immunomodulatory effects. Immunity. 2016;45:931‐943. [DOI] [PubMed] [Google Scholar]
- 111. Wang S, Campos J, Gallotta M, et al. Intratumoral injection of a CpG oligonucleotide reverts resistance to PD‐1 blockade by expanding multifunctional CD8+ T cells. Proc Natl Acad Sci USA. 2016;113:E7240‐E7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Yin P, Liu X, Mansfield AS, et al. CpG‐induced antitumor immunity requires IL‐12 in expansion of effector cells and down‐regulation of PD‐1. Oncotarget. 2016;7:70223‐70231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107‐118. [DOI] [PubMed] [Google Scholar]
- 114. Ichinohe T, Pang IK, Kumamoto Y, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci USA. 2011;108:5354‐5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Das S, Suarez G, Beswick EJ, Sierra JC, Graham DY, Reyes VE. Expression of B7–H1 on gastric epithelial cells: its potential role in regulating T cells during Helicobacter pylori infection. J immunol. 2006;176:3000‐3009. [DOI] [PubMed] [Google Scholar]
- 116. Wu YY, Lin CW, Cheng KS, et al. Increased programmed death‐ligand‐1 expression in human gastric epithelial cells in Helicobacter pylori infection. Clin Exp Immunol. 2010;161:551‐559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Crabtree JE, Shallcross TM, Heatley RV, Wyatt JI. Mucosal tumour necrosis factor alpha and interleukin‐6 in patients with Helicobacter pylori associated gastritis. Gut. 1991;32:1473‐1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Yadegar A, Mobarez AM, Zali MR. Genetic diversity and amino acid sequence polymorphism in Helicobacter pylori CagL hypervariable motif and its association with virulence markers and gastroduodenal diseases. Cancer Med. 2019;8:1619‐1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Bodger K, Wyatt JI, Heatley RV. Gastric mucosal secretion of interleukin‐10: relations to histopathology, Helicobacter pylori status, and tumour necrosis factor‐alpha secretion. Gut. 1997;40:739‐744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Perri F, Clemente R, Festa V, et al. Serum tumour necrosis factor‐alpha is increased in patients with Helicobacter pylori infection and CagA antibodies. Ital J Gastroenterol Hepatol. 1999;31:290‐294. [PubMed] [Google Scholar]
- 121. Xie Z, Chen Y, Zhao S, et al. Intrahepatic PD‐1/PD‐L1 up‐regulation closely correlates with inflammation and virus replication in patients with chronic HBV infection. Immunol Invest. 2009;38:624‐638. [DOI] [PubMed] [Google Scholar]
- 122. Mason LG, Palmer B, Klarquist J, Mengshol JA, Castelblanco N, Rosen HR. Upregulation of PD‐1 expression on circulating and intrahepatic hepatitis C virus‐specific CD8+ T cells associated with reversible immune dysfunction. J Virol. 2007;81:9249‐9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Yang W, Song Y, Lu YL, Sun JZ, Wang HW. Increased expression of programmed death (PD)‐1 and its ligand PD‐L1 correlates with impaired cell‐mediated immunity in high‐risk human papillomavirus‐related cervical intraepithelial neoplasia. Immunology. 2013;139:513‐522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Fang W, Zhang J, Hong S, et al. EBV‐driven LMP1 and IFN‐γ up‐regulate PD‐L1 in nasopharyngeal carcinoma: Implications for oncotargeted therapy. Oncotarget. 2014;5:12189‐12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Bai R, Lv Z, Xu D, Cui J. Predictive biomarkers for cancer immunotherapy with immune checkpoint inhibitors. Biomark Res. 2020;8:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Shen H, Yang ES‐H, Conry M, et al. Predictive biomarkers for immune checkpoint blockade and opportunities for combination therapies. Genes Dis. 2019;6:232‐246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Yang X, Kui L, Tang M, et al. High‐throughput transcriptome profiling in drug and biomarker discovery. Front Genet. 2020;11:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Lightbody G, Haberland V, Browne F, et al. Review of applications of high‐throughput sequencing in personalized medicine: barriers and facilitators of future progress in research and clinical application. Brief Bioinform. 2019;20:1795‐1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Gong J, Raffle AC, Hickok VP, Guan M, Hendifar A, Salgia R. The gut microbiome and response to immune checkpoint inhibitors: preclinical and clinical strategies. Clin Transl Med. 2019;8:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Shui L, Yang X, Li J, Yi C, Sun Q, Zhu H. Gut Microbiome as a potential factor for modulating resistance to cancer immunotherapy. Front Immunol. 2020;10:2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Ingham AC, Kielsen K, Cilieborg MS, et al. Specific gut microbiome members are associated with distinct immune markers in pediatric allogeneic hematopoietic stem cell transplantation. Microbiome. 2019;7:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Wang Y, Wiesnoski DH, Helmink BA, et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor‐associated colitis. Nat Med. 2018;24:1804‐1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Pushalkar S, Hundeyin M, Daley D, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 2018;8:403‐416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Malczewski AB, Navarro S, Coward JI, Ketheesan N. Microbiome‐derived metabolome as a potential predictor of response to cancer immunotherapy. J Immunother Cancer. 2020;8:e001383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Bultman SJ. The microbiome and its potential as a cancer preventive intervention. Semin Oncol. 2016;43:97‐106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Zhang J, Hong Y, Harman NJ, Das A, Ebner PD. Genome sequence of a Salmonella phage used to control salmonella transmission in Swine. Genome Announc. 2014;2:e521‐e614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev. 2017;279:70‐89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Krajicek E, Fischer M, Allegretti JR, Kelly CR. Nuts and bolts of fecal microbiota transplantation. Clin Gastroenterol Hepatol. 2019;17:345‐352. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


