Abstract
Therapeutic options for SARS-CoV-2 are limited merely to the symptoms or repurposed drugs and non-specific interventions to promote the human immune system. In the present study, chromatographic and in silico approaches were implemented to identify bioactive compounds which might play pivotal role as inhibitor for SARS-CoV-2 and human immunomodulator (TGF-β and TNF-α). Tinospora cordifolia (Willd.) Miers was evaluated for phenolic composition and explored for bioactive compounds by high-performance thin layer chromatography (HPTLC). Furthermore, the bioactive compounds such as cordifolioside, berberine, and magnoflorine were appraised as human immunomodulatory and potent inhibitor against Main Protease (Mpro) of SARS-CoV-2 through multiple docking strategies. Cordifolioside formed six stable H-bonds with His41, Ser144, Cys145, His163, His164, and Glu166 of Mpro of SARS-CoV-2, which displayed a significant role in the viral replication/transcription during infection acting towards the common conserved binding cleft among all strains of coronavirus. Overall, the study emphasized that the proposed cordifolioside might use for future investigations, which hold as a promising scaffold for developing anti-COVID-19 drug and reduce human cytokine storm.
Keywords: COVID-19, SARS-CoV-2, Main protease, Tinospora cordifolia, HPTLC, Sequence analysis, Molecular docking, Dynamics simulations
Introduction
The novel COronaVIrus Disease-2019 (COVID-19) outbreak has become a zoonotic pandemic worldwide since its first outbreak in December 2019. Officially, on 11th February 2020, it has been named by the International Committee on Taxonomy of Viruses (ICTV) as Severe Acute Respiratory Syndrome CoronaVirus-2 (SARS-CoV-2) (Rosendaal 2020). The virus has turned into a deadly disease from mild symptoms to chronic respiratory disorder since its appearance as it spreads rapidly from human to human (Anand et al. 2003; Hilgenfeld 2014). The lack of drugs or vaccines provoked the research communities and become tremendously important to understand the underlying mechanism of this virus its control. The use of viral proteases and the protease inhibitors halt the viral replication in turn preventing the disease progression. Owing to the importance of viral proteases, one of the attractive drug targets among coronaviruses known as main protease (Mpro, also called 3CLpro) was marked as a therapeutic target in the current study due to its significant role in processing the polyproteins that are translated from the viral Ribonucleic acid (RNA) (Anand et al. 2003; Hilgenfeld 2014). Furthermore, the Mpro also been recognized as non-homologous to human proteases with inhibitors that are unlikely to be toxic (Naik et al. 2020). However, several synthetic drugs which have shown some promising results to a certain extent against SARS-CoV-2 are remdesivir (used to control Ebola virus and other RNA viruses), chloroquine (antimalarial, anti-viral, and immunomodulator), hydroxychloroquine (Wang et al. 2020; Naik et al. 2020). The treatment options with limited effect have posed a challenge to many researchers in most of the countries to find out a valid and proper solution for this universal pandemic.
Bioactive compounds from natural resources have been into use since ancient times for treating several health ailments, which also include viral infections (Ganjhu et al. 2015). Development of novel anti-viral drugs has been guided by several purified natural compounds (Mani et al. 2020), as during 1981 and 2014, most of the drugs which have been approved (approx. 50%) have been derived from or may have mimicked from natural compounds (Newman and Cragg 2016). With proved anti-viral activity (Kusumoto et al. 1995; Asres et al. 2005), considering the present pandemic (COVID-19), several therapeutic systems with natural compounds are proposed, some being proved in treating several viruses mediated (virus-mediated) diseases such as herpes simplex virus, influenza virus, human immunodeficiency virus, SARS and MERS (middle east respiratory syndrome) (Xu et al. 2000; Du et al. 2003; Cinatl et al. 2003; Lin et al. 2017; Kannan and Kolandaivel 2017). A recent study reported an effective virtual screening protocol of natural compounds as multi-target molecules for therapeutic option against SARS-CoV-2 infection, including some biological compounds which might be possible therapeutic agents against COVID-19 (Liang et al. 2020; Shree et al. 2020; Jiménez-Alberto et al. 2020; Meyer-Almes 2020; Chen 2020; Chen et al. 2020a; Chen et al. 2020b; Qin et al. 2020; Pitsillou et al. 2020).
To gaze at, Tinospora cordifolia (Willd.) Miers ex Hook. f. & Thomas. (Menispermaceae) has an important place in Indian Ayurvedic traditional medicine due to its predominant role in treating several health disorders and multi-faceted therapeutic properties as an anti-inflammatory, antimalarial (Sharma et al. 2012), antiallergic (Sunanda and Ainapure 2020), antipyretic (Ashok et al. 2010), antiarthritic (Rao et al. 2008), antidiabetic (Wadood et al. 1992), and antihepatotoxic (Sharma et al. 2012). The active constituents of the plant have not been explored for its potential, indeed, it has been used at the household level for treating several infections traditionally in the Indian system (Dahanukar et al. 1999; Panchabhai et al. 2008). The bioactive compounds of the T. cordifolia have been reported (Jyoti et al. 2015), and well documented for the immunomodulatory effect due to one of the active constituent (Supe and Purandare 2007; Aranha et al. 2012).
In the present study, an attempt was made to identify natural bioactive compounds with dual activity, immunomodulator and potential inhibitor against SARS-CoV-2 of Mpro. Thus, T. cordifolia explored for the characterization of bioactive components with different solvent extractions and characterized for phenolic components using high-performance thin layer chromatography (HPTLC). Furthermore, the molecular interactions of identified compounds through extensive in silico approaches of molecular docking and dynamics simulations was investigated against SARS-CoV-2 inhibitory and as human immune modulator.
Materials and methods
Sample and chemicals
HPLC grade reference standards of cordifolioside, magnoflorine, berberine chloride, gallic acid and rutin were purchased from Sigma-Aldrich Co. St.Louis, USA. Analytical grade chemicals; 2,2′-diphenyl-1-picrylhydrazyl (DPPH), 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS+), 2,4,6-tris(2-pyridyl)-s-triazine (TPTZ), and trolox were procured from Sigma-Aldrich Co. St.Louis, USA. Folin–ciocalteu reagent and other chemicals used for analysis were of analytical grade, which were acquired from Merck, Mumbai, India. HPTLC plates, Silica gel 60 F254 were purchased from Merck KGaA, Darmstadt, Germany. T. cordifolia plant powder sample was procured from the local market with commercial suppliers in Dharwad, Karnataka, India.
Extraction of phenolic compounds
The extraction of phenolic compounds was carried out by previously examined methods (Goudar and Sathisha 2016), with slight modification. In brief, extraction was carried out with four different solvents: water, acetone, ethyl alcohol, and methanol. 2 g of the sample extracted with 50 mL of each solvent, vortexed and incubated for 30 min under sonication, and centrifuged for 4000 RPM maintained at 4 °C (REMI, Mumbai, India). The supernatant obtained was decanted and the residue was re-extracted using fresh solvents by reiterating the extraction step. The obtained supernatant pooled and concentrated using a rotary evaporator (Rotavapor R-300, Buchi, Switzerland). The residue attained was dissolved in 5 mL of methanol and used for further analysis.
Total phenolic content (TPC)
TPC in the sample extract was determined using the Folin-ciocalteu reagent method (Singleton and Rossi 1965). Sample (200 mg) was extracted with acidified methanol (4 mL) for 1 h at room temperature. 200 μL of sample extract was mixed with 1.5 mL of diluted Folin–Ciocalteu reagent (tenfold). Aliquot was incubated for 5 min, then 1.5 mL of sodium carbonate solution (6%). Further incubated for 60 min under dark conditions at room temperature and the absorbance was recorded at 725 nm (Cary 50, Varian, Middelburg, Netherlands). The results were expressed as gallic acid equivalent milligram per 100 g (mg GAE/100 g).
Total flavonoid content (TFC)
TFC of the sample extracts were analyzed by an applied method, which was reported in the earlier study (Goudar et al. 2018). Sample extracts (250 µL) were made to react with sodium nitrite (5%) and aluminium chloride (10%), incubated after the addition of sodium hydroxide (1 M). The absorbance of the solutions was recorded at 415 nm by UV–Visible Spectrophotometer (Cary 50, Varian, Middelburg, Netherlands). Rutin was used as standard reference and the values were expressed as milligram of catechin equivalent per 100 g (mg CE/100 g).
Antioxidant activity
Antioxidant activity was investigated and evaluated by different methods explained previously viz, 2,2′-diphenyl-1-picrylhydrazyl (DPPH) radical scavenging method (Brand-Williams et al. 1995), 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS+) assay (Re et al. 1999), and ferric reducing antioxidant power (FRAP) assay (Benzie and Strain 1996).
In DPPH method, the sample extracts (100 µL) were mixed with 3.9 mL of 6 × 10–5 mol/L of DPPH solution in methanol, then the absorbance of the solutions along with blank was recorded at 515 nm at 0 and 30 min and the antioxidant activity (%) was calculated by the equation:
For ABTS analysis, the sample extracts (100 μL) combined with ABTS solution, and the absorbance was marked at 734 nm. The absorbance of the samples was compared with the Trolox standard and results were reported as micromole Trolox equivalent per gram (µmTE/g).
In FRAP method, 100 μL of sample extracts were reacted with FRAP reagent prepared as explained in the method. The absorbance of the solutions was recorded at 593 nm and results were expressed as µmol Fe(II)/g.
Chromatographic analysis by HPTLC
Chromatographic fingerprint was developed for the sample extracts as per the previously described method (Goudar and Sathisha 2016). The aluminium backed TLC plates bonded with silica gel 60 F254 of size 20 × 10 cm were used for investigation. The sample extracts along with reference standards were applied with Hamilton syringe (10 µL) using a sample applicator (Linomat V, CAMAG, Switzerland). The TLC plates were developed inside the glass chamber with a size of 20 × 10 cm after pre-saturation with a mobile phase which was the combination of solvents: ethyl acetate: methanol: water: formic acid in the ratio (80:10:5:1, v/v). The development of the plates was carried out until the solvent reach up to 90 mm, then removed, air-dried for 10 min and heated inside a hot air oven at 60 °C for 10 min to remove the solvent vapors. The scanning of the plates was performed using the TLC scanner III (Camag, Switzerland) and the bioactive compounds were recognized and quantified by comparing the Rf values of the standards with the Rf values achieved for sample extracts.
Sequence analysis
The SARS-CoV-2 belongs to the family of Coronaviridae, which consists of a single-stranded, non-segmented positive-sense large RNA genome of size ~ 26–32 kb. To date, several human coronaviruses (HCoVs) have been identified namely HCoV-229E, HCoV-NL63, HCoV-OC43, HCoV-HKU1, Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV), and Middle East respiratory syndrome coronavirus (MERS-CoV). Except for SARS-CoV and MERS-CoV, the other four viruses were globally disseminated in the human population and causes common cold infections among one-third of the human population (Killerby et al. 2018). Other, two life-threatening coronavirus disease outbreaks occurred in the past two decades. SARS-CoV in 2003 originated in Guangdong Province, China, and spread the disease to more than 30 countries including all major continents, resulting in 8098 human infections and 774 deaths (https://www.cdc.gov/sars/about/fs-sars.html). The second includes MERS-CoV in 2012 originated in Saudi Arabia and genetically different from SARS-CoV but infected 2249 people in 27 countries with 35% case fatality (https://www.who.int/news-room/fact-sheets/detail/middle-east-respiratory-syndrome-coronavirus-(mers-cov). The current SARS-CoV-2 made the globe into lockdown mode with the case fatality rates, inclined economic downfall and sky-scraping infection speed changing exponentially at every minute (Lu 2020; Liu et al. 2020; Yu et al. 2020; Yang et al. 2020). The beginning of SARS-CoV-2 was linked to a cluster of patients with pneumonia of unknown etiology connected to a local Huanan South China Seafood Market in Wuhan, Hubei Province, China in December 2019 (Huang et al. 2020; Wu et al. 2020).
In the present study, Mpro protein sequence of seven coronaviruses retrieved from the National Center for Biotechnology Information (NCBI) viral genome sequence database (https://www.ncbi.nlm.nih.gov/genome/viruses/) for the evolutionary relationship and conserved common binding pocket analysis was explored by the MEGA-X and CLUSTAL X tool (Higgins and Sharp 1988; Prathima et al. 2017; Munikumar et al. 2018). The conserved common binding pocket further utilized for the grid generation in molecular docking protocol.
Mpro protein structure analysis
The Mpro encoded by open reading frame 1 (ORF1) as a non-structural protein 5 (nsp5) with no less than 11 cleavage sites in the polyproteins flanked by the Nsp4 and Nsp6 responsible for processing the huge polyproteins (pp1a and pp1ab). Along with part of Nsp3, anchor the replication and transcription complex to double-membrane vesicles that are derived from the endoplasmic reticulum membrane during the infection (Hilgenfeld 2014). The co-crystal structure of SARS-CoV-2 Mpro complexed with an nucleotide peptide inhibitor N3 (n-[(5-methylisoxazol-3-yl)carbonyl]alanyl-l-valyl-n~1~-((1r,2z)-4-(benzyloxy)-4-oxo-1-{[(3r)-2oxopyrrolidin-3-yl]methyl}but-2-enyl)-l-leucinamide) was acquired from the protein data bank (PDB) encoded with the ID:6LU7 with crystal resolution 1.5 Ȧ (Jin et al. 2020). The co-crystal inhibitor of N3 peptide (positive control) formed nine H-bonds with amino acid residues Phe140, Gly143, Cys145, His163, His164, Glu166 (2), Gln189, and Thr190 with the bond lengths of 3.13, 2.87, 2.05, 2.37, 2.8, 2.98, 2.83, 2.89, and 2.85 Ȧ, respectively, in the native cleft located between domain I and domain II of Mpro. The Mpro residues namely, Thr24, Thr25, Thr26, Thr26, His41, Phe140, Leu141, Asn142, Gly143, Ser144, Cys145, His163, His164, Met165, Glu166, Pro168, His172, Arg188, Gln189, Thr190, Ala191, and Gln192 formed strong Van der Waals (VdW) interactions that are in attendance within 4 Ȧ region of N3 peptide (Supplementary Fig. 1) (Jin et al. 2020). Besides, additional tools like PDBsum and PyMol were utilized for the validation of binding site prediction. Thus, the residues positioned around the N3 peptide considered as crucial molecular interactive cleft towards the Mpro and utilized for the grid generation in molecular docking analysis.
Ligand preparation
The selected compounds of Cordifolioside (PubChem ID: 45359937), Berberine (PubChem ID: 2353), and Magnoflorine (PubChem ID: 73337) were obtained from the PubChem database and prepared for the docking experiment by BIOVIA Discovery Studio v20.1 and AutoDock Vina 1.1.2. For Discovery Studio, the “prepare ligand” wizard was utilized to correct the three-dimensional (3D) coordinates through the addition of hydrogen atoms, neutralization of the charged groups, generation of ionization states, clean geometry, tautomerization states, and finally, the conversion of the yields into MOL2 format. For AutoDock Vina, the 3D coordinates of Structure Data File (SDF) format and each selected compound were transformed to PDBQT file format for docking study with the help of PyMol and ADT tool of molecular docking.
Molecular docking
Molecular docking is a modern and robotic-modeling technique for the molecule level interactions of two or more, i.e., protein–ligand, protein–protein, protein–nucleotide, and drug–drug in the rational drug design and discovery process. The outcomes obtained from the molecular docking approaches can be utilized to predict the binding energy, free energy, and stability of complexes (Pradhan et al. 2014; Khandare et al. 2018; Manne et al. 2018; Munikumar et al. 2019). In the present study, two different docking strategies, Discovery Studio and AutoDock Vina are employed for the molecular interactions between Mpro and selected bioactive compounds of T. cordifolia.
For Discovery Studio, the preparation of the Mpro protein, all HOH, and co-crystallized N3 peptide removed and hydrogens were added. Protein was processed with the ‘protein preparation’ wizard to model missing loop regions, calculate protein optimization, and protonate the protein structure. The prepared Mpro protein was defined as a receptor with a binding site defined from N3 peptide. The first binding site sphere (− 11.74, 13.63, 70.63, 8.1) of Mpro was selected for molecular docking analysis. T. cordifolia compounds and N3 peptide were docked with Mpro by employing the LibDock module and binding energies are calculated for the individual compounds. The highest LibDock score and least binding energy with the best-docked pose of each compound was examined with the aid of the molecular interaction module.
For AutoDock Vina, all HOH molecules were removed, and Auto Dock Tool (ADT) software was used to prepare the required files for AutoDock Vina by assigning hydrogen polarities, calculating Gasteiger charges to protein structures, and converting protein structures from the Mpro. PDB file format to Mpro. PDBQT format. The prepared T. cordifolia compounds and N3 peptide were docked on an individual basis to the Mpro of SARS-CoV-2 with grid coordinates (grid center) and grid boxes of certain sizes for N3 peptide. The compounds were set to be flexible when interacting with the macromolecule of Mpro under rigid conditions. The configuration file was engaged by opening notepad to run AutoDock Vina (Sliwoski et al. 2014; Kodchakorn et al. 2020). ADT was required to prepare the input PDBQT file for Mpro and to set the size and the center of the grid box. Kollman charges and polar hydrogen atoms were added in the Mpro structure (Morris et al. 2009; Azam and Abbasi 2013). The grid size was set at 25 × 25 × 25 (x, y, and z) points, and the grid center was designated at x, y, and z dimensions of − 11.171, 16.853, and 66.519, respectively, with a grid spacing of 1000 Å. The prepared Mpro file was saved in the PDBQT format. Compound-binding affinities were predicted as negative Gibbs free energy (∆G) scores (kcal/mol), which were calculated based on the AutoDock Vina scoring function (Trott and Olson 2009).
Post-docking analyses were visualized using the Discovery Studio, which showed the sizes and locations of the binding sites, H-bond, hydrophobic interactions, and bonding distances as interaction radii of 5 Å from the position of the docked compounds with the binding site of Mpro. Subsequently, the binding poses of each compound examined and their interactions with the protein characterized, and the best and most energetically favorable conformations of a compound from two different docking strategies picked for the molecular dynamics (MD) simulations utilizing Desmond v2019-4 at 100 ns stipulated run time.
Molecular dynamics simulations
MD simulations for the topmost best binding conformation compound with Mpro docked complex was carried out using the Desmond v2019-4 program, an explicit solvent MD package (Desmond Molecular Dynamics System; D. E. Shaw Research, New York, NY, USA) with integral optimized potentials for liquid simulation (OPLS-2005) force field. The correctness of the chemical structures provided to Desmond ensured using the Protein Preparation Wizard (macro models). The system was set up for simulation using a predefined water model (TIP3P) as a solvent in a cubic box with periodic boundary conditions specifying the shape and size of the box as 10 Å × 10 Å × 10 Å distance. After building the solvated system, the protein–ligand complex minimization and relaxation were performed under the NPT ensemble using minimization and relaxation protocol that consists of nine stages (Vilar et al. 2011). The natural ions of 0.15 M NaCl (physiological concentration of monovalent ions) added in 10 Å buffer using the system-built option. The final production run was carried out for 100 ns, and the trajectory sampling was done at an interval of 4.0 ps (Leimkuhler and Sweet 2004; Munikumar et al. 2018).
Statistical analysis
All the wet laboratory vetting were performed in triplicate and expressed as mean ± standard deviation. Statistical analyses for one-way analysis of variance (ANOVA) and significant difference (p < 0.05) between means were analyzed using SPSS software (SPSS Institute Inc., Cary, NC, USA).
Results and discussion
Phenolic composition
TPC of the T. cordifolia extracts analyzed in ethyl alcohol, methanol, acetone, and water solvents showed variability in the values compared among different solvents (Table 1). Ethyl alcohol and methanol extracts accounted for higher TPC with 571 and 518 mg GAE/100 g, respectively, which was approximately double in comparison to the acetone and water extracts, which were 245 and 216 mg GAE/100 g, respectively. A similar study declared TPC in T.cordofolia leaf extracts of ethyl alcohol, methanol, and water with 264, 301, and 465 mg GAE/g, respectively (Jyoti et al. 2015). Yadav and Agarwala (2011), affirmed for TPC in leaf and stem extracts T.cordofolia with 40.8 and 12.8 mg GAE/g, respectively. The outcome in the present study for TPC was strongly corroborated with the reported values from the reputed previous published studies.
Table 1.
Total phenolic content (TPC), total flavonoid content (TFC) and antioxidant properties in T. cordifolia solvent extracts
| Extraction solvent | Phenolic components | Antioxidant properties | |||
|---|---|---|---|---|---|
| TPC (mgGAE/100 g) | TFC (mgRE/100 g) | DPPH method (%) | ABTS method (µmTE/g) | FRAP method (µmol Fe(II)/g) | |
| Acetone | 245.87 ± 6.24c | 228.48 ± 8.99c | 118.85 ± 7.06b | 136.17 ± 5.00c | 0.96 ± 0.01c |
| Ethyl alcohol | 571.35 ± 5.65a | 448.97 ± 8.70a | 162.11 ± 4.67a | 172.77 ± 1.88a | 1.15 ± 0.06a |
| Methanol | 518.32 ± 5.68b | 374.61 ± 4.88b | 161.35 ± 4.02a | 153.50 ± 3.51b | 1.03 ± 0.01b |
| Water | 216.82 ± 6.35d | 186.75 ± 6.34d | 109.34 ± 4.06b | 119.61 ± 2.73d | 0.78 ± 0.01d |
Mean values in the table followed by different superscript letter (a–d) are significantly different (p < 0.05) compared within the column for different solvent extracts
GAE gallic acid equivalent, RE rutin equivalent
TFC of the T. cordifolia extracts was comparatively downbeat in comparison to TPC in ethyl alcohol, methanol, acetone, and water solvents. Whereas when compared among solvents, ethyl alcohol and methanol had a higher content of TFC compared with acetone and water (Table 1). Hydro-alcoholic extract of T. cordifolia plant was reported for TFC with 18.91 mg QE/g, whereas TFC of T. cordifolia stem extracts in ethyl alcohol, petroleum ether, dichloromethane, n-butanol, and aqueous solvents were reported between 0.17 and 0.98 mg QE/g (Polu et al. 2017).
Antioxidant activity
The antioxidant activity in different solvent extracts of T. cordifolia is reported in Table 1. Antioxidant activities analyzed by ABTS and FRAP method showed note-worthy variation among different solvent extracts (p < 0.05). The DPPH method displayed antioxidant activity, which was ranging from 109.34 to 162.11%. Another study reported DPPH activity for different solvent extracts of T. cordifolia as IC50 ranging from 14.18 to 183.47 µg/mL (Polu et al. 2017). The antioxidant activity analyzed by ABTS method in different extracts were ranging from 119.61 to 172.77 µmTE/g, wherein the highest activity was found in the ethyl alcohol and the least in the water extract. A similar study by Polu et al. (2017), reported antioxidant activity by ABTS method in T. cordifolia which was between 28.24 and 132.4 IC50µg/mL, with exception of petroleum ether extract with more than 1000 IC50µg/mL. The results for the FRAP method of antioxidant activity in different extracts were found to be between 0.78 and 1.15 mmol Fe(II)/g, wherein ethyl alcohol and methanol depicted the highest content in comparison to acetone and water extracts. These results obtained will prove that all types of solvents showed antioxidant activity with varying concentrations, which might help combat several oxidants produced during physiological processes inside the cells.
Chromatographic analysis
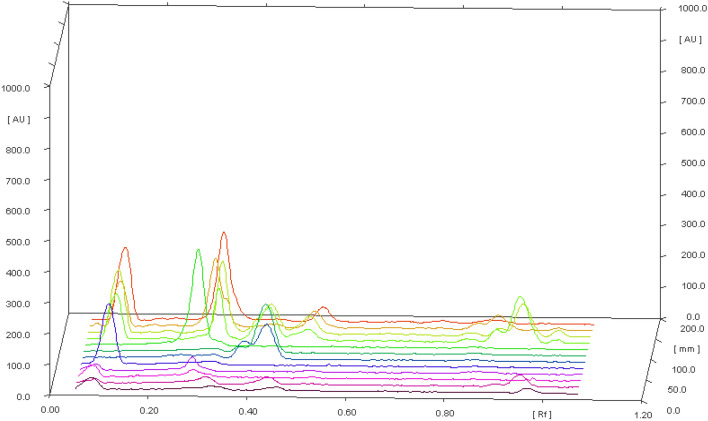
The bioactive compounds cordifolioside, magnoflorine, and berberine were analyzed by HPTLC method and the results acquired are reported in Table 2. Momentous variation was found among the bioactive compounds analyzed among different solvent extracts (p < 0.05). Cordifolioside was predominantly found in the ethyl alcohol extract (432.51 mg/100 g) in comparison to other solvent extracts analyzed. Whereas, magnoflorine was detected with highest content in methanol extract (554.08 mg/100 g) followed by ethyl alcohol (455.33 mg/100 g), acetone (96.90 mg/100 g), and water (71.16 mg/100 g) extracts (Fig. 1).
Table 2.
HPTLC evaluation of T. cordifolia bioactive compounds in different solvent extracts
| Extraction solvent | Cordifolioside (mg/100 g) | Magnoflorine (mg/100 g) | Berberine (mg/100 g) |
|---|---|---|---|
| Acetone | 37.78 ± 1.43c | 96.90 ± 1.76c | 10.82 ± 1.43c |
| Ethyl alcohol | 432.51 ± 2.66a | 455.33 ± 4.92a | 326.37 ± 2.89a |
| Methanol | 293.97 ± 3.59b | 554.08 ± 5.19b | 221.65 ± 3.94b |
| Water | 21.61 ± 0.67d | 71.16 ± 1.35d | 65.53 ± 2.91d |
Mean values in the table followed by different superscript letter (a–d) are significantly different (p < 0.05) compared within the column for different solvent extracts
Fig. 1.
HPTLC chromatogram of T. cordifolia extracts and standards; cordifolioside, magnoflorine, and berberine
Compared to cordifolioside, and magnoflorine; berberine was detected with less quantity in ethyl alcohol, methanol, and acetone extracts. These bioactive compounds of T. cordifolia are known to impart and have been involved in several health beneficial effects such as anti-HIV, antidiabetic, antistress, hypolipidemic, antiosteoporotic, wound healing, anti-complement and immunomodulating activity (Shanbhag et al. 2005; Kalikar et al. 2008; Patel and Mishra 2012; Shivananjappa and Muralidhara 2012; Ali and Dixit 2013; Sharma et al. 2019). Hence, these compounds are evaluated at the molecular level for stable binding energy, H-bond, and hydrophobic interactions with the drug target of SARS-CoV-2 and properties of immune modulation.
Sequence and conserved binding site analysis
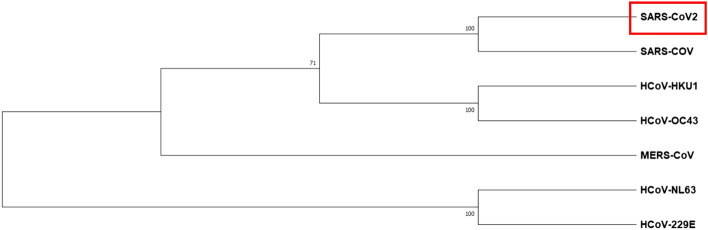
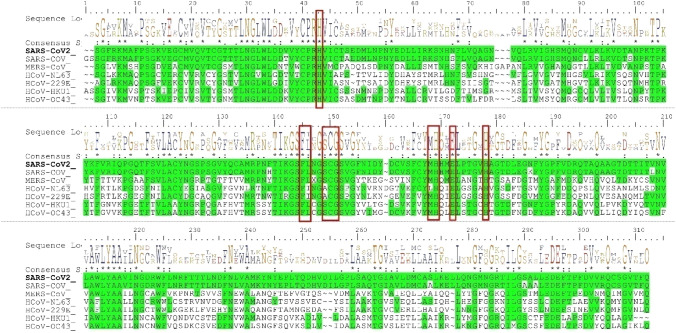
Initially, the SARS-CoV-2 Mpro primary sequence (identity and similarity) and the phylogenetic tree constructed between the closely related seven human coronaviruses using MEGA-X. Figure 2 shows the cladogram representation of the phylogenetic tree for the Mpro for the seven coronaviruses, i.e., HCoV-229E, HCoV-NL63, HCoV-HKU1, HCoV-OC43, MERS-CoV, SARS-CoV, and SARS-CoV-2. The SARS-CoV is the closest strain to the newly emerged SARS-CoV-2 and distantly related to HCoV-229E of Mpro coronavirus. The MSA of the Mpro from the different strains of the corona family shown in Fig. 3. The SARS-CoV-2 of Mpro, the percent of sequence identity against SARS-CoV, MERS-CoV, HCoV-HKU1, HCoV-OC43, HCoV-NL63, and HCoV-229E of strains were determined 96.08, 50.65, 49.17, 48.04, 44.30, and 41.04%, respectively. The multiple sequence alignment (MSA) results projected that conserved binding site residues viz, His41, Phe140, Cys145, Gly146, His164, Glu166, and His172 were 100% identical, and the rest of the residues; Ile141, Ser144, and Met163 were similar in highlighted (red color box) region marked in seven strains of corona family (Fig. 3).
Fig. 2.
Phylogenetic tree analysis of the corona family. The Mpro of SARS-CoV-2 evolutionary associated with SARS-CoV and distantly related to H-CoV-229E
Fig. 3.
Conserved Mpro-binding site analysis (red-colored box) in the corona family. The residues of His41, Phe140, Cys145, Gly146, His164, Glu166, and His172 have 100% identity with the family of corona and Ile141, Ser144, and Met163 has physicochemically similar
Validation of docking protocol
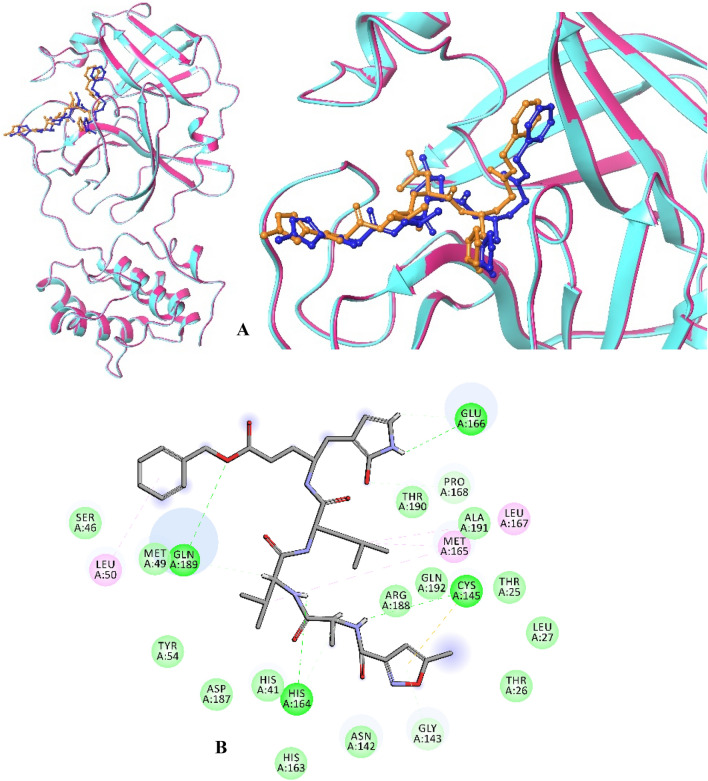
Re-docking, scoring, and ranking methods have been commonly reported for the validating the docking protocols. The pose section by the re-dock into the target-binding site with a known conformation and orientation typically measured, root mean square deviation (RMSD) value from the experimentally derived structure (co-crystal) is considered to have performed successfully. In the present study, the molecular docking analysis was performed through two different docking protocols; Discovery Studio (LibDock) and AutoDock Vina (Figs. 4, 5).
Fig. 4.
Molecular interactions of the native and docked complex of Mpro with N3 peptide by the Discovery Studio of LibDock. a The native co-crystal structure of Mpro (pink) with N3 peptide (blue) and docked structure of Mpro (green) with N3 peptide (red) showed RMSD with 1.53 Å. b Interactions of docked N3 peptide within 4 Ȧ of Mpro
Fig. 5.
Molecular interactions of the native and docked complex of Mpro with N3 peptide by the AutoDock Vina. a The native complex of Mpro (pink) with N3 peptide (blue) has RMSD of 1.84 Å when associated with the docked complex of Mpro(cyan) with N3 peptide (orange). b Interactions of docked N3 peptide within 4 Ȧ of Mpro
Initially, docking protocols were validated with the native structure of Mpro with a co-crystal inhibitor of N3 peptide to reproduce the orientation and position of the N3 observed in the crystal structure (Figs. 4, 5). After docking, the N3 peptide is bound firmly and formed identical six H-bond with His163, Glu166 (3), and Gln189 (2) of Mpro of SARS-CoV-2. Additionally, Ser46, Met49, Phe140, Met165 (3), Pro168, and His172 formed eight H-bond interactions within 4 Ȧ region of N3 peptide (Figs. 4b, 5b), which are significantly replaced with native complex in two different docking approaches. The RMSD between the native and docked structures through LibDock and AutoDock Vina has 1.53 and 1.84 Å, respectively (Figs. 4a, 5a). Thus, the upshot outcome of the identical binding site and superimposed RMSD revealed that the docked complex predominantly supplanted with co-crystal structure N3 peptide with Mpro of SARS-CoV-2 (Figs. 4, 5). Therefore, the validated docking protocols have proceeded further in molecular docking of the Mpro with cordifolioside, berberine, and magnoflorine compounds of T. cordifolia.
Compound-binding analysis
To study the T. cordifolia selected compound-binding mechanisms, cordifolioside, berberine, magnoflorine and N3 (positive control) were docking obsessed by the 3D structure of Mpro of SARS-CoV-2 by utilizing two different docking protocols; LibDock module of Discovery Studio and AutoDock Vina. Remarkably, identical molecular interactions were observed in both the subjects of the protocol. The selected compounds have constituted good docking scores, binding energies, and molecular interactions in comparison to the positive control (Table 3). Among, the cordifolioside has best docking score (LibDock: 142.20; Binding energy: − 21.09 kcal/mol; AutoDock Vina: − 7.0 kcal/mol) in the different protocols when equaled, berberine (LibDock: 95.54; binding energy: − 17.92 kcal/mol; AutoDock Vina: − 6.8 kcal/mol), magnoflorine (LibDock: 84.56; binding energy: − 14.18 kcal/mol; AutoDock Vina: − 6.4 kcal/mol) (Table 4) and N3 peptide (LibDock: 80.84; Binding energy: − 13.20 kcal/mol; AutoDock Vina: − 4.8 kcal/mol) (Table 3).
Table 3.
Molecular docking score, interactions, and bond length of the native and docked complex of Mpro (6LU7) with N3 peptide in the two different docking strategies
| Name of the compound | 2D structure | Discovery studio | AutoDock Vina | |||||
|---|---|---|---|---|---|---|---|---|
| LibDock | Binding energy (kcal/mol) | H-bonds | Bond length (Ȧ) | Docking score (kcal/mol) | H-bonds | Bond length (Ȧ) | ||
| N3 peptide |
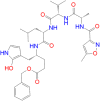
|
80.84 | − 13.20 | Leu50 ← π–π | 5.15 | − 4.8 | Ser46: HG → O8 | 2.02 |
| Gly143: → π–π | 2.56 | Met49 → π | 4.98 | |||||
| Cys145: SG → π–π | 2.54 | Phe140: O ← H89 | 2.76 | |||||
| His164: O ← H59 | 2.62 | His163: HE2 → H88 | 1.79 | |||||
| His164: O → H55 | 2.52 | Met165 ← C26 | 5.03 | |||||
| Met165 ← C18 | 5.29 | Met165: HA → O24 | 2.44 | |||||
| Met165 ← C26 | 4.10 | Met165: SD → π | 4.49 | |||||
| Glu166: OE1 ← H88 | 3.01 | Glu166: OE2 → H89 | 2.43 | |||||
| Glu166: OE1 ← H87 | 2.47 | Glu166: OE2 ← H89 | 2.78 | |||||
| Leu167 ← C26 | 5.29 | Glu166: HN → O24 | 2.02 | |||||
| Pro168: HA → O39 | 2.67 | Pro168 → π | 5.39 | |||||
| Gln189: OE1 ← H60 | 2.41 | His172: HD2 → O3 | 2.82 | |||||
| Gln189: HE21 → O43 | 3.02 | Gln189: OE1 ← H68 | 2.50 | |||||
| Gln189: OE1 → H69 | 2.32 | |||||||
→ ← H-bond donor and acceptor
Table 4.
Molecular docking score, interactions, and bond length of T. cordifolia compound with Mproof SARS-CoV-2 (6LU7)
| Name of the compound | 2D structure | LibDock | Binding energy (kcal/mol) | Auto dock score (kcal/mol) | H-bonds | Bond length (Ȧ) |
|---|---|---|---|---|---|---|
| Cordifolioside |
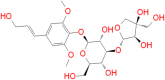
|
142.20 | − 21.09 | − 7.0 | His41 → π | 4.04 |
| His41 → C33 | 3.42 | |||||
| Tyr54: OH → H42 | 1.92 | |||||
| Phe140: O ← H41 | 3.05 | |||||
| Leu141: O ← H55 | 2.90 | |||||
| Ser144: HG → O10 | 2.80 | |||||
| Cys145 ← C33 | 3.53 | |||||
| His163: HE2 → O10 | 2.16 | |||||
| Met165 ← C32 | 5.04 | |||||
| Met165 → π | 4.56 | |||||
| Glu166: HN → O11 | 2.59 | |||||
| Glu166: O ← H40 | 2.87 | |||||
| Glu166: OE1 ← H49 | 2.30 | |||||
| Glu166: O ← H52 | 2.62 | |||||
| Glu166: O ← H61 | 2.72 | |||||
| Leu167: O ← H52 | 2.85 | |||||
| Leu167: O ← H53 | 3.00 | |||||
| Pro168: HA → O9 | 2.43 | |||||
| Asp187: O ← H66 | 2.55 | |||||
| Berberine |

|
95.54 | − 17.92 | − 6.8 | Thr26: O ← H35 | 2.51 |
| Thr26: O ← H36 | 2.93 | |||||
| Cys145 ← π | 4.72 | |||||
| Cys145 ← π | 5.01 | |||||
| Met165 ← C25 | 3.82 | |||||
| Met165 ← π | 4.37 | |||||
| Glu166: O ← H38 | 2.44 | |||||
| Arg188: O ← H42 | 2.66 | |||||
| Gln189: HA → O4 | 2.36 | |||||
| Magnoflorine |
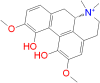
|
84.56 | − 14.18 | − 6.4 | His41 → C25 | 4.48 |
| His41 → π | 5.08 | |||||
| Met49 ← C25 | 4.29 | |||||
| Met165: SD → O1 | 3.09 | |||||
| Pro168 ← C24 | 4.46 | |||||
| Arg188: O ← H26 | 2.82 | |||||
| Gln189: OE1 ← H28 | 2.63 | |||||
| Thr190: O ← H46 | 2.37 |
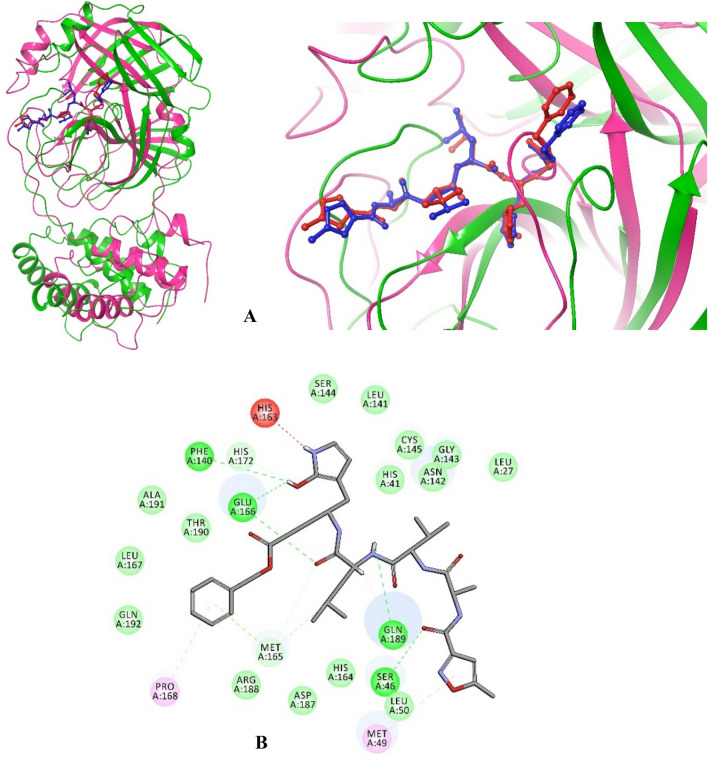
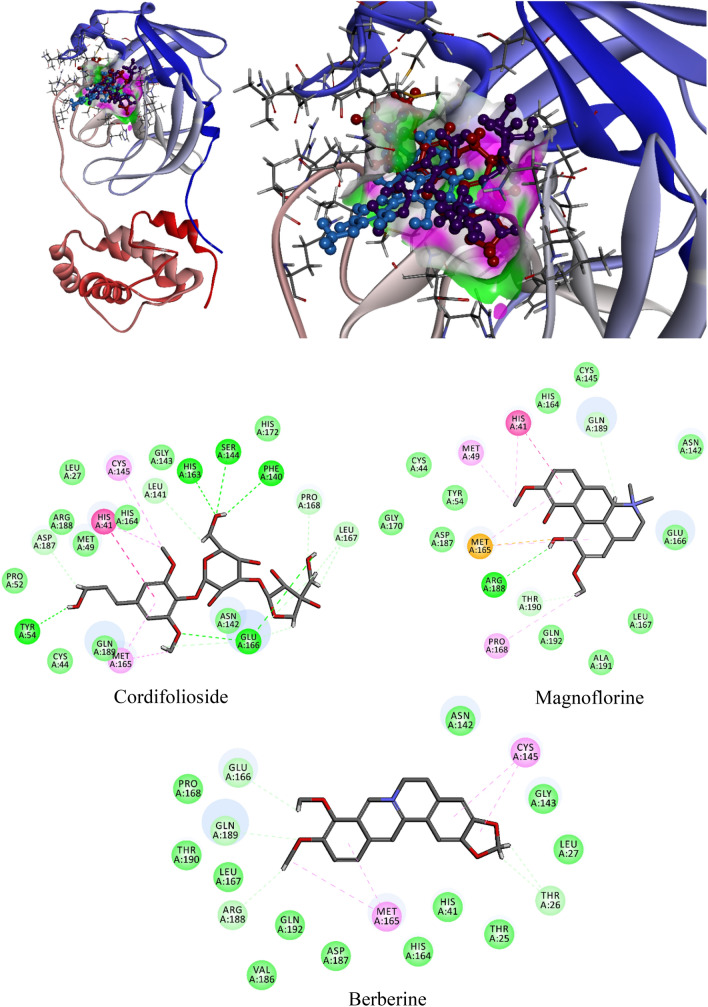
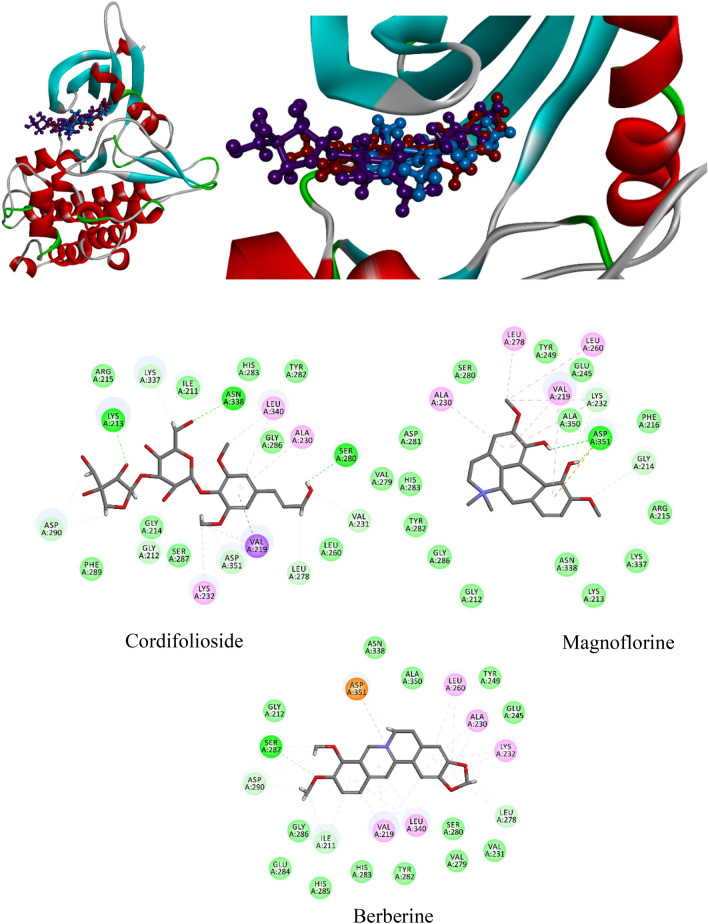
When analyzed for molecular interactions; cordifolioside formed 15 H-bonds with His41, Tyr54, Phe140, Leu141, Ser144, Cys145, His163, Met165, Glu166 (4), Leu167 (2), and Asp187 of Mpro and additionally, His41 and Met165 formed two π–π interactions within 4 Ȧ region (Table 4; Fig. 6). The berberine formed six h-bonds with Thr26 (2), Met165, Glu166, Arg188, Gln189, and three π–π interactions with Cys145 (2) and Met165 of Mpro of SARS-CoV-2. Eventually, the residues of His41, Met49, Met165, Pro168, Arg188, Gln189, and Thr190 formed seven H-bonds and His41 formed one π–π interaction with 4 Å region of magnoflorine (Table 4; Fig. 6). Interestingly, spotted residues were significantly identical to the native complex of Mpro-N3. Thus, the results displayed that, different docking approaches of docking scores and significant molecular interactions with T. cordifolia selected compound were substantial inhibitors or placebo against Mpro of SARS-CoV-2.
Fig. 6.
Molecular interaction of cordifolioside (purple), berberine (red), and magnoflorine (blue) with Mpro of SARS-CoV-2
Molecular dynamics simulations
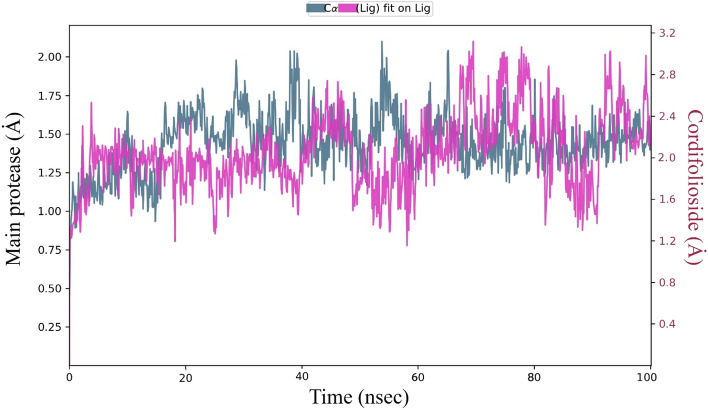
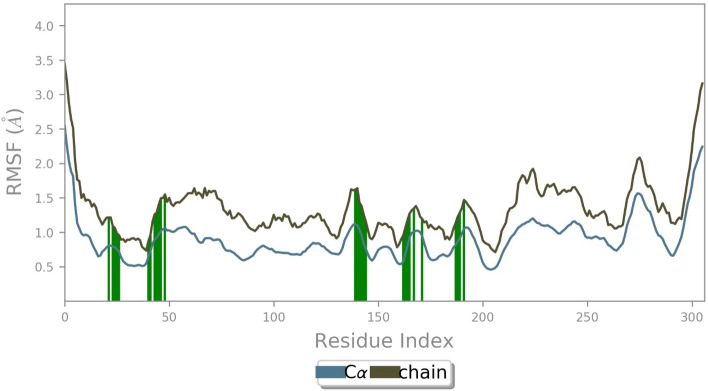
The molecular dynamic simulations were examined with the physical movement of atoms, molecules, and dynamics simulations based on protein and ligand properties of RMSD, root mean square fluctuation (RMSF), molecular interactions, cordifolioside torsion profile, and radius of gyration (rGyr) values as a function of time at 1000 ns period. The RMSD is a quantitative parameter to determine the stability of the Mpro–cordifolioside complex equilibrium was noticed by values of 1.4 and 2.05 Ȧ, respectively, with the stable state throughout the simulation (Fig. 7). The RMSF provides the fluctuation of Mpro residues of Cα-atoms and side chains have an average of 1.44 and 2.61 Ȧ, respectively, in the overall simulation (Fig. 8). RMSF was calculated for Mpro of binding site residues with the cordifolioside, which defined unsteady fluctuation (green lines).
Fig. 7.
RMSD of Cα-atoms of Mpro (greenish-blue) with cordifolioside (pink) complex at 100 ns molecular simulations run
Fig. 8.
RMSF of Mpro of Cα-atoms and side-chain with binding site residues (green) at 100 ns molecular simulations run
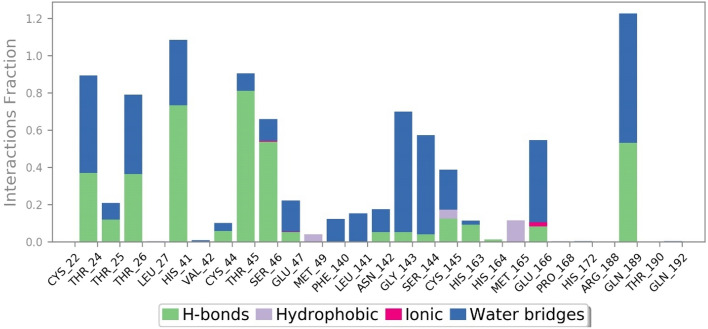
The post dynamics simulation of Mpro of Thr24, Thr25, Thr26, His41, Val42, Cys44, Thr45, Ser46, Glu47, Asn142, Gly143, Ser144, Cys145, His163, His164, Glu166, and Gln189 residues formed H-bond and HOH intermediated interactions within 4 Ȧ of cordifolioside (Fig. 9). Amongst, Leu27, Met49, Cys145, Met165, and Pro168 also formed hydrophobic and metal ion interaction with Glu166 with the compound throughout the simulation time (Fig. 9). Thus, it was predicted that the H-bond, HOH, and metal ion interactions play a significant role in drug design because of their strong influence on drug specificity, metabolization, and adsorption.
Fig. 9.
Binding site residues of SARS-CoV-2 Mpro displaying H-bonds, Hydrophobic, Ionic, and water-associated interactions with cordifolioside
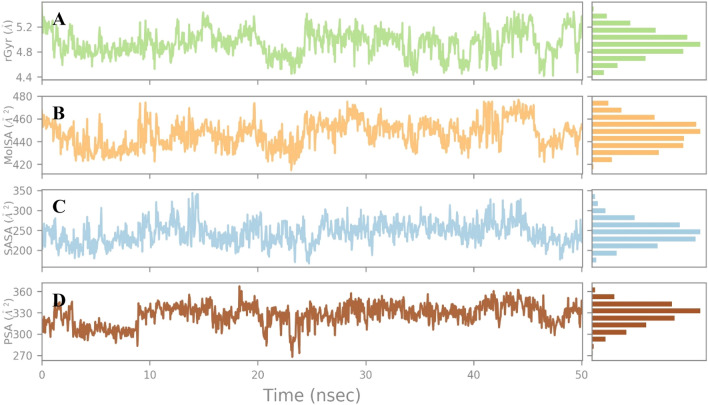
The rGyr is the radial distance to a point, which would measure the compactness of the ligand with the moment of inertia, and the total mass of the ligand was concentrated. The cordifolioside showed the equilibrium around 4.9 Å (Fig. 10a) which is stable at simulations runtime. The MolSA is a surface calculation with the default value of 1.4 Å probe radius, which is equivalent to a van der Waals surface area of the compound. The MolSA of the cordifolioside range of 420–480 Å2 and the equilibrium around 446.91 Å2, which is constant throughout the total simulation run time of 100 ns (Fig. 10b). The surface area of a cordifolioside molecule was accessible by an HOH molecule, the results portrayed that, it ranged around 200–350 Å2, and the equilibrium around 243.48 Å2 (Fig. 10c). The PSA denotes the surface area in a molecule contributed only by oxygen and nitrogen atoms of the cordifolioside, which depicts the range between 270 and 360 Å2 and the equilibrium around 326.94 Å2 (Fig. 10d). Remarkably, the torsion angle of the cordifolioside of each bond was observed during the simulation time, which is stable during the MD run. Hence, the cordifolioside properties by the MD simulations confirm that the stability of the ligand in the cleft between the two-domain region of the Mpro (Fig. 11).
Fig. 10.
Cordifolioside properties of average a rGyr: 4.9, b MolSA: 446.9, c SASA: 243.4 and d PSA: 326.9 were stable at MD simulation at 100 ns simualtions
Fig. 11.
Depiction of cordifolioside and Mpro SARS-CoV-2 complex involved in torsion angle graph plotted at 100 ns simulation time
Depicted from the different molecular docking approaches and dynamics simulations results presented that, Mpro residues His41, Ser144, Cys145, His163, His164, and Glu166 identically formed H-bonds with cordifolioside. Moreover, the natural compound also formed significant H-bond towards the common conserved binding site residues His41, Ser144, Cys145, His163, His164, and Glu166 among all corona family strains.
Concurrently, we also performed molecular docking approaches for the human anti-inflammatory and pro-inflammatory at molecular level analysis for the bioactive compounds of T. cordifolia.
Anti-inflammatory and pro-inflammatory analysis
In the majority of the patients with SARS-CoV-2, mortality is due to lung failure by acute respiratory distress syndrome (ARDS) in which fluid fills up the air sacs in the lungs (Manne et al. 2018). The syndrome is attributed for elevated cytokine concentration level on account of the over-stimulated immune response of inflammatory and pro-inflammatory characterized by a cytokine storm, edema, and fibrosis in the lungs at the end stages (Manne et al. 2018; Singleton and Rossi 1965; Sliwoski et al. 2014; Sunanda and Ainapure 2020). Considering innate immunity at the gene level, a recent study on targeting stimulated hub genes has identified for significant interaction with agonists for COVID-19 (Prasad et al. 2020).
In the majority of the patients with SARS-CoV-2, it has been observed that transforming growth factor-beta (TGF-β) (Xu et al. 2000; Liu et al. 2020; Leng et al. 2020) and tumor necrosis factor-alpha (TNF-α) (O’Connell et al. 2019; Wan et al. 2020; Jin et al. 2020) levels were elevated with more severe diseases. Sharma et al., Aranha et al. and Kalikar et al. stated that the bioactive compounds mainly cordifolioside, berberine, and magnoflorine of T. cordifolia are having beneficial immune-modulator activity (Kalikar et al. 2008; Aranha et al. 2012; Sharma et al. 2012). In the current study, we performed multiple molecular docking strategies against cordifolioside, berberine, and magnoflorine with TGF-β and TNF-α to check the molecular interactions (Figs. 12, 13). The co-crystal structure of TGF-β with inhibitor of PY1 (4-(3-pyridin-2-yl-1 h-pyrazol-4-yl))quinolone (PDB ID: 1PY5) (Sawyer et al. 2004) and TNF-α with the small molecule inhibitor of (R)-{1-[(2,5-dimethylphenyl)methyl]-6-(1-methyl-1H-pyrazol-4-yl)-1H-benzimidazol-2-yl}(pyridin-4-yl)methanol (PDB ID: 6OP0) (O’Connell et al. 2019) were obtained from PDB and implemented with two docking approaches (Figs. 12, 13).
Fig. 12.
Binding modes and molecular interactions of T. cordifolia selected compounds with anti-inflammatory biomarkers of TGF-β (1PY5) by a molecular docking simulation study
Fig. 13.
Binding mode and interactive residues of T. cordifolia selected compound with a pro-inflammatory biomarker of TNF-α (6OP0) obtained by molecular docking simulation
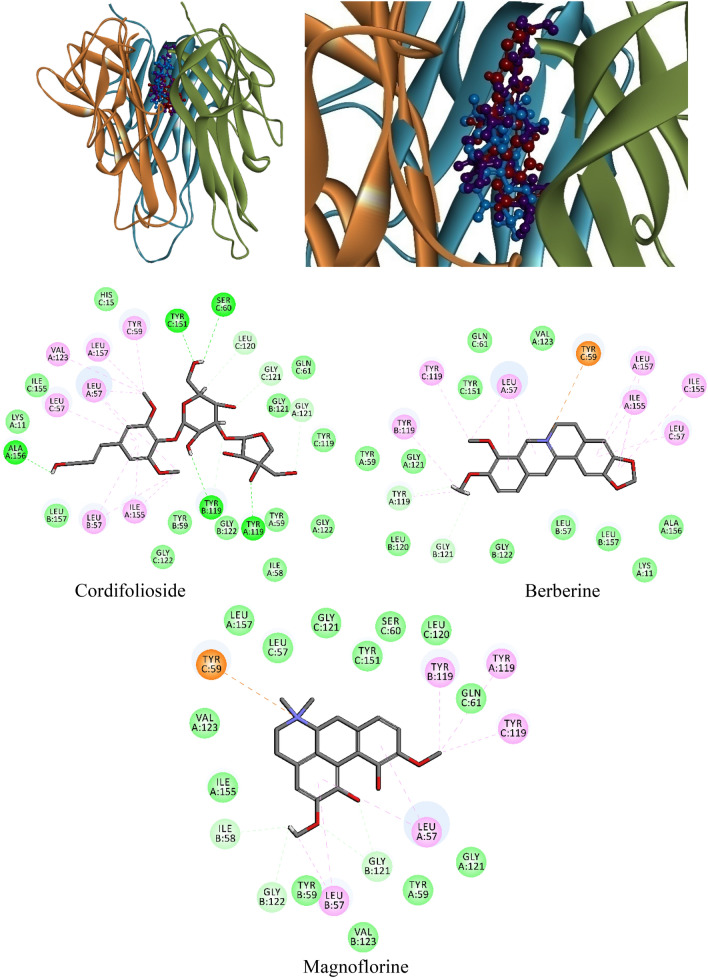
In the native structure of TGF-β, the residues His283 and Asp351 formed H-bond with the co-crystal inhibitor of PY1 and Ile211, Val219, Ala230, Val231, Lys232, Leu278, Ser280, Asp281, Tyr282, Gly286, and Leu340, which was involved in VdW within 4Å region. Another native structure of TNF-α (homomer) which consists of three chains (A, B, and C) formed three H-bond with chain-A: Ser60, and Tyr119, and chain-C of Tyr151. Whereas, chain-A: Leu57, Tyr59, Val123, Ile155 and Leu157, chain-B: Leu57, Tyr119, Gly121 and chain-C residues of Leu57, Tyr59, Ser60, Tyr119, Leu120, and Ile155 formed Vdw interactions within 4 Å region of the small inhibitor of A7A. Therefore, a grid box was generated in accordance with co-crystal inhibitors (positive control), where interactive residues fall within 4 Å region for molecular docking analysis implemented with two different docking strategies. Remarkably, cordifolioside, berberine, and magnoflorine had good docking scores, binding energies, and binding interactions with TGF-β and TNF-α, respectively, in comparison to the positive control. In both the docking approaches, cordifolioside had defined with the best docking score, binding energy, and molecular interactions with human TGF-β and TNF-α (Tables 5, 6). The cordifolioside formed 16 H-bond with residues Gly212, Lys213 (4), Val219, Val231, Lys232, Leu278, Ser280, Asp290 (2), Lys337, Asn338, Leu340, and Asp351 of human TGF-β, followed by Val219, Ala230 and Leu340 residues formed π–π interactions within 4 Å region (Fig. 12). The homomer human-TNF-α active site residue of chain-A: Leu57 (2), Tyr119, Gly121, Val123, Ile155, Ala156, Leu157, chain-B: Leu57, Tyr119 (2), and chain-C: Tyr59, Ser60, Leu120, Gly121, and Tyr151 residues formed 16 H-bonds and additionally, chain-A: Leu57, Ile155, chain-B: Leu57 and chain-C of Leu57 were significantly involved in π–π interactions (Fig. 13).
Table 5.
Molecular docking score, interactions and bond length of T. cordifolia selected compound with anti-inflammatory biomarker of TGF-β (1PY5)
| Name of the compound | 2D structure | LibDock | Binding energy (kcal/mol) | Auto dock score (kcal/mol) | H-bonds | Bond length (Ȧ) |
|---|---|---|---|---|---|---|
| Cordifolioside |
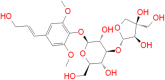
|
98.19 | − 14.54 | − 9.12 | Gly212: HA1 → O7 | 2.4 |
| Lys213: HN → O5 | 1.95 | |||||
| Lys213: HZ1 → O5 | 3.09 | |||||
| Lys213: O ← H46 | 2.46 | |||||
| Lys213: HN–O5 | 1.95 | |||||
| Val219: HG23 → π | 2.58 | |||||
| Val219: O ← C33 | 4.77 | |||||
| Ala230 ← π | 5.39 | |||||
| Val231: HA ← O13 | 2.99 | |||||
| Lys232 ← C33 | 4.76 | |||||
| Leu278: O ← H67 | 2.92 | |||||
| Ser280: OG → H42 | 2.16 | |||||
| Asp290: OD1 ← H49 | 2.61 | |||||
| Asp290: OD2 ← H52 | 2.48 | |||||
| Lys337: O ← H55 | 2.41 | |||||
| Asn338: HD21 ← O10 | 2.16 | |||||
| Leu340 ← C32 | 4.66 | |||||
| Leu340 ← π | 5.36 | |||||
| Asp351: OD1 ← H64 | 2.73 | |||||
| Berberine |
|
95.5439 | -17.92 | − 6.8 | Gly214: HA1 ← O4 | 2.94 |
| Val219 → π | 5.36 | |||||
| Val219 → π | 4.46 | |||||
| Ala230 → π | 5.36 | |||||
| Lys232 → π | 5.26 | |||||
| Lys232 ← C24 | 4.16 | |||||
| Lys232: HE1 ← O3 | 2.8 | |||||
| Leu260 → C24 | 5.26 | |||||
| Leu278 → C24 | 4.46 | |||||
| Asp351: OD1 → H26 | 1.9 | |||||
| Asp351: OD1 → H27 | 2.01 | |||||
| Asp351: OD2 → π | 3.91 | |||||
| Magnoflorine |
|
84.5607 | − 14.18 | − 6.4 | Ile211: O ← H42 | 2.25 |
| Ile211 ← C25 | 5.36 | |||||
| Ile211 ← π | 4.81 | |||||
| Val219 ← π | 5.2 | |||||
| Val219 ← π | 4.18 | |||||
| Val219 ← π | 4.86 | |||||
| Ala230 ← π | 4.73 | |||||
| Ala230 ← π | 4.94 | |||||
| Lys232 ← π | 4.24 | |||||
| Lys232 ← π | 4.47 | |||||
| Leu260 ← π | 5.48 | |||||
| Leu260 ← π | 5.2 | |||||
| Leu278: O ← H35 | 2.2 | |||||
| Leu278 ← π | 4.94 | |||||
| Ser287: HN → O4 | 2.39 | |||||
| Ser287: HB1 → O3 | 2.66 | |||||
| Asp290: OD2 ← H43 | 2.35 | |||||
| Asp290: OD2 ← H38 | 2.8 | |||||
| Leu340 ← π | 5.31 | |||||
| Leu340 ← π | 4.92 | |||||
| Asp351: OD2 ← N5 | 4.14 | |||||
| Asp351: OD2 ← H26 | 2.38 |
Table 6.
Molecular docking scores, molecular interactions, and bond length of T. cordifolia selected compound with a pro-inflammatory biomarker of TNF-α (6OP0)
| Name of the compound | 2D structure | LibDock | Binding energy (kcal/mol) | Auto dock score (kcal/mol) | H-bonds | Bond length (Ȧ) |
|---|---|---|---|---|---|---|
| Cordifolioside |
|
173.92 | − 27.26 | − 7.0 | A: Leu57 ← π | 4.82 |
| A: Leu57 ← C33 | 4.66 | |||||
| A: Leu57 ← C32 | 4.52 | |||||
| A: Tyr119: HH → O4 | 1.83 | |||||
| A: Gly121: HA1 → O9 | 2.27 | |||||
| A: Val123 ← C32 | 4.67 | |||||
| A: Ile155 ← π | 4.61 | |||||
| A: Ile155 ← C33 | 4.54 | |||||
| A: Ala156: O ← H42 | 1.75 | |||||
| A: Leu157 ← C32 | 4.17 | |||||
| B: Leu57 ← C33 | 4.33 | |||||
| B: LEU57 ← π | 5.28 | |||||
| B: Tyr119: OH ← H43 | 2.54 | |||||
| B: Tyr119: OH ← H39 | 2.87 | |||||
| C: Leu57 ← π | 5.32 | |||||
| C: Tyr59 → C32 | 3.38 | |||||
| C: Ser60: O ← H41 | 2.85 | |||||
| C: Leu120: O ← H48 | 2.58 | |||||
| C: Gly121: HA1 → O6 | 2.14 | |||||
| C: Tyr151: HH → O10 | 1.89 | |||||
| Berberine |
|
113.54 | − 25.86 | − 6.8 | A: Leu57 ← π | 4.77 |
| A: Leu57 ← π | 4.21 | |||||
| A: Leu57 ← C24 | 5.21 | |||||
| A: Tyr119: OH ← H43 | 2.48 | |||||
| A: Tyr119 → C25 | 5.00 | |||||
| A: Ile155 ← π | 4.86 | |||||
| A: Ile155 ← π | 5.03 | |||||
| A: Leu157 ← π | 5.39 | |||||
| A: Leu157 → π | 5.1 | |||||
| B: Tyr119 → C25 | 4.59 | |||||
| B: Gly121: O ← H41 | 2.86 | |||||
| C: Leu57 ← π | 5.07 | |||||
| C: Tyr59 ← N5 | 4.89 | |||||
| C: Tyr119 → C24 | 4.85 | |||||
| C: Ile155 ← π | 5.49 | |||||
| Magnoflorine |
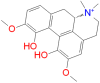
|
84.56 | − 14.18 | − 6.4 | A: Leu57 ← π | 4.74 |
| A: Leu57 ← π | 5.19 | |||||
| A: Tyr119 → C25 | 4.57 | |||||
| B: Leu57 ← C24 | 3.68 | |||||
| B: Leu57 ← π | 5.09 | |||||
| B: Ile58: O ← H46 | 2.12 | |||||
| B: Tyr119 → C25 | 4.39 | |||||
| B: Gly121: HA1 → O1 | 2.56 | |||||
| B: Gly121: HA1 → O2 | 2.64 | |||||
| B: Gly122: O ← H46 | 2.29 | |||||
| C: Tyr59 ← N5 | 4.15 | |||||
| C: Tyr119 → C25 | 3.64 |
Thus, the bioactive compound, cordifolioside from T. cordifolia exhibited a higher degree of interaction with the Mpro accompanied by the lowest binding energy and consistently stable conformation, which might perceive as a potential antagonist against SARS-CoV-2, including beneficial immunomodulator distinct toxic effects for candidates. However, additional exploration is inevitable for the current findings of the inherent use in vivo activity.
Conclusion
An attempt was made in the present study to identify natural bioactive compound which perhaps effective against SARS-CoV-2 with immune-modulator activity, such studies are scarcely observed. The bioactive compounds cordifolioside, berberine, and magnoflorine established in the study varied significantly among different solvent extracts and portrayed antioxidant activity for phenolic compounds. The MD simulation investigations confirmed that the cordifolioside proposed in the study confer stable confirmations, indicating high inhibitory activity towards the binding pocket of Mpro. The exploited sequence to structure and dynamics simulations study revealed that, cordifolioside as an attractive inhibitor against Mpro of SARS-CoV-2 which can majorly act as an immune modulator on human TGF-β and TNF-α. The compound also paved a strong groundwork for other computational and practical drug discovery methods to determine the active stabilized inhibitors in deteriorating the symptoms and spread of SARS-CoV-2 that can block its activity. Thus, the findings provided constructive insight and evidence for cordifolioside from T. cordifolia might use as a natural bioactive compound in treating COVID-19 causing infection. However, preclinical studies would afford evidence for these budding molecules to perceive long-term drug design techniques to understand the mechanism of actions and drugability of the bioactive compound in the anti-viral treatment regimen allied with COVID-19 infection.
Acknowledgements
MM highly thankful to TATA trust for providing facilities at the NIN-TATA Centre of Excellence in Public Health and Nutrition, ICMR-National Institute of Nutrition, Hyderabad, Telangana State. GG highly thankful for the financial support from RKVY RAFTAAR, DAC and FW, Ministry of Agriculture, Government of India, New Delhi, India and KRISHIK ABI, University of Agricultural Sciences, Dharwad, Karnataka, India for carrying out the research work (Sanction No. RKVY-RAFTAAR/K-ABI/71/2020-21).
Author contributions
MM: in silico analysis, conceptualization, methodology, writing–original draft preparation; GG: in vitro analysis, conceptualization, data curation, writing–original draft preparation; SRV: validation, writing–reviewing and editing; MCK: in vitro validation, writing–reviewing and editing; HK: in silico validation, writing–reviewing and editing; PN: in silico validation, writing–reviewing and editing; MDU: statistical analysis and table preparation; VAY: in vitro validation, writing–reviewing and editing; SC: writing–reviewing and editing; PD: in vitro validation, writing–reviewing and editing; TDSS: writing–reviewing and editing and SP: writing–reviewing and editing; VP: writing–reviewing and editing. All authors revised and reviewed the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare no conflicts and competing interest over the work.
Footnotes
Munikumar Manne and Giridhar Goudar contributed equally.
Contributor Information
Munikumar Manne, Email: mannemunikumar.bioinfo@gmail.com.
Giridhar Goudar, Email: giri1411@rediffmail.com.
References
- Ali H, Dixit S. Extraction optimization of Tinospora cordifolia and assessment of the anticancer activity of its alkaloid palmatine. Sci World J. 2013 doi: 10.1155/2013/376216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K, Ziebuhr J, Wadhwani P, et al. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science (80-) 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- Aranha I, Clement F, Venkatesh YP. Immunostimulatory properties of the major protein from the stem of the ayurvedic medicinal herb, guduchi (Tinospora cordifolia) J Ethnopharmacol. 2012;139:366–372. doi: 10.1016/j.jep.2011.11.013. [DOI] [PubMed] [Google Scholar]
- Ashok B, Ravishankar B, Prajapati P, Bhat S. Antipyretic activity of guduchi ghrita formulations in albino rats. AYU (Int Q J Res Ayurveda) 2010;31:367. doi: 10.4103/0974-8520.77162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asres K, Seyoum A, Veeresham C, et al. Naturally derived anti-HIV agents. Phyther Res. 2005;19:557–581. doi: 10.1002/ptr.1629. [DOI] [PubMed] [Google Scholar]
- Azam SS, Abbasi SW. Molecular docking studies for the identification of novel melatoninergic inhibitors for acetylserotonin-O-methyltransferase using different docking routines. Theor Biol Med Model. 2013;10:63. doi: 10.1186/1742-4682-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzie IFF, Strain JJ. The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”: The FRAP assay. Anal Biochem. 1996;239:70–76. doi: 10.1006/abio.1996.0292. [DOI] [PubMed] [Google Scholar]
- Brand-Williams W, Cuvelier ME, Berset C. Use of a free radical method to evaluate antioxidant activity. LWT Food Sci Technol. 1995;28:25–30. doi: 10.1016/S0023-6438(95)80008-5. [DOI] [Google Scholar]
- Chen J. Pathogenicity and transmissibility of 2019-nCoV—a quick overview and comparison with other emerging viruses. Microbes Infect. 2020;22:69–71. doi: 10.1016/j.micinf.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Wu D, Guo W, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Han W, Wang G, Zhao X. Application prospect of polysaccharides in the development of anti-novel coronavirus drugs and vaccines. Int J Biol Macromol. 2020;164:331–343. doi: 10.1016/j.ijbiomac.2020.07.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinatl J, Morgenstern B, Bauer G, et al. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet. 2003;361:2045–2046. doi: 10.1016/S0140-6736(03)13615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahanukar SA, Thatte UM, Rege NN. Immunomodulatory agents from plants. Basel: Birkhäuser; 1999. Immunostimulants in ayurveda medicine; pp. 289–323. [Google Scholar]
- Du J, He ZD, Jiang RW, et al. Antiviral flavonoids from the root bark of Morus alba L. Phytochemistry. 2003;62:1235–1238. doi: 10.1016/S0031-9422(02)00753-7. [DOI] [PubMed] [Google Scholar]
- Ganjhu RK, Mudgal PP, Maity H, et al. Herbal plants and plant preparations as remedial approach for viral diseases. VirusDisease. 2015;26:225–236. doi: 10.1007/s13337-015-0276-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudar G, Sathisha GJ. Effect of processing on ferulic acid content in foxtail millet (Setaria italica) grain cultivars evaluated by HPTLC. Orient J Chem. 2016;32:2251–2258. doi: 10.13005/ojc/320458. [DOI] [Google Scholar]
- Goudar G, Khetagoudar MC, Patil SR. Evaluation of hydroxycinnamic acids in finger millet (Eleusine coracana) grain cultivars by HPTLC. Int J Chem Stud. 2018;6:328–332. [Google Scholar]
- Higgins DG, Sharp PM. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Alberto A, Ribas-Aparicio RM, Aparicio-Ozores G, Castelán-Vega JA. Virtual screening of approved drugs as potential SARS-CoV-2 main protease inhibitors. Comput Biol Chem. 2020 doi: 10.1016/j.compbiolchem.2020.107325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Du X, Xu Y, et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Jyoti VV, Giridhar G, Shameembanu AB, et al. Identification of bio-active components in leaf extracts of Aloe vera, Ocimum tenuiflorum (Tulasi) and Tinospora cordifolia (Amrutballi) J Med Plants Res. 2015;9:764–770. doi: 10.5897/jmpr2013.5197. [DOI] [Google Scholar]
- Kalikar M, Thawani V, Varadpande U, et al. Immunomodulatory effect of Tinospora cordifolia extract in human immuno-deficiency virus positive patients. Indian J Pharmacol. 2008;40:107–110. doi: 10.4103/0253-7613.42302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan S, Kolandaivel P. Antiviral potential of natural compounds against influenza virus hemagglutinin. Comput Biol Chem. 2017;71:207–218. doi: 10.1016/j.compbiolchem.2017.11.001. [DOI] [PubMed] [Google Scholar]
- Khandare AL, Validandi V, Manne M, et al. Tamarind fruit extract ameliorates fluoride toxicity by upregulating carbonic anhydrase II: a mechanistic study. Fluoride. 2018;51:137–152. [Google Scholar]
- Killerby ME, Biggs HM, Haynes A, et al. Human coronavirus circulation in the United States 2014–2017. J Clin Virol. 2018;101:52–56. doi: 10.1016/j.jcv.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodchakorn K, Poovorawan Y, Suwannakarn K, Kongtawelert P. Molecular modelling investigation for drugs and nutraceuticals against protease of SARS-CoV-2. J Mol Graph Model. 2020 doi: 10.1016/j.jmgm.2020.107717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumoto IT, Nakabayashi T, Kida H, et al. Screening of various plant extracts used in ayurvedic medicine for inhibitory effects on human immunodeficiency virus type 1 (HIV-1) protease. Phyther Res. 1995;9:180–184. doi: 10.1002/ptr.2650090305. [DOI] [Google Scholar]
- Leimkuhler BJ, Sweet CR. The canonical ensemble via symplectic integrators using Nosé and Nosé-Poincaré chains. J Chem Phys. 2004;121:108–116. doi: 10.1063/1.1740753. [DOI] [PubMed] [Google Scholar]
- Leng Z, Zhu R, Hou W, et al. Transplantation of ACE2-mesenchymal stem cells improves the outcome of patients with COVID-19 pneumonia. Aging Dis. 2020;11:216. doi: 10.14336/AD.2020.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Pitsillou E, Karagiannis C, et al. Interaction of the prototypical α-ketoamide inhibitor with the SARS-CoV-2 main protease active site in silico: molecular dynamic simulations highlight the stability of the ligand-protein complex. Comput Biol Chem. 2020 doi: 10.1016/j.compbiolchem.2020.107292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Ho CT, Chuo WH, et al. Effective inhibition of MERS-CoV infection by resveratrol. BMC Infect Dis. 2017 doi: 10.1186/s12879-017-2253-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Yang Y, Zhang C, et al. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci China Life Sci. 2020;63:364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV) Biosci Trends. 2020 doi: 10.5582/BST.2020.01020. [DOI] [PubMed] [Google Scholar]
- Mani JS, Johnson JB, Steel JC, et al. Natural product-derived phytochemicals as potential agents against coronaviruses: a review. Virus Res. 2020;284:197989. doi: 10.1016/j.virusres.2020.197989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manne M, Validandi V, Khandare AL. Reduction of fluoride toxicity by tamarind components: an in silico study. Fluoride. 2018;51:122–136. [Google Scholar]
- Meyer-Almes FJ. Repurposing approved drugs as potential inhibitors of 3CL-protease of SARS-CoV-2: virtual screening and structure based drug design. Comput Biol Chem. 2020 doi: 10.1016/j.compbiolchem.2020.107351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GM, Ruth H, Lindstrom W, et al. Software news and updates AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munikumar M, Krishna VS, Reddy VS, et al. In silico design of small peptides antagonist against leptin receptor for the treatment of obesity and its associated immune-mediated diseases. J Mol Graph Model. 2018;82:20–36. doi: 10.1016/j.jmgm.2018.04.002. [DOI] [PubMed] [Google Scholar]
- Munikumar M, Natarajan P, Amineni U, Radha Krishna KV. Discovery of potential lumazine synthase antagonists for pathogens involved in bacterial meningitis: In silico study. Inform Med Unlocked. 2019 doi: 10.1016/j.imu.2019.100187. [DOI] [Google Scholar]
- Naik VR, Munikumar M, Ramakrishna U, et al. Remdesivir (GS-5734) as a therapeutic option of 2019-nCOV main protease–in silico approach. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1781694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod. 2016;79:629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- O’Connell J, Porter J, Kroeplien B, et al. Small molecules that inhibit TNF signalling by stabilising an asymmetric form of the trimer. Nat Commun. 2019 doi: 10.1038/s41467-019-13616-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchabhai TS, Kulkarni UP, Rege NN. Validation of therapeutic claims of Tinospora cordifolia: a review. Phyther Res. 2008;22:425–441. doi: 10.1002/ptr.2347. [DOI] [PubMed] [Google Scholar]
- Patel MB, Mishra SM. Magnoflorine from Tinospora cordifolia stem inhibits α-glucosidase and is antiglycemic in rats. J Funct Foods. 2012;4:79–86. doi: 10.1016/j.jff.2011.08.002. [DOI] [Google Scholar]
- Pitsillou E, Liang J, Karagiannis C, et al. Interaction of small molecules with the SARS-CoV-2 main protease in silico and in vitro validation of potential lead compounds using an enzyme-linked immunosorbent assay. Comput Biol Chem. 2020 doi: 10.1016/j.compbiolchem.2020.107408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polu PR, Nayanbhirama U, Khan S, Maheswari R. Assessment of free radical scavenging and anti-proliferative activities of Tinospora cordifolia Miers (Willd) BMC Complement Altern Med. 2017 doi: 10.1186/s12906-017-1953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan D, Priyadarshini V, Munikumar M, et al. Para-(benzoyl)-phenylalanine as a potential inhibitor against LpxC of Leptospira spp.: homology modeling, docking, and molecular dynamics study. J Biomol Struct Dyn. 2014;32:37–41. doi: 10.1080/07391102.2012.758056. [DOI] [PubMed] [Google Scholar]
- Prasad K, Khatoon F, Rashid S, et al. Targeting hub genes and pathways of innate immune response in COVID-19: a network biology perspective. Int J Biol Macromol. 2020;163:1–8. doi: 10.1016/j.ijbiomac.2020.06.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prathima P, Venkaiah K, Pavani R, et al. α-Lipoic acid inhibits oxidative stress in testis and attenuates testicular toxicity in rats exposed to carbimazole during embryonic period. Toxicol Rep. 2017;4:373–381. doi: 10.1016/j.toxrep.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C, Zhou L, Hu Z, et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SK, Rao PS, Rao BN. Preliminary investigation of the radiosensitizing activity of guduchi (Tinospora cordifolia) in tumor-bearing mice. Phyther Res. 2008;22:1482–1489. doi: 10.1002/ptr.2508. [DOI] [PubMed] [Google Scholar]
- Re R, Pellegrini N, Proteggente A, et al. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic Biol Med. 1999;26:1231–1237. doi: 10.1016/S0891-5849(98)00315-3. [DOI] [PubMed] [Google Scholar]
- Rosendaal FR. Review of: “Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial Gautret et al 2010. Int J Antimicrob Agents. 2020;56:106063. doi: 10.1016/j.ijantimicag.2020.106063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer JS, Beight DW, Britt KS, et al. Synthesis and activity of new aryl- and heteroaryl-substituted 5,6-dihydro-4H-pyrrolo[1,2-b]pyrazole inhibitors of the transforming growth factor-β type I receptor kinase domain. Bioorgan Med Chem Lett. 2004;14:3581–3584. doi: 10.1016/j.bmcl.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Shanbhag T, Shenoy S, Rao MC. Wound healing profile of Tinospora cordifolia. Indian Drugs. 2005;42:217–221. [Google Scholar]
- Sharma U, Bala M, Kumar N, et al. Immunomodulatory active compounds from Tinospora cordifolia. J Ethnopharmacol. 2012;141:918–926. doi: 10.1016/j.jep.2012.03.027. [DOI] [PubMed] [Google Scholar]
- Sharma P, Dwivedee BP, Bisht D, et al. The chemical constituents and diverse pharmacological importance of Tinospora cordifolia. Heliyon. 2019;5:e02437. doi: 10.1016/j.heliyon.2019.e02437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivananjappa MM, Muralidhara Abrogation of maternal and fetal oxidative stress in the streptozotocin-induced diabetic rat by dietary supplements of Tinospora cordifolia. Nutrition. 2012;28:581–587. doi: 10.1016/j.nut.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Shree P, Mishra P, Selvaraj C, et al. Targeting COVID-19 (SARS-CoV-2) main protease through active phytochemicals of ayurvedic medicinal plants—Withania somnifera (Ashwagandha), Tinospora cordifolia (Giloy) and Ocimum sanctum (Tulsi)—a molecular docking study. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1810778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton VL, Rossi JA. Colorimetry of total phenolics with phosphomolybdic-phosphotungstic acid reagents. Am J Enol Vitic. 1965;16:144–158. [Google Scholar]
- Sliwoski G, Kothiwale S, Meiler J, Lowe EW. Computational methods in drug discovery. Pharmacol Rev. 2014;66:334–395. doi: 10.1124/pr.112.007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunanda DN, Ainapure S. Antiallergic properties of Tinospora cordifolia in animal models. Indian J Pharmacol. 2020;18:250. [Google Scholar]
- Supe A, Purandare H. Immunomodulatory role of Tinospora cordifolia as an adjuvant in surgical treatment of diabetic foot ulcers: a prospective randomized controlled study. Indian J Med Sci. 2007;61:347. doi: 10.4103/0019-5359.32682. [DOI] [PubMed] [Google Scholar]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2009 doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilar S, Karpiak J, Berk B, Costanzi S. In silico analysis of the binding of agonists and blockers to the β2-adrenergic receptor. J Mol Graph Model. 2011;29:809–817. doi: 10.1016/j.jmgm.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadood N, Wadood A, Shah SAW. Effect of Tinospora cordifolia on blood glucose and total lipid levels of normal and alloxan-diabetic rabbits. Planta Med. 1992;58:131–136. doi: 10.1055/s-2006-961414. [DOI] [PubMed] [Google Scholar]
- Wan S, Yi Q, Fan S, et al. Characteristics of lymphocyte subsets and cytokines in peripheral blood of 123 hospitalized patients with 2019 novel coronavirus pneumonia (NCP) New York: Cold Spring Harbor Laboratory Press; 2020. [Google Scholar]
- Wang M, Cao R, Zhang L, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Li J, Zhu G, et al. Clinical features of maintenance hemodialysis patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. Clin J Am Soc Nephrol. 2020 doi: 10.2215/cjn.04160320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu HX, Ming DS, Hui D, But PPH. A new anti-HIV triterpene from Geum japonicum. Chem Pharm Bull. 2000;48:1367–1369. doi: 10.1248/cpb.48.1367. [DOI] [PubMed] [Google Scholar]
- Yadav RNS, Agarwala M. Phytochemical analysis of some medicinal plants. J Phytol. 2011;3:10–14. [Google Scholar]
- Yang Y, Peng F, Wang R, et al. The deadly coronaviruses: the 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J Autoimmun. 2020;109:102434. doi: 10.1016/j.jaut.2020.102434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Du L, Ojcius DM, et al. Measures for diagnosing and treating infections by a novel coronavirus responsible for a pneumonia outbreak originating in Wuhan, China. Microbes Infect. 2020;22:74–79. doi: 10.1016/j.micinf.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]