Abstract
Middle East respiratory syndrome coronavirus (MERS-CoV) is the causative agent of a severe respiratory disease with a high mortality of ~ 35%. The lack of approved treatments for MERS-CoV infection underscores the need for a user-friendly system for rapid drug screening. In this study, we constructed a MERS-CoV replicon containing the Renilla luciferase (Rluc) reporter gene and a stable luciferase replicon-carrying cell line. Using this cell line, we showed that MERS-CoV replication was inhibited by combined application of lopinavir and ritonavir, indicating that this cell line can be used to screen inhibitors of MERS-CoV replication. Importantly, the MERS-replicon cell line can be used for high-throughput screening of antiviral drugs without the need for live virus handling, providing an effective and safe tool for the discovery of antiviral drugs against MERS-CoV.
Electronic supplementary material
The online version of this article (10.1007/s12250-020-00341-z) contains supplementary material, which is available to authorized users.
Keywords: Middle East respiratory syndrome coronavirus (MERS-CoV), Replicon cell line, Antiviral screening, Luciferase reporter gene
Introduction
Middle East respiratory syndrome coronavirus (MERS-CoV) is the causative agent of a severe respiratory syndrome that initially emerged in the Middle East in 2012 and continues to cause epidemic outbreaks globally (Zaki et al. 2012; de Wit et al. 2016). At the end of November 2019, 2494 laboratory-confirmed cases of MERS and 858 deaths were reported in 27 countries, with a mortality rate of ~ 35% (WHO 2019). Camels are direct sources of MERS-CoV and have been carrying the virus for at least 30 years (Muller et al. 2015). Close contact between humans and camels contributes to the ongoing MERS-CoV epidemic in humans (Azhar et al. 2014; Dudas et al. 2018). However, there are currently no approved antiviral drugs or vaccines against MERS-CoV infection, underscoring the urgent need for an efficient antiviral inhibitor screening method.
MERS-CoV, a member of the family Coronaviridae, is an enveloped, positive-sense, single-stranded RNA virus. The 5′ end of the genome harbors two large open reading frames (ORFs), ORF1a and ORF1b, which encode polyproteins pp1a and pp1ab. Two viral proteins, 3C-like protease and papain-like protease, process polyproteins to generate 16 mature non-structural proteins. These non-structural proteins are essential for MERS-CoV RNA replication and transcription. At the 3′ end of the genome, several ORFs encode four structural proteins (spike [S], envelope [E], membrane [M], and nucleocapsid [N]) and five accessory proteins (ORF3, ORF4a, ORF4b, ORF5, ORF8b) (van Boheemen et al. 2012). The structural proteins of coronaviruses are related to virus entry, assembly, and release (Siu et al. 2008; Kuo et al. 2016). Furthermore, the N protein has been proven to be essential for coronavirus transcription (Almazan et al. 2004; Zúñiga et al. 2010; Hurst et al. 2013). The accessory proteins mediate virus–host interaction and contribute to viral pathogenicity (Yang et al. 2013; Lee et al. 2019).
Antiviral assays based on infectious MERS-CoV require a high-level biosafety laboratory and are time-consuming. The virus replicon system contains the necessary elements for genome transcription and replication and is non-infectious, as it lacks viral structural genes. A stable replicon cell line that carries replicon RNA and expresses a reporter gene would allow easy screening for an antiviral inhibition based on luminescent or fluorescent signals (Hertzig et al. 2004; Ge et al. 2008). Thus, the replicon system may be a convenient and safe tool for antiviral drug discovery.
In this study, we constructed a MERS-CoV replicon cDNA clone harboring the Renilla luciferase (Rluc) reporter gene and we developed a stable cell line containing the MERS-CoV replicon with the aim to develop a user-friendly tool for rapid screening of antiviral inhibitors against MERS-CoV. We demonstrate that the MERS-CoV stable cell line can be used for the screening of MERS-CoV replication inhibitors based on Rluc activity detection.
Materials and Methods
Cells and Virus
BHK21 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Gibco). BHK21-MERS-Rep cells were maintained in DMEM supplemented with 10% FBS and 0.3 mg/mL G418 (G418 sulfate, Gibco, USA). MERS-CoV strain EMC/2012 viral RNA was used as a template for cDNA synthesis.
MERS-CoV Replicon Plasmid Construction
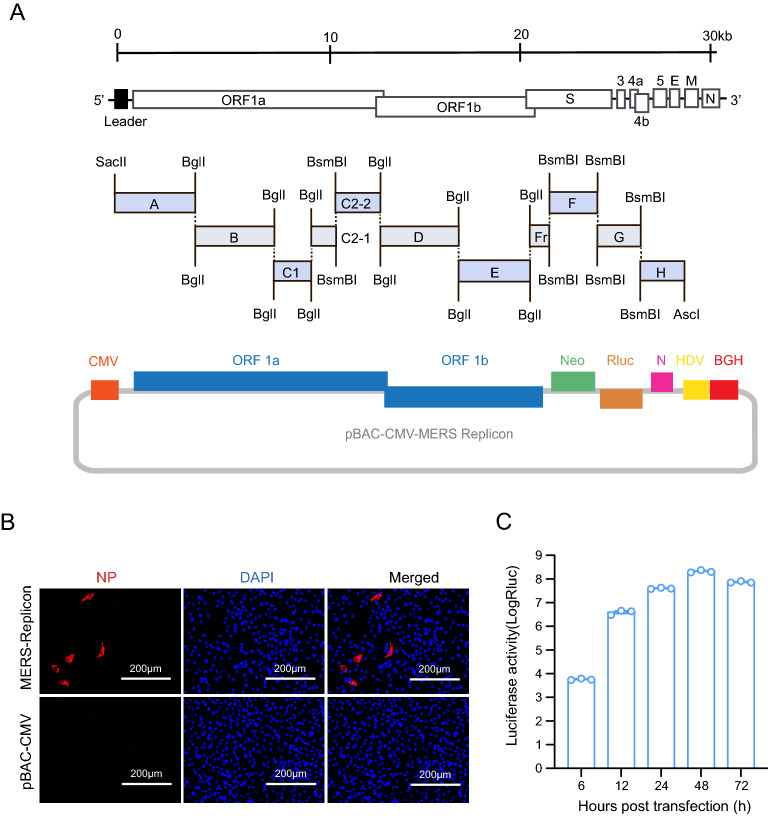
The MERS-CoV replicon mainly contains genes encoding the polyproteins pp1a and pp1ab and the N protein, a neomycin phosphotransferase gene (neo), and the Rluc-coding gene (Fig. 1A). The MERS-CoV 5′ UTR and the region containing the polyprotein genes were divided into eight contiguous fragments (fragment A–Fr). The N protein gene under the control of its transcription-regulating sequence (TRS) together with the 3′ UTR was regarded fragment H. Neo and Rluc under the control of TRSs of the E gene and M gene, respectively, were regarded fragments F and G. The cDNA fragments were amplified by PCR using specific primers (primer sequences are available upon request) to introduce BglI and BsmBI restriction cleavage sites to allow unidirectional ligation of all contiguous cDNA fragments. The amplicons were cloned into pGEM-T Easy (Promega). SacII and AscI sites were introduced at the 5′ end of fragment A and the 3′ end of fragment H, respectively. A poly(A) sequence (25 nt) was added to the 3′ end of fragment H. The cDNA clone was constructed as described previously (Zeng et al. 2016). Briefly, to construct a BAC cDNA clone, subclones A–H were digested with restriction enzyme and then ligated into pBAC-CMV vector (Zeng et al. 2016) and transformed into DH10B competent cells. The clones were screened by PCR and verified by restriction enzyme cleavage and sequencing, and the correct clone was designated as pBAC-CMV-MERS replicon.
Fig. 1.
Construction and characterization of the MERS-CoV replicon. A Schematic diagram of MERS-CoV replicon cDNA clone construction. The structural genes encoding E and M were replaced with a neomycin resistance gene and an Rluc gene, respectively. B Detection of MERS-CoV N protein. BHK21 cells transfected with the MERS-CoV replicon were analyzed by IFA using an anti-N protein rabbit polyclonal antibody as the primary antibody and a Cy3-conjugated goat anti-rabbit IgG as the secondary antibody. C Luciferase activity assay. BHK21 cells were transiently transfected with the MERS-CoV replicon cDNA clone. Luciferase activity was measured at the indicated time points
Detection of the MERS-CoV Replicon in BHK21 Cells
An amount of 1.5 μg of pBAC-CMV-MERS replicon was transfected into BHK21 cells that had been grown to 70% confluence in a 24-well plate. The same amount of empty pBAC-CMV vector was transfected as a negative control. Transfection was conducted using Lipofectamine 3000 (Invitrogen) in Opti-MEM Medium (Gibco) according to the manufacturer’s instructions. After incubation for 4 h, the culture medium was replaced with fresh DMEM containing 10% FBS.
MERS-CoV replicon in transfected BHK21 cells was detected by immunofluorescence assay (IFA). Briefly, at 24 h post transfection, cells were fixed with 4% paraformaldehyde at room temperature for 20 min. The fixed cells were incubated with a house-made rabbit polyclonal antibody against the MERS-CoV N protein. A Cy3-conjugated goat anti-rabbit antibody was used as a secondary antibody. Images were captured under a fluorescence microscope (Invitrogen).
To characterize the replication of the MERS-CoV replicon in BHK21 cells, BHK21 cells in a 24-well plate were transfected with pBAC-CMV-MERS replicon. The transfected cells were lysed at various time points, and Rluc activity was measured. For the Rluc assay, cells were lysed with 1 × Rluc lysis buffer (Promega) at room temperature for 15 min. Rluc activity was measured by mixing 20 μL of cell lysate with 100 μL of substrate (Promega), using a GloMax luminometer (Promega).
Generation and Selection of a MERS-CoV Replicon BHK21 Cell Line
Approximately 5 × 105 BHK21 cells were seeded in a 35-mm dish. After 24 h of incubation, the cells were transfected with 6 μg pBAC-CMV-MERS replicon as described above. At 48 h post transfection, G418 was added to the cell culture medium at a final concentration of 0.3 mg/mL for selection. The culture medium was changed every 3 days. On day 12, the cells were trypsinized and pooled in a T75 flask for further expansion under G418 selection. When the cells had grown into a confluent monolayer, they were trypsinized for establishing individual cell clones by gradient dilution. Individual cell clones were initially expanded in individual wells of 96-well plates and were then transferred to individual wells of 24-well plates for further expansion. All expansion steps were under G418 selection. Cells of passage 5 were aliquoted and stored in a liquid nitrogen.
Analysis of BHK21-MERS-Rep Cells
The cloned BHK21-MERS-Rep cells were seeded in 24-well plates and incubated for 24 h. Then, cell lysates were prepared for Western blotting and luciferase assay. For Western blotting, the cells were washed twice with PBS and then lysed. MERS-CoV N protein was detected using a house-made mouse monoclonal antibody and HRP-conjugated goat anti-mouse antibody. Rluc activity was measured as described above.
Drug Inhibition of MERS-CoV Replication
BHK21-MERS-Rep cells were seeded in 96-well plates at 1.2 × 104 cells/well and were cultured in DMEM containing 10% FBS and 0.3 mg/mL G418 for 16 h. Lopinavir (Selleck) and ritonavir (Selleck) were a gift from Prof. Gengfu Xiao. The compounds dissolved in DMSO were serially diluted in DMEM containing 10% FBS medium in two-fold increments and were then added to the cells. After 48 h of incubation, the cell culture medium was removed. The cells were washed thrice with PBS and Rluc activity was measured as described above. The 50% effective concentration (IC50) was determined as the concentration at which there was a 50% decrease in MERS-CoV replication by setting untreated cells as 0% inhibition, using GraphPad 8 (GraphPad).
After treatment with the compounds at different concentration for 48 h, the inhibition of MERS-CoV replication was confirmed by reverse-transcription quantitative PCR (RT-qPCR). BHK21-MERS-Rep cells were harvested and total cellular RNA was isolated using the RNeasy Mini Kit (Qiagen). The RNA was subjected to SYBR green one-step real-time RT-PCR (Vazyme) with primers targeting MERS-CoV N protein gene (forward, 5′-GGGTGTACCTCTTAATGCCAATTC-3′, reverse, 5′-TCTGTCCTGTCTCCGCCAAT-3′). The replicon RNA copy number was determined by interpolation using a standard curve generated using a MERS-CoV infectious clone plasmid.
Cytotoxicity Assay in BHK21-MERS-Rep Cells
The cytotoxicity of compounds was measured using CCK-8 assays. BHK21-MERS-Rep cells were seeded in a 96-well plate and cultured for 16 h. Then, the culture medium was removed and fresh medium containing the compounds at different concentrations was added. After 48 h, 10 μL of CCK-8 reagent (MedChem Express) was added to each well and the cells were incubated for another 1 h. The absorbance was read using an Infinite 200 PRO plate reader (Tecan) with a 450-nm filter.
Statistical Analysis
The experiments involved in statistical analysis were performed in triplicate for two or three times. Each data point represents the group mean, and error bars define the SEM. Statistical analysis was performed with GraphPad 8.
Results
Construction and Characterization of MERS-CoV Replicon
The MERS-CoV replicon plasmid was constructed as described in Materials and Methods (Fig. 1A). To characterize the replication of the MERS-CoV replicon, BHK21 cells were transfected with pBAC-CMV-MERS replicon and then analyzed by IFA and Rluc assay. N protein expression was observed in MERS-CoV replicon cDNA clone-transfected BHK21 cells (Fig. 1B), indicating correct transcription and translation of the MERS CoV polyprotein and N genes. Rluc activity in BHK21 cells was measured at different time points. Rluc signal increased as of 6 h post transfection and reached a peak at 48 h post transfection (Fig. 1C), demonstrating replication of the MERS-CoV replicon. Collectively, these findings indicated that MERS-CoV replicon was successfully constructed.
Establishment of the Stable BHK21-MERS-Rep Cell Line Carrying MERS-CoV Replicon RNA
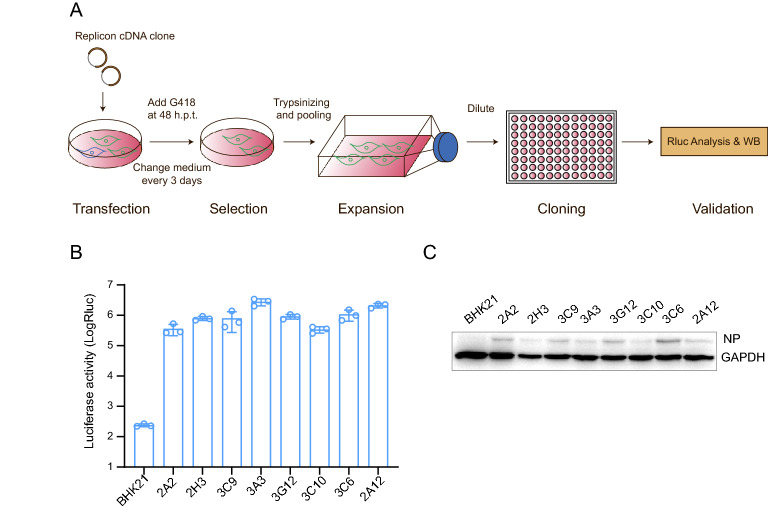
A stable cell line containing MERS-CoV replicon was established as shown in the flowchart in Fig. 2A. BHK21 cells were transfected with pBAC-CMV-MERS replicon and selected with G418. After 12 days of G418 selection, all surviving cells were trypsinized, pooled, and expanded. Subsequently, the expanded cells were diluted and plated in 96-well plates to obtain single cell clones. Finally, eight G418-resistant single colonies were obtained and expanded for analysis of the presence of MERS-CoV replicon.
Fig. 2.
Development and characterization of MERS-CoV replicon-carrying cells. A Flowchart of the development of a replicon-carrying cell line. B Luciferase activity assay of BHK21-MERS-Rep cells. Individual clones of BHK21-MERS-Rep cells were plated in a 24-well plate and were lysed after 24 h for luciferase activity measurement. C MERS-CoV N protein detection in BHK21-MERS-Rep cells. BHK21-MERS-Rep cells were detected using an anti-N protein mouse monoclonal antibody and an HRP-conjugated goat anti-mouse IgG as primary and secondary antibody, respectively
Rluc activity and viral protein expression were determined in different colonies of MERS-CoV replicon-transfected BHK21 cells. All eight single cell colonies produced 105–106 Rluc signal, which was approximately 103–104 fold higher than the signal produced by naive BHK21 cells (Fig. 2B). MERS-CoV N protein expression was detected in all cell clones by Western blotting, and the cell clone 3C6 exhibited the highest protein levels (Fig. 2C). Then the cell clone 3C6 was chosen to use as BHK21-MERS-Rep cell line. Together, these data indicated that a BHK21 cell line stably carrying the MERS-CoV replicon was successfully constructed.
Antiviral Screening Assay with the BHK21-MERS-Rep Cell Line
To test whether BHK21-MERS-Rep cell line can be used for antiviral screening, the antiviral activity of lopinavir/ritonavir was determined in BHK21-MERS-Rep cells. Lopinavir/ritonavir is currently under evaluation in the MIRACLE trial (Arabi et al. 2018; Arabi et al. 2020). Rluc activity decreased upon lopinavir/ritonavir (molar ratio, 4.6:1) treatment in a dose-dependent manner, with a calculated IC50 of 8.34 μmol/L. No obvious cytotoxicity was observed at concentrations below 50 μmol/L (Fig. 3A). Accordingly, the cell numbers in each test group were further normalized. MERS-CoV replicon replication was confirmed by RT-qPCR. MERS-CoV replicon RNA levels decreased in a dose-dependent manner after treatment with lopinavir/ritonavir (Fig. 3B). These data indicated that BHK21-MERS-Rep cell line can be used for antiviral screening based on Rluc signal measurement.
Fig. 3.
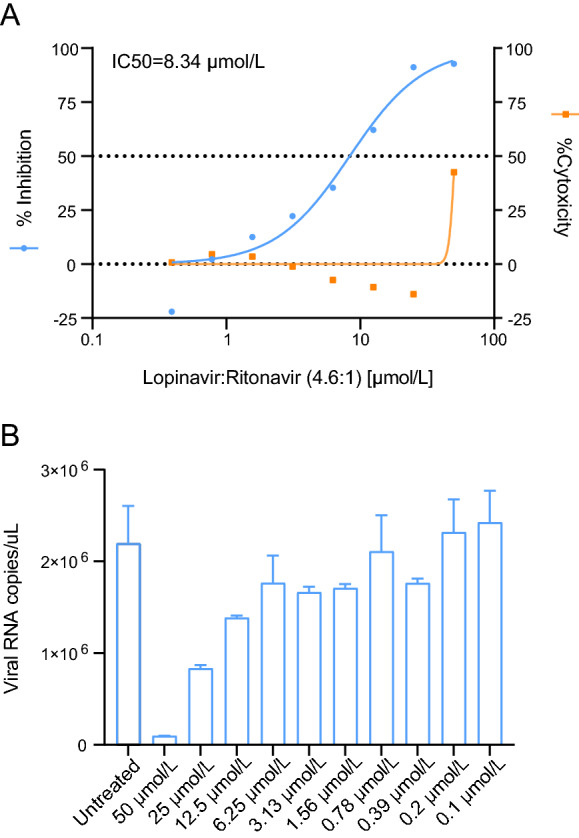
Antiviral assay based on the BHK21-MERS-Rep cell line. BHK21-MERS-Rep cells were treated with lopinavir/ritonavir at the indicated concentrations, and luciferase activity and cytotoxicity (A) and viral RNA levels (B) were measured at 48 h post treatment
Discussion
The replicon cell line is a safe and convenient tool for antiviral screening. As no infectious virus is used, it can be used in low biosafety-level laboratories. Additionally, the replicon cell line can be used for high-throughput screening. Replicons of coronaviruses, such as SARS-CoV-2, SARS-CoV, and 229E (Hertzig et al. 2004; Almazan et al. 2006; Ge et al. 2008; Zhang et al. 2021), as well as of flaviviruses (Xie et al. 2016; Kato et al. 2019) and hepatitis C virus (Camus et al. 2018) have been widely used.
The replicon we developed and described here can be used for antiviral screening of inhibitors target viral proteases and host cellular factors that are involved in viral replication and transcription. As the MERS-CoV replicon does not contain the structural proteins S, E, and M, this cell line cannot be used for antiviral assays related to virus entry, assembly, and release, as well as the function associated with the accessory proteins such as ORF3, ORF4a, ORF4b and ORF5.
The MERS-CoV replicon expresses the Rluc reporter gene and neomycin resistant gene and needs neomycin selection for the maintenance in cells. In our study, a single clone of replicon-containing was purified and could continually produce the replicon after 20 passages without obvious variations (Supplementary Fig. S1).
In summary, we successfully constructed a MERS-CoV replicon cDNA clone and we developed a stable MERS-CoV replicon cell line. The MERS-CoV replicon cDNA clone can be used for the study of MERS-CoV transcription and replication. The stable MERS-CoV replicon cell is an effective and safe tool for high-throughput screening of antivirals against MERS-CoV.
Supplementary Information
Supplementary Figure S1 Replication stability of the BHK21-MERS-Rep cell line. Luciferease activities of BHK21-MERS-Rep cells at different passage numbers were measured. Supplementary material 1 (pdf 66 kb)
Acknowledgements
This study was supported by the strategic priority research program of the Chinese academy of sciences (XDB29010101). We thank our lab members and colleagues for helpful supports during this course of work and Professor Gengfu Xiao for providing Lopinavir and Ritonavir inhibitors.
Author Contributions
JC and Z-LS designed the experiments; JC, BJH, KZ, YL and HFL performed experiments; JC performed data analysis; JC and Z-LS wrote the manuscript. Z-LS finalized the article. All authors read and approved the final manuscript.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Animal and Human Rights Statement
This article does not contain any studies with human and animal subjects performed by any of the authors.
References
- Almazan F, Galan C, Enjuanes L. The nucleoprotein is required for efficient coronavirus genome replication. J Virol. 2004;78:12683–12688. doi: 10.1128/JVI.78.22.12683-12688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazan F, DeDiego ML, Galan C, Escors D, Alvarez E, Ortego J, Sola I, Zuniga S, Alonso S, Moreno JL, Nogales A, Capiscol C, Enjuanes L. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J Virol. 2006;80:10900–10906. doi: 10.1128/JVI.00385-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi YM, Alothman A, Balkhy HH, Al-Dawood A, AlJohani S, Al Harbi S, Kojan S, Al Jeraisy M, Deeb AM, Assiri AM, Al-Hameed F, AlSaedi A, Mandourah Y, Almekhlafi GA, Sherbeeni NM, Elzein FE, Memon J, Taha Y, Almotairi A, Maghrabi KA, Qushmaq I, Al Bshabshe A, Kharaba A, Shalhoub S, Jose J, Fowler RA, Hayden FG, Hussein MA, The Mtg Treatment of Middle East Respiratory Syndrome with a combination of lopinavir-ritonavir and interferon-beta1b (MIRACLE trial): study protocol for a randomized controlled trial. Trials. 2018;19:81. doi: 10.1186/s13063-017-2427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi YM, Asiri AY, Assiri AM, Balkhy HH, Al Bshabshe A, Al Jeraisy M, Mandourah Y, Azzam MHA, Bin Eshaq AM, Al Johani S, Al Harbi S, Jokhdar HAA, Deeb AM, Memish ZA, Jose J, Ghazal S, Al Faraj S, Al Mekhlafi GA, Sherbeeni NM, Elzein FE, Al-Hameed F, Al Saedi A, Alharbi NK, Fowler RA, Hayden FG, Al-Dawood A, Abdelzaher M, Bajhmom W, AlMutairi BM, Hussein MA, Alothman A. Interferon Beta-1b and Lopinavir-Ritonavir for Middle East Respiratory Syndrome. N Engl J Med. 2020;383:1645–1656. doi: 10.1056/NEJMoa2015294. [DOI] [PubMed] [Google Scholar]
- Azhar EI, El-Kafrawy SA, Farraj SA, Hassan AM, Al-Saeed MS, Hashem AM, Madani TA. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med. 2014;370:2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- Camus G, Xu S, Han B, Lu J, Dvory-Sobol H, Yu M, Cheng G, Miller MD, Doehle BP, Mo H. Establishment of robust HCV genotype 4d, 5a, and 6a replicon systems. Virology. 2018;514:134–141. doi: 10.1016/j.virol.2017.11.003. [DOI] [PubMed] [Google Scholar]
- de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudas G, Carvalho LM, Rambaut A, Bedford T (2018) MERS-CoV spillover at the camel-human interface. Elife 7:e31257 [DOI] [PMC free article] [PubMed]
- Ge F, Xiong S, Lin FS, Zhang ZP, Zhang XE. High-throughput assay using a GFP-expressing replicon for SARS-CoV drug discovery. Antiviral Res. 2008;80:107–113. doi: 10.1016/j.antiviral.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzig T, Scandella E, Schelle B, Ziebuhr J, Siddell SG, Ludewig B, Thiel V. Rapid identification of coronavirus replicase inhibitors using a selectable replicon RNA. J Gen Virol. 2004;85:1717–1725. doi: 10.1099/vir.0.80044-0. [DOI] [PubMed] [Google Scholar]
- Hurst KR, Koetzner CA, Masters PS. Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex. J Virol. 2013;87:9159–9172. doi: 10.1128/JVI.01275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato F, Nio Y, Yagasaki K, Suzuki R, Hijikata M, Miura T, Miyazaki I, Tajima S, Lim CK, Saijo M, Takasaki T, Hishiki T. Identification of inhibitors of dengue viral replication using replicon cells expressing secretory luciferase. Antiviral Res. 2019;172:104643. doi: 10.1016/j.antiviral.2019.104643. [DOI] [PubMed] [Google Scholar]
- Kuo L, Hurst-Hess KR, Koetzner CA, Masters PS. Analyses of Coronavirus Assembly Interactions with Interspecies Membrane and Nucleocapsid Protein Chimeras. J Virol. 2016;90:4357–4368. doi: 10.1128/JVI.03212-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Bae S, Myoung J. Middle East respiratory syndrome coronavirus-encoded ORF8b strongly antagonizes IFN-β promoter activation: its implication for vaccine design. J Microbiol. 2019;57:803–811. doi: 10.1007/s12275-019-9272-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller MA, Meyer B, Corman VM, Al-Masri M, Turkestani A, Ritz D, Sieberg A, Aldabbagh S, Bosch BJ, Lattwein E, Alhakeem RF, Assiri AM, Albarrak AM, Al-Shangiti AM, Al-Tawfiq JA, Wikramaratna P, Alrabeeah AA, Drosten C, Memish ZA. Presence of Middle East respiratory syndrome coronavirus antibodies in Saudi Arabia: a nationwide, cross-sectional, serological study. Lancet Infectious Diseases. 2015;15:559–564. doi: 10.1016/S1473-3099(15)70090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu YL, Teoh KT, Lo J, Chan CM, Kien F, Escriou N, Tsao SW, Nicholls JM, Altmeyer R, Peiris JS, Bruzzone R, Nal B. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J Virol. 2008;82:11318–11330. doi: 10.1128/JVI.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Osterhaus AD, Haagmans BL, Gorbalenya AE, Snijder EJ, Fouchier RA. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3:e00473–e00512. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2019) Middle East respiratory syndrome coronavirus (MERS-CoV). https://www.who.int/emergencies/mers-cov/en/
- Xie X, Zou J, Shan C, Yang Y, Kum DB, Dallmeier K, Neyts J, Shi PY. Zika Virus Replicons for Drug Discovery. EBioMedicine. 2016;12:156–160. doi: 10.1016/j.ebiom.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhang L, Geng H, Deng Y, Huang B, Guo Y, Zhao Z, Tan W. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell. 2013;4:951–961. doi: 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zeng LP, Gao YT, Ge XY, Zhang Q, Peng C, Yang XL, Tan B, Chen J, Chmura AA, Daszak P, Shi ZL. Bat Severe Acute Respiratory Syndrome-Like Coronavirus WIV1 Encodes an Extra Accessory Protein, ORFX, Involved in Modulation of the Host Immune Response. J Virol. 2016;90:6573–6582. doi: 10.1128/JVI.03079-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Song W, Chen S, Yuan Z, Yi Z. A bacterial artificial chromosome (BAC)-vectored noninfectious replicon of SARS-CoV-2. Antiviral Res. 2021;185:104974. doi: 10.1016/j.antiviral.2020.104974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zúñiga S, Cruz JL, Sola I, Mateos-Gómez PA, Palacio L, Enjuanes L. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J Virol. 2010;84:2169–2175. doi: 10.1128/JVI.02011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1 Replication stability of the BHK21-MERS-Rep cell line. Luciferease activities of BHK21-MERS-Rep cells at different passage numbers were measured. Supplementary material 1 (pdf 66 kb)