Abstract
Background & Aims
RING finger protein 43 (RNF43) is a tumor suppressor that frequently is mutated in gastric tumors. The link between RNF43 and modulation of Wingless-related integration site (WNT) signaling has not been shown clearly in the stomach. Because mutations in RNF43 are highly enriched in microsatellite-unstable gastric tumors, which show defects in DNA damage response (DDR), we investigated whether RNF43 is involved in DDR in the stomach.
Methods
DDR activation and cell viability upon γ-radiation was analyzed in gastric cells where expression of RNF43 was depleted. Response to chemotherapeutic agents 5-fluorouracil and cisplatin was analyzed in gastric cancer cell lines and xenograft tumors. In addition, involvement of RNF43 in DDR activation was analyzed upon Helicobacter pylori infection in wild-type and Rnf43ΔEx8 mice. Furthermore, a cohort of human gastric biopsy specimens was analyzed for RNF43 expression and mutation status as well as for activation of DDR.
Results
RNF43 depletion conferred resistance to γ-radiation and chemotherapy by dampening the activation of DDR, thereby preventing apoptosis in gastric cells. Upon Helicobacter pylori infection, RNF43 loss of function reduced activation of DDR and apoptosis. Furthermore, RNF43 expression correlated with DDR activation in human gastric biopsy specimens, and RNF43 mutations found in gastric tumors conferred resistance to DNA damage. When exploring the molecular mechanisms behind these findings, a direct interaction between RNF43 and phosphorylated H2A histone family member X (γH2AX) was observed.
Conclusions
We identified a novel function for RNF43 in the stomach as a regulator of DDR. Loss of RNF43 function in gastric cells confers resistance to DNA damage-inducing radiotherapy and chemotherapy, suggesting RNF43 as a possible biomarker for therapy selection.
Keywords: RNF43, DNA Damage Response (DDR), Gastric Cancer, Helicobacter pylori
Abbreviations used in this paper: ATM, ataxia-telangiectasia mutated; ATR, ATM-and Rad3-Related; CHK, checkpoint kinase; CRISPR/Cas9, Clustered related interspaced short palindromic repeats/CRISPR associated 9; D196, deletion at aspartic acid 196; DDR, DNA damage response; DSB, double-strand break; GC, gastric cancer; H2AX, H2A histone family member X; MSI, microsatellite unstable; MSI-high, microsatellite instability-high; PMSS1, pre-mouse SS1; RNF43, RING finger protein 43; RNP, ribonucleoprotein; SDS, sodium dodecyl sulfate; shControl, short hairpin control; shRNF43, short hairpin RING finger protein 43; SNP, single-nucleotide polymorphism; WNT, Wingless-related integration site; WT, wild-type
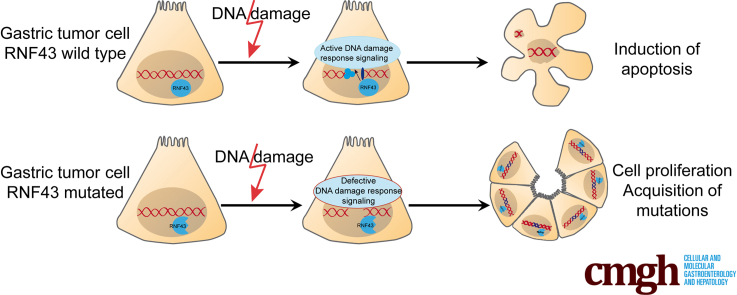
Graphical abstract
Summary.
The tumor-suppressor RING finger protein 43 modulates DNA damage response in gastric cells. The RING finger protein 43 mutational status might be used as a biomarker for therapy selection.
Gastric cancer (GC) is the fifth most common malignancy and the third leading cause of cancer-related death worldwide (Global Cancer Observatory (GLOBOCAN) 2018). Several attempts have been made to classify gastric tumors to guide treatment selection.1, 2, 3 Despite these numerous attempts, surgery is the only curative treatment available to date. Although the addition of chemotherapy or radiotherapy can improve outcomes, prognosis remains poor, with a median survival of 10–12 months and a 5-year survival rate of <10%.4 GC patients show a high interindividual as well as intra-individual heterogeneity, making the identification of novel biomarkers indispensable for patient stratification. To date, not many biomarkers for therapy response exist, except for Human epidermal growth factor receptor 2 (HER2) expression or Programmed death-ligand 1 (PD-L1) and microsatellite instability (MSI) levels.4, 5, 6
Mutations in well-known tumor suppressors, as well as oncogenes such as TP53 or KRAS, have been identified as drivers of GC.3,7,8 Mutations in the E3 ubiquitin ligase RING finger protein 43 (RNF43) also have frequently been reported to occur in microsatellite instability (MSI)-high tumors, suggesting an important role of RNF43 in gastric carcinogenesis.3,8, 9, 10, 11, 12 Recently, we observed that loss of RNF43 function enhances the tumorigenic potential of GC cells in vitro and in vivo,13 supporting a tumor-suppressor function of RNF43 in the stomach. The tumor-suppressive function of RNF43 has been shown to be related to its capacity to negatively regulate Wingless-related integration site (WNT) signaling by 2 different mechanisms. Located in the cell membrane, RNF43 targets Frizzled receptors for ubiquitin-mediated internalization and degradation, thereby decreasing WNT activity.14 When expressed in the nucleus, we could show that RNF43 negatively regulates WNT signaling downstream of β-catenin and Adenomatous polyposis coli (APC) by sequestering T-cell factor 4 to the nuclear membrane, thereby suppressing its transcriptional activity.15 For this inhibitory activity, the presence of an intact RING domain, where the ubiquitin ligase function resides, is essential. Notably, the WNT inhibitory capacity of RNF43 in the stomach has not been completely demonstrated. Our previous results suggested a WNT-independent function of RNF43. Thus, Rnf43ΔEx8 mice carrying an inactivating deletion of the RING domain did not show alterations in the gastric expression of important WNT target genes such as Axin2 or Lgr5. However, we observed thickening of the gastric mucosa, hyperplasia, and cellular atypia in these mice, confirming an important function of RNF43 in gastric homeostasis.13 Nevertheless, the molecular mechanisms involved remained unknown.
Interestingly, ubiquitination of the main players of the DNA damage response (DDR) is a key event for the activation of this signaling cascade, and several E3 ubiquitin ligases have been reported to be involved.16, 17, 18, 19 The DDR is critical for maintaining genomic stability commonly lost in tumors. Two main kinase-signaling pathways, ataxia-telangiectasia mutated (ATM)–checkpoint kinase 2 (CHK2) and ATM- and Rad3-Related (ATR)–CHK1, coordinate cellular responses to DNA damage. ATM-CHK2 signaling is activated by radiation and genotoxins inducing double-strand breaks (DSBs), while ATR-CHK1 is activated when replication is impeded.20 Mutations in ATR or CHK1 were found in gastric tumors with MSI,21 and mutated ATM was associated strongly with MSI,22 indicating that inhibition of DDR might be important for the development of gastric tumors harboring MSI.
Helicobacter pylori infection, one of the major risk factors for the development of GC, has been linked to DDR. H pylori elicits an immune response that leads to the production of reactive oxygen and nitrogen species, which can induce DNA damage.23,24 In addition, H pylori has been shown to induce DSBs directly in host cells.25,26 Furthermore, H pylori infection induces epigenetic modifications leading to the up-regulation of ATM.27 Thus, induction of DNA damage by infection and lack of functional repair mechanisms can highly contribute to gastric carcinogenesis.
Considering the high mutation rate of RNF43 in gastric tumors showing MSI and our previous data showing that in vivo loss of RNF43 function leads to gastric pathology independent of alterations in WNT signaling,13 we sought to determine whether RNF43 could be involved in DDR in the stomach and thereby influence response to DNA damage-inducing cancer therapy.
Results
Loss of RNF43 Function Confers Resistance to DNA Damage-Induced Cell Death
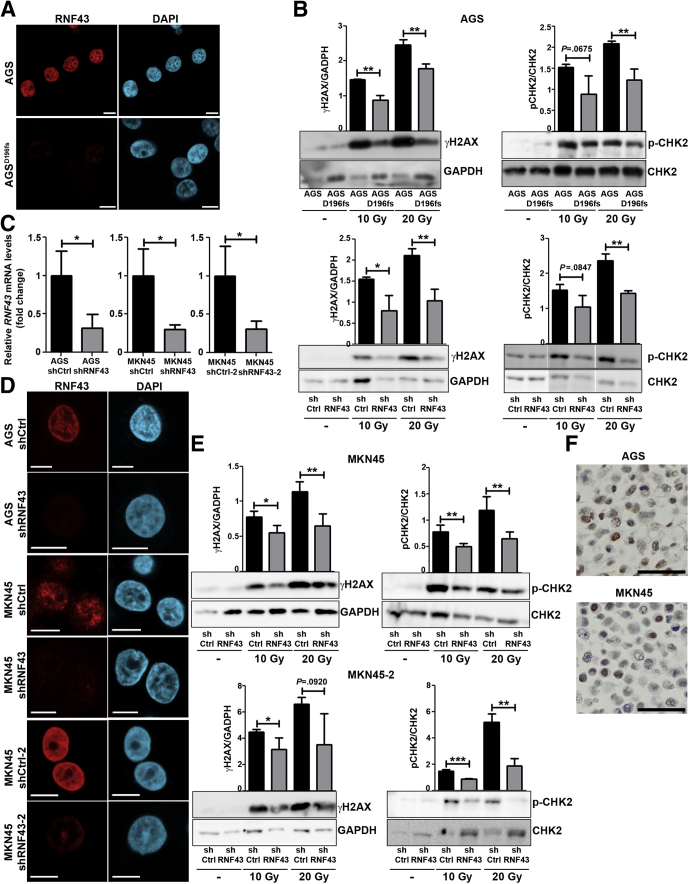
To explore whether RNF43 is involved in DDR, we first analyzed the levels of phosphorylated H2A histone family member X (γH2AX) and CHK2 in AGS control and in AGS cells where expression of RNF43 had been depleted by CRISPR/Cas9 (AGSdeletion at aspartic acid 196 [D196]fs) (Figure 1A). Cellular damage caused by γ-radiation highly induced H2AX and CHK2 phosphorylation in cells expressing RNF43 (Figure 1B). Depletion of RNF43 expression resulted in reduced activation of DDR because lower levels of γH2AX and phosphorylated CHK2 were detected in AGSD196fs cells (Figure 1B).
Figure 1.
RNF43 is involved in DDR. (A) Representative immunofluorescence images of RNF43 (red) in control and AGSD196fs cells. Scale bars: 10 μm. (B) Western blot analysis and quantification of γH2AX and CHK2 expression in WT AGS, AGSD196fs, control AGS (shCtrl), and RNF43 knockdown AGS (shRNF43) cells after γ-radiation. GAPDH was used as a protein loading control (N = 3). (C) RNF43 mRNA levels in control and RNF43 knockdown AGS and MKN45 cells. Cycle threshold (CT)values were normalized to GAPDH and fold change was calculated over control cells. (D) Representative immunofluorescence images of RNF43 (red) in control and RNF43 knockdown AGS and MKN45 cells. Scale bars: 10 μm. (E) Western blot analysis and quantification of γH2AX and CHK2 expression in control MKN45 (shCtrl) and RNF43 knockdown MKN45 (shRNF43) cells after γ-radiation. GAPDH was used as a protein loading control (N = 3). (F) Endogenous RNF43 expression in AGS and MKN45 cells detected by immunocytochemistry. Scale bar: 50 μm. Error bars indicate SD. ∗P ≤ .05, ∗∗P ≤ .01, ∗∗∗P ≤ .001, 2-tailed unpaired t test. DAPI, 4′,6-diamidino-2-phenylindole; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; mRNA, messenger RNA.
No single clones could be obtained for MKN45 cells after transfection of the guide RNAs. Therefore, we depleted RNF43 by lentiviral transduction of specific short hairpin RNAs (Figure 1C and D). Similar to our observations in AGSD196fs cells, impaired activation of DDR was observed in MKN45–short hairpin RNF43 (shRNF43) and AGS–shRNF43 cells (Figure 1E and B, respectively). These observations indicate that RNF43 is important for the induction of DDR in gastric cells.
It has to be noted that AGS and MKN45 cells do not show mutations in RNF43 (Catalogue Of Somatic Mutations In Cancer (Cosmic) ID: COSS906790 and COSS925340, respectively), and therefore express wild-type (WT) protein in the nucleus (Figure 1F).
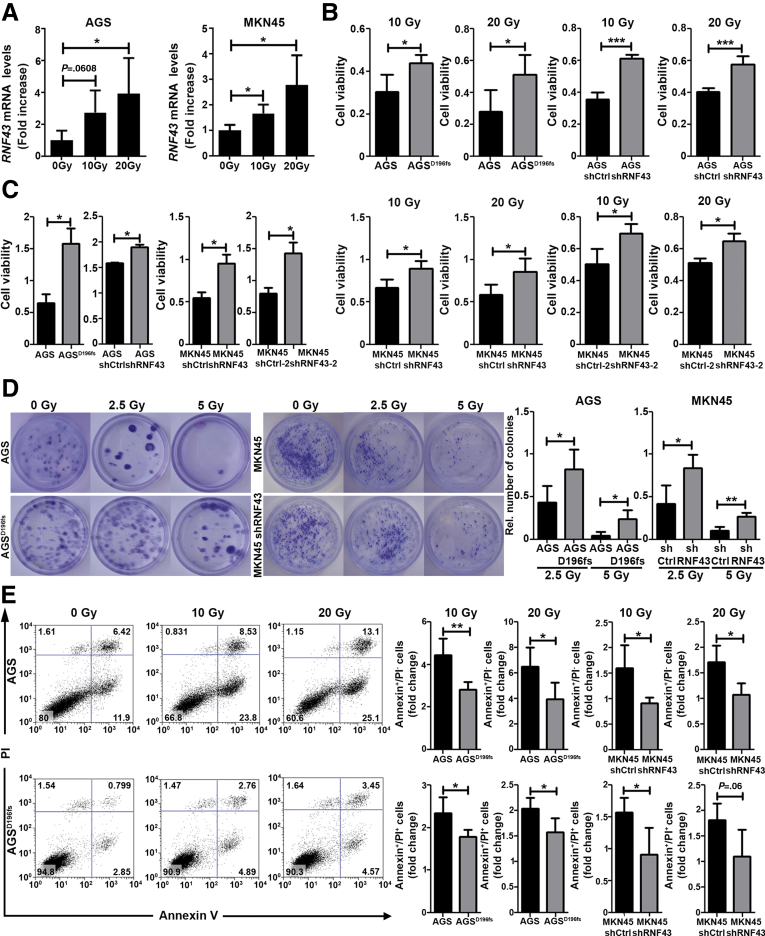
Interestingly, increased levels of RNF43 messenger RNA were detected in AGS and MKN45 GC cells after applying different doses of γ-radiation (Figure 2A), suggesting that DNA damage induces the expression of RNF43.
Figure 2.
Loss of RNF43 function confers resistance to DNA damage-induced cell death. (A) RNF43 mRNA levels in AGS and MKN45 gastric cancer cells upon increasing doses of γ-radiation. Cycle threshold (CT) values were normalized to GAPDH and fold change was calculated over untreated cells (N = 4). (B) Cell viability of WT AGS, AGSD196fs, control MKN45 (shCtrl), and RNF43 knockdown MKN45 (shRNF43) cells after γ-radiation. Values were normalized over untreated cells (N = 4). (C) Cell viability of AGS, AGSD196fs cells, as well as control AGS and MKN45 (shCtrl) and RNF43 knockdown AGS and MKN45 (shRNF43) cells under basal conditions (N = 3). (D) Representative images and quantification of cell colonies after γ-radiation (N = 4). (E) Flow cytometry analysis of Annexin V and propidium iodide (PI)-positive AGS and MKN45 cells after irradiation. Values were normalized over untreated cells (N = 4). Error bars indicate SD. ∗P ≤ .05, ∗∗P ≤ .01, ∗∗∗P ≤ .001, 2-tailed unpaired t test. mRNA, messenger RNA.
We next measured cell viability upon induction of DNA damage through γ-radiation. AGSD196fs cells showed enhanced cell viability after irradiation (Figure 2B). RNF43 knock-down MKN45 and RNF43 knock-down AGS cells showed increased cell viability compared with short hairpin control (shControl)-transduced cells upon treatment with ionizing radiation (Figure 2B). Notably, depletion of RNF43 either by Clustered related interspaced short palindromic repeats (CRISPR)/ CRISPR associated 9 (Cas9) or short hairpin RNAs already induced changes in cell proliferation under basal conditions (Figure 2C), which were accounted for when comparing cell viability of WT and RNF43-depleted cells after γ-radiation.
Cell survival after radiation also was assessed in clonogenic assays. Loss of RNF43 enhanced clonal proliferative capacity of gastric cells because more colonies of AGSD196fs and MKN45–shRNF43 compared with respective controls were detected after exposing the cells to γ-radiation (Figure 2D).
Activation of DDR results in cellular apoptosis when the repair mechanisms cannot cope with the cellular damage inflicted. Thus, we next evaluated whether enhanced viability of cells lacking RNF43 expression upon γ-radiation was the result of reduced cellular apoptosis. We observed that a higher percentage of AGS- and MKN45-expressing WT RNF43 underwent early (Annexin V–positive) and late apoptosis (Annexin V/propidium iodide–double-positive) compared with AGSD196fs and MKN45–shRNF43 cells (Figure 2E).
Together, our results indicate that RNF43 is involved in DDR in GC cells and depletion of its expression confers resistance to DNA damage-induced apoptosis.
RNF43 Influences Susceptibility to DNA Damage-Inducing Chemotherapeutics
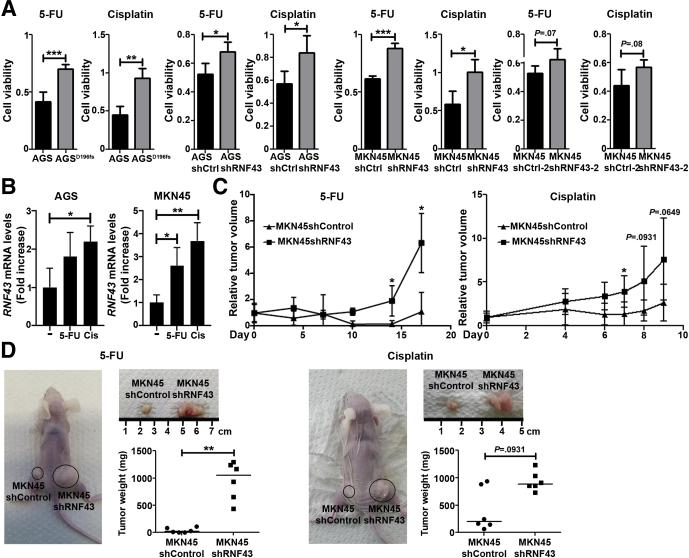
Current GC therapeutic treatments are based on the use of DNA damage-inducing chemotherapeutics, such as cisplatin or 5-fluorouracil.28 To analyze whether depletion of RNF43 influenced the response of GC cells to DNA damage-inducing chemotherapeutics, we analyzed cell viability in AGS control and AGSD196fs cells upon treatment with lethal doses 50 (LD50) of 5-fluorouracil and cisplatin. AGSD196fs showed higher cell viability after treatment when compared with control cells (Figure 3A), indicating that depletion of RNF43 confers resistance to chemotherapy. Similar results were observed for MKN45–shRNF43 and AGS–shRNF43 cells (Figure 3A).
Figure 3.
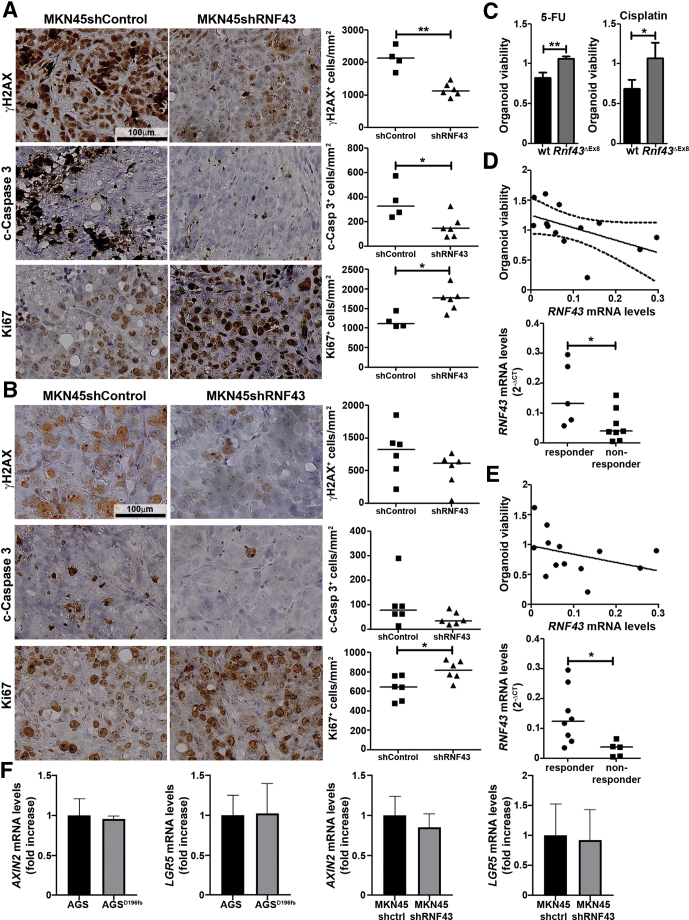
RNF43 influences susceptibility to DNA damage-inducing chemotherapeutics. (A) Cell viability of WT AGS, AGSD196fs, control MKN45 and AGS (shCtrl), and RNF43 knockdown MKN45 and AGS (shRNF43) cells after treatment with 5-fluorouracil (5-FU) or cisplatin. Values were normalized over untreated cells (N = 3). (B) RNF43 mRNA levels in AGS and MKN45 GC cells after treatment with 5-FU or cisplatin (Cis) for 48 hours. Cycle threshold (CT) values were normalized to GAPDH and fold change was calculated over untreated cells (N = 3). (C) Relative volume of xenograft tumors derived from control MKN45 (shCtrl) and RNF43 knockdown MKN45 (shRNF43) cells after treatment of mice with 5-FU or cisplatin. Relative volumes were calculated over tumor volume before treatment (N = 6/group). (D) Representative images of xenograft tumors derived from control MKN45 (shCtrl) and RNF43 knockdown MKN45 (shRNF43) cells after treatment of mice with 5-FU or cisplatin. Size and weight of the resected tumors are shown (n = 6/group). Error bars indicate SD. Horizontal lines represent the median values. ∗P ≤ .05, ∗∗P ≤ .01, and ∗∗∗P ≤ .001, (A and B) 2-tailed unpaired t test, (C and D) Mann–Whitney test.
The expression of RNF43 was increased in AGS and MKN45 cells treated with 5-fluorouracil and cisplatin (Figure 3B), confirming up-regulation of RNF43 in response to DNA damage.
Resistance to chemotherapy after depletion of RNF43 expression was explored further in a tumor xenograft model. When tumors originating from MKN45–shControl and MKN45–shRNF43 cells reached 40–70 mm3, mice were treated with 5-fluorouracil or cisplatin. Tumors derived from MKN45–shRNF43 cells grew bigger compared with tumors that originated from MKN45–shControl cells, as previously reported.13 Therefore, tumor growth upon treatment with chemotherapeutics was calculated relative to the initial tumor size to account for the growth-promoting effect of RNF43 loss. Tumors derived from MKN45–shRNF43 cells showed enhanced resistance to chemotherapy in vivo because bigger tumors were observed after treatment (Figure 3C and D), confirming the results observed in vitro. In addition, we evaluated activation of the DDR, apoptosis, and proliferation by staining the tumors for γH2AX, cleaved–caspase 3, and Ki67, respectively, at the end point of treatment with 5-fluorouracil (Figure 4A) or cisplatin (Figure 4B). Tumors derived from MKN45–shRNF43 cells showed reduced levels of γH2AX upon treatment with 5-fluorouracil, indicating lower activation of DDR (Figure 4A). Furthermore, reduced apoptosis and increased proliferation were observed in MKN45–shRNF43–derived tumors (Figure 4A) compared with tumors that originated from MKN45–shControl cells. Likewise, after treatment with cisplatin, tumors derived from MKN45–shRNF43 cells showed reduced DDR activation and apoptosis (Figure 4B). These tumors also were more proliferative compared with tumors originating from MKN45–shControl cells (Figure 4B). Together, these results suggest that, in the absence of RNF43, GC cells become resistant to chemotherapeutics.
Figure 4.
RNF43 influences susceptibility to DNA damage-inducing chemotherapeutics. Representative images showing γH2AX, cleaved caspase 3 (c-caspase 3), and Ki67 detected by immunohistochemistry in xenograft tumors that originated from control MKN45 (shCtrl) and RNF43 knockdown MKN45 (shRNF43) cells after treatment with (A) 5-fluorouracil or (B) cisplatin. Quantification of positive cells per square millimeter is shown. Each dot represents 1 mouse. (C) Viability of gastric organoids from WT or Rnf43ΔEx8 mice after treatment with 5-fluorouracil (5-FU) or cisplatin. Values were normalized over untreated (N = 3). Spearman correlation between RNF43 mRNA expression levels in human gastric tissue samples (N = 13), and viability of organoids generated from the same tissue samples after treatment with (D) 5-fluorouracil or (E) cisplatin. RNF43 mRNA levels normalized to GAPDH also are shown. (F) AXIN2 and LGR5 mRNA levels in AGS, AGSD196fs, and MKN45 control and RNF43 knockdown cells. Cycle threshold (CT)values were normalized to GAPDH and fold change was calculated over control cells (N = 3). Error bars indicate SD. Horizontal lines represent the median values. ∗P ≤ .05, ∗∗P ≤ .01, (C and F) 2-tailed unpaired t test, (A, B, D, and E) Mann–Whitney test. mRNA, messenger RNA.
To further substantiate resistance to DNA damage-inducing chemotherapeutics after loss of RNF43 function, we generated gastric organoids from WT and Rnf43ΔEx8 mice.13 Organoids were treated with 5-fluorouracil or cisplatin, and cell viability was measured after 4 days. Organoids derived from Rnf43ΔEx8 mice were more resistant to treatment with chemotherapeutics inflicting DNA damage (Figure 4C), confirming the results observed in vitro and in the xenograft model.
Finally, we assessed whether levels of RNF43 correlate with response to chemotherapy using human gastric organoids. Organoids were generated from stomach biopsy specimens and treated with 5-fluorouracil or cisplatin for 5 days, after which cell viability was measured. An inverse correlation between response to 5-fluorouracil treatment and RNF43 expression was observed (Spearman rs = -6264; P = .0220) (Figure 4D). RNF43 expression was higher in organoids responding to 5-fluorouracil than in organoids not responding (Figure 4D). Similar results were detected when human gastric organoids were treated with cisplatin (Figure 4E).
These results suggest that cells lacking RNF43 function become resistant to DNA damage-inducing chemotherapeutics.
Because RNF43 has been described to inhibit WNT signaling in different cellular models, we analyzed whether the effects induced by depleting RNF43 expression depended on alterations in WNT. We assessed the expression levels of the WNT target genes AXIN2 and LGR5 in AGS control and AGSD196fs and MKN45–shControl and MKN45–shRNF43. No differences in the expression of these genes were detected (Figure 4F), indicating that the effects are independent of changes in WNT signaling.
RNF43 Is Involved in H pylori–Induced DNA Damage Response
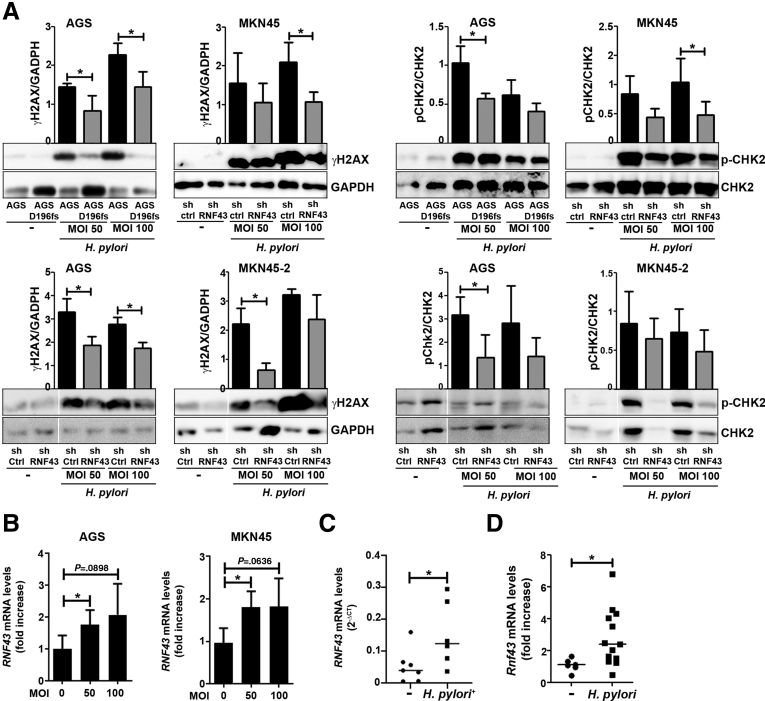
H pylori has been described to induce DNA damage in gastric epithelial cells.25 We analyzed whether activation of DDR in response to H pylori infection is compromised in cells with suppressed RNF43 expression. Indeed, we observed that activation of DDR, as assessed by H2AX and CHK2 phosphorylation, was reduced in AGSD196fs cells (Figure 5A), indicating that RNF43 is involved in DDR elicited by H pylori. Likewise, impairment of DDR in response to H pylori infection was detected in MKN45–shRNF43 and AGS–shRNF43 cells (Figure 5A).
Figure 5.
RNF43 is involved in H pylori–induced DDR. (A) Western blot analysis and quantification of γH2AX and CHK2 expression in WT AGS, AGSD196fs, control MKN45 and AGS (shCtrl), and RNF43 knockdown MKN45 and AGS (shRNF43) cells upon H pylori PMSS1 infection at different multiplicity of infection (MOI). GAPDH was used as a loading control (N = 4). (B) RNF43 mRNA levels after H pylori infection of AGS and MKN45 cells. Cycle threshold (CT)values were normalized to GAPDH and fold change was calculated over uninfected cells (N = 4). (C) RNF43 mRNA levels normalized to GAPDH in gastric biopsy specimens from uninfected (n = 6) and H pylori–infected (n = 6) subjects. (D) Rnf43 mRNA levels in the stomach of WT mice (n = 13) after 3-month H pylori PMSS1 infection. CT values were normalized to Gapdh and fold change was calculated over uninfected mice (n = 6). Error bars indicate SD. ∗P ≤ .05, (A and B) 2-tailed unpaired t test, (C and D) Mann–Whitney test. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; mRNA, messenger RNA.
H pylori infection up-regulated the expression of RNF43 in AGS and MKN45 cells (Figure 5B). In addition, we observed that the expression of RNF43 was higher in gastric biopsy specimens from H pylori–infected patients compared with noninfected individuals (Figure 5C).
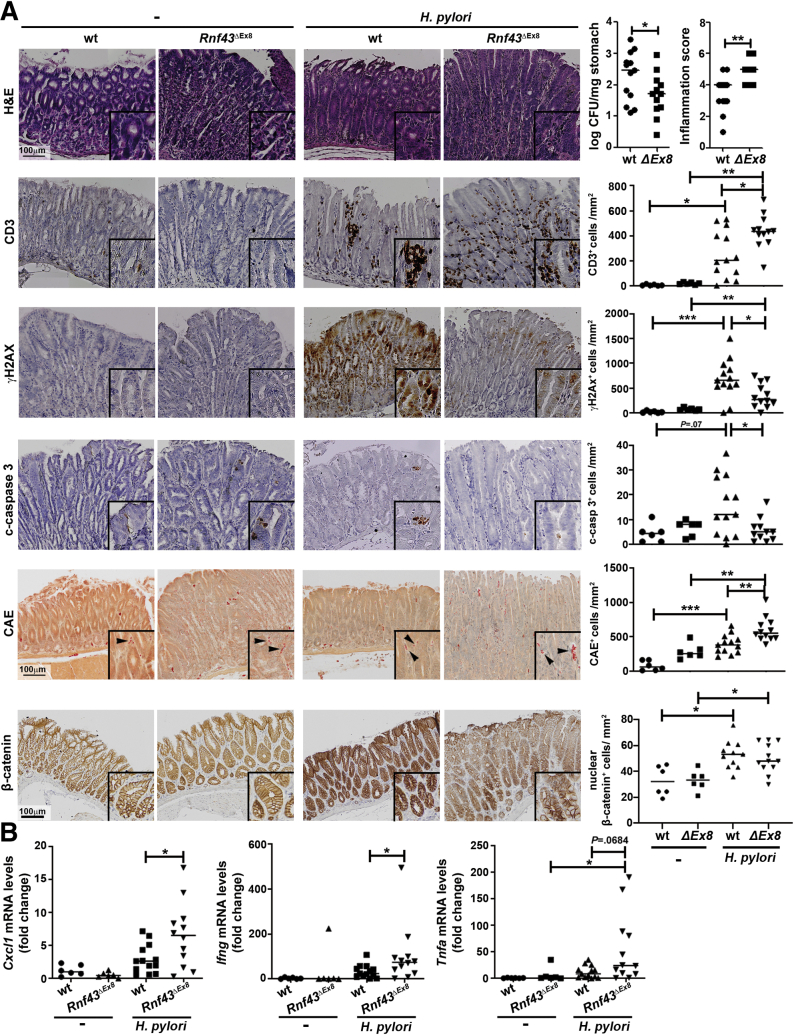
To further explore the involvement of RNF43 in DDR upon H pylori infection in vivo, we infected WT or Rnf43ΔEx8 mice with the H pylori pre-mouse SS1 (PMSS1) strain and killed them after 3 months. As previously reported, at this age, Rnf43ΔEx8 mice did not show pathologic changes in the stomach.13 H pylori infection up-regulated the expression of Rnf43 in the stomach of WT mice (Figure 5D). Notably, Rnf43ΔEx8 mice were colonized at lower levels than WT mice (Figure 6A) and showed more severe gastric inflammation (Figure 6A). Increased recruitment of CD3+ cells into the stomach was observed in Rnf43ΔEx8 mice when compared with WT infected mice (Figure 6A). In addition, increased infiltration of neutrophils and mast cells was detected in Rnf43ΔEx8 mice (Figure 6A). Concomitantly, Rnf43ΔEx8 mice showed increased expression levels of the proinflammatory cytokines Cxcl1 (murine homologue of interleukin 8), Ifng, and Tnfa (Figure 6B).
Figure 6.
RNF43 is involved in H pylori–induced DDR. (A) Colony forming units (CFU)/mg of stomach of WT (n = 13) and Rnf43ΔEx8 mice (n = 12), and inflammatory score after H pylori infection. Activity and chronicity were evaluated in antrum and corpus according to the updated Sydney system. Representative images and quantification of CD3, γH2AX, cleaved-caspase 3 (c-casp3), chloroacetate esterase (CAE)-positive cells (neutrophils and mast cells), and nuclear β-catenin–positive cells per square millimeter of tissue stained by immunohistochemistry in murine gastric tissue samples are shown. Arrowheads indicate positive cells. (B) Cxcl1, Ifng, and Tnfa mRNA levels in the stomach after 3-month H pylori PMSS1 infection. Cycle threshold (CT)values were normalized to Gapdh and fold change was calculated over uninfected mice. Horizontal lines represent the median values. ∗P ≤ .05, ∗∗P ≤ .01, and ∗∗∗P ≤ .001, Mann–Whitney test. mRNA, messenger RNA.
We next assessed activation of DDR response upon H pylori infection by analyzing γH2AX-positive cells in the stomach of WT and Rnf43ΔEx8 mice. H pylori infection resulted in increased levels of γH2AX-positive cells in WT mice. In contrast, no increase in the induction of DDR was detected in infected Rnf43ΔEx8 mice, which showed lower numbers of γH2AX-positive cells when compared with infected WT mice (Figure 6A). In addition, Rnf43ΔEx8 mice showed reduced apoptosis compared with WT mice, as shown by fewer cleaved caspase 3–positive cells in the stomach after infection (Figure 6A).
We also analyzed whether the changes observed in Rnf43ΔEx8 mice could be related to WNT activation by staining stomach tissue samples of naïve and infected mice for β-catenin. β-catenin expression was up-regulated upon infection in WT as well as in Rnf43 mutant mice, confirming activation of WNT signaling by H pylori. However, no differences between wild-type and Rnf43ΔEx8 mice were observed (Figure 6A).
Together, these results indicate that loss of Rnf43 aggravates H pylori–induced inflammation, but at the same time results in reduced DDR. In addition, these effects are independent of alterations in the WNT pathway.
RNF43 Expression Varies During Gastric Carcinogenesis
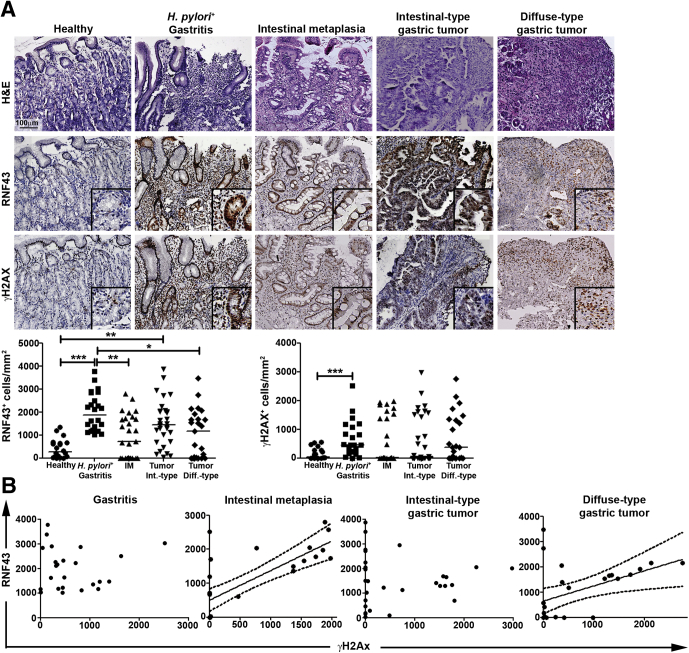
Our results indicate that H pylori infection induces RNF43 expression, which relates to induction of DDR. To further confirm these observations in human tissue, we analyzed human gastric biopsy specimens from healthy, as well as H pylori–infected subjects presenting with gastritis (Table 1). A higher number of RNF43 and γH2AX-positive cells was detected in H pylori–positive gastritis compared with healthy mucosa (Figure 7A), confirming that H pylori infection up-regulates RNF43 expression and induces DNA damage in gastric cells. We further analyzed intestinal metaplasia and gastric tumor samples (Table 1), the latter restricted to intestinal- and diffuse-type tumors according to Lauren’s1 classification. Increased expression of RNF43 was observed in all gastric lesions analyzed (Figure 7A). Interestingly, expression of RNF43 correlated to γH2AX levels in intestinal metaplasia (Spearman rs = 0.7103; P < .0001) as well as in diffuse-type gastric tumors (Spearman rs = 0.4426; P = .0267) (Figure 7B), although no correlation could be observed in gastritis or intestinal-type gastric tumors (Figure 7B).
Table 1.
List of Human Gastric Biopsy Specimens Classified by Gastric Pathology Observed
| Sample ID | Sex | Age, y | H pylori status |
|---|---|---|---|
| Healthy stomach | |||
| H1 | M | 76 | Negative |
| H2 | M | 55 | Negative |
| H3 | F | 90 | Negative |
| H4 | F | 80 | Negative |
| H5 | F | 39 | Negative |
| H6 | M | 72 | Negative |
| H7 | F | 84 | Negative |
| H8 | M | 59 | Negative |
| H9 | M | 69 | Negative |
| H10 | M | 27 | Negative |
| H11 | F | 62 | Negative |
| H12 | M | 59 | Negative |
| H13 | M | 72 | Negative |
| H14 | M | 33 | Negative |
| H15 | F | 50 | Negative |
| H16 | F | 24 | Negative |
| H17 | F | 27 | Negative |
| H18 | F | 41 | Negative |
| H19 | F | 22 | Negative |
| H20 | F | 27 | Negative |
| Gastritis | |||
| G1 | M | 60 | Positive |
| G2 | M | 57 | Positive |
| G3 | M | 59 | Positive |
| G4 | F | 60 | Positive |
| G5 | F | 32 | Positive |
| G6 | M | 58 | Positive |
| G7 | F | 85 | Positive |
| G8 | F | 73 | Positive |
| G9 | M | 90 | Positive |
| G10 | F | 83 | Positive |
| G11 | F | 72 | Positive |
| G12 | F | 78 | Positive |
| G13 | F | 82 | Positive |
| G14 | M | 60 | Positive |
| G15 | M | 90 | Positive |
| G16 | M | 91 | Positive |
| G17 | M | 47 | Positive |
| G18 | F | 48 | Positive |
| G19 | F | 32 | Positive |
| G20 | M | 74 | Positive |
| G21 | F | 81 | Positive |
| G22 | F | 52 | Positive |
| G23 | M | 90 | Positive |
| G24 | M | 68 | Positive |
| Intestinal metaplasia | |||
| IM1 | M | 76 | Eradicated |
| IM2 | M | 81 | Negative |
| IM3 | F | 67 | Eradicated |
| IM4 | F | 80 | Negative |
| IM5 | F | 68 | Eradicated |
| IM6 | M | 55 | Eradicated |
| IM7 | M | 31 | Eradicated |
| IM8 | M | 63 | Eradicated |
| IM9 | F | 80 | Eradicated |
| IM10 | M | 40 | Eradicated |
| IM11 | M | 67 | Eradicated |
| IM12 | F | 84 | Positive |
| IM13 | F | 85 | Eradicated |
| IM14 | F | 66 | Eradicated |
| IM15 | M | 75 | Eradicated |
| IM16 | M | 75 | Eradicated |
| IM17 | M | 64 | Negative |
| IM18 | F | 82 | Eradicated |
| IM19 | F | 52 | Eradicated |
| IM20 | M | 86 | Positive |
| IM21 | F | 64 | Eradicated |
| IM22 | M | 76 | Positive |
| IM23 | F | 54 | Positive |
| IM24 | F | 33 | Eradicated |
| IM25 | M | 61 | Negative |
| IM26 | M | 86 | Eradicated |
| IM27 | M | 68 | Eradicated |
| Intestinal-type gastric cancer | |||
| GCi1 | F | 80 | Negative |
| GCi2 | F | 57 | Positive |
| GCi3 | M | 71 | Eradicated |
| GCi4 | M | 87 | Eradicated |
| GCi5 | F | 78 | Negative |
| GCi6 | M | 66 | Negative |
| GCi7 | M | 53 | Positive |
| GCi8 | F | 82 | ND |
| GCi9 | F | 79 | Positive |
| GCi10 | M | 72 | Negative |
| GCi11 | M | 86 | Negative |
| GCi12 | F | 79 | Eradicated |
| GCi13 | M | 88 | Negative |
| GCi14 | F | 77 | Eradicated |
| GCi15 | F | 82 | ND |
| GCi16 | M | 70 | 0 |
| GCi17 | M | 70 | Positive |
| GCi18 | F | 95 | Positive |
| GCi19 | M | 88 | Negative |
| GCi20 | F | 98 | ND |
| GCi21 | M | 86 | Negative |
| GCi22 | M | 62 | ND |
| GCi23 | F | 58 | ND |
| GCi24 | M | 86 | Eradicated |
| GCi25 | M | 73 | Eradicated |
| GCi26 | F | 80 | Negative |
| GCi27 | F | 98 | Positive |
| GCi28 | M | 82 | ND |
| GCi29 | F | 95 | Eradicated |
| GCi30 | F | 84 | Eradicated |
| Diffuse-type gastric cancer | |||
| GCd1 | F | 86 | Negative |
| GCd2 | F | 77 | Eradicated |
| GCd3 | M | 71 | Eradicated |
| GCd4 | F | 75 | Eradicated |
| GCd5 | M | 69 | Negative |
| GCd6 | M | 77 | Eradicated |
| GCd7 | F | 81 | Negative |
| GCd8 | F | 82 | Negative |
| GCd9 | F | 58 | ND |
| GCd10 | M | 98 | ND |
| GCd11 | F | 83 | ND |
| GCd12 | M | 82 | Negative |
| GCd13 | F | 94 | ND |
| GCd14 | M | 89 | Positive |
| GCd15 | F | 87 | Eradicated |
| GCd16 | F | 86 | Eradicated |
| GCd17 | M | 76 | Positive |
| GCd18 | M | 34 | Eradicated |
| GCd19 | M | 85 | ND |
| GCd20 | M | 51 | Positive |
| GCd21 | M | 72 | Negative |
| GCd22 | M | 51 | ND |
| GCd23 | F | 85 | Negative |
| GCd24 | F | 91 | Eradicated |
| GCd25 | F | 81 | Negative |
| GCd26 | F | 78 | Positive |
ND, not determined.
Figure 7.
RNF43 expression varies during gastric carcinogenesis. (A) Representative images and quantification of RNF43 and γH2AX-positive cells per square millimeter of human gastric tissue of healthy individuals (n = 20), and patients with gastritis (n = 24), intestinal metaplasia (n = 27), and intestinal-type (n = 20) or diffuse (n = 25) gastric cancer. Horizontal lines represent the median values. Each dot represents 1 sample. ∗P ≤ .05, ∗∗P ≤ .01, and ∗∗∗P ≤ .001, 1-way analysis of variance multiple comparisons. (B) Spearman correlation between the number of RNF43 and γH2AX-positive cells per square millimeter of human gastric tissue. Diff, diffuse; IM, intestinal metaplasia; Int, intestinal.
RNF43 Mutations Upstream of the RING Domain Confer Resistance to DNA Damage
RNF43 was observed to be mutated frequently in gastric tumors. To determine whether our samples carried WT or mutated RNF43, we sequenced a subset of intestinal metaplasia (n = 12), intestinal-type (n = 15), and diffuse-type (n = 13) gastric tumors. No mutations in RNF43 were detected in intestinal metaplasia samples. Three of the 12 samples analyzed harbored mutations in APC (p.A1471Gfs∗14; p.R1463fs; p.R1450X). In 2 cases, APC mutation co-occurred with a mutation in BRAF (p.N581S) and KRAS (p.G13D), respectively. Mutated ERBB4 (p.L713V) was observed in 1 of the intestinal metaplasia cases (Table 2).
Table 2.
List of Mutations Found in Human Gastric Tissue Samples
| Sample | RNF43 SNPs | RNF43 mutations | Other mutations | MSI status |
|---|---|---|---|---|
| Intestinal metaplasia | ||||
| IM1 | p.I47V p.R117H p.P686R |
– | APC p.A1471Gfs∗14 BRAF p.N581S |
|
| IM2 | p.I47V p.R117H |
– | APC p.R1445fs KRAS p.G13D |
|
| IM3 | p.R117H p.P686R |
– | – | |
| IM4 | – | – | ERBB4 p.L713V | |
| IM5 | p.I47V p.R117H p.L418M |
– | APC p.R1432X | |
| IM6 | p.R117H p.P686R |
– | – | |
| IM7 | p.R117H | – | – | |
| IM8 | p.I47V p.L418M p.P686R |
– | – | |
| IM9 | p.R117H p.L418M |
– | – | |
| IM10 | p.I47V | – | – | |
| IM11 | p.R117H | – | – | |
| IM12 | p.I47V p.R117H p.L418M |
– | – | |
| Intestinal-type gastric cancer | ||||
| GCi1 | p.I47V p.R117H p.L418M |
– | PTPRT p.P1075Rfs PIK3CA p.E542K TLR4 p.L10M |
|
| Gci2 | p.P686R | – | ERBB2 p.V842I | MSS |
| Gci3 | p.R117H p.P760P |
– | MDM2 amplification | |
| Gci4 | – | – | Sox2 p.E36G APC p.R1432X PREX2 p.L50V TP53 p.G66R |
|
| Gci5 | p.I47V p.R117H p.L418M |
– | TP53 p.E204X | |
| Gci6 | p.I47V p.L418M p.P686R |
– | TP53 p.R116Q TP53 p.Y88H CDKN2A p.Q50H |
|
| Gci7 | p.I47V p.R117H |
– | TLR4 p.L298V PTPRT p.E911G |
|
| Gci8 | p.I47V p.L418M p.P686R |
– | ARID1A p.P1467fs PTEN M134T TP53 R151H |
|
| Gci9 | WT | – | – | |
| Gci10 | p.I47V p.L418M |
– | – | MSS |
| Gci11 | p.I47V p.R343H p.L418M |
– | CDKN2A p.Q70X PTEN p.R130X TGFBR2 p.D405N |
|
| Gci12 | R117H | – | MSS | |
| Gci13 | p.I47V p.L418M |
– | TP53 p.G134E APC p.R1432X |
|
| Gci14 | p.I47V p.R117H |
p.R371R p.D628G |
CCND1 p.S257P | MSS |
| Gci15 | – | p.R132X | FBXW7 p.A508D RHOA p.Y42C ARID1A p.P2095fs |
MSI |
| Diffuse-type gastric cancer | ||||
| GCd1 | p.I47V p.L418M |
– | PREX2 p.R186Q | |
| GCd2 | p.I47V p.R117H p.P686R |
– | – | |
| GCd3 | – | p.R584fs | ERBB2 p.R678Q CDH1 p.E429G |
MSS |
| GCd4 | p.I47V p.L418M |
– | – | MSS |
| GCd5 | p.L418M | – | CDH1 p.D254H | |
| GCd6 | p.I47V p.L418M |
– | – | |
| GCd7 | – | p.R132X | TP53 p.R117M | MSS |
| GCd8 | p.I47V p.R343H p.L418M |
– | ARID1A p.R2158L CDH1 c.531+1G>T |
MSS |
| GCd9 | p.I47V p.L418M |
– | CDH1 p.E243K | MSS |
| GCd10 | p.I47V p.L418M |
– | KRAS p.G13D TP53 p.R150W PIK3CA p.N345K PIK3CA p.A1066V MTOR p.R2317I |
|
| GCd11 | p.P686R | – | – | |
| GCd12 | p.I47V p.L418M |
– | – | |
| GCd13 | p.I47V p.L418M |
– | PIK3CA p.H1047R | |
MSS, microsatellite stable.
Mutations in RNF43 were found only in gastric tumors. Two of the intestinal-type gastric tumors carried mutated RNF43 (p.D628G and p.R371R; p.R132X). Interestingly, 5 of the 15 cases analyzed presented mutations in TP53. RNF43 was mutated in 2 diffuse-type tumors (p.R584fs; p.R132X). In this subset of tumors, mutations in CDH1 were the most frequent (4 of 13 cases), as expected for this type of stomach cancer. A summary of the mutations can be found in Table 2. RNF43 mutations have been found more frequently in MSI-high tumors. Mutated diffuse-type tumors were genomically stable, as expected.3 In contrast, 1 of the intestinal-type tumors carrying mutated RNF43 was MSI-high (Table 2).
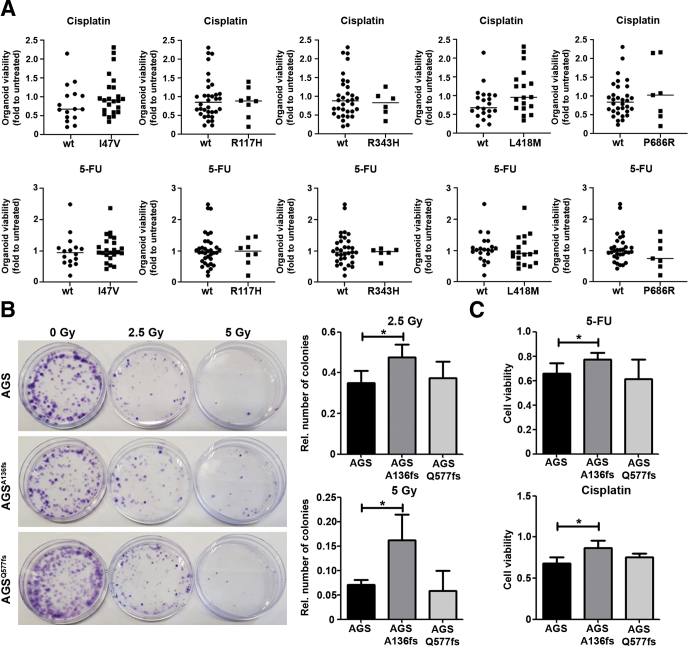
In most of the cases analyzed we found single-nucleotide polymorphism (SNP) variants of RNF43, namely I47V (rs3744093), R117H (rs2257205), L418M (rs2526374), and P686R (rs9652855) (Table 2). It recently was shown that a SNP of the RNF43 X117 site is associated with overall survival of colorectal cancer patients.29 To determine whether this could be related to an altered response to DNA damage-inducing chemotherapy, we sequenced human gastric organoids to identify SNPs and analyzed their response to 5-fluorouracil and cisplatin. Five SNPs were found frequently. The I47V variant was found in 23 of the 40 cases (57.5%). Nine cases carried the R117H variant (22.5%). The L418M variant was observed in 18 cases (45%), and the P686R variant was observed in 7 cases (17.5%). In 6 cases (15%) we found the variant R343H (rs34523089), which was detected in only 2 tumor samples (Table 2). No difference in response to chemotherapy was detected between samples harboring WT RNF43 and samples presenting any of the aforementioned SNPs (Figure 8A).
Figure 8.
RNF43 mutations upstream of the RING domain confer resistance to DNA damage. (A) Viability of gastric organoids (n = 40) carrying different SNP variants of RNF43 after treatment with 5-fluorouracil (5-FU) or cisplatin. Fold change was calculated over untreated organoids. Horizontal lines represent the median values. Mann–Whitney test. (B) Representative images and quantification of cell colonies after γ-radiation. (n = 4). (C) Cell viability after treatment with 5-FU or cisplatin. Values were normalized over untreated cells (n = 4). Error bars indicate SD. ∗P ≤ .05, 2-tailed unpaired t test. Rel, relative.
To determine whether mutations occurring in tumors conferred resistance to γ-radiation, we selected 2 mutations observed in our tumor cohort and generated AGS cells carrying these mutations by CRISPR/CAS9 editing. We selected mutations occurring before (p.R132X) or downstream of the RING domain (p.R584fs). After transfection of guide RNAs we could not introduce the indicated mutations but obtained cells carrying the frameshift mutations A136fs and Q577fs, respectively. Because these mutations give rise to similar proteins, we used the generated AGSA136fs and AGSQ577fs in further experiments. AGSA136fs cells showed resistance to γ-radiation as more colonies were observed (Figure 8B). In contrast, AGSQ577fs cells were as sensitive to γ-radiation as WT cells. Similarly, the frameshift mutation A136fs conferred increased resistance to chemotherapy in AGS cells (Figure 8C), while the viability of cells expressing the Q577fs mutation after 5-fluorouracil or cisplatin treatment was similar to the viability of AGS WT cells.
These findings show that mutations upstream of the RING domain of RNF43 in gastric cells confer resistance to DNA damage-inducing agents, and confirm the importance of the RING domain for proper function of the protein.
RNF43 Regulates DNA Damage Response by Ubiquitinating γH2AX
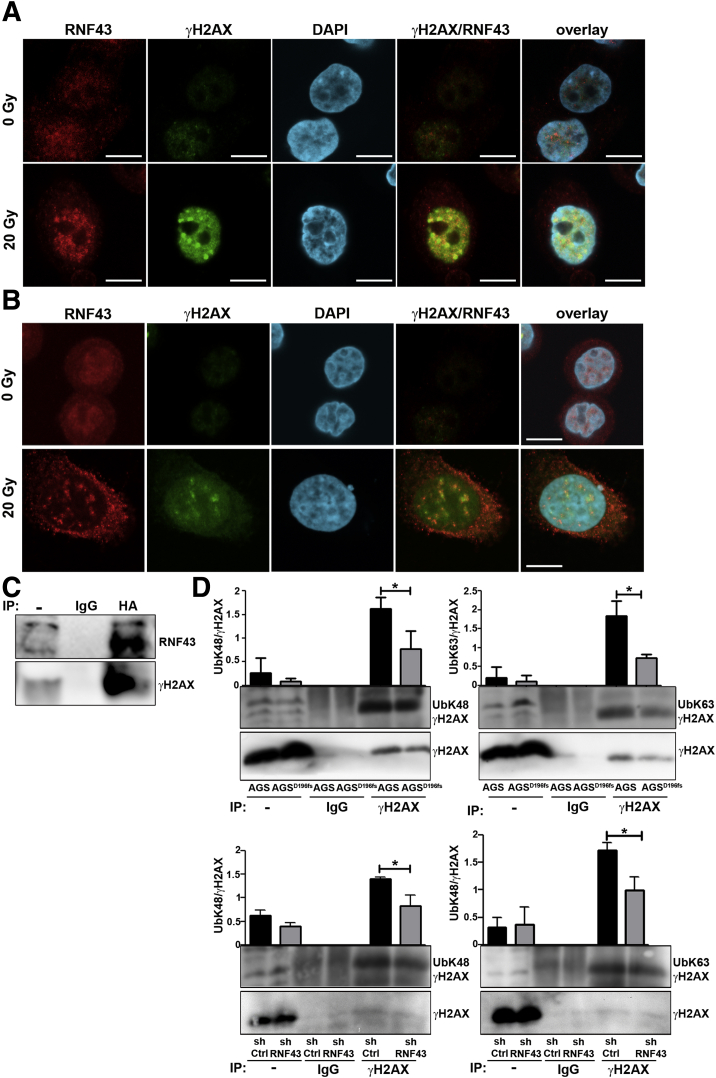
We finally explored the molecular mechanism by which RNF43 regulates DDR. We observed that upon induction of DNA damage, RNF43 co-localized with γH2AX in the nucleus of irradiated cells (Figure 9A and B). Moreover, direct interaction between RNF43 and γH2AX was detected in co-immunoprecipitation experiments after irradiation of AGS cells (Figure 9C).
Figure 9.
RNF43 is up-regulated in response to DNA damage and it is recruited to sites of DNA damage. Representative immunofluorescence images of RNF43 (red) and γH2AX (green) in control and irradiated (A) AGS and (B) MKN45 cells. Scale bars: 10 μm. (C) Immunoprecipitation of RNF43 (human influenza hemagglutinin, HA) from AGS cells transfected with HA-tagged RNF43 upon γ-radiation (20 Grey), followed by Western blot analysis (n = 3; 1 representative experiment is shown). (D) Immunoprecipitation of γH2AX from AGS and MKN45 cells after radiation (20 Grey), followed by Western blot analysis. -, whole lysate. Quantification of ubiquitinated γH2AX from total γH2AX is shown (n = 3). Error bars indicate SD. ∗P ≤ .05, 2-tailed unpaired t test. DAPI, 4′,6-diamidino-2-phenylindole; IP, immunoprecipitation.
We next analyzed whether RNF43 influenced γH2AX ubiquitination status. To this end, we immunoprecipitated γH2AX in AGSD196fs and MKN45shRNF43 cells and detected K48 and K63 ubiquitination by Western blot. We observed that ubiquitination of γH2AX was reduced in RNF43-depleted cells (Figure 9D), indicating that RNF43 is involved in DDR by interacting and ubiquitinating γH2AX.
Discussion
Several studies have shown that RNF43 is mutated in tumors of the gastrointestinal tract, including colorectal, pancreatic, and GC. RNF43 mutations found in GC were considered driving mutations of gastric carcinogenesis,8 occurring early during transition of adenomas to carcinomas.30 Functionally, the importance of RNF43 for tissue homeostasis derives from its role as an inhibitor of the WNT signaling pathway.14,15 However, our previous data suggest that the tumor-suppressor activity of RNF43 in gastric cells is independent of WNT. In a xenograft model using human gastric epithelial cells with lentiviral-mediated silencing of RNF43, we observed no changes in the activation of WNT signaling. In addition, gastric alterations detected in Rnf43ΔEx8 mice were not related to changes in the expression of WNT target genes,13 indicating that RNF43 might have an alternative function in the stomach. Interestingly, various RING finger proteins have been reported to play important roles in DDR by ubiquitinating the central histone H2AX. For instance, a complex of RNF2 and RNF51 ubiquitinates γH2AX at the lysine residues K118/K119,31 while RNF8 and RNF168 attach further K63-linked ubiquitin chains to this histone.17,18,32 RNF168 also has been described to ubiquitinate γH2AX at the lysine residues K13/K15, which has been shown to be important for DDR signaling.33 In our study, we observed that loss of RNF43 function resulted in reduced levels of K63- and K48-linked γH2AX ubiquitination upon irradiation, which translated into impaired DDR activation and enhanced cellular survival. These results indicate an important role of RNF43 in DDR because ubiquitination of γH2AX has been described extensively as a central post-translational modification for the initiation19,34 and maintenance17,18,33 of DDR signaling.
The expression of RNF43 was found to be increased upon DNA damage. Top transcription factor binding sites described in the RNF43 gene promoter include aryl hydrocarbon receptor (AhR), Activator protein 1 (AP-1), Aryl Hydrocarbon Receptor Nuclear Translocator (Arnt), c-Fos Proto-Oncogene, AP-1 Transcription Factor Subunit (c-Fos), c-Jun Proto-Oncogene, AP-1 Transcription Factor Subunit (c-Jun), Forkhead Box O4 (FOXO4), and Peroxisome Proliferator Activated Receptor Gamma (PPAR-γ) (GeneCards). All of these transcription factors have been implicated in different aspects of the DDR, and therefore could be regulating RNF43 expression upon DNA damage. The identification of the transcription factors involved and the regulatory mechanisms behind it require further investigation.
H pylori infection is the main risk factor for GC development. Previous studies have indicated that H pylori can induce DNA damage directly to host cells through interaction of the type IV secretion system with integrin β1. This interaction would lead to the activation of nuclear factor-κB signaling and thereby to the activation of nucleotide excision repair endonucleases responsible for inducing DSBs.26 In contrast, the involvement of other virulence factors such as vacuolating cytotoxin A (VacA) in the induction of DSBs was excluded.25 Whether these virulence factors play a role in the RNF43-mediated DDR therefore deserves further investigations.
Current treatment of GC patients includes adjuvant chemotherapy with cisplatin or 5-fluorouracil, as well as irradiation.35 The efficacy of these treatments to eliminate tumor cells is based on their ability to induce DNA damage. However, not all tumors respond to adjuvant therapy, and biomarkers that could predict response still are lacking. In a previous study, overexpression of RNF43 was shown to render gastric cells more susceptible to oxaliplatin and 5-fluorouracil, but the molecular mechanisms involved remained unclear.36 In line with this observation, we also found that loss of RNF43 function confers resistance to DNA damage-inducing chemotherapy. More importantly, we identified the mechanism responsible, namely the direct role of RNF43 in DDR.36
Furthermore, we observed a correlation between response to chemotherapy and RNF43 expression levels in human gastric organoids, indicating that not only loss of functional protein but also its down-regulation or up-regulation might modify resistance to DNA damage-inducing chemotherapeutics. We postulate that treating patients expressing low levels of RNF43, or carrying mutations affecting its functional domain, with DNA-damaging drugs and radiation therapy (as recommended in the guidelines28) would lead to detrimental effects. Thus, cells lacking RNF43 not only would resist radiotherapy or chemotherapy-induced apoptosis, but eventually would accumulate additional mutations induced by the treatments themselves, contributing to tumor progression and thereby worsening prognosis. Along these lines, decreased RNF43 expression has been associated with distant metastasis, TNM stage, and poorer overall survival of GC patients,36 supporting our hypothesis. It also is relevant to note that RNF43 mutations are highly enriched in MSI gastric tumors,8,11,12,30 which are characterized by a high mutation burden in tandem repeats37 caused by deficient DNA repair machinery.38 Therefore, it is plausible that acquired mutations in RNF43 may contribute to the increased mutation rate in MSI tumors by further impairing DDR activation.
Our data suggest that mutations downstream of the catalytic domain of RNF43 still would retain functionality. This observation is in line with recent data showing that, for instance, the most common RNF43 mutation in gastric tumors (G659Vfs∗41) is still fully functional in terms of WNT inhibition.39
In light of our findings, the inclusion of RNF43 in GC gene panels for the screening of patients should be considered to select therapeutic regimens based on mutation status. This, together with the examination of protein expression levels of WT RNF43, may serve as a biomarker for therapy selection for GC patients. Moreover, screening of RNF43 mutations and expression also could be considered for other types of tumors such as colorectal or pancreatic cancers, where mutations in RNF43 have been observed frequently. In fact, and as mentioned earlier, a possible role of RNF43 in DDR in pancreatic cells already has been suggested.40 As such, further investigations in other tissues will be important to confirm a general function of RNF43 as a modulator of DDR.
Materials and Methods
Cell Culture and Cell Treatments
AGS (ATCC CRL-1739) and MKN45 (RCB1001) cells were cultured in Dulbecco’s modified Eagle medium (Invitrogen, Carlsbad, CA) containing 10% fetal calf serum (Sigma-Aldrich, St. Louis, MO) at 37°C in a humidified atmosphere (5% CO2). Cells were tested routinely for Mycoplasma contamination. Short tandem repeat analysis was performed for authentication of the cell lines used. AGS and MKN45 shControl and shRNF43 cells were generated using lentiviruses as previously described.13 Sequences are listed in Table 3.
Table 3.
DNA-Oligo Sequences for Lentiviral Knockdown and CRISPR/Cas9 Editing
| Oligo | Forward | Reverse |
|---|---|---|
| shRNF43 1 | GGAGAAAGCTATTGCACAGAA | TTCTGTGCAATAGCTTTCTCC |
| shRNF43 2 | CGCGTCCCCTTCTTGGTAAGATC | CGATTTCCAAAAATTCTTGGTAAGAT |
| GAGAGTTCAAGAGACTCTCGATCTT | CGAGAGTCTCTTGAACTCTCGATCT | |
| ACCAAGAATTTTTGGAAAT | TACCAAGAA GGGGA | |
| shctrl 1 | GCAACTTCAGCTATATCATTT | AAATGATATAGCTGAAGTTGC |
| shctrl 2 | CGCGTCCCCGTACAGCCGCCTCAATTCT | CGATTTCCAAAAAGTACAGCCGCCTCA |
| TTCAAGAGAAGAATTGAGGCGGCTGT | ATTCTTCTCTTGAAAGAATTGAGGCGGCT | |
| ACTTTTTGGAAAT | GTACGGGGA | |
| Guide RNA CRISPR/Cas9 AGSD196fs | CCACACATCATAATCTGGCT | AGCCAGATTATGATGTGTGG |
| Guide RNA CRISPR/Cas9 AGSR132X | GATGTCAAAGAGGACAGCAC | – |
| Guide RNA CRISPR/Cas9 AGSR584fs | AAACCGGAGTCCCCCAGTCC | – |
AGSD196fs, AGSA136fs, and AGSQ577fs cells were generated by CRISPR/Cas9. To generate AGSD196fs cells, we used guide RNAs targeting exon 6 of RNF43 to introduce a 4-bp deletion at aspartic acid 196. The resulting frameshift mutation leads to a premature stop codon 5 amino acids downstream, deleting the functional RING domain. Guide RNA sequences (Table 3) were designed using www.tools.genome-engineering.org, annealed, and cloned into a pX330 vector (42230; Addgene, Watertown, Massachusetts, USA) containing green fluorescent protein (GFP). Cells were transfected with Lipofectamine 2000 (Invitrogen), sorted for GFP, and grown as single-cell colonies. Genomic DNA was extracted using the PureLink Genomic DNA Mini Kit (Invitrogen), and mutations in RNF43 and selected off targets were checked by Sanger sequencing.
For targeted mutations, Cas9 ribonucleoproteins (RNPs) were transfected together with an oligonucleotide containing the desired mutations and homology arms flanking 30 bp of the site of interest on both sides. Guide RNA and repair oligonucleotides were planned using Benchling (www.benchling.com), and guide RNAs, ATTO 550-labeled tracer RNA, and Cas9 protein were purchased from Integrated DNA Technologies (Coralville, IA). Repair oligonucleotides were purchased as single-stranded DNA, as follows:
R132X: TGCCTCTGCAGGCTCGGATGGCGGGTGAGTGAGGAGCAAGTGCTGTCCTCTTTGACATCA;
R584fs: CAGAAACCGGAGTCCCCCAGTCCAGACCTCCTATTCCTCGACACAGCCCCAGCCAGAGCCACCTTCTCCT.
RNP assembly and transfection were performed as described by Integrated DNA Technologies. AGS cells (5∗105) were electroporated with RNPs using a Nucleofector 4D (Kit X, pulse code DS-135; Lonza, Basel, Switzerland). To increase homology-directed repair, Nu7026 (20 μmol/L; Sigma-Aldrich) and Trichostatin A (0.01 μmol/L; Sigma-Aldrich) were added. At 24 hours after electroporation, cells were sorted for ATTO 550 and seeded as single-cell clones for selection.
Lipofectamine 2000 (Invitrogen) was used for plasmid transfections. Transfected cells were cultured for 36 hours before being subjected to γ-radiation and subsequent co-immunoprecipitation.
γ-radiation was performed using a Cs137 radiation source (Buchler, Braunschweig, Germany). For chemotherapeutic treatment, cells were seeded in a 96-well plate (7500 cells/well) and treated with 5-fluorouracil (2 μg/mL) or cisplatin (10 μg/mL) for 48 hours, followed by measurement of cell viability using Cell-counting kit 8 (Sigma-Aldrich).
Cell Viability and Clonogenicity Assay
AGS and MKN45 cells were seeded in a 96-well plate (2500 cells/well) after γ-radiation, and serum-starved for 24 hours. After addition of serum-containing medium, cells were allowed to grow for 48 hours and cell viability was measured using Cell-counting kit 8 (Sigma-Aldrich).
To assess clonal expansion, AGS and MKN45 cells were irradiated and seeded at low density. After 14 days, single-cell colonies were fixed and stained with 6% glutaraldehyde containing Crystal Violet.
Annexin V Staining
Four days (AGS) or 6 days (MKN45) after γ-radiation, cells were stained with Annexin V Pacific Blue conjugate (Invitrogen) and propidium iodide (Sigma-Aldrich). Fluorescence was measured in a CyAn ADP flow cytometer (Beckmann Coulter, Brea, CA) and analyzed with FlowJo (Becton, Dickinson & Company, Franklin Lakes, NJ).
H pylori Infection
H pylori strain PMSS141 was cultured on Wilkins-Chalgren Dent agar plates in a microaerophilic atmosphere.
Cells were infected at a multiplicity of infection (MOI) of 50 and 100 (OD600 1 = 2∗108 bacteria/mL) for 24 hours, and lysed in sodium dodecyl sulfate (SDS) sample buffer or RNA lysis buffer.
Quantitative Polymerase Chain Reaction
GenElute Mammalian Total RNA Miniprep Kit (Sigma-Aldrich) was used to isolate RNA. Mouse tissue was homogenized using a Precellys (Bertin Instruments, Montigny-le-Bretonneux, France) lysing kit. MMLV Reverse Transcriptase RNase H−Point Mutant (Promega, Madison, WI) was used for reverse transcription. Transcript abundance was assessed using the GoTaq quantitative polymerase chain reaction Mastermix (Promega) and a CFX384 system (Bio-Rad, Hercules, CA). Primer sequences are included in Table 4. Gene expression was analyzed with the comparative relative expression (ΔΔCT) method.
Table 4.
Sequences of the Primers Used in This Study
| Primer | Human |
Murine |
||
|---|---|---|---|---|
| Forward | Reverse | Forward | Reverse | |
| GAPDH | GAAGGTGAAGGTCGGAGT | GAAGATGGTGATGGGATTTC | GCCTTCTCCATGGTGGTGAA | GCACAGTCAAGGCCGAGAAT |
| RNF43 (qPCR) | GAGTGTGCTCCAGATGTGTT | AGTCCTCTTCCA GTCCTTCTA | GGGGCAAACTATGACGTGTG | CTGCTGAAGAGGATCCGGTC |
| AXIN2 | CTC CCC ACC TTG AAT GAA GA | GTT TCC GTG GAC CTC ACA CT | ||
| LGR5 | TGA TGA CCA TTG CCT ACA C | GTA AGG TTT ATT AAA GAG GAG AAG | ||
| Ifng | – | – | TCAAGTGGCATAGATGTGGAAGAA | TGGCTCTGCAGGATTTTCATG |
| Cxcl1 | – | – | TGCACCCAAACCGAAGTCAT | TTGTCAGAAGCCAGCGTTCAC |
| TNFα | – | – | CGATGGGTTGTACCTTGTC | CGGACTCCGCAAAGTCTAAG |
Immunofluorescence
Cells were seeded on glass coverslips either untreated or immediately after being subjected to γ-radiation (20 Gγ). Immunofluorescence staining was performed as previously described.13 The monoclonal antibody GGTRNF 8D6 (IgG2b/k) was raised in rats against amino acid residues 329–348 (SRSYQEPGRRLHLIRQHPGH) of human RNF43. Staining was analyzed using a FluoView FV10i confocal microscope (Olympus, Shinjuku, Japan).
Co-immunoprecipitation and Western blot
Cells were lysed in RIPA buffer (50 mmol/L Tris-HCl, pH 7.4, 1% Nonidet P-40 (NP-40), 150 mmol/L NaCl, 0.25% Deoxycholate (DOC), 1 mmol/L ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid) containing phosphatase and protease inhibitors and precleared using 1 μg appropriate IgG and agarose conjugate. Lysates were centrifuged at 3000 rpm for 1 minute at 4°C, and incubated with primary antibody or IgG overnight at 4°C while rotating. Protein A/G agarose beads (Roche, Basel, Switzerland) were added to the lysates and incubated for 4 hours at 4°C. After washing with phosphate-buffered saline, beads were collected by centrifugation, diluted in SDS sample buffer for Western blot, and boiled for 5 minutes at 95°C.
Protein lysates were subjected to SDS–polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. After blocking with 5% low-fat milk in Tris-buffered saline-tween (TBS-T), membranes were incubated in primary antibody (Table 5) overnight at 4°C. Membranes were washed in TBS-T followed by incubation with horseradish peroxidase–conjugated secondary antibodies for 1 hour at room temperature. Signal was visualized using Clarity ECL Western substrate (Bio-Rad) and an Intas chemiluminescence detection system (Intas, Göttingen, Germany).
Table 5.
Antibodies Used in This Study
| Target | Clone | Assay | Dilution | Company | Reactivity |
|---|---|---|---|---|---|
| Annexin V, Pacific Blue conjugate | ----------- | Flow cytometry | 1:100 | Thermo Fisher | H, M |
| (non-p-)β-catenin | D2U8Y | IHC | 1:1000 | Cell Signaling (Danvers, MA) | H, M |
| CD3 | SP7 | IHC | 1:150 | Thermo Fisher | H, M |
| Chk2 | 1C12 | WB | 1:1000 | Cell Signaling | H |
| Cleaved caspase 3 (Asp175) | 9661 | IHC | 1:300 | Cell Signaling | H, M |
| GAPDH | 14C10 | WB | 1:1000 | Cell Signaling | H, M |
| HA | H6908 | IP | 1:100 | Sigma | – |
| HA | H3663 | WB | 1:1000 | Sigma | – |
| Ki67 | D2H10 | IHC | 1:400 | Cell Signaling | H |
| p-Chk2 (Thr68) | C13C1 | WB | 1:1000 | Cell Signaling | H |
| RNF43 | HPA008079 | IHC | 1:1000 | Invitrogen (ATLAS) | H |
| RNF43 | 8D6 | IF | 1:150 | – | H |
| K48-linked Ubiquitin | D9D5 | WB | 1:1000 | Cell Signaling | H |
| K63-linked Ubiquitin | D7A11 | WB | 1:1000 | Cell Signaling | H |
| γH2Ax | ab81299 | WB, IHC, IF | 1:10000 (WB) | Abcam | H, M |
| 1:5000 (IHC) | |||||
| 1:300 (IF) | |||||
| γH2Ax | 5438 | IP | 1:50 | Cell Signaling | H, M |
GAPDH, glyceraldehyde-3-phosphate dehydrogenase; H, human; IF, immunofluorescence; IHC, immunohistochemistry; IP, immunoprecipitation; M, mouse; WB, Western blot.
Gastric Organoids
Gastric organoid culture was established as previously described.42 There were 100 (48-well plate) or 300 (24-well plate) gastric glands seeded per well.
After 1 week, organoids were incubated with 5-fluorouracil (2 μg/mL) or cisplatin (10 μg/mL) for 4 (murine) or 5 (human) days. Viability was analyzed using Cell Titer Glo 3D (Promega). Luminescence was measured using a SpectraMax plate reader (Molecular Devices, San José, CA). Viability was normalized to untreated controls. For human organoids, an organoid viability normalized to control greater than 95% was defined as a nonresponder, while organoids with a viability of less than 95% were defined as responders.
Animal Experiments
Friend Virus B NIH Jackson (FVBN) WT and Rnf43ΔEx8 mice13 were used for in vivo experiments. Rnf43ΔEx8 mice were generated by introducing a 57-bp deletion in exon 8 of the genomic sequence of Rnf43 using CRISPR/Cas9 technology. This deletion led to a systemic loss of the functional RING domain of Rnf43. Mice were co-housed under specific pathogen-free conditions in individually ventilated cages (Tecniplast, Hohenpeißenberg, Germany) containing enrichment material. Mice were fed a rodent diet (Envigo, Indianapolis, IN) ad libitum.
Eight- to 10-week-old male and female FVBN WT or Rnf43ΔEx8 mice were infected twice with 2∗108 H pylori PMSS1 diluted in 200 μL Brain heart infusion (BHI) 20% fetal calf serum by oral gavage. Mice were killed after 3 months.
Xenograft tumors were established as previously described.13 Tumor volume was measured with a caliper and calculated as follows: volume = length∗width∗height/2. When tumors reached a volume of 40–70 mm3, mice were injected with 20 mg/kg 5-fluorouracil or 10 mg/kg cisplatin twice a week. When tumors reached a size of 500 mm3, mice were killed and tumors were resected.
Immunohistochemistry
Mouse gastric samples were fixed in 4% formaldehyde and embedded in paraffin. Heat-induced antigen retrieval was performed using 10 mmol/L sodium citrate (pH 6). Primary antibodies were applied overnight (Table 5), and bound secondary antibodies were detected with diaminobenzidine. Slides were scanned and analyzed using an Olympus Virtual Slide Imaging System.
Sequencing
H&E-stained sections of gastric biopsy specimens were used to select the lesion to be analyzed. Sections (10-μm) were used to extract DNA with a GeneRead DNA FFPE Kit (Qiagen, Hilden, Germany). DNA concentrations were determined by a QuiBit 2.0 fluorometer (ThermoFisher, Waltham, MA) and the amplifiable genomic DNA was quantified using TaqMan RNase P detection kit (ThermoFisher). Barcoded libraries were generated using the Ion Xpress Barcode Adapters (ThermoFisher). Samples were purified using the Agencourt AMPure XP kit (Beckman Coulter) and quantified with the Ion Library TaqMan quantitation kit (ThermoFisher). Libraries were loaded on the Ion 530 chip kit (ThermoFisher) and sequenced using an Ion S5XL sequencer (ThermoFisher).
Statistics
Data were first tested for normality using the Shapiro–Wilk test. Normally distributed data then were analyzed by the Student t test. The Mann–Whitney U test or the Kruskal–Wallis test for multiple comparisons was used to compare not normally distributed data. Results of at least 3 independent experiments were analyzed. Statistical significance was defined when P < .05.
Study Approval
Experiments with human gastric organoids were approved by the Ethics Committee of Klinikum Rechts der Isar (116/17 S).
All animal experiments were conducted in compliance with European guidelines for the care and use of laboratory animals and were approved by the Regierung von Oberbayern (AZ 55.2-1-55-2532-239-15 and AZ 55.2-1-54-2532-196-2016).
Formalin-fixed, paraffin-embedded human gastric samples (Table 1) were obtained from the tissue bank of the Institut für Pathologie at Klinikum Bayreuth (Germany) after approval of the local ethics committee (Project number 243_19Bc).
All authors had access to the study data and reviewed and approved the final manuscript.
Acknowledgments
The authors thank Martina Grandl, Karin Mink, Raphaela P. Semper, Andreas Wanisch, and Nicolai Buse for their invaluable technical assistance, and Claudia Crowell for proofreading the manuscript.
CRediT Authorship Contributions
Victoria Neumeyer (Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Writing: Lead; Formal analysis: Lead)
Anna Brutau-Abia (Investigation: Supporting)
Michael Allgäuer (Investigation: Supporting)
Nicole Pfarr (Formal analysis: Supporting; Methodology: Supporting)
Wilko Weichert (Resources: Supporting)
Christina Falkeis-Veits (Formal analysis: Supporting)
Elisabeth Kremmer (Methodology: Supporting)
Michael Vieth (Formal analysis: Supporting)
Markus Gerhard (Supervision: Supporting)
Raquel Mejías Luque (Conceptualization: Lead; Formal analysis: Lead; Supervision: Lead; Writing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by Deutsche Forschungsgemeinschaft grant GE2042 12-1.
References
- 1.Lauren P. Histogenesis of intestinal and diffuse types of gastric carcinoma. Scand J Gastroenterol Suppl. 1991;180:160–164. [PubMed] [Google Scholar]
- 2.Correa P. Human gastric carcinogenesis: a multistep and multifactorial process--First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992;52:6735–6740. [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orditura M., Galizia G., Sforza V., Gambardella V., Fabozzi A., Laterza M.M., Andreozzi F., Ventriglia J., Savastano B., Mabilia A., Lieto E., Ciardiello F., De Vita F. Treatment of gastric cancer. World J Gastroenterol. 2014;20:1635–1649. doi: 10.3748/wjg.v20.i7.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aichler M., Luber B., Lordick F., Walch A. Proteomic and metabolic prediction of response to therapy in gastric cancer. World J Gastroenterol. 2014;20:13648–13657. doi: 10.3748/wjg.v20.i38.13648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim S.T., Cristescu R., Bass A.J., Kim K.M., Odegaard J.I., Kim K., Liu X.Q., Sher X., Jung H., Lee M., Lee S., Park S.H., Park J.O., Park Y.S., Lim H.Y., Lee H., Choi M., Talasaz A., Kang P.S., Cheng J., Loboda A., Lee J., Kang W.K. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med. 2018;24:1449–1458. doi: 10.1038/s41591-018-0101-z. [DOI] [PubMed] [Google Scholar]
- 7.Lianos G.D., Glantzounis G.K., Bali C.D., Katsios C., Roukos D.H. Identification of novel genes by whole-exome sequencing can improve gastric cancer precision oncology. Future Oncol. 2017;13:883–892. doi: 10.2217/fon-2016-0430. [DOI] [PubMed] [Google Scholar]
- 8.Wang K., Yuen S.T., Xu J., Lee S.P., Yan H.H., Shi S.T., Siu H.C., Deng S., Chu K.M., Law S., Chan K.H., Chan A.S., Tsui W.Y., Ho S.L., Chan A.K., Man J.L., Foglizzo V., Ng M.K., Chan A.S., Ching Y.P., Cheng G.H., Xie T., Fernandez J., Li V.S., Clevers H., Rejto P.A., Mao M., Leung S.Y. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–582. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 9.Maruvka Y.E., Mouw K.W., Karlic R., Parasuraman P., Kamburov A., Polak P., Haradhvala N.J., Hess J.M., Rheinbay E., Brody Y., Koren A., Braunstein L.Z., D'Andrea A., Lawrence M.S., Bass A., Bernards A., Michor F., Getz G. Analysis of somatic microsatellite indels identifies driver events in human tumors. Nat Biotechnol. 2017;35:951–959. doi: 10.1038/nbt.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho J., Ahn S., Son D.S., Kim N.K., Lee K.W., Kim S., Lee J., Park S.H., Park J.O., Kang W.K., An J.Y., Choi M.G., Lee J.H., Sohn T.S., Bae J.M., Kim S., Kim K.M. Bridging genomics and phenomics of gastric carcinoma. Int J Cancer. 2019;145:2407–2417. doi: 10.1002/ijc.32228. [DOI] [PubMed] [Google Scholar]
- 11.Cho J., Chang Y.H., Heo Y.J., Kim S., Kim N.K., Park J.O., Kang W.K., Lee J., Kim K.M. Four distinct immune microenvironment subtypes in gastric adenocarcinoma with special reference to microsatellite instability. ESMO Open. 2018;3 doi: 10.1136/esmoopen-2018-000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung S.H., Kim S.Y., An C.H., Lee S.H., Jung E.S., Park H.C., Kim M.S., Chung Y.J., Lee S.H. Clonal structures of regionally synchronous gastric adenomas and carcinomas. Clin Cancer Res. 2018;24:4715–4725. doi: 10.1158/1078-0432.CCR-18-0345. [DOI] [PubMed] [Google Scholar]
- 13.Neumeyer V., Grandl M., Dietl A., Brutau-Abia A., Allgauer M., Kalali B., Zhang Y., Pan K.F., Steiger K., Vieth M., Anton M., Mejias-Luque R., Gerhard M. Loss of endogenous RNF43 function enhances proliferation and tumour growth of intestinal and gastric cells. Carcinogenesis. 2019;40:551–559. doi: 10.1093/carcin/bgy152. [DOI] [PubMed] [Google Scholar]
- 14.Koo B.K., Spit M., Jordens I., Low T.Y., Stange D.E., van de Wetering M., van Es J.H., Mohammed S., Heck A.J., Maurice M.M., Clevers H. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488:665–669. doi: 10.1038/nature11308. [DOI] [PubMed] [Google Scholar]
- 15.Loregger A., Grandl M., Mejias-Luque R., Allgauer M., Degenhart K., Haselmann V., Oikonomou C., Hatzis P., Janssen K.P., Nitsche U., Gradl D., van den Broek O., Destree O., Ulm K., Neumaier M., Kalali B., Jung A., Varela I., Schmid R.M., Rad R., Busch D.H., Gerhard M. The E3 ligase RNF43 inhibits Wnt signaling downstream of mutated beta-catenin by sequestering TCF4 to the nuclear membrane. Sci Signal. 2015;8:ra90. doi: 10.1126/scisignal.aac6757. [DOI] [PubMed] [Google Scholar]
- 16.Bohgaki M., Bohgaki T., El Ghamrasni S., Srikumar T., Maire G., Panier S., Fradet-Turcotte A., Stewart G.S., Raught B., Hakem A., Hakem R. RNF168 ubiquitylates 53BP1 and controls its response to DNA double-strand breaks. Proc Natl Acad Sci U S A. 2013;110:20982–20987. doi: 10.1073/pnas.1320302111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D.H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J., Lukas J., Lukas C. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 18.Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 19.Pan M.R., Peng G., Hung W.C., Lin S.Y. Monoubiquitination of H2AX protein regulates DNA damage response signaling. J Biol Chem. 2011;286:28599–28607. doi: 10.1074/jbc.M111.256297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith J., Tho L.M., Xu N., Gillespie D.A. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res. 2010;108:73–112. doi: 10.1016/B978-0-12-380888-2.00003-0. [DOI] [PubMed] [Google Scholar]
- 21.Menoyo A., Alazzouzi H., Espin E., Armengol M., Yamamoto H., Schwartz S., Jr. Somatic mutations in the DNA damage-response genes ATR and CHK1 in sporadic stomach tumors with microsatellite instability. Cancer Res. 2001;61:7727–7730. [PubMed] [Google Scholar]
- 22.Kim J.W., Im S.A., Kim M.A., Cho H.J., Lee D.W., Lee K.H., Kim T.Y., Han S.W., Oh D.Y., Lee H.J., Kim T.Y., Yang H.K., Kim W.H., Bang Y.J. Ataxia-telangiectasia-mutated protein expression with microsatellite instability in gastric cancer as prognostic marker. Int J Cancer. 2014;134:72–80. doi: 10.1002/ijc.28245. [DOI] [PubMed] [Google Scholar]
- 23.Touati E., Michel V., Thiberge J.M., Wuscher N., Huerre M., Labigne A. Chronic Helicobacter pylori infections induce gastric mutations in mice. Gastroenterology. 2003;124:1408–1419. doi: 10.1016/s0016-5085(03)00266-x. [DOI] [PubMed] [Google Scholar]
- 24.Butcher L.D., den Hartog G., Ernst P.B., Crowe S.E. Oxidative stress resulting from Helicobacter pylori infection contributes to gastric carcinogenesis. Cell Mol Gastroenterol Hepatol. 2017;3:316–322. doi: 10.1016/j.jcmgh.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toller I.M., Neelsen K.J., Steger M., Hartung M.L., Hottiger M.O., Stucki M., Kalali B., Gerhard M., Sartori A.A., Lopes M., Muller A. Carcinogenic bacterial pathogen Helicobacter pylori triggers DNA double-strand breaks and a DNA damage response in its host cells. Proc Natl Acad Sci U S A. 2011;108:14944–14949. doi: 10.1073/pnas.1100959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartung M.L., Gruber D.C., Koch K.N., Gruter L., Rehrauer H., Tegtmeyer N., Backert S., Muller A.H. pylori-induced DNA strand breaks are introduced by nucleotide excision repair endonucleases and promote NF-kappaB target gene expression. Cell Rep. 2015;13:70–79. doi: 10.1016/j.celrep.2015.08.074. [DOI] [PubMed] [Google Scholar]
- 27.Santos J.C., Gambeloni R.Z., Roque A.T., Oeck S., Ribeiro M.L. Epigenetic mechanisms of ATM activation after Helicobacter pylori infection. Am J Pathol. 2018;188:329–335. doi: 10.1016/j.ajpath.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Smyth E.C., Verheij M., Allum W., Cunningham D., Cervantes A., Arnold D., ESMO Guidelines Committee Gastric cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2016;27(Suppl 5):v38–v49. doi: 10.1093/annonc/mdw350. [DOI] [PubMed] [Google Scholar]
- 29.Wen D., Wang G., Huang Z., Cui X., Song J., Zhu Z., Cui L. Reduced frequency and prognostic significance of ring finger protein 43 nucleotide polymorphisms in a Chinese colorectal cancer cohort. DNA Cell Biol. 2019;38:541–548. doi: 10.1089/dna.2019.4645. [DOI] [PubMed] [Google Scholar]
- 30.Min B.H., Hwang J., Kim N.K., Park G., Kang S.Y., Ahn S., Ahn S., Ha S.Y., Lee Y.K., Kushima R., Van Vrancken M., Kim M.J., Park C., Park H.Y., Chae J., Jang S.S., Kim S.J., Kim Y.H., Kim J.I., Kim K.M. Dysregulated Wnt signalling and recurrent mutations of the tumour suppressor RNF43 in early gastric carcinogenesis. J Pathol. 2016;240:304–314. doi: 10.1002/path.4777. [DOI] [PubMed] [Google Scholar]
- 31.Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R.S., Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 32.Wang B., Elledge S.J. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci U S A. 2007;104:20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mattiroli F., Vissers J.H., van Dijk W.J., Ikpa P., Citterio E., Vermeulen W., Marteijn J.A., Sixma T.K. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell. 2012;150:1182–1195. doi: 10.1016/j.cell.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 34.Wu C.Y., Kang H.Y., Yang W.L., Wu J., Jeong Y.S., Wang J., Chan C.H., Lee S.W., Zhang X., Lamothe B., Campos A.D., Darnay B.G., Lin H.K. Critical role of monoubiquitination of histone H2AX protein in histone H2AX phosphorylation and DNA damage response. J Biol Chem. 2011;286:30806–30815. doi: 10.1074/jbc.M111.257469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim I.H. Current status of adjuvant chemotherapy for gastric cancer. World J Gastrointest Oncol. 2019;11:679–685. doi: 10.4251/wjgo.v11.i9.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao Y., Cai A., Xi H., Li J., Xu W., Zhang Y., Zhang K., Cui J., Wu X., Wei B., Chen L. Ring finger protein 43 associates with gastric cancer progression and attenuates the stemness of gastric cancer stem-like cells via the Wnt-beta/catenin signaling pathway. Stem Cell Res Ther. 2017;8:98. doi: 10.1186/s13287-017-0548-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamoto H., Imai K. Microsatellite instability: an update. Arch Toxicol. 2015;89:899–921. doi: 10.1007/s00204-015-1474-0. [DOI] [PubMed] [Google Scholar]
- 38.Jackson S.P., Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tu J., Park S., Yu W., Zhang S., Wu L., Carmon K., Liu Q.J. The most common RNF43 mutant G659Vfs∗41 is fully functional in inhibiting Wnt signaling and unlikely to play a role in tumorigenesis. Sci Rep. 2019;9:18557. doi: 10.1038/s41598-019-54931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gala M.K., Mizukami Y., Le L.P., Moriichi K., Austin T., Yamamoto M., Lauwers G.Y., Bardeesy N., Chung D.C. Germline mutations in oncogene-induced senescence pathways are associated with multiple sessile serrated adenomas. Gastroenterology. 2014;146:520–529. doi: 10.1053/j.gastro.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arnold I.C., Lee J.Y., Amieva M.R., Roers A., Flavell R.A., Sparwasser T., Muller A. Tolerance rather than immunity protects from Helicobacter pylori-induced gastric preneoplasia. Gastroenterology. 2011;140:199–209. doi: 10.1053/j.gastro.2010.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartfeld S., Clevers H. Organoids as model for infectious diseases: culture of human and murine stomach organoids and microinjection of Helicobacter pylori. J Vis Exp. 2015;105:53359. doi: 10.3791/53359. [DOI] [PMC free article] [PubMed] [Google Scholar]