Abstract
Background & Aims
The functions of the liver and the intestine are closely tied in both physiological and pathologic conditions. The gut microbiota (GM) often cause deleterious effects during hepatic pathogenesis. Autophagy is essential for liver homeostasis, but the impact of hepatic autophagy function on liver-gut interaction remains unknown. Here we investigated the effect of hepatic autophagy deficiency (Atg5Δhep) on GM and in turn the effect of GM on the liver pathology.
Methods
Fecal microbiota were analyzed by 16S sequencing. Antibiotics were used to modulate GM. Cholestyramine was used to reduce the enterohepatic bile acid (BA) level. The functional role of fibroblast growth factor 15 (FGF15) and ileal farnesoid X receptor (FXR) was examined in mice overexpressing FGF15 gene or in mice given a fibroblast growth factor receptor-4 (FGFR4) inhibitor.
Results
Atg5Δhep causes liver injury and alterations of intestinal BA composition, with a lower proportion of tauro-conjugated BAs and a higher proportion of unconjugated BAs. The composition of GM is significantly changed with an increase in BA-metabolizing bacteria, leading to an increased expression of ileal FGF15 driven by FXR that has a higher affinity to unconjugated BAs. Notably, antibiotics or cholestyramine treatment decreased FGF15 expression and exacerbated liver injury. Consistently, inhibition of FGF15 signaling in the liver enhances liver injury.
Conclusions
Deficiency of autophagy function in the liver can affect intestinal environment, leading to gut dysbiosis. Surprisingly, such changes provide an adaptive protection against the liver injury through the FGF15-FGFR4 signaling. Antibiotics use in the condition of liver injury may thus have unexpected adverse consequences via the gut-liver axis.
Keywords: Autophagy, Gut Dysbiosis, Liver Injury, FGF15
Abbreviations used in this paper: ABX, antibiotics; ALP, alkaline phosphatase; ALT, alanine transaminase; ASBT (SLC10A2), apical sodium–bile acid transporter; AST, aspartate transaminase; ATG, autophagy-related gene; BA, bile acid; BAS, bile acid sequestrants; BLU, Blu-9931; BSH, bile salt hydrolase; CA, cholic acid; CDCA, chenodeoxycholic acid; CK19, cytokeratin 19; CYP7A1, cytochrome p450 7a1; CYP8B1, cytochrome p450 family 8b1; DCA, deoxycholic acid; ERK, extracellular signal-regulated kinase; FEX, fexaramine; FGF15, fibroblast growth factor 15; FGFR4, fibroblast growth factor receptor 4; FXR, farnesoid X receptor; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GM, gut microbiota; IBABP, ileal bile acid-binding protein; MCA, muricholic acid; NAFLD, nonalcoholic fatty liver disease; NRF2, nuclear factor erythroid 2-related factor 2; OST-α, organic solute transporter subunit-α; OST-β, organic solute transporter subunit-β; PBS, phosphate-buffered saline; PCoA, principal coordinates analysis; qRT-PCR, quantitative real-time polymerase chain reaction; SE, standard error; SHP, small heterodimer partner; SQSTM1, sequestosome-1; TBA, total bile acid; TCA, taurocholic acid; TCDCA, taurochenodeoxycholic acid; TDCA, taurodeoxycholic acid; TLCA, taurolithocholic acid; TMCA, tauromuricholic acid; TUDCA, tauroursodeoxycholic acid
Graphical abstract
Summary.
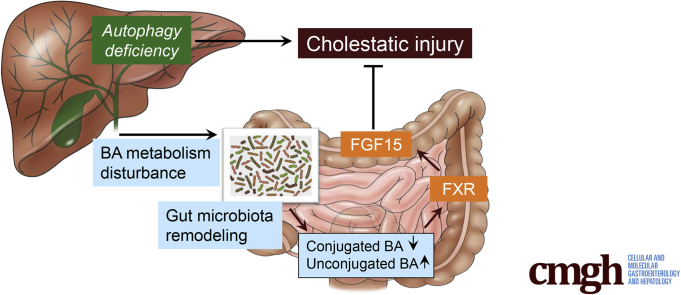
Autophagy deficiency in the liver can cause alterations of intestinal bile acid (BA) composition and gut microbiota with a significantly higher proportion of BA-metabolizing bacteria. Gut dysbiosis provides an adaptive protection to the liver via the FGF15-FGFR4 signaling pathway.
Gut microbiota (GM) consist of a diverse community of symbiotic bacteria and have a complex interplay with the host.1 Interactions between GM and the host result in production of metabolites by microbes, including secondary bile acids (BAs).1 Alteration of GM has been associated with multiple diseases, including fatty liver disease. Gut dysbiosis is found in patients with nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis.2 In patients with alcohol-related liver disease, the microbiota composition and function are varied in association with the severity of liver condition and whether the patients are active drinkers.3 On the other hand, gut dysbiosis may contribute to hepatic pathogenesis by translocation of microbial-associated molecular patterns and bacteria that can cross the gut barrier.4 However, evidence of beneficial effects through a defined mechanism is scarce.
BAs are produced in hepatocytes through 2 major biosynthetic pathways, the classic pathway, which converts cholesterol to 7α-hydroxycholesterol by cytochrome P450 7A1 (CYP7A1), and the alternative pathway, which converts cholesterol to 27-hydroxycholesterol by cytochrome P450 27A1.5,6 Eventually the classic pathway leads to the synthesis of cholic acid (CA) and chenodeoxycholic acid (CDCA), whereas the alternative pathway only leads to the synthesis of CDCA in mice.5,6 In the human liver, CDCA is an end product; however, CDCA is further converted to muricholic acids (MCAs) by cytochrome P450 2C70 in the mouse liver.5,6 Primary BAs are conjugated after synthesis and secreted to the intestine, where they are converted by GM into secondary BAs and reabsorbed by the liver through the portal circulation.5,6 Dysfunction of BA metabolism and gut dysbiosis can be associated with each other, often in the context of liver diseases.4,6 Gut dysbiosis has been found in patients with primary biliary cholangitis or primary sclerosing cholangitis and in a mouse model of cholestasis.4,7 It is not known whether gut dysbiosis would in turn affect liver pathogenesis in the context of dysregulated BA metabolism.
Macroautophagy, hereafter simply referred to as autophagy, is an evolutionarily conserved degradation process and is critical for hepatic homeostasis.8 Deficiency of key autophagy-related genes (Atg) in the liver, eg, Atg5 and Atg7, causes severe liver injury, fibrosis, and tumorigenesis.9,10 Our previous study demonstrates that autophagy deficiency–induced liver injury is accompanied with altered hepatic BA metabolism, cholestatic injury, and impaired farnesoid X receptor (FXR) activity in the liver.11 It is not known whether hepatic autophagy function can be important for the gut-liver interaction and exert its impact to the GM.
Our study shows that hepatic autophagy deficiency leads to an increased proportion of BA-metabolizing bacteria and the alteration of BA composition in the intestine. The significance of these changes is the enhanced activation of ileal FXR and an increased expression of fibroblast growth factor 15 (FGF15). Unexpectedly, we found an exacerbated liver injury and ductular reaction in autophagy-deficient livers after GM removal or blockage of the FGF15–fibroblast growth factor receptor 4 (FGFR4) signaling pathway. Our findings thus indicate that gut dysbiosis can be beneficial in protecting the liver from further injury, and the mechanism is medicated by a gut-liver signaling axis. Furthermore, the findings suggest that antibiotics (ABX) use in the condition of liver injury should be cautious, considering the potential adverse effect on the liver via the gut-liver signaling.
Results
Liver-Specific Deletion of Atg5 Altered the Composition of GM
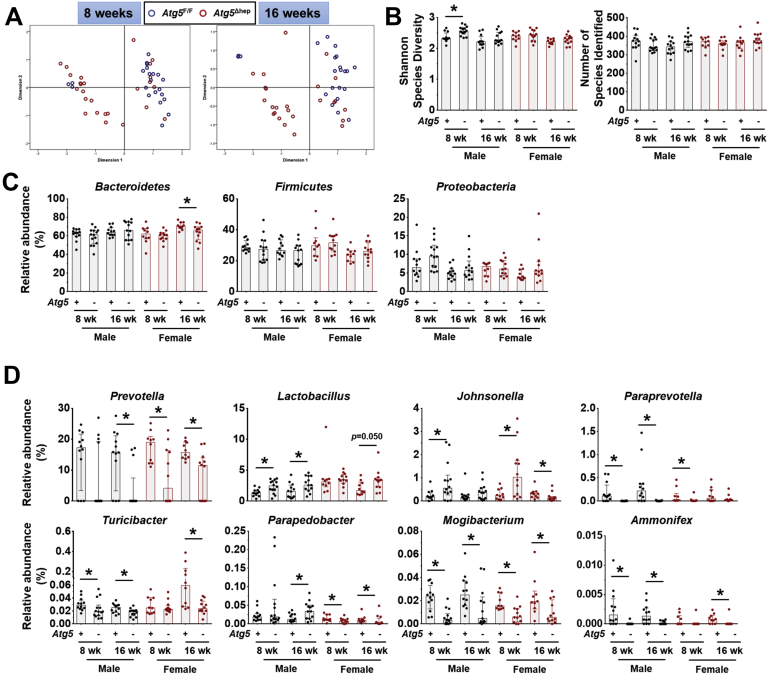
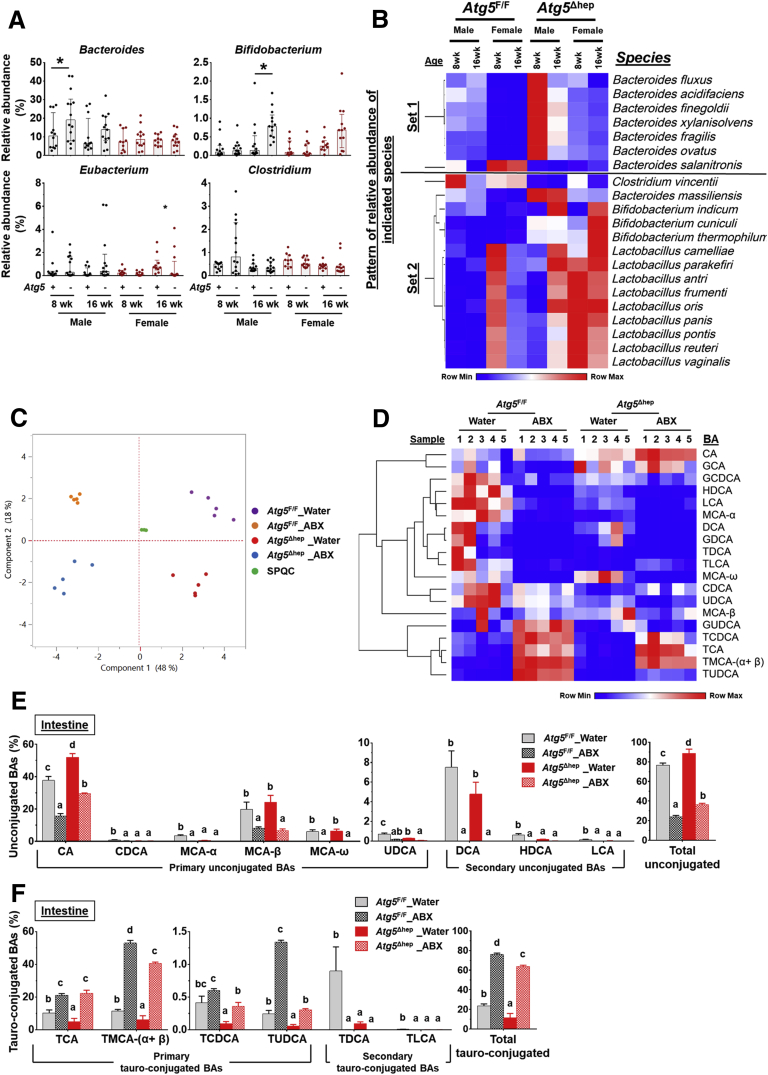
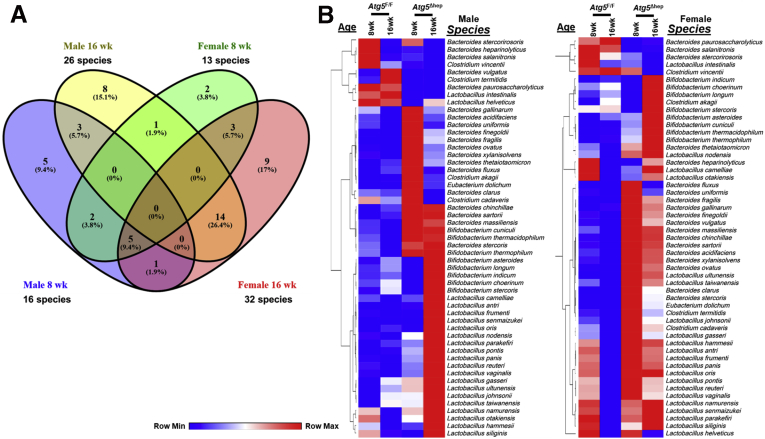
Hepatic autophagy deficiency due to the deletion of a key autophagy gene, Atg5 or Atg7, causes significant liver injury, which is dependent on the nuclear factor erythroid 2-related factor 2 (NRF2) activation.9,12,13 To investigate the impact of hepatic autophagy deficiency on GM, we first assessed the composition of GM in fecal samples from Atg5F/F and Atg5Δhep mice by 16S sequencing. Principal coordinates analysis (PCoA) showed that the composition of GM was noticeably separated between Atg5F/F and Atg5Δhep mice at the age of 8 or 16 weeks (Figure 1A). However, the diversity and the number of species were comparable between Atg5F/F and Atg5Δhep mice (Figure 1B). The most abundant bacteria at the phylum level were Bacteroidetes, Firmicutes, and Proteobacteria, and their proportions were in general comparable between Atg5F/F and Atg5Δhep mice (Figure 1C). These results suggest that hepatic Atg5 deletion alters the proportion of bacterial species rather than the diversity of GM. Analysis at the genus level showed significant disproportions of 8 bacteria between Atg5F/F and Atg5Δhep mice (Figure 1D). A higher proportion of Lactobacillus but a lower proportion of Prevotella, Paraprevotella, Turicibacter, Mogibacterium, and Ammonifex were observed in Atg5Δhep mice. The proportion of Johnsonella was also increased in most of the Atg5Δhep groups except the 16-week-old female group. In addition, Parapedobacter seemed enriched only in the male Atg5Δhep mice but impoverished in the females.
Figure 1.
Liver-specific deletion of Atg5 altered the composition of GM. (A) PCoA based on relative abundance at species level shows a distinguishable profile of GM between 8- and 16-week-old Atg5F/F and Atg5Δhep mice. Data from mice of both sexes were plotted. (B) Shannon species diversity and number of species identified were similar among different groups of mice. (C) Proportion of 3 most abundant bacteria at phylum level. (D) Proportion of bacteria that showed significant changes in at least 3 comparisons between the age- and sex- matched Atg5F/F and Atg5Δhep mice at the genus level. Data are shown as median with interquartile range, n = 10/group. Mann-Whitney analysis was used to determine significance, ∗P < .05.
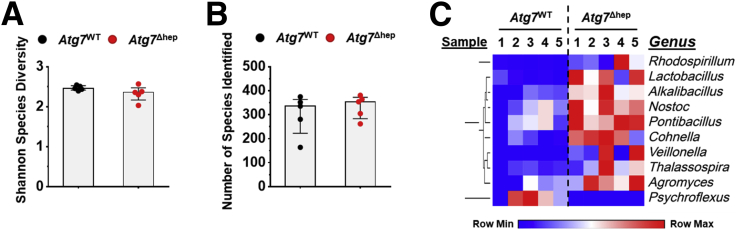
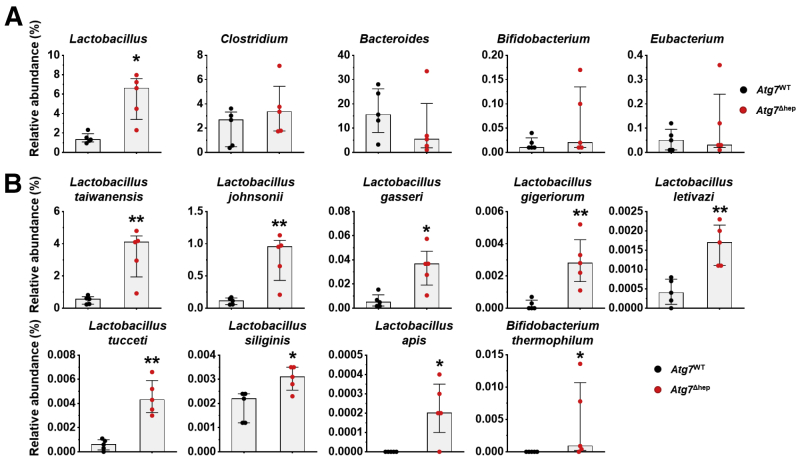
To understand whether the disproportion of GM was correlated with the autophagy deficiency, we analyzed GM in Atg7Δhep mice, another hepatic autophagy-deficiency mouse model. As observed in the Atg5Δhep mice, the diversity and the number of identified species were comparable between Atg7Δhep mice and their controls (Figure 2A and B). However, proportions of 9 genera were changed in Atg7Δhep mice (Figure 2C), including Lactobacillus. These results indicate that hepatic autophagy deficiency can cause gut dysbiosis.
Figure 2.
Liver-specific deletion of Atg7 caused gut dysbiosis. Shannon species diversity (A) and the number of species identified (B) were analyzed in each group of mice. (C) Bacteria with disproportionate representation in Atg7Δhep mice at genus level. Heatmap was generated, and values in the heatmap were mapped to colors using the minimum and maximum of each row independently. The hierarchical cluster of different genus was constructed using one minus Pearson correlation method. Male and female mice were 6–26 weeks old when fecal samples were collected (n = 5). Data are shown as median with interquartile range. Mann-Whitney analysis did not show statistical significance between the groups.
ABX Treatment Aggravated Atg5 Deficiency-Induced Liver Injury
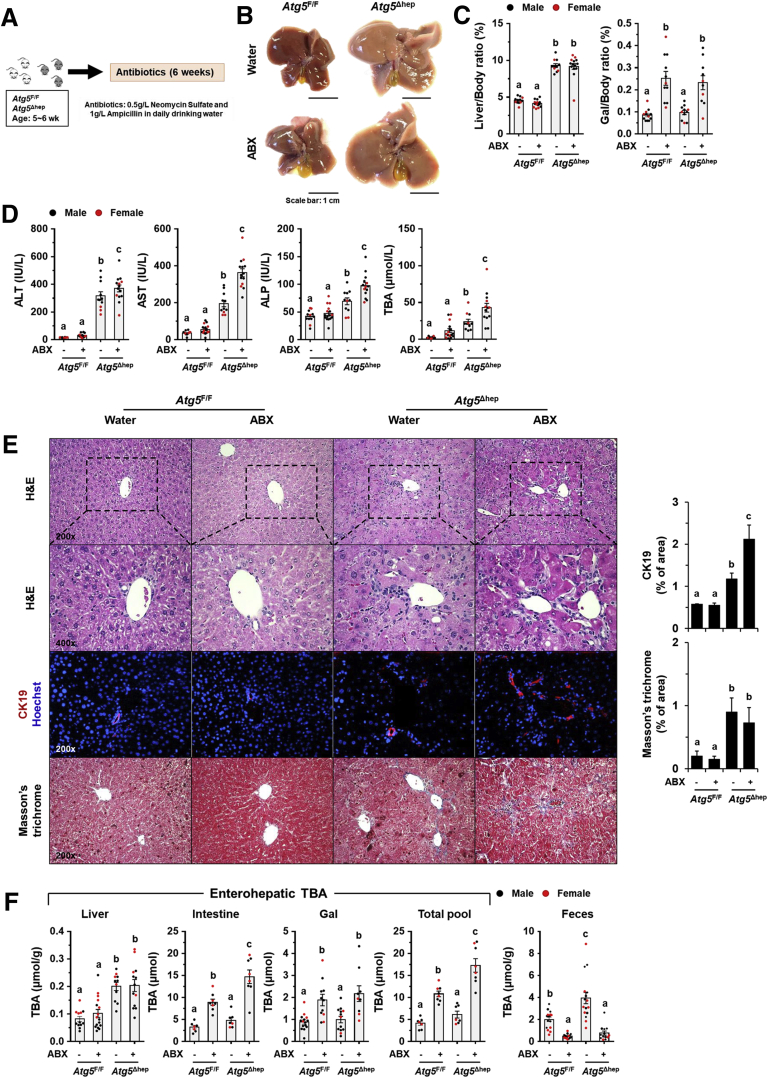
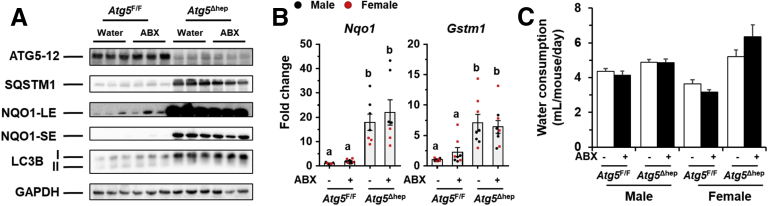
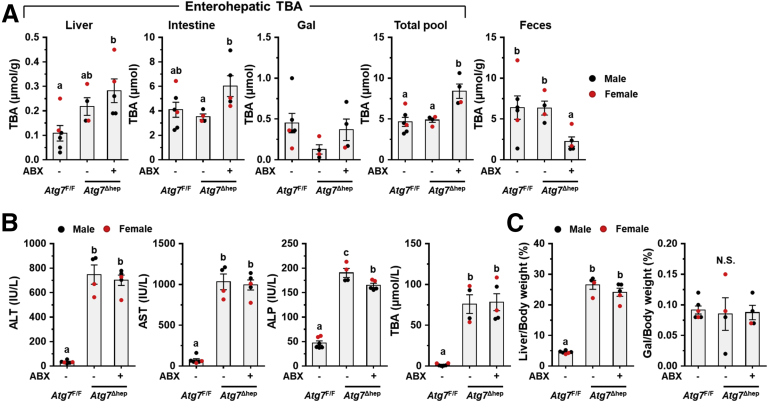
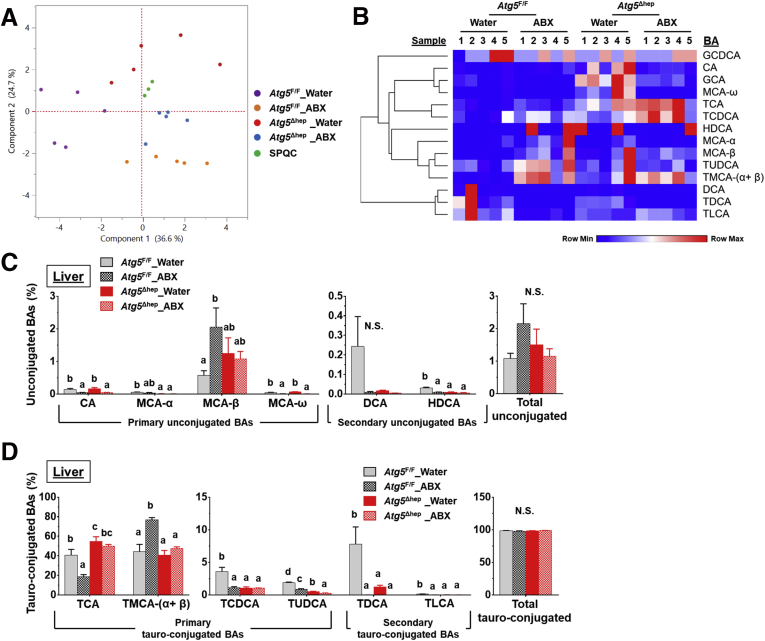
To investigate the potential impact of GM dysbiosis on the pathogenesis of hepatic autophagy deficiency, mice were given ABX for 6 weeks (Figure 3A). ABX treatment did not affect either hepatic NRF2 activity (Figure 4A and B), which could affect liver injury,12 or water consumption (Figure 4C). The hepatomegaly in Atg5Δhep mice remained unchanged after ABX treatment (Figure 3B and C). Nevertheless, gallbladders were enlarged after ABX treatment (Figure 3B and C), which is consistent with previous findings in germ-free mice.14 Unexpectedly, serum levels of alanine transaminase (ALT), aspartate transaminase (AST), alkaline phosphatase (ALP), and total BA (TBA) were significantly increased in Atg5Δhep mice but not significantly changed in Atg5F/F mice after ABX treatment (Figure 3D). Consistently, ABX treatment enhanced ductular reaction but not fibrotic reaction as shown by H&E, cytokeratin 19 (CK19), and Masson’s trichrome staining in Atg5-deficient livers (Figure 3E). Autophagy deficiency causes liver injury accompanied with cholestasis.11 Consistently, Atg5Δhep mice presented increased TBA mainly in the liver and the feces (Figure 3F). ABX treatment caused further elevations in TBA levels mainly in the intestine and the gallbladder, and thus the TBA pool elevated in Atg5F/F mice but more so in Atg5Δhep mice (Figure 3F). Similar to germ-free mice, accumulation of TBA in the intestine was accompanied with decreased fecal levels of TBA after ABX treatment (Figure 3F). Overall, ABX treatment enhanced cholestatic liver injury of Atg5Δhep mice.
Figure 3.
ABX treatment aggravated Atg5 deficiency-induced liver injury. (A) Scheme of the ABX treatment. Mice were given neomycin sulfate and ampicillin sodium salt mixture in daily drinking water for 6 weeks. (B) Representative gross anatomy of livers of indicated genotypes and treatments. (C) Liver weight and gallbladder (Gal) weight were determined as percentages of the body weight (n = 10–16/group). (D) Serum levels of ALT, AST, ALP, and TBA in mice (n = 10–16/group). (E) Liver sections were subjected to H&E, anti-CK19, or Masson’s trichrome staining. Percentage of positive area was quantified with ImageJ (anti-CK19 staining quantification, n = 3–4/group; Masson’s trichrome staining quantification, n = 8–12/group). (F) TBA levels in indicated compartments were measured (n = 7–16/group). Data are shown as means ± SE. Groups with different lowercase letters had significant differences (P < .05).
Figure 4.
ABX treatment did not affect NRF2 activity in Atg5 deficiency livers. (A) Immunoblotting analysis of hepatic samples. (B) The mRNA level of Nqo1 and Gstm1 was determined by qRT-PCR (n = 7–9). (C) Average daily water consumption per mouse was determined. Data are shown as means ± SE. Groups with different lowercase letters had significant differences (P < .05). Gstm1, glutathione s-transferase mu 1; LC3B, microtubule associated protein 1 light chain 3 beta; Nqo1, NAD(P)H quinone dehydrogenase 1.
In Atg7Δhep mice, ABX treatment increased TBA in the intestine and the total pool while decreased fecal excretion of TBA (Figure 5A), the changes of which are similar to Atg5Δhep mice. However, hepatic enzyme levels in the blood were not further elevated in ABX-treated Atg7Δhep mice (Figure 5B). This could be due to the well-documented more severe liver injury seen in Atg7Δhep mice (Figure 5B and C vs Figure 3C and D). The more severe phenotype of Atg7-deficient livers may mask the effects of ABX treatment.
Figure 5.
ABX treatment suppressed ileal FXR activity and altered enterohepatic TBA levels in Atg7Δhepmice. (A) TBA levels in the indicated compartments were measured (n = 4–6). (B) Serum levels of ALT, AST, ALP, and TBA in Atg7Δhep mice after ABX treatment (n = 4–6). (C) Liver weight and gallbladder (Gal) weight were shown as percentage of body weight (n = 4–6). Data are shown as means ± SE. Groups with different lowercase letters had significant differences (P < .05).
BA-Metabolizing Bacteria Were Enriched in Atg5Δhep Mice
Because GM are critical for BA metabolism in the intestine,15,16 we examined whether the disproportion of GM affected BA metabolism in Atg5Δhep mice. The major BA-metabolizing bacteria are those that express bile salt hydrolase (BSH) and/or 7α/β-dehydroxylation activity, which include Lactobacillus, Bacteroides, Bifidobacterium, Eubacterium, and Clostridium.1 Of note, unlike BSH activity, only a small number of bacteria belonging to the class Clostridia have 7α/β-dehydroxylation activity.17 A higher proportion of Lactobacillus (Figure 1E) but not Clostridium or Eubacterium (Figure 6A) was consistently found in Atg5Δhep mice. The elevation of Bacteroides was only observed in male Atg5Δhep mice, and that of Bifidobacterium was seen only in 16-week-old Atg5Δhep mice (Figure 6A). Thus, there were different levels of increments in BA-metabolizing bacteria in Atg5Δhep mice. In addition, we also observed an enrichment of Lactobacillus at genus level in Atg7Δhep mice (Figure 7A), suggesting that hepatic autophagy deficiency altered proportions of BA-metabolizing bacteria.
Figure 6.
Hepatic autophagy deficiency affects intestinal BA composition in correlation with gut dysbiosis. (A) Hepatic autophagy deficiency affected the proportion of bacteria with the BSH and/or 7α/β-dehydroxylation activities at the genus level (Lactobacillus is shown in Figure 1E). Data are shown as median with interquartile range, n = 10/group. (B) Heatmap shows the BA-metabolizing bacteria that are disproportionate in Atg5Δhep mice at the species level. Heatmap was generated, and values in the heatmap were mapped to colors using the minimum and maximum of each row independently. The hierarchical cluster of different species was constructed using one minus Pearson correlation method. Proportion of bacteria in Set 1 was significantly changed in both male and female Atg5Δhep mice at 8 weeks old. Proportion of bacteria in Set 2 was significantly changed in both male and female Atg5Δhep mice at 16 weeks old. (C) PCoA of BAs in the intestine data (log2-scaled μmol/L). (D) Heatmap was generated, and values in the heatmap were mapped to colors using the minimum and maximum of each row independently. Heatmap shows the cluster of indicated BA species in the intestine of different groups of mice. The hierarchical cluster of different BAs was constructed using one minus Pearson correlation method. (E and F) Intestinal levels of unconjugated (E) and tauro-conjugated (F) BAs in male mice. Data are shown as percentage of TBA level (means ± SE), n = 5/group. Groups with different lowercase letters or indicated by asterisk had significant differences (P < .05). HDCA, hyodeoxycholic acid; LCA, lithocholic acid; UDCA, ursodeoxycholic acid.
Figure 7.
Alterations in the proportion of BA-metabolizing bacteria in Atg7Δhepmice. (A) Proportion of bacteria with BSH and/or 7α/β-dehydroxylation activity at the genus level in floxed Atg7 or in Atg7Δhep mice. Mice were 6–26 weeks old. Both male and female mice were included. (B) The bacterial species with BSH and/or 7α/β-dehydroxylation activity that were disproportionate in Atg7Δhep mice. Data are shown as median with interquartile range, n = 5. Mann-Whitney analysis, ∗P < .05, ∗∗P < .01.
Further heterogeneity at the species levels within each of these 5 genera could be observed in terms of the enrichment in Atg5Δhep mice. Because the variations could be related to the sex and/or the age, only a few species were overlapped between the cross-age/sex comparisons (Figure 8A). Two heatmaps based on the gender were then generated to include all disproportionate species, which presented the overall disproportion of the BA-metabolizing bacteria in Atg5Δhep mice (Figure 8B). As expected, most of these species with higher proportions were in both male and female Atg5Δhep mice, albeit with variations. We then focused on species that were disproportionately altered in both age-matched male and female Atg5Δhep mice. We found that among the 7 species that were altered in 8-week-old Atg5Δhep mice, 6 of them were enriched (Set 1 in Figure 6B). Similar enrichment of these species was seen in 16-week-old Atg5Δhep mice, although it was not statistically significant (Figure 6B). On the other hand, in the 16-week-old Atg5Δhep mice, 14 other species were disproportionate, and 13 of them were enriched (Set 2 in Figure 6B). We observed similar changes in Atg7Δhep mice. At species level, proportion of 9 species, including Bifidobacterium thermophilum and another 8 from genus Lactobacillus, was enriched in Atg7Δhep mice (Figure 7B). Overall, our results indicate that BA-metabolizing bacteria are enriched in hepatic autophagy-deficient mice at both genus and species levels, despite that there are age- and sex-related variations.
Figure 8.
Hepatic autophagy deficiency caused changes in the proportion of BA-metabolizing bacteria. (A) Venn diagram shows the number of bacterial species with BSH and/or 7α/β-dehydroxylation activity, which were disproportionate in Atg5Δhep mice compared with the sex- and age- matched Atg5F/F mice. (B) Disproportionate bacteria at the species level were segregated on the basis of gender of the mouse. Heatmap was generated, and values in the heatmap were mapped to colors using the minimum and maximum of each row independently. The hierarchical cluster of different species was constructed using one minus Pearson correlation method.
Liver-Specific Deletion of Atg5 Altered the Composition of Intestinal BAs
Because BA-metabolizing bacteria were enriched in Atg5Δhep mice, we interrogated the composition of BAs in the intestine and the liver. We found that the composition of intestinal BAs was distinctively separated among different groups (Figure 6C and D). In the intestine, the total level of unconjugated BAs was elevated, with a significant increase in CA in Atg5Δhep mice (Figure 6E). Correspondingly, most of the tauro-conjugated BAs, either primary or secondary, were significantly reduced in Atg5Δhep mice (Figure 6F). The composition of intestinal BAs suggested a stronger capacity of deconjugation in Atg5Δhep mice. These observations are consistent with the higher proportion of BA-metabolizing bacteria in Atg5Δhep mice, which seemed to be more capable of deconjugation but not necessarily dehydroxylation. After ABX treatment, we observed that the proportion of unconjugated BAs was dramatically increased whereas the proportion of conjugated BAs was decreased in both Atg5F/F and Atg5Δhep mice (Figure 6E and F), the pattern of which is similar to the finding in germ-free mice.14 Interestingly, Atg5 deletion in the liver did not change the effects of ABX on intestinal composition of BAs, which nondiscriminately eliminated BA-metabolizing bacteria for both deconjugation and dehydroxylation capability.
We then examined what potential impact of Atg5 deficiency and ABX treatment on hepatic BA compositions as comparisons. BA compositions in the liver were distinctly separable among the groups (Figure 9A and B). As expected, the major BA species in the liver were the primary conjugated BAs (Figure 9C and D). Whereas total levels of both tauro-conjugated and unconjugated BAs did not seem to be affected significantly, individual BA composition showed changes that were associated with Atg5 deletion and/or ABX treatment. We observed a significant increase of taurocholic acid (TCA), but not tauromuricholic acid (TMCA), in Atg5-deficient livers (Figure 9D). Hepatic levels of taurochenodeoxycholic acid (TCDCA), tauroursodeoxycholic acid (TUDCA), taurodeoxycholic acid (TDCA), and taurolithocholic acid (TLCA) were all decreased by Atg5 deletion (Figure 9D). After ABX treatment, hepatic levels of TCA and TCDCA were significantly decreased in Atg5F/F but not in Atg5Δhep mice (Figure 9D). Hepatic levels of TMCA-(α+β) and MCA-β were significantly increased in Atg5F/F mice, which is similar to the case of germ-free mice,14 whereas the change was not observed in Atg5Δhep mice (Figure 9C and D). These results suggest that hepatic composition of BAs is altered but is less sensitive to ABX treatment in Atg5Δhep mice.11
Figure 9.
BA composition in the liver after ABX treatment. (A) PCoA of BAs in the liver (log2-scaled μmol/L). (B) Heatmap was generated, and values in the heatmap were mapped to colors using the minimum and maximum of each row independently. The hierarchical cluster of different BAs was constructed using one minus Pearson correlation method. (C and D) Hepatic levels of unconjugated BAs (C) and hepatic levels of tauro-conjugated bile acids (D) in male mice (n = 5). Data are shown as means ± SE. Groups with different lowercase letters had significant differences (P < .05). HDCA, hyodeoxycholic acid; LCA, lithocholic acid; UDCA, ursodeoxycholic acid.
Ileal FXR Activation and FGF15 Expression Were Up-regulated in Atg5Δhep Mice in a Manner Dependent on GM
Intestinal level of TMCA was significantly reduced in Atg5Δhep mice, which meanwhile was significantly affected by GM as indicated by the robust elevation after ABX treatment (Figure 6F). Indeed, TMCA-β in the intestine is known to particularly inhibit ileal FXR, which is activated by TCA.16 Accumulation of TMCA-β was found in germ-free mice and in conventional mice given ABX treatment, which dramatically inhibited ileal FXR, which thereby reduced expression of FGF15.14 FGF15 is secreted into portal circulation to function as a hormone.18 In the liver, FGF15 can inhibit BA synthesis when binding to its receptor FGFR4, which is critical for BA homeostasis.19, 20, 21, 22, 23
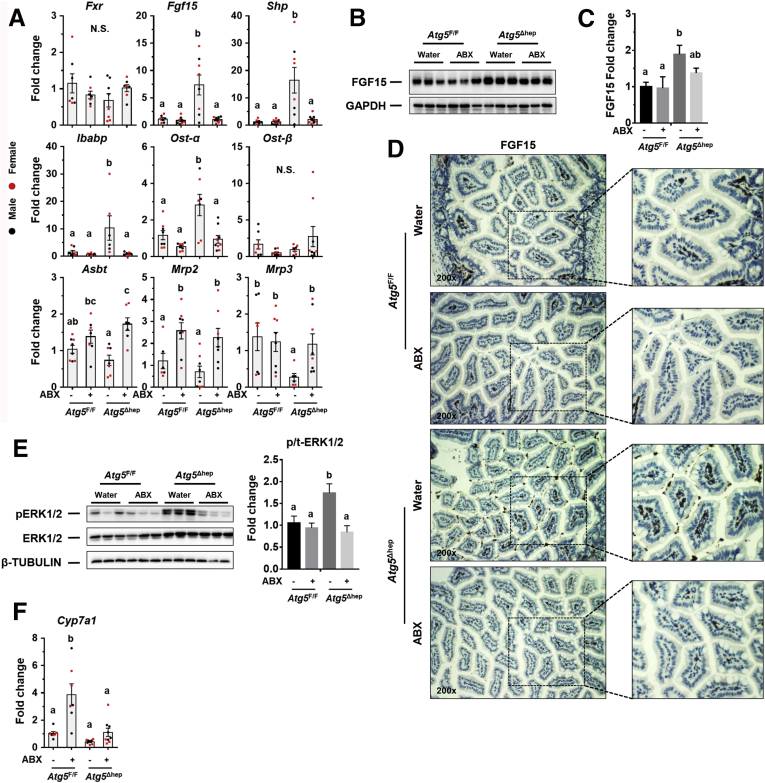
We therefore hypothesized that the lower level of intestinal TMCA in Atg5Δhep mice might allow a higher level of ileal FXR activation and a higher level of FGF15 expression. As expected, we found a significant increase of ileal expression of Fgf15 and small heterodimer partner (Shp) in Atg5Δhep mice, which is compromised by ABX treatment (Figure 10A), implying the involvement of TMCA. Fxr expression itself was not affected. In addition to Fgf15 and Shp, expression of intestinal BA transporters, which is regulated by FXR,24 is also affected by hepatic autophagy deficiency. In Atg5Δhep mice, ileal expression of both ileal bile acid-binding protein (Ibabp) and organic solute transporter subunit-α (Ost)-α, but not Ost-β, was up-regulated, but compromised by ABX treatment (Figure 10A). Ileal apical sodium–bile acid transporter (Asbt) expression was noticeably decreased in Atg5Δhep mice (Figure 10A). After ABX treatment, expression of Mrp2 and Asbt was induced in mice of both genotypes (Figure 10A). The pattern of gene expression indicates that the activation of ileal FXR in Atg5Δhep mice can be compromised by ABX treatment, suggesting a potential association of these changes with GM.
Figure 10.
GM regulates ileal FXR activation and FGF15 expression in Atg5Δhepmice. (A) mRNA level of indicated genes in the ileum was analyzed by qRT-PCR (n = 7–9/group). (B and C) Ileal FGF15 protein levels were examined by immunoblotting assay (B) and quantified with densitometry (C, n = 3/group). (D) Representative images of anti-FGF15 immunohistochemistry staining in the ileum of indicated genotypes and treatments. (E) ERK1/2 level in the liver was analyzed by immunoblotting assay and quantified by densitometry. Phosphorylation level of ERK1/2 was normalized by the total protein level and expressed as the fold change of the Atg5F/F control group (n = 5–7/group). (F) Expression of Cyp7a1 was analyzed by qRT-PCR in the liver (n = 6–9/group). Data are shown as means ± SE. Groups with different lowercase letters had significant differences (P < .05). Mrp2 (Abcc2), multidrug resistance-associated protein 2; Mrp3 (Abcc3), multidrug resistance-associated protein 3; N.S., no statistical significance.
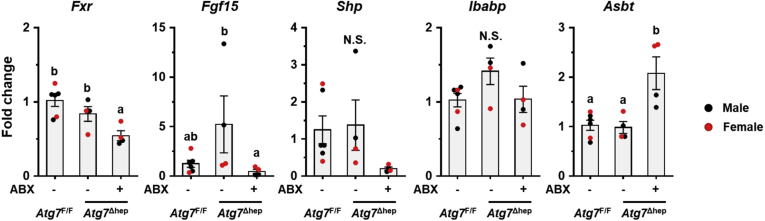
The elevation of ileal FGF15 expression was further confirmed by immunoblotting (Figure 10B and C) and immunochemistry (Figure 10D), both of which were increased in Atg5Δhep mice in a manner dependent on GM. Interestingly, ileal expression of Fgf15 and Ibabp was induced in Atg7Δhep mice, which was also compromised after ABX treatment (Figure 11), suggesting that the modulation of ileal FXR activity by hepatic autophagy deficiency was not dependent on specific autophagy-related genes.
Figure 11.
ABX treatment suppressed ileal FXR activity and reduced Fgf15 expression in Atg7Δhepmice. Ileal expression of indicated genes was analyzed by qRT-PCR (n = 4–6). Data are shown as means ± SE. Groups with different lowercase letters had significant differences (P < .05).
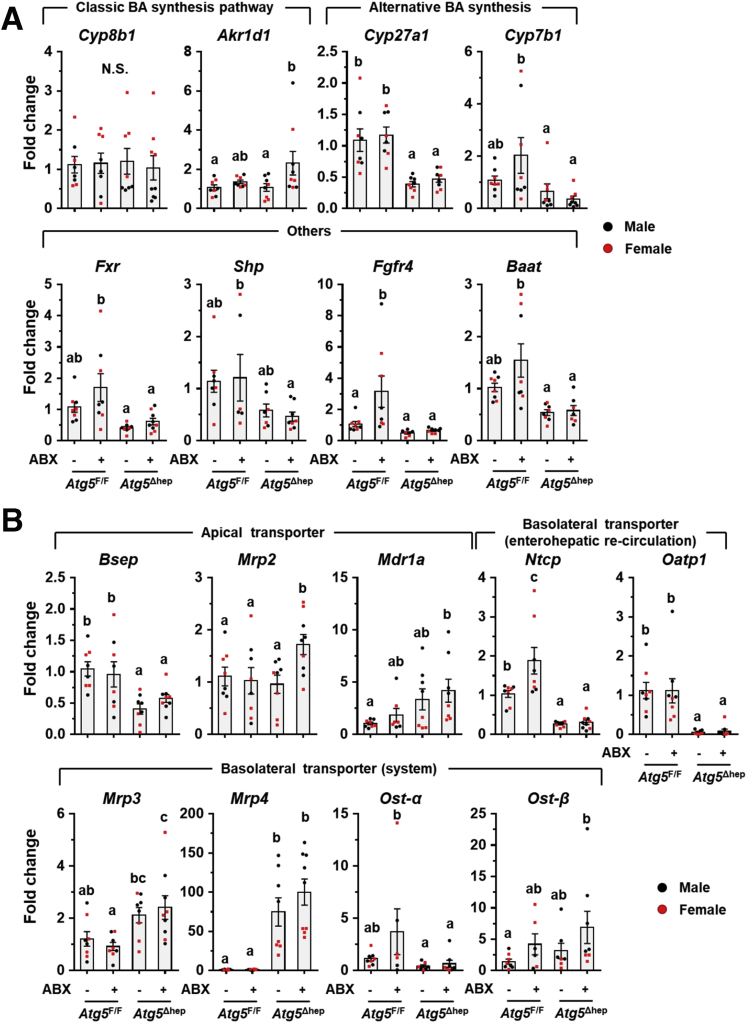
To examine the consequent impacts of FGF15 on Atg5-deficient livers, we analyzed FGF15-related pathways. In livers, FGF15 can bind to FGFR4 and activate the extracellular signal-regulated kinase (ERK) pathway.19 As expected, phosphorylation level of ERK1/2 in Atg5-deficient livers was significantly induced but compromised after ABX treatment (Figure 10E). Cyp7a1 gene that encodes the rate-limiting enzyme in the classic BA synthesis pathway is negatively regulated by FGF15 signaling.19 Indeed, whereas Cyp7a1 mRNA level is low in Atg5Δhep livers, its expression was noticeably elevated after ABX treatment (Figure 10F), which is similar to the case in the germ-free mice14 and consistent with the alteration in the FGF15 signaling. Expression of Fxr and most of its target genes was suppressed in Atg5-deficient liver, as shown in our previous study,11 but not affected by ABX treatment (Figure 12A and B).
Figure 12.
The mRNA level of hepatic genes related to BA metabolism after ABX treatment. (A) Expression of BA metabolism-related genes was analyzed by qRT-PCR in the liver (n = 6–9). (B) Hepatic expression of BA transporters was analyzed by qRT-PCR (n = 6–9). Data are shown as means ± SE. Groups with different lowercase letters had significant differences. Akr1d1, aldo-keto reductase family 1 member d1; Baat, bile acid-coa:amino acid n-acyltransferase; Bsep (Abcb11), bile salt export pump; Cyp7b1, cytochrome P450 7b1; Cyp27a1, cytochrome P450 27a1; Mdr1a (Abcb1), multidrug resistance protein 1; Mrp2 (Abcc2), multidrug resistance-associated protein 2; Mrp3 (Abcc3), multidrug resistance-associated protein 3; Mrp4 (Abcc4), multidrug resistance-associated protein 4; Ntcp (Slc10a1), Na/Taurocholate cotransporting polypeptide; Oatp1 (Slco1a1), organic anion-transporting polypeptide 1.
Taken together, ABX treatment affects the intestinal composition of BA and eliminates the relative advantage of Atg5Δhep mice in producing more ileal FGF15 because of a lower level of TMCA in the intestine.
Bile Acid Sequestrants Reduced TBA Pool but Also Ileal FGF15 Production, Leading to an Enhanced Atg5 Deficiency-Induced Liver Injury
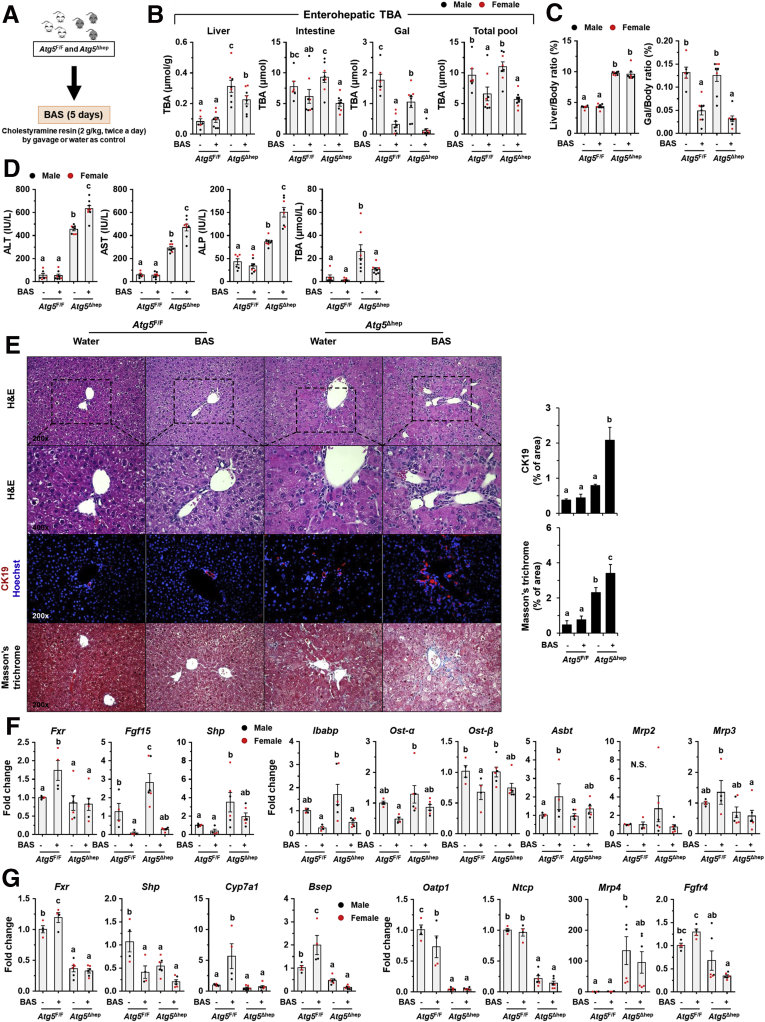
Bile acid sequestrants (BAS) are large polymers that bind negatively charged BAs in the small intestine, which can prevent reabsorption of BAs in the gut, increase their fecal excretion, and finally disrupt enterohepatic circulation.25 BAS are efficient to reduce TBA pool; however, evidence also has shown that they can consequently inhibit ileal FXR and reduce expression of FGF15 in mice26 and FGF19 in humans.19 To determine the impact of enterohepatic TBA on autophagy-deficient livers, mice were given cholestyramine resin treatment (Figure 13A). As expected, a significant decrease of TBA levels in livers and enterohepatic circulation (Figure 13B) and reduced gallbladder size (Figure 13C) were observed in mice after BAS treatment. Although hepatomegaly in Atg5Δhep mice was not further enhanced (Figure 13C), serum levels of ALT, AST, and ALP were all increased in Atg5Δhep mice after BAS treatment (Figure 13D). H&E staining showed an increased number of oval cells around the periportal areas in Atg5-deficient livers after BAS treatment (Figure 13E). The increased positive areas of CK19 and Masson’s trichrome staining indicate a more severe ductular reaction in Atg5-deficient livers after BAS treatment (Figure 13E). These results suggest that BAS treatment in Atg5Δhep mice paradoxically enhances liver injury.
Figure 13.
BAS reduced TBA pool but enhanced Atg5 deficiency-induced liver injury. (A) Scheme of the BAS treatment. Water was given as control. (B) TBA levels in the indicated compartments were measured (n = 6–8/group). (C) Liver weight and gallbladder (Gal) weight were determined as percentage of body weight (n = 6–8/group). (D) Serum levels of ALT, AST, ALP, and TBA in mice after BAS treatment (n = 6–8/group). (E) Liver sections were subjected to H&E, anti-CK19, or Masson’s trichrome staining. Percentage of positive area was quantified with ImageJ (anti-CK19 staining quantification, n = 3–4/group; Masson's trichrome staining quantification, n = 3–5/group). (F and G) Expression of genes related to BA metabolism in ileums (F) or in livers (G) was analyzed by qRT-PCR (n = 4–6/group). Data are shown as means ± SE. Groups with different lowercase letters had significant differences (P < .05). Bsep (Abcb11), bile salt export pump; Cyp7a1, cytochrome P450 7a1; Mrp2 (Abcc2), multidrug resistance-associated protein 2; Mrp3 (Abcc3), multidrug resistance-associated protein 3; Mrp4 (Abcc4), multidrug resistance-associated protein 4; Ntcp (Slc10a1), Na/Taurocholate cotransporting polypeptide; Oatp1 (Slco1a1), organic anion-transporting polypeptide 1.
To identify potential mechanisms in which BAS treatment contributed to enhanced liver injury in Atg5Δhep mice, we examined expression of FXR-regulated genes in both ileums and livers. Despite an increase of Fxr gene expression in Atg5F/F mice after BAS treatment, ileal expression of FXR-promoted genes, including Fgf15, Shp, Ibabp, Ost-α, and Ost-β, was significantly reduced in mice after BAS treatment (Figure 13F). The ileal level of Asbt mRNA, an FXR-suppressed gene, was increased after BAS treatment (Figure 13F). These results suggested that ileal FXR activity was strongly inhibited by BAS treatment and consequently decreased expression of ileal Fgf15. In the liver, BAS treatment suppressed hepatic FXR activity in Atg5F/F mice but did not significantly change the expression of FXR targets in Atg5Δhep mice (Figure 13G), perhaps because the FXR activity was already reduced in autophagy-deficient livers. Taken together, these results suggest that BAS treatment in Atg5Δhep mice reduces ileal FXR activation and Fgf15 expression, which may contribute to the paradoxical effects on autophagy deficiency–induced liver injury.
Intestine-Specific FXR Agonist Activated Ileal FXR in Atg5F/F Mice but not in Atg5Δhep Mice
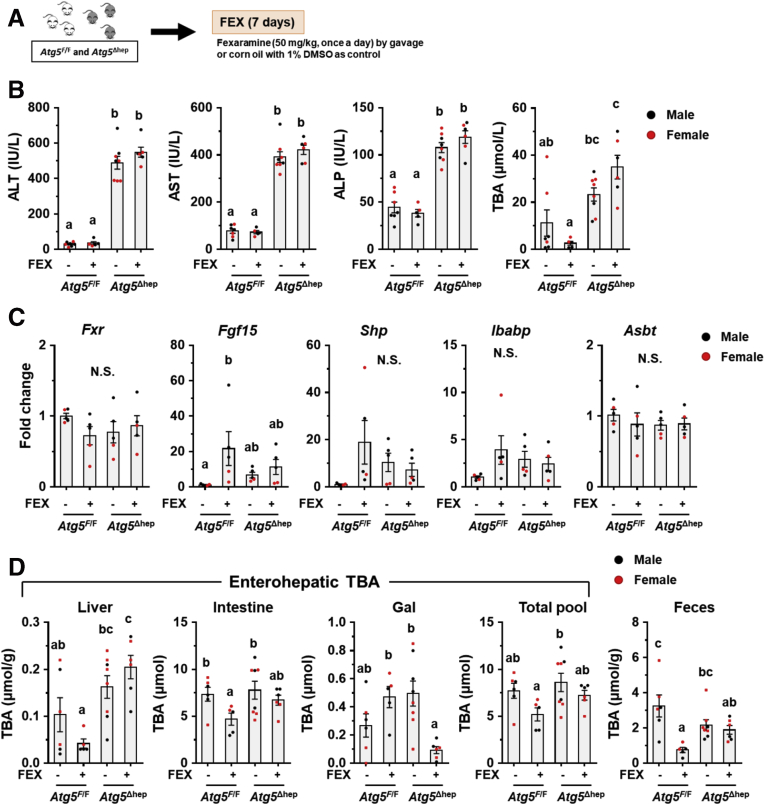
Because both ABX and BAS treatments exacerbate liver injury in Atg5Δhep mice accompanied with reduced ileal FXR activity, we asked whether further activated ileal FXR can improve Atg5 deficiency-induced liver injury. Hence, mice were given an intestine-specific FXR agonist, fexaramine (FEX), for 7 days (Figure 14A). Unexpectedly, FEX treatment did not change serum levels of ALT, AST, ALP, and TBA in Atg5Δhep mice, whereas serum level of TBA was decreased in Atg5F/F mice (Figure 14B). Analysis of the expression of ileal Fxr and its target genes suggested an activation of FXR in Atg5F/F mice but not in Atg5Δhep mice after FEX treatment (Figure 14C). The TBA pool and fecal TBA level were both significantly reduced in Atg5F/F mice after FEX treatment, but not so much in Atg5Δhep mice (Figure 14D). We suspect that the lack of the effect of FEX in Atg5Δhep mice may be due to the altered GM and saturated FXR with existing BA agonists. Consistently, a prior study had also shown that GM could influence the efficacy of FEX treatment.27
Figure 14.
Intestine-specific FXR agonist activated ileal FXR in Atg5F/Fbut not in Atg5Δhepmice. (A) Scheme of the study using an intestine-specific FXR agonist, FEX. Solvent (1% dimethyl sulfoxide in corn oil) was given as the control. (B) Serum levels of ALT, AST, ALP, and TBA in mice after FEX treatment (n = 5–8). (C) Ileal expression of indicated genes was analyzed by qRT-PCR (n = 5). (D) TBA levels in the indicated compartments were measured (n = 5–8). Data are shown as means ± SE. Groups with different lowercase letters had significant differences (P < .05).
Overexpression of FGF15 Attenuated Pathologic Features in Atg5Δhep Livers
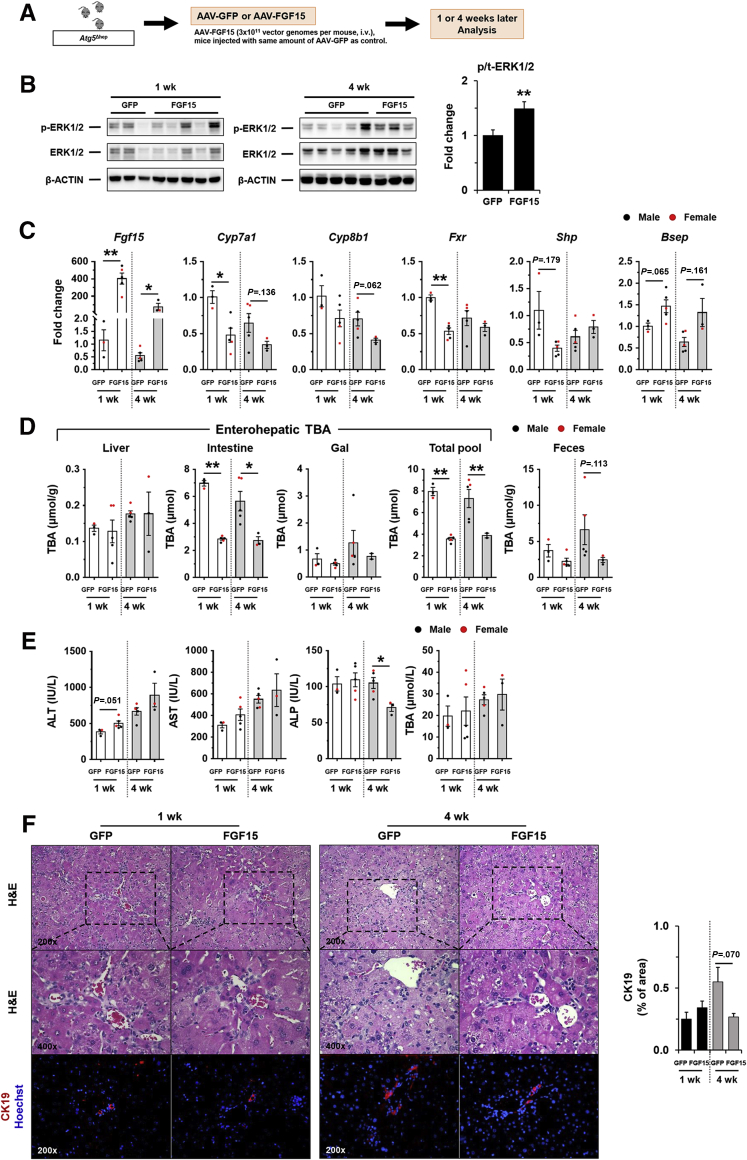
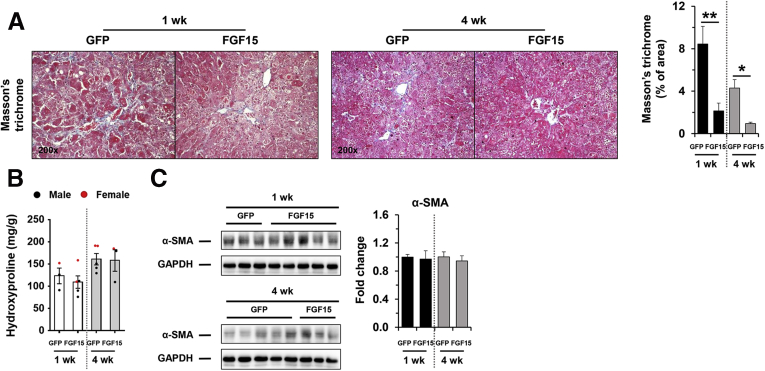
FGF15 was significantly induced in Atg5Δhep mice but was reduced after treatments with ABX or BAS. We thus hypothesized that an elevated level of FGF15 could play a protective role in Atg5 deficiency-induced liver injury. Because recombinant FGF15 does not seem stable in the circulation for long-term experimentation,28,29 AAV8-FGF15 was given to Atg5Δhep mice (Figure 15A). The level of phosphorylated ERK1/2 in the liver was elevated after AAV8-FGF15 injection 1 week and 4 weeks later (Figure 15B), suggesting that FGF15 signaling was enhanced in the liver as supported by a higher hepatic expression of Fgf15 mRNA (Figure 15C). The protein level of FGF15 overexpression could not be defined because of lack of proper antibodies (data not shown). Consequently, hepatic levels of 2 classic targets of FGF15 signaling, Cyp7a1 and cytochrome p450 family 8b1 (Cyp8b1), were reduced after FGF15 overexpression (Figure 15C). Paradoxically, hepatic Fxr and Shp expression was not elevated after FGF15 overexpression, although the expression of bile salt export pump (Bsep), a direct target of FXR, was induced (Figure 15C), suggesting FXR-independent effects of FGF15 on hepatic gene expression. Consistent with the reduced expression of Cyp7a1 and Cyp8b1, the total TBA pool, mainly contributed by the intestinal level of BA, and the fecal TBA level were significantly reduced (Figure 15D).
Figure 15.
Overexpression of FGF15 attenuated pathologic features inAtg5Δheplivers. (A) Scheme of the FGF15 overexpression study in mouse livers. (B) Expression of ERK1/2 in the liver was analyzed by immunoblotting assay and quantified by densitometry. Phosphorylation levels of ERK1/2 were normalized to those of the total protein levels and expressed as fold change of GFP group (n = 8/group). (C) Hepatic expression of indicated genes was analyzed by qRT-PCR (n = 3–5/group). Data were expressed as fold change of GFP for the 1-week group. (D) TBA levels in the indicated compartments were measured (n = 3–5/group). (E) Serum levels of ALT, AST, ALP, and TBA in mice after AAV injection (n = 3–5/group). (F) Liver sections were subjected to H&E, anti-CK19, or Masson’s trichrome staining. Percentage of positive area was quantified with ImageJ (n = 3–5/group). Data are shown as means ± SE. ∗P < .05, ∗∗P < .01. AAV, adeno-associated virus; Bsep (Abcb11), bile salt export pump; Cyp7a1, cytochrome P450 7a1; GFP, green fluorescence protein.
Serum biochemistry analysis suggested FGF15 overexpression in the liver reduced ALP but not ALT, AST, and TBA (Figure 15E) in Atg5Δhep mice, suggesting a potential improvement in biliary injury. Consistently, the H&E staining and CK19 staining showed a reduced ductular reaction (Figure 15F). On the other hand, the parameters of fibrosis (Figure 16) showed a variable improvement to FGF15 overexpression, suggesting that the short treatment had a minor impact on this process.
Figure 16.
Effects of FGF15 on fibrosis in mouse livers. (A) Liver sections were subjected to Masson’s trichrome staining. Percentage of positive area was quantified with ImageJ (n = 3–5). (B) Hepatic levels of hydroxyproline (n = 3–5). (C) Protein level of α-SMA in the liver was analyzed by immunoblotting assay and quantified by densitometry (n = 3–5). Data are shown as means ± SE. ∗P < .05, ∗∗P < .01. α-SMA, α-smooth muscle actin. GFP, green fluorescence protein.
Inhibition of FGFR4 Aggravated Liver Injury in Atg5Δhep Mice
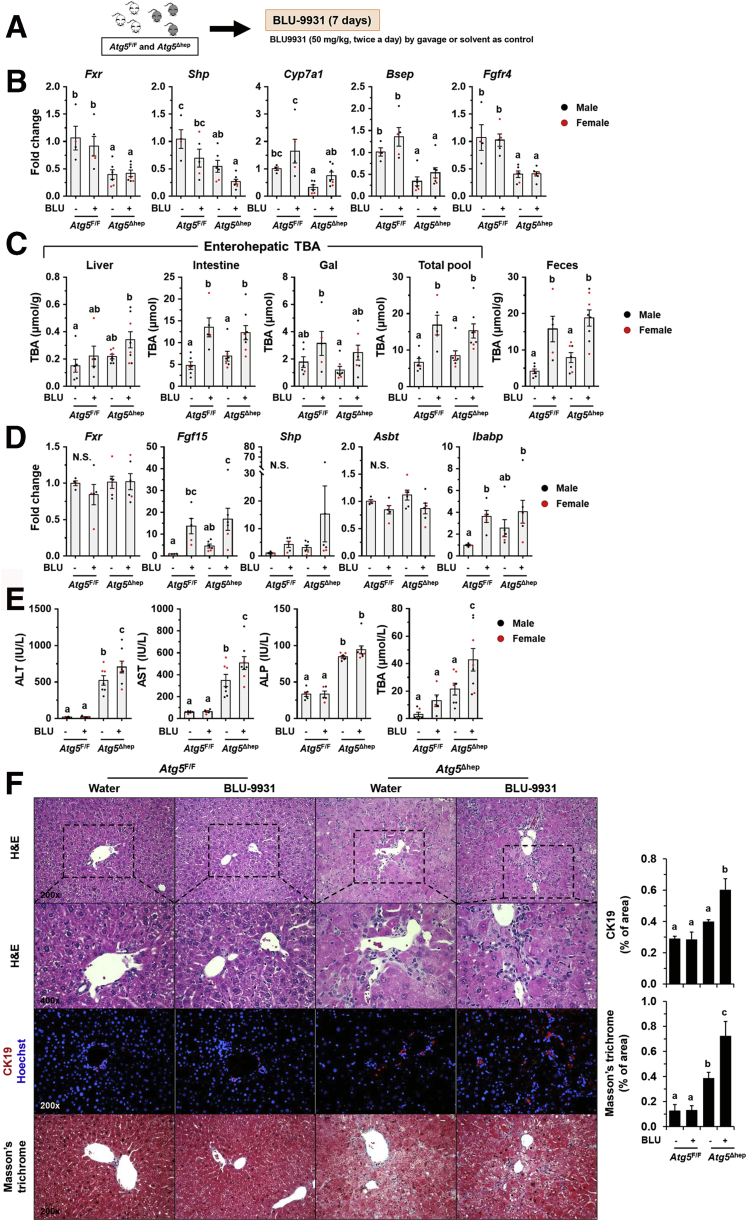
FGFR4 is the receptor that mediates FGF15 signaling in the liver.19 To determine the role of FGF15-FGFR4 signaling in autophagy-deficient livers, we treated mice with Blu-9931 (BLU), a novel small molecular that selectively inhibits FGFR4,30 to block FGFR4 activity (Figure 17A).
Figure 17.
Inhibition of FGFR4 aggravated liver injury in Atg5Δhepmice. (A) Scheme of treatment with FGFR4 inhibitor, Blu-9931 (BLU). Solvent (0.5% methylcellulose/1% Tween 80) was given as control. (B) Hepatic expression of indicated genes was analyzed by qRT-PCR (n = 4–7/group). (C) TBA levels in indicated compartments were measured (n = 6–8/group). (D) Ileal expression of indicated genes was analyzed by qRT-PCR (n = 4–6/group). (E) Serum levels of ALT, AST, ALP, and TBA in mice after BLU treatment (n = 6–8/group). (F) Liver sections were subjected to H&E, anti-CK19, or Masson’s trichrome staining. Percentage of positive area was quantified with ImageJ (CK19 staining quantification, n = 3–6/group; Masson’s trichrome staining quantification, n = 4–5/group). Data are shown as means ± SE. Groups with different lowercase letters had significant differences (P < .05). Bsep (Abcb11), bile salt export pump; N.S., no statistical significance.
In both Atg5F/F and Atg5Δhep mice, BLU treatment reduced Shp expression, whereas it induced Cyp7a1 expression in the liver (Figure 17B). Consequently, the TBA pool and fecal TBA level were significantly increased after BLU treatment (Figure 17C), indicating an increase of BA synthesis in the liver. Consistently, with the inhibition of hepatic FGFR4, ileal expression of Fgf15 and Ibabp was remarkably induced after BLU treatment (Figure 17D), which is possibly attributed to the increase of intestinal TBA.
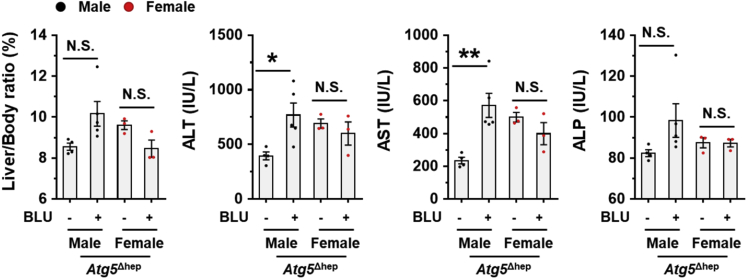
After BLU treatment, liver injury was enhanced in Atg5Δhep mice as indicated by the significantly elevated serum levels of ALT, AST, and TBA (Figure 17E). Notably, the male mice were more susceptible to BLU treatment for the serum enzyme activation (Figure 18). We did not see significant pathologic changes in livers of Atg5F/F mice (Figure 17F), suggesting that inhibition of FGFR4 was not toxic in healthy livers. However, the ductular reaction around the periportal areas was further enhanced in Atg5-deficient livers after BLU treatment as indicated by H&E staining and the positive areas of CK19 staining (Figure 17F). Positive staining of Masson’s trichrome was also significantly increased in Atg5-deficient livers after BLU treatment (Figure 17F). Although we found a noticeable sexual disparity in serum enzyme changes, changes in histologic studies were comparable between male and female Atg5Δhep mice. Taken together, these results suggest that FGF15-FGFR4 signaling protects livers from further injury in Atg5-deficient mice.
Figure 18.
Effects of Blu-9931 on mouse livers. Measurement of liver weight and serum ALT, AST, and ALP levels in Atg5-deficient livers with and without Blu-9931 (BLU) treatment. Results showed more significant effect of BLU treatment in male mice. Data are shown as means ± SE. ∗P <.05, ∗∗P < .01. N.S., no statistical significance.
Discussion
Interaction Between GM and Hepatic Autophagy Deficiency and BA Metabolism
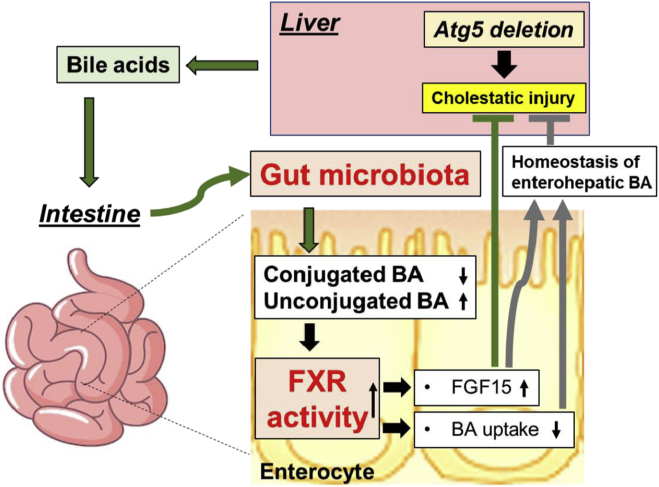
In this study, we showed that autophagy deficiency in the liver led to the alteration of intestinal BA composition and gut dysbiosis with a significantly higher proportion of BA-metabolizing bacteria. Unexpectedly, ABX treatment increased enterohepatic level of BAs and exacerbated the pathology in autophagy-deficient livers. Together with other evidence, we demonstrate that enhanced activation of ileal FXR-FGF15 signaling, because of the effects of altered BA metabolism and GM, accounted for the protection of the autophagy-deficient liver from further injury (Figure 19). Therefore gut dysbiosis in liver diseases can be an adaptive response to mitigate the injury via a gut-liver signaling pathway.
Figure 19.
GM-dependent FXR-FGF15 signaling improves hepatic presentation in Atg5-deficient mice.Atg5 deficiency in the liver impairs autophagy process and BA homeostasis, which causes liver injury accompanied with cholestasis. Disruption of hepatic BA homeostasis altered the composition of BAs in the intestine and GM , which in turn affects BA composition in the intestine. The changes in intestinal BA composition activate ileal FXR and consequently induce FGF15 expression and reduce BA uptake in the intestine, both of which can improve the homeostasis of enterohepatic BA. Importantly, FGF15 is beneficial to the improvement of the liver injury induced by autophagy deficiency.
Autophagy deficiency in the liver causes hepatomegaly, chronic injury, and tumorigenesis.9,10 Mechanically, consistent activation of NRF2 by sequestosome-1 (SQSTM1) is critical for pathologic changes induced by hepatic Atg5 or Atg7 deletion,9,12,13 yet how SQSTM1-NRF2 signaling leads to hepatocyte death remains unclear. Our previous study shows a compromised FXR activity and altered BA homeostasis in autophagy-deficient livers,11 in which activation of FXR in the liver can ameliorate autophagy deficiency–induced liver injury, suggesting that reduced hepatic FXR expression and disrupted BA homeostasis may at least partially contribute to injury in autophagy-deficient livers.
In this study we have defined a unique interaction between GM and liver injury in the context of autophagy deficiency, which affects BA metabolism and gut-liver signaling. First, we show that dysfunction of BA metabolism induced by Atg5 or Atg7 deficiency in the liver alters GM and leads to a significant enrichment of BA-metabolizing bacteria with BSH activity. GM is critical for BA metabolism by deconjugation of BAs and conversion of primary BAs into secondary BAs.6 Consistent with enriched BA-metabolizing bacteria, we observed a lower level of tauro-conjugated BAs but a higher level of unconjugated BAs in the intestine. However, levels of most secondary BAs in the intestine are lower in Atg5Δhep intestines except deoxycholic acid (DCA). In mice, DCA is converted from TCA, whereas other secondary BAs are converted from TCDCA.31 Interestingly, TCDCA level is decreased in Atg5Δhep livers, suggesting a reduced capability of CDCA synthesis in these mice, which may contribute to the decreased levels of non-DCA secondary BAs in the intestine. Overall, the evidence here suggests that autophagy deficiency in the liver alters hepatic BA metabolism, which generates a different BA profile and favors the growth of a specific set of GMs. Thus the altered BA metabolism is the link connecting hepatic autophagy deficiency to gut dysbiosis.
Interestingly there are several studies indicating that GM are altered in patients with cholestatic liver diseases.32, 33, 34 Several bacteria are consistently enriched in patients with primary biliary cholangitis or primary sclerosing cholangitis, including Veillonella, Streptococcus, Enterococcus, Lactobacillus, Haemophilus, and Fusobacterium, whereas the proportion of some other bacteria are decreased in these patients, such as Phascolarctobacterium, Faecalibacterium, and Blautia. These findings indicate that some GM can be altered by cholangitis regardless of etiologies. In comparison, our data showed that Lactobacillus was elevated consistently in multiple groups of mice across the age and the sex at the genus level (Figures 1D, 2C, 7A) and at the species level (Figures 6B, 7B). In addition, Streptococcus and Enterococcus had shown some changes in some groups of Atg5Δhep mice, although not as consistently as Lactobacillus across the age and the sex (data not shown). Overall there is a notable similarity between the human cholestatic injury and the autophagy deficiency–induced cholestatic injury in terms of gut dysbiosis.
Second, we found that altered GM maintain the enterohepatic BA level. We have reported cholestatic injury in autophagy-deficient livers, with a significant increase of TBA levels in serum and livers.11 Here we not only confirmed our previous findings but also found that despite the increase of hepatic TBA level, the total TBA pool is comparable between the autophagy-deficient mice and the control mice (Figures 3F, 5A). This may be due to increased fecal excretion, which is reduced after ABX treatment, suggesting that GM maintained the total enterohepatic level of TBA in autophagy-deficient mice by an increased BA excretion from the intestine. The mechanism of this increased excretion may be related to the elevation of the BA-metabolizing bacteria because most excreted BA species are secondary and/or unconjugated BAs, which are produced through the action of bacteria.
Third, we present evidence that GM-mediated FXR activation in the ileum can induce FGF15 expression, thereby protecting mice from further liver damage caused by autophagy deficiency in the liver. Both the ABX and BAS treatments lead to dramatic inhibition of ileal FXR activity and significant decrease of FGF15 expression. Evidence from overexpression of FGF15 and inhibition of FGFR4 in the Atg5-deficient liver suggests that GM-mediated FGF15 expression at least partially protected Atg5Δhep mice from further liver damage by a FGF15-FGFR4 feedback signaling pathway.
We previously found that Atg7 deletion induced more severe pathologic changes than Atg5 deletion in the liver, which leads to different response to alcohol treatment.35 In this study, we also observed that ABX treatment enhanced liver injury in Atg5Δhep but not obviously in Atg7Δhep mice, although increased BA pool and reduced ileal FXR activity after ABX treatment were observed in both Atg5Δhep and Atg7Δhep mice. It is possible that the protection effect from gut dysbiosis is overcome by additional hepatic phenotypes exerted by the more severe damage in the absence of Atg7, which sits on the upstream of Atg5 in the autophagy signaling pathway.
Gut Dysbiosis Can Be an Adaptive Response to Liver Injury
A number of studies had shown that gut dysbiosis contributes to the progress of liver disease, and correction of the dysbiosis may improve pathologic changes in the liver.36, 37, 38, 39, 40 Among the detrimental effects of GM, “invasion” of the liver by the product of GM because of increased gut permeability is thought to be the major one.4 Although the detrimental role of gut dysbiosis seems to be widely recognized, there is also evidence indicating a beneficial impact of GM on acute liver injury in mice. Enrichment of intestinal Lactobacillus was found in mice with liver injury induced by acute concanavalin A treatment, which can prevent further liver inflammation through activation of interleukin 22 production.41 Furthermore, conflicting evidence supporting either a detrimental42,43 or beneficial14,44 effect of GM can be found for the liver injury in the adenosine triphosphate–binding cassette, subfamily B (MDR/TAP), member 4 (Mdr2) knockout (Mdr2-/-) mice.
In the present study, we have found GM are altered in Atg5Δhep mice. Surprisingly, ABX treatment enhanced Atg5 deficiency-induced liver injury, clearly indicating a protective role of GM in Atg5Δhep mice. We further identified an increase of FXR activity and ileal FGF15 expression in Atg5Δhep mice, which is associated with the altered intestinal BA composition and the dysbiosis status of GM. Our findings demonstrate that FGF15 can be a beneficial feedback signal from gut dysbiosis attributed to hepatic autophagy deficiency.
In mice, FGF15 is induced by FXR activation in the ileum, and its human ortholog is FGF19.18 FGF15/19 is required for the efficiency of SHP-mediated CYP7A1 repression and plays a critical role in repressing BA synthesis.18 Conversely, decrease of intestinal level of BA by BAS can reduce ileal FGF15 expression in mice26 and serum FGF19 levels in healthy humans.45 Animal experiments have shown that FGF15 is essential for hepatic homeostasis, and overexpression of FGF15/19 in the liver has beneficial effects on multiple liver diseases including sclerosing cholangitis,20 alcoholic fatty liver,22 and NAFLD.21,29 In addition to its functions in hepatic metabolism, FGF15 has also been shown to contribute to liver regeneration.23,46,47 In humans, circulating FGF19 has been found to be increased in patients with biliary cirrhosis48 and in patients with alcoholic hepatitis,49 which is accompanied with inhibition of BA synthesis. Nevertheless, the function and cause of increased levels of circulating FGF19 remain unclear. Our current finding of the beneficial effects of FGF15-FGFR4 signaling in autophagy-deficient livers provides evidence that increase of FGF15/19 level in the setting of liver diseases can potentially be a protective mechanism via the gut-liver interaction.
Our study suggests that GM can adapt to metabolic changes in the liver and consequently activate feedback signaling, such as FXR-FGF15 signaling, in the gut to protect the liver from further damage. The present study also suggests caution should be exercised in the use of ABX during specific liver diseases to avoid potential detrimental effects, not because of reduced hepatic drug metabolism but because of disruption of beneficial gut-liver signaling.
Conclusion
In summary, the findings in our studies indicate a primary liver disease can lead to alteration of GM, which then activates FXR-FGF15 feedback signaling through modulating the composition of intestinal BAs. Our results suggest that ABX treatment can exacerbate hepatic pathogenesis in Atg5-deficient livers by the reduction of FGF15 expression. Taken together, our present study demonstrates a protective role of gut dysbiosis in liver injury, which is associated with the FXR-FGF15-FGFR4 feedback signaling.
Methods
Animals and Treatments
Atg5F/F mice (B6.129S-Atg5tm1Myok)10 and Atg7F/F mice12 had been reported in previous studies. Atg5Δhep and Atg7Δhep mice were created by cross Atg5F/F or Atg7F/F with the Alb:Cre transgenic mice (The Jackson Laboratory, Bar Harbor, ME), respectively. Mice were maintained on a 12-hour dark/12-hour light cycle with free access to food and water. Both male and female mice were used in the studies if not further addressed. Age- and sex-matched mice were randomly assigned to treatment or control group. For ABX treatment, mice (5 to 6 weeks old) were given ABX (0.5 g/L neomycin sulfate and 1 g/L ampicillin) in daily drinking water for 6 weeks. For BAS treatment, cholestyramine resin (2 g/kg) was mixed in water and given to 8- to 12-week-old mice twice a day by oral gavage for 5 days. Control groups were given a same volume of water. For FEX treatment, FEX was dissolved in dimethyl sulfoxide and further diluted in corn oil according to manufacturer’s protocol. Mice (8 to 12 weeks old) were given 50 mg/kg FEX daily by oral gavage for 7 days. Control mice were given the same volume of corn oil containing the same amount of dimethyl sulfoxide. For adeno-associated virus (AAV8)-mediated overexpression of FGF15, Atg5Δhep mice (6 to 8 weeks old) were given 3 × 1011 vector genomes of AAV8-FGF15 per mouse by intravenous injection. Control mice were given AAV8-GFP. Mice were euthanized for further analysis 1 or 4 weeks later. For FGFR4 inhibitor treatment, BLU was dissolved in 0.5% methylcellulose/1% Tween 80 solution according to a previous study.30 Mice (8 to 12 weeks old) were given 50 mg/kg BLU twice per day by oral gavage for 7 days. Control mice were given the same volume of solvent. All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of Indiana University.
Antibodies and Chemicals
Antibodies and polymerase chain reaction primers used in this study are listed in Tables 1 and 2, respectively.
Table 1.
Antibody List
| Antibody name | Company | Catalog # | Host |
|---|---|---|---|
| ATG12 | Cell Signaling | 2011 | Rabbit |
| β-ACTIN | Cell Signaling | 3700 | Mouse |
| CK19 | DSHB | TROMA-III | Rat |
| Phospho-ERK1/2 (Thr202/Tyr204) | Cell Signaling | 4370 | Rabbit |
| ERK1/2 | Cell Signaling | 9102 | Rabbit |
| FGF15 | Santa Cruz | sc-514647 | Mouse |
| GAPDH | Novus biologicals | NB 300-221 | Mouse |
| LC3B | Sigma | L7543 | Rabbit |
| NQO1 | Abcam | ab34173 | Rabbit |
| P62/SQSTM1 | Abnova | H00008878-M01 | Mouse |
| α-SMA | Thermo | PA5-19465 | Rabbit |
| β-TUBULIN | Cell Signaling | 86298 | Mouse |
Table 2.
Primer List
| Gene name | equence (forward) | Sequence (reverse) |
|---|---|---|
| Actin | 5′-ACTATTGGCAACGAGCGGTT-3′ | 5′-CAGGATTCCATACCCAAGAAGGA-3′ |
| Akr1d1 | 5′-CTCATTGGGCTTGGAACCTA-3′ | 5′-CATTGATGGGACATGCTCTG-3′ |
| Asbt (Slc10a2) | 5′-CGACATGGACCTCAGTGTTAG-3′ | 5′-CAACCCACATCTTGGTGTAGA-3′ |
| Baat | 5′-GTCCTTTTCCAGGGGTCATT-3′ | 5′-CCAGAGCTAAGGTGGCAAAG -3′ |
| Bsep (Abcb11) | 5′-CTGCCAAGGATGCTAATGCA-3′ | 5′-CGATGGCTACCCTTTGCTTCT-3′ |
| Cyp27a1 | 5′-GCCTCACCTATGGGATCTTCA-3′ | 5′-TCAAAGCCTGACGCAGATG-3′ |
| Cyp7a1 | 5′-AACAACCTGCCAGTACTAGATAGC-3′ | 5′-GTGTAGAGTGAAGTCCTCCTTAGC-3′ |
| Cyp7b1 | 5′-CAGCTATGTTCTGGGCAATG-3′ | 5′-TCGGATGATGCTGGAGTATG-3′ |
| Cyp8b1 | 5′-AGTACACATGGACCCCGACATC-3′ | 5′-GGGTGCCATCCGGGTTGAG-3′ |
| Fgfr4 | 5′-CTGCCAGAGGAAGACCTCAC-3′ | 5′-GTAGTGGCCACGGATGACTT-3′ |
| Fgf15 | 5′-ATGGCGAGAAAGTGGAACG -3′ | 5′-CTGACACAGACTGGGATTGCT-3′ |
| Fxr (Nr1h4) | 5′-GGCCTCTGGGTACCACTACA-3′ | 5′-TGTACACGGCGTTCTTGGTA-3′ |
| Gstm1 | 5′-ACTTGATTGATGGGGCTCAC-3′ | 5′-TCTCCAAAATGTCCACACGA-3′ |
| Ibabp | 5′-CCCCAACTATCACCAGACTTC-3′ | 5′-ACATCCCCGATGGTGGAGAT-3′ |
| Mdr1a (Abcb1) | 5′-AAAGGCTCTACGACCCCCTA-3′ | 5′-CCTGACTCACCACACCAATG-3′ |
| Mrp2 (Abcc2) | 5′-GCACTGTAGGCTCTGGGAAG-3′ | 5′-TGCTGAGGGACGTAGGCTAT-3′ |
| Mrp3 (Abcc3) | 5′-GGACTTCCAGTGCTCAGAGG-3′ | 5′-AGCTGTGGCCTCGTCTAAAA-3′ |
| Mrp4 (Abcc4) | 5′-TGTTTGATGCACACCAGGAT-3′ | 5′-GACAAACATGGCACAGATGG-3′ |
| Nqo1 | 5′-GCACTGATCGTACTGGCTCA-3′ | 5′-CATGGCATAGAGGTCCGACT-3′ |
| Ntcp (Slc10a1) | 5′-CACCATGGAGTTCAGCAAGA-3′ | 5′-CCAGAAGGAAAGCACTGAGG-3′ |
| Ost-α (Slc51A) | 5′-GTCTCAAGTGATGAACTGCCA-3′ | 5′-TTGAGTGCTGAGTCCAGGTC-3′ |
| Ost-β (Slc51B) | 5′-GTATTTTCGTGCAGAAGATGCG-3′ | 5′-TTTCTGTTTGCCAGGATGCTC-3′ |
| Oatp1 (Slco1a1) | 5′-ATCCAGTGTGTGGGGACAAT-3′ | 5′-GCAGCTGCAATTTTGAAACA-3′ |
| Shp | 5′-CTGGTTGAGCGCCTGAGAC-3′ | 5′-CTGCCTGGATGCCCTTTATC -3′ |
Fecal 16S rRNA Sequencing of Gut Microbial Communities
Mice were 6–26 weeks old when fecal samples were collected. For Atg5Δhep mice, fecal samples were collected from Atg5F/F and Atg5Δhep mice at 8 or 16 weeks old. For Atg7Δhep mice, fecal samples were collected from floxed Atg7 mice (heterozygous or homozygous, Atg7WT) and Atg7Δhep mice. Both male and female mice were used and equally distributed in different genotypes. Fecal DNA was extracted from frozen fecal samples by using the E.Z.N.A. Stool DNA Kit (Omega Bio-Tek, Inc, Norcross, GA). All DNA samples were stored at –80°C before sequencing, which was performed by SeqMatic LLC (Fremont, CA) using Illumina (San Diego, CA) sequencing libraries. FASTQ data were processed by using the Qiime pipeline on Illumina’s BaseSpace servers.
For 16S sequencing analysis, relative abundance of each bacteria was calculated. PCoA was performed by using multidimensional scaling function based on relative abundance at species level using SPSS for Windows 17.0 Software (SPSS, Inc, Chicago, IL). Heatmaps were generated by using Morpheus (https://software.broadinstitute.org/morpheus), and values in the heatmap were mapped to colors using the minimum and maximum of each row independently. The hierarchical cluster of each heatmap was constructed using one minus Pearson correlation method.
Serum Biochemistry Analysis
Serum levels of ALT, AST, and ALP were measured by using kits from Pointe Scientific (Canton, MI) according to the manufacturer’s protocol. Serum TBAs were measured using the TBAs assay kit from Diazyme Laboratories, Inc (Poway, CA).
Tissue and Fecal TBA Content Analysis
Sample preparation and BA quantification were performed as in previous studies with modifications.27 For the liver tissue, samples (100 mg) were homogenized in 1 mL of 90% ethanol and incubated at 55°C overnight. The lysates were centrifuged at 10,000 rpm for 10 minutes. Supernatant was used to measure BA concentration. The whole intestine (with its content) was homogenized in 5 mL water and was incubated at 55°C overnight after addition of 45 mL ethanol. One milliliter of the lysates was removed for centrifugation at 10,000 rpm for 10 minutes. An aliquot of the supernatant was diluted 5 times and measured for BA concentration. For BAs in the gallbladder, the entire organ was put in 1 mL of 90% ethanol. After the gallbladder was cut to release the bile, the samples were incubated at 55°C overnight. The lysates were centrifuged at 10,000 rpm for 10 minutes. An aliquot of the supernatant was diluted 50 times to measure the BA concentration. For fecal BA measurement, overnight-dried fecal samples (150–250 mg) were admixed with 1 mL of 95% EtOH and incubated at 55°C overnight. The lysates were centrifuged at 10,000 rpm for 10 minutes. An aliquot of the supernatant was diluted 10 times for BA quantification. The BA concentration of each diluted supernatants was measured by using the TBAs assay kit from Diazyme Laboratories, Inc.
Liver and Intestinal BA Composition Analysis
Liver and intestinal samples from male mice were used for BA profile analysis. BA analysis was by using a Biocrates Bile Acids Kit (Biocrates Life Science AG, Innsbruck, Austria). Samples were prepared according to the manufacturer’s protocol with modifications. At least 30 mg of liver tissue was homogenized in 3-fold volume of extraction buffer (ethanol/phosphate buffer, 85:15 v/v). For intestine samples, they were homogenized in 3 mL phosphate-buffered saline (PBS), and then 17 mL of 100% ethanol was added to extract BAs. All samples were probe-sonicated for 3 bursts with 10 seconds each. Samples were chilled in an ice bath for at least 60 seconds between bursts. Homogenized samples were stored at –80°C. Further BA extraction and profile analysis were performed according to manufacturer’s protocol in Center for Genomic and Computational Biology, Duke University. Sample pool quality controls were created using equal volumes of all liver and intestine samples respectively. BAs were identified by using Biocrates MetIDQ software. A PCoA was performed for BAs using JMP Pro v14.0 software (SAS, Cary, NC).
For BA composition analysis, results of analytes with more than 40% missing values were removed before statistical analysis. Missing values of the remaining results were then replaced with the limit of detection. Results from liver samples were calculated as nmoL/g or percentage of TBA level and were calculated as nmoL/whole intestine or percentage of TBA level from intestine samples. Heatmaps were generated using Morpheus (https://software.broadinstitute.org/morpheus), and values in the heatmap were mapped to colors using the minimum and maximum of each row independently. The hierarchical cluster of each heatmap was constructed using one minus Pearson correlation method.
Immunoblotting Analysis
The liver samples were homogenized in the radio immunoprecipitation assay buffer containing a protease cocktail and phosphatase inhibitors. Supernatant was collected after centrifugation at 12,000 rpm for 12 minutes. Protein concentration was determined by using BCA protein assay kit (Thermo Fisher Scientific-Pierce, Waltham, MA). The proteins were separated on a sodium dodecyl sulfate polyacrylamide gel electrophoresis. Proteins were transferred onto polyvinylidene fluoride membranes, which were then blocked with 5% bovine serum albumin or 5% skim milk for 1 hour at room temperature. Membranes were incubated with the appropriate primary antibody overnight and then washed with TBST (Tris-buffered saline, 0.1% Tween 20) before being incubated with the horseradish peroxidase–coupled secondary antibodies at room temperature. Protein bands were detected by using enhanced chemiluminescence kit (Thermo Fisher Scientific-Pierce). The images were taken digitally with a BioRad ChemiDoc Image System (BioRad, Hercules, CA). Densitometry was measured by using the companion software, and the values were normalized to those of β-tubulin, β-actin, or glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which were then converted to fold change of the control.
RNA Isolation and Quantitative Real-Time PCR Analysis
Total RNA was prepared from liver samples using GeneTET RNA purification kit (Thermo Fisher Scientific, Grand Island, NY) according to manufacturer’s protocols. Complementary DNA (cDNA) was synthesized using Oligo dT primers and an M-MLV reverse transcriptase system (Life Technologies-Thermo Fisher Scientific). Quantitative real-time polymerase chain reaction (qRT-PCR) analysis was performed on a QuantStudio 3 Real-Time PCR System (Life Technologies-Applied Biosystems, Waltham, MA) using SYBR Green master mixes (Life Technologies-Applied Biosystems). All qRT-PCR results were normalized to the level of β-actin, and the gene expression was calculated using the 2-ΔΔCt method.
Histologic Study
Liver or ileum samples were rinsed with PBS and fixed in 10% formalin overnight. Samples were further fixed in 70% ethanol and processed as paraffin-embedded blocks. The paraffin-embedded liver tissues were sectioned and stained with H&E or Mason’s trichrome C. The paraffin-embedded ileum tissues were sectioned, and immunohistochemical staining was performed with FGF15 antibody. Photomicrographs were taken using a Nikon (Tokyo, Japan) Eclipse E200 light microscope equipped with a SPOT RT Slider color digital camera (Diagnostic Instruments, Inc, Sterling Heights, MI). Area with positive Mason’s trichrome C staining was quantified by using ImageJ software (National Institutes of Health, Bethesda, MD); at least 4 random fields of each section from each mouse liver were used for quantification.50
Immunofluorescence Microscopy
Paraffin sections were subjected to antigen retrieval treatment using the Citrate buffer (0.01 mol/L, pH 6.0) after deparaffinization. Slides were blocked with 5% goat serum in PBS containing 0.1% Triton X (PBS-Tx) for 1 hour and then incubated with primary antibodies diluted in 1% bovine serum albumin/PBS-Tx overnight at 4°C. Sections were washed with PBS, followed by incubation with fluorochrome-conjugated secondary antibodies. Hoechst 33342 was used for nucleus staining. Images were obtained by using a Nikon Eclipse TE 200 epi-immunofluorescence microscope and the companion NIS-Elements AR3.2 software. Quantification was performed by using ImageJ software; at least 4 random fields of each section from each mouse liver were analyzed.
Statistical Analysis
The 16S sequencing data were represented as median with interquartile range. All the other data were represented as means with standard errors (SEs). For 16S sequencing data, Mann-Whitney test was performed to identify bacteria with significantly different proportions between Atg5Δhep and sex- and age-matched Atg5F/F mice. For all other data, to determine statistical significance, Student t test was used to determine differences between 2 groups. Differences among more than 2 treatment groups were determined by using one-way analysis of variance followed by Duncan’s post hoc test. Results were considered statistically significant for P value <.05. Statistical analyses were performed by using SPSS for Windows 17.0 Software (SPSS, Inc).
Acknowledgments
CRediT Authorship Contributions
Shengmin Yan (Conceptualization: Supporting; Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Writing – original draft: Lead; Writing – review & editing: Equal),
Bilon Khambu (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting),
Xiaoyun Chen (Investigation: Supporting; Methodology: Supporting)
Zheng Dong (Investigation: Supporting; Methodology: Supporting)
Grace Guo (Investigation: Supporting; Resources: Supporting; Writing – review & editing: Supporting)
Xiao-Ming Yin, MD, PhD (Conceptualization: Lead; Data curation: Support; Formal analysis: Support; Funding acquisition: Lead; Project administration: Lead; Resources: Lead; Supervision: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported in part by the USA National Institutes of Health (NIH) grants DK116605 (to X.-M. Yin).
References
- 1.Nicholson J.K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 2.Schnabl B., Brenner D.A. Interactions between the intestinal microbiome and liver diseases. Gastroenterology. 2014;146:1513–1524. doi: 10.1053/j.gastro.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bajaj J.S. Alcohol, liver disease and the gut microbiota. Nat Rev Gastroenterol Hepatol. 2019;16:235–246. doi: 10.1038/s41575-018-0099-1. [DOI] [PubMed] [Google Scholar]
- 4.Tripathi A., Debelius J., Brenner D.A., Karin M., Loomba R., Schnabl B., Knight R. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol. 2018;15:397–411. doi: 10.1038/s41575-018-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li T., Chiang J.Y. Bile acids as metabolic regulators. Curr Opin Gastroenterol. 2015;31:159–165. doi: 10.1097/MOG.0000000000000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jia W., Xie G., Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol. 2018;15:111–128. doi: 10.1038/nrgastro.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sabino J., Vieira-Silva S., Machiels K., Joossens M., Falony G., Ballet V., Ferrante M., Van Assche G., Van der Merwe S., Vermeire S., Raes J. Primary sclerosing cholangitis is characterised by intestinal dysbiosis independent from IBD. Gut. 2016;65:1681–1689. doi: 10.1136/gutjnl-2015-311004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine B., Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komatsu M., Waguri S., Koike M., Sou Y.S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., Hamazaki J., Nishito Y., Iemura S., Natsume T., Yanagawa T., Uwayama J., Warabi E., Yoshida H., Ishii T., Kobayashi A., Yamamoto M., Yue Z., Uchiyama Y., Kominami E., Tanaka K. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 10.Takamura A., Komatsu M., Hara T., Sakamoto A., Kishi C., Waguri S., Eishi Y., Hino O., Tanaka K., Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khambu B., Li T., Yan S., Yu C., Chen X., Goheen M., Li Y., Lin J., Cummings O.W., Lee Y.A., Friedman S., Dong Z., Feng G.S., Wu S., Yin X.M. Hepatic autophagy deficiency compromises farnesoid X receptor functionality and causes cholestatic injury. Hepatology. 2019;69:2196–2213. doi: 10.1002/hep.30407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y.S., Ueno I., Sakamoto A., Tong K.I., Kim M., Nishito Y., Iemura S., Natsume T., Ueno T., Kominami E., Motohashi H., Tanaka K., Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 13.Ni H.M., Woolbright B.L., Williams J., Copple B., Cui W., Luyendyk J.P., Jaeschke H., Ding W.X. Nrf2 promotes the development of fibrosis and tumorigenesis in mice with defective hepatic autophagy. J Hepatol. 2014;61:617–625. doi: 10.1016/j.jhep.2014.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sayin S.I., Wahlstrom A., Felin J., Jantti S., Marschall H.U., Bamberg K., Angelin B., Hyotylainen T., Oresic M., Backhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Ridlon J.M., Kang D.J., Hylemon P.B. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 16.Wahlstrom A., Sayin S.I., Marschall H.U., Backhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016;24:41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Urdaneta V., Casadesus J. Interactions between bacteria and bile salts in the gastrointestinal and hepatobiliary tracts. Front Med (Lausanne) 2017;4:163. doi: 10.3389/fmed.2017.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kliewer S.A., Mangelsdorf D.J. Bile acids as hormones: the FXR-FGF15/19 pathway. Dig Dis. 2015;33:327–331. doi: 10.1159/000371670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gadaleta R.M., Moschetta A. Metabolic messengers: fibroblast growth factor 15/19. Nature Metabolism. 2019;1:588–594. doi: 10.1038/s42255-019-0074-3. [DOI] [PubMed] [Google Scholar]
- 20.Zhou M., Learned R.M., Rossi S.J., DePaoli A.M., Tian H., Ling L. Engineered fibroblast growth factor 19 reduces liver injury and resolves sclerosing cholangitis in Mdr2-deficient mice. Hepatology. 2016;63:914–929. doi: 10.1002/hep.28257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou M., Luo J., Chen M., Yang H., Learned R.M., DePaoli A.M., Tian H., Ling L. Mouse species-specific control of hepatocarcinogenesis and metabolism by FGF19/FGF15. J Hepatol. 2017;66:1182–1192. doi: 10.1016/j.jhep.2017.01.027. [DOI] [PubMed] [Google Scholar]
- 22.Hartmann P., Hochrath K., Horvath A., Chen P., Seebauer C.T., Llorente C., Wang L., Alnouti Y., Fouts D.E., Starkel P., Loomba R., Coulter S., Liddle C., Yu R.T., Ling L., Rossi S.J., DePaoli A.M., Downes M., Evans R.M., Brenner D.A., Schnabl B. Modulation of the intestinal bile acid/farnesoid X receptor/fibroblast growth factor 15 axis improves alcoholic liver disease in mice. Hepatology. 2018;67:2150–2166. doi: 10.1002/hep.29676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong B., Sun R., Huang M., Chow M.D., Zhong X.B., Xie W., Lee Y.H., Guo G.L. Fibroblast growth factor 15-dependent and bile acid-independent promotion of liver regeneration in mice. Hepatology. 2018;68:1961–1976. doi: 10.1002/hep.30041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dawson P.A. Role of the intestinal bile acid transporters in bile acid and drug disposition. Handb Exp Pharmacol. 2011;201:169–203. doi: 10.1007/978-3-642-14541-4_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Out C., Groen A.K., Brufau G. Bile acid sequestrants: more than simple resins. Curr Opin Lipidol. 2012;23:43–55. doi: 10.1097/MOL.0b013e32834f0ef3. [DOI] [PubMed] [Google Scholar]
- 26.Fuchs C.D., Paumgartner G., Mlitz V., Kunczer V., Halilbasic E., Leditznig N., Wahlstrom A., Stahlman M., Thuringer A., Kashofer K., Stojakovic T., Marschall H.U., Trauner M. Colesevelam attenuates cholestatic liver and bile duct injury in Mdr2(-/-) mice by modulating composition, signalling and excretion of faecal bile acids. Gut. 2018;67:1683–1691. doi: 10.1136/gutjnl-2017-314553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pathak P., Xie C., Nichols R.G., Ferrell J.M., Boehme S., Krausz K.W., Patterson A.D., Gonzalez F.J., Chiang J.Y.L. Intestine farnesoid X receptor agonist and the gut microbiota activate G-protein bile acid receptor-1 signaling to improve metabolism. Hepatology. 2018;68:1574–1588. doi: 10.1002/hep.29857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo J., Ko B., Elliott M., Zhou M., Lindhout D.A., Phung V., To C., Learned R.M., Tian H., DePaoli A.M., Ling L. A nontumorigenic variant of FGF19 treats cholestatic liver diseases. Sci Transl Med. 2014;6:247ra100. doi: 10.1126/scitranslmed.3009098. [DOI] [PubMed] [Google Scholar]
- 29.Alvarez-Sola G., Uriarte I., Latasa M.U., Fernandez-Barrena M.G., Urtasun R., Elizalde M., Barcena-Varela M., Jimenez M., Chang H.C., Barbero R., Catalan V., Rodriguez A., Fruhbeck G., Gallego-Escuredo J.M., Gavalda-Navarro A., Villarroya F., Rodriguez-Ortigosa C.M., Corrales F.J., Prieto J., Berraondo P., Berasain C., Avila M.A. Fibroblast growth factor 15/19 (FGF15/19) protects from diet-induced hepatic steatosis: development of an FGF19-based chimeric molecule to promote fatty liver regeneration. Gut. 2017;66:1818–1828. doi: 10.1136/gutjnl-2016-312975. [DOI] [PubMed] [Google Scholar]
- 30.Hagel M., Miduturu C., Sheets M., Rubin N., Weng W., Stransky N., Bifulco N., Kim J.L., Hodous B., Brooijmans N., Shutes A., Winter C., Lengauer C., Kohl N.E., Guzi T. First selective small molecule inhibitor of FGFR4 for the treatment of hepatocellular carcinomas with an activated FGFR4 signaling pathway. Cancer Discov. 2015;5:424–437. doi: 10.1158/2159-8290.CD-14-1029. [DOI] [PubMed] [Google Scholar]
- 31.Chiang J.Y.L., Ferrell J.M. Bile acids as metabolic regulators and nutrient sensors. Annu Rev Nutr. 2019;39:175–200. doi: 10.1146/annurev-nutr-082018-124344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kummen M., Hov J.R. The gut microbial influence on cholestatic liver disease. Liver Int. 2019;39:1186–1196. doi: 10.1111/liv.14153. [DOI] [PubMed] [Google Scholar]
- 33.Little R., Wine E., Kamath B.M., Griffiths A.M., Ricciuto A. Gut microbiome in primary sclerosing cholangitis: a review. World J Gastroenterol. 2020;26:2768–2780. doi: 10.3748/wjg.v26.i21.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen W., Wei Y., Xiong A., Li Y., Guan H., Wang Q., Miao Q., Bian Z., Xiao X., Lian M., Zhang J., Li B., Cao Q., Fan Z., Zhang W., Qiu D., Fang J., Gershwin M.E., Yang L., Tang R., Ma X. Comprehensive analysis of serum and fecal bile acid profiles and interaction with gut microbiota in primary biliary cholangitis. Clin Rev Allergy Immunol. 2020;58:25–38. doi: 10.1007/s12016-019-08731-2. [DOI] [PubMed] [Google Scholar]
- 35.Yan S., Zhou J., Chen X., Dong Z., Yin X.M. Diverse consequences in liver injury in mice with different autophagy functional status treated with alcohol. Am J Pathol. 2019;189:1744–1762. doi: 10.1016/j.ajpath.2019.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Minicis S., Rychlicki C., Agostinelli L., Saccomanno S., Candelaresi C., Trozzi L., Mingarelli E., Facinelli B., Magi G., Palmieri C., Marzioni M., Benedetti A., Svegliati-Baroni G. Dysbiosis contributes to fibrogenesis in the course of chronic liver injury in mice. Hepatology. 2014;59:1738–1749. doi: 10.1002/hep.26695. [DOI] [PubMed] [Google Scholar]
- 37.Jena P.K., Sheng L., Liu H.X., Kalanetra K.M., Mirsoian A., Murphy W.J., French S.W., Krishnan V.V., Mills D.A., Wan Y.Y. Western diet-induced dysbiosis in farnesoid X receptor knockout mice causes persistent hepatic inflammation after antibiotic treatment. Am J Pathol. 2017;187:1800–1813. doi: 10.1016/j.ajpath.2017.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferrere G., Wrzosek L., Cailleux F., Turpin W., Puchois V., Spatz M., Ciocan D., Rainteau D., Humbert L., Hugot C., Gaudin F., Noordine M.L., Robert V., Berrebi D., Thomas M., Naveau S., Perlemuter G., Cassard A.M. Fecal microbiota manipulation prevents dysbiosis and alcohol-induced liver injury in mice. J Hepatol. 2017;66:806–815. doi: 10.1016/j.jhep.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 39.Grander C., Adolph T.E., Wieser V., Lowe P., Wrzosek L., Gyongyosi B., Ward D.V., Grabherr F., Gerner R.R., Pfister A., Enrich B., Ciocan D., Macheiner S., Mayr L., Drach M., Moser P., Moschen A.R., Perlemuter G., Szabo G., Cassard A.M., Tilg H. Recovery of ethanol-induced Akkermansia muciniphila depletion ameliorates alcoholic liver disease. Gut. 2018;67:891–901. doi: 10.1136/gutjnl-2016-313432. [DOI] [PubMed] [Google Scholar]
- 40.Chen P., Starkel P., Turner J.R., Ho S.B., Schnabl B. Dysbiosis-induced intestinal inflammation activates tumor necrosis factor receptor I and mediates alcoholic liver disease in mice. Hepatology. 2015;61:883–894. doi: 10.1002/hep.27489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamoto N., Amiya T., Aoki R., Taniki N., Koda Y., Miyamoto K., Teratani T., Suzuki T., Chiba S., Chu P.S., Hayashi A., Yamaguchi A., Shiba S., Miyake R., Katayama T., Suda W., Mikami Y., Kamada N., Ebinuma H., Saito H., Hattori M., Kanai T. Commensal Lactobacillus controls immune tolerance during acute liver injury in mice. Cell Rep. 2017;21:1215–1226. doi: 10.1016/j.celrep.2017.10.022. [DOI] [PubMed] [Google Scholar]
- 42.Tedesco D., Thapa M., Chin C.Y., Ge Y., Gong M., Li J., Gumber S., Speck P., Elrod E.J., Burd E.M., Kitchens W.H., Magliocca J.F., Adams A.B., Weiss D.S., Mohamadzadeh M., Grakoui A. Alterations in intestinal microbiota lead to production of interleukin 17 by intrahepatic gammadelta T-cell receptor-positive cells and pathogenesis of cholestatic liver disease. Gastroenterology. 2018;154:2178–2193. doi: 10.1053/j.gastro.2018.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao L., Schneider K.M., Galvez E.J.C., Frissen M., Marschall H.U., Su H., Hatting M., Wahlstrom A., Haybaeck J., Puchas P., Mohs A., Peng J., Bergheim I., Nier A., Hennings J., Reissing J., Zimmermann H.W., Longerich T., Strowig T., Liedtke C., Cubero F.J., Trautwein C. Intestinal dysbiosis augments liver disease progression via NLRP3 in a murine model of primary sclerosing cholangitis. Gut. 2019;68:1477–1492. doi: 10.1136/gutjnl-2018-316670. [DOI] [PubMed] [Google Scholar]
- 44.Tabibian J.H., O’Hara S.P., Trussoni C.E., Tietz P.S., Splinter P.L., Mounajjed T., Hagey L.R., LaRusso N.F. Absence of the intestinal microbiota exacerbates hepatobiliary disease in a murine model of primary sclerosing cholangitis. Hepatology. 2016;63:185–196. doi: 10.1002/hep.27927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lundasen T., Galman C., Angelin B., Rudling M. Circulating intestinal fibroblast growth factor 19 has a pronounced diurnal variation and modulates hepatic bile acid synthesis in man. J Intern Med. 2006;260:530–536. doi: 10.1111/j.1365-2796.2006.01731.x. [DOI] [PubMed] [Google Scholar]
- 46.Uriarte I., Fernandez-Barrena M.G., Monte M.J., Latasa M.U., Chang H.C., Carotti S., Vespasiani-Gentilucci U., Morini S., Vicente E., Concepcion A.R., Medina J.F., Marin J.J., Berasain C., Prieto J., Avila M.A. Identification of fibroblast growth factor 15 as a novel mediator of liver regeneration and its application in the prevention of post-resection liver failure in mice. Gut. 2013;62:899–910. doi: 10.1136/gutjnl-2012-302945. [DOI] [PubMed] [Google Scholar]
- 47.Kong B., Huang J., Zhu Y., Li G., Williams J., Shen S., Aleksunes L.M., Richardson J.R., Apte U., Rudnick D.A., Guo G.L. Fibroblast growth factor 15 deficiency impairs liver regeneration in mice. Am J Physiol Gastrointest Liver Physiol. 2014;306:G893–G902. doi: 10.1152/ajpgi.00337.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Z., Lin B., Lin G., Wu Y., Jie Y., Li X., Ko B., Chong Y., Luo J. Circulating FGF19 closely correlates with bile acid synthesis and cholestasis in patients with primary biliary cirrhosis. PLoS One. 2017;12 doi: 10.1371/journal.pone.0178580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brandl K., Hartmann P., Jih L.J., Pizzo D.P., Argemi J., Ventura-Cots M., Coulter S., Liddle C., Ling L., Rossi S.J., DePaoli A.M., Loomba R., Mehal W.Z., Fouts D.E., Lucey M.R., Bosques-Padilla F., Mathurin P., Louvet A., Garcia-Tsao G., Verna E.C., Abraldes J.G., Brown R.S., Jr., Vargas V., Altamirano J., Caballeria J., Shawcross D., Starkel P., Ho S.B., Bataller R., Schnabl B. Dysregulation of serum bile acids and FGF19 in alcoholic hepatitis. J Hepatol. 2018;69:396–405. doi: 10.1016/j.jhep.2018.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kennedy D.J., Vetteth S., Periyasamy S.M., Kanj M., Fedorova L., Khouri S., Kahaleh M.B., Xie Z., Malhotra D., Kolodkin N.I., Lakatta E.G., Fedorova O.V., Bagrov A.Y., Shapiro J.I. Central role for the cardiotonic steroid marinobufagenin in the pathogenesis of experimental uremic cardiomyopathy. Hypertension. 2006;47:488–495. doi: 10.1161/01.HYP.0000202594.82271.92. [DOI] [PubMed] [Google Scholar]