Abstract
Background & Aims
Patients with ulcerative colitis have low concentrations of the major membrane lipid phosphatidylcholine (PC) in gastrointestinal mucus, suggesting that defects in colonic PC metabolism might be involved in the development of colitis. To determine the precise role that PC plays in colonic barrier function, we examined mice with intestinal epithelial cell (IEC)-specific deletion of the rate-limiting enzyme in the major pathway for PC synthesis: cytidine triphosphate:phosphocholine cytidylyltransferase-α (CTαIKO mice).
Methods
Colonic tissue of CTαIKO mice and control mice was analyzed by histology, immunofluorescence, electron microscopy, quantitative polymerase chain reaction, Western blot, and thin-layer chromatography. Histopathologic colitis scores were assigned by a pathologist blinded to the experimental groupings. Intestinal permeability was assessed by fluorescein isothiocyanate–dextran gavage and fecal microbial composition was analyzed by sequencing 16s ribosomal RNA amplicons. Subsets of CTαIKO mice and control mice were treated with dietary PC supplementation, antibiotics, or 4-phenylbutyrate.
Results
Inducible loss of CTα in the intestinal epithelium reduced colonic PC concentrations and resulted in rapid and spontaneous colitis with 100% penetrance in adult mice. Colitis development in CTαIKO mice was traced to a severe and unresolving endoplasmic reticulum stress response in IECs with altered membrane phospholipid composition. This endoplasmic reticulum stress response was linked to the necroptotic death of IECs, leading to excessive loss of goblet cells, formation of a thin mucus barrier, increased intestinal permeability, and infiltration of the epithelium by microbes.
Conclusions
Maintaining the PC content of IEC membranes protects against colitis development in mice, showing a crucial role for IEC phospholipid equilibrium in colonic homeostasis. SRA accession number: PRJNA562603.
Keywords: Phosphatidylcholine, Inflammation, Colon, Mouse Model, Lipid
Abbreviations used in this paper: ATF6, activating transcription factor 6; CDP, cytidine diphosphate; CTα, CTP:phosphocholine cytidylyltransferase-α; CTαIKO, intestinal epithelial cell–specific deletion of CTP:phosphocholine cytidylyltransferase-α; ER, endoplasmic reticulum; FITC, fluorescein isothiocyanate; GM-CSF, granulocyte-macrophage colony-stimulating factor; IBD, inflammatory bowel diseases; IEC, intestinal epithelial cell; IL, interleukin; IRE1α, inositol-requiring enzyme 1-α; LIF, leukemia inhibitory factor; MCP-1, monocyte chemoattractant protein-1; mRNA, messenger RNA; PBA, 4-phenyl butyrate; PC, phosphatidylcholine; PCR, polymerase chain reaction; PE, phosphatidylethanolamine; PERK, protein kinase R-like ER kinase; RIP, receptor-interacting serine/threonine-protein kinase; rRNA, ribosomal RNA; TNF-α, tumor necrosis factor-α; TUNEL, terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling; UC, ulcerative colitis; UPR, unfolded protein response; XBP1, X-box binding protein 1
Graphical abstract
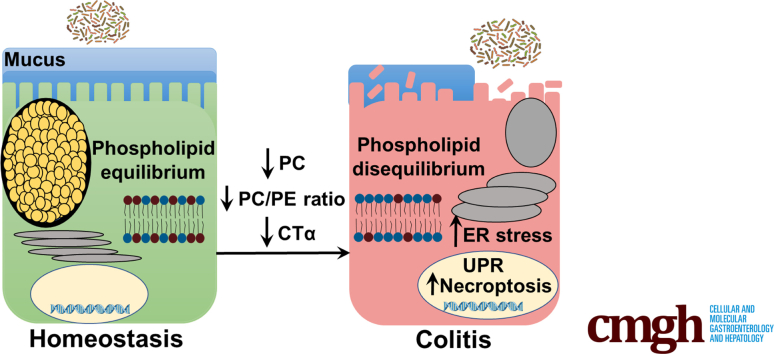
Summary.
Using mice with impaired intestinal de novo phosphatidylcholine (PC) synthesis, we found that maintaining the PC content of intestinal epithelial cell membranes is crucial to prevent endoplasmic reticulum stress and colitis development, showing an important role for PC in colonic function.
Inflammatory bowel diseases (IBDs), comprising Crohn’s disease and ulcerative colitis (UC), are increasing in incidence and prevalence worldwide.1 Crohn’s disease can occur in any area of the gastrointestinal tract and is characterized histologically by transmural inflammation, noncaseating granulomas, and a thickened submucosa.2 UC, on the other hand, occurs primarily in the colon and is characterized by superficial damage to the mucosa, cryptitis, and crypt abscesses.2 A unique histologic feature of UC is the loss of goblet cell mucus granules from the colonic epithelium,3 although the mechanisms underlying this loss of mucus granules remain unclear. Interestingly, gastrointestinal mucus samples from UC patients show low levels of the major membrane lipid phosphatidylcholine (PC) as compared with mucus from patients with Crohn’s disease or people without IBD.4,5 Furthermore, human clinical trials designed to restore colonic PC concentrations in UC patients have shown promising results.6, 7, 8 However, despite these important clinical links between PC and UC, the precise molecular mechanisms linking changes to intestinal PC concentrations (and membrane lipid composition) to intestinal inflammation and features of UC pathology in vivo remain unclear.
PC is produced primarily by the cytidine diphosphate (CDP)–choline pathway in mammalian tissues.9 The rate-limiting step of the CDP–choline pathway, the conversion of phosphocholine to CDP–choline, is catalyzed by cytidine triphosphate (CTP):phosphocholine cytidylyltransferase-α (CTα; encoded by Pcyt1a). An adequate supply of PC is required for the prevention and resolution of endoplasmic reticulum (ER) stress in a variety of cell types,10,11 and might be particularly important in intestinal epithelial cells (IECs) owing to their high secretory activity and constant exposure to environmental antigens. Furthermore, a relatively low molar ratio of PC to phosphatidylethanolamine (PE) is associated with nonalcoholic steatohepatitis in human beings,12 suggesting that perturbations to membrane lipid composition might influence the initiation or progression of inflammatory diseases. Consistent with an anti-inflammatory role for PC in IECs, exogenous delivery of PC, but not PE, to Caco2 cells after treatment with tumor necrosis factor-α (TNF-α) dampens the induction of proinflammatory transcripts.13 To date, the link between PC and intestinal inflammation has not yet been examined thoroughly in vivo. In addition to de novo synthesis, IECs of the small intestine can obtain PC from the diet, bile, and circulating lipoproteins. However, because dietary and biliary PC is absorbed primarily in the proximal small intestine,14,15 the colon is reliant on de novo PC synthesis to maintain membrane lipid composition and thus might be particularly sensitive to dietary or environmental factors that disrupt membrane lipid homeostasis.
To determine the role that PC plays in mucosal barrier function, and to gain insight into the mechanisms by which colonic PC depletion is linked to inflammation in UC patients, we examined mice with IEC-specific deletion of CTα (CTαIKO) mice.16 We found that inducible loss of CTα in the intestinal epithelium reduces colonic PC concentrations and results in rapid and spontaneous colitis with 100% penetrance in adult mice that is characterized by crypt abscesses, goblet cell depletion, and immune cell infiltration. Colitis development after IEC PC depletion is initiated by a severe and unresolving ER stress response. This ER stress response is linked to the induction of receptor-interacting serine/threonine-protein kinase (RIP)3 and the death of IECs by necroptosis, leading to loss of goblet cells, formation of a thin mucus barrier, infiltration of the epithelium by microbes, and the induction of an array of proinflammatory cytokines. Taken together, our data show that maintaining membrane lipid composition in IECs is crucial for normal colonic barrier function.
Results
CTαIKO Mice Have Altered Colonic Phospholipid Concentrations
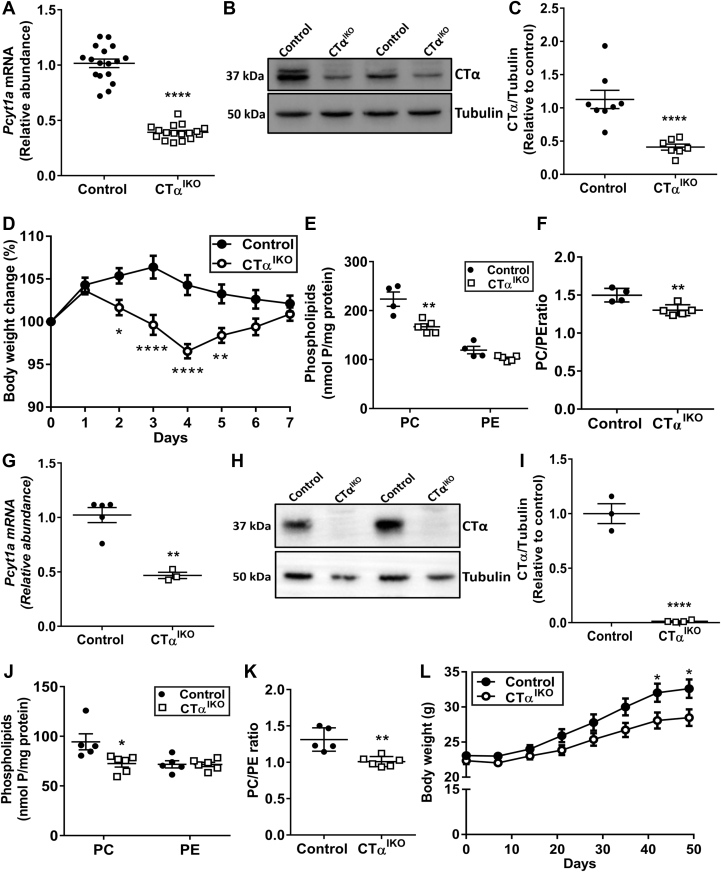
Induction of Cre recombinase with tamoxifen resulted in the generation of adult CTαIKO mice with approximately 60% lower Pcyt1a messenger RNA (mRNA) (Figure 1A) and approximately 75% lower CTα protein abundance (Figure 1B and C) in the colon compared with control mice. Residual CTα protein levels likely were owing to the presence of nonepithelial cell types including muscle and infiltrating immune cells. Consistent with our previous report,16 CTαIKO mice experienced rapid body weight loss of varying severity upon Cre induction, whereas control mice did not experience body weight loss (Figure 1D). One of 17 CTαIKO mice experienced severe wasting with more than 20% body weight loss and was euthanized on day 5 after Cre induction. Most CTαIKO mice began to regain body weight on day 5 and were a similar body weight to controls by day 7 (Figure 1D). CTαIKO mice had lower PC concentrations in epithelial cells isolated from the colon 7 days after Cre induction compared with epithelial cells isolated from control mice (Figure 1E). PE concentrations in colonic epithelial cells were comparable between groups (Figure 1E), resulting in a significantly lower ratio of PC/PE in epithelial cells of CTαIKO mice compared with control mice (Figure 1F).
Figure 1.
CTαIKO mice have low colonic PC concentrations and experience acute body weight loss. (A) The mRNA abundance of Pcyt1a in colonic tissue of control mice and CTαIKO mice 7 days after the end of tamoxifen treatment (n = 17/group). (B) Western blot and (C) quantification of CTα relative to tubulin in colonic epithelial cells isolated from control mice and CTαIKO mice 7 days after the end of tamoxifen treatment (n = 8/group). (D) Body mass change relative to body mass at the end of tamoxifen treatment in control mice and CTαIKO mice (n = 17/group). (E) PC and PE concentrations in colonic epithelial cells isolated from control mice and CTαIKO mice 7 days after the end of tamoxifen treatment (n = 4–5/group). (F) The ratio of PC/PE in colonic epithelial cells of control mice and CTαIKO mice 7 days after the end of tamoxifen treatment (n = 4–5/group). (G) The mRNA abundance of Pcyt1a in colonic tissue of control mice and CTαIKO mice 7 weeks after the end of tamoxifen treatment (n = 3–5/group). (H) Western blot and (I) quantification of CTα relative to tubulin in ileal epithelial cells isolated from control mice and CTαIKO mice 7 weeks after the end of tamoxifen treatment (n = 3–4/group). (J) PC and PE concentrations in ileal epithelial cells isolated from control mice and CTαIKO mice 7 weeks after the end of tamoxifen treatment (n = 5–6/group). (K) The ratio of PC/PE in ileal epithelial cells of control mice and CTαIKO mice 7 weeks after the end of tamoxifen treatment (n = 5–6/group). (L) Body mass in control mice and CTαIKO mice over 7 weeks after Cre induction (n = 18–20/group). Mice killed because of excessive weight loss (2 of 35 CTαIKO mice) were not included in any analyses. Values are means ± SEM. ∗P < .05, ∗∗P < .01, and ∗∗∗∗P < .0001.
Colonic Pcyt1a mRNA remained repressed in the colons of CTαIKO mice at 7 weeks after Cre induction (Figure 1G). In addition, CTα protein levels remained absent in epithelial cells isolated from the ileums of CTαIKO mice after 7 weeks (Figure 1H and I). CTαIKO mice also had lower PC concentrations in ileal epithelial cells after 7 weeks compared with control mice, while PE concentrations were similar (Figure 1J), resulting in a lower ratio of PC to PE in ileal epithelial cells (Figure 1K) at 7 weeks after Cre induction. Furthermore, CTαIKO mice gained less body weight compared with controls over the 7-week follow-up period (Figure 1L), but no additional morbidity was observed after day 8 when 1 further CTαIKO mouse was killed because of excessive body weight loss.
CTαIKO Mice Develop Spontaneous Colitis
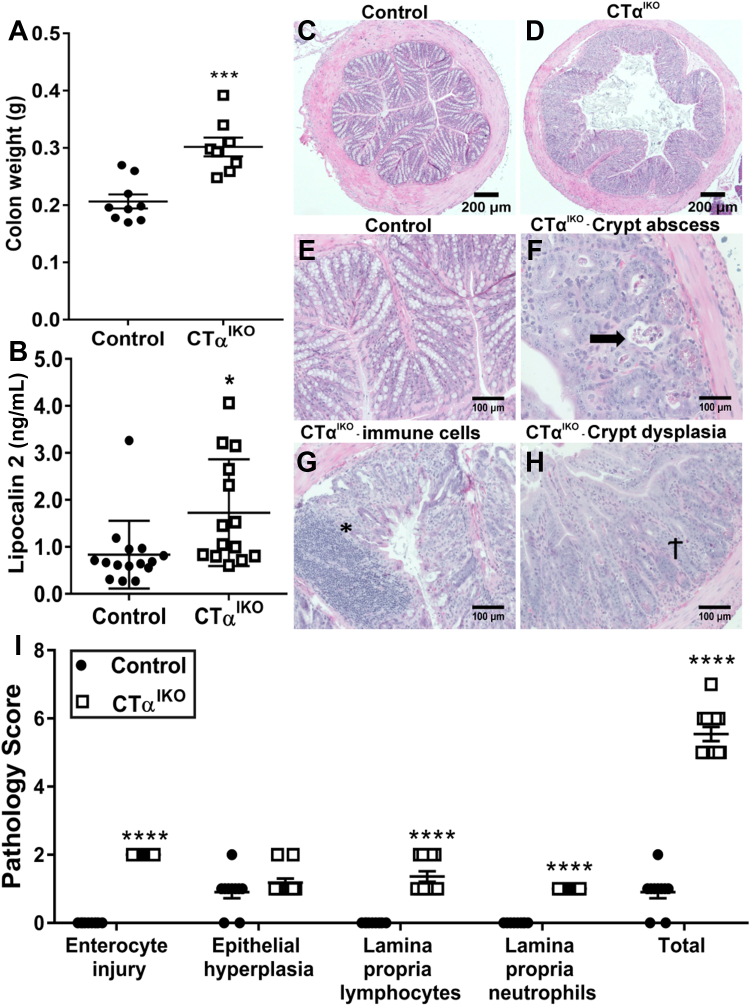
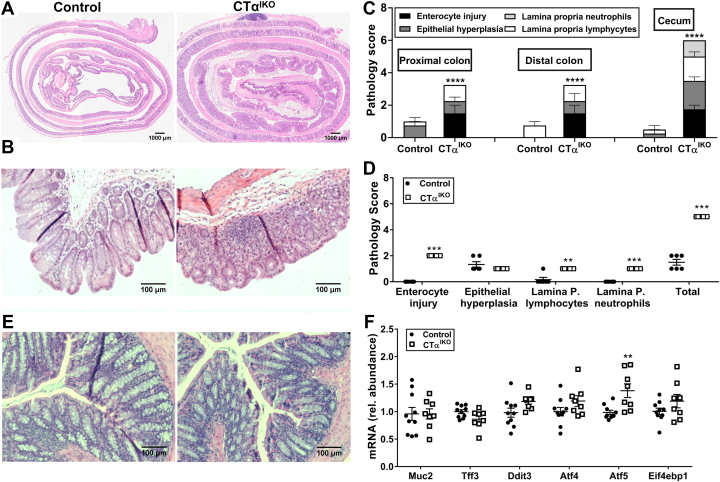
Seven days after Cre induction, the colons of CTαIKO mice weighed approximately 50% more than those of controls (Figure 2A). Furthermore, red blood cells, hemoglobin, and hematocrit were significantly lower in CTαIKO mice, while blood reticulocyte concentrations were higher, which together is indicative of anemia and suggested that CTαIKO mice might have lost blood through the injured bowel (Table 1). In addition, circulating concentrations of the bacteriostatic protein lipocalin 2 were increased in CTαIKO mice compared with control mice (Figure 2B).
Figure 2.
CTαIKO mice develop spontaneous colitis. (A) Colon weight in control mice and CTαIKO mice 7 days after the end of tamoxifen treatment (n = 8–9/group). (B) Plasma lipocalin 2 concentrations in control mice and CTαIKO mice 4 days after the end of tamoxifen treatment (n = 14–15/group). (C and D) Representative H&E-stained distal colon sections from control mice and CTαIKO mice 4 days after the end of tamoxifen treatment. (E) Representative H&E-stained distal colon section from control mice 4 days after the end of tamoxifen treatment. (F and H) Representative H&E-stained distal colon sections from CTαIKO mice 4 days after the end of tamoxifen treatment. (F) Arrow indicates crypt abscess. (G) ∗Immune cell infiltration. (H) †Crypt dysplasia. (I) Pathology scores of control mice and CTαIKO mice 4 days after the end of tamoxifen treatment (n = 10–11/group). Values are means ± SEM. ∗P < .05, ∗∗∗P < .001.
Table 1.
Complete Blood Cell Count Data
| Complete blood cell count | Control | CTαIKO |
|---|---|---|
| Hematocrit, % | 54.00 ± 1.31 | 50.32 ± 0.48∗ |
| Red blood cells, ×10E12 cells/L | 10.38 ± 0.26 | 9.72 ± 0.11∗ |
| Hemoglobin, g/L | 156.70 ± 3.06 | 147.80 ± 1.8∗ |
| Reticulocytes, ×10E09 cells/L | 149.40 ± 20.05 | 295.80 ± 29.50∗∗ |
| Reticulocytes, % | 1.46 ± 0.22 | 3.05 ± 0.31∗∗ |
| White blood cell count peroxidase method, ×10E09 cells/L | 2.85 ± 0.88 | 2.33 ± 1.10 |
| White blood cell count basophile method, ×10E09 cells/L | 3.21 ± 1.03 | 2.29 ± 0.99 |
| Mean corpuscular volume, fL | 52.02 ± 0.17 | 51.78 ± 0.29 |
| Mean corpuscular hemoglobin, pg | 15.12 ± 0.10 | 15.23 ± 0.80 |
| Mean corpuscular hemoglobin concentration, g/L | 290.30 ± 1.86 | 293.80 ± 1.99 |
| Platelets, ×10E09 cells/L | 701.00 ± 11.85 | 796.20 ± 44.19 |
| Neutrophils, % | 9.90 ± 1.83 | 11.38 ± 2.51 |
| Lymphocytes, % | 80.32 ± 2.20 | 79.13 ± 1.86 |
| Monocytes, % | 2.90 ± 0.68 | 3.32 ± 0.36 |
| Eosinophils, % | 2.55 ± 0.51 | 3.15 ± 0.61 |
| Large unstained cells, % | 3.95 ± 1.28 | 2.57 ± 0.71 |
| Basophils, % | 0.47 ± 0.071 | 0.47 ± 0.1 |
| Neutrophils, ×10E09 cells/L | 0.25 ± 0.50 | 0.21 ± 0.06 |
| Lymphocytes, ×10E09 cells/L | 2.65 ± 0.95 | 1.88 ± 0.87 |
| Monocytes, ×10E09 cells/L | 0.07 ± 0.02 | 0.06 ± 0.2 |
| Eosinophils, ×10E09 cells/L | 0.06 ± 0.01 | 0.05 ± 0.11 |
| Large unstained cells, ×10E09 cells/L | 0.15 ± 0.07 | 0.07 ± 0.04 |
| Basophils, ×10E09 cells/L | 0.02 ± 0.01 | 0.01 ± 0.01 |
NOTE. Data are means ± SEM.
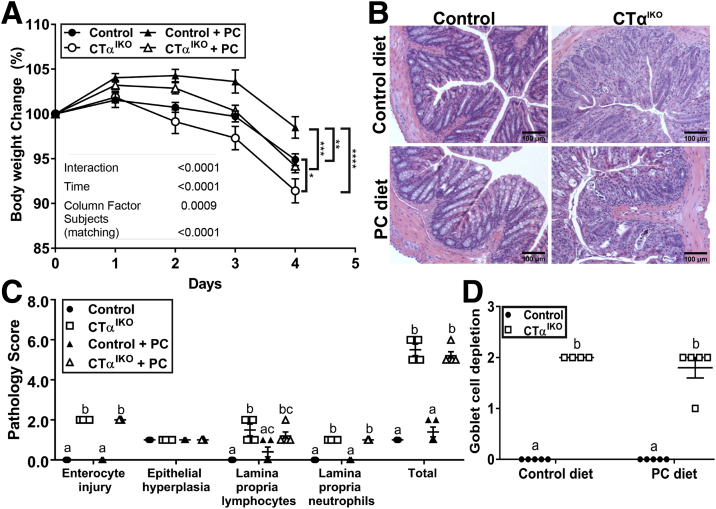
There was extensive damage to the colonic epithelium of CTαIKO mice compared with controls (Figure 2C–H), including crypt abscesses (Figure 2F), lymphocyte infiltration to the lamina propria (Figure 2D), and crypt dysplasia (Figure 2H). Pathology scoring of H&E-stained distal colon sections showed that 100% of CTαIKO mice developed spontaneous colitis by day 4 after Cre induction (Figure 2I). Dietary PC supplementation was unable to rescue body weight loss (Figure 3A) or colitis development (Figure 3B) in CTαIKO mice, as indicated by assessment of pathology scores (Figure 3C) and goblet cell depletion (Figure 3D). The inability of exogenous dietary PC to rescue any metrics of disease pathology in CTαIKO mice likely reflects that dietary PC is efficiently hydrolyzed and absorbed in the proximal small intestine and that very little dietary PC reaches the colon.15
Figure 3.
Dietary PC supplementation does not rescue body weight loss or colitis development in CTαIKO mice. (A) Body weight change relative to body weight at the end of tamoxifen treatment in control mice and CTαIKO mice treated either with or without supplementary PC in the diet (n = 6–7/group). (B) Representative H&E-stained distal colon sections in control mice and CTαIKO mice treated either with or without supplementary PC in the diet. (C) Pathology scores for control mice and CTαIKO mice treated either with or without supplementary PC in the diet (n = 4–5/group). (D) Goblet cell depletion score in control mice and CTαIKO mice treated either with or without supplementary PC in the diet (n = 4–5/group). (A) ∗Statistical significance in body weight on day 4 after the end of tamoxifen treatment based on 2-way analysis of variance followed by the Tukey post hoc test. (C) Columns that do not share a letter (a, b, or c) are significantly different (α = .05). Values are means ± SEM. ∗P < .05, ∗∗P < .01,∗∗∗P < .001, and ∗∗∗∗P < .0001.
Regional analysis of colon Swiss rolls showed similarly high levels of enterocyte injury, epithelial hyperplasia, and lymphocyte infiltration to the lamina propria of the proximal compared with the distal colon of CTαIKO mice, and pathology scores also were higher in the cecum of CTαIKO mice compared with controls (Figure 4A–C). Although we initially hypothesized that the weight regain observed in CTαIKO mice on day 5 after Cre induction (Figure 1D) might be linked to improved metrics of disease severity, pathology scoring showed that colitis had not improved by day 7 (Figure 4D), even after most CTαIKO mice were a similar body weight to control mice. Assessment of colon histology 7 weeks after Cre induction showed improved disease pathology relative to 4 days and 7 days after Cre induction in CTαIKO mice (Figure 4E), with infrequent immune cell infiltration, absence of crypt abscesses, and partial restoration of goblet cells. Consistent with the restoration of goblet cells and improvement to the colonic epithelium after 7 weeks in CTαIKO mice, mRNA levels of the goblet cell marker Muc2 were comparable between groups (Figure 4F), while Tff3 levels tended to be modestly lower in CTαIKO mice but did not reach statistical significance (P = .055). The ER stress marker Atf5 was significantly higher in the colons of CTαIKO mice after 7 weeks, while Ddit3 (P = .063), Atf4 (P = .127), and Eif4bp1 (P = .161) also tended to be higher but did not reach statistical significance. These observations suggest that epithelial cells of CTαIKO mice activate compensatory pathways to promote mucosal healing after the initial inflammatory response but remain modestly stressed relative to control mice.
Figure 4.
Acute inflammation extends to all parts of the colon and cecum in CTαIKO mice but disease severity improves by 7 weeks after Cre induction. (A) Representative H&E-stained colon Swiss rolls from control mice and CTαIKO mice 4 days after the end of tamoxifen treatment. (B) Representative H&E-stained cecum sections from control mice and CTαIKO mice 4 days after the end of tamoxifen treatment. (C) Pathology scores in proximal colon Swiss roll sections, distal colon Swiss roll sections, and cecum sections of control mice and CTαIKO mice 4 days after the end of tamoxifen treatment (n = 4/group). (D) Pathology scores of control mice and CTαIKO mice 7 days after the end of tamoxifen treatment (n = 6–7/group). Representative H&E-stained colon sections from control mice and CTαIKO mice 7 weeks after the end of tamoxifen treatment. (E) The mRNA abundance of Muc2, Tff3, Ddit3, Atf4, Atf5, and Eif4ebp1 in the colons of control mice and CTαIKO mice 7 weeks after the end of tamoxifen treatment (n = 8–10/group). Values are means ± SEM. ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. P., propria; rel., relative.
CTαIKO Mice Acutely Lose Goblet Cell Mucus Granules and Have Ultrastructural Damage to Theca in Goblet Cells
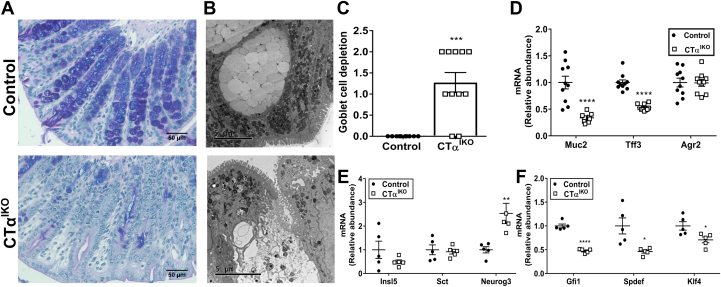
There was a marked decrease in Alcian blue/periodic acid–Schiff staining, which identifies goblet cell mucus granules, in the colons of CTαIKO mice compared with control mice on day 4 after Cre induction (Figure 5A). Electron microscopy showed that the ultrastructural integrity of mucus-containing theca in CTα-deficient goblet cells was compromised, with loss of mucus granules and infiltration of cellular debris (Figure 5B). This atypical appearance of mucus granules has been observed previously in UC patients.17 Loss of mucus granules in CTαIKO mice resulted in a significantly higher pathology score for goblet cell depletion compared with control mice (Figure 5C).
Figure 5.
Loss of mucus granules and ultrastructural damage to theca in goblet cells of CTαIKO mice. (A) Representative Alcian blue/periodic acid–Schiff staining in colons from control mice and CTαIKO mice. (B) Representative transmission electron micrographs of goblet cell theca in the colons of control mice and CTαIKO mice. (C) Pathology score for goblet cell depletion in the colons of control mice and CTαIKO mice (n = 10–11/group). (D) The mRNA abundance of Muc2, Tff3, and Agr2 in the colons of control mice and CTαIKO mice (n = 10/group). (E) The mRNA abundance of enteroendocrine cell markers in the colons of control mice and CTαIKO mice (n = 5/group). (F) The mRNA abundance goblet cell maturation factors in the colons of control mice and CTαIKO mice (n = 5/group). Values are means ± SEM. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001, ∗∗∗∗P < .0001.
The mRNA abundance of the goblet cell markers Muc2 and Tff3 were lower in the colons of CTαIKO mice compared with control mice (Figure 5D). However, the mRNA abundance of Agr2, which encodes a protein disulfide isomerase that is expressed in Paneth cells and enteroendocrine cells in addition to goblet cells,18 was not different between groups (Figure 5D). Furthermore, enteroendocrine cell markers (Insl5 and Sct) were comparable between groups, except for Neurog3, which was higher in CTαIKO mice (Figure 5E). These data together suggest that goblet cells might be particularly sensitive to loss of CTα, possibly owing to their high secretory activity.19 Further consistent with the lower abundance of goblet cells, the mRNA levels of Gfi1, Spdef, and Klf4, which are involved in goblet cell maturation, were lower in the colons of CTαIKO mice compared with control mice (Figure 5F).
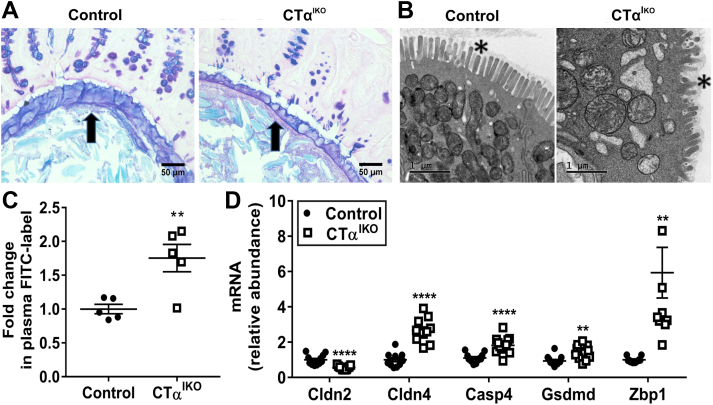
CTαIKO Mice Have a Thin Mucus Layer and Enhanced Intestinal Permeability
The mucus layer of CTαIKO mice was thinner than that of control mice (Figure 6A), and electron microscopy showed that there was extensive damage to the apical brush border of colonic epithelial cells on day 4 after Cre induction (Figure 6B). CTαIKO mice also had a striking increase in intestinal permeability, as assessed by the appearance of fluorescein isothiocyanate (FITC)-labeled dextran in circulation after oral administration (Figure 6C). Altered expression of tight junction components is linked to increased intestinal permeability in UC.20 Accordingly, CTαIKO mice had a lower abundance of Cldn2 transcripts and a higher abundance of Cldn4 transcripts in the colon compared with control mice (Figure 6D). Pyroptotic cell death is triggered in response to gram-negative bacterial infections and thus plays a crucial role in antibacterial innate immune defense.17,21, 22, 23, 24 The mRNA levels of Casp4 (encodes caspase-11) and Gsdmd (encodes gasdermin D), which are markers of pyroptosis, were higher in the colonic tissue of CTαIKO mice (Figure 6D), and mRNA levels of the cytosolic bacterial DNA sensor Zbp1 also strongly were induced (Figure 6D), suggesting that microbes had infiltrated the colonic epithelium after breakdown of the mucosal barrier.
Figure 6.
Impaired mucus layer integrity and increased intestinal permeability in response to impaired de novo PC synthesis in IECs. (A) Representative light microscope images of the colonic mucus layer (arrow) in control mice and CTαIKO mice after Carnoy’s fixation and Alcian blue/periodic acid–Schiff staining. (B) Representative electron micrographs of the colonic microvilli (asterisk) in control mice and CTαIKO mice. (C) Relative fluorescence in plasma of control mice and CTαIKO mice 2 hours after an oral gavage of FITC-labeled dextran (n = 5/group). (D) The mRNA abundance of Cldn2, Cldn4, Casp4, Gsdmd, and Zbp1 in the colons of control mice and CTαIKO mice (n = 10–12/group). Values are means ± SEM. ∗∗P < .01, ∗∗∗∗P < .001.
Antibiotics Dampen Inflammatory Cytokine Secretion but Do Not Prevent Colitis Development in CTαIKO Mice
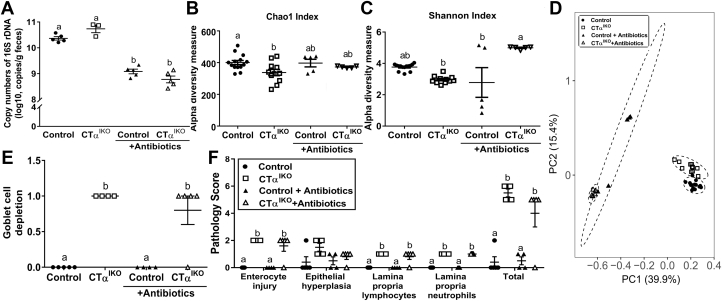
To comprehensively examine the interplay between microbes, the colonic epithelium, and the intestinal immune system of CTαIKO mice, we sequenced the fecal microbiome and profiled colonic cytokine and chemokine concentrations under both untreated and antibiotic-treated conditions on day 4 after Cre induction. Antibiotic treatment reduced fecal bacterial loads in both control mice and CTαIKO mice compared with untreated mice (12%–18% decrease in 16S ribosomal DNA copy numbers) (Figure 7A). In line with previous studies showing a decrease in gut microbial diversity in both human beings25 and mice26 with UC, CTαIKO mice had lower richness of the fecal microbiota compared with control mice as indicted by chao1 index, and antibiotic treatment ameliorated these differences between groups (Figure 7B). The Shannon index also indicated that loss of intestinal CTα tended to reduce the richness and evenness of the microbiome, although this did not reach statistical significance (Figure 7C). Although there was a clear clustering of samples according to antibiotic treatment status (Figure 7D), there was no difference in β diversity between CTαIKO mice and control mice under untreated (R2 = 0.703, P = .090) or antibiotic-treated (R2 = 0.380, P = .822) conditions. However, CTαIKO mice showed a marked expansion of Proteobacteria, including the genus unclassified Enterobacteriaceae (Table 2), which has been linked previously to intestinal inflammation.27 In addition, the genus Akkermansia was increased significantly, while anaerobic bacteria belonging to the family Ruminococcaceae were reduced, in CTαIKO mice compared with control mice (Table 2). Antibiotic treatment prevented the expansion of Enterobacteriaceae and Akkermansia in CTαIKO mice (Table 2). Thus, CTαIKO mice have changes to gut microbes that reflect increased intestinal inflammation including lower richness of fecal microbiome and increased abundance of Enterobacteriaceae.
Figure 7.
Loss of intestinal CTα changes the microbiome but depletion of gut microbes with antibiotics does not prevent colitis development in CTαIKO mice. (A) Copy number of 16S rRNA in feces of control mice and CTαIKO mice treated with and without antibiotics (n = 3–5/group). (B) Chao1 and (C) Shannon indexes of gut microbiota from control mice and CTαIKO mice treated with and without antibiotics (control, n = 14; CTαIKO, n = 11; control + antibiotics, n = 5; CTαIKO + antibiotics, n = 5). (D) Principle component analysis plots of the bacterial communities based on the Bray–Curtis distance matrix. Each point represents an individual mouse (control, n = 14; CTαIKO, n = 11; control + antibiotics, n = 5; CTαIKO + antibiotics, n = 5). (E) Pathology score for goblet cell depletion in the colons of control mice and CTαIKO mice treated with and without antibiotics (n = 4–5/group). (F) Pathology scores of control mice and CTαIKO mice treated with and without antibiotics (n = 4–5/group). Columns that do not share a letter (a, b, or c) are significantly different (α = .05).
Table 2.
Relative Abundance of Predominant Fecal Bacterial Phyla and Genera Under Untreated or Antibiotic-Treated Conditions
| Control | CTαIKO | Control + antibiotics | CTαIKO + antibiotics | SEM | P value | FDR_P value | |
|---|---|---|---|---|---|---|---|
| Phylum | |||||||
| p__Actinobacteria | 0.44 | 0.43 | 0.24 | 0.44 | 0.05 | .385 | .405 |
| p__Bacteroidetes | 41.12a,b | 43.53a | 14.98c | 33.20b,c | 2.14 | .001 | .001 |
| p__Firmicutes | 33.06b | 11.42c | 64.92a | 43.43a,b | 3.43 | <.001 | <.001 |
| p__Proteobacteria | 11.18a | 16.79a | 10.25a | 7.74b | 0.98 | .003 | .003 |
| p__Verrucomicrobia | 13.05b | 26.35a | 0.46c | 0.06c | 2.03 | <.001 | <.001 |
| p__Tenericutes | 0.17c | 0.24b,c | 1.54a | 0.66a,b | 0.15 | .002 | .003 |
| p__Deferribacteres | 0.48b | 0.93a | 0.04c | 0.04c | 0.13 | <.001 | <.001 |
| p__TM7 | 0.21a | 0.04b | 0.08a,b | 0.20a,b | 0.03 | .021 | .025 |
| p__Spirochaetes | 0.02b | 0.01c | 3.43a | 8.59a | 0.57 | <.001 | <.001 |
| p__Lentisphaerae | NDb | NDb | 0.42a | 1.00a | 0.12 | .008 | .012 |
| Actinobacteria | |||||||
| g__Bifidobacterium | 0.33a | 0.22a | NDb | 0.01b | 0.05 | <.001 | <.001 |
| f_Coriobacteriaceae;g_ | 0.01b | 0.10a,b | 0.06a,b | 0.17a | 0.02 | .010 | .017 |
| g__Adlercreutzia | 0.10a | 0.10a | NDb | NDb | 0.01 | <.001 | <.001 |
| g__Collinsella | NDb | NDb | 0.16a | 0.22a | 0.02 | <.001 | <.001 |
| Bacteroidetes | |||||||
| o__Bacteroidales;f__;g__ | 5.74 | 4.17 | 2.97 | 6.84 | 0.48 | .086 | .120 |
| g__Bacteroides | 4.59b | 14.06a | 0.24c | 0.16c | 1.25 | <.001 | <.001 |
| g__Parabacteroides | 4.71a | 8.21a | 1.36b | 3.46a,b | 0.82 | .002 | .004 |
| g__Prevotella | 0.08b | 0.03c | 5.72a | 11.93a | 0.83 | <.001 | <.001 |
| g__[Prevotella] | 1.41 | 2.48 | 1.63 | 3.60 | 0.37 | .202 | .220 |
| f__Rikenellaceae;g__ | 7.21a | 7.00a | 0.05b | 0.01b | 0.65 | <.001 | <.001 |
| g_AF12 | 0.33a | 0.19a | NDb | NDb | 0.04 | <.001 | <.001 |
| f_S24-7;g_ | 10.60a | 3.71b | 1.16b | 3.58b | 0.77 | <.001 | <.001 |
| g__Odoribacter | 6.44a | 3.67a | 0.01b | NDb | 0.57 | <.001 | <.001 |
| Firmicutes | |||||||
| f__Bacillaceae;g__ | 0.01 | 0.03 | 0.04 | ND | 0.01 | .093 | .128 |
| g__Enterococcus | 0.01b | 0.06a,b | 0.35a | 0.59a | 0.04 | <.001 | <.001 |
| g__Lactobacillus | 1.50b | 1.43b | 1.76a,b | 3.90a | 0.23 | .015 | .023 |
| g__Lactococcus | 1.12 | 0.16 | 45.62 | 3.92 | 3.62 | .054 | .078 |
| g__Turicibacter | 0.26a | 0.01b | 0.03b | 0.04b | 0.03 | <.001 | <.001 |
| o__Clostridiales;f__;g__ | 13.36a | 3.67b | 3.53b | 7.05a,b | 1.05 | <.001 | .001 |
| f__Christensenellaceae;g__ | 0.01b | 0.01b | 1.02a | 2.50a | 0.17 | <.001 | <.001 |
| f__Clostridiaceae;g__ | 0.02b | 0.14a,b | 0.25a | 0.55a | 0.04 | .003 | .006 |
| g__Clostridium | 0.03b | 0.02b | 0.04a,b | 0.10a | 0.01 | .015 | .024 |
| g__Dehalobacterium | 0.33a | 0.08a | NDb | 0.02a,b | 0.03 | <.001 | <.001 |
| f__Lachnospiraceae;g__ | 1.80a | 0.64b | 0.84a,b | 1.66a | 0.13 | <.001 | .001 |
| g__Coprococcus | 0.69a | 0.12b | 0.40a,b | 0.63a | 0.06 | <.001 | .001 |
| g__Dorea | 0.21a | 0.03b | 0.21a,b | 0.40a | 0.03 | <.001 | <.001 |
| g__[Ruminococcus] | 0.89a | 0.20b | NDc | NDc | 0.08 | <.001 | <.001 |
| f__Peptococcaceae;g__ | 0.07a | 0.02a,b | NDb | 0.01b | 0.01 | <.001 | <.001 |
| f__Ruminococcaceae;Other | 1.21a | 0.53b | 0.01c | 0.01c | 0.10 | <.001 | <.001 |
| f__Ruminococcaceae;g__ | 4.28b | 1.28c | 5.23a,b,c | 11.17a | 0.67 | <.001 | .001 |
| g__Oscillospira | 5.31a | 1.23b | 0.56b | 1.41b | 0.40 | <.001 | <.001 |
| g__Ruminococcus | 0.94a | 0.40b | 0.36b | 0.63a,b | 0.06 | <.001 | .001 |
| g__Megasphaera | NDb | 0.01b | 0.31a | 0.78a | 0.05 | <.001 | <.001 |
| g__Anaerovorax | 0.04a | 0.04a | NDb | NDb | 0.00 | <.001 | <.001 |
| f__Erysipelotrichaceae;g__ | 0.17 | 0.06 | 0.04 | 0.21 | 0.03 | .124 | .141 |
| g__Allobaculum | 0.70a | 1.13a | 0.01b | NDb | 0.14 | <.001 | <.001 |
| g__[Eubacterium] | NDb | 0.01b | 0.15a | 0.15a | 0.02 | <.001 | <.001 |
| Proteobacteria | |||||||
| o__RF32;f__;g__ | 0.59a,b | 2.83a | 0.28b | 0.59a,b | 0.53 | .013 | .020 |
| g__Sutterella | 0.02b | 3.25a | 3.40a | 0.20a,b | 0.37 | <.001 | <.001 |
| f__Desulfovibrionaceae;g__ | 8.46a | 0.65a,b | 0.19b | NDb | 0.78 | <.001 | <.001 |
| g__Desulfovibrio | 0.10 | 0.09 | 0.05 | 0.17 | 0.01 | .069 | .098 |
| g__Campylobacter | 0.01b | NDb | 1.50a | 3.22a | 0.22 | <.001 | <.001 |
| g__Flexispira | 1.04 | 1.55 | 0.18 | 0.23 | 0.40 | .634 | .649 |
| g__Helicobacter | 0.38a | 0.05b | 0.23a | 0.25a | 0.04 | .001 | .001 |
| f__Enterobacteriaceae;g__ | 0.16c | 6.88a | 2.75b | 0.64b | 0.76 | .001 | .003 |
| Verrucomicrobia | |||||||
| g__Akkermansia | 13.05b | 26.35a | 0.42c | 0.02c | 2.03 | <.001 | <.001 |
| Tenericutes | |||||||
| f__Mycoplasmataceae;g__ | 0.03a,b | 0.02a,b | 1.31a | NDb | 0.14 | .012 | .020 |
| Deferribacteres | |||||||
| g__Mucispirillum | 0.48b | 0.93a | 0.04c | 0.04c | 0.13 | <.001 | <.001 |
| TM7 | |||||||
| f__F16;g__ | 0.21a | 0.08b | 0.08a,b | 0.20a | 0.03 | .021 | .032 |
| Spirochaetes | |||||||
| g__Treponema | 0.01b,c | 0.01c | 2.76a,b | 7.28a | 0.48 | <.001 | <.001 |
| Lentisphaerae | |||||||
| f__Victivallaceae;g__ | NDb | NDb | 0.10a | 0.39a | 0.03 | <.001 | <.001 |
| f__R4-45B;g__ | NDb | NDb | 0.32a | 0.61a | 0.05 | <.001 | <.001 |
NOTE. The relative abundance data (%) are presented as means ± pooled SEM. The nonparametric Kruskall–Wallis test with the Dwass, Steel, Critchlow–Fligner multiple comparisons post hoc procedure was used to compare the differences between treatment groups. The P value and FDR-adjusted P values are shown. Control, n = 14; CTαIKO, n = 11; control + antibiotics, n = 5; CTαIKO + antibiotics, n = 5. α = 0.05.
FDR, false discovery rate; ND, not detected.
Means that do not share a common letter are significantly different.
We hypothesized that a thinner mucus layer allowed microbes to infiltrate the intestinal epithelium of CTαIKO mice, and that decreasing the abundance of gut microbes with antibiotics might reduce microbial infiltration and associated inflammation. The master inflammatory cytokines interleukin (IL)1β, IL1α, and interferon-γ (P = .06) were higher in the colons of untreated CTαIKO mice compared with control mice, and antibiotics partially ameliorated their induction (Table 3). Furthermore, the proinflammatory factors granulocyte-macrophage colony-stimulating factor (GM-CSF) and leukemia inhibitory factor (LIF) were strongly induced, while monocyte chemoattractant protein-1 (MCP-1) and TNF-α also tended to be higher, in the colons of CTαIKO mice compared with control mice (Table 3). Antibiotic treatment partially blunted LIF induction, did not influence the induction of GM-CSF or TNF-α (Table 3), and further stimulated the secretion of MCP-1 in CTαIKO mice. Colonic concentrations of IL10, and a panel of other cytokines and chemokines, were not different between groups (Table 3). Therefore, changes to colonic PC concentrations induce the secretion of a specific array of inflammatory factors, and antibiotics can dampen the induction of some, but not all, of these factors. Notably, antibiotic treatment did not improve the loss of goblet cells (Figure 7E) or histopathologic colitis scores in CTαIKO mice (Figure 7F), suggesting that although luminal bacteria can exacerbate inflammation in CTαIKO mice, nonbacterial inflammatory stimuli (eg, inflammatory signaling cascades initiated within the IECs as a result of perturbations to membrane phospholipid composition) are likely the primary drivers of inflammation in these mice.
Table 3.
Colonic Cytokines and Chemokines in Control Mice and CTαIKO With and Without Antibiotics
| Protein name | Control, pg/mg protein | CTαIKO, pg/mg protein | Control + antibiotics, pg/mg protein | CTαIKO + antibiotics, pg/mg protein | P genotype | P treatment | P interaction |
|---|---|---|---|---|---|---|---|
| IL1β | 3.00 ± 1.45a | 24.49 ± 6.32b | 5.87 ± 1.42a | 13.27 ± 1.72a,b | .0002 | .18 | .03 |
| IL1α | 12.53 ± 4.09a | 32.98 ± 4.64b | 10.52 ± 3.53a | 23.41 ± 4.48a,b | .001 | .19 | .38 |
| IFNγ | 0.88 ± 0.06 | 2.86 ± 0.88 | 1.31 ± 0.36 | 2.27 ± 0.49 | .009 | .87 | .32 |
| GM-CSF | 1.02 ± 0.1a | 2.23 ± 0.26b | 1.02 ± 0.1a | 4.64 ± 0.44b | <.0001 | .001 | .001 |
| LIF | 1.14 ± 0.33a | 20.69 ± 7.69b | 1.38 ± 0.42a | 8.44 ± 1.25a,b | .001 | .08 | .07 |
| MCP-1 | 4.68 ± 1.71a | 10.23 ± 1.72a,b | 5.18 ± 1.19a | 15.6 ± 2.32b | .0005 | .12 | .2 |
| TNF-α | 0.35 ± 0.09a | 0.78 ± 0.17a,b | 0.35 ± 0.06a | 0.96 ± 0.13b | .0004 | .39 | .53 |
| IL10 | 3.82 ± 1.42 | 6.04 ± 1.07 | 5.27 ± 1.4 | 7.15 ± 0.94 | .12 | .32 | .89 |
| IL2 | 2.26 ± 0.69a | 0.78 ± 0.21a,b | 0.44 ± 0.21b | 0.29 ± 0.11b | .06 | .01 | .12 |
| IL3 | 0.46 ± 0.23 | 0.66 ± 0.21 | 0.68 ± 0.16 | 0.83 ± 0.12 | .53 | .46 | .8 |
| IL5 | 0.29 ± 0.12 | 0.38 ± 0.07 | 0.22 ± 0.07 | 1.42 ± 0.88 | .19 | .32 | .26 |
| IL6 | 1.37 ± 0.54 | 1.88 ± 0.54 | 1.48 ± 0.4 | 2.29 ± 0.29 | .16 | .57 | .74 |
| IL9 | 16.03 ± 8.58 | 3.23 ± 2.88 | 0.50 ± 0.13 | 0.30 ± 0.1 | .19 | .07 | .2 |
| IL12 (p40) | 6.40 ± 1.18 | 6.81 ± 2.23 | 7.12 ± 1.16 | 12.00 ± 2.94 | .21 | .16 | .29 |
| IL12 (p70) | 2.41 ± 1.29 | 5.19 ± 1.42 | 4.08 ± 0.79 | 6.99 ± 2.15 | .08 | .27 | .97 |
| MIP-1α | 7.00 ± 2.81 | 12.29 ± 2.04 | 8.07 ± 1.44 | 12.95 ± 1.85 | .03 | .69 | .92 |
| MIP-2 | 32.40 ± 4.48 | 50.31 ± 5.72 | 38.48 ± 3.14 | 63.55 ± 16.13 | .04 | .32 | .71 |
| RANTES | 1.60 ± 0.77 | 1.24 ± 0.43 | 0.93 ± 0.27 | 0.65 ± 0.10 | .51 | .2 | .94 |
| G-CSF | 0.85 ± 0.09 | 1.29 ± 0.22 | 0.96 ± 0.07 | 1.97 ± 0.52 | .03 | .2 | .35 |
| M-CSF | 1.63 ± 0.72 | 5.78 ± 3.02 | 1.08 ± 0.26 | 3.03 ± 1.11 | .051 | .27 | .46 |
| Eotaxin | 4.47 ± 2.62 | 3.94 ± 3.47 | 1.25 ± 0.79 | 2.10 ± 0.82 | .94 | .24 | .75 |
NOTE. Data are means ± SEM. A 2-way analysis of variance with the Tukey post-test was used. Samples that were lower than the limit of detection were assigned a value of half the limit of detection (the lowest point obtained from the standard curve).
G-CSF, granulocyte colony stimulating factor; IFN, interferon; M-CSF, macrophage colony-stimulating factor; MIP, macrophage inflammatory protein; RANTES, Regulated upon Activation, Normal T Cell Expressed and Presumably Secreted.
Columns that do not share a letter are significantly different (α = .05).
PC Depletion in IECs Leads to ER Stress and Unfolded Protein Response Activation
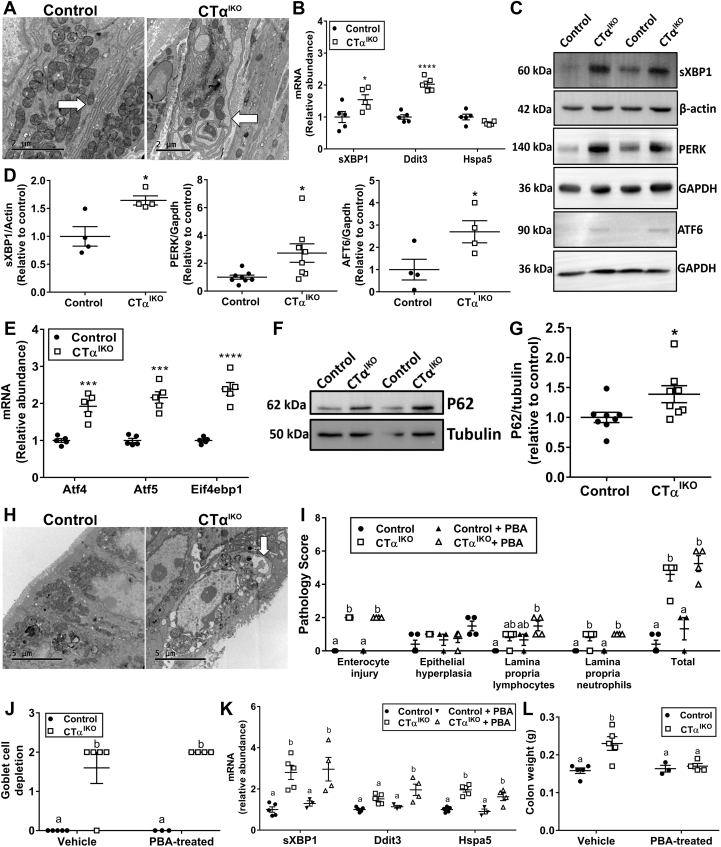
Electron microscopy showed that the ER in colonic IECs of CTαIKO mice appeared dilated and distended (Figure 8A). The unfolded protein response (UPR) can be activated independently of unfolded proteins in the ER by perturbations to membrane lipid composition resulting in lipid bilayer stress.28, 29, 30, 31 Accordingly, both mRNA (Figure 8B) and protein levels (Figure 8C and D) of spliced X-box binding protein 1 (XBP1), the major downstream target of the UPR initiator inositol-requiring enzyme 1-α (IRE1α), were higher in the colons of CTαIKO mice compared with control mice. Furthermore, protein levels of the ER stress sensors protein kinase R-like ER kinase (PERK) and activating transcription factor 6 (ATF6) were increased significantly in the colons of CTαIKO mice (Figure 8C and D). The mRNA levels the downstream UPR targets Ddit3, Atf4, Atf5, and Eif4ebp1 also were robustly higher in the colons of CTαIKO mice (Figure 8B and E), while Hspa5 levels were higher in some, but not all, experiments (Figure 8B and I). Furthermore, colonic epithelial cells of CTαIKO mice had higher P62 protein levels than those of control mice (Figure 8F and G) and contained numerous autophagic vesicles (Figure 8H), as has been reported previously during pathologic UPR activation.32 Thus, perturbations to membrane phospholipid composition results in ER stress and activation of the UPR in IECs of CTαIKO mice.
Figure 8.
Phosphatidylcholine depletion in IECs leads to ER stress and UPR activation. (A) Representative electron micrographs of the ER in colonic epithelial cells of control mice and CTαIKO mice. Arrow indicates ER. (B) The mRNA abundance of sXBP1, Ddit3, and Hspa5 in the colons of control mice and CTαIKO mice (n = 5/group). (C) Representative Western blots and (D) quantification of spliced XBP1, PERK, and ATF6 relative to loading controls in the colons of control mice and CTαIKO mice (n = 4–8/group). (E) The mRNA abundance of Atf4, Atf5, and Eif4ebp1 in the colons of control mice and CTαIKO mice (n = 5/group). (F) Representative Western blot and (G) quantification of P62 relative to tubulin in the colons of control mice and CTαIKO mice (n = 8/group). (H) Representative electron micrographs of autophagic vesicles in colonic epithelial cells of control mice and CTαIKO mice. Arrow indicates autophagosome. (I) Pathology scores for control mice and CTαIKO mice treated with and without PBA (n = 3–5/group). (J) Goblet cell depletion scores for control mice and CTαIKO mice treated with and without PBA (n = 3–5/group). (K) The mRNA abundance of sXBP1, Ddit3, and Hspa5 in the colons of control mice and CTαIKO mice treated with and without PBA (n = 3–5/group). (L) Colon weight of control mice and CTαIKO mice treated with and without PBA (n = 3–5/group). Values are means ± SEM. (I–L) Columns that do not share a letter (a or b) are significantly different (α = .05). ∗P < .05, ∗∗∗P < .001, and ∗∗∗∗P < .0001. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; sXBP1, spliced XBP1.
We next treated mice with 4-phenyl butyrate (PBA), a chemical chaperone that alleviates ER stress driven by misfolded protein accumulation33 but that fails to alleviate ER stress arising from perturbations to membrane lipid composition.29,34 PBA failed to improve total pathology scores (Figure 8I), goblet cell depletion (Figure 8J), or the colonic ER stress markers spliced X-box binding protein 1 (sXBP1), Ddit3, or Hspa5 (Figure 8K) in CTαIKO. PBA also further increased the levels of GM-CSF, decreased IL10 and IL12 (p40) concentrations, and did not affect colonic TNF-α or LIF in CTαIKO mice compared with control mice (Table 4). However, colonic IL1β concentrations, which were 10-fold higher in untreated CTαIKO mice compared with control mice, were normalized with PBA treatment (Table 4). Furthermore, MCP-1 concentrations, which were increased in CTαIKO mice compared with controls, were equalized between groups after PBA treatment (Table 4). Surprisingly, PBA treatment completely prevented the increase in colon weight observed in untreated CTαIKO mice (Figure 8L). Together, these data show that PBA is largely ineffective at improving colitis pathology in CTαIKO mice, which is consistent with a primary role for lipid bilayer stress, as opposed to protein misfolding, in the initiation of ER stress in CTαIKO mice. However, impaired protein folding, likely occurring secondary to changes in ER function, appears to exacerbate inflammation after perturbation to IEC phospholipid composition because PBA treatment blunted the induction of IL1β and MCP-1 in the colons of CTαIKO mice.
Table 4.
Colonic Cytokines and Chemokines in Control Mice and CTαIKO With and Without PBA
| Protein | Control, pg/mg protein | CTαIKO, pg/mg protein | Control + PBA, pg/mg protein | CTαIKO + PBA, pg/mg protein | P genotype | P treatment | P interaction |
|---|---|---|---|---|---|---|---|
| IL1β | 6.94 ± 2.93a | 48.86 ± 15.41b | 9.16 ± 4.35a | 8.32 ± 2.92a | .03 | .4 | .2 |
| MCP-1 | 2.13 ± 1.28a | 12.02 ± 2.4b | 6.57 ± 3.24a | 6.88 ± 2.3a | .04 | .88 | .05 |
| GM-CSF | 0.58 ± 0.0a | 2.53 ± 0.75a | 0.87 ± 0.29a | 5.13 ± 0.6b | <.0001 | .01 | .04 |
| IL10 | 2.86 ± 0.97a,b | 6.26 ± 1.37a | 6.74 ± 0.44a | 2.21 ± 0.41b | .57 | .93 | .002 |
| IL12 (p40) | 5.33 ± 2.28a,b | 8.31 ± 2.80a,b | 12.50 ± 0.78a | 1.38 ± 0.45b | .07 | .95 | .006 |
| TNF-α | 0.28 ± 0.13a | 1.75 ± 0.49b | 0.45 ± 0.17a | 1.86 ± 0.25b | .0004 | .63 | .91 |
| LIF | 0.71 ± 0.3a | 7.45 ± 1.65b | 0.58 ± 0.12a | 5.27 ± 0.53b | <.0001 | .23 | .28 |
| IL12 (p70) | 0.64 ± 0.48 | 4.17 ± 2.19 | 6.41 ± 3.18 | 0.1 ± 0.02 | .41 | .61 | .01 |
| IL6 | 0.85 ± 0.41 | 2.18 ± 0.70 | 2.76 ± 1.12 | 1.18 ± 0.33 | .84 | .47 | .04 |
| IL2 | 2.88 ± 0.66 | 2.11 ± 0.66 | 1.52 ± 0.68 | 4.21 ± 0.64 | .18 | .59 | .03 |
| IL3 | 0.24 ± 0.15 | 0.53 ± 0.19 | 0.73 ± 0.31 | 0.09 ± 0.01 | .34 | .88 | .02 |
| M-CSF | 0.43 ± 0.22 | 1.36 ± 0.33 | 0.94 ± 0.38 | 0.52 ± 0.02 | .35 | .54 | .03 |
| MIP-1α | 2.99 ± 0.75 | 4.07 ± 0.92 | 2.44 ± 1.01 | 5.36 ± 0.42 | .03 | .65 | .27 |
| Eotaxin | 5.07 ± 2.51 | 22.15 ± 10.87 | 0.61 ± 0.26 | 19.3 ± 3.73 | .01 | .56 | .9 |
| IFNγ | 0.41 ± 0.08 | 0.96 ± 0.19 | 0.77 ± 0.37 | 0.62 ± 0.10 | .29 | .98 | .07 |
| G-CSF | 1.23 ± 0.33 | 1.91 ± 0.43 | 1.07 ± 0.25 | 1.50 ± 0.23 | .13 | .42 | .72 |
| IL1α | 20.14 ± 1.90 | 26.57 ± 2.89 | 22.04 ± 7.77 | 25.99 ± 5.38 | .26 | .88 | .78 |
| IL5 | 0.38 ± 0.12 | 0.20 ± 0.09 | 0.08 ± 0.05 | 0.28 ± 0.10 | .92 | .32 | .08 |
| IL9 | 16.91 ± 4.77 | 8.00 ± 2.83 | 11.62 ± 9.76 | 18.17 ± 6.27 | .84 | .68 | .21 |
| MIP-2 | 28.70 ± 8.55 | 59.90 ± 11.34 | 42.53 ± 3.31 | 41.02 ± 1.86 | .15 | .98 | .11 |
| RANTES | 1.57 ± 0.87 | 2.73 ± 0.92 | 1.22 ± 0.21 | 2.26 ± 0.53 | .18 | .65 | .99 |
NOTE. Data are means ± SEM. A 2-way analysis of variance with the Tukey post-test was used. Samples that were lower than the limit of detection were assigned a value of half the limit of detection (the lowest point obtained from the standard curve).
G-CSF, granulocyte colony stimulating factor; IFN, interferon; M-CSF, macrophage colony-stimulating factor; MIP, macrophage inflammatory protein; RANTES, Regulated upon Activation, Normal T Cell Expressed and Presumably Secreted.
Columns that do not share a letter are significantly different (α = .05).
ER Stress Induced by Altered Membrane Lipid Composition Drives IEC Necroptosis
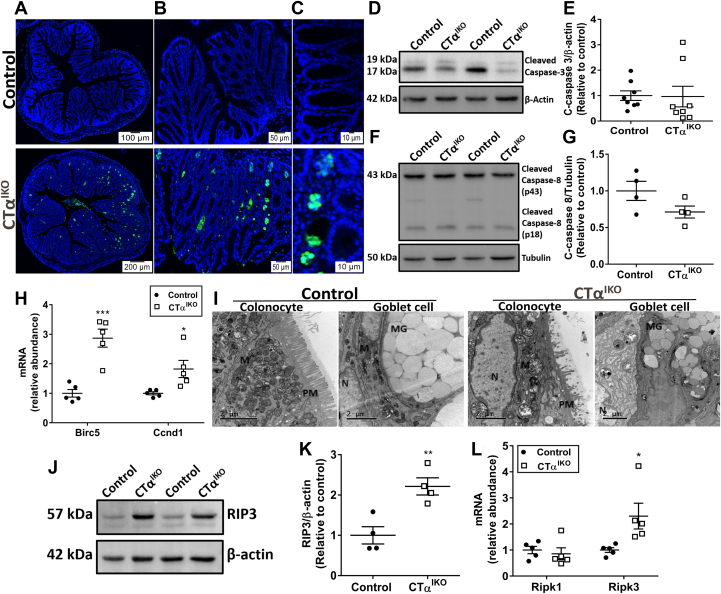
Terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling (TUNEL) staining, which detects DNA fragmentation caused by apoptosis, necroptosis, or pyroptosis, was more intense in the colons of CTαIKO mice compared with control mice (Figure 9A–C). TUNEL staining was especially prominent in colonic crypts, which typically are filled with goblet cells (Figure 9C). Pyroptotic cell death induced after microbial invasion of IECs likely contributes to TUNEL-positive staining in the colons of CTαIKO mice (Figure 6D). However, because makers of pyroptosis were increased only modestly in CTαIKO mice, we investigated whether other forms of cell death were involved in the loss of goblet cells before microbial invasion of the epithelium. Surprisingly, protein levels of both cleaved caspase-3 and cleaved caspase-8, critical mediators of apoptosis, tended to be lower in the colons of most CTαIKO mice compared with controls (Figure 9D–G). Furthermore, mRNA levels of the anti-apoptotic (prosurvival) factors Birc5 (encodes survivin) and Ccnd1 (encodes cyclin D1) were higher in the colons of CTαIKO mice compared with controls (Figure 9H). These data suggest that apoptosis is not a major contributor to TUNEL-positive staining in the colons of CTαIKO mice.
Figure 9.
ER stress induced by altered membrane lipid composition drives IEC necroptosis in CTαIKO mice. (A–C) Representative TUNEL staining in the colons of control mice and CTαIKO mice. (D) Representative Western blot of cleaved caspase-3 and (E) quantification of cleaved caspase-3 relative to β-actin in the colons of control mice and CTαIKO mice (n = 4/group). (F) Representative Western blot of cleaved caspase-8 and (G) quantification of cleaved caspase-8 relative to tubulin in the colons of control mice and CTαIKO mice (n = 8/group). (H) The mRNA abundance of Birc5 and Ccnd1 in the colons of control mice and CTαIKO mice (n = 5/group). (I) Representative electron micrographs of necroptotic features in colonocytes and goblet cells of CTαIKO mice compared with control mice. Note swollen mitochondria (M), ruptured plasma membrane (PM), and nuclei (N) without chromatin condensation. (J) Representative Western blot of RIP3 and (K) quantification of RIP3 in the colons of control mice and CTαIKO mice (n = 4/group). (L) The mRNA abundance of Ripk1 and Ripk3 in the colons of control mice and CTαIKO mice (n = 5/group). Values are means ± SEM. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. MG, _____.
Necroptosis, a form of programmed cell death that shares the subcellular characteristics of necrosis, is regulated negatively by caspases, and is initiated by RIP3 (encoded by Ripk3), has been implicated recently in human IBD.35,36 Hallmarks of necroptosis were immediately evident in IECs of CTαIKO mice by electron microscopy (Figure 9I), including swollen mitochondria and ER, disrupted plasma membranes, and many cytoplasmic vacuoles. The lack of chromatin condensation in the nuclei of cells with swollen mitochondria further suggested that the cells were necroptotic as opposed to apoptotic.37 Importantly, the mRNA and protein levels of the major mediator of necroptosis, RIP3, were strongly induced in the colons of CTαIKO mice compared with control mice (Figure 9J–L). Swollen mitochondria and cytoplasmic vacuoles were prominent in cells containing damaged mucus granules (Figures 9I). Together, these data show that phospholipid imbalance in IECs induces an ER stress response that promotes IEC death by necroptosis, loss of barrier function, microbial infiltration, and spontaneous colitis in mice.
Discussion
The present study shows that disturbances to membrane phospholipid composition in IECs leads to an ER stress response that drives spontaneous colitis in mice. The spontaneous nature of colitis development in CTαIKO mice (ie, in the absence of aggravating factors such as dextran sodium sulfate) points to a particularly important role for membrane PC content in maintaining gut barrier function. CTαIKO mice experience severe colonic ER stress that results in IEC necroptosis, a form of cell death that recently was implicated in human IBD.35,36,38,39 IEC death is linked to goblet cell depletion, crypt abscess formation, increased intestinal permeability, microbial invasion of the intestinal epithelium, immune cell infiltration, and augmented inflammatory cytokine secretion in CTαIKO mice. Thus, phospholipid disequilibrium in murine IECs leads to a form of colitis that closely resembles that seen in human beings.2 An important role for PC in maintaining goblet cell function correlates with human clinical observations because UC patients typically have low colonic PC concentrations with goblet cell mucus granule depletion.3, 4, 5
Impaired ER homeostasis can arise for many reasons including viral infections, drug toxicity, impaired calcium or redox balance, and lipid accumulation or depletion.10,40 Eukaryotes have evolved the UPR to relay information regarding stressful conditions in the ER to the nucleus, where adaptive changes to cellular gene expression or cell fate decisions occur.40 Accumulation of unfolded or misfolded proteins in the lumen of the ER is a well-characterized activator of the UPR, as its name suggests.40 In the colon, accumulation of misfolded MUC2 promotes ER stress and UC-like inflammation in mice.19 Furthermore, slowing the accumulation of misfolded proteins with the chemical chaperones PBA or tauroursodeoxycholic acid reduces colitis severity in several genetically modified mouse models.33 In addition to misfolded protein accumulation, the UPR can be activated directly by perturbations to membrane lipid composition, resulting in lipid bilayer stress.28, 29, 30, 31 Our experiments show that perturbations to IEC phospholipid composition lead to ultrastructural changes to the ER; activation of PERK, ATF6, and IRE1α; and colitis development. These data highlight the physiological consequences of lipid bilayer stress in the colon. The ER stress sensors IRE1α and PERK sense unfolded proteins with their luminal domains, but sense perturbations to membrane lipid composition by an alternative mechanism involving their transmembrane domains.28,29 Furthermore, the UPR activator ATF6 responds to proteotoxic and lipotoxic stress by distinct mechanisms.34 Importantly, PBA is largely ineffective at resolving the form of ER stress that arises due to perturbations to membrane lipid composition.29,34 PBA did not improve colitis scores or rescue goblet cell depletion in CTαIKO mice, consistent with a primary role for lipid bilayer stress in colitis development after IEC PC depletion. However, the accumulation of misfolded proteins secondary to changes in ER structure might exacerbate inflammation in CTαIKO mice because PBA ameliorated the induction of a subset of proinflammatory cytokines including IL1β. PBA treatment also prevented the increase in colon mass observed in CTαIKO mice, an effect that might be linked to its decreasing of colonic levels of the prosurvival chemokine MCP-1.41 Antihypertrophic effects of PBA have been reported previously in pressure overload–induced myocardial hypertrophy.42
Consistent with our results highlighting the importance of maintaining membrane lipid equilibrium in IECs, pancreatic β cells loaded with saturated fatty acids,43 macrophages loaded with cholesterol,44 and the livers of mice fed a high-fat diet45 have high levels of ER stress. Lipids that increase membrane saturation (ie, reduce membrane fluidity), in particular, have been shown to promote UPR activation.28,43,46,47 For example, loading cells with palmitate increases the content of saturated fatty acid–containing phospholipids in cellular membranes before induction of ER stress response pathways and cell death.48 Similarly, CTαIKO mice have a lower ratio of PC/PE in IEC membranes (a change predicted to increase membrane saturation) relative to control mice, which is linked to a strong induction of the UPR and cell death. Overexpression of spliced XBP1 in fibroblasts increases CTα activity and PC synthesis, linking XBP1 to PC synthesis for membrane expansion during ER stress.11 In CTαIKO mice, impaired PC synthesis triggers XBP1 splicing, but CTα deficiency in these mice does not allow subsequent membrane expansion. The uncontrolled inflammation that develops in CTαIKO mice highlights the importance of PC synthesis for the prevention and resolution of colonic ER stress. Although immune cell–derived cytokines likely exacerbate ER stress in CTαIKO mice,49 the use of a villin promotor traces colonic pathology in CTαIKO mice to IECs. Furthermore, although our data show that dietary phospholipid treatment might not be a viable way to improve colitis in CTαIKO mice, likely owing to phospholipid absorption in the proximal small intestine, future studies should examine the effects of local phospholipid delivery on ER stress and associated inflammation in UC.
We found using H&E staining, Alcian Blue staining, electron microscopy, quantitative polymerase chain reaction (PCR) of goblet cell markers, and assessment of the mucus layer that CTαIKO mice lose goblet cells from the colonic epithelium by 4 days after Cre induction. A primary reason for this loss of goblet cells appears to be induction of IEC necroptosis, as indicated by TUNEL-positive staining localized to colonic crypts, electron micrographs of necroptotic goblet cells, and the induction of RIP3 without induction of cleaved caspase-3 in colonic epithelial cells of CTαIKO mice. Pyroptosis also might contribute to IEC death after breakdown of the mucosal barrier and microbial infiltration of the epithelium17,21, 22, 23, 24 because mRNA levels of Casp4 and Gsdmd were increased modestly in the colons CTαIKO mice. It traditionally has been reported that unresolved ER stress leads to cell death by activation of the intrinsic mitochondrial apoptotic pathway.40 However, it increasingly is appreciated that severe ER stress also can induce necroptosis.50,51 Importantly, necroptosis is active in human IBD.35,36,38,39 It should be noted that goblet cells are not the only IEC type to undergo necroptosis in the colons of CTαIKO mice because electron microscopy also showed necroptotic features in IECs without mucus granules. However, loss of goblet cells is predicted to be particularly detrimental to mucosal barrier function owing to their role in mucus production and secretion of various protective factors including trefoil factor 3 (TFF3).52 Electron microscopy of goblet cells that remained in the colons of CTαIKO mice showed that the integrity of mucus-containing theca appeared compromised, with loss of mucus granule structure and infiltration of cellular debris and vacuoles. Reasons for subcellular damage to goblet cell theca in CTαIKO mice might include impaired intracellular membrane integrity or swelling of organelles upon initiation of necroptosis. It is conceivable that impairments to ER function might be especially detrimental to goblet cells because they must continuously produce, fold, and secrete massive quantities of the MUC2 glycoprotein, as has been suggested previously.30 Furthermore, CTαIKO mice appear unable to replace mature goblet cells owing to transcriptional repression of the goblet cell maturation factors Gfi1, Klf4, and Spdef. An inability to produce mature IECs is a feature of colitis-associated cancer53 that, when also considering the 50% increase in colon mass within 7 days of Cre induction, might be owing to high mucosal concentrations of the prosurvival factors LIF and MCP-1 in CTαIKO mice.41,54
Given that UC onset typically occurs in young adulthood, the use of a tamoxifen-inducible Cre-recombinase system allowing us to delete CTα in young adult mice was an advantage in these experiments. CTαIKO mice rapidly lose weight upon Cre induction before beginning to regain body weight on day 5 and arriving at a similar body weight to controls by day 7, although pathology scoring showed that severe colitis remained in CTαIKO mice on day 7. The colonic epithelium of CTαIKO mice, however, was substantially less damaged at 7 weeks after Cre induction when compared 4 days or 7 days after Cre induction. These observations suggest that epithelial cells of CTαIKO mice activate compensatory pathways to promote mucosal healing after the initial inflammatory response. A similar pattern of acute and rapid weight loss followed by weight regain and colonic restitution has been observed previously in mice infected by Citrobacter rodentium, a murine pathogen that can subvert the colonic mucus barrier and cause colitis.55 Although some of the epithelial repair mechanisms induced after Citrobacter infection, such as production of anti-inflammatory or antimicrobial factors,55,56 might apply to CTαIKO mice, a rewiring of cellular lipid metabolism likely also is required. An example of a cellular adaptation to limited PC supply was reported previously with the slowing of PC catabolism in the brains of mice fed a diet deficient in choline, the dietary precursor of PC.57 The course of severe and debilitating colonic inflammation followed by gradual epithelial restitution observed in CTαIKO mice is comparable with that seen in human IBD and will make CTαIKO mice a useful model for studying mechanisms of epithelial repair and return to homeostasis after bouts of colitis.
We reported in our previous article16 that the small intestines of CTαIKO mice look overtly normal and, unlike the colons of CTαIKO mice, do not have obvious structural damage. Severe colonic pathology with overtly normal small intestinal morphology also was observed previously in MUC2-deficient mice.1 It is conceivable that the colons of CTαIKO mice are affected more severely by disturbances to PC synthesis than their small intestines because there is a constant supply of PC to the small intestine in bile, whereas very little biliary PC reaches the colon.14,15 We also reported previously that CTαIKO mice have small intestinal lipid malabsorption.16 There is evidence from other murine models that intestinal inflammation and lipid malabsorption are linked. For example, mice lacking B cells (which secrete IgA to restrict bacterial interaction with the epithelium) have enhanced interferon-related immune responses in the gut, which is linked to lipid malabsorption.58 Furthermore, enteric infection of mice with C rodentium has been shown to induce diarrhea and water efflux to promote pathogen clearance.59 Impaired dietary lipid absorption also has been described in patients with UC.60, 61, 62 Future studies will investigate the mechanisms underlying the relationship between intestinal inflammation and dietary lipid malabsorption in CTαIKO mice.
Although most CTαIKO mice experience body weight loss upon Cre induction, a minority of mice also experience severe wasting that requires euthanasia. This striking response to colonic PC depletion might be owing in part to loss of blood through the injured bowel, as indicated by low circulating levels of red blood cells, hemoglobin, and hematocrit. However, additional inflammatory stimuli likely also play a role. First, necroptotic cells undergo plasma membrane permeabilization and release their cellular contents (including cytokines such as IL1α) to amplify local inflammation and recruit neutrophils and lymphocytes to affected tissues.63 Furthermore, ER stress has been shown to independently activate inflammatory signaling pathways and proinflammatory cytokine secretion.49 Blockage of protein translation by induction of Eif4ebp1 (likely leading to lower secretion of mucus components) also could contribute to disease pathogenesis in CTαIKO mice. High concentrations of colonic proinflammatory cytokines promote tight junction dysfunction, as indicated by altered expression of claudins and substantially increased gut permeability to FITC-labeled dextran in CTαIKO mice. Increased intestinal permeability, combined with a thin mucus layer, allows microbes to infiltrate the epithelium, as indicated by higher circulating levels of lipocalin 246,47 and high colonic concentrations of the cytosolic bacterial DNA sensor ZBP1 in CTαIKO mice, generating uncontrolled inflammation and further immune cell recruitment to the colon. Consistent with a role for microbes in exacerbating inflammation in CTαIKO mice, microbe depletion by antibiotic treatment reduces colonic IL1β and LIF concentrations in CTαIKO mice without affecting goblet cell depletion, total pathology scores, or the induction of GM-CSF or MCP-1. Therefore, ER stress, necroptotic signals, cytokines, enhanced intestinal permeability, microbes, and mucus layer depletion promote inflammation in CTαIKO mice.
In conclusion, the present study shows that de novo PC synthesis is required to maintain the intestinal mucosal barrier by protecting IECs against ER stress and subsequent nonapoptotic cell death. CTαIKO mice will be a useful model for studying aspects of UC pathogenesis.
Materials and Methods
Mice
Mice were housed in a temperature-controlled room with a 12-hour light–dark cycle and free access to food and water. Generation of Pcyt1aLoxP/LoxP;villin-CreERT2 and Pcyt1aLoxP/LoxP mice has been described previously.16 Cre was induced in age-matched 8- to 20-week-old female Pcyt1aLoxP/LoxP;villin-CreERT2 (CTαIKO) mice fed a chow diet (5001; Lab Diet, St. Louis, MO) by intraperitoneal injection of tamoxifen (1 mg/d in sunflower oil for 5 days), while tamoxifen-treated Pcyt1aLoxP/LoxP mice were used as controls. Twenty-four hours after the end of tamoxifen treatment (time 0), mice were placed on a semipurified diet (40% fat, 20% protein, 40% carbohydrate16) for either 4 days, 7 days, or 7 weeks until termination, as indicated. Colon sections, colon Swiss rolls, or cecum sections were fixed in 10% neutral-buffered formalin for histology. Colonic epithelial cells and the overlying mucus layer were collected as previously described.64 Briefly, colons were flushed with a solution containing 0.154 mol/L NaCl and 1 mmol/L dithiothreitol to remove contents. Colons were next ligated, filled with phosphate-buffered saline, and incubated at 37°C for 15 minutes. The phosphate-buffered saline then was replaced with phosphate-buffered saline containing 1.5 mmol/L ethylenediaminetetraacetic acid and 0.5 mmol/L dithiothreitol before being incubated for a further 30 minutes at 37°C. After 30 minutes, 1 ligature was removed, and colonic epithelial cells were collected. Samples were frozen at -80°C before being used for phospholipid analysis or Western blot. Whole blood was collected in ethylenediaminetetraacetic acid–coated tubes containing a protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO) for the measurement of complete blood counts on a Siemens ADVIA 2120i Hematology System (Washington, DC), or for the measurement of plasma lipocalin 2 (R&D Systems, Minneapolis, MN). A group of control mice and CTαIKO mice were given an antibiotic cocktail (500 mg/L bacitracin, 1 g/L neomycin, and 500 mg/L vancomycin) in the drinking water or no antibiotics from the time of first tamoxifen injection until death (10 days), as described previously.65 A separate group of CTαIKO mice and control mice were administered PBA (500 mg/kg body weight; Scandinavian Formulas, Sellersville, PA), dissolved in phosphate-buffered saline or vehicle twice daily by oral gavage from the time of first tamoxifen injection until death (10 days), as described previously.33 A third group of CTαIKO mice and control mice were fed either a diet containing 4 times the recommended level of choline as PC or a control diet that was matched to the experimental diet for total calories and total choline content (Table 5). The University of Alberta’s Institutional Animal Care Committee approved all animal procedures, which were in accordance with guidelines of the Canadian Council on Animal Care.
Table 5.
PC-Supplemented Diet and Control Diet Ingredients
| Ingredients (grams) | Control diet | PC diet |
|---|---|---|
| Caseina | 270 | 270 |
| Corn starcha | 170.65 | 170.65 |
| Sucroseb | 195.35 | 195.35 |
| Cellulosea | 80 | 80 |
| AIN-93-VX vitamin mixa | 19 | 19 |
| Bernhart–Tomarelli mineral mixa | 50 | 50 |
| Calcium phosphate dibasicb | 3.4 | 3.4 |
| Myo-inositolb | 6.3 | 6.3 |
| L-cystineb | 1.8 | 1.8 |
| Choline bitartrateb | 10.0 | 2.5 |
| Crisco vegetable oilc | 32 | 23 |
| Mazola corn oild | 10 | 10 |
| Larde | 155 | 127 |
| DHAscof | 1.5 | 1.5 |
| Arascof | 1.5 | 1.5 |
| PC (soy lecithin)g | 0 | 90 |
Harlan Teklad (Indianapolis, IN).
Sigma.
Crisco J.M. Smucker Company (Orrville, OH).
Mazola ACH Food Companies, Inc (Oakbrook Terrace, IL).
TenderFlake (Chicago, IL).
DSM Nutritional Products, Inc (Heerlen, The Netherlands).
General Nutrition Company (product number 005648; Pittsburgh, PA).
All authors had access to the study data and reviewed and approved the final manuscript.
Microscopy
Formalin-fixed, paraffin-embedded tissue slices (5 μm) were stained with H&E or Alcian blue/periodic acid–Schiff and visualized with a light microscope (Zen, AxioCamMR3; Zeiss, Dublin, CA). TUNEL staining was performed using the In Situ Cell Death Detection Kit (Sigma-Aldrich) and images were obtained with a fluorescence microscope (Olympus, Markham, Ontario, Canada) with Surveyor (Objective Imaging Inc, Kansasville, WI) and Image-Pro Plus (Media Cybernetics Inc, Rockville, MD) software. For electron microscopy, 2-cm colonic rings were fixed with 2.5% glutaraldehyde in 0.1 mol/L phosphate buffer (pH 7.2) and 2% paraformaldehyde. Sections were cryosectioned with an ultramicrotome (Ultracut E; Reichert-Jung, Buffalo Grove, IL) and images were obtained using a Philips 410 transmission electron microscope (Eindhoven, The Netherlands), as previously described.16
Histopathologic Scoring of Colitis
H&E-stained distal colon sections were assessed by an experienced pathologist who was blinded to the experimental groupings. The pathologist assigned a colitis severity score based on a previously established protocol66 that included assessment of epithelial hyperplasia, enterocyte injury, and the presence of lymphocytes and neutrophils in the lamina propria, as outlined in Table 6. The pathologist also assigned a score for goblet cell depletion using the following scale: 0, no goblet cell depletion; 1, modest goblet cell depletion; and 2, substantial goblet cell depletion.
Table 6.
Histopathologic Colitis Scoring System
| Group | Description | Score |
|---|---|---|
| Enterocytes | ||
| Normal | Rare epithelial lymphocytes | 0 |
| Mild | Intraepithelial neutrophils | 1 |
| Moderate | Mucosal necrosis and/or luminal pus | 2 |
| Severe | Necrosis muscularis mucosa | 3 |
| Epithelial hyperplasia | ||
| Normal | 0 | |
| Mild | 1 | |
| Moderate | 2 | |
| Pseudopolyps | 3 | |
| Lamina propria mononuclear infiltrate | ||
| Normal | 1 small lymphoid aggregate | 0 |
| Slightly increased | >1 small aggregate | 1 |
| Markedly increased | Large aggregates and/or greatly increased single cells | 2 |
| Lamina propria neutrophil infiltrate | ||
| Normal | 0 | |
| Slightly increased | 1 | |
| Markedly increased | 2 |
Lipid Measurements
The protein content of colonic epithelial cells was determined by bicinchoninic acid assay (Thermo Scientific, Waltham, MA), and lipids were extracted from homogenates by the method of Folch et al.67 PC and PE were separated on silica plates (EM1.05721.0001; VWR, Radnor, PA) by thin-layer chromatography using the solvent system of chloroform:methanol:acetic acid:water (50:30:8:4). Plates were exposed to iodine for visualization, PC and PE bands were scraped into glass tubes, and lipids were liberated from the silica by heating with perchloric acid (77230; Sigma) for 60 minutes at 180°C. Subsequently, 0.5 mL ammonium molybdate (431346; Sigma) and L-ascorbic acid (255564; Sigma) was added to the samples before heating at 95°C for 15 minutes. The phosphorous content of samples was determined by measuring the absorbance of the samples at 820 nm on a spectrophotometer (Spectra Max Pro; Molecular Devices, San Jose, CA) and comparing the sample absorbance with the absorbance of a phosphorous standard curve (53139; Sigma), as described previously.68
Cytokine and Chemokine Concentrations
Sections of the distal colon were homogenized in buffer containing a protease inhibitor cocktail (Sigma-Aldrich) and dithiothreitol (Sigma-Aldrich). The protein concentration of the supernatant was determined by bicinchoninic acid assay (Thermo Scientific) after centrifugation at 10,000 rpm for 10 minutes to remove debris. Samples were adjusted to 3 mg protein/mL homogenate, and cytokine and chemokine concentrations were determined using Multiplex LASER Bead Technology (MD31; Eve Technology, Calgary, Canada).
mRNA Isolation and Quantitation by PCR
Total RNA was isolated from frozen colonic tissue using TRIzol (Invitrogen, Carlsbad, CA), as described previously.16 Superscript II (Invitrogen) was used to reverse-transcribe isolated RNA. Quantitative PCR was run for 40 cycles on a StepOne Plus system (Applied Biosystems, Waltham, MA) using Power SYBR Green PCR Master Mix (Thermo Fisher Scientific, Waltham, MA). Quantitation was performed using the standard curves method. Relative mRNA expression was normalized to Rplp0. Primer sequences and gene names are listed in Table 7.
Table 7.
Quantitative PCR Primer Sets
| Gene symbol | Gene name | Forward primer sequence | Reverse primer sequence |
|---|---|---|---|
| Mouse | |||
| Muc2 | Mucin 2 | CCATTGAGTTTGGGAACATGC | TTCGGCTCGGTGTTCAGAG |
| Tff3 | Trefoil factor 3 | CTGGGATAGCTGCAGATTACG | CATTTGCCGGCACCATAC |
| Agr2 | Anterior gradient 2, protein disulfide isomerase family member | CCTCAACCTGGTCTATGAAACA | ACCGTCAGGGATGGGTCT |
| Gfi1 | Growth factor independent 1 transcriptional repressor | ATGTGCGGCAAGACCTTC | ACAGTCAAAGCTGCGTTCCT |
| Spdef | SAM pointed domain containing ETS transcription factor | GATGTACTGCATGCCCACCT | GGAGGCGCAGTAGTGAAGG |
| Klf4 | Kruppel-like factor 4 | CCGTCCTTCTCCACGTTC | GAGTTCCTCACGCCAACG |
| Zbp1 | Z-DNA binding protein 1 | CAGGAAGGCCAAGACATAGC | GACAAATAATCGCAGGGGACT |
| Cldn2 | Claudin 2 | TGTGAATGAACTGAAGGAAAGC | ATCCTGCACCCAGCTGTATT |
| Cldn4 | Claudin 4 | TTTTGTGGTCACCGACTTTG | TGTAGTCCCATAGACGCCATC |
| Xbp1 (spliced) | X-box binding protein 1 (spliced isoform) | GAGTCCGCAGCAGGTG | GTG TCA GAG TCC ATG GGA |
| Ddit3 | DNA damage inducible transcript 3 | GCGACAGAGCCAGAATAACA | GATGCACTTCCTTCTGGAACA |
| Atf4 | Activating transcription factor 4 | CTCAGACACCGGCAAGGA | TCATCCAACGTGGTCAAGAG |
| Atf5 | Activating transcription factor 5 | GCAGCACCTAGGGTACAGGT | CGCTGGAGACAGACGTACAC |
| Hspa5 | Heat shock protein family A (Hsp70) member 5 | CTGAGGCGTATTTGGGAAAG | TCATGACATTCAGTCCAGCAA |
| Eif4ebp1 | Eukaryotic translation initiation factor 4E binding protein 1 | GATGAGCCTCCCATGCAA | AATGTCCATCTCAAATTGTGACTC |
| Birc5 | Baculoviral IAP repeat-containing 5 | TGATTTGGCCCAGTGTTTTT | CAGGGGAGTGCTTTCTATGC |
| Ccnd1 | Cyclin D1 | GCACAACGCACTTTCTTTCC | TCCAGAAGGGCTTCAATCTG |
| Rplp0 | Ribosomal protein lateral stalk subunit P0 | ACTGGTCTAGGACCCGAGAAG | CTCCCACCTTGTCTCCAGTC |
| Neurog3 | Neurogenin 3 | ACTGCTGCTTGTCACTGACTG | ATGGTGAGCGCATCCAAG |
| Sct | Secretin | GCTGTGGTCGAACACTCAGA | GAGACAGGGACCCATCCAG |
| Insl5 | Insulin-like 5 | GCATTTCCACTCTCAACAAGC | GATGGCTCGTGCCTGTCTA |
| Pcyt1a | Phosphate cytidylyltransferase 1, choline, α isoform | GCTAAAGTCAATTCGAGGAA | CATAGGGCTTACTAAAGTCAACT |
| Casp4 | Caspase 4 (caspase 11) | GTGGTGAAAGAGGAGCTTACAGC | GCACCAGGAATGTGCTGTCTGA |
| Target | |||
|---|---|---|---|
| Gsdmd | Gasdermin D | GGTGCTTGACTCTGGAGAACTG | GCTGCTTTGACAGCACCGTTGT |
| Bacteria | |||
| UniF340/UniR514 | All bacteria | ACTCCTACGGGAGGCAGCAGT | ATTACCGCGGCTGCTGGC |
ETS, _; IAP, __; SAM, ___; Z-DNA, ____.
Western Blot
Colonic epithelial cells containing 40–50 μg of protein were resolved on a sodium dodecyl sulfate–polyacrylamide gel before being transferred to a polyvinylidene difluoride membrane and probed with antibodies against CTα (gift from Dr R. K. Mallampalli), spliced XBP1 (D2C1F, 12782; Cell Signaling Technology, Danvers, MA), PERK (C33E10, 3192; Cell Signaling Technology), ATF6 (D4Z8V, 65880S; Cell Signaling Technology), sequestosome 1 (p62, Ab56416; Abcam, Cambridge, MA), RIP3 (AHP1797; Bio-Rad Laboratories, Hercules, CA), cleaved caspase 3 (9661; Cell Signaling Technology), cleaved caspase 8 (8592; Cell Signaling Technology), β-actin (4967; Cell Signaling Technology), glyceraldehyde-3-phosphate dehydrogenase (ab8245; Abcam), and α-tubulin (T6199; Sigma-Aldrich). Immunoreactive proteins were detected with ECL Western Blot Reagent (GE Healthcare, Amersham, UK), and images were obtained with a Chemi-Doc MP Imager (Bio-Rad Laboratories, CA).
Assessment of Paracellular Permeability
Mice were fasted for 12 hours, weighed, and orally gavaged with 4 kilodaltons FITC–dextran (FD4, 0.44 mg/g mouse; Sigma). Blood was collected by cardiac puncture after 2 hours before centrifuging at 2000 × g for 5 minutes to obtain plasma. Plasma was diluted in water (1:1) before fluorescence was measured with an excitation of 485 nm and an emission wavelength of 528 nm on an EnVision Multilabel plate reader (Perkin Elmer, Waltham, MA). The appearance of the nondigestible FITC–dextran in plasma after oral administration is a measure of paracellular permeability.69
Microbial Analysis
DNA was extracted from 2 fecal pellets collected aseptically from nonantibiotic (control, n = 14; CTαIKO, n = 11) and antibiotic-treated groups (control + antibiotics, n = 5; CTαIKO + antibiotics, n = 5) as previously described.70 Real-time PCR was performed to quantify the total bacterial load in feces using primer set UniF340/UniR514 (Table 7). The PCR reaction was performed on an ABI StepOne real-time System (Applied Biosystems, Foster City, CA) using PerfeCTa SYBR Green Supermix (Quantabio, Gaithersburg, MD). The amplification program contained an initial denaturation step at 95°C for 3 minutes, followed by 40 cycles of denaturation at 95°C for 10 seconds, and annealing at 60°C for 30 seconds. The genomic DNA from a gut commensal Escherichia coli strain with a genome size of 5,190,098 bp was chosen to create an 8-log-fold standard curve for direct quantification of the total bacteria.1 The 16S ribosomal RNA (rRNA) gene copies were calculated using the following formula: ([([(16S rRNA gene copies in the genome)/(genome size of the E coli strain)]∗(DNA concentration at the first serial dilution)]/(average mass of 1 bp dsDNA)] ∗ (Avogadro’s number)]/(1∗109 ng/g). The cycle threshold value was associated with 16S rRNA gene copies (log copy number) to construct a function for quantification of all samples. The total bacterial load was expressed as 16S rRNA gene copies per gram of feces on a base 10 logarithmic scale. Microbial composition was assessed by 16S rRNA gene amplicon sequencing on an Illumina MiSeq platform (San Diego, CA). Amplicon library construction, paired-end sequencing targeting the V3–V4 region of the 16S rRNA gene, and data analysis have been published previously.70
Statistics, Data Analysis, and Visualization
Data are expressed as means ± SEM using GraphPad Prism (version 7, San Diego, CA). Data were analyzed with a Student t test or 2-way analysis of variance with the Tukey post-test. A Mann–Whitney test was used to compare histopathologic colitis scores and goblet cell depletion scores between control and CTαIKO mice. For microbiota analysis, permutational multivariate analysis of variance of the weighted UniFrac distance was conducted to identify the difference in overall microbial structure between groups, using the adonis function in the vegan package71 with 999 permutations (R v3.4.4). The principal coordinate analysis based on the Bray–Curtis dissimilarity metric was plotted using the phyloseq package (R v3.4.4).72 Comparison of individual taxa/operational taxonomic units between treatments was performed using the Kruskal–Wallis test with the Dwass, Steel, Critchlow–Fligner multiple comparisons post hoc procedure (SAS v9.4, Cary, NC). Raw sequences of the 16S rRNA gene amplicon data are available through the Sequence Read Archive (SRA) with accession number PRJNA562603.
Acknowledgments
The authors thank Kelly-Ann Leonard and Nicole Coursen for technical support, Dr Ethan Wine for helpful comments on the manuscript, Dr Xuejun Sun and Priscilla Guo of the Cross-Cancer Institute for help with electron microscopy, and Lynette Elder of the Alberta Diabetes Institute Histology Core.
CRediT Authorship Contributions
John P. Kennelly (Conceptualization: Supporting; Data curation: Lead; Formal analysis: Lead; Funding acquisition: Supporting; Investigation: Lead; Methodology: Lead; Writing – original draft: Lead; Writing – review & editing: Supporting)
Stephanie Carlin (Data curation: Supporting; Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Tingting Ju (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Jelske van der Veen (Conceptualization: Supporting; Data curation: Supporting; Formal analysis: Supporting; Methodology: Supporting; Supervision: Supporting; Writing – review & editing: Supporting)
Randal Nelson (Investigation: Supporting; Methodology: Supporting; Supervision: Supporting; Writing – review & editing: Supporting)
Jean Buteau (Methodology: Supporting; Supervision: Supporting; Writing – review & editing: Supporting)
Aducio Thiesen (Data curation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Caroline Richard (Investigation: Supporting; Methodology: Supporting; Supervision: Supporting; Writing – review & editing: Supporting)
Ben Willing (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting; Supervision: Supporting; Writing – review & editing: Supporting)
Rene Jacobs (Conceptualization: Lead; Formal analysis: Lead; Funding acquisition: Lead; Supervision: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by Canadian Institutes of Health Research Grant 156243 (R.L.J.), the Alberta Diabetes Institute (R.L.J. and J.P.K.), and Alberta Innovates–Technology Futures (J.P.K.).
References
- 1.Ng S.C., Shi H.Y., Hamidi N., Underwood F.E., Tang W., Benchimol E.I., Panaccione R., Ghosh S., Wu J.C.Y., Chan F.K.L., Sung J.J.Y., Kaplan G.G. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2017;390:2769–2778. doi: 10.1016/S0140-6736(17)32448-0. [DOI] [PubMed] [Google Scholar]
- 2.Khor B., Gardet A., Xavier R.J. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strugala V., Dettmar P.W., Pearson J.P. Thickness and continuity of the adherent colonic mucus barrier in active and quiescent ulcerative colitis and Crohn's disease. Int J Clin Pract. 2008;62:762–769. doi: 10.1111/j.1742-1241.2007.01665.x. [DOI] [PubMed] [Google Scholar]
- 4.Braun A., Treede I., Gotthardt D., Tietje A., Zahn A., Ruhwald R., Schoenfeld U., Welsch T., Kienle P., Erben G., Lehmann W.D., Fuellekrug J., Stremmel W., Ehehalt R. Alterations of phospholipid concentration and species composition of the intestinal mucus barrier in ulcerative colitis: a clue to pathogenesis. Inflamm Bowel Dis. 2009;15:1705–1720. doi: 10.1002/ibd.20993. [DOI] [PubMed] [Google Scholar]
- 5.Ehehalt R., Wagenblast J., Erben G., Lehmann W.D., Hinz U., Merle U., Stremmel W. Phosphatidylcholine and lysophosphatidylcholine in intestinal mucus of ulcerative colitis patients. A quantitative approach by nanoElectrospray-tandem mass spectrometry. Scand J Gastroenterol. 2004;39:737–742. doi: 10.1080/00365520410006233. [DOI] [PubMed] [Google Scholar]
- 6.Stremmel W., Merle U., Zahn A., Autschbach F., Hinz U., Ehehalt R. Retarded release phosphatidylcholine benefits patients with chronic active ulcerative colitis. Gut. 2005;54:966–971. doi: 10.1136/gut.2004.052316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stremmel W., Ehehalt R., Autschbach F., Karner M. Phosphatidylcholine for steroid-refractory chronic ulcerative colitis: a randomized trial. Ann Intern Med. 2007;147:603–610. doi: 10.7326/0003-4819-147-9-200711060-00004. [DOI] [PubMed] [Google Scholar]
- 8.Karner M., Kocjan A., Stein J., Schreiber S., von Boyen G., Uebel P., Schmidt C., Kupcinskas L., Dina I., Zuelch F., Keilhauer G., Stremmel W. First multicenter study of modified release phosphatidylcholine "LT-02" in ulcerative colitis: a randomized, placebo-controlled trial in mesalazine-refractory courses. Am J Gastroenterol. 2014;109:1041–1051. doi: 10.1038/ajg.2014.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Veen J.N., Kennelly J.P., Wan S., Vance J.E., Vance D.E., Jacobs R.L. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochim Biophys Acta. 2017;1859:1558–1572. doi: 10.1016/j.bbamem.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Ho N., Xu C., Thibault G. From the unfolded protein response to metabolic diseases – lipids under the spotlight. J Cell Sci. 2018;131 doi: 10.1242/jcs.199307. [DOI] [PubMed] [Google Scholar]
- 11.Sriburi R., Jackowski S., Mori K., Brewer J.W. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol. 2004;167:35–41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z., Agellon L.B., Allen T.M., Umeda M., Jewell L., Mason A., Vance D.E. The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis. Cell Metab. 2006;3:321–331. doi: 10.1016/j.cmet.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Treede I., Braun A., Sparla R., Kühnel M., Giese T., Turner J.R., Anes E., Kulaksiz H., Füllekrug J., Stremmel W., Griffiths G., Ehehalt R. Anti-inflammatory effects of phosphatidylcholine. J Biol Chem. 2007;282:27155–27164. doi: 10.1074/jbc.M704408200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nilsson A. Intestinal absorption of lecithin and lysolecithin by lymph fistula rats. Biochim Biophys Acta. 1968;152:379–390. doi: 10.1016/0005-2760(68)90047-7. [DOI] [PubMed] [Google Scholar]
- 15.Parthasarathy S., Subbaiah P.V., Ganguly J. The mechanism of intestinal absorption of phosphatidylcholine in rats. Biochem J. 1974;140:503–508. doi: 10.1042/bj1400503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kennelly J.P., van der Veen J.N., Nelson R.C., Leonard K.A., Havinga R., Buteau J., Kuipers F., Jacobs R.L. Intestinal de novo phosphatidylcholine synthesis is required for dietary lipid absorption and metabolic homeostasis. J Lipid Res. 2018;59:1695–1708. doi: 10.1194/jlr.M087056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aachoui Y., Leaf I.A., Hagar J.A., Fontana M.F., Campos C.G., Zak D.E., Tan M.H., Cotter P.A., Vance R.E., Aderem A., Miao E.A. Caspase-11 protects against bacteria that escape the vacuole. Science. 2013;339:975–978. doi: 10.1126/science.1230751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park S.W., Zhen G., Verhaeghe C., Nakagami Y., Nguyenvu L.T., Barczak A.J., Killeen N., Erle D.J. The protein disulfide isomerase AGR2 is essential for production of intestinal mucus. Proc Natl Acad Sci U S A. 2009;106:6950–6955. doi: 10.1073/pnas.0808722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heazlewood C.K., Cook M.C., Eri R., Price G.R., Tauro S.B., Taupin D., Thornton D.J., Png C.W., Crockford T.L., Cornall R.J., Adams R., Kato M., Nelms K.A., Hong N.A., Florin T.H., Goodnow C.C., McGuckin M.A. Aberrant mucin assembly in mice causes endoplasmic reticulum stress and spontaneous inflammation resembling ulcerative colitis. PLoS Med. 2008;5:e54. doi: 10.1371/journal.pmed.0050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lameris A.L., Huybers S., Kaukinen K., Mäkelä T.H., Bindels R.J., Hoenderop J.G., Nevalainen P.I. Expression profiling of claudins in the human gastrointestinal tract in health and during inflammatory bowel disease. Scand J Gastroenterol. 2013;48:58–69. doi: 10.3109/00365521.2012.741616. [DOI] [PubMed] [Google Scholar]
- 21.Kayagaki N., Stowe I.B., Lee B.L., O’Rourke K., Anderson K., Warming S., Cuellar T., Haley B., Roose-Girma M., Phung Q.T., Liu P.S., Lill J.R., Li H., Wu J., Kummerfeld S., Zhang J., Lee W.P., Snipas S.J., Salvesen G.S., Morris L.X., Fitzgerald L., Zhang Y., Bertram E.M., Goodnow C.C., Dixit V.M. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 22.Broz P., Ruby T., Belhocine K., Bouley D.M., Kayagaki N., Dixit V.M., Monack D.M. Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature. 2012;490:288–291. doi: 10.1038/nature11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kayagaki N., Warming S., Lamkanfi M., Walle L.V., Louie S., Dong J., Newton K., Qu Y., Liu J., Heldens S., Zhang J., Lee W.P., Roose-Girma M., Dixit V.M. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 24.Rathinam Vijay A.K., Vanaja Sivapriya K., Waggoner L., Sokolovska A., Becker C., Stuart Lynda M., Leong John M., Fitzgerald Katherine A. TRIF licenses caspase-11-dependent NLRP3 inflammasome activation by gram-negative bacteria. Cell. 2012;150:606–619. doi: 10.1016/j.cell.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T., Mende D.R., Li J., Xu J., Li S., Li D., Cao J., Wang B., Liang H., Zheng H., Xie Y., Tap J., Lepage P., Bertalan M., Batto J.-M., Hansen T., Le Paslier D., Linneberg A., Nielsen H.B., Pelletier E., Renault P., Sicheritz-Ponten T., Turner K., Zhu H., Yu C., Li S., Jian M., Zhou Y., Li Y., Zhang X., Li S., Qin N., Yang H., Wang J., Brunak S., Doré J., Guarner F., Kristiansen K., Pedersen O., Parkhill J., Weissenbach J., Antolin M., Artiguenave F., Blottiere H., Borruel N., Bruls T., Casellas F., Chervaux C., Cultrone A., Delorme C., Denariaz G., Dervyn R., Forte M., Friss C., van de Guchte M., Guedon E., Haimet F., Jamet A., Juste C., Kaci G., Kleerebezem M., Knol J., Kristensen M., Layec S., Le Roux K., Leclerc M., Maguin E., Melo Minardi R., Oozeer R., Rescigno M., Sanchez N., Tims S., Torrejon T., Varela E., de Vos W., Winogradsky Y., Zoetendal E., Bork P., Ehrlich S.D., Wang J., Meta H.I.T.C. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wohlgemuth S., Haller D., Blaut M., Loh G. Reduced microbial diversity and high numbers of one single Escherichia coli strain in the intestine of colitic mice. Environ Microbiol. 2009;11:1562–1571. doi: 10.1111/j.1462-2920.2009.01883.x. [DOI] [PubMed] [Google Scholar]
- 27.Lupp C., Robertson M.L., Wickham M.E., Sekirov I., Champion O.L., Gaynor E.C., Finlay B.B. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of enterobacteriaceae. Cell Host Microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 28.Volmer R., van der Ploeg K., Ron D. Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc Natl Acad Sci U S A. 2013;110:4628–4633. doi: 10.1073/pnas.1217611110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho N., Yap W.S., Xu J., Wu H., Koh J.H., Goh W.W.B., George B., Chong S.C., Taubert S., Thibault G. Stress sensor Ire1 deploys a divergent transcriptional program in response to lipid bilayer stress. J Cell Biol. 2020;219 doi: 10.1083/jcb.201909165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halbleib K., Pesek K., Covino R., Hofbauer H.F., Wunnicke D., Hänelt I., Hummer G., Ernst R. Activation of the unfolded protein response by lipid bilayer stress. Mol Cell. 2017;67:673–684.e8. doi: 10.1016/j.molcel.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 31.Thibault G., Shui G., Kim W., McAlister G.C., Ismail N., Gygi S.P., Wenk M.R., Ng D.T. The membrane stress response buffers lethal effects of lipid disequilibrium by reprogramming the protein homeostasis network. Mol Cell. 2012;48:16–27. doi: 10.1016/j.molcel.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee H., Noh J.-Y., Oh Y., Kim Y., Chang J.-W., Chung C.-W., Lee S.-T., Kim M., Ryu H., Jung Y.-K. IRE1 plays an essential role in ER stress-mediated aggregation of mutant huntingtin via the inhibition of autophagy flux. Hum Mol Genet. 2011;21:101–114. doi: 10.1093/hmg/ddr445. [DOI] [PubMed] [Google Scholar]
- 33.Cao S.S., Zimmermann E.M., Chuang B.M., Song B., Nwokoye A., Wilkinson J.E., Eaton K.A., Kaufman R.J. The unfolded protein response and chemical chaperones reduce protein misfolding and colitis in mice. Gastroenterology. 2013;144:989–1000.e6. doi: 10.1053/j.gastro.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tam A.B., Roberts L.S., Chandra V., Rivera I.G., Nomura D.K., Forbes D.J., Niwa M. The UPR activator ATF6 responds to proteotoxic and lipotoxic stress by distinct mechanisms. Dev Cell. 2018;46:327–343.e7. doi: 10.1016/j.devcel.2018.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pierdomenico M., Negroni A., Stronati L., Vitali R., Prete E., Bertin J., Gough P.J., Aloi M., Cucchiara S. Necroptosis is active in children with inflammatory bowel disease and contributes to heighten intestinal inflammation. Am J Gastroenterol. 2014;109:279–287. doi: 10.1038/ajg.2013.403. [DOI] [PubMed] [Google Scholar]
- 36.Wang R., Li H., Wu J., Cai Z.-Y., Li B., Ni H., Qiu X., Chen H., Liu W., Yang Z.-H., Liu M., Hu J., Liang Y., Lan P., Han J., Mo W. Gut stem cell necroptosis by genome instability triggers bowel inflammation. Nature. 2020;580:386–390. doi: 10.1038/s41586-020-2127-x. [DOI] [PubMed] [Google Scholar]
- 37.Gunther C., Neumann H., Neurath M.F., Becker C. Apoptosis, necrosis and necroptosis: cell death regulation in the intestinal epithelium. Gut. 2013;62:1062–1071. doi: 10.1136/gutjnl-2011-301364. [DOI] [PubMed] [Google Scholar]
- 38.Gunther C., Martini E., Wittkopf N., Amann K., Weigmann B., Neumann H., Waldner M.J., Hedrick S.M., Tenzer S., Neurath M.F., Becker C. Caspase-8 regulates TNF-alpha-induced epithelial necroptosis and terminal ileitis. Nature. 2011;477:335–339. doi: 10.1038/nature10400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Negroni A., Colantoni E., Pierdomenico M., Palone F., Costanzo M., Oliva S., Tiberti A., Cucchiara S., Stronati L. RIP3 AND pMLKL promote necroptosis-induced inflammation and alter membrane permeability in intestinal epithelial cells. Dig Liver Dis. 2017;49:1201–1210. doi: 10.1016/j.dld.2017.08.017. [DOI] [PubMed] [Google Scholar]
- 40.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 41.McClellan J.L., Davis J.M., Steiner J.L., Enos R.T., Jung S.H., Carson J.A., Pena M.M., Carnevale K.A., Berger F.G., Murphy E.A. Linking tumor-associated macrophages, inflammation, and intestinal tumorigenesis: role of MCP-1. Am J Physiol Gastrointest Liver Physiol. 2012;303:G1087–G1095. doi: 10.1152/ajpgi.00252.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo T., Chen B., Wang X. 4-PBA prevents pressure overload-induced myocardial hypertrophy and interstitial fibrosis by attenuating endoplasmic reticulum stress. Chem Biol Interact. 2015;242:99–106. doi: 10.1016/j.cbi.2015.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cunha D.A., Hekerman P., Ladrière L., Bazarra-Castro A., Ortis F., Wakeham M.C., Moore F., Rasschaert J., Cardozo A.K., Bellomo E., Overbergh L., Mathieu C., Lupi R., Hai T., Herchuelz A., Marchetti P., Rutter G.A., Eizirik D.L., Cnop M. Initiation and execution of lipotoxic ER stress in pancreatic beta-cells. J Cell Sci. 2008;121:2308–2318. doi: 10.1242/jcs.026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng B., Yao P.M., Li Y., Devlin C.M., Zhang D., Harding H.P., Sweeney M., Rong J.X., Kuriakose G., Fisher E.A., Marks A.R., Ron D., Tabas I. The endoplasmic reticulum is the site of cholesterol-induced cytotoxicity in macrophages. Nat Cell Biol. 2003;5:781–792. doi: 10.1038/ncb1035. [DOI] [PubMed] [Google Scholar]
- 45.Fu S., Yang L., Li P., Hofmann O., Dicker L., Hide W., Lin X., Watkins S.M., Ivanov A.R., Hotamisligil G.S. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature. 2011;473:528–531. doi: 10.1038/nature09968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kitai Y., Ariyama H., Kono N., Oikawa D., Iwawaki T., Arai H. Membrane lipid saturation activates IRE1alpha without inducing clustering. Genes Cells. 2013;18:798–809. doi: 10.1111/gtc.12074. [DOI] [PubMed] [Google Scholar]
- 47.Ariyama H., Kono N., Matsuda S., Inoue T., Arai H. Decrease in membrane phospholipid unsaturation induces unfolded protein response. J Biol Chem. 2010;285:22027–22035. doi: 10.1074/jbc.M110.126870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borradaile N.M., Han X., Harp J.D., Gale S.E., Ory D.S., Schaffer J.E. Disruption of endoplasmic reticulum structure and integrity in lipotoxic cell death. J Lipid Res. 2006;47:2726–2737. doi: 10.1194/jlr.M600299-JLR200. [DOI] [PubMed] [Google Scholar]
- 49.Zhang K., Kaufman R.J. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454:455–462. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saveljeva S., Mc Laughlin S.L., Vandenabeele P., Samali A., Bertrand M.J.M. Endoplasmic reticulum stress induces ligand-independent TNFR1-mediated necroptosis in L929 cells. Cell Death Dis. 2015;6:e1587. doi: 10.1038/cddis.2014.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu P., Hu S., Jin Q., Li D., Tian F., Toan S., Li Y., Zhou H., Chen Y. Ripk3 promotes ER stress-induced necroptosis in cardiac IR injury: a mechanism involving calcium overload/XO/ROS/mPTP pathway. Redox Biol. 2018;16:157–168. doi: 10.1016/j.redox.2018.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johansson M.E., Hansson G.C. Immunological aspects of intestinal mucus and mucins. Nat Rev Immunol. 2016;16:639–649. doi: 10.1038/nri.2016.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reya T., Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 54.Yu H., Yue X., Zhao Y., Li X., Wu L., Zhang C., Liu Z., Lin K., Xu-Monette Z.Y., Young K.H., Liu J., Shen Z., Feng Z., Hu W. LIF negatively regulates tumour-suppressor p53 through Stat3/ID1/MDM2 in colorectal cancers. Nat Commun. 2014;5:5218. doi: 10.1038/ncomms6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mundy R., MacDonald T.T., Dougan G., Frankel G., Wiles S. Citrobacter rodentium of mice and man. Cell Microbiol. 2005;7:1697–1706. doi: 10.1111/j.1462-5822.2005.00625.x. [DOI] [PubMed] [Google Scholar]
- 56.Buffie C.G., Pamer E.G. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Z., Agellon L.B., Vance D.E. Choline redistribution during adaptation to choline deprivation. J Biol Chem. 2007;282:10283–10289. doi: 10.1074/jbc.M611726200. [DOI] [PubMed] [Google Scholar]
- 58.Shulzhenko N., Morgun A., Hsiao W., Battle M., Yao M., Gavrilova O., Orandle M., Mayer L., Macpherson A.J., McCoy K.D., Fraser-Liggett C., Matzinger P. Crosstalk between B lymphocytes, microbiota and the intestinal epithelium governs immunity versus metabolism in the gut. Nat Med. 2011;17:1585–1593. doi: 10.1038/nm.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsai P.Y., Zhang B., He W.Q., Zha J.M., Odenwald M.A., Singh G., Tamura A., Shen L., Sailer A., Yeruva S., Kuo W.T., Fu Y.X., Tsukita S., Turner J.R. IL-22 upregulates epithelial claudin-2 to drive diarrhea and enteric pathogen clearance. Cell Host Microbe. 2017;21:671–681.e4. doi: 10.1016/j.chom.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salem S.N., Truelove S.C. Small-intestinal and gastric abnormalities in ulcerative colitis. Br Med J. 1965;1:827–831. doi: 10.1136/bmj.1.5438.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chakravarti K.R., Sehgal A.K., Chakravarti R.N., Chnuttani P.N. A study of intestinal function and morphology in nonspecific ulrcerative colitis in acute phase and remission in India. Am J Dig Dis. 1973;18:191–198. doi: 10.1007/BF01071972. [DOI] [PubMed] [Google Scholar]
- 62.Andersson H., Dotevall G., Gillberg R., Jagenburg R., Kock N.G. Absorption studies in patients with Crohn's disease and in patients with ulcerative colitis. Acta Med Scand. 1971;190:407–410. doi: 10.1111/j.0954-6820.1971.tb07450.x. [DOI] [PubMed] [Google Scholar]
- 63.Kaczmarek A., Vandenabeele P., Krysko D.V. Necroptosis: the release of damage-associated molecular patterns and its physiological relevance. Immunity. 2013;38:209–223. doi: 10.1016/j.immuni.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 64.Nenci A., Becker C., Wullaert A., Gareus R., van Loo G., Danese S., Huth M., Nikolaev A., Neufert C., Madison B., Gumucio D., Neurath M.F., Pasparakis M. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 2007;446:557–561. doi: 10.1038/nature05698. [DOI] [PubMed] [Google Scholar]
- 65.Out C., Patankar J.V., Doktorova M., Boesjes M., Bos T., de Boer S., Havinga R., Wolters H., Boverhof R., van Dijk T.H., Smoczek A., Bleich A., Sachdev V., Kratky D., Kuipers F., Verkade H.J., Groen A.K. Gut microbiota inhibit Asbt-dependent intestinal bile acid reabsorption via Gata4. J Hepatol. 2015;63:697–704. doi: 10.1016/j.jhep.2015.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Madsen K., Cornish A., Soper P., McKaigney C., Jijon H., Yachimec C., Doyle J., Jewell L., De Simone C. Probiotic bacteria enhance murine and human intestinal epithelial barrier function. Gastroenterology. 2001;121:580–591. doi: 10.1053/gast.2001.27224. [DOI] [PubMed] [Google Scholar]
- 67.Folch J., Lees M., Sloane Stanley G.H. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 68.Zhou X., Arthur G. Improved procedures for the determination of lipid phosphorus by malachite green. J Lipid Res. 1992;33:1233–1236. [PubMed] [Google Scholar]
- 69.Woting A., Blaut M. Small intestinal permeability and gut-transit time determined with low and high molecular weight fluorescein isothiocyanate-dextrans in C3H mice. Nutrients. 2018;10:685. doi: 10.3390/nu10060685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ju T., Shoblak Y., Gao Y., Yang K., Fouhse J., Finlay B.B., So Y.W., Stothard P., Willing B.P. Initial gut microbial composition as a key factor driving host response to antibiotic treatment, as exemplified by the presence or absence of commensal Escherichia coli. Appl Environ Microbiol. 2017;83:e01107–e01117. doi: 10.1128/AEM.01107-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Oksanen J, Kindt R, Legendre P, et al. The Vegan Package. 2007. Available: http://cran.r-project.org/, http://vegan.r-forge.r-project.org/.
- 72.McMurdie P.J., Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]