Abstract
In kidney transplantation, short-term allograft survival has improved due to improvements in acute rejection episodes without corresponding improvements in long-term survival. While current organ allocation algorithms take into account human leukocyte antigen (HLA) matching to reduce anti-donor allo-immune responses, it is likely that genomic variation at non-HLA loci (i.e. non-HLA donor-recipient (D-R) pair mismatches) play a role in the “non-self” responses and ultimately affect long-term allograft survival. Existing data from both animal models and human studies suggest an association between non-HLA D-R mismatches and kidney allograft outcomes. In this minireview, we examine existing and emerging data and discuss putative mechanisms on the role of non-HLA D-R mismatches on long-term allograft outcomes in kidney transplantation.
Introduction
Allo-immune responses that cause kidney allograft damage arise from T-lymphocytic “non-self” recognition, when recipient T-cells recognize donor antigens via the direct pathway (donor major histocompatibility complex (MHC) plus peptide on donor cells), indirect pathway (donor-derived antigens presented by recipient antigen presenting cells (APC)), or the semi-direct pathway (presentation of self-peptides by donor MHC on recipient APC via membrane transfer). Fundamentally, alloreactivity is based on specific peptide/MHC differences between the host (recipient) and donor cells. At the level of the genome, the processes that recognize the donor organ as non-self and culminate in acute rejection (AR) are largely determined by the human leukocyte antigen (HLA) region of the donor-recipient (D-R) pair. AR itself has been repeatedly shown to be associated with decreased allograft survival (1,2). In current organ allocation algorithms and clinical care, we attempt to take into account HLA mismatching at the A- B- and DR-loci. Though other HLA loci (i.e. DP, DQ) are not factored into organ allocation, mismatches at these loci have also been shown to associate with kidney transplant outcomes (3,4). In spite of these and other measures, improved long-term allograft survival remains an elusive goal in kidney transplantation - though acute cellular rejection episodes have been significantly reduced (5).
Putative Mechanisms of Non-HLA Loci in Transplantation
Less is known whether variations at non-HLA regions of D-R genomes impact allograft survival independent of or additive to HLA variation. Variants in non-HLA regions could impact outcomes depending on their presence in donor organs, recipients, or their presence as “mismatches” between given D-R pairs. Hypothesis-based, targeted analyses have primarily identified single nucleotide polymorphisms (SNPs) that have been associated with predefined phenotypes (e.g. acute allograft rejection, graft survival) (6,7). Recent unbiased examinations of non-HLA genomic sequence variations via genome wide association studies (GWAS) in donors or recipients have reported novel loci associated with graft outcomes (8–11). Of note, only one of these three studies included both donor and recipient variation (8) while the other two included only kidney transplant recipients. Besides SNPs, the relevance of inter-individual non-HLA variations from copy number variants (CNVs) which can span exons or entire genes has been previously reported in bone marrow transplantation (BMT) (12).
Intronic or intergenic variants may impact regulation of gene expression or splicing without directly altering protein sequences. In fact, a summary of GWASs for complex traits identified that most GWAS loci localize to non-coding regions (13). Results from the Encyclopedia of DNA elements (ENCODE) project have attributed regulatory functions to such GWAS-identified non-coding loci within the human genome, annotating these SNPs as expression quantitative trait loci (eQTL)(14). Cis- or trans- eQTLs in the recipient or donor genomes could therefore alter expression of proinflammatory cytokines or profibrotic proteins by acting as regulatory elements and promoting inflammation or histologic damage. Unfortunately, the identified loci from these GWASs have not been independently validated for the tested transplant phenotypes (rejection or graft survival) (8), and the altered expression of cytokines remains speculative. The lack of validation of these loci could relay heterogenous ancestral background in study populations or lack of uniform reporting of phenotypes (e.g. cellular vs humoral rejection, subclinical vs clinical rejection).
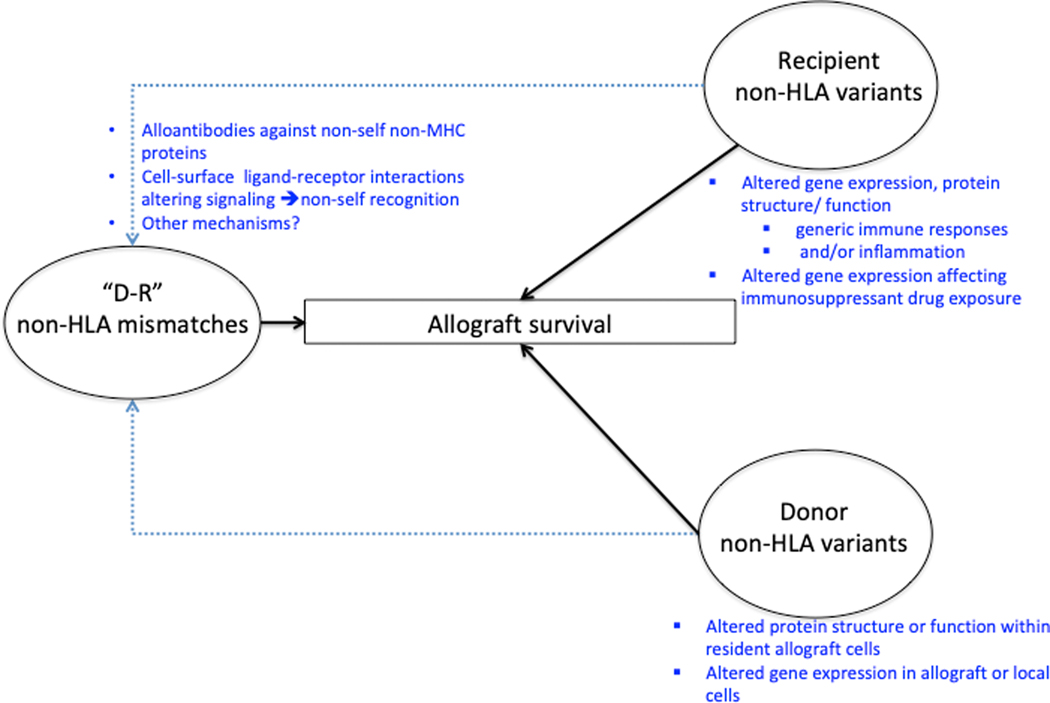
Aside from individual donor or recipient variants that have been associated with allograft function, “D-R mismatches” at non-HLA loci or “minor antigen” mismatches are known to influence non-self responses to allografts in experimental models (15–18). In humans, non-self responses directed at targeted loci have been reported in rejection phenotypes independent of HLA (19–21). Non-self responses also arise from D-R mismatches in specific CNVs in kidney transplantation (22). Recent work has interrogated non-HLA mismatches in human transplantation in a quantitative yet genome-wide basis (19,23). These data have identified global non-HLA mismatch signals that significantly impact allograft rejection phenotypes and survival, independent of HLA. Thus, non-HLA variation in donors and recipients may have donor-organ- or recipient-specific effects respectively, while combined donor-recipient mismatches could impact non-self responses in transplantation (19,24) (Figure 1). As several excellent reviews have tabulated associations of donor and recipient SNPs with allograft outcomes (1,25), we focus our review on recent literature regarding non-HLA “mismatches” reported in D-R pairs and their associations with allograft rejection and survival.
Figure 1.
Mechanisms of Non-HLA variation and impact on transplant outcomes
Donor-Recipient non-MHC “minor antigen” mismatches in kidney transplantation:
Minor antigen mismatch in animal models of transplantation:
Animal models have repeatedly demonstrated the importance of non-MHC (or “minor”) antigen mismatch in transplantation. A role for the male antigen (H-Y) in mice was evident as early as 1955, where males tolerated female skin grafts while females rejected skin homografts from males (26). The significance of Y-chromosome coded antigens has also been confirmed in subsequent rodent models (27). In multiple inbred strains of rats, skin grafts were transplanted into syngeneic but sex-mismatched recipients. While female skin grafts in female recipients survived, male grafts were rejected by female recipients. In humans, small series have detected anti-H-Y antibody responses in male-to-female transplants (28), and larger epidemiologic investigations suggest worse outcomes for such allografts compared to other D-R gender combinations (29).
In an aortic graft transplant model of mice the following D-R combinations were examined: Animals which were disparate at 1) both MHC and non-MHC loci, 2) MHC loci only, and 3) non-MHC loci only. Only female animals were used to control for Y-chromosome coded antigens. Grafts from fully disparate (combination 1) or non-MHC (combination 3) disparate donors showed more severe intimal lesions (and allograft arteriosclerosis) than grafts disparate at MHC only (combination 2) at all time points, highlighting the role of non-MHC antigens in this histologic response (15). In a miniature swine transplantation model, MHC-matched, minor-antigen mismatched recipients rejected both skin and heart allografts at 11 and 35 days, respectively (16). Interestingly, minor-antigen mismatched corneal transplants in mice presented more of a barrier to successful transplant than MHC mismatches (17). Bone marrow transplant models in miniature swine have also supported the importance of non-MHC-antigen mismatch (18).
Non A/B/DR HLA-mismatches in clinical renal transplantation:
HLA-DQ and DP genes are expressed at lower levels on the cell surface and are less polymorphic, although shown to be important in graft-vs-host disease and mortality in allogenic bone marrow transplant (30). In BMT, HLA DQ and DP mismatches impacted mortality only when at least one HLA-A, B, C or DR mismatch coexisted between D-R pairs. In kidney transplantation, analysis of deceased donors from the Australian and New Zealand Transplant Registry (ANZDATA) showed an association between DQ-mismatches and all rejection episodes, antibody mediated rejection (ABMR) episodes, 1- and 5-year graft function, and overall graft survival. These associations were independent of HLA-A and B mismatches and clinico-demographic factors, but additive to the presence of DR-mismatches (31). Recent analysis from the US UNOS reaffirmed the importance of DQ mismatches independent of A-, B-, and DR-mismatches for acute rejection (<1-year) and graft loss (3). We recently showed from UNOS data that among ~ 96,000 deceased donor kidney transplants, HLA-matching better correlated with kidney graft survival and rejection phenotype than HLA-mismatching (4). Examination of DQ loci among D-R pairs in this cohort showed that HLA-matching at A-, B-, and DR-loci was a better surrogate of matching at DQ-loci (by linkage) than mismatching. HLA-DP mismatches have been associated with increased risk of graft loss in re-transplants, only suggesting the need for prior allo-sensitization and antibody formation to have occurred for such mismatches to impact clinical outcomes (32). These data also suggested that mismatches at loci outside of routinely measured A-, B-, and DR-loci could impact graft outcomes in clinical kidney transplantation.
Non-HLA mismatches in human kidney transplantation:
In human kidney transplantation involving HLA-identical D-R pairs, clinical observations have reported the occurrence of graft losses from antibody mediated rejection (33,34), implying development of antibodies directed at proteins coded outside HLA or arising from non-HLA D-R “mismatches”. Relevance of antibodies against non-HLA proteins have also emerged in heart (35) and lung (36) transplantation as an extension of auto-immune phenomena. However, when D-R mismatches exist at non-HLA loci, non-HLA antibody development is an allo-immune response, and not part of auto-immunity (37). In either case, non-HLA antibody development and maturation must require a T-cell mediated immune response providing B-cell help, and therefore amenable to generic immunosuppression.
The impact of these antibodies against non-HLA antigens have been best reported in two instances in kidney transplantation – MHC class I–related chain A (MICA) and Angiotensin-II type-1 receptor (AT1R). MICA is polymorphic in humans (allowing D-R mismatches), expressed on endothelial cells but not on lymphocytes, and involved in innate immune activation (38). Anti-endothelial antibodies including anti-MICA are not picked up on routine cross matching if the antigens are cell-specific and not expressed on B- or T-cells. Especially in HLA-matched transplants, the presence of anti-MICA antibodies was associated with poorer 1-year graft survival (20), while the combined presence of antibodies against HLA and MICA were associated with the lowest 3-month estimated glomerular filtration rate (eGFR) (39). However, other work has suggested anti-MICA antibodies in recipient sera may not be specific to donor MICA antigens, and ~36% recipients had anti-self MICA antibodies (40). Antibodies directed against extracellular epitopes of AT1R were associated with C4d-negative ABMR with a unique phenotype – malignant hypertension, anemia, and response to angiotensin receptor blockade (ARB) therapy (21). While MICA is highly polymorphic, only few polymorphisms in AT1R have been associated with anti-AT1R antibodies (41). Such pre-transplant antibodies must form as a result of prior sensitizing events (e.g. anti-AT1R via pregnancy) or heterologous immunity and cross-reactivity with the donor. In the latter circumstance, mismatches between donor and recipient sequences at these non-HLA loci allow allo-reactivity without auto-immunity.
Genome-wide non-HLA mismatches in kidney transplantation:
Aside from targeted examinations of individual antigens, the relevance of mismatches between D-R pairs at polymorphic non-HLA sequences and their impact on transplantation has recently been reported on a genome-wide basis. Leveraging whole-exome sequencing of 28 D-R pairs and simultaneous gene expression data, Pineda et al, identified that non-HLA loci mismatches were significantly enriched in rejection phenotype or AR (T-cell mediated rejection (TCMR) or ABMR) vs no rejection, while mismatches at 28 variants were associated specifically with ABMR (23). Using known cell-specific gene expression data, as well GTEX eQTL database, the authors identified that high proportions of predictive SNPs associated with ABMR were annotated as non-synonymous or as eQTLs in genes expressed in kidney or vascular endothelial cells. In this study, while HLA-mismatches correlated with non-HLA mismatches in D-R pairs, a significant association between HLA matching and AR was not identified. On the other hand, whether ABMR-associated non-HLA mismatches were also associated with anti-HLA donor-specific antibodies (DSA) or other anti-donor antibodies was not investigated. A more recent thorough analysis of 477 deceased-donor D-R pairs of European ethnicity from Austria & Czech republic, identified that genome-wide non-HLA mismatches associated with death-censored graft survival (19). The authors annotated the mismatched SNPs as intronic & exonic (the latter as synonymous and non-synonymous variants), and further using Uniprot database to annotate variants within genes that encode transmembrane or secreted proteins vs all other cellular locations. Most interestingly in adjusted analysis, only the normalized total number of mismatched SNPs that were non-synonymous and within transmembrane/secreted proteins had significant impact on death-censored graft survival. Finally, in a subset of transplants with chronic ABMR, recipient serum showed significantly increased non-HLA anti-donor antibodies detected using specially designed peptide arrays based on mismatched loci. Notably, this association identified was with chronic ABMR, suggesting a smoldering injury from non-MHC mismatches. Mesnard et al calculated an “allogenomics mismatch score” (AMS), which reflected the number of amino acid D-R mismatches in transmembrane proteins. In this exome sequencing study of 34 living donors and 34 kidney transplant recipients, the AMS was significantly associated with eGFR up to 3 years after transplantation (42). These data suggest that in modern transplantation non-HLA mismatches may drive alloimmune phenomena leading to ABMR, renal function, and survival.
An exciting study led by Kiryluk et al, revealed a novel association between allograft rejection and D-R mismatch at LIMS1 gene locus (rs894304). The authors discovered,rs894304 from a genome-wide survey of CNVs through CNV-tagging SNPs (i.e. SNPs in high linkage disequilibrium with nearby CNVs). The SNP rs894304 tags a 1.5 kilobase intronic deletion downstream of the LIMS1 gene, and was identified as an eQTL of LIMS1 in multiple datasets. At rs894304, genotype corresponding to homozygous deletion in recipients was associated with acute kidney rejection (i.e. ABMR or TCMR), in a time-to-event analysis. LIMS1 protein was shown as expressed across multiple renal cell types. In D-R pairs across 3 validation cohorts (n=2004), the authors then identified that the effect sizes became larger when accounting for donor genotypes or when the locus existed a “D-R mismatch”. Such a D-R mismatch at this locus was then shown to be associated with allograft rejection as well as the production of anti-LIMS1 immunoglobulins. Hence, LIMS1 deletion in the recipient may reduce the expression of this surface protein, which is associated with focal adhesion complexes, and predispose them to an anti-LIMS1 immune response when encountering a high-expression donor (22). It should also be noted that this multinational study included multiethnic D-R cohorts as opposed to the restricted Caucasian-to-Caucasian, deceased-donor cohort in the Austrian study. On the other hand, the LIMS1-locus mismatch associated with any-AR, which is a heterogenous outcome, while the aggregated global D-R mismatch of non-synonymous SNPs in genes coding transmembrane and secreted proteins was associated with the hard outcome of graft loss.
The discrepancy between acute rejection episodes and long-term allograft survival encountered in the current era could reflect that distinct pathogenetic events may be contributing to these two outcomes. For instance, a common histological entity, allograft fibrosis or interstitial fibrosis and tubular atrophy (IF/TA) of unclear etiology has accounted for 30–40% of cases of allograft loss in large studies(43,44). In our own analysis from the GoCAR study using a multiethnic mixed deceased and live-donor cohort, genome-wide D-R differences expressed quantitatively (as a continuous variable) were associated with development of chronic allograft damage (Ci, Ct, and Cv scores, IF/TA) after excluding the MHC region in its entirety (45). These global non-HLA differences were not associated with cellular rejection (i, t scores). Very interestingly, non-HLA global differences in our data were best associated with vascular intimal fibrosis scores, similar to prior experimental data (15). Ongoing efforts are validating these findings in independent D-R cohorts. Together, these interesting findings raise the need to promote genome-wide efforts to understand the impact of non-HLA D-R differences in allograft outcomes in the current era. Potential clinical implications for such non-HLA “compatibility” data could include improved donor selection (such as for recipients with >1 potential live donors) or in post transplant risk stratification (categorizing high or low level mismatched D-Rs) and management (surveillance biopsy strategies or immunosuppression levels).
In the analysis by Pineda et al, non HLA D-R differences that were associated with ABMR also correlated with ancestral differences between given D-R pairs, which they adjusted for in subsequent analysis (23). In our analysis genome-wide D-R differences using quantitatively derived genetic ancestry (based on the 1000-genome project) were also associated with graft survival (45). Similar data wherein quantitative genome-wide SNP-based ancestry measures associated with graft loss have been subsequently reported (46). These data also highlight the relevance of non-MHC loci informative of ancestry and D-R mismatches thereof may impact graft survival.
Alternate mechanisms of non-self responses independent of HLA:
Aside from eliciting a classical allo-response via T- and B-cells, innate immune cells have recently been shown to have non-self recognition capacities (4). In elegant data from mice, non-self recognition via innate immune mechanisms were also shown to involve unique monocyte-APC, ligand with non-self-receptor, activating recipient monocytes and promoting T-cell mediated rejection when encountering the donor cell surface antigen (24,47). In this report in a bone marrow plug transplantation model, activating interaction between recipient-monocyte and donor-APC was independent of MHC mismatches and dependent on a single locus mismatch (e.g. CD47 (receptor) & SIRPα (ligand), both of which are cell-surface proteins in the D-R combination). Both recent published human D-R data identified enrichment of non-HLA mismatched loci that localized within cell-surface proteins (19,23). Such unique ligand-receptor interaction between surface-expressed antigens altering cell-signaling in specific D-R pairs is a further putative mechanism for the impact of non-HLA mismatches on transplantation. For instance, SIRPα gene is polymorphic in humans, and similar mechanisms should be explored in kidney transplantation. However, since several data point to the association of global D-R differences with graft survival, it is less likely that any single locus mismatch will account for the impact of non-HLA loci in the current clinical paradigm.
Conclusions:
Existing and emerging data in D-R pairs provide evidence that taking into account HLA mismatches alone does not completely account for current long-term allograft outcomes. The identification of increased prevalence of non-HLA antibodies in the presence of global or locus-specific non-HLA mismatches is suggestive that ABMR may mediate the effect of D-R mismatches via classical adaptive allo-immune responses. Intriguing, albeit preliminary, human data also suggest a dichotomous impact of MHC mismatches – leading on to AR, vs non-MHC mismatches – reflecting chronic allograft damage and vascular fibrosis as a manifestation of chronic rejection. Further mechanistic data are warranted to examine exact mechanisms of selected D-R mismatch loci or groups of such loci that impact allograft rejection, histology, function and survival. Identifying ancestry from genome-wide data are likely to be relevant in customizing future testing for unique variants that may be relevant exclusively in certain D-R combinations.
Further human studies recruiting heterogeneous populations and larger numbers of patients are required to better elucidate how information regarding non-HLA D-R mismatches may be incorporated to inform organ allocation, personalization of immunosuppression and surveillance. The global non-HLA signals identified in human data are a reminder of the genetic variation governing complexity of the non-self responses in humans which lie enroute to achieving optimal allograft survival.
Acknowledgements:
The authors would like to acknowledge Dr Barbara Murphy, PI of the GoCAR study [NIH 5U01AI070107-03]. MCM acknowledges Pilot funding from Clinical Trials in Organ Transplantation-19 – 2U01AI063594 – 11 Anciliary studies for “Non-HLA Dependent Donor-Recipient Genomic Distance Impacts Long-Term Renal Allograft Outcomes.” We acknowledge Dr Peter Heeger for critical feedback.
Footnotes
Disclosure
The authors of this manuscript have no conflicts to disclose as described by the American Journal of Transplantation.
Data Sharing Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- 1.Sankaran D, Asderakis A, Ashraf S et al. Cytokine gene polymorphisms predict acute graft rejection following renal transplantation. Kidney International 1999; 56: 281. [DOI] [PubMed] [Google Scholar]
- 2.Sayegh MH, Watschinger B, Carpenter CB. Mechanisms Of T Cell Recognition Of Alloantigen: The Role of Peptides. Transplantation 1994; 57: 1295. [DOI] [PubMed] [Google Scholar]
- 3.Leeaphorn N, Pena JRA, Thamcharoen N, Khankin EV, Pavlakis M, Cardarelli F. HLA-DQ Mismatching and Kidney Transplant Outcomes. Clinical Journal of the American Society of Nephrology 2018; 13: 763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yacoub R, Nadkarni GN, Cravedi P et al. Analysis of OPTN/UNOS registry suggests the number of HLA matches and not mismatches is a stronger independent predictor of kidney transplant survival. Kidney International 2018; 93: 482. [DOI] [PubMed] [Google Scholar]
- 5.Meier-Kriesche H-U, Schold JD, Kaplan B. Long-term renal allograft survival: have we made significant progress or is it time to rethink our analytic and therapeutic strategies? American Journal of Transplantation: Official Journal of the American Society of Transplantation and the American Society of Transplant Surgeons 2004; 4: 1289. [DOI] [PubMed] [Google Scholar]
- 6.Grinyó J, Vanrenterghem Y, Nashan B et al. Association of four DNA polymorphisms with acute rejection after kidney transplantation. Transplant International: Official Journal of the European Society for Organ Transplantation 2008; 21: 879. [DOI] [PubMed] [Google Scholar]
- 7.Park H, Lee N, In JW et al. Association of Foxp3 Polymorphism With Allograft Outcome in Kidney Transplantation. Annals of Laboratory Medicine 2017; 37: 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hernandez-Fuentes MP, Franklin C, Rebollo-Mesa I et al. Long- and short-term outcomes in renal allografts with deceased donors: A large recipient and donor genome-wide association study. American Journal of Transplantation: Official Journal of the American Society of Transplantation and the American Society of Transplant Surgeons 2018; 18: 1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Brien RP, Phelan PJ, Conroy J et al. A genome-wide association study of recipient genotype and medium-term kidney allograft function. Clinical Transplantation 2013; 27: 379. [DOI] [PubMed] [Google Scholar]
- 10.Ghisdal L, Baron C, Lebranchu Y et al. Genome‐Wide Association Study of Acute Renal Graft Rejection. American Journal of Transplantation 2017; 17: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oetting WS, Wu B, Schladt DP et al. Genome-wide association study identifies the common variants in CYP3A4 and CYP3A5 responsible for variation in tacrolimus trough concentration in Caucasian kidney transplant recipients. The Pharmacogenomics Journal 2018; 18: 501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCarroll SA, Bradner JE, Turpeinen H et al. Donor-recipient mismatch for common gene deletion polymorphisms in graft-versus-host disease. Nature Genetics 2009; 41: 1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hindorff LA, Sethupathy P, Junkins HA et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences of the United States of America 2009; 106: 9362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koulack J, McAlister VC, MacAulay MA, Bitter-Suermann H, MacDonald AS, Lee TDG. Importance of Minor Histocompatibility Antigens in the Development of Allograft Arteriosclerosis. Clinical Immunology and Immunopathology 1996; 80: 273. [DOI] [PubMed] [Google Scholar]
- 16.Ovid: Histocompatible miniature swine: an inbred large-animal model1. [Internet]. [cited 2019 Mar 22] Available from: https://ovidsp.tx.ovid.com/sp-3.33.0b/ovidweb.cgi?QS2=434f4e1a73d37e8cb17da02d43bbd96c9542092274d51f248f643dc30c45c68467f6fab082b6670784fe7d0eb5201311bd3aff8351ab28ee78644fe5b296d29a62b6161dcd27b2b0e240d554760840b604358fde3d701e07738e274231e2cbfe9e042612ffb85e7cbc64b317fe7cbd345040f7510c0a89586a2e6b2c8b5af7ae94ed16ffb2be7d1aabc9c1d75e4681643c6c7435c4edc9a7160943a59298143cebd8dc625fcf3f4048e676914e5f6fa3a460e1972d0e1080b4b515a3b91ba36e42fa95f23cd3389052eb24d038aa57d59b9856069e6b533d5d9a6367c6380858e31fbf06a55f74cc7e5bfce27576f5b1d3c875b24be60127ec8f225362ee153f
- 17.Sano Y, Ksander BR, J Wayne S. Minor H, rather than MHC, alloantigens offer the greater barrier to successful orthotopic corneal transplantation in mice. Transplant Immunology 1996; 4: 53. [DOI] [PubMed] [Google Scholar]
- 18.Guzzetta PC, Sundt TM, Suzuki T, Mixon A, Rosengard BR, Sachs DH. Induction of kidney transplantation tolerance across major histocompatibility complex barriers by bone marrow transplantation in miniature swine. Transplantation 1991; 51: 862. [DOI] [PubMed] [Google Scholar]
- 19.Reindl-Schwaighofer R, Heinzel A, Kainz A et al. Contribution of non-HLA incompatibility between donor and recipient to kidney allograft survival: genome-wide analysis in a prospective cohort. Lancet (London, England) 2019; 393: 910. [DOI] [PubMed] [Google Scholar]
- 20.Zou Y, Stastny P, Süsal C, Döhler B, Opelz G. Antibodies against MICA antigens and kidney-transplant rejection. The New England Journal of Medicine 2007; 357: 1293. [DOI] [PubMed] [Google Scholar]
- 21.Dragun D, Müller DN, Bräsen JH et al. Angiotensin II type 1-receptor activating antibodies in renal-allograft rejection. The New England Journal of Medicine 2005; 352: 558. [DOI] [PubMed] [Google Scholar]
- 22.Steers NJ, Li Y, Drace Z et al. Genomic Mismatch at LIMS1 Locus and Kidney Allograft Rejection. New England Journal of Medicine 2019; 380: 1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pineda S, Sigdel TK, Chen J, Jackson AM, Sirota M, Sarwal MM. Novel Non-Histocompatibility Antigen Mismatched Variants Improve the Ability to Predict Antibody-Mediated Rejection Risk in Kidney Transplant. Frontiers in Immunology 2017; 8: 1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai H, Friday AJ, Abou-Daya KI et al. Donor SIRPα polymorphism modulates the innate immune response to allogeneic grafts. Science Immunology 2017; 2: eaam6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chand S, McKnight AJ, Borrows R. Genetic polymorphisms and kidney transplant outcomes. Current Opinion in Nephrology and Hypertension 2014; 23: 605. [DOI] [PubMed] [Google Scholar]
- 26.Eichwald EJ, Silmser CR. Skin. Transplantation Bulletin 1955; 2: 148. [PubMed] [Google Scholar]
- 27.Muramatsu K, Kurokawa Y, Ihara K, Sakamoto S, You-Xin S, Kawai S. Behavior of male-specific minor histocompatibility antigen in skin and limb transplantation. Journal of Surgical Research 2003; 115: 106. [DOI] [PubMed] [Google Scholar]
- 28.Tan JC, Wadia PP, Coram M et al. H-Y Antibody Development Associates With Acute Rejection in Female Patients With Male Kidney Transplants. Transplantation 2008; 86: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gratwohl A, Döhler B, Stern M, Opelz G. H-Y as a minor histocompatibility antigen in kidney transplantation: a retrospective cohort study. Lancet (London, England) 2008; 372: 49. [DOI] [PubMed] [Google Scholar]
- 30.Fernández-Viña MA, Klein JP, Haagenson M et al. Multiple mismatches at the low expression HLA loci DP, DQ, and DRB3/4/5 associate with adverse outcomes in hematopoietic stem cell transplantation. Blood 2013; 121: 4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim WH, Chapman JR, Coates PT et al. HLA-DQ Mismatches and Rejection in Kidney Transplant Recipients. Clinical journal of the American Society of Nephrology: CJASN 2016; 11: 875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mytilineos J, Deufel A, Opelz G. Clinical relevance of HLA-DPB locus matching for cadaver kidney retransplants: a report of the Collaborative Transplant Study. Transplantation 1997; 63: 1351. [DOI] [PubMed] [Google Scholar]
- 33.Grafft CA, Cornell LD, Gloor JM et al. Antibody-mediated rejection following transplantation from an HLA-identical sibling. Nephrology, Dialysis, Transplantation: Official Publication of the European Dialysis and Transplant Association - European Renal Association 2010; 25: 307. [DOI] [PubMed] [Google Scholar]
- 34.Crespo M, Redondo D, Butler C et al. Antibody-Mediated Rejection With and Without HLA Donor-Specific Antibodies in Kidney. Transplantation 2018; 102: S211. [Google Scholar]
- 35.Kalache S, Dinavahi R, Pinney S, Mehrotra A, Cunningham MW, Heeger PS. Anticardiac myosin immunity and chronic allograft vasculopathy in heart transplant recipients. Journal of immunology (Baltimore, Md. : 1950) 2011; 187: 1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reinsmoen NL, Mirocha J, Ensor CR et al. A 3-Center Study Reveals New Insights Into the Impact of Non-HLA Antibodies on Lung Transplantation Outcome. Transplantation 2017; 101: 1215. [DOI] [PubMed] [Google Scholar]
- 37.Dinavahi R, George A, Tretin A et al. Antibodies reactive to non-HLA antigens in transplant glomerulopathy. Journal of the American Society of Nephrology: JASN 2011; 22: 1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zwirner NW, Marcos CY, Mirbaha F, Zou Y, Stastny P. Identification of MICA as a new polymorphic alloantigen recognized by antibodies in sera of organ transplant recipients. Human Immunology 2000; 61: 917. [DOI] [PubMed] [Google Scholar]
- 39.Sánchez-Zapardiel E, Castro-Panete MJ, Mancebo E et al. Early renal graft function deterioration in recipients with preformed anti-MICA antibodies: partial contribution of complement-dependent cytotoxicity. Nephrology, Dialysis, Transplantation: Official Publication of the European Dialysis and Transplant Association - European Renal Association 2016; 31: 150. [DOI] [PubMed] [Google Scholar]
- 40.Sapák M, Chreňová S, Tirpáková J et al. Donor non-specific MICA antibodies in renal transplant recipients. Immunobiology 2014; 219: 109. [DOI] [PubMed] [Google Scholar]
- 41.Zhang G, Wang H, Wang F et al. Gene polymorphisms of the renin-angiotensin-aldosterone system and angiotensin II type 1-receptor activating antibodies in renal rejection. The Tohoku Journal of Experimental Medicine 2007; 213: 203. [DOI] [PubMed] [Google Scholar]
- 42.Mesnard L, Muthukumar T, Burbach M et al. Exome Sequencing and Prediction of Long-Term Kidney Allograft Function. PLoS computational biology 2016; 12: e1005088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.El-Zoghby ZM, Stegall MD, Lager DJ et al. Identifying specific causes of kidney allograft loss. American Journal of Transplantation: Official Journal of the American Society of Transplantation and the American Society of Transplant Surgeons 2009; 9: 527. [DOI] [PubMed] [Google Scholar]
- 44.De Vusser K, Lerut E, Kuypers D et al. The predictive value of kidney allograft baseline biopsies for long-term graft survival. Journal of the American Society of Nephrology: JASN 2013; 24: 1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.American Society of Nephrology | Kidney Week - Abstract Details [Internet]. [cited 2019 Mar 28] Available from: https://www.asn-online.org/education/kidneyweek/2017/program-abstract.aspx?controlId=2783427
- 46.American Society of Nephrology | Kidney Week - Abstract Details [Internet]. [cited 2019 Mar 30] Available from: https://www.asn-online.org/education/kidneyweek/2018/program-abstract.aspx?controlId=3022505
- 47.Oberbarnscheidt MH, Zeng Q, Li Q et al. Non-self recognition by monocytes initiates allograft rejection. The Journal of Clinical Investigation 2014; 124: 3579. [DOI] [PMC free article] [PubMed] [Google Scholar]