Abstract
Hypercapnia, the elevation of CO2 in blood and tissues, commonly occurs in severe acute and chronic respiratory diseases and is associated with increased risk of death. Recent studies have shown that hypercapnia inhibits expression of select innate immune genes and suppresses host defence against bacterial and viral pneumonia in mice. In the current study, we evaluated the effect of culture under conditions of hypercapnia (20% CO2) versus normocapnia (5% CO2), both with normoxia, on global gene transcription in human THP-1 and mouse RAW 264.7 macrophages stimulated with lipopolysaccharide (LPS). We found that hypercapnia selectively downregulated transcription of LPS-induced genes associated with innate immunity, antiviral response, type I interferon signalling, cytokine signalling and other inflammatory pathways in both human and mouse macrophages. Simultaneously, hypercapnia increased expression of LPS-downregulated genes associated with mitosis, DNA replication and DNA repair. These CO2-induced changes in macrophage gene expression help explain hypercapnic suppression of antibacterial and antiviral host defence in mice and reveal a mechanism that may underlie, at least in part, the high mortality of patients with severe lung disease and hypercapnia.
Keywords: hypercapnia, CO2, lipopolysaccharide, macrophage, gene expression, innate immunity
1. Introduction
Hypercapnia, that is, elevation of the partial pressure of CO2 in blood and tissues, commonly develops in patients with severe acute and chronic pulmonary disorders, and is associated with an increased risk of death in chronic obstructive pulmonary disease [1–4], cystic fibrosis [5], community-acquired pneumonia [6] and adenoviral lung infection [7]. The possibility of a mechanistic link between hypercapnia and poor clinical outcomes was first suggested by reports that elevated CO2 inhibited expression of cytokines and chemokines that are important for host defence in macrophages and other cultured cells [8–12]. Our own studies showed that hypercapnia inhibited expression of tumour necrosis factor (TNF) and interleukin-6 (IL-6) in human and mouse alveolar macrophages, and in macrophage cell lines from both species [11]. We further showed that elevated CO2 inhibited cytokine expression at the level of gene transcription, and that the inhibition was non-cytotoxic, reversible and independent of changes in extracellular or intracellular pH [11]. Hypercapnia also inhibited macrophage phagocytosis, respiratory burst activity and autophagy-mediated bacterial killing [11,13].
Beyond its effects on cultured cells, hypercapnia suppresses innate immunity and host defence in vivo. We showed that exposure of mice to 10% CO2/21% O2, resulting in normoxic hypercapnia, reduced lung cytokine expression, suppressed neutrophil function and increased the mortality from Pseudomonas pneumonia in mice [14]. As in our studies with cultured macrophages, the effects of hypercapnia in the murine pneumonia model were not attributable to acidosis. More recently, we found that hypercapnia inhibited the antiviral response, enhanced viral replication and increased the mortality of influenza A virus (IAV) infection in mice [15]. The adverse effects of elevated CO2 on IAV infection were largely due to suppression of antiviral activity in lung macrophages, mediated by activation of Akt1 [15].
Because of its pleiotropic effects on macrophage function, and the adverse impact of hypercapnia on antibacterial and antiviral host defence, in the current study we evaluated the effect of elevated CO2 on global gene transcription in stimulated human and murine macrophages. We chose lipopolysaccharide (LPS) as the stimulus because it broadly induces expression of innate immune and pro-inflammatory genes that are critical to the outcome of bacterial, viral and other infections, as well as non-infectious insults to the lung and other tissues. We used human THP-1 and mouse RAW 264.7 macrophages for the investigation, since responses of the cell lines and primary human and mouse alveolar macrophages to hypercapnia were closely matched in our previous studies [11,13,15]. We show that elevated CO2 selectively inhibits LPS-induced expression of innate immune, pro-inflammatory and antiviral genes, while selectively attenuating LPS-induced suppression of genes required for DNA replication and cell division. These results have important implications for pulmonary host defence in patients who develop hypercapnia associated with severe lung disease.
2. Methods
2.1. Cells
Human monocytic leukaemia THP-1 cells (American Type Culture Collection (ATCC)) were cultured in RPMI 1640, supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 1 mM sodium pyruvate, 20 µM 2-mercaptoethanol, 100 U ml−1 penicillin and 100 µg ml−1 streptomycin and differentiated to a macrophage phenotype by exposure to 5 nM phorbol myristate acetate (PMA) for 48 h [11]. Mouse monocyte–macrophage RAW 264.7 cells (ATCC) were cultured in Dulbecco's modified Eagle's medium (supplemented with 10% heat-inactivated FBS, 100 U ml−1 penicillin and 100 µg ml−1 streptomycin). THP-1 and RAW 264.7 cells were expanded and THP-1 cells were differentiated under a standard culture atmosphere of humidified 5% CO2/95% air.
2.2. Hypercapnia and LPS exposure
For analysis of gene expression by microarray, THP-1 and RAW 264.7 macrophages were stimulated with LPS in humidified 20% CO2/21% O2/59% N2 (hypercapnia) or maintained in humidified 5% CO2/95% air (normocapnia), for 0.5, 1.5 or 3 h. Cells were exposed to hypercapnia in an environmental chamber (C-174; BioSpherix) contained within the same incubator where control cultures were simultaneously exposed to normocapnia. At time zero, culture media were removed and replaced with new media that had been pre-saturated for 4 h with 5% or 20% CO2, as appropriate, prior to the addition to the cells. Immediately following the medium change, ultrapure Escherichia coli K12 LPS (1 ng ml−1; InvivoGen) was added to the cells. The partial pressure of CO2 (PCO2) and pH of the pre-saturated media were measured using a pHOx Plus Blood Gas Analyzer (Nova Biomedical Corp.). For the normocapnia- and hypercapnia-equilibrated media, the PCO2 values were 44 and 112 mmHg, and the corresponding pH values were 7.4 and 7.1, respectively. We previously showed that exposure to 20% CO2 and stimulation with LPS for up to 24 h has no adverse effect on the viability of the macrophage cell lines [11].
In a separate set of experiments for polymerase chain reaction (PCR) validation of microarray results for selected genes, PMA-differentiated THP-1 cells were exposed for 3 h to a hypercapnic atmosphere of 15% CO2/21% O2/64% N2, or normocapnia as control, in a manner analogous to the experiment described above.
2.3. RNA isolation and Illumina geneChip hybridization
Total RNA was isolated using the RNeasy Mini Kit (Qiagen). The quality and quantity of each RNA sample were assessed using a 2100 BioAnalyzer (Agilent). Whole-genome microarray analysis was performed on LPS-stimulated THP-1 and RAW 264.7 cells using Illumina HumanRef-8 v3 Expression BeadChips and MouseRef-8 v2 Expression BeadChips, respectively.
2.4. Microarray data analysis
The criteria for differential gene expression were a fold-change cut-off of greater than or equal to 1.35 and false discovery rate of less than 0.05. K-means clustering and heat map visualization for differential gene expression induced by LPS in cells exposed to normocapnia and hypercapnia were performed using the Morpheus web tool (https://software.broadinstitute.org/morpheus). Enrichment analysis of gene ontology (GO) terms from the biological processes of all genes downregulated or upregulated by LPS and hypercapnia were separately analysed using the Gene Ontology Analysis InnateDB tool [16], which uses a manually curated knowledge base of genes, proteins, interactions and signalling pathways involved in mammalian innate immune responses. Results from the InnateDB analysis were confirmed using GeneGo Metacore (Thomson Reuter), a separately curated database and pathway analysis tool. From the GO biological term results, the four to six most enriched processes were selected, and interaction networks were constructed for each set of differentially expressed genes associated with these terms using the GeneMANIA plug-in [17] of Cytoscape 3.8.0 software [18]. GeneMANIA can find other genes that are related to the set of input genes and produce a functional association network based on their relationships, such as pathways, co-expression, co-localization, genetic interaction, physical interaction, shared protein domains and so on, based on the published literature.
2.5. Quantitative real-time PCR
RNA was extracted using an RNeasy Mini Kit (Qiagen) and reverse transcribed to cDNA using an iScript cDNA synthesis kit (Bio-Rad). PCR amplification was performed using a CFX Connect real-time system (Bio-Rad) and the TaqMan (Applied Biosystems) or PrimeTime®Predesigned (IDT) gene expression assays with FAM-labelled probes. The following primer/probe sets were used, from TaqMan: CCL2 (Hs00234140), IL6 (Hs00174031), ICAM1 (Hs00164932), EGR1 (Hs00152928); and from PrimeTime: NFKB1 (Hs.PT.58.21008943), EBI2 (Hs.PT.58.2335925) and EEF1A1 (Hs.PT.58.15621992.g) as reference. Relative expression was calculated by the comparative CT method (ΔΔCT) [19].
3. Results
3.1. LPS increases expression of immune response genes and decreases expression of genes involved in DNA replication and mitosis in human and mouse macrophages
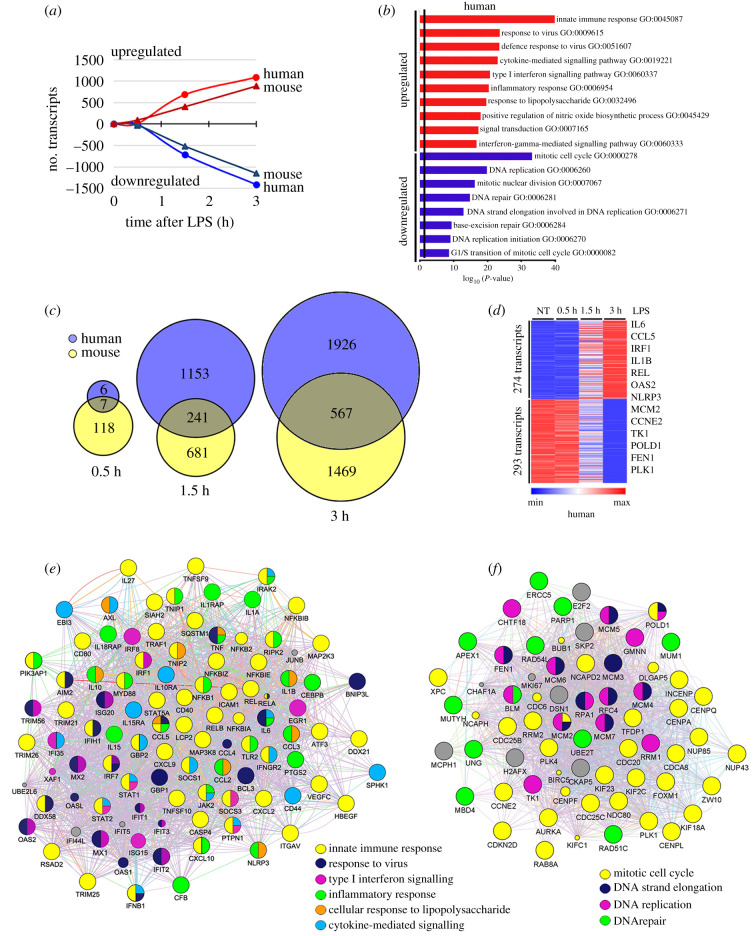
The transcriptomic response to LPS under normocapnic conditions was assessed in human THP-1 and mouse RAW 264.7 macrophages cultured under 5% CO2/95% air (PCO2 44 mmHg) and stimulated with LPS (1 ng ml−1) for 0.5, 1.5 or 3 h, followed by analysis of global gene expression on Illumina microarrays. In THP-1 macrophages cultured in normocapnia, LPS increased expression of 9, 683 and 1085 genes at 0.5, 1.5 and 3 h, respectively, and decreased expression of 4, 711 and 1408 genes at the same time points (figure 1a). In RAW 264.7 macrophages cultured in normocapnia, LPS increased expression of 89, 402 and 886 genes and decreased expression of 35, 519 and 1149 genes, at 0.5, 1.5 and 3 h, respectively. All these genes were changed by greater than or equal to 1.35-fold, among 14 320 genes mapped in the human array and 17 572 genes in the mouse array (figure 1a).
Figure 1.
LPS induces transcriptional changes in human and mouse macrophages. Global gene expression was assessed on Illumina microarrays in human THP-1 and mouse RAW 264.7 cells stimulated with LPS (1 ng ml−1) in normocapnia (5% CO2, PCO2 44 mmHg) for 0.5, 1.5 or 3 h. Transcripts downregulated and upregulated by greater than or equal to 1.35-fold at 0.5, 1.5 or 3 h after LPS treatment (a). Bars represent the top GO biological processes of downregulated (blue) and upregulated (red) genes induced by LPS after 3 h in THP-1 cells (b). Venn diagrams of human and mouse common genes changed by LPS at 0.5, 1.5 or 3 h (c). Heat map of common genes changed by LPS over time in human macrophages (d). Networks of GO biological processes upregulated (e) and downregulated (f) by LPS after 3 h of treatment.
GO analysis of the LPS response in human THP-1 cells showed that, for LPS-upregulated genes, the most enriched processes were innate immune and inflammatory responses including response to virus, cytokine-mediated signalling, type I interferon signalling, nitric oxide biosynthesis, interferon-γ signalling and, as expected, response to LPS (figure 1b). Among LPS-downregulated genes, the most enriched processes involved mitosis, DNA replication and repair, and G1/S cell cycle transition (figure 1b). Not surprisingly, analysis of genes differentially expressed in RAW 264.7 cells stimulated with LPS documented highly similar GO processes to those seen in human macrophages (electronic supplementary material, figure S1A), confirming that the response to LPS is conserved from mice to humans. Since the GO processes were so similar, we identified genes regulated by LPS in common in human and mouse macrophages. Figure 1c shows that 7, 241 and 567 common genes were differentially expressed following LPS stimulation for 0.5, 1.5 or 3 h, respectively. These 567 common genes were grouped by K-means clustering, and heatmaps were generated to illustrate the LPS response in human and mouse macrophages. In human macrophages the LPS-upregulated cluster comprises 274 genes and the LPS-downregulated cluster comprises 293 genes (figure 1d), while in mouse macrophages LPS upregulated 285 genes and downregulated 282 genes (electronic supplementary material, figure S1B). Interestingly, when gene networks were generated for the commonly upregulated and downregulated clusters (figure 1e,f), the GO biological processes represented were nearly identical to those obtained when the LPS responses of human and mouse macrophages were analysed separately (figure 1b and electronic supplementary material, figure S1B). As shown in figure 1e, for the common LPS-upregulated genes, the most enriched processes were all related to inflammation and innate immunity, comprising genes from the NF-κB pathway (REL, RELA, RELB, NFKB1, NFKB2 and others), response to virus (INFB1, IRF7, DDX58, IFIH1, GBP1, BNIP3L and others); type I interferon signalling (IRF1, IRF7, IRF8, STAT1, STAT2, SOCS3, EGR1 and others), inflammatory response (IL1A, IL6, IL15, IL18, CXCL10, CCL2, CCL5, IRAK2, MYD88, CEBPB, NLRP3, PTGS2, CFB and others), cytokine-mediated signalling (IL10RA, IL15RA, CD44, GBP2, IFNGR2, EBI3, IRAK2, PTPN1 and others); mitogen-activated protein (MAP) kinases (MAP2K3, MAP3K8); and other LPS response genes (including ICAM1, CD40, CXCL9, ATF3, PI3KAP1 and TNIP2). For the common LPS-downregulated genes, the most enriched processes were mitotic cell cycle (RRM2, CDC20, AURKA, KIF2C, FOXM1, CCNE2, CENPA, INCENP, PLK1, NUP43 and others); DNA replication and elongation (FEN1, MCM2, MCM3, MCM4, MCM5, GMNN, CHTF18 and others) and DNA repair (APEX1, MUM1, PARP1, UNG, RAD51C, RAD54L, MBD4 and others; figure 1f).
3.2. Hypercapnia selectively modulates LPS-regulated gene transcription changes in human and mouse macrophages
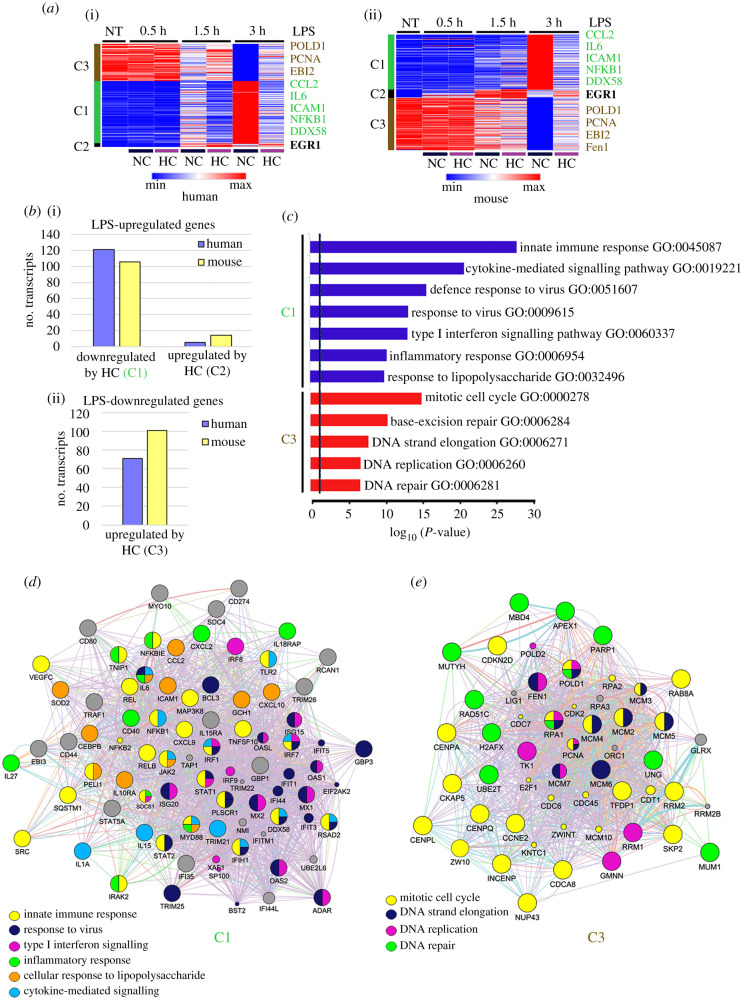
The effect of hypercapnia on LPS-regulated gene expression was assessed by stimulating THP-1 and RAW 264.7 macrophages with LPS (1 ng ml−1) for 0.5, 1.5 or 3 h under 20% CO2/21% O2/59% N2 (PCO2 112 mmHg). The cells exposed to hypercapnia were from the same passages and were stimulated with LPS simultaneously with those in normocapnia whose results are described above. Microarray analysis showed that hypercapnia modulated expression of many genes regulated by LPS in both THP-1 and RAW 264.7 macrophages. Relative to normocapnia, hypercapnia downregulated expression of 742 genes and upregulated expression of 434 genes by greater than or equal to 1.35-fold in THP-1 cells stimulated with LPS for 3 h. Similarly, hypercapnia downregulated 611 genes and upregulated 508 genes by greater than or equal to 1.35-fold in RAW 264.7 cells stimulated with LPS for 3 h. Complete lists of all genes downregulated and upregulated by hypercapnia after 0.5, 1.5 and 3 h of LPS stimulation in THP-1 and RAW 264.7 macrophages are available in the electronic supplementary material, tables S1 and S2.
The heatmaps in figure 2a depict changes in expression of the LPS-regulated genes that were commonly modulated by hypercapnia in human THP-1 (i) and mouse RAW 264.7 (ii) macrophages. In each case, K-means analysis generated three clusters: genes that were upregulated by LPS in normocapnia and relatively downregulated by hypercapnia (cluster 1, C1), genes that were upregulated by LPS in normocapnia and further upregulated by hypercapnia (cluster 2, C2) and genes that were downregulated by LPS in normocapnia and relatively upregulated by hypercapnia (cluster 3, C3). (No genes that were downregulated by LPS in normocapnia were further downregulated by hypercapnia.) Among the genes commonly upregulated by LPS in normocapnia, hypercapnia downregulated 121 of 274 (44%) in THP-1 macrophages and 106 of 285 (37%) in RAW 264.7 macrophages (C1, figure 2b(i)). Of these LPS-upregulated genes in normocapnia, hypercapnia further upregulated only 6 (2%) in the human and 14 (5%) in the mouse macrophages (C2; figure 2b(i)). Among the genes commonly downregulated by LPS in normocapnia, hypercapnia upregulated 71 of 293 (24%) and 101 of 282 (36%) in the human and mouse macrophages (C3; figure 2b(ii)).
Figure 2.
Hypercapnia alters expression of LPS-upregulated and -downregulated genes in human and mouse macrophages. Global gene expression was assessed on Illumina microarrays in human THP-1 and mouse RAW 264.7 cells stimulated with LPS (1 ng ml−1) in normocapnia (5% CO2, PCO2 44 mmHg) or hypercapnia (20% CO2, PCO2 112 mmHg) for 0.5, 1.5 or 3 h. K-means clustering of common differentially expressed genes changed by LPS and then by hypercapnia in human THP-1 (i) and mouse RAW 264.7 (ii) macrophages are presented as a heatmap (a). K-means generate three clusters C1, C2 and C3. Bars show the number of transcripts upregulated by LPS in normocapnia that are uniquely and commonly downregulated (C1) or further upregulated (C2) by hypercapnia (i), and transcripts downregulated by LPS in normocapnia that are uniquely and commonly upregulated (C3) by hypercapnia (ii) in human and mouse (b). GO biological processes of C1 and C3 (c). Global network analysis of C1 (d) and C3 (e).
Major GO biological processes represented by the LPS-upregulated genes whose expression was decreased by hypercapnia belonging to cluster 1 include innate immune response, cytokine-mediated signalling, response to virus, type I interferon signalling, inflammatory response and response to LPS (figure 2c). The LPS-downregulated genes whose expression was upregulated by elevated CO2 in cluster 3 involve biological processes related to the mitotic cell cycle, and to DNA replication, elongation and repair (figure 2c). Genes that were upregulated by LPS and further increased by hypercapnia in cluster 2 were too few in number to map to GO processes; these include ATF3, EGR1, ERRFI1, IFNB1, NLRP3 and ZFP36.
3.3. Hypercapnia downregulates LPS-induced genes associated with innate immunity, response to virus, type I interferon signalling, inflammatory response and cytokine signalling
Figure 2d shows the network of human genes belonging to cluster 1, i.e. genes whose expression was upregulated by LPS and relatively downregulated by hypercapnia. Genes central to all of the major biological processes related to innate immunity and inflammation represented by the transcriptional response to LPS in normocapnia (figure 1b,e) were downregulated by hypercapnia. These include NF-κB pathway genes (NFKB1, NFKB2, NFKBIE, MAP3K8, REL, RELB and others), antiviral and type I interferon signalling genes (STAT1, STAT2, DDX58, IRF1, IRF2, IRF7, IFIT1, ISG20, MX1, MX2, OAS2, RSAD2 and others); cytokine signalling genes (IL1A, IL6, IL15, JAK2, TRIM21 and others); and other inflammatory and LPS response genes (IRAK2, CEBPB, PELI1, TLR2, ICAM1, CXCL2, CXCL9, CXCL10, CD40, IL27 and others). Taken together, these results indicate that hypercapnia broadly suppresses induction of innate immune, antiviral and inflammation-related gene transcription in human and mouse macrophages.
3.4. Hypercapnia increases expression of LPS-downregulated genes associated with mitosis and DNA replication
Figure 2e shows the network of top human genes belonging to cluster 3, i.e. genes that were downregulated by LPS and relatively upregulated by hypercapnia. Genes with roles in each of the biological processes suppressed by LPS were counter-regulated by hypercapnia. These include genes involved in the mitotic cell cycle (CCNE2, CDKN2D, CENPA, CENPL, CENPQ, INCENP, CKAP5, NUP43, SKP2 and others); and DNA strand elongation, replication and repair (FEN1, MCM2, MCM4, MCM5, MCM6, POLD1, RPA1, PARP1, UNG, RAD51C, MBD4 and others). Thus, hypercapnia attenuates LPS-induced suppression of genes required for mitosis and DNA replication and repair.
3.5. Validation of microarray results for selected genes by quantitative PCR
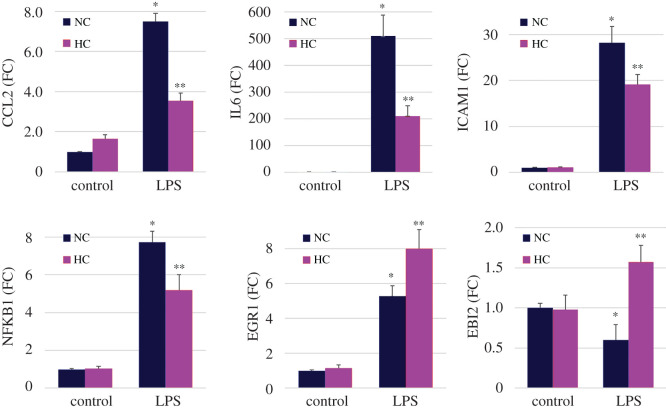
To validate LPS- and CO2-regulated changes in gene expression demonstrated by microarray, we performed a set of independent experiments in which THP-1 macrophages were stimulated with LPS for 3 h under normocapnic and hypercapnic conditions. As shown in figure 3, hypercapnia significantly downregulated the LPS-induced increases in mRNA expression of the cluster 1 genes, CCL2, IL6, ICAM1 and NFKB1, as determined by quantitative PCR (qPCR). Similarly, analysis by qPCR confirmed that hypercapnia further upregulated the LPS-induced increase in expression of the cluster 2 gene EGR1 and reversed LPS-induced suppression of the cluster 3 gene EBI2 (figure 3). Thus, we were able to confirm the directionality of both LPS- and CO2-regulated expression of representative genes in each of the three clusters from the microarrays in independent experiments analysed by qPCR.
Figure 3.
qPCR confirmation of selected immunoregulatory genes. THP-1 cells stimulated with or without LPS (1 ng ml−1) in normocapnia (5% CO2; NC) or hypercapnia (15% CO2; HC) for 3 h. CCL2, IL-6, ICAM1, NFKB1, EBI1 and EGR1 mRNA expression assessed by qPCR. Means ± s.e.m., n = 4–6 from at least four independent experiments; *p < 0.01 versus control in NC, **p < 0.05 versus LPS in NC.
4. Discussion
The transcriptional responses to LPS we observed in human THP-1 and mouse RAW 264.7 macrophages are similar to LPS responses in the two cell lines and in primary macrophages reported by others previously, including rapid induction of a wide range of innate immune and inflammatory genes, and downregulation of many genes involved in DNA replication and cell division [20–23]. The LPS-triggered changes in gene expression we documented were broad, robust and highly similar in the human and mouse macrophage lines. These factors, plus the fact that LPS has been shown to induce a core macrophage transcriptional response that aligns closely with responses to a range of pathogens and other microbe-related agonists [24], support the approach of using LPS stimulation as a model to interrogate the effects of hypercapnia on gene expression related to immunity, host defence and other critical cell functions in the macrophage.
The principal finding of our study is that hypercapnia broadly downregulated LPS-induced expression of innate immune and inflammatory genes in both human and mouse macrophages. The hypercapnia-downregulated genes are important for host defence against bacterial, viral and fungal pathogens. The results corroborate and extend our previous finding that elevated CO2 inhibited LPS-induced macrophage expression of TNF and IL-6 [11]. Broad suppression by elevated CO2 of innate immune gene expression in macrophages is also consistent with our observation that hypercapnia inhibited lung cytokine gene expression and increased the mortality of mice with Pseudomonas pneumonia [14]. Likewise, hypercapnic suppression of antiviral and type I interferon pathway gene expression in the macrophage cell lines coincides with our recent report that hypercapnia suppresses IAV-induced antiviral gene and protein expression in alveolar macrophages and other lung cells, and increases the mortality of IAV infection in mice [15]. Of note, we have also shown that hypercapnia downregulates innate immune gene expression in human bronchial epithelial cells [25]. Thus, hypercapnic suppression of immune gene expression is not restricted to macrophages, indicating that the adverse impact of elevated CO2 on host defence against bacterial and viral pulmonary infections in vivo probably results from impacts of hypercapnia on multiple cell types in the lung.
Interestingly, we have also shown that hypercapnia inhibits expression of antimicrobial peptides and other immune genes in Drosophila [26]. As in the mouse, exposure to elevated CO2 increased the mortality from bacterial infection in the fly [26]. Further, in a genome-wide RNAi screen in Drosophila cells, we identified a number of genes whose expression is required for hypercapnic inhibition of antimicrobial peptide gene expression, and confirmed that one of these, the zinc finger homeobox transcription factor, zfh2, mediates CO2-induced suppression of antibacterial host defence in vivo [27]. Thus, hypercapnic suppression of innate immune gene expression is conserved from Drosophila to mammals. To explore the basis of this conservation, we are now studying the role of Zfhx3, a mammalian orthologue of zfh2, as a possible mediator of hypercapnic immunosuppression in mice.
A second major result of our analysis is that hypercapnia countered LPS-induced downregulation of multiple mitosis-related and DNA replication and repair genes. Suppression of these gene programmes underlies the inhibition of DNA synthesis and cell cycle progression first observed in LPS-treated macrophages many years ago [28,29]. More recently, it has been shown that LPS reprogrammes macrophage metabolism to a glycolytic phenotype [23,30], which, in combination with the block in proliferation, optimizes the cell for antimicrobial activity. On the other hand, attenuation by hypercapnia of LPS-induced downregulation of mitosis-associated and DNA replication genes might be expected to divert cellular resources away from antimicrobial functions and towards proliferative pathways. In this way, CO2-induced upregulation of non-immune, proliferation-related genes might further impair macrophage innate immune activity and contribute to the host defence defect caused by hypercapnia.
The molecular mechanism(s) by which hypercapnia alters gene expression remain to be determined. Given the large number and variety of genes whose expression is altered, it seems likely that multiple transcriptional regulators may be involved. One possibility is that the activity of transcription factors or their upstream regulators could be altered directly by elevated CO2 via carbamylation [31]. Alternatively, signalling proteins and/or transcription factors could be targeted for other activity-modifying, post-translational modifications triggered by signals from yet-to-be-discovered upstream CO2 sensor(s). In line with the latter possibility, hypercapnia has been shown to activate and signal via AMP-activated protein kinase in alveolar epithelial and skeletal muscle cells [32,33], miR-183 and isocitrate dehydrogenase 2 in alveolar epithelial cells and lung fibroblasts [34], caspase-7, miR-133a and RhoA in airway smooth muscle [35], Wnts in several tissues [36], and heat shock factor 1 [37] and Akt1 [15] in macrophages.
In conclusion, using global gene profiling we have shown that hypercapnia selectively downregulates a broad array of innate immune and inflammatory genes, while upregulating genes involved in cell division and DNA replication in LPS-stimulated human and mouse macrophages. These results align with our previous finding that hypercapnia inhibits macrophage TNF and IL-6 mRNA and protein expression in a non-cytotoxic, reversible and pH-independent manner [11]. They also help explain the immune defects that underlie the hypercapnia-induced increase in mortality of mice with Pseudomonas and IAV pneumonia [14,15]. Furthermore, our results reveal a mechanism by which elevated CO2 may contribute, at least in part, to the high mortality of patients with severe acute and chronic lung disease complicated by hypercapnia.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Jacob I. Sznajder for helping to motivate the work and thoughtful discussions.
Data accessibility
Primary microarray data are provided as electronic supplementary material.
Authors' contributions
S.M.C.-M., N.W., K.L.G., G.J.B. and P.H.S.S. conceptualized the study. S.M.C.-M., N.W. and P.H.S.S. designed the experiments. S.M.C.-M., N.W. and A.N. performed the experiments. S.M.C.-M. and S.B. performed the bioinformatics analysis. S.M.C.-M., S.B. and P.H.S.S. analysed and interpreted the data. S.M.C.-M. and P.H.S.S. wrote the manuscript. All authors reviewed the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by the National Institutes of Health (grant nos. R01 HL107629 to P.H.S.S and G.J.B., and R01 HL131745 to P.H.S.S.).
References
- 1.Moser KM, Shibel EM, Beamon AJ. 1973. Acute respiratory failure in obstructive lung disease. Long-term survival after treatment in an intensive care unit. JAMA 225, 705–707. ( 10.1001/jama.1973.03220340019004) [DOI] [PubMed] [Google Scholar]
- 2.Martin TR, Lewis SW, Albert RK. 1982. The prognosis of patients with chronic obstructive pulmonary disease after hospitalization for acute respiratory failure. Chest 82, 310–314. ( 10.1378/chest.82.3.310) [DOI] [PubMed] [Google Scholar]
- 3.Groenewegen KH, Schols AM, Wouters EF. 2003. Mortality and mortality-related factors after hospitalization for acute exacerbation of COPD. Chest 124, 459–467. ( 10.1378/chest.124.2.459) [DOI] [PubMed] [Google Scholar]
- 4.Mohan A, Premanand R, Reddy LN, Rao MH, Sharma SK, Kamity R, Bollineni S. 2006. Clinical presentation and predictors of outcome in patients with severe acute exacerbation of chronic obstructive pulmonary disease requiring admission to intensive care unit. BMC Pulm. Med. 6, 27 ( 10.1186/1471-2466-6-27) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belkin RA, et al. 2006. Risk factors for death of patients with cystic fibrosis awaiting lung transplantation. Am. J. Respir. Crit. Care Med. 173, 659–666. ( 10.1164/rccm.200410-1369OC) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sin DD, Man SF, Marrie TJ. 2005. Arterial carbon dioxide tension on admission as a marker of in-hospital mortality in community-acquired pneumonia. Am. J. Med. 118, 145–150. ( 10.1016/j.amjmed.2004.10.014) [DOI] [PubMed] [Google Scholar]
- 7.Murtagh P, Giubergia V, Viale D, Bauer G, Pena HG. 2009. Lower respiratory infections by adenovirus in children. Clinical features and risk factors for bronchiolitis obliterans and mortality. Pediatr. Pulmonol. 44, 450–456. ( 10.1002/ppul.20984) [DOI] [PubMed] [Google Scholar]
- 8.West MA, Baker J, Bellingham J. 1996. Kinetics of decreased LPS-stimulated cytokine release by macrophages exposed to CO2. J. Surg. Res. 63, 269–274. ( 10.1006/jsre.1996.0259) [DOI] [PubMed] [Google Scholar]
- 9.Takeshita K, et al. 2003. Hypercapnic acidosis attenuates endotoxin-induced nuclear factor-κB activation. Am. J. Respir. Cell Mol. Biol. 29, 124–132. ( 10.1165/rcmb.2002-0126OC) [DOI] [PubMed] [Google Scholar]
- 10.Lang CJ, Dong P, Hosszu EK, Doyle IR. 2005. Effect of CO2 on LPS-induced cytokine responses in rat alveolar macrophages. Am. J. Physiol. Lung Cell. Mol. Physiol. 289, L96–L103. ( 10.1152/ajplung.00394.2004) [DOI] [PubMed] [Google Scholar]
- 11.Wang N, Gates KL, Trejo H, Favoreto S Jr, Schleimer RP, Sznajder JI, Beitel GJ, Sporn PH. 2010. Elevated CO2 selectively inhibits interleukin-6 and tumor necrosis factor expression and decreases phagocytosis in the macrophage. FASEB J. 24, 2178–2190. ( 10.1096/fj.09-136895) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cummins EP, et al. 2010. NF-κB links CO2 sensing to innate immunity and inflammation in mammalian cells. J. Immunol. 185, 4439–4445. ( 10.4049/jimmunol.1000701) [DOI] [PubMed] [Google Scholar]
- 13.Casalino-Matsuda SM, Nair A, Beitel GJ, Gates KL, Sporn PH. 2015. Hypercapnia inhibits autophagy and bacterial killing in human macrophages by increasing expression of Bcl-2 and Bcl-xL. J. Immunol. 194, 5388–5396. ( 10.4049/jimmunol.1500150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gates KL, Howell HA, Nair A, Vohwinkel CU, Welch LC, Beitel GJ, Hauser AR, Sznajder JI, Sporn PH. 2013. Hypercapnia impairs lung neutrophil function and increases mortality in murine Pseudomonas pneumonia. Am. J. Respir. Cell Mol. Biol. 49, 821–828. ( 10.1165/rcmb.2012-0487OC) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casalino-Matsuda SM, et al. 2020. Hypercapnia suppresses macrophage antiviral activity and increases mortality of influenza A infection via Akt1. J. Immunol. 205, 489–501. ( 10.4049/jimmunol.2000085) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breuer K, et al. 2013. InnateDB: systems biology of innate immunity and beyond—recent updates and continuing curation. Nucleic Acids Res. 41(Database issue), D1228–D1233. ( 10.1093/nar/gks1147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warde-Farley D, et al. 2010. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38(Web Server issue), W214–W220. ( 10.1093/nar/gkq537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. ( 10.1101/gr.1239303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Casalino-Matsuda SM, Forteza RM, Monzon ME. 2008. Hyaluronan fragments induce MUC5B expression through a monocyte chemoattractant protein-1/ CCR2 dependent mechanism. Am. J. Respir. Crit. Care Med. 177, A994. [Google Scholar]
- 20.Nilsson R, et al. 2006. Transcriptional network dynamics in macrophage activation. Genomics 88, 133–142. ( 10.1016/j.ygeno.2006.03.022) [DOI] [PubMed] [Google Scholar]
- 21.Huang H, Park CK, Ryu JY, Chang EJ, Lee Y, Kang SS, Kim HH. 2006. Expression profiling of lipopolysaccharide target genes in RAW264.7 cells by oligonucleotide microarray analyses. Arch. Pharm. Res. 29, 890–897. ( 10.1007/BF02973911) [DOI] [PubMed] [Google Scholar]
- 22.Sharif O, Bolshakov VN, Raines S, Newham P, Perkins ND. 2007. Transcriptional profiling of the LPS induced NF-κB response in macrophages. BMC Immunol. 8, 1 ( 10.1186/1471-2172-8-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu L, et al. 2016. Proinflammatory signal suppresses proliferation and shifts macrophage metabolism from Myc-dependent to HIF1alpha-dependent. Proc. Natl Acad. Sci. USA 113, 1564–1569. ( 10.1073/pnas.1518000113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenner RG, Young RA. 2005. Insights into host responses against pathogens from transcriptional profiling. Nat. Rev. Microbiol. 3, 281–294. ( 10.1038/nrmicro1126) [DOI] [PubMed] [Google Scholar]
- 25.Casalino-Matsuda SM, et al. 2018. Hypercapnia alters expression of immune response, nucleosome assembly and lipid metabolism genes in differentiated human bronchial epithelial cells. Sci. Rep. 8, 13508 ( 10.1038/s41598-018-32008-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helenius IT, Krupinski T, Turnbull DW, Gruenbaum Y, Silverman N, Johnson EA, Sporn PH, Sznajder JI, Beitel GJ. 2009. Elevated CO2 suppresses specific Drosophila innate immune responses and resistance to bacterial infection. Proc. Natl Acad. Sci. USA 106, 18 710–18 715. ( 10.1073/pnas.0905925106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helenius IT, Haake RJ, Kwon YJ, Hu JA, Krupinski T, Casalino-Matsuda SM, Sporn PH, Sznajder JI, Beitel GJ. 2016. Identification of Drosophila Zfh2 as a mediator of hypercapnic immune regulation by a genome-wide RNA interference screen. J. Immunol. 196, 655–667. ( 10.4049/jimmunol.1501708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cocks BG, Vairo G, Bodrug SE, Hamilton JA. 1992. Suppression of growth factor-induced CYL1 cyclin gene expression by antiproliferative agents. J. Biol. Chem. 267, 12 307–12 310. [PubMed] [Google Scholar]
- 29.Nagao S, Ikegami S, Tanaka A. 1984. Inhibition of macrophage DNA synthesis by immunomodulators. II. Characterization of the suppression by muramyl dipeptide or lipopolysaccharide [3H]thymidine incorporation into macrophages. Cell. Immunol. 89, 427–438. ( 10.1016/0008-8749(84)90344-7) [DOI] [PubMed] [Google Scholar]
- 30.Ubanako P, Xelwa N, Ntwasa M. 2019. LPS induces inflammatory chemokines via TLR-4 signalling and enhances the Warburg effect in THP-1 cells. PLoS ONE 14, e0222614 ( 10.1371/journal.pone.0222614) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linthwaite VL, Janus JM, Brown AP, Wong-Pascua D, O'Donoghue AC, Porter A, Treumann A, Hodgson DR, Cann MJ. 2018. The identification of carbon dioxide mediated protein post-translational modifications. Nat. Commun. 9, 3092 ( 10.1038/s41467-018-05475-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vadasz I, et al. 2008. AMP-activated protein kinase regulates CO2-induced alveolar epithelial dysfunction in rats and human cells by promoting Na,K-ATPase endocytosis. J. Clin. Invest. 118, 752–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaitovich A, et al. 2015. High CO2 levels cause skeletal muscle atrophy via AMP-activated kinase (AMPK), FoxO3a protein, and muscle-specific Ring finger protein 1 (MuRF1). J. Biol. Chem. 290, 9183–9194. ( 10.1074/jbc.M114.625715) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vohwinkel CU, Lecuona E, Sun H, Sommer N, Vadasz I, Chandel NS, Sznajder JI. 2011. Elevated CO2 levels cause mitochondrial dysfunction and impair cell proliferation. J. Biol. Chem. 286, 37 067–37 076. ( 10.1074/jbc.M111.290056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shigemura M, et al. 2018. Hypercapnia increases airway smooth muscle contractility via caspase-7-mediated miR-133a-RhoA signaling. Science Transl. Med. 10, eaat1662 ( 10.1126/scitranslmed.aat1662) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shigemura M, et al. 2019. Elevated CO2 regulates the Wnt signaling pathway in mammals, Drosophila melanogaster and Caenorhabditis elegans. Sci. Rep. 9, 18251 ( 10.1038/s41598-019-54683-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu Z, Casalino-Matsuda SM, Nair A, Buchbinder A, Budinger GRS, Sporn PHS, Gates KL. 2018. A role for heat shock factor 1 in hypercapnia-induced inhibition of inflammatory cytokine expression. FASEB J. 32, 3614–3622. ( 10.1096/fj.201701164R) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Primary microarray data are provided as electronic supplementary material.