Abstract
Background and Aims
Gut bacteria-derived short-chain fatty acids (SCFAs) play crucial roles in the maintenance of intestinal homeostasis. However, how SCFAs regulate epithelial turnover and tissue repair remain incompletely understood. In this study, we investigated how the SCFA propionate regulates cell migration to promote epithelial renewal and repair.
Methods
Mouse small intestinal epithelial cells (MSIE) and human Caco-2 cells were used to determine the effects of SCFAs on gene expression, proliferation, migration, and cell spreading in vitro. Video microscopy and single cell tracking were used to assess cell migration kinetically. 5-bromo-2’-deoxyuridine (BrdU) and hydroxyurea were used to assess the effects of SCFAs on migration in vivo. Lastly, an acute colitis model using dextran sulfate sodium (DSS) was used to examine the effects of SCFAs in vivo.
Results
Using video microscopy and single cell tracking, we found that propionate promoted intestinal epithelial cell migration by enhancing cell spreading and polarization, which led to increases in both cell speed and persistence. This novel function of propionate was dependent on inhibition of class I histone deacetylases (HDAC) and GPR43 and required signal transducer and activator of transcription 3 (STAT3). Furthermore, using 5-bromo-2’-deoxyuridine (BrdU) and hydroxyurea in vivo, we found that propionate enhanced cell migration up the crypt-villus axis under homeostatic conditions, while also protecting against ulcer formation in experimental colitis.
Conclusion
Our results demonstrate a mechanism by which propionate stimulates cell migration in an HDAC inhibition, GPR43, and STAT3 dependent manner, and suggest that propionate plays an important role in epithelial migration independent of proliferation.
Keywords: HDAC, IEC, Propionate, Migration, STAT3
Abbreviations used in this paper: ANOVA, analysis of variance; DMEM, Dulbecco’s modified Eagle’s medium; DSS, dextran sulfate sodium; ECAR, extracellular acidification rate; EGF, endothelial growth factor; FBS, fetal bovine serum; gRNA, guide RNA; HDAC, histone deacetylase; HDACi, histone deacetylase inhibitor; IEC, intestinal epithelial cell; IFNγ, interferon gamma; IL, interleukin; KO, knockout; MEK, mitogen-activated protein kinase; MSD, mean-squared displacement; MSIE, mouse small intestinal epithelial cell; mTOR, mammalian target of rapamycin; OCR, oxygen consumption rate; SCFA, short-chain fatty acid; STAT3, signal transducer and activator of transcription 3; TGFβ, transforming growth factor beta; TNFα, tumor necrosis factor alpha; WT, wild-type
Graphical abstract
Summary.
In this study, we demonstrate that the short-chain fatty acid propionate stimulates intestinal epithelial cell spreading and polarization, which leads to an increase in cell speed and persistence. These effects lead to increases in epithelial cell migration in vitro and in vivo.
The gut epithelium, which separates the host from the external environment and forms the first line of defense to enteric infection, comprises a single layer of epithelial cells that renew themselves constantly, turning over every 3–5 days.1,2 In order to maintain homeostasis, the epithelial layer depends on a host of growth signals and energy sources to proliferate, differentiate, and migrate. This begins with stem cells in the crypt base dividing every 2–3 hours driven by the secretion of growth factors such as Wnt from underlying mesenchymal fibroblast.3 Following division, stem cells give rise to transient amplifying cells, which then differentiate in response to both host and microbial factors including short-chain fatty acids (SCFAs).4 These cells migrate toward the villus axis, where they are extruded into the gut lumen. It has been shown that gut microbiota plays a crucial role in epithelial maturation and turnover; however, the mechanisms remain poorly understood.
Intestinal epithelial restitution is a complex process that is important for tissue regeneration in inflammatory bowel disease (IBD).5 It has been shown that during the first several hours following injury, cells from adjacent crypts migrate into the wound bed to form a temporary barrier to protect the underlying stem cells from exposure to luminal contents.6, 7, 8 This process is independent of cellular proliferation, as seen in other organs such as the skin. Several signals have been described to play a role in epithelial restitution including prostaglandins, transforming growth factor beta (TGFβ), trefoil factors, and gut microbiota–derived SCFAs such as propionate.9, 10, 11, 12 However, the mechanisms behind how these signals contribute to the restitution remain unclear.
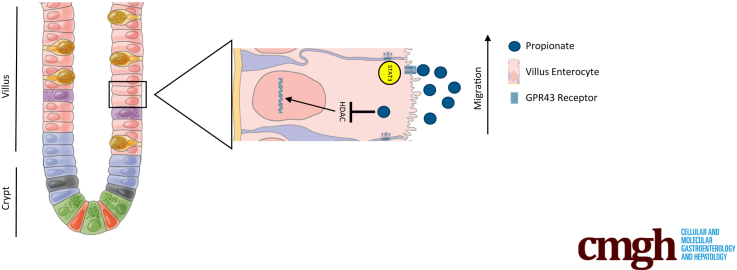
SCFAs are bacterial metabolites from the fermentation of dietary fibers by microbes in the gut lumen.13, 14, 15 There are 5 SCFAs, with acetate, propionate, and butyrate making up the large majority of SCFAs in the gut.16 SCFAs are vital for intestinal homeostasis, where they affect both the epithelial cells as wells as immune cells in the underlying lamina propria.17,18 SCFAs are known to enhance mucus production by promoting goblet cell differentiation and mucus production.19, 20, 21 Previously, we and several other groups have shown that SCFAs are important for stimulating intestinal epithelial cell (IEC) antimicrobial peptide production to protect against enteric infection.22 It has been demonstrated that an SCFA gradient exists in the gut, with high levels of SCFAs exerting an apoptotic effect on cells via inhibition of histone deacetylases (HDACs), and low concentration allowing for histone acetyltransferase activity and increased stem cell proliferation.23 Furthermore, it has been shown that SCFAs such as propionate can enhance epithelial restitution in vitro, and promote epithelial turnover by promoting proliferation in the crypt.12,24 Previously, it was thought that epithelial migration that occurs during epithelial turnover was a passive process as a result of the force generated from dividing cells in the crypt.25 However, recent data suggest that actin polymerization, but not proliferation, is the major driving force for cell migration on the villus.26
Propionate, a major SCFA present at high levels in the gut lumen, has been shown to contribute to intestinal homeostasis through promoting epithelial proliferation and migration.12,16,23,27 However, how propionate drives epithelial cell proliferation and migration and the mechanisms involved, are still largely unclear. In the current study, we utilized video microscopy to track individual cells to analyze cell speed and persistence using the classic scratch assay (wound healing model) and DiPer in vitro.28,29 Additionally, we used BrdU (5-bromo-2’-deoxyuridine) and hydroxyurea to investigate the effects of propionate on IEC migration in vivo.26 We report here that propionate increases both cell speed and persistence through inhibition of HDAC and activation of GPR43 and STAT3 to promote epithelial cell migration and wound closure, which enhances epithelial turnover and protection against ulcer formation.
Results
Propionate Promotes the Migration of IECs
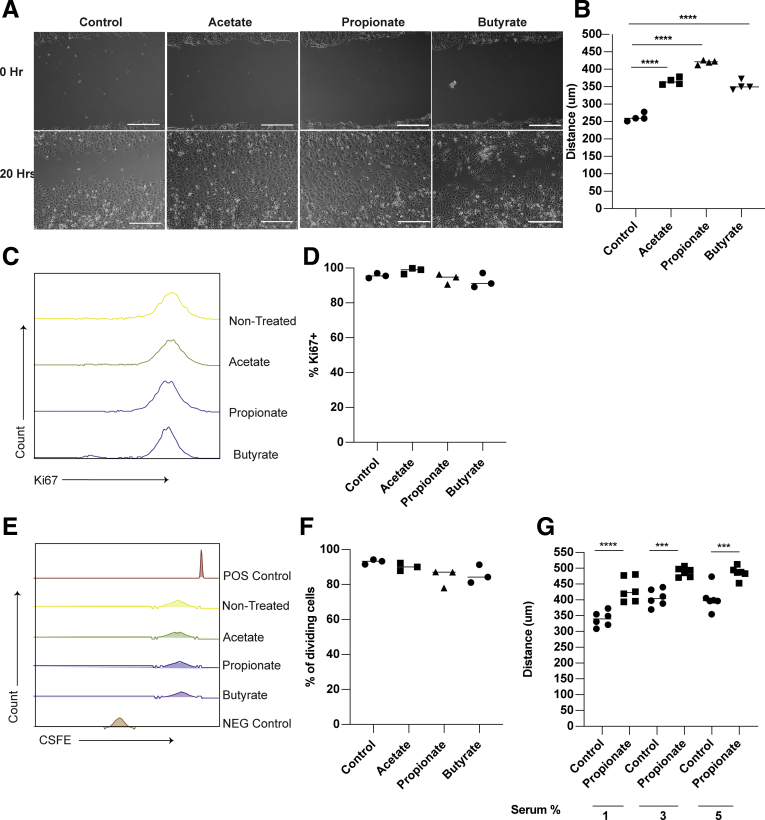
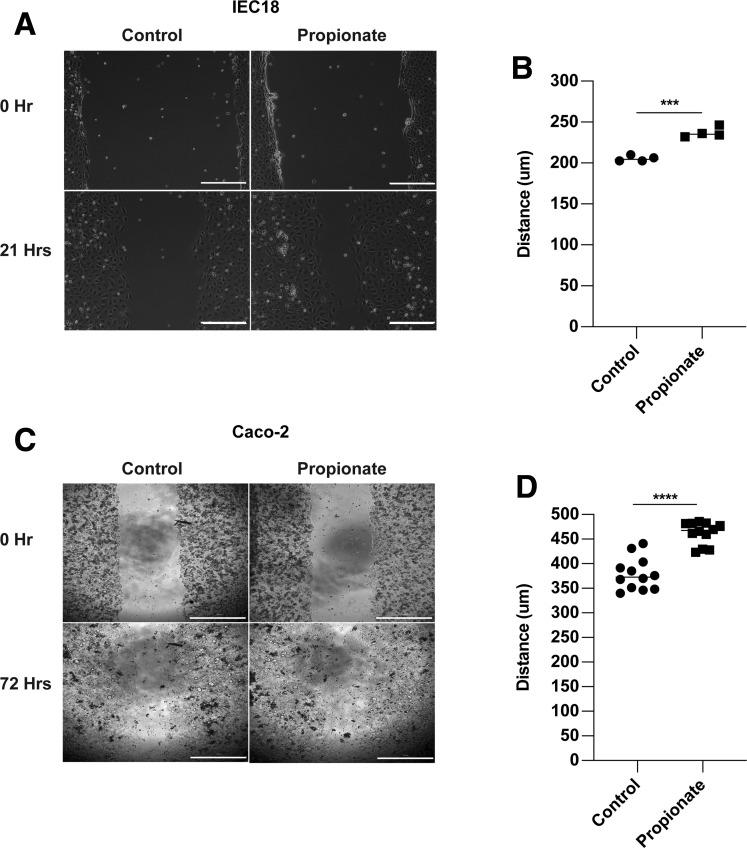
To determine whether propionate stimulates epithelial migration, we used a classic in vitro wound healing model to monitor the movement of cells into a denuded area.29 Mouse small intestinal epithelial cells (MSIEs), an immortalized nontransformed epithelial cell line that retains properties of primary IECs,30 were used. MSIEs treated with propionate enhanced cell movement into the denuded area over 20 hours (Figure 1A and B). Acetate and butyrate also functioned similarly in promoting cell movement into the denuded area (Figure 1A and B), which is consistent with previous reports.12 To determine if proliferation was a driving factor behind the enhanced wound closure, MSIEs were treated with acetate, propionate, and butyrate for 24 hours and stained for Ki67 to measure the proliferating cells. Cells treated with SCFAs did not affect proliferation, as evidenced by no difference in Ki67 expression between SCFA-treated cells and untreated cells (Figure 1C and D). To confirm the Ki67 data, MSIEs were labeled with the fluorescent dye CFSE (carboxyfluorescein diacetate succinimidyl ester) and treated for 24 hours. No differences were seen between control and SCFA-treated cells, with most cells undergoing 1 division within 24 hours, which is similar to the average doubling time of MSIEs (Figure 1E and F). These data indicated that the ability for SCFA-treated cells to fill the denuded area is due to cell migration but not proliferation. Next, to ensure that performing the migration experiments in serum-reduced media was not the reason for the difference between control and SCFA-treated cells, we performed a migration experiment using propionate under various concentrations of fetal bovine serum (FBS). Although cells at baseline migrated faster with higher concentrations of FBS, propionate was able to significantly enhance migration over control cells under each scenario (Figure 1G). Finally, to ensure that SCFA stimulates IEC migration occurred in different species, IEC18, a nontransformed rat intestinal epithelial cell line, and Caco-2, a human colorectal cell line, were treated with or without propionate. Propionate treatment of both IEC18 and Caco-2 enhanced cell migration (Figure 2A–D). These data suggest that SCFAs promotion of IEC migration is conserved across several species.
Figure 1.
Propionate promotes the migration of intestinal epithelial cells. (A, B) MSIEs were wounded and treated with SCFAs. (A) Representative phase contrast images of MSIEs. Scale bar = 300 μm. (B) Quantification of average migration distance of 3 samples per treatment. (C, D) MSIEs were treated with SCFAs, stained for Ki67, and analyzed via flow cytometry. (C) Representative graphs and (D) quantification of Ki67 from 3 samples per treatment. (E, F) MSIEs were labeled with CSFE, treated with SCFAs, and analyzed via flow cytometry. (E) Representative graphs and (F) quantification of proliferation from 3 samples per treatment. (G) MSIEs were wounded and treated with propionate in increasing concentrations of FBS. Quantification of average migration with 6 samples per treatment. ∗P < .05, ∗∗∗P < .001 ∗∗∗∗P < .0001 by 1-way ANOVA with Tukey’s posttest.
Figure 2.
Propionate promotes the migration of rat and human epithelial cells. (A, B) IEC-18 cells were wounded and treated with propionate. (A) Representative phase contrast images. Scale bar = 300 μm. (B) Quantification of average migration distance with 4 samples per treatment. (C, D) Caco-2 cells were wounded and treated with propionate. (C) Representative phase contrast images. Scale bar = 1000 μm. (D) Quantification of average migration distance with 12 samples per treatment group. ∗∗∗P < .001, ∗∗∗∗P < .0001 by 2 tailed Student’s t-test.
Propionate Promotes IEC Spreading and Polarization
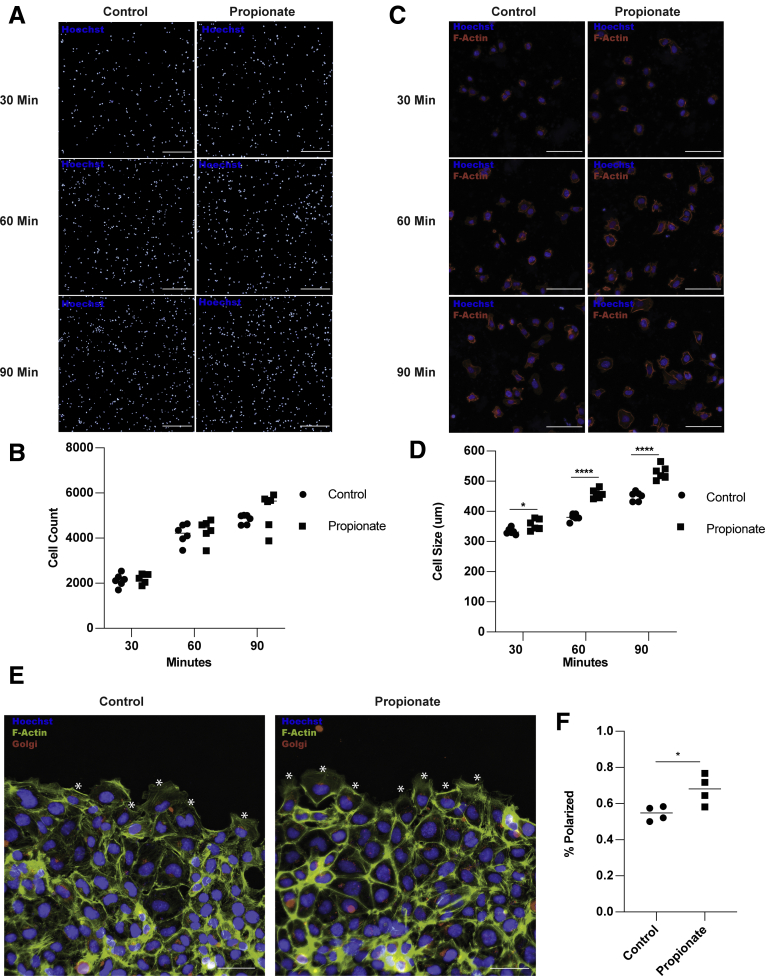
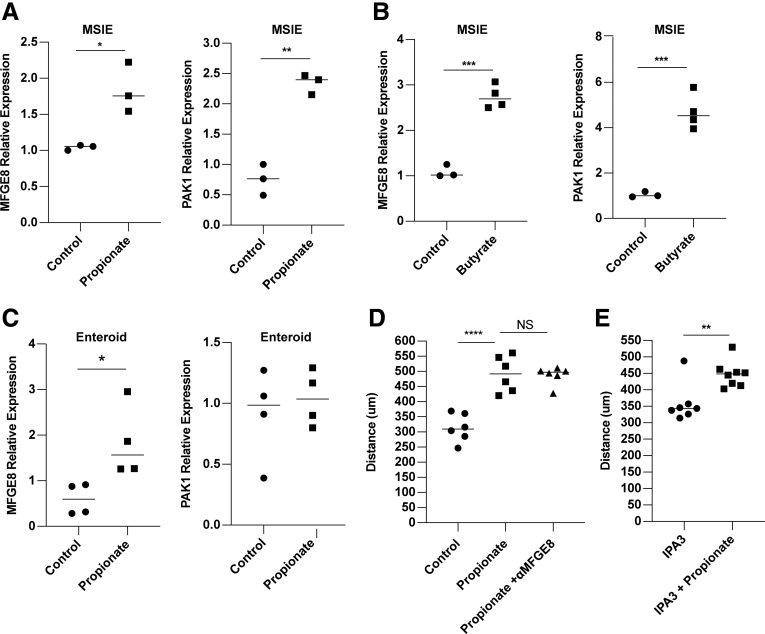
Epithelial migration requires continuous actin remodeling to allow for extension and retraction of cell lamellipodia, filopodia, and adhesions.31,32 This process is critical for epithelial turnover and wound healing.26,33 To investigate whether propionate promotes actin remodeling, we pretreated MSIEs for 16 hours with or without propionate. After 16 hours, cells were gently dissociated and replated onto Matrigel-coated plates for 30, 60, or 90 minutes, followed by actin staining and quantification for attachment and cell spreading. No differences were observed in cell attachment (Figure 3A and B). However, there was a significant increase in cell size in MSIEs treated with propionate, with many cells spreading rapidly by 60 minutes postplating (Figure 3C and D). To investigate if propionate could enhance cell polarization, which is important for cell directionality,34 we pretreated MSIEs with or without propionate for 16 hours before wounding the monolayers with a scratch. Two hours postscratch, cells were stained for golgin-97 to visualize the Golgi complex, which relocates between the nucleus and leading edge of polarized cells, and is indicative of cell polarization.34 Propionate was able to increase the number of cells polarized along the wound margins, with many cells positioning the Golgi between the nucleus and forming lamellipodia as compared with control cells, in which Golgi staining was more often equally distributed perinuclear with smaller lamellipodia (Figure 3E and F). Additionally, propionate treatment upregulated the expression of P21-associated kinase 1 (Pak1), a downstream target of CDC42, important for cell spreading and lamellipodia dynamics in MSIEs,35 and milkfat-globule epidermal growth factor 8 (Mfge8) in MSIE, which has also been shown to play a role in cell spreading with cells taking on a type II migration phenotype (Figure 4A).36 This finding was also extended to butyrate in MSIEs (Figure 4B). Propionate treatment also upregulated MFGE8 but not PAK1 expression in enteroids (Figure 4C). To determine whether propionate induces cell spreading and polarization through PAK1 and MFGE8, we included either anti-MFGE8 or the PAK1 inhibitor IPA-3 in scratch assays. However, neither neutralization of MFGE8 nor inhibition of PAK1 was able to decrease propionate-induced cell migration (Figure 4D and E). Together, these data suggest that propionate could increase cell migration by promoting cell spreading and Golgi positioning independent of PAK1 and MFGE8.
Figure 3.
Propionate promotes IEC spreading and polarization. (A–D) MSIEs were pretreated with propionate for 16 hours and then replated onto Matrigel-coated plates for 30, 60, or 90 minutes to assess attachment and spreading. (A) Representative images of cells stained for Hoechst (blue). Scale bar = 300 μm. (B) Quantification of cell attachment with 6 samples per treatment. (C) Representative images of the cells stained for phalloidin (red) or Hoechst (blue). Scale bar = 20 μm. (D) Quantification of average cell area per sample with 6 samples per treatment. (E, F) MSIEs were pretreated with propionate for 16 hours, wounded, and stained for phalloidin (green), golgin-97 (red), and Hoechst (blue) 2 hours postwounding. (E) Representative immunofluorescent images with (∗) indicating polarized cells. Scale bar = 100 μm. (F) Quantification of cell polarization with 4 samples per treatment. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001 by 1-way ANOVA with Tukey’s posttest for groups of 3 or more and by Student’s t test for groups of 2.
Figure 4.
Propionate-induced migration is independent of MFGE8 and PAK1. (A, B) MSIEs were treated for 24 hours with propionate or butyrate and analyzed for messenger RNA expression of MFGE8 and PAK1. (C) Jejunal enteroids were treated for 24 hours with propionate and analyzed for messenger RNA expression of MFGE8 and PAK1. Relative expression of MFGE8 and PAK1 with 3–4 samples per treatment group. (D, E) MSIEs wounded and treated with propionate with or without inhibitors. (D) Quantification of average migration distance of cells treated with MFGE8 neutralizing antibody with 7 samples per treatment. (E) Quantification of average migration distance of cells treated with the PAK1 inhibitor IPA3 with 7 samples per treatment. ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001 by 1-way ANOVA with Tukey’s posttest for groups of 3 or more and by Student’s t test for groups of 2.
Propionate Promotes IEC Speed and Persistence
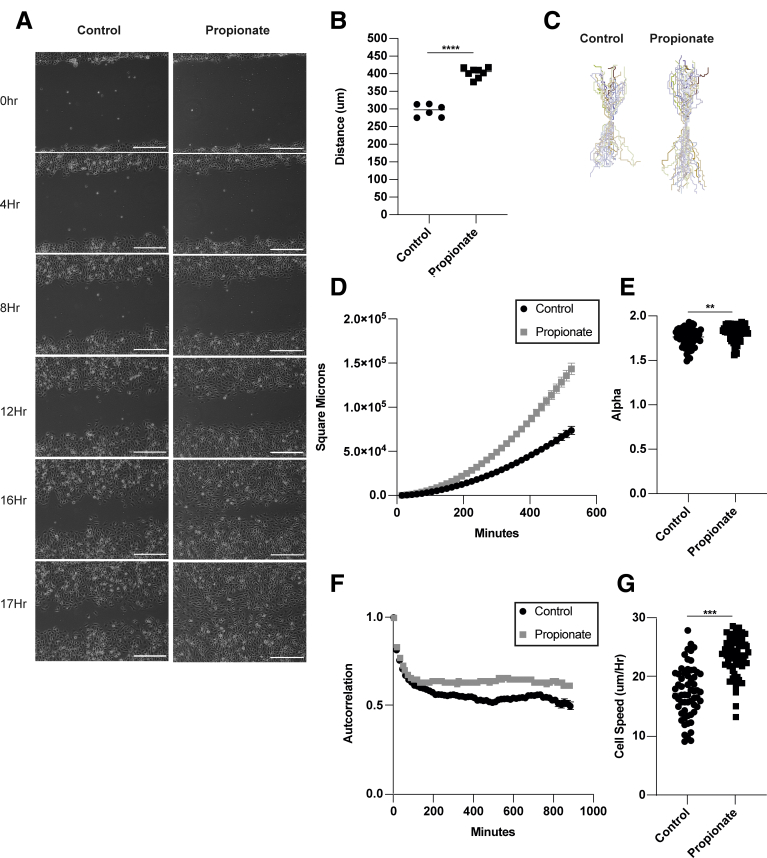
Appropriate cell migration is critical to the development and homeostasis of tissues.37 It has been shown that there is a universal coupling between cell speed and cell persistence, and that faster cells move in straighter lines.38 However, cell persistence can also be affected by the physical constraints of neighboring cells during sheet migration, which constantly remodels their junctions to uniformly migrate together.39 To investigate whether SCFA treatment affects cell speed and persistence, we performed video microscopy. MSIEs were treated with or without propionate, and cell movement was recorded every 15 minutes for up 24 hours (Supplementary Movie 1). Treatment with propionate enhanced cell migration (Figure 5A and B), which was seen from approximately 8–12 hours posttreatment of propionate (Figure 5A). Cell motion was analyzed by tracking the centroid position of cells that moved the furthest during the assay. Plot at origins revealed that cells treated with propionate moved further and straighter than nontreated cells (Figure 5C). Analysis via DiPER showed a significant increase in mean-squared displacement (MSD), which has been shown to take into account both cell speed and persistence, when treated with propionate (Figure 5D). To determine cell persistence, we calculated alpha values, indicative of more persistent movement toward the wounded area, by taking the log slope of the MSD curve. Alpha values range from 1 to 2, with 1 being completely random motion and 2 indicating nonrandom directed motion. Propionate treatment increased alpha values (Figure 5E). To confirm this finding, we also calculated cell autocorrelation, which measures persistence independent of speed by using only the angles of the vector tangential to the cells trajectory.40 This calculation revealed increased cell persistence in response to propionate treatment (Figure 5F). Last, we calculated cell speed by averaging the instantaneous speed of cells every 15 minutes. This revealed that propionate treatment significantly increased cell speed (Figure 5G). Taken together, these data indicate that by promoting cell spreading and polarization, propionate treatment increases both cell speed and persistence, which are critical for enhanced cell movement.
Figure 5.
Propionate promotes IEC speed and persistence. (A–G) MSIEs were wounded and video microscopy was performed. Videos were analyzed by tracking the centroid position of 15–20 cells per sample that moved the furthest during the assay. (A) Representative phase contrast images of MSIEs at 0, 4, 8, 12, 16, and 17 hours. Scale bar = 300 μm. (B) Quantification of average migration distance with 5–6 samples per treatment. (C) Plot at origin graphs of 58 cells from 3 samples per treatment. (D) MSD of 58 cells from 3 samples per treatment. (E) Alpha values of 58 cells from 3 samples per treatment. (F) Autocorrelation of cells over time representative of 58 cells from 3 samples per treatment. (G) Average cell speed per hour of 58 cell from 3 samples per treatment. ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001 by 1-way ANOVA with Tukey’s posttest for groups of 3 or more and by Student’s t test for groups of 2.
HDAC Inhibition and GPR43 Mediate the Effects of Propionate on IEC Migration
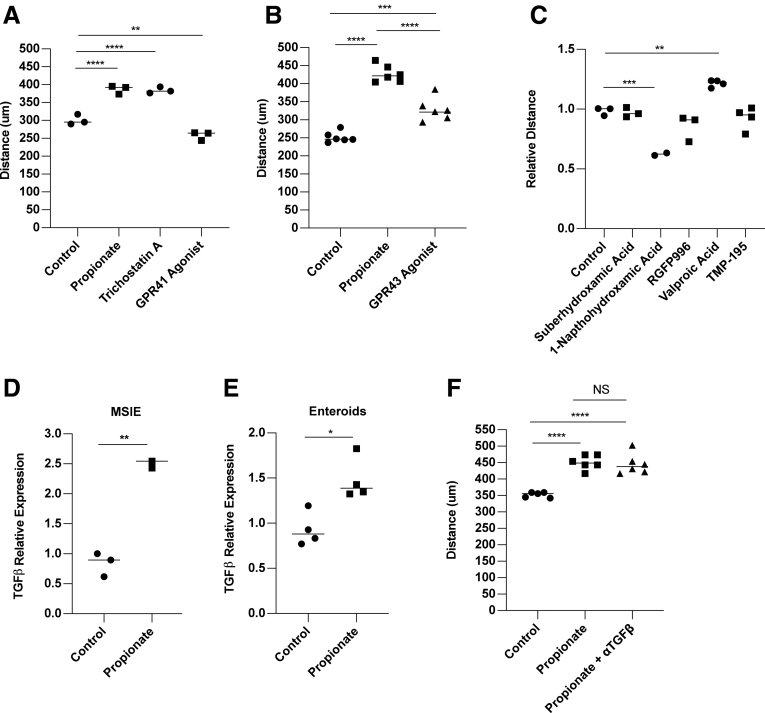
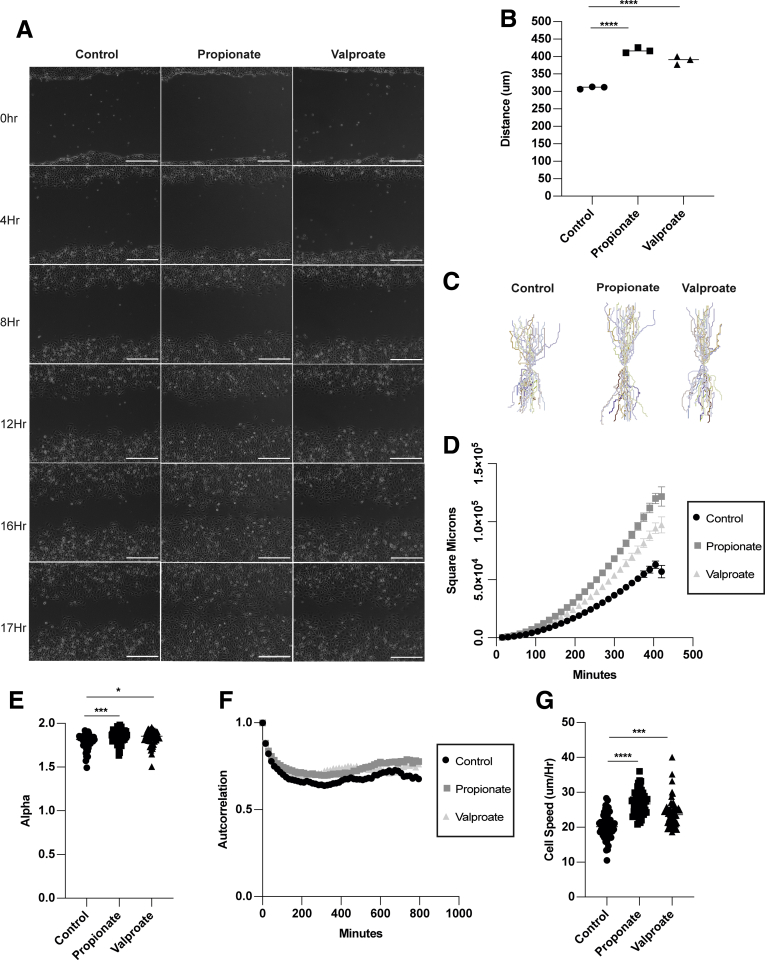
It has been shown that SCFAs function through binding their receptors GPR41, GPR43, and GPR109, and through inhibition of HDAC.41, 42, 43, 44, 45 HDAC inhibitors (HDACis) are global regulators of gene transcription due to their ability to module histones.46 HDACis have been shown to stimulate cell migration through a TGFβ-dependent pathway and enhance wound healing in vivo.47 Additionally, GPR43 is known to promote neutrophil migration into the gut during inflammation.48 To determine whether HDAC inhibition and GPR stimulation mediate propionate induction of IEC migration, we treated MSIEs with the global HDACi trichostatin A to mimic the HDAC inhibitory function of propionate, as well as ligands for GPR41 and GPR43, the receptors for propionate. We found that trichostatin A was able to significantly enhance MSIE cell migration, with GPR43 agonist also playing a role and GPR41 having an inhibitory effect (Figure 6A and B). HDACs are divided into 3 classes: type I, IIA, and IIB. Next, we investigated the specificity of propionate-mediated HDACi in MSIEs. To determine which HDACs were important for MSIE migration, we performed an HDACi screen with various inhibitors specific for different classes of HDAC, including TMP195a (a class IIa inhibitor), SBHA (a HDAC1 and HDAC3 inhibitor), 1-napthohydroxamic acid (a HDAC8, HDAC1, HDAC6 inhibitor), RGFP966 (a HDAC3 inhibitor), and valproate (an inhibitor of class I HDAC and proteasomal degradation of HDAC2).49 We found that only valproate was able to recapitulate the effects seen with propionate (Figure 6C). Next, we investigated the role of TGFβ in this process, as HDACi have been shown to promote TGFβ expression and TGFβ-dependent cell migration.10,47 We confirmed that propionate increased TGFβ expression in MSIEs (Figure 6D) and enteroids (Figure 6E) as previously reported.50 We then performed a scratch assay with propionate in the presence of a TGFβ-neutralizing antibody. However, TGFβ neutralization had little effect on migrating MSIEs treated with propionate (Figure 6F). To determine whether valproate could enhance both cell speed and persistence in a similar manner to propionate, we performed video microscopy with MSIEs and analyzed cell motion in response to valproate treatment (Supplementary Movie 2). Valproate treatment was able to enhance cell migration similar to propionate treatment (Figure 7A and B). However, valproate-treated cells did not close the wound gap as fast as propionate-treated cells, with a difference occurring with propionate-treated cells for 8–12 hours vs 12–14 hours with valproate (Figure 7A). Plot at origins showed that propionate and valproate increased the distance and straightness of cell migration (Figure 7C). Plotting MSD showed that both propionate and valproate increased displacement but to different extents (Figure 7D). Analysis of cell persistence by calculating alpha values revealed that both propionate and valproate increased cell persistence, albeit to different degrees (Figure 7E), whereas autocorrelation found that both valproate and propionate enhanced cell persistence to a similar extent (Figure 7F). Furthermore, both treatments with propionate and valproate enhanced cell speed (Figure 7G). Together, these data indicate that class I HDACis can recapitulate many of the effects of propionate-induced IEC migration, and that this mechanism is independent of TGFβ.
Figure 6.
GPR43 and inhibition of class I HDACs mediate propionate promotion of IEC migration. (A–C and E) MSIEs were wounded, then cultured with propionate, HDACis, or anti-TGFβ antibody. Quantification of average migration distance with (A) propionate, trichostatin A, and GPR41 agonist with 3 per treatment; (B) propionate and GPR43 agonist with 6 per treatment; and (C) HDAC inhibitors with 2–3 per treatment. (D) MSIEs or (E) jejunal enteroids were treated for 24 hours with propionate and analyzed for messenger RNA expression. Relative expression of TGFβ with 3 samples per treatment group. (F) Quantification of average migration distance with propionate with or without anti-TGFβ antibody. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001 by 1-way ANOVA with Tukey’s posttest for groups of 3 or more and by Student’s t test for groups of 2.
Figure 7.
Inhibition of HDAC mediates the effects of propionate on IEC migration. MSIEs were wounded and treated with propionate or valproate. Video microscopy was performed with images taken every 15 minutes. Videos were analyzed by tracking the centroid position of 15–20 cells per sample that moved the furthest during the assay. (A) Representative phase contrast images of MSIEs at 0, 4, 8, 12, 16, and 17 hours. Scale bar = 300 μm. (B) Quantification of average migration distance with 3 samples per treatment. (C) Plot at origin graphs of 52, 52, and 49 cells for control, propionate-treated, or valproate-treated samples from 3 samples per treatment. (D) MSD of 52, 52, and 49 cells for control, propionate-treated, or valproate-treated samples from 3 samples per treatment. (E) Alpha values of 52, 52, and 49 cells for control, propionate-treated, or valproate-treated samples from 3 samples per treatment. (F) Autocorrelation of cells over time representative of 52, 52, and 49 cells for control, propionate-treated, or valproate-treated samples from 3 samples per treatment. (G) Average cell speed per hour of 52, 52, and 49 cells for control, propionate-treated, or valproate-treated samples from 3 samples per treatment. ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001 by 1-way ANOVA with Tukey’s posttest for groups of 3 or more and by Student’s t test for groups of 2.
STAT3 Is Critical for Propionate Induction of Cell Persistence
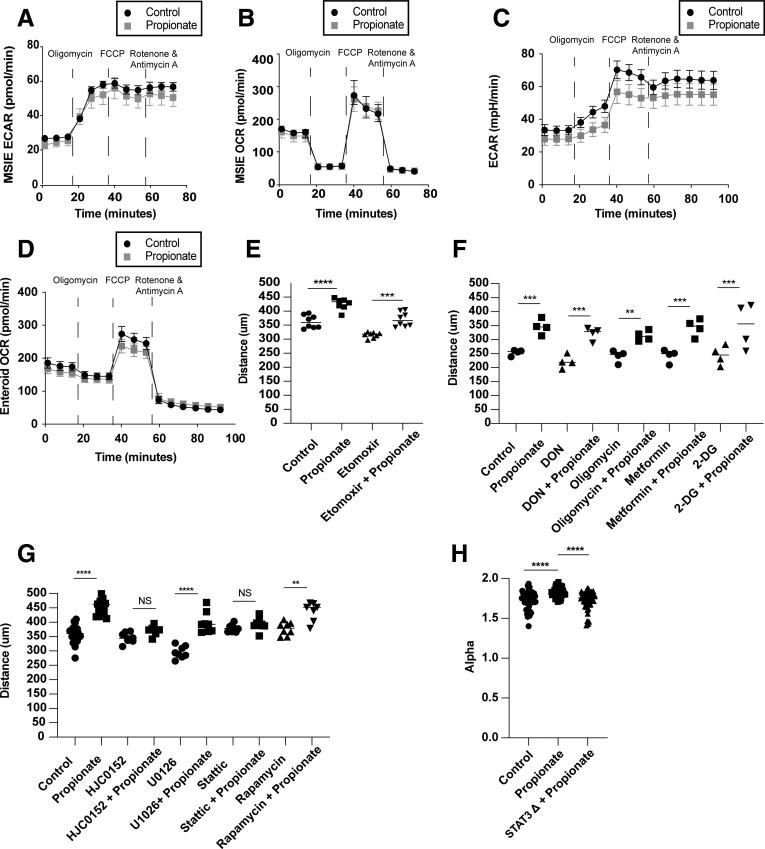
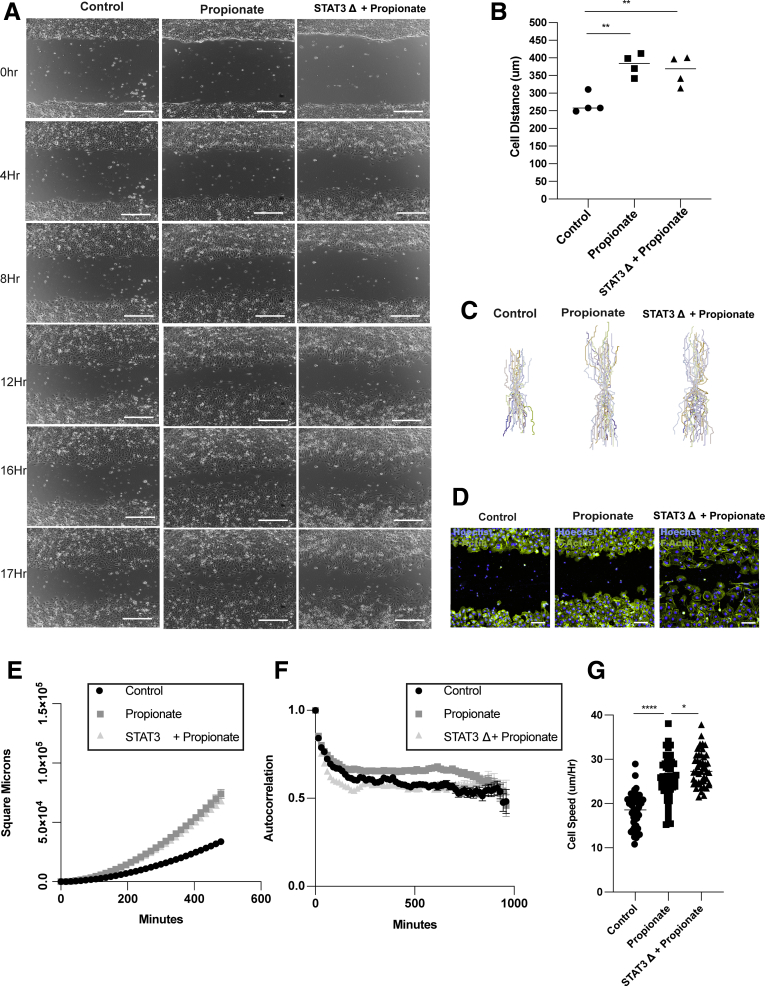
SCFAs are known to act as an energy source for intestinal epithelial cells.45 Additionally, SCFAs affect cell metabolism of epithelial cells by promoting oxidative phosphorylation.51,52 To investigate whether propionate affects IEC metabolism, MSIEs or enteroid monolayers were treated with or without propionate for 8 hours and subjected to an extracellular flux Seahorse analyzer to measure the oxygen consumption rate (OCR), which is primarily attributed to mitochondrial oxidation, and the extracellular acidification rate (ECAR) that represents glycolysis. There were no significant differences in oxygen consumption or extracellular acidification rate, indicating that propionate at this early time point did not affect cell metabolism (Figure 8A–D). To verify this result, we performed scratch assays in the presence of various metabolic inhibitors for glycolysis and mitochondrial oxidation, including etomoxir (a fatty acid oxidation inhibitor), DON (6-Diazo-5-oxo-L-norleucine) (a glutamine inhibitor), oligomycin (a complex 5 inhibitor), metformin (a TCA inhibitor), and 2-deoxyglucose (a glycolysis inhibitor). All metabolic inhibitors tested had no effect on propionate-induced migration (Figure 8E and F). We have previously shown that SCFAs in a GPR43-dependent manner promoted activation of STAT3, mammalian target of rapamycin (mTOR), and mitogen-activated protein kinase (MEK).22 Because GPR43 agonist increased cell migration of MSIE (Figure 6B), we investigated whether any of these pathways had an effect on propionate-induced IEC migration. We performed a screen using U0126 (a MEK inhibitor), HJC0152 and Stattic (STAT3 inhibitors), and rapamycin (an mTOR inhibitor) with or without propionate. We found that both Stattic and HJC0152, but not other inhibitors, attenuated propionates ability to induce migration (Figure 8G). To examine the effects of STAT3 on propionate-induced migration, we performed video microscopy with wild-type (WT) and STAT3 knockout (KO) MSIEs, as previously described,53 in the presence or absence of propionate (Supplementary Movie 3). Interestingly, deficiency in STAT3 had no effect on total migration distance in response to propionate treatment (Figure 9A and B). However, further analysis revealed major differences between WT and STAT3 KO MSIEs. Plots at origins revealed that although propionate-treated STAT3 KO cells moved a similar distance as propionate-treated WT cells, the movement was more random (Figure 9C). During migration experiments, the leader cells at the front edge of the wound margins in STAT3 KO cells tended to separate from the rest of the monolayer (Figure 9A). To confirm this finding, we performed phalloidin staining for F-actin. Propionate treatment promoted sheet migration in WT cells but often led to dissociation of the epithelial sheet in STAT3 KO cells (Figure 9D). Plotting MSD confirmed similarities in the total migration distance, as there was no difference between propionate-treated WT and STAT3 KO cells (Figure 9E). However, calculating alpha values indicated large differences in cell persistence between propionate-treated WT and STAT3 KO cells (Figure 8H), which was further confirmed by autocorrelation (Figure 9F). Furthermore, although propionate significantly increased cell speed over WT cells, propionate treatment further enhanced STAT3 KO cell speed (Figure 9G). Collectively, these data indicated that STAT3 is critical for propionate-induced directional persistence but is dispensable for the enhanced speed seen with propionate treatment. This is important as it has been shown that conditional STAT3 KO in IECs leads to severe experimental colitis and aberrant wound healing.54
Figure 8.
Propionate induced migration is independent of metabolism and dependent on STAT3. (A, B) MSIEs or (C, D) jejunal enteroids monolayers were treated for 8 hours with propionate followed by a Mito Stress Test. (A, C) ECAR and (B, D) OCR over time with 3 samples per treatment for panels A and B and 4–6 samples per treatment for panels C and D. MSIEs were wounded and treated with propionate with or without (E) etomoxir or (F) DON, oligomycin, metformin, or 2-DG. Quantification of average migration distance with 3–8 samples per treatment group. (G) Quantification of average migration distance of MSIEs treated with propionate with or without HJC0152, U0126, Stattic, or rapamycin with 3 or more samples per treatment. (H) Alpha values of 45, 48, and 47 cells for control, propionate-treated, or STAT3 KO–treated samples from 3 samples per treatment. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001 by 1-way ANOVA with Tukey’s posttest for groups of 3 or more and by Student’s t test for groups of 2.
Figure 9.
STAT3 is critical for propionate induction of cell persistence. WT and STAT3 KO MSIEs were wounded and video microscopy was performed. Videos were analyzed by tracking the centroid position of 15–20 cells per sample that moved the furthest during the assay. (A) Representative phase contrast images of MSIEs at 0, 4, 8, 12, 16, and 17 hours. Scale bar = 300 μm. (B) Quantification of average migration distance with 4 samples per treatment. (C) Plot at origin graphs of 45, 48, and 47 cells for control, propionate-treated, or STAT3 KO–treated samples from 3 samples per treatment. (D) MSIEs were wounded and phalloidin (green) and Hoechst (blue) staining was performed 16 hours postwounding. Scale bar = 100 μm. (E) MSD of 45, 48, and 47 cells for control, propionate-treated, or STAT3 KO–treated samples from 3 samples per treatment. (F) Autocorrelation of cells over time representative of 45, 48, and 47 cells for control, propionate-treated, or STAT3 KO–treated samples from 3 samples per treatment. (G) Average cell speed per hour of 45, 48, and 47 cells for control, propionate-treated, or STAT3 KO–treated samples from 3 samples per treatment. ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001 by 1-way ANOVA with Tukey’s posttest for groups of 3 or more and by Student’s t test for groups of 2.
Propionate Stimulates Migration of IECs In Vivo
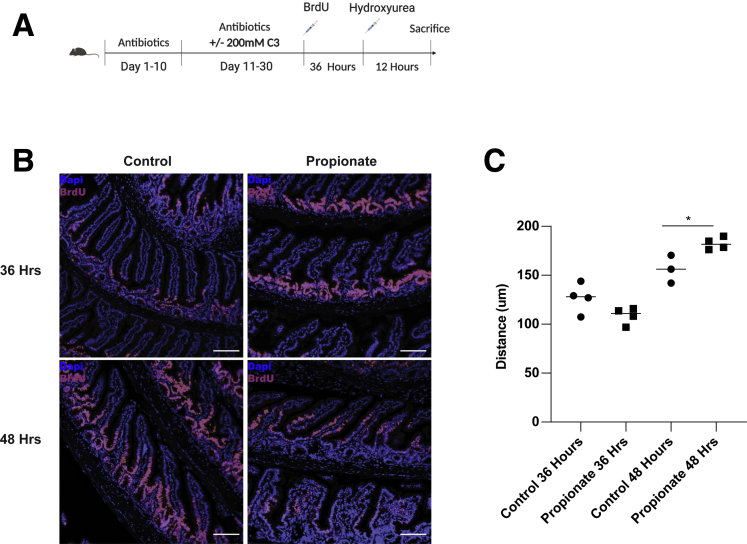
It has been shown that SCFAs are pertinent for epithelial turnover.24 However, until recently, it was not known that epithelial cells actively migrate up the villus independent of crypt proliferation.25 To quantitatively assess whether propionate could stimulate migration of cells up the crypt villus axis in vivo, we employed a protocol as previously reported using hydroxyurea.26 WT mice were fed a cocktail of antibiotics including vancomycin, metronidazole, ampicillin, and kanamycin for 10 days as we previously described.55 After 10 days, mice were continuedly fed the antibiotic cocktail with or with 200-mM propionate for an additional 21 days. All groups of mice were intraperitoneally injected with 100 mg/kg of BrdU. Thirty-six hours postinjection, some of the mice were sacrificed and used for baseline measurements of epithelial migration. The remaining mice received an injection of 50 mg/kg hydroxyurea to halt crypt proliferation and sacrificed 12 hours later (Figure 10A). Thirty-six– and 48-hour time points were used based on data from a previous report that found that proliferation in the crypts affects villus migration the least when cells were at the midway point of the villus.26 To further protect against the effect of proliferation, low-dose hydroxyurea was also used. We found that the BrdU front in control and propionate-treated mice moved a similar distance 36 hours postinjection (Figure 10B and C). However, by 48 hours post–BrdU injection, the BrdU front in propionate-treated mice was significantly increased compared with control mice, with propionate-treated cells moving an average of 73.6 μm (95% confidence interval, 49.61–97.71 μm) compared with 29.3 μm (95% confidence interval, 3.32-55.28 μm) in control cells (Figure 10B and C). Taken together, these data indicate that propionate stimulates the migration of IECs along the villus independent of proliferation.
Figure 10.
Propionate stimulates IEC migration in vivo. (A) Schematic of measuring epithelial migration in vivo. (B) Representative images of BrdU (red/pink) and DAPI (blue) in the ileum of control and propionate-treated mice at 36 and 48 hours post–BrdU injection. Scale bar = 100 μm. (C) Quantification of BrdU height from the base of the crypt to the uppermost positive BrdU cell with 3–4 samples per group. ∗P < .01 by 2-way ANOVA with Tukey posttest.
Propionate Reduces Ulceration in Experimental Colitis
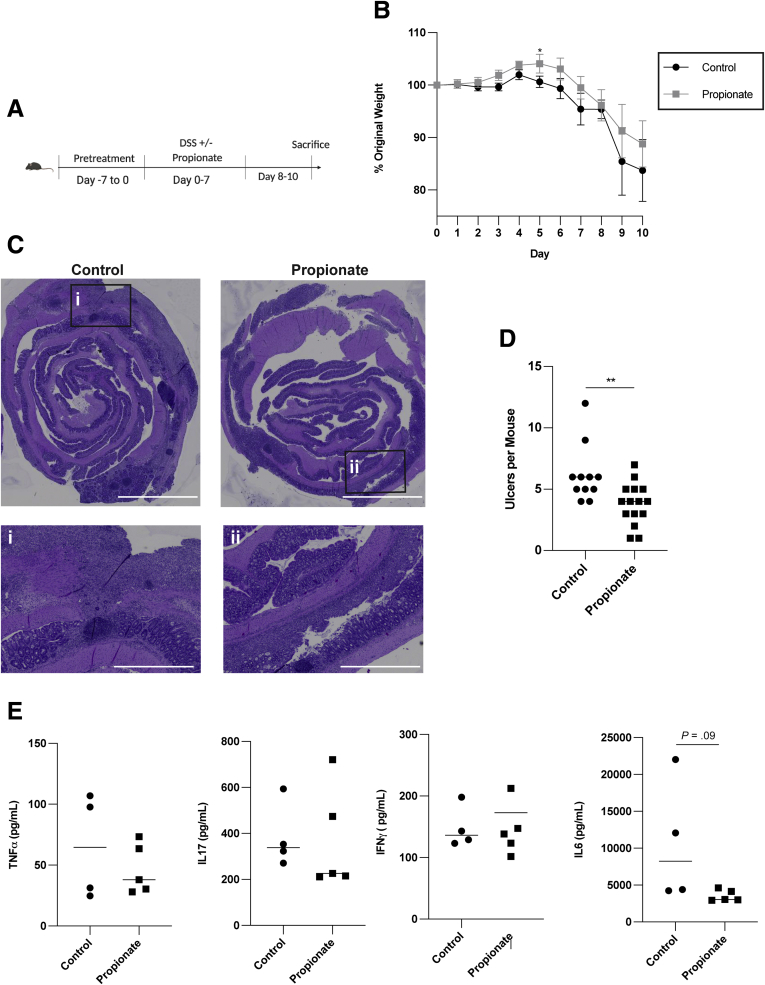
Finally, we examined if propionate could protect mice against the development of colitis in experimental colitis upon dextran sulfate sodium (DSS) insult (Figure 11A). WT mice were pretreated for 7 days with or without 200 mM propionate in their drinking water. Then mice were given DSS containing water with or without 200 mM propionate for 7 days before being switched to regular drinking water with or without propionate for an additional 3 days. Mice were monitored daily for weight changes. Mice were sacrificed 10 days postadministration of DSS. There were no differences in weight loss between either group (Figure 11B). However, propionate-treated mice had a significant reduction in ulcer development in the colon (Figure 11C and D). There were no differences in inflammatory cytokine production, including tumor necrosis factor alpha (TNFα), interleukin (IL)-17, interferon gamma (IFNγ), and IL-6, between control and propionate-treated mice (Figure 11E). These results indicate that propionate mainly affects intestinal epithelial cell migration and wound healing but not inflammatory responses.
Figure 11.
Propionate reduces ulceration in experimental colitis. (A) Schematic of DSS colitis model. (B) Percent of original weight with 4–5 mice per treatment. (C) Representative hematoxylin and eosin images of control and propionate-treated mice. Scale bar = 4000 μm for whole mount images and 1000 μm for zoomed regions. (D) Total ulcers per mouse for 12 control and 15 propionate-treated mice pooled from 3 independent experiments. (E) Colonic organ culture for inflammatory cytokines with 4–5 samples per treatment. (D) ∗P < .05, ∗∗P < .01 by 2-way ANOVA with Tukey posttest and (B) by multiple t test corrected for multiple comparisons using the Holm-Sidak method.
Discussion
SCFAs are the primary gut bacterial products of fermentation of soluble fibers contributed from foods such as fruits, vegetables, and grains, which are known to be protective against colorectal cancer and potentially beneficial for patients with inflammatory disorders such as IBD.56, 57, 58, 59 However, the mechanisms by which they protect the intestinal epithelium are still being unraveled. In this study, we found that propionate and other SCFAs stimulated IEC migration that was independent of cell proliferation. The propionate-enhanced IEC migration was collective rather than single cell in nature, which is pertinent for processes such as epithelial turnover and wound healing, in which neighboring junctions with other cells remain intact.37,39 Importantly, propionate drives intestinal epithelial migration through regulating cell speed and persistence in an HDAC inhibition–, GPR43-, and STAT3-dependent manner.
It has been shown that migrating cells require 3 processes for effective migration: attachment, polarization, and actin polymerization. In the gut, actin polymerization was shown to be a major driving factor for cell migration on the villus.26 We found that propionate upregulated genes involved with actin reorganization such as Pak1 and Mfge8.36,60, 61, 62, 63 However, although propionate drives cell spreading and polarization without affecting attachment, this process was independent of both Pak1 and MFGE8, despite these proteins being linked to this process. These differences suggest that each of these genes is dispensable for this function possibly via the redundant actions of downstream proteins. Using video microscopy, we found that propionate enhanced both cell speed and persistence, most likely due to its ability to increase cell spreading and enhance polarization, which is pertinent for both cell speed and directionality. Thus, our findings support the argument that both cell speed and persistence are linked to cell migration.38
Propionate exerts its actions on cells via G-coupled protein receptors and by acting as an inhibitor for HDAC.41,44 Here we found that HDAC inhibition and GPR43 mediated propionate-induced cell speed and persistence. This indicates that modulation of gene transcription is most likely pertinent to propionate-induced migration, although we were not able to identify a single gene that contributed to this process. We also found that inhibition of class I HDACs via valproate could recapitulate the effects of propionate on cell migration. Valproate has been shown to be beneficial in ameliorating colitis in experimental colitis, which raises the possibility that valproate is stimulating epithelial migration to allow for recovery in these mice.64 Additionally, our finding is in line with a previous report, which found that the HDAC inhibition could stimulate the migration of intestinal epithelial cells.47 However, in our model, TGFβ was not responsible for the induction of migration. This is most likely due to the duration of the migration experiment. Epithelial restitution is a fast process, occurring within 24–48 hours. Thus, we chose to assay over a shorter time period, whereas the previous report chose to assess migration over 6 days, where TGFβ secretion into the supernatant would most likely have a major impact on migration as is well known.10
SCFAs are known to alter the metabolism of IECs, driving them toward oxidative phosphorylation and β-oxidation.45,51,65 In our study, we found that propionate was unable to change the metabolic activity in IECs in the early stages. We assessed metabolic activity 8 hours posttreatment with propionate, whereas other groups in previous reports have examined this effect at 24 hours with SCFAs such as butyrate.51,52 Thus, the difference in incubation, SCFA used, and dosage could be a significant factor affecting the results. However, this time point was used due to the significant changes in cell migration between nontreatment and propionate treatment at the 8- to 12-hour time frame. Additionally, the use of etomixir, a β-oxidation inhibitor, alongside propionate further confirmed that propionate was not being metabolized at this crucial time frame, but rather exerting its functions via alteration of gene expression. However, our data do not exclude a possible metabolic regulation of IECs for SCFAs at later stages of tissue repair.
SCFAs have been shown to activate many enzymes including MEK, mTOR, and STAT3 to exert their functions.22 We found that inhibition of STAT3, but not MEK and mTOR, could affect propionate-induced cell persistence. This is consistent with a previous report, which found that STAT3 was important for cell persistence in fibroblast, with STAT3 KO cells unable to effectively polarize and move collectively.66 Furthermore, we found that STAT3 KO IECs dissociated from one another and moved individually instead of as a collective sheet. Thus, although propionate-treated STAT3 KO IEC moved as far as propionate-treated WT IEC, their migration was disorganized. This is critical, as collective migration is required for epithelium tissue organization and wound healing. This is in line with a previous report that VilCre Stat3fl/fl mice, in which STAT3 is deficient specifically in IEC, have defective wound healing with acute DSS injury leading to severe disease and aberrant wound healing.54
Given that propionate was able to induce cell migration in vitro via promoting cell spreading and polarization, we assessed its ability to promote migration in vivo. To eliminate the effect of existing bacteria produced propionate in the intestinal lumen, we treated mice with an antibiotic cocktail. BrdU time points of 36 and 48 hours postinjection were used to minimize the effect of cell proliferation on migration as found in a previous report.26 Additionally, to further reduce the proliferative effect, we injected mice with hydroxyurea, an S-phase inhibitor. We found that propionate induced migration of cells on the villus. This finding suggests that propionate is a major regulator of epithelial turnover, enhancing both cell proliferation and cell migration, which would allow the coordinated renewal of the intestinal epithelium. Of note, we did not find that propionate significantly enhanced proliferation at the 36-hour time point, as previously reported.24 However, our finding does not contradict theirs, as they assessed proliferation at a 48-hour time point and used a cocktail of SCFAs, rather than a single SCFA propionate as in our study. Based on our findings that propionate induces IEC migration, and previous results indicating that HDAC inhibition ameliorated colitis,47,64 we expected that propionate would protect against colitis. Indeed, our results demonstrated that propionate was able to reduce ulceration of the epithelium.
In summary, we demonstrate that propionate promotes epithelial migration to drive epithelial turnover and repair, which depends on HDAC inhibition of class I HDACs, GPR43, and activation of STAT3, allowing for collective cell migration. This suggests that at the base of the crypt, where HDAC inhibitory activity is low, SCFAs are able to promote stem cell proliferation.23 However, as SCFA concentrations rise as cells move out of the crypt, HDAC inhibition enhances cell migration by promoting cell spreading to coordinate the movement of cells out of the crypt with active migration of cells up the villus and eventual cell extrusion into the lumen. Our findings thus suggest a novel mechanism for how SCFAs contribute to intestinal homeostasis.
Materials and Methods
Animals and Models
C57BL/6 WT mice were purchased from the Jackson Laboratory (Bar Harbor, ME). All mice were housed in the specific pathogen-free animal facility in the Animal Resource Center at the University of Texas Medical Branch. All described animal experiments were performed in accordance with protocols reviewed and approved by the Institutional Animal Care and Use Committees of the University of Texas Medical Branch.
For DSS model of colitis, WT mice were fed water ± 200 mM propionate ad libitum for 7 days. Mice were then fed water containing 1.9% DSS (Cat# DS1004; Gojira FC, Cleveland, OH) w/v ± 200 mM propionate for 7 days. A 3-day washout was performed in which mice were fed water ± 200 mM propionate. Mouse weights were monitored daily.
For epithelial migration in vivo, mice were fed water ad libitum containing an antibiotic cocktail containing 1g/L of metronidazole (Cat# 443-48-1; Acros Organics, Geel, Belgium), ampicillin (Cat# BP1760-5; Thermo Fisher Scientific, Waltham, MA), kanamycin (Cat# BP906-5; Thermo Fisher Scientific), and 0.5 g/L of vancomycin (Cat# 1404-93-9; Acros Organics) for 10 days. Mice were then treated with water containing the antibiotic cocktail ± 200 mM propionate for an additional 21 days. Mice were intraperitoneally injected with 100 mg/kg of BrdU (Cat# ab142567; Abcam, Cambridge, United Kingdom). Half of the mice were sacrificed 36 hours post-BrdU injection. The remaining mice were injected with 50 mg/kg of the S phase Inhibitor hydroxyurea (Cat# 127-07-1; Acros Organics) to inhibit crypt proliferation, and sacrificed 12 hours post–hydroxyurea injection.
Reagents
Recombinant murine IFNγ (Cat# 575306) was purchased from BioLegend (San Diego, CA). Culture medium RPMI 1640 (Cat# SH30027.01), penicillin/streptomycin (Cat# SV30010), were purchased from GE Healthcare (Milwaukee, WI. ITS (Cat# 354350) and Matrigel (Cat# 356231) were purchased from Corning (Corning, NY). Dulbecco’s modified Eagle’s medium (DMEM) (Cat# 30-2002) was purchased from ATCC (Manassas, VA). Advanced DMEM/F12 (advanced DMEM) was purchased from Gibco (Gaithersburg, MD). Recombinant mouse endothelial growth factor (EGF) (Cat# 2028-EG), human WNT-3A (Cat# 5036-WN), mouse Noggin (Cat# 1967-NG), and mouse R-Spondin (Cat# 3474-RS) were purchased from R&D Systems (Minneapolis, MN). The ROCK inhibitor Y-27632 dihydrochloride (Cat# 1254) was purchased from Tocris Bioscience (Bristol, United Kingdom). STAT3 inhibitor HJC0152 was synthesized by Dr. Jia Zhou’s laboratory at the University of Texas Medical Branch at Galveston following their reported procedures.67 STAT3 Inhibitor Stattic (Cat# S7024), PAK1 inhibitor IPA3 (Cat# S7093), HDACI TMP195 (Cat# S8502), valproate (Cat# S1168), RGFP966 (Cat# S7229), and 1-naphthohydroxamic acid were purchased from Selleckchem (Houston, TX). MEK inhibitor U0126 (Cat# U120) butyrate (Cat# 303410), acetate (Cat# S5636), propionate (Cat# P1880), TSA (Cat# t8552), sodium dichloroacetate (Cat# 347795), DON (Cat# D2141), Bz-423 (Cat# SML1944), metformin hydrochloride (Cat# PHR1084), 2-DG (2-deoxy-D-glucose) (Cat# D6134), and etomoxir (Cat# E1905) were purchased from Sigma-Aldrich (St. Louis, MO). Primary antibody against BrdU (Cat# ab6326) and Texas red goat anti-rat secondary (Cat# T-6392) were purchased from Abcam. Primary antibody against golgin-97 (Cat# A-21270), Texas red goat anti-mouse secondary (Cat# T-6390), phalloidin-488 (Cat# A12379), phalloidin-Texas Red (Cat# T7471), Hoechst 33342 (Cat# 62249) and Prolong Diamond Antifade Mount containing DAPI (Cat# P36970) were purchased from Thermo Fisher Scientific. The following enzyme-linked immunosorbent assay kits were purchased from BioLegend: mouse IFN-γ, IL-17, TNF, IL-6, and human IL-10 antibodies (Cat#: 431414, 430804, 432504, 430904, 431304, and 430604, respectively). For flow cytometry, Ki67 was purchased from BioLegend. Anti-MFGE8 monoclonal antibody (Cat# D199-3) was purchased from MBL International Corporation (Woburn, MA).
Knockout of STAT3 Using CRISPR
Knockout of STAT3 was performed as previously described.68 LentiCRISPR vector (plasmid no. 52961; Addgene, Cambridge, MA) was used to knockdown STAT3 in MSIEs. The design and cloning of the target guide RNA (gRNA) sequences were performed using the Zhang laboratory’s protocol (http://www.genome-engineering.org).69 Briefly, the suitable target sites for the gRNA sequence against STAT3 were established using the CRISPR design tool software (http://crispr.mit.edu). Cas9 target sequences for the indicated genes were designed in the Zhang laboratory (http://www.genome-engineering.org). Then, gRNA (Integrated DNA Technologies, Coralville, IA) were synthesized and subcloned into the lentiCRISPR-v2 vector. The newly constructed lentiCRISPR plasmids were then transfected into MSIEs. Following antibiotic positive selection, transfected cells were established as a stable cell line (STAT3 forward: 5′ACCGCGATTACCTGCACTCGCTTC3′, reverse: 5′ AAACGAAGCGAGTGCAGGTAATCGC3′).
Epithelial Cell Culture
MSIEs were cultured in RPMI 1640 medium supplemented with 5-U/mL IFNγ, 5% FBS, ITS (5 μg/mL insulin, 5 μg /mL transferrin, and 5 ng/mL selenous acid), and 100-U/mL penicillin/streptomycin at the permissive temperature of 33°C. Before treatment with SCFAs, cells were starved in RPMI 1640 medium with 0.5% or indicated FBS for 16 hours at 37°C. Caco-2 and IEC-18 cells were cultured in DMEM supplemented with 10% FBS and 100 U/mL penicillin/streptomycin at 37°C and 5% CO2. Before treatment with SCFAs, Caco-2 and IEC-18 cells were starved overnight in medium with 1% FBS at 37°C.
Enteroid Culture
Enteroids were generated as previously described.22 Briefly. The jejunum was dissected from the mouse, minced, and rocked for 30 minutes at 4°C with 2 mM EDTA. The tissue was then treated with PBS containing 43.3 mM sucrose and 54.9 mM sorbitol and rocked for 2 minutes. Supernatant was filtered through a 70-μm cell strainers. The pellet containing detached crypts was resuspended in a 50% Matrigel plug and overlayed with LWRN conditioned media (ATCC) and cultured at 37°C as previously described.70 For the first 2 days of culture, cells were additionally supplemented with 10 μM of the ROCK inhibitor Y-27632. After 2 days, the media was changed to LWRN media without Y-27632. On days 3–4, spheroids were gently dissociated and passaged using TrypLE (Thermo Fisher Scientific). For treatment of cells for RNA analysis or Seahorse XF Cell Mitro Stress Test, cells were washed twice with Advanced DMEM F12 containing 4 mM glutamine, 10 mM HEPES, and 100-U/mL penicillin/streptomycin. Treatment media was composed of advanced DMEM/F12 with 4 mM glutamine, 10 mM HEPES, 10-ng/mL WNT3A, 71-ng/mL Noggin, 75-g/mL RSPO, and 50-g/mL EGF as described previously.71
Epithelial Migration Assay
MSIE, IEC-18, and Caco-2 were seeded in either 24- or 96-well plates and allowed to attach for 24 hours. After serum starving, a scratch was made in the monolayer with a 200/L pipette tip and cells placed in their respective treatments. The cells were then placed onto the microscope stage of a Nikon Eclipse TI (Nikon, Melwille, NY) located inside of an Okolab cage incubator (Okolab, San Francisco, CA) and imaged at 10× at 37°C and 5% CO2 or into the Biotek Cytation 5 (Biotek Instruments, Winooski, VT), and imaged at 4× or 10× at 37°C and 5% CO2. For single time point experiments, wound closure was calculated as the The widths of the wounds were measured using the MRI Wound Healing Tool macro for FIJI software (National Institutes of Health, Bethesda, MD; http://dev.mri.cnrs.fr/attachments/download/1992/MRI_Wound_Healing_Tool.ijm) and manually checked for accuracy. When quantification could not be done automatically, the Fiji line tool was used to measure the area of the wound. For some experiments, cells were stained for F-actin using phalloidin-488 (Cat# A12379; Invitrogen, Carlsbad, CA) to assess the adhesion of the monolayers. For time-lapse experiments, cells were imaged continuously every 15 minutes. Single cell tracking was performed by tracking the centroid position of 15–20 cells per sample that moved the furthest using Fiji.72 Data were uploaded and analyzed via DiPER for autocorrelation (cell persistence), cell speed, MSD, and plots at origin.28 Alpha values were calculated by taking the at each time point and averaged together over the course of the entire experiment.
Epithelial Cell Spreading
The 96-well plates were coated with 60-μL of a (1:50) dilution of Matrigel in ice-cold DPBS for 1 hour at 37°C and 5% CO2. Following a 16-hour pretreatment, MSIEs were harvested using TrypLE Express (Cat# 12604021; Thermo Fisher Scientific), and plated in fresh media at 5000 cells per well onto Matrigel-coated plates. Following incubation, cells were washed to remove cells that did not adhere, fixed in 4% paraformaldehyde, blocked in 10% goat serum, and incubated with phalloidin-Texas Red-X (1:40) and Hoechst 33342 (1:1000) overnight. Cells were washed with phosphate-buffered saline with Tween 20 and imaged using the Biotek Cytation 5 (Biotek Instruments, Winooski, VT). For cell counts, the entire well was image automatically and automatically quantified using Gen5.3 (Biotek Instruments). For cell spreading, a 5 × 5 grid, which encompassed approximately 75% of the well was imaged automatically at 10×. Cell size was quantified automatically by Gen5.3 by calculating the total area of phalloidin staining. Images were checked manually for accuracy. To calculate cell spreading, the average cell size and SEM were determined for each sample.
Epithelial Polarization
MSIEs were seeded in 96-well plates and serum starved overnight. MSIEs were then pretreated with propionate for 16 hours and scratched as described previously. Two hours postscratching, cells were fixed in 4% paraformaldehyde, permeabilized with 0.1% Triton-×100, blocked in 10% goat serum, and incubated overnight incubation with golgin-97 (1:100). The following day, cells were incubated in goat anti-mouse Texas Red secondary (1:500), washed, and then incubated overnight in phalloidin-488 (1:40) and Hoechst 33342 (1:1000). Cells were imaged using the Biotek Cytation 5 at 40× in 4 similar locations across samples. Cells at the leading edge were then quantified for the extent of polarization indicated by the majority of the Golgi being located between the nucleus and the leading edge. Between 35 and 55 cells were analyzed per sample, and the percent of cells polarized was calculated as .
Flow Cytometry Analysis
For analysis of proliferation, MSIEs were plated in a 24-well plate. Following serum starvation, cells were treated with CFSE and treated for 24 hours with SCFAs. Cells were then fixed with 1% paraformaldehyde. For Ki67 staining, serum-starved MSIEs were treated for 24 hours with SCFAs. Cells were fixed and permeabilized followed by fluorochrome-conjugated anti-mouse Ki67 (1:100). Cells were then fixed in 1% paraformaldehyde. Quantification was performed with a LSRII/Fortessa and FACSDiva software (Becton Dickinson, Mountain View, CA). Data were analyzed with FlowJo (v10.7, FlowJo LLC, Ashland, OR).
BrdU Staining
Five-micrometer formalin-fixed ileal tissue was deparaffinized with CitriSolv (Cat# 04-355-121; Thermo Fisher Scientific) and rehydrated using a series of ethanol washes. Antigen retrieval was performed with sodium citrate (pH 6.8) at 100°C. Tissue was permeabilized using 0.2% Triton-x100, blocked in 10% goat serum, and incubated overnight in rat anti-BrdU (1:200). The next day, slides were incubated with rabbit anti-rat 588 (1:500). Slides were mounted using ProLong Diamond Antifade Mount containing DAPI (Thermo Fisher, Scientific). Images were acquired at 20× with a Biotek Cytation 5. The BrdU front was measured using Gen 5.3.72 A total of 15–30 villi were quantified per sample to obtain a BrdU front average and SEM.
Enzyme-Linked Immunosorbent Assays
Enzyme-linked immunosorbent assays were performed according to the manufacturer’s protocols for IL-6, IFN, TNFα, and IL17A using culture supernatant derived from organ cultures. High-affinity 96-well plates were coated with capture antibody at a (1:200) dilution and incubated overnight at 4°C. After blocking, supernatants were incubated for with detection antibody at a (1:200) followed by streptavidin conjugated to horseradish peroxidase at (1:1000) prior to the addition of TMB substrate. Absorbance was measured using a plate reader.
Quantitative Real-Time Polymerase Chain Reaction
Total RNA was extracted with TRIzol reagent (Life Technologies, Carlsbad, CA), quantified via nanodrop, and used for cDNA synthesis. Quantitative real-time polymerase chain reaction was performed by using SYBR Green Gene Expression Assays (Bio-Rad, Hercules, CA). GAPDH was used as the endogenous reference gene. The relative gene by normalizing to GAPDH (GAPDH Forward: 5′TCAACAGCAACTCCCACTCTTCCA3′, Reverse: 5′ACCCTGTTGTAGCCGTATTCA3; TFGβ Forward:5′TGACGTCACTGGAGTTGTACGG3′, Reverse: 5′GGTTCATGTCATGGATGGTGC3′; Mfge8 Forward: 5′CGCACAGGATCGTCAAT3′, Reverse: 5′CGCAGAAGGTTCACCTGGAT3′; Pak1 Forward: 5′GTGTCTGAGACCCCAGCAGTA3′, Reverse: 5′GTGGTTCAATCACAGATCGTGT3′).
Cell Metabolism Measurement
MSIEs were seeded at 8000 cells per well into a 96-well Seahorse plate. Following serum starvation, cells were treated for 8 hours with propionate and subjected to the Seahorse XF Cell Mito Stress Test (Cat# 103708-100; Agilent, Santa Clara, CA) to determine OCR and ECAR. For enteroid monolayers, spheroids grown in LWRN were dissociated and plated at 350 crypt pieces per well onto a 96-well Seahorse plated coated with 30 L of 10% Matrigel (1:10 dilution into DPBS). Cells were overlayed with LWRN conditioned media for 2 days supplemented with Y-27632. After 2 days, cells were treated in treatment media as described above with or without propionate. Following 8 hours of treatment, cells were subjected to the Seahorse XF Cell Mito Stress Test to determined OCR and ECAR.
Histopathological Assessment
At necropsy, the colon was swiss-rolled and fixed in 10% neutral buffered formalin for 24 hours. Tissue was then paraffin embedded and 5 μm sections were prepared and stained with hematoxylin and eosin. The severity of the disease was calculated based upon epithelial architecture and inflammatory. A score between 0 and 4 was given for each category and summed together to give a total pathology score between 0 and 8. Additionally, the total number of ulcers were quantified per sample. All slides were read by a blinded pathologist.
Statistical Analysis
All results were presented as mean or mean ± SEM. Student’s t test was used to test for differences between the means of 2 groups. One-way analysis of variance (ANOVA) was used to assess the differences between the means of 3 or more groups. Post hoc pairwise comparisons were assessed if more than 2 groups were present in an experiment with adjustment for multiple comparisons using the Tukey method. When data were compiled from multiple experiments, 2-Way ANOVAs assessing the treatment and experiment effects were conducted. We also conducted a post hoc test of the statistical interaction assessing if the treatment effect differed across the experiments, but this was not significant (P > .05). We reported our 2-way ANOVA results without the statistical interaction. We also conducted pairwise comparisons between the means of the treatment and control groups at prespecified time points adjusting for multiple comparisons with the Tukey method. For experiments with multiple time points comparing 2 treatments, multiple t tests were performed with each row analyzed individually without assuming a consistent SD, corrected for multiple comparisons using the Holm-Sidak method. All the statistical analysis was performed using Prism 8 (GraphPad Software, San Diego, CA). Significance was set a priori at P < .05.
CRediT Authorship Contributions
Anthony J. Bilotta, BS (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Validation: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Chunyan Ma, PhD (Investigation: Supporting; Writing – review & editing: Supporting)
Wenjing Yang, MD, PhD (Investigation: Supporting; Writing – review & editing: Supporting)
Yanbo Yu, MD (Investigation: Supporting; Writing – review & editing: Supporting)
Yu Yu, MM (Investigation: Supporting; Writing – review & editing: Supporting)
Xiaojing Zhao, MM (Investigation: Supporting; Writing – review & editing: Supporting)
Zheng Zhou, MS (Investigation: Supporting; Writing – review & editing: Supporting)
Suxia Yao, MD (Investigation: Supporting; Writing – review & editing: Supporting)
Sara M Dann, PhD (Investigation: Supporting; Resources: Supporting; Writing – review & editing: Supporting)
Yingzi Cong, PhD (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Funding acquisition: Lead; Investigation: Lead; Methodology: Lead; Project administration: Lead; Resources: Lead; Supervision: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by National Institutes of Health grants DK105585, DK112436, DK125011, AI150210, and DK124132 and the University of Texas System STARs award (to Yingzi Cong), and F30 DK120212 (to Anthony J. Bilotta).
Supplementary Material
Propionate promotes intestinal epithelial cell (IEC) cell speed and persistence. Mouse small intestinal epithelial cells (MSIEs) were wounded and treated with the short-chain fatty acid (SCFA) propionate, and video microscopy was performed with images taken every 15 minutes. Representative phase contrast images of MSIEs taken every 15 minutes and stacked into a movie. Scale bar = 300 μm.
Inhibition of histone deacetylase (HDAC) mediates the effects of propionate on IEC migration. MSIEs were wounded and treated with the SCFA propionate or valproate, and video microscopy was performed with images taken every 15 minutes. Representative phase contrast images of MSIEs taken every 15 minutes and stacked into a movie.
STAT3 is critical for propionate induction of cell persistence. Puromycin control and STAT3 knockout (KO) cells were wounded and treated with the SCFA propionate, and video microscopy was performed with images taken every 15 minutes. Representative phase contrast images of MSIEs taken every 15 minutes and stacked into a movie.
References
- 1.Creamer B., Shorter R.G., Bamforth J. The turnover and shedding of epithelial cells. I. The turnover in the gastro-intestinal tract. Gut. 1961;2:110–118. doi: 10.1136/gut.2.2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Potten C.S., Kellett M., Roberts S.A., Rew D.A., Wilson G.D. Measurement of in vivo proliferation in human colorectal mucosa using bromodeoxyuridine. Gut. 1992;33:71–78. doi: 10.1136/gut.33.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinto D., Gregorieff A., Begthel H., Clevers H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17:1709–1713. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siavoshian S., Segain J.P., Kornprobst M., Bonnet C., Cherbut C., Galmiche J.P., Blottière H.M. Butyrate and trichostatin a effects on the proliferation/differentiation of human intestinal epithelial cells: Induction of cyclin D3 and p21 expression. Gut. 2000;46:507–514. doi: 10.1136/gut.46.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sturm A., Dignass A.U. Epithelial restitution and wound healing in inflammatory bowel disease. World J Gastroenterol. 2008;14:348–353. doi: 10.3748/wjg.14.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lacy E.R. Epithelial restitution in the gastrointestinal tract. J Clin Gastroenterol. 1988;10(Suppl 1):S72–S77. doi: 10.1097/00004836-198812001-00012. [DOI] [PubMed] [Google Scholar]
- 7.Allaire J.M., Crowley S.M., Law H.T., Chang S.Y., Ko H.J., Vallance B.A. The intestinal epithelium: central coordinator of mucosal immunity. Trends Immunol. 2018;39:677–696. doi: 10.1016/j.it.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Nusrat A., Delp C., Madara J.L. Intestinal epithelial restitution. Characterization of a cell culture model and mapping of cytoskeletal elements in migrating cells. J Clin Invest. 1992;89:1501–1511. doi: 10.1172/JCI115741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyoshi H., VanDussen K.L., Malvin N.P., Ryu S.H., Wang Y., Sonnek N.M., Lai C.W., Stappenbeck T.S. Prostaglandin E2 promotes intestinal repair through an adaptive cellular response of the epithelium. EMBO J. 2017;36:5–24. doi: 10.15252/embj.201694660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciacci C., Lind S.E., Podolsky D.K. Transforming growth factor beta regulation of migration in wounded rat intestinal epithelial monolayers. Gastroenterology. 1993;105:93–101. doi: 10.1016/0016-5085(93)90014-4. [DOI] [PubMed] [Google Scholar]
- 11.Dignass A., Lynch-Devaney K., Kindon H., Thim L., Podolsky D.K. Trefoil peptides promote epithelial migration through a transforming growth factor beta-independent pathway. J Clin Invest. 1994;94:376–383. doi: 10.1172/JCI117332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson A.J., Gibson P.R. Short-chain fatty acids promote the migration of colonic epithelial cells in vitro. Gastroenterology. 1997;113:487–496. doi: 10.1053/gast.1997.v113.pm9247468. [DOI] [PubMed] [Google Scholar]
- 13.Binder H.J., Mehta P. Short-chain fatty acids stimulate active sodium and chloride absorption in vitro in the rat distal colon. Gastroenterology. 1989;96:989–996. doi: 10.1016/0016-5085(89)91614-4. [DOI] [PubMed] [Google Scholar]
- 14.Louis P., Young P., Holtrop G., Flint H.J. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ Microbiol. 2010;12:304–314. doi: 10.1111/j.1462-2920.2009.02066.x. [DOI] [PubMed] [Google Scholar]
- 15.Reichardt N., Duncan S.H., Young P., Belenguer A., McWilliam L.C., Scott K.P., Flint H.J., Louis P. Phylogenetic distribution of 3 pathways for propionate production within the human gut microbiota. ISME J. 2014;8:1323–1335. doi: 10.1038/ismej.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pomare E.W., Branch H.W.J., Naylor C.P.E., Macfarlane T. Short chain fatty acids in human large intestine , portal , hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bilotta A.J., Cong Y. Gut microbiota metabolite regulation of host defenses at mucosal surfaces: implication in precision medicine. Precis Clin Med. 2019;2:110–119. doi: 10.1093/pcmedi/pbz008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorburn A.N., Macia L., Mackay C.R. Diet, metabolites, and ‘Western-lifestyle’ inflammatory diseases. Immunity. 2014;40:833–842. doi: 10.1016/j.immuni.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 19.Finnie I.A., Dwarakanath A.D., Taylor B.A., Rhodes J.M. Colonic mucin synthesis is increased by sodium butyrate. Gut. 2007;36:93–99. doi: 10.1136/gut.36.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatayama H., Iwashita J., Kuwajima A., Abe T. The short chain fatty acid, butyrate, stimulates MUC2 mucin production in the human colon cancer cell line, LS174T. Biochem Biophys Res Commun. 2007;356:599–603. doi: 10.1016/j.bbrc.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 21.Burger-van Paassen N., Vincent A., Puiman P.J., van der Sluis M., Bouma J., Boehm G., van Goudoever J.B., van Seuningen I., Renes I.B. The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: implications for epithelial protection. Biochem J. 2009;420:211–219. doi: 10.1042/BJ20082222. [DOI] [PubMed] [Google Scholar]
- 22.Zhao Y., Chen F., Wu W., Sun M., Bilotta A.J., Yao S., Xiao Y., Huang X., Eaves-Pyles T.D., Golovko G., Fofanov Y., D'Souza W., Zhao Q., Liu Z., Cong Y. GPR43 mediates microbiota metabolite SCFA regulation of antimicrobial peptide expression in intestinal epithelial cells via activation of mTOR and STAT3. Mucosal Immunol. 2018;11:752–762. doi: 10.1038/mi.2017.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donohoe D.R., Collins L.B., Wali A., Bigler R., Sun W., Bultman S.J. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol Cell. 2012;48:612–626. doi: 10.1016/j.molcel.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park J.H., Kotani T., Konno T., Setiawan J., Kitamura Y., Imada S., Usui Y., Hatano N., Shinohara M., Saito Y., Murata Y., Matozaki T. Promotion of intestinal epithelial cell turnover by commensal bacteria: role of short-chain fatty acids. PLoS One. 2016;11 doi: 10.1371/journal.pone.0156334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker A., Maclaren O.J., Fletcher A.G., Muraro D., Kreuzaler P.A., Byrne H.M., Maini P.K., Watson A.J., Pin C. Cell proliferation within small intestinal crypts is the principal driving force for cell migration on villi. FASEB J. 2017;31:636–649. doi: 10.1096/fj.201601002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krndija D., El Marjou F., Guirao B., Richon S., Leroy O., Bellaiche Y., Hannezo E., Matic Vignjevic D. Active cell migration is critical for steady-state epithelial turnover in the gut. Science. 2019;365:705–710. doi: 10.1126/science.aau3429. [DOI] [PubMed] [Google Scholar]
- 27.Gibson P.R., Rosella O., Wilson A.J., Mariadason J.M., Rickard K., Byron K., Barkla D.H. Colonic epithelial cell activation and the paradoxical effects of butyrate. Carcinogenesis. 1999;20:539–544. doi: 10.1093/carcin/20.4.539. [DOI] [PubMed] [Google Scholar]
- 28.Gorelik R., Gautreau A. Quantitative and unbiased analysis of directional persistence in cell migration. Nat Protoc. 2014;9:1931–1943. doi: 10.1038/nprot.2014.131. [DOI] [PubMed] [Google Scholar]
- 29.Liang C.-C., Park A.Y., Guan J.-L. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007;2:329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 30.Whitehead R.H., Robinson P.S. Establishment of conditionally immortalized epithelial cell lines from the intestinal tissue of adult normal and transgenic mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;296:G455–G460. doi: 10.1152/ajpgi.90381.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mogilner A., Oster G. Cell motility driven by actin polymerization. Biophys J. 1996;71:3030–3045. doi: 10.1016/S0006-3495(96)79496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Webb D.J., Parsons J.T., Horwitz A.F. Adhesion assembly, disassembly and turnover in migrating cells – over and over and over again. Nat Cell Biol. 2002;4:E97–E100. doi: 10.1038/ncb0402-e97. [DOI] [PubMed] [Google Scholar]
- 33.Martin P. Wound healing--aiming for perfect skin regeneration. Science. 1997;276:75–81. doi: 10.1126/science.276.5309.75. [DOI] [PubMed] [Google Scholar]
- 34.Kupfer A., Louvard D., Singer S.J. Polarization of the Golgi apparatus and the microtubule-organizing center in cultured fibroblasts at the edge of an experimental wound. Proc Natl Acad Sci U S A. 1982;79:2603–2607. doi: 10.1073/pnas.79.8.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zegers M.M., Friedl P. Rho GTPases in collective migration. Small GTPases. 2014;5 doi: 10.4161/sgtp.28997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bu H.-F., Zuo X.L., Wang X., Ensslin M.A., Koti V., Hsueh W., Raymond A.S., Shur B.D., Tan X.D. Milk fat globule-EGF factor 8/lactadherin plays a crucial role in maintenance and repair of murine intestinal epithelium. J Clin Invest. 2007;117:3673–3683. doi: 10.1172/JCI31841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scarpa E., Mayor R. Collective cell migration in development. J Cell Biol. 2016;212:143–155. doi: 10.1083/jcb.201508047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maiuri P., Rupprecht J.F., Wieser S., Ruprecht V., Bénichou O., Carpi N., Coppey M., De Beco S., Gov N., Heisenberg C.P., Lage Crespo C., Lautenschlaeger F., Le Berre M., Lennon-Dumenil A.M., Raab M., Thiam H.R., Piel M., Sixt M., Voituriez R. Actin flows mediate a universal coupling between cell speed and cell persistence. Cell. 2015;161:374–386. doi: 10.1016/j.cell.2015.01.056. [DOI] [PubMed] [Google Scholar]
- 39.Rorth P. Collective cell migration. Annu Rev Cell Dev Biol. 2009;25:407–429. doi: 10.1146/annurev.cellbio.042308.113231. [DOI] [PubMed] [Google Scholar]
- 40.Vedel S., Tay S., Johnston D., Bruus H., Quake S. Migration of cells in a social context. Proc Natl Acad Sci USA. 2013;110:129–134. doi: 10.1073/pnas.1204291110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le Poul E., Loison C., Struyf S., Springael J.Y., Lannoy V., Decobecq M.E., Brezillon S., Dupriez V., Vassart G., Van Damme J., Parmentier M., Detheux M. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J Biol Chem. 2003;278:25481–25489. doi: 10.1074/jbc.M301403200. [DOI] [PubMed] [Google Scholar]
- 42.Thangaraju M., Cresci G.A., Liu K., Ananth S., Gnanaprakasam J.P., Browning D.D., Mellinger J.D., Smith S.B., Digby G.J., Lambert N.A., Prasad P.D., Ganapathy V. GPR109A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res. 2009;69:2826–2832. doi: 10.1158/0008-5472.CAN-08-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waldecker M., Kautenburger T., Daumann H., Busch C., Schrenk D. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. J Nutr Biochem. 2008;19:587–593. doi: 10.1016/j.jnutbio.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 44.Brown A.J., Goldsworthy S.M., Barnes A.A., Eilert M.M., Tcheang L., Daniels D., Muir A.I., Wigglesworth M.J., Kinghorn I., Fraser N.J., Pike N.B., Strum J.C., Steplewski K.M., Murdock P.R., Holder J.C., Marshall F.H., Szekeres P.G., Wilson S., Ignar D.M., Foord S.M., Wise A., Dowell S.J. The orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003;278:11312–11319. doi: 10.1074/jbc.M211609200. [DOI] [PubMed] [Google Scholar]
- 45.Donohoe D.R., Garge N., Zhang X., Sun W., O'Connell T.M., Bunger M.K., Bultman S.J. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13:517–526. doi: 10.1016/j.cmet.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glozak M.A., Sengupta N., Zhang X., Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 47.Friedrich M., Gerbeth L., Gerling M., Rosenthal R., Steiger K., Weidinger C., Keye J., Wu H., Schmidt F., Weichert W., Siegmund B., Glauben R. HDAC inhibitors promote intestinal epithelial regeneration via autocrine TGFβ1 signalling in inflammation. Mucosal Immunol. 2019;12:656–667. doi: 10.1038/s41385-019-0135-7. [DOI] [PubMed] [Google Scholar]
- 48.Sina C., Gavrilova O., Förster M., Till A., Derer S., Hildebrand F., Raabe B., Chalaris A., Scheller J., Rehmann A., Franke A., Ott S., Häsler R., Nikolaus S., Fölsch U.R., Rose-John S., Jiang H.P., Li J., Schreiber S., Rosenstiel P. G protein-coupled receptor 43 is essential for neutrophil recruitment during intestinal inflammation. J Immunol. 2009;183:7514–7522. doi: 10.4049/jimmunol.0900063. [DOI] [PubMed] [Google Scholar]
- 49.Schulthess J., Pandey S., Capitani M., Rue-Albrecht K.C., Arnold I., Franchini F., Chomka A., Ilott N.E., Johnston D.G.W., Pires E., McCullagh J., Sansom S.N., Arancibia-Cárcamo C.V., Uhlig H.H., Powrie F. The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity. 2019;50:432–445.e7. doi: 10.1016/j.immuni.2018.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin-Gallausiaux C., Béguet-Crespel F., Marinelli L., Jamet A., Ledue F., Blottière H.M., Lapaque N. Butyrate produced by gut commensal bacteria activates TGF-beta1 expression through the transcription factor SP1 in human intestinal epithelial cells. Sci Rep. 2018;8:9742. doi: 10.1038/s41598-018-28048-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaiko G.E., Ryu S.H., Koues O.I., Collins P.L., Solnica-Krezel L., Pearce E.J., Pearce E.L., Oltz E.M., Stappenbeck T.S. The colonic crypt protects stem cells from microbiota-derived metabolites. Cell. 2016;165:1708–1720. doi: 10.1016/j.cell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blouin J.M., Penot G., Collinet M., Nacfer M., Forest C., Laurent-Puig P., Coumoul X., Barouki R., Benelli C., Bortoli S. Butyrate elicits a metabolic switch in human colon cancer cells by targeting the pyruvate dehydrogenase complex. Int J Cancer. 2011;128:2591–2601. doi: 10.1002/ijc.25599. [DOI] [PubMed] [Google Scholar]
- 53.Xiao Y., Huang X., Zhao Y., Chen F., Sun M., Yang W., Chen L., Yao S., Peniche A., Dann S.M., Sun J., Golovko G., Fofanov Y., Miao Y., Liu Z., Chen D., Cong Y. Interleukin-33 promotes REG3γ expression in intestinal epithelial cells and regulates gut microbiota. Cell Mol Gastroenterol Hepatol. 2019;8:21–36. doi: 10.1016/j.jcmgh.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pickert G., Neufert C., Leppkes M., Zheng Y., Wittkopf N., Warntjen M., Lehr H.A., Hirth S., Weigmann B., Wirtz S., Ouyang W., Neurath M.F., Becker C. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med. 2009;206:1465–1472. doi: 10.1084/jem.20082683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu W., Sun M., Chen F., Cao A.T., Liu H., Zhao Y., Huang X., Xiao Y., Yao S., Zhao Q., Liu Z., Cong Y. Microbiota metabolite short-chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Mucosal Immunol. 2017;10:946–956. doi: 10.1038/mi.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scheppach W., Sommer H., Kirchner T., Paganelli G.M., Bartram P., Christl S., Richter F., Dusel G., Kasper H. Effect of butyrate enemas on the colonic mucosa in distal ulcerative colitis. Gastroenterology. 1992;103:51–56. doi: 10.1016/0016-5085(92)91094-k. [DOI] [PubMed] [Google Scholar]
- 57.Harig J.M., Soergel K.H., Komorowski R.A., Wood C.M. Treatment of diversion colitis with short-chain-fatty acid irrigation. N Engl J Med. 1989;320:23–28. doi: 10.1056/NEJM198901053200105. [DOI] [PubMed] [Google Scholar]
- 58.Breuer R.I., Buto S.K., Christ M.L., Bean J., Vernia P., Paoluzi P., Di Paolo, MC, Caprilli R. Rectal irrigation with short-chain fatty acids for distal ulcerative colitis. Preliminary report. Dig Dis Sci. 1991;36:185–187. doi: 10.1007/BF01300754. [DOI] [PubMed] [Google Scholar]
- 59.Ben Q., Sun Y., Chai R., Qian A., Xu B., Yuan Y. Dietary fiber intake reduces risk for colorectal adenoma: a meta-analysis. Gastroenterology. 2013;146:689–699.e6. doi: 10.1053/j.gastro.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 60.Wakatsuki T., Wysolmerski R.B., Elson E.L. Mechanics of cell spreading: role of myosin II. J Cell Sci. 2003;116:1617–1625. doi: 10.1242/jcs.00340. [DOI] [PubMed] [Google Scholar]
- 61.Ridley A.J. Rho GTPase signalling in cell migration. Curr Opin Cell Biol. 2015;36:103–112. doi: 10.1016/j.ceb.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Delorme-Walker V.D., Peterson J.R., Chernoff J., Waterman C.M., Danuser G., DerMardirossian C., Bokoch G.M. Pak1 regulates focal adhesion strength, myosin IIA distribution, and actin dynamics to optimize cell migration. J Cell Biol. 2011;193:1289–1303. doi: 10.1083/jcb.201010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sells M.A., Boyd J.T., Chernoff J. p21-Activated kinase 1 (Pak1) regulates cell motility in mammalian fibroblasts. J Cell Biol. 1999;145:837–849. doi: 10.1083/jcb.145.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Glauben R., Batra A., Fedke I., Zeitz M., Lehr H.A., Leoni F., Mascagni P., Fantuzzi G., Dinarello C.A., Siegmund B. Histone hyperacetylation is associated with amelioration of experimental colitis in mice. J Immunol. 2006;176:5015–5022. doi: 10.4049/jimmunol.176.8.5015. [DOI] [PubMed] [Google Scholar]
- 65.Roediger W.E. Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut. 1980;21:793–798. doi: 10.1136/gut.21.9.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Teng T.S., Lin B., Manser E., Ng D.C.H., Cao X. Stat3 promotes directional cell migration by regulating Rac1 activity via its activator βPIX. J Cell Sci. 2009;122:4150–4159. doi: 10.1242/jcs.057109. [DOI] [PubMed] [Google Scholar]
- 67.Chen H., Yang Z., Ding C., Chu L., Zhang Y., Terry K., Liu H., Shen Q., Zhou J. Discovery of O-alkylamino-tethered niclosamide derivatives as potent and orally bioavailable anticancer agents. ACS Med Chem Lett. 2013;4:180–185. doi: 10.1021/ml3003082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu Y., Yang W., Bilotta A.J., Yu Y., Zhao X., Zhou Z., Yao S., Xu J., Zhou J., Dann S.M., Li Y., Cong Y. STING controls intestinal homeostasis through promoting antimicrobial peptide expression in epithelial cells. FASEB J. 2020;34:15417–15430. doi: 10.1096/fj.202001524R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miyoshi H., Stappenbeck T.S. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc. 2013;8:2471–2482. doi: 10.1038/nprot.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y., DiSalvo M., Gunasekara D.B., Dutton J., Proctor A., Lebhar M.S., Williamson I.A., Speer J., Howard R.L., Smiddy N.M., Bultman S.J., Sims C.E., Magness S.T., Allbritton N.L. Self-renewing monolayer of primary colonic or rectal epithelial cells. Cell Mol Gastroenterol Hepatol. 2017;4:165–182.e7. doi: 10.1016/j.jcmgh.2017.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Propionate promotes intestinal epithelial cell (IEC) cell speed and persistence. Mouse small intestinal epithelial cells (MSIEs) were wounded and treated with the short-chain fatty acid (SCFA) propionate, and video microscopy was performed with images taken every 15 minutes. Representative phase contrast images of MSIEs taken every 15 minutes and stacked into a movie. Scale bar = 300 μm.
Inhibition of histone deacetylase (HDAC) mediates the effects of propionate on IEC migration. MSIEs were wounded and treated with the SCFA propionate or valproate, and video microscopy was performed with images taken every 15 minutes. Representative phase contrast images of MSIEs taken every 15 minutes and stacked into a movie.
STAT3 is critical for propionate induction of cell persistence. Puromycin control and STAT3 knockout (KO) cells were wounded and treated with the SCFA propionate, and video microscopy was performed with images taken every 15 minutes. Representative phase contrast images of MSIEs taken every 15 minutes and stacked into a movie.