Abstract
Background
Cholangiocarcinoma (CCA) is the second most common liver malignant tumor. CircRNA hsa_circ_0005230 (circDNM3OS) has been reported to exert an oncogenic role in CCA. However, the mechanisms related to circDNM3OS in CAA progression have not been fully elucidated.
Methods
The expression of circDNM3OS, microRNA (miR)-145-5p, and MORC2 (MORC Family CW-Type Zinc Finger 2) mRNA were analyzed by quantitative real-time polymerase chain reaction (qRT-PCR). Cell proliferation, colony formation, migration, invasion, and apoptosis were evaluated by Cell Counting Kit-8 (CCK-8), colony formation, transwell, wound-healing, and flow cytometry assays. The levels of glutamine, α-KG (α-ketoglutarate), and ATP (adenosine triphosphate) were detected using commercial kits. The relationship between circDNM3OS or MORC2 and miR-145-5p was verified by dual-luciferase reporter and/or RNA immunoprecipitation (RIP) assays. Protein level of MORC2 was measured by Western blotting. The role of circDNM3OS in CCA growth was verified by xenograft experiment.
Results
CircDNM3OS and MORC2 were upregulated while miR-145-5p was downregulated in CCA tissues and cells. Inhibition of circDNM3OS reduced xenograft tumor growth in vivo and constrained proliferation, colony formation, migration, invasion, induced apoptosis, and reduced glutamine metabolism of CCA cells in vitro. CircDNM3OS sponged miR-145-5p to elevate MORC2 expression. MiR-145-5p silencing overturned circDNM3OS knockdown-mediated influence on malignancy and glutamine metabolism of CCA cells. Also, MORC2 overexpression reversed the repressive impact of miR-145-5p mimic on malignancy and glutamine metabolism of CCA cells.
Conclusion
CircDNM3OS facilitates CCA growth and glutamine metabolism by regulating the miR-145-5p/MORC2 pathway, offering a novel mechanism to understand the progression of CCA.
Keywords: CCA, circDNM3OS, miR-145-5p, MORC2, glutamine
Introduction
Cholangiocarcinoma (CCA), which originates from bile duct epithelial cells, is the second most common liver malignant tumor following hepatocellular cancer.1 Radical surgery is the best choice for the treatment of CCA patients, but most patients cannot be treated with surgery due to metastasis and lymph node invasion.2,3 Also, there is a higher risk of recurrence after radical surgery.4 Furthermore, more than 90% of patients die within 5 years, and most patients survive for less than 12 months after diagnosis.5 Thus, exploring the potential mechanisms related to the occurrence and progression of CCA is indispensable for the development of new therapeutic measures.
Glutamine, a non-essential amino acid, can be converted into α-ketoglutarate (α-KG) and enter the tricarboxylic acid (TCA) cycle under the action of glutamine dehydrogenase and glutamate dehydrogenase.6 Glutamine plays an important role in maintaining redox homeostasis, regulating signaling pathways, and supporting macromolecular biosynthesis.7 Glutamine metabolism plays a promoting impact on tumor growth.8 Furthermore, targeting glutamine metabolism has been shown as a promising anti-cancer therapy.9 Thus, it is indispensable to survey the molecular mechanisms associated with CCA growth and glutamine metabolism.
Circular RNAs (circRNAs), a group of endogenous biomolecules with a closed-loop structure, are generated by pre-mRNA via variable shear processing.10 They exert important biological functions via serving as protein or microRNA (miR) sponges.11 CircRNAs have become a research hotspot because their disorders are closely related to the development of diseases.12 For example, circWBSCR17 elevated fibrosis and inflammation of human kidney tubular cells through regulating the miR-185-5p/SOX6 regulatory axis in human kidney tubular cells in diabetic nephropathy.13 Furthermore, circZNF652 sponged miR-205 to contribute to epithelial-mesenchymal transition and proliferation of renal carcinoma cells.14 Also, some circRNAs have been uncovered to participate in the tumorigenesis of CCA. CircRNA circ-0000673 played an oncogenic role in distal CCA.15 High circRNA SMARCA5 expression constrained malignancy and elevated cisplatin sensitivity of intrahepatic CCA cells.16 Moreover, circRNA CDR1as promoted CCA progression by activating the AKT/mTOR pathway through adsorbing miR-641.17 CircRNA hsa_circ_0005230 (circDNM3OS), located at chr1:172,109,619–172,113,577, is formed by splicing from the DNM3OS (DNM3 opposite strand/antisense RNA) gene. Previous research revealed that circDNM3OS promoted CCA cell metastasis and growth.18 Nevertheless, the mechanism by which circDNM3OS regulates the growth of CCA is still unclear.
Herein, circDNM3OS accelerated cell malignancy and glutaminolysis through elevating MORC2 (MORC Family CW-Type Zinc Finger 2) expression by absorbing miR-145-5p in CCA.
Materials and Methods
Case Selection
The study protocols were approved by the Ethics Committee of Jiangxi Provincial Cancer Hospital. 35 paired CCA tissues and surrounding normal tissues were acquired from CCA patients who underwent radical surgery at Jiangxi Provincial Cancer Hospital. The clinical characteristics of CCA patients were shown in Table 1. The research has been carried out in accordance with the World Medical Association Declaration of Helsinki. Written informed consent was obtained from all patients recruited in the study.
Table 1.
Association Between circDNM3OS Expression and Clinicopathologic Features of CCA Patients
| Clinical Characteristics | Total (n=35) | circDNM3OS Expression | P value | |
|---|---|---|---|---|
| High | Low | |||
| Age | 0.685 | |||
| ≤ 60 years | 19 | 10 | 9 | |
| > 60 years | 16 | 8 | 8 | |
| Gender | 0.549 | |||
| Male | 18 | 9 | 9 | |
| Female | 17 | 9 | 8 | |
| Tumor size | 0.041* | |||
| < 3cm | 20 | 12 | 8 | |
| ≥ 3cm | 15 | 6 | 9 | |
| TNM stage | 0.001* | |||
| I–II | 12 | 3 | 9 | |
| III | 23 | 15 | 8 | |
| Histologic differentiation | 0.398 | |||
| Well | 10 | 4 | 6 | |
| Moderate | 18 | 10 | 8 | |
| Poor | 7 | 4 | 3 | |
| Lymph node invasion | 0.004* | |||
| Present | 21 | 13 | 8 | |
| Absent | 14 | 5 | 9 | |
Note: *P < 0.05.
Cell Culture
CCA cells (HuCCT1 and HuH28), human intrahepatic biliary epithelial cells (HIBEC), and 293T were obtained from Biovector Science Lab, Inc., (Beijing, China) and cultured in Roswell Park Memorial Institute (RPMI)-1640 medium Procell (Wuhan, China) supplemented with 10% fetal bovine serum (Procell). The growth of these cells was kept in a moist atmosphere with 5% CO2 at 37°C.
Transient Transfection
Small interfering (si) RNA against circDNM3OS (si-circDNM3OS: 5ʹ-ATACTCTTTGTTTTGCACACT-3ʹ) and matching negative control (NC) (si-NC: 5ʹ-TTCTCCGAACGTGTCACGT-3ʹ), short hairpin (sh) RNA targeting circ-circDNM3OS (sh-circDNM3OS:5ʹ-CACCATACTCTTTGTTTTGCACACTCTCGAGAGTGTGCAAAACAAAGAGTA-3ʹ) and its NC (sh-NC: 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ), miR-145-5p mimic (miR-145-5p) and corresponding NC (miR-NC), and miR-145-5p inhibitor (in-miR-145-5p) and matching NC (in-miR-NC) were acquired from GenePharma (Shanghai, China). The pcDNA-MORC2 (MORC2) vectors were constructed using the pcDNA vectors (Biovector Science Lab, Inc.). Transient transfection was executed using Lipofectamine 3000 reagent (Thermo Fisher Scientific, Waltham, MA, USA).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was isolated with the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). Reverse transcription was performed using the miRNA Reverse Transcription Kit (Qiagen) or GoScript Reverse Transcription System (Promega, Madison, WI, USA). The obtained complementary DNA was used with the SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) for qRT-PCR in the ABI7500 system (Applied Biosystems). All primer sequences were listed in Table 2. Relative expression was figured by the 2−ΔΔCt method and normalized to β-actin or U6 small nuclear RNA (U6).
Table 2.
Primer Sequences for RT-qPCR
| Genes | Primer Sequences (5ʹ-3ʹ) |
|---|---|
| circDNM3OS | Forward (F): 5ʹ-CTGCTGAGAAAAGACTGCCGA-3ʹ |
| Reverse (R): 5ʹ-ACCTAGGTTCCCTTGGTCACA-3ʹ | |
| MORC2 | F: 5ʹ-GGAGGTTCCTTCTCCCAAAGTC-3ʹ |
| R: 5ʹ-CAGAAACTGCGACACTCCGCTT-3ʹ | |
| miR-145-5p | F: 5ʹ-GTCCAGTTTTCCCAGGA-3ʹ |
| R: 5ʹ-GAACATGTCTGCGTATCTC-3ʹ | |
| DNM3OS | F: 5ʹ-GGTCCTAAATTCATTGCCAGTTC-3ʹ |
| R: 5ʹ-ACTCAAGGGCTGTGATTTCC-3ʹ | |
| β-actin | F: 5ʹ-AAATCTGGCACCACACCTTC-3ʹ |
| R: 5ʹ-GGGGTGTTGAAGGTCTCAAA-3ʹ | |
| U6 | F: 5ʹ-GCTCGCTTCGGCAGCACA-3ʹ |
| R: 5ʹ-GAGGTATTCGCACCAGAGGA-3ʹ |
Subcellular Fractionation Assay
The cytoplasmic RNA and nuclear RNA of HuCCT1 and HuH28 cells were separated using the PARIS™ kit (Thermo Fisher Scientific). Relative expression of circDNM3OS in the cytoplasmic RNA and nuclear RNA was estimated by qRT-PCR. U6 and β-actin were used as controls for nuclear RNA and cytoplasmic RNA.
RNase R Treatment
The circular structure of circDNM3OS was validated by RNase R treatment. In short, total RNA from HuCCT1 and HuH28 cells was digested with DEPC-treated water (Sigma, St Louis, MO, USA) or RNase R (Epicentre Technologies, Madison, WI, USA) at 37°C for 30 min. Relative expression of circDNM3OS and DNM3OS mRNA was detected by qRT-PCR. Primer for DNM3OS was listed in Table 2.
Cell Proliferation Assessment
Cell proliferation was analyzed in the light of the manufacturer’s directions of the Cell Counting Kit-8 (CCK-8) kit (Solarbio, Beijing, China). Briefly, transfected HuCCT1 and HuH28 cells (1.5 × 103) were seeded into 96-well plates. After culture for a period of time, CCK-8 solution (10 μL) was added to each well and incubated for 2 h. The optical density (OD) value at 450 nm was measured using a microplate reader (PerkinElmer, Waltham, MA, USA).
Colony Formation Assay
Briefly, 1 mL growth medium containing 0.6% agarose was tiled to 6-well plates. 20 min later, HuCCT1 and HuH28 cells (4 × 102) in 1 mL growth medium containing 0.3% agarose were seeded into the upper layer. 14 days later, the colonies were stained with 0.01% crystal violet solution (Sigma). The number of colonies (> 50 cells) was counted using a microscope (Nikon Eclipse E600, Nikon, Melville, NY, USA) at 40 × magnification.
Cell Migration Analysis
In short, 600 μL RPMI-1640 medium containing 10% FBS was placed to the lower chamber of the 24-well transwell chamber (Corning, Corning, NY, USA). Then, transfected HuCCT1 and HuH28 cells (1×105 cells/chamber) in 200 μL RPMI-1640 medium containing 1% bovine serum albumin (Sigma) were tiled to the upper chamber. 24 h later, the cells in the low surface were fixed by 4% paraformaldehyde (Sigma) and stained with 0.25% crystal solution (Sigma). The number of migrated cells was figured using a microscope (Nikon Eclipse E600, Nikon) at 100 × magnification.
Wound-Healing Assay
The invasion of HuCCT1 and HuH28 cells was assessed by wound-healing assay. After transfection for 24 h, a linear scratch was made on the cell layer with a 200 μL pipette tip. Then, the cells were washed with PBS (phosphate buffer saline) and then cultured in FBS-free medium for 24 h. The wounds were photographed at 0 h and 24 h using a Nikon Eclipse E600 microscope (Nikon). The widths of the wounds were measured with Image-Pro Plus 6.0 software (Media Cybernetics, Rockville, MD, USA) at 40 × magnification.
Flow Cytometry Assay
Cell apoptosis was evaluated using an Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (Becton Dickinson, San Jose, CA, USA) in light of the manufacturer’s instructions. In short, the cells were digested with trypsin (Sigma) and then re-suspended in 1× binding buffer. Thereafter, the cells (1.0 × 105) were incubated with Annexin V-FITC (5 μL) for 10 min and then stained with PI (5 μL) for 5 min. Live cells, early apoptotic cells, dead cells, and late apoptotic cells were determined using a FACS Calibur Flow Cytometer (Becton Dickinson) with a CellQuest software program (Becton Dickinson).
Measurement of Glutamine, α-KG, and ATP (Adenosine Triphosphate) Levels
After transfection, the levels of glutamine, α-KG, and ATP in HuCCT1 and HuH28 cells were analyzed with a Glutamine/Glutamate Determination Kit (Sigma), α-KG Assay Kit (Sigma), or ATP Colorimetric/Fluorometric Assay Kit (Sigma).
Dual-Luciferase Reporter Assay
The sequence of circDNM3OS or MORC2 3ʹ untranslated regions (UTR) complementary to miR-145-5p was predicted using the circInteractome or microT_CDS databases. The wild type (WT) sequences of circDNM3OS (circDNM3OS WT) and MORC2 3ʹ UTR (MORC2 3ʹUTR WT) and their mutant (MUT) sequences (circDNM3OS MUT and MORC2 3ʹUTR MUT) were synthesized and inserted into the pGL3 promoter vector (Promega), respectively. HuCCT1 and HuH28 cells were co-transfected with a luciferase reporter plasmid, pRL-SV40 (renilla luciferase vector) (Promega), and miR-145-5p or miR-NC using Lipofectamine 3000 reagent (Thermo Fisher Scientific). The luciferase activities in cell lysates were analyzed using the luciferase reporter assay kit (Promega).
RNA Immunoprecipitation (RIP) Assay
RIP was performed using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, Massachusetts, USA). HuCCT1 and HuH28 cells were lysed using RIPA lysis buffer (Sigma). After centrifugation, the supernatant was incubated with magnetic beads conjugated with Anti-IgG (ab133470, Abcam, Cambridge, MA, USA) or Anti-AGO2 (ab32381, Abcam) antibodies. After removing the beads, the enrichment of circDNM3OS and miR-145-5p in the purified RNA was evaluated with the qRT-PCR.
Western Blotting
Total protein was extracted using the RIPA lysis buffer (Sigma). After quantification with a BCA assay kit (Pierce, Rockford, IL, USA), the extracted total protein was isolated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to polyvinylidene fluoride (PVDF) membranes (Sigma). Thereafter, the membranes were blocked in tris buffered saline tween buffer containing 5% skim milk. The membranes were incubated with primary anti-MORC2 (#PA5-49,339, 1:1000, Thermo Fisher Scientific) or anti-β-actin (#PA1-16,889, 1:5000, Thermo Fisher Scientific) antibodies, β-actin was deemed as a loading control. Then, the membranes were incubated with a goat anti-rabbit IgG secondary antibody (#31,460, 1:10,000, Thermo Fisher Scientific). The blots were detected by enhanced chemiluminescence (ECL) substrates (Thermo Fisher Scientific).
Lentivirus Transduction
The pLKO.1 vectors (Sigma) carrying sh-NC or sh-circDNM3OS were transfected into 293T cells using polyethylenimine (Ploysciencs, Warrington, PA, USA). The lentiviral particles from the supernatant of 293T cells were transduced into HuH28 cells and selected with puromycin (5 μg/mL, Solarbio).
Animal Xenograft Experiment
The experiment was approved by the Animal Ethics Committee of Jiangxi Provincial Cancer Hospital. Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. All animals received humane care according to the National Institutes of Health (USA) guidelines. 10 BALB/c nude mice (5-week-old) were bought from Vital River Laboratory (Beijing, China) and randomly divided into 2 groups and managed as follows: One group of mice was injected with HuH28 cells carrying sh-NC (5×106/0.2 mL PBS), and the other group of mice was injected with HuH28 cells carrying sh-circDNM3OS (5×106/0.2 mL PBS). 28 days later, all mice were killed by cervical dislocation under anesthesia. Tumor volume was measured once a week and calculated based on the following equation: Volume = (length ×width2)/2. All xenograft tumors were excised after the mouse heart stopped beating.
Statistical Analysis
All data are expressed as mean ± standard deviation and obtained from three repeated experiments. Statistical analysis was executed using GraphPad Prism 7.0 (Graph-Pad Software, La Jolla, CA). The correlation among circDNM3OS, miR-145-5p, and MORC2 in CCA tissues was estimated by Pearson’s correlation analysis. Statistical significance was accepted when P < 0.05. The difference between 2 groups was analyzed using paired Student’s t-test (CCA tissues and surrounding normal tissues) or unpaired Student’s t-test. The differences among 3 or more groups were analyzed using one-way variance analysis followed by Turkey’s post hoc test.
Results
circDNM3OS Was Highly Expressed in CCA Tissues and Cells
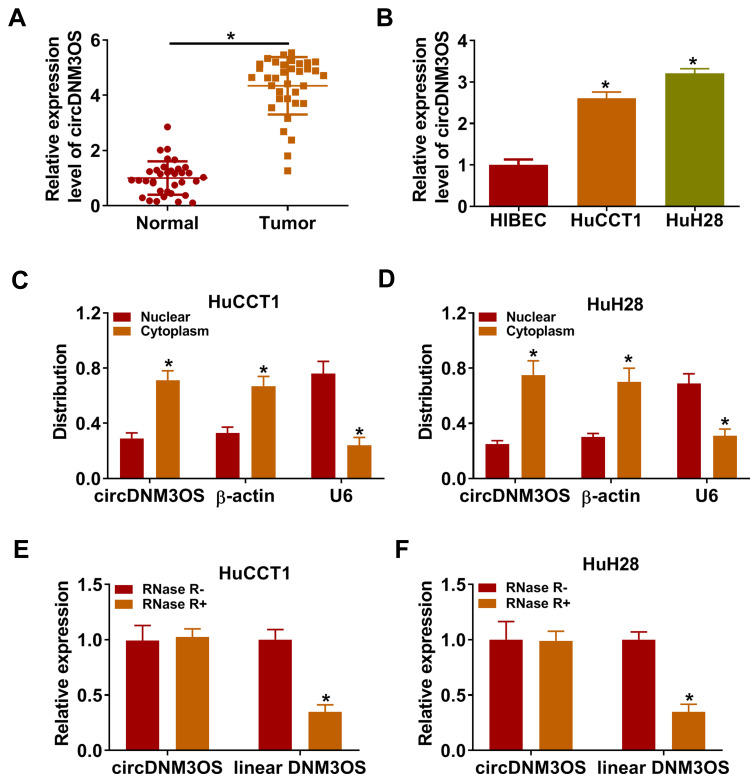
To validate the differential expression of circDNM3OS in CCA, we carried out qRT-PCR analysis. In contrast to surrounding normal tissues, circDNM3OS expression was signally increased in CCA tissues (Figure 1A). High circDNM3OS expression was associated with tumor size, TNM stage, and lymph node invasion in CCA patients (Table 1). Consistently, circDNM3OS was highly expression in CCA cells (HuCCT1 and HuH28) than that in HIBEC cells (Figure 1B). We also surveyed the distribution of circDNM3OS in CCA cells. QRT-PCR presented that circDNM3OS was preferentially distributed in the cytoplasm of HuCCT1 and HuH28 cells (Figure 1C and D). Moreover, RNase R treatment was performed to further verify the circular structure of circDNM3OS. As displayed in Figure 1E and F, the expression of linear DNM3OS was observably decreased after RNase R treatment, but circDNM3OS expression did not change. Collectively, these results manifested that circDNM3OS was upregulated in CCA.
Figure 1.
Expression pattern of circDNM3OS in CCA tissues and cells. (A, B) The expression of circDNM3OS in CCA tissues and cells (HuCCT1 and HuH28) was assessed by qRT-PCR, and surrounding normal tissues and HIBEC cells were used as controls. (C, D) After isolating cytoplasmic RNA and nuclear RNA, the distribution of circDNM3OS in HuCCT1 and HuH28 was evaluated by qRT-PCR. (E, F) After RNase R treatment, the levels of circDNM3OS and DNM3OS mRNA were detected by qRT-PCR. *P < 0.05.
circDNM3OS Knockdown Repressed Malignancy and Glutamine Metabolism of CCA Cells
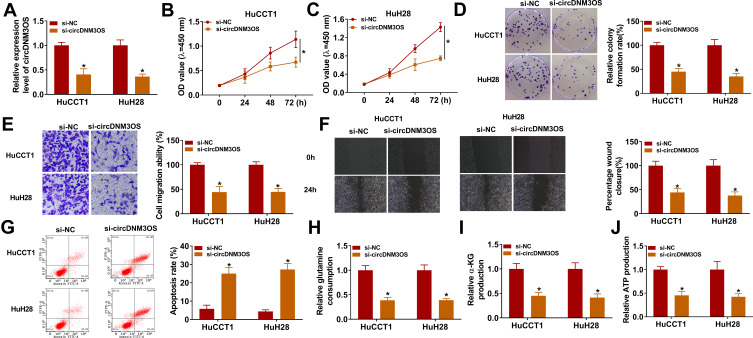
To investigate the biological function of circDNM3OS in CCA, we conducted loss-of-function experiments. CircDNM3OS expression was observably decreased in HuCCT1 and HuH28 cells after transfection with si-circDNM3OS (Figure 2A). However, the expression of linear DNM3OS did not change in si-circDNM3OS-transfected HuCCT1 and HuH28 cells (Supplementary Figure 1A and B). CCK-8 assay presented that the downregulation of circDNM3OS repressed the proliferation of HuCCT1 and HuH28 cells (Figure 2B and C). Colony formation assay showed that circDNM3OS inhibition reduced the ability of HuCCT1 and HuH28 cells to form clones (Figure 2D). Also, circDNM3OS silencing decreased the migratory number of HuCCT1 and HuH28 cells in transwell assay (Figure 2E). Wound-healing assay displayed that circDNM3OS silencing constrained the invasion capacity of HuCCT1 and HuH28 cells (Figure 2F). Flow cytometry assay presented that the apoptotic rate of HuCCT1 and HuH28 cells was elevated after circDNM3OS inhibition (Figure 2G). Additionally, we observed that the inhibition of circDNM3OS reduced glutamine consumption, α-KG production, and ATP level in HuCCT1 and HuH28 cells, implying that circDNM3OS silencing repressed the conversion of glutamine to α-KG, thereby inhibiting the TCA and reducing the production of ATP. (Figure 2H–J). Together, these findings indicated that the inhibition of circDNM3OS constrained malignancy and glutamine metabolism of CCA cells.
Figure 2.
Influence of circDNM3OS inhibition on malignancy and glutamine metabolism of CCA cells. (A) QRT-PCR was carried out to verify the knockdown efficiency of circDNM3OS in HuCCT1 and HuH28 cells. (B–G) The proliferation, colony formation, migration, invasion, and apoptosis of HuCCT1 and HuH28 cells transfected with si-NC or si-circDNM3OS were assessed by CCK-8 assay (B, C), colony formation assay (D), transwell assay (E), wound-healing assay (F), or flow cytometry assay (G). (H–J) The levels of glutamine, α-KG, and ATP in HuCCT1 and HuH28 cells transfected with si-NC or si-circDNM3OS were detected using a corresponding commercial kit. *P < 0.05.
circDNM3OS Was Verified as a Sponge for miR-145-5p in CCA Cells
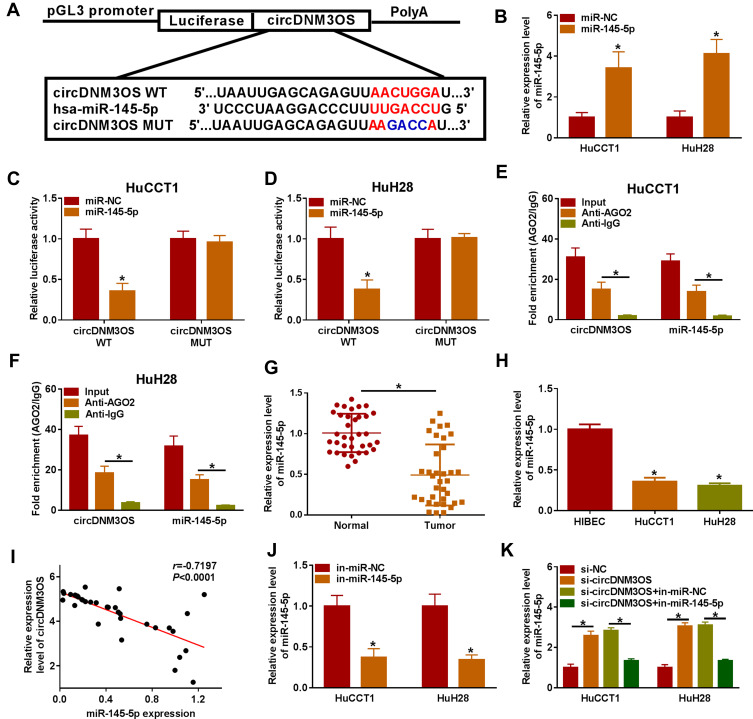
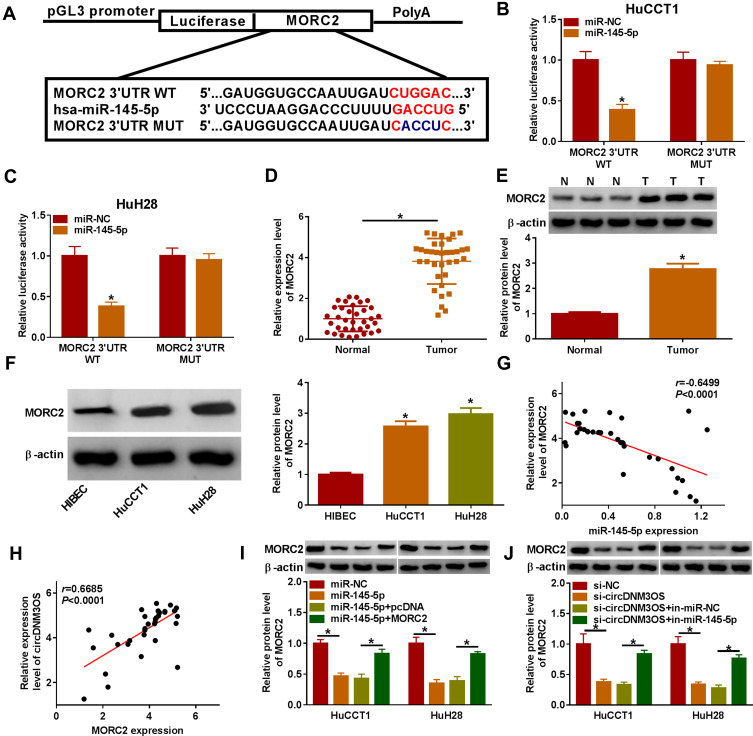
Given that circDNM3OS was preferentially distributed in the cytoplasm of HuCCT1 and HuH28 cells, we further explored the regulatory mechanism of circDNM3OS in CCA progression. Bioinformatics prediction (circinteractome database) showed that there were 14 miRs that might interact with circDNM3OS through prediction using the circinteractome database. We randomly selected 6 candidate miRs (miR-767-3p, miR-640, miR-618, miR-532-3p, miR-1208, and miR-145-5p) for analysis and found that circDNM3OS overexpression repressed the expression of miR-640, miR-618, miR-532-3p, miR-1208, and miR-145-5p in CCA cells (Supplementary Figure 2A). In addition, miR-145 has been reported to be associated with glutamine metabolism.19,20 Therefore, miR-145-5p was selected for further analysis and the predicted binding sites of circDNM3OS in miR-145-5p were presented in Figure 3A. The overexpression efficiency of miR-145-5p was presented in Figure 3B. Dual-luciferase reporter assay displayed that miR-145-5p overexpression reduced the luciferase activity of the luciferase reporter carrying circDNM3OS WT, but the luciferase activity of the luciferase reporter carrying circDNM3OS MUT did not change (Figure 3C and D). RIP assay showed that circDNM3OS and miR-145-5p were markedly enriched in Anti-AGO2 complex rather than Anti-IgG complex (Figure 3E and F). Moreover, an apparent decrease of miR-145-5p was observed in CCA tissues and cells (Figure 3G and H). Pearson’s correlation analysis presented that there was a negative correlation between circDNM3OS and miR-145-5p in CCA tissues (Figure 3I). Furthermore, miR-145-5p was markedly downregulated in HuCCT1 and HuH28 cells after in-miR-145-5p transfection (Figure 3J). Also, circDNM3OS silencing elevated miR-145-5p expression in HuCCT1 and HuH28 cells, but this influence was restored after miR-145-5p knockdown (Figure 3K). These results suggested that circDNM3OS served as a sponge for miR-145-5p in CCA cells.
Figure 3.
CircDNM3OS acted as a sponge for miR-145-5p in CCA cells. (A) Schematic diagram of circDNM3OS sequence complementary to miR-145-5p. (B) Analysis of the expression of miR-145-5p in HuCCT1 and HuH28 cells transfected with miR-145-5p mimic or miR-NC by qRT-PCR. (C, D) The luciferase activity in HuCCT1 and HuH28 cells was determined by dual-luciferase reporter assay. (E, F) The enrichment of miR-145-5p and circDNM3OS in Anti-AGO2 complex and Anti-IgG complex was detected by qRT-PCR. (G, H) Relative expression of miR-145-5p in CCA tissues and cells (HuCCT1 and HuH28) was tested using qRT-PCR. (I) The correlation between circDNM3OS and miR-145-5p expression in CCA tissues was determined by Pearson’s correlation analysis. (J) Relative expression of miR-145-5p in HuCCT1 and HuH28 transfected with in-miR-NC or in-miR-145-5p was evaluated by qRT-PCR. (K) HuCCT1 and HuH28 were transfected with si-NC, si-circDNM3OS, si-circDNM3OS+in-miR-NC, or si-circDNM3OS+in-miR-145-5p. Relative expression of miR-145-5p in HuCCT1 and HuH28 cells was detected using qRT-PCR. *P < 0.05.
circDNM3OS Sponged miR-145-5p to Regulate Malignancy and Glutamine Metabolism of CCA Cells
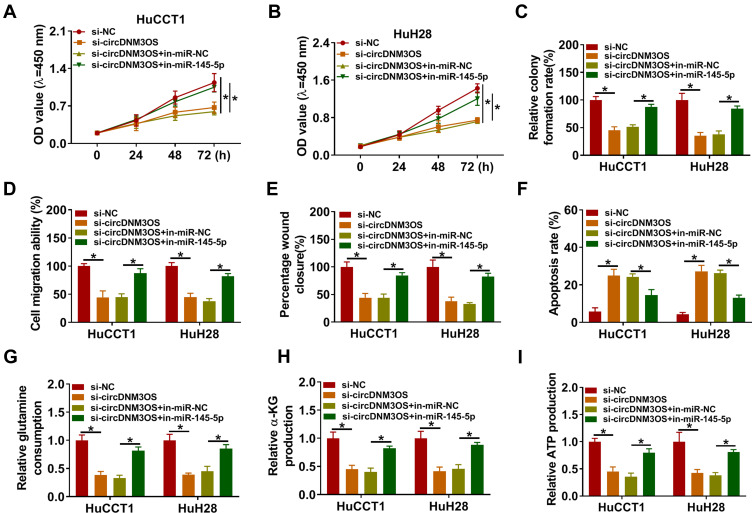
To ascertain that circDNM3OS modulated CCA advancement by sponging miR-145-5p, we carried out rescue experiments. As showed in Figure 4A–E, the repressive influence of circDNM3OS knockdown on proliferation, colony formation, migration, and invasion of HuCCT1 and HuH28 cells was reversed after in-miR-145-5p introduction. Moreover, the downregulation of miR-145-5p in circDNM3OS-inhibited HuCCT1 and HuH28 cells obviously reduced the number of apoptotic cells (Figure 4F). Also, the decrease of glutamine consumption, α-KG production, and ATP level in circDNM3OS-silenced HuCCT1 and HuH28 cells were overturned by miR-145-5p inhibition (Figure 4G–I). Together, these dada suggested that circDNM3OS accelerated malignancy and glutamine metabolism of CCA cells through absorbing miR-145-5p.
Figure 4.
CircDNM3OS regulated malignancy and glutamine metabolism of CCA cells through sponging miR-145-5p. (A–I) HuCCT1 and HuH28 cells were co-transfected with si-NC or si-circDNM3OS and in-miR-NC or in-miR-145-5p. (A–F) Assessment of the proliferation, colony formation, migration, invasion, apoptosis of HuCCT1 and HuH28 cells using CCK-8 assay (A, B), colony formation assay (C), transwell assay (D), wound-healing assay (E), or flow cytometry assay (F). (G–I) The levels of glutamine, α-KG, and ATP in HuCCT1 and HuH28 cells were measured using commercial kits. *P < 0.05.
circDNM3OS Sponged miR-145-5p to Regulate MORC2 Expression
To investigate the involvement of miR-145-5p in circDNM3OS-mediated malignancy and glutamine metabolism of CCA cells, we randomly selected 6 candidate targets of miR-145-5p (ULK1, SMAD5, HMGA2, TLR4, SIRT3 and MORC2) for analysis after prediction in the microT_CDS database. Also, miR-145-5p constrained the mRNA expression levels of SMAD5, HMGA2, TLR4, SIRT3 and MORC2 in CCA cells, and MORC2 was inhibited most significantly (Supplementary Figure 2B). As showed in Figure 5A, MORC2 might be a target of miR-145-5p. The luciferase intensity of the luciferase reporter containing MORC2 3ʹUTR WT was reduced in HuCCT1 and HuH28 cells transfected with miR-145-5p mimic, but there was no obvious difference in the luciferase reporter containing MORC2 3ʹUTR MUT (Figure 5B and C). We observed an overt upregulation of MORC2 mRNA in CCA tissues (Figure 5D). The level of MORC2 protein was also increased in CCA tissues and cells (Figure 5E and F). Moreover, the expression of MORC2 mRNA was negatively correlated with miR-145-5p and positively correlated with circDNM3OS in CCA tissues (Figure 5G and H). Also, the downregulation of MORC2 protein in miR-145-5p-elevated HuCCT1 and HuH28 cells was restored after MORC2 introduction (Figure 5I). Additionally, the silence of miR-145-5p reversed the downregulation of MORC2 protein in HuCCT1 and HuH28 cells mediated by circDNM3OS inhibition (Figure 5J). Collectively, these results proved that circDNM3OS elevated MORC2 expression by absorbing miR-145-5p in CCA cells.
Figure 5.
CircDNM3OS regulated MORC2 expression by sponging miR-145-5p in CCA cells. (A) Schematic diagram of the complementary sequence between MORC2 and miR-145-5p. (B, C) Dual-luciferase reporter assay was performed to verify the relationship between MORC2 and miR-145-5p. (D) Analysis of the relative expression of MORC2 mRNA in CCA tissues by qRT-PCR. (E, F) Relative protein level of MORC2 in CCA tissues and cells was analyzed by Western blotting. (G) The correlation between MORC2 mRNA and miR-145-5p or circDNM3OS in CCA tissues was estimated by Pearson’s correlation analysis. (I) Relative protein level of MORC2 in HuCCT1 and HuH28 cells transfected with miR-NC, miR-145-5p, miR-145-5p+pcDNA, or miR-145-5p+MORC2 was determined by Western blotting. (J) Relative protein level of MORC2 in HuCCT1 and HuH28 cells transfected with si-NC, si-circDNM3OS, si-circDNM3OS+in-miR-NC, or si-circDNM3OS+in-miR-145-5p was detected using Western blotting. *P < 0.05.
miR-145-5p Repressed Malignancy and Glutamine Metabolism of CCA Cells by Targeting MORC2
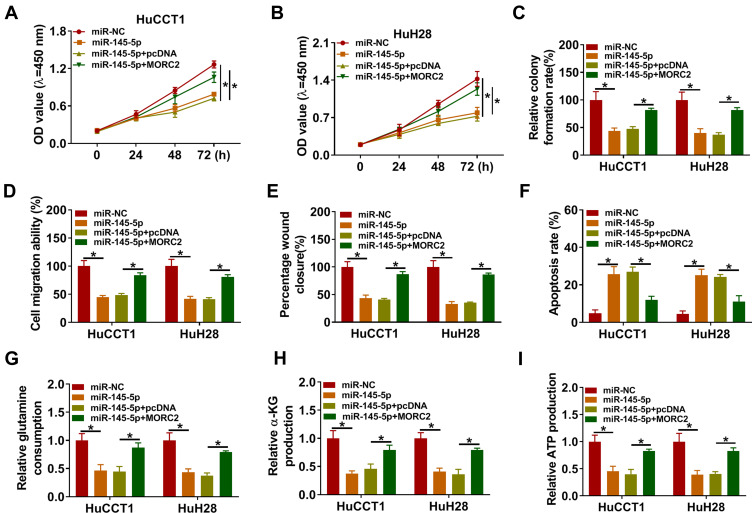
To determine that miR-145-5p regulated malignancy and glutamine metabolism of CCA cells by targeting MORC2, we performed rescue experiments. The results presented that miR-145-5p overexpression repressed proliferation, colony formation, migration, invasion, and accelerated apoptosis of HuCCT1 and HuH28 cells, but these impacts were overturned by forcing MORC2 expression (Figure 6A–F). Furthermore, the elevation of miR-145-5p reduced glutamine consumption, α-KG production, and ATP level in HuCCT1 and HuH28, but this influence was abolished after MORC2 overexpression (Figure 6G–I). Together, these data indicated that miR-145-5p modulated malignancy and glutamine metabolism of CCA cells through targeting MORC2.
Figure 6.
MiR-145-5p targeted MORC2 to modulate malignancy and glutamine metabolism of CCA cells. (A–I) HuCCT1 and HuH28 cells were transfected with miR-NC, miR-145-5p, miR-145-5p+pcDNA, or miR-145-5p+MORC2. (A–F) Assessment of the proliferation, colony formation, migration, invasion, and apoptosis of HuCCT1 and HuH28 cells by CCK-8 assay (A, B), colony formation assay (C), transwell assay (D), wound-healing assay (E), or flow cytometry assay (F). (G–I) Detection of the levels of glutamine, α-KG, and ATP in HuCCT1 and HuH28 cells using commercial kits. *P < 0.05.
Inhibition of circDNM3OS Reduced the Growth of Xenograft Tumor
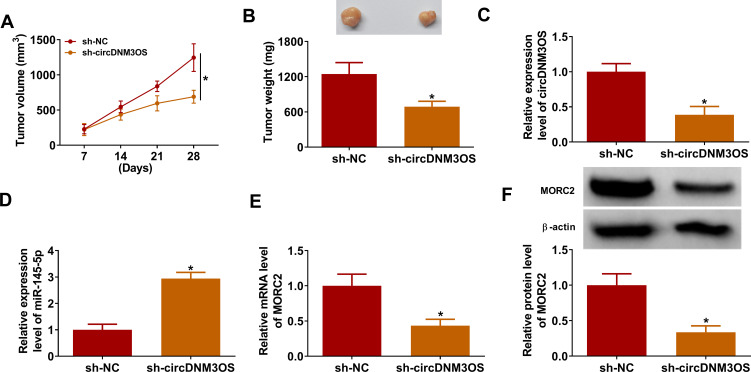
In view of the above findings, we further validated the role of circDNM3OS in CCA progression by xenograft assay. Compared to the control group, xenograft tumor volume and weight in mice injected with HuH28 cells carrying sh-circDNM3OS were markedly reduced (Figure 7A and B). Moreover, the relative expression of circDNM3OS was reduced in xenograft tumors in the sh-circDNM3OS group, but miR-145-5p expression had an opposing trend (Figure 7C and D). Also, the levels of MORC2 mRNA and protein were also decreased in xenograft tumors in the sh-circDNM3OS group (Figure 7E and F). Collectively, these results indicated that circDNM3OS silencing reduced CCA growth in vivo.
Figure 7.
CircDNM3OS knockdown repressed the growth of CCA in mouse model. 10 BALB/c nude mice were subcutaneously injected with HuH28 cells carrying sh-circDNM3OS or sh-NC. (A) Tumor volume of the sh-circDNM3OS or sh-NC groups was estimated once a week. (B) Tumor weight of the sh-circDNM3OS or sh-NC groups was shown. (C–F) The levels of circDNM3OS, miR-145-5p, MORC2 mRNA, and MORC2 protein in xenograft tumors in the sh-circDNM3OS or sh-NC groups were assessed by qRT-PCR or Western blotting. *P < 0.05.
Discussion
Mounting evidence has revealed that the abnormal expression of circRNAs is implicated in the progression of CCA.17,21,22 Moreover, whole-transcriptome sequencing has revealed that there are 110 differentially expressed circRNAs in CCA tissues in contrast to adjacent normal tissues.23 Previous study uncovered that circRNA circ_0000284 aroused the malignant phenotype of CCA cells.21 Also, circRNA circCDR1as exerted a promoting impact on the malignancy of CCA cells through sponging miR-641.17 Furthermore, circRNA circCCAC1 accelerated CCA development by sequestering miR-514a-5p and elevating YY1 expression.22 In this study, circDNM3OS had a higher expression in CCA tissues and cells. Moreover, circDNM3OS downregulation decreased xenograft tumor growth in vivo and constrained glutamine metabolism and malignancy of CCA cells in vitro. Xu et al also revealed that circDNM3OS played an oncogenic function in CCA through absorbing miR-1299 and miR-1238.18 In breast cancer, circDNM3OS overexpression contributed to invasion and proliferation via the miR-618/CBX8 axis.24 Accordingly, we concluded that circDNM3OS exerted a promoting influence on tumor growth and glutamine metabolism in CCA.
Based on the ceRNA hypothesis, circRNAs act as miR sponges by competitively binding miR response elements, thereby reducing the inhibitory effect of miRs on their targets.25 Herein, we uncovered that circDNM3OS was a sponge of miR-145-5p. Many researches had revealed that miR-145-5p exerted an inhibitory impact on a series of tumor growth.26–28 Zhou et al marked that miR-145-5p repressed cell invasion, migration, and proliferation of gastric cancer cells through targeting ANGPT2 and modulating the NOD_LIKE_RECEPTOR pathway.26 Moreover, circRNA circ_0058063 sponged miR-145-5p to elevate CDK6 expression, thereby accelerating cell migration and growth in bladder cancer.27 Furthermore, miR-145-5p played a suppressive effect on hepatocellular cancer growth.29 Zeng et al discovered that lncRNA TUG1 promoted malignancy and glutamine metabolism of intrahepatic CCA cells by regulating the miR-145/Sirt3/GDH pathway.19 Herein, miR-145-5p had a lower expression in CCA tissues and cells. The repressive effect of circDNM3OS silencing on the malignancy and glutamine metabolism of CCA cells was overturned by miR-145-5p inhibitor. Thus, we inferred that circDNM3OS promoted malignancy and glutamine metabolism of CCA cells through sponging miR-145-5p. Given that circDNM3OS facilitated migration and growth via sponging miR-1299 and miR-1238 in CCA,18 we found that MORC2 may be the target of miR-1238 and miR-1299 through online prediction (microT_CDS), implying that circDNM3OS might regulate CCA progression by regulating MORC2 expression by sponging miR-1238, miR-1299, and miR-145-5p. Unfortunately, we have not confirmed this, which can be discussed in depth in the future.
MiRs, a type of short non-coding RNA molecules, control the expression of mRNA associated with various biological processes.30 In our study, MORC2 was validated as a target for miR-145-5p. MORC2 is a member of the MORC ATPase superfamily, and it plays vital roles in diverse biological processes, such as lipogenesis, DNA damage repair, and chromatin remodeling.31–33 MORC2 had been uncovered as an oncogene in liver cancer.34 Moreover, miR-186-5p targeted MORC2 to curb metastasis and growth of CCA cells.35 Herein, MORC2 had a higher expression in CCA tissues and cells. Overexpression of MORC2 overturned miR-145-5p elevation-mediated influence on glutamine metabolism and malignancy of CCA cells. Also, circDNM3OS sponged miR-145-5p to modulate MORC2 expression. Accordingly, we concluded that circDNM3OS facilitated the malignancy and glutamine metabolism of CCA cells through upregulating MORC2 via sponging miR-145-5p.
In summary, circDNM3OS accelerated tumor growth and glutamine metabolism in CCA. Furthermore, circDNM3OS elevated MORC2 expression through absorbing miR-145-5p, thereby facilitating the malignancy and glutamine metabolism of CCA cells, providing a novel mechanism to comprehend the advancement of CCA.
Funding Statement
There is no funding to report.
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
- 1.Li G, Liu T, Zhang B, Chen W, Ding Z. Genome-wide identification of a competing endogenous RNA network in cholangiocarcinoma. J Cell Biochem. 2019;120(11):18995–19003. doi: 10.1002/jcb.29222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet. 2014;383(9935):2168–2179. doi: 10.1016/S0140-6736(13)61903-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fitzmaurice C, Allen C, Barber RM, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 Cancer Groups, 1990 to 2015: a systematic analysis for the Global Burden of Disease Study. JAMA Oncol. 2017;3(4):524–548. doi: 10.1001/jamaoncol.2016.5688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bridgewater J, Galle PR, Khan SA, et al. Guidelines for the diagnosis and management of intrahepatic cholangiocarcinoma. J Hepatol. 2014;60(6):1268–1289. [DOI] [PubMed] [Google Scholar]
- 5.Yeh C-N, Chen M-H, Chang Y-C, et al. Over-expression of TNNI3K is associated with early-stage carcinogenesis of cholangiocarcinoma. Mol Carcinog. 2019;58(2):270–278. doi: 10.1002/mc.22925 [DOI] [PubMed] [Google Scholar]
- 6.Michalak KP, Maćkowska-Kędziora A, Sobolewski B, Woźniak P. Key roles of glutamine pathways in reprogramming the cancer metabolism. Oxid Med Cell Longev. 2015;2015:964321. doi: 10.1155/2015/964321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao D, Zeng L, Yao K, Kong X, Wu G, Yin Y. The glutamine-alpha-ketoglutarate (AKG) metabolism and its nutritional implications. Amino Acids. 2016;48(9):2067–2080. doi: 10.1007/s00726-016-2254-8 [DOI] [PubMed] [Google Scholar]
- 8.Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123(9):3678–3684. doi: 10.1172/JCI69600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer. 2016;16(10):619–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J, Li D, Luo H, Zhu X. Circular RNAs: the star molecules in cancer. Mol Aspects Med. 2019;70:141–152. doi: 10.1016/j.mam.2019.10.006 [DOI] [PubMed] [Google Scholar]
- 11.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi: 10.1038/s41576-019-0158-7 [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. 2018;34:267–274. doi: 10.1016/j.ebiom.2018.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li G, Qin Y, Qin S, Zhou X, Zhao W, Zhang D. Circ_WBSCR17 aggravates inflammatory responses and fibrosis by targeting miR-185-5p/SOX6 regulatory axis in high glucose-induced human kidney tubular cells. Life Sci. 2020;259:118269. doi: 10.1016/j.lfs.2020.118269 [DOI] [PubMed] [Google Scholar]
- 14.Zhang L, Guo Y. Silencing circular RNA-ZNF652 represses proliferation and EMT process of renal carcinoma cells via raising miR-205. Artif Cells Nanomed Biotechnol. 2020;48(1):648–655. doi: 10.1080/21691401.2020.1725532 [DOI] [PubMed] [Google Scholar]
- 15.Zhao X, Zhang X, Zhang Z, et al. Comprehensive circular RNA expression profiling constructs a ceRNA network and identifies hsa_circ_0000673 as a novel oncogene in distal cholangiocarcinoma. Aging (Albany NY). 2020;12(22):23251–23274. doi: 10.18632/aging.104099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu Q, Fang T. Circular RNA SMARCA5 correlates with favorable clinical tumor features and prognosis, and increases chemotherapy sensitivity in intrahepatic cholangiocarcinoma. J Clin Lab Anal. 2020;34(4):e23138. doi: 10.1002/jcla.23138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li D, Tang Z, Gao Z, Shen P, Liu Z, Dang X. Circular RNA CDR1as exerts oncogenic properties partially through regulating MicroRNA 641 in cholangiocarcinoma. Mol Cell Biol. 2020;40:15. doi: 10.1128/MCB.00042-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Yao Y, Liu Y, et al. Elevation of circular RNA circ_0005230 facilitates cell growth and metastasis via sponging miR-1238 and miR-1299 in cholangiocarcinoma. Aging (Albany NY). 2019;11(7):1907–1917. doi: 10.18632/aging.101872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng B, Ye H, Chen J, et al. LncRNA TUG1 sponges miR-145 to promote cancer progression and regulate glutamine metabolism via Sirt3/GDH axis. Oncotarget. 2017;8(69):113650–113661. doi: 10.18632/oncotarget.21922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Li X, Wu L, Pei M, Li H, Jiang Y. miR-145 inhibits glutamine metabolism through c-myc/GLS1 pathways in ovarian cancer cells. Cell Biol Int. 2019;43(8):921–930. doi: 10.1002/cbin.11182 [DOI] [PubMed] [Google Scholar]
- 21.Wang S, Hu Y, Lv X, et al. Circ-0000284 arouses malignant phenotype of cholangiocarcinoma cells and regulates the biological functions of peripheral cells through cellular communication. Clin Sci. 2019;133(18):1935–1953. doi: 10.1042/CS20190589 [DOI] [PubMed] [Google Scholar]
- 22.Xu Y, Leng K, Yao Y, et al. A novel circular RNA, circ-CCAC1, contributes to CCA progression, induces angiogenesis, and disrupts vascular endothelial barriers. Hepatology. 2020. doi: 10.1002/hep.31493 [DOI] [PubMed] [Google Scholar]
- 23.Chu K-J, Ma Y-S, Jiang X-H, et al. Whole-transcriptome sequencing identifies key differentially expressed mRNAs, miRNAs, lncRNAs, and circRNAs associated with CHOL. Mol Ther Nucleic Acids. 2020;21:592–603. doi: 10.1016/j.omtn.2020.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y, Yao Y, Leng K, et al. Increased expression of circular RNA circ_0005230 indicates dismal prognosis in breast cancer and regulates cell proliferation and invasion via miR-618/CBX8 signal pathway. Cell Physiol Biochem. 2018;51(4):1710–1722. doi: 10.1159/000495675 [DOI] [PubMed] [Google Scholar]
- 25.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou K, Song B, Wei M, Fang J, Xu Y. MiR-145-5p suppresses the proliferation, migration and invasion of gastric cancer epithelial cells via the NOD_LIKE_RECEPTOR axis. Cancer Cell Int. 2020;20:416. doi: 10.1186/s12935-020-01483-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun M, Zhao W, Chen Z, et al. Circ_0058063 regulates CDK6 to promote bladder cancer progression by sponging miR-145-5p. J Cell Physiol. 2019;234(4):4812–4824. doi: 10.1002/jcp.27280 [DOI] [PubMed] [Google Scholar]
- 28.Chen Z-L, Li X-N, Ye C-X, Chen H-Y, Wang Z-J. Elevated levels of circRUNX1 in colorectal cancer promote cell growth and metastasis via miR-145-5p/IGF1 signalling. Onco Targets Ther. 2020;13:4035–4048. doi: 10.2147/OTT.S254133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang B, Dong W, Li X. miR-145-5p acts as a novel tumor suppressor in hepatocellular carcinoma through targeting RAB18. Technol Cancer Res Treat. 2019;18:1533033819850189. doi: 10.1177/1533033819850189 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Chen L, Heikkinen L, Wang C, Yang Y, Sun H, Wong G. Trends in the development of miRNA bioinformatics tools. Brief Bioinform. 2019;20(5):1836–1852. doi: 10.1093/bib/bby054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie H-Y, Zhang T-M, Hu S-Y, Shao Z-M, Li D-Q. Dimerization of MORC2 through its C-terminal coiled-coil domain enhances chromatin dynamics and promotes DNA repair. Cell Commun Signal. 2019;17(1):160. doi: 10.1186/s12964-019-0477-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sánchez-Solana B, Li D-Q, Kumar R. Cytosolic functions of MORC2 in lipogenesis and adipogenesis. Biochim Biophys Acta. 2014;1843(2):316–326. doi: 10.1016/j.bbamcr.2013.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao X-H, Zhang Y, Dong W-J, Shao Z-M, Li D-Q. Chromatin remodeling protein MORC2 promotes breast cancer invasion and metastasis through a PRD domain-mediated interaction with CTNND1. Oncotarget. 2017;8(58):97941–97954. doi: 10.18632/oncotarget.18556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan Z, Ding Q, Guo Q, et al. MORC2, a novel oncogene, is upregulated in liver cancer and contributes to proliferation, metastasis and chemoresistance. Int J Oncol. 2018;53(1):59–72. doi: 10.3892/ijo.2018.4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao G, Liu X, Wu D, et al. MORC2 promotes cell growth and metastasis in human cholangiocarcinoma and is negatively regulated by miR-186-5p. Aging (Albany NY). 2019;11(11):3639–3649. doi: 10.18632/aging.102003 [DOI] [PMC free article] [PubMed] [Google Scholar]