Abstract
Purpose
To explore the molecular mechanism and search for candidate lncRNA and mRNA associated with pyroptosis in the gene expression profile of LPS-induced acute lung injury (ALI).
Methods
We investigated lncRNA and mRNA expression in lipopolysaccharide (LPS)-induced ALI at an early stage. RNA sequencing (RNA-Seq) was carried out to analyze lncRNA and mRNA expression profiles between the LPS-induced and control groups. We used bioinformatics analysis to predict target genes of early differential lncRNAs among obtained the differential mRNAs.
Results
A total of 78 lncRNAs and 248 mRNAs were upregulated at 2 hours and downregulated at 9 hours, and 21 lncRNAs and 107 mRNAs were downregulated at 2 and upregulated at 9 hours in early ALI models. We predicted 7 cis-and trans-regulated target genes of the top 20 lncRNAs. Gene Ontology (GO) analysis indicated that the target genes for the screened lncRNAs were most enriched in three-terms: regulation of protein serine/threonine kinase activity, pertussis, and cellular response to LPS. Additionally, target genes of lncRNAs were the top three enriched in pertussis, osteoclast differentiation, and cAMP signaling pathways with Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. We also identified vital mRNAs and lncRNAs. Protein-protein interaction (PPI) network analysis suggested that Tnf, Jun, and Atf3 were the top three key genes. Hub lncRNA4344 (NONRATT004344.2) and cis-regulated target mRNA (NLRP3) were validated in vitro. Finally, luciferase assay results confirmed that lncRNA4344 sponged miR‐138-5p to promote pyroptosis in inflammatory responses to LPS‐induced acute lung injury by targeting NLRP3.
Conclusion
Based on analysis of lncRNA and mRNA expression profiles by RNA-Seq and experimental verification, this study is the first to reveal that lncRNA4344 sponged miR‐138-5p to promote pyroptosis in inflammatory responses of LPS‐induced acute lung injury by targeting NLRP3. These newly identified lncRNA, miRNA, and mRNA might be novel potential targets for early treatment and prevention in early ALI.
Keywords: acute lung injury, alveolar macrophage, ceRNA network long noncoding RNAs, pyroptosis, RNA sequencing
Introduction
Acute lung injury (ALI) progresses to acute respiratory distress syndrome (ARDS) and is a frequently occurring clinical syndrome with erupted uncontrolled lung inflammation, which leads to high mortality rates due to a lack of effective treatment.1 A recent global study of 29,144 patients in the ICU population found that ARDS has a high prevalence. ARDS patients represented 10.4% of total ICU admission, and 23.4% of these patients required mechanical ventilation and were hospitalized for more than four weeks.2 Clinicians have achieved considerable progress in understanding pathogenesis and advances in supportive treatments such as ventilatory support therapy or other medications; however, mortality rates from ALI and ARDS remain at approximately 40% globally.3 Although medical professionals have been looking for new treatments or drugs that inhibit the excessive inflammatory response to ALI/ARDS, the results have been minimal.4,5 Many high-quality research results suggest that other pathogeneses may be involved in ALI/ARDS. Since the pathophysiological processes of ALI/ARDS have not been elucidated at the genetic and transcriptional levels, novel therapeutic targets and directions for ALI/ARDS urgently need to be developed.
Long noncoding RNAs (lncRNAs) with no protein-coding ability have been considered noise in the transcription process in recent years. However, a growing body of research shows that lncRNAs play critical roles in regulating different gene expression levels, such as chromatin formation, transcription, and posttranscriptional translation.6,7 Accumulating evidence indicates that lncRNAs are involved in various processes of cellular activities, such as adipogenesis, apoptosis, pyroptosis, cell differentiation, epigenetic modification, and tumorigenesis and modulation.8–13 They regulate genomic expression and posttranscription in cis or trans by interacting with chromatin, protein, and RNA in the nucleus or cytoplasm. Li et al14 reported that lncRNA-MALAT1 can suppress inflammatory responses by increasing miR-146a expression in lipopolysaccharide (LPS) -treated ALI. Similarly, Wang et al15 also reported that lncRNA expression profiles were changed in the LPS- stimulated mouse ALI model, which suggested that lncRNAs and their target gene mRNAs may be closely associated with ALI and ARDS. LncRNA-HOTAIR was also observed to activate NF-κB, promoting the release of cytokines in the macrophages of LPS-induced sepsis mice, which showed that lncRNAs might be intimately related to ALI.16 However, the potential roles of lncRNAs have not been adequately elaborated in ALI; our study aimed to profile both lncRNAs and mRNAs in AM of ALI rats at an early stage and tried to determine their roles through bioinformatics analysis and cell study in vitro.
Materials and Methods
Alveolar Macrophage (AM) Modeling for ALI and RNA Isolation of Samples
We purchased NR8383 AMs from the Chinese Academy of Sciences Cell Bank (Shanghai, China) for ALI modeling. NR8383 AMs were cultured in Ham’s F-12K containing 15% (v/v) FBS and differentiated into macrophages by treatment with 25 ng/mL phorbol 12-myristate 13-acetate (PMA) (Sigma) overnight before use. Then, the medium was replaced with fresh non-PMA fresh medium. NR8383 AMs were cultured in Ham’s F-12K containing 15% (v/v) FBS. Then, NR8383 AMs were to induce an inflammatory response by stimulation with 1 µg/µL LPS (Escherichia coli 055:B5, Sigma-Aldrich) unless otherwise specified. Rat NR8383 AMs were randomly divided into three groups: LPS-stimulated 0 h group, LPS-stimulated 2 h group, and LPS-stimulated 9 h group, each group contained three samples. We used TRIzol reagent (Invitrogen, Carlsbad, USA) to isolate and purify the total RNA of rat NR8383 AMs. The quantity and quality of RNA and RNA integrity were evaluated and determined using the standard spectrophotometric and electrophoretic methods, as described in a previous study.17
Enzyme‐Linked Immunosorbent Assay (ELISA)
ELISA was performed according to the product manual. In brief, AMs were seeded into 24‐well plates at a density of 2×104 cells/mL. Supernatant macrophages of each group were harvested from cultured Petri dishes at preprogrammed time points. Expression levels of the inflammatory cytokines IL-6, IL‐18, IL‐1β, and TNF-α levels in the culture supernatant (100 μL) were analyzed using Boster Biotech Systems ELISA kits (Boster BioTECH, EK0412, EK0592, EK0526, and EK0393, Wuhan, China) according to the product manufacturer’s protocol. The absorbance at 450 nm (A450) was measured to calculate their concentration by regression analysis of a standard curve.
RNA Sample Collection and Library Preparation
We selected 3 μg of RNA from each sample for RNA sample preparations. First, we used an Epicentre Ribo-zero™ rRNA Removal Kit (RiBOBiTech, Guangzhou, China) to remove ribosomal RNA (rRNA) and clean residual free RNA with ethanol precipitation. Second, we used rRNA-depleted RNA (a NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina®, San Diego, CA, USA) to establish twelve sequencing libraries. The prepared RNA-Seq libraries were finally quantified. Finally, the diluted libraries were loaded onto the reagent cartridge and subjected to sequencing using an Illumina NextSeq 500 system (Illumina, San Diego, CA, USA) with a NextSeq 500/550 V2 kit (Illumina, San Diego, CA, USA).
RNA-Seq Analysis
We analyzed raw sequencing data generated from the Illumina NextSeq 500 system that passes the Illumina chastity filter. All raw sequencing data were subjected to strict quality control and screened to remove those low-quality reads to obtain high quality and clean sequencing data. After complete removal of the remaining rRNA reads, bioinformatics analysis was used to analyze the clean reads mapped to an rRNA reference using the short-read alignment software SOAPaligner/SOAP2. The mapped reads for each sample were independently assembled using Cufflinks (2.1.1). We calculated and screened the expression profiles and differentially expressed mRNAs and lncRNAs with data from statistical alignment analysis.
Bioinformatics Prediction and Analysis
Gene Ontology (GO) analysis was used to calculate the functions of selected differentially expressed genes, consisting of biological processes, molecular functions, and cellular components. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was used to map genes to the main signaling pathway. We used Fisher’s exact tests for statistical analyses and set 0.05 as statistically significant.
Construction of the Protein-Protein Interaction Network
We used the STRING (version 10.5; https://string-db.org/) database with confidence > 0.4 to identify the protein-protein interaction (PPI) pairs between differentially expressed mRNAs. Then, the PPI network was established by Cytoscape (Version 3.8.0; https://cytoscape.org/). The hub nodes in the PPI net were those that scored highly as network topology property indicators and were analyzed by Moncode in Cytoscape for factors including Betweenness centrality, Closeness centrality, Degree centrality, Edge percolated component (EPC), Radiality, and EcCentricity. In general, the high indicator score of the topology property network indicates a vital role in the network.
LncRNA-mRNA Coexpression Network
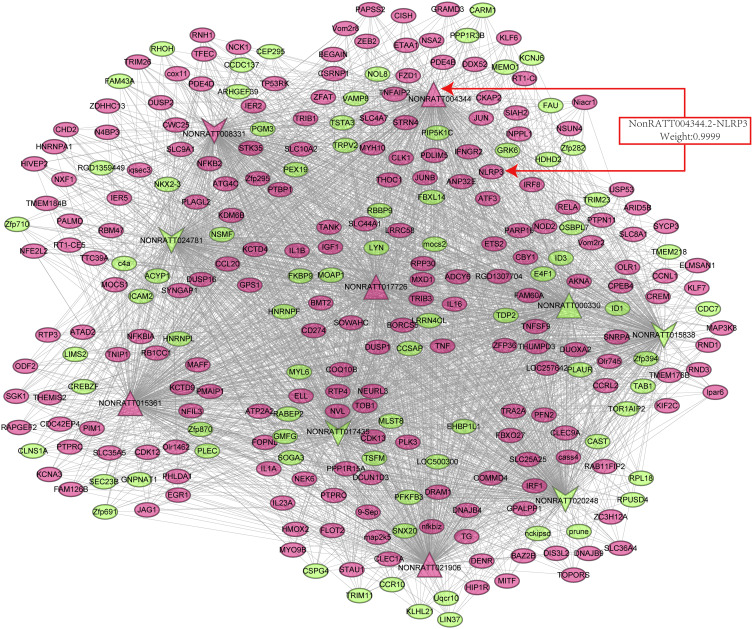
We established a coding-noncoding gene coexpression (CNC) network analysis to explore the correlation between differentially expressed mRNA and lncRNA. The correlations between lncRNAs and mRNAs pairs were differentiated with Pearson’s correlation coefficients (PCCs) of no less than 0.99.18 Finally, the lncRNA-mRNA coexpression network graphic was constructed using Cytoscape software 3.8.0 (http://www.cytoscape.org/) (Figure 1).
Figure 1.
Coexpression network between key differentially expressed lncRNAs and mRNAs. Dots represent mRNAs, and the arrows represent lncRNAs. The red arrows represent the five upregulated and the green arrow represents five downregulated differentially expressed lncRNAs and mRNAs. The weighting factor of lncRNA4344 (NONRATT004344.2) and the NLRP3 Coexpression network is 0.9999.
Abbreviation: NLRP3, Nod-like receptor protein-3.
Cis- and Trans-Regulated Gene Analysis
Cis-acting lncRNAs can suppress and activate genes within or near them on the same chromosome. In contrast, the regulatory functions of trans-acting lncRNAs are often limited at independent loci. All lncRNAs were identified after their gene positions on the chromosomes were determined. Potential “cis-regulated mRNAs” were identified, which were situated within the 300 kbp range upstream and downstream of the selected lncRNAs in the lncRNA-mRNA CNCs (r > 0.99 or r < −0.99, P < 0.01). “trans-regulated mRNAs” with the potential coding ability of trans-regulated proteins located beyond 100 kb in genomic distance from dysregulated lncRNAs in CNCs. To analyze the functions of the potential “cis-regulated mRNAs” or “trans-regulated mRNAs,” GO enrichment and pathway analysis were also performed.
Cell Culture and Cell Transfection with siRNA
NR8383 AMs were cultured in Ham’s F12K medium supplemented with 15% fetal bovine serum (Excell Bio, Shanghai, China) in a humidified atmosphere with 5% CO2 at 37°C. Then, all of the cells were plated in a six-well plate for further experiments. The small interfering RNA (siRNA) duplexes for lncRNA4344 (NONRATT004344.2, sequence: 5ʹ-GCAGCAGAAGTCACTTATA-3ʹ) were commercially synthesized by RIBOBIO (RIBOBIO Co., Guangzhou, China). We used siRNA-lncRNA4344 and siRNA-scrambled to transfect NR8383 AMs at 70% confluence following Lipofectamine 2000 referencing the product instructions. After transfection for thirty-six hours, NR8383 AMs were exposed to LPS (1µg/ µL) (Sigma Chemical Co., USA) for 6 h. Total RNA was isolated and purified following the manufactory protocol. PCR analysis for lncRNAs and mRNAs was performed. The primer sequences for PCR are shown in Supplementary Table 1. miRNA quantification: Bulge-loopTM miRNA qRT-PCR Primer Sets (one RT primer and a pair of qPCR primers for each set) specific for miR-138-5p and U6 were designed by RiboBio (RiboBio BioTech, Guangzhou, China).
Construction of the ceRNA Regulatory Network
The interactions between miRNAs and NLRP3 were predicted using miranda, TargetScan, and miRWalk, and the overlapping miRNA–mRNA pairs predicted by these three methods were selected. Additionally, miRNAs targeting lncRNA4344 were predicted using miRanda with default parameters. Next, mRNA–lncRNA–miRNA ceRNA regulatory networks were constructed.
Luciferase Assay
The wild-type and mutant fragments in the 3′-UTR of lncRNA4344 and NLRP3 related to the miR‐138-5p binding site were cloned into the pmirGLO vector (Promega, CA, USA) according to the product manufacturer′s instructions. Then 293T cells transfected with lncRNA4344-Wt or lncRNA4344-Mut were cotransfected with miR-138-5p mimics or miR138‐5p NC. After 48 h, the dual-luciferase reporter assay system (Promega, CA, USA) was used to measure the luciferase activities according to the manufacturer’s standard protocol.
Western Blot Analysis
Cells were collected and lysed in M-PER Mammalian Protein Extraction Reagent. All samples were normalized by their protein concentrations, separated in 10% SDS-PAGE gels, and then transferred to membranes (Washington, NY) using a wet transfer blotting system (Hercules, CA). The antibodies used for Western blotting were: anti-NLRP3 (Abcam), anti-caspase-1 (Abcam), and anti-GAPDH (Boster, Wuhan, China), at a dilution of 1:500. Protein bands were developed using ECL reagents, whose images were acquired using the BIO-RAD Imaging system. Western blot examination was repeated three times.
Statistical Analysis
Normalization (reads per kilobase pair per million mapped reads, RPKM, and counts per million mapped reads, CPM) analysis was used to control the quality of the sequence data of mRNAs and lncRNAs in our study. The negative binomial generalized linear model was used to calculate the CPM values of the control group and the LPS-induced group. We used R 4.02 software to analyze the differential expression of mRNAs and lncRNAs. The criteria for selecting genes were |Fold change| ≥ 2 and P ≤ 0.05. The qRT-PCR results are expressed as the mean ± SD in our study. Statistical comparisons of the data were made using SPSS software (SPSS 24, Chicago, IL, USA). Multiple comparisons among all groups were performed by one-way analysis of variance (ANOVA) followed by post hoc testing. Comparisons between two groups were performed by the Student–Newman–Keuls test. P <0.05 was defined as a statistically significant difference.
Results
Establishment of Early ALI Model
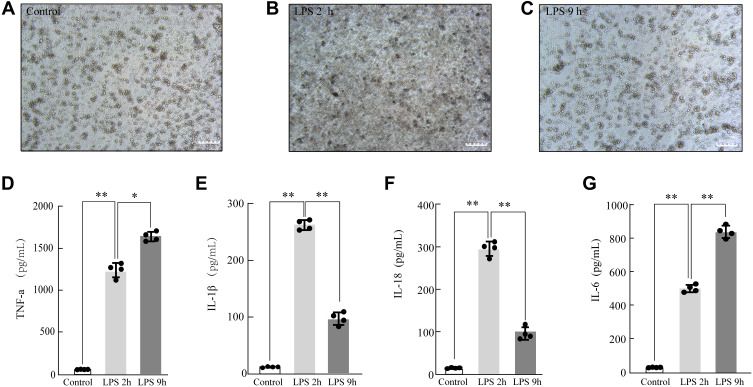
After LPS stimulation, the microscopy results showed that LPS dramatically increased the leukocyte infiltration of rat NR8383 AMs. In a normal state, NR8383 AMs were small, round, and bright, and became larger and pseudopods extended wider after LPS stimulation for 2 h. After 9 hours of LPS stimulation, their shapes became irregular with rough surfaces and were crumpled, and smaller (Figure 2A–C). A significant inflammatory response was observed in ALI AM models. The expression of early inflammatory factors (IL-18, IL-1β, and TNF-α) was significantly increased after LPS stimulation (P < 0.05). The expression of IL-18 and IL-1β increased at 2 hours but decreased at 9 hours; the TNF-α expression continued to increase for 9 hours (Figure 2D–F), suggesting the successful establishment of the early ALI AM model.
Figure 2.
Modeling and validation of LPS-treated acute lung injury (ALI). The morphological changes of LPS-treated rat NR8383 alveolar macrophage (AM)(200×) Control (A), LPS 2 h (B), LPS 9 h (C); LPS-induced AM modeling for ALI was validated by measuring the inflammatory cytokine expression levels of TNF-α (D), IL-1β (E), IL-18 (F), and IL-6 (G) by ELISA. Each value represents the mean ± SD of three separate sets of experiments. *p < 0.05; **p < 0.01
Abbreviations: ALI, acute lung injury; ELISA, enzyme‐linked immunosorbent assay; IL‐1β, interleukin (IL)‐1β; IL‐6, interleukin (IL)‐6; IL-18, interleukin (IL)-18; LPS, lipopolysaccharides; TNF-α, tumor necrosis factor-α; NS, no statistically significant difference.
LncRNAs and mRNAs Expression Profiles in Early ALI
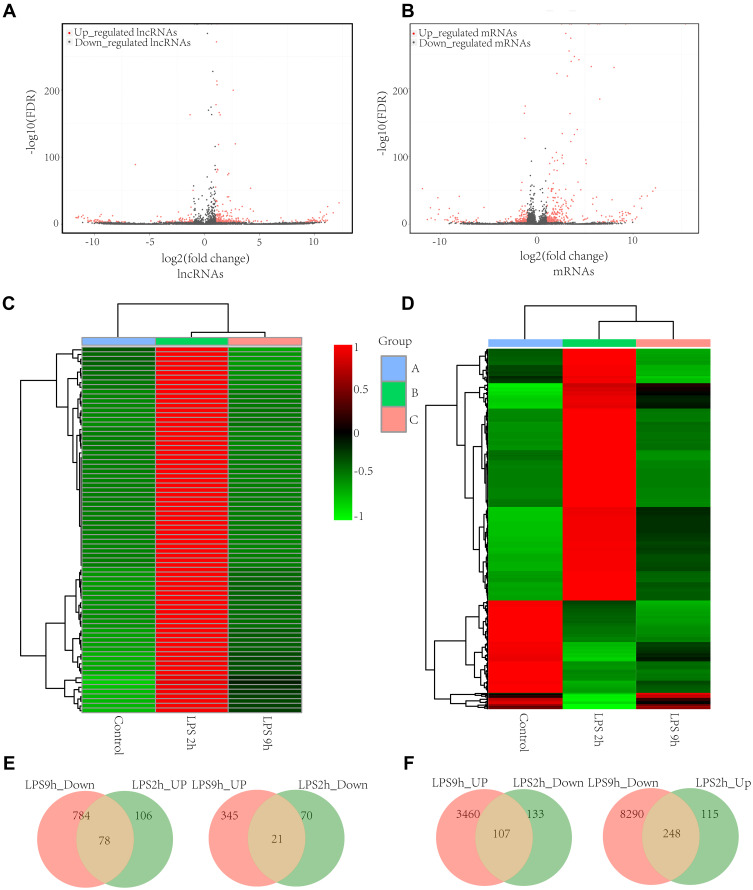
We identified 1503 differentially expressed lncRNAs in LPS-induced ALI models, with 78 lncRNAs upregulated at 2 hours and downregulated at 9 hours, and 21 lncRNAs downregulated at 2 hours and upregulated at 9 hours (P-value < 0.05 and |fold-change| > 2.0), (Figure 3A, C, and E). NONRATT008331.2 (fold-change: 2.80, P=2.80E-123) and NONRATT000330.2 (fold-change: −6.28, P=2.69E-92) were the most upregulated and downregulated lncRNAs among the identified lncRNAs, respectively. The top twenty differentially upregulated and downregulated expressed lncRNAs were identified by RNA-Seq (Table 1). A total of 248 mRNAs were upregulated at 2 hours and were downregulated at 9 hours, and 107 mRNAs downregulated at 2 h and upregulated at 9 h in LPS-induced ALI (P-value < 0.05 and |fold-change| > 2.0) (Figure 3B, D, and F). NFKBIA (fold change: 3.40, P=0.00) and PLAUR (fold change: −11.07, P=6.12E-27) were the top significantly upregulated and downregulated mRNAs, respectively. Table 2 summarizes the top 10 upregulated and 10 downregulated differentially expressed mRNAs selected among the identified mRNAs via RNA-Seq.
Figure 3.
The distinct lncRNA and mRNA expression profiles between LPS-induced AM and control AM. Volcano plots of differentially expressed lncRNAs (A) and mRNAs (B); The heatmap represents hierarchical clustering for differentially expressed lncRNAs (C) and mRNAs (D) at 0 h, 2h, and 9 h; lncRNAs (E) and mRNAs (F) that are significantly upregulated and downregulated at 2 h compared to 9 h. were shown by the Venn diagram between LPS-stimulated AM and control AM. The range of colors from green to red indicates expression value from relatively low to high, respectively (n=3 for the LPS-stimulated group and the control group, respectively).
Abbreviations: AM, alveolar macrophages; LPS, lipopolysaccharides.
Table 1.
The Detailed Information of the Top Ten Up-Regulated and Down-Regulated LncRNAs
| Transcript ID | New ID | Log2.Ratio | Regulation | P-value | FDR | Gene Length | Chrom |
|---|---|---|---|---|---|---|---|
| NONRATT008331.2 | Shykoy.aSep08-unspliced | 2.798165 | Up | 2.81E-123 | 1.54E-120 | 519 | Chr13 |
| NONRATT004344.2 | Nlrp3.cSep08 | 4.172958 | Up | 1.08E-56 | 3.11E-54 | 1033 | Chr10 |
| NONRATT017726.2 | Flady.aSep08-unspliced | 3.416925 | Up | 2.78E-29 | 3.49E-27 | 491 | Chr20 |
| NONRATT021906.2 | Porzoy.aSep08-unspliced | 2.802134 | Up | 1.86E-25 | 2.05E-23 | 1497 | Chr4 |
| NONRATT015361.2 | Veyblaw.aSep08-unspliced | 3.144573 | Up | 2.26E-24 | 2.36E-22 | 689 | Chr2 |
| NONRATT009244.2 | Ccni.cSep08 | 10.50581 | Up | 1.52E-20 | 1.30E-18 | 1374 | Chr14 |
| NONRATT004883.2 | STAT_alpha.0.aSep08 | 11.67904 | Up | 1.08E-19 | 8.81E-18 | 608 | Chr10 |
| NONRATT020829.2 | LOC500300.aSep08-unsplice | 9.79604 | Up | 4.82E-12 | 2.23E-10 | 1283 | Chr4 |
| NONRATT014868.2 | Glrx1.bSep08-unspliced | 6.93178 | Up | 9.26E-12 | 4.16E-10 | 308 | Chr2 |
| NONRATT017346.2 | Glady.aSep08-unspliced | 2.859657 | Up | 5.62E-11 | 2.33E-09 | 2732 | Chr20 |
| NONRATT000330.2 | Koflor.aSep08-unspliced | −6.2754 | Down | 2.69E-92 | 1.12E-89 | 361 | Chr1 |
| NONRATT017435.2 | Cdc2l6.bSep08 | −11.0293 | Down | 9.28E-16 | 5.84E-14 | 735 | Chr20 |
| NONRATT024781.2 | Cox7a2l.cSep08 | −3.96677 | Down | 1.91E-13 | 1.00E-11 | 821 | Chr6 |
| NONRATT015838.2 | Sep15.cSep08 | −10.6119 | Down | 6.32E-12 | 2.87E-10 | 736 | Chr2 |
| NONRATT020248.2 | Tmem106b.bSep08 | −10.3685 | Down | 8.99E-08 | 2.42E-06 | 562 | Chr4 |
| NONRATT020695.2 | LOC500251.cSep08 | −2.9189 | Down | 3.12E-07 | 7.74E-06 | 421 | Chr4 |
| NONRATT016007.2 | Fcho2.aSep08 | −8.56224 | Down | 8.17E-07 | 1.87E-05 | 1588 | Chr2 |
| NONRATT020531.2 | Gng12.bSep08 | −10.5651 | Down | 3.56E-06 | 7.14E-05 | 395 | Chr4 |
| NONRATT004734.2 | Tmem49.eSep08 | −9.04439 | Down | 3.23E-05 | 0.000514 | 898 | Chr10 |
| NONRATT004074.2 | Rnps1.cSep08 | −9.23362 | Down | 6.74E-05 | 0.000986 | 735 | Chr10 |
Table 2.
The Detailed Information of the Top Ten Up-Regulated and Down-Regulated mRNAs
| Gene ID | New ID | Log2.Ratio | Regulation | P-value | FDR | Gene Length | Chrom |
|---|---|---|---|---|---|---|---|
| NFKBIA | ENSRNOG00000007390 | 3.403453 | Up | 0 | 0 | 1566 | Chr6 |
| NFKBIZ | ENSRNOG00000031163 | 6.54208 | Up | 0 | 0 | 2990 | Chr11 |
| CCRL2 | ENSRNOG00000033234 | 5.349875 | Up | 0 | 0 | 1979 | Chr8 |
| NLRP3 | ENSRNOG00000003170 | 3.594497 | Up | 0 | 0 | 3857 | Chr10 |
| TNF | ENSRNOG00000055156 | 15.45128 | Up | 0 | 0 | 1687 | Chr20 |
| OLR1 | ENSRNOG00000056219 | 5.408867 | Up | 0 | 0 | 3751 | Chr4 |
| JUNB | ENSRNOG00000042838 | 3.002839 | Up | 8.93E-285 | 1.37E-281 | 1785 | Chr19 |
| DUSP2 | ENSRNOG00000013862 | 3.518036 | Up | 1.02E-277 | 1.49E-274 | 1634 | Chr3 |
| IL1A | ENSRNOG00000004575 | 4.363508 | Up | 4.54E-246 | 5.37E-243 | 1946 | Chr3 |
| IL1B | ENSRNOG00000004649 | 8.020804 | Up | 1.65E-234 | 1.75E-231 | 1314 | Chr3 |
| MYL6 | ENSRNOG00000054140 | −1.65249 | Down | 3.96E-53 | 9.59E-51 | 677 | Chr7 |
| PLEC | ENSRNOG00000023781 | −8.07682 | Down | 7.18E-44 | 1.39E-41 | 15085 | Chr7 |
| CARM1 | ENSRNOG00000031129 | −10.2992 | Down | 5.93E-42 | 1.10E-39 | 3131 | Chr8 |
| TRPV2 | ENSRNOG00000003104 | −1.54239 | Down | 3.28E-29 | 4.08E-27 | 2720 | Chr10 |
| CAST | ENSRNOG00000010286 | −5.50553 | Down | 2.36E-27 | 2.76E-25 | 4441 | Chr2 |
| PLAUR | ENSRNOG00000037931 | −11.0701 | Down | 6.12E-27 | 7.08E-25 | 1201 | Chr1 |
| THUMPD3 | ENSRNOG00000006941 | −10.1737 | Down | 2.20E-24 | 2.31E-22 | 1982 | Chr4 |
| LYN | ENSRNOG00000008180 | −1.28847 | Down | 1.69E-22 | 1.63E-20 | 3367 | Chr5 |
| HNRNPF | ENSRNOG00000014562 | −1.50434 | Down | 1.97E-22 | 1.89E-20 | 2188 | Chr4 |
| PFKFB3 | ENSRNOG00000018911 | −1.55032 | Down | 9.20E-20 | 7.61E-18 | 2148 | Chr17 |
Abbreviation: FDR, false discovery rate.
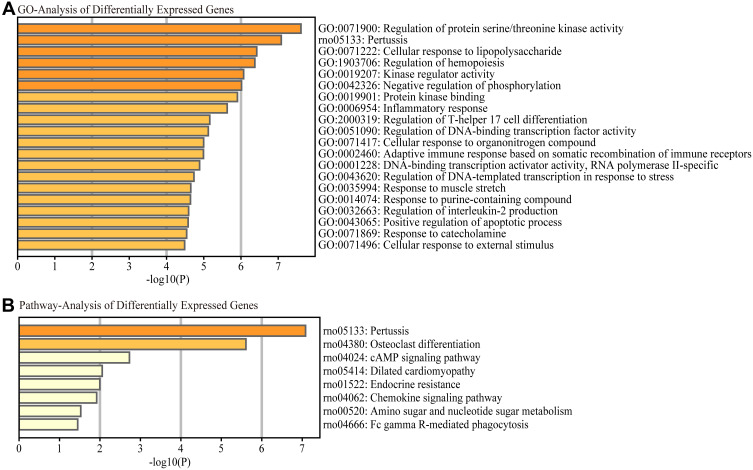
GO and KEGG Analyses
We investigated the probable mechanisms and latent functions of the most differentially expressed lncRNAs; GO analyses were performed to predict target genes. The top three enriched functional terms of differentially expressed genes were: regulation of protein serine/threonine kinase activity (GO: 0071900), pertussis (rno: 05133), and cellular response to LPS (GO: 0071222) (Figure 4A). Simultaneously, we investigated the probable mechanisms and possible functions of the most differentially expressed lncRNAs; KEGG analysis was performed to predict target genes. We found that pertussis, osteoclast differentiation, and the cAMP signaling pathway were the top pathways (Figure 4B).
Figure 4.
Functional enrichment analysis of differentially expressed mRNAs. The most significantly differentially expressed mRNAs involved in (A) biological process, cellular component and molecular function were identified by Gene Ontology (GO) analysis and (B) signaling pathways by Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis.
PPI Network Analysis of mRNAs
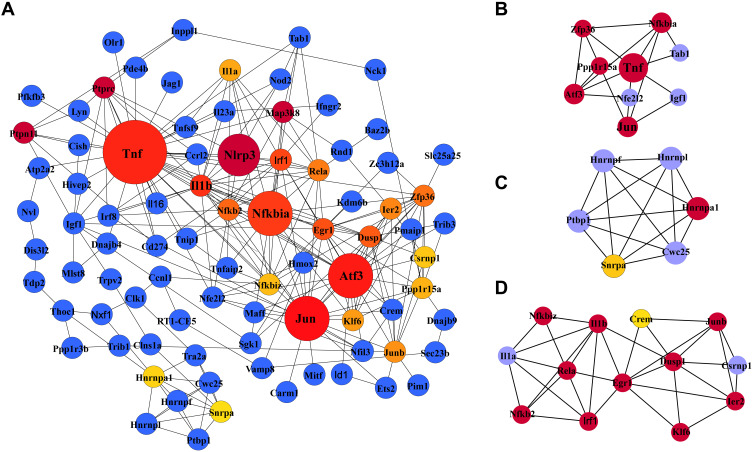
The PPI network containing 282 nodes and 307 edges was established from differentially expressed mRNAs (Figure 5). According to the ranking of network topology property indicators of Betweenness centrality, Closeness centrality, Degree centrality, EPC, Radiality, and EcCentricity, and the top 30 nodes were separately identified as hub nodes (Table 3). Tnf, Jun, and Atf3 were the central hub nodes among these hub nodes, which have more neighborhood connections to other mRNAs (Figure 5).
Figure 5.
Protein-protein interaction (PPI) network of differentially expressed mRNAs (A) and the top three clusters (B–D) of PPIs. (A) The PPI network consists of 282 nodes and 307 edges. Dot size represents the MCC of nodes. (B–D) The top three highly MCC connected clusters are composed of nine hub genes, six hub genes, and eleven hub genes.
Abbreviations: PPI, protein-protein interaction network; MCC, maximal clique centrality.
Table 3.
The Hub Nodes in the Protein-Protein Interaction Network According to the Score of Network Topology Property Indicators
| Rank | Node_Name | Degree | EPC | EcCentricity | Closeness | Radiality | Betweenness |
|---|---|---|---|---|---|---|---|
| 1 | Tnf | 35 | 86.702 | 0.08918 | 81.26349 | 12.58823 | 13682.09 |
| 2 | Jun | 23 | 86.702 | 0.08107 | 72.18337 | 12.33418 | 3286.734 |
| 3 | Atf3 | 21 | 86.702 | 0.08107 | 70.08456 | 12.24086 | 2937.374 |
| 4 | IL1b | 19 | 86.702 | 0.08107 | 69.00956 | 12.25641 | 1016.724 |
| 5 | Nfkbia | 17 | 86.702 | 0.08107 | 67.12504 | 12.18901 | 730.6439 |
| 6 | Egr1 | 16 | 86.702 | 0.08107 | 68.88337 | 12.30307 | 3120.393 |
| 7 | Zfp36 | 14 | 86.702 | 0.08107 | 66.7917 | 12.25641 | 1685.765 |
| 8 | Dusp1 | 14 | 86.702 | 0.08107 | 65.33456 | 12.14754 | 676.7042 |
| 9 | Rela | 13 | 86.526 | 0.08107 | 64.40837 | 12.14754 | 667.0527 |
| 10 | Nfkb2 | 12 | 86.617 | 0.08107 | 62.83456 | 12.04384 | 332.3751 |
| 11 | Junb | 12 | 86.702 | 0.07431 | 59.70678 | 11.91423 | 2575.122 |
| 12 | Irf1 | 12 | 86.702 | 0.08107 | 62.86075 | 12.04384 | 498.7108 |
| 13 | Ppp1r15a | 11 | 85.836 | 0.07431 | 58.68416 | 11.83128 | 2288.497 |
| 14 | Ptprc | 11 | 86.702 | 0.08107 | 59.73575 | 11.93497 | 671.6867 |
| 15 | Igf1 | 10 | 86.471 | 0.08107 | 64.18337 | 12.19938 | 3147.178 |
| 16 | Nfkbiz | 10 | 86.322 | 0.08107 | 62.80004 | 12.16827 | 3664.288 |
| 17 | Ptpn11 | 9 | 85.634 | 0.08107 | 59.23575 | 11.94015 | 938.0699 |
| 18 | Ier2 | 9 | 86.27 | 0.07431 | 56.69131 | 11.78461 | 577.7848 |
| 19 | IL1a | 8 | 86.31 | 0.08107 | 57.57742 | 11.87794 | 49.35081 |
| 20 | Hnrnpa1 | 8 | 39.866 | 0.07431 | 38.554 | 10.12035 | 841.2823 |
| 21 | Klf6 | 8 | 86.41 | 0.07431 | 56.6175 | 11.77424 | 34.7899 |
| 22 | Map3k8 | 8 | 85.95 | 0.08107 | 58.81789 | 11.97126 | 1494.478 |
| 23 | Nfe2l2 | 8 | 85.165 | 0.08107 | 60.44408 | 12.01792 | 831.0779 |
| 24 | Nlrp3 | 7 | 85.119 | 0.08107 | 57.51075 | 11.87275 | 1074.296 |
| 25 | Snrpa | 7 | 39.897 | 0.0686 | 39.47342 | 10.35366 | 1497.442 |
| 26 | Cwc25 | 7 | 39.353 | 0.0686 | 36.427 | 9.85594 | 420.5 |
| 27 | Crem | 7 | 85.706 | 0.07431 | 54.01988 | 11.64981 | 360.8152 |
| 28 | Cish | 7 | 80.231 | 0.08107 | 58.48575 | 11.93497 | 1814.863 |
| 29 | Fbxo27 | 7 | 76.674 | 0.07431 | 48.42821 | 11.32837 | 528.1243 |
| 30 | Sgk1 | 6 | 83.203 | 0.07431 | 55.45916 | 11.78461 | 848.2421 |
Abbreviation: EPC, edge percolated component.
CNC Network Analysis of mRNAs and LncRNAs
The CNC network was established to analyze the correlation between mRNAs and lncRNAs, selected differentially expressed lncRNAs and mRNAs, whose PCC value was more than 0.99 are shown in Figure 1. Table 4 listed the top 20 lncRNA–mRNA interactions. We selected the top ten of those lncRNAs marked as red up arrows and green down arrows in Figure 1. Additionally, one lncRNA was commonly coexpressed with multiple mRNAs and lncRNAs.
Table 4.
Top Twenty Rank Pairs Interactions in the LncRNA–mRNA Network
| Number | Gene1 | Biotype1 | Gene2 | Biotype2 | Interaction | Type | P-value |
|---|---|---|---|---|---|---|---|
| 1 | NONRATT008331.2 | lncRNA | IER5 | mRNA | 1.0 | Up | 4.85E-09 |
| 2 | NONRATT004344.2 | lncRNA | NLRP3 | mRNA | 0.998 | Up | 7.03E-06 |
| 3 | NONRATT017726.2 | lncRNA | ARID5B | mRNA | 0.999 | Up | 8.48E-07 |
| 4 | NONRATT021906.2 | lncRNA | BORCS5 | mRNA | 0.996 | Up | 2.03E-05 |
| 5 | NONRATT015361.2 | lncRNA | RAPGEF2 | mRNA | 0.998 | Up | 4.90E-06 |
| 6 | NONRATT009244.2 | lncRNA | JUNB | mRNA | 0.999 | Up | 1.46E-08 |
| 7 | NONRATT004883.2 | lncRNA | KCNA3 | mRNA | 1.0 | Up | 3.12E-10 |
| 8 | NONRATT020829.2 | lncRNA | CCRL2 | mRNA | 1.0 | Up | 1.70E-07 |
| 9 | NONRATT014868.2 | lncRNA | TNF | mRNA | 1.0 | Up | 9.26E-11 |
| 10 | NONRATT017346.2 | lncRNA | DUSP2 | mRNA | 0.998 | Up | 3.90E-06 |
| 11 | NONRATT000330.2 | lncRNA | CBY1 | mRNA | −1.0 | Down | 1.23E-08 |
| 12 | NONRATT017435.2 | lncRNA | KCNA3 | mRNA | −1.0 | Down | 1.77E-11 |
| 13 | NONRATT024781.2 | lncRNA | NFKBIZ | mRNA | −1.0 | Down | 4.15E-11 |
| 14 | NONRATT015838.2 | lncRNA | MTCONS_00034514 | mRNA | −1.0 | Down | 7.38E-11 |
| 15 | NONRATT020248.2 | lncRNA | MTCONS_00013625 | mRNA | −1.0 | Down | 1.16E-10 |
| 16 | NONRATT020695.2 | lncRNA | SGK1 | mRNA | −0.999 | Down | 1.33E-06 |
| 17 | NONRATT016007.2 | lncRNA | MTCONS_00014018 | mRNA | −0.999 | Down | 1.40E-06 |
| 18 | NONRATT020531.2 | lncRNA | NCK1 | mRNA | −1.0 | Down | 1.24E-07 |
| 19 | NONRATT004734.2 | lncRNA | PFN2 | mRNA | −0.999 | Down | 7.22E-07 |
| 20 | NONRATT004074.2 | lncRNA | MTCONS_00047512 | mRNA | −0.999 | Down | 5.76E-07 |
The Cis- and Trans-Regulated mRNAs Analysis
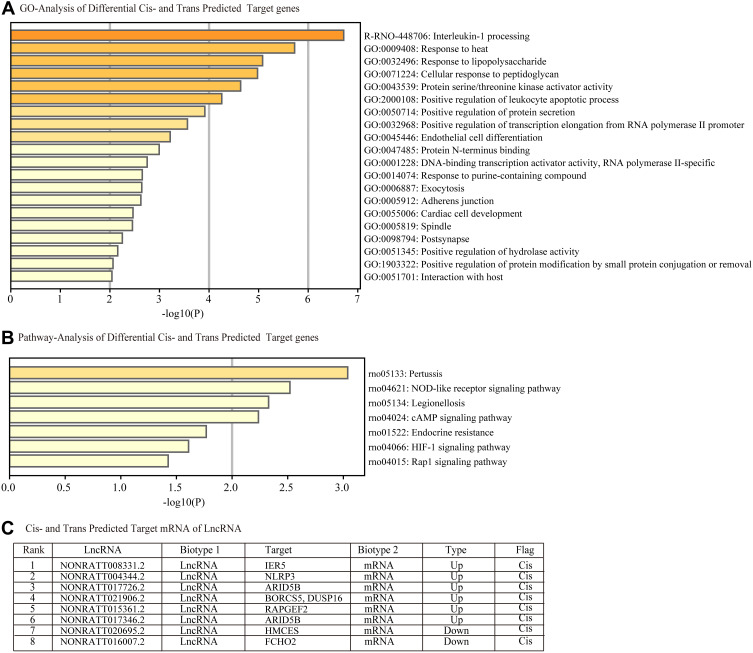
Figure 6A summarizes the predicted cis- and trans-regulated mRNAs of lncRNAs by GO analysis. The top three functional terms are interleukin-1 processing, response to heat, and response to LPS. We found that pertussis, NOD-like receptor signaling pathway, and legionellosis were the common pathways for cis- and trans-regulated mRNAs (Figure 6B) by KEGG analysis. We predicted 7 regulated target genes via cis- and trans- methods in the top 20 lncRNAs (Figure 6C).
Figure 6.
Functional enrichment analysis of differentially expressed predicted cis-and trans-regulated genes of lncRNAs. (A) Cis- and trans- predicted differentially expressed target genes involved in (A) biological processes, cellular components and molecular functions were identified by Gene Ontology (GO) analysis and (B) signaling pathways by Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. (C) Table C lists the cis-and trans- regulated mRNAs of lncRNAs.
Construction of the LncRNA–miRNA-mRNA ceRNA Regulatory Network
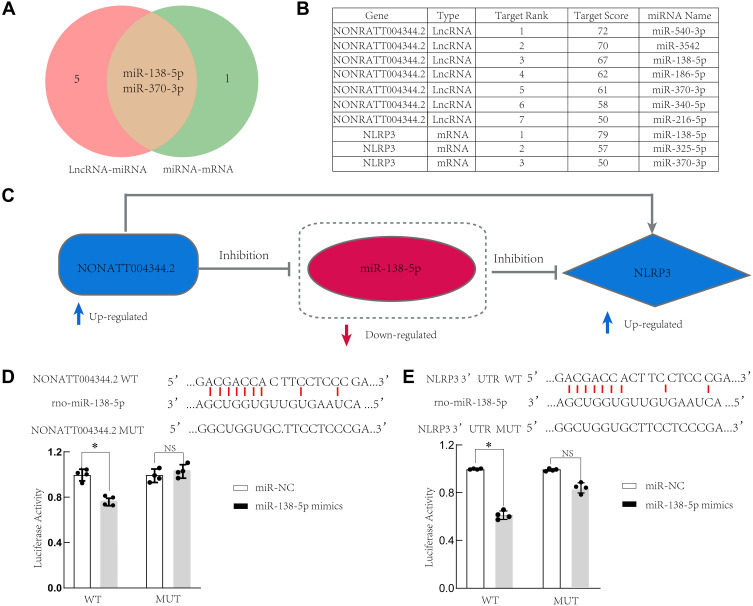
After predicting NLRP3-miRNA and lncRNA4344- miRNA interaction pairs, the pairs were integrated to construct the lncRNA–miRNA-mRNA ceRNA regulatory network. Among the ceRNA relationships, two predicted target miRNAs were identified, namely, rno-miR-138-5p and rno-miR-370-3p (Figure 7A–C). We selected rno-miR-138-5p and validated the lncRNA4344-miR-138-5p-NLRP3 ceRNA network by luciferase assay (Figure 7D and E). We found that lncRNA4344 regulated the expression level of the NLRP3 gene by competitively binding to the same miRNA rno-miR-138-5p. Corresponding regulative relationships are presented in Figure 8. Additionally, to characterize the functions of genes in our ceRNA network, we established the lncRNA4344-miR-138-5p-NLRP3 pathway network.
Figure 7.
LncRNA4344 (NONRATT004344.2), rno-miR-138-5p, and NLRP3 ceRNA network. (A) miRNAs that are interact with both lncRNA4344 and NLRP3 are shown in the Venn diagram. (B) The table shows the details of the miRNAs targeted by both lncRNA4344 and NLRP3 in the miRDB database. (C) The regulatory relationship map among lncRNA4344, miR-138-5p, and NLRP3. (D) A firefly luciferase reporter containing either wild‐type or mutant lncRNA4344 was cotransfected into A549 cells with miR‐NC or miR‐138-5p mimics. (E). A firefly luciferase reporter containing either wild‐type or mutant NLRP3 was cotransfected into A549 cells with miR‐NC or miR‐138-5p mimics. The luciferase activity of miR‐NC‐transfected cells was supposed to be 1. Each experiment was repeated three times. *p < 0.05; NS, no statistically significant difference.
Abbreviations: qRT-PCR, quantitative real-time reverse transcriptase-PCR; NLRP3, Nod-like receptor protein-3; IL‐1β, interleukin (IL)‐1β; IL‐6, interleukin (IL)‐6; IL-18, interleukin (IL)-18; LPS, lipopolysaccharides; TNF-α, tumor necrosis factor-α.
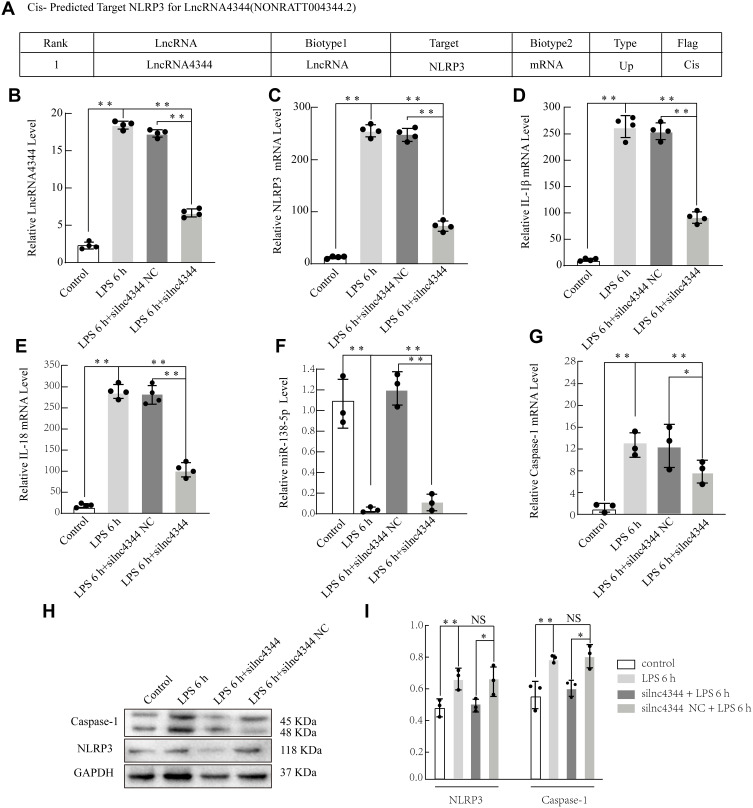
Figure 8.
Effect of lncRNA siRNA on early LPS-induced cytokine production in NR8383 alveolar macrophages (AMs). Information about lncRNA4344 (NONRATT004344.2) and the cis-predicted target gene NLRP3 (A). silnc4344 significantly decreased the expression of lncRNA4344 in NR8383 cells after stimulation with LPS for 6 h (B). siRNA-lnc4344 significantly affected the expression of NLRP3 (C), IL-1β (D), IL-1β (E), miR-138-5p (F), and Caspase-1 (G) in NR8383 cells after LPS stimulation for 6 h. (H and I) Expression levels of NLRP3 and caspase-1 proteins in LPS-induced AMs measured using Western blot analysis. *p < 0.05; **p < 0.01; NS, no statistically significant difference.
Abbreviations: qRT-PCR, quantitative real-time reverse transcriptase-PCR; NLRP3, Nod-like receptor protein-3; IL‐1β, interleukin (IL)‐1β; IL‐6, interleukin (IL)‐6; IL-18, interleukin (IL)-18; LPS, lipopolysaccharides; NLRP3, Nod-like receptor protein-3; TNF-α, tumor necrosis factor-α.
Experimental Validation of Selected LncRNAs and Target Genes
Our study selected lncRNA4344 and its predicted target gene - pyrin domain containing 3 (NLRP3) by bioinformatics to further explore its function (Figure 8A). After silencing lncRNA4344, its expression was significantly compared with that of the control group, and siRNA-lncRNA4344 also significantly decreased the expression of NLRP3, IL-18, Caspase-1, and IL-1β in NR8383 AMs after LPS stimulation for 6 h. In contrast, miR-138-5p was increased (Figure 8B–G). The protein expression levels of NLRP3 and Caspase-1 protein were decreased in siRNA-lncRNA4344 samples pretreated with LPS-induced AMs compared with control AMs (Figure 8H and I).
Discussion
Progress in microarray and RNA sequencing research has allowed us to discover many unknown lncRNAs across many species in the occurrence and development of various diseases.7,9,10 Numerous studies have focused on the potential function of different lncRNAs during the development of ALI. However, kinds of literature regarding possible roles and mechanisms of lncRNAs in early ALI is still restricted. In the present study, we explored the mRNA and lncRNA expression profiles in LPS-induced ALI in the early stage; in total, 355 differentially expressed mRNAs, and 99 lncRNAs were identified via RNA-seq. We analyzed the function of the regulated target genes of the top 20 differentially expressed lncRNAs. An in vitro study confirmed that lncRNAs might function in early ALI treatment by regulating the inflammatory response. Our research found that lncRNAs are involved in the pathophysiological process of early ALI.
Several previous studies have found that various lncRNAs are involved in multiple biological processes of diseases via various regulated mechanisms at different levels, including chromatin remodeling, transcription, posttranscription, protein modification, and epigenetic regulation.19,20 GO analysis suggested that the early differentially expressed mRNAs are mostly associated with the regulation of protein serine/threonine kinase activity, pertussis, and cellular response to LPS. It has been widely accepted that changes in response to external stimuli are involved in early ALI/ARDS; responding to infection and LPS includes various cellular metabolism and structural modifications that coordinate responses to multiple stimuli.21 LncRNAs may play critical roles in epigenetic and transcriptional regulation and have demonstrated great potential as key regulatory molecules of early inflammatory factor expression programs in response to microbial-derived clues.22 Previous studies have shown that lncRNAs regulate the LPS-stimulated inflammatory response in rat lung tissue, and that lncRNAs are essential regulators of the rat innate immune response.15 Our data reveal lncRNAs involved in functional and structural changes in AMs that occur in the early stages of ALI, indicating lncRNA-mediated changes in the LPS-induced ALI model and may provide insights for early intervention of ALI through lncRNAs.
KEGG analysis found that most differentially expressed mRNAs were related to pertussis, osteoclast differentiation, and cAMP signaling pathways. Pertussis has been found to provoke the release of LPS in the external cell wall, thus further mediating the innate immune response and inflammatory process in ALI.23 Osteoclast differentiation leads to the fusion of mononuclear macrophages formed from the differentiation of myeloid progenitor cells into each other and the formation of multinucleated giant cells under LPS stimulation.24 Growing studies have confirmed that the cAMP signaling pathway has a protective effect on the whole pathological progression of LPS-induced ALI.25 These three signaling pathways are involved in the early cellular response to LPS stimulation, which supported our bioinformatics results.
Based on evidence from current research, most lncRNAs perform functions locally to interfere with their neighboring or overlapping genes.26 Thus, the potential cis- or trans-target genes for differentially expressed lncRNAs were explored for the possible significance of investigating these lncRNAs in the lung response to LPS stimulation. Furthermore, GO analysis found that interleukin-1 processing, response to heat, and response to LPS are involved in early ALI pathogenesis, as confirmed by previous publications.27,28 KEGG pathway analysis indicated that trans-and cis- predicted regulated mRNAs are involved in pertussis, the NOD-like receptor signaling pathway, and the legionellosis signaling pathway. Based on previous findings, these pathways are critical for the development of early ALI to external stimulus,29–31 suggesting that altered lncRNAs may play a role in ALI/ARDS through such signaling pathways enriched by cis-or trans-target genes.
Additionally, we performed a CNC and PPI net analysis to investigate lncRNAs and mRNAs’ early ALI influences. We identified three key hub genes, namely, including Tnf, Jun, and Atf3, through the PPI network. Some studies have confirmed that both c-Jun N-terminal kinase (JNK) and nuclear factor kappa B (NF-KB) activation can be induced by upregulated ATF3 expression via the TLR4 signaling pathway.32 The CNC network showed that most lncRNAs are closely related to the functional expression of multiple protein-coding genes via the regulation of different molecules. Altered genes were differentially correlated with the same lncRNAs. Integrated analysis of CNCs, PPIs, and cis- or trans-prediction results, revealed that lncRNA4344 and NLRP3 were positively related to early inflammatory processing factors (IL-18 and IL-1β) in early ALI. The activated NLRP3 releases IL-1β and then mediates neutrophilic airway pyroptosis.33,34 Herein, our data implied that these lncRNAs might play different roles in early ALI immune responses. Accurately, lncRNA4344 and the predicted target gene NLRP3 would promote the onset of inflammatory responses; in contrast, we did not validate those predicted early downregulated lncRNAs by bioinformatics in ALI-related pyroptosis. Our findings indicated that lncRNAs played crucial roles in the pyroptosis response and innate immunity in early LPS-induced ALI. CNC analysis suggests that most lncRNAs are involved in the coexpression of multiple mRNAs and lncRNAs, suggesting various trans- and cis-regulatory mechanisms. Further studies should be conducted to explore the possible molecular mechanisms of these altered lncRNAs.
To further confirm the functional roles and mechanisms of lncRNAs in early ALI, the NLRP3, lncRNA4344, and rno-miR-138-5p networks revealed ceRNA regulatory relationships among the transcripts and non-coding RNAs, which was validated by luciferase assay. Finally, we performed a vitro study to investigate whether interventions targeting lncRNAs play a role in early ALI progression. We used siRNA-lncRNA4344 and observed that interference with lncRNA4344 increased the expression of NLRP3 and LPS-induced IL-18 and IL-1β production, which is considered a critical role in early ALI.35,36 Western Blot results also confirmed the NLRP3 and Caspase-1 expression levels were decreased under siRNA-lncRNA4344 conditions. We proposed that therapy targeting lncRNAs related to ceRNA regulatory mechanisms may provide a different direction for ALI/ARDS treatment. Moreover, more in vivo studies and clinical research should be performed to investigate the therapeutic potential of lncRNAs in ALI/ARDS.
Conclusion
In conclusion, most lncRNAs and mRNAs in AMs were differentially expressed in the early-stage after LPS stimulation. Bioinformatics analyses revealed that different lncRNAs exhibit diverse potential functions that are related to differentially expressed genes. The PPI network analysis suggested that Tnf, Jun, and Atf3 were the top three key genes, and they may represent potential biomarkers or clinical targets for early ALI. The in vitro study suggested that lncRNA4344 sponged rno-miR‐138-5p to promote pyroptosis in the inflammatory responses of LPS‐induced acute lung injury by targeting NLRP3. However, further studies need to be performed to investigate the intergenic regulatory mechanisms and biological functions among NLRP3, lncRNA4344, and rno-miR-138-5p. They could be potential targets for early ALI intervention.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81560306 and 81460292), and the Graduate Student Innovation Special Fund Project of Jiangxi Province (YC2020-B033). No financial relationship between a commercial entity interested in our article and the authors was found. All data generated or analyzed in our study can be obtained from the corresponding author.
Disclosure
The authors declare that they have no competing interests.
References
- 1.Butt Y, Kurdowska A, Allen TC. Acute lung injury: a clinical and molecular review. Arch Pathol Lab Med. 2016;140:345–350. doi: 10.5858/arpa.2015-0519-RA [DOI] [PubMed] [Google Scholar]
- 2.Bellani G, Laffey JG, Pham T, et al. Epidemiology, patterns of care, and mortality for patients with acute respiratory distress syndrome in intensive care units in 50 countries. JAMA. 2016;315:788–800. doi: 10.1001/jama.2016.0291 [DOI] [PubMed] [Google Scholar]
- 3.Confalonieri M, Salton F, Fabiano F. Acute respiratory distress syndrome. Eur Respir Rev. 2017;26:160116. doi: 10.1183/16000617.0116-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howell MD, Davis AM. Management of ARDS in adults. JAMA. 2018;319:711–712. doi: 10.1001/jama.2018.0307 [DOI] [PubMed] [Google Scholar]
- 5.Standiford TJ, Ward PA. Therapeutic targeting of acute lung injury and acute respiratory distress syndrome. Transl Res. 2016;167:183–191. doi: 10.1016/j.trsl.2015.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen LL. Linking long noncoding RNA localization and function. Trends Biochem Sci. 2016;41:761–772. doi: 10.1016/j.tibs.2016.07.003 [DOI] [PubMed] [Google Scholar]
- 7.de Oliveira JC, Oliveira LC, Mathias C, et al. Long non‐coding RNAs in cancer: another layer of complexity. J Gene Med. 2019;21:e3065. doi: 10.1002/jgm.3065 [DOI] [PubMed] [Google Scholar]
- 8.Tang S, Zhou J, Jing H, et al. HanbingWang, functional roles of lncRNAs and its potential mechanisms in neuropathic pain. Clin Epigenetics. 2019;11:78. doi: 10.1186/s13148-019-0671-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma C, Luo H, Liu B, Li F, Tschope C, Fa X. Long noncoding RNAs: a new player in the prevention and treatment of diabetic cardiomyopathy? Diabetes Metab Res Rev. 2018;34:e3056. doi: 10.1002/dmrr.3056 [DOI] [PubMed] [Google Scholar]
- 10.Geng H, Tan X-D. Functional diversity of long non-coding RNAs in immune regulation. Genes Dis. 2016;3:72–81. doi: 10.1016/j.gendis.2016.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bach DH, Lee SK. Long noncoding RNAs in cancer cells. Cancer Lett. 2018;419:152–166. doi: 10.1016/j.canlet.2018.01.053 [DOI] [PubMed] [Google Scholar]
- 12.Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics. 2016;14:42–54. doi: 10.1016/j.gpb.2015.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo D, Liu F, Zhang J, et al. Functional crosstalk between long non-coding RNAs and the NLRP3 inflammasome in the regulation of diseases. Mol Immunol. 2021. doi: 10.1016/j.molimm.2020.12.038 [DOI] [PubMed] [Google Scholar]
- 14.Dai L, Zhang G, Cheng Z, et al. Knockdown of LncRNA MALAT1 contributes to the suppression of inflammatory responses by up-regulating miR-146a in LPS-induced acute lung injury. Connect Tissue Res. 2018;59:581–592. doi: 10.1080/03008207.2018.1439480 [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Shen YC, Chen ZN, et al. Microarray profiling of lung long non-coding RNAs and mRNAs in lipopolysaccharide-induced acute lung injury mouse model. Biosci Rep. 2019;39:1–12. doi: 10.1042/BSR20181634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Obaid M, Udden SN, Deb P, Shihabeddin N, Zaki MH, Mandal SS. LncRNA HOTAIR regulates lipopolysaccharide-induced cytokine expression and inflammatory response in macrophages. Sci Rep. 2018;8:1–18. doi: 10.1038/s41598-018-33722-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forsgren E, Locke B, Semberg E, Laugen AT, de Miranda JR. Sample preservation, transport and processing strategies for honeybee RNA extraction: influence on RNA yield, quality, target quantification and data normalization. J Virol Methods. 2017;246:81–89. doi: 10.1016/j.jviromet.2017.04.010 [DOI] [PubMed] [Google Scholar]
- 18.Pacholewska A, Sung M-H. lncRNA expression predicts mRNA abundance. Epigenomics. 2019;11:1121–1128. doi: 10.2217/epi-2019-0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gheitasi R, Manoochehri H, Gharib D, Dalil N, Gharib A. LncRNAs: a new trend in molecular biology of diseases; A review. Chronic Dis J. 2019;7:195–206. doi: 10.22122/cdj.v7i3.418 [DOI] [Google Scholar]
- 20.Yao R-W, Wang Y, Chen -L-L. Cellular functions of long noncoding RNAs. Nat Cell Biol. 2019;21:542–551. doi: 10.1038/s41556-019-0311-8 [DOI] [PubMed] [Google Scholar]
- 21.Hu Q, Hao C, Tang S. From sepsis to acute respiratory distress syndrome (ARDS): emerging preventive strategies based on molecular and genetic researches. Biosci Rep. 2020;40. doi: 10.1177/1753465815585716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scacalossi KR, van Solingen C, Moore KJ. Long non-coding RNAs regulating macrophage functions in homeostasis and disease. Vasc Pharmacol. 2019;114:122–130. doi: 10.1016/j.vph.2018.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Summers C, Rankin SM, Condliffe AM, Singh N, Peters AM, Chilvers ER. Neutrophil kinetics in health and disease. Trends Immunol. 2010;31:318–324. doi: 10.1016/j.it.2010.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soysa NS, Neil A, Aoki K, Ohya K. Osteoclast formation and differentiation: an overview. J Med Dent Sci. 2012;59:65–74. doi: 10.11480/jmds.590301 [DOI] [PubMed] [Google Scholar]
- 25.Wang X-F, Li Y-J, Qiang Hu Z, et al. Protective effect of quercetin in LPS-induced murine acute lung injury mediated by cAMP-Epac pathway. Inflammation. 2018;41:1093–1103. doi: 10.1128/IAI.01346-09 [DOI] [PubMed] [Google Scholar]
- 26.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10 [DOI] [PubMed] [Google Scholar]
- 27.Lutay N, Ambite I, Hernandez JG, et al. Bacterial control of host gene expression through RNA polymerase II. J Clin Invest. 2013;123:2366–2379. doi: 10.1172/JCI66451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhatia M, Zemans RL, Jeyaseelan S. Role of chemokines in the pathogenesis of acute lung injury. Am J Respir Cell Mol Biol. 2012;46:566–572. doi: 10.1165/rcmb.2011-0392TR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arpaia N, Green JA, Moltedo B, et al. A distinct function of regulatory T cells in tissue protection. Cell. 2015;162:1078–1089. doi: 10.1016/j.cell.2015.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mata AF, Sarnaik AA. Bronchoscopy with N-acetylcysteine lavage in severe respiratory failure from pertussis infection. Pediatrics. 2013;132:e1418–e1423. doi: 10.1542/peds.2013-0912 [DOI] [PubMed] [Google Scholar]
- 31.Mukherjee T, Hovingh ES, Foerster EG, Abdel-Nour M, Philpott DJ, Girardin SE. NOD1 and NOD2 in inflammation, immunity and disease. Arch Biochem Biophys. 2019;670:69–81. doi: 10.1542/peds.2013-0912 [DOI] [PubMed] [Google Scholar]
- 32.Eunyoung K, Young SH, Jooyoung K, et al. ATF3 plays a key role in Kdo2-lipid A-induced TLR4-dependent gene expression via NF-κB activation. PLoS One. 2010;5:e14181. doi: 10.1371/journal.pone.0014181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong Y, Lu Y, Yang X, et al. The roles of NLRP3 inflammasome in bacterial infection. Mol Immunol. 2020;122:80–88. doi: 10.1016/j.molimm.2020.03.020 [DOI] [PubMed] [Google Scholar]
- 34.Alarcón MML, Ruocco JF, Ferreira F, et al. Toll-like receptor 4 and NLRP3 caspase 1- interleukin-1β-axis are not involved in colon ascendens stent peritonitis-associated heart disease. Shock. 2018;50:483–492. doi: 10.1097/SHK.0000000000001059 [DOI] [PubMed] [Google Scholar]
- 35.Jia X, Cao B, An Y, Zhang X, Wang C. Rapamycin ameliorates lipopolysaccharide-induced acute lung injury by inhibiting IL-1β and IL-18 production. Int Immunopharmacol. 2019;67:211–219. doi: 10.1016/j.intimp.2018.12.017 [DOI] [PubMed] [Google Scholar]
- 36.Cao F, Tian X, Li Z, et al. Suppression of NLRP3 inflammasome by erythropoietin via the EPOR/JAK2/STAT3 pathway contributes to attenuation of acute lung injury in mice. Front Pharmacol. 2020;11:306. doi: 10.3389/fphar.2020.00306 [DOI] [PMC free article] [PubMed] [Google Scholar]