Abstract
Rapid wound detection by distant leukocytes is essential for antimicrobial defense and post-infection survival 1. The reactive oxygen species hydrogen peroxide and the polyunsaturated fatty acid arachidonic acid are among the earliest known mediators of this process 2-4. It is unknown whether or how these highly conserved cues collaborate to achieve wound detection over distances of several hundreds of microns within a few minutes. To investigate this, we locally applied arachidonic acid and skin permeable peroxide by micropipette perfusion to unwounded zebrafish tail fins. As in wounds, arachidonic acid rapidly attracted leukocytes through dual oxidase (Duox) and 5-lipoxygenase (Alox5a). Peroxide promoted chemotaxis to arachidonic acid without being chemotactic on its own. Intravital biosensor imaging showed that wound peroxide and arachidonic acid converged on half-millimeter long lipid peroxidation gradients that promoted leukocyte attraction. Our data suggest that lipid peroxidation functions as spatial redox relay that enables long-range detection of early wound cues by immune cells, outlining a beneficial role for this otherwise toxic process.
Hydrogen peroxide (H2O2) is a relatively stable reactive oxygen species (ROS) produced by NADPH oxidase or mitochondria in cells. At wounds, it rapidly forms concentration gradients proposed to provide paracrine regulation of wound healing, inflammation, and regeneration throughout phylae 2,5. H2O2 can bias immune cell motility in vitro 3,6, and promote leukocyte recruitment to injury sites, at least in part through redox-activation of Lyn 3,7. Quantitative analysis of wound peroxide gradients in live zebrafish, however, suggests that these patterns do not reach far enough to directly alter redox signaling in cells farther than ~30 μm from the wound margin 8. Notably, in the same model, leukocytes are rapidly summoned from distances of up to ~ 300 μm. Thus, some relay mechanism must exist that extends the chemotactic signaling range of the wound peroxide gradient by roughly an order of magnitude.
Together with peroxide, arachidonic acid (AA) is locally released at wounds by a mechanosensitive phospholipase (cPLA2) in response to calcium signals and nuclear envelope stretch 4,9. Whereas conventional chemotaxis receptors for peroxide remain elusive, AA fuels the eicosanoid cascade, a metabolic pathway that oxidizes AA into potent lipid chemoattractants, such as the G-protein coupled receptor (GPCR) ligands leukotriene B4, and 5-oxoETE. Bathing wounded zebrafish larvae in exogenous AA, when wound-induced AA release is osmotically prevented, reconstitutes rapid leukocyte recruitment to injury sites 4. Yet, like for wound peroxide, it is unclear whether AA just amplifies chemotaxis to other wound signals, for example by promoting leukotriene-mediated relay signaling between leukocytes 10, or whether it can act as primary wound chemoattractant itself.
Zebrafish larvae are a powerful model for innate immune responses 11. Their tail fin tissue is not vascularized; hence, interstitial and vascular damage detection mechanisms can be separately studied. In healthy, unwounded tail fins, neutrophils are largely confined to the caudal hematopoietic region. Thus, fin injury can be applied at secure distance (100-300 μm) from a leukocyte, minimizing the risk of inadvertently injuring and activating it.
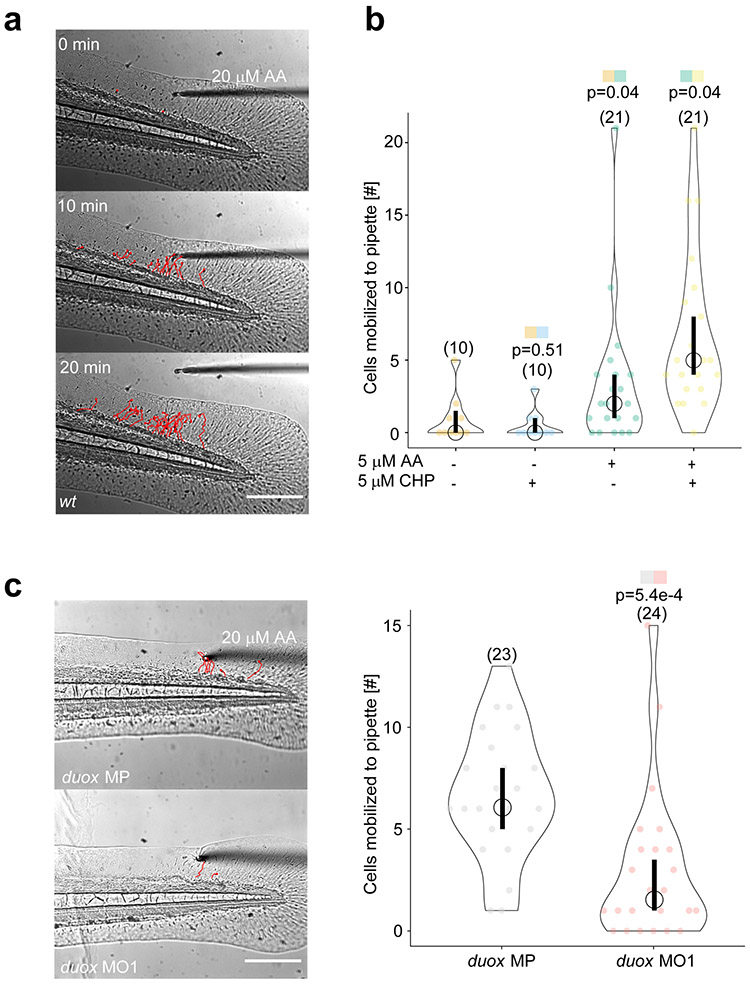
To test whether local AA- or peroxide-application suffices to attract leukocytes in vivo, we patched a custom forged, polished micropipette (tip diameter ~ 10-20 μm) filled with AA onto unwounded zebrafish tail fins using mild, non-damaging suction applied by a syringe. We then monitored rapid leukocyte responses by transmitted light imaging (Figure 1a, Supplementary Video 1). Native AA is not a leukocyte chemoattractant, and intact larval tail fins typically lack interstitial leukocytes that could oxidize AA into leukotrienes. However, AA microperfusion caused massive leukocyte recruitment to the pipette tip at concentrations ≥ 5 μM. Like a wound, microperfused AA mobilized leukocytes from up to ~300 μm towards the signal source within ~10 min. Different commercial AA preparations gave similar responses (Extended Data Figure 1a). The number of responsive leukocytes depended on AA dose: 5 μM AA (Figure 1b) mobilized about half as many leukocytes as 20 μM (Figure 1c). These concentrations are within the range of free AA measured in normal or inflamed human skin 12. After detachment of the pipette, leukocytes immediately returned to the vascular region (Supplementary Video 1), reminiscent of reverse leukocyte migration previously described for zebrafish wounds 13. Next, we tested whether oxidative stress alone could initiate leukocyte recruitment. At zebrafish wounds, the epithelial NADPH oxidase Duox generates extracellular/luminal H2O2 by consuming cytoplasmic NADPH 2. As H2O2 does not penetrate intact larval skin 14, we instead microperfused cumene hydroperoxide (CHP), a lipophilic peroxide often used to trigger lipid peroxidation. CHP treatment elevated tissue H2O2 levels (Extended Data Figure 1b). Although CHP was not chemotactic on its own, it augmented leukocyte mobilization to AA (Figure 1b). Conversely, local Duox inhibition by co-perfusion of the flavoprotein inhibitor diphenyleneiodonium (DPI), or global duox knockdown with an established antisense morpholino 2,3 suppressed directional migration or responsiveness of leukocytes, respectively, towards microperfused AA (Figure 1c, Extended Data Figure 4, Supplementary Video 2) as previously observed for wounds 2. At zebrafish wounds, AA metabolites hence act as primary, redox-regulated attractants, transcending their known intermediary role in relay chemotaxis and leukocyte swarming.
Figure 1. Oxidative stress enhances wound detection through the arachidonic acid pathway.
(a) Rapid leukocyte migration to 20 μM arachidonic acid (AA) released by micropipette patched onto an intact, larval zebrafish tail fin. Note, leukocytes immediately turn back to the vasculature after the pipette is removed. Red, leukocyte tracks at indicated times after patching. Representative of 3 biologically independent animal experiments. (b) Intact fins perfused with dimethyl sulfoxide (DMSO) carrier control, 5 μM cumene hydroperoxide (CHP), 5 μM AA alone or in combination with CHP. Violin plot depicting the number of leukocytes per animal mobilized towards the pipette tip within 20 min after patching. Data aggregated from n=10 (AA−/CHP−), n=10 (AA−/CHP+), n=21 (AA+/CHP−), and n=21 (AA+/CHP+) biologically independent animal experiments. P-value, two-sided, heteroscedastic Student’s t-test for pairwise comparison of the indicated color-coded samples. Circle, median. Bar, 95% confidence interval. (c) Intact fins of control (duox MP) and duox morphant (duox MO1) larvae perfused with 20 μM AA. Left panel: Representative images of control (duox MP) or duox morphant (duox MO1) tail fins at 20 min after patching. Red, leukocyte tracks. Right panel: Violin plot depicting the number of leukocytes per animal mobilized towards the pipette tip within 20 min after patching. Circle, median. Bar, 95% confidence interval. Data aggregated from n=23 (duox MP), and n=24 (duox MO1) biologically independent animal experiments. P-value, two-sided, heteroscedastic Student’s t-test for pairwise comparison of the indicated color-coded samples. Parentheses, number of different animals. Scale bars, 200 μm. Statistical source data can be found at Source data figure 1.
The synthesis of AA-derived leukocyte chemoattractants is initiated by stereospecific peroxidation of AA by 5- and 12-lipoxygenase into 5(S)- and 12(S)-hydroperoxyeicosatetraenoic acid (HpETE), respectively. Lipoxygenases are intrinsically redox-sensitive: Their catalytic iron must be oxidized from ferrous (Fe2+) to ferric (Fe3+) by long chain fatty acid hydroperoxides 15,16, which may be generated by lipoxygenases or non-enzymatic lipid peroxidation. Reduction of H2O2 or other organic peroxides by Fe2+ provides the radicals (HO∙, etc.) that start and propel lipid autooxidation. Autooxidation continuously generates lipid peroxides that, with Fe2+, form new radicals and can activate lipoxygenases. If the process slips out of control, this chain reaction can trigger ferroptotic cell death 17. Ferroptosis can move through multicellular systems as a wave in vitro and ex vivo 18-20. Intriguingly, in two out of our ~200 micropipette experiments (i.e., ~1%), we observed a reminiscent in vivo phenomenon: A rapid wave of tissue deformation (Extended Data Figure 1c, Supplementary Video 3) surged out from the AA patch site. Its transmitted light features (e.g., tissue swelling/collapse, cell opaqueness at patch site) at least seemed consistent with some form of necrotic tissue degeneration. Peroxidation of polyunsaturated fatty acids (PUFAs), such as AA, can drive ferroptosis 21. It is intriguing to speculate that ferroptotic ‘death waves’, as those reported in Riegman et al.20 in this issue, can occur in vivo, perhaps, when antioxidant depletion coincides with the availability of reactive iron and a ferroptotic driver PUFA, such as AA. More work is needed to test this idea.
Interestingly, it was shown that α-tocopherol depletion of zebrafish larvae promotes the formation of the 5-lipoxygenase metabolite 5-HETE 22, suggesting that lipid peroxidation regulates this enzyme in vivo. But the physiological purpose, if any, of redox-sensing by 5-lipoxygenase has remained elusive.
We probed for lipid peroxidation at wounds by ratiometric C11-BODIPY581/591 (C11B)-imaging. C11B is a fluorescent fatty acid analog with redox sensitive double bonds. When those double bonds react with lipid radicals, C11B shifts its fluorescence emission from red to green 23. Hence, it acts as fluorescent proxy for radical mediated PUFA oxidation during lipid peroxidation. Its ratiometric readout allows normalization of sensor signals to local sensor concentration. We imaged lipid peroxidation in live larvae ~15-30 min after tail fin tip amputation, that is, when the wound-induced H2O2 gradient is at peak amplitude 2,8. For better comparison, we plotted the C11B green/red pixel ratios against wound distance (Wound Ratio) after internal normalization by the median baseline ratio (Base Ratio, insets). The Wound Ratio highlights wound-induced, spatial changes in lipid peroxidation, whereas the Base Ratio reports on global lipid peroxidation shifts.
C11B-imaging of live zebrafish larvae revealed extensive lipid peroxidation gradients extending ~ half a millimeter from the wound margin into the unwounded tissue (Figure 2a, b). Like the H2O2 gradient 2, and rapid leukocyte recruitment to AA and wounds, these patterns were inhibited by DPI and duox knockdown (Figure 2a, Extended Data Figure 1d, e). Unlike the wound H2O2 gradient 4,8, but just like leukocyte recruitment 4, lipid peroxidation gradients were completely suppressed by bath isotonicity (Figure 2b), which prevents osmotic release of AA, and perhaps other PUFAs, by blocking mechanosensitive phospholipases, such as cPla2 4,9. Conversely, bath application of unsaturated fatty acids to unwounded larvae induced lipid peroxidation (Figure 2c). The PUFAs linoleic (18:2), linolenic (18:3), and arachidonic acid (20:4) caused more lipid peroxidation than single-unsaturated oleic acid (18:1). Saturated arachidic acid (20:0) induced no lipid peroxidation at all. Unsaturated fatty acids can activate NADPH oxidases 24. Thereby, AA alone (e.g., patched onto intact fins) seems to generate enough oxidative stress to drive its chemotactic conversion, even in the absence of other, wound-derived ROS sources. Linoleic acid, which causes as much lipid peroxidation as AA, caused little leukocyte responses (Figure 2d, Extended Data Figure 4; Supplementary Video 4). Again, this underlines that oxidative stress, at least at non-cytotoxic doses, ineffectively initiates inflammatory responses on its own. Instead, it acts through the AA pathway.
Figure 2. Lipid peroxidation gradients integrate oxidative and osmotic wound cues.
(a) C11B-imaging of control (duox MP) and duox knockdown (duox MO1) zebrafish larvae ~15-30 min after tail fin tip amputation. Upper panel: Representative C11B ratio images. Lower panel: Wound ratio, mean C11B ratio normalized to median baseline ratio ± SEM plotted against wound distance. Inset, boxplots of baseline ratios normalized to control. Central mark, median. Box edges, 25th/75th percentiles. Whiskers, most extreme, non-outlier data points. +, outliers. Data aggregated from n=59 (duox MP), and n=45 (duox MO1) biologically independent animal experiments. (b) C11B-imaging of zebrafish larvae ~15-30 min after tail fin tip amputation in hypotonic (Hypo) or isotonic (Iso: Hypo +135 mM NaCl) bathing solution. Upper panel: Representative C11B ratio images. Lower panel: Wound ratio, mean C11B ratio normalized to median baseline ratio ± SEM plotted against wound distance. Inset, boxplots of baseline ratios normalized to control. Central mark, median. Box edges, 25th/75th percentiles. Whiskers, most extreme, non-outlier data points. +, outliers. Data aggregated from n=19 (Hypo), and n=19 (Iso) biologically independent animal experiments. (c) Violin plot of C11B ratios in unwounded tail fins ~60 min after exposure to 20 μM of the indicated free fatty acids or DMSO carrier control. Circle, median. Bar, 95% confidence interval. P-value, two-sided, heteroscedastic Student’s t-test for pairwise comparison of the indicated color-coded samples. Data aggregated from n=15 (Control), n=19 (Arachidonic), n=18 (Arachidic), n=17 (Linoleic), n=18 (Linolenic), and n=18 (Oleic) biologically independent animal experiments. (d) Left panel: Representative images of wild type (wt) tail fins patched with 20 μM arachidonic (AA) or linoleic (Lin) acid at 20 min after patching. Red, leukocyte tracks. Right panel: Violin plot depicting the number of leukocytes per animal mobilized towards the pipette tip within 20 min after patching. Data aggregated from n=10 (Arachidonic), and n=10 (Linoleic) biologically independent animal experiments. P-value, two-sided, heteroscedastic Student’s t-test for pairwise comparison of the indicated color-coded samples. Circle, median. Bar, 95% confidence interval. Parentheses, number of different animals. Scale bars, 200 μm. Statistical source data can be found at Source data figure 2.
In the presence of excess thiols (e.g., in vivo), the selenium-containing compound ebselen (Ebs) acts as glutathione peroxidase mimetic. It antagonizes lipid peroxidation by reducing lipid hydroperoxides to lipid alcohols as opposed to radical scavenging 25. Like duox knockdown, ebselen attenuated the lipid peroxidation gradient with little effect on baseline peroxidation levels (Figure 3a). In contrast, the lipid radical scavenger and ferroptosis inhibitor liproxstatin-1 (Lpx) 26 primarily affected baseline lipid peroxidation levels (Figure 3b). This indicates that wound-induced peroxides initialize the gradient, and lipid radicals isometrically amplify the prepattern through perpetuating lipid autooxidation. Ebselen, liproxstatin-1, and the lipid radical scavenger α-tocopherol inhibited rapid leukocyte recruitment to wounds, suggesting that lipid peroxidation promotes wound detection (Figure 3c, d; Supplementary Video 5, 6).
Figure 3. Inhibiting lipid peroxidation inhibits wound detection.
(a) C11B-imaging of control larvae and larvae treated with the GPX mimetic ebselen (Ebs, 3 μM) ~15-30 min after tail fin tip amputation. Upper panel: Representative C11B ratio images. Lower panel: Wound ratio, mean C11B ratio normalized to median baseline ratio ± SEM plotted against wound distance. Inset, boxplots of baseline ratios normalized to control. Central mark, median. Box edges, 25th/75th percentiles. Whiskers, most extreme, non-outlier data points. +, outliers. Data aggregated from n=17 (Ctr), and n=20 (Ebs) biologically independent animal experiments. (b) C11B-imaging of control larvae and larvae treated with the lipid radical scavenger liproxstatin-1 (Lpx, 20 μM) ~15-30 min after tail fin tip amputation. Upper panel: Representative C11B ratio images. Lower panel: Wound ratio, mean C11B ratio normalized to median baseline ratio ± SEM plotted against wound distance. Inset, boxplots of baseline ratios normalized to control. Central mark, median. Box edges, 25th/75th percentiles. Whiskers, most extreme, non-outlier data points. +, outliers. Data aggregated from n=17 (Ctr), and n=12 (Lpx) biologically independent animal experiments. (c) Left panel: Representative images of wounded tail fins treated with (Ebs, 3 μM) or DMSO control (Ctr) at 40 min after injury. Red, leukocyte tracks. Right panel: Violin plot depicting the number of leukocytes per animal recruited to a tail fin wound within 40 min after injury. Circles, median. Bars, 95% confidence intervals. Data aggregated from n=55 (Ctr), and n=39 (Ebs) biologically independent animal experiments. P-value, two-sided, heteroscedastic Student’s t-test for pairwise comparison of the indicated color-coded samples. (d) Left panel: Representative images of wounded fish treated with liproxstatin-1 (Lpx, 20 μM), or α-tocopherol (αTOC, 100 μM), or DMSO control (Ctr) at 40 min after injury. Red, leukocyte tracks. Right panel: Violin plot depicting the number of leukocytes per animal recruited to a tail fin wound within 40 min after injury. Circles, median. Bars, 95% confidence intervals. Data aggregated from n=118 (Ctr), n=135 (Lpx), and n=33 (αTOC) biologically independent animal experiments. P-value, two-sided, heteroscedastic Student’s t-test for pairwise comparison of the indicated color-coded samples. Parentheses, number of different animals. Scale bars, 200 μm. Statistical source data can be found at Source data figure 3.
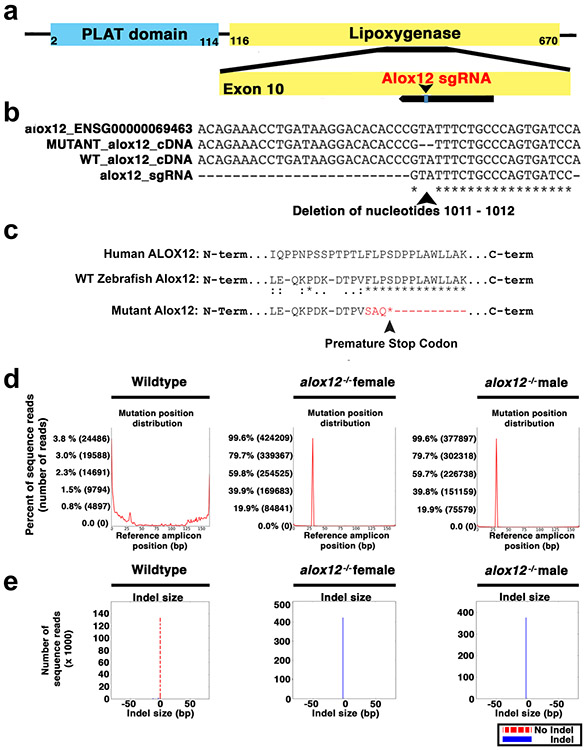
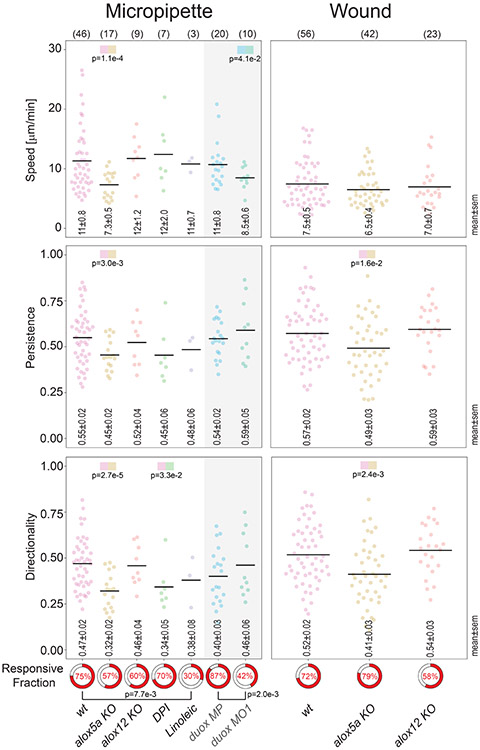
As discussed above, lipid peroxidation may exert its effects through redox regulation of lipoxygenases. Judging from mRNA and their (HETE-)product levels, alox12 is the predominant lipoxygenase in zebrafish larvae, followed by alox5a 22,27. Like their mammalian orthologs, zebrafish 5- and 12-lipoxygenases (Alox5a, Alox12) generate 5(S)-HpETE and 12(S)-HpETE, respectively 27,28. To test which, if any, of these enzymes is involved in wound detection, we generated CRISPR/Cas9 frameshift KO mutants for both (Extended Data Figure 2, 3). Homozygous mutants grew to larval and adult stages without obvious developmental phenotypes. Leukocyte migration to microperfused AA and wounds, respectively, was strongly (72% and 61% by average, see Source Data for the respective figures) inhibited in alox5a mutant larvae, while overall leukocyte abundance in the tail fin was largely unaffected by the mutation (Figure 4a, b; Supplementary Video 7, 8; Extended Data Figure 3e). Alox5a KO affected speed, persistence and directionality of leukocyte recruitment (Extended Data Figure 4). By contrast, Alox12 mutant larvae showed only slightly (22% and 17% by average) attenuated leukocyte responses to microperfused AA or wounds, respectively. Relative to H2O2 inhibition by duox knockdown (Figure 2a), lipoxygenase KO had relatively little effect on lipid peroxidation gradients (Figure 4c), suggesting that their pre-patterning occurs upstream of lipoxygenases and downstream of Duox. As compared to liproxstatin-1 (Figure 3b), alox12 mutants only showed a moderate reduction of baseline lipid peroxidation (69% vs. 29% by average). Together with the previously noticed redox-sensitivity of 5-lipoxygenase product formation in vitro and in vivo 15,16,22, the above data collectively argue that lipid peroxidation gradients instruct wound detection through redox-sensing by the 5-lipoxygenase arm of the eicosanoid cascade. In humans, co-expression of cPLA2, ALOX5, and DUOX1 occurs in the mucosal linings of the lower urinary tract 29, raising the possibility that this damage detection pathway is evolutionary conserved.
Figure 4. Arachidonic acid initiates wound detection through 5-lipoxygenase.
(a) Left panel: Representative images of wild type (wt), alox12 KO, and alox5a KO tail fins patched with 20 μM AA at 20 min after patching. Right panel: Violin plot depicting number of leukocytes per animal mobilized towards the pipette tip within 20 min for each genotype. Circles, median. Bars, 95% confidence intervals. Data aggregated from n=42 (wt), n=16 (alox12 KO), and n=31 (alox5a KO) biologically independent animal experiments. P-value, two-sided, heteroscedastic Student’s t-test for pairwise comparison of the indicated color-coded samples. (b) Left panel: Representative images of wounded wt, alox12 KO, and alox5a KO tail fins at 40 min after injury. Right panel: Violin plot depicting number of leukocytes per animal recruited towards the tail fin wound within 40 min after injury for each genotype. Data aggregated from n=246 (wt), n=192 (alox12 KO), and n=71 (alox5a KO) biologically independent animal experiments. P-value, two-sided, heteroscedastic Student’s t-test for pairwise comparison of the indicated color-coded samples. Circles, median. Bars, 95% confidence interval. (c) C11B ratios of wt, alox12 KO, and alox5a KO larvae ~15-30 min after tail fin amputation. Wound ratio, mean C11B ratio normalized to median baseline ratio ± SEM plotted against wound distance. Inset, boxplots of baseline ratios normalized to control. Central mark, median. Box edges, 25th/75th percentiles. Whiskers, most extreme, non-outlier data points. +, outliers. Data aggregated from n=149 (wt), n=77 (alox12 KO), and n=74 (alox5a KO) biologically independent animal experiments. (d) Superposition of the calibrated wound H2O2 gradient (blue, published in Figure 3D and 4 of Jelcic & Enyedi, 2017 8) and the lipid peroxidation gradient (red, duox MP, Figure 2a) at ~ 15-30 min after tail fin tip amputation. Data presented as mean values ± SEM. Parentheses, number of different animals. Scale bars, 200 μm. Statistical source data can be found at Source data figure 4.
It is conceivable that redox-sensitive wound chemoattractant production acts in concert with redox switches in leukocytes, such as Lyn 3,7, to first initiate and later amplify rapid chemotaxis to zebrafish wounds. We cannot exclude that additional redox-sensitivity exists downstream of 5-lipoxygenase. For example, synthesis of the chemotactic AA metabolite 5-oxoETE, which we previously implicated in the early wound response 4, is putatively driven by the hypothetical enzyme 5-HEDH, which requires NADP+ as cofactor 30. Testing the contribution of 5-HEDH to wound redox sensing must await its genetic identification. Regardless, our work identifies eicosanoid metabolism as instructive redox sensor of wound detection with AA functioning akin to an alarmin 1. Through redox activation of Alox5a, lipid peroxidation gradients may impinge on long-range production gradients of lipid chemoattractants (e.g., of 5-oxoETE4) that directly guide distant leukocytes to wounds via conventional GPCR-mediated signaling. Likewise, there could be local Alox5a activation at the wound site and subsequent long-range diffusion of lipid chemoattractants, or any mixture of the above mechanisms, which may complement each other.
Currently, ROS signaling is mostly attributed to reversible cysteine oxidation of kinases and phosphatases, although physiologic roles for lipid peroxidation are starting to emerge 31. In zebrafish larvae, cytoplasmic antioxidants are predicted to confine H2O2-mediated cysteine oxidation to the immediate wound margin vicinity (<30 μm) 8. Thus, it is difficult to envision how wound margin H2O2 can directly affect redox signaling in leukocytes 100-300 μm away. But lipid peroxidation gradients reach much deeper into the tissue (Figure 4d) and hence can act as tissue scale relay—potentially for both, inflammatory cues and cell death (Extended Data Figure 5).
Methods
General fish procedures
Adult wild type and mutant Casper 32 zebrafish, and larvae were maintained as previously described 33 and according to the institutional animal healthcare guidelines with the approval of the Memorial Sloan Kettering Cancer Center (MSKCC) Institutional Animal Care and Use Committee. For experiments, 2.5-4 days post-fertilization (2.5-4 dpf) larvae were anaesthetized in E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) containing 0.2 mg/ml ethyl 3-aminobenzoate methanesulfonate (Sigma, E10521). Sex was indeterminate at the stages of development that were used for the experiments in this paper.
Imaging leukocyte responses to wounds and micropipettes
For imaging leukocyte responses to wounds or micropipettes, anesthetized 2.5-4 dpf larvae were embedded in ~0.5 ml 1% isotonic low-melting agarose (Goldbio, A-204-100) in a glass-bottom dish (Matek Corporation, P35G-1.5-20C). For wounding experiments, the caudal tail fin was incised with a tungsten needle (Fine Science Tools, 10130-20) during agarose solidification. Transmitted light images were acquired at room temperature (~25 °C) using NIS-Elements (Nikon, 64bit 3.22.15 Build 738) on a Nikon Eclipse Ti microscope equipped with a 10x, plan-apochromat NA 0.45 air objective lens, and an Andor Clara CCD camera. Multiple larvae were imaged in parallel using a motorized stage.
In some cases, leukocyte recruitment after pharmacologic treatment was assessed. To this end, larvae were preincubated with 3 μM ebselen (Sigma, E3520), 20 μM liproxstatin-1 (Sigma, SML1414), or 100 μM α-tocopherol (Sigma, 258024) in E3 for 30-60 min by bathing at 28°C. Compound concentrations were maintained throughout the entire wounding/imaging experiment.
For micropipette experiments, the agarose covering the tail fin was removed to allow tail fin-patching. Glass capillaries (World Precision Instruments, TW100-3) were cut, polished, and bent (~30°) using a microforge (Narishige, MF-900) to yield a tip diameter of ~10-20 μm. The micropipettes were filled with solutions using a syringe needle (World Precision Instruments, MF28G-5) and connected to a plastic syringe filled with the same solution. The following solutions were used: 5 or 20 μM arachidonic acid (Sigma, A3611 or Cayman, 90010), 20 μM linoleic acid (Sigma, L1376) in E3 medium, either alone or in combination with 5 μM cumene hydroperoxide (CHP; Invitrogen, included in Image-iT Lipid Peroxidation Kit, C10445), or 50 μM diphenyleneiodonium chloride (DPI; Sigma, D2926). The micropipette was mounted onto the microscope using course-, and fine-micromanipulators (Narishige, MN-4 & MMO-4), and lowered onto the caudal tail fin for attachment via mild negative pressure manually applied with the syringe. Transmitted light images were acquired every 15-30s for 20 min at room temperature as described above. Leukocytes that approaches the pipette tip within 20 min of patching were counted as “Cells mobilized to pipette”.
Image-based measurement of lipid peroxidation
Anesthetized 2.5-3 dpf larvae were incubated in a solution of 10 μM BODIPY® 581/591 C11 (C11B; Invitrogen, Image-iT Lipid Peroxidation Kit, C10445) in E3 medium for 30 min at 28°C. Larvae were transferred to a glass bottom dish, and tail fin tips were amputated using a needle blade (Fine Science Tools, 10318-14) in the same C11B solution. Larvae were incubated another 15 min at 28°C, and then washed three times with E3. After mounting them in 1% low melting agarose, C11B fluorescence was excited with an LED light source (LumenCor 7, Light Engine) with internal 549/15 and 475/28 bandpass filters, respectively, combined with a multispectral dichroic filter (Chroma, 89021bs). Emission of reduced C11B was sampled with a 632/60 bandpass filter (Chroma, ET632/60m), and emission of oxidized C11B was sampled with a 525/50 bandpass filter (Chroma, 525/50m). Transmitted light and C11B images were captured at room temperature using NIS-Elements AR on a Nikon Eclipse Ti microscope equipped with a 10x, plan-apochromat NA 0.45 air objective lens, and an Andor Clara CCD camera.
In some cases, the impact of pharmacologic treatments on lipid peroxidation was assessed. To this end, larvae were co-incubated with 3 μM ebselen (Sigma, E3520) or 20 μM liproxstatin-1 (Sigma, SML1414), or 20-100 μM DPI together with 10 μM C11B in E3 medium for 30-60 min at 28°C. C11B was washed off, but compound concentrations were maintained throughout the imaging experiment.
Image analysis was primarily conducted in MATLAB (MathWorks, R2019b) using the custom scripts/functions ND2ratio, ND2SUM, NormalizeWoundProfiles (https://github.com/niethamp/KatikaneniAndJelcic2020). Briefly, ND2-files were imported into MATLAB using the Bioformats Toolbox for MATLAB (v5.9, OME) function bfGetReader. In the transmitted light image, the wound margin and the notochord region were manually marked for calculating wound distances of pixels and for masking out the thick notochord region for quantitative C11B ratio analysis. Images were smoothed using a gaussian filter if necessary (e.g., at low camera binning). Modal background fluorescence was subtracted from the smoothed image. Green fluorescence emission was divided by red fluorescence emission. Thus, C11B ratios increase with lipid peroxidation. C11B ratios were calculated pixel-by-pixel together with their Euclidian distance from the wound margin. The median foreground C11B pixel ratios at 650-750 μm from the wound margin were defined as baseline C11B ratio. All pixels of an animal (excluding the notochord region) were divided by this baseline ratio for normalization and plotted against their wound distance to yield the depicted normalized Wound ratio profile. The baseline ratios (insets) for each experimental group were normalized by the median baseline ratio of all control animals.
For measuring fatty acid-induced lipid peroxidation, uninjured and anesthetized 2.5-3 dpf larvae were embedded in ~0.5 ml 1% isotonic low-melting agarose in a glass-bottom dish. After solidification, the agarose surrounding the tail fins was removed with a needle blade to allow for better topical compound application. Larvae were preincubated for 60 min in 10 μM C11B in E3 at 28°C. The C11B solution was replaced with carrier control (0.4% DMSO), 20 μM arachidonic acid (Sigma, A3611), arachidic acid (Sigma, 10930), linolenic acid (Sigma, L2376), linoleic acid (Sigma, L1376), or oleic acid (Sigma, O1008) in E3, and incubated for 45 min at 28°C. C11B images were acquired as described above. Image analysis was performed in MATLAB essentially as detailed above, with the exception that a random polygonal region of the unwounded caudal tail fin (excluding the notochord) was selected from the transmitted light image for ratiometric C11B analysis.
Image-based measurement of tissue H2O2 production
Anesthetized, unwounded larvae were embedded in 1% low melting agarose in glass bottom dishes. After solidification, the agarose covering the caudal tail fin part of the larvae was removed, and 10 μM acetyl-pentafluorobenzene sulphonyl fluorescein (APSF, Calbiochem, 386794) 34 solution in E3 was added into the resulting well. Tail fins were exposed to the dye solution for ~40 min at room temperature before being washed three times with E3 supplemented with 5 μM CHP or DMSO carrier control (Ctr). Either 5 μM CHP or DMSO (Ctr) in E3 was then added to the wells. APSF fluorescence was excited with an LED light source with an internal 475/28 bandpass filter combined with a multispectral dichroic filter (Chroma, ET89021bs). APSF emission was sampled with a 525/50 bandpass filter (Chroma, ET525/50m). Transmitted light and APSF images were captured at room temperature using NIS-Elements on a Nikon Eclipse Ti microscope equipped with a 10x, plan-apochromat NA 0.45 air objective lens, and an Andor Clara CCD camera with variable camera binning. Both Ctr and CHP samples were acquired in parallel using a motorized stage with double dish-holder.
Analysis was conducted with the custom MATLAB script ND2Int (https://github.com/niethamp/KatikaneniAndJelcic2020). Briefly, ND2-files were imported using the Bioformats Toolbox for MATLAB function bfGetReader. In the transmitted light image, a background region close to the fin, and the tail fin region caudal to the notochord tip were manually marked for calculating background corrected APSF signal in the caudal fin. Background-corrected APSF tail fin signals It at each time point t were normalized by calculating the relative fluorescence ΔIt increase over the initial signal It0 with ΔIt=(It-It0)/It0.
Leukocyte tracking analysis
Leukocytes were tracked in the ventral part of the caudal tail fin from transmitted light time lapse movies using the MTrackJ 35 (v1.5.1, imagescience.org) plugin of FIJI 36 (ImageJ v1.5.2) by manually marking their position with the mouse cursor. Only clearly distinct positions were marked (i.e., no clicking in place, marks were not placed in every frame). MDF-files containing the tracking data were imported into MATLAB and processed with the custom scripts/functions AnalyzeMDF and AnalyzeMTrackJ (https://github.com/niethamp/KatikaneniAndJelcic2020) to obtain average cell speed, persistence and directionality as previously described 2,4 for animals with at least two tracks >50 μm. Such animals were categorized as “responsive”. Animals with less than two tracks >50 μm were categorized as non-responsive and excluded from the aggregated tracking analysis (Extended Data Figure 4). The fraction of responsive versus non-responsive animals was calculated as separate parameter and represented as pie chart in Extended Data Figure 4, and statistical testing was conducted using the MATLAB fishertest function.
Sudan black staining of leukocytes
Leukocyte staining by Sudan black B (Sigma, 199664) was essentially performed as previously described 37 on euthanized 2.5-3 dpf wild type or alox5a −/− larvae.
Morpholino injection
One-cell stage embryos were injected with 2.3 nL of 250-500 μM duox MO1 (5'-AAGCGTCACTTACTATAATGTTGGA-3', Gene Tools) or MP (5'-TCCCTTTTAGAATTTACCTTGCCGA-3', Gene Tools)2 together with 200 μM p53 morpholino (5'-ATGCTCAACTATAATGTTGGACATT-3', Gene Tools)38 diluted in water. P53 morpholino co-injection with duox MO1/MP served to suppress potential, pleiotropic morpholino-associated toxicity as previously described 2,38.
To assess the efficacy of the injected morpholino, ~100 larvae (2 dpf) were dissociated in 500 μL of TRIzol reagent (Invitrogen, 15596026) using RNase-free disposable pellet pestles (Fisherbrand, 12-141-364). Total RNA was extracted as per manufacturer’s instructions. Contaminating DNA was removed using the TURBO DNA-free Kit (Invitrogen, AM1907). Knockdown was confirmed by using the OneStep Ahead RT-PCR Kit (Qiagen, 220211) with the following primers: duox forward 5’-ACACATGTGACTTCATATCCAG-3’ and duox reverse 5’-ATTATTAACTCATCCACATCCAG-3’.
Generation of alox12−/− zebrafish
Cas9 was cloned into the pCS2 vector to generate pCS2-Cas9, capped mRNAs were transcribed using mMESSAGE mMACHINE SP6 Transcription kit (Thermo Fisher Scientific, AM1340), DNase (Invitrogen, AM1907) treated, and precipitated with lithium chloride. To generate templates for sgRNA transcription, gene specific oligonucleotides against zebrafish alox12 (ENSDARG00000069463) were designed using CHOPCHOP (https://chopchop.cbu.uib.no). A Sp6 consensus sequence was placed upstream of the targeting region of the alox12 sgRNA for in vitro transcription of the sgRNA (Sp6: ATTTAGGTGACACTATA promoter sequence), the 20 base target site of alox12 (alox12: GGATCACTGGGCAGAAATAC), were annealed to a constant oligonucleotide encoding the reverse-complement of the tracrRNA tail (5′-AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC-3’). The ssDNA overhangs were filled in with T4 DNA polymerase (NEB, M0203), and the resulting sgRNA template were purified using QIAquick columns (Qiagen, 28115). sgRNAs were transcribed using the MEGAscript SP6 Transcription kit (Invitrogen, AM1330). All sgRNAs were DNase treated and precipitated with 3 M ammonium acetate / 70 % ethanol.
One-cell stage Casper zebrafish zygotes were collected and the cytoplasm of the cell was injected with a single mix of 25 pg sgRNA and 100 pg Cas9 mRNA.
For sequencing, total RNA was isolated with TRIzol reagent from > 50 F3 alox12 −/− mutant zebrafish at 3 dpf. Each single mutant was reverse transcribed using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, K1622). The alox12 gene was amplified with forward: 5’-TTGAACAGAAACCTGATAAGGACA-3’ and reverse: 5’-TGGCCACACAGATCACTTCT - 3’ primers spanning intron / exon boundaries to ensure mRNA was specifically amplified. The resulting amplicon was TA–cloned with the StrataClone PCR Cloning kit (Aligent, 240205). Single colonies were isolated, and Sanger-sequenced.
Genomic DNA was isolated from the tail fins of adult F2 alox12 −/− zebrafish. HOT-SHOT gDNA isolation procedure was utilized. Briefly, 100 μL of 50 mM NaOH was added to each tail fin and incubated at 95 °C for 10 min. The gDNA was then placed on ice and 10 μL of 1 M Tris–HCl (pH 8.0) was added. The tube was spun down to remove debris and the resulting gDNA was used to amplify the region targeting alox12 (see above). The resulting 164 base pair amplicon for wildtype and control samples was collected and sent to the Memorial Sloan Kettering Cancer Center (MSKCC) Integrated Genomics Core (iGO). The resulting amplicon had NGS tails ligated onto the product and was sequenced on an Illumina NGS at a respective read-depth of greater than one-hundred thousand reads per sample. CRISPResso analysis was performed by the MSKCC Bioinformatics Core (http://crispresso.pinellolab.partners.org).
Generation of alox5a−/− zebrafish
An alox5a (ENSDARG00000057273) CRISPR RNA (crRNA) designed from the following 20 base sequence (5’-TGGGTGCCGCCAAGTACTGA-3’) was selected from a predesigned list of alox5a targeting crRNA’s generated by a propriety algorithm from Integrated DNA Technologies (IDT) and was ordered as an Alt-R® CRISPR-Cas9 crRNA from IDT. The alox5a ALT-R® crRNA and transactivating crRNA (tracrRNA) (IDT, 1072532) were solubilized in Nuclease Free IDTE pH 7.5 solution (IDT, 11-01-02-02) to final concentrations of 100 μM each. Solubilized alox5a crRNA and tracrRNA were mixed with IDT Nuclease Free Duplex Buffer (IDT, 11-01-03-01) to final concentrations of 3 μM each. The mixture was subsequently heated at 95 °C for 5 min, removed from heat, and allowed to cool to RT on the bench top to form the gRNA solution. ALT-R® S.p. Cas9 Nuclease (IDT, 1081058) was diluted to 1 mg mL−1 in 150 mM KCl, 20 mM HEPES pH 7.5 solution. Prior to injection, the Cas9-gRNA ribonucleoprotein (RNP) complex was assembled by combining the gRNA and Cas9 protein solutions 1:1 (v/v), then incubating at 37 °C for 10 min and allowing it to cool to RT on the bench top. 2.3 nL of the Cas9-gRNA RNP complex solution was subsequently injected into the cytoplasm of 1-cell stage Casper zebrafish embryos and injected F0 larvae were grown to sexual maturity (2-3 months post fertilization). Individual F0 adults were crossed to wild type Casper zebrafish and F1 progeny were collected. At 2-3 days post fertilization (dpf) genomic DNA (gDNA) was isolated from 4-6 individual F1 larvae for genotyping. Briefly, euthanized 2.5-3 dpf were separately suspended in 100 μL of 50 mM NaOH and incubated at 95 °C for 10 min. Samples were then cooled on ice, neutralized with 10 μL of 1 M Tris-HCl pH 8 and centrifuged at 16,000 g to remove debris. The alox5a sequence of interest was PCR amplified from the gDNA samples with the following primers designed using CHOPCHOP: alox5a geno fwd: (5’-GCTGTAATCCAGTGGTCATCAA-3’) and alox5a geno rev: (5’-TGATCTCACTGGAGACTGGAGA-3’) 39. Next, PCR products were incubated with the FastDigest ScaI (Thermo Fisher Scientific, FD0434) restriction enzyme for ~2 h at 37 °C and then samples were separated by agarose gel electrophoresis. F1 heterozygous larvae were identified by the presence of three double stranded DNA products at ~1102, 693, and 409 base pairs (bp). The ~1102 bp product signifies a mutant allele for which the ScaI restriction site has been destroyed as a result of a Cas9-induced double strand break and subsequent DNA damage repair. F1 homozygous wild type larvae were identified by the presence of only the 693 and 409 bp products, which are the result of complete digest of the amplified wild type allele by ScaI. An F0 alox5a KO founder adult was identified by the presence of F1 heterozygous alox5a progeny. Once an F0 alox5a KO founder adult was identified, the fish was once again crossed to a wild type Casper. At 3 dpf, progeny tail fin tips were amputated, and genomic DNA was isolated by suspending the tail fin tip in 20 μL of 50 mM NaOH then heating at 95 °C for 10 min. Samples were then cooled on ice, neutralized with 2 μL of 1 M Tris-HCl pH 8, and centrifuged at 16,000 g to remove debris. Genotyping was conducted by PCR amplification followed by ScaI digest. Heterozygous alox5a F1 larvae were identified and grown to sexual maturity. F1 adults were tail fin clipped to isolate gDNA and genotyped by PCR amplification and ScaI digest. The mutated alleles were sequenced by Sanger-sequencing. F1 heterozygous male and female adults containing the frameshift mutation of interest (8 bp deletion; Extended Data Figure 3) were crossed to generate F2 homozygous mutant alox5a progeny (alox5a −/−).
Statistics and Reproducibility
Sample sizes were not predetermined by statistical methods and based on our previous experience with biological variability in this system. Each group plotted in the graphs typically contains n=9 or more biologically independent animal experiments often aggregated from different experimental days (as indicated in the Source data for the respective figures). For micropipette experiments, we sequentially imaged animals leading to lower experimental numbers as compared to wounding experiments, where several wounded animals could be imaged in parallel using a motorized microscope stage. The group sizes in some of the leukocyte tracking experiments are below n=9 (but at least n=3), because leukocytes in non-responsive animals could not be tracked (Extended Data Figure 4). The experiments were not randomized, and investigators were not blinded to allocation during experiments and outcome assessment with the following exceptions: Part of the wounding and patching experiments summarized in in Figure 4a and 4b were blinded by AK prior to experiment and analysis by PN to establish the principle mutant phenotypes at highest experimental rigor. Given its considerable logistic effort, blinding was later not consequently performed for the sake of increasing experimental throughput. During computational C11B and APSF image analysis in MATLAB, PN was selecting measurement regions based on the transmitted light images. Those images contained no result-relevant information, excluding the possibility of biased region-picking based on lipid peroxidation signal intensity. Data were represented as dot plot, violin plot, or boxplot using the PlotsOfData 40 website (https://huygens.science.uva.nl/PlotsOfData/), or MATLAB, respectively. One dot in a graph represents n=1 biologically independent animal experiment (with a single larva). Number in parentheses represent the total n per group. Animals were never reused for different experiments. In the violin plot, the center of the open circle indicates the median value, and the vertical bar the 95% confidence interval. In the dot plots of Extended Data Figure 4, the mean is denoted as horizontal bar. Boxplots were generated with the MATLAB boxplot function. The central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles. Red crosses indicate outliers. Profile plots with shaded error regions were generated in MATLAB using the shadedErrorBar function (v1.3.1, https://www.github.com/raacampbell/shadedErrorBar). The midline indicates the mean and the shaded region the standard error of mean (SEM). P-values for pairwise comparisons of samples were calculated using the Excel (Microsoft Office 365, Build 11929.20606) TTEST function (unpaired, two-tailed distribution, two sample unequal variance). Two-tailed Fisher’s exact tests in the Extended Data Figure 4 were performed in MATLAB.
The wave-like tissue degeneration phenomenon depicted in Extended Data Figure 1c was observed two times out of a total (incl. morphants and mutants) of the 241 reported AA-patching experiments (i.e., at ~1% frequency).
Code availability
Custom MATLAB analysis code can be found on https://github.com/niethamp/KatikaneniAndJelcic2020.
Data availability
All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
Extended Data
Extended Data Fig. 1. Additional data supporting Figure 1 and 2.
(a) Leukocyte mobilization to different AA preparations. Circles, median. Bars, 95% confidence interval. Data aggregated from n=10 (Cayman AA), and n=10 (Sigma AA) biologically independent animal experiments. No significant difference (p=0.51) detected between groups by two-sided, heteroscedastic Student’s t-test. (b) CHP stimulates H2O2. Zebrafish tail fins of wild type larvae were loaded with the H2O2 sensor dye acetyl-pentafluorobenzene sulphonyl fluorescein, and then exposed to 5 μM CHP or DMSO control (Ctr). Top panel, representative dye emission images at 0 min and 45 min after CHP exposure. Lower panel, plots (mean ± SEM) of normalized dye fluorescence over 45 min after CHP exposure. Data aggregated from n=15 (Ctr), and n=18 (CHP) biologically independent animal experiments (c) Time-lapse montage of a wave-like tissue degeneration emerging from an AA (20 μM) microperfusion patch site. Pink shade highlights the affected region. Time, hh:mm:ss. Images representative of the 2 out of 241 biologically independent animal experiments with observable waves. (d) Confirmation of duox knockdown by RT-PCR in 2 dpf embryos. Representative for three independent analyses. (e) C11B-imaging of control larvae (Ctr) and larvae treated with 20 μM or 100 μM DPI ~15-30 min after tail fin tip amputation. Upper panel: Representative C11B ratio images. Lower panel: Wound ratio, mean C11B ratio normalized to median baseline ratio ± SEM plotted against wound distance. Inset, boxplots of baseline ratios normalized to control. Central mark, median. Box edges, 25th/75th percentiles. Whiskers, most extreme, non-outlier data points. +, outliers. Data aggregated from n=17 (Ctr, 0 μM DPI), n=9 (20 μM DPI), and n=9 (100 μM DPI) biologically independent animal experiments. Parentheses, number of different animals. Scale bar, 200 μm. Statistical source data can be found at Source data Extended Data Figure 1.
Extended Data Fig. 2. Characterization of the alox12 KO mutant.
(a) Schematic representation of the protein domains of zebrafish 12-lipxoygenase (Alox12). The lipoxygenase domain of Alox12 responsible for enzymatic function was targeted with a short guide RNA (sgRNA). (b) Sanger sequence analysis of cDNA identified a two-base pair deletion at nucleotide position 1011-1012 within exon 10 of alox12. (c) The targeted region of the lipoxygenase domain of zebrafish Alox12 is well conserved compared to human ALOX12. The frameshift mutant alox12 allele contains a premature stop codon in the coding sequence. Asterisks, fully conserved residues. Colons, conservation of strongly similar residues. Periods, conservation of weakly similar residues. (d) Next-Gen Sequencing and CRISPResso analysis of the amplicon containing the sgRNA cut site was performed on genomic DNA from tail fin clips of F2 adult alox12-targeted zebrafish. Alox12-targeted F2 male and female zebrafish exhibited a 2 bp mutation positioned at the proposed sgRNA cut site in greater than 99% of sequence reads over a depth of more than 300 and 400 thousand reads, respectively. Wild type zebrafish displayed background sequence abnormalities below 1% in more than 140 thousand total reads. (e) CRISPResso analysis of F2 male and female zebrafish targeted with the sgRNA against alox12 identifies a single 2 bp indel. An indel is denoted by a blue solid bar and no indel is represented by a red dashed bar. Wildtype zebrafish display no indels in most of the sequence reads.
Extended Data Fig. 3. Characterization of the alox5a KO mutant.
(a) Schematic representation of zebrafish Alox5a. A single guide RNA (sgRNA) was designed to target alox5a for gene disruption within the lipoxygenase domain at exon 7 (bold text). Successful target disruption is predicted to occur at a ScaI restriction enzyme recognition site (AGT^ACT) (highlighted red text) within the genomic DNA (gDNA) sequence, potentially destroying the sequence upon DNA repair. (b) alox5a wild type and knockout mutant alleles. The mutant alox5a allele contains an 8-base pair (bp) deletion resulting in a frameshift and the destruction of the ScaI restriction enzyme recognition site. Red text, mutant alox5a sequence that differs from the wild type sequence. (c) The targeted region of the lipoxygenase domain of zebrafish Alox5a is highly conserved compared to human ALOX5. The mutant alox5a allele contains a premature stop codon in the coding sequence, resulting in a truncated Alox5a. Asterisks, fully conserved residues. Colons, conservation of strongly similar residues. Periods, conservation of weakly similar residues. Red text, mutant Alox5a sequence that differs from the wild type sequence. (d) Heterozygous alox5a larvae were crossed and gDNA from isolated progeny was PCR amplified, ScaI-digested, and analyzed by agarose gel electrophoresis for genotyping. PCR product from the wild type alox5a allele is cleaved into two smaller products upon incubation with ScaI (409 and 693 bp products). The mutant alox5a allele is not cleaved by ScaI (1094 bp product only). Heterozygotes are identified by the combined presence of a cleaved wild type allele and non-cleaved mutant allele (409, 693, and 1094 bp products). Representative of three independent analyses. (e) Sudan black-stained neutrophils in the tail region of zebrafish larvae at 2.5-3 days post fertilization. Shown are three representative animals from a total of 20 different larvae stained per genotype. Scale bar, 200 μm.
Extended Data Fig. 4. Aggregated leukocyte tracking analysis.
Left panels: leukocyte speed, migration persistence, directionality, and responsiveness towards microperfused 20 μM AA tracked over 20 min after patching. Right panels: leukocyte speed, migration persistence, directionality, and responsiveness towards a tail fin wound tracked over 40 min after tail fin injury. Leukocyte speed, migration persistence, directionality was calculated as previously described (see Methods). The Responsive Fractions (red color, pie charts) denote the fractions of animals for which at least two tracks > 50 μm could be reliably measured in the caudal tail fin below the notochord line. Tracking data are only aggregated for responsive animals. Only clearly distinct positions were marked for tracking; stationary leukocytes were not tracked. The tracking analysis was performed on the time lapse data underlying Figure 1c, 2d, 4a, 4b, and an additional AA/DPI pipette experiment, in which leukocyte migration to a micropipette filled with 20 μM AA + 50 μM DPI (DPI, green dots) was tracked. Gray shaded region, duox MO1 morphants were compared to duox MP animals instead of wild type animals, as both samples are also co-morphants for p53 (see Methods). Horizontal bars, mean. Data aggregated from n=46 (wt, Micropipette ), n=17 (alox5a KO, Micropipette), and n=9 (alox12 KO, Micropipette), n=7 (DPI, Micropipette), n=3 (Linoleic, Micropipette), n=20 (duox MP, Micropipette), n=10 (duox MO1, Micropipette), n=56 (wt, Wound), n=42 (alox5a KO, Wound), and n=23 (alox12 KO, Wound) biologically independent animal experiments. P-values with color code, two-sided, heteroscedastic Student’s t-test for pairwise comparison of the indicated color-coded samples. P-value near brackets, two-tailed Fisher’s exact test between Responsive Fractions indicated by brackets. Parentheses, number of different animals. Statistical source data can be found at Source data Extended Data Figure 4.
Extended Data Fig. 5. Cartoon scheme of proposed mechanism.
Wounding activates Duox to produce H2O2 and causes hypotonic (Hypo) shock, which leads to release of PUFAs (including AA). H2O2 reacts with Fe2+ to generate HO∙ by Fenton chemistry. HO∙ reacts with PUFAs/AA to generate long-chain lipid ROS (PUFA-OO∙, PUFA-OOH) that further promote lipid peroxidation, and activate Alox5a to produce leukocyte chemoattractants. If lipid peroxidation slips out of control, it may cause cell death.
Supplementary Material
Supplementary Video 4 Microperfusion of zebrafish tail fins with 20 μM arachidonic acid (AA, upper panel) or 20 μM linoleic acid (Lin, lower panel). Red, leukocyte tracks. Time, hh:mm:ss. Scale bar, 200 μm.
Supplementary Video 5 Leukocyte recruitment to zebrafish tail fin wounds treated with DMSO carrier control (Ctr, upper panel) or 3 μM ebselen (Ebs, lower panel). Red, leukocyte tracks. Time, hh:mm:ss. Scale bar, 200 μm.
Supplementary Video 6 Leukocyte recruitment to zebrafish tail fin wounds treated with DMSO carrier control (upper panel), 20 μM liproxstatin-1 (Lpx, middle panel), or 100 μM α-tocopherol (a-TOC, lower panel). Red, leukocyte tracks. Time, hh:mm:ss. Scale bar, 200 μm.
Supplementary Video 7 Microperfusion of wild type (wt, upper panel), alox12 KO (middle panel), or alox5a KO (lower panel) tail fins with 20 μM arachidonic acid. Red, leukocyte tracks. Time, hh:mm:ss. Scale bar, 200 μm.
Supplementary Video 1 Microperfusion of a wild type zebrafish tail fin with 20 μM arachidonic acid. Note, leukocytes immediately return to the vasculature once the pipette is removed. Time, hh:mm:ss. Scale bar, 200 μm.
Supplementary Video 2 Microperfusion of control (duox MP, upper panel) and duox knockdown (duox MO1, lower panel) tail fins with 20 μM arachidonic acid. Red, leukocyte tracks. Time, hh:mm:ss. Scale bar, 200 μm.
Supplementary Video 8 Leukocyte recruitment to wounds in wild type (wt, upper panel), alox12 KO (middle panel), or alox5a KO (lower panel) tail fins. Red, leukocyte tracks. Time, hh:mm:ss. Scale bar, 200 μm.
Supplementary Video 3 Microperfusion of a zebrafish tail fin with 20 μM arachidonic acid. Note, wave emerging at 00:36:45. Time, hh:mm:ss (after patching). Scale bar, 200 μm.
Acknowledgements
Research was supported by the NIH/NIGMS grants R01GM099970 and R01GM127356 to PN, the MSKCC Functional Genomics Initiative, and in part through the NIH/NCI Cancer Center Support grant P30CA008748. The authors thank the Integrated Genomics Operation Core and Bioinformatics Core at MSKCC for their assistance.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Huang C & Niethammer P Tissue damage signaling is a prerequisite for protective neutrophil recruitment to microbial infection in zebrafish. Immunity 48, 1006–1013.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niethammer P, Grabher C, Look AT & Mitchison TJ A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature 459, 996–999 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoo SK, Starnes TW, Deng Q & Huttenlocher A Lyn is a redox sensor that mediates leukocyte wound attraction in vivo. Nature 480, 109–112 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enyedi B, Kala S, Nikolich-Zugich T & Niethammer P Tissue damage detection by osmotic surveillance. Nat. Cell Biol 15, 1123–1130 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enyedi B & Niethammer P Mechanisms of epithelial wound detection. Trends Cell Biol. 25, 398–407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klyubin IV, Kirpichnikova KM & Gamaley IA Hydrogen peroxide-induced chemotaxis of mouse peritoneal neutrophils. Eur. J. Cell Biol 70, 347–351 (1996). [PubMed] [Google Scholar]
- 7.Evans IR, Rodrigues FSLM, Armitage EL & Wood W Draper/CED-1 mediates an ancient damage response to control inflammatory blood cell migration in vivo. Curr. Biol 25, 1606–1612 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jelcic M, Enyedi B, Xavier JB & Niethammer P Image-Based Measurement of H2O2 Reaction-Diffusion in Wounded Zebrafish Larvae. Biophys. J 112, 2011–2018 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enyedi B, Jelcic M & Niethammer P The Cell Nucleus Serves as a Mechanotransducer of Tissue Damage-Induced Inflammation. Cell 165, 1160–1170 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lämmermann T et al. Neutrophil swarms require LTB4 and integrins at sites of cell death in vivo. Nature 498, 371–375 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trede NS, Langenau DM, Traver D, Look AT & Zon LI The use of zebrafish to understand immunity. Immunity 20, 367–379 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Hammarström S et al. Increased concentrations of nonesterified arachidonic acid, 12L-hydroxy-5,8,10,14-eicosatetraenoic acid, prostaglandin E2, and prostaglandin F2alpha in epidermis of psoriasis. Proc. Natl. Acad. Sci. USA 72, 5130–5134 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathias JR et al. Resolution of inflammation by retrograde chemotaxis of neutrophils in transgenic zebrafish. J. Leukoc. Biol 80, 1281–1288 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Gault WJ, Enyedi B & Niethammer P Osmotic surveillance mediates rapid wound closure through nucleotide release. J. Cell Biol 207, 767–782 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riendeau D, Denis D, Choo LY & Nathaniel DJ Stimulation of 5-lipoxygenase activity under conditions which promote lipid peroxidation. Biochem. J 263, 565–572 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rouzer CA & Samuelsson B The importance of hydroperoxide activation for the detection and assay of mammalian 5-lipoxygenase. FEBS Lett. 204, 293–296 (1986). [DOI] [PubMed] [Google Scholar]
- 17.Conrad M et al. Regulation of lipid peroxidation and ferroptosis in diverse species. Genes Dev. 32, 602–619 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linkermann A et al. Synchronized renal tubular cell death involves ferroptosis. Proc. Natl. Acad. Sci. USA 111, 16836–16841 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim SE et al. Ultrasmall nanoparticles induce ferroptosis in nutrient-deprived cancer cells and suppress tumour growth. Nat. Nanotechnol 11, 977–985 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riegman M et al. Ferroptosis occurs through an osmotic mechanism and propagates independently of cell rupture. Nat. Cell Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang WS et al. Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc. Natl. Acad. Sci. USA 113, E4966–75 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lebold KM et al. Novel liquid chromatography-mass spectrometry method shows that vitamin E deficiency depletes arachidonic and docosahexaenoic acids in zebrafish (Danio rerio) embryos. Redox Biol 2, 105–113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pap EHW et al. Ratio-fluorescence microscopy of lipid oxidation in living cells using C11-BODIPY581/591. FEBS Lett. 453, 278–282 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Brash AR Arachidonic acid as a bioactive molecule. J. Clin. Invest 107, 1339–1345 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noguchi N, Yoshida Y, Kaneda H, Yamamoto Y & Niki E Action of ebselen as an antioxidant against lipid peroxidation. Biochem. Pharmacol 44, 39–44 (1992). [DOI] [PubMed] [Google Scholar]
- 26.Zilka O et al. On the Mechanism of Cytoprotection by Ferrostatin-1 and Liproxstatin-1 and the Role of Lipid Peroxidation in Ferroptotic Cell Death. ACS Cent. Sci 3, 232–243 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adel S, Heydeck D, Kuhn H & Ufer C The lipoxygenase pathway in zebrafish. Expression and characterization of zebrafish ALOX5 and comparison with its human ortholog. Biochim. Biophys. Acta 1861, 1–11 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Haas U et al. Targeted knock-down of a structurally atypical zebrafish 12S-lipoxygenase leads to severe impairment of embryonic development. Proc. Natl. Acad. Sci. USA 108, 20479–20484 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uhlén M et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell Proteomics 4, 1920–1932 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Erlemann K-R et al. Airway epithelial cells synthesize the lipid mediator 5-oxo-ETE in response to oxidative stress. Free Radic. Biol. Med 42, 654–664 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Albadri S et al. Redox signaling via lipid peroxidation regulates retinal progenitor cell differentiation. Dev. Cell 50, 73–89.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 32.White RM et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2, 183–189 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nüsslein-Volhard C & Dahm R Zebrafish: a practical approach. 303 (Oxford University Press, 2002). [Google Scholar]
- 34.Maeda H et al. Fluorescent probes for hydrogen peroxide based on a non-oxidative mechanism. Angew. Chem. Int. Ed. Engl 43, 2389–2391 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Meijering E, Dzyubachyk O & Smal I Methods for cell and particle tracking. Meth. Enzymol 504, 183–200 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosowski EE et al. Macrophages inhibit Aspergillus fumigatus germination and neutrophil-mediated fungal killing. PLoS Pathog. 14, e1007229 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robu ME et al. p53 activation by knockdown technologies. PLoS Genet. 3, e78 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Labun K, Montague TG, Gagnon JA, Thyme SB & Valen E CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 44, W272–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Postma M & Goedhart J PlotsOfData-A web app for visualizing data together with their summaries. PLoS Biol. 17, e3000202 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Video 4 Microperfusion of zebrafish tail fins with 20 μM arachidonic acid (AA, upper panel) or 20 μM linoleic acid (Lin, lower panel). Red, leukocyte tracks. Time, hh:mm:ss. Scale bar, 200 μm.
Supplementary Video 5 Leukocyte recruitment to zebrafish tail fin wounds treated with DMSO carrier control (Ctr, upper panel) or 3 μM ebselen (Ebs, lower panel). Red, leukocyte tracks. Time, hh:mm:ss. Scale bar, 200 μm.
Supplementary Video 6 Leukocyte recruitment to zebrafish tail fin wounds treated with DMSO carrier control (upper panel), 20 μM liproxstatin-1 (Lpx, middle panel), or 100 μM α-tocopherol (a-TOC, lower panel). Red, leukocyte tracks. Time, hh:mm:ss. Scale bar, 200 μm.
Supplementary Video 7 Microperfusion of wild type (wt, upper panel), alox12 KO (middle panel), or alox5a KO (lower panel) tail fins with 20 μM arachidonic acid. Red, leukocyte tracks. Time, hh:mm:ss. Scale bar, 200 μm.
Supplementary Video 1 Microperfusion of a wild type zebrafish tail fin with 20 μM arachidonic acid. Note, leukocytes immediately return to the vasculature once the pipette is removed. Time, hh:mm:ss. Scale bar, 200 μm.
Supplementary Video 2 Microperfusion of control (duox MP, upper panel) and duox knockdown (duox MO1, lower panel) tail fins with 20 μM arachidonic acid. Red, leukocyte tracks. Time, hh:mm:ss. Scale bar, 200 μm.
Supplementary Video 8 Leukocyte recruitment to wounds in wild type (wt, upper panel), alox12 KO (middle panel), or alox5a KO (lower panel) tail fins. Red, leukocyte tracks. Time, hh:mm:ss. Scale bar, 200 μm.
Supplementary Video 3 Microperfusion of a zebrafish tail fin with 20 μM arachidonic acid. Note, wave emerging at 00:36:45. Time, hh:mm:ss (after patching). Scale bar, 200 μm.
Data Availability Statement
All other data supporting the findings of this study are available from the corresponding author upon reasonable request.