Abstract
The coronavirus disease 2019 (COVID-19) pandemic has highlighted the cardinal importance of rapid and accurate diagnostic assays. Since the early days of the outbreak, researchers with different scientific backgrounds across the globe have tried to fulfill the urgent need for such assays, with many assays having been approved and with others still undergoing clinical validation. Molecular diagnostic assays are a major group of tests used to diagnose COVID-19. Currently, the detection of SARS-CoV-2 RNA by reverse transcription polymerase chain reaction (RT-PCR) is the most widely used method. Other diagnostic molecular methods, including CRISPR-based assays, isothermal nucleic acid amplification methods, digital PCR, microarray assays, and next generation sequencing (NGS), are promising alternatives. In this review, we summarize the technical and clinical applications of the different COVID-19 molecular diagnostic assays and suggest directions for the implementation of such technologies in future infectious disease outbreaks.
Keywords: Coronavirus, SARS-CoV-2, COVID-19, molecular diagnostic techniques, nucleic acid amplification techniques
Introduction
The recent coronavirus disease 2019 (COVID-19) outbreak caused by SARS-CoV-2, which initially originated in Wuhan, Hubei province of China in December 2019, has since spread globally. The causative agent, SARS-CoV-2, is a single-stranded RNA virus that is spread primarily via droplet transmission but that can also display airborne transmission under certain circumstances [1,2]. This infection has a variable clinical presentation ranging from asymptomatic carrier state to fulminant respiratory failure. Dry cough, fever, and shortness of breath are the most common symptoms of COVID-19 [3]. It has been shown that asymptomatic and pre-symptomatic carriers play an important role in the transmission of SARS-CoV-2 [4,5]. This highlights the importance of the widespread availability of accurate and efficient diagnostic assays to identify those infected so as to help control further spread of infection.
Currently, two major categories of diagnostic assays are commercially available for diagnosing SARS-CoV-2. The first group of assays identifies the viral RNA using molecular techniques that are based mostly on polymerase chain reaction (PCR) or nucleic acid hybridization. The second group are immunological assays that detect either antibodies that are produced in response to the infection or antigenic proteins. Laboratory-based SARS-CoV-2 molecular assays are currently the reference standard for the diagnosis of this infection [6]. However, point-of-care diagnostic tests and technologies are emerging. In this review, we provide an overview of the molecular diagnostic assays used to diagnose COVID-19, describe their importance in the control of this pandemic, and highlight the application of such assays in potential future infectious disease outbreaks.
SARS-CoV-2 genome and structure
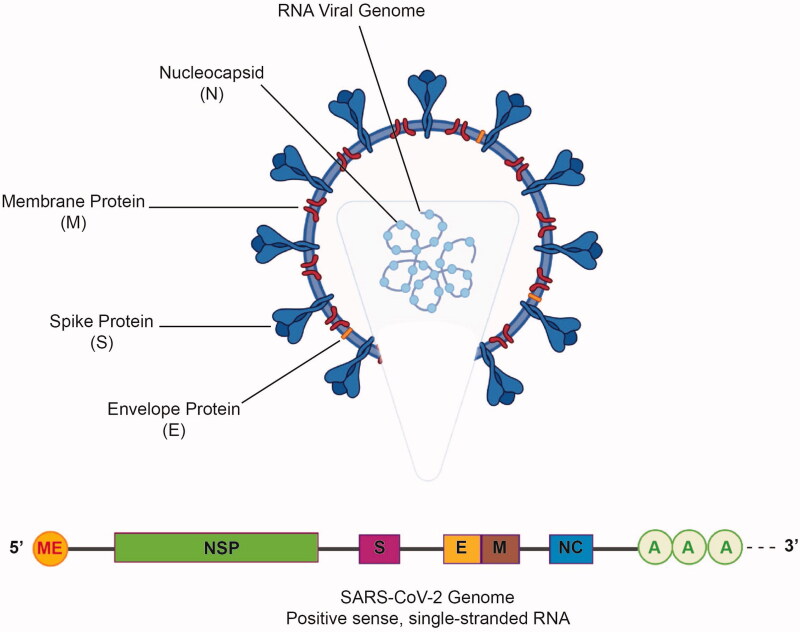
SARS-CoV-2 belongs to the Coronaviridae family of the Nidovirales order [1]. The viral genome was first sequenced using deep meta-transcriptomic sequencing of the bronchoalveolar lavage fluid of a 41-year-old man who was admitted to the Central Hospital of Wuhan due to pneumonia of unknown etiology (Genbank: MN908947). The viral genome, which is approximately 30 kb in size, is a positive single-stranded RNA with a 5′-cap and a 3′-poly-A tail [7]. It contains 14 open reading frames (ORFs) encoding different replication, structural and nonstructural accessory proteins. At the 5′-terminal region of the genome, ORF1 and ORF2 encode 15 nonstructural proteins responsible for viral replication while the structural proteins, nucleocapsid (N), membrane protein (M), envelope protein (E), and spike protein (S), in addition to eight accessory proteins, are encoded by the 3′-terminal region [8,9] (Figure 1).
Figure 1.
The general structure of SARS-CoV-2. Major structural proteins, namely, the spike protein (S), membrane protein (M), and envelope protein (E), are present on the viral envelope. The nucleocapsid protein (N) along with the genomic RNA is present inside the viral envelope (above). SARS-CoV-2 RNA genome has a 5’ methylated cap and a 3’ poly-A tail. The positions of the genes encoding the nonstructural proteins (NSP) and spike (S), membrane (M), envelope (E), and nucleocapsid (NC) proteins are shown (below).
Coronaviruses are equipped with genetic proofreading mechanisms [10]. This has, in turn, led to low sequence diversity among different SARS-CoV-2 strains [11]. However, potentially favorable mutations conferring increased infectivity and immunologic resistance may accumulate with continued human-to-human transmission over time. One such example is the emergence of the D614G S protein variant, which disrupts an inter-protomer contact and alters the conformation of the S protein, and which is associated with increased infectivity [12,13]. Furthermore, recent findings highlight the emergence of novel SARS-CoV-2 variants in different regions across the globe. On December 14, 2020, the British authorities reported to the World Health Organization (WHO) a variant containing 23 nucleotide substitutions, which was phylogenetically unrelated to the variant commonly circulating in the UK at that time [14]. This novel variant, SARS-CoV-2 VOC 202012/01 (Variant of Concern, year 2020, month 12, variant 01), which is also named B.1.1.7, is found to be 75% more transmissible than previous variants [15]. On December 18, 2020, the authorities at South Africa reported a novel variant with the N501Y mutation that was spreading in three provinces of the country. This variant, called 501Y.V2 by the South African authorities, is associated with a greater viral load and with potentially higher transmissibility according to preliminary studies [14].
Clinical specimens
Upper or lower respiratory tract specimens - nasopharyngeal (NP) specimens, oropharyngeal (OP) specimens, nasal mid-turbinate swabs, anterior nares (nasal swab) specimens, NP wash/aspirates, or nasal wash/aspirate (NW) specimens - collected by healthcare providers are the recommended specimens for SARS-CoV-2 diagnostic testing [16]. It is possible to use lower respiratory tract specimens in patients with productive cough (sputum) or those receiving mechanical ventilation (bronchoalveolar lavage or lower respiratory tract aspirates) [16]. The virus also has been detected in other clinical specimens (e.g. blood, feces) [17].
Molecular assays
Reverse transcription-polymerase chain reaction (RT-PCR)
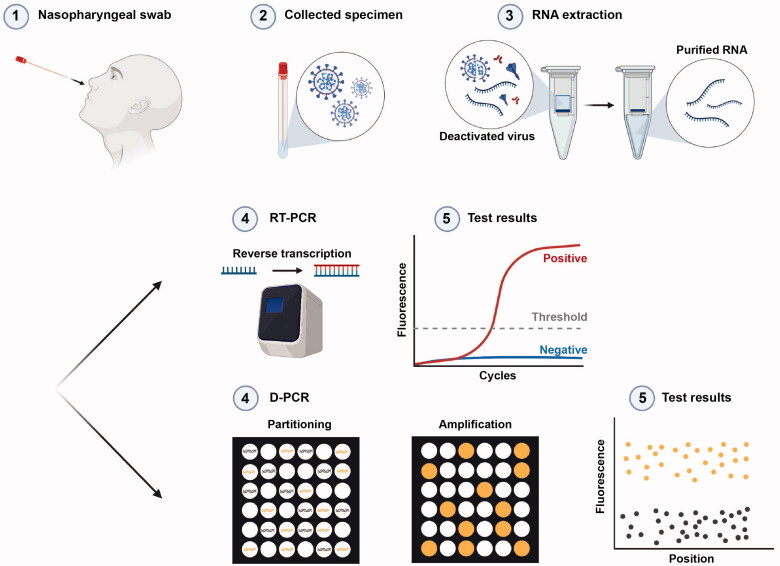
Within a few days after the start of the outbreak, the full genomic sequence of the virus was made publicly available [18]. This, in turn, enabled biotechnological companies and research groups around the globe to develop different molecular diagnostic assays through designing specific primers and probes. These primer sets have been optimized thanks to ongoing global efforts by different researchers [19]. RT-PCR, the gold standard method for SARS-CoV-2 diagnosis, amplifies a small segment of virus genetic material. In this method, the extracted viral RNA from the clinical specimen is converted to cDNA, which subsequently undergoes amplification [20]. DNA amplification is monitored in real-time using a fluorescent dye or a combination of a quencher molecule and a sequence-specific DNA probe labeled with a fluorescent molecule [21] (Figure 2).
Figure 2.
Schematic representation of RT-PCR and digital PCR procedures used to detect SARS-CoV-2. In both assays, appropriate specimens are collected and viral RNA is extracted. In RT-PCR, the relative or absolute concentration of the target of interest is assessed by measuring the fluorescent signal that shows the amplification in each cycle. In digital PCR, the absolute concentration of the target nucleic acid is determined based on the number of partitions that are either positive or negative for amplification based on fluorescent signals.
In general, the specific genetic regions selected as the target in RT-PCR diagnostic assays are very important. Assays targeting the E gene, which has been identified to be similar to that of other coronaviridae strains, have been shown to have the highest sensitivity [20]. On the other hand, the low homology of the RdRp, N, and S genes in SARS-CoV-2 with those in other bat-related viruses makes these genes specific targets [20,22–24]. Multiplexed assays targeting multiple genes simultaneously or detecting different regions in the same target gene have been used in various laboratories to increase the sensitivity of detection [25,26].
Considering the overwhelming number of cases, the diagnostic laboratory infrastructure in different regions, particularly in those with limited resources, has not been able to cope with the very large number of clinical specimens that are submitted. Pooled sample testing (i.e. mixing a number of samples together in a batch and then treating the batch as one specimen) has been suggested to be a potential solution for this problem [27,28]. However, due to the dilution of the genetic material as a result of pooling, the likelihood of false-negative results increases. Therefore, it is recommended to perform pooled sample testing only in settings with a low prevalence of cases [29].
SARS-CoV-2 RT-PCR assays have been shown to have a detection limit below 10 copies/reaction [23]. Low viral loads at the very early or very late stages of infection can lead to false-negative results. Mutations in primer target regions in the virus genome can also lead to false-negative results. Furthermore, despite the high specificity of RT-PCR assays, false-positive results due to sample contamination can occur; negative control templates may be helpful in identifying these cases [30]. Additionally, it has been shown that patients recovering from COVID-19 can test positive for the virus for a long period of time after complete resolution of symptoms [31]. Although the significance of this observation remains elusive, this highlights the inability of RT-PCR to discriminate intact whole virus particles from viral RNA.
Overall, RT-PCR is the most commonly used method to diagnose COVID-19 due to its high sensitivity and specificity and also to its ability to process large numbers of samples. However, its widespread use is hindered by its requirement for expensive laboratory instruments and skilled laboratory personnel.
Digital PCR
In quantitative PCR, the quantity of the template of interest is inferred based on the intensity of the fluorescent signal emitted during the amplification phase of a single PCR reaction compared with an internal or external calibrator that is amplified simultaneously. On the other hand, in digital PCR, the template is isolated into single molecules by subdividing the reaction mixture into thousands of microscopic partitions with the ultimate goal of each partition containing, on average, less than a single copy of the template of interest. The quantity of the template in the sample is subsequently calculated using Poisson statistics based on the overall number of compartments that are either amplification-positive or -negative (Figure 2) [32,33].
The advantages of digital PCR over quantitative PCR include quantification without the need for calibration curves, higher precision, and less susceptibility to artifacts that may arise from sub-optimal amplification efficacy because of PCR inhibitors or primer/template mismatch [34–37]. In addition, digital PCR has a higher analytical sensitivity due to its ability to partition the samples; this leads to decreased competition between various targets for the amplification reagents [32]. It also has a higher multiplexing capability compared with quantitative PCR [38]. Nonetheless, the complicated workflow, which requires more expensive instruments and consumables and also longer hands-on time and higher staff costs, is a major disadvantage of this method that results in higher per-test cost compared with quantitative PCR [39].
In a study of 77 suspected COVID-19 patients, digital PCR was shown to have a higher sensitivity (94% vs 40%), negative predictive value (63% vs 16%), and accuracy (95% vs 47%) compared with RT-PCR [40]. This advantage of digital PCR over RT-PCR was corroborated by other independent studies [41,42]. An investigation of 55 suspected COVID-19 cases who had had previous negative RT-PCR test results showed evidence of SARS-CoV-2 genome in the NP samples of 35% of the tested individuals when they were retested with digital PCR [43].
Isothermal nucleic acid amplification
In contrast to RT-PCR, which requires thermal cycling, this method amplifies the target sequence at a constant temperature and therefore enables nucleic acid amplification without the need for expensive thermal cycling equipment [44].
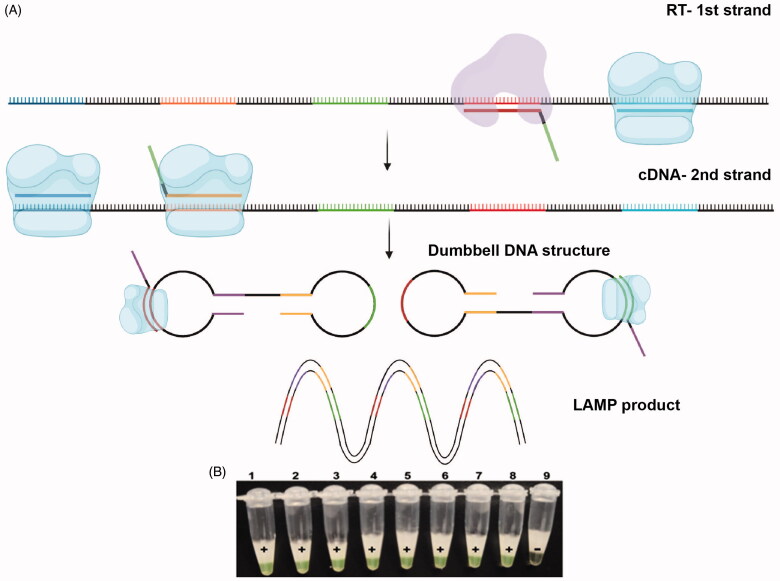
Reverse transcription loop-mediated isothermal amplification (RT-LAMP), which uses four to six sets of primers specifically designed to bind to distinct target regions, allows the identification of RNA sequences of interest [45,46] (Figure 3). The higher number of primers used in this method leads to an overall increase in the specificity of this assay [45]. Detection methods include measurement of turbidity caused by magnesium pyrophosphate precipitation during the reaction or use of fluorescent dyes [47]. RT-LAMP, which was used for the detection of the previous Middle East respiratory syndrome coronavirus (MERS-CoV) and severe acute respiratory syndrome coronavirus (SARS-CoV) global outbreaks, has also been shown to be an efficient diagnostic assay for SARS-CoV-2 [48–50]. In a study that analyzed 130 clinical specimens of individuals suspected to be infected with SARS-CoV-2 and compared the diagnostic accuracy of RT-LAMP with RT-PCR as the reference standard, RT-LAMP had a sensitivity of 100% (95% confidence interval [CI] 92.3-100%) and specificity of 100% (95% CI 93.7–100%); notably, the RT-LAMP assay detected the virus in a mean duration of 26.28 min while the RT-PCR assay required 1–2 h after viral RNA extraction [48].
Figure 3.
Schematic view of reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay. (A) Initially, the primer and reverse transcriptase (shown in purple) convert RNA to cDNA while at the same time the primer and DNA polymerase with strand displacing activity (shown in blue) make the second cDNA strand and release the first cDNA strand. The displaced single-stranded cDNA subsequently acts as a template for further extension reactions by other specific primers and DNA polymerases with strand displacing activity. The product subsequently self-anneals and forms dumbbell-shaped structures leading to subsequent rounds of exponential amplification. Reproduced and modified with permission from ref [46]. (B) The amplification process can be visually monitored by the color change of the fluorescent calcein from orange to green that indicates a positive result. Reprinted from [48] with permission from Elsevier.
Other techniques based on isothermal amplification include transcription-mediated amplification, rolling circle amplification, and circle-to-circle amplification [51–53]. The latter assay was able to detect sub-femtomolar synthetic SARS-CoV-2 complementary DNA within 100 min [54].
CRISPR-based assays
Clustered regularly interspaced short palindromic repeats (CRISPR) were initially discovered in Escherichia coli [55]. This system, which is the only known adaptive immune system identified in prokaryotes, recognizes and cleaves foreign RNA or DNA material in a sequence-specific way [56]. It is comprised of one or more CRISPR-associated endonuclease (Cas) systems that need base pairing between guide RNA or CRISPR RNA (crRNA) and the target sequence (DNA or RNA) located next to a proto-spacer adjacent motif for the identification and cleavage of the target nucleic acids [57]. Although the CRISPR-Cas system is widely known as a powerful tool for gene editing, the application of RNA-guided, RNA-targeting enzymes, namely CRISPR-Cas12a and CRISPR-Cas13a for recognition of specific sequences, has paved the way for the development of CRISPR-based diagnostic assays [58–60].
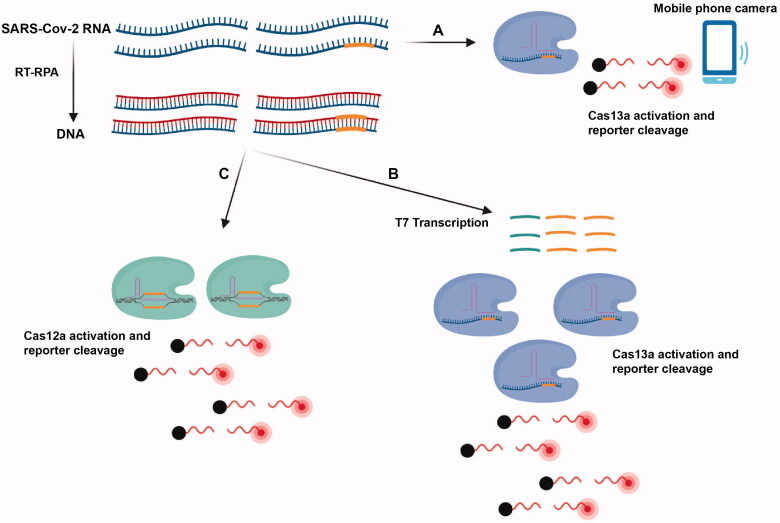
A platform termed Specific High Sensitivity Enzymatic Reporter UnLOCKing (SHERLOCK), which is based on the amplification of the target DNA or RNA followed by Cas13-mediated detection using colorimetric and fluorescent readouts, has ushered in rapid diagnostic assays for the detection of infectious agents in clinical specimens [61,62] (Figure 4). One assay with a fluorescent read-out and a viral RNA detection limit of 42 copies/reaction was clinically validated in a study of 154 NP and OP specimens and showed a sensitivity and specificity of 100% [63], while the sensitivity and specificity of the assay with a lateral flow readout were 97% and 100%, respectively [63].
Figure 4.
Overview of the CRISPR-based assays. (A) Amplification-free detection of the SARS-CoV-2 RNA using the Cas13a- crRNA complex and a mobile phone camera. In both the SHERLOCK (B) and DETECTR (C) assays, the viral RNA is converted to dsDNA using RT-recombinase polymerase amplification. Subsequently, in SHERLOCK, complementary RNA generated from this DNA template by T7 transcription is subsequently detected by Cas13: RNA complexes binding to the target sequence, which leads to the cleavage of fluorescent RNA molecules. In the DETECTR method, T7 transcription does not occur and target sequences on RT-RPA reaction products are detected directly by Cas12: RNA complexes.
SHERLOCK testing in a one pot (STOPCovid.v2) method, which combines magnetic bead purification for RNA extraction with isothermal amplification and Cas12b-mediated detection of SARS-CoV-2, has been shown to detect one-thirtieth of the viral load detected by the Centers for Disease Control and Prevention RT-qPCR test [64].
The SARS-CoV-2 DNA endonuclease-targeted CRISPR trans reporter (DETECTR) assay consists of pre-amplification of the E gene and N2 region of the N gene using RT-LAMP and subsequent cleavage of reporter RNA by Cas12a and visualization by lateral flow strip or a fluorescent reader [65]. In addition, another CRISPR-based assay that, unlike most CRISPR assays, does not depend on nucleic acid amplification has been developed. This assay, which was able to detect ∼100 copies/µL of viral SARS-CoV-2 in 30 min using CRISPR-Cas13a and a mobile phone camera, offers promise for rapid point of care (POC) SARS-CoV-2 CRISPR-based diagnostics (Figure 4) [66].
Nucleic acid microarray assays
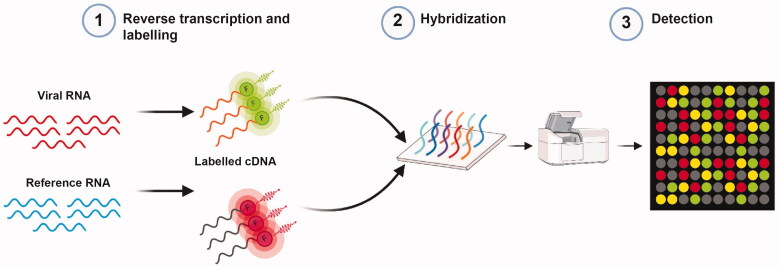
Microarray technologies combine robust nucleic acid amplification with the high throughput screening potential of microarray techniques and therefore enable detection of different microbial agents with high sensitivity and specificity [67]. Microarray techniques can be used to detect coronaviruses by generating cDNA from the viral RNA using reverse transcription and subsequent labeling with specific probes. It is then loaded into microarray wells coated with oligonucleotides that signal the presence of viral nucleic acid [68] (Figure 5).
Figure 5.
Overview of microarray assay workflow. Viral and reference RNA undergo reverse transcription to generate cDNA and differential fluorescent labeling. They are then mixed and transferred to microarray wells coated with highly specific probes that enable the fluorescent scan to provide the image for analysis.
Microarray assays have shown promising results in the detection of previous coronaviruses. A 60mer oligonucleotide microarray was able to detect the SARS-CoV genome [69]. A single nucleotide polymorphism (SNP) DNA microarray was shown to detect SNPs associated with the SARS-CoV S gene, which was related to its virus pathogenicity, with 100% accuracy [70]. Furthermore, a portable microarray-based diagnostic tool, mobile analysis platform (MAP), was able to detect respiratory viruses, including influenza A, influenza B, respiratory syncytial virus, and MERS-CoV, effectively [71].
Although the high cost of microarray assays has generally been the most significant impediment to their widespread use, nonfluorescent low-density microarray assays, which are less expensive and have been shown to have a sensitivity equal to that of RT-PCR in detecting multiple coronavirus strains, offer a glimmer of hope [72]. Considering the diversity and evolving nature of SARS-CoV-2 and emerging viral strains, microarray-based methods could be a useful tool in surveillance and timely identification of emerging strains [73,74].
Next generation sequencing
Next generation sequencing (NGS) has enabled genetic sequencing with a wide range of applications in different areas of medical research at an entirely unprecedented scale [75–79]. NGS allows comprehensive characterization and analysis of viral genetic material. Thanks to global sequencing efforts made possible by this method, over 380,000 SARS-CoV-2 genome sequences have so far been shared on the GISAID initiative (global initiative promoting rapid open-access sharing of the genetic sequencing data of influenza viruses and SARS-CoV-2), paving the way for real-time surveillance and monitoring of the pandemic as well as offering profound insight into the virus evolution and genomic epidemiology [80].
The major advantage of NGS is that it can screen for and identify viral agents without the need for previous knowledge of the causative agent [81,82]. Shotgun metagenomics (i.e. untargeted sequencing of all microbial genomes in a sample) facilitates the identification of viral strains that cannot otherwise be detected due to highly mutagenic regions that hinder the proper function of assays such as RT-PCR [83]. A study of 75 patients under investigation for SARS-CoV-2 that used RNA metagenomics next generation sequencing to assess other viral etiologies showed the enormous capability of this method in identifying other respiratory viruses in the setting of co-infection or infection due to other etiologies; in four patients who were infected with human coronavirus NL63, respiratory syncytial virus and human metapneumovirus, the causative agent was not detected by routine clinical testing for respiratory infections [84].
Despite the great advantages of metagenomics next generations sequencing, the massive amount of data generated, which in turn requires advanced data storage and processing infrastructure, limits its widespread use in the setting of an outbreak [85]. In addition, NGS platforms are much more expensive compared to other molecular diagnostic assays and involve intricate sample preparation and library construction protocols that require significant expertise. Furthermore, most current NGS assays cannot compete with other molecular diagnostic assays with respect to speed; it takes more than 18 h to perform the sequencing run alone on a standard Illumina sequencing platform (Illumina, San Diego CA) [86]. An alternative, focused approach is target enrichment [87]. Target enrichment using the Illumina respiratory virus oligo panel, one of the latest developments in this field, is designed to detect a wide range of respiratory viruses including SARS-CoV-2 [88].
Swab-Seq, another method that applies next generation sequencing to the diagnosis of SARS-COV-2, employs unique molecular barcodes embedded in RT-PCR primers to enable parallel sequencing of thousands of samples using very short reads in a single run [89].
Quality control
Although an increase in COVID-19 testing capacity is indispensable for containing the pandemic, the reliability and validity of the tests used are of utmost importance [90]. In the absence of adequate quality control in COVID-19 molecular testing, beginning from the test design stages up to their use in the clinical settings, the reliability of such test results and the subsequent clinical management of patients would be jeopardized. Important test quality parameters commonly assessed by regulatory agencies include sensitivity, specificity, positive predictive value and negative predictive value [91].
The quality and amount of viral RNA present in the samples are important factors that determine the analytical sensitivity of the test (i.e. lowest amount of analytical sample measurable by the assay) [92]. This in turn depends largely on the sample type and the site of collection [93]. In a meta-analysis of 8136 clinical specimens, the highest positive rate (91.8%) was seen in broncho-alveolar lavage fluid specimens. This was followed by rectal swabs and sputum specimens with positive rates of 87.8% and 68.1%, respectively. Notably in this study, NP swab specimens, which are the most widely used samples, had a positive rate of only 45.5% [94].
Another important consideration in COVID-19 molecular diagnostic assays is novel strains. Laboratories and diagnostic assay manufacturers must be vigilant about the emergence of novel virus strains that could affect the efficacy of their diagnostic assays. This goal can be attained by periodic evaluation of the primer sets against publicly available viral genome sequence databases to ensure their efficacy. Any mutation in the viral genome that would compromise the expected efficacy of molecular diagnostic assays should lead to their re-design and subsequent re-validation. A recent example is the 69/70del mutation found in the VOC 202012/01 variant that has been shown to affect the performance of PCR assays that target the S gene [95].
Discussion and conclusions
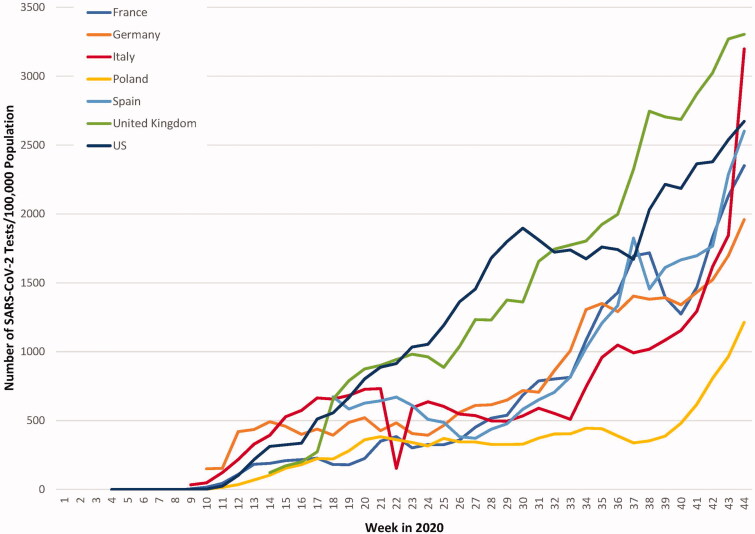
Timely and accurate identification of patients infected with SARS-CoV-2 plays a major role in the management of this pandemic. The appropriate use of testing in different clinical settings and in the various stages of infection can provide critical information necessary for clinical decision making and formulation of healthcare policy. Furthermore, beyond the clinical setting, molecular diagnostic assays have far-reaching applications in population-wide COVID-19 surveillance [96]. The SARS-CoV-2 RNA concentration in sewage has been shown to correlate with COVID-19 prevalence, highlighting the utility of sewage surveillance as a sensitive monitoring tool [97]. Despite the substantial increase in testing capacities in developed countries, there are still glaring disparities among these countries (Figure 6). The situation is even worse in developing countries that face shortages of equipment, advanced laboratory infrastructure, and trained personnel to carry out tests [98,99]. This has prompted the WHO to ask researchers to mobilize their efforts for the development of POC diagnostic assays that could be used at the community level [100].
Figure 6.
Weekly SARS-CoV-2 testing rates per 100,000 population. Although the weekly number of tests per 100,000 population for all the countries presented has increased during this period, there is a considerable disparity among countries. (Data extracted from The COVID Tracking Project COVID Tracking Project [https://covidtracking.com/data/national] and European Center for Disease Prevention and Control [https://www.ecdc.europa.eu/en/publications-data/covid-19-testing]).
Many rapid POC immunologic and molecular assays have been developed to address this urgent need. Overall, in different studies, rapid antigen assays have been shown to have considerable variation in sensitivity [average sensitivity 56.2%, 95% CI 29.5-79.8%)], while molecular assays were shown to have consistently higher sensitivity [average sensitivity 95.2%, 95% CI 86.7–98.3%)] [101]. However, even though serologic assays often require only basic equipment and are rapid, the long period between the onset of symptoms and the development of detectable antibody response (days to weeks) limits the utility of such assays in the diagnosis of acute SARS-CoV-2 infection [102]. This highlights the influential role of the molecular diagnostic assays discussed in this review.
At this time, RT-PCR is the most widely used molecular assay to diagnose COVID-19. However, because these assays require the transfer of clinical specimens to a laboratory with expert staff and because of the associated costs and slow turnaround times, researchers around the globe have been motivated to explore other molecular diagnostic assays that avoid these issues. Although isothermal amplification methods such as RT-LAMP seem attractive alternatives to RT-PCR because of their lower cost and rapid analysis time, nonspecific amplification due to the absence of temperature gating mechanisms may lead to false-positive results, raising potential concerns over the widespread implementation of such methods [103].
Prolonged detection of viral RNA after the resolution of symptoms has been reported by many researchers. However, it should be noted that detectable viral RNA does not necessarily indicate the presence of an infectious virus. A study of nine young- to middle-aged COVID-19 patients with no underlying disorders highlighted the importance of viral load; infectious virus could not be identified in clinical specimens with a viral RNA load of less than 106 copies/mL [104]. Considering the inability of RT-PCR to differentiate degraded noninfectious viral RNA from intact infectious virus, a focus on molecular assays with the potential to distinguish between these two entities could have wide-ranging implications, for the adverse economic consequences of keeping individuals in quarantine who test positive but are no longer infectious are irrefutable. Although the superior precision of digital PCR may not be clinically relevant in the context of quantification of SARS-CoV-2 RNA, the ability of droplet digital PCR to ascertain the integrity of the viral genome could be very useful in this regard [105,106].
In June 2020, the US Food and Drug Administration granted Emergency Use Authorization for the first COVID-19 NGS diagnostic assay [107]. Notwithstanding that the advanced technical requirements of NGS assays have been the major bottleneck in their widespread availability, exploring the untapped potential of NGS could pave the way for their use not only to detect SARS-CoV-2 but also to gauge the patient’s immune response to the infection. Recent studies have shed light on transcriptomic findings in patients infected with SARS-CoV-2 [108–110]. Considering the enormous number of human inflammatory cells in different clinical specimens (e.g. NP specimen), profiling the host transcriptome response using shotgun NGS could be used to tailor clinical management.
Another major obstacle in the wide-scale use of molecular diagnostic assays is the viral RNA extraction step; the supply chain has not been able to meet the surging demand for reagents and extraction kits, which has led to global shortages. Although many investigators have tried to come up with protocols to tackle this issue, this area remains a lively area of investigation [64,111,112]. Nanotechnology methods could have far-reaching applications in improving the efficiency of SARS-CoV-2 diagnostic assays [113–115]. Magnetic nanoparticles show promise for high sensitivity detection of viruses without the need for purification [116,117]. Nanotechnology can also facilitate the development of POC molecular testing assays. Gold nanoparticles, which have been previously used in CRISPR-based detection assays for other viral pathogens, can be used in SARS-CoV-2 POC molecular calorimetric assays [118]. This highlights the importance of nanotechnology in the development of cost-effective and efficient SARS-CoV-2 molecular diagnostic assays.
Overall, the delay in test result reporting that accompanies centralized laboratory SARS-CoV-2 testing has been the most significant challenge confronting healthcare systems globally.
Although the development of high quality laboratory assays is necessary to provide accurate diagnoses, this should not hinder the development of rapid diagnostic tests that are of cardinal importance during the early days of an outbreak. It should be noted that even tests that are less accurate and precise, yet are rapid and inexpensive, can play a substantial role in the fight against an outbreak during the initial days when high standard laboratory assays are under development [119]. Considering the wide range of POC molecular diagnostic assays available, they should play an important role in containing the pandemic, particularly in areas with limited resources. However, it should be noted that the reallocation of scarce resources in these areas should not undermine the surveillance efforts for other pathogens such as human immunodeficiency virus, Mycobacterium tuberculosis and malaria [120]. Collaboration between the clinicians, scientists and public health administrators in a multidisciplinary effort can undoubtedly achieve this goal.
The COVID-19 pandemic has led to unprecedented investment in the development of molecular diagnostic assays, particularly POC tests. Building on this invaluable experience, the scientific community can address a wide range of human diseases beyond coronavirus infection through further development of such assays.
Acknowledgments
We would like to thank Dr. Milad Sharifpour (Emory School of Medicine), and Dr. Paul Rutland (retired researcher, University College London), for their invaluable comments. The schematic representations used in the figures were created using BioRender (BioRender.com).
Glossary
Abbreviations
- CI
confidence interval
- COVID-19
coronavirus disease 2019
- Cas
CRISPR-associated endonuclease
- CRISPR
clustered regularly interspaced short palindromic repeats
- crRNA
CRISPR RNA
- DETECTR
DNA endonuclease-targeted CRISPR trans reporter
- MAP
mobile analysis platform
- MERS-CoV
Middle East respiratory syndrome coronavirus
- NGS
next generation sequencing
- NP
nasopharyngeal
- NW
nasal wash
- OP
oropharyngeal
- ORF
open reading frame
- PCR
polymerase chain reaction
- POC
point of care
- RT-LAMP
reverse transcription loop-mediated isothermal amplification
- RT-PCR
reverse transcription polymerase chain reaction
- SARS-CoV
severe acute respiratory syndrome coronavirus
- SHERLOCK
Specific High Sensitivity Enzymatic Reporter UnLOCKing
- SNP
single nucleotide polymorphism
- WHO
World Health Organization
Disclosure statement
The authors report no conflict of interest.
References
- 1.Habibzadeh P, Stoneman EK.. The novel coronavirus: a bird's eye view. Int J Occup Environ Med. 2020;11(2):65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scientific Brief: SARS-CoV-2 and Potential Airborne Transmission: CDC; 2020. [cited 2020 Nov 2]. Available from: https://www.cdc.gov/coronavirus/2019-ncov/more/scientific-brief-sars-cov-2.html
- 3.Wiersinga WJ, Rhodes A, Cheng AC, et al. . Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324(8):782–793. [DOI] [PubMed] [Google Scholar]
- 4.Arons MM, Hatfield KM, Reddy SC, et al. . Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N Engl J Med. 2020. May 28;382(22):2081–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai Y, Yao L, Wei T, et al. . Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020;323(14):1406–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.CDC’s Diagnostic Test for COVID-19 Only and Supplies: CDC; 2020. [updated 2020 Jul 15; 2020 Sep 30]. Available from: https://www.cdc.gov/coronavirus/2019-ncov/lab/virus-requests.html
- 7.Wu F, Zhao S, Yu B, et al. . A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu A, Peng Y, Huang B, et al. . Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malik YS, Sircar S, Bhat S, et al. . Emerging novel coronavirus (2019-nCoV)-current scenario, evolutionary perspective based on genome analysis and recent developments. Vet Q. 2020;40(1):68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sevajol M, Subissi L, Decroly E, et al. . Insights into RNA synthesis, capping, and proofreading mechanisms of SARS-coronavirus. Virus Res. 2014;194:90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fauver JR, Petrone ME, Hodcroft EB, et al. . Coast-to-coast spread of SARS-CoV-2 during the early epidemic in the United States. Cell. 2020;181(5):990.e5–996.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korber B, Fischer WM, Gnanakaran S, et al. . Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yurkovetskiy L, Wang X, Pascal KE, et al. . SARS-CoV-2 spike protein variant D614G increases infectivity and retains sensitivity to antibodies that target the receptor binding domain. bioRxiv. 2020. DOI: 10.1101/2020.07.04.187757 [DOI] [Google Scholar]
- 14.WHO. SARS-CoV-2 Variants: WHO ; 2020. Available from: https://www.who.int/csr/don/31-december-2020-sars-cov2-variants/en/
- 15.Leung K, Shum MH, Leung GM, et al. . Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom. Euro Surveill. 2021;26:2002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Interim guidelines for collecting, handling, and testing clinical specimens for COVID-19: C DC; 2020. [updated 2020 Jul 8]. Available from: https://www.cdc.gov/coronavirus/2019-nCoV/lab/guidelines-clinical-specimens.html
- 17.Wang W, Xu Y, Gao R, et al. . Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen J. Chinese researchers reveal draft genome of virus implicated in Wuhan pneumonia outbreak. Science; 2020. Available from: https://www.sciencemag.org/news/2020/01/chinese-researchers-reveal-draft-genome-virus-implicated-wuhan-pneumonia-outbreak
- 19.Park M, Won J, Choi BY, et al. . Optimization of primer sets and detection protocols for SARS-CoV-2 of coronavirus disease 2019 (COVID-19) using PCR and real-time PCR. Exp Mol Med. 2020;52(6):963–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corman VM, Landt O, Kaiser M, et al. . Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.VanGuilder HD, Vrana KE, Freeman WM.. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques. 2008;44(5):619–626. [DOI] [PubMed] [Google Scholar]
- 22.Zhou P, Yang XL, Wang XG, et al. . A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chu DKW, Pan Y, Cheng SMS, et al. . Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem. 2020;66(4):549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi J, Han D, Zhang R, et al. . Molecular and serological assays for SARS-CoV-2: insights from genome and clinical characteristics. Clin Chem. 2020;66(8):1030–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan JF, Yip CC, To KK, et al. . Improved molecular diagnosis of COVID-19 by the Novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J Clin Microbiol. 2020;58(5):e00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Accelerated Emergency Use Authorization (EUA) Summary Orig3n 2019 Novel Coronavirus (COVID-19) Test (ORIG3N, INC.) FDA; 2020. Available from: https://www.fda.gov/media/136873/download
- 27.Lohse S, Pfuhl T, Berko-Gottel B, et al. . Pooling of samples for testing for SARS-CoV-2 in asymptomatic people. Lancet Infect Dis. 2020;20(11):P1231–P1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garg J, Singh V, Pandey P, et al. . Evaluation of sample pooling for diagnosis of COVID-19 by Real time PCR- A resource saving combat strategy. J Med Virol. 2021;93(3):1526–1531. [DOI] [PubMed] [Google Scholar]
- 29.Pooled Sample Testing and Screening Testing for COVID-19: FDA; 2020. Available from: https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/pooled-sample-testing-and-screening-testing-covid-19
- 30.Tahamtan A, Ardebili A.. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev Mol Diagn. 2020. May;20(5):453–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Habibzadeh P, Sajadi MM, Emami A, et al. . Rate of re-positive RT-PCR test among patients recovered from COVID-19. Biochem Med (Zagreb). 2020;30(3):030401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quan P-L, Sauzade M, Brouzes E.. A technology review. Sensors. 2018;18(4):1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salipante SJ, Jerome KR.. Digital PCR-an emerging technology with broad applications in microbiology. Clin Chem. 2020;66(1):117–123. [DOI] [PubMed] [Google Scholar]
- 34.Hindson CM, Chevillet JR, Briggs HA, et al. . Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 2013;10(10):1003–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svec D, Tichopad A, Novosadova V, et al. . How good is a PCR efficiency estimate: recommendations for precise and robust qPCR efficiency assessments. Biomol Detect Quantif. 2015;3:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dingle TC, Sedlak RH, Cook L, et al. . Tolerance of droplet-digital PCR vs real-time quantitative PCR to inhibitory substances. Clin Chem. 2013;59(11):1670–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoffman NG, Cook L, Atienza EE, et al. . Marked variability of BK virus load measurement using quantitative real-time PCR among commonly used assays. J Clin Microbiol. 2008;46(8):2671–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whale AS, Huggett JF, Tzonev S.. Fundamentals of multiplexing with digital PCR. Biomol Detect Quantif. 2016;10:15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuypers J, Jerome KR.. Applications of digital PCR for clinical microbiology. J Clin Microbiol. 2017;55(6):1621–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suo T, Liu X, Feng J, et al. . ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg Microbes Infect. 2020;9(1):1259–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Falzone L, Musso N, Gattuso G, et al. . Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection. Int J Mol Med. 2020;46(3):957–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu X, Feng J, Zhang Q, et al. . Analytical comparisons of SARS-COV-2 detection by qRT-PCR and ddPCR with multiple primer/probe sets. Emerg Microbes Infect. 2020;9(1):1175–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alteri C, Cento V, Antonello M, et al. . Detection and quantification of SARS-CoV-2 by droplet digital PCR in real-time PCR negative nasopharyngeal swabs from suspected COVID-19 patients. PLoS One. 2020;15(9):e0236311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Craw P, Balachandran W.. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: a critical review. Lab Chip. 2012;12(14):2469–2486. [DOI] [PubMed] [Google Scholar]
- 45.Notomi T, Okayama H, Masubuchi H, et al. . Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kellner MJ, Ross JJ, Schnabl J, et al. . A rapid, highly sensitive and open-access SARS-CoV-2 detection assay for laboratory and home testing. bioRxiv. 2020. DOI: 10.1101/2020.06.23.166397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mori Y, Nagamine K, Tomita N, et al. . Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun. 2001;289(1):150–154. [DOI] [PubMed] [Google Scholar]
- 48.Yan C, Cui J, Huang L, et al. . Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin Microbiol Infect. 2020;26(6):773–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shirato K, Yano T, Senba S, et al. . Detection of Middle East respiratory syndrome coronavirus using reverse transcription loop-mediated isothermal amplification (RT-LAMP). Virol J. 2014;11:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hong TC, Mai QL, Cuong DV, et al. . Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J Clin Microbiol. 2004;42(5):1956–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kacian DL, Fultz TJ. inventors; Gen-Probe Inc, assignee. Kits for nucleic acid sequence amplification methods. United States patent US5888779A. 1999. Available from: https://patentimages.storage.googleapis.com/02/21/aa/e06896e20b88c1/US5888779.pdf
- 52.Ali MM, Li F, Zhang Z, et al. . Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine. Chem Soc Rev. 2014;43(10):3324–3341. [DOI] [PubMed] [Google Scholar]
- 53.Dahl F, Baner J, Gullberg M, et al. . Circle-to-circle amplification for precise and sensitive DNA analysis. Proc Natl Acad Sci USA. 2004;101(13):4548–4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tian B, Gao F, Fock J, et al. . Homogeneous circle-to-circle amplification for real-time optomagnetic detection of SARS-CoV-2 RdRp coding sequence. Biosens Bioelectron. 2020;165:112356. [DOI] [PubMed] [Google Scholar]
- 55.Ishino Y, Shinagawa H, Makino K, et al. . Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169(12):5429–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hille F, Charpentier E.. CRISPR-Cas: biology, mechanisms and relevance. Philos Trans R Soc Lond B Biol Sci. 2016;371(1707):20150496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pickar-Oliver A, Gersbach CA.. The next generation of CRISPR-Cas technologies and applications. Nat Rev Mol Cell Biol. 2019;20(8):490–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gootenberg JS, Abudayyeh OO, Lee JW, et al. . Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen JS, Ma E, Harrington LB, et al. . CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Myhrvold C, Freije CA, Gootenberg JS, et al. . Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360(6387):444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kellner MJ, Koob JG, Gootenberg JS, et al. . SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc. 2019;14(10):2986–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang F, Abudayyeh OO, Gootenberg JS. A protocol for detection of COVID-19 using CRISPR diagnostics (v.20200321); 2020. Available from: https://www.broadinstitute.org/files/publications/special/COVID-19%20detection%20(updated).pdf
- 63.Patchsung M, Jantarug K, Pattama A, et al. . Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat Biomed Eng. 2020;4(12):1140–1149. [DOI] [PubMed] [Google Scholar]
- 64.Joung J, Ladha A, Saito M, et al. . Detection of SARS-CoV-2 with SHERLOCK one-pot testing. N Engl J Med. 2020;383(15):1492–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Broughton JP, Deng X, Yu G, et al. . CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38(7):870–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fozouni P, Son S, Díaz de León Derby M, et al. . Direct detection of SARS-CoV-2 using CRISPR-Cas13a and a mobile phone. medRxiv. 2020. DOI: 10.1101/2020.09.28.20201947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vora GJ, Meador CE, Stenger DA, et al. . Nucleic acid amplification strategies for DNA microarray-based pathogen detection. Appl Environ Microbiol. 2004;70(5):3047–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen Q, Li J, Deng Z, et al. . Comprehensive detection and identification of seven animal coronaviruses and human respiratory coronavirus 229E with a microarray hybridization assay. Intervirology. 2010;53(2):95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shi R, Ma W, Wu Q, et al. . Design and application of 60mer oligonucleotide microarray in SARS coronavirus detection. Chin Sci Bull. 2003;48(12):1165–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guo X, Geng P, Wang Q, et al. . Development of a single nucleotide polymorphism DNA microarray for the detection and genotyping of the SARS coronavirus. J Microbiol Biotechnol. 2014;24(10):1445–1454. [DOI] [PubMed] [Google Scholar]
- 71.Hardick J, Metzgar D, Risen L, et al. . Initial performance evaluation of a spotted array Mobile Analysis Platform (MAP) for the detection of influenza A/B, RSV, and MERS coronavirus. Diagn Microbiol Infect Dis. 2018;91(3):245–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Souza Luna LK, Heiser V, Regamey N, et al. . Generic detection of coronaviruses and differentiation at the prototype strain level by reverse transcription-PCR and nonfluorescent low-density microarray. J Clin Microbiol. 2007;45(3):1049–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pachetti M, Marini B, Benedetti F, et al. . Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J Transl Med. 2020;18(1):179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Benedetti F, Snyder GA, Giovanetti M, et al. . Emerging of a SARS-CoV-2 viral strain with a deletion in nsp1. J Transl Med. 2020;18(1):329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Habibzadeh P, Honarvar B, Silawi M, et al. . Association between rs2303861 polymorphism in CD82 gene and non-alcoholic fatty liver disease: a preliminary case-control study. Croat Med J. 2019;60(4):361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Habibzadeh P, Inaloo S, Silawi M, et al. . A novel TTC19 mutation in a patient with neurological, psychological, and gastrointestinal impairment. Front Neurol. 2019;10:944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Habibzadeh P, Silawi M, Dastsooz H, et al. . Clinical and molecular characterization of a patient with mitochondrial Neurogastrointestinal Encephalomyopathy. BMC Gastroenterol. 2020;20(1):142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Muhamad Rizal NS, Neoh HM, Ramli R, et al. . Advantages and limitations of 16S rRNA next-generation sequencing for pathogen identification in the diagnostic microbiology laboratory: perspectives from a middle-income country. Diagnostics (Basel). 2020;10(10):816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Habibzadeh P, Tabatabaei Z, Inaloo S, et al. . Case report: expanding the genetic and phenotypic spectrum of autosomal recessive spastic ataxia of Charlevoix-Saguenay. Front Genet. 2020;11:585136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.GISAID; 2020. [updated 2021 Jan 16]. Available from: https://www.gisaid.org/
- 81.Hayes S, Mahony J, Nauta A, et al. . Metagenomic approaches to assess bacteriophages in various environmental niches. Viruses. 2017;9(6):127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nooij S, Schmitz D, Vennema H, et al. . Overview of virus metagenomic classification methods and their biological applications. Front Microbiol. 2018;9:749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Quince C, Walker AW, Simpson JT, et al. . Shotgun metagenomics, from sampling to analysis. Nat Biotechnol. 2017;35(9):833–844. [DOI] [PubMed] [Google Scholar]
- 84.Babiker A, Bradley HL, Stittleburg VD, et al. . Metagenomic sequencing to detect respiratory viruses in persons under investigation for COVID-19. J Clin Microbiol. 2020. DOI: 10.1128/JCM.02142-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hillmann B, Al-Ghalith GA, Shields-Cutler RR, et al. . Evaluating the Information content of shallow shotgun metagenomics. mSystems. 2018. DOI: 10.1128/mSystems.00069-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chiu CY, Miller SA.. Clinical metagenomics. Nat Rev Genet. 2019;20(6):341–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kiselev D, Matsvay A, Abramov I, et al. . Current trends in diagnostics of viral infections of unknown etiology. Viruses. 2020;12(2):211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Detection and characterization of respiratory viruses, including SARS-CoV-2, using Illumina RNA Prep with Enrichment: Illumina; 2020. [cited 2020]. Available from: https://emea.illumina.com/content/dam/illumina/gcs/assembled-assets/marketing-literature/illumina-rna-enrichment-coronavirus-app-note-470-2020-005/illumina-rna-enrichment-coronavirus-app-note-470-2020-005.pdf
- 89.Bloom JS, Jones EM, Gasperini M, et al. . Swab-Seq: a high-throughput platform for massively scaled up SARS-CoV-2 testing. medRxiv. 2020. DOI: 10.1101/2020.08.04.20167874 [DOI] [Google Scholar]
- 90.Bohn MK, Mancini N, Loh TP, et al. . IFCC Interim guidelines on molecular testing of SARS-CoV-2 Infection. Clin Chem Lab Med. 2020;58(12):1993–2000. [DOI] [PubMed] [Google Scholar]
- 91.Habibzadeh F, Habibzadeh P, Yadollahie M, et al. . On the information hidden in a classifier distribution. Sci Rep. 2021;11(1):917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vandenberg O, Martiny D, Rochas O, et al. . Considerations for diagnostic COVID-19 tests. Nat Rev Microbiol. 2020. DOI: 10.1038/s41579-020-00461-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lieberman JA, Pepper G, Naccache SN, et al. . Comparison of commercially available and laboratory-developed assays for in vitro detection of SARS-CoV-2 in clinical laboratories. J Clin Microbiol. 2020. DOI: 10.1128/JCM.00821-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bwire GM, Majigo MV, Njiro BJ, et al. . Detection profile of SARS-CoV-2 using RT-PCR in different types of clinical specimens: a systematic review and meta-analysis. J Med Virol. 2021;93(2):719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mutant coronavirus in the United Kingdom sets off alarms, but its importance remains unclear: Science magazine ; 2020. Available from: https://www.sciencemag.org/news/2020/12/mutant-coronavirus-united-kingdom-sets-alarms-its-importance-remains-unclear
- 96.Daughton CG. Wastewater surveillance for population-wide Covid-19: the present and future. Sci Total Environ. 2020;736:139631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Medema G, Heijnen L, Elsinga G, et al. . Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ Sci Technol Lett. 2020;7(7):511–516. [DOI] [PubMed] [Google Scholar]
- 98.Giri AK, Rana DR.. Charting the challenges behind the testing of COVID-19 in developing countries: Nepal as a case study. Biosaf Health. 2020;2(2):53–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Abdullahi IN, Emeribe AU, Akande AO, et al. . Roles and challenges of coordinated public health laboratory response against COVID-19 pandemic in Africa. J Infect Dev Ctries. 2020;14(7):691–695. [DOI] [PubMed] [Google Scholar]
- 100.COVID-19 Public Health Emergency of International Concern (PHEIC) global research and innovation forum towards a research roadmap: WHO; 2020. [cited 2020 Oct 30]. Available from: https://www.who.int/publications/m/item/covid-19-public-health-emergency-of-international-concern-(pheic)-global-research-and-innovation-forum
- 101.Dinnes J, Deeks JJ, Adriano A, et al. . Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev. 2020;(8):CD013705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang W, Du RH, Li B, et al. . Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9(1):386–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rolando JC, Jue E, Barlow JT, et al. . Real-time kinetics and high-resolution melt curves in single-molecule digital LAMP to differentiate and study specific and non-specific amplification. Nucleic Acids Res. 2020;48(7):e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wolfel R, Corman VM, Guggemos W, et al. . Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. [DOI] [PubMed] [Google Scholar]
- 105.Hindson BJ, Ness KD, Masquelier DA, et al. . High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83(22):8604–8610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Regan JF, Kamitaki N, Legler T, et al. . A rapid molecular approach for chromosomal phasing. PLoS One. 2015;10(3):e0118270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Regan JF, Kamitaki N, Legler T, et al. . First NGS-based COVID-19 diagnostic. Nat Biotechnol. 2020;38(7):777. [DOI] [PubMed] [Google Scholar]
- 108.Butler DJ, Mozsary C, Meydan C, et al. . Shotgun transcriptome and isothermal profiling of SARS-CoV-2 infection reveals unique host responses, viral diversification, and drug interactions. bioRxiv. 2020. DOI: 10.1101/2020.04.20.048066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu M, Chen Y, Xia H, et al. . Transcriptional and proteomic insights into the host response in fatal COVID-19 cases. Proc Natl Acad Sci USA. 2020;117(45):28336–28343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xiong Y, Liu Y, Cao L, et al. . Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect. 2020;9(1):761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Miranda JP, Osorio J, Videla M, et al. . Analytical and clinical validation for RT-qPCR detection of SARS-CoV-2 without RNA extraction. Front Med. 2020;7:567572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fomsgaard AS, Rosenstierne MW.. An alternative workflow for molecular detection of SARS-CoV-2 – escape from the NA extraction kit-shortage, Copenhagen, Denmark. Euro Surveill. 2020;25(14):2000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Munir S, Ahmed S, Ibrahim M, et al. . A spellbinding interplay between biological barcoding and nanotechnology. Front Bioeng Biotechnol. 2020;8:883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen L, Liang J.. An overview of functional nanoparticles as novel emerging antiviral therapeutic agents. Mater Sci Eng C Mater Biol Appl. 2020;112:110924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cheong J, Yu H, Lee CY, et al. . Fast detection of SARS-CoV-2 RNA via the integration of plasmonic thermocycling and fluorescence detection in a portable device. Nat Biomed Eng. 2020;4(12):1159–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Perez JM, Josephson L, Weissleder R.. Use of magnetic nanoparticles as nanosensors to probe for molecular interactions. Chembiochem. 2004;5(3):261–264. [DOI] [PubMed] [Google Scholar]
- 117.Ganganboina AB, Chowdhury AD, Khoris IM, et al. . Hollow magnetic-fluorescent nanoparticles for dual-modality virus detection. Biosens Bioelectron. 2020;170:112680. [DOI] [PubMed] [Google Scholar]
- 118.Zhou R, Li Y, Dong T, et al. . A sequence-specific plasmonic loop-mediated isothermal amplification assay with orthogonal color readouts enabled by CRISPR Cas12a. Chem Commun (Camb). 2020;56(24):3536–3538. [DOI] [PubMed] [Google Scholar]
- 119.Ramdas K, Darzi A, Jain S. 'Test, re-test, re-test': using inaccurate tests to greatly increase the accuracy of COVID-19 testing. Nat Med. 2020;26(6):810–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Venkatesan P. COVID-19 diagnostics-not at the expense of other diseases. Lancet Microbe. 2020;1(2):e64. [DOI] [PMC free article] [PubMed] [Google Scholar]