Abstract
Microglia are specialized macrophages of the central nervous system (CNS) and first to react to pathogens or injury. Over the last decade, transcriptional profiling of microglia significantly contributed to our understanding of their functions. In the case of human CNS samples, either potential CNS pathology in the case of surgery samples, or a postmortem delay (PMD) due to the time needed for tissue access and collection, are potential factors that affect gene expression profiles. To determine the effect of PMD on the microglia transcriptome, we first analyzed mouse microglia, where genotype, antemortem conditions and PMD can be controlled. Microglia were isolated from mice after different PMDs (0, 4, 6, 12, and 24 hr) using fluorescence‐activated cell sorting (FACS). The number of viable microglia significantly decreased with increasing PMD, but even after a 12 hr PMD, high‐quality RNA could be obtained. PMD had very limited effect on mouse microglia gene expression, only 50 genes were differentially expressed between different PMDs. These genes were related to mitochondrial, ribosomal, and protein binding functions. In human microglia transcriptomes we previously generated, 31 of the 50 PMD‐associated mouse genes had human homologs, and their relative expression was also affected by PMD. This study provides a set of genes that shows relative expression changes in relation to PMD, both in mouse and human microglia. Although the gene expression changes detected are subtle, these genes need to be accounted for when analyzing microglia transcriptomes generated from samples with variable PMDs.
Keywords: gene expression profiling, human, microglia, mouse, postmortem delay
Main Points
Postmortem delay affects microglia gene expression to a limited extent.
The effect of postmortem delay on the mouse and human microglia transcriptome is comparable.
Affected genes decrease in expression with increasing postmortem delay.
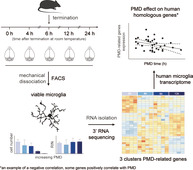
1. INTRODUCTION
Postmortem human brain tissue is a valuable resource to study the etiology and pathology of neurological disorders and neurodegenerative diseases. However, when dealing with such human brain tissue, variations in antemortem conditions and postmortem delay (PMD) are inevitable. As degradation of RNA was shown to result in false positive observations in RNA sequencing (Gallego Romero, Pai, Tung, & Gilad, 2014; Sigurgeirsson, Emanuelsson, & Lundeberg, 2014), successfully isolating high quality mRNA is of paramount importance.
Previous research showed conflicting results on how PMD affects the stability of RNA in brain tissue. In some studies, total RNA was shown to be well‐preserved in postmortem brain tissue, and RNA degradation did not correlate with PMD (Cummings, Strum, Yoon, Szymanski, & Hulette, 2001; Ervin et al., 2007; Johnson, Morgan, & Finch, 1986; Kobayashi et al., 1990; Robinson et al., 2016), but rather with other factors such as tissue handling, brain pH (homogenate) and agonal state (Durrenberger et al., 2010; Harrison et al., 1995; Preece & Cairns, 2003; Robinson et al., 2016). In addition, degradation of selective gene transcripts was monitored from brain tissues with different postmortem intervals by PCR, and no correlation between degradation of specific mRNAs and postmortem interval were detected for the PMDs investigated (Heinrich, Matt, Lutz‐Bonengel, & Schmidt, 2007; Kobayashi et al., 1990; Koppelkamm, Vennemann, Lutz‐Bonengel, Fracasso, & Vennemann, 2011).
However, these studies were all based on the analysis of a restricted set of transcripts. Therefore, it is essential to address the effect of PMD on mRNAs genome‐wide with microarrays or RNA‐Seq, to determine whether PMD affects a subset of gene transcripts that are potentially more susceptible to PMD‐related degradation. In a microarray study of PMD influence on mRNA degradation in mouse brain tissue, a subgroup of mRNAs was identified with a 3′ untranslated region (3′UTR) AUUUA motif that was more susceptible to PMD‐related RNA degradation (Catts et al., 2005). In another microarray study, a PMD less than 4 hr at room temperature (RT) resulted in a gene expression profile that highly correlated with that from mouse brains without PMD. However, a PMD longer than 8 hr at RT was shown to reduce this correlation of gene expression (Trotter, Brill, & Bennett, 2002). For human brain tissue, a PCR‐based array was used to determine gene expression levels in 79 frozen cerebellar cortex samples with different postmortem intervals. The expression level of a great proportion of genes decreased with PMD (65 out of 89) (Birdsill, Walker, Lue, Sue, & Beach, 2011). In contrast, several microarray studies showed that, at least for the studied intervals, PMD had no significant effect on RNA integrity and the expression profile of human brain tissues (Popova, Mennerich, Weith, & Quast, 2008; Tomita et al., 2004). It is postulated that PMD‐related mRNA degradation is tissue‐specific, gene‐specific, and even genotype‐dependent (Ferreira et al., 2018; Zhu, Wang, Yin, & Yang, 2017). Although the specific reason is unknown, PMD has limited impact on the transcriptomes of human brain compared to other tissues (Ferreira et al., 2018; Zhu et al., 2017).
Microglia are the tissue‐resident macrophages of the central nervous system (CNS) and the first to respond during CNS dysfunction and disease. Fluorescence‐activated cell sorting (FACS) enables isolation of microglia from fresh postmortem brain tissue. Over the last decade, gene expression profiling of microglia has greatly contributed to our understanding and characterization of these cells, both under normal and disease conditions (Gerrits, Heng, Boddeke, & Eggen, 2020). However, it is unknown whether PMD affects microglia transcriptomes, and it is important to determine how faithfully microglia isolated from samples with PMD reflect acutely isolated microglia. Here, we isolated microglia using FACS from mice after different postmortem intervals (0, 4, 6, 12, and 24 hr). PMD led to an extensive reduction in the number of viable (DAPIneg) microglia that could be FACS‐sorted from mouse brains. However, high quality RNA was still isolated from the remaining microglia, irrespective of PMD. Overall, PMD had a limited effect on microglia gene expression in mice, but the relative abundance of a set of genes was changed after PMD. This mouse PMD‐related microglia gene set was further investigated in our previously generated postmortem human microglia transcriptome dataset (Galatro et al., 2017). We observed that PMD also affected the expression of a large proportion of these genes in the human microglia transcriptome.
2. MATERIALS AND METHODS
2.1. Animals
Male C57BL/6 mice (22–25 g, Envigo, The Netherlands) between 8 and 10 weeks of age were used in all experiments. Mice were raised on a 12 hr light/dark cycle with food and water ad libitum and were housed in groups of four per cage. All experiments were performed in the Central Animal Facility (CDP) of the UMCG, approved by the National Central Authority for Scientific Procedures on Animals (CCD, The Netherlands) and the Animal Care and Use Committee (DEC) of the University of Groningen. Mice were terminated by cervical dislocation and left at RT for 0, 4, 6, 12, and 24 hr.
2.2. Microglia isolation from mouse brain
Microglia were isolated from adult mouse brain using the protocol as described before (Galatro, Vainchtein, Brouwer, Boddeke, & Eggen, 2017). Briefly, brains were isolated and triturated using a tissue homogenizer. Homogenized brain samples were passed through a 70 μM cell strainer to obtain single cell suspensions. Cells were centrifuged at 220g for 10 min at 4°C and the pellets were resuspended in 24% Percoll (GE Healthcare, 17‐0891‐01) gradient buffer. Of note, 3 ml PBS was pipetted onto the gradient buffer and myelin was removed by centrifuging at 950g for 20 min at 4°C (accelerate 4 and brake 0). Cell pellets were incubated with the antibodies CD11b‐PE (eBioscience, 12‐0112‐82), CD45‐FITC (eBioscience, 11‐0451‐85), and Ly6C‐APC (Biolegend, 128025). Microglia were FACS‐sorted as DAPIneg CD11bhigh CD45int Ly6Cneg events.
2.3. Library preparation
Total RNA was isolated from sorted microglia using the RNeasy Plus Micro Kit (Qiagen, 74034). RNA quantity and quality were analyzed using an Experion electrophoresis system (Bio‐Rad Laboratories, Hercules, CA). Sequencing libraries were prepared with the QuantSeq 3′ mRNA‐Seq Library Prep Kit FWD (Lexogen, 015.96).
2.4. RNA‐Seq data analysis
Read trimming was first performed with Trim Galore (version 0.4.5). Next, trimmed reads were aligned to the mouse genome (GRCm38.92) using HISAT2 (version 2.1.0; Kim, Langmead, & Salzberg, 2015; Kim, Paggi, Park, Bennett, & Salzberg, 2019). FeatureCounts of the Subread package (version 1.6.2) was used for reads summarization (Liao, Smyth, & Shi, 2014). Raw read counts were imported into R and normalized using the Bioconductor package DESeq2 package (Love, Huber, & Anders, 2014). Lowly expressed genes (rowSums <1) were filtered out. Since mouse samples were generated from two separated isolations, data was normalized using ComBat (Johnson, Li, & Rabinovic, 2007). This method uses an empirical Bayesian framework that can used to reduce known batch effects. Next, differentially expressed genes were identified with the rowttests function from the genefilter package (version 1.66.0), a function that compares two conditions and returns the mean difference and respective p‐value. The function qvalue (version 2.16.0) was performed to calculate the q‐value from the obtained p‐values. Genes with a q‐value <0.1 were considered to be significant and were used to be visualized in a heatmap. Hierarchical clustering on the subset was used to determine the number of clusters. Gene ontology (GO) term enrichment analysis was performed using Metascape (http://www.metascape.org/).
2.5. Statistical analysis
Statistical significance was determined by either a one‐way ANOVA followed by Bonferroni correction or an unpaired t‐test, as indicated in the legends. The analyses of the correlation between PMD time and median counts of PMD‐related genes (excluding MT‐CYTB and MT‐ND1), as well as the correlation between PMD time and average counts of MT‐CYTB and MT‐ND1, were done using Pearson correlation using Graphpad Prism v.6 (Graphpad Software, La Jolla, CA). The significance level for all tests was set at p < .05.
2.6. Data availability
All next‐generation sequencing data can be viewed at NCBI GEO under accession number GSE162209.
3. RESULTS
3.1. The effect of PMD on mouse microglia numbers and RNA quality
To determine the effect of PMD on the mouse microglia transcriptome, microglia were FACS‐sorted as Ly6Cneg CD11bhigh CD45intDAPIneg events from mice with PMDs of 0, 4, 6, 12, and 24 hr (Figure 1a). The number of FACS‐isolated viable (DAPIneg) microglia significantly decreased with increasing PMD (Figure 1b). The number of microglia isolated from mice after a 24 hr PMD was so low (average number of 3,970), that this group was not included in subsequent gene expression profiling and analysis. RNA was isolated from 0, 4, 6, and 12 hr PMD microglia. Remarkably, up to 12 hr, PMD had no effect on the integrity of the RNA isolated from DAPIneg Ly6Cneg CD11bhigh CD45int microglia (Figure 1c).
FIGURE 1.
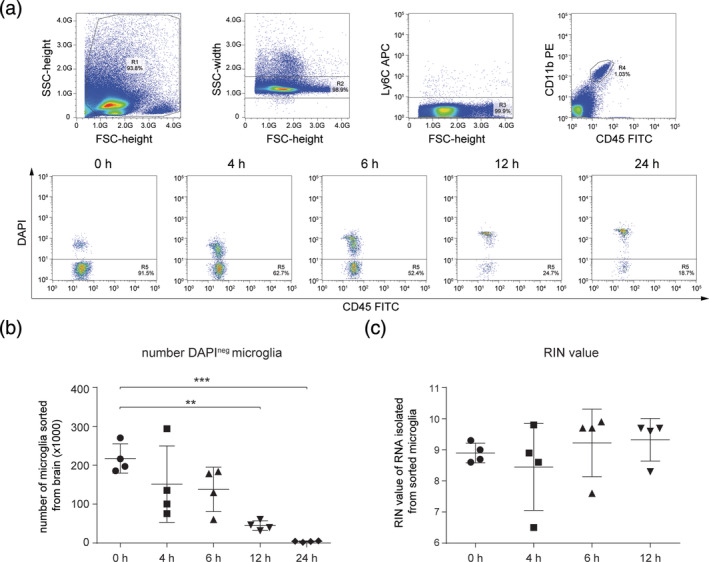
The effects of postmortem delay (PMD) on viable fluorescence‐activated cell sorting (FACS)‐sorted mouse microglia numbers and RNA quality. (a) FACS plots depicting the sorting strategy of DAPIneg Ly6Cneg CD11bhigh CD45int microglia and the PMD induced decrease in the percentage of DAPIneg microglia in gate R5. (b) Dot plot depicting the number of DAPIneg microglia sorted from entire mouse brains at different PMD times (n = 4 mice per PMD). (c) Dot plot depicting RIN values of RNA extracted from sorted DAPIneg Ly6Cneg CD11bhigh CD45int microglia at different PMD times (n = 4 mice per PMD). A one‐way ANOVA followed by a Bonferroni correction for multiple comparisons was performed to assess significance. **, p < .01; ***, p < .001
3.2. The effect of PMD on the mouse microglia transcriptome
To determine the effect of PMD on microglia gene expression, we performed RNA‐Seq on microglia RNA samples with four postmortem time intervals: 0, 4, 6, and 12 hr. Principal component analysis (PCA) indicated that gene expression profiles of samples with a PMD of 0 and 4 hr were similar, but the 6 and 12 hr PMD samples clearly segregated, indicative of an altered transcriptome (Figure 2a).
FIGURE 2.
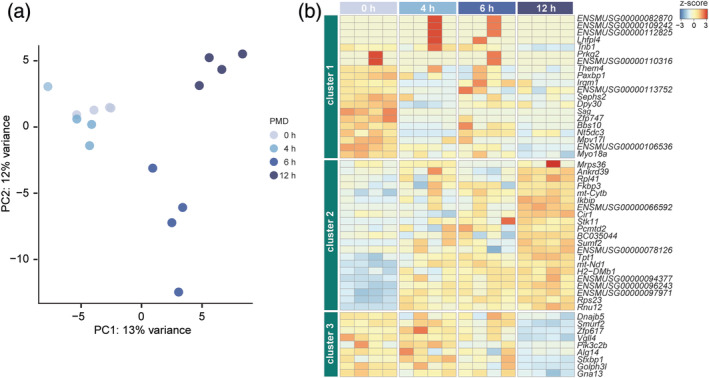
The effect of postmortem delay (PMD) on the mouse microglia transcriptome. (a) PCA analysis of microglia samples with different PMD. Each dot represents a mouse (n = 4 per PMD interval). (b) Heatmap depicting row z‐scores of 50 PMD‐related genes in mouse microglia identified by pairwise comparisons with a cutoff q‐value < .1. Unsupervised hierarchical clustering resulted in three gene clusters
Next, differential gene expression analysis by pairwise comparisons was performed. Genes that were differentially expressed (q‐value < 0.1) in at least one of the comparisons were regarded as PMD‐related genes. In total, out of 15,554 detected genes, we identified 50 PMD‐related genes (Table S1). To identify groups of genes with similar expression patterns, unsupervised hierarchical clustering of these 50 genes was performed, and three clusters were identified (Figure 2b). Genes in Cluster 1 were most abundantly expressed in the 0 hr PMD samples (Figure 2b). GO analysis indicated these genes were related to membrane organization (GO:0061024). In addition, the relative expression of Them4, Irgm1, Dpy30, Zfp747, and Bbs10 decreased with increasing PMD and these genes are related to protein binding (GO:0005515). Sag is related to phosphoprotein binding (GO:0051219) and opsin binding (GO:0002046). Myo18a is related to actin filament binding (GO:0003779). Decreased relative abundance of these genes suggested PMD influenced microglia membrane organization. The relative expression of genes in Cluster 2 increased with PMD (Figure 2b). This cluster consisted of several genes that are related to mitochondrial and ribosomal functions, such as Mrps36, Rpl41, mt‐Cytb, mt‐Nd1, and Rps23. Interestingly, several genes related to protein binding (GO:0005515) were also identified in this cluster but their relative expression increased with PMD: Ankrd39, Fkbp3, Ikbip, Cir1, Stk11, and Tpt1. Genes in Cluster 3 also showed a relative decrease in relative expression with increasing PMD, and this decrease was most prominent at 12 hr PMD (Figure 2b). Six out of eight genes in this cluster were related to protein binding (GO:0005515): Dnajb5, Smurf2, Vgll4, Pik3c2b, Stxbp1, Golph3l, and Gna13.
In summary, the relative expression level of several genes was altered in microglia with increasing PMD. The expression of genes related to mitochondrial and ribosomal functions showed a relative increase with increasing PMD. Genes involved in protein binding were also significantly affected, and their relative expression levels either increased or decreased with longer PMD.
3.3. The effect of PMD on human microglia transcriptome
A previously published RNA‐Seq dataset was used to determine whether the relative expression level of these PMD‐associated genes was altered in human microglia transcriptomes generated from postmortem samples (Galatro, Holtman, et al., 2017). Donor information is provided in Table S2. Of the 50 PMD‐associated genes in mouse microglia, 31 have a known human homolog (Table S3). Hierarchical clustering of these 31 genes in the human microglia gene expression data resulted in segregation of the samples into two clusters (Figure 3a). Samples in Cluster 1 showed significantly higher abundance of PMD‐related genes compared to Cluster 2 (Figure 3b) with the exception of MT‐CYTB and MT‐ND1 of which relative expression was significantly lower in Cluster 1 and higher in Cluster 2 samples (Figure 3c). Changes in the relative expression levels of genes in these clusters did not associate with the age of the donors (Figure 3a). Cluster 1 contained significantly more samples with shorter PMD than Cluster 2 (Figure 3a,d), suggesting that in human microglia, the relative abundance of these genes is also affected by PMD.
FIGURE 3.
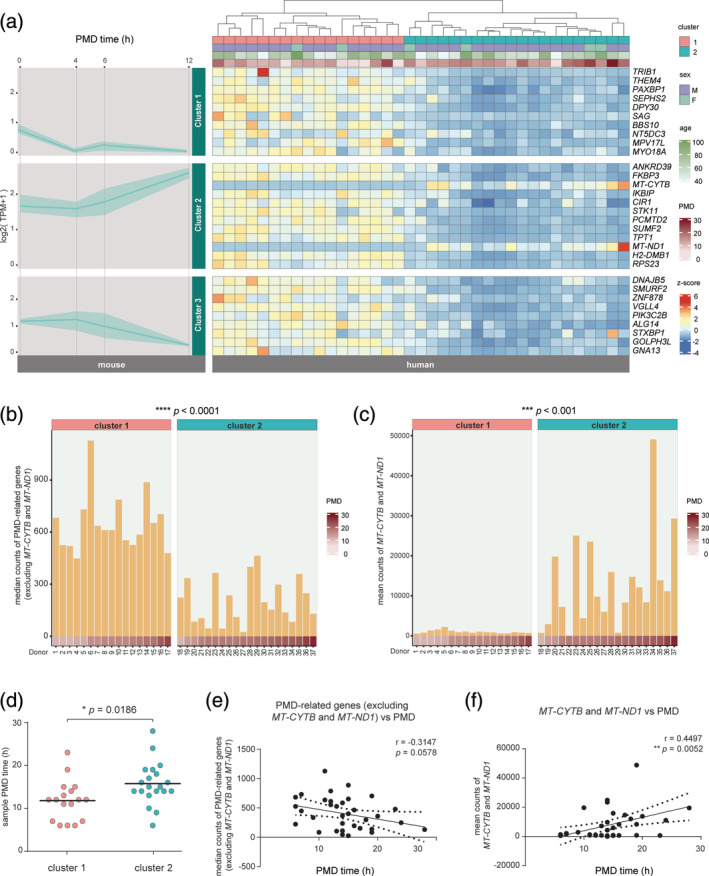
The effects of postmortem delay (PMD) on the human microglia transcriptome. (a) Line and ribbon graphs depict expression pattern of mouse genes with a human homologue, divided into three gene clusters that were previously identified in mouse microglia (left). The y‐axis indicates expression values, and postmortem time is depicted on the x‐axis. The line depicts the mean expression values of the genes in their respective cluster, the ribbon illustrates the highest and lowest gene expression values of the given PMD. A heatmap of 31 human homologs of PMD‐related genes identified in mouse (right). Human samples clustered into two groups based on the relative gene expression of PMD‐related homologs. (b,c) Bar plots depicting the median gene expression of PMD‐related genes excluding MT‐CYTB and MT‐ND1 (b) and mean expression of MT‐CYTB and MT‐ND1 (c) for each individual human sample. The PMD for each human sample is indicated. An unpaired t‐test was performed to assess significance. ***, p < .001; ****, p < .0001. (d) Dot plot depicting the PMD time of the samples in Cluster 1 and 2. An unpaired t‐test was used to determine the significance *, p < .05. (e, f) Linear regression analysis for median counts of PMD‐related genes excluding MT‐CYTB and MT‐ND1 (e) and mean counts of MT‐CYTB and MT‐ND1 (f) to PMD time. Results show linear regression lines with 95% confidence limits and Pearson r values with p values indicating degree of significance. **, p < .01
However, the directionality of these effects was only partly overlapping between human and mouse microglia. Homologous genes in mouse Cluster 1 and 3 had similar gene expression patterns in human microglia, with a lower expression level in samples with relatively longer PMDs (Figure 3a). The relative expression level of homologous genes in mouse Cluster 2, with the exception of MT‐CYTB and MT‐ND1, was lower in samples with relatively longer PMDs in humans, which is different from what was observed in mice (Figure 3a).
Next, a correlation analysis between PMD time and the expression level of PMD‐related genes in the human dataset was performed. The MT‐CYTB and MT‐ND1 genes were analyzed separately from the other PMD‐related genes in view of their reciprocal expression patterns (Figure 3a). A near‐significant (p = .0578) negative correlation between the median counts of PMD‐related genes (except MT‐CYTB and MT‐ND1) and PMD time was observed (Figure 3e). In contrast, a significant (p = .0052) positive correlation was observed for MT‐CYTB and MT‐ND1 (Figure 3f). These data show that the relative abundance of genes affected by PMD in mice were also altered with PMD in human microglia.
4. DISCUSSION
Microglia can be isolated from postmortem human brain samples or from tissues obtained during surgery, but when placed in culture, expression of microglia‐specific genes is rapidly lost (Gosselin et al., 2017). Cultured microglia lack brain‐specific signals that are necessary to maintain microglia gene expression signatures (Gosselin et al., 2017). Therefore, profiling of acutely isolated microglia is of vital importance for understanding microglia transcriptional signatures both in health and disease. Acutely isolated microglia from human tissue are challenging to investigate, due to limited sample sizes, antemortem factors (like medical history, comorbidities, medication, nutrition) and technical variations in sample processing. Additionally, samples obtained from surgery or with PMD are confounded by several factors. Surgery obtained tissue is generally taken from an area of the brain in which pathology is present. As the procedure is pathology driven, often the tissue is obtained from various regions, rather than a predefined area of interest. Samples with a PMD are challenging due to the different circumstances leading up to the death of the donor, as well as postmortem effects. Here, we were able to determine PMD effects on microglia gene expression in mice, where antemortem conditions and PMD can be tightly controlled. To minimize ex vivo microglial activation and related transcriptional changes, we performed the mechanical dissociation and Percoll gradient separation at low temperature (Galatro, Vainchtein, et al., 2017).
Surprisingly, viable microglia (DAPIneg Ly6Cneg CD11bhigh CD45int) could be isolated from mouse brains even 24 hr after death. But the number of viable microglia (DAPIneg) decreased sharply with increasing PMD. In our human cases, the number of FACS‐sorted viable microglia did not correlate with PMD (data not shown). This is also supported by a recent study demonstrating that PMD does not affect viable primary microglia yield from human postmortem brain tissue (Mizee et al., 2017).
Although in mice, the number of isolated microglia decreased with increasing PMD, high quality RNA was still obtained from these cells, even after 12 hr PMD, which could be explained by the selection of viable (DAPIneg) microglia. Gene expression in DAPIneg Ly6Cneg CD11bhigh CD45int microglia was only moderately affected by PMD, as out of 15,554 genes detected, only 50 genes displayed significant PMD‐associated relative expression changes. Of these 50 genes, 31 had human homologs, which were investigated in microglia transcriptomes generated from human postmortem samples (Galatro, Holtman, et al., 2017). The relative expression of human homologs MT‐CYTB and MT‐ND1 was positively correlated with increasing PMD. However, for the remaining 29 human homologs, their relative expression was not altered in human microglia in relation to PMD. The observed discrepancies between human and mouse microglia in terms of PMD effects could be caused by unavoidable confounding factors of human postmortem brain, different isolation methods, or intrinsic differences between human and mouse microglia. In human microglia, PMD might affect more genes than we identified, but larger datasets are required to correct for PMD, donor age, antemortem conditions and other relevant parameters in order to reliably detect these genes.
Although the effects of PMD on the microglia transcriptome have not been studied before, some studies showed that PMD had a limited effect on the brain transcriptome. Sobue et al. used microarrays to compare gene expression profiles of mouse brain samples with three different PMD conditions: immediately after death, after 3 hr at RT, and after storage at 4°C for 18 hr. They found that 87.6% of the transcripts (total 45,141 genes/probes) were not affected by PMD (Sobue et al., 2016). In another study, mouse brain transcriptomes were analyzed immediately after death and with increasing PMD. Moderate changes were detected after 8–24 hr PMD at RT: 365 of the 588 genes detected by the microarrays were unaffected by PMD (Trotter et al., 2002). Two other studies investigated the effects of PMD on gene expression in various human tissues and detected no effects of PMD on the cerebellum and very limited influence on the cerebral cortex; only 105 genes were differentially expressed with a cutoff FDR < 0.05 in the cerebral cortex (Ferreira et al., 2018; Zhu et al., 2017). Similar to these previous studies in brain tissues, we also observed that the effects of PMD on microglia transcriptomes were limited.
In a previous study, relative Ikbip expression was shown to be increased, and relative Zfp617 expression was shown to be decreased with PMD in mouse brain tissue (Sobue et al., 2016). We observed similar changes in mouse microglia in this study. For humans, Zhu et al. reported that the expression of SEPHS2 (in aorta artery), NT5DC3 (in thyroid), MYO18A (in lung and skeletal muscle), FKBP3 (in esophageal mucosa), IKBIP (in whole blood), TPT1 (in skeletal muscle), GNA13 (in skeletal muscle) was decreased with increasing PMD (Zhu et al., 2017). These genes also showed decreased relative expression with increasing PMD in human microglia in this study. However, no overlap was observed between our human microglia PMD‐associated genes and their human brain PMD‐associated genes (Zhu et al., 2017).
Postmortem human brain tissue is invaluable for the study of CNS diseases. Over the last decade, transcriptional analysis of purified human microglia from postmortem brain tissues has greatly contributed to our understanding and characterization of these cells (Gerrits et al., 2020). Thus, understanding the common postmortem changes in gene expression, as presented in this study, is of importance for identifying biological changes rather than PMD effects on microglia gene expression profiles. This study provides evidence that PMD has limited effects on the microglia transcriptome, and suggests that transcriptional profiling microglia isolated from postmortem brain tissues provides reliable results.
Supporting information
Table S1 Mouse PMD‐related genes
Table S2 Donor information
Table S3 Human homologs of mouse PMD‐related genes
ACKNOWLEDGMENTS
The authors thank Xiaoming Zhang, Qiong Jiang and Nieske Brouwer for technical support and Geert Mesander, Johan Teunis and Theo Bijma of the UMCG FACS facility. The authors thank Susanne Kooistra for critical proofreading of the manuscript. This work was supported by a China Scholarship Council fellowship to Yang Heng.
Heng Y, Dubbelaar ML, Marie SKN, Boddeke EWGM, Eggen BJL. The effects of postmortem delay on mouse and human microglia gene expression. Glia. 2021;69:1053–1060. 10.1002/glia.23948
Yang Heng and Marissa L. Dubbelaar contributed equally to this work.
DATA AVAILABILITY STATEMENT
All next‐generation sequencing data can be viewed at NCBI GEO under accession number GSE162209.
REFERENCES
- Birdsill, A. C. , Walker, D. G. , Lue, L. , Sue, L. I. , & Beach, T. G. (2011). Postmortem interval effect on RNA and gene expression in human brain tissue. Cell and Tissue Banking, 12(4), 311–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catts, V. S. , Catts, S. V. , Fernandez, H. R. , Taylor, J. M. , Coulson, E. J. , & Lutze‐Mann, L. H. (2005). A microarray study of post‐mortem mRNA degradation in mouse brain tissue. Brain Research. Molecular Brain Research, 138(2), 164–177. [DOI] [PubMed] [Google Scholar]
- Cummings, T. J. , Strum, J. C. , Yoon, L. W. , Szymanski, M. H. , & Hulette, C. M. (2001). Recovery and expression of messenger RNA from postmortem human brain tissue. Modern Pathology, 14(11), 1157–1161. [DOI] [PubMed] [Google Scholar]
- Durrenberger, P. F. , Fernando, S. , Kashefi, S. N. , Ferrer, I. , Hauw, J. J. , Seilhean, D. , … Reynolds, R. (2010). Effects of antemortem and postmortem variables on human brain mRNA quality: A BrainNet Europe study. Journal of Neuropathology and Experimental Neurology, 69(1), 70–81. [DOI] [PubMed] [Google Scholar]
- Ervin, J. F. , Heinzen, E. L. , Cronin, K. D. , Goldstein, D. , Szymanski, M. H. , Burke, J. R. , … Hulette, C. M. (2007). Postmortem delay has minimal effect on brain RNA integrity. Journal of Neuropathology and Experimental Neurology, 66(12), 1093–1099. [DOI] [PubMed] [Google Scholar]
- Ferreira, P. G. , Munoz‐Aguirre, M. , Reverter, F. , Sa Godinho, C. P. , Sousa, A. , Amadoz, A. , … Guigo, R. (2018). The effects of death and post‐mortem cold ischemia on human tissue transcriptomes. Nature Communications, 9(1), 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galatro, T. F. , Holtman, I. R. , Lerario, A. M. , Vainchtein, I. D. , Brouwer, N. , Sola, P. R. , … Eggen, B. J. L. (2017). Transcriptomic analysis of purified human cortical microglia reveals age‐associated changes. Nature Neuroscience, 20(8), 1162–1171. [DOI] [PubMed] [Google Scholar]
- Galatro, T. F. , Vainchtein, I. D. , Brouwer, N. , Boddeke, E. , & Eggen, B. J. L. (2017). Isolation of microglia and immune infiltrates from mouse and primate central nervous system. Methods in Molecular Biology, 1559, 333–342. [DOI] [PubMed] [Google Scholar]
- Gallego Romero, I. , Pai, A. A. , Tung, J. , & Gilad, Y. (2014). RNA‐seq: Impact of RNA degradation on transcript quantification. BMC Biology, 12, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrits, E. , Heng, Y. , Boddeke, E. , & Eggen, B. J. L. (2020). Transcriptional profiling of microglia; current state of the art and future perspectives. Glia, 68(4), 740–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin, D. , Skola, D. , Coufal, N. G. , Holtman, I. R. , Schlachetzki, J. C. M. , Sajti, E. , … Glass, C. K. (2017). An environment‐dependent transcriptional network specifies human microglia identity. Science, 356(6344), eaal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, P. J. , Heath, P. R. , Eastwood, S. L. , Burnet, P. W. , McDonald, B. , & Pearson, R. C. (1995). The relative importance of premortem acidosis and postmortem interval for human brain gene expression studies: Selective mRNA vulnerability and comparison with their encoded proteins. Neuroscience Letters, 200(3), 151–154. [DOI] [PubMed] [Google Scholar]
- Heinrich, M. , Matt, K. , Lutz‐Bonengel, S. , & Schmidt, U. (2007). Successful RNA extraction from various human postmortem tissues. International Journal of Legal Medicine, 121(2), 136–142. [DOI] [PubMed] [Google Scholar]
- Johnson, S. A. , Morgan, D. G. , & Finch, C. E. (1986). Extensive postmortem stability of RNA from rat and human brain. Journal of Neuroscience Research, 16(1), 267–280. [DOI] [PubMed] [Google Scholar]
- Johnson, W. E. , Li, C. , & Rabinovic, A. (2007). Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics, 8(1), 118–127. [DOI] [PubMed] [Google Scholar]
- Kim, D. , Langmead, B. , & Salzberg, S. L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nature Methods, 12(4), 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Paggi, J. M. , Park, C. , Bennett, C. , & Salzberg, S. L. (2019). Graph‐based genome alignment and genotyping with HISAT2 and HISAT‐genotype. Nature Biotechnology, 37(8), 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, H. , Sakimura, K. , Kuwano, R. , Sato, S. , Ikuta, F. , Takahashi, Y. , … Tsuji, S. (1990). Stability of messenger RNA in postmortem human brains and construction of human brain cDNA libraries. Journal of Molecular Neuroscience, 2(1), 29–34. [DOI] [PubMed] [Google Scholar]
- Koppelkamm, A. , Vennemann, B. , Lutz‐Bonengel, S. , Fracasso, T. , & Vennemann, M. (2011). RNA integrity in post‐mortem samples: Influencing parameters and implications on RT‐qPCR assays. International Journal of Legal Medicine, 125(4), 573–580. [DOI] [PubMed] [Google Scholar]
- Liao, Y. , Smyth, G. K. , & Shi, W. (2014). featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics, 30(7), 923–930. [DOI] [PubMed] [Google Scholar]
- Love, M. I. , Huber, W. , & Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biology, 15(12), 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizee, M. R. , Miedema, S. S. , van der Poel, M. , Adelia, Schuurman , K. G., van Strien, M. E. , … Huitinga, I. (2017). Isolation of primary microglia from the human post‐mortem brain: Effects of ante‐ and post‐mortem variables. Acta Neuropathologica Communications, 5(1), 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova, T. , Mennerich, D. , Weith, A. , & Quast, K. (2008). Effect of RNA quality on transcript intensity levels in microarray analysis of human post‐mortem brain tissues. BMC Genomics, 9, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preece, P. , & Cairns, N. J. (2003). Quantifying mRNA in postmortem human brain: Influence of gender, age at death, postmortem interval, brain pH, agonal state and inter‐lobe mRNA variance. Brain Research. Molecular Brain Research, 118(1–2), 60–71. [DOI] [PubMed] [Google Scholar]
- Robinson, A. C. , Palmer, L. , Love, S. , Hamard, M. , Esiri, M. , Ansorge, O. , … Mann, D. M. (2016). Extended post‐mortem delay times should not be viewed as a deterrent to the scientific investigation of human brain tissue: A study from the Brains for Dementia Research Network Neuropathology Study Group, UK. Acta Neuropathologica, 132(5), 753–755. [DOI] [PubMed] [Google Scholar]
- Sigurgeirsson, B. , Emanuelsson, O. , & Lundeberg, J. (2014). Sequencing degraded RNA addressed by 3′ tag counting. PLoS One, 9(3), e91851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobue, S. , Sakata, K. , Sekijima, Y. , Qiao, S. , Murate, T. , & Ichihara, M. (2016). Characterization of gene expression profiling of mouse tissues obtained during the postmortem interval. Experimental and Molecular Pathology, 100(3), 482–492. [DOI] [PubMed] [Google Scholar]
- Tomita, H. , Vawter, M. P. , Walsh, D. M. , Evans, S. J. , Choudary, P. V. , Li, J. , … Bunney, W. E., Jr. (2004). Effect of agonal and postmortem factors on gene expression profile: Quality control in microarray analyses of postmortem human brain. Biological Psychiatry, 55(4), 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter, S. A. , Brill, L. B. , & Bennett, J. P. (2002). Stability of gene expression in postmortem brain revealed by cDNA gene array analysis. Brain Research, 942(1–2), 120–123. [DOI] [PubMed] [Google Scholar]
- Zhu, Y. , Wang, L. , Yin, Y. , & Yang, E. (2017). Systematic analysis of gene expression patterns associated with postmortem interval in human tissues. Scientific Reports, 7(1), 5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Mouse PMD‐related genes
Table S2 Donor information
Table S3 Human homologs of mouse PMD‐related genes
Data Availability Statement
All next‐generation sequencing data can be viewed at NCBI GEO under accession number GSE162209.
All next‐generation sequencing data can be viewed at NCBI GEO under accession number GSE162209.


