Abstract
How nonspore haploid Saccharomyces cells choose sites of budding and polarize towards pheromone signals in order to mate has been a subject of intense study. Unlike nonspore haploids, sibling spores produced via meiosis and sporulation by a diploid cell are physically interconnected and encased in a sac derived from the old cell wall of the diploid, called the ascus. Nonspore haploids bud adjacent to previous sites of budding, relying on stable cortical landmarks laid down during prior divisions, but because spore membranes are made de novo, it was assumed that, as is known for fission yeast, Saccharomyces spores break symmetry and polarize at random locations. Here, we show that this assumption is incorrect: Saccharomyces cerevisiae spores are born prepolarized to outgrow, prior to budding or mating, away from interspore bridges. Consequently, when spores bud within an intact ascus, their buds locally penetrate the ascus wall, and when they mate, the resulting zygotes adopt a unique morphology reflective of repolarization towards pheromone. Long‐lived cortical foci containing the septin Cdc10 mark polarity sites, but the canonical bud site selection programme is dispensable for spore polarity, thus the origin and molecular composition of these landmarks remain unknown. These findings demand further investigation of previously overlooked mechanisms of polarity establishment and local cell wall digestion and highlight how a key step in the Saccharomyces life cycle has been historically neglected.
Keywords: cell wall, fungal, life cycle stages, Saccharomyces, septins, spores
When budding yeast spores germinate, they outgrow away from each other, following cortical foci of the septin protein Cdc10. If spores then bud, and old cell wall of the mother diploid cell still surrounds them, the buds locally penetrate that wall. If spores instead mate with each other, their outward prepolarization leads to a unique zygote morphology. Prepolarization does not require Rsr1, a key component of the canonical bud site selection pathway in nonspore cells.
1. INTRODUCTION
Most steps in the Saccharomyces cerevisiae life cycle have been described in great detail at the cellular and molecular levels. For example, in the YeastBook series of reviews published by the journal GENETICS, 43 articles to date ‘span the breadth of Saccharomyces biology’ (https://www.genetics.org/content/yeastbook). Here and in other sources can be found numerous mechanistic insights into the ways that budding yeast cells choose bud sites by positioning polarity factors according to mating‐type‐specific cortical landmarks; how a potent extrinsic signal, mating pheromone, can override these landmarks; and how cells use hydrolytic enzymes to remodel their cell walls upon budding and cell fusion. The unique morphology yeast cells adopt as they grow chemotropically toward a pheromone source even garnered an enduring and endearing name, the shmoo, based on its resemblance to a comic strip character from the mid‐1900s. Indeed, early studies of the signal transduction cascades underlying the pheromone response and the ways that yeast cells repolarize to track pheromone gradients were foundational in our general understanding of eukaryotic signal transduction and polarity determination.
Accordingly, we find it remarkable how little is known about a step of the Saccharomyces life cycle for which polarity, chemotropism and cell wall remodelling are crucial. Germination is the process by which spores awaken from a nearly dormant state, break out of a rigid, specialized cell wall, and grow in a single direction by producing new membrane and cell wall. Only a single review dedicated to germination has been published, over a decade ago (Geijer, Joseph‐Strauss, Simchen, Barkai, & Hohmann, 2010).
Most natural isolates of S. cerevisiae are diploid. Hence, most spores are haploid, and germinating spores are capable of mating immediately (without a prior budding event) if a suitable mating partner is sufficiently close by (Joseph‐Strauss, Zenvirth, Simchen, & Barkai, 2007). The assumption from the literature is that mating between spores follows the same rules as what has been worked out for nonspore (‘vegetative’) haploids, but this has not been rigorously tested.
Alternatively, and often regardless of the proximity of a mating partner (McClure, Jacobs, Zyla, & Lew, 2018), following outgrowth haploid spores can enter S phase and bud from the tip of the outgrowth. The most recent comprehensive review of yeast cell polarity states that ‘in yeasts, germinating spores lack obvious positional cues and appear to break symmetry to initiate polar growth’ (Chiou, Balasubramanian, & Lew, 2017). This statement is clearly true for the fission yeast Schizosaccharomyces pombe, where an elegant study demonstrated that polarity factors, including active Cdc42, wander randomly around the cortex of growing spores until the outer spore wall breaks, whereupon polarity is stabilized at the site of wall rupture (Bonazzi et al., 2014). In S. cerevisiae, on the other hand, a single polarity site is already apparent in dormant spores, prior to germination; the only known marker of this site is the septin protein Cdc10 (Joseph‐Strauss et al., 2007). Polarization of the actin cytoskeleton, a prerequisite for outgrowth, occurs only after germination begins (Kono et al., 2005). In nonspore cells, septins are recruited to the site of future budding by active Cdc42 and Cdc42‐interacting proteins (Iwase et al., 2006); it is not known how Cdc10 is deposited during sporulation at a single site on each spore. During sporulation, the spore cortex is synthesized de novo, precluding any influence on bud site selection by persistent cortical landmarks produced by previous budding events.
Yeast sporulation takes place within a single (usually diploid) cell and generates (usually four) spores encased within the original cell wall of the sporulating cell, called the ascus (Neiman, 2011). Whereas in S. pombe, the ascus wall is globally digested by cell wall hydrolases immediately upon the successful completion of meiosis (Dekker, van Rijssel, Distel, & Hochstenbach, 2007; Encinar del Dedo, Dueñas, Arnáiz, del Rey, & Vázquez de Aldana, 2009; Guo & King, 2013), in Saccharomyces the ascus wall remains intact throughout sporulation. In the absence of external factors, such as those found in insect guts (Coluccio, Rodriguez, Kernan, & Neiman, 2008), the Saccharomyces ascus wall breaks down slowly during germination, often persisting long enough for spores to mate and bud following germination. Saccharomyces spores (but not spores of S. pombe) are also interconnected by cell wall junctions called interspore bridges that persist following ascus wall digestion (Coluccio & Neiman, 2004). Outgrowth by germinating spores first requires local break down of a rigid outer spore wall that confers stress resistance via layers of chitosan and polymerized dityrosine (in Saccharomyces; Briza, Ellinger, Winkler, & Breitenbach, 1990) or a proteinaceous coat (in S. pombe; Fukunishi et al., 2014). Subsequent budding or mating requires local breakdown of the newly emerged cell wall at the site of budding or fusion, respectively. If Saccharomyces spores bud when the ascus wall is still intact, those buds can locally penetrate the ascus wall, as visualized by electron microscopy many decades ago (Hashimoto, Conti, & Naylor, 1958; Rij, 1978; Sando, Oguchi, Nagano, & Osumi, 1980). A more recent study using light microscopy marked bud necks with fluorescently tagged septins to verify that the protrusions were buds (McMurray & Thorner, 2008). The mechanisms by which germinating Saccharomyces spores digest cell walls during outgrowth, budding and mating have not been investigated.
Here, we describe how the sites where spores bud and fuse upon mating, and the localization of Cdc10 with relation to the position of other haploid spores produced by the same diploid mother, demonstrate that each S. cerevisiae spore is born prepolarized to direct outgrowth away from its ‘sibling’ spores. Spore buds are thereby positioned to penetrate the ascus wall.
2. MATERIALS AND METHODS
2.1. Yeast strains, plasmid and media
All strains used in the new experiments described here are of the S288C strain background, specifically derived from the ‘designer deletion’ strains BY4741 (MAT a his3∆1 leu2∆0 met15∆0 ura3∆0), BY4742 (MATα his3∆1 leu2∆0 lys2∆0 ura3∆0) and BY4743 (the diploid formed by mating BY4741 and BY4742). Unless specified otherwise in the figure legend, all experiments were done with a diploid strain made by mating FY2742 (MATα his3∆1 leu2∆0 lys2∆0 ura3∆0 TAO1 MKT1 RME1) and FY2839 (MAT a his3∆1 leu2∆0 lys2∆0 ura3∆0 TAO1 MKT1 RME1), two haploid strains that carry at three loci (TAO1, MKT1 and RME1) dominant alleles from the efficiently sporulating SK‐1 strain background (Kloimwieder & Winston, 2011). JTY3985 carries CDC10‐GFP integrated at the CDC10 locus using the URA3 selectable marker and is otherwise isogenic to BY4741 (Johnson, Weems, Brewer, Thorner, & McMurray, 2015). JTY3992 carries CDC10‐mCherry integrated at the CDC10 locus using the kanMX selectable marker and is otherwise isogenic to BY4742 (McMurray et al., 2011). MMY0341 (gas1∆::kanMX/gas1∆::kanMX), MMY0286 (acf2∆::kanMX, acf2∆::kanMX) and MMY0291 (rsr1∆::kanMX, rsr1∆::kanMX) were retrieved from the homozygous diploid deletion collection derived from BY4743 (Giaever et al., 2002). Previously published Gas1‐GFP localization data reproduced here used a diploid strain of the SK‐1 background in which both genomic copies of GAS1 were deleted and Gas1‐GFP was expressed from a high‐copy plasmid (Rolli et al., 2011). The low‐copy (ARS‐CEN) plasmid pZL02 (Heasley, Garcia, & McMurray, 2014) carries the URA3 selectable marker and CDC10‐mCherry under control of the CDC10 promoter.
Haploid strains were mated together using sterile toothpicks by mixing approximately equal amounts on the surface of YPD agar (per litre: 10 g of yeast extract, 20 g of peptone, 20 g of dextrose, 2% of agar) in a petri dish. After overnight incubation at 30°C, cells from the mixture were streaked with a toothpick to agar media that was either selective for the diploid, or non‐selective (YPD). In the latter case, diploid clones were identified as individual colonies containing cells that were able to sporulate. The plasmid pZL02 was introduced by transformation using the Frozen‐EZ Yeast Transformation II kit (Zymo Research, Irvine, CA).
To induce sporulation, in most cases, diploid cells were cultured in 5 ml of liquid YPD (per litre: 10 g of yeast extract, 20 g of peptone, 20 g of dextrose) in glass culture tubes rotated in a roller drum overnight to near‐saturation. For analysis of rsr1∆/rsr1∆ cells in ‘old’ rich medium, the culture time in YPD was extended for six additional days. A 200‐μl aliquot of these cells was washed with 5 ml of sterile water and resuspended in 2.5 ml of sporulation medium (1% of potassium acetate, 0.05% of glucose, 20 mg/L of leucine and 40 mg/L of uracil) in a new tube to an optical density at 600 nm of approximately 0.5. These cell suspensions were rotated at 22°C for at least 4 days, after which time the percentage of cells that had formed mature asci did not noticeably increase. For confocal microscopy, sporulation was induced by spreading cells with a sterile toothpick from YPD agar to solid sporulation medium (same composition as above but with added 2% agar). To induce germination, aliquots of cells from sporulation cultures were pelleted and resuspended in 1 ml of YPD in 1.7‐ml microcentrifuge tubes and rotated at 22°C or 30°C for 4–6 h (or as indicated in figure legends). To digest the ascus wall, a 50‐μl aliquot of cells from sporulation culture was pelleted and resuspended in 1 mg/ml of Zymolyase‐20T (#320921, MP Biomedicals) dissolved in water and incubated at 30°C for 10 min.
2.2. Fluorescence labelling
FM™ 4–64 dye (N‐(3‐triethylammoniumpropyl)‐4‐(6‐(4‐(diethylamino) phenyl) hexatrienyl) Pyridinium Dibromide, from Molecular Probes, Inc. #T3166) was dissolved in dimethyl sulfoxide to make a 1.6 μM of stock solution. An aliquot of cells from a sporulation culture that had been in sporulation medium for 18 h was pelleted and resuspended in 50 μl of ice‐cold YPD to which 1 μl of the dye stock was then added. After 20 min on ice in the dark, the cells were pelleted again and washed twice by resuspension in 1 ml of ice‐cold water each time. Finally, the cells were suspended in 50 μl of water and visualized by microscopy using the Texas red LED filter cube.
Calcofluor white M2R was dissolved in water at a concentration of 10 mg/ml. One microlitre was added to a 100‐μl aliquot of cells that had been in sporulation medium for 4 days, after which the cells were washed three times with water by pelleting and resuspension. After the third wash, the cells were resuspended in 1 ml of YPD and incubated at 30°C for 6 h, then pelleted and resuspended in 50 μl before imaging.
2.3. Microscopy and imaging
Aliquots of cells from sporulation cultures were imaged directly on agarose pads made with 1% agarose in water or, in the case of cells expressing Cdc10‐GFP, before and after a 3.5‐h interval of incubation at 30°C on an agarose pad made with 1% agarose in sporulation medium. Germinating cells were pelleted and resuspended in water before applying to agarose pads. Most images were captured on an EVOSfl all‐in‐one microscope (ThermoFisher Scientific, Waltham, MA) with a 1.42 NA Olympus 60 × Plan‐Apo oil objective. Filter cubes were as follows: GFP (AMEP4651, excitation 470/22 nm, emission 510/42 nm), Texas red (AMEP4655, excitation 585 nm, emission 624 nm), RFP (AMEP4652, excitation 531/40 nm, emission 593/40 nm) and DAPI (AMEP4650, excitation 357/44 nm, emission 447/60 nm). z‐stacks of images of Cdc10‐mCherry foci were collected on a Nikon Ti‐E microscope (Nikon Instruments Inc., Melville, NY) equipped with a 1.45 NA 100 × CFI Plan Apo objective, piezo electric stage (PhysikInstrumente, Auburn, MA), spinning disk confocal scanner unit (CSU10; Yokogawa Corporation of America, Sugar Land, TX), 561‐nm laser (Agilent Technologies, Santa Clara, CA), and an EMCCD camera (iXon Ultra 897, Andor Technology, Belfast, UK) using NIS Elements software (Nikon). Laser power was 92%, 500‐ms exposure time, 10‐μm stack and 250‐nm separation. Images were cropped and adjusted (always the same way for each image of the same type from the same experiment) and inverted in Photoshop (Adobe, San Jose, CA) or Fiji (Schindelin et al., 2012). Three‐dimensional projections and rotations were performed in Fiji.
3. RESULTS
3.1. Stable cortical foci of the septin Cdc10 at the ascus periphery
That single S. cerevisiae spores are born prepolarized was known from the discrete localization of the septin Cdc10 to a single site on the spore cortex from which cell wall outgrowth occurs (Joseph‐Strauss et al., 2007). However, it was not known how the site of spore prepolarization relates to the spatial relationship between the spores, the interspore bridges and the ascus wall. If spore buds commonly penetrate the ascus wall, then spores should be polarized to outgrow and bud away from the interspore bridges at the centre of the ascus. To test this prediction, we visualized Cdc10‐GFP in spores within intact, mature asci. Fluorescent protein fusions slightly compromise Cdc10 function (McMurray, 2016), and diploid cells homozygous for fluorescently tagged alleles of CDC10 sporulated poorly and often displayed abnormal morphology (data not shown). To minimize these issues, we used diploid strains carrying CDC10‐GFP or CDC10‐mCherry and one untagged allele. We also tried boosting Cdc10‐mCherry signal by introducing additional copies of CDC10‐mCherry on a low‐copy plasmid, but there was no obvious benefit (not shown). As is true of diverse fluorescently tagged proteins, strong signal was observed in the areas between the spores (Figure 1a,b). Presumably, this signal reflects Cdc10 molecules that were outside the prospore membranes as they closed and thus remained in the ascal cytoplasm, which becomes concentrated between the spores as the ascus wall compresses tightly to surround the spores during the final stages of ascus maturation. Cdc10‐GFP signal is not found at these locations in spores that are isolated from asci (Joseph‐Strauss et al., 2007). Apart from this signal, Cdc10 puncta, usually one per spore, were found in many (but not all) spores (Figure 1a,b); the failure to observe a punctum in every spore may reflect the fact that only one allele of CDC10 in the diploid strain expresses the fluorescently tagged fusion. While it is known that all spores from such CDC10‐GFP/CDC10 heterozygous diploids inherit some pre‐existing Cdc10‐GFP protein, regardless of their haploid genotype, those that fail to inherit the CDC10‐GFP allele have fainter signal (Joseph‐Strauss et al., 2007). When they were visible, Cdc10 puncta were found in locations consistent with a mode of outgrowth upon germination that precedes spore budding through an intact ascus wall (Figure 1a–c and data not shown).
FIGURE 1.
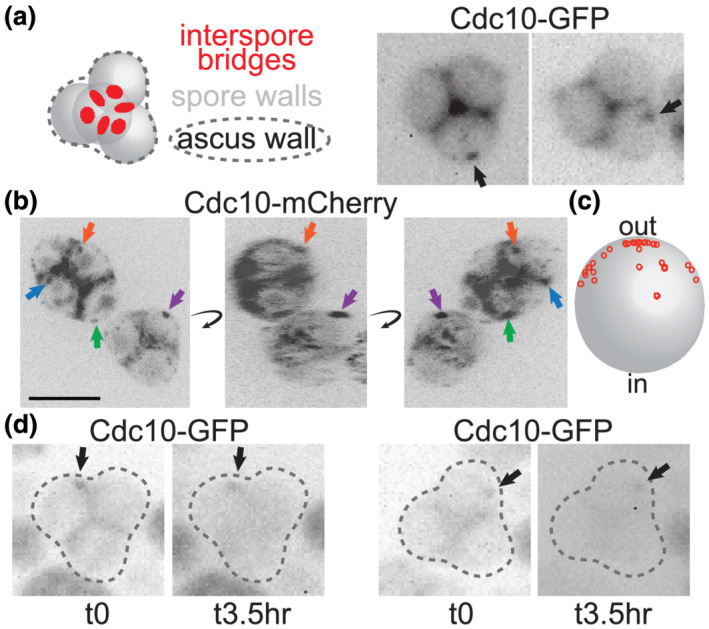
Stable cortical foci of the septin Cdc10 persist on the periphery of asci. (a) At left is an illustration of a typical pyramidal four‐spored ascus in which the three spores at the ‘base’ of the pyramid are in the same focal plane. Red circles indicate interspore bridges. At right, Cdc10‐GFP fluorescence in intact asci from sporulation culture. Arrows indicate cortical foci. Cells are of a diploid strain made by mating haploid strains JTY3985 and FY2742. (b) 2D images from 3D reconstruction of images captured via confocal microscopy of Cdc10‐mCherry fluorescence in asci from sporulation medium. The 3D reconstruction was rotated (curved arrows) to improve visualization of cortical signal versus signal in between spores, which we assume to be in the residual ascal cytoplasm. Arrows point to cortical foci and are color‐coded to track the same foci through the rotation. Scale bar, 5 μm. Cells are of a diploid strain made by mating haploid strains JTY3992 and FY2839. (c) The approximate positions of 35 foci visualized as in (b) were plotted as small red circles onto a large grey circle representing a spore, where ‘out’ refers to the periphery of the ascus and ‘in’ refers to the centre. All foci were on the spore periphery; red circles not at the edge of the grey circle would project into or out of the plane of the image. These data also include foci from diploid cells as in (b) but carrying the CDC10‐mCherry plasmid pZL02. (d) As in (a), but the same asci were visualized before and after a 3.5‐h interval on solid sporulation medium
Spores can survive for long periods in nutrient‐free conditions and then, once nutrients are provided, germinate efficiently. We see no obvious effect on the site of budding during germination of the length of time between sporulation and germination (unpublished observation); indeed, the images in Figure 1a were taken of asci that had been in sporulation medium for over 7 days, with the vast majority of asci in these cultures having visibly completed maturation by day 4. If Cdc10 foci mark the location of spore prepolarization, then, these cortical foci must not diffuse to any great extent over time. Alternatively, they may be able to diffuse on regions of the cortex far from the interspore bridges but are prevented from diffusing into the cortical areas near interspore bridges. To distinguish between these possibilities, we visualized Cdc10‐GFP foci before and after an interval of 3.5 h. In every case, the position of the focus at the later time was indistinguishable from its starting position (Figure 1b). While these experiments cannot exclude the possibility that the foci moved in between the imaging time points, we interpret these data as evidence that diffusion of the cortical Cdc10‐GFP foci is limited, potentially allowing spores to maintain cortical polarity for long periods of time.
3.2. Ascus wall penetration upon spore budding points to prepolarization of spores away from interspore bridges
If spores are prepolarized to outgrow upon germination towards the periphery of the ascus, then when germinating spores bud while they are still within the ascus, they should locally penetrate the ascus wall, such that the buds protrude from the ascus surface. If, instead, spore prepolarization is random, then spore budding should frequently result in budding in between the spores, within the ascus, without penetrating the ascus wall. We exploited the weak red autofluorescence of the post‐germination spore wall (Joseph‐Strauss et al., 2007) to identify trapped buds (nonfluorescent cell wall outgrowth) within the crowded environment of an intact ascus full of germinating spores. Among hundreds of such asci, we have only seen a single likely case of a bud ‘trapped’ within the ascus (Figure 2a).
FIGURE 2.
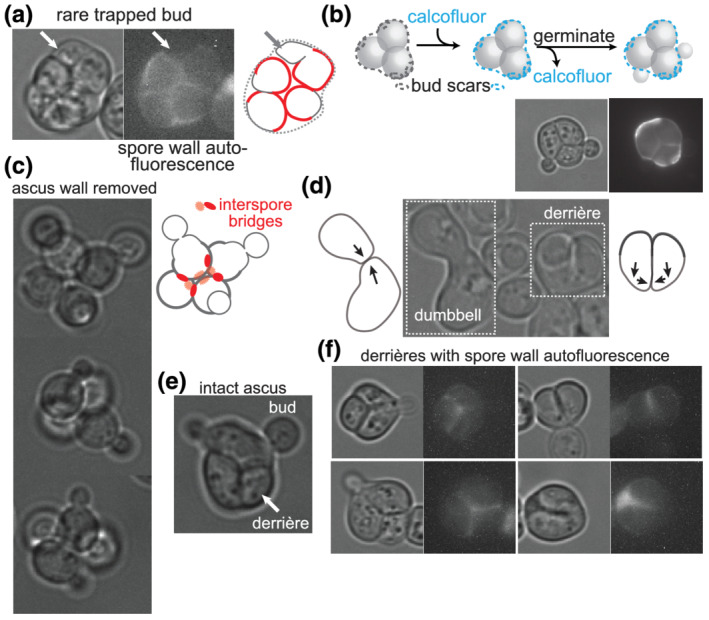
Saccharomyces spores outgrow away from interspore bridges upon germination. (a) Germinating ascus as viewed by transmitted light and with autofluorescence of the spore wall visualized with an RFP filter. (b) According to the illustration, asci were exposed to the chitin‐binding dye calcofluor white and then, after washing away free dye, allowed to germinate. Pre‐existing bud scars on the ascus wall were deposited during diploid budding events prior to sporulation. Calcofluor fluorescence and transmitted light are shown. (c) Tetrads for which the ascus wall was removed by exposure to Zymolyase prior to germination. In the illustration, red circles are interspore bridges, and the spore wall is thicker than the new, vegetative cell wall. (d) Image taken several hours after asci were allowed to germinate, showing dumbbell‐shaped zygote and derrière‐shaped zygote, with illustrations of presumptive directions of outgrowth prior to fusion. (e) Germinating ascus showing two buds penetrating the ascus wall and the other two spores fused into a derrière. (f) As in (a), after 7.75 h of germination, showing localization of the autofluorescent spore wall with regard to the shape of the derrière
As an independent way to visualize bud penetration of the ascus wall, we performed a pulse‐chase with calcofluor white, which fluorescently labels cell wall chitin (Cabib & Bowers, 1975). The ascus wall in mature asci was labelled with calcofluor, and then excess dye was washed away prior to the induction of germination by addition of rich (YPD) medium (Figure 2b). New cell wall synthesized in the absence of calcofluor white is nonfluorescent. Spore buds emerged from holes in the ascus wall, which otherwise remained intact (Figure 2b). Thus, budding by germinating spores is directed away from the interspore bridges that connect spores to each other and towards the ascus wall that surrounds them. Budding away from interspore bridges was also apparent for interconnected spores in which the ascus wall was enzymatically removed (Figure 2c).
3.3. Unique zygote morphology provides independent evidence of spore prepolarization
We noticed independent evidence of spore prepolarization in the morphology of zygotes produced by spores that mated within the ascus. Figure 2d shows an image of cells from a population of germinating asci in which a zygote formed by mating between nonspore haploid cells (or possibly a spore with a nonspore cell) is adjacent to a zygote formed by mating between what we assume to be sibling spores. (See Figure 2f for unambiguous sister‐spore matings.) The dumbbell morphology of the zygote formed by nonspore mating is consistent with the morphology established in the literature for mating between wild‐type haploids (Sena, Radin, & Fogel, 1973), where prior to cell fusion each partner grows directly towards the other, following a pheromone gradient. The zygote formed from intra‐ascus mating between spores is distinctly different, as if both mating partners initially grew in approximately the same direction, roughly perpendicular to a line directly connecting them, and then redirected growth towards each other prior to fusion (Figure 2d). Similar to the use of the term ‘shmoo’ to refer to the unique morphology adopted by nonspore cells just prior to mating, we sought a new term to refer to the unique morphology of zygotes produced by mating between sibling spores. Inspired by the inescapable resemblance of one side of the resulting shape to the shape of the human buttocks, we propose the term ‘derrière’. Derrières were also seen within intact asci after germination (Figure 2e).
The rigid outer spore wall is likely a barrier to outgrowth, which requires cell wall breakdown at the site of fusion. Cell wall expansion upon germination is thus restricted to the site of outer spore wall breakdown, and the residual spore wall changes little upon germination and thereafter. If spores break down the outer spore wall at sites approximately opposite from the interspore bridges, then two adjacent spores within an ascus should grow along vectors that do not converge and instead are parallel or, more likely, divergent (Figure 2d). If redirected growth towards a pheromone source is only possible for vegetative cell wall, where polarized exocytosis targets cell wall synthesis enzymes to specific sites, then some amount of cell wall outgrowth is presumably required before subsequent cell wall outgrowth can be redirected towards the pheromone source. This model predicts derriére‐shaped zygotes with spore wall in the twin bulges. To test this model, we visualized spore wall autofluorescence within derrières. As expected, spore wall fluorescence was restricted to the twin bulges of derrières (Figure 2f). Buds then emerged from the site of fusion (Figure 2d,f).
3.4. The old plasma membrane of the sporulating diploid cell disappears prior to germination
In addition to local digestion via budding, the ascus wall breaks down globally during germination, albeit on a slower time scale. In S. pombe, two hydrolytic enzymes, the α‐1,3‐glucanase Agn2 and the endo‐β‐1,3‐glucanase Eng2, reside in the ascal cytoplasm because they lack signal sequences to drive secretion (Dekker et al., 2007; Encinar del Dedo et al., 2009). It has been speculated that after completion of spore wall synthesis, the old plasma membrane of the diploid fission yeast cell ‘may disintegrate through an unknown mechanism’ (Dekker et al., 2007) allowing the two enzymes access to their substrates.
In S. cerevisiae, if spore buds frequently penetrate the ascus wall, we wondered if in doing so they also penetrate the old plasma membrane of the sporulating cell, or if, as is thought for fission yeast, that membrane has already been destroyed. Late in sporulation, vacuole lysis releases hydrolytic enzymes that destroy, among other things, nuclei that were not protected by prospore membrane engulfment (Eastwood, Cheung, Lee, Moffat, & Meneghini, 2012). The functional integrity of old plasma membrane becomes compromised at the same time, suggesting that this membrane may also be destroyed by vacuole lysis (Eastwood & Meneghini, 2015). A direct examination of plasma membrane persistence during sporulation has not, to our knowledge, been reported. To label the plasma membrane in cells undergoing sporulation, we exposed an asynchronously sporulating culture to FM4–64, a lipophilic dye (Vida & Emr, 1995). As can be seen in Figure 3, very early in sporulation, when spore walls had not yet been made, only the plasma membrane was labelled. Later, when spore walls had been made but the ascus wall had not yet compressed around the spores, the old plasma membrane was labelled, and in some cases, the spore membranes were also labelled. In mature asci, in which the ascus wall had compressed tightly around the spores, only spore membrane labelling was visible. We interpret these results as an intact plasma membrane ‘shielding’ the spore membranes from labelling early in sporulation, then losing structural integrity later in sporulation, and finally, disappearing altogether in mature asci. If our interpretation is correct, when spores germinate and bud within an intact ascus, they only penetrate an old cell wall, and not an old membrane, in order to exit the ascus.
FIGURE 3.
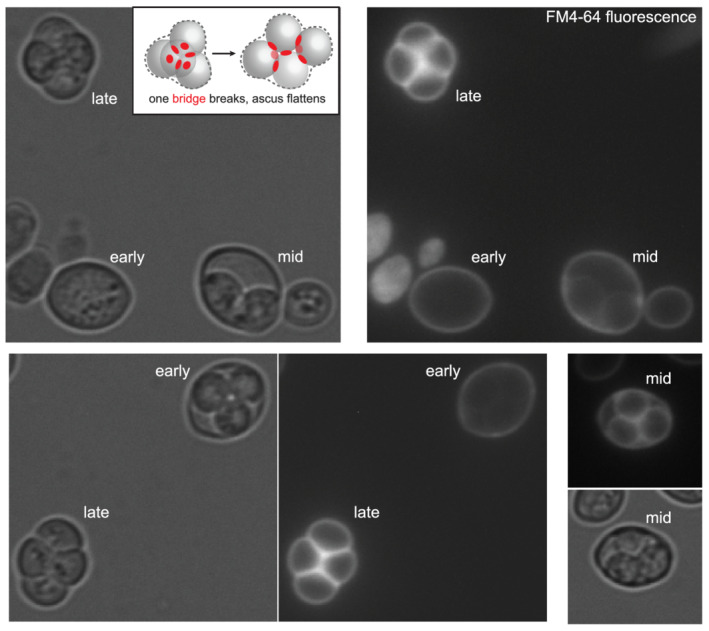
Gradual loss of the old plasma membrane during sporulation. An asynchronously sporulating culture was exposed briefly to the lipophilic dye FM™4–64. The stage of sporulation is labelled and was estimated based on spore and ascus wall appearance by transmitted light. Inset, an illustration of the arrangement of spores in a tetrahedral ascus with all six interspore bridges (red circles) intact and in a rhomboid ascus in which one of the bridges has broken (dashed red circle) as an ascus flattens under a coverslip
3.5. Global ascus wall digestion does not require the putative cell wall hydrolase Acf2
In the S. cerevisiae genome, the ACF2 gene (alias ENG2) encodes a homologue of S. pombe Eng2, one of the enzymes responsible for global ascus wall digestion (Encinar del Dedo et al., 2009). Like S. pombe Eng2, Acf2 lacks a predicted signal sequence. In S. pombe, the absence of either Agn2 or Eng2 is sufficient to almost completely prevent global ascus wall digestion, resulting in mature spores trapped within an ascus wall (Dekker et al., 2007; Encinar del Dedo et al., 2009). If in S. cerevisiae Acf2 digests the ascus wall during germination, we predicted that it should either be translated near the end of sporulation and then secreted into the ascal cytoplasm upon germination, or both translated and secreted immediately upon germination. No translation data are available for germination, but existing ribosome profiling data from synchronously sporulating cells (Brar et al., 2012) demonstrate that Acf2 translation spikes at the last stage of sporulation, just before mature spores are produced (Figure 4a). If gradual Acf2‐mediated digestion gradually thins the ascus wall during germination, we predicted that in asci lacking Acf2, the ascus wall might remain too thick and inhibit penetration by spore buds. However, we saw no discernible difference in the frequency or morphology of instances in which the buds of acf2∆ spores penetrated the ascus wall upon germination (Figure 4b). In S. cerevisiae, either Acf2 performs a different function than Egn2, or loss of a single digestive enzyme is not enough to toughen the ascus wall to an extent that is impenetrable by buds.
FIGURE 4.
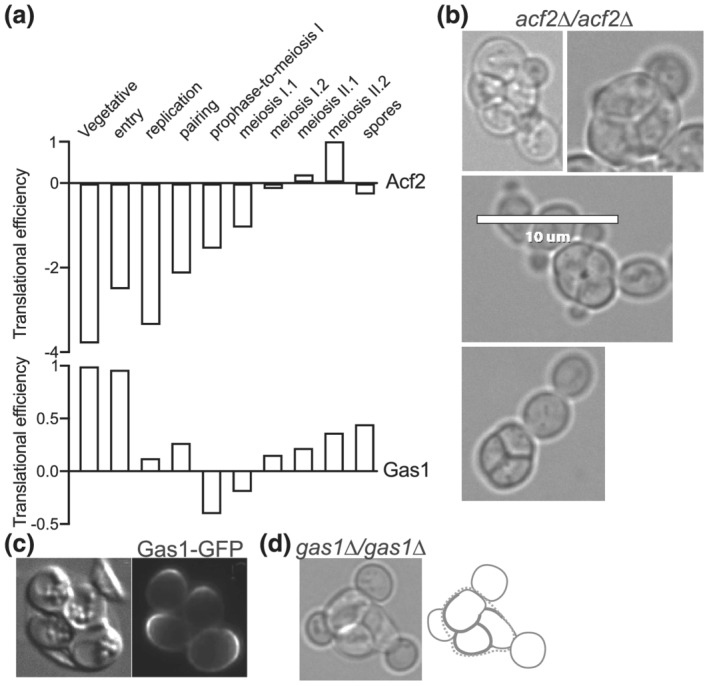
Penetration of the ascus wall by spore budding does not require Gas1 or Acf2. (a) Translational efficiencies (a measure of ribosome occupancy per mRNA) for Acf2 and Gas1 at various time points during sporulation. Data are from Brar et al. (2012). (b) Germinating asci of acf2∆/acf2∆ strain MMY0286 showing penetration of the ascus wall by spore budding. Scale bar, 10 μm. (c) Localization of Gas1‐GFP to sites of spore outgrowth upon germination. Reproduced from Rolli et al. (2011). (d) A germinating ascus of gas1∆/gas1∆ strain MMY0341
3.6. Local ascus wall digestion does not require the putative cell wall hydrolase Gas1
To identify candidate enzymes that may mediate local digestion of the ascus wall during germination, we searched the literature for published localization patterns of cell wall hydrolytic enzymes, with the logic that a protein responsible for local wall digestion should localize at the site of digestion. Translation of the α‐1,3‐glucanosyltransferase Gas1 translation increases at the end of sporulation (Figure 4a), and although this pattern was not specifically noted by the authors, examination of a GFP‐tagged allele of Gas1 in spores germinating within an intact ascus revealed that Gas1‐GFP clearly localizes to a broad region of the spore cortex opposite from interspore bridges (Rolli et al., 2011) (Figure 4c). To ask if Gas1 is required for the ability of buds to penetrate the ascus wall, we monitored germination by gas1∆/gas1∆ mutant diploid cells. Consistent with the phenotype reported in the literature for nonspore haploid gas1∆ cells (Watanabe, Watanabe, Nogami, Morishita, & Ohya, 2009), the buds produced by gas1∆ spores were oddly shaped, but they had no problems penetrating the ascus wall (Figure 4d). Thus, deletion of GAS1 is insufficient to prevent local ascus wall digestion by germinating spores.
3.7. The canonical bud site selection pathway does not drive polarity in spores
The Ras‐family GTPase Rsr1 is required for the canonical bud site selection pathways in haploid and diploid cells (Bender & Pringle, 1989). We noticed that RSR1 is transcriptionally induced early in germination (Joseph‐Strauss et al., 2007). To ask if the same pathway that controls budding in vegetative yeast cells also controls spore polarization, we sporulated rsr1∆/rsr1∆ diploids and monitored budding upon germination.
Analysis of budding was complicated by the fact that many of the mutant cells had multiple buds even prior to germination (Figure 5a). Unlike mitotic DNA replication, premeiotic DNA replication is usually uncoupled from bud emergence. The few singly‐budded asci found in nominally wild‐type strain backgrounds presumably arise from diploid cells that were in a small window of early S phase when sporulation began and proceeded directly into meiosis upon completion of DNA replication (Croes, Dodemont, & Stumm, 1976). In the rsr1∆/rsr1∆ mutants, however, either buds formed during multiple prior cell cycles failed to separate from the mother, or multiple buds formed simultaneously upon premeiotic S phase entry. We favour the latter interpretation, considering that simultaneous multiple budding is a known phenotype of rsr1∆ haploid cells expressing a synthetic fusion of the polarity scaffold protein Bem1 to the v‐SNARE Snc2 and that this phenotype is exacerbated when cells are cultured in minimal, as opposed to rich, medium (Howell et al., 2009). The starvation conditions induced by sporulation medium may bypass the need for the Bem1‐Snc2 fusion in driving multiple budding events. Indeed, we found that starving rsr1∆/rsr1∆ diploid cells by prolonged culture (1 week) in rich medium, which does not induce sporulation, was sufficient to induce a bi‐budded phenotype (Figure 5b).
FIGURE 5.
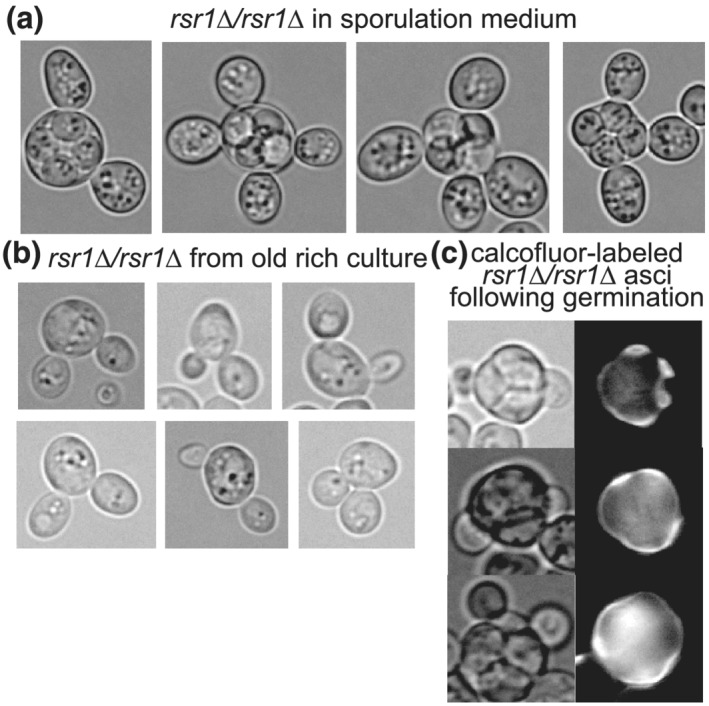
Cells lacking the polarity factor Rsr1 form multiple buds upon starvation but spore buds are able to penetrate the ascus wall. rsr1∆/rsr1∆ strain MMY0291 was cultured in (a) sporulation medium, (b) rich medium for 1 week or (c) sporulation medium, followed by pulse labelling with calcofluor white, and then rich medium to induce germination. Fluorescence images show calcofluor fluorescence
To unambiguously identify buds produced during germination, we applied the calcofluor pulse‐chase method. The vast majority of rsr1∆/rsr1∆ asci that were not budded prior to germination showed buds penetrating the ascus upon germination (Figure 5c and data not shown). We conclude from these results that spore polarity is not determined by the canonical pathway that drives polarity during budding by vegetative cells.
4. DISCUSSION
The evidence for prepolarization of S. cerevisiae spores has been hiding in plain sight for decades. For every published image we could find in which the orientation of a germinating spore relative to its sibling spores is discernible, the direction of outgrowth, budding and/or mating is consistent with prepolarization away from the interspore bridges. Ascus wall penetration by buds has also been documented previously (Hashimoto et al., 1958; Rij, 1978; Sando et al., 1980), but the implications for spore polarity were not considered. The mechanism of local ascus digestion upon spore budding also remains unknown. Budding itself requires cell wall digestion at a single site on an unbudded cell. Thus, when a spore buds inside an ascus, two walls are locally digested: the wall of the spore and the ascus wall. Another step in the yeast life cycle also requires local digestion of two walls: cell fusion upon mating. Here, it is thought that targeted secretion of enzymes followed by cell‐contact‐limited diffusion restricts digestion to a narrow pore (Huberman & Murray, 2014). Intra‐ascus mating events do not locally digest the ascus wall because repolarization towards the mating partner directs the digestive enzymes elsewhere. During an inter‐ascus mating event, on the other hand, four walls must be digested to allow fusion: one wall for each of the spores and two ascus walls. In the simplest scenario, the same enzymes that digest two walls during mating between vegetative cells also digest all four walls during inter‐ascus mating between spores. We thus find it likely that they also mediate ascus wall penetration during budding. Our results show that, despite localizing to the right place at the right time, Gas1 is dispensable for this process; other enzymes may act redundantly.
By placing the localization of Cdc10 in the context of spore positioning within an ascus, our results build on the prior identification of the septin protein Cdc10 as a marker of the sites of spore prepolarization. For other septin functions, for example, cytokinesis, Cdc10 acts together with other septin proteins as a stable hetero‐oligomeric protein complex that act as diffusion barriers and scaffolds for the recruitment and cortical retention of other proteins (Oh & Bi, 2011). Polymerization of hetero‐oligomeric septin complexes into filaments is required for cytokinesis (McMurray et al., 2011); no single septin is sufficient. Hence, although we have not yet asked if other septins colocalize with Cdc10 to cortical foci in spores, we speculate that they do. What specific function, if any, Cdc10 might perform at these foci is an intriguing question for future inquiry. In nonspore cells, septins encircle active Cdc42 at both the site of bud formation (Okada et al., 2013) and the shmoo (Kelley et al., 2015), but the cortical puncta in spores are not obviously ring‐shaped, and it is not known if Cdc42 (and/or another Rho‐family GTPase) is also there.
How does Cdc10 arrive at this location? Cdc10 and other septins localize around the spindle pole bodies at the end of meiosis metaphase II, where the prospore membrane first appears, and then localize as bars and horseshoes associated with the prospore membrane as it grows (Pablo‐Hernando et al., 2008). The nuclear envelope and prospore membrane are connected via the spindle pole body until the meiosis‐specific components of the ‘meiotic outer plaque’, Mpc54 and Mpc70, are destroyed just after meiosis II (Knop & Strasser, 2000). The exocyst complex, which targets exocytic vesicle docking and fusion, is found (along with septins) at the meiotic outer plaque (Mathieson et al., 2010), at the shmoo tip (Kelley et al., 2015) and at the site of future budding by non‐spore cells (Guo, Tamanoi, & Novick, 2001). Meiotic anaphase II pushes the spindle pole bodies toward the periphery of the ascus. If septins and/or the exocyst persist on the spore membrane at the former site of contact with the meiotic outer plaque, these ‘landmarks’ will be near the periphery of the ascus, with the sites of prospore membrane fusion (and interspore bridges) in the centre. Provided cortical diffusion of the landmarks is limited in mature spores, these sites would correspond to the Cdc10 localization patterns and sites of outgrowth that we observe upon germination.
Although future work will be required to test this model, there are several precedents for polarity factors persisting at cortical locations that were, like the meiotic outer plaque, sites of targeted vesicle trafficking. In the dumbbell‐shaped zygotes formed by vegetative cells, polarity factors at the former shmoo sites diffuse slowly from the cortex at the site of cell fusion, biasing the first diploid budding events to occur nearby (Zapanta Rinonos et al., 2014). [Incidentally, we note that in every instance we have observed, the first diploid bud produced by a derrière is also ‘medially’ positioned (Figure 2d,f and data not shown). If the derrière was still within an intact ascus, the bud penetrated the ascus wall.] Conversely, in budded vegetative cells exposed to pheromone, polarity factors persist at the bud site following cytokinesis and bias shmooing to occur nearby, independent of the direction of the pheromone gradient (Vasen, Dunayevich, & Colman‐Lerner, 2020). Polarization in both these cases does not require Rsr1 (Vasen et al., 2020; Zapanta Rinonos et al., 2014), like the polarization we observe in spores (Figure 5c).
In S. pombe spores, lack of prepolarization has been clearly demonstrated (Bonazzi et al., 2014), and the assumption that S. cerevisiae is the same is understandable, given the many similarities with regard to mechanisms of sporulation. Several key differences are worth noting, however. Fission yeast spores lack interspore bridges, and ascus wall breakdown is a programmed event taking place immediately following successful completion of meiosis (Guo & King, 2013). Hence, S. pombe spores are designed for dispersal, consistent with a mostly haploid lifestyle in this species, whereas Saccharomyces species appear to have evolved to prioritize return to diploidy following meiosis. From this perspective, why prepolarize a spore away from its meiotic siblings, at least two‐thirds of which will be of compatible mating type? A simple explanation may be that interspore bridges represent a physical barrier to outgrowth, analogous to the difficulties that nonspore yeast cells encounter when mutations drive them to rebud through a chitin‐rich bud scar (Tong et al., 2007). We propose another model. In a crowded four‐spored ascus, navigating gradients of multiple pheromones in order to identify a compatible partner is nontrivial (Rappaport & Barkai, 2012). If germinating spores grew into the centre of the ascus, the situation would become even more complex, especially if spores budded and then underwent mating type switching, a process available to most wild isolates but only after during the G1 phase following the spore's first budding event (Haber, 2012). Hence, prepolarization away from the centre may facilitate pheromone sensing by simplifying the gradients of signals spores must navigate to find a mate.
From a broader perspective, studies of polarity determination, pheromone detection and mating by isolated nonspore Saccharomyces cells in the laboratory setting have provided numerous insights into basic biology. However, considering what we now know about the Saccharomyces life cycle outside the lab (Tsai, Bensasson, Burt, & Koufopanou, 2008), budding and mating by vegetative haploids may represent a kind of backup plan for circumstances in which a spore is physically separated from—or chooses to ignore (McClure et al., 2018)—its meiotic siblings. Instead, budding and mating by spores and their immediate progeny may be the context in which evolution has had the most opportunities to sculpt these processes. With this idea in mind, our findings provide new lenses through which to view key cellular events (see Figure 6). First, by placing two cells of one mating type adjacent to two cells of opposite mating type, axial budding by haploids has been proposed to promote a rapid return to diploidy following mating‐type switching by an isolated spore (Gimeno & Fink, 1992). We further propose that axial budding helps ensure that if a spore is still within an intact ascus when it buds a second time, the second bud also penetrates the ascus wall (Figure 6). Second, it was recently discovered that unbudded vegetative daughter cells exposed to pheromone are biased to polarize away from the sites of separation from their mothers (their distal poles) (Vasen et al., 2020). Those authors speculated that a distal bias for daughter cell polarization may have evolved as a way to promote outbreeding, by disfavouring mating between a daughter and its mother following mating‐type switching (Vasen et al., 2020). We propose that, even if it has been locally digested by budding, an otherwise intact ascus wall surrounding the former connection between a spore and its first daughter may also inhibit mating between those two cells (Figure 6). Our work thus highlights outstanding questions and lays a foundation for future studies.
FIGURE 6.
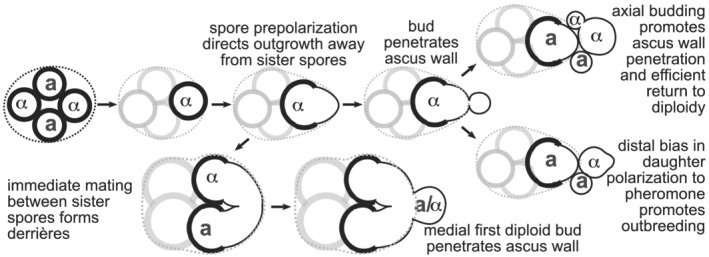
Illustration of possible roles for polarity decisions in key steps of the S. cerevisiae life cycle. Thicker lines represent the spore wall. Dashed line, ascus wall. Mating types are indicated with ‘a’ or ‘α’
ACKNOWLEDGEMENTS
We thank Jeff Moore (Univ. Colorado Anschutz Medical Campus) for use of his confocal microscope. This work was supported by the National Science Foundation, Directorate of Biological Sciences (award 1928900 to M.M.) and by the National Institute of General Medical Sciences of the National Institutes of Health (award T32GM008730 to L.R.H., as part of the Molecular Biology PhD Program). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Science Foundation or the National Institutes of Health.
Heasley LR, Singer E, Cooperman BJ, McMurray MA. Saccharomyces spores are born prepolarized to outgrow away from spore–spore connections and penetrate the ascus wall. Yeast. 2021;38:90–101. 10.1002/yea.3540
REFERENCES
- Bender, A. , & Pringle, J. R. (1989). Multicopy suppression of the cdc24 budding defect in yeast by CDC42 and three newly identified genes including the ras‐related gene RSR1. Proceedings of the National Academy of Sciences of the United States of America, 86(24), 9976–9980. 10.1073/pnas.86.24.9976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonazzi, D. , Julien, J.‐D. , Romao, M. , Seddiki, R. , Piel, M. , Boudaoud, A. , & Minc, N. (2014). Symmetry breaking in spore germination relies on an interplay between polar cap stability and spore wall mechanics. Developmental Cell, 28(5), 534–546. 10.1016/j.devcel.2014.01.023 [DOI] [PubMed] [Google Scholar]
- Brar, G. A. , Yassour, M. , Friedman, N. , Regev, A. , Ingolia, N. T. , & Weissman, J. S. (2012). High‐resolution view of the yeast meiotic program revealed by ribosome profiling. Science (New York, N.Y.), 335(6068), 552–557. 10.1126/science.1215110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briza, P. , Ellinger, A. , Winkler, G. , & Breitenbach, M. (1990). Characterization of a DL‐dityrosine‐containing macromolecule from yeast ascospore walls. The Journal of Biological Chemistry, 265(25), 15118–15123. [PubMed] [Google Scholar]
- Cabib, E. , & Bowers, B. (1975). Timing and function of chitin synthesis in yeast. Journal of Bacteriology, 124(3), 1586–1593. 10.1128/JB.124.3.1586-1593.1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou, J.‐G. , Balasubramanian, M. K. , & Lew, D. J. (2017). Cell polarity in yeast. Annual Review of Cell and Developmental Biology, 33, 77–101. 10.1146/annurev-cellbio-100616-060856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluccio, A. , & Neiman, A. M. (2004). Interspore bridges: A new feature of the Saccharomyces cerevisiae spore wall. Microbiology (Reading, England), 150(Pt 10), 3189–3196. 10.1099/mic.0.27253-0 [DOI] [PubMed] [Google Scholar]
- Coluccio, A. E. , Rodriguez, R. K. , Kernan, M. J. , & Neiman, A. M. (2008). The yeast spore wall enables spores to survive passage through the digestive tract of Drosophila . PLoS ONE, 3(8) e2873. 10.1371/journal.pone.0002873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croes, A. F. , Dodemont, H. J. , & Stumm, C. (1976). Induction of meiosis in Saccharomyces cerevisiae at different points in the mitotic cycle. Planta, 130(2), 131–136. 10.1007/BF00384409 [DOI] [PubMed] [Google Scholar]
- Dekker, N. , van Rijssel, J. , Distel, B. , & Hochstenbach, F. (2007). Role of the alpha‐glucanase Agn2p in ascus‐wall endolysis following sporulation in fission yeast. Yeast (Chichester, England), 24(4), 279–288. 10.1002/yea.1464 [DOI] [PubMed] [Google Scholar]
- Eastwood, M. D. , Cheung, S. W. T. , Lee, K. Y. , Moffat, J. , & Meneghini, M. D. (2012). Developmentally programmed nuclear destruction during yeast gametogenesis. Developmental Cell, 23(1), 35–44. 10.1016/j.devcel.2012.05.005 [DOI] [PubMed] [Google Scholar]
- Eastwood, M. D. , & Meneghini, M. D. (2015). Developmental coordination of gamete differentiation with programmed cell death in sporulating yeast. Eukaryotic Cell, 14(9), 858–867. 10.1128/EC.00068-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encinar del Dedo, J. , Dueñas, E. , Arnáiz, Y. , del Rey, F. , & Vázquez de Aldana, C. R. (2009). {beta}‐glucanase Eng2 is required for ascus wall endolysis after sporulation in the fission yeast Schizosaccharomyces pombe . Eukaryotic Cell, 8(8), 1278–1286. 10.1128/EC.00148-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunishi, K. , Miyakubi, K. , Hatanaka, M. , Otsuru, N. , Hirata, A. , Shimoda, C. , & Nakamura, T. (2014). The fission yeast spore is coated by a proteinaceous surface layer comprising mainly Isp3. Molecular Biology of the Cell, 25(10), 1549–1559. 10.1091/mbc.e13-12-0731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geijer, C. , Joseph‐Strauss, D. , Simchen, G. , Barkai, N. , & Hohmann, S. (2010). Saccharomyces cerevisiae spore germination In Lubzens E., Cerda J. & Clark M. (Eds.), Dormancy and resistance in harsh environments (pp. 29–41). Berlin Heidelberg: Springer; 10.1007/978-3-642-12422-8_3 [DOI] [Google Scholar]
- Giaever, G. , Chu, A. M. , Ni, L. , Connelly, C. , Riles, L. , Véronneau, S. , … Johnston, M. (2002). Functional profiling of the Saccharomyces cerevisiae genome. Nature, 418(6896), 387–391. 10.1038/nature00935 [DOI] [PubMed] [Google Scholar]
- Gimeno, C. , & Fink, G. (1992). The logic of cell division in the life cycle of yeast. Science, 257(5070), 626 10.1126/science.1496375 [DOI] [PubMed] [Google Scholar]
- Guo, H. , & King, M. C. (2013). A quality control mechanism linking meiotic success to release of ascospores. PLoS ONE, 8(12) e82758. 10.1371/journal.pone.0082758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, W. , Tamanoi, F. , & Novick, P. (2001). Spatial regulation of the exocyst complex by Rho1 GTPase. Nature Cell Biology, 3(4), 353–360. 10.1038/35070029 [DOI] [PubMed] [Google Scholar]
- Haber, J. E. (2012). Mating‐type genes and MAT switching in Saccharomyces cerevisiae . Genetics, 191(1), 33–64. 10.1534/genetics.111.134577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, T. , Conti, S. F. , & Naylor, H. B. (1958). Fine structure of microorganisms. III. Electron microscopy of resting and germinating ascospores of Saccharomyces cerevisiae . Journal of Bacteriology, 76(4), 406–416. 10.1128/JB.76.4.406-416.1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasley, L. R. , Garcia, G. , & McMurray, M. A. (2014). Off‐target effects of the septin drug forchlorfenuron on nonplant eukaryotes. Eukaryotic Cell, 13(11), 1411–1420. 10.1128/EC.00191-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, A. S. , Savage, N. S. , Johnson, S. , Bose, I. , Wagner, A. W. , Zyla, T. R. , … Lew, D. J. (2009). Singularity in polarization: Rewiring yeast cells to make two buds. Cell, 139, 731–743. 10.1016/j.cell.2009.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman, L. B. , & Murray, A. W. (2014). A model for cell wall dissolution in mating yeast cells: Polarized secretion and restricted diffusion of cell wall remodeling enzymes induces local dissolution. PLoS ONE, 9(10) e109780. 10.1371/journal.pone.0109780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase, M. , Luo, J. , Nagaraj, S. , Longtine, M. , Kim, H. B. , Haarer, B. K. , … Bi, E. (2006). Role of a Cdc42p effector pathway in recruitment of the yeast septins to the presumptive bud site. Molecular Biology of the Cell, 17(3), 1110–1125. 10.1091/mbc.E05-08-0793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, C. R. , Weems, A. D. , Brewer, J. M. , Thorner, J. , & McMurray, M. A. (2015). Cytosolic chaperones mediate quality control of higher‐order septin assembly in budding yeast. Molecular Biology of the Cell, 26, 1323–1344. 10.1091/mbc.E14-11-1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph‐Strauss, D. , Zenvirth, D. , Simchen, G. , & Barkai, N. (2007). Spore germination in Saccharomyces cerevisiae: Global gene expression patterns and cell cycle landmarks. Genome Biology, 8(11), R241 10.1186/gb-2007-8-11-r241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, J. B. , Dixit, G. , Sheetz, J. B. , Venkatapurapu, S. P. , Elston, T. C. , & Dohlman, H. G. (2015). RGS proteins and septins cooperate to promote chemotropism by regulating polar cap mobility. Current Biology: CB, 25(3), 275–285. 10.1016/j.cub.2014.11.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloimwieder, A. , & Winston, F. (2011). A screen for germination mutants in Saccharomyces cerevisiae. G3: Genes, genomes . Genetics, 1(2), 143–149. 10.1534/g3.111.000323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop, M. , & Strasser, K. (2000). Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. The EMBO Journal, 19(14), 3657–3667. 10.1093/emboj/19.14.3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono, K. , Matsunaga, R. , Hirata, A. , Suzuki, G. , Abe, M. , & Ohya, Y. (2005). Involvement of actin and polarisome in morphological change during spore germination of Saccharomyces cerevisiae . Yeast (Chichester, England), 22(2), 129–139. 10.1002/yea.1205 [DOI] [PubMed] [Google Scholar]
- Mathieson, E. M. , Suda, Y. , Nickas, M. , Snydsman, B. , Davis, T. N. , Muller, E. G. D. , & Neiman, A. M. (2010). Vesicle docking to the spindle pole body is necessary to recruit the exocyst during membrane formation in Saccharomyces cerevisiae . Molecular Biology of the Cell, 21(21), 3693–3707. 10.1091/mbc.E10-07-0563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure, A. W. , Jacobs, K. C. , Zyla, T. R. , & Lew, D. J. (2018). Mating in wild yeast: Delayed interest in sex after spore germination. Molecular Biology of the Cell, 29(26), 3119–3127. 10.1091/mbc.E18-08-0528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray, M. A. (2016). Assays for genetic dissection of septin filament assembly in yeast, from de novo folding through polymerization. Methods in Cell Biology, 136, 99–116. 10.1016/bs.mcb.2016.03.012 [DOI] [PubMed] [Google Scholar]
- McMurray, M. A. , Bertin, A. , Garcia, G. 3rd , Lam, L. , Nogales, E. , & Thorner, J. (2011). Septin filament formation is essential in budding yeast. Developmental Cell, 20(4), 540–549. 10.1016/j.devcel.2011.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray, M. A. , & Thorner, J. (2008). Septin stability and recycling during dynamic structural transitions in cell division and development. Current Biology: CB, 18(16), 1203–1208. 10.1016/j.cub.2008.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman, A. M. (2011). Sporulation in the budding yeast Saccharomyces cerevisiae . Genetics, 189(3), 737–765. 10.1534/genetics.111.127126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, Y. , & Bi, E. (2011). Septin structure and function in yeast and beyond. Trends in Cell Biology, 21(3), 141–148. 10.1016/j.tcb.2010.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, S. , Leda, M. , Hanna, J. , Savage, N. S. , Bi, E. , & Goryachev, A. B. (2013). Daughter cell identity emerges from the interplay of Cdc42, septins, and exocytosis. Developmental Cell, 26(2), 148–161. 10.1016/j.devcel.2013.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pablo‐Hernando, M. E. , Arnaiz‐Pita, Y. , Tachikawa, H. , del Rey, F. , Neiman, A. M. , & Vázquez de Aldana, C. R. (2008). Septins localize to microtubules during nutritional limitation in Saccharomyces cerevisiae . BMC Cell Biology, 9, 55 10.1186/1471-2121-9-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport, N. , & Barkai, N. (2012). Disentangling signaling gradients generated by equivalent sources. Journal of Biological Physics, 38(2), 267–278. 10.1007/s10867-011-9240-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rij, N. J. W. K.‐V. (1978). Electron microscopy of germinating ascospores of Saccharomyces cerevisiae . Archives of Microbiology, 117(1), 73–77. 10.1007/BF00689354 [DOI] [PubMed] [Google Scholar]
- Rolli, E. , Ragni, E. , de Medina‐Redondo, M. , Arroyo, J. , de Aldana, C. R. V. , & Popolo, L. (2011). Expression, stability, and replacement of glucan‐remodeling enzymes during developmental transitions in Saccharomyces cerevisiae . Molecular Biology of the Cell, 22(9), 1585–1598. 10.1091/mbc.E10-03-0268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sando, N. , Oguchi, T. , Nagano, M. , & Osumi, M. (1980). Morphological changes in ascospores of Saccharomyces cerevisiae during aerobic and anaerobic germination. The Journal of General and Applied Microbiology, 26(6), 403–412. 10.2323/jgam.26.403 [DOI] [Google Scholar]
- Schindelin, J. , Arganda‐Carreras, I. , Frise, E. , Kaynig, V. , Longair, M. , Pietzsch, T. , … Cardona, A. (2012). Fiji: An open‐source platform for biological‐image analysis. Nature Methods, 9(7), 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena, E. , Radin, D. , & Fogel, S. (1973, May). Synchronous mating in yeast. Proceedings of the National Academy of Sciences of the United States of America, 70, 1373–1377. 10.1073/pnas.70.5.1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, Z. , Gao, X.‐D. , Howell, A. S. , Bose, I. , Lew, D. J. , & Bi, E. (2007). Adjacent positioning of cellular structures enabled by a Cdc42 GTPase‐activating protein‐mediated zone of inhibition. The Journal of Cell Biology, 179(7), 1375–1384. 10.1083/jcb.200705160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, I. J. , Bensasson, D. , Burt, A. , & Koufopanou, V. (2008). Population genomics of the wild yeast Saccharomyces paradoxus: Quantifying the life cycle. Proceedings of the National Academy of Sciences of the United States of America, 105(12), 4957–4962. 10.1073/pnas.0707314105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasen, G. , Dunayevich, P. , & Colman‐Lerner, A. (2020). Mitotic and pheromone‐specific intrinsic polarization cues interfere with gradient sensing in Saccharomyces cerevisiae . Proceedings of the National Academy of Sciences of the United States of America, 117(12), 6580–6589. 10.1073/pnas.1912505117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida, T. , & Emr, S. D. (1995, March). A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. The Journal of Cell Biology, 128, 779–792. 10.1083/jcb.128.5.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, M. , Watanabe, D. , Nogami, S. , Morishita, S. , & Ohya, Y. (2009). Comprehensive and quantitative analysis of yeast deletion mutants defective in apical and isotropic bud growth. Current Genetics, 55(4), 365–380. 10.1007/s00294-009-0251-0 [DOI] [PubMed] [Google Scholar]
- Zapanta Rinonos, S. , Rai, U. , Vereb, S. , Wolf, K. , Yuen, E. , Lin, C. , & Tartakoff, A. M. (2014). Sequential logic of polarity determination during the haploid‐to‐diploid transition in Saccharomyces cerevisiae . Eukaryotic Cell, 13(11), 1393–1402. 10.1128/EC.00161-14 [DOI] [PMC free article] [PubMed] [Google Scholar]


