Significance Statement
This technical advance introduces a robust and reliable disease assay to monitor bacterial growth of Pseudomonas syringae in agroinfiltrated tissues using an antibiotics cocktail to select against Agrobacterium tumefaciens.
Keywords: Agrobacterium, Nicotiana benthamiana, plant immunity, Pseudomonas syringae, disease assay, technical advance
Summary
The lengthy process to generate transformed plants is a limitation in current research on the interactions of the model plant pathogen Pseudomonas syringae with plant hosts. Here we present an easy method called agromonas, where we quantify P. syringae growth in agroinfiltrated leaves of Nicotiana benthamiana using a cocktail of antibiotics to select P. syringae on plates. As a proof of concept, we demonstrate that transient expression of PAMP receptors reduces bacterial growth, and that transient depletion of a host immune gene and transient expression of a type‐III effector increase P. syringae growth in agromonas assays. We show that we can rapidly achieve structure−function analysis of immune components and test the function of immune hydrolases. The agromonas method is easy, fast and robust for routine disease assays with various Pseudomonas strains without transforming plants or bacteria. The agromonas assay offers a reliable approach for further comprehensive analysis of plant immunity.
INTRODUCTION
Understanding the plant immune system and microbial pathogenicity is essential to improve plant biotechnologies and crop protection. To evaluate the level of resistance of a plant or the virulence of bacterial pathogens, the routine method relies on infection assays that quantify bacterial growth (i.e. colony count assays). Colony count assays are usually performed on stable transformant plants. However, generation of stable transgenic lines is time and resource consuming, and is limited to plant species that are amenable to genetic transformation. Therefore, there is a need for faster disease assays particularly in the studies of the model plant pathogen Pseudomonas syringae, which causes important economic damages in many plant species (Mansfield et al., 2012).
Rapid overexpression and transcript depletion of various exogenous and endogenous genes is facilitated by Agrobacterium tumefaciens‐mediated transient expression (agroinfiltration). Agroinfiltration is used throughout plant science to study protein localisation and for their biochemical characterisation. Agroinfiltrated leaves are routinely used to study the interaction between the model plant Nicotiana benthamiana and the potato blight pathogen Phytophthora infestans (Chaparro‐Garcia et al., 2011; Bozkurt et al., 2014; Dagdas et al., 2018) and other Phytophthora species. However, agroinfiltrated N. benthamiana leaves are not routinely used for disease assays with P. syringae. One problem is that selective isolation of P. syringae from agroinfiltrated tissue is challenging because there is an overlap of endogenous or introduced antibiotics resistance. For example, rifampicin is commonly used to select for antibiotic‐resistance genes in the genome of A. tumefaciens and P. syringae strains, whereas resistance to kanamycin is frequently used to maintain plasmids in both bacteria and therefore these antibiotics are not useful for selective isolation in co‐inoculated tissue.
A solution to this problem is to use selection for endogenous bacterial resistance to antibiotics. The combination of 10 µg ml−1 cetrimide, 10 µg ml−1 fucidin and 50 µg ml−1 cephaloridine (CFC; Figure 1a) permits the selection of Pseudomonas species (Mead and Adams, 1977). CFC is used for the selective isolation of Pseudomonas species during the microbiological examination of environmental, clinical, food and plant samples (Krueger and Sheikh, 1987; Hill et al., 2005; Pantazi et al., 2008; Fones et al., 2010; Straub et al., 2018), but has not yet been exploited in combination with agroinfiltrated leaves.
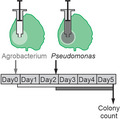
Figure 1.
Selection against Agrobacterium tumefaciens.
(a) Chemical structures of cephaloridine, fucidin and cetrimide (CFC).
(b) Pseudomonas syringae grows on CFC selection, Agrobacterium does not. Nicotiana benthamiana leaves were infiltrated with 1 × 106 CFU ml−1 P. syringae or 1 × 108 CFU ml−1 AtumGV3101, and bacterial populations were determined 3 days later using colony count method using Luria−Bertani (LB) plates containing CFC or not. Error bars represent SE of n = 3 biological replicates. Student’s t‐test statistics (***P < 0.001). CFU, colony‐forming units.
Here, we use CFC selection to establish a rapid and easy disease assay to quantify growth of P. syringae from agroinfiltrated leaves. We tested whether immunity can be studied in agroinfiltrated leaves even though these leaves contain A. tumefaciens. The method we developed is called ‘agromonas’ because it is based on agroinfiltration followed by inoculation of Pseudomonas syringae by both infiltration and spray inoculation. We demonstrate that the agromonas assay can be applied to different P. syringae strains, and demonstrate its adaptability to study the impact of immune components and bacterial effectors on P. syringae growth in planta.
RESULTS
CFC facilitates Pseudomonas syringae selection from agroinfiltrated tissues
To confirm that CFC facilitates P. syringae selection, we tested three different P. syringae strains that are pathogenic on N. benthamiana. We tested P. syringae pv. tomato DC3000, the causative agent of the bacterial speck disease of tomato lacking the type III effector gene hopQ1‐1 [PtoDC3000(∆hQ)]; P. syringae pv. tabaci 6605 (Pta6605), the causative agent for wildfire disease in tobacco; and P. syringae pv. syringae B728a (PsyB728a), the causative agent of bacterial brown spot of bean. We also included A. tumefaciens GV3101 (AtumGV3101), the non‐oncogenic strain that is routinely used for agroinfiltration. These strains were infiltrated into N. benthamiana leaves and, at 3 days post‐infiltration (3 dpi), leaf extracts were generated, diluted in water and plated out on Luria−Bertani (LB) medium with or without CFC selection. All tested P. syringae pathovars grew equally well on LB medium supplemented with or without CFC (Figure 1b), demonstrating that CFC does not affect P. syringae growth. By contrast, AtumGV3101 did not grow at all on plates containing CFC (Figure 1b). Likewise, CFC also blocks growth of the non‐oncogenic AtumC58C1 as well as all tested Xanthomonas strains (Table S1), confirming the selectivity of CFC.
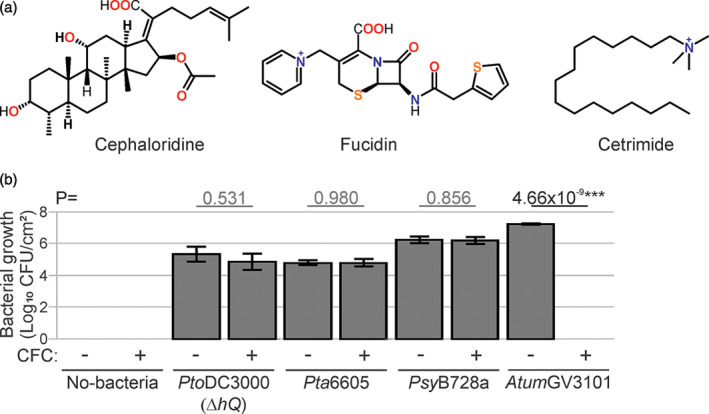
To apply CFC selection to facilitate the selective isolation of P. syringae from agroinfiltrated leaves, N. benthamiana leaves were first infiltrated with AtumGV3101. Two days after agroinfiltration, each of the three different P. syringae strains were infiltrated into the agroinfiltrated regions. Three days later, leaf homogenates were plated onto LB plates containing CFC or gentamicin (Figure 2a). While P. syringae strains were specifically isolated on CFC plates, AtumGV3101 was isolated on plates containing gentamicin (Figure 2b,c). No colonies with Agrobacterium morphology were detected on CFC plates (Figure 2b), demonstrating that A. tumefaciens cannot grow on CFC plates, even when P. syringae is growing. These data demonstrate that CFC‐containing medium facilitates the selection of living Pseudomonas spp. from agroinfiltrated leaves. Comparison with the samples taken immediately upon P. syringae infiltration (0 dpi) shows that P. syringae populations grow at least 10‐fold in agroinfiltrated tissues (Figure 2c).
Figure 2.
Concept of agromonas assay.
(a) Experimental procedure for agromonas assay. Two days after agroinfiltration, agroinfiltrated leaves are infiltrated with Pseudomonas syringae bacteria. Bacterial growth is measured 3 days later (3 days post‐infiltration, 3 dpi) by a classic colony count on Luria−Bertani (LB) agar plates containing cephaloridine, fucidin and cetrimide (CFC).
(b) CFC selects P. syringae from agroinfiltrated leaves. Agroinfiltrated leaves were infiltrated with 1 × 106 CFU ml−1 PtoDC3000(∆hQ) and 3 days later leaf extracts were diluted, and each dilution was plated onto medium supplemented with or without CFC. Pictures were taken 48 h later.
(c) Selective isolation of Pseudomonas spp. from agroinfiltrated leaves. Agroinfiltrated leaves were infiltrated with 1 × 106 CFU ml−1 P. syringae and, at 0 and 3 dpi, leaf extracts were plated on medium containing CFC or gentamicin to select P. syringae or Agrobacterium, respectively. Error bars represent SE of n = 3 biological replicates.
While comparing unmixed infection with mixed infection (Figures 1b and 2c), we observed that the presence of A. tumefaciens suppresses P. syringae growth, as previously reported (Rico et al., 2010). We compared the growth of PtoDC3000(∆hQ) in the presence and absence of AtumGV3101. PtoDC3000(∆hQ) grew ninefold less in N. benthamiana leaves infiltrated with AtumGV3101 as compared with a non‐agroinfiltrated sample (Figure S1), confirming that A. tumefaciens reduces P. syringae growth in planta. Therefore, it is essential to use agroinfiltrated leaves expressing the empty vector (EV) as control in agromonas assays.
PAMP receptors reduce bacterial growth in agromonas assay
To demonstrate that the agromonas assay can be used to study genes that confer immunity, we used two pattern recognition receptors (PRRs) that are absent in N. benthamiana. We tested tomato FLS3 (flagellin‐sensing 3) and Arabidopsis EFR (EF‐Tu receptor), which recognize the flgII‐28 epitope of flagellin and the elf18 epitope of EF‐Tu, respectively (Kunze et al., 2004; Zipfel et al., 2006; Cai et al., 2011; Hind et al., 2016).
To confirm the functionality of FLS3 and EFR upon agroinfiltration, we transiently expressed FLS3 and EFR in N. benthamiana leaves and measured the production of reactive oxygen species (ROS) upon treatment with flgII‐28 and elf18. Leaves transiently expressing FLS3 were able to release a ROS burst upon flgII‐28 treatment, whereas EFR and EV expressing leaves remained unresponsive to flgII‐28 (Figure 3a). Likewise, leaves that transiently express EFR were able to release an oxidative burst upon elf18 treatment, whereas FLS3 and EV expressing leaves remained unresponsive to elf18 (Figure 3a). These results show that FLS3 and EFR are functional in N. benthamiana, consistent with previous studies (Lacombe et al., 2010; Hind et al., 2016).
Figure 3.
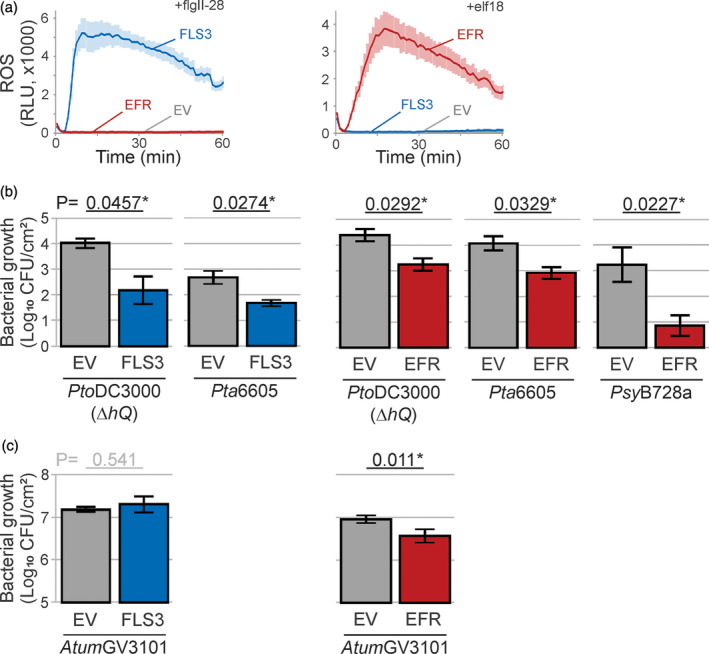
PAMP receptors reduce bacterial growth in the agromonas assay.
(a) Transient expression of tomato FLS3 and Arabidopsis EFR in Nicotiana benthamiana confers flgII‐28 and elf18 responsiveness, respectively. Leaf discs from agroinfiltrated leaves expressing FLS3 (blue), EFR (red) or empty vector (EV; grey) were treated with 100 nm flgII‐28 or elf18, and reactive oxygen species (ROS) was measured in relative light units (RLU). Error intervals (shaded regions) represent SE of n = 12 biological replicates.
(b) Transient expression of FLS3 or EFR reduces Pseudomonas syringae growth. Two days after agroinfiltration, agroinfiltrated leaves expressing FLS3 (blue), EFR (red) or EV (grey) were spray‐inoculated with the indicated strains of P. syringae (at 1 × 108 CFU ml−1), and bacterial growth was measured 3 days later using cephaloridine, fucidin and cetrimide (CFC) selection. Error bars represent SE of n = 3 biological replicates. Student’s t‐test statistics (*P < 0.05).
(c) Transient expression of EFR, but not FLS3, affect Agrobacterium growth. Bacterial growth of AtumGV3101 was measured by plating the leaf extracts described in (b) on medium containing gentamicin. Error bars represent SE of n = 3 replicates. Student’s t‐test statistics (* P < 0.05).
We next tested whether agroinfiltrated leaves expressing FLS3 have enhanced resistance to P. syringae upon infection. Agroinfiltrated leaves of N. benthamiana plants transiently expressing FLS3 showed reduced bacterial growth of both PtoDC3000(∆hQ) and Pta6605 strains compared with leaves expressing the EV control (Figure 3b). Similarly, EFR transient expression caused a strong reduction in the growth of PtoDC3000(∆hQ), Pta6605 and PsyB728a (Figure 3b). Altogether these data demonstrate that agroinfiltration of FLS3 and EFR increases immunity to P. syringae in N. benthamiana.
We also measured bacterial growth of AtumGV3101 in the same extracts using gentamicin selection. While no effect on AtumGV3101 growth was detected in leaves transiently expressing EV and FLS3 (Figure 3c), transient expression of EFR in N. benthamiana reduced AtumGV3101 growth (Figure 3c), consistent with a previous study using EFR transgenic plants (Lacombe et al., 2010).
Depletion of host immunity gene increases bacterial growth in agromonas assay
To test if we could also promote bacterial growth by depleting a host immune component in agromonas assays, we depleted NbFLS2 by RNAi using hairpin (hp) constructs (Yan et al., 2012). We monitored ROS production after flg22 treatment to confirm NbFLS2 depletion. Agroinfiltration of hpFLS2 had no effect on flg22‐induced ROS production 3 days after agroinfiltration, but suppressed the response 10 days after agroinfiltration in contrast to hpGFP (Figure 4a,b), confirming the selective depletion of NbFLS2.
Figure 4.
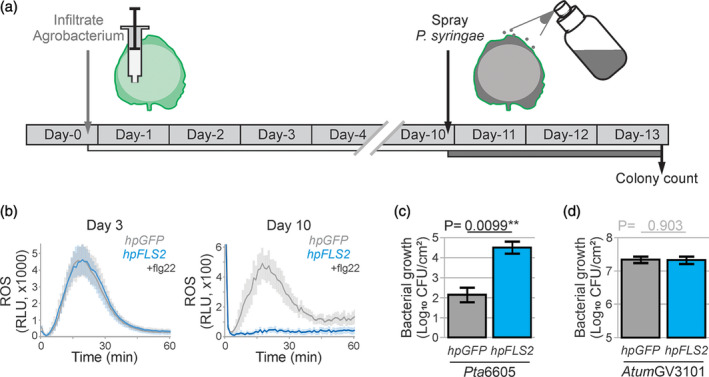
Depletion of host immune gene increases Pseudomonas syringae growth in the agromonas assay.
(a) Experimental procedure for studying the role of endogenous immune components in agromonas assay. Ten days after agroinfiltration, agroinfiltrated leaves are spray‐inoculated with P. syringae bacteria. Bacterial growth is measured 3 days later by a classic colony count on Luria−Bertani (LB) agar plates containing cephaloridine, fucidin and cetrimide (CFC).
(b) FLS2 depletion reduces reactive oxygen species (ROS) production upon flg22 treatment. Leaves were agroinfiltrated (OD600 = 0.2) with hpGFP (grey) or hpFLS2 (blue) and, at 3 and 10 dpi, leaf discs were treated with 100 nm flg22. Error intervals represent SE of n = 12 replicates.
(c) FLS2 depletion increases P. syringae growth. Agroinfiltrated leaves expressing hpGFP (grey) or hpFLS2 (blue) were spray‐inoculated at 10 dpi with 1 × 108 CFU ml−1 Pta6605 and bacterial growth was measured 3 days later using CFC selection. Error bars represent SE of n = 3 biological replicates. Student’s t‐test statistics (**P < 0.01).
(d) FLS2 depletion does not affect Agrobacterium growth. Bacterial growth of AtumGV3101 was measured by plating the leaf extracts described in (c) on medium containing gentamicin. Error bars represent SE of n = 3 replicates. Student’s t‐test statistics.
We next inoculated these agroinfiltrated leaves depleted for NbFLS2 with Pta6605 to measure plant immunity to bacteria. NbFLS2 depletion using hpFLS2 resulted in significantly more P. syringae growth compared with the hpGFP control (Figure 4c). These data are consistent with the reported role of NbFLS2 in immunity to P. syringae (Segonzac et al., 2011), demonstrating that depletion with hairpin constructs can be used in agromonas assays to study the role of endogenous immune components. By contrast, AtumGV3101 grew equally well in both hpGFP and hpFLS2 expressing leaves (Figure 4d), consistent with the absence of immunogenic sequences in flagellin of A. tumefaciens that are recognized by NbFLS2 (Hann and Rathjen, 2007).
Rapid functional analysis of immune components in agromonas assay
To illustrate that the agromonas assay can be used for fast functional analysis (Figure 5a), we generated the non‐phosphorylatable mutant EFRY836F known to be inactive in elf18‐induced signalling (Macho et al., 2014). Indeed, agroinfiltrated leaves expressing EFRY836F were unable to mount an elf18‐induced ROS burst, unlike wild‐type EFR (Figure 5b), confirming the non‐functionality of this EFRY836F mutant. Consequently, leaves expressing EFRY836F were more susceptible to PtoDC3000(ΔhQ), in contrast to leaves expressing wild‐type EFR (Figure 5c). Consistent with a role for EFR in conferring resistance to A. tumefaciens, growth of AtumGV3101 was also reduced by the functional EFR receptor, but not in the presence of EFRY836F (Figure 5d). These data are consistent with the reported crucial role of Y836 of EFR in immunity to P. syringae (Macho et al., 2014), and illustrate that the agromonas assay can be used for quick and robust functional analysis of immune components.
Figure 5.
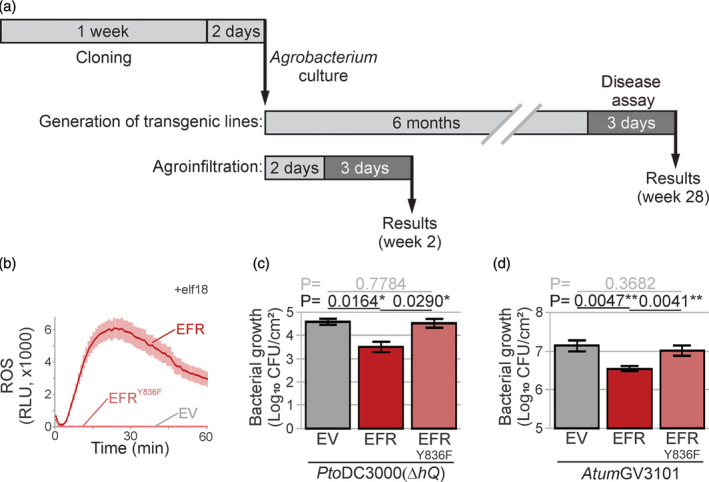
Rapid functional analysis of immune components.
(a) Time scale for functional analysis by generation transgenic plants and by agroinfiltration (agromonas assay).
(b) Phosphomutant EFRY836F is unable to trigger reactive oxygen species (ROS) burst upon elf18 treatment. Leaves were agroinfiltrated with EFR (red), EFRY836F (light red) or empty vector (EV; grey), and the ROS burst was measured at 3 dpi in leaf discs treated with 100 nm elf18. Error intervals represent SE of n = 12 replicates.
(c) Phosphomutant EFRY836F is blocked in elf18‐triggered immunity. Two days after agroinfiltration, agroinfiltrated leaves expressing EFR, EFRY836F or EV were spray‐inoculated with 1 × 108 CFU ml−1 PtoDC3000(∆hQ) and bacterial growth was measured 3 days later using cephaloridine, fucidin and cetrimide (CFC) selection. Error bars represent SE of n = 3 biological replicates. Student’s t‐test statistics (*P < 0.05).
(d) Agroinfiltration of EFR, but not EFRY836F, reduces Agrobacterium growth. Bacterial growth of AtumGV3101 was measured by plating the leaf extracts described in (c) on medium containing gentamicin. Error bars represent SE of n = 3 biological replicates. Student’s t‐test statistics (** P < 0.01).
T3 effector increases bacterial growth in agromonas assay
To demonstrate that bacterial growth can be increased in agromonas assays by transient expression of microbial effectors, we tested the type III (T3) effector AvrPto, which is a kinase inhibitor blocking PRRs (Xiang et al., 2008). As expected, expression of AvrPto blocked the ROS burst induced by flgII‐28 when co‐expressed with FLS3 (Figure 6a), consistent with an earlier study (Hind et al., 2016). In addition, AvrPto expression also blocked the ROS burst induced by flg22 (Figure 6b), consistent with an earlier study (Xing et al., 2007). Consequently, agroinfiltration of AvrPto increased growth of PtoDC3000(ΔhQ) in leaves transiently expressing FLS3 (Figure 6c), demonstrating that agroinfiltration of pathogenic microbial effector suppresses host defence. By contrast, AtumGV3101 grew equally well on leaves agroinfiltrated with AvrPto (Figure 6d), indicating that AvrPto does not affect A. tumefaciens growth by blocking PRRs.
Figure 6.
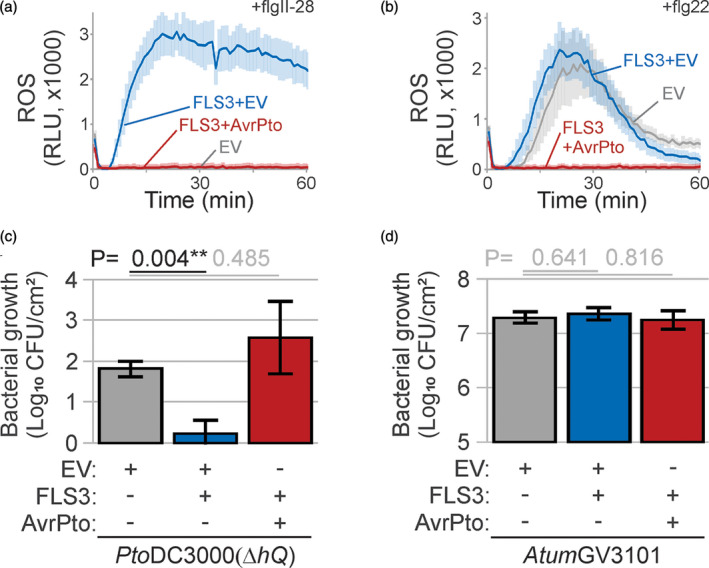
T3 effector suppresses immunity in the agromonas assay.
(a,b) Expression of AvrPto blocks reactive oxygen species (ROS) production upon flgII‐28 and flg22 treatment. Leaf discs from agroinfiltrated leaves expressing FLS3 with empty vector (EV; blue), FLS3 with AvrPto (red) or EV alone (grey) were treated with 100 nm flgII‐28 (a) or flg22 (b) and the ROS burst was measured in RLU. Error intervals represent SE of n = 12 replicates.
(c) Agroinfiltration of AvrPto increases Pseudomonas syringae growth in Nicotiana benthamiana and suppresses FLS3‐mediated immunity. Two days after agroinfiltration, agroinfiltrated leaves expressing FLS3 in combination with either AvrPto (red) or EV (blue) were spray‐inoculated with 1 × 108 CFU ml−1 PtoDC3000(∆hQ) and bacterial growth was measured 3 days later using cephaloridine, fucidin and cetrimide (CFC) selection. Error bars represent SE of n = 3 biological replicates. Student’s t‐test statistics (**P < 0.01).
(d) AvrPto does not affect growth of AtumGV3101. Bacterial growth of AtumGV3101 was measured by plating the leaf extracts described in (c) on medium containing gentamicin. Error bars represent SE of n = 3 biological replicates. Student’s t‐test statistics.
Secreted immune hydrolases reduce bacterial growth in agromonas assay
We recently discovered that the β‐galactosidase NbBGAL1 contributes to FLS2‐mediated immunity by initiating the hydrolytic release of flagellin elicitors, presumably by removing the terminal glycan from the flagellin polymer (Buscaill et al., 2019). To test whether immunity triggered by NbBGAL1 can be detected in the agromonas assay, we agroinfiltrated NbBGAL1. Consistent with previous studies (Buscaill et al., 2019; Kriechbaum et al., 2020), apoplastic fluids from leaves of N. benthamiana bgal1 mutant plants transiently overexpressing NbBGAL1 had strong β‐galactosidase activity as NbBGAL1 can cleave galactose from FDG (fluorescein di‐β‐d‐galactopyranoside) and no such activity was detected in the EV control (Figure 7a). These EV and NbBGAL1 expressing leaves were spray‐inoculated with Pta6605, which carries BGAL1‐sensitive glycans. Leaves overexpressing NbBGAL1 had reduced bacterial growth as compared with EV control leaves (Figure 7b), demonstrating that transient expression of NbBGAL1 in N. benthamiana increases resistance to Pta6605. We also inoculated agroinfiltrated leaves with PsyB728a, which carries BGAL1‐insensitive glycans. Bacterial growth of PsyB728a was not affected by NbBGAL1 when compared with EV expressing leaves (Figure 7c), consistent with the fact that NbBGAL1 acts in immunity only against strains carrying sensitive glycans.
Figure 7.
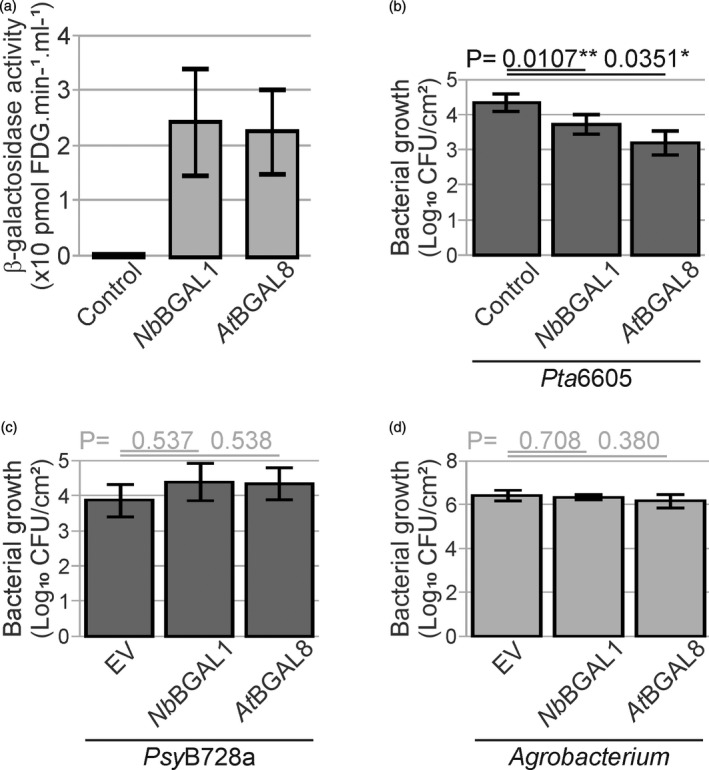
β‐Galactosidases reduce bacterial growth of BGAL‐sensitive strains in agromonas assay.
(a) NbBGAL1 and AtBGAL8 have β‐galactosidase activity. FDG‐hydrolysing activity was measured in apoplastic fluids isolated from bgal1 mutant leaves transiently expressing NbBGAL1 or AtBGAL8. Error bars represent SE of n = 3 biological replicates.
(b) Agroinfiltration of NbBGAL1 and AtBGAL8 reduce Pta6605 growth. Two days after agroinfiltration, agroinfiltrated leaves expressing NbBGAL1 or AtBGAL8 were spray‐inoculated with 1 × 108 CFU ml−1 Pta6605 and bacterial growth was measured 3 days later using cephaloridine, fucidin and cetrimide (CFC) selection. Error bars represent SE of n = 6 biological replicates; t‐test P‐values (* P < 0.05).
(c) NbBGAL1 or AtBGAL8 do not reduce PsyB728a growth. Two days after agroinfiltration, agroinfiltrated leaves expressing NbBGAL1 or AtBGAL8 were spray‐inoculated with 1 × 108 CFU ml−1 PsyB728a and bacterial growth was measured 3 days later using CFC selection. Error bars represent SE of n = 3 biological replicates; t‐test P‐values.
(d) NbBGAL1 and AtBGAL8 do not affect Agrobacterium growth. Bacterial growth of AtumGV3101 was measured by plating the leaf extracts described in (c) on medium containing gentamicin. Error bars represent SE of n = 3 biological replicates. Student’s t‐test statistics.
AtumGV3101 grew equally well on both plants agroinfiltrated with NbBGAL1 and EV (Figure 7d), indicating that NbBGAL1 does not affect Agrobacterium growth, consistent with the absence of immunogenic flagellin peptides triggering NbFLS2 (Hann and Rathjen, 2007).
Finally, we tested whether the NbBGAL1 orthologue in Arabidopsis, AtBGAL8, can provide immunity to strains carrying NbBGAL1‐sensitive glycans. NbBGAL1 and AtBGAL8 share 72% amino acid identity, and AtBGAL8 carries the catalytic residues (Figure S2). Similarly to NbBGAL1, AtBGAL8 has β‐galactosidase activity when produced by agroinfiltration in N. benthamiana bgal1 mutant plants (Figure 7a) and, as with NbBGAL1, agroinfiltration of AtBGAL8 reduces growth of Pta6605 (Figure 7b) but did not affect the growth of PsyB728a (Figure 7c). Thus, similarly to NbBGAL1, AtBGAL8 can confer specific immunity to P. syringae strains carrying BGAL1‐sensitive glycans. By contrast, AtumGV3101 grew equally well on agroinfiltrated leaves expressing AtBGAL8 (Figure 7d), indicating that AtBGAL8 does not affect Agrobacterium growth. These data demonstrate that the agromonas assay can be used to study diverse components of the immune system.
DISCUSSION
Agromonas is an easy, robust and simple assay to monitor P. syringae growth in agroinfiltrated leaf tissues both upon infiltration and spray inoculation of P. syringae. Using well‐established PAMP receptors, a T3 effector and immune hydrolases, we demonstrated that the agromonas assay can be used to study components that enhance immunity (e.g. FLS3, EFR and BGAL1) and reduce immunity (e.g. hpFLS2 and AvrPto). This assay is now routinely used in our lab to study various components of plant immunity, perform structure−function analysis, and study putative roles of effectors in immune suppression. This manuscript sets examples and parameters for this assay so it can be used widely by the research community.
Four practical considerations to take away
This manuscript establishes the methodology of the agromonas assays. As for all disease assays, experimental conditions are of fundamental importance. There are four essential parameters that should be considered for agromonas assays.
First, it is essential to use EV and hpGFP controls in overexpression and depletion assays, respectively. Agrobacterium suppresses P. syringae growth directly and/or indirectly, so a mock control (i.e. buffer infiltration) is not very useful. The control should be based on the same Agrobacterium strain carrying the same vector and at the same final OD600.
Second, it is worth simultaneously monitoring growth of Agrobacterium simply using gentamicin selection, when the P. syringae strain being used is not gentamicin resistant. Occasionally, Agrobacterium growth is also affected by modulation of host immunity and this may affect protein expression. However, in the cases described here, Agrobacterium growth was reduced by EFR consistent with the literature (Kunze et al., 2004), but immunity to P. syringae was still enhanced, indicating that P. syringae growth was suppressed by EFR not by reduced Agrobacterium levels.
Third, it is crucial to allow sufficient protein accumulation in the agroinfiltrated leaves prior to P. syringae infection. This may differ between proteins. Likewise, depletion of endogenous proteins by hpRNAi may need time. For instance, 3 days upon agroinfiltration of hpFLS2 was not enough to deplete endogenous NbFLS2.
Fourth, we recommend analysing at least 3 (ideally 6) plants per condition per experiment, and repeating experiments at least three times. This is normal practice in P. syringae infections, and will undoubtedly display the high reproducibility of the agromonas assay.
Five limitations of agromonas assays
Despite the broad versatility of the agromonas assay, we would like to point out five limitations of the assay that should be considered for future experiments.
First, excessively high expression levels could cause artefacts, but this problem can be mitigated by using different promoters (Grefen et al., 2010). We like to note that many microscopy studies on fluorescent tagged proteins show the expected subcellular localisations (Bally et al., 2018), indicating that N. benthamiana usually delivers proteins at the intended site. Overexpression of host proteins may also mitigate the functions of specific effectors, for example, when these suppress this host protein.
Second, A. tumefaciens strains employed for agroinfiltration are non‐oncogenic but they still trigger host immunity. Indeed, agroinfiltration into Nicotiana tabacum leaves elicits a low level of callose deposition (Rico et al., 2010), and the csp22 epitope of Cold Shock Protein (CSP) of A. tumefaciens is recognised by the receptor CORE (cold shock protein receptor) of N. benthamiana (Felix and Boller, 2003; Wang et al., 2016). CSP recognition increases in adult plants because CORE is expressed higher in 6‐week‐old plants (Wang et al., 2016), but we found that agromonas assays work in both young (3 weeks old) and adult plants (6 weeks old). Increased host immunity is visible in reduced P. syringae growth, and may be a limitation to study specific immune components.
Third, some immune responses are suppressed by agroinfiltration. For instance, agroinfiltration into N. tabacum leaves results in reduced abscisic acid levels and salicylic acid production (Rico et al., 2010). Likewise, A. tumefaciens strain GV3101(pMP90) used for our experiments is known to produce cytokinin through the trans‐zeatin synthase encoded by the Ti plasmid, which induces stromules and changes the position of chloroplasts, a phenomenon that is reduced when using strain LBA4404 (Erickson et al., 2014).
Fourth, despite the fact that N. benthamiana is commonly used as a model for plant−pathogen interactions (Goodin et al., 2008) and has the PRR co‐receptors SOBIR1 and BAK1 (Heese et al., 2007; Liebrand et al., 2013), it may not possess all supporting components when testing immune components from other plant species. For example, N. benthamiana has ZAR1, but lacks ZED1 for the recognition of the bacterial effector HopZ1 (Baudin et al., 2017).
Fifth, quantification of bacterial growth by colony count assays remains a bottle neck for high‐throughput screening. Alternative pathogen infection assays have been developed to increase throughput. For instance, bacterial DNA can be quantified by real‐time polymerase chain reaction (PCR; Ross and Somssich, 2016), but this technique does not distinguish between living and dead bacteria, overestimating the titres of living bacteria (Rooney et al., 2020). An alternative approach for monitoring bacterial density is using bioluminescence (Fan et al., 2008), but this method requires the transformation of each bacterial strain with the luxCDABE operon (Fan et al., 2008). Also, bioluminescence reflects the metabolic state rather than bacterial viability, and cannot be used to detect low titres. However, bioluminescence might be a powerful approach to increase throughput in specific contexts.
Six opportunities of using agromonas assays
We believe that the agromonas assay can be applied to a wide range of applications. There are at least six main opportunities for a wider application of the agromonas assay.
First, the agromonas assay is a rapid and easy method to subject plant and pathogen proteins in immunity for structure−function analysis, without the need of transgenic plants. For instance, we tested the non‐phosphorylatable mutant EFRY836F (Macho et al., 2014) to demonstrate that agromonas assay facilitates rapid structure−function analysis of immune components.
Second, because CFC selects Pseudomonas spp., the agromonas assay works without genetic manipulation of P. syringae, allowing the study of the available repertoire of P. syringae mutants and strains. For instance, PtoDC3000 polymutants (Wei et al., 2015) and PsyB728a mutants (Vinatzer et al., 2006) can be tested in agromonas assays. However, phenotypes associated with effector functions may not always be visible when the effector target is overexpressed by agroinfiltration.
Third, although in this study we only used N. benthamiana, we anticipate that the agromonas assay can be easily adapted to plant species suitable for agroinfiltration, such as potato (Du et al., 2014), tobacco (Van der Hoorn et al., 2000), pea (Guy et al., 2016), Medicago (Picard et al., 2013), grapefruit (Figueiredo et al., 2011), lettuce (Chen et al., 2016), tomato (Wroblewski et al., 2009), flax (Dodds et al., 2006), cassava (Zeng et al., 2019), strawberry (Guidarelli and Baraldi, 2015) and Mucuna bracteate (Abd‐Aziz et al., 2020). This facilitates studies of immunity in other plant species using corresponding P. syringae pathovars.
Fourth, for plant species that are not amenable to agroinfiltration, N. benthamiana remains an excellent heterologous expression system to study proteins from various organisms (plants, microbes and animals). We demonstrate this by studying EFR and AtBGAL8 from Arabidopsis and FLS3 from tomato. We anticipate that agromonas assays can be used to study more immune‐related genes or quantitative trait locus (QTL) from various plants and microbes. For instance, PAMPs receptors LORE and LYM1/3 (Willmann et al., 2011; Ranf et al., 2015) are absent in N. benthamiana and should reduce bacterial growth in agromonas assays.
Fifth, P. syringae growth can be monitored in agromonas assay using both infiltration and spray inoculation, testing both post‐ and pre‐invasive immunity. Here, we used the method of inoculation described in literature for each immune component tested (Xiang et al., 2008; Segonzac et al., 2011; Macho et al., 2014; Buscaill et al., 2019). We anticipate that leaf‐dipping and vacuum inoculation should also be applicable in agromonas assays.
Sixth, agromonas assays can be used to study depletion of host immunity genes. We demonstrated this by depleting NbFLS2 using hpFLS2, which resulted in a significant enhancement of P. syringae growth. Similar results were observed with depletion of NbFLS2 by virus‐induced gene silencing (VIGS; Segonzac et al., 2011). However, VIGS is at least 4 weeks slower than hpRNAi technique, and VIGS requires approval for work with modified tobacco rattle virus. We anticipate that agromonas assays can also be used to study the depletion of additional endogenous immune components, such as mitogen‐activated protein kinases, calcium‐dependent protein kinases, transcription factors, PAMP receptors, and nucleotide‐binding/leucine‐rich repeats. However, depletion of these components with transient hpRNAi will depend on the stability of the protein.
In conclusion, the ability to characterise immune components from plants and microbes using agromonas assays will speed up our understanding of the plant immune system and microbial colonisation, and generate promising strategies for crop protection.
EXPERIMENTAL PROCEDURES
Plants
Nicotiana benthamiana plants were grown in a growth chamber at 21°C and ~60% relative humidity with a 16 h photoperiod and a light intensity of 2000 cd sr m−2.
Molecular cloning
All constructs were generated using standard molecular biology procedures. All vectors used in this study are listed in Table S2. EFR (At5g20480) and At BGAL8 (At2g28470) were amplified from Arabidopsis thaliana ecotype Col‐0 complementary DNA (cDNA), and Sl FLS3 (LOC101248095/Solyc04g009640) was amplified from Solanum lycopersicum cv. Rio Grande genomic DNA (gDNA) using primers listed in Table S3. BGAL1 was amplified from N. benthaminana cDNA using primers listed in Table S3. The PCR products were combined with pICH51288 (Engler and Marillonnet, 2014), pICH41414 (Engler and Marillonnet, 2014) and pJK001c (Paulus et al., 2020) in a BsaI GoldenGate reaction to generate pPB069 (pL2M‐2x35S::EFR), pJK668 (pL2M‐2x35S::FLS3), pJK646 (pL2M‐2x35S::NbBGAL1) and pJK645 (pL2M‐2x35S::AtBGAL8), respectively. The EFR tyrosine mutant EFRY836F was generated by site directed mutagenesis using primers listed in Table S3. All binary plasmids were transformed into A. tumefaciens GV3101 (pMP90) by freeze‐thawing, and transformants were selected by kanamycin resistance.
Silencing by RNA interference (RNAi)
An intron‐containing hairpin RNA (ihpRNA) construct targeting a conserved region in FLS2a/b was designed to silence both FLS2 homologues (i.e. NbD013936.1 and NbD024362.1) detected in the NbDE database (Kourelis et al., 2019). The 300‐bp fragment of GFP (SeqA; Table S3) and the 300‐bp fragment of FLS2a/b (SeqB; Table S3) used for RNAi were commercially synthesized (Invitrogen, Carlsbad, CA, USA). The fragments were cloned into the pRNAiGG vector (Yan et al., 2012) using BsaI restriction sites resulting in vector pPB070 and pPB072, respectively (Table S2). The binary constructs were transformed into AtumGV3101 (pMP90) by freeze‐thawing, and transformants were selected by kanamycin resistance. Three‐week‐old N. benthamiana leaves were agroinfiltrated with the hairpin silencing construct at a final OD600 = 0.2. Further experiments (ROS production and spray infection) were performed 10 days after agroinfiltration.
Agroinfiltration
For transient expression of proteins in N. benthamiana, overnight cultures of A. tumefaciens GV3101 (AtumGV3101) carrying binary vectors were harvested by centrifugation. Cells were resuspended in induction buffer (10 mm MgCl2, 10 mm MES pH5.0, and 150 µm acetosyringone) and mixed (1:1) with bacteria carrying silencing inhibitor P19 at OD600 = 0.5. After 1 h at 21°C, cells were infiltrated with a needleless syringe into the abaxial side of three leaves of 4‐week‐old N. benthamiana. Leaves were harvested and processed at the indicated days after agroinfiltration.
Bacterial strains
The bacterial strains used in this study are listed in Table S4. Pseudomonas and Xanthomonas strains were grown in LB medium at 28°C. For the infection assays, bacteria were cultured in LB medium containing 10 mm MgCl2 at 28°C. Agrobacterium tumefaciens strains were grown in LB medium containing 50 μg ml−1 rifampicin, 10 μg ml−1 gentamicin and 50 μg ml−1 kanamycin at 28°C.
Oxidative burst assays
The generation of ROS was measured by a luminol‐based assay on leaf discs adapted from Smith and Heese (2014). Luminol (Sigma‐Aldrich, Saint Louis, MO, USA) was dissolved in dimethyl sulphoxide at a concentration of 10 mg ml−1 and kept in the dark. Horseradish peroxidase (HRP; Thermo Fisher Scientific, Waltham, MA, USA) was dissolved in water at a concentration of 10 mg ml−1. Leaf discs (6 mm diameter), were incubated overnight in water in Petri dishes. Leaf discs from agroinfiltrated leaves were deposed in a 96‐well plates, one leaf disc per well (Costar, Kennebunk, ME, USA); 100 µl of 25 ng µl−1 luminol, 25 ng µl−1 HRP and 100 nm elf18 (Kunze et al., 2004) or 100 nm flg22 (Felix et al., 1999) or 100 nm flgII‐28 (Cai et al., 2011) was added and chemiluminescence was recorded immediately in relative light units (RLU) using an Infinite M200 plate reader (Tecan, Mannedorf, Switzerland). Measurements were taken every minute for 1 h. Standard errors were calculated at each time point and for each treatment. Experiments were repeated at least three times.
Bacterial growth upon inoculation
For syringe inoculations, an overnight culture was washed and resuspended in sterile water to a density of 1 × 106 CFU ml−1 and infiltrated into agroinfiltrated leaves using a blunt syringe via the abaxial side of the leaves. For spray inoculations, an overnight culture was washed and resuspended in sterile water to a density of 1 × 108 CFU ml−1 and sprayed onto adaxial surfaces of agroinfiltrated leaves. Before inoculation, plants were covered with a humidified dome for 1 day. After infection, plants were re‐covered with the dome and kept for 3 days in a growth cabinet at 21°C. For determination of in planta bacterial growth, three leaf discs (1 cm diameter) were excised 3 days after inoculation from inoculated leaves. Each leaf disc was soaked in 15% H2O2 for 2 min to sterilise the leaf surface. Leaf discs were washed twice in sterile water and dried under sterile conditions for 30 min. Leaf discs were then ground in sterile water for 5 min using the tissue‐lyser and metal beads (Biospec Products, Bartelsville, OK, USA). Serial dilutions of the homogenate were plated onto LB agar supplemented with either gentamicin (10 µg ml−1) for selection of AtumGV3101 or CFC (Oxoid™ C‐F‐C Supplement) at 1× concentration, prepared according to the manufacturer’s instructions for selection of P. syringae strains and incubated at 28°C. Colonies were counted after 36 h incubation at 28°C. The P‐value was calculated using the two‐tailed Student’s t‐test to binary compare bacterial growth between agroinfiltrated plants.
Statistics
All values shown are mean values, and the error intervals shown represent standard error of the mean (SE), unless otherwise indicated. P‐values were calculated using the two‐tailed Student’s t‐test. All experiments have been reproduced and representative datasets are shown.
Protein alignment
Sequences were aligned using Clustal Omega (Sievers et al., 2011). Alignment was visualised and analysed using Jalview (Waterhouse et al., 2009), and edited using CorelDraw (Corel Corporation, Ottawa, Ontario, Canada).
Author contributions
PB, NS, GP and RH conceived the project; PB, NS, YL and JK performed experiments; PB and RH wrote the manuscript with input from all authors.
Conflict of interest
The authors declare no competing interests.
ACKNOWLEDGEMENTS
The authors thank Alan Collmer for providing the PtoDC3000(∆hQ) mutant; Yuki Ichinose for providing the Pta6605 strain; Tolga Bozkurt for providing the pRNAiGG vector; Nicola J. Patron and Sylvestre Marillonnet for providing Golden Gate plasmids; Ursula Pyzio for plant care; Sarah Rodgers and Caroline O’Brien for technical support. ERC Consolidator grant 616447 ‘GreenProteases’ (PB, RH); BBSRC grants BB/R017913/1 (PB, RH); the Royal Thai Government Scholarship (NS); the Clarendon foundation (JK); the Oxford Interdisciplinary Bioscience DTP (BB/M011224/1 to NS, GP).
Data Availability Statement
All data are available in the manuscript, the supplementary materials, and the cited references.
REFERENCES
- Abd‐Aziz, N. , Tan, B.C. , Rejab, N.A. , Othman, R.Y. and Khalid, N. (2020) A new plant expression system for producing pharmaceutical proteins. Mol. Biotechnol. 62, 240–251. [DOI] [PubMed] [Google Scholar]
- Bally, J. , Jung, H. , Mortimer, C. , Naim, F. , Philips, J.G. , Hellens, R. , Bombarely, A. , Goodin, M.M. and Waterhouse, P.M. (2018) The rise and rise of Nicotiana benthamiana: a plant for all reasons. Annu. Rev. Phytopathol. 56, 405–426. [DOI] [PubMed] [Google Scholar]
- Baudin, M. , Hassan, J.A. , Schreiber, K.J. and Lewis, J.D. (2017) Analysis of the ZAR1 immune complex reveals determinants for immunity and molecular interactions. Plant Physiol. 174, 2038–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt, T.O. , Richardson, A. , Dagdas, Y.F. , Mongrand, S. , Kamoun, S. and Raffaele, S. (2014) The plant membrane‐associated REMORIN1.3 accumulates in discrete perihaustorial domains and enhances susceptibility to Phytophthora infestans . Plant Physiol. 165, 1005–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaill, P. , Chandrasekar, B. , Sanguankiattichai, N. et al (2019) Glycosidase and glycan polymorphism control hydrolytic release of immunogenic flagellin peptides. Science, 364, eaav0748. [DOI] [PubMed] [Google Scholar]
- Cai, R. , Lewis, J. , Yan, S. et al (2011) The plant pathogen Pseudomonas syringae pv. tomato is genetically monomorphic and under strong selection to evade tomato immunity. PLoS Pathog. 7, e1002130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro‐Garcia, A. , Wilkinson, R.C. , Gimenez‐Ibanez, S. , Findlay, K. , Coffey, M.D. , Zipfel, C. , Rathjen, J.P. , Kamoun, S. and Schornack, S. (2011) The receptor‐like kinase SERK3/BAK1 is required for basal resistance against the late blight pathogen phytophthora infestans in Nicotiana benthamiana . PLoS One, 6, e16608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Q. , Dent, M. , Hurtado, J. , Stahnke, J. , McNulty, A. , Leuzinger, K. and Lai, H. (2016) Transient protein expression by agroinfiltration in lettuce. Methods Mol. Biol. 1385, 55–67. [DOI] [PubMed] [Google Scholar]
- Dagdas, Y.F. , Pandey, P. , Tumtas, Y. et al (2018) Host autophagy machinery is diverted to the pathogen interface to mediate focal defense responses against the Irish potato famine pathogen. eLife, 7, e37476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N. , Lawrence, G.J. , Catanzariti, A.M. , Teh, T. , Wang, C.I. , Ayliffe, M.A. , Kobe, B. and Ellis, J.G. (2006) Direct protein interaction underlies gene‐for‐gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl Acad. Sci. USA, 103, 8888–8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, J. , Rietman, H. and Vleeshouwers, V.G. (2014) Agroinfiltration and PVX agroinfection in potato and Nicotiana benthamiana . J. Vis. Exp. e50971 10.3791/50971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler, C. and Marillonnet, S. (2014) Golden gate cloning. Methods Mol. Biol. 1116, 119–131. [DOI] [PubMed] [Google Scholar]
- Erickson, J.L. , Ziegler, J. , Guevara, D. , Abel, S. , Klösgen, R.B. , Mathur, J. , Rothstein, S.J. and Schattat, M.H. (2014) Agrobacterium‐derived cytokinin influences plastid morphology and starch accumulation in Nicotiana benthamiana during transient assays. BMC Plant Biol. 14, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, J. , Crooks, C. and Lamb, C. (2008) High‐throughput quantitative luminescence assay of the growth in planta of Pseudomonas syringae chromosomally tagged with Photorhabdus luminescens luxCDABE . Plant J. 53, 393–399. [DOI] [PubMed] [Google Scholar]
- Felix, G. and Boller, T. (2003) Molecular sensing of bacteria in plants. The highly conserved RNA‐binding motif RNP‐1 of bacterial cold shock proteins is recognized as an elicitor signal in tobacco. J. Biol. Chem. 278, 6201–6208. [DOI] [PubMed] [Google Scholar]
- Felix, G. , Duran, J.D. , Volko, S. and Boller, T. (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276. [DOI] [PubMed] [Google Scholar]
- Figueiredo, J.F. , Römer, P. , Lahaye, T. , Graham, J.H. , White, F.F. and Jones, J.B. (2011) Agrobacterium‐mediated transient expression in citrus leaves: a rapid tool for gene expression and functional gene assay. Plant Cell Rep. 30, 1339–1345. [DOI] [PubMed] [Google Scholar]
- Fones, H. , Davis, C.A. , Rico, A. , Fang, F. , Smith, J.A. and Preston, G.M. (2010) Metal hyperaccumulation armors plants against disease. PLoS Pathog. 6, e1001093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodin, M.M. , Zaitlin, D. , Naidu, R.A. and Lommel, S.A. (2008) Nicotiana benthamiana: its history and future as a model for plant‐pathogen interactions. Mol. Plant Microbe Interact. 21, 1015–1026. [DOI] [PubMed] [Google Scholar]
- Grefen, C. , Donald, N. , Hashimoto, K. , Kudla, J. , Schumacher, K. and Blatt, M.R. (2010) A ubiquitin‐10 promoter‐based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 64, 355–365. [DOI] [PubMed] [Google Scholar]
- Guidarelli, M. and Baraldi, E. (2015) Transient transformation meets gene function discovery: the strawberry fruit case. Front. Plant Sci. 6, 444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy, E. , Boulain, H. , Aigu, Y. , Le Pennec, C. , Chawki, K. , Morlière, S. , Schädel, K. , Kunert, G. , Simon, J.C. and Sugio, A. (2016) Optimization of agroinfiltration in Pisum sativum provides a new tool for studying the salivary protein functions in the pea aphid complex. Front. Plant Sci. 7, 1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann, D.R. and Rathjen, J.P. (2007) Early events in the pathogenicity of Pseudomonas syringae on Nicotiana benthamiana . Plant J. 49, 607–618. [DOI] [PubMed] [Google Scholar]
- Heese, A. , Hann, D.R. , Gimenez‐Ibanez, S. , Jones, A.M. , He, K. , Li, J. , Schroeder, J.I. , Peck, S.C. and Rathjen, J.P. (2007) The receptor‐like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl Acad. Sci. USA, 104, 12217–12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, D. , Rose, B. , Pajkos, A. et al (2005) Antibiotic susceptabilities of Pseudomonas aeruginosa isolates derived from patients with cystic fibrosis under aerobic, anaerobic, and biofilm conditions. J. Clin. Microbiol. 43, 5085–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hind, S.R. , Strickler, S.R. , Boyle, P.C. et al (2016) Tomato receptor FLAGELLIN‐SENSING 3 binds flgII‐28 and activates the plant immune system. Nat. Plants, 2, 16128. [DOI] [PubMed] [Google Scholar]
- Kourelis, J. , Kaschani, F. , Grosse‐Holz, F.M. , Homma, F. , Kaiser, M. and van der Hoorn, R.A.L. (2019) A homology‐guided, genome‐based proteome for improved proteomics in the alloploid Nicotiana benthamiana. BMC Genom., 20, 722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriechbaum, R. , Ziaee, E. , Grünwald‐Gruber, C. , Buscaill, P. , van der Hoorn, R.A.L. and Castilho, A. (2020) BGAL1 depletion boosts the level of β‐galactosylation of N‐ and O‐glycans in Nicotiana benthamiana . Plant Biotechnol. J. 18, 1537–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger, C.L. and Sheikh, W. (1987) A new selective medium for isolating Pseudomonas spp. from water. Appl. Environ. Microbiol. 53, 895–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze, G. , Zipfel, C. , Robatzek, S. , Niehaus, K. , Boller, T. and Felix, G. (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell, 16, 3496–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe, S. , Rougon‐Cardoso, A. , Sherwood, E. et al (2010) Interfamily transfer of a plant pattern‐recognition receptor confers broad‐spectrum bacterial resistance. Nat. Biotechnol. 28, 365–369. [DOI] [PubMed] [Google Scholar]
- Liebrand, T.W. , van den Berg, G.C. , Zhang, Z. et al (2013) Receptor‐like kinase SOBIR1/EVR interacts with receptor‐like proteins in plant immunity against fungal infection. Proc. Natl Acad. Sci. USA, 110, 10010–10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho, A.P. , Schwessinger, B. , Ntoukakis, V. et al (2014) A bacterial tyrosine phosphatase inhibits plant pattern recognition receptor activation. Science, 343, 1509–1512. [DOI] [PubMed] [Google Scholar]
- Mansfield, J. , Genin, S. , Magori, S. et al (2012) Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead, G.C. and Adams, B.W. (1977) A selective medium for the rapid isolation of Pseudomonads associated with poultry meat spoilage. Br. Poult. Sci. 18, 661–670. [DOI] [PubMed] [Google Scholar]
- Pantazi, D. , Papavergou, A. , Pournis, N. , Kontominas, M.G. and Savvaidis, I.N. (2008) Shelf‐life of chilled fresh Mediterranean swordfish (Xiphias gladius) stored under various packaging conditions: microbiological, biochemical and sensory attributes. Food Microbiol. 25, 136–143. [DOI] [PubMed] [Google Scholar]
- Paulus, J.K. , Kourelis, J. , Ramasubramanian, S. et al (2020) Extracellular proteolytic cascade in tomato activates immune protease Rcr3. Proc. Natl Acad. Sci. USA, 117, 17409–17417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard, K. , Lee, R. , Hellens, R. and Macknight, R. (2013) Transient gene expression in Medicago truncatula leaves via agroinfiltration. Methods Mol. Biol. 1069, 215–226. [DOI] [PubMed] [Google Scholar]
- Ranf, S. , Gisch, N. , Schäffer, M. et al (2015) A lectin S‐domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana . Nat. Immunol. 16, 426–433. [DOI] [PubMed] [Google Scholar]
- Rico, A. , Bennett, M.H. , Forcat, S. , Huang, W.E. and Preston, G.M. (2010) Agroinfiltration reduces ABA levels and suppresses Pseudomonas syringae‐elicited salicylic acid production in Nicotiana tabacum . PLoS One, 5, e8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney, W.M. , Grinter, R.W. , Correia, A. , Parkhill, J. , Walker, D.C. and Milner, J.J. (2020) Engineering bacteriocin‐mediated resistance against the plant pathogen Pseudomonas syringae . Plant Biotechnol. J. 18, 1296–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, A. and Somssich, I.E. (2016) A DNA‐based real‐time PCR assay for robust growth quantification of the bacterial pathogen. Plant Methods, 12, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonzac, C. , Feike, D. , Gimenez‐Ibanez, S. , Hann, D.R. , Zipfel, C. and Rathjen, J.P. (2011) Hierarchy and roles of pathogen‐associated molecular pattern‐induced responses in Nicotiana benthamiana . Plant Physiol. 156, 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers, F. , Wilm, A. , Dineen, D. et al (2011) Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J.M. and Heese, A. (2014) Rapid bioassay to measure early reactive oxygen species production in Arabidopsis leave tissue in response to living Pseudomonas syringae . Plant Methods, 10, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub, C. , Colombi, E. , Li, L. , Huang, H. , Templeton, M.D. , McCann, H.C. and Rainey, P.B. (2018) The ecological genetics of Pseudomonas syringae from kiwifruit leaves. Environ. Microbiol. 20, 2066–2084. [DOI] [PubMed] [Google Scholar]
- van der Hoorn, R.A.L. , Laurent, F. , Roth, R. and De Wit, P.J.G.M. (2000) Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf‐9‐induced and Avr4/Cf‐4‐induced necrosis. Mol. Plant Microbe Interact. 13, 439–446. [DOI] [PubMed] [Google Scholar]
- Vinatzer, B.A. , Teitzel, G.M. , Lee, M.W. , Jelenska, J. , Hotton, S. , Fairfax, K. , Jenrette, J. and Greenberg, J.T. (2006) The type III effector repertoire of Pseudomonas syringae pv. syringae B728a and its role in survival and disease on host and non‐host plants. Mol Microbiol. 62, 26–44. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Albert, M. , Einig, E. , Fürst, U. , Krust, D. and Felix, G. (2016) The pattern‐recognition receptor CORE of Solanaceae detects bacterial cold‐shock protein. Nat. Plants, 2, 16185. [DOI] [PubMed] [Google Scholar]
- Waterhouse, A.M. , Procter, J.B. , Martin, D.M. , Clamp, M. and Barton, G.J. (2009) Jalview Version 2 ‐ a multiple sequence alignment editor and analysis workbench. Bioinformatics, 25, 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, H.L. , Chakravarthy, S. , Mathieu, J. , Helmann, T.C. , Stodghill, P. , Swingle, B. , Martin, G.B. and Collmer, A. (2015) Pseudomonas syringae pv. tomato DC3000 type III secretion effector polymutants reveal an interplay between HopAD1 and AvrPtoB. Cell Host Microbe, 17, 752–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann, R. , Lajunen, H.M. , Erbs, G. et al (2011) Arabidopsis lysin‐motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc. Natl Acad. Sci. USA, 49, 19824–19829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewski, T. , Caldwell, K.S. , Piskurewicz, U. et al (2009) Comparative large‐scale analysis of interactions between several crop species and the effector repertoires from multiple pathovars of Pseudomonas and Ralstonia . Plant Physiol. 150, 1733–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, T. , Zong, N. , Zou, Y. et al (2008) Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr. Biol. 18, 74–80. [DOI] [PubMed] [Google Scholar]
- Xing, W. , Zou, Y. , Liu, Q. et al (2007) The structural basis for activation of plant immunity by bacterial effector protein AvrPto. Nature, 449, 243–247. [DOI] [PubMed] [Google Scholar]
- Yan, P. , Shen, W. , Gao, X. , Li, X. , Zhou, P. and Duan, J. (2012) High‐throughput construction of intron‐containing hairpin RNA vectors for RNAi in plants. PLoS One, 7, e38186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, H. , Xie, Y. , Liu, G. , Wei, Y. , Hu, W. and Shi, H. (2019) Agrobacterium‐mediated gene transient overexpression and Tobacco Rattle Virus (TRV)‐based gene silencing in Cassava. Int. J. Mol. Sci. 20, 3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C. , Kunze, G. , Chinchilla, D. , Caniard, A. , Jones, J.D.G. , Boller, T. and Felix, G. (2006) Perception of the bacterial PAMP EF‐Tu by the receptor EFR restricts Agrobacterium‐mediated transformation. Cell, 125, 749–760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are available in the manuscript, the supplementary materials, and the cited references.


