Abstract
Background:
The molecular drivers of human papillomavirus-related head and neck squamous cell carcinoma (HPV+HNSCC) are not entirely understood. This study evaluated the relationship between HPV integration, expression of E6/E7, and patient outcomes in p16+ HNSCCs.
Methods:
HPV type was determined by HPV PCR-MassArray, and integration was called using Detection of Integrated Papillomavirus Sequences (DIPS) PCR. We investigated whether fusion transcripts were produced by RT-PCR. E6/E7 expression was assessed by qRT-PCR. We assessed if there was a relationship between integration and E6/E7 expression, clinical variables, or patient outcomes.
Results:
Most samples demonstrated HPV integration, which sometimes resulted in a fusion transcript. HPV integration was positively correlated with age at diagnosis and E6/E7 expression. There was a significant difference in survival between patients with versus without integration.
Conclusions:
Contrary to previous reports, HPV integration was associated with improved patient survival. Therefore, HPV integration may act as a molecular marker of good prognosis.
Keywords: head and neck squamous cell carcinoma (HNSCC), oropharynx, human papillomavirus (HPV), integration, survival
INTRODUCTION
Human papillomavirus (HPV)-induced head and neck squamous cell carcinoma (HPV+HNSCC) represents a growing public health concern due to its rapidly increasing incidence worldwide. The incidence rate of HPV+HNSCC in the United States is 4.62 per 100,000 persons.1 This cancer type most frequently presents in the oropharynx (HPV+OPSCC) but can also arise in other anatomic subsites of the head and neck region.2 HPV+HNSCC is clinically distinguished from HPV-negative HNSCC (HPV- HNSCC) by p16 status, which acts as a surrogate immunohistochemical marker for HPV positivity. Currently, HPV+ and HPV- HNSCCs are treated in a similar manner, but HPV+ patients have a significantly better outcome.3, 4 Despite this improved outcome, still 20–30% of these patients recur or fail to respond to initial therapies.5 Therefore, it is essential to understand the molecular drivers of this disease to help identify patients at high-risk of recurrence and to develop alternate therapy regimens.
The process of HPV integration into the human genome is of particular interest as a potential driver of HPV+HNSCC. HPV has been reported to be integrated in a large proportion of cervical, head and neck, and other anogenital tumors with estimates ranging from ~50–70%.6–12 This process has been most heavily investigated in cervical cancers, but there is a growing body of literature implicating integration as a potentially useful biomarker in head and neck cancer. It has been debated whether integration is a stochastic process that occurs randomly throughout the genome or whether it is a targeted process. Some studies have reported that integration occurs into/near genes or other genomic hotspots more frequently than expected by chance and that this can lead to functional alteration of critical genes.6, 12, 13
In addition to altering cellular gene expression, integration has also been thought to contribute to oncogenesis by increasing HPV oncogene levels within the cell by a variety of mechanisms, including disruption of viral E2.14 E2 is frequently, but not always, disrupted as a result of integration, which results in increased E6/E7 due to the role of E2 as a negative transcriptional regulator.15 Integration of HPV has also been reported to be associated with increased expression of shorter, spliced transcripts of E6 known as E6*I and E6*II16, which have been shown to be associated with dysregulation of key cancer pathways and worse outcomes for HPV+HNSCC patients.17 Additionally, integration into cellular genes can lead to the generation of viral-host fusion transcripts, and it has been reported that these transcripts may be more stable than episomally-derived HPV transcripts that then allows for the HPV oncogenes to persist longer.18 Some have reported that E6/E7 levels are increased in HNSCC cell lines and tumors with integrated HPV19, 20, but others have reported this is not necessarily true in every case.12, 21 Therefore, the relationship between HPV integration and E6/E7 levels is not entirely clear.
Due to its impact on both viral and cellular gene expression, it has been of great interest whether integration status can be used clinically as a prognostic marker of poor outcome. A handful of studies have attempted to elucidate the relationship between HPV integration and patient outcomes with conflicting results. Some studies of integration, as measured by loss of E2, revealed that patients with integrated HPV had worse outcomes than those with episomal HPV22–26, but others reported no significant difference between these two patient groups.27, 28 Another group recently compared the survival of patients with and without viral-cellular fusion transcripts and found that patients with these transcripts had a significantly worse survival.29 We recently examined the integration sites in patients who were responsive versus non-responsive to treatment and found that most responsive patients had integration into intergenic regions of the genome, whereas non-responsive patients had integrations into cellular genes.30 This suggests that integration site may be an important factor in whether integration impacts cellular behavior leading to altered survival.
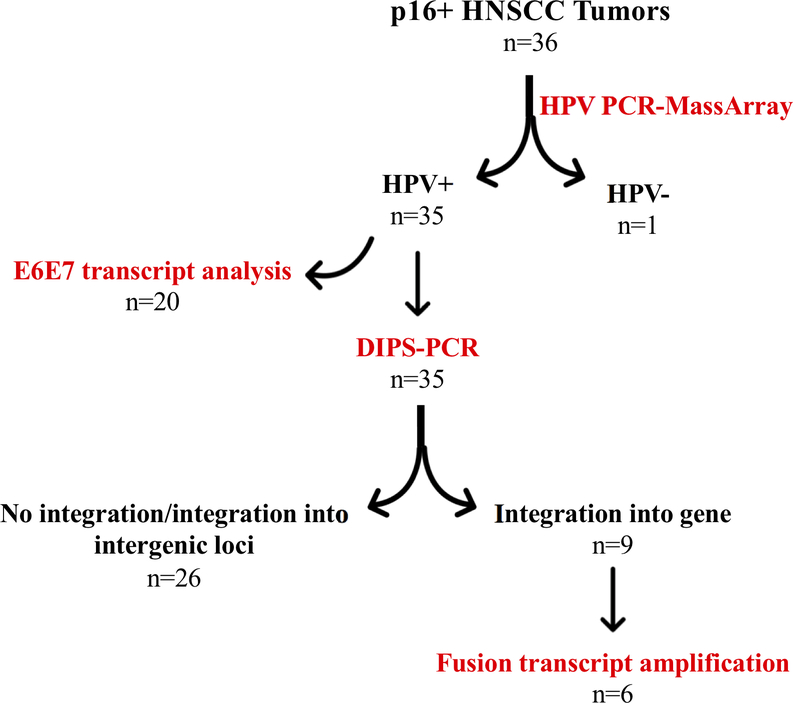
Due to this conflicting literature, we sought to clarify the relationship between E6/E7 expression and HPV integration, as well the potential impact of integration status and site on patient outcomes. Here we present an analysis of HPV types, HPV integration, and oncogene expression in thirty-six p16+ HNSCCs (Figure 1). We found that HPV integrated at a similar frequency (60%) in our cohort as previous studies, and sometimes resulted in the generation of a viral-cellular fusion transcript. There was a significant positive correlation between HPV integration status and E6/E7 expression level, and contrary to what others have reported, we found that patients with tumors containing HPV integration had a significantly improved disease-specific survival (DSS).
Figure 1:
Analysis of p16+ HNSCC tumors
MATERIALS AND METHODS
Tumor Specimens:
Thirty-six p16+ HNSCC tumors were obtained from the Beaumont Hospital BioBank (n=21, fresh frozen) and the Head and Neck Cancer SPORE Biorepository at the University of Michigan (n=15, formalin-fixed, paraffin embedded (FFPE) pre-treatment biopsies/surgical specimens for DNA analysis only. In four of these cases, frozen tissue was available for RNA analysis). Written informed consent to investigate their tissue was obtained from patients under studies approved by the Institutional Review Board at each institution. To reduce selection bias, p16+ HNSCC samples were acquired consecutively.
DNA/RNA Isolation:
Tumor tissue was identified by a head and neck pathologist and was subsequently microdissected from 10μm sections of FFPE tissue blocks from the University of Michigan. Following microdissection, DNA was extracted from the tissue using the NucleoSpin DNA FFPE kit (Macherey-Nagel, Duren, Germany) according to the manufacturer’s protocol. Briefly, paraffin was dissolved with xylene, and the tissue was lysed with lysis buffer and Proteinase K overnight at 56° C. Following overnight digestion, DNA was de-crosslinked, loaded onto the NucleoSpin DNA columns, washed and then eluted in water. DNA concentration was measured using the QUBIT 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA).
RNA was extracted using the RNeasy Mini Kit (Qiagen, Hilden, Germany); RNA isolation was only performed from samples with fresh frozen tissue (n=20). RNA concentration was measured using the QUBIT 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). cDNA was prepared from the resulting RNA using SuperScript III (Thermo Fisher Scientific, Waltham, MA, USA).
Viral Testing:
HPV PCR-MassArray was performed as previously described.31 In brief, this method detects and identifies fifteen high-risk HPV subtypes (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68, and 73), two low-risk subtypes (6 and 11), and HPV90, considered to be a possible high-risk subtype. The test included interrogation of human GAPDH as a control for sample DNA quality and assay validity. Type-specific, multiplex, competitive PCR was performed to amplify the E6 region of HPV, followed by probe-specific single base extension to discriminate between naturally occurring HPV present in the sample and the synthetic competitors included in the reaction. Matrix-assisted laser desorption/ionization time of flight mass spectroscopy was used for separation of products on a matrix-loaded silicon chip array. Samples were run in quadruplicate with appropriate positive and negative controls.
Detection of Integrated Papillomavirus Sequences (DIPS-PCR):
DIPS-PCR was performed to identify the sites of HPV integration in the genome of the tumors, as previously described.32 For each tumor, 0.75μg DNA was digested with one of two restriction enzymes, either TaqA1 or Sau3AI (New England Biolabs, Ipswich, MA, USA). Adapters complementary to the unique overhangs created by restriction digestion were annealed to digested DNA. Linear PCR was performed on each sample using multiple viral primers to amplify viral fragments. Following linear PCR, exponential PCR using nested viral primers and an adapter-specific primer was performed. All DIPS-PCR primer sequences are listed in Table S1. Products of the exponential PCR reactions were separated by gel electrophoresis (3% agarose gel). Bands were excised from the gel and were purified using the Qiaquick Gel Extraction Kit (Qiagen, Hilden, Germany). Sanger sequencing of the isolated products was performed by the University of Michigan Advanced Genomics Core, and the results were mapped to the human and HPV genomes using NCBI-BLAST.
Integration Site Transcript Analysis:
RT-PCR assays were designed to amplify predicted viral-cellular transcripts in cases where RNA was available and integration took place within a cellular gene (n=6). The designed primers are listed in Table S2. All successfully amplified transcripts were sequenced for verification.
Viral Transcript Analysis:
Samples with RNA available (n=20) were tested for HPV E6 and E7 transcripts by both quantitative RT-PCR (qRT-PCR) and RT-PCR. qRT-PCR was performed using QuantiTect SYBR Green PCR kit (Qiagen, Hilden, Germany) with GAPDH as an endogenous control. Relative gene expression was calculated using the ΔΔCt method compared to UM-SCC-47 (E6 and E7 expression in UM-SCC-47 were each set to 1). RT-PCR was performed using primers spanning the entire HPV16, HPV18 and/or HPV33 E6E7 region as appropriate; products were separated by gel electrophoresis (1.5% agarose gel). Primer sequences are listed in Table S3.
Statistical Analysis:
Censored Kaplan Meier curves were generated using GraphPad Prism 8; survival curves were compared using log-rank testing (Mantel-Cox). Associations between integration status and clinical variables were analyzed by Spearman’s rank correlation testing. P values of 0.05 or lower were considered significant.
RESULTS
Clinical Summary
Two cohorts of p16+ HNSCC patients were analyzed from either Beaumont Hospital (n=21) or Michigan Medicine (n=15). The patients from Beaumont Hospital were collected as part of a retrospective study; patients were diagnosed between 2005–2012. Patients from Michigan Medicine were collected prospectively and were recently diagnosed (2015 onward). Tumor information, patient sex, age, smoking history, year of diagnosis, treatment, and outcomes are summarized in Tables 1 and S4. We included thirty-four oropharyngeal SCCs, as well as one SCC from the oral cavity and one from the nasopharynx. As expected, there was a higher proportion of males included in this study (79% males, 21% females). Age at diagnosis ranged from 46 to 87 with an average age of 63. The majority of patients were at one time regular smokers (45% former smokers and 15% current smokers) with an average of 22 pack years. The remaining 40% of patients identified as never smokers. Only a small number of patients had history of heavy alcohol use (18%) defined as 8 or more drinks per week for females or 15 or more drinks per week for males; most patients identified as either never, light, or social drinkers.
Table 1.
Clinical information summary.
| Variable | HNSCC Patients n=33* |
|---|---|
| Av. Age at Diagnosis | 62.9 [46–87] |
| Sex | |
| Male | 26 (79%) |
| Female | 7 (21%) |
| Smoking Status | |
| Current | 5 (15%) |
| Former | 15 (45%) |
| Never | 13 (40%) |
| Av. Pack Years | 22 [0–100] |
| Drinking History | |
| Never | 14 (42%) |
| Social | 5 (15%) |
| Light | 8 (24%) |
| Heavy | 6 (18%) |
| Tumor Site | |
| Oropharynx | 31 (94%) |
| Oral Cavity | 1 (3%) |
| Nasopharynx | 1 (3%) |
| T Classification† | |
| T1 | 5 (15%) |
| T2 | 12 (36%) |
| T3 | 7 (21%) |
| T4 | 7 (21%) |
| Recurrence | 2 (6%) |
| Treatment | |
| CRT | 22 (67%) |
| CRT + Immunotherapy | 1 (3%) |
| RT | 2 (6%) |
| Surgery | 4 (12%) |
| Surgery + RT | 2 (6%) |
| Surgery + CRT | 2 (6%) |
| Disease Progression | |
| No LRF or DM | 22 (67%) |
| LRF and DM | 3 (9%) |
| LRF only | 4 (12%) |
| DM only | 3 (9%) |
| Unknown | 1 (3%) |
| Survival | |
| Alive, NED | 21 (64%) |
| Died of disease | 9 (27%) |
| Died, unrelated cause | 3 (9%) |
Excludes 3 patients (n=1, HPV-negative. n=2, data unavailable).
AJCC 7th edition.
Abbreviations: CRT, chemoradiation. RT, radiation therapy. LRF, locoregional failure. DM, distant metastasis. NED, no evidence of disease.
Patients presented with tumors across the TNM classifications (AJCC 7th edition). The most frequently reported T classification was T2 (36%), but there were patients with T1, T3, and T4 tumors as well. The majority of patients (71%) had some level of nodal involvement (26% N1, 3% N2, 23% N2b, 19% N2c). Only one patient had distant metastasis at diagnosis. The majority of patients were treated with chemoradiation alone or in combination with surgery (73%). A variety of chemotherapy agents were used, including erbitux, cisplatin, carboplatin, taxol, fluorouracil, docetaxel, and gemcitabine. Other treatments included surgery alone (12%), radiation alone (6%), and surgery plus radiation (6%). Patients who developed local recurrences or metastases were treated initially with chemoradiation, followed by different chemotherapy regimens or immunotherapy.
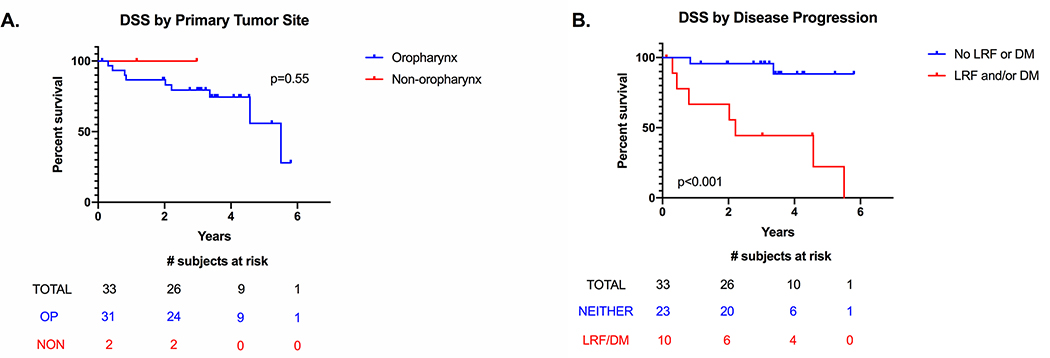
We were able to collect at least two years of follow-up on the majority of this cohort with a median follow-up time of 3.25 years; four patients were lost to follow-up before the two-year mark. Only three patients (9%) developed both locoregional failure (LRF) and distant metastases (DM); four patients developed only LRF (12%), and three patients developed only DM (9%). Nine patients (27%) died of their disease; the average time to death was 1.5 years with a range of 3 months to 3.2 years. The majority of patients who died of disease did so within 2 years of diagnosis. The 3-year disease-specific survival (DSS) of the OPSCC patients was 80% and did not differ significantly from the non-oropharyngeal patients (Figure 2A). We compared the survival curves of patients who developed LRF and/or DM versus those who didn’t, and as expected, patients whose tumors progressed had a significant worse DSS (Figure 2B). We also examined the influence of age, smoking and drinking histories, and T and N classification, but none of these variables showed significant differences in survival (Figure S1).
Figure 2:
Kaplan-Meier censored disease-specific survival (DSS) curves. A, Separated by primary tumor site (oropharynx vs nonoropharynx). B, Separated by disease progression (patients with vs without locoregional failure (LRF) and/or distant metastases (DM),includes both oropharynx and nonoropharynx patients
Viral Genotypes
We tested the HPV genotypes present in thirty-six p16+ HNSCCs by HPV PCR-MassArray (Table 2). The majority of samples were positive for a single HPV type; thirty samples (83%) were HPV16+ and one sample (3%) was HPV18+. Four additional samples were positive for multiple HPV types; three samples were HPV16+ HPV33+ (8%) and one sample was HPV16+ HPV18+ (3%). Only one sample (3%) was negative for all HPV types and was excluded from further analysis.
Table 2.
HPV PCR-MassArray results.
| HPV Result | No. of patients (%) by HPV type |
|---|---|
| HPV16 | 30 (83%) |
| HPV16 + HPV33 | 3 (8%) |
| HPV16 + HPV18 | 1 (3%) |
| HPV18 | 1 (3%) |
| Negative | 1 (3%) |
| TOTAL | 36 |
Viral Integration
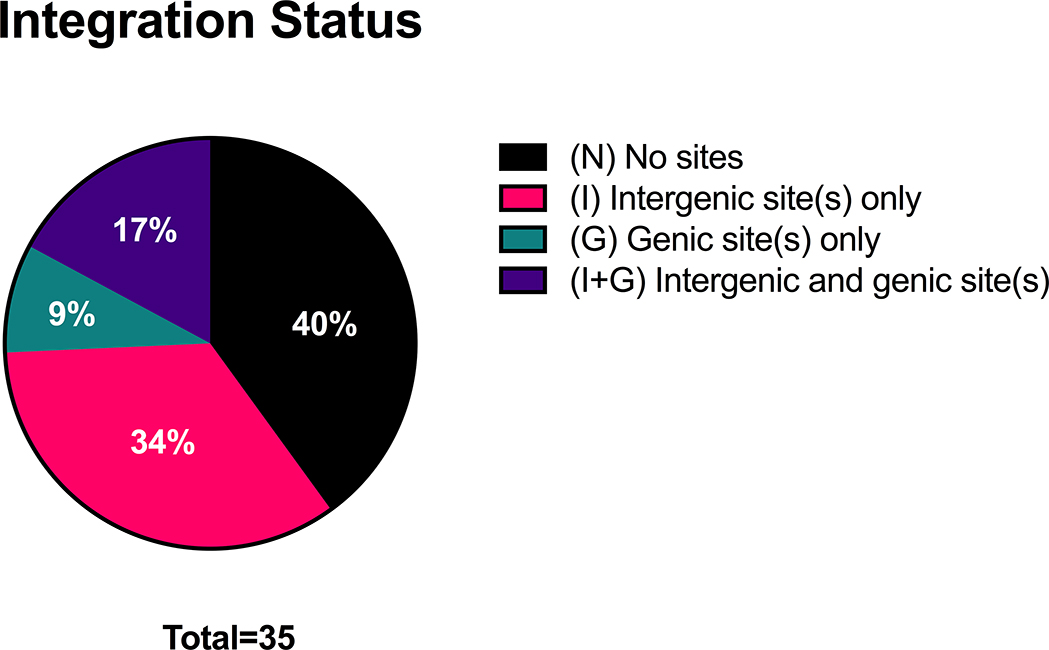
We tested thirty-five samples for HPV16 and/or HPV18 viral integration as appropriate by DIPS-PCR. We discovered at least one integration site in the majority of samples (60%) but were unable to find any integration sites in fourteen out of thirty-five samples (40%). Interestingly, the sample that was positive for both HPV16 and HPV18 (UM-3898) showed integration of both HPV types into different loci. Of the twenty-one samples with HPV integration, the median number of sites we discovered in each was 1, ranging from 1 to 4.
By Sanger Sequencing, we were able to determine that the vast majority of cellular loci affected by integration were gene-poor intergenic regions of the genome; we discovered a total of thirty-five integration sites and only eight of them involved cellular genes (Table 3). Of the samples with HPV integration, the majority had integration into intergenic sites only (n=12) (Figure 3). Some samples had integration into both intergenic and genic regions (n=6), and a few samples (n=3) had integration into genic regions only.
Table 3.
Integration status and site descriptions.
| Sample ID | HPV Type | HPV Integration Status | HPV/Human Region(s) Involved |
|---|---|---|---|
| BMT-396 | Negative | - | - |
| BMT-8 | 16 | N | - |
| BMT-56 | 16 | N | - |
| BMT-280 | 16+33 | N | - |
| BMT-403 | 16 | N | - |
| BMT-412 | 16 | N | - |
| BMT-700 | 16+33 | N | - |
| BMT-1327 | 16 | N | - |
| UM-3884 | 18 | N | - |
| UM-3917 | 16 | N | - |
| UM-3955 | 16 | N | - |
| UM-3962 | 16 | N | - |
| UM-3989 | 16 | N | - |
| UM-4028 | 16 | N | - |
| UM-4093 | 16 | N | - |
| BMT-233 | 16 | I | E1: SCAF |
| E1: SCAF | |||
| E2: SCAF | |||
| L1: SCAF | |||
| BMT-319 | 16 | I | E1: SCAF |
| L1: Chrom 13 | |||
| BMT-322 | 16 | I | E1: SCAF |
| BMT-344 | 16 | I | E1: SCAF |
| BMT-400 | 16 | I | E1: SCAF |
| BMT-402 | 16 | I | E2: SCAF |
| L2: SCAF | |||
| L1: SCAF | |||
| BMT-404 | 16+33 | I | E2: Chrom 4 |
| BMT-411 | 16 | I | E1: SCAF |
| BMT-427 | 16 | I | E1: SCAF |
| UM-3940 | 16 | I | E2: Chrom 17q21 |
| UM-3948 | 16 | I | L1: Chrom 13q14 |
| UM-4067 | 16 | I | L1: Chrom 13q14 |
| BMT-251 | 16 | I+G | E1: SCAF |
| E2: SCAF | |||
| L2: SGCZ | |||
| BMT-323 | 16 | I+G | E1: Chrom 2q |
| L2: UTP18 | |||
| L1: Chrom 4 | |||
| BMT-1159 | 16 | I+G | E1: SCAF |
| L1: KIF21B | |||
| UM-3898 | 16+18 | I+G | L1: Chrom 13q14 |
| (HPV18) E1: NDST1 | |||
| UM-3938 | 16 | I+G | L2: YIPF1 |
| L1: Chrom 6q21 | |||
| UM-3954 | 16 | I+G | E1: Chrom 3p25 |
| L1: DNAI1-L1: NPAS3 | |||
| BMT-331 | 16 | G | E1: Chrom 1q21: SCN1B |
| UM-4011 | 16 | G | E1: PTPRN2 |
| UM-4068 | 16 | G | L1: RLN1 |
Abbreviations: N, no sites. I, intergenic sites. G, genic sites. SCAF, genomic scaffold region.
Figure 3:
Integration status of HPV+ HNSCCs. HNSCC, head and neck squamous cell carcinoma; HPV, human papillomavirus
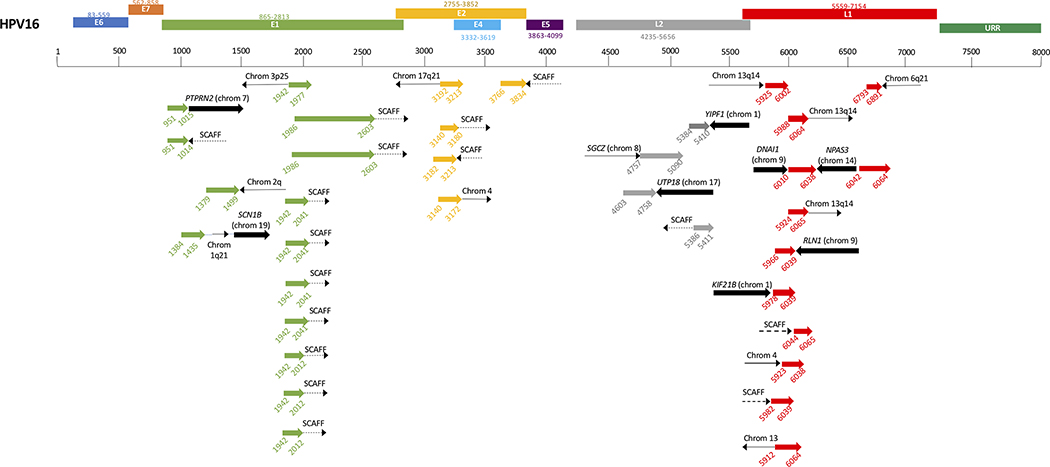
A large number of integrations occurred in unplaced genomic scaffold regions of the genome (14/35 events) (Figure 4). The most frequently affected chromosome was chromosome 13 (4/35 events).The cellular genes involved in the integration sites we found included PTPRN2, SCN1B, YIPF1, SGCZ, DNAI1, NPAS3, UTP18, RLN1, and KIF21B. Integration most frequently involved the HPV genes E1 (n=14) and L1 (n=11). A few integrations also involved E2 (n=5) and L2 (n=4).
Figure 4:
HPV integration sites aligned to HPV genome. Corresponding colors represent HPV gene (green = E1, etc). Black arrows indicate human sequence. Wide black arrow, cellular gene. Thin solid black line, intergenic region. Dashed black line labeled SCAFF,genomic scaffold region
Viral-cellular Fusion Transcript Expression
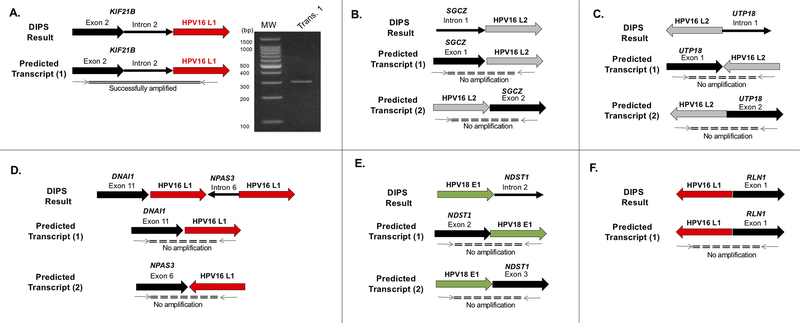
We were interested whether those integration sites involving cellular genes led to the generation of viral-cellular fusion transcripts that have been reported in many HNSCC samples. Of the nine samples with integration into a gene, RNA was available for fusion transcript analysis for six samples. We attempted to amplify the predicted fusion transcripts with primers designed spanning the junction site discovered by DIPS-PCR (Figure 5). In BMT-1159, we detected an integration of HPV16 L1 into intron 2 of KIF21B by DIPS-PCR and were able to amplify a fusion transcript across this junction as shown in Fig 5A. This amplicon was sequenced by Sanger sequencing to confirm its identity, and the resulting sequence matched correctly to KIF21B and L1. In BMT-251, HPV16 L2 integrated into intron 1 of SGCZ; we attempted to amplify junctions up and downstream of L2, but no amplicons were generated. We performed similar amplifications in BMT-323 (UTP18:HPV16 L2), UM-3954 (DNAI1:HPV16 L1:NPAS3:HPV16 L1), UM-3898 (HPV18 E1-NDST1) and UM-4068 (HPV16:RLN1) with no amplification of any of the predicted fusion transcripts.
Figure 5:
HPV integration sites aligned to HPV genome. Corresponding colors represent HPV gene (green = E1, etc). Black arrows indicate human sequence. Wide black arrow, cellular gene. Thin solid black line, intergenic region. Dashed black line labeled SCAFF,genomic scaffold region
Viral E6E7 Transcripts
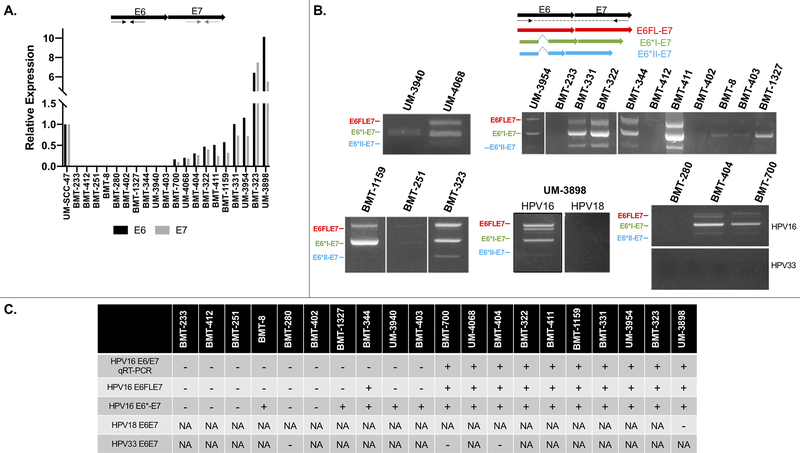
We assessed expression of HPV E6 and E7 in samples with available RNA by qRT-PCR and RT-PCR (Figure 6). Of twenty samples tested for HPV16 by qRT-PCR, ten (50%) expressed E6 and E7 transcripts at varying levels relative to expression in UM-SCC-47 which very strongly expresses these transcripts. The remaining ten samples (50%) did not express detectable levels of HPV16 transcripts, despite testing HPV16+ at the DNA level. However, upon assessment of the expression of HPV16 E6-E7 alternate transcripts by RT-PCR, we found that five of these samples showed expression of one or more transcript. We found that the majority of samples expressed both full-length (E6FLE7) and spliced E6* transcripts (n=10), and a small number of samples (n=4) only expressed E6* transcripts. Samples positive for more than one HPV type (HPV16/18+ or HPV16/33+) were tested for transcripts of both HPV types; three samples expressed HPV16 transcripts but not HPV18 (UM-3898) or HPV33 (BMT-700 and BMT-404) transcripts. A fourth sample (BMT-280) did not express HPV16 or HPV33 transcripts. There was no significant difference in survival between patients who expressed any E6/E7 transcripts versus those who didn’t, and there was also no significant difference in survival between patients who expressed only E6* transcripts versus both E6FL and E6* transcripts. (Figure S2).
Figure 6:
HPV16 E6 and E7 transcript expression. A, Top: qRT-PCR primer design, Bottom: relative expression of E6 and E7,compared to UM-SCC-47. B, Top: RT-PCR primer design to amplify alternate HPV16/18/33 transcripts, Bottom: expression of alternate transcripts. C, Summary table of results. +, positive result. −, negative result; HPV, human papillomavirus; NA, not applicable
Association with Clinical Variables
We tested whether there was an association between HPV genomic integration and other variables gathered during this study by Spearman’s rank correlation (Table 4). We tested for a correlation between HPV integration and age, smoking history, drinking history, T classification, nodal involvement, E6/E7 expression level by qRT-PCR, and expression of E6FL or E6*. Of these, only age (r=0.453, p=0.008), E6/E7 expression level by qRT-PCR (r=0.480, p=0.038) and E6FL expression (r=.459, p=0.048) demonstrated a significant positive correlation with HPV integration. This indicates that patients with integration were more likely to be older and had higher expression of the HPV oncogenes, specifically the full-length E6 transcript.
Table 4.
Correlation between HPV integration and other relevant variables.
| HPV Integration vs… | Spearman’s r | p value |
|---|---|---|
| Age | 0.453 | 0.008* |
| Smoking | 0.112 | 0.537 |
| Heavy drinking | 0.219 | 0.220 |
| T classification | −0.213 | 0.251 |
| Nodal involvement | −0.215 | 0.229 |
| E6/E7 qRT-PCR expression | 0.480 | 0.038* |
| E6FLE7 expression | 0.459 | 0.048* |
| E6*-E7 expression | 0.186 | 0.447 |
Significant p-value.
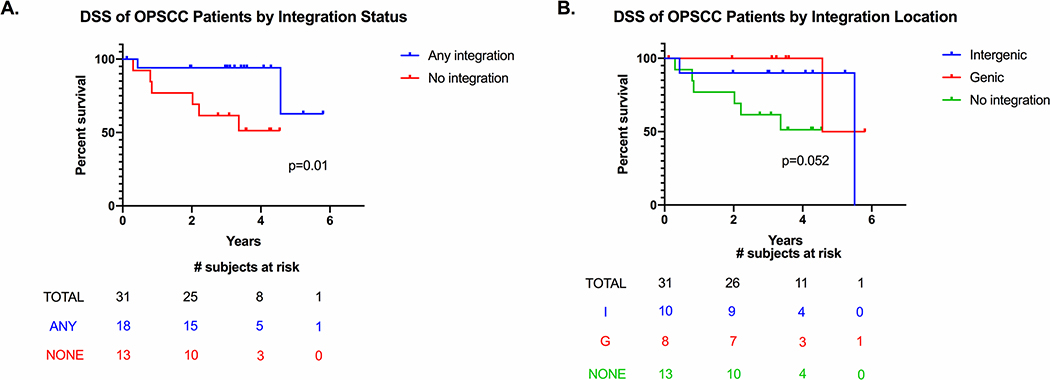
We were interested in whether HPV integration influenced patient outcomes. There was no significant association between HPV integration and locoregional failure (p=0.676) or distant metastasis (p=0.659) as assessed by Fisher’s exact test, although the number of events in each group was likely too small to power this analysis. The DSS curves of the oropharynx patients separated by integration status and site are shown in Figure 7. Integration positive OPSCC patients had a significantly improved DSS compared to integration negative patients (p=0.01). When we separated integration positive patients by site of integration (intergenic sites only vs any genic sites), there was no significant difference in the survival curves.
Figure 7:
Kaplan-Meier curves of oropharynx patients separated by integration status, A, and integration subsite, B, censored
DISCUSSION
HPV+ HNSCC, particularly HPV+OPSCC, has been increasing in incidence rapidly over the past few decades.33–35 Despite improved outcomes compared to HPV- HNSCC, still 20–30% of patients fail to respond to initial therapies or recur5, and the factors that contribute to the progression of this disease are not well understood. Given the high morbidity of HNSCC treatment, there is a push in the field to de-escalate treatment for patients at low risk of disease recurrence.36 However, the biomarkers for response to treatment are not well developed yet, which makes stratifying patients difficult. Studies of treatment de-escalation are ongoing based on clinical risk factors37–39, but there is still a need to investigate the molecular drivers of this disease in order to understand what distinguishes high versus low risk patients.
One such process that has been investigated as a potential driver of HPV+ HNSCC is the process of viral integration. Viral integration has been well characterized in cervical cancer as a marker of disease progression.40 Studies in cervical cancer and HNSCC have shown that integration into the genome can have a variety of effects on both the cellular and viral genomes, including large scale rearrangements, amplifications, deletions, alterations in gene expression and generation of viral-cellular fusion transcripts.6–8, 11–13, 19 Others have attempted to characterize the relationship between HPV integration and E6/E7 expression as well as between HPV integration and patient outcomes with mixed results.16, 22–31
Here we have presented an analysis of integration sites, HPV oncogene expression and associations with clinical variables in a cohort of p16+ HNSCCs. Only one patient tested negative for all HPV types by HPV PCR-MassArray and was excluded from further analysis. Of the thirty-five patients tested for HPV16 and/or HPV18 integration by DIPS-PCR, we found at least one integration site in 60% of samples and were unable to find integration in 40%. We considered samples without HPV integration sites to be “integration-negative”, although it is theoretically possible sites of integration were missed by DIPS-PCR. However, previous studies of HPV integration using a variety of methods reported similar proportions, ranging from 30–50% integration negative.6–12 The use of different HPV integration detection methods likely accounts for the variability seen between studies.
The use of DIPS-PCR allows us to identify the number and location of HPV integration sites within each sample. The majority of samples contained only one integration site, although there were samples in which we were able to identify more than one. Of particular interest was UM-3898, which contained integrations for both HPV16 and HPV18; it is unclear how integration of more than one HPV type might affect the progression of tumorigenesis. E1 was the HPV gene most frequently involved in integration (40% of sites), which is in agreement with previous studies.12, 41 Even though there were a limited number of integration sites detected (n=35), we were able to determine that integration events took place across eleven different chromosomes (chromosomes 1, 2, 3, 4, 6, 7, 8, 9, 13, 14, 17). Of the integration sites detected, only eight (23%) were within cellular genes. Previous studies have proposed that integration is a directed process that occurs preferentially in/near genes or other genomic features, such as miRNAs or lncRNAs6, 13, 40, 42, but our results show more of a stochastic pattern given the wide range of chromosomes affected and low percentage involving genes. However, the number of events we detected is relatively small, and therefore it is challenging to detect predilections for a specific type of location or chromosomal hotspots. Furthermore, the limiting size of the genomic segments in the SCAF insertions detected by this method prohibits precise identification of the actual locus affected.
We further investigated the integration sites that occurred within cellular genes at the transcript level. Viral-host fusion transcripts have been reported by other groups to increase E6/E7 expression.18–20 Previous work from our group has shown that viral-cellular fusion transcripts may or may not form depending on the location of the integration site within the gene (within an intron vs exon).20, 30, 43 It is possible that some integrations within introns are spliced out and therefore do not produce a fusion transcript, while others may alter splice acceptor/donor sites such that they are retained at the transcript level. We attempted to amplify the predicted fusion transcripts based on the DNA-level information we obtained from DIPS-PCR in six samples but were only successful in amplifying the fusion in one sample (BMT-1159). This fusion involved HPV16 L1 integrating into intron 2 of the cellular gene KIF21B, which encodes for a microtubule-dependent motor protein. In this case, we were able to amplify a transcript that included KIF21B exon 2–KIF21B intron 2–HPV16 L1, indicating this integration resulted in alteration of splice sites such that intron 2 was retained in the transcript. KIF21B and other kinesin superfamily proteins have been implicated in the progression of many solid tumors via dysregulation of mitosis44–46; therefore, it is of great interest to discover how this fusion may have played a role in the carcinogenesis in this case.
We performed a similar analysis on the other five samples, three of which involved integration into introns and two involved gene exons, but we were unable to amplify any of the predicted fusion transcripts. It is not necessarily surprising that these integration sites did not yield fusion transcripts, but it is possible that the site is more complicated than we expect, resulting in a false negative. Another open question is whether these fusion transcripts are being driven off of a cellular or HPV promoter, which is difficult to address with the relatively short sequences obtained during DIPS-PCR. Gathering more sequence surrounding the site may be helpful in the future to amplify these transcripts.
We also assessed expression of the E6 and E7 oncogenes within tumors with available RNA (n=20) by qRT-PCR, which showed varying levels of expression compared to UM-SCC-47, an HPV+ HNSCC cell line we showed previously has high E6/E7 expression.20 Interestingly, half of the samples showed no expression of E6 or E7. However, analysis of these samples by RT-PCR using primers designed to amplify alternate E6E7 transcripts revealed that they did in fact express one or more E6E7 transcripts. It is unclear why they lacked expression by qRT-PCR, but it is possible they were below the threshold of detection for this assay. There were still five samples which showed no expression of E6E7, which is curious given that they were p16+ by IHC and HPV16+ at the DNA level. E6/E7 are negatively regulated by E2, which is frequently reported to be disrupted by the process of HPV integration; therefore, some have proposed that HPV integration leads to increased E6/E7 levels.15 In this cohort, we saw a significant positive correlation between HPV integration and E6/E7 expression levels, which supports this idea. However, it is not a perfect correlation; some samples with HPV integration still have no expression of E6/E7. This aligns with those who have published that E2 is not always disrupted during integration, and therefore not all integrated samples will have increased E6/E7 levels.12, 21 Alternatively, E6/E7 expression could be altered due to methylation of the E2 binding sites in the upstream regulatory region (URR) of HPV16 rather than loss of E2 itself.47, 48
We assessed the expression of alternate E6* transcripts; these transcripts are thought to contribute to a more aggressive phenotype, resulting in larger tumors and worse patient prognosis.17 We found that the majority of samples expressed both E6FLE7 and alternate E6* transcripts with a few samples only expressing E6* transcripts. Three out of four samples that contained multiple HPV types only expressed HPV16 transcripts but not from other HPV types. There was a significant positive correlation between HPV integration and E6FL expression, but not between HPV integration and E6* expression. This contrasts with reports that E6* variants are more common in tumors with integrated HPV16; however, it is possible our results differed due to our relatively small sample size.
We assessed the association of HPV integration with clinical variables, including age, smoking and drinking histories, and T/N classification, to further examine this process. Of these, only age showed a significant positive correlation with HPV integration, indicating that older patients were more likely to have integrated HPV. It is unclear why this may be; one explanation could be that HPV integration occurs more frequently in older patients because DNA damage accumulates in aging tissue, as it has been previously proposed that HPV integration occurs at sites of unresolved DNA damage.49
We compared the survival of OPSCC patients with versus without integration and found that integration-positive patients had a significantly improved disease-specific survival over integration-negative patients. This contrasts with what others have previously reported; studies either reported no significant difference between the two groups or that integration-negative patients had a survival advantage over integration-positive patients.23–29 It has been hypothesized that integration acts as an additional oncogenic driver through its various effects on the human and viral genomes. The reason for the discrepancy between our findings and previous reports is unclear, but it could be due to different methods of detecting HPV integration. These previous studies measured integration indirectly by assessing loss of E2 DNA22–27 or mRNA.28 Another study based integration status on the presence of fusion transcripts.29 However, given that E2 is not always lost due to integration and not every integration results in a fusion transcript, our preferred method to detect integration is DIPS-PCR. We have used DIPS-PCR previously to assess integration sites in a small cohort of responsive vs non-responsive patients and found that non-responsive patients were more likely to have integration into genes rather than intergenic loci.30 The underlying mechanism behind the improved survival we reported here in integration positive patients is unclear and requires further investigation. One possible hypothesis is that the process of HPV integration generates tumor neoantigens which can then be recognized as non-self by the host immune system and enhance antitumor immune response. HPV+ OPSCC patients with higher levels of infiltrating CD8+ T cells, which are involved in recognizing tumor antigens, have been shown to have improved outcomes50, but it is currently unknown if integration-positive vs integration negative patients have differential immune infiltration patterns and whether they can present these neoantigens for immune recognition.
There are two major limitations of this study that could be addressed in future research. First, our study population was relatively small, which limited our ability to examine the relationships between HPV integration status/site and LRF or DM given that so few patients experienced these events. Secondly, we used DIPS-PCR as our preferred method of detecting integration sites because it is highly specific, but some of the amplicons we generated were too short to provide enough context for us to be able to place them at a specific locus and therefore had to be denoted as “genomic scaffold”. DIPS-PCR alone is also unable to distinguish between samples with only integrated HPV and samples that contain a mixture of integrated and episomal HPV, although sometimes episomal HPV copies may appear as 6–8 kb bands upon gel electrophoresis. It is unclear how these two samples types may differ in terms of HPV-related genetic or epigenetic changes. In the future, we will focus on pairing DIPS-PCR with long-range sequencing technologies, such as Nanopore sequencing, in order to better define the complex structural rearrangements caused by HPV integration19 and explain the structural basis of local amplification at integration sites.12 Comprehensive investigation of HPV integration sites and how they impact the course of HNSCC is necessary to provide insight for the development of alternate therapies for non-responsive tumors. Overall, this study shows that HPV integration influences patient outcomes, which we feel warrants the implementation of viral integration analysis in the clinic.
Supplementary Material
ACKNOWLEDGEMENTS:
The authors acknowledge the assistance of the University of Michigan Advanced Genomics core for carrying out Sanger sequencing and Lila Peters who helped with the isolation of RNA from frozen tissue. This work was supported by R01 CA194536. L.M. Pinatti was supported by R01 CA194536 and the Cancer Biology Training Grant NIH-NCI T32 CA009676.
REFERENCES
- 1.Mahal BA, Catalano PJ, Haddad RI, et al. Incidence and Demographic Burden of HPV-Associated Oropharyngeal Head and Neck Cancers in the United States. Cancer Epidemiol Biomarkers Prev 2019. October;28(10):1660–7. [DOI] [PubMed] [Google Scholar]
- 2.Gillison ML, Koch WM, Capone RB, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst 2000. May 3;92(9):709–20. [DOI] [PubMed] [Google Scholar]
- 3.Fakhry C, Westra WH, Li S, et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst 2008. February 20;100(4):261–9. [DOI] [PubMed] [Google Scholar]
- 4.Licitra L, Perrone F, Bossi P, et al. High-risk human papillomavirus affects prognosis in patients with surgically treated oropharyngeal squamous cell carcinoma. Journal of Clinical Oncology. 2006. December 20;24(36):5630–6. [DOI] [PubMed] [Google Scholar]
- 5.Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. The New England journal of medicine. 2010. July 01;363(1):24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodelon C, Untereiner ME, Machiela MJ, Vinokurova S, Wentzensen N. Genomic characterization of viral integration sites in HPV-related cancers. Int J Cancer. 2016. November 01;139(9):2001–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao G, Wang J, Kasperbauer JL, et al. Whole genome sequencing reveals complexity in both HPV sequences present and HPV integrations in HPV-positive oropharyngeal squamous cell carcinomas. BMC Cancer. 2019. April 11;19(1):352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holmes A, Lameiras S, Jeannot E, et al. Mechanistic signatures of HPV insertions in cervical carcinomas. NPJ Genom Med 2016;1:16004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalantari M, Villa LL, Calleja-Macias IE, Bernard HU. Human papillomavirus-16 and −18 in penile carcinomas: DNA methylation, chromosomal recombination and genomic variation. Int J Cancer. 2008. October 15;123(8):1832–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morel A, Neuzillet C, Wack M, et al. Mechanistic Signatures of Human Papillomavirus Insertions in Anal Squamous Cell Carcinomas. Cancers (Basel). 2019. November 22;11(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nulton TJ, Olex AL, Dozmorov M, Morgan IM, Windle B. Analysis of The Cancer Genome Atlas sequencing data reveals novel properties of the human papillomavirus 16 genome in head and neck squamous cell carcinoma. Oncotarget 2017. March 14;8(11):17684–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parfenov M, Pedamallu CS, Gehlenborg N, et al. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc Natl Acad Sci U S A. 2014. October 28;111(43):15544–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Z, Zhu D, Wang W, et al. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat Genet 2015. February;47(2):158–63. [DOI] [PubMed] [Google Scholar]
- 14.McBride AA, Warburton A. The role of integration in oncogenic progression of HPV-associated cancers. PLoS Pathog 2017. April;13(4):e1006211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McBride AA. The papillomavirus E2 proteins. Virology. 2013. October;445(1–2):57–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Koneva LA, Virani S, et al. Subtypes of HPV-Positive Head and Neck Cancers Are Associated with HPV Characteristics, Copy Number Alterations, PIK3CA Mutation, and Pathway Signatures. Clin Cancer Res 2016. September 15;22(18):4735–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin T, Koneva LA, Liu Y, et al. Significant association between host transcriptome-derived HPV oncogene E6* influence score and carcinogenic pathways, tumor size, and survival in head and neck cancer. Head Neck. 2020. September;42(9):2375–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeon S, Lambert PF. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc Natl Acad Sci U S A. 1995. February 28;92(5):1654–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akagi K, Li J, Broutian TR, et al. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res 2014. February;24(2):185–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walline HM, Goudsmit CM, McHugh JB, et al. Integration of high-risk human papillomavirus into cellular cancer-related genes in head and neck cancer cell lines. Head Neck. 2017. May;39(5):840–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olthof NC, Speel EJ, Kolligs J, et al. Comprehensive analysis of HPV16 integration in OSCC reveals no significant impact of physical status on viral oncogene and virally disrupted human gene expression. Plos One. 2014;9(2):e88718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ibragimova M, Tsyganov M, Shpileva O, et al. HPV status and its genomic integration affect survival of patients with cervical cancer. Neoplasma 2018. March 14;65(3):441–8. [DOI] [PubMed] [Google Scholar]
- 23.Kiseleva VI, Mkrtchyan LS, Ivanov SA, et al. The Presence of Human Papillomavirus DNA Integration is Associated with Poor Clinical Results in Patients with Third-Stage Cervical Cancer. Bull Exp Biol Med 2019. November;168(1):87–91. [DOI] [PubMed] [Google Scholar]
- 24.Shin HJ, Joo J, Yoon JH, Yoo CW, Kim JY. Physical status of human papillomavirus integration in cervical cancer is associated with treatment outcome of the patients treated with radiotherapy. Plos One. 2014;9(1):e78995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anayannis NV, Schlecht NF, Ben-Dayan M, et al. Association of an intact E2 gene with higher HPV viral load, higher viral oncogene expression, and improved clinical outcome in HPV16 positive head and neck squamous cell carcinoma. Plos One. 2018;13(2):e0191581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nulton TJ, Kim NK, DiNardo LJ, Morgan IM, Windle B. Patients with integrated HPV16 in head and neck cancer show poor survival. Oral Oncol 2018. May;80:52–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim MY, Dahlstrom KR, Sturgis EM, Li G. Human papillomavirus integration pattern and demographic, clinical, and survival characteristics of patients with oropharyngeal squamous cell carcinoma. Head Neck. 2016. August;38(8):1139–44. [DOI] [PubMed] [Google Scholar]
- 28.Vojtechova Z, Sabol I, Salakova M, et al. Analysis of the integration of human papillomaviruses in head and neck tumours in relation to patients’ prognosis. Int J Cancer. 2016. January 15;138(2):386–95. [DOI] [PubMed] [Google Scholar]
- 29.Koneva LA, Zhang Y, Virani S, et al. HPV Integration in HNSCC Correlates with Survival Outcomes, Immune Response Signatures, and Candidate Drivers. Mol Cancer Res 2017. September 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walline HM, Komarck CM, McHugh JB, et al. Genomic Integration of High-Risk HPV Alters Gene Expression in Oropharyngeal Squamous Cell Carcinoma. Mol Cancer Res 2016. October;14(10):941–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walline HM, Komarck CM, McHugh JB, et al. High-risk human papillomavirus detection in oropharyngeal, nasopharyngeal, and, oral cavity cancers: Comparison of multiple methods. JAMA Otolaryngology. 2013. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luft F, Klaes R, Nees M, et al. Detection of integrated papillomavirus sequences by ligation-mediated PCR (DIPS-PCR) and molecular characterization in cervical cancer cells. Int J Cancer. 2001. April 01;92(1):9–17. [PubMed] [Google Scholar]
- 33.Lundberg M, Leivo I, Saarilahti K, Makitie AA, Mattila PS. Increased incidence of oropharyngeal cancer and p16 expression. Acta Otolaryngol 2011. September;131(9):1008–11. [DOI] [PubMed] [Google Scholar]
- 34.Wittekindt C, Wagner S, Bushnak A, et al. Increasing Incidence rates of Oropharyngeal Squamous Cell Carcinoma in Germany and Significance of Disease Burden Attributed to Human Papillomavirus. Cancer Prev Res (Phila). 2019. June;12(6):375–82. [DOI] [PubMed] [Google Scholar]
- 35.Chaturvedi AK, Engels EA, Pfeiffer RM, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol 2011. November 10;29(32):4294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bigelow EO, Seiwert TY, Fakhry C. Deintensification of treatment for human papillomavirus-related oropharyngeal cancer: Current state and future directions. Oral Oncol 2020. April 2;105:104652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marur S, Li S, Cmelak AJ, et al. E1308: Phase II Trial of Induction Chemotherapy Followed by Reduced-Dose Radiation and Weekly Cetuximab in Patients With HPV-Associated Resectable Squamous Cell Carcinoma of the Oropharynx- ECOG-ACRIN Cancer Research Group. J Clin Oncol 2017. February 10;35(5):490–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Misiukiewicz K, Gupta V, Miles BA, et al. Standard of care vs reduced-dose chemoradiation after induction chemotherapy in HPV+ oropharyngeal carcinoma patients: The Quarterback trial. Oral Oncol 2019. August;95:170–7. [DOI] [PubMed] [Google Scholar]
- 39.Seiwert TY, Foster CC, Blair EA, et al. OPTIMA: a phase II dose and volume de-escalation trial for human papillomavirus-positive oropharyngeal cancer. Ann Oncol 2019. October 1;30(10):1673. [DOI] [PubMed] [Google Scholar]
- 40.Tian R, Cui Z, He D, et al. Risk stratification of cervical lesions using capture sequencing and machine learning method based on HPV and human integrated genomic profiles. Carcinogenesis. 2019. October 16;40(10):1220–8. [DOI] [PubMed] [Google Scholar]
- 41.Liu L, Ying C, Zhao Z, et al. Identification of reliable biomarkers of human papillomavirus 16 methylation in cervical lesions based on integration status using high-resolution melting analysis. Clin Epigenetics. 2018;10:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitz M, Driesch C, Jansen L, Runnebaum IB, Durst M. Non-random integration of the HPV genome in cervical cancer. Plos One. 2012;7(6):e39632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pinatti LM, Walline HM, Carey TE, Klussman JP, Huebbers CU. Viral Integration Analysis Reveals Likely Common Clonal Origin of Bilateral HPV16-Positive, p16-Positive Tonsil Tumors. Archives of Clinical and Medical Case Reports. 2020;4:680–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J, Li S, Zhou S, et al. Kinesin superfamily protein expression and its association with progression and prognosis in hepatocellular carcinoma. J Cancer Res Ther 2017;13(4):651–9. [DOI] [PubMed] [Google Scholar]
- 45.Sun ZG, Pan F, Shao JB, Yan QQ, Lu L, Zhang N. Kinesin superfamily protein 21B acts as an oncogene in non-small cell lung cancer. Cancer Cell Int 2020;20:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao HQ, Dong BL, Zhang M, et al. Increased KIF21B expression is a potential prognostic biomarker in hepatocellular carcinoma. World J Gastrointest Oncol 2020. March 15;12(3):276–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reuschenbach M, Huebbers CU, Prigge ES, et al. Methylation status of HPV16 E2-binding sites classifies subtypes of HPV-associated oropharyngeal cancers. Cancer-Am Cancer Soc 2015. June 15;121(12):1966–76. [DOI] [PubMed] [Google Scholar]
- 48.von Knebel Doeberitz M, Prigge ES. Role of DNA methylation in HPV associated lesions. Papillomavirus Res 2019. June;7:180–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wallace NA, Khanal S, Robinson KL, Wendel SO, Messer JJ, Galloway DA. High-Risk Alphapapillomavirus Oncogenes Impair the Homologous Recombination Pathway. J Virol 2017. October 15;91(20). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oguejiofor K, Hall J, Slater C, et al. Stromal infiltration of CD8 T cells is associated with improved clinical outcome in HPV-positive oropharyngeal squamous carcinoma. Br J Cancer. 2015. September 15;113(6):886–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.