Abstract
Background
The prevalence of age-related macular degeneration (AMD) varies from 6.8 to 18.3% for all forms of AMD and from 0.6 to 2.6% for late AMD according to race, suggesting the existence of genetic differences among races. The purpose of this study was to determine the genetic causes of differences in the prevalence of AMD among individuals of different races.
Methods
We collected 138 AMD-associated single nucleotide polymorphisms (SNPs) from a genome-wide association studies catalog. Their population-level allele frequencies were derived based on the 1000 Genomes Project and Korean Reference Genome Database. We used Fisher’s exact tests to assess whether the effect allele at a given SNP was significantly enriched or depleted in the database.
Results
European, American, and South Asian populations showed similar heatmap patterns, whereas East Asian, and Korean populations had distinct patterns. Korean populations exhibited patterns that were different from those of the other groups; rs5754227 (SYN3), rs1626340 (TGFBR1/COL15A1), rs3750846(ARMS2/HTRA1), and rs9564692 (B3GALTL) were enriched, whereas rs2230199 (C3) and rs73036519 (EXOC3L2/MARK4) were depleted in Koreans; these SNPs are associated with late AMD. The genetic risk score calculated from allele frequencies was not less in East Asians than in Europeans.
Conclusion
The prevalence of AMD is lower in Asians than in Europeans. However, our study showed that genetic risk scores in East Asians were similar to those in Europeans, which may explain why the global projected number of people with AMD by 2040 is in largest for East Asians, including Koreans.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12886-021-01830-9.
Keywords: Age-related macular degeneration, Allele frequency, Single nucleotide polymorphism, Genetic risk scores, Prevalence
Background
Age-related macular degeneration (AMD) is the leading cause of visual impairment in developed countries [1, 2], particularly in people older than 60 years. AMD includes early AMD and late AMD; early AMD is characterized by drusen and/or pigmentary changes [3, 4], whereas late AMD is can show two subtypes, i.e., geographic atrophy (GA) and choroidal neovascularization (CNV). According to a study by Wong et al., the global prevalence of all types of AMD is approximately 8.7% worldwide [5]. In Europe, the prevalence of early AMD increases from 3.5% in those aged 55–59 years to 17.9% in those aged 85 years; for late AMD, the prevalence increases from 0.1 to 9.8% [6]. For Asian AMD, the prevalence rates of early and late AMD were found to range from 1.4 to 17.3% and 0.1 to 7.3%, respectively, with a higher prevalence in elderly groups [7, 8]. Importantly, the number of people affected is expected to increase dramatically during the coming decades as Asian populations age, and it is estimated that there will be more Asians with AMD than in the rest of the world combined by 2040 [2].
AMD (both early and late) is common among non-Hispanic white and Hispanic populations, but is less common in non-Hispanic black populations; in contrast, late AMD is less common in Hispanics than in non-Hispanic whites [9–12]. These differences suggest that genetic factors could be important determinants of AMD. In recent years, large consortiums using genome-wide association studies (GWASs) have revealed 52 independently associated variants spanning 34 loci, including the complement factor H (CFH), complement component 3 (C3), ARMS2/HTRA1, and complement factor I (CFI) genes [13]. Additional meta-analyses have shown that several single nucleotide polymorphisms (SNPs) and variants located in extracellular matrix-related genes, such as COL15A1, COL8A1, MMP19, and CTRB2/CTRB1, were significantly associated with late AMD [13–15]. Thus, by combining these results and data from the GWAS catalog of the National Human Genome Research Institute-European Bioinformatics Institute (NHGRI-EBI), it may be possible to identify SNPs related to any AMD (i.e., both early and late) and to late AMD specifically.
Accordingly, in this study, we compiled a comprehensive set of SNPs associated AMD and assessed differences in allele frequencies in various populations by utilizing phase 3 data from the 1000 Genome Project [16], which identifies genetic variants among 26 worldwide populations. Because this database does not include Korean genome data, it was possible to assess allele frequency pattern-related AMD in Koreans using the Korean Reference Genome Database (KRGDB) [17], which consists of 1722 whole-genome sequencing data from healthy Koreans produced by the Korea National Institute of Health in 2016. The rationale for conducting this study was to apply whole-genome sequencing data of healthy subjects to aging-related disease, which suggested by previous researchers [18]. Hence, we aimed to gain insights into genetic causes of allele frequencies differences among races for SNPs related to AMD and to compare composite genetic risk scores and polygenetic risk scores using SNPs related to AMD and AMD prevalence for different ethnic groups, including Koreans.
Methods
Ethical considerations
This study was approved and monitored by the Institutional Review Board (IRB) of the Veterans Health Service Medical Center, Korea (IRB no. 2019–07-008).
Comparison of AMD-related SNPs in the global population and east Asians
AMD can be classified as early or late AMD. Early AMD is characterized by yellow subretinal deposits or retinal pigment epithelial irregularities, whereas late AMD is characterized by leakage of the choroidal neovascular membrane fluid or blood into the subretinal space or GA. “Any AMD” included both types of AMD. We searched the NHGRI-EBI GWAS catalog (https://www.ebi.ac.uk/gwas/home, December 2019) for SNPs that were associated with AMD traits (EFO_0001365). The catalog included 28 studies, which revealed 232 associations. After eliminating repetitive SNPs and removal of information not found in the 1000 Genome Projects database, 138 AMD-associated SNPs from the GWAS catalog were used for analysis of allele frequencies associated with any AMD (Supplementary Tables 1and 2).
Among SNPs associated with AMD-related traits, we determined AMD risk by examining beta-coefficients and odds ratios for the effect allele. We also read the text descriptions in the primary GWAS reports. The details and advantages of this method have been described elsewhere [19]. Briefly, the population-level allele frequencies of SNPs were derived based on the 1000 Genomes Project phase 3 (n = 2504) and KRGDB (n = 1722). The 1000 Genomes Project surveyed genetic variations among 2504 individuals from 26 worldwide populations, grouped into African, East Asian, European, South Asian, and American populations based on their geographical locations and ancestries [16]; the data were downloaded from ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/ (last accessed: January 15, 2020). The variant coordinates were based on the human genome assembly GRCh37. Because the East Asian data in the 1000 Genomes Project did not include data from Korean populations, we compared the data from five continents and East Asian countries in the 1000 Genome Projects with data extracted from KRGDB, which included whole-genome sequencing data for 1722 Korean individuals [17]. Data on the population frequencies of the SNPs were downloaded from the web-based database (http://152.99.75.168:9090/KRGDB/menuPages/download.jsp/, last accessed: January 15, 2020). For comparison of the distributions of individual risk alleles of the Korean population, individual genotyping results from the second phase of KRGDB (n = 1099) were obtained from the National Human Resource Bank of Korea.
Comparison of SNPs related to late AMD in the global population and east Asians
Late AMD in this study included CNV and GA. Although a previous study revealed the SNPs related to each type of late AMD, it is difficult to select only one phenotype because the SNPs are often related to both CNV and GA [13]. SNPs related to late AMD are shown in Table 1, and these data were obtained from overlapping SNPs from GWAS catalog data and a study by the International AMD Genomics Consortium [13, 14]. The population-level allele frequencies of these SNPs were derived as described above.
Table 1.
Effect allele frequencies (EAFs) of 31 late age-related macular degeneration related single nucleotide polymorphisms in continental groups, including Koreans
| SNP ID | Chr | Position | Function | Ref allele | Alt allele | Nearby/containing Gene | Global EAF | AMR EAF | AMR log10 P | AFR EAF | AFR log10 P | EAS EAF | EAS log10 P | SAS EAF | SAS log10 P | EUR EAF | EUR log10 P | KOR EAF | KOR log10 P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs10033900 | chr4 | 110,659,067 | intergenic | T | C | CFI | 0.49 | 0.53 | 1.06 | 0.66 | 27.44 | 0.39 | −8.01 | 0.31 | −23.92 | 0.54 | 2.30 | 0.34 | −43.57 |
| rs10781182 | chr9 | 76,617,720 | intergenic | T | G | MIR6130/RORB | 0.46 | 0.54 | 3.26 | 0.15 | −101.56 | 0.49 | 1.04 | 0.53 | 3.66 | 0.70 | 42.77 | 0.41 | −4.43 |
| rs10922109 | chr1 | 196,704,632 | intronic | C | A | CFH | 0.51 | 0.55 | 1.06 | 0.56 | 2.79 | 0.44 | −4.05 | 0.56 | 2.15 | 0.43 | −5.16 | 0.47 | −3.33 |
| rs11080055 | chr17 | 26,649,724 | intronic | A | C | TMEM97/VTN | 0.58 | 0.58 | 0.00 | 0.61 | 1.23 | 0.72 | 16.01 | 0.48 | −7.41 | 0.49 | −6.53 | 0.76 | 64.55 |
| rs1142 | chr7 | 104,756,326 | downstream | C | T | KMT2E/SRPK2 | 0.31 | 0.33 | 0.31 | 0.27 | −2.23 | 0.37 | 3.55 | 0.23 | −5.81 | 0.36 | 2.56 | 0.34 | 1.89 |
| rs11884770 | chr2 | 228,086,920 | ncRNA_intronic | T | C | COL4A3 | 0.67 | 0.72 | 1.62 | 0.49 | −31.04 | 0.78 | 11.41 | 0.73 | 3.30 | 0.71 | 1.76 | 0.76 | 20.03 |
| rs12019136 | chr19 | 5,835,677 | intronic | G | A | C3 | 0.15 | 0.05 | −14.85 | 0.32 | 39.45 | 0.14 | −0.34 | 0.14 | −0.31 | 0.04 | −24.78 | 0.08 | −23.98 |
| rs12357257 | chr10 | 24,999,593 | intronic | G | A | ARHGAP21 | 0.14 | 0.17 | 1.07 | 0.11 | −2.37 | 0.05 | −15.48 | 0.16 | 0.79 | 0.25 | 15.64 | 0.03 | −65.27 |
| rs13081855 | chr3 | 99,481,539 | ncRNA_intronic | G | T | COL8A1/FILIP1L | 0.07 | 0.06 | −0.31 | 0.03 | −4.92 | 0.05 | −1.23 | 0.10 | 3.36 | 0.10 | 3.01 | 0.03 | −15.11 |
| rs1626340 | chr9 | 101,923,372 | intergenic | G | A | TGFBR1/COL15A1 | 0.29 | 0.25 | −1.07 | 0.29 | 0.00 | 0.46 | 23.83 | 0.24 | −2.47 | 0.21 | −6.58 | 0.51 | 95.30 |
| rs181705462 | chr6 | 31,947,027 | intronic | G | T | C2/CFB/SKIV2L | 0.01 | 0.01 | 0.00 | 0.01 | 0.18 | 0.00 | −1.52 | 0.02 | 0.66 | 0.01 | 0.38 | 0.00 | −4.86 |
| rs2043085 | chr15 | 58,680,954 | intergenic | T | C | AQP9;LIPC | 0.49 | 0.62 | 8.95 | 0.36 | −16.28 | 0.50 | 0.23 | 0.44 | −2.15 | 0.63 | 15.08 | 0.48 | −0.30 |
| rs2230199 | chr19 | 6,718,387 | exonic | G | C | C3 | 0.09 | 0.10 | 0.31 | 0.03 | −11.70 | 0.00 | −30.53 | 0.10 | 0.66 | 0.22 | 28.35 | 0.00 | −97.67 |
| rs2740488 | chr9 | 107,661,742 | intronic | A | C | ABCA1 | 0.33 | 0.28 | −1.62 | 0.43 | 10.40 | 0.24 | −7.72 | 0.38 | 2.29 | 0.25 | −6.05 | 0.26 | −12.12 |
| rs3138141 | chr12 | 56,115,778 | ncRNA_intronic | C | A | RDH5/CD63/MMP19 | 0.15 | 0.11 | −1.86 | 0.01 | −61.21 | 0.12 | −1.76 | 0.38 | 53.57 | 0.19 | 2.57 | 0.07 | −32.36 |
| rs3750846 | chr10 | 124,215,565 | intronic | T | C | ARMS2/HTRA1 | 0.29 | 0.25 | −1.07 | 0.26 | −1.44 | 0.41 | 12.35 | 0.34 | 2.47 | 0.19 | −10.25 | 0.42 | 32.52 |
| rs429358 | chr19 | 45,411,941 | exonic | T | C | APOE | 0.15 | 0.10 | −2.81 | 0.27 | 21.51 | 0.09 | −7.38 | 0.09 | −6.61 | 0.16 | 0.37 | 0.09 | −14.75 |
| rs55975637 | chr3 | 99,419,853 | ncRNA_intronic | G | A | COL8A1 | 0.08 | 0.08 | −0.04 | 0.05 | −2.62 | 0.06 | −1.52 | 0.09 | 0.40 | 0.12 | 4.31 | 0.03 | −20.71 |
| rs570618 | chr1 | 196,657,064 | intronic | T | G | CFH | 0.78 | 0.78 | 0.00 | 0.81 | 1.71 | 0.95 | 43.09 | 0.71 | −4.95 | 0.64 | −18.54 | 0.92 | 72.90 |
| rs5754227 | chr22 | 33,105,817 | intronic | T | C | SYN3/TIMP3 | 0.37 | 0.39 | 0.31 | 0.56 | 33.60 | 0.61 | 42.65 | 0.12 | −57.77 | 0.13 | −53.79 | 0.67 | 167.56 |
| rs61818925 | chr1 | 196,815,450 | intergenic | T | G | CFHR1 | 0.59 | 0.52 | −2.60 | 0.72 | 17.39 | 0.41 | −24.37 | 0.69 | 7.77 | 0.58 | −0.26 | 0.50 | −15.15 |
| rs61985136 | chr14 | 68,769,199 | intronic | C | T | RAD51B | 0.45 | 0.60 | 11.79 | 0.13 | −110.82 | 0.45 | 0.00 | 0.63 | 23.38 | 0.58 | 12.87 | 0.42 | −2.45 |
| rs62247658 | chr3 | 64,715,155 | ncRNA_intronic | C | T | ADAMTS9-AS2 | 0.28 | 0.29 | 0.15 | 0.13 | −30.69 | 0.24 | −1.92 | 0.22 | −3.58 | 0.56 | 60.51 | 0.20 | −15.79 |
| rs6565597 | chr17 | 79,526,821 | intronic | C | T | NPLOC4/TSPAN10 | 0.24 | 0.23 | −0.15 | 0.16 | −9.47 | 0.21 | −1.28 | 0.26 | 0.66 | 0.36 | 13.46 | 0.21 | −2.48 |
| rs67538026 | chr19 | 1,031,438 | intronic | C | T | CNN2 | 0.38 | 0.33 | −1.54 | 0.15 | −60.48 | 0.52 | 15.07 | 0.49 | 8.86 | 0.45 | 4.28 | 0.50 | 24.75 |
| rs72802342 | chr16 | 75,234,872 | intergenic | C | A | CTRB2/CTRB1 | 0.06 | 0.06 | 0.00 | 0.01 | −21.05 | 0.13 | 11.80 | 0.04 | −1.35 | 0.08 | 1.56 | 0.11 | 15.95 |
| rs73036519 | chr19 | 45,748,362 | intergenic | G | C | EXOC3L2/MARK4 | 0.23 | 0.17 | −2.80 | 0.36 | 19.67 | 0.00 | −98.90 | 0.29 | 3.66 | 0.29 | 4.05 | 0.00 | − 284.04 |
| rs7803454 | chr7 | 99,991,548 | intronic | C | T | PILRB/PILRA | 0.09 | 0.11 | 1.00 | 0.03 | −16.28 | 0.03 | −11.57 | 0.11 | 1.42 | 0.19 | 18.37 | 0.02 | −37.14 |
| rs8135665 | chr22 | 38,476,276 | intronic | C | T | SLC16A8 | 0.25 | 0.31 | 2.43 | 0.37 | 16.33 | 0.14 | −14.06 | 0.20 | −2.76 | 0.22 | −1.32 | 0.10 | −65.11 |
| rs943080 | chr6 | 43,826,627 | intergenic | C | T | VEGFA | 0.67 | 0.52 | −12.42 | 0.83 | 30.61 | 0.73 | 3.60 | 0.67 | 0.00 | 0.51 | −20.22 | 0.67 | 0.14 |
| rs9564692 | chr13 | 31,821,240 | intronic | C | T | B3GALTL | 0.50 | 0.51 | 0.15 | 0.45 | −2.79 | 0.71 | 33.55 | 0.54 | 1.42 | 0.33 | −22.06 | 0.73 | 101.51 |
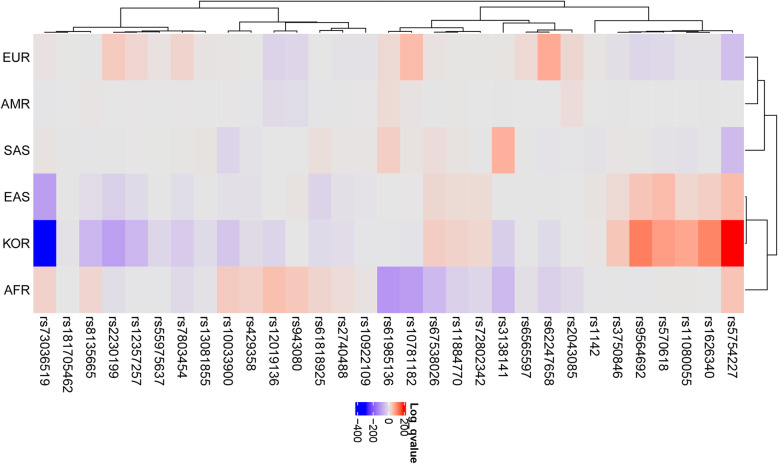
Chr chromosome, EAF effect allele frequency, ref. allele reference allele, alt allele alterative allele, AMR Americans, AFR Africans, EAS East Asians, SAS South Asians, EUR Europeans, KOR Koreans P-value: adjusted Fischer’s test assuming 138 hypotheses, statistical significance was set at P < 0.05
Calculation of genetic risk scores using SNPs related to any AMD and late AMD
To compare the composite genetic risk of AMD, we adopted the following equation described by Mao et al [19]:
where “I” refers to the number of AMD-related SNPs, and “Xi” refers to copies of risk alleles (Xi ∈ {0,1,2}) at the ith SNP. In one extreme case, if a person has two copies of the risk allele at each AMD-related SNP, then the person’s risk score was set as 1. In contrast, if a person has no copies of risk alleles at any AMD-related SNP, then the person’s risk score was set as 0. A person with a composite score of 1 had the highest possible genetic risk for AMD, whereas a person with a score of 0 had the lowest possible genetic risk. If copies of effect alleles (0/1/2) were randomly assigned to each SNP, the expected value of the risk score was set at 0.5. In addition, polygenic risk score (PRS) examine the cumulative effect of genetic variants on a disease or trait by aggregating the individual genetic effects into a single measurement [20]. To compare the PRS, we adopted the following equation:
Where "βi ” refers to an average odd ratio of the ith SNP reported in the GWAS studies.
SNPs with frequency differences of more than 10% between the total (n = 1722) and second phase (n = 1099) data of KRGDB were excluded from the genetic score calculation. We used the average composite genetic risk scores for populations to determine correlations with country-wise AMD prevalence data. For subgroup analysis of late AMD, the relationships between the composite genetic risk score using late AMD-related SNPs and continental-wise late AMD prevalence data were analyzed. Because the prevalence of AMD is affected by age, the prevalence was analyzed for individuals ages greater than or equal to 40 years old or greater than or equal to 65 years old. The prevalence data for any AMD and late AMD were evaluated based in the results of studies on Europeans [2, 6, 10, 21], East Asians [2, 7, 21, 22], Americans [2, 10] and Koreans [8, 23, 24]. Studies on prevalence among Japanese people were sufficient; thus, the prevalence data for Japanese populations were used to represent East Asians. Data on the prevalence of AMD in Africans and South Asians were limited; therefore, these populations were not included in this analysis.
Data analyses
We used Fisher’s exact test to assess whether the effect allele at a given SNP was significantly enriched or depleted compared with the global population frequency in the 1000 Genomes Project database. Since we compared 138 SNPs, we calculated adjusted P values assuming 138 hypotheses and the P values were first log10-transformed. In the heatmap generated to visualize allele enrichment or depletion patterns in different populations, red and purple colors indicated higher and lower frequencies than the global average, respectively. If the effect allele of an SNP was enriched in a population, then the negative of log10 of the enrichment P value (a positive number) was used to represent the SNP associated with that population in the heatmap. In contrast, if the allele of an SNP was depleted in a population, the value of log10 of the depletion P value (a negative number) was used to represent the SNP for that population in the heatmap. Statistical analyses were performed using R software version 3.6.0 (R Foundation, Vienna, Austria). Statistical significance was set at P < 0.05.
Results
Patterns of any AMD-related SNPs in the global population and in east Asians
We collected 232 AMD-associated SNPs from 24 GWASs from the NHGRI-EBI GWAS catalog. The SNPs were identified in 17 European populations, four East Asian populations, and three South Asian populations; three studies were performed in mixed ethnic populations. Clearly, populations other than Europeans were understudied, particularly African and American populations. However, there were no significant differences in the SNP frequencies among these populations (Fig. 1). This result suggested that many SNPs found in Europeans were also applicable to other populations. After collecting 138 AMD-associated SNPs, we determined the effect allele frequencies (EAFs) for each of the continental groups and for Koreans based on genotype information from the 1000 Genomes Project and KRGDB (Supplementary Table S1). Heatmap analysis showed how significantly the effect allele was enriched or depleted across the Korean and continental groups (Supplementary Fig. 1). For Koreans, 55 AMD-related SNPs were significantly enriched, 74 SNPs were depleted, and nine SNPs were similar to the global EAFs. Additionally, heatmap analysis enabled visualization of the proportion of effect alleles in each continental group compared with the global average. For example, rs5754227 (SYN3), rs1626340 (TGFBR1/COL15A1), rs3750846 (ARMS2/HTRA1), and rs9564692 (B3GALTL) were enriched, whereas rs2230199 (C3) and rs73036519 (EXOC3L2/MARK4) were depleted in Koreans. The hierarchical clustering tree showed differences among the populations; Europeans, Americans, and South Asians were in one cluster, whereas Africans, East Asians, and Koreans were in another cluster.
Fig. 1.

Comparison of the frequency of age-related macular degeneration (AMD)-related single nucleotide polymorphisms (SNPs) according to super population. AMR: American, EUR: European, SAS: South Asian, AFR: African, EAS: East Asian, KOR: Korean
Next, we compared the EAFs of East Asians and Koreans (Supplementary Table S2and Fig. 2). Although the EAF did not differ much among East Asians, 21 SNPs were enriched, 30 SNPs were depleted, and 87 SNPs were similar to those in Koreans. Moreover, the heatmap clearly showed that the main allele frequency pattern in East Asians in the 1000 Genomes Project was similar to that in Koreans; in contrast, few alleles showed the opposite allele frequency patterns between East Asians and Koreans.
Fig. 2.
Heatmap generated using single nucleotide polymorphisms related to late age-related macular degeneration in the global population. Each row shows an SNP, and each column shows a continent. Red color indicates that the effect allele is enriched, whereas purple color indicates that the effect allele is depleted. AMR: American, EUR: European, SAS: South Asian, AFR: African, EAS: East Asian, KOR: Korean
Patterns of late AMD-related SNPs in the global population and in east Asians
Thirty-one SNPs related to late AMD were selected from 138 AMD-related SNPs (Table 1), and a heatmap showing how significantly the EAF was enriched or depleted across the Koreans and continental groups (Fig. 2) with a log scale among 31 late AMD-related SNPs was generated. For Koreans, 10 AMD-related SNPs were significantly enriched, 19 SNPs were depleted, and two SNPs were similar to global EAFs. For example, rs11080055, located in the intronic region of the TMEM97 gene, which encodes transmembrane protein 97, had A > C alleles; the C allele was detected in 49% of Europeans and 76% of Koreans. Additionally, rs1626340, located in the intergenic region of TGFBR1/COL15A1, which encodes collagen type XV alpha 1 chain, had G > A alleles; the A allele was detected in 21% of Europeans and 51% of Koreans. This SNP has been reported to be associated with various diseases, including colon cancer [25, 26]. rs5754227, located in the intronic region of SYN3 which encodes synapsin-3 (a neuronal phosphoprotein associated with the cytoplasmic surface of synaptic vesicles), had T > C alleles; the C allele frequencies were 13% for Europeans and 67% for Koreans. Some SNPs were depleted in Koreans. For example, rs73036519, located in the intergenic region of the EXOC3L2/MARK4 gene, which encodes a member of the microtubule affinity-regulating kinase family, had G > C alleles; the C allele frequencies were 29% for Europeans and 0% for Koreans. Hierarchical clustering tree analysis showed differences among the populations; Europeans, Americans, and South Asians were in one cluster, whereas Africans, East Asians, and Koreans were in another cluster (Fig. 2).
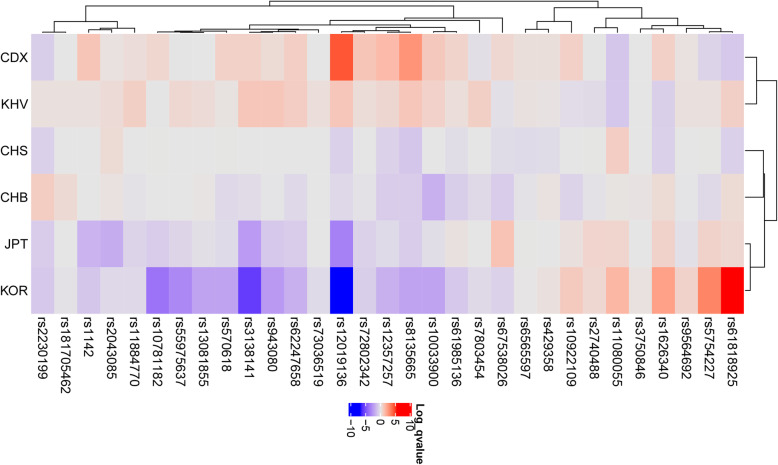
In addition, heatmap analysis showed differences in EAFs across Koreans and East Asians (Table 2, Fig. 3). The rs61818925, located in the intergenic region of the CFHR1 gene, which encodes complement factor H, had T > G alleles; the G allele frequencies were 33% for Chinese populations and 50% for Koreans. Notably, some SNPs were depleted in Koreans; for example, rs12019136, located in the intronic region of the C3 gene, which encodes complement C3, had G > A alleles; the A allele frequencies were 21% for Vietnamese populations and 8% for Koreans. However, the EAFs did not differ substantially among East Asians; for Koreans, four SNPs were enriched, 11 SNPs were depleted, and 16 SNPs were similar to those in global East Asian populations.
Table 2.
Effect allele frequencies (EAFs) of 31 late age-related macular degeneration related single nucleotide polymorphisms in East Asian groups including Koreans
| SNP ID | Chr | Position | Function | Ref allele | Alt allele |
Nearby/containing Gene | Global East Asian EAF | CHS EAF | CHS log10 P | CDX EAF | CDX log10 P | KHV EAF | KHV log10 P | CHB EAF | CHB log10 P | JPT EAF | JPT log10 P | KOR EAF | KOR log10 P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs10033900 | chr4 | 110,659,067 | intergenic | T | C | CFI | 0.39 | 0.39 | 0.00 | 0.45 | 0.92 | 0.43 | 0.65 | 0.31 | −1.72 | 0.37 | −0.24 | 0.34 | −2.17 |
| rs10781182 | chr9 | 76,617,720 | intergenic | T | G | MIR6130/RORB | 0.49 | 0.51 | 0.01 | 0.54 | 0.50 | 0.49 | 0.02 | 0.50 | 0.00 | 0.41 | −0.81 | 0.41 | −3.69 |
| rs10922109 | chr1 | 196,704,632 | intronic | C | A | CFH | 0.44 | 0.46 | 0.00 | 0.51 | 0.67 | 0.40 | −0.27 | 0.38 | −0.57 | 0.47 | 0.19 | 0.47 | 0.89 |
| rs11080055 | chr17 | 26,649,724 | intronic | A | C | TMEM97/VTN | 0.72 | 0.80 | 0.77 | 0.63 | −1.03 | 0.63 | −0.99 | 0.74 | 0.08 | 0.77 | 0.54 | 0.76 | 1.50 |
| rs1142 | chr7 | 104,756,326 | downstream | C | T | KMT2E/SRPK2 | 0.37 | 0.35 | 0.00 | 0.47 | 1.03 | 0.39 | 0.20 | 0.37 | 0.00 | 0.26 | −1.61 | 0.34 | −1.03 |
| rs11884770 | chr2 | 228,086,920 | ncRNA_intronic | T | C | COL4A3 | 0.78 | 0.78 | 0.00 | 0.81 | 0.28 | 0.84 | 0.72 | 0.76 | −0.13 | 0.73 | −0.56 | 0.76 | −0.43 |
| rs12019136 | chr19 | 5,835,677 | intronic | G | A | C3 | 0.14 | 0.08 | −0.69 | 0.29 | 4.20 | 0.21 | 0.99 | 0.10 | −0.42 | 0.04 | −3.29 | 0.08 | −6.61 |
| rs12357257 | chr10 | 24,999,593 | intronic | G | A | ARHGAP21 | 0.05 | 0.02 | −0.65 | 0.11 | 1.37 | 0.08 | 0.47 | 0.02 | −0.79 | 0.03 | −0.35 | 0.03 | −1.70 |
| rs13081855 | chr3 | 99,481,539 | ncRNA_intronic | G | T | COL8A1/FILIP1L | 0.05 | 0.04 | −0.01 | 0.05 | 0.00 | 0.07 | 0.35 | 0.05 | 0.06 | 0.03 | −0.22 | 0.03 | −2.17 |
| rs1626340 | chr9 | 101,923,372 | intergenic | G | A | TGFBR1/COL15A1 | 0.46 | 0.38 | −0.69 | 0.53 | 0.67 | 0.38 | −0.72 | 0.50 | 0.37 | 0.52 | 0.60 | 0.51 | 2.17 |
| rs181705462 | chr6 | 31,947,027 | intronic | G | T | C2/CFB/SKIV2L | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.20 | 0.01 | 0.40 | 0.00 | 0.00 | 0.00 | −0.13 |
| rs2043085 | chr15 | 58,680,954 | intergenic | T | C | AQP9;LIPC | 0.50 | 0.55 | 0.35 | 0.52 | 0.13 | 0.55 | 0.35 | 0.52 | 0.13 | 0.38 | −1.85 | 0.48 | −0.43 |
| rs2230199 | chr19 | 6,718,387 | exonic | G | C | C3 | 0.00 | 0.00 | −0.67 | 0.00 | −0.75 | 0.01 | 0.20 | 0.01 | 0.79 | 0.00 | −0.81 | 0.00 | − 0.93 |
| rs2740488 | chr9 | 107,661,742 | intronic | A | C | ABCA1 | 0.24 | 0.26 | 0.01 | 0.25 | 0.02 | 0.20 | −0.35 | 0.22 | −0.13 | 0.29 | 0.55 | 0.26 | 0.45 |
| rs3138141 | chr12 | 56,115,778 | ncRNA_intronic | C | A | RDH5/CD63/MMP19 | 0.12 | 0.12 | 0.00 | 0.16 | 0.66 | 0.19 | 0.99 | 0.09 | −0.33 | 0.04 | −2.42 | 0.07 | −5.43 |
| rs3750846 | chr10 | 124,215,565 | intronic | T | C | ARMS2/HTRA1 | 0.41 | 0.39 | −0.01 | 0.40 | 0.00 | 0.41 | 0.00 | 0.43 | 0.08 | 0.40 | 0.00 | 0.42 | 0.14 |
| rs429358 | chr19 | 45,411,941 | exonic | T | C | APOE | 0.09 | 0.06 | −0.29 | 0.10 | 0.22 | 0.09 | 0.08 | 0.10 | 0.13 | 0.08 | 0.00 | 0.09 | 0.18 |
| rs55975637 | chr3 | 99,419,853 | ncRNA_intronic | G | A | COL8A1 | 0.06 | 0.06 | 0.00 | 0.06 | 0.02 | 0.09 | 0.45 | 0.05 | 0.00 | 0.03 | −0.53 | 0.03 | −3.03 |
| rs570618 | chr1 | 196,657,064 | intronic | T | G | CFH | 0.95 | 0.96 | 0.01 | 0.98 | 0.66 | 0.96 | 0.11 | 0.93 | −0.42 | 0.93 | −0.35 | 0.92 | −2.17 |
| rs5754227 | chr22 | 33,105,817 | intronic | T | C | SYN3/TIMP3 | 0.61 | 0.63 | 0.01 | 0.55 | −0.57 | 0.65 | 0.21 | 0.56 | −0.40 | 0.67 | 0.62 | 0.67 | 2.97 |
| rs61818925 | chr1 | 196,815,450 | intergenic | T | G | CFHR1 | 0.41 | 0.33 | −0.69 | 0.32 | −0.92 | 0.48 | 0.70 | 0.46 | 0.37 | 0.46 | 0.46 | 0.50 | 5.24 |
| rs61985136 | chr14 | 68,769,199 | intronic | C | T | RAD51B | 0.45 | 0.40 | −0.26 | 0.51 | 0.59 | 0.49 | 0.27 | 0.38 | −0.75 | 0.47 | 0.16 | 0.42 | −0.93 |
| rs62247658 | chr3 | 64,715,155 | ncRNA_intronic | C | T | ADAMTS9-AS2 | 0.24 | 0.25 | 0.00 | 0.31 | 0.75 | 0.31 | 0.78 | 0.19 | −0.42 | 0.17 | − 0.81 | 0.20 | −1.70 |
| rs6565597 | chr17 | 79,526,821 | intronic | C | T | NPLOC4/TSPAN10 | 0.21 | 0.17 | −0.36 | 0.24 | 0.24 | 0.23 | 0.15 | 0.18 | −0.15 | 0.22 | 0.01 | 0.21 | 0.03 |
| rs67538026 | chr19 | 1,031,438 | intronic | C | T | CNN2 | 0.52 | 0.47 | −0.29 | 0.57 | 0.46 | 0.49 | −0.20 | 0.44 | −0.79 | 0.62 | 1.12 | 0.50 | −0.64 |
| rs72802342 | chr16 | 75,234,872 | intergenic | C | A | CTRB2/CTRB1 | 0.13 | 0.12 | 0.00 | 0.20 | 1.03 | 0.16 | 0.30 | 0.11 | −0.13 | 0.09 | −0.65 | 0.11 | −0.75 |
| rs73036519 | chr19 | 45,748,362 | intergenic | G | C | EXOC3L2/MARK4 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.27 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | −0.35 |
| rs7803454 | chr7 | 99,991,548 | intronic | C | T | PILRB/PILRA | 0.03 | 0.03 | 0.00 | 0.02 | −0.23 | 0.06 | 0.72 | 0.01 | −0.40 | 0.03 | 0.00 | 0.02 | −0.40 |
| rs8135665 | chr22 | 38,476,276 | intronic | C | T | SLC16A8 | 0.14 | 0.09 | −1.02 | 0.23 | 2.55 | 0.19 | 0.99 | 0.10 | −0.79 | 0.10 | −0.81 | 0.10 | −2.17 |
| rs943080 | chr6 | 43,826,627 | intergenic | C | T | VEGFA | 0.73 | 0.73 | 0.00 | 0.76 | 0.37 | 0.79 | 0.99 | 0.72 | −0.06 | 0.67 | −0.92 | 0.67 | −2.46 |
| rs9564692 | chr13 | 31,821,240 | intronic | C | T | B3GALTL | 0.71 | 0.71 | 0.00 | 0.73 | 0.14 | 0.73 | 0.20 | 0.70 | −0.06 | 0.69 | −0.20 | 0.73 | 0.63 |
CHS Southern Han Chinese, China, CDX Chinese Dai in Xishuangbanna, China, KHV Kinh in Ho Chi Minh City, Vietnam, CHB: Han Chinese in Beijing, China, JPT Japanese in Tokyo in 1000 genome project, KOR Korean Reference Genome data base, Chr chromosome, EAF effect allele frequency, ref. allele reference allele, alt allele alterative allele, P-value: adjusted Fischer’s test assuming 138 hypotheses, statistical significance was set at P < 0.05
Fig. 3.
Heatmap generated using single nucleotide polymorphisms related to late age-related macular degeneration in the East Asian population. Each row shows an SNP, and each column shows a continent. Red color indicates that the effect allele is enriched, whereas purple color indicates that the effect allele is depleted. CDX: Chinese Dai in Xishuangbanna; CHB: Han Chinese in Beijing, China; CHS: Southern Han Chinese, China; JPT: Japanese in Tokyo, Japan; KOR: Korean in the Republic of Korea; KHV: Kinh in Ho Chi Minh City, Vietnam
Genetic risk scores calculated using SNPs related to any AMD and late AMD
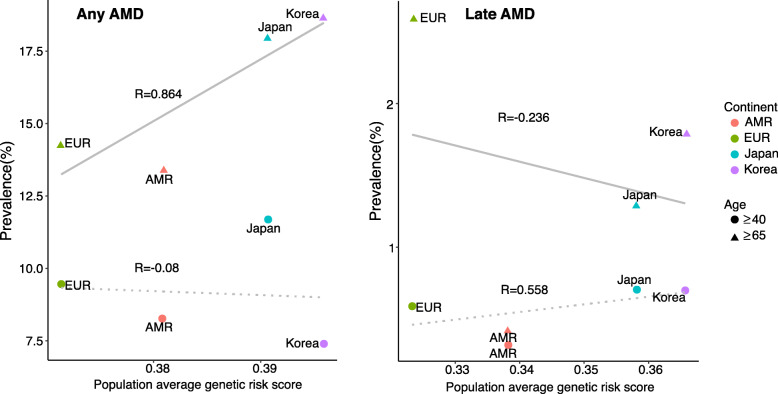
Next, we calculated the composite genetic risk scores based on copies of effect alleles at AMD-associated SNPs, with the assumption that allelic associations from most GWAS-identified variants could be replicated in non-European populations [27]. The genetic risk score of any AMD was highest in East Asians, followed by South Asians, Americans, and Europeans (Fig. 4). The prevalence of any AMD in individuals 65 years old or older was correlated with the population average genetic risk score (R = 0.864; Fig. 5). In addition, the genetic risk score of late AMD was highest in East Asians, followed by Europeans, Americans, and South Asians (Fig. 4). However, differences in genetic risk scores of late AMD were not greater than those of any AMD, as supported by meta-analysis data indicating that the prevalence of late AMD was similar among Europeans and Asians [7]. The prevalence of late AMD in individuals 40 years old or older was correlated with the population average genetic risk score (R = 0.558; Fig. 5). In addition, the PRS of any AMD or late AMD in East Asians was similar among Europeans and Asians (Supplementary Fig. S3). The prevalence of any AMD in individuals 65 years old or older was correlated with the population PRS (R = 0.846; Supplementary Fig. S3) and the prevalence of late AMD in individuals 40 years old or older was correlated with the population PRS (R = 0.572; Supplementary Fig. S3).
Fig. 4.
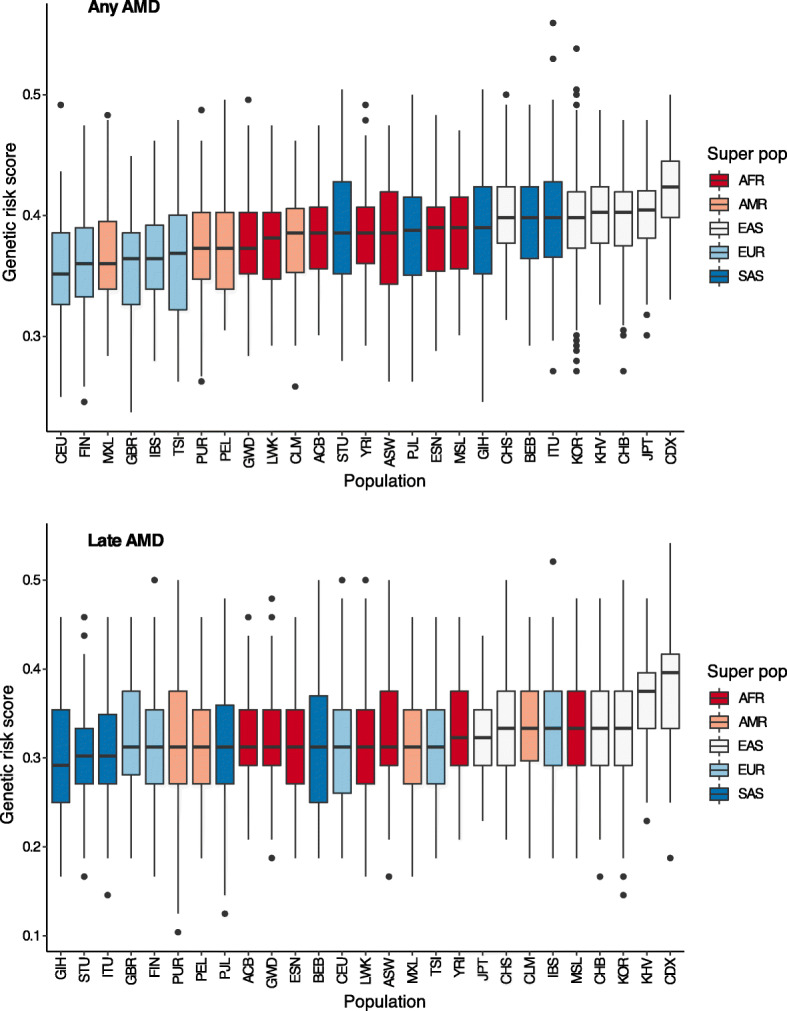
Genetic risk score calculations of age-related macular degeneration for any AMD or late AMD using related single nucleotide polymorphisms. ACB: African Caribbean in Barbados; ASW: African ancestry in the Southwest USA; BEB: Bengali in Bangladesh; CDX: Chinese Dai in Xishuangbanna; CEU: Utah residents with Northern and Western European ancestry; CHB: Han Chinese in Beijing, China; CHS: Southern Han Chinese, China; CLM: Colombian in Medellin, Colombia; ESN: Esan in Nigeria; FIN: Finnish in Finland; GBR: British in England and Scotland; GIH: Gujarati Indian in Houston, TX, USA; GWD: Gambian in Western Division, Gambia; IBS: Iberian populations in Spain; ITU: Indian Telugu in the UK; JPT: Japanese in Tokyo, Japan; KOR: Korean in the Republic of Korea; KHV: Kinh in Ho Chi Minh City, Vietnam; LWK: Luhya in Webuye, Kenya; MSL: Mende in Sierra Leone; MXL: Mexican ancestry in Los Angeles, CA, USA; PEL: Peruvian in Lima, Peru; PJL: Punjabi in Lahore, Pakistan; PUR: Puerto Rican in Puerto Rico; STU: Sri Lankan Tamil in the UK; TSI: Toscani in Italy; YRI: Yoruba in Ibadan, Nigeria
Fig. 5.
Correlation plots of the prevalence of age-related macular degeneration (any or late) and genetic risk score using related single nucleotide polymorphisms. The graphs on the left and right indicate correlations of prevalence and genetic risk scores with any AMD and late AMD, respectively. The dotted line indicates the relationship between genetic risk score and prevalence (circular shape) for people with AMD who are 40 years old or older, and the solid line indicates the relationship between genetic risk score and prevalence of AMD (triangular shape) for people 65 years of age or older. AMR: American, EUR: European
Discussion
The etiology of AMD is not fully understood; however, several hypotheses have demonstrated the involvement of genetics, inflammation, complement, lipids, and oxidative stress [28]; aging and family history are known risk factors for AMD [29]. Since the discovery of the CFH gene as a major AMD susceptibility gene in 2005, several studies using GWASs have reported various genetic risk loci [30–34]. Complement-related genes, such as complement H (CFH), complement factor I (CFI), and complement components C2, C3, and C9, are related to the pathogenesis of AMD [30]. ARMS2/HTRA1 genes (rs10490924 and rs11200638) are risk factors for late AMD according to results of a Korean study [35]. In our study, we found that these SNPs showed higher prevalence rates in Koreans than in other races. Importantly, because the affected allele frequencies were high in their study, there were no problems with interpreting significance. The multi-hit threshold model of AMD pathogenesis [30] suggests that genes related to the complement pathway, immune response, lipid transport, extracellular matrix remodeling, angiogenesis, and cell survival are related to biologically relevant pathways. Therefore, race is expected to be one of important factor affecting AMD prevalence.
AMD prevalence differs among racial and ethnic groups [36]. According to a study by Wong and colleagues, early AMD occurs more frequently in individuals of European ancestry (11.2%) than in Asian individuals (6.8%), and any AMD was more common in populations of European ancestry (12.3%) than in those of Asian ancestry (7.4%) [2]. The occurrence of any AMD was markedly less common in populations of African ancestry [2, 21]. In a meta-analysis of AMD in Asians, the age-specific prevalence of late AMD in Asians was found to be comparable with that in Europeans; however, early AMD was found to be less common among Asians (6.8%) than among Europeans (8.8%) [7]. In our study, genetic risk scores of AMD were higher in Asians than in Europeans. Accordingly, our findings showed that the prevalence of AMD may be increased owing to genetic predisposition in East Asians, including Koreans. However, environmental factors, such as lifespan and eating habits, as well as differences in the penetrance of AMD-related genes may explain these results. For example, a study of Japanese immigrants in Brazil showed that the prevalence rates for early and late AMD were 13.4 and 1.3%, respectively, suggesting that genetic predisposition was affected by environmental factors [37]. In the Korea National Health and Nutrition Examination Survey, the prevalence rates of AMD were found to be 6.62% for any AMD, 6.02% for early AMD, and 0.6% for late AMD, similar to the prevalence rate of pooled Asian and European populations [23, 24]. In addition, according to a study of the projected number of people with AMD by region [2], global projected cases of any AMD in 2040 was the highest for Asian populations.
In a previous study, Fritsche and colleagues showed that the ARMS2/HTRA1 gene plays a larger role in Asian populations than in European populations based on an analysis of the effects of allele frequencies of known AMD risk variants from 1000 Genomes data [30]. Thus, differences in genetic risk scores in Asians may favor late AMD, which could explain in prevalence of late AMD in Asian populations [7]. Any AMD was more prevalent in European populations than in Asian populations; however, Asian individuals may be more likely to develop late AMD than white individuals [36], consistent with the results of our study. Indeed, in our study, the country-wise AMD prevalence and genetic risk scores showed positive or negative correlations according age groups, suggesting that aging and environmental risk factors, such as smoking, may also play substantial roles in the development of AMD.
A major strength of our study was the inclusion of the large Korean whole-genome dataset (n = 1722), which reflected the allele frequency of SNPs related to any stage of AMD and to late AMD. Additionally, we did not systematically organize the new AMD cohort and analyze the effects; instead, we compared the 1000 Genomes Project data with AMD-related SNP data from the GWAS catalog. However, there were a few limitations to this study. First, the GWAS catalog contained data for which the risk allele was not clearly defined according to the minor allele frequency (MAF). However, we did not exclude these from in our study because the majority of MAFs were likely to be risk alleles; thus, removing all of the undefined alleles could result in inaccurate subgroup analysis. Further studies are needed for data curation of 50 undefined SNPs. To solve this problem, risk allele curation is necessary for the GWAS catalog based on the results of additional large population studies using AMD cohorts. Second, the statistical significance of EAF in Koreans was high and should be interpreted with caution because the Korean reference genome number was 1722 (whole genome data was 1099), which was very high; the genome number of populations belonging to the 1000 Genomes Project was approximately 100 (range: 61–113). In addition, there is a possibility that the difference in sequencing depth (30X in KRGDB vs. 4X in 1000 Genome Project) affects coverage. However, the authors expect fewer missing SNPs since the population frequency can be accurately found up to 1% through the combined calling method in the 1000 Genome Projects and the microarray consist of common SNPs. Third, our study analyzed data based on current knowledge; we did not detect new genetic loci or perform pathway analysis using cell culture and animal studies. Additionally, our findings should be interpreted while considering the fact that penetrance is variable, even when causative SNPs are present in specific individuals. Nevertheless, our results are expected to improve our understanding of the genetic etiology of AMD for ophthalmologists.
Conclusion
Our study showed substantial population differentiation in allele frequencies for SNPs related to any AMD and late AMD. Based on the allele frequencies of these SNPs, the composite risk scores for AMD and late AMD for 26 ethnic groups in the 1000 Genomes Project and Koreans showed that East Asians, including Koreans, had a higher risk than Europeans for any AMD and late AMD. Finally, we observed differences in allele frequencies associated with SNPs related to AMD between Koreans and other races, which may explain the increased prevalence of AMD, predominantly in East Asians.
Supplementary Information
Additional file 1 Supplemental Figure 1 Heatmap generated using single nucleotide polymorphisms related to age-related macular degeneration in the global population. Each row shows an SNP, and each column shows a continent. Red color indicates that the effect allele is enriched, whereas purple color indicates that the effect allele is depleted. AMR: American, EUR: European, SAS: South Asian, AFR: African, EAS: East Asian, KOR: Korean.
Additional file 2 Supplemental Figure 2 Heatmap generated using single nucleotide polymorphisms related to age-related macular degeneration in East Asian populations. Each row shows an SNP, and each column shows a country. Red color indicates that the effect allele is enriched, whereas purple color indicates that the effect allele is depleted. CDX: Chinese Dai in Xishuangbanna; CHB: Han Chinese in Beijing, China; CHS: Southern Han Chinese, China; JPT: Japanese in Tokyo, Japan; KOR: Korean in the Republic of Korea; KHV: Kinh in Ho Chi Minh City, Vietnam.
Additional file 3 Supplemental Figure 3 Polygenic risk score calculations of age-related macular degeneration for any AMD or late AMD using related single nucleotide polymorphisms and correlation plots of the prevalence of age-related macular degeneration (any or late) and polygenic risk score
Additional file 4 Supplemental Table 1 Effect allele frequencies (EAFs) of any age-related macular degeneration related single nucleotide polymorphisms in continental groups, including Koreans.
Additional file 5 Supplemental Table 2 Effect allele frequencies (EAFs) of any age-related macular degeneration-related single nucleotide polymorphisms in East Asian groups, including Koreans.
Acknowledgments
This study was conducted with bio resources from the National Biobank of Korea, the Center for Disease Control and Prevention, Republic of Korea (KBN-2019-053).
Ethical approval
This study was approved and monitored by the IRB of the Veterans Health Service Medical Center, Korea (IRB no. 2019–07-008).
Availability of data and material
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AMD
Age-related macular degeneration
- CNV
Choroidal neovascularization
- C3
Complement component 3
- CFH
Complement factor H
- CFI
Complement factor I
- EAFs
Effect allele frequencies
- GWASs
Genome-wide association studies
- GA
Geographic atrophy
- IRB
Institutional Review Board
- KRGDB
Korean Reference Genome Database
- MAF
Minor allele frequency
- NHGRI-EBI
National Human Genome Research Institute-European Bioinformatics Institute
- PRS
Polygenic risk score
- SNPs
Single nucleotide polymorphisms
Authors’ contributions
HTS collected data, analysis and reviewed/edited the manuscript, BWY researched data collection and were involved in the analysis and interpretation of data, JHS made study design, reviewed data collection, and wrote the manuscript. All the authors approved the final version of the manuscript to be published. JHS is the guarantor of this work. The authors read and approved the final manuscript.
Funding
This study was supported by a VHS Medical Center Research Grant (grant no.: VHSMC19033). The sponsor or funding organization had no role in the design or conduct of this study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hyun-Tae Shin and Byung Woo Yoon contributed equally to this work.
References
- 1.Lim LS, Mitchell P, Seddon JM, Holz FG, Wong TY. Age-related macular degeneration. Lancet. 2012;379(9827):1728–1738. doi: 10.1016/S0140-6736(12)60282-7. [DOI] [PubMed] [Google Scholar]
- 2.Wong WL, Su X, Li X, Cheung CM, Klein R, Cheng CY, Wong TY. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Health. 2014;2(2):e106–e116. doi: 10.1016/S2214-109X(13)70145-1. [DOI] [PubMed] [Google Scholar]
- 3.Gottlieb JL. Age-related macular degeneration. JAMA. 2002;288(18):2233–2236. doi: 10.1001/jama.288.18.2233. [DOI] [PubMed] [Google Scholar]
- 4.de Jong PT. Age-related macular degeneration. N Engl J Med. 2006;355(14):1474–1485. doi: 10.1056/NEJMra062326. [DOI] [PubMed] [Google Scholar]
- 5.Wong TY, Chakravarthy U, Klein R, Mitchell P, Zlateva G, Buggage R, Fahrbach K, Probst C, Sledge I. The natural history and prognosis of neovascular age-related macular degeneration: a systematic review of the literature and meta-analysis. Ophthalmology. 2008;115(1):116–126. doi: 10.1016/j.ophtha.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Colijn JM, Buitendijk GHS, Prokofyeva E, Alves D, Cachulo ML, Khawaja AP, Cougnard-Gregoire A, Merle BMJ, Korb C, Erke MG, et al. Prevalence of age-related macular degeneration in Europe: the past and the future. Ophthalmology. 2017;124(12):1753–1763. doi: 10.1016/j.ophtha.2017.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawasaki R, Yasuda M, Song SJ, Chen SJ, Jonas JB, Wang JJ, Mitchell P, Wong TY. The prevalence of age-related macular degeneration in Asians: a systematic review and meta-analysis. Ophthalmology. 2010;117(5):921–927. doi: 10.1016/j.ophtha.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Park SJ, Kwon KE, Choi NK, Park KH, Woo SJ: Prevalence and incidence of exudative age-related macular degeneration in South Korea: a Nationwide population-based study. Ophthalmology 2015, 122(10):2063–2070 e2061. [DOI] [PubMed]
- 9.Klein R, Rowland ML, Harris MI. Racial/ethnic differences in age-related maculopathy. Third National Health and nutrition examination survey. Ophthalmology. 1995;102(3):371–381. doi: 10.1016/S0161-6420(95)31012-3. [DOI] [PubMed] [Google Scholar]
- 10.Klein R, Chou CF, Klein BE, Zhang X, Meuer SM, Saaddine JB. Prevalence of age-related macular degeneration in the US population. Arch Ophthalmol. 2011;129(1):75–80. doi: 10.1001/archophthalmol.2010.318. [DOI] [PubMed] [Google Scholar]
- 11.Sommer A, Tielsch JM, Katz J, Quigley HA, Gottsch JD, Javitt JC, Martone JF, Royall RM, Witt KA, Ezrine S. Racial differences in the cause-specific prevalence of blindness in East Baltimore. N Engl J Med. 1991;325(20):1412–1417. doi: 10.1056/NEJM199111143252004. [DOI] [PubMed] [Google Scholar]
- 12.Varma R, Fraser-Bell S, Tan S, Klein R, Azen SP. Los Angeles Latino eye study G: prevalence of age-related macular degeneration in Latinos: the Los Angeles Latino eye study. Ophthalmology. 2004;111(7):1288–1297. doi: 10.1016/j.ophtha.2004.01.023. [DOI] [PubMed] [Google Scholar]
- 13.Fritsche LG, Igl W, Bailey JN, Grassmann F, Sengupta S, Bragg-Gresham JL, Burdon KP, Hebbring SJ, Wen C, Gorski M, et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48(2):134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeAngelis MM, Owen LA, Morrison MA, Morgan DJ, Li M, Shakoor A, Vitale A, Iyengar S, Stambolian D, Kim IK, et al. Genetics of age-related macular degeneration (AMD) Hum Mol Genet. 2017;26(R1):R45–R50. doi: 10.1093/hmg/ddx228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Domalpally A, Agron E, Pak JW, Keenan TD, Ferris FL, 3rd, Clemons TE, Chew EY. Prevalence, risk, and genetic Association of Reticular Pseudodrusen in age-related macular degeneration: age-related eye disease study 2 report 21. Ophthalmology. 2019;126(12):1659–1666. doi: 10.1016/j.ophtha.2019.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Genomes Project C. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J, Weber JA, Jho S, Jang J, Jun J, Cho YS, Kim HM, Kim H, Kim Y, Chung O, et al. KoVariome: Korean National Standard Reference Variome database of whole genomes with comprehensive SNV, indel, CNV, and SV analyses. Sci Rep. 2018;8(1):5677. doi: 10.1038/s41598-018-23837-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinese M, Lacaze P, Rath EM, Stone A, Brion MJ, Ameur A, Nagpal S, Puttick C, Husson S, Degrave D, et al. The medical genome reference Bank contains whole genome and phenotype data of 2570 healthy elderly. Nat Commun. 2020;11(1):435. doi: 10.1038/s41467-019-14079-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mao L, Fang Y, Campbell M, Southerland WM. Population differentiation in allele frequencies of obesity-associated SNPs. BMC Genomics. 2017;18(1):861. doi: 10.1186/s12864-017-4262-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domingue BW, Belsky DW, Harris KM, Smolen A, McQueen MB, Boardman JD. Polygenic risk predicts obesity in both white and black young adults. PLoS One. 2014;9(7):e101596. doi: 10.1371/journal.pone.0101596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jonas JB, Cheung CMG, Panda-Jonas S. Updates on the epidemiology of age-related macular degeneration. Asia Pac J Ophthalmol (Phila) 2017;6(6):493–497. doi: 10.22608/APO.2017251. [DOI] [PubMed] [Google Scholar]
- 22.Wong CW, Yanagi Y, Lee WK, Ogura Y, Yeo I, Wong TY, Cheung CMG. Age-related macular degeneration and polypoidal choroidal vasculopathy in Asians. Prog Retin Eye Res. 2016;53:107–139. doi: 10.1016/j.preteyeres.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Park SJ, Lee JH, Woo SJ, Ahn J, Shin JP, Song SJ, Kang SW, Park KH. Epidemiologic survey Committee of the Korean Ophthalmologic S: age-related macular degeneration: prevalence and risk factors from Korean National Health and nutrition examination survey, 2008 through 2011. Ophthalmology. 2014;121(9):1756–1765. doi: 10.1016/j.ophtha.2014.03.022. [DOI] [PubMed] [Google Scholar]
- 24.Cho BJ, Heo JW, Kim TW, Ahn J, Chung H. Prevalence and risk factors of age-related macular degeneration in Korea: the Korea National Health and nutrition examination survey 2010-2011. Invest Ophthalmol Vis Sci. 2014;55(2):1101–1108. doi: 10.1167/iovs.13-13096. [DOI] [PubMed] [Google Scholar]
- 25.Pasche B, Wisinski KB, Sadim M, Kaklamani V, Pennison MJ, Zeng Q, Bellam N, Zimmerman J, Yi N, Zhang K, et al. Constitutively decreased TGFBR1 allelic expression is a common finding in colorectal cancer and is associated with three TGFBR1 SNPs. J Exp Clin Cancer Res. 2010;29:57. doi: 10.1186/1756-9966-29-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baas AF, Medic J, Van 't Slot R, de Kovel CG, Zhernakova A, Geelkerken RH, Kranendonk SE, van Sterkenburg SM, Grobbee DE, Boll AP, et al. Association of the TGF-beta receptor genes with abdominal aortic aneurysm. Eur J Hum Genet. 2010;18(2):240–244. doi: 10.1038/ejhg.2009.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carlson CS, Matise TC, North KE, Haiman CA, Fesinmeyer MD, Buyske S, Schumacher FR, Peters U, Franceschini N, Ritchie MD, et al. Generalization and dilution of association results from European GWAS in populations of non-European ancestry: the PAGE study. PLoS Biol. 2013;11(9):e1001661. doi: 10.1371/journal.pbio.1001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchell P, Liew G, Gopinath B, Wong TY. Age-related macular degeneration. Lancet. 2018;392(10153):1147–1159. doi: 10.1016/S0140-6736(18)31550-2. [DOI] [PubMed] [Google Scholar]
- 29.Klein R, Cruickshanks KJ, Nash SD, Krantz EM, Nieto FJ, Huang GH, Pankow JS, Klein BE. The prevalence of age-related macular degeneration and associated risk factors. Arch Ophthalmol. 2010;128(6):750–758. doi: 10.1001/archophthalmol.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fritsche LG, Fariss RN, Stambolian D, Abecasis GR, Curcio CA, Swaroop A. Age-related macular degeneration: genetics and biology coming together. Annu Rev Genomics Hum Genet. 2014;15:151–171. doi: 10.1146/annurev-genom-090413-025610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rivera A, Fisher SA, Fritsche LG, Keilhauer CN, Lichtner P, Meitinger T, Weber BH. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005;14(21):3227–3236. doi: 10.1093/hmg/ddi353. [DOI] [PubMed] [Google Scholar]
- 32.Chen W, Stambolian D, Edwards AO, Branham KE, Othman M, Jakobsdottir J, Tosakulwong N, Pericak-Vance MA, Campochiaro PA, Klein ML, et al. Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc Natl Acad Sci U S A. 2010;107(16):7401–7406. doi: 10.1073/pnas.0912702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holliday EG, Smith AV, Cornes BK, Buitendijk GH, Jensen RA, Sim X, Aspelund T, Aung T, Baird PN, Boerwinkle E, et al. Insights into the genetic architecture of early stage age-related macular degeneration: a genome-wide association study meta-analysis. PLoS One. 2013;8(1):e53830. doi: 10.1371/journal.pone.0053830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sobrin L, Ripke S, Yu Y, Fagerness J, Bhangale TR, Tan PL, Souied EH, Buitendijk GH, Merriam JE, Richardson AJ, et al. Heritability and genome-wide association study to assess genetic differences between advanced age-related macular degeneration subtypes. Ophthalmology. 2012;119(9):1874–1885. doi: 10.1016/j.ophtha.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woo SJ, Ahn J, Morrison MA, Ahn SY, Lee J, Kim KW, DeAngelis MM, Park KH. Analysis of genetic and environmental risk factors and their interactions in Korean patients with age-related macular degeneration. PLoS One. 2015;10(7):e0132771. doi: 10.1371/journal.pone.0132771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klein R, Klein BE, Knudtson MD, Wong TY, Cotch MF, Liu K, Burke G, Saad MF, Jacobs DR., Jr Prevalence of age-related macular degeneration in 4 racial/ethnic groups in the multi-ethnic study of atherosclerosis. Ophthalmology. 2006;113(3):373–380. doi: 10.1016/j.ophtha.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 37.Oguido AP, Casella AM, Matsuo T, Ramos Filho EH, Berbel R, Silva RM. Prevalence of age-related macular degeneration in Japanese immigrants and their descendants living in Londrina (PR) - Brazil. Arq Bras Oftalmol. 2008;71(3):375–380. doi: 10.1590/S0004-27492008000300013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Supplemental Figure 1 Heatmap generated using single nucleotide polymorphisms related to age-related macular degeneration in the global population. Each row shows an SNP, and each column shows a continent. Red color indicates that the effect allele is enriched, whereas purple color indicates that the effect allele is depleted. AMR: American, EUR: European, SAS: South Asian, AFR: African, EAS: East Asian, KOR: Korean.
Additional file 2 Supplemental Figure 2 Heatmap generated using single nucleotide polymorphisms related to age-related macular degeneration in East Asian populations. Each row shows an SNP, and each column shows a country. Red color indicates that the effect allele is enriched, whereas purple color indicates that the effect allele is depleted. CDX: Chinese Dai in Xishuangbanna; CHB: Han Chinese in Beijing, China; CHS: Southern Han Chinese, China; JPT: Japanese in Tokyo, Japan; KOR: Korean in the Republic of Korea; KHV: Kinh in Ho Chi Minh City, Vietnam.
Additional file 3 Supplemental Figure 3 Polygenic risk score calculations of age-related macular degeneration for any AMD or late AMD using related single nucleotide polymorphisms and correlation plots of the prevalence of age-related macular degeneration (any or late) and polygenic risk score
Additional file 4 Supplemental Table 1 Effect allele frequencies (EAFs) of any age-related macular degeneration related single nucleotide polymorphisms in continental groups, including Koreans.
Additional file 5 Supplemental Table 2 Effect allele frequencies (EAFs) of any age-related macular degeneration-related single nucleotide polymorphisms in East Asian groups, including Koreans.
Data Availability Statement
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.