Abstract
The assembly of the bipolar mitotic spindle requires the careful orchestration of a myriad of enzyme activities like protein posttranslational modifications. Among these, phosphorylation has arisen as the principle mode for spatially and temporally activating the proteins involved in early mitotic spindle assembly processes. Here, we review key kinases, phosphatases, and phosphorylation events that regulate critical aspects of these processes. We highlight key phosphorylation substrates that are important for ensuring the fidelity of centriole duplication, centrosome maturation, and the establishment of the bipolar spindle. We also highlight techniques used to understand kinase–substrate relationships and to study phosphorylation events. We conclude with perspectives on the field of posttranslational modifications in early mitotic spindle assembly.
Keywords: centriole, centromere, centrosome, kinase, kinetochore, microtubules, mitotic spindle
1. INTRODUCTION
The faithful congression and segregation of chromosomes during mitosis requires the assembly and regulation of a large and complex microtubule‐based structure called the mitotic spindle. Numerous proteins with structural and signaling roles coordinate to assemble and regulate the mitotic spindle (Prosser & Pelletier, 2017). Importantly, these spindle components are often regulated by posttranslational modifications such as phosphorylation and ubiquitylation that allow for precise spatial and temporal control over their activity (Ong & Torres, 2019). The preparations for mitotic spindle assembly initiate during S phase where centrioles must duplicate and recruit and assemble the appropriate factors to mature into centrosomes (Nigg & Holland, 2018). The centrosomes then disjoin and move to opposite ends of the cell where they will serve as the spindle poles for the bipolar spindle. Subsequently, in early mitosis, the spindle poles nucleate and form the microtubule spindle, which promotes kinetochore‐microtubule attachments and the alignment of the chromosomes at the cell mid‐plane for cell division (Prosser & Pelletier, 2017). The synchronization and execution of these processes is highly regulated and necessary for faithful cell division, and their dysregulation can lead to chromosomal instability (Maniswami et al., 2018), apoptosis (Steegmaier et al., 2007; Torres et al., 2011), and aneuploidy (Kang et al., 2006; Tan, Caro, Potnis, Lanza, & Slawson, 2013; Yasui et al., 2004). Understanding how centrosomes, microtubule‐associated proteins, and the mitotic spindle are regulated are important avenues toward understanding diseases like aging (Fu et al., 2008; Macedo et al., 2018) and cancer (Gordon, Resio, & Pellman, 2012).
Kinases are critical for regulating proteins that have essential roles in early spindle assembly through their ability to transfer phosphate groups onto their substrates to modulate protein activity (Arquint & Nigg, 2016; Johnson & Hunter, 2005; Joukov & De Nicolo, 2018). In particular, phosphorylation regulates centriole duplication and procentriole elongation in S phase, centrosome maturation, disjunction, and separation in G2 phase, and microtubule nucleation and spindle assembly in late G2 and early M phase (Figure 1) (Arquint & Nigg, 2016; Carmena, Wheelock, Funabiki, & Earnshaw, 2012; Johmura et al., 2011; Mardin et al., 2010; Prosser & Pelletier, 2017). Here, we review the activation and roles of the key kinases Plk4, Plk1, Aurora A, Aurora B, and Cdk1, the key phosphatases PP1 and PP2A, and substrates of these enzymes as they function to promote mitotic spindle assembly from S phase to early M phase (Table 1). We discuss useful techniques for understanding the kinase–substrate relationship and identifying and characterizing phosphorylation sites and conclude with perspectives on future avenues into understanding the roles of phosphorylation and other posttranslational modifications on spindle assembly.
FIGURE 1.

Kinases regulate early mitotic spindle assembly. Key kinases, including Plk4, Aurora A, Plk1, and Aurora B regulate aspects of centriole duplication, centrosome maturation, and spindle assembly throughout the cell cycle [Color figure can be viewed at wileyonlinelibrary.com]
TABLE 1.
Summary of kinases, substrates, and phosphatases in mitotic spindle assembly
| Kinase | Key substrates | Substrate function | Consequence of phosphorylation | Relevant phosphatases |
|---|---|---|---|---|
|
Centriole duplication and procentriole elongation |
||||
| Plk4 | Plk4 (in trans) | Master kinase involved in centriole duplication and elongation in G1/S | Promotes localization, phase separation, degradation by β‐TrCP (Park et al., 2019; Rogers, Rusan, Roberts, Peifer, & Rogers, 2009) | PP2A‐Twins (Drosophila) (Brownlee, Klebba, Buster, & Rogers, 2011), Cdc14B (mammalian) (Wu et al., 2008) |
| STIL | Marks site of procentriole | Promotes STIL localization to centriole and binding of CPAP and Sas6 to STIL to promote centriole elongation (Moyer, Clutario, Lambrus, Daggubati, & Holland, 2015; Moyer & Holland, 2019; Ohta et al., 2014) | ||
| Sas6 | Forms nine‐spoked cartwheel at core of centriole | Promotes centriole formation (in Caenorhabditis elegans) (Kitagawa, Busso, Flückiger, & Gönczy, 2009) | ||
| GCP6 | Component of γ‐TuRC | Promotes centriole duplication (Bahtz et al., 2012) | ||
| Cep152 | Centrosome‐associated protein | Unknown (Hatch, Kulukian, Holland, Cleveland, & Stearns, 2010) | ||
| Centrosome maturation and early spindle pole assembly | ||||
| Aurora A | Aurora A | Master G2/M kinase | Autophosphorylation promotes Aurora A kinase activity (Zorba et al., 2014) | PP6 (Zeng, Bastos, Barr, & Gruneberg, 2010) |
| TPX2 | Nucleator for branched microtubules | Promotes TPX2‐CLASP1 interaction and proper spindle length (Fu et al., 2015) | ||
| TACC3 | Microtubule‐associated protein; forms TACC3/XMAP215/clathrin complex in mitosis | Promotes TACC3‐clathrin binding, TACC3 binding to microtubules, and TACC3/XMAP215/clathrin complex localization to mitotic spindle (Barros, Kinoshita, Hyman, & Raff, 2005; Burgess et al., 2018; Hood et al., 2013; Lin, Hu, & Shih, 2010) | ||
| NDEL1 | Microtubule‐associated protein | Promotes NDEL1 localization to centrosomes (Mori et al., 2007) | ||
| Hice1 | Component of Augmin complex, microtubule nucleator | Weakens interaction between Hice1 and microtubules (Tsai et al., 2011) | ||
| Plk1 | Cep192 | Centrosome‐associated protein | Promotes γ‐TuRC recruitment to centrosome and centrosome maturation (in Xenopus laevis) (Joukov, de Nicolo, Rodriguez, Walter, & Livingston, 2010) | |
| PCNT | Principle component of centrosomal matrix | Promotes subsequent microtubule nucleation activity via recruitment of PCM proteins (Lee & Rhee, 2011) | ||
| Nedd1 | Localizes γ‐TuRC to centrosome | Phosphorylation and binding by Plk1 requires priming phosphorylation by Cdk1 (Johmura et al., 2011; Zhang et al., 2009); promotes γ‐TuRC recruitment to centrosome and phosphorylation of Hice1 (Johmura et al., 2011; Zhang et al., 2009) | ||
| Hice1 | Component of Augmin complex, microtubule nucleator | Promotes Augmin localization to microtubule spindle and microtubule nucleation activity (Johmura et al., 2011) | ||
| Kizuna | Centrosomal protein involved in centrosome cohesion | Promotes spindle bistability, centrosome cohesion (Oshimori, Ohsugi, & Yamamoto, 2006) | Cdc25B (Thomas et al., 2014) | |
| LRRK1 | Kinase with various functions | Phosphorylation first by Plk1, then by Cdk1, activates LRRK1 kinase activity; LRRK1 phosphorylates CDK5RAP2, activating its microtubule‐nucleating capacity (Hanafusa et al., 2015) | ||
| NEK9 | Kinase with mitotic roles | Phosphorylation by Cdk1 and Plk1 promote NEK9 kinase activity; NEK9 phosphorylates Nedd1 to promote recruitment of Nedd1 to centrosomes (Bertran et al., 2011; Sdelci et al., 2012) | ||
| Centrosome disjunction and separation | ||||
| Plk1 | Cep85 | Centrosome‐associated protein; suppresses NEK2A activity | Disrupts Cep85‐NEK2A complex and promotes NEK2A kinase activity (Chen et al., 2019) | |
| Mst2 (STK3) | Hippo pathway kinase | Phosphorylation of Mst2 causes dissociation of PP1γ from Mst2‐NEK2A‐PP1γ complex and promotes NEK2A activity and localization to centrosome (Mardin, Agircan, Lange, & Schiebel, 2011; Mardin et al., 2010) | PP1γ (Mardin et al., 2010, 2011) | |
| NEK9 | Kinase with mitotic roles | Phosphorylation by Cdk1 and Plk1 promote NEK9 kinase activity; activated NEK9 phosphorylates NEK6/7 (Belham et al., 2003) | ||
| NEK2A | NEK2A | Kinase with mitotic roles | Autophosphorylation activates NEK2A kinase activity (Rellos et al., 2007) | PP1γ (Meraldi & Nigg, 2001) |
| C‐Nap1, Rootletin, LRRC45, β‐catenin | Form a protein network that connects the duplicated centrosomes | Dissociates protein network and promotes centrosome disjunction (Bahe, Stierhof, Wilkinson, Leiss, & Nigg, 2005; Bahmanyar et al., 2008; Hardy et al., 2014; He et al., 2013) | ||
| NEK9 | NEK9 | Kinase with mitotic roles | Autophosphorylation activates NEK9 kinase activity (Roig, Groen, Caldwell, & Avruch, 2005) | |
| NEK6/7 | Kinases with mitotic roles | Activates NEK6/7 kinase activity (Belham et al., 2003) | ||
| NEK6/7 | Eg5 | Major kinesin involved in centrosome separation and spindle bipolarity | Promotes Eg5‐TPX2 binding (Eibes et al., 2018) and consequently Eg5 localization to centrosome (Bertran et al., 2011; Rapley et al., 2008) | |
| Cdk1 | Promotes Eg5‐MT binding (Cahu et al., 2008; Slangy et al., 1995) | |||
| SRC kinases | Phosphorylation by c‐Src decreases Eg5 motor activity (Bickel et al., 2017) | |||
| Aurora A (in X. laevis) | Unknown (Giet, Uzbekov, Cubizolles, Le Guellec, & Prigent, 1999) | |||
| Spindle positioning | ||||
| Cdk1 | NuMA | Links mitotic spindle poles to cell membrane | NuMA not phosphorylated at Thr2055 localizes to cell cortex (Kotak, Busso, & Gönczy, 2013) | PP1‐Repo‐Man (B. H. Lee, Schwager, Meraldi, & Gotta, 2018), PP2A‐B55γ (Keshri, Rajeevan, & Kotak, 2020) |
| APC/C | Promotes mitotic progression | Regulates APC/C activity in early mitosis by anchoring it to spindle poles via END network (Ban et al., 2007) | PP2A‐B55α (Torres, Ban, & Jackson, 2010) | |
| Early spindle assembly—centromere | ||||
| Aurora B kinase | Aurora B | Master mitotic kinase | Autophosphorylation in trans activates Aurora B kinase activity (Yasui et al., 2004) | PP2A, PP1 |
| INCENP | Core component of CPC | Phosphorylation of and binding to INCENP activates Aurora B kinase activity (Honda, Körner, & Nigg, 2003) | ||
KMN network
|
Bind kinetochore proteins and microtubules to form kinetochore‐MT attachments | Phosphorylation of Knl1, Dsn1, and Hec1 reduces the binding affinity of the KMN network with microtubules (Cheeseman, Chappie, Wilson‐Kubalek, & Desai, 2006; DeLuca et al., 2006; Welburn et al., 2010) | ||
| Ska complex (Ska1‐3) | Stabilizes kinetochore‐MT attachments | Phosphorylation of Ska1 and Ska3 decreases Ska‐KMN network affinity (Chan, Jeyaprakash, Nigg, & Santamaria, 2012) | ||
| Cdt1 | Origin licensing in DNA replication; in mitosis, stabilizes kinetochore‐MT attachments | Phosphorylation decreases Cdt1‐MT affinity (Agarwal et al., 2018) | ||
| Early mitotic spindle assembly—kinetochore | ||||
| Aurora A | Plk1 | Master G2/M kinase involved in centrosome maturation | Phosphorylation activates Plk1 kinase activity (Macůrek et al., 2008) | |
| Cdk1 | Mypt1 | Regulatory unit of PP1 | Phosphorylation promotes Mypt1 localization to kinetochores and binding to Plk1 to antagonize Plk1 activity (Dumitru, Rusin, Clark, Kettenbach, & Compton, 2017; Yamashiro et al., 2008) | |
| Plk1 | PBIP1 (CENP‐U) | Centrosome‐associated protein | Phosphorylation promotes PBIP1‐Plk1 binding and localization to kinetochores (Dumitru et al., 2017; Yamashiro et al., 2008) | PP2A, PP1, Mypt1‐PP1 (Dumitru et al., 2017; Yamashiro et al., 2008) |
| NudC | Dynein‐associated protein | Phosphorylation promotes NudC‐Plk1 binding and localization to kinetochores (Nishino et al., 2006) | ||
| Bub1 | Kinetochore protein with roles in promoting spindle assembly checkpoint | Phosphorylation and binding by Plk1 requires priming phosphorylation by Cdk1; phosphorylation promotes Plk1 localization to kinetochores and possibly to enhance Plk1 phosphorylation of other substrates | ||
| CLASP2 | MT plus‐end tracking protein | Phosphorylation and binding by Plk1 requires priming phosphorylation by Cdk1; phosphorylation promotes Plk1 localization to kinetochores and formation of kinetochore‐MT attachments | ||
| CENP‐F | In mitosis, recruits other proteins to kinetochores | Phosphorylation possibly promotes CENP‐F localization to kinetochores | ||
| Sgt1 | Cochaperone for heat‐shock proteins | Phosphorylation promotes KMN complex localization at kinetochores | ||
| CLIP‐170 | MT plus‐end tracking protein | Phosphorylation promotes CLIP‐170 localization at kinetochores and kinetochore‐MT attachments | ||
| BubR1 | Component of mitotic checkpoint complex; promotes spindle assembly checkpoint | Phosphorylation and binding by Plk1 requires priming phosphorylation by Cdk1; recruits phosphatase PP2A‐B56ɑ to kinetochores to antagonize Aurora B (Wang et al., 2016) | ||
Abbreviations: APC/C, anaphase promoting complex/cyclosome; CPC, chromosomal passenger complex; MT, microtubules; PP2A, protein phosphatase 2A; γ‐TuRC, γ‐tubulin ring complex.
2. CENTRIOLE DUPLICATION AND PROCENTRIOLE ELONGATION
The centrioles are rod‐shaped microtubule‐based structures that form the core of the future centrosome (Nigg & Holland, 2018). Before S phase, a cell has two centrioles connected by a protein linker (Bahe et al., 2005). During S phase, an emerging procentriole will form at the base of each centriole, eventually forming two centriole pairs (Ohta et al., 2014). Each centriole pair will subsequently serve as one of the two poles of the bipolar spindle (Mardin et al., 2011). Plk4 is a key regulator of centriole biogenesis (Nigg & Holland, 2018) and is recruited to nascent centrioles through its binding to the centriolar proteins Cep152 and Cep192 (Kim et al., 2013; Sonnen, Gabryjonczyk, Anselm, Nigg, & Stierhof, 2013) (Figure 2). Although Plk4 phosphorylates Cep152, which residue(s) it phosphorylates and the importance of this modification is unknown (Hatch et al., 2010). At the centrioles, Plk4 binds STIL, a protein marker for the site of procentriole elongation, activating Plk4 kinase activity via autophosphorylation of its T‐loop on Thr170 (Moyer et al., 2015). Once activated, Plk4 phosphorylates STIL at Ser1108 and Ser1116 (Moyer et al., 2015; Moyer & Holland, 2019). These modifications are required for maintaining STIL localization at the centriole (Moyer et al., 2015) and for downstream recruitment of Sas6 (Ohta et al., 2014), which forms the symmetric core of the centriole. Plk4 also phosphorylates STIL at Ser428 (Dzhindzhev et al., 2017; Moyer & Holland, 2019), which promotes the binding of STIL with CPAP, a protein involved in procentriole elongation, and connects the growing microtubule cartwheel to the centriole wall (Moyer & Holland, 2019). While Plk4 is necessary upstream of Sas6, it remains unknown whether Plk4 modifies Sas6 in mammals. In Caenorhabditis elegans, Zyg‐1 (the ortholog of Plk4) phosphorylates Sas6 at Ser123, to promote centriole formation (Kitagawa et al., 2009). However, this residue is not conserved in the human sequence of Sas6. Beyond its roles in centriole biogenesis, Plk4 also has roles in recruiting the γ‐tubulin ring complex (γ‐TuRC), a key protein involved in microtubule nucleation, to the centrosome. GCP6 is a member of the γ‐TuRC complex and a substrate of Plk4 (Bahtz et al., 2012). Plk4 phosphorylates GCP6 on at least 10 residues throughout the sequence, and these modifications may be required for centriole duplication (Bahtz et al., 2012).
FIGURE 2.

Plk4 kinase activity regulates centriole duplication and procentriole elongation. Plk4 binds to STIL, a marker of the site of procentriole formation, and auto‐phosphorylates itself in trans to activate its kinase activity. Once activated, Plk4 phosphorylates STIL. Sas6 binds to STIL and promotes procentriole elongation. Once autophosphorylated, Plk4 binds to β‐TrCP (substrate adaptor for SCF ubiquitin ligase complex), promoting Plk4 degradation [Color figure can be viewed at wileyonlinelibrary.com]
Plk4 protein levels are precisely regulated by posttranslational modifications (Rogers et al., 2009) and misregulation of Plk4 is associated with tumorigenesis (Maniswami et al., 2018). Overexpression of Plk4 leads to centriole over‐duplication, whereas loss of Plk4 leads to a reduction in centriole number (Arquint & Nigg, 2016; Habedanck, Stierhof, Wilkinson, & Nigg, 2005; Kleylein‐Sohn et al., 2007). Both overexpression and underexpression of Plk4 are characteristic of numerous types of cancers. Plk4 autophosphorylates itself at Ser698, Ser700, Thr704, and Thr707 to promote the formation of phase separated condensates (Park et al., 2019). These condensates localize and concentrate Plk4 and exclude its ubiquitin ligase, β‐TrCP, promoting Plk4's stability (Gouveia et al., 2019; Park et al., 2019). Moreover, Plk4 autophosphorylates itself in trans at Ser285 and Thr289 (Cunha‐Ferreira et al., 2013; Guderian, Westendorf, Uldschmid, & Nigg, 2010). Phosphorylation of these residues generates a phosphodegron recognized by β‐TrCP and promotes Plk4 degradation in S and G2 phases (Cunha‐Ferreira et al., 2013; Guderian et al., 2010). In Drosophila melanogaster, Slimb (β‐TrCP ortholog) binding to Plk4 at the centrosome is antagonized by the phosphatase PP2A‐Twins, as PP2A‐Twins dephosphorylates Plk4, presumably removing the phosphodegron and promoting Plk4 stability (Brownlee et al., 2011). There is no obvious mammalian ortholog of the Drosophila PP2A regulatory B subunit Twins in mammalian systems. However, Cdc14B is a likely candidate as a regulator of Plk4 activity, as it localizes to the centrosomes and loss of Cdc14B activity leads to centriole amplification (Wu et al., 2008). As Plk4 localization and concentration both drive Plk4 kinase activity and degradation (autophosphorylation in trans both increases kinase activity and recognition by β‐TrCP), Plk4 levels and activity are tightly regulated. For example, lysine acetylation on Plk4 at Lys45 and Lys46 by acetyltransferases KAT2A and KAT2B are thought to induce structural changes that inactivate Plk4's kinase activity (Fournier et al., 2016).
3. CENTROSOME MATURATION AND EARLY SPINDLE POLE ASSEMBLY
To become competent for nucleating mitotic spindle microtubules, centrosomes must undergo a maturation process where they accumulate pericentriolar material (PCM) and recruit the γ‐TuRC. These two factors nucleate new microtubules and recruit microtubule‐associated proteins for microtubule organization, growth, and stability. The Aurora A kinase regulates many proteins with roles in centrosome maturation, spindle pole assembly, and mitotic spindle integrity (Brittle & Ohkura, 2005) (Figure 3). However, prior to doing so, Aurora A must be activated, a process dependent on phosphorylation and protein–protein interactions. First, Aurora A kinase activity is activated by phosphorylation of Thr288 on its T‐loop (Littlepage et al., 2002), a modification that may be carried out by another kinase, such as PAK1 (Zhao, Lim, Ng, Lim, & Manser, 2005), or via autophosphorylation (Zorba et al., 2014). Phosphorylation at Thr288 is opposed by phosphatases, such as PP6 (Zeng et al., 2010). Second, Aurora A activity is further promoted by binding to branched microtubule nucleator TPX2 (Bayliss, Sardon, Vernos, & Conti, 2003; Zorba et al., 2014), a regulatory event that causes a conformational shift in Aurora A toward an open conformation. Moreover, the binding of TPX2 to Aurora A may protect Aurora A from dephosphorylation at Thr288 (Bayliss et al., 2003) and ubiquitin‐mediated degradation (Giubettini et al., 2011). Similarly, Aurora A binding to TACC3, a microtubule‐associated protein that generally stabilizes microtubules (Peset & Vernos, 2008), promotes activation of Aurora A kinase activity (Burgess et al., 2015). Additionally, Aurora A activity may also be enhanced by incorporation into phase separated condensates via binding to microtubule‐associated protein BuGZ at the centromeres (Huang et al., 2018) and by binding to Cep192 (Joukov et al., 2010).
FIGURE 3.

Aurora A regulates centrosome maturation and early spindle assembly. Aurora A activates Plk1 at the centrosome and phosphorylates microtubule‐associated proteins like TPX2 and TACC3. These microtubule‐associated proteins promote centrosome maturation and microtubule organizing center activity, and consequently serve to assemble and stabilize the microtubule spindle [Color figure can be viewed at wileyonlinelibrary.com]
Once activated, Aurora A phosphorylates TPX2 at Ser121 and Ser125, promoting an interaction between TPX2 and CLASP1, a microtubule stabilizing protein (Lawrence, Zanic, & Rice, 2020), and consequently normal spindle length (Fu et al., 2015). Similarly, Aurora A phosphorylates NDEL1, a microtubule‐associated protein, on Ser251 (Mori et al., 2007). This modification promotes NDEL1 localization to the centrosomes and may be required for ubiquitin‐mediated degradation of NDEL1 (Mori et al., 2007). Moreover, this modification was not required for the binding of NDEL1 to TACC3 but nonetheless may promote the localization of TACC3 and γ‐tubulin to the centrosome, suggesting that phosphorylation on Ser251 of NDEL1 serves to recruit other proteins to the centrosome (Mori et al., 2007). Aurora A phosphorylates Ser34, Ser552, and Ser558 on TACC3, a regulator of microtubules and a component of the TACC3/XMAP215/clathrin complex (Kinoshita et al., 2005). Phosphorylation of TACC3 on Ser558 is not necessary for localization to the centrosomes or recruitment of XMAP215 (also known as ch‐TOG), a key microtubule nucleator and polymerase (Thawani, Kadzik, & Petry, 2018), to the centrosomes (Barros et al., 2005). Rather, this phosphorylation event promotes TACC3 binding to the minus ends of microtubules, stabilizes astral microtubules (Barros et al., 2005) and promotes the binding of TACC3 to clathrin (Burgess et al., 2018; Hood et al., 2013; Lin et al., 2010). Phosphorylation at Ser552 and Ser558 of TACC3 by Aurora A also promotes TACC3 localization to the centrosome, mitotic spindle, and mitotic spindle assembly (Fu et al., 2010). Similarly, TACC3 binding with Aurora A is necessary for TACC3 targeting to the mitotic spindle (Burgess et al., 2015). Although TACC3 can bind XMAP215 without Aurora A kinase activity (Thakur et al., 2014), TACC3 binding to XMAP215 stimulates XMAP215 microtubule nucleation activity (Kinoshita et al., 2005). Together, the TACC3/XMAP215/clathrin complex localizes to the mitotic spindle and promotes spindle stability (Hood et al., 2013; Lin et al., 2010).
Cep192 binds Aurora A and promotes Aurora A phosphorylation at Thr288 (Joukov, Walter, & De Nicolo, 2014). Once activated, Aurora A phosphorylates Plk1 at Thr210 (Joukov et al., 2014; Macůrek et al., 2008; Seki, Coppinger, Jang, Yates, & Fang, 2008). Plk1 binds to Cep192 via recognition of a phosphorylated Thr46 on Cep192 in Xenopus laevis (Thr44 in humans), though whether this modification comes from Plk1 or from another kinase is uncertain (Joukov et al., 2014). Nevertheless, in Xenopus egg extracts, once bound to Cep192, Plk1 phosphorylates Ser481, Ser507, Ser509, Ser919, and Ser923 (amino acid numbering refers to X. laevis Cep192 sequence), promoting γ‐TuRC recruitment and centrosome maturation (Joukov et al., 2014). In human cells, Plk1 also binds Cep192 through the conserved phospho‐Thr44 (Meng et al., 2015). However, whether Plk1 also modifies Cep192 for the purpose of γ‐TuRC recruitment in humans is unclear, as the phosphorylated serine residues in X. laevis are not conserved in humans.
Besides Cep192, Plk1 phosphorylates the long isoform of PCNT, a protein necessary for PCM expansion and centrosome maturation, at Thr1209, Thr1221, Ser1235, and Ser1241 (Lee & Rhee, 2011). While these modifications are not necessary for PCNT targeting to the centrosome, they are required for subsequent microtubule nucleation activity (Lee & Rhee, 2011). In particular, phosphorylation of Ser1235 and Ser1241 promotes bipolar spindle formation and recruitment of PCM proteins including Cep192, Aurora A, Nedd1, Plk1, and γ‐tubulin, proteins which are all involved in γ‐TuRC recruitment and microtubule nucleation (Lee & Rhee, 2011). Nedd1, the anchor for the γ‐TuRC complex in the centrosome, is first phosphorylated on Thr550 by Cdk1, allowing for the binding of Plk1 and subsequent phosphorylation by Plk1 on Thr382, Ser397, Ser426, and Ser637 (Zhang et al., 2009). These modifications promote γ‐TuRC recruitment to the centrosome (Zhang et al., 2009). Moreover, Cdk1 also phosphorylates Nedd1 on Ser460, which again promotes Plk1 binding to Nedd1 (Johmura et al., 2011). The Plk1‐Nedd1 complex phosphorylates the Hice1 subunit of Augmin, a noncentrosomal microtubule nucleator, at 17 unique sites (Johmura et al., 2011). These modifications drive Augmin binding to the microtubule spindle, microtubule nucleation, and spindle stability (Johmura et al., 2011). Interestingly, Aurora A also phosphorylates Hice1 at a number of residues in its N‐terminal microtubule binding region, including Thr17, Ser19, Ser20, and Ser20 (Tsai et al., 2011). In contrast to Plk1 phosphorylation, which promotes Hice1‐microtubule binding, Aurora A phosphorylation of Hice1 weakened the affinity of Hice1 for microtubules (Tsai et al., 2011). Plk1 phosphorylation of Kizuna, a centrosomal protein involved in centrosome cohesion, on Thr379 does not affect localization to the centrosomes, but does promote spindle bistability and centrosome cohesion (Oshimori et al., 2006). This modification is antagonized by the phosphatase Cdc25B (Thomas et al., 2014).
Plk1 also activates a number of kinases that regulate processes in centrosome maturation (Figure 4). Plk1 phosphorylates LRRK1, a kinase involved in cytoskeletal regulation and endosome trafficking (Hanafusa et al., 2019; Kedashiro et al., 2015), at Ser1790, then Cdk1 phosphorylates LRRK1 at Thr1400 in the T‐loop to activate LRRK1 kinase activity (Hanafusa et al., 2015). Loss of LRRK1 phosphorylation at this residue results in spindle orientation defects and deficiencies in the formation of astral microtubules (Hanafusa et al., 2015). Activated LRRK1 phosphorylates Thr102 and Ser140 of CDK5RAP2 (Hanafusa et al., 2015), a protein involved in maintaining centrosome cohesion (Barrera et al., 2010). While these modifications do not affect the recruitment of CDK5RAP2 or PCNT to centrosomes, they promote CDK5RAP2 binding to γ‐tubulin and subsequent microtubule nucleation (Hanafusa et al., 2015). The NEK family of kinases regulate many aspects of cell division and cell cycle progression (Moniz, Dutt, Haider, & Stambolic, 2011). Cdk1 phosphorylates NEK9 on Ser869 (Bertran et al., 2011; Sdelci et al., 2012). This modification serves as a priming phosphorylation that allows for subsequent Plk1 binding and NEK9 activation via phosphorylation on Thr210 in the T‐loop of NEK9 (Bertran et al., 2011; Sdelci et al., 2012). Whether Plk1 is necessary to activate NEK9 is uncertain, as phosphorylation of this residue can also occur via autophosphorylation (Roig et al., 2005). Nevertheless, once activated, NEK9 phosphorylates Nedd1 on Ser337 to promote the recruitment of Nedd1 and subsequent recruitment of γ‐tubulin to the centrosome (Sdelci et al., 2012).
FIGURE 4.
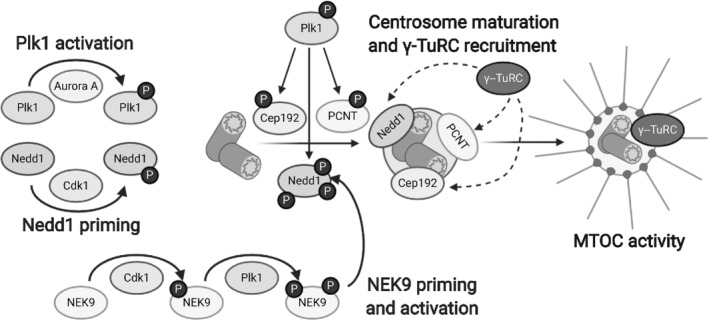
Plk1 regulates centrosome maturation and microtubule organizing center (MTOC) activity. Once activated by Aurora A, Plk1 phosphorylates centrosomal proteins like Cep192, PCNT, and Nedd1. Cdk1 primes NEK9 for activation by Plk1, and NEK9 also phosphorylates Nedd1. These and other centrosomal proteins play roles in centrosome maturation, pericentriolar material expansion, and γ‐TuRC recruitment to the centrosome, allowing the centrosome to serve as a MTOC [Color figure can be viewed at wileyonlinelibrary.com]
4. CENTROSOME DISJUNCTION AND SEPARATION
Plk1 activates a number of kinases that regulate the process of centrosome disjunction (separation of mother and daughter centrioles) (Figure 5) and centrosome separation (the separation of one centrosome from another to establish a bipolar spindle) (Hinchcliffe & Sluder, 2001; Wang, Jiang, & Zhang, 2014) (Figure 6). Downstream of Plk1 is NEK2A, the longer isoform of NEK2 that plays a role in centrosome disjunction (Hames & Fry, 2002). Centrosomal protein Cep85 binds to NEK2A to suppress NEK2A kinase activity (Chen et al., 2019). Plk1 binds to the Cep85‐NEK2A complex and subsequently phosphorylates Cep85 (Chen et al., 2019). Once phosphorylated, Cep85 has a lower binding affinity for NEK2A, freeing NEK2A and promoting NEK2A kinase activity (Chen et al., 2019). NEK2A activity is influenced by multiple autophosphorylation sites in its T‐loop, modifications which can either increase or decrease its kinase activity (Rellos et al., 2007). Plk1 also phosphorylates an upstream regulator of NEK2A, Mst2 (also known as STK3) (Mardin et al., 2011). NEK2A activity is antagonized by phosphatases (Meraldi & Nigg, 2001), including PP1γ. Conversely, NEK2A binds to and phosphorylates PP1γ at Thr307 and Thr318 to reduce its phosphatase activity (Helps, Luo, Barker, & Cohen, 2000). NEK2A forms a complex with PP1γ and Hippo‐pathway kinase Mst2 (Mardin et al., 2010, 2011). Mst2 phosphorylates NEK2A at Ser356, Ser365, Thr406, and Ser438 (Mardin et al., 2010). These modifications do not affect NEK2A kinase activity but are necessary for NEK2A localization to the centrosome (Mardin et al., 2010). To regulate NEK2A activity, Plk1 phosphorylates Mst2 at Ser15, Ser18, and Ser316, disrupting the binding of PP1γ to the NEK2A‐PP1γ‐Mst2 complex and promoting NEK2A activity (Mardin et al., 2011).
FIGURE 5.
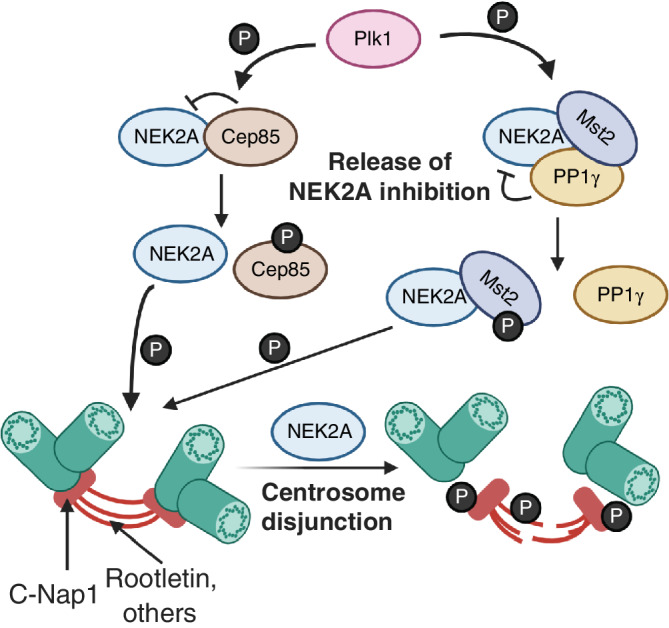
Plk1 regulates NEK2A in centrosome disjunction. Plk1 phosphorylates Cep85 to free Cep85 inhibition of NEK2A. Similarly, Plk1 phosphorylates Mst2 to disrupt the PP1γ from antagonizing the activity of NEK2A. Once activated, NEK2A phosphorylates a number of linker proteins at the centrioles, including C‐Nap1 and Rootletin, to free the two centrosomes from each other [Color figure can be viewed at wileyonlinelibrary.com]
FIGURE 6.

Kinases regulate Eg5 in centrosome separation and bipolar spindle formation. Once the centrosomes have disjoined, they need to separate to opposite ends of the cell to form a bipolar spindle. NEK9 activates NEK6 and NEK7, which in turn phosphorylate Eg5, localizing it to the centrosome and promoting centrosome separation and bipolar spindle formation. Other kinases also regulate Eg5 activity [Color figure can be viewed at wileyonlinelibrary.com]
During G2 phase, the two pairs of centrioles are linked by a network of filaments that must be removed for centrosome disjunction and separation (Paintrand, Moudjou, Delacroix, & Bornens, 1992). While the composition of these filaments is not entirely known, a number of candidate proteins have been identified, including C‐Nap1 and Rootletin (Hinchcliffe & Sluder, 2001; Wang et al., 2014). When activated, NEK2A phosphorylates 27 residues in the C‐terminus of C‐Nap1, a protein that resides at the proximal end of centrioles and may serve as the point of attachment for the protein linker network (Hardy et al., 2014). These modifications weaken the affinity of C‐Nap1 for Cep135, presumably via electrostatic interactions, and promote the release of C‐Nap1 from the centrosome, freeing the pairs of centrioles from each other and promoting centrosome disjunction (Hardy et al., 2014) (Figure 5). Rootletin is a protein that forms fibers that bridge the proximal ends of centrioles and binds to C‐Nap1 and is similarly phosphorylated by NEK2A at many sites to promote centrosome disjunction (Bahe et al., 2005). A similar mechanism may apply for NEK2A phosphorylation of protein fiber LRRC45 at Ser661 (He et al., 2013). β‐catenin also forms a complex with Rootletin at the centrosomes, is phosphorylated by NEK2A, and plays a role in centrosome separation (Bahmanyar et al., 2008). Whereas C‐Nap1 and Rootletin do not localize at the centrosomes during mitosis, β‐catenin is found at mitotic centrosomes, suggesting additional roles for β‐catenin in centrosome function (Bahmanyar et al., 2008).
Once centrosomes have been disjoined, they must separate to opposite ends of the cell to form a bipolar spindle. Eg5 is a tetrameric bipolar kinesin that acts on antiparallel microtubules to generate forces that separate centrosomes and promote spindle bistability (Kapitein et al., 2005; Shimamoto, Forth, & Kapoor, 2015) (Figure 6). NEK9 is a downstream kinase of Plk1 that phosphorylates NEK6 at Ser206 and NEK7 at Ser195 at the T‐loop to activate NEK6/7 (Belham et al., 2003). NEK6/7 phosphorylate Eg5 at Ser1033, promoting Eg5 binding to TPX2 (Eibes et al., 2018), inducing Eg5 localization to the centrosome, and ultimately promoting centrosome separation (Bertran et al., 2011; Rapley et al., 2008). Cdk1 also phosphorylates Eg5 on Thr926 (Slangy et al., 1995), increasing the affinity of Eg5 for microtubules (Cahu et al., 2008).The PP2A‐B55α complex dephosphorylates this residue to regulate Eg5 activity (Liu et al., 2017). SRC kinases phosphorylate Eg5 on Tyr125, Tyr211, and Tyr231 (Bickel et al., 2017). These modifications decrease Eg5 motor activity (Bickel et al., 2017). Of interest, Aurora A has been shown to phosphorylate Eg5 in X. laevis, but the site of phosphorylation and the consequence of this modification is unknown, and there are no reports of Aurora A phosphorylating Eg5 in human cells (Giet et al., 1999).
5. EARLY MITOTIC SPINDLE ASSEMBLY—CENTROMERE
Critical to chromosome congression are kinetochore‐microtubule attachments. Centromeres are regions of DNA that recruit centromeric proteins (CENPs, such as CENP‐A) and function to physically link the chromosomes to the microtubule spindle via the kinetochore (Fukagawa & Earnshaw, 2014). At the centromere, the chromosomal passenger complex (CPC), consisting of Aurora B kinase, INCENP, Survivin, and Borealin, regulates many aspects of mitosis, including early mitotic spindle assembly (Carmena et al., 2012). To localize the CPC at the centromere, kinase Haspin phosphorylates histone H3 at Thr3, leading to recruitment of the CPC to the centromere via Survivin (Kelly et al., 2010). At the centromere, Aurora B kinase activity is activated by two main modifications: Aurora B phosphorylation of INCENP at three consecutive residues (TSS 893–895) (Honda et al., 2003) and Aurora B autophosphorylation in trans at Thr232 in the T‐loop of its kinase domain (Yasui et al., 2004) are thought to result in structural changes that fully activate the kinase (Sessa et al., 2005).
Once activated, Aurora B phosphorylates a number of substrates with the general purpose of destabilizing erroneous kinetochore‐microtubule attachments (Figure 7, bottom). The KMN network, a protein structure composed of the KNL1 complex, the Mis12 (or MIND) complex, and the Ndc80 complex, is the core protein complex that connects microtubules to the kinetochores (Cheeseman et al., 2006). Centromeric Aurora B phosphorylates and regulates many members of this complex in order to monitor proper spindle assembly. Aurora B phosphorylates the N‐terminus of Hec1, one of the key members of the Ndc80 complex, at Ser5, Ser15, Thr49, Ser55, Ser69, and possibly Ser44 (DeLuca et al., 2006). In microtubule binding assays with C. elegans proteins, these phosphorylation events led to a weaker binding between Ndc80 and microtubules, suggesting that Aurora B phosphorylation of Hec1 decreases the affinity of Hec1 for microtubules (Cheeseman et al., 2006). In a similar manner, Aurora B also phosphorylates KNL1 at Ser25 and Ser60 and phosphorylates Dsn1, a member of the Mis12 complex, at Ser28, Ser78, Ser109, and possibly Ser100 (Welburn et al., 2010). Similar to phosphorylation of Hec1, these phosphorylation events reduced the affinity of these complexes for microtubules (Welburn et al., 2010). Thus, Aurora B phosphorylation of the KMN complex serves to promote the disruption of kinetochore‐microtubule attachments.
FIGURE 7.

Kinetochore‐microtubule (MT) dynamics are regulated by phosphorylation. At the kinetochores, Cdk1 and Plk1 (top) phosphorylate a number of proteins that serve to promote stable kinetochore‐microtubule attachments. At the centromere, Aurora B (bottom) phosphorylates proteins, including the microtubule binding KMN complex, to discourage untimely or erroneous kinetochore‐microtubule attachments [Color figure can be viewed at wileyonlinelibrary.com]
While the KMN complex is a crucial component of kinetochore‐microtubule attachments, other proteins also support kinetochore‐microtubule attachments. Notably, the Ska complex, comprising of Ska1‐3, directly binds Ndc80 (Zhang et al., 2017) and microtubules (Welburn et al., 2009), adding an additional microtubule‐binding modality at the kinetochores. In a similar manner, Cdt1, a protein canonically known for its roles in replication origin licensing in G1 phase, also binds Ndc80 (Varma et al., 2012) and microtubules (Agarwal et al., 2018). In both cases, Ska 1/3 and Cdt1 are phosphorylated by Aurora B (Agarwal et al., 2018; Chan et al., 2012). Expression of nonphosphorylatable mutants of Ska1/3 (Chan et al., 2012) or Cdt1 (Agarwal et al., 2018) both led to hyper‐stability of kinetochore‐microtubule attachments, whereas expression of phosphomimetic mutants led to a decrease in kinetochore‐fiber microtubule stability. In particular, phosphorylation of Ska1/3 decreases the Ska complex's affinity for the KMN complex, indirectly causing weaker kinetochore‐microtubule attachments (Chan et al., 2012) whereas phosphorylation of Cdt1 decreases its affinity for microtubules themselves (Agarwal et al., 2018). For both proteins, loss of the correct phosphorylation patterns led to increases in mitotic timing and the presence of mitotic failures.
Interestingly, the loss of the Ska complex led to loss of Aurora B kinase activity in cells assayed with an FRET reporter for Aurora B activity, and the addition of Ska1 into in vitro Aurora B kinase assay increased Aurora B kinase activity, suggesting that the Ska complex also increases Aurora B activity (Redli, Gasic, Meraldi, Nigg, & Santamaria, 2016). These experiments suggest a feedback loop between the Ska complex and Aurora B whereby properly localized Ska limits its own enrichment and microtubule‐binding activity at the kinetochore by maintaining high levels of active Aurora B, keeping kinetochore‐microtubule attachments in a dynamic state (Redli et al., 2016).
MCAK is another important microtubule motor at the centromere that serves to depolymerize microtubules (Maney, Hunter, Wagenbach, & Wordeman, 1998). Aurora B phosphorylates MCAK, at Ser95, Ser109, Ser111, Ser115, and Ser192 (amino acid numbering uses the short isoform of MCAK, UniProt identifier: Q99661‐2) (Andrews et al., 2004). Phosphorylation of MCAK inhibits its microtubule depolymerization activity (Andrews et al., 2004). Further studies in X. laevis demonstrated that phosphorylation of Ser196 (Ser192 in humans) is responsible for the inhibition of microtubule depolymerization activity (Lan et al., 2004), whereas other residues are responsible for the localization of MCAK. Aurora B phosphorylation of X. laevis Thr95 (Ser95 in human MCAK) and Ser196 (Ser192 in humans) inhibits MCAK binding to centromeres, whereas Aurora B phosphorylation at X. laevis Ser110 (Ser111 in humans) promotes MCAK localization to centromeres (Zhang, Lan, Ems‐McClung, Stukenberg, & Walczak, 2007).
6. EARLY SPINDLE ASSEMBLY—KINETOCHORE
Key to ensuring the timing and fidelity of chromosome movements are kinetochore‐microtubule attachments. Plk1 kinase activity is necessary for mitotic spindle formation and kinetochore‐microtubule attachments (Lera et al., 2016; Liu, Davydenko, & Lampson, 2012) and is tightly regulated by modifications (Figure 7, top). Plk1 kinase activity is activated via phosphorylation of its T‐loop on Thr210 by Aurora A kinase (Macůrek et al., 2008; Seki et al., 2008) and possibly by other kinases (Paschal, Maciejowski, & Jallepalli, 2012). Methylation on Lys191 of Plk1 by methyltransferases SET7/9 inhibits Plk1 kinase activity (Yu et al., 2020). This modification presumably allows for fine‐tuning of Plk1 activity at the kinetochores, though a demethylase has not yet been identified. Moreover, phosphorylation of Plk1 Thr210 is also regulated by phosphatases. Cdk1/Cyclin A phosphorylates Mypt1, a binding partner and localization subunit for phosphatase PP1 at Ser473 (Dumitru et al., 2017). Phosphorylation at Ser473 promotes Mypt1 localization to the kinetochores (Dumitru et al., 2017) and subsequent binding to Plk1 (Dumitru et al., 2017; Yamashiro et al., 2008). Once bound to Plk1, the Mypt‐PP1 complex antagonizes Plk1 function by dephosphorylating Thr210 (Dumitru et al., 2017).
Plk1 localization to the kinetochores is mediated by several protein interactions. Plk1 phosphorylates kinetochore protein PBIP1 at a number of residues, and phosphorylation at Thr78 generates a consensus polo‐box domain‐binding motif that is necessary for Plk1‐PBIP1 binding (Kang et al., 2006). This interaction is required for Plk1 targeting to the kinetochores (Kang et al., 2006). Similarly, NudC, a dynein‐associated protein, is phosphorylated by Plk1 at Ser274 and Ser326, and, in the absence of NudC, Plk1 localization at the kinetochores is weakened (Nishino et al., 2006). Cdk1 phosphorylation of kinetochore protein Bub1 at Thr609 also promotes Plk1 binding to Bub1 and localization of Plk1 to the kinetochores (Qi, Tang, & Yu, 2006). Plk1 phosphorylates Bub1, but which residues are phosphorylated and the consequence of the modification are not fully understood (Qi et al., 2006). One possible downstream application of Plk1‐Bub1 binding may be to enhance Plk1 phosphorylation of Cdc20 at Ser92, a modification that inhibits the E3 ubiquitin ligase anaphase promoting complex/cyclosome (APC/C) and regulates mitotic progression (Jia, Li, & Yu, 2016). Another consequence of Plk1‐Bub1 binding may be that Bub1 serves as a scaffold for Plk1 to phosphorylate other substrates, such as the Mps1 kinase (Ikeda & Tanaka, 2017).
At the kinetochores, Plk1 phosphorylation activates a number of microtubule‐associated proteins with the purpose of promoting stable kinetochore‐microtubule attachments (Lera et al., 2016; Liu, Davydenko, & Lampson, 2012). Cdk1 phosphorylates CLASP2 at Ser1233, Ser1234, and Ser1250, with Ser1234 being the main modification priming CLASP2 for recognition and subsequent phosphorylation by Plk1 at Ser12348, Ser1255, Ser1274, and Ser1313 (Maia et al., 2012). These modifications are necessary for maintenance of spindle bipolarity and proper kinetochore‐microtubule attachments (Maia et al., 2012). CENP‐F is a large microtubule‐associated protein with roles in stabilizing kinetochore‐microtubule attachments (Auckland, Roscioli, Coker, & McAinsh, 2020). Plk1 phosphorylates CENP‐F on at least eight residues, and these phosphorylation marks may be important for CENP‐F localization at the kinetochores (Santamaria et al., 2011).
Similarly, Plk1 phosphorylates Sgt1, a co‐chaperone for heat‐shock proteins, at Ser331, and this phosphorylation is necessary for Sgt1 localization at kinetochores and enhances subsequent localization of the KMN microtubule‐kinetochore complex (Liu, Song, et al., 2012). In a similar manner, CLIP‐170, a major component of the microtubule plus end‐binding proteins (Bieling et al., 2008), was phosphorylated by Plk1 at Ser195 and kinase CK2 at Ser1318 (Li et al., 2010). Phosphorylation at Ser195 does not affect CLIP‐170 binding to microtubules but enhances the association of kinase CK2 with CLIP‐170, CLIP‐170 localization to the kinetochores, and the formation of microtubule‐kinetochore attachments (Li et al., 2010). After Cdk1 primes kinetochore protein BubR1 by phosphorylating Thr620, Plk1 phosphorylates BubR1 at Ser676 (Elowe, Hümmer, Uldschmid, Li, & Nigg, 2007) and at Thr680 (Suijkerbuijk, Vleugel, Teixeira, & Kops, 2012). Together with other phosphorylation marks by kinases like Mps1 (Huang et al., 2008), these modifications recruit PP2A‐B56α which may balance Aurora B activity at the kinetochore (Suijkerbuijk et al., 2012; Xu, Raetz, Kitagawa, Virshup, & Lee, 2013).
7. PHOSPHATASES IN MITOTIC SPINDLE ASSEMBLY
Phosphatases antagonize the activity of protein kinases. While they play a role in suppressing kinase activity, the timely activation of protein phosphatases is also necessary to promote timely cell cycle progression and spindle assembly. Here, we focus on the two main classes of phosphatases in mitotic spindle assembly: PP1 and PP2A. The PP1 holoenzyme generally consists of the catalytic PP1 subunit and one or more substrate‐recognition and/or localization co‐factors (Bertolotti, 2018), whereas the PP2A holoenzyme generally consists of a scaffolding A subunit, a catalytic C subunit, and a regulatory, substrate‐recognition B subunit (Sents, Ivanova, Lambrecht, Haesen, & Janssens, 2013).
In mitosis, PP2A functions to promote proper kinetochore‐microtubule attachments at the kinetochores and centromeres. At the kinetochores, PP2A‐B56γ binds to Cdk1‐ and Plk1‐phosphorylated BubR1 (Kruse et al., 2013; Wang et al., 2016). Binding of PP2A to BubR1 may be promoted by the presence of centromeric protein Sgo1 (Vallardi, Allan, Crozier, & Saurin, 2019). At the centromere, PP2A‐B56α localization is dependent on Sgo2, not on Sgo1 (Vallardi et al., 2019). Interestingly, however, Sgo1 also binds the CPC via Borealin, and a Borealin‐Sgo1‐PP2AC complex was reconstituted in vitro from recombinant proteins (Bonner et al., 2020). Together, these data suggest that Sgo1 may coordinate to allow PP2A‐B56 to antagonize CPC‐based Aurora B activity at the centromere (Meppelink, Kabeche, Vromans, Compton, & Lens, 2015; Vallardi et al., 2019) and inner kinetochore (Suijkerbuijk et al., 2012; Xu et al., 2013). Altogether, PP2A regulates kinetochore‐microtubule attachments, as loss of PP2A leads to a loss of stable k‐fibers and chromosome attachment, regulating both Aurora B and Plk1 activity (Foley, Maldonado, & Kapoor, 2011).
In contrast to PP2A, which has roles at both the centromere and kinetochores, PP1 functions mainly at the kinetochores. PP1 binds competitively to kinetochore protein KNL1 at the same location as KNL1's microtubule‐binding site, suggesting that KNL1 could either bind microtubules or PP1 (Bajaj, Bollen, Peti, & Page, 2018). Once bound to KNL1, PP1 dephosphorylates KNL1, causing dissociation of spindle assembly checkpoint kinetochore proteins from KNL1, thus silencing spindle assembly checkpoint signaling (Bajaj et al., 2018). PP1 is also recruited to the kinetochores via binding to Ska1 (Sivakumar et al., 2016). At the kinetochores, both PP1 and PP2A dephosphorylate and inactivate Msp1, a master kinase of spindle assembly checkpoint signaling, shutting down the spindle assembly checkpoint (Hayward et al., 2019; Moura et al., 2017). Thus, the proper localization and accumulation of PP1 and PP2A at the kinetochores serve to antagonize Aurora B activity and quench spindle assembly checkpoint signaling.
Besides regulating kinetochore‐microtubule attachments, PP1 and PP2A also function to regulate spindle orientation and stability during mitosis. NuMA is a microtubule‐binding protein that anchors the mitotic spindle to the cortex via the plasma membrane (Kotak, Busso, Gonczy, & Gönczy, 2014). Cdk1 phosphorylates Thr2055 of NuMA; this phosphorylation event inhibits NuMa localization at the cortex, such that only NuMA not phosphorylated at this residue localizes to the cortex (Kotak et al., 2013). The proper extent of NuMA localization at the cortex is regulated by PP2A‐B55γ (Keshri et al., 2020) and by PP1‐Repo‐Man (Lee et al., 2018). Expression of a construct of NuMA that could not be phosphorylated on Thr2055 resulted in spindle oscillations (i.e., the mitotic spindle wobbled unsteadily) (Kotak et al., 2013), and inhibition of either phosphatase complex resulted in improper metaphase spindle positioning (Keshri et al., 2020; Kotak et al., 2013). During anaphase, when Cdk1 activity is low, these phosphatases promote increased NuMA localization to the cortex, generating dynein‐dependent forces that result in spindle elongation and progression through anaphase (Keshri et al., 2020; Kotak et al., 2013). At the spindle poles, NuMA also forms the END (Emi1, NuMA, dynein‐dynactin) network, a complex that localizes the APC/C at the spindle poles and inhibits its activity, allowing for proper Cdk1‐mediated spindle assembly in early mitosis (Ban et al., 2007). Loss of PP2A‐B55α led to the mislocalization of the APC/C from the spindle poles during mitotic spindle assembly, the formation of multipolar spindles, and failures in chromosome congression, suggesting that PP2A antagonizes Cdk1‐regulation of APC/C localization at the spindle poles in early mitosis (Torres et al., 2010).
8. TECHNIQUES IN IDENTIFYING AND STUDYING PHOSPHORYLATION
Numerous methods of identifying and studying phosphorylation and the relationship between kinase and substrate have been developed (Johnson & Hunter, 2005; Xue & Tao, 2013). A traditional method for characterizing a kinase–substrate pair has been an in vitro kinase assay, where the isolated kinase and putative substrate are incubated with radioactive γ‐32P ATP and the transfer of the radioactive phosphoryl group onto the substrate is monitored by western blotting and radiometry. If the substrate is phosphorylated, then the reaction is repeated using nonradioactive ATP and the sites of phosphorylation on the substrate are identified via mass spectrometry, as the addition of a phosphate group leads to a detectable mass increase of ~80 Da. These types of approaches have been used to identify numerous Plk1 substrates including the StarD9 protein that is important for PCM cohesion at the centrosome (Senese et al., 2015). Similar experiments can be performed in cells either in the presence or absence of kinase inhibitors or by overexpressing or knocking down/knocking out the kinase of interest and assessing for differential phosphorylation on potential substrates using immunoblotting (with a phospho‐specific antibody) or mass spectrometry‐based approaches, which have also been used to identify Plk1 substrates (Santamaria et al., 2011).
Other popular approaches include classical yeast two‐hybrid approaches or affinity proteomics approaches, which can also identify potential kinase–substrate relationships (Johnson & Hunter, 2005; Xue & Tao, 2013). Once a phosphorylation site has been mapped, a common approach to study the effect of the modification is to mutate the site(s) of phosphorylation to a nonphosphorylatable residue (usually Ser/Thr to Ala and Tyr to Phe) or a phosphomimetic residue (usually to Asp or Glu, though sometimes these phosphomimetic residues do not recapitulate the phosphorylated state of the protein) (Johnson & Hunter, 2005; Xue & Tao, 2013) and analyze the cellular consequences of the modification.
In some cases, understanding what motif the kinase will phosphorylate is useful for identifying additional substrates of that kinase. Computational approaches, such as ScanSite (Obenauer, Cantley, & Yaffe, 2003) or PhosphoPredict (Song et al., 2017) have been developed that analyze phosphosites and their motifs and predict potential substrates, such as the prediction and validation of SPICE1 as an Aurora A and Aurora B substrate (Deretic, Kerr, & Welburn, 2019). Phage display approaches or peptide‐library based approaches have also been used to define kinase phosphorylation motifs. In the case of peptide‐library based approaches, a microchip with a peptide library is allowed to react with the kinase of interest, such as with the NEK family of kinases (van de Kooij et al., 2019). Phosphorylated peptides are identified and computationally analyzed to identify a consensus phosphorylation motif. In the case of phage‐based approaches, phages are modified to display a library of peptides on their surface. The kinase is allowed to react with the peptides, and phages that are phosphorylated are enriched, usually via binding to a phospho‐antibody. After sequential rounds of enrichment, the selected phages have their DNA sequenced, and again the selected peptides are computationally analyzed to identify a consensus phosphorylation motif. Such a technique was used to expand the phosphorylation motif for Plk1 (Santamaria et al., 2011).
While these classic approaches have been useful in understanding kinase–substrate relationships, these techniques have difficulty addressing important biological questions. First, in vitro kinase assays are low‐throughput and biased, requiring a guess as to what a kinase's substrate may be. Moreover, while numerous techniques have been used to assay kinase activity in vitro, assessing kinase activity in cells has been a challenge. Finally, chemical inhibitors are useful agents in understanding a kinase's role, but chemical inhibitors lack spatial specificity that make it difficult to study a kinase with more than one localization. Here, we highlight some innovative techniques used for understanding kinase–substrate relationships and kinase activity in cells. We focus on “bump‐hole” technology for orthogonal identification of kinase substrates, FRET sensors for in situ readouts of kinase activity in cells, and strategies for localized inhibition of kinases for targeted kinase inhibition. Strategies for studying phosphatases are similar, including techniques in mass spectrometry and the use of FRET‐ or fluorescence‐based assays, and are reviewed elsewhere (Fahs, Lujan, & Köhn, 2016).
8.1. Bump‐hole
While mass spectrometry can identify phosphorylation sites that increase or decrease in abundance upon expression or inhibition of a kinase, these modifications may come about from the activation or inactivation of downstream kinases. For example, many Cdk1 phosphorylation events serve as priming modifications for later binding and phosphorylation by Plk1 (Elowe et al., 2007; Maia et al., 2012; Qi et al., 2006; Zhang et al., 2009). Thus, if mass spectrometry is performed on cell lysates where Cdk1 is overexpressed, the identified phosphorylation sites may not simply be Cdk1 sites but also Plk1 sites. While in vitro kinase assays can confirm whether or not those sites are due to Cdk1, such assays are time‐consuming and/or expensive. A high‐throughput, unbiased, and more direct method of identifying kinase substrates employs “bump‐hole” technology (Figure 8a′). First, an analogue of ATP is synthesized that is sterically larger than unmodified ATP. This increased steric size creates a “bump” in ATP that renders it unable to bind to unmodified kinases or ATPases (Figure 8a′, Circle 1) (Liu, Shah, Yang, Witucki, & Shokat, 1998; Shah, Liu, Deirmengian, & Shokat, 1997). The gamma phosphate of the modified ATP analogue (i.e., the phosphate group that is transferred to the substrate) is also modified to contain a thiophosphate group for later detection (Blethrow, Glavy, Morgan, & Shokat, 2008; Hengeveld et al., 2012). Mutation of a bulky “gatekeeper” residue in the ATP binding pocket to an amino acid with a smaller side chain, such as glycine or alanine, introduces a “hole” into the kinase of interest. This “hole” allows the mutated kinase of interest, but not other kinases or ATPases, to bind to the “bumpy” substrate (Figure 8a′, Circle 2) (Liu et al., 1998; Shah et al., 1997). Since the mutated kinase can bind the analogue of ATP, these kinases are often referred to as “analog sensitive” enzymes. Importantly, the “hole” also decreases the affinity of the mutated enzyme for unmodified ATP, allowing the mutated enzyme to bind to and use “bumpy” ATP with reasonable selectivity for its enzymatic reactions (Figure 8a′, Circle 3). Proteins modified by the “bumpy” substrate can subsequently be isolated via enrichment for the thiophosphate tag and analyzed via mass spectrometry, allowing for high‐throughput and unbiased identification of substrates of a particular kinase (Figure 8a") (Blethrow et al., 2008; Hengeveld et al., 2012). Such an approach has been used to identify substrates of Aurora B (Hengeveld et al., 2012) and Cdk1 (Blethrow et al., 2008) via mass spectrometry. One limitation of this approach is that due to poor cell permeability of the ATP analogue, these experiments are usually performed in cell extracts, not in whole cells. However, some cell‐permeable inhibitors have been identified, allowing for more physiological labeling conditions (Bishop et al., 1998, 1999, 2000). Additionally, further developments have allowed the generation of otherwise cell‐impermeable ATP analogues within cells (Hertz et al., 2013) and for the use of ATP analogues in animal models (Cibrián Uhalte et al., 2012; Soskis et al., 2012).
FIGURE 8.

Selected techniques in determining kinase function. (a) Bump‐hole technology. (a') A “bump” is chemically added to ATP (brown) to make it sterically bulky (red) and unable to bind to a wild‐type kinase (left, purple) (Circle 1). A corresponding “hole” is made in a modified kinase (right, blue) that allows it to bind to and use the modified ATP analogue (Circle 2). The catalytic efficiency of the modified kinase for unmodified ATP is usually weakened (Circle 3). (a") The modified kinase selectively incorporates the ATP analogue onto its substrates. These substrates are then enriched and identified via mass spectrometry‐based approaches. (b) Fluorescence resonance energy transfer (FRET) sensors. When the kinase is inactive (left), the FRET donor and acceptor are not within FRET distance and the FRET signal is low. When the kinase is active (right), the kinase phosphorylates a phosphosite (dark blue) in the linker sequence, causing the linker sequence to bind to the phospho‐binding domain (dark brown),which brings the FRET donor and acceptor close to each other, producing an FRET signal. (c) Localized kinase inhibition (LoKI). A kinase inhibitor (purple triangle) has poor location specificity, inhibiting its target kinase (red circular sector) at all locations and prohibiting the study of the kinase at a specific location (left, centrosome and kinetochores used as examples). In an LoKI construct (right), the inhibitor is covalently bound via an SNAP‐tag (brown) to a protein targeting domain (orange; centrosome‐targeting sequence used as example). The LoKI construct promotes inhibition of the kinase only at that domain (here, only at the centrosome and not at the kinetochores) [Color figure can be viewed at wileyonlinelibrary.com]
8.2. FRET sensors
In vitro kinase assays are useful for assessing kinase activity. These assays typically employ a means of measuring ATP hydrolysis over time (e.g., with colorimetric assays) or of measuring phosphorylation of the substrate over time (e.g., via intensity of the phosphoprotein band by western blotting or via radiolabeled phosphate incorporation on the substrate) (Johnson & Hunter, 2005; Xue & Tao, 2013). However, a read‐out of kinase activity within living cells had been largely intractable until the recent development of fluorescence resonance energy transfer (FRET)‐based sensors. From N‐terminus to C‐terminus, an FRET construct for kinase activity consists of an FRET donor (CFP), a phospho‐amino acid binding domain, a linker peptide with a phosphorylation motif for the kinase of interest, and an FRET acceptor (YFP) (Zhang, Ma, Taylor, & Tsien, 2001). When the kinase is inactive, excitation of the FRET donor results in a low or no FRET signal, as the FRET donor is usually not within the vicinity of the FRET acceptor for an FRET signal to occur (Figure 8b, left). When the kinase is active, however, it phosphorylates the phosphorylation motif within the linker peptide. The phosphorylated residue binds to the phospho‐amino acid binding domain, bringing the FRET acceptor closer to the FRET donor and increasing the FRET signal (Figure 8b, right). Assessing the FRET signal over time and distance allows for an understanding of when and where a kinase is active in situ. Such a reporter was used to determine the spatial and temporal activity of Aurora B (Fuller et al., 2008).
8.3. Localized inhibition
While small molecule inhibitors have been useful tools to elucidate kinase function, they lack precise temporal or spatial precision. For example, Plk1 is an enzyme with prominent roles at both the centrosome and the kinetochores. If one has an interest in the role of Plk1 at the centrosome, adding a Plk1 inhibitor will disturb Plk1 signaling at both the centrosome and the kinetochores, causing unwanted artifacts and complicating subsequent analysis (Figure 8c, left). Conversely, if one has an interest in the role of Plk1 at the kinetochore, a Plk1 inhibitor will disturb the function of Plk1 at the centrosome, inhibiting downstream mitotic spindle assembly and potentially introducing artifacts into the analysis of the kinetochore. One recent technological advancement that provides for localized inhibition of kinases employs the use of localized kinase inhibition (LoKI) (Bucko et al., 2019). An LoKI construct consists of two self‐labeling protein SNAP‐tags followed by a localization domain, such as the PACT domain for centrosome localization (Bucko et al., 2019) (Figure 8c, right; only one SNAP‐tag is shown in Figure 8c for clarity). The inhibitor of interest is chemically modified such that it is compatible with the self‐labeling protein tag (i.e., it will form a covalent bond with the protein tag) while still retaining its abilities to inhibit the kinase of interest. Adding the modified inhibitor to a cell expressing the LoKI construct will localize the inhibitor to the region of interest, allowing for acute and region‐specific inhibition of a kinase (Figure 8c, right). The original LoKI construct could be targeted to centrosomes, kinetochores, mitochondria, and the plasma membrane. In theory, LoKI can be targeted to any cellular location, as long as an appropriate localization signal is used. Similarly, while Plk1 and Aurora A inhibitors have been used for this purpose, any kinase inhibitor can be used with the LoKI system as long as the inhibitor can be chemically adapted to be compatible with the self‐labeling protein tag without the loss of function.
While the original LoKI construct used two SNAP‐tags, in theory, other self‐labeling protein tags, such as the HaloTag or CLIP‐tag, could be used as well (Gautier et al., 2008; Stagge, Mitronova, Belov, Wurm, & Jakobs, 2013). These tags could be used in the same LoKI construct to fine‐tune control of enzyme activity for downstream analyses. For example, a construct may consist of an LoKI construct with an SNAP‐tag and a HaloTag, used in conjunction with an SNAP‐modified Plk1 inhibitor and a Halo‐modified APC/C inhibitor. In this way, one LoKI construct will contain two (or more, if multiple self‐labeling protein domains are used) inhibitors, allowing for localized and specific inhibition of two (or more) biological processes within one region.
Other localization‐based kinase inhibition strategies have also been developed. In one example, Lera et al. generated two alleles of Plk1 within one cell (Lera et al., 2016). The first allele coded for an analogue sensitive construct of Plk1 with mutations that rendered it sensitive to the “bumpy” inhibitor 3‐MB‐PP1 but insensitive to the Plk1 ATP‐competitive inhibitor BI‐2536. This allele retained the wild‐type localization of Plk1. The other allele coded for a construct of Plk1 that was wild‐type in the ATP binding pocket but with varying C‐terminal localization domains that localized it to chromatin, the inner centromere and outer kinetochore, or the inner kinetochore. The localization‐specific construct of Plk1 was sensitive to the inhibitor BI‐2536 but insensitive to the inhibitor 3‐MB‐PP1. Thus, by using different combinations of inhibitors and localization constructs, the authors were able to selectively inhibit Plk1 at different cellular structures to understand the different functions of Plk1 at each structure. For example, the authors found that Plk1 localized to the inner centromere and outer kinetochore, but not Plk1 localized to chromatin or the inner kinetochore, was responsible for chromosome alignment in metaphase. These experiments were performed under 3‐MB‐PP1 inhibition (suppressing the activity of the analog sensitive Plk1 with wild‐type localization) to allow for precise control of Plk1 localization. Furthermore, the authors showed that the ability of Plk1 to localize to the inner centromere and outer kinetochore to promote chromosome alignment was due to Plk1 activity, as the phenotype was sensitive to BI‐2536 treatment.
9. CONCLUSIONS AND PERSPECTIVES
Although this review has focused on the phosphoregulation of mitotic spindle assembly, many other posttranslational modifications have key roles in regulating various aspects of early mitotic spindle assembly. The APC/C, for example, promotes mitotic progression by ubiquitylating and degrading substrates throughout mitosis (Pines, 2011; Primorac & Musacchio, 2013). Misregulation of the APC/C can lead to defects in spindle assembly and in cyclin B distribution on the mitotic spindle (Torres et al., 2010). Moreover, tubulin itself is heavily modified in mitosis. Besides phosphorylation, tubulin is also modified via detyrosination, acetylation, and polyglutamylation (Barisic & Maiato, 2016; Ferreira, Figueiredo, Orr, Lopes, & Maiato, 2018; Janke & Magiera, 2020). In some cases, these modifications promote or direct motor localization or activity, but understanding how these modifications “cross‐talk” and how the ensemble of these modifications gives rise to the complex dynamics of mitosis remains an open question (Barisic & Maiato, 2016; Ferreira et al., 2018; Janke & Magiera, 2020). Other modifications, such as the addition and removal of O‐linked N‐acetylglucosamine (O‐GlcNAc) to serine or threonine residues of nuclear or cytoplasmic proteins is essential for cell cycle progression. Misregulation of O‐GlcNAc cycling leads to defects in spindle size and shape (Slawson et al., 2005), in part due to misregulation of Cdk1 and Aurora B (Tan et al., 2013). Most of the identified O‐GlcNAc sites in mitosis are in close proximity or lie within known phosphorylation sites, causing altered phosphorylation of proteins associated with the mitotic spindle (Wang et al., 2010). Other modifications, including farnesylation (Moudgil et al., 2015) and myristoylation (Timm, Titus, Bernd, & Barroso, 1999), promote the localization of microtubule‐associated proteins to the mitotic spindle.
Thus, while phosphorylation and ubiquitylation have been heavily studied as regulators of the events leading to mitotic spindle assembly (Ong & Torres, 2019), other posttranslational modifications have received less attention, perhaps because of the difficulty of studying these modifications in a high‐throughput manner with mass spectrometry (An, Froehlich, & Lebrilla, 2009; Tate, Kalesh, Lanyon‐Hogg, Storck, & Thinon, 2015). Nonetheless, improvements in computation and analytical techniques have emerged to aid our understanding of the role of these modifications in spindle assembly (Klamer, Hsueh, Ayala‐Talavera, & Haab, 2019). Altogether, understanding how the myriad of modifications coordinate as a signaling network to give rise to the dynamic process of mitotic spindle assembly will be a prime future goal for the field of cell division.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Michelle C. Bradley, Jorge Z. Torres: Conceptualization. Joseph Y. Ong, Michelle C. Bradley: Writing – original draft. Joseph Y. Ong, Michelle C. Bradley, and Jorge Z. Torres: Writing – review and editing. Jorge Z. Torres: Supervision.
ACKNOWLEDGMENTS
The authors apologize to colleagues whose work could not be cited due to space constraints. This material is based upon work supported by the National Institutes of Health NIGMS grant number R01GM117475 and National Science Foundation grant number MCB1912837 to J. Z. T. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Institutes of Health or the National Science Foundation. This work was also supported by an NIH‐NIGMS Ruth L. Kirschstein National Research Service Award GM007185 and a Whitcome Pre‐doctoral Fellowship in Molecular Biology from the UCLA MBI to J. Y. O and M. C. B; and a National Science Foundation Graduate Research Fellowship DGE‐1650604 to J. Y. O. Figures 1, 2, 3, 4, 5, 6, 7 and parts of Figure 8c were created with BioRender.com.
Ong JY, Bradley MC, Torres JZ. Phospho‐regulation of mitotic spindle assembly. Cytoskeleton. 2020;77:558–578. 10.1002/cm.21649
Funding information Division of Graduate Education, Grant/Award Number: DGE‐1650604; Division of Molecular and Cellular Biosciences, Grant/Award Number: MCB1912837; National Institute of General Medical Sciences, Grant/Award Numbers: GM007185, R01GM117475
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- Agarwal, S. , Smith, K. P. , Zhou, Y. , Suzuki, A. , McKenney, R. J. , & Varma, D. (2018). Cdt1 stabilizes kinetochore‐microtubule attachments via an Aurora B kinase‐dependent mechanism. Journal of Cell Biology, 217(10), 3446–3463. 10.1083/JCB.201705127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, H. J. , Froehlich, J. W. , & Lebrilla, C. B. (2009). Determination of glycosylation sites and site‐specific heterogeneity in glycoproteins. Current Opinion in Chemical Biology, 13(4), 421–426. 10.1016/j.cbpa.2009.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, P. D. , Ovechkina, Y. , Morrice, N. , Wagenbach, M. , Duncan, K. , Wordeman, L. , & Swedlow, J. R. (2004). Aurora B regulates MCAK at the mitotic centromere. Developmental Cell, 6(2), 253–268. 10.1016/S1534-5807(04)00025-5 [DOI] [PubMed] [Google Scholar]
- Arquint, C. , & Nigg, E. A. (2016). The PLK4‐STIL‐SAS‐6 module at the core of centriole duplication. Biochemical Society Transactions, 44, 1253–1263. 10.1042/BST20160116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auckland, P. , Roscioli, E. , Coker, H. L. E. , & McAinsh, A. D. (2020). CENP‐F stabilizes kinetochore‐microtubule attachments and limits dynein stripping of corona cargoes. The Journal of Cell Biology, 219(5). 10.1083/jcb.201905018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahe, S. , Stierhof, Y. D. , Wilkinson, C. J. , Leiss, F. , & Nigg, E. A. (2005). Rootletin forms centriole‐associated filaments and functions in centrosome cohesion. Journal of Cell Biology, 171(1), 27–33. 10.1083/jcb.200504107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahmanyar, S. , Kaplan, D. D. , DeLuca, J. G. , Giddings, T. H. , O'Toole, E. T. , Winey, M. , … Barth, A. I. M. (2008). β‐Catenin is a Nek2 substrate involved in centrosome separation. Genes and Development, 22(1), 91–105. 10.1101/gad.1596308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahtz, R. , Seidler, J. , Arnold, M. , Haselmann‐Weiss, U. , Antony, C. , Lehmann, W. D. , & Hoffmann, I. (2012). GCP6 is a substrate of Plk4 and required for centriole duplication. Journal of Cell Science, 125(2), 486–496. 10.1242/jcs.093930 [DOI] [PubMed] [Google Scholar]
- Bajaj, R. , Bollen, M. , Peti, W. , & Page, R. (2018). KNL1 binding to PP1 and microtubules is mutually exclusive. Structure, 26(10), 1327–1336.e4. 10.1016/j.str.2018.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban, K. H. , Torres, J. Z. , Miller, J. J. , Mikhailov, A. , Nachury, M. V. , Tung, J. J. , … Jackson, P. K. (2007). The END network couples spindle pole assembly to inhibition of the anaphase‐promoting complex/cyclosome in early mitosis. Developmental Cell, 13(1), 29–42. 10.1016/j.devcel.2007.04.017 [DOI] [PubMed] [Google Scholar]
- Barisic, M. , & Maiato, H. (2016). The tubulin code: A navigation system for chromosomes during mitosis. Trends in Cell Biology, 26(10), 766–775. 10.1016/j.tcb.2016.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera, J. A. , Kao, L. R. , Hammer, R. E. , Seemann, J. , Fuchs, J. L. , & Megraw, T. L. (2010). CDK5RAP2 regulates centriole engagement and cohesion in mice. Developmental Cell, 18(6), 913–926. 10.1016/j.devcel.2010.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros, T. P. , Kinoshita, K. , Hyman, A. A. , & Raff, J. W. (2005). Aurora A activates D‐TACC‐Msps complexes exclusively at centrosomes to stabilize centrosomal microtubules. Journal of Cell Biology, 170(7), 1039–1046. 10.1083/jcb.200504097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss, R. , Sardon, T. , Vernos, I. , & Conti, E. (2003). Structural basis of Aurora‐A activation by TPX2 at the mitotic spindle. Molecular Cell, 12(4), 851–862. 10.1016/S1097-2765(03)00392-7 [DOI] [PubMed] [Google Scholar]
- Belham, C. , Roig, J. , Caldwell, J. A. , Aoyama, Y. , Kemp, B. E. , Comb, M. , & Avruch, J. (2003). A mitotic cascade of NIMA family kinases: Nercc1/Nek9 activates the Nek6 and Nek7 kinases. Journal of Biological Chemistry, 278(37), 34897–34909. 10.1074/jbc.M303663200 [DOI] [PubMed] [Google Scholar]
- Bertolotti, A. (2018). The split protein phosphatase system. Biochemical Journal, 475(23), 3707–3723. 10.1042/BCJ20170726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertran, M. T. , Sdelci, S. , Regué, L. , Avruch, J. , Caelles, C. , & Roig, J. (2011). Nek9 is a Plk1‐activated kinase that controls early centrosome separation through Nek6/7 and Eg5. EMBO Journal, 30(13), 2634–2647. 10.1038/emboj.2011.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel, K. G. , Mann, B. J. , Waitzman, J. S. , Poor, T. A. , Rice, S. E. , & Wadsworth, P. (2017). Src family kinase phosphorylation of the motor domain of the human kinesin‐5, Eg5. Cytoskeleton, 74(9), 317–330. 10.1002/cm.21380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling, P. , Kandels‐Lewis, S. , Telley, I. A. , Van Dijk, J. , Janke, C. , & Surrey, T. (2008). CLIP‐170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulinbinding sites. Journal of Cell Biology, 183(7), 1223–1233. 10.1083/jcb.200809190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, A. C. , Kung, C. Y. , Shah, K. , Witucki, L. , Shokat, K. M. , & Liu, Y. (1999). Generation of monospecific nanomolar tyrosine kinase inhibitors via a chemical genetic approach. Journal of the American Chemical Society, 121(4), 627–631. 10.1021/ja983267v [DOI] [Google Scholar]
- Bishop, A. C. , Shah, K. , Liu, Y. , Witucki, L. , Kung, C. Y. , & Shokat, K. M. (1998). Design of allele‐specific inhibitors to probe protein kinase signaling. Current Biology, 8(5), 257–266. 10.1016/S0960-9822(98)70198-8 [DOI] [PubMed] [Google Scholar]
- Bishop, A. C. , Ubersax, J. A. , Pøtsch, D. T. , Matheos, D. P. , Gray, N. S. , Blethrow, J. , … Shokat, K. M. (2000). A chemical switch for inhibitor‐sensitive alleles of any protein kinase. Nature, 407(6802), 395–401. 10.1038/35030148 [DOI] [PubMed] [Google Scholar]
- Blethrow, J. D. , Glavy, J. S. , Morgan, D. O. , & Shokat, K. M. (2008). Covalent capture of kinase‐specific phosphopeptides reveals Cdk1‐cyclin B substrates. Proceedings of the National Academy of Sciences of the United States of America, 105(5), 1442–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner, M. K. , Haase, J. , Saunders, H. , Gupta, H. , Li, B. I. , & Kelly, A. E. (2020). The Borealin dimerization domain interacts with Sgo1 to drive Aurora B–mediated spindle assembly. Molecular Biology of the Cell, 31(20), 2207–2218. 10.1091/mbc.E20-05-0341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brittle, A. L. , & Ohkura, H. (2005). Centrosome maturation: Aurora lights the way to the poles. Current Biology, 15, R880–R882. 10.1016/j.cub.2005.10.022 [DOI] [PubMed] [Google Scholar]
- Brownlee, C. W. , Klebba, J. E. , Buster, D. W. , & Rogers, G. C. (2011). The protein phosphatase 2A regulatory subunit twins stabilizes Plk4 to induce centriole amplification. Journal of Cell Biology, 195(2), 231–243. 10.1083/jcb.201107086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucko, P. J. , Lombard, C. K. , Rathbun, L. , Garcia, I. , Bhat, A. , Wordeman, L. , … Scott, J. D. (2019). Subcellular drug targeting illuminates local kinase action. eLife, 8 10.7554/eLife.52220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, S. G. , Mukherjee, M. , Sabir, S. , Joseph, N. , Gutiérrez‐Caballero, C. , Richards, M. W. , … Bayliss, R. (2018). Mitotic spindle association of TACC3 requires Aurora‐A‐dependent stabilization of a cryptic α‐helix. The EMBO Journal, 37(8). 10.15252/embj.201797902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, S. G. , Peset, I. , Joseph, N. , Cavazza, T. , Vernos, I. , Pfuhl, M. , … Bayliss, R. (2015). Aurora‐A‐dependent control of TACC3 influences the rate of mitotic spindle assembly. PLoS Genetics, 11(7), e1005345 10.1371/journal.pgen.1005345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahu, J. , Olichon, A. , Hentrich, C. , Schek, H. , Drinjakovic, J. , Zhang, C. , … Surrey, T. (2008). Phosphorylation by Cdk1 increases the binding of Eg5 to microtubules in vitro and in Xenopus egg extract spindles. PLoS One, 3(12), e3936 10.1371/journal.pone.0003936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena, M. , Wheelock, M. , Funabiki, H. , & Earnshaw, W. C. (2012). The chromosomal passenger complex (CPC): From easy rider to the godfather of mitosis. Nature Reviews Molecular Cell Biology, 13(12), 789–803. 10.1038/nrm3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, Y. W. , Jeyaprakash, A. A. , Nigg, E. A. , & Santamaria, A. (2012). Aurora B controls kinetochore‐microtubule attachments by inhibiting Ska complex‐KMN network interaction. Journal of Cell Biology, 196(5), 563–571. 10.1083/jcb.201109001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I. M. , Chappie, J. S. , Wilson‐Kubalek, E. M. , & Desai, A. (2006). The conserved KMN network constitutes the Core microtubule‐binding site of the kinetochore. Cell, 127(5), 983–997. 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Chen, C. , Xu, Z. , Zhang, T. , Lin, L. , Lu, M. , Xie, C. , & Yu, X. (2019). Cep85 relays Plk1 activity to phosphorylated Nek2A for its timely activation in centrosome disjunction. IScience, 11, 114–133. 10.1016/j.isci.2018.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibrián Uhalte, E. , Kirchner, M. , Hellwig, N. , Allen, J. J. , Donat, S. , Shokat, K. M. , … Abdelilah‐Seyfried, S. (2012). In vivo conditions to identify Prkci phosphorylation targets using the analog‐sensitive kinase method in zebrafish. PLoS One, 7(6), e40000 10.1371/journal.pone.0040000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha‐Ferreira, I. , Bento, I. , Pimenta‐Marques, A. , Jana, S. C. , Lince‐Faria, M. , Duarte, P. , … Bettencourt‐Dias, M. (2013). Regulation of autophosphorylation controls PLK4 self‐destruction and centriole number. Current Biology, 23(22), 2245–2254. 10.1016/j.cub.2013.09.037 [DOI] [PubMed] [Google Scholar]
- DeLuca, J. G. , Gall, W. E. , Ciferri, C. , Cimini, D. , Musacchio, A. , & Salmon, E. D. (2006). Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell, 127(5), 969–982. 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- Deretic, J. , Kerr, A. , & Welburn, J. P. I. (2019). A rapid computational approach identifies SPICE1 as an Aurora kinase substrate. Molecular Biology of the Cell, 30(3), 312–323. 10.1091/mbc.E18-08-0495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitru, A. M. G. , Rusin, S. F. , Clark, A. E. M. , Kettenbach, A. N. , & Compton, D. A. (2017). Cyclin a/Cdk1 modulates Plk1 activity in prometaphase to regulate kinetochore‐microtubule attachment stability. eLife, 6 10.7554/eLife.29303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev, N. S. , Tzolovsky, G. , Lipinszki, Z. , Abdelaziz, M. , Debski, J. , Dadlez, M. , & Glover, D. M. (2017). Two‐step phosphorylation of Ana2 by Plk4 is required for the sequential loading of Ana2 and Sas6 to initiate procentriole formation. Open Biology, 7(12), 170247 10.1098/rsob.170247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eibes, S. , Gallisà‐Suñé, N. , Rosas‐Salvans, M. , Martínez‐Delgado, P. , Vernos, I. , & Roig, J. (2018). Nek9 phosphorylation defines a new role for TPX2 in Eg5‐dependent centrosome separation before nuclear envelope breakdown. Current Biology, 28(1), 121–129.e4. 10.1016/j.cub.2017.11.046 [DOI] [PubMed] [Google Scholar]
- Elowe, S. , Hümmer, S. , Uldschmid, A. , Li, X. , & Nigg, E. A. (2007). Tension‐sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore‐microtubule interactions. Genes and Development, 21(17), 2205–2219. 10.1101/gad.436007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahs, S. , Lujan, P. , & Köhn, M. (2016). Approaches to study phosphatases. ACS Chemical Biology, 11(11), 2944–2961. 10.1021/acschembio.6b00570 [DOI] [PubMed] [Google Scholar]
- Ferreira, L. T. , Figueiredo, A. C. , Orr, B. , Lopes, D. , & Maiato, H. (2018). Dissecting the role of the tubulin code in mitosis. Methods in Cell Biology, 144, 33–74. 10.1016/bs.mcb.2018.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley, E. A. , Maldonado, M. , & Kapoor, T. M. (2011). Formation of stable attachments between kinetochores and microtubules depends on the B56‐PP2A phosphatase. Nature Cell Biology, 13(10), 1265–1271. 10.1038/ncb2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier, M. , Orpinell, M. , Grauffel, C. , Scheer, E. , Garnier, J. M. , Ye, T. , … Tora, L. (2016). KAT2A/KAT2B‐targeted acetylome reveals a role for PLK4 acetylation in preventing centrosome amplification. Nature Communications, 7(1), 1–16. 10.1038/ncomms13227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, J. , Bian, M. , Xin, G. , Deng, Z. , Luo, J. , Guo, X. , … Zhang, C. (2015). TPX2 phosphorylation maintains metaphase spindle length by regulating microtubule flux. Journal of Cell Biology, 210(3), 373–383. 10.1083/jcb.201412109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, W. , Tao, W. , Zheng, P. , Fu, J. , Bian, M. , Jiang, Q. , … Zhang, C. (2010). Clathrin recruits phosphorylated TACC3 to spindle poles for bipolar spindle assembly and chromosome alignment. Journal of Cell Science, 123(21), 3645–3651. 10.1242/jcs.075911 [DOI] [PubMed] [Google Scholar]
- Fu, Z. , Malureanu, L. , Huang, J. , Wang, W. , Li, H. , van Deursen, J. M. , … Chen, J. (2008). Plk1‐dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nature Cell Biology, 10(9), 1076–1082. 10.1038/ncb1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa, T. , & Earnshaw, W. C. (2014). The centromere: Chromatin foundation for the kinetochore machinery. Developmental Cell, 30(5), 496–508. 10.1016/j.devcel.2014.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller, B. G. , Lampson, M. A. , Foley, E. A. , Rosasco‐Nitcher, S. , Le, K. V. , Tobelmann, P. , … Kapoor, T. M. (2008). Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature, 453(7198), 1132–1136. 10.1038/nature06923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier, A. , Juillerat, A. , Heinis, C. , Corrêa, I. R. , Kindermann, M. , Beaufils, F. , & Johnsson, K. (2008). An engineered protein tag for multiprotein labeling in living cells. Chemistry and Biology, 15(2), 128–136. 10.1016/j.chembiol.2008.01.007 [DOI] [PubMed] [Google Scholar]
- Giet, R. , Uzbekov, R. , Cubizolles, F. , Le Guellec, K. , & Prigent, C. (1999). The Xenopus laevis aurora‐related protein kinase pEg2 associates with and phosphorylates the kinesin‐related protein XlEg5. Journal of Biological Chemistry, 274(21), 15005–15013. 10.1074/jbc.274.21.15005 [DOI] [PubMed] [Google Scholar]
- Giubettini, M. , Asteriti, I. A. , Scrofani, J. , De Luca, M. , Lindon, C. , Lavia, P. , & Guarguaglini, G. (2011). Control of Aurora‐A stability through interaction with TPX2. Journal of Cell Science, 124(1), 113–122. 10.1242/jcs.075457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, D. J. , Resio, B. , & Pellman, D. (2012). Causes and consequences of aneuploidy in cancer. Nature Reviews Genetics, 13(3), 189–203. 10.1038/nrg3123 [DOI] [PubMed] [Google Scholar]
- Gouveia, S. M. , Zitouni, S. , Kong, D. , Duarte, P. , Gomes, B. F. , Sousa, A. L. , … Bettencourt‐Dias, M. (2019). PLK4 is a microtubule‐associated protein that self‐assembles promoting de novo MTOC formation. Journal of Cell Science, 132(4), jcs219501 10.1242/jcs.219501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guderian, G. , Westendorf, J. , Uldschmid, A. , & Nigg, E. A. (2010). Plk4 trans‐autophosphorylation regulates centriole number by controlling βTrCP‐mediated degradation. Journal of Cell Science, 123(13), 2163–2169. 10.1242/jcs.068502 [DOI] [PubMed] [Google Scholar]
- Habedanck, R. , Stierhof, Y. D. , Wilkinson, C. J. , & Nigg, E. A. (2005). The polo kinase Plk4 functions in centriole duplication. Nature Cell Biology, 7(11), 1140–1146. 10.1038/ncb1320 [DOI] [PubMed] [Google Scholar]
- Hames, R. S. , & Fry, A. M. (2002). Alternative splice variants of the human centrosome kinase Nek2 exhibit distinct patterns of expression in mitosis. Biochemical Journal, 361(1), 77–85. 10.1042/0264-6021:3610077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanafusa, H. , Kedashiro, S. , Tezuka, M. , Funatsu, M. , Usami, S. , Toyoshima, F. , & Matsumoto, K. (2015). PLK1‐dependent activation of LRRK1 regulates spindle orientation by phosphorylating CDK5RAP2. Nature Cell Biology, 17(8), 1024–1035. 10.1038/ncb3204 [DOI] [PubMed] [Google Scholar]
- Hanafusa, H. , Yagi, T. , Ikeda, H. , Hisamoto, N. , Nishioka, T. , Kaibuchi, K. , … Matsumoto, K. (2019). LRRK1 phosphorylation of Rab7 at S72 links trafficking of EGFR‐containing endosomes to its effector RILP. Journal of Cell Science, 132(11), jcs228809 10.1242/jcs.228809 [DOI] [PubMed] [Google Scholar]
- Hardy, T. , Lee, M. , Hames, R. S. , Prosser, S. L. , Cheary, D. M. , Samant, M. D. , … Fry, A. M. (2014). Multisite phosphorylation of C‐Nap1 releases it from Cep135 to trigger centrosome disjunction. Journal of Cell Science, 127(11), 2493–2506. 10.1242/jcs.142331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch, E. M. , Kulukian, A. , Holland, A. J. , Cleveland, D. W. , & Stearns, T. (2010). Cep152 interacts with Plk4 and is required for centriole duplication. Journal of Cell Biology, 191(4), 721–729. 10.1083/jcb.201006049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward, D. , Bancroft, J. , Mangat, D. , Alfonso‐Pérez, T. , Dugdale, S. , McCarthy, J. , … Gruneberg, U. (2019). Checkpoint signaling and error correction require regulation of the MPS1 T‐loop by PP2A‐B56. Journal of Cell Biology, 218(10), 3188–3199. 10.1083/JCB.201905026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, R. , Huang, N. , Bao, Y. , Zhou, H. , Teng, J. , & Chen, J. (2013). LRRC45 is a centrosome linker component required for centrosome cohesion. Cell Reports, 4(6), 1100–1107. 10.1016/j.celrep.2013.08.005 [DOI] [PubMed] [Google Scholar]
- Helps, N. R. , Luo, X. , Barker, H. M. , & Cohen, P. T. W. (2000). NIMA‐related kinase 2 (Nek2), a cell‐cycle‐regulated protein kinase localized to centrosomes, is complexed to protein phosphatase 1. Biochemical Journal, 349(2), 509–518. 10.1042/0264-6021:3490509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengeveld, R. C. C. , Hertz, N. T. , Vromans, M. J. M. , Zhang, C. , Burlingame, A. L. , Shokat, K. M. , & Lens, S. M. A. (2012). Development of a chemical genetic approach for human Aurora B kinase identifies novel substrates of the chromosomal passenger complex. Molecular & Cellular Proteomics, 11(5), 47–59. 10.1074/mcp.M111.013912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz, N. T. , Berthet, A. , Sos, M. L. , Thorn, K. S. , Burlingame, A. L. , Nakamura, K. , & Shokat, K. M. (2013). A neo‐substrate that amplifies catalytic activity of Parkinson's‐disease‐related kinase PINK1. Cell, 154(4), 737–747. 10.1016/j.cell.2013.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe, E. H. , & Sluder, G. (2001). “It takes two to tango”: Understanding how centrosome duplication is regulated throughout the cell cycle. Genes and Development, 15(10), 1167–1181. 10.1101/gad.894001 [DOI] [PubMed] [Google Scholar]
- Honda, R. , Körner, R. , & Nigg, E. A. (2003). Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Molecular Biology of the Cell, 14(8), 3325–3341. 10.1091/mbc.E02-11-0769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood, F. E. , Williams, S. J. , Burgess, S. G. , Richards, M. W. , Roth, D. , Straube, A. , … Royle, S. J. (2013). Coordination of adjacent domains mediates TACC3‐ch‐TOG‐clathrin assembly and mitotic spindle binding. Journal of Cell Biology, 202(3), 463–478. 10.1083/jcb.201211127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H. , Hittle, J. , Zappacosta, F. , Annan, R. S. , Hershko, A. , & Yen, T. J. (2008). Phosphorylation sites in BubR1 that regulate kinetochore attachment, tension, and mitotic exit. Journal of Cell Biology, 183(4), 667–680. 10.1083/jcb.200805163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y. , Li, T. , Ems‐McClung, S. C. , Walczak, C. E. , Prigent, C. , Zhu, X. , … Zheng, Y. (2018). Aurora A activation in mitosis promoted by BuGZ. Journal of Cell Biology, 217(1), 107–116. 10.1083/jcb.201706103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda, M. , & Tanaka, K. (2017). Plk1 bound to Bub1 contributes to spindle assembly checkpoint activity during mitosis. Scientific Reports, 7(1), 1–15. 10.1038/s41598-017-09114-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke, C. , & Magiera, M. M. (2020). The tubulin code and its role in controlling microtubule properties and functions. In Nature Reviews Molecular Cell Biology. Nature Research., 21, 307–326. 10.1038/s41580-020-0214-3 [DOI] [PubMed] [Google Scholar]
- Jia, L. , Li, B. , & Yu, H. (2016). The Bub1–Plk1 kinase complex promotes spindle checkpoint signalling through Cdc20 phosphorylation. Nature Communications, 7, 10818 10.1038/NCOMMS10818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johmura, Y. , Soung, N. K. , Park, J. E. , Yu, L. R. , Zhou, M. , Bang, J. K. , … Lee, K. S. (2011). Regulation of microtubule‐based microtubule nucleation by mammalian polo‐like kinase 1. Proceedings of the National Academy of Sciences of the United States of America, 108(28), 11446–11451. 10.1073/pnas.1106223108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, S. A. , & Hunter, T. (2005). Kinomics: Methods for deciphering the kinome. Nature Methods, 2(1), 17–25. 10.1038/nmeth731 [DOI] [PubMed] [Google Scholar]
- Joukov, V. , & de Nicolo, A. (2018). Aurora‐PLK1 cascades as key signaling modules in the regulation of mitosis. Science Signaling, 11(543), eaar4195 10.1126/scisignal.aar4195 [DOI] [PubMed] [Google Scholar]
- Joukov, V. , de Nicolo, A. , Rodriguez, A. , Walter, J. C. , & Livingston, D. M. (2010). Centrosomal protein of 192 kDa (Cep192) promotes centrosome‐driven spindle assembly by engaging in organelle‐specific Aurora A activation. Proceedings of the National Academy of Sciences of the United States of America, 107(49), 21022–21027. 10.1073/pnas.1014664107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joukov, V. , Walter, J. C. , & De Nicolo, A. (2014). The Cep192‐organized Aurora A‐Plk1 Cascade is essential for centrosome cycle and bipolar spindle assembly. Molecular Cell, 55(4), 578–591. 10.1016/j.molcel.2014.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, Y. H. , Park, J. E. , Yu, L. R. , Soung, N. K. , Yun, S. M. , Bang, J. K. , … Lee, K. S. (2006). Self‐regulated Plk1 recruitment to kinetochores by the Plk1‐PBIP1 interaction is critical for proper chromosome segregation. Molecular Cell, 24(3), 409–422. 10.1016/j.molcel.2006.10.016 [DOI] [PubMed] [Google Scholar]
- Kapitein, L. C. , Peterman, E. J. G. , Kwok, B. H. , Kim, J. H. , Kapoor, T. M. , & Schmidt, C. F. (2005). The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature, 435(7038), 114–118. 10.1038/nature03503 [DOI] [PubMed] [Google Scholar]
- Kedashiro, S. , Pastuhov, S. I. , Nishioka, T. , Watanabe, T. , Kaibuchi, K. , Matsumoto, K. , & Hanafusa, H. (2015). LRRK1‐phosphorylated CLIP‐170 regulates EGFR trafficking by recruiting p150Glued to microtubule plus ends. Journal of Cell Science, 128(2), 385–396. 10.1242/jcs.161547 [DOI] [PubMed] [Google Scholar]
- Kelly, A. E. , Ghenoiu, C. , Xue, J. Z. , Zierhut, C. , Kimura, H. , & Funabiki, H. (2010). Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science, 330(6001), 235–239. 10.1126/science.1189505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshri, R. , Rajeevan, A. , & Kotak, S. (2020). PP2A–B55γ counteracts Cdk1 and regulates proper spindle orientation through the cortical dynein adaptor NuMA. Journal of Cell Science, 133(14), jcs243857 10.1242/jcs.243857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T. S. , Park, J. E. , Shukla, A. , Choi, S. , Murugan, R. N. , Lee, J. H. , … Lee, K. S. (2013). Hierarchical recruitment of Plk4 and regulation of centriole biogenesis by two centrosomal scaffolds, Cep192 and Cep152. Proceedings of the National Academy of Sciences of the United States of America, 110(50), E4849–E4857. 10.1073/pnas.1319656110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, K. , Noetzel, T. L. , Pelletier, L. , Mechtler, K. , Drechsel, D. N. , Schwager, A. , … Hyman, A. A. (2005). Aurora A phosphorylation of TACC3/maskin is required for centrosome‐dependent microtubule assembly in mitosis. Journal of Cell Biology, 170(7), 1047–1055. 10.1083/jcb.200503023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa, D. , Busso, C. , Flückiger, I. , & Gönczy, P. (2009). Phosphorylation of SAS‐6 by ZYG‐1 is critical for centriole formation in C. elegans embryos. Developmental Cell, 17(6), 900–907. 10.1016/j.devcel.2009.11.002 [DOI] [PubMed] [Google Scholar]
- Klamer, Z. , Hsueh, P. , Ayala‐Talavera, D. , & Haab, B. (2019). Deciphering protein glycosylation by computational integration of on‐chip profiling, glycan‐array data, and mass spectrometry. Molecular and Cellular Proteomics, 18(1), 28–40. 10.1074/mcp.RA118.000906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein‐Sohn, J. , Westendorf, J. , Le Clech, M. , Habedanck, R. , Stierhof, Y. D. , & Nigg, E. A. (2007). Plk4‐induced centriole biogenesis in human cells. Developmental Cell, 13(2), 190–202. 10.1016/j.devcel.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Kotak, S. , Busso, C. , & Gönczy, P. (2013). NuMA phosphorylation by CDK1 couples mitotic progression with cortical dynein function. The EMBO Journal, 32(18), 2517–2529. 10.1038/emboj.2013.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak, S. , Busso, C. , Gonczy, P. , & Gönczy, P. (2014). NuMA interacts with phosphoinositides and links the mitotic spindle with the plasma membrane. The EMBO Journal, 33(16), 1815–1830. 10.15252/embj.201488147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse, T. , Zhang, G. , Larsen, M. S. Y. , Lischetti, T. , Streicher, W. , Nielsen, T. K. , … Nilsson, J. (2013). Direct binding between BubR1 and B56‐PP2A phosphatase complexes regulate mitotic progression. Journal of Cell Science, 126(5), 1086–1092. 10.1242/jcs.122481 [DOI] [PubMed] [Google Scholar]
- Lan, W. , Zhang, X. , Kline‐Smith, S. L. , Rosasco, S. E. , Barrett‐Wilt, G. A. , Shabanowitz, J. , … Stukenberg, P. T. (2004). Aurora B phosphorylates Centromeric MCAK and regulates its localization and microtubule Depolymerization activity. Current Biology, 14(4), 273–286. 10.1016/j.cub.2004.01.055 [DOI] [PubMed] [Google Scholar]
- Lawrence, E. J. , Zanic, M. , & Rice, L. M. (2020). CLASPs at a glance. Journal of Cell Science, 133(8), jcs243097 10.1242/jcs.243097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B. H. , Schwager, F. , Meraldi, P. , & Gotta, M. (2018). p37/UBXN2B regulates spindle orientation by limiting cortical NuMA recruitment via PP1/repo‐man. Journal of Cell Biology, 217(2), 483–593. 10.1083/jcb.201707050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K. , & Rhee, K. (2011). PLK1 phosphorylation of pericentrin initiates centrosome maturation at the onset of mitosis. Journal of Cell Biology, 195(7), 1093–1101. 10.1083/jcb.201106093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lera, R. F. , Potts, G. K. , Suzuki, A. , Johnson, J. M. , Salmon, E. D. , Coon, J. J. , & Burkard, M. E. (2016). Decoding polo‐like kinase 1 signaling along the kinetochore‐centromere axis. Nature Chemical Biology, 12(6), 411–418. 10.1038/nchembio.2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Liu, X. S. , Yang, X. , Wang, Y. , Wang, Y. , Turner, J. R. , & Liu, X. (2010). Phosphorylation of CLIP‐170 by Plk1 and CK2 promotes timely formation of kinetochore‐microtubule attachments. EMBO Journal, 29(17), 2953–2965. 10.1038/emboj.2010.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C. H. , Hu, C. K. , & Shih, H. M. (2010). Clathrin heavy chain mediates TACC3 targeting to mitotic spindles to ensure spindle stability. Journal of Cell Biology, 189(7), 1097–1105. 10.1083/jcb.200911120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlepage, L. E. , Wu, H. , Andresson, T. , Deanehan, J. K. , Amundadottir, L. T. , & Ruderman, J. V. (2002). Identification of phosphorylated residues that affect the activity of the mitotic kinase Aurora‐A. Proceedings of the National Academy of Sciences of the United States of America, 99(24), 15440–15445. 10.1073/pnas.202606599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D. , Davydenko, O. , & Lampson, M. A. (2012). Polo‐like kinase‐1 regulates kinetochore‐microtubule dynamics and spindle checkpoint silencing. Journal of Cell Biology, 198(4), 491–499. 10.1083/jcb.201205090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. S. , Song, B. , Tang, J. , Liu, W. , Kuang, S. , & Liu, X. (2012). Plk1 phosphorylates Sgt1 at the kinetochores to promote timely kinetochore‐microtubule attachment. Molecular and Cellular Biology, 32(19), 4053–4067. 10.1128/mcb.00516-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Zhang, Z. , Liang, H. , Zhao, X. , Liang, L. , Wang, G. , … Yin, Y. (2017). Protein phosphatase 2A (PP2A) regulates EG5 to control mitotic progression. Scientific Reports, 7(1), 1–13. 10.1038/s41598-017-01915-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Shah, K. , Yang, F. , Witucki, L. , & Shokat, K. M. (1998). Engineering Src family protein kinases with unnatural nucleotide specificity. Chemistry and Biology, 5(2), 91–101. 10.1016/S1074-5521(98)90143-0 [DOI] [PubMed] [Google Scholar]
- Macedo, J. C. , Vaz, S. , Bakker, B. , Ribeiro, R. , Bakker, P. L. , Escandell, J. M. , … Logarinho, E. (2018). FoxM1 repression during human aging leads to mitotic decline and aneuploidy‐driven full senescence. Nature Communications, 9(1), 2834 10.1038/s41467-018-05258-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macůrek, L. , Lindqvist, A. , Lim, D. , Lampson, M. A. , Klompmaker, R. , Freire, R. , … Medema, R. H. (2008). Polo‐like kinase‐1 is activated by Aurora A to promote checkpoint recovery. Nature, 455(7209), 119–123. 10.1038/nature07185 [DOI] [PubMed] [Google Scholar]
- Maia, A. R. R. , Garcia, Z. , Kabeche, L. , Barisic, M. , Maffini, S. , Macedo‐Ribeiro, S. , … Maiato, H. (2012). Cdk1 and Plk1 mediate a CLASP2 phospho‐switch that stabilizes kinetochore‐microtubule attachments. Journal of Cell Biology, 199(2), 285–301. 10.1083/jcb.201203091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney, T. , Hunter, A. W. , Wagenbach, M. , & Wordeman, L. (1998). Mitotic centromere‐associated kinesin is important for anaphase chromosome segregation. Journal of Cell Biology, 142(3), 787–801. 10.1083/jcb.142.3.787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniswami, R. R. , Prashanth, S. , Karanth, A. V. , Koushik, S. , Govindaraj, H. , Mullangi, R. , … Jegatheesan, S. K. (2018). PLK4: A link between centriole biogenesis and cancer. Expert Opinion on Therapeutic Targets, 22(1), 59–73. 10.1080/14728222.2018.1410140 [DOI] [PubMed] [Google Scholar]
- Mardin, B. R. , Agircan, F. G. , Lange, C. , & Schiebel, E. (2011). Plk1 controls the Nek2A‐PP1γ antagonism in centrosome disjunction. Current Biology, 21(13), 1145–1151. 10.1016/j.cub.2011.05.047 [DOI] [PubMed] [Google Scholar]
- Mardin, B. R. , Lange, C. , Baxter, J. E. , Hardy, T. , Scholz, S. R. , Fry, A. M. , & Schiebel, E. (2010). Components of the hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nature Cell Biology, 12(12), 1166–1176. 10.1038/ncb2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, L. , Park, J.‐E. , Kim, T.‐S. , Lee, E. H. , Park, S.‐Y. , Zhou, M. , … Lee, K. S. (2015). Bimodal interaction of mammalian polo‐like kinase 1 and a Centrosomal scaffold, Cep192, in the regulation of bipolar spindle formation. Molecular and Cellular Biology, 35(15), 2626–2640. 10.1128/mcb.00068-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meppelink, A. , Kabeche, L. , Vromans, M. J. M. , Compton, D. A. , & Lens, S. M. A. (2015). Shugoshin‐1 balances Aurora B kinase activity via PP2A to promote chromosome bi‐orientation. Cell Reports, 11(4), 508–515. 10.1016/j.celrep.2015.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi, P. , & Nigg, E. A. (2001). Centrosome cohesion is regulated by a balance of kinase and phosphatase activities. Journal of Cell Science, 114(Pt 20), 3749–3757. [DOI] [PubMed] [Google Scholar]
- Moniz, L. , Dutt, P. , Haider, N. , & Stambolic, V. (2011). Nek family of kinases in cell cycle, checkpoint control and cancer. Cell Division, 6, 18 10.1186/1747-1028-6-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, D. , Yano, Y. , Toyo‐oka, K. , Yoshida, N. , Yamada, M. , Muramatsu, M. , … Hirotsune, S. (2007). NDEL1 phosphorylation by Aurora‐A kinase is essential for Centrosomal maturation, separation, and TACC3 recruitment. Molecular and Cellular Biology, 27(1), 352–367. 10.1128/mcb.00878-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moudgil, D. K. , Westcott, N. , Famulski, J. K. , Patel, K. , Macdonald, D. , Hang, H. , & Chan, G. K. T. (2015). A novel role of farnesylation in targeting a mitotic checkpoint protein, human spindly, to kinetochores. Journal of Cell Biology, 208(7), 881–896. 10.1083/jcb.201412085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura, M. , Osswald, M. , Leça, N. , Barbosa, J. , Pereira, A. J. , Maiato, H. , … Conde, C. (2017). Protein phosphatase 1 inactivates Mps1 to ensure efficient spindle assembly checkpoint silencing. eLife, 6 10.7554/eLife.25366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer, T. C. , Clutario, K. M. , Lambrus, B. G. , Daggubati, V. , & Holland, A. J. (2015). Binding of STIL to Plk4 activates kinase activity to promote centriole assembly. Journal of Cell Biology, 209(6), 863–878. 10.1083/jcb.201502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer, T. C. , & Holland, A. J. (2019). Plk4 promotes centriole duplication by phosphorylating stil to link the procentriole cartwheel to the microtubule wall. eLife, 8 10.7554/eLife.46054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg, E. A. , & Holland, A. J. (2018). Once and only once: Mechanisms of centriole duplication and their deregulation in diseases. Nature Reviews Molecular Cell Biology, 19(5), 297–312. 10.1038/nrm.2017.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino, M. , Kurasawa, Y. , Evans, R. , Lin, S. H. , Brinkley, B. R. , & Yu‐Lee, L. y. (2006). NudC is required for Plk1 targeting to the kinetochore and chromosome Congression. Current Biology, 16(14), 1414–1421. 10.1016/j.cub.2006.05.052 [DOI] [PubMed] [Google Scholar]
- Obenauer, J. C. , Cantley, L. C. , & Yaffe, M. B. (2003). Scansite 2.0: Proteome‐wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Research, 31(13), 3635–3641. 10.1093/nar/gkg584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, M. , Ashikawa, T. , Nozaki, Y. , Kozuka‐Hata, H. , Goto, H. , Inagaki, M. , … Kitagawa, D. (2014). Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nature Communications, 5, 5267 10.1038/ncomms6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong, J. Y. , & Torres, J. Z. (2019). Dissecting the mechanisms of cell division. Journal of Biological Chemistry, 294(30), 11382–11390. 10.1074/jbc.AW119.008149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshimori, N. , Ohsugi, M. , & Yamamoto, T. (2006). The Plk1 target Kizuna stabilizes mitotic centrosomes to ensure spindle bipolarity. Nature Cell Biology, 8(10), 1095–1101. 10.1038/ncb1474 [DOI] [PubMed] [Google Scholar]
- Paintrand, M. , Moudjou, M. , Delacroix, H. , & Bornens, M. (1992). Centrosome organization and centriole architecture: Their sensitivity to divalent cations. Journal of Structural Biology, 108(2), 107–128. 10.1016/1047-8477(92)90011-X [DOI] [PubMed] [Google Scholar]
- Park, J.‐E. , Zhang, L. , Bang, J. K. , Andresson, T. , DiMaio, F. , & Lee, K. S. (2019). Phase separation of polo‐like kinase 4 by autoactivation and clustering drives centriole biogenesis. Nature Communications, 10(1), 4959 10.1038/s41467-019-12619-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschal, C. R. , Maciejowski, J. , & Jallepalli, P. V. (2012). A stringent requirement for PlK1 T210 phosphorylation during K‐fiber assembly and chromosome congression. Chromosoma, 121(6), 565–572. 10.1007/s00412-012-0375-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peset, I. , & Vernos, I. (2008). The TACC proteins: TACC‐ling microtubule dynamics and centrosome function. Trends in Cell Biology, 18(8), 379–388. 10.1016/j.tcb.2008.06.005 [DOI] [PubMed] [Google Scholar]
- Pines, J. (2011). Cubism and the cell cycle: The many faces of the APC/C. Nature Reviews Molecular Cell Biology, 12(7), 427–438. 10.1038/nrm3132 [DOI] [PubMed] [Google Scholar]
- Primorac, I. , & Musacchio, A. (2013). Panta rhei: The APC/C at steady state. The Journal of Cell Biology, 201(2), 177–189. 10.1083/jcb.201301130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser, S. L. , & Pelletier, L. (2017). Mitotic spindle assembly in animal cells: A fine balancing act. Nature Reviews Molecular Cell Biology, 18(3), 187–201. 10.1038/nrm.2016.162 [DOI] [PubMed] [Google Scholar]
- Qi, W. , Tang, Z. , & Yu, H. (2006). Phosphorylation‐ and polo‐box‐dependent binding of Plk1 to Bub1 is required for the kinetochore localization of Plk1. Molecular Biology of the Cell, 17(8), 3705–3716. 10.1091/mbc.E06-03-0240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapley, J. , Nicolàs, M. , Groen, A. , Regué, L. , Bertran, M. T. , Caelles, C. , … Roig, J. (2008). The NIMA‐family kinase Nek6 phosphorylates the kinesin Eg5 at a novel site necessary for mitotic spindle formation. Journal of Cell Science, 121(23), 3912–3921. 10.1242/jcs.035360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redli, P. M. , Gasic, I. , Meraldi, P. , Nigg, E. A. , & Santamaria, A. (2016). The Ska complex promotes Aurora B activity to ensure chromosome biorientation. Journal of Cell Biology, 215(1), 77–93. 10.1083/jcb.201603019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rellos, P. , Ivins, F. J. , Baxter, J. E. , Pike, A. , Nott, T. J. , Parkinson, D. M. , … Smerdon, S. J. (2007). Structure and regulation of the human Nek2 centrosomal kinase. Journal of Biological Chemistry, 282(9), 6833–6842. 10.1074/jbc.M609721200 [DOI] [PubMed] [Google Scholar]
- Rogers, G. C. , Rusan, N. M. , Roberts, D. M. , Peifer, M. , & Rogers, S. L. (2009). The SCFSlimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. Journal of Cell Biology, 184(2), 225–239. 10.1083/jcb.200808049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig, J. , Groen, A. , Caldwell, J. , & Avruch, J. (2005). Active Nercc1 protein kinase concentrates at centrosomes early in mitosis and is necessary for proper spindle assembly. Molecular Biology of the Cell, 16(10), 4827–4840. 10.1091/mbc.E05-04-0315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria, A. , Wang, B. , Elowe, S. , Malik, R. , Zhang, F. , Bauer, M. , … Nigg, E. A. (2011). The Plk1‐dependent Phosphoproteome of the early mitotic spindle. Molecular & Cellular Proteomics, 10(1), M110.004457 10.1074/mcp.M110.004457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sdelci, S. , Schütz, M. , Pinyol, R. , Bertran, M. T. , Regué, L. , Caelles, C. , … Roig, J. (2012). Nek9 phosphorylation of NEDD1/GCP‐WD contributes to Plk1 control of γ‐tubulin recruitment to the mitotic centrosome. Current Biology, 22(16), 1516–1523. 10.1016/j.cub.2012.06.027 [DOI] [PubMed] [Google Scholar]
- Seki, A. , Coppinger, J. A. , Jang, C.‐Y. , Yates, J. R. , & Fang, G. (2008). Bora and the kinase Aurora A cooperatively activate the kinase Plk1 and control mitotic entry. Science, 320(5883), 1655–1658. 10.1126/science.1157425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senese, S. , Cheung, K. , Lo, Y.‐C. , Gholkar, A. A. , Xia, X. , Wohlschlegel, J. A. , & Torres, J. Z. (2015). A unique insertion in STARD9's motor domain regulates its stability. Molecular Biology of the Cell, 26(3), 440–452. 10.1091/mbc.E14-03-0829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sents, W. , Ivanova, E. , Lambrecht, C. , Haesen, D. , & Janssens, V. (2013). The biogenesis of active protein phosphatase 2A holoenzymes: A tightly regulated process creating phosphatase specificity. FEBS Journal, 280(2), 644–661. 10.1111/j.1742-4658.2012.08579.x [DOI] [PubMed] [Google Scholar]
- Sessa, F. , Mapelli, M. , Ciferri, C. , Tarricone, C. , Areces, L. B. , Schneider, T. R. , … Musacchio, A. (2005). Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Molecular Cell, 18(3), 379–391. 10.1016/j.molcel.2005.03.031 [DOI] [PubMed] [Google Scholar]
- Shah, K. , Liu, Y. , Deirmengian, C. , & Shokat, K. M. (1997). Engineering unnatural nucleotide specificity for Rous sarcoma virus tyrosine kinase to uniquely label its direct substrates. Proceedings of the National Academy of Sciences of the United States of America, 94(8), 3565–3570. 10.1073/pnas.94.8.3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto, Y. , Forth, S. , & Kapoor, T. M. (2015). Measuring pushing and braking forces generated by ensembles of kinesin‐5 crosslinking two microtubules. Developmental Cell, 34(6), 669–681. 10.1016/j.devcel.2015.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar, S. , Janczyk, P. , Qu, Q. , Brautigam, C. A. , Stukenberg, P. T. , Yu, H. , & Gorbsky, G. J. (2016). The human SKA complex drives the metaphase‐anaphase cell cycle transition by recruiting protein phosphatase 1 to kinetochores. eLife, 5 10.7554/eLife.12902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slangy, A. , Lane, H. A. , d'Hérin, P. , Harper, M. , Kress, M. , & Niggt, E. A. (1995). Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin‐related motor essential for bipolar spindle formation in vivo. Cell, 83(7), 1159–1169. 10.1016/0092-8674(95)90142-6 [DOI] [PubMed] [Google Scholar]
- Slawson, C. , Zachara, N. E. , Vosseller, K. , Cheung, W. D. , Lane, M. D. , & Hart, G. W. (2005). Perturbations in O‐linked β‐N‐acetylglucosamine protein modification cause severe defects in mitotic progression and cytokinesis. Journal of Biological Chemistry, 280(38), 32944–32956. 10.1074/jbc.M503396200 [DOI] [PubMed] [Google Scholar]
- Song, J. , Wang, H. , Wang, J. , Leier, A. , Marquez‐Lago, T. , Yang, B. , … Daly, R. J. (2017). PhosphoPredict: A bioinformatics tool for prediction of human kinase‐specific phosphorylation substrates and sites by integrating heterogeneous feature selection. Scientific Reports, 7(1), 6862 10.1038/s41598-017-07199-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen, K. F. , Gabryjonczyk, A. M. , Anselm, E. , Nigg, E. A. , & Stierhof, Y. D. (2013). Human cep192 and cep152 cooperate in plk4 recruitment and centriole duplication. Journal of Cell Science, 126(14), 3223–3233. 10.1242/jcs.129502 [DOI] [PubMed] [Google Scholar]
- Soskis, M. J. , Ho, H. Y. H. , Bloodgood, B. L. , Robichaux, M. A. , Malik, A. N. , Ataman, B. , … Greenberg, M. E. (2012). A chemical genetic approach reveals distinct EphB signaling mechanisms during brain development. Nature Neuroscience, 15(12), 1645–1654. 10.1038/nn.3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagge, F. , Mitronova, G. Y. , Belov, V. N. , Wurm, C. A. , & Jakobs, S. (2013). Snap‐, CLIP‐ and halo‐tag labelling of budding yeast cells. PLoS One, 8(10), e78745 10.1371/journal.pone.0078745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steegmaier, M. , Hoffmann, M. , Baum, A. , Lénárt, P. , Petronczki, M. , Krssák, M. , … Rettig, W. J. (2007). BI 2536, a potent and selective inhibitor of polo‐like kinase 1, inhibits tumor growth in vivo. Current Biology, 17(4), 316–322. 10.1016/j.cub.2006.12.037 [DOI] [PubMed] [Google Scholar]
- Suijkerbuijk, S. J. E. , Vleugel, M. , Teixeira, A. , & Kops, G. J. P. L. (2012). Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore‐microtubule attachments. Developmental Cell, 23(4), 745–755. 10.1016/j.devcel.2012.09.005 [DOI] [PubMed] [Google Scholar]
- Tan, E. P. , Caro, S. , Potnis, A. , Lanza, C. , & Slawson, C. (2013). O‐linked N‐acetylglucosamine cycling regulates mitotic spindle organization. Journal of Biological Chemistry, 288(38), 27085–27099. 10.1074/jbc.M113.470187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate, E. W. , Kalesh, K. A. , Lanyon‐Hogg, T. , Storck, E. M. , & Thinon, E. (2015). Global profiling of protein lipidation using chemical proteomic technologies. Current Opinion in Chemical Biology, 24, 48–57. 10.1016/j.cbpa.2014.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur, H. C. , Singh, M. , Nagel‐Steger, L. , Kremer, J. , Prumbaum, D. , Fansa, E. K. , … Piekorz, R. P. (2014). The centrosomal adaptor TACC3 and the microtubule polymerase chTOG interact via defined C‐terminal subdomains in an Aurora‐A kinase‐independent manner. Journal of Biological Chemistry, 289(1), 74–88. 10.1074/jbc.M113.532333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thawani, A. , Kadzik, R. S. , & Petry, S. (2018). XMAP215 is a microtubule nucleation factor that functions synergistically with the γ‐tubulin ring complex. Nature Cell Biology, 20(5), 575–585. 10.1038/s41556-018-0091-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, Y. , Peter, M. , Mechali, F. , Blanchard, J. M. , Coux, O. , & Baldin, V. (2014). Kizuna is a novel mitotic substrate for CDC25B phosphatase. Cell Cycle, 13(24), 3867–3877. 10.4161/15384101.2014.972882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm, S. , Titus, B. , Bernd, K. , & Barroso, M. (1999). The EF‐hand Ca2+‐binding protein p22 associates with microtubules in an N‐myristoylation‐dependent manner. Molecular Biology of the Cell, 10(10), 3473–3488. 10.1091/mbc.10.10.3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, J. Z. , Ban, K. H. , & Jackson, P. K. (2010). A specific form of phospho protein phosphatase 2 regulates anaphase‐promoting complex/cyclosome association with spindle poles. Molecular Biology of the Cell, 21(6), 897–904. 10.1091/mbc.e09-07-0598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, J. Z. , Summers, M. K. , Peterson, D. , Brauer, M. J. , Lee, J. , Senese, S. , … Jackson, P. K. (2011). The STARD9/Kif16a kinesin associates with mitotic microtubules and regulates spindle pole assembly. Cell, 147(6), 1309–1323. 10.1016/J.CELL.2011.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, C. Y. , Ngo, B. , Tapadia, A. , Hsu, P. H. , Wu, G. , & Lee, W. H. (2011). Aurora‐A phosphorylates augmin complex component hice1 protein at an N‐terminal serine/threonine cluster to modulate its microtubule binding activity during spindle assembly. Journal of Biological Chemistry, 286(34), 30097–30106. 10.1074/jbc.M111.266767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallardi, G. , Allan, L. A. , Crozier, L. , & Saurin, A. T. (2019). Division of labour between pp2a‐b56 isoforms at the centromere and kinetochore. eLife, 8 10.7554/eLife.42619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Kooij, B. , Creixell, P. , van Vlimmeren, A. , Joughin, B. A. , Miller, C. J. , Haider, N. , … Yaffe, M. B. (2019). Comprehensive substrate specificity profiling of the human nek kinome reveals unexpected signaling outputs. eLife, 8, 8 10.7554/eLife.44635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma, D. , Chandrasekaran, S. , Sundin, L. J. R. , Reidy, K. T. , Wan, X. , Chasse, D. A. D. , … Cook, J. G. (2012). Recruitment of the human Cdt1 replication licensing protein by the loop domain of Hec1 is required for stable kinetochore‐microtubule attachment. Nature Cell Biology, 14(6), 593–603. 10.1038/ncb2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G. , Jiang, Q. , & Zhang, C. (2014). The role of mitotic kinases in coupling the centrosome cycle with the assembly of the mitotic spindle. Journal of Cell Science, 127(19), 4111–4122. 10.1242/jcs.151753 [DOI] [PubMed] [Google Scholar]
- Wang, J. , Wang, Z. , Yu, T. , Yang, H. , Virshup, D. M. , Kops, G. J. P. L. , … Rao, Z. (2016). Crystal structure of a PP2A B56‐BubR1 complex and its implications for PP2A substrate recruitment and localization. Protein & Cell, 7(7), 516–526. 10.1007/s13238-016-0283-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Udeshi, N. D. , Slawson, C. , Compton, P. D. , Sakabe, K. , Cheung, W. D. , … Hart, G. W. (2010). Extensive crosstalk between O‐GlcNAcylation and phosphorylation regulates cytokinesis. Science Signaling, 3(104), ra2 10.1126/scisignal.2000526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn, J. P. I. , Grishchuk, E. L. , Backer, C. B. , Wilson‐Kubalek, E. M. , Yates, J. R. , & Cheeseman, I. M. (2009). The human kinetochore Ska1 complex facilitates microtubule depolymerization‐coupled motility. Developmental Cell, 16(3), 374–385. 10.1016/j.devcel.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn, J. P. I. , Vleugel, M. , Liu, D. , Yates, J. R. , Lampson, M. A. , Fukagawa, T. , & Cheeseman, I. M. (2010). Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore‐microtubule Interface. Molecular Cell, 38(3), 383–392. 10.1016/j.molcel.2010.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , Cho, H. P. , Rhee, D. B. , Johnson, D. K. , Dunlap, J. , Liu, Y. , & Wang, Y. (2008). Cdc14B depletion leads to centriole amplification, and its overexpression prevents unscheduled centriole duplication. Journal of Cell Biology, 181(3), 475–483. 10.1083/jcb.200710127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, P. , Raetz, E. A. , Kitagawa, M. , Virshup, D. M. , & Lee, S. H. (2013). BUBR1 recruits PP2A via the B56 family of targeting subunits to promote chromosome congression. Biology Open, 2(5), 479–486. 10.1242/bio.20134051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, L. , & Tao, W. A. (2013). Current technologies to identify protein kinase substrates in high throughput. Frontiers in Biology, 8(2), 216–227. 10.1007/s11515-013-1257-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro, S. , Yamakita, Y. , Totsukawa, G. , Goto, H. , Kaibuchi, K. , Ito, M. , … Matsumura, F. (2008). Myosin phosphatase‐targeting subunit 1 regulates mitosis by antagonizing polo‐like kinase 1. Developmental Cell, 14(5), 787–797. 10.1016/j.devcel.2008.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui, Y. , Urano, T. , Kawajiri, A. , Nagata, K. I. , Tatsuka, M. , Saya, H. , … Inagaki, M. (2004). Autophosphorylation of a newly identified site of Aurora‐B is indispensable for cytokinesis. Journal of Biological Chemistry, 279(13), 12997–13003. 10.1074/jbc.M311128200 [DOI] [PubMed] [Google Scholar]
- Yu, R. , Wu, H. , Ismail, H. , Du, S. , Cao, J. , Wang, J. , … Yao, X. (2020). Methylation of PLK1 by SET7/9 ensures accurate kinetochore–microtubule dynamics. Journal of Molecular Cell Biology, 12(6), 462–476. 10.1093/jmcb/mjz107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, K. , Bastos, R. N. , Barr, F. A. , & Gruneberg, U. (2010). Protein phosphatase 6 regulates mitotic spindle formation by controlling the T‐loop phosphorylation state of Aurora A bound to its activator TPX2. Journal of Cell Biology, 191(7), 1315–1332. 10.1083/jcb.201008106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Ma, Y. , Taylor, S. S. , & Tsien, R. Y. (2001). Genetically encoded reporters of protein kinase a activity reveal impact of substrate tethering. Proceedings of the National Academy of Sciences of the United States of America, 98(26), 14997–15002. 10.1073/pnas.211566798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , Sivakumar, S. , Chen, Y. , Gao, H. , Yang, L. , Yuan, Z. , … Liu, H. (2017). Ska3 phosphorylated by Cdk1 binds Ndc80 and recruits Ska to kinetochores to promote mitotic progression. Current Biology, 27(10), 1477–1484.e4. 10.1016/j.cub.2017.03.060 [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Chen, Q. , Feng, J. , Hou, J. , Yang, F. , Liu, J. , … Zhang, C. (2009). Sequential phosphorylation of Nedd1 by Cdk1 and Plk1 is required for targeting of the γTuRC to the centrosome. Journal of Cell Science, 122(13), 2240–2251. 10.1242/jcs.042747 [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Lan, W. , Ems‐McClung, S. C. , Stukenberg, P. T. , & Walczak, C. E. (2007). Aurora B phosphorylates multiple sites on mitotic centromere‐associated kinesin to spatially and temporally regulate its function. Molecular Biology of the Cell, 18(9), 3264–3276. 10.1091/mbc.E07-01-0086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Z. S. , Lim, J. P. , Ng, Y. W. , Lim, L. , & Manser, E. (2005). The GIT‐associated kinase PAK targets to the centrosome and regulates Aurora‐A. Molecular Cell, 20(2), 237–249. 10.1016/j.molcel.2005.08.035 [DOI] [PubMed] [Google Scholar]
- Zorba, A. , Buosi, V. , Kutter, S. , Kern, N. , Pontiggia, F. , Cho, Y. J. , & Kern, D. (2014). Molecular mechanism of Aurora A kinase autophosphorylation and its allosteric activation by TPX2. eLife, 2014(3). 10.7554/eLife.02667.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


