Summary
Plant hormones play important roles in plant growth and development and physiology, and in acclimation to environmental changes. The hormone signaling networks are highly complex and interconnected. It is thus important to not only know where the hormones are produced, how they are transported and how and where they are perceived, but also to monitor their distribution quantitatively, ideally in a non‐invasive manner. Here we summarize the diverse set of tools available for quantifying and visualizing hormone distribution and dynamics. We provide an overview over the tools that are currently available, including transcriptional reporters, degradation sensors, and luciferase and fluorescent sensors, and compare the tools and their suitability for different purposes.
Keywords: plant hormone, biosensor, imaging, quantification
Significance Statement
Sensors are a valuable tool for understanding how the spatial and temporal distribution of plant hormones is coordinated, and how these changes trigger diverse responses. This review summarizes the diverse tools available to quantify and visualize the distribution and dynamics of hormones, provides an overview of the tools currently available, including transcription reporters, degradation sensors, and luciferase and fluorescent sensors, and compares the tools and their suitability for different purposes.
INTRODUCTION
Plant growth regulators, or plant hormones, are chemical messengers that play crucial roles in the control of plant growth and development. A wide range of unique hormones has been characterized in plants, i.e., abscisic acid, auxins, brassinosteroids, cytokinins, ethylene, gibberellins, jasmonic acid, salicylic acid, strigolactone and small peptides (Verma et al., 2016; Stührwohldt and Schaller, 2019; Bowman et al., 2019). The concentration of the hormones can vary between different plant tissues, developmental stages and environmental conditions (Swarup et al., 2007); however, it is not entirely understood how the spatial and temporal distribution of plant hormones is coordinated, nor how these changes trigger diverse responses. In the past, traditional biochemical analyses have been used to gain insight into the distribution and dynamics of plant hormones. For instance, immunohistochemistry using anti‐hormone antibodies has been used to determine the distribution of hormones in plant cells and tissues (Forestan and Varotto, 2013; Schlicht et al., 2006; Nishimura et al., 2011). The need to fix and section the material limits the spatial and temporal resolution, however. Mass spectrometry combined with gas or liquid chromatography has been used to identify and quantify plant hormones at high accuracy and sensitivity (Okamoto et al., 2009; Owen and Abrams, 2009; Gemperline et al., 2016). Although rapid sampling can provide temporal resolution for chromatographic assays, the extracts reflect an average over many cells in an organ, such as a leaf, and thus cannot detect spatial differences, such as gradients. Over the past two decades, advances in the development of fluorescence microscopy technologies and the engineering of biosensors has provided new tools for monitoring plant hormones with minimal invasion and cellular or even subcellular resolution (Walia et al., 2018; Waadt, 2020). Moreover, genetically encoded biosensors are able to detect rapid changes in the concentration and distribution of plant hormones in living cells. In this review, we introduce the current imaging technologies available for plant hormone detection (Tables 1 and 2), and we discuss possible routes to expand and improve biosensors for plant hormones.
Table 1.
Biosensors for plant hormones
| Analyte | Biosensor | Sensor type | Range of ligand concentrations applied endogenously or exogenously | Reference |
|---|---|---|---|---|
| ABA | 6xABRE | Transcriptional | approx. 1–25 µm | (Wu et al., 2018) |
| ABACUS | FRET based | approx. 2–80 µm | (Jones et al., 2014) | |
| ABAleon2.1 | FRET based | approx. 100 nm | (Waadt et al., 2014) | |
| ABAleon2.15 | FRET based | approx. 100 nm | (Waadt et al., 2014) | |
| ABAleonSD1‐3L21 | FRET based | approx. 100 nm | (Waadt et al., 2020) | |
| SNACS | FRET based | 20 µm ABA (exogenous) | (Zhang et al., 2020) | |
| Auxin | DR5:reporter | Transcriptional | 10–1000 nm (DR5) | (Ulmasov et al., 1997; Liao et al., 2015) |
| 3–1000 nm (DR5v2) | ||||
| pIAAmotif | Transcriptional | 10 nm –1 µm 2,4‐D (exogenous) | (Lieberman‐Lazarovich et al., 2019) | |
| DII‐VENUS | Degron based | 1–1000 nm | (Brunoud et al., 2012) | |
| R2D2 | Degron based | 1 µm IAA (exogenous) | (Liao et al., 2015) | |
| L2min17‐Luc | Degron based | 10–1000 nm | (Wend et al., 2013) | |
| AuxSen | FRET based | 5–50 µm | (Herud‐Sikimic et al., 2020) | |
| Brassinosteroid | BZR1‐YFP | Transcriptional | 100 µm brassinolide (exogenous) | (Chaiwanon and Wang, 2015) |
| Cytokinin | ARR5 | Transcriptional | 10–1000 nm | (D'Agostino et al., 2000; Romanov et al., 2002) |
| TCS | Transcriptional | 1–1000 nm | (Müller & Sheen, 2007) | |
| TCSn | Transcriptional | 1–1000 nm | (Zürcher et al., 2013) | |
| Ethylene | EIN3‐GFP | Degron based | 50 µm ACC (exogenous) | (Guo and Ecker, 2003) |
| EIL1‐GFP | Degron based | 100 µm ACC (exogenous) | (An et al., 2010) | |
| FP‐EBF 3’UTR | Translational | 10 µm ACC (exogenous) | (Merchante et al., 2015) | |
| FP‐6x EPU | Translational | 10 µm ACC (exogenous) | (Li et al., 2015) | |
| EBS:GUS | Transcriptional | 10 µM ACC (exogeneous) | (Stepanova et al., 2007) | |
| AEP | Artificial metalozyme | 100 µM ACC (exogenous) | (Vong et al., 2019) | |
| Gibberellin | GFP‐RGA | Degron based | 100 µM GA3 (exogenous) | (Silverstone et al., 2001) |
| GPS1 | FRET based | 0.03–1 µM GA1 | (Rizza et al., 2017) | |
| 0.1–2 µM GA3 | ||||
| 0.005–0.2 µM GA4 | ||||
| Jasmonic acid | Jas9‐Venus | Degron based | 50 nm−10 µM coronatine | (Larrieu et al., 2015) |
| JAI3‐FP | Degron based | 50 µM jasmonate (exogenous) | (Chini et al., 2007) | |
| Karrikin | DLK2:LUC | Transcriptional | 1 µM < KAR1 (exogenous) | (Sun et al., 2016) |
| 10 nm < KAR2 (exogenous) | ||||
| pRATIO‐SMAX1 | Degron based | approx. 200 nm KAR1 (exogenous) | (Khosla et al., 2020) | |
| Salicylic acid | NPR1‐FP | Transcriptional | 0.5 mM INA (exogenous) | (Mou et al., 2003) |
| Strigolactone | D53‐GFP | Degron based | 5 µM rac‐GR24 (exogeneous) | (Zhou et al., 2013) |
| SMXL6‐YFP | Degron based | 5 µM rac‐GR24 (exogeneous) | (Bennett et al., 2016) | |
| SMXL7‐YFP | Degron based | 5 µM rac‐GR24 (exogeneous) | (Liang et al., 2016) | |
| StrigoQuant | Degron based | 10 pM–1 nm | (Samodelov et al., 2016) | |
| rDAD2cpGFP | cpFP based | 50–500 nm rec‐GR24 (exogenous) | (Chesterfield et al., 2020) | |
| rShHTL7cpGFP | cpFP based | approx. 10–100 nm rec‐GR24 (exogenous) | (Chesterfield et al., 2020) |
Table 2.
Comparison of FRET‐based direct biosensors
| Biosensor | Analyte | K D | Dynamic range | Ref. |
|---|---|---|---|---|
| ABACUS1‐2µ | ABA | approx. 2 µm | +60% (500 µm ABA) | (Jones et al., 2014) |
| ABACUS1‐80µ | ABA | approx. 80 µm | +160% (500 µm ABA) | (Jones et al., 2014) |
| ABAleon2.1 | ABA | approx. 79 nm | –8.98% | (Waadt et al., 2014) |
| ABAleon2.15 | ABA | approx. 600 nm | –10.09% | (Waadt et al., 2014) |
| ABAleonSD1‐3L21 | ABA | approx. 938 nm | n.d. | (Waadt et al., 2020) |
| GPS1 | GA1 | approx. 110 nm | +60% (200 nm GA1) | (Rizza et al., 2017) |
| GA3 | approx. 240 nm | +40% (200 nm GA3) | (Rizza et al., 2017) | |
| GA4 | approx. 24 nm | +90% (200 nm GA4) | (Rizza et al., 2017) |
DETECTION OF PLANT HORMONES IN LIVING PLANT MATERIALS
To understand the physiological effects of plant hormones, it is important to determine the distributions and to quantify the concentrations of plant hormones with high spatiotemporal resolution, which is important for understanding the physiological responses of plant hormones. Fluorescence is an excellent tool for the minimally invasive monitoring of the distribution of bioactive molecules within intact cells, tissues and organs. Fluorescence can be effectively quantified and has femto‐ and nanosecond excitation and emission rates that allow fluorescence‐based probes to faithfully record rapid processes with subcellular resolution. Combined with the development of increasingly sophisticated and sensitive imaging technologies, fluorescence can be used to non‐destructively and directly visualize rapid cellular processes. Such fluorescence microscopy provides data with spatial and temporal resolution that is not possible with traditional biochemical methods. The ways that fluorescence can be used to detect plant hormones are discussed below.
FLUORESCENT TAGS AND ANALOGS OF PLANT HORMONES
Organic chemistry has contributed to the progress in plant hormone research (Rigal et al., 2014). Fluorescent dyes can be conjugated to standard forms of plant hormones, and some analogs are naturally fluorescent. Fluorescent conjugates of auxin, 7‐nitro‐2,1,3‐benzoxadiazole (NBD)‐naphthalene‐1‐acetic acid (NAA) and NBD‐indole‐3‐acetic acid (IAA) enabled the visualization of auxin transport dynamics and subcellular auxin distribution in Nicotiana tabacum (tobacco) BY‐2 cells and Arabidopsis thaliana seedlings (Hayashi et al., 2014). Although these fluorescent‐labeled auxin analogs are not functional with respect to auxin signaling or metabolism, a fluorescent‐labeled brassinosteroid (BR), Alexa Fluor 647‐castasterone (AFCS), has been shown to be bioactive. AFCS was used to visualize endocytosis of the BR receptor in living Arabidopsis cells (Irani et al., 2012). Bioactive fluorescein‐labeled gibberellic acids (GA‐fls) accumulated in the endodermal cells of the Arabidopsis root elongation zone, and the GA‐Fl accumulation was inhibited by ethylene signaling (Shani et al., 2013). The strigolactone‐agonist probe Yoshimulactone Green (YLG), which has activity similar to strigolactone, becomes fluorescent when hydrolyzed by the hydrolase activity of the strigolactone receptor, thereby enabling the visualization of the perception of applied strigolactone by Striga embryos (Tsuchiya et al., 2015).
Peptide hormones have also been successfully labeled with fluorescent dyes to track their localization and uptake. For example, the elicitor‐induced peptide pep1 of Arabidopsis was N‐terminally tagged with tetramethylrhodamine (TAMRA) and used to monitor binding to the PEPR receptors followed by clathrin‐mediated endocytosis of peptide‐receptor complexes (Ortiz‐Morea et al., 2016). In a few cases, peptide hormones were monitored by genetically encoded peptide‐FP fusions. Using such tools, the shoot‐to‐root translocations of C‐TERMINALLY ENCODED PEPTIDE DOWNSTREAM 1 (CEPD1) and CEPD‐like 2 (CEPDL2) were observed using GFP‐CEPD1 and GFP‐CEPDL2 reporters (Ohkubo et al., 2017; Ota et al., 2020), revealing the signaling pathway that integrates nitrate uptake in Arabidopsis. Large fluorescent moieties often interfere with peptide–receptor interactions and reduce bioactivity, however, or are unstable and are processed from the peptide.
The use of synthetic chemistry is an excellent tool that can provide new insights into plant hormone dynamics in living plants. It is noteworthy that these small molecules need to be applied exogenously and are not likely to represent the actual distribution or dynamics of the endogenous hormones, especially because of the differential recognition by transporters, receptors and degradation enzymes in the plant. In addition, often some chemical compounds cannot pass the cuticular layer at the plant surface, and it is difficult to force the entry of compounds into tissues by infiltration in a uniform manner, thereby limiting the reliable quantification of the compounds in the plant.
GENETICALLY ENCODED FLUORESCENT PROTEIN BIOSENSORS
Genetically encoded biosensors have been developed for better quantitative and less invasive imaging of specific plant hormones in genetically transformable plants such as Arabidopsis. Genetically encoded biosensors can be categorized as indirect and direct biosensors (Walia et al., 2018). The responses of indirect biosensors is dependent upon changes in gene expression or protein degradation in response to changes in analyte (plant hormone) concentration (Walia et al., 2018; Waadt, 2020). On the other hand, the responses of direct biosensors are dependent on conformational change of the biosensor protein itself (Okumoto et al., 2012). In this section, we introduce different types of biosensors for plant hormone detection and general aspects regarding how they respond to the respective analytes.
Indirect biosensors include two major types: transcriptional reporter and degron‐based reporter proteins (Figure 1). Transcriptional reporters consist of a promoter sequence with a conserved motif that is indirectly regulated by analyte concentration, through the activation of a transcription factor that acts within the hormone‐dependent signal transduction cascade and controls the expression of a quantifiable reporter gene (Waadt, 2020). Although promoters generally respond to many different cues (Juven‐Gershon and Kadonaga, 2010), using specific hormone‐responsive cis‐elements as components of artificial promoters can be effective in achieving high specificity for the hormone of interest. In many cases, a fragment of the cauliflower mosaic virus (CaMV) 35S promoter (minimal CaMV 35S promoter) will be combined with enhancer sequences and specific transcription factor (TF) binding motifs that bind hormone‐controlled TFs (Waadt, 2020). Reporter proteins, such as β‐glucuronidase (GUS), luciferase (LUC) and green fluorescent protein (GFP), as well as other fluorescent proteins (FPs), can be used to visually represent promoter activity in response to hormone levels (Walia et al., 2018). The read‐out of transcriptional reporters is affected by multiple parameters: (i) analyte‐dependent activation/deactivation of transcription factors, and (ii) subsequent induction/attenuation of reporter gene expression. The temporal resolution of transcriptional reporters is in the range of minutes to hours (D’Agostino et al., 2000; Brunoud et al., 2012), as multiple steps, including transcription, translation, FP maturation and turnover of reporters affect the actual output. The spatial resolution of transcriptional reporters provides cellular‐level resolution but not subcellular‐level resolution.
Figure 1.
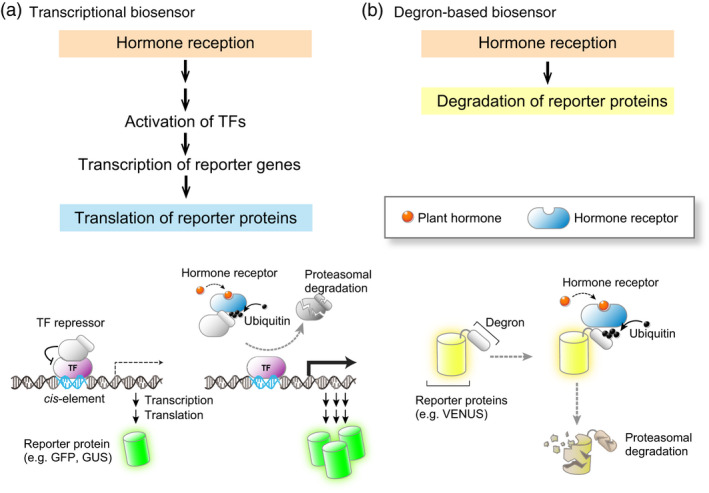
Models of indirect biosensors. Indirect sensors comprise transcriptional or degradation‐based biosensors. (a) Typical mechanism commandeered for transcriptional reporters. Plant hormone perception by its receptor induces the proteasomal degradation of repressors for specific transcription factors (TFs). The activated TFs bind to specific sequence motifs (cis‐elements) and induce the transcription of reporter genes such as GFP, GUS or LUC. The read‐out of the transcriptional reporters depends on hormone reception, proteasomal degradation of the TF repressors, transcription, translation, and folding and activity of reporter proteins. Reporter activity can remain even after transcription returns to base levels after hormone levels drop, limiting the temporal resolution of transcriptional reporters; however, transcriptional sensors can be considered memory reporters as they will also report past activity. (b) Typical mechanism commandeered for degron‐based biosensors. A reporter protein is fused with a degron. The reporter is active in the absence of hormone, but is rapidly ubiquitinated and degraded by the activities of a specific hormone receptor and the proteasome, respectively. The reporter–degron fusion protein is typically expressed under the control of constitutive and ubiquitous promoters. Therefore, the temporal resolution of the degron‐based biosensors is more accurate compared with transcriptional reporters.
Degron‐based biosensors consist of a reporter protein, such as an FP, and a specific domain, the so‐called degron. Degron domains undergo ubiquitination, mediated by specific plant hormone receptors that are associated with SKP1‐Cullin‐F‐Box (SCF) complexes and subsequent degradation by the proteasome (Santner and Estelle, 2010). Constitutive promoters such as CaMV 35S, UBQ10 or RPS5A are typically employed for expressing degron‐based biosensors across many cells and stages (Brunoud et al., 2012; Wend et al., 2013; Larrieu et al., 2015; Liao et al., 2015; Samodelov et al., 2016). Therefore, degron‐based biosensors typically have a faster temporal resolution (minutes until a degron‐sensor response is detectable) compared to transcriptional reporters (estimated to >2 h until FP is formed) (Brunoud et al., 2012). The temporal resolution of degron‐based biosensors is determined by not only the transcription rate but also by the FP maturation time and degradation rate of the reporter protein. To maximize the temporal resolution, fast‐maturing FPs such as VENUS (with a maturation time of approx. 18 min in Escherichia coli; Balleza et al., 2018) are often chosen as reporters for degron‐based biosensors.
Direct biosensors consist of a sensory domain that binds directly to a specific plant hormone and two spectral FP variants that function as Förster resonance energy transfer (FRET) donors and acceptors, respectively (Figure 2). The fluorescence read‐out generated by direct biosensors is the result of conformational rearrangements in the sensory domain that affect the FRET of the sensor and not the abundance of the sensor protein (Walia et al., 2018). Thus, direct biosensors provide rapidly reversible read‐outs of plant hormone concentrations with high spatiotemporal resolution. As the read‐out of direct biosensors relies on conformational protein dynamics, such sensors are not suitable for analyses of fixed plant specimens. For the engineering of direct biosensors, both endogenous plant hormone receptors and modified synthetic enzymes have been used for the sensory domain (Waadt, 2020). Specific examples are given in the next section.
Figure 2.
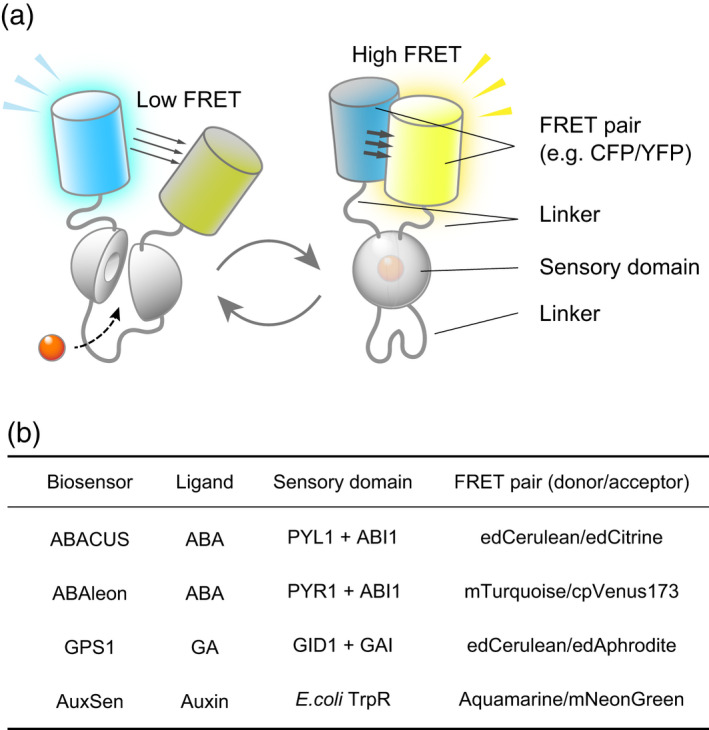
A model of FRET‐based direct biosensors. ‘Direct biosensors’ directly bind the plant hormone and report this binding. (a) The typical structure of a FRET‐based direct biosensor. The sensor generally consists of a sensory domain sandwiched by two fluorescent proteins (FPs) that differ in emission spectra and that act as a Förster resonance energy transfer (FRET) pair. Ligand binding causes a conformational rearrangement. The conformational change physically alters the distance and orientation between the two FPs and results in a readily detectable change in the emission ratio of the two FPs. Importantly, the FPs and the sensory domains are connected by flexible linkers, which influence the sensor activity. (b) A short list of FRET‐based biosensors for plant hormones.
PLANT HORMONE SENSORS
Abscisic acid
Abscisic acid (ABA) is a monocyclic sesquiterpene that plays key roles in diverse processes, such as seed development and dormancy, stomatal closure, abscission and responses to abiotic stresses in plants (Cutler et al., 2010; Yoshida et al., 2019). In the absence of ABA, protein phosphatase 2C (PP2C) represses the activity of SnRK2‐type kinases via dephosphorylation. ABA signaling is initiated by the binding of ABA to a member of the PYL/PYR/RCAR family and co‐receptor PP2C‐type protein phosphatases. The binding leads to the activation of a third protein, the SnRK2 protein kinase. SnRK2 then phosphorylates downstream proteins, including ABA‐responsive element (ABRE)‐binding transcription factors and ion channels. The distribution of ABA has successfully been detected by matrix‐assisted laser desorption/ionization time‐of‐flight mass spectrometry (MALDI‐TOF‐MS) imaging (Shiono et al., 2017) and transcriptional biosensors (Wu et al., 2018). Transcriptional biosensors were developed that consisted of synthetic promoters, based on repeats of the ABRE elements, upstream of GFP fused with an endoplasmic reticulum (ER)‐retention signal (erGFP) (Wu et al., 2018). These 6xABRE reporters were used to characterize ABRE‐mediated gene expression in the root under stress conditions. Induction of 6xABRE reporters was detected 4 h after treatment with 10 µm ABA in essentially all tissues. 6xABRE reporters have a detection range of at least 1 µm up to 25 µm of exogenously applied ABA. 6xABRE reporters were also induced by osmotic stress (50–150 mm NaCl or 100–300 mm mannitol) (Wu et al., 2018).
To visualize the dynamics of ABA in living tissues, FRET‐based direct biosensors, namely ABACUS and ABAleon, were generated using the ABA perception machinery (Jones et al., 2014; Waadt et al., 2014; Waadt et al., 2020). The sensory domain of ABACUS consists of PYL1 fused to the minimal ABA interaction domain of ABI1 (PP2C) as a recognition element that is sandwiched between the reporter elements edCerulean and edCitrine, as FRET donor and acceptor, respectively (Figure 2). The sensor was developed by testing a large number of variants (Jones et al., 2014). Affinity variants with equilibrium dissociation constant (K D) values of 2 µm and 80 µm have been engineered (Table 2). ABAleon2.1 uses PYR1 fused to a truncated ABI1 as the sensory domain linked to mTurquoise and circularly permutated Venus (cpVenus) as FRET donor and acceptor, respectively. The detection range of ABAleon2.1 was 100–600 nm with a K D of 79 nm (Table 2). ABACUS showed an increasing emission ratio when ABA levels increased, whereas ABAleon was characterized by high FRET in the absence of ABA and a decreased ratio when ABA was bound. It is noteworthy that the transgenic Arabidopsis lines expressing the ABA biosensors showed ABA‐related phenotypes, however. The primary root growth and germination of ABACUS1 lines were hypersensitive to ABA. As the degree of sensitivity and the binding affinity of the ABACUS1 sensors were correlated, it is likely that the sensor interacts with the endogenous ABA signaling pathway, with effects similar to those found in PYL1 overexpression lines (Jones et al., 2014). Although ABAleon2.1 is sensitive enough to detect changes in endogenous ABA levels induced by abiotic stress, for example, the Arabidopsis lines that expressed high levels of ABAleon2.1 and produced bright fluorescence were ABA hyposensitive. The expression level of ABAleon correlated with reduced sensitivity, indicating that the sensor scavenged cytosolic ABA (Waadt et al., 2014). ABAleon has been improved through the screening of deletion variants of ABAleon2.15 and optimizing FRET pairs, sensory domains and linkers (Waadt et al., 2020). The improved ABAleon, ABAleonSD1‐3L21, used the amino acids LD and T as linkers between the FPs and sensory domains, instead of the amino acid pairs GP and PG in ABAleon2.15. When Arabidopsis seedlings expressing ABA indicators were treated with 10 µm ABA, ABAleon2.15 and ABAleonSD1‐3L21 showed faster responses with a half‐life (t 1/2) of approx. 15 min, whereas ABACUS1‐2µ responded at a slower rate (t 1/2 ~ 29 min).
Recently, another type of FRET sensor, an SnRK2 activity sensor (SNACS) was developed and used to monitor ABA accumulation (Zhang et al., 2020). SNACS is composed of a 48‐amino‐acid sensory domain surrounding Serine‐30 of the Arabidopsis ABA‐RESPONSIVE KINASE SUBSTRATE 1 transcription factor (AKS1) and the full‐length 14‐3‐3 protein GF14phi. In SNACS, the binding of the 14‐3‐3 protein to AKS1, which is phosphorylated by SnRK2, causes a conformational change and changes the FRET efficiency. The emission ratio of SNACS in Arabidopsis guard cells increased in a time‐dependent manner after the addition of 20 µm ABA. ABAleon2.15 and SNACS were both used to test whether CO2 elevation directly activates ABA signaling; however, neither sensor showed any detectable change in emission ratio in guard cells in response to increasing CO2 concentrations (Hsu et al., 2018; Zhang et al., 2020).
AUXIN
Auxins, with their key player indole acetic acid (IAA), are structurally related to the amino acid tryptophan and have roles in a wide range of developmental processes in plants, including morphogenesis, cell proliferation and organogenesis. The predominant endogenous auxin IAA is synthesized in proliferating tissues, such as embryos, shoot meristems and young leaves, from where it is transported to roots by polar auxin transport. Auxin gradients play central roles in many phases of plant pattern formation and development (Leyser, 2005; Möller and Weijers, 2009).
Transcriptional reporters, degron‐based biosensors and FRET‐based direct biosensors have been developed for monitoring auxin (Table 1). The widely used transcriptional reporter DR5 was the first sensor to be generated, and DR5‐based transcriptional reporters are still widely used today. The DR5 promoter contains multiple auxin‐response elements (AuxREs), which were first identified by promoter deletion analysis (Ulmasov et al., 1997). The improved version, DR5v2, carries AuxREs with a higher affinity for auxin response factors (ARFs), and is thus more sensitive (Liao et al., 2015). DR5 and DR5v2 reporters enabled the visualization of the distribution of auxin in embryos, in post‐embryonic root cells such as the quiescent center (QC), in the columella root cap and protoxylem cells, in the shoot apical meristem and in many other tissues. The two artificial DR5 and DR5v2 promoters responded to exogenously added auxin concentrations as low as 3 nm, but the reporter amplitude is higher for DR5v2 compared with DR5. This difference in sensitivity may explain why DR5v2 can detect auxin in a wider range of cell types, including the root metaxylem, pericycle, lateral root cap and root epidermal cells (Liao et al., 2015). In addition to fluorescent proteins such as GFP, the DR5 promoter has also been combined with GUS and LUC as reporters (Ulmasov et al., 1997; Moreno‐Risueno et al., 2010; Van Norman et al., 2013). DR5‐based transcriptional reporters have been used in a variety of plant species (Pattison and Catalá, 2012; Galli et al., 2015). Based on the conservation of AuxRE sequences (TGTCGG/CC/TC), in particular the pIAAmotif, highly sensitive synthetic reporters have been engineered (Lieberman‐Lazarovich et al., 2019).
Auxin transport plays an important role in the control of cellular auxin levels. The auxin influx carrier AUX1 and the PIN‐family of auxin efflux carriers, which are regulated by protein kinases, determine the level and direction of auxin transport. Therefore, auxin biosensors must have rapid kinetics to be able to follow rapid changes in auxin dynamics, e.g., for the analysis of auxin relocation during blue light phototropic or gravitropic responses. To visualize dynamic changes of auxin, the degron‐based auxin reporter DII‐VENUS was developed (Brunoud et al., 2012). DII‐VENUS uses domain II (DII) of the AUX/IAA repressor IAA28 fused to a variant yellow fluorescent protein (VENUS), which carries nuclear localization sequences (NLSs). DII is ubiquitinated by the auxin receptor TIR1/AFB in the presence of auxin, and this ubiquitination targets AUX/IAA for proteasomal degradation. The CaMV 35S RNA promoter has been used for constitutive expression of the DII‐VENUS, such that the expression level of the biosensor should not be affected by auxin concentration changes. An auxin‐insensitive variant, mDII‐VENUS, with impaired TIR1/AFB interaction, can serve as a negative control.
To enable auxin detection in embryos and meristems, a similar approach was used to engineer the ratiometric auxin reporter R2D2. The R2D2 sensor places both RPS5A:DII‐n3xVENUS and RPS5A:mDII‐ntdTomato in a single transgene (Liao et al., 2015). As an alternative for efficient quantification, DII of AtIAA17 fused to Firefly was fused to Renilla luciferase via the self‐cleaving 2A peptide for normalization, named min17‐Luc (Wend et al., 2013). The sensor is ratiometric, as the relative luminescence of Firefly to Renilla luciferase enabled the quantification of the auxin levels in Arabidopsis protoplasts. Three types of sensor modules derived from three different Aux/IAAs provide a wide range of sensitivities and binding affinities for IAA and NAA.
Although degron‐based sensors are likely to be faster compared with transcriptional sensors and to integrate auxin signals over shorter time scales, sensors that transform a binding‐induced conformational rearrangement into a change in fluorescence can be orders of magnitude faster and enable subcellular analyses. As the bacterial periplasmic binding protein superfamily covers a wide range of ions and metabolites, this class of proteins is widely used for engineering such sensors (Fukami‐Kobayashi et al., 1999). Our group could not identify periplasmic binding proteins that could recognize plant hormones but did make use of repressors that bind tryptophan. As tryptophan and auxin are structurally related, we engineered a sensor for tryptophan as a first step towards building an auxin sensor by subsequent modification of the substrate specificity (Kaper et al., 2007). The Jürgens lab then used the tryptophan sensor FLIPW as a starting point for the screening of mutants that can recognize auxin with high selectivity. The resulting auxin sensor, AuxSen, was then used to measure auxin levels in protoplasts and roots of Arabidopsis (Herud‐Sikimic et al., 2020). A major advantage of using heterologous proteins as binding domains (here the bacterial Trp repressor) over plant endogenous proteins is that it is unlikely that a fully orthogonal protein domain interferes with plant signaling pathways.
BRASSINOSTEROIDS
Brassinosteroids (BRs) are polyhydroxylated sterol derivatives. BRs are involved in cell elongation, cell division, cell proliferation, seed germination and stress responses, in combination with other plant hormones (Zhu et al., 2013). BR signaling is initiated by the binding of BR to the extracellular domain of the receptor kinase BRI1. Activated BRI1 transiently interacts with BAK1 and then disassociates. In the presence of BR, the GSK3‐like kinase BIN1 is inactivated, which leads to the dephosphorylation of BRASSINAZOLE RESISTANT 1 (BZR1)‐type transcription factors. Dephosphorylated BZR1 translocates to the nucleus, where it regulates BR‐responsive gene expression. Therefore, it is assumed that BZR1 accumulation in the nucleus reports active BR signaling. To monitor the distribution of BR, BZR1‐YFP fusions were expressed either from the BZR1 or the CaMV 35S promoter (Chaiwanon and Wang, 2015). BZR1‐YFP accumulated to low levels in the nuclei of the stem cell region and at higher levels in the nuclei of epidermal cells in the transition and elongation zone of roots, which indicated a close correlation between rapid cell elongation and the level of BZR1 expression (Table 1). To our knowledge, direct BR biosensors have not been engineered or published yet.
CYTOKININS
Cytokinins are nucleobase or nucleoside analogs that play important roles in cell division and shoot and root morphogenesis. Cytokinin distribution can be visualized by mass spectrometry‐based imaging (Shiono et al., 2017). Cytokinins are perceived by transmembrane two‐component receptors that phosphorylate downstream mobile phospho‐transmitters (To et al., 2007). In turn, the phospho‐transmitters phosphorylate nuclear transcription factors (such as the ARRs) that induce or repress cytokinin‐responsive gene expression. To create a cytokinin reporter, the promoter region of ARR5 was fused to GUS or to fluorescent proteins (D’Agostino et al., 2000). Although cytokinin signaling was known at this time to play essential roles in post‐embryonic growth and development, its role during early embryogenesis was unclear. To visualize the cytokinin distribution in Arabidopsis embryos, a synthetic reporter, two‐component‐output‐sensor (TCS), as well as an improved derivative, TCS‐new (TCSn), were engineered. TCSn consists of 24 repeats of DNA‐binding sites [(A/G)GAT(C/T)] for cytokinin‐responsive transcription factors followed by a fluorescent reporter (Zürcher et al., 2016) that is induced in Arabidopsis roots by external addition of trans‐zeatin or isopentenyl‐adenine (Liu and Müller, 2017). TCSn:GFP, TCSn:GUS and TCSn:VENUS:H2B have been used for the analysis of cytokinin responses and cytokinin distribution in Arabidopsis, Oryza sativa (rice), and Hordeum vulgare (barley), respectively (Tao et al., 2017; Kirschner et al., 2018). Although transcriptional cytokinin sensors are highly useful for monitoring cytokinin, a FRET sensor that directly detects the dynamics of cytokinins, and that may be able to differentiate between different cytokinins, might be useful in the future.
ETHYLENE
Ethylene is a gaseous hormone synthesized from methionine that affects plant growth, flower and fruit development, and senescence. Ethylene signaling is initiated by binding to one of the five ETR receptors in Arabidopsis, leading to the inactivation of the cytosolic CTR1 kinase, which derepresses EIN2, an ER‐bound transmembrane protein. EIN2 is proteolytically cleaved upon ethylene perception, and the C‐terminal fragment of EIN2 enters the nucleus to regulate EIN3/EILs transcription factor activities, resulting in the activation or repression of ethylene‐responsive gene expression (An et al., 2010; Qiao et al., 2012). EIN3 is a transcriptional activator of ethylene‐responsive genes. EIN3‐GFP and EIL1‐GFP were developed as sensors for ethylene. EIN3‐GFP and EIL1‐GFP accumulate in Arabidopsis seedlings in response to treatment with the ethylene precursor ACC (An et al., 2010). To monitor the distribution of ethylene in plants, a reporter using GUS driven by the synthetic EIN3‐responsive reporter EIN3‐binding site (EBS) was developed (EBS:GUS) (Stepanova et al., 2007). The distribution of ethylene was reported in the roots of Arabidopsis expressing EBS:GUS treated with ACC (Stepanova et al., 2007).
In the absence of ethylene, EIN3 is degraded by proteolysis mediated by two F‐box proteins, EBF1 and EBF2 (Guo and Ecker, 2003), whereas in the presence of ethylene, EIN3 is stabilized by the degradation of EBF1 and EBF2 induced by ethylene signaling (An et al., 2010). Degradation of EBF1/2 upon ethylene signaling is induced through the recognition of its 3′ untranslated region (3′‐UTR) being recognized by the C terminus of EIN2 (Merchante et al., 2015; Li et al., 2015). Using the 590‐bp 3′‐UTR of EBF2 (3′EBF2) and an ethylene‐responsive RNA element containing poly‐uridylates (EPU) from the 3′‐UTR of EBF1, translational regulation probes such as FP‐3′EBF2 and FP‐6x EPU were developed (Merchante et al., 2015; Li et al., 2015), enabling the monitoring of ethylene dynamics in plant tissues (Fernandez‐Moreno and Stepanova, 2020).
As an alternative ethylene detection technology, the enzyme‐based chemical biosensor artificial‐metalloenzyme ethylene probe (AEP) was developed (Vong et al., 2019). AEP was designed to have an albumin scaffold that would solubilize and protect a metal complex. The metal complex consists of the fluorophore 7‐diethylaminocoumarin (DEAC), the second‐generation Hoveyda–Grubbs (HG) catalyst and the olefin‐containing DABCYL quencher. In the absence of ethylene, the DABCYL quencher suppresses DEAC fluorescence by FRET interaction. When the AEP is exposed to ethylene, cross‐metathesis catalyzed by HG releases DABCYL, yielding DEAC fluorescence. This has been applied to the detection of ethylene gas in fruits and Arabidopsis leaves (Vong et al., 2019).
GIBBERELLINS
Gibberellic acids or gibberellins (GAs) belong to the diterpene family. GAs come in many different varieties: some are precursors, whereas others are active or inactive. GAs are involved in seed germination, stem elongation, fruit growth and floral development. GA signaling is initiated through the perception of GA by GID1 accompanied by DELLA proteins in Arabidopsis (Murase et al., 2008). DELLA proteins, such as GAI and RGA, act as repressors of GA‐responsive gene expression. When interacting with GA, the GID1–DELLA complex recruits an E3 ubiquitin ligase to target the complex for degradation, which induces the expression of GA‐responsive genes. This ubiquitin‐dependent degradation mechanism, which is analogous to the auxin receptor, was used to engineer the degron‐based GA biosensor, GFP‐RGA (Silverstone et al., 2001). GFP‐RGA carrying a nuclear targeting sequence and under the control of the CaMV 35S promoter was expressed, and the GFP‐RGA sensor showed decreased fluorescence 30 min after the addition of 100 µm GA3 (Table 1).
The FRET‐based GA sensor GPS1 was developed for the high‐resolution quantification of spatiotemporal GA distribution (Rizza et al., 2017). GPS1 is composed of domains from the Arabidopsis GID1 receptor AtGAI as sensory domains. The GA biosensor was optimized by a screening of ligand sensory domains fused to pairs of FRET donor/acceptor variants. Three Arabidopsis GID1 proteins (AtGID1A, AtGID1B or AtGID1C) and two DELLA family proteins (AtGAI or AtRGA) were tested as sensory domains with five different flexible linkers and CFP and YFP variants (Figure 3). The optimized GPS1 consists of edAphrodite (edAFP) at the N terminus and edCelurean at the C terminus as FRET donor and acceptor, and with the truncated 74‐amino‐acid AtGAI (D28–D101) and the AtGID1C linked via a 12‐amino‐acid linker (L12) as the sensory domains. GPS1 responds to nanomolar concentrations of GA4. The K D values of GPS1 for GA1, GA3 and GA4 were 110, 240 and 24 nm, respectively (Tables 1 and 2). The binding to GPS1 was partially reversible for GA1 and GA3, but not for GA4. To assess the GA distribution in planta, a nuclear‐targeted variant of GPS1 (nlsGPS1) was stably expressed under the control of the ubiquitous constitutive p16 promoter in Arabidopsis. GPS1 emission was detected in individual cells of both roots and shoots. In seedling roots, a gradient of GA was observed, with a low nlsGPS1 emission ratio in the apical cell division zone that transitioned to increasing nlsGPS1 emission ratio in the elongation zone. nlsGPS1 also showed a higher concentration of GA in the elongation zone of dark‐grown hypocotyls. Furthermore, the sensor was successfully used to evaluate hormone levels in Arabidopsis mutants. The FRET‐based GPS1 sensor was successfully used to monitor GA levels and investigate the quantitative relationship between the distribution of gibberellin and cell elongation (Rizza et al., 2017).
Figure 3.
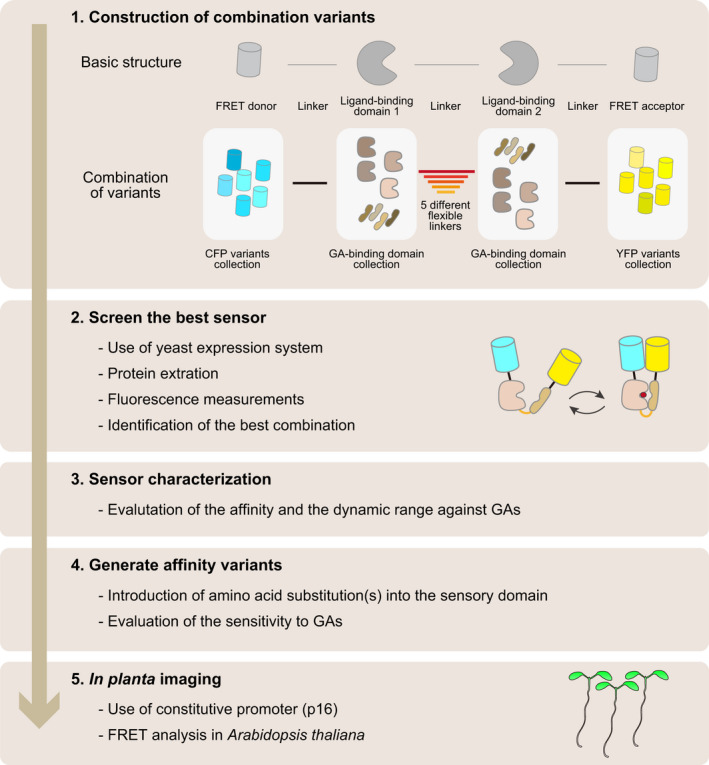
Flow chart for biosensor engineering. We present the engineering procedure for a plant hormone biosensor, in this instance the gibberellin (GA) sensor GPS1 (Jones et al., 2014; Rizza et al., 2017). The biosensor was created by combining domains of different bipartite GA receptors through an artificial linker and flanking the ligand‐binding domains with potential Förster resonance energy transfer (FRET) pairs. Linkers and collections of fluorescent protein (FP) variants were tested as FRET pairs and compared in high‐throughput yeast expression assays. Candidate GA‐binding domains were selected from the receptors AtGID1A, AtGID1B and AtGID1C, and truncated AtGAI or AtRGA domains (DELLA protein domain). The GA‐binding domains were linked to each other by different linkers. The combinatorial library was constructed using Gateway® cloning technology, introduced into a protease‐deficient yeast strain, expressed and isolated. The best five‐way combination was screened by in vitro fluorescence measurements of the sensors isolated from yeast. Affinity mutants with reduced or no response to GAs were engineered by rational design. The affinity mutants can be used as negative controls during in vivo analysis to exclude artifacts such as those caused by pH changes. GPS1 and the affinity mutant GPS1‐NR were expressed in Arabidopsis plants under the control of a constitutive promoter (p16).
JASMONIC ACID
Jasmonic acid (JA) is a product of the octadecanoid pathway (Turner et al., 2002). The amino acid conjugate Ja‐Ile is the bioactive form responsible for signaling (Ruan et al., 2019). JA is involved in plant responses to abiotic/biotic stresses, plant growth and development (Turner et al., 2002). Similar to the case of the auxin and gibberellin signaling pathways, JA is also perceived by a two‐component receptor that is part of an SCF complex and is involved in the degradation of downstream effectors like JAZ. At low JA levels, JAZ represses transcription factors that affect JA‐responsive genes. JA‐Ile promotes the binding of JAZ to the F‐box protein COI1, leading in turn to the ubiquitination of JAZ and the degradation of the complex. As a result, transcription factors of JA‐responsive genes are released from repression (Chini et al., 2007; Thines et al., 2007). A JA degron sensor was developed on the basis of the JA‐dependent degradation of JAZ protein (Larrieu et al., 2015). A specific Jas motif in the JAZ proteins is responsible for its targeting for degradation. The Jas motif of AtJAZ9 protein was fused to Venus‐NLS driven by the CaMV 35S promoter (Jas9‐VENUS). The degradation of Jas9‐VENUS depends on COI1. Jas9‐VENUS was used to measure fluorescence in roots after leaf injury and to investigate the long‐distance JA signaling in Arabidopsis seedling (Larrieu et al., 2015)(Table 1).
STRIGOLACTONES
Strigolactones (SLs) are a diverse group of plant hormones that share a common core: a tricyclic lactone coupled to a hydroxymethyl butanolide (Waters et al., 2017). SLs are involved in branching inhibition and root development, and also function as signaling molecules that induce symbiosis and parasitic interactions. SL signaling is initiated by binding to the α/β‐hydrolase D14/AtD14 (Umehara et al., 2008). SL‐binding to D14 leads to the formation of a co‐receptor complex comprised of the F‐box protein D3/MAX2 and target regulator proteins in the D53/SMXL family (Zhou et al., 2013; Waters et al., 2017). D3/MAX2 engages an SCF complex, leading to the proteolytic degradation of the D53 protein (Waters et al., 2017). Using this SL‐triggered degradation mechanism, degron‐based sensors were developed. In rice roots, D53‐GFP enabled the visualization of D53 degradation in response to the addition of 5 µm of the synthetic SL rac‐GR24 (Zhou et al., 2013). In Arabidopsis roots, the degradation of SMXL6‐YFP and SMXL7‐YFP were also induced by rac‐GR24 treatment (Bennett et al., 2016; Liang et al., 2016). Another degron‐based SL sensor is the ratiometric StrigoQuant (Samodelov et al., 2016). StrigoQuant was developed with the same structure as min17‐Luc, the ratiometric sensor for auxin, using SMXL6, a homolog of rice D53, as the sensor module instead of the auxin‐responsive DII. Other ratiometric SL biosensors, namely rDAD2cpGFP and rShHTL7cpGFP, incorporate circularly permutated GFP (cpGFP) with the SL receptors DAD2 from Petunia hybrida (D14 in rice) or HTL7 from Striga hermonthica (Chesterfield et al., 2020). The cpGFP can report conformational rearrangements induced by the ligand that cause a change in fluorescence intensity. LSSmOrange was fused to the C terminus of DAD2cpGFP and ShHTL7cpGFP via a rigid linker as reference controls. These ratiometric sensors have been used in tobacco protoplasts (Chesterfield et al., 2020).
PEPTIDE HORMONES
Peptide hormones are small signaling peptides such as phytosulfokine (PSK) and CLAVATA 3 (CLV3) (Murphy et al., 2012). Most known peptide hormones interact with receptors that assemble into heteromeric complexes upon peptide addition. Such induced receptor interactions can be visualized by FRET between appropriately engineered fusions of receptors and FPs. The use of a peptide receptor as a sensor was conceptionally shown using the well‐established interaction of the immune receptor FLS2 with its co‐receptor BAK1, which is triggered by the presence of the flg22 peptide. Upon peptide addition to Nicotiana benthamiana leaf cells expressing FLS2‐mCherry and BAK1‐GFP, rapid changes in fluorescence lifetime and fluorescence anisotropy indicated the peptide‐dependent induction of the assembly of heteromeric and homomeric receptor complexes with high spatial resolution (Somssich et al., 2015). The functionality of this approach has not yet been confirmed in transgenic Arabidopsis plants, however, and improvements in the design of such complex sensor systems may be required to achieve the necessary sensitivity. Furthermore, many peptides do not induce receptor complex assembly but complex rearrangements or redistribution into nanoclusters at the membrane, thus generating a technically more demanding output for further analysis (Bücherl et al., 2017).
OTHER HORMONES
Karrikins (KARs) are butenolides found in wildfire smoke that stimulate seed germination (Nelson et al., 2009). KARs are also involved in photomorphogenesis, root hair development, root density and drought responses. The KAR signaling mechanism is apparently highly similar to that for SLs. The putative KAR receptor, KARRIKIN INSENSITIVE 2 (KAI2), is a homolog of D14 (Waters et al., 2012). In Arabidopsis, KAR signaling is mediated by KAI2, MAX2, SMAX1 and SMXL2, which increase the expression of D14‐LIKE2 (DLK2). The first developed transcriptional KAR reporter is DLK2:LUC (Sun et al., 2016). Arabidopsis seeds carrying DLK2:LUC are incubated with bioactive KARs and the cell lysate can be used for a LUC assay. In this system, the application of 10 nm KAR2 and 1 µm KAR1 led to a fourfold increase in luciferase activity after 24 h (Table 1). Recently, a degron‐based ratiometric KAR/KL biosensor was developed based on the SMAX1 degradation mechanism (Khosla et al., 2020).
The development of biosensors for other plant hormones, such as salicylic acid (SA) and nitric oxide (NO), is also of major interest. For the detection of SA, a bacterial reporter strain was engineered by inserting a luciferase gene into the chromosome between salA and salR (Acinetobacter sp. ADPWH_lux). The expression of salA‐luxCDABE‐salR responds to SA. This reporter strain was successfully used to visualize the spatiotemporal distribution of SA in leaves of plants during virus infection (Huang et al., 2006). NPR1‐GFP, which is known to migrate from the cytoplasm to the nucleus in response to SA accumulation (Mou et al., 2003), has also been used as an SA reporter. The reporter indicated that defense responses induced by lipopolysaccharides result in SA accumulation in the leaves (Sun et al., 2012). For NO, several FP‐based sensors, including those using FRET, have been developed for mammalian cells, but none has been tested in plants thus far (Eroglu et al., 2018).
HORMONE TRANSPORTER ACTIVITY REPORTERS
The FRET and cpFP‐based sensors make use of conformational rearrangements in a binding protein or receptor. We hypothesized that as transport proteins undergo complex rearrangements during the transport cycle, the concepts used for engineering binding protein‐based sensors could be used to build reporters for the activity of specific transporters (or enzymes). The approach was successfully used to construct reporters for ammonium and nitrate transport (AmTrac, Amtryoshka and NiTrac) (De Michele et al., 2013; Ho and Frommer, 2014; Ast et al., 2017). Evidence has been provided that members of the CHL1/NRT1/NPF family, which include the nitrate transporter used to build NiTrac, are able to transport a wide range of hormones (Krouk et al., 2010; Chiba et al., 2015; Michniewicz et al., 2019). It is therefore conceivable that NiTrac could also respond to IAA, and that other hormone transporters could be used to engineer transport activity reporters.
FUTURE IMPROVEMENTS THROUGH ENGINEERING THE BIOSENSORS
Sensors that exploit FRET between two spectral variants were the first reporters used to create sensors for calcium and many metabolites (Okumoto et al., 2012). In the simplest form a binding protein was sandwiched between the two fluorophores, as in the prototype maltose and glucose sensors (Fehr et al., 2002; Fehr et al., 2003; Okumoto et al., 2005). Notably, in these sensors, bacterial proteins were used that have a low likelihood of interacting with endogenous plant proteins, thus avoiding artifacts that could occur when using endogenous proteins as recognition elements for sensor construction. Ligand‐induced conformational rearrangements change the relative emission of chimeric sensors. Typical FRET sensors have detection ranges of about two orders of magnitude and a rather limited signal‐to‐noise ratio (SNR). A major advantage of using bacterial periplasmic binding proteins rather than endogenous receptors is that periplasmic binding proteins typically have a high affinity for the ligands, allowing for the rapid development of a series of affinity mutants to cover a broad detection range. Miyawaki and Tsien’s original calcium sensor used an endogenous calmodulin combined with an endogenous calcium‐calmodulin binding protein as a means to increase the conformational rearrangement (Miyawaki et al., 1997). Later on, they improved the sensor by introducing orthogonal mutations to avoid interaction with the endogenous machinery. A second approach, also developed by Tsien’s lab, was the use of a circularly permutated fluorescent protein (cpFP) as a reporter (Baird et al., 1999; Akerboom et al., 2009). These intensiometric sensors often have a much higher sensitivity than other FRET sensors, yet they are sensitive to the changes in sensor levels that arise from promoter activity variability or differential degradation. Recently, Matryoshka technology was developed to convert the intensiometric sensors into ratiometric sensors (Figure 4) (Ast et al., 2017). As an alternative, the fusion of a second FP was used to achieve a comparable result, although specific advantages such as simultaneous excitation and single‐step insertion are not possible in this case (Waadt et al., 2017).
Figure 4.
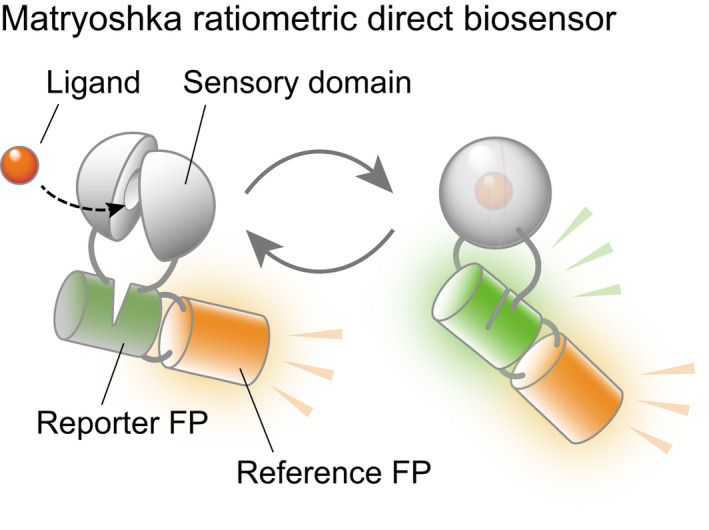
Further optimization of genetically encoded hormone biosensors. A model of a ratiometric direct biosensor. The sensory domain is linked to an environmentally sensitive circularly permutated fluorescent protein (cpFP) that carries an inserted reference FP (like a Matryoshka or nested doll) (Ast et al., 2017). Ligand binding brings the reporter FP components back together and increases the fluorescence. A green–orange (GO) Matryoshka cassette composed of cpsfGFP and LSSmOrange can be used for engineering ratiometric sensors that make use of the large change in intensity caused by analyte binding in the cpFP.
As all these sensors make use of fluorescent proteins and operate on the principle of conformational changes in a recognition element, it is worth noting that the conformation of the recognition element and the fluorescent proteins are sensitive to many other parameters, such as ionic strength, pH and redox status (Hochreiter et al., 2015). The maturation time of donor and acceptor fluorescent proteins can also vary the fluorescent read‐out (Liu et al., 2018). Affinity mutants are thus important and useful controls for excluding artifacts. Surprisingly, affinity mutants have so far been rarely used in studies and only in the animal/medical field. The selectivity of ligands for a sensory domain should also be considered, because binding proteins have often been tested with only a few analytes. This is especially important for hormones such as GA that exist as many different analogs. We could expect that hormone sensors could be fine‐tuned for each individual analog. Mutations used to create such variants are often in the binding pocket and can generate both high‐affinity and low‐affinity sensors (Jones et al., 2014), which may impart ligand selectivity to the sensors. An extensive structure–activity relationship (SAR) analysis enables a better interrogation of the ligand–receptor binding machinery (Rigal et al., 2014). If the ligand affinity is too high, the binding protein may interfere with endogenous signaling machinery. This might be avoided if orthologous systems are employed. Although biosensors represent minimally invasive tools, they can have effects on the cell, either by acting as scavengers or by interacting with other cellular components. They thus essentially represent an ‘observer effect’ problem (Buks et al., 1998). From another perspective, such a drawback can be turned into an advantage. The binding protein can be deployed intentionally as a molecular ‘sponge’ that sequesters a ligand (Armbruster et al., 2020), providing an effective tool for the functional analysis of signal transduction. In the absence of even less‐invasive technologies, it will be important to keep this in mind and to perform proper controls. For example, when a glucose sensor was used in yeast, the analysis of growth curves and even flux analysis did not reveal any significant impact of the sensors on metabolism (Bermejo et al., 2010), whereas some surface‐displayed glutamate sensors clearly impacted processes that led to defects in growth and development (Castro‐Rodríguez et al., 2020), and some others did not have any clear impact (Toyota et al., 2018). Affinity mutants are ideal controls, as they often differ only in a single amino acid, and should respond differently if the detected ratio change is caused by a change in the analyte concentration.
Genetically encoded sensors can be targeted to specific compartments, or even attached to specific proteins. For example, a calcium sensor can be attached to a calcium channel to obtain insights into calcium dynamics in the vicinity of the channel. Although a cytosolic biosensor does not require a targeting sequence, it is not always easy to visualize analytes and detect them separately in each cell, as many plant cells carry large vacuoles, squeezing the cytoplasm towards the cell periphery. Nuclear targeting can help with the identification and tracking of individual cells (Rizza et al., 2017), which provides an option for multidimensional imaging.
Transcriptional sensors report the result of a treatment and thus report on past activity. As some reporter molecules, like GUS, are extremely stable in living cells, the reporter integrates over comparatively long periods, which can increase sensitivity. By contrast, most genetically encoded binding‐dependent sensors report analyte changes with high temporal and even subcellular spatial resolution. For instance, a new type of genetically encoded sensor was developed that provides activity snapshots within the brain (Fosque et al., 2015). The prototype for such activity snapshot sensors is CaMPARI. CaMPARI carries a photoswitchable FP. When calcium levels increase in a particular cell, the FP in CaMPARI can be red‐shifted only when a calcium elevation coincides with user‐controlled violet light. CaMPARI has been used successfully in zebrafish, flies and mice. No such sensor for hormones has been reported so far, but the generation of hormone sensors that make use of photoswitchable FPs or Matryoshka variants may provide a promising starting point for engineering sensors that can take hormone activity snapshots.
IMAGING TECHNOLOGIES FOR THE QUANTIFICATION OF SENSOR OUTPUT
Initially, epifluorescence microscopy in conjunction with high‐quality electron‐multiplying CCD (EMCCD) cameras and simple perfusions systems were used for the quantitative imaging of sensor responses. These systems use either fast switching between multiple excitation and multiple emission wavelengths or devices for image splitting or multiple cameras. Extensive protocols have been published for FRET sensor quantification in yeast and animal cells and contain important information for establishing such imaging systems (Hou et al., 2011; Bermejo et al., 2011). Although confocal microscopy is generally considered to be more advanced, it bears some risks, such as when treatment with an osmotically active substance affects the cell diameter in the z‐axis and causes a shift of the focal plane to a different region of the cell. By contrast, epifluorescence integrates over the whole z‐axis and can provide information if z‐stacks are generated. There are possible artifacts that arise from differences in the thickness of the organs. For example, in roots, one can observe single cells in a single layer in the periphery, but there are multiple tissues overlaying each other in the z‐axis in the center. The adoption of nuclear‐targeting biosensors may overcome these difficulties.
Plants sense and respond to mechano‐stimulation by synthesizing plant hormones and other chemicals (Chehab et al., 2008). As a similar response occurs in plants stimulated by more subtle mechanical cues, such as touch, technological advances for eliminating physical contact are needed. The initial development of a new imaging platform made use of small baths created with Play‐Doh and peristaltic pumps (Deuschle et al., 2006; Chaudhuri et al., 2008; Chaudhuri et al., 2011). Such systems can provide substantial throughput, but are typically limited to single seedlings and require substantial set‐up time for each seedling. The RootChip, a microfluidic chip platform, allows root growth in a controlled environment on the microscope (Grossmann et al., 2011; Grossmann et al., 2012). Combined imaging using FRET sensors with the RootChip can achieve the non‐invasive real‐time detection of metabolites such as cytosolic zinc and calcium levels and fluxes in Arabidopsis roots (Lanquar et al., 2014; Stanley et al., 2018). Although RootChip can provide opportunities to observe the distribution and dynamics of plant hormones in roots, similar imaging platforms must be developed for other tissues.
Another major goal of plant hormone research is to achieve four‐dimensional imaging, which can analyze dynamics in an entire cell or tissue over time. Over the last 10 years, light‐sheet fluorescence microscopy (LSFM) has contributed substantially to the emerging field of real‐time visualization of complex developmental processes. As a result of the low photo‐toxicity and high‐speed multiview acquisition, LSFM has proven to be a powerful tool for the study of organ morphogenesis and function in zebrafish, Drosophila and other model organisms. Although still in its infancy, LSFM is increasingly being used in plant science (Grossmann et al., 2018). Recently, FRET sensors were used with LSFM to monitor the dynamics of MgATP2– in root hairs (De Col et al., 2017) and intracellular calcium release during root or root hair growth (Costa et al., 2013; Candeo et al., 2017). LSFM does still have some limitations in sample conditioning, however, as a result of the spatial arrangement between the lens that produces the single‐plane illumination and the objective lens that is used for imaging. Despite these limitations, LSFM imaging technology has the potential to significantly advance the real‐time quantitative imaging of hormones in living plants.
OUTLOOK
Plant hormones are highly dynamic and interact in complex ways with each other. The relationships between these dynamics and their physiological phenotypes regulated by plant hormone signaling are not fully understood. Different approaches to follow this dynamism have been undertaken. Imaging mass spectrometry has been developed as a powerful method to detect the distribution of multiple hormones simultaneously (Shiono et al., 2017; Shiono and Taira, 2020), but, for now, the spatial resolution of imaging mass spectrometry is comparatively low. As a complementary technology, biosensors that provide extremely high spatiotemporal information are in great demand. In animal research, the trend is shifting to biosensors that are based on fluorescence lifetime changes (Greenwald et al., 2018). Although not intuitive, this requires the complete redesign of sensors for optimization to the lifetime of fluorescence. Although many ‘FRET sensors’ appear to carry out non‐radiative energy transfer from the donor to the acceptor fluorophore, as evidenced by the emission of the FP with the longer wavelength that is not excited directly, the change seen in the ratio is not necessarily a result of FRET. For example, for the FLIPglu FRET glucose sensors, we have not detected a glucose‐induced change in donor FP lifetime. Thus, we do not recommend using terms such as FRET channel or axis labels such as FRET change, as scientists rarely measure absolute FRET values, but instead measure ratios. NiTrac is a good example of an unexpected although useful effect, i.e., a change in donor quenching or absorption in the sensor caused by the addition of nitrate substrate (Ho and Frommer, 2014). Pioneering work in animal systems conducted by the Zhang lab has enabled the multiplexing and simultaneous recording of many processes, in part through differential subcellular targeting and in part through the development of fluorescence‐lifetime imaging (FLIM) sensors (Greenwald et al., 2018). Recently, multiparametric imaging analysis has been achieved by 2‐in‐1 genetically encoded fluorescence indicators (GEFIs), consisting of two GEFIs fused via a 14‐amino‐acid linker or the self‐cleaving 22‐amino‐acid P2A linker (Waadt et al., 2020). The type of biosensor, it’s ligand affinity and the dynamic range should be selected based on the characteristics of the phenomenon to be observed in the plants. The recent intensely active development of plant hormone sensors shows the importance of biosensors, but this field is still at the frontier. Matryoshka technology may help to advance the detection of hormone dynamics in plant cells. Understanding complex hormonal networks with temporal and spatial information is the critical step to a comprehensive understanding of plant hormone science.
ACKNOWLEDGEMENTS
This research was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany´s Excellence Strategy (EXC‐2048/1, project ID 390686111), the Alexander von Humboldt Professorship and a Japan Society for the Promotion of Science (JSPS) grant (19H000932) to WBF, and the Human Frontier Science Program to MN. ITbM is supported by the World Premier International Research Center Initiative (WPI), Japan.
AUTHOR CONTRIBUTIONS
RI, WBF and MN planned the article. All authors wrote the article and prepared the figures.
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
Contributor Information
Wolf B. Frommer, Email: frommew@hhu.de.
Masayoshi Nakamura, Email: mnakamu@itbm.nagoya-u.ac.jp.
References
- Akerboom, J. , Rivera, J.D. , Guilbe, M.M. et al (2009) Crystal structures of the GCaMP calcium sensor reveal the mechanism of fluorescence signal change and aid rational design. J. Biol. Chem. 284, 6455–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, F. , Zhao, Q. , Ji, Y. et al (2010) Ethylene‐induced stabilization of ETHYLENE INSENSITIVE3 and EIN3‐LIKE1 Is mediated by proteasomal degradation of EIN3 binding F‐Box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell, 22, 2384–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster, M. , Dulla, C.G. and Diamond, J.S. (2020) Effects of fluorescent glutamate indicators on neurotransmitter diffusion and uptake J. Huguenard, ed. eLife, 9, e54441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ast, C. , Foret, J. , Oltrogge, L.M. , Michele, R.D. , Kleist, T.J. , Ho, C.‐H. and Frommer, W.B. (2017) Ratiometric Matryoshka biosensors from a nested cassette of green‐ and orange‐emitting fluorescent proteins. Nat. Comm. 8, 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird, G.S. , Zacharias, D.A. and Tsien, R.Y. (1999) Circular permutation and receptor insertion within green fluorescent proteins. Proc. Natl Acad. Sci. USA, 96, 11241–11246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balleza, E. , Kim, J.M. and Cluzel, P. (2018) Systematic characterization of maturation time of fluorescent proteins in living cells. Nat. Methods, 15, 47–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, T. , Liang, Y. , Seale, M. , Ward, S. , Müller, D. and Leyser, O. (2016) Strigolactone regulates shoot development through a core signalling pathway. Biol Open, 5, 1806–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo, C. , Haerizadeh, F. , Takanaga, H. , Chermak, D. and Frommer, W.B. (2010) Dynamic analysis of cytosolic glucose and ATP levels in yeast using optical sensors. Biochemical J. 432, 399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo, C. , Haerizadeh, F. , Takanaga, H. , Chermak, D. and Frommer, W.B. (2011) Optical sensors for measuring dynamic changes of cytosolic metabolite levels in yeast. Nat. Protoc. 6, 1806–1817. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L. , Briginshaw, L.N. , Fisher, T.J. and Flores‐Sandoval, E. (2019) Something ancient and something neofunctionalized—evolution of land plant hormone signaling pathways. Curr. Opin Plant Biol. 47, 64–72. [DOI] [PubMed] [Google Scholar]
- Brunoud, G. , Wells, D.M. , Oliva, M. et al (2012) A novel sensor to map auxin response and distribution at high spatio‐temporal resolution. Nature, 482, 103–106. [DOI] [PubMed] [Google Scholar]
- Bücherl, C.A. , Jarsch, I.K. , Schudoma, C. , Segonzac, C. , Mbengue, M. , Robatzek, S. , MacLean, D. , Ott, T. and Zipfel, C. (2017) Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. eLife, 6, e25114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buks, E. , Schuster, R. , Heiblum, M. , Mahalu, D. and Umansky, V. (1998) Dephasing in electron interference by a ‘which‐path’ detector. Nature, 391, 871–874. [Google Scholar]
- Candeo, A. , Doccula, F.G. , Valentini, G. , Bassi, A. and Costa, A. (2017) Light sheet fluorescence microscopy quantifies calcium oscillations in root hairs of Arabidopsis thaliana . Plant Cell Physiol. 58, 1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro‐Rodríguez, V. , Kleist, T.J. , Gappel, N.M. , Okumoto, S. , Machado, M. , Denyer, T. , Timmermans, M.C.P. , Frommer, W.B. and Wudick, M.M. (2020) Noxious effects of cell surface display glutamate sensors on plant growth and development. bioRxiv, 2020.03.24.005223.
- Chaiwanon, J. and Wang, Z.‐Y. (2015) Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Curr. Biol. 25, 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri, B. , Hörmann, F. and Frommer, W.B. (2011) Dynamic imaging of glucose flux impedance using FRET sensors in wild‐type Arabidopsis plants. J. Exp. Bot. 62, 2411–2417. [DOI] [PubMed] [Google Scholar]
- Chaudhuri, B. , Hörmann, F. , Lalonde, S. , Brady, S.M. , Orlando, D.A. , Benfey, P. and Frommer, W.B. (2008) Protonophore‐ and pH‐insensitive glucose and sucrose accumulation detected by FRET nanosensors in Arabidopsis root tips. Plant J., 56, 948–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehab, E.W. , Kaspi, R. , Savchenko, T. , Rowe, H. , Negre‐Zakharov, F. , Kliebenstein, D. and Dehesh, K. (2008) Distinct roles of jasmonates and aldehydes in plant‐defense responses. PLoS One, 3, e1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesterfield, R.J. , Whitfield, J.H. , Pouvreau, B. , Cao, D. , Beveridge, C.A. and Vickers, C.E. (2020) Rational design of novel fluorescent enzyme biosensors for direct detection of strigolactones. bioRxiv, 2020.03.10.986562. [DOI] [PubMed]
- Chiba, Y. , Shimizu, T. , Miyakawa, S. , Kanno, Y. , Koshiba, T. , Kamiya, Y. and Seo, M. (2015) Identification of Arabidopsis thaliana NRT1/PTR FAMILY (NPF) proteins capable of transporting plant hormones. J. Plant Res. 128, 679–686. [DOI] [PubMed] [Google Scholar]
- Chini, A. , Fonseca, S. , Fernández, G. et al (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature, 448, 666–671. [DOI] [PubMed] [Google Scholar]
- Costa, A. , Candeo, A. , Fieramonti, L. , Valentini, G. and Bassi, A. (2013) Calcium dynamics in root cells of Arabidopsis thaliana visualized with selective plane illumination microscopy. PLoS One, 8, e75646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler, S.R. , Rodriguez, P.L. , Finkelstein, R.R. and Abrams, S.R. (2010) Abscisic acid: emergence of a core signaling network. Annu Rev. Plant Biol. 61, 651–679. [DOI] [PubMed] [Google Scholar]
- D’Agostino, I.B. , Deruere, J. and Kieber, J.J. (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 124, 1706–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Col, V. , Fuchs, P. , Nietzel, T. et al (2017) ATP sensing in living plant cells reveals tissue gradients and stress dynamics of energy physiology. eLife, 6, e26770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Michele, R. , Ast, C. , Loque, D. et al (2013) Fluorescent sensors reporting the activity of ammonium transceptors in live cells. eLife, 2, e00800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuschle, K. , Chaudhuri, B. , Okumoto, S. , Lager, I. , Lalonde, S. and Frommer, W.B. (2006) Rapid metabolism of glucose detected with FRET glucose nanosensors in epidermal cells and intact roots of Arabidopsis RNA‐silencing mutants. Plant Cell, 18, 2314–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eroglu, E. , Charoensin, S. , Bischof, H. , Ramadani, J. , Gottschalk, B. , Depaoli, M.R. , Waldeck‐Weiermair, M. , Graier, W.F. and Malli, R. (2018) Genetic biosensors for imaging nitric oxide in single cells. Free Radic. Biol. Med. 128, 50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr, M. , Frommer, W.B. and Lalonde, S. (2002) Visualization of maltose uptake in living yeast cells by fluorescent nanosensors. Proc. Natl. Acad. Sci. USA, 99, 9846–9851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr, M. , Lalonde, S. , Lager, I. , Wolff, M.W. and Frommer, W.B. (2003) In vivo imaging of the dynamics of glucose uptake in the cytosol of COS‐7 cells by fluorescent nanosensors. J. Biol. Chem. 278, 19127–19133. [DOI] [PubMed] [Google Scholar]
- Fernandez‐Moreno, J.‐P. and Stepanova, A.N. (2020) Monitoring Ethylene in Plants: genetically encoded Reporters and biosensors. Small Meth., 4, 1900260. [Google Scholar]
- Forestan, C. and Varotto, S. (2013) Auxin immunolocalization in plant tissues I De Smet, ed., Totowa, NJ: Humana Press; https://link.springer.com/protocol/10.1007%2F978‐1‐62703‐221‐6_15 [Accessed October 20, 2020]. [DOI] [PubMed] [Google Scholar]
- Fosque, B.F. , Sun, Y. , Dana, H. et al (2015) Neural circuits. Labeling of active neural circuits in vivo with designed calcium integrators. Science, 347, 755–760. [DOI] [PubMed] [Google Scholar]
- Fukami‐Kobayashi, K. , Tateno, Y. and Nishikawa, K. (1999) Domain dislocation: a change of core structure in periplasmic binding proteins in their evolutionary history. J. Mol. Biol. 286, 279–290. [DOI] [PubMed] [Google Scholar]
- Galli, M. , Liu, Q. , Moss, B.L. et al (2015) Auxin signaling modules regulate maize inflorescence architecture. Proc. Natl. Acad. Sci. USA, 112, 13372–13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemperline, E. , Keller, C. , Jayaraman, D. , Maeda, J. , Sussman, M.R. , Ané, J.‐M. and Li, L. (2016) Examination of endogenous peptides in Medicago truncatula using mass spectrometry imaging. J. Proteome Res. 15, 4403–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald, E.C. , Mehta, S. and Zhang, J. (2018) Genetically Encoded Fluorescent Biosensors Illuminate the Spatiotemporal Regulation of Signaling Networks. Chem. Rev. 118, 11707–11794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann, G. , Guo, W.J. , Ehrhardt, D.W. , Frommer, W.B. , Sit, R.V. , Quake, S.R. and Meier, M. (2011) The RootChip: an integrated microfluidic chip for plant science. Plant Cell, 23, 4234–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann, G. , Krebs, M. , Maizel, A. , Stahl, Y. , Vermeer, J.E.M. and Ott, T. (2018) Green light for quantitative live‐cell imaging in plants. J Cell Sci, 131. Available at: https://jcs.biologists.org/content/131/2/jcs209270 [Accessed June 22, 2020]. [DOI] [PubMed]
- Grossmann, G. , Meier, M. , Cartwright, H.N. , Sosso, D. , Quake, S.R. , Ehrhardt, D.W. and Frommer, W.B. (2012) Time‐lapse fluorescence imaging of Arabidopsis root growth with rapid manipulation of the root environment using the RootChip. J. Visualized Experiments : JoVE. Available at http://www.ncbi.nlm.nih.gov/pubmed/22805296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H. and Ecker, J.R. (2003) Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)‐dependent proteolysis of EIN3 transcription factor. Cell, 115, 667–677. [DOI] [PubMed] [Google Scholar]
- Hayashi, K. , Nakamura, S. , Fukunaga, S. et al (2014) Auxin transport sites are visualized in planta using fluorescent auxin analogs. Proc. Natl. Acad. Sci. USA, 111, 11557–11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herud‐Sikimic, O. , Stiel, A.C. , Ortega‐Perez, M. , Shanmugaratnam, S. , Höcker, B. and Jürgens, G. (2020) Design of a biosensor for direct visualisation of auxin. bioRxiv, 2020.01.19.911735.
- Ho, C.‐H. and Frommer, W.B. (2014) Fluorescent sensors for activity and regulation of the nitrate transceptor CHL1/NRT1.1 and oligopeptide transporters. Elife, 3, e01917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochreiter, B. , Garcia, A.P. and Schmid, J.A. (2015) Fluorescent proteins as genetically encoded FRET biosensors in life sciences. Sensors, 15, 26281–26314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, B.H. , Takanaga, H. , Grossmann, G. et al (2011) Optical sensors for monitoring dynamic changes of intracellular metabolite levels in mammalian cells. Nat. Protoc. 6, 1818–1833. [DOI] [PubMed] [Google Scholar]
- Hsu, P.‐K. , Takahashi, Y. , Munemasa, S. , Merilo, E. , Laanemets, K. , Waadt, R. , Pater, D. , Kollist, H. and Schroeder, J.I. (2018) Abscisic acid‐independent stomatal CO2 signal transduction pathway and convergence of CO2 and ABA signaling downstream of OST1 kinase. Proc. Natl. Acad. Sci. USA, 115, E9971–E9980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W.E. , Huang, L. , Preston, G.M. , Naylor, M. , Carr, J.P. , Li, Y. , Singer, A.C. , Whiteley, A.S. and Wang, H. (2006) Quantitative in situ assay of salicylic acid in tobacco leaves using a genetically modified biosensor strain of Acinetobacter sp. ADP1. Plant J., 46, 1073–1083. [DOI] [PubMed] [Google Scholar]
- Irani, N.G. , Di Rubbo, S. , Mylle, E. et al (2012) Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat. Chem. Biol. 8, 583–589. [DOI] [PubMed] [Google Scholar]
- Jones, A.M. , Danielson, J.A. , Manojkumar, S.N. , Lanquar, V. , Grossmann, G. and Frommer, W.B. (2014) Abscisic acid dynamics in roots detected with genetically encoded FRET sensors. eLife, 3, e01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven‐Gershon, T. and Kadonaga, J.T. (2010) Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 339, 225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper, T. , Looger, L.L. , Takanaga, H. , Platten, M. , Steinman, L. and Frommer, W.B. (2007) Nanosensor detection of an immunoregulatory tryptophan influx/kynurenine efflux cycle. PLoS Biol. 5, e257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla, A. , Morffy, N. , Li, Q. et al (2020) Structure‐function analysis of SMAX1 reveals domains that mediate its Karrikin‐Induced proteolysis and interaction with the receptor KAI2. Plant Cell, Available at: http://www.plantcell.org/content/early/2020/05/20/tpc.19.00752 [Accessed June 22, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner, G.K. , Stahl, Y. , Imani, J. , von Korff, M. and Simon, R. (2018) Fluorescent reporter lines for auxin and cytokinin signalling in barley (Hordeum vulgare). PLoS One, 13, e0196086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk, G. , Lacombe, B. , Bielach, A. et al (2010) Nitrate‐regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell, 18, 927–937. [DOI] [PubMed] [Google Scholar]
- Lanquar, V. , Grossmann, G. , Vinkenborg, J.L. , Merkx, M. , Thomine, S. and Frommer, W.B. (2014) Dynamic imaging of cytosolic zinc in Arabidopsis roots combining FRET sensors and RootChip technology. New Phytol. 202, 198–208. [DOI] [PubMed] [Google Scholar]
- Larrieu, A. , Champion, A. , Legrand, J. et al (2015) A fluorescent hormone biosensor reveals the dynamics of jasmonate signalling in plants. Nat. Comm. 6, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser, O. (2005) Auxin distribution and plant pattern formation: how many angels can dance on the point of PIN? Cell, 121, 819–822. [DOI] [PubMed] [Google Scholar]
- Li, W. , Ma, M. , Feng, Y. , Li, H. , Wang, Y. , Ma, Y. , Li, M. , An, F. and Guo, H. (2015) EIN2‐directed translational regulation of ethylene signaling in Arabidopsis. Cell, 163, 670–683. [DOI] [PubMed] [Google Scholar]
- Liang, Y. , Ward, S. , Li, P. , Bennett, T. and Leyser, O. (2016) SMAX1‐LIKE7 signals from the nucleus to regulate shoot development in Arabidopsis via partially EAR motif‐independent mechanisms. Plant Cell, 28, 1581–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, C.‐Y. , Smet, W. , Brunoud, G. , Yoshida, S. , Vernoux, T. and Weijers, D. (2015) Reporters for sensitive and quantitative measurement of auxin response. Nat. Methods, 12, 207–210 2 p following 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman‐Lazarovich, M. , Yahav, C. , Israeli, A. and Efroni, I. (2019) Deep conservation of cis‐element variants regulating plant hormonal responses. Plant Cell, 31, 2559–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B. , Mavrova, S.N. , van den Berg, J. , Kristensen, S.K. , Mantovanelli, L. , Veenhoff, L.M. , Poolman, B. and Boersma, A.J. (2018) Influence of fluorescent protein maturation on FRET measurements in living cells. ACS Sens. 3, 1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. and Müller, B. (2017) Imaging TCSn:GFP, a synthetic cytokinin reporter, in Arabidopsis thaliana. Methods Mol. Biol., 1497, 81–90. [DOI] [PubMed] [Google Scholar]
- Merchante, C. , Brumos, J. , Yun, J. et al (2015) Gene‐specific translation regulation mediated by the hormone‐signaling molecule EIN2. Cell, 163, 684–697. [DOI] [PubMed] [Google Scholar]
- Michniewicz, M. , Ho, C.‐H. , Enders, T.A. et al (2019) TRANSPORTER OF IBA1 links auxin and cytokinin to influence root architecture. Dev. Cell, 50, 599–609.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki, A. , Llopis, J. , Heim, R. , McCaffery, J.M. , Adams, J.A. , Ikura, M. and Tsien, R.Y. (1997) Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature, 388, 882–887. [DOI] [PubMed] [Google Scholar]
- Möller, B. and Weijers, D. (2009) Auxin control of embryo patterning. Cold Spring Harb Perspect Biol. 1, a001545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno‐Risueno, M.A. , Van Norman, J.M. , Moreno, A. , Zhang, J. , Ahnert, S.E. and Benfey, P.N. (2010) Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science, 329, 1306–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou, Z. , Fan, W. and Dong, X. (2003) Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell, 113, 935–944. [DOI] [PubMed] [Google Scholar]
- Murase, K. , Hirano, Y. , Sun, T. and Hakoshima, T. (2008) Gibberellin‐induced DELLA recognition by the gibberellin receptor GID1. Nature, 456, 459–463. [DOI] [PubMed] [Google Scholar]
- Murphy, E. , Smith, S. and Smet, I.D. (2012) Small signaling peptides in Arabidopsis development: how cells communicate over a short distance. Plant Cell, 24, 3198–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D.C. , Riseborough, J.A. , Flematti, G.R. , Stevens, J. , Ghisalberti, E.L. , Dixon, K.W. and Smith, S.M. (2009) Karrikins discovered in smoke trigger Arabidopsis seed germination by a mechanism requiring gibberellic acid synthesis and light. Plant Physiol. 149, 863–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, T. , Toyooka, K. , Sato, M. , Matsumoto, S. , Lucas, M.M. , Strnad, M. , Baluška, F. and Koshiba, T. (2011) Immunohistochemical observation of indole‐3‐acetic acid at the IAA synthetic maize coleoptile tips. Plant Signal Behav. 6, 2013–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo, Y. , Tanaka, M. , Tabata, R. , Ogawa‐Ohnishi, M. and Matsubayashi, Y. (2017) Shoot‐to‐root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nature Plants, 3, 1–6. [DOI] [PubMed] [Google Scholar]
- Okamoto, M. , Hanada, A. , Kamiya, Y. , Yamaguchi, S. and Nambara, E. (2009) Measurement of abscisic acid and gibberellins by gas chromatography/mass spectrometry. Methods Mol. Biol. 495, 53–60. [DOI] [PubMed] [Google Scholar]
- Okumoto, S. , Jones, A. and Frommer, W.B. (2012) Quantitative imaging with fluorescent biosensors. Annu Rev Plant Biol. 63, 663–706. [DOI] [PubMed] [Google Scholar]
- Okumoto, S. , Looger, L.L. , Micheva, K.D. , Reimer, R.J. , Smith, S.J. and Frommer, W.B. (2005) Detection of glutamate release from neurons by genetically encoded surface‐displayed FRET nanosensors. Proc. Natl. Acad. Sci. USA, 102, 8740–8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz‐Morea, F.A. , Savatin, D.V. , Dejonghe, W. et al (2016) Danger‐associated peptide signaling in Arabidopsis requires clathrin. Proc Natl Acad Sci USA, 113, 11028–11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota, R. , Ohkubo, Y. , Yamashita, Y. , Ogawa‐Ohnishi, M. and Matsubayashi, Y. (2020) Shoot‐to‐root mobile CEPD‐like 2 integrates shoot nitrogen status to systemically regulate nitrate uptake in Arabidopsis. Nat. Comm. 11, 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen, S.J. and Abrams, S.R. (2009) Measurement of plant hormones by liquid chromatography‐mass spectrometry. Methods Mol. Biol. 495, 39–51. [DOI] [PubMed] [Google Scholar]
- Pattison, R.J. and Catalá, C. (2012) Evaluating auxin distribution in tomato (Solanum lycopersicum) through an analysis of the PIN and AUX/LAX gene families. Plant J. 70, 585–598. [DOI] [PubMed] [Google Scholar]
- Qiao, H. , Shen, Z. , Huang, S.S.C. , Schmitz, R.J. , Urich, M.A. , Briggs, S.P. and Ecker, J.R. (2012) Processing and subcellular trafficking of ER‐tethered EIN2 control response to ethylene gas. Science, 338, 390–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigal, A. , Ma, Q. and Robert, S. (2014) Unraveling plant hormone signaling through the use of small molecules. Front Plant Sci, 5. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4115670/ [Accessed June 20, 2020]. [DOI] [PMC free article] [PubMed]
- Rizza, A. , Walia, A. , Lanquar, V. , Frommer, W.B. and Jones, A.M. (2017) In vivo gibberellin gradients visualized in rapidly elongating tissues. Nat. Plants, 3, 803–813. [DOI] [PubMed] [Google Scholar]
- Ruan, J. , Zhou, Y. , Zhou, M. , Yan, J. , Khurshid, M. , Weng, W. , Cheng, J. and Zhang, K. (2019) Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 20, 2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samodelov, S.L. , Beyer, H.M. , Guo, X. et al (2016) StrigoQuant: a genetically encoded biosensor for quantifying strigolactone activity and specificity. Sci. Adv. 2, e1601266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner, A. and Estelle, M. (2010) The ubiquitin‐proteasome system regulates plant hormone signaling. Plant J. 61, 1029–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicht, M. , Strnad, M. , Scanlon, M.J. , Mancuso, S. , Hochholdinger, F. , Palme, K. , Volkmann, D. , Menzel, D. and Baluska, F. (2006) Auxin immunolocalization implicates vesicular neurotransmitter‐like mode of polar auxin transport in root apices. Plant Signal Behav. 1, 122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani, E. , Weinstain, R. , Zhang, Y. , Castillejo, C. , Kaiserli, E. , Chory, J. , Tsien, R.Y. and Estelle, M. (2013) Gibberellins accumulate in the elongating endodermal cells of Arabidopsis root. Proc. Natl. Acad. Sci. U.S.A., 110, 4834–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiono, K. , Hashizaki, R. , Nakanishi, T. et al (2017) Multi‐imaging of cytokinin and abscisic acid on the roots of rice (Oryza sativa ) using Matrix‐Assisted Laser Desorption/Ionization mass spectrometry. J. Agr. Food Chem. 65, 7624–7628. [DOI] [PubMed] [Google Scholar]
- Shiono, K. and Taira, S. (2020) Imaging of multiple plant hormones in roots of rice (Oryza sativa) using nanoparticle‐assisted laser desorption/Ionization mass spectrometry. J. Agric. Food Chem. 68, 6770–6775. [DOI] [PubMed] [Google Scholar]
- Silverstone, A.L. , Jung, H.‐S. , Dill, A. , Kawaide, H. , Kamiya, Y. and Sun, T. (2001) Repressing a repressor: gibberellin‐induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell, 13, 1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich, M. , Ma, Q. , Weidtkamp‐Peters, S. , Stahl, Y. , Felekyan, S. , Bleckmann, A. , Seidel, C.A.M. and Simon, R. (2015) Real‐time dynamics of peptide ligand–dependent receptor complex formation in planta. Sci. Signal. 8, ra76–ra76. [DOI] [PubMed] [Google Scholar]
- Stanley, C.E. , Shrivastava, J. , Brugman, R. , Heinzelmann, E. , van Swaay, D. and Grossmann, G. (2018) Dual‐flow‐RootChip reveals local adaptations of roots towards environmental asymmetry at the physiological and genetic levels. New Phytol. 217, 1357–1369. [DOI] [PubMed] [Google Scholar]
- Stepanova, A.N. , Yun, J. , Likhacheva, A.V. and Alonso, J.M. (2007) Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell, 19, 2169–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stührwohldt, N. and Schaller, A. (2019) Regulation of plant peptide hormones and growth factors by post‐translational modification. Plant Biol. 21, 49–63. [DOI] [PubMed] [Google Scholar]
- Sun, A. , Nie, S. and Xing, D. (2012) Nitric oxide‐mediated maintenance of redox homeostasis contributes to NPR1‐dependent plant innate immunity triggered by lipopolysaccharides. Plant Physiol, 160, 1081–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y.K. , Flematti, G.R. , Smith, S.M. and Waters, M.T. (2016) Reporter gene‐facilitated detection of compounds in Arabidopsis leaf extracts that activate the karrikin signaling pathway. Front. Plant Sci, 7. Available at: https://www.frontiersin.org/articles/10.3389/fpls.2016.01799/full [Accessed June 26, 2020]. [DOI] [PMC free article] [PubMed]
- Swarup, R. , Perry, P. , Hagenbeek, D. , Van Der Straeten, D. , Beemster, G.T.S. , Sandberg, G. , Bhalerao, R. , Ljung, K. and Bennett, M.J. (2007) Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell, 19, 2186–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, J. , Sun, H. , Gu, P. , Liang, Z. , Chen, X. , Lou, J. , Xu, G. and Zhang, Y. (2017) A sensitive synthetic reporter for visualizing cytokinin signaling output in rice. Plant Methods, 13, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines, B. , Katsir, L. , Melotto, M. et al (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature, 448, 661–665. [DOI] [PubMed] [Google Scholar]
- To, J.P.C. , Deruère, J. , Maxwell, B.B. , Morris, V.F. , Hutchison, C.E. , Ferreira, F.J. , Schaller, G.E. and Kieber, J.J. (2007) Cytokinin regulates type‐A Arabidopsis response regulator activity and protein stability via two‐component phosphorelay. Plant Cell, 19, 3901–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota, M. , Spencer, D. , Sawai‐Toyota, S. , Jiaqi, W. , Zhan, T. , Koo, A.J. , Howe, G.A. and Gilroy, S. (2018) Glutamate triggers long‐distance, calcium‐based plant defense signaling. Science, 361, 1112–1115. [DOI] [PubMed] [Google Scholar]
- Tsuchiya, Y. , Yoshimura, M. , Sato, Y. et al (2015) Probing strigolactone receptors in Striga hermonthica with fluorescence. Science, 349, 864–868. [DOI] [PubMed] [Google Scholar]
- Turner, J.G. , Ellis, C. and Devoto, A. (2002) The jasmonate signal pathway. Plant Cell, 14, S153–S164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T. , Murfett, J. , Hagen, G. and Guilfoyle, T.J. (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell, 9, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara, M. , Hanada, A. , Yoshida, S. et al (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature, 455, 195–200. [DOI] [PubMed] [Google Scholar]
- Van Norman, J.M. , Xuan, W. , Beeckman, T. and Benfey, P.N. (2013) To branch or not to branch: the role of pre‐patterning in lateral root formation. Development, 140, 4301–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma, V. , Ravindran, P. and Kumar, P.P. (2016) Plant hormone‐mediated regulation of stress responses. BMC Plant Biol. 16, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vong, K. , Eda, S. , Kadota, Y. , Nasibullin, I. , Wakatake, T. , Yokoshima, S. , Shirasu, K. and Tanaka, K. (2019) An artificial metalloenzyme biosensor can detect ethylene gas in fruits and Arabidopsis leaves. Nat. Comm. 10, 5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt, R. (2020) Phytohormone signaling mechanisms and genetic methods for their modulation and detection. Curr. Opin Plant Biol. 57, 31–40. [DOI] [PubMed] [Google Scholar]
- Waadt, R. , Hitomi, K. , Nishimura, N. , Hitomi, C. , Adams, S.R. , Getzoff, E.D. and Schroeder, J.I. (2014) FRET‐based reporters for the direct visualization of abscisic acid concentration changes and distribution in Arabidopsis. Elife, 3, e01739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt, R. , Köster, P. , Andrés, Z. , Waadt, C. , Bradamante, G. , Lampou, K. , Kudla, J. and Schumacher, K. (2020) Dual‐reporting transcriptionally linked genetically encoded fluorescent indicators resolve the spatiotemporal coordination of cytosolic abscisic acid and second messenger dynamics in Arabidopsis. Plant Cell, 32, 2582–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt, R. , Krebs, M. , Kudla, J. and Schumacher, K. (2017) Multiparameter imaging of calcium and abscisic acid and high‐resolution quantitative calcium measurements using R‐GECO1‐mTurquoise in Arabidopsis. New Phytol. 216, 303–320. [DOI] [PubMed] [Google Scholar]
- Walia, A. , Waadt, R. and Jones, A.M. (2018) Genetically encoded biosensors in plants: pathways to discovery. Annu Rev Plant Biol, 69, 497–524. [DOI] [PubMed] [Google Scholar]
- Waters, M.T. , Gutjahr, C. , Bennett, T. and Nelson, D.C. (2017) Strigolactone signaling and evolution. Annu. Rev. Plant Biol. 68, 291–322. [DOI] [PubMed] [Google Scholar]
- Waters, M.T. , Nelson, D.C. , Scaffidi, A. , Flematti, G.R. , Sun, Y.K. , Dixon, K.W. and Smith, S.M. (2012) Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development, 139, 1285–1295. [DOI] [PubMed] [Google Scholar]
- Wend, S. , Dal Bosco, C. , Kämpf, M.M. , Ren, F. , Palme, K. , Weber, W. , Dovzhenko, A. and Zurbriggen, M.D. (2013) A quantitative ratiometric sensor for time‐resolved analysis of auxin dynamics. Sci. Rep. 3, 2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, R. , Duan, L. , Pruneda‐Paz, J.L. , Oh, D. , Pound, M. , Kay, S. and Dinneny, J.R. (2018) The 6xABRE Synthetic promoter enables the spatiotemporal analysis of ABA‐mediated transcriptional regulation. Plant Physiol. 177, 1650–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, T. , Christmann, A. , Yamaguchi‐Shinozaki, K. , Grill, E. and Fernie, A.R. (2019) Revisiting the basal role of ABA – roles outside of stress. Trends Plant Sci. 24, 625–635. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Takahashi, Y. , Hsu, P.‐K. , Kollist, H. , Merilo, E. , Krysan, P.J. and Schroeder, J.I. (2020) FRET kinase sensor development reveals SnRK2/OST1 activation by ABA but not by MeJA and high CO2 during stomatal closure D. C. Bergmann, C. S. Hardtke, and J. Leung, eds. eLife, 9, e56351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, F. , Lin, Q. , Zhu, L. et al (2013) D14–SCFD3‐dependent degradation of D53 regulates strigolactone signalling. Nature, 504, 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.‐Y. , Sae‐Seaw, J. and Wang, Z.‐Y. (2013) Brassinosteroid signalling. Development, 140, 1615–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zürcher, E. , Liu, J. , di Donato, M. , Geisler, M. and Müller, B. (2016) Plant development regulated by cytokinin sinks. Science, 353, 1027–1030. [DOI] [PubMed] [Google Scholar]


