Tumoral cells secrete an antagonist that attenuates insulin signaling in neurons. It induces mitochondrial defects and synapse loss; restoring neuronal insulin activity rescues neurodegeneration.
Abstract
Cell to cell communication facilitates tissue development and physiology. Under pathological conditions, brain tumors disrupt glia-neuron communication signals that in consequence, promote tumor expansion at the expense of surrounding healthy tissue. The glioblastoma is one of the most aggressive and frequent primary brain tumors. This type of glioma expands and infiltrates into the brain, causing neuronal degeneration and neurological decay, among other symptoms. Here, we describe in a Drosophila model how glioblastoma cells produce ImpL2, an antagonist of the insulin pathway, which targets neighboring neurons and causes mitochondrial disruption as well as synapse loss, both early symptoms of neurodegeneration. Furthermore, glioblastoma progression requires insulin pathway attenuation in neurons. Restoration of neuronal insulin activity is sufficient to rescue synapse loss and to delay the premature death caused by glioma. Therefore, signals from glioblastoma to neuron emerge as a potential field of study to prevent neurodegeneration and to develop anti-tumoral strategies.
Significance Statement
Glioblastoma (GB) is among the most aggressive type of brain tumors, and currently there is no cure or effective treatment. Patients course with neurological decay and previous data in animal models indicate that GB causes a neurodegenerative process. In a Drosophila model, we describe here a molecule named ImpL2 that is produced by GB cells and impacts on neighboring neurons. ImpL2 is an antagonist of the insulin pathway, and insulin signaling reduction in neurons causes mitochondrial defects and synapse loss. These mechanisms underlying GB-induced neurodegeneration play a central role in the premature death caused by this tumor. Restoration of insulin signaling in neurons prevents tumor progression and rescues the lethality caused by GB in Drosophila models.
Introduction
Cancer is one of the leading causes of mortality worldwide and is expected to be responsible for 15 million deaths in 2020 (65% in less developed countries) according to the World Health Organization. Notwithstanding recent advances in health treatments and extended lifespan of patients, some tumors still remain incurable. Among them, GB stands out because it is the most frequent and a very aggressive primary brain tumor. It is originated from glial cells and causes death within the first year after diagnosis (Bi & Beroukhim, 2014), despite standard treatments such as resection, radiotherapy, and chemotherapy. This is accompanied by broad neurological dysfunctions (Messaoudi et al, 2015). Brain tumors cause cognitive decline and neuronal dysfunction (reviewed in Gehrke et al [2013] and Bergo et al [2016]). These cognitive defects are consistent with typical neurodegenerative-associated symptoms such as synapse loss and mitochondrial alterations (Granholm et al, 2010; Levenson et al, 2014).
Drosophila melanogaster, the fruit fly, has emerged as a reliable animal system to mimic human diseases such as cancer (Sonoshita & Cagan, 2017). The aim is to model cellular and molecular mechanisms of human diseases, to identify targets for eventual diagnosis and treatments of patients. The power of Drosophila genetics allows genetic and pharmacological screens that may be translated to medicine, particularly for neurodegenerative disorders (Casas-Tinto et al, 2011; Dar et al, 2012; Burke et al, 2017; Held et al, 2019; Portela et al, 2019a). In fact, a Drosophila GB model that recapitulates most of the human disease features has been developed and validated (Read et al, 2009, 2013; Read, 2011; Portela et al, 2019a, 2019b). This model is based on two of the most frequent mutations in patients, a constitutively active forms of the epidermal growth factor receptor (EGFR) and the phosphatidylinositol-3 kinase (PI3K) catalytic subunit p110α (PI3K92E) driven by the glial specific repo-Gal4 (Read et al, 2009). This animal model has brought novel understandings into GB molecular mechanisms (Read, 2011; Read et al, 2013; Weng et al, 2017; Portela et al, 2019a, 2019b).
miRNAs are short noncoding RNAs that control gene activity mainly through post-transcriptional mechanisms. Recently, they have been linked to almost all biological processes and diseases, particularly cancer (Peng & Croce, 2016; Sander & Herranz, 2019). Concerning glioma, the miR-200 family (which includes miR-200, miR-141, and miR-429) plays central roles in GB development, metastasis, therapeutic response, and prognosis (reviewed in Peng et al [2018]). Low levels of miR-200 are indicative of poor prognosis in GB (Men et al, 2014). In colorectal cancer and GB, low expression levels of miRNAs correlates with up-regulation of insulin-like growth binding protein 7 (IGFBP7) (Jones et al, 2015). Likewise, in GB there is a transforming growth factor beta-2 (TGFB2)–dependent increase in IGFBP7 protein levels (Pen et al, 2008). However, the mechanisms involved in IGFBP7 influence on GB progression and its regulation by miR-200 remains unsolved. In Drosophila, miR-200 and IGFBP7 are represented by miR-8 and Imaginal morphogenesis protein-late 2 (ImpL2), respectively (Honegger et al, 2008). In juvenile stages, miR-8 has been found to regulate glial cell growth and to promote synaptic growth at the neuromuscular junction (Morante et al, 2013; Loya et al, 2014).
In contrast, Drosophila ImpL2 induces cachexia, a systemic effect characterized by anorexia and metabolic alterations induced by other malignant tumors (Petruzzelli & Wagner, 2016). Secreted ImpL2 from epithelial tumor cells induces systemic organ wasting and insulin resistance by antagonizing insulin signaling (Figueroa-Clarevega & Bilder, 2015; Kwon et al, 2015). Interestingly, PI3K and Drosophila Ras homolog enriched in brain (dRheb), two members of the insulin pathway, induce the formation of synapses between neurons (a process known as synaptogenesis) in the Drosophila larval brain (Martín-Peña et al, 2006). Actually, it has been shown that AKT, also involved in insulin signaling, acts as a pro-synaptogenic element (Jordán-Álvarez et al, 2017). These data strongly support a role for insulin signaling in the regulation of neuronal synaptic activity in Drosophila. In mammals, a similar effect of insulin signaling on synaptic plasticity has been described (Knafo & Esteban, 2017). Notably, synapse loss is an early step in neurodegeneration (Sephton & Yu, 2015; Henstridge et al, 2016). We have recently re-evaluated GB as a neurodegenerative disease, showing that GB reduces the number of synapsis through wingless/frizzled 1 (wg/fz1) signaling (Portela et al, 2019b), equivalent to mammalian WNT pathway (Arnés & Casas Tintó, 2017). However, whether tumoral glial cells are able to modify insulin signaling in contiguous neurons, and consequently alter the number of synapses, is yet unknown.
To study the mechanisms of communication among malignant glial cells and neurons, we used a previously well-characterized Drosophila GB model that reproduces the oncogenic transformation of glial cells and lethal glial neoplasia in post-embryonic larval (Read et al, 2009, 2013) or adult brains (Portela et al, 2019a), leading to lethal glial neoplasia. We previously reported a reduction in the number of synapses in the neuromuscular junction (NMJ) of adult flies caused by GB progression (Portela et al, 2019b). This neurodegenerative process is consequence of genetic modifications caused in glial cells. This phenomenon suggests that signals originated in the glial tumor can impact on neighboring neurons. Synaptogenesis is tightly regulated by PI3K, a main player in insulin signaling pathway (Jordán-Álvarez et al, 2017). Moreover, GB progression correlates with high levels of secreted molecules that decrease insulin pathway activity, such as ImpL2 (IGFBP7 in humans) (Pen et al, 2008; Jones et al, 2015).
In this report, we showed that secreted ImpL2 from glial-derived tumoral cells antagonizes insulin signaling in neighboring neurons, inducing a reduction in synapse number and consequently promoting neurodegeneration. ImpL2 expression in GB cells is regulated by miR-8, thus linking functionally miRNA pathway with insulin signaling in a GB model. We described the function of ImpL2 as a mediator in GB–neuron communication, responsible for the reduction in synapse number and neurological defects. Indeed, we propose the insulin pathway as a core signal in GB progression and neurological decay. Finally, we propose neurodegeneration as a relevant factor in the lethality induced by GB.
Results
ImpL2 mediates GB progression and the associated neurodegeneration
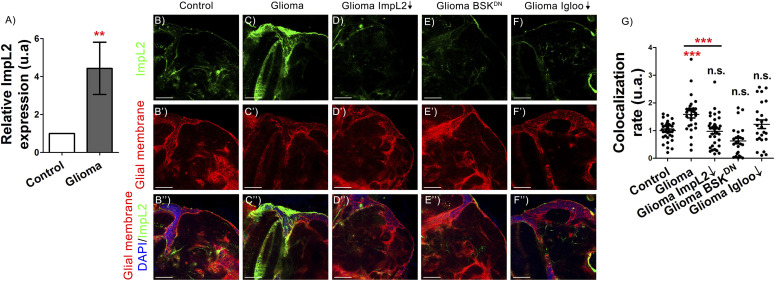
To determine ImpL2 mRNA expression levels in GB we performed qRT-PCR experiments. ImpL2 mRNA showed an increase in GB samples as compared with control brains (Fig 1A). To discriminate between ImpL2 expression in neuronal or glial (GB) cells, we used a MIMIC GFP reporter that reproduced faithfully ImpL2 expression (Nagarkar-Jaiswal et al, 2015). Consistently, GB cells showed higher reporter GFP levels than control glial cells, which are restored to control levels upon ImpL2 knockdown (Fig 1B–D and G). Therefore, these results validate the ImpL2 RNAi tool and indicate that ImpL2 is up-regulated in GB cells.
Figure 1. ImpL2 is up-regulated in glioma cells.
(A) qRT-PCR of 7-d-old adult brain extracts from repo>UAS-LacZ (Control) and repo>UAS-dEGFRλ, UAS-dp110CAAX (Glioma) flies shows an up-regulation of ImpL2 in gliomas (t test). (B, C, D, E, F) Confocal microscopy images of 7-d-old adult brains from (B) repo>UAS-LacZ (Control), (C) repo>UAS-dEGFRλ, UAS-dp110CAAX (Glioma), (D) repo>UAS-dEGFRλ, UAS-dp110CAAX, UAS-ImpL2 RNAi (Glioma ImpL2↓), (E) repo>UAS-dEGFRλ, UAS-dp110CAAX, UAS-BSKDN (Glioma BSKDN), and (F) repo>UAS-dEGFRλ, UAS-dp110CAAX, UAS-igloo RNAi (Glioma Igloo↓) animals, in all cases combined with an ImpL2-MIMIC GFP transgene (scale bar, 50 μm). (B′, C′, D′, E′, F′) Glial membrane is marked in red. (B″, C″, D″, E″, F″) Merge and DAPI staining. (G) Quantification and statistical analysis of co-localization rate between ImpL2-MIMIC GFP and glial membrane in at least N = 10 per genotype (ANOVA, post hoc Bonferroni) (**P-value < 0.01, ***P-value < 0.001).
In a previous work, we established that tumoral progression depended on the formation of a network of protrusions (i.e., an expansion of the membrane surface) named tumor microtubules (TMs), similarly to human GB (Osswald et al, 2015; Portela et al, 2019b). Besides, in mammals and flies, the TM network required the GAP43 and igloo gene functions, respectively (Osswald et al, 2015; Portela et al, 2019b). We also showed that GB progression requires c-Jun N-terminal Kinase (JNK) pathway activity (Portela et al, 2019b). The Drosophila JNK homolog Basket (Bsk) plays a central role in JNK signaling in normal and tumoral conditions (Fahey-Lozano et al, 2019). To determine if ImpL2 up-regulation in glial cells requires TMs network expansion, or JNK pathway activity in GB cells, we down-regulated igloo expression to prevent TMs formation, or overexpressed a dominant negative form of bsk (BSKDN) to block JNK activity. ImpL2 expression levels were reverted to normal levels in both cases, indicating that glial ImpL2 expression was dependent on TMs and JNK pathway activity, but not transcriptionally regulated by activated EGFR or Dp110 (Fig 1E–G).
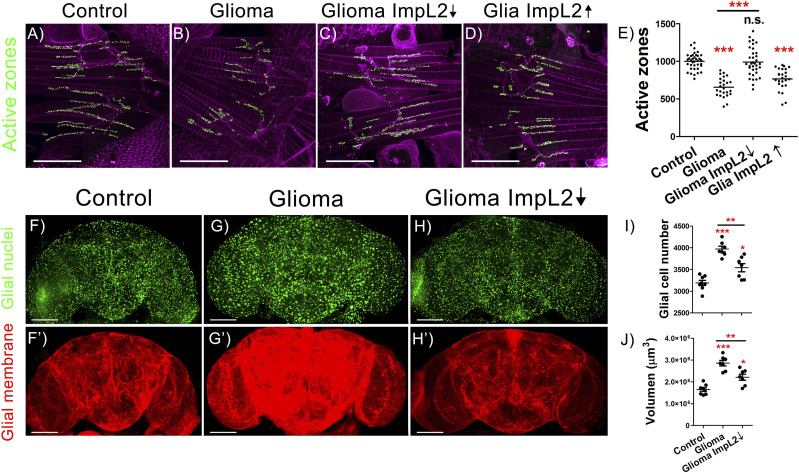
To study the neurodegeneration associated to GB progression, we quantified the number of synapses in the neuromuscular junction (NMJ) of the adult flies with GB. NMJ is a stereotyped structure that allows counting the number of synapses in the synaptic buttons of the motor neurons by using anti-bruchpilot, a specific antibody that recognizes synapses unambiguously (see the Materials and Methods section for details). To determine the contribution of glial ImpL2 to synapse loss, we knocked-down ImpL2 in GB cells and counted the number of synapses in adult NMJs. The results showed that Impl2 reduction in GB cells counteracted the reduction in the number of synapses observed in GB samples (Fig 2A–C and E). Next, to determine if ImpL2 is sufficient to decrease the number of synapses or it requires further features of GB cells, we overexpressed ImpL2 in wild-type (wt) glial cells and quantified the number of synapses. The results show a decreased number of synapses in NMJs (Fig 2D and E), consistent with our previous results, and suggest that ImpL2 expressed in glial cells is sufficient to cause synapse loss in neurons.
Figure 2. ImpL2 down-regulation in glioma cells rescues neurodegeneration and reduces tumor progression.
(A, B, C, D) Confocal images of adult neuromuscular junction (NMJ) at 29°C from (A) repo>UAS-LacZ (Control), (B) repo>UAS-dEGFRλ, UAS-dp110CAAX (Glioma), (C) repo>UAS-dEGFRλ, UAS-dp110CAAX, UAS-ImpL2 RNAi (Glioma ImpL2↓), and (D) repo>ImpL2 (Glia ImpL2↑) 7-d-old animals. Active zones are marked in green (anti-NC82) (scale bar, 50 μm). (E) Quantification and statistical analysis of active zones in at least N = 13 per genotype (ANOVA, post hoc Bonferroni). (F, G, H) Confocal microscopy images of adult brains from (F) repo>UAS-LacZ, (G) repo>UAS-dEGFRλ, UAS-dp110CAAX and (H) repo>UAS-dEGFRλ, UAS-dp110CAAX, UAS-ImpL2 RNAi flies after 7 d at 29°C with the glial nuclei marked in green. (F′, G′, H′) Glial membrane is shown in red (scale bar, 100 μm). (I, J) Quantification of (I) glial cell number and (J) glial membrane volume for at least N = 7 per genotype (ANOVA, post hoc Bonferroni) (*P-value < 0.05, **P-value < 0.01, ***P-value < 0.001).
Besides, we studied the two additional and typical features of GB such as the increase in the number of glial cells and the expansion of the TM network (Fig 2F, G, I, and J). The confocal images and quantifications showed that the down-regulation of ImpL2 RNA levels in GB caused a striking reduction in the number of glial cells (Fig 2H and I) and in the total tumor volume (Fig 2J). Thus, we concluded that GB cells up-regulated and secreted ImpL2, a necessary step to induce tumoral expansion and a reduction of the synapse number in surrounding neurons. These results suggest that GB progression and neurodegeneration are coupled events.
miRNAs levels inversely correlate with ImpL2 expression and GB progression
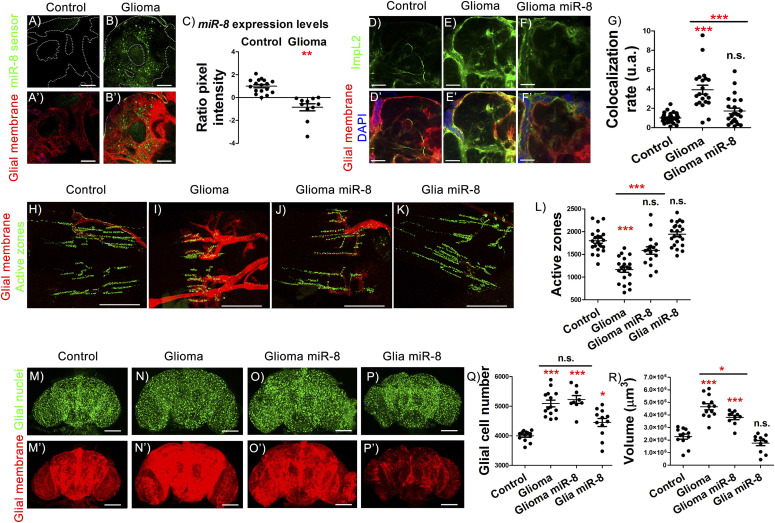
It has been described that low levels of miRNAs correlated with high levels of ImpL2 homolog in human GB, suggesting that ImpL2 regulation might be mediated by miRNAs (Jones et al, 2015). Accordingly, there is a down-regulation of the miRNA miR-200 family in GB samples (reviewed in Peng et al [2018]). Given that miR-8, the Drosophila homolog of miR-200 family, negatively regulates ImpL2 mRNA stability in the fat body (Lee et al, 2015), we hypothesized that miR-8 might play a role in GB progression and ImpL2 levels regulation. We used a miR-8 sensor to monitor miR-8 activity in GB brains. It includes miR-8 binding sites in the 3′UTR of the GFP gene (Kennell et al, 2012). Thus, high levels of GFP indicate low levels of miR-8 activity and vice versa (see the Materials and Methods section). GFP signal was increased in GB cells (Fig 3A–C), indicating that miR-8 levels were reduced. To determine if the increase in ImpL2 in GB depended on miR-8, we analyzed ImpL2 sensor upon miR-8 overexpression in that context. We observed a significant reduction in ImpL2 expression in GB cells in vivo upon miR-8 overexpression (Fig 3D–G). Consistently, miR-8 gain-of-function in GB rescued the loss of synapses, recapitulating the effect of ImpL2 loss of function in GB conditions (Fig 3H–J and L). These results are consistent with the effect of ImpL2 down-regulation in GB that also rescued the synapse number (Fig 2C and E), and suggest that miR-8 regulates ImpL2 expression in GB.
Figure 3. Increasing miR-8 levels in gliomas impairs ImpL2 overexpression and synapse loss.
(A, B) Confocal microscopy images of adult brain showing the expression pattern of miR-8 in repo>UAS-LacZ control animals compared with repo>UAS-dEGFRλ, UAS-dp110CAAX flies using miR-8 sensor (see the Materials and Methods section). (A′, B′) Glial membrane is marked in red. (C) Quantification and statistical analysis of miR-8 expression based on the GFP pixel intensity in at least N = 3 per genotype (ANOVA, post hoc Bonferroni) (scale bar, 50 μm). GFP control levels were normalized to one and GFP increase in glioma brains was transformed into negative values to indicate the relation between the increase in GFP and the reduction in miR-8 levels of expression. (D, E, F) Confocal microscopy images of 7-d-old adult brains from the following genotypes: (D) repo>UAS-LacZ (Control), (E) repo>UAS-dEGFRλ, UAS-dp110CAAX (Glioma), (F) repo>UAS-dEGFRλ, UAS-dp110CAAX, UAS-miR-8 (Glioma miR-8) animals, in all cases combined with ImpL2-MIMIC GFP transgene (scale bar, 50 μm). (D′, E′, F′) Merge with glial membrane in red and DAPI staining. (G) Quantification and statistical analysis of co-localization rate between ImpL2-MIMIC GFP and glial membrane in at least N = 10 per genotype (ANOVA, post hoc Bonferroni). (H, I, J, K) Adult neuromuscular junction (NMJ) from (H) repo>UAS-LacZ (controI), (I) repo>UAS-dEGFRλ, UAS-dp110CAAX (Glioma), (J) repo>UAS-dEGFRλ, UAS-dp110CAAX, UAS-miR-8 (Glioma miR-8), and (K) repo>miR-8 (Glia miR-8), active zones marked in green. (L) Quantification and statistical analysis of active zones in at least N = 10 per genotype (ANOVA, post hoc Bonferroni) (scale bar, 50 μm). (M, N, O, P) Confocal microscopy images of adult brains from 7-d-old flies of (M) repo>UAS-LacZ, (N) repo>UAS-dEGFRλ, UAS-dp110CAAX, (O) repo>UAS-dEGFRλ, UAS-dp110CAAX, UAS-miR-8 and (P) repo>UAS-miR-8 genotypes, glial nuclei marked in green. (M′, N′, O′, P′) Glial membrane is shown in red. (Q, R) Quantification of (Q) glial cells and (R) glial membrane volume for at least N = 9 per genotype (ANOVA, post hoc Bonferroni) (scale bar, 100 μm) (*P-value < 0.05**P-value < 0.01, ***P-value < 0.001).
Furthermore, miR-8 overexpression in wt glial cells (with low ImpL2 levels) did not alter the number of synapses (Fig 3K and L). Intriguingly, GB cell number expansion was not prevented by miR-8 overexpression (Fig 3M–O and Q), and consistently miR-8 overexpression increased glial cell number in wild type conditions (Fig 3P and Q). However, we did observe a reduction in GB membrane volume upon miR-8 up-regulation, something that did not occur in normal glial cells (Fig 3R). Altogether, these results showed an inverse correlation between miR-8 and ImpL2 expression in GB cells and suggested that ImpL2 levels are regulated by miR-8 in vivo.
GB secreted ImpL2 reduces insulin signaling in neurons
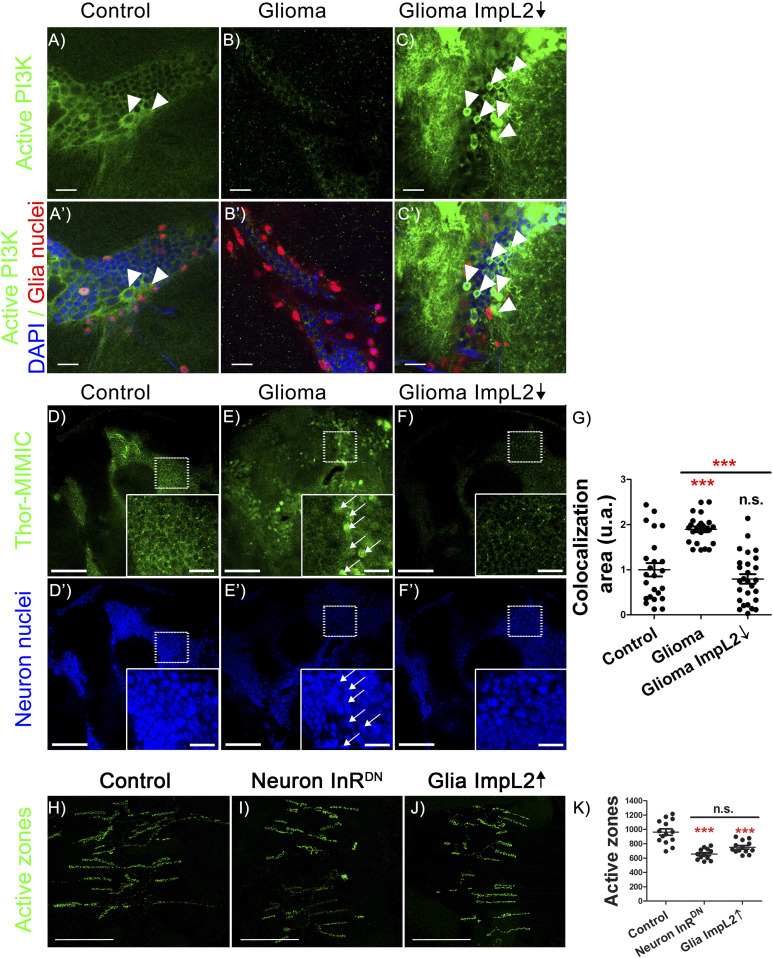
To evaluate the impact of GB on insulin signaling in neurons, we used a fluorescent reporter (tGPH, composed by the fusion of a pleckstrin homology domain plus green fluorescent protein under the control of the tubulin promoter) that reports the activity of PI3K and, thus, is widely used as a monitor of insulin pathway (Britton et al, 2002). tGPH reporter activity in neurons is strongly reduced (although still detectable) upon GB induction compared with control neurons (Fig 4A and B). In addition, ImpL2 knockdown specifically in GB cells prevents tGPH reporter reduction in neurons, suggesting a normalization of insulin signaling levels (Fig 4C).
Figure 4. Glioma-secreted ImpL2 inhibits insulin pathway activity in neurons.
(A, B, C) Confocal microscopy images of adult brain of 7-d-old flies from (A) repo>UAS-LacZ (Control), (B) repo>UAS-dEGFRλ, UAS-dp110CAAX (Glioma) and (C) repo>UAS-dEGFRλ, UAS-dp110CAAX, UAS-ImpL2 RNAi (Glioma ImpL2↓) genotypes, combined with tGPH transgene reporter of PI3K activity (green). (A′, B′, C′) Merge with glial nuclei in red and DAPI staining (scale bar 10 μm). White arrows point to non-glial cells with PI3K activity. The images are representative of at list N = 6 per genotype. (D, E, F) Confocal microscopy images of 7-d-old adult fly brains of the genotypes: (D) repo>UAS-LacZ, (E) repo>UAS-dEGFRλ, UAS-dp110CAAX and (F) repo>UAS-dEGFRλ, UAS-dp110CAAX, UAS-ImpL2 RNAi, in all the cases combined with ThorMI09732 line. (D′, E′, F′) Neuronal nuclei are marked in blue. White arrows indicate nuclear localization of ThorGFP signal. (G) Quantification and statistical analysis of co-localization area in N = 9 per genotype (ANOVA, post hoc Bonferroni) (scale bar, 50/10 μm). (H, I, J, K) Adult neuromuscular junction (NMJ) from the genotypes: (H) D42>UAS-LacZ (Control), (I) D42>UAS-InRDN (Neuron InRDN), and (J) repo>UAS-ImpL2 (Glia ImpL2↑) animals after 7 d at 29°C (active zones shown in green). D42 is expressed in motor neurons. (K) Quantification and statistical analysis of active zones in at least N = 6 per genotype (ANOVA, post hoc Bonferroni) (scale bar, 50 μm) (***P-value < 0.001).
To further analyze insulin-dependent FOXO activity, we used the ThorMI09732 line that bears a MIMIC transgene inserted in the genomic region corresponding to the gene Thor. The Thor gene encodes for a protein that is involved in translational control. It is regulated by FOXO, and its expression can hence be used as a surrogate of FOXO activity. In normal conditions, Thor transcription remains at low but detectable levels (Teleman et al, 2005). However, when insulin activity is compromised, Thor is highly transcribed, as reflected by LacZ or MIMIC lines (Galagovsky et al, 2014). Neurons exposed to GB had reduced insulin signaling, as shown by ThorMI09732 GFP expression. This effect on insulin pathway was restored by down-regulating ImpL2 in GB cells (Fig 4D–G). All these results together suggested that ImpL2 up-regulation in GB cells decreased the activity of insulin pathway in neurons, which is compatible with neurodegeneration.
However, the central function for insulin signaling pathway in synaptogenesis was described mainly in larval NMJ synapses (Martín-Peña et al, 2006). Whether or not insulin signaling plays a similar role in the central adult nervous system has not been evaluated yet. We expressed a dominant negative form of the insulin receptor (InRDN) in adult motor neurons and quantified the number of synapses. The quantification shows that InRDN expression in neurons caused a reduction in the number of synapses (Fig 4H–K). Moreover, the overexpression of ImpL2 in glial cells also provoked a reduction in the synapse number. Thus, these results together suggest that the ImpL2 effect on synapsis was due to a deregulation of the insulin signaling in neurons.
Insulin signaling in neurons rescues partially neurodegeneration and mortality in GB
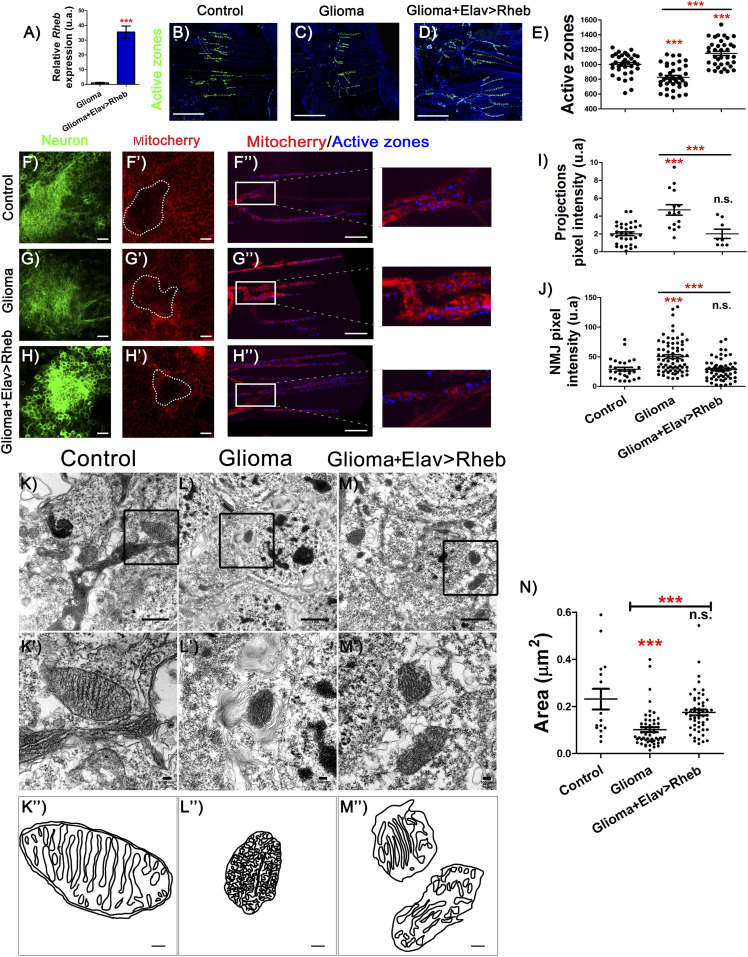
Our results suggest that in a GB, ImpL2 overexpression causes an effective decrease in neuronal insulin pathway activity which in turn induces neurodegeneration. In consequence, we proposed that restoring insulin signaling specifically in neurons should prevent GB-induced neurodegeneration. However, to manipulate insulin pathway activity in neuronal population simultaneously with GB induction in glial cells, we needed to use the LexA/lexAop system (see the Materials and Methods section). We overexpressed Rheb (lexAop-dRheb), to activate insulin signaling in neurons under the control of an elav-LexA line. dRheb mRNA levels increase significantly upon LexA/lexAop system activation, therefore validating the lexAop-dRheb tool (Fig 5A).
Figure 5. Rheb expression in neurons rescues active zone loss and mitochondrial alterations.
(A) qRT-PCR of 7-d-old adult brains from repo>UAS-dEGFRλ, UAS-dp110CAAX; Elav-LexA>lexAop-CD8GFP (Glioma) and repo>UAS-dEGFRλ, UAS-dp110CAAX; elav-LexA>lexAop-CD8GFP, lexAop-Rheb (Glioma+Elav>Rheb) flies show Rheb expression in glioma brains, and Rheb up-regulation by ectopic expression of Rheb in neurons (t test). (B, C, D) Adult neuromuscular junction (NMJ) from (B) repo>UAS-LacZ; elav-LexA (C) repo>UAS-dEGFRλ, UAS-dp110CAAX; elav-LexA and (D) repo>UAS-dEGFRλ, UAS-dp110CAAX; elav-LexA lexAop-Rheb after 7 d at 29°C (active zones are marked in green). (E) Quantification and statistical analysis of active zones in at least N = 20 per genotype (ANOVA, post hoc Bonferroni) (scale bar, 50 μm). (F, F′, G, G′, H, H′) Confocal microscopy images of adult brains (detail of Kenyon cells) from (F) repo>UAS-LacZ; elav-LexA (G) repo>UAS-dEGFRλ, UAS-dp110CAAX; elav-LexA and (H) repo>UAS-dEGFRλ, UAS-dp110CAAX; elav-LexA lexAop-Rheb 7 d at 29°C in all the cases combined with lexAop-mito-Cherry transgene (red), and neural membrane shown in green and (F′, G′, H′), mitochondrial membrane in red (scale bar, 10 μm). (F, F″, G, G″, H, H″) NMJ confocal images of the same genotypes as in (F, G, H) show mitochondrial marked in red and active zones in blue (scale bar, 10 μm). (I, J) Quantification and statistical analysis of pixel intensity in (I) projections and (J) NMJ in at least N = 15 and N = 6, respectively per genotype (ANOVA, post hoc Bonferroni). (K, K′, K″, L, L′, L″, M, M′, M″) Electron microscopy images of neurons from adult brains of (K) repo>UAS-LacZ; elav-LexA, (L) repo>UAS-dEGFRλ, UAS-dp110CAAX; elav-LexA and (M) repo>UAS-dEGFRλ, UAS-dp110CAAX; elav-LexA lexAop-Rheb (scale bar, 1 μm K′, L′, M′), an amplification from black square of mitochondria in detail (scale bar, 100 nm) and cristae (K″, L″, M″) a schematic representation of the mitochondrial (scale bar, 100 nm). (N) Quantification and statistical analysis of mitochondrial area (***P-value < 0.001).
The quantifications of adult NMJs showed an increase in the synapse number in NMJ when compared with GB and even to control genotypes, thus showing a similar effect (although slightly stronger) to GB with low ImpL2 levels (Fig 5B–D and E). In conclusion, the larval synaptogenic pathway regulated by insulin signaling members was conserved in adult brains, at least for InR pathway and dRheb.
Insulin signaling mediates glucose metabolism and mitochondrial physiology (Cheng et al, 2010). In addition, mitochondrial alterations lead to synapse dysfunction and neurodegeneration (Chen et al, 2019; Zhao et al, 2019). In particular, the transport of mitochondria through the axons is altered in other neurodegenerative diseases (Vanhauwaert et al, 2019). Therefore, we investigated if GB progression affects neuronal mitochondria.
We used a lexAop-mito-Cherry reporter transgene to quantify the distribution of mitochondria in axon terminals. Pixel intensity quantification showed that GB causes a significant increase in mitochondria in the neuronal projections of Kenyon cells in the mushroom body (Fig 5F–H, F′–H′, and I), compatible with a neurodegenerative process (Chevalier-Larsen & Holzbaur, 2006). In line with this, we observed an increase in mitochondria accumulated in NMJ boutons that correlated with lower number of synaptic boutons (Fig 5F″–H″ and J). Both effects in neuronal projections and NMJs were prevented upon insulin pathway signaling activation in the neuronal population of a GB brain (Fig 5H, H′, and H″). Consistently, electron microscopy images of neuronal mitochondria in GB samples showed defects in the cristae, a typical feature of nonfunctional mitochondria (Long et al, 2015; Miyazono et al, 2018). This mitochondrial defective morphology was also reverted upon dRheb overexpression in neurons (Fig 5K–M). In addition, we quantified the size of the mitochondria in neurons and observed that, in GB samples, mitochondria area is significantly reduced, and dRheb overexpression prevents this morphological defect (Fig 5N). Altogether, these results indicated that neurons showed mitochondrial disruption as a consequence of low insulin signaling. Increasing insulin pathway activity in neurons exposed to GB might be sufficient to recover functional organelles, as suggested by the synapse number.
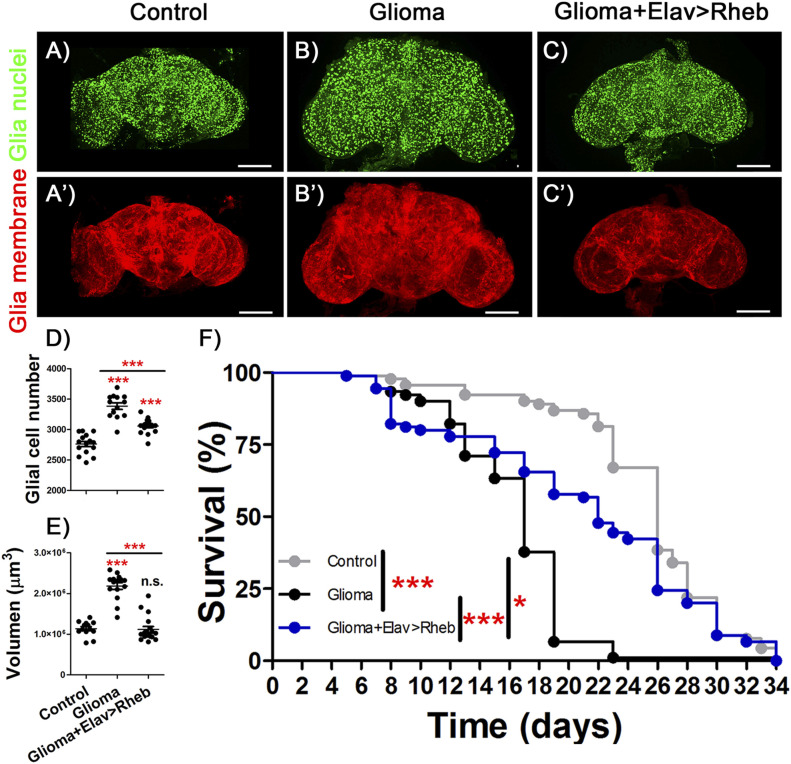
GB brains with high insulin signaling in neurons also showed a reduction in the number of glial cells and in tumor volume (Fig 6A–E). More importantly, GB caused premature death, an effect that was significantly rescued by overexpressing dRheb in neurons (Fig 6F). These results suggested that restoration of neuronal insulin pathway activity improved the lifespan in animals with GB, thus linking synaptogenesis to a slower disease progression and functional protection. In conclusion, the communication between GB cells and neurons is proposed as a novel field of study for GB progression and provides a possible novel therapeutic target.
Figure 6. Overexpression of Rheb in neurons protect against glioma effects.
(A, B, C) Confocal microscopy images of adult brains from (A) repo>UAS-LacZ; elav-LexA (Control), (B) repo>UAS-dEGFRλ, UAS-dp110CAAX; elav-LexA (Glioma) and (C) repo>UAS-dEGFRλ, UAS-dp110CAAX; elav-LexA, lexAop-Rheb (Glioma+Elav>Rheb) after 7 d at 29°C with glial nuclei marked in green. (A′, B′, C′) Glial membrane is shown in red. (D, E) Glial cells number and (E) glial membrane volume quantification and statistical analysis for at least N = 13 per genotype (ANOVA, post hoc Bonferroni) (scale bar, 100 μm). (F) Graph shows a survival assay of repo>UAS-LacZ; elav-LexA (gray), repo>UAS-dEGFRλ, UAS-dp110CAAX; elav-LexA (black) and repo>UAS-dEGFRλ, UAS-dp110CAAX, elav-LexA, lexAop-Rheb (blue) male flies and statistical analysis in N = 90 (Mantel–Cox test) (*P-value < 0.05,**P-value < 0.01, ***P-value < 0.001).
Discussion
GB is one of the most aggressive type of brain tumor (Bi & Beroukhim, 2014). During GB progression, tumoral cells extend a network of membrane projections that contribute to brain infiltration and results in poor prognosis for the patient (Osswald et al, 2015). GB courses with a neurological decay that includes sleep disturbances, speech difficulties and other typical symptoms of neurodegeneration (Bergo et al, 2016). For decades, the origin of this decay was attributed to the high pressure caused by the GB solid mass and the associated edema. Despite the solid mass of the GB being removed after surgery, the neurodegenerative process continues, more likely because of the diffuse GB progression. This indicates that mechanisms underlying neurological decay are not restricted to the intracranial pressure and edema.
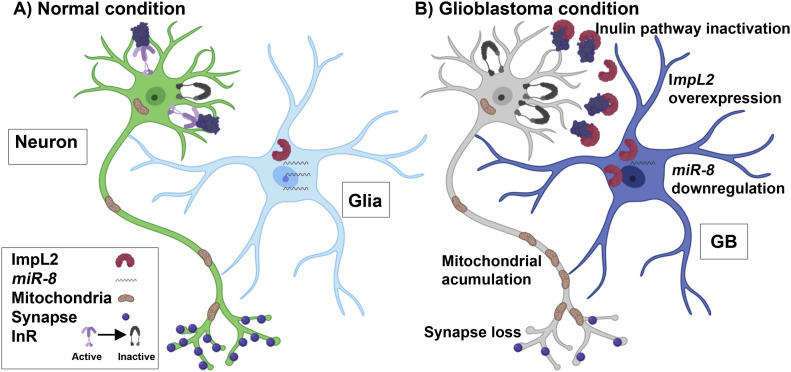
Recent publications suggested an active communication between GB cells and the surrounding healthy tissue, including neurons. Experiments performed with human GB cells in mice xenografts revealed a physical interaction between GB cells and neurons as electrical and chemical synapses (Venkataramani et al, 2019; Venkatesh et al, 2019). In this case, neurons act as presynaptic structures whereas GB tumoral cells are postsynaptic elements. These so-called “synapses” are required for GB progression. Besides, we have recently described cellular mechanisms for GB to deplete Wingless (Wg)/WNT from neurons. GB cells project TMs that enwrap neurons and accumulate Frizzled1 (Fz1) receptor to vampirize Wg from neighboring neurons. This unidirectional mechanism facilitates GB proliferation and causes a loss of synapses in the neurons (Portela et al, 2019b). In addition to this, here we also describe a bidirectional communication system between GB cells and neurons. In contrast to what happen with Wg/Fz1, ImpL2 protein is originated in GB cells and target healthy neurons, but not vice versa. ImpL2 binds InR ligands and act as an antagonist of the pathway. In consequence, insulin signaling might be reduced in neurons which in turn caused synapse loss and lethality (Fig 7). Neuronal insulin signaling can be restored via dRheb up-regulation, and this is sufficient to extend the lifespan of GB animals. It is tempting to propose that ImpL2 impacts both tumor expansion (GB membrane) and neurodegeneration. The rescue of the neurodegenerative phenotype by ImpL2 knockdown could be caused by an autonomous GB signaling that impairs tumoral progression, or by the restoration of insulin signaling in neurons. However, dRheb up-regulation in neurons prevented GB cells number increase, suggesting that GB progression is impaired when neuronal insulin signaling is active. These results suggest that reducing insulin signaling (and the subsequent neurodegeneration) is critical for GB progression and invasion, and ultimately for the lethality caused by GB. In conclusion, the GB is able to alter neuronal regular functions actively by at least two ways: vampirizing a required growth factor from neurons (like Wg) or secreting an antagonist of the pathway (such as Impl2). In both cases, the consequence is similar; a signaling pathway essential for synaptogenesis in the healthy neuronal population is down-regulated.
Figure 7. Schematic representation of the effect of glioblastoma (GB) on neuronal insulin pathway.
(A) In normal condition glial cells show physiological levels of miR-8 and ImpL2. (B) In GB condition, miR-8 expression is down-regulated, and ImpL2 levels increase, which in turn causes an inactivation of the insulin pathway signaling in neurons. This effect triggers several changes in neurons such as synapse loss and mitochondrial accumulation throughout the axon. The three strategies addressed in this work to revert the tumoral phenotype consist in the up-regulation of miR-8 expression in GB cells, the down-regulation of ImpL2 also in GB cells and the activation of the insulin pathway in the surrounding neurons by means of Rheb overexpression.
The regulation of ImpL2 expression in GB cells seems to depend on miRNAs, at least in part. In cancer, miRNAs have emerged as general regulators key for tumoral progression, including GB (Sander & Herranz, 2019). Interestingly, miR-200 family (miR-8 in Drosophila) plays a key role in human GB. We have described a correlation between miR-8 levels, ImpL2 expression and GB progression. However, there are no predicted miR-8 binding sites in ImpL2 sequence (STarMirSoftware for Statistical Folding of Nucleic Acids and Studies of Regulatory RNAs- (Rennie et al, 2014) or TargetSCAnFly- http://www.targetscan.org/); therefore, it is unlikely that miR-8 regulates directly ImpL2 mRNA stability. One possibility is the existence of mediator proteins that depend directly on miR-8. The most evident possibility is a transcription factor whose mRNA stability is sensitive to miR-8 and acts as a transcriptional regulator of ImpL2. However, the regulation of ImpL2 and its association to miRNAs is a matter for future studies. Another intriguing point is the differential effects that both miR-8 overexpression and ImpL2 down-regulation have on GB growth. Whereas an excess of miR-8, which in turn reduces ImpL2, is unable to reduce GB cell number, direct down-regulation of Impl2 significantly reduces the growth of GB cells. Nevertheless, it is known that most of miRNAs control several mRNAs, thus miR-8 overexpression might affect to different extent other mRNAs than just ImpL2 mRNA, which may account for such differences.
Altogether, our data suggest that the progression of brain tumors in Drosophila depends not only on the intrinsic properties of the tumoral cells, but also on the physiological condition of the surrounding cells (Fig 7). The potential relationship between Wg/WNT and insulin pathway has been proposed under physiological or tumoral conditions (Desbois-Mouthon et al, 2001; Yi et al, 2008; Palsgaard et al, 2012) and represent a potential issue of interest to study in GB-host biology. We described recently (Portela et al, 2019b) the positive feedback loop established with wingless/JNK/MMPs and tumor microtubes that promote GB progression. We do not have evidences that miR-8/ImpL2 regulation is directly controlled by EGFR and/or PI3K signaling pathways. However, the results included in Fig 1 suggest that a reduction in JNK pathway (BSKDN) or the knockdown of igloo (prevention of TMs formation) reduces ImpL2 expression. It has been described that miR-8-mutant animals activate JNK signaling, but there is no evidence that JNK pathway can regulate miR-8. However, miRNA regulation has the tendency to establish reciprocal feedback loops and networks (Herranz & Cohen, 2010), so it might be plausible that JNK signaling and miR-8 would have such a reciprocal regulation. These results suggest that ImpL2 up-regulation is sensitive to TMs formation and JNK, and one could speculate that it might be also dependent on Wg/WNT signaling pathway. Our observations in Drosophila suggest that both pathways (insulin and Wg) participate in the equilibrium between GB cells and neurons. The relations among all different pathways and the mutual regulation should be matter of study of future projects.
GB patients respond differently to the progression of the GB: some patients survive for a few months, whereas others survive for years. If we accept that the coordinated effect in GB and neurons result in differential tumor progression, the vast differences in how patients respond to GB could be, in part, dependent on genetic or epigenetic conditions related to InR signaling genes in neurons, and probably other pathways such as WNT or Hedgehog.
Materials and Methods
Fly stocks and genetics
All fly stocks were maintained at 25°C (unless otherwise specified) on a 12/12 h light/dark cycles at constant humidity in a standard medium. The stocks used from Bloomington Stock Center were tub-Gal80ts (BL-7019), Repo-Gal4 (BL-7415), D42-Gal4 (BL-8816), UAS-InR DN (Bl-2852), UAS-myr-RFP (BL-7119) UAS-LacZ (BL-8529), UAS-ImpL2 RNAi (BL-55855), ImpL2-MIMIC (BL-59246), lexAop-mito-Cherry (BL-66530), tGPH (BL-8164), and ThorMI09732 (BL-53159). Other fly stocks used were miR-8 sensor (Kennell et al, 2012), UAS-miR-8-RFP (Lu et al, 2014), Elav-LexA (BL52676), UAS-HRP::CD2 (gifted by L Luo), UAS-dEGFRλ;UAS-dp110CAAX (gift from R Read, Read et al, 2009) UAS-ImpL2 (gift from Hugo Stocker), and lexAop-Rheb (gift from Nuria Romero).
The glioma-inducing line contains the UAS-dEGFRλ, UAS-dp110CAAX transgenes that encodes for the constitutively active forms of the human orthologs PI3K and EGFR, respectively (Read et al, 2009). Repo-Gal4 line drives the Gal4 expression to glial cells and precursors (Lee & Jones, 2005; Casas-Tintó et al, 2017) and combined with the UAS-dEGFRλ, UAS-dp110CAAX line allow us to generate a glioma thanks to the Gal4 system (Brand & Perrimon, 1993). Elav-LexA line drives the expression to neurons, allowing us to manipulate neurons in a glioma combining LexA and Gal4 expression systems (Lai & Lee, 2006).
Gal80TS is a repressor of the Gal4 activity at 18°C, although at 29°C is inactivated (McGuire et al, 2003). The tub-Gal80ts construct was used in all the crosses to avoid the lethality caused by the glioma development during the larval stage. The crosses were kept at 17°C until the adult flies emerged. To inactivate the Gal80ts protein and activate the Gal4/UAS system to allow the expression of our genes of interest, the adult flies were maintained at 29°C for 7 d except in the survival assay (flies were at 29°C until death).
Because of the inherent variability of tumor growth and the use of different reporters, we used the appropriate control and glioma genotypes that include them and performed the experiment in parallel for each grouped panel (at least three times): Figs 1B–F, 2, 3A, B, D–F, and –P, 4A–C and F–H, 5A–E and J–L, and 6.
Immunostaining and image acquisition
Adult brains were dissected and fixed with 4% formaldehyde in phosphate-buffered saline for 20 min whereas adult NMJ were fixed 10 min; in both cases, samples were washed 3 × 15 min with PBS+0.4% triton, blocked for 1 h with PBS+0.4% triton+ BSA 5%, incubated overnight with primary antibodies, washed 3 × 15 min, incubated with secondary antibodies for 2 h, and mounted in Vectashield mounting medium, with DAPI in the case of the brains. The primary antibodies used were anti-repo mouse (1/200; DSHB) to recognize glial nuclei, anti-bruchpilot-NC82-mouse (1/50; DSHB) to recognize the presynaptic protein bruchpilot, anti-HRP rabbit (1/400; Cell Signalling) to recognize membranes, anti-GFP rabbit (1:500; DSHB) and anti-Elav (1:100; DSHB) to recognize neuron nuclei. The secondary antibodies used were Alexa 488 or 647 (1/500; Life Technologies). Images were taken by a Leica SP5 confocal microscopy.
RNA extraction, reverse transcription and qRT-PCR
For RNA extraction, 1- to 4-d-old male adults were entrained to a 12:12 h LD cycle for 7 d at 29°C and then collected on dry ice at ZT 6. Total RNA was extracted from 30 heads of adult males of the Control (repo>LacZ), Glioma (repo>UAS-dEGFRλ, UAS-dp110CAAX), and repo>UAS-dEGFRλ, UAS-dp110CAAX, elav-LexA, lexAop-Rheb genotypes after 7 d of glioma development. RNA was extracted with TRIzol and phenol chloroform. Total RNA concentration was measured by using NanoDrop ND-1000. cDNA was synthetized from 1 mg of total RNA using M-MLV RT (Invitrogen). cDNA samples from 1:5 dilutions were used for real-time PCR reactions. Transcription levels were determined in a 14-ml volume in duplicate using SYBR Green (Applied Biosystem) and 7500 qPCR (Thermo Fisher Scientific). We analyzed transcription levels of ImpL2, dRheb, and Rp49 as housekeeping gene reference.
Sequences of primers were RP49 F: GCATACAGGCCCAAGATCGT, Rp49 R: AACCGATGTTGGGCATCAGA, ImpL2 F: CCGAGATCACCTGGTTGAAT, ImpL2 R: AGGTATCGGCGGTATCCTTT, dRheb F:CGACGTAATGGGCAAGAAAT, and dRheb R: CAAGACAACCGCTCTTCTCC.
After completing each real-time PCR run, outlier data were analyzed using 7500 software (Applied Biosystems). Ct values of duplicates from three biological samples were analyzed calculating 2DDCt and comparing the results using a t test with GraphPad (GraphPad Software).
Viability and survival assays
Lifespan was determined under 12:12 h LD cycles at 29°C conditions. Three replicates of 30 1- to 4-d-old male adults were collected in vials containing standard Drosophila media and transferred every 2–3 d to fresh Drosophila media.
Electron microscopy
Adult brains of repo>LacZ, repo>UAS-dEGFRλ, UAS-dp110CAAX, and repo>UAS-dEGFRλ, UAS-dp110CAAX, elav-LexA>lexAop-Rheb animals expressing CD2-HRP in glial membranes were dissected after 7 d of glioma development and fixed with 4% formaldehyde in phosphate-buffered saline for 30 min. The samples were washed twice with PBS and incubate with R.T.U. VECTASTAIN kit (VECTOR) for 30 min at RT and washed once with PBS. Followed by an incubation in dark with SIGMA FAST 3,3′-Diaminobenzidine Tablet SETS (Sigma-Aldrich) for 75 min at RT, washed once with PBT, and incubate with 4% formaldehyde + 2% glutaraldehyde for 1 h and stored at 4°C. Following fixation samples were washed three times in 0.1 M phosphate buffer. Post-fixation was performed in 1% osmium tetroxide + 1% potassium ferrocyanide for 1 h at 4°C, three washes in H2O2dd and incubated in PBS 0.1M + 0.15% tanic acid for 1 min, washed once in PBS 0.1M, and twice in H2O2dd. After incubation in 2% uranil acetate, it was incubated for 1 h at RT in darkness and washesd three times in H2O2dd. Dehydration was done in ethanol series (30%, 50%, 70%, 90%, and 3 × 100%). The samples were infiltrated with increasing concentrations of epoxy resin TAAB-812 (TAAB Laboratories) in propilenoxid and encapsulated in BEEM capsules to polymerize 48 h at 60°C. Ultrathin sections of 70–80 nm were cut using Ultracut E microtome (Leica) and stained with 2% uranyl acetate solution in water and lead Reynols citrate. Grids were examined with JeolJEM1400Flash electron microscope at 80 kV. Images were taken with a OneView (4K × 4K) CMOS camera (Gatan).
Quantification
Fluorescent reporter-relative ImpL2 and Thor signals within brains were determined from images taken at the same confocal settings avoiding saturation. For the analysis of co-localization rates, “co-localization” tool from LAS AF Lite software (Leica) was used taking the co-localization rate data for the statistics analyzing the co-localization between green signal (both cases) and signal coming from glial tissue (for ImpL2 levels) or neuronal nuclei (for Thor levels) from three slices per brain in similar positions of the z axis.
Average pixel intensity from miR-8 sensor and mito-Cherry was measured using measurement log tool from Adobe Photoshop CS5.1. Average pixel intensity of miR-8 sensor was analyzed quantifying the green sensor signaling glial tissue compared with green signal that did not overlap with glial cells to generate a ratio. Measurements were taken from similar localizations in three slices per brain with equivalent positions of the z axis. Average pixel intensity of mito-Cherry was analyzed taking measurements in the red signal from the reporter from at least five points of the NMJ.
Glial network was marked by a UAS-myristoylated-RFP reporter specifically expressed under the control of repo-Gal4. The total volume was quantified using Imaris surface tool (Imaris 6.3.1 software). Glial nuclei were marked by staining with the anti-Repo (DSHB). The number of Repo+ cells and number of synapses (anti-nc82; DSHB) were quantified by using the spots tool in Imaris 6.3.1 software. We selected a minimum size and threshold for the spot in the control samples of each experiment: 0.5 μm for active zones and 2 μm for glial cell nuclei. Then we applied the same conditions to the analysis the corresponding experimental sample.
For electron microscopy images quantification, we used FIJI (ImageJ 1.52v) software. After manually selecting the perimeter of each mitochondrion, we measured the area, major and minor axis. All measurements were taken blind. In total, at least 15 mitochondria were measured from three different animals per genotype.
Statistics
The results were analyzed using the GraphPad Prism 5 software (www.graphpad.com). Quantitative parameters were divided into parametric and nonparametric using the D’Agostino and Pearson omnibus normality test, and the variances were analyzed with F test. t test and ANOVA test with Bonferroni’s post hoc were used in parametric parameters, using Welch’s correction when necessary. To the nonparametric parameters, Mann–Whitney test and Kruskal–Wallis test with Dunn’s post hoc were used. The survival assays were analyzed with Mantel–Cox test. The P limit value for rejecting the null hypothesis and considering the differences between cases as statistically significant was P < 0.05 (*). Other P-values are indicated as ** when P < 0.01 and *** when P < 0.001.
Supplementary Material
Acknowledgements
We thank Professor Alberto Ferrús for helpful discussions. We are grateful to R Read, the Bloomington Drosophila stock Centre and the Developmental Studies Hybridoma Bank for supplying fly stocks and antibodies, and FlyBase for its wealth of information. We acknowledge the support of the Confocal Microscopy unit and Molecular Biology unit at the Cajal Institute for their help with this project. We want to thank the continuous support from Emirates Khalifa Capital to this project. Research has been funded by grant BFU2015-65685P and PGC2018-094630-B-I00 (Ministerio de Innovacion y Ciencia [MICINN]). FA Martín is a recipient of a Ramon y Cajal Contract (RyC-2014–14961).
Author Contributions
P Jarabo: formal analysis, funding acquisition, validation, investigation, methodology, and writing—original draft, review, and editing.
C de Pablo: formal analysis, investigation, and methodology.
H Herranz: formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, and writing—original draft, review, and editing.
FA Martín: conceptualization, resources, formal analysis, supervision, funding acquisition, validation, investigation, methodology, project administration, and writing—original draft, review, and editing.
S Casas-Tintó: conceptualization, resources, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, project administration, and writing—original draft, review, and editing.
Conflict of Interest Statement
The authors declare that they have no conflict of interest.
References
- Arnés M, Tintó SC (2017) Aberrant Wnt signaling: A special focus in CNS diseases. J Neurogenet 31: 216–222. 10.1080/01677063.2017.1338696 [DOI] [PubMed] [Google Scholar]
- Bergo E, Lombardi G, Pambuku A, Della Puppa A, Bellu L, D’avella D, Zagonel V (2016) Cognitive rehabilitation in patients with gliomas and other brain tumors: State of the art. Biomed Res Int 2016: 3041824 10.1155/2016/3041824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi WL, Beroukhim R (2014) Beating the odds: Extreme long-term survival with glioblastoma. Neuro Oncol 16: 1159–1160. 10.1093/neuonc/nou166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Britton JS, Lockwood WK, Li L, Cohen SM, Edgar BA (2002) Drosophila’s insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev Cell 2: 239–249. 10.1016/s1534-5807(02)00117-x [DOI] [PubMed] [Google Scholar]
- Burke C, Trinh K, Nadar V, Sanyal S (2017) AxGxE: Using flies to interrogate the complex etiology of neurodegenerative disease. Curr Top Dev Biol 121: 225–251. 10.1016/bs.ctdb.2016.07.007 [DOI] [PubMed] [Google Scholar]
- Casas-Tinto S, Zhang Y, Sanchez-Garcia J, Gomez-Velazquez M, Rincon-Limas DE, Fernandez-Funez P (2011) The ER stress factor XBP1s prevents amyloid-β neurotoxicity. Hum Mol Genet 20: 2144–2160. 10.1093/hmg/ddr100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Tintó S, Arnés M, Ferrús A (2017) Drosophila enhancer-Gal4 lines show ectopic expression during development. R Soc Open Sci 4: 170039 10.1098/rsos.170039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Turnbull DM, Reeve AK (2019) Mitochondrial dysfunction in Parkinson’s disease—cause or consequence? Biology (Basel) 8: 38 10.3390/biology8020038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Tseng Y, White MF (2010) Insulin signaling meets mitochondria in metabolism. Trends Endocrinol Metab 21: 589–598. 10.1016/j.tem.2010.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier-Larsen E, Holzbaur ELF (2006) Axonal transport and neurodegenerative disease. Biochim Biophys Acta 1762: 1094–1108. 10.1016/j.bbadis.2006.04.002 [DOI] [PubMed] [Google Scholar]
- Dar AC, Das TK, Shokat KM, Cagan RL (2012) Chemical genetic discovery of targets and anti-targets for cancer polypharmacology. Nature 486: 80–84. 10.1038/nature11127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbois-Mouthon C, Cadoret A, Blivet-Van Eggelpoël MJ, Bertrand F, Cherqui G, Perret C, Capeau J (2001) Insulin and IGF-1 stimulate the β-catenin pathway through two signalling cascades involving GSK-3β inhibition and Ras activation. Oncogene 20: 252–259. 10.1038/sj.onc.1204064 [DOI] [PubMed] [Google Scholar]
- Fahey-Lozano N, La Marca JE, Portela M, Richardson HE (2019) Drosophila models of cell polarity and cell competition in tumourigenesis. Adv Exp Med Biol 1167: 37–64. 10.1007/978-3-030-23629-8_3 [DOI] [PubMed] [Google Scholar]
- Figueroa-Clarevega A, Bilder D (2015) Malignant drosophila tumors interrupt insulin signaling to induce cachexia-like wasting. Dev Cell 33: 47–55. 10.1016/j.devcel.2015.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagovsky D, Katz MJ, Acevedo JM, Sorianello E, Glavic A, Wappner P (2014) The Drosophila insulin-degrading enzyme restricts growth by modulating the PI3K pathway in a cell-autonomous manner. Mol Biol Cell 25: 916–924. 10.1091/mbc.E13-04-0213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke AK, Baisley MC, Sonck ALB, Wronski SL, Feuerstein M (2013) Neurocognitive deficits following primary brain tumor treatment: Systematic review of a decade of comparative studies. J Neurooncol 115: 135–142. 10.1007/s11060-013-1215-2 [DOI] [PubMed] [Google Scholar]
- Granholm A-C, Boger H, Emborg M (2010) Mood, memory and movement: An age-related neurodegenerative complex? Curr Aging Sci 1: 133–139. 10.2174/1874609810801020133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held A, Major P, Sahin A, Reenan RA, Lipscombe D, Wharton KA (2019) Circuit dysfunction in SOD1-ALS model first detected in sensory feedback prior to motor neuron degeneration is alleviated by BMP signaling. J Neurosci 39: 2347–2364. 10.1523/JNEUROSCI.1771-18.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henstridge CM, Pickett E, Spires-Jones TL (2016) Synaptic pathology: A shared mechanism in neurological disease. Ageing Res Rev 28: 72–84. 10.1016/j.arr.2016.04.005 [DOI] [PubMed] [Google Scholar]
- Herranz H, Cohen SM (2010) MicroRNAs and gene regulatory networks: Managing the impact of noise in biological systems. Genes Dev 24: 1339–1344. 10.1101/gad.1937010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honegger B, Galic M, Köhler K, Wittwer F, Brogiolo W, Hafen E, Stocker H (2008) Imp-L2, a putative homolog of vertebrate IGF-binding protein 7, counteracts insulin signaling in Drosophila and is essential for starvation resistance. J Biol 7: 10 10.1186/jbiol72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Jeyapalan JN, Forshew T, Tatevossian RG, Lawson ARJ, Patel SN, Doctor GT, Mumin MA, Picker SR, Phipps KP, et al. (2015) Molecular analysis of pediatric brain tumors identifies microRNAs in pilocytic astrocytomas that target the MAPK and NF-κB pathways. Acta Neuropathol Commun 3: 86 10.1186/s40478-015-0266-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordán-Álvarez S, Santana E, Casas-Tintó S, Acebes Á, Ferrús A (2017) The equilibrium between antagonistic signaling pathways determines the number of synapses in Drosophila. PLoS One 12: e0184238 10.1371/journal.pone.0184238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennell JA, Cadigan KM, Shakhmantsir I, Waldron EJ (2012) The microRNA miR-8 is a positive regulator of pigmentation and eclosion in Drosophila. Dev Dyn 241: 161–168. 10.1002/dvdy.23705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knafo S, Esteban JA (2017) PTEN: Local and global modulation of neuronal function in health and disease. Trends Neurosci 40: 83–91. 10.1016/j.tins.2016.11.008 [DOI] [PubMed] [Google Scholar]
- Kwon Y, Song W, Droujinine IA, Hu Y, Asara JM, Perrimon N (2015) Systemic organ wasting induced by localized expression of the secreted Insulin/IGF antagonist ImpL2. Dev Cell 33: 36–46. 10.1016/j.devcel.2015.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SL, Lee T (2006) Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci 9: 703–709. 10.1038/nn1681 [DOI] [PubMed] [Google Scholar]
- Lee BP, Jones BW (2005) Transcriptional regulation of the Drosophila glial gene repo. Mech Dev 122: 849–862. 10.1016/j.mod.2005.01.002 [DOI] [PubMed] [Google Scholar]
- Lee GJ, Jun JW, Hyun S (2015) MicroRNA miR-8 regulates multiple growth factor hormones produced from Drosophila fat cells. Insect Mol Biol 24: 311–318. 10.1111/imb.12156 [DOI] [PubMed] [Google Scholar]
- Levenson RW, Sturm VE, Haase CM (2014) Emotional and behavioral symptoms in neurodegenerative disease: A model for studying the neural bases of psychopathology. Annu Rev Clin Psychol 10: 581–606. 10.1146/annurev-clinpsy-032813-153653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q, Zhao D, Fan W, Yang L, Zhou Y, Qi J, Wang X, Liu X (2015) Modeling of mitochondrial donut formation. Biophys J 109: 892–899. 10.1016/j.bpj.2015.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loya CM, McNeill EM, Bao H, Zhang B, Van Vactor D (2014) miR-8 controls synapse structure by repression of the actin regulator enabled. Development 141: 1864–1874. 10.1242/dev.105791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CS, Zhai B, Mauss A, Landgraf M, Gygi S, Van Vactor D (2014) MicroRNA-8 promotes robust motor axon targeting by coordinate regulation of cell adhesion molecules during synapse development. Philos Trans R Soc B Biol Sci 369: 20130517 10.1098/rstb.2013.0517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Peña A, Acebes A, Rodríguez JR, Sorribes A, De Polavieja GG, Fernández-Fúnez P, Ferrús A (2006) Age-independent synaptogenesis by phosphoinositide 3 kinase. J Neurosci 26: 10199–10208. 10.1523/JNEUROSCI.1223-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL (2003) Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302: 1765–1768. 10.1126/science.1089035 [DOI] [PubMed] [Google Scholar]
- Men D, Liang Y, Chen L (2014) Decreased expression of microRNA-200b is an independent unfavorable prognostic factor for glioma patients. Cancer Epidemiol 38: 152–156. 10.1016/j.canep.2014.01.003 [DOI] [PubMed] [Google Scholar]
- Messaoudi K, Clavreul A, Lagarce F (2015) Toward an effective strategy in glioblastoma treatment. Part I: Resistance mechanisms and strategies to overcome resistance of glioblastoma to temozolomide. Drug Discov Today 20: 899–905. 10.1016/j.drudis.2015.02.011 [DOI] [PubMed] [Google Scholar]
- Miyazono Y, Hirashima S, Ishihara N, Kusukawa J, Nakamura KI, Ohta K (2018) Uncoupled mitochondria quickly shorten along their long axis to form indented spheroids, instead of rings, in a fission-independent manner. Sci Rep 8: 350 10.1038/s41598-017-18582-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morante J, Vallejo DM, Desplan C, Dominguez M (2013) Conserved miR-8/miR-200 defines a glial niche that controls neuroepithelial expansion and neuroblast transition. Dev Cell 27: 174–187. 10.1016/j.devcel.2013.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarkar-Jaiswal S, Lee PT, Campbell ME, Chen K, Anguiano-Zarate S, Gutierrez MC, Busby T, Lin WW, He Y, Schulze KL, et al. (2015) A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. Elife 4: e05338 10.7554/eLife.05338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osswald M, Jung E, Sahm F, Solecki G, Venkataramani V, Blaes J, Weil S, Horstmann H, Wiestler B, Syed M, et al. (2015) Brain tumour cells interconnect to a functional and resistant network. Nature 528: 93–98. 10.1038/nature16071 [DOI] [PubMed] [Google Scholar]
- Palsgaard J, Emanuelli B, Winnay JN, Sumara G, Karsenty G, Kahn CR (2012) Cross-talk between insulin and Wnt signaling in preadipocytes: Role of Wnt co-receptor low density lipoprotein receptor-related protein-5 (LRP5). J Biol Chem 287: 12016–12026. 10.1074/jbc.M111.337048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pen A, Moreno MJ, Durocher Y, Deb-Rinker P, Stanimirovic DB (2008) Glioblastoma-secreted factors induce IGFBP7 and angiogenesis by modulating Smad-2-dependent TGF-β signaling. Oncogene 27: 6834–6844. 10.1038/onc.2008.287 [DOI] [PubMed] [Google Scholar]
- Peng Y, Croce CM (2016) The role of microRNAs in human cancer. Signal Transduct Target Ther 1: 15004 10.1038/sigtrans.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Fu J, Ming Y (2018) The miR-200 family: Multiple effects on gliomas. Cancer Manag Res 10: 1987–1992. 10.2147/CMAR.S160945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruzzelli M, Wagner EF (2016) Mechanisms of metabolic dysfunction in cancer-associated cachexia. Genes Dev 30: 489–501. 10.1101/gad.276733.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portela M, Segura-Collar B, Argudo I, Sáiz A, Gargini R, Sánchez-Gómez P, Casas-Tintó S (2019. a) Oncogenic dependence of glioma cells on kish/TMEM167A regulation of vesicular trafficking. Glia 67: 404–417. 10.1002/glia.23551 [DOI] [PubMed] [Google Scholar]
- Portela M, Venkataramani V, Fahey-Lozano N, Seco E, Losada-Perez M, Winkler F, Casas-Tintó S (2019. b) Glioblastoma cells vampirize WNT from neurons and trigger a JNK/MMP signaling loop that enhances glioblastoma progression and neurodegeneration. PLoS Biol 17: e3000545 10.1371/journal.pbio.3000545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read RD (2011) Drosophila melanogaster as a model system for human brain cancers. Glia 59: 1364–1376. 10.1002/glia.21148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read RD, Cavenee WK, Furnari FB, Thomas JB (2009) A Drosophila model for EGFR-Ras and PI3K-dependent human glioma. PLoS Genet 5: e1000374 10.1371/journal.pgen.1000374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read RD, Fenton TR, Gomez GG, Wykosky J, Vandenberg SR, Babic I, Iwanami A, Yang H, Cavenee WK, Mischel PS, et al. (2013) A kinome-wide RNAi screen in Drosophila glia reveals that the RIO kinases mediate cell proliferation and survival through TORC2-Akt signaling in glioblastoma. PLoS Genet 9: e1003253 10.1371/journal.pgen.1003253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennie W, Liu C, Carmack CS, Wolenc A, Kanoria S, Lu J, Long D, Ding Y (2014) STarMir: A web server for prediction of microRNA binding sites. Nucleic Acids Res 42: W114–W118. 10.1093/nar/gku376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M, Herranz H (2019) MicroRNAs in drosophila cancer models. Adv Exp Med Biol 1167: 157–173. 10.1007/978-3-030-23629-8_9 [DOI] [PubMed] [Google Scholar]
- Sephton CF, Yu G (2015) The function of RNA-binding proteins at the synapse: Implications for neurodegeneration. Cell Mol Life Sci 72: 3621–3635. 10.1007/s00018-015-1943-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoshita M, Cagan RL (2017) Modeling human cancers in Drosophila. Curr Top Dev Biol 121: 287–309. 10.1016/bs.ctdb.2016.07.008 [DOI] [PubMed] [Google Scholar]
- Teleman AA, Chen YW, Cohen SM (2005) Drosophila melted modulates FOXO and TOR activity. Dev Cell 9: 271–281. 10.1016/j.devcel.2005.07.004 [DOI] [PubMed] [Google Scholar]
- Vanhauwaert R, Bharat V, Wang X (2019) Surveillance and transportation of mitochondria in neurons. Curr Opin Neurobiol 57: 87–93. 10.1016/j.conb.2019.01.015 [DOI] [PubMed] [Google Scholar]
- Venkataramani V, Tanev DI, Strahle C, Studier-Fischer A, Fankhauser L, Kessler T, Körber C, Kardorff M, Ratliff M, Xie R, et al. (2019) Glutamatergic synaptic input to glioma cells drives brain tumour progression. Nature 573: 532–538. 10.1038/s41586-019-1564-x [DOI] [PubMed] [Google Scholar]
- Venkatesh HS, Morishita W, Geraghty AC, Silverbush D, Gillespie SM, Arzt M, Tam LT, Espenel C, Ponnuswami A, Ni L, et al. (2019) Electrical and synaptic integration of glioma into neural circuits. Nature 573: 539–545. 10.1038/s41586-019-1563-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng YL, An R, Cassin J, Joseph J, Mi R, Wang C, Zhong C, Jin SG, Pfeifer GP, Bellacosa A, et al. (2017) An intrinsic epigenetic barrier for functional axon regeneration. Neuron 94: 337–346.e6. 10.1016/j.neuron.2017.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F, Sun J, Lim GE, Fantus IG, Brubaker PL, Jin T (2008) Cross talk between the insulin and Wnt signaling pathways: Evidence from intestinal endocrine L cells. Endocrinology 149: 2341–2351. 10.1210/en.2007-1142 [DOI] [PubMed] [Google Scholar]
- Zhao XY, Lu MH, Yuan DJ, Xu DE, Yao PP, Ji WL, Chen H, Liu WL, Yan CX, Xia YY, et al. (2019) Mitochondrial dysfunction in neural injury. Front Neurosci 13: 30 10.3389/fnins.2019.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.