Abstract
Key points
The electrogenic Na+/HCO3 −cotransporter NBCe1‐B is widely expressed in many tissues, including pancreas, submandibular gland, brain, heart, etc. NBCe1‐B has very low activity under basal condition due to auto‐inhibition, but can be fully activated by protein interaction with the IP3R‐binding protein released with inositol 1,4,5‐trisphosphate (IRBIT).
The structural components of the auto‐inhibition domain and the IRBIT‐binding domain of NBCe1‐B are finely characterized based on systematic mutations in the present study and data from previous studies.
Reducing negative charges on the cytosol side of the transmembrane domain greatly decreases the magnitude of the auto‐inhibition of NBCe1‐B.
We propose that the auto‐inhibition domain functions as a brake module that inactivates NBCe1‐B by binding to, via electrostatic attraction, the transmembrane domain; IRBIT activates NBCe1‐B by releasing the brake from the transmembrane domain via competitive binding to the auto‐inhibition domain.
Abstract
The electrogenic Na+/HCO3 − cotransporter NBCe1‐B is widely expressed in many tissues in the body. NBCe1‐B exhibits only basal activity due to the action of the auto‐inhibition domain (AID) in its unique amino‐terminus. However, NBCe1‐B can be activated by interaction with the IP3R‐binding protein released with inositol 1,4,5‐trisphosphate (IRBIT). Here, we investigate the molecular mechanism underlying the auto‐inhibition of NBCe1‐B and its activation by IRBIT. The IRBIT‐binding domain (IBD) of NBCe1‐B spans residues 1−52, essentially consisting of two arms, one negatively charged (residues 1−24) and the other positively charged (residues 40−52). The AID mainly spans residues 40−85, overlapping with the IBD in the positively charged arm. The magnitude of auto‐inhibition of NBCe1‐B is greatly decreased by manipulating the positively charged residues in the AID or by replacing a set of negatively charged residues with neutral ones in the transmembrane domain. The interaction between IRBIT and NBCe1‐B is abolished by mutating a set of negatively charged Asp/Glu residues (to Asn/Gln) plus a set of Ser/Thr residues (to Ala) in the PEST domain of IRBIT. However, this interaction is not affected by replacing the same set of Ser/Thr residues in the PEST domain with Asp. We propose that: (1) the AID, acting as a brake, binds to the transmembrane domain via electrostatic interaction to slow down NBCe1‐B; (2) IRBIT activates NBCe1‐B by releasing the brake from the transmembrane domain.
Keywords: auto‐inhibition, bicarbonate transporter, electrostatic interaction, IRBIT, NBCe1, SLC4A4
Key points
The electrogenic Na+/HCO3 −cotransporter NBCe1‐B is widely expressed in many tissues, including pancreas, submandibular gland, brain, heart, etc. NBCe1‐B has very low activity under basal condition due to auto‐inhibition, but can be fully activated by protein interaction with the IP3R‐binding protein released with inositol 1,4,5‐trisphosphate (IRBIT).
The structural components of the auto‐inhibition domain and the IRBIT‐binding domain of NBCe1‐B are finely characterized based on systematic mutations in the present study and data from previous studies.
Reducing negative charges on the cytosol side of the transmembrane domain greatly decreases the magnitude of the auto‐inhibition of NBCe1‐B.
We propose that the auto‐inhibition domain functions as a brake module that inactivates NBCe1‐B by binding to, via electrostatic attraction, the transmembrane domain; IRBIT activates NBCe1‐B by releasing the brake from the transmembrane domain via competitive binding to the auto‐inhibition domain.
Introduction
IRBIT (the IP3R binding protein released with inositol 1,4,5‐trisphosphate) can interact with a wide array of proteins with distinct biological functions. IRBIT was originally identified as a binding partner that inhibits the activity of inositol 1,4,5‐trisphosphate receptor (IP3R), a Ca2+ channel expressed in the endoplasmic reticulum membrane (Ando et al. 2003, 2006). IRBIT can stimulate a series of acid‐base transporters, such as Na+/HCO3 − cotransporter NBCe1 (Shirakabe et al. 2006), NBCn1 (Hong et al. 2013; Wang et al. 2020 a), NBCn2 (Wang et al. 2020 a), Na+/H+ exchanger NHE3 (He et al. 2008; He et al. 2010), Cl−/HCO3 − exchanger SLC26A6 (Park et al. 2013), Na+/dicarboxylate cotransporter SLC13A3 (Khamaysi et al. 2019). IRBIT can also regulate the calcium calmodulin‐dependent kinase II alpha (Kawaai et al. 2015), ribonucleotide reductase involved in regulation of DNA synthesis (Arnaoutov & Dasso, 2014), the cleavage and polyadenylation specificity factor involved in RNA splicing (Kiefer et al. 2009), and apoptosis regulator Bcl‐2 (Bonneau et al. 2016). Dysfunction of AHCYL1 encoding IRBIT is associated with abnormal embryonic development (Cooper et al. 2006), altered life‐span expectation (Parkhitko et al. 2016), disturbance in catecholamine homeostasis (Kawaai et al. 2015), carcinogenesis (Jeong et al. 2012), drug‐ and chemo‐resistance in cancers (Wittig et al. 2002; Nakazawa et al. 2019), and increased suicidality (Niculescu et al. 2017).
NBCe1, a member of the SLC4 family, was first discovered and then cloned from the proximal tubule of salamander (Boron & Boulpaep, 1983; Romero et al. 1997). NBCe1 is widely distributed in diverse tissues in the body, playing an essential role in maintaining the acid‐base and electrolyte homeostasis on both cellular and systemic levels. NBCe1 dysfunction causes severe proximal tubular acidosis accompanied by many clinical manifestations, such as familial migraine, mental retardation, growth retardation, ocular abnormalities and dentition defects (Igarashi et al. 1999; Suzuki et al. 2010; Salerno et al. 2019; for reviews, see Parker & Boron, 2013; Kurtz, 2014; Parker, 2018).
SLC4A4 expresses five NBCe1 variants: NBCe1‐A to ‐E. NBCe1‐A is predominantly expressed at basolateral proximal tubule, responsible for reabsorbing ∼80% of filtered HCO3 −, a house‐keeping function of the kidney (Boron & Boulpaep, 1983; Soleimani et al. 1987; Romero et al. 1997; Maunsbach et al. 2000). NBCe1‐B is broadly expressed in many tissues, including but not limited to, exocrine gland tissues such as pancreas (Abuladze et al. 1998; Satoh et al. 2003), parotid gland (Kim et al. 2003) and submandibular gland (Luo et al. 2001), enamel (Lacruz et al. 2010; Jalali et al. 2014), heart (Choi et al. 1999), cornea and the central nervous system (Parker & Boron, 2013). In the exocrine gland tissues, NBCe1‐B mediates uptake of Na+ and HCO3 − from the interstitial space, contributing to the secretion of HCO3 −‐rich fluid that is stimulated by both hormone and vagus nerve. NBCe1‐C is abundantly expressed in astrocytes in the central nervous system (Bevensee et al. 2000). NBCe1‐D and ‐E are two minor variants identified from mouse reproductive tracts (Liu et al. 2011).
NBCe1‐A contains an auto‐stimulatory domain (ASD) in its unique amino‐terminus (residues 1−41, designated as NtA). NBCe1‐B and NBCe1‐C contain an auto‐inhibitory domain (AID) in their unique amino‐terminus (residues 1−85, designated as NtB). The activity of NBCe1‐A is severalfold greater than those of NBCe1‐B and ‐C (McAlear et al. 2006; Lee et al. 2012; Hong et al. 2013; Shcheynikov et al. 2015; Vachel et al. 2018). NBCe1‐B and ‐C can be fully activated by removing the NtB (McAlear et al. 2006; Lee et al. 2012), or by protein interaction with IRBIT (Shirakabe et al. 2006; Thornell et al. 2010; Lee et al. 2012; Hong et al. 2013; Shcheynikov et al. 2015). The basal activities of NBCe1‐B and ‐C are probably due to activation by endogenous IRBIT (Kawaai et al. 2017; Wang et al. 2020 b).
Uniquely, NtB contains a ‘basic cluster’ (sequence ‘RRRRRHKRK’) in residues 40−48. Deleting region 40−62 or even just the ‘basic cluster’ fully activates NBCe1‐B (Shcheynikov et al. 2015; Lee & Boron, 2018), suggesting that the ‘basic cluster’ is a central component of the AID.
NtB also contains an IRBIT‐binding domain (IBD). Removing the initial acidic motif ‘EDE’ in NtB almost completely abolishes NBCe1‐B activation by IRBIT (Shirakabe et al. 2006; Lee et al. 2012), indicating that ‘EDE’ is a key component of the IBD. A yeast two‐hybrid study suggests that region 37−62 also contains a structural component essential for IRBIT binding (Shirakabe et al. 2006).
What are the molecular mechanisms underlying the auto‐inhibition of NBCe1‐B and its activation by IRBIT? To address these issues, we introduced systematic mutations to NtB of NBCe1‐B. We find that AID comprises components in region 40−85 of NtB, whereas IBD comprises components in region 1−52. AID overlaps with IBD in the ‘basic cluster’. Interestingly, manipulating a set of negatively charged residues (Asp→Asn, Glu→Gln) in the transmembrane domain (TMD) could increase by 3‐ to 4‐fold the basal activity of NBCe1‐B without IRBIT, while having little effect on the IRBIT‐stimulated activity. Moreover, changing a set of negatively charged residues (Asp→Asn, Glu→Gln) plus Ser/Thr residues (to Ala) in the PEST (Pro‐Glu‐Ser‐Thr) domain of IRBIT completely abolishes its capacity to activate NBCe1‐B. In contrast, replacing the Ser/Thr residues with Asp has little effect. We hypothesize that (1) the AID acts as a brake module to bind to the transmembrane domain of NBCe1 via electrostatic attraction, resulting in inhibition of NBCe1‐B; (2) IRBIT binds to the IBD via electrostatic attraction to release the brake, resulting in the activation of NBCe1‐B.
Methods
Ethical approval
Xenopus laevis (clawed frogs) were purchased from Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). The frogs were housed in a fish tank at 18°C and were routinely fed every three days. All procedures regarding the usage of frogs were carried out according to guidelines from the Institutional Committee on Animal Care and Use at Huazhong University of Science and Technology, and conformed to the principles and regulations of The Journal of Physiology (Grundy, 2015).
Structural modelling
For the generation of the Nt model for NBCe1, a pair‐wise alignment of the Nt region (residues 82–381) of mouse NBCe1‐B (NCBI accession NP_061230.2) and the homologous portion of human NDCBE Nt (NCBI accession AAY79176) was generated by SWISS‐MODEL (Waterhouse et al. 2018; Studer et al. 2020). The overall identity is 54.55% between the two sequences. The alignment was used for structural modelling with the crystal structure of human NDCBE Nt (PDB ID: 5JHO) as the template. Structural assessment shows that the simulated model of NBCe1 Nt had the following values: QMEAN −3.25, Cβ −3.11, solvation −0.62, torsion −2.54, and scored 1.86 by using a MolProbity approach (Chen et al. 2010). The resulting PDB file for the structure model of the Nt domain of mouse NBCe1‐B can be accessed at the open Zenodo repository at https://doi.org/10.5281/zenodo.4281509 (https://zenodo.org).
The structure model of the transmembrane domain of mouse NBCe1 was generated by SWISS‐MODEL with the cryo‐EM structure of human NBCe1 (PDB ID: 6CAA) as the template. Residues 445−1007 of mouse NBCe1‐B were aligned with the homologous region of human NBCe1 using SWISS‐MODEL. The sequence identity between the two sequences is 96.09%. Structural assessment shows that the simulated model of mouse NBCe1 had the following values: QMEAN −6.86, Cβ −3.57, solvation −1.80, torsion −5.49, and scored 1.49 by MolProbity approach. The relatively low QMEAN value was probably due to the presence of the poorly folded large extracellular loops, e.g. EL3, in the transmembrane domain of NBCe1. Removing EL3, which was lacking in the template structure of human NBCe1, modestly improved the QMEAN value of the simulated structure. In the template structure of human NBCe1, the intracellular loop IL6 is well folded and completely displayed, whereas IL5 only is partially displayed. The electrostatic surface of NBCe1 transmembrane domain was generated by using the PyMOL Molecular Graphics System (Version 2.0 Schrödinger, LLC). The resulting PDB file for the structure model of the transmembrane domain of mouse NBCe1‐B can be accessed at Zenodo at https://doi.org/10.5281/zenodo.4281682.
Molecular biology
Mouse NBCe1‐A (accession no. HQ018820.1), NBCe1‐B (accession no. NM_018760.2), NBCe1‐D (accession no. HQ285250.1), NBCe1‐E (accession no. HQ204220.1), and mouse IRBIT (accession no. NM_145542.4) were used for the present study. The cDNA of NBCe1 or IRBIT was inserted into pGH19, a Xenopus laevis oocyte expression vector, by using regular molecular cloning techniques. All NBCe1 constructs were tagged with EGFP at the carboxyl‐terminal (Ct) end. Constructs of IRBIT were tagged with myc (EQKLISEEDL) at the Nt. All constructs were confirmed by full‐length DNA sequencing.
Plasmids containing the cDNA of NBCe1 or IRBIT were linearized by restrictive digestion. Capped cRNAs were transcribed from the linearized plasmids with T7 mMessage mMachine kit (Ambion, Austin, TX, USA) according to the manufacturer's instructions.
Oocyte preparation and cRNA injection
For oocyte isolation, a female Xenopus laevis was anaesthetized by immersion in 0.2% ethyl 3‐aminobenzoate methanesulfonate or tricaine (catalogue no. A5040, Sigma‐Aldrich, St Louis, MO, USA). An ovary lobe was extracted, cut into small pieces, and digested with 2 mg ml−1 collagenase Type 1A (Sigma‐Aldrich) in Ca2+‐free NRS solution for 90 min at room temperature. Stage V−VI oocytes were selected for usage.
Oocytes were injected with 25 ng cRNA of NBCe1. Some other oocytes were simultaneously injected with 8 ng cRNA of IRBIT. Control blank oocytes were injected with H2O only. Injected oocytes were incubated at 18°C in OR3 medium for 4−5 days prior to electrophysiology measurement.
Yeast two‐hybrid assay
A yeast two‐hybrid assay was performed with the Matchmaker Gold Yeast Two‐Hybrid System (catalogue no. PT4084‐1, Clontech Takara, Mountain View, CA, USA) according to the manufacturer's instructions. Briefly, the cDNA encoding mouse IRBIT was inserted into pGBKT7 containing the DNA‐binding domain of the transcription factor GAL4 and transformed into haploid yeast stain Y2HGold. The cDNA encoding fragments of NBCe1‐B were inserted into pGADT7 containing the activation domain of GAL4 and transformed into haploid yeast stain Y187. Y2HGold and Y187 were mated to form diploid yeast and then grown on double dropout (DDO) medium or quadruple dropout (QDO) medium containing X‐α‐gal at 30°C for 3−4 days. The strength of protein interaction was judged based on the number and colour lightness of blue colonies.
Two‐electrode voltage clamp
Two‐electrode voltage clamp was performed with an OC‐725C oocyte clamp (Warner Instruments, Hamden, CT, USA) controlled by pCLAMP10.2 (Molecular Devices, San Jose, CA, USA). Electrodes were pulled from thin‐walled borosilicate glass capillaries with outer diameter of 2.0 mm and inner diameter of 1.56 mm (Sutter Instruments, catalogue no. BF100‐78‐10, Navato, CA, USA) and filled with 3 m KCl. The electrodes had a resistance of 0.5−2.0 MΩ. For voltage clamp recording, an oocyte was placed in a chamber containing standard ND96 solution and impaled with two microelectrodes, one for sensing membrane potential (V m) and the other for passing current. The cell was superfused with ND96 until the V m was stable. The current−voltage (I‐V) relationship was generated by voltage clamp using a protocol stepping the potential from −160 mV to +60 mV with 20‐mV increments. Each step was recorded for a duration of 100 ms followed by an interval of 100 ms at a holding potential close to the spontaneous V m of the oocyte. The I‐V relationship for CO2/HCO3 − was generated immediately after the hyperpolarization was maximized upon the switch from ND96 to 5% CO2/33 mm HCO3 −.
Solutions for physiological experiments
Ca2+‐free NRS (in mm): 82 NaCl, 2 KCl, 20 MgCl2, 5 Hepes, pH 7.50.
OR3 medium: one packet of L‐15 medium was dissolved in 1.5 l of H2O, followed by the addition of 100 ml of penicillin/streptomycin (10,000 units ml−1 of penicillin, 10,000 μg ml−1 of streptomycin; Invitrogen, Carlsbad, CA, USA) and 5 mm Hepes, pH 7.50.
Standard ND96 (in mm): 96 NaCl, 2 KCl, 1 MgCl2, 1.8 CaCl2 and 5 Hepes, pH7.50.
5% CO2/33 mm HCO3 − (in mm): 63 NaCl, 2 KCl, 1 MgCl2, 1.8 CaCl2 and 5 Hepes, followed by titration to pH 7.50 and the addition of 33 NaHCO3. The solution was then bubbled with 5% CO2 balanced with N2.
Protein preparation and western blotting
For membrane preparation, a batch of 20 oocytes were placed in 1 ml protein isolation buffer (in mm: 7.5 NaH2PO4, 250 sucrose, 5 EDTA,5 EGTA, pH 7.0) containing 1% protease inhibitor cocktail (catalogue no. P8340, Sigma‐Aldrich) and homogenized. The crude homogenate was centrifuged at 3000 g for 10 min at 4°C to remove the cell debris, and then centrifuged at 100,000 g for 1 h at 4°C to collect the membrane proteins. The pellet was dissolved in 200 μl of protein resuspension buffer (20 mm Tris, 5 mm EDTA, 5% sodium dodecyl sulfate or SDS, pH 8.0). The membrane preparations were mixed with SDS sample buffer, separated by SDS polyacrylamide gel electrophoresis (SDS‐PAGE), and transferred onto polyvinylidene difluoride (PVDF) membrane for western blotting.
Biotinylation was performed with the Cell Surface Protein Isolation Kit (catalogue no. 89881, Pierce, Rockford, IL, USA) according to manufacturer's instructions but adapted for usage with Xenopus oocytes (Liu et al. 2013). Briefly, 15 oocytes were pooled and incubated at 4°C for 1 h in phosphate‐buffered saline (diluted to 200 mOsm for usage with Xenopus oocytes) containing 240 μg ml−1 Sulfo‐NHS‐SS‐biotin. The reaction was stopped by adding 200 μl of Quenching Solution. The cells were saved and washed with tris‐buffered solution (TBS), then disrupted by trituration with a pipette in 200 μl of TBST (TBS with 1% Triton X‐100) containing 1% Protease Inhibitor Cocktail (catalogue no. P8340, Sigma‐Aldrich). The crude lysate was centrifuged at 3000 rpm at 4°C for 10 min. A total of 40 μl of the resultant supernatant was saved, representing the ‘total proteins’. The rest was incubated with 120 μl Immobilized NeutrAvidin (Pierce, Rockford, IL, USA) at room temperature for 1 h and then washed 5 times with TBST. The bound proteins were collected, representing the ‘surface proteins’, by elution with 100 μl of SDS‐sample buffer containing 50 mm dithiothreitol. Finally, the samples were separated by SDS‐PAGE for western blotting analysis. Each lane was loaded with 10 μl of ‘total proteins’ or 20 μl of ‘surface proteins’. The proteins were then blotted onto a PVDF membrane for western blotting.
For western blotting, the blot was incubated with 5% milk in 1 × TBST (1 mm Tris, 150 mm NaCl, 0.1% Tween20, pH 7.4) for 1 h at room temperature, incubated with primary antibody in 1 × TBST at 4°C overnight, washed 5 times with 1 × TBST, and then incubated with HRP‐conjugated secondary antibody 1 h at RT. After 5 washes with 1 × TBST, the blot was incubated with SuperSignal West Pico PLUS chemiluminescent substrate (catalogue no. 34 580; Thermo Scientific, Rockford, IL, USA) prior to X‐ray exposure. Densitometry was performed by ImageJ (National Institutes of Health, USA).
Anti‐EGFP was purchased from Proteintech (catalogue no. 66 002, Proteintech Group, Inc., Wuhan, China). Anti‐Myc was purchased from Abclonal (catalogue no. AE010, Abclonal Technology, Wuhan, China). Goat‐anti‐muse HRP‐conjugated secondary antibody was purchased from Beyotime (catalogue no. A0216, Beyotime Biotechnology, Shanghai, China).
Statistical analysis
All bars in graphs represent the mean ± SD. For multiple comparisons, one‐way ANOVA followed by post hoc Tukey's comparison was performed with Minitab (Minitab, State College, PA, USA). A confidence interval of 95% was used for the ANOVA analysis. The exact numerical values for the bars and results of detailed statistical analyses are provided as Supporting information in the Statistical summary document.
Results
Effect of cassette I on NBCe1 activity
Figure 1 A shows structural diagrams of NBCe1‐A to ‐E. The activities of NBCe1‐D and ‐E, both lacking cassette I (9 amino acids or aa), remain to be characterized. Figure 1 B shows a model of the 3‐dimension structure of the Ntcore of NBCe1 simulated based on the crystal structure of the Nt of human NDCBE (Alvadia et al. 2017). Cassette I is located in a poorly folded loop. To examine the effect of cassette I on transport activity, NBCe1‐A, ‐B, ‐D and ‐E, all tagged with EGFP at the carboxyl‐terminus (Ct), were each expressed in Xenopus oocytes. For electrophysiology recording, an oocyte was superfused with nominally HCO3 −‐free ND96 and then with 5% CO2/33 mm HCO3 −.
Figure 1. Functional comparison of NBCe1 variants in Xenopus oocytes.
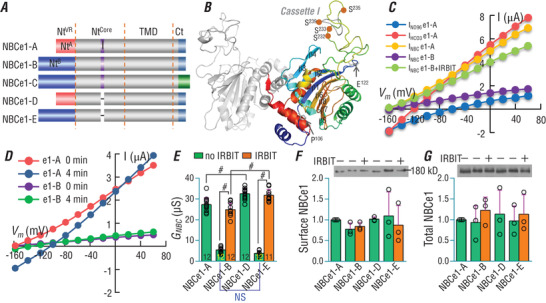
A, diagram showing primary structure of NBCe1 variants. NtVR: variable region of the initial amino terminus (Nt). NtA: unique Nt of NBCe1‐A (red) containing the auto‐stimulatory domain (ASD). NtB: unique Nt of NBCe1‐B (blue) containing the auto‐inhibitory domain (AID). Ntcore: conserved core region of the Nt. TMD: transmembrane domain. Ct: carboxyl terminus. The purple indicates the optionally spliced cassette I. B, 3‐dimensional model of Ntcore domain of NBCe1. The model was simulated based on the crystal structure of human NDCBE Nt (PDB ID: 5JHO). Cassette I (indicated in grey in the coloured monomer) is located in a poorly folded loop that is rich in Ser and Thr residues. C, representative I‐V curves of an oocyte expressing NBCe1‐A or NBCe1‐B ± IRBIT in ND96 and 5% CO2/33 mm HCO3 −. D, comparison of electrophysiology recordings for day‐matched oocytes expressing NBCe1‐A or ‐B at time 0 and 4 min after switch from ND96 to CO2/HCO3 −. Shown here are representative of four independent recordings. E, summary of G NBC of NBCe1 with or without IRBIT. G NBC represents the slope conductance of I NBC between ±40 mV calculated from experiments like those shown in panel C. F and G, expression and abundance of NBCe1 in surface (F) and total fractions (G) of Xenopus oocytes. NBCe1 was probed with anti‐EGFP. The protein abundance was normalized to NBCe1‐A. The numerals on the bars indicate the number of individual oocytes. Panels F and G represent the average of three experiments. Green bars: without IRBIT. Orange bars: with IRBIT. #Statistically significant. NS: not significant. One‐way ANOVA followed by post hoc Tukey's comparison was performed for statistical analysis. Numerical values for all bars and detailed results of statistical tests are provided as Supporting information in Statistical Summary Tables 1E−1G.
Figure 1 C shows an example of the I‐V curves in ND96 (I ND96) and CO2/HCO3 − solution (I HCO3) for an oocyte expressing NBCe1‐A. Here, the electrophysiology recordings for CO2/HCO3 − were carried out immediately after the maximal hyperpolarization (time 0) upon the switch from ND96 to CO2/HCO3 −. I NBC was obtained by subtracting I ND96 from I HCO3, representing the net HCO3 −‐dependent current mediated by NBCe1. The slope conductance (G NBC) of I NBC between ±40 mV provides an index for NBCe1 activity. For comparison, we also show the I NBC for NBCe1‐B ± IRBIT in Fig. 1 C. Note that the reversal potential (V rev) of I NBC is virtually identical for NBCe1‐A and NBCe1‐B ± IRBIT, suggesting that the initial intracellular [Na+] and [HCO3 −] were very similar in cells expressing NBCe1‐A vs. NBCe1‐B ± IRBIT.
Figure 1 D shows the net I NBC curves of a cell expressing either NBCe1‐A or NBCe1‐B, obtained from recordings either at time 0 or after 4 min of continuous perfusion in CO2/HCO3 − solution. Again, the V rev of NBCe1‐A and NBCe1‐B at time 0 is virtually identical. However, the V rev of both NBCe1‐A and NBCe1‐B is shifted rightward after 4‐min perfusion in CO2/HCO3 −. Similar observations were reported in a very recent study (Moss & Boron, 2020). The rightward shift in V rev probably reflects an increase in the intracellular [Na+] and [HCO3 −] due to the activity of NBCe1. However, this effect of perfusion with CO2/HCO3 − on intracellular [Na+] and [HCO3 −] would vary depending on the degree of NBCe1 activity. Note that, the shift in V rev is much greater for NBCe1‐A (∼60 mV), which is highly active, than for NBCe1‐B (∼20 mV), which contains only basal activity without IRBIT. Interestingly, the perfusion in CO2/HCO3 − also increases the apparent G NBC of NBCe1‐A more than NBCe1‐B. In the following experiments, to minimize the effect of CO2/HCO3 − perfusion, the electrophysiology recordings for CO2/HCO3 − solution were carried out immediately after the maximal hyperpolarization upon the switch from ND96 to CO2/HCO3 − solution.
Figure 1 E summarizes the activities of different NBCe1 variants. The apparent activity of NBCe1‐A is ∼5 times greater than that of NBCe1‐B, whereas the apparent activity of NBCe1‐D is ∼8 times greater than that of NBCe1‐E. Both NBCe1‐B and ‐E are fully activated by IRBIT, indicating the presence of auto‐inhibition in NBCe1‐B and ‐E. Neither the surface nor the total expression of the NBCe1 variants is significantly different from each other (Fig. 1 F and G). Thus, the difference in the G NBC values probably reflects the difference in the intrinsic activity, i.e. the single molecular activity, of NBCe1 variants.
The G NBC of NBCe1‐D is higher by ∼20% than that of NBCe1‐A. The G NBC of IRBIT‐activated NBCe1‐E is higher by ∼30% than that of IRBIT‐activated NBCe1‐B. These two comparisons suggest that cassette I is modestly inhibitory to the activity of NBCe1.
AID is dominant over ASD when both are present simultaneously
We generated a series of NBCe1 chimeras by fusing regions of NBCe1‐B Nt containing the AID to wild‐type (WT) NBCe1‐A (AWT hereafter), or by fusing NtA containing the ASD to WT NBCe1‐B (BWT hereafter) at the Nt or Ct end (Fig. 2 A). For example, N85B‐A was created by attaching N85B (residues 1−85 of NBCe1‐B) to the Nt of AWT via lnk20, a linker of 20aa consisting of four repeats of ‘GGGGS’ ([GGGGS]4). A‐lnk40‐N106B was created by attaching N106B (residues 1−106 of NBCe1‐B) with lnk40 ([GGGGS]8) to the Ct of AWT.
Figure 2. AID is inhibitory at both Nt and Ct ends, but requires covalent linkage to NBCe1.
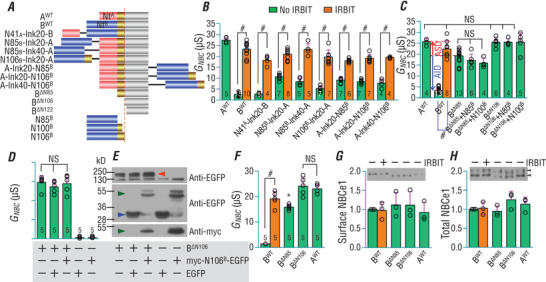
A, diagram showing the design of constructs. Red: NtA, the unique Nt of NBCe1‐A (red) containing the ASD; blue: NtB, the unique Nt of NBCe1‐B (blue) containing the AID; gold: region 86−106; grey: residues 107 to Ct end (amino‐acid numbering according to NBCe1‐B). B, IRBIT stimulates NBCe1 variants simultaneously containing AID and ASD. C, isolated AID‐containing Nt fragments of NBCe1‐B have no effect on the functional expression of BΔN85 and BΔN106. The blue arrow indicates the AID effect. The red arrow indicates the ASD effect. D, Myc‐N106B‐EGFP has no effect on the functional expression of BΔN106. E, western blot showing the expression of BΔN106 (indicated by red triangle) and Myc‐N106B‐EGFP (indicated by green triangle). Blue triangle indicates EGFP. F, comparison of G NBC of BWT, BΔN85, BΔN106 and AWT. G and H, expression and relative abundance of AWT, BWT, BΔN85 and BΔN106 in surface and total fractions of Xenopus oocytes. The protein abundance was normalized to BWT without IRBIT. The two bands indicated by triangles in the total fraction presumably represent glycosylated vs. non‐glycosylated NBCe1 (Choi et al. 2003). The numerals on the bars in panels B−D and F indicate the number of individual oocytes. Panels G and H each represent the average of three experiments. Green bars: without IRBIT. Orange bars: with IRBIT. #Significantly different from the corresponding bar without IRBIT (two‐tailed unpaired Student's t test). *BΔN85 is significantly different from BΔN106 and AWT. NSBars in the group not significant from each other. For multiple comparisons, one‐way ANOVA followed by post hoc Tukey's comparison was performed. In panel E, red triangle indicates BΔN106; blue triangle indicates EGFP; green triangle indicates myc‐N106B‐EGFP. For voltage clamp recordings, NBCe1 variants were always analysed with day‐matched BWT. Numerical values for all bars and detailed results of statistical tests are provided as Supporting information in Statistical Summary Tables 2B−D and 2F−H.
The activities of all AID‐containing constructs are substantially lower than that of AWT, but are greatly enhanced by IRBIT (Fig. 2 B), indicating the presence of auto‐inhibition in these constructs. Our data demonstrate that: (1) the AID is able to inhibit NBCe1‐A activity at both the Nt and Ct ends; (2) when present simultaneously, the AID is dominant over the ASD.
Auto‐inhibition requires covalent attachment of the AID to NBCe1
We generated three Nt‐truncated variants of BWT (Fig. 2 A): BΔN85 (2−85 removed), BΔN106 (2−106 removed), and BΔN122 (2−122 removed). Note that, BΔN85 is equivalent to AΔN41 (NBCe1‐A variant without ASD).
Without IRBIT, the G NBC of both BΔN85 and BΔN106 is 5−6 times greater than that of BWT (Fig. 2 C). BΔN122 is completely inactive, presumably due to a major folding defect in the Nt domain of NBCe1 (data not shown). Note that region 107−122 represents a central β‐strand in the Ntcore (Fig. 1 B). The difference in activities between BΔN85 and BWT represents the AID effect, whereas the difference between BΔN85 (equivalent of AΔN41) and AWT represents the ASD effect.
To examine the effect of isolated AID on NBCe1, we attempted to co‐express isolated N85B or N100B (residues 1−100 of NBCe1‐B) with BΔN85 or BΔN106 in oocytes. As shown in Fig. 2 C, neither N85B nor N100B changes the activities of BΔN85 and BΔN106. The isolated N85B and N100B were presumably degraded in oocytes as no protein could be detected by western blotting (data not shown). As shown in Fig. 2 D, the activity of BΔN106 was not affected by myc‐N106B‐EGFP – N106B tagged with myc at Nt and EGFP at Ct. The expression of myc‐N106B‐EGFP was verified by western blotting with both anti‐Myc and anti‐EGFP (Fig. 2 E). Together, these findings show that auto‐inhibition requires covalent linkage of AID to NBCe1.
Interestingly, the G NBC of BΔN106 is consistently higher than that of BΔN85, but not different from that of AWT (Fig. 2 C and F). Neither the surface nor the total expression is significantly different among BWT, BΔN85, BΔ106 and AWT (Fig. 2 G and H). Taken together, our data suggest that region 86−106 is modestly inhibitory to NBCe1 activity.
Mutations in motifs ‘EDE’, ‘DEEEVE’ and the ‘basic cluster’ in NtB abolish IRBIT binding
In the following sections, we aim to characterize the key structural elements in NtB that are important for NBCe1‐B auto‐inhibition and the binding of IRBIT. Figure 3 A shows the primary sequence of NtB. We employed a yeast two‐hybrid assay to characterize the structural elements in NtB essential for IRBIT binding. The results are summarized in Fig. 3 B.
Figure 3. Identification and characterization of structural elements in NtB essential for IRBIT binding.
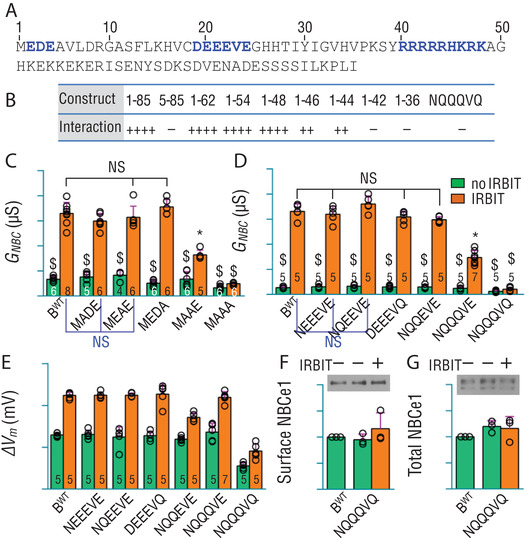
A, primary amino‐acid sequence of NtB. The blue indicates elements essential for IRBIT binding. B, summary of yeast two‐hybrid results for protein interaction between NtB fragments and IRBIT. The construct name indicates regions of NtB used for yeast two‐hybrid assay. NQQQVQ: full‐length NtB variant with ‘DEEEVE’ replaced by ‘NQQQVQ’. ‘++++’: strong protein interaction; ‘++’: weak interaction; ‘−’: no detectable interaction by yeast two‐hybrid assay. The strength of protein interaction between NtB fragments and IRBIT was judged based on the number and colour‐intensity of blue colonies on quadruple‐dropout medium (QDO). C, effect of perturbing residues in ‘MEDE’ on NBCe1‐B stimulation by IRBIT. D, effect of perturbing residues in DEEEVE on NBCe1‐B stimulation by IRBIT. E, summary of maximum hyperpolarization of oocytes expressing NBCe1 variants. F and G, expression and relative abundance of BWT, NQQQVQ ± IRBIT in surface and total fractions of Xenopus oocytes. The protein abundance was normalized to BWT without IRBIT. For voltage clamp measurements, NBCe1 mutants were analysed with day‐matched BWT. The numerals on the bars indicate the number of individual oocytes. Panels F and G each represent the average of three experiments. Green bars: without IRBIT. Orange bars: with IRBIT. $Bars are not significantly different from BWT. NSBars in the group are not significantly different from each other. *Bar is significantly different from all other bars. One‐way ANOVA followed by post hoc Tukey's comparison was performed for statistical analysis. Numerical values for all bars and detailed results of statistical tests are provided as Supporting information in Statistical Summary Tables 3C−3G.
EDE
Our yeast two‐hybrid data show that the initial ‘EDE’ motif is essential for IRBIT binding to NtB (Fig. 3 B), consistent with the previous observation that deleting ‘EDE’ virtually abolishes NBCe1‐B activation by IRBIT (Lee et al. 2012). NBCe1‐B variants were generated by replacing the acidic residues in ‘EDE’ with Ala. The G NBC of these variants without IRBIT is not significantly different from that of non‐activated BWT (Fig. 3 C), indicating full auto‐inhibition in all variants. With IRBIT, the G NBC values of MADE, MEAE and MEDA are not different from IRBIT‐activated BWT, whereas that of MAAE is just ∼50% of that of IRBIT‐activated BWT. Finally, IRBIT only slightly stimulates MAAA (P = 0.005, Student's t test for MAAA with vs. without IRBIT). Interestingly, adding a 10‐aa linker ([GGGGS]2) following ‘MEDE’ has no effect on IRBIT‐NBCe1 interaction (data not shown).
Together, our data show that (1) motif ‘EDE’ is essential for IRBIT binding to NBCe1‐B; (2) two acidic residues are minimal in this motif for full activation of NBCe1‐B by IRBIT.
DEEEVE
Using a yeast two‐hybrid assay (Fig. 3 B), construct NQQQVQ (NtB variant with ‘DEEEVE’ → ‘NQQQVQ’) elicits no interaction with IRBIT. NBCe1‐B mutants were generated by mutating acidic residues in ‘DEEEVE’. Without IRBIT, the basal G NBC of all mutants is not different from that of BWT (by ANOVA), indicating full auto‐inhibition. As shown in Fig. 3 D, the G NBC of IRBIT‐activated mutants NEEEVE, NQEEVE, NQQEVE and DEEEVQ is not significantly different from that of IRBIT‐stimulated BWT, indicating full activation. However, the G NBC of NQQQVE with IRBIT is just ∼55% of that of IRBIT‐activated BWT. Finally, IRBIT has no effect on mutant NQQQVQ. NQQQVQ ± IRBIT produced modest hyperpolarization upon the addition of CO2/HCO3 − (Fig. 3 E), characteristic of electrogenic NBC activity (Romero et al. 1997). Note that, without IRBIT, the basal G NBC of NQQQVQ is somewhat lower than that of BWT (P = 0.0002 by Student's t test). Neither the surface nor the total expression of NQQQVQ ± IRBIT is significantly different from that of BWT (Fig. 3 F and G). Thus, the activity of NQQQVQ is probably intrinsically lower than BWT.
Together, our data show that (1) DEEEVE is essential for IRBIT binding to NBCe1‐B; (2) two acidic residues are minimal in this motif for full activation of NBCe1‐B by IRBIT.
The ‘basic cluster’ (RRRRRHKRK)
Yeast two‐hybrid analysis shows that all fragments containing region 1−48 elicit strong interactions with IRBIT (Fig. 3 B). Fragments 1−46 and 1−44 retain only weak interaction, whereas 1−42 and 1−36 show no interaction with IRBIT. Our yeast two‐hybrid data indicate that region 1−48 is sufficient for IRBIT binding. Moreover, the ‘basic cluster’ is essential for IRBIT binding to NBCe1‐B.
Altogether, NtB contains three structural elements essential for IRBIT binding to NBCe1‐B. Later in the Discussion section, we will propose that these three elements probably represent the minimal structural requirement for IBD.
Effect of perturbing Ct portions of NtB on auto‐inhibition and activation of NBCe1‐B by IRBIT
The previous study found that, starting from the Nt end, deleting portions of the NtB progressively increases NBCe1 activity without IRBIT (Lee et al. 2012). To examine the effect of deleting Ct portions of NtB on NBCe1‐B, Nt fragments of NtB are attached to BΔN85‐Nt via GS‐linkers, the length of which matches the deleted portions of NtB (Fig. 4 A). For example, N80B‐BΔN85 was generated by attaching region 1−80 of NtB to BΔN85 with a 5aa linker [GGGGS].
Figure 4. Minimum structural requirements for IRBIT binding and effect of mutation to ‘basic cluster’ on the magnitude of auto‐inhibition of NBCe1‐B.
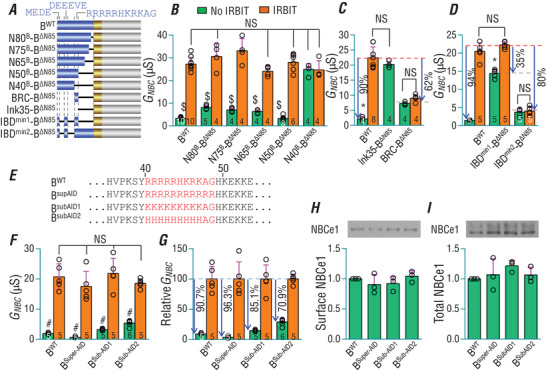
A, diagram showing the design of NBCe1‐B variants. Blue boxes represent elements of NtB. Gold boxes represent region 86−106 of NBCe1‐B. Lines indicate GS‐linkers (repeats of ‘GGGGS’). The length of the GS‐linker matches the length of the replaced region of NBCe1‐B. B−D, summary of G NBC of NBCe1‐B variants (with different parts of NtB) ± IRBIT. E, mutation to ‘basic cluster’ of NBCe1‐B. F, summary of G NBC of ‘basic‐cluster’ variants ± IRBIT. G, relative G NBC of ‘basic‐cluster’ variants as percentiles of the corresponding stimulated ones. To obtain the relative G NBC, the G NBC of each individual oocyte contributed to the bars in panel F was normalized to the average G NBC of the same variant activated by IRBIT. The percentile difference between the non‐activated vs. the IRBIT‐activated G NBC indicates the magnitude of auto‐inhibition. H and I, expression and relative abundance of BWT, BSuper‐AID, BSub‐AID1 and BSub‐AID2 in the surface and total fractions of Xenopus oocytes. The protein abundance was normalized to BWT. NBCe1 was probed with anti‐EGFP. For voltage clamp recordings, mutants were always analysed with day‐matched BWT. The numerals on the bars indicate the number of individual oocytes. Panels H and I each represent the average of three experiments. Green bars: without IRBIT. Orange bars: with IRBIT. NSThese groups are not significantly different from each other. $Groups are not significantly different from each other. *The group is significantly different from all other groups. #These four groups are significantly different from each other. One‐way ANOVA followed by post hoc Tukey's comparison was performed for statistical analysis. Numerical values for all bars and detailed results of statistical tests are provided as Supporting information in Statistical Summary Tables 4B−4D and 4F−4I.
Without IRBIT, the basal G NBC values of N80B‐BΔN85 (P < 0.0001 by Student's t test), N75B‐BΔN85 (P < 0.0001 by Student's t test), and N65B‐BΔN85 (P < 0.0001 by Student's t test) are increased by ∼1‐ to 2‐fold compared to BWT. However, the basal G NBC of N50B‐BΔN85 is not different from that of BWT (Fig. 4 B). Like BWT, all these four variants can be fully activated by IRBIT. With or without IRBIT, the G NBC of N40B‐BΔN85, which lacks the ‘basic cluster’ is not significantly different from that of IRBIT‐activated BWT, indicating absence of auto‐inhibition in N40B‐BΔN85. Together, the data indicate that the ‘basic cluster’ is essential for NBCe1‐B auto‐inhibition, consistent with previous observations (Shcheynikov et al. 2015; Lee & Boron, 2018).
To further investigate the role of the ‘basic cluster’ in auto‐inhibition, we generate BRC‐BΔN85 by attaching just the ‘basic cluster’ (BRC) to BΔN85 via a 35aa linker. Not surprisingly, BRC‐BΔN85 cannot be activated by IRBIT due to lacking ‘EDE’ and ‘DEEEVE’ essential for IRBIT binding (Fig. 4 C). Without IRBIT, the G NBC of BRC‐BΔN85 is lower by ∼62% than that of lnk35‐BΔN85, indicating that the ‘basic cluster’ alone can produce profound auto‐inhibition.
Based on BRC‐BΔN85, we created IBDmin1‐BΔN85 by restoring ‘EDE’ and ‘DEEEVE’, and IBDmin2‐BΔN85 by restoring ‘EDE’, ‘DEEEVE’, and region 51−85. As shown in Fig. 4 D, IBDmin1‐BΔN85 retains ∼35% of auto‐inhibition and can be fully activated by IRBIT. IBDmin2‐BΔN85 retains ∼80% of auto‐inhibition. Surprisingly, this IBDmin2‐BΔN85 cannot be stimulated by IRBIT, although it contains all three elements, i.e. EDE, DEEEVE, and the ‘basic cluster’, that are essential for IRBIT binding. The data indicate that these three elements are not sufficient for IRBIT to activate NBCe1 in the context of IBDmin2‐BΔN85. Later in Discussion (section ‘Explanation for no stimulation of IBDmin2‐BΔN85 by IRBIT’), we will address how this observation makes sense.
Mutation to charged residues in ‘basic cluster’ alters the magnitude of auto‐inhibition in NBCe1‐B
The ‘basic cluster’ of NBCe1‐B contains 9 alkaline residues, including 6Arg+2Lys+1His, and thus would be highly positively charged at near‐neutral physiological pH. Consider the pK a (−log of acid dissociation constant) of the side chains (6.00 for His, 10.53 for Lys, and 12.48 for Arg), the tendency to be positively charged would be Arg > Lys > His. Starting from BWT, we created BSuper‐AID, BSub‐AID1 and BSub‐AID2 by manipulating residues in the ‘basic cluster’ (Fig. 4 E). In principal, at physiological pH, the tendency to be positively charged of these ‘basic‐cluster’ variants would be BSuper‐AID > BWT > BSub‐AID1 > BSub‐AID2.
As summarized in Fig. 4 F, the G NBC values of the IRBIT‐activated variants are not significantly different from each other. Without IRBIT, the G NBC progressively increases in the following order: BSuper‐AID < BWT < BSub‐AID1 < BSub‐AID2.
We normalized the non‐activated G NBC to the IRBIT‐activated G NBC of the same variant. The percentile difference in the relative G NBC between the non‐activated and IRBIT‐activated NBCe1 represents the auto‐inhibition magnitude. As summarized in Fig. 4 G, the auto‐inhibition magnitude progressively decreases in the order: BSuper‐AID (96.3%) > BWT (90.7%) > BSub‐AID1 (85.1%) > BSub‐AID2 (70.9%), consistent with the order for tendency of the ‘basic cluster’ to be positively charged. Finally, neither the surface level nor the total level of NBCe1 is significantly different among BWT, BSuper‐AID, BSub‐AID1 and BSub‐AID2 (Fig. 4 H−I).
Together, our data indicate that the magnitude of auto‐inhibition of NBCe1 is correlated to the tendency to be positively charged of the ‘basic cluster’.
Mutating residues in the transmembrane domain greatly decreases the magnitude of auto‐inhibition in NBCe1‐B
Given the highly positively charged nature of the ‘basic cluster’, could it inhibit NBCe1 activity by binding to the TMD via electrostatic attraction? If so, then mutating the binding sites for the ‘basic cluster’ in TMD would disrupt this electrostatic interaction, thereby increasing the basal activity of NBCe1‐B without IRBIT.
Potential binding sites for the ‘basic cluster’ in TMD could be: (1) acidic residues, i.e. Asp and Glu, that could form a salt bridge with positively charged residues; (2) Phe and Tyr residues with aromatic rings that could be powerfully attractive, via cation‐π interaction, to the positive charges (Kennedy et al. 2016). Based upon a careful examination into the electrostatic surface model of NBCe1 TMD, we identified D853, E858, E860, E866 and E875 on intracellular loop (IL5), as well as D924, D938, F939 on IL6, for further testing (Fig. 5 A and B). Figure 5 C shows the localization of the acidic and aromatic residues in a side‐view of the 3‐dimensional structure of NBCe1 TMD.
Figure 5. Screening for potential AID‐binding targets in the transmembrane domain of NBCe1.
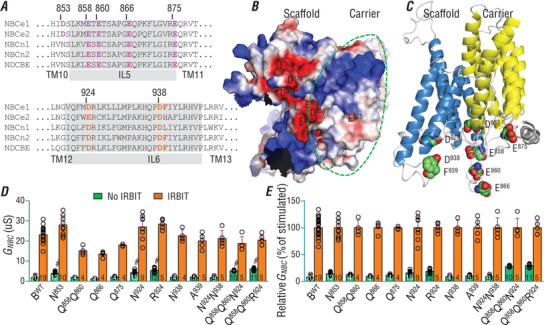
A, sequence alignment of intracellular loop IL5 and IL6 of NBCe1, NBCe2, NBCn1, NBCn2 and NDCBE. Purple in IL5 and orange in IL6 of NBCe1 indicate the negatively charged or aromatic residues manipulated in the present study. Most of these residues are conserved in other NBC members. B, bottom view of 3‐dimension structure of NBCe1 transmembrane domain showing the electrostatic surface of the intracellular side. Red indicates negatively charged surface area. Blue indicates positively charged surface area. The region surrounded by a dashed green line represents the carrier domain. C, side view of 3‐dimensional structure of NBCe1 transmembrane domain. Yellow represents the carrier domain. Blue represents the scaffold domain. Shown as spheres are the acidic and aromatic residues in the IL5 and IL6 that are presumably involved in the binding of AID. Red spheres = O, green spheres = C and blue spheres = N. D, functional expression of different NBCe1 constructs with and without IRBIT in Xenopus oocytes. E, relative G NBC as percentile of the stimulated NBCe1. The 3‐dimensional model of mouse NBCe1 was simulated based on the cryo‐EM structure of human NBCe1 (PDB ID 6CAA) by using SWISS‐MODEL and visualized using the PyMOL Molecular Graphics System (Version 2.0 Schrödinger, LLC). For voltage clamp recordings, NBCe1 mutants were always analysed with day‐matched oocytes expressing BWT. The relative G NBC was calculated as in Fig. 4 G. Green bars: without IRBIT. Orange bars: with IRBIT. The numerals on the bars indicate the number of individual oocytes. #Significantly different from BWT by one‐way ANOVA followed by post hoc Tukey's comparison among the groups without IRBIT. Numerical values for all bars and detailed results of statistical tests are provided as Supporting information in Statistical Summary Tables 5D and 5E. [Correction made on 6 January 2021, after first online publication: The label of the y axis in Figure 5D was changed from ‘GHCO3 (uS)’ to ‘GNBC (uS)’ to be consistent with the bar graphs of the rest of the figures.]
As shown in Fig. 5 D and E, all NBCe1‐B variants with single or double mutations to these residues retain normal basal activity and can be substantially activated by IRBIT. Single mutants, including N853 (Asp853→Asn), N924 (Asp924→Asn) and R924 (Asp924→Arg), elicit a slightly lower degree of auto‐inhibition compared to BWT. Strikingly, NBCe1‐B variants with triple or more mutations, e.g. Q858Q860R924 (Fig. 5 D), NQQNNF and NQQNNA (Fig. 6 A), elicit significantly increased basal activity without IRBIT. Specifically, the basal activity of non‐activated NQQNNA (without IRBIT) is ∼4.5 times higher than that of non‐activated BWT, corresponding to a decrease in the auto‐inhibition magnitude by 31% (92% in BWT − 61% in NQQNNA; Fig. 6 B).
Figure 6. Manipulating residues in TMD decreases the magnitude of NBCe1‐B auto‐inhibition.
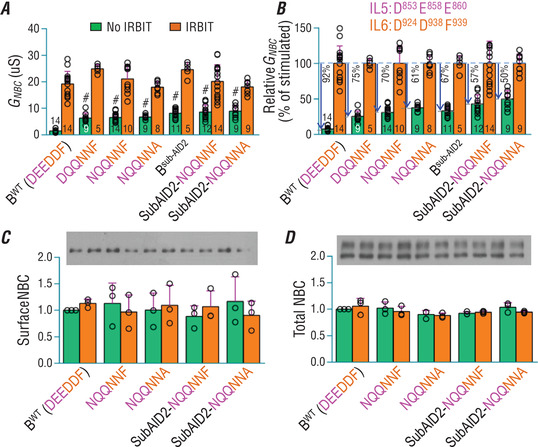
A, functional expression of different NBCe1 constructs with and without IRBIT in Xenopus oocytes. B, relative G NBC as a percentile of the stimulated NBCe1. For voltage clamp recordings, NBCe1 mutants were always analysed with day‐matched oocytes expressing BWT. The relative G NBC was calculated as in Fig. 4 G. The numerals on the bars indicate the number of individual oocytes. #Significantly different from BWT and all orange bars. C and D, expression and relative abundance of selected NBCe1 variants in surface (C) and total fractions (D) of Xenopus oocytes. The protein abundance was normalized to BWT without IRBIT. The numerals on the bars indicate the number of individual oocytes. Panels C and D each represent the average of three experiments. Green bars: without IRBIT. Orange bars: with IRBIT. ANOVA followed by post hoc Tukey's comparison was performed for statistical analysis. Numerical values for all bars and detailed results of statistical tests are provided as Supporting information in Statistical Summary Tables 6A−6D.
Interestingly, a combination of subAID2 – mutation to ‘basic cluster’ which decreases auto‐inhibition magnitude by ∼30%, as shown in Fig. 4 E – and NQQNNA (mutation to the TMD) just decreases auto‐inhibition magnitude by 42% (92% − 50% = 42%; Fig. 6 B). This observation indicates that the combination of the two sets of mutations in the ‘basic cluster’ and TMD has no direct additive effect of decreasing the auto‐inhibition magnitude. Thus, the effects of mutations in the ‘basic cluster’ and those in the TMD on auto‐inhibition are probably part of the same continuum.
Selected NBCe1 constructs, including BWT, NQQNNF, NQQNNA, subAID2‐NQQNNF and subAID2‐NQQNNA, were examined for expression in the surface as well as the total fraction of oocytes (Fig. 6 C and D). Neither the surface nor the total protein levels are significantly different among these NBCe1 variants ± IRBIT. Thus, the decrease in the auto‐inhibition magnitude is not an effect of changes in the expression level of NBCe1.
Altogether, our data show that mutations in the TMD can greatly reduce the auto‐inhibition magnitude of NBCe1, consistent with the hypothesis that the ‘basic cluster’ interacts with the TMD to inhibit NBCe1 activity.
Mutations to acid residues and Thr/Ser residues in PEST domain of IRBIT attenuate its interaction with NBCe1‐B
We hypothesize that IRBIT binds to the ‘basic cluster’, releasing the AID from TMD, and thus activating NBCe1‐B. If so, mutating the binding sites for the ‘basic cluster’ in IRBIT could disrupt IRBIT‐NBCe1 interaction and therefore abolish the stimulation of NBCe1‐B by IRBIT.
What are the ‘basic‐cluster’ binding sites in IRBIT? Given the highly positively charged nature of the ‘basic cluster’, we speculate that the ‘basic‐cluster’ binding sites in IRBIT would be negatively charged. As shown in Fig. 7 A, the unique Nt of IRBIT contains a so‐called PEST (Pro‐Glu‐Ser‐Thr) domain containing five acidic residues, i.e. Asp73, Asp83, Asp86, Asp87 and Glu88 plus 15 Ser/Thr/Tyr residues that could be potentially phosphorylated ((Devogelaere et al. 2008)). Phosphorylation of the PEST domain plays an essential role in the interaction of IRBIT with IP3R (Ando et al. 2006) and NBCe1‐B (Shirakabe et al. 2006). The acidic and phosphorylated residues would cause the PEST domain to be highly negatively charged.
Figure 7. Effects of mutation to charged residues and potential phosphorylation sites in PEST domain of IRBIT on its interaction with NBCe1‐B.
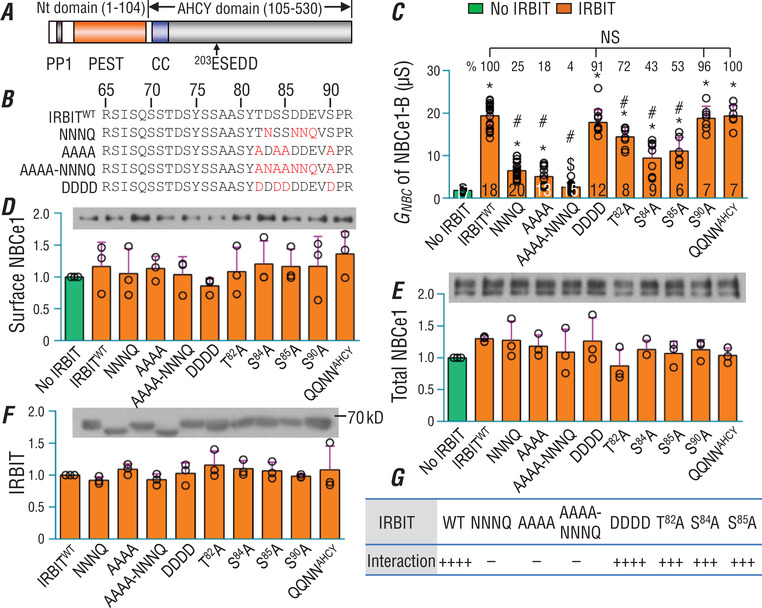
A, diagram to show the module structure of IRBIT. IRBIT contains a unique Nt domain and an AHCY domain that is homologous to the adenosylhomocysteine hydrolase. The PEST domain, a core portion in the unique Nt, is rich in acidic residues and potential phosphorylation sites. PP1: putative protein phosphatase 1 binding site. CC: coiled‐coil domain. B, alignment to show mutations to the PEST domain in IRBIT variants. C, stimulatory effects of IRBIT variants on NBCe1‐B activity. The percentages above the orange bars indicate the stimulatory effect of IRBIT variants on NBCe1‐B relative to that of WT IRBIT, computed by (G NBC mutant IRBIT – G NBC No IRBIT)/(G NBC WT IRBIT – G NBC No IRBIT). D and E, expression and abundance of NBCe1‐B in surface (D) and total (E) fractions of Xenopus oocytes relative to control ‘No IRBIT’ (i.e. BWT only). F, expression of IRBIT in total fraction of Xenopus oocytes relative to wild‐type (WT) IRBIT. G, summary of results of a yeast two‐hybrid assay for protein interaction between NtB and IRBIT variants. The numerals on the bars indicate the number of individual oocytes. Panels D−F each represent the average of three experiments. Green bars: without IRBIT. Orange bars: with IRBIT. $Not significantly different from the control ‘No IRBIT’. *Bars significantly different from the control ‘No IRBIT’. #Bars significantly different from wild‐type IRBIT. NS: bars in this group are not significantly different from each other. One‐way ANOVA followed post hoc Tukey's comparison was performed for statistical analysis. The number of ‘+’ signs in panel G indicates the relative strength of protein interaction. ‘−’ signs indicate no detectable protein interaction by yeast two‐hybrid assay. The strength of protein interaction between NtB fragments and IRBIT was judged based on the number and colour‐intensity of blue colonies on the quadruple‐dropout (QDO) medium. Numerical values for all bars and detailed results of statistical tests are provided as Supporting information in Statistical Summary Tables 7C−7F.
Here, we generated IRBIT mutants by disturbing the acidic residues Asp83, Asp86, Asp87 and Glu88 plus the potential phosphorylation sites Thr82, Ser84, Ser85 and Ser90 in the Ct half of the PEST domain (Fig. 7 B). Compared to wild‐type IRBIT, the compound mutation in NNNQ and AAAA greatly reduces by about 75−80%, whereas the single mutation in T82A, S84A and S85A reduces by about 30−60%, the stimulatory effect of IRBIT on NBCe1‐B activity (Fig. 7 C). Combination mutation to the acidic and the Ser/Thr residues in construct AAAA‐NNNQ completely abolishes the stimulatory effect of IRBIT on NBCe1‐B. We generated mutant DDDD by replacing Thr82, Ser84, Ser85 and Ser90 all with Asp to mimic phosphorylation. This DDDD remains almost as powerful as wild‐type IRBIT at activating NBCe1‐B. Finally, replacing an acidic‐residue cluster in the AHCY domain (construct ‘QQNNAHCY’ with 203ESEDD→QSQNN in the AHCY domain) has no effect on the stimulatory effect of IRBIT.
Both the surface (Fig. 7 D) and the total expression levels (Fig. 7 E) are similar for NBCe1‐B co‐expressed with different IRBIT variants. The protein expression levels are similar for all IRBIT variants (Fig. 7 F). The protein expression data rule out the possibility that, compared to wild‐type IRBIT, the decreased stimulatory effect of specific IRBIT mutants on NBCe1‐B is due to a decrease in the surface expression of NBCe1‐B or a decrease in the expression of IRBIT proteins in oocytes.
Finally, we examined by yeast two‐hybrid assay the protein interaction of NtB with different IRBIT variants. As summarized in Fig. 7 G, wild‐type IRBIT, DDDD, T82A, S84A, and S85A all elicit strong protein interaction with NtB, consistent with the functional data in Fig. 7 C. However, NNNQ, AAAA and AAAA‐NNNQ elicit no detectable interaction with NtB by yeast two‐hybrid assay. Note that, the activities of NBCe1‐B in the presence of NNNQ and AAAA are slightly higher than the control without IRBIT, indicating a modest stimulatory effect of these IRBIT variants on NBCe1‐B. The apparent discrepancy between the yeast two‐hybrid data and the functional data presumably reflects differential sensitivities of the two approaches to detecting protein interaction.
Taken together, the above data indicate that the acidic residues as well as the four Thr/Ser residues in the PEST domain play an important role in the interaction of IRBIT with NBCe1‐B. It is likely that the phosphorylation of at least some of Thr82, Ser84, Ser85 and Ser90 is necessary for the binding of IRBIT to NBCe1‐B. Our data suggest that NBCe1‐B activation by IRBIT requires the negative charges attributed by the acidic residues and the presumably phosphorylated Ser/Thr residues in the PEST domain.
Discussion
Structural consideration of the unique Nt of NBCe1‐B
In proteins, intrinsically disordered regions (IDRs) are usually rich in uncharged polar or charged residues, and thus do not maintain a stable globular conformation due to a lack of long‐range hydrophobic interactions. IDRs are widely involved in protein‐protein interaction, protein‐complex assembly, and protein‐RNA interaction (Habchi et al. 2014; Babu, 2016). The SLC4 transporters are believed to function as a dimer based upon structural studies on the Nt and the transmembrane domains (Zhang et al. 2000; Kao et al. 2008; Shnitsar et al. 2013; Chang et al. 2014; Arakawa et al. 2015; Alvadia et al. 2017; Huynh et al. 2018). Based on the crystal structure of AE1 Nt (Zhang et al. 2000), a structural modelling study shows that the conserved Ntcore of NBCe1 is well folded with a typical α + β fold containing central anti‐parallel β‐sheets surrounded by two layers of α helices (Chang et al. 2008). However, the NtB of NBCe1‐B is likely to be intrinsically disordered, as supported by the following arguments.
Firstly, NtB is highly rich in polar and charged residues. In the total of 85 residues of NtB, 18 are acidic (12 Glu + 6 Asp) and would be negatively charged at physiological pH, and 24 are alkaline (10 Lys + 8 Arg + 6 His) and therefore could be positively charged. In addition, NtB contains 13 uncharged polar residues (Ser, Thr and Asn). The remaining 22 hydrophobic residues are scattered in the hydrophilic strings. Thus, it is unlikely that NtB forms a hydrophobic core essential for a well‐folded globular structure.
Secondly, in the present study, no protein expression could be detected for the isolated NtB in Xenopus oocytes, suggesting that NtB is sensitive to protease degradation, consistent with the notion that NtB is intrinsically disordered.
Thirdly, deleting the Nt half of NtB (Lee et al. 2012) or replacing the Ct half with GS linkers (present study) retains considerable or even full auto‐inhibition, suggesting that the structure of NtB is highly dynamic, consistent with the notion that NtB is intrinsically disordered.
Figure 8 A shows the sequence of NtB. In view of the distribution of charged residues, Arm N (residues 1–24) containing two acidic motifs ‘EDE’ and ‘DEEEVE’ would be highly negatively charged at physiological pH. Region 25−39 would be slightly positively charged. Arm P (residues 40−52) containing the ‘basic cluster’ would be highly positively charged. Finally, Arm M (residues 53−85) would be mixed with positive and negative charges.
Figure 8. Proposed mechanisms for the auto‐inhibition and activation of NBCe1‐B by IRBIT and high‐activity of NBCe1‐A.
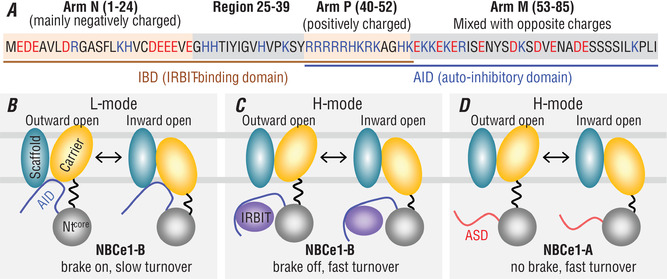
A, functional modules of unique Nt of NBCe1‐B. IRBIT‐binding domain (IBD) consists of structural elements in the Nt half. The two acidic motifs ‘EDE’ and ‘DEEEVE’ in negatively charged Arm N and the ‘basic cluster’ in positively charged Arm P are essential for IRBIT binding. Region 25−39 also contains elements important for IRBIT binding. The auto‐inhibitory domain (AID) consists of elements in the Ct half including Arm P and Arm M (mixed with opposite charges). Red indicates residues that could be negatively charged, whereas blue indicates residues that could be positively charged. B, NBCe1‐B functions in low activity mode (L‐mode) in the absence of IRBIT. Here, the AID functions as a brake module by binding to the MTD of NBCe1‐B, and thus slows down the turnover rate of the transporter. In the TMD, both the scaffold domain and the carrier domain presumably contain binding sites for the AID. C, NBCe1‐B functions in high activity mode (H‐mode) in the presence of IRBIT. The binding of IRBIT to IBD releases the brake from the TMD, resulting in full turnover rate of the transporter. D, NBCe1‐A constitutively functions in H‐mode due to lacking a brake module.
Arms P and M, but not N, are responsible for auto‐inhibition
Arm N
Perturbing motif ‘EDE’ (i.e. ΔN4, Fig. 6 in Lee et al. 2012); Ala‐substitution in Fig. 3 C, present study) or motif ‘DEEEVEE’ (Fig. 3 E, present study) has little effect on NBCe1‐B auto‐inhibition inasmuch as the basal activity of the mutants without IRBIT is not considerably different from that of BWT. Moreover, in the background BWT, deleting the first 16 residues has little effect on NBCe1‐B basal activity (construct ΔN16, Fig. 6 in Lee et al. 2012). We conclude that Arm N is not essential for NBCe1‐B auto‐inhibition.
Arm P
Arm P is a key component of the AID. Firstly, deleting region 40−62 (largely corresponding to Arm P) completely abolishes NBCe1‐B auto‐inhibition in HeLa cells (Shcheynikov et al. 2015). Secondly, Ala‐substitution or deletion of the 9 alkaline residues in Arm P completely abolishes NBCe1‐B auto‐inhibition in Xenopus oocytes (Lee & Boron, 2018). Here, we show that Arm P alone decreases NBCe1 activity by ∼60% (construct BRC‐BΔN85 in Fig. 4 C).
We propose that Arm P accounts for one‐half of auto‐inhibition in BWT. The potency of Arm P to inhibit NBCe1 activity depends on the number of positive‐charge residues in Arm P. Firstly, in the previous study (Fig. 5C in Lee & Boron, 2018), while deleting region 40−48 abolishes NBCe1‐B auto‐inhibition, restoring Arg progressively increases the magnitude of auto‐inhibition. Secondly, in the present study, the auto‐inhibition magnitude of NBCe1 with mutations to the ‘basic cluster’ increases in the following order: BSub‐AID2 (9 His in the ‘basic cluster’) < BSub‐AID1 (8 Lys + 1 His) < BWT (6 Arg + 2 Lys +1 His) < BSuper‐AID (11 Arg) (Fig. 4 G). Thirdly, we note that, similar to our data for BSuper‐AID, in the previous study (Construct ‘9R’ in Fig. 5D in Lee & Boron, 2018), replacing the native ‘basic cluster’ with 9 Arg virtually eliminates the basal activity of NBCe1, indicating super auto‐inhibition in construct ‘9R’.
Arm M
Arm M is also an important component of the AID. Firstly, in the background of BWT, deleting region 1−59 eliminates just ∼50% of auto‐inhibition (construct ΔN59, Fig. 6 in Lee et al. 2012), suggesting that region 60−85 contains elements of AID. Secondly, in the present study, the magnitude of auto‐inhibition of IBDmin2‐BΔN85 containing both Arm P and Arm M is much greater than that of IBDmin1‐BΔN85 containing just Arm P. We propose that Arm M accounts for the other half of auto‐inhibition in BWT.
Taking together, we conclude that the elements of AID are predominantly distributed in Arm P and Arm M of NBCe1‐B. In BWT, these two arms presumably each contribute about half of the auto‐inhibition effect. What is unusual is that, in the background of BWT, removing just Arm P (Δ940‐48, Fig. 3D in Lee & Boron, 2018) or Arm P plus the very initial part of Arm M (Δ40−62, Fig. 2B in Shcheynikov et al. 2015) completely abolishes the auto‐inhibition. We will address this apparent inconsistency below in the section on ‘Potential local interactions in NtB affecting AID binding to TMD’.
Arm N, region 25−39, and Arm P, but not Arm M, are responsible for IRBIT binding
Arm N: EDE
A previous yeast two‐hybrid study showed that region 1−19 of NtB contains structural elements essential for IRBIT binding (Shirakabe et al. 2006). Here, our yeast two‐hybrid data (Fig. 3 B) show that region 1−48 is sufficient for IRBIT binding. Moreover, our yeast two‐hybrid study shows that the initial acidic ‘EDE’ motif is essential for IRBIT binding, consistent with the functional studies showing that deleting ‘EDE’ motif (construct ‘ΔN4’, Fig. 6 in (Lee et al. 2012) or replacing with Ala (Fig. 3 C, present study) greatly reduces NBCe1‐B stimulation by IRBIT.
Arm N: DEEEVE
Here, we show for the first time by yeast two‐hybrid and functional studies (Fig. 3) that motif ‘DEEEVE’ is essential for IRBIT binding.
Region 25−39
Region 25−39 between Arm N and Arm P probably also contains a structural component important for IRBIT interaction. Supporting this hypothesis are the previous observations that construct Δ1228‐39 with region 28−39 deleted exhibits very low basal activity—indicating the presence of full auto‐inhibition as in BWT, but cannot be stimulated by IRBIT (Fig. 3 in Lee & Boron, 2018). In the section on ‘Explanation for no stimulation of Δ1228‐39 by IRBIT’ below, we will address why Δ1228‐39 is insensitive to IRBIT.
Arm P
The ‘basic cluster’ (‘RRRRRHKRK’) represents the core element of Arm P. Previous studies using a yeast two‐hybrid assay (Shirakabe et al. 2006) and co‐immunoprecipitation (Shcheynikov et al. 2015) showed that region 37−62 of NtB contains structural elements essential for IRBIT binding. Our yeast two‐hybrid study shows that Arm P accounts for this essentiality (Fig. 3 B).
Arm M
In the present study, constructs that retain Arm N and Arm P but lack Arm M (e.g. N50B‐BΔN85, Fig. 4 B) can be fully stimulated by IRBIT, demonstrating that Arm M is not essential for IRBIT‐binding.
Taking together, we conclude that the IBD predominantly consists of elements in Arm N and Arm P of NBCe1‐B. Motifs ‘EDE’ and ‘DEEEVE’ in Arm N, as well as the ‘basic cluster’ in Arm P, the three essential components of IBD, probably represent the minimal structural requirement for IRBIT to activate NBCe1, given that construct ‘IBDmin1‐BΔN85’ can be fully activated by IRBIT (Fig. 4 D).
Binding site for NBCe1‐B in IRBIT
A previous study using pull‐down showed that neither the Nt nor the AHCY domain of IRBIT alone can bind to NBCe1‐B, suggesting that both the Nt and AHCY domains contain elements essential for NBCe1‐B binding (Shirakabe et al. 2006). Our data (Fig. 7) indicate that the negative charge in the PEST domain is a key factor determining the interaction between IRBIT and NBCe1‐B. We propose that the PEST domain is the binding site for the positively charged Arm P in the IBD of NBCe1‐B. The binding site for the negatively charged Arm N is probably located in the AHCY domain of IRBIT. This hypothesis requires testing in future studies.
Proposed model for molecular mechanism underlying NBCe1‐B auto‐inhibition and its stimulation by IRBIT
Membrane transporters employ an alternating‐access mechanism to translocate substrate across the plasma membrane (Jardetzky, 1966; Drew & Boudker, 2016). Three different alternating‐access models have emerged: rocker switch, rocking bundle and elevator mechanism (Drew & Boudker, 2016). The members of the SLC4, SLC23 and SLC26 families share a very similar overall fold for their TMD (Lu et al. 2011; Arakawa et al. 2015; Geertsma et al. 2015; Thurtle‐Schmidt & Stroud, 2016; Chang & Geertsma, 2017; Huynh et al. 2018; Wang et al. 2019). The TMD of these transporters comprises 14 transmembrane α helices (TMs), forming a carrier domain (aka core domain) and a scaffold domain (aka gate domain). Evidences from structural studies suggest that these transporters probably employ an elevator‐like mechanism for ion translocation (Thurtle‐Schmidt & Stroud, 2016; Ficici et al. 2017; Yu et al. 2017; Chang et al. 2019; Wang et al. 2019).
According to the alternating‐access mechanism, a complete transport cycle includes three steps. First, the substrate binds to the transporter. Then the transporter must undergo a global conformation transition from the outward‐open state to the inward‐open one or vice versus. Finally, the substrate releases from the transporter. In theory, any of these three steps could be rate‐limiting for the transporter. It has been shown that neither the Na+ affinity nor the HCO3 − affinity of NBCe1‐A is considerably different from those of NBCe1‐B (Sciortino & Romero, 1999; McAlear et al. 2006). Thus, the difference in the activities of NBCe1‐A vs. non‐activated NBCe1‐B cannot be accounted for by substrate binding. Could it be accounted for by the global‐conformation‐transition rate and/or substrate releasing?
In a previous study (Lee & Boron, 2018), the authors assumed that the full activity of NBCe1 requires an interaction between Ntcore (the so‐called ‘gNt’) and TMD of NBCe1. To explain the auto‐inhibition of NBCe1‐B, the authors proposed that the putative interaction between Ntcore and the TMD is interrupted either by electrostatic repulsion between the positively charged Arm P (the so‐called ‘cationic cluster’) and a positively charged area in TMD (model A), or by electrostatic attraction between the positively charged Arm P and the negatively charged plasma membrane (model B).
The above model A appears inconsistent with the observations in the present study that mutating the negatively charged residues in the TMD decreases the magnitude of the auto‐inhibition of NBCe1‐B, e.g. specific constructs in Figs 5 and 6.
The above model B also appears inconsistent with the following two observations. Firstly, the AID at the Ct end inhibits the activity of NBCe1, i.e. constructs A‐lnk20‐N85B, A‐lnk20‐N106B and A‐lnk40‐N106B in the present study (Fig. 2). In these cases, the AID, if interacting with the plasma membrane, would have no direct effect on the assumed interaction between the Ntcore and the TMD. One might argue that the Ct domain of NBCe1 might interact with the Ntcore. Thus, the Ct‐attached AID, when interacting with the plasma membrane, could pull Ntcore off the TMD indirectly. If this is the case, then increasing the length of the linker between the Ct of NBCe1‐A and the AID would probably attenuate the pulling to the Ntcore, thus decreasing the magnitude of NBCe1 auto‐inhibition. However, a very similar magnitude of auto‐inhibition was observed for A‐lnk20‐N85B, A‐lnk20‐N106B and A‐lnk40‐N106B, although the linker in the last construct is twice as long as that in the other two. Secondly, just the region 60−85 of NtB, which would be largely negatively charged, inhibits NBCe1 activity by ∼50% (Lee et al. 2012).
We propose that the AID binds to the TMD of NBCe1‐B, blocking the global conformation turnover of TMD during the transport cycle, resulting in the low activity of NBCe1‐B (L‐mode, Fig. 8 B). The binding of the AID to the TMD is presumably mainly mediated by electrostatic attraction. IRBIT competitively binds to the IBD via electrostatic attraction – note the overlap of IBD and AID in Arm P, releasing the AID from the TMD, resulting in activation of NBCe1‐B (H‐mode, Fig. 8 C). Thus, the AID acts as a brake module to tune down the transport rate of NBCe1‐B. NBCe1‐A lacks the brake, and thus is constitutively active (Fig. 8 D).
The TMD of NBCe1 presumably contains the binding sites for both Arm P and Arm M in the NtB. The binding surface for the positively charged Arm P could be contributed by negatively charged acidic residues and/or aromatic residues that could form cation‐π interaction with positively charged residues. Consistent with this idea, manipulating a set of residues, including D853, E858, and E860 in intracellular loop IL5 of the carrier domain, as well as D924, D938, and F939 in IL6 of the scaffold domain, could increase by ∼3‐fold the basal activity of NBCe1, while having little effect on the IRBIT‐activated activity (e.g. construct ‘NQQNNA’, Fig. 6 A). As shown in the 3‐dimensional structure model of NBCe1 (Fig. 5 C), these residues are well lined up along IL5 and IL6 in a large cleft between the carrier domain and the scaffold domain. We propose that the flexible positively charged region of the AID bends into the cleft and binds to IL5 and IL6, therefore blocking the movement of the carrier domain relative to the scaffold domain, and thus decreasing the rate of global confirmation‐turnover of the transporter.
Note that it is probably not possible to completely eliminate the auto‐inhibition by mutating residues in the TMD, because some of AID‐binding sites in the TMD might also be essential for the transport process per se. For example, such residues might directly contribute to or indirectly affect the formation of the substrate‐binding pocket of NBCe1. Disturbing such residues might inactivate the transporter. In our present study, replacing Glu866 plus Glu875 with Gln in the background of ‘NQQNNA’ completely inactivates NBCe1 (data not shown).
It is interesting that, although the isolated IBD in the fusion protein Myc‐N106‐EGFP could completely eliminate the stimulatory effect of IRBIT on NBCe1‐B (Wang et al. 2020 b), the isolated AID in this fusion protein has no inhibitory effect on NBCe1 activity (Fig. 2 D). This last observation indicates that the AID must be covalently attached to NBCe1 to act as a brake to inhibit NBCe1 activity. It appears that the AID and the TMD must be tethered in close proximity to enable their interaction.
Finally, in the present study, we cannot rule out the possibility that the low activity of NBCe1‐B could be due to an altered rate of substrate release compared to NBCe1 with full activity, e.g. NBCe1‐A or IRBIT‐activated NBCe1‐B. The substrate‐releasing step during a transport cycle could indeed be rate‐limiting, as reported for the serotonin transporter (Hasenhuetl et al. 2016).
Explanation for no stimulation of IBDmin2‐BΔN85 by IRBIT
IBDmin2‐BΔN85 contains a nearly full AID, i.e. Arm P and Arm M. Not surprisingly, IBDmin2‐BΔN85 retains almost full auto‐inhibition (Fig. 4 D). However, IBDmin2‐BΔN85 retains a minimized IBD comprising of just the three essential elements for IRBIT‐binding. Therefore, the strength of IRBIT interaction with IBDmin2‐BΔN85 should be greatly attenuated compared to its interaction with BWT. This attenuated interaction is probably not powerful enough to disrupt the strong interaction of the full AID with TMD. Thus, IBDmin2‐BΔN85 cannot be stimulated by IRBIT.
IBDmin1‐BΔN85 contains the same mini IBD as IBDmin2‐BΔN85. However, IBDmin1‐BΔN85 contains a reduced AID comprising just Arm P. Thus, the interaction of the reduced AID with TMD should be attenuated. Consistently, the basal activity of IBDmin1‐BΔN85 in the absence of IRBIT is greatly increased compared to BWT (Fig. 4 D). In turn, the interaction of IRBIT with the mini IBD should be powerful enough to disrupt the interaction of the reduced AID with TMD.
Explanation for no stimulation of Δ1228‐39 by IRBIT
Similar to the case of IBDmin2‐BΔN85 in the present study, construct Δ1228‐39 in the previous study (Fig. 3 in Lee & Boron, 2018) contains a reduced IBD due by lacking the sequence between Arm N and Arm P, and a nearly full AID. Not surprisingly, Δ1228‐39 retains full auto‐inhibition but cannot be activated by IRBIT, because the attenuated interaction between the reduced IBD of Δ1228‐39 and IRBIT is not be powerful enough to disrupt the strong interaction between the full AID and TMD.
Potential local interactions in NtB affecting AID binding to TMD
As discussed above, the NtB is likely to be intrinsically disordered and lacks stable tertiary structure. However, it is possible that different elements in the NtB can locally interact with each other. Such local interaction might affect the interaction of the AID with TMD, therefore modulating the magnitude of NBCe1 auto‐inhibition.
Note that the basal activity of NQQQVQ is lower by ∼50% than that of BWT, suggesting super auto‐inhibition in NQQQVQ (Fig. 3 D). Similarly, BRC‐BΔN85 elicits super auto‐inhibition compared to IBDmin1‐BΔN85 (Fig. 4 C and D). Compared to BWT, mutant NQQQVQ loses five negative charges in Arm N. Compared to IBDmin1‐BΔN85, BRC‐BΔN85 lacks the two negatively charged motifs in Arm N. We presume that the negatively charged Arm N somehow interacts with the positively charged Arm P, thereby imposing an effect on the binding of AID to the TMD. Reducing the number of negative charges in Arm N might abolish this putative interaction, thereby enhancing the interaction between Arm P and the TMD, resulting in super auto‐inhibition.
As we have concluded above, Arm M contributes about 50% of auto‐inhibition in NBCe1‐B. One would expect that deleting elements in Arm M would attenuate the magnitude of auto‐inhibition of NBCe1. Indeed, in the background of BWT, removing the carboxyl‐terminal portion of Arm M attenuates the auto‐inhibition in N80B‐BΔN85, N75B‐BΔN85 and N65B‐BΔN85 compared to BWT. Surprisingly, removing the entire Arm M restores full auto‐inhibition in N50B‐BΔN85 (Fig. 4 B). Even more surprisingly, in the background of BWT, manipulating the amino‐terminal portion of Arm M leads to super auto‐inhibition in 14NA49‐62 and Δ1449‐62 (Fig. 6 D, Lee & Boron, 2018). We presume that Arm M locally interacts with other structural elements, e.g. Arm N and/or Arm P. The variation in the auto‐inhibition magnitude of the above variants with Arm M modified could reflect different patterns of interaction among elements in NtB.
Finally, in the background of BWT, removing just Arm P (Δ940‐48, Fig. 3D in Lee & Boron, 2018) or Arm P plus the very initial portion of Arm M (Δ40−62, Fig. 2B, in Shcheynikov et al. 2015) completely abolishes the auto‐inhibition. We presume that, in the absence of Arm P, Arm M interacts with a specific structural element in NtB, e.g. Arm N, thereby neutralizing the inhibitory effect of Arm M on NBCe1.
The structure of NtB would be highly dynamic under physiological conditions. The hypothetical interaction among elements in NtB, if any, might be very complicated and highly dynamic. The above hypotheses about the local interactions in NtB require testing in future studies with approaches capable of monitoring the structural dynamics of NtB.
Consideration of the role of cassette I in NBCe1
Due to the ASD effect vs. the AID effect, the activity of NBCe1‐A is several fold greater than those of NBCe1‐B and ‐C (McAlear et al. 2006; Lee et al. 2012). Here, we show that the same is true for NBCe1‐D vs. NBCe1‐E, both lacking cassette I. Moreover, two comparisons (NBCe1‐D vs. ‐A and NBCe1‐E + IRBIT vs. ‐B + IRBIT, Fig. 1 D) indicate that cassette I imposes a modest inhibition on the activity of NBCe1 in the high‐activity mode.
The mechanism whereby cassette I affects NBCe1 activity remains to be investigated. In the 3‐dimension model of NBCe1 Ntcore (Fig. 1 B), cassette I (‘RMFSNPDNG’, from 236 to 244’) is located in an intrinsically disordered loop that is rich in Ser and Thr. As examined in HEK293, a likely phosphorylation of Ser232/Ser233/Ser235 right before cassette I switches NBCe1‐B from high‐activity state to low‐activity state (Vachel et al. 2018). Note that cassette I contains a potential phosphorylation site. It is possible that optional inclusion of cassette I alters the phosphorylation of the loop structure in the Ntcore.
Concluding remarks
The AID of NBCe1‐B is mainly defined by elements from residues 40−85 of the Nt, with the ‘basic cluster’ being a central component. The IBD of NBCe1‐B for IRBIT binding is mainly defined by elements in the region 1−52, with the acidic motifs ‘EDE’ and ‘DEEEVE’ and the ‘basic cluster’ being the three essential components. The AID acts as a brake module to bind to the TMD via electrostatic attraction, and therefore slows down the global turnover rate of NBCe1‐B. IRBIT activates NBCe1‐B by releasing the brake from the TMD by competitively binding to the IBD via electrostatic attraction. The balance between the two interactions, i.e. the one between the AID and TMD, the other between the IBD and IRBIT, determines the activity state of NBCe1‐B.
Our findings have general implications in a broad physiological context given the wide distribution of NBCe1‐B. For example, in the resting condition in exocrine gland tissues such as the pancreatic duct, the parotid gland, or the submandibular gland, NBCe1‐B would be inhibited with the brake on. Upon stimulation triggered by hormonal and/or vagus nerve signals, NBCe1‐B would be activated due to the release of the brake by the interaction with IRBIT. The findings from our study of NBCe1‐B also provide insight into the molecular mechanism underlying the regulation of NBCe1‐C. Both NBCe1‐C and IRBIT are highly expressed in the central nervous system. Here, the activity of NBCe1‐C might be highly regulated. Finally, our findings also shed light on understanding the molecular mechanisms underlying the activation of other membrane transporters such as NBCn1, NBCn2, and NDCBE and presumably the regulation of membrane channels by IRBIT.
Additional information
Competing interests
The authors declare that no competing interests exist.
Authors contributions
P.S., Y.L. and L.‐M.C designed the study. P.S., H.W., M.W. and L.C. performed the experiments and data collection. P.S., Y.L. and L.‐M.C. analysed the data and wrote the manuscript. All authors have approved the final version of the manuscript and agree to be accountable for all aspects of the work. All persons designated as authors qualify for authorship, and all those who qualify for authorship are listed.
Funding
This work was supported by NSFC grant nos 31771294 (L.‐M.C.), 31571201 (Y.L.), and 81571388 (L.‐M.C.), and a grant to L.‐M.C. from Research Centre for Innovative Education and Critical Thinking at HUST (grant no. 2019CT008).
Supporting information
Statistical Summary Document
Acknowledgements
We thank the reviewers for their careful reading of the manuscript and critical comments which helped us greatly to improve the manuscript.
Biography
Pan Su received his BSc degree in Biological Technology from Anyang Institute of Technology, Henan, China, in 2014. He then joined the School of Life Science and Technology at Huazhong University of Science and Technology, Wuhan, China, in 2014. He is now a PhD candidate, expecting his PhD degree in 2020. He is broadly interested in the physiology, biophysics and pathology of acid‐base transporters. His PhD study focuses on the structure‐function relationship and functional regulation of the electrogenic Na+/HCO3 − cotransporter NBCe1 of SLC4 family.

Edited by: Peying Fong & Rajini Rao
Contributor Information
Ying Liu, Email: liuying@hust.edu.cn.
Li‐Ming Chen, Email: liming.chen@hust.edu.cn.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- Abuladze N, Lee I, Newman D, Hwang J, Boorer K, Pushkin A & Kurtz I (1998). Molecular cloning, chromosomal localization, tissue distribution, and functional expression of the human pancreatic sodium bicarbonate cotransporter. J Biol Chem 273, 17689–17695. [DOI] [PubMed] [Google Scholar]
- Alvadia CM, Sommer T, Bjerregaard‐Andersen K, Damkier HH, Montrasio M, Aalkjaer C & Morth JP (2017). The crystal structure of the regulatory domain of the human sodium‐driven chloride/bicarbonate exchanger. Sci Rep 7, 12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando H, Mizutani A, Kiefer H, Tsuzurugi D, Michikawa T & Mikoshiba K (2006). IRBIT suppresses IP3 receptor activity by competing with IP3 for the common binding site on the IP3 receptor. Mol Cell 22, 795–806. [DOI] [PubMed] [Google Scholar]
- Ando H, Mizutani A, Matsu‐ura T & Mikoshiba K (2003). IRBIT, a novel inositol 1,4,5‐trisphosphate (IP3) receptor‐binding protein, is released from the IP3 receptor upon IP3 binding to the receptor. J Biol Chem 278, 10602–10612. [DOI] [PubMed] [Google Scholar]
- Arakawa T, Kobayashi‐Yurugi T, Alguel Y, Iwanari H, Hatae H, Iwata M, Abe Y, Hino T, Ikeda‐Suno C, Kuma H, Kang D, Murata T, Hamakubo T, Cameron AD, Kobayashi T, Hamasaki N & Iwata S (2015). Crystal structure of the anion exchanger domain of human erythrocyte band 3. Science 350, 680–684. [DOI] [PubMed] [Google Scholar]
- Arnaoutov A & Dasso M (2014). Enzyme regulation. IRBIT is a novel regulator of ribonucleotide reductase in higher eukaryotes. Science 345, 1512–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu MM (2016). The contribution of intrinsically disordered regions to protein function, cellular complexity, and human disease. Biochem Soc Trans 44, 1185–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevensee MO, Schmitt BM, Choi I, Romero MF & Boron WF (2000). An electrogenic Na/HCO3 cotransporter (NBC) with a novel C terminus, cloned from rat brain. Am J Physiol Cell Physiol 278, C1200‐C1211. [DOI] [PubMed] [Google Scholar]
- Bonneau B, Ando H, Kawaai K, Hirose M, Takahashi‐Iwanaga H & Mikoshiba K (2016). IRBIT controls apoptosis by interacting with the Bcl‐2 homolog, Bcl2l10, and by promoting ER‐mitochondria contact. eLife 5, e19896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boron WF & Boulpaep EL (1983). Intracellular pH regulation in the renal proximal tubule of the salamander: basolateral HCO3 − transport. J Gen Physiol 81, 53–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MH, Chen AP & Romero MF (2014). NBCe1A dimer assemble visualized by bimolecular fluorescence complementation (BiFC). Am J Physiol Renal Physiol 306, F672–F680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MH, DiPiero J, Sonnichsen FD & Romero MF (2008). Entry to “HCO3 tunnel” revealed by SLC4A4 human mutation and structural model. J Biol Chem 283, 18402–18410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YN & Geertsma ER (2017). The novel class of seven transmembrane segment inverted repeat carriers. Biol Chem 398, 165–174. [DOI] [PubMed] [Google Scholar]
- Chang YN, Jaumann EA, Reichel K, Hartmann J, Oliver D, Hummer G, Joseph B & Geertsma ER (2019). Structural basis for functional interactions in dimers of SLC26 transporters. Nat Commun 10, 2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS & Richardson DC (2010). MolProbity: all‐atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallog 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi I, Hu L, Rojas JD, Schmitt BM & Boron WF (2003). Role of glycosylation in the renal electrogenic Na+‐HCO3 − cotransporter (NBCe1). Am J Physiol Renal Physiol 284, F1199–F1206. [DOI] [PubMed] [Google Scholar]
- Choi I, Romero MF, Khandoudi N, Bril A & Boron WF (1999). Cloning and characterization of a human electrogenic Na+‐HCO3 − cotransporter isoform (hhNBC). Am J Physiol 276, C576–C584. [DOI] [PubMed] [Google Scholar]
- Cooper BJ, Key B, Carter A, Angel NZ, Hart DN & Kato M (2006). Suppression and overexpression of adenosylhomocysteine hydrolase‐like protein 1 (AHCYL1) influences zebrafish embryo development: a possible role for AHCYL1 in inositol phospholipid signaling. J Biol Chem 281, 22471–22484. [DOI] [PubMed] [Google Scholar]
- Devogelaere B, Sammels E & De Smedt H (2008). The IRBIT domain adds new functions to the AHCY family. BioEssays 30, 642–652. [DOI] [PubMed] [Google Scholar]
- Drew D & Boudker O (2016). Shared molecular mechanisms of membrane transporters. Ann Rev Biochem 85, 543–572. [DOI] [PubMed] [Google Scholar]
- Ficici E, Faraldo‐Gomez JD, Jennings ML & Forrest LR (2017). Asymmetry of inverted‐topology repeats in the AE1 anion exchanger suggests an elevator‐like mechanism. J Gen Physiol 149, 1149–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geertsma ER, Chang YN, Shaik FR, Neldner Y, Pardon E, Steyaert J & Dutzler R (2015). Structure of a prokaryotic fumarate transporter reveals the architecture of the SLC26 family. Nat Struct Mol Biol 22, 803–808. [DOI] [PubMed] [Google Scholar]
- Grundy D (2015). Principles and standards for reporting animal experiments in The Journal of Physiology and Experimental Physiology . J Physiol 593, 2547–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habchi J, Tompa P, Longhi S & Uversky VN (2014). Introducing protein intrinsic disorder. Chem Rev 114, 6561–6588. [DOI] [PubMed] [Google Scholar]
- Hasenhuetl PS, Freissmuth M & Sandtner W (2016). Electrogenic binding of intracellular cations defines a kinetic decision point in the transport cycle of the human serotonin transporter. J Biol Chem 291, 25864–25876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P, Klein J & Yun CC (2010). Activation of Na+/H+ exchanger NHE3 by angiotensin II is mediated by inositol 1,4,5‐triphosphate (IP3) receptor‐binding protein released with IP3 (IRBIT) and Ca2+/calmodulin‐dependent protein kinase II. J Biol Chem 285, 27869–27878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P, Zhang H & Yun CC (2008). IRBIT, inositol 1,4,5‐triphosphate (IP3) receptor‐binding protein released with IP3, binds Na+/H+ exchanger NHE3 and activates NHE3 activity in response to calcium. J Biol Chem 283, 33544–33553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JH, Yang D, Shcheynikov N, Ohana E, Shin DM & Muallem S (2013). Convergence of IRBIT, phosphatidylinositol (4,5) bisphosphate, and WNK/SPAK kinases in regulation of the Na+‐HCO3 − cotransporters family. Proc Natl Acad Sci USA 110, 4105–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh KW, Jiang J, Abuladze N, Tsirulnikov K, Kao L, Shao X, Newman D, Azimov R, Pushkin A, Zhou ZH & Kurtz I (2018). CryoEM structure of the human SLC4A4 sodium‐coupled acid‐base transporter NBCe1. Nat Commun 9, 900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi T, Inatomi J, Sekine T, Cha SH, Kanai Y, Kunimi M, Tsukamoto K, Satoh H, Shimadzu M, Tozawa F, Mori T, Shiobara M, Seki G & Endou H (1999). Mutations in SLC4A4 cause permanent isolated proximal renal tubular acidosis with ocular abnormalities. Nat Genet 23, 264–266. [DOI] [PubMed] [Google Scholar]
- Jalali R, Guo J, Zandieh‐Doulabi B, Bervoets TJM, Paine ML, Boron WF, Parker MD, Bijvelds MJC, Medina JF, DenBesten PK & Bronckers ALJJ (2014). NBCe1 (SLC4A4) a potential pH regulator in enamel organ cells during enamel development in the mouse. Cell Tissue Res 358, 433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardetzky O (1966). Simple allosteric model for membrane pumps. Nature 211, 969–970. [DOI] [PubMed] [Google Scholar]
- Jeong W, Kim J, Ahn SE, Lee SI, Bazer FW, Han JY & Song G (2012). AHCYL1 is mediated by estrogen‐induced ERK1/2 MAPK cell signaling and microRNA regulation to effect functional aspects of the avian oviduct. PLoS One 7, e49204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao L, Sassani P, Azimov R, Pushkin A, Abuladze N, Peti‐Peterdi J, Liu W, Newman D & Kurtz I (2008). Oligomeric structure and minimal functional unit of the electrogenic sodium bicarbonate cotransporter NBCe1‐A. J Biol Chem 283, 26782–26794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaai K, Ando H, Satoh N, Yamada H, Ogawa N, Hirose M, Mizutani A, Bonneau B, Seki G & Mikoshiba K (2017). Splicing variation of Long‐IRBIT determines the target selectivity of IRBIT family proteins. Proc Natl Acad Sci U S A 114, 3921–3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaai K, Mizutani A, Shoji H, Ogawa N, Ebisui E, Kuroda Y, Wakana S, Miyakawa T, Hisatsune C & Mikoshiba K (2015). IRBIT regulates CaMKIIalpha activity and contributes to catecholamine homeostasis through tyrosine hydroxylase phosphorylation. Proc Natl Acad Sci USA 112, 5515–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy CR, Lin S & Jacobsen EN (2016). The cation‐π interaction in small‐molecule catalysis. Angew Chem 55, 12596–12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khamaysi A, Anbtawee‐Jomaa S, Fremder M, Eini‐Rider H, Shimshilashvili L, Aharon S, Aizenshtein E, Shlomi T, Noguchi A, Springer D, Moe OW, Shcheynikov N, Muallem S & Ohana E (2019). Systemic succinate homeostasis and local succinate signaling affect blood pressure and modify risks for calcium oxalate lithogenesis. J Am Soc Nephrol 30, 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer H, Mizutani A, Iemura S, Natsume T, Ando H, Kuroda Y & Mikoshiba K (2009). Inositol 1,4,5‐triphosphate receptor‐binding protein released with inositol 1,4,5‐triphosphate (IRBIT) associates with components of the mRNA 3' processing machinery in a phosphorylation‐dependent manner and inhibits polyadenylation. J Biol Chem 284, 10694–10705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YB, Yang BH, Piao ZG, Oh SB, Kim JS & Park K (2003). Expression of Na+/HCO3 − cotransporter and its role in pH regulation in mouse parotid acinar cells. Biochem Biophys Res Commun 304, 593–598. [DOI] [PubMed] [Google Scholar]
- Kurtz I (2014). NBCe1 as a model carrier for understanding the structure‐function properties of Na+‐coupled SLC4 transporters in health and disease. Pflugers Arch 466, 1501–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacruz RS, Nanci A, White SN, Wen X, Wang H, Zalzal SF, Luong VQ, Schuetter VL, Conti PS, Kurtz I & Paine ML (2010). The sodium bicarbonate cotransporter (NBCe1) is essential for normal development of mouse dentition. J Biol Chem 285, 24432–24438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK & Boron WF (2018). Exploring the autoinhibitory domain of the electrogenic Na+/HCO3 − transporter NBCe1‐B, from residues 28 to 62. J Physiol 596, 3637–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Boron WF & Parker MD (2012). Relief of autoinhibition of the electrogenic Na‐HCO3 cotransporter NBCe1‐B: role of IRBIT vs. amino‐terminal truncation. Am J Physiol Cell Physiol 302, C518–C526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Qin X, Wang DK, Guo YM, Gill HS, Morris N, Parker MD, Chen LM & Boron WF (2013). Effects of optional structural elements, including two alternative amino termini and a new splicing cassette IV, on the function of NBCn1 (SLC4A7). J Physiol 591, 4983–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Xu JY, Wang DK, Wang L & Chen LM (2011). Cloning and identification of two novel NBCe1 splice variants from mouse reproductive tract tissues: A comparative study of NCBT genes. Genomics 98, 112–119. [DOI] [PubMed] [Google Scholar]
- Lu F, Li S, Jiang Y, Jiang J, Fan H, Lu G, Deng D, Dang S, Zhang X, Wang J & Yan N (2011). Structure and mechanism of the uracil transporter UraA. Nature 472, 243–246. [DOI] [PubMed] [Google Scholar]
- Luo X, Choi JY, Ko SB, Pushkin A, Kurtz I, Ahn W, Lee MG & Muallem S (2001). HCO3 − salvage mechanisms in the submandibular gland acinar and duct cells. J Biol Chem 276, 9808–9816. [DOI] [PubMed] [Google Scholar]
- McAlear SD, Liu X, Williams JB, McNicholas‐Bevensee CM & Bevensee MO (2006). Electrogenic Na/HCO3 cotransporter (NBCe1) variants expressed in Xenopus oocytes: functional comparison and roles of the amino and carboxy termini. J Gen Physiol 127, 639–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunsbach AB, Vorum H, Kwon TH, Nielsen S, Simonsen B, Choi I, Schmitt BM, Boron WF & Aalkjær C (2000). Immunoelectron microscopic localization of the electrogenic Na/HCO3 cotransporter in rat and Ambystoma kidney. J Am Soc Nephrol 11, 2179–2189. [DOI] [PubMed] [Google Scholar]
- Moss FJ & Boron WF (2020). Carbonic anhydrases enhance activity of endogenous Na‐H exchangers and not the electrogenic Na/HCO3 cotransporter NBCe1‐A, expressed in Xenopus oocytes. J Physiol 598, 5821–5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa N, Ogata K, Yokobori T, Ide M, Baatar S, Ubukata Y, Kimura A, Kogure N, Sohda M, Kuwano H, Saeki H & Shirabe K (2019). Low IRBIT levels are associated with chemo‐resistance in gastric cancer patients. Anticancer Res 39, 4111–4116. [DOI] [PubMed] [Google Scholar]
- Niculescu AB, Le‐Niculescu H, Levey DF, Phalen PL, Dainton HL, Roseberry K, Niculescu EM, Niezer JO, Williams A, Graham DL, Jones TJ, Venugopal V, Ballew A, Yard M, Gelbart T, Kurian SM, Shekhar A, Schork NJ, Sandusky GE & Salomon DR (2017). Precision medicine for suicidality: from universality to subtypes and personalization. Mol Psychiatry 22, 1250–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Shcheynikov N, Hong JH, Zheng C, Suh SH, Kawaai K, Ando H, Mizutani A, Abe T, Kiyonari H, Seki G, Yule D, Mikoshiba K & Muallem S (2013). Irbit mediates synergy between Ca2+ and cAMP signaling pathways during epithelial transport in mice. Gastroenterology 145, 232–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MD (2018). Mouse models of SLC4‐linked disorders of HCO3 −‐transporter dysfunction. Am J Physiol Cell Physiol 314, C569–C588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MD & Boron WF (2013). The divergence, actions, roles, and relatives of sodium coupled bicarbonate transporters. Physiol Rev 93, 803–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhitko AA, Binari R, Zhang N, Asara JM, Demontis F & Perrimon N (2016). Tissue‐specific down‐regulation of S‐adenosyl‐homocysteine via suppression of dAhcyL1/dAhcyL2 extends health span and life span in Drosophila. Genes Dev 30, 1409–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero MF, Hediger MA, Boulpaep EL & Boron WF (1997). Expression cloning and characterization of a renal electrogenic Na+/HCO3 − cotransporter. Nature 387, 409–413. [DOI] [PubMed] [Google Scholar]
- Salerno EE, Patel SP, Marshall A, Marshall J, Alsufayan T, Mballo CSA, Quade BN & Parker MD (2019). Extrarenal signs of proximal renal tubular acidosis persist in nonacidemic Nbce1b/c‐null mice. J Am Soc Nephrol 30, 979–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh H, Moriyama N, Hara C, Yamada H, Horita S, Kunimi M, Tsukamoto K, Iso O, Inatomi J, Kawakami H, Kudo A, Endou H, Igarashi T, Goto A, Fujita T & Seki G (2003). Localization of Na+‐HCO3 − cotransporter (NBC‐1) variants in rat and human pancreas. Am J Physiol Cell Physiol 284, C729–C737. [DOI] [PubMed] [Google Scholar]
- Sciortino CM & Romero MF (1999). Cation and voltage dependence of rat kidney electrogenic Na+‐HCO3 − cotransporter, rkNBC, expressed in oocytes. Am J Physiol 277, F611–F623. [DOI] [PubMed] [Google Scholar]
- Shcheynikov N, Son A, Hong JH, Yamazaki O, Ohana E, Kurtz I, Shin DM & Muallem S (2015). Intracellular Cl− as a signaling ion that potently regulates Na+/HCO3− transporters. Proc Natl Acad Sci USA 112, E329‐E337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakabe K, Priori G, Yamada H, Ando H, Horita S, Fujita T, Fujimoto I, Mizutani A, Seki G & Mikoshiba K (2006). IRBIT, an inositol 1,4,5‐trisphosphate receptor‐binding protein, specifically binds to and activates pancreas‐type Na+/HCO3 − cotransporter 1 (pNBC1). Proc Natl Acad Sci U S A 103, 9542–9547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shnitsar V, Li J, Li X, Calmettes C, Basu A, Casey JR, Moraes TF & Reithmeier RA (2013). A substrate access tunnel in the cytosolic domain is not an essential feature of the solute carrier 4 (SLC4) family of bicarbonate transporters. J Biol Chem 288, 33848–33860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleimani M, Grassl SM & Aronson PS (1987). Stoichiometry of Na+‐HCO3 − cotransport in basolateral membrane vesicles isolated from rabbit renal cortex. J Clin Invest 79, 1276–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer G, Rempfer C, Waterhouse AM, Gumienny R, Haas J & Schwede T (2020). QMEAND is co‐distance constraints applied on model quality estimation. Bioinformatics 36, 1765–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Van Paesschen W, Stalmans I, Horita S, Yamada H, Bergmans BA, Legius E, Riant F, De Jonghe P, Li Y, Sekine T, Igarashi T, Fujimoto I, Mikoshiba K, Shimadzu M, Shiohara M, Braverman N, Al Gazali L, Fujita T & Seki G (2010). Defective membrane expression of the Na+‐HCO3 − cotransporter NBCe1 is associated with familial migraine. Proc Natl Acad Sci U S A 107, 15963–15968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornell IM, Wu J & Bevensee MO (2010). The IP3 receptor‐binding protein IRBIT reduces phosphatidylinositol 4,5‐bisphosphate (PIP2) stimulation of Na/bicarbonate cotransporter NBCe1 variants expressed in Xenopus laevis oocytes. FASEB J 24, 815.816. [Google Scholar]
- Thurtle‐Schmidt BH & Stroud RM (2016). Structure of Bor1 supports an elevator transport mechanism for SLC4 anion exchangers. Proc Natl Acad Sci U S A 113, 10542–10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vachel L, Shcheynikov N, Yamazaki O, Fremder M, Ohana E, Son A, Shin DM, Yamazaki‐Nakazawa A, Yang CR, Knepper MA & Muallem S (2018). Modulation of Cl− signaling and ion transport by recruitment of kinases and phosphatases mediated by the regulatory protein IRBIT. Sci Signal 11, eaat5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CC, Sun B, Zhang X, Huang XW, Zhang MH, Guo H, Chen X, Huang F, Chen TY, Mi HL, Yu F, Liu LN & Zhang P (2019). Structural mechanism of the active bicarbonate transporter from cyanobacteria. Nat Plants 5, 1184–1193. [DOI] [PubMed] [Google Scholar]
- Wang JL, Wang XY, Wang DK, Parker MD, Musa‐Aziz R, Popple J, Guo YM, Min TX, Xia T, Tan M, Liu Y, Boron WF & Chen LM (2020a). Multiple acid‐base and electrolyte disturbances upregulate NBCn1, NBCn2, IRBIT and L‐IRBIT in the mTAL. J Physiol 598, 3395–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wu H, Liu Y & Chen L‐M (2020b). Activation of mouse NBCe1‐B by Xenopus laevis and mouse IRBITs: role of the variable Nt appendage of IRBITs. Biochim Biophys Acta Biomembr 1862, 183240. [DOI] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R & Schwede T (2018). SWISS‐MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46, W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig R, Nessling M, Will RD, Mollenhauer J, Salowsky R, Munstermann E, Schick M, Helmbach H, Gschwendt B, Korn B, Kioschis P, Lichter P, Schadendorf D & Poustka A (2002). Candidate genes for cross‐resistance against DNA‐damaging drugs. Cancer Res 62, 6698–6705. [PubMed] [Google Scholar]
- Yu X, Yang G, Yan C, Baylon JL, Jiang J, Fan H, Lu G, Hasegawa K, Okumura H, Wang T, Tajkhorshid E, Li S & Yan N (2017). Dimeric structure of the uracil:proton symporter UraA provides mechanistic insights into the SLC4/23/26 transporters. Cell Res 27, 1020–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Kiyatkin A, Bolin JT & Low PS (2000). Crystallographic structure and functional interpretation of the cytoplasmic domain of erythrocyte membrane band 3. Blood 96, 2925–2933. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Statistical Summary Document
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


