Abstract
Purpose
To estimate for each distinct fiber population within voxels containing multiple brain tissue types.
Methods
A diffusion‐ correlation experiment was carried out in an in vivo human brain using tensor‐valued diffusion encoding and multiple repetition times. The acquired data were inverted using a Monte Carlo algorithm that retrieves nonparametric distributions of diffusion tensors and longitudinal relaxation rates . Orientation distribution functions (ODFs) of the highly anisotropic components of were defined to visualize orientation‐specific diffusion‐relaxation properties. Finally, Monte Carlo density‐peak clustering (MC‐DPC) was performed to quantify fiber‐specific features and investigate microstructural differences between white matter fiber bundles.
Results
Parameter maps corresponding to ’s statistical descriptors were obtained, exhibiting the expected contrast between brain tissue types. Our ODFs recovered local orientations consistent with the known anatomy and indicated differences in between major crossing fiber bundles. These differences, confirmed by MC‐DPC, were in qualitative agreement with previous model‐based works but seem biased by the limitations of our current experimental setup.
Conclusions
Our Monte Carlo framework enables the nonparametric estimation of fiber‐specific diffusion‐ features, thereby showing potential for characterizing developmental or pathological changes in within a given fiber bundle, and for investigating interbundle differences.
Keywords: diffusion‐relaxation correlation, fiber‐specific microstructure, inverse Laplace transform, multivariate distribution, orientation distribution function, tensor‐valued diffusion encoding
1. INTRODUCTION
While diffusion MRI 1 , 2 , 3 , 4 , 5 has provided enhanced sensitivity to tissue microstructure in vivo by capturing the translational motion of water molecules, diffusion‐relaxation MRI (drMRI) 6 , 7 , 8 , 9 , 10 , 11 , 12 , 13 , 14 additionally reports on the local chemical composition of the aqueous phase. For instance, the longitudinal relaxation time is mainly determined in vivo by cross relaxation, magnetization transfer, and spin diffusion with macromolecules in general, 15 , 16 , 17 by myelin lipids in particular, 18 , 19 , 20 , 21 , 22 and by the interplay between relaxation and diffusion. 23 However, microstructural studies have been hindered by the fact that the measured drMRI signal is only sensitive to the voxel‐averaged diffusion‐relaxation profile, with typical cubic‐millimeter voxels comprising multiple cell types and the extracellular space. 24 , 25 , 26 , 27 , 28
Three strategies were explored to alleviate the lack of specificity of the drMRI signal. First, multiple models and signal representations have been developed to relate either the diffusion‐ 29 , 30 , 31 , 32 or diffusion‐ 7 , 8 , 33 MRI signal to the voxel content. However, these approaches rely on compartmental/functional assumptions and/or on model‐selection strategies that may disagree with the underlying tissue microstructure. 34 , 35 , 36 Second, while nonparametric inversion of the diffusion‐relaxation NMR signal is already common practice in the porous media field, 37 , 38 , 39 , 40 nonparametric inversion techniques of the drMRI signal have also been developed, 14 , 41 , 42 with applications ranging from porous media 6 to biological tissues such as the heart, 43 spinal cord, 44 , 45 placenta, 11 and brain. 46 However, these techniques have so far only been employed to retrieve 2D or 3D diffusivity–relaxation distributions. 14 , 42 Their lack of specificity in terms of diffusion orientation thus render them unable to isolate subparts of the distribution belonging to distinct subvoxel anisotropic diffusion environments, for example, white matter (WM) fiber populations. Third, “tensor‐valued” diffusion‐encoding gradient waveforms have enhanced the specificity of the data itself by targeting specific features of the intravoxel diffusion profile 47 , 48 , 49 , 50 , 51 , 52 , 53 , 54 via an axisymmetric encoding tensor introduced in Refs. [3, 55, 56]. Tensor‐valued diffusion acquisition schemes have since been used to further investigate signal representations 50 , 57 , 58 , 59 and models. 32 , 60 , 61 , 62 , 63 , 64
The advent of tensor‐valued diffusion measurements has resulted in the development of nonparametric Monte Carlo signal inversion algorithms of the 2D diffusion 65 and 6D diffusion‐‐ 12 NMR signals in porous media, and of the 4D diffusion 66 and 5D diffusion‐ 13 MRI signals in the in vivo brain. Although noise sensitive, 36 these algorithms do not rely on any compartmental/functional assumption regarding the voxel content, nor on any regularization 67 , 68 , 69 , 70 narrowing the space of suitable solutions to the inverse problem. They are also not limited by constraints regarding data compression, or restricted to dense acquisition sampling schemes that are difficult to extend to higher dimensions, unlike previous works in Ref. [71] and Refs. [41, 45, 72, 73, 74], respectively. Enhanced by methods aiming to visualize and quantify fiber‐specific properties, Monte Carlo signal inversions have recently provided critical sensitivity and specificity to fiber‐specific values. 75 , 76 , 77 However, this work has yet to be extended to fiber‐specific ‐values, which are of particular interest to evaluate changes in bundle‐specific myelin contents 78 relevant to the study of neurodevelopment, plasticity, aging, and neurological disorders. 79 , 80 , 81
In this work, we leverage nonparametric distributions of diffusion tensors "/> and longitudinal relaxation rates obtained via Monte Carlo inversion of a 5D diffusion‐ weighted in vivo human brain dataset to resolve subvoxel diffusion‐ components. We first estimate parameter maps of ’s statistical descriptors and extract orientation‐resolved values within the pool of highly anisotropic components output by the Monte Carlo inversion algorithm. These values are then color‐mapped onto nonparametric orientation distribution functions (ODFs) 75 , 76 and quantified in terms of median value and precision using Monte Carlo density‐peak clustering (MC‐DPC). 77 In particular, we identify significant differences with respect to relaxation between major WM bundles without relying on limiting assumptions, albeit in a single healthy volunteer.
After describing how our in vivo human brain data was acquired in Section 2.1, we lay down the theory underlying the Monte Carlo inversion algorithm and the statistical descriptors of in Section 2.2, and detail our ODF and MC‐DPC procedures in Section 2.3. We then present our results in Section 3 and discuss them in Section 4, and conclude in Section 5.
2. METHODS
2.1. In vivo human brain data
Data collection was approved by the Spectrum Medical Imaging local ethics committee. A healthy volunteer was scanned on a 3T GE 750w equipped with a 32‐channel receiver head and neck GEM coils (only 12‐16 channels used for head) using a prototype GE multidimensional diffusion (MDD) spin‐echo sequence with EPI readout, echo time ms, FOV = 240 × 240 × 12 , voxel size = 3 × 3 × 3 , fat saturation pulses, 82 and ASSET acceleration factor=2, customized for tensor‐valued diffusion encoding, 57 , 83 and variable repetition time . Tensor‐valued diffusion encoding was performed with numerically optimized 84 Maxwell‐compensated 85 waveforms whose gradient power spectra share similar frequency contents. 86 , 87 , 88 The same tensor‐valued diffusion‐weighted sequence, including linear, planar, and spherical b‐tensors with a maximal b‐value of , was repeated for , 2 and 5 seconds. The five dimensions of the resulting 20‐minute 363‐point acquisition scheme, shown in Figure 1, encode information allowing estimation of the 5D distribution . The signal‐to‐noise ratio (SNR) of this dataset, estimated across voxels of the corona radiata by computing the mean‐to‐standard‐deviation ratio of the spherically encoded diffusion signal at b = 0.1 ms/ and seconds (see Supporting Information of Ref. [83]), equals 40. As indicated above, only 4 axial slices were acquired in order to limit the acquisition time. While an inversion‐recovery slice‐shuffling sequence could drastically reduce acquisition time, 9 , 10 our prototype sequence is currently limited to sequential slices and lacks eddy current nulling. 89
FIGURE 1.
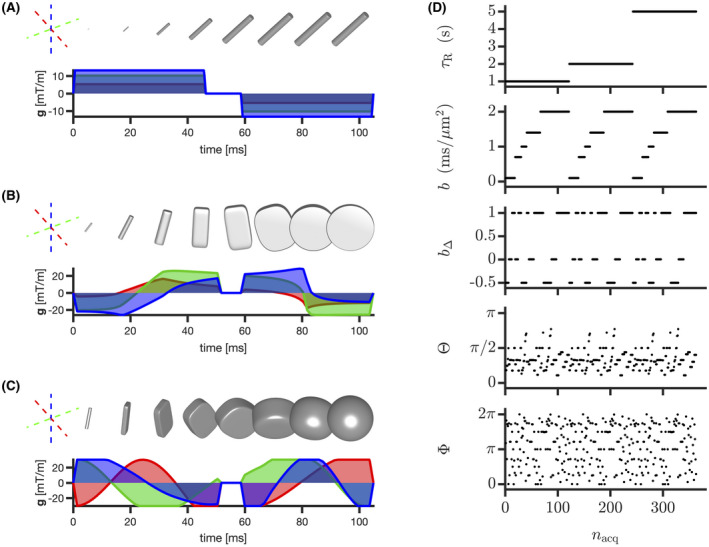
Visualization of our acquisition scheme. (A, B, and C) Gradient waveforms g(t) yielding linear, planar, and spherical diffusion encoding, respectively. Each color relates each gradient component to a given orthogonal axis of the spatial frame of reference. Tensor glyphs represent the b‐tensor progressively acquiring its final size (trace) , shape (normalized anisotropy) and orientation at the end of a given waveform. 49 , 54 D, Acquisition parameters as a function of sorted acquisition point index . denotes the varying repetition time of our acquisition scheme
2.2. Nonparametric Monte Carlo inversion
2.2.1. Signal fitting and bootstrapping
We modified the 5D Monte Carlo inversion algorithm found in Ref. [13] and pioneered by Ref. [37] to analyze the diffusion‐ dataset described in Section 2.1. Let us consider axisymmetric diffusion tensors, parameterized by their axial diffusivity , radial diffusivity , and orientation . An alternative parameterization includes the isotropic diffusivity and normalized anisotropy . 49 , 54 , 90 , 91 Our Monte Carlo inversion technique retrieves nonparametric intravoxel 5D distributions by fitting diffusion‐ weighted signals with a weighted sum of components , with . Given that the ‐weighting of the dataset detailed in Section 2.1 is provided through a spin‐echo sequence with constant echo time and variable repetition time , the algorithm inverts the following discretized signal equation 92 :
| (1) |
where is the mth acquired signal, is the weight of the th component (normalized so that ), is the diffusion‐encoding tensor (b‐tensor), 47 , 48 , 49 , 50 , 51 , 54 and “:” is the Frobenius inner product. The Monte Carlo inversion algorithm randomly samples components within the following ranges, , , and , and estimates the weights quantifying the components’ propensity to fit the acquired signals via non‐negative least‐squares fitting 69 , 70 , 71 , 93 , 94 of Equation (1). This process is repeated iteratively following a quasi‐genetic filtering detailed in Refs. [12, 13, 65, 95]. Using the same wording as these references, we used initial components, proliferation steps, mutation/extinction steps, and output components. Embracing the inherent ill‐conditioning of Laplace inversion problems, we performed bootstrapping with replacement 96 , 97 on the data and estimated for each voxel an ensemble of plausible sets of components, also called “bootstrap solutions,” each denoted by .
2.2.2. Statistical descriptors and binning
The final solution of the Monte Carlo inversion algorithm, , can be understood as the median of all bootstrap solutions. Following previous works, 13 , 36 we quantified the main features of this final solution by computing the median across bootstrap solutions of the per‐bootstrap means , variances , and covariances of , with . The median operator acts across bootstrap solutions, and , and denote the per‐bootstrap average, variance, and covariance over bootstrap solution , respectively. For simplicity, we implicitly omit the median operator when addressing a statistical descriptor, thereby writing averages, variances, and covariances as “,” “,” and “,” respectively.
Tissue‐specific statistical descriptors can be extracted by subdividing the 5D configuration space of into multiple bins. For instance, the “thin,” “thick,” and “big” bins introduced in Refs. [13, 54] aim to isolate the signal contributions from WM, gray matter (GM), and cerebrospinal fluid (CSF), respectively. The bin boundaries, illustrated in the panels C, D, and E of Figure 2, were defined as follows:
FIGURE 2.
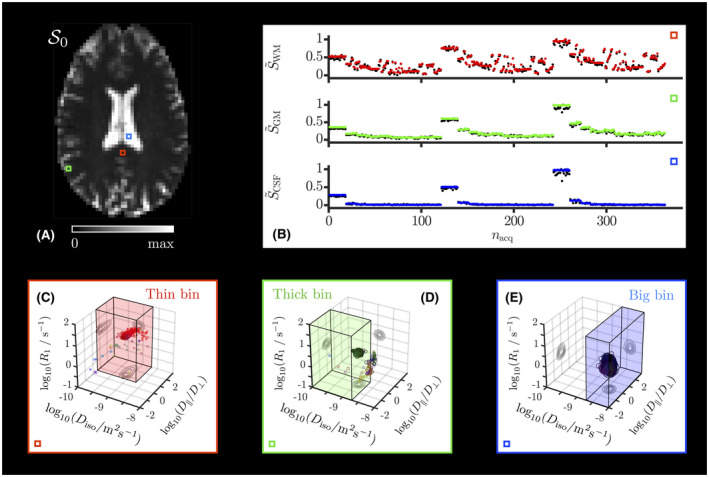
Monte Carlo fitted signal and retrieved 5D distributions in typical voxels. A, map estimated by the Monte Carlo inversion. The colored squares delineate typical WM (red), GM (green), and CSF (blue) voxels. B, Normalized signal measured (black points) and fitted (colored points) in the archetypal voxels of panel A as a function of the sorted acquisition point index of Figure 1. (C, D, E) Nonparametric distributions estimated for the archetypal voxels of panel A and reported as scatter plots in a 3D space of the logarithms of the longitudinal relaxation rate , isotropic diffusivity , and axial‐radial diffusivity ratio . Diffusion orientations are color‐coded according to . Symbol area is proportional to the statistical weight of the corresponding component n. The contour lines on the sides of the plots represent projections of the 5D distributions onto the respective 2D planes. The “thin,” “thick,” and “big” bins defined in Section 2.2.2 are illustrated as colored boxes in the panels where they are most relevant. The colors bounding panels C, D, and E match those of the highlighted voxels in panel A
“thin” bin within , and .
“thick” bin within , and .
“big” bin within , and .
Bin‐specific statistical descriptors were estimated following the above process for the retrieved components specifically falling into each bin. Note that this manual binning consists merely in a preliminary attempt to comprehend the rich information contained in , and is as such a limitation that could be mitigated by automatic clustering methods or by higher dimensional versions of data‐driven techniques such as those of Refs. [46, 98].
2.3. Orientation distribution functions and Monte Carlo density‐peak clustering
Orientation distribution functions were defined from the thin‐bin components output by the Monte Carlo inversion of Section 2.2.1 using the procedure detailed in Refs. [75, 76, 95]. This procedure consists in mapping the set of thin‐bin components onto the nodes of a dense spherical mesh , building up the ODF radii from the thin‐bin component weights and orientations . In addition, it enables to compute per‐bootstrap orientation‐specific diffusion‐relaxation means , with . Besides coloring ODFs according to local orientation, this mapping allows to color ODFs according to the local value , which we used to visualize orientation‐specific diffusion‐relaxation quantities. For simplicity, the short‐hand notation “” is now retained for .
MC‐DPC, 77 that is, a combination of the Monte Carlo inversion algorithm of Section 2.2.1 and density‐peak clustering, 99 was used to quantify the median value and precision of orientation‐resolved means of across bootstrap solutions, which we employed to detect differences between subvoxel fiber populations. Briefly, MC‐DPC first delineates subvoxel clusters as orientational aggregates of thin‐bin solutions, which are interpreted as orientational regions of interest associated with subvoxel fiber populations. MC‐DPC then computes per‐bootstrap cluster‐specific (orientation‐resolved) means , where denotes the cluster index. Finally, one extracts cluster‐specific medians and interquartile ranges of across bootstrap solutions. For simplicity, the short‐hand notation “” is now used to describe the collection of orientation‐resolved means originating from all bootstrap solutions and all clusters . Also, and were computed separately, as both quantities are commonly found in the MRI literature and does not generally equal .
The ODF and MC‐DPC procedures are detailed for the diffusion‐ case in Supporting Information. In addition, an in silico evaluation of these techniques is provided in Supporting Information Figures S1 and S2, demonstrating their accuracy in capturing relaxation‐based differences across distinct subvoxel fiber populations at intermediate‐to‐high SNR levels.
3. RESULTS
Figure 2 presents the fitted signals and distributions estimated by our Monte Carlo inversion algorithm in typical voxels associated with WM in the corpus callosum (CC), cortical GM, and CSF in the ventricles. Figure 3 displays typical axial maps of ’s global and bin‐specific statistical descriptors. Figure 4 shows orientation‐colored and ‐colored ODFs in a typical axial slice. Figure 5 investigates possible microstructural differences between subvoxel fiber populations by leveraging MC‐DPC in regions of interest that target specific fiber crossings, namely the crossing between the CC and the cingulum (CING), and the crossing between the corpus callosum, the arcuate fasciculus (AF), and the corticospinal tract (CST) in the posterior corona radiata.
FIGURE 3.
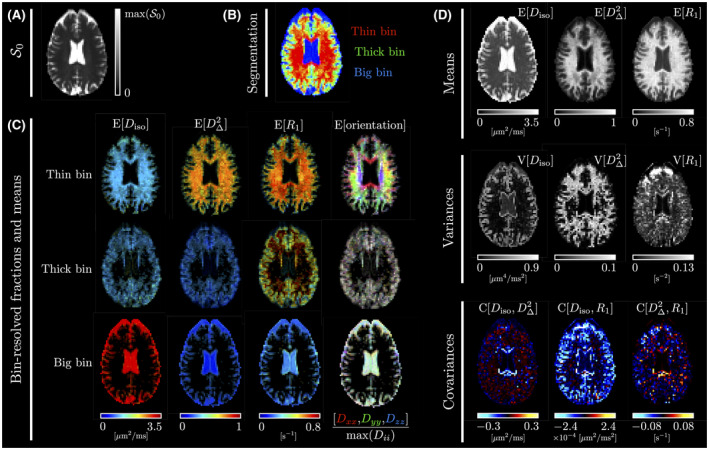
Typical axial maps of the statistical descriptors described in Section 2.2.2. While panel (A) shows the non‐diffusion‐weighted and non‐‐weighted signal , panel (B) presents a segmentation map of the brain into the thin, thick, and big bins defined in Section 2.2.2, colored according to . Panel (C) features maps of the bin‐specific means of , , and orientation. The bin‐specific averaged subvoxel orientation is color‐coded for orientation according to , where the average diffusivities are associated with the directions corresponding to the “left‐right,” “anterior‐posterior,” and “superior‐inferior” directions, respectively. For a given bin, the intensity of the bin‐specific maps equals the voxel‐wise average fraction of components belonging to this bin. Finally, panel (D) contains the global means, variances, and covariances of , , and
FIGURE 4.
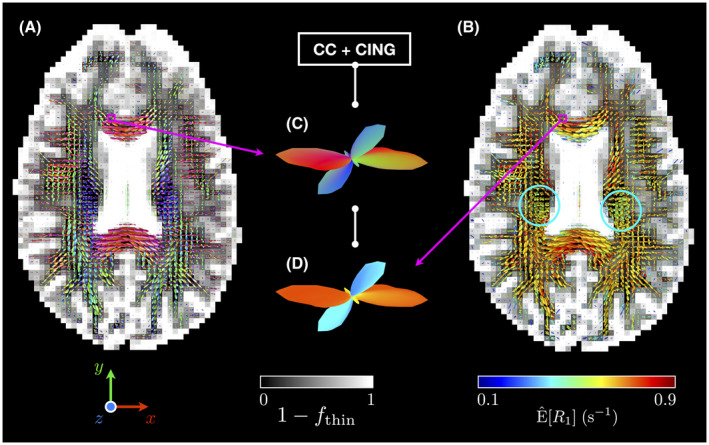
Axial gray‐scale maps of the fraction of non thin‐bin components with superimposed ODFs colored by (A) local orientation (with , and corresponding to the “left‐right,” “anterior‐posterior,” and “superior‐inferior” directions, respectively) and by (B) (see Section 2.3). The middle insets zoom on a voxel containing a fiber crossing between the corpus callosum (CC) and the cingulum (CING), and presents the estimated (C) orientation‐colored and (D) ‐colored ODFs for this voxel. While differences in seem to exist between CC and CING, such differences may also exist in the regions where the CST’s pyramidal tracts are located (blue circles), as indicated by the greener ‐colored ODFs therein
FIGURE 5.
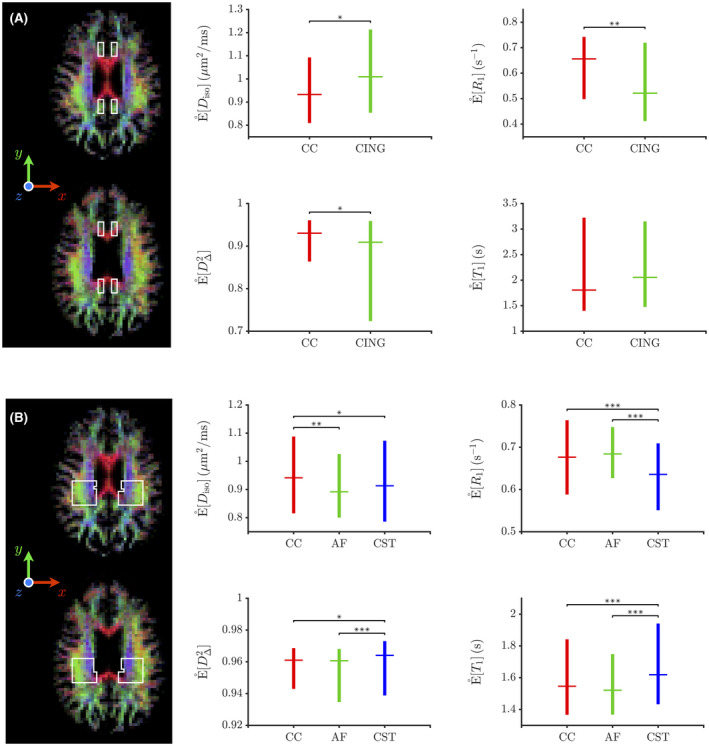
Boxplots of the medians of the orientation‐resolved means obtained from MC‐DPC (see Section 2.3) within hand‐drawn regions of interest (ROIs), represented as white‐lined boxes over axial slices of the orientation‐colored average fraction of thin‐bin components . For a given boxplot, the horizontal line and whiskers indicate the median and the range between the first and third quartiles of the medians of the orientation‐resolved means , respectively. While panel A’s ROIs focus on crossing areas between the corpus callosum (CC) and the cingulum (CING), those of panel B focus on crossing areas between the corpus callosum (CC), the arcuate fasciculus (AF) and the corticospinal tract (CST) in the posterior corona radiata. Each MC‐DPC cluster (and associated orientation‐resolved means) is robustly assigned to one of these bundles depending on whether its median orientation is closer to the “left‐right” direction (CC), to the “anterior‐posterior” direction (CING or AF), or to the “superior‐inferior” direction (CST). The asterisks report the results of nonparametric Mann‐Whitney ‐tests assessing whether or not two orientation‐resolved means assigned to distinct bundles are sampled from identically shaped non‐median‐shifted continuous distributions (null hypothesis ). The ‐values resulting from these tests inform on the acceptance or rejection of at a certain significance level: (*), (**) and (***)
4. DISCUSSION
As observed in previous works, 13 , 66 , 95 Figure 2 demonstrates that the Monte Carlo inversion algorithm yields distributions consistent with the features of measured raw signals in various voxels pertaining to WM, cortical GM, and CSF in the ventricles. In particular, the diffusion isotropy of GM and CSF implies ‐ and b‐tensor orientation‐independent measured signals, and the ‐dependence of the signals shows that increases when going from CSF to GM, and from GM to WM. In addition, the three bins capture these distinct environments in accordance with their original design.
Figure 3 shows that the Monte Carlo inversion algorithm can estimate maps of ’s statistical descriptors. In particular, it retrieves non‐‐related maps that are consistent with those thoroughly discussed in Ref. [13]. Let us thus mainly discuss the ‐related maps. The bin‐specific maps of Figure 3C present a clear contrast between our tissue‐specific bins, due to high‐ WM, intermediate‐ GM, and low‐ CSF. In Figure 3D, the global map resembles an expected low‐resolution conventional map, that is, bright in WM, slightly darker in GM, and very dark in CSF. The map resembles a noisier version of the map, as both give high values in mixed CSF‐WM/GM voxels. The noise in could be reduced by adding more repetition times in the acquisition scheme. The map is negative at the interface between CSF and either WM or cortical GM. Indeed, upon entering CSF from WM/GM, increases and decreases rapidly. Finally, the map exhibits no specific pattern. We emphasize the fact that although and are often correlated in the brain, they nevertheless report on different properties, namely microstructure and chemical composition. Therefore, it may still be useful to map them separately (means and variances) and jointly (covariance), especially in pathological cases.
Figure 4 features nonparametric ODFs capturing local orientations that are consistent with the known anatomy. Regarding ‐colored ODFs (see Section 2.3), they change colors when approaching tissue interfaces with CSF. This gradual change in may originate from molecular exchange and/or magnetization transfer between tissues and CSF. Importantly, Figure 4 shows that potential differences in relaxation may exist between major fiber bundles, namely CC and CING (insets in Figure 4), and CC, AF, and CST (greener ‐colored ODFs in the CST’s pyramidal tracts).
These potential microstructural differences are quantified in Figure 5. Focusing on relaxation‐based differences, Figure 5A shows that CC and CING exhibit significant differences in that are qualitatively consistent with those found in Refs. [8, 33], that is, tends to be lower in CING compared to CC. As for Figure 5B, it shows that CST features significant differences in and with CC and AF, with no statistically significant differences between CC and AF. These differences are qualitatively consistent with those identified for CST in Ref. [8], that is, tends to be higher in CST compared to CC and AF. These differences justify the need for a 5D inversion of the drMRI signal. Indeed, while a 3D inversion of the powder‐averaged drMRI signal is possible, it would only be useful if were independent of orientation, which is often not the case.8, 33
Quantitatively, the values estimated by in Figure 5 (around 1.5‐2 seconds) are overestimated compared to those of Ref. [8] (around 0.9‐1 second) and Ref. [33] (around 0.7 second). This discrepancy can be explained by the following factors. First, the acquisition scheme described in Section 2. 1 does not maximize the amount of diffusion‐relaxation correlations built into the inversion kernel of Equation (1), because the same diffusion‐weighting block was repeated for each acquired repetition time. Similar problems have been suggested to lead to a loss of accuracy for the Monte Carlo inversion. 36 Besides, the presence of a similar overestimation at SNR = 40 in the in silico evaluation of MC‐DPC presented in Supporting Information Figure S2 indicates that our acquisition sampling scheme is a limiting factor of this present work. Second, the use of saturation recovery with a spoiled spin echo for encoding is very sensitive to flip‐angle inaccuracies caused by both inhomogeneity across the subject and slice‐profile imperfections. Saturation‐recovery based mapping is also sensitive to magnetization‐transfer effects, especially in the present setup comprising an additional refocusing pulse and a fat‐saturation pulse. 100 , 101 , 102 These technical limitations should be mitigated upon developing a sequence that includes inversion preparation for enhanced sensitivity and slice shuffling for optimized time efficiency. 9 , 10
5. CONCLUSIONS
Diffusion‐ weighted datasets incorporating multiple b‐tensor shapes can be inverted without relying on limiting assumptions to obtain nonparametric distributions using the Monte Carlo inversion algorithm. The main features of the retrieved distributions can be visualized as maps of global and bin‐specific statistical descriptors related to means, variances, and covariances of diffusion‐relaxation properties. In particular, the bin‐specific maps exhibit the expected contrast between WM, GM, and CSF. Further insight into WM microstructure is provided by the “thin bin,” which isolates highly anisotropic components that should report on WM tissues. From these thin‐bin components, visualization of fiber‐specific information is improved upon defining ODFs that can be color‐mapped with respect to local orientation or diffusion‐relaxation features. 75 While ‐colored ODFs hint at possible differences between fiber bundles, MC‐DPC enables their quantification in terms of fiber‐specific diffusion‐relaxation measures. 77
Importantly, significant relaxation‐based differences are detected between the CC and the CING, and between the CST and the CC and AF. These differences, qualitatively consistent with those found in previous works, 8 , 33 offer a first proof of concept for the potential of our Monte Carlo framework in terms of nonparametric fiber‐specific relaxometry. Such approach would be practical in identifying differences in between distinct subvoxel fiber populations, characterizing developmental or pathological changes in within a given subvoxel fiber population, and measuring the angular dependence of longitudinal relaxation times in WM with respect to the main MRI magnetic field . 103 , 104 Moreover, fiber‐specific values could be relevant for microstructure‐informed tractography. 105 , 106 , 107
Nevertheless, we emphasize that this work can be improved in multiple ways. First, our acquisition setup could be optimized in terms of speed 9 , 10 and sensitivity. 89 , 108 , 109 , 110 Second, our manual binning could be made data‐driven upon using automatic clustering techniques similar to that of Ref. [77], or upon extending recent works to higher dimensions. 46 , 98 Third, better matching between the output of our Monte Carlo framework and plausible WM tracts would be obtained by integrating tractography 111 , 112 , 113 , 114 , 115 , 116 , 117 , 118 into our analysis pipeline. Finally, our framework remains to be applied to more subjects to assess its consistency across samples and studies.
CONFLICT OF INTEREST
D. Topgaard owns shares in Random Walk Imaging AB (Lund, Sweden, http://www.rwi.se/), holding patents related to the described methods.
Supporting information
FIGURE S1 Subvoxel orientations retrieved for the in silico data described in Section S3 using the Monte Carlo inversion for various numbers of bootstrap solutions and various SNR levels. While the ODFs were obtained via the process detailed in Section S1, the orientational clusters, here represented on the unit sphere, were extracted via MC‐DPC according to Section S2. denotes the angular deviation, computed for a given orientational cluster as the shortest angle between either the cluster geometric median orientation (circles, see Equation S7) or the corresponding ODF peak (squares, see Section S1), and the closest ground‐truth anisotropic component orientation. The color mapped onto the ODF codes for local orientation according to [red, green, blue] ≡ [|x|, |y|, |z|]/max([|x|, |y|, |z|]). As for the clusters, while opacity codes for the weight of the intra‐cluster averaged components (see Equation S6), color codes for the geometric median orientation of each cluster (see Equation S7). The conditions of the in vivo study presented in the main body of the paper are closest to the case FIGURE S2 Orientation‐resolved means [χ] (see Equation S5) and weights (see Equation S6) associated with the MC‐DPC clusters of Figure S1. While ground truth is shown as horizontal lines, the circles and whiskers represent the medians and interquartile ranges of the orientation‐resolved means across bootstrap solutions, respectively. Squares correspond to the estimated ODF‐peak metrics. Colors match those of the orientational clusters/ODF peaks presented in Figure S1. In the rightmost panels, cluster weights were normalized so that the sum of all median weights across clusters equals one. Their ODF‐peak equivalents were simply obtained by taking the mesh‐projected component weights (ie, ODF radii) along the peaks of a given ODF (see Section S1). These ODF‐peak weights were then normalized to sum up to one, for easier comparison with normalized cluster weights. The conditions of the in vivo study presented in the main body of the paper are closest to the case
ACKNOWLEDGMENTS
This work was financially supported by the Swedish Foundation for Strategic Research (ITM17‐0267) and the Swedish Research Council (2018‐03697).
Reymbaut A, Critchley J, Durighel G, et al. Toward nonparametric diffusion‐ characterization of crossing fibers in the human brain. Magn Reson Med. 2021;85:2815–2827. 10.1002/mrm.28604
DATA AVAILABILITY STATEMENT
The code and in silico data that support the findings of this study are openly available in a GitHub repository at https://github.com/areymbaut (last accessed: 22nd of November 2020).
REFERENCES
- 1. Le Bihan D. Diffusion/perfusion MR imaging of the brain: from structure to function. Radiology. 1990;177:328‐329. [DOI] [PubMed] [Google Scholar]
- 2. Le Bihan D, Turner R, Douek P, Patronas N. Diffusion MR imaging: clinical applications. Am J Roentgenol. 1992;159:591‐599. [DOI] [PubMed] [Google Scholar]
- 3. Basser PJ, Mattiello J, LeBihan D. MR diffusion tensor spectroscopy and imaging. Biophys J. 1994;66:259‐267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jones DK. Diffusion MRI. Oxford: Oxford University Press; 2010. [Google Scholar]
- 5. Topgaard D. (Ed.). Advanced diffusion encoding methods in MRI New Developments in NMR. London: The Royal Society of Chemistry; 2020. [Google Scholar]
- 6. Zhang Y, Blümich B. Spatially resolved D–T 2 correlation NMR of porous media. J Magn Reson (San Diego, Calif.: 1997). 2014;242:41‐48. [DOI] [PubMed] [Google Scholar]
- 7. De Santis S, Barazany D, Jones DK, Assaf Y. Resolving relaxometry and diffusion properties within the same voxel in the presence of crossing fibres by combining inversion recovery and diffusion‐weighted acquisitions. Magn Reson Med. 2016;75:372‐380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Santis S, Assaf Y, Jeurissen B, Jones DK, Roebroeck A. T 1 relaxometry of crossing fibres in the human brain. NeuroImage. 2016;141:133‐142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hutter J, Slator PJ, Christiaens D, et al. Integrated and efficient diffusion‐relaxometry using ZEBRA. Sci Rep. 2018;8:15138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Park DJ, Witzel T, Leppert I, et al. Rapid multi‐inversion SMS‐EPI integrated with gradient‐echo, spin‐echo and diffusion‐weighted EPI acquisitions. Proceedings of the International Society for Magnetic Resonance Imaging, Paris, France, 2018. Abstract #4229. [Google Scholar]
- 11. Slator PJ, Hutter J, Palombo M, et al. Combined diffusion‐relaxometry MRI to identify dysfunction in the human placenta. Magn Reson Med. 2019;82:95‐106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Almeida Martins JP, Topgaard D. Multidimensional correlation of nuclear relaxation rates and diffusion tensors for model‐free investigations of heterogeneous anisotropic porous materials. Sci Rep. 2018;8:2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Almeida Martins JP, Tax CMW, Szczepankiewicz F, Jones DK, Westin CF, Topgaard D. Transferring principles of solid‐state and Laplace NMR to the field of in vivo brain MRI. Magn Reson. 2020;1:27‐43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tax CMW. Chapter 7 estimating chemical and microstructural heterogeneity by correlating relaxation and diffusion In: Advanced Diffusion Encoding Methods in MRI. London: The Royal Society of Chemistry; 2020:186‐227. [PubMed] [Google Scholar]
- 15. Edzes HT, Samulski ET. Cross relaxation and spin diffusion in the proton NMR of hydrated collagen. Nature. 1977;265:521‐523. [DOI] [PubMed] [Google Scholar]
- 16. Halle B. Molecular theory of field‐dependent proton spin‐lattice relaxation in tissue. Magn Reson Med. 2006;56:60‐72. [DOI] [PubMed] [Google Scholar]
- 17. Rooney WD, Johnson G, Li X, et al. Magnetic field and tissue dependencies of human brain longitudinal 1H2O relaxation in vivo. Magn Reson Med. 2007;57:308‐318. [DOI] [PubMed] [Google Scholar]
- 18. Mottershead JP, Schmierer K, Clemence M, et al. High field MRI correlates of myelincontent and axonal density in multiple sclerosis. J Neurol. 2003;250:1293‐1301. [DOI] [PubMed] [Google Scholar]
- 19. Bjarnason T, Vavasour I, Chia C, MacKay A. Characterization of the NMR behavior of white matter in bovine brain. Magn Reson Med. 2005;54:1072‐1081. [DOI] [PubMed] [Google Scholar]
- 20. De Santis S, Drakesmith M, Bells S, Assaf Y, Jones DK. Why diffusion tensor MRI does well only some of the time: variance and covariance of white matter tissue microstructure attributes in the living human brain. NeuroImage. 2014;89:35‐44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lutti A, Dick F, Sereno MI, Weiskopf N. Using high‐resolution quantitative mapping of R1 as an index of cortical myelination. NeuroImage. 2014;93:176‐188. In‐vivo Brodmann mapping of the human brain. [DOI] [PubMed] [Google Scholar]
- 22. Stüber C, Morawski M, Schäfer A, et al. Myelin and iron concentration in the human brain: a quantitative study of MRI contrast. NeuroImage. 2014;93:95‐106. [DOI] [PubMed] [Google Scholar]
- 23. Brownstein K, Tarr C. Spin‐lattice relaxation in a system governed by diffusion. J Magn Reson (1969). 1977;26:17‐24. [Google Scholar]
- 24. Stanisz GJ, Wright GA, Henkelman RM, Szafer A. An analytical model of restricted diffusion in bovine optic nerve. Magn Reson Med. 1997;37:103‐111. [DOI] [PubMed] [Google Scholar]
- 25. Norris DG. The effects of microscopic tissue parameters on the diffusion weighted magnetic resonance imaging experiment. NMR Biomed. 2001;14:77‐93. [DOI] [PubMed] [Google Scholar]
- 26. Sehy JV, Ackerman JJ, Neil JJ. Evidence that both fast and slow water ADC components arise from intracellular space. Magn Reson Med. 2002;48:765‐770. [DOI] [PubMed] [Google Scholar]
- 27. Minati L, Weglarz WP. Physical foundations, models, and methods of diffusion magnetic resonance imaging of the brain: a review. Con Magn Reson Part A. 2007;30A:278‐307. [Google Scholar]
- 28. Mulkern RV, Haker SJ, Maier SE. On high b diffusion imaging in the human brain: ruminations and experimental insights. Magn Reson Imaging. 2007;30A:278‐307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Veraart J, Novikov DS, Fieremans E. TE dependent diffusion imaging (TEdDI) distinguishes between compartmental T 2 relaxation times. NeuroImage. 2018;182:360‐369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lemberskiy G, Fieremans E, Veraart J, Deng FM, Rosenkrantz AB, Novikov DS. Characterization of prostate microstructure using water diffusion and NMR relaxation. Front Phys. 2018;6:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ning L, Gagoski B, Szczepankiewicz F, Westin C, Rathi Y. Joint relaxation‐diffusion imaging moments to probe neurite microstructure. IEEE Trans Med Imaging. 2020;39:668‐677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lampinen B, Szczepankiewicz F, Mårtensson J, et al. Towards unconstrained compartment modeling in white matter using diffusion‐relaxation MRI with tensor‐valued diffusion encoding. Magn Reson Med. 2020;84:1605‐1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Andrews DA, Campbell JSW, Leppert IR, et al. Efficient whole‐brain tract‐specific T 1 mapping with slice‐shuffled inversion‐recovery diffusion‐weighted imaging at 3T. Proceedings of the International Society for Magnetic Resonance Imaging, Montreal, Québec, Canada, 2019. Abstract #0941. [Google Scholar]
- 34. Jelescu IO, Budde MD. Design and validation of diffusion MRI models of white matter. Front Phys. 2017;5:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Novikov DS, Kiselev VG, Jespersen SN. On modeling. Magn Reson Med. 2018;79:3172‐3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reymbaut A, Mezzani P, de Almeida Martins JP, Topgaard D. Accuracy and precision of statistical descriptors obtained from multidimensional diffusion signal inversion algorithms. NMR Biomed. 2020;33:e4267. [DOI] [PubMed] [Google Scholar]
- 37. Prange M, Song YQ. Quantifying uncertainty in NMR T 2 spectra using Monte Carlo inversion. J Magn Reson. 2009;196:54‐60. [DOI] [PubMed] [Google Scholar]
- 38. Galvosas P, Callaghan PT. Multi‐dimensional inverse Laplace spectroscopy in the NMR of porous media. Comptes Rendus Phys. 2010;11:172‐180. Multiscale NMR and relaxation. [Google Scholar]
- 39. Bernin D, Topgaard D. NMR diffusion and relaxation correlation methods: new insights in heterogeneous materials. Curr Opin Colloid Interface Sci. 2013;18:166‐172. [Google Scholar]
- 40. Song YQ, Venkataramanan L, Kausik R, Heaton N. Chapter 4 two‐dimensional NMR of diffusion and relaxation. London: The Royal Society of Chemistry; 2017:111‐155. [Google Scholar]
- 41. Benjamini D, Basser PJ. Multidimensional correlation MRI. NMR Biomed. 2020;33:e4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Benjamini D. Chapter 10 nonparametric inversion of relaxation and diffusion correlation data Advanced Diffusion Encoding Methods in MRI. London: The Royal Society of Chemistry; 2020:278‐316. [Google Scholar]
- 43. Seland JG, Bruvold M, Anthonsen H, et al. Determination of water compartments in rat myocardium using combined D–T 1 and T 1–T 2 experiments. Magn Reson Imaging. 2005;23:353‐354. Proceedings of the Seventh International Conference on Recent Advances in MR Applications to Porous Media. [DOI] [PubMed] [Google Scholar]
- 44. Benjamini D, Basser PJ. Magnetic resonance microdynamic imaging reveals distinct tissue microenvironments. NeuroImage. 2017;163:183‐196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim D, Doyle EK, Wisnowski JL, Kim JH, Haldar JP. Diffusion‐relaxation correlation spectroscopic imaging: a multidimensional approach for probing microstructure. Magn Reson Med. 2017;78:2236‐2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pas K, Komlosh ME, Perl DP, Basser PJ, Benjamini D. Retaining information from multidimensional correlation MRI using a spectral regions of interest generator. Sci Rep. 2020;10:3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Eriksson S, Lasic S, Topgaard D. Isotropic diffusion weighting in PGSE NMR by magic‐angle spinning of the q‐vector. J Magn Reson. 2013;226:13‐18. [DOI] [PubMed] [Google Scholar]
- 48. Westin CF, Szczepankiewicz F, Pasternak O, et al. Measurement Tensors in Diffusion MRI: generalizing the concept of diffusion encoding In: Golland P., Hata N., Barillot C., Hornegger J., Howe R. (Eds.), Medical Image Computing and Computer‐Assisted Intervention—MICCAI 2014—MICCAI. Cham: Springer International Publishing; 2014:209‐216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Eriksson S, Lasic S, Nilsson M, Westin CF, Topgaard D. NMR diffusion‐encoding with axial symmetry and variable anisotropy: distinguishing between prolate and oblate microscopic diffusion tensors with unknown orientation distribution. J Chem Phys. 2015;142:104201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Westin CF, Knutsson H, Pasternak O, et al. Q‐space trajectory imaging for multidimensional diffusion MRI of the human brain. NeuroImage. 2016;135:345‐362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Topgaard D. Multidimensional diffusion MRI. J Magn Reson. 2017;275:98‐113. [DOI] [PubMed] [Google Scholar]
- 52. Topgaard D. Multiple dimensions for random walks. J Magn Reson. 2019;306:150‐154. [DOI] [PubMed] [Google Scholar]
- 53. Lundell H, Lasič, S. Chapter 2 diffusion encoding with general gradient waveforms Advanced Diffusion Encoding Methods in MRI. London: The Royal Society of Chemistry; 2020:12‐67. [Google Scholar]
- 54. Reymbaut A. Chapter 3 diffusion anisotropy and tensor‐valued encoding In: Advanced Diffusion Encoding Methods in MRI. London: The Royal Society of Chemistry; 2020:68‐102. [Google Scholar]
- 55. Mattiello J, Basser P, Lebihan D. Analytical expressions for the b matrix in NMR diffusion imaging and spectroscopy. J Magn Reson Ser A. 1994;108:131‐141. [Google Scholar]
- 56. Mattiello J, Basser PJ, Le Bihan D. The b matrix in diffusion tensor echo‐planar imaging. Magn Reson Med. 1997;37:292‐300. [DOI] [PubMed] [Google Scholar]
- 57. Lasi S, Szczepankiewicz F, Eriksson S, Nilsson M, Topgaard D. Microanisotropy imaging: quantification of microscopic diffusion anisotropy and orientational order parameter by diffusion MRI with magic‐angle spinning of the q‐vector. Front Phys. 2014;2:11. [Google Scholar]
- 58. Cottaar M, Szczepankiewicz F, Bastiani M, et al. Improved fibre dispersion estimation using b‐tensor encoding. NeuroImage. 2020;116832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Reymbaut A. Matrix moments of the diffusion tensor distribution. arXiv e‐prints. 2020.
- 60. Lampinen B, Szczepankiewicz F, Mårtensson J, Westen VD, Sundgren PC, Nilsson M. Neurite density imaging versus imaging of microscopic anisotropy in diffusion MRI: a model comparison using spherical tensor encoding. NeuroImage. 2017;147:517‐531. [DOI] [PubMed] [Google Scholar]
- 61. Coelho S, Pozo JM, Jespersen SN, Jones DK, Frangi AF. Resolving degeneracy in diffusion MRI biophysical model parameter estimation using double diffusion encoding. Magn Reson Med. 2019;82:395‐410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Coelho S, Pozo JM, Jespersen SN, Frangi AF. Optimal experimental design for biophysical modelling in multidimensional diffusion MRI In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2019. Cham: Springer International Publishing; 2019;617‐625. [Google Scholar]
- 63. Reisert M, Kiselev VG, Dhital B. A unique analytical solution of the white matter standard model using linear and planar encodings. Magn Reson Med. 2019;81:3819‐3825. [DOI] [PubMed] [Google Scholar]
- 64. Reymbaut A, Valcourt Caron A, Gilbert G, et al. Magic DIAMOND: multi‐fascicle diffusion compartment imaging with tensor distribution modeling and tensor‐valued diffusion encoding. arXiv e‐prints, 2020. [DOI] [PubMed]
- 65. de Almeida Martins JP, Topgaard D. Two‐dimensional correlation of isotropic and directional diffusion using NMR. Phys Rev Lett. 2016;116:087601. [DOI] [PubMed] [Google Scholar]
- 66. Yon M, dJP Almeida Martins, Bao Q, Budde MD, Frydman L, Topgaard D. Diffusion tensor distribution imaging of an in vivo mouse brain at ultrahigh magnetic field by spatiotemporal encoding. NMR Biomed. 2020;33:e4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Provencher SW. A constrained regularization method for inverting data represented by linear algebraic or integral equations. Comput Phys Commun. 1982;27:213‐227. [Google Scholar]
- 68. Kroeker RM, Henkelman RM. Analysis of biological NMR relaxation data with continuous distributions of relaxation times. J Magn Reson (1969). 1986;69):218‐235. [Google Scholar]
- 69. Whittall KP, MacKay AL. Quantitative interpretation of NMR relaxation data. J Magn Reson (1969). 1989;84:134‐152. [Google Scholar]
- 70. Mitchell J, Chandrasekera T, Gladden L. Numerical estimation of relaxation and diffusion distributions in two dimensions. Prog Nucl Magn Reson Spectrosc. 2012;62:34‐50. [DOI] [PubMed] [Google Scholar]
- 71. Venkataramanan L, Song Yi‐Qiao, Hurlimann MD. Solving Fredholm integrals of the first kind with tensor product structure in 2 and 2.5 dimensions. IEEE Trans Signal Process. 2002;50:1017‐1026. [Google Scholar]
- 72. Benjamini D, Basser PJ. Use of marginal distributions constrained optimization (MADCO) for accelerated 2D MRI relaxometry and diffusometry. J Magn Reson. 2016;271:40‐45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Benjamini D, Basser PJ. Towards clinically feasible relaxation‐diffusion correlation MRI using MADCO. Microporous Mesoporous Mater. 2018;269:93‐96. Proceedings of the 13th International Bologna Conference on Magnetic Resonance in Porous Media (MRPM13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kim D, Wisnowski JL, Nguyen CT, Haldar JP. Multidimensional correlation spectroscopic imaging of exponential decays: from theoretical principles to in vivo human applications. NMR Biomed. 2020;33:e4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. de Almeida Martins JP, Tax CMW, Reymbaut A, Szczepankiewicz F, Chamberland M, Jones DK, et al. Computing and visualising intra‐voxel orientation‐specific relaxation–diffusion features in the human brain. Hum Brain Mapp. 2020. 10.1002/hbm.25224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. de Almeida Martins JP. Pushing diffusion MRI towards new dimensions. [PhD thesis]. Lund, Sweden: Lund University, Faculty of Science, Department of Chemistry, Division of Physical Chemistry; 2020. [Google Scholar]
- 77. Reymbaut A, de Almeida Martins JP, Tax CMW, Szczepankiewicz F, Jones DK, Topgaard D. Resolving orientation‐specific diffusion‐relaxation features via Monte‐Carlo density‐peak clustering in heterogeneous brain tissue. arXiv e‐prints, 2020.
- 78. Liu H, Rubino C, Dvorak AV, et al. Myelin water atlas: a template for myelin distribution in the brain. J Neuroimaging. 2019;29:699‐706. [DOI] [PubMed] [Google Scholar]
- 79. Heuvel VDMP, Mandl RCW, Stam CJ, Kahn RS, Hulshoff Pol HE. Aberrant frontal and temporal complex network structure in schizophrenia: a graph theoretical analysis. J Neurosci. 2010;30:15915‐15926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Caeyenberghs K, Metzler‐Baddeley C, Foley S, Jones DK. Dynamics of the human structural connectome underlying working memory training. J Neurosci. 2016;36:4056‐4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mancini M, Giulietti G, Dowell N, et al. Introducing axonal myelination in connectomics: a preliminary analysis of g‐ratio distribution in healthy subjects. NeuroImage. 2018;182:351‐359. [DOI] [PubMed] [Google Scholar]
- 82. Haase A, Frahm J, Hanicke W, Matthaei D. 1H NMR chemical shift selective (CHESS) imaging. Phys Med Biol. 1985;30:341‐344. [DOI] [PubMed] [Google Scholar]
- 83. Szczepankiewicz F, Sjölund J, Ståhlberg F, Lätt J, Nilsson M. Tensor‐valued diffusion encoding for diffusional variance decomposition (DIVIDE): technical feasibility in clinical MRI systems. PLoS ONE. 2019;14:1‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sjölund J, Szczepankiewicz F, Nilsson M, Topgaard D, Westin CF, Knutsson H. Constrained optimization of gradient waveforms for generalized diffusion encoding. J Magn Reson. 2015;261:157‐168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Szczepankiewicz F, Westin CF, Nilsson M. Maxwell‐compensated design of asymmetric gradient waveforms for tensor‐valued diffusion encoding. Magn Reson Med. 2019;82:1424‐1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Callaghan PT, Stepisnik J. Frequency‐domain analysis of spin motion using modulated‐gradient NMR. J Magn Reson Ser A. 1995;117:118‐122. [Google Scholar]
- 87. Lundell H, Nilsson M, Dyrby TB, et al. Multidimensional diffusion MRI with spectrally modulated gradients reveals unprecedented microstructural detail. Sci Rep. 2019;9:9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Szczepankiewicz F, Westin CF, Nilsson M. Gradient waveform design for tensor‐valued encoding in diffusion MRI. ArXiv e‐prints. 2020. [DOI] [PMC free article] [PubMed]
- 89. Yang G, McNab JA. Eddy current nulled constrained optimization of isotropic diffusion encoding gradient waveforms. Magn Reson Med. 2019;81:1818‐1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Haeberlen U. High Resolution NMR in Solids: Selective Averaging. New York: Academic Press; 1976. [Google Scholar]
- 91. Conturo TE, McKinstry RC, Akbudak E, Robinson BH. Encoding of anisotropic diffusion with tetrahedral gradients: a general mathematical diffusion formalism and experimental results. Magn Reson Med. 1996;35:399‐412. [DOI] [PubMed] [Google Scholar]
- 92. Perman W, Hilal S, Simon H, Maudsley A. Contrast manipulation in NMR imaging. Magn Reson Imaging. 1984;2:23‐32. Second Annual Meeting of the Society for Magnetic Resonance Imaging. [DOI] [PubMed] [Google Scholar]
- 93. Lawson CL, Hanson RJ. Solving least squares problems. Classics in Applied Mathematics. Philadelphia, USA: Soc Indust Appl Math; 1995. [Google Scholar]
- 94. English AE, Whittall KP, Joy MLG, Henkelman RM. Quantitative two‐dimensional time correlation relaxometry. Magn Reson Med. 1991;22:425‐434. [DOI] [PubMed] [Google Scholar]
- 95. Topgaard D. Diffusion tensor distribution imaging. NMR Biomed. 2019;32:e4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Efron B. Bootstrap methods: another look at the jackknife. Ann Statist. 1979;7:1‐26. [Google Scholar]
- 97. Kort DDW, Duynhoven VJPM, Hoeben FJM, Janssen HM, Van As H. NMR nanoparticle diffusometry in hydrogels: enhancing sensitivity and selectivity. Anal Chem. 2014;86:9229‐9235. [DOI] [PubMed] [Google Scholar]
- 98. Slator P, Hutter J, Marinescu R, et al. InSpect: iNtegrated SPECTral component estimation and mapping for multi‐contrast microstructural MRI In: Bao S., Gee J., Yushkevich P., Chung A. (Eds.), Information Processing in Medical Imaging–26th International Conference, IPMI 2019. Proceedings–Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). Information Processing in Medical Imaging. Vol. 11492 Cham: Springer; 2019:755‐766. [Google Scholar]
- 99. Rodriguez A, Laio A. Clustering by fast search and find of density peaks. Science. 2014;344:1492‐1496. [DOI] [PubMed] [Google Scholar]
- 100. Wolff SD, Balaban RS. Magnetization transfer contrast (MTC) and tissue water proton relaxation in vivo. Magn Reson Med. 1989;10:135‐144. [DOI] [PubMed] [Google Scholar]
- 101. Teixeira RP, Malik SJ, Hajnal JV. Fast quantitative MRI using controlled saturation magnetization transfer. Magn Reson Med. 2019;81:907‐920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Teixeira RP, Neji R, Wood TC, Baburamani AA, Malik SJ, Hajnal JV. Controlled saturation magnetization transfer for reproducible multivendor variable flip angle T 1 and T 2 mapping. Magn Reson Med. 2020;84:221‐236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Henkelman RM, Stanisz GJ, Kim JK, Bronskill MJ. Anisotropy of NMR properties of tissues. Magn Reson Med. 1994;32:592‐601. [DOI] [PubMed] [Google Scholar]
- 104. Knight MJ, Damion RA, Kauppinen RA. Observation of angular dependence of T 1 in the human white matter at 3T. Biomed Spectroscopy Imaging. 2018;7:125‐133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Daducci A, Dal Palú A, Descoteaux M, Thiran JP. Microstructure informed tractography: pitfalls and open challenges. Front Neurosci. 2016;10:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Girard G, Daducci A, Petit L, et al. AxTract: toward microstructure informed tractography. Hum Brain Mapp. 2017;38:5485‐5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Schiavi S, Ocampo‐Pineda M, Barakovic M, et al. A new method for accurate in vivo mapping of human brain connections using microstructural and anatomical information. Sci Adv. 2020;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Song YQ, Venkataramanan L, Burcaw L. Determining the resolution of Laplace inversion spectrum. J Chem Phys. 2005;122:104104. [DOI] [PubMed] [Google Scholar]
- 109. Bates A, Daducci A, Sadeghi P, Caruyer E. A 4D basis and sampling scheme for the tensor encoded multi‐dimensional diffusion MRI signal. IEEE Signal Process Lett. 2020;27:790‐794. [Google Scholar]
- 110. Song YQ, Xiao L. Optimization of multidimensional MR data acquisition for relaxation and diffusion. NMR Biomed. 2020;33:e4238. [DOI] [PubMed] [Google Scholar]
- 111. Mori S, Crain BJ, Chacko VP, Van Zijl PCM. Three‐dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann Neurol. 1999;45:265‐269. [DOI] [PubMed] [Google Scholar]
- 112. Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A. In vivo fiber tractography using DT‐MRI data. Magn Reson Med. 2000;44:625‐632. [DOI] [PubMed] [Google Scholar]
- 113. Morris DM, Embleton KV, Parker GJ. Probabilistic fibre tracking: differentiation of connections from chance events. NeuroImage. 2008;42:1329‐1339. [DOI] [PubMed] [Google Scholar]
- 114. Reisert M, Mader I, Anastasopoulos C, Weigel M, Schnell S, Kiselev V. Global fiber reconstruction becomes practical. NeuroImage. 2011;54:955‐962. [DOI] [PubMed] [Google Scholar]
- 115. Christiaens D, Reisert M, Dhollander T, Sunaert S, Suetens P, Maes F. Global tractography of multi‐shell diffusion‐weighted imaging data using a multi‐tissue model. NeuroImage. 2015;123:89‐101. [DOI] [PubMed] [Google Scholar]
- 116. Neher PF, Côté MA, Houde JC, Descoteaux M, Maier‐Hein KH. Fiber tractography using machine learning. NeuroImage. 2017;158:417‐429. [DOI] [PubMed] [Google Scholar]
- 117. Konopleva L, Il’yasov KA, Skibbe H, et al. Modelfree global tractography. NeuroImage. 2018;174:576‐586. [DOI] [PubMed] [Google Scholar]
- 118. Poulin P, Jörgens D, Jodoin PM, Descoteaux M. Tractography and machine learning: current state and open challenges. Magn Reson Imaging. 2019;64:37‐48. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Subvoxel orientations retrieved for the in silico data described in Section S3 using the Monte Carlo inversion for various numbers of bootstrap solutions and various SNR levels. While the ODFs were obtained via the process detailed in Section S1, the orientational clusters, here represented on the unit sphere, were extracted via MC‐DPC according to Section S2. denotes the angular deviation, computed for a given orientational cluster as the shortest angle between either the cluster geometric median orientation (circles, see Equation S7) or the corresponding ODF peak (squares, see Section S1), and the closest ground‐truth anisotropic component orientation. The color mapped onto the ODF codes for local orientation according to [red, green, blue] ≡ [|x|, |y|, |z|]/max([|x|, |y|, |z|]). As for the clusters, while opacity codes for the weight of the intra‐cluster averaged components (see Equation S6), color codes for the geometric median orientation of each cluster (see Equation S7). The conditions of the in vivo study presented in the main body of the paper are closest to the case FIGURE S2 Orientation‐resolved means [χ] (see Equation S5) and weights (see Equation S6) associated with the MC‐DPC clusters of Figure S1. While ground truth is shown as horizontal lines, the circles and whiskers represent the medians and interquartile ranges of the orientation‐resolved means across bootstrap solutions, respectively. Squares correspond to the estimated ODF‐peak metrics. Colors match those of the orientational clusters/ODF peaks presented in Figure S1. In the rightmost panels, cluster weights were normalized so that the sum of all median weights across clusters equals one. Their ODF‐peak equivalents were simply obtained by taking the mesh‐projected component weights (ie, ODF radii) along the peaks of a given ODF (see Section S1). These ODF‐peak weights were then normalized to sum up to one, for easier comparison with normalized cluster weights. The conditions of the in vivo study presented in the main body of the paper are closest to the case
Data Availability Statement
The code and in silico data that support the findings of this study are openly available in a GitHub repository at https://github.com/areymbaut (last accessed: 22nd of November 2020).


