Abstract
G‐quadruplex (G4), a non‐canonical higher‐order structure formed by guanine‐rich nucleic acid sequences, affects various genetic events in cis, including replication, transcription and translation. Whereas up‐regulation of innate immune/interferon‐stimulated genes (ISGs) is implicated in cancer progression, G4‐forming oligonucleotides that mimic telomeric repeat‐containing RNA suppress ISG induction in three‐dimensional (3D) culture of cancer cells. However, it is unclear how G4 suppresses ISG expression in trans. In this study, we found that G4 binding to splicing factor 3B subunit 2 (SF3B2) down‐regulated STAT1 phosphorylation and ISG expression in 3D‐cultured cancer cells. Liquid chromatography‐tandem mass spectrometry analysis identified SF3B2 as a G4‐binding protein. Either G4‐forming oligonucleotides or SF3B2 knockdown suppressed ISG induction, whereas Phen‐DC3, a G4‐stabilizing compound, reversed the inhibitory effect of G4‐forming oligonucleotides on ISG induction. Phen‐DC3 inhibited SF3B2 binding to G4 in vitro. SF3B2‐mediated ISG induction appeared to occur independently of RNA splicing because SF3B2 knockdown did not affect pre‐mRNA splicing under the experimental conditions, and pharmacological inhibition of splicing by pladienolide B did not repress ISG induction. These observations suggest that G4 disrupts the ability of SF3B2 to induce ISGs in cancer. We propose a new mode for gene regulation, which employs G4 as an inhibitory trans‐element.
Keywords: cancer, G‐quadruplex, innate immune gene, interferon‐stimulated gene, SF3B2, splicing factor, telomere
We demonstrate that a splicing factor SF3B2 mediates interferon‐stimulated gene (ISG) expression in cancer cells. G‐quadruplex‐forming oligonucleotides bind to SF3B2 and suppress ISG up‐regulation. The findings provide a mechanism for the function of G‐quadruplex as a trans‐element of gene regulation.
![]()
1. INTRODUCTION
DNA typically forms a right‐handed double helix under physiological conditions, whereas it can also form non‐canonical higher‐order structures such as a G‐quadruplex (G4; Sen & Gilbert, 1988). G4s are formed by guanine‐rich single‐stranded DNA and RNA sequences, GnNxGnNyGnNzGn, in which four guanine residues are held together by Hoogsteen hydrogen bonds in the same plane to form a G‐tetrad (also called a G‐quartet), and π–π stacking of at least two G‐tetrads constitutes a G4 (Zhang et al., 2014). A monovalent cation, such as K+ and Na+, is located at the center of a G‐tetrad and stabilizes G4. Chromatin‐immunoprecipitation‐seq analysis with a G4‐specific antibody has indicated that there are about 10,000 G4s in human chromatin in a genome‐wide manner (Hänsel‐Hertsch et al., 2016). In particular, G4 is present in highly transcribed genetic loci such those in the promoters and 5′‐untranslated regions of cancer‐associated genes. A small molecule that stabilizes G4 (G4 ligand) in the promoter region of the c‐MYC oncogene exerts an anticancer effect (Siddiqui‐Jain et al., 2002). G4 ligands also induce DNA damage response in telomeres because telomeric TTAGGG repeats are the most typical G4‐forming sequence in the genome (Gomez et al., 2006; Hasegawa et al., 2016; Miyazaki et al., 2012; Nakamura et al., 2017; Tahara et al., 2006).
Telomeres are specialized structures at the ends of linear chromosomes in eukaryotes (Seimiya, 2020). Telomeric loop structures formed by self‐strand invasion are required to protect chromosome ends from being recognized as DNA strand breaks. However, in human somatic cells, telomeres gradually shorten with each cell division because of the so‐called end replication problem. Eventually, excessive telomere shortening disrupts the loop structure and fails to protect the chromosome ends from the DNA damage response, which results in cellular senescent or apoptosis. This is one of the fundamental mechanisms for tumor suppression (Feldser & Greider, 2007; Greenberg et al., 1999). Conversely, one of the hallmarks of cancer is the limitless reproductive potential of the cells (Hanahan & Weinberg, 2000). Mechanistically, most human cancer cells activate the telomere‐synthesizing enzyme telomerase through gain‐of‐function mutations, methylation of the promoter or amplification of the catalytic subunit TERT, which maintain their telomeres to overcome the end replication problem (Seimiya, 2020).
Considering that cancer cells with shorter telomeres are more susceptible to the deleterious effect of telomerase inhibitors (Chiappori et al., 2015; Fujiwara et al., 2018), it would be expected that longer telomeres are more advantageous for the cell growth and survival. Intriguingly, however, cancer cells often retain shorter telomeres than normal cells (Barthel et al., 2017; Sommerfeld et al., 1996). One possible reason for this paradoxical phenomenon is that short telomeres tend to cause genomic instability, which would be advantageous for cancer evolution (Artandi et al., 2000; Okamoto & Seimiya, 2019). To further address the rationale of telomere shortening in cancer, we have previously elongated telomeres in various human cancer cell lines using a Cre‐loxP on/off system for exogenous TERT expression (Hirashima et al., 2013). The study revealed that telomere elongation in cancer cells causes tumor tissue reorganization with duct‐like structure formation in mouse xenograft models. Furthermore, compared with the parental cells, telomere‐elongated cancer cells exhibit altered gene expression in a genome‐wide manner. Particularly in three‐dimensional (3D) cultures, which mimic tumor microenvironment, or in xenografted tumors of parental cancer cells, innate immune/interferon‐stimulated genes (ISGs) are up‐regulated, such as ISG15, STAT1 and OAS3, whereas telomere‐elongated tumors fail to induce these genes (Hirashima et al., 2013; Hirashima & Seimiya, 2015).
The ubiquitin family protein ISG15 is a critical component of the host defense system against viral infection (Morales & Lenschow, 2013), but it is overexpressed in various cancers and contributes to cancer malignancy (Hirashima et al., 2013). In the tumor microenvironment of pancreatic ductal adenocarcinoma, secreted ISG15 enhances the cancer stem cell phenotype (Alcala et al., 2020). These observations suggest that shorter telomeres allow ISG15 induction, which is involved in tumor progression. Mechanistically, telomere‐elongated cells increase UUAGGG tracts in telomeric repeat‐containing RNA (TERRA), a long non‐coding RNA transcribed from telomeres (Hirashima & Seimiya, 2015). Because TERRA can form G4s and a TERRA‐mimicking oligonucleotide and another G4‐forming oligonucleotide suppress ISG expression in 3D‐cultured cancer cells (Hirashima & Seimiya, 2015), telomere‐elongated tumors may repress ISG expression through TERRA G4. In fact, transcriptome analysis has revealed that telomere‐elongated tumors and 3D‐cultured cancer cells treated with G4‐forming oligonucleotides commonly down‐regulate ISG expression. Nevertheless, how G4‐forming oligonucleotides suppress ISG expression remains elusive.
In this study, we found that G4‐forming oligonucleotides bind SF3B2, subunit 2 of the splicing factor 3b protein complex. Our results show that SF3B2 is required for ISG induction in 3D‐cultured cancer cells, and SF3B2 binding to G4‐forming oligonucleotides is associated with loss of ISG induction. These observations provide new insights into the biological functions of G4, especially as a trans‐element for gene suppression, in cancer.
2. RESULTS
2.1. G4‐forming oligonucleotides inhibit STAT1 activation and ISG induction
We have previously demonstrated that telomeric G‐strand and AS1411 (Girvan et al., 2006) oligonucleotides, both of which can form G4, repress ISG induction in 3D‐cultured cancer cells (Hirashima & Seimiya, 2015). However, whether this gene repression is also induced by other G4‐forming oligonucleotides and whether the oligonucleotides need to be single stranded remain unknown. Therefore, we first investigated the effects of various oligonucleotides (Table 1) on ISG induction in 3D‐cultured PC3 prostate cancer cells. As shown in Figure 1a,b, the cells under non‐adherent 3D culture conditions (no oligo), which mimic tumor microenvironment, up‐regulated the expression for ISGs, such as ISG15, STAT1 and OAS3, 2 to 5 times compared with the two‐dimensional (2D) culture conditions. This gene up‐regulation was significantly inhibited by transfection of G4‐forming oligonucleotides, (ttaggg)4, and (ttaggg)25 (4 and 25 repeats of the telomeric sequence, respectively), hras‐1 G4 (derived from the HRAS promoter containing a G4‐forming sequence; Cogoi et al., 2014), and center G4 that can form a G4 consisting of two G‐tetrads in the center position of the oligonucleotide. Conversely, oligonucleotides that do not form G4, including (ttaccc)4, (ttaccc)25, hras‐1 non‐G4 (complementary sequence to hras‐1 G4) and center non‐G4, which has the same guanine content as center G4, did not repress ISG induction or repressed only marginally. Western blot analysis indicated that 3D culture of the cells induced activating phosphorylation at Tyr701 and Ser727 residues of STAT1, a master regulator of ISG expression and ISG15 up‐regulation at protein levels (Figure 1c). These responses were specifically inhibited by G4‐forming oligonucleotides, but not the control oligonucleotides, irrespective of the percentage of guanine contents or the oligonucleotide length. G4‐forming oligonucleotide‐mediated suppression of ISG15 induction was also observed in A549 lung adenocarcinoma cells at both RNA and protein levels (Figure S1).
Table 1.
Oligonucleotides used in this study

Guanine base (G) in magenta indicates a component of the G‐quartet in G4. G‐score was calculated by QGRS Mapper (https://bioinformatics.ramapo.edu/QGRS/index.php). TelG indicates (ttaggg)n telomere sequence.
The complementary strand has no G4‐forming sequence.
Figure 1.
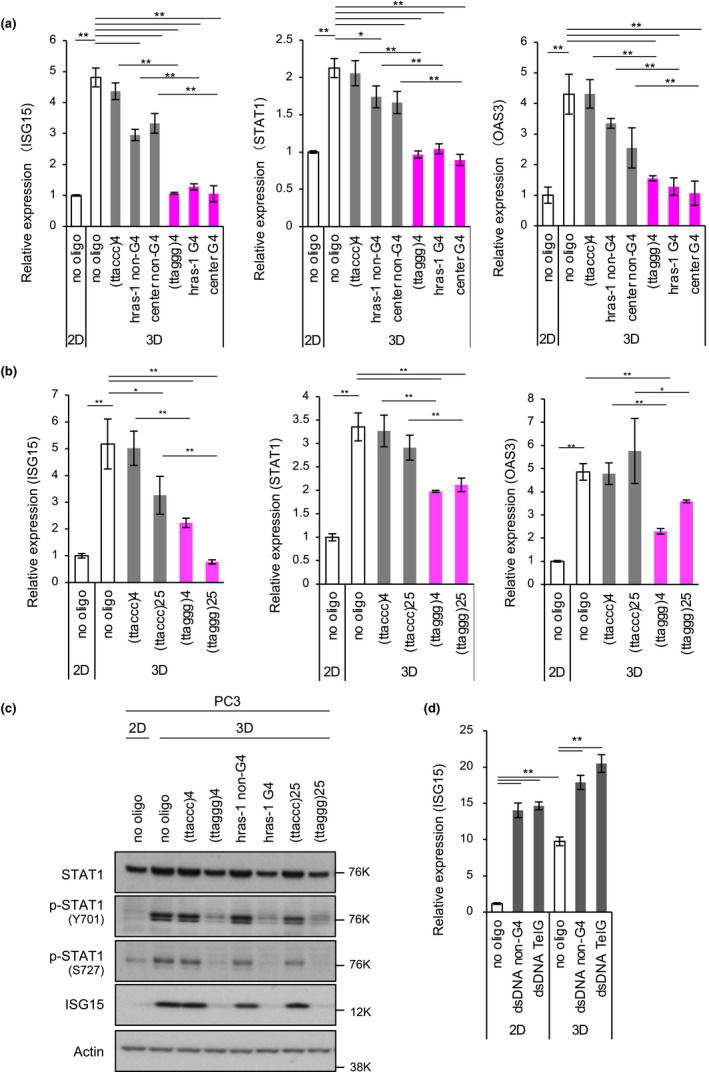
G4‐forming oligonucleotides repress STAT1 signaling in 3D‐cultured PC3 prostate cancer cells. (a) Effects of G4‐forming oligonucleotides on induction of interferon‐stimulated genes (ISGs) in 3D‐cultured PC3 cells. Cells were transfected with the indicated oligonucleotides and incubated under 2D or 3D conditions for 72 hr. Expression levels of ISG15, STAT1 and OAS3 mRNAs were analyzed by RT‐qPCR and normalized to GPI mRNA levels. A relative expression value of 1 was defined as expression in cells without oligonucleotides under 2D culture conditions. Error bars indicate standard deviation (n = 3). *p < .05, **p < .01 as determined using the Tukey–Kramer test. (b) Effects of different lengths of G4‐forming oligonucleotides on ISG induction. Cells were transfected with the indicated oligonucleotides and then subjected to RT‐qPCR as described in (a). (c) Effect of G4‐forming oligonucleotides on STAT1 signaling. PC3 cells were treated as described in (a), and then, Western blot analysis was performed with the indicated primary antibodies. (d) Effect of double‐stranded DNA (dsDNA) on ISG15 induction. PC3 cells were transfected with dsDNAs under 2D or 3D culture conditions for 72 hr and then subjected to RT‐qPCR to quantitate ISG15 expression. Error bars indicate standard deviation (n = 3). *p < .05, **p < .01 as determined using the Tukey–Kramer test
Because only single‐stranded nucleic acids can form G4, double‐stranded DNA (dsDNA) may not repress ISG expression. To test this hypothesis, we used dsDNA sequences with G4‐forming telomeric or non‐G4 sequences. As expected, dsDNA regardless of the sequence did not suppress ISG15 expression (Figure 1d). Conversely, these DNA fragments significantly induced ISG15 expression under the 2D culture conditions and to lesser extents under the 3D culture conditions, where were consistent with the previous observations that cytoplasmic dsDNA causes ISG15 accumulation via the cGAS‐STING pathway (Bianco & Mohr, 2017). These observations indicate that single‐stranded G4‐forming oligonucleotides selectively inhibit STAT1 activation and ISG induction in 3D‐cultured cancer cells.
2.2. SF3B2 binds to G4‐forming oligonucleotides and is required for ISG15 induction in 3D‐cultured PC3 cells
To gain insights into the molecular mechanism for G4‐forming oligonucleotide‐mediated suppression of ISG induction, we next analyzed proteins that bound to G4‐forming oligonucleotides. Whole A549 cell extracts were incubated with the biotin‐conjugated G4‐forming oligonucleotide, TelG [(ttaggg)12, total G‐score for G4 formation = 84, calculated by QGRS Mapper (max length: 30, min G‐group size: 2, loop size: from 0 to 36; https://bioinformatics.ramapo.edu/QGRS/index.php)]. As the second G4‐forming oligonucleotide, we also used a biotin‐conjugated non‐telomeric sequence, AS1411 [(GGTGGTGGTGGTTGTGGTGGTGGTGG)3, total G‐score = 126], which represses ISG induction in 3D‐cultured cancer cells (Hirashima & Seimiya, 2015). Furthermore, biotin‐conjugated control oligonucleotide [(GCACGCGTATCTTTGGCGCAGGTG)3, total G‐score = 18] was used in the experiments. These oligonucleotides were incubated with the whole cell extracts and pulled down with avidin beads. Co‐precipitated proteins were then analyzed by liquid chromatography‐tandem mass (LC‐MS/MS) spectrometry. Table 2 shows the proteins that preferentially bound to TelG and AS1411 oligonucleotides. They included well‐known G4‐binding proteins, such as heterogeneous nuclear ribonucleoprotein components (hnRNPA2/B1 and FUS), RNA helicases (DDX17 and DDX1) and an RNA‐binding protein (EWSR1). Various ribosomal proteins (RPL36L, RPS18, RPL17, RPS23, RPS15, RPS9, RPL27A, RPL21 and RPS10) were also identified. Gene ontology (GO) analysis of these proteins identified RNA processing‐related terms such as mRNA metabolic process [false discovery rate (FDR) = 1.9 × 10−16], RNA catabolic process (FDR = 1.5 × 10−12) and RNA splicing (FDR = 2.2 × 10−8; Figure 2a). Furthermore, STRING analysis categorized these proteins into four blocks associated with (a) translational initiation and ribosome, (b) mRNA splicing via spliceosome, (c) tRNA‐splicing ligase complex and (d) nuclear speckle (Figure 2b).
Table 2.
G4‐binding proteins identified by LC‐MS/MS spectrometry
| Accession no. | Gene | Description | TelG | AS1411 | ||||
|---|---|---|---|---|---|---|---|---|
| Coverage (%) | Peptides | Area | Coverage (%) | Peptides | Area | |||
| P22626 | HNRNPA2B1 | Heterogeneous nuclear ribonucleoproteins A2/B1 | 80.17 | 45 | 5.30E + 08 | 76.49 | 32 | 5.34E + 07 |
| P35637 | FUS | RNA‐binding protein FUS | 24.90 | 12 | 2.82E + 07 | 26.05 | 14 | 4.45E + 07 |
| P31942 | HNRNPH3 | Heterogeneous nuclear ribonucleoprotein H3 | 62.14 | 14 | 1.62E + 07 | 63.01 | 15 | 1.22E + 07 |
| P84090 | ERH | Enhancer of rudimentary homolog | 40.38 | 4 | 1.05E + 07 | 45.19 | 5 | 6.30E + 06 |
| Q9Y2W1 | THRAP3 | Thyroid hormone receptor‐associated protein 3 | 21.68 | 17 | 3.26E + 06 | 34.66 | 36 | 8.47E + 06 |
| Q9Y3I0 | RTCB | tRNA‐splicing ligase RtcB homolog | 31.09 | 10 | 3.17E + 06 | 42.18 | 19 | 6.90E + 06 |
| Q92841 | DDX17 | Probable ATP‐dependent RNA helicase DDX17 | 12.21 | 3 | 2.64E + 06 | 37.45 | 19 | 8.70E + 06 |
| Q13310 | PABPC4 | Polyadenylate‐binding protein 4 | 10.71 | 1 | 2.61E + 06 | 37.27 | 11 | 1.42E + 07 |
| Q7Z417 | NUFIP2 | Nuclear fragile × mental retardation‐interacting protein 2 | 17.41 | 9 | 2.59E + 06 | 54.53 | 36 | 9.23E + 06 |
| Q9BY77 | POLDIP3 | Polymerase delta‐interacting protein 3 | 42.76 | 11 | 2.46E + 06 | 53.44 | 18 | 5.07E + 06 |
| Q92499 | DDX1 | ATP‐dependent RNA helicase DDX1 | 19.59 | 9 | 2.28E + 06 | 46.08 | 29 | 4.83E + 06 |
| Q13435 | SF3B2 | Splicing factor 3B subunit 2 | 12.40 | 8 | 1.40E + 06 | 10.84 | 7 | 8.81E + 05 |
| Q9Y224 | RTRAF | UPF0568 protein C14orf166 | 45.90 | 7 | 1.26E + 06 | 58.61 | 13 | 2.71E + 06 |
| Q9NYF8 | BCLAF1 | Bcl‐2‐associated transcription factor 1 | 7.61 | 6 | 1.01E + 06 | 23.80 | 22 | 4.00E + 06 |
| P83881 | RPL36A | 60S ribosomal protein L36a | 12.26 | 2 | 9.58E + 05 | 22.64 | 4 | 5.93E + 05 |
| P62269 | RPS18 | 40S ribosomal protein S18 | 9.87 | 2 | 9.18E + 05 | 44.08 | 11 | 3.01E + 06 |
| Q01844 | EWSR1 | RNA‐binding protein EWS | 2.13 | 1 | 7.66E + 05 | 16.01 | 7 | 5.63E + 06 |
| Q99873 | PRMT1 | Protein arginine N‐methyltransferase 1 | 8.59 | 2 | 7.61E + 05 | 12.47 | 3 | 6.35E + 05 |
| P18621 | RPL17 | 60S ribosomal protein L17 | 5.98 | 1 | 7.60E + 05 | 30.98 | 7 | 2.49E + 06 |
| Q8NCA5 | FAM98A | Protein FAM98A | 2.31 | 1 | 6.11E + 05 | 16.76 | 6 | 1.39E + 06 |
| P62266 | RPS23 | 40S ribosomal protein S23 | 8.39 | 1 | 5.15E + 05 | 29.37 | 4 | 1.14E + 06 |
| P62841 | RPS15 | 40S ribosomal protein S15 | 33.79 | 2 | 3.95E + 05 | 36.55 | 4 | 4.48E + 05 |
| P46781 | RPS9 | 40S ribosomal protein S9 | 8.76 | 2 | 3.94E + 05 | 27.84 | 7 | 1.42E + 06 |
| P46776 | RPL27A | 60S ribosomal protein L27a | 7.43 | 1 | 3.71E + 05 | 25.00 | 4 | 9.00E + 05 |
| P46778 | RPL21 | 60S ribosomal protein L21 | 9.38 | 1 | 3.07E + 05 | 25.00 | 3 | 5.65E + 05 |
| P46783 | RPS10 | 40S ribosomal protein S10 | 5.45 | 1 | 2.40E + 05 | 5.45 | 1 | 4.33E + 05 |
A549 cell lysates were incubated with the indicated G4‐forming oligonucleotides, and streptavidin bead‐bound proteins were identified as described in Section 4.6.
Figure 2.
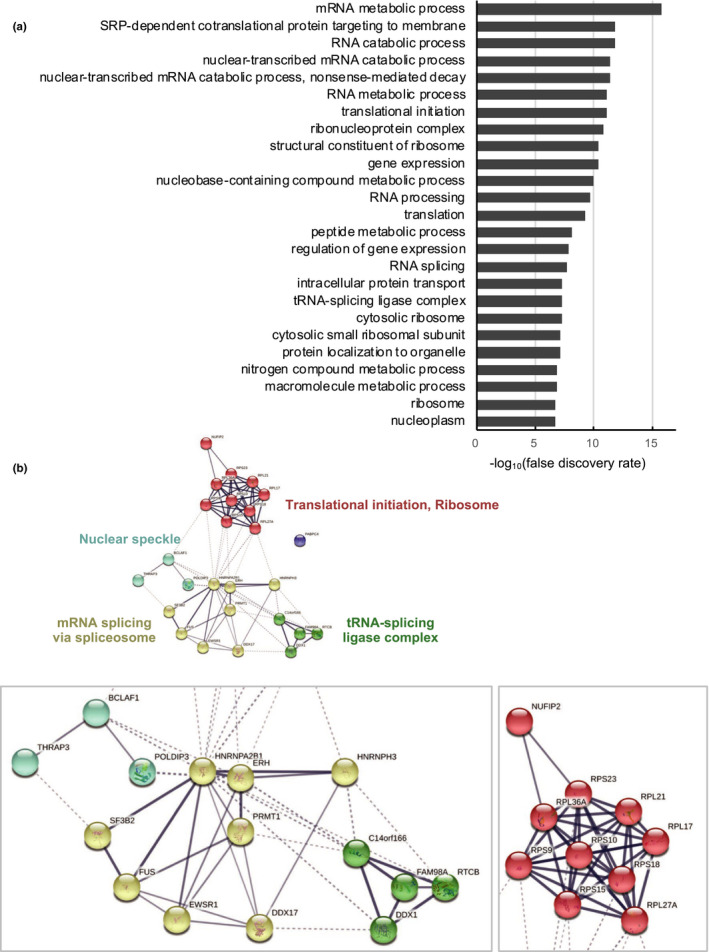
Gene ontology and clustering analyses of G4‐binding proteins. (a) Gene ontology analysis of genes listed in Table 2 by STRING (https://string‐db.org). Ontology terms with the 25 lowest false discover rates (FDR) in Biological Process, Molecular Function and Cellular Component are shown. (b) Genes listed in Table 2 were clustered by STRING with the k‐means clustering algorithm (numbers of cluster = 5). Line thickness indicates the strength of data support. The minimum required interaction score was 0.4. Active interaction sources include text mining, experiments, databases, coexpression, neighborhood, gene fusion and co‐occurrence
To examine whether these proteins affected ISG expression in 3D‐cultured PC3 cells, we transfected the cells with siRNAs against the genes listed in Table 2. After 72 hr of 3D culture, we evaluated the effect of each siRNA on ISG induction (K.M., K.O. and H.S., unpublished observations). As a result, we found that two different siRNAs for the splicing factor 3B subunit 2 (SF3B2; also known as SF3B145 or SAP145) suppressed ISG15 and STAT1 induction, although the efficiency of STAT1 suppression was not higher than that of ISG15 suppression (Figure 3a,b). Knockdown efficiencies of SF3B2 siRNAs were >80%, and introduction of (ttaccc)25 or (ttaggg)25 had no effect on SF3B2 expression (Figure 3a). Consistently, ISG15 protein accumulation in 3D‐cultured PC3 cells was also inhibited by SF3B2 siRNAs, although the reduction in STAT1 phosphorylation by siSF3B2 #2 was only marginal (Figure 3c). These observations indicate that SF3B2 is required for ISG induction in 3D‐cultured PC3 cells.
Figure 3.
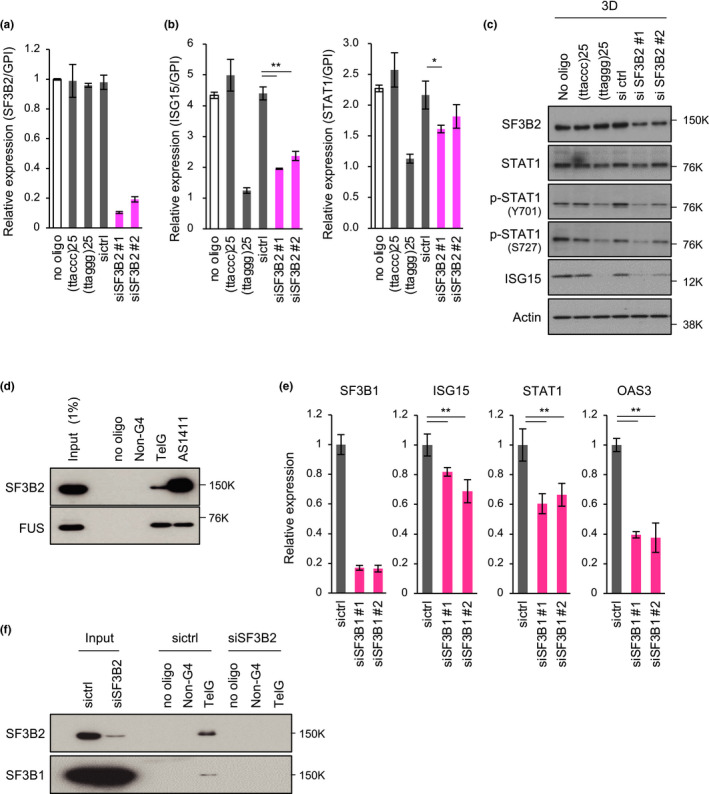
SF3B2 binds to G4 and promotes ISG induction in 3D‐cultured PC3 cells. (a) RT‐qPCR analysis to confirm knockdown efficiency of SF3B2. PC3 cells were transfected with the indicated oligonucleotides or siRNAs and cultured under 3D conditions for 72 hr. RNAs were prepared, and SF3B2 transcripts were quantitated by RT‐qPCR and normalized to GPI expression. Error bars indicate standard deviation (n = 3). (b) Effect of SF3B2 depletion on ISG induction. PC3 cells were transfected with the indicated oligonucleotides or siRNAs and cultured under 3D conditions for 72 hr. Expression levels of ISG15 and STAT1 mRNAs were analyzed by RT‐qPCR and normalized to GPI mRNA levels. Error bars indicate standard deviation (n = 3). *p < .05, **p < .01 as determined using the Tukey–Kramer test. (c) Western blot analysis of 3D‐cultured PC3 cells. Cells were treated as described in (b), and cell lysates were subjected to Western blotting with the indicated primary antibodies. (d) In vitro pull‐down assay. Whole PC3 cell extracts were incubated with biotin‐conjugated oligonucleotides and pulled down with Dynabeads M‐280 Streptavidin. The precipitated proteins were subjected to Western blot analysis. FUS served as a positive control of a G4‐binding protein. (e) Effect of SF3B1 depletion on ISG induction. Cells were transfected with siRNAs and cultured as described in (b). SF3B1 and ISG expression was measured by RT‐qPCR analysis. (f) In vitro pull‐down assay with lysates of SF3B2‐depleted cells. Cells were transfected with the indicated siRNAs, and cell lysates were subjected to Western blot analysis of SF3B2 and SF3B1
To validate the binding of SF3B2 to G4‐forming oligonucleotides, we performed a pull‐down assay of whole PC3 cell extracts with the biotin‐conjugated oligonucleotides. As shown in Figure 3d, SF3B2 was pulled down by TelG and AS1411, but not the control oligonucleotide without the G4‐forming sequence [(GCACGCGTATCGTTGCGCAGTGTG)3, total G‐score = 0]. As a positive control of G4‐binding proteins, FUS also bound to these G4‐forming oligonucleotides.
SF3B2 is a component of U2 small nuclear ribonucleoprotein particle (snRNP) and forms a complex with SF3B1. Therefore, we examined whether SF3B1 knockdown also affected ISG expression. As shown in Figure 3e, SF3B1 was efficiently knocked down by two different siRNAs that caused repression of ISG induction in 3D‐cultured PC3 cells. Although our LC‐MS/MS analysis did not identify SF3B1 as a G4‐binding protein (Table 2), we found marginal binding of SF3B1 to the TelG oligonucleotide (Figure 3f). When lysates of SF3B2‐depleted cells were pulled down, TelG‐bound SF3B1 was not detected, which suggested that SF3B1 indirectly bound to G4 thorough an interaction with SF3B2. Taken together, these observations suggest that SF3B2 mediates ISG induction in 3D‐cultured PC3 cells and that G4‐forming oligonucleotides inhibit ISG induction through an interaction with SF3B2.
2.3. Phen‐DC3, a G4 ligand, prevents G4 binding to SF3B2 and promotes ISG15 expression
To investigate whether the interaction between SF3B2 and G4 was involved in repression of ISG induction, we next examined the effect of Phen‐DC3 (De Cian et al., 2007), a G4 ligand, on ISG15 expression in 3D‐cultured PC3 cells. As shown in Figure 4a, whereas (ttaggg)4 oligonucleotide inhibited ISG15 induction upon 3D culture, treatment with Phen‐DC3 at 1 or 3 µM for 72 hr enhanced ISG15 expression compared with DMSO treatment. Because G4 stabilization in the telomeric G‐strand prevents telomerase‐mediated telomere synthesis, G4 ligands have a telomerase‐inhibitory activity. BIBR1532, which inhibits telomerase activity but does not stabilize G4 (Pascolo et al., 2002), did not affect ISG15 expression. These observations suggest that Phen‐DC3‐mediated enhancement of ISG15 expression is caused by G4 stabilization, but not telomerase inhibition. Importantly, (ttaggg)4 oligonucleotide‐mediated repression of ISG15 expression was reversed by Phen‐DC3 in a dose‐dependent manner (Figure 4b). These results prompted us to speculate that Phen‐DC3 inhibits G4 binding with SF3B2. To test this hypothesis, we performed a pull‐down assay of SF3B2 with the G4‐forming oligonucleotide. The biotin‐conjugated (ttaggg)12 oligonucleotide was incubated by either DMSO or Phen‐DC3 (4 µM) and mixed with the whole PC3 cell extract. Then, the oligonucleotide‐bound proteins were subjected to Western blot analysis. As shown in Figure 4c, Phen‐DC3 inhibited SF3B2 binding to the G4‐forming oligonucleotide. These observations suggest that G4 binds to SF3B2 and blocks the ability of SF3B2 to promote ISG15 expression. Furthermore, Phen‐DC3 might disrupt the SF3B2‐G4 interaction, which results in derepression of SF3B2 functions for ISG15 expression.
Figure 4.
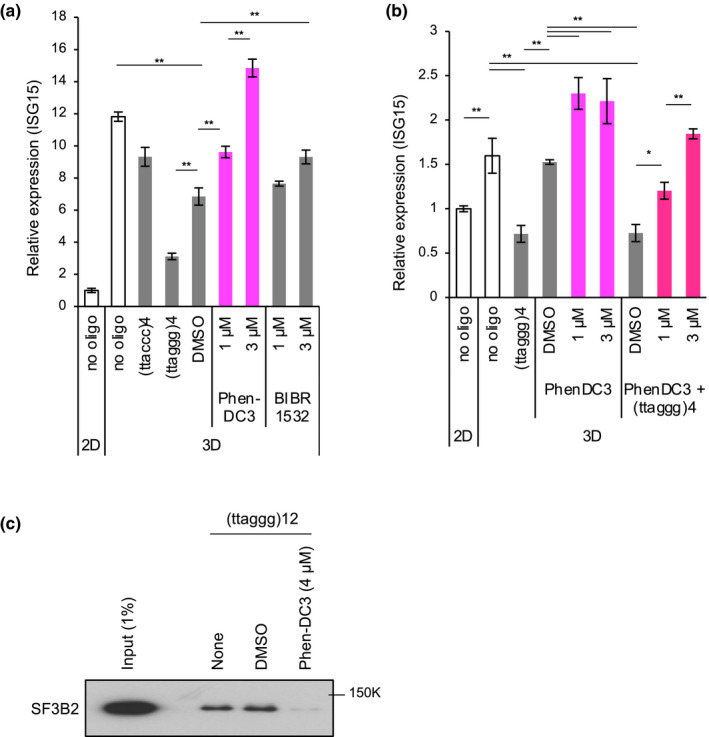
G4 ligand counteracts G4‐forming oligonucleotides and enhances ISG15 induction. (a) PC3 cells were transfected with oligonucleotides or treated with Phen‐DC3 or BIBR1532 for 72 hr under 3D culture conditions. Then, RT‐qPCR was performed to quantitate ISG15 expression. The data were normalized to GPI mRNA levels. A relative expression value of 1 was defined as the expression in cells cultured under 2D conditions. Error bar indicates standard deviation (n = 3). **p < .01 as determined by the Tukey–Kramer test. (b) Effect of Phen‐DC3 on G4‐forming oligonucleotide‐mediated repression of ISG15 induction. PC3 cells were treated with Phen‐DC3 in the presence or absence of (ttaggg)4 oligonucleotide. After 72 hr, RNAs were prepared and subjected to RT‐qPCR as described in (a). (c) Effect of Phen‐DC3 on the interaction of SF3B2 with G4‐forming oligonucleotide. Biotin‐conjugated (ttaggg)12 was incubated with whole PC3 cell extracts in the presence of DMSO or Phen‐DC3 and pulled down by Dynabeads M‐280 Streptavidin. The bead‐bound proteins were subjected to Western blot analysis of SF3B2
2.4. SF3B2 knockdown suppresses ISG expression in a splicing‐independent manner
Because SF3B2 is a component of the U2 snRNP subcomplex in the spliceosome, it is possible that ISG suppression by SF3B2 knockdown is due to dysfunction of pre‐mRNA splicing. However, SF3B2 knockdown at least in our experimental conditions did not affect the splicing of DNAJB1 pre‐mRNA, which was assessed using the PCR‐based splicing detection assay (Figure 5a). We selected DNAJB1 because this gene was used in the splicing assay in a previous literature (Kotake et al., 2007). Furthermore, neither (ttaccc)25 nor (ttaggg)25 oligonucleotides inhibited the splicing. Therefore, we next examined whether pharmacological inhibition of pre‐mRNA splicing suppressed ISG expression in 3D‐cultured PC3 cells. The cells were treated for 6–24 hr with pladienolide B, a pre‐mRNA splicing inhibitor that targets the SF3b complex, and then, total RNAs were subjected to the splicing detection assay. As shown in Figure 5b, pladienolide B inhibited DNAJB1 pre‐mRNA splicing. After 24 hr of incubation with pladienolide B, 0.5 µM was sufficient for maximal accumulation of the unspliced transcript, although the spliced transcript was still detectable. Under these conditions of incomplete pre‐mRNA splicing, pladienolide B did not repress STAT1 phosphorylation or ISG15 accumulation (Figure 5c).
Figure 5.
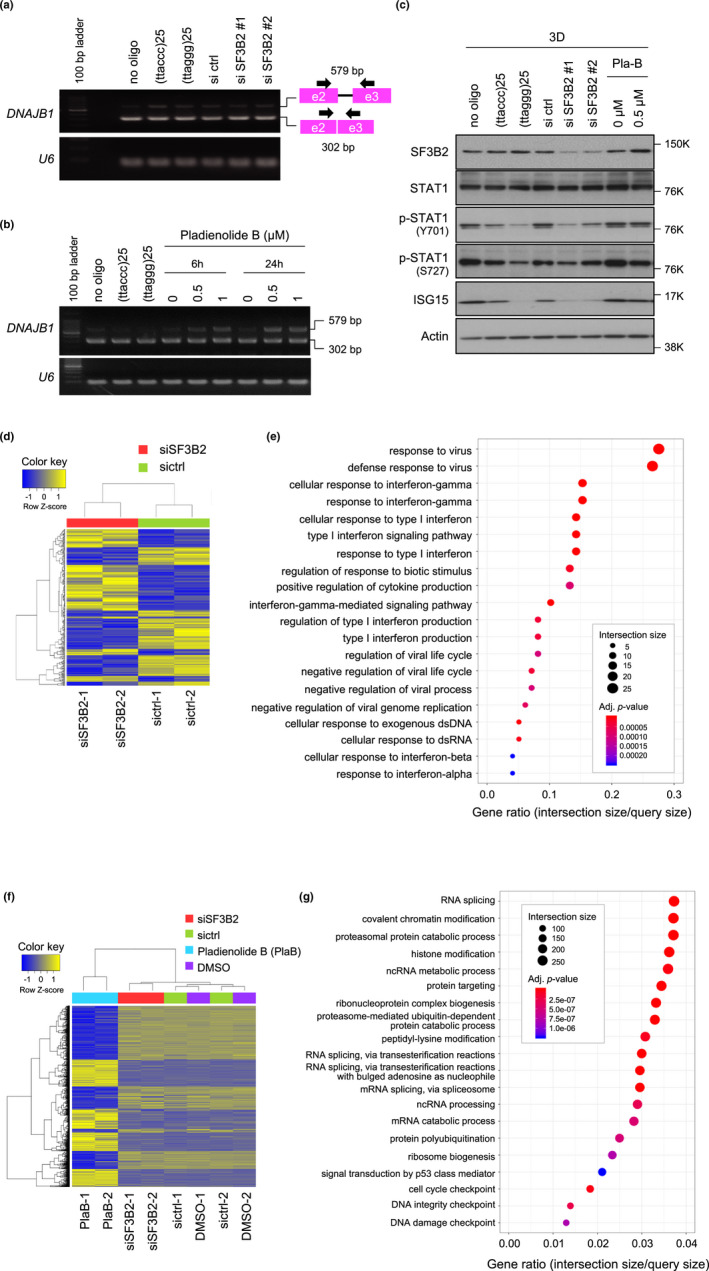
Pharmacological inhibition of RNA splicing is not associated with repression of STAT1 signaling. (a) Effect of SF3B2 knockdown on pre‐mRNA splicing. PC3 cells were transfected with SF3B2 siRNAs and cultured for 72 hr. RNAs were prepared, and RT‐PCR was performed to analyze the splicing state of DNAJB1 transcripts. Because the PCR primers were designed to span two exons, unspliced mRNA products were larger than the spliced mRNA. U6 RNA is shown as an internal control. Arrow and square indicate the PCR primer and exon, respectively. (b) Effect of pladienolide B on pre‐mRNA splicing. PC3 cells were treated with 0.5 or 1 µM pladienolide B for 6 or 24 hr. After 72 hr of culture, RT‐PCR was performed as described in (a). (c) Effect of pladienolide B on STAT1 signaling. PC3 cells were treated with the indicated concentrations of pladienolide B, and then, cell lysates were subjected to Western blot analysis with the indicated primary antibodies. (d) Hierarchical cluster analysis (Euclidean method, complete linkage) of RNA‐seq data from PC3 cells transfected with control or SF3B2 siRNAs. A heat map was drawn using 285 genes with a fold change of >2 and exactTest raw p‐value of <.05. (e) Gene ontology (GO) analysis of down‐regulated genes in SF3B2‐depleted cells in (d). The top 20 GO categories of biological processes identified by g:Profiler are indicated. (f) Hierarchical cluster analysis of RNA‐seq data from PC3 cells transfected with control or SF3B2 siRNAs or treated with 0.5 µM pladienolide B or DMSO for 24 hr. A heat map was drawn using the top 3,000 genes of 11,306 genes with a fold change of >2 and exactTest raw p‐value of <.05 listed in more than at least one of the comparison pairs: SF3B2 siRNA versus control siRNA; pladienolide B versus DMSO; SF3B2 siRNA versus pladienolide B. (g) GO analysis of genes with altered expression in pladienolide B‐treated PC3 cells in (f)
To further evaluate the difference between SF3B2 knockdown and pladienolide B treatment, we performed RNA‐seq analysis of the 3D‐cultured PC3 cells treated with SF3B2 siRNA or pladienolide B (0.5 µM, 24 hr). First, we compared the gene signatures of control siRNA‐ and SF3B2 siRNA‐treated cells (Figure 5d, |fc| >= 2, exactTest raw p‐value < .05). In particular, we focused on down‐regulated genes in SF3B2‐depleted cells. GO analysis identified GO terms related to innate immunity, such as the immune system process, defense response, response to other organisms, innate immune response, cellular response to type I interferon, and type I interferon signaling pathway (Figure 5e; Table S2). Of note, these GO terms were very similar to those related to the G4‐induced suppression signature (G4SS) that is commonly observed in G4‐forming oligonucleotide‐treated cells and xenografted tumors formed by telomere‐elongated cancer cells (i.e., cancer cells with up‐regulated TERRA signals; Hirashima & Seimiya, 2015). In fact, SF3B2 knockdown down‐regulated most G4SS genes (13 out of 18 genes) by more than 2‐fold, which included CMPK2, DDX58, DDX60, IFIT1, IFIT2, IFIT3, OAS2, RSAD2, USP18, IFI27, IFITM1, OAS1 and OAS3, although the levels of down‐regulation for seven genes were not statistically significant (Table S3). These observations support that G4‐forming oligonucleotides suppress the gene expression facilitated by SF3B2. Next, we analyzed the gene expression signature of pladienolide B‐treated cells (Figure 5f). GO analysis of genes with altered expression in pladienolide B‐treated cells identified GO terms related to RNA splicing, spliceosome and RNA processing (Figure 5g). Importantly, innate immunity‐related GO terms identified by down‐regulated genes in SF3B2‐depleted cells were not related to the down‐regulated genes in pladienolide B‐treated cells (Table S4). Taken together, these observations support that SF3B2 knockdown represses ISG expression in a splicing‐independent manner.
3. DISCUSSION
In the present study, we employed LC‐MS/MS spectrometry analysis and identified SF3B2 among G4‐binding proteins. Either G4‐forming oligonucleotides or SF3B2 depletion repressed ISG induction upon 3D culture of PC3 cells, whereas Phen‐DC3, a G4 ligand, blocked the G4‐SF3B2 interaction and abolished the G4‐forming oligonucleotide‐induced repression of ISG induction. These observations have revealed a novel mechanism for specific gene regulation in which G4‐forming nucleic acids are involved in trans.
In our previous study (Hirashima & Seimiya, 2015), we demonstrated that only two types of G4‐forming oligonucleotides, telomeric repeats (both DNA and RNA) and AS1411, suppress ISG induction in 3D‐cultured cancer cells. Here, we expanded these analyses with various types of oligonucleotides and confirmed that suppression of ISG induction was specifically mediated by G4‐forming oligonucleotides. Of note, dsDNA up‐regulated, but not down‐regulated, ISG15 expression under 2D and 3D culture conditions irrespective of the presence or absence of G4‐forming sequences. This would be caused by the cGAS‐STING cytosolic DNA sensing pathway that induces type I interferon production and consequential ISG expression for host defense (Cai et al., 2014; Wesoly et al., 2007). Conversely, the single‐stranded G4‐forming oligonucleotides did not affect STING phosphorylation at Ser366, a hallmark of STING inactivation (Konno et al., 2013; K.M., K.O. and H.S., unpublished observation). Our results demonstrate a unique feature of single‐stranded G4‐forming nucleic acids, which exert a completely opposite effect on ISG induction.
Among the G4‐binding proteins identified by LC‐MS/MS analysis, we focused on SF3B2 because its knockdown by different siRNAs suppressed ISG induction. It has been reported that G4 binds to the arginine‐ and glycine‐rich RGG motif of proteins (Kondo et al., 2018; Takahama et al., 2011). Consistent with this finding, we identified several G4‐binding proteins with RGG motifs, including FUS, DDX17 and EWSR1 (Table 2) (Thandapani et al., 2013). Although both SF3B2 and SF3B1 have one RGG sequence, these RGGs may not be sufficient for G4 binding because typical RGG motifs have multiple arginine and glycine residues (Thandapani et al., 2013; Vasilyev et al., 2015). Furthermore, SF3B3, which is complexed with SF3B2 (Das et al., 1999), has no RGG motif. Instead of the typical RGG motif, SF3B2 has the N‐terminal SAP domain, a potential DNA‐binding domain (Aravind & Koonin, 2000). Interestingly, we found that Phen‐DC3 inhibited SF3B2 binding to the G4‐forming oligonucleotide. Because Phen‐DC3 interacts with G4 thorough π stacking with the surface G‐tetrad (Chung et al., 2014), it is possible that SF3B2 interacts with the surface G‐tetrad of G4. Meanwhile, SF3B4, which directly interacts with SF3B2 (Champion‐Arnaud & Reed, 1994), has two N‐terminal RNA‐recognition motifs. Whether SF3B2 directly recognizes G4 or indirectly associates with G4 via other proteins remains to be determined.
Our RNA‐seq analysis demonstrated that SF3B2 depletion widely suppressed innate immune/ISGs under the conditions that did not significantly disrupt pre‐mRNA splicing (Figure 5a). Conversely, pladienolide B treatment under the conditions that disrupted pre‐mRNA splicing did not affect the innate immune/ISGs, which suggested that SF3B2 depletion down‐regulates these gene expression in a splicing‐independent manner. Because pladienolide B only partially inhibited the pre‐mRNA splicing (Figure 5b), it is possible that complete inhibition of pre‐mRNA splicing may repress STAT1 phosphorylation or ISG accumulation. However, it is difficult to examine this possibility because complete inhibition of pre‐mRNA splicing will cause significant cytotoxicity.
Recently, it has been reported that SF3B2 promotes androgen receptor splice variant‐7 (AR‐V7) expression in prostate cancer cells and correlates with aggressive phenotypes of the cancer (Kawamura et al., 2019). However, the PC3 cells used in this study do not express AR, and it is unlikely that SF3B2‐mediated ISG induction is caused by AR‐V7. Recently, Wang et al. reported that SF3b promotes mRNA export in a manner independent of the splicing complex of U2 snRNP (Wang et al., 2019). They demonstrated that knockdown of either SF3B1 or SF3B2 inhibits mRNA export from nuclei to the cytoplasm. In this study, we demonstrated that not only SF3B2, but also SF3B1 knockdown suppressed ISG expression (Figure 3e). It is of interest to speculate that G4 may inhibit mRNA export thorough SF3B2 dysfunction. However, even in this scenario, it remains unclear why G4 preferentially targets ISGs.
Originally, G4SS was supposed to emerge as a cancer cell response to excessive telomere elongation (Hirashima et al., 2013; Okamoto & Seimiya, 2019). Mechanistically, telomere elongation increases the number of TTAGGG repeats of telomeric DNA, which increases the number of UUAGGG repeats of TERRA (Hirashima & Seimiya, 2015). This means that telomere elongation mimics introduction of G4‐forming oligonucleotides, at least, in terms of G4 accumulation in the cells. In fact, GO terms associated with down‐regulated genes by telomere elongation are related to innate immune/defense responses, which is similar to those in G4‐forming oligonucleotide‐treated cells (Hirashima et al., 2013; Hirashima & Seimiya, 2015). Although we used DNA oligonucleotides in this study, we have demonstrated that TERRA‐mimicking G4‐forming RNA oligonucleotide, (uuaggg)4, also inhibits ISG up‐regulation in cancer cells (Hirashima & Seimiya, 2015). It is of interest to transfect or express longer TERRA RNA as a future experimental plan.
In general, innate immune genes/ISGs are important for host defense against viruses and other organisms. However, these genes are up‐regulated in many types of cancer and have bidirectional functions in carcinogenesis and their malignant progression. In the pathological aspect, the innate immune/defense response is implicated in the therapy resistance and metastatic potential of cancer (Khodarev et al., 2009; Weichselbaum et al., 2008). Furthermore, the low expression of innate immune genes/ISGs, such as STAT1, IFI44, ISG15, IFIT1, MX1, OAS1 and USP18, predicts a good prognosis of glioblastoma multiforme (Duarte et al., 2012). Considering that cancer cells often exhibit shorter telomeres than normal cells in spite of telomerase reactivation (Barthel et al., 2017; Chiba et al., 2017), it is possible that cancer cells keep their telomeres shorter to prevent accumulation of TERRA G4 and allow SF3B2‐mediated ISG expression.
In conclusion, our present findings suggest a novel function of G4 as an extrachromosomal modulator of SF3B2‐mediated gene expression (Figure 6). Although we used TERRA‐mimicking oligonucleotides in the experiments, it is possible that not only TERRA, but also other endogenous G4‐forming nucleic acids are involved in specific gene regulation in a similar fashion.
Figure 6.
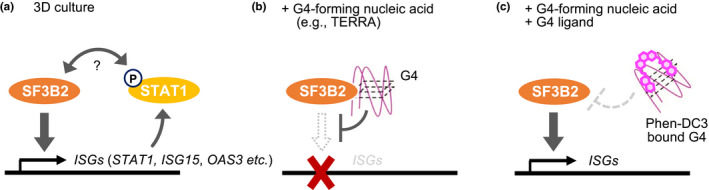
Schematic model for SF3B2‐mediated ISG induction and its inhibition by G4‐forming oligonucleotides. (a) Under the tumor‐mimicking 3D culture conditions, SF3B2 induces ISG expression through a mechanism that involves STAT1 phosphorylation. (b) G4‐forming nucleic acids, such as TERRA, bind to SF3B2 and repress the ISGs induction. (c) Phen‐DC3, a G4 ligand, binds to the G4‐forming nucleic acids, which in turn disrupts the interaction between SF3B2 and G4, resuming the ISG expression
4. EXPERIMENTAL PROCEDURES
4.1. Cell culture and chemicals
Human prostate cancer PC3 and lung cancer A549 cells were authenticated by short tandem repeat analysis (BEX) and maintained as described previously (Jang et al., 2020; Okamoto et al., 2018) in Dulbecco's modified Eagle's medium (DMEM; Nacalai Tesque) containing 10% heat‐inactivated fetal bovine serum (FBS) and 0.1 mg/ml kanamycin in a humidified atmosphere with 5% CO2. Cells were routinely tested for mycoplasma contamination by PCR with the primers 5′‐CACCATCTGTCACTCTGTTAACC‐3′ and 5′‐GGAGCAAACAGGATTAGATACCC‐3′. Pladienolide B, Phen‐DC3 and BIBR1532 were purchased from Santa Cruz Biotechnology, Polysciences and Selleck Chemicals, respectively.
4.2. Transfection of oligonucleotides, siRNAs and DNAs
Oligonucleotides (Table 1) were synthesized by Fasmac. We have already confirmed that more than four TTAGGG repeats and AS1411 form G4s (Hirashima & Seimiya, 2015; Nakamura et al., 2017; S.O. and H.S., unpublished observations). The center G4 oligonucleotide contains the same G4‐forming sequence as AS1411. G4 formation in hras‐1 oligonucleotide was previously reported (Cogoi et al., 2014; Membrino et al., 2011). Silencer select siRNAs [SF3B2 (#1: s21642, #2: s225464), SF3B1 (#1: s23851, #2: s223598) and negative control (#1)] were purchased from Thermo Fisher Scientific. Double‐stranded DNAs (non‐G4 and TelG) were amplified by PCR using plasmids containing the non‐G4 or TelG sequences (Table 1) as templates. PCR products were electrophoresed on an agarose gel and purified with a QIAquick Gel Extraction Kit (Qiagen). Transfection was performed using Lipofectamine RNAiMAX (Invitrogen) by the reverse transfection method in accordance with the manufacturer's instructions. In brief, 0.5 × 105 cells for 2D adherent culture or 3.0 × 105 cells for 3D culture were transfected with 1 µg oligonucleotides, 5 nmol siRNA or 500 ng of dsDNA. Cells were then incubated in a 12‐well flat‐bottomed plate (Iwaki) for 2D culture or a 24‐well EZ‐BindShutII plate (Iwaki) for 3D culture in 1 ml DMEM containing 3% heat‐inactivated FBS and then harvested at 72 hr after transfection.
4.3. Reverse transcription‐quantitative PCR (RT‐qPCR)
Total RNA was extracted using an RNeasy Mini kit (Qiagen) or FastGene RNA Basic Kit (Nippon Genetics). cDNA was synthesized using SuperScript III First‐Strand Synthesis SuperMix for RT‐qPCR (Thermo Fisher Scientific) or ReverTra Ace qPCR RT Master Mix (Toyobo). RT‐qPCR was performed using LightCycler 480 Probes Master (Roche) or FastStart Essential DNA Probe Master Mix (Nippon Genetics) with a LightCycler 96 (Roche). Universal ProbeLibrary probes (Roche) were used to detect target genes and a reference gene (GPI or GUSB) as an internal control to normalize differences in the amounts of RNA in each sample. All primers and probes (Table S1) were designed or selected in accordance with the Universal ProbeLibrary Assay Design Center (https://qpcr.probefinder.com/organism.jsp).
4.4. Western blot analysis
Cell lysates were prepared with buffer consisting of 50 mM NaCl, 1% Nonidet P‐40, 50 mM Tris‐HCl (pH 8.0), 125 mM dithiothreitol and 1/50 volume of protease inhibitor cocktail (Nacalai Tesque). Western blot analysis was performed as described previously (Jang et al., 2020) with the following primary antibodies: rabbit anti‐ISG15 (10 µg/ml, ab14374, Abcam), rabbit anti‐Tyr701‐STAT1 (1:1,000, 9167S, Cell Signaling Technology), rabbit anti‐Ser727‐STAT1 (1:1,000, 9177S, Cell Signaling Technology), rabbit anti‐STAT1 (0.67 or 0.13 µg/ml, ab118638, Abcam), mouse anti‐actin (1:20,000, A5441, Sigma), mouse anti‐FUS (1 µg/ml, sc‐47711, Santa Cruz Biotechnology), mouse anti‐SF3B2/SAP145 (0.4 µg/ml, sc‐514930, Santa Cruz Biotechnology) or mouse anti‐SF3B1/SAP155 (0.2 µg/ml, sc‐514655, Santa Cruz Biotechnology).
4.5. In vitro pull‐down assay
A549 cells (4 × 106 cells) were resuspended in 150 µl lysis buffer consisting of 10 mM Tris‐HCl (pH 7.5), 400 mM KCl, 0.5% Triton X‐100 and 1 mM EDTA. After incubation for 15 min on ice, 250 µl dilution buffer [10 mM Tris‐HCl (pH 7.5), 0.5% Triton X‐100 and 1 mM EDTA] was added, followed by centrifugation at 20,400 g to obtain the supernatant as the cell lysate. The lysates were precleared using Dynabeads M‐280 Streptavidin (Veritas) for 1 hr at 4°C. Then, the oligonucleotides were heated to 95°C for 5 min, cooled down slowly at the rate of 0.5°C/30 s to 4°C, followed by 0.25 µM pre‐annealed biotin‐conjugated oligonucleotides (Fasmac) were added to the precleared lysates and the mixtures were rotated for 1 hr at 4°C. Then, Dynabeads M‐280 Streptavidin was added to the mixtures, followed by rotation for 1 hr at 4°C. The beads were washed with wash buffer consisting of 10 mM Tris‐HCl (pH 7.5), 150 mM KCl, 0.5% Triton X‐100 and 1 mM EDTA three times and TE buffer (10 mM Tris‐HCl and 1 mM EDTA) twice. The bead‐bound proteins were eluted with elution buffer (95% formamide and 10 mM EDTA) for 10 min at 65°C. In the case of PC3 cells, lysates were prepared from 1.5 × 106 cells with 400 µl lysis buffer. Pre‐annealed biotin oligonucleotides were incubated with or without Phen‐DC3 and then added to the precleared cell lysates. Then, the mixtures were rotated for 1 hr at 4°C, followed by addition of Dynabeads M‐280 Streptavidin and rotation for 1 hr at 4°C. The beads were washed with the wash buffer three times, and the bead‐bound proteins were eluted with 1× sample buffer (50 mM Tris pH 6.8, 2% sodium dodecyl sulfate [SDS], 10% glycerol, 0.05% bromophenol blue and 2% 2‐mercaptoethanol) for 5 min at 95°C. The eluted proteins were subjected to Western blot analysis as described above.
4.6. LC‐MS/MS
A549 pull‐down samples were prepared as described above and dissolved and reduced in 1× Laemmli's sample buffer with 10 mM Tris (2‐carboxyethyl)phosphine at 100°C for 10 min. Following alkylation with 50 mM iodoacetamide at ambient temperature for 45 min, protein samples were subjected to SDS‐PAGE. The electrophoresis was stopped at a migration distance of 2 mm from the top edge of the separation gel. After Coomassie Brilliant Blue staining, protein bands were excised, destained and cut finely prior to in‐gel digestion with Trypsin/Lys‐C Mix (Promega) at 37°C for 12 hr. The resulting peptides were extracted from gel fragments and analyzed with an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific) combined with UltiMate 3000 RSLC nano‐flow HPLC (Thermo Fisher Scientific). Peptides were enriched with μ‐Precolumn (0.3 mm i.d. × 5 mm, 5 μm, Thermo Fisher Scientific) and separated on an AURORA column (0.075 mm i.d. × 250 mm, 1.6 μm, Ion Opticks Pty Ltd) using a two‐step gradient: 2%–40% acetonitrile for 110 min, followed by 40%–95% acetonitrile for 5 min in the presence of 0.1% formic acid. The analytical parameters of the Orbitrap Fusion Lumos were set as follows: resolution of full scans = 50,000, scan range (m/z) = 350–1500, maximum injection time of full scans = 50 ms, AGC target of full scans = 4 × 105, dynamic exclusion duration = 30 s, cycle time of data‐dependent MS/MS acquisition = 2 s, activation type = HCD, detector of MS/MS = Ion trap, maximum injection time of MS/MS = 35 ms, AGC target of MS/MS = 1 × 104. The MS/MS spectra were searched against the Homo sapiens protein sequence database (20,366 entries) in SwissProt using Proteome Discoverer 2.4 software (Thermo Fisher Scientific), in which peptide identification filters were set at a false discovery rate of <1%. Label‐free relative quantification analysis of the identified proteins was performed with the default parameters of Minora Feature Detector node, Feature Mapper node and Precursor Ions Quantifier node in Proteome Discoverer 2.4 software.
4.7. PCR‐based detection of RNA splicing
Total RNA was prepared as described above, and potential DNA contamination was eliminated using a TURBO DNA‐free Kit (Thermo Fisher Scientific). cDNA was synthesized using ReverTra Ace qPCR RT Master Mix (Toyobo). PCR amplification of cDNAs derived from DNAJB1 mRNA and U6 snRNA (Kotake et al., 2007; O'Brien et al., 2008) was performed with the following primers (DNAJB1‐Fw: 5′‐GAACCAAAATCACTTTCCCCAAGGAAGG‐3′; DNAJB1‐Rv: 5′‐AATGAGGTCCCCACGTTTCTCGGGTGT‐3′; U6‐Fw: 5′‐CGCTTCGGCAGCACATATAC‐3′; U6‐Rv: 5′‐GAATTTGCGTGTCATCCTTG‐3′) and KOD‐Plus‐Neo (Toyobo) at 94°C for 5 min, 32 cycles of (94°C for 30 s, 60°C for 30 s and 72°C for 1 min) and then 72°C for 7 min. Products were analyzed by 2% agarose gel electrophoresis with SYBR Safe DNA Gel Stain (Thermo Fisher Scientific).
4.8. RNA‐seq analysis
Total RNA was prepared and treated with TURBO DNase as described above. Handling of the RNA‐seq library construction, next‐generation sequencing and differential expressed gene analysis was conducted by Macrogen Japan. Transcriptome libraries were prepared using a TruSeq Stranded mRNA LT Sample Prep Kit (Illumina), following the manufacturer's instructions. Briefly, poly‐A‐containing mRNAs were purified using poly‐T oligo‐attached magnetic beads. The resulting mRNA was fragmented into small pieces and reverse transcribed into cDNA. The cDNA fragments were ligated with adaptors on both ends. The final cDNA library was created by purifying the products and subjecting them to PCR (Martin & Wang, 2011). Each transcriptome library was sequenced on an Illumina NextSeq 6000 as paired‐end 101‐bp reads. Raw sequence reads were trimmed to remove adaptor sequences, contaminant DNA and PCR duplicates, and those with a quality lower than Q20 were removed using Trimmomatic 0.38 (Bolger et al., 2014). The sequence reads were aligned to the GRCh38 reference genome using HISAT2 version 2.1.0, Bowtie2 2.3.5.1(Kim et al., 2015). All reads were assembled with StringTie software version 2.1.3b using annotated genes from the NCBI database (Pertea et al., 2015) (https://www.ncbi.nlm.nih.gov/genome/annotation_euk/Homo_sapiens/109/ver.109_20200522). Low‐quality transcripts were filtered. Then, TMM normalization was performed. Statistical analysis was performed using fold change (fc) exactTest with edgeR per comparison pair. Significant results were selected on the basis of |fc| >= 2 and exactTest raw p‐value < .05. The gene expression data have been deposited in the Gene Expression Omnibus and are accessible through accession number GSE157725. The data will be released on 30 September 2021.
4.9. GO analysis
Proteins identified by LC‐MS/MS spectrometry as both Tel‐G4 and AS1411 oligonucleotide‐bound proteins, but not as non‐G4 oligonucleotide‐bound proteins, were defined as G4‐specific binding proteins. Furthermore, among the proteins that were bound to all three oligonucleotides, proteins with more than a 10‐fold higher frequency of identification among both Tel‐G4 and AS1411 oligonucleotide‐bound proteins than non‐G4 oligonucleotide‐bound proteins were also included as G4‐binding proteins. In both criteria, the blank bead‐bound proteins were excluded from the list. GO analysis of these proteins was performed by STRING (https://string‐db.org). For RNA‐seq data, an enrichment test based on the Gene Ontology database (http://geneontology.org/) was conducted with significant gene list using g:Profiler tool (https://biit.cs.ut.ee/gprofiler/; Raudvere et al., 2019). The p‐value for each GO term reflects the enrichment in frequency of that GO term in the input probe list relative to all probes in the background list.
4.10. Statistical analysis
Statistical analysis was performed using Prism 8 (GraphPad Software) or R (https://cran.ism.ac.jp/index.html). Unpaired Student's t tests (two‐sided) were performed to examine differences between two experimental groups. One‐way analysis of variance (Tukey's test) was performed to examine every combination of multiple experimental groups.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
Supporting information
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Seimiya laboratory, especially Myung‐Kyu Jang and Kyotaro Hirashima, for invaluable discussions. We also thank Mitchell Arico from Edanz Group (https://en‐author‐services.edanzgroup.com/ac) for editing a draft of this manuscript. This work was supported in part by a Grants‐in‐Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS) [Grant‐in‐Aid for Scientific Research on Innovative Areas, Frontier Research on Chemical Communications (20H04789 to H.S.), Grant‐in‐Aid for Challenging Research (Exploratory, 20K21555 to H.S.) and Grant‐in‐Aid for Scientific Research (C) (18K07252 to K.O.)], Translational Research Program, Strategic Promotion for Practical Application of Innovative Medical Technology, Japan Agency for Medical Research and Development (AMED) to H.S., and a grant from Nippon Foundation to H.S.
Matsumoto K, Okamoto K, Okabe S, et al. G‐quadruplex‐forming nucleic acids interact with splicing factor 3B subunit 2 and suppress innate immune gene expression. Genes Cells.2021;26:65–82. 10.1111/gtc.12824
Communicated by: Hisao Masai
REFERENCES
- Alcalá, S. , Sancho, P. , Martinelli, P. , Navarro, D. , Pedrero, C. , Martín‐Hijano, L. , Valle, S. , Earl, J. , Rodríguez‐Serrano, M. , Ruiz‐Cañas, L. , Rojas, K. , Carrato, A. , García‐Bermejo, L. , Fernández‐Moreno, M. Á. , Hermann, P. C. , & Sainz, B. (2020). ISG15 and ISGylation is required for pancreatic cancer stem cell mitophagy and metabolic plasticity. Nature Communications, 11(1), 2682 10.1038/s41467-020-16395-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind, L. , & Koonin, E. V. (2000). SAP—A putative DNA‐binding motif involved in chromosomal organization. Trends in Biochemical Sciences, 25(3), 112–114. 10.1016/s0968-0004(99)01537-6 [DOI] [PubMed] [Google Scholar]
- Artandi, S. E. , Chang, S. , Lee, S. L. , Alson, S. , Gottlieb, G. J. , Chin, L. , & DePinho, R. A. (2000). Telomere dysfunction promotes non‐reciprocal translocations and epithelial cancers in mice. Nature, 406(6796), 641–645. 10.1038/35020592 [DOI] [PubMed] [Google Scholar]
- Barthel, F. P. , Wei, W. , Tang, M. , Martinez‐Ledesma, E. , Hu, X. , Amin, S. B. , Akdemir, K. C. , Seth, S. , Song, X. , Wang, Q. , Lichtenberg, T. , Hu, J. , Zhang, J. , Zheng, S. , & Verhaak, R. G. W. (2017). Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nature Genetics, 49(3), 349–357. 10.1038/ng.3781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco, C. , & Mohr, I. (2017). Restriction of human cytomegalovirus replication by ISG15, a host effector regulated by cGAS‐STING double‐stranded‐DNA sensing. Journal of Virology, 91(9), e02483‐16 10.1128/jvi.02483-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger, A. M. , Lohse, M. , & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 30(15), 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, X. , Chiu, Y. H. , & Chen, Z. J. (2014). The cGAS‐cGAMP‐STING pathway of cytosolic DNA sensing and signaling. Molecular Cell, 54(2), 289–296. 10.1016/j.molcel.2014.03.040 [DOI] [PubMed] [Google Scholar]
- Champion‐Arnaud, P. , & Reed, R. (1994). The prespliceosome components SAP 49 and SAP 145 interact in a complex implicated in tethering U2 snRNP to the branch site. Genes & Development, 8(16), 1974–1983. 10.1101/gad.8.16.1974 [DOI] [PubMed] [Google Scholar]
- Chiappori, A. A. , Kolevska, T. , Spigel, D. R. , Hager, S. , Rarick, M. , Gadgeel, S. , Blais, N. , Von Pawel, J. , Hart, L. , Reck, M. , Bassett, E. , Burington, B. , & Schiller, J. H. (2015). A randomized phase II study of the telomerase inhibitor imetelstat as maintenance therapy for advanced non‐small‐cell lung cancer. Annals of Oncology, 26(2), 354–362. 10.1093/annonc/mdu550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba, K. , Lorbeer, F. K. , Shain, A. H. , McSwiggen, D. T. , Schruf, E. , Oh, A. , & Hockemeyer, D. (2017). Mutations in the promoter of the telomerase gene TERT contribute to tumorigenesis by a two‐step mechanism. Science, 357(6358), 1416–1420. 10.1126/science.aao0535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, W. J. , Heddi, B. , Hamon, F. , Teulade‐Fichou, M. P. , & Phan, A. T. (2014). Solution structure of a G‐quadruplex bound to the bisquinolinium compound Phen‐DC(3). Angewandte Chemie, 53(4), 999–1002. 10.1002/anie.201308063 [DOI] [PubMed] [Google Scholar]
- Cogoi, S. , Shchekotikhin, A. E. , & Xodo, L. E. (2014). HRAS is silenced by two neighboring G‐quadruplexes and activated by MAZ, a zinc‐finger transcription factor with DNA unfolding property. Nucleic Acids Research, 42(13), 8379–8388. 10.1093/nar/gku574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, B. K. , Xia, L. , Palandjian, L. , Gozani, O. , Chyung, Y. , & Reed, R. (1999). Characterization of a protein complex containing spliceosomal proteins SAPs 49, 130, 145, and 155. Molecular and Cellular Biology, 19(10), 6796–6802. 10.1128/mcb.19.10.6796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cian, A. , Delemos, E. , Mergny, J. L. , Teulade‐Fichou, M. P. , & Monchaud, D. (2007). Highly efficient G‐quadruplex recognition by bisquinolinium compounds. Journal of the American Chemical Society, 129(7), 1856–1857. 10.1021/ja067352b [DOI] [PubMed] [Google Scholar]
- Duarte, C. W. , Willey, C. D. , Zhi, D. , Cui, X. , Harris, J. J. , Vaughan, L. K. , Mehta, T. , McCubrey, R. O. , Khodarev, N. N. , Weichselbaum, R. R. , & Gillespie, G. Y. (2012). Expression signature of IFN/STAT1 signaling genes predicts poor survival outcome in glioblastoma multiforme in a subtype‐specific manner. PLoS One, 7(1), e29653 10.1371/journal.pone.0029653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldser, D. M. , & Greider, C. W. (2007). Short telomeres limit tumor progression in vivo by inducing senescence. Cancer Cell, 11(5), 461–469. 10.1016/j.ccr.2007.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara, C. , Muramatsu, Y. , Nishii, M. , Tokunaka, K. , Tahara, H. , Ueno, M. , Yamori, T. , Sugimoto, Y. , & Seimiya, H. (2018). Cell‐based chemical fingerprinting identifies telomeres and lamin A as modifiers of DNA damage response in cancer cells. Scientific Reports, 8(1), 14827 10.1038/s41598-018-33139-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girvan, A. C. , Teng, Y. , Casson, L. K. , Thomas, S. D. , Juliger, S. , Ball, M. W. , & Bates, P. J. (2006). AGRO100 inhibits activation of nuclear factor‐kappaB (NF‐kappaB) by forming a complex with NF‐kappaB essential modulator (NEMO) and nucleolin. Molecular Cancer Therapeutics, 5(7), 1790–1799. 10.1158/1535-7163.MCT-05-0361 [DOI] [PubMed] [Google Scholar]
- Gomez, D. , O'Donohue, M. F. , Wenner, T. , Douarre, C. , Macadre, J. , Koebel, P. , & Riou, J. F. (2006). The G‐quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFP‐POT1 dissociation from telomeres in human cells. Cancer Research, 66(14), 6908–6912. 10.1158/0008-5472.CAN-06-1581 [DOI] [PubMed] [Google Scholar]
- Greenberg, R. A. , Chin, L. , Femino, A. , Lee, K.‐H. , Gottlieb, G. J. , Singer, R. H. , Greider, C. W. , & DePinho, R. A. (1999). Short dysfunctional telomeres impair tumorigenesis in the INK4aΔ2/3 cancer‐prone mouse. Cell, 97(4), 515–525. 10.1016/s0092-8674(00)80761-8 [DOI] [PubMed] [Google Scholar]
- Hanahan, D. , & Weinberg, R. A. (2000). The hallmarks of cancer. Cell, 100(1), 57–70. 10.1016/s0092-8674(00)81683-9 [DOI] [PubMed] [Google Scholar]
- Hänsel‐Hertsch, R. , Beraldi, D. , Lensing, S. V. , Marsico, G. , Zyner, K. , Parry, A. , Di Antonio, M. , Pike, J. , Kimura, H. , Narita, M. , Tannahill, D. , & Balasubramanian, S. (2016). G‐quadruplex structures mark human regulatory chromatin. Nature Genetics, 48(10), 1267–1272. 10.1038/ng.3662 [DOI] [PubMed] [Google Scholar]
- Hasegawa, D. , Okabe, S. , Okamoto, K. , Nakano, I. , Shin‐ya, K. , & Seimiya, H. (2016). G‐quadruplex ligand‐induced DNA damage response coupled with telomere dysfunction and replication stress in glioma stem cells. Biochemical and Biophysical Research Communications, 471(1), 75–81. 10.1016/j.bbrc.2016.01.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirashima, K. , Migita, T. , Sato, S. , Muramatsu, Y. , Ishikawa, Y. , & Seimiya, H. (2013). Telomere length influences cancer cell differentiation in vivo. Molecular and Cellular Biology, 33(15), 2988–2995. 10.1128/MCB.00136-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirashima, K. , & Seimiya, H. (2015). Telomeric repeat‐containing RNA/G‐quadruplex‐forming sequences cause genome‐wide alteration of gene expression in human cancer cells in vivo. Nucleic Acids Research, 43(4), 2022–2032. 10.1093/nar/gkv063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, M. K. , Mashima, T. , & Seimiya, H. (2020). Tankyrase inhibitors target colorectal cancer stem cells via AXIN‐dependent downregulation of c‐KIT tyrosine kinase. Molecular Cancer Therapeutics, 19(3), 765–776. 10.1158/1535-7163.MCT-19-0668 [DOI] [PubMed] [Google Scholar]
- Kawamura, N. , Nimura, K. , Saga, K. , Ishibashi, A. , Kitamura, K. , Nagano, H. , Yoshikawa, Y. , Ishida, K. , Nonomura, N. , Arisawa, M. , Luo, J. , & Kaneda, Y. (2019). SF3B2‐mediated RNA splicing drives human prostate cancer progression. Cancer Research, 79(20), 5204–5217. 10.1158/0008-5472.CAN-18-3965 [DOI] [PubMed] [Google Scholar]
- Khodarev, N. N. , Roach, P. , Pitroda, S. P. , Golden, D. W. , Bhayani, M. , Shao, M. Y. , Darga, T. E. , Beveridge, M. G. , Sood, R. F. , Sutton, H. G. , Beckett, M. A. , Mauceri, H. J. , Posner, M. C. , & Weichselbaum, R. R. (2009). STAT1 pathway mediates amplification of metastatic potential and resistance to therapy. PLoS One, 4(6), e5821 10.1371/journal.pone.0005821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Langmead, B. , & Salzberg, S. L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nature Methods, 12(4), 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo, K. , Mashima, T. , Oyoshi, T. , Yagi, R. , Kurokawa, R. , Kobayashi, N. , Nagata, T. , & Katahira, M. (2018). Plastic roles of phenylalanine and tyrosine residues of TLS/FUS in complex formation with the G‐quadruplexes of telomeric DNA and TERRA. Scientific Reports, 8(1), 2864 10.1038/s41598-018-21142-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno, H. , Konno, K. , & Barber, G. N. (2013). Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell, 155(3), 688–698. 10.1016/j.cell.2013.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake, Y. , Sagane, K. , Owa, T. , Mimori‐Kiyosue, Y. , Shimizu, H. , Uesugi, M. , Ishihama, Y. , Iwata, M. , & Mizui, Y. (2007). Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nature Chemical Biology, 3(9), 570–575. 10.1038/nchembio.2007.16 [DOI] [PubMed] [Google Scholar]
- Martin, J. A. , & Wang, Z. (2011). Next‐generation transcriptome assembly. Nature Reviews Genetics, 12(10), 671–682. 10.1038/nrg3068 [DOI] [PubMed] [Google Scholar]
- Membrino, A. , Cogoi, S. , Pedersen, E. B. , & Xodo, L. E. (2011). G4‐DNA formation in the HRAS promoter and rational design of decoy oligonucleotides for cancer therapy. PLoS One, 6(9), e24421 10.1371/journal.pone.0024421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki, T. , Pan, Y. , Joshi, K. , Purohit, D. , Hu, B. , Demir, H. , & Nakano, I. (2012). Telomestatin impairs glioma stem cell survival and growth through the disruption of telomeric G‐quadruplex and inhibition of the proto‐oncogene, c‐Myb. Clinical Cancer Research, 18(5), 1268–1280. 10.1158/1078-0432.CCR-11-1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales, D. J. , & Lenschow, D. J. (2013). The antiviral activities of ISG15. Journal of Molecular Biology, 425(24), 4995–5008. 10.1016/j.jmb.2013.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, T. , Okabe, S. , Yoshida, H. , Iida, K. , Ma, Y. , Sasaki, S. , Yamori, T. , Shin‐ya, K. , Nakano, I. , Nagasawa, K. , & Seimiya, H. (2017). Targeting glioma stem cells in vivo by a G‐quadruplex‐stabilizing synthetic macrocyclic hexaoxazole. Scientific Reports, 7(1), 3605 10.1038/s41598-017-03785-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien, K. , Matlin, A. J. , Lowell, A. M. , & Moore, M. J. (2008). The biflavonoid isoginkgetin is a general inhibitor of Pre‐mRNA splicing. Journal of Biological Chemistry, 283(48), 33147–33154. 10.1074/jbc.M805556200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, K. , Ohishi, T. , Kuroiwa, M. , Iemura, S. I. , Natsume, T. , & Seimiya, H. (2018). MERIT40‐dependent recruitment of tankyrase to damaged DNA and its implication for cell sensitivity to DNA‐damaging anticancer drugs. Oncotarget, 9(88), 35844–35855. 10.18632/oncotarget.26312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, K. , & Seimiya, H. (2019). Revisiting telomere shortening in cancer. Cells, 8(2), 107 10.3390/cells8020107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascolo, E. , Wenz, C. , Lingner, J. , Hauel, N. , Priepke, H. , Kauffmann, I. , Garin‐Chesa, P. , Rettig, W. J. , Damm, K. , & Schnapp, A. (2002). Mechanism of human telomerase inhibition by BIBR1532, a synthetic, non‐nucleosidic drug candidate. Journal of Biological Chemistry, 277(18), 15566–15572. 10.1074/jbc.M201266200 [DOI] [PubMed] [Google Scholar]
- Pertea, M. , Pertea, G. M. , Antonescu, C. M. , Chang, T. C. , Mendell, J. T. , & Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA‐seq reads. Nature Biotechnology, 33(3), 290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudvere, U. , Kolberg, L. , Kuzmin, I. , Arak, T. , Adler, P. , Peterson, H. , & Vilo, J. (2019). g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Research, 47(W1), W191–W198. 10.1093/nar/gkz369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seimiya, H. (2020). Crossroads of telomere biology and anticancer drug discovery. Cancer Science, 111, 3089–3099. 10.1111/cas.14540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen, D. , & Gilbert, W. (1988). Formation of parallel four‐stranded complexes by guanine‐rich motifs in DNA and its implications for meiosis. Nature, 334(6180), 364–366. 10.1038/334364a0 [DOI] [PubMed] [Google Scholar]
- Siddiqui‐Jain, A. , Grand, C. L. , Bearss, D. J. , & Hurley, L. H. (2002). Direct evidence for a G‐quadruplex in a promoter region and its targeting with a small molecule to repress c‐MYC transcription. Proceedings of the National Academy of Sciences USA, 99(18), 11593–11598. 10.1073/pnas.182256799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommerfeld, H. J. , Meeker, A. K. , Piatyszek, M. A. , Bova, G. S. , Shay, J. W. , & Coffey, D. S. (1996). Telomerase activity: A prevalent marker of malignant human prostate tissue. Cancer Research, 56(1), 218–222. [PubMed] [Google Scholar]
- Tahara, H. , Shin‐Ya, K. , Seimiya, H. , Yamada, H. , Tsuruo, T. , & Ide, T. (2006). G‐Quadruplex stabilization by telomestatin induces TRF2 protein dissociation from telomeres and anaphase bridge formation accompanied by loss of the 3′ telomeric overhang in cancer cells. Oncogene, 25(13), 1955–1966. 10.1038/sj.onc.1209217 [DOI] [PubMed] [Google Scholar]
- Takahama, K. , Kino, K. , Arai, S. , Kurokawa, R. , & Oyoshi, T. (2011). Identification of Ewing's sarcoma protein as a G‐quadruplex DNA‐ and RNA‐binding protein. FEBS Journal, 278(6), 988–998. 10.1111/j.1742-4658.2011.08020.x [DOI] [PubMed] [Google Scholar]
- Thandapani, P. , O'Connor, T. R. , Bailey, T. L. , & Richard, S. (2013). Defining the RGG/RG motif. Molecular Cell, 50(5), 613–623. 10.1016/j.molcel.2013.05.021 [DOI] [PubMed] [Google Scholar]
- Vasilyev, N. , Polonskaia, A. , Darnell, J. C. , Darnell, R. B. , Patel, D. J. , & Serganov, A. (2015). Crystal structure reveals specific recognition of a G‐quadruplex RNA by a beta‐turn in the RGG motif of FMRP. Proceedings of the National Academy of Sciences USA, 112(39), E5391–5400. 10.1073/pnas.1515737112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K. E. , Yin, C. , Du, X. , Chen, S. , Wang, J. , Zhang, L. I. , Wang, L. , Yu, Y. , Chi, B. , Shi, M. , Wang, C. , Reed, R. , Zhou, Y. U. , Huang, J. , & Cheng, H. (2019). A U2‐snRNP‐independent role of SF3b in promoting mRNA export. Proceedings of the National Academy of Sciences USA, 116(16), 7837–7846. 10.1073/pnas.1818835116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichselbaum, R. R. , Ishwaran, H. , Yoon, T. , Nuyten, D. S. A. , Baker, S. W. , Khodarev, N. , Su, A. W. , Shaikh, A. Y. , Roach, P. , Kreike, B. , Roizman, B. , Bergh, J. , Pawitan, Y. , van de Vijver, M. J. , & Minn, A. J. (2008). An interferon‐related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proceedings of the National Academy of Sciences USA, 105(47), 18490–18495. 10.1073/pnas.0809242105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesoly, J. , Szweykowska‐Kulinska, Z. , & Bluyssen, H. A. (2007). STAT activation and differential complex formation dictate selectivity of interferon responses. Acta Biochimica Polonica, 54(1), 27–38. 10.18388/abp.2007_3266 [DOI] [PubMed] [Google Scholar]
- Zhang, S. , Wu, Y. , & Zhang, W. (2014). G‐quadruplex structures and their interaction diversity with ligands. ChemMedChem, 9(5), 899–911. 10.1002/cmdc.201300566 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


