Abstract
Glucose metabolism is tightly regulated and disrupting glucose homeostasis is a hallmark of many diseases. Caloric restriction (CR), periodic fasting, and circadian rhythms are interlinked with glucose metabolism. Here, we directly investigated if CR depends on periodic fasting and circadian rhythms to improve glucose metabolism. CR was implemented as two‐meals per day (2M‐CR), provided at 12‐hour intervals, and compared with one meal per day CR, mealtime (MT), and ad libitum (AL) feeding. The 2M‐CR impacted the circadian rhythms in blood glucose, metabolic signaling, circadian clock, and glucose metabolism gene expression. 2M‐CR significantly reduced around the clock blood glucose and improved glucose tolerance. Twenty‐four‐hour rhythms in mTOR signaling and gene expression observed under AL, MT, and CR, became 12‐hour rhythms in 2M‐CR. The 12‐hour rhythms in behavior, gene expression, and signaling persisted in fasted mice, implicating some internal regulation. The study highlights that the reduction in caloric intake rather than meal frequency and duration of fasting is essential for metabolic reprograming and improvement in glucose metabolism and provides evidence on food‐entrained molecular pacemaker, which can be uncoupled from the light‐entrained circadian clock and rhythms.
Keywords: aging, metabolism, diet, circadian rhythms, gene expression, glucose homeostasis, 12‐hour rhythms
Abbreviations
- 2M‐CR
Two‐meals per day caloric restriction
- AL
Ad libitum
- ANOVA
Analysis of variance
- AUC
Area Under the Curve
- Bmal1
Brain and Muscle ARNT‐Like 1
- CR/1M‐CR
Caloric restriction
- Cry1/2
Cryptochrome
- EDTA
Ethylenediaminetetraacetic acid
- EGTA
Ethylene glycol tetraacetic acid
- FAA
Food Anticipatory Activity
- Fbp1
Fructose‐1,6‐bisphosphatase 1
- G6pc
Glucose‐6‐phosphatase, catalytic subunit
- GAPDH
Glyceraldehyde 3‐phosphate dehydrogenase
- Gck
Glucokinase
- GTT
Intraperitoneal Glucose Tolerance Test
- HF
High‐fat
- HRP
Horseradish peroxidase
- IF
Intermittent fasting
- IgG
Immunoglobulin G
- mRNA
messenger ribonucleic acid
- MT
Mealtime feeding
- mTOR
Mechanistic target of rapamycin
- PBS
Phosphate Buffer Saline
- Pck1
Phosphoenolpyruvate carboxykinase 1
- PCR
Polymerase Chain Reaction
- Per1/2
Period
- Pfk1
Phosphofructokinase‐1
- PVDF
Polyvinylidene difluoride
- Rev‐erbα
Nuclear receptor subfamily 1 group D member 1
- RNA
Ribonucleic acid
- Rorγ
RAR‐related orphan receptor gamma
- rRNA
Ribosomal ribonucleic acid
- SD
Standard deviation
- TBST
Tris Buffered Saline + Tween 20
- TRF
Time‐restricted feeding
- ZT
Zeitgeber
1. INTRODUCTION
It is common knowledge, coming from different cultures, that diet and health are connected. There is multiple evidence that some diets have negative effects and others have positive effects on physiology and metabolism. In addition to the composition of the diet, the meal size, frequency of meal, and time of meal are recognized as contributing factors. 1 Experimental dietary interventions that explore periodic fasting are growing in popularity for proposed health benefits. The periodic fasting diets include four major variants. First, intermittent fasting (IF) or intermittent energy restriction involves fasting or reduced food intake on some days while eating ad libitum (AL) on other days. 2 Second, time‐restricted feeding (TRF) limits food consumption to a restricted time window. 3 While the amount of food is not restricted during TRF, the restriction of the time window to several hours might also lead to a reduction in intake. 4 Third, mealtime feeding (MT), 100% of calculated daily AL food intake is provided as a single meal once per day, the animals typically consume the entire meal in several hours and fast for some time. 5 Fourth, caloric restriction (CR), in which the food intake is reduced by 20%‐40%. 6 The most common meal frequency in CR experiments is once per day. Other variants such as providing the meal every second day or more than one meal per day are also known. One meal per day CR animals consume the provided food in about 2‐3 hours and fast for the rest of the day, 7 , 8 , 9 in other words, CR is a severe form of MT or self‐implemented TRF. CR, IF, and MT improve health and increase longevity in different organisms including mammals. 2 , 5 , 6 TRF restores circadian rhythms disrupted by the high‐fat diet and ameliorates negative effects of the high‐fat diet on physiology. 3 , 10 TRF might also be beneficial in prediabetic and obese individuals. 11 Little is known if TRF has metabolic benefits on regular chow. CR is the most well documented dietary intervention with multiple health benefits. 12 Some of these benefits are common between CR and other above‐discussed diets and all these diets involve periodic fasting. Together, this leads to a hypothesis that some of the metabolic benefits of CR are due to periodic fasting and can be achieved without a reduction in caloric intake.
Periodic fasting/feeding cycle is linked with the circadian clocks and rhythms. 13 , 14 Twenty‐four‐hour circadian rhythms are generated by the circadian clocks that are present in different tissues and synchronize organism physiology and metabolism with the 24‐hour periodicity of the daily cycle. Several transcriptional regulators form negative and positive feedback loops, interact with cellular transcriptional/translational machinery to drive rhythmic gene expression. 15 , 16 , 17 Circadian clocks regulate rhythmic transcription of at least 10% of the genes in a tissue‐specific manner. 18 These rhythms are believed to be important for the optimization of many processes in an organism. The disruption of circadian rhythms, through an unhealthy diet or desynchronization with the environment, is associated with negative effects on physiology and metabolism. As a result, the circadian disruption increases the risk of many diseases. 19 , 20 Moreover, robust circadian rhythms are associated with improved organism fitness, which is characteristic of CR and contributes to the metabolic benefits. 8 , 21 , 22
Improvement in glucose homeostasis is one of the well‐documented effects of CR. 23 CR reduces blood glucose and improves glucose tolerance in mammals. CR also reduces blood insulin and improves insulin sensitivity. 6 IF mice also have reduced blood glucose even when the food intake was not reduced. 2 We recently found that TRF, wherein animals have unlimited access to the regular chow during 12‐hours, improves insulin sensitivity, but does not reduce the blood glucose and does not improve glucose tolerance. 24 The duration of fasting is different between CR and TRF in the above experiments: indeed, CR mice fast for about 22 hours 7 , 8 , 9 and TR mice fast only for 12 hours. This raises a question if the duration of fasting is an important factor that might influence the outcome. To establish if the duration of periodic fasting in CR is essential for the improvement in glucose homeostasis in the present study, 30% CR was implemented as two‐meals per day (2M‐CR). The metabolic effects, with a focus on glucose metabolism and circadian rhythms, were compared between 2M‐CR, one meal per day CR (1M‐CR), MT, and AL feeding. We found that 2M‐CR improved glucose homeostasis as efficient as 1M‐CR. The improvement was achieved even when the circadian rhythms in gene expression and signaling were disrupted by 2M‐CR. Interestingly, 24‐hour rhythms in metabolism were replaced with 12‐hour rhythms suggesting high plasticity in metabolic reprograming and adjustment with meal frequency.
2. MATERIALS AND METHODS
2.1. Animals
All animal experiments were approved and conducted according to the guidelines of the Institutional Animal Care and Use Committee (IACUC) at Cleveland State University. C57BL/6J male mice were maintained under controlled conditions under 12‐hour light: 12‐hour dark cycle (LD 12:12) with lights on at 7 AM (ZT0) and lights off at 7 PM (ZT12). Mice were fed 2018 Teklad Global 18% protein diet (Envigo (formerly Harlan) Catalog# 2018) and allowed ab libitum access to water.
2.2. Study design/animal experiments
Mice were provided an AL amount of food prior to the start of the experiment. Three‐month‐old mice were randomly assigned to four feeding regimens. Ad libitum (AL)‐fed mice have unlimited access to the food around the clock. AL mice were single‐caged and group‐caged, no significant difference between single‐ and group‐caged AL mice were observed. Three other groups were subject to different forms of food restriction. All food‐restricted mice were single‐caged. Mealtime (MT)‐fed mice received 100% of their food intake, 5 g per day on average, calculated from their AL intake in the week before the experiment, once per day at ZT14, 2 hours after light was off. Two other groups were on 30% CR. Caloric restriction was introduced gradually: 10% CR on week 1, followed by 20% restriction on week 2, and 30% restriction from week 3 until the end of the experiment. One CR group (1M‐CR) received food as a single meal, 3.5 gram per day on average, at ZT14. Another CR group (2M‐CR) received the food as two‐meals per day: first meal, 1.75 grams per meal on average, at ZT2 and second meal, 1.75 grams per meal on average, at ZT14. Body weight measurements were performed at ZT14 before the feeding once per week. Tissues were collected at the end of 8 weeks, snap‐frozen on dry ice, and stored at −80°C until further analysis. Tissues for AL and 2M‐CR were collected around the clock at 2‐hour intervals from ZT2 to ZT24, whereas 1M‐CR tissues were collected at 4‐hour intervals from ZT2 to ZT22. Additional groups were used as experimental controls for GTT experiments: time‐restricted fed mice, this group received food at ZT14 and food was withdrawn at ZT2.
2.3. In‐cage locomotor activity
Mice were on the diets for at least 5 weeks before behavioral experiments were started. Single‐caged mice were placed in their home cage and their in‐cage locomotor activity was evaluated using PAS Home Cage Photobeam System (San Diego Instruments). Locomotor activity was continuously monitored and automatically recorded every 60 minutes for 48 hours. The data were analyzed using the PAS software.
2.4. Blood glucose and intraperitoneal glucose tolerance test
Around the clock blood glucose was collected via tail vein nick. Mice in all diet groups were not fasted prior to blood collection. Glucose tolerance test was performed on mice after at least 5 weeks on the diets. MT, TRF, 1M‐CR, and 2M‐CR mice were not additionally fasted before GTT. AL mice were fasted for 12 hours before the experiment. Mice were intraperitoneally injected with glucose (0.4 g/kg body weight) in PBS. Blood glucose was collected via tail vein nick at intervals indicated in the figure. Blood Glucose was measured using CVS Health Advanced Blood Glucose Meter and CVS Health Advanced Glucose Meter Test Strips (CVS).
2.5. RNA isolation and real time––PCR analysis
Total RNA extraction was performed from frozen liver tissues using TRIzol method according to the manufacturer's protocols. RNA quantification was done using Nanodrop‐2000 (Thermo Fisher Scientific). Reverse transcription of total RNA was performed using SuperScript IV Reverse Transcriptase (InvitrogenTM). Real‐time quantitative PCR was performed on CFX Connect real‐time PCR detection instrument using iTaq Universal SYBR Green Supermix (Bio‐Rad) and gene‐specific primers. Changes in mRNA expression were determined by ΔΔCt method and the expression levels were normalized using 18s rRNA gene.
Genes and sequences used were as follows: Bmal1 (Fwd 5ʹ CACTGTCCCAGGCATTCCA 3ʹ; Rev 5ʹ TTCCTCCGCGATCATTCG 3ʹ), Per1 (Fwd 5ʹ AGGTGGCTTTCGTGTTGG 3ʹ; Rev 5ʹ CAATCGATGGATCTGCTCTGAG 3ʹ), Per2 (Fwd 5ʹ AGGCACCTCCAACATGCAA 3ʹ; Rev 5ʹ GGATGCCCCGCTTCTAGAC 3ʹ), Cry1 (Fwd 5ʹ CGTCTGTTTGTGATTCGGGG 3ʹ; Rev 5ʹ ATTCACGCCACAGGAGTTGC 3ʹ), Cry2 (Fwd 5ʹ ACCGATGGAGGTTCCTACTG 3ʹ; Rev 5ʹ AGCCTTGGGAACACATCAG 3ʹ), Rev‐erbα (Fwd 5ʹ TGGCCTCAGGCTTCCACTATG 3ʹ; Rev 5ʹ CCGTTGCTTCTCTCTCTTGGG 3ʹ), Rorγ (Fwd 5ʹ ACTACGGGGTTATCACCTGTGAG 3ʹ; Rev 5ʹ GTGCAGGAGTAGGCCACATTAC 3ʹ), Gck (Fwd 5ʹ CACAATGATCTCCTGCTACT 3ʹ; Rev 5ʹ TTCTGCATCTCCTCCATGTA 3ʹ), Pfk1 (Fwd 5ʹ AGAGGACCTTTGTTTTGGAG 3ʹ; Rev 5ʹ TCTGCGATGATGATGATGTT 3ʹ), Fbp1 (Fwd 5ʹ TGACCTGGTGATCAATATGC 3ʹ; Rev 5ʹ CAAAAATGGTTCCGATGGAC 3ʹ), Pck1 (Fwd 5ʹ TGAGATCTAGGAGAAAGCCA 3ʹ; Rev 5ʹ CCTTGAAGTGGAACCAAAAC 3ʹ), G6pc1 (Fwd 5ʹ CTAAAGCCTCTGAAACCCAT 3ʹ; Rev 5ʹ ATGACTCAGTTTCCAGCATT 3ʹ), and 18s rRNA (Fwd 5ʹ GCTTAATTTGACTCAACACGGGA 3ʹ; Rev 5ʹ AGCTATCAATCTGTCAATCCTGTC 3ʹ).
2.6. Protein expression analysis
Total proteins were extracted from liver tissues using Cell Signaling Lysis Buffer (1M of Tris base pH 7.5, 5M of NaCl, 0.5M of EGTA, 0.5M of EDTA, Triton‐X, 0.1M of Na4P2O7, 1M of β‐glycerophosphate, 1M of Na3VO4) with protease and phosphatase inhibitor cocktails. Protein concentration was quantified using Bradford method as per the manufacturer's instructions. The protein lysates were run on 4%‐12% Bis‐Tris NUPAGE gels (Thermo Fisher Scientific) and transferred on PVDF membrane (Thermo Fisher Scientific). Membranes were blocked with 5% nonfat dry milk for 1 hour prior to overnight incubation at 4°C with primary antibodies as indicated below. Next day, membranes were washed with 1X TBST and incubated with the corresponding secondary antibody. The signal was detected using ECL Substrate solution. GAPDH was used for normalization. Image quantification was performed using Image Studio Lite software (LI‐COR Biosciences). Antibodies used include: Cell Signaling Technology: Phospho‐p70 S6K (Thr389) (Cat# 9205), Phospho‐S6 Ribosomal Protein (Ser235/236) (Cat# 4858S), p70 S6K (Cat# 9202), Anti‐rabbit IgG, HRP‐linked (Cat# 7074), Anti‐mouse IgG, HRP‐linked (Cat# 7076), and GAPDH (D16H11) XP® Rabbit monoclonal (Cat #5174); Santa Cruz Biotechnology: Ribosomal Protein S6 Antibody (C‐8) (Cat# sc‐74459).
2.7. Statistics and rhythms analysis
Statistical analyses were performed using GraphPad Prism 5.0 (San Diego, CA). Data for all diets were analyzed using either one‐way or two‐way ANOVA. Post hoc analysis was done using Bonferroni method. Number of biological replicas used in each experiment is indicated in the Figure legends. Data are represented as Mean ± SD. Statistical significance was set at P ≤ .05. Circadian analysis of gene and protein expression for each diet was performed using JTK‐Cycle (Hughes et al 2010). The adjusted P‐value for circadian rhythmicity was set at ≤.05.
3. RESULTS
3.1. Effect of two‐meals per day CR on daily behavior rhythms
Three‐month‐old mice were randomly assigned to four feeding regimens: Ad libitum (AL) and three different forms of food restriction (Figure 1A). Food‐restricted mice received meals at the same time every day. Mealtime (MT)‐fed mice received 100% of their daily AL food intake at ZT14. CR mice received 70% of their daily AL food intake either as single meal per day at ZT14 (referred to as 1M‐CR throughout the text); or as two equal‐size meals per day (referred to as 2M‐CR throughout the text). In agreement with previous observations, 8 , 9 1M‐CR mice consumed all the provided food in 1‐2 hours. Similar to 1M‐CR, 2M‐CR mice consumed all provided food in about 1 hour. MT mice consume a variable amount of food immediately after the feeding, but typically they have some food in the cages for at least 12‐15 hours. 5 AL mice typically consume the food as two major meals: first after the light is off, around ZT12‐ZT14 and second before the light is on around ZT22‐ZT24, but they also eat around the clock. 24 Therefore, based on these observations, we expect the following duration of fasting in every diet; 1M‐CR mice fast continuously for about 21‐22 hours. 2M‐CR mice fast continuously for about 11 hours twice a day. MT mice might fast for several hours, and AL mice might fast for a few hours when lights are on or do not fast at all.
FIGURE 1.
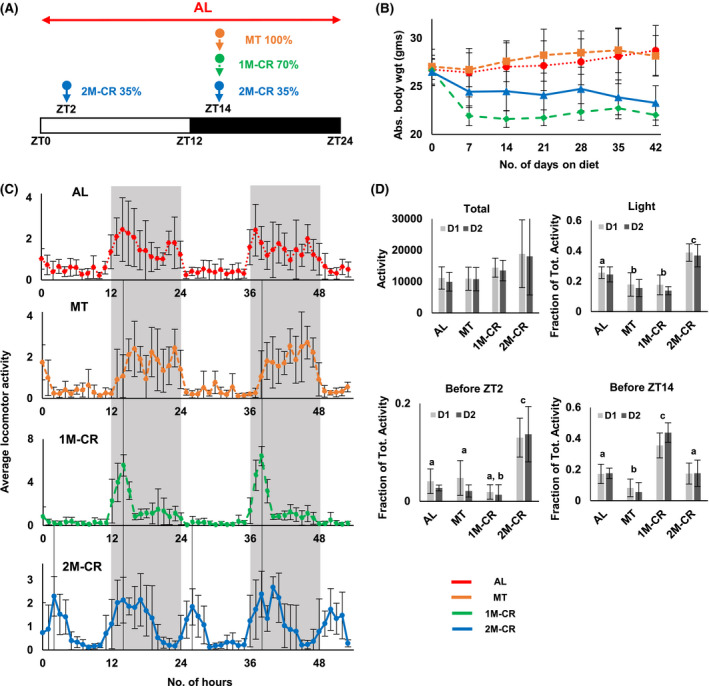
Effect of different diets on mouse daily rhythms in locomotor activity. A, Design of the experiment. B, Body weight of mice on different diets: AL––dotted red line, N = 13; MT––small‐dashed brown line, N = 6; 1M‐CR––large‐dashed green line, N = 6; and 2M‐CR––solid blue line, N = 22. C, Average in‐cage locomotor activity for mice on indicated diets. The activity was recorded for two random days. The activity for each mouse was normalized to average hourly activity, after that average activity and standard deviations were calculated for mice on every diet. Black line indicted the time when the food was provided to mice. D, In‐cage activity on Day 1 (light gray) and Day 2 (dark gray): total activity––upper left panel; the activity during the light phase of the day, shown as a fraction of total daily activity, which was set up as 1.0––upper right panel; the activity in 2 hours before ZT2, data are shown as a fraction of total daily activity, which was set up as 1.0––lower left panel; and the activity in 2 hours before ZT14, data are shown as a fraction of total daily activity, which was set up as 1.0––lower right panel. N = 6 per diet, Bars with different letters are significantly different from each other. The light was on at ZT0 and off at ZT12. Gray panels indicate dark phase of the day
Body weight of mice was assayed once per week (Figure 1B). In agreement with previous data, there was no difference in body weight between AL and MT mice. 1M‐CR resulted in reduced body weight with a significant difference observed after the first week. 2M‐CR also significantly reduced the body weight compared with AL and MT. 2M‐CR were heavier compared with 1M‐CR, detailed analysis revealed that the difference between two CR groups was due to fast changes in body weight around the feeding time. Indeed, CR mice consumed the provided food in a matter of 1‐2 hours after the food is provided. The body weight of mice presented in Figure 1B was taken at ZT14, before the feeding. After that the mice received the food: 1M‐CR––3.5 grams on average and 2M‐CR––1.75 grams on average. If the body weight was taken at ZT16, no significant difference between 1M‐CR and 2M‐CR was detected.
The feeding restriction affects daily locomotor activity and might induce Food Anticipatory Activity (FAA) in rodents. 25 Locomotor activity was assayed around the clock for mice on all four diets and the representative locomotion is shown in Figure 1C and Figures [Link], [Link]. Total activity was not different between AL and MT mice, the activity was significantly higher in 1M‐CR and 2M‐CR compared with AL and showed a tendency, but did not reach significance, compared with MT (Figure 1D, first panel). AL mice were more active during the night and have some activity during the light phase of the day (Figure 1D, second panel). The pattern of night locomotor activity of MT mice was similar with AL mice, but these mice have a significantly reduced light phase activity (Figure 1D). CR significantly changed the patterns of daily behavior (Figure 1B and Figure S2). 1M‐CR mice demonstrated high level of activity around the feeding time and little activity through the rest of the day (Figure 1D and Figure S2). About 40% of daily activity was at ZT12‐ZT13, 1‐2 hour before the feeding time (Figure 1C,D), in agreement with the expected high level of FAA. 25 Interestingly, MT and 1M‐CR mice demonstrated comparable level of light phase activity (Figure 1D, second panel), but MT did not demonstrate any anticipation peak in activity before the meal (Figure 1C, second panel). The pattern of daily locomotor activity of 2M‐CR mice was significantly different from AL, MT, and 1M‐CR. These mice demonstrated two distinct peaks of activity: about 13% of daily activity before ZT2 feeding and about 17% before ZT14 feeding (Figure 1D, third and fourth panel). Thus, 2M‐CR mice demonstrated the ability to anticipate two‐meals per day. In summary, all three experimental diets significantly impact daily pattern of locomotor activity compared with AL. Both CRs have the strongest effect by increasing the total activity and activity around feeding time.
3.2. Effect of two‐meals per day CR on circadian clock gene expression
Circadian clock gene expression in the liver is affected by the diets. 3 , 18 , 26 , 27 , 28 We compared the effect of 2M‐CR on the expression of several core clock genes (Figure 2A), which were highly rhythmic in AL liver (Table S1). 1M‐CR significantly increased the amplitude of expression of five genes: Bmal1, Per1, Per2, Rorγ, and Rev‐erbα. The expression of Cry2 gene was reduced and became arrhythmic. There was no effect on Cry1 expression. 2M‐CR significantly changed the expression in a gene‐specific manner. The mRNAs of four genes: Bmal1, Per1, Rorγ, and Rev‐erbα were rhythmic with period 24 hours (Figure 2B, Table S1), but with reduced amplitude. The expression of three genes: Per2, Cry1, and Cry2 lost circadian rhythmicity. The rhythms were analyzed for 12‐hour period (Table S2). Twelve‐hour rhythms were not detected in AL or 1M‐CR. Four genes: Per2, Cry1, Per1, and Rev‐erbα were rhythmic with a 12‐hour period and demonstrated two peaks within a 24‐hour period. Cry1, Rev‐erbα, and Per1 mRNAs demonstrated one high amplitude peak, which coincides with the peak in AL or 1M‐CR, and a second low amplitude peak. Per2 demonstrated two distinct high amplitude peaks. Two peaks in Per2 expression are in line with a proposed role for Per2 in the regulation of food anticipation, 29 and two peaks in locomotor activity were observed in 2M‐CR mice (Figure 1C). Interestingly, Rev‐erbα and Per1 mRNAs were rhythmic with both 12‐ and 24‐hour periods. Thus, 2M‐CR disturbed circadian rhythms in the expression of circadian clock genes and the effect of 2M‐CR was significantly different from the 1M‐CR.
FIGURE 2.
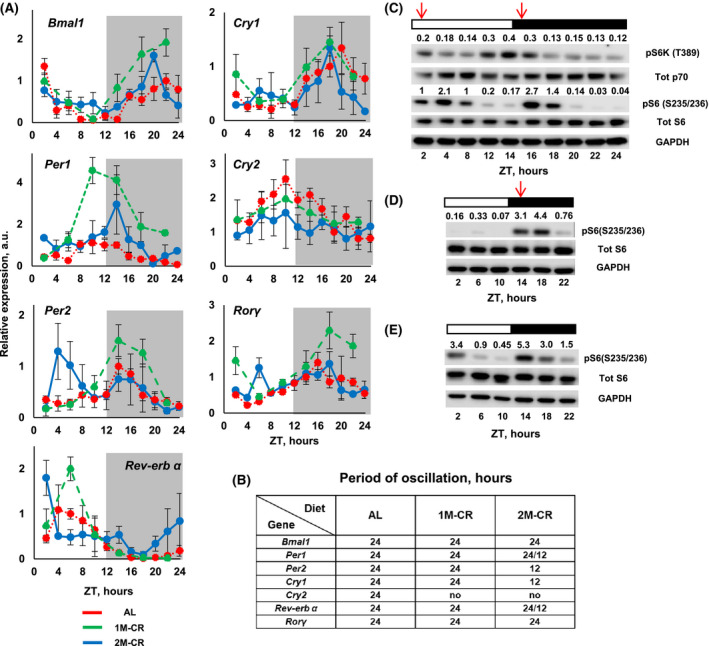
Effect of one and two‐meal CR on circadian rhythms in the liver. A, The expression of indicated circadian clock genes in the liver of mice: AL––dotted red line; 1M‐CR––large‐dashed green line; and 2M‐CR––solid blue line. N = 3 per time point per diet. Gray panels indicate dark phase of the day. B, Results of rhythmic analysis indicate the period of oscillation for mRNA expression for the genes in (A). The detailed analysis is presented in Tables S1 and S2. C, Representative western blotting for mTORC1 signaling in the liver of 2M‐CR mice. For every line the lysates from three different animals were pooled together. D, Representative western blotting for mTORC1 signaling in the liver of 1M‐CR mice. For every line the lysates from three different animals were pooled together. E, Representative western blotting for mTORC1 signaling in the liver of AL mice. For every line, the lysates from three different animals were pooled together. Arrows indicate the feeding time. Open and black bars indicate the light and dark time of the day
3.3. Effect of two‐meals per day CR on mTORC1 signaling
Mechanistic target of rapamycin (mTOR) is an important regulator of liver metabolism. 30 , 31 mTOR complex 1 (mTORC1) signaling is implicated in the regulation of longevity and mechanisms of CR. 32 mTORC1 activity in the liver is under circadian clock control and is highly sensitive to feeding/fasting. 33 , 34 , 35 , 36 The activity is reduced during fasting and increased upon re‐feeding. 37 CR reduces mTORC1 signaling in the liver in a time‐of‐the‐day‐dependent manner. 38 Therefore, we decided to check what will be the effect of 2M‐CR on mTORC1 signaling in the liver. mTORC1 activity was assayed by the phosphorylation of p70‐S6K1 protein kinase on Thr389 (direct target of mTORC1) and by the phosphorylation of ribosomal protein S6 on Ser235/236 (indirect target of mTORC1). The phosphorylation of both proteins demonstrated high amplitude oscillation around the day with two peaks: one around ZT2 and another around ZT14 (Figure 2C and Figure S4A,B). mTORC1 was highly circadian in both AL and 1M‐CR. In AL liver, the activity was high between ZT14 and ZT2, during the feeding period in AL mice, and low at ZT6‐ZT10 (Figure 2E and Figure S4B). In CR liver, the activity was low through most of the day with strong peak at ZT14‐ZT18, around the feeding time (Figure 2D and Figure S4B). The analysis of rhythms confirmed 24‐hour period for AL and 1M‐CR and 12‐hour period for 2M‐CR (Figure 2B, Tables S10 and S11). The pattern of daily mTORC1 activity under all diets was highly correlated with feeding/fasting cycle. The activity cycles were similar for AL and 1M‐CR mice and significantly different from the mTORC1 cycle in 2M‐CR liver, which demonstrated two distinct peaks. Thus, similar to the effect on circadian clock gene expression, 2M‐CR disrupted the circadian rhythms in mTORC1 signaling in the liver.
3.4. Two‐meals per day CR improved glucose homeostasis
Improvement of glucose homeostasis by CR is well documented. 6 We assayed blood glucose in AL, MT, 1M‐CR, and 2M‐CR. Glucose was assayed across the day at multiple time points and animals were not fasted before the blood collection. Across the day average blood glucose, assayed at multiple time points around the clock, is shown in Figure 3A. AL and MT mice have similar blood glucose levels. 1M‐CR have significantly reduced blood glucose compared with AL or MT. 2M‐CR resulted in significant reduction in blood glucose compared with AL and MT, but it was higher compared with 1M‐CR. Blood glucose around the day is presented in Figure 3B. AL blood glucose was slightly fluctuated. There were peaks in blood glucose before the feeding for three other diets: at ZT14 in MT mice, at ZT10 in 1M‐CR mice and two peaks at ZT2, and ZT12 in 2M‐CR. Thus, mice on different diets have different daily profiles of blood glucose. The rhythm analysis was performed for 12‐hour and 24‐hour period (Table S3). Blood glucose was circadian in MT and 1M‐CR. It was rhythmic with 12‐hour period in 2M‐CR (Table S3) and it was arrhythmic in AL.
FIGURE 3.
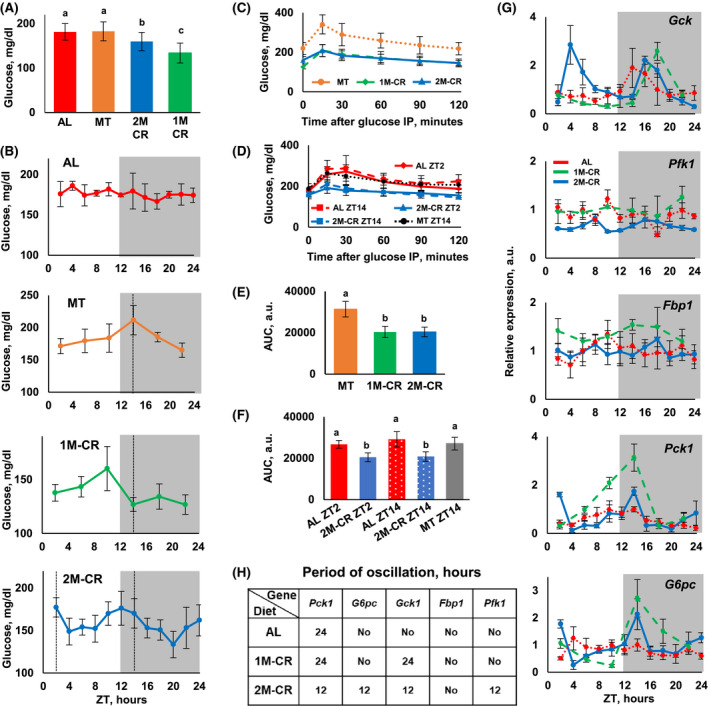
2M‐CR improves glucose homeostasis and reprograms the expression of rate‐limiting glucose metabolism genes. A, Daily average blood glucose level for mice on different diets. One‐way ANOVA was used for the statistical analysis; the statistical results are presented in Table S14. Bars that have the same letter are not different from each other, and bars with different letters are significantly different form each other indicating statistical difference (P < .05) between groups. AL (N = 13), MT (N = 6), 1M‐CR (N = 6), and 2M‐CR (N = 6). B, Blood glucose around the clock in mice on indicated diets. N = 6 per time point per diet. C, Glucose tolerance test (GTT) was performed at ZT14 with the mice: MT––dotted brown line, N = 8; 1M‐CR––dashed green line, N = 10; and 2M‐CR––solid blue line, N = 8. D, GTT was performed with the mice at ZT2––solid or ZT14 dashed lines, N = 8; AL––red line, N = 8; 2M‐CR––blue line, N = 8; and Time‐restricted feeding (TRF)––dotted gray line, N = 8. E, Area Under the Curve (AUC) for GTT experiment in (C). F, AUC for the experiment in (D). One‐way ANOVA was used for the statistical analysis; the statistical results are presented in Tables S15 and S16. Bars that have the same letter are not different from each other, and bars with different letters are significantly different form each other indicating statistical difference (P < .05) between groups. G, The expression of indicated glycolysis and gluconeogenesis genes in the liver of mice: AL––dotted red line; 1M‐CR––dashed green line; and 2M‐CR––solid blue line. N = 3 per time point per diet. Gray panels indicate dark phase of the day. (H) Results of rhythmic analysis indicate the period of oscillation for mRNA expression for the genes in (G). The detailed analysis is presented in Tables S4 and S5
To further investigate the effect of 2M‐CR on glucose homeostasis we performed GTT with MT, 1M‐CR, and 2M‐CR mice. Mice in all three groups received the food at ZT14, 2M‐CR mice also received the food at ZT2, and GTT was performed at ZT14 before the next feeding. The kinetic of GTT experiments is presented in Figure 3C and Area Under the Curve (AUC) in Figure 3E. Both 1M‐CR and 2M‐CR significantly improve glucose tolerance compared with MT mice. No difference was detected between one and two‐meal CR mice. Importantly, 1M‐CR mice were fasted for about 22 hours and 2M‐CR were fasted for only about 11 hours, thus, the duration of fasting before the GTT did not significantly affect the glucose tolerance. There is some evidence that daytime feeding might have negative effect on rodent glucose homeostasis, 39 one of two‐meals for 2M‐CR mice was at ZT2, the beginning of the light cycle. Therefore, we compared GTT for 2M‐CR mice at ZT2 and ZT14. AL mice were used as controls, the food was removed at ZT2 for GTT performed at ZT14, and at ZT14 for GTT performed at ZT2, thus the AL mice fasted for about 12 hours before GTT. Time‐restricted feeding (TRF) mice, which have unlimited food access between ZT14 and ZT2 and no food between ZT2 and Z14 for 2 months served as additional control. TRF mice consumed a comparable amount of food as AL mice, therefore, they were not restricted in caloric intake. GTT was performed with TRF mice at ZT14. Thus, in all five groups in this experiment the mice fasted for comparable time. The kinetics is shown in Figure 3D and Area Under the Curve (AUC) in Figure 3F. 2M‐CR have significantly improved glucose tolerance at both time points compared with fasted AL and TRF mice. There was no significant difference in glucose tolerance at ZT2 and at ZT14 for 2M‐CR mice. There was also no difference between AL and TRF mice in agreement with previously reported data. 24 Importantly, starting blood glucose levels in GTT experiments were different between the groups, MT and TRF mice have higher blood glucose compared with fasted AL and CR mice, which have about the same levels. These results suggest that the improved glucose tolerance in CR mice was not a direct consequence of the difference in starting blood glucose. Together results in Figure 3A‐F support the hypothesis that the improvement in glucose homeostasis was mostly a consequence of a reduced caloric intake and the duration of fasting was less significant.
CR affects the expression of glucose metabolism enzymes. 7 , 24 , 40 mRNA expression of Gck and Pfk1, which encode committed step glycolytic enzymes and Pck1, G6pc, and Fbp1, which encode committed step gluconeogenic enzymes across the day are shown in Figure 3G. The results of rhythms analysis are shown in Figure 3H and Tables S4 and S5. In AL liver, only Pck1 was rhythmic whereas G6pc and Gck demonstrated a tendency, but it did not reach a statistical significance. Pck1 was rhythmic with increased amplitude and Gck became rhythmic in 1M‐CR liver, of note G6pc expression was almost rhythmic in 1M‐CR. Twenty‐four‐hour rhythms in expression were disrupted in 2M‐CR liver for all glucose metabolism genes. Instead, with exception for Fbp1 that was not rhythmic under any diet, the expression of other four genes became rhythmic with 12‐hour period. Importantly for all dietary interventions the phases of expression of glucose metabolism genes were in agreement with their expected role in glucose metabolism. Pck1 and G6pc peaked before the feeding and Gck after the meal suggesting that liver rhythmic transcription was adjusted to better fit metabolic needs of the organism.
3.5. Fasting restored 24‐hour rhythms in circadian clock gene expression but not the rhythms in behavior or glucose metabolism
Two‐meals per day induced 12‐hour rhythms in behavior, gene expression, and metabolism. We asked if these 12‐hour rhythms represented some reprogramed metabolic adaptation or they are just a simple reflection of two‐meal feeding. In the case of reprograming one would expect that the rhythms would persist even without feeding. To test this hypothesis, 2M‐CR animals were fed at ZT14 and animals were without food for the next 48 hours. Locomotor activity was similar between fed and fasted 2M‐CR mice (compare Figure 4A and Figure S4C with Figure 1C and Figure S2). Fasted 2M‐CR mice demonstrated activity peaks before ZT2 and ZT14, the times when they expected the food, similar with fed mice. There was a tendency for the increase in total activity (Figure 4B), but it did not reach statistical significance. Fasting restored the circadian rhythms in the expression of clock genes (Figure 4C, Figure S3, Tables S6 and S8). However, as fasting progressed, some clock genes lost their 24‐hour rhythms and some retained. Thus, 12‐hour rhythms in clock gene expression were a direct consequence of the feeding pattern rather than a metabolic reprograming. Without food as an external cue the expressions of circadian clock genes were regulated by an internal mechanism, which has a 24‐hour period.
FIGURE 4.
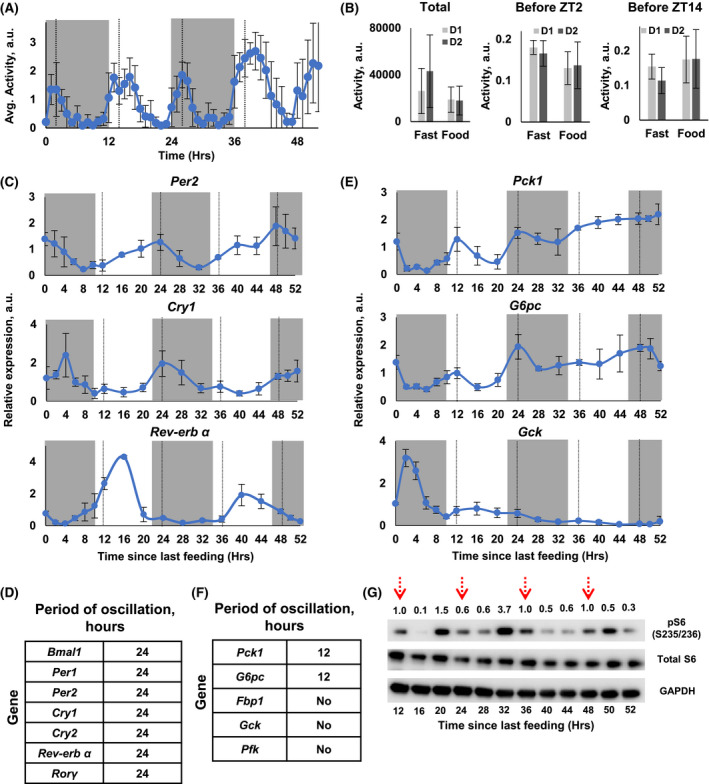
Effect of fasting on circadian and 12‐hour rhythms in 2M‐CR mice. 2M‐CR mice were fed at ZT14, indicated by solid black line, and were not fed for the next 48 hours, dotted black line indicate expected time of feeding. A, Average in‐cage locomotor activity for mice. B, In‐cage activity on Day 1 (light gray) and Day 2 (dark gray): total activity––left panel; the activity in 2 hours before ZT2, data are shown as a fraction of total daily activity, which was set up as 1.0––center panel; the activity in 2 hours before ZT14, data are shown as a fraction of total daily activity, which was set up as 1.0––right panel. C, The expression of circadian clock genes in fasted 2M‐CR mice, N = 3 per time point. D, Results of rhythmic analysis indicate the period of oscillation for mRNA expression for the genes in (C). The detailed analysis is presented in Tables S6 and S8. E, The expression of glucose metabolism genes in fasted 2M‐CR mice, N = 3 per time point. F, Results of rhythmic analysis indicate the period of oscillation for mRNA expression for the genes in (E). The detailed analysis is presented in Tables S7 and S9. G, Representative western blotting for S6 protein phosphorylation on Ser235/236 in the liver of fasted 2M‐CR mice. For every line, the lysates from three animals were pooled together. Red dotted arrows indicate time when mice were supposed to receive food instead were fasted
Rhythms in glucose metabolism genes demonstrated different patterns in the liver of fasted 2M‐CR mice (Figure 4D, Figure S3, Tables S7 and S9). Pck1 and G6pc still demonstrated robust 12‐hour rhythms and the peaks coincided with the expected time of feeding. As duration of fasting increased, the expression of both genes gradually increased, and the amplitude of oscillation decreased. Gck expression decreased with duration of fasting, the single peak in the expression was detected only after the feeding. Thus, rhythms in Pck1 and G6pc expression were a result of metabolic reprograming and they were maintained through some internal mechanism. Moreover, the 12‐hour rhythmic expression of Pfk1 and Gck was a direct reflection of rhythmic feeding. Finally, phosphorylation of ribosomal protein S6 demonstrated robust oscillations with 12‐hour period in fasted liver (Figure 4E, Figure S3 and Tables S12 and S13) suggesting that it was regulated by some internal mechanism.
4. DISCUSSION
Improvement in glucose homeostasis by CR is implicated in increased longevity. 6 In mammals, CR is a combination of reduced food intake and self‐implemented periodic fasting. 7 , 8 , 9 The relative contribution of each of these components is not known. It was proposed that periodic fasting diets: IF and TRF can provide some metabolic benefits, including effect on blood glucose, without reduction in food intake. 1 IF without reduction in caloric intake reduces blood glucose on regular chow. 2 However, it is unclear if the blood glucose was assayed on the day of fasting or the day of AL feeding. Importantly, the reduction in blood glucose is not necessarily an indication of improved glucose homeostasis. For example, blood glucose level is reduced with the duration of fasting and 24‐hour fasting reduces blood glucose to a level comparable with CR or lower, however, random fasting does not improve glucose tolerance. 24 GTT was not performed in IF study, 2 therefore, the effect of IF on glucose homeostasis still needs to be assayed. TRF reduces blood glucose and improves glucose tolerance in mice with metabolic syndrome induced by high‐fat diet. 3 TRF with the same duration of fasting, 12 hours, does not improve glucose metabolism in healthy young mice on regular chow. 24 MT––a self‐implemented periodic fasting was also reported not to improve glucose homeostasis, 5 as in this study. 2M‐CR mice were fasted for not more than 12 hours, but their glucose homeostasis was improved to the level comparable with 1M‐CR (Figure 3). Thus, neither periodic fasting without reduction in caloric intake nor the reduction in caloric intake without periodicity were able to improve glucose homeostasis, but the combination of both was sufficient.
What might be the importance of the periodic component in CR? Acute fasting induces some changes in signaling and gene expression, but these changes can be easily reversed. For example: the blood glucose gradually reduced with duration of fasting and rapidly returned back to AL level in 1 hour after refeeding. 24 In contrast to this, the blood glucose in CR mice is tightly regulated and maintained near stable level independently from the duration of fasting. We propose that the periodicity of fasting might be important for reprograming of transcriptome/translatome in the liver and, probably, other tissues.
Periodic feeding/fasting cycle is interlinked with the circadian clock. 19 , 20 The circadian clock is a master regulator of metabolism and clock disruption is associated with metabolic syndrome and increased risk of cardiometabolic diseases. 19 For example, HF diet disrupts the circadian rhythms in gene expression and metabolism and these disruptions might contribute to HF‐induced metabolic syndrome. 26 In agreement with this, TRF restores the circadian rhythms and ameliorates metabolic abnormalities induced by HF diet, and it is hypothesized that the restoration of the rhythms plays a major role in the improved metabolism. 3 Restoration of the rhythms by TRF in a disease state is a question of great clinical importance due to increased obesity and cardiometabolic diseases throughout the world. However, there is another compelling question; could the circadian clock and rhythms be involved in the improvement of metabolism in a healthy state? There is a significant body of evidence that dietary interventions known to improve metabolism in healthy animals such as CR or ketogenic diet also significantly impact the clock. 21 , 41 , 42 In line with that, full metabolic benefits of CR, including the effect on longevity, cannot be achieved in circadian clock mutants. The effect is conserved in mice and flies, 8 , 22 but in flies the outcome might be circadian clock gene specific. 43 Pharmacological intervention that increases the robustness of the clock also improves metabolism and longevity. 44 Therefore, improvement in glucose homeostasis when circadian rhythms are disrupted is somewhat surprising. This suggests that the interaction between the circadian rhythms and metabolism in CR is complicated and improvement in glucose homeostasis can be uncoupled from the circadian clocks and rhythms. Interestingly, TRF improves HF diet disrupted metabolism even in circadian clock mutant mice. 45 In Drosophila, both the overexpression 46 and deletion 43 of clock genes might increase life span. Thus, a connection between the clock, metabolism, and longevity is complicated.
The plasticity of circadian metabolic rhythms is well documented. Peripheral circadian clocks can be phase shifted by restricted feeding, 27 and it is believed that this shift contributes to metabolic adaptation. 2M‐CR did not have uniform effects on the expression of canonical clock genes (Figure 2). Some were only slightly affected while others adapted to 12‐hour rhythms. Interestingly, feeding around the clock, with 3‐hour interval, also have differential effect on clock gene expression: the expressions of Bmal1, Per1, or Cry1 are not significantly affected and the amplitudes of Per2 and Rev‐erbα are reduced. 13 The same study also illustrates the fact that most of 24‐hour rhythms in liver transcriptome are driven by periodic feeding rather than an internal clock. Thus, meal frequency dictates changes in clock gene expression. Replacement of 24‐hour rhythms with 12‐hour rhythms might be essential for metabolic adaptation to 12‐hour periodicity in food supply. Interestingly, 12‐hour rhythms in gene expression can be detected for AL mice, but the physiological significance of these rhythms and their mechanism is unknown. 14 The changes in the expression of several rate‐limiting glucose metabolism enzymes (Figure 3) were in agreement with the hypothesis on metabolic adaptation. Low amplitude oscillations in blood glucose in both 1M‐CR and 2M‐CR strongly correlated with feeding patterns: 24‐hour in 1M‐CR and 12‐hour in 2M‐CR. Importantly, the peaks in blood glucose were detected before the feeding and coincided with the peaks in mRNA expression for Pck1 and G6pc, committed step gluconeogenic enzymes, and can be an indication of the increased gluconeogenesis. We also cannot exclude the possibility that the peaks were caused by reduced glucose consumption in skeletal muscles and/or adipose tissues. Furthermore, detailed studies on kinetics of glucose production and consumption in CR animals will help to answer this question. mTOR signaling in the liver and other tissues is rhythmic and can be entrained by the food. 34 , 36 , 47 It was proposed that rhythms in mTOR signaling might be important for circadian metabolism. This study demonstrated that mTOR rhythms can be entrained to 12‐hour periodicity, again in agreement with metabolic adaptation to meal frequency. There is a cross talk between the circadian clock and mTOR signaling. 33 , 35 BMAL1 and PER2 regulate the rhythms in mTORC1 activity. 48 , 49 In turn, mTORC1 feeds back to regulate circadian rhythms. 33 , 50 The current study suggests that meal frequency can uncouple rhythms in mTORC1 and expression of clock genes. mTOR rhythms can be detected even in circadian clock mutants, 34 which supports a relative independence of these oscillators. Importantly, mTOR is a nutrient sensor, it is expected that mTOR activity in the liver is mostly dictated by hormones such as insulin and the level of nutrients such as amino acids. 30 , 51 Twelve‐hour rhythms in mTORC1 activity were not surprising under 12‐hour meal frequency. Moreover, 12‐hour rhythms in mTORC1 signaling persisted in fasting suggesting some internal oscillatory mechanism. Light‐entrained circadian clock, driven by a dozen of transcriptional regulators, which form molecular transcriptional‐translational feedback loops is well described. Recent evidence suggests the existence of circadian rhythms, which do not depend on of the transcriptional‐translational loop and canonical clock genes. The examples are 24‐hour redox rhythms in red blood cells, 52 periodic transcriptome and proteome in Bmal1 deficient cells, 53 and robust food anticipatory rhythms in circadian clock mutants. 54 Several interesting questions are raised as a result of our study: are the same or different clock‐like mechanisms responsible for 12‐hour rhythms in behavior, glucose metabolism, and mTORC1 signaling, and do they interact with other clocks such as the light‐entrained circadian clock, transcription independent redox clock, and Bmal1 independent circadian molecular oscillator?
In summary, two‐meals per day CR‐induced 12‐hour metabolic rhythms, suggesting reprograming and metabolic adaptation to meal frequency. Some of these rhythms were a direct reflection of feeding, while others persisted in fasting for 48 hours suggesting the existence of some internal clock‐like mechanisms. These oscillators, most likely, are different from canonical circadian clocks that generate 24‐hour rhythms. CR robustly increases longevity in contrast to a modest effect of MT, which correlates with the effect of these diets on glucose homeostasis. Our data supported the importance of caloric intake and demonstrated a high plasticity of metabolic adaptation to meal frequency under CR.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
RK and NV designed the experiments. NV, VM, and AP performed the experiments. RK, NV, and VM analyzed the data. RK, NV, and VM prepared the figures and wrote the manuscript. All authors reviewed and approved the manuscript.
Supporting information
Fig S1
Fig S2
Fig S3
Fig S4
Table S1‐S16
ACKNOWLEDGMENTS
This work was supported by NIH grant 5RO1‐AG039547 and internal support by the GRHD Department at CSU. The authors thank the members of Kondratov's laboratory for technical assistance.
Velingkaar N, Mezhnina V, Poe A, Kondratov RV. Two‐meal caloric restriction induces 12‐hour rhythms and improves glucose homeostasis. The FASEB Journal. 2021;35:e21342 10.1096/fj.202002470R
Funding information
This work was supported by NIH grant 5RO1‐AG039547 and internal support by the GRHD Department at CSU.
Contributor Information
Nikkhil Velingkaar, Email: n.velingkaar@vikes.csuohio.edu.
Roman V. Kondratov, Email: r.kondratov@csuohio.edu.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in Mendeley Data at http://dx.doi.org/10.17632/txz8y4dsn8.1
REFERENCES
- 1. Mattson MP, Allison DB, Fontana L, et al. Meal frequency and timing in health and disease. Proc Natl Acad Sci USA. 2014;111:16647‐16653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anson RM, Guo Z, de Cabo R, et al. Intermittent fasting dissociates beneficial effects of dietary restriction on glucose metabolism and neuronal resistance to injury from calorie intake. Proc Natl Acad Sci USA. 2003;100:6216‐6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hatori M, Vollmers C, Zarrinpar A, et al. Time‐restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high‐fat diet. Cell Metab. 2012;15:848‐860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sherman H, Genzer Y, Cohen R, Chapnik N, Madar Z, Froy O. Timed high‐fat diet resets circadian metabolism and prevents obesity. FASEB J. 2012;26:3493‐3502. [DOI] [PubMed] [Google Scholar]
- 5. Mitchell SJ, Bernier M, Mattison JA, et al. Daily Fasting Improves Health and Survival in Male Mice Independent of Diet Composition and Calories. Cell Metab. 2018;29:221‐228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Longo VD, Antebi A, Bartke A, et al. Interventions to slow aging in humans: are we ready? Aging Cell. 2015;14:497‐510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bruss MD, Khambatta CF, Ruby MA, Aggarwal I, Hellerstein MK. Calorie restriction increases fatty acid synthesis and whole body fat oxidation rates. Am J Physiol ‐ Endocrinol Metab. 2010;298:E108‐E116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Patel SA, Chaudhari A, Gupta R, Velingkaar N, Kondratov RV. Circadian clocks govern calorie restriction‐mediated life span extension through BMAL1‐ and IGF‐1‐dependent mechanisms. FASEB J. 2016;30:1634‐1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Acosta‐Rodríguez VA, de Groot MHM, Rijo‐Ferreira F, Green CB, Takahashi JS. Mice under Caloric Restriction Self‐Impose a Temporal Restriction of Food Intake as Revealed by an Automated Feeder System. Cell Metab. 2017;26:267‐277.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chaix A, Zarrinpar A, Phuong Miu SP. Time‐restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab. 2014;20:991‐1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sutton EF, Beyl R, Early KS, Cefalu WT, Ravussin E, Peterson CM. Early time‐restricted feeding improves insulin sensitivity, blood pressure, and oxidative stress even without weight loss in men with prediabetes. Cell Metab. 2018;27:1212‐1221.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Taormina G, Mirisola MG. Calorie restriction in mammals and simple model organisms. Biomed Res Int. 2014;2014:308690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Greenwell BJ, Trott AJ, Beytebiere JR, et al. Rhythmic food intake drives rhythmic gene expression more potently than the hepatic circadian clock in mice. Cell Rep. 2019;27:649‐657. [DOI] [PubMed] [Google Scholar]
- 14. Atger F, Gobet C, Marquis J, et al. Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc Natl Acad Sci USA. 2015;112:E6579‐E6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nakahata Y, Grimaldi B, Sahar S, Hirayama J, Sassone‐Corsi P. Signaling to the circadian clock: plasticity by chromatin remodeling. Curr Opin Cell Biol. 2007;19:230‐237. [DOI] [PubMed] [Google Scholar]
- 16. Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445‐462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aguilar‐Arnal L, Sassone‐Corsi P. Chromatin landscape and circadian dynamics: Spatial and temporal organization of clock transcription. Proc Natl Acad Sci USA. 2015;112:6863‐6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eckel‐Mahan KL, Patel VR, De Mateo S, et al. Reprogramming of the circadian clock by nutritional challenge. Cell. 2013;155:1464‐1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bass J. Circadian topology of metabolism. Nature. 2012;491:348‐356. [DOI] [PubMed] [Google Scholar]
- 20. Asher G, Sassone‐Corsi P. Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell. 2015;161:84‐92. [DOI] [PubMed] [Google Scholar]
- 21. Sato S, Solanas G, Peixoto FO, et al. Circadian reprogramming in the liver identifies metabolic pathways of aging. Cell. 2017;170:664‐677.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Katewa SD, Akagi K, Bose N, et al. Peripheral circadian clocks mediate dietary restriction‐dependent changes in lifespan and fat metabolism in drosophila. Cell Metab. 2016;23:143‐154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Piper MD, Bartke A. Diet and aging. Cell Metab. 2008;8:99‐104. [DOI] [PubMed] [Google Scholar]
- 24. Velingkaar N, Mezhnina V, Poe A, Makwana K, Tulsian R, Kondratov RV. Reduced caloric intake and periodic fasting independently contribute to metabolic effects of caloric restriction. Aging Cell e13138. 2020;19(4):13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mistlberger RE. Neurobiology of food anticipatory circadian rhythms. Physiol Behav. 2011;104:535‐545. [DOI] [PubMed] [Google Scholar]
- 26. Kohsaka A, Laposky AD, Ramsey KM, et al. High‐fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414‐421. [DOI] [PubMed] [Google Scholar]
- 27. Damiola F, Le Minli N, Preitner N, Kornmann B, Fleury‐Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950‐2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Astafev AA, Patel SA, Kondratov RV. Calorie restriction effects on circadian rhythms in gene expression are sex dependent. Sci Rep. 2017;7(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chavan R, Feillet C, Costa SSF, et al. Liver‐derived ketone bodies are necessary for food anticipation. Nat. Commun. 2016;7:10580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kennedy BK, Lamming DW. The mechanistic target of rapamycin: the grand conductor of metabolism and aging. Cell Metab. 2016;23:990‐1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471‐484. [DOI] [PubMed] [Google Scholar]
- 32. Lamming DW, Sabatini DM. A radical role for TOR in longevity. Cell Metab. 2011;13:617‐618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cao R, Robinson B, Xu H, et al. Translational control of entrainment and synchrony of the suprachiasmatic circadian clock by mTOR/4E‐BP1 signaling. Neuron. 2013;79:712‐724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Khapre RV, Patel S, Kondratova AA, et al. Metabolic clock generates nutrient anticipation rhythms in mTOR signaling. Aging (Albany. NY). 2014;6:675‐689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lipton JO, Yuan ED, Boyle LM, et al. The circadian protein BMAL1 regulates translation in response to S6K1‐mediated phosphorylation. Cell. 2015;161:1138‐1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jouffe C, Cretenet G, Symul L, et al. The circadian clock coordinates ribosome biogenesis. PLoS Biol. 2013;11:e1001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960‐976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tulsian R, Velingkaar N, Kondratov R. Caloric restriction effects on liver mTOR signaling are time‐of‐day dependent. Aging (Albany, NY). 2018;10:1640‐1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mukherji A, Kobiita A, Chambon P. Shifting the feeding of mice to the rest phase creates metabolic alterations, which, on their own, shift the peripheral circadian clocks by 12 hours. Proc Natl Acad Sci USA. 2015;112(48):E6683–E6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hagopian K, Ramsey JJ, Weindruch R. Influence of age and caloric restriction on liver glycolytic enzyme activities and metabolite concentrations in mice. Exp. Gerontol. 2003;38:253‐266. [DOI] [PubMed] [Google Scholar]
- 41. Solanas G, Peixoto FO, Perdiguero E, et al. Aged stem cells reprogram their daily rhythmic functions to adapt to stress. Cell. 2017;170:678‐692.e20. [DOI] [PubMed] [Google Scholar]
- 42. Tognini P, Murakami M, Liu Y, et al. Distinct circadian signatures in liver and gut clocks revealed by ketogenic diet. Cell Metab. 2017;26:523‐538. [DOI] [PubMed] [Google Scholar]
- 43. Ulgherait M, Chen A, McAllister SF, et al. Circadian regulation of mitochondrial uncoupling and lifespan. Nat. Commun. 2020;11:1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nohara K, Mallampalli V, Nemkov T, et al. Nobiletin fortifies mitochondrial respiration in skeletal muscle to promote healthy aging against metabolic challenge. Nat. Commun. 2019;10:3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chaix A, Lin T, Le HD, Chang MW, Panda S. Time‐restricted feeding prevents obesity and metabolic syndrome in mice lacking a circadian clock. Cell Metab. 2019;29:303‐319.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Solovev I, Shegoleva E, Fedintsev A, Shaposhnikov M, Moskalev A. Circadian clock genes’ overexpression in Drosophila alters diet impact on lifespan. Biogerontology. 2019;20:159‐170. [DOI] [PubMed] [Google Scholar]
- 47. Cao R, Anderson FE, Jung YJ, Dziema H, Obrietan K. Circadian regulation of mammalian target of rapamycin signaling in the mouse suprachiasmatic nucleus. Neuroscience. 2011;181:79‐88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Khapre RV, Kondratova AA, Patel S, et al. BMAL1‐dependent reulation of the mTOR signaling pathway delays aging. Aging (Albany NY). 2014;6:48‐57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu R, Dang F, Li P, et al. The circadian protein period2 suppresses mTORC1 activity via recruiting Tsc1 to mTORC1 complex. Cell Metab. 2019;29:653‐657. [DOI] [PubMed] [Google Scholar]
- 50. Cao R, Lee B, Cho HY, Saklayen S, Obrietan K. Photic regulation of the mTOR signaling pathway in the suprachiasmatic circadian clock. Mol Cell Neurosci. 2008;38:312‐324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274‐293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469:498‐503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ray S, Valekunja UK, Stangherlin A, Howell SA, Snijders AP, Damodaran G, Reddy AB. (2020) Circadian rhythms in the absence of the clock gene Bmal1. Science. 367:800–806. [DOI] [PubMed] [Google Scholar]
- 54. Patton DF, Mistlberger RE. Circadian adaptations to meal timing: Neuroendocrine mechanisms. Front. Neurosci. 2013;7:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Fig S4
Table S1‐S16
Data Availability Statement
The data that support the findings of this study are openly available in Mendeley Data at http://dx.doi.org/10.17632/txz8y4dsn8.1


