Summary
Plant hormones are essential for regulating the interactions between plants and their complex biotic and abiotic environments. Each hormone initiates a specific molecular pathway and these different hormone pathways are integrated in a complex network of synergistic, antagonistic and additive interactions. This inter‐pathway communication is called hormone crosstalk. By influencing the immune network topology, hormone crosstalk is essential for tailoring plant responses to diverse microbes and insects in diverse environmental and internal contexts. Crosstalk provides robustness to the immune system but also drives specificity of induced defense responses against the plethora of biotic interactors. Recent advances in dry‐lab and wet‐lab techniques have greatly enhanced our understanding of the broad‐scale effects of hormone crosstalk on immune network functioning and have revealed underlying principles of crosstalk mechanisms. Molecular studies have demonstrated that hormone crosstalk is modulated at multiple levels of regulation, such as by affecting protein stability, gene transcription and hormone homeostasis. These new insights into hormone crosstalk regulation of plant defense are reviewed here, with a focus on crosstalk acting on the jasmonic acid pathway in Arabidopsis thaliana, highlighting the transcription factors MYC2 and ORA59 as major targets for modulation by other hormones.
Keywords: hormone crosstalk, defense, network, jasmonic acid, salicylic acid, abscisic acid, ethylene, MYC2, ORA59
Significance Statement
Plant hormone pathways interact with each other in a complex network, which manages synergistic, antagonistic and additive effects between different sectors of the network, thereby providing robustness and specificity to the immune system. Here we review hormone crosstalk regulation at the network, protein, gene expression and hormone homeostasis levels.
INTRODUCTION
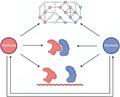
Plants in nature and agriculture are constantly interacting with their biotic and abiotic environment. To ensure their survival in different and often hostile conditions they evolved a sophisticated and flexible environmental signaling network that is steered by plant hormones. This elaborate hormone‐controlled network finetunes the plants’ responses according to highly dynamic and heterogeneous circumstances. Immune signaling is part of this overarching network and can be activated and tweaked by the intricate molecular communication between the plant and the microbe or insect that it encounters. The intertwinement of the immune network with other stress and internal networks allows for adjustments in plant defense responses according to the abiotic conditions, plant developmental stage and time of day (Atkinson and Urwin, 2012; Lu et al., 2017; Nobori and Tsuda, 2019; Figure 1).
Figure 1.
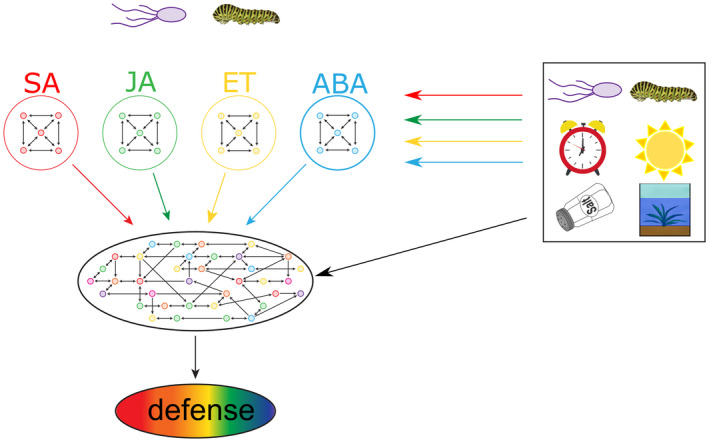
Schematic overview of integration of hormone networks involved in plant defense. Microbes and insects elicit the accumulation of specific blends of hormones. The main hormones involved in the regulation of plant defense responses are salicylic acid (SA), jasmonic acid (JA), ethylene (ET) and abscisic acid (ABA). Each hormone regulates its own pathway, but also influences other hormone pathways in a complex mix of synergistic, antagonistic and additional interactions, a phenomenon known as hormone crosstalk. Moreover, accumulation of these hormones and the responsiveness to them can be further modulated by (i) light quality, (ii) time of day, (iii) abiotic stresses such as drought, flooding and salt stress and (iv) prior or simultaneous interactions with other microbes or insects. Integration of the different hormone networks shapes the defense response leading to elimination or accommodation of the microbe or insect in diverse environmental and internal contexts.
Plant hormones are central regulators of plant immunity. Depending on the type of attacker different hormones accumulate in the plant, whereby each hormone regulates its own core pathway in the immune network (Figure 1). The two most studied defense pathways are those regulated by jasmonic acid (JA) and salicylic acid (SA), which form the backbone of the hormone‐regulated part of the immune system (Wasternack and Song, 2017; Zhang and Li, 2019). The JA pathway can be subdivided into two branches (Pieterse et al., 2012). The ERF branch of the JA pathway is co‐regulated by ethylene (ET) and is activated by infection with pathogens with a necrotrophic lifestyle. The MYC branch of the JA pathway is co‐regulated by abscisic acid (ABA) and generally provides protection against chewing insects. The SA pathway is considered to be mostly directed against pathogens with a biotrophic lifestyle. So, the infection or infestation strategy of the attacker determines which hormones accumulate and which pathways the plant activates to express the appropriate defense responses to the attacker at hand. Moreover, hormone homeostasis is greatly influenced by the status of the plant, being internal, for example age, or being external, for example experiencing other stresses (Berens et al., 2019; Nobori and Tsuda, 2019). Overall, the final hormone balance and responsiveness is a cumulative result of the activation of plant immunity and the context in the plant (Figure 1).
The plant immune system is built on two layers, and hormone signaling is essential for both layers. In the first layer, plants recognize small conserved microbe‐ or insect‐derived molecules, called microbe/pathogen‐associated molecular patterns (M/PAMPs) or herbivore‐associated molecular patterns (HAMPs). If there is damage caused by an attacker, plant‐derived small molecules called damage‐associated molecular patterns (DAMPs) are released and these can also be recognized. P/M/HAMPs and DAMPs trigger immune signaling resulting in pattern‐triggered immunity (PTI), which wards off most of the non‐adapted microbes and insects (Dangl et al., 2013; Erb and Reymond, 2019). However, successful pathogens and insects, which can be pathogenic, beneficial or neutral to the plant, can secrete variable effectors into the host plant to suppress PTI signaling. This is known as effector‐triggered susceptibility (ETS) and commonly is established by repression of effective defense hormone pathways (Han and Kahmann, 2019). Resistant plants recognize these effectors or their action, setting off a second layer of immunity called effector‐triggered immunity (ETI). In the case of plant interactions with biotrophic pathogens ETI often results in a hypersensitive response (HR), which arrests the invading pathogen (Cui et al., 2015). During PTI, ETS and ETI, plant hormones trigger extensive transcriptional reprogramming and thereby tightly regulate defense responses (Berens et al., 2017). This ultimately leads to elimination of harmful microbes and insects and accommodation of beneficial microbes and insects, which can occur simultaneously in the plant.
It is important for plant health and long‐term survival that defense responses are finetuned to turn on effective defenses but switch off ineffective defenses. Moreover, defense responses need to be balanced with general housekeeping and responses to other stresses (Vos et al., 2013a; Vos et al., 2015; Berens et al., 2017; Van Butselaar and Van den Ackerveken, 2020). To this end, different hormone networks interact in a complex interplay of synergistic, antagonistic and additive interactions, a phenomenon known as hormone crosstalk (Figure 1). Hormone crosstalk is an important component of the architecture of the immune signaling network. Besides finetuning and balancing of responses, antagonistic interactions can also serve to provide robustness to the response. For example, two sectors can positively regulate the same immune response, but negatively regulate each other. That means that if one sector is compromised (for example by manipulation by a pathogen) the other sector is derepressed and can take over the function of the first sector. The classical example of crosstalk in defense regulation is that between the SA and JA pathways. Antagonism between these two pathways is the most studied and prevalent form, although large‐scale additive and synergistic interactions have been described as well (Hickman et al., 2019). Additionally, the ERF branch and the MYC branch of the JA pathway have been reported to repress each other (Pieterse et al., 2012; Gimenez‐Ibanez and Solano, 2013; Wasternack and Hause, 2013).
A molecular‐ and systems‐level understanding of hormone crosstalk will improve our predictions of effects that disruption or overactivation of parts of the network have on the overall plant response. Implementation of this knowledge can help breeders to engineer crops with a strengthened immune response without undesired traits like enhanced susceptibility to other attackers or decreased plant growth and yield. Here, we review recent advances in hormone crosstalk within the immune network. Different levels of regulation, from network and genome scale to single gene and protein scale, are described. We focus on crosstalk in the JA pathway in Arabidopsis thaliana (hereafter Arabidopsis), as a showcase for the multiple regulation levels of pathway interference in hormone defense signaling.
CROSSTALK AT THE NETWORK LEVEL
Studying crosstalk at the network level enables the investigation of crosstalk without defining beforehand all the individual components. This coarse‐grain overview can reveal the overall architecture of the hormone‐regulated plant immune system. Furthermore, it can provide hypotheses about crosstalk at more fine‐grain levels, which can be validated experimentally.
A network approach encompasses gathering information on a genome‐wide scale. As RNA sequencing (RNA‐seq) is the most widely available genome‐wide technique, information on the transcriptome is most commonly used for hormone network‐scale analyses until now. However, newly emerging technologies also allow gathering information on other regulation levels such as the proteome and translatome (Lee and Bailey‐Serres, 2019; Zander et al., 2020). In addition to these molecular data, relatively simple phenotypical readouts can be used for network analysis. The different types of data are usually gathered from leaves of plants that are given a stimulus such as hormone application or pathogen infection. Comparisons can be made between effects that one stimulus has on various mutants that are impaired in hormone signaling sectors. Alternatively, effects of different hormone treatments on one plant genotype (wild type) can be compared.
Network modeling using hormone mutants
A good example of network‐level research is a series of papers that describe how different hormone sectors interact to regulate PTI and ETI (Tsuda et al., 2009; Kim et al., 2014; Hillmer et al., 2017; Mine et al., 2017). The researchers used single and higher‐order mutants of key regulators of the SA, JA and ET pathways to understand how each pathway contributes to PTI and ETI. A mutant of PAD4 was added because although PAD4 expression is known to be induced by SA and SA response‐eliciting pathogens, PAD4 itself can regulate both SA‐dependent and ‐independent responses (Jirage et al., 1999; Glazebrook et al., 2003). Using bacterial growth as a readout, they found that each of the four sectors alone positively contributes to both PTI and ETI (see also Figure 1). However, interactions between the sectors differ between the PTI and ETI responses (Tsuda et al., 2009). The PTI response involves both synergistic and antagonistic interactions (Tsuda et al., 2009; Kim et al., 2014) and gene expression in this response is almost always influenced by one or multiple interactions between sectors (Hillmer et al., 2017). Robustness during PTI is mostly provided by the JA and ET sectors. This was demonstrated by the finding that in a mutated JA or ET background knocking out another sector had much more impact on MAMP‐induced immunity levels against two Pseudomonas syringae strains than in the wild‐type background (Kim et al., 2014). In contrast, during ETI all of the sectors act antagonistically and can (partially) take over the response if one of the sectors is inactive. This crosstalk mechanism ensures that the ETI response is robust against manipulation or dysfunction of one of the involved network sectors caused by an attacker or another stimulus (Tsuda et al., 2009).
An example of how such network robustness can be achieved was elegantly demonstrated in a follow‐up paper by Mine et al. (2017). They provided evidence for a robust regulation of SA biosynthesis by interactions between transcriptional regulators of the JA, SA and PAD4 sectors. These regulators form a so‐called incoherent type 4 feed‐forward loop, in which two components positively regulate one target, but one of these two components also negatively regulates the other component (Mangan and Alon, 2003). In this case, both PAD4 and the JA master regulator MYC2 (Kazan and Manners, 2013) positively regulate EDS5 (Mine et al., 2017), a gene essential for SA biosynthesis (Rekhter et al., 2019), but MYC2 represses PAD4 (Mine et al., 2017). These interactions provide robustness to the system, which was demonstrated by the following. In wild‐type plants PAD4 positively contributed to basal and flg22‐induced EDS5 expression, whereas the JA sector had no effect on EDS5 expression and in fact contributed negatively to SA accumulation (Mine et al., 2017). However, when PAD4 was compromised, which in nature could result from activity of a pathogen effector or high temperature, MYC2 was able to activate EDS5 and hence, the JA pathway could positively influence SA biosynthesis. This was demonstrated using the dde2 pad4 mutant (impaired in JA signaling and the PAD4 sector), which, compared to the pad4 single mutant, exhibited lower levels of flg22‐induced EDS5 expression and free SA (Mine et al., 2017). So, in the case of SA biosynthesis the JA sector can functionally replace the PAD4 sector when the latter is compromised. Moreover, a direct interaction between the MYC2 and PAD4 proteins has been shown to affect free SA accumulation (Cui et al., 2018), as described in the section ‘Crosstalk at the hormone homeostasis level’. Vice versa, the SA sector stimulates JA biosynthesis during ETI (Liu et al., 2016a), which is described in the section ‘Crosstalk by modulation of protein stability’.
Network modeling using time series of hormone treatments of wild‐type plants
A complementary approach to the above‐described systems biology network studies using mutants as conducted by the Tsuda and Katagiri groups is using hormone applications to wild‐type plants. A high‐throughput time series set‐up can provide extra power to the analysis, as this unveils regulatory connections between different components that shape the dynamic architecture of the network. Such an approach facilitates our understanding of the temporal information flow through the different sectors of the individual hormone regulatory networks, including the interactions with sectors of other hormone networks. This approach was taken for JA (Hickman et al., 2017; Zander et al., 2020), the JA mimic phytotoxin (of P. syringae) coronatine (Attaran et al., 2014), SA (Hickman et al., 2019), ET (Chang et al., 2013) and ABA (Song et al., 2016), which followed up on seminal time series papers studying responses to pathogen infection (Windram et al., 2012; Lewis et al., 2015).
Hickman et al. (2017) built a gene regulatory network model of the JA response based on a transcriptome study of a time series of 14 time points within 16 after a one‐time treatment of mature Arabidopsis with aqueous methyl jasmonate (MeJA), which is converted to JA in the plant. The majority of the differentially expressed genes were detected as such within 2 h after treatment. Correlation analysis of expression at different time points showed that there were six distinct phases of upregulation and four distinct phases of downregulation. Each phase was enriched for different processes and contained specific transcription factors (TFs) that were predicted to regulate genes in subsequent phases, based on enrichment of TF binding motifs in genes differentially expressed during these phases. Intersections of the JA network with other hormone networks were also observed. For example, genes related to the SA pathway were downregulated in early phases (1–2 h) and genes related to the growth hormone auxin were downregulated in later phases (3–4 h).
Zander et al. (2020) integrated more data types to elucidate the JA response in etiolated seedlings that had received continuous treatment with gaseous MeJA for up to 24 h. They focused on the role of MYC2 (Boter et al., 2004; Lorenzo et al., 2004; Dombrecht et al., 2007) and MYC3 (Fernández‐Calvo et al., 2011) as master regulators of the JA response by conducting chromatin immunoprecipitation–sequencing (ChIP‐seq) analysis of these TFs, and ChIP‐seq or DNA affinity purification–sequencing (DAP‐seq) (basically in vitro ChIP‐seq) of five of their targets. In addition, six other known JA‐related TFs were included in their ChIP‐seq/DAP‐seq experiments. They also generated proteome and phosphoproteome data, and integrated these with the other data types to infer a regulatory network. This network contained known and new components of the JA regulatory network and pointed to known and novel nodes of crosstalk of the JA pathway with other hormone pathways. An important role for MYC2 and MYC3 in modulation of other hormone pathways was demonstrated by the finding that 37–59% of genes that are annotated as being part of other hormone signaling pathways were bound by MYC2 and MYC3 and that their transcription responded to the MeJA treatment. Furthermore, the JA‐induced transcriptional repressor STZ was predicted to suppress genes from several other hormone pathways, including the SA, gibberellin (GA) and brassinosteroid (BR) pathways (Hickman et al., 2019; Zander et al., 2020).
In a follow‐up paper of the Hickman et al. (2017) paper on individual MeJA treatment, another part of the same experiment was reported, for which plants received a single SA treatment or a combination treatment of MeJA with SA (Hickman et al., 2019). The single SA treatment had a greater impact on the transcriptome than the single MeJA treatment, affecting the expression of more genes and having more prolonged effects. Validation of the built SA gene regulatory network model confirmed that specific TFs regulate specific paths in the network that are biologically relevant for defense against biotrophic pathogens. Comparison of the individual SA and MeJA treatments showed that there is a high level of interplay between the SA and JA networks (see also Figure 1). Of the MeJA‐responsive genes 69% was also modulated by the individual SA treatment, and of the SA‐responsive genes (which was a greater set) 26% was modulated by MeJA. Contrary to the paradigm of SA/JA antagonism, only half of the overlapping genes were regulated in an opposite manner (upregulated by SA and downregulated by MeJA or vice versa), while the other half of the genes were regulated in a similar direction by the two hormones. Noteworthily, hormone biosynthesis and pathways regulators like LOX2, MYC2, EDS1 and PAD4 were generally upregulated by the respective hormone treatment but downregulated by the other hormone treatment (Figure 2a, see also the section ‘Crosstalk at the hormone homeostasis level’). Moreover, many of the SA‐ and MeJA‐co‐upregulated genes were canonical JA and SA pathway genes, like GRX480, ANAC019, ANAC055, some JAZs and RAP2.6. Many of these genes are also associated with ET and ABA signaling, hinting to their responsiveness to a broad range of hormone‐inducing environmental stimuli. The combined SA and MeJA treatment showed that 68% of the MeJA‐responsive genes changed its expression when SA was added to the treatment, while this was the case for only 12% of SA‐responsive genes. While antagonistic and synergistic effects of the dual treatment were observed, the vast majority of the effects were just additive. Short‐term effects by MeJA were overridden by SA effects over time, resulting in a dominance of the SA profile over the MeJA profile (Hickman et al., 2019).
Figure 2.
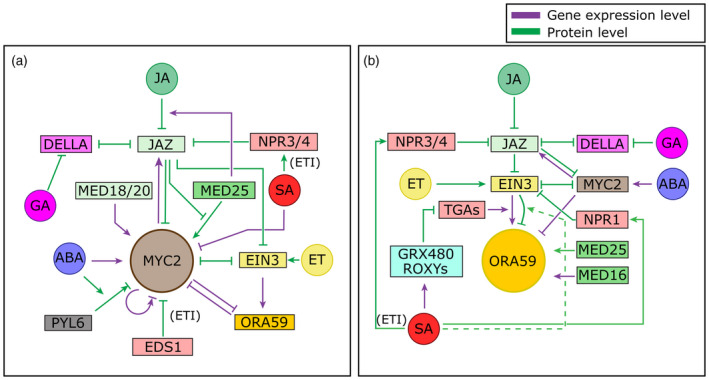
Schematic overview of hormone crosstalk acting on two key transcription factors, MYC2 and ORA59, of the two branches of the jasmonic acid (JA) pathway.
(a) Crosstalk acting on the MYC branch master regulator MYC2. In the context of defense, MYC2 mostly regulates anti‐insect responses. MYC2 is repressed by interacting JAZ repressors, and MYC2 itself can induce transcription of these JAZs. JA induces the breakdown of JAZs, thus leading to activation of MYC2. MED25 promotes MYC2 transcriptional activity, but JAZs prevent binding of MED25 to MYC2. MED25 also promotes JAZ breakdown by recruiting COI1, and alters JAZ splicing and thereby JAZ sensitivity to JA. MED18 and MED20 promote transcription of MYC2. EIN3 is activated by JA‐mediated breakdown of JAZ proteins and ethylene (ET)‐mediated stabilization. It binds to and represses MYC2 and vice versa. EIN3 also transcriptionally activates ORA59 and ORA59 can repress MYC2 transcription and vice versa (either directly or indirectly, see also panel b). MYC2 can enhance its own transcription in the short term but represses it in the long term. During AvrRps4‐induced ETI, EDS1 can repress MYC2. Furthermore, SA can promote degradation of JAZs via NPR3 and NPR4 during ETI. Generally, SA is an inhibitor of MYC2 transcription. Abscisic acid (ABA) directly activates transcription of MYC2 and enhances binding of the ABA receptor PYL6 to MYC2, modulating transcriptional activity of MYC2, which differentially acts on the JAZ6 and JAZ8 promoters (leading to repression versus activation). DELLA proteins bind JAZs and thereby JAZs and DELLAs prevent each other from binding to their respective target transcription factors (TFs). Gibberellin (GA) induces breakdown of DELLAs and thus indirectly represses MYC2. (b) Crosstalk acting on the ERF branch master regulator ORA59. ORA59 mostly regulates defense against necrotrophic pathogens. ORA59 is indirectly regulated by both JA and ET through their action on EIN3: JA releases EIN3 from its repression by JAZ and ET stabilizes EIN3. When released from repression and degradation, EIN3 activates transcription of ORA59. TGA TFs are also needed for this activation. GRX480 and other ROXYs are induced by SA and repress the transcriptional activity of these TGAs, leading to reduced transcription of ORA59. EIN3 also mediates degradation of ORA59. Because this only leads to reduced ORA59 functioning under high SA levels and not under high ET levels we propose that SA specifically modulates EIN3 activity such that it leads to ORA59 degradation (dotted line). SA activates NPR1’s activity as a co‐transcriptional regulator. NPR1 can interact with EIN3, leading to repression of EIN3 transcriptional activity. We propose that it is unlikely that NPR1 modulates EIN3‐mediated ORA59 activation during SA/JA crosstalk (see the section ‘Crosstalk by modulation of protein stability’). EIN3 is further repressed by MYC2 through direct binding and this also occurs the other way around. MYC2 is repressed by interaction of JAZ repressors and itself activates transcription of these JAZs. It is also transcriptionally activated by ABA. ORA59 expression is inhibited by MYC2, both directly and possibly indirectly via inhibition of EIN3 by MYC2. DELLAs bind to JAZs and thereby they inhibit each other from binding to target TFs in their respective pathways. GA leads to breakdown of DELLAs, thus indirectly repressing ORA59. MED25 interacts with ORA59 and promotes its transcriptional activity, and MED16 promotes ORA59 transcription. Modulation of JAZ breakdown and JAZ RNA splicing by MED25 is not shown here (see panel a). During ETI SA can promote degradation of JAZs via NPR3 and NPR4. Note that in most cases where EIN3 is mentioned, EIL1 likely has the same function. However, in most research only EIN3 is extensively characterized.
Mechanisms acting on the gene expression level are colored purple and mechanisms acting on the protein level are colored green. Arrows and bar‐headed lines indicate positive and negative effects, respectively. Mechanisms acting downstream of MYC2 and ORA59 or at the hormone homeostasis level are not shown.
Chang et al. (2013) combined ChIP‐seq of the ET master regulator TF EIN3 with RNA‐seq at five time points within 24 h following continuous ET treatment. ET was found to influence many other hormone pathways besides the JA pathway (Lorenzo et al., 2003; Anderson et al., 2004), as the GA, auxin, BR, ABA, SA and cytokinin pathways were also affected by ET treatment. A study by Song et al. (2016) investigated the ABA network based on RNA‐seq time series and ChIP‐seq experiments with 21 ABA‐responsive TFs in the presence of ABA. A complex network of regulation by multiple master regulators, including extensive feedback regulation, was revealed. A thousand genes involved in other hormone pathways were bound by at least one of the investigated ABA‐responsive TFs. However, the genes that were bound by a large number of TFs and/or by TFs that showed increased binding after ABA treatment usually belonged to the ABA pathway itself.
CROSSTALK AT THE PROTEIN LEVEL
Proteins can modulate the functioning of other proteins in their own pathway or in other hormone pathways through various mechanisms, such as co‐activation, repression, competitive binding to multiple targets and chemical modification (e.g., phosphorylation, ubiquitination, sumoylation, nitrosylation or sulfonylation). Several molecular players in crosstalk have been demonstrated to modulate proteins that act in other hormone pathways. These include hormone receptors and their interactors. In addition, TFs have been implicated as crosstalk mediators at the protein level, with leading roles for the bHLH TF MYC2 and the ERF TF ORA59 as important targets for crosstalk in the JA pathway (Figure 2).
Crosstalk by protein–protein interactions
Protein–protein interactions are of major importance for hormone pathway crosstalk. Recently, an extensive network of protein–protein interactions between members of all hormone pathways in Arabidopsis was revealed using yeast two‐hybrid with 1226 genes with probable or genetically demonstrated functions in plant hormone signaling (Altmann et al., 2020). Not only was there high connectivity within each single hormone pathway, but also many inter‐pathway contact points were uncovered. Validation of a subset of these inter‐pathway contact points suggested that many of these interactions indeed likely represented crosstalk mechanisms. This was demonstrated by the finding that a mutant of one interaction partner influenced the plant phenotype that correlated with the hormone‐associated function of the other interaction partner. It should be noted that such validation does not explicitly show that the convergence of two pathways depends on the detected protein interaction. Alternatively, it could be regulated by another factor that acts downstream in the pathway of the mutated gene.
Hormone receptors were especially often found to interact with proteins that were not involved in the canonical hormone pathway of the receptor (Altmann et al., 2020). This suggests that signaling by hormone receptors through non‐canonical pathways has a more prominent role in integrated hormone signaling than previously anticipated. One such example was previously shown for the ABA receptor PYL6, which interacts with the JA master regulator MYC2 (Aleman et al., 2016; Figure 2a). In the presence of ABA, the binding of PYL6 to MYC2 is enhanced, which alters the transcriptional specificity of MYC2 from promoting JAZ6 expression to JAZ8 expression. The genome‐wide implications of this mechanism have yet to be determined. Another example of an interaction between hormone receptors and key components of non‐canonical hormone pathways is that of the SA receptors NPR3 and NPR4 with JAZ repressors. This is discussed in the section ‘Crosstalk by modulation of protein stability’.
MYC branch/ERF branch antagonistic crosstalk in the JA defense pathway is likely also (partly) regulated by protein–protein interactions. MYC2 can suppress the ERF branch by directly binding to the TF EIN3, which causes reduced binding of EIN3 to the promoter of a target gene (Song et al., 2014; Zhang et al., 2014; Figure 2). This effect was only shown for the promoter of HLS1, a gene involved in the formation of the apical hook in etiolated seedlings (Song et al., 2014; Zhang et al., 2014). It is thus unclear if this mechanism also underlies MYC2‐mediated antagonism on JA‐responsive defense genes in the ERF branch. Vice versa, binding of EIN3 and EIL1 to MYC2 represses the transcriptional activity of MYC2 (Figure 2). This could likely explain the enhanced level of MYC2‐regulated VSP2 expression in the ein3 eil1 mutant and the reduced growth of a caterpillar that feeds on this mutant (Song et al., 2014). The reciprocal inhibition between MYC2 and ERF branch signaling components is not only regulated at the level of protein–protein interactions but also at the level of transcriptional regulation. This was demonstrated by the direct positive effect of MYC2 on expression of the F‐box protein‐coding gene EBF1, whose protein product targets EIN3 for degradation (Zhang et al., 2014), thus further suppressing ET signaling through a combined effect of MYC2 on ET signaling via transcription, protein stability and protein–protein interaction.
Crosstalk by modulation of protein stability
Modulation of stability of activators and especially repressors is an important regulatory mechanism in many different pathways. The most famous example from hormone defense signaling is that of JAZ proteins, which form a co‐receptor complex with the E3 ubiquitin ligase F‐box protein COI1 for JA‐Ile, the active form of JA (Fonseca et al., 2009; Sheard et al., 2010). JAZ proteins inhibit transcription within the MYC and ERF branches through direct binding to key TFs, recruitment of co‐repressors and inhibition of the interaction of the Mediator subunit MED25 with MYCs (see the section ‘Crosstalk by the Mediator complex’). Upon perception of JA‐Ile, JAZs are degraded by the 26S proteasome, which releases the previously bound TFs and initiates the JA response (Chini et al., 2007; Thines et al., 2007; Figure 2). The key function of JAZ proteins is substantiated by experiments that identified JAZs as targets for interference of immune signaling by pathogen and insect effectors. For example, HARP1, an effector of the chewing cotton bollworm (Helicoverpa armigera) was recently shown to bind to multiple JAZs in Arabidopsis, cotton (Gossypium hirsutum) (its preferred host) and tobacco (Nicotiana benthamiana) (Chen et al., 2019). This led to an increase in stability of JAZs, likely because it prevented JA‐Ile‐induced binding of JAZs with COI1. Via this mechanism, HARP1 reduced wound‐induced defense signaling and increased plant susceptibility to the insect (Chen et al., 2019). HopX1, an effector from the hemi‐biotrophic bacterial pathogen P. syringae pv. tabaci 11528, was also found to interact with JAZs but in contrast this led to a decrease (rather than an increase) in their stability, in a COI1‐independent manner. The resulting activation of the JA pathway in turn led to repression of the SA pathway and thus increased susceptibility to this pathogen (Gimenez‐Ibanez et al., 2014). Similarly, the effector HopZ1a from P. syringae pv. syringae A2 causes degradation of JAZs and consequent activation of the JA pathway and repression of SA signaling. However, in contrast to the effect of HopX1, JAZ degradation by HopZ1a depends on COI1 and likely involves acetylation of JAZs (Jiang et al., 2013). The most studied example of an effector that causes degradation of JAZs, the JA‐Ile mimic coronatine, is discussed in the section ‘Crosstalk at the hormone homeostasis level’.
Degradation of JAZ proteins is essential for synergistic effects between the JA pathway and the ET, ABA, SA and BR pathways, as we will outline in this paragraph. Synergism between the JA and ET pathways drives activation of the ERF branch of defense. JAZ proteins can physically interact with and repress the ET response TFs EIN3 and EIL1 by recruitment of the chromatin modifier HDA6 as a co‐repressor (Zhu et al., 2011). In the presence of JA, the JAZs are degraded and thereby the interaction between HDA6 and EIN3/EIL1 is reduced and thus EIN3/EIL1 transcriptional activity is enhanced. In combination with ET’s activity to stimulate EIN3/EIL1 protein accumulation, the de‐repression of EIN3/EIL1 by JA enhances transcription of ERF1 and ORA59 (Zhu et al., 2011; Figure 2), which are key TFs in the ERF branch of the JA pathway (Pré et al., 2008; Pieterse et al., 2012). ABA/JA synergistic crosstalk is also partly regulated through stability of a JAZ protein. This is mediated by the RING E3 ligase KEG, which is promoted for self‐ubiquitination and subsequent degradation by ABA. KEG normally decreases the COI1‐mediated degradation of JAZ12, but this is prevented under high ABA conditions due to degradation of KEG, leading to reduced JAZ12 levels (Pauwels et al., 2015). This was associated with an enhanced expression level of the MYC branch marker gene VSP2 under basal conditions in a keg knockdown line (Pauwels et al., 2015). No evidence was found for a role of JAZ (de)stabilization in antagonistic SA/JA crosstalk on JA‐responsive gene expression when plants were exogenously treated with SA and/or MeJA (Van der Does et al., 2013; Liu et al., 2016a). However, a role for SA‐mediated JAZ degradation was reported in synergistic SA/JA crosstalk occurring during ETI triggered by P. syringae pv. maculicola (Psm) ES4326 carrying the effector AvrRpt2 (Liu et al., 2016a). JAZ proteins were shown to bind to the SA response regulators NPR3 and NPR4, and this binding was enhanced by SA. Being substrate adaptors for Cullin 3 (Cul3) ubiquitin E3 ligases, NPR3 and NPR4 target the JAZs for degradation (Liu et al., 2016a; Figure 2). This results in increased JA signaling, which is necessary for a full HR response. It is unclear why SA‐mediated breakdown of JAZ proteins would occur only during ETI triggered by Psm ES4326 AvrRpt2 but not after exogenous SA application. In rice (Oryza sativa), BR/JA crosstalk is regulated through modulation of OsJAZ4 stability (He et al., 2020). This is mediated by OsGSK2, a kinase that itself is negatively regulated by BR in Arabidopsis through dephosphorylation and degradation (Peng et al., 2008; Kim et al., 2009; He et al., 2020). OsGSK2 was shown to interact with and phosphorylate OsJAZ4, which subsequently leads to disruption of the OsJAZ4–OsNINJA and OsJAZ4–OsJAZ11 interactions and to degradation of OsJAZ4 in an OsCOI1‐dependent manner (He et al., 2020). In accordance, high BR levels lead to reduced OsGSK2 levels and activity, which in turn leads to higher OsJAZ4 levels and thus decreased JA signaling, resulting in enhanced susceptibility to the rice black‐streaked dwarf virus (He et al., 2017).
SA/JA antagonistic crosstalk is partly regulated through modulation of the stability of the TF ORA59, a master regulator in the ERF branch of the JA pathway (Pré et al., 2008). SA treatment leads to breakdown of ORA59 (Van der Does et al., 2013; He et al., 2017) but not in an ein3 eil1 mutant (He et al., 2017). Furthermore, SA increases EIN3 protein abundance and co‐transfection of EIN3 and ORA59 in N. benthamiana leads to degradation of ORA59 unless a proteasome inhibitor is added (He et al., 2017). Also, EIN3 interacts with ORA59 (He et al., 2017). Hence, the SA‐mediated degradation of ORA59 depends on the interaction of ORA59 with EIN3 and likely also its homolog EIL1 (Figure 2b). However, the underlying molecular mechanism by which SA can induce EIN3‐mediated breakdown of ORA59 is not completely clear. It is unlikely that the SA‐increased protein abundance of EIN3 can explain SA‐mediated ORA59 degradation, since ET also increases EIN3 protein abundance (Dolgikh et al., 2019) but has a positive effect on ORA59 functioning in the ERF branch of defense (Pré et al., 2008). We propose that SA modulates the activity of EIN3 such that it specifically causes degradation of ORA59 (Figure 2b, dotted line). Recently, it was found that the SA master transcriptional regulator NPR1 can bind to EIN3 and repress its transcriptional activity in the regulation of apical hook formation (Huang et al., 2020; Figure 2b). Hypothetically, an NPR1–EIN3 interaction may also be required for EIN3‐mediated breakdown of ORA59 during SA/JA crosstalk. In contrast, under high ET conditions, which would increase EIN3 levels, the SA/JA crosstalk was shown to be independent of NPR1 (Leon‐Reyes et al., 2009), making this hypothesis very unlikely. This suggests that another protein that functions in the SA pathway must be the missing link for SA/JA crosstalk via EIN3‐mediated ORA59 degradation.
Crosstalk by competitive binding of proteins to multiple other proteins
A regulatory protein can be held inactive if binding to its downstream target protein is prevented due to its bound status to another protein. An example of such a crosstalk mechanism that is based on competitive binding to multiple proteins is provided by the interaction between JAZs and DELLAs, which are repressors of the JA response and the GA response, respectively (Hou et al., 2010). When they are bound to each other, JAZs compete for binding of DELLAs to growth‐promoting PIF TFs, and DELLAs compete for binding of JAZs to the JA master regulator MYC2 (Hou et al., 2010; Hou et al., 2013; Figure 2). When plants are attacked by an insect the JA levels rise, causing degradation of JAZs and thus release of MYC2, so that JA‐responsive transcription is initiated. At the same time more DELLAs can bind to PIFs and elongation growth is inhibited. Other JA pathway‐regulating TFs that physically associate with JAZs, such as EIN3 and EIL1, are likely also indirectly affected by DELLAs, which may impact transcription of ORA59 (Hou et al., 2013; Figure 2). In contrast, if GA levels are high, such as under far‐red light conditions (Franklin, 2008), DELLAs are degraded, thereby releasing PIFs and thus leading to elongation growth, while the JAZs can now bind more MYC2, leading to repression of JA‐mediated defense responses (Hou et al., 2013; Figure 2). This crosstalk mechanism between JA and GA signaling is traditionally viewed as important for regulation of the defense–growth trade‐off (Hou et al., 2013). However, recently, conflicting results on the role of the JAZ–DELLA interaction in the JA‐mediated growth effects were reported and other interaction points of the JA pathway with growth signaling have been pinpointed (Chakraborty et al., 2019; Liu et al., 2019; Major et al., 2020; Ortigosa et al., 2020).
Crosstalk by redox regulation and ROXY glutaredoxins
Redox status is an important determinant of protein functioning and as such plays a role in plant defense hormone signaling, among which hormone crosstalk. The defense hormone SA itself induces cycles of oxidation and reduction in the cell, leading to an increase in the amount of the antioxidant glutathione and changing the ratio between the oxidized and reduced states of glutathione (Spoel and Loake, 2011). The changes in redox potential and the change in glutathione state as a result of SA elevation has consequences for oxidation or reduction of cellular proteins and thereby modulates their function. The increase in glutathione levels was found to coincide with the time frame in which SA could suppress JA signaling. That is, SA treatment prior to MeJA treatment led to a reduction of MeJA‐induced PDF1.2 expression only if MeJA was applied in the time frame when glutathione levels were increased by SA. Additionally, chemical inhibition of glutatione biosynthesis severely diminished the ability of SA to suppress MeJA‐induced PDF1.2 expression (Koornneef et al., 2008). This suggests that the SA‐induced shift in redox potential is involved in SA/JA antagonistic crosstalk.
Glutaredoxins are small oxidoreductases that are involved in reduction of oxidative modifications using glutathione (Ströher and Millar, 2012). Four of the CC‐type glutaredoxins, which are also known as ROXYs and include GRX480, are induced by SA and can suppress induction of ORA59 by EIN3, making them potential candidates for SA‐mediated crosstalk on the ERF branch (Zander et al., 2012; Figure 2b). Several lines of evidence indicate that under high ET levels group II TGA TFs regulate ORA59 induction, but that the TGAs recruit ROXYs under high SA levels (Ndamukong et al., 2007; Zander et al., 2010; Zander et al., 2012; Zander et al., 2014). It was believed that this would lead to redox modification of the TGAs, which would cause a decrease of their transcriptional activity. However, the ROXYs were recently shown to recruit the co‐repressor TPL through the same motif that was shown to be essential for repression of ORA59 transcription (Uhrig et al., 2017). This suggests that the effect of GRX480 and other ROXYs on SA/ERF branch crosstalk via suppression of TGA‐mediated transcription of ORA59 is caused by recruitment of a transcriptional co‐repressor rather than by redox modification.
CROSSTALK AT THE GENE EXPRESSION LEVEL
Regulation of gene transcription is a major component of hormone crosstalk. In fact, most of the regulation at the protein level that is discussed above eventually leads to altered transcription of downstream target genes. In this section we discuss that crosstalk can act across multiple regulatory scales of gene expression, from binding of a TF to the promoter of a gene to translation of mRNA to protein.
Crosstalk by binding of TFs to promoters
Most TFs have affinity for binding to a specific DNA motif (Franco‐Zorrilla et al., 2014; Weirauch et al., 2014; O’Malley et al., 2016). TF–DNA binding in the promoter region of a gene largely determines activation or repression of transcription. A gene may be subject to crosstalk if, for example, TFs from different hormone pathways compete for binding to the same DNA motif in the promoter of a gene. Also binding of different TFs to different DNA motifs or cooperative binding of different TFs to the promoter of a gene may modulate the expression of that gene by multiple hormones. The increasing amount of available ChIP‐seq and DAP‐seq data (of currently >500 Arabidopsis TFs) greatly facilitates the identification of DNA motifs and their trans‐acting TFs (O’Malley et al., 2016; Zander et al., 2020). Besides DNA sequence, many other factors determine where a TF binds to DNA, as demonstrated by the fact that TFs from the same family often bind to very similar motifs, but regulate divergent genes (O’Malley et al., 2016). This can for example be determined by the chromatin structure, which can be more compact or relaxed, depending on, for example, acetylation/methylation/ubiquitination of the histones. This local chromatin status of genomic DNA influences the exposure of cis‐regulatory DNA elements for proteins and consequently transcription. Additional important factors that determine transcriptional activity are the 3D structure of the DNA (Muiño et al., 2014; Mathelier et al., 2016), methylation of the DNA (O’Malley et al., 2016) and spacing between adjacent DNA motifs (Krawczyk et al., 2002; O’Malley et al., 2016). Although to our knowledge these types of information have not been implicated in hormone crosstalk research yet, they provide an enormous potential to uncover cross‐regulatory mechanisms based on differential TF–DNA binding.
Investigations with overexpression and/or knockout lines implied a role for several SA‐activated WRKY TFs in SA/JA crosstalk (reviewed by Caarls et al. (2015)). The role of the WRKY‐bound W‐box motif in SA/JA crosstalk was investigated using RNA‐seq data derived from time course experiments with MeJA, SA and MeJA + SA combination treatments (see also the section ‘Network modeling using time series of hormone treatments of wild‐type plants’). The W‐box was, as expected, significantly enriched in SA‐upregulated genes as well as in MeJA‐downregulated genes (Hickman et al., 2017; Hickman et al., 2019). However, there was no enrichment of the W‐box in MeJA‐upregulated genes that were antagonized by SA in the combination treatment (Hickman et al., 2019). This is in contrast to a previously reported microarray‐based study of a single, relatively late time point (28 h), in which the W‐box was enriched in a small number of ERF branch response genes that were upregulated by MeJA, but antagonized by the combination with SA (Van der Does et al., 2013). This difference may be explained by the fact that compared to the above‐mentioned RNA‐seq study the ERF branch was activated to a higher extent by the MeJA treatment in the latter experiment. Thus, WRKY TFs may regulate SA/JA crosstalk through binding to a subset of promoters of MeJA‐inducible genes in the ERF branch to repress their expression under certain conditions, but likely do not play such a role in the entire JA pathway, which is in line with the finding that in contrast to the ERF branch response genes, the MYC branch response genes, which are antagonized by SA, were not enriched in the W‐box (Van der Does et al., 2013).
Two other motifs, the GCC‐box and the G‐box, which are mostly known for their role in JA and ET signaling, may also be important in SA‐mediated crosstalk. These motifs are bound by ERF TFs, among which ORA59, and bHLH TFs, among which MYC2, respectively. Both motifs are enriched in MeJA‐induced genes that are suppressed by SA (Van der Does et al., 2013; Hickman et al., 2019). Moreover, a synthetic promoter only containing four repeats of the GCC‐box was demonstrated to be inducible by a single MeJA treatment but repressed by a combination treatment of MeJA and SA (Van der Does et al., 2013). A study by Caarls et al. (2017) investigated if this was caused by repressive SA‐induced ERFs or EAR domain‐containing ERFs that may compete with JA‐induced ERF activators for binding to target genes in the ERF branch. Gene expression analyses of 16 erf mutants showed that the tested ERFs are not required for SA/JA crosstalk. To the best of our knowledge there are also no known potential JA pathway‐repressing bHLH TFs from the SA pathway. It is worth noting that there is a clade of bHLH TFs that repress the JA pathway, consisting of JAM1, JAM2 and JAM3, but they are described as JA‐responsive rather than SA‐responsive (Sasaki‐Sekimoto et al., 2013). Together, these results suggest that it is unlikely that the JA pathway is antagonized through large‐scale binding of SA‐responsive TFs directly to the promoters of JA‐activated genes. Instead, SA may antagonize the JA pathway by inhibiting the transcriptional activity of certain key activator TFs of the JA pathway such as ORA59 and MYC2. In agreement with this hypothesis, SA treatment is known to cause reduced transcription of the ORA59 gene and degradation of the ORA59 protein (see the sections ‘Crosstalk by modulation of protein stability’ and ‘Crosstalk by redox regulation and ROXY glutaredoxins’ and Figure 2b).
There is ample evidence for extensive regulation of ORA59 and MYC2 by several proteins that are involved in hormone crosstalk (see previous and subsequent sections, Figure 2) but the exact underlying mechanisms have not always been elucidated. For example, Verhage et al. (2011) showed that basal and caterpillar‐induced ORA59 mRNA levels were higher in a myc2 mutant and caterpillar‐induced MYC2 levels were higher in an ORA59 RNAi line. This suggests that MYC2 and ORA59 directly or indirectly repress each other’s transcription (Figure 2), in addition to the reciprocal inhibitory effects of binding between MYC2 and EIN3, the upstream regulator of ORA59 (Figure 2 and the section ‘Crosstalk by protein–protein interactions’). Later, it was shown that repression of ORA59 by MYC2 was mediated by direct binding of MYC2 to a G‐box in the ORA59 promoter (Zhai et al., 2013), but to the best of our knowledge the mechanism underlying the repressive effect of ORA59 on the transcription of MYC2 has not been elucidated yet. Another example is the finding that MYC2 is transcriptionally upregulated by both JA and ABA and can itself regulate genes in both JA and ABA signaling (Abe et al., 1997; Abe et al., 2003; Hickman et al., 2017; Figure 2). As such, MYC2 is both an integrator and a regulator of JA and ABA signaling. Possibly, the enhanced MYC2 expression by ABA treatment is related to ABA‐enhanced JA biosynthesis (see the section ‘Crosstalk at the hormone homeostasis level’), which not only activates MYC2 transcriptional activity but also enhances transcription of MYC2 through auto‐regulation (Wang et al., 2019; Figure 2).
Crosstalk by the Mediator complex
The multiprotein Mediator complex forms the molecular bridge that relays signals from DNA‐binding TFs to the transcription machinery (Zhai and Li, 2019). It plays an important role in hormone signaling pathways, including hormone crosstalk. MED25 is the most studied Mediator subunit in defense hormone signaling. It interacts with the TFs MYC2, MYC3, MYC4, ORA59 and ERF1 as well as with the JA receptor component COI1. In doing so MED25 is involved in the transcriptional activity of these TFs in both the MYC and the ERF branch (Çevik et al., 2012; Chen et al., 2012; An et al., 2017; Figure 2). Additionally, MED25 promotes JAZ breakdown by recruiting COI1 to target promoters (An et al., 2017). Vice versa, JAZ proteins inhibit the MED25–MYC interaction (Zhang et al., 2015; Figure 2a).
MED25 promotes transcription in the JA pathway via various mechanisms such as recruitment of RNA polymerase II, histone acyltransferase HAC1 and JA‐related enhancers to promoters that are targeted by for example MYC2 and ORA59 (Çevik et al., 2012; Chen et al., 2012; An et al., 2017; Wang et al., 2019; You et al., 2019; Figure 2). Moreover, MED25 has a role in alternative splicing of JAZ proteins, which determines their sensitivity to JA (Wu et al., 2020; Figure 2a). This multifaceted role makes MED25 an obvious candidate player in crosstalk regulation. Indeed, med25 mutant studies and interaction studies of MED25 with MYC2 and ABI5 (key regulators of JA and ABA signaling, respectively) suggest that it plays a positive role in JA signaling but has a negative role in ABA signaling (Chen et al., 2012).
Other Mediator subunits have also been implicated in defense hormone crosstalk. This was mostly based on mutant studies. MED14, MED15 and MED16 were found to be involved in suppression of MYC branch marker genes by SA and ET (Wang et al., 2015; Wang et al., 2016). This suggests that these three Mediator subunits are required for SA/JA and ERF branch/MYC branch crosstalk. In another study, MED18 and MED20 were found to be involved in the activation of transcription of MYC2 and the MYC branch marker gene VSP1 by Fusarium oxysporum and in the repression of SA pathway marker genes (Fallath et al., 2017; Figure 2a). It is important to note that in all experiments the Mediator subunits were not only found to suppress a certain pathway, but also to activate another pathway that is known to antagonize the suppressed pathway. Therefore, it is possible that the repression by Mediator subunits is not a direct effect on that pathway but rather an indirect effect resulting from activation of a repressor derived from the antagonizing pathway. For example, MED16 was not only found to suppress MYC branch marker gene expression, but also to activate ORA59 expression (Wang et al., 2015; Figure 2b). ORA59 represses MYC2 expression (see the section ‘Crosstalk by binding of TFs to promoters’ and Figure 2a), which may explain how MED16 causes suppression of MYC branch response genes.
Crosstalk affecting mRNA maturation and translation to protein
Above we described the different steps in initiation of transcription. The subsequent steps in gene expression are also potential points of crosstalk. For example, crosstalk may act through modulation of alternative splicing, stability of mRNA, retention of mRNA in the nucleus or translation efficiency of mRNA into protein in the cytosol. In ET and JA signaling extensive regulation of these regulatory steps is suggested by the findings that only a subset of genes that are bound by key TFs show alterations in mRNA levels (Chang et al., 2013; Zander et al., 2020) and that transcript and protein abundance do not match up after MeJA treatment (Zander et al., 2020).
Some JAZ proteins undergo alternative splicing, potentially making them insensitive to JA‐Ile‐induced breakdown mediated by COI1 (Chung et al., 2010). So far, no evidence for selective favoring of undegradable JAZ isoforms during SA/JA crosstalk has been found (Van der Does et al., 2013). The myc2 mutant was shown to affect phosphorylation of proteins that act in the spliceosome (Zander et al., 2020). In agreement with this, the isoforms of 151 transcripts were switched after MeJA treatment (Zander et al., 2020). Only two of these genes were related to JA, while the rest was related to other processes, including ABA signaling. This suggests that MYC2 can influence other signaling pathways by modulation of transcript splicing. However, the importance of this observed JA‐induced alternative splicing and the role of MYC2 in this mechanism need further investigation.
Besides the stability of proteins, the stability of mRNA molecules may also serve as a way for hormones to influence each other’s pathway activities. A role for RNA‐binding proteins and small RNAs (Narsai et al., 2007) in determining mRNA stability during plant immune responses and root nodule symbiosis has been indicated (Staiger et al., 2013; Zanetti et al., 2020). For proper responsiveness to the hormone ET, the mRNA of the F‐box protein‐coding gene EBF2 is targeted to decay in P‐bodies (Merchante et al., 2015). However, to the best of our knowledge the role of mRNA stability in hormone crosstalk has not been explored yet.
Mature mRNAs can be temporarily disengaged from the translation process by retaining them in the nucleus. There is recent evidence for a role of selective nuclear retention of mRNAs in the control of gene expression activity during adaptation to hypoxia, an abiotic stress. After reaeration the mRNAs are quickly released to the cytosol to be translated into protein (Lee and Bailey‐Serres, 2019). However, there is no evidence yet that regulation of plant immunity or hormone crosstalk acts on temporary retention of mRNAs in the nucleus in order to divert all the plant’s molecular attention to the most critical response.
The final step in gene expression is that of translation from mRNA to protein. Translation efficiency is influenced by different features of the mRNA (Merchante et al., 2017). The literature on translational regulation of plant immunity, although scarce, points to translational control of specific mRNAs via upstream, short open reading frames (uORFs) during defense activation by the pathogen elicitors AvrRpm1 and elf18 in Arabidopsis (Pajerowska‐Mukhtar et al., 2012; Meteignier et al., 2017; Xu et al., 2017). The elicitor treatments transiently alleviate the repressive effect of the uORFs on expression of the main ORF, the heat shock factor gene TBF1, so that ribosomes can engage in its translation, leading to activation of the immune system. Furthermore, it has been shown that in the establishment of root nodule symbiosis, small RNAs are crucial by determining stability and translatability of mRNAs (Zanetti et al., 2020). If and how control at the translation level can affect crosstalk between different hormone pathways in defense is not known yet.
CROSSTALK AT THE HORMONE HOMEOSTASIS LEVEL
The previous sections focused on crosstalk by hormones via their interference with responsiveness to other hormones, namely downstream of these other hormones. Here, we describe effects that hormones have on the levels of other hormones. For example, ABA is known to enhance the biosynthesis of JA (Adie et al., 2007; Fan et al., 2009; Wang et al., 2018). This is correlated with the ABA‐induced expression of PLIP2 and PLIP3, which encode lipases that catalyze the release of polyunsaturated fatty acids (PUFAs) (Wang et al., 2018), which can be further metabolized to form JA (Wasternack and Hause, 2013). In accordance, overexpression lines of PLIP2 and PLIP3 show enhanced JA signaling (Wang et al., 2018). The ERF TF gene ORA47 is upregulated via MYC2 by JA treatment (Zander et al., 2020) and directly targets promoters of JA and ABA biosynthesis genes, which leads to enhanced JA levels, and upon wounding also to enhanced ABA levels (Pauwels et al., 2008; Chen et al., 2016; Hickman et al., 2017; Zander et al., 2020). Hence, the canonical JA pathway regulator MYC2 acts at multiple levels as an integrator of JA and ABA signaling: MYC2 is itself positively regulated by JA and ABA at the protein and gene expression level (see the sections ‘Crosstalk by protein–protein interactions’ and ‘Crosstalk by binding of TFs to promoters’) and subsequently, MYC2 regulates JA and ABA levels. Apart from activating JA biosynthesis genes, MYC2 also activates transcription of JAZ repressors, whose protein products in the long term attenuate the JA response via repression of MYC2 and other JA master regulators (Chini et al., 2007). This form of short‐term self‐activation and long‐term self‐inhibition of MYC2 is reinforced by MED25. This Mediator subunit promotes looping of a MYC2 enhancer, which is also bound by MYC2 itself, to the MYC2 promoter. For unknown reasons this leads to self‐activation of the MYC2 promoter during short‐term JA responses but inhibition of the MYC2 promoter during long‐term JA responses (Wang et al., 2019). Besides JA, other hormones also activate or repress transcription of different JAZ genes, which potentially modulates JA responsiveness, resulting in synergism, antagonism or reestablishment of the basal situation when both pathways are elicited in the same cell (Hickman et al., 2019).
SA and JA can also influence each other’s levels. RNA‐seq analyses after MeJA treatment pointed to repression of JA and SA biosynthesis genes by the respective reciprocal treatments with SA and MeJA (Hickman et al., 2017; Hickman et al., 2019). Several underlying mechanisms for this antagonism in transcriptional activity and the resultant decrease in hormone levels have been elucidated. For example, SA inhibits activity of the catalase CAT2, which leads to reduced activity of the acyl‐CoA oxidases ACX2 and ACX3, which are enzymes involved in JA biosynthesis. This effect of SA leads to lower JA levels and reduced defense against Botrytis cinerea (Yuan et al., 2017). Additionally, WRKY51, which is transcriptionally activated by SA, inhibits JA biosynthesis by repressing transcription of the JA biosynthesis gene AOS (Yan et al., 2018). This repression is mediated by a complex containing WRKY51, JAV1 and JAZ8. However, during wounding JAV1 is degraded, leading to de‐repression of AOS and increased JA biosynthesis (Yan et al., 2018). Vice versa, JA also has the potential to reduce free SA levels. This is exploited by biotrophic pathogens to reduce effective plant defense responses. For example, P. syringae pv tomato (Pst) DC3000 produces the JA‐Ile mimic coronatine (COR), which, via MYC2, MYC3 and MYC4, activates transcription of three NAC TFs, ANAC019, ANAC055 and ANAC072 (Zheng et al., 2012; Gimenez‐Ibanez et al., 2017). These NAC TFs repress expression of the SA biosynthesis gene ICS1 and activate expression of the SA methyltransferase gene BSMT1 (Zheng et al., 2012). This leads to lower levels of free SA and reduced defense against Pst DC3000. The bacterial effectors HopX1 and HopZ1a (see the section ‘Crosstalk by modulation of protein stability’) are likely to have the same effect on free SA levels as COR has. This was investigated for HopZ1a, which reduces the transcription level of ICS1 (Jiang et al., 2013). Another study found that the negative effect of MYC2 on SA accumulation is antagonized by EDS1 during ETI that is triggered by the pathogen effector AvrRps4 (Cui et al., 2018; Figure 2a). This antagonism by EDS1 involves the competitive binding of EDS1 to PAD4, which otherwise binds to MYC2. This results in reduced binding of MYC2 to the ANAC019 promoter and less BSMT1 expression, and thus enhanced SA levels (Cui et al., 2018; Bhandari et al., 2019).
Research on the effect of crosstalk on hormone levels has been restricted mostly to measurements of (proven) active compounds, like JA or JA‐Ile. However, the concentrations of hormone derivates, resulting from effects on biosynthesis or catabolism, will also change and these may also modulate plant responses. For example, a mutation in the JA biosynthesis gene OPR3 that completely blocked the canonical JA biosynthesis pathway and led to accumulation of the JA precursors OPDA and dn‐ODPDA enabled research on the role of these compounds in the absence of JA (Chini et al., 2018). It was demonstrated that these JA precursors promote thermotolerance through a mechanism independent of COI1 in Arabidopsis as well as in a bryophyte and a charophyte alga (Monte et al., 2020). Whether hormone derivates are targeted by other hormones and whether the derivates themselves could affect other hormone pathways remains to be investigated.
PERSPECTIVES
This review reports on several molecular components in hormone crosstalk that regulate plant defense responses. In most cases their regulation at the transcriptional or protein level has been demonstrated, but the exact mechanisms underlying their role in hormone crosstalk have often not been fully elucidated yet. The integration of data derived from different technologies aiming to address different regulation levels has the potential to unveil these crosstalk mechanisms in different internal and external contexts of the plant.
The network‐level understanding of defense hormone crosstalk as a whole is still rudimentary. Until now, most research has been restricted to the use of hormone mutants, single hormone applications, or TF–DNA binding studies under control conditions. These systems approaches gave us a little glimpse of regulatory nodes in hormone signaling networks and their possible role in hormone crosstalk. However, addition of multiple hormones and the integration of multiple data types regarding different levels of regulation are crucial to unveil new crosstalk mechanisms. Such multiomics research is now possible thanks to modern wet‐lab technologies as well as advanced data analysis and modeling tools (Zander et al., 2020). A network‐level understanding of crosstalk is also necessary to ultimately grasp the impact of hormone signal integration under different conditions for the plant. Therefore, we need to learn not only the ‘how’, but also the ‘when’ and ‘where’ of hormone crosstalk. In addition to interactions with microbes and insects, other environmental conditions of the plant and the internal context in the plant determine its hormone balance and hormone responsiveness (Figure 1). Indeed, the extent of crosstalk between hormone pathways during immune responses has been shown to be influenced by additional stresses, location of the stimulus, and plant age. Likely, different crosstalk mechanisms, such as described above and below in this review, are engaged in different situations and are regulated in a spatiotemporal manner.
The order in which sequential stresses occur and the nature of the stresses determine whether hormone crosstalk is effective. For example, for primed expression of JA‐mediated defenses in systemic tissue that expresses MYC2 after local herbivory by caterpillars of Pieris rapae, the ABA pathway needs to be activated by a secondary infestation (Vos et al., 2013b). In contrast, ABA can inhibit the JA pathway in tissue that is primed for dehydration stress: A first experience of dehydration stress leads to induction of the JA pathway, but this does not occur during a second dehydration stress (Ding et al., 2013), which is likely due to a lack of MYC2 activation during the second stress if ABA levels had already increased previously (Liu et al., 2016b; Avramova, 2019). An RNA‐seq experiment studying responsiveness to the sequential stresses drought, herbivory and infection showed that the transcriptome profiles during the sequential stresses rapidly change to largely resemble those of the last stress (Coolen et al., 2016). This shows that many crosstalk mechanisms can be overridden by a second, dominating stress. Nevertheless, it was also found that the first stress leaves a relatively small expression signature, which is enriched for hormone signaling genes, suggesting a lasting effect of induced hormone signaling by the first stress.
A spatial separation between SA and JA signaling has been demonstrated when a local hypersensitive response (HR) is activated during ETI as a result of infection with the avirulent pathogen Pst DC3000 carrying AvrRpt2. The zone around the HR‐induced cell death is surrounded by a small layer where the SA marker gene PR1 is highly expressed, followed by a region where the JA marker gene VSP1 is highly expressed. This demands differential prioritization of SA versus JA antagonism mechanisms. In the SA zone SA‐mediated defenses against Pst DC3000 AvrRpt2 can be activated, while in the JA zone runaway cell death and secondary infections by necrotrophic pathogens are limited, which could otherwise take advantage of the dead tissue generated by the HR response (Betsuyaku et al., 2018).
Leaf age is another factor that can influence hormone crosstalk. Biotic and abiotic stress responses are differently balanced in older leaves compared to younger leaves (Berens et al., 2019). Abiotic stress suppresses immune responses in older leaves through ABA. This antagonistic effect on immunity is blocked in young leaves, which is dependent on NPR1 as well as on the SA biosynthesis component PBS3, but independent of ICS1. This suggests an SA‐independent function of NPR1 and PBS3 in regulating leaf age‐dependent crosstalk (Berens et al., 2019).
The above examples illustrate that knowing the ‘when’ and ‘where’ of hormone pathway integration in immune networks is important to predict the outcome of immune signaling. To advance our knowledge, future research should focus on the different levels of hormone network regulation under different internal and external conditions. For such biological experimental systems, it would be even more meaningful to use single‐cell methods instead of bulk analyses. This will provide a better spatiotemporal resolution, which is particularly powerful when studying plant–microbe interactions, where relevant molecular events are often restricted to localized cell populations (of specific cell types), ranging from being infected themselves, to residing in the same leaf or in distant tissue and not (yet) being infected. Moreover, information on the single‐cell level will increase the accuracy of network predictions, as the measured responses are derived from one cell and not diluted by bulk analyses of multiple cells. This will further increase our knowledge of the ‘how’ of hormone pathway integration. Together, this molecular‐ and systems‐level knowledge is crucial to design crops with a strengthened immune response without undesired side effects like enhanced sensitivity to other stresses or decreased plant growth and yield.
AUTHOR CONTRIBUTIONS
NA and SCMW wrote the manuscript and designed the figures. MPM helped design the figures and substantially contributed to conception of the paper.
Acknowledgments
The graphical abstract was created using BioRender. The authors have no conflicts of interest to declare. This work was supported by the Netherlands Organization for Scientific Research (ALWGS.2016.005) and by the CAPES Foundation, Ministry of Education of Brazil (DF 70040‐020 to MPM).
REFERENCES
- Abe, H. , Urao, T. , Ito, T. , Seki, M. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell, 15, 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe, H. , Yamaguchi‐Shinozaki, K. , Urao, T. , Iwasaki, T. , Hosokawa, D. and Shinozaki, K. (1997) Role of Arabidopsis MYC and MYB homologs in drought‐ and abscisic acid‐regulated gene expression. Plant Cell, 9, 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adie, B.A.T. , Perez‐Perez, J. , Perez‐Perez, M.M. , Godoy, M. , Sanchez‐Serrano, J.J. , Schmelz, E.A. and Solano, R. (2007) ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell, 19, 1665–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleman, F. , Yazaki, J. , Lee, M. , Takahashi, Y. , Kim, A.Y. , Li, Z. , Kinoshita, T. , Ecker, J.R. and Schroeder, J.I. (2016) An ABA‐increased interaction of the PYL6 ABA receptor with MYC2 transcription factor: a putative link of ABA and JA signaling. Sci. Rep. 6, 28941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann, M. , Altmann, S. , Rodriguez, P.A. et al (2020) Extensive signal integration by the phytohormone protein network. Nature, 583(7815), 271–276. [DOI] [PubMed] [Google Scholar]
- An, C. , Li, L. , Zhai, Q. et al (2017) Mediator subunit MED25 links the jasmonate receptor to transcriptionally active chromatin. Proc. Natl. Acad. Sci. USA, 114, E8930–E8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J.P. , Badruzsaufari, E. , Schenk, P.M. , Manners, J.M. , Desmond, O.J. , Ehlert, C. , Maclean, D.J. , Ebert, P.R. and Kazan, K. (2004) Antagonistic interaction between abscisic acid and jasmonate‐ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell, 16, 3460–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson, N.J. and Urwin, P.E. (2012) The interaction of plant biotic and abiotic stresses: from genes to the field. J. Exp. Bot. 63, 3523–3543. [DOI] [PubMed] [Google Scholar]
- Attaran, E. , Major, I.T. , Cruz, J.A. , Rosa, B.A. , Koo, A.J.K. , Chen, J. , Kramer, D.M. , He, S.Y. and Howe, G.A. (2014) Temporal dynamics of growth and photosynthesis suppression in response to jasmonate signaling. Plant Physiol. 165, 1302–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avramova, Z. (2019) Defence‐related priming and responses to recurring drought: Two manifestations of plant transcriptional memory mediated by the ABA and JA signalling pathways. Plant Cell Environ. 42, 983–997. [DOI] [PubMed] [Google Scholar]
- Berens, M.L. , Berry, H.M. , Mine, A. , Argueso, C.T. and Tsuda, K. (2017) Evolution of hormone signaling networks in plant defense. Annu. Rev. Phytopathol. 55, 401–425. [DOI] [PubMed] [Google Scholar]
- Berens, M.L. , Wolinska, K.W. , Spaepen, S. et al (2019) Balancing trade‐offs between biotic and abiotic stress responses through leaf age‐dependent variation in stress hormone cross‐talk. Proc. Natl. Acad. Sci. USA, 116, 2364–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betsuyaku, S. , Katou, S. , Takebayashi, Y. , Sakakibara, H. , Nomura, N. and Fukuda, H. (2018) Salicylic acid and jasmonic acid pathways are activated in spatially different domains around the infection site during effector‐triggered immunity in Arabidopsis thaliana. Plant Cell Physiol. 59, 8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari, D.D. , Lapin, D. , Kracher, B. , von Born, P. , Bautor, J. , Niefind, K. and Parker, J.E. (2019) An EDS1 heterodimer signalling surface enforces timely reprogramming of immunity genes in Arabidopsis. Nat. Commun. 10, 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boter, M. , Ruiz‐Rivero, O. , Abdeen, A. and Pratt, S. (2004) Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18, 1577–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caarls, L. , Pieterse, C.M.J. and Van Wees, S.C.M. (2015) How salicylic acid takes transcriptional control over jasmonic acid signaling. Front. Plant Sci. 6, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caarls, L. , Van der Does, D. , Hickman, R. , Jansen, W. , Van Verk, M.C. , Proietti, S. , Lorenzo, O. , Solano, R. , Pieterse, C.M.J. and Van Wees, S.C.M. (2017) Assessing the role of ETHYLENE RESPONSE FACTOR transcriptional repressors in salicylic acid‐mediated suppression of jasmonic acid‐responsive genes. Plant Cell Physiol. 58, 266–278. [DOI] [PubMed] [Google Scholar]
- Çevik, V. , Kidd, B.N. , Zhang, P. et al (2012) MEDIATOR25 acts as an integrative hub for the regulation of jasmonate‐responsive gene expression in Arabidopsis. Plant Physiol. 160, 541–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty, M. , Gangappa, S.N. , Maurya, J.P. et al (2019) Functional interrelation of MYC2 and HY5 plays an important role in Arabidopsis seedling development. Plant J. 99, 1080–1097. [DOI] [PubMed] [Google Scholar]
- Chang, K.N. , Zhong, S. , Weirauch, M.T. , Hon, G. , Pelizzola, M. , Li, H. , Huang, S.‐S.‐C. , Schmitz, R.J. , Urich, M.A. and Kuo, D. (2013) Temporal transcriptional response to ethylene gas drives growth hormone cross‐regulation in Arabidopsis . Elife, 2, e00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.Y. , Liu, Y.Q. , Song, W.M. et al (2019) An effector from cotton bollworm oral secretion impairs host plant defense signaling. Proc. Natl. Acad. Sci. USA, 116, 14331–14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H.Y. , Hsieh, E.J. , Cheng, M.C. , Chen, C.Y. , Hwang, S.Y. and Lin, T.P. (2016) ORA47 (octadecanoid‐responsive AP2/ERF‐domain transcription factor 47) regulates jasmonic acid and abscisic acid biosynthesis and signaling through binding to a novel cis‐element. New Phytol. 211, 599–613. [DOI] [PubMed] [Google Scholar]
- Chen, R. , Jiang, H. , Li, L. et al (2012) The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell, 24, 2898–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini, A. , Fonseca, S. , Fernandez, G. et al (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature, 448, 666–671. [DOI] [PubMed] [Google Scholar]
- Chini, A. , Monte, I. , Zamarreño, A.M. , Hamberg, M. , Lassueur, S. , Reymond, P. , Weiss, S. , Stintzi, A. , Schaller, A. and Porzel, A. (2018) An OPR3‐independent pathway uses 4,5‐didehydrojasmonate for jasmonate synthesis. Nat. Chem. Biol. 14, 171. [DOI] [PubMed] [Google Scholar]
- Chung, H.S. , Cooke, T.F. , Depew, C.L. , Patel, L.C. , Ogawa, N. , Kobayashi, Y. and Howe, G.A. (2010) Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 63, 613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolen, S. , Proietti, S. , Hickman, R. et al (2016) Transcriptome dynamics of Arabidopsis during sequential biotic and abiotic stresses. Plant J. 86, 249–267. [DOI] [PubMed] [Google Scholar]
- Cui, H. , Qiu, J. , Zhou, Y. , Bhandari, D.D. , Zhao, C. , Bautor, J. and Parker, J.E. (2018) Antagonism of transcription factor MYC2 by EDS1/PAD4 complexes bolsters salicylic acid defense in Arabidopsis effector‐triggered immunity. Mol. Plant, 11, 1053–1066. [DOI] [PubMed] [Google Scholar]
- Cui, H. , Tsuda, K. and Parker, J.E. (2015) Effector‐triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L. , Horvath, D.M. and Staskawicz, B.J. (2013) Pivoting the plant immune system from dissection to deployment. Science, 341, 746–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Y. , Liu, N. , Virlouvet, L. , Riethoven, J.‐J. , Fromm, M. and Avramova, Z. (2013) Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Biol. 13, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgikh, V.A. , Pukhovaya, E.M. and Zemlyanskaya, E.V. (2019) Shaping ethylene response: the role of EIN3/EIL1 transcription factors. Front. Plant Sci. 10, 1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht, B. , Xue, G.P. , Sprague, S.J. et al (2007) MYC2 differentially modulates diverse jasmonate‐dependent functions in Arabidopsis . Plant Cell, 19, 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb, M. and Reymond, P. (2019) Molecular interactions between plants and insect herbivores. Annu. Rev. Plant Biol. 70, 527–557. [DOI] [PubMed] [Google Scholar]
- Fallath, T. , Kidd, B.N. , Stiller, J. , Davoine, C. , Björklund, S. , Manners, J.M. , Kazan, K. and Schenk, P.M. (2017) MEDIATOR18 and MEDIATOR20 confer susceptibility to Fusarium oxysporum in Arabidopsis thaliana . PLoS One, 12, e0176022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, J. , Hill, L. , Crooks, C. , Doerner, P. and Lamb, C. (2009) Abscisic acid has a key role in modulating diverse plant‐pathogen interactions. Plant Physiol. 150, 1750–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández‐Calvo, P. , Chini, A. , Fernández‐Barbero, G. et al (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell, 23, 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca, S. , Chini, A. , Hamberg, M. , Adie, B. , Porzel, A. , Kramell, R. , Miersch, O. , Wasternack, C. and Solano, R. (2009) (+)‐7‐iso‐Jasmonoyl‐L‐isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5, 344–350. [DOI] [PubMed] [Google Scholar]
- Franco‐Zorrilla, J.M. , López‐Vidriero, I. , Carrasco, J.L. , Godoy, M. , Vera, P. and Solano, R. (2014) DNA‐binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. USA, 111, 2367–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin, K.A. (2008) Shade avoidance. New Phytol. 179, 930–944. [DOI] [PubMed] [Google Scholar]
- Gimenez‐Ibanez, S. , Boter, M. , Fernandez‐Barbero, G. , Chini, A. , Rathjen, J.P. and Solano, R. (2014) The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis . PLoS Biol. 12, e1001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez‐Ibanez, S. , Boter, M. , Ortigosa, A. , García‐Casado, G. , Chini, A. , Lewsey, M.G. , Ecker, J.R. , Ntoukakis, V. and Solano, R. (2017) JAZ2 controls stomata dynamics during bacterial invasion. New Phytol. 213, 1378–1392. [DOI] [PubMed] [Google Scholar]
- Gimenez‐Ibanez, S. and Solano, R. (2013) Nuclear jasmonate and salicylate signaling and crosstalk in defense against pathogens. Front. Plant Sci. 4, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J. , Chen, W. , Estes, B. , Chang, H. , Nawrath, C. , Métraux, J. , Zhu, T. and Katagiri, F. (2003) Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 34, 217–228. [DOI] [PubMed] [Google Scholar]
- Han, X. and Kahmann, R. (2019) Manipulation of phytohormone pathways by effectors of filamentous plant pathogens. Front. Plant Sci. 10, 822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X. , Jiang, J. , Wang, C.Q. and Dehesh, K. (2017) ORA59 and EIN3 interaction couples jasmonate‐ethylene synergistic action to antagonistic salicylic acid regulation of PDF expression. J. Integr. Plant Biol. 59, 275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y. , Hong, G. , Zhang, H. et al (2020) The OsGSK2 kinase integrates brassinosteroid and jasmonic acid signaling by interacting with OsJAZ4. Plant Cell, 32, 2806–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman, R. , Mendes, M.P. , Van Verk, M.C. , Van Dijken, A.J.H. , Di Sora, J. , Denby, K. , Pieterse, C.M.J. and Van Wees, S.C.M. (2019) Transcriptional dynamics of the salicylic acid response and its interplay with the jasmonic acid pathway. bioRxiv, 742742. [Google Scholar]
- Hickman, R. , Van Verk, M.C. , Van Dijken, A.J.H. et al (2017) Architecture and dynamics of the jasmonic acid gene regulatory network. Plant Cell, 29, 2086–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillmer, R.A. , Tsuda, K. , Rallapalli, G. , Asai, S. , Truman, W. , Papke, M.D. , Sakakibara, H. , Jones, J.D. , Myers, C.L. and Katagiri, F. (2017) The highly buffered Arabidopsis immune signaling network conceals the functions of its components. PLoS Genet. 13, e1006639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, X. , Ding, L. and Yu, H. (2013) Crosstalk between GA and JA signaling mediates plant growth and defense. Plant Cell Rep. 32, 1067–1074. [DOI] [PubMed] [Google Scholar]
- Hou, X. , Lee, L.Y.C. , Xia, K. , Yen, Y. and Yu, H. (2010) DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell, 19, 884–894. [DOI] [PubMed] [Google Scholar]
- Huang, P. , Dong, Z. , Guo, P. , Zhang, X.C. , Qiu, Y. , Li, B. , Wang, Y. and Guo, H.J. (2020) Salicylic acid suppresses apical hook formation via NPR1‐mediated repression of EIN3 and EIL1 in Arabidopsis. Plant Cell, 32, 612–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, S. , Yao, J. , Ma, K.‐W. , Zhou, H. , Song, J. , He, S.Y. and Ma, W. (2013) Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLoS Pathog. 9, e1003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage, D. , Tootle, T.L. , Reuber, T.L. , Frost, L.N. , Feys, B.J. , Parker, J.E. , Ausubel, F.M. and Glazebrook, J. (1999) Arabidopsis thaliana PAD4 encodes a lipase‐like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA, 96, 13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan, K. and Manners, J.M. (2013) MYC2: the master in action. Mol. Plant, 6, 686–703. [DOI] [PubMed] [Google Scholar]
- Kim, T.‐W. , Guan, S. , Sun, Y. , Deng, Z. , Tang, W. , Shang, J.‐X. , Sun, Y. , Burlingame, A.L. and Wang, Z.‐Y. (2009) Brassinosteroid signal transduction from cell‐surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11, 1254–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y. , Tsuda, K. , Igarashi, D. , Hillmer, R.A. , Sakakibara, H. , Myers, C.L. and Katagiri, F. (2014) Mechanisms underlying robustness and tunability in a plant immune signaling network. Cell Host Microbe, 15, 84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, A. , Leon‐Reyes, A. , Ritsema, T. , Verhage, A. , Den Otter, F.C. , Van Loon, L.C. and Pieterse, C.M.J. (2008) Kinetics of salicylate‐mediated suppression of jasmonate signaling reveal a role for redox modulation. Plant Physiol. 147, 1358–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk, S. , Thurow, C. , Niggeweg, R. and Gatz, C. (2002) Analysis of the spacing between the two palindromes of activation sequence‐1 with respect to binding to different TGA factors and transcriptional activation potential. Nucleic Acids Res. 30, 775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T.A. and Bailey‐Serres, J. (2019) Integrative analysis from the epigenome to translatome uncovers patterns of dominant nuclear regulation during transient stress. Plant Cell, 31, 2573–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon‐Reyes, A. , Spoel, S.H. , De Lange, E.S. , Abe, H. , Kobayashi, M. , Tsuda, S. , Millenaar, F.F. , Welschen, R.A.M. , Ritsema, T. and Pieterse, C.M.J. (2009) Ethylene modulates the role of NONEXPRESSOR OF PATHOGENESIS‐RELATED GENES1 in cross talk between salicylate and jasmonate signaling. Plant Physiol. 149, 1797–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, L.A. , Polanski, K. , de Torres‐Zabala, M. et al (2015) Transcriptional dynamics driving MAMP‐triggered immunity and pathogen effector‐mediated immunosuppression in Arabidopsis leaves following infection with Pseudomonas syringae pv tomato DC3000. Plant Cell, 27, 3038–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L. , Sonbol, F.‐M. , Huot, B. , Gu, Y. , Withers, J. , Mwimba, M. , Yao, J. , He, S.Y. and Dong, X. (2016a) Salicylic acid receptors activate jasmonic acid signalling through a non‐canonical pathway to promote effector‐triggered immunity. Nat. Commun. 7, 13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, N. , Staswick, P.E. and Avramova, Z. (2016b) Memory responses of jasmonic acid‐associated Arabidopsis genes to a repeated dehydration stress. Plant Cell Environ. 39, 2515–2529. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Wei, H. , Ma, M. et al (2019) Arabidopsis FHY3 and FAR1 regulate the balance between growth and defense responses under shade conditions. Plant Cell, 31, 2089–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo, O. , Chico, J.M. , Sanchez‐Serrano, J.J. and Solano, R. (2004) JASMONATE‐INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate‐regulated defense responses in Arabidopsis . Plant Cell, 16, 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo, O. , Piqueras, R. , Sánchez‐Serrano, J.J. and Solano, R. (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell, 15, 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, H. , McClung, C.R. and Zhang, C. (2017) Tick tock: circadian regulation of plant innate immunity. Annu. Rev. Phytopathol. 55, 287–311. [DOI] [PubMed] [Google Scholar]
- Major, I.T. , Guo, Q. , Zhai, J. , Kapali, G. , Kramer, D.M. and Howe, G.A. (2020) A phytochrome B‐independent pathway restricts growth at high levels of jasmonate defense. Plant Physiol. 183, 733–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan, S. and Alon, U. (2003) Structure and function of the feed‐forward loop network motif. Proc. Natl. Acad. Sci. USA, 100, 11980–11985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier, A. , Xin, B. , Chiu, T.‐P. , Yang, L. , Rohs, R. and Wasserman, W.W. (2016) DNA shape features improve transcription factor binding site predictions in vivo. Cell Syst. 3, 278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchante, C. , Brumos, J. , Yun, J. , Hu, Q. , Spencer, K.R. , Enríquez, P. , Binder, B.M. , Heber, S. , Stepanova, A.N. and Alonso, J.M. (2015) Gene‐specific translation regulation mediated by the hormone‐signaling molecule EIN2. Cell, 163, 684–697. [DOI] [PubMed] [Google Scholar]
- Merchante, C. , Stepanova, A.N. and Alonso, J.M. (2017) Translation regulation in plants: an interesting past, an exciting present and a promising future. Plant J. 90, 628–653. [DOI] [PubMed] [Google Scholar]
- Meteignier, L.‐V. , El Oirdi, M. , Cohen, M. , Barff, T. , Matteau, D. , Lucier, J.‐F. , Rodrigue, S. , Jacques, P.‐E. , Yoshioka, K. and Moffett, P. (2017) Translatome analysis of an NB‐LRR immune response identifies important contributors to plant immunity in Arabidopsis. J. Exp. Bot. 68, 2333–2344. [DOI] [PubMed] [Google Scholar]
- Mine, A. , Nobori, T. , Salazar‐Rondon, M.C. , Winkelmüller, T.M. , Anver, S. , Becker, D. and Tsuda, K. (2017) An incoherent feed‐forward loop mediates robustness and tunability in a plant immune network. EMBO Rep. 18, 464–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte, I. , Kneeshaw, S. , Franco‐Zorrilla, J.M. , Chini, A. , Zamarreño, A.M. , García‐Mina, J.M. and Solano, R. (2020) An ancient COI1‐independent function for reactive electrophilic oxylipins in thermotolerance. Curr. Biol. 30, 962–971.e963. [DOI] [PubMed] [Google Scholar]
- Muiño, J.M. , Smaczniak, C. , Angenent, G.C. , Kaufmann, K. and Van Dijk, A.D. (2014) Structural determinants of DNA recognition by plant MADS‐domain transcription factors. Nucleic Acids Res. 42, 2138–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai, R. , Howell, K.A. , Millar, A.H. , O'Toole, N. , Small, I. and Whelan, J. (2007) Genome‐wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana . Plant Cell, 19, 3418–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndamukong, I. , Abdallat, A.A. , Thurow, C. , Fode, B. , Zander, M. , Weigel, R. and Gatz, C. (2007) SA‐inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA‐responsive PDF1.2 transcription. Plant J. 50, 128–139. [DOI] [PubMed] [Google Scholar]
- Nobori, T. and Tsuda, K. (2019) The plant immune system in heterogeneous environments. Curr. Opin. Plant Biol. 50, 58–66. [DOI] [PubMed] [Google Scholar]
- O’Malley, R.C. , Huang, S.S.C. , Song, L. , Lewsey, M.G. , Bartlett, A. , Nery, J.R. , Galli, M. , Gallavotti, A. and Ecker, J.R. (2016) Cistrome and epicistrome features shape the regulatory DNA landscape. Cell, 165, 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortigosa, A. , Fonseca, S. , Franco‐Zorrilla, J.M. , Fernández‐Calvo, P. , Zander, M. , Lewsey, M.G. , García‐Casado, G. , Fernández‐Barbero, G. , Ecker, J.R. and Solano, R. (2020) The JA‐pathway MYC transcription factors regulate photomorphogenic responses by targeting HY5 gene expression. Plant J. 102, 138–152. [DOI] [PubMed] [Google Scholar]
- Pajerowska‐Mukhtar, K.M. , Wang, W. , Tada, Y. , Oka, N. , Tucker, C.L. , Fonseca, J.P. and Dong, X. (2012) The HSF‐like transcription factor TBF1 is a major molecular switch for plant growth‐to‐defense transition. Curr. Biol. 22, 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels, L. , Morreel, K. , De Witte, E. , Lammertyn, F. , Van Montagu, M. , Boerjan, W. , Inzé, D. and Goossens, A. (2008) Mapping methyl jasmonate‐mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. USA, 105, 1380–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels, L. , Ritter, A. , Goossens, J. et al (2015) The RING E3 ligase KEEP ON GOING modulates JASMONATE ZIM‐DOMAIN12 stability. Plant Physiol. 169, 1405–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, P. , Yan, Z. , Zhu, Y. and Li, J. (2008) Regulation of the Arabidopsis GSK3‐like kinase BRASSINOSTEROID‐INSENSITIVE 2 through proteasome‐mediated protein degradation. Mol. Plant, 1, 338–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M.J. , Van der Does, D. , Zamioudis, C. , Leon‐Reyes, A. and Van Wees, S.C.M. (2012) Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. [DOI] [PubMed] [Google Scholar]
- Pré, M. , Atallah, M. , Champion, A. , De Vos, M. , Pieterse, C.M.J. and Memelink, J. (2008) The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147, 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekhter, D. , Lüdke, D. , Ding, Y. , Feussner, K. , Zienkiewicz, K. , Lipka, V. , Wiermer, M. , Zhang, Y. and Feussner, I. (2019) Isochorismate‐derived biosynthesis of the plant stress hormone salicylic acid. Science, 365, 498–502. [DOI] [PubMed] [Google Scholar]
- Sasaki‐Sekimoto, Y. , Jikumaru, Y. , Obayashi, T. , Saito, H. , Masuda, S. , Kamiya, Y. , Ohta, H. and Shirasu, K. (2013) Basic Helix‐Loop‐Helix transcription factors JA‐ASSOCIATED MYC2‐LIKE 1 (JAM1), JAM2 and JAM3 are negative regulators of jasmonate responses in Arabidopsis . Plant Physiol. 163, 291–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard, L.B. , Tan, X. , Mao, H.B. et al (2010) Jasmonate perception by inositol‐phosphate‐potentiated COI1‐JAZ co‐receptor. Nature, 468, 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, L. , Huang, S.‐S.‐ C. , Wise, A. , Castanon, R. , Nery, J.R. , Chen, H. , Watanabe, M. , Thomas, J. , Bar‐Joseph, Z. and Ecker, J.R. (2016) A transcription factor hierarchy defines an environmental stress response network. Science, 354, aag1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, S. , Huang, H. , Gao, H. et al (2014) Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis . Plant Cell, 26, 263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel, S.H. and Loake, G.J. (2011) Redox‐based protein modifications: the missing link in plant immune signalling. Curr. Opin. Plant Biol. 14, 358–364. [DOI] [PubMed] [Google Scholar]
- Staiger, D. , Korneli, C. , Lummer, M. and Navarro, L. (2013) Emerging role for RNA‐based regulation in plant immunity. New Phytol. 197, 394–404. [DOI] [PubMed] [Google Scholar]
- Ströher, E. and Millar, A.H. (2012) The biological roles of glutaredoxins. Biochem. J. 446, 333–348. [DOI] [PubMed] [Google Scholar]
- Thines, B. , Katsir, L. , Melotto, M. , Niu, Y. , Mandaokar, A. , Liu, G.H. , Nomura, K. , He, S.Y. , Howe, G.A. and Browse, J. (2007) JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature, 448, 661–665. [DOI] [PubMed] [Google Scholar]
- Tsuda, K. , Sato, M. , Stoddard, T. , Glazebrook, J. and Katagiri, F. (2009) Network properties of robust immunity in plants. PLoS Genet. 5, e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrig, J.F. , Huang, L.‐J. , Barghahn, S. , Willmer, M. , Thurow, C. and Gatz, C. (2017) CC‐type glutaredoxins recruit the transcriptional co‐repressor TOPLESS to TGA‐dependent target promoters in Arabidopsis thaliana . Biochim. Biophys. Acta Gene Regul. Mech. 1860, 218–226. [DOI] [PubMed] [Google Scholar]
- Van Butselaar, T. and Van den Ackerveken, G. (2020) Salicylic acid steers the growth–immunity tradeoff. Trends Plant Sci. 25, 566–576. [DOI] [PubMed] [Google Scholar]
- Van der Does, D. , Leon‐Reyes, A. , Koornneef, A. et al (2013) Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1‐JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell, 25, 744–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage, A. , Vlaardingerbroek, I. , Raaymakers, C. , Van Dam, N. , Dicke, M. , Van Wees, S.C.M. and Pieterse, C.M.J. (2011) Rewiring of the jasmonate signaling pathway in Arabidopsis during insect herbivory. Front. Plant Sci. 2, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos, I.A. , Moritz, L. , Pieterse, C.M.J. and Van Wees, S.C.M. (2015) Impact of hormonal crosstalk on plant resistance and fitness under multi‐attacker conditions. Front. Plant Sci. 6, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos, I.A. , Pieterse, C.M.J. and Van Wees, S.C.M. (2013a) Costs and benefits of hormone‐regulated plant defences. Plant Pathol. 62, 43–55. [Google Scholar]
- Vos, I.A. , Verhage, A. , Schuurink, R.C. , Watt, L.G. , Pieterse, C.M.J. and Van Wees, S.C.M. (2013b) Onset of herbivore‐induced resistance in systemic tissue primed for jasmonate‐dependent defenses is activated by abscisic acid. Front. Plant Sci. 4, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , Du, X. and Mou, Z. (2016) The mediator complex subunits MED14, MED15, and MED16 are involved in defense signaling crosstalk in Arabidopsis . Front. Plant Sci. 7, 1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , Yao, J. , Du, X. , Zhang, Y. , Sun, Y. , Rollins, J.A. and Mou, Z. (2015) The Arabidopsis mediator complex subunit16 is a key component of basal resistance against the necrotrophic fungal pathogen Sclerotinia sclerotiorum . Plant Physiol. 169, 856–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Li, S. , Li, Y.A. et al (2019) MED25 connects enhancer–promoter looping and MYC2‐dependent activation of jasmonate signalling. Nat. Plants, 5, 616–625. [DOI] [PubMed] [Google Scholar]
- Wang, K. , Guo, Q. , Froehlich, J.E. , Hersh, H.L. , Zienkiewicz, A. , Howe, G.A. and Benning, C. (2018) Two abscisic acid‐responsive plastid lipase genes involved in jasmonic acid biosynthesis in Arabidopsis thaliana . Plant Cell, 30, 1006–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C. and Hause, B. (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111, 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C. and Song, S. (2017) Jasmonates: biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 68, 1303–1321. [DOI] [PubMed] [Google Scholar]
- Weirauch, M.T. , Yang, A. , Albu, M. et al (2014) Determination and inference of eukaryotic transcription factor sequence specificity. Cell, 158, 1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windram, O. , Madhou, P. , McHattie, S. et al (2012) Arabidopsis defense against Botrytis cinerea: Chronology and regulation deciphered by high‐resolution temporal transcriptomic analysis. Plant Cell, 24, 3530–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F. , Deng, L. , Zhai, Q. , Zhao, J. , Chen, Q. and Li, C. (2020) Mediator subunit MED25 couples alternative splicing of JAZ genes with fine‐tuning of jasmonate signaling. Plant Cell, 32, 429–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, G. , Greene, G.H. , Yoo, H. , Liu, L. , Marqués, J. , Motley, J. and Dong, X. (2017) Global translational reprogramming is a fundamental layer of immune regulation in plants. Nature, 545, 487–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, C. , Fan, M. , Yang, M. , Zhao, J. , Zhang, W. , Su, Y. , Xiao, L. , Deng, H. and Xie, D. (2018) Injury activates Ca2+/calmodulin‐dependent phosphorylation of JAV1‐JAZ8‐WRKY51 complex for jasmonate biosynthesis. Mol. Cell, 70, 136–149. [DOI] [PubMed] [Google Scholar]
- You, Y. , Zhai, Q. , An, C. and Li, C. (2019) LEUNIG_HOMOLOG mediates MYC2‐dependent transcriptional activation in cooperation with the coactivators HAC1 and MED25. Plant Cell, 31, 2187–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, H.M. , Liu, W.C. and Lu, Y.T. (2017) CATALASE2 coordinates SA‐mediated repression of both auxin accumulation and JA biosynthesis in plant defenses. Cell Host Microbe, 21, 143–155. [DOI] [PubMed] [Google Scholar]
- Zander, M. , Chen, S. , Imkampe, J. , Thurow, C. and Gatz, C. (2012) Repression of the Arabidopsis thaliana jasmonic acid/ethylene‐induced defense pathway by TGA‐interacting glutaredoxins depends on their C‐terminal ALWL motif. Mol. Plant, 5, 831–840. [DOI] [PubMed] [Google Scholar]
- Zander, M. , La Camera, S. , Lamotte, O. , Métraux, J.‐P. and Gatz, C. (2010) Arabidopsis thaliana class‐II TGA transcription factors are essential activators of jasmonic acid/ethylene‐induced defense responses. Plant J. 61, 200–210. [DOI] [PubMed] [Google Scholar]
- Zander, M. , Lewsey, M.G. , Clark, N.M. et al (2020) Integrated multi‐omics framework of the plant response to jasmonic acid. Nat. Plants, 6, 290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander, M. , Thurow, C. and Gatz, C. (2014) TGA transcription factors activate the salicylic acid‐suppressible branch of the ethylene‐induced defense program by regulating ORA59 expression. Plant Physiol. 165, 1671–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti, M.E. , Blanco, F. , Reynoso, M. and Crespi, M. (2020) To keep or not to keep: mRNA stability and translatability in root nodule symbiosis. Curr. Opin. Plant Biol. 56, 109–117. [DOI] [PubMed] [Google Scholar]
- Zhai, Q. and Li, C. (2019) The plant Mediator complex and its role in jasmonate signaling. J. Exp. Bot. 70, 3415–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, Q. , Yan, L. , Tan, D. , Chen, R. , Sun, J. , Gao, L. , Dong, M.‐Q. , Wang, Y. and Li, C. (2013) Phosphorylation‐coupled proteolysis of the transcription factor MYC2 is important for jasmonate‐signaled plant immunity. PLoS Genet. 9, e1003422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F. , Yao, J. , Ke, J. et al (2015) Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature, 525, 269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Zhu, Z. , An, F. , Hao, D. , Li, P. , Song, J. , Yi, C. and Guo, H. (2014) Jasmonate‐activated MYC2 represses ETHYLENE INSENSITIVE3 activity to antagonize ethylene‐promoted apical hook formation in Arabidopsis . Plant Cell, 26, 1105–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. and Li, X. (2019) Salicylic acid: biosynthesis, perception, and contributions to plant immunity. Curr. Opin. Plant Biol. 50, 29–36. [DOI] [PubMed] [Google Scholar]
- Zheng, X.‐Y. , Spivey, N.W. , Zeng, W. , Liu, P.‐P. , Fu, Z.Q. , Klessig, D.F. , He, S.Y. and Dong, X. (2012) Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe, 11, 587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Z. , An, F. , Feng, Y. et al (2011) Derepression of ethylene‐stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis . Proc. Natl. Acad. Sci. USA, 108, 12539–12544. [DOI] [PMC free article] [PubMed] [Google Scholar]


