Abstract
Aims
In long QT syndrome (LQTS) patients, modifier genes modulate the arrhythmic risk associated with a disease-causing mutation. Their recognition can improve risk stratification and clinical management, but their discovery represents a challenge. We tested whether a cellular-driven approach could help to identify new modifier genes and especially their mechanism of action.
Methods and results
We generated human-induced pluripotent stem cell-derived cardiomyocytes (iPSC-CM) from two patients carrying the same KCNQ1-Y111C mutation, but presenting opposite clinical phenotypes. We showed that the phenotype of the iPSC-CMs derived from the symptomatic patient is due to impaired trafficking and increased degradation of the mutant KCNQ1 and wild-type human ether-a-go-go-related gene. In the iPSC-CMs of the asymptomatic (AS) patient, the activity of an E3 ubiquitin-protein ligase (Nedd4L) involved in channel protein degradation was reduced and resulted in a decreased arrhythmogenic substrate. Two single-nucleotide variants (SNVs) on the Myotubularin-related protein 4 (MTMR4) gene, an interactor of Nedd4L, were identified by whole-exome sequencing as potential contributors to decreased Nedd4L activity. Correction of these SNVs by CRISPR/Cas9 unmasked the LQTS phenotype in AS cells. Importantly, the same MTMR4 variants were present in 77% of AS Y111C mutation carriers of a separate cohort. Thus, genetically mediated interference with Nedd4L activation seems associated with protective effects.
Conclusion
Our finding represents the first demonstration of the cellular mechanism of action of a protective modifier gene in LQTS. It provides new clues for advanced risk stratification and paves the way for the design of new therapies targeting this specific molecular pathway.
Keywords: Long QT syndrome, Induced pluripotent stem cells, Arrhythmias, Variants, Nedd4L, MTMR4
1. Introduction
One of the most puzzling questions in arrhythmic disorders of genetic origin concerns why the clinical manifestations in two siblings carrying the same disease-causing mutations may vary from benign to highly malignant. Genetic variants acting as ‘modifier genes’ are the commonly accepted explanation.1 The congenital long QT syndrome (LQTS) is caused by mutations of the genes encoding ion channels or excitation-contraction coupling proteins,2 has a prevalence of 1 in 2000,3 is associated with life-threatening arrhythmias,4 shows a good genotype–phenotype correlation,5 and is regarded as a paradigm for sudden cardiac death.6
LQT1, the most common type of LQTS, is caused by loss-of-function mutations on the KCNQ1 gene encoding for the repolarizing current IKs.2,5 As IKs mutations impair QT shortening during heart rate increase, most arrhythmic events among LQT1 patients are triggered by sympathetic activation during exercise and emotions.5
Here, we performed cellular electrophysiology in patient-specific induced pluripotent stem cell (iPSC)-derived cardiomyocytes (iPSC-CMs)7,8 and whole-exome sequencing to identify potential mechanisms by which modifier genes may exert their action in LQTS. We focused on the common mutation KCNQ1-Y111C (henceforth, Y111C) characterized by a generally benign phenotype.9,10 Y111C is associated with channel trafficking defects11 and accelerated degradation by the proteasome.12 By studying a family whose members carry the same Y111C mutation but have distinct clinical severity of LQTS, we discovered two protective single-nucleotide variants (SNVs) on the same gene present in the asymptomatic (AS) Y111C carriers and unravelled their mechanism of action.
2. Methods
An expanded method section is available in the Supplementary material online, Appendix.
2.1 Generation of human iPSCs and iPSC-CMs
The study was approved by the ethics committee of the Fondazione IRCCS Policlinico San Matteo (Pavia, Italy). The study conformed to the principles outlined in the Declaration of Helsinki and informed written consent was given prior to the inclusion of subjects in the study. Human dermal fibroblasts were isolated from skin biopsies and reprogrammed to iPSCs by using an established protocol.13 These and control (CTR) iPSC lines were then differentiated into iPSC-CMs with a standard protocol.14
2.2 Electrophysiology
Extracellular field potentials (FPs) were recorded in spontaneously beating embryoid bodies (EBs) by Multi-Electrode Arrays (MEA). QT intervals normalized to the intrinsic beating rate (Bazett’s correction) were used as a surrogate of the QTc. Action potentials (APs) were recorded by whole-cell patch-clamp in isolated iPSC-CMs paced at 0.5, 1, 2, and 3.33 Hz during Tyrode’s superfusion. AP parameters were measured from the same cell in the absence (‘native’) and in the presence of injected IK1 (by dynamic clamp). Under native conditions, iPSC-CMs had partially depolarized diastolic potentials (Ediast −49.5 ± 3.4 mV at 0.5 Hz), a likely consequence of low IK1 expression in immature isolated cardiomyocytes.15 To overcome this problem, numerically modelled IK1 (see Supplementary material online, Appendix) was injected in iPSC-CMs by dynamic clamp,16 which was customized as previously reported.17IKs and IKr were isolated in voltage-clamped iPSC-CMs as HMR1556- and E4031-sensitive currents, respectively. All measurements were performed at physiological temperature (36.5°C) (see Supplementary material online, Appendix).
2.3 Protein interaction
Nedd4-like, E3 ubiquitin-protein ligase (Nedd4L)-KCNQ1 and Nedd4L-Myotubularin-related protein 4 (MTMR4) protein interactions were investigated in a 293T-based heterologous system (see Supplementary material online, Appendix). Whole-exome sequencing was performed using Illumina HiSeq with the SeqCap EZ Human Exome Library v3.0 (see Supplementary material online, Appendix).18
2.4 Statistical analysis
All the results are expressed as mean ± Standard Error of the Mean (SEM). Statistical comparisons were performed using Student’s t-test for paired observations. One-way or two-ways analysis of variance (ANOVA) were performed when multiple independent groups were compared. Post hoc comparison between individual means was performed by Tukey’s method and P-values have been corrected for multiple testings. A P-value of <0.05 was considered statistically significant.
3. Results
3.1 Characterization of patients, iPSCs, and iPSC-CMs
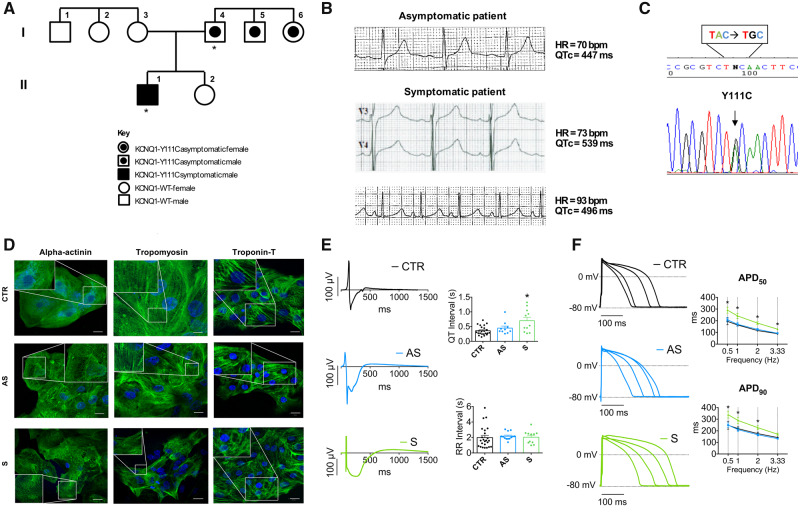
The two subjects studied are father and son and their family pedigree is shown in Figure 1A. The father, our AS carrier, has never experienced symptoms despite no treatment; his son, our symptomatic (S) proband, is a 16-year-old boy who experienced a prolonged syncope with sphincter release during combined emotional and physical stress at age 9 and became AS on β-blocker therapy (propranolol 2.5 mg/kg). His 12-lead electrocardiogram recorded before initiation of propranolol showed a consistently prolonged corrected QT interval (QTc) (often >550 ms), whereas the AS father had borderline values (Figure 1B). The mutation screening showed that the heterozygous LQTS mutation consisted in a single base exchange (332A→G) in the KCNQ1 gene, resulting in the Y111C missense mutation previously shown to be associated with LQT1 (Figure 1C).11,12 Compound heterozygosity and/or double mutations in the S proband were excluded since no additional mutations in all known LQTS genes were detected.
Figure 1.
Generation of patient-specific iPSC-CMs. (A) Pedigree of the family segregating the KCNQ1-Y111C mutation. The asterisks indicate the two subjects enrolled in the study: the symptomatic (S) proband is a 16-year-old boy (II-1) with an episode of syncope with sphincter release during a combined emotional and physical stress, while the father (I-4) has never experienced symptoms, i.e. he is an asymptomatic carrier (AS). Other two family members, one uncle (I-5) and one aunt (I-6) of the S proband also carry the same heterozygous mutation, but their QT interval is normal and they have never experienced symptoms. (B) Electrocardiogram tracings. Representative electrocardiograms of the AS individual and of the S patient at age 8 prior to institution of β-blocker therapy. Rate corrected QT interval (QTc, Bazett’s formula) are indicated at the top of the tracings. (C) DNA sequencing. The analysis shows the presence of the heterozygous 332 A>G mutation in the KCNQ1 coding region, resulting, at protein level, in the substitution of the tyrosine (Y) in position 111 with a cysteine (C). (D) iPSC-CMs. Cardiac differentiation was demonstrated by expression of alpha-actinin, cardiac troponin-T, and tropomyosin (green) organized to form cross-striations (insets). Nuclei were stained with Hoechst 33258 (blue). Scale bar = 20 μm. (E) Multi-Electrode array analysis. Representative field potential recordings (left) of CTR-, AS-, and S-iPSC-CMs. Average values for QT- and RR interval durations recorded in spontaneously beating EBs from CTR-, AS-, and S-iPSC-CMs (right) (*P < 0.05; n = 10–23). (F) Action potentials. APs recorded with IK1 injection by dynamic clamp in CTR-, AS-, and S-iPSC-CMs paced at 0.5, 1, 2, and 3.33 Hz (*P < 0.05 vs. S; n = 7–13). One-way ANOVA for unpaired measurements was applied to MEA and patch-clamp data. Post hoc comparison among individual means was performed with Tukey’s method.
Fibroblasts of the S and AS mutation carrier and a CTR cell line were successfully transformed into iPSC clones able to maintain pluripotent features and to differentiate into beating EBs (Supplementary material online, Appendix and Figure S1). The iPSC-CMs stained positive for several cardiac-specific myofilament proteins (Figure 1D) and were able to beat spontaneously. To assess the electrophysiological phenotype of Y111C-mutated iPSC-CMs, we first measured the electrical activity (FP) of spontaneously beating EBs by MEA. The mean QT interval was longer in the S-iPSC-CMs compared with both AS and CTR iPSC-CMs, while the mean RR interval was similar among all groups (Figure 1E). Patch-clamp data showed that, under dynamic clamp condition, the AP duration (APD) measured at 50% (APD50) and 90% of the repolarization phase (APD90) on single iPSC-CMs was longer in S-iPSC-CMs compared with AS- and CTR-iPSC-CMs at all pacing rates (Figure 1F). Albeit with larger variability, cells studied under native conditions showed a qualitatively similar pattern (Supplementary material online, Figure S2).
3.2 Expression of KCNQ1
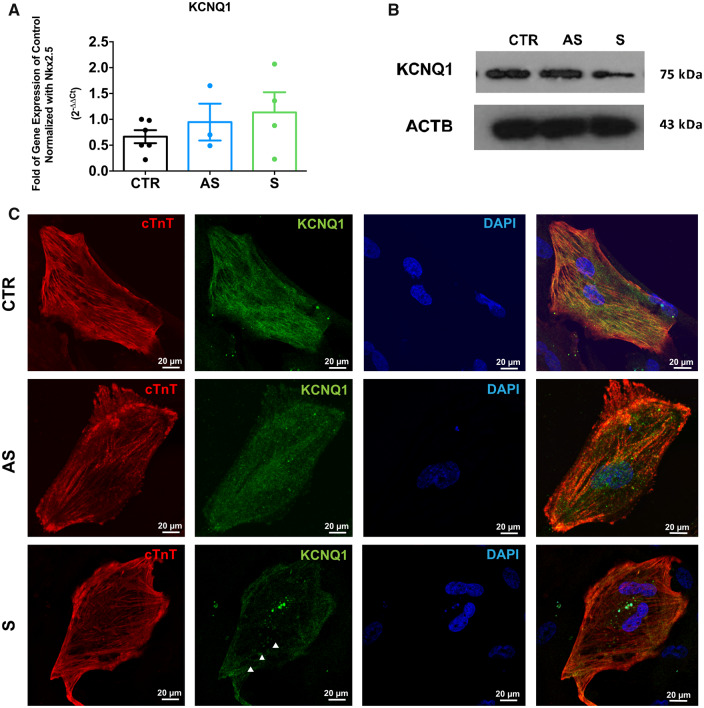
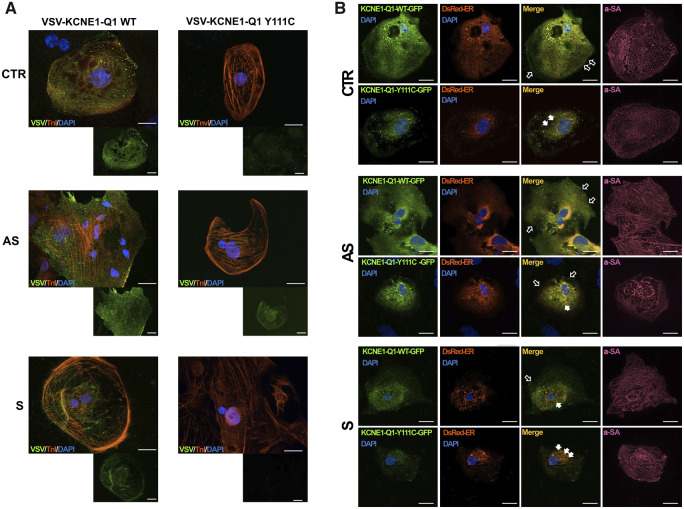
We then explored differences between AS- and S-iPSC-CMs in terms of KCNQ1 levels and/or distribution inside the iPSC-CMs. First, we quantified mRNA levels of KCNQ1 without finding any difference among CTR-, AS-, and S-iPSC-CMs (Figure 2A). In contrast, the KCNQ1 protein level was lower in S-iPSC-CMs compared with both CTR- and AS-iPSC-CMs (Figure 2B). Moreover, immunocytochemistry showed reduced expression of KCNQ1 on the cell membrane of S-iPSC-CMs compared with AS- and CTR-iPSC-CMs (Figure 2C). To better discriminate differences in cellular localization of KCNQ1 between S and AS subjects, we transfected the iPSC-CMs with a plasmid over-expressing wild-type (WT) or mutant KCNQ1 proteins fused with a VSV tag located on the extracellular N-terminal domain.11 VSV-positive staining of WT KCNQ1 was present in S-, AS-, and CTR-iPSC-CMs but the signal intensity was lower in S-iPSC-CMs compared with AS- and CTR-iPSC-CMs (Figure 3A). Conversely, the mutant fusion protein was undetectable on the membrane of both S- and CTR-iPSC-CMs while it was present in AS-iPSC-CMs (Figure 3A).
Figure 2.
Expression of endogenous KCNQ1 in Y111C iPSC-CMs. (A) Quantification of KCNQ1 mRNA levels by RT-qPCR and ΔΔCt method. Nkx2.5 was used as reference gene. Data were expressed as fold increase vs. CTR (n = 3, **P < 0.005). (B) Quantification of KCNQ1 protein levels by SDS-PAGE and immunoblot in CTR, S, and AS iPSC-CMs. (C) Immunostaining of the endogenous KCNQ1 in CTR-, AS-, and S-iPSC-CMs. KCNQ1 was stained in green, cardiac troponin-T (cTnT) in red, and the nuclei in blue.
Figure 3.
Trafficking defect of KCNQ1 in iPSC-CMs. (A) Trafficking defect of the VSV-KCNE1-KCNQ1 (VSV-E1-Q1) Y111C fusion protein. Representative images of CTR-, AS-, and S-iPSC-CMs acquired 72 h after transfection with WT or Y111C VSV-E1-Q1 plasmid. Anti-VSV staining (green) was performed on live cells. Then, the cells were fixed, permeabilized, and co-stained with an anti-Troponin-I (TnI) antibody, used as a marker specific for cardiomyocytes (red). Nuclei were stained with DAPI (blue). Scale bar = 20 μm. (B) Retention of Y111C VSV-E1-Q1-GFP fusion protein in endoplasmic reticulum. Images of CTR-, AS-, and S-iPSC-CMs co-transfected with WT or Y111C VSV-E1-Q1-GFP plasmids (green), and pDsRed2-ER plasmid (red). Cardiac cells were visualized by α-sarcomeric actin (α-SA) staining (purple). Nuclei were stained with DAPI (blue). Hollow and filled arrows indicate membrane and ER localization, respectively. Scale bar = 20 μm.
The absence of channel surface expression is commonly associated with increased retention of misfolded channels in the endoplasmic reticulum (ER), followed by proteasome degradation through ubiquitination.19 To verify if this could be reproducible also in our model, cells were co-transfected with WT or mutant VSV fusion proteins tagged with green fluorescent protein (GFP) and with a pDsRed2-ER plasmid enabling red fluorescent labelling of the ER. After co-transfection with WT protein, exogenous KCNQ1 was detected at the membrane level in both CTR- and AS-iPSC-CMs, while part of the WT channels in S-iPSC-CM was sequestered at the ER level, suggesting the presence of an intrinsic trafficking defect in the S patient affecting also the transport of exogenous KCNQ1 (Figure 3B). Vice-versa, the mutant KCNQ1 protein was retained at the ER level in both CTR- and S-iPSC-CMs, while it was partly present at the membrane level in the AS-iPSC-CMs (Figure 3B).
To summarize, the Y111C mutation was associated with increased protein turnover and failure of trafficking of mutant KCNQ1 to the membrane in S-iPSC-CMs and CTR-iPSC-CMs, while in the AS-iPSC-CMs residual, although limited, trafficking of mutant KCNQ1 was observed. The presence of endogenous mutant KCNQ1 impaired also the trafficking of exogenous WT KCNQ1 protein exclusively in S-iPSC-CMs but not in the AS-iPSC-CMs. Taken together, these observations suggest the presence of a compensatory mechanism in the AS-iPSC-CMs that may correct, at least partially, the trafficking defect associated with KCNQ1.
3.3 Functional studies
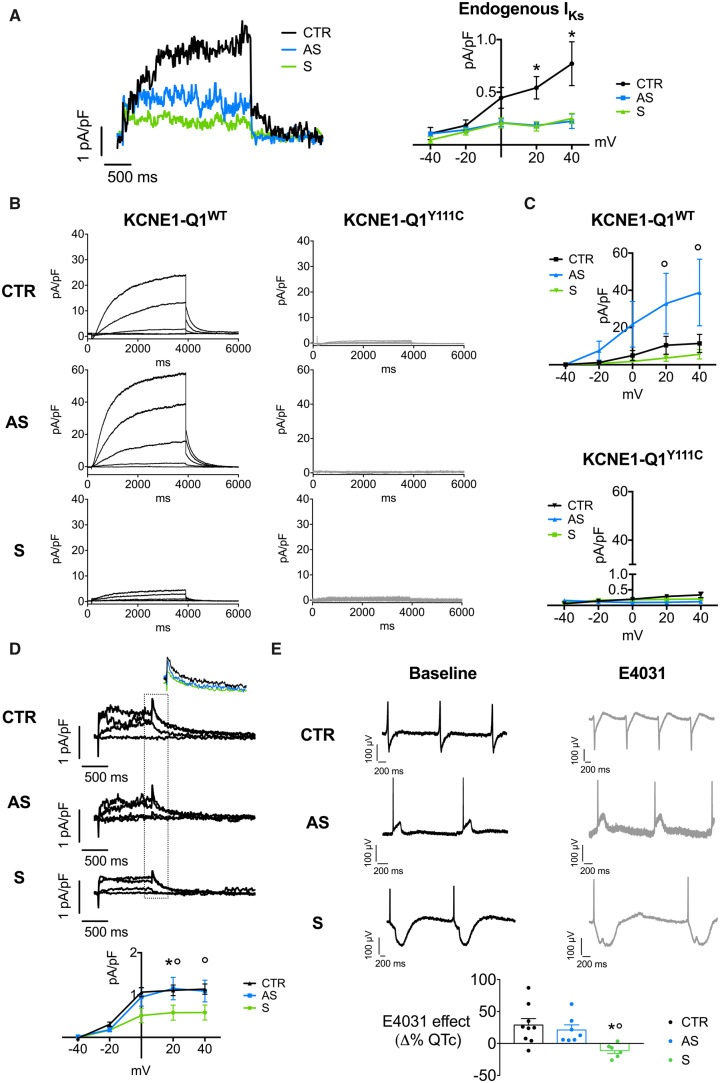
Next, we verified whether the molecular pattern observed led to functional differences by measuring IKs. Endogenous IKs was weak but detectable in CTR-iPSC-CMs, while it was almost undetectable in both S- and AS-iPSC-CMs (Figure 4A). To test the role of trafficking defects in mutant iPSC-CMs, IKs was measured also after transfection with WT or Y111C VSV-KCNE1-Q1-GFP fusion constructs. Surprisingly, transfection with the WT construct increased IKs density in AS-iPSC-CMs more than in CTR-iPSC-CMs, while a very limited increase was recorded in S-iPSC-CMs (Figure 4B and C). In contrast, transfection with the Y111C construct failed to restore (or to increase above the endogenous level) IKs in iPSC-CMs from all subjects. This indicates that, while the mutant channel was dysfunctional in all cells, trafficking of the WT channel was depressed in S-iPSC-CMs but even enhanced in AS-iPSC-CMs. Thus, in the heterozygous condition, AS patients might have a higher membrane density of WT KCNQ1 channels as compared with S ones, a factor that may contribute to their benign clinical phenotype.
Figure 4.
Electrophysiological characterization of LQT-iPSC-CMs. (A) Voltage-clamp recordings of endogenous IKs in CTR- and Y111C-iPSC-CMs. Representative traces (left) and average current–voltage (I–V) relationships (right) of endogenous IKs from CTR- (black), AS- (blue), S-iPSC-CMs (green). (B and C) Voltage-clamp recordings of heterologous IKs current in CTR- and LQT-iPSC-CMs transiently transfected with VSV-E1-Q1-GFP WT or Y111C. Representative patch-clamp recordings (B) and average I–V relationships (C) of heterologous IKs from CTR-, AS-, and S-iPSC-CMs transfected with VSV-E1-Q1-GFP WT or Y111C plasmids. (D) Voltage-clamp recordings of endogenous IKr. Representative IKr recordings (top panel) and average data (bottom panel) for CTR-, AS-, and S-iPSC-CMs. IKr tail currents have been magnified and overlaid in the inset. (E) Response to IKr blocking. Representative field potentials recorded with MEA in CTR-, AS-, and S-iPSC-CMs at baseline (top left, black) and after IKr block with E4031 (top right, grey). Average data (bottom panel) are expressed as delta percentage of the effect vs. the respective baseline (n = 14–19 for patch-clamp data; n = 6–9 for MEA data; *P < 0.05 CTR vs. S; °P < 0.05 AS vs. S).
An alternate possibility could be that the differences between S- and AS-iPSC-CMs might also reside in changes in membrane expression of ion channels other than KCNQ1 and consequently in changes in other currents contributing to the repolarization reserve. Accordingly, we determined whether there were differences in endogenous IKr, a repolarizing current functionally interacting with IKs with a compensatory role. Patch-clamp analysis showed that IKr density was similar between CTR- and AS-iPSC-CMs, but was significantly reduced in S-iPSC-CMs (Figure 4D). In addition, MEA demonstrated that the selective IKr blocker E4031 prolonged the QT interval in CTR- and AS-beating EBs but not in S-beating EBs (Figure 4E). The functional demonstration of reduced IKr was matched by the finding that the human ether-a-go-go-related gene (hERG) protein levels (Supplementary material online, Figure S3A) and membrane localization (Supplementary material online, Figure S3B) were lower in S-iPSC-CMs compared with CTR- and AS-iPSC-CMs. A lower IKr density implies a reduced repolarization reserve, which facilitates the unmasking of the abnormality caused by mutation-induced IKs deficit.
3.4 Proteasome activity
The demonstration of imbalanced expression of KCNQ1 and hERG in S-iPSC-CMs prompted us to explore the mechanism of their cellular turnover. Both KCNQ1 and hERG localization and turnover are physiologically regulated by Nedd4L,20,21 which is involved in protein tagging and ubiquitination, eventually promoting degradation via proteasome.22 Accordingly, we tested if this protein was involved in the cellular mechanism behind the different phenotypes displayed by the S- and AS-iPSC-CMs.
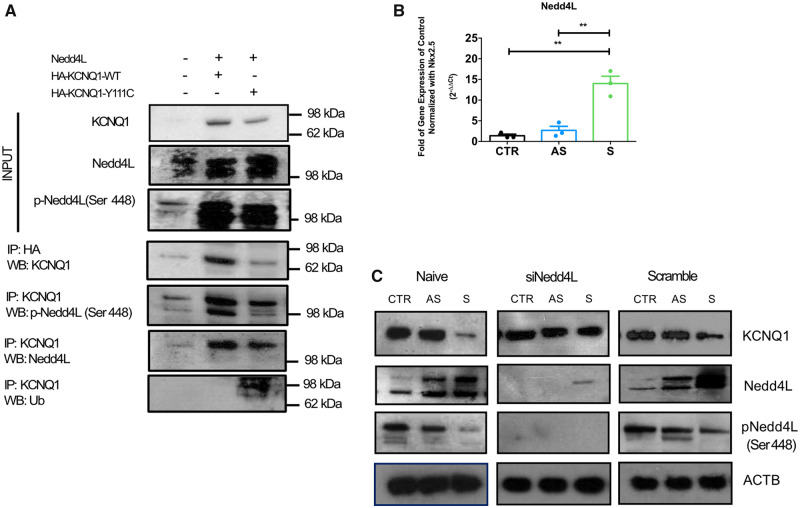
First, we verified whether the physiological Nedd4L-mediated KCNQ1 ubiquitination might be amplified in the presence of the Y111C mutation. Hence, we studied the interactions of Nedd4L with WT or Y111C KCNQ1 by immunoprecipitation in a 293T-based heterologous system. The concomitant over-expression of Nedd4L and WT KCNQ1 resulted in a normal degradation of KCNQ1. The presence of the Y111C mutation further magnified this effect (Figure 5A). We also detected higher levels of the enzymatically active form of Nedd4L (dephosphorylated at Ser448) bound to mutant KCNQ1 compared with WT KCNQ1, confirming that the Y111C mutation stimulates KCNQ1 degradation through the Nedd4L pathway.
Figure 5.
Quantification of Nedd4L in iPSC-CMs and interaction with KCNQ1. (A) KCNQ1–Nedd4L protein interaction. Nedd4L–KCNQ1 protein interactions were investigated in the 293T heterologous system by co-immunoprecipitation. 293T were co-transfected with pcDNA3.1-hNedd4L-WT and pCB6-HA-KCNQ1 expressing the WT or the Y111C KCNQ1. Forty-eight hours after transfection, whole-cell extracts were collected and subjected to immunoprecipitation and western blot with the indicated antibodies (Supplementary material online, Table S2). The upper row represents a western blot of the immunoprecipitated (IP) HA-KCNQ1 proteins and shows depleted level of HA-KCNQ1-Y111C in the presence of Nedd4L over-expression. The middle row represents the immunoblot of phosphorylated-Nedd4L (Ser448) proteins co-immunoprecipitated with HA-KCNQ1-WT or -Y111C and shows dephosphorylation of Nedd4L interacting with HA-KCNQ1-Y111C. The lower row confirms equivalent level of basal Nedd4L interacting either with HA-KCNQ1-WT or -Y111C. The activated Nedd4L in Y111C group ubiquitinated the mutated channel, KCNQ1-Y111C. (B) Nedd4L mRNA levels were quantified by RT-qPCR and ΔΔCt method. Nkx2.5 was used as reference gene (n = 3, **P < 0.005). (C) KCNQ1 and Nedd4L in CTR- and diseased iPSC-CMs. Evaluation of KCNQ1, Nedd4L, phospho-Nedd4L (p-Nedd4L) protein levels by western blot in naïve CTR-, AS-, and S-iPSC-CMs, after transfection with Nedd4L siRNA or after transfection with scramble siRNA.
Next, we investigated the correlation between Nedd4L activity and KCNQ1 in iPSC-CMs. First, we found a significant up-regulation of Nedd4L at both mRNA and protein levels in S-iPSC-CMs compared with AS- and CTR-iPSC-CMs (Figure 5B and C and Supplementary material online, Figure S4); the level of the inactive (phosphorylated) Nedd4L form was lower in S-iPSC-CMs (Figure 5C). Consequently, it is logical to assume that the active Nedd4L form was higher in S-iPSC-CMs compared with the other cells analysed. When matched with the reduced levels of KCNQ1 and hERG (Figure 5C and Supplementary material online, Figure S3A), these data suggested that the Y111C mutation triggered Nedd4L-mediated KCNQ1 and hERG degradation. On the other hand, even in the presence of the same ubiquitination-triggering KCNQ1 mutation, AS-iPSC-CMs failed to display Nedd4L up-regulation and KCNQ1 or hERG reduction (Figure 5C and Supplementary material online, Figure S3A); this suggests a differential upstream regulation of Nedd4L activation in AS- vs. S-iPSC-CMs. To confirm that differences in Nedd4L activation determine the molecular and functional differences observed in AS- and S-iPSC-CMs, Nedd4L expression was knocked-down by siRNA. As expected, silencing of Nedd4L in S-iPSC-CMs restored KCNQ1 protein levels (Figure 5C).
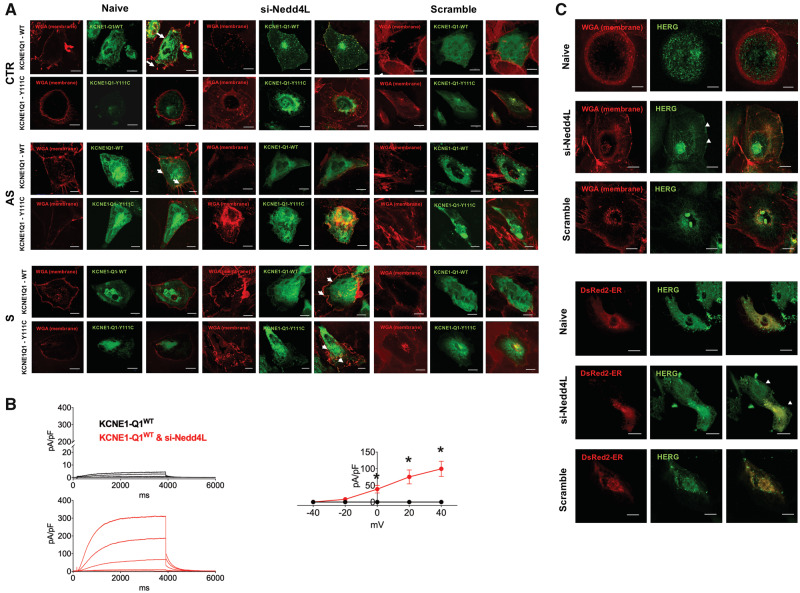
Importantly, silencing of Nedd4L restored trafficking of both WT and Y111C VSV-KCNE1-Q1 in S-iPSC-CMs (Figure 6A); moreover, it increased IKs density in S-iPSC-CMs transfected with WT VSV-KCNE1-Q1 (Figure 6B). Finally, immunocytochemistry analysis showed that silencing of Nedd4L restored the membrane expression of hERG in S-iPSC-CMs (Figure 6C).
Figure 6.
Regulation of KCNQ1 and hERG by Nedd4L in Y111C iPSC-CMs. (A) Nedd4L knockdown restores trafficking of KCNQ1-Y111C. CTR-, AS-, and S-iPSC-CMs were co-transfected with WT or mutated VSV-E1-Q1-GFP fusion protein, and with Nedd4L siRNA or scramble. To confirm the localization of KCNQ1, cell membrane was stained with wheat germ agglutinin (WGA). Arrows indicate the cell membrane. (B) Nedd4L knockdown rescued the IKs current in S-iPSC-CMs. Representative patch-clamp recordings (top panels) and average current–voltage (I–V) relationships (bottom panel) of S-iPSC-CMs transfected with the WT VSV-E1-Q1-GFP fusion protein alone (black) or in combination with the Nedd4L siRNA (red) (n = 11, 9, *P < 0.05 vs. KCNE1-Q1 WT). (C) Nedd4L knockdown restored hERG trafficking in S-iPSC-CMs. S-iPSC-CMs were transfected with scramble or Nedd4L siRNA. Expression of endogenous hERG was assessed by immunostaining (green). Surface localization was confirmed by wheat germ agglutinin (WGA)-Texas Red membrane co-staining (red). Arrows indicate the cell membrane.
3.5 MTMR4 variants
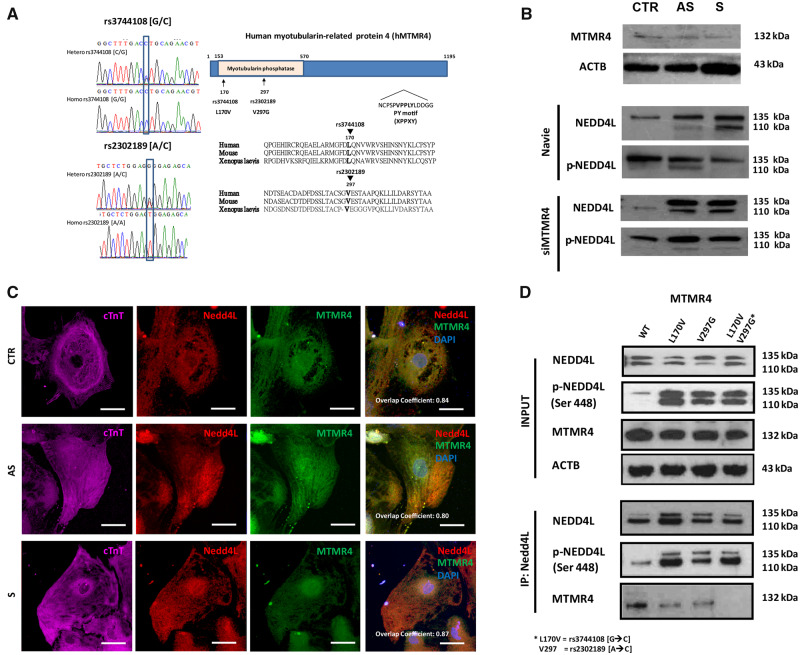
To discover putative modifier genes able to regulate the activity of Nedd4L, we performed whole-exome sequencing that led to the identification of 17 missense discordant SNVs between S and AS (Supplementary material online, Table S2). Among them, two variants at the MTMR4 locus potentially had functional relevance for the activity of Nedd4L. MTMR4 protein is an inositol phosphatase known to be an interactor of Nedd4L.23,24 The two SNVs identified were rs2302189 (c.890T>G p. Val297Gly) and rs3744108 (c.508C>G p. Leu170Val) with minor allele frequency of 0.3848 and 0.3774, respectively. These two SNVs lie on the conserved phosphatase region of MTMR4 ranging from the 153rd to 570th amino acids (Figure 7A). The AS patient and his two siblings, who are also AS carriers, carry the minor alleles of both MTMR4 SNVs in heterozygosis. Conversely, the S patient and his unaffected mother are homozygous for the major alleles (Supplementary material online, Figure S5). In our iPSC-CMs, the MTMR4 protein was expressed (Figure 7B) and, upon its inhibition with siRNA, we documented an increase in pNedd4L in S-iPSC-CMs (Figure 7B).
Figure 7.
Role of MTMR4. (A) Sequencing results. The analysis identified two interesting SNVs: rs2302189 (V297G), with either wild-type (A/A) or heterozygous variant (A/C) and rs2302189 (L170V), with either wild-type (G/G) or heterozygous variant (G/C). The illustration on the right illustrates the location of the two SNVs V297G and L170V on the conserved phosphatase region of the MTMR4 protein. (B) MTMR4 protein expression revealed by SDS-PAGE and immunoblot in CTR-, S-, and AS iPSC-CMs. Knock-down of MTMR4 by siRNA confirmed the role of MTMR4 on dephosphorylation of Nedd4L in iPSC-CM. (C) Co-localization of MTMR4 and Nedd4L in CTR and S cells. Confocal images of CTR-, AS-, and S-iPSC-CMs visualized with cTnT co-staining (purple). Nedd4L and MTMR4 were shown in red and green channel and the nuclei in blue (DAPI). Scale bar = 20 μm. (D) MTMR4–Nedd4L protein interaction investigated in 293T heterologous system. HEK cells transfected either with pCMV6-XL4-hMTMR4-WT, L170V (rs3744108 G to C single-nucleotide variant), V297G (rs2302189 A to C single-nucleotide variant), or double SNVs together with pcDNA3.1-hNedd4L. Forty-eight hours after transfection, whole-cell extracts were collected and subjected to immunoprecipitation and western blot with the indicated antibodies.
Next, we asked if the MTMR4 protein could dephosphorylate Nedd4L, and if MTMR4 dephosphorylation activity might be blunted by the presence of the SNVs. Accordingly, we studied the interactions of Nedd4L with WT or MTMR4 variants by immunoprecipitation in a 293T heterologous system. WT-MTMR4 co-immunoprecipitated with Nedd4L and reduced the levels of Nedd4L phosphorylation (Figure 7D). The two MTMR4 minor alleles V297G and L170V likely reduced MTMR4 dephosphorylation activity, with L170V showing the strongest effect. When tested in combination, the minor alleles reduced both MTMR4 activity and binding of MTMR4 to Nedd4L. These results confirm the presence of a link between MTMR4 and Nedd4L regulation.
3.6 Phenotype of the isogenic AS-MTMR4Cor-iPSC-CMs
To confirm that the presence of both minor alleles in AS-iPSC-CMs can reduce MTMR4 dephosphorylation activity, thus blunting the proteasomal degradation of KCNQ1 and hERG mediated by Nedd4L, we generated MTMR4 corrected AS-iPSCs (AS-MTMR4Cor-iPSCs), using CRISPR/Cas9 technology. The gene correction strategy is described in detail in the Supplementary material online, Result section and Figures S6 and S7.
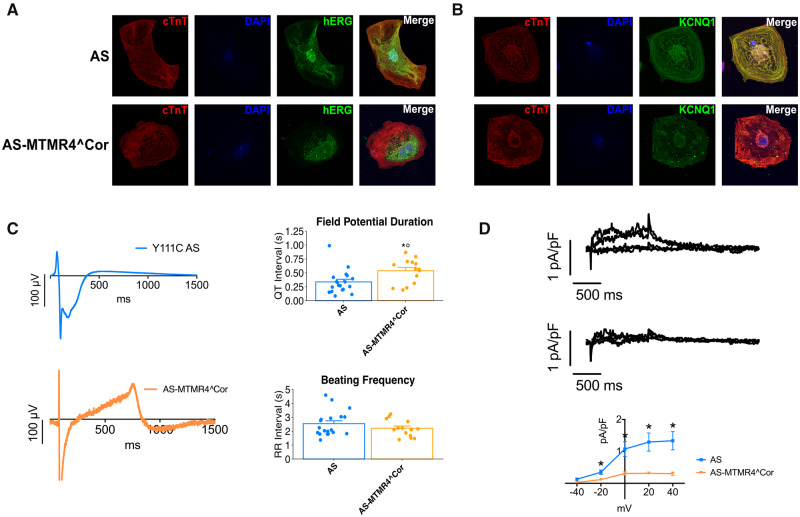
KCNQ1 mRNA levels were unchanged compared with AS-iPSC-CMs, but protein levels decreased suggesting an increase of KCNQ1 protein degradation in the corrected line (Supplementary material online, Figure S8). Immunostaining analysis showed trafficking defects of KCNQ1 similar to that observed in S cells (Figure 8A). Also, hERG protein was expressed at lower levels in the AS-MTMR4Cor-iPSC-CMs compared with AS-CMs (Supplementary material online, Figure S8) and not properly localized at plasma membrane (Figure 8B).
Figure 8.
Isogenic AS-MTMR4Cor-iPSC-CMs. (A) Immunostaining of the endogenous KCNQ1 in AS- and AS-MTMR4Cor-iPSC-CM. KCNQ1 was stained in green, cTnT in red, and nuclei in blue. Scale bar 20 µm. (B) Immunostaining for the hERG channel in AS- and AS-MTMR4Cor-iPSC-CM. hERG was stained in green, cTnT in red, and nuclei in blue. Scale bar =20 µm. (C) Representative field potential recordings of AS- and AS-MTMR4Cor groups. Average values for QT- and RR interval durations recorded in spontaneously beating EBs from AS and AS-MTMR4Cor groups. *P < 0.05 vs. AS (D). Representative IKr recordings and average data from AS and AS-MTMR4Cor groups. *P < 0.05 vs. AS.
Gene editing did not affect MTMR4 expression levels but altered only its activity. MTMR4 protein was similarly expressed in AS and AS-MTMR4Cor-iPSC-CMs (Supplementary material online, Figure S9) but MTMR4 activity and interaction with Nedd4L were resumed upon correction of the two SNVs, as indicated by the increased dephosphorylation of Nedd4L and co-localization with Nedd4L in AS-MTMR4Cor-iPSC-CMs (Supplementary material online, Figure S9).
These molecular changes were also visible on the functional phenotype of AS-MTMR4Cor-iPSC-CMs. Specifically, the correction of the two MTMR4 SNVs induced a prolongation of the QT interval recorded with the MEA to levels comparable with those recorded in S cells, suggesting that the protective role of the MTMR4 SNVs in the presence of Y111C mutation is lost when the two SNVs are corrected (Figure 8C). Moreover, patch-clamp analysis showed that IKr density, previously normal in AS-iPSC-CMs, was significantly reduced in AS-MTMR4Cor-iPSC-CMs (Figure 8D), further confirming the protective role of these SNVs.
3.7 Presence of MTMR4 SNVs in additional Y111C carriers
Following our cell study, we identified 13 additional AS Y111C carriers belonging to different families and documented that 10 out of 13 (77%) were also carriers of the MTMR4 rs2302189 (V297G) and rs3744108 (L170V) variants in heterozygosis. Interestingly, the average QT interval of the 10 patients carrying the MTMR4 variants was shorter (464 ± 20 ms) compared with that of the three non-carriers (502 ± 35 ms) P = 0.03.
4. Discussion
Our findings represent the first biological evidence of a genetic mechanism that, by specifically interfering with the effect of an LQTS-causing mutation, provides antiarrhythmic protection. By comparing patient-specific iPSC-CMs derived from symptomatic and AS LQT1 patients carrying the same mutation, we demonstrated that the clinical penetrance of the disease is determined by differences in trafficking and degradation of the two potassium channels (IKs and IKr) encoded by KCNQ1 and hERG. We also showed that the activation status of Nedd4L accounts for these biological differences. More specifically, genetic variants of MTMR4, an upstream regulator of Nedd4L, partly correct the defect in potassium channel turnover, thus mitigating the clinical manifestations of the disease in the AS patient. The present study suggests a mechanism for protective action of common genetic variants and, in addition to the recent demonstration of a worsening effect,25 contributes to the concept of the important role of iPSC in the identification of new ‘modifier genes’.1 It is likely that this or similar mechanisms may be relevant to other LQTS-causing mutations and possibly to other arrhythmogenic disorders related to channel trafficking.
Some LQT1 mutations are associated with ion channel trafficking defects,26 and Y111C is among these.11,12,27 Trafficking defects usually lead to malignant phenotype, but most Y111C mutation carriers present a low incidence of life-threatening cardiac events.9,10 Using patient-derived iPSC-CMs, we demonstrated that the Y111C mutation triggers an increased activity of Nedd4L that amplifies the proteasome degradation of both WT and mutant KCNQ1 protein. In S-iPSC-CMs, increased Nedd4L activity also enhances the degradation of the hERG protein, likely by targeting the same PY-motif expressed by KCNQ1.20,28 The loss of compensatory IKr due to the ‘bystander’ degradation of hERG channel by Nedd4L plays an important role in the pathophysiology of LQT1 in S-iPSC-CMs (Figure 9).
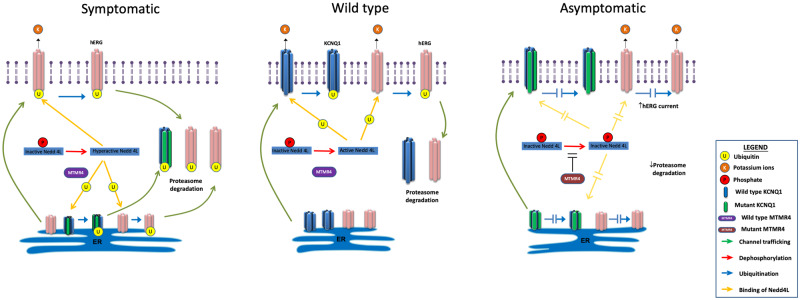
Figure 9.
Schematic illustration of the cellular mechanisms regulated by MTMR4. Cardiomyocytes of normal subject (middle panel): KCNQ1 and hERG are ion channels conducting the IKs and IKr currents, respectively. They are synthetized in the endoplasmic reticulum (ER) from where they traffick to the plasma membrane. Their steady-state expression level at cell surface is physiologically regulated by the ubiquitin ligase Nedd4L. The active form of Nedd4L ubiquitinates ‘old’ KCNQ1 and hERG channels which can then be degraded via the proteasome complex. We have proved that MTMR4 interacts with Nedd4L in iPSC-CMs. Cardiomyocytes of symptomatic patient (left panel): Mutant KCNQ1-Y111C channels are recognized as defective and retained in the ER. This triggers an increased activation of Nedd4L that amplifies the proteasome degradation not only of mutant KCNQ1, but also of WT hERG. Both IKs and IKr are reduced, and repolarization reserve compromised. LQT1 phenotype is manifested. Cardiomyocytes of asymptomatic patient (right panel): SNVs on MTMR4 inhibit Nedd4L activation. The total amount of KCNQ1 and hERG on the plasma membrane is increased compared with cardiomyocytes of the symptomatic patient. IKs deficit is partly compensated by higher IKr expression. LQT1 phenotype is mitigated.
Our observations in the AS-iPSC-CMs provide insights into the mechanisms for the incomplete penetrance and relative benign clinical course in LQT1 patients carrying the Y111C mutation. In AS-iPSC-CMs, the mutant KCNQ1 apparently failed to trigger Nedd4L activation. The lack of QT prolongation in AS-iPSC-CMs might result from enhanced trafficking of residual WT KCNQ1 (heterozygosity) and/or of hERG proteins, the latter functionally concealing IKs deficit by robust IKr expression.29 Because of mutations of channels post-transcriptionally assembled as multimers (as KCNQ1) are characterized by strong negative-dominance,30 the latter mechanism is likely to prevail.
Overall, the primary factor limiting the phenotype severity may be represented by blunted Nedd4L response to the mutant protein. By using whole-exome sequencing, we identified in AS-iPSC-CMs the presence of common SNVs in MTMR4 affecting its phosphatase function on Nedd4L that consequently remains inactive.22,23 This reduction of Nedd4L active form resulted in less efficient proteasome degradation not only of both mutant and WT KCNQ1, but also of hERG, with the consequent shortening of QTc (Figure 9). The results observed in iPSC-CMs derived from CRISPR/Cas9 corrected AS-iPSC seem to confirm this interpretation.
Post-translational modification by ubiquitination mediated via Nedd4L is crucial in the targeted recycling, degradation or stabilization of many ion channels, including KCNQ1 and hERG; and loss of function of Nedd4L is associated with a broad range of diseases pathologies.31 On the other hand, the common variants of MTMR4 identified in this study are highly prevalent in the general population and only modified the Nedd4L activity during hyperactivation. Our findings suggest that these MTMR4 variants may account for the incomplete penetrance and relative benign clinical course in LQT1 due to KCNQ1-Y111C mutation. Indeed, genotyping of other 13 AS Y111C mutation carriers showed that 77% of them carried the MTMR4 protective variants.
In conclusion, our study strongly supports the use of iPSCs as a system to model LQTS and other channelopathies17,32–35 suggesting that this technology has the potential of fine-tuning risk stratification and of guiding the design of new drugs for the development of personalized therapies aimed at correcting the consequences of disease-causing mutations.36,37 The conceptual implications of our finding are clear. The practical relevance, however, would dramatically increase if the protective impact of these SNVs would affect not only the carriers of the Y111C mutation but also the much larger group of patients with different mutations causing LQT1 and LQT2. Accordingly, we are currently testing this possibility.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Supplementary Material
Acknowledgements
The authors are grateful to Pinuccia De Tomasi, BS, for expert editorial support.
Conflict of interest: none declared.
Funding
This work was supported by the Italian Ministry of Education, University and Research (MIUR) (PRIN 2010BWY8E9 to M.G., P.J.S., and L.C.), and a grant to the Department of Molecular Medicine of the University of Pavia under the initiative ‘Dipartimenti di Eccellenza’ (2018–2022); Italian Ministry of Health (GR-2010-2305717 to M.G. and L.C.); Leducq Foundation for Cardiovascular Research (18CVD05) ‘Towards Precision Medicine with Human iPSCs for Cardiac Channelopathies’ (to M.G., P.J.S., L.C., L.S., and M.M.); Hong Kong Research Grant Council: Theme-Based Research Scheme T12-705/11 (to H.F.T.). This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 795209 (to L.S.).
Time for primary review: 7 days
Translational perspective
KCNQ1-Y111C penetrance in the asymptomatic patient is incomplete for the presence of common variants on the MTMR4 gene, which attenuates Nedd4L-mediated proteasome degradation not only of wild type and mutant KCNQ1 but also of hERG channels, thus restoring repolarization reserve and maintaining a normal QTc. This is the first evidence of a protective modifier gene documented biologically in the context of inherited arrhythmogenic diseases and represents a proof-of-concept for the possibility of identifying the mechanisms of action of protective ‘modifier genes’ using patient-specific iPSCs. Our finding paves the way to the development of novel approaches to stratify the risk in LQTS patients and to implement personalized therapies.
References
- 1. Schwartz PJ, Crotti L, George AL Jr.. Modifier genes for sudden cardiac death. Eur Heart J 2018;39:3925–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schwartz PJ, Ackerman MJ, George AL Jr, Wilde AA.. Impact of genetics on the clinical management of channelopathies. J Am Coll Cardiol 2013;62:169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schwartz PJ, Stramba-Badiale M, Crotti L, Pedrazzini M, Besana A, Bosi G, Gabbarini F, Goulene K, Insolia R, Mannarino S, Mosca F, Nespoli L, Rimini A, Rosati E, Salice P, Spazzolini C.. Prevalence of the congenital long-QT syndrome. Circulation 2009;120:1761–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schwartz PJ, Ackerman MJ.. The long QT syndrome: a transatlantic clinical approach to diagnosis and therapy. Eur Heart J 2013;34:3109–3116. [DOI] [PubMed] [Google Scholar]
- 5. Schwartz PJ, Priori SG, Spazzolini C, Moss AJ, Vincent GM, Napolitano C, Denjoy I, Guicheney P, Breithardt G, Keating MT, Towbin JA, Beggs AH, Brink P, Wilde AA, Toivonen L, Zareba W, Robinson JL, Timothy KW, Corfield V, Wattanasirichaigoon D, Corbett C, Haverkamp W, Schulze-Bahr E, Lehmann MH, Schwartz K, Coumel P, Bloise R.. Genotype-phenotype correlation in the long-QT syndrome: gene-specific triggers for life-threatening arrhythmias. Circulation 2001;103:89–95. [DOI] [PubMed] [Google Scholar]
- 6. Schwartz PJ, Crotti L.. Long and short QT syndromes In Zipes DP, Lalife J (eds). Cardiac Electrophysiology: From Cell to Bedside. 6th ed.Philadelphia: Elsevier/Saunders; 2014. pp. 935–946. [Google Scholar]
- 7. Gnecchi M, Schwartz PJ.. The unstoppable attraction for induced pluripotent stem cells: are they the magic bullet for modeling inherited arrhythmogenic diseases? J Am Coll Cardiol 2012;60:1001–1004. [DOI] [PubMed] [Google Scholar]
- 8. Gnecchi M, Stefanello M, Mura M.. Induced pluripotent stem cell technology: toward the future of cardiac arrhythmias. Int J Cardiol 2017;237:49–52. [DOI] [PubMed] [Google Scholar]
- 9. Winbo A, Diamant UB, Rydberg A, Persson J, Jensen SM, Stattin EL.. Origin of the Swedish long QT syndrome Y111C/KCNQ1 founder mutation. Heart Rhythm 2011;8:541–547. [DOI] [PubMed] [Google Scholar]
- 10. Winbo A, Diamant UB, Stattin EL, Jensen SM, Rydberg A.. Low incidence of sudden cardiac death in a Swedish Y111C type 1 long-QT syndrome population. Circ Cardiovasc Genet 2009;2:558–564. [DOI] [PubMed] [Google Scholar]
- 11. Dahimene S, Alcolea S, Naud P, Jourdon P, Escande D, Brasseur R, Thomas A, Baro I, Merot J.. The N-terminal juxtamembranous domain of KCNQ1 is critical for channel surface expression: implications in the Romano-Ward LQT1 syndrome. Circ Res 2006;99:1076–1083. [DOI] [PubMed] [Google Scholar]
- 12. Peroz D, Dahimene S, Baro I, Loussouarn G, Merot J.. LQT1-associated mutations increase KCNQ1 proteasomal degradation independently of Derlin-1. J Biol Chem 2009;284:5250–5256. [DOI] [PubMed] [Google Scholar]
- 13. Lai WH, Ho JC, Lee YK, Ng KM, Au KW, Chan YC, Lau CP, Tse HF, Siu CW.. ROCK inhibition facilitates the generation of human-induced pluripotent stem cells in a defined, feeder-, and serum-free system. Cell Reprogram 2010;12:641–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, Palecek SP.. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat Protoc 2013;8:162–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Doss MX, Di Diego JM, Goodrow RJ, Wu Y, Cordeiro JM, Nesterenko VV, Barajas-Martinez H, Hu D, Urrutia J, Desai M, Treat JA, Sachinidis A, Antzelevitch C.. Maximum diastolic potential of human induced pluripotent stem cell-derived cardiomyocytes depends critically on IKr. PLoS One 2012;7:e40288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Meijer van Putten RM, Mengarelli I, Guan K, Zegers JG, van Ginneken AC, Verkerk AO, Wilders R.. Ion channelopathies in human induced pluripotent stem cell derived cardiomyocytes: a dynamic clamp study with virtual IK1. Front Physiol 2015;6:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rocchetti M, Sala L, Dreizehnter L, Crotti L, Sinnecker D, Mura M, Pane LS, Altomare C, Torre E, Mostacciuolo G, Severi S, Porta A, De Ferrari GM, George AL Jr, Schwartz PJ, Gnecchi M, Moretti A, Zaza A.. Elucidating arrhythmogenic mechanisms of long-QT syndrome CALM1-F142L mutation in patient-specific induced pluripotent stem cell-derived cardiomyocytes. Cardiovasc Res 2017;113:531–541. [DOI] [PubMed] [Google Scholar]
- 18. Tse HF, Ho JC, Choi SW, Lee YK, Butler AW, Ng KM, Siu CW, Simpson MA, Lai WH, Chan YC, Au KW, Zhang J, Lay KW, Esteban MA, Nicholls JM, Colman A, Sham PC.. Patient-specific induced-pluripotent stem cells-derived cardiomyocytes recapitulate the pathogenic phenotypes of dilated cardiomyopathy due to a novel DES mutation identified by whole exome sequencing. Hum Mol Genet 2013;22:1395–1403. [DOI] [PubMed] [Google Scholar]
- 19. Andersen MN, Krzystanek K, Petersen F, Bomholtz SH, Olesen SP, Abriel H, Jespersen T, Rasmussen HB.. A phosphoinositide 3-kinase (PI3K)-serum- and glucocorticoid-inducible kinase 1 (SGK1) pathway promotes Kv7.1 channel surface expression by inhibiting Nedd4-2 protein. J Biol Chem 2013;288:36841–36854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Albesa M, Grilo LS, Gavillet B, Abriel H.. Nedd4-2-dependent ubiquitylation and regulation of the cardiac potassium channel hERG1. J Mol Cell Cardiol 2011;51:90–98. [DOI] [PubMed] [Google Scholar]
- 21. Jespersen T, Membrez M, Nicolas CS, Pitard B, Staub O, Olesen SP, Baro I, Abriel H.. The KCNQ1 potassium channel is down-regulated by ubiquitylating enzymes of the Nedd4/Nedd4-like family. Cardiovasc Res 2007;74:64–74. [DOI] [PubMed] [Google Scholar]
- 22. Boase NA, Kumar S.. NEDD4: the founding member of a family of ubiquitin-protein ligases. Gene 2015;557:113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Laporte J, Bedez F, Bolino A, Mandel JL.. Myotubularins, a large disease-associated family of cooperating catalytically active and inactive phosphoinositides phosphatases. Hum Mol Genet 2003;12:R285–R292. [DOI] [PubMed] [Google Scholar]
- 24. Plant PJ, Correa J, Goldenberg N, Bain J, Batt J.. The inositol phosphatase MTMR4 is a novel target of the ubiquitin ligase Nedd4. Biochem J 2009;419:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chai S, Wan X, Ramirez-Navarro A, Tesar PJ, Kaufman ES, Ficker E, George AL, Deschênes I.. Physiological genomics identifies genetic modifiers of long QT syndrome type 2 severity. J Clin Invest 2018;128:1043–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wilson AJ, Quinn KV, Graves FM, Bitner-Glindzicz M, Tinker A.. Abnormal KCNQ1 trafficking influences disease pathogenesis in hereditary long QT syndromes (LQT1). Cardiovasc Res 2005;67:476–486. [DOI] [PubMed] [Google Scholar]
- 27. Seebohm G, Strutz-Seebohm N, Ureche ON, Henrion U, Baltaev R, Mack AF, Korniychuk G, Steinke K, Tapken D, Pfeufer A, Kääb S, Bucci C, Attali B, Merot J, Tavare JM, Hoppe UC, Sanguinetti MC, Lang F.. Long QT syndrome-associated mutations in KCNQ1 and KCNE1 subunits disrupt normal endosomal recycling of IKs channels. Circ Res 2008;103:1451–1457. [DOI] [PubMed] [Google Scholar]
- 28. Bongiorno D, Schuetz F, Poronnik P, Adams DJ.. Regulation of voltage-gated ion channels in excitable cells by the ubiquitin ligases Nedd4 and Nedd4-2. Channels (Austin) 2011;5:79–88. [DOI] [PubMed] [Google Scholar]
- 29. Varro A, Balati B, Iost N, Takacs J, Virag L, Lathrop DA, Csaba L, Talosi L, Papp JG.. The role of the delayed rectifier component IKs in dog ventricular muscle and Purkinje fibre repolarization. J Physiol 2000;523:67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Demolombe S, Baro I, Pereon Y, Bliek J, Mohammad-Panah R, Pollard H, Morid S, Mannens M, Wilde A, Barhanin J, Charpentier F, Escande D.. A dominant negative isoform of the long QT syndrome 1 gene product. J Biol Chem 1998;273:6837–6843. [DOI] [PubMed] [Google Scholar]
- 31. Manning JA, Kumar S.. Physiological functions of Nedd4-2: Lessons from knockout mouse models. Trends Biochem Sci 2018;43:635–647. [DOI] [PubMed] [Google Scholar]
- 32. Sala L, Gnecchi M, Schwartz PJ.. Long QT syndrome modelling with cardiomyocytes derived from human-induced pluripotent stem cells. Arrhythm Electrophysiol Rev 2019;8:105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu JC, Garg P, Yoshida Y, Yamanaka S, Gepstein L, Hulot JS, Knollmann BC, Schwartz PJ.. Towards precision medicine with human iPSCs for cardiac channelopathies. Circ Res 2019;125:653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mura M, Mehta A, Ramachandra CJ, Zappatore R, Pisano F, Ciuffreda MC, Barbaccia V, Crotti L, Schwartz PJ, Shim W, Gnecchi M.. The KCNH2-IVS9-28A/G mutation causes aberrant isoform expression and hERG trafficking defect in cardiomyocytes derived from patients affected by Long QT Syndrome type 2. Int J Cardiol 2017;240:367–371. [DOI] [PubMed] [Google Scholar]
- 35. Musunuru K, Sheikh F, Gupta RM, Houser SR, Maher KO, Milan DJ, Terzic A, Wu JC; American Heart Association Council on Functional Genomics and Translational Biology; Council on Cardiovascular Disease in the Young; and Council on Cardiovascular and Stroke Nursing. Induced pluripotent stem cells for cardiovascular disease modeling and precision medicine: a scientific statement from the American Heart Association. Circ Genom Precis Med 2018;11:e000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mehta A, Ramachandra CJA, Singh P, Chitre A, Lua CH, Mura M, Crotti L, Wong P, Schwartz PJ, Gnecchi M, Shim W.. Identification of a targeted and testable antiarrhythmic therapy for long-QT syndrome type 2 using a patient-specific cellular model. Eur Heart J 2018;39:1446–1455. [DOI] [PubMed] [Google Scholar]
- 37. Schwartz PJ, Gnecchi M, Dagradi F, Castelletti S, Parati G, Spazzolini C, Sala L, Crotti L.. From patient-specific induced pluripotent stem cells to clinical translation in long QT syndrome Type 2. Eur Heart J 2019;40:1832–1836. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.