Abstract
Many pathogens are capable of disrupting autophagy within host cells. In this issue of Developmental Cell, Miao et al. discover that the SARS-CoV-2 protein ORF3a inhibits autophagosome-lysosome fusion by dysregulating the HOPS complex.
Many pathogens are capable of disrupting autophagy within host cells. In this issue of Developmental Cell, Miao et al. discover that the SARS-CoV-2 protein ORF3a inhibits autophagosome-lysosome fusion by dysregulating the HOPS complex.
Main text
As the world continues to wrestle with the coronavirus disease 2019 (COVID-19) pandemic, caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), research on the virus presses on, with the hope of developing therapies and informing public health policy. Clarifying the interplay between SARS-CoV-2 and the cell it has infected can contribute to our understanding of COVID-19 pathogenesis.
Autophagy is one of the major defense mechanisms a cell employs against pathogens. It is the cell’s bulk degradation pathway by which material that is large in either size or quantity gets engulfed by the autophagosome and delivered to the lysosomal lumen after the autophagosome fuses with lysosomes. When marked by autophagy adaptors, pathogens can be eliminated in a similar manner. However, many pathogens have evolved ways to evade autophagy or even turn the pathway to their own advantage (Levine et al., 2011).
Coronaviruses are known to interact with the autophagy pathway (Carmona-Gutierrez et al., 2020; Delorme-Axford and Klionsky, 2020; Miller et al., 2020). Although core autophagy genes don’t seem to be required for coronavirus infection (Zhao et al., 2007; Schneider et al., 2021; Hoffmann et al., 2020), the nonlipidated form of LC3 (LC3-I) appears to be important for viral replication in the case of mouse hepatitis virus infection (Reggiori et al., 2010). LC3 is one of the proteins marking the membranes of autophagosomes, which viral replication complexes strongly resemble; both are derived from the ER, and both are double-membrane vesicles. The formation of these two vesicular structures may share the same mechanism, as suggested by recent findings of autophagy-essential proteins on the ER, TMEM41B and VMP1, shown to serve as host factors for SARS-CoV-2 and other coronaviruses (Schneider et al., 2021; Hoffmann et al., 2020). In this issue of Developmental Cell, Miao et al. reveal another link between SARS-CoV-2 infection and autophagy: SARS-CoV-2 can prevent autophagy progression by hindering autophagosome-lysosome fusion (Miao et al., 2020).
The authors began the study by expressing SARS-CoV-2 proteins, one by one, in human HeLa cells and evaluating the effect of this expression on autophagy activity. They found that the expression of ORF3a, ORF7a, M, or NSP6 resulted in the accumulation of structures positive for autophagosome markers. Such structures also accumulated in SARS-CoV-2-infected cells. As ORF3a expression displayed the strongest effect, the authors chose to focus on this protein.
To understand how ORF3a, a multi-spanning membrane protein, was interfering with autophagy, the authors started by identifying the accumulated structures. After a series of experiments, the structures were found to be closed autophagosomes and amphisomes (the latter referring to autophagosomes that have fused with late endosomes) that were positive for the autophagosomal SNARE STX17 and yet devoid of lysosomal markers. This finding indicated that ORF3a suppresses autophagosome-lysosome fusion.
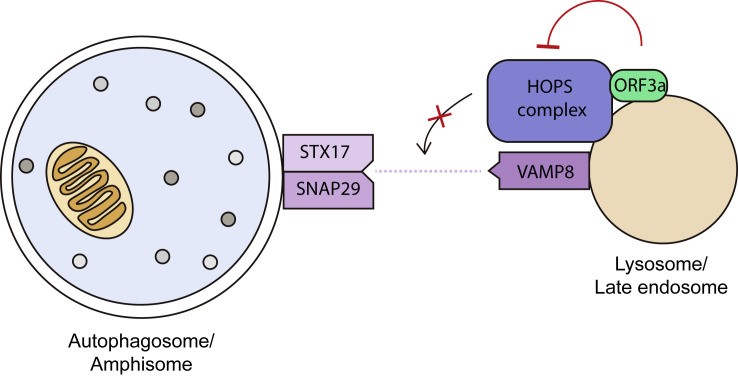
Miao et al. then systematically tested the interactions between ORF3a and the collection of tethering factors and SNARE complexes coordinating autophagosome-lysosome fusion. They found that ORF3a consistently displayed a strong interaction with VPS39, a component of the HOPS complex, one of the tethering factors essential for autophagosome-lysosome fusion (Zhao and Zhang, 2019). ORF3a sequesters the HOPS complex (or part of the complex) to ORF3a-positive endosomes and lysosomes (Figure 1 ). Furthermore, the binding of ORF3a to VPS39 was shown to negatively impact HOPS complex assembly and even the formation of the STX17-SNAP29-VAMP8 SNARE complex that is essential for autophagosome-lysosome fusion. This ability to disrupt the fusion step of autophagy is unique to the ORF3a of SARS-CoV-2, as the highly similar ORF3a of SARS-CoV was found to be unable to interact with the HOPS complex and had no effect on autophagy. This difference in ORF3a function should be taken into account when explaining the difference in pathogenicity and infectivity of these two genetically similar viruses.
Figure 1.
ORF3a of SARS-CoV-2 blocks fusion between autophagosomes/amphisomes and endolysosomes
SARS-CoV-2’s ORF3a localizes to late endosomes and lysosomes, where it binds to VPS39 of the HOPS complex. The resulting HOPS complex is unable to mediate STX17-SNAP29-VAMP8 SNARE complex formation. As this SNARE complex mediates autophagosome-lysosome fusion, autophagosomes or amphisomes (autophagosomes that have fused with late endosomes) are unable to mature to autolysosomes in cells with ORF3a.
It is intriguing that the ORF3a-bound HOPS complex cannot mediate autophagosome-lysosome fusion, despite being in the right place (on late endosomes and lysosomes) at the right time (after autophagosomes have formed). ORF3a might disrupt the arrangement of proteins that make up the HOPS complex. Although Miao et al. did not characterize the other three SARS-CoV-2 proteins (ORF7a, M, and NSP6) found to inhibit autophagy, it appears likely that they, too, work toward preventing autophagosome-lysosome fusion, suggesting that doing so is important to SARS-CoV-2’s replication. However, what remains unclear is whether SARS-CoV-2 proteins are actively targeted for degradation by autophagy. If they are, blocking fusion would allow SARS-CoV-2 to evade lysosomal degradation and avoid degradation products from being used in antigen presentation to T cells (Levine et al., 2011). If not, the production of autophagosomes might somehow be beneficial to viral replication. Further studies of SARS-CoV-2 activity in autophagy-deficient cells or the activity of SARS-CoV-2 deprived of ORF3a may provide answers that contribute to our understanding of the virus’s replication cycle.
Acknowledgments
Declaration of interests
The authors declare no competing interests.
References
- Carmona-Gutierrez D., Bauer M.A., Zimmermann A., Kainz K., Hofer S.J., Kroemer G., Madeo F. Digesting the crisis: autophagy and coronaviruses. Microb. Cell. 2020;7:119–128. doi: 10.15698/mic2020.05.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme-Axford E., Klionsky D.J. Highlights in the fight against COVID-19: does autophagy play a role in SARS-CoV-2 infection? Autophagy. 2020;16:2123–2127. doi: 10.1080/15548627.2020.1844940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann H.H., Schneider W.M., Rozen-Gagnon K., Miles L.A., Schuster F., Razooky B., Jacobson E., Wu X., Yi S., Rudin C.M., et al. TMEM41B is a pan-flavivirus host factor. Cell. 2020;184:133–148. doi: 10.1016/j.cell.2020.12.005. Published online December 9, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Mizushima N., Virgin H.W. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao G., Zhao H., Li Y., Ji M., Chen Y., Shi Y., Bi Y., Wang P., Zhang H. ORF3a of the COVID-19 virus SARS-CoV-2 blocks HOPS complex-mediated assembly of the SNARE complex required for autolysosome formation. Dev. Cell. 2020;56 doi: 10.1016/j.devcel.2020.12.010. Published online December 16, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K., McGrath M.E., Hu Z., Ariannejad S., Weston S., Frieman M., Jackson W.T. Coronavirus interactions with the cellular autophagy machinery. Autophagy. 2020;16:2131–2139. doi: 10.1080/15548627.2020.1817280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Monastyrska I., Verheije M.H., Calì T., Ulasli M., Bianchi S., Bernasconi R., de Haan C.A.M., Molinari M. Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7:500–508. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W.M., Luna J.M., Hoffmann H.H., Sánchez-Rivera F.J., Leal A.A., Ashbrook A.W., Le Pen J., Ricardo-Lax I., Michailidis E., Peace A., et al. Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks. Cell. 2021;184:120–132. doi: 10.1016/j.cell.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.G., Zhang H. Autophagosome maturation: an epic journey from the ER to lysosomes. J. Cell Biol. 2019;218:757–770. doi: 10.1083/jcb.201810099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Thackray L.B., Miller B.C., Lynn T.M., Becker M.M., Ward E., Mizushima N.N., Denison M.R., Virgin H.W., 4th Coronavirus replication does not require the autophagy gene ATG5. Autophagy. 2007;3:581–585. doi: 10.4161/auto.4782. [DOI] [PubMed] [Google Scholar]