Abstract
This paper proposes a mixture of linear dynamical systems model for quantifying the heterogeneous progress of Parkinson’s disease from DaTscan Images. The model is fitted to longitudinal DaTscans from the Parkinson’s Progression Marker Initiative. Fitting is accomplished using robust Bayesian inference with collapsed Gibbs sampling. Bayesian inference reveals three image-based progression subtypes which differ in progression speeds as well as progression trajectories. The model reveals characteristic spatial progression patterns in the brain, each pattern associated with a time constant. These patterns can serve as disease progression markers. The subtypes also have different progression rates of clinical symptoms measured by MDS-UPDRS Part III scores.
Keywords: Parkinson’s disease, disease progression model, DaTscans, linear dynamical system, centrosymmetric matrix, t-distribution
I. Introduction
Parkinson’s disease (PD) is a neurodegenerative disease characterized by the loss of dopaminergic neurons in the substantia nigra. Different individuals with PD progress along different disease trajectories. This variability is called progression heterogeneity, or simply, heterogeneity. Heterogeneity is understood in terms of progression subtypes, each subtype being a prototypical progression trajectory.
Another characteristic of PD progression is that it exhibits specific spatial patterns in the brain. These patterns, called Braak stages [1], have mostly been analyzed by histology of deceased PD patient brains. Spatial progression patterns in living PD patients have not yet been reported using DaTscans.
The goal of this paper is to propose a mathematical model and a Bayesian analysis method to (i) quantify PD heterogeneity by identifying progression subtypes, and (ii) to identify spatial progression patterns and their time constants in living PD patients using longitudinal analysis of DaTscan images, or DaTscans. DaTscans are the commercial name for SPECT imaging with 123I-FP-CIT. DaTscans measure the local presynaptic dopamine transporter (DAT) density. DAT density decreases as dopaminergic neurons are lost in PD, and this manifests as signal loss in DaTscan images [2]. In early-stage PD, signal loss is most significant in the striatum [1], hence we study the dynamics of PD progression using the striatal binding ratio (SBR) [3] in the caudates and putamina. The SBR at voxel v is defined as SBRv = (Iv − μ)/μ where Iv is the intensity in voxel v and μ is the mean (or median) intensity in a reference region, such as the occipital lobe, that does not have specific ligand binding. The SBR normalizes for radioligand dose as well as compensates for the amount of nonspecific radioligand binding.
The model we propose is a mixture of linear dynamical systems (MLDS). In this model, PD subjects are assigned to different progression subtypes, where each subtype is defined by a multivariate linear dynamical system (LDS). The eigenvectors of the transition matrix of the dynamical system give spatial progression patterns of DAT loss in the brain. The corresponding eigenvalues give time constants of disease progression along these patterns. The data used to fit the model comes from the Parkinson’s Progression Marker Initiative (PPMI) (https://www.ppmi-info.org/).
This paper introduces several novel techniques for PD DaTscan image analysis, and we briefly summarize them here: First, we model coupled progression of the disease in several regions of interest. Second, our model is specifically designed to capture progression heterogeneity. This is in contrast to most previous PD SPECT or PET image analyses, which only model a single region-of-interest (ROI) at a time (e.g. [4]) and do not model heterogeneity. Third, we identify a new constraint called population mirror symmetry. A justification for the constraint, based on DaTscan data, is presented in Section II-E. Finally, we use Bayesian analysis with a robust t-distribution to model the residues. Using the t-distribution makes the parameter estimates robust to outliers [5], [6].
The paper is organized as follows: We begin in Section II by a brief review of disease progression literature and of PD progression. The MLDS model is explained in Section III. Bayesian inference for the model is in Section IV. The results of fitting the model to the data are reported in Section V. Section VI contains a discussion, while Section VII concludes the paper. Preliminary work using a Gaussian distribution to model the noise was reported in [7].
II. Progression Models, PD, and the PPMI Dataset
A. Disease Progression Models
Most disease progression models (DPM) reported in the literature are for Alzheimer’s disease. These DPMs model the temporal progress of biomarkers such as brain MRI regional volumes, cerebrospinal fluid measures, and clinical scores. DPMs can be categorized as either event-based, explicit function of time-based, or differential equation-based.
Event-based models are discrete in time; they define various disease stages and model transitions from one disease state to another. An example is [8], where transitions from normal to severe atrophy in different brain regions are defined as events. The model finds a consistent ordering of these events in a group of subjects. Enhanced versions of this basic model, with more events and applied to the ADNI dataset are in [9], and with different orderings for different groups of subjects and subject specific orderings in [10], [11].
In contrast to event-based models, explicit function models characterize the continuous longitudinal progress of biomarkers by a parametric or a non-parametric function of time and other variates. An example of parametric modeling is [12] which regresses covariates such as time, baseline age, brain regional volume with cognitive scores [12]. A similar scheme with subject specific time shift is used with the PPMI data in [13]. Non-linear models with logistic [14] or sigmoidal [15] functions are also used. For high-dimensional data, clustering is used to reduce the number of parameters [16]. An example of a non-parametric model is [17] where disease trajectories are modeled with a group-wise monotonic Gaussian process trajectory plus an individual trajectory. In the above models, time explicitly enters the regression. In contrast are models where image features at different time points are regressed to clinical scores [18], [19].
Differential equation models use a differential equation to model the longitudinal trajectories of biomarkers, e.g. [20], [21]. Neurodegenerative diseases progress by toxic protein transmission along neuronal pathways [22]. This suggests that modeling neuronal pathways as edges in a graph can lead to using diffusion on the graph as a model for disease progression [23], [24]. An extension adds regional sporadic stimulus [25] to the model. Recently, a graph-based differential equation has been applied to MRI images of PD relating atrophy patterns to diffusion seeded at the substantia nigra [26].
Bayesian analysis has been used with neurodegenerative DPMs before [8], [10], [12], [13]. However, to the best of our knowledge, Bayesian modeling of a mixture of linear dynamical systems has not been reported with PD DaTscans.
As mentioned above, most of the above methods are designed for Alzheimer’s disease, and they predominantly use MRI images. In contrast, our goal is to model Parkinson’s disease progression using DaTscans. DaTscans do not provide any connectivity information.
B. Early Stage Parkinson’s Disease
PD progression in DaTscans is quantified by ROI analysis which shows that the mean SBR in the putamen and caudate decreases exponentially with time [27], [28]. Exponential decrease is also observed with PET (non-DaTscan) imaging tracers [4]. The rates of SBR decrease vary widely, from 5% to 13% per annum, indicating strong heterogeneity [27]. Because the putamen is affected before the caudate in the early stages [1], the difference between the mean SBR in the putamen and the caudate is taken as an indicator of disease progression [29].
Early-stage PD is also asymmetric; one brain hemisphere is affected more than the other [30]. Asymmetry is caused by a complex interplay of hereditary and environmental factors [31]. Initially, either brain hemisphere may be affected with almost equal probability, but the disease becomes more symmetric as it progresses.
C. Parkinson’s Disease Subtypes
In the PD literature, subtypes are usually derived from clinical examination, i.e. from the Movement Disorders Society’s Unified Parkinson’s Disease Rating Scale (MDS-UPDRS) scores, resulting in subtypes such as akinetic/rigid-dominant or tremor-dominant [32]. Typically, clinical progression subtypes are found by clustering the baseline clinical scores and comparing the progression rates of these clusters [33], [34]. A review of these methods is available in [32]. Recently, a complex combination of neural networks, dynamic time warping, t-SNE embedding and k-means was used to cluster the PPMI data [35] into subtypes. To the best of our knowledge progression subtypes have not been found so far using DaTscans.
D. The PPMI Dataset
The PPMI DaTscan dataset has 449 early-stage PD subjects. Their demographics are as follows: 65% of the subjects are male, 35% are female. Their ages at the time of entry into the study are 34 − 85 years, with a median age of 63 years. The subjects are scanned at baseline, and then approximately at 1, 2, 4, and 5 years from baseline (the imaging protocol for the PPMI DaTscans is documented in http://www.ppmi-info.org/wp-content/uploads/2013/02/PPMI-Protocol-AM5-Final-27Nov2012v6–2.pdf). Not all subjects have a scan for all of these time points, and the scan times for different subjects are not exactly at 1, 2, 4, 5 years.
The PPMI dataset also has longitudinal MDS-UPDRS scores for the subjects. We relate the image subtypes to Part III of the scores.
E. Population-level Mirror Symmetry
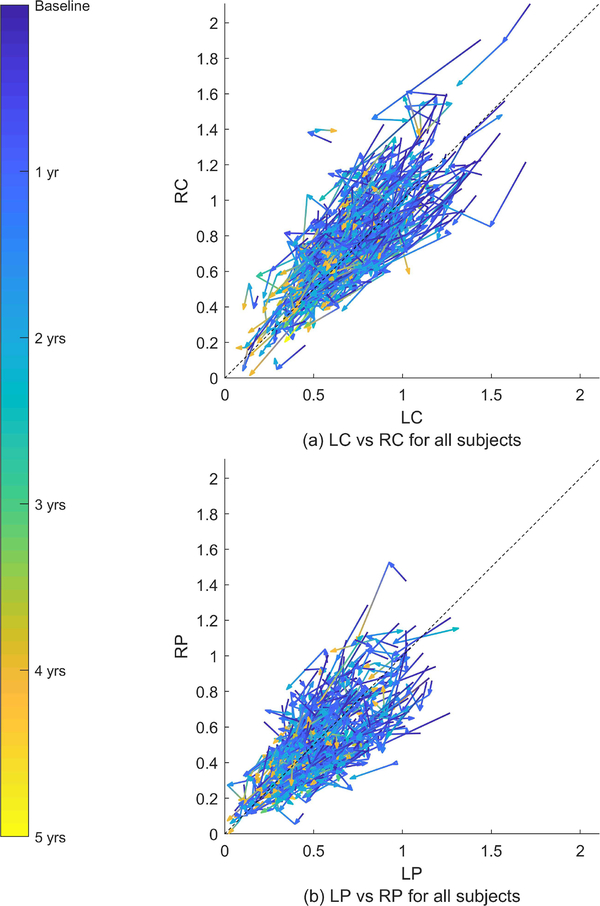
There is a population-level symmetry in the PD DaTscans. For every PD patient whose brain is asymmetrically affected in one direction, there is another patient whose brain is asymmetrically affected in the reverse direction. Moreover these subjects progress in a mirror image fashion. Fig. 1 illustrate this. The subfigures plot the time series of the mean SBR in the bilateral caudates and putamina for the PPMI subjects. The time series for every subject is plotted as a sequence of vectors, each vector pointing from a time point to the subsequent time point. The vectors are rendered with continuously changing colors that denote time from the baseline DaTscan. The relation between color and time is in the colorbar on the left. A 45 degree line is also shown. Any departure from this line represents asymmetry. Note that the spread of the data exhibits mirror symmetry around the 45 degree line. That is, given a time series for a subject, its mirror image across the 45 degree line is also a valid time series. This implies that if we were to use a single model to describe all of the trajectories in a population, then the model should remain invariant if we swapped the right and left hemispheres for all subjects. We call this property population mirror symmetry.
Fig. 1.
Time series of the mean SBR in the left caudate (LC), right caudate (RC), left putamen (LP), and right putamen (RP) for 365 subjects from the PPMI dataset. The arrows are rendered with continuously changing colors that correspond to the 5 year period indicated by the colorbar on the left side. The asymmetry of the disease is observable at baseline; in approximately 45% of the subjects, the right hemisphere is more severely affected at baseline. The left hemisphere is more severely affected in the rest.
III. The MLDS Model
Leaving aside the issue of subtypes for now, Fig. 1 suggests a single multivariate linear dynamical system (LDS) as a model for all disease trajectories. Suppose that the mean SBR in the left caudate (LC), left putamen (LP), right putamen (RP), right caudate (RC) are arranged in a vector x = [LC, LP, RP, RC]T, then the time evolution of x can be modeled as the LDS dx/dt = Ax, where A is a D × D transition matrix (D = 4). This model is coupled as long as A is not a diagonal matrix. The solution of the LDS is x(t) = eAtx(0), where eAt is the matrix exponential and x(0) is the initial condition. The solution has interesting, and well-known, properties:
The semi-group property: Suppose x(t) is a time series that satisfies dx(t)/dt = Ax(t). Further suppose that we observe this time series starting from some later point in time, i.e. suppose y(t) = x(t + T), T > 0. Then y(t) continues to follow the same differential equation without time shift, i.e. dy(t)/dt = Ay(t), an equation that is independent of T.
For a given A, the initial condition x(0) determines the entire trajectory. Since different PD patients have different initial conditions, the trajectory determined by the differential equation is automatically subject-specific.
These properties suggest that as far as fitting a matrix A to a time series goes, it is not essential to know, or to model, the exact starting time for every patient.
Different progression subtypes can be described by different LDSs (with different transition matrices A). This leads to the mixture of linear dynamical systems (MLDS) model. We make two comments about this model before we give mathematical details:
First, a subtype in this model does not correspond to a single speed of progress, neither does the model cluster the time series by progression speeds. The differential equation dx/dt = Ax expresses a relation between the values of the SBRs (at any point in time) to the rates of change of SBRs at that point in time. The values of SBR and its rate (speed) can be arbitrary; all that the equation requires is that the relation between the two be similar for subjects to be modeled by the same equation.
Second, different subtypes modeled in this way cannot be seen as early or late stages of a single trajectory. Suppose dx1(t) = A1x1(t) and dx2(t)/dt = A2x2(t) with A1 ≠ A2. Then, excluding trivial initial conditions similar to x1(0) = x2(0) = 0, there is no time shift T ≠ 0 (positive or negative) such that x1(t) = x2(t + T) for all t.
We now give mathematical details of this model beginning with the constraint on A due to population mirror symmetry.
A. Dynamical System with Population Mirror Symmetry
Given the arrangement of x that we use (i.e. LC, LP, RP, RC), population mirror symmetry is mathematically equivalent to saying that the differential equation dx/dt = Ax remains invariant under a permutation that swaps the right SBRs with the left SBRs.
Definition 1.
is a symmetric permutation if (π(x))i = (x)D−i+1 for i = 1, · · · ,D, for any , where (x)k refers to the kth component of the vector x.
Because of the way the SBRs are arranged in the vector x, population mirror symmetry corresponds to applying a symmetric permutation to x. Applying this permutation to both sides of dx/dt = Ax gives: π(dx/dt) = dπ(x)/dt = π(Ax). Population mirror symmetry requires dπ(x)/dt = Aπ(x), i.e. π(Ax) = Aπ(x). It is easy to check that this constraint is mathematically equivalent to the pre-processing procedure in the clinical literature that relabels the two hemispheres of all the subjects to dominant/non-dominant for analysis (hence ignores the left/right difference and assumes a mirror-like progression pattern) [4].
Population mirror symmetry is equivalent to A being a centrosymmetric matrix:
Definition 2.
A D × D matrix A is centrosymmetric if π (Ax) = Aπ (x) for all , where π is a symmetric permutation [36].
In terms of elements of A, centrosymmetry means that (A)i,j = (A)D−i+1,D−j+1. Loosely speaking, centrosymmetry means that elements which are located on the same line through the center of the matrix, but which are on opposite sides of the center, are equal.
Definition 3.
The dynamical system dx/dt = Ax has population mirror symmetry if A is a centrosymmetric matrix.
From now on we assume that our dynamical system has mirror symmetry. The following properties of centrosymmetric matrices are important for developing and interpreting our model:
The set of all D × D centrosymmetric matrices is a subspace of the vector space of D × D matrices. This subspace has dimension ⌈D2/2].
If the eigenvalues of a centrosymmetric matrix are distinct, then the corresponding eigenvectors are either symmetric or anti-symmetric [36].
The first property indicates that the number of parameters used in fitting a centrosymmetric matrix is reduced by half for even numbered D. The second property has implications for interpreting the differential equation dx/dt = Ax.
To see the significance of interpreting a differential equation with a centrosymmetric transition matrix, note that the solution of the differential equation can be written as
| (1) |
where {vi : i = 1, …, D} are the eigenvectors of A and {λi} are the corresponding eigenvalues, and [c1, …, cD]T = V−1x(0) where V = [v1, …, vD]. The eigenvectors v1, …, vD are linearly independent but are not guaranteed to be orthonormal. Hence the coefficients of x along the eigenvectors are found by taking the inner product of x with the dual basis of v1, …, vD. Suppose {u1, …, uD} is such a dual basis, i.e. UTV = I where U = [u1, …, uD]. We have an exponential function for projected SBR values along each ui, .
The dual basis of V are eigenvectors of AT with the same eigenvalues as that of A. Since AT is also centrosymmetric, the dual basis vectors are also either symmetric or antisymmetric. The symmetry/anti-symmetry of the dual basis of the eigenvectors of the transition matrix has an interesting interpretation. If ui is symmetric, i.e. ui = [α, β, β, α]T, then the projection is the linear combination α × LC + β × LP + β × RP + α × RC, i.e. the projection is a symmetric measurement across the brain hemispheres. If ui is anti-symmetric, then the projection is an asymmetric measurement. Thus projecting on the dual basis tells us how symmetric and asymmetric parts of the SBR vector evolve – they evolve with the corresponding λi as time constants. Note however, that because the dual basis is not orthonormal, the orthogonal projection is not the component of x(t) along ui. Rather it is the component of x(t) along vi.
B. Discretization and Probabilistic Formulation
Suppose that SBRs are available for N subjects and the ith subject has SBRs at time points , where Ti is the total number of time points and the time points are not assumed to be evenly spaced. Then, the time series for the ith subject can be modeled by a discrete version of the linear differential equation dx/dt = Ax as:
| (2) |
where Δtij = τi,j+1 − τij, and ϵij is the model residue. The residue is assumed to follow a Student’s t-distribution, i.e. where σ2ID is the scale matrix and ν is the degree of freedom.
Letting denote the entire time series for subject i,
| (3) |
where we assume that the probability distribution of the first element of the time series is , and the conditional probability distribution is
| (4) |
The form of the conditional distribution follows from (2) and the t-distribution. The distribution is the same for every subject. It models the “spread” of initial data xi1, which we take to be independent of A, σ2, ν.
Directly expressing p(xi,j+1|xij, A, σ2, ν) as the t-distribution causes technical problems in Bayesian inference – there are no conjugate priors for A, σ2, ν. However, a standard modification makes it possible to create conjugate priors for A, σ2, ν [37], [38]. The modification follows from the observation that a t-distributed random variable can be generated by first sampling a scalar random variable from a Gamma distribution, and then sampling from a Gaussian distribution with a covariance matrix scaled by the Gamma distribution sample, i.e.
| (5) |
where w is the scale parameter. To use this formulation, we introduce latent scale variables {wij : i = 1, …, N, j = 1, …, Ti − 1} with , and write (4) with a normal distribution on the right hand side:
| (6) |
With this modification, we can rewrite (3) as
| (7) |
where and . Note that according to (5), combining p(xi|wi,A, σ2) and p(wi|ν) with wi integrated out gives the time series distribution in (3). This is the discretized probabilistic version of dx/dt = Ax with t-distributed model residues.
C. The Mixture Model
All subjects that have the same transition matrix belong to the same subtype. To extend the model to K distinct progression subtypes, we allow each subtype to have its own transition matrix Ak and model residue σk. Let zi be a latent random variable taking values in {1,2, …, K} and indicating the subtype of the ith subject. Given zi, the probability density of the time series xi of the ith subject is the density of (7) with and :
| (8) |
The latent variable zi has a categorical distribution:
| (9) |
where π = (π1,· · · ,πK), such that πl ≥ 0 for all l and . Also if the argument of is true and zero otherwise.
Finally, let X = {x1, …, xN}, W = {w1, …, wN}, and z = (z1, …, zN) denote the time series, the latent variables for the t-distribution, and the latent variables for the class labels. Setting gives
| (10) |
| (11) |
as the complete model. Note that integrating out the latent variables W and z gives the mixture model
where is defined in (3). We want to infer the parameters θ and ν from the observed data X.
IV. Bayesian Inference
Our Bayesian inference methodology is to use Gibbs sampling, and to keep the sampling scheme tractable we use priors that are conjugate to the conditional densities in (10) and (11):
| (12) |
| (13) |
| (14) |
where Dir(·) is the Dirichlet distribution, NIG(·) is the normal-inverse-gamma distribution, , and α = (α/K, …, α/K), γ = {ξ0, τ0}, β = {μ0, Λ0, ν0, κ0} are hyperparameters. We define the prior on Ak using its coordinates on a basis for centrosymmetric matrices, i.e. vec(Ak) = Eak where the jth column of E has the form […, 1, …, 1, … ]T where 1 only appears at the jth position and the (D2 − j + 1)th position and the others are zero. The rationale for choosing (14) for p(X|z,W, θ) and (13) for p(W|ν) is provided in Supplementary Section II.
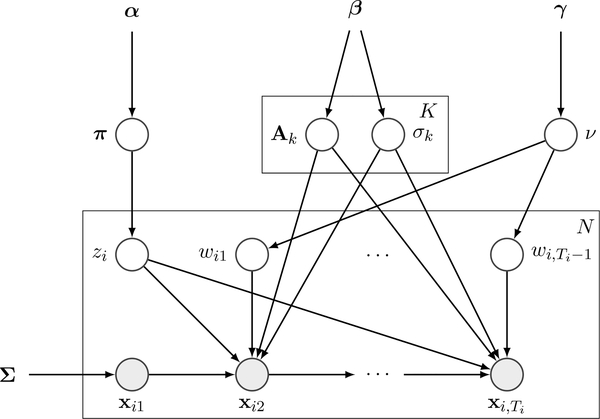
With these priors, the probabilistic graphical model is shown in Fig. 2. We can infer the parameters by drawing samples from the posterior p(z, θ,W, ν|X, α, β, γ).
Fig. 2.
Probabilistic graphical model of the MLDS with t-distributed residues. It features a standard structure of four layers: hyperparameters (α, β, γ), parameters to infer , latent variables (z = (z1, …, zN), W = {w1, …, wN}), and observed data (X = {x1, …, xN}).
A. The Gibbs Sampler
A detailed derivation of the Gibbs sampler is available in Supplementary Section III. Here we briefly describe the salient points of the sampler. The sampler works by sampling z, θ and W, ν in sequence conditioned on the remaining random variables. The sampling proceeds as below:
1. Sample p(z, θ|X,α, β,W) as follows:
1.1. Sample z from p(z|X,α, β,W) with θ integrated out. This is known as collapsed Gibbs sampling [39]. Sampling z corresponds to sequentially sampling each zi given the rest {zj : j ≠ i}.
1.2. Sample θ from p(θ|z,X,α, β,W) by sampling π from a Dirichlet distribution and Ak, from a NIG distribution.
2. Sample p(W, ν|X, γ, z, θ) as follows:
2.1. Sample W from p(wij|X, γ, z, θ, ν) by independently sampling each wij from a Gamma distribution.
2.2. Sample ν from p(ν|W, γ) using adaptive rejection sampling (ARS) [40].
A detailed version of this algorithm is in Supplementary Section III-B. The above steps are iterated till the chain converges and provides sufficient samples for parameter estimation.
We set the hyperparameters to α = K, , γ = (10−3, 10−3), which corresponds to having weak priors. The initialization of each sampling chain is done by assigning each zi ∈ {1, …, K} randomly, wij = 1, ∀i, j, ν = 30. Following Section 11.4 of [41], we run 5 chains (1500 iterations each) with random initialization, discard the first half of each chain as burn-in samples, and split the remaining samples to calculate a ratio of between-sequence variance and within-sequence variance of log p (X|θ, ν) to check convergence. For any combination of chains, if this ratio is close to one, we conclude that this combination converges to the same distribution. We pick samples from one chain of a converged combination for analysis.
Following standard Bayesian methodology, we take the parameter estimates to be the averages of the post-burn-in samples of the converged chain. The estimates of the transition matrices Ak are of particular importance, since they define the progression subtypes. We estimate Ak by the mixture estimator [42]:
where L is the number of post-burn-in samples. The means on the right hand side are directly available when we sample Ak, . The above estimator has a lower variance than the empirical estimator (i.e. averaging the samples) according to the Rao-Blackwell theorem (see Section 2.4.4 in [43]).
B. Model Selection
The Gibbs sampler described above estimates model parameters, once the number of subtypes (the number of components of the model) are known. To find the number of subtypes, we use cross validation and Bayesian model selection [44], [45]. For cross validation, we divide the dataset into 10 subsets (10-fold cross-validation). Using each subset as test set, we use the remaining data as training set to infer the parameters θ, ν. Then, the log-likelihood of each test set is evaluated and the sum of these log-likelihood values is considered for each K ∈ {1, …, Kmax}, where Kmax is the maximum number of components considered.
For Bayesian model selection, we denote ηK = {α, β, γ} for the hyperparameters with K components, and let . Assuming p(K) ∝ constant on K = 1, …, Kmax, we have . Finding the optimal is equivalent to finding the maximum of p(X|ηK) which can be evaluated by the integral
| (15) |
Since the Gibbs sampler has already generated samples from p(z, θ,W, ν|X, ηK), we use importance sampling with the proposal distribution p(θ, ν|X, ηK) to calculate the integral. The details are in Supplementary Section IV.
V. Results
A. Data Preparation
The PPMI DaTscan dataset was described earlier in Section II-D. The DICOM headers for PPMI DaTscan images reveal that the images have a size of 109 × 91 × 91 voxels, with 2 mm3 isotropic voxels. The images are distributed by PPMI and already registered in standard Montreal Neurological Institute (MNI) space. However, we did find some misregistered images in the data. These were eliminated in the preprocessing step described below. After elimination, the mean SBRs in the caudates and the putamina were obtained using the MNI atlas, which is also explained below.
Image Pre-processing:
We pre-processed the image data in two steps. First, we eliminated all subjects that had only one scan, since time series information cannot be gleaned from a single scan. This led to 382 remaining subjects. Next, we eliminated all subjects that had misregistered images. Misregistered images were found by taking the image sequence for every subject and calculating the correlation coefficient of all voxels outside the striatum between every pair of images in the series. The smallest correlation coefficient in this set was taken as the indicator of misregistration. If this indicator was less than the median minus three times the mean absolute deviation of correlation coefficients of all subjects, then the entire sequence for the subject was removed. This step eliminated 17 such subjects, leaving 365 subjects, which entered the analysis (id’s of the eliminated subjects are available in the supplementary material). Of these subjects, 45 had 2 scans, 190 had 3 scans, 127 had 4 scans and 3 had 5 scans.
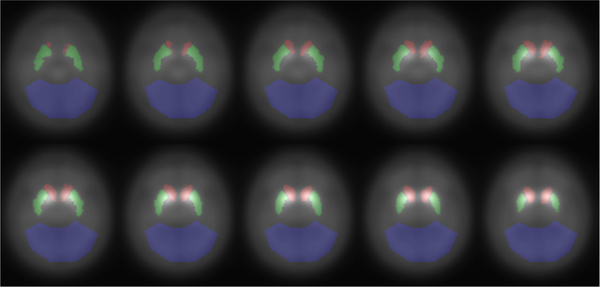
Next the SBR feature vectors xij were extracted from the images by using a set of 3D masks for the two caudates, two putamina, and the occipital lobe. Fig. 3 shows the masks overlaid on a subset of the axial slices of the mean baseline image of all subjects. The masks for the caudates and putamina were taken from the MNI atlas, dilated by 1 voxel and smoothed by a Gaussian filter with σ = 0.5 pixels to capture the partial volume effects. The occipital lobe mask was created manually, and is similar to the mask in [29]. Then, the median of the occipital lobe was used as the denominator to calculate the SBR at each voxel, and mean SBRs in the caudates and putamina were organized as xij as described in Section III-A. PPMI also provides imaging dates for all subjects, and these dates were used to calculate the time intervals Δtij.
Fig. 3.
Masks for the left and right caudates (red), left and right putamina (green), and the occipital lobe (blue). The background shows the 33rd - 42nd slices of the mean baseline image.
B. Simulation on Synthetic Dataset
Because Bayesian analysis is new to PD DaTscan image analysis, we first evaluated its accuracy – especially clustering accuracy – by creating a synthetic dataset with known class labels. To create a synthetic dataset that is close to real DaTscan data, we used our algorithm on the real PPMI dataset with 3 clusters (see Section V-C) to obtain estimates of the model parameters ({, , }, ). Using these estimated parameters, we created a low, medium and large noise dataset (the noise σk’s were set to with λ = 2, 1, 0) by keeping existing {xi1} and {Δtij} and generating the remaining data according to (4). We also set πk = 1/K to ensure that the data are evenly distributed across different classes.
We divided this synthetic dataset randomly into 10 subsets where 9 subsets were retained for training and 1 subset for testing (rotated over all subsets). This procedure was repeated 10 times and Gibbs sampling was run on the 10 by 10 training sets. For comparison, we also ran two other algorithms: 1. An EM algorithm that maximizes the log-likelihood of p(X|θ, ν) of Section III-C, but with Gaussian noise. 2. A simplified Gibbs sampling with Gaussian noise and the centrosymmetric constraint [7].
We used three measures to compare clustering accuracies of the algorithms. The first measure is purity, which measures the percentage of overlap of estimated and true class labels. The second measure is Rand index, which measures the proportion of data point pairs that are in agreement with the true labels in terms of falling in the same class or different classes [46]. Purity and Rand index have range 0 – 1, where 1 represents perfect clustering. The third measure is prediction error. We use the MLDS model to predict the SBR values for time points ≥ 3 and take the prediction error to be the difference (L1 norm) between the prediction and true values (details in Supplementary Section V).
Table I shows the mean purity and mean Rand index over the 10 by 10 training sets, and mean prediction error over the 10 by 10 test sets, for the three methods. We see that the Gibbs sampling algorithms outperform the EM algorithm for all performance measures. The two Gibbs samplers perform very similarly except for the large noise case, where the t-distribution version has a higher purity and Rand index and lower prediction error. This analysis of synthetic data justifies the use of Bayesian analysis with t-distributions over maximum-likelihood methods.
Table I.
CLUSTERING AND PREDICTION ACCURACIES OF GIBBS SAMPLING VS. THE EM-ALGORITHM USING A SYNTHETIC DATASET.
| Noise Level | 0.01 | 0.1 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Purity | Rd Id | Pr Er | Purity | Rd Id | Pr Er | Purity | Rd Id | Pr Er | |
| Gibbs-tb | 1.000a | 1.000 | 0.004 | 0.997 | 0.996 | 0.037 | 0.853 | 0.644 | 0.393 |
| Gibbs | 1.000 | 1.000 | 0.004 | 0.997 | 0.996 | 0.037 | 0.781 | 0.642 | 0.401 |
| EM c | 0.988 | 0.938 | 0.022 | 0.988 | 0.957 | 0.045 | 0.758 | 0.629 | 0.408 |
The best entry in each category is boldfaced.
Gibbs-t / Gibbs is the proposed Gibbs sampling with t-distributed / normal-distributed model residues.
EM is the version that maximizes the likelihood without the centrosymmetric constraint.
C. Fitting MLDS to the PPMI Data
Having established the superiority of Gibbs sampling with synthetic data, we turn to analyzing real PPMI data. We first determined the number of subtypes using cross-validation and Bayesian model selection as described in Section IV-B, and then used Bayesian analysis to explore the posterior distribution of the parameters.
1). Determining the number of subtypes:
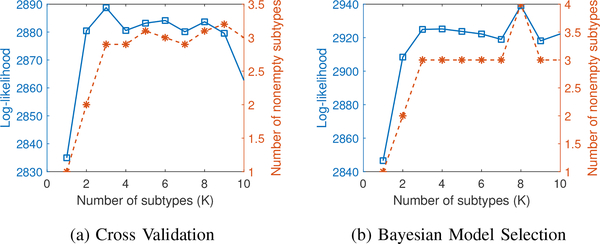
The results of using cross-validation and Bayesian model selection are shown in Fig. 4. The number of subtypes explored was between 1 and 10 (Kmax = 10). The blue solid curve in Fig. 4(a) shows the log-likelihood of the test sets as a function of the number of subtypes. As the number of subtypes increases, several subtypes turn out to be empty, i.e. no subjects are assigned to that subtype. The orange dashed curve in Fig. 4(a) shows the mean number of nonempty subtypes. Fig. 4(b) shows the same two quantities for Bayesian model selection.
Fig. 4.
Model selection using cross validation (a) and Bayesian (b). The y-axis has two scales corresponding to log-likelihood value (blue solid square) and final number of nonempty subtypes (orange dashed star) respectively. The number of nonempty subtypes is averaged over 10 folds for cross validation.
Fig. 4 clearly shows that the log-likelihood values for cross-validation and Bayesian model selection behave similarly. The log-likelihood increases monotonically from 1 to 3 subtypes and then appears to saturate. The number of nonempty subtypes found by both methods is similar as well. In the final model, we chose 3 subtypes (K = 3) for further analysis.
2). Parameter estimation and interpretation:
The MLDS model with three subtypes (K = 3) was fit to the PPMI dataset using Bayesian analysis. The clustering results are shown in Fig. 5. The subtype label is created by calculating p(zi|X,α, β, γ) from the samples, and assigning argmaxk p(zi = k|X,α, β, γ) to subject i. This gives us 46, 257, and 62 subjects in subtypes 1, 2, and 3 respectively. As the estimated trajectories show, different subtypes progress with different speeds with subtype 1 being the fastest and subtype 3 being the slowest. The mean and standard deviation of the posterior distribution of the parameters are shown in the top half of Table II for each subtype. The bottom half of Table II (rows indicated by λ, v, u) shows the eigenvalues, eigenvectors and dual basis of the eigenvectors of the mean transition matrix for each subtype.
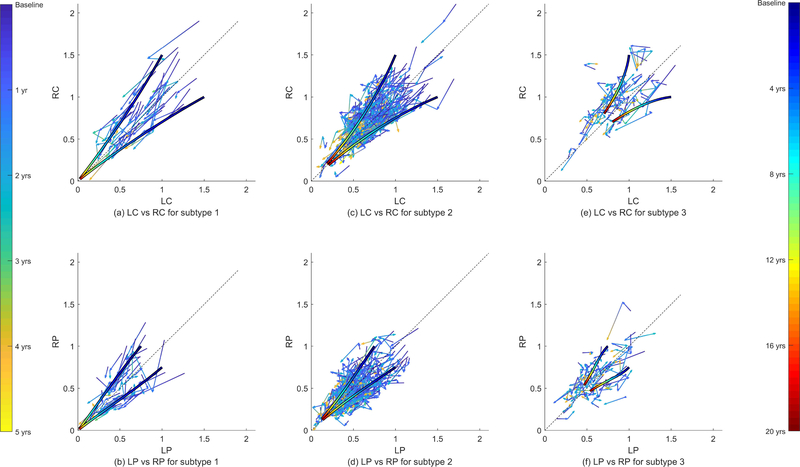
Fig. 5.
Clustered time series of the mean SBR for all the PPMI subjects. The columns show the subtypes discovered by the proposed approach. The arrows are rendered with continuously changing colors that correspond to the 5 year period indicated by the colorbar on the left side. We can see that different subtypes exhibit different progression rates. We also show our estimated trajectory starting from a fixed point (x = [1, 0.75, 1, 1.5]T ) and its reflection (duration indicated by the colorbar on the right side).
TABLE II.
PARAMETERS AND EIGENSTRUCTURE ESTIMATED BY BAYESIAN INFERENCE.
| Estimated Parameters (mean
(std))a | |||||||||||||
| Subtype | k = 1 | k = 2 | k = 3 | ||||||||||
| ν | 2.85 (0.21) | ||||||||||||
| πk | 0.146 (0.036) | 0.612 (0.081) | 0.242 (0.076) | ||||||||||
| σk | 0.064 (0.007) | 0.059 (0.003) | 0.067 (0.005) | ||||||||||
| Ak | −0.33 (0.06) | 0.16 (0.07) | −0.10 (0.06) | 0.09 (0.05) | −0.21 (0.02) | 0.12 (0.03) | −0.01 (0.03) | 0.04 (0.02) | −0.09 (0.04) | 0.05 (0.05) | −0.08 (0.05) | 0.08 (0.04) | |
| 0.15 (0.06) | −0.44 (0.08) | 0.09 (0.06) | −0.05 (0.05) | 0.06 (0.02) | −0.22 (0.03) | 0.07 (0.02) | −0.02 (0.02) | 0.13 (0.03) | −0.24 (0.05) | 0.03 (0.05) | −0.01 (0.04) | ||
| −0.05 (0.05) | 0.09 (0.06) | −0.44 (0.08) | 0.15 (0.06) | −0.02 (0.02) | 0.07(0.02) | −0.22 (0.03) | 0.06 (0.02) | −0.01 (0.04) | 0.03 (0.05) | −0.24 (0.05) | 0.13 (0.03) | ||
| 0.09 (0.05) | −0.10 (0.06) | 0.16 (0.07) | −0.33 (0.06) | 0.04 (0.02) | −0.01 (0.03) | 0.12 (0.03) | −0.21 (0.02) | 0.08 (0.04) | −0.08 (0.05) | 0.05 (0.05) | −0.09 (0.04) | ||
| Estimated
Eigenstructureb | |||||||||||||
| λ | −0.71 | −0.39 | −0.23 | −0.20 | −0.37 | −0.22 | −0.16 | −0.09 | −0.37 | −0.18 | −0.08 | −0.02 | |
| v | LC | 0.46 | 0.26 | 0.58 | 0.59 | 0.50 | 0.64 | 0.60 | 0.59 | 0.40 | 0.13 | 0.58 | 0.59 |
| LP | −0.54 | −0.66 | 0.40 | 0.39 | −0.50 | −0.30 | 0.38 | 0.39 | −0.58 | 0.70 | 0. 41 | 0.39 | |
| RP | 0.54 | −0.66 | −0.40 | 0.39 | 0.50 | −0.30 | −0.38 | 0.39 | 0.58 | 0.70 | −0.41 | 0.39 | |
| RC | −0.46 | 0.26 | −0.58 | 0.59 | −0.50 | 0.64 | −0.60 | 0.59 | −0.40 | 0.13 | −0.58 | 0.59 | |
| u | LC | 0.41 | 0.40 | 0.54 | 0.67 | 0.39 | 0.46 | 0.51 | 0.35 | 0.41 | −0.54 | 0.58 | 0.97 |
| LP | −0.58 | −0.60 | 0.46 | 0.27 | −0.61 | −0.70 | 0.51 | 0.75 | −0.58 | 0.82 | 0.40 | −0.18 | |
| RP | 0.58 | −0.60 | −0.46 | 0.27 | 0.61 | −0.70 | −0.51 | 0.75 | 0.58 | 0.82 | −0.40 | −0.18 | |
| RC | −0.41 | 0.40 | −0.54 | 0.67 | −0.39 | 0.46 | −0.51 | 0.35 | −0.41 | −0.54 | −0.58 | 0.97 | |
π is the fraction of the PD subjects that are contained in each subtype. Ak and are the transition matrix and the unscaled variance for each subtype.
The bottom half shows the eigenvalues (λ), the eigenvectors (v) and the dual basis of the eigenvectors (u) of the mean transition matrices.
The main characteristics of Table II are: Row πk (mixing coefficient) in Table II indicates that subtype 2 has the highest occupancy; a little over half of the subjects are contained in this subtype. Subtype 3 and 1 have sequentially smaller occupancy. All subtypes have similar values for σk, suggesting that all subtypes have similar model residues.
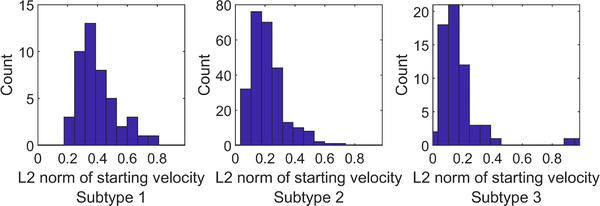
Different subtypes have different progression rates and progression trajectories, i.e. the MLDS model has successfully captured PD heterogeneity. The variability in progression rates is apparent in the eigenvalues of the transition matrices in Table II (row λ). All eigenvalues are real, distinct, and negative. Checking the magnitude of the eigenvalues, we see that subtype 1 is the fastest progressing subtype, followed by subtype 2 and then by subtype 3. Further evidence for the relative speeds of the subtypes can be directly found in Fig. 6, which shows histograms of starting speeds (i.e. the magnitude of ) of all subjects in each subtype (initial changes are the largest and therefore present the clearest evidence in presence of noise).
Fig. 6.
Histograms of the initial speeds for subtypes 1, 2, and 3. The median velocities are 0.36, 0.19, and 0.13 SBR/year respectively.
The subtypes differ not only in speed but also in the shape of the SBR trajectories as well. This is apparent in Fig. 5 which shows the SBR trajectories of subjects in each subtype. The figure also shows model trajectories (smooth curves overlaid on raw trajectories) for two initial points. These trajectories clearly have different speed, extent, and shape.
The spatial patterns of progression as evident in the dual basis of the eigenvectors of the transition matrices are especially interesting. Recall from Section III-A that a symmetric or anti-symmetric dual basis vector can be interpreted as representing the symmetry or asymmetry of the disease across the two brain hemispheres. Since all eigenvalues are real and negative, symmetric/anti-symmetric dual basis vectors capture how the symmetry/asymmetry of dopamine transporter concentration (i.e. the mean/difference of α×Caudate+β×Putamen between both hemispheres) changes as the disease progresses.
The leading dual basis vector in every subtype in Table II (row v) is anti-symmetric, with α, β having opposite signs. This implies that the loss of asymmetry in the disease is the fastest spatial progression pattern among all linear combinations of SBRs. The last dual basis vector in subtype 1 and 2 is symmetric with α, β having the same sign. This dynamical mode clearly represents the “mean” of all four regions. Since this mode has the smallest eigenvalue, the mean SBR is the slowest index of disease progression in early-stage PD. Thus Table II suggests that the rate of asymmetry change is several times faster than the change in the mean SBR.
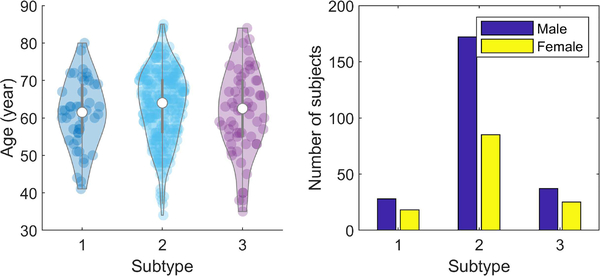
3). Relation to Demographics:
Fig. 7 shows violin plots of the age and sex distribution of PD subjects in the three subtypes. The 95-percentile age range in the three subtypes is 61.0 ± 18.1, 63.2 ± 19.3, 61.5 ± 22.2 years respectively. A t-test with a null hypothesis of equality of means of the ages of subtype 1 vs 2, 2 vs 3, 1 vs 3 gives p-values of 0.14, 0.26, 0.82. Thus the null hypothesis cannot be rejected, suggesting that the mean ages in the subtypes are equal. The male population is distributed in the three subtypes as 11.8%, 72.6%, 15.6%. The female population is distributed as 14.1%, 66.4%, 19.5%. A chi-square test, evaluating the null hypothesis that these distributions are equal, gives a p-value of 0.46, suggesting that the null hypothesis cannot be rejected again. In spite of that, the female population is slightly more dispersed with subtypes 1 and 3 having larger fractions of the population.
Fig. 7.
Age and sex distribution in image-based subtypes.
D. Model Validation and Results Sensitivity
Finally, we turn to evaluating other aspects of the model: the use of t-distributions for model residues, the train-test consistency of the model residue, and the sensitivity of the result to caudate and putamen templates.
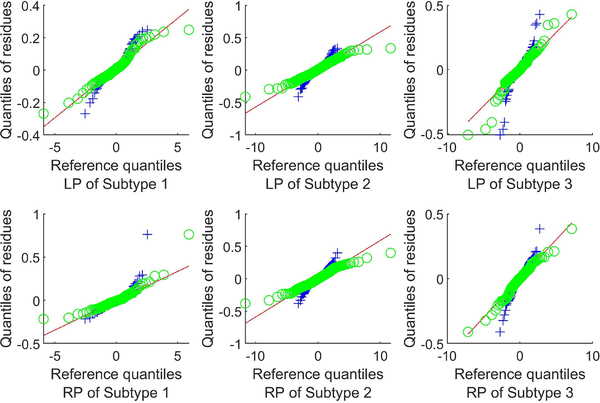
1). Validating the residue distribution:
We validate the model residue distribution by using a Q-Q plot, i.e. by plotting the quantiles of the model residues against that of a normal distribution and a t-distribution for each region and each subtype. Fig. 8 shows Q-Q plots of the putamen residues (the caudate residues show a similar trend and are omitted to save space) and fitted lines representing a perfect fit to a residue model. A Q-Q plot crossing the line at a steeper slope implies that the data have heavier tails than the assumed distribution. It is clear from Fig. 8 that the residues in all three subtypes have significantly heavier tails than the normal distribution. And the t-distribution assumption appears to be a significant improvement over the normal distribution assumption in every subtype.
Fig. 8.
Q-Q plots of the model residue vs normal (blue cross) and vs t-distribution (green circle) for the putamina.
2). Train-test consistency of the model residue:
To test whether our algorithm overfits the data, we also evaluated the train-test consistency of the model residue with 10-fold cross validation. Specifically, nine folds were taken as the training set with K = 3 to estimate parameters , . These parameters were applied to the subjects in the remaining fold (test set) to predict their subtypes (via using Eq. (S20) in Supplementary Section V) and the norms of the model residues ‖ϵij‖ were calculated from the subtype according to Eq. (2).
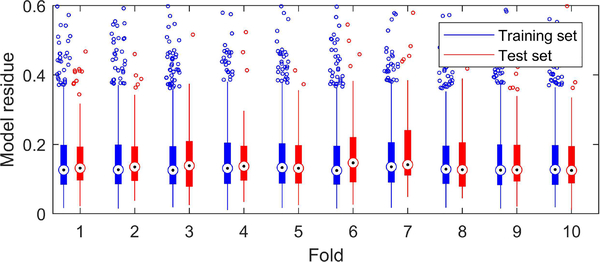
Fig. 9 shows box plots of training and test set residues. For clarity, only the scatter of the residues in the extreme quantiles is shown. Fig. 9 shows that our algorithm does not overfit the data (overfitting would lead to substantially larger test errors).
Fig. 9.
Box plots of model residues from training sets and test sets in the 10-fold cross validation. The whiskers correspond to ±2.7σ and 99.3% coverage if the residues are normally distributed. Residues beyond the limit of y-axis are not shown.
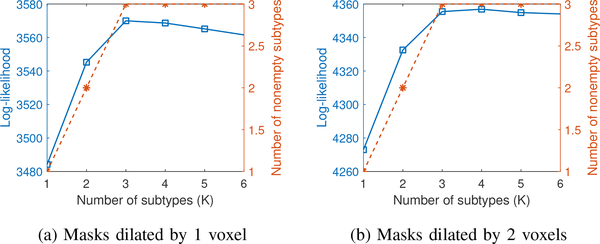
3). Sensitivity to caudate and putamen masks:
Partial voluming and small variations in subject-specific anatomy can potentially affect the estimated model parameters. To test the sensitivity to these, we further dilated the caudate and putamen masks by 1 and 2 voxels and then filtered them with a Gaussian filter having σ = 0.5 pixels. We re-estimated the parameters using Bayesian analysis with these dilated masks. Dilating by 1 voxel increases the volume of the mask by 55% for caudate and 41% for putamen (133% and 101% when dilating 2 voxels). Even with such large changes to the masks, the number of subtypes (see Fig. 10) as well as the parameter estimates were similar to the original estimates. The number of subtypes remained at 3, and the relative changes in the transition matrices of the three subtypes, calculated as , were 0.04, 0.08, 0.11 for masks dilated by 1 voxel and 0.47, 0.19, 1.67 for masks dilated by 2 voxels. We also calculated the Rand index between the subtype labels in Section V-C and the labels estimated using the dilated masks. For masks dilated by 1 voxel, the Rand index was 0.92, while for masks dilated by 2 voxels, the Rand index was 0.73. This implies that the progression pattern and subtyping are not sensitive to partial volume effects or anatomical variations.
Fig. 10.
Bayesian model selection for dilated masks. The y-axis has two scales corresponding to log-likelihood value (blue solid square) and final number of nonempty subtypes (orange dashed star) respectively.
E. Correlation with MDS-UPDRS scores
Finally, we sought correlation between DaTscan-based progression subtypes and clinical movement scores as present in the Part III of the MDS-UPDRS exam. In PPMI, longitudinal scores of each patient are sampled more frequently than DaTscan images: at 3 months intervals for the first year, at 6 months intervals for the next 4 years, and at 1 year intervals for the following 3 years. We retained only those scores that corresponded to the imaging times. Part III scores can be influenced by medication, but PPMI provides scores for subjects in the off-medication state. We only used the off-medication scores.
MDS-UPDRS Part III has 36 scores of which we retained the first 33 for every subject for every imaging time. The last three ratings (“were dyskinesias present?”, “did these movements interfere with your ratings?”, “Hoehn and Yahr stage”) were discarded either because they were non-informative (all subjects scored the same score) or because they could be considered a summary of other ratings (e.g. “Hoehn and Yahr stage”).
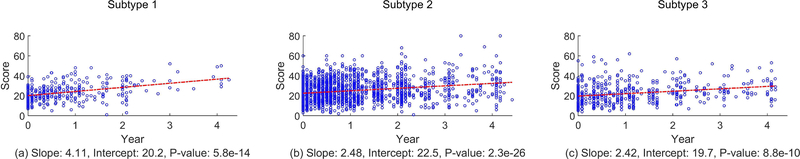
In the PD literature, Part III scores are added to create a single number which summarizes the state of movement disorder for the patient. This summed score is called Total Movement Score (TMS) [47]. Larger values of TMS reflect worse PD symptoms. Fig. 11 shows the scatter plots of TMS for all subjects in the three progression subtypes. The plots in Fig. 11 also show the best-fit linear time regression to the scores. A slope, intercept and a p-value are calculated from the best-fit line. The p-value corresponds to testing the null hypothesis that the slope of the best-fit line is 0. A p-value less than 0.05 indicates that the slope is not zero, at a significance level of 0.05.
Fig. 11.
Linear regression to the MDS-UPDRS Part III total scores for each subtype. The p-value is for the null hypothesis that the slope is 0.
Fig. 11 shows that regressing TMS linearly with time for all subtypes gives a positive slope with a p-value significantly less than 0.05. Moreover, the slope for subtype 1 is bigger than the slopes for subtype 2 and subtype 3, which is consistent with image-based progression – subtype 1 has the fastest progression. The slope for subtype 2 is only slightly bigger than that for subtype 3, implying that the difference in terms of clinical symptom progression rate of TMS between subtype 2 and subtype 3 is smaller. Thus, TMS progression is consistent with DaTscan progression subtypes.
VI. Discussion
A. The MLDS Model
MLDS models the mean SBR from the caudates and the putamina. This is consistent with almost all of the PD DaTscan analysis literature, e.g. [48], [47]. In spite of its popularity, we readily acknowledge that there are limitations to this paradigm: the posterior-to-anterior anatomical gradient of disease progression in the striatum cannot be captured by mean SBRs. Moreover, extra-striatal structures such as the globus pallidus and the thalamus are also affected by PD [49], [50], but they are not included in the model. Clearly, what is needed is a finer grained model which also takes striatal subregions and extra-striatal regions into account. We plan to address this in the future.
The MLDS model can be generalized to other longitudinal datasets as long as the underlying progression satisfies a linear differential equation. For example, it can be naturally extended to high dimensional features (e.g. voxelwise SBR) from DaTscan images by imposing a low-rank constraint on the transition matrices. Another example is the graph diffusion equation for modeling the progression of misfolded protein in the brain’s connectivity network [23], where the connectivity information can be encoded in the prior to constrain the transition matrices.
B. Inferred Subtypes and Progression Patterns
The subtypes found by Bayesian inference clearly capture the progression heterogeneity as it manifests in DaTscans. Evidence, presented in Supplementary Section VI-B, shows that the subtypes do not represent time delayed versions of a single progression prototype.
The subtypes have different progression speeds with subtype 1 having the fastest progression, subtype 2 having a more moderate progression, and subtype 3 having the slowest progression. The values for πk in Table II suggest that slightly more than half of the PD subjects belong to subtype 2, the remaining divided between subtype 1 and 3. Thus, one interpretation of the subtypes is that subtype 2 represents typical progression, while subtypes 1 and 3 represent the extremes of progression.
The presence of three progression subtypes in the PPMI dataset is also supported by other machine learning approaches. For example, Zhang et al. combine image and non-image features in the PPMI dataset (SBR, clinical scores, biospecimen exams) in a deep learning framework to find moderate, mild, and rapid progression subtypes [35]. However, that analysis does not reveal any eigenvectors or time constants. And our analysis only uses DaTscans.
It is remarkable that the eigenvector with the fastest time constant in all three subtypes corresponds to conversion from asymmetry to symmetry. The decrease of asymmetry in (non-DaTscan) PET images has been noted in the previous research [4], [51]. What MLDS reveals is that the decrease in asymmetry has the fastest possible time constant amongst all linear combinations of LC, LP, RP, RC.
The correlation between image-based subtypes and MDS-UPDRS Part III scores shows that at a group level, the progression rates measured by DaTscans reflect the progression rates of clinical symptoms. Note that in the PD literature, the reported correlations between DaTscans and UPDRS scores are usually quite small, ranging in magnitude from 0.1 to 0.3. The PD literature also suggests that correlations between changes in DaTscans and MDS-UPDRS are not significant [47]. However, these studies do not take subtypes into account. Our results show that TMS changes in the subtypes are similar to the subtype progression rates.
VII. Conclusions
This paper introduced a new longitudinal model and a Bayesian inference methodology for identifying progression subtypes and for finding disease progression patterns and their time constants for PD. The model is a mixture of linear dynamical systems, and is based on identifying key properties of PD progression. The model introduces several new ideas to PD modeling: coupled progression of multiple regions with population mirror symmetry, t-distributed model residues, mixtures of dynamical systems for heterogeneity, and a proper Bayesian analysis using Gibbs sampling.
Three image-based progression subtypes are found, differing in progression speeds. Each subtype displays characteristic spatial progression patterns with associated time constants. The fastest progression pattern in all subtypes is the loss of hemispheric asymmetry, while the slowest progression pattern is the change in the mean SBR. This finding has implications for clinical trials that assess the effectiveness of disease modifying therapies. The DaTscan-based subtypes also have different TMS progression rates.
Supplementary Material
Acknowledgements
This research was supported by the NIH grant R01NS107328. The data used in the preparation of this article was obtained from the Parkinson’s Progression Markers Initiative (PPMI) database (up-to-date information is available at www.ppmi-info.org). PPMI – a public-private partnership – is funded by the Michael J. Fox Foundation for Parkinson’s Research and multiple funding partners. The full list of PPMI funding partners can be found at ppmiinfo.org/fundingpartners.
The work is partially supported by NIH grant R01NS107328.
Footnotes
Supplementary downloadable material is available at http://ieeexplore.ieee.org, provided by the authors.
Contributor Information
Yuan Zhou, Dept. of Radiology and Biomedical Imaging, Yale University, New Haven, CT, USA.
Sule Tinaz, Dept. of Neurology, Yale University, New Haven, CT, USA.
Hemant D. Tagare, Dept. of Radiology and Biomedical Imaging, Dept. of Biomedical Engineering, Dept. of Statistics and Data Science, Yale University, New Haven, CT, USA.
References
- [1].Braak H, Tredici KD, Rub U, de Vos RA, Steur ENJ, and Braak E, “Staging of brain pathology related to sporadic Parkinson’s disease,” Neurobiology of Aging, vol. 24, no. 2, pp. 197–211, 2003. [DOI] [PubMed] [Google Scholar]
- [2].Booij J et al. , “Practical benefit of [123I]FP-CIT SPET in the demonstration of the dopaminergic deficit in Parkinson’s disease,” European Journal of Nuclear Medicine, vol. 24, no. 1, pp. 68–71, January 1997. [DOI] [PubMed] [Google Scholar]
- [3].Innis RB et al. , “Consensus nomenclature for in vivo imaging of reversibly binding radioligands,” Journal of Cerebral Blood Flow & Metabolism, vol. 27, no. 9, pp. 1533–1539, 2007. [DOI] [PubMed] [Google Scholar]
- [4].Nandhagopal R et al. , “Longitudinal progression of sporadic Parkinson’s disease: a multi-tracer positron emission tomography study,” Brain, vol. 132, no. 11, pp. 2970–2979, 08 2009. [DOI] [PubMed] [Google Scholar]
- [5].Hampel FR, Ronchetti E, Rousseeuw P, and Stahel WA, Robust Statistics: The Approach Based on Influence Functions, 01 1986, vol. 29. [Google Scholar]
- [6].Lange KL, Little RJA, and Taylor JMG, “Robust statistical modeling using the t distribution,” Journal of the American Statistical Association, vol. 84, no. 408, pp. 881–896, 1989. [Google Scholar]
- [7].Zhou Y and Tagare HD, “Bayesian longitudinal modeling of early stage Parkinson’s disease using DaTscan images,” in International Conference on Information Processing in Medical Imaging Springer, 2019, pp. 405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fonteijn HM et al. , “An event-based model for disease progression and its application in familial Alzheimer’s disease and Huntington’s disease,” NeuroImage, vol. 60, no. 3, pp. 1880–1889, 2012. [DOI] [PubMed] [Google Scholar]
- [9].Young AL et al. , “A data-driven model of biomarker changes in sporadic alzheimer’s disease,” Brain, vol. 137, no. 9, pp. 2564–2577, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].——, “Uncovering the heterogeneity and temporal complexity of neurodegenerative diseases with subtype and stage inference,” Nature communications, vol. 9, no. 1, pp. 1–16, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Venkatraghavan V, Bron EE, Niessen WJ, Klein S, Initiative ADN et al. , “Disease progression timeline estimation for Alzheimer’s disease using discriminative event based modeling,” NeuroImage, vol. 186, pp. 518–532, 2019. [DOI] [PubMed] [Google Scholar]
- [12].Li K and Luo S, “Dynamic predictions in Bayesian functional joint models for longitudinal and time-to-event data: An application to Alzheimer’s disease,” Statistical methods in medical research, vol. 28, no. 2, pp. 327–342, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Iddi S et al. , “Estimating the evolution of disease in the Parkinson’s Progression Markers Initiative,” Neurodegenerative Diseases, vol. 18, pp. 173–190, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Donohue MC et al. , “Estimating long-term multivariate progression from short-term data,” Alzheimer’s & Dementia, vol. 10, pp. S400–S410, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jedynak BM et al. , “A computational neurodegenerative disease progression score: method and results with the Alzheimer’s disease Neuroimaging Initiative cohort,” Neuroimage, vol. 63, no. 3, pp. 1478–1486, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Marinescu RV et al. , “DIVE: A spatiotemporal progression model of brain pathology in neurodegenerative disorders,” NeuroImage, vol. 192, pp. 166–177, 2019. [DOI] [PubMed] [Google Scholar]
- [17].Lorenzi M et al. , “Probabilistic disease progression modeling to characterize diagnostic uncertainty: application to staging and prediction in alzheimer’s disease,” NeuroImage, 2017. [DOI] [PubMed] [Google Scholar]
- [18].Zhou J, Liu J, Narayan VA, and Ye J, “Modeling disease progression via fused sparse group lasso,” in Proceedings of the 18th ACM SIGKDD international conference on Knowledge discovery and data mining ACM, 2012, pp. 1095–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang H et al. , “High-order multi-task feature learning to identify longitudinal phenotypic markers for Alzheimer’s disease progression prediction,” in Advances in Neural Information Processing Systems, 2012, pp. 1277–1285. [Google Scholar]
- [20].Oxtoby NP et al. , “Learning imaging biomarker trajectories from noisy Alzheimer’s disease data using a Bayesian multilevel model,” in Bayesian and grAphical Models for Biomedical Imaging, Cardoso MJ, Simpson I, Arbel T, Precup D, and Ribbens A, Eds. Cham: Springer International Publishing, 2014, pp. 85–94. [Google Scholar]
- [21].——, “Data-driven models of dominantly-inherited Alzheimer’s disease progression,” Brain, vol. 141, no. 5, pp. 1529–1544, 03 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Seeley WW, Crawford RK, Zhou J, Miller BL, and Greicius MD, “Neurodegenerative diseases target large-scale human brain networks,” Neuron, vol. 62, no. 1, pp. 42–52, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Raj A, Kuceyeski A, and Weiner M, “A network diffusion model of disease progression in dementia,” Neuron, vol. 73, no. 6, pp. 1204–1215, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Raj A and Powell F, “Models of network spread and network degeneration in brain disorders,” Biological Psychiatry: Cognitive Neuroscience and Neuroimaging, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hu C, Hua X, Ying J, Thompson PM, Fakhri GE, and Li Q, “Localizing sources of brain disease progression with network diffusion model,” IEEE journal of selected topics in signal processing, vol. 10, no. 7, pp. 1214–1225, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Pandya S et al. , “Predictive model of spread of Parkinson’s pathology using network diffusion,” NeuroImage, vol. 192, pp. 178–194, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Au WL, Adams JR, Troiano A, and Stoessel AJ, “Parkinson’s disease: in vivo assessment of disease progression using positron emission tomography,” Mol. Brain Research, vol. 134, pp. 24–33, 2005. [DOI] [PubMed] [Google Scholar]
- [28].Hilker R et al. , “Nonlinear progression of Parkinson disease as determined by serial positron emission tomographic imaging of striatal fluorodopa F 18 activity,” Archives of Neurology, vol. 62, no. 3, pp. 378–382, 2005. [DOI] [PubMed] [Google Scholar]
- [29].Zubal IG, Early M, Yuan O, Jennings D, Marek K, and Seibyl JP, “Optimized, automated striatal uptake analysis applied to SPECT brain scans of Parkinson’s disease patients,” The Journal of Nuclear Medicine, vol. 48, no. 6, pp. 857–864, 2007. [DOI] [PubMed] [Google Scholar]
- [30].Hoehn MM and Yahr MD, “Parkinsonism: onset, progression and mortality,” Neurology, vol. 17, pp. 427–442, 1967. [DOI] [PubMed] [Google Scholar]
- [31].Riederer P, Jellinger K, Kolber P, Hipp G, Sian-Hülsmann J, and Krüger R, “Lateralisation in Parkinson disease,” Cell and tissue research, vol. 373, no. 1, pp. 297–312, 2018. [DOI] [PubMed] [Google Scholar]
- [32].Thenganatt MA and Jankovic J, “Parkinson disease subtypes,” JAMA neurology, vol. 71, no. 4, pp. 499–504, 2014. [DOI] [PubMed] [Google Scholar]
- [33].Fereshtehnejad S-M, Romenets SR, Anang JB, Latreille V, Gagnon J-F, and Postuma RB, “New clinical subtypes of parkinson disease and their longitudinal progression: a prospective cohort comparison with other phenotypes,” JAMA neurology, vol. 72, no. 8, pp. 863–873, 2015. [DOI] [PubMed] [Google Scholar]
- [34].Lawton M et al. , “Developing and validating Parkinson’s disease subtypes and their motor and cognitive progression,” J Neurol Neurosurg Psychiatry, vol. 89, no. 12, pp. 1279–1287, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhang X et al. , “Data-driven subtyping of Parkinson’s disease using longitudinal clinical records: a cohort study,” Scientific reports, vol. 9, no. 1, p. 797, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Weaver JR, “Centrosymmetric (cross-symmetric) matrices, their basic properties, eigenvalues, and eigenvectors,” The American Mathematical Monthly, vol. 92, no. 10, pp. 711–717, 1985. [Google Scholar]
- [37].Fröhwirth-Schnatter S and Kaufmann S, “Model-based clustering of multiple time series,” Journal of Business & Economic Statistics, vol. 26, no. 1, pp. 78–89, 2008. [Google Scholar]
- [38].Peel D and McLachlan GJ, “Robust mixture modelling using the t distribution,” Statistics and computing, vol. 10, no. 4, pp. 339–348, 2000. [Google Scholar]
- [39].Murphy KP, Machine learning: a probabilistic perspective. MIT press, 2012. [Google Scholar]
- [40].Gilks WR and Wild P, “Adaptive rejection sampling for gibbs sampling,” Applied Statistics, pp. 337–348, 1992. [Google Scholar]
- [41].Gelman A, Stern HS, Carlin JB, Dunson DB, Vehtari A, and Rubin DB, Bayesian data analysis. Chapman and Hall/CRC, 2013. [Google Scholar]
- [42].Liu JS, Wong WH, and Kong A, “Covariance structure of the gibbs sampler with applications to the comparisons of estimators and augmentation schemes,” Biometrika, vol. 81, no. 1, pp. 27–40, 1994. [Google Scholar]
- [43].Sudderth EB, “Graphical models for visual object recognition and tracking,” Ph.D. dissertation, Massachusetts Institute of Technology, 2006. [Google Scholar]
- [44].Kass RE and Raftery AE, “Bayes factors,” Journal of the American Statistical Association, vol. 90, no. 430, pp. 773–795, 1995. [Google Scholar]
- [45].McLachlan GJ and Rathnayake S, “On the number of components in a Gaussian mixture model,” Wiley Interdisciplinary Reviews: Data Mining and Knowledge Discovery, vol. 4, no. 5, pp. 341–355, 2014. [Google Scholar]
- [46].Hubert L and Arabie P, “Comparing partitions,” Journal of classification, vol. 2, no. 1, pp. 193–218, 1985. [Google Scholar]
- [47].Simuni T et al. , “Longitudinal change of clinical and biological measures in early Parkinson’s disease: Parkinson’s progression markers initiative cohort,” Movement Disorders, vol. 33, no. 5, pp. 771–782, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Prashanth R, Roy SD, Mandal PK, and Ghosh S, “Automatic classification and prediction models for early Parkinson’s disease diagnosis from SPECT imaging,” Expert Systems with Applications, vol. 41, no. 7, pp. 3333–3342, 2014. [Google Scholar]
- [49].Rajput A, Sitte H, Rajput A, Fenton M, Pifl C, and Hornykiewicz O, “Globus pallidus dopamine and Parkinson motor subtypes: clinical and brain biochemical correlation,” Neurology, vol. 70, no. 16 Part 2, pp. 1403–1410, 2008. [DOI] [PubMed] [Google Scholar]
- [50].Remy P, Doder M, Lees A, Turjanski N, and Brooks D, “Depression in Parkinson’s disease: loss of dopamine and noradrenaline innervation in the limbic system,” Brain, vol. 128, no. 6, pp. 1314–1322, 2005. [DOI] [PubMed] [Google Scholar]
- [51].Fu JF et al. , “Joint pattern analysis applied to PET DAT and VMAT2 imaging reveals new insights into Parkinson’s disease induced presynaptic alterations,” NeuroImage: Clinical, vol. 23, p. 101856–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.