Introduction
The use of rodent models has been the standard for preclinical pain research for decades. It is obvious that animal models offer an invaluable means to explore the functional, morphological and molecular properties of the sensory system. However, differences in nociceptor biology between rodent and humans exist and almost certainly underlie some of the limited success in the translatability of preclinical analgesic strategies into effective clinical treatments [7; 33; 37]. Such differences may arise from intra-species variances in the expression of drug targets in the peripheral sensory system (i.e. in nociceptors) or in CNS regions. Many failures of analgesics in human clinical trials have been blamed on a lack of reproducibility and experimental rigor in rodent studies; but the primary translatability issue might be an insufficient base of knowledge on expression similarities and differences between mouse and human nociceptors.
Dorsal root ganglion sensory neurons are molecularly defined by their mRNA-expression signatures. Classically, three broad populations of neurons have been delineated in mouse DRG which include the CGRP-positive, peptidergic nociceptors, the P2X3R and/or isolectin B4-binding (IB4), non-peptidergic nociceptors, and the large diameter mechanoreceptors that express neurofilament-200 (NF200). Although there is some overlap between these markers, their exclusivity as unique subpopulations in mouse DRG is widely recognized and has been confirmed by single cell RNA sequencing [49]. However, long-standing evidence supports these markers may not exclusively demarcate DRG neuronal subpopulations in other species. For instance, studies in rats have shown that there is a large overlap in the expression of CGRP and IB4-positive neurons in DRG [38] and NF200 marks all sensory neurons in human DRG [41]. Several studies have found important differences using bulk RNA-seq, physiological or histochemical measures between mouse and human DRG [20] but relatively few studies have quantitatively investigated the expression and population distribution for a broad number of mRNA targets in DRG tissues from this species in parallel. The goal of the present work was to define DRG mRNA expression similarities or differences between mouse and human for (A) genes that are important population markers, (B) genes that are specifically expressed in DRG, (C) genes that are existing or potential drug targets, or (D) genes that have clear links to human pain states.
We used multiplexed RNAscope in situ hybridization to quantify cellular mRNA expression for Trp-channels, cholinergic receptors, potassium channels, sodium channels, immune and other markers in combination with known nociceptor population markers (CGRP and P2X3R) in mouse and human DRG. We also assessed population distribution, cell diameters, and species differences. While we found some strong similarities between mouse and human, there were multiple mRNAs that showed substantial differences in the level of expression, cell diameter distribution and cellular population profile. The most striking difference was the substantial overlap of peptidergic and non-peptidergic markers in human DRG, raising the question of whether this commonly utilized demarcation exists among the human nociceptor population. Our findings highlight the importance of the use of human sensory tissues as a translational bridge from preclinical model to clinical testing of new pain therapeutics.
Materials and Methods
Tissue preparation
All experiments were approved by the Institutional Animal Care and Use Committee at the University of Texas at Dallas and conducted in accordance with the National Institutes of Health and International Association for the Study of Pain guidelines. 8-week old male C57BL/6 mouse dorsal root ganglion tissue (bilateral L3-L5) was dissected after gross hemi-section of the spinal column, embedded in OCT together in a cryomold and flash frozen immediately over dry ice. Human dorsal root ganglion (L5) tissue from donors was frozen in dry ice at the time of extraction and shipped on dry ice by Anabios. The pain history of 2 of the donors was unremarkable but one had a history of low back pain, which was likely discogenic. Because the lumbar discs are innervated largely by L1 and L2 DRG [14] and not by L5 DRG, which we used in this study, we did not consider this DRG to have a clear pain phenotype, although this possibility cannot be excluded. The study was certified as exempt by the University of Texas at Dallas Institutional Review Board because the human DRGs were obtained from organ donors and no identifying information was shared with the research team. Upon arrival, the human DRGs were gradually embedded with OCT in a cryomold by adding small volumes of OCT over dry ice to avoid thawing. All tissues were cryostat sectioned at 20 μm onto SuperFrost Plus charged slides. Sections were only briefly thawed in order to adhere to the slide but were immediately returned to the −20°C cryostat chamber until completion of sectioning. The slides were then immediately utilized for histology.
RNAscope in situ hybridization
RNAscope in situ hybridization (ish) multiplex version 1 was performed as instructed by Advanced Cell Diagnostics (ACD). Slides were removed from the cryostat and immediately transferred to cold (4°C) 10% formalin for 15 minutes. The tissues were then dehydrated in 50% ethanol (5 min), 70% ethanol (5 min) and 100% ethanol (10 min) at room temperature. The slides were air dried briefly and then boundaries were drawn around each section using a hydrophobic pen (ImmEdge PAP pen; Vector Labs). When hydrophobic boundaries had dried, protease IV reagent was added to each section until fully covered. The protease IV incubation period was optimized as recommended by ACD. A 2-minute protease digestion was used for all mouse experiments in order to preserve protein epitope for IHC label. IHC was not performed in human experiments (as NF200 is non-specific for all sensory neurons in human [41]) so the maximum 30 min protease IV digestion was used for all human DRG experiments. Slides were washed briefly in 1X phosphate buffered saline (PBS, pH 7.4) at room temperature. Each slide was then placed in a prewarmed humidity control tray (ACD) containing dampened filter paper and a mixture of Channel 1, Channel 2, and Channel 3 probes (50:1:1 dilution, as directed by ACD due to stock concentrations) was pipetted onto each section until fully submerged. This was performed one slide at a time to avoid liquid evaporation and section drying. The humidity control tray was placed in a HybEZ oven (ACD) for 2 hours at 40°C. All experiments involved a target of interest in Channel 1 in combination with CALCA (CGRP) probe in channel 2, and P2RX3 (P2X3R) in channel 3. A table of all the probes used is shown in Table 1 (note: some targets were not assessed in mouse due to the availability of expression patterns in the literature). Following probe incubation, the slides were washed two times in 1X RNAscope wash buffer and returned to the oven for 30 minutes after submersion in AMP-1 reagent. Washes and amplification were repeated using AMP-2, AMP-3 and AMP-4 reagents with a 15-min, 30-min, and 15-min incubation period, respectively. AMP-4 ALT B (Channel 1 = Atto 550, Channel 2 = Alexa 488, Channel 3 = Atto 647) was used for all mouse experiments and AMP-4 ALT C (Channel 1 = Atto 550, Channel 2 = Atto 647, Channel 3 = Alexa 488) was used for all human experiments. Slides were then washed two times in 0.1M phosphate buffer (PB, pH7.4). Mouse slides were then processed for immunohistochemistry, while human slides were incubated in 1:5000 DAPI in 0.1M PB for 1 min before being washed, air dried, and cover-slipped with Prolong Gold Antifade mounting medium.
Table 1.
Summary table of Advanced Cell Diagnostics (ACD) RNAscope probes.
| ACD Probe Cat No. | |||
|---|---|---|---|
| mRNA | Gene name | Mouse | Human |
| CD68 | Macrophage Antigen CD68 | 316611 | 560591 |
| CALCA | Calcitonin gene-related peptide | 417961 | 605551 |
| CHRNA6 | Cholinergic Receptor Nicotinic Alpha 6 | 485191 | 516981 |
| CHRNA9 | Cholinergic Receptor Nicotinic Alpha 9 | 525921 | 489681 |
| CHRNA10 | Cholinergic Receptor Nicotinic Alpha 10 | 525911 | 516971 |
| CRLF2 | Cytokine Receptor Like Factor 2 | 525931 | 445261 |
| HCN1 | Hyperpolarization Activated Cyclic Nucleotide Gated Potassium Channel 1 | 423651 | 516991 |
| HCN2 | Hyperpolarization Activated Cyclic Nucleotide Gated Potassium Channel 2 | 427001 | 517021 |
| HDAC6 | Histone Deacetylase 6 | 422871 | 485141 |
| KCNB1 | Potassium Voltage-Gated Channel Subfamily B Member 1; Kv2.1 | 517001 | |
| KCNS1 | Potassium Voltage-Gated Channel Modifier Subfamily S Member 1; Kv9.1 | 525941 | 487141 |
| P2RX3 | Purinergic Receptor P2X3 | 521611 | 406301 |
| SCN9A | Sodium Voltage-Gated Channel Alpha Subunit 9; Nav1.7 | 562251 | |
| SCN10A | Sodium Voltage-Gated Channel Alpha Subunit 10; Nav1.8 | 406291 | |
| TRPA1 | Transient Receptor Potential Cation Channel Subfamily A Member 1 | 400211 | 311951 |
| TRPV1 | Transient Receptor Potential Cation Channel Subfamily V Member 1 | 313331 | 415381 |
Immunohistochemistry (IHC)
Given the non-specificity for NF200 in human DRG [41], IHC for NF200 was only conducted on mouse tissues. After completion of RNAscope in situ hybridization, mouse slides were incubated in blocking buffer (10% Normal Goat Serum, 0.3% Triton-X 100 in 0.1M PB) for 1 hour at room temperature while being shielded from light. Slides were placed in a light-protected humidity-controlled tray and incubated in primary antibody (mouse-anti-Neurofilament 200; clone N52; Sigma) at 1:500 in blocking buffer overnight at 4°C. The next day, slides were washed two times in 0.1M PB, and then incubated in secondary antibody (goat-anti-mouse H&L 405; 1:2000) for 1 hour at room temperature. Sections were washed two times in 0.1M PB, air dried, and cover-slipped with Prolong Gold Antifade mounting medium.
In a single experiment, IHC was conducted on fresh-frozen human DRG tissue for Nav1.7 protein. The tissues were sectioned, fixed and dehydrated as described above. Following PAP-pen labeling, IHC was performed as described above but using a knockout-validated [19] mouse monoclonal antibody (mouse-anti-Nav1.7; cloneN68/6; NeuroMab; RRID:AB_10672976; 1:250) and with secondary antibody (goat-anti-mouse IgG1 555; 1:2000).
Tissue Quality Check
All tissues were checked for RNA quality by using a positive control probe cocktail (ACD) which contains probes for high, medium and low-expressing mRNAs that are present in all cells (ubiquitin C > Peptidyl-prolyl cis-trans isomerase B > DNA-directed RNA polymerase II subunit RPB1). Only tissues that showed signal for all 3 positive controls were used for experiments. Sample sizes for human are 2–3 and for mouse it is 3. Human donor information is provided in Table 2. Although this is a small sample size, the mRNA is high quality with little-to-no variability between samples in all experiments. This speaks to the importance and robustness of the RNA quality check included with the RNAscope technology. RNA quality is not usually considered or assessed in standard in situ hybridization studies. A negative control probe against the bacterial DapB gene (ACD) was used to check for non-specific/background label.
Table 2.
Human donor information.
| Sex | Age | Cause of Death | Patient history |
|---|---|---|---|
| Male | 22 | Asphyxiation | Depression; heavy drinker; recreational drug use |
| Male | 56 | Stroke | Cardiac disease; hypertension; type 1 diabetes; recreational drug use, tobacco smoker, chronic low back pain due to severe stenosis of L3–4 and moderate stenosis of L4–5. |
| Female | 48 | Head Trauma | Depression; GERD; past tobacco use; recreational drug use; alcohol use |
Image Analysis
DRG sections were imaged on an Olympus FV3000 confocal microscope at 20X magnification. One image was acquired of each mouse DRG section, and 3 sections were imaged per mouse (total: ~9 images/experiment for an n=3). Two-to-three images were acquired of each human DRG section, and 3 sections were imaged per human (total: ~18 images/experiment for an n=2; or 27 images for n=3). The sections imaged were chosen at random, but preference was given to sections that did not have any sectioning artifact, or sections that encompassed the entire DRG bulb. As such, the mouse analysis is a mix of L3-L5 DRG sections. The acquisition parameters were set based on guidelines for the FV3000 provided by Olympus. In particular, the gain was kept at the default setting 1, HV ≤ 600, offset = 4 (based on HI-LO settings; doesn’t change between experiments), and laser power ≤ 10% (but generally the laser power was ≤ 5% for our experiments). The raw image files were brightened and contrasted in Olympus CellSens software (v1.18), and then analyzed manually one cell at a time for expression of Calca and P2rx3 with the target of interest. Cell diameters were measured using the polyline tool. Total neuron counts for mouse were assessed by counting all of the NF200, CALCA, P2RX3, and target of interest positive-neurons. There were two mouse experiments (Chrna6 and Trpa1) where the NF200-IHC staining did not work; most likely due to the protease-digestion required for the ish label. For these experiments, we over-brightened the image to reveal background autofluorescence in order to count the total number of neurons in the image and to measure cell diameters. Total neuron counts for human samples were acquired by counting all of the CALCA/P2RX3-positive cells and all cells that were clearly outlined by DAPI (satellite cell) signal and contained lipofuscin in the overlay image. Each human DRG experiment was quality checked to account for variability in image representation of the nociceptor / neuronal populations. This was performed by assessing the distribution of each neuronal population for each experiment: the total nociceptor (human CALCA, P2RX3, CALCA/P2RX3 populations), total non-nociceptor (human neurons that were not positive for CALCA nor P2RX3), and total neuronal populations (Supplementary tables 1–3).
Large globular structures and/or signal that auto-fluoresced in all 3 channels (488, 550, and 647; appears white in the overlay images) was considered to be background lipofuscin and was not analyzed. Aside from adjusting brightness/contrast, we performed no digital image processing to subtract background. We attempted to optimize automated imaging analysis tools for our purposes, but these tools were designed to work with fresh, low background rodent tissues, not human samples taken from older organ donors. As such, we chose to implement a manual approach in our imaging analysis in which we used our own judgement of the negative/positive controls and target images to assess mRNA label. Images were not analyzed in a blinded fashion.
IHC for Nav1.7 was analyzed by drawing regions of interest (ROIs) around each neuron (not including the lipofuscin) and measuring its fluorescence intensity in CellSens. Neurons were considered positive for Nav1.7 if they showed a fluorescence intensity value above background signal. The background signal was assessed by drawing ROIs around all neurons (not including lipofuscin) in the negative control which was only exposed to the secondary antibody and averaging that value.
Given that each target of interest was assessed for colocalization with CALCA and P2RX3, we combined all of the CALCA/P2RX3 data from each experiment to create the CALCA/P2RX3 data figure. The sample size for all mouse experiments was 12 (analysis of 36 lumbar DRG sections for the CALCA/P2RX3 datasheet) and the sample size for all human experiments was 2–3 (analysis of 35, 37 or 18 sections, respectively for the CALCA/P2RX3 datasheet). Experiments that utilized sections from the same mouse and/or human were averaged together to generate the final values in the CALCA/P2RX3 data figure. N values shown represent biological replicates. The CALCA/P2RX3 histograms are the combination of all CALCA/P2RX3-positive cells that were co-positive for the target of interest in each experiment and a single experiment for mouse and human where the diameters of all CALCA and P2RX3 neurons were measured.
Data Analysis and Statistics
Graphs and statistical analyses were generated using GraphPad Prism version 7.01 (GraphPad Software, Inc. San Diego, CA USA). Statistical differences between groups were assessed using two-way ANOVA with Bonferroni or unpaired t-tests as indicated in the text and figure captions. Statistical analyses were only conducted for datasets that had an n=3 or greater for both mouse and human. All statistics including F and exact p-values are shown in Supplementary tables 4. All data are represented as mean ± SEM, with p <0.05 considered significant. Sample sizes are displayed in the graphs of each figure (mouse n=3, human n=2–3). All pie-charts were generated by averaging the population distributions from each mouse (n=3, 3 sections/mouse) or human (n=2- or 3, 2–3 sections/human). The total number of neurons assessed between all subjects is indicated on the pie-chart; however, the pie-charts were generated based on population averages as noted above. Relative frequency distribution histograms with a Gaussian distribution curve were generated using the diameters of all target-positive neurons. Raw histogram data is provided in supplementary file 1.
Results
CALCA (CGRP) and P2RX3 (P2X3R) label distinct neuronal subpopulations in mouse, but not human, DRG
We conducted RNAscope in situ hybridization on mouse and human DRGs to assess the population distribution of the standard nociceptor subpopulation markers, CALCA (CGRP) and P2RX3 (P2X3R). We selected P2RX3 mRNA expression rather than IB4-binding because IB4-binding glycoproteins do not exist in primate DRG [10]. While antibodies against proteins for these markers label distinct neuronal subpopulations in mouse, these markers show substantial overlap in protein labeling in the rat [38]. We observed significantly more CALCA mRNA-positive cells that were co-positive for P2RX3-mRNA in humans than in mouse (Fig 1A–D). Interestingly, these differences stemmed from a significant increase in CALCA mRNA-expressing neurons in human DRG (Fig 1D). In mouse DRG, 32.9% and 52.4% of the neurons expressed Calca and P2rx3, respectively (Fig 1D). However, in human DRG, the percentage of CALCA-expressing neurons increased to 59.3% while the percentage of P2RX3 remained similar to mouse at 52.6% (Fig 1D). These data indicate that there are more CALCA mRNA-positive neurons in human DRG. We observed that CALCA mRNA expression was most abundant in neurons that were ~35–40um but was moderately to weakly positive in other neurons suggesting a gradient of expression within this entire population. However, we did not formally quantify this gradient of expression as we did not base our analyses on signal intensity. The Calca and P2rx3 mRNA-positive cells in mouse ranged from ~12 – 59 μm in diameter with (Fig 1E; supplementary file 1). The human CALCA and P2RX3 mRNA-positive populations ranged from 27 – 125 μm (Fig 1F; supplementary file 1).
Figure 1. Distribution of peptidergic (CALCA; CGRP) and non-peptidergic (P2RX3; P2X3R) populations in mouse and human DRG.
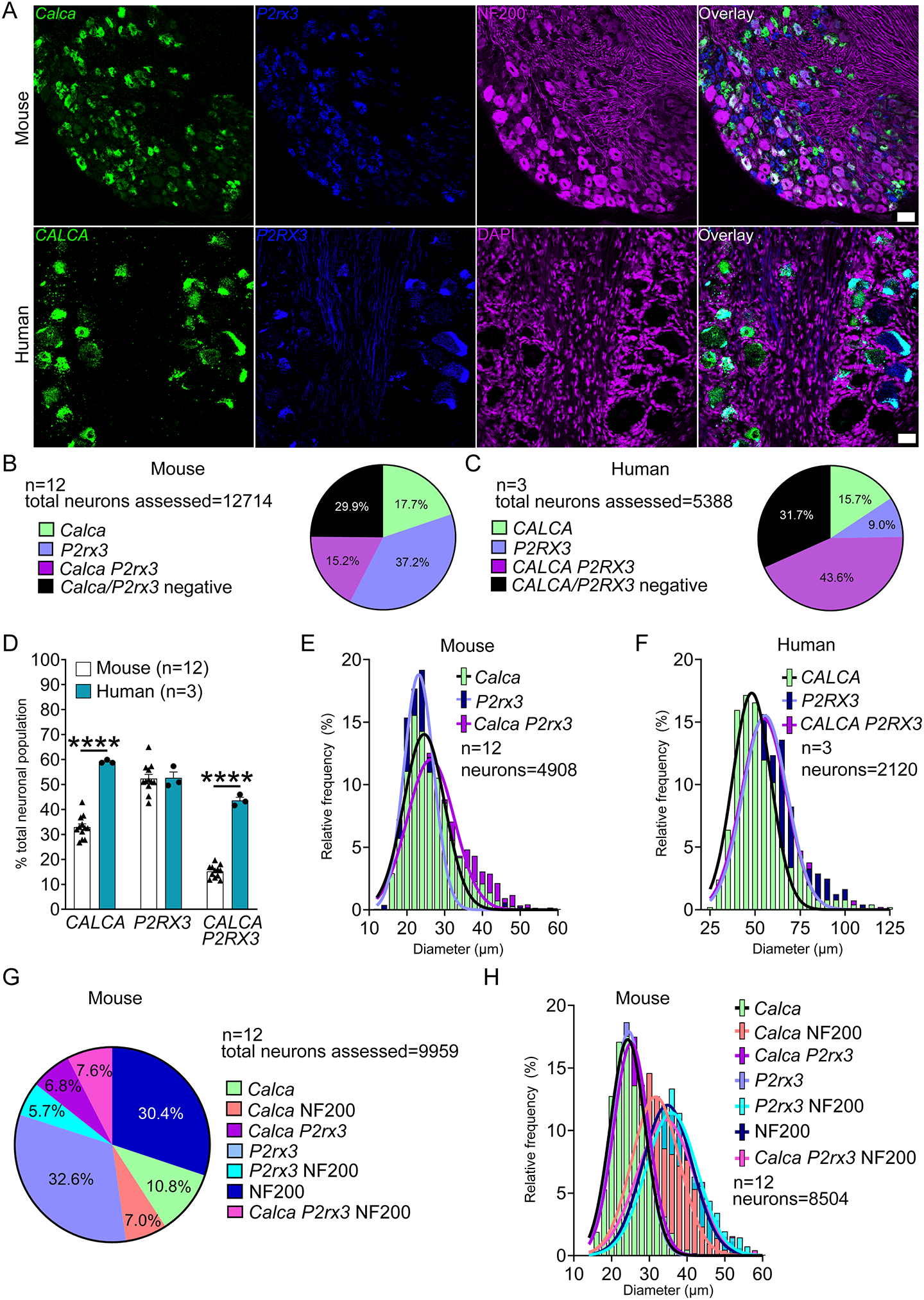
A) Representative 20x images of mouse and human DRG labeled with RNAscope in situ hybridization for CALCA (green) and P2RX3 (blue) mRNA. Mouse and human DRG were costained for NF200 protein (purple) and DAPI (purple), respectively. B) Pie chart representation of CALCA and P2RX3 subpopulations in mouse and C) human DRG. D) The total number of CALCA-expressing neurons (all CALCA, and CALCA/P2RX3 neurons) was significantly increased in human DRG compared to mouse, but the population of P2RX3-expressing neurons (all P2RX3 and CALCA/P2RX3 neurons) was unchanged. The percentage of CALCA/P2RX3 co-expressing neurons was significantly increased in human DRG compared to mouse. E) Histogram with Gaussian distribution displaying the diameters of CALCA, P2RX3, and CALCA/P2RX3 co-expressing neurons in mouse and F) human DRG. G) Sensory neuron subpopulations were further divided in mouse DRG based on expression with Neurofilament-200 (NF200). H) Histogram with Gaussian distribution displaying the size distribution of Calca, P2rx3, and NF200 sensory neuron subpopulations in mouse DRG. Two way ANOVA with Bonferroni ****p<0.0001. Scale bar = 50 μm.
Approximately 29.9% and 31.7% of the neurons in mouse and human, respectively, were negative for CALCA and/or P2RX3 mRNA expression. These neurons in mice were NF200-positive neurons (Fig 1G). In mice NF200 was expressed in an exclusively NF200-positive population as well as subpopulations that coexpressed Calca and P2rx3 (Fig 1G, supplementary file 1). However, because NF200 antibodies label all DRG neurons in humans [41], we were unable to label these cells with a clear neurochemical marker initially. Further analyses of other mRNA targets in human revealed that these CALCA/P2RX3 mRNA-negative neurons were large in size, potentially fitting the size profile for an A-β mechanoreceptor which are known to be large cells, and expressing some specific neurochemical markers (for example KCNS1) (Supplementary Fig 1A–B).
Voltage-gated sodium channels
We focused on SCN10A and SCN9A, which encode the Nav1.8 and Nav1.7 proteins, respectively. We chose these two targets based on their relative specificity for expression in the DRG and genetic evidence linking these channels to human pain disorders or pain insensitivities [13; 21]. SCN10A, (Nav1.8) is a nociceptor-specific marker in rodent DRG at the protein and mRNA levels [1; 12; 42] and has been the gene-promotor of choice in creating nociceptor-specific transgenic lines [44]. In human DRG, we found that SCN10A mRNA is exclusively expressed in CALCA and P2RX3 mRNA-positive neurons with no expression in the CALCA/P2RX3-negative neuronal population (Fig 2A–C). SCN10A positive neurons ranged in size from 31 – 90 μm (Fig 2D; supplementary file 1). This suggests that Nav1.8 is specifically expressed in the nociceptor population in human DRG neurons, much as it is in mice and rats. We did not analyze SCN10A expression in mice given the large literature on this target [50].
Figure 2. Distribution of SCN10A (Nav1.8) mRNA in human DRG.
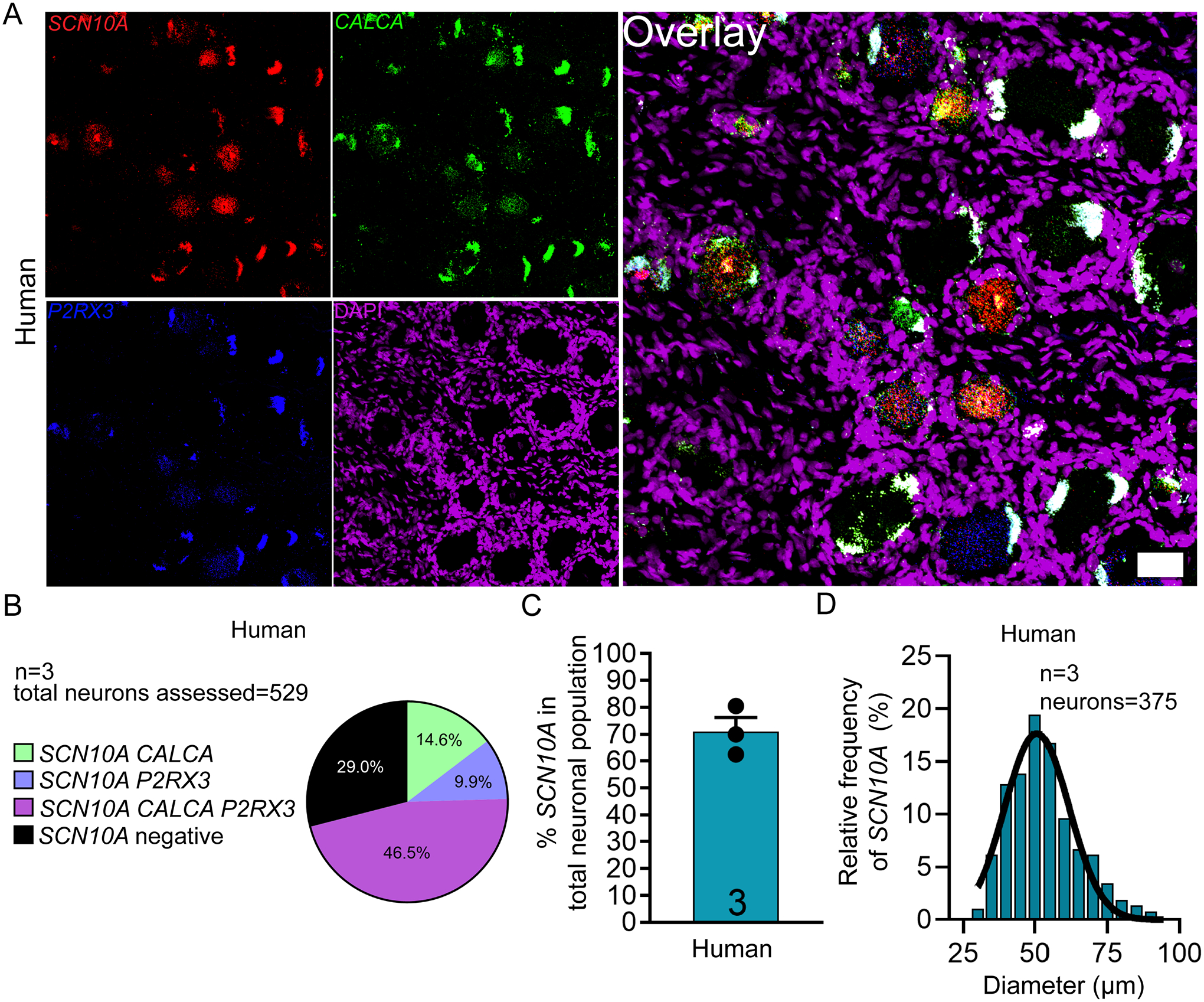
A) Representative 20x images of human DRG labeled with RNAscope in situ hybridization for CALCA (green) and P2RX3 (blue), SCN10A (red), and DAPI (purple). B) Pie chart representation of all SCN10A-positive sensory neuron subpopulations in human DRG. SCN10A was expressed exclusively in neurons that co-expressed CALCA and P2RX3. C) SCN10A was expressed in 71% of all sensory neurons in human DRG. D) Histogram with Gaussian distribution displaying the size profile of all SCN10A-positive neurons in human DRG. Scale bar = 50 μm.
At the protein level, Nav1.7 (SCN9A) has been shown to be expressed in the majority of small diameter nociceptors and a small subset of large diameter NF200-positive neurons in rodent [6; 11; 28]. In human, these Nav1.7-protein positive nociceptors co-express TRPV1 and CGRP [28]. While the protein seems to be nociceptor exclusive, single-cell sequencing of mouse DRG demonstrates that SCN9A mRNA is expressed in all sensory neurons [49]. We found that SCN9A mRNA is not exclusive to nociceptors in human but is expressed in 96.0% of neurons in human DRG (Fig 3A–C) which ranged in size from 31 – 143 μm (Fig 3D; supplementary file 1). At the protein level, using a knockout mouse-validated antibody, we observed that Nav1.7 protein is also expressed in a large population of sensory neurons (~90.5%) (Fig 3E–F) which ranged in size from 32 – 130 μm (Fig 3G; supplemental file 1). Consistent with previous reports, we also observed SCN9A mRNA and Nav1.7 protein present in the axons of DRG neurons (supplementary Fig 2A–B)) supporting the notion that Nav1.7 mRNA and protein is found along the length of DRG axons [6] across species.
Figure 3. Distribution of SCN9A mRNA and Nav1.7 protein in human DRG.
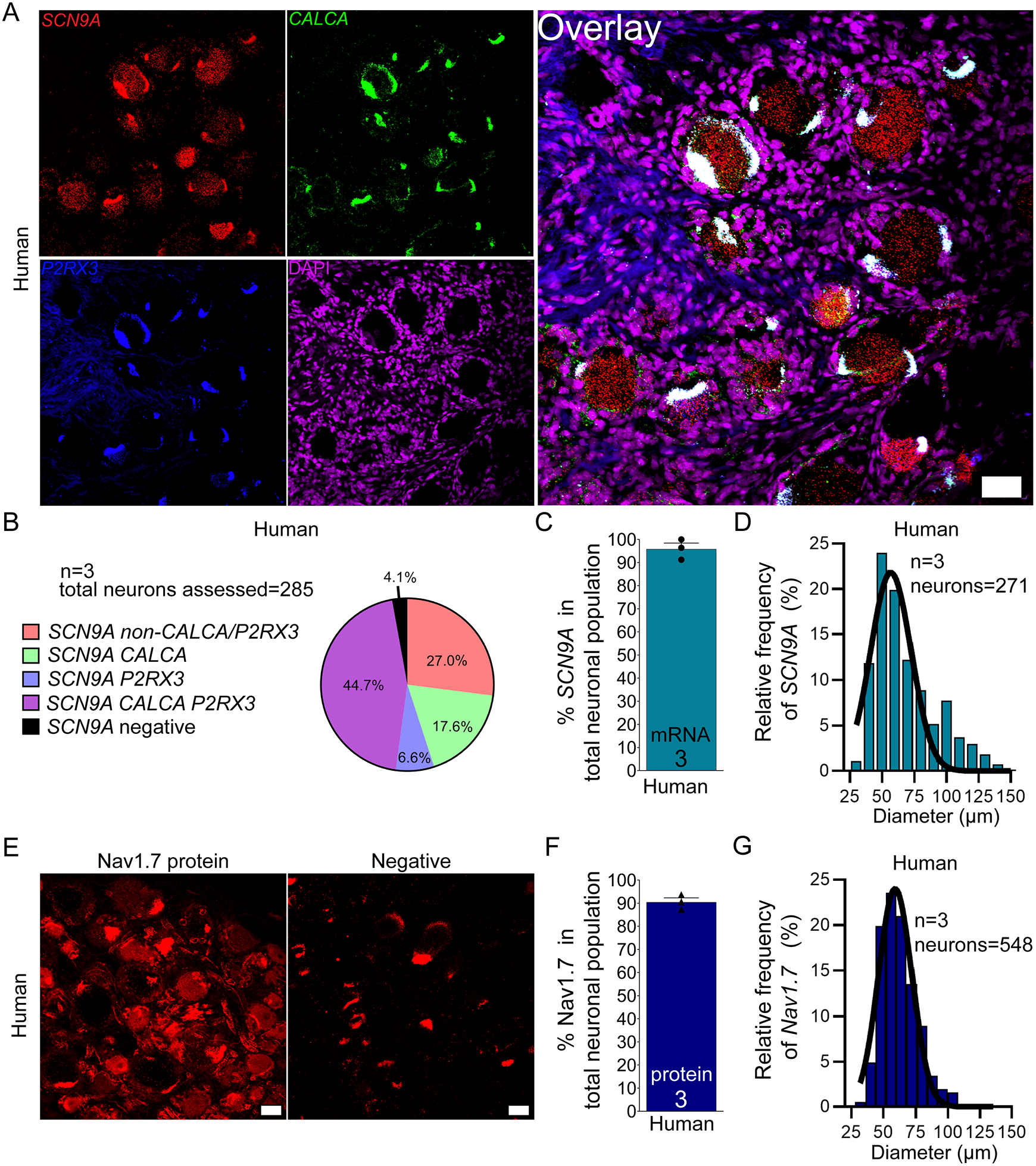
A) Representative 20x images of human DRG labeled with RNAscope in situ hybridization for CALCA (green) and P2RX3 (blue), SCN9A (red), and DAPI (purple). B) Pie chart representation of all Scn9a-positive sensory neuron subpopulations in human DRG. C) SCN9A was expressed in 96% of sensory neurons in human DRG. D) Histogram with Gaussian distribution displaying the size profile of all SCN9A -positive neurons in human DRG. E) Representative 20x images of Nav1.7 (red) protein staining using a knockout-validated monoclonal antibody. The negative control was imaged at the same settings (no primary with secondary antibody). The negative control shows background lipofuscin (large globular structures) and myelin autofluorescence. F) Nav1.7 protein was expressed in 90.5% of sensory neurons in human DRG. G) Histogram with Gaussian distribution displaying the size profile of all Nav1.7 protein-positive neurons in human DRG. 20x scale bar = 50 μm.
Voltage-gated potassium channels
We focused on two voltage-gated potassium channels with links to neuropathic pain. KCNS1 encodes an accessory subunit (Kv9.1) for many voltage gated potassium channels that is downregulated after nerve injury [47]. KCNS1 mRNA is expressed in virtually all large diameter A-beta mechanoreceptors that express NF200 in rat [47]. We found that KCNS1 is expressed in 55.5% and 42.4% of sensory neurons in mouse and human DRG, respectively (Fig 4A–E). KCNS1 neurons ranged in size from 16 – 58 μm in mouse, and 54 – 140 μm in human (Fig 4F–G; supplementary file 1). 75.2% of NF200-expressing neurons in mouse and 79.4% of CALCA/P2XR3-negative neurons in human coexpressed KCNS1 (Table 4). Kcns1 may be an excellent molecular marker for the A-beta neuronal population in human DRG.
Figure 4. Distribution of KCNS1 (Kv9.1) mRNA in mouse and human DRG.
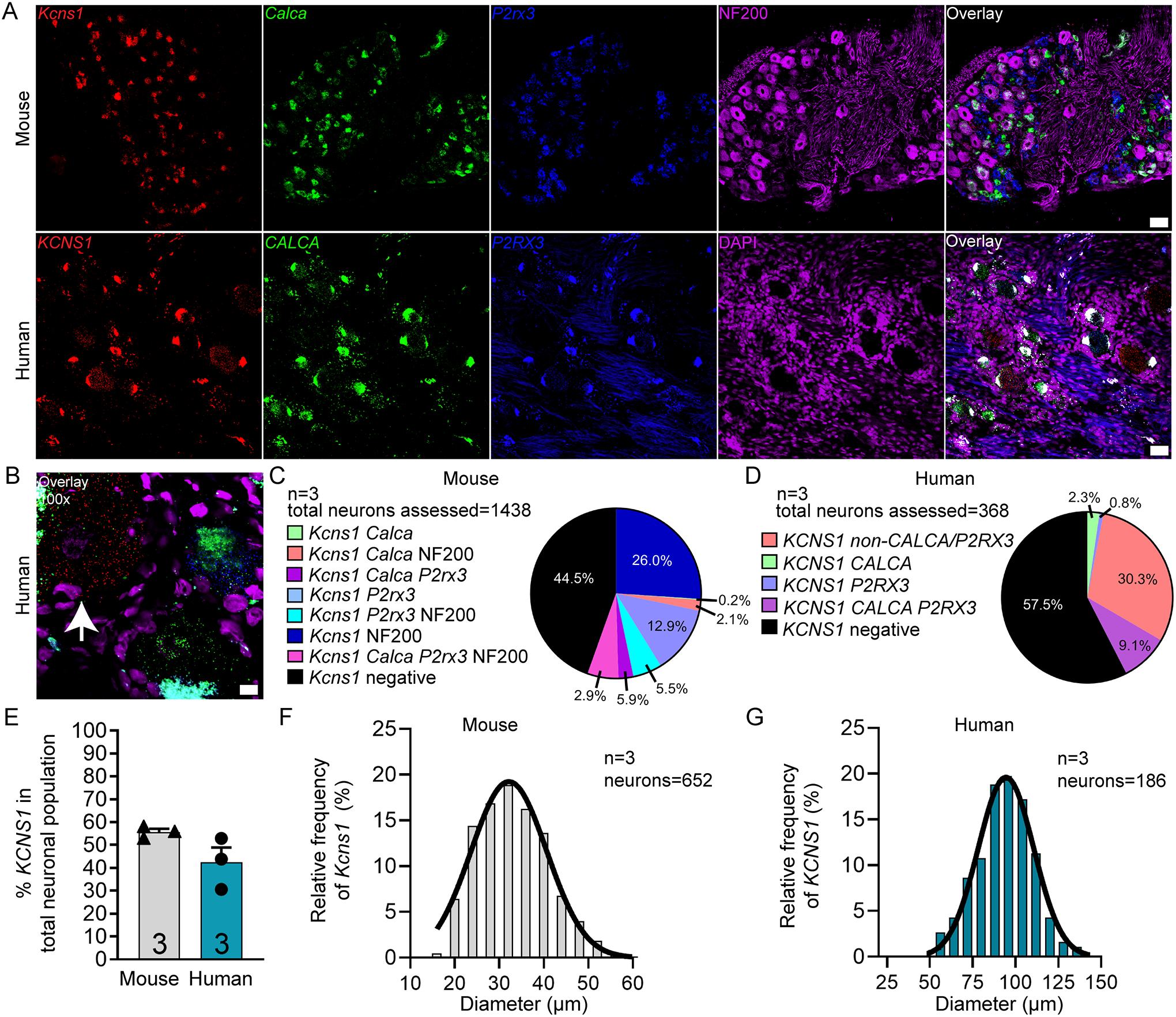
A) Representative 20x images of mouse and human DRG labeled with RNAscope in situ hybridization for CALCA (green), P2RX3 (blue), and KCNS1 (red) mRNA. Mouse and human DRG were costained for NF200 protein (purple) and DAPI (purple), respectively. B) Representative 100x overlay image showing CALCA (green), P2RX3 (blue), KCNS1 (red) and DAPI (purple) signal in human DRG. White arrow points toward a KCNS1-positive neuron. C) Pie chart representation of all KCNS1-positive sensory neuron subpopulations in mouse and D) human DRG. E) KCNS1 was expressed in 55.5% of sensory neurons in mouse DRG and 42.4% in human DRG. F) Histogram with Gaussian distribution displaying the size profile of all KCNS1-positive neurons in mouse and G) human DRG. Unpaired t-test, p>0.5. 20x scale bar = 50 μm. 100x scale bar = 10 μm.
Table 4.
Summary table showing the percentage of CALCA, P2RX3, NF200 and/or non-CALCA/P2RX3 neuronal populations that co-express the target of interest. ND = not detected
| CALCA population | P2RX3 population | NF200 population | Non-CALCA/P2RX3 population | Total # neurons assessed | ||||
|---|---|---|---|---|---|---|---|---|
| Target | Mouse | Human | Mouse | Human | Mouse | Human | Mouse | Human |
| CD68 | non-neuronal | non-neuronal | non-neuronal | non-neuronal | non-neuronal | non-neuronal | ||
| CALCA | 100 | 100 | 29.6 | 82.5 | 29.2 | 0 | 12714 | 5388 |
| CHRNA9 | ND | 40.6 | ND | 38.6 | ND | 1.2 | 468 | |
| CHRNA10 | 3.2 | 5.7 | 2.0 | 5.5 | 2.0 | 2.0 | 1682 | 366 |
| CRLF2 | 78.0 | 6.1 | 73.1 | 6.5 | 71.2 | 2.3 | 1348 | 360 |
| HCN1 | 76.1 | 99.0 | 60.8 | 99.6 | 89.0 | 84.9 | 1483 | 334 |
| HCN2 | 69.5 | 59.3 | 67.8 | 56.4 | 86.1 | 50.3 | 1266 | 331 |
| HDAC6 | 84.4 | 77.1 | 87.9 | 73.9 | 89.7 | 29.1 | 1344 | 326 |
| KCNB1 | 96.2 | 97.2 | 64.8 | 605 | ||||
| KCNS1 | 35.6 | 21.3 | 48.6 | 20.6 | 75.2 | 79.4 | 1176 | 368 |
| P2RX3 | 45.9 | 73.7 | 100 | 100 | 26.6 | 0 | 12714 | 5388 |
| SCN9A | 100 | 100 | 85.6 | 285 | ||||
| SCN10A | 98.9 | 98.1 | 0 | 529 | ||||
| TRPV1 | 64.6 | 88.6 | 29.2 | 89.5 | 6.2 | 34.4 | 1438 | 423 |
| CALCA population | P2RX3 population | Non-CALCA/P2RX3 population | Total # neurons assessed | |||||
| Target | Mouse | Human | Mouse | Human | Mouse | Human | Mouse | Human |
| CHRNA6 | 32.9 | 56.3 | 52.4 | 52.8 | 6.7 | 27.6 | 1194 | 410 |
| TRPA1 | 33.7 | 27.2 | 75.7 | 20.6 | 10.6 | 0.80 | 1606 | 583 |
KCNB1 encodes the Kv2.1 protein which is also known to be downregulated after nerve injury [48]. Because this channel contributes to afterhyperpolarizations, its absence supports high frequency firing of DRG neurons, potentially contributing to neuropathic pain. KCNB1 mRNA is present in a large, mixed population of small, medium and large diameter neurons in rat DRG [48]. In human DRG we similarly observed KCNB1 in 85.4% of all sensory neurons which ranged in size from 32 – 130 μm (Supplementary Fig 3A–D; supplementary file 1)). KCNB1 mRNA was expressed in virtually all neurons that co-expressed CALCA (96.2%) or P2RX3 (97.2%) and was also present in a majority of neurons that were negative for CALCA/P2RX3(64.8%) (Table 4).
Hyperpolarization Activated Cyclic Nucleotide Gated Potassium and Sodium (HCN) Channels
HCN channels are expressed in excitable tissues where they contribute to rebound depolarizations and are regulated by cyclic nucleotide signaling. HCN2, in particular, has been implicated in chronic pain states using knockout mouse models [16; 29; 45], but HCN1 and 2 are both expressed in DRG. We found that HCN1 mRNA was expressed in 70.9% of sensory neurons in mouse DRG, and 94.4.% in human (Fig 5A–E). These neurons ranged in size from 15 – 57 μm in mouse, and 27 – 138 μm in human (Fig 5F–G).
Figure 5. Distribution of HCN1 mRNA in mouse and human DRG.
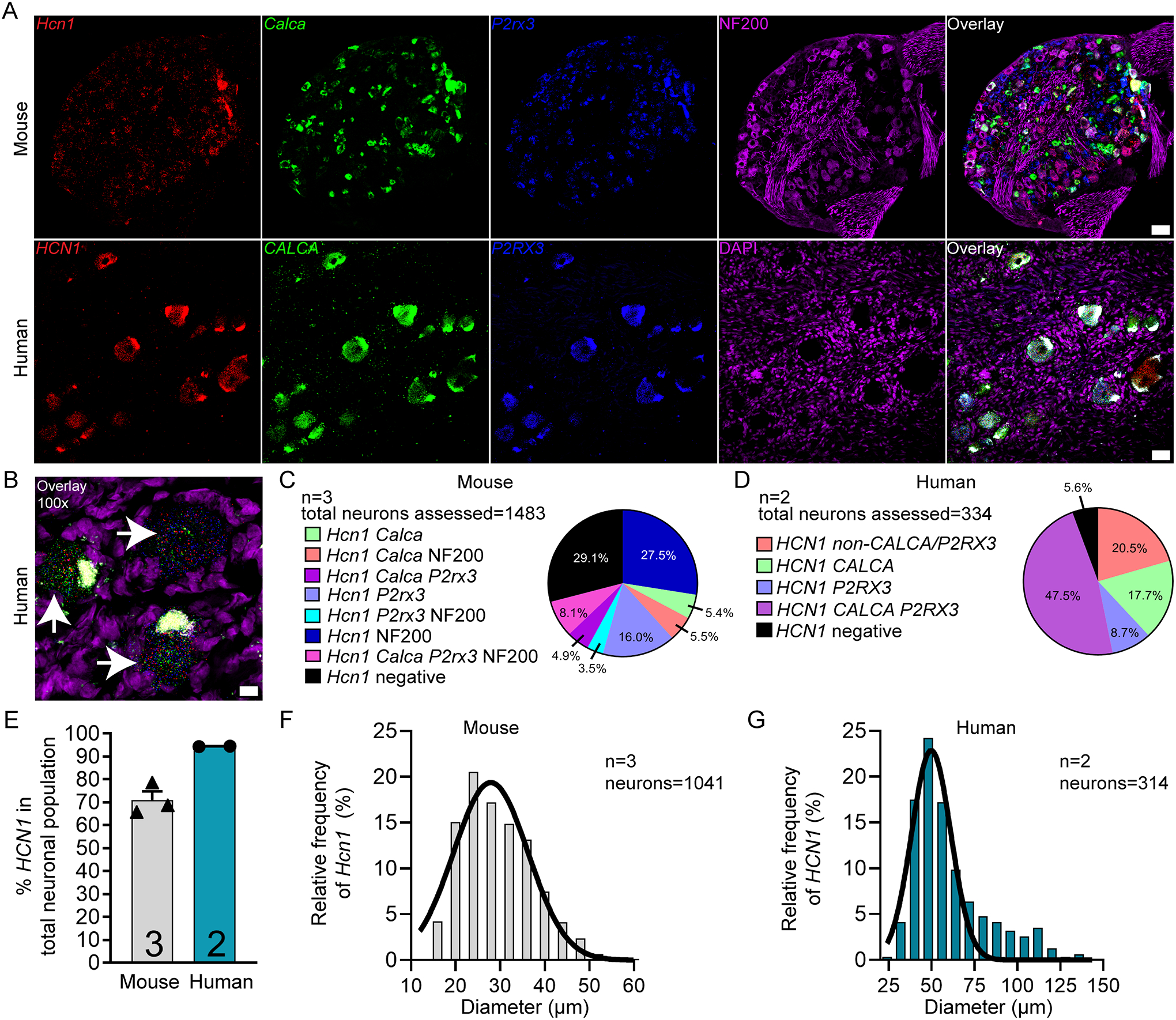
A) Representative 20x images of mouse and human DRG labeled with RNAscope in situ hybridization for CALCA (green), P2RX3 (blue), and HCN1 (red) mRNA. Mouse and human DRG were costained for NF200 protein (purple) and DAPI (purple), respectively. B) Representative 100x overlay image showing CALCA (green), P2RX3 (blue), HCN1 (red) and DAPI (purple) signal in human DRG. White arrows point toward HCN1-positive neurons. C) Pie chart representation of all HCN1-positive sensory neuron subpopulations in mouse and D) human DRG. E) HCN1 was expressed in 70.9% of sensory neurons in mouse DRG and 94.4% in human DRG. F) Histogram with Gaussian distribution displaying the size profile of all HCN1-positive neurons in mouse and G) human DRG. 20x scale bar = 50 μm. 100x scale bar = 10 μm.
HCN2 mRNA was found in 71.5% and 44.0% of all sensory neurons in mouse and human DRG, respectively (Supplementary Fig 4 A–E)). These neurons ranged in size from 16 – 58 μm in mouse, and 48 – 155 μm in human (Supplementary Fig 4F–G; supplementary file 1). These marked species differences may have important implications for the role of different HCN isoforms in pain states between mouse models and human patients.
Nicotinic acetylcholine receptors
Nicotinic receptors are prominently expressed in DRG neurons and are known to play a role in sensory neuron excitability [35]. We focused on several subunits, α 6, 9 and 10, that show prominent species differences in DRG mRNA expression or specificity based on RNA sequencing experiments [39]. CHRNA6 (nicotinic acetyl choline receptor alpha 6; nAChRα6) mRNA was present in 30.2% and 43.8% of all mouse and human sensory neurons, respectively (Supplementary Fig 5A–E). Consistent with previous reports, ~ 33% of Calca expressing neurons in mouse also expressed Chrna6 (~ 35% previously reported for the protein [51]) while 52.4% of all P2rx3-expressing neurons co-expressed Chrna6 mRNA (58% previously reported for the protein in the IB4 population [51]) (Table 4). Similar to the protein [51], CHRNA6 was expressed in neurons that ranged from 15 – 44 μm in mouse and 41 – 123 μm in human (Supplementary Fig 5F–G; supplementary file 1).
CHRNA9 (nicotinic acetyl choline receptor alpha 9; nAChRα9) was expressed in 25.6% of all sensory neurons in human DRG but was not detected in the mouse DRG (Fig 6A–D). This finding parallels RNA sequencing results [51]. CHRNA9 mRNA-positive neurons ranged in size from 40 – 109 μm in human DRG (Fig 6E; supplementary file 1)). nAChRα9 can form functional channels with nAChRα10 (CHRNA10; nicotinic acetyl choline receptor alpha 10). CHRNA10 was only found in only 1.6% and 3.7% of mouse and human sensory neurons, respectively (Supplemental Fig 6A–E)). There were very few neurons positive for CHRNA10 mRNA in mouse and human; however, the size of these neurons ranged from 21 – 50 μm in mouse and 44 – 101 μm in human (Supplemental Fig 6F–G). nAChRα9 - nAChRα10 heteromers would only be possible in human DRG, and there they would likely be limited to a small population of nociceptors. Our findings suggest that homomeric nAChRα9 channels would be the most likely functional channel in nociceptors, but only in human DRG.
Figure 6. Distribution of CHRNA9 mRNA in mouse and human DRG.
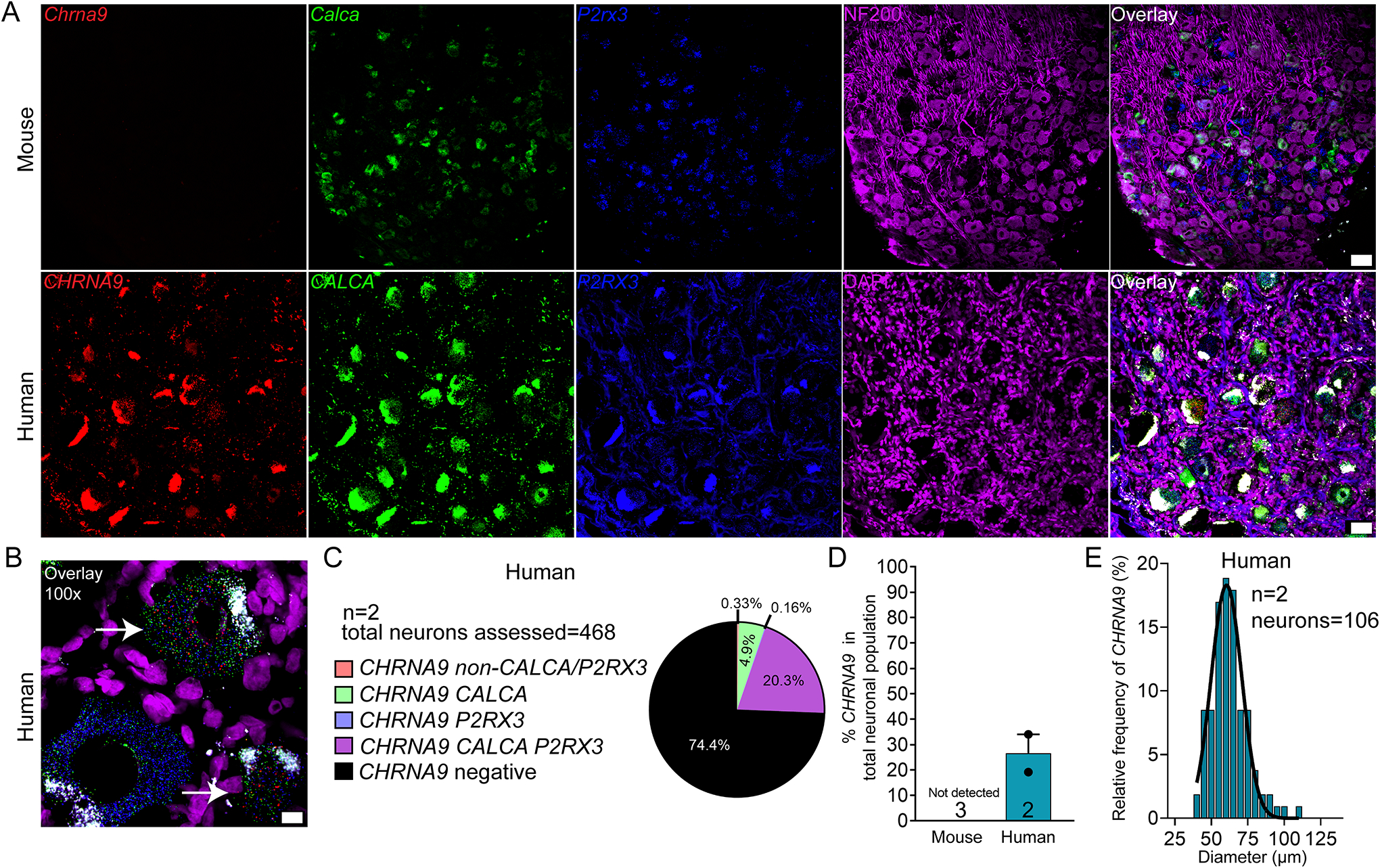
A) Representative 20x images of mouse and human DRG labeled with RNAscope in situ hybridization for CALCA (green), P2RX3 (blue), and CHRNA9 (red) mRNA. Mouse and human DRG were costained for NF200 protein (purple) and DAPI (purple), respectively. B) Representative 100x overlay image showing CALCA (green), P2RX3 (blue), CHRNA9 (red) and DAPI (purple) signal in human DRG. White arrows point toward CHRNA9-positive neurons. C) Pie chart representation of all CHRNA9-positive sensory neuron subpopulations in mouse and human DRG. D) CHRNA9 mRNA was not detected in mouse DRG but was expressed in 25.6% of sensory neurons in human DRG. E) Histogram with Gaussian distribution displaying the size profile of all CHRNA 9-positive neurons in human DRG. 20x scale bar = 50 μm. 100x scale bar = 10 μm.
Transient receptor potential cation (TRP) channels
We focused on TRPV1 (transient receptor potential cation channel subfamily v member 1) and TRPA1 (transient receptor potential cation channel subfamily a member 1) as both channels are strongly implicated in acute and chronic pain, and both channels are intensely studied as therapeutic targets [4; 24]. We found that TRPV1 mRNA was expressed in significantly more sensory neurons in human (74.7%) than in mouse (30.3%) (Fig 7A–E). TRPV1--positive neurons ranged in size from 17 – 45 μm in mouse, and 28 – 122 μm in human (Fig 7F–G; supplementary file 1). However, unlike in mouse, TRPV1 mRNA was expressed in almost all CALCA (88.6%) and P2RX3 (89.5%) co-expressing neurons in human DRG (Table 4).
Figure 7. Distribution of TRPV1 mRNA in mouse and human DRG.
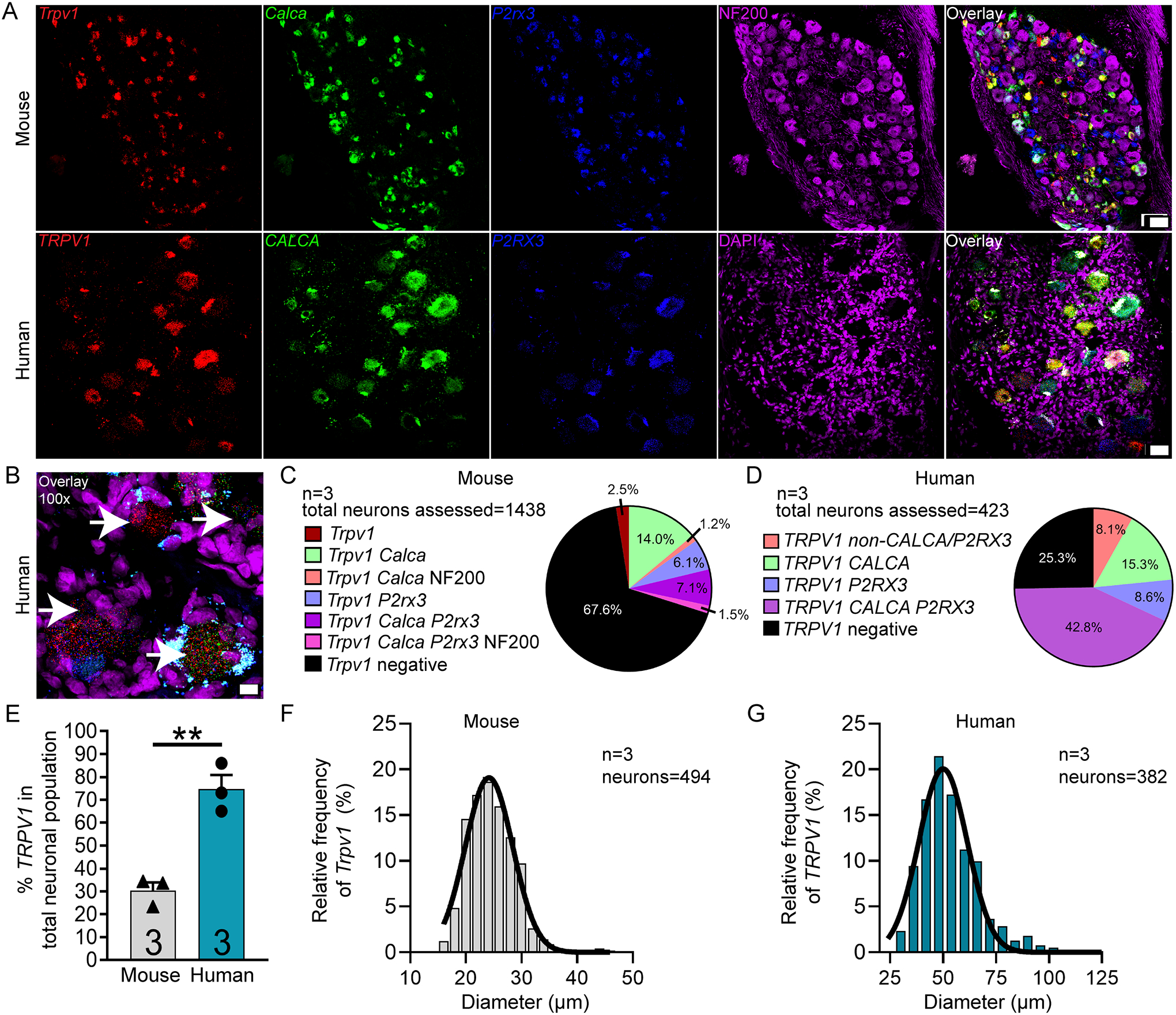
A) Representative 20x images of mouse and human DRG labeled with RNAscope in situ hybridization for CALCA (green), P2RX3 (blue), and TRPV1 (red) mRNA. Mouse and human DRG were costained for NF200 protein (purple) and DAPI (purple), respectively. B) Representative 100x overlay image showing CALCA (green), P2RX3 (blue), TRPV1 (red) and DAPI (purple) signal in human DRG. White arrows point toward Trpv1-positive neurons. C) Pie chart representation of all TRPV1-positive sensory neuron subpopulations in mouse and D) human DRG. E) TRPV1 was expressed in significantly more neurons in human DRG (74.7%) than in mouse DRG (32.4%). F) Histogram with Gaussian distribution displaying the size profile of all TRPV1-positive neurons in mouse and G) human DRG. Unpaired t-test, **p<0.01. 20x scale bar = 50 μm. 100x scale bar = 10 μm.
Conversely, TRPA1 mRNA was significantly more abundant in mouse DRG (53.2%) than in human (16.3%) (Fig 8A–E), consistent with published findings [3]. Trpa1 mRNA was expressed in 75.7% of P2rx3-expressing neurons in mouse (Table 4). TRPA1 positive neurons ranged in size from 12 – 43 μm in mouse and 29 – 77 μm in human (Fig 8F–G; Supplementary file 11).
Figure 8. Distribution of TRPA1 mRNA in mouse and human DRG.
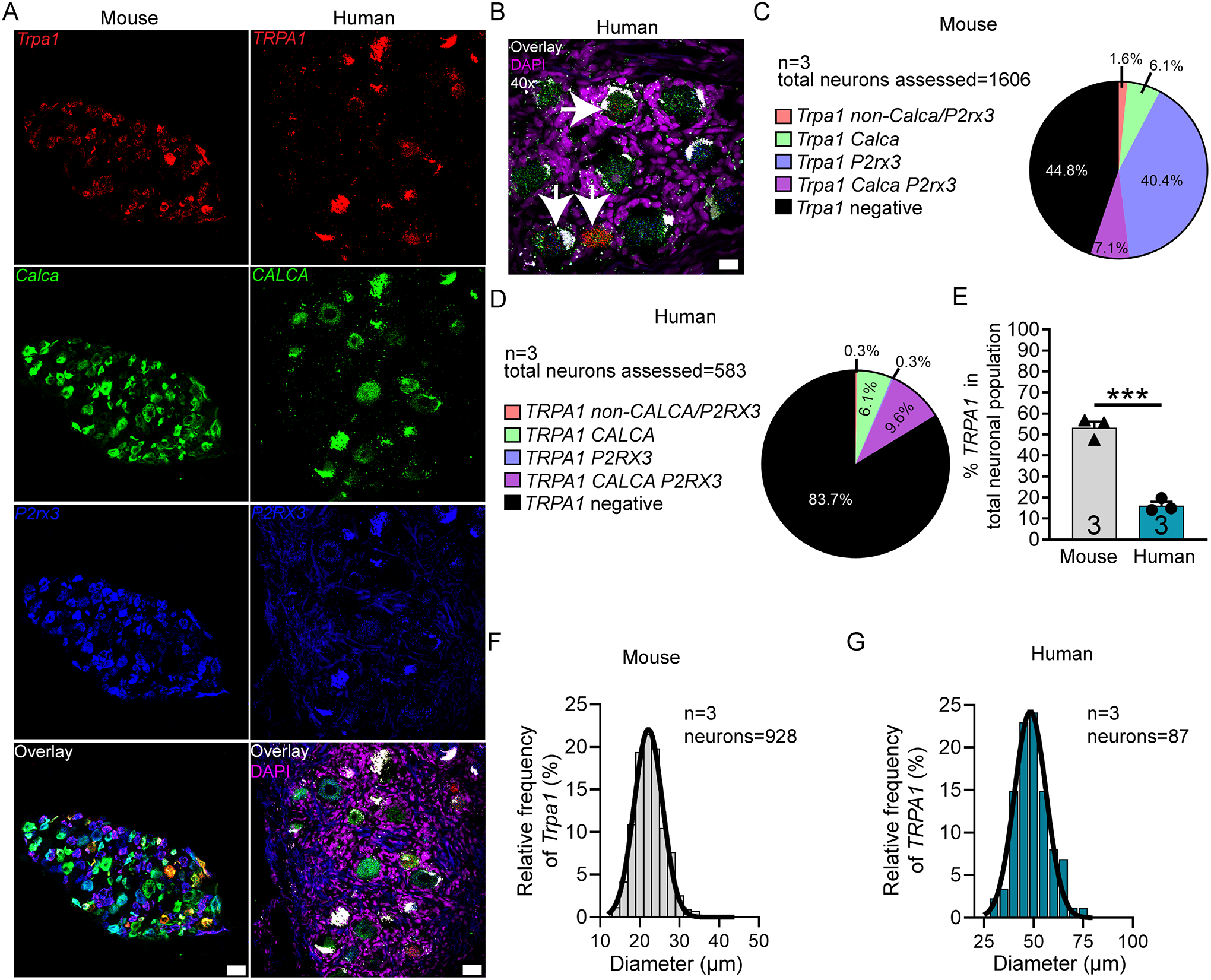
A) Representative 20x images of mouse and human DRG labeled with RNAscope in situ hybridization for CALCA (green), P2RX3 (blue), and TRPA1 (red) mRNA. Human DRG was costained for DAPI (purple). B) Representative 40x overlay image showing CALCA (green), P2RX3 (blue), Trpa1 (red) and DAPI (purple) signal in human DRG. White arrows point toward TRPA1-positive neurons. C) Pie chart representation of all TRPA1-positive sensory neuron subpopulations in mouse and D) human DRG. E) TRPA1 was expressed in significantly more neurons in mouse DRG (55.2%) than in human DRG (16.3%). F) Histogram with Gaussian distribution displaying the size profile of all TRPA1-positive neurons in mouse and G) human DRG. Unpaired t-test, ***p<0.001. 20x scale bar = 50 μm. 40x scale bar = 20 μm.
Immune and other markers
CD68 is a classic marker for macrophages but is also expressed in other cells such as lymphocytes, fibroblasts and endothelial cells [9]. Macrophages are resident to the DRG in rodent and primates and they have been associated with the development of chronic pain in model systems and in patients with neuropathic pain [2; 36; 40; 43]. We did observe abundant CD68 mRNA expression in both species, which was confined to the extra-neuronal space particularly the epineurium and nerve (Supplemental Fig 7A–C). In both mouse and human DRG, the majority, if not all, of the CD68 mRNA signal appeared to be non-neuronal. In human, CD68 mRNA was present in satellite glial cells. While we were not convinced that Cd68 mRNA was present in neurons, we cannot rule this out as scarce Cd68 mRNA puncta was observed in a few neurons along the outside perimeter which could be neuronal or satellite glial cell in origin given the proximity of these cells to the neuron border.
CRLF2 (Cytokine receptor like factor 2) is a co-receptor for cytokine thymic stromal lymphopoietin (TSLP) that has been implicated in itch in mouse models [52]. Our previous RNA sequencing experiments demonstrate a large difference in expression for the CRLF2 gene between mouse and human DRG [39]. Consistent with those RNA sequencing results, we found CRLF2 mRNA expression in 70.4% and 4.0% of all sensory neurons in mouse and human DRG, respectively (Supplemental Fig 8A–E). In mouse, Crlf2 neurons ranged in size from 16 – 59 μm (Supplemental Fig 8F; supplementary file 1). In human, there were very few positive neurons for CRLF2, but these neurons ranged in size from 45 – 118 μm (Supplemental Fig 8G; supplementary file 1).
Finally, we assessed HDAC6 (Histone deacetylase 6) due its recent implication in a variety of neuropathic pain states [25; 30; 31]. HDAC6 had a unique subcellular localization in the mouse and human DRG where the mRNA was almost exclusively confined to the nucleus (Supplemental Fig 9A–B). We observed HDAC6 signal in both neurons and non-neuronal cell types in human, particularly satellite glial cells (Supplemental Fig 9B). HDAC6 mRNA was present in 86.6% and 59.6% of sensory neurons in mouse and human DRG, respectively (Supplemental Fig 9C–E). In mouse, the majority of Calca (84.4%), P2rx3 (87.9%), and NF200-positive neurons (89.7%) also expressed Hdac6 (Table 4). In human, the majority of CALCA (77.1%) and P2RX3-positive neurons (73.9%) also co-expressed HDAC6, but it was less abundant in CALCA/P2RX3-negative cells (29.1%) (Table 4). HDAC6-positive neurons ranged in size from 15 – 56 μm in mouse and 36 – 133 μm in human (Supplemental Fig 9F–G; supplementary file 1).
Discussion
Herein, we conducted RNAscope in situ hybridization to elucidate the distribution of sensory neuron populations in mouse and human DRG. We did this to investigate not only intraspecies homology in expression patterns, but also to begin classifying human DRG neurons based on their gene expression profiles, cell body diameter, and function. We observed species divergence and similarities for a variety of pain processing and/or drug targets, consistent with a growing body of evidence highlighting such difference [20; 38; 41; 54]. These data affirm the importance of confirming target homology between rodent models and humans to ascertain translational potential and therapeutic efficacy.
Classically, the field has used NF200 to label large diameter Aβ mechanoreceptors, and markers such as CGRP and P2X3R/IB4 to label peptidergic and non-peptidergic Aδ and C-nociceptors, respectively. However, recent work suggests that these markers are not-specific for these neuronal subpopulations in some mammals. For example, NF200 is expressed in all sensory neurons in human DRG [41] and CGRP and IB4 significantly overlap in rat DRG [38]. Similarly, we identified a large population of CGRP and P2X3R co-expressing neurons in human DRG that was not present in mouse. One of the major contributors to this difference was a large increase in the number of CGRP-expressing neurons in human (59.3% in human compared to 32.9% in mouse). Indeed, others have reported CALCA mRNA expression in 50–70% of human DRG neurons [17; 26], although these findings have not yet been validated at the protein level using a knockout validated antibody. CGRP is well-known to be involved in mouse and human pain neurobiology [23], however, its higher abundance in human may help explain why monoclonal antibodies against CGRP are so effective for the treatment of migraine [15]. It remains to be seen if CGRP can be targeted to treat other types of pain in humans. Nevertheless, our results highlight an important species difference that is at the core of pain anatomy classification terminology and this difference may have important functional consequences.
We also observed strong species divergence in the expression of CHRNA9 mRNA. Multiple sources including bulk and single-cell RNA sequencing in addition to our RNAscope data have confirmed that CHRNA9 is not expressed in mouse DRG [39; 49]. However, CHRNA9 is found in human DRG [39] and is expressed in 25.6% of all human sensory neurons. It is unknown if this species difference exists within the Rodentia order. The data for CHRNA9 expression in rat has been inconclusive with some reporting the mRNA as undetectable [22], while others have reported protein expression in every sensory neuron using antibody-based methods [8]. It is likely that other human-specific molecular markers exist in the human DRG, indicating that potential therapeutic targets may be overlooked by exclusive studies in rodent. Bulk [39] and single-cell sequencing of human DRG will be instrumental to the identification of these molecular targets.
We identified species differences in key molecular targets for pain sensory neurobiology. For example, TRPV1 has been studied for decades and is considered to be an essential component of the nociceptive sensory system [4; 24]. However, we detected TRPV1 mRNA in almost the entire nociceptor population in human (77.7% of all neurons) compared to only 32.4% in mouse. TRPV1 antagonism has shown therapeutic efficacy for acute pain in human individuals [32], but these compounds have side-effects that may preclude their clinical use [5]. It remains to be seen if enhanced efficacy will be found in clinical trials compared to mouse model findings but this might be predicted given the greater distribution of expression. Contrastingly, we found significantly less TRPA1 mRNA expressing neurons in human DRG (16.3%) than mouse (55.2%). These drastic species differences affirm the importance of identifying target expression homology in humans for even long-standing therapeutic targets.
Over the last 30 years, the field has shifted towards almost exclusively using mice in pain research [33]. This transition came around the time of the advent of the Cre-LoxP system and the increasing availability of transgenic mouse lines. While transgenic animals offer an invaluable means to explore the function of a specific gene/protein, this transition may not have been the best step forward for clinical translatability. As we have shown, the human sensory system is different between mouse and human DRG in some very fundamental ways, offering insight into one of the main challenges for clinical translation. We observed SCN9A mRNA in virtually all neurons in human DRG while the protein was present in most neurons (86.3%) but was absent from very large diameter cells. These data corroborate clinical reports of human patients with loss-of-function mutations in Nav1.7 who are insensitive to pain but have normal proprioception [18]. Importantly, these findings do point out that mRNA expression does not necessarily predict protein distribution and warrant that further investigations at the protein-level are necessary to identify functional significance. This issue is further highlighted by a recent study showing discordance between functional opioid receptor signaling and receptor mRNA expression in acutely dissociated human DRG neurons [34].
A recent clinical trial tested a non-specific HCN blocker in an acute pain condition in which topical capsaicin was applied to the skin. However, this therapeutic failed to show efficacy in reducing pain [27]. This same compound, ivabradine, causes robust analgesia in mouse models of neuropathic pain [45; 46; 53]. Interestingly, HCN2 shows substantially higher expression in nociceptors and large diameter neurons in mouse (71.5% of all mouse sensory neurons) compared to its lower expression in mainly larger-diameter neurons in human (44% of all human neurons). HCN1 shows higher expression in human DRG neurons than in mice. Our findings suggest that specifically targeting HCN1 in chronic pain states in humans may be a better strategy than focusing on HCN2, as mouse models have indicated. This raises an important observation that we extend from previous RNAseq experiments [36; 39], that expression of gene families tends to be consistent from species to species within the DRG, but specific genes within these families often have quite divergent expression patterns. It is possible that HCN expression changes in pathological pain, thereby ivabradine may be more effective in neuropathic pain patients. We propose validation of therapeutic targets in human DRG using single-cell sequencing, which has notably not been done on human DRG neurons, and RNAscope in situ hybridization assays as critical tools for clinical translation.
Supplementary Material
Table 3.
Summary table of mRNA expression for each target in mouse vs human DRG. These data summarize all data shown in the pie charts of all figures. ND = not detected
| % total neuronal population | % nociceptors (CGRP and/or P2X3R positive) | % other neurons (CGRP and/or P2X3R-negative) | Total # neurons assessed | |||||
|---|---|---|---|---|---|---|---|---|
| Target | Mouse | Human | Mouse | Human | Mouse | Human | Mouse | Human |
| CD68 | non-neuronal | non-neuronal | non-neuronal | non-neuronal | non-neuronal | non-neuronal | ||
| CALCA | 32.9 | 59.1 | 32.9 | 59.1 | 0 | 0 | 12714 | 5388 |
| CHRNA6 | 30.3 | 43.8 | 27.7 | 34.4 | 2.6 | 9.40 | 1194 | 410 |
| CHRNA9 | ND | 25.6 | ND | 25.3 | ND | 0.30 | 468 | |
| CHRNA10 | 1.65 | 3.68 | 1.32 | 3.01 | 0.33 | 0.67 | 1682 | 366 |
| CRLF2 | 70.4 | 4.57 | 52.1 | 3.83 | 18.3 | 0.74 | 1348 | 360 |
| HCN1 | 70.9 | 94.4 | 43.4 | 73.9 | 27.5 | 20.5 | 1483 | 334 |
| HCN2 | 71.5 | 44.0 | 44.2 | 24.9 | 27.3 | 19.1 | 1266 | 331 |
| HDAC6 | 86.6 | 58.0 | 60.2 | 48.1 | 26.4 | 9.90 | 1344 | 326 |
| KCNB1 | 85.4 | 65.2 | 20.2 | 605 | ||||
| KCNS1 | 55.5 | 42.4 | 29.4 | 12.1 | 26.1 | 30.3 | 1176 | 368 |
| P2RX3 | 66.3 | 52.6 | 66.3 | 52.6 | 0 | 0 | 12714 | 5388 |
| SCN9A | 96.0 | 69.0 | 27.0 | 285 | ||||
| SCN10A | 71.0 | 71.0 | 0 | 529 | ||||
| TRPA1 | 55.2 | 16.3 | 53.6 | 16.0 | 1.60 | 0.30 | 1606 | 583 |
| TRPV1 | 32.4 | 74.7 | 29.8 | 66.6 | 2.54 | 8.10 | 1438 | 423 |
ACKNOWLEDGEMENTS:
This work was supported by Merck Sharp and Dohme.
Footnotes
The authors declare no conflicts of interest
References
- [1].Akopian AN, Sivilotti L, Wood JN. A tetrodotoxin-resistant voltage-gated sodium channel expressed by sensory neurons. Nature 1996;379(6562):257–262. [DOI] [PubMed] [Google Scholar]
- [2].Albrecht DS, Ahmed SU, Kettner NW, Borra RJH, Cohen-Adad J, Deng H, Houle TT, Opalacz A, Roth SA, Melo MFV, Chen L, Mao J, Hooker JM, Loggia ML, Zhang Y. Neuroinflammation of the spinal cord and nerve roots in chronic radicular pain patients. Pain 2018;159(5):968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Anand U, Otto WR, Facer P, Zebda N, Selmer I, Gunthorpe MJ, Chessell IP, Sinisi M, Birch R, Anand P. TRPA1 receptor localisation in the human peripheral nervous system and functional studies in cultured human and rat sensory neurons. Neurosci Lett 2008;438(2):221–227. [DOI] [PubMed] [Google Scholar]
- [4].Basbaum AI, Bautista DM, Scherrer G, Julius D. Cellular and molecular mechanisms of pain. Cell 2009;139(2):267–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bevan S, Quallo T, Andersson DA. Trpv1. Handb Exp Pharmacol 2014;222:207–245. [DOI] [PubMed] [Google Scholar]
- [6].Black JA, Frezel N, Dib-Hajj SD, Waxman SG. Expression of Nav1.7 in DRG neurons extends from peripheral terminals in the skin to central preterminal branches and terminals in the dorsal horn. Mol Pain 2012;8:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Burma NE, Leduc-Pessah H, Fan CY, Trang T. Animal models of chronic pain: Advances and challenges for clinical translation. J Neurosci Res 2017;95(6):1242–1256. [DOI] [PubMed] [Google Scholar]
- [8].Butts KA, Weinberg J, Young AH, Phillips AG. Glucocorticoid receptors in the prefrontal cortex regulate stress-evoked dopamine efflux and aspects of executive function. Proc Natl Acad Sci U S A 2011;108(45):18459–18464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chistiakov DA, Killingsworth MC, Myasoedova VA, Orekhov AN, Bobryshev YV. CD68/macrosialin: not just a histochemical marker. Lab Invest 2017;97(1):4–13. [DOI] [PubMed] [Google Scholar]
- [10].Davidson S, Copits BA, Zhang J, Page G, Ghetti A, Gereau RW. Human sensory neurons: Membrane properties and sensitization by inflammatory mediators. Pain 2014;155(9):1861–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dib-Hajj SD, Cummins TR, Black JA, Waxman SG. Sodium channels in normal and pathological pain. Annu Rev Neurosci 2010;33:325–347. [DOI] [PubMed] [Google Scholar]
- [12].Dib-Hajj SD, Tyrrell L, Cummins TR, Black JA, Wood PM, Waxman SG. Two tetrodotoxin-resistant sodium channels in human dorsal root ganglion neurons. FEBS Lett 1999;462(1–2):117–120. [DOI] [PubMed] [Google Scholar]
- [13].Eberhardt MJ, Leffler A. [Pain and analgesia : Mutations of voltage-gated sodium channels]. Schmerz 2017;31(1):14–22. [DOI] [PubMed] [Google Scholar]
- [14].Edgar MA. The nerve supply of the lumbar intervertebral disc. J Bone Joint Surg Br 2007;89(9):1135–1139. [DOI] [PubMed] [Google Scholar]
- [15].Edvinsson L The CGRP Pathway in Migraine as a Viable Target for Therapies. Headache 2018;58 Suppl 1:33–47. [DOI] [PubMed] [Google Scholar]
- [16].Emery EC, Young GT, Berrocoso EM, Chen L, McNaughton PA. HCN2 ion channels play a central role in inflammatory and neuropathic pain. Science 2011;333(6048):1462–1466. [DOI] [PubMed] [Google Scholar]
- [17].Giaid A, Gibson SJ, Ibrahim BN, Legon S, Bloom SR, Yanagisawa M, Masaki T, Varndell IM, Polak JM. Endothelin 1, an endothelium-derived peptide, is expressed in neurons of the human spinal cord and dorsal root ganglia. Proc Natl Acad Sci U S A 1989;86(19):7634–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Goldberg YP, MacFarlane J, MacDonald ML, Thompson J, Dube MP, Mattice M, Fraser R, Young C, Hossain S, Pape T, Payne B, Radomski C, Donaldson G, Ives E, Cox J, Younghusband HB, Green R, Duff A, Boltshauser E, Grinspan GA, Dimon JH, Sibley BG, Andria G, Toscano E, Kerdraon J, Bowsher D, Pimstone SN, Samuels ME, Sherrington R, Hayden MR. Loss-of-function mutations in the Nav1.7 gene underlie congenital indifference to pain in multiple human populations. Clin Genet 2007;71(4):311–319. [DOI] [PubMed] [Google Scholar]
- [19].Grubinska B, Chen L, Alsaloum M, Rampal N, Matson DJ, Yang C, Taborn K, Zhang M, Youngblood B, Liu D, Galbreath E, Allred S, Lepherd M, Ferrando R, Kornecook TJ, Lehto SG, Waxman SG, Moyer BD, Dib-Hajj S, Gingras J. Rat NaV1.7 loss-of-function genetic model: Deficient nociceptive and neuropathic pain behavior with retained olfactory function and intra-epidermal nerve fibers. Mol Pain 2019;15:1744806919881846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Haberberger RV, Barry C, Dominguez N, Matusica D. Human Dorsal Root Ganglia. Front Cell Neurosci 2019;13:271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hameed S Nav1.7 and Nav1.8: Role in the pathophysiology of pain. Mol Pain 2019;15:1744806919858801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hone AJ, Meyer EL, McIntyre M, McIntosh JM. Nicotinic acetylcholine receptors in dorsal root ganglion neurons include the alpha6beta4* subtype. FASEB J 2012;26(2):917–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Iyengar S, Ossipov MH, Johnson KW. The role of calcitonin gene-related peptide in peripheral and central pain mechanisms including migraine. Pain 2017;158(4):543–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Julius D TRP channels and pain. Annu Rev Cell Dev Biol 2013;29:355–384. [DOI] [PubMed] [Google Scholar]
- [25].Krukowski K, Ma J, Golonzhka O, Laumet GO, Gutti T, van Duzer JH, Mazitschek R, Jarpe MB, Heijnen CJ, Kavelaars A. HDAC6 inhibition effectively reverses chemotherapy-induced peripheral neuropathy. Pain 2017;158(6):1126–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Landry M, Aman K, Dostrovsky J, Lozano AM, Carlstedt T, Spenger C, Josephson A, Wiesenfeld-Hallin Z, Hokfelt T. Galanin expression in adult human dorsal root ganglion neurons: initial observations. Neuroscience 2003;117(4):795–809. [DOI] [PubMed] [Google Scholar]
- [27].Lee MC, Bond S, Wheeler D, Scholtes I, Armstrong G, McNaughton P, Menon D. A randomised, double-blind, placebo-controlled crossover trial of the influence of the HCN channel blocker ivabradine in a healthy volunteer pain model: an enriched population trial. Pain 2019;160(11):2554–2565. [DOI] [PubMed] [Google Scholar]
- [28].Li Y, North RY, Rhines LD, Tatsui CE, Rao G, Edwards DD, Cassidy RM, Harrison DS, Johansson CA, Zhang H, Dougherty PM. DRG Voltage-Gated Sodium Channel 1.7 Is Upregulated in Paclitaxel-Induced Neuropathy in Rats and in Humans with Neuropathic Pain. J Neurosci 2018;38(5):1124–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Liu X, Zhang L, Jin L, Tan Y, Li W, Tang J. HCN2 contributes to oxaliplatin-induced neuropathic pain through activation of the CaMKII/CREB cascade in spinal neurons. Mol Pain 2018;14:1744806918778490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ma J, Huo X, Jarpe MB, Kavelaars A, Heijnen CJ. Pharmacological inhibition of HDAC6 reverses cognitive impairment and tau pathology as a result of cisplatin treatment. Acta Neuropathol Commun 2018;6(1):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ma J, Trinh RT, Mahant ID, Peng B, Matthias P, Heijnen CJ, Kavelaars A. Cell-specific role of histone deacetylase 6 in chemotherapy-induced mechanical allodynia and loss of intraepidermal nerve fibers. Pain 2019;160(12):2877–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Manitpisitkul P, Brandt M, Flores CM, Kenigs V, Moyer JA, Romano G, Shalayda K, Mayorga AJ. TRPV1 antagonist JNJ-39439335 (mavatrep) demonstrates proof of pharmacology in healthy men: a first-in-human, double-blind, placebo-controlled, randomized, sequential group study. Pain Rep 2016;1(4):e576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mogil JS. Animal models of pain: progress and challenges. Nat Rev Neurosci 2009;10(4):283–294. [DOI] [PubMed] [Google Scholar]
- [34].Moy JK, Hartung JE, Duque MG, Friedman R, Nagarajan V, Loeza-Alcocer E, Koerber HR, Christoph T, Schroder W, Gold MS. Distribution of functional opioid receptors in human dorsal root ganglion neurons. Pain 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Naser PV, Kuner R. Molecular, Cellular and Circuit Basis of Cholinergic Modulation of Pain. Neuroscience 2018;387:135–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].North RY, Li Y, Ray P, Rhines LD, Tatsui CE, Rao G, Johansson CA, Zhang H, Kim YH, Zhang B, Dussor G, Kim TH, Price TJ, Dougherty PM. Electrophysiological and transcriptomic correlates of neuropathic pain in human dorsal root ganglion neurons. Brain 2019;142(5):1215–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Percie du Sert N, Rice AS. Improving the translation of analgesic drugs to the clinic: animal models of neuropathic pain. Br J Pharmacol 2014;171(12):2951–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Price TJ, Flores CM. Critical evaluation of the colocalization between calcitonin gene-related peptide, substance P, transient receptor potential vanilloid subfamily type 1 immunoreactivities, and isolectin B4 binding in primary afferent neurons of the rat and mouse. J Pain 2007;8(3):263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ray P, Torck A, Quigley L, Wangzhou A, Neiman M, Rao C, Lam T, Kim JY, Kim TH, Zhang MQ, Dussor G, Price TJ. Comparative transcriptome profiling of the human and mouse dorsal root ganglia: an RNA-seq-based resource for pain and sensory neuroscience research. Pain 2018;159(7):1325–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Rosen S, Ham B, Mogil JS. Sex differences in neuroimmunity and pain. J Neurosci Res 2017;95(1–2):500–508. [DOI] [PubMed] [Google Scholar]
- [41].Rostock C, Schrenk-Siemens K, Pohle J, Siemens J. Human vs. Mouse Nociceptors - Similarities and Differences. Neuroscience 2018;387:13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sangameswaran L, Delgado SG, Fish LM, Koch BD, Jakeman LB, Stewart GR, Sze P, Hunter JC, Eglen RM, Herman RC. Structure and function of a novel voltage-gated, tetrodotoxin-resistant sodium channel specific to sensory neurons. J Biol Chem 1996;271(11):5953–5956. [DOI] [PubMed] [Google Scholar]
- [43].Shepherd AJ, Copits BA, Mickle AD, Karlsson P, Kadunganattil S, Haroutounian S, Tadinada SM, de Kloet AD, Valtcheva MV, McIlvried LA, Sheahan TD, Jain S, Ray PR, Usachev YM, Dussor G, Krause EG, Price TJ, Gereau RWt, Mohapatra DP. Angiotensin II Triggers Peripheral Macrophage-to-Sensory Neuron Redox Crosstalk to Elicit Pain. J Neurosci 2018;38(32):7032–7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Stirling LC, Forlani G, Baker MD, Wood JN, Matthews EA, Dickenson AH, Nassar MA. Nociceptor-specific gene deletion using heterozygous NaV1.8-Cre recombinase mice. Pain 2005;113(1–2):27–36. [DOI] [PubMed] [Google Scholar]
- [45].Tsantoulas C, Lainez S, Wong S, Mehta I, Vilar B, McNaughton PA. Hyperpolarization-activated cyclic nucleotide-gated 2 (HCN2) ion channels drive pain in mouse models of diabetic neuropathy. Sci Transl Med 2017;9(409):eaam6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tsantoulas C, Mooney ER, McNaughton PA. HCN2 ion channels: basic science opens up possibilities for therapeutic intervention in neuropathic pain. Biochem J 2016;473(18):2717–2736. [DOI] [PubMed] [Google Scholar]
- [47].Tsantoulas C, Zhu L, Shaifta Y, Grist J, Ward JP, Raouf R, Michael GJ, McMahon SB. Sensory neuron downregulation of the Kv9.1 potassium channel subunit mediates neuropathic pain following nerve injury. J Neurosci 2012;32(48):17502–17513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Tsantoulas C, Zhu L, Yip P, Grist J, Michael GJ, McMahon SB. Kv2 dysfunction after peripheral axotomy enhances sensory neuron responsiveness to sustained input. Exp Neurol 2014;251:115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Usoskin D, Furlan A, Islam S, Abdo H, Lonnerberg P, Lou D, Hjerling-Leffler J, Haeggstrom J, Kharchenko O, Kharchenko PV, Linnarsson S, Ernfors P. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci 2015;18(1):145–153. [DOI] [PubMed] [Google Scholar]
- [50].Waxman SG, Zamponi GW. Regulating excitability of peripheral afferents: emerging ion channel targets. Nat Neurosci 2014;17(2):153–163. [DOI] [PubMed] [Google Scholar]
- [51].Wieskopf JS, Mathur J, Limapichat W, Post MR, Al-Qazzaz M, Sorge RE, Martin LJ, Zaykin DV, Smith SB, Freitas K, Austin JS, Dai F, Zhang J, Marcovitz J, Tuttle AH, Slepian PM, Clarke S, Drenan RM, Janes J, Al Sharari S, Segall SK, Aasvang EK, Lai W, Bittner R, Richards CI, Slade GD, Kehlet H, Walker J, Maskos U, Changeux JP, Devor M, Maixner W, Diatchenko L, Belfer I, Dougherty DA, Su AI, Lummis SC, Imad Damaj M, Lester HA, Patapoutian A, Mogil JS. The nicotinic alpha6 subunit gene determines variability in chronic pain sensitivity via cross-inhibition of P2X2/3 receptors. Sci Transl Med 2015;7(287):287ra272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wilson SR, The L, Batia LM, Beattie K, Katibah GE, McClain SP, Pellegrino M, Estandian DM, Bautista DM. The epithelial cell-derived atopic dermatitis cytokine TSLP activates neurons to induce itch. Cell 2013;155(2):285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Young GT, Emery EC, Mooney ER, Tsantoulas C, McNaughton PA. Inflammatory and neuropathic pain are rapidly suppressed by peripheral block of hyperpolarisation-activated cyclic nucleotide-gated ion channels. Pain 2014;155(9):1708–1719. [DOI] [PubMed] [Google Scholar]
- [54].Zhang X, Priest BT, Belfer I, Gold MS. Voltage-gated Na(+) currents in human dorsal root ganglion neurons. Elife 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


