Abstract
Single cell sequencing (SCS) has impacted many areas of cancer research and improved our understanding of intratumor heterogeneity, the tumor microenvironment, metastasis and therapeutic resistance. The development and refinement of SCS technologies has led to massive reductions in costs, increased cell throughput and improved reproducibility, paving the way for clinical applications. However, before translational applications can be realized, there are a number of logistical and technical challenges that must be overcome. This review discusses past cancer research studies, emerging technologies and future clinical applications that are bound to transform cancer medicine.
Introduction
The field of single cell genomics has progressed rapidly over the last 10 years, since the development of the first single cell DNA sequencing (scDNA-seq) method (Navin et al., 2011) and single cell RNA sequencing (scRNA-seq) method (Tang et al., 2009) for mammalian cells. While initial studies were limited to sequencing only a few cells at a time, the development of high-throughput systems including nanowells (Fan et al., 2015a; Gierahn et al., 2017), microdroplets (Lan et al., 2017; Macosko et al., 2015) and microfluidic platforms (Wang et al., 2012) have made it possible to sequence thousands of cells in parallel, while reducing costs to less than $1 per cell (Svensson et al., 2018). Developments in whole-genome amplification (WGA) and whole-transcriptome-amplification (WTA) chemistries, as well as the use of unique molecular identifiers (UMIs) (Islam et al., 2014) have greatly improved SCS data quality and reproducibility (Gawad et al., 2016; Wang and Navin, 2015). Over the last decade, a major shift in the field has been the transfer of these technologies from a few expert laboratories, into the hands of cancer research groups around the world (Stuart and Satija, 2019; Tanay and Regev, 2017). The democratization of SCS methods has been facilitated both by the open sharing of protocols and the commercialization of technologies (eg. 10X Genomics, Illumina, Mission Bio, Takara Biosciences). SCS methods offer many advantages over traditional ‘bulk’ DNA-seq and RNA-seq approaches, that are limited to providing a mixed signal that represents many cell types in the microenvironment or an amalgamation of tumor clones with different genotypes. Although computational methods exist to deconvolute bulk sequencing data, these approaches can only provide ‘rough estimates’ of cell type mixtures or tumor clones present in a tumor sample. Although SCS has been used widely in cancer research studies, implementation in clinical studies remains limited, where issues such as costs, throughput and reproducibility are paramount (Navin and Hicks, 2011). This review will discuss logistical issues related to clinical SCS, as well as emerging technologies and past research studies that have impacted many areas of cancer research over the last 10 years.
Current and Emerging Technologies
The recent commercialization of SCS now provides stable platforms that can be applied widely to cancer research and clinical applications (Table 1). Microdroplet systems have emerged as the most widely used platform for high-throughput 3’ scRNA-seq (eg. 10X Genomics) and are capable of profiling up to 10,000 cells in a single experiment (Klein et al., 2015; Macosko et al., 2015; Zheng et al., 2017b). However for research applications, alternative platforms such as nanowells (Fan et al., 2015b; Gao et al., 2017; Gierahn et al., 2017), combinatorial indexing (Cao et al., 2017) and high-throughput FACS (Patel et al., 2014) can provide additional features, including cell imaging, cell selection, reduced costs and multi-step chemistry. For example, to perform full-length mRNA profiling in single cells, FACS based methods using smart-seq2 (Ramskold et al., 2012) or nanowell-based platforms (Goldstein et al., 2017) can be used to study alternative splicing. Other platforms such as combinatorial indexing (sci-RNA-seq) can achieve very high scalability (millions of cells) for detecting rare cell subpopulations (Cao et al., 2017; Cao et al., 2019) such as circulating tumor cells (CTCs), cancer stem cells or minimal residual disease. Overall, scRNA-seq approaches represent the most mature SCS methods, however they still have several technical limitations including high transcript drop-outs for low expressed genes, low total gene counts per cell and high bias for 3’ coverage.
Table 1 -.
Technologies for High-Throughput Single Cell Sequencing
| Method | Technology | Name | Platform | Cost | Chemistry | References | |
|---|---|---|---|---|---|---|---|
| scRNA-seq | microdroplets | 10X Genomics RNA | commercial | 10K | $$$$ | 3' or 5' | Zheng et al. 2017 |
| microdroplets | Drop-seq | research | 10K | $$ | 3' | Macosko et al. 2015 | |
| microdroplets | Indrop | research | 10K | $$ | 3' | Klein et al. 2015 | |
| nanowells | Seq-Well | research | 10K | $ | 3' | Gierahn et al. 2017 | |
| nanowells | Takara Wafergen | commercial | 1.8K | $$$$ | 3', full-length | Goldstein et al. 2017 | |
| nanowells | cytoseq | research | 100K | $$ | 3' | Fodor et al. 2015 | |
| FACS | smart-seq2 | research | 384 | $ | full-length | Ramskold et al. 2012 | |
| FACS | sci-RNA-seq | research | 100K | $$ | 3' | Cao et al. 2017 | |
| scEpigenomics | microdroplets | 10X Chromium ATAC | commercial | 10K | $$$$ | tagmentation | Satpathy et al. 2019 |
| FACS | ATAC tagmentation | research | 384 | $ | tagmentation | Buenrostro et al. 2015 | |
| FACS | dscATAC-seq | research | 100K | $ | tagmentation | Cusavonich et al. 2015 | |
| micromanipulation | scRBBS | research | 100 | $$ | RBBS | Guo et al. 2013 | |
| scDNA-seq | microdroplets | 10X Chromium CNV | commercial | 10K | $$$$ | MDA | Andor et al. 2018 |
| microdroplets | Mission Bio Tapestri | commercial | 10K | $$$ | amplicon PCR | Lan et al. 2017 | |
| nanowells | Wafergen Takara | commercial/res | 1.8K | $$$$ | tagmentation | Laks et al. 2019 | |
| Microfluidics | tagmentation | research | 200 | $$ | tagmentation | Zahn et al. 2017 | |
| FACS | sci-seq | research | 100K | $$ | tagmentation | Adey et al. 2017 | |
For epigenomic profiling, scATAC-seq has become the most widely used assay to measure chromatin accessibility of single cells. In early versions of scATAC-seq technologies, experiments were performed using FACS and tagmentation chemistry (Buenrostro et al., 2015) or microfluidic platforms (eg. Fluidigm) at low cell throughput (96 cells). However the development of microdroplet platforms for scATAC-seq (10X Genomics) have emerged as the most popular platform for profiling over 10K cells in a single experiment (Satpathy et al., 2019). The scATAC-seq methods have also been adopted for combinatorial indexing (dscATAC-seq) to achieve very high scalability to profile hundreds of thousands of cells at low cost (Cusanovich et al., 2015; Lareau et al., 2019). A major advantage of scATAC-seq compared to scRNA-seq, is that it can provide deeper insights into gene regulation and transcription factors or other, as well as more information on cell lineages and identity. However scATAC-seq methods are still challenged by technical errors, including low library complexities and high sensitivity to tissue dissociation. Another approach is to measure cytosine methylation of DNA in single cells, using methods such as single-cell bisulfite sequencing (scBS-seq) and single-cell reduced-representation bisulfite sequencing (scRRBS) (Guo et al., 2013; Mooijman et al., 2016; Smallwood et al., 2014; Zhu et al., 2017). While DNA methylation in single cells is highly desirable for epigenomic profiling and lineage tracing, current methods remain challenged by technical issues including low cell throughput and poor genomic coverage. Thus, these scEpigenomic methods still represent the least mature platforms for profiling single cancer cells, but they are under active development.
For high-throughput scDNA-seq several commercial platforms have been developed (10X Genomics, Mission Bio) using microdroplet systems (Andor et al., 2018; Lan et al., 2017). Additionally, a number of research platforms including high-density FACS assays (Baslan et al., 2015; Leung et al., 2016), microfluidic platforms (Zahn et al., 2017a), nanowell systems (Laks et al., 2019) and combinatorial indexing methods (Vitak et al., 2017; Yin et al., 2019) have also been developed and offer higher scalability, enable cell selection, have lower costs and provide more flexibility for custom chemistry steps. Currently, the most common application of scDNA-seq is copy number aberration (CNA) profiling. While high-throughput systems such as microdroplets (10X Genomics) and combinatorial indexing can achieve very high-throughput (up to 10K cells) for single cell CNA profiling, they are challenged by lower data quality and limited genomic resolution. In contrast, nanowells, FACS and microfluidic platforms that utilize tagmentation chemistry (Laks et al., 2019; Zahn et al., 2017a) can provide very high-quality copy number data at single molecule resolution, but have modest throughput (hundreds to one thousand cells). Another major application of scDNA-seq is mutation detection, which requires higher coverage depth of the mutation sites of interest. While initial studies performed whole genome or exome sequencing of single cells using whole-genome amplification (WGA) methods (Hou et al., 2012; Wang et al., 2014b; Zong et al., 2012), these studies were limited to profiling a small number of cells due to high costs. To reduce costs and increase throughput, subsequent approaches have focused on sequencing targeted regions of the genome, such as cancer gene panels (Leung et al., 2017a; Leung et al., 2016) to increase depth and reduce costs. To further scale up scDNA-seq to over 10K cells for mutation detection, a two-step microdroplet approach was developed (Lan et al., 2017) and commercialized (Mission Bio) that performs PCR amplicon profiling of single cells at hundreds of targeted genomic regions. These highly targeted approaches are ideal for clinical applications but have limited applications in research for unbiased discovery and inferring cancer evolution. Technical errors that must still be overcome include high allelic dropout rates for mutational profiling (10-20%), high false-positive error rates for unbiased detection and non-uniform coverage depth due to WGA over and under amplification (Filbin et al., 2018; Gawad et al., 2016).
Several emerging technologies are on the horizon and have potential to transform the field of SCS. Multiomic SCS methods are designed to interrogate multiple layers of molecular information from the same single cells, such as DNA & RNA, RNA & ATAC, or all three layers combined (‘triple-omics’) (Stuart and Satija, 2019). These methods can provide deeper insights into the genotype-phenotype relationship and the regulation of gene expression by epigenomics. While initial technologies have demonstrated the technical feasibility of sequencing DNA and RNA in the same cell (Dey et al., 2015; Macaulay et al., 2015), they are currently low-throughput, expensive and labor intensive. The future development of these methods using nanowell systems, microdroplet platforms and combinatorial indexing (Yin et al., 2019) hold promise for overcoming many of these technical obstacles, which may lead to their widespread adoption in cancer research in the near future.
Another emerging technology is spatially-resolved SCS. A major limitation of standard SCS methods is that they require cell suspensions as input materials, and therefore inherently lose all spatial information of the location of the cell in its native tissue context during dissociation. To preserve spatial information, Laser Capture Microdissection (LCM) can be combined with scDNA-seq (Casasent et al., 2018) or scRNA-seq (Nichterwitz et al., 2016). However LCM methods are low-throughput (< 100 cells) and labor intensive. Other methods such as Spatial Transcriptomic (ST) microarrays (Stahl et al., 2016) and Slide-Seq (Rodriques et al., 2019) are high-throughput and can profile small groups of cells (eg. 10-100 cells) across thousands of spatial locations, but do not have single cell resolution (they require a tissue lysis step). In contrast in situ sequencing technologies, have single cell resolution, including FISSEQ (Lee et al., 2014), MERFISH (Chen et al., 2015) and seqFISH (Shah et al., 2017) but can only image limited spatial areas and are restricted to smaller sets of gene targets. The further development of these spatial technologies will greatly improve our understanding of cancer biology and are expected to revolutionize clinical pathology, by linking qualitative and morphological features of tissues to genomic data at single cell resolution.
Cancer Research Studies
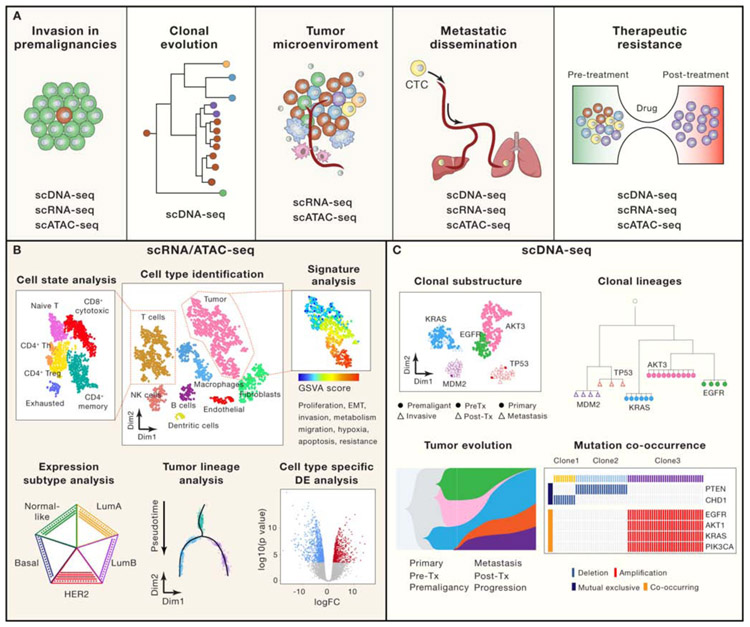
Cancer research studies published to date using SCS can roughly be categorized into five main areas: (1) invasion in premalignant disease, (2) clonal evolution and intratumor heterogeneity (ITH) in primary tumors, (3) reprogramming of the tumor microenvironment, (4) metastatic dissemination, and (5) therapeutic resistance (Figure 1A). In these studies scRNA-seq and scATAC-seq methods have often been used to resolve cell types and cell states (i.e. expression/epigenomic programs) in the TME, as well as the expression programs and subtypes of tumor cells. Analysis of cell states in the TME can reveal how different stromal and immune cell types are reprogrammed, reflecting different biological functions that may promote or suppress tumor growth (Figure 1B). Similarly, scRNA-seq methods can provide insight into the phenotypic diversity of tumor cells through the analysis of gene signatures for proliferation, stemness, hypoxia, EMT, metabolism and other cancer hallmarks. An advantage of scRNA-seq over bulk RNA-seq is the ability to perform cell-type specific differential expression (DE) analysis, to determine if cell types in the TME have different genes expressed under conditions such as treatment or progression. Other analyses may include reconstructing differentiation lineages or identifying tumor expression subtypes (eg. PAM50) at single cell resolution (Figure 1B). Several groups have also shown that it is possible to infer low-resolution genomic information from scRNA-seq data by inferring DNA copy number information to distinguish aneuploid tumor cells from the TME (Durante et al., 2020; Tickle T, 2019), and that prior knowledge of mutation sites (eg. from exome sequencing) can be used to genotype full-length mRNA data in single cells (Filbin et al., 2018; Petti et al., 2019). However these data are highly sparse and CNAs detected in RNA data often do not correlate with DNA data, hence the best application of the RNA copy number approach is for classifying tumor cells from normal cells, rather than inferring clonal substructure in tumors. Another layer of genomic information that can be gained from the 5’ or full length single cell RNA data is to sequence the clonotype of the T-cell receptor (TCR), which has important applications in understanding the expansion of T-cells in tumors(Azizi et al., 2018; Stubbington et al., 2016) and this approach has recently been commercialized (10X Genomics). In contrast, scDNA-seq methods can be used to resolve clonal substructure and reconstruct clonal lineages during tumor evolution in the context of premalignant disease, metastasis and therapeutic resistance (Figure 1C). Methods for scDNA-seq are particularly useful for resolving mutation co-occurrence and mutual exclusivity in different tumor clones, which are difficult to resolve in bulk sequencing data using deconvolution methods that provide only a rough estimates of the clonal subpopulations (Leung et al., 2017b; Wang et al., 2014b). Below we discuss several areas of cancer research in which SCS approaches have been used to illuminate cancer biology.
Figure 1 – Cancer Research Applications and Analyses.
(A) Cancer research applications of SCS include investigating invasion in premalignant cancers, clonal evolution in primary tumors, the tumor microenvironment, metastatic dissemination and therapeutic resistance. (B) Biological analyses that can be performed using scRNA-seq and scATAC-seq methods include cell type identification, cell state analysis, gene signature and pathway analysis, expression subtype analysis, tumor lineage inference and cell-type specific differential expression (DE) analysis. (C) Biological analysis that can be performed using scDNA-seq data for copy number aberrations or mutations, include delineating clonal substructure, inferring clonal lineages, reconstructing tumor evolution and identifying mutation co-occurrence or mutual exclusivity. Matched samples for these analyses may include premalignant and invasive tumors, pre-treatment and post-treatment samples, or primary and metastatic tumors.
Insights into Premalignant Disease
A major question is how premalignant cancers progress to invasive malignancies. Two studies using scDNA-seq have focused on understanding this problem in the most common form premalignant breast cancer, called ductal carcinoma in situ (DCIS). By developing a spatially resolved scDNA-seq method called Topographic Single Cell Sequencing (TSCS), genomic copy number aberrations (CNAs) were linked to spatial information in 10 cases of synchronous DCIS-IDC tissues, which showed that multiple clones co-migrated from the ducts into the invasive regions (Casasent et al., 2018). In a smaller study of four DCIS-IDC patients, scDNA-seq data showed that in situ tumor cells underwent a population bottleneck during invasion leading to the expansion of a selected genotype (Martelotto et al., 2017b). While both studies showed evidence for direct genomic lineages from the DCIS to IDC regions, they reported different results on the selection of clonal genotypes during invasion. In Barrett’s oesophagus and early gastric cancers, two studies used scRNA-seq to identify marker genes in premalignant tumor cells that were associated with progression (Owen et al., 2018; Zhang et al., 2019). In another study, scRNA-seq was used to compare the TME between premalignant and advanced pancreatic cancer, which showed a loss of pro-inflammatory immune cells and gain of reprogrammed myofibroblasts during progression (Bernard et al., 2019a). These studies highlight the utility of using SCS to understand the progression of premalignant to invasive disease, in which tumor cells are often rare populations that cannot easily be profiled with bulk genomic methods.
Decoding the Tumor Microenvironment
The TME is largely divided into the immune and stromal components, which can both be resolved using scRNA-seq and scATAC-seq methods. In the stromal TME, fibroblasts have been a cell type of considerable interest since they are often reprogrammed as carcinoma associated fibroblasts (CAFs) that have been shown to interact with tumor cells to promote or suppress tumor growth (Kalluri, 2016). In several cancers, scRNA studies have revealed heterogenous CAFs subtypes with different functions in matrix construction, angiogenesis and immune-regulation (Bartoschek et al., 2018; Elyada et al., 2019; Lambrechts et al., 2018; Puram et al., 2017). Another stromal cell type of interest is endothelial cells, which can be reprogrammed as tumor endothelial cells (TECs) that can promote tumor progression by recruiting vasculature and regulating immune cells (Hida et al., 2018). Methods for scRNA-seq can resolve subtypes of endothelial cells and reveal signaling pathways that can be targeted (Goveia et al., 2020; Lambrechts et al., 2018; Zhao et al., 2018). The immune TME is another area of intense interest in solid tumors, due to the growing use of immunotherapy in cancer treatment (Sharma and Allison, 2015). Several studies have used scRNA-seq to profile T-cells and show that a suppressive immune microenvironment is correlated with poor prognosis, in which increased T-cell exhaustion signatures and decreased activated T-cells were associated with progression in a variety of human cancers (Guo et al., 2018; Peng et al., 2019; Savas et al., 2018; Tirosh et al., 2016a; Zheng et al., 2017a). In addition, scRNA-seq methods have implicated tissue-resident memory (TRM) T-cells as having higher cytotoxic activity in breast tumors and correlated with better prognosis for triple-negative breast cancer (TNBC) patients (Savas et al., 2018). Myeloid cells are another component of the TME that are associated with patient outcome in several cancer types (Engblom et al., 2016). Tumor-associated macrophages (TAMs) have traditionally been classified as M1 (inflammatory) or M2 (tumor promoting); however, scRNA-seq data has shown that there is a continuum of macrophage expression programs, with a large diversity of cell states, challenging this simple classification (Azizi et al., 2018; Song et al., 2019). Other myeloid subtypes identified by scRNA-seq include myeloid derived suppressor cells (MDSCs) and monocytes (Azizi et al., 2018; Cassetta et al., 2019; Song et al., 2019; Wang et al., 2019). Notably, two glioma studies using scRNA-seq showed that increased peripheral macrophage expression programs relative to microglia in the TME were associated with progression and poor survival in glioma patients (Muller et al., 2017; Venteicher et al., 2017).
Diversity of Tumor Cell Phenotypes
Tumor cells can exhibit diverse phenotypes in proliferation, stemness, EMT, invasion, migration, metabolism, immune evasion, apoptosis and hypoxia (Hanahan and Weinberg, 2011). This diversity may play an important role in progression, therapeutic response, invasion and metastasis. In contrast to bulk genomic methods, scRNA-seq and scATAC seq have the ability to resolve the phenotypic diversity and plasticity of tumor cells. In glioblastoma (GBM), scRNA-seq identified plasticity in EMT and stemness signatures, showing that most tumors consisted of co-existing cells with different GBM subtype signatures. Similarly, in TNBC snRNA-seq showed that while a basal-like (PAM50) expression subtype usually dominated each tumor, there were also many cancer cells with other expression signatures co-existing in the tumor mass (Gao et al., 2017) that did not change in response to chemotherapy (Kim et al., 2018). Variation in EMT states have also been identified using scRNA-seq in head and neck cancers, where a partial-EMT program correlated with invasion and metastatic dissemination (Puram et al., 2017). In breast cancers, scRNA-seq has identified phenotypic variation in EMT, angiogenesis and stemness across patients (Chung et al., 2017; Karaayvaz et al., 2018). In TNBC, scRNA-seq analysis identified a glycosphingolipid metabolism signature that correlated with treatment resistance and metastasis, that predicted patient outcome (Karaayvaz et al., 2018). Methods such as scRNA-seq can also be used to infer cell hierarchies, lineage plasticity and developmental trajectories in human tumors. Using scRNA-seq methods in brain tumors has provided insight into differentiation trajectories of tumor cells in early stage gliomas (astrocytic, oligodendrocytic) as well as dynamic plasticity between cell states in invasive GBM (Filbin et al., 2018; Neftel et al., 2019; Tirosh et al., 2016b). By spatially sampling GBM tumors, another scRNA-seq study showed less hypoxia, lower proliferation and less adhesion in the invasive tumor cells near the tumor margins compared to the inner cores (Darmanis et al., 2017). Single cell multiomics studies have also revealed connections between genetic mutations and transcriptional reprogramming. Using Genotyping of Transcriptomes (GoT) investigators identified unfolded protein response in CALR mutated myeloproliferative neoplasms by comparing wildtype and mutant clones (Nam et al., 2019). Another study of primary and metastatic paired samples in colorectal cancer used single cell ‘tripleomics’ (DNA, RNA, methylation) to show that DNA methylation levels were found to be diverse across different genetic lineages, while relatively stable during metastasis (Bian et al., 2018).
Clonal Evolution
During the expansion of the primary tumor mass, tumor cells undergo Darwinian evolution and form divergent clonal lineages in response to selective pressures. Bulk methods have limited ability to resolve ITH and clonal lineages in tumors, compared to scDNA-seq approaches (Gawad et al., 2014; Navin et al., 2011). These data can provide insight into general models of tumor evolution (Davis et al., 2017). A notable evolutionary model discovered by scDNA-seq is punctuated copy number evolution (PCNE) in breast cancer, which showed that early bursts of genome instability give rise to hundreds of genomic rearrangements, that re-stabilize and undergo stable clonal expansions, challenging the paradigm of gradual evolution (Gao et al., 2016; Navin et al., 2011). Consistent studies using single cell copy number analysis in breast cancers, xenografts and ovarian cancers have also shown that CNAs are highly stable across individual tumor cells (Laks et al., 2019; Zahn et al., 2017b). In contrast to copy number data, scDNA-seq of mutations have often supported branching evolution, by showing that mutations occur more gradually and that multiple lineages and clones often co-exist in the tumor at the same point in time (Gawad et al., 2014; Li et al., 2012; Wang et al., 2017; Wang et al., 2014b; Yu et al., 2014). Methods for scDNA-seq data can be used to identify combinations and mutual exclusivity of driver mutations in clones to provide a guidance for clinical intervention. For example, in childhood acute lymphoblastic leukemia (ALL) targeted scDNA-seq identified late oncogenic mutations involved in proliferation that could be targeted to treat the disease (Gawad et al., 2014). In acute myeloid leukemia (AML) a microdroplet scDNA-seq approach identified concurrent mutations in NRAS, KRAS, and FLT3 that emerged after treatment with a FLT3 inhibitor, providing additional targets for treatment (McMahon et al., 2019). By applying both scDNA-seq and scRNA-seq, a study of chronic lymphocytic leukemia (CLL) identified driver mutations in LCP1 and WNK1, and showed that different genetic lineages can adopt similar expression programs (Wang et al., 2017). Another CLL study applied scRRBS-seq and scRNA-seq to reconstruct B-cell lineages and showed that after ibrutinib treatment a specific lineage of B-cells had upregulation of Toll-like receptor pathway in post treatment cells (Gaiti et al., 2019).
Metastasis and CTC profiling
Metastatic disease is highly correlated with morbidity and mortality in cancer patients. Gaps in our knowledge of metastasis include understanding which clones in the primary tumor are capable of dissemination, how many times cancer cells disseminate to distant organ sites, and whether the TME plays a role in the metastatic niche. SCS methods can resolve ITH and the TME in the primary and metastatic tumors, as well as the key intermediates, circulating tumor cells (CTCs), to provide insight into these questions. In matched primary colorectal cancers and liver metastases, scDNA-seq in two patients identified a late-dissemination model to the liver, in which most of the driver events were acquired in primary tumor clones prior to the first seeding events (Leung et al., 2017b). In a breast cancer PDX, scRNA-seq identified stem-like cells that initiated metastasis, and implicated MYC expression in high burden disease (Lawson et al., 2015). Another PDX study of renal cell carcinomas (RCC) used scRNA-seq to identify metastasis signatures, which included genes such as EGFR, Scr, BRAF/MEK that provide potential therapeutic targets (Kim et al., 2016). In human head & neck cancers, scRNA-seq of tumor cells identified invasive expression programs, including cell cycle, stress, hypoxia, differentiation and partial-EMT programs, that promoted metastasis (Puram et al., 2017).
SCS methods have also been applied to profile the genomes and transcriptomes of CTCs in metastatic disease. Studies using scDNA-seq have identified a high concordance of mutations and CNAs in CTCs when compared to primary and metastatic tumors (Lohr et al., 2014; Ni et al., 2013). The transcriptomes of CTCs and CTC clusters have also been studied in human blood samples using scRNA-seq to identify genes involved in dissemination. In pancreatic cancer scRNA-seq identified metastatic signatures with low proliferation, enriched stem cell genes and stromal-derived extracellular matrix (ECM) genes (Ting et al., 2014). In breast cancer scRNA-seq of CTCs identified plakoglobin as a critical cell junction gene in CTC cluster formation that increased dissemination efficiency (Aceto et al., 2014). CTC gene expression signatures identified by scRNA-seq have correlated with therapeutic response and metastasis risk in lung cancer (Su et al., 2019), breast cancer (Kwan et al., 2018) and prostate cancer (Miyamoto et al., 2018), paving the way for clinical applications.
Therapeutic resistance
Drug resistance is a major obstacle in the treatment of human cancers. While therapies, including chemotherapy, hormonal therapy, targeted therapy and immunotherapy are often initially effective, many patients develop resistance and progress to metastatic disease. Key areas for investigation include understanding the mechanisms of resistance, identifying predictive biomarkers of response, and elucidating the evolution of resistant disease. SCS methods can profile ITH and the TME from pre-treatment samples with clinical outcome data, or longitudinal time-point samples collected before and after treatment to address these topics. Several studies have applied scRNA-seq to identify signatures of therapeutic response and resistance using tissue samples and CTCs. In ER+/HER2− breast cancer, scRNA-seq analysis identified differences in HER2-signatures in CTCs during chemotherapy that were associated with resistance (Jordan et al., 2016). In melanoma, two studies using scRNA-seq reported MITF, AXL, and DCT signatures that correlated with RAF/MEK-inhibitor resistance (Ho et al., 2018; Rambow et al., 2018; Tirosh et al., 2016a). In prostate cancer, scRNA-seq analysis of CTCs identified activated noncanonical Wnt signaling in resistance of androgen receptor inhibitors (Miyamoto et al., 2015). Cell lines treated with drugs and analyzed by scRNA-seq have also provided insights into resistance. In endocrine resistant breast cancer, cell lines profiled with scRNA-seq showed that KDM5 inhibitor resistance was due to an acquired epigenetic state with high expression of KDM5 and ITH in ER+ cells that were resistant to antiestrogens (Hinohara et al., 2018). In another breast cancer study, scRNA-seq in cell lines treated with chemotherapy identified upregulation of EMT and stemness genes and down-regulation of cell cycle genes in the resistant cells (Prieto-Vila et al., 2019) In melanoma cell lines treated with BRAF inhibitors, a rare pre-resistant cell state was identified, which required further epigenetic reprogramming and loss of SOX10 to achieve a fully resistant state (Shaffer et al., 2017). In other cell line studies in oral carcinomas treated with chemotherapy, scRNA-seq analysis showed that phenotypic heterogeneity favored selection of pre-existing clones, while homogenous populations led to epigenomic reprogramming through increased SOX9 expression and loss of SOX2 (Sharma et al., 2018).
To understand the genomic evolution of drug resistance several studies have used scDNA-seq methods to understand whether resistant clones are pre-existing in the tumor mass and selected after therapy (adaptive resistance), or alternatively have resistance mutations induced by treatment (acquired resistance). In a study of CTCs from SCLC patients, analysis of blood samples before and after chemotherapy showed consistent CNA profiles after treatment, indicating an intrinsic or adaptive resistance model (Carter et al., 2017). In another study of neoadjuvant chemotherapy resistance in TNBC, combined profiling using scDNA-seq and scRNA-seq showed that resistant genotypes were pre-existing in the tumor mass and adaptively selected, followed by further transcriptional reprogramming to acquire a fully resistant phenotype (Kim et al., 2018). In castration-resistant prostate cancer, scDNA-seq of CTCs showed that MYC and AR amplifications were selected in subclones during resistance (Dago et al., 2014). In AML patients treated with FLT3 inhibitors, scDNA-seq identified rare subclones that emerged upon relapse with mutations in DNMT3A or combinations of IDH2, AXSL1 and NRAS following an adaptive resistance model (Pellegrino et al., 2018).
TME reprogramming in resistant disease can also be investigated using scRNA-seq and scATAC-seq methods. In melanoma scRNA-seq identified a TCF7+ memory-like state in the cytotoxic T cell population that was associated with therapeutic response (Sade-Feldman et al., 2018). In basal cell carcinoma (BCC) scATAC-seq was used to analyze biopsies pre and post-treatment with PD-1 blockade, which showed that exhausted T-cells were highly expanded after therapy, suggesting that PD-1 blockade impacts both CD4+ and CD8+ cell states in the TME (Satpathy et al., 2019). These studies highlight the potential of SCS approaches to uncover the role of the immune TME in therapeutic response and resistance.
Sample Processing Logistics for SCS in the Clinic
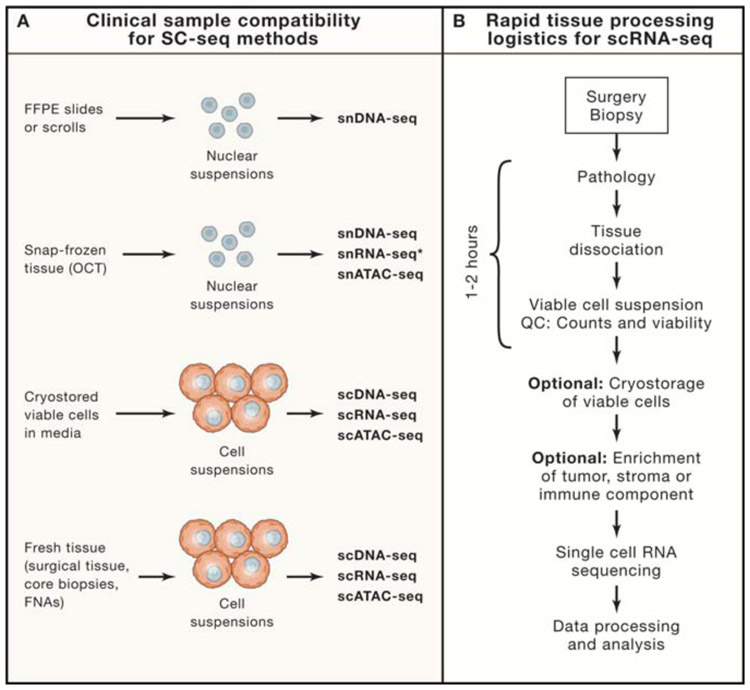
The collection and processing of fresh tissues from the clinic for SCS presents unique logistical and technical challenges. While scDNA-seq and scATAC-seq methods can be applied to archival materials (snap-frozen tissues (OCT) and even FFPE), methods such as scRNA-seq require viable cell suspension as input material (Figure 2A). The problem is that snap freezing of tissue (without freezing media) leads to the rupture of the cellular membrane, and therefore cannot provide intact cells for subsequent dissociation and running scRNA-seq. Fortunately, the nuclear membrane remains intact during freeze-thaw cycles, protecting the DNA, chromatin and even RNA in the nucleus (Baslan et al., 2012). This unique property has allowed researchers to isolate nuclear suspensions from snap-frozen or OCT tissues for single nucleus sequencing, including snDNA-seq (Navin et al., 2011; Wang et al., 2014a), snATAC-seq (Fujiwara et al., 2019) and snRNA-seq (Gao et al., 2017; Habib et al., 2016) (Figure 2A). However while this strategy works well for scDNA assays, the profiling of nuclear RNA instead of cytoplasmic RNA can result in lower gene counts, lower UMIs and differences in the expression of many genes and pathways (Gao et al., 2017; Habib et al., 2017). Therefore, in most research studies and clinical applications, scRNA-seq methods are the preferred approach and requires implementing a rapid tissue dissociation (RTD) program, which currently do not exist in routine pathology at most cancer hospitals.
Figure 2 – Compatibility of Clinical Tissues and Processing Logistics for SCS.
(A) Clinical samples types for SCS may include FFPE tissues, snap-frozen tumors, cryostored viable cells and fresh tissues, and their compatibility with single cell and single nucleus sequencing assays is listed below. Asterisk indicates that single-nucleus RNA-seq of frozen tissues is highly dependent on the quality of the tissues. (B) Rapid tissue collection (RTC) program to procure fresh tissues collected after surgery or biopsy procedures for SCS. Optional steps include cryostorage of viable cell suspensions in freezing media and enrichment of cell types prior to running scRNA-seq assays.
To procure fresh clinical tissues in a rapid time-frame for scRNA-seq, an RTD program must be established that involves close coordination between the oncologists, surgeons, pathologists and researchers (Figure 2B). In an RTD program, the surgeon must deliver the excised tumor tissues to pathologists in a rapid time frame, after which the pathologist must identify regions of high tumor purity for macrodissection (surgical specimens) and place the tissue into cell culture media, ideally in less than 1 hour. For core biopsy or fine-needle-aspirate (FNA) samples this time can be cut down substantially, since the radiologist can place the sample directly into a tube of media and provide the pathologist with a different core sample. Once the tissue is submerged in media, the research team can transfer the sample back to the laboratory for dissociation to generate single cell suspensions. Ideally, a cancer hospital would setup a dedicated RTD facility that can centralize the processing of incoming tissue samples to generate viable cell suspensions from tissues after they are collected from surgeries or biopsy procedures. However a challenge for RTD is that each tissue type (eg. breast, pancreas, liver) requires a different dissociation protocol, that may vary in dissociation time, digestion enzymes, red blood cell lysis and other steps. Fortunately, collaborative efforts such as the Human Cell Atlas (HCA) are developing open source protocols for many tissue types (Regev et al., 2017) that are publicly accessible online (www.protocols.io) to help researchers. After dissociation, the cells must undergo quality check (QC) for cell viability (ideally >70%) and cell counts (ideally >100K total cells). Fortunately after this step, the viable cells can be cryostored in freezing media for an extended period of time and batched to run many scRNA-seq experiments together (Wohnhaas et al., 2019). However running samples from fresh cell suspensions without cryostorage leads to the best data quality, when it is logistically feasible. Another optional step involves enriching cells of interest using FACS or antibody columns, to enrich immune cells (eg. CD45), stromal cells or tumor cells (eg. EpCAM, Cytokeratins) (Nguyen et al., 2018). The final cell suspensions are then used as input material for SCS methods followed by next-generation sequencing (NGS).
Although FFPE blocks represent an abundant source of clinical tissue with long-term clinical outcome information, these materials are not compatible with standard SCS methods. The main problem is that FFPE processing leads to the fragmentation and degradation of RNA and DNA when the tissue is placed in formalin. While scRNA and scATAC methods may never possible in FFPE materials, developments in scDNA-seq methods may make the analysis of CNAs and targeted mutational profiling feasible in the near future. Encouragingly, initial success was shown through the addition of DNA damage repair enzymes prior to WGA for single cell CNA profiling in FFPE blocks from premalignant breast cancer tissues (Martelotto et al., 2017a) . Other strategies designed to PCR small targeted regions may also be feasible for FFPE tissue in which the DNA is fragmented. Therefore, the development of future technologies for scDNA-seq may open up the doors for FFPE tissues and H&E slides that are commonly used in clinical pathology.
Applications in Cancer Medicine
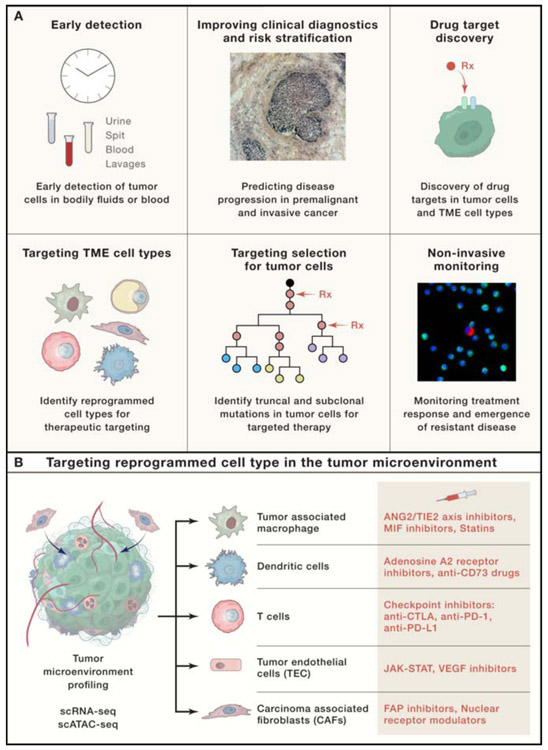
While most SCS studies published to date have been research focused, the development and refinement of SCS technologies provides new opportunities for clinical translation. We expect that several areas of cancer medicine will be transformed by SCS including early detection, diagnostics & risk stratification, drug target discovery, targeting the microenvironment, targeting tumor clones, and non-invasive monitoring (Figure 3A). We discuss initial progress in these areas and remaining challenges for clinical implication that lie ahead.
Figure 3 – Clinical Applications of Single Cell Genomics.
(A) Clinical areas that may benefit from SCS include early detection, improving clinical diagnostics & risk stratification, drug target discovery, targeting the tumor microenvironment, target selection for tumor cells and non-invasive monitoring. Red arrows in the tumor targeting panel indicate targeting of clonal or subclonal mutations (B) An example of profiling a patient’s tumor to identify reprogrammed cell types in the TME using scRNA-seq or scATAC-seq, followed by selecting specific drugs that target individual reprogrammed cell types in the TME to treat the patient.
Early Detection
Early detection is paramount in modern oncology, since it can lead to intervention and reduced morbidity in patients. However, the detection of early cancers still relies heavily on imaging techniques and pathological analysis. Disseminated tumors cells (DTCs) can be detected by cytopathology in many bodily fluids that are proximal to the cancer origin, such as urine for bladder and prostate carcinomas (Nawroth et al., 2014); peritoneal washings for ovarian (Naz et al., 2015), endometrial and pancreatic carcinomas (Yamada et al., 2007); and saliva for nasopharyngeal cancer(Chang et al., 2003). These samples provide a unique opportunity for SCS methods to asses risk of progression. Early detection is of particular importance in people that have a high risk of cancer development, including patients with germline mutations in tumor suppressors (BRCA, APC mutations), acid reflux, inflammatory bowel disease, and a history of heavy smoking. Although most studies to date have focused on using imaging methods or pathological staining to identify early cancer cells in bodily fluids, SCS methods can provide far richer information on whether these cells are likely to present a risk for progression to the patient. In one of the first studies, scRNA-seq was used to study 40 multiple myeloma patients and 11 healthy controls, which identified the expression programs of rare invasive plasma cells in several patients with asymptomatic disease (Ledergor et al., 2018). We expect that future applications of scDNA-seq and scRNA-seq in combination with early screening and diagnostic procedures will hold great promise to improve early detection and assess the risk of progression to malignant disease.
Improving Clinical Diagnostics & Risk Stratification
The clinical diagnosis of invasive cancers relies heavily on histopathological evaluations that often have discrepancies. SCS methods have proven to be very effective at profiling premalignant cells in DCIS (Casasent et al., 2018), pancreatic premalignancies (Bernard et al., 2019b), prostate intraductal neoplasia (PIN) (Barros-Silva et al., 2018) and atypical adenomatous hyperplasia (AAH) (Allin et al., 2018) in the research setting. In principle this data can be used to asses risk of progression, in larger clinical studies that compare patients that progress to invasive disease to patients that remain indolent. In invasive prostate cancer, aggressive clonal populations detected by single cell analysis from tissue biopsies were shown to correlate very well with surgical Gleason scores, thus guiding surgery decisions based on pathological diagnosis (Alexander et al., 2018). In another study, the subtyping of colorectal cancer by SCS has been used to identify the best treatment options for individual patients (Bian et al., 2018). Another clinical opportunity involves the diagnosis of cancer of unknown primary (CUP) origin, which represents an aggressive cancer without a clear organ site of origin, where scRNA-seq and scATAC-seq methods may provide insight into the initial organ sites and thus have implications for treatment (Varadhachary et al., 2004).
Risk stratification is another important area for personalized therapy planning at multiple times during cancer treatment including: (1) at diagnosis, (2) after the completion of local treatment, and (3) at the time of metastatic recurrence. Current risk stratification tools rely on histopathological analysis, cytogenetic markers, germline mutations or gene expression panels (eg. oncotypeDX, mammaprint, cologuard). However predictive gene expression panels using bulk RNA-seq analysis and qPCR are challenged by the mixtures of tumor, stromal and immune cells, which vary across patients. Instead of a precise prediction for individual patients, current gene expression panels provide population based prediction (e.g., the percentage of recurrence for the group of patients with same score). These assays could potentially be improved by scRNA-seq methods to obtain pure cell-type specific expression signatures from tumor cells or the TME to predict the risk of progression, metastasis or therapeutic resistance. Thus SCS methods have the potential to improve diagnostics and risk assessment to determine which patients will require more aggressive treatment strategies.
Drug Target Discovery
SCS methods provide powerful tools for unbiased discovery of new drug targets in the tumor cells and TME in primary, metastatic or therapy-resistant disease. For example, in prostate cancer, scRNA-seq analysis of CTCs identified activated noncanonical Wnt signaling in resistance to androgen receptor inhibitors, providing a new therapeutic target for treating advanced disease (Miyamoto et al., 2015). In breast cancer, scRNA-seq identified inversely correlated expression of NOTCH1 and HER2 in patient-derived CTCs, where HER2-negative CTCs showed reduced sensitivity to docetaxel, but were sensitive to Notch inhibitors that could potentially be exploited for treatment (Jordan et al., 2016). In a pre-clinical study, scRNA-seq of docetaxel sensitive and resistant MCF7 breast cancer cell lines identified upregulated EMT and stemness-related genes and downregulated cell cycle genes that were regulated by LEF1 in the resistant cells, providing a potential drug target to overcome resistance (Prieto-Vila et al., 2019). Longitudinal sampling of tumors on therapy can also identify the emergence of resistant clones and potential drug targets. In chemoresistant TNBC patients, snRNA-seq of longitudinal biopsies identified signatures for EMT, AKT1 signaling, hypoxia, CDH1 and angiogenesis that could potentially be targeted to treat resistant disease (Kim et al., 2018). Methods for scRNA-seq and scATAC-seq can be used to delineate cell types in the TME and identify drug targets in the stroma and immune cell types that contribute to progression, metastasis or therapeutic resistance (Azizi et al., 2018; Elyada et al., 2019; Lambrechts et al., 2018). In a breast cancer mouse model treated with anti-Her2 and CDK4/6 inhibitors, scRNA-seq identified an enrichment of Gr1+ immature myeloid cells (IMCs) that were sensitive to cabozantinib, providing a new therapeutic target to validate in human patients (Wang et al., 2019). In another breast cancer study, scRNA seq revealed serial adaptive changes by transcriptomic reprogramming and copy number changes in patients undergoing endocrine therapy (Hong et al., 2019). Thus, we expect that SCS methods will lead to a new era of drug development, by providing new drug targets in the TME and tumor cells to improve the treatment of cancer patients.
Targeting the Tumor Microenvironment
Reprogrammed cell types in the TME including CAFs, TECs, TAMs and tumor adipocytes (Altshuler-Keylin et al., 2016) contribute significantly to tumor progression, metastasis or therapeutic resistance (McAllister and Weinberg, 2014). In the research setting, scRNA-seq or scATAC-seq methods have proven to be powerful tools to delineate the TME and identify reprogrammed immune and stromal cell types in tumors that either promote or inhibit tumor growth or therapy resistance (Elyada et al., 2019; Puram et al., 2017; Tirosh et al., 2016a). These data can in principle be used to select targeted therapies against the aberrant cell types individually in a clinical setting (Figure 3B). For example, tumors that harbor TAMs could be treated with ANG2/TIE2 axis inhibitor (Rebastinib), MIF inhibitor (anti-CD74 antibody), or statins to indirectly regulate the function of TAM (simvastatin, atorvastatin). The tumors with reprogrammed dendritic cells could be treated with dual inhibitor of Adenosine A2A and A2B receptors (AB-928) or anti-CD73 (BMS-986179). T-cells could be treated with checkpoint inhibitors: anti-CTLA (ipilimumab), anti-PD1 (pembrolizumab, durvalumab), anti-PD-L1(atezolizumab, Avelumab). Similarly, tumors with reprogrammed stromal cell types, such as TECs could be treated with inhibitors against JAK-STAT (Ruxolitinib) and VEGF (bevacizumab, cabozantinib). CAFs could be treated with FAP inhibitors (PT-100, sibrotuzumab) or Nuclear receptor modulators (MORAb2, WYC-209). These SCS tools could greatly improve our ability to target reprogrammed cell types in the TME to improve the efficacy of cancer therapies (Abbosh et al., 2017).
Targeting Tumor Cells
ITH is common in solid tumors and therefore one single gene target detected by bulk sequencing may not be effective in all of the clones in the tumor mass (Alizadeh et al., 2015; McGranahan and Swanton, 2015). ITH can be resolved with scDNA-seq to reconstruct clonal lineages and identify mutations that are truncal (in all tumor cells), subclonal (shared in lineages) or private (exclusive to one clone) in the tumor lineages. Importantly, scDNA-seq can reveal the combination of mutations in clones in the tumor mass, to know if certain mutations are mutually exclusive or concurrent in the same tumor cells. Oncologists can use this data to guide treatment decisions on targeting mutations that are truncal, or specific to subpopulations that may have a higher risk for metastasis, therapeutic resistance or progression (Figure 3A). In AML, high-throughput scDNA-seq using a micro-droplet system was used to identify clonal remodeling in the context of therapy to determine relevant pathologic clones for therapeutic targeting in recurrent disease (Pellegrino et al., 2018). Methods such as scRNA-seq of tumor cells can also play an important role in guiding therapy selection by providing phenotypic information on tumor cells and their signaling pathways to investigate heterogeneity in processes like EMT, proliferation, migration and apoptosis, (Dentro et al., 2018; Patel et al., 2014). Thus, SCS methods provide powerful tools to resolve clonal substructure in tumors and guide treatment decisions based on mutation co-occurrence and aberrant expression programs.
Non-invasive monitoring
The most advanced translational application of SCS methods has been in genomic analysis of CTCs to perform non-invasive monitoring of disease progression, detect the emergence of resistant clones and track minimal residual disease (MRD). In contrast to metastatic tissue biopsies, CTC analysis can provide a more holistic view of genomic aberrations across many organ sites and micrometastases. Further, these methods allow longitudinal measurements to track genomic changes within tumors cells in ‘real time’ during disease progression and treatment without invasive biopsies. In contrast to circulating tumor DNA (ctDNA) methods, CTCs can also resolve clonal diversity in the blood and provide information on RNA gene expression. In prostate cancer, the use of scDNA-seq methods to profile CTCs over time has been shown to be useful in detecting clonal and subclonal changes of CTCs that provide information on progression, regression, and the emergence of resistant disease (Alberter et al., 2016; Dago et al., 2014). CTCs from single time-point samples also have utility for predicting therapeutic response and clinical outcome. In a breast cancer study, scRNA-seq of CTCs identified over 17 breast cancer specific RNA signatures that were used to generate a score to predict clinical outcome and risk of progression (Kwan et al., 2018). Similarly, in prostate cancer two CTC scores for metastatic and localized sites were established using a gene signature that correlated with poor overall survival and early dissemination (Miyamoto et al., 2018). Thus, non-invasive monitoring and risk prediction of CTCs using SCS methods hold great potential for translation, where previous translational work has largely focused on CTC enumeration.
Conclusions
In closing, SCS technologies have already transformed many areas of cancer research and are poised to make an even bigger impact in the clinic. In the same way that NGS technologies have transformed modern oncology over the last decade, we expect that SCS methods will impact many areas of cancer medicine and will become a common tool in cancer hospitals. While the biggest barrier to implementing SCS studies broadly in hospitals is likely to be the implementation of RTD programs, the wealth of information that can be obtained from SCS analysis can easily justify these efforts. Emerging technologies including SCS spatial and multiomic methods will further expand the capabilities of SCS methods over the next few years, and have a major impact on cancer research and clinical pathology. We expect that the implementation of SCS in cancer medicine over the next decade will lead to vast improvements in the diagnosis and treatment of cancer patients.
Acknowledgements
This work was supported by grants to N.E.N. from the American Cancer Society (129098-RSG-16-092-01-TBG), the National Cancer Institute (RO1CA240526, RO1CA236864), the Chan-Zuckerberg Initiative (HCA-A-1704-01668), the Emerson Collective Cancer Research Fund and the CPRIT Single Cell Genomics Center (RP180684). N.E.N and B.L. are supported by the MD Anderson Moonshot Program. B.L. is supported by the SWOG Hope Foundation grant and the Morgan Welch Inflammatory Breast Cancer Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, Le Quesne J, Moore DA, Veeriah S, Rosenthal R, et al. (2017). Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545, 446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, Yu M, Pely A, Engstrom A, Zhu H, et al. (2014). Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158, 1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberter B, Klein CA, and Polzer B (2016). Single-cell analysis of CTCs with diagnostic precision: opportunities and challenges for personalized medicine. Expert Rev Mol Diagn 16, 25–38. [DOI] [PubMed] [Google Scholar]
- Alexander J, Kendall J, McIndoo J, Rodgers L, Aboukhalil R, Levy D, Stepansky A, Sun GL, Chobardjiev L, Riggs M, et al. (2018). Utility of Single-Cell Genomics in Diagnostic Evaluation of Prostate Cancer. Cancer Research 78, 348–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizadeh AA, Aranda V, Bardelli A, Blanpain C, Bock C, Borowski C, Caldas C, Califano A, Doherty M, Elsner M, et al. (2015). Toward understanding and exploiting tumor heterogeneity. Nature Medicine 21, 846–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allin DM, Shaikh R, Carter P, Thway K, Sharabiani MTA, Gonzales-de-Castro D, O'Leary B, Garcia-Murillas I, Bhide S, Hubank M, et al. (2018). Circulating tumour DNA is a potential biomarker for disease progression and response to targeted therapy in advanced thyroid cancer. Eur J Cancer 103, 165–175. [DOI] [PubMed] [Google Scholar]
- Altshuler-Keylin S, Shinoda K, Hasegawa Y, Ikeda K, Hong H, Kang Q, Yang Y, Perera RM, Debnath J, and Kajimura S (2016). Beige adipocyte maintenance is regulated by autophagy-induced mitochondrial clearance. Cell metabolism 24, 402–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andor N, Lau BT, Catalanotti C, Kumar V, Sathe A, Belhocine K, Wheeler TD, Price AD, Song M, Stafford D, et al. (2018). Joint single cell DNA-Seq and RNA-Seq of gastric cancer reveals subclonal signatures of genomic instability and gene expression. bioRxiv, 445932. [Google Scholar]
- Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, Nainys J, Wu K, Kiseliovas V, Setty M, et al. (2018). Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 174, 1293–1308 e1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros-Silva JD, Linn DE, Steiner I, Guo G, Ali A, Pakula H, Ashton G, Peset I, Brown M, and Clarke NW (2018). Single-cell analysis identifies LY6D as a marker linking castration-resistant prostate luminal cells to prostate progenitors and cancer. Cell reports 25, 3504–3518. e3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoschek M, Oskolkov N, Bocci M, Lövrot J, Larsson C, Sommarin M, Madsen CD, Lindgren D, Pekar G, and Karlsson G (2018). Spatially and functionally distinct subclasses of breast cancer-associated fibroblasts revealed by single cell RNA sequencing. Nature communications 9, 5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baslan T, Kendall J, Rodgers L, Cox H, Riggs M, Stepansky A, Troge J, Ravi K, Esposito D, Lakshmi B, et al. (2012). Genome-wide copy number analysis of single cells. Nat Protoc 7, 1024–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baslan T, Kendall J, Ward B, Cox H, Leotta A, Rodgers L, Riggs M, D'Italia S, Sun G, Yong M, et al. (2015). Optimizing sparse sequencing of single cells for highly multiplex copy number profiling. Genome Res 25, 714–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard V, Semaan A, Huang J, San Lucas FA, Mulu FC, Stephens BM, Guerrero PA, Huang Y, Zhao J, and Kamyabi N (2019a). Single-Cell Transcriptomics of Pancreatic Cancer Precursors Demonstrates Epithelial and Microenvironmental Heterogeneity as an Early Event in Neoplastic Progression. Clinical Cancer Research 25, 2194–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard V, Semaan A, Huang J, San Lucas FA, Mulu FC, Stephens BM, Guerrero PA, Huang Y, Zhao J, Kamyabi N, et al. (2019b). Single-Cell Transcriptomics of Pancreatic Cancer Precursors Demonstrates Epithelial and Microenvironmental Heterogeneity as an Early Event in Neoplastic Progression. Clin Cancer Res 25, 2194–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian S, Hou Y, Zhou X, Li X, Yong J, Wang Y, Wang W, Yan J, Hu B, Guo H, et al. (2018). Single-cell multiomics sequencing and analyses of human colorectal cancer. Science 362, 1060–1063. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, and Greenleaf WJ (2015). Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, Furlan SN, Steemers FJ, et al. (2017). Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, et al. (2019). The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter L, Rothwell DG, Mesquita B, Smowton C, Leong HS, Fernandez-Gutierrez F, Li Y, Burt DJ, Antonello J, Morrow CJ, et al. (2017). Molecular analysis of circulating tumor cells identifies distinct copy-number profiles in patients with chemosensitive and chemorefractory small-cell lung cancer. Nat Med 23, 114–119. [DOI] [PubMed] [Google Scholar]
- Casasent AK, Schalck A, Gao R, Sei E, Long A, Pangburn W, Casasent T, Meric-Bernstam F, Edgerton ME, and Navin NE (2018). Multiclonal Invasion in Breast Tumors Identified by Topographic Single Cell Sequencing. Cell 172, 205–217 e212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassetta L, Fragkogianni S, Sims AH, Swierczak A, Forrester LM, Zhang H, Soong DYH, Cotechini T, Anur P, Lin EY, et al. (2019). Human Tumor-Associated Macrophage and Monocyte Transcriptional Landscapes Reveal Cancer-Specific Reprogramming, Biomarkers, and Therapeutic Targets. Cancer Cell 35, 588–602 e510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HW, Chan A, Kwong DLW, Wei WI, Sham JST, and Yuen APW (2003). Detection of hypermethylated RIZ1 gene in primary tumor, mouth, and throat rinsing fluid, nasopharyngeal swab, and peripheral blood of nasopharyngeal carcinoma patient. Clinical cancer research 9, 1033–1038. [PubMed] [Google Scholar]
- Chen KH, Boettiger AN, Moffitt JR, Wang S, and Zhuang X (2015). RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W, Eum HH, Lee HO, Lee KM, Lee HB, Kim KT, Ryu HS, Kim S, Lee JE, Park YH, et al. (2017). Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat Commun 8, 15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Daza R, Adey A, Pliner HA, Christiansen L, Gunderson KL, Steemers FJ, Trapnell C, and Shendure J (2015). Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348, 910–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dago AE, Stepansky A, Carlsson A, Luttgen M, Kendall J, Baslan T, Kolatkar A, Wigler M, Bethel K, Gross ME, et al. (2014). Rapid phenotypic and genomic change in response to therapeutic pressure in prostate cancer inferred by high content analysis of single circulating tumor cells. PLoS One 9, e101777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Croote D, Mignardi M, Chernikova S, Samghababi P, Zhang Y, Neff N, Kowarsky M, and Caneda C (2017). Single-cell RNA-seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell reports 21, 1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A, Gao R, and Navin N (2017). Tumor evolution: Linear, branching, neutral or punctuated? Biochim Biophys Acta Rev Cancer 1867, 151–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentro SC, Leshchiner I, Haase K, Tarabichi M, Wintersinger J, Deshwar AG, Yu K, Rubanova Y, Macintyre G, and Vazquez-Garcia I (2018). Portraits of genetic intra-tumour heterogeneity and subclonal selection across cancer types. BioRxiv, 312041. [Google Scholar]
- Dey SS, Kester L, Spanjaard B, Bienko M, and van Oudenaarden A (2015). Integrated genome and transcriptome sequencing of the same cell. Nat Biotechnol 33, 285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durante MA, Rodriguez DA, Kurtenbach S, Kuznetsov JN, Sanchez MI, Decatur CL, Snyder H, Feun LG, Livingstone AS, and Harbour JW (2020). Single-cell analysis reveals new evolutionary complexity in uveal melanoma. Nat Commun 11, 496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elyada E, Bolisetty M, Laise P, Flynn WF, Courtois ET, Burkhart RA, Teinor JA, Belleau P, Biffi G, Lucito MS, et al. (2019). Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discov 9, 1102–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engblom C, Pfirschke C, and Pittet MJ (2016). The role of myeloid cells in cancer therapies. Nat Rev Cancer 16, 447–462. [DOI] [PubMed] [Google Scholar]
- Fan HC, Fu GK, and Fodor SP (2015a). Expression profiling. Combinatorial labeling of single cells for gene expression cytometry. Science 347, 1258367. [DOI] [PubMed] [Google Scholar]
- Fan HC, Fu GK, and Fodor S. P. a. (2015b). Combinatorial labeling of single cells for gene expression cytometry supplement. Science 347, 1258367–1258367. [DOI] [PubMed] [Google Scholar]
- Filbin MG, Tirosh I, Hovestadt V, Shaw ML, Escalante LE, Mathewson ND, Neftel C, Frank N, Pelton K, Hebert CM, et al. (2018). Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science 360, 331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S, Baek S, Varticovski L, Kim S, and Hager GL (2019). High Quality ATAC-Seq Data Recovered from Cryopreserved Breast Cell Lines and Tissue. Sci Rep 9, 516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiti F, Chaligne R, Gu H, Brand RM, Kothen-Hill S, Schulman RC, Grigorev K, Risso D, Kim K-T, and Pastore A (2019). Epigenetic evolution and lineage histories of chronic lymphocytic leukaemia. Nature 569, 576–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Davis A, McDonald TO, Sei E, Shi X, Wang Y, Tsai PC, Casasent A, Waters J, Zhang H, et al. (2016). Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nat Genet 48, 1119–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Kim C, Sei E, Foukakis T, Crosetto N, Chan LK, Srinivasan M, Zhang H, Meric-Bernstam F, and Navin N (2017). Nanogrid single-nucleus RNA sequencing reveals phenotypic diversity in breast cancer. Nat Commun 8, 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawad C, Koh W, and Quake SR (2014). Dissecting the clonal origins of childhood acute lymphoblastic leukemia by single-cell genomics. Proc Natl Acad Sci U S A 111, 17947–17952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawad C, Koh W, and Quake SR (2016). Single-cell genome sequencing: current state of the science. Nat Rev Genet 17, 175–188. [DOI] [PubMed] [Google Scholar]
- Gierahn TM, Wadsworth MH 2nd, Hughes TK, Bryson BD, Butler A, Satija R, Fortune S, Love JC, and Shalek AK (2017). Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat Methods 14, 395–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein LD, Chen YJ, Dunne J, Mir A, Hubschle H, Guillory J, Yuan W, Zhang J, Stinson J, Jaiswal B, et al. (2017). Massively parallel nanowell-based single-cell gene expression profiling. BMC Genomics 18, 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goveia J, Rohlenova K, Taverna F, Treps L, Conradi LC, Pircher A, Geldhof V, de Rooij L, Kalucka J, Sokol L, et al. (2020). An Integrated Gene Expression Landscape Profiling Approach to Identify Lung Tumor Endothelial Cell Heterogeneity and Angiogenic Candidates. Cancer Cell 37, 21–36 e13. [DOI] [PubMed] [Google Scholar]
- Guo H, Zhu P, Wu X, Li X, Wen L, and Tang F (2013). Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome research 23, 2126–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Zhang Y, Zheng L, Zheng C, Song J, Zhang Q, Kang B, Liu Z, Jin L, Xing R, et al. (2018). Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med 24, 978–985. [DOI] [PubMed] [Google Scholar]
- Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M, Choudhury SR, Aguet F, Gelfand E, Ardlie K, et al. (2017). Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods 14, 955–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib N, Li Y, Heidenreich M, Swiech L, Avraham-Davidi I, Trombetta JJ, Hession C, Zhang F, and Regev A (2016). Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science 353, 925–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, and Weinberg RA (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. [DOI] [PubMed] [Google Scholar]
- Hida K, Maishi N, Annan DA, and Hida Y (2018). Contribution of Tumor Endothelial Cells in Cancer Progression. Int J Mol Sci 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinohara K, Wu H-J, Vigneau S, McDonald TO, Igarashi KJ, Yamamoto KN, Madsen T, Fassl A, Egri SB, and Papanastasiou M (2018). KDM5 histone demethylase activity links cellular transcriptomic heterogeneity to therapeutic resistance. Cancer cell 34, 939–953. e939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho YJ, Anaparthy N, Molik D, Mathew G, Aicher T, Patel A, Hicks J, and Hammell MG (2018). Single-cell RNA-seq analysis identifies markers of resistance to targeted BRAF inhibitors in melanoma cell populations. Genome Res 28, 1353–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SP, Chan TE, Lombardo Y, Corleone G, Rotmensz N, Bravaccini S, Rocca A, Pruneri G, McEwen KR, Coombes RC, et al. (2019). Single-cell transcriptomics reveals multi-step adaptations to endocrine therapy. Nat Commun 10, 3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu X, Li F, Wu K, Liang J, Shao D, et al. (2012). Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell 148, 873–885. [DOI] [PubMed] [Google Scholar]
- Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, Lonnerberg P, and Linnarsson S (2014). Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods 11, 163–166. [DOI] [PubMed] [Google Scholar]
- Jordan NV, Bardia A, Wittner BS, Benes C, Ligorio M, Zheng Y, Yu M, Sundaresan TK, Licausi JA, and Desai R (2016). HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature 537, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R (2016). The biology and function of fibroblasts in cancer. Nat Rev Cancer 16, 582–598. [DOI] [PubMed] [Google Scholar]
- Karaayvaz M, Cristea S, Gillespie SM, Patel AP, Mylvaganam R, Luo CC, Specht MC, Bernstein BE, Michor F, and Ellisen LW (2018). Unravelling subclonal heterogeneity and aggressive disease states in TNBC through single-cell RNA-seq. Nature communications 9, 3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Gao R, Sei E, Brandt R, Hartman J, Hatschek T, Crosetto N, Foukakis T, and Navin NE (2018). Chemoresistance Evolution in Triple-Negative Breast Cancer Delineated by Single-Cell Sequencing. Cell 173, 879–893 e813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KT, Lee HW, Lee HO, Song HJ, Jeong da E, Shin S, Kim H, Shin Y, Nam DH, Jeong BC, et al. (2016). Application of single-cell RNA sequencing in optimizing a combinatorial therapeutic strategy in metastatic renal cell carcinoma. Genome Biol 17, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, and Kirschner MW (2015). Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan TT, Bardia A, Spring LM, Giobbie-Hurder A, Kalinich M, Dubash T, Sundaresan T, Hong X, LiCausi JA, Ho U, et al. (2018). A Digital RNA Signature of Circulating Tumor Cells Predicting Early Therapeutic Response in Localized and Metastatic Breast Cancer. Cancer Discov 8, 1286–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laks E, McPherson A, Zahn H, Lai D, Steif A, Brimhall J, Biele J, Wang B, Masud T, Ting J, et al. (2019). Clonal Decomposition and DNA Replication States Defined by Scaled Single-Cell Genome Sequencing. Cell 179, 1207–1221 e1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts D, Wauters E, Boeckx B, Aibar S, Nittner D, Burton O, Bassez A, Decaluwe H, Pircher A, Van den Eynde K, et al. (2018). Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med 24, 1277–1289. [DOI] [PubMed] [Google Scholar]
- Lan F, Demaree B, Ahmed N, and Abate AR (2017). Single-cell genome sequencing at ultra-high-throughput with microfluidic droplet barcoding. Nat Biotechnol 35, 640–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau CA, Duarte FM, Chew JG, Kartha VK, Burkett ZD, Kohlway AS, Pokholok D, Aryee MJ, Steemers FJ, Lebofsky R, and Buenrostro JD (2019). Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nat Biotechnol 37, 916–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson DA, Bhakta NR, Kessenbrock K, Prummel KD, Yu Y, Takai K, Zhou A, Eyob H, Balakrishnan S, Wang CY, et al. (2015). Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature 526, 131–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledergor G, Weiner A, Zada M, Wang S-Y, Cohen YC, Gatt ME, Snir N, Magen H, Koren-Michowitz M, and Herzog-Tzarfati K (2018). Single cell dissection of plasma cell heterogeneity in symptomatic and asymptomatic myeloma. Nature medicine 24, 1867–1876. [DOI] [PubMed] [Google Scholar]
- Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Yang JL, Ferrante TC, Terry R, Jeanty SS, Li C, Amamoto R, et al. (2014). Highly multiplexed subcellular RNA sequencing in situ. Science 343, 1360–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung ML, Davis A, Gao R, Casasent A, Wang Y, Sei E, Sanchez E, Maru D, Kopetz S, and Navin NE (2017a). Single cell DNA sequencing reveals a late-dissemination model in metastatic colorectal cancer. Genome Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung ML, Davis A, Gao R, Casasent A, Wang Y, Sei E, Vilar E, Maru D, Kopetz S, and Navin NE (2017b). Single-cell DNA sequencing reveals a late-dissemination model in metastatic colorectal cancer. Genome Res 27, 1287–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung ML, Wang Y, Kim C, Gao R, Jiang J, Sei E, and Navin NE (2016). Highly multiplexed targeted DNA sequencing from single nuclei. Nat Protoc 11, 214–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xu X, Song L, Hou Y, Li Z, Tsang S, Li F, Im KM, Wu K, Wu H, et al. (2012). Single-cell sequencing analysis characterizes common and cell-lineage-specific mutations in a muscle-invasive bladder cancer. Gigascience 1, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr JG, Adalsteinsson VA, Cibulskis K, Choudhury AD, Rosenberg M, Cruz-Gordillo P, Francis JM, Zhang C-Z, Shalek AK, and Satija R (2014). Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nature biotechnology 32, 479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay IC, Haerty W, Kumar P, Li YI, Hu TX, Teng MJ, Goolam M, Saurat N, Coupland P, Shirley LM, et al. (2015). G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat Methods. [DOI] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. (2015). Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelotto LG, Baslan T, Kendall J, Geyer FC, Burke KA, Spraggon L, Piscuoglio S, Chadalavada K, Nanjangud G, and Ng CK (2017a). Whole-genome single-cell copy number profiling from formalin-fixed paraffin-embedded samples. Nature medicine 23, 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelotto LG, Baslan T, Kendall J, Geyer FC, Burke KA, Spraggon L, Piscuoglio S, Chadalavada K, Nanjangud G, Ng CK, et al. (2017b). Whole-genome single-cell copy number profiling from formalin-fixed paraffin-embedded samples. Nat Med 23, 376–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister SS, and Weinberg RA (2014). The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nature cell biology 16, 717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGranahan N, and Swanton C (2015). Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 27, 15–26. [DOI] [PubMed] [Google Scholar]
- McMahon CM, Ferng T, Canaani J, Wang ES, Morrissette JJ, Eastburn DJ, Pellegrino M, Durruthy-Durruthy R, Watt CD, and Asthana S (2019). Clonal selection with Ras pathway activation mediates secondary clinical resistance to selective FLT3 inhibition in acute myeloid leukemia. Cancer discovery, CD-18–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto DT, Lee RJ, Kalinich M, LiCausi JA, Zheng Y, Chen T, Milner JD, Emmons E, Ho U, and Broderick K (2018). An RNA-based digital circulating tumor cell signature is predictive of drug response and early dissemination in prostate cancer. Cancer discovery 8, 288–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, Desai R, Fox DB, Brannigan BW, and Trautwein J (2015). RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science 349, 1351–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooijman D, Dey SS, Boisset JC, Crosetto N, and van Oudenaarden A (2016). Single-cell 5hmC sequencing reveals chromosome-wide cell-to-cell variability and enables lineage reconstruction. Nat Biotechnol 34, 852–856. [DOI] [PubMed] [Google Scholar]
- Muller S, Kohanbash G, Liu SJ, Alvarado B, Carrera D, Bhaduri A, Watchmaker PB, Yagnik G, Di Lullo E, Malatesta M, et al. (2017). Single-cell profiling of human gliomas reveals macrophage ontogeny as a basis for regional differences in macrophage activation in the tumor microenvironment. Genome Biol 18, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam AS, Kim KT, Chaligne R, Izzo F, Ang C, Taylor J, Myers RM, Abu-Zeinah G, Brand R, Omans ND, et al. (2019). Somatic mutations and cell identity linked by Genotyping of Transcriptomes. Nature 571, 355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin N, and Hicks J (2011). Future medical applications of single-cell sequencing in cancer. Genome Med 3, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. (2011). Tumour evolution inferred by single-cell sequencing. Nature 472, 90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawroth R, Weckermann D, and Retz M (2014). Prostate and bladder cancer: detection of disseminated tumor cells in bone marrow. Der Urologe Ausg A 53, 514–518. [DOI] [PubMed] [Google Scholar]
- Naz S, Hashmi AA, Ali R, Faridi N, Hussian SD, Edhi MM, and Khan M (2015). Role of peritoneal washing cytology in ovarian malignancies: correlation with histopathological parameters. World J Surg Oncol 13, 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, Richman AR, Silverbush D, Shaw ML, Hebert CM, et al. (2019). An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 178, 835–849 e821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen QH, Pervolarakis N, Nee K, and Kessenbrock K (2018). Experimental Considerations for Single-Cell RNA Sequencing Approaches. Front Cell Dev Biol 6, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni X, Zhuo M, Su Z, Duan J, Gao Y, Wang Z, Zong C, Bai H, Chapman AR, and Zhao J (2013). Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proceedings of the National Academy of Sciences 110, 21083–21088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichterwitz S, Chen G, Aguila Benitez J, Yilmaz M, Storvall H, Cao M, Sandberg R, Deng Q, and Hedlund E (2016). Laser capture microscopy coupled with Smart-seq2 for precise spatial transcriptomic profiling. Nat Commun 7, 12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen RP, White MJ, Severson DT, Braden B, Bailey A, Goldin R, Wang LM, Ruiz-Puig C, Maynard ND, Green A, et al. (2018). Single cell RNA-seq reveals profound transcriptional similarity between Barrett's oesophagus and oesophageal submucosal glands. Nat Commun 9, 4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al. (2014). Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino M, Sciambi A, Treusch S, Durruthy-Durruthy R, Gokhale K, Jacob J, Chen TX, Geis JA, Oldham W, Matthews J, et al. (2018). High-throughput single-cell DNA sequencing of acute myeloid leukemia tumors with droplet microfluidics. Genome Res 28, 1345–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Sun B-F, Chen C-Y, Zhou J-Y, Chen Y-S, Chen H, Liu L, Huang D, Jiang J, and Cui G-S (2019). Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell research 29, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petti AA, Williams SR, Miller CA, Fiddes IT, Srivatsan SN, Chen DY, Fronick CC, Fulton RS, Church DM, and Ley TJ (2019). A general approach for detecting expressed mutations in AML cells using single cell RNA-sequencing. Nat Commun 10, 3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Vila M, Usuba W, Takahashi RU, Shimomura I, Sasaki H, Ochiya T, and Yamamoto Y (2019). Single-Cell Analysis Reveals a Preexisting Drug-Resistant Subpopulation in the Luminal Breast Cancer Subtype. Cancer Res 79, 4412–4425. [DOI] [PubMed] [Google Scholar]
- Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, Rodman C, Luo CL, Mroz EA, and Emerick KS (2017). Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 171, 1611–1624. e1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambow F, Rogiers A, Marin-Bejar O, Aibar S, Femel J, Dewaele M, Karras P, Brown D, Chang YH, Debiec-Rychter M, et al. (2018). Toward Minimal Residual Disease-Directed Therapy in Melanoma. Cell 174, 843–855 e819. [DOI] [PubMed] [Google Scholar]
- Ramskold D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, et al. (2012). Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol 30, 777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, Bodenmiller B, Campbell P, Carninci P, Clatworthy M, et al. (2017). The Human Cell Atlas. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriques SG, Stickels RR, Goeva A, Martin CA, Murray E, Vanderburg CR, Welch J, Chen LM, Chen F, and Macosko EZ (2019). Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, Lieb DJ, Chen JH, Frederick DT, Barzily-Rokni M, et al. (2018). Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell 175, 998–1013 e1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy AT, Granja JM, Yost KE, Qi Y, Meschi F, McDermott GP, Olsen BN, Mumbach MR, Pierce SE, Corces MR, et al. (2019). Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat Biotechnol 37, 925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas P, Virassamy B, Ye C, Salim A, Mintoff CP, Caramia F, Salgado R, Byrne DJ, Teo ZL, Dushyanthen S, et al. (2018). Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat Med 24, 986–993. [DOI] [PubMed] [Google Scholar]
- Shaffer SM, Dunagin MC, Torborg SR, Torre EA, Emert B, Krepler C, Beqiri M, Sproesser K, Brafford PA, and Xiao M (2017). Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 546, 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, Lubeck E, Zhou W, and Cai L (2017). seqFISH Accurately Detects Transcripts in Single Cells and Reveals Robust Spatial Organization in the Hippocampus. Neuron 94, 752–758 e751. [DOI] [PubMed] [Google Scholar]
- Sharma A, Cao EY, Kumar V, Zhang X, Leong HS, Wong AML, Ramakrishnan N, Hakimullah M, Teo HMV, Chong FT, et al. (2018). Longitudinal single-cell RNA sequencing of patient-derived primary cells reveals drug-induced infidelity in stem cell hierarchy. Nat Commun 9, 4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, and Allison JP (2015). Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell 161, 205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood SA, Lee HJ, Angermueller C, Krueger F, Saadeh H, Peat J, Andrews SR, Stegle O, Reik W, and Kelsey G (2014). Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nature methods 11, 817–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q, Hawkins GA, Wudel L, Chou PC, Forbes E, Pullikuth AK, Liu L, Jin G, Craddock L, Topaloglu U, et al. (2019). Dissecting intratumoral myeloid cell plasticity by single cell RNA-seq. Cancer Med 8, 3072–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl PL, Salmen F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, Giacomello S, Asp M, Westholm JO, Huss M, et al. (2016). Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82. [DOI] [PubMed] [Google Scholar]
- Stuart T, and Satija R (2019). Integrative single-cell analysis. Nat Rev Genet 20, 257–272. [DOI] [PubMed] [Google Scholar]
- Stubbington MJT, Lonnberg T, Proserpio V, Clare S, Speak AO, Dougan G, and Teichmann SA (2016). T cell fate and clonality inference from single-cell transcriptomes. Nat Methods 13, 329–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Ni X, Duan J, Gao Y, Zhuo M, Li R, Zhao J, Ma Q, Bai H, and Chen H (2019). Inferring the Evolution and Progression of Small-Cell Lung Cancer by Single-Cell Sequencing of Circulating Tumor Cells. Clinical Cancer Research, clincanres. 3571.2018. [DOI] [PubMed] [Google Scholar]
- Svensson V, Vento-Tormo R, and Teichmann SA (2018). Exponential scaling of single-cell RNA-seq in the past decade. Nat Protoc 13, 599–604. [DOI] [PubMed] [Google Scholar]
- Tanay A, and Regev A (2017). Scaling single-cell genomics from phenomenology to mechanism. Nature 541, 331–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, et al. (2009). mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 6, 377–382. [DOI] [PubMed] [Google Scholar]
- Tickle T,TI, Georgescu C, Brown M, Haas B (2019). inferCNV of the Trinity CTAT Project. [Google Scholar]
- Ting DT, Wittner BS, Ligorio M, Jordan NV, Shah AM, Miyamoto DT, Aceto N, Bersani F, Brannigan BW, and Xega K (2014). Single-cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell reports 8, 1905–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. (2016a). Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K, Fisher JM, Rodman C, Mount B, Filbin MG, et al. (2016b). Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539, 309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadhachary GR, Abbruzzese JL, and Lenzi R (2004). Diagnostic strategies for unknown primary cancer. Cancer-Am Cancer Soc 100, 1776–1785. [DOI] [PubMed] [Google Scholar]
- Venteicher AS, Tirosh I, Hebert C, Yizhak K, Neftel C, Filbin MG, Hovestadt V, Escalante LE, Shaw ML, Rodman C, et al. (2017). Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitak SA, Torkenczy KA, Rosenkrantz JL, Fields AJ, Christiansen L, Wong MH, Carbone L, Steemers FJ, and Adey A (2017). Sequencing thousands of single-cell genomes with combinatorial indexing. Nat Methods 14, 302–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Fan HC, Behr B, and Quake SR (2012). Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell 150, 402–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Fan J, Francis JM, Georghiou G, Hergert S, Li S, Gambe R, Zhou CW, Yang C, and Xiao S (2017). Integrated single-cell genetic and transcriptional analysis suggests novel drivers of chronic lymphocytic leukemia. Genome research 27, 1300–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Guldner IH, Golomb SM, Sun L, Harris JA, Lu X, and Zhang S (2019). Single-cell profiling guided combinatorial immunotherapy for fast-evolving CDK4/6 inhibitor-resistant HER2-positive breast cancer. Nat Commun 10, 3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, and Navin NE (2015). Advances and applications of single-cell sequencing technologies. Mol Cell 58, 598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X, Chen K, Scheet P, Vattathil S, and Liang H (2014a). Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature 512, 155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X, Chen K, Scheet P, Vattathil S, Liang H, et al. (2014b). Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature 512, 155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohnhaas CT, Leparc GG, Fernandez-Albert F, Kind D, Gantner F, Viollet C, Hildebrandt T, and Baum P (2019). DMSO cryopreservation is the method of choice to preserve cells for droplet-based single-cell RNA sequencing. Scientific reports 9, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Takeda S, Fujii T, Nomoto S, Kanazumi N, Sugimoto H, Kasuya H, Kodera Y, Nagasaka T, and Morita S (2007). Clinical implications of peritoneal cytology in potentially resectable pancreatic cancer: positive peritoneal cytology may not confer an adverse prognosis. Annals of surgery 246, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Jiang Y, Lam KG, Berletch JB, Disteche CM, Noble WS, Steemers FJ, Camerini-Otero RD, Adey AC, and Shendure J (2019). High-Throughput Single-Cell Sequencing with Linear Amplification. Molecular cell 76, 676–690 e610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Yu J, Yao X, Wu WK, Lu Y, Tang S, Li X, Bao L, Li X, Hou Y, et al. (2014). Discovery of biclonal origin and a novel oncogene SLC12A5 in colon cancer by single-cell sequencing. Cell Res 24, 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn H, Steif A, Laks E, Eirew P, VanInsberghe M, Shah SP, Aparicio S, and Hansen CL (2017a). Scalable whole-genome single-cell library preparation without preamplification. Nat Methods 14, 167–173. [DOI] [PubMed] [Google Scholar]
- Zahn H, Steif A, Laks E, Eirew P, VanInsberghe M, Shah SP, Aparicio S, and Hansen CL (2017b). Scalable whole-genome single-cell library preparation without preamplification. Nature methods 14, 167. [DOI] [PubMed] [Google Scholar]
- Zhang P, Yang M, Zhang Y, Xiao S, Lai X, Tan A, Du S, and Li S (2019). Dissecting the Single-Cell Transcriptome Network Underlying Gastric Premalignant Lesions and Early Gastric Cancer. Cell Rep 27, 1934–1947 e1935. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Eichten A, Parveen A, Adler C, Huang Y, Wang W, Ding Y, Adler A, Nevins T, Ni M, et al. (2018). Single-Cell Transcriptome Analyses Reveal Endothelial Cell Heterogeneity in Tumors and Changes following Antiangiogenic Treatment. Cancer Res 78, 2370–2382. [DOI] [PubMed] [Google Scholar]
- Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, Guo X, Kang B, Hu R, Huang JY, Zhang Q, et al. (2017a). Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 169, 1342–1356 e1316. [DOI] [PubMed] [Google Scholar]
- Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. (2017b). Massively parallel digital transcriptional profiling of single cells. Nature communications 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Gao Y, Guo H, Xia B, Song J, Wu X, Zeng H, Kee K, Tang F, and Yi C (2017). Single-cell 5-formylcytosine landscapes of mammalian early embryos and ESCs at single-base resolution. Cell Stem Cell 20, 720–731. e725. [DOI] [PubMed] [Google Scholar]
- Zong C, Lu S, Chapman AR, and Xie XS (2012). Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science 338, 1622–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]