Abstract
The COVID-19 pandemic in Egypt is a part of the worldwide global crisis of coronavirus 2 (SARS-CoV-2). The contagious life-threatening condition causes acute respiratory syndrome. The present study aimed to assess the compounds identified by LC-MS of the methanolic leaves extracts from three conifers trees cultivated in Egypt (Araucaria bidwillii, Araucaria. cunninghamii and Araucaria heterophylla) via docking technique as potential inhibitor of COVID-19 virus on multiple targets; viral main protease (Mpro, 6LU7), non-structural protein-16 which is a methyl transferase (nsp16, 6W4H) and RNA dependent RNA polymerase (nsp12, 7BV2). Among the three targets, nsp16 was the best target recognized by the tested compounds as can be deduced from docking studies. Moreover, the methanolic extract of A. cunninghamii showed the highest radical-scavenging activity using (DPPH test) with 53.7 µg/mL comparable to ascorbic acid with IC50 = 46 µg/mL The anti-inflammatory potential carried using enzyme linked immunoassay showed the highest activity for A. cunninghamii and A. bidwillii followed by A. heterophylla with IC50 = 23.20 ± 1.17 µg/mL, 82.83 ± 3.21 µg/mL and 221.13 ± 6.7 µg/mL, respectively (Celecoxib was used as a standard drug with IC50 = 141.92 ± 4.52 µg/mL). Moreover, a molecular docking study was carried for the LC-MS annotated metabolites to validate their anti-inflammatory inhibitory effect using Celecoxib as a reference compound and showed a high docking score (−7.7 kcal/mol) for Octadecyl (E) P-coumarate and (−7.3 kcal/mol) for secoisolariciresinol rhamnoside.
Communicated by Ramaswamy H. Sarma
Keywords: A. heterophylla, A. cunninghamii, A. bidwillii, Araucariaceae, COVID-19, Molecular Docking, Anti-inflammatory, DPPH
1. Introduction
The COVID-19 pandemic in Egypt is a part of the worldwide global crisis of coronavirus 2 (SARS-CoV-2). It is a contagious condition causes acute respiratory syndrome (Chiappetta et al., 2020; Zabetakis et al., 2020). The first case reported with that disease was a women in local market in Wuhan province, China in December 2019, the scientists believe it’s acquired from animals and started spreading between humans (https://africacdc.org/covid-19/). The danger of that disease is the rapid spreading as it’s highly contagious and its vigorous symptoms which might lead to death (Schett et al., 2020). Up till now is available only a supportive treatment as a strategy in the treatment protocol, WHO welcomed the satisfactory results of using dexamethasone as a preliminary treatment for COVID-19 (World Health Organiztion, 2020) as it reduces the risk of mortality in infected patients for its role as anti-inflammatory reducing the immune-system cytokine storm reducing the exaggerated immune response caused by the COVID-19 (Mahmudpour et al., 2020)
Inflammation is a biological body response as a result of exposure to stimuli which provokes the immune system to eliminate the harmful stimuli. Prostaglandins are responsible for the generation of any inflammatory response (Martínez-Sánchez et al., 2020). Prostaglandins biosynthesis is markedly increased in inflamed tissue, arachidonic acid biosynthesis to prostaglandin H2 is catalyzed by cyclooxygenase (Lee et al., 2017). As a result of the human defense mechanism during the inflammation, there is an excessive production of reactive oxygen species by phagocytic leucocytes (Jayawardena et al., 2020), here appears the role of antioxidants to scavenge the reactive species, these auxiliary process aids tissue repair and healing (Ik et al., 2013).
Plants used medicinally considered as a huge source for constituents with biological various activities. So, it was a substantial importance to seek a plant origin drug that might provide a safer therapy to control the covid-19 symptoms or as a curative treatment. Therefore, three Araucaria species were subjected for the following study to investigate their anti-inflammatory and antioxidant potential activities. Anti-inflammatory activity for that genus was reported before using different techniques, exerting a significant results (Aslam et al., 2013; Elshamy et al., 2020) which was interesting to carry further investigations for the anti-inflammatory activity using different and accurate assay; as the enzyme immunoassay ELISA method which is performed here in the current research comparing three species.
It is well known now the role of inflammation in the pathogenesis of SARS-CoV-2 which suggests a promising effect of anti-inflammatory drugs as adjuvant therapy in COVID-19 patients to suppress the cytokine storm. Furthermore, dexamethasone was recently used to control SARS-CoV-2 progression (World Health Organiztion, 2020). This information has encouraged us to virtually investigate our compounds to see if any of them could have a potential effect on the new COVID-19 virus possible targets. The active sites of three targets, viral main protease (Mpro, 6LU7), non-structural protein-16 which is a methyl transferase (nsp16, 6W4H) and RNA dependent RNA polymerase (nsp12, 7BV2) using a published recently high-resolution structure of COVID-19 main protease (Mpro ) (Jin et al., 2020; Zhang et al., 2020) allowed the chance for the creation of an inhibitor with an important role affecting the transcription and replication of virus (Aly, 2020). The importance of Mpro in the life cycle of coronavirus in contrast it is not present as an identical human homologues, introduced Mpro as favoured antiviral targeted drug designed (Pillaiyar et al., 2016).
2. Materials and methods
2.1. Plant material
A. bidwillii, A. cunninghamii and A. heterophylla leaves were collected and shed dried. The plants identity was confirmed at the Department of Flora and Phytotaxonomy, ARC, Cairo, Egypt.
2.2. Extracts preparation
The powdered leaves dried in the air of A. bidwillii, A. cunninghamii and A. heterophylla weighing 200 g for each species, was separately extracted with methanol by percolation. methanolic extracts were subjected for evaporation under reduced pressure at 45 °C to a constant weight then were subjected to lyophilization and kept in the dark at 4° C until analysis.
2.3. Chemical reagents
Analytical grade reagents were purchased and used with no additional purification. Organic solvents: methanol, (Adwic, Nasr Pharma, Egypt), DPPH, standardized extract of Ascorbic acid., (Memphis Co., Egypt). (2, 2-diphenyl-1-picrylhydrazyl) free radical (Sigma Co., USA), COX-1 (human) Inhibitor Screening Assay Kit., by (Caymanchem, USA), Celecoxib (Pfizer Inc., Egypt).
2.4. Metabolic profiling of A. bidwillii, A. cunninghamii and A. heterophylla
Metabolic profiling was performed on methanolic extracts of A. bidwillii, A. heterophylla and A. cunninghamii according to (Sobeh et al., 2017). HPLC-PDA-MS/MS was carried at Institute of Pharmacy and Molecular Biotechnology, Heidelberg University, Germany using a ThermoFinnigan LC system (Thermo Electron Corporation, Austin, TX, USA). A Zorbax Eclipse (XDB-C18), Rapid resolution of 4.6 × 150 mm, 3.5 µm column was used (Agilent, Santa Clara, CA, USA). Operating mass parameters were used in the negative mode. A combination of gradient water and acetonitrile (ACN), 0.1% formic acid have been used, through 60 min in 1 mL/min flow rate and 1:1 splitted before the ESI source, acetonitrile was raised from 5% → 30%. The samples were injected separately using the autosampler. For MS analysis, LCQ-Duo ion trap with a ThermoQuest ESI source has been used. The Xcalibur software was used (Xcalibur™ 2.0.7, Thermo Scientific, Waltham, MA, USA) for system controlling.
2.5. Antioxidant activity evaluation by DPPH free radical scavenging assay
The plant extract ability to quench DPPH free radicals was evaluated by a standard method (Takao et al., 1994) accredited with slight modifications (Kumarasamy et al., 2007). Methanolic extracts of A. bidwillii, A. cunninghamii and A. heterophylla were dissolved in methanol to accomplish 15 mg/mL concentrations. Dilutions were carried to have the concentrations of 500, 250, 125, 62.5, 31.25 μg/mL. (100 μL each) of the solutions after dilution were added to 3 mL of methanolic solution of DPPH with the concentration of 0.002%. As, the standard ascorbic acid stock solution was dissolved in methanol for achieving a 1.5 mg/mL concentration, dilutions were carried to achieve concentrations 50, 25, 12.5, 6.25, 3.125 μg/mL incubation in the dark for 30 min, the absorbances were observed. The reaction undergoes between the antioxidant and free radical to produce 1,1-diphenyl-2-picryl hydrazine. The capacity to quench the free radical, DPPH was recorded at the absorbance of 517 nm.
2.6. Anti-inflammatory
The biosynthesis of AA to PGH2 is catalyzed by Cyclooxygenase. PGH2 produces PGF2α, via stannous chloride reduction to be assessed by enzyme immunoassay (ELISA) (Pradelles et al., 1985). This is a competitive method among a PG tracer (PG-acetylcholinesterase conjugate) and PGs for the limited quantity of PG antiserum. The PG tracer amount that can bind to the PG antiserum is indirectly proportional to the amount of PGs in the wells, the PG tracer concentration is kept constant, but PG concentration varies. This complex of antibody-PG conjugate to an antibody (anti-IgG) which was before linked to the well. The buffer and the acetylcholinesterase substrate (Ellman’s reagent) poured to the well. The reaction enzymatically produces a yellow colour which is measured via a spectrophotometer in a Microplate special Reader (BioRad, Japan) at 412 nm. The technique to have 100% COX activity was carried out in the presence and absence of DMSO as a solvent control. The inhibitory experiment was carried for extracts at different concentrations starting from 1 ending with 16 μg total phenolic compound/mL) or of the celecoxib drug as a standard anti-inflammatory. After adding the enzyme, a pre-incubation period was allowed of 10 min before adding the inhibitor and 2 min of incubation in case of AA presence at 37 °C. inactivation of COXs by keeping them in water and allowed to boil for 3 min as Enzyme control. 29 PG/mL was the detection limit and the coefficients of variations intra and interassay were 5 and 10%, respectively. The anti-inflammatory activity of the tested extracts was assessed by determining the percent of inhibition of PGE2 produced and the concentration of the test compounds leading to 50% inhibition of the release of PGE2 (IC50) was detected from the curve of concentration-inhibition response by analytical regression.
2.7. Molecular docking
Docking took place in the compounds active sites of the corresponding proteins including COX-1 (1EQG), main protease of SARS-CoV-2 (6LUV7), nsp16 (6W4H) and nsp12 (7BV2). Ligands and proteins were prepared as reported earlier (Ebada et al., 2020). In short, structures were either drawn or used from the PubChem (www.pubchem.ncbi.nlm.nih.gov). Minimization for Structures via one thousand steps of steepest descent after that a minimization through one thousand steps of gradient conjugate. Available Proteins were used from the protein data bank (www.rcsb.org) then hydrogens were added and merged, water and non-bonded fragments were removed, and Gasteiger charges were calculated. AutoDock vina was used for carrying docking (Trott & Olson, 2010) with a grid box of 25 × 25 × 25 Å3 centered on the internal ligand using exhaustiveness of 16. PyMOL was responsible for generating images (Schrodinger, 2010).
2.8. Molecular dynamics simulation
Amber ff14SB force field was used to implement Amber18 to make the ligand-protein complexes in water. The module of antechamber was used to adjust the Gasteiger atomic partial charges (Case et al., 2020). For preparation of the general Amber GAFF force field and the force field parameters (Wang et al., 2004), Addition of hydrogen atoms was to target proteins via the tleap program of the AmberTools18 package (Case et al., 2020). First, the complexes of ligand-protein were each laid in a box with cubical shape with conditions of periodic boundary, TIP3P water as a solvent was included in the box for the MD simulations of MD. (Jorgensen et al., 1983), Cl− and Na+ ions supplied a salt of 0.10 M concentration. the system was set up, minimization of energy was carried via five hundred steps of steepest descent algorithm and five hundred steps of gradient conjugation method. The essential atoms for complex clustering were fixed by the additional harmonic potential with constant force of 2.0 kcal/mol subjecting the system to the phase of equilibration. Equilibration of the system was performed in three steps: (1) gradual heating of the system using a Langevin thermostat with a collision frequency of 2.0 ps−1 heated from 0 K to 310 K for 1 ns in NVT ensemble; (2) equilibration of pressure was done at 1.0 bar in NPT for 1 ns ensemble via Berendsen barostat with a 2.0 ps (Case et al., 2020); (3) equilibration was achieved at 310 K over 0.5 ns in conditions of constant volume and the complex assembly constraints were eliminated. Once the equilibration was accomplished, the simulations of MD were performed at temperature T = 310 K and p = 1 bar and for 150 ns in NPT ensemble. SHAKE algorithm was used to constrained bonds participating in hydrogen atoms (Ryckaert, 1977) for accomplishing the integration time-step of 2 ps. Calculations of the electrostatic Long-range interactions were made via Particle Mesh Ewald algorithm (Essmann et al., 1995). Truncations at 8 Å of van der Waals and Coulomb interactions and the calculations of energy values of binding were made with Amber18 (Case et al., 2020) via the MM/GBSA method (Genheden & Ryde, 2015). 750 snapshots were calculated produced from the final 150 ns of the MD trajectories, by maintaining the snapshots every 0.2 ns. Computations of the energies of the polar solvation in continuum solvent via Poisson-Boltzmann continuum-solvation model with ionic strength of 0.10. The non-polar terms were evaluated via solvent accessible surface areas. Application of the Nmode module in Amber 18 was performed for the calculation of the entropy for the binding free energy (Case et al., 2020). MD trajectories analysis was carried by the CPPTRAJ module of AmberTools 18 (Case et al., 2020).
3. Results and discussion
3.1. A. Cunninghamii; A. bidwillii and A. heterophylla methanolic extracts metabolic profiling
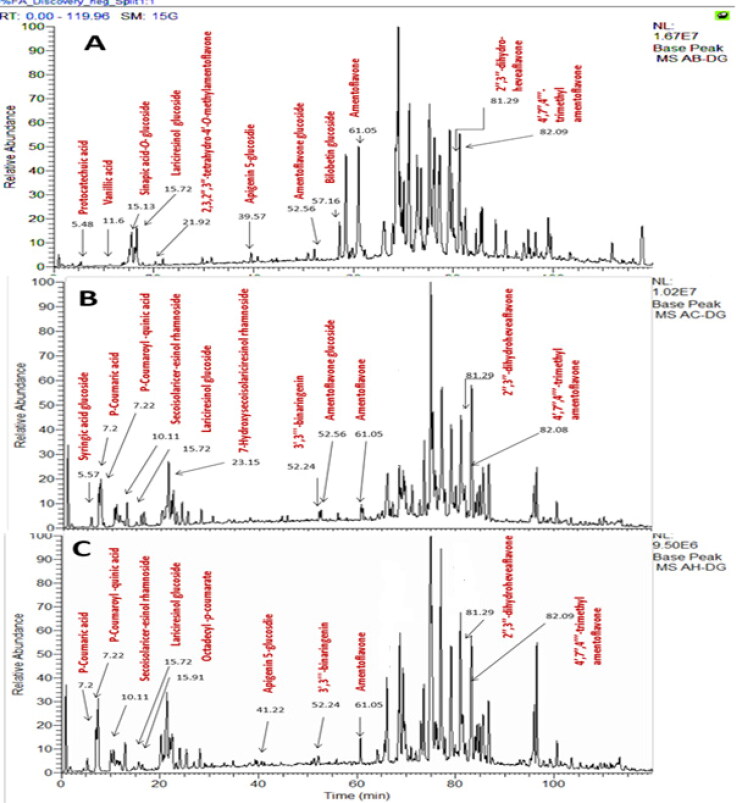
Secondary metabolites of crude methanolic extract of A. bidwillii, A. cunninghamii, and A. heterophylla dereplication revealed the annotation of various compounds of different classes. The annotated compounds detect the presence of diverse phenolic acids, biflavonoids and lignans Table 1 and Figure 1.
Table 1.
Annotated compounds in the methanolic extracts of A. cunninghamii, A. bidwillii and A. heterophylla.
|
m/z (M-H) |
R.T (min.) |
M.wt. | Name | Molecular formula |
Araucaria |
References | ||
|---|---|---|---|---|---|---|---|---|
| bid. | Cun. | Het. | ||||||
| 153.10 | 5.48 | 154.02 | Protocatechuic acid | C7H6O4 | + | − | − | Fang et al. (2002) |
| 359.21 | 5.57 | 360.11 | Syringic acid glucoside | C15H20O10 | − | + | − | Fang et al. (2002) |
| 163.01 | 7.20 | 164.04 | P-Coumaric acid | C9H8O3 | − | + | + | Fang et al. (2002) |
| 337.99 | 7.22 | 338.31 | P-Coumaroyl-quinic acid | C16H18O8 | − | + | + | Fang et al. (2002) |
| 521.26 | 10.11 | 522.25 | Secoisolariceresinol rhamnoside | C27H38O10 | − | + | + | Eklund et al. (2008) |
| 167.06 | 11.16 | 168.04 | Vanillic acid | C8H8O4 | + | − | − | Fang et al. (2002) |
| 385.05 | 15.13 | 386.12 | Sinapic acid-O-glucoside | C17H22O10 | + | − | − | Sánchez‐Rabaneda et al. (2004) |
| 521.00 | 15.72 | 522.15 | Lariciresinol glucoside | C26H34O11 | + | + | + | Ağalar et al. (2017) |
| 415.00 | 15.91 | 416.32 | Octadecyl-p-coumarate | C27H44O3 | + | + | Purev et al. (1988) | |
| 555.14 | 21.92 | 556.52 | (2S,2′′S)-2,3,2′′,3′′-tetrahydro-4′- O-methyl amentoflavone | C31H24O10 | + | − | − | Moawad et al. (2010) |
| 523.00 | 23.15 | 524.13 | 7-Hydroxy seco-isolariciresinol rhamnoside | C26H36O11 | − | + | − | Eklund et al. (2008) |
| 431.24 | 39.57 | 432.38 | Apigenin 5-glucosdie | C21H20O10 | + | − | − | Carini et al. (2001) |
| 415.21 | 41.22 | 416.11 | Apigenin 5-O-rhamnoside | C21H20O9 | − | + | + | El-Hawary et al. (2017) |
| 541.23 | 52.24 | 542.21 | 3′,3′''-binaringenin | C30H22O10 | − | + | + | Yao et al. (2017) |
| 699.15 | 52.56 | 700.14 | Amentoflavone glucoside | C36H28O15 | + | + | − | Yao et al. (2017) |
| 713.00 | 57.16 | 714.10 | Bilobetin glucoside | C31H20O10 | + | + | + | Yao et al. (2017) |
| 537.32 | 61.05 | 538.09 | Amentoflavone | C30H18O10 | + | + | + | Yao et al. (2017) |
| 581.15 | 81.29 | 582.56 | 2′’,3′’-dihydroheveaflavone | C33H26O10 | + | + | + | Yao et al. (2017) |
| 579.19 | 82.08 | 580.12 | Kayaflavone (4′,7′’,4′’’-trimethyl amentoflavone | C33H24O10 | + | + | + | Yao et al. (2017) |
R.T: retention time, bid: bidwillii, cun: cunninghamii, het: heterophylla.
Figure 1.
Chromatogram LC-MS of methanolic extracts of A. bidwillii (A), A. cunninghamii (B), A. heterophylla (C).
3.2. Anti-inflammatory activity
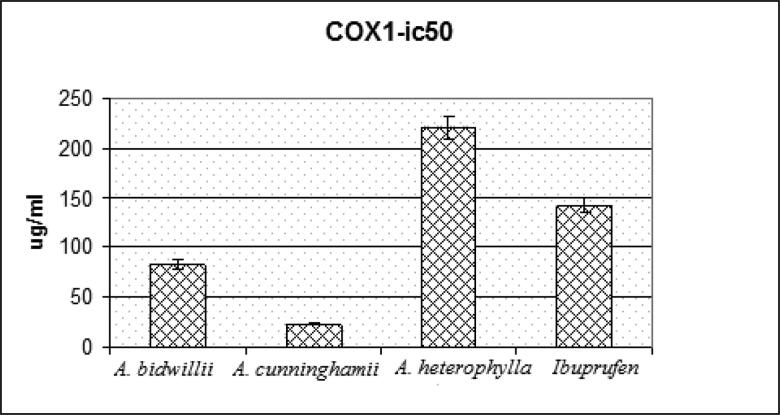
The concentration inhibiting 50% of the release of PGE2 (IC50) of the extracts under investigation, A.bidwillii, A.cunninghamii and A. heterophylla to examine the extracts anti-inflammatory activity was determined from the curve of concentration-inhibition response through regression analysis. The reported values are the means of three experiments and represented in Table 2 and Figure 2.
Table 2.
IC50 of A. cunninghamii, A. bidwillii, A. heterophylla and Celecoxib.
| Methanolic extracts | IC50 µg/mL |
|---|---|
| A. bidwillii | 82.83 ± 3.21 |
| A. cunninghamii | 23.20 ± 1.17 |
| A. heterophylla | 221.13 ± 6.7 |
| Celecoxib | 141.92 ± 4.52 |
Figure 2.
IC50 of A. cunninghamii, A. bidwillii, A. heterophylla and Celecoxib.
3.3. Antioxidant activity
The DPPH and its % inhibition of methanolic extracts showed IC50 values 93.32 μg/mL, 53.7 μg/mL and 120.226 μg/mL for A. bidwillii, A. cunninghamii and A. heterophylla respectively. The reference drug used was ascorbic acid which showed 46 μg/mL Among these results methanolic extract of A. cunninghamii exhibited a potent antioxidant activity than the other extracts followed by A. bidwillii and A. heterophylla extracts. The total results of inhibition percentage as represented in Table 3, respective to IC50 values.
Table 3.
IC50 of A. cunninghamii, A. bidwillii, A. heterophylla and ascorbic acid.
| Methanolic extracts | IC50 µg/mL |
|---|---|
| A. bidwillii | 93.32 |
| A. cunninghamii | 53.7 |
| A. heterophylla | 120.226 |
| Ascorbic acid | 46 |
3.4. Statistical analysis
Standard Deviation (SD) ± means data set of n = 3 was determined through the subsequent equation, whilst the values of IC50 were calculated from plotting inhibition percentage against concentration, via a non-linear regression algorithm. Presenting data as mean values ± of (n = 3) standard deviation.
3.5. Molecular docking
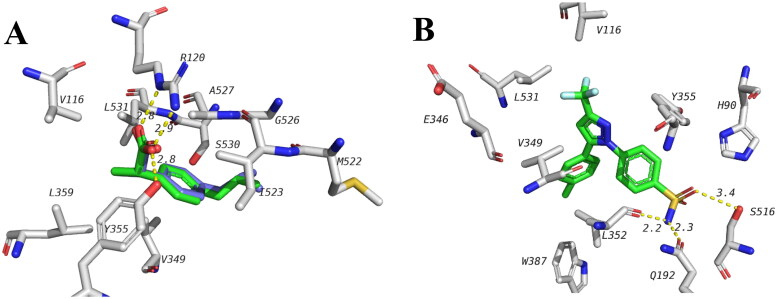
The plant extracts of A. bidwillii, A. cunninghamii, and A. heterophylla showed anti-inflammatory effects as shown by their ability to inhibit the release of PGE2. The possible inhibitory effect of compounds in the extracts for COX-1 was studied using docking. The docking study was done using 1EQG PDB file which has co-crystalized ibuprofen. To establish the docking procedure, ibuprofen was redocked in its active site and the correct pose was predicted with high accuracy (RMSD of 0.381) and the docking energy was found to be −7.8 kcal/mol (Figure 3A). This indicates the validity of docking procedure to predict correct pose. Ibuprofen (and most of the anti-inflammatory non-steroidal drugs, NSAIDS) forms 2 hydrogen bonds with R120 and Y355. The hydrophobic part of ibuprofen extends in a hydrophobic pocket formed around I345, V349, L359, I523. Selective COX-2 inhibitors, on the other hand, do not inhibit COX-1 to the same extent. Although that is the case, a selective COX-2 inhibitor, celecoxib, is known to bind to one of two subunits of COX-1 and the interference of the action of low dose aspirin (Rimon et al., 2010). Binding site of celecoxib is close but different from that of known NSAIDs such as celecoxib (Figure 3B). Celecoxib sulphamoyl moiety forms hydrogen bonds with S516, Q192 and L352 while hydrophobic interactions are seen with V116, L531 and V349. Redocking of celecoxib in COX-1 resulted in an energy of −5.0 kcal/mol.
Figure 3.
A docked ibuprofen (green) in the crystal structure of COX-1 active site (1EQG) overlaid with the ibuprofen from the crystal structure (blue) with RMSD of 0.381. Key interactions are shown. B, Celecoxib binding with only one unit in the COX-1 dimer as seen in 3KK6 PDB file.
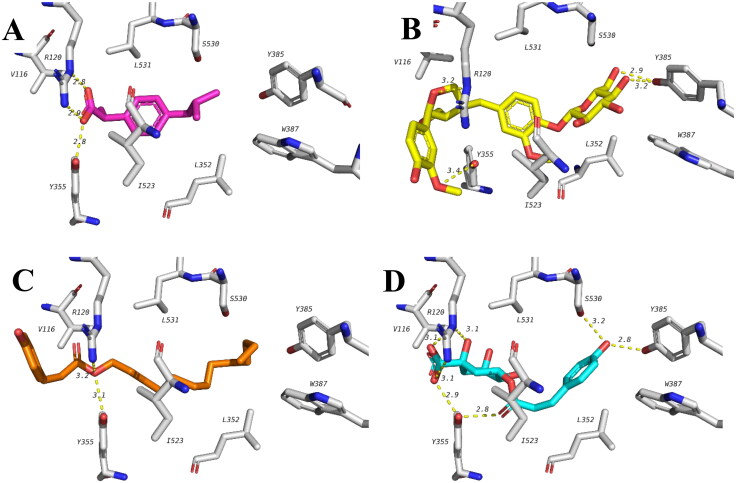
Among the tested compounds lariciresinol glucoside, octadecyl (E)-p-coumarate and p-coumaroylquinic acid showed best docking scores which is comparable or even slightly higher than that of ibuprofen (Table 4). All the three compounds can form hydrogen bonds with the same residues similar to ibuprofen including R120 and Y355 as well as multiple hydrophobic interactions. In addition, lariciresinol glucoside and p-coumaroylquinic acid both formed hydrogen bonds with Y385 and p-coumaroylquinic acid has an extra hydrogen bond with S530 which is in close proximity to the binding pocket of celecoxib reported earlier. Proposed binding modes of these three compounds with COX-1 are illustrated in Figure 4.
Table 4.
Docking score of the annotated compounds.
| Compound | Cox-1 (1EQG) | Covid-19 targets |
|||
|---|---|---|---|---|---|
| Mpro (6LU7) | nsp16 (6W4H) |
nsp12 (7BV2) |
|||
| 1 | 2,3,2′',3′'-tetrahydrobilobetin | −3.5 | −9.5 | −9.6 | −8.9 |
| 2 | 2′',3′'-dihydroheveaflavone | −3.4 | −8.0 | −7.9 | −8.1 |
| 3 | 3′,3′''-binaringenin | −5.8 | −9.1 | −9.5 | −8.0 |
| 4 | 4′,7,'',4′'' tri-o-methyl amentoflavone (kayaflavone) | −3.2 | −9.1 | −10.1 | −8.5 |
| 5 | 7-Hydroxy-secoisolaricersinol | −7.3 | −6.6 | −7.8 | −5.7 |
| 6 | Amentoflavone Glucoside | −6.1 | −9.3 | −10.0 | −9.5 |
| 7 | Amentoflavone | −3.7 | −9.8 | −10.1 | −8.5 |
| 8 | Apigenin 5-O-rhamnoside | −6.5 | −8.3 | −8.8 | −6.9 |
| 9 | Apigenin 5-glucoside | −6.7 | −8.4 | −8.8 | −7.4 |
| 10 | Bilobetin glucoside | −6.3 | −9.7 | −10.1 | −9.0 |
| 11 | Lariciresinol glucoside | −8.0 | −7.7 | −9.0 | −7.6 |
| 12 | Octadecyl (E)-p-coumarate | −7.7 | −4.9 | −6.2 | −4.5 |
| 13 | p-coumaric acid | −6.3 | −5.1 | −6.0 | −5.2 |
| 14 | p-coumaroylquinic acid | −7.5 | −7.1 | −7.8 | −6.8 |
| 15 | Protocatechuic acid | −6.1 | −5.4 | −6.5 | −5.7 |
| 16 | Secoisolariciresinol rhamnoside | −7.3 | −7.1 | −8.5 | −5.9 |
| 17 | Sinapic acid-O-glucoside | −6.3 | −6.9 | −7.6 | −6.7 |
| 18 | Syringic acid glucoside | −5.8 | −7.4 | −6.4 | −5.8 |
| 19 | Vanillic acid | −6.0 | −5.1 | −6.3 | −5.5 |
| Co-crystalized Ligand | −7.8 (ibuprofen) | −7.9 (N3) |
−8.2 (SAM) | −6.6 (Remdesivir) | |
Figure 4.
Docking poses of celecoxib and top ranked compounds with COX-1 showing important hydrogen bonds and hydrophobic interactions. (A) Ibuprofen (pink), (B) lariciresinol glucoside (yellow), (C) octadecyl (E)-p-coumarate (orange) and (D) p-coumaroylquinic acid (blue).
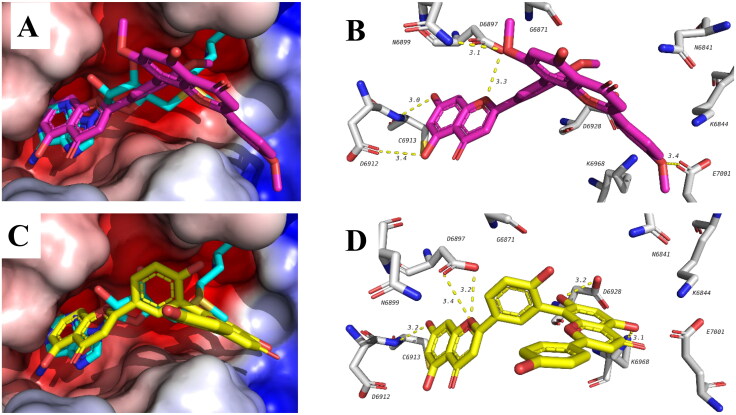
A recent study has shown that the anti-inflammatory corticosteroid dexamethasone has positive effect in improving survival rate of COVID-19 hospitalized patients as demonstrated by randomized clinical trials (Abdolahi et al., 2020). This suggests promising effect of anti-inflammatory drugs as adjuvant therapy in COVID-19 patients for challenging the storm of cytokine which is a state of release of excessive of a diverse of inflammatory cytokines. The molecular mechanism of the cytokine storm is still unexplored. We were encouraged to theoretically study possible effect of our identified compounds against multiple targets of SARS-CoV-2. The compounds identified were docked in 3 targets active sites, viral main protease (Mpro, 6LU7), non-structural protein-16 which is a methyl transferase (nsp16, 6W4H) and RNA dependent RNA polymerase (nsp12, 7BV2). Table 4 shows the docking results of tested compounds against the 3 viral targets. In general, bioflavonoids have shown better scores compared to other compounds in all the three targets. Among the three targets, nsp16 was the best target recognized by the tested compounds as can be deduced from docking energies of each compound with the 3 targets (Table 4). Three biflavonoids were among the top compounds bound to nsp16 with energies better than S-adenosylmethionine (SAM) which is the co-crystalized ligand. These biflavonoids include kayaflavone, amentoflavone and Bilobetin glucoside. A representative example of the binding of kayaflavone and amentoflavone is shown in Figure 5. Rings A and B of biflavonoids overlapped with the purine ring of the SAM and form similar hydrogen bonds with D6912 and C6913. Other hydrogen bonds are also formed with D6897. The other side of the bioflavonoids extends into the KDKE methyl transferase motif and forms hydrogen bonds with E7001 in case of kayaflavone and with K6968 for amentoflavone. These results suggest possible effect of these compounds against SARS-CoV-2 and worth further investigation.
Figure 5.
Binding mode and interactions of kayaflavone (top, pink) and amentoflavone (bottom, yellow) in the active site of nsp16. Compounds are overlapped with SAM (blue).
3.6. Molecular dynamics simulations
So, the molecular docking data revealed that all selected compounds effectively block the target proteins and that is confirmed by the binding energy values lower compared with those predicted for the molecules used as a positive control in the calculations. However, it is well-known that estimating the binding affinity profiles using molecular docking studies involves a number of approximations, resulting in the Lack of trustworthiness on the capability of scoring functions to produce accurate values of binding free energy (Meng et al., 2011; Salmaso & Moro, 2018). In this context, the parameters of thermodynamic depending on the MD simulations are much more credible than those predicted from the analysis of the static docking models. contrary of the computational approaches, MD simulation moves each atom individually through the field of the rested atoms showing the flexibility of both the ligand and protein which is more effective than other algorithms (Meng et al., 2011; Salmaso & Moro, 2018).
Nevertheless, analyzed compounds to the target proteins resulted from the post-modeling analysis of the static ligand/protein structures their data, is confirmed by the MD simulations the high binding affinity. Analysis of the dynamic ligand/protein models shows that, in both cases of interest, within the MD simulations, these complexes are relatively stable, as proved by the averages of binding free energies and the corresponding standard deviations (Table 5). With the Given MM/GBSA method errors of 1–3 kcal/mol (Genheden & Ryde, 2015), we can propose that the dynamic ligand/protein structures having the averages of binding free energy lower than those calculated for the reference compounds using the same computational protocols (Table 5). The exception is compound B' targeting the NSP16-10 protein of SARS-Cov-2 and exhibiting the binding affinity comparable with that obtained for the control molecule SAM (Table 5).
Table 5.
Values mean of binding free energy < ΔG > for the compounds analyzed and their standard deviations (ΔG)STD.
| COX-1 | |||
|---|---|---|---|
| Compound | <ΔH> ± (ΔH)STD (kcal/mol) |
<TΔS> ±(TΔS)STD (kcal/mol) | <ΔG> ± (ΔG)STD (kcal/mol) |
| A | −64.39 ± 3.39 | −24.69 ± 10.76 | −39.71 ± 11.74 |
| B | −54.04 ± 3.69 | −24.15 ± 10.09 | −29.90 ± 10.97 |
| C | −50.41 ± 4.50 | −18.92 ± 6.05 | −31.49 ± 7.28 |
| IBP | −33.20 ± 2.07 | −15.24 ± 8.26 | −17.95 ± 8.72 |
| NSP16-10 | |||
| A' | −38.93 ± 4.02 | −23.60 ± 5.42 | −15.33 ± 6.44 |
| B' | −27.84 ± 7.01 | −19.21 ± 3.29 | −8.63 ± 7.10 |
| C' | −44.14 ± 3.26 | −24.69 ± 5.97 | −19.45 ± 5.84 |
| SAM | −27.30 ± 4.69 | −18.60 ± 7.19 | −8.70 ± 8.51 |
The compounds are designated as follows: A—3-Lariciresinol-glucoside, B—14-Octadecyl (E)-p-coumarate, C—16-p-coumaroylquinic acid, IBP—Ibuprofen. A'—5-Kayaflavon, B'—8-Amentoflavone, C'—11-Bilobetin-glucoside, SAM—S-Adenosylmethionine.
Values of mean of enthalpic and entropic components of free energy are <ΔH > and < TΔS>, respectively; standard deviations corresponding to these values are (ΔH)STD and (TΔS)STD
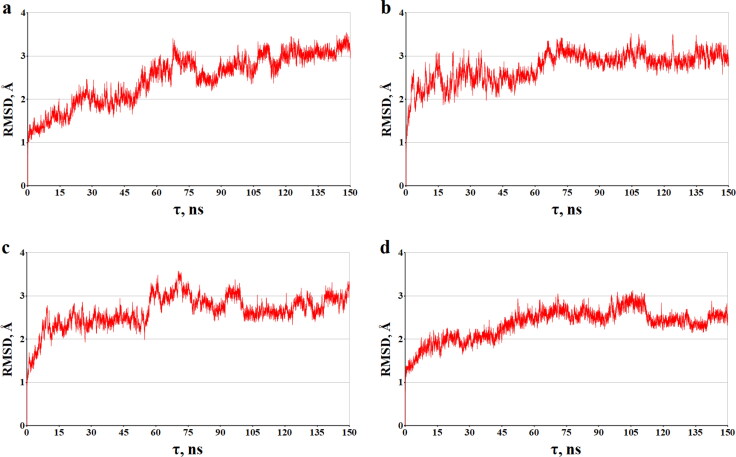
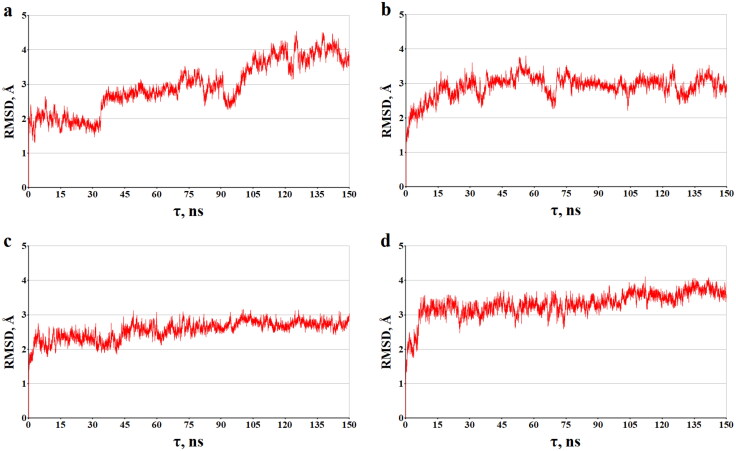
Regarding the observed stability of the dynamic ligand/protein structures is also an evidence for the data depending on the time of the root-mean square deviations (RMSD) for the atomic positions of the static and dynamic models of the analyzed compounds bounding to the target proteins (Figures 6 and 7). Analysis of Figures 6 and 7 reveals that these complexes do not support significant structural rearrangements on the MD trajectories, and this is confirmed by the calculated RMSD averages of the dynamic models of molecules A, B, C and A, B, C in the complexes with COX-1 and NSP16-10, respectively. For the ligand/COX-1 complexes, the RMSD mean values and standard deviations, which are 2.48 ± 0.58 Å (compound A), 2.74 ± 0.35 Å (compound B), 2.64 ± 0.37 (compound C), are close to those of 2.34 ± 0.36 Å calculated for the COX-1 inhibitor IBP (Figure 6). In the case of the ligand/NSP16-10 complexes, these averages are 2.94 ± 0.74 Å (compound A'), 2.90 ± 0.33 Å (compound B'), 2.55 ± 0.26 Å (compound C'), and 3.31 ± 0.37 Å (SAM).
Figure 6.
The time dependences calculated of the RMSD (Å) between all of the MD ligand/COX-1 structures and their starting models. The backbone atoms of COX-1 were used in the calculations. The graphs shown correspond to the following ligand/COX-1 complexes: (a) A/COX-1, (b) B/COX-1, (c) C/COX-1, and (D) IBP/COX-1.
Figure 7.
The time dependences of the RMSD (Å) calculated between all of the MD ligand/NSP16-10 structures and their starting models. The backbone atoms of NSP16-10 were used in the calculations. The graphs shown correspond to the following ligand/COX-1 complexes: (a) A'/NSP16-10, (b) B'/ NSP16-10, (c) C'/ NSP16-10, and (D) SAM/ NSP16-10.
Comparison of the MD structures between every subsequent dynamic model with the previous one results in the averages of RMSD which are also evidence of the relative stability of the ligand/protein complexes at MD simulations. For the compounds bound to COX-1, these averages equal to 0.77 ± 0.05 Å (compound A), 0.78 ± 0.05 Å (compound B) and 0.78 ± 0.05 Å (compound C) are very close to those calculated for the control molecule IBP (0.76 ± 0.05 Å). Compounds A', B', C' show the mean values of RMSD in the complexes with NSP16-10 which are respectively 0.84 ± 0.06 Å, 0.84 ± 0.06 Å, 0.83 ± 0.06 Å and very close to the SAM/NSP16-10 structure (0.85 ± 0.06 Å).
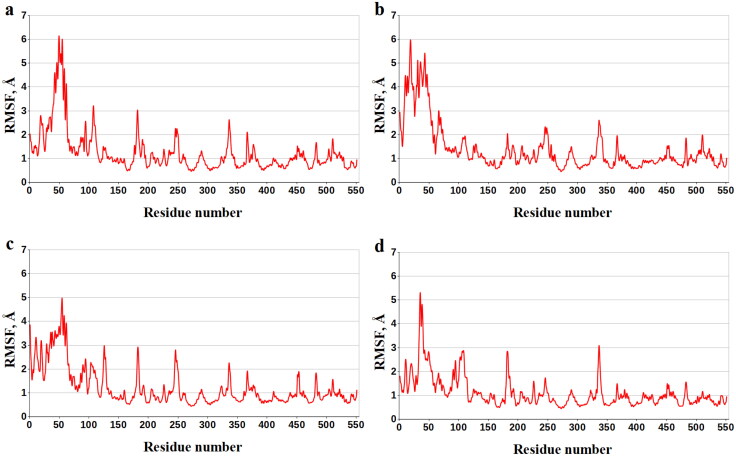
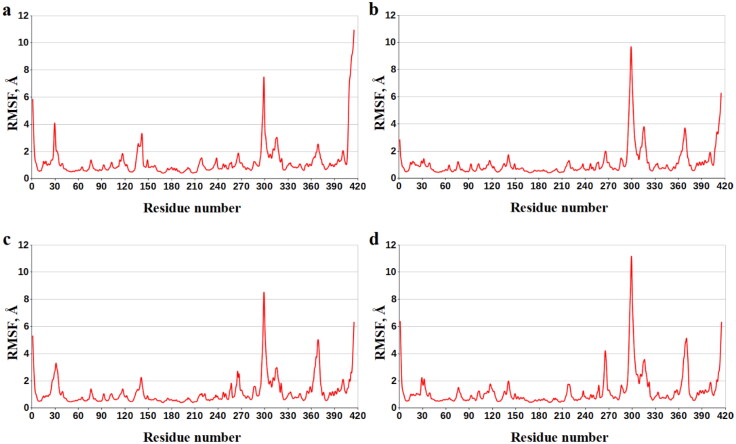
Enthalpy components decomposition of the binding energy to the contributions of individual COX-1 amino acids exposes the residues dominating the ligand/COX-1 interactions in all cases of interest (Table 6). Analysis of Table 6 shows that these COX-1 residues are Val-116, Val-349, Leu-352, Tyr-355, Ile-523, and Ala-527. The data obtained suggest that there are a number of the basic anchoring residues of COX-1 that provide strong attachment of the analyzed compounds at the active site of this protein. This supposition is supported by the values of the data of root-mean-square fluctuations (RMSF) for residues of COX-1 designating the flexibility of each amino acid throughout the MD simulations (Figure 8, Table 7). Analysis of Table 7 shows that, according to the values of RMSF, the key anchoring residues Val-116, Val-349, Leu-352, Tyr-355, Ile-523, and Ala-527 of COX-1 their position are restrained on the MD trajectories, in agreement with the data on their contributions into the binding enthalpy (Table 6).
Table 6.
Averages of the binding enthalpy for the amino-acid residues of COX-1 bound to compounds A, B and Ca,2,3.
| Compound | |||
|---|---|---|---|
| Residue of COX-1 | A | B | C |
| Residue Contribution to the Binding Enthalpy (kcal/mol) | |||
| ILE-89 | −1.17 ± 0.50 | – | – |
| LEU-93 | −0.90 ± 0.51 | −0.85 ± 0.37 | – |
| LEU-112 | −0.59 ± 0.48 | −1.02 ± 0.42 | – |
| MET-113 | – | −1.52 ± 0.74 | – |
| VAL-116 | −1.44 ± 0.35 | −1.52 ± 0.40 | −0.74 ± 0.31 |
| LEU-117 | – | −0.80 ± 0.47 | −0.69 ± 0.32 |
| ARG-120 | – | – | −0.54 ± 0.36 |
| ILE-345 | – | – | −0.60 ± 0.50 |
| VAL-349 | −1.90 ± 0.31 | −1.32 ± 0.34 | −1.67 ± 0.34 |
| LEU-352 | −1.86 ± 0.39 | −1.22 ± 0.36 | −1.48 ± 0.33 |
| SER-353 | −0.88 ± 0.33 | −0.70 ± 0.25 | −0.76 ± 0.28 |
| TYR-355 | −1.59 ± 0.68 | −1.15 ± 0.38 | −0.72 ± 0.31 |
| LEU-357 | −0.70 ± 0.52 | −1.09 ± 0.45 | – |
| LEU-359 | −0.48 ± 0.24 | −1.39 ± 0.53 | −0.94 ± 0.27 |
| LEU-384 | – | – | −0.57 ± 0.28 |
| TRP-387 | −0.47 ± 0.24 | – | – |
| PHE-518 | −0.93 ± 0.26 | −0.52 ± 0.26 | −0.87 ± 0.28 |
| MET-522 | – | – | −0.56 ± 0.33 |
| ILE-523 | −2.26 ± 0.64 | −1.16 ± 0.35 | −1.59 ± 0.47 |
| GLY-526 | −0.83 ± 0.26 | −0.50 ± 0.24 | −0.89 ± 0.24 |
| ALA-527 | −1.90 ± 0.33 | −1.34 ± 0.31 | −1.63 ± 0.31 |
| SER-530 | −0.46 ± 0.38 | – | −0.47 ± 0.41 |
| LEU-531 | −0.61 ± 0.27 | −0.56 ± 0.27 | −1.51 ± 0.55 |
aData for the COX-1 residues with the binding enthalpy ≤ −0.4 kcal/mol are presented.
bThe averages of the residue contributions to the binding enthalpy and corresponding standard deviations are given.
cThe COX-1 residues dominating the ligand/protein interaction are highlighted by bold.
Figure 8.
Values of RMSF (Å) for each residue along the COX-1 amino-acid sequence. Numbering of the protein amino acids corresponds to an original PDB numbering in the following manner: 1-551 − 33-583.
Table 7.
Values of RMSF for the COX-1 residues contributing to the binding enthalpy.
| Compound | |||
|---|---|---|---|
| Residue of COX-1 | A | B | C |
| Values of RMSF (Å) for the individual residues of COX-1 | |||
| ILE-89 | 3.47 | – | – |
| LEU-93 | 3.07 | 1.19 | – |
| LEU-112 | 1.23 | 1.46 | – |
| MET-113 | – | 1.35 | – |
| VAL-116 | 1.53 | 1.47 | 1.22 |
| LEU-117 | – | 1.32 | 1.34 |
| ARG-120 | – | – | 1.44 |
| ILE-345 | – | – | 0.70 |
| VAL-349 | 0.62 | 0.75 | 0.69 |
| LEU-352 | 0.73 | 0.80 | 0.97 |
| SER-353 | 0.77 | 0.89 | 1.06 |
| TYR-355 | 1.08 | 1.08 | 1.29 |
| LEU-357 | 0.91 | 1.06 | – |
| LEU-359 | 0.82 | 0.99 | 0.88 |
| LEU-384 | – | – | 0.63 |
| TRP-387 | 0.63 | – | – |
| PHE-518 | 0.87 | 0.73 | 0.97 |
| MET-522 | – | – | 0.73 |
| ILE-523 | 0.80 | 1.19 | 0.71 |
| GLY-526 | 0.80 | 0.91 | 0.78 |
The COX-1 residues presenting the dominant contributors to the ligand/protein interaction are highlighted by bold.
The data on decomposition of binding energy into the contributions of individual NSP16-10 amino acids are given in Table 8, and the data on the values of RMSF for the individual residues of this protein are presented in Figure 9 and Table 9.
Table 8.
Averages of the binding enthalpy for the amino-acid residues of NSP16-10 bound to compounds A', B' and C'a,b,c.
| Compound | |||
|---|---|---|---|
| Residue of NSP16-10 | A' | B' | C' |
| Residue Contribution to the Binding Enthalpy (kcal/mol) | |||
| GLY-6869 | – | – | −0.55 ± 0.24 |
| LEU-6898 | −3.91 ± 0.89 | −0.98 ± 1.14 | −2.10 ± 0.80 |
| ASN-6899 | −1.06 ± 0.80 | – | – |
| CYS-6913 | −0.64 ± 0.35 | – | −0.56 ± 0.36 |
| MET-6929 | −1.97 ± 0.40 | −0.65 ± 0.91 | −2.08 ± 0.47 |
| TYR-6930 | – | −2.39 ± 0.94 | −1.54 ± 0.59 |
| ASP-6931 | −0.60 ± 0.53 | – | −1.05 ± 0.51 |
| PRO-6932 | −1.51 ± 0.87 | −2.54 ± 0.74 | −1.08 ± 0.78 |
| LYS-6933 | – | −0.54 ± 0.83 | – |
| THR-6934 | – | – | −0.73 ± 0.44 |
| LYS-6935 | – | −0.76 ± 1.03 | – |
| LYS-6944 | – | – | −0.63 ± 0.25 |
| PHE-6947 | −1.59 ± 0.89 | – | −2.01 ± 0.61 |
| PHE-6948 | – | – | −0.79 ± 0.27 |
| LEU-7093 | −0.71 ± 0.91 | – | – |
aData for the NSP16-10 residues with the binding enthalpy ≤ −0.4 kcal/mol are presented.
bThe averages of the residue contributions to the binding enthalpy and corresponding standard deviations are given.
cThe NSP16-10 residues dominating the ligand/protein interaction are highlighted by bold.
Figure 9.
Values of RMSF (Å) for each residue along the NSP16-10 amino-acid sequence. Numbering of the protein amino acids along the X-axis corresponds to an original PDB numbering in the following manner: 1-299 − 6798-7096 and 300-415 − 4271-4386.
Table 9.
RMSF values for the NSP16-10 residues in contribution to the binding enthalpy.
| Compound | |||
|---|---|---|---|
| Residue of NSP16-10 | A' | B' | C' |
| RMSF values (Å) for the individual residues of NSP16-10 | |||
| GLY-6869 | – | – | 0.65 |
| LEU-6898 | 0.90 | 0.87 | 0.97 |
| ASN-6899 | 1.12 | – | – |
| CYS-6913 | 1.74 | – | 1.23 |
| MET-6929 | 0.63 | 0.67 | 0.79 |
| TYR-6930 | – | 0.80 | 0.94 |
| ASP-6931 | 1.29 | – | 1.16 |
| PRO-6932 | 1.70 | 1.07 | 1.24 |
| LYS-6933 | – | 1.06 | – |
| THR-6934 | – | – | 1.32 |
| LYS-6935 | – | 0.90 | – |
| LYS-6944 | – | – | 0.68 |
| PHE-6947 | 0.91 | – | 0.70 |
| PHE-6948 | – | – | 0.63 |
| LEU-7093 | 3.12 | – | – |
The NSP16-10 residues presenting the dominant contributors to the ligand/protein interaction are highlighted by bold.
Intermolecular calculations of hydrogen bonds appeared in the MD trajectories of the ligand/COX-1 complexes indicate (Table 10) that the concerned compounds form hydrogen bonds with Arg-120COX-1 (compound A), Leu-117COX-1 (compound B), Ala-527COX-1 and Leu-531COX-1 (compound C). Notably, as compound C, the control molecule IBP is also involved in hydrogen bonding with Leu-531COX-1 (Table 10). Analysis of the dynamic ligand/NSP16-10 models reveals hydrogen bonds between compound A' and Asn-6899NSP16-10, compound B' and Lys-6933NSP16-10, as well as compound C' and Lys-6930NSP16-10, Asp-6931NSP16-10, Met-6929 NSP16-10, and Cys-6913 NSP16-10. At the same time, the reference compound SAM forms hydrogen bonds with the NSP16-10 residues Thr-4292, Asn-4293, Val-4295 (Table 10). Among these H-bonds, it should be specially noted hydrogen bonds C···Ala-527COX-1 and C'···Lys-6930NSP16-10 demonstrating the relatively high percentage occupancies on the MD trajectories (Table 10).
Table 10.
Intermolecular hydrogen bonds in the dynamic ligand/protein structures.
| Hydrogen Bonds | |||
|---|---|---|---|
| Ligand | COX-1 | Ligand | NSP16-10 |
| A | O…HN[Arg-120; 10.83%] | A' | O…HN[Asn-6899; 29.48%] |
| B | O…HN[Leu-117; 28.08%] | B' | O…HN[Lys-6933; 12.43%] |
| C | O…HN[Ala-527; 68.37%] O…HN[Leu-531; 18.63%] | C' | O…HN[Lys-6930; 59.96%], O…HN[Asp-6931; 45.91%], O…HN[Met-6929; 25.15%] O…HN[Cys-6913; 17.69%] |
| IBP | O…HN[Leu-531; 10.19%] | SAM | NH2…O[Thr-4292; 17.16%], NH1…O[Thr-4292; 17.16%], NH2…O[Asn-4293; 12.65%], NH1…O[Asn-4293; 12.65%], O…HN[Val-4295; 11.03%] |
Donors and acceptors of the hydrogen bonds relating to the ligands are shown first, followed by the corresponding functional groups of the protein amino acids. The protein residues and percentage occupancies of hydrogen bonds are indicated in square brackets.
The data on the hydrogen bonds exhibiting percentage occupancies >10%are given.
Thus, the data of molecular modeling indicate that the analyzed compounds show strong attachment to the target proteins. This follows from the calculation of low binding free energy values for the ligand/protein complexes in terms of the scoring functions of molecular docking and molecular dynamics. In both cases, the predicted values of this thermodynamic parameter testify to the higher binding affinity of the identified compounds to their targets compared with the reference molecules IBP and SAM.
4. Conclusions
In light of the results of the current investigation, we propose that the extracts of A. bidwillii, A. cunninghamii and A. heterophylla cultivated in Egypt, could be a possible source of potent natural anti-inflammatory and antioxidant metabolites. Those activities are of extensive importance as medicated agents for inhibiting and regress the progression of inflammation correlated with oxidative stress-relevant to viral infection and trying to avoid the side effects accompanied by the administration of dexamethasone such as elevated blood pressure, oedema and hyperglycemia. Those results were deduced from the virtual study carried for the structure relationship of LC-MS annotated compounds and the receptor interactions in three different binding sites of proteolytic enzyme in the COVID-19 as all the tested exhibited significant binding stability as observed for the amentoflavone and bilobetin glucoside with significant low binding energy. Our data suggested possible effect of these compounds against SARS-CoV-2 and worth further investigation. Several pathological conditions for instance as processes of inflammation are accompanied with reactive oxygen species generation. Thus, inhibitory activity on COX-1 enzyme of these plant extracts probably due to their capacity to reduce oxidative stress. Finally, we believe that the potential anti-COVID effects of the reported compounds required and worth further investigations.
Acknowledgments
The authors would like to extend their sincere appreciation to the central laboratory at Jouf University for support this study. M.M. Al-Sanea also extends his appreciation to the Korea Institute of Science and Technology (KIST) for funding this work through the Grant name “2021 KIST School Partnership Project”.
Funding Statement
The authors extend their appreciation to the Deputyship for Research and Innovation, Ministry of Education in Saudi Arabia for funding this work through the project number “375213500”
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Abdolahi, N., Kaheh, E., Golsha, R., Khodabakhshi, B., Norouzi, A., Khandashpoor, M., Besharat, S., Tavassoli, S., Livani, S., Azimi, S. A., Gharib, M. H., Peivandi, B., Fazel, A., Shirzad-Aski, H., & Roshandel, G. (2020). Letter to the editor: Efficacy of different methods of combination regimen administrations including dexamethasone, intravenous immunoglobulin, and interferon-beta to treat critically ill COVID-19 patients: A structured summary of a study protocol for a randomized controlled trial. Trials, 21(1), 1–3. 10.1186/s13063-020-04499-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ağalar, H. G., Çiftçi, G. A., Göger, F., & Kırımer, N. (2017). Activity guided fractionation of Arum italicum miller tubers and the LC/MS-MS profiles. Records of Natural Products, 12(1), 64–75. 10.25135/rnp.06.17.05.089 [DOI] [Google Scholar]
- Aly, O. M. (2020). Molecular docking reveals the potential of aliskiren, dipyridamole, mopidamol, rosuvastatin, rolitetracycline and metamizole to inhibit COVID-19 virus main protease. [DOI] [PMC free article] [PubMed]
- Aslam, M. S., Choudhary, B. A., Uzair, M., & Ijaz, A. S. (2013). Phytochemical and ethno-pharmacological review of the genus Araucaria–review. Tropical Journal of Pharmaceutical Research, 12(4), 651–659. 10.4314/tjpr.v12i4.31 [DOI] [Google Scholar]
- Carini, M., Aldini, G., Furlanetto, S., Stefani, R., & Facino, R. M. (2001). LC coupled to ion-trap MS for the rapid screening and detection of polyphenol antioxidants from Helichrysum stoechas. Journal of Pharmaceutical and Biomedical Analysis, 24(3), 517–526. 10.1016/S0731-7085(00)00431-3 [DOI] [PubMed] [Google Scholar]
- Case, D. A., Belfon, K., Ben-Shalom, I., Brozell, S. R., Cerutti, D., Cheatham, T., & Giambasu, G. (2020). Amber 2020.
- Chiappetta, S., Sharma, A. M., Bottino, V., & Stier, C. (2020). COVID-19 and the role of chronic inflammation in patients with obesity. International Journal of Obesity (2005), 44(8), 1790–1793. 10.1038/s41366-020-0597-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebada, S. S., Al-Jawabri, N. A., Youssef, F. S., Albohy, A., Aldalaien, S. M., Disi, A. M., & Proksch, P. (2020). In vivo antiulcer activity, phytochemical exploration, and molecular modelling of the polyphenolic-rich fraction of Crepis sancta extract. Inflammopharmacology, 28(1), 321–331. 10.1007/s10787-019-00637-x [DOI] [PubMed] [Google Scholar]
- Eklund, P. C., Backman, M. J., Kronberg, L. Å., Smeds, A. I., & Sjöholm, R. E. (2008). Identification of lignans by liquid chromatography-electrospray ionization ion-trap mass spectrometry. Journal of Mass Spectrometry: JMS, 43(1), 97–107. 10.1002/jms.1276 [DOI] [PubMed] [Google Scholar]
- El-Hawary, S., Hammouda, F., Tawfik, W., Kassem, H., Abdelshafeek, K., & El-Shamy, S. (2017). Investigation of some chemical constituents, cytotoxicity and antioxidant activities of Beta Vulgaris var. altissima cultivated in Egypt. Rasayan Journal of Chemistry, 10(4), 1391-1401. 10.7324/RJC.2017.1041936 [DOI]
- Elshamy, A. I., Ammar, N. M., Hassan, H. A., Al-Rowaily, S. L., Ragab, T. I., El Gendy, A. E.-N G., & Abd-ElGawad, A. M. (2020). Essential oil and its nanoemulsion of Araucaria heterophylla resin: Chemical characterization, anti-inflammatory, and antipyretic activities. Industrial Crops and Products, 148, 112272. 10.1016/j.indcrop.2020.112272 [DOI] [Google Scholar]
- Essmann, U., Perera, L., Berkowitz, M. L., Darden, T., Lee, H., & Pedersen, L. G. (1995). A smooth particle mesh Ewald method. The Journal of Chemical Physics, 103(19), 8577–8593. 10.1063/1.470117 [DOI] [Google Scholar]
- Fang, N., Yu, S., & Prior, R. L. (2002). LC/MS/MS characterization of phenolic constituents in dried plums. Journal of Agricultural and Food Chemistry, 50(12), 3579–3585. 10.1021/jf0201327 [DOI] [PubMed] [Google Scholar]
- Genheden, S., & Ryde, U. (2015). The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opinion on Drug Discovery, 10(5), 449–461. 10.1517/17460441.2015.1032936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ik, A., Fleischer, T., Annan, K., Dickson, R., Mensah, A., & Sarpong, F. (2013). Anti-inflammatory, antioxidant and antimicrobial activity of the stem bark extract and fractions of Ficus exasperata Vahl.(Moraceae). Journal of Pharmacognosy and Phytochemistry, 2(3), 38–44. [Google Scholar]
- Jayawardena, R., Sooriyaarachchi, P., Chourdakis, M., Jeewandara, C., & Ranasinghe, P. (2020). Enhancing immunity in viral infections, with special emphasis on COVID-19: A review. Diabetes & Metabolic Syndrome: Clinical Research & Reviews, 14(4), 367–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., & Duan, Y. (2020). Structure-based drug design, virtual screening and high-throughput screening rapidly identify antiviral leads targeting COVID-19. BioRxiv. 10.1101/2020.02.26.964882 [DOI]
- Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W., & Klein, M. L. (1983). Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics, 79(2), 926–935. 10.1063/1.445869 [DOI] [Google Scholar]
- Kumarasamy, Y., Byres, M., Cox, P. J., Jaspars, M., Nahar, L., & Sarker, S. D. (2007). Screening seeds of some Scottish plants for free radical scavenging activity. Phytotherapy Research: PTR, 21(7), 615–621. 10.1002/ptr.2129 [DOI] [PubMed] [Google Scholar]
- Lee, S. R., Lee, S., Moon, E., Park, H.-J., Park, H. B., & Kim, K. H. (2017). Bioactivity-guided isolation of anti-inflammatory triterpenoids from the sclerotia of Poria cocos using LPS-stimulated Raw264.7 cells. Bioorganic Chemistry, 70, 94–99. 10.1016/j.bioorg.2016.11.012 [DOI] [PubMed] [Google Scholar]
- Mahmudpour, M., Roozbeh, J., Keshavarz, M., Farrokhi, S., & Nabipour, I. (2020). COVID-19 cytokine storm: The anger of inflammation. Cytokine, 133, 155151. 10.1016/j.cyto.2020.155151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Sánchez, G., Schwartz, A., & Donna, V. D. (2020). Potential cytoprotective activity of ozone therapy in SARS-CoV-2/COVID-19. Antioxidants, 9(5), 389. 10.3390/antiox9050389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moawad, A., Hetta, M., Zjawiony, J. K., Jacob, M. R., Hifnawy, M., Marais, J. P., & Ferreira, D. (2010). Phytochemical investigation of Cycas circinalis and Cycas revoluta leaflets: Moderately active antibacterial biflavonoids. Planta Medica, 76(8), 796–802. 10.1055/s-0029-1240743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillaiyar, T., Manickam, M., Namasivayam, V., Hayashi, Y., & Jung, S.-H. (2016). An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: Peptidomimetics and small molecule chemotherapy. Journal of Medicinal Chemistry, 59(14), 6595–6628. 10.1021/acs.jmedchem.5b01461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradelles, P., Grassi, J., & Maclouf, J. (1985). Enzyme immunoassays of eicosanoids using acetylcholine esterase as label: An alternative to radioimmunoassay. Analytical Chemistry, 57(7), 1170–1173. 10.1021/ac00284a003 [DOI] [PubMed] [Google Scholar]
- Purev, O., Pospíšil, F., & Motl, O. (1988). Flavonoids from Ephedra sinica stapf. Collection of Czechoslovak Chemical Communications, 53(12), 3193–3196. 10.1135/cccc19883193 [DOI] [Google Scholar]
- Rimon, G., Sidhu, R. S., Lauver, D. A., Lee, J. Y., Sharma, N. P., Yuan, C., Frieler, R. A., Trievel, R. C., Lucchesi, B. R., & Smith, W. L. (2010). Coxibs interfere with the action of aspirin by binding tightly to one monomer of cyclooxygenase-1. Proceedings of the National Academy of Sciences of the United States of America, 107(1), 28–33. 10.1073/pnas.0909765106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckaert, J. (1977). Ciccotti+, G. & Berendsen, HJC numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of N-alkanes. Journal of Computational Physics, 23, 321–341. [Google Scholar]
- Sánchez‐Rabaneda, F., Jauregui, O., Lamuela‐Raventós, R. M., Viladomat, F., Bastida, J., & Codina, C. (2004). Qualitative analysis of phenolic compounds in apple pomace using liquid chromatography coupled to mass spectrometry in tandem mode. Rapid Communications in Mass Spectrometry: RCM, 18(5), 553–563. 10.1002/rcm.1370 [DOI] [PubMed] [Google Scholar]
- Schett, G., Manger, B., Simon, D., & Caporali, R. (2020). COVID-19 revisiting inflammatory pathways of arthritis. Nature Reviews. Rheumatology, 16(8), 465–466. 10.1038/s41584-020-0451-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger, L. (2010). The PyMOL molecular graphics system, Version 1.3r1, vol. 5.
- Sobeh, M., Mahmoud, M., Sabry, O., Adel, R., Dmirieh, M., El-Shazly, A., & Wink, M. (2017). HPLC-PDA-MS/MS characterization of bioactive secondary metabolites from Turraea fischeri bark extract and its antioxidant and hepatoprotective activities in vivo. Molecules, 22(12), 2089. 10.3390/molecules22122089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao, T., Kitatani, F., Watanabe, N., Yagi, A., & Sakata, K. (1994). A simple screening method for antioxidants and isolation of several antioxidants produced by marine bacteria from fish and shellfish. Bioscience, Biotechnology, and Biochemistry, 58(10), 1780–1783. 10.1271/bbb.58.1780 [DOI] [Google Scholar]
- Trott, O., & Olson, A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A., & Case, D. A. (2004). Development and testing of a general amber force field. Journal of Computational Chemistry, 25(9), 1157–1174. 10.1002/jcc.20035 [DOI] [PubMed] [Google Scholar]
- World Health Organiztion . (2020). https://www.who.int/news-room/detail/16-06-2020-who-welcomes-preliminary-results-about-dexamethasone-use-in-treating-critically-ill-covid-19-patients.
- Yao, H., Chen, B., Zhang, Y., Ou, H., Li, Y., Li, S., Shi, P., & Lin, X. (2017). Analysis of the total biflavonoids extract from Selaginella doederleinii by HPLC-QTOF-MS and its in vitro and in vivo anticancer effects. Molecules, 22(2), 325. 10.3390/molecules22020325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabetakis, I., Lordan, R., Norton, C., & Tsoupras, A. (2020). COVID-19: The inflammation link and the role of nutrition in potential mitigation. Nutrients, 12(5), 1466. 10.3390/nu12051466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (New York, N.Y.), 368(6489), 409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, X.-Y., Zhang, X.-H., Mezei, M., & Cui, M. (2011). Molecular docking: A powerful approach for structure-based drug discovery. Current Computer-Aided Drug Design, 7(2), 146–157. 10.2174/157340911795677602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmaso, V., & Moro, S. (2018). Bridging molecular docking to molecular dynamics in exploring ligand-protein recognition process: An overview. Frontiers in Pharmacology, 9, 923. 10.3389/fphar.2018.00923 [DOI] [PMC free article] [PubMed] [Google Scholar]